The bacterial phytotoxin coronatine mimics a plant hormone, jasmonic acid, and thus antagonizes signaling by another plant hormone, salicylic acid, important for plant defense. This study demonstrates that coronatine also suppresses SA-independent host defense, including the production of defense-promoting indole glucosinolates and has a target other than the jasmonic acid receptor.
Abstract
The phytotoxin coronatine (COR) promotes various aspects of Pseudomonas syringae virulence, including invasion through stomata, growth in the apoplast, and induction of disease symptoms. COR is a structural mimic of active jasmonic acid (JA) conjugates. Known activities of COR are mediated through its binding to the F-box–containing JA coreceptor CORONATINE INSENSITIVE1. By analyzing the interaction of P. syringae mutants with Arabidopsis thaliana mutants, we demonstrate that, in the apoplastic space of Arabidopsis, COR is a multifunctional defense suppressor. COR and the critical P. syringae type III effector HopM1 target distinct signaling steps to suppress callose deposition. In addition to its well-documented ability to suppress salicylic acid (SA) signaling, COR suppresses an SA-independent pathway contributing to callose deposition by reducing accumulation of an indole glucosinolate upstream of the activity of the PEN2 myrosinase. COR also suppresses callose deposition and promotes bacterial growth in coi1 mutant plants, indicating that COR may have multiple targets inside plant cells.
INTRODUCTION
Successful bacterial pathogens overcome plant immune responses. Contributions to defense suppression come from phytotoxins and type III effectors, which are bacterial proteins injected into host cells via the type III secretion system (Büttner and Bonas, 2002; He et al., 2004; Chisholm et al., 2006). Coronatine (COR) is a phytotoxin produced by several strains of Pseudomonas syringae, including P. syringae pv tomato strain DC3000 (Pto). Two moieties, coronafacic acid and coronamic acid, are conjugated by an amide linkage to form COR (Brooks et al., 2004), which is the predominantly active molecule in plant tissues (Uppalapati et al., 2005). COR is a structural and functional mimic of JA-Ile, the bioactive conjugate of jasmonic acid (JA) and Ile (Fonseca et al., 2009). JA signaling regulates plant growth and development and also plays essential roles in plant defense (Ballaré, 2011). The F-box protein CORONATINE INSENSITIVE1 (COI1) is a key component of the JA signaling pathway (Feys et al., 1994; Xie et al., 1998). JA-Ile interacts with a complex of COI1 and a jasmonate ZIM-domain (JAZ) transcriptional repressors protein and promotes the SCFCOI1 ubiquitin E3 ligase complex to induce proteasome-mediated degradation of the JAZ protein (Thines et al., 2007; Sheard et al., 2010). JA-responsive gene expression is activated by elimination of inhibitory JAZ proteins (Chini et al., 2007). Remarkably, COR is ∼1000 times more active than JA-Ile, in vitro, at stabilizing interactions between JAZ proteins and tomato (Solanum lycopersicum) COI1 (Katsir et al., 2008).
COR promotes multiple aspects of P. syringae virulence, including reopening of stomata to facilitate invasion, proliferation in the apoplast, and development of disease symptoms (Bender et al., 1987, 1999; Brooks et al., 2004, 2005; Cui et al., 2005; Melotto et al., 2006; Uppalapati et al., 2008; Freeman and Beattie, 2009; Ishiga et al., 2009). The reduced virulence of Pto mutants unable to produce COR (hereafter referred to as Ptocor-) correlates with enhanced defense responses of the plant. The defense suppressing activity of COR, as well as endogenous JA conjugates, is at least partially dependent on their ability to antagonize salicylic acid (SA) signaling via COI1 activation. COR inhibits SA accumulation by differentially regulating the transcription of genes involved in its biosynthesis and metabolism (Zheng et al., 2012). coi1 mutant plants, which display enhanced resistance against Pto and other biotrophs, accumulate more SA at early stages of Pto infection (Kloek et al., 2001). Also, the impaired growth of Ptocor- is restored in SA signaling–deficient mutant plants (Brooks et al., 2005; Melotto et al., 2006; Zeng and He, 2010). COR also may function independent of suppressing SA signaling. The COR-dependent symptoms induced by Pto are not restored when Ptocor- infects tomato plants deficient in SA signaling (Brooks et al., 2005; Uppalapati et al., 2005). Also, pathogen-associated molecular pattern (PAMP)–induced callose deposition in Arabidopsis thaliana roots, which does not require SA signaling, is suppressed by COR (Millet et al., 2010).
COR may promote bacterial virulence through regulation of secondary metabolism. When Arabidopsis is challenged by Pto, COR affects the expression of genes involved in the synthesis of anthocyanin as well as Trp- and Met-derived glucosinolates (Thilmony et al., 2006). Trp-derived indole glucosinolates contribute to the elicitation of Arabidopsis defenses in response to PAMPs and non-host-adapted fungi, with the hydrolysis of 4-methoxy-indol-3ylmethylglucosinolate (4MI3G) by the atypical myrosinase, PENETRATION2 (PEN2), playing a key role (Bednarek et al., 2009; Clay et al., 2009). PEN2 is required for callose deposition induced by flg22 (a peptide PAMP from bacterial flagellin) in the cotyledons of liquid-grown Arabidopsis seedlings, and, notably, exogenous JA suppresses the response (Clay et al., 2009).
In addition to COR, Pto also deploys type III effectors to promote its virulence. HopM1 and AvrE1 are key type III effector genes within the conserved effector locus (CEL), a region flanking the genes encoding the structural components of the type III pilus (Alfano et al., 2000). A Pto mutant lacking the CEL (PtoΔCEL) displays reduced virulence on tomato and Arabidopsis (DebRoy et al., 2004; Badel et al., 2006). PtoΔCEL grows less well and induces stronger cell wall reinforcement, as measured by callose deposition, than wild-type Pto on Arabidopsis plants competent in SA signaling. These defenses are suppressed by either HopM1 or AvrE1, indicating that HopM1 and AvrE1 proteins suppress SA-dependent defense (DebRoy et al., 2004). Curiously, despite the ability of COR to suppress SA signaling, PtoΔCEL still elicits SA-dependent defense. Thus, in the context of a PtoΔCEL infection, COR may promote virulence independent of suppressing SA signaling.
Here, we analyzed defense responses elicited when Pto or Pto mutants lacking the CEL, unable to produce COR, or both are introduced into the leaves of soil-grown Arabidopsis plants with compromised defense signal transduction, including single and multiple mutants disrupting SA production, SA signaling, JA perception, and glucosinolate metabolism (see Supplemental Table 1 online). To focus on postinvasive defense, the experiments were done following infiltration of bacteria into the interior of plant leaves. Our results indicate that the activity of COR can be obscured by partially overlapping functions of type III effectors of the CEL. By examining the activity of COR in the PtoΔCEL strain, we show that, in addition to suppressing SA signaling, COR also suppresses callose deposition and promotes bacterial growth in a manner independent of suppressing SA signaling. We show that COR inhibits accumulation of an indole glucosinolate involved in the callose response. Additionally, we show that COR inhibits callose deposition in a coi1 mutant, indicating a COI1-independent function of COR. These findings significantly extend our understanding of the relationship between COR and type III effectors in suppressing plant immunity and indicate that COR is a multifunctional toxin that likely has a plant target(s) in addition to COI1.
RESULTS
COR Suppresses SA-Dependent and SA-Independent Defense Responses
COR suppresses SA accumulation via COI1 activation (Kloek et al., 2001; Uppalapati et al., 2005, 2007), and yet the PtoΔCEL mutant, which produces COR, elicits SA-dependent defense responses that promote callose deposition and restrict bacterial growth (DebRoy et al., 2004). To understand the effect of COR on SA-dependent and SA-independent defense responses against Pto, we examined the interaction of Arabidopsis with Pto, PtoΔCEL (Alfano et al., 2000), Ptocor- (Brooks et al., 2004), and a PtoΔCEL cor- double mutant (see Supplemental Figure 1A online).
We first measured expression of the PATHOGENESIS RELATED1 (PR-1) gene, which is a marker of SA-dependent defense. PR1 transcript and protein accumulation was monitored after infiltration of Columbia-0 (Col-0) plants with a high-titer suspension of all four strains (see Supplemental Figure 2 online). While all four strains induced significantly more PR-1 transcript than buffer, Pto and PtoΔCEL induced less transcript than Ptocor- and PtoΔCEL cor-. Additionally, PtoΔCEL cor- induced more PR-1 protein accumulation than did PtoΔCEL after 48 and 72 h. Collectively, these data confirm previous findings that COR suppresses Pto-induced PR-1 expression, an effect that has been attributed to COR mimicking JA-Ile and antagonizing SA signaling. Free SA levels were not higher after challenge with the COR-deficient strains than the COR-producing strains (see Supplemental Figure 3 online). Thus, COR may suppress Pto-induced PR-1 expression independent of suppressing SA levels.
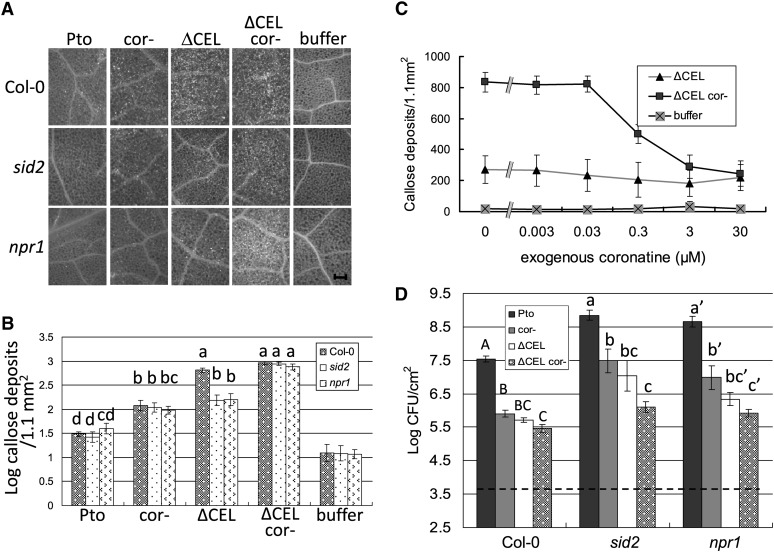
Next, we examined the elicitation of callose in wild-type Col-0 and plants deficient in SA production and SA signaling (Figures 1A and 1B). SA-induction deficient-2 (SID2) encodes an isochorismate synthase required for defense-associated SA production (Wildermuth et al., 2001). In sid2 mutant plants, neither Pto nor any of our mutant strains induced detectable levels of SA (see Supplemental Figure 3 online). Nonexpressor of pathogenesis related genes 1 (NPR1) encodes a protein that makes key contributions to SA signaling, including the activation of PR-1 expression (Cao et al., 1997). PtoΔCEL elicited high levels of callose deposition in Col-0 plants and reduced levels in sid2 and npr1 mutant plants, consistent with the findings of DebRoy et al. (2004) in other Arabidopsis mutant plants that do not accumulate SA (eds5 mutant and nahG expressing). Thus, although COR suppresses SA-dependent PR-1 expression elicited by PtoΔCEL, it fails to suppress SA-dependent callose deposition.
Figure 1.
COR Promotes Bacterial Virulence in SA Signaling–Deficient Plants.
(A) Callose deposition in Col-0, sid2, and npr1 Arabidopsis leaves after infiltration with the indicated bacterial strains or buffer. Shown are representative fluorescence microscopy images of aniline blue–stained leaves. Bar = 0.2 mm.
(B) Quantification of callose deposits following treatments as in (A). Shown are the mean and se of combined data from two independent biological replicates. Statistical analyses of log-transformed data of the indicated samples were by one-way ANOVA and Tukey HSD test with significant differences (P < 0.05) indicated by lowercase letters.
(C) Effect of exogenous COR on callose deposition elicited by ΔCEL cor- in sid2 mutant plants. Shown are the mean and sd of combined data from two independent biological replicates.
(D) Growth of the indicated strains 4 d after inoculation into Col-0, sid2, and npr1 Arabidopsis leaves. The dashed line indicates the starting inoculum of bacteria. Shown are the mean and se of four biological replicates. Different letter types (uppercase, lowercase, and lowercase’) indicate significant differences (P < 0.05) by one-way ANOVA and Tukey HSD test of comparisons between the different bacterial strains on individual plant genotypes.
COR did suppress SA-independent callose deposition during Pto infection. Ptocor- elicited more callose than Pto in Col-0 and sid2 plants (Figure 1B). Thus, the inhibition of callose deposition by COR cannot be accounted for by suppression of SA signaling. This observation was reinforced by the observation that PtoΔCEL cor- elicited more callose than PtoΔCEL in sid2 and npr1 plants. Thus, COR suppresses a pathway that contributes to callose deposition in plants deficient in SA accumulation or signaling.
To determine if the increased callose response against COR-deficient strains of Pto resulted from their inability to produce COR, we tested for complementation by exogenous COR. PtoΔCEL or PtoΔCEL cor- alone or in combination with various concentrations of COR were infiltrated into the leaves of sid2 plants (Figure 1C). The increased callose deposition elicited by PtoΔCEL cor- relative to PtoΔCEL was inhibited by exogenous COR in a dose-dependent manner, with partial suppression by 0.3 μM COR and full suppression by 3 and 30 μM COR. Thus, COR suppresses callose deposition in response to Pto.
The results for PR-1 expression and callose induction indicate that COR suppresses both SA-dependent and SA-independent Arabidopsis defense responses. Next, we measured the contribution of COR to Pto growth in both wild-type and SA signaling–deficient plants. Col-0, sid2, and npr1 plants were infiltrated with a low-titer inoculum of Pto or Ptocor-, and bacterial numbers were measured after 4 d (Figure 1D). Consistent with earlier reports that COR promotes Pto growth when the bacteria are directly infiltrated into the apoplast at a low concentration (Zeng and He, 2010), we found that Ptocor- grew to lower levels than Pto in Col-0. And, as expected, sid2 and npr1 plants showed enhanced disease susceptibility (eds) relative to Col-0 plants. Notably, Pto grew to significantly higher levels than Ptocor- in sid2, npr1, and sid2 npr1 mutant plants. (Figure 1D; see Supplemental Figure 4B online). Thus, COR promotes Pto growth in SA signaling–deficient plants.
COR and Type III Effectors of the CEL Make Overlapping Contributions to the Virulence of Pto
We also measured the growth of PtoΔCEL and PtoΔCEL cor- in wild-type and SA signaling–deficient plants. The Ptocor- and PtoΔCEL single mutant strains grew to similar levels to one another and both strains grew significantly less than Pto in both Col-0 and SA signaling–deficient mutants (Figure 1D). The PtoΔCEL cor- double mutant strain consistently grew less than either the PtoΔCEL or Ptocor- single mutant. Although not supported by analysis of variance (ANOVA) on the composite data set in Figure 1D, the reduced growth of PtoΔCEL cor- relative to PtoΔCEL was significant (P < 0.05) by two-tailed t test in 11 of 12 comparisons of the two bacteria in Col-0, sid2, or npr1 from the four biological replicates comprising the data. Thus, COR and type III effectors from the CEL promote bacterial growth in the apoplastic space in a manner that (1) is at least partially independent of suppressing SA signaling and (2) is overlapping but not entirely redundant.
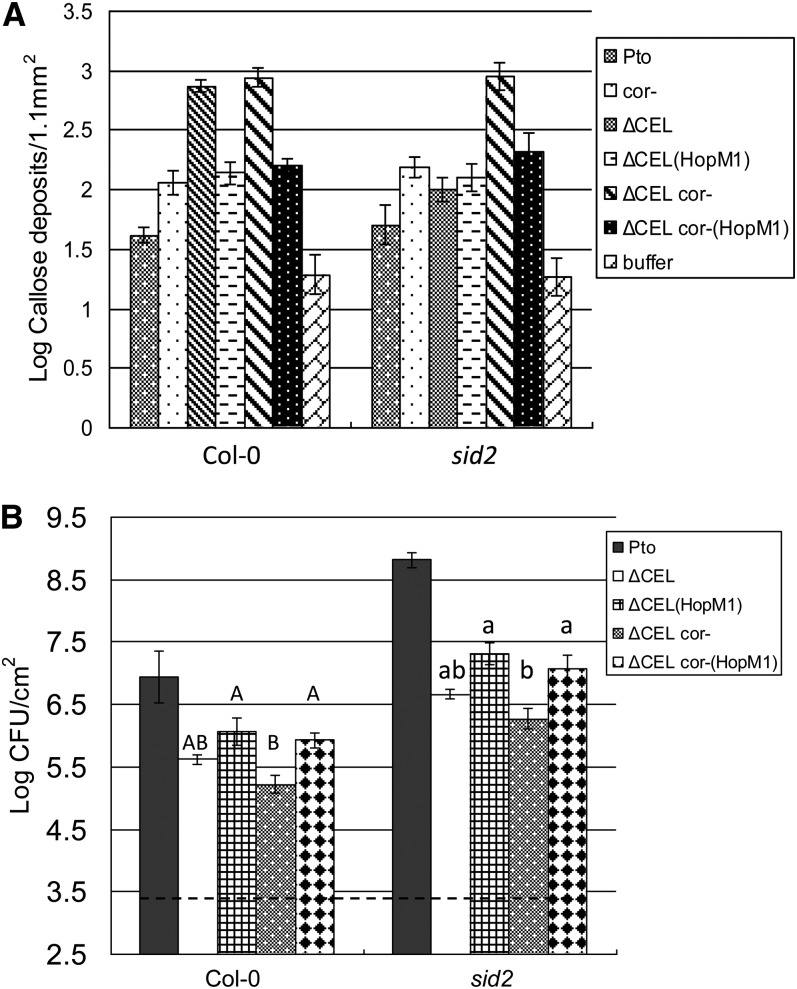
DebRoy et al. (2004) found that HopM1 suppresses SA-dependent defense signaling elicited by PtoΔCEL. Our results indicate that (1) both SA-dependent and SA-independent pathways contribute to callose elicitation by PtoΔCEL and (2) COR suppresses the SA-independent pathway. To examine the defense suppressing activity of HopM1 in the context of our findings, we analyzed strains of PtoΔCEL and PtoΔCEL cor- expressing HopM1 from a plasmid, called PtoΔCEL (HopM1) and PtoΔCEL cor- (HopM1), respectively. Callose induction by and growth of these strains were measured in Col-0 and sid2 plants. HopM1 suppressed callose deposition induced by PtoΔCEL and PtoΔCEL cor- in Col-0 as well as that induced by PtoΔCEL cor- in sid2 (Figure 2A). Consistent with the findings of DebRoy et al. (2004), PtoΔCEL (HopM1) grew to higher levels than PtoΔCEL (Figure 2B). Though this conclusion is not supported by ANOVA, the differences in Col-0 and sid2 were significant (P < 0.05) by two-tailed t test. Notably, we also observed that PtoΔCEL cor- (HopM1) grew significantly better than PtoΔCEL cor- in both plant backgrounds. The ability of HopM1 to suppress callose deposition and enhance bacterial growth in sid2 plants indicates that its virulence activity is not mediated by suppression of SA signaling per se.
Figure 2.
HopM1 Promotes Bacterial Virulence in SA Signaling–Deficient Plants.
(A) Quantification of callose deposits following infiltration of the indicated strains into Col-0 and sid2 leaves. Shown are the mean and se of combined data from two independent biological replicates.
(B) Growth of the indicated strains 4 d after inoculation into Col-0 and sid2 plants. The dashed line indicates the starting inoculum of bacteria. Shown are the mean and se of four biological replicates. Different letter types indicate significant differences (P < 0.05) by one-way ANOVA and Tukey HSD test of comparisons between the indicated bacterial strains on individual plant genotypes.
COI1 Suppresses SA-Dependent and SA-Independent Defense Responses Effective against Pto
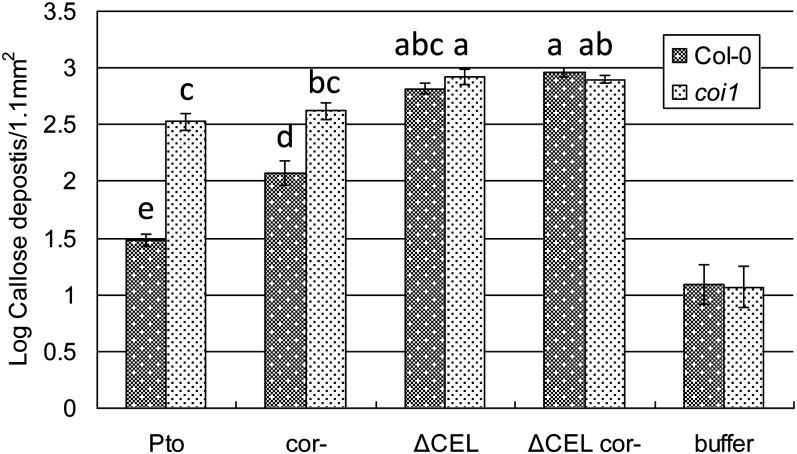
We examined the role of COI1 in Arabidopsis defense against Pto and activity of COR. We tested if suppression of callose deposition by COR is dependent on COI1 (Figure 3A). Ptocor- elicited more callose than did Pto in wild-type Col-0. However, although overall levels of callose were elevated, Pto and Ptocor- elicited similar amounts of callose in coi1 plants. These results indicate that the ability of COR to suppress callose induction by Pto is dependent on COI1 (at least in plants with intact SA signaling; see below). Furthermore, these results indicate that, even if the bacteria do not produce COR, COI1-dependent signaling suppresses the levels of callose deposition, likely by responding to increases in plant-derived JA conjugates. Callose levels are similar in Col-0 and coi1 plants following challenge with PtoΔCEL and PtoΔCEL cor-, indicating that the lack of COR or the coi1 mutation does not significantly increase callose deposition beyond the high level induced by PtoΔCEL in Col-0.
Figure 3.
COR Promotes Bacterial Virulence by Targeting COI1.
Quantification of callose deposits following infiltration of the indicated strains into Col-0 and coi1 leaves. Shown are the mean and se of combined data from two independent biological replicates. Statistical analyses of log-transformed data of indicated samples were by one-way ANOVA and Tukey HSD test (P < 0.05).
Next, we tested the role of COI1 in mediating the growth enhancement provided by COR to Pto. Consistent with other coi1 alleles (Feys et al., 1994; Kloek et al., 2001), we observed that coi1-16 plants displayed an enhanced disease resistance (edr) phenotype against wild-type Pto (Figure 4C; see Supplemental Figure 4B online). The significant reduction in growth of Ptocor- relative to Pto in Col-0 plants was not apparent in coi1, and, similarly, growth of PtoΔCEL cor- was not reduced relative to PtoΔCEL in coi1 plants (Figure 4C; see Supplemental Figure 4B online). Thus, COR fails to enhance the growth of Pto in coi1 plants, indicating that, in SA signaling–competent plants, COR requires COI1 to promote Pto growth.
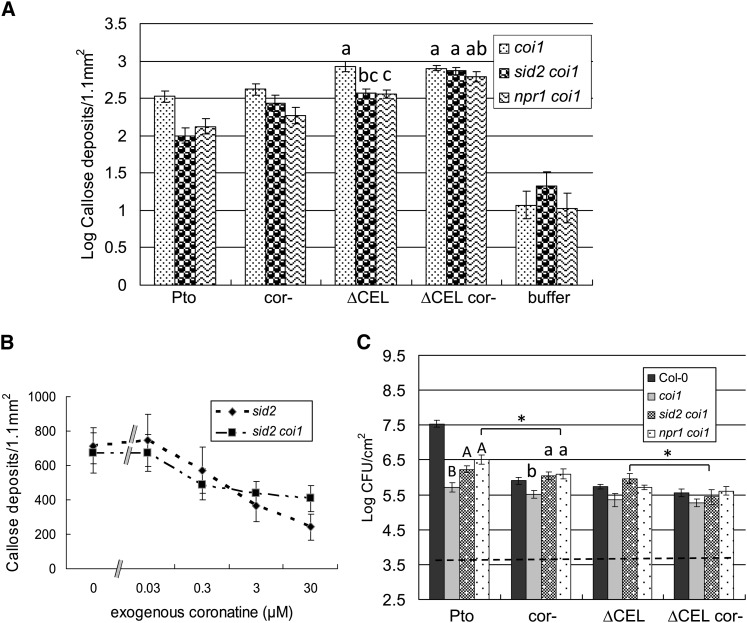
Figure 4.
COR Promotes Bacterial Virulence in SA Signaling–Deficient Plants Independent of Targeting COI1.
(A) Quantification of callose deposits following infiltration of the indicated strains into coi1, sid2 coi1, and npr1 coi1. Shown are the mean and se of combined data from two independent biological replicates. Statistical analyses of log transformed data of indicated samples were by one-way ANOVA and Tukey HSD test (P < 0.05). See also Supplemental Figure 4 online.
(B) Effect of exogenous COR on callose deposition elicited by PtoΔCEL cor- in sid2 and sid2 coi1 mutant plants. Shown are the mean and sd of combined data from two independent biological replicates.
(C) Growth of the indicated strains 4 d after inoculation into Col-0, coi1, sid2 coi1, and npr1 coi1 plants. The dashed line indicates the starting inoculum of bacteria. Shown are the mean and se of four biological replicates. Different letter types indicate significant differences (P < 0.05) by one-way ANOVA and Tukey HSD test of comparisons between the indicated plant genotypes with individual bacterial strains. Asterisks indicate significant differences between indicated samples as determined by two-tailed t test with * P < 0.05. See also Supplemental Figure 4 online.
The edr phenotype of coi1 mutant plants to Pto is hypothesized to result from elevated SA signaling due to the lack of inhibitory crosstalk from COI1 (Feys et al., 1994; Kloek et al., 2001). However, our findings that COR also suppresses SA-independent defense raises the possibility that COI1 suppresses both SA-dependent and SA-independent defense responses. To explore this possibility, we generated sid2 coi1 and npr1 coi1 double mutant and sid2 npr1 coi1 triple mutant plants. We observed that each of our Pto strains induced elevated SA accumulation in coi1 relative to Col-0 and that this increase was statistically significant for all but wild-type Pto (see Supplemental Figure 3 online). No SA accumulation is detected in the sid2 coi1 double mutant, excluding the possibility that the coi1 mutation stimulates SID2-independent SA production. Thus, the sid2 and sid2 coi1 mutants are SA accumulation deficient in response to the examined Pto strains.
Pto strains grew better in multiple mutant plants deficient in both SA and JA signaling than in coi1 plants deficient in just JA signaling (Figure 4C; see Supplemental Figure 4B online). Thus, COI1-mediated suppression of SA signaling does contribute to restricting Pto growth. However, Pto strains grew less well in the SA and JA signaling–deficient plants than in sid2, npr1, and sid2 npr1 plants only deficient in SA signaling (Figures 1D and 4C; see Supplemental Figure 4B online). Thus, the edr phenotype of coi1 is apparent in SA signaling–deficient plants; therefore, COI1 also suppresses SA-independent defenses important for restricting the growth of Pto.
COR Promotes Pto Virulence Independent of Targeting COI1
From the results described so far, an activity of COR independent of targeting COI1 is not apparent. However, we speculated that the edr phenotype of coi1 might be dominant over the virulence promoting function of COR because (1) SA-mediated defense is activated in the coi1 mutant, (2) COR suppresses the SA-dependent defenses through its interaction with COI1, and (3) the SA-mediated defense is dominant over the putative COI1-independent virulence activity of COR. Thus, to test for COI1-independent activity of COR, we examined COR function in coi1 plants that are also deficient in SA signaling. We observed that PtoΔCEL cor- elicits more callose than PtoΔCEL in sid2 coi1, npr1 coi1, and sid2 npr1 coi1 plants (Figure 4A; see Supplemental Figure 4A online). This finding is consistent with COR suppressing callose deposition independent of targeting COI1. However, an alternate possibility is that coi1-16 (a missense mutation in the Leu-rich repeats of COI1) is leaky and the potent activity of COR, relative to JA-Ile, overcomes a reduction of COI1 protein activity in coi1-16. To examine this possibility, we inoculated PtoΔCEL cor- along with various concentrations of COR into sid2 and sid2 coi1 plants (Figure 4B). The dose response for suppression of callose by COR was similar in both backgrounds, indicating that leakiness of the coi1-16 allele is unlikely to account for the COI1-independent suppression of callose deposition by COR. Furthermore, Pto grew to significantly higher levels than Ptocor- in npr1 coi1 (Figure 4C) and sid2 npr1 coi1 (see Supplemental Figure 4B online) and PtoΔCEL grew to significantly higher levels than PtoΔCEL cor- in sid2 coi1 (Figure 4C). Collectively, these results reveal a COI1-independent function of COR that suppresses callose deposition and promotes bacterial multiplication. A target of COR, other than COI1, has not been described.
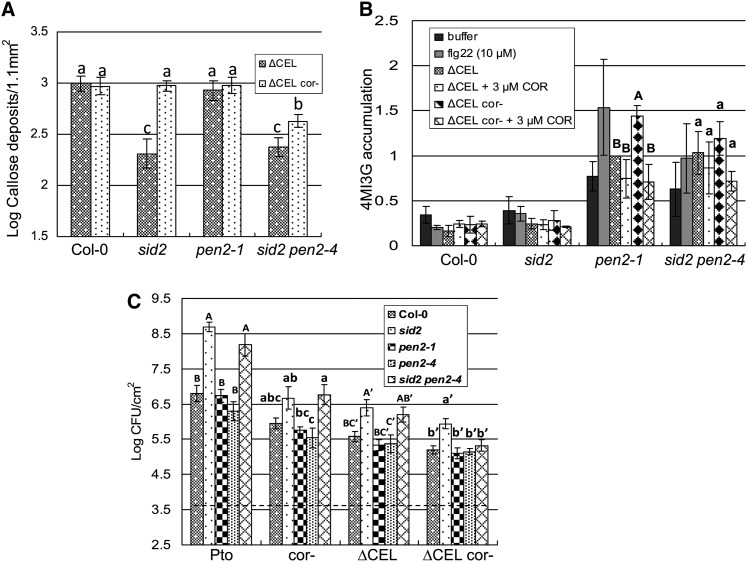
COR Inhibits the PEN2-Dependent Branch of a Pathway Contributing to Callose Deposition in Response to Pto
The atypical myrosinase PEN2 (Bednarek et al., 2009) is required for flg22-induced callose deposition in the cotyledons and roots of liquid-grown Arabidopsis seedlings (Clay et al., 2009; Millet et al., 2010). However, the pen2 mutant did not differ from wild-type Col-0 in flg22-induced callose deposition in the leaves of soil-grown plants (see Supplemental Figure 5A online), although we did reproduce the finding that flg22-induced callose deposition is dependent on pen2 and is not dependent on sid2 in liquid-grown seedlings (see Supplemental Figure 5B online). In the liquid-grown seedling assay, pretreatment with exogenous SA rescued callose deposition in pen2 mutant plants (Clay et al., 2009). Based on these results, we hypothesize that (1) in the liquid-grown seedling assay, diffusion of endogenous SA renders the plants effectively SA accumulation deficient and thus dependent on PEN2; and (2) in soil-grown plants, the PAMP-induced accumulation of SA (Tsuda et al., 2008) permits callose deposition in the pen2 mutant background.
We further hypothesized that COR suppresses callose deposition elicited by PtoΔCEL in SA signaling–deficient plants by targeting the PEN2-dependent pathway. To examine the relationship between COR, PEN2-dependent signaling, and SA-dependent signaling, we compared callose deposition induced by PtoΔCEL and PtoΔCEL cor- in wild-type, sid2, pen2, and sid2 pen2 mutant plants (Figure 5A). In Col-0 and pen2 mutant plants, callose deposition was strong in response to both strains. In sid2 single mutant plants, callose deposition was strong in response to PtoΔCEL cor- and was reduced in response to PtoΔCEL. In sid2 pen2 double mutant plants, callose deposition was reduced in response to both PtoΔCEL and PtoΔCEL cor-. Thus, the high level of callose elicited by PtoΔCEL cor- in sid2 mutant plants is PEN2 dependent and COR suppresses the PEN2-dependent pathway. Interestingly, PtoΔCEL cor- elicited more callose than PtoΔCEL in the sid2 pen2 double mutant plants. This result indicates that the pen2 mutant is not completely penetrant, perhaps due to other myrosinases that can substitute for PEN2, and that COR suppresses the residual activity in the pen2 mutant background.
Figure 5.
COR Inhibits Indole Glucosinolate Accumulation Upstream of PEN2.
(A) Quantification of callose deposits following infiltration of PtoΔCEL or PtoΔCEL cor- into Col-0, sid2, pen2-1, and sid2 pen2-4 leaves. Shown are the mean and se of combined data from two independent biological replicates. Statistical analyses of log-transformed data were by one-way ANOVA and Tukey HSD test (P < 0.05).
(B) Accumulation of 4MI3G after infiltration of buffer, flg22, or the indicated bacterial strains, with or without 3 μM COR, into Col-0, sid2, pen2-1, and sid2 pen2-4 leaves. Quantities of 4MI3G were calculated relative to sinigrin (spiked into each sample) and normalized with the amount elicited by PtoΔCEL in pen2-1 set to 1. Shown are the means and se from three biological replicates. Different letter types indicate significant differences (P < 0.05) by one-way ANOVA and Tukey HSD test of comparisons between the indicated bacterial strains on individual plant genotypes. See also Supplemental Figure 6 online.
(C) Growth of the indicated strains 4 d after inoculation into Col-0, sid2, pen2-1, pen2-4, and sid2 pen2-4 plants. The dashed line indicates the starting inoculum of bacteria. Shown are the mean and se of five biological replicates, except pen2-4 data, which are from two biological replicates. Different letter types indicate significant differences (P < 0.05) by one-way ANOVA and Tukey HSD test of comparisons between plant genotypes with individual bacterial strains.
COR Suppresses Indole Glucosinolate Metabolism Upstream of PEN2
Next, we sought to ascertain the effect of COR on the PEN2-dependent pathway. Pathogen or PAMP elicitation induces expression of numerous enzymes, including cytochrome P450s, that mediate the multistep conversion of Trp into 4MI3G (Bednarek et al., 2009; Clay et al., 2009). PEN2 then hydrolyzes 4MI3G into an as yet unidentified substrate that is crucial for the eventual deposition of callose. 4MI3G levels do not fluctuate significantly following defense elicitation in wild-type plants, presumably because it is efficiently processed by PEN2. However, 4MI3G levels increase following elicitation in pen2 mutant plants. We reasoned that COR would prevent the accumulation of 4MI3G in pen2 mutant plants if it functions upstream of PEN2 enzyme activity, but not if it inhibits PEN2 activity or downstream events. Infiltration of flg22, PtoΔCEL, or PtoΔCEL cor- each elicited elevated accumulation of 4MI3G in pen2 mutant plants relative to Col-0 (Figure 5B). Notably, in pen2, PtoΔCEL cor- elicited significantly more 4MI3G than did PtoΔCEL. Also, exogenous COR (3 μM) suppressed 4MI3G accumulation elicited by PtoΔCEL cor-. The precursor indol-3ylmethylglucosinolate (I3G) is first hydroxylated then converted by a methyltransferase into 1-methoxy-indol-3ylmethylglucosinolate (1MI3G) and 4MI3G (Pfalz et al., 2011). Infiltration with flg22 also induced elevated accumulation of 1MI3G in pen2 relative to Col-0 plants (see Supplemental Figure 6A online). And similar to 4MI3G, 1MI3G accumulation induced by PtoΔCEL cor- was suppressed by exogenous COR. Though not supported by ANOVA, this difference was significant (P < 0.05) by two-tailed t test. Levels of I3G in pen2 plants did not vary significantly with any of the tested infiltrations (see Supplemental Figure 6B online). Collectively, these data indicate that COR suppresses glucosinolate metabolism upstream of 1MI3G and 4MI3G accumulation.
We sought to determine the relationship between SA signaling and the suppression of glucosinolate metabolism by COR. In each of three biological replicates, flg22 elicited significantly higher levels of 4MI3G and 1MI3G in pen2 than sid2 pen2 plants (composite data shown in Figure 5B and Supplemental Figure 6A online). This observation, along with the elicitation of SA by flg22 (Tsuda et al., 2008), indicates that SA promotes indole glucosinolate metabolism. On the contrary, PtoΔCEL elicited similar levels of 4MI3G and 1MI3G in both pen2 and sid2 pen2. Thus, COR may limit glucosinolate metabolism upstream of PEN2 function by inhibiting SA signaling. However, COR also suppresses glucosinolate metabolism independent of suppressing SA signaling. In sid2 pen2 plants, accumulation of 4MI3G and 1MI3G induced by PtoΔCEL cor- was suppressed by exogenous COR. Though not supported by ANOVA, this conclusion is in both cases significant (P < 0.05) by two-tailed t test. Thus, in SA signaling–deficient plants, COR suppresses both glucosinolate metabolism and callose deposition.
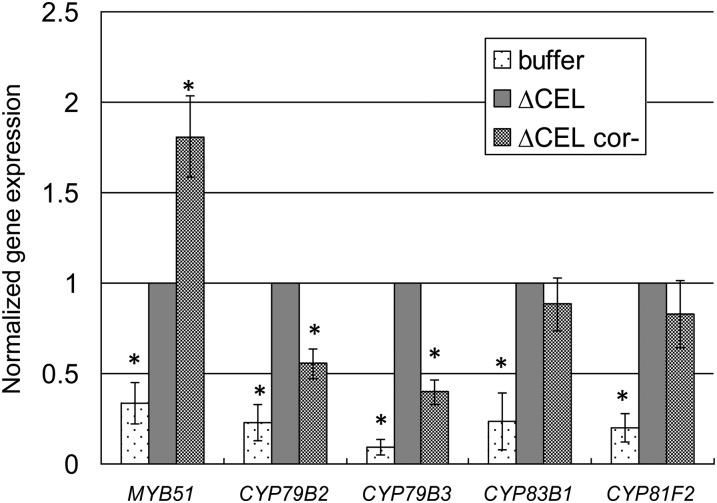
COR might suppress 4MI3G and 1MI3G accumulation by preventing the expression of enzymes required for glucosinolate metabolism. In roots, COR suppresses the expression of MYB51 (Millet et al., 2010), which is an R2R3 MYB family transcription factor that promotes the expression of numerous cytochrome P450 monooxygenases involved in glucosinolate metabolism (Gigolashvili et al., 2007) and is required for PAMP-induced callose deposition in liquid-grown seedlings (Clay et al., 2009). CYP79B2 and CYP79B3 encode cytochrome P450 monooxygenases that convert Trp into indol-3ylacetaldoxime (Hull et al., 2000). CYP83B1 and CYP81F2 participate in the multistep conversion of indol-3ylacetaldoxime into 4MI3G (Bak et al., 2001; Bednarek et al., 2009). PAMP-induced expression of CYP79B2, CYP79B3, and CYP83B1, but not CYP81F2, are MYB51 dependent in liquid-grown seedlings (Bednarek et al., 2009; Clay et al., 2009). In soil-grown plants, PtoΔCEL induced expression of all five genes between approximately threefold and ∼10-fold relative to a buffer infiltration (Figure 6). The complex effect of COR on expression of these genes was revealed by comparing PtoΔCEL to PtoΔCEL cor-. Relative to PtoΔCEL cor-, PtoΔCEL induced less expression of MYB51, comparable expression of CYP83B1 and CYP81F2, and increased expression of CYP79B2 and CYP79B3. Thus, bacterially produced COR has variable effects on the expression of genes involved in indole glucosinolates metabolism. MYB51 may act as a master regulator responsible for the ultimate reduction in 4MI3G accumulation (Clay et al., 2009), but how it does so remains unclear.
Figure 6.
Bacterially Produced COR Variably Affects Expression of Genes Involved in Indole Glucosinolate Metabolism.
qRT-PCR analysis of MYB51, CYP79B2, CYP79B3, CYP83B1, and CYP81F2 expression 6 h after infiltration of Col-0 leaves with buffer, PtoΔCEL, or PtoΔCEL cor-. Shown are the average and se of normalized data from of four (MYB51) or three (CYP79B2, CYP79B3, CYP83B1, and CYP81F2) biological repeats with the level of each transcript induced by PtoΔCEL set to 1. Asterisks indicate significant differences between the indicated samples and ΔCEL sample as determined by two-tailed t test with *P < 0.005.
PEN2 is important for antifungal defense (Bednarek et al., 2009). To test whether PEN2 contributes to defense against Pto, we tested bacterial growth in Col-0, sid2, pen2, and sid2 pen2 mutant plants (Figure 5C). The growth of Pto, Ptocor-, PtoΔCEL, and PtoΔCEL cor- does not differ significantly between Col-0 and pen2, indicating that PEN2 does not play a major role in defense against these strains of Pto. Interestingly, the eds phenotype of sid2 pen2 relative to pen2, which is apparent for Pto, Ptocor-, and PtoΔCEL, is not apparent for PtoΔCEL cor-. Thus, PEN2 contributes to the susceptibility of SA-deficient plants to Pto severely compromised by the lack of key type III effectors from the CEL and an inability to produce COR.
DISCUSSION
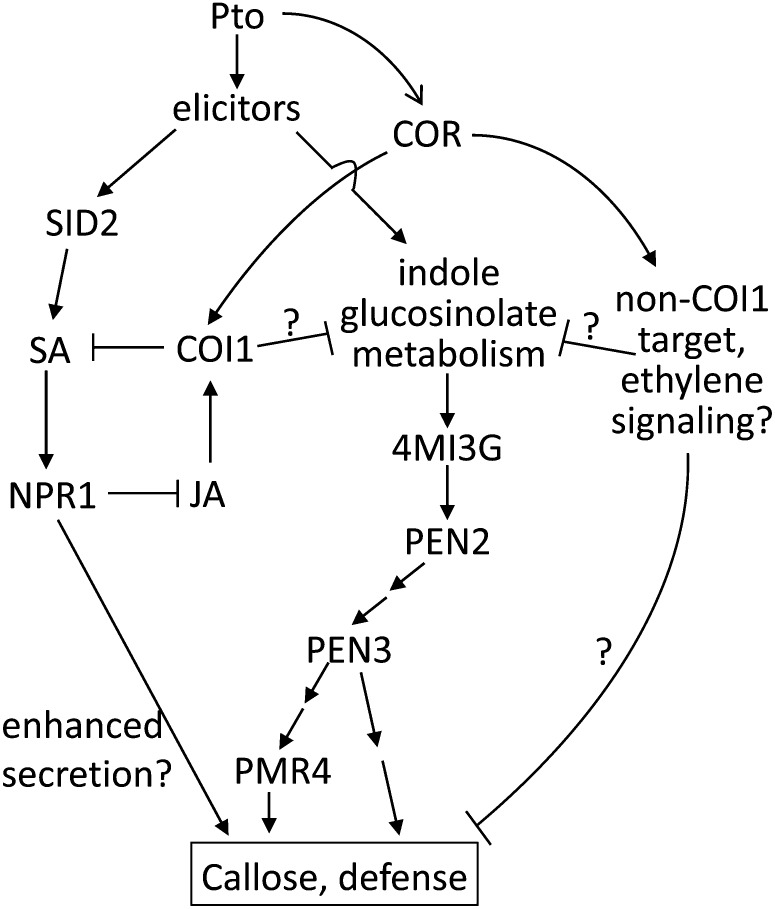
Our results support a model integrating the contributions of SA signaling, JA signaling, and indole glucosinolate metabolism and the suppressive activities of COR to Arabidopsis defense against Pto (Figure 7). COR and type III effectors from the CEL both make significant contributions to suppressing callose and promoting Pto growth. Although these virulence activities are largely overlapping with respect to bacterial growth, COR and HopM1 differ in their modes of action. COR suppresses callose deposition mediated by a PEN2-dependent pathway but cannot block callose deposition in SA signaling–competent plants. HopM1, on the other hand, suppresses callose deposition mediated through the SA-dependent and PEN2-dependent pathways. Thus, HopM1 does not suppress SA signaling per se, but instead may suppress a signaling step upstream or downstream of the requirements for SA and PEN2.
Figure 7.
Model: Suppression of the Arabidopsis Immune Response by COR.
Unknown elicitors from Pto (PAMPs and/or T3Es) activate SA signaling and indole glucosinolate metabolism. SA signaling and JA signaling are antagonistic, with COR activating COI1 to suppress SA accumulation. COR also suppresses indole glucosinolate metabolism upstream of 4MI3G accumulation. SA signaling promotes indole glucosinolate metabolism, perhaps by suppressing JA accumulation, and also promotes callose deposition independent of COI1 through an unknown pathway. COR suppresses defense in a COI1-independent manner through an unknown pathway.
COR suppresses SA signaling. The ability of COR to suppress SA-dependent PR-1 expression has been shown here and elsewhere (Kloek et al., 2001) and is thought to occur through suppression of SA accumulation by activated COI1 (Spoel and Dong, 2008). However, SA measurements did not reveal increased SA accumulation in response to COR-deficient strains. Furthermore, COR fails to suppress SA-mediated callose deposition elicited by PtoΔCEL. Thus, differences may exist between the quantitative requirement for and qualitative nature of SA signaling in callose deposition versus PR-1 expression. SA signaling promotes various aspects of Arabidopsis defense against P. syringae, including activation of PR genes by NPR1 in association with TGA transcription factors and genes involved in vesicle secretion by NPR1 through TL1 elements in their promoters (Fan and Dong, 2002; Wang et al., 2005). Notably, vesicle secretion is required for efficient cell wall fortification, including callose deposition (Assaad et al., 2004; Hardham et al., 2007). Thus, the activation of secretion by SA could contribute to its support of callose deposition, perhaps downstream or independent of PEN2. The reduced accumulation of 4MI3G and 1MI3G following PAMP elicitation in sid2 pen2, compared with pen2, indicates that SA signaling also supports callose deposition by promoting indole glucosinolate metabolism upstream of PEN2. This activity of SA may result from its ability to suppress the accumulation of JA, which, similar to COR, may antagonize 4MI3G accumulation. 4MI3G accumulation was not compromised in SA signaling–deficient plants in response to aphid feeding, perhaps because JA signaling is already strongly induced (Kim and Jander, 2007).
COR also suppresses Arabidopsis defense independent of suppressing SA signaling. The enhanced callose deposition elicited by and the reduced growth of COR-deficient strains was apparent in SA signaling–deficient plants. In fact, the contribution of COR to suppressing callose induction by PtoΔCEL is only apparent in SA signaling–deficient mutants. Defense suppressing activities of COR in SA signaling–deficient mutant plants are apparent in both COI1 and coi1 backgrounds. Of particular note is the ability of COR to suppress callose deposition and promote the growth of P. syringae in coi1 plants. An activity of COR, other than targeting COI1, has not been described. One possibility is that, in functioning as a JA mimic, COR stimulates COI1-independent responses. JA induces COI1-independent transcriptional reprogramming in Arabidopsis (Devoto et al., 2005). Alternatively, COR may have a function other than acting as a JA mimic. The coronamic acid component of COR is a structural mimic of ACC and COR has been shown to elicit de novo production of ethylene from Met (Ferguson and Mitchell, 1985; Kenyon and Turner, 1992). Ethylene signaling contributes to the expression of MYB51 and plays a key role in callose deposition (Clay et al., 2009; Millet et al., 2010). Thus, it is reasonable to speculate that COR perturbs ethylene signaling.
COR regulates secondary metabolism. Glucosinolates are a class of thioglucosides produced by the Capparales, including Arabidopsis, which play significant roles in defense against insects and microorganisms (Halkier and Gershenzon, 2006; Bednarek et al., 2009; Clay et al., 2009). Trp-derived indole glucosinolates, including 4MI3G, are key in defense against microorganisms. Our and previous findings indicate that COR activates the expression of CYP79B2 and CYP79B3, the gene products of which convert Trp into indol-3ylacetaldoxime (Zhao et al., 2002; Thilmony et al., 2006). In another interesting result, spraying Arabidopsis leaves with 5 μM COR induces the expression of ST5a, which synthesizes I3G (Piotrowski et al., 2004), which is subsequently converted by CYP81F2 and indole glucosinolate methyltransferases into 4MI3G and 1MI3G (Bednarek et al., 2009; Pfalz et al., 2011). We observed here that bacterially produced and/or exogenous COR suppressed MYB51 expression 6 h and 4MI3G and 1MI3G accumulation upstream of the PEN2 myrosinase 9 h after bacterial challenge. Thus, the effects of COR on gene expression and glucosinolate metabolism are complex.
SA signaling and COI1-dependent signaling are antagonistic to one another (Spoel and Dong, 2008). Here, we demonstrate that, in addition to antagonizing one another, both regulate Arabidopsis defense against P. syringae independent of the other. The eds phenotype of SA signaling mutants is still apparent in the coi1 background, and the edr phenotype of the coi1 mutant is still apparent in SA signaling mutant backgrounds. The virulence activity of COR can be mediated in three ways: (1) activation of COI1 to suppress SA signaling, (2) activation of COI1 to suppress SA-independent defense responses, and (3) through an unknown mechanism that is independent of targeting COI1 and may be independent of mimicking JA. Thus, COR is a multifunctional suppressor of plant immunity.
METHODS
Plants
Arabidopsis thaliana plants used in this work were of the Col-0 ecotype and were grown under 8 h (115 μmol m−2 s−1), 23°C days, and 16 h, 16°C nights. The following mutants were used: sid2-2 has a deletion from amino acid 439 to 455 (Wildermuth et al., 2001); npr1-1 has a mutation of conserved His into Tyr at amino acid 334 (Cao et al., 1997); coi1-16 has a mutation of Leu to Phe at amino acid 245 (Ellis and Turner, 2002); pen2-4 has a mutation of Gly to Asp at amino acid 150 (Westphal et al., 2008); pen2-1 has a mutation at amino acid 48 Trp to stop codon (Lipka et al., 2005). The original line carrying coi1-16 also harbored the pen2-4 mutant allele (Westphal et al., 2008). The coi1-16 and pen2-4 single mutants and the various double and triple mutants described in the work were produced by crossing and marker-assisted selection (see Supplemental Figures 1B to 1E and Supplemental Tables 1 and 2 online).
Bacteria
Pto and mutant strains were grown at 28°C on King's B (KB) plates containing the appropriate antibiotics for selection. Mutant strains used are PtoΔCEL (Alfano et al., 2000) and Ptocor- (DB4G3) (Brooks et al., 2004). To construct the PtoΔCEL cor- double mutant, homologous recombination was used to introduce the cfa6 mutation into PtoΔCEL. Plasmid pDB29, which is a derivative of pRK415 (Tcr) containing a 7.8-kb genomic fragment from strain DB4G3 including the 3.2-kb Tn5 insertion in cfa6 and 4.5-kb of flanking DNA (cfa6::Tn5 uidA Kmr) (Brooks et al., 2004), was transformed into PtoΔCEL (Sm/Spr). Selection of kanamycin and tetracycline resistance clones isolated cells carrying pDB29. These were then propagated for 4 d in liquid KB media (diluting the saturated culture 1:1000 into fresh media every day) containing kanamycin (50 μg/mL) but lacking tetracycline to allow: (1) homologous recombination between the wild type, chromosomal cfa6, and plasmid-borne, cfa6::Tn5 and (2) subsequent loss of the pRK415 plasmid. The final bacterial culture was diluted and plated on KB containing kanamycin. Individual clones were then replica plated onto Sp Km and Sp Km Tc to identify Spr Kmr Tcs clones as candidate pRK415-free double mutants. PCR screening of individual clones for insertion of Tn5 into cfa6 identified the PtoΔCEL cor- double mutant. Primers used for screening are shown in Supplemental Table 2 online. The plasmid pORF43, which carries HopM1-ShcM (DebRoy et al., 2004), was transformed into PtoΔCEL and PtoΔCEL cor- by electroporation to generate PtoΔCEL (HopM1) and PtoΔCEL cor- (HopM1).
Bacterial growth assays were conducted by infiltrating 105 colony-forming units (CFU)/mL (OD600 = 0.0002) in 10 mM MgCl2 into the underside of the leaves of 5-week-old plants using a needleless 1cc syringe. After the infiltrated leaves were dry, plants were returned to the growth room. After 4 d, nine leaf discs were separated into three technical replicates containing three leaf discs each. The bacterial titer in each technical replicate was determined by grinding leaf discs to homogeneity in 10 mM MgCl2, serially diluting the samples in a 96-well plate, and transferring with a multiplating tool onto KB plates with appropriate selection (Kim and Mackey, 2008). Colonies were counted and used to calculate the mean CFU/cm2 for each treatment, and the values were log transformed. The log-transformed means from individual biological replicates, as single data points, were then combined from multiple independent biological replicates and used to calculate the mean and standard error. Minitab 16 statistical software was used to determine significant differences by one-way ANOVA and Tukey honestly significant difference (HSD) test (P < 0.05), either between bacteria in the same plant background or between plants infiltrated with the same bacterial strain. Where indicated by brackets, the two-tailed t test was used to compare the indicated pairs of treatments.
Protein Immunoblotting
Five-week-old leaves of Col-0 plants were hand-infiltrated with 108 CFU/mL (OD600 = 0.2) of bacteria in 10 mM MgCl2. After infiltrated leaves were dry, the plants were covered with the transparent lid for the reminder of the experiment. Protein extracts were prepared and quantified as previously described (Kim et al., 2005). Samples containing 20 μg of protein were resolved on 12% SDS-PAGE gels, transferred to polyvinylidene difluoride membrane, and blotted with Anti-PR-1 sera (Kliebenstein et al., 1999) at 1:10,000.
Callose Staining
Callose detection was conducted as previously described (Kim and Mackey, 2008). Briefly, leaves of 4-week-old plants were hand-infiltrated with 108 CFU/mL of bacteria in 10 mM MgCl2 or 30 μM flg22. At 15 h after infiltration, leaves were collected, cleared with lactophenol, washed with 50% ethanol and then with water, stained with 0.01% aniline blue dissolved in 150 mM K2HPO4, pH 9.5, mounted on slides in 50% glycerol, and examined with a Nikon Eclipse 80i epifluorescent microscope. At least four to five individual leaves were analyzed for each treatment. Images were captured from the similar middle area of each leaf, and the number of callose deposits was calculated using ImageJ software (http://rsbweb.nih.gov/ij). Except where noted in Supplemental Figure 5 online, similar results for all callose experiments were observed in three or more independent biological replicates. Statistical analyses of log-transformed data were by one-way ANOVA and Tukey HSD test (P < 0.05) using Minitab 16 statistical software.
Callose staining of 10-d-old liquid-grown seedlings was done after seedlings were exposed to 1 μM flg22 or water for 18 h. Cotyledons were cleared and stained as described (Clay et al., 2009).
Quantitative Real-Time PCR
MYB51, CYP79B2, CYP79B3, CYP83B1, CYP81F2, and PR1 transcript levels were measured using quantitative real-time PCR (qRT-PCR). Leaves of 5-week-old Col-0 plants were hand-infiltrated with buffer or 108 CFU/mL of PtoΔCEL or PtoΔCEL cor-. At 6 h after infiltration, leaves were frozen in liquid nitrogen and ground by mortar and pestle. Total RNA was prepared using the Plant RNeasy Mini Prep Kit (Qiagen). One microgram of total RNA was treated with DNaseI (Invitrogen) and then cDNA synthesis was done using the reverse transcription system (Promega). For individual biological replicates, each cDNA sample was tested by three technical replicates. qRT-PCR reactions were set up using iQ SYBR green supermix (Bio-Rad) and run in an iQ5 real-time PCR detection system (Bio-Rad). Gene expression data were analyzed using the ΔCT (cycle threshold) method with the relative quantification to ACTIN2 as the reference gene. The specificity of PCR products was verified on 1.5% agarose gels and by melt curves in the iQ5. Primers used for RT-PCR are shown in Supplemental Table 3 online. Statistical analysis for the combined data from biological replicates was done using one-way ANOVA and Tukey HSD test (P < 0.05) by Minitab 16 statistical software.
SA Extraction and Quantification
Leaves of 5-week-old plants were hand-infiltrated with buffer or 108 CFU/mL of bacteria in 10 mM MgCl2. At 15 h after infiltration, leaves were collected and ground in liquid N2 and 0.2 g of grindate was extracted twice overnight in 350 μL of 100% methanol in the dark at 4°C. The supernatants were removed after centrifugation at 13K rpm for 10 min, pooled, and stored at −20°C until HPLC analyses. HPLC fluorescence analyses were performed using an Alliance 2690 separation module (Waters) equipped with an autosampler and a 474 fluorescence detector (Waters). The autosampler and column temperatures were set to 4 and 30°C, respectively. Chromatographic separation of methanolic extracts was performed using a Waters Xterra RP18 analytical column (3.9 μm) coupled with a 3.0 × 20-mm guard column. The binary mobile phase consisted of water/acetic acid (A) (98:2, v/v) and methanol/acetic acid (B) (98:2, v/v) with a flow rate of 1 mL/min. The gradient was as follows (percentages refer to proportions of eluant B): 0 to 10% (0 to 2 min), 10 to 50% (20 to 30 min), 50 to 80% (30 to 32 min), and 80 to 100% (32 to 34 min). The injection volume for all samples was 15 μL. Quantification of SA was achieved using fluorescence detection set to λem = 400 nm. Identification of SA was done by matching chromatographic profiles of individual samples to an external standard of SA. Individual peak areas of SA were quantified against an external standard of SA. Statistical analysis for the combined data from three biological replicates was done using one-way ANOVA and Fisher LSD test (P < 0.05) by Minitab 16 statistical software.
Glucosinolate Extraction and Measurement
Leaves of 5-week-old plants were infiltrated with buffer, 10 μM flg22, or 108 CFU/mL of bacteria in 10 mM MgCl2 with or without 3 μM COR. After 9 h, 100 mg of leaves was frozen in liquid nitrogen and ground by mortar and pestle. The ground tissue was combined with 600 μL 80% HPLC-grade methanol, vortexed well, and spiked with 30 μL of 1.25 mM sinigrin (Sigma-Aldrich) as an internal standard. After incubating at 75°C for 15 min to deactivate myrosinases, the extraction mixture was centrifuged at 15K rpm for 10 min at 4°C and 400 μL of supernatant was collected.
4MI3G, 1MI3G, and I3G were detected as previously described (Cataldi et al., 2007). Then, 30-μL samples were run on liquid chromatograph–mass spectrometer (Varian) in a negative ion electrospray mode. The samples were separated by HPLC fitted with a C18-A column (A2001150X046). The mobile phases were A - water and B - 90% acetonitrile (Fisher Scientific) at room temperature. Column linear gradient was 0 to 18 min, 85% A, 15% B; 18 to 25 min, 45% A, 55% B; 25 to 28 min, 100% B; 28 to 30 min, 85% A, 15% B, with a flow rate of 300 μL/min. The internal standard sinigrin, 4MI3G, 1MI3G and I3G were observed as mass-to-charge ratios of 358, 477, 477, and 447, respectively. For all four compounds, the retention times as well as masses of daughter ions match published values (Cataldi et al., 2007). Statistical analysis for the combined data from three biological replicates was done using one-way ANOVA and Tukey HSD test (P < 0.05) by Minitab 16 statistical software.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: SID2 (AT1G74710), NPR1 (AT1G64280), COI1 (AT2G39940), PEN2 (AT2G44490), ACTIN2 (AT3G18780), MYB51 (AT1G18570), CYP79B2 (AT4G39950), CYP79B3 (AT2G22330), CYP83B1 (AT4G31500), CYP81F2 (AT5G57220), and PR-1 (AT2G14610).
Supplemental Data
The following materials are available in the online version of this article:
Supplemental Figure 1. DNA Analysis of Mutant Bacteria and Plants.
Supplemental Figure 2. COR Inhibits PR-1 Expression.
Supplemental Figure 3. SA Accumulation after Bacterial Infiltration.
Supplemental Figure 4. COR Promotes Bacterial Virulence in sid2 npr1 Plants Independent of Targeting COI1.
Supplemental Figure 5. PEN2 Is Not Required for flg22-Induced Callose Deposition in Soil-Grown Plants.
Supplemental Figure 6. COR Inhibits Indole Glucosinolate Accumulation Upstream of PEN2.
Supplemental Table 1. Phenotype of Arabidopsis Types Used in This Study.
Supplemental Table 2. Primers Used for Screening Mutant Plants and Bacteria.
Supplemental Table 3. Primers Used for qRT-PCR.
Acknowledgments
We thank Barbara Kunkel for Ptocor- (DB4G3) and plasmid pDB29, John Turner for coi1-16 seed, Kristin Mercer for advice on statistics, and the National Science Foundation (MCB-0718882), the USDA (NIFA 2008-35319-04506), the Ohio Agricultural Research and Development Center of The Ohio State University, and the Korean RDA Next-Generation BioGreen Program (SSAC and PJ009088) for support.
AUTHOR CONTRIBUTIONS
X.G. and D.M. designed the research. X.G. and A.G. performed the research. X.G., A.G., J.C., and D.M. analyzed data. X.G. and D.M. wrote the article.
Glossary
- COR
Coronatine
- Pto
P. syringae pv tomato strain DC3000
- JA
jasmonic acid
- SA
salicylic acid
- PAMP
pathogen-associated molecular pattern
- 4MI3G
4-methoxy-indol-3ylmethylglucosinolate
- CEL
conserved effector locus
- Col-0
Columbia-0
- ANOVA
analysis of variance
- I3G
indol-3ylmethylglucosinolate
- KB
to be defined
- CFU
colony-forming units
- HSD
to be defined
- qRT-PCR
quantitative real-time PCR
- 1MI3G
to be defined
References
- Alfano J.R., Charkowski A.O., Deng W.L., Badel J.L., Petnicki-Ocwieja T., van Dijk K., Collmer A. (2000). The Pseudomonas syringae Hrp pathogenicity island has a tripartite mosaic structure composed of a cluster of type III secretion genes bounded by exchangeable effector and conserved effector loci that contribute to parasitic fitness and pathogenicity in plants. Proc. Natl. Acad. Sci. USA 97: 4856–4861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assaad F.F., Qiu J.L., Youngs H., Ehrhardt D., Zimmerli L., Kalde M., Wanner G., Peck S.C., Edwards H., Ramonell K., Somerville C.R., Thordal-Christensen H. (2004). The PEN1 syntaxin defines a novel cellular compartment upon fungal attack and is required for the timely assembly of papillae. Mol. Biol. Cell 15: 5118–5129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badel J.L., Shimizu R., Oh H.S., Collmer A. (2006). A Pseudomonas syringae pv. tomato avrE1/hopM1 mutant is severely reduced in growth and lesion formation in tomato. Mol. Plant Microbe Interact. 19: 99–111 [DOI] [PubMed] [Google Scholar]
- Bak S., Tax F.E., Feldmann K.A., Galbraith D.W., Feyereisen R. (2001). CYP83B1, a cytochrome P450 at the metabolic branch point in auxin and indole glucosinolate biosynthesis in Arabidopsis. Plant Cell 13: 101–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballaré C.L. (2011). Jasmonate-induced defenses: A tale of intelligence, collaborators and rascals. Trends Plant Sci. 16: 249–257 [DOI] [PubMed] [Google Scholar]
- Bednarek P., Pislewska-Bednarek M., Svatos A., Schneider B., Doubsky J., Mansurova M., Humphry M., Consonni C., Panstruga R., Sanchez-Vallet A., Molina A., Schulze-Lefert P. (2009). A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 323: 101–106 [DOI] [PubMed] [Google Scholar]
- Bender C.L., Alarcón-Chaidez F., Gross D.C. (1999). Pseudomonas syringae phytotoxins: Mode of action, regulation, and biosynthesis by peptide and polyketide synthetases. Microbiol. Mol. Biol. Rev. 63: 266–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender C.L., Stone H.E., Cooksley D.A. (1987). Reduced pathogen fitness of Pseudomonas syringae pv. tomato Tn5 mutants defective in coronatine production. Physiol. Mol. Plant Pathol. 30: 273–283 [Google Scholar]
- Brooks D.M., Bender C.L., Kunkel B.N. (2005). The Pseudomonas syringae phytotoxin coronatine promotes virulence by overcoming salicylic acid-dependent defences in Arabidopsis thaliana. Mol. Plant Pathol. 6: 629–639 [DOI] [PubMed] [Google Scholar]
- Brooks D.M., Hernández-Guzmán G., Kloek A.P., Alarcón-Chaidez F., Sreedharan A., Rangaswamy V., Peñaloza-Vázquez A., Bender C.L., Kunkel B.N. (2004). Identification and characterization of a well-defined series of coronatine biosynthetic mutants of Pseudomonas syringae pv. tomato DC3000. Mol. Plant Microbe Interact. 17: 162–174 [DOI] [PubMed] [Google Scholar]
- Büttner D., Bonas U. (2002). Getting across—Bacterial type III effector proteins on their way to the plant cell. EMBO J. 21: 5313–5322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H., Glazebrook J., Clarke J.D., Volko S., Dong X.N. (1997). The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88: 57–63 [DOI] [PubMed] [Google Scholar]
- Cataldi T.R., Rubino A., Lelario F., Bufo S.A. (2007). Naturally occurring glucosinolates in plant extracts of rocket salad (Eruca sativa L.) identified by liquid chromatography coupled with negative ion electrospray ionization and quadrupole ion-trap mass spectrometry. Rapid Commun. Mass Spectrom. 21: 2374–2388 [DOI] [PubMed] [Google Scholar]
- Chini A., Fonseca S., Fernández G., Adie B., Chico J.M., Lorenzo O., García-Casado G., López-Vidriero I., Lozano F.M., Ponce M.R., Micol J.L., Solano R. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671 [DOI] [PubMed] [Google Scholar]
- Chisholm S.T., Coaker G., Day B., Staskawicz B.J. (2006). Host-microbe interactions: Shaping the evolution of the plant immune response. Cell 124: 803–814 [DOI] [PubMed] [Google Scholar]
- Clay N.K., Adio A.M., Denoux C., Jander G., Ausubel F.M. (2009). Glucosinolate metabolites required for an Arabidopsis innate immune response. Science 323: 95–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Bahrami A.K., Pringle E.G., Hernandez-Guzman G., Bender C.L., Pierce N.E., Ausubel F.M. (2005). Pseudomonas syringae manipulates systemic plant defenses against pathogens and herbivores. Proc. Natl. Acad. Sci. USA 102: 1791–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DebRoy S., Thilmony R., Kwack Y.B., Nomura K., He S.Y. (2004). A family of conserved bacterial effectors inhibits salicylic acid-mediated basal immunity and promotes disease necrosis in plants. Proc. Natl. Acad. Sci. USA 101: 9927–9932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devoto A., Ellis C., Magusin A., Chang H.S., Chilcott C., Zhu T., Turner J.G. (2005). Expression profiling reveals COI1 to be a key regulator of genes involved in wound- and methyl jasmonate-induced secondary metabolism, defence, and hormone interactions. Plant Mol. Biol. 58: 497–513 [DOI] [PubMed] [Google Scholar]
- Ellis C., Turner J.G. (2002). A conditionally fertile coi1 allele indicates cross-talk between plant hormone signalling pathways in Arabidopsis thaliana seeds and young seedlings. Planta 215: 549–556 [DOI] [PubMed] [Google Scholar]
- Fan W., Dong X. (2002). In vivo interaction between NPR1 and transcription factor TGA2 leads to salicylic acid-mediated gene activation in Arabidopsis. Plant Cell 14: 1377–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson I.B., Mitchell R.E. (1985). Stimulation of ethylene production in bean leaf discs by the pseudomonad phytotoxin coronatine. Plant Physiol. 77: 969–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys B., Benedetti C.E., Penfold C.N., Turner J.G. (1994). Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6: 751–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca S., Chico J.M., Solano R. (2009). The jasmonate pathway: The ligand, the receptor and the core signalling module. Curr. Opin. Plant Biol. 12: 539–547 [DOI] [PubMed] [Google Scholar]
- Freeman B.C., Beattie G.A. (2009). Bacterial growth restriction during host resistance to Pseudomonas syringae is associated with leaf water loss and localized cessation of vascular activity in Arabidopsis thaliana. Mol. Plant Microbe Interact. 22: 857–867 [DOI] [PubMed] [Google Scholar]
- Gigolashvili T., Yatusevich R., Berger B., Müller C., Flügge U.I. (2007). The R2R3-MYB transcription factor HAG1/MYB28 is a regulator of methionine-derived glucosinolate biosynthesis in Arabidopsis thaliana. Plant J. 51: 247–261 [DOI] [PubMed] [Google Scholar]
- Halkier B.A., Gershenzon J. (2006). Biology and biochemistry of glucosinolates. Annu. Rev. Plant Biol. 57: 303–333 [DOI] [PubMed] [Google Scholar]
- Hardham A.R., Jones D.A., Takemoto D. (2007). Cytoskeleton and cell wall function in penetration resistance. Curr. Opin. Plant Biol. 10: 342–348 [DOI] [PubMed] [Google Scholar]
- He S.Y., Nomura K., Whittam T.S. (2004). Type III protein secretion mechanism in mammalian and plant pathogens. Biochim. Biophys. Acta 1694: 181–206 [DOI] [PubMed] [Google Scholar]
- Hull A.K., Vij R., Celenza J.L. (2000). Arabidopsis cytochrome P450s that catalyze the first step of tryptophan-dependent indole-3-acetic acid biosynthesis. Proc. Natl. Acad. Sci. USA 97: 2379–2384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiga Y., Uppalapati S.R., Ishiga T., Elavarthi S., Martin B., Bender C.L. (2009). The phytotoxin coronatine induces light-dependent reactive oxygen species in tomato seedlings. New Phytol. 181: 147–160 [DOI] [PubMed] [Google Scholar]
- Katsir L., Schilmiller A.L., Staswick P.E., He S.Y., Howe G.A. (2008). COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc. Natl. Acad. Sci. USA 105: 7100–7105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon J.S., Turner J.G. (1992). The stimulation of ethylene synthesis in Nicotiana-Tabacum leaves by the phytotoxin coronatine. Plant Physiol. 100: 219–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.H., Jander G. (2007). Myzus persicae (green peach aphid) feeding on Arabidopsis induces the formation of a deterrent indole glucosinolate. Plant J. 49: 1008–1019 [DOI] [PubMed] [Google Scholar]
- Kim M.G., da Cunha L., McFall A.J., Belkhadir Y., DebRoy S., Dangl J.L., Mackey D. (2005). Two Pseudomonas syringae type III effectors inhibit RIN4-regulated basal defense in Arabidopsis. Cell 121: 749–759 [DOI] [PubMed] [Google Scholar]
- Kim M.G., Mackey D. (2008). Measuring cell-wall-based defenses and their effect on bacterial growth in Arabidopsis. Methods Mol. Biol. 415: 443–452 [DOI] [PubMed] [Google Scholar]
- Kliebenstein D.J., Dietrich R.A., Martin A.C., Last R.L., Dangl J.L. (1999). LSD1 regulates salicylic acid induction of copper zinc superoxide dismutase in Arabidopsis thaliana. Mol. Plant Microbe Interact. 12: 1022–1026 [DOI] [PubMed] [Google Scholar]
- Kloek A.P., Verbsky M.L., Sharma S.B., Schoelz J.E., Vogel J., Klessig D.F., Kunkel B.N. (2001). Resistance to Pseudomonas syringae conferred by an Arabidopsis thaliana coronatine-insensitive (coi1) mutation occurs through two distinct mechanisms. Plant J. 26: 509–522 [DOI] [PubMed] [Google Scholar]
- Lipka V., et al. (2005). Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science 310: 1180–1183 [DOI] [PubMed] [Google Scholar]
- Melotto M., Underwood W., Koczan J., Nomura K., He S.Y. (2006). Plant stomata function in innate immunity against bacterial invasion. Cell 126: 969–980 [DOI] [PubMed] [Google Scholar]
- Millet Y.A., Danna C.H., Clay N.K., Songnuan W., Simon M.D., Werck-Reichhart D., Ausubel F.M. (2010). Innate immune responses activated in Arabidopsis roots by microbe-associated molecular patterns. Plant Cell 22: 973–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfalz M., Mikkelsen M.D., Bednarek P., Olsen C.E., Halkier B.A., Kroymann J. (2011). Metabolic engineering in Nicotiana benthamiana reveals key enzyme functions in Arabidopsis indole glucosinolate modification. Plant Cell 23: 716–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotrowski M., Schemenewitz A., Lopukhina A., Müller A., Janowitz T., Weiler E.W., Oecking C. (2004). Desulfoglucosinolate sulfotransferases from Arabidopsis thaliana catalyze the final step in the biosynthesis of the glucosinolate core structure. J. Biol. Chem. 279: 50717–50725 [DOI] [PubMed] [Google Scholar]
- Sheard L.B., et al. (2010). Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468: 400–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel S.H., Dong X. (2008). Making sense of hormone crosstalk during plant immune responses. Cell Host Microbe 3: 348–351 [DOI] [PubMed] [Google Scholar]
- Thilmony R., Underwood W., He S.Y. (2006). Genome-wide transcriptional analysis of the Arabidopsis thaliana interaction with the plant pathogen Pseudomonas syringae pv. tomato DC3000 and the human pathogen Escherichia coli O157:H7. Plant J. 46: 34–53 [DOI] [PubMed] [Google Scholar]
- Thines B., Katsir L., Melotto M., Niu Y., Mandaokar A., Liu G., Nomura K., He S.Y., Howe G.A., Browse J. (2007). JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448: 661–665 [DOI] [PubMed] [Google Scholar]
- Tsuda K., Sato M., Glazebrook J., Cohen J.D., Katagiri F. (2008). Interplay between MAMP-triggered and SA-mediated defense responses. Plant J. 53: 763–775 [DOI] [PubMed] [Google Scholar]
- Uppalapati S.R., Ayoubi P., Weng H., Palmer D.A., Mitchell R.E., Jones W., Bender C.L. (2005). The phytotoxin coronatine and methyl jasmonate impact multiple phytohormone pathways in tomato. Plant J. 42: 201–217 [DOI] [PubMed] [Google Scholar]
- Uppalapati S.R., Ishiga Y., Wangdi T., Kunkel B.N., Anand A., Mysore K.S., Bender C.L. (2007). The phytotoxin coronatine contributes to pathogen fitness and is required for suppression of salicylic acid accumulation in tomato inoculated with Pseudomonas syringae pv. tomato DC3000. Mol. Plant Microbe Interact. 20: 955–965 [DOI] [PubMed] [Google Scholar]
- Uppalapati S.R., Ishiga Y., Wangdi T., Urbanczyk-Wochniak E., Ishiga T., Mysore K.S., Bender C.L. (2008). Pathogenicity of Pseudomonas syringae pv. tomato on tomato seedlings: Phenotypic and gene expression analyses of the virulence function of coronatine. Mol. Plant Microbe Interact. 21: 383–395 [DOI] [PubMed] [Google Scholar]
- Wang D., Weaver N.D., Kesarwani M., Dong X. (2005). Induction of protein secretory pathway is required for systemic acquired resistance. Science 308: 1036–1040 [DOI] [PubMed] [Google Scholar]
- Westphal L., Scheel D., Rosahl S. (2008). The coi1-16 mutant harbors a second site mutation rendering PEN2 nonfunctional. Plant Cell 20: 824–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildermuth M.C., Dewdney J., Wu G., Ausubel F.M. (2001). Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414: 562–565 [DOI] [PubMed] [Google Scholar]
- Xie D.-X., Feys B.F., James S., Nieto-Rostro M., Turner J.G. (1998). COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280: 1091–1094 [DOI] [PubMed] [Google Scholar]
- Zeng W., He S.Y. (2010). A prominent role of the flagellin receptor FLAGELLIN-SENSING2 in mediating stomatal response to Pseudomonas syringae pv tomato DC3000 in Arabidopsis. Plant Physiol. 153: 1188–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y.D., Hull A.K., Gupta N.R., Goss K.A., Alonso J., Ecker J.R., Normanly J., Chory J., Celenza J.L. (2002). Trp-dependent auxin biosynthesis in Arabidopsis: Involvement of cytochrome P450s CYP79B2 and CYP79B3. Genes Dev. 16: 3100–3112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X.Y., Spivey N.W., Zeng W., Liu P.P., Fu Z.Q., Klessig D.F., He S.Y., Dong X. (2012). Coronatine promotes Pseudomonas syringae virulence in plants by activating a signaling cascade that inhibits salicylic acid accumulation. Cell Host Microbe 11: 587–596 [DOI] [PMC free article] [PubMed] [Google Scholar]