Upon accumulation of unfolded proteins in the endoplasmic reticulum (ER), cells activate an ER stress response to enable plants to tolerate these conditions. This work shows that one facet of this response is the activation of the autophagy pathway for degradation of ER fragments in the vacuole, which is regulated by the IRE1b splicing factor.
Abstract
In this article, we show that the endoplasmic reticulum (ER) in Arabidopsis thaliana undergoes morphological changes in structure during ER stress that can be attributed to autophagy. ER stress agents trigger autophagy as demonstrated by increased production of autophagosomes. In response to ER stress, a soluble ER marker localizes to autophagosomes and accumulates in the vacuole upon inhibition of vacuolar proteases. Membrane lamellae decorated with ribosomes were observed inside autophagic bodies, demonstrating that portions of the ER are delivered to the vacuole by autophagy during ER stress. In addition, an ER stress sensor, INOSITOL-REQUIRING ENZYME-1b (IRE1b), was found to be required for ER stress–induced autophagy. However, the IRE1b splicing target, bZIP60, did not seem to be involved, suggesting the existence of an undiscovered signaling pathway to regulate ER stress–induced autophagy in plants. Together, these results suggest that autophagy serves as a pathway for the turnover of ER membrane and its contents in response to ER stress in plants.
INTRODUCTION
In eukaryotic cells, the endoplasmic reticulum (ER) is a central organelle for protein folding and maturation. To ensure correct protein function, polypeptide chains that enter the ER need to be successfully folded and exported to their destination. However, under severe environmental stress conditions, the load of unfolded or misfolded proteins in the ER can exceed the capacity of the cellular folding machinery, resulting in the accumulation of unfolded or misfolded proteins, which causes ER stress (Ron and Walter, 2007). To overcome this problem, the cell transmits a signal from ER to nucleus to activate the unfolded protein response (UPR) (Ron and Walter, 2007). The UPR usually involves the transcriptional upregulation of some ER-resident proteins, such as the binding protein (BiP) family of molecular chaperones, to assist in proper folding of the misfolded proteins, and the attenuation of protein translation to reduce the load of nascent proteins entering the ER. The UPR-targeted genes can also activate the ER-associated degradation (ERAD) machinery to eliminate the misfolded proteins by delivery to the cytoplasm for proteasomal degradation (Mori, 2000; Ron and Walter, 2007). If ER homeostasis cannot be reestablished, cell death is triggered (Ron and Walter, 2007).
The first example of an ER-to-nucleus signaling pathway triggering the UPR was discovered in yeast, where the ER-resident transmembrane protein inositol-requiring enzyme-1 (IRE1) binds to unfolded proteins (Gardner and Walter, 2011), thus sensing the ER stress, and splices the basic domain/leucine zipper (bZIP) transcription factor homologous to Atf/Creb1 (HAC1) mRNA (Cox et al., 1993; Mori et al., 1993, 1996; Cox and Walter, 1996). The translation product from the spliced HAC1 mRNA regulates the transcription of targeted genes that possess UPR cis-activating regulatory elements in their promoter regions (Mori et al., 1992; Kohno et al., 1993). Subsequently, three different classes of ER stress transducers were identified in mammalian systems, all of which are ER transmembrane proteins that sense ER stress through their lumenal domain and transduce the signal to the nucleus (Ron and Walter, 2007). These include (1) the mammalian homolog of IRE1, which splices the mRNA of the bZIP-like transcription factor X-box binding protein 1 (XBP1) (Tirasophon et al., 1998; Yoshida et al., 2001; Calfon et al., 2002); (2) the activating transcription factor-6 (ATF6), which is transported to the Golgi to be processed by site 1 and site 2 proteases (S1P and S2P), followed by movement to the nucleus to activate targeted genes (Haze et al., 1999; Ye et al., 2000; Yoshida et al., 2000); and (3) the RNA-activated protein kinase–like ER kinase (PERK), which regulates translation initiation factor-2a, thus attenuating translation (Harding et al., 2000). In plants, IRE1 homologs and two other ER stress sensors, bZIP28 and bZIP60, have been identified and/or characterized (Koizumi et al., 2001; Noh et al., 2002; Okushima et al., 2002; Iwata and Koizumi, 2005; Liu et al., 2007; Iwata et al., 2008, 2009; Tajima et al., 2008; Deng et al., 2011; Nagashima et al., 2011; Chen and Brandizzi, 2012). In Arabidopsis thaliana, the IRE1 gene family has two members, IRE1a and IRE1b; both are located in the perinuclear ER (Koizumi et al., 2001). IRE1b is widely expressed throughout the plant, whereas IRE1a is restricted to embryos and seeds (Koizumi et al., 2001; Noh et al., 2002). Like their yeast and animal counterparts, the plant IRE1s also possess kinase and endoribonuclease domains (Koizumi et al., 2001). In response to ER stress, IRE1 splices the mRNA encoding bZIP60, which is a bZIP transcription factor (Deng et al., 2011; Nagashima et al., 2011; Chen and Brandizzi, 2012), in a manner similar to that of Hac1 in yeast or XBP1 in animals. The spliced bZIP60 mRNA is translated and activates the BINDING PROTEIN3 gene (Deng et al., 2011). bZIP17 and -28 are additional transcription factors that localize to the ER membrane in unstressed conditions. In response to stress, they are cleaved and translocated to the nucleus, thus allowing activation of transcriptional responses (Liu et al., 2007); whether these proteins directly sense ER stress is not known.
While some aspects of the UPR signaling pathways have been elucidated in plants, morphological changes to the ER resulting from the UPR remain unclear (Urade, 2007, 2009; Moreno and Orellana, 2011). Previous research suggests that unfolded proteins can be degraded by both cytoplasmic ubiquitin-proteasome system (UPS)-dependent pathways and UPS-independent pathways (Urade, 2007). However, the molecular mechanisms of UPS-independent ERAD-like pathways in plants are still unknown.
Autophagy functions as a degradation process in the recycling of cellular cytoplasmic contents and the removal of damaged proteins or organelles under unfavorable growth conditions. Upon induction of autophagy, a double-membrane structure termed an autophagosome forms around cytoplasmic components. The autophagosome then delivers this cargo to the vacuole for degradation by hydrolases (Yang and Klionsky, 2009). After fusion of the outer membrane of the autophagosome with the vacuolar membrane, autophagic bodies, consisting of the inner membrane enclosing the cargo, are released into the vacuole, degraded, and the degradation products recycled. The autophagy pathway is largely conserved from yeast to animals and plants. Studies in yeast cells demonstrated that autophagy is triggered by ER stress, and the IRE1-HAC1 signaling pathway is required for ER stress–induced autophagy (Yorimitsu et al., 2006). Electron microscopy analyses have shown that autophagosomes contain ER lamellar membrane structures during ER stress (Bernales et al., 2006). Mammalian cells also showed the induction of autophagy under ER stress, and IRE1 is required for this process (Ogata et al., 2006). However, XBP1 mRNA splicing by the IRE1 endoribonuclease activity is not involved in autophagy (Ogata et al., 2006). Instead, the IRE1 kinase activity-mediated c-Jun N-terminal kinase pathway seems to be required for autophagy induction (Urano et al., 2000; Ogata et al., 2006). In addition, the ER was found to be one of the membrane sources for autophagosome formation (Hayashi-Nishino et al., 2009; Ylä-Anttila et al., 2009). In plants, autophagy is involved in responses to nutrient deprivation conditions, oxidative stress, salt and drought stresses, pathogen infection, and senescence (Hanaoka et al., 2002; Liu et al., 2005, 2009; Xiong et al., 2005, 2007b; Liu and Bassham, 2012). Recent studies also suggest that autophagy can selectively degrade certain plant organelles, such as ribosomes (Hillwig et al., 2011) and chloroplasts (Ishida et al., 2008; Wada et al., 2009). However, whether autophagy contributes to ER membrane turnover and homeostasis in plants has not been addressed.
In this study, the potential involvement of autophagy and the morphological changes in ER structure during ER stress in plants were investigated. Tunicamycin (TM) and DTT were found to trigger autophagy in Arabidopsis. Confocal and electron microscopy showed that portions of the ER are engulfed by autophagosomes and delivered to the vacuole, most likely for degradation. Moreover, one of the ER stress sensors, IRE1b, but not its homolog IRE1a, was found to be required for autophagy induction by ER stress, although its bZIP60 mRNA splicing activity does not seem to be involved. Our results suggest that autophagy is involved in mitigating ER stress in plants and that an alternative signaling pathway involving IRE1 activates autophagy in response to ER stress.
RESULTS
ER Stress Induces Autophagy in Arabidopsis
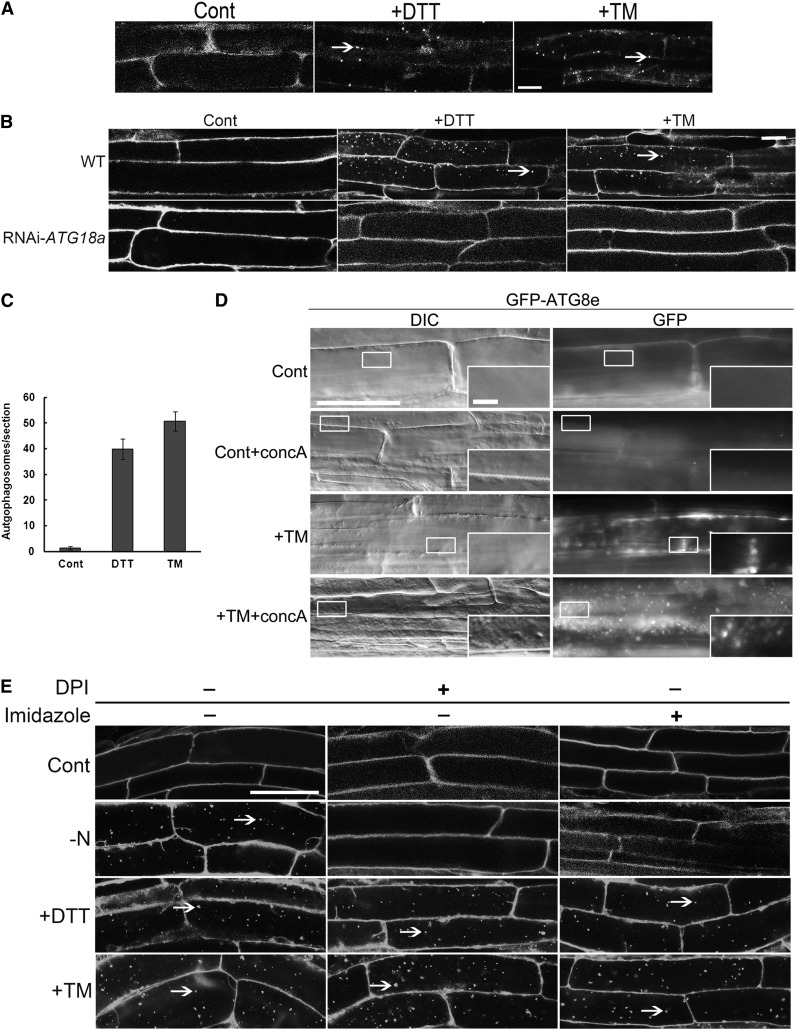
The effect of ER stress on autophagy in Arabidopsis was tested by treating wild-type seedlings or transgenic seedlings expressing green fluorescent protein (GFP)-AUTOPHAGY-RELATED 8e (ATG8e) with TM or DTT for 8 h. ATG8e is an autophagosome membrane-associated protein that is conjugated to phosphatidylethanolamine and is used as an autophagosome marker (Yoshimoto et al., 2004; Contento et al., 2005; Thompson et al., 2005). The markers used herein label both autophagosomes (in the cytoplasm) and autophagic bodies (in the vacuole); the term autophagosome is used to denote both types of structure unless further specified. Autophagosomes and autophagic bodies were detected in transgenic plants by visualization of GFP-ATG8e (Figure 1A) or in wild-type plants by staining with the autophagosome-selective dye monodansylcadaverine (MDC) (Figure 1B) (Contento et al., 2005; Xiong et al., 2007b). In control conditions, autophagosomes were rarely seen, whereas numerous autophagosomes were visualized after TM or DTT treatment. To quantify the extent of autophagy activation, autophagosome numbers per root section were analyzed (Figure 1C). Compared with control conditions, significantly more autophagosomes were detected upon TM and DTT treatments. These data suggest that autophagy is induced by both TM and DTT treatment and, therefore, most likely by ER stress.
Figure 1.
Autophagy Is Activated in Response to ER Stress in Arabidopsis Roots.
(A) Seven-day-old GFP-ATG8e transgenic plants were transferred to MS liquid medium supplemented with 5 µg/mL TM, 2 mM DTT, or DMSO (Cont) for 8 h. GFP-ATG8e–labeled autophagosomes in root epidermal cells were visualized by confocal microscopy. Arrows indicate GFP-labeled autophagosomes or autophagic bodies. Bar = 10 μm.
(B) Seven-day-old wild-type (WT) or RNAi-ATG18a seedlings were transferred to MS liquid medium supplemented with 5 µg/mL TM or 2 mM DTT, or an equivalent amount of DMSO, for 8 h, followed by MDC staining, and roots were observed using confocal microscopy. Arrows indicate MDC-stained autophagosomes/autophagic bodies. Bar = 10 μm.
(C) The number of MDC-stained autophagosomes per root section was counted in control conditions or after DTT and TM treatment as above and the average number determined for 20 seedlings per treatment. Error bars represent se.
(D) Seven-day-old GFP-ATG8e seedlings were transferred to liquid MS medium plus DMSO (Cont), plus 1 µM concA (+concA), plus 5 µg/mL TM, or plus 5 µg/mL TM with 1 µM concA (TM+concA) for 12 h, followed by both fluorescence and DIC microscopy. Insets show enlargement of indicated boxes. Bars = 50 µm for main figure and 5 µm for insets.
(E) Seven-day-old seedlings were transferred to liquid MS medium plus 2 mM DTT plus or minus 20 mM imidazole/20 µM DPI for 8 h, plus 5 µg/mL TM plus or minus 20 mM imidazole/20 µM DPI for 8 h, or transferred to plates lacking nitrogen (−N) for 4 d and then to liquid MS medium plus or minus 20 mM imidazole/20 µM DPI for 8 h. Arrows indicate autophagosomes/autophagic bodies. Bar = 25 µm.
Previously, ATG18a was identified as an essential gene for autophagy induction under several abiotic stresses. Thus, RNA interference (RNAi)-ATG18a transgenic plants showed an autophagy-defective phenotype (Xiong et al., 2005, 2007b; Liu et al., 2009). To investigate whether ATG18a is required for ER stress–induced autophagy, 7-d-old RNAi-ATG18a seedlings were subjected to ER stress as described above (Figure 1B). No autophagosomes were detected under control conditions or upon TM or DTT treatment; this indicates that ER stress–induced autophagy is dependent on the function of ATG18a and, therefore, most likely occurs via the classical autophagy pathway.
There are two possibilities to explain the increased number of autophagosomes seen under ER stress; either the ER stress leads to an increased rate of formation of autophagosomes, or the ER stress inhibits the delivery to or degradation of autophagosomes in the vacuole. To distinguish between these possibilities, the vacuolar H+-ATPase inhibitor concanamycin A (concA) was used to inhibit the degradation of autophagic bodies in the vacuole (Dröse et al., 1993; Yoshimoto et al., 2004). ConcA raises the vacuolar pH and inhibits trafficking of vacuolar proteins, preventing vacuolar protein degradation (Matsuoka et al., 1997; Yoshimoto et al., 2004). Autophagic bodies therefore accumulate in vacuoles after treatment with concA instead of being degraded (Yoshimoto et al., 2004). Seven-day-old GFP-ATG8e seedlings grown on Murashige and Skoog (MS) plates were transferred to liquid MS medium containing 1 µM concA or DMSO as a solvent control, 5 µg/mL TM, or 5 µg/mL TM + 1 µM concA for 12 h, then observed by both fluorescence microscopy using a fluorescein isothiocyanate filter to visualize the GFP signal and differential interference contrast (DIC) microscopy to image autophagic bodies directly (Figure 1D). In control conditions, the GFP signal was diffuse and the vacuole rarely contained any spherical structures in the DIC images. In the control+concA condition, GFP-ATG8e–labeled puncta and spherical structures were sometimes detected in the vacuole in certain cells since plants usually have a basal level of housekeeping autophagy (Inoue et al., 2006; Xiong et al., 2007a). After adding TM, several GFP-ATG8e–labeled puncta and spherical structures were observed, indicating the formation of autophagosomes. Compared with the control+concA sample, the majority of cells treated with both TM and concA showed a large number of GFP-ATG8e–labeled puncta and spherical structures in the vacuole, which have been shown previously to be autophagic bodies (Yoshimoto et al., 2004). This indicates that after adding TM and concA, autophagic bodies accumulate inside the vacuole instead of being degraded. Identical results were obtained using DTT instead of TM (see Supplemental Figure 1 online). Together, these results demonstrate that the increased number of autophagosomes present upon TM treatment is due to the increased formation of autophagosomes, rather than decreased degradation because of a deficiency in delivery to or fusion between autophagosomes and vacuoles.
ER Stress–Induced Autophagy Is Regulated by an NADPH Oxidase-Independent Pathway
Autophagy is regulated by either NADPH oxidase–dependent or –independent pathways, depending on the stress conditions (Liu et al., 2009). To determine whether ER stress–induced autophagy is dependent on NADPH oxidase, the NADPH oxidase inhibitors diphenyleneiodonium (DPI) and imidazole were used. Seven-day-old wild-type plants grown on MS plates were transferred to liquid MS medium plus DTT or TM, plus or minus imidazole or DPI, for 8 h (Figure 1E). As demonstrated previously, autophagy could be induced by nitrogen starvation (−N treatment), and imidazole or DPI inhibited this induction (–N + imidazole/DPI treatment) (Figure 1E) (Liu et al., 2009). Compared with control conditions, wild-type plants had substantially more autophagosomes when treated with DTT or TM as shown above. The addition of imidazole or DPI together with DTT/TM did not block autophagy induction, in contrast with the inhibition seen during nitrogen starvation. This result implies that ER stress–induced autophagy is regulated by an NADPH oxidase–independent pathway.
ER Is Delivered to the Vacuole during ER Stress
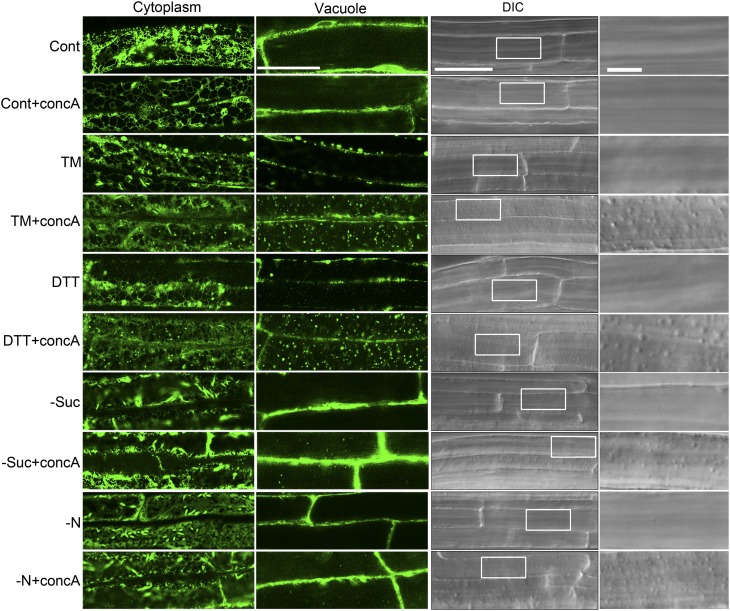
One role of autophagy during ER stress might be to degrade regions of the ER, thereby turning over some of the ER membrane and contents. To investigate the physical relationship between the autophagy pathway and the ER, the subcellular localization of both ER and autophagosome markers was analyzed in response to ER stress. GFP fused with the ER retention signal HDEL was used to label the ER (Batoko et al., 2000). To prevent vacuolar degradation, 1 µM concA or DMSO as carrier control was added to the corresponding stress-inducing liquid MS medium, followed by incubation for 12 h in the dark. Seedlings were imaged by confocal microscopy, with two focal planes shown for each sample: one through the cortical cytoplasm and one through the vacuole. In the control, confocal microscopy showed typical ER networks in the cytoplasm, and the vacuole was relatively devoid of any GFP signal (Figure 2, left panels). Likewise, few spherical structures were observed in DIC images of the vacuole in the controls (Figure 2, right panels). The concA-treated sample also showed typical ER patterns in the cytoplasm and no GFP signal in the vacuole, although in some cells small numbers of spherical structures were observed in the DIC images.
Figure 2.
The Localization of GFP-HDEL Changes during ER Stress in Arabidopsis Roots.
Seven-day-old GFP-HDEL transgenic plants were transferred to liquid MS medium with or without 1 µM concA for 12 h (control conditions). To induce ER stress, 7-d-old GFP-HDEL seedlings were transferred to liquid MS medium plus 2 mM DTT or 5 µg/mL TM with or without 1 µM concA for 12 h. To induce starvation stress, seedlings were first transferred to MS plates lacking Suc or nitrogen for 4 d, then transferred to MS liquid medium lacking Suc (−Suc) or nitrogen (−N), with or without 1 µM concA for 12 h. DMSO was used as a solvent control for all experiments. The plants were then observed with both confocal microscopy (left panels) and DIC microscopy (right panels). Far-right panels show enlarged boxes. Bars = 50 µm for main figures and 10 µm for insets.
[See online article for color version of this figure.]
To induce ER stress, 7-d-old GFP-HDEL transgenic Arabidopsis seedlings were treated with TM or DTT for 8 h. With either treatment, the typical ER structure was still observed in the cytoplasm, although some punctate structures were occasionally seen. Vacuoles were still largely devoid of any GFP signal, and spherical structures were rarely seen in vacuoles by DIC imaging, similar to the control. However, upon TM+concA or DTT+concA treatment, some of the signal from GFP-HDEL appeared as numerous dots, and GFP punctate structures and spherical structures in the DIC images, which morphologically resemble autophagic bodies, greatly increased in number. For comparison, 7-d-old GFP-HDEL plants grown on MS plates were transferred to MS plates lacking Suc (−Suc) or nitrogen (−N) for 4 d to cause starvation stress, a well-characterized autophagy-activating condition. In the –Suc and –N conditions, the GFP signal also appeared as an ER network in the cytoplasm and the spherical structures were rarely seen in the vacuole. Interestingly, in –Suc+concA and –N+concA conditions, increased numbers of spherical structures were observed in the DIC images because autophagy is induced under starvation conditions (Contento et al., 2005), whereas the GFP signal mainly showed an ER pattern in the cytoplasm and only a few GFP punctate structures were observed in the vacuole. These observations suggest that ER is transported to the vacuole via an autophagy-like pathway during ER stress but not largely transported to the vacuole during nutrient starvation conditions.
ER Is a Substrate of the Autophagy Pathway during ER Stress
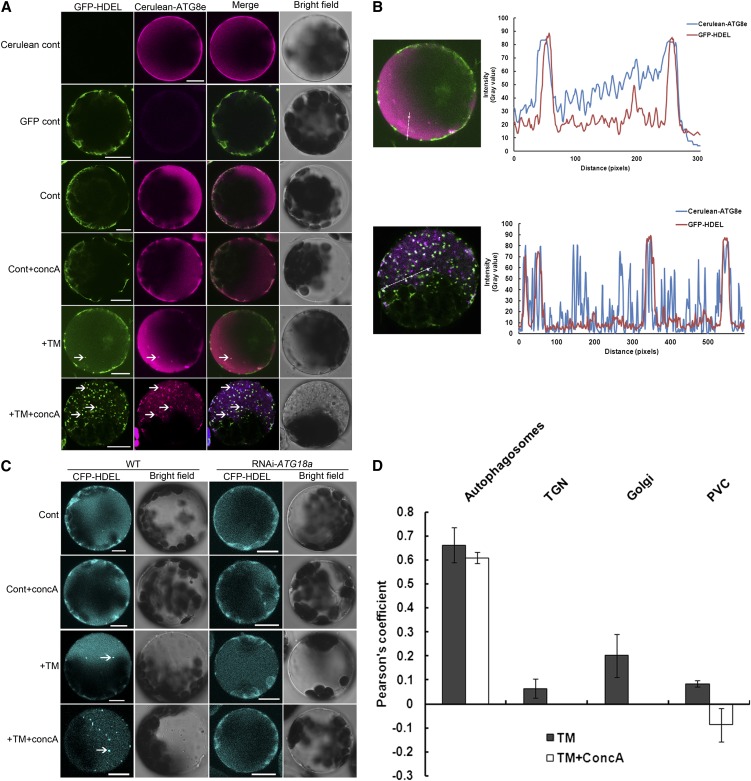
To test whether the spherical structures that deliver ER to the vacuole are autophagosomes, leaf protoplasts obtained from 4-week-old GFP-HDEL plants were transformed with a cerulean-ATG8e fusion construct, then incubated in the dark for 12 h to allow gene expression. Confocal microscopy was performed to visualize the subcellular localization of both GFP-HDEL and cerulean-ATG8e (Figure 3A). (Note: The plant protoplasts expressing only GFP-HDEL or cerulean-ATG8e displayed little fluorescence contamination between these two individual signals.) In the controls, GFP-HDEL showed a typical cytoplasmic ER pattern, and the cerulean-ATG8e gave a diffuse cytosolic signal. In the control+concA condition, most cells also showed the GFP-labeled ER network and a cytosolic cerulean signal, although a few cells had GFP and cerulean puncta in the vacuole likely due to a basal level of autophagy. In the presence of TM, both the cerulean- and the GFP-labeled puncta appeared in most of the cells, and some of the cerulean puncta and GFP puncta colocalized with each other. After incubation with both TM and ConcA, in most of the cells, numerous GFP puncta appeared as shown previously in planta, and the cerulean-ATG8e was also found in multiple puncta. Many of the structures labeled with these two markers colocalized, and in the bright-field images, these puncta corresponded to small vesicles in the vacuole.
Figure 3.
Autophagy Delivers ER to the Vacuole during ER Stress in Arabidopsis Leaf Protoplasts.
(A) Leaf protoplasts obtained from 4-week-old GFP-HDEL plants were transformed with a cerulean-ATG8e fusion construct. For control conditions, the protoplasts were incubated in W5 solution with or without 1 µM concA for 12 h. DMSO was used as a solvent control for all experiments. To induce ER stress, protoplasts were incubated in W5 solution plus 5 µg/mL TM with or without 1 µM concA for 12 h. Confocal microscopy was used to visualize the GFP and cerulean fluorescence. GFP-HDEL protoplasts lacking the cerulean-ATG8e construct were used as a GFP fluorescence control; wild-type protoplasts transformed with the cerulean-ATG8e construct were used as a cerulean fluorescence control. Arrows indicate autophagic bodies labeled with both GFP-HDEL and cerulean-ATG8e. Bar = 10µm.
(B) A line fluorescence tracing from images +TM and +TM+concA treatment in (A).
(C) Leaf protoplasts obtained from 4-week-old wild-type (WT) or RNAi-ATG18a plants were transformed with a CFP-HDEL fusion construct. For the control, the protoplasts were incubated in W5 solution with or without 1 µM concA for 12 h. To induce ER stress, the protoplasts were incubated in W5 solution plus 5 µg/mL TM with or without 1 µM concA for 12 h. Confocal microscopy was used to visualize CFP fluorescence. Arrows indicate CFP-HDEL–labeled structures inside the vacuole. Bars = 10µm.
(D) Pearson’s colocalization coefficient for GFP-HDEL and cerulean-ATG8e (autophagosomes), CFP-HDEL and ST-GFP (Golgi), CFP-HDEL and GFP-VHA1 (TGN), and CFP-HDEL and YFP-RHA1 (PVC). Pearson’s coefficient was derived from three independent experiments. Error bars represent se.
To analyze further and quantify the colocalization between the cerulean-ATG8e and the GFP-HDEL signals, the fluorescence patterns of the two signals were analyzed using the ImageJ software (Abramoff et al., 2004) (Figure 3B). The patterns obtained from both TM and TM+concA treatment showed significant overlap, and the Pearson coefficient relating the patterns (Figure 3D) also suggested that these two patterns positively correlate with each other. Together, these data suggest that during ER stress, ER is delivered to the vacuole through an ATG8e-containing vesicle, presumably an autophagosome.
Delivery of ER to the Vacuole Requires the Autophagy-Related Gene ATG18a
To test further the role of the autophagy pathway in delivering ER to the vacuole, similar experiments as described above were performed comparing wild-type and RNAi-ATG18a leaf protoplasts transiently expressing a cyan fluorescent protein (CFP)-HDEL fusion construct as an ER marker (Liu et al., 2007) (Figure 3C). Autophagosome formation is defective in RNAi-ATG18a plants, which thus can be used to test whether the loss of the autophagy pathway blocks ER transport to the vacuole during ER stress. In both wild-type and RNAi-ATG18a protoplasts, the CFP signal labeled an ER membrane network in control conditions. After addition of concA, most of the wild-type protoplasts showed an ER pattern and a few showed some CFP puncta, but almost all RNAi-ATG18a protoplasts observed displayed an ER pattern without CFP puncta. After adding TM, CFP puncta were observed in a majority of wild-type protoplasts, but not in RNAi-ATG18a protoplasts. After adding both TM and concA to the medium, most of the wild-type protoplasts observed accumulated numerous CFP puncta inside the vacuole; however, RNAi-ATG18a protoplasts still displayed the ER pattern, suggesting that delivery of ER to the vacuole is blocked when autophagy is defective. These results indicate that the delivery of ER to the vacuole is dependent on the autophagy-related gene ATG18a and this, when taken together with the colocalization between the ER marker and the autophagosome marker, suggests that the ER is delivered in autophagosomes.
ER Marker Does Not Colocalize with Other Endomembrane Markers during ER Stress
Two major vacuolar trafficking routes have been identified in plants: the autophagy pathway and the biosynthetic pathway, which involves the Golgi, the trans-Golgi network (TGN), and the prevacuolar compartment (PVC) (Sanderfoot et al., 1998; Törmäkangas et al., 2001). To investigate whether the ER structures identified in the vacuole can also be transported through the biosynthetic pathway, the subcellular colocalization between the ER marker and the Golgi marker ST-GFP (Boevink et al., 1998) (see Supplemental Figure 2 online), the TGN marker GFP-VHA1 (Dettmer et al., 2006) (see Supplemental Figure 3 online), and the PVC marker YFP-RHA1 (Preuss et al., 2004) (see Supplemental Figure 4 online) were also analyzed using Arabidopsis leaf protoplasts as described above. Both the confocal images and the Pearson coefficient (Figure 3D) suggested that the punctate ER fluorescence signal does not colocalize with the Golgi, the TGN, and the PVC structures. The TM+concA condition was not analyzed for colocalization with ST-GFP or GFP-VHA1 because concA disrupts Golgi and TGN structures (Dettmer et al., 2006). These results indicate that the ER structures identified in the vacuole during ER stress are not transported via the Golgi, TGN, and the PVC pathway.
Structure of ER Stress–Induced Autophagosomes
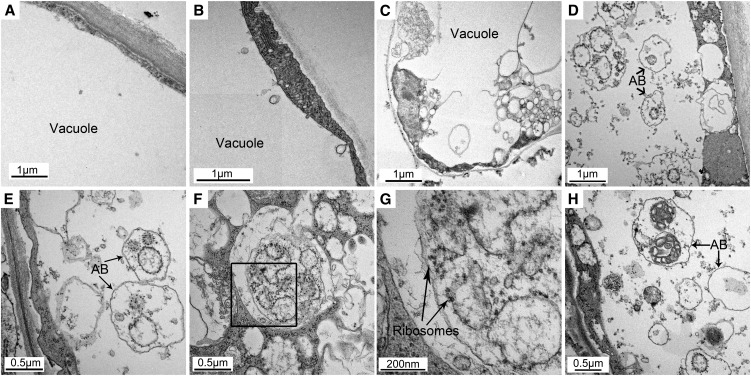
Although it can be interpreted that the colocalization of ER puncta with an autophagosome marker indicates that ER is transported to the vacuole by autophagosomes, previous studies in animals showed that the autophagosome membrane can be derived from ER membrane (Hayashi-Nishino et al., 2009; Ylä-Anttila et al., 2009) in which case one might observe a similar pattern of colocalization. To clarify whether ER is delivered by autophagosomes or is a component of the autophagosome membrane during ER stress, electron microscopy was performed to examine the detailed structures of ER stress–induced autophagosomes (Figure 4). In the controls, a few small vesicles were observed in both the vacuoles and the cytoplasm (Figure 4A). In the control+concA condition, there were greater numbers of vesicles observed in the vacuole due to the inhibition of vacuolar degradation (Figure 4B). In response to TM treatment, numerous vesicles appeared in the cytoplasm, but the contents of the vesicles were difficult to identify (Figure 4C). This might be due to the fusion of autophagosomes with a smaller lysosome-like or endosome-like organelle, leading to the degradation of the contents in the autophagosomes before fusion with the vacuole (Rose et al., 2006; Toyooka et al., 2006; Bassham, 2007). Following treatment with TM+concA, a multitude of small vesicles were observed in both the cytoplasm and the vacuole (Figure 4D). Since concA was used to inhibit vacuolar degradation, the contents inside the autophagic bodies could be identified. These small vesicles contained a variety of cargos, some with unidentified cytoplasmic contents (Figure 4H), whereas many had membrane structures decorated with electron-dense ribosomes (Figures 4E to 4G), typical of ER. Not all autophagosomes or autophagic bodies contained ER, consistent with the partial colocalization seen by confocal microscopy. From this we conclude that ER membranes had been engulfed by autophagosomes. Together, these results imply that ER is transported to the vacuole for degradation via the autophagy pathway during ER stress.
Figure 4.
ER Membranes Are Engulfed by Autophagosomes during ER Stress.
Seven-day-old wild-type seedlings were transferred to MS liquid medium without (A) or with (B) 1 µM concA or MS liquid medium supplemented with 5 µg/ml TM without (C) or with ([D] to [H]) 1 µM concA for 12 h, followed by electron microscopy analysis of root cells. AB, autophagic bodies.
(E) and (F) Autophagic bodies with ribosome-decorated membranes inside.
(G) Enlargement of a section indicted in (F).
(H) Autophagic bodies with unidentified content inside.
IRE1b Is Required for ER Stress–Induced Autophagy
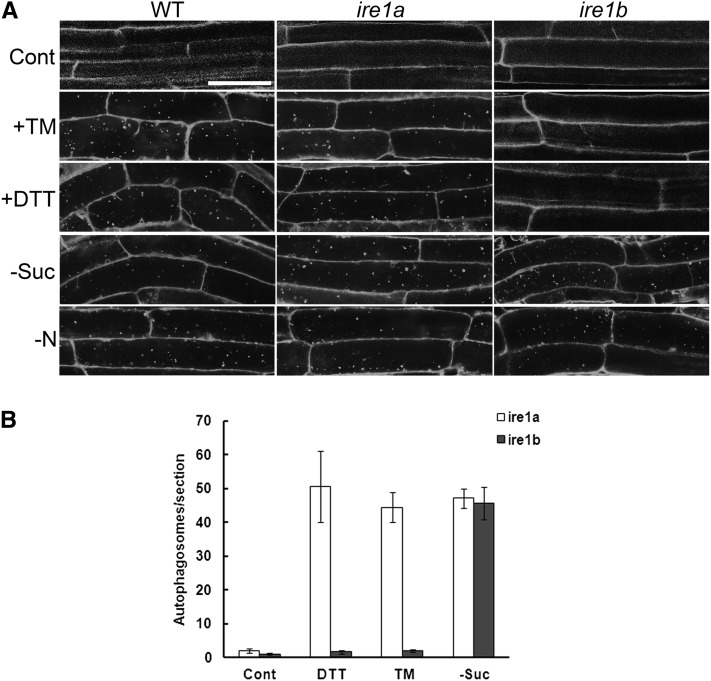
In Arabidopsis, IRE1 has been identified as an ER stress sensor. IRE1 senses ER stress and splices the mRNA encoding bZIP60, which is a bZIP-containing transcription factor implicated in the UPR in plants (Deng et al., 2011). There are two members of the IRE1 gene family in Arabidopsis, IRE1a and IRE1b (Koizumi et al., 2001), and IRE1b is most highly expressed in seedlings (http://bbc.botany.utoronto.ca/efp/cgi-bin/efpWeb.cgi?dataSource=Developmental_Map) and is primarily responsible for bZIP60 splicing in seedlings (Deng et al., 2011). To investigate whether ER stress–induced autophagy is activated via IRE1 genes, the induction of autophagy was examined in ire1a or ire1b null mutants (Humbert et al., 2012) compared with wild-type plants. Seven-day-old ire1a, ire1b, and wild-type plants grown on MS plates were transferred to MS liquid medium plus DMSO for 8 h as a control, MS liquid medium supplemented with 5 µg/mL TM, or 1 mM DTT for 8 h to induce ER stress, or MS plates lacking Suc (−Suc) or nitrogen (−N) for 4 d to induce starvation stresses, followed by MDC staining (Figure 5A). In control conditions, all three genotypes showed very few autophagosomes. Both wild-type and ire1a plants showed elevated autophagy during TM or DTT treatment and starvation conditions (–Suc and −N). Autophagy was also induced in ire1b during starvation conditions (−Suc and −N), but autophagy induction was not observed in response to DTT or TM treatment.
Figure 5.
ER Stress–Induced Autophagy Is Dependent on IRE1b Function in Arabidopsis Roots.
(A) Seven-day-old wild-type (WT), ire1a, and ire1b plants were transferred to MS liquid medium plus DMSO for 8 h as a control, MS liquid medium supplemented with 5 µg/mL TM or 2 mM DTT for 8 h to induce ER stress, or MS plates lacking Suc (−Suc) or nitrogen (−N) for 4 d to induce starvation stress, followed by MDC staining and confocal microscopy. Bar = 50µm.
(B) The number of MDC-stained autophagosomes per root section was counted after DTT and TM treatment as above and the average number determined for 20 seedlings per treatment. Error bars represent se.
To quantify these observations, autophagosome numbers were analyzed per root section for both ire1a and ire1b mutants (Figure 5B). The ire1a mutant responded to both ER stress (DTT or TM) treatment and the starvation stress, whereas the ire1b mutant responded to the starvation stress but showed significantly lower autophagosome numbers during ER stress treatment. To confirm further the MDC staining result, concA was used to prevent vacuolar degradation of autophagic bodies, which were visualized by DIC microscopy. Seven-day-old ire1a, ire1b, and wild-type plants were transferred to ER stress (+TM and +DTT) or starvation (−Suc and −N) conditions as described above, plus concA or an equal volume of DMSO as a solvent control, followed by DIC image analysis (see Supplemental Figure 5 online). In the absence of concA, ire1a, ire1b, and wild-type plants all displayed few spherical structures in the vacuole in almost all conditions tested. In the control+concA condition, all three types of plants showed a small number of spherical structures, predominantly corresponding to autophagic bodies, in the vacuole in some cells as described above. In the TM+concA or DTT+concA conditions, a substantial increase in spherical structures was observed in both wild-type and ire1a plants, but not in ire1b plants, which had a similar number as in control+concA conditions. In −Suc+concA or –N+concA conditions, all three types of plants showed an increased level of spherical structure accumulation compared with the control+concA condition. The DIC image analysis together with the MDC staining results indicate that IRE1b, and not IRE1a, is required for ER stress–induced autophagy but not for starvation-induced autophagy in roots.
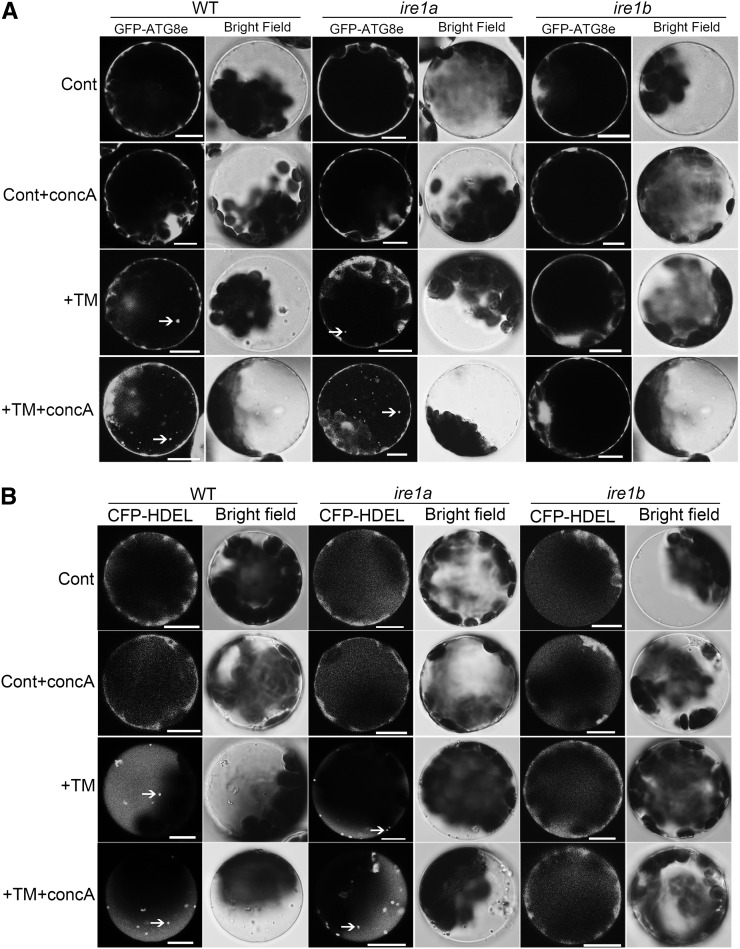
To confirm further the MDC and DIC results obtained in seedling roots, 4-week-old leaf protoplasts from wild-type, ire1a, and ire1b plants were transformed with a GFP-ATG8e fusion construct to visualize the induction of autophagy under ER stress by confocal microscopy (Figure 6A). In the control, the fluorescence from GFP-ATG8e was diffuse in all three types of plant protoplasts. In the control+concA condition, most of the cells observed displayed a diffuse signal and occasionally GFP puncta were detected. In response to TM treatment, most of the wild-type and ire1a plants contained GFP-ATG8e labeled puncta, indicating the induction of autophagy, whereas the GFP signal was diffuse in ire1b, indicating no autophagosome formation. In the TM+concA condition, most of the wild-type and ire1a protoplasts observed showed substantial accumulation of GFP puncta, but ire1b plants behaved similarly as in the control+concA condition. These results again suggest that the ER stress–induced autophagy is dependent on IRE1b.
Figure 6.
ER Stress–Induced Autophagy Is Dependent on IRE1b Function in Arabidopsis Leaf Protoplasts.
Leaf protoplasts obtained from 4-week-old wild-type (WT), ire1a, and ire1b plants were transformed with GFP-ATG8e (A) or CFP-HDEL (B) fusion constructs. As a control, protoplasts were incubated in W5 solution with or without 1 µM concA for 12 h. To induce ER stress, the protoplasts were incubated in W5 solution plus 5 µg/mL TM with or without 1 µM concA for 12 h. DMSO was used as a solvent control. Confocal microscopy was used to visualize the GFP and CFP fluorescence. Arrows indicate autophagic bodies containing GFP-ATG8e or CFP-HDEL. Bars = 10 µm.
IRE1b Is Required for Transport of ER to the Vacuole
Next, the role of IRE1b in delivery of ER to the vacuole upon ER stress was analyzed using wild-type, ire1a, and ire1b leaf protoplasts transiently expressing a CFP-HDEL fusion construct (Figure 6B). In control conditions, the ER pattern was typical in all three types of plant protoplasts. In the control+concA treatment, most of the cells displayed an ER pattern and occasionally CFP-labeled puncta were detected. In the presence of TM, most of the wild-type and ire1a plants contained CFP-labeled puncta, whereas ire1b plants mainly showed a typical ER labeling pattern. In the TM+concA samples, most of the wild-type and ire1a plants observed showed substantial accumulation of CFP-HDEL puncta, but ire1b plants did not, behaving similarly as in control+concA conditions. These results imply that the delivery of ER to the vacuole is dependent on IRE1b but not on IRE1a. Together, the findings both in planta and in protoplasts suggest that IRE1b is required for ER stress–induced autophagy.
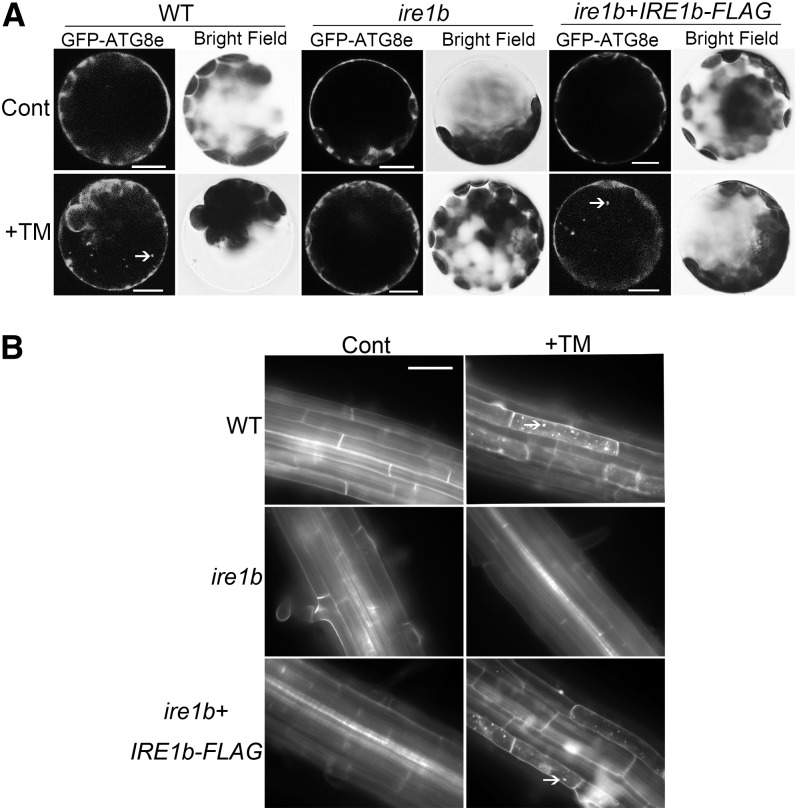
To confirm that the loss of autophagy induction during ER stress in the ire1b mutant was actually due to the lack of IRE1b gene function, autophagy induction was tested in both ire1b leaf protoplasts transiently expressing a FLAG-tagged IRE1b construct (Figure 7A) and transgenic lines expressing the IRE1b cDNA (IRE1b-FLAG) in the ire1b mutant background (Figure 7B; see Supplemental Figure 6 online). Wild-type leaf protoplasts transiently expressing GFP-ATG8e displayed a diffuse GFP signal in control conditions, and GFP puncta were observed in the presence of TM as expected (Figure 7A). ire1b leaf protoplasts transiently expressing GFP-ATG8e showed a diffuse GFP signal in both the control and after TM treatment. However, upon transformation of ire1b leaf protoplasts with both IRE1b-FLAG and GFP-ATG8e constructs, the GFP signal was diffuse in the control conditions, but GFP puncta were seen in the presence of TM, similar to wild-type protoplasts. To confirm these results in planta, 7-d-old wild-type, ire1b, and ire1b/IRE1b-FLAG complementation lines (see Supplemental Figure 6 online) were transferred to liquid MS medium supplemented with TM or DMSO, followed by MDC staining (Figure 7B). With TM treatment, both the wild type and IRE1b complemented plants contained significantly more MDC-labeled autophagosomes compared with control conditions, whereas in the ire1b mutant, few autophagosomes were detected in both control and in +TM conditions. Together, these results indicate that the defect for autophagy induction in ire1b in response to ER stress can be attributed to the loss of IRE1b gene function, rather than other defects in the autophagy pathway. This again suggests that ER stress–induced autophagy is dependent on the IRE1b gene.
Figure 7.
Defects in Autophagy Induction in ire1b during ER Stress Can Be Attributed to the Loss of IRE1b Gene Function.
(A) Leaf protoplasts obtained from 4-week-old wild-type (WT) or ire1b plants were transformed with the GFP-ATG8e fusion construct, or ire1b leaf protoplasts were transformed with both GFP-ATG8e and IRE1b-FLAG fusion constructs. For controls, the protoplasts were incubated in W5 solution. To induce ER stress, protoplasts were incubated in W5 solution plus 5 µg/mL TM. Arrows indicate GFP-ATG8e–labeled autophagic bodies.
(B) Seven-day-old wild-type, ire1b, and ire1b/IRE1b-FLAG seedlings were transferred to MS liquid medium supplemented with 5 µg/mL TM or DMSO as a solvent control for 8 h, followed by MDC staining and fluorescence microscopy. Arrows indicate MDC-stained autophagosomes and autophagic bodies.
bZIP60 Is Not Involved in Regulating ER Stress–Induced Autophagy
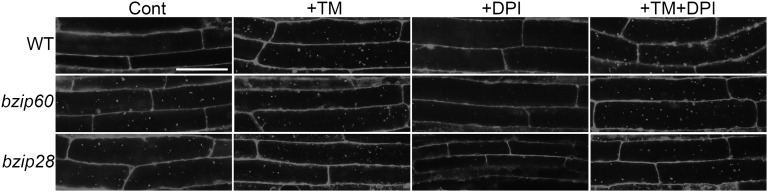
As discussed above, IRE1b splices bZIP60 mRNA to produce an active transcription factor, thus upregulating the UPR genes in plants (Deng et al., 2011). To test whether regulation of ER stress–induced autophagy by IRE1b occurs via IRE1b splicing of bZIP60, the induction of autophagy was examined in a bzip60 T-DNA insertion mutant (Deng et al., 2011). Seven-day-old wild-type and bzip60 plants grown on MS plates were transferred to MS liquid medium supplemented with TM or DSMO as a solvent control, followed by MDC staining (Figure 8). Unexpectedly, the bzip60 mutant showed constitutive autophagy even under control conditions. One explanation for the constitutive autophagy in the bzip60 mutant is that the loss of bZIP60 function causes constitutive ER stress, thus inducing autophagy. Alternatively, the loss in bZIP60 function may lead to general cellular stress, causing an increased level of basal autophagy. This complicated the testing of whether ER stress induces autophagy in bzip60, as autophagy seen upon TM treatment in bzip60 could either be increased basal autophagy or a mixture of the basal autophagy and TM induced autophagy.
Figure 8.
bZIP60 and bZIP28 Are Not Involved in Regulating ER Stress–Induced Autophagy.
Seven-day-old wild-type (WT), bzip60, and bzip28 plants grown on MS plates were transferred to MS liquid medium supplemented with 5 µg/mL TM, 20 µM DPI, or 5 µg/mL TM together with 20 µM DPI, with DMSO used as a solvent control. MDC staining of roots was performed to visualize autophagosomes. Bar = 50 µm.
To distinguish between these two possibilities, the NADPH oxidase inhibitor DPI was used to inhibit the general starvation and salt stress–induced autophagy pathway (Liu et al., 2009). As shown in Figure 1, the addition of an NADPH oxidase inhibitor does not block ER stress–induced autophagy. In the presence of DPI, no autophagy was seen in bzip60 (Figure 8), indicating that the constitutive autophagy observed in bzip60 is inhibited by DPI and is therefore most likely a general stress response and unrelated to ER stress. After adding both DPI and TM to the medium, wild-type plants still showed autophagy induction. Autophagosomes were also present in the bzip60 mutant in the presence of DPI and TM, which suggests that after DPI inhibition of the enhanced basal autophagy, an alternative pathway for activation of ER stress–induced autophagy was still active. These data indicate that autophagy can still be induced by ER stress in the bzip60 mutant and, therefore, that bZIP60 is not required for ER stress–induced autophagy. Thus, ER stress–induced autophagy is regulated by IRE1b but is not dependent on the downstream factor bZIP60.
bZIP28 Is Not Involved in Regulating ER Stress–Induced Autophagy
Animal cells contain another two ER stress sensors, ATF6 and PERK, in addition to IRE1. Cells lacking ATF6 or PERK are capable of autophagy induction in response to ER stress (Ogata et al., 2006). In plants, bZIP28 may be functionally equivalent to ATF6, whereas PERK signaling has not been demonstrated in plants (Liu et al., 2007; Tajima et al., 2008). To investigate whether bZIP28 is involved in ER stress–induced autophagy, a bzip28-2 knockout mutant (Liu et al., 2007; Che et al., 2010) was tested for autophagy induction during ER stress. Similar experiments as described above for bzip60 were performed with 7-d-old bzip28-2 plants (Figure 8). The bzip28-2 plants displayed constitutive autophagy even in control conditions. This constitutive autophagy was inhibited by the addition of the NADPH oxidase inhibitor DPI, indicating that the constitutive autophagy seen in the bzip28-2 mutant was most likely a general stress response and unrelated to ER stress. After adding both DPI and TM to the medium, autophagosomes were present in bzip28-2, indicating that after the inhibition of general stress-induced autophagy, autophagy can still be induced by ER stress in the bzip28-2 mutant. These data imply that, like bZIP60, bZIP28 is not required for ER stress–induced autophagy.
DISCUSSION
Although a number of studies have focused on UPR signaling pathways in plants, little is understood about ER morphology changes in response to ER stress or as mediated by the UPS-independent ERAD pathway (Urade, 2007, 2009; Moreno and Orellana, 2011). Previously, plant autophagy had been shown to be involved in senescence, nutrient deprivation, oxidative stress, salt and drought stresses, and pathogen infection (Doelling et al., 2002; Hanaoka et al., 2002; Liu et al., 2005, 2009; Xiong et al., 2005, 2007b). In this article, we demonstrate that autophagy is activated in the response to ER stress in plants. MDC staining and GFP-ATG8e transgenic plants showed autophagy induction after TM or DTT treatment. In addition, portions of the ER are engulfed by autophagosomes and delivered to the vacuole for degradation. Together, this evidence implicates autophagy in ER turnover in response to ER stress. To investigate the upstream signaling pathway that activates ER stress–induced autophagy, a mutant lacking one of the ER stress sensors, IRE1b, was tested for autophagy induction upon ER stress. Leaf protoplasts transiently expressing CFP-HDEL or GFP-ATG8e indicated that IRE1b is required for ER stress–induced autophagy. To characterize further the IRE1b-dependent autophagy pathway, a mutant lacking the splicing target of IRE1b, bZIP60, was also analyzed. The bzip60 mutant was capable of inducing autophagy in response to ER stress, suggesting that ER stress–induced autophagy does not rely on the splicing activity of IRE1b.
Our data identified IRE1b as an upstream component of ER stress–induced autophagy in Arabidopsis seedlings. However, we cannot exclude the possibility that IRE1a could also be involved in autophagy. According to the eFP Browser (http://bbc.botany.utoronto.ca/efp/cgi-bin/efpWeb.cgi?dataSource=Developmental_Map), the expression of IRE1a is quite limited in vegetative tissues. Thus, by analyzing root tissues and protoplasts, we may not have been able to assess the contribution by IRE1a simply because it is not highly expressed in roots. IRE1a plays newly recognized roles in plant defense responses, so it will be interesting to determine whether those responses also involve autophagy (Moreno et al., 2012). There is some discrepancy in the literature about the extent to which the roles of IRE1a and IRE1b overlap, which may be due to allelic differences in the mutants used in the different studies (Deng et al., 2011; Nagashima et al., 2011; Chen and Brandizzi, 2012).
The detailed molecular mechanism of regulation of ER stress–induced autophagy is yet to be determined. In yeast, ER stress–induced autophagy is regulated through the IRE1 endoribonuclease activity toward HAC1 mRNA (Yorimitsu et al., 2006). In animals, IRE1 is also required for autophagy induction; however, the IRE1 kinase activity-mediated c-Jun N-terminal kinase pathway, which is absent in plants, rather than the splicing activity toward XBP1 seems to control autophagy induction (Urano et al., 2000; Ogata et al., 2006). Our results showed that in plants, ER stress–induced autophagy is dependent on IRE1b, suggesting a conserved role for the IRE1 gene during autophagy induction from yeast to animals and plants. Similar to animals, autophagy does not depend on the IRE1b downstream splicing target, which in the case of Arabidopsis is bZIP60 (Deng et al., 2011). As bZIP60 is the only known target of IRE1b ribonuclease activity, this suggests that either (1) IRE1b has additional splicing targets that have yet to be discovered that regulate autophagy activation or (2) other activities associated with IRE1b may be responsible for the autophagy induction in response to ER stress, rather than its splicing activity. However, other functions of IRE1b in addition to the splicing of bZIP60 mRNA have not been discovered to date (Deng et al., 2011). This suggests that a distinct, previously undiscovered, signaling pathway functions in activation of autophagy during ER stress in Arabidopsis. To elucidate further the role of IRE1 in autophagy induction, more experiments are needed to identify its substrates and downstream signals. Intriguingly, the loss of XBP1 in Drosophila melanogaster causes constitutive autophagy (Arsham and Neufeld, 2009), similar to that of the bzip60 and bzip28 mutants observed here. The authors suggest that the absence of XBP1 activity may lead to accumulation of unfolded proteins, triggering XBP1-independent UPR signaling. Whether this happens in plant cells still needs to be determined.
Target of rapamycin (TOR) has been shown to be a negative regulator of autophagy from yeast to animals and plants (Díaz-Troya et al., 2008; Liu and Bassham, 2010). TOR regulates the downstream ATG1 kinase complex, recently characterized in Arabidopsis (Suttangkakul et al., 2011), which in turn regulates the initiation of autophagy (Díaz-Troya et al., 2008). Several studies have shown an interplay between ER stress and mTOR signaling in animals; for example, constitutive activation of mTOR leads to ER stress (Ozcan et al., 2008) and chaperone availability controls mTOR signaling (Qian et al., 2010). It was also suggested that ER stress induces autophagy through the inactivation of mTOR (Qin et al., 2010). However, whether TOR is associated with the control of autophagy during ER stress in plants is still unknown. A recent study in Chlamydomonas reinhardtii reported that the phosphorylation state of the BiP chaperone is regulated by TOR (Díaz-Troya et al., 2011). The authors showed that under ER stress when increased chaperone levels are needed, BiP protein is dephosphorylated, resulting in its activation. When protein synthesis was inhibited by downregulating TOR activity, BiP was phosphorylated to its inactive form (Díaz-Troya et al., 2011). These results indicate a potential TOR function in its interaction with the ER stress signal, thereby regulating both protein synthesis and the autophagy degradation pathway. However, how exactly TOR senses ER stress, and whether IRE1 fits into this pathway, still needs to be determined.
Generally, autophagy is a nonselective process; however, organelle-specific autophagy has been identified in both yeast and animals (Reumann et al., 2010). For example, the selective degradation of peroxisomes (pexophagy) (Hutchins et al., 1999), mitochondria (mitophagy) (Kim et al., 2007), ribosomes (ribophagy) (Kraft et al., 2008), and ER (ER-phagy) (Bernales et al., 2006) have been reported. In plants, organelle-specific autophagy has not been studied extensively. Nevertheless, increasing evidence is emerging for organelle-specific autophagy in plants, such as the degradation of ribosomes (Hillwig et al., 2011) and bodies containing ribulose-1,5-bisphosphate carboxylase/oxygenase (Ishida et al., 2008; Wada et al., 2009). However, whether the engulfment of ER by autophagosomes is a selective process is unknown. One possibility is that during ER stress, the ER begins to fragment, allowing it to be incorporated into autophagosomes nonselectively. Another possibility is that the autophagosome can recognize ER fragments containing misfolded proteins (Yorimitsu and Klionsky, 2007), therefore sequestering both the misfolded proteins and the ER membranes. Recent studies in animals suggest that p62/SEQUESTOSOME1 and NBR1 (for neighbor of BRCA1 gene) function as selective autophagy cargo receptors (Pankiv et al., 2007; Ichimura et al., 2008; Kirkin et al., 2009). Both p62 and NBR1 harbor an Atg8-interacting motif (AIM domain), a WXXL amino acid sequence that can be recognized by Atg8/LC3, and a UBA (for Ub associate) domain, which can interact with ubiquitinated proteins (Noda et al., 2010; Johansen and Lamark, 2011). It was proposed that upon ubiquitination of protein aggregates, the UBA domains of p62 and NBR1 recognize and bind to the ubiquitinated protein aggregates, then the AIM domains of p62 and NBR1 recruit Atg8/LC3, followed by the formation of autophagosomes (Johansen and Lamark, 2011). Similar mechanisms have been reported in plants. NBR1 homologs in both Arabidopsis and tobacco (Nicotiana tabacum) have been identified, and they both interact with ATG8 (Svenning et al., 2011; Zientara-Rytter et al., 2011). Therefore, it is possible that the autophagosome sequesters ER through identifying ER fragments containing protein aggregates or containing surface proteins tagged by ubiquitylation, as has been seen in the case of mitophagy (Ashrafi and Schwarz, 2012); however, more evidence is required before this conclusion can be drawn.
In this study, we provide another link between organelle degradation and autophagy by showing that the ER is a target of autophagy during ER stress in plants. Activities other than the splicing of bZIP60 by IRE1b may function as upstream events to regulate ER stress–induced autophagy. However, future experiments are needed to determine the downstream targets of IRE1b and the detailed regulation mechanisms in the ER stress–induced autophagy pathway.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana Columbia ecotype seeds were surface sterilized with 0.1% (v/v) Triton X-100 and 33% (v/v) bleach solution for 20 min, followed by cold treatment for at least 2 d. Arabidopsis seedlings were grown under long-day conditions (16 h light) at 22°C on nutrient solid MS medium (Murashige-Skoog vitamin and salt mixture [Caisson, MSPA0910], 1% [w/v] Suc, 2.4 mM MES, pH 5.7, and 0.8% [w/v] phytagar). T-DNA insertion mutants used in this study were ire1a (salk_018112), ire1b (sail_238_F07), bzip60-1 (salk_050203), and bzip28-2 (salk_132285). Transgenic plants used in this study have been described previously as follows: RNAi-ATG18a (Xiong et al., 2005), GFP-ATG8e (Xiong et al., 2007b), GFP-HDEL (Batoko et al., 2000), CFP-HDEL (Liu et al., 2007), GFP-VHA1 (Dettmer et al., 2006), YFP-RHA1 (Preuss et al., 2004), and ST-GFP (Boevink et al., 1998).
For TM and DTT treatment, 7-d-old seedlings grown on solid MS plates were transferred to liquid MS medium supplemented with 5 µg/mL TM or 2 mM DTT (Liu et al., 2007), or DMSO as a solvent control for the indicated times in the dark.
For starvation treatment, 7-d-old seedlings grown on solid MS plates were transferred to MS plates lacking Suc or nitrogen for an additional 4 d. Plants grown on Suc starvation plates were incubated in the dark. If concA treatment (see below) was also required, the seedlings were then transferred to liquid MS medium lacking Suc or nitrogen plus concA for 12 h in the dark.
For imidazole and DPI treatment, seedlings grown on solid MS plates were transferred to MS liquid medium plus or minus 20 mM imidazole or 20 µM DPI for the indicated times. The solvent for DPI was DMSO; an equivalent volume of DMSO was added to controls.
For concA treatment, seedlings grown on MS plates were transferred to MS liquid medium containing 1 µM concA or DMSO as a solvent control for 12 h in the dark. The roots were mounted in water and then observed by confocal, fluorescence, and DIC microscopy.
MDC Staining and Microscopy
Arabidopsis seedlings were stained with MDC as previously described (Contento et al., 2005). Seedlings were incubated with 0.05 mM MDC for 10 min, washed three times with PBS, and observed using a Zeiss Axioplan II compound microscope equipped with Axio Cam HRC digital imaging system (Carl Zeiss). MDC fluorescence was visualized using a 4′,6-diamidino-2-phenylindole–specific filter, and GFP fluorescence was visualized using a fluorescein isothiocyanate–specific filter. Confocal microscopy was performed with a Leica confocal microscope using a ×63 Leica oil immersion objective.
Generation of Cerulean-ATG8e Construct
The ATG8e cDNA was synthesized by RT-PCR from total RNA from 7-d-old seedlings grown on MS plates, using gene-specific primers (see Supplemental Table 1 online). The cDNA was sequenced for verification and ligated into the pAN578 vector using BglII and NotI restriction sites (Rizzo et al., 2004). Protoplasts transformed with the Cerulean-ATG8e construct were observed with a CFP-optimized filter.
Transient Transformation of Leaf Protoplasts
Arabidopsis leaf protoplasts were prepared and transformed according to Sheen (2002). Twenty micrograms of plasmid DNA was used for each transformation. Protoplasts were incubated at room temperature in darkness for 12 h, with 40 rpm orbital shaking. Confocal microscopy was performed with a Leica confocal microscope using a ×63 Leica oil immersion objective.
Pearson’s colocalization coefficients were derived using ImageJ software (Abramoff et al., 2004) with the “sync windows” plugin and the “plot profile” function. All Pearson’s coefficients were derived from three completely independent experiments.
Generation of IRE1b-FLAG Construct and Complementation of Transgenic Plants
IRE1b coding sequence was amplified from Columbia-0 cDNA using gene-specific primers. A 3XFLAG tag was added after the transmembrane domain of IRE1b by overlapping PCR. Primers used are listed in Supplemental Table 1 online. IRE1b-N primers were used to amplify the first half of the IRE1b gene up to and including the transmembrane domain, IRE1b-C primers were used to amplify the second half of the IRE1b gene after the transmembrane domain, and FLAG primers were used for the 3XFLAG tag. The IRE1b-3XFLAG DNA fragments were then ligated into the pSKM36 vector using AscI-SpeI restriction sites (Ikeda et al., 2006). This construct was introduced into Agrobacterium tumefaciens by electroporation (Mersereau et al., 1990) and then into ire1b plants by Agrobacterium-mediated transformation using the floral dipping method (Clough and Bent, 1998). Transgenic plants were identified by kanamycin resistance. Individuals from the T2 generation were used for further studies.
Electron Microscopy Analysis
Electron microscopy was performed at the Iowa State University Microscopy and NanoImaging Facility. For transmission electron microscopy, samples were collected and fixed with 2% glutaraldehyde (w/v) and 2% paraformaldehyde (w/v) in 0.1 M cacodylate buffer, pH 7.2, for 48 h at 4°C. Samples were rinsed three times in 0.1 M cacodylate buffer and then postfixed in 1% osmium tetroxide in 0.1 M cacodylate buffer for 1 h (room temperature). The samples were rinsed in deionized distilled water and enbloc stained with 2% aqueous uranyl acetate for 30 min, dehydrated in a graded ethanol series, cleared with ultra-pure acetone, infiltrated, and embedded using Spurr’s epoxy resin (Electron Microscopy Sciences). Resin blocks were polymerized for 48 h at 65°C. Ultrathin sections were made using a Reichert UC6 ultramicrotome (Leeds Precision Instruments), followed by collection onto copper grids and counterstaining with 2% uranyl acetate in deionized distilled water for 30 min. Images were captured using a JEOL 2100 scanning and transmission electron microscope (Japan Electron Optic Laboratories).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: At2g17520 (IRE1a), At5g24360 (IRE1b), At3g10800 (bZIP28), At1g42990 (bZIP60), and At3g62770 (ATG18a).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Autophagy Is Activated in the Presence of DTT.
Supplemental Figure 2. CFP-HDEL Does Not Colocalize with Golgi Structures during ER Stress.
Supplemental Figure 3. CFP-HDEL Does Not Colocalize with a TGN Marker during ER Stress.
Supplemental Figure 4. CFP-HDEL Does Not Colocalize with a PVC Marker during ER Stress.
Supplemental Figure 5. Autophagosomes Do Not Accumulate in ire1b Roots in Response to ER Stress.
Supplemental Figure 6. 35S-IRE1b-Flag Partially Complements an ire1b Mutant.
Supplemental Table 1. Primers Used for Generation of the Cerulean-ATG8e and IRE1b-FLAG Constructs and for PCR.
Acknowledgments
We thank Harry (Jack) T. Horner, Randall Den Adel, and Tracey M. Pepper for assistance with the fluorescence, DIC, and the electron microscopy and Margaret Carter for assistance with confocal microscopy. We also thank Ian Moore for the GFP-HDEL transgenic plant, Karin Schumacher, Chris Hawes, and Erik Nielsen for constructs, and Anthony L. Contento for generating the cerulean-ATG8e construct. This research was supported by Grants IOB-0515998 and MCB-1051818 from the National Science Foundation to D.C.B.
AUTHOR CONTRIBUTIONS
Y.L., S.H.H., and D.C.B. designed research. Y.L., J.S.B., Y.D., and R.S. performed research. Y.L., S.H.H., and D.C.B. analyzed data. Y.L., S.H.H., and D.C.B. wrote the article.
Glossary
- ER
endoplasmic reticulum
- UPR
unfolded protein response
- ERAD
ER-associated degradation
- bZIP
basic domain/leucine zipper
- PERK
protein kinase–like ER kinase
- UPS
ubiquitin-proteasome system
- TM
tunicamycin
- GFP
green fluorescent protein
- MDC
monodansylcadaverine
- RNAi
RNA interference
- concA
concanamycin A
- MS
Murashige and Skoog
- DIC
differential interference contrast
- DPI
diphenyleneiodonium
- CFP
cyan fluorescent protein
- TGN
trans-Golgi network
- PVC
prevacuolar compartment
References
- Abramoff M., Magalhaes P., Ram S. (2004). Image Processing with ImageJ. Biophotonics International 11: 36–42 [Google Scholar]
- Arsham A.M., Neufeld T.P. (2009). A genetic screen in Drosophila reveals novel cytoprotective functions of the autophagy-lysosome pathway. PLoS ONE 4: e6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashrafi G., Schwarz T.L. (June 29, 2012). The pathways of mitophagy for quality control and clearance of mitochondria. Cell Death Differ. http://dx.doi.org/.10.1038/cdd.2012.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassham D.C. (2007). Plant autophagy—More than a starvation response. Curr. Opin. Plant Biol. 10: 587–593 [DOI] [PubMed] [Google Scholar]
- Batoko H., Zheng H.Q., Hawes C., Moore I. (2000). A rab1 GTPase is required for transport between the endoplasmic reticulum and Golgi apparatus and for normal Golgi movement in plants. Plant Cell 12: 2201–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernales S., McDonald K.L., Walter P. (2006). Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol. 4: e423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boevink P., Oparka K., Santa Cruz S., Martin B., Betteridge A., Hawes C. (1998). Stacks on tracks: The plant Golgi apparatus traffics on an actin/ER network. Plant J. 15: 441–447 [DOI] [PubMed] [Google Scholar]
- Calfon M., Zeng H., Urano F., Till J.H., Hubbard S.R., Harding H.P., Clark S.G., Ron D. (2002). IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415: 92–96 [DOI] [PubMed] [Google Scholar]
- Che P., Bussell J.D., Zhou W., Estavillo G.M., Pogson B.J., Smith S.M. (2010). Signaling from the endoplasmic reticulum activates brassinosteroid signaling and promotes acclimation to stress in Arabidopsis. Sci. Signal. 3: ra69. [DOI] [PubMed] [Google Scholar]
- Chen Y., Brandizzi F. (2012). AtIRE1A/AtIRE1B and AGB1 independently control two essential unfolded protein response pathways in Arabidopsis. Plant J. 69: 266–277 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Contento A.L., Xiong Y., Bassham D.C. (2005). Visualization of autophagy in Arabidopsis using the fluorescent dye monodansylcadaverine and a GFP-AtATG8e fusion protein. Plant J. 42: 598–608 [DOI] [PubMed] [Google Scholar]
- Cox J.S., Shamu C.E., Walter P. (1993). Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell 73: 1197–1206 [DOI] [PubMed] [Google Scholar]
- Cox J.S., Walter P. (1996). A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell 87: 391–404 [DOI] [PubMed] [Google Scholar]
- Deng Y., Humbert S., Liu J.X., Srivastava R., Rothstein S.J., Howell S.H. (2011). Heat induces the splicing by IRE1 of a mRNA encoding a transcription factor involved in the unfolded protein response in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 7247–7252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettmer J., Hong-Hermesdorf A., Stierhof Y.D., Schumacher K. (2006). Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell 18: 715–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Troya S., Pérez-Pérez M.E., Florencio F.J., Crespo J.L. (2008). The role of TOR in autophagy regulation from yeast to plants and mammals. Autophagy 4: 851–865 [DOI] [PubMed] [Google Scholar]
- Díaz-Troya S., Pérez-Pérez M.E., Pérez-Martín M., Moes S., Jeno P., Florencio F.J., Crespo J.L. (2011). Inhibition of protein synthesis by TOR inactivation revealed a conserved regulatory mechanism of the BiP chaperone in Chlamydomonas. Plant Physiol. 157: 730–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doelling J.H., Walker J.M., Friedman E.M., Thompson A.R., Vierstra R.D. (2002). The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J. Biol. Chem. 277: 33105–33114 [DOI] [PubMed] [Google Scholar]
- Dröse S., Bindseil K.U., Bowman E.J., Siebers A., Zeeck A., Altendorf K. (1993). Inhibitory effect of modified bafilomycins and concanamycins on P- and V-type adenosinetriphosphatases. Biochemistry 32: 3902–3906 [DOI] [PubMed] [Google Scholar]
- Gardner B.M., Walter P. (2011). Unfolded proteins are Ire1-activating ligands that directly induce the unfolded protein response. Science 333: 1891–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanaoka H., Noda T., Shirano Y., Kato T., Hayashi H., Shibata D., Tabata S., Ohsumi Y. (2002). Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol. 129: 1181–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding H.P., Zhang Y., Bertolotti A., Zeng H., Ron D. (2000). Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol. Cell 5: 897–904 [DOI] [PubMed] [Google Scholar]
- Hayashi-Nishino M., Fujita N., Noda T., Yamaguchi A., Yoshimori T., Yamamoto A. (2009). A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat. Cell Biol. 11: 1433–1437 [DOI] [PubMed] [Google Scholar]
- Haze K., Yoshida H., Yanagi H., Yura T., Mori K. (1999). Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol. Biol. Cell 10: 3787–3799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillwig M.S., Contento A.L., Meyer A., Ebany D., Bassham D.C., Macintosh G.C. (2011). RNS2, a conserved member of the RNase T2 family, is necessary for ribosomal RNA decay in plants. Proc. Natl. Acad. Sci. USA 108: 1093–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humbert S., Zhong S., Deng Y., Howell S.H., Rothstein S.J. (2012). Alteration of the bZIP60/IRE1 pathway affects plant response to ER stress in Arabidopsis thaliana. PLoS ONE 7: e39023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins M.U., Veenhuis M., Klionsky D.J. (1999). Peroxisome degradation in Saccharomyces cerevisiae is dependent on machinery of macroautophagy and the Cvt pathway. J. Cell Sci. 112: 4079–4087 [DOI] [PubMed] [Google Scholar]
- Ichimura Y., Kumanomidou T., Sou Y.S., Mizushima T., Ezaki J., Ueno T., Kominami E., Yamane T., Tanaka K., Komatsu M. (2008). Structural basis for sorting mechanism of p62 in selective autophagy. J. Biol. Chem. 283: 22847–22857 [DOI] [PubMed] [Google Scholar]
- Ikeda Y., Banno H., Niu Q.W., Howell S.H., Chua N.-H. (2006). The ENHANCER OF SHOOT REGENERATION 2 gene in Arabidopsis regulates CUP-SHAPED COTYLEDON 1 at the transcriptional level and controls cotyledon development. Plant Cell Physiol. 47: 1443–1456 [DOI] [PubMed] [Google Scholar]
- Inoue Y., Suzuki T., Hattori M., Yoshimoto K., Ohsumi Y., Moriyasu Y. (2006). AtATG genes, homologs of yeast autophagy genes, are involved in constitutive autophagy in Arabidopsis root tip cells. Plant Cell Physiol. 47: 1641–1652 [DOI] [PubMed] [Google Scholar]
- Ishida H., Yoshimoto K., Izumi M., Reisen D., Yano Y., Makino A., Ohsumi Y., Hanson M.R., Mae T. (2008). Mobilization of rubisco and stroma-localized fluorescent proteins of chloroplasts to the vacuole by an ATG gene-dependent autophagic process. Plant Physiol. 148: 142–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata Y., Fedoroff N.V., Koizumi N. (2008). Arabidopsis bZIP60 is a proteolysis-activated transcription factor involved in the endoplasmic reticulum stress response. Plant Cell 20: 3107–3121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata Y., Koizumi N. (2005). An Arabidopsis transcription factor, AtbZIP60, regulates the endoplasmic reticulum stress response in a manner unique to plants. Proc. Natl. Acad. Sci. USA 102: 5280–5285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata Y., Yoneda M., Yanagawa Y., Koizumi N. (2009). Characteristics of the nuclear form of the Arabidopsis transcription factor AtbZIP60 during the endoplasmic reticulum stress response. Biosci. Biotechnol. Biochem. 73: 865–869 [DOI] [PubMed] [Google Scholar]
- Johansen T., Lamark T. (2011). Selective autophagy mediated by autophagic adapter proteins. Autophagy 7: 279–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I., Rodriguez-Enriquez S., Lemasters J.J. (2007). Selective degradation of mitochondria by mitophagy. Arch. Biochem. Biophys. 462: 245–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkin V., et al. (2009). A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell 33: 505–516 [DOI] [PubMed] [Google Scholar]
- Kohno K., Normington K., Sambrook J., Gething M.J., Mori K. (1993). The promoter region of the yeast KAR2 (BiP) gene contains a regulatory domain that responds to the presence of unfolded proteins in the endoplasmic reticulum. Mol. Cell. Biol. 13: 877–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi N., Martinez I.M., Kimata Y., Kohno K., Sano H., Chrispeels M.J. (2001). Molecular characterization of two Arabidopsis Ire1 homologs, endoplasmic reticulum-located transmembrane protein kinases. Plant Physiol. 127: 949–962 [PMC free article] [PubMed] [Google Scholar]
- Kraft C., Deplazes A., Sohrmann M., Peter M. (2008). Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat. Cell Biol. 10: 602–610 [DOI] [PubMed] [Google Scholar]
- Liu J.X., Srivastava R., Che P., Howell S.H. (2007). An endoplasmic reticulum stress response in Arabidopsis is mediated by proteolytic processing and nuclear relocation of a membrane-associated transcription factor, bZIP28. Plant Cell 19: 4111–4119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Bassham D.C. (2010). TOR is a negative regulator of autophagy in Arabidopsis thaliana. PLoS ONE 5: e11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Bassham D.C. (2012). Autophagy: Pathways for self-eating in plant cells. Annu. Rev. Plant Biol. 63: 215–237 [DOI] [PubMed] [Google Scholar]
- Liu Y., Schiff M., Czymmek K., Tallóczy Z., Levine B., Dinesh-Kumar S.P. (2005). Autophagy regulates programmed cell death during the plant innate immune response. Cell 121: 567–577 [DOI] [PubMed] [Google Scholar]
- Liu Y., Xiong Y., Bassham D.C. (2009). Autophagy is required for tolerance of drought and salt stress in plants. Autophagy 5: 954–963 [DOI] [PubMed] [Google Scholar]
- Matsuoka K., Higuchi T., Maeshima M., Nakamura K. (1997). A vacuolar-type H+-ATPase in a nonvacuolar organelle is required for the sorting of soluble vacuolar protein precursors in tobacco cells. Plant Cell 9: 533–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mersereau M., Pazour G.J., Das A. (1990). Efficient transformation of Agrobacterium tumefaciens by electroporation. Gene 90: 149–151 [DOI] [PubMed] [Google Scholar]
- Moreno A.A., Mukhtar M.S., Blanco F., Boatwright J.L., Moreno I., Jordan M.R., Chen Y., Brandizzi F., Dong X., Orellana A., Pajerowska-Mukhtar K.M. (2012). IRE1/bZIP60-mediated unfolded protein response plays distinct roles in plant immunity and abiotic stress responses. PLoS ONE 7: e31944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno A.A., Orellana A. (2011). The physiological role of the unfolded protein response in plants. Biol. Res. 44: 75–80 [DOI] [PubMed] [Google Scholar]
- Mori K. (2000). Tripartite management of unfolded proteins in the endoplasmic reticulum. Cell 101: 451–454 [DOI] [PubMed] [Google Scholar]
- Mori K., Kawahara T., Yoshida H., Yanagi H., Yura T. (1996). Signalling from endoplasmic reticulum to nucleus: Transcription factor with a basic-leucine zipper motif is required for the unfolded protein-response pathway. Genes Cells 1: 803–817 [DOI] [PubMed] [Google Scholar]
- Mori K., Ma W., Gething M.J., Sambrook J. (1993). A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell 74: 743–756 [DOI] [PubMed] [Google Scholar]
- Mori K., Sant A., Kohno K., Normington K., Gething M.J., Sambrook J.F. (1992). A 22 bp cis-acting element is necessary and sufficient for the induction of the yeast KAR2 (BiP) gene by unfolded proteins. EMBO J. 11: 2583–2593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagashima Y., Mishiba K., Suzuki E., Shimada Y., Iwata Y., Koizumi N. (2011). Arabidopsis IRE1 catalyses unconventional splicing of bZIP60 mRNA to produce the active transcription factor. Sci. Rep. 1: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda N.N., Ohsumi Y., Inagaki F. (2010). Atg8-family interacting motif crucial for selective autophagy. FEBS Lett. 584: 1379–1385 [DOI] [PubMed] [Google Scholar]
- Noh S.J., Kwon C.S., Chung W.I. (2002). Characterization of two homologs of Ire1p, a kinase/endoribonuclease in yeast, in Arabidopsis thaliana. Biochim. Biophys. Acta 1575: 130–134 [DOI] [PubMed] [Google Scholar]
- Ogata M., et al. (2006). Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol. Cell. Biol. 26: 9220–9231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y., Koizumi N., Yamaguchi Y., Kimata Y., Kohno K., Sano H. (2002). Isolation and characterization of a putative transducer of endoplasmic reticulum stress in Oryza sativa. Plant Cell Physiol. 43: 532–539 [DOI] [PubMed] [Google Scholar]
- Ozcan U., Ozcan L., Yilmaz E., Düvel K., Sahin M., Manning B.D., Hotamisligil G.S. (2008). Loss of the tuberous sclerosis complex tumor suppressors triggers the unfolded protein response to regulate insulin signaling and apoptosis. Mol. Cell 29: 541–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv S., Clausen T.H., Lamark T., Brech A., Bruun J.A., Outzen H., Øvervatn A., Bjørkøy G., Johansen T. (2007). p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282: 24131–24145 [DOI] [PubMed] [Google Scholar]
- Preuss M.L., Serna J., Falbel T.G., Bednarek S.Y., Nielsen E. (2004). The Arabidopsis Rab GTPase RabA4b localizes to the tips of growing root hair cells. Plant Cell 16: 1589–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian S.B., Zhang X., Sun J., Bennink J.R., Yewdell J.W., Patterson C. (2010). mTORC1 links protein quality and quantity control by sensing chaperone availability. J. Biol. Chem. 285: 27385–27395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin L., Wang Z., Tao L., Wang Y. (2010). ER stress negatively regulates AKT/TSC/mTOR pathway to enhance autophagy. Autophagy 6: 239–247 [DOI] [PubMed] [Google Scholar]
- Reumann S., Voitsekhovskaja O., Lillo C. (2010). From signal transduction to autophagy of plant cell organelles: Lessons from yeast and mammals and plant-specific features. Protoplasma 247: 233–256 [DOI] [PubMed] [Google Scholar]
- Rizzo M.A., Springer G.H., Granada B., Piston D.W. (2004). An improved cyan fluorescent protein variant useful for FRET. Nat. Biotechnol. 22: 445–449 [DOI] [PubMed] [Google Scholar]
- Ron D., Walter P. (2007). Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 8: 519–529 [DOI] [PubMed] [Google Scholar]
- Rose T.L., Bonneau L., Der C., Marty-Mazars D., Marty F. (2006). Starvation-induced expression of autophagy-related genes in Arabidopsis. Biol. Cell 98: 53–67 [DOI] [PubMed] [Google Scholar]
- Sanderfoot A.A., Ahmed S.U., Marty-Mazars D., Rapoport I., Kirchhausen T., Marty F., Raikhel N.V. (1998). A putative vacuolar cargo receptor partially colocalizes with AtPEP12p on a prevacuolar compartment in Arabidopsis roots. Proc. Natl. Acad. Sci. USA 95: 9920–9925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J. (2002). A transient expression assay using Arabidopsis mesophyll protoplasts. http://genetics.mgh.harvard.edu/sheenweb/. Accessed November 19, 2012
- Suttangkakul A., Li F., Chung T., Vierstra R.D. (2011). The ATG1/ATG13 protein kinase complex is both a regulator and a target of autophagic recycling in Arabidopsis. Plant Cell 23: 3761–3779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svenning S., Lamark T., Krause K., Johansen T. (2011). Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic adapters NBR1 and p62/SQSTM1. Autophagy 7: 993–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima H., Iwata Y., Iwano M., Takayama S., Koizumi N. (2008). Identification of an Arabidopsis transmembrane bZIP transcription factor involved in the endoplasmic reticulum stress response. Biochem. Biophys. Res. Commun. 374: 242–247 [DOI] [PubMed] [Google Scholar]
- Thompson A.R., Doelling J.H., Suttangkakul A., Vierstra R.D. (2005). Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol. 138: 2097–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirasophon W., Welihinda A.A., Kaufman R.J. (1998). A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev. 12: 1812–1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Törmäkangas K., Hadlington J.L., Pimpl P., Hillmer S., Brandizzi F., Teeri T.H., Denecke J. (2001). A vacuolar sorting domain may also influence the way in which proteins leave the endoplasmic reticulum. Plant Cell 13: 2021–2032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyooka K., Moriyasu Y., Goto Y., Takeuchi M., Fukuda H., Matsuoka K. (2006). Protein aggregates are transported to vacuoles by a macroautophagic mechanism in nutrient-starved plant cells. Autophagy 2: 96–106 [DOI] [PubMed] [Google Scholar]
- Urade R. (2007). Cellular response to unfolded proteins in the endoplasmic reticulum of plants. FEBS J. 274: 1152–1171 [DOI] [PubMed] [Google Scholar]
- Urade R. (2009). The endoplasmic reticulum stress signaling pathways in plants. Biofactors 35: 326–331 [DOI] [PubMed] [Google Scholar]
- Urano F., Wang X., Bertolotti A., Zhang Y., Chung P., Harding H.P., Ron D. (2000). Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science 287: 664–666 [DOI] [PubMed] [Google Scholar]
- Wada S., Ishida H., Izumi M., Yoshimoto K., Ohsumi Y., Mae T., Makino A. (2009). Autophagy plays a role in chloroplast degradation during senescence in individually darkened leaves. Plant Physiol. 149: 885–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., Contento A.L., Bassham D.C. (2005). AtATG18a is required for the formation of autophagosomes during nutrient stress and senescence in Arabidopsis thaliana. Plant J. 42: 535–546 [DOI] [PubMed] [Google Scholar]
- Xiong Y., Contento A.L., Bassham D.C. (2007a). Disruption of autophagy results in constitutive oxidative stress in Arabidopsis. Autophagy 3: 257–258 [DOI] [PubMed] [Google Scholar]
- Xiong Y., Contento A.L., Nguyen P.Q., Bassham D.C. (2007b). Degradation of oxidized proteins by autophagy during oxidative stress in Arabidopsis. Plant Physiol. 143: 291–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Klionsky D.J. (2009). An overview of the molecular mechanism of autophagy. Curr. Top. Microbiol. Immunol. 335: 1–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J., Rawson R.B., Komuro R., Chen X., Davé U.P., Prywes R., Brown M.S., Goldstein J.L. (2000). ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol. Cell 6: 1355–1364 [DOI] [PubMed] [Google Scholar]
- Ylä-Anttila P., Vihinen H., Jokitalo E., Eskelinen E.L. (2009). 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy 5: 1180–1185 [DOI] [PubMed] [Google Scholar]
- Yorimitsu T., Klionsky D.J. (2007). Eating the endoplasmic reticulum: Quality control by autophagy. Trends Cell Biol. 17: 279–285 [DOI] [PubMed] [Google Scholar]
- Yorimitsu T., Nair U., Yang Z., Klionsky D.J. (2006). Endoplasmic reticulum stress triggers autophagy. J. Biol. Chem. 281: 30299–30304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H., Matsui T., Yamamoto A., Okada T., Mori K. (2001). XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107: 881–891 [DOI] [PubMed] [Google Scholar]
- Yoshida H., Okada T., Haze K., Yanagi H., Yura T., Negishi M., Mori K. (2000). ATF6 activated by proteolysis binds in the presence of NF-Y (CBF) directly to the cis-acting element responsible for the mammalian unfolded protein response. Mol. Cell. Biol. 20: 6755–6767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto K., Hanaoka H., Sato S., Kato T., Tabata S., Noda T., Ohsumi Y. (2004). Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 16: 2967–2983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zientara-Rytter K., Lukomska J., Moniuszko G., Gwozdecki R., Surowiecki P., Lewandowska M., Liszewska F., Wawrzyńska A., Sirko A. (2011). Identification and functional analysis of Joka2, a tobacco member of the family of selective autophagy cargo receptors. Autophagy 7: 1145–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]