Hemicellulose can retain a large amount of Al. This study demonstrates that an important component of hemicellulose, xyloglucan, can bind Al, and knockout of XTH31 increases Al resistance in Arabidopsis by decreasing xyloglucan endotransglucosylase action and the accumulation of xyloglucan in hemicellulose, which in turn reduces the retention of Al in the cell wall, thus excluding Al from roots.
Abstract
Xyloglucan endohydrolase (XEH) and xyloglucan endotransglucosylase (XET) activities, encoded by xyloglucan endotransglucosylase-hydrolase (XTH) genes, are involved in cell wall extension by cutting or cutting and rejoining xyloglucan chains, respectively. However, the physiological significance of this biochemical activity remains incompletely understood. Here, we find that an XTH31 T-DNA insertion mutant, xth31, is more Al resistant than the wild type. XTH31 is bound to the plasma membrane and the encoding gene is expressed in the root elongation zone and in nascent leaves, suggesting a role in cell expansion. XTH31 transcript accumulation is strongly downregulated by Al treatment. XTH31 expression in yeast yields a protein with an in vitro XEH:XET activity ratio of >5000:1. xth31 accumulates significantly less Al in the root apex and cell wall, shows remarkably lower in vivo XET action and extractable XET activity, has a lower xyloglucan content, and exhibits slower elongation. An exogenous supply of xyloglucan significantly ameliorates Al toxicity by reducing Al accumulation in the roots, owing to the formation of an Al-xyloglucan complex in the medium, as verified by an obvious change in chemical shift of 27Al-NMR. Taken together, the data indicate that XTH31 affects Al sensitivity by modulating cell wall xyloglucan content and Al binding capacity.
INTRODUCTION
Al toxicity is a major constraint for crop production on acid soils worldwide (Foy, 1988; Kochian, 1995). Al rapidly inhibits root growth, along with the uptake of water and nutrients, which ultimately results in the loss of crop production (Kochian, 1995). The earliest and most dramatic visual symptom of Al toxicity is the inhibition of root elongation, but the underlying physiological and molecular mechanisms are still not well understood (Zheng and Yang, 2005; Horst et al., 2010).
Both cell division and cell expansion contribute to root elongation. But as the inhibition of root elongation is observed within 30 min in an Al-sensitive cultivar (Llugany et al., 1995), it is now generally accepted that Al inhibition of cell expansion is the main cause of the inhibition of root elongation. Cell expansion, as well as other developmental processes, requires the modification of plant primary cell walls. The cell wall is a dynamic architecture composed of cellulose embedded in a matrix of hemicellulosic and pectic polysaccharides plus structural proteins (Hayashi, 1989; Carpita and Gibeaut, 1993). The wall plays important roles in not only the regulation of growth and development but also the perception and manifestation of Al toxicity. When plants suffer Al toxicity, the cell wall is the major site for Al accumulation. For instance, 85 to 90% of the total Al accumulated by barley (Hordeum vulgare) roots is tightly bound to the cell walls (Clarkson, 1967), and almost 90% of the total Al is associated with the cell walls of cultured tobacco (Nicotiana tabacum) cells (Chang et al., 1999). The major Al binding site in the cell wall was generally considered to be the pectic polysaccharides, since their negatively charged carboxylic groups have a particularly high affinity for Al3+ (Blamey et al., 1990; Chang et al., 1999). However, recent evidence has shown that cell wall hemicellulose metabolism is more susceptible to Al stress. For example, the most significant Al-induced change in cell wall components involves the hemicellulose fraction in wheat (Triticum aestivum; Tabuchi and Matsumoto, 2001), triticale (X Triticosecale Wittmack; Liu et al., 2008), and rice (Oryza sativa; Yang et al., 2008), especially in Al-sensitive cultivars. Moreover, we found that in Arabidopsis thaliana, hemicellulose is not only susceptible to Al stress but is also the principal binding site for Al; the amount of Al accumulated in hemicellulose is much more than that in pectin (Yang et al., 2011), although how hemicellulose can bind Al is unclear. Thus, the role of not only pectin but also hemicellulose in Al toxicity/resistance and its underlying physiological and molecular mechanisms need to be further explored.
Hemicelluloses, synthesized in the Golgi, are a group of neutral or slightly acidic polysaccharides, such as xyloglucan, arabinoxylan, and mannan, that are bound tightly to the surface of cellulose to form a strong yet resilient network (Carpita and Gibeaut, 1993; Cosgrove, 2005). There is a remarkable difference in primary cell wall hemicellulose composition between dicotyledons and poalean monocotyledons (e.g., cereals). In the cereals, such as rice and maize (Zea mays), there are large amounts of glucuronoarabinoxylan and, at least in some tissues, the taxonomically restricted polymer (1-3),(1-4)-β-d-glucan (mixed-linked glucan), while xyloglucan is a minor component (Carpita and Gibeaut, 1993; Carpita, 1996, Hazen et al., 2002). However, in the nonpoalean monocotyledons and dicotyledons, such as Arabidopsis, xyloglucan is the major primary cell wall hemicellulose. Xyloglucan has been widely proposed to tether cellulose microfibrils by means of hydrogen bonds (Hayashi, 1989; Fry, 1989), forming one of three coextensive frameworks of the cell wall (the cellulose/xyloglucan, pectic, and extensin networks). The key architectural role of the cellulose/xyloglucan network was challenged by Cavalier et al. (2008), who reported that a xyloglucan-deficient mutant of Arabidopsis grew more or less normally, suggesting that, in the absence of xyloglucan, pectins and arabinoxylans assume a larger role in cell wall biomechanics (Park and Cosgrove, 2012). Recently, through analysis of the xxt1 xxt2 xxt5 triple mutant, Zabotina et al. (2012) found that cell walls undergo rearrangements in polysaccharide interactions in the absence of xyloglucan without substantially increasing the synthesis of any other wall component. Nevertheless, it remains true that in normal plants the presence of xyloglucan is particularly important for the process of cell wall extension (creep) induced by α-expansins during acid growth, and xyloglucan itself indeed strengthens the primary cell wall (judged from rapid stress/strain assays) (Park and Cosgrove, 2012). Furthermore, Van Sandt et al. (2007) demonstrated the effects of an exogenous xyloglucan-modifying enzyme on wall extensibility. The common backbone of xyloglucan is (1-4)-linked β-d-glucopyranosyl residues, a large proportion of which are substituted with α-d-xylopyranosyl residues at O-6. In the standard nomenclature for xyloglucan structures, unsubstituted Glc residues are represented by G, while X, L, and F indicate Glc residues that are 6-O-substituted with α-d-Xylp, β-d-Galp-(1-2)-α-d-Xylp, and α-l-Fucp-(1-2)-β-d-Galp-(1-2)-α-d-Xylp side chains, respectively (Fry et al., 1993). Treatment of Arabidopsis xyloglucan with an endoglucanase (XEG) that attacks unsubstituted Glc residues yields an oligosaccharide mixture that includes XXXG, XXLG, XLXG, XXG, GXXG, XLLG, XXFG, and XLFG (the sequences are shown with the reducing end of the molecule positioned to the right; Madson et al., 2003; Obel et al., 2009; Sampedro et al., 2012).
Modifications of the cell wall network are catalyzed by several enzymes, including the xyloglucan endotransglucosylase/hydrolase (XTH) family (Nishitani and Vissenberg, 2007). XTHs either cut and rejoin xyloglucan chains through xyloglucan endotransglucosylase (XET) activity (Fry et al., 1992; Nishitani and Tominaga, 1992; Thompson and Fry, 2001) or catalyze the hydrolysis of xyloglucan through xyloglucan endohydrolase (XEH) activity, thus contributing to cell wall extension (Van Sandt et al., 2007). In our previous report, we found that Al inhibits XET action and, among the XTH genes expressed in roots, XTH14, 15, and 31 expression is significantly downregulated by Al in Arabidopsis (Yang et al., 2011); however, the functions of individual XTH genes in terms of root growth regulation at toxic Al concentrations are still unclear.
XTH enzymes are generally encoded by a large multigene family, for example, there are 41 members in poplar (Populus spp; Geisler-Lee et al., 2006), 25 in tomato (Solanum lycopersicum ; Saladié et al., 2006), 29 in rice (Yokoyama et al., 2004) and 22 in barley (Strohmeier et al., 2004). Of all 33 identified XTH genes in Arabidopsis (Yokoyama and Nishitani, 2001), one-third occurs as clusters resulting from genome duplication (Blanc et al., 2000). Different XTHs have diverse and distinct expression patterns in terms of organ specificity and in response to developmental and environmental stimuli. Yokoyama and Nishitani (2001) revealed that at least 10 XTH genes are predominantly expressed in roots. Later, Becnel et al. (2006) found that a subset of XTHs has robust root expression, especially XTH14, 15, and 31, in Arabidopsis. Recently, we suggested that XTH31 might make a bigger contribution to this XET action than any other enzyme does, which was inhibited by Al stress in the root tip (Yang et al., 2011). In this study, we found that a T-DNA insertional mutant of XTH31 gains the function of Al resistance. We characterized XTH31, including the enzymological activities of XTH31 in vitro and found that XTH31 modulates XET action in roots, which may thereby regulate the content of xyloglucan, a hemicellulose that can bind Al in Arabidopsis, accumulated in the cell wall.
RESULTS
An XTH31 T-DNA Insertional Mutant Has Increased Al Resistance
In our previous study, we found that the majority of Al in roots is bound to the cell wall hemicellulose and that the expression of one gene, XTH31, correlates with hemicellulose modification. XTH31 expression is suppressed by Al stress together with declining Arabidopsis XET action as monitored by in vivo assays with a fluorescent acceptor substrate (Yang et al., 2011). To determine whether XTH31 function is required for Al stress responses, we characterized a T-DNA insertion line (Salk_046167). The T-DNA is inserted into the second exon region of XTH31 (211 bp from the translation initiator ATG codon; Figure 1A), and XTH31 transcripts were not detected in the homozygous line (Figure 1B), indicating that the T-DNA insertion may lead to the loss of XTH31 function. The roots (Figures 1C and 1D) and shoots (see Supplemental Figure 1 online) of xth31 were shorter than those of the wild type, showing that XTH31 function is required for normal growth (defined as the irreversible increase in volume). Decreased growth, so defined, necessarily involves decreased cell expansion. To gain insight into the cellular basis for the shorter roots in the mutant, we used propidium iodide staining to visualize the cell size. Cellular shape and size were similar in the mutant and the wild type (see Supplemental Figure 2 online). Therefore, cell division paralleled cell expansion during the establishment of the short root phenotype under normal growth conditions. The findings are consistent with the conclusion that XTH31 is not essential for cell shape determination but instead may have an essential function in the regulation and/or processes of cell expansion.
Figure 1.
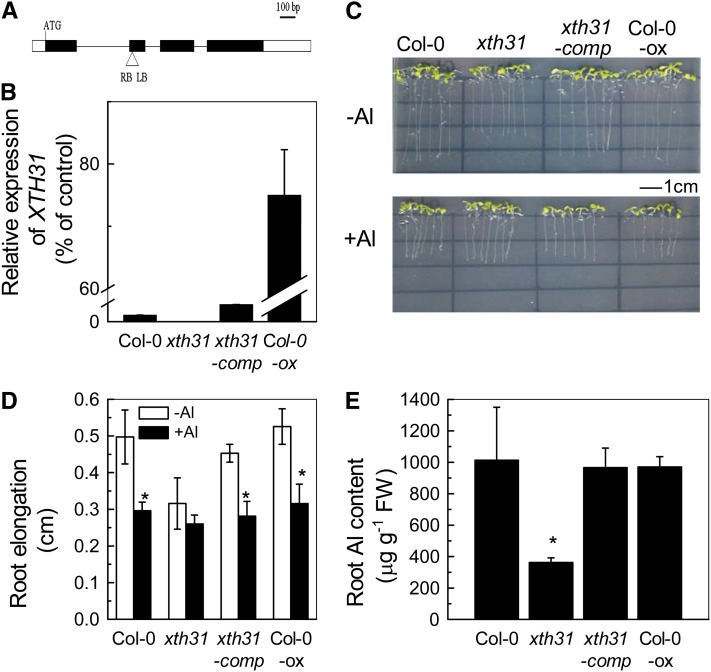
Phenotype of the xth31 Mutant, Complementation, and Overexpression of XTH31.
(A) Schematic structure of the xth31 mutant carrying a single copy of the T-DNA insert in the second exon. The black and white boxes represent the coding and untranslated regions, respectively.
(B) Quantitative RT-PCR study of XTH31 expression in the whole roots of Arabidopsis. Values are mean ± sd (n = 3).
(C) Col-0 and xth31, xth31-comp (complementation), and Col-0 overexpression (ox) lines grown on Murashige and Skoog plates in the presence or absence of 50 µM Al3+. Seedlings were treated with Al when the roots were ∼1 cm long.
(D) Root elongation of Col-0 and xth31, xth31-comp, and Col-0 overexpression lines in the presence or absence of 50 µM Al3+. Plants with ∼1-cm roots were transferred to agar medium containing 0 or 50 µM Al and grown for a further 24 h. Root length was measured before and after treatment. Data are means ± sd (n = 10). The asterisk shows a significant effect of Al at P < 0.05 by Student’s t test.
(E) Root Al content of Col-0 and xth31, xth31-comp, and Col-0 overexpression lines. Data are means ± sd (n = 4). The asterisk shows a significant difference between xth31 and the others at P < 0.05 by Student’s t test. FW, fresh weight.
Furthermore, although the growth of wild-type seedling roots was inhibited by ∼40% when the seedlings were grown on agar medium containing 50 μM Al3+ for 7 d, no significant Al-dependent growth change occurred in xth31 (Figures 1C and 1D). Strikingly, Al-exposed xth31 roots accumulate less than half the Al that is found in wild-type roots grown under similar conditions (Figure 1C). Root morin staining also showed much less Al accumulation in the root tip of xth31 than in Columbia-0 (Col-0) (see Supplemental Figure 3 online). One possibility is that XTH31 function is required for Al accumulation in wild-type roots and that Al accumulation is required for the Al-dependent root growth inhibition.
To confirm that the growth and Al resistance phenotypes of xth31 are caused by XTH31 loss of function, we performed a complementation test by transforming the xth31 mutant with the full-length coding region of XTH31. Transgene expression was confirmed by quantitative RT-PCR (Figure 1B). Complemented xth31 plants showed Al sensitivity and overall growth similar to those of the wild type (Figures 1C to 1E; see Supplemental Figure 1 online), verifying that the reported xth31 mutant phenotypes are the consequence of the loss of XTH31 function. We also generated transgenic lines that overexpress XTH31 in the wild type (Figure 1B); these Col-0–overexpressing lines showed no detectable phenotypic differences from the wild type in the absence or presence of Al stress (Figures 1C and 1D). In addition, overexpression of XTH31 did not affect root Al accumulation (Figure 1E). One possible explanation for the lack of overt phenotypic changes due to XTH31 overexpression is that wild-type XTH31 expression levels are not limiting for XTH31 function and the amount of synthesized xyloglucan may be the limiting factor.
Sequence Analysis of XTH31 and the Cellular Localization of the Protein
According to a phylogenetic analysis of the Arabidopsis XTH family, XTH31 (At3g44990) is classified, together with XTH27, XTH28, XTH29, XTH30, XTH32, XTH33, in subgroup 3, while XTH1 to 11 are in subgroup 1 and XTH12 to 26 are in subgroup 2 (Rose et al., 2002). Similar to other reported XTH members, XTH31 also contains a sequence encoding a potential signal peptide at the beginning of the first exon. The deduced XTH31 sequence also contains four Cys residues in the C-terminal portion of the protein that could form disulfide bridges (Figure 2A); these four Cys residues are found in all known XETs (Xu et al., 1995), although the position of the second Cys residue varies slightly.
Figure 2.
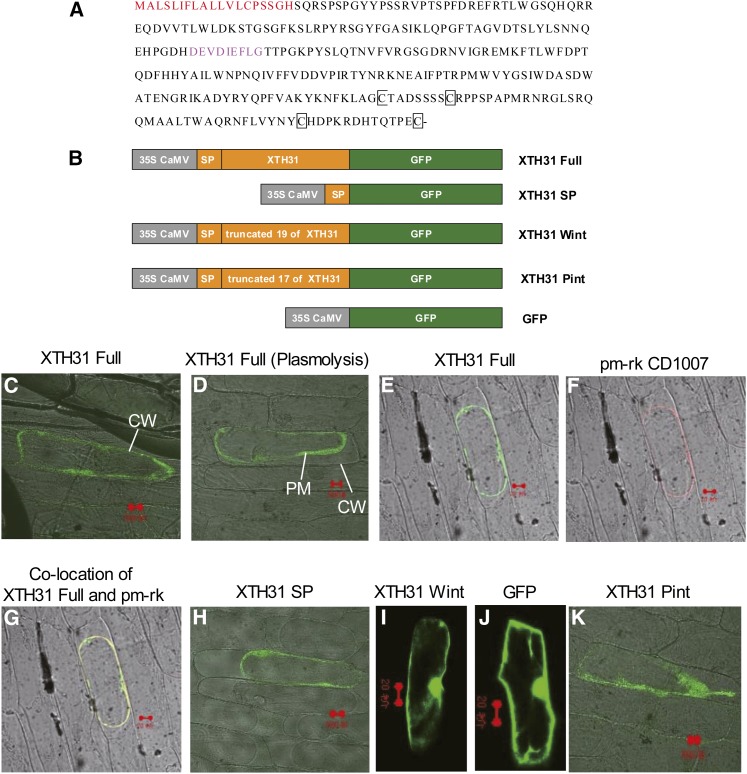
The Amino Acid Sequence of XTH31 and Its Subcellular Localization by Transient Expression in Onion Cells.
(A) The signal peptide at the N terminus, shown in red, is the putative wall/membrane targeting signal, the sequences in pink show the conserved GH16_XET domain, and conserved Cys residues (C) are in black squares.
(B) Schematic representation of XTH31 constructs fused to GFP. SP, signal peptide (19 amino acids at the N terminus of the full XTH31 coding sequence; see Methods for details). XTH31 Wint, the full XTH31 coding sequence (XTH31 Full) without the 19 putative SP residues; XTH31 Pint, the full XTH31 sequence without the first 17 of the 19 putative signal peptide residues. CaMV, Cauliflower mosaic virus.
(C) XTH31 Full-GFP localization. CW, cell wall.
(D) and (E) XTH31 Full-GFP localization in plasmolysed cells. PM, plasma membrane.
(F) pm-rk CD-1007 (plasma membrane marker) localization in the same plasmolysed cells as in (E).
(G) XTH31 Full-GFP colocalized with pm-rk CD-1007.
(H) XTH31 SP-GFP localization in plasmolysed cells.
(I) XTH31 Wint-GFP localization in nonplasmolysed cells.
(J) 35S:GFP localization in nonplasmolysed cells.
(K) XTH31 Pint-GFP localization in plasmolysed cells.
Red bars = 20 µm.
Consistent with the overall amino acid similarities between XTH31 and other XTHs, XTH31 harbors a potential signal peptide at the N terminus. This region lacks charged amino acids and is rich in hydrophobic residues, and cleavage of the potential signal sequence is predicted to occur after His-19 (Figure 2A). To elucidate the potential subcellular localization of XTH31, we produced three different constructs fused to green fluorescent protein (XTH31 Full-GFP, XTH31 SP-GFP, and XTH31 Wint-GFP; Figure 2B) and used these to transform onion (Allium cepa) cells. The full-length XTH31 fusion protein (XTH31 Full-GFP) was found around the cell periphery, which was consistent with the interpretation of plasma membrane localization (Figures 2C and 2D). Moreover, when a plasma membrane–localized marker (pm-rk) was coapplied during transformation, XTH31 Full-GFP and pm-rk staining patterns were indistinguishable (Figures 2E to 2G). A similar result was obtained when only the first 19 amino acids of XTH31 were fused to GFP (XTH31 SP-GFP; Figure 2H), indicating that these N-terminal amino acids are sufficient to confer plasma membrane localization of GFP and therefore likely function, as predicted, as a plasma membrane–targeted signal peptide. Consistent with this conclusion, the putative signal peptide is required for plasma membrane localization because a truncated XTH31-GFP fusion protein that lacks the whole putative signal peptide (XTH31 Wint-GFP) showed localization similar to that of GFP alone (Figures 2I and 2J). These results are consistent with the interpretation that XTH31 is most likely targeted to the membrane by an N-terminal signal peptide.
The Tissue-Specific Localization of XTH31 Expression
The expression pattern of XTH31 mRNA was first examined with quantitative real-time PCR of isolated organs. According to the RT-PCR data, XTH31 transcripts accumulate in the roots as well as in the leaves, stems, flowers, and siliques (see Supplemental Figure 4A online); roots have the highest expression, whereas flowers have relatively low expression.
The in vivo tissue-specific localization of XTH31 was further investigated with β-glucuronidase (GUS) staining. A DNA fragment consisting of 2.3 kb of sequences located upstream of the coding region was used to drive the expression of the GUS reporter gene, and the construct containing the reporter fusion was introduced into Arabidopsis wild-type plants by transformation. GUS analysis in the transgenic plants revealed that the XTH31 upstream sequences result in GUS accumulation in both roots and shoots (see Supplemental Figure 4B online). In the roots, XTH31:GUS was predominantly expressed in the root tips, including the elongation zone (see Supplemental Figures 4E and 4F online). In the shoots, high GUS activity was observed in developing young leaves (see Supplemental Figure 4D online), whereas the activity was greatly reduced in fully expanded leaves (see Supplemental Figure 4C online).
The Dose Response of XTH31 Expression Levels to Aluminum
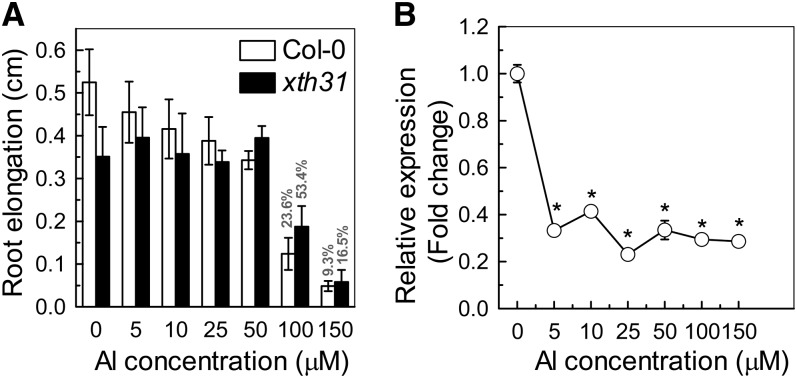
In our previous report, a time-course experiment indicated that XTH31 transcription is suppressed by 50 µM Al and a significant change can be detected even within 30 min of Al treatment (Yang et al., 2011). To investigate whether inhibition of XTH31 expression correlates with the phenotype of shortened roots, we conducted a dose–response experiment. We found that root elongation decreased with increasing external Al concentrations (Figure 3A). In Col-0, relative root elongation (100 × root elongation in Al/root elongation without Al) decreased to 23.6 and 9.3% at 100 μΜ and 150 μΜ Al, respectively, whereas in xth31, it was reduced only to 53.4 and 16.5% by 100 and 150 μΜ Al, respectively (Figure 3A). However, XTH31 expression in the wild type was substantially inhibited even at an Al concentration as low as 5 µM (Figure 3B). Therefore, XTH31 expression is very responsive to Al stress, with significant reductions in expression at Al levels lower than that required for root growth inhibition. As the knockout of XTH31 results in increased Al resistance (Figures 1C and 1D) and expression of XTH31 is decreased by Al treatment, it is possible that XTH31 function impairs Al resistance and decreased XTH31 expression somehow confers protection against Al toxicity in plants.
Figure 3.
Dose Response of Seedlings to Al and the Relative Expression of XTH31.
(A) One-centimeter-long seedlings were selected and grown in agar medium containing 0 to 150 µM Al3+ for 24 h. Root length was measured before and after treatment. The percentages shown in the last two bars are the root elongation of the wild type and mutant relative to the respective Al-untreated controls. Data are means ± sd (n = 10).
(B) Quantitative RT-PCR analysis of XTH31 expression in Col-0 roots exposed to 0 to 150 µM Al3+ for 24 h. The y axis shows XTH31 RNA levels normalized to that of the control (0 µM Al3+). Values are mean ± sd (n = 3). The asterisks show significant differences between control and Al treatments at P < 0.05 by Student’s t test.
To determine whether XTH31 has an Al sensitive promoter, we examined GUS activity in XTH31:GUS transgenics subjected to Al stress. We found that GUS activity derived from the expression of XTH31:GUS indeed decreased in response to Al stress (see Supplemental Figures 5A and 5B online), suggesting the existence of an Al-sensitive promoter. XTH31:GUS activity was also reduced when the transgenics were subjected to cold or salt stress (see Supplemental Figures 5C and 5D online), suggesting the inhibition of XTH31 expression is not specific to Al.
XTH31 Contributes to in Vivo XET Action and in Vitro XEH/XET Activity
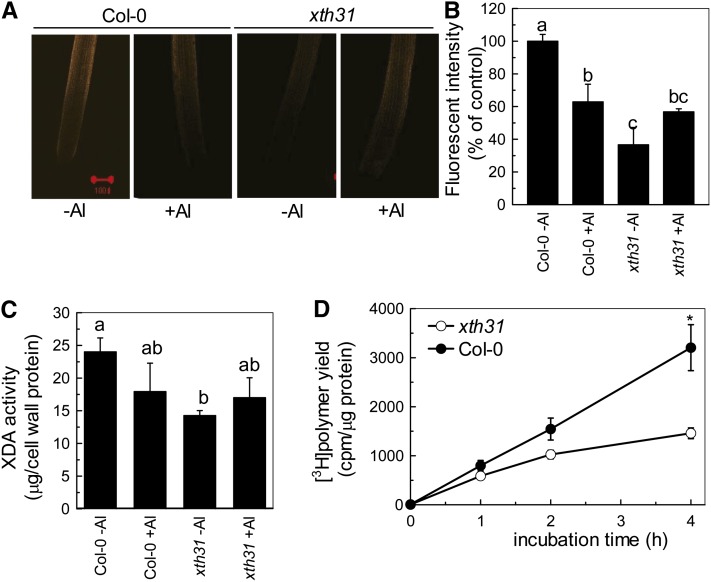
It is valuable to distinguish between XET/XEH enzyme action occurring in vivo (with endogenous donor substrate) and enzyme activity assayed in vitro (Fry, 2004). In our previous study, we demonstrated that XET action was inhibited remarkably and quickly by Al stress (Yang et al., 2011). We extended these findings in this work and found that in vivo XET action, using endogenous xyloglucan as donor substrate, in the xth31 mutant was sharply decreased under normal growth conditions (Figures 4A and 4B), suggesting that XTH31 is in some way responsible for a major proportion of the assayable XET action in the root tip. Furthermore, the loss of XET action on endogenous donor substrate that occurs in Col-0 upon Al stress (Figures 4A and 4B; Yang et al., 2011) was not apparent in Al-stressed xth31 (Figures 4A and 4B). This lack of inhibition of XET action by Al in xth31 is in accordance with the lack of inhibition of root growth by Al in xth31.
Figure 4.
Effect of Al on XET Action, Extractable XET Activity, and Extractable XDA in Col-0 and the xth31 Mutant.
Incubation with or without Al for 24 h corresponds to the panels “+Al” and “-Al,” respectively.
(A) Seedlings with ∼1-cm roots were grown on plates containing 0 or 50 µM Al for 24 h. Roots were then subjected to cytochemical assays of XET action for 1 h. Images reveal XET action as orange fluorescence in representative roots. Red bar = 100 µm.
(B) XET action expressed as fluorescence relative to untreated wild type. Data are means ± sd, n = 6. Different letters indicate significant differences at P < 0.05 by Student’s t test.
(C) Plants were treated with or without 50 µM Al for 24 h. Root extracts were subjected to in vitro assays of XDA. Data are means ± sd, n = 4. Different letters indicate significant differences at P < 0.05 by Student’s t test.
(D) Radiochemical assay of XET activity in root extracts. Buffer-extractable enzymes from the roots of Col-0 and xth31 seedlings were assayed for XET activity with tamarind xyloglucan and [3H]XXLGol as donor and acceptor substrates, respectively. Data are the mean ± se of six independent extracts. The asterisk shows a significant difference between xth31 and Col-0 at P < 0.05 by Student’s t test.
To further characterize the potential mechanisms that underlie the phenotypes in the xth31 mutant, we measured extractable xyloglucan degrading activity (XDA; i.e., XET and/or XEH activity as well as the hydrolytic activity of nonspecific endoglucanases) by an in vitro assay using tamarind xyloglucan as donor and/or hydrolyzable substrate according to Sasidharan et al. (2010). XDA is a measure of the degradation of xyloglucan by XTHs and/or cellulases that are present in a crude enzyme extract of the root. The values of plant-extractable XDA in vitro showed trends, both in response to Al stress and as the consequence of the loss of XTH31 function, similar to the in vivo XET action assay data (Figure 4C). Similarly, the plant extractable XET activity, assayed radiochemically, was significantly lower in xth31 roots than in wild-type roots (Figure 4D).
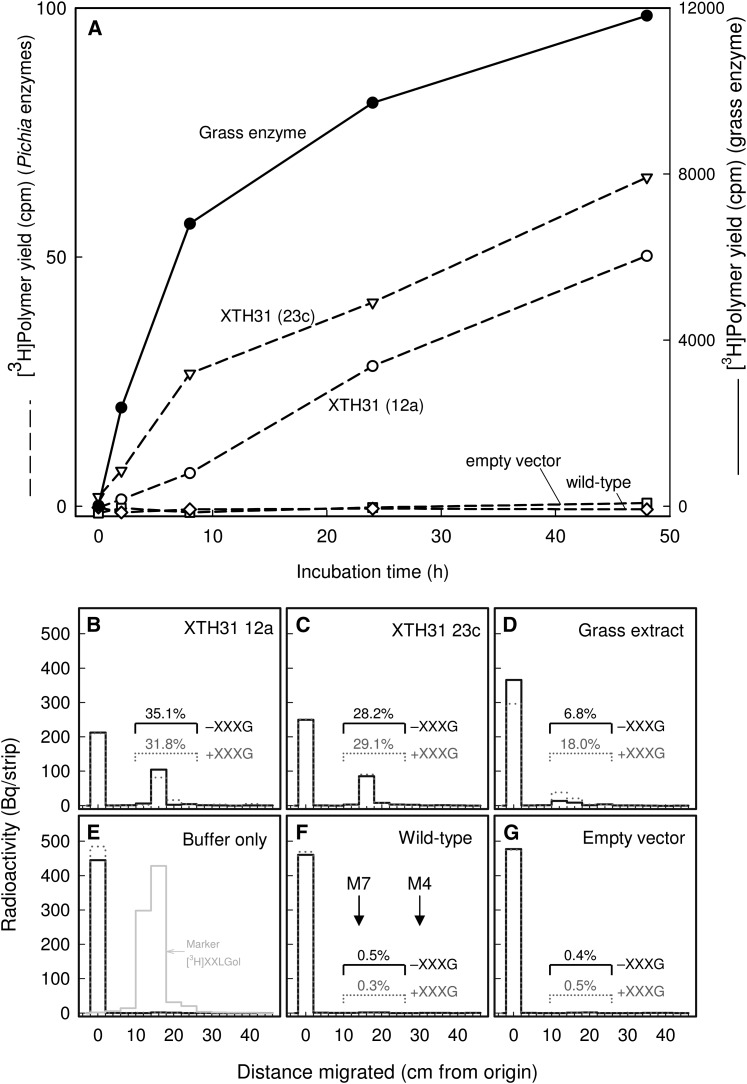
The XDA assay, as used in Figure 4C, does not distinguish XET from XEH; however, XTH31 is in a phylogenetic group of enzymes known or predicted to have XEH activity. To investigate the enzymological activities of XTH31, we employed the heterologous Pichia pastoris expression system to produce XTH31. P. pastoris–generated XTH31 exhibited slight XET activity in a highly sensitive radiochemical assay that is specific for XET, with nonradioactive xyloglucan as donor substrate and [3H]XXLGol as acceptor (Figure 5A). The XET activity of XTH31 was ∼1000-fold lower than that of a crude Holcus lanatus extract (Figure 5A).
Figure 5.
P. pastoris Cultures Expressing XTH31 Produce Slight XET but High XEH Activity.
(A) Standard radiochemical XET assay. Proteins secreted by XTH31-transformed P. pastoris cells (lines 12a and 23c), wild-type cells, or cells transformed with empty vector were tested by a sensitive radiochemical assay for XET activity (dashed lines). A crude extract of the grass H. lanatus was tested under identical conditions (solid line; right-hand scale).
(B) to (G) Novel assay for XEH and XET activity. The same five protein preparations ([B], XTH31 from P. pastoris line 12a; [C], XTH31 from P. pastoris line 23c; [D], H. lanatus; [F], wild-type P. pastoris; [G], P. pastoris transformed with empty vector), and a buffer-only control (E) were incubated with [reducing-end-3H]xyloglucan in the absence (solid line) or presence (dotted line) of 200 µM nonradioactive XXXG. After 48 h of incubation, the products were analyzed by paper chromatography, revealing any free 3H-oligosaccharides generated by the enzyme (∼15 cm from origin) and the remaining [3H]xyloglucan (0 cm from origin). The yields of 3H-oligosaccharides as a percentage of total 3H are indicated above each enzyme’s profile. (E) also shows the profile of authentic free [3H]XXLGol (gray histogram), which is expected to be the major oligosaccharide released. (F) shows the positions of the external markers maltoheptaose (M7) and maltotetraose (M4).
To test whether XTH31 also has XEH activity, we designed a new radiochemical assay for both XEH and XET activities. The donor (and/or hydrolyzable) substrate was [reducing-end-3H]xyloglucan. XEH activity will hydrolyze this substrate at G units. If the G selected for cleavage is the one nearest the reducing end, then free [3H]XXLGol will be released, detectable by paper chromatography. Supplementing the reaction mixture with nonradioactive XXXG is expected to have little effect on XEH activity. The release of [3H]XXLGol by XET activity in the absence of XXXG, on the other hand, may be a rare occurrence because the only acceptor substrate available is the [3H]xyloglucan itself, present at ∼7 µM. However, with the addition of 200 µM XXXG (an almost saturating acceptor substrate concentration; Rose et al., 2002), XET activity will operate at close to its maximum rate. In summary, XEH releases [3H]XXLGol at its maximum rate in the presence and absence of XXXG, whereas XET is expected to release [3H]XXLGol at its maximum rate only in the presence of cold XXXG.
P. pastoris–produced XTH31 released ∼30% of the radioactivity from [reducing-end-3H]xyloglucan as [3H]XXLGol in 48 h, and the addition of 200 µM nonradioactive XXXG had little effect, indicating the occurrence predominantly XEH activity (Figures 5B and 5C). By contrast, the slight [3H]XXLGol release by the H. lanatus enzyme was enhanced 2.6-fold by supplemental XXXG and thus was predominantly due to XET rather than XEH activity (Figure 5D). The 6.8% released in the absence of XXXG (Figure 5D) is an upper limit on the possible H. lanatus XEH activity because of the likelihood that H. lanatus XET activity can appreciably catalyze some polysaccharide-to-polysaccharide transglycosylation in the absence of added XXXG. In conclusion, the Holcus:XTH31 ratio for XEH activity is <0.2 (Figures 5B to 5D), whereas Holcus:XTH31 ratio for XET activity is ∼1000 (Figure 5A). Therefore, XTH31 had slight XET activity in vitro but >5000-fold greater XEH activity.
In the light of the above observations, it was surprising that XEH activity, assayed in vitro in root extracts and with [reducing-end-3H]xyloglucan as the substrate, was almost identical in the wild-type and xth31 (Table 1).
Table 1. In Vitro XEH Activity in Root Extracts.
| Enzyme Incubation Time (h) | Yield of [3H]Octasaccharide
(cpm/µg Protein) a |
|
|---|---|---|
| The Wild Type | xth31 | |
| 12 | 4.27 ± 0.90 | 3.08 ± 0.74 |
| 24 | 7.30 ± 0.90 | 7.92 ± 1.40 |
Mean ± se for six independent extracts; data are zeroed against a 0-h control.
Reduced Hemicellulose Content in xth31 Roots
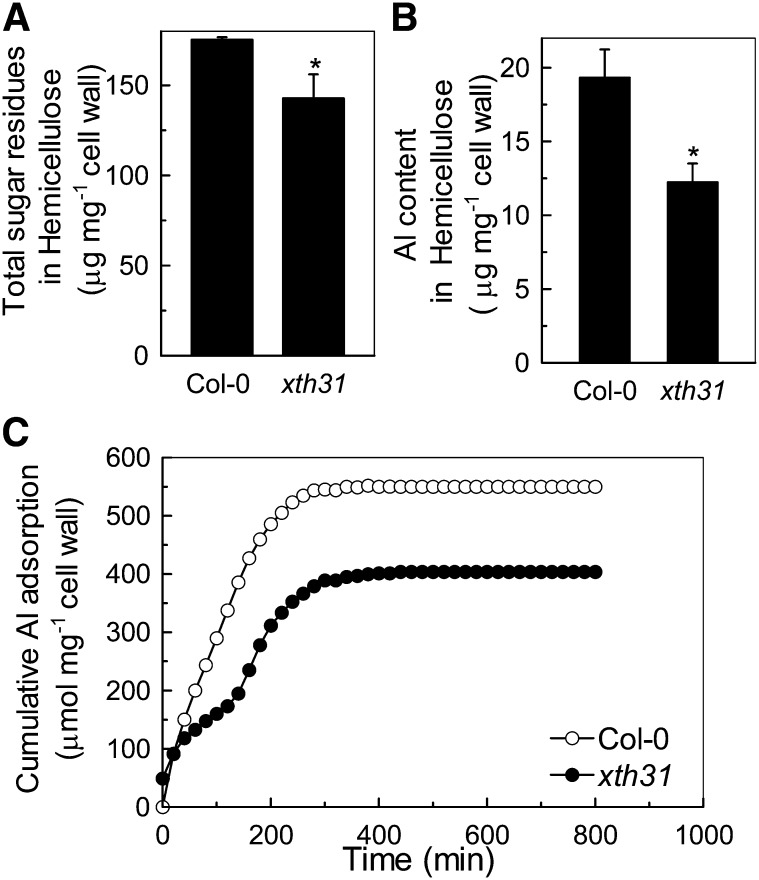
Since XTHs are involved in hemicellulose modulation (Fry et al., 1992), hemicellulose is the major Al binding site in Arabidopsis (Yang et al., 2011), and xth31 accumulates less Al in the roots when grown on Al (Figure 1C; see Supplemental Figure 3 online), we examined the hemicellulose content of xth31. Total sugar residues and Al content in hemicellulose was significantly lower in xth31 (Figures 6A and 6B). The in vitro Al adsorption kinetics also showed that the root cell walls of xth31 adsorbed significantly less Al than did those of Col-0 (Figure 6C).
Figure 6.
Total Sugar Residues, Al Content in Hemicellulose, and Cell Wall Al Adsorption Kinetics.
(A) Total sugar residues in extractable hemicellulose of Col-0 and xth31 mutant. Cell wall material from Al-untreated roots was fractionated into different polysaccharide classes. Data are means ± sd. n = 4; the asterisk shows a significant difference between Col-0 and the xth31 mutant at P < 0.05 by Student’s t test.
(B) Al content in the hemicellulose of Col-0 and the xth31 mutant. Cell wall hemicellulose from Al-treated roots was assayed for Al. Data are means ± sd. n = 4. The asterisk shows a significant difference between Col-0 and the xth31 mutant at P < 0.05 by Student’s t test.
(C) Al adsorption kinetics of cell walls from Col-0 and xth31. Cell wall material from Al-untreated roots was placed into a 2-mL column and Al adsorption kinetics were monitored as previously described (Zheng et al., 2004).
Reduced Xyloglucan Content in xth31 Cell Walls
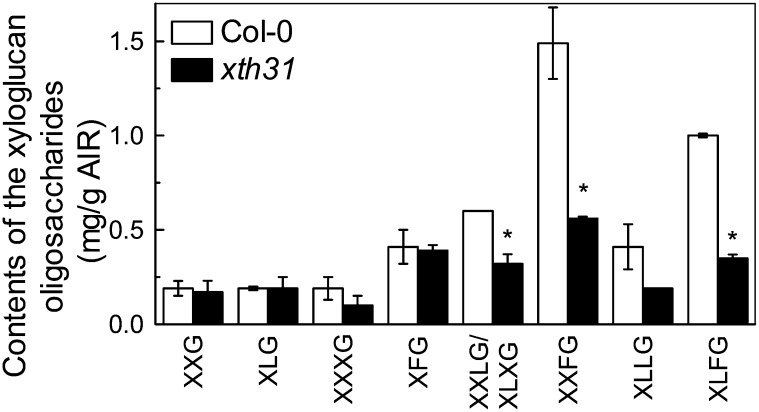
As xyloglucan is a major component of the cell wall hemicellulose and is the substrate for XET and XEH activities, we analyzed the xyloglucan content in root cell walls after xyloglucanase digestion and matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF) analysis. The content of total xyloglucanase-accessible xyloglucan repeat units was significantly reduced in xth31, especially XXFG, XLFG, and XXLG (and/or XLXG) (Figure 7).
Figure 7.
MALDI-TOF MS Analysis of the Relative Abundance of Xyloglucan Oligosaccharides Released by Xyloglucanase.
Cell wall material was extracted from Col-0 and xth31 roots not treated with Al and digested with XEG. The oligosaccharides obtained were analyzed by MALDI-TOF MS. Data are means ± sd; n = 2. The asterisk shows a significant difference between xth31 and Col-0 at P < 0.05 by Student’s t test.
Xyloglucan Can Bind Al
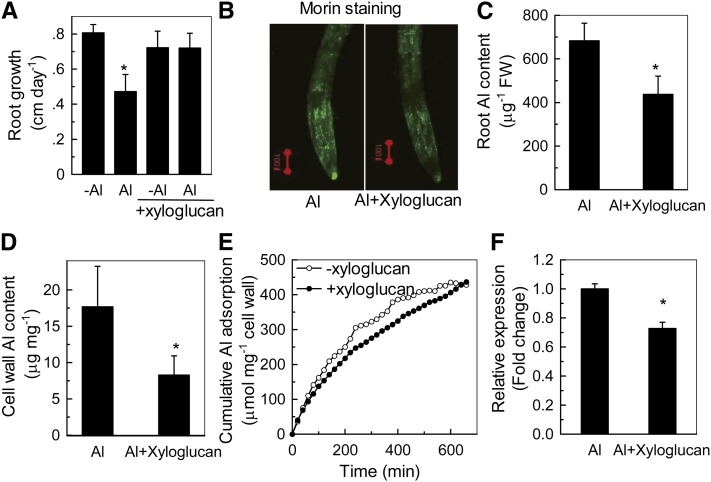
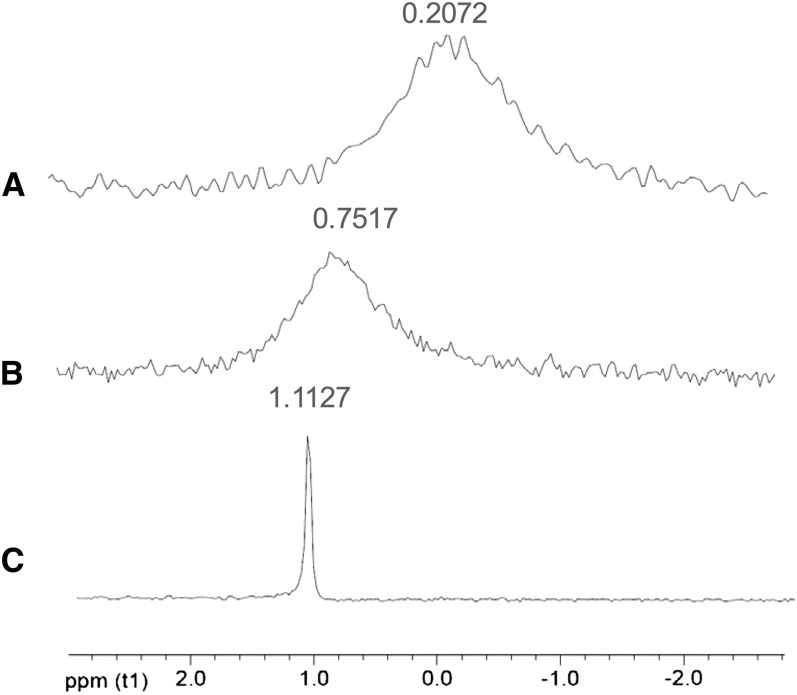
How may xyloglucan content be related to Al sensitivity? To address this question, we applied xyloglucan to the culture medium of Col-0, and, to our surprise, xyloglucan treatment significantly alleviated Al-induced root inhibition (Figure 8A). Furthermore, exogenous application of xyloglucan decreased Al accumulation in the roots (Figures 8B and 8C) and cell wall (Figures 8D and 8E) and caused a decrease in XTH31 transcript accumulation (Figure 8F). The role of xyloglucan in Al accumulation was further verified in the xxt1 xxt2 double mutant, which has hardly any detectable xyloglucan (Cavalier et al., 2008). The double mutant contained lower levels of Al in the root and accumulated and adsorbed less Al in the root cell wall (see Supplemental Figure 6 online). Together, these data provide strong evidence that xyloglucan may be a key factor affecting Al sensitivity. However, the question remained of whether xyloglucan could form a complex with Al. To test this possibility, we produced 27Al-NMR spectra of AlCl3 without or with polysaccharides or citrate. Changes in the 27Al chemical shift represent the formation of complexes between Al and organic compounds in the solution. Citrate is known to chelate Al to form stable complexes with altered chemical shifts in NMR spectra (Ma et al., 1997). Chemical shifts of 27Al from pure AlCl3 and its complexes with xyloglucan and citrate were all clearly distinct (Figure 9), while no resonances were observed outside the chemical shift range shown, demonstrating the formation of a distinct kind of complex between xyloglucan and Al.
Figure 8.
Alleviation of Al Toxicity by Xyloglucan in Col-0.
(A) Root growth in the presence (Al) or absence (-Al) of Al treated with or without xyloglucan. Seedlings with a root length of ∼1 cm were transferred to plates containing 0 or 50 µM Al, with or without 2 µg mL−1 xyloglucan, and grown for a further 24 h. Root length was measured before and after treatment. Data are means ± sd (n = 10). The asterisk shows a significant effect of Al at P < 0.05 by Student’s t test.
(B) Morin staining of the roots treated with or without xyloglucan in the presence of Al according to (A). Red bar = 100 µm.
(C) Al content in roots treated with or without xyloglucan in the presence of Al. The asterisk shows a significant difference at P < 0.05 by Student’s t test. FW, fresh weight.
(D) Al content in the cell wall treated with or without xyloglucan in the presence of Al. The asterisk shows a significant difference at P < 0.05 by Student’s t test.
(E) Al adsorption kinetics of isolated cell walls treated with or without xyloglucan. Cell wall material was extracted from Al-untreated roots that had grown for 24 h in the presence or absence of xyloglucan. Dry cell wall material was placed in a 2-mL column and kinetics were monitored as previously described (Zheng et al., 2004).
(F) Relative expression of XTH31 in the Col-0 root treated with or without xyloglucan in the presence of Al, as determined by RT-PCR. The asterisk shows a significant difference at P < 0.05 by Student’s t test. Data are means ± sd. n = 3.
Figure 9.
27Al-NMR Profile of AlCl3 in the Presence of Citrate and Xyloglucan.
No resonances were observed outside the chemical shift range shown. AlCl3 alone (A), AlCl3 + xyloglucan (∼1:6, Al3+:sugar residue molar ratio) (B), and AlCl3 + citrate (1:1 molar ratio) (C).
DISCUSSION
XTH31 Is Required for Al Sensitivity in Arabidopsis
Recently, we found that hemicellulose in the root cell wall binds much more Al than pectin, and XET action, along with the expression of some XTH genes, especially XTH31, is significantly inhibited by Al stress (Yang et al., 2011); thus, we proposed that xyloglucans and enzymes that act on them hold the key to understanding why Al3+ is toxic to Arabidopsis. In this study, we further investigated the function of XTH31 in its relationship with Al resistance and demonstrated that XTH31 function somehow increases xyloglucan accumulation in the cell wall, which contributes to the Al binding capacity of the cell wall, such that knockout of XTH31 results in a lower wall xyloglucan content and less Al accumulation in the cell wall (Figures 6B and 7). The xth31 mutant had short roots, and root growth was little further inhibited by 5 to 50 µM Al. By contrast, in the wild type and the complemented line of xth31 (i.e., when XTH31 is present), roots elongated rapidly and their growth was strongly inhibited by Al3+. This could be attributed to unique features of the XTH31-dependent element of growth, whereas the XTH31-independent element of growth is only slightly affected by Al3+ (Figure 1). These observations are consistent with the possibility that Al3+ may modify xyloglucan chains in the cell wall in such a way as to prevent XTH31 from promoting cell expansion. This would explain why it may be biologically economical for the plant to respond to elevated Al3+ by downregulating the expression of XTH31 (Figure 3B; see Supplemental Figure 5 online). Moreover, there exists an Al-responsive promoter in XTH31 as indicated by GUS staining (see Supplemental Figure 5 online). All of these observations support the conclusion that XTH31, a gene related to the modification of cell wall composition, directly affects Al sensitivity in Arabidopsis.
XTH31 Is Involved in Cell Wall Modification and Cell Elongation
XTH31 and XTH32 are the only two XTH members for which XEH activity has been predicted. They have thus been placed in group IIIA, and this activity is predicted to have evolved as a gain of function from the ancestral XET activity (Baumann et al., 2007). XTH31 has XET-like conserved sequences similar to those of bacterial β-glucanases, especially the sequence DEIDF/IEFLG, though the first Ile is replaced by Val in XTH31 (Figure 2). This change conserves the apolar, uncharged nature of this residue and most likely would not change the protein’s structure. It was also shown that the alteration of Phe to Ile in a nasturtium (Tropaeolum majus) seed XTH caused no change in XET activity (de Silva et al., 1993). In Bacillus β-glucanases, the first E of the DEIDIEFLG sequence is proposed to fall within the active site (Borriss et al., 1990) and therefore may be a critical amino acid for the cleavage of (1-4)-β-glucosyl linkages (de Silva et al., 1993; Okazawa et al., 1993; Xu et al., 1995). Thus, XTH31 is likely to be capable of modifying the structure of xyloglucan in the cell wall.
XTH31:GFP fusions localized at the plasma membrane, where XTH31 may modify recently secreted xyloglucans. We found that when only the XTH31 putative signal peptide is fused to GFP, the subcellular localization pattern is indistinguishable from that obtained with the entire XTH31 molecule (i.e., both variants colocalize with a plasma membrane marker) (Figures 2G and 2H), indicating that the signal peptide is functional, in accordance with that of maize XTH1 (Genovesi et al., 2008). Since xyloglucan is synthesized in the Golgi and transported via exocytosis to undergo transglycosylation immediately upon release into the wall (Thompson and Fry, 2001), this membrane-localized XTH31 may be well positioned for catalyzing either this process or the partial hydrolysis of newly secreted xyloglucans. XTH33 has also been reported to be plasma membrane localized (Ndamukong et al., 2009). Here, we further demonstrated that the localization of XTH31 was totally dependent on the signal peptide (Figure 2). The signal peptide may target the protein to the plasma membrane underlying the cell wall in a process that is possibly related to the deposition of released xyloglucan within that domain. It is interesting that fusions with truncated XTH31, lacking 17 amino acids at the N terminus, were still localized at the plasma membrane (Figure 2K), whereas fusions were no longer located at the plasma membrane when truncated by 19 amino acids at the N terminus (Figure 2I). The biological function of these two amino acids needs further investigation.
XTHs with XET activity have been proposed to function in the process of cell expansion (Fry et al., 1992; Van Sandt et al., 2007) and, in many circumstances, extractable XET activity correlates well with growth rate (Fry et al., 1992; Hetherington and Fry, 1993; Potter and Fry, 1993, 1994; Pritchard et al., 1993). In line with this observation, the xth31 mutant was inhibited in growth (Figures 1C and 1D; see Supplemental Figure 1 online). Those XTHs that continue to be expressed in the xth31 mutant (e.g., XTH5, 14, 15, 18, 18, 19, 20, and 26), which contribute to the residual XET activity, in combination with other potentially growth-promoting agents, such as expansins (Cosgrove, 1998), could be responsible for the element of growth that continues, and is Al3+ insensitive, in the xth31 mutant.
Finally, the organ-specific localization of XTH31 transcripts gives further insight into the possible developmental functions of the XTH31-encoded enzyme. XTH31 expression appears to be prominent in regions where cellular expansion is likely to occur, as XTH31-GUS is strongly expressed in the elongation zones of roots (which is consistent with the XET activity) and the young expanding leaves (see Supplemental Figure 4 online), further correlating XTH31 expression with cell expansion.
In Vivo XET Action and in Vitro XEH/XET Activity
In this study, we found that XTH31 produced heterologously in P. pastoris showed high XEH activity and low XET activity in vitro (Figure 5). This result agrees with the sequence-based predictions that as a member of group IIIA, XTH31 has XEH activity (Baumann et al., 2007). Thus, it was unexpected that, relative to the wild type, the xth31 mutant root (in the absence of Al3+) has low in vivo XET action (indicating the colocalization of xyloglucan with an XET-active protein; Figures 4A and 4B), low extractable XDA (indicating extractable XET- and/or XEH-active XTH protein concentration; Figure 4C), and low extractable XET activity (Figure 4D), but normal extractable XEH activity (Table 1). The apparent contradiction that the xth31 mutant was compromised in in vivo XET action and in extractable XET activity but not in extractable XEH activity may be due to the fact that the main action of XTH31 in planta is XET and that the in vitro activity of the heterologously produced enzyme is misleading: For example, (1) there may be differences between P. pastoris- and Arabidopsis-produced XTH31 protein structure (additional peptide tags and differences in posttranslational modification); (2) the innate activity of the enzyme as observed in free solution may be changed in the local environment of the plant wall/membrane interface (Fry, 2004); (3) XTH31 in the root may undergo synergistic or direct protein–protein interactions with other XTHs; (4) the pH of the medium (pH 4.5) that must be used for cultivation of plants in Al3+ resistance tests may be suboptimal for the XET activity of XTH31 (other XTHs have pH optima for XET activity of 5 to 7.5; Maris et al., 2009, 2011); and (5) it is also possible that XTH31, because it is a membrane-bound protein, was not readily solubilized and that any extracted XTH31 was exceeded by other extractable XEH-active proteins (e.g., XTH32 or nonspecific endoglucanases [cellulases] that may also be capable of hydrolyzing xyloglucan). Unfortunately, no method is currently available for specifically assaying the in vivo XEH action of inextractable XTHs on endogenous wall-bound xyloglucan (cf. the test for putative inextractable heterotransglucosylases; Mohler et al., 2012), which would have offered an informative comparison with the in vivo XET action seen in Figure 4A.
Whichever is the correct explanation for the effect of the XTH31 mutation, it remains true that wild-type XTH31 has at least four distinct effects in the root: (1) it increases plant extractable XET activity and in vivo observable XET action (Figure 4); (2) it promotes the elongation of roots (Figures 1C, 1D, and 3A) and stems (see Supplemental Figure 1 online); (3) it increases the accumulation of hemicelluloses (Figure 6A), especially XEG-accessible xyloglucan (Figure 7), in the cell wall; and (4) presumably as a consequence of 3 it increases the Al3+ binding capacity of the roots and their walls (Figures 1C, 6B, and 6C; see Supplemental Figure 3 online). Thus, as an alternative to 1 to 5 above, it may also be plausible that knocking out of XTH31 indirectly diminishes the action and activity of some or all of the 32 other XTH proteins of Arabidopsis. Although the knockout has little effect on, or slightly increases, the steady state mRNA concentrations of eight other XTHs tested, it does pleiotropically diminish the expression of XTH12 and XTH13 in Arabidopsis roots (see Supplemental Figure 7 online); thus, such a side effect of XTH31 may be imposed at the transcriptional level, but this needs to be investigated in future work.
Possible Mechanism of Xyloglucan Involvement in Al Resistance
Our findings raise the question of how XTH31 could affect root Al3+ resistance. The high XET action observed in situ in the xyloglucan-rich roots expressing wild-type XTH31 is significantly inhibited by added Al3+ (Figure 4B; Yang et al., 2011), and this inhibition is accompanied by diminished accumulation of XTH31 mRNA (Figure 3B), thus phenocopying the xth31 mutation. We propose that the in vivo XET action of XTH31 at or near the plasma membrane modifies newly secreted xyloglucan in such a way as to enhance further xyloglucan accumulation. This effect could be related to that observed by Paull and Jones (1978), who showed that elevated concentrations of extracellular xyloglucan suppress further synthesis and secretion of fucosylated polysaccharides (presumably mainly xyloglucan). We thus suggest that the high xyloglucan concentrations due to wild-type XTH31 expression will result in more Al3+ binding sites and, hence, more total Al3+ accumulation within the root elongation zone. This conclusion is based on two experimental results: First, xyloglucan applied exogenously can significantly alleviate Al toxicity in Arabidopsis (Figure 8), presumably by trapping Al outside the root, while the xyloglucan-deficient double mutant xxt1 xxt2 accumulates significantly less Al in its cell walls and roots (see Supplemental Figure 6 online). Second, there is also a chemical shift in the 27Al-NMR spectrum when Al interacts with xyloglucan (Figure 9). Other polysaccharides, such as homogalacturonan, glucuronoxylan, glucomannan, xylan, and mannan (but not cellulose), can also bind with Al, as indicated by changes in the 27Al-NMR chemical shift (see Supplemental Figure 8 online), though these are less relevant to the question of the roles of XTH31. The substrate of XTH is xyloglucan, and we detected a significant decrease of (XEG-accessible) xyloglucan in the xth31 cell walls (Figure 7). It should be noted that treatment with XEG and determination of subunits by MALDI-TOF gives information only about the xyloglucan domains that are accessible to XEG and that the proportion accessible might vary between xth31 and the wild type. Thus, the lower xyloglucan content in the cell walls of both xth31 and xxt1 xxt2 mutants results in a lower proportion of the Al that enters the root being bound in their elongation zone cell walls; thus, less Al-mediated wall tightening occurs.
In conclusion, our results indicate that XTH31 confers Al sensitivity in Arabidopsis by encoding XEH/XET activity and secondarily promoting XET action and xyloglucan accumulation, which leads to the elevated binding of xyloglucan to Al and consequently the inhibition of wall loosening in the elongation zone.
METHODS
Plant Culture and Treatments
All wild-type, mutant, and transgenic Arabidopsis thaliana plants used were in the Col-0 wild-type background. Seeds were surface-sterilized and germinated on an agar-solidified nutrient medium in Petri dishes. The nutrient medium was based on Murashige and Skoog salts (Murashige and Skoog, 1962) containing the following macronutrients (in mM): KNO3, 6.0; Ca(NO3)2, 4.0; MgSO4, 1; and NH4H2PO4, 0.1; and the following micronutrients (in μM): Fe(III)-EDTA, 50; H3BO3, 12.5; MnSO4, 1; CuSO4, 0.5; ZnSO4, 1; H2MoO4, 0.1; and NiSO4, 0.1. The final pH was adjusted to 4.5. The seeds were vernalized at 4°C for 1 d. Petri dishes were placed into a growth chamber, positioned vertically and kept under controlled environmental conditions at 24°C, 140 µmol m−2 s−1 and a 16/8-h day/night rhythm. It is noteworthy that as the xxt1 xxt2 mutant cannot grow well under pH 4.5, these plants were first cultivated at pH 5.7 for 3 weeks and then transplanted to nutrient solution of a pH of 4.5 for 1 week for acclimation before Al treatment, as was done for the Col-0 plants with which they were compared.
For the tissue expression pattern experiment, seedlings were sampled from the root, stem, leave, flower, and silique. For the dose–response experiment, seedlings were exposed to 0.5 mM CaCl2 medium, pH 4.5, containing 0, 5, 10, 25, 50, 100, or 150 μM AlCl3 for 24 h. For the xyloglucan applied exogenously experiments, 2 μg mL−1 xyloglucan was mixed. For the analysis of promoter to other stress, T3 generation of XTH31-GUS transgenic lines with a root length of 1 cm were exposed to 0.5 mM CaCl2 medium, pH 4.5, containing 50 μM AlCl3 and 100 mM NaCl or frozen with ice for cold treatment for 24 h before GUS staining.
Effect of Al on Root Growth
Seedlings with a root length of 1 cm (4 d old) were selected and transferred to Petri dishes containing agar-solidified CaCl2 (0.5 mM) medium with 50 µM Al in the form of AlCl3. Root morphology was recorded using a digital camera connected to a computer. Data were quantified and analyzed using Photoshop 7.0 (Adobe Systems). Plants grew vertically for an additional 7 d, at which point root elongation was determined. For the dose–response experiment, 1-cm long seedlings were exposed to 0.5 mM CaCl2 medium, pH 4.5, containing 0, 5, 10, 25, 50, 100, or 150 μM AlCl3 for 24 h.
Microscopy Observations
Localization of Al ions in roots was determined by staining with morin (C15H10O7; Sigma-Aldrich), according to the method described by Tice et al. (1992). The roots were rinsed with double-deionized water and stained with a 100 µM aqueous solution of morin for 30 min. Then, the roots were washed twice with double-deionized water for 5 min each. Samples were mounted on glass slides and inspected under a confocal laser scanning microscope (LSM 510; Zeiss).
For propidium iodide staining, roots treated with or without Al were floated in a 2 mg mL −1 propidium iodide solution for 1 min before being mounted on glass slides and inspected by confocal laser scanning microscopy (LSM 510; Zeiss).
Cytochemical Assay
The XET action of XTH was determined according to Vissenberg et al. (2000). In brief, roots were incubated in a 6.5 µM sulforhodamine-labeled xyloglucan oligosaccharides (XGO-SRs) mixture for 1 h according to Yang et al. (2011). The assay was followed by a 10-min wash in ethanol/formic acid/water (15:1:4, v/v/v) to remove remaining unreacted xyloglucan oligosaccharide SRs; a further incubation overnight in 5% formic acid removed apoplastic, non-wall-bound xyloglucan-SR. Samples were mounted on glass slides and inspected under a confocal laser scanning microscope (LSM 510; Zeiss) using an excitation light of 540 nm.
Gene Expression Analysis
Total RNA was isolated from root apices (0 to 10 mm) using TRIzol (Invitrogen). cDNA was prepared from 1 µg of total RNA using the PrimeScript RT reagent kit (Takara). For real-time RT-PCR analysis, 1 μL of a 10-fold dilution of cDNA was used for the quantitative analysis of gene expression performed with SYBR Premix ExTaq (Takara) with the following pairs of gene-specific primers (for tubulin: forward, 5′-AAGTTCTGGGAAGTGGTT-3′; reverse, 5′-CTCCCAATGAGTGACAAA-3′; and for XTH31: forward, 5′-TGTCACTCTTTGGCTCG-3′; reverse, 5′- ACCTCATCGTGGTCTCC-3′). Each cDNA sample was run in triplicate. Expression data were normalized with the expression level of the tubulin gene.
Root Cell Wall Extraction and Fractionation
Extraction of root crude cell wall materials and subsequent fractionation of cell wall components were performed according to Zhong and Lauchli (1993) with minor modifications according to Zhu et al. (2012). Roots were ground with a mortar and pestle in liquid nitrogen and then homogenized with 75% ethanol for 20 min in an ice-cold water bath. The sample was then centrifuged at 8000 rpm for 10 min, and the supernatant was removed. The pellets were homogenized and washed with acetone, methanol:chloroform at a ratio of 1:1, and methanol, respectively, for 20 min each, with each supernatant being removed after centrifugation between the washes. The remaining pellet (i.e., the cell wall material) was dried and stored at 4°C for further use.
Pectin was extracted three times by hot water (100°C) for 1 h each and the extracts were combined. Then, hemicellulose fraction 1 (hemicellulose 1) was extracted twice with 4% (w/v) KOH containing 0.02% (w/v) KBH4 at room temperature for 12 h. Hemicellulosic material in the remaining solid residue (hemicellulose 2) was extracted with a solution containing 24% (w/v) KOH and 0.02% (w/v) KBH4 in a similar manner. The hemicellulose 1 plus the hemicellulose 2 fractions were referred to as hemicellulose material.
Determination of Total Sugar Residues
The content of total sugar residues in the hemicellulosic fractions was determined by the phenol–sulfuric acid method (Dubois et al., 1956) and expressed as Glc equivalents. Briefly, 200 μL of hemicellulose extracts was incubated with 1 mL of 98% H2SO4 and 10 μL of 80% phenol at room temperature for 15 min and then incubated at 100°C for 15 min. After cooling, the absorbance at 490 nm was measured spectrophotometrically.
MALDI-TOF Mass Spectrometry Analysis of Xyloglucan Oligosaccharides
The alcohol-insoluble residues (AIRs) were generated from roots of Col-0 and xth31 and destarched with α-amylase (Bacillus sp). The xyloglucan-enriched KOH-soluble fraction was prepared by treating 50 mg of destarched AIRs in 4 n KOH solution, after neutralization and dialysis, finally lyophilized. Then, 0.5 mg of AIRs or KOH fraction was incubated in 100 μL of 50 mM ammonium formate, pH 5.0, with one unit of xyloglucanase (E-XEGP; Megazyme) for 18 h at 37°C. The supernatants were recovered, and 1 μL of aqueous sample plus 10 ng xylopentaose was spotted with an equal volume of matrix solution (10 mg mL−1 2,5-dihydroxbenzoic acid). After being dried on the MALDI target plate, spectra were analyzed on a Bruker Autoflex MALDI-TOF mass spectrometry (MS) instrument (Bruker) in the positive reflection mode with an acceleration voltage of 20 kV. The relative height of each generated oligosaccharide ion peak was counted to determine their relative abundance as described by Zhang et al. (2012).
Measurement of XDA
Four-week-old Arabidopsis Col-0 and xth31 mutant plants were used for this experiment. Seedlings of a similar size were selected and treated in a 0.5 mM CaCl2 solution with or without 50 µM Al for 24 h. After treatment, roots were harvested and enzyme extracts were prepared as described by Soga et al. (1999). Briefly, roots were homogenized in ice-cold sodium phosphate buffer (10 mM, pH 7) and then centrifuged. The remaining cell wall pellet was washed three times with sodium phosphate buffer (10 mM, pH 7), after which the wall pellet was resuspended in sodium phosphate buffer (10 mM, pH 6) containing 1 M NaCl. The walls were extracted in this buffer for at least 24 h at 4°C before centrifugation. This supernatant was then used as a crude enzyme extract to measure XDA as described by Sulová et al. (1995). The reaction mixture contained 100 μL of the enzyme extract (containing ∼40 µg protein), 0.4 mg mL−1 xyloglucan (Megazyme International), 0.2 mg mL−1 (∼150 µM) xyloglucan oligosaccharides (Megazyme International), and 0.1 M sodium phosphate buffer, pH 6, in a final volume of 200 µL. Xyloglucan oligosaccharides act as additional glycosyl acceptors so that both XET and XEH activity are recorded. The reaction was terminated after 60 min at 37°C via the addition of 0.1 mL of 1 n HCl. The remaining xyloglucan was then assayed by the iodine staining method as below. The XDA activity was calculated as the proportion of the xyloglucan (total 40 µg) degraded by 1 µg of cell wall protein in 60 min. Protein estimation was performed using the Bradford assay using a commercially available Bradford Reagent (Bio-Rad). Each experiment had four biological replicates. Experiments were repeated twice.
Xyloglucan was determined by Kooiman’s iodine staining method with a slight modification (Kooiman 1960; Nishitani and Masuda, 1981). A 100-μL solution was mixed with 0.25 mL of an aqueous solution containing 0.5% I2 and 1.0% KI followed by 2.0 mL of an aqueous solution of 20% sodium sulfate. After the reaction mixture was kept for 1 h at 4°C in darkness, A640 was read. Xyloglucan contents were calculated and tamarind xyloglucan (Megazyme) was used as a standard. Xyloglucan was dissolved at room temperature with continuous stirring for 24 h and then kept at room temperature for two more days for complete dissolution.
Construction of the Expression Vector
The cDNA of each of the XTH31 proteins was amplified without its own predicted secretion signal sequence and ligated into a pMD18-T vector using primers that contained XhoI and XbaI restriction sites. After sequencing, the XhoI-XbaI fragment from this plasmid was ligated into the pPICZαC vector in frame with the vector’s α-factor secretion signal sequence for production and secretion in the Pichia pastoris expression system (Invitrogen). This vector also provides a c-myc epitope and a His tag at the C terminus of the protein.
Recombinant Protein Production
The pPICZα-XTH expression vector was linearized with PmeI and introduced into the P. pastoris strain X33 by electroporation according to the manufacturer’s protocol. Multiple-copy integrants were selected on YPDS plates (yeast/peptone/dextrose/sorbitol regeneration medium) containing 100 µg mL−1 zeocin according to Maris et al. (2009). Colony-PCR on selected colonies confirmed the presence of the expression construct in the yeast genome. Two selected and verified colonies (12a and 23c, which expressed the construct with XTH31; the empty vector expressed the construct without XTH31) were grown overnight in liquid buffered complex medium containing glycerol (BMGY) and 100 µg mL−1 zeocin at 28°C on a shaker at a speed of 250 rpm, and the same was done for the P. pastoris strain X33 wild type, except for the absence of 100 µg mL−1 zeocin. Then, 100 mL of BMGY medium in a 500-mL culture flask was inoculated with 80 μL of this culture and grown overnight under the same conditions until OD600 reached 2.0 to 6.0. Yeast cells were harvested by centrifugation (2000g for 5 min) and diluted to OD600 = 1 in buffered complex medium containing methanol (BMMY). Induction was performed in 1.0-liter flasks containing 200 mL of culture at 28°C, and the samples were cultivated on a shaker at 250 rpm for 4 d with the addition of methanol to a final concentration ∼1% every 24 h. At the end of the induction period, yeast cells were discarded by centrifugation (5000g for 10 min), and proteins were recovered from the medium by precipitation with 40% (w/v) ammonium sulfate and centrifugation (10,000g for 45 min). Pellets were redissolved in 0.5 M NaCl, 20 mM Tris-HCl, and 5 mM imidazole, pH 7.9, and dialyzed overnight against 8.0 liters of the same buffer.
Assay of XEH and XET Activities of P. pastoris–Produced XTH31 in Vitro
For a highly sensitive assay of XET activity, a radiochemical method based on that of Fry et al. (1992) was used. The reaction mixture (final volume 60 µL) contained (final concentrations) 1.7 mg/mL nonradioactive tamarind xyloglucan, 1.26 µM [1-3H]XXLGol (containing a small proportion of [1-3H]XLXGol; specific activity 66 MBq/µmol; synthesized by reacting tamarind xyloglucan oligosaccharides with NaB3H4 and purified by preparative thin layer chromatography), 67 mM succinate (Na+, pH 5.5), 0.5% chlorobutanol, and 20 μL enzyme preparation (added last; XTH31 from P. pastoris, protein from wild-type or empty vector P. pastoris, a crude grass extract [extracted by the method of Franková and Fry (2011) from young shoots of the grass Holcus lanatus], or protein-free blank). At timed intervals, 10-μL aliquots were stopped with 10 μL formic acid, the acidified solution was dried onto Whatman 3MM paper, washed in running tap water for 24 h and redried, and any 3H-polysaccharide remaining on the paper was assayed in OptiScint HiSafe (Perkin-Elmer).
XEH and XET activities were measured by a new method using [reducing-end-3H]xyloglucan as substrate. This substrate was prepared by incubating a reaction mixture (final volume 600 µL) containing 50 kBq [1-3H]XXLGol, 1.7 mg/mL tamarind xyloglucan, 67 mM succinate (Na+, pH 5.5), and 200 μL of the XET-rich H. lanatus enzyme preparation at 20°C for 24 h. Products were then heated at 100°C for 1 h, denatured proteins were pelleted by centrifugation, and the [3H]xyloglucan in the supernatant was freed of buffer and oligosaccharides by dialysis twice against 0.5% chlorobutanol; yield = 35 kBq (specific activity ∼12.5 kBq/mg; median Mr ∼2 × 105).
In the novel assay for XEH, the reaction mixture (final volume 70 µL) contained 1.3 kBq (1.5 mg/mL) [reducing-end-3H]xyloglucan, 57 mM succinate (Na+, pH 5.5), 0.5% chlorobutanol, and 20 μL enzyme (as listed above; added last). For an assay of XET activity under comparable conditions, the reaction mixture was supplemented with 200 µM XXXG. In both assays, after 48 h at 20°C, the products were analyzed by chromatography on Whatman 3MM paper in ethyl acetate/acetic acid/water (10:5:6) for 48 h in the presence of external markers (maltoheptaose, maltotetraose, and [3H]XXLGol). Strips of the dried chromatogram were assayed for 3H in OptiScint HiSafe, which has a counting efficiency of ∼7% for oligosaccharides and ∼30% for xyloglucan. Nonradioactive markers were stained with AgNO3 (Fry, 2000).
Radiochemical Assay of XET and XEH Activities in Root Extracts
Mutant and wild-type roots were frozen, placed in ice-cold 3.2 M (NH4)2SO4, and stored at 4°C. Surplus (NH4)2SO4 solution was then removed, and roots (10 to 20 mg wet weight) were homogenized with a pestle in 250 μL of ice-cold 0.3 M succinate (Na+, pH 5.5) containing 10 mM CaCl2 and 20 mM ascorbate. The homogenate was left at 0°C for 2.5 h with occasional shaking and then centrifuged for 5 min at 13,000g. The supernatant enzyme solution (protein concentration 0.12 to 0.69 µg/μL by the Bradford assay) was aliquoted and stored at –80°C. XEH activity was assayed in a reaction mixture containing the buffered enzyme solution (64% v/v), 11.6 Bq/μL [reducing-end-3H]xyloglucan, and 0.18% (w/v) chlorobutanol at 20°C for 12 and 24 h. The [3H]octasaccharide released by hydrolysis (from 232 Bq substrate) was assayed by paper chromatography as above. XET activity was assayed in a reaction mixture containing buffered enzyme solution (50%, v/v), 2.5 µg/μL tamarind xyloglucan, and 114 Bq/μL [1-3H]XXLGol at 20°C. At intervals (0 to 4 h), 3H-polysaccharide (formed from 1140 Bq acceptor substrate) was assayed by paper binding as above.
Al Content Measurement
The Al content in each pellet was extracted by 2 n HCl for 24 h with occasional shaking. For total Al determination, roots were harvested and digested with HNO3:HClO4 (4:1, v/v). Al concentrations in the extracts were determined by inductively coupled plasma–atomic emission spectrometry (IRIS/AP optical emission spectrometer).
Adsorption Kinetics
To determine the ability of different cell wall components to adsorb Al, a total of 5 mg dried cell wall material was placed in a 2-mL column equipped with a filter at the bottom. The cell wall material was rehydrated for 2 h in 0.5 mM CaCl2 at pH 4.5. The adsorption solution consisted of 20 µM AlCl3 in 0.5 mM CaCl2 at pH 4.5. The solution was passed through the bed of cell walls by a peristaltic pump at 12 mL h−1. The eluate was collected in 4-mL aliquots, which were assayed for Al spectrophotometrically with pyrocatechol violet according to Kerven et al. (1989) with some modification (Zheng et al., 2004). The kinetics study was performed twice independently, and one set of adsorption curves is presented in Results.
Measurement of 27Al-NMR
For 27Al -NMR, 50 μL of 10 mM AlCl3 in 99.9% D2O was mixed either with 450 μL of 10 mg mL−1 polysaccharide solution or suspension (xyloglucan, glucomannan, mannan, polygalacturonic acid, glucuronoxylan, cellulose, or xylan, giving sugar residues:AlCl3 molar ratios of between 5.1:1 and 6.8:1) or with 400 μL 99.9% D2O and 50 μL of 10 mM citric acid (giving residues:AlCl3 molar ratios of 1:1) to a final volume of 500 µL. The 27Al-NMR spectra were obtained with a Bruker Advanced DMX 500 NMR. The parameters used were as follows: frequency range, 104.2 kHz; acquisition time, 0.31 s. Aluminum chloride (0.2 mM AlCl3 in 0.1 M HCl) was used as an external reference for calibration of the chemical shift.
Generation of XTH31-GUS Transgenic Lines
To assess the regulation of expression of Arabidopsis XTH31, we took the approach of generating transgenic plants expressing the GUS-encoding reporter gene under the control of the potential regulatory regions lying upstream of the XTH genes. Using gene-specific PCR primers (forward, 5′-GCAAGCTTGTTGATTTTGATTGGTTAT-3′; reverse, 5′-CGGATCCTTTTGAGTGAAGTAAAACCT-3′), we were able to generate DNA products corresponding to an ∼2.3-kb region found upstream from the translational start site of the XTH31. These PCR fragments were verified by sequencing and ligated upstream of the GUS reporter gene in the pBI 121 vector, which is capable of propagating in Agrobacterium tumefaciens and being transformed into plant cells.
35S:XTH31-GFP Expression Constructs, Transient Onion Transformation, and Plasmolysis
The full XTH31 coding sequence (XTH31 Full), the same without the whole signal peptide (XTH31 Wint), tXTH31 Full without the first 17 of the 19 putative SP residues (XTH31 Pint), and only the XTH31 signal peptide (XTH31 SP) were cloned in the pBI 221 vector under the control of a cauliflower mosaic virus 35S promoter and fused in the 3′ region with the GFP according to Genovesi et al. (2008). Onion (Allium cepa) cells were bombarded at 900 p.s.i. with 5 µg of DNA plasmids for expression of the fusion with or without plasma membrane marker pm-rk CD-1007 or GFP alone as a control using a Biolistic PSD-1000/He particle delivery system (Bio-Rad). After particle bombardment, the samples were incubated for 24 to 60 h at 25°C in the dark. Samples were mounted on glass slides and inspected under a confocal laser scanning microscope (LSM 510; Zeiss). When indicated, cells were plasmolysed in saturated Suc for 15 min.
Arabidopsis Transformation
The cDNA sequence of XTH31 was PCR amplified (forward, 5′-ACCCGGGATGGCTTTGTCTCTTATC-3′, reverse, 5′-CTCTAGAACATTCTGGTGTTTGGGTATG-3′), sequenced, subcloned into the pCAMBIA2300 vector, which was predigested with SmaI and XbaI, and then transferred into Agrobacterium (GV3101 strain) by heat shock. Arabidopsis plants were transformed by the vacuum infiltration method described by Bechtold et al. (1993). T1 transgenic lines were selected on plates containing 0.8% (w/v) agar and 50 µg mL−1 kanamycin. The kanamycin-resistant seedlings were transferred to soil and grown to maturation to harvest the seeds.
Statistical Analysis
Each experiment was repeated independently at least two times, and one set of data is shown in Results. Data were analyzed by one-way analysis of variance, and the means were compared by Student’s t test. Different letters and asterisks on the histograms indicate statistical differences at the P < 0.05 level.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers:XTH31: At3g44990.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. The Phenotypes of Col-0, xth31 Mutant, xth31 Complemented, and Overexpression Lines.
Supplemental Figure 2. Photographs Are of Representative Col-0 and xth31 Roots Stained with Propidium Iodide.
Supplemental Figure 3. Morin Staining of Col-0 and xth31.
Supplemental Figure 4. Quantitative Real-Time RT-PCR Analysis and GUS Analysis of XTH31 Expression in Various Tissues.
Supplemental Figure 5. GUS Staining of XTH31 Expression in Stress.
Supplemental Figure 6. Al Content in Root, Cell Wall, and Cell Wall Al Adsorption Kinetics.
Supplemental Figure 7. Quantitative RT-PCR Analysis of Other XTH Gene Expression in Roots of xth31 Compared with Col-0.
Supplemental Figure 8. 27Al-NMR Profile of AlCl3 in the Presence of Polysaccharide.
Acknowledgments
We thank Zhao Pu Liu (Nanjing Agriculture University) and Yong Quan Li (Zhejiang University) for assisting with the yeast heterologous expression system; Qing Shao and Jian Yang Pang (College of Medicine) and Shi Hua Wu (College of Life Science of Zhejiang University) for assisting with NMR analysis; Han Ming Chen and She long Zhang (College of Life Science of Zhejiang University) for assisting with confocal analysis; Lenka Franková, Thomas Simmons, and Janice Miller (University of Edinburgh) for help with the radiochemical assays; and Kenneth Keegstra (Michigan State University) for the gift of xxt1 xxt2 mutant seeds. This research was supported by a grant from Natural Science Foundation of China (30830076), a Changjiang Scholarship, the Program for Innovative Research Team in Universities (IRT1185), the Fundamental Research Funds for the Central Universities, the UK Biotechnology and Biological Sciences Research Council, and the U.S. National Science Foundation (MCB 0817976 to J.B.).
AUTHOR CONTRIBUTIONS
X.F.Z., Y.Z.S., G.J.L., T.J., and X.Y.X. performed research. X.F.Z. analyzed data and wrote the draft. S.C.F. designed the new XEH assay, supervised the radiochemical analyses, and revised the article. B.C.Z. and Y.H.Z. measured xyloglucan content and revise the article. J.B. developed the T-DNA mutant and revised the article. C.Z.M. was involved in genetic analysis. Y.J.P. was involved in 27Al-NMR analysis. J.L.Y., P.W., and S.J.Z. designed the research. S.J.Z. wrote the article.
Glossary
- XEG
xyloglucanase
- XTH
xyloglucan endotransglucosylase/hydrolase
- XET
xyloglucan endotransglucosylase
- XEH
xyloglucan endohydrolase
- Col-0
Columbia-0
- GFP
green fluorescent protein
- GUS
β-glucuronidase
- XDA
xyloglucan degrading activity
- MALDI-TOF
to be defined
- AIR
alcohol-insoluble residue
- MS
mass spectrometry
References
- Baumann M.J., Eklöf J.M., Michel G., Kallas Å.M., Teeri T.T., Czjzek M., Brumer H., III (2007). Structural evidence for the evolution of xyloglucanase activity from xyloglucan endo-transglycosylases: Biological implications for cell wall metabolism. Plant Cell 19: 1947–1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold N., Ellis J., Pelletier G. (1993). In-planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. CR. Acad. Bulg. Sci. 316: 1194–1199 [Google Scholar]
- Becnel J., Natarajan M., Kipp A., Braam J. (2006). Developmental expression patterns of Arabidopsis XTH genes reported by transgenes and Genevestigator. Plant Mol. Biol. 61: 451–467 [DOI] [PubMed] [Google Scholar]
- Blamey F.P.C., Edmeades D.C., Wheeler D.M. (1990). Role of cation-exchange capacity in differential aluminum tolerance of Lotus species. J. Plant Nutr. 13: 729–744 [Google Scholar]
- Blanc G., Barakat A., Guyot R., Cooke R., Delseny M. (2000). Extensive duplication and reshuffling in the Arabidopsis genome. Plant Cell 12: 1093–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borriss R., Buettner K., Maentsaelae P. (1990). Structure of the β-l,3-1,4-glucanase gene of Bacillus macerans: Homologies to other beta-glucanases. Mol. Gen. Genet. 222: 278–283 [DOI] [PubMed] [Google Scholar]
- Carpita N.C. (1996). Structure and biogenesis of the cell walls of grasses. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47: 445–476 [DOI] [PubMed] [Google Scholar]
- Carpita N.C., Gibeaut D.M. (1993). Structural models of primary cell walls in flowering plants: Consistency of molecular structure with the physical properties of the walls during growth. Plant J. 3: 1–30 [DOI] [PubMed] [Google Scholar]
- Cavalier D.M., Lerouxel O., Neumetzler L., Yamauchi K., Reinecke A., Freshour G., Zabotina O.A., Hahn M.G., Burgert I., Pauly M., Raikhel N.V., Keegstra K. (2008). Disrupting two Arabidopsis thaliana xylosyltransferase genes results in plants deficient in xyloglucan, a major primary cell wall component. Plant Cell 20: 1519–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y.C., Yamamoto Y., Matsumoto H. (1999). Accumulation of aluminium in the cell wall pectin in cultured tobacco (Nicotiana tabacum L.) cells treated with a combination of aluminium and iron. Plant Cell Environ. 22: 1009–1017 [Google Scholar]
- Clarkson D.T. (1967). Interactions between aluminum and phosphorus on root surfaces and cell wall material. Plant Soil 27: 347–356 [Google Scholar]
- Cosgrove D.J. (1998). Cell wall loosening by expansins. Plant Physiol. 118: 333–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove D.J. (2005). Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 6: 850–861 [DOI] [PubMed] [Google Scholar]
- de Silva J., Jarman C.D., Arrowsmith D.A., Stronach M.S., Chengappa S., Sidebottom C., Reid J.S.G. (1993). Molecular characterization of a xyloglucan-specific endo-(1—>4)-β-D-glucanase (xyloglucan endo-transglycosylase) from nasturtium seeds. Plant J. 3: 701–711 [PubMed] [Google Scholar]
- Dubois M., Gilles K.A., Hamilton J.K., Rebers P.A., Smith F. (1956). Colorimetric method for determination of sugars and related substances. Anal. Chem. 28: 350–356 [Google Scholar]
- Foy C.D. (1988). Plant adaptation to acid, aluminum-toxic soils. Commun. Soil Sci. Plant Anal. 19: 959–987 [Google Scholar]
- Franková L., Fry S.C. (2011). Phylogenetic variation in glycosidases and glycanases acting on plant cell wall polysaccharides, and the detection of transglycosidase and trans-β-xylanase activities. Plant J. 67: 662–681 [DOI] [PubMed] [Google Scholar]
- Fry S.C. (1989). Cellulases, hemicelluloses and auxin-stimulated growth: A possible relationship. Physiol. Plant. 75: 532–536 [Google Scholar]
- Fry S.C. (2000). The Growing Plant Cell Wall: Chemical and Metabolic Analysis, Reprint Edition. (Caldwell, NJ: The Blackburn Press)
- Fry S.C. (2004). Tansley Review: Primary cell wall metabolism: Tracking the careers of wall polymers in living plant cells. New Phytol. 161: 641–675 [DOI] [PubMed] [Google Scholar]
- Fry S.C., Smith R.C., Renwick K.F., Martin D.J., Hodge S.K., Matthews K.J. (1992). Xyloglucan endotransglycosylase, a new wall-loosening enzyme activity from plants. Biochem. J. 282: 821–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry S.C., et al. (1993). An unambiguous nomenclature for xyloglucan-derived oligosaccharides. Physiol. Plant. 89: 1–3 [Google Scholar]
- Geisler-Lee J., et al. (2006). Poplar carbohydrate-active enzymes. Gene identification and expression analyses. Plant Physiol. 140: 946–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genovesi V., Fornalé S., Fry S.C., Ruel K., Ferrer P., Encina A., Sonbol F.M., Bosch J., Puigdomènech P., Rigau J., Caparrós-Ruiz D. (2008). ZmXTH1, a new xyloglucan endotransglucosylase/hydrolase in maize, affects cell wall structure and composition in Arabidopsis thaliana. J. Exp. Bot. 59: 875–889 [DOI] [PubMed] [Google Scholar]
- Hayashi T. (1989). Xyloglucans in the primary cell wall. Annu. Rev. Plant Physiol. Plant Mol. Biol. 40: 139–168 [Google Scholar]
- Hazen S.P., Scott-Craig J.S., Walton J.D. (2002). Cellulose synthase-like genes of rice. Plant Physiol. 128: 336–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetherington P.R., Fry S.C. (1993). Xyloglucan endotransglycosylase activity in carrot cell suspensions during cell elongation and somatic embryogenesis. Plant Physiol. 103: 987–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horst W.J., Wang Y., Eticha D. (2010). The role of the root apoplast in aluminum-induced inhibition of root elongation and in aluminum resistance of plants: A review. Ann. Bot. (Lond.) 106: 185–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerven G.L., Edwards D.G., Asher C.J., Halman P.S., Kokot S. (1989). Aluminium determination in soil solution. II. Short-term colorimetric procedures for the measurement of inorganic monomeric aluminium in the presence of organic ligands. Aust. J. Soil Res. 27: 91–102 [Google Scholar]
- Kochian L.V. (1995). Cellular mechanisms of aluminum toxicity and resistance in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 46: 237–260 [Google Scholar]
- Kooiman P. (1960). A method for the determination of amyloid in plant seeds. Recl. Trav. Chim. Pays-Bas. 79: 675–678.
- Liu Q., Yang J.L., He L.S., Li Y.Y., Zheng S.J. (2008). Effect of aluminum on cell wall, plasma membrane, antioxidants and root elongation in triticale. Biol. Plant. 52: 87–92 [Google Scholar]
- Llugany M., Poschenrieder C., Barceló J. (1995). Monitoring of aluminium-induced inhibition of root elongation in four maize cultivars differing in resistance to aluminium and proton toxicity. Physiol. Plant. 93: 265–271 [Google Scholar]
- Ma J.F., Hiradate S., Nomoto K., Iwashita T., Matsumoto H. (1997). Internal detoxification mechanism of Al in hydrangea-Identification of Al form in the leaves. Plant Physiol. 113: 1033–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madson M., Dunand C., Li X., Verma R., Vanzin G.F., Caplan J., Shoue D.A., Carpita N.C., Reiter W.D. (2003). The MUR3 gene of Arabidopsis encodes a xyloglucan galactosyltransferase that is evolutionarily related to animal exostosins. Plant Cell 15: 1662–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maris A., Kaewthai N., Eklöf J.M., Miller J.G., Brumer H., Fry S.C., Verbelen J.-P., Vissenberg K. (2011). Differences in enzymic properties of five recombinant xyloglucan endotransglucosylase/hydrolase (XTH) proteins of Arabidopsis thaliana. J. Exp. Bot. 62: 261–271 [DOI] [PubMed] [Google Scholar]
- Maris A., Suslov D., Fry S.C., Verbelen J.P., Vissenberg K. (2009). Enzymic characterization of two recombinant xyloglucan endotransglucosylase/hydrolase (XTH) proteins of Arabidopsis and their effect on root growth and cell wall extension. J. Exp. Bot. 60: 3959–3972 [DOI] [PubMed] [Google Scholar]
- Mohler K.E., Simmons T.J., Fry S.C. (October 19, 2012). Mixed-linkage glucan:xyloglucan endotransglucosylase (MXE) re-models hemicelluloses in Equisetum shoots but not in barley shoots or Equisetum callus. New Phytol. http://dx.doi.org/10.1111/j.1469-8137.2012.04371.x. [DOI] [PubMed]
- Murashige T., Skoog F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15: 473–496 [Google Scholar]
- Ndamukong I., Chetram A., Saleh A., Avramova Z. (2009). Wall-modifying genes regulated by the Arabidopsis homolog of trithorax, ATX1: Repression of the XTH33 gene as a test case. Plant J. 58: 541–553 [DOI] [PubMed] [Google Scholar]
- Nishitani K., Masuda Y. (1981). Auxin-induced changes in the cell wall structure: Changes in the sugar compositions, intrinsic viscosity and molecular weight distribution of matrix polysaccharides of the epicotyl cell wall of Vigna angularis. Physiol. Plant. 52: 482–494 [Google Scholar]
- Nishitani K., Tominaga R. (1992). Endo-xyloglucan transferase, a novel class of glycosyltransferase that catalyzes transfer of a segment of xyloglucan molecule to another xyloglucan molecule. J. Biol. Chem. 267: 21058–21064 [PubMed] [Google Scholar]
- Nishitani K., Vissenberg K. (2007). Roles of the XTH protein family in the expanding cell. In The Expanding Cell. Plant Cell Monographs, Vol. 5, J.P. Verbelen and K. Vissenberg, eds (Berlin, Heidelberg, New York: Springer), 89–116.
- Obel N., Erben V., Schwarz T., Kühnel S., Fodor A., Pauly M. (2009). Microanalysis of plant cell wall polysaccharides. Mol. Plant 2: 922–932 [DOI] [PubMed] [Google Scholar]
- Okazawa K., Sato Y., Nakagawa T., Asada K., Kato I., Tomita E., Nishitani K. (1993). Molecular cloning and cDNA sequencing of endoxyloglucan transferase, a novel class of glycosyltransferase that mediates molecular grafting between matrix polysaccharides in plant cell walls. J. Biol. Chem. 268: 25364–25368 [PubMed] [Google Scholar]
- Park Y.B., Cosgrove D.J. (2012). Changes in cell wall biomechanical properties in the xyloglucan-deficient xxt1/xxt2 mutant of Arabidopsis. Plant Physiol. 158: 465–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull R.E., Jones R.L. (1978). Regulation of synthesis and secretion of fucose-containing polysaccharides in cultured sycamore cells. Aust. J. Plant Physiol. 5: 457–467 [Google Scholar]
- Potter I., Fry S.C. (1993). Xyloglucan endotransglycosylase activity in pea internodes. Effects of applied gibberellic acid. Plant Physiol. 103: 235–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter I., Fry S.C. (1994). Changes in xyloglucan endotransglycosylase (XET) activity during hormone-induced growth in lettuce and cucumber hypocotyls and spinach cell suspension cultures. J. Exp. Bot. 45: 1703–1710 [Google Scholar]
- Pritchard J., Hetherington P.R., Fry S.C., Tomos A.D. (1993). Xyloglucan endotransglycosylase activity, microfibril orientation and the profites of cell wall properties along growing regions of maize roots. J. Exp. Bot. 44: 1281–1289 [Google Scholar]
- Rose J.K.C., Braam J., Fry S.C., Nishitani K. (2002). The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: Current perspectives and a new unifying nomenclature. Plant Cell Physiol. 43: 1421–1435 [DOI] [PubMed] [Google Scholar]
- Saladié M., Rose J.K.C., Cosgrove D.J., Catalá C. (2006). Characterization of a new xyloglucan endotransglucosylase/hydrolase (XTH) from ripening tomato fruit and implications for the diverse modes of enzymic action. Plant J. 47: 282–295 [DOI] [PubMed] [Google Scholar]
- Sampedro J., Gianzo C., Iglesias N., Guitián E., Revilla G., Zarra I. (2012). AtBGAL10 is the main xyloglucan β-galactosidase in Arabidopsis, and its absence results in unusual xyloglucan subunits and growth defects. Plant Physiol. 158: 1146–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasidharan R., Chinnappa C.C., Staal M., Elzenga J.T.M., Yokoyama R., Nishitani K., Voesenek L.A.C.J., Pierik R. (2010). Light quality-mediated petiole elongation in Arabidopsis during shade avoidance involves cell wall modification by xyloglucan endotransglucosylase/hydrolases. Plant Physiol. 154: 978–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soga K., Wakabayashi K., Hoson T., Kamisaka S. (1999). Hypergravity increases the molecular mass of xyloglucans by decreasing xyloglucan-degrading activity in azuki bean epicotyls. Plant Cell Physiol. 40: 581–585 [DOI] [PubMed] [Google Scholar]
- Strohmeier M., Hrmova M., Fischer M., Harvey A.J., Fincher G.B., Pleiss J. (2004). Molecular modeling of family GH16 glycoside hydrolases: Potential roles for xyloglucan transglucosylases/hydrolases in cell wall modification in the poaceae. Protein Sci. 13: 3200–3213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulová Z., Lednická M., Farkaš V. (1995). A colorimetric assay for xyloglucan-endotransglycosylase from germinating seeds. Anal. Biochem. 229: 80–85 [DOI] [PubMed] [Google Scholar]
- Tabuchi A., Matsumoto H. (2001). Changes in cell-wall properties of wheat (Triticum aestivum) roots during aluminum-induced growth inhibition. Physiol. Plant. 112: 353–358 [DOI] [PubMed] [Google Scholar]
- Thompson J.E., Fry S.C. (2001). Restructuring of wall-bound xyloglucan by transglycosylation in living plant cells. Plant J. 26: 23–34 [DOI] [PubMed] [Google Scholar]
- Tice K.R., Parker D.R., Demason D.A. (1992). Operationally defined apoplastic and symplastic aluminum fractions in root tips of aluminum-intoxicated wheat. Plant Physiol. 100: 309–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Sandt V.S., Suslov D., Verbelen J.-P., Vissenberg K. (2007). Xyloglucan endotransglucosylase activity loosens a plant cell wall. Ann. Bot. (Lond.) 100: 1467–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissenberg K., Martinez-Vilchez I.M., Verbelen J.-P., Miller J.G., Fry S.C. (2000). In vivo colocalization of xyloglucan endotransglycosylase activity and its donor substrate in the elongation zone of Arabidopsis roots. Plant Cell 12: 1229–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W., Purugganan M.M., Polisensky D.H., Antosiewicz D.M., Fry S.C., Braam J. (1995). Arabidopsis TCH4, regulated by hormones and the environment, encodes a xyloglucan endotransglycosylase. Plant Cell 7: 1555–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J.L., Li Y.Y., Zhang Y.J., Zhang S.S., Wu Y.R., Wu P., Zheng S.J. (2008). Cell wall polysaccharides are specifically involved in the exclusion of aluminum from the rice root apex. Plant Physiol. 146: 602–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J.L., Zhu X.F., Peng Y.X., Zheng C., Li G.X., Liu Y., Shi Y.Z., Zheng S.J. (2011). Cell wall hemicellulose contributes significantly to aluminum adsorption and root growth in Arabidopsis. Plant Physiol. 155: 1885–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama R., Nishitani K. (2001). A comprehensive expression analysis of all members of a gene family encoding cell-wall enzymes allowed us to predict cis-regulatory regions involved in cell-wall construction in specific organs of Arabidopsis. Plant Cell Physiol. 42: 1025–1033 [DOI] [PubMed] [Google Scholar]
- Yokoyama R., Rose J.K.C., Nishitani K. (2004). A surprising diversity and abundance of xyloglucan endotransglucosylase/hydrolases in rice. Classification and expression analysis. Plant Physiol. 134: 1088–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabotina O.A., Avci U., Cavalier D., Pattathil S., Chou Y.H., Eberhard S., Danhof L., Keegstra K., Hahn M.G. (2012). Mutations in multiple XXT genes of Arabidopsis reveal the complexity of xyloglucan biosynthesis. Plant Physiol. 159: 1367–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S.J., Song X.Q., Yu B.S., Zhang B.C., Sun C.Q., Knox J.P., Zhou Y.H. (2012). Identification of quantitative trait loci affecting hemicellulose characteristics based on cell wall composition in a wild and cultivated rice species. Mol. Plant 5: 162–175. [DOI] [PubMed]
- Zheng S.J., Lin X.Y., Yang J.L., Liu Q., Tang C. (2004). The kinetics of aluminum adsorption and desorption by root cell walls of an aluminum resistance wheat (Triticum aestivum L.) cultivar. Plant Soil 261: 85–90 [Google Scholar]
- Zheng S.J., Yang J.L. (2005). Target sites of aluminum phytotoxicity. Biol. Plantarum 49: 321–331 [Google Scholar]
- Zhong H., Lauchli A. (1993). Changes of cell wall component and polymer size in primary roots of cotton seedlings under high salinity. J. Exp. Bot. 44: 773–778 [Google Scholar]
- Zhu X.F., Lei G.J., Jiang T., Liu Y., Li G.X., Zheng S.J. (2012). Cell wall polysaccharides are involved in P-deficiency-induced Cd exclusion in Arabidopsis thaliana. Planta 236: 989–997 [DOI] [PubMed] [Google Scholar]