This work shows that the Magnaporthe oryzae effector AvrPiz-t enters into rice cells to target the RING E3 ubiquitin ligase APIP6 for suppression of PAMP-triggered immunity in rice. It also describes that APIP6 degrades AvrPiz-t in planta and positively regulates basal defense to M. oryzae.
Abstract
Although the functions of a few effector proteins produced by bacterial and oomycete plant pathogens have been elucidated in recent years, information for the vast majority of pathogen effectors is still lacking, particularly for those of plant-pathogenic fungi. Here, we show that the avirulence effector AvrPiz-t from the rice blast fungus Magnaporthe oryzae preferentially accumulates in the specialized structure called the biotrophic interfacial complex and is then translocated into rice (Oryza sativa) cells. Ectopic expression of AvrPiz-t in transgenic rice suppresses the flg22- and chitin-induced generation of reactive oxygen species (ROS) and enhances susceptibility to M. oryzae, indicating that AvrPiz-t functions to suppress pathogen-associated molecular pattern (PAMP)-triggered immunity in rice. Interaction assays show that AvrPiz-t suppresses the ubiquitin ligase activity of the rice RING E3 ubiquitin ligase APIP6 and that, in return, APIP6 ubiquitinates AvrPiz-t in vitro. Interestingly, agroinfection assays reveal that AvrPiz-t and AvrPiz-t Interacting Protein 6 (APIP6) are both degraded when coexpressed in Nicotiana benthamiana. Silencing of APIP6 in transgenic rice leads to a significant reduction of flg22-induced ROS generation, suppression of defense-related gene expression, and enhanced susceptibility of rice plants to M. oryzae. Taken together, our results reveal a mechanism in which a fungal effector targets the host ubiquitin proteasome system for the suppression of PAMP-triggered immunity in plants.
INTRODUCTION
Plants have evolved multiple layers of preformed and induced defenses to protect themselves against pathogens. Upon pathogen infection, the recognition of pathogen-associated molecular patterns (PAMPs) by the pattern recognition receptors (PRRs) triggers the first layer of defense response, which is referred to as PAMP-triggered immunity (PTI) and includes the accumulation of reactive oxygen species (ROS) and the deposition of phenolic compounds (Jones and Dangl, 2006). However, pathogens deliver effector proteins to target PRRs or key components in the PTI signaling pathway to suppress the defense triggered by PAMPs. Some of the effectors, referred to as avirulence (Avr) proteins, can interact either directly or indirectly with the cognate resistance (R) gene products or bind to the promoter of the cognate R genes, triggering a second layer of defense called effector-triggered immunity (ETI). ETI is a more robust and effective defense than PTI and is often associated with a hypersensitive response (HR) at the infection site that inhibits pathogen proliferation (Chisholm et al., 2006; Jones and Dangl, 2006). However, several recent studies have challenged the distinction between PTI and ETI and have provided evidence for a continuum between the two types of immunities (Lee et al., 2009; Thomma et al., 2010).
Ubiquitination is one of the important protein posttranslational modifications in eukaryotic cells. The ubiquitin-activating enzymes (E1), ubiquitin-conjugating enzymes (E2), and ubiquitin ligases (E3) are three main enzymes in the ubiquitin proteasome system (UPS) (Vierstra, 2003). Ubiquitination-related proteins are much more abundant in plant genomes than in Saccharomyces cerevisiae and Drosophila melanogaster genomes, reflecting the importance of ubiquitination in plants. Ubiquitination is involved in many biological processes in plants, such as the cell cycle, circadian rhythm control, hormone signaling, growth, and development (Smalle and Vierstra, 2004; Santner and Estelle, 2010). Recent research also suggests that ubiquitination is involved in disease resistance and in suppression of host plant innate immunity by pathogen (Zeng et al., 2006; Shirsekar et al., 2010). For example, the F-box–containing GALA effector proteins in Ralstonia solanacearum interact with Arabidopsis thaliana SKP1-like proteins, a component of the SCF-type E3 ubiquitin ligases (Angot et al., 2006). In a second example, the Pseudomonas syringae effector HopM1 does not contain any known conserved domain associated with the UPS but mediates the proteasome-dependent degradation of an Arabidopsis protein MIN7 (HopM interactor 7) that is required for cell wall–mediated defense (Nomura et al., 2006). Blocking the degradation of trans-Golgi network/early endosome-associated MIN7 by P. syringae is an essential part of the ETI mechanism for counteracting bacterial suppression of PTI (Nomura et al., 2011). A third example concerns AvrPtoB, an avirulence factor produced by P. syringae pv tomato (Abramovitch and Martin, 2005). Instead of targeting the UPS in plants, AvrPtoB contains a U-box–like domain in its C-terminal region and suppresses cell death triggered by Pto and Fen. The E3 ubiquitin ligase activity of AvrPtoB is required for the degradation of the tomato protein kinase Fen, which together with the nucleotide binding site–Leu-rich repeat (NBS-LRR) R protein Prf signals in defense against the pathogen (Rosebrock et al., 2007). A recent study showed that Pto is resistant to AvrPtoB-mediated degradation because it inactivates AvrPtoB E3 ubiquitin ligase activity (Ntoukakis et al., 2009), demonstrating a novel host immunity strategy that inactivates the enzymatic activity of a bacterial effector. The involvement of the UPS in the interactions between plants and oomycetes is emerging. A recent study with potato (Solanum tuberosum) reported that the Phytophthora infestans effector AVR3a interacts with and stabilizes host U-box E3 ubiquitin ligase CMPG1, which is required for the INF1-triggered cell death (Bos et al., 2010). CMPG1 degradation depends on the UPS, and the degradation is prevented by the modifications of CMPG1 E3 ubiquitin ligase activity by AVR3a. However, it is not clear whether fungal effectors and host E3 ubiquitin ligases have a similar relationship or not during plant–fungus interactions.
Rice blast, caused by the fungal pathogen Magnaporthe oryzae, is a devastating disease that causes huge economic losses in epidemic years (Dean et al., 2005). The disease has become an advanced model pathosystem for understanding the molecular basis of plant–fungal interactions because of the availability of both host and pathogen genome sequences, the availability of a wealth of genetic resources, and the feasibility of molecular manipulation of both species (Valent, 1990; Shimamoto and Kyozuka, 2002; Caracuel-Rios and Talbot, 2007). To date, a total of 18 rice (Oryza sativa) R genes, two rice blast quantitative trait loci, and nine Avr genes in M. oryzae have been cloned (J. Liu et al., 2010a; Takahashi et al., 2010; Chen et al., 2011; Okuyama et al., 2011; Yuan et al., 2011; Zhai et al., 2011; Gupta et al., 2012). With the exception of Pi-d2, which encodes a B-lectin kinase protein, all rice R genes encode NBS-LRR proteins. With regard to the pathogen, putative molecular functions of the identified Avr genes vary greatly (Valent and Khang, 2010). The ACE1 gene corresponding to the Pi33 resistance gene encodes a putative hybrid between a polyketide syntase and a nonribosomal peptide synthetase and is a member of an infection-specific gene cluster involved in microbial secondary metabolism (Böhnert et al., 2004; Collemare et al., 2008). The deduced amino acid sequence of Avr-Pita has high similarity to fungal neutral metalloproteases (Orbach et al., 2000). The other seven Avr effectors do not have known functions or identifiable functional domains. Some M. oryzae Avr effectors (i.e., Avr-Pita, PWL1, and PWL2) and biotrophy-associated secreted (BAS) proteins are secreted into biotrophic interface complexes (BICs), and so far only PWL2 and BAS1 have been demonstrated to be translocated into living rice cells (Khang et al., 2010).
We cloned three broad-spectrum R genes, Pi2, Pi9, and Piz-t (Qu et al., 2006; Zhou et al., 2006). Each of these three allelic genes confers resistance to a diverse set of blast isolates and encodes an NBS-LRR protein with high sequence similarity to the other proteins. Sequence comparisons among them revealed a disproportionate number of sequence variations located within the LRR region, and eight amino acid changes differentiating Pi2 from Piz-t are confined to three consecutive LRR repeats that determine resistance specificity (Zhou et al., 2006, 2007). We also cloned AvrPiz-t, the cognate Avr gene for the R gene Piz-t (Li et al., 2009). AvrPiz-t encodes a predicted 108–amino acid polypeptide with a secretion signal at the N terminus. The deduced amino acid sequence of the AvrPiz-t protein shows no sequence similarity to any known proteins. In this study, we demonstrated that AvrPiz-t is secreted into the BIC and translocated into rice cells. Ectopic expression of the AvrPiz-t gene in the Piz-t-lacking rice plants reduces the flg22- and chitin-triggered increase in ROS and enhances susceptibility to a virulent rice blast strain. AvrPiz-t interacts with the E3 ubiquitin ligase AvrPiz-t Interacting Protein 6 (APIP6) and suppresses its E3 ligase activity in vitro. Interestingly, APIP6 also ubiquitinates AvrPiz-t in vitro and promotes degradation of the AvrPiz-t protein in vivo. Silencing of APIP6 reduced the flg22-triggered ROS generation and enhanced the susceptibility to a virulent blast strain. In contrast with previous reports, our study has discovered that a fungal effector can target a host E3 ubiquitin ligase for degradation to suppress PTI in plants. In return, the host E3 ubiquitin ligase can also degrade the fungal effector to reduce its interference on host PTI.
RESULTS
AvrPiz-t Is Translocated into Living Rice Cells during Rice Blast Infection
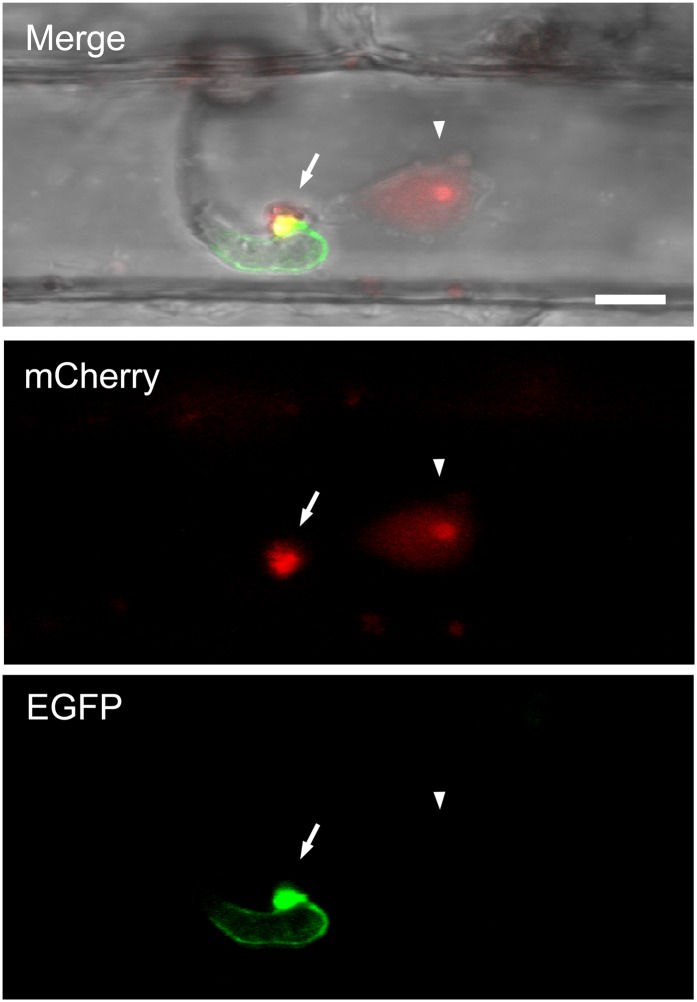
Functional recognition of pathogen Avr proteins by plant intracellular NBS-LRR R proteins leads to a rapid HR and defense response; this process is also known to require the translocation of Avr proteins into plant cells (Houterman et al., 2009; Rafiqi et al., 2010). To assess whether AvrPiz-t is translocated into rice cells, we used a live-cell imaging approach that was successfully used to localize M. oryzae effectors in rice sheath epidermal cells (Khang et al., 2010). The AvrPiz-t gene with its native promoter was engineered to express a C-terminal fusion of mCherry with a nuclear localization signal (AvrPiz-tpro:AvrPiz-t:mCherry:NLS) to facilitate visualization of translocated fluorescent protein by concentrating the AvrPiz-t:mCherry:NLS fusion protein in the rice nucleus. This fusion construct was cloned together with a fusion construct expressing a green fluorescent protein (GFP)–labeled version of the putative extracellular matrix protein BAS4 (BAS4pro:BAS4:EGFP) in a single T-DNA region and transformed into the blast strain O-137. BAS4:EGFP fusion protein was used as a control for the presence of intact host-derived membrane around invasive hyphae (Khang et al., 2010). The transformed strain was inoculated in rice sheaths, and fluorescence was observed by confocal microscopy 30 h after inoculation (Khang et al., 2010). We observed 10 independent fungal transformants that expressed both AvrPiz-t:mCherry:NLS and BAS4:EGFP fusion proteins. All transformants exhibited accumulation of mCherry red fluorescence in BICs (n > 60), and there was no detectable fluorescence inside invasive hyphae. This indicates successful secretion of the fluorescently labeled AvrPiz-t to BICs. To demonstrate the translocation of AvrPiz-t into host cells, we focused on three independent transformants that showed bright fluorescence of AvrPiz-t:mCherry:NLS. At successful infection sites where BAS4:EGFP was not spilled into host cells (n = 22) (Figure 1, EGFP, bottom panel), AvrPiz-t-associated red fluorescence was observed in the nuclei of invaded host cells (n = 21) (Figure 1, mCherry, middle panel) and in BIC (Figure 1, Merge, upper panel). This result indicated that AvrPiz-t:mCherry:NLS had been translocated into host cells. We could not detect red fluorescence in the nuclei of adjoining cells that did not contain IH.
Figure 1.
Translocation of AvrPiz-t into Rice Cells during Rice Blast Infection.
The fungal transformant KV129 expressing AvrPiz-t:mCherry:NLS and putative interfacial matrix protein BAS4:EGFP at 30 h after infection in the sheath cells of rice cultivar YT16 rice is shown as a projection of confocal optical sections over a depth of 4 μm. “Merge” shows bright-field, mCherry (red), and enhanced GFP (green). Arrows indicate BIC, arrowheads indicate rice nucleus, and yellow indicates overlapping mCherry and enhanced GFP fluorescence signals. Pinhole settings were 1 airy unit for enhanced GFP and 12.5 airy units for mCherry. mCherry fluorescence occurred in the BIC and in the nucleus of the invaded rice cell with brighter fluorescence in the presumed nucleolus. Bar = 5 μm.
Ectopic Expression of AvrPiz-t Reduces Rice Basal Defense against M. oryzae
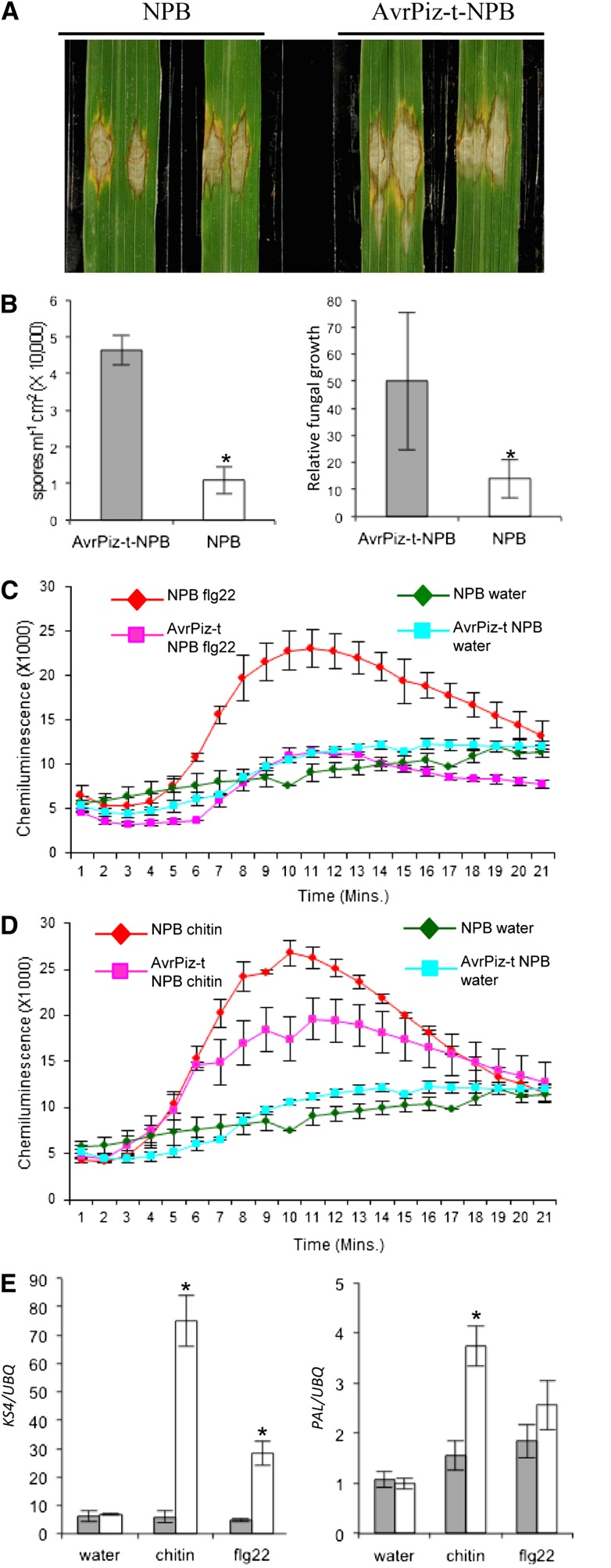
To investigate the function of AvrPiz-t in rice cells, we generated stable transgenic rice expressing the truncated AvrPiz-t gene (without the N-terminal signal peptide sequence) with a C-terminal fusion of Myc tag (AvrPiz-t:Myc) under the maize (Zea mays) ubiquitin promoter in the Piz-t-lacking cultivar Nipponbare (NPB). We obtained over 20 independent transgenic lines and used three homozygous T3 lines for the following experiments. The expression of AvrPiz-t:Myc in the transgenic lines was confirmed by immunoblot analysis (see Supplemental Figure 1 online). The transgenic plants grew normally in our growth chambers (i.e., growth was similar to that of the segregated wild-type plants). With standard spray inoculation, avirulent and virulent strains caused similar blast symptoms on both transgenic and wild-type control plants without noticeable difference in disease severity in terms of lesion size and number (see Supplemental Figure 2 online). To investigate whether expression of AvrPiz-t affected rice basal resistance to M. oryzae, we used the punch inoculation method, which is more suitable to measure the basal resistance levels of the transgenic plants than the standard spraying inoculation method as previously described (Ono et al., 2001). Punch inoculation with the virulent strain RB22 showed that disease lesions were larger on the AvrPiz-t transgenic plants than on the control plants (Figures 2A). To further confirm the observation, we counted spore number and estimated the relative fungal biomass in the infected leaves by DNA-based quantitative PCR (qPCR) (Berruyer et al., 2006). The assays showed that more spores were produced in the lesions of the transgenic plants than in those of the control plants (Figure 2B, left panel), corroborating the lesion size result. Moreover, the relative fungal biomass, as indicated by the ratio of the M. oryzae Pot2 gene versus the rice ubiquitin gene (determined by DNA-based qPCR), was greater in the AvrPiz-t plants than in the control plants (Figure 2B, right panel). These results suggested that ectopic expression of AvrPiz-t in transgenic rice leads to enhanced susceptibility of rice plants to M. oryzae.
Figure 2.
Response of AvrPiz-t Transgenic Rice to Blast Infection, flg22, and Chitin Treatments.
(A) Punch inoculation of the AvrPiz-t transgenic and the segregated wild-type (NPB) plants. Rice leaves of 6-week-old plants were inoculated with the virulent isolate RB22. Leaves were photographed 10 DAI.
(B) Sporulation (left) and relative fungal growth [2 [CT(OsUBQ) − CT(MoPot2)] × 10,000] (right) on the inoculated leaves. Samples were taken for the assays 10 DAI. Values are the means of three replications, and error bars represent the se (n = 8, *P value < 0.05).
(C) Flg22-induced ROS burst in the AvrPiz-t and NPB plants. Rice leaf disks were treated with 100 nM flg22 and water. ROS were detected with a luminol-chemiluminescence assay. Error bars represent the se (n = 3).
(D) Chitin-induced ROS burst in the AvrPiz-t and NPB plants. Rice leaf disks were treated with 8 nM chitin (hexa-N-acetyl-chitohexaose) and water. ROS were detected with a luminol-chemiluminescence assay. Error bars represent the se (n = 3).
(E) Induction of the defense-related genes KS4 and PAL at 1 h after incubation in water, chitin, or flg22. Gray bars indicate the AvrPiz-t transgenic plants, and white bars indicate NPB control plants. qRT-PCR was performed with gene-specific primers. Values are the means of three replications, and error bars represent the se (n = 3).
AvrPiz-t Suppresses PTI Signaling in Rice
A variety of plant pathogen effectors suppress the induction of ROS in infected tissues as a virulence function to perturb PTI (Boller and He, 2009; Block and Alfano, 2011; Dou and Zhou, 2012). Recent studies showed that rice cells can perceive PAMP elicitor flg22 through the PRR FLS2 (Takai et al., 2008) and can perceive chitin through the PRRs CEBiP and CERK1 (Kaku et al., 2006; Shimizu et al., 2010). To examine whether expression of AvrPiz-t in rice affected ROS generation after PAMP elicitor treatment, we collected leaves from the AvrPiz-t transgenic and segregating wild-type plants and measured the ROS level after flg22 and chitin treatments using a ROS inhibition assay (Schwacke and Hager, 1992). Like other plants, such as Arabidopsis (Felix et al., 1999; Gómez-Gómez et al., 1999; Kunze et al., 2004; Miya et al., 2007), tissues of 4-week-old rice plants also exhibited a ROS burst when they were exposed to flg22 or chitin (Figures 2C and 2D). The flg22-induced hydrogen peroxide accumulation was completely inhibited and the chitin-induced hydrogen peroxide accumulation was reduced by 40 to 50% in the AvrPiz-t transgenic plants relative to the control plants (Figures 2C and 2D). Quantitative RT-PCR (qRT-PCR) revealed that the expression of the PTI-related genes, such as KS4, was significantly suppressed after 1 h in both the chitin-treated and flg22-treated AvrPiz-t plants (Figure 2E, left panel). By contrast, the expression of PAL was significantly suppressed in the chitin-treated but not in the flg22-treated AvrPiz-t plants (Figure 2E, right panel). Together with the results from the above inoculations and defense gene expression analysis, these data suggested that AvrPiz-t contributes to the virulence of M. oryzae by suppressing rice basal defense during the compatible interaction.
AvrPiz-t Interacts with APIP6 H58Y in Vitro and in Vivo
To elucidate the molecular mechanism underlying AvrPiz-t–mediated suppression of host PTI, we performed a yeast two-hybrid screen to identify potential host targets of AvrPiz-t. With the AvrPiz-t without the signal peptide sequence as the bait, a total of 12 distinct putative interactors, named AvrPiz-t interacting protein [APIP], were isolated from a rice cDNA library (Vega-Sánchez et al., 2008). Interestingly, four APIPs (APIP2, 6, 8, and 10; in bold type in Supplemental Table 1 online) are likely involved in the UPS because APIP2, 6, and 10 are putative C3H4-type RING E3 ubiquitin ligases, and APIP8 is a homolog of the yeast UBIQUITIN FUSION DEGRADATION1 protein, which is responsible for the retrotranslocation of ubiquitinated proteins with their partners (Ye et al., 2003). This result suggested that AvrPiz-t may manipulate the host UPS during blast infection.
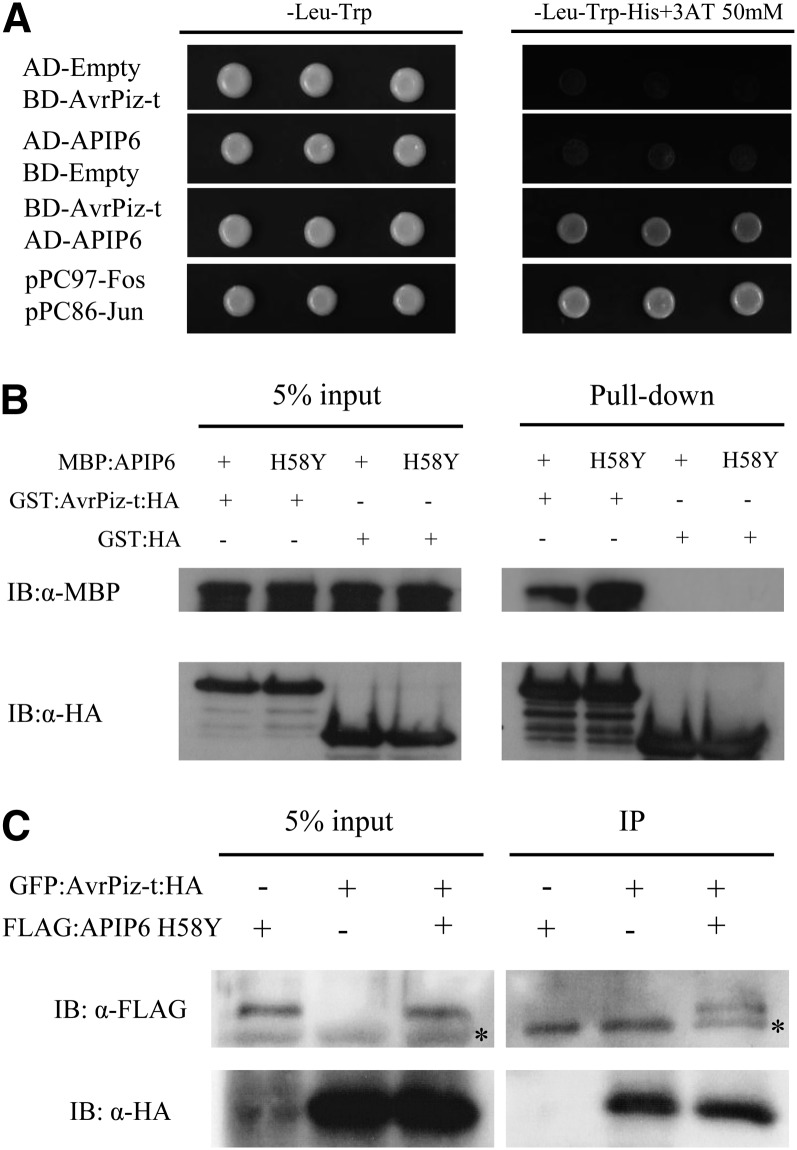
We analyzed the function of APIP6 first because its interaction with AvrPiz-t in yeast was strong in our preliminary screens. We confirmed the AvrPiz-t-APIP6 interaction in yeast using three selections (Vega-Sánchez et al., 2008) (Figure 3A). A glutathione S-transferase (GST) pull-down assay showed that AvrPiz-t binds APIP6 in vitro (Figure 3B). To validate the interaction result in yeast, we expressed both AvrPiz-t (without the signal peptide sequence) and APIP6 in Nicotiana benthamiana using the agroinfection method (Goodin et al., 2002). As a preliminary experiment, Agrobacterium tumefaciens carrying FLAG:APIP6 was infiltrated into N. benthamiana leaves, and the tissues were harvested 1 to 3 d after agroinfection (DAI) to monitor whether the APIP6 protein can be efficiently expressed in N. benthamiana leaves. Surprisingly, APIP6 was degraded completely within 1 to 3 DAI (see Supplemental Figure 3A online, top panel and lanes 1 to 3), and the degradation was not inhibited by MG132, an inhibitor of the 26S proteasome pathway (see Supplemental Figure 3A online, top panel and lane 4). We then mutated the His residue at the amino acid position 58 to Tyr (H58Y) in the APIP6 RING finger domain. Interestingly, the FLAG:APIP6 H58Y mutant protein was not degraded in the agroinfected leaves (see Supplemental Figure 3A online, top panel and lanes 5 to 7). Both immunoblot with the peroxidase antiperoxidase antibody and RT-PCR confirmed the expression of the APIP6 and APIP6 H58Y genes in the agroinfected tissues (see Supplemental Figure 3A online, second to fourth panel). To exclude the possibility that the FLAG tag and APIP6 were not in frame, we expressed FLAG:APIP6 and FLAG:APIP6 H58Y in Escherichia coli as MBP or GST fusion proteins. These proteins were detected with correct size by the immunoblot analysis with the anti-FLAG antibody (see Supplemental Figure 3B online).
Figure 3.
Interaction between AvrPiz-t and APIP6 in Vitro and in Vivo.
(A) Yeast two-hybrid assay between BD-AvrPiz-t (without the signal peptide sequence) and AD-APIP6. Cells were plated on -Leu-Trp-His media containing 50 mM 3-amino-1,2,4-triazole (3AT), a competitive inhibitor of the His3p enzyme.
(B) GST pull-down of MBP:APIP6 and MBP:APIP6 H58Y by GST:AvrPiz-t:HA. GST:HA was used as a negative control
(C) Co-IP analysis of FLAG:APIP6 H58Y and GFP:AvrPiz-t:HA (without the signal peptide sequence) in vivo. The GFP:AvrPiz-t:HA and FLAG:APIP6 H58Y genes were expressed in N. benthamiana using agroinfection. The Co-IP experiment was performed with the anti-HA antibody, and the isolated protein was analyzed by immunoblot using anti-FLAG antibody to detect APIP6 and anti-HA antibody to detect AvrPiz-t. Asterisk indicates nonspecific bands (left panel) or IgG bands from anti-HA antibody used for Co-IP (right panel).
Since the FLAG:APIP6 protein was not detectable in the agroinfected tissue, we coinfiltrated the Agrobacterium strains carrying the plasmids of FLAG:APIP6 H58Y and GFP:AvrPiz-t:HA in N. benthamiana leaves to confirm the AvrPiz-t-APIP6 interaction in vivo. Protein extracts isolated from the N. benthamiana leaves agroinfected with both Agrobacterium strains were subjected to a coimmunoprecipitation (Co-IP) assay. When the GFP:AvrPiz-t:HA fusion protein was immunoprecipitated from the plant extract using the anti-HA antibody, FLAG:APIP6 H58Y was detected in the immunocomplex of GFP:AvrPiz-t:HA using the anti-FLAG antibody (Figure 3C, third lane of the right panel). By contrast, no signal was visible in the reactions where only GFP:AvrPiz-t:HA (Figure 3C, first lane of the right panel) or FLAG:APIP6 (Figure 3C, second lane of the right panel) was expressed in N. benthamiana. Moreover, we performed bimolecular fluorescence complementation (BiFC) analysis by coinfiltrating the cYFP:AvrPiz-t and APIP6 H58Y:nYFP constructs into N. benthamiana leaves. Reconstituted fluorescence signals due to the interaction between AvrPiz-t:cYFP and APIP6 H58Y:nYFP were observed in the entire cell of the agroinfected tissue with stronger signals in the nucleus (see Supplemental Figure 4A online). By contrast, there were no signals in the cells agroinfected with two negative control combinations (see Supplemental Figures 4B and 4C online). To confirm the above result, we coexpressed APIP6 H58Y:nYFP and AvrPiz-t:cYFP along with red fluorescent protein (RFP):SPIN1, a nuclear protein related to flowering (Vega-Sánchez et al., 2008). Reconstituted fluorescence signals were predominantly observed in the nucleus and colocalized with the nuclear protein RFP:SPIN1 (see Supplemental Figure 4D online). Together these results indicated that AvrPiz-t:cYFP and APIP6 H58Y:nYFP interact in plant cells with more accumulation in the nucleus.
APIP6 Is a Functional RING Finger E3 Ubiquitin Ligase
The open reading frame of APIP6 is 1320 bp long, and the deduced protein contains 439 amino acids. BLAST searches against the rice genome database revealed that APIP6 is a single-copy gene in the rice genome and that the deduced protein belongs to a large family of conserved and ubiquitous RING finger proteins. The conserved C3H4 RING finger domain is located at the N terminus of APIP6 (35 to 80 amino acids; see Supplemental Figure 5A online). To determine whether APIP6 possesses E3 ubiquitin ligase activity, we performed an in vitro E3 ubiquitin ligase assay that included a series of negative controls lacking E1, E2, or ubiquitin. To investigate whether the E3 ligase activity depends on its RING finger domain, we included the APIP6 H58Y mutant protein with a His-to-Tyr mutation in the RING domain (see Supplemental Figure 5B online) in the activity assay. The MBP:APIP6 or MBP:APIP6 H58Y fusion protein purified from E. coli was incubated with wheat (Triticum aestivum) E1, Arabidopsis E2 (UBC10), ubiquitin (Ub), and ATP in the reactions. The immunoblot analysis showed that the signal of polyubiquitin products with high molecular weight was detected only in the presence of all the components (see Supplemental Figure 5C online, lane 6), suggesting that APIP6 is an active E3 ubiquitin ligase. In the reaction with the APIP6 H58Y mutant protein, however, the E3 ubiquitin ligase activity was completely abolished (see Supplemental Figure 5C online, lane 7), suggesting that an intact RING finger domain is required for the E3 ubiquitin ligase activity of APIP6 in vitro.
AvrPiz-t Is Ubiquitinated by APIP6 and Suppresses APIP6-Mediated Ubiquitination in Vitro
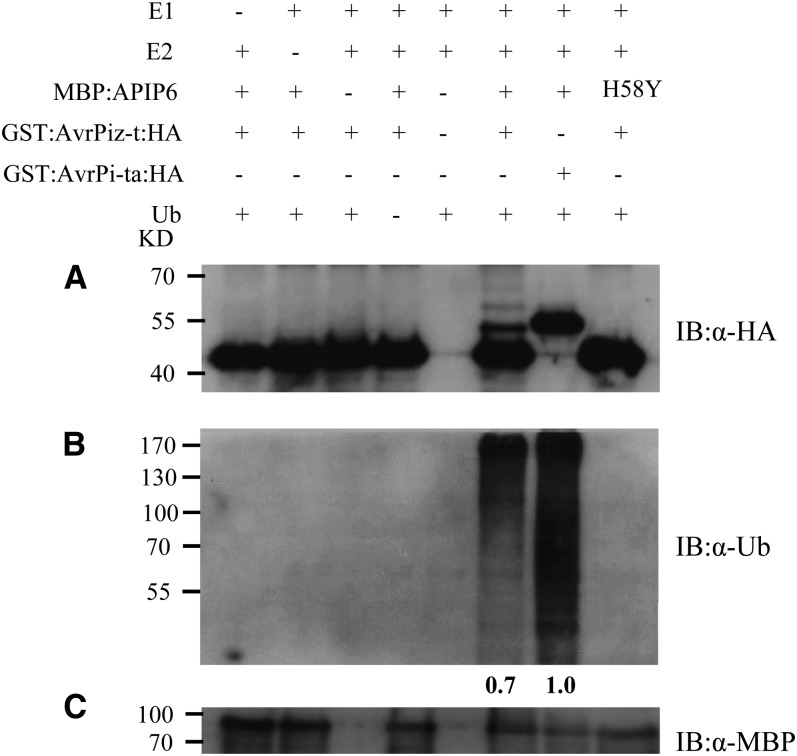
Because APIP6 has E3 ubiquitin ligase activity in vitro and interacts with AvrPiz-t in yeast, we speculated that AvrPiz-t might be a substrate of APIP6. To test this hypothesis, we performed a similar ubiquitination assay as described earlier for APIP6 E3 ubiquitin ligase activity (see Supplemental Figure 5C online) but added the GST:AvrPiz-t:HA fusion protein in the reaction. Strong signals of two high molecular weight bands of ubiquitinated products were detected by the anti-HA antibody only in the reaction with GST:AvrPiz-t:HA (Figure 4A, lane 6), indicating that AvrPiz-t is ubiquitinated by APIP6. By contrast, no high molecular weight band was observed in the control reaction in which the unrelated Avr-Pita:HA fusion protein was added (Figure 4A, lane 7) (Jia et al., 2000). However, the APIP6 H58Y mutant protein lacking E3 ubiquitin ligase activity was not able to ubiquitinate GST:Avr-Piz-t:HA (Figure 4A, lane 8). Surprisingly, when the signal on the same immunoblot was detected using the antiubiquitin antibody, the ubiquitination signal was significantly weaker in the reaction that included the GST:AvrPiz-t:HA fusion protein (∼30% signal reduction; Figure 4B, lane 6) than in the reaction that also included the control protein GST:Avr-Pita:HA (Figure 4B, lane 7; see Supplemental Figure 6B online, lane 3). Also, APIP6 showed reduced E3 ligase activity in the presence of AvrPiz-t (see Supplemental Figure 6B online, lane 2) compared with that in the absence of AvrPiz-t (see Supplemental Figure 6B online, lane 1). Consistent with the E3 ligase assay, the ubiquitination ability of the APIP6 H58Y mutant protein was completely abolished (Figure 4B, lane 8; see Supplemental Figure 6B online, lane 4). Similar results were obtained when the GST tag was replaced by the MBP tag in the AvrPiz-t fusion construct. The E3 ligase activity of APIP6 was significantly reduced when the MBP:AvrPiz-t fusion protein was included in the reactions (see Supplemental Figure 7A online). Furthermore, multiple high molecular weight bands above the MBP:AvrPiz-t protein were detected by immunoblot analysis with anti-MBP antibody in the presence of the MBP:AvrPiz-t recombinant protein (see Supplemental Figure 7B online). These results clearly demonstrated that APIP6 ubiquitinates AvrPiz-t, and in return, AvrPiz-t suppresses APIP6 E3 ligase activity in vitro.
Figure 4.
Ubiquitination of AvrPiz-t by APIP6 and Suppression of APIP6 E3 Ligase Activity by AvrPiz-t in Vitro.
(A) In vitro ubiquitination assay of GST:AvrPiz-t:HA by the MBP:APIP6 fusion protein. Ubiquitination of AvrPiz-t by APIP6 was detected by immunoblot with the anti-HA antibody. GST:Avr-Pita:HA was used as a negative control for determining the specificity of AvrPiz-t ubiquitination by APIP6.
(B) Suppression of APIP6 E3 ligase activity by AvrPiz-t. E3 ligase activity of APIP6 in the presence of AvrPiz-t or AvrPi-ta was determined by immunoblot with the antiubiquitin antibody. Relative E3 ligase activity was calculated by comparison to the control (lane 7) using ImageJ software.
(C) Immunoblot with the anti-MBP antibody to quantify the MBP:APIP6 or MBP:APIP6 H58Y protein in each lane.
AvrPiz-t and APIP6 Promote Degradation of Each Other in N. benthamiana Leaves
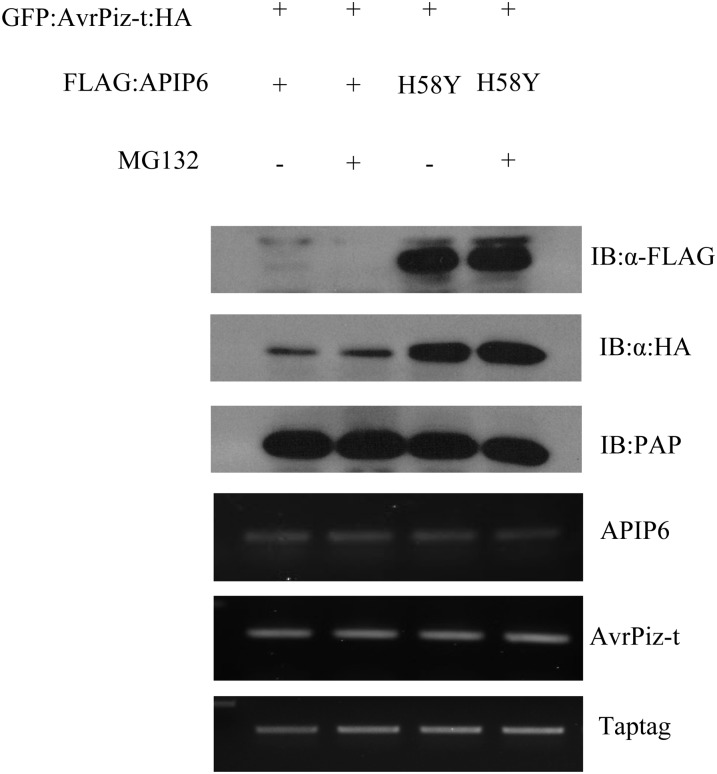
In vitro ubiquitination of AvrPiz-t by APIP6 prompted us to investigate whether the fungal effector AvrPiz-t can be degraded in planta in the presence of the rice E3 ubiquitin ligase APIP6. We coinfiltrated the Agrobacterium strain carrying the GFP:AvrPiz-t:HA construct with that containing the FLAG:APIP6 or FLAG:APIP6 H58Y construct into N. benthamiana leaves and extracted total protein from the treated leaves at 2 DAI. The immunoblot analysis revealed that the protein level of GFP:AvrPiz-t:HA was significantly lower in the tissue where APIP6 was coexpressed (Figure 5, lanes 1 and 2 in the second panel) than in the control where APIP6 H58Y was coexpressed (Figure 5, lanes 3 and 4 in the second panel). Pretreatment of the leaves with MG132 did not result in any obvious change in the protein levels of GFP:AvrPiz-t:HA (Figure 5, lanes 2 and 4 in the second panel), suggesting that AvrPiz-t degradation may not be effectively inhibited by MG132 or through a non-26S proteasome degradation pathway in N. benthamiana. Nevertheless, these results demonstrated that APIP6 promotes the degradation of AvrPiz-t and that its E3 ligase activity is essential for the degradation.
Figure 5.
Degradation of AvrPiz-t in the Presence of APIP6 in N. benthamiana.
FLAG:APIP6 or FLAG:APIP6 H58Y (H58Y) was coexpressed with GFP:AvrPiz-t:HA (without the signal peptide sequence) by agroinfection, and the tissues were harvested 2 DAI. MG132 (50 μM) was infiltrated with DMSO 18 h before sampling. The TAP tag gene was expressed as an internal control, and the protein was detected by immunoblot with the peroxidase antiperoxidase (PAP) antibody. The transcriptional level of each gene was determined by RT-PCR.
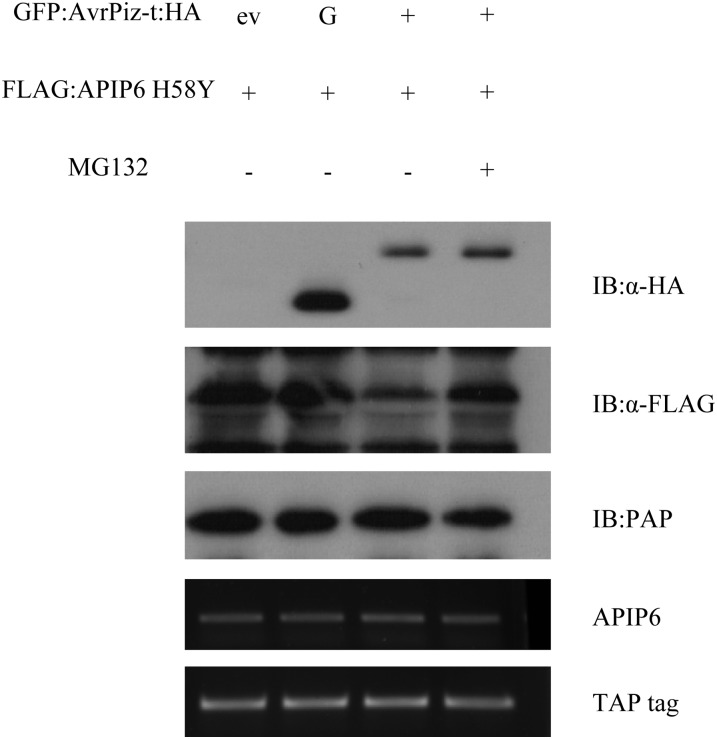
The oomycete effector Avr3a targets and stabilizes the host E3 ligase CMPG1 to potentially prevent host cell death during the biotrophic phase of infection (Bos et al., 2010). Because we found that AvrPiz-t suppresses APIP6 E3 ubiquitin ligase activity in vitro, we postulated that AvrPiz-t might affect the accumulation or stability of APIP6 in plant cells. To confirm this speculation, we coinfiltrated the plasmids containing GFP:AvrPiz-t:HA and FLAG:APIP6 H58Y into N. benthamiana leaves because the wild-type APIP6 was not detectable (see Supplemental Figure 3A online) and extracted total protein from the treated leaves 3 DAI. In the coagroinfection assay, the GFP:HA protein was used as a negative control. As shown in the first panel of Figure 6, a GFP-HA band was detected in the second lane and a GFP:AvrPiz-t:HA band was detected in the third and fourth lanes using the anti-HA antibody. The immunoblot analysis with the anti-FLAG antibody also showed that the protein level of FLAG:APIP6 H58Y was significantly less when coexpressed with GFP:AvrPiz-t:HA than in the control containing an empty vector (ev) or GFP:HA (Figure 6, lane 3 in the second panel). Interestingly, treatment of the leaf tissue with MG132 inhibited the degradation of the APIP6 H58Y protein (Figure 6, lane 4 in the second panel). These results demonstrated that AvrPiz-t promotes degradation of APIP6 H58Y and possibly its wild-type protein, APIP6, via the 26S proteasome system in plant cells.
Figure 6.
Degradation of APIP6 by AvrPiz-t in N. benthamiana.
Agrobacterium carrying FLAG:APIP6 H58Y construct was coinfiltrated into N. benthamiana leaves with either an empty vector (ev), GFP:HA (G), or GFP:AvrPiz-t:HA (without the signal peptide sequence). The agroinfected tissues were harvested 3 d after treatment. MG132 (50 μM) was infiltrated with DMSO 18 h before sampling. The transcriptional level of each gene was determined by RT-PCR.
Knockdown of APIP6 Compromises Basal Defense in Rice
To determine the function of APIP6 in Piz-t–mediated resistance, we constructed an APIP6 RNA interference (RNAi) construct targeting the region between 470 and 739 bp after the ATG codon (see Supplemental Figure 5A online). To determine the specificity of APIP6 silencing, we performed BLAST searches (Altschul et al., 1997) with the rice genome databases and found that the RNAi fragment matched 100% to the DNA sequence of APIP6 (Os05g0154600) and 80% to Os01g0350900 (see Supplemental Figures 8A and 8B online). Window pane analysis of the RNAi fragment revealed that all the 21 nucleotides only 100% matched to the APIP6 sequence (see Supplemental Table 2 online), indicating a high efficacy of the RNAi construct on the silencing of APIP6. The RNAi construct was transformed into the Piz-t–carrying cultivar Toride (TRD) (Li et al., 2009) using the Agrobacterium-mediated transformation method (Qu et al., 2006). Over 20 independent lines were obtained and self-pollinated to produce seeds in growth chambers. In both the T1 (primary transformants) and T2 generations, the RNAi plants grew normally and did not show any changes in growth and development. Three homozygous lines of the T3 generation with significant reduction of the APIP6 transcript were chosen for the phenotypic analyses. To exclude the possibility of silencing Os01g0350900 as an off-target in the APIP6 RNAi lines, we measured the expression level of APIP6 and Os01g0350900 in the segregated wild type and the three independent RNAi lines by qRT-PCR. We found that, while the APIP6 expression was reduced significantly compared with the wild type, the expression of Os01g0350900 in the transgenic lines was not changed compared with that in the wild type (see Supplemental Figure 9 online).
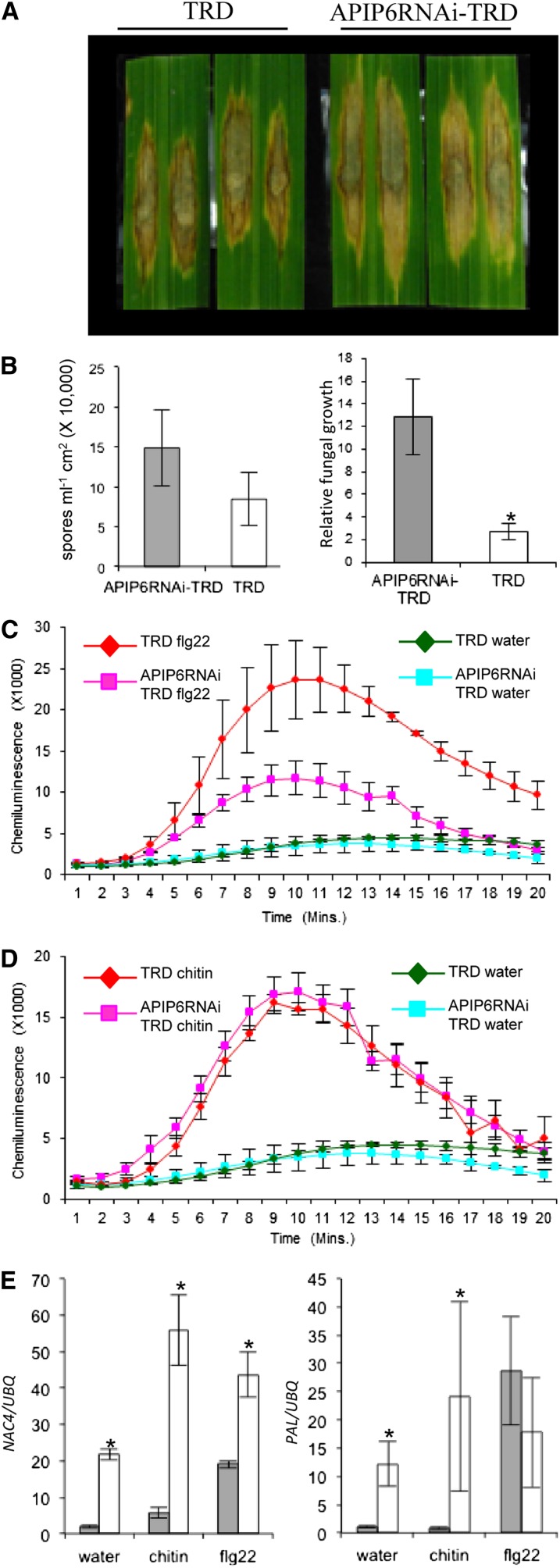
We first tested whether silencing of APIP6 affected Piz-t resistance by inoculating the RNAi plants with the AvrPiz-t–carrying strain RO1-1. Blast inoculations showed that the APIP6 RNAi plants were as resistant as the wild-type Piz-t plants (TRD) to the isolate, suggesting that APIP6 is not required for the Piz-t–mediated resistance to the avirulent strain. Next, we evaluated the resistance of the APIP6 RNAi plants to the virulent strain RB22. Punch inoculation assays showed that lesions were larger on the APIP6 RNAi plants than on the segregated wild-type plants (Figure 7A). More spores were produced on the infected leaves of the APIP6 RNAi plants than on the control plants (Figure 7B, left panel). In addition, the fungal biomass (as indicated by the relative amount of the Pot2 gene per infected leaf area) was greater in the RNAi plants than in the control plants (Figure 7B, right panel). These results demonstrated that knockdown of APIP6 compromises basal defense against M. oryzae in rice.
Figure 7.
Responses of APIP6 RNAi Plants to Blast Infection and PAMP Elicitor Treatments.
(A) Punch inoculation of the APIP6 RNAi and segregated wild-type TRD plants. Leaves of 6-week-old rice plants were inoculated with the virulent isolate RB22. The leaves were photographed 10 DAI.
(B) Sporulation (left) and relative fungal growth [2 [CT(OsUBQ) − CT(MoPot2)] × 100] (right) were measured 10 d after punch inoculation. Values are the means of three replications, and error bars represent the se (n = 8; *P value < 0.05).
(C) Measurement of the flg22-induced ROS burst. Leaf disks from the APIP6 RNAi and the control plants were treated with 100 nM flg22 and water. ROS were detected with a luminol-chemiluminescence assay. Error bars represent the se (n = 3).
(D) Measurement of chitin-induced ROS burst. Leaf disks from the APIP6 RNAi and the control plants were treated with 8 nM chitin (hexa-N-acetyl-chitohexaose) and water. ROS were detected with a luminol-chemiluminescence assay. Error bars represent the se (n = 3).
(E) Induction of the defense-related genes NAC4 (1 h after incubation) and PAL (3 h after incubation) in water, chitin, or flg22. Gray bars indicate APIP6 RNAi plants, and white bars indicate the control plants. qPCR was performed with gene-specific primers. Values are the means of three replications, and error bars represent the se (n = 3; *P value < 0.05).
To determine the molecular basis of the reduced resistance in the APIP6 RNAi plants, we first analyzed the induction of ROS after flg22 and chitin treatments in both the RNAi and segregated wild-type plants. As observed with the AvrPiz-t overexpression plants, knockdown of APIP6 resulted in ∼60% suppression of the ROS generation in the rice leaves within 10 min after the flg22 treatment compared with that in control plants (Figure 7C). However, the suppression of APIP6 expression in the APIP6 RNAi plants did change ROS production induced by chitin (Figure 7D), suggesting that the reduction of basal resistance in the APIP6 RNAi plants is independent of the chitin-induced rapid and transient ROS burst in rice. Real-time qPCR analysis revealed that the expression of the NAC4 gene was significantly reduced 1 h after the chitin and flg22 treatments in the RNAi plants compared with the control plants (Figure 7E, left panel). Similarly, the expression of the PAL gene was also markedly reduced in the RNAi plants but the suppression of PAL occurred 3 h after the chitin treatment (Figure 7E, right panel). However, the expression of PAL was similar in the RNAi and control plants even 3 h after the flg22 treatment (Figure 7E, right panel). Together, these results demonstrated that APIP6 in rice is an important component in the PTI pathway in response to M. oryzae and may regulate the expression of the basal defense genes via the UPS.
DISCUSSION
AvrPiz-t Is a Cytoplasmic Effector and Suppresses PTI in Rice Cells
Although some M. oryzae effector proteins are translocated into rice cells during infection, presumably via the BIC (Khang et al., 2010), it is unclear whether avirulence proteins in M. oryzae are also delivered into rice cells via the BIC. In this study, we used a live-cell imaging approach developed by Khang et al. (2010) to determine whether AvrPiz-t accumulated in BICs and was translocated into rice cells. Imaging analysis showed that AvrPiz-t:mCherry:NLS fusion protein is evident not only in the BIC area but also in the rice nucleus, indicating that AvrPiz-t:mCherry:NLS is indeed translocated from invasive hyphae into rice cells. Unlike PWL2, which is present even in uninfected neighboring cells (Khang et al., 2010), AvrPiz-t:mCherry:NLS is absent in the nuclei of adjoining cells that lack invasive hyphae, indicating that AvrPiz-t might not move ahead of hyphae into uninfected neighboring cells. In plant pathogenic oomycetes, the N-terminal region of a subset of cytoplasmic effector proteins contain the RXLR motif, which is required for the translocation of these effectors into host plant cells (Whisson et al., 2007). Similarly, some effector proteins from M. oryzae also contain the LxAR-like motif but their function in translocation is unclear (Yoshida et al., 2009). Further research will focus on identifying the region that is required for the translocation of the AvrPiz-t protein into rice cells.
Some avirulence proteins of plant pathogens are known to have dual activities (Göhre and Robatzek, 2008). On the one hand, in the presence of its cognate resistance protein, an avirulence protein can trigger a rapid HR that is often associated with localized cell death. On the other hand, an avirulence protein may suppress PTI and thereby enhance pathogenesis in the host cells in the absence of its cognate resistance protein (Göhre and Robatzek, 2008). We previously found that AvrPiz-t suppresses BAX-mediated programmed cell death in N. benthamiana (Li et al., 2009). In this study, we expressed the AvrPiz-t gene without the signal peptide sequence in non-Piz-t backgrounds. The AvrPiz-t transgenic plants showed a nearly complete suppression of ROS generation triggered by the PAMP elicitors, flg22 and chitin, and a significant reduction of defense gene expression. In addition, the AvrPiz-t transgenic plants showed enhanced susceptibility to the virulent M. oryzae isolate. These results clearly demonstrated that AvrPiz-t enhances virulence by suppressing early defense responses to blast infection.
Function of Ubiquitin-Mediated Degradation in the Interaction between AvrPiz-t and APIP6
In the in vitro ubiquitination assays, we found that AvrPiz-t is ubiquitinated by the RING finger E3 ligase APIP6 (Figure 4A). This prompted us to test the stability of AvrPiz-t in rice cells. When the GFP:AvrPiz-t-HA fusion construct was expressed in rice protoplasts, no signal was detected in immunoblots when the anti-GFP antibody was used. We suspect that there are many host proteins that can target AvrPiz-t for degradation in rice cells. For example, we identified three RING-finger E3 ligases that interact with AvrPiz-t in our Y2H screens. However, the GFP:AvrPiz-t:HA fusion protein is stably expressed in N. benthamiana when agroinfected. This discrepancy might result from the difference between the ubiquitination systems in rice and N. benthamiana, and there is a lack of the components essential for the degradation of AvrPiz-t in N. benthamiana. Because the GFP:AvrPiz-t:HA fusion gene can be expressed in N. benthamiana, we were able to coexpress it with FLAG:APIP6 to test whether the expression of the APIP6 protein can affect the stability of AvrPiz-t in N. benthamiana leaves. Interestingly, we found that APIP6 interacts with AvrPiz-t and targets it for degradation and that MG132 does not inhibit the degradation. Although we believe that a one-to-one interaction assay between AvrPiz-t and APIP6 in N. benthamiana is a suitable system to test the interaction between these two proteins in planta, we should find a suitable assay to test whether the degradation of AvrPiz-t in rice occurs via the 26S UPS.
It is intriguing that APIP6 is degraded after agroinfection in N. benthamiana (see Supplemental Figure 3A online). However, the APIP6 H58Y mutant protein with a mutation in the RING finger domain is relatively stable, suggesting that APIP6 may be self-ubiquitinated in N. benthamiana as observed for other plant E3 ligases (Jackson et al., 2000; Vaux and Silke, 2005). Using the APIP6 H58Y mutant protein, we found that AvrPiz-t promotes the degradation of the APIP6 H58Y protein in N. benthamiana and that the degradation is significantly inhibited by MG132 (Figure 6), suggesting that the APIP6 degradation depend on the UPS. Because a suitable in vivo assay system is lacking in rice, we were unable to test whether AvrPiz-t affects the stability of APIP6 in rice. However, we found that ectopic expression of AvrPiz-t and silencing of APIP6 leads to similar phenotypes, such as enhanced susceptibility to rice blast and suppression of the PTI-related defense gene expression in transgenic rice. These results indicate that AvrPiz-t might interact with and destabilize APIP6 in rice during blast infection. This finding is contrary to the previous report that the oomycete effector AVR3a interacts with and stabilizes the U-box E3 ligase CMPG1, which is required for INF1-triggered cell death (Bos et al., 2010). The biochemical mechanism for the degradation of APIP6 in rice cells remains to be elucidated.
Function of APIP6 in Basal Resistance to M. oryzae
Knockdown of APIP6 in the Piz-t background does not lead to any cell death or change in resistance to an AvrPiz-t–carrying blast strain, suggesting that APIP6 is not required for Piz-t–mediated disease resistance. Instead, we found that APIP6 RNAi plants show reduced basal resistance to the virulent blast strain, significant suppression of defense gene expression, and significant suppression of ROS generation after flg22 treatment. Interestingly, the suppression of ROS generation occurred after flg22 treatment but not to chitin treatment. By contrast, reduced defense gene expression was observed in the APIP6 RNAi plants in response to both chitin and flg22 treatments (Figure 7E). Previous research showed that a ROS burst can act in multiple ways to affect basal defense in plants: ROS bursts can act as a potent antibiotic, can enhance cell wall cross linking, and can serve as a secondary stress signal to activate different defense responses (Apel and Hirt, 2004; Boller and Felix, 2009). Expression of defense genes can also be independent of the oxidative burst induced by different elicitors. In parsley (Petroselinum crispum), for example, mitogen-activated protein kinases play an important role in oxidative burst–independent expression of pathogenesis-related genes (Kroj et al., 2003). In Arabidopsis, the oligogalacturonide-induced oxidative burst is not involved in the activation of the defense response to Botrytis cinerea (Galletti et al., 2008). Therefore, we speculate that APIP6 might regulate gene expression downstream of or independent of the initial rapid ROS burst in the chitin-induced defense signaling pathway. Alternatively, APIP6 might only affect the flg22-mediated defense response, and AvrPiz-t might affect the chitin signaling pathway via another APIP in rice. Therefore, functional analysis of other APIPs may identify a target of AvrPiz-t for the suppression of the chitin-mediated defense pathway in rice. Similarly, comparison of transcriptome profiles of AvrPiz-t transgenic and APIP6 RNAi plants before and after chitin treatment may identify chitin-associated defense genes in rice. The relationship between the APIP6-mediated ubiquitination pathway and the FLS2-mediated PTI pathway is another interesting area for further study. A recent study in Arabidopsis showed that the phosphorylated U-box E3 ligase PUB13 interacts with FLS2 and promotes the degradation of FLS2 after flg22 treatment (Lu et al., 2011). It will be interesting to investigate whether rice FLS2 or its downstream protein(s) is the substrate of APIP6 during rice blast infection.
Based on the results obtained in this study, we propose a working model to illustrate the functions of AvrPiz-t and APIP6 in rice PTI. When M. oryzae infects rice plants, the secreted AvrPiz-t targets the RING finger E3 ubiquitin ligase APIP6 for degradation through an unknown mechanism. Because of the positive function of APIP6 in PTI, APIP6 degradation leads to reduced ROS generation and increased susceptibility of rice plants to M. oryzae.
METHODS
Rice Blast Inoculations and Disease Resistance Evaluations
The Magnaporthe oryzae isolates used in the study were grown on oatmeal agar for 2 weeks in the dark at 28 to 30°C and then exposed to fluorescent lights for 1 week at room temperature for sporulation. Spore concentration was adjusted to 5 × 105 spores mL−1 with a hemacytometer before spores were applied by spraying (Qu et al., 2006) or punch inoculation (Ono et al., 2001).
Rice (Oryza sativa) seeds were sterilized by immersion in 75% ethanol for 1 min followed by immersion in 2% sodium hypochlorite for 40 min. After they were washed with sterile water, seeds were germinated on half-strength Murashige and Skoog medium for 1 week and then transferred to soil. Rice plants were maintained in a growth chamber at 26°C and 80% relative humidity with a 12-h light/dark photoperiod. Three-week-old seedlings were spray inoculated with spores following the procedure described by Qu et al. (2006). The resistance of 6- to 8-week-old rice plants to M. oryzae was evaluated by the punch inoculation method with slight modification (Ono et al., 2001). In brief, rice leaves were lightly wounded with a mouse ear punch, and 10 μL of spore suspension (5 × 105 spores mL−1) was added to the wound. Both sides of the inoculated area were sealed with Scotch tape to hold the spore suspension, and inoculated plants were returned to the growth chamber. Lesions were photographed 9 DAI, and lesion area was measured by analyzing the photographs with Adobe Photoshop software; the number of pixels with and without lesion was determined in a 300-mm2 area. For determination of the blast sporulation rate on the lesions, a portion of the lesion (∼3 × 1 cm) was cut from the leaf and placed in a microcentrifuge tube containing 100 μL of distilled water with 1% Tween 20. The samples were vigorously mixed in a vortex mixer for 2 min to dislodge the spores, and the number of spores mL−1 was determined with a microscope and a hemacytometer. The fungal biomass in infected rice leaf tissue was quantified using a slight modification of a previously described method (Kawano et al., 2010). In brief, a small piece of infected rice tissue (3 × 1 cm) was cut for DNA extraction using the standard Cetyltrimethyl ammonium bromide extraction protocol. The DNA was treated with 1 μL RNase A (10 mg mL−1) to remove RNA. DNA-based qPCR was performed using the iQ5 real-time PCR detection system (Bio-Rad). Relative fungal growth was calculated using the threshold cycle value (CT) of M. oryzae Pot2 DNA against the CT of rice genomic ubiquitin DNA. The CTs of Os-Ubq and Mo-Pot2 were measured, and the CT of Ubq was subtracted from the CT of Pot2. Relative fungal growth was then calculated as a ratio (Mo-Pot2/Os-Ubq) represented by the equation ECT(Os-UBQ) − CT(Mo-Pot2), in which the amplification efficiency, E, is 2 for the primer pairs designed for the respective genes (data not shown).
Rice Transformation
The AvrPiz-t overexpression plasmid, named pCXUN-DSAPZ, was constructed in pCXUN in which the AvrPiz-t gene without its signal peptide sequence was under the control of the maize (Zea mays) ubiquitin promoter (Chen et al., 2009). The APIP6 RNAi construct, pCXUNAP6i-2, was made in pCXUN in which the region between 470 and 739 bp after the ATG code in APIP6 was cloned into the vector using an overlapping PCR method as described previously (Chen et al., 2009).
Rice callus was induced from the embryos of mature seeds of the Japonica rice cultivars NPB and TRD. Rice transformation was conducted using the Agrobacterium tumefaciens–mediated method described previously (Yin et al., 2000; Qu et al., 2006).
Live-Cell Imaging Analysis of AvrPiz-t in Rice Cells
For the PAvr-Piz-t:AvrPiz-tCDS:mCherry:NLS construct, the AvrPiz-t promoter and its entire 324-bp sequence was PCR amplified with the primer pair AvrPiz-t-FPro-MfeI/AvrPiz-t-R-BamHI. The amplified fragment was then digested with MfeI and BamHI and cloned into the EcoRI-SalI site of the pBV436 vector along with the mCherry:NLS fragment released from pBV591 by enzyme digestion with BamHI and SalI (Khang et al., 2010). The AvrPiz-t:mCherry:NLS-BAS:EGFP plasmid was transformed into blast isolate O-137 (Valent et al., 1991) using Agrobacterium-mediated transformation (Khang et al., 2006). Leaf sheath inoculation was performed by incubating fungal spores (3 × 104 spores/mL in 0.25% gelatin) in the hollow interior of detached leaf sheaths of rice cultivar YT16. The inner epidermal layer was excised immediately before it was examined with a Zeiss Axiovert 200M confocal microscope equipped with a Zeiss LSM 510 META system. A C-Apochromat ×40/1.2–numerical aperture water immersion objective was used, and excitation/emission wavelengths were 488 nm/505 to 550 nm for enhanced GFP and 543 nm/560 to 615 nm for mCherry.
Plasmid Construction
Plasmids were constructed following the standard procedures described by Sambrook and Russell (2001). The primers used in plasmid construction and qPCR are listed in Supplemental Table 3 online. For the AvrPiz-t construct, AvrPiz-t that was PCR amplified with the primer pair AvrPiz-t-HAF1/AvrPiz-t-HAR was cloned into pXUN-CHA, which is a derivative of pXUN (Chen et al., 2009), which contains the HA sequence in the C terminus of the cloning site. Then, the AvrPiz-t-CHA fragment was amplified with the primer pair AvrPiz-t-HAF1/AvrPiz-t-HAR2 and digested with BamHI and XhoI. The digested product was inserted into the BamHI-SalI site of pGex-6p-1 or into the BglII-SalI site of pGDG for expression in Escherichia coli and Nicotiana benthamiana, respectively. For the cloning of APIP6 into the pMAL-c2x vector to express protein in E. coli, the APIP6 fragment was amplified with a primer pair, APIP6-ProF/APIP6-ProR. The amplicon digested with EcoRI and SalI was inserted into the EcoRI-SalI site of pMAL-c2x. To generate the APIP6 H58Y mutation, the RING finger domain fragment was mutated by PCR with the primer pair AP6mutF/AP6mutR, and the purified fragment was digested with PstI and HindIII. The digested fragment was cloned into PstI-HindIII site of pMAL-c2x. For pGD-FLAG:APIP6 and pGD-FLAG:APIP6 H58Y constructs to be used for agroinfection in N. benthamiana, either amplified APIP6 or APIP6 H58Y fragment was cloned into pXUN-FLAG vector (Chen et al., 2009). From these two vectors, the BamHI and SalI fragments containing FLAG:APIP6 or FLAG:APIP6 H58Y were inserted into the BglII-SalI site of pGD vector. The same BamHI and SalI fragment was also cloned into either pGex-6p-1 or pMAL-c2x for expression in E. coli. For the TAP tag construct, the TAP tag fragment amplified from the plasmid pubi.nc1300.ntapintron.new (Wang et al., 2006) with the primer pair of Taptag-F/Taptag-R was cloned into pGD for the expression in N. benthamiana as an internal control. To measure the expression of each gene after agroinfection, total RNA was extracted using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions followed by DNase I treatment for 15 min at room temperature. First-strand cDNA was prepared from 2 μg of RNA using the reverse transcription system (Promega) according to the manufacturer’s protocol. RT-PCR was performed using 25 cycles of amplification, and the PCR products were separated on a 1.4% agarose gel. The amplification of the TAP tag gene was used as a control for an equal amount of the first-strand cDNA being used for each PCR reaction. The primers used are listed in Supplemental Table 3 online.
Agroinfection Assay in N. benthamiana
Agrobacterium strain GV3101 carrying different constructs was grown at 28°C in a shaking incubator at 200 rpm. After 18 h, bacterial cells were spun down at 3200g for 20 min and resuspended in MES buffer (10 mM MgCl2 and 10 mM MES, pH 5.6) augmented with acetosyringone (final concentration 150 μm) to a final OD600 of 1.5 for all constructs except the p19 and the TAP tag constructs, which had final ODs of 1.0, and 0.25, respectively. The bacterial suspensions were incubated for 3 h, prior to agroinfection into N. benthamiana leaves (L. Liu et al., 2010b; Ning et al., 2011). For the BiFC assay, constructs expressing the N- and C-terminal halves of GFP were mixed with the p19 expressing construct in the ratio of 1:1:1 (v/v).
In Vivo Co-IP and BiFC
For the in vivo Co-IP assay, Agrobacterium strain GV3101 carrying the expression vectors FLAG:APIP6, FLAG:APIP6 H58Y, or GFP:AvrPiz-t:HA was infiltrated into N. benthamiana leaves. Two days after agroinfection, the leaf tissue was harvested for extraction of total proteins with a native buffer that was composed of 50 mM Tris-MES, pH 8.0, 0.5 M Suc, 1 mM MgCl2, 10 mM EDTA, 5 mM DTT, and plant protease inhibitor cocktail (1 mM 4′-2′-aminoethyl benzene sulfonyl fluoride, 1 µM bestatin, 1 µM pepstatin A, 100 µM leupeptin, 5 µM E-64, and 10 mM phenanthroline in DMSO) (L. Liu et al., 2010b). A 10-μL volume of anti-HA agarose suspension (Sigma-Aldrich) was added to the protein samples, and the mixtures were kept at 4°C with slow shaking overnight. The samples were washed three times using 1× IP buffer according to the manufacturer’s instructions (Sigma-Aldrich). After 50 μL of 1× SDS loading buffer was added, the sample was heated to 95°C for 5 min. About 15 μL of each sample was loaded on the protein gel for immunoblot analysis using the anti-HA or anti-FLAG antibody.
For BiFC, we followed the procedure previously published by Waadt et al. (2008) and used the vector hygII-SPYNE173 for cloning of the APIP6 H58Y fragment and the vector kanII-SPYCE (M) for cloning of the AvrPiz-t fragment (without the signal peptide sequence). Leaf disks were collected 48 h after coagroinfection. Live-cell imaging was performed using a Nikon C2 confocal microscope equipped with the NIS Elements software suite (Nikon). A ×20 oil immersion objective lens was used to observe fluorescence. All images were taken with the same settings of exposure time, laser strength, and signal gain (excitation, 488 nm; emission, 515 nm). For RFP detection, the excitation was 553 nm and emission was 574 nm.
Protein Purification from E. coli and E3 Ubiquitin Ligase Activity Assay
Full-length cDNAs of APIP6 and AvrPiz-t (lacking the signal peptide sequence and having the HA tag sequence at the C terminus) were expressed as N-terminal MBP and GST-tagged proteins, respectively, in the Rosetta2 (DE3) strain of E. coli. The two proteins were affinity purified with maltose (NEB) and with glutathione matrix (Sigma-Aldrich) respectively. In vitro ubiquitination reactions were performed by adding 1 µg of MBP:APIP6, 1 µg of GST:AvrPiz-t:HA, 40 ng of wheat (Triticum aestivum) E1, 100 ng of Arabidopsis thaliana E2 (UBC10), 1 μg of ubiquitin, and 1.5 μL of 20× reaction buffer (1 M Tris HCl, pH 7.5, 40 mM ATP, 100 mM MgCl2, 40 mM DTT, 600 mM creatine phosphate, and 1 mg mL−1 creatine phosphokinase). The reaction was incubated at 30°C for 1.5 h in a 30-μL reaction volume before it was stopped with the SDS sample loading buffer and heated to 100°C for 5 min. Samples of the reactions were then separated on a 10% SDS-PAGE gel. Ubiquitination signals were detected by immunoblotting with the antiubiquitin antibody (Biomol) followed by chemiluminescence with the ECL kit (Promega).
GST Pull-Down
GST pull-down was conducted by following the procedures described by Sambrook and Russell (2001). About 50 μg of glutathione-agarose beads were mixed with ∼10 μg of GST:HA or GST:AvrPiz-t:HA. After the incubation at 4°C for 2 h with end-over-end mixing, supernatant was removed and beads were washed four times with 1 mL of ice-cold PBS buffer. Next, ∼10 μg of MBP:APIP6 or MBP:APIP6 H58Y was added into above beads for the incubation at 4°C for 2 h with end-over-end mixing. After four times of washing with 1× PBST, the SDS sample loading buffer was added to beads and heated to 100°C for 5 min for the immunoblot analysis.
Protein Extraction from Rice Plants and Immunoblot Analysis
Total protein from 4-week-old rice plants was extracted. Briefly, ∼4 cm2 of leaf tissues was ground in liquid nitrogen and subsequently dissolved in protein extraction buffer (50 mM Tris-MES, pH 8.0, 0.5 M Suc, 1 mM MgCl2, 10 mM EDTA, 5 mM DTT, 100 µM MG132, and plant protease inhibitor cocktail). Insoluble debris was pelleted by centrifugation at 13,000 rpm for 10 min at 4°C. Protein concentration was measured by Bio-Rad protein assay reagent. Tricine sample buffer was added, and samples were incubated at 65°C for 2 min. Thirty micrograms of each sample was separated on a 10 to 20% Tris-Tricine Mini-Protean gel (Bio-Rad). Protein was then transferred onto Immobilon-PSQ PVDF membrane (Millipore) in CAPS buffer, pH 11.0, for 40 min (350 mA, 80 V). Immunoblots were done using standard protocols. Anti-Myc tag antibody (Abcam) and horseradish peroxidase–conjugated goat anti-rabbit antibody (Jackson ImmunoResearch) were used at dilutions of 1:3000 and 1:10,000, respectively. Chemiluminescene was detected using Pierce ECL substrate (Thermo Scientific) and the ChemiDoc XRS system (Bio-Rad).
Yeast Two-Hybrid Screening
The ProQuest two-hybrid system (Invitrogen) was used for screening of the AvrPiz-t interacting proteins. The AvrPiz-t coding region (without the signal peptide sequence) was cloned in frame into the bait vector pDBlue. pDBlue-AvrPiz-t was transformed into yeast strain Mav203. Yeast cells carrying pDBlue-AvrPiz-t were transformed with DNA isolated from a rice cDNA library, which was constructed into the prey vector pPC86 using mRNA isolated from the seedlings of rice line 75-1-127 (Vega-Sánchez et al., 2008). Candidate clones growing on the SD/-Leu-Trp-His+50 mM +3AT medium were subjected to β-galactosidase assays for confirmation following the manufacturer’s protocol (Invitrogen).
Measurement of ROS
Leaf disks from 4-week-old plants were cut and preincubated overnight in sterile distilled water. ROS generation after elicitor treatment in the leaf disks was monitored using the luminol chemiluminescence assay (Schwacke and Hager, 1992). Three leaf disks per sample were placed in a microcentrifuge tube containing 100 μL of luminol (Bio-Rad Immun-Star horseradish peroxidase substrate 170-5040), 1.0 μL of horseradish peroxidase (Jackson ImmunoResearch), and elicitor (100 nM flg22, 8 nM hexa-N-acetyl-chitohexaose, or water as control). Immediately after the treatment, luminescence was measured continuously at 10-s intervals for 21 min with a Glomax 20/20 luminometer (Promega). Three replications were performed for each sample and treatment. Standard errors were calculated for each treatment.
qPCR for Analysis of Defense Marker Gene Expression
Leaf disks were collected as described for the measurement of ROS and were treated with elicitors (100 nM flg22, 8 nM hexa-N-acetyl-chitohexaose, or water as control) for 1, 3, and 6 h. Total RNA was extracted and first-strand cDNA was synthesized as described above for RT-PCR. qPCR was performed using the iQ5 real-time PCR detection system (Bio-Rad). Primer pairs used for qPCR of the genes NAC4, PAL, and KS4 were synthesized based on the information reported in a previous study (Shimizu et al., 2010) and are listed in Supplemental Table 3 online.
Accession Numbers
Sequence data from this article can be found in Rice Genome Project website (http://rice.plantbiology.msu.edu/) or the Magnaporthe comparative database website (http://www.broadinstitute.org/annotation/genome/magnaporthe_comparative/MultiHome.html) under the following accession numbers: APIP6, Os05g06270; KS4, Os04g10060; NAC4, Os01g60020; PAL, Os02g41680; Ubq, Os03g13170, AvrPiz-t, MGG_18041; and Pot2, MGG_13294.6.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Detection of the AvrPiz-t:Myc Protein Expression in Transgenic Rice by Immunoblot Analysis.
Supplemental Figure 2. Disease Reaction of 3-Week-Old AvrPiz-t Transgenic Plants to M. oryzae Isolates RB22 (Virulent) and C9240 (Avirulent).
Supplemental Figure 3. Expression of the FLAG:APIP6 and FLAG:APIP6 H58Y Proteins in N. benthamiana and E. coli.
Supplemental Figure 4. BiFC Assay for the APIP6 H58Y and AvrPiz-t Interaction in N. benthamiana Using Agrobacterium-Mediated Transient Expression System.
Supplemental Figure 5. Structure and E3 Ubiquitin Ligase Activity of APIP6.
Supplemental Figure 6. Ubiquitination of AvrPiz-t by APIP6 and Suppression of APIP6 E3 Ligase Activity by AvrPiz-t in Vitro.
Supplemental Figure 7. Suppression of APIP6 E3 Ligase Activity by AvrPiz-t (without the Signal Peptide Sequence) and Ubiquitination of AvrPiz-t by APIP6.
Supplemental Figure 8. BLASTN Results of APIP6 Silencing Fragment.
Supplemental Figure 9. Specificity of APIP6 Silencing.
Supplemental Table 1. The List of APIPs Identified by the Yeast Two-Hybrid Screen.
Supplemental Table 2. Window Pane Analysis for APIP6 RNAi Target Region Specificity.
Supplemental Table 3. Primers Used in This Study.
Acknowledgments
This project is supported by grants from The National Science Foundation–Integrative Organismal Systems (1120949) and the 973 Project (2012CB114005) of the Ministry of Science and Technology China to G.-L.W., by National Research Initiative Competitive Grants Program Grant 2010-65108-20538 from the USDA National Institute of Food and Agriculture to B.V. and C.H.K., and by grants from Natural Science Foundation of China to B.Z. (30971878) and Y.S.N. (31101405). We thank Qi Xie for providing Arabidopsis E2 (AtUBC10). This is contribution number 13-141-J from the Agricultural Experiment Station at Kansas State University. We thank Christopher DeFraia for his technical assistance with the BiFC analysis and Keith Slotkin for the use of his confocal microscope.
AUTHOR CONTRIBUTIONS
C.-H.P., S.C., G.S., B.Z., B.V., and G.-L.W. designed the research. C.-H.P., S.C., G.S., B.Z., C.H.K., P.S., A.J.A., Y.N., R.W., and M.B. performed the experiments and analyzed the data. C.-H.P., S.C., G.S, B.V., and G.-L.W wrote the article.
Glossary
- PAMP
pathogen-associated molecular pattern
- PRR
pattern recognition receptor
- PTI
PAMP-triggered immunity
- ROS
reactive oxygen species
- ETI
effector-triggered immunity
- HR
hypersensitive response
- UPS
ubiquitin proteasome system (
- NBS-LRR
nucleotide binding site–Leu-rich repeat
- BIC
biotrophic interface complex
- GFP
green fluorescent protein
- NPB
Nipponbare
- qPCR
quantitative PCR
- qRT-PCR
quantitative RT-PCR
- GST
glutathione S-transferase
- DAI
days after agroinfection
- Co-IP
coimmunoprecipitation
- BiFC
bimolecular fluorescence complementation
- RFP
red fluorescent protein
- RNAi
RNA interference
- TRD
Toride
- CT
cycle threshold
References
- Abramovitch R.B., Martin G.B. (2005). AvrPtoB: a bacterial type III effector that both elicits and suppresses programmed cell death associated with plant immunity. FEMS Microbiol. Lett. 245: 1–8 [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angot A., Peeters N., Lechner E., Vailleau F., Baud C., Gentzbittel L., Sartorel E., Genschik P., Boucher C., Genin S. (2006). Ralstonia solanacearum requires F-box-like domain-containing type III effectors to promote disease on several host plants. Proc. Natl. Acad. Sci. USA 103: 14620–14625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apel K., Hirt H. (2004). Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55: 373–399 [DOI] [PubMed] [Google Scholar]
- Berruyer R., Poussier S., Kankanala P., Mosquera G., Valent B. (2006). Quantitative and qualitative influence of inoculation methods on in planta growth of rice blast fungus. Phytopathology 96: 346–355 [DOI] [PubMed] [Google Scholar]
- Block A., Alfano J.R. (2011). Plant targets for Pseudomonas syringae type III effectors: Virulence targets or guarded decoys? Curr. Opin. Microbiol. 14: 39–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhnert H.U., Fudal I., Dioh W., Tharreau D., Notteghem J.L., Lebrun M.H. (2004). A putative polyketide synthase/peptide synthetase from Magnaporthe grisea signals pathogen attack to resistant rice. Plant Cell 16: 2499–2513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T., He S.Y. (2009). Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science 324: 742–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T., Felix G. (2009). A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60: 379–406 [DOI] [PubMed] [Google Scholar]
- Bos J.I., Armstrong M.R., Gilroy E.M., Boevink P.C., Hein I., Taylor R.M., Zhendong T., Engelhardt S., Vetukuri R.R., Harrower B., Dixelius C., Bryan G., et al. (2010). Phytophthora infestans effector AVR3a is essential for virulence and manipulates plant immunity by stabilizing host E3 ligase CMPG1. Proc. Natl. Acad. Sci. USA 107: 9909–9914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caracuel-Rios Z., Talbot N.J. (2007). Cellular differentiation and host invasion by the rice blast fungus Magnaporthe grisea. Curr. Opin. Microbiol. 10: 339–345 [DOI] [PubMed] [Google Scholar]
- Chen J., Shi Y., Liu W., Chai R., Fu Y., Zhuang J., Wu J. (2011). A Pid3 allele from rice cultivar Gumei2 confers resistance to Magnaporthe oryzae. J. Genet. Genomics 38: 209–216 [DOI] [PubMed] [Google Scholar]
- Chen S., Songkumarn P., Liu J., Wang G.L. (2009). A versatile zero background T-vector system for gene cloning and functional genomics. Plant Physiol. 150: 1111–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm S.T., Coaker G., Day B., Staskawicz B.J. (2006). Host-microbe interactions: Shaping the evolution of the plant immune response. Cell 124: 803–814 [DOI] [PubMed] [Google Scholar]
- Collemare J., Pianfetti M., Houlle A.E., Morin D., Camborde L., Gagey M.J., Barbisan C., Fudal I., Lebrun M.H., Böhnert H.U. (2008). Magnaporthe grisea avirulence gene ACE1 belongs to an infection-specific gene cluster involved in secondary metabolism. New Phytol. 179: 196–208 [DOI] [PubMed] [Google Scholar]
- Dean R.A., Talbot N.J., Ebbole D.J., Farman M.L., Mitchell T.K., Orbach M.J., Thon M., Kulkarni R., Xu J.R., Pan H., Read N.D., Lee Y.H., et al. (2005). The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 434: 980–986 [DOI] [PubMed] [Google Scholar]
- Dou D., Zhou J.-M. (2012). Phytopathogen effectors subverting host immunity: Different foes, similar battleground. Cell Host Microbe 12: 484–495 [DOI] [PubMed] [Google Scholar]
- Felix G., Duran J.D., Volko S., Boller T. (1999). Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 18: 265–276 [DOI] [PubMed] [Google Scholar]
- Galletti R., Denoux C., Gambetta S., Dewdney J., Ausubel F.M., De Lorenzo G., Ferrari S. (2008). The AtrbohD-mediated oxidative burst elicited by oligogalacturonides in Arabidopsis is dispensable for the activation of defense responses effective against Botrytis cinerea. Plant Physiol. 148: 1695–1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göhre V., Robatzek S. (2008). Breaking the barriers: microbial effector molecules subvert plant immunity. Annu. Rev. Phytopathol. 46: 189–215 [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez L., Felix G., Boller T. (1999). A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana. Plant J. 18: 277–284 [DOI] [PubMed] [Google Scholar]
- Goodin M.M., Dietzgen R.G., Schichnes D., Ruzin S., Jackson A.O. (2002). pGD vectors: Versatile tools for the expression of green and red fluorescent protein fusions in agroinfiltrated plant leaves. Plant J. 31: 375–383 [DOI] [PubMed] [Google Scholar]
- Gupta S.K., Rai A.K., Kanwar S.S., Chand D., Singh N.K., Sharma T.R. (2012). The single functional blast resistance gene Pi54 activates a complex defence mechanism in rice. J. Exp. Bot. 63: 757–772 [DOI] [PubMed] [Google Scholar]
- Houterman P.M., Ma L., van Ooijen G., de Vroomen M.J., Cornelissen B.J., Takken F.L., Rep M. (2009). The effector protein Avr2 of the xylem-colonizing fungus Fusarium oxysporum activates the tomato resistance protein I-2 intracellularly. Plant J. 58: 970–978 [DOI] [PubMed] [Google Scholar]
- Jackson P.K., Eldridge A.G., Freed E., Furstenthal L., Hsu J.Y., Kaiser B.K., Reimann J.D. (2000). The lore of the RINGs: Substrate recognition and catalysis by ubiquitin ligases. Trends Cell Biol. 10: 429–439 [DOI] [PubMed] [Google Scholar]
- Jia Y., McAdams S.A., Bryan G.T., Hershey H.P., Valent B. (2000). Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 19: 4004–4014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J.D., Dangl J.L. (2006). The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Kaku H., Nishizawa Y., Ishii-Minami N., Akimoto-Tomiyama C., Dohmae N., Takio K., Minami E., Shibuya N. (2006). Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc. Natl. Acad. Sci. USA 103: 11086–11091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano Y., Akamatsu A., Hayashi K., Housen Y., Okuda J., Yao A., Nakashima A., Takahashi H., Yoshida H., Wong H.L., Kawasaki T., Shimamoto K. (2010). Activation of a Rac GTPase by the NLR family disease resistance protein Pit plays a critical role in rice innate immunity. Cell Host Microbe 7: 362–375 [DOI] [PubMed] [Google Scholar]
- Khang C.H., Park S.Y., Rho H.S., Lee Y.H., Kang S. (2006). Filamentous Fungi (Magnaporthe grisea and Fusarium oxysporum). Methods Mol. Biol. 344: 403–420 [DOI] [PubMed] [Google Scholar]
- Khang C.H., Berruyer R., Giraldo M.C., Kankanala P., Park S.Y., Czymmek K., Kang S., Valent B. (2010). Translocation of Magnaporthe oryzae effectors into rice cells and their subsequent cell-to-cell movement. Plant Cell 22: 1388–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroj T., Rudd J.J., Nürnberger T., Gäbler Y., Lee J., Scheel D. (2003). Mitogen-activated protein kinases play an essential role in oxidative burst-independent expression of pathogenesis-related genes in parsley. J. Biol. Chem. 278: 2256–2264 [DOI] [PubMed] [Google Scholar]
- Kunze G., Zipfel C., Robatzek S., Niehaus K., Boller T., Felix G. (2004). The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell 16: 3496–3507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.W., Han S.W., Sririyanum M., Park C.J., Seo Y.S., Ronald P.C. (2009). A type I-secreted, sulfated peptide triggers XA21-mediated innate immunity. Science 326: 850–853 [DOI] [PubMed] [Google Scholar]
- Li W., Wang B., Wu J., Lu G., Hu Y., Zhang X., Zhang Z., Zhao Q., Feng Q., Zhang H., Wang Z., Wang G., et al. (2009). The Magnaporthe oryzae avirulence gene AvrPiz-t encodes a predicted secreted protein that triggers the immunity in rice mediated by the blast resistance gene Piz-t. Mol. Plant Microbe Interact. 22: 411–420 [DOI] [PubMed] [Google Scholar]
- Liu J., Wang X., Mitchell T., Hu Y., Liu X., Dai L., Wang G.L. (2010a). Recent progress and understanding of the molecular mechanisms of the rice-Magnaporthe oryzae interaction. Mol. Plant Pathol. 11: 419–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Zhang Y., Tang S., Zhao Q., Zhang Z., Zhang H., Dong L., Guo H., Xie Q. (2010b). An efficient system to detect protein ubiquitination by agroinfiltration in Nicotiana benthamiana. Plant J. 61: 893–903 [DOI] [PubMed] [Google Scholar]
- Lu D., Lin W., Gao X., Wu S., Cheng C., Avila J., Heese A., Devarenne T.P., He P., Shan L. (2011). Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science 332: 1439–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miya A., Albert P., Shinya T., Desaki Y., Ichimura K., Shirasu K., Narusaka Y., Kawakami N., Kaku H., Shibuya N. (2007). CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 104: 19613–19618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning Y., Jantasuriyarat C., Zhao Q., Zhang H., Chen S., Liu J., Liu L., Tang S., Park C.H., Wang X., Liu X., Dai L., et al. (2011). The SINA E3 ligase OsDIS1 negatively regulates drought response in rice. Plant Physiol. 157: 242–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura K., Debroy S., Lee Y.H., Pumplin N., Jones J., He S.Y. (2006). A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science 313: 220–223 [DOI] [PubMed] [Google Scholar]
- Nomura K., Mecey C., Lee Y.N., Imboden L.A., Chang J.H., He S.Y. (2011). Effector-triggered immunity blocks pathogen degradation of an immunity-associated vesicle traffic regulator in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 10774–10779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntoukakis V., Mucyn T.S., Gimenez-Ibanez S., Chapman H.C., Gutierrez J.R., Balmuth A.L., Jones A.M., Rathjen J.P. (2009). Host inhibition of a bacterial virulence effector triggers immunity to infection. Science 324: 784–787 [DOI] [PubMed] [Google Scholar]
- Okuyama Y., Kanzaki H., Abe A., Yoshida K., Tamiru M., Saitoh H., Fujibe T., Matsumura H., Shenton M., Galam D.C., Undan J., Ito A., et al. (2011). A multifaceted genomics approach allows the isolation of the rice Pia-blast resistance gene consisting of two adjacent NBS-LRR protein genes. Plant J. 66: 467–479 [DOI] [PubMed] [Google Scholar]
- Ono E., Wong H.L., Kawasaki T., Hasegawa M., Kodama O., Shimamoto K. (2001). Essential role of the small GTPase Rac in disease resistance of rice. Proc. Natl. Acad. Sci. USA 98: 759–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orbach M.J., Farrall L., Sweigard J.A., Chumley F.G., Valent B. (2000). A telomeric avirulence gene determines efficacy for the rice blast resistance gene Pi-ta. Plant Cell 12: 2019–2032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu S., Liu G., Zhou B., Bellizzi M., Zeng L., Dai L., Han B., Wang G.L. (2006). The broad-spectrum blast resistance gene Pi9 encodes a nucleotide-binding site-leucine-rich repeat protein and is a member of a multigene family in rice. Genetics 172: 1901–1914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafiqi M., Gan P.H., Ravensdale M., Lawrence G.J., Ellis J.G., Jones D.A., Hardham A.R., Dodds P.N. (2010). Internalization of flax rust avirulence proteins into flax and tobacco cells can occur in the absence of the pathogen. Plant Cell 22: 2017–2032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosebrock T.R., Zeng L., Brady J.J., Abramovitch R.B., Xiao F., Martin G.B. (2007). A bacterial E3 ubiquitin ligase targets a host protein kinase to disrupt plant immunity. Nature 448: 370–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Russell D.W. (2001). Molecular cloning: A laboratory manual. (Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press)
- Santner A., Estelle M. (2010). The ubiquitin-proteasome system regulates plant hormone signaling. Plant J. 61: 1029–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacke R., Hager A. (1992). Fungal elicitors induce a transient release of active oxygen species from cultured spruce cells that is dependent on Ca<sup>2+</sup> and protein-kinase activity. Planta 187: 136–141 [DOI] [PubMed] [Google Scholar]
- Shimamoto K., Kyozuka J. (2002). Rice as a model for comparative genomics of plants. Annu. Rev. Plant Biol. 53: 399–419 [DOI] [PubMed] [Google Scholar]
- Shimizu T., Nakano T., Takamizawa D., Desaki Y., Ishii-Minami N., Nishizawa Y., Minami E., Okada K., Yamane H., Kaku H., Shibuya N. (2010). Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 64: 204–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirsekar G., Dai L., Hu Y., Wang X., Zeng L., Wang G.-L. (2010). Role of Ubiquitination in Plant Innate Immunity and Pathogen Virulence. J. Plant Biol. 53: 10–18 [Google Scholar]
- Smalle J., Vierstra R.D. (2004). The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 55: 555–590 [DOI] [PubMed] [Google Scholar]
- Takahashi A., Hayashi N., Miyao A., Hirochika H. (2010). Unique features of the rice blast resistance Pish locus revealed by large scale retrotransposon-tagging. BMC Plant Biol. 10: 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai R., Isogai A., Takayama S., Che F.S. (2008). Analysis of flagellin perception mediated by flg22 receptor OsFLS2 in rice. Mol. Plant Microbe Interact. 21: 1635–1642 [DOI] [PubMed] [Google Scholar]
- Thomma B.P., Nürnberger T., Joosten M.H. (2011). Of PAMPs and effectors: The blurred PTI-ETI dichotomy. Plant Cell 23: 4–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valent B. (1990). Rice blast as a model system for plant pathology. Phytopathology 80: 33–36 [Google Scholar]
- Valent B., Khang C.H. (2010). Recent advances in rice blast effector research. Curr. Opin. Plant Biol. 13: 434–441 [DOI] [PubMed] [Google Scholar]
- Valent B., Farrall L., Chumley F.G. (1991). Magnaporthe grisea genes for pathogenicity and virulence identified through a series of backcrosses. Genetics 127: 87–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaux D.L., Silke J. (2005). IAPs, RINGs and ubiquitylation. Nat. Rev. Mol. Cell Biol. 6: 287–297 [DOI] [PubMed] [Google Scholar]
- Vega-Sánchez M.E., Zeng L., Chen S., Leung H., Wang G.L. (2008). SPIN1, a K homology domain protein negatively regulated and ubiquitinated by the E3 ubiquitin ligase SPL11, is involved in flowering time control in rice. Plant Cell 20: 1456–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra R.D. (2003). The ubiquitin/26S proteasome pathway, the complex last chapter in the life of many plant proteins. Trends Plant Sci. 8: 135–142 [DOI] [PubMed] [Google Scholar]
- Waadt R., Schmidt L.K., Lohse M., Hashimoto K., Bock R., Kudla J. (2008). Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J. 56: 505–516 [DOI] [PubMed] [Google Scholar]
- Wang Y.S., Pi L.Y., Chen X., Chakrabarty P.K., Jiang J., De Leon A.L., Liu G.Z., Li L., Benny U., Oard J., Ronald P.C., Song W.Y. (2006). Rice XA21 binding protein 3 is a ubiquitin ligase required for full Xa21-mediated disease resistance. Plant Cell 18: 3635–3646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whisson S.C., Boevink P.C., Moleleki L., Avrova A.O., Morales J.G., Gilroy E.M., Armstrong M.R., Grouffaud S., van West P., Chapman S., Hein I., Toth I.K., et al. (2007). A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature 450: 115–118 [DOI] [PubMed] [Google Scholar]
- Ye Y., Meyer H.H., Rapoport T.A. (2003). Function of the p97-Ufd1-Npl4 complex in retrotranslocation from the ER to the cytosol: dual recognition of nonubiquitinated polypeptide segments and polyubiquitin chains. J. Cell Biol. 162: 71–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Z., Chen J., Zeng L., Goh M., Leung H., Khush G.S., Wang G.L. (2000). Characterizing rice lesion mimic mutants and identifying a mutant with broad-spectrum resistance to rice blast and bacterial blight. Mol. Plant Microbe Interact. 13: 869–876 [DOI] [PubMed] [Google Scholar]
- Yoshida K., Saitoh H., Fujisawa S., Kanzaki H., Matsumura H., Yoshida K., Tosa Y., Chuma I., Takano Y., Win J., Kamoun S., Terauchi R. (2009). Association genetics reveals three novel avirulence genes from the rice blast fungal pathogen Magnaporthe oryzae. Plant Cell 21: 1573–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan B., Zhai C., Wang W., Zeng X., Xu X., Hu H., Lin F., Wang L., Pan Q. (2011). The Pik-p resistance to Magnaporthe oryzae in rice is mediated by a pair of closely linked CC-NBS-LRR genes. Theor. Appl. Genet. 122: 1017–1028 [DOI] [PubMed] [Google Scholar]
- Zeng L.R., Vega-Sánchez M.E., Zhu T., Wang G.L. (2006). Ubiquitination-mediated protein degradation and modification: An emerging theme in plant-microbe interactions. Cell Res. 16: 413–426 [DOI] [PubMed] [Google Scholar]
- Zhai C., Lin F., Dong Z., He X., Yuan B., Zeng X., Wang L., Pan Q. (2011). The isolation and characterization of Pik, a rice blast resistance gene which emerged after rice domestication. New Phytol. 189: 321–334 [DOI] [PubMed] [Google Scholar]
- Zhou B., Dolan M., Sakai H., Wang G.L. (2007). The genomic dynamics and evolutionary mechanism of the Pi2/9 locus in rice. Mol. Plant Microbe Interact. 20: 63–71 [DOI] [PubMed] [Google Scholar]
- Zhou B., Qu S., Liu G., Dolan M., Sakai H., Lu G., Bellizzi M., Wang G.L. (2006). The eight amino-acid differences within three leucine-rich repeats between Pi2 and Piz-t resistance proteins determine the resistance specificity to Magnaporthe grisea. Mol. Plant Microbe Interact. 19: 1216–1228 [DOI] [PubMed] [Google Scholar]