Abstract
Protease-Activated Receptor-1 (PAR1) is the prototypical member of a family of G protein-coupled receptors that mediate cellular responses to thrombin and related proteases. Thrombin irreversibly activates PAR1 by cleaving the N-terminal exodomain of the receptor, which exposes a tethered peptide ligand that binds the receptor’s heptahelical bundle to effect G protein-activation. Here we report a 2.2Å resolution crystal structure of human PAR1 bound to vorapaxar, a PAR1 antagonist. The structure reveals an unusual mode of drug binding that explains how a small molecule binds virtually irreversibly to inhibit receptor activation by PAR1’s tethered ligand. In contrast to deep, solvent-exposed binding pockets observed in other peptide-activated GPCRs, the vorapaxar-binding pocket is superficial but has little surface exposed to the aqueous solvent. PARs are important targets for drug development. The structure reported here will aid development of improved PAR1 antagonists and discovery of antagonists to other members of this receptor family.
Introduction
Protease-activated receptors (PARs) are G protein-coupled receptors (GPCRs) that mediate cellular responses to specific proteases1,2. The coagulation protease thrombin activates the prototypical PAR, PAR1, by specific cleavage of the receptor’s N-terminal exodomain to generate a new N-terminus. This new N-terminus then functions as a tethered peptide agonist that binds intramolecularly to the seven-transmembrane helix bundle of the receptor to effect G protein activation (Fig. 1a)1,3–8. In adult mammals, the four members of the PAR family link tissue injury and local generation of active coagulation proteases to cellular responses that help orchestrate hemostasis and thrombosis, inflammation, and perhaps tissue repair2,9. PARs may also participate in the progression of specific cancers10,11.
Figure 1. PAR1 activation and overall structure of human PAR1 complex with antagonist vorapaxar.
a, Thrombin cleaves PAR1 N-terminus and exposes a new N-terminal peptide SFLLRN, which can bind to and activate the transmembrane core of PAR1. PAR1 can activate several G proteins including Gi, G12/13 and Gq. b, Overall view of the human PAR1 structure and the extracellular surface. The receptor is shown in blue ribbon and vorapaxar is shown as green spheres. Monoolein is shown in orange, water in red. The disulfide bond is shown as a yellow stick. c, Surface view of the ligand-binding pocket viewed from two different perspectives. The vorapaxar binding pocket is close to the extracellular surface but not well exposed to the extracellular solvent.
In contrast to a typical receptor-agonist binding interaction, the interaction of PAR1 with its activator, thrombin, is that of a protease substrate, with thrombin binding transiently to the receptor, cleaving it, then dissociating1,3–7,12. Proteolytic unmasking of the receptor’s tethered peptide agonist is irreversible, and although a free synthetic hexapeptide with the amino acid sequence of the tethered agonist (SFLLRN) can activate the receptor with EC50 in the 3–10 µM range, the local concentration of the tethered agonist peptide is estimated to be about 0.4 mM. Accordingly, PAR signaling must be actively terminated13–15 and, unlike most other GPCRs that can go though many rounds of activation by reversible diffusible hormones and neurotransmitters, PARs are degraded after a single activation6,13–17. Identification of effective PAR antagonists has been challenging because low molecular weight compounds must compete with the very high local concentration of the tethered agonist generated by proteolytic cleavage.
Vorapaxar is a highly specific, virtually irreversible PAR1 antagonist18 (Supplementary Figure 1). In a Phase 3 trial, vorapaxar protected patients against recurrent myocardial infarction at a cost of increased bleeding19,20. Given the latter, an antagonist that is reversible in the setting of bleeding might be desirable. While the very slow dissociation rate of vorapaxar from PAR1 likely accounts for its ability to inhibit receptor activation by its tethered agonist peptide, it may be possible to develop a drug with an off rate slow enough to block signaling but fast enough to allow useful reversal after cessation of drug.
In an effort to advance our understanding of PAR1 structure and function and to provide a foundation for discovery of new agents to advance the pharmacology of PARs, we obtained a crystal structure of vorapaxar-bound human PAR1.
Crystallization of the human PAR1 receptor
To facilitate crystallogenesis, T4 lysozyme was inserted in intracellular loop 3 (ICL3) in human PAR1, the N-linked glycosylation sites in ECL2 were mutated21, and the N-terminal exodomain was removed by site-specific cleavage at a Tobacco Etch Virus protease site introduced between amino acids 85 and 864 (Supplementary Figure 2). The structure of human PAR1-T4L bound to vorapaxar was determined to 2.2 Å by merging diffraction data sets from 18 crystals grown in lipidic cubic phase (Supplementary Figures 3 and 4). Details of data collection and structure refinement are listed in Supplementary Table 1.
PAR1 has the expected seven-transmembrane segment (TM) bundle (Fig. 1b). There are several lipid molecules assigned as monoolein from lipidic cubic phase in the structure (Fig. 1b), but no ordered cholesterol molecules were observed. The remaining N-terminal fragment A86-E90 and a part of the intracellular loop 2 from Q209 to W213 are not modeled in the structure because of the weak electron density. There is no clear electron density for residues after C378, and no helix 8 is observed after transmembrane segment TM7 in the structure. Whether this reflects a lack of a helix 8 in PAR1 in its native state or conditions in the crystal is not known.
C1753.25 in helix III and C254 in extracellular loop 2 (ECL2) form a conserved disulfide (Figs. 1b and 2a). Amino terminal to C254, ECL2 loops outward in two anti-parallel beta strands. This structural feature is found in other peptide receptors including the CXCR4 receptor and the opioid receptors22–25 despite absence of amino acid sequence homology among these receptors in ECL2 (Supplementary Figure 5). In contrast to the open, solvent-exposed binding pocket observed in the µ-opioid receptor (MOR) and other peptide receptors, access to the vorapaxar-binding pocket is restricted by the central location of ECL2 (Fig. 1, Supplementary Figures 5 and 6), which almost completely covers the extracellular-facing surface of vorapaxar. ECL2 is anchored in this position by hydrogen bonds between H255 in ECL2 and Y3537.35 in TM7, and between D256 in ECL2 and Y95 in the amino terminus (Supplementary Figure 6), and by extensive interactions with vorapaxar (Fig. 2). The covered vorapaxar binding pocket in PAR1 more closely resembles rhodopsin and the lipid-activated sphingosine-1-phosphate receptor than other peptide-activated GPCRs (Supplementary Figure 5).
Figure 2. Binding interactions of vorapaxar with human PAR1.
a, b, Ligand binding pocket viewed from extracellular surface (a) and from side of transmembrane helix bundle (b). ECL2 is colored in orange in panels a and b. Ligand vorapaxar is shown as green sticks. Water molecules are shown as red spheres. Hydrogen bonds are shown as black dotted lines. c, Two residues L262 and L263 in ICL2 (shown as dot surface), which pack against residues H255, F2715.39 and Y3376.59 (shown in CPK representation), may contribute to the selectivity of vorapaxar for human PAR1. Also shown are F2745.42 and F2785.46 in TM5 (shown as dot surface), which may indirectly influence vorapaxar binding by packing interactions with F2715.39.
Divergence of PAR1 from other Family A GPCRs
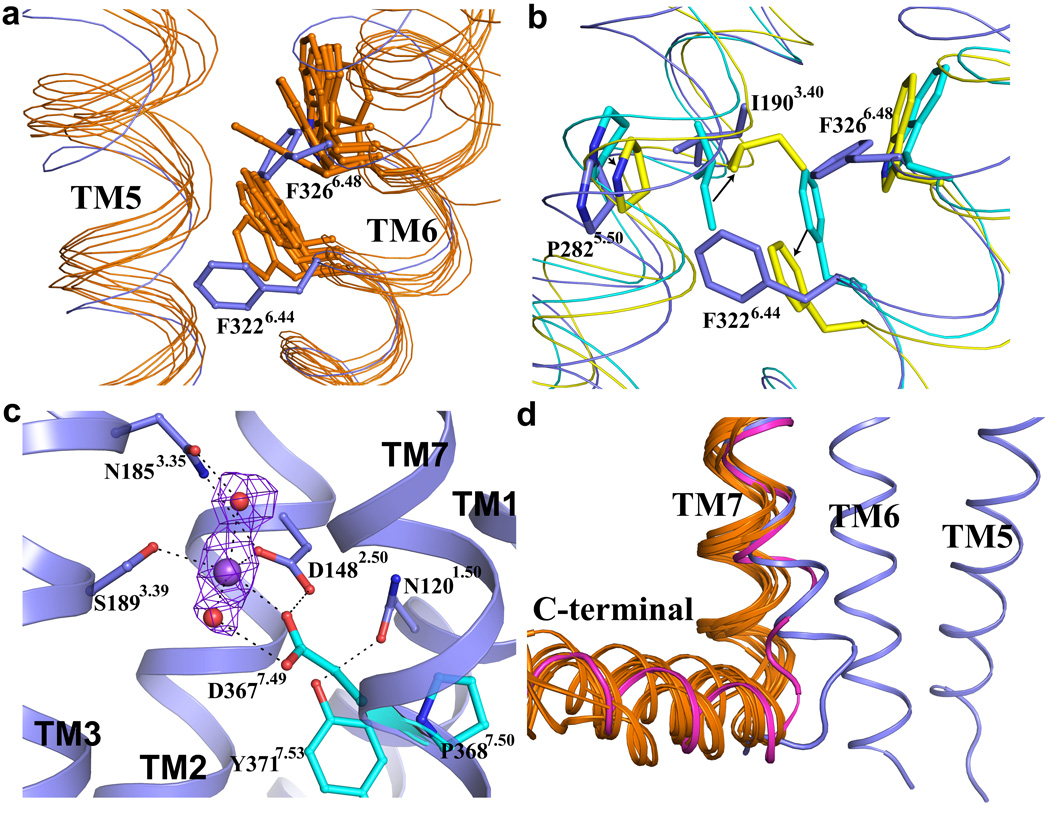
Members of the Family A GPCRs share a set of conserved amino acids that are thought to be important in signal transduction26,27. Specific residues in the highly conserved FxxCWxP motif in TM6, and the NPxxY motif in TM7 undergo structural rearrangements during activation of the β2-adrenergic receptor (β2AR) (Fig. 3b, d). However, based on a phylogenetic analysis of amino acid sequences28, PAR1 is a more distant relative of the family A GPCRs that have been crystallized thus far. PAR1 belongs to the δ subfamily, which includes the glycoprotein receptors, the purinergic receptors and the olfactory receptors28. The tryptophan residue in FxxCW6.48xP proposed to act as a toggle switch during activation in some GPCRs28 is replaced by F6.48 in all PARs (Fig. 3a). F6.44, also highly conserved in Family A GPCRs, is F6.44 in PAR1, but Y6.44 in PAR2 and A6.44 in PAR4. When comparing inactive and active states of the β2-adrenergic receptor, changes in packing interactions involving P5.50, I3.40 and F6.44 appear to play a role in structural changes needed to accommodate G protein binding29–31. Packing interactions of the corresponding residues P2825.50, I1903.40 and F3226.44 in the PAR1 differ from those in both active and inactive β2AR structures (Fig. 3b). Taken together, these differences suggest that the PAR1 may differ from other Family A GPCRs in the mechanism by which signals propagate from the extracellular peptide-binding interface to the cytoplasmic domains that interact with G proteins and other signaling molecules.
Figure 3. Structure motifs in PAR1 compared with other Family A GPCRs.
a, Superimposition of TM5 and TM6 of human PAR1 (in blue) with those of other GPCRs including β2- and β1-adrenergic receptors (AR), A2A adenosine receptor, dopamine D3 receptor, M2 muscarinic receptor, histamine H1 receptor, µ-opioid receptor, S1P1 receptor and CXCR4 (all in orange). F3266.48 and F3226.44 in the F6.44xxCF6.48xP motif in PAR1 are shown as sticks. This motif is FxxCW6.48xP in most other Family A GPCRs. F3266.48 and F3226.44 are both in different conformations compared to their counterparts in other GPCRs. b, In the β2AR, rearrangements of three residues, P5.50, I3.40 and F6.44, are associated with receptor activation. Black arrows indicate changes of these residues in going from inactive (cyan) to active (yellow) β2AR structures. The counterparts in the inactive state structure of PAR1 (P2825.50, I1903.40 and F3226.44) are shown in blue. c, DP7.50xxY motif in TM7 and sodium binding site in PAR1. Residues D3677.49, P3687.50 and Y3717.53 in DP7.50xxY motif are shown as cyan sticks. This motif is normally NPxxY in most other Family A GPCRs. Sodium is shown as a purple sphere and water molecules are shown as red spheres. Polar interactions are shown as black dash lines. An Fo-Fc omit electron density map for the putative sodium ion and water molecules contoured at 4σ is shown as purple mesh. d, Superimposition of the C-terminal part of TM7 in the structure of human PAR1 (blue), in the inactive structures of other GPCRs (all in orange) mentioned in panel a and in the active structure of β2AR (in magenta). The C-terminal part of TM7 in PAR1 adopts a conformation more similar to that observed in the active state of the β2AR.
The NP7.50xxY motif at the end of helix VII observed in most Family A GPCRs is DP7.50xxY in PAR1. This region undergoes structural rearrangement upon activation of the β2AR. In PAR1, D3677.49 and Y3717.53 form hydrogen bonds with residues in TM2 and TM1 (Fig. 3c). The hydrogen-bonding network associated with D3677.49 is extensive and includes several water molecules and a putative sodium ion. Na+, rather than a water molecule, was assigned to this region of electron density as it has five oxygen neighbors and short distances to its oxygen ligands (average refined distance 2.4 Å), both consistent with known Na+-oxygen interactions32, and it interacts with two acidic side chains that, assuming deprotonated states, would repel one another without charge neutralization provided by the Na+. The sodium ion also interacts with a conserved D1482.50 in TM2 and S1893.39 in TM3, with two water molecules nearby (Fig 3c). Na+ is an allosteric modulator for several family A GPCRs such as the α2A adrenergic receptor, A2A adenosine receptor, µ- and δ-opioid receptors and D2 dopamine receptor33–37. The conserved D2.50 is necessary for sodium sensitivity of α2A adrenergic receptor37 and D2 dopamine receptor36,38. In PAR1, D3677.49 might be expected to form a stronger hydrogen-bonding network and sodium coordination site than asparagine residues found in most other family A GPCRs. This more stable network may contribute to the unusual position of the cytoplasmic end of TM7 that is displaced inward towards TM2. This position is more similar to the active β2AR bound with either nanobody 80 or heterotrimeric G protein29,31(Fig. 3d).
Structural insights into vorapaxar binding properties
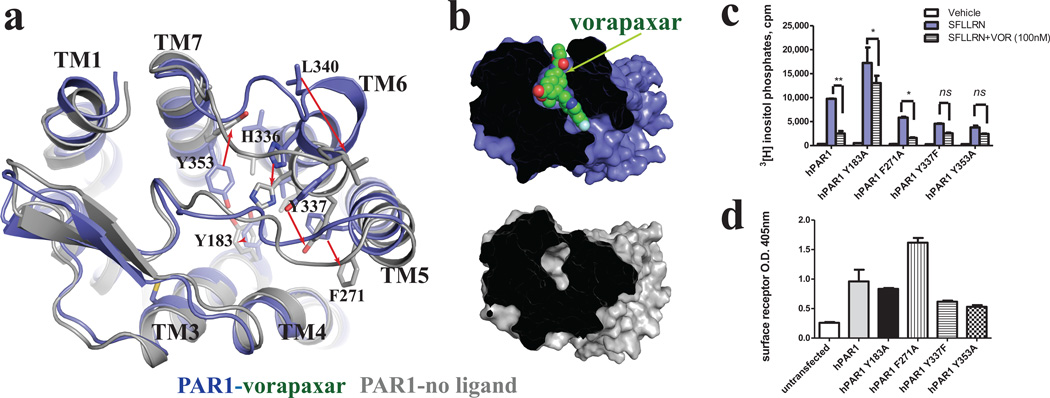
Vorapaxar binds in an unusual location very close to the extracellular surface of PAR1. In contrast, ligands for other GPCRs penetrate more deeply into the transmembrane core (Fig. 1, Supplementary Figures 5 and 7). The vorapaxar-binding pocket, composed of residues from TMs 3, 4, 5, 6 and 7 as well as ECL2 and ECL3, forms a tunnel across the receptor with one end open between TM4 and TM5 and the other between TM6 and TM7 occupied by the ethyl carbamate tail of vorapaxar (Figs. 1 and 2). There is only a small opening in the extracellular surface between ECL2 and ECL3. Details of interactions between vorapaxar and PAR1 are illustrated in Fig. 2 and Supplementary Figure 8.
Vorapaxar shows high selectivity for human PAR1 over human PAR2 and PAR4, and mouse PAR1 in functional assays (Supplementary Figure 9A and B). The structural basis for this selectivity is not readily apparent from the crystal structure. Nearly all the residues that interact with vorapaxar in human PAR1 are conserved in human PAR2, human PAR4 and mouse PAR1 (Supplementary Figure 10). Residues L262 and L263, which are involved in weak hydrophobic interactions with vorapaxar in human PAR1, are alanine and asparagine, respectively, in human PAR4, and L263 is a methionine in mouse PAR1 (Fig. 2c, Supplementary Figure 10). These differences by themselves would not be expected to explain the high selectivity of vorapaxar. However, L262 and L263 pack against other amino acids that have more extensive interactions with vorapaxar. L262 interacts with H255 in ECL2 and L263 interacts with F2715.39 at the top of TM5 and Y3376.59 at the top of TM6 (Fig. 2c). These interactions may influence ligand-binding selectivity indirectly by contributing to the overall structure and stability of the binding pocket. Amino acid differences between PAR1 and PAR2 and PAR4 more distant from the ligand-binding pocket may also contribute to subtype-specific binding of vorapaxar. F2745.42 is Leu in PAR2 and 4, and F2785.46 is Val in PAR2 and Gly in PAR4. While neither F2745.42 nor F2785.46 directly contact vorapaxar, F2785.46 packs against F2745.42, which in turn packs against F2715.39 in the binding pocket (Fig. 2c).
In human PAR2 and human PAR4, ECL3 connecting TM6 and TM7 is one residue shorter than it is in human PAR1 (Supplementary Figure 9C). Y3376.59 at the C-terminal end of TM6 forms a strong hydrogen bond with vorapaxar (Fig. 2 and Supplementary Figure 9D). Another residue Y3537.35, which forms the base of the ligand-binding pocket together with Y1833.33, is located at the N-terminal end of TM7 (Fig. 2 and Supplementary Figure 9D). A shorter ECL3 in human PAR2 and PAR4 may change the relative position of these amino acids thereby altering the overall geometry of the binding pocket. While the length of ECL3 in mouse and human PAR1 is the same, 4 of the 8 amino acids are different (Supplementary Figure 9C). These differences may effect the structure of ECL3 and thereby influence interactions between vorapaxar in Y3376.59 and Y3537.35. Alternatively, these differences could have an effect on the mechanism by which vorapaxar gains access to the binding pocket.
Structural insights into vorapaxar inhibition of PAR1 activation
While this structure is compatible with the very slow dissociation rate of vorapaxar, it does not provide insight into the mechanism by which vorapaxar or an agonist peptide gains access to the binding pocket. None of the three openings to the vorapaxar-binding pocket are large enough to accommodate the passage of the ligand. We thus wondered whether the unliganded receptor might have a more open structure, similar to that observed for opioid receptors, with unique interactions between vorapaxar and PAR1 causing an otherwise open binding pocket to close upon vorapaxar binding. To investigate this issue, we performed long-timescale molecular dynamics (MD) simulations of PAR1 with and without vorapaxar bound. Intriguingly, removal of the ligand led not to a more open binding pocket, but to one that was even more closed (Figs. 4a and b, andSupplementary Figure 11, Supplementary Table 2). The extracellular end of TM6 moved about 4Å inward toward TM4, bringing ECL3 in full contact with ECL2 and completely occluding the binding pocket. By contrast, in a similar study on the MOR, the binding pocket remained open when the ligand was removed (Supplementary Fig 11). The collapse of the vorapaxar binding pocket may reflect the fact that both vorapaxar and its binding pocket are uncharged, while in the opioid receptors and many other family A GPCRs, the charged residue D3.32 helps keep the binding pocket hydrated after the ligand is removed.
Figure 4. Collapse of ligand-binding pocket in long-timescale molecular dynamics (MD) simulations of unliganded PAR1.
MD simulations were performed on PAR1 from which vorapaxar had been removed. The vorapaxar-bound PAR1 crystal structure is shown in blue and the unliganded structure obtained from MD simulation is shown in gray. a, The largest differences between vorapaxar-bound and unliganded PAR1 are at the extracellular end of TM6 and in ECL3. Residues involved in vorapaxar binding are shown as sticks. b, Surface view showing collapse of the ligand-binding pocket during MD simulation in the absence of vorapaxar. c Signaling and d cell surface expression for wild-type human PAR1 and PAR1 binding site-mutants. Cos7 cells expressing the indicated receptor constructs were labeled with [3H] myoinositol, pretreated with vehicle or 100nM vorapaxar in DMEM medium containing 0.1% BSA, 20mM HEPES, 0.2% β-hydroxy cyclodextrin (to retain vorapaxar in solution) for 1h, then incubated with vehicle or PAR1 agonist (100µM SFLLRN) for 1h at 37°C. Total [3H] inositol-phosphate accumulation was measured. Surface expression of receptors in cells transfected in parallel was assessed by measuring binding of anti-FLAG antibody to an epitope displayed at the receptor's N terminus. Results are representative of three separate experiments.
It is interesting to speculate that vorapaxar, a highly lipophilic molecule, may access the binding pocket through the lipid bilayer, possibly between TMs 6 and 7. This is similar to the binding mode proposed for retinal to rhodopsin and the lipid S1P to the S1P1 receptor39,40 (Supplementary Fig 5).
In an effort to understand the ability of vorapaxar to inhibit agonist binding and activation, we examined the functional consequences of mutating four aromatic amino acids that form strong interactions with vorapaxar: Y1833.33, Y3537.35, F2715.39 and Y3376.59 (Figs. 2a and b and Supplementary Fig 3C). Three of these (Y3537.35, F2715.39 and Y3376.59) assume substantially different positions in simulations of unliganded PAR1 (Fig. 4a). Tyrosine residues Y1833.33 in TM3 and Y3537.35 in TM7 are linked to each other by a hydrogen bond and form the base of the binding “tunnel”, and they are part of a hydrophobic cage that surrounds the ligand. Y3537.35 also forms a hydrogen bond with H255, the most deeply buried amino acid in ECL2 (Fig. 2a and Supplementary Figure 6). This interaction of H255 with Y3537.35 contributes to the closed conformation of ECL2 over the ligand-binding pocket. F2715.39 interacts with the fluoro-phenyl ring and Y3376.59 forms a strong hydrogen bond with the pyridine ring of vorapaxar.
Mutation of Y3376.59 to phenylalanine and Y3537.35 to alanine led to a reduction in cell surface expression, making it difficult to interpret the associated reduction in agonist peptide activation (Fig. 4c and d). Mutation of F2715.39 to alanine was associated with enhanced cell surface expression, but reduced activation by agonist peptide. There was little effect of this mutation on maximal inhibition by vorapaxar. While not conclusive, this result suggests that F2715.39 may play a role in both peptide and vorapaxar binding. Of interest, mutation of Y1833.33 exhibited enhanced response to the agonist peptide and loss of inhibition by vorapaxar (Fig. 4c). This result suggests a possible role for Y1833.33 in maintaining the receptor in an inactive state and implies that interactions between Y1833.33 and vorapaxar may further stabilize an inactive conformation.
Activation of PAR1 by the agonist peptide
Our structure is consistent with data from mutagenesis studies that suggest that the PAR1 tethered agonist peptide may activate the receptor's heptahelical bundle by interacting with superficial structures rather than penetrating deeply into the transmembrane core8,41–45. E260, a solvent-exposed residue in ECL2 in both vorapaxar-bound and unliganded PAR1 (Fig. 5), is of particular interest, as evidence from mutagenesis studies suggests an interaction with R46 in the tethered peptide SFLLRN8,44. Substitution of E260 with R dramatically reduces activation of PAR1 by a peptide with the native tethered ligand sequence (SFLLRN) but facilitates activation by SFLLEN. R substitution of E264, which is surface-exposed and near E260 in the structure, also facilitates activation by SFLLEN.
Figure 5. Residues important for agonist peptide binding and receptor activation.
a, Mutations of residues E260, D256, L96 and E3477.29 near the extracellular surface have been shown to reduce activation of PAR1 by the free agonist peptide. Among them only E260 is completely exposed to the solvent, while D256 is the most deeply buried, forming H-bond with residue Y95. While none of these amino acids form part of the vorapaxar binding pocket, D256 forms a hydrogen bond with Y95 that may stabilize ECL2 over the vorapaxar binding pocket. Vorapaxar is shown in green. b, c, Superimposition of the unliganded MD simulation model (gray) with the ligand-bound crystal structure (blue). In b, Residues E260, D256, L96 and E3477.29, which are important for agonist peptide signaling, are in similar positions in both structures. c, The positions of residues that differ between human and Xenopus PAR1 in ECL2. Substitution of these residues in human PAR1 with corresponding residues from Xenopus PAR1 results in increased basal activity.
Mutation of other residues near the extracellular surface including L96A (amino terminus), D256A (ECL2) and E3477.29A/Q 45 (Fig. 5a), dramatically reduces activation of PAR1 by the peptide agonist. However, these mutations have only a small effect on agonist peptide binding, with only D256A resulting in a more than ten-fold loss of binding affinity43. The positions of these residues do not change substantially when comparing vorapaxar-bound and unliganded PAR1 (Fig. 5b). Of interest, only E3477.29 is surface exposed (Fig. 5a and b), suggesting that L96 and D256 may not interact directly with the agonist peptide or that these amino acids are more exposed than would appear from the MD model of the unliganded receptor. In the inactive structure, D256 forms a hydrogen bond with Y95 and helps stabilize interactions between the C-terminal end of the amino terminus and ECL2 (Fig. 5a).
Interestingly, substitution of human PAR1 sequence N259-A268, the region of ECL2 implicated in tethered ligand binding, with the cognate Xenopus ECL2 sequence results in an approximately 10-fold increase in basal activity46. Figure 5c shows the position of amino acids that differ between human and Xenopus receptors in ECL2 in both the crystal structure and the unliganded MD simulation. The superficial location of these activating mutations suggest that very superficial interactions between the tethered agonist peptide and the extracellular loops may be sufficient to activate PAR1. Taken together these mutagenesis studies suggest that the agonist peptide of PAR1 may activate by binding more superficially than do agonist peptides for opioid receptors. Alternatively, the tethered peptide may bind in a sequential manner, initially to the extracellular loops but penetrating more deeply into the core of the receptor through a sequence of conformational intermediates.
Conclusion
The unusual mode of activation and the paucity of pharmacological tools have made PAR1 one of the more challenging GPCRs to characterize and a difficult target for drug development. The crystal structure offers insights into the very high affinity interaction with the antagonist vorapaxar. This structure will provide a template for the development of PAR1 antagonists with better drug properties and the development of antagonists for other PAR subtypes to probe their biological roles. The mechanism of activation of PAR1 remains poorly understood. MD simulations of an unliganded receptor together with the location of amino acids known to influence agonist peptide activity suggest that activation of PAR1 by its agonist peptide may involve superficial interactions with extracellular loops. Future efforts will focus on an active-state structure of PAR1 bound to its tethered agonist peptide.
METHODS
PAR1-T4L expression and purification
To facilitate crystallogenesis, a human PAR1 construct was generated with several modifications. A TEV protease recognition site was introduced after residue P85, two N-linked glycosylation sites in ECL2 were removed by mutation (N250G and N259S), and the carboxyl terminus was truncated after residue S395. T4 lysozyme residues 2–16148 were inserted in the third intracellular loop between residue A301 and A303, with only one residue V302 removed. To facilitate purification, an N-terminal FLAG epitope was inserted after a signal peptide and a C-terminal deca-histidine tag was introduced. The final crystallization construct PAR1-T4L is shown in Supplementary Figure 2.
The modified PAR1 was expressed in Sf9 cells using the pFastBac baculovirus system (Invitrogen). The ligand vorapaxar was added at 100nM to the cells during expression. The cells were infected with baculovirus at 27°C for 48 hour before harvest. To purify the receptor, infected cells were lysed by osmotic shock in low-salt buffer containing 10mM Tris-HCl pH 7.5, 1mM EDTA, 100nM vorapaxar and 2mg/ml iodoacetamide. Iodoacetamide was used to alkylate reactive cysteines to prevent non-specific oligomerization. The protein was further extracted from cell membranes using a glass dounce homogenizer in buffer containing 20mM Tris-HCl pH7.5, 350mM NaCl, 1% dodecyl maltoside (DDM), 0.03% cholesterol hemisuccinate (CHS), 0.2% sodium cholate, 10% glycerol, 2mg/ml iodoacetamide and 100nM vorapaxar. Cell debris was removed by high-speed centrifugation. From this point, 1µM vorapaxar was added in all the following buffers used for purification. Nickel-NTA agarose resin was added to the supernatant after homogenization and stirred for 1h at 4°C. The resin was then washed 3 times in batch with buffer comprised of 20mM Hepes pH 7.5, 350mM NaCl, 0.1% DDM, 0.02% CHS and 1µM vorapaxar and transferred to a glass column. The bound receptor was eluted with buffer containing 300mM imidazole and loaded onto an anti-FLAG M1 affinity column. After extensive washing with buffer comprised of 20mM Hepes pH7.5, 350mM NaCl, 0.1% DDM, 0.02% CHS, 1µM vorapaxar and 2mM Ca2+, the receptor was eluted from M1 resin using the same buffer without Ca2+ but with 200ug/ml FLAG peptide and 5mM EDTA. To remove extra N-terminal residues and the FLAG epitope, TEV protease was added to the receptor and the cleavage reaction run at room temperature overnight. Size exclusion chromatography was used to obtain the final monodisperse receptor preparation. Purified PAR1-T4L was concentrated to 40–50mg/ml using 100kDa cutoff Vivaspin concentrators for crystallization.
Crystallization
As for other T4 lysozyme fused GPCRs crystallized so far, in meso crystallization was used to obtain PAR1-T4L crystals49,50. The protein was mixed with monoolein and cholesterol (10:1 by mass) using the two syringe mixing method by weight of 1:1.5 (protein: lipid). After a clear lipidic cubic phase formed, the mix was dispensed onto glass plates in 20–40nl drops overlaid with 700nl precipitant solution using a Gryphon LCP robot. Crystals appeared in two days in 0.1–0.2M sodium chloride, 100mM sodium phosphate pH6.0–6.5, 25%–35% PEG300, and grew to full size after one week (Supplementary Figure 3).
Data collection and structure determination
Crystals were harvested and frozen in liquid nitrogen. Date collection was performed at beamline 23-ID of GM/CA@APS at the Advanced Photon Source. Microbeams of 10- or 20-µM diameter were used to acquire all diffraction data. Due to radiation damage, only 5–20 degrees of rotation data were collected from each crystal. All data were processed with the HKL2000 package51. A 2.2 Å data set was obtained by merging diffraction data from 18 crystals. The space group was determined to be P212121. Molecular replacement was performed using the program Phaser52 in Phenix53, with the CXCR4 structure (PDB code 3ODU) as the search model. The 7-transmembrane helices without any loops, and the T4 lysozyme in the CXCR4 structure, were used as independent search models. The initial structure model was completed and improved through iterative refinement in Phenix53 and manual rebuilding of all the loops and several parts in the trans-membrane region in Coot54. Model refinement in Phenix and manual adjustment in Coot was performed to improve the model. The final structure was determined at 2.2Å resolution. The quality of the structure was assessed using Molprobity55. Data processing and structure refinement statistics are shown in Supplementary Table 1. Figures were prepared with PyMol56.
Phosphoinositide hydrolysis assays and cell surface expression level
The Quick Change Site Directed Mutagenesis kit (Agilent) was used to generate human Par1 mutants and all mutants were fully sequenced. Cos7 cells were transiently transfected using Fugene HD with either vehicle or wild-type human Par1 and mutants in the mammalian expression vector pBJ1 and signaling assays were performed as described in ref. 46. Briefly, Cos7 cells expressing the indicated hPar1 wild-type and mutants were labeled with [3H]-myoinositol, then incubated with vehicle or 100nM vorapaxar in DMEM medium containing 0.1% BSA, 20mM HEPES, 0.2% 2-hydroxypropyl-β-cyclodextrin (to retain vorapaxar in solution) for 1h at 37°C. Agonist (100µM SFLLRN or 10nM thrombin for PAR1 or other PAR agonists as indicated) was added and incubation continued for 1h. The total amount of accumulated [3H]-inositol phosphates accumulated was determined as in ref. 46. Cos7 cells transfected with empty vector had little response to PAR agonists, and treatment with vorapaxar alone did not affect phosphoinositide hydrolysis (Supplementary Figure 9A).
Cos7 cells were transiently transfected with empty pBJ1 or pBJ1 directing expression of N-terminal FLAG-tagged versions of wild-type human Par1 or mutants, and surface expression of receptors was measured as described in ref. 13 and 16. Briefly, cells were washed once with serum-free medium containing 0.1% BSA and 20mM HEPES, then incubated with 3µg/ml FLAG M1 antibody (Sigma) for 1h at 4°C in the same medium. The cells were then washed twice with PBS containing Ca2+ and Mg2+ to remove unbound antibody and fixed with 2% paraformaldehyde for 5 min. The cells were then washed twice with PBS with Ca2+ and Mg2+, and incubated with goat-anti mouse HRP-conjugated secondary antibody, washed and developed with one-step ABTS HRP substrate (Pierce). The absorbance at 405 nm was measured as indication of cell surface receptor expression levels.
Platelet signaling assays
Washed human platelets were prepared and PAR1-dependent responses were measured as described in ref. 57. Briefly, acid-citrate-dextrose (ACD) anti-coagulated human blood samples (60mL per donor) were obtained from AllCells, LLC (Emeryville, CA, USA). Blood was centrifuged without braking at 250×g at 37°C for 15 minutes. The upper platelet-rich plasma (PRP) phase was collected, incubated at 37°C for 10 minutes in the presence of prostacyclin (PGI2, 0.5µM), and centrifuged at 2200×g for 15 minutes. The pellet was resuspended in complete Tyrode’s solution (134mM NaCl, 12mM NaHCO3, 2.9mM KCl, 0.34mM Na2HPO4, 1.0mM MgCl2, 10mM HEPES, 0.9 % (w/v) dextrose, pH 7.4) containing 2mM CaCl2, 0.35% (w/v) bovine serum albumin (BSA), 10U/mL heparin, and 0.5µM PGI2. The platelet suspension was incubated for 10 minutes at 37°C then centrifuged at 1900×g for 8 minutes. This wash step was repeated and the final pellet resuspended in Tyrode’s buffer supplemented with BSA and 0.02U/mL apyrase. Platelets were incubated at 37 °C for 30 minutes to allow recovery from the effects of PGI2, counted using a Hemavet FS950 (Drew Scientific, Oxford, CT) and diluted to 300,000 cells/µL in Tyrode’s solution.
To antagonize PAR1, vorapaxar or vehicle (2% w/v 2-hydroxypropyl-β-cyclodextrin in DMSO) were added to platelet suspensions that were then incubated for one hour at 37°C before addition of agonists. The final concentrations of 2-hydroxypropyl-β-cyclodextrin and DMSO in platelet suspensions were 0.002% and 0.1%, respectively. Where reversibility was evaluated, platelets were washed twice with Tyrode’s buffer containing BSA and PGI2 after vorapaxar treatment then diluted for cell activation assays as above.
For flow cytometric analysis of platelet activation, platelets suspended in Tyrode’s solution containing 2mM CaCl2, 0.35% BSA and 0.02U/mL apyrase were incubated with agonist and antibody simultaneously. Fluor-conjugated antibodies directed against human P-selectin (PE-conjugate of AK-4; Ebiosciences; 1:25 dilution) and the activated conformation of human integrin αIIbβ3(FITC-conjugate of PAC-1, BD Biosciences; 1:25 dilution) were used. After 15 minutes at 37°C, the platelet suspension was diluted with phosphate-buffered saline and platelet-bound antibody measured using an Accuri C6 flow cytometer (Accuri; Ann Arbor, MI). Samples from at least three different donors were analyzed, each in triplicate.
Molecular dynamics simulation methods
In all simulations, the receptor was embedded in a hydrated lipid bilayer with all atoms, including those in the lipids and water, represented explicitly. Simulations were performed on Anton47, a special-purpose computer designed to accelerate molecular dynamics simulations by orders of magnitude.
System set-up and simulation protocol
Simulations of PAR1 were based on the crystal structure of the PAR1–vorapaxar complex. The crystallized construct has T4L inserted into ICL3 in place of residue 302. For the simulations, the T4L portion was omitted, and residue 302 was modeled in. The unresolved segment of ICL2 (residues 209–213) was also modeled in. Residues 209 and 213 were added manually, and residues 210 through 212 were modeled in using Prime (Schrödinger LLC). The Refine Loops tool in Prime, with default settings, was then used to refine residues 209–213.
The simulation of the µ-opioid receptor (MOR) dimer was based on the crystal structure of MOR bound to the irreversible antagonist β-funaltrexamine (PDB code 4DKL). Both monomers of the crystallographic dimer were included in the simulation, but β-funaltrexamine was deleted from the binding pocket. As with PAR1, the T4L sequence was omitted in our simulations. Side chains for residue M651.29, T671.31, K260ICL3, and R263ICL3 were not fully resolved in the crystal structure, so they were modeled in by hand, with rotamers chosen to avoid any clashes with resolved residues.
For both PAR1 and MOR, hydrogens were added to the crystal structures using Maestro (Schrödinger LLC), as described in previous work58. Histidines were singly protonated on the epsilon nitrogen. All other titratable residues were left in their dominant protonation state for pH 7.0, except for D3677.49 in PAR1 and D1142.50 in MOR, which were protonated, and D1482.50 in PAR1, which was protonated in certain simulations. PAR1 was simulated both with and without the crystallographic sodium ion by D1482.50 (Supplementary Table 2); D1482.50 was not protonated in simulations that included this ion, but was protonated otherwise. The conserved aspartate at position 2.50 is known to be protonated in rhodopsin59, and the residue at position 7.49 is most often an (uncharged) asparagine residue in family A GPCRs (the “N” of the NPxxY motif).
Prepared protein structures were inserted into an equilibrated POPC bilayer as described previously32. Sodium and chloride ions were added to neutralize the net charge of the system and to create a 150 mM solution.
Simulations of the PAR1 receptor initially measured 88.9 × 88.9 × 88.7 Å and contained 174 lipid molecules, and approximately 13,152 water molecules, for a total of ~67,500 atoms. When the crystallographic sodium ion near D1482.50 was included, the simulation contained 32 sodium ions and 36 chloride ions. When the crystallographic sodium ion was not included, the system contained 31 sodium ions and 36 chloride ions. To simulate the unliganded PAR1 receptor, vorapaxar was deleted from the binding pocket. Simulations of the MOR dimer initially measured 100.0 × 100.0 × 89.0 Å and contained 204 lipid molecules, 19 sodium ions, 43 chloride ions, and approximately 16,654 water molecules, for a total of ~86,700 atoms.
All simulations were equilibrated using Anton in the NPT ensemble at 310 K (37 °C) and 1 bar with 5 kcal mol−1 Å−2 harmonic position restraints applied to all non-hydrogen atoms of the protein and the ligand; these restraints were tapered off linearly over 50 ns. All bond lengths to hydrogen atoms were constrained using m-shake60. A RESPA integrator61 was used with a time step of 2 fs, and long-range electrostatics were computed every 6 fs. Production simulations were initiated from the final snapshot of the corresponding equilibration runs, with velocities sampled from the Boltzmann distribution at 310 K, using the same integration scheme, long-range electrostatics method, temperature and pressure. For PAR1, Van der Waals and short-range electrostatic interactions were cut off at 10.3 Å and long-range electrostatic interactions were computed using the k-space Gaussian Split Ewald method62 with a 32 × 32 × 32 grid, σ =2.27 Å, and σs=1.59 Å. For MOR, van der Waals and short-range electrostatic interactions were cut off at 10.16 Å and long-range electrostatic interactions were computed using the k-space Gaussian Split Ewald method with a 64 × 64 × 64 grid, σ=2.25 Å, and σs=1.55 Å.
We performed two vorapaxar-bound PAR1 simulations and four unliganded PAR1 simulations, and results were consistent across each set. The two receptors in our MOR dimer simulation also exhibited consistent behavior. The simulation protocol we followed has been validated in previous simulations of GPCRs63,64. Nevertheless, it is possible that different behavior might have been observed in even longer simulations, with different force field parameters, or with a different choice of simulation conditions.
Force field parameters
We used the CHARMM27 parameter set for protein molecules and salt ions, with the CHARMM TIP3P water model65; protein parameters incorporated CMAP terms66 and modified charges on the Asp, Glu, and Arg side chains67. We utilized a modified CHARMM lipid force field68. Force field parameters for vorapaxar were obtained from the CHARMM ParamChem web server69, version 0.9.6 β.
Analysis protocols
Trajectory snapshots, each containing a record of all atom positions at a particular instant in time, were saved every 180 ps during production simulations. Time series data shown in Supplementary Figure 11 were smoothed by applying a 9.9-ns (55-snapshot) running average. VMD was used to visualize trajectories70.
Supplementary Material
Acknowledgements
We acknowledge support from the National Institutes of Health Grants NS028471 (B.K.K), and HL44907 and HL65590 (S.R.C), and from the Mathers Foundation (B.K.K. and W.I.W.).
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contribution C.Z. optimized construct, expressed and purified human PAR1-T4L for crystallization; developed the purification procedure; performed crystallization trials, optimized crystallization conditions, collected diffraction data, solved and refined the structure. Y.S. helped design and make constructs for baculoviral expression, and executed and analyzed cell-based functional assays of wild-type and mutant PAR1. D.H.A. designed, performed, and analyzed MD simulations. J.J.F developed the initial expression and purification protocol for PAR1. D.P. helped design, executed and analyzed platelet function studies. Y.Z. helped design and make constructs for baculoviral PAR expression. H.F.G. performed and analyzed MD simulations; assisted with manuscript preparation. A.P. made vorapaxar. R.O.D. oversaw, designed, and analyzed MD simulations; assisted with manuscript preparation. D.E.S. oversaw MD simulations and analysis. W.I.W. assisted with X-ray diffraction data processing and crystal structure refinement. S.R.C. and B.K.K. initiated project, planned and analyzed experiments, supervised the research and wrote the manuscript with C.Z.
Author Information Coordinates and structure factors for PAR1 are deposited in the Protein Data Bank (accession code 3VW7). Reprints and permission information is available at www.nature.com/reprints. The authors declare no competing financial interests. Readers are welcome to comment on the online version of this article at www.nature.com/nature.
REFERENCES
- 1.Vu TK, Hung DT, Wheaton VI, Coughlin SR. Molecular cloning of a functional thrombin receptor reveals a novel proteolytic mechanism of receptor activation. Cell. 1991;64:1057–1068. doi: 10.1016/0092-8674(91)90261-v. [DOI] [PubMed] [Google Scholar]
- 2.Coughlin SR. Thrombin signalling and protease-activated receptors. Nature. 2000;407:258–264. doi: 10.1038/35025229. [DOI] [PubMed] [Google Scholar]
- 3.Vu TK, Wheaton VI, Hung DT, Charo I, Coughlin SR. Domains specifying thrombin-receptor interaction. Nature. 1991;353:674–677. doi: 10.1038/353674a0. [DOI] [PubMed] [Google Scholar]
- 4.Chen J, Ishii M, Wang L, Ishii K, Coughlin SR. Thrombin receptor activation. Confirmation of the intramolecular tethered liganding hypothesis and discovery of an alternative intermolecular liganding mode. J Biol Chem. 1994;269:16041–16045. [PubMed] [Google Scholar]
- 5.Liu LW, Vu TK, Esmon CT, Coughlin SR. The region of the thrombin receptor resembling hirudin binds to thrombin and alters enzyme specificity. J Biol Chem. 1991;266:16977–16980. [PubMed] [Google Scholar]
- 6.Ishii K, Hein L, Kobilka B, Coughlin SR. Kinetics of thrombin receptor cleavage on intact cells. Relation to signaling. J Biol Chem. 1993;268:9780–9786. [PubMed] [Google Scholar]
- 7.Mathews II, et al. Crystallographic structures of thrombin complexed with thrombin receptor peptides: existence of expected and novel binding modes. Biochemistry. 1994;33:3266–3279. doi: 10.1021/bi00177a018. [DOI] [PubMed] [Google Scholar]
- 8.Gerszten RE, et al. Specificity of the thrombin receptor for agonist peptide is defined by its extracellular surface. Nature. 1994;368:648–651. doi: 10.1038/368648a0. [DOI] [PubMed] [Google Scholar]
- 9.Coughlin SR. Protease-activated receptors in hemostasis, thrombosis and vascular biology. J Thromb Haemost. 2005;3:1800–1814. doi: 10.1111/j.1538-7836.2005.01377.x. [DOI] [PubMed] [Google Scholar]
- 10.Garcia-Lopez MT, Gutierrez-Rodriguez M, Herranz R. Thrombin-activated receptors: promising targets for cancer therapy? Curr Med Chem. 2010;17:109–128. doi: 10.2174/092986710790112639. [DOI] [PubMed] [Google Scholar]
- 11.Ramachandran R, Noorbakhsh F, Defea K, Hollenberg MD. Targeting proteinase-activated receptors: therapeutic potential and challenges. Nat Rev Drug Discov. 2012;11:69–86. doi: 10.1038/nrd3615. [DOI] [PubMed] [Google Scholar]
- 12.Ishii K, et al. Determinants of thrombin receptor cleavage. Receptor domains involved, specificity, and role of the P3 aspartate. J Biol Chem. 1995;270:16435–16440. doi: 10.1074/jbc.270.27.16435. [DOI] [PubMed] [Google Scholar]
- 13.Shapiro MJ, Trejo J, Zeng D, Coughlin SR. Role of the thrombin receptor's cytoplasmic tail in intracellular trafficking. Distinct determinants for agonist-triggered versus tonic internalization and intracellular localization. J Biol Chem. 1996;271:32874–32880. doi: 10.1074/jbc.271.51.32874. [DOI] [PubMed] [Google Scholar]
- 14.Trejo J, Coughlin SR. The cytoplasmic tails of protease-activated receptor-1 and substance P receptor specify sorting to lysosomes versus recycling. J Biol Chemh. 1999;274:2216–2224. doi: 10.1074/jbc.274.4.2216. [DOI] [PubMed] [Google Scholar]
- 15.Trejo J, Hammes SR, Coughlin SR. Termination of signaling by protease-activated receptor-1 is linked to lysosomal sorting. Proc Natl Acad Sci U S A. 1998;95:13698–13702. doi: 10.1073/pnas.95.23.13698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shapiro MJ, Coughlin SR. Separate signals for agonist-independent and agonist-triggered trafficking of protease-activated receptor 1. J Biol Chem. 1998;273:29009–29014. doi: 10.1074/jbc.273.44.29009. [DOI] [PubMed] [Google Scholar]
- 17.Hein L, Ishii K, Coughlin SR, Kobilka BK. Intracellular targeting and trafficking of thrombin receptors. A novel mechanism for resensitization of a G protein-coupled receptor. J Biol Chem. 1994;269:27719–27726. [PubMed] [Google Scholar]
- 18.Chackalamannil S, et al. Discovery of potent orally active thrombin receptor (protease activated receptor 1) antagonists as novel antithrombotic agents. J Med Chem. 2005;48:5884–5887. doi: 10.1021/jm0502236. [DOI] [PubMed] [Google Scholar]
- 19.Morrow DA, et al. Vorapaxar in the secondary prevention of atherothrombotic events. N Engl J Med. 2012;366:1404–1413. doi: 10.1056/NEJMoa1200933. [DOI] [PubMed] [Google Scholar]
- 20.Kosoglou T, et al. Pharmacodynamics and pharmacokinetics of the novel PAR-1 antagonist vorapaxar (formerly SCH 530348) in healthy subjects. Eur J Clin Pharmacol. 2012;68:249–258. doi: 10.1007/s00228-011-1120-6. [DOI] [PubMed] [Google Scholar]
- 21.Soto AG, Trejo J. N-linked glycosylation of protease-activated receptor-1 second extracellular loop: a critical determinant for ligand-induced receptor activation and internalization. J Biol Chem. 2010;285:18781–18793. doi: 10.1074/jbc.M110.111088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu B, et al. Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science. 2010;330:1066–1071. doi: 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manglik A, et al. Crystal structure of the micro-opioid receptor bound to a morphinan antagonist. Nature. 2012;485:321–326. doi: 10.1038/nature10954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu H, et al. Structure of the human kappa-opioid receptor in complex with JDTic. Nature. 2012;485:327–332. doi: 10.1038/nature10939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Granier S, et al. Structure of the delta-opioid receptor bound to naltrindole. Nature. 2012;485:400–404. doi: 10.1038/nature11111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smit MJ, et al. Pharmacogenomic and structural analysis of constitutive g protein-coupled receptor activity. Annu Rev Pharmacol Toxicol. 2007;47:53–87. doi: 10.1146/annurev.pharmtox.47.120505.105126. [DOI] [PubMed] [Google Scholar]
- 27.Schwartz TW, Frimurer TM, Holst B, Rosenkilde MM, Elling CE. Molecular mechanism of 7TM receptor activation--a global toggle switch model. Annu Rev Pharmacol Toxicol. 2006;46:481–519. doi: 10.1146/annurev.pharmtox.46.120604.141218. [DOI] [PubMed] [Google Scholar]
- 28.Fredriksson R, Lagerstrom MC, Lundin LG, Schioth HB. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol Pharmacol. 2003;63:1256–1272. doi: 10.1124/mol.63.6.1256. [DOI] [PubMed] [Google Scholar]
- 29.Rasmussen SG, et al. Structure of a nanobody-stabilized active state of the beta(2) adrenoceptor. Nature. 2011;469:175–180. doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosenbaum DM, et al. Structure and function of an irreversible agonist-beta(2) adrenoceptor complex. Nature. 2011;469:236–240. doi: 10.1038/nature09665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rasmussen SG, et al. Crystal structure of the beta2 adrenergic receptor-Gs protein complex. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harding MM. Metal-ligand geometry relevant to proteins and in proteins: sodium and potassium. Acta Crystallogr D Biol Crystallogr. 2002;58:872–874. doi: 10.1107/s0907444902003712. [DOI] [PubMed] [Google Scholar]
- 33.Horstman DA, et al. An aspartate conserved among G-protein receptors confers allosteric regulation of alpha 2-adrenergic receptors by sodium. J Biol Chem. 1990;265:21590–21595. [PubMed] [Google Scholar]
- 34.Costa T, Lang J, Gless C, Herz A. Spontaneous association between opioid receptors and GTP-binding regulatory proteins in native membranes: specific regulation by antagonists and sodium ions. Mol Pharmacol. 1990;37:383–394. [PubMed] [Google Scholar]
- 35.Gao ZG, Ijzerman AP. Allosteric modulation of A(2A) adenosine receptors by amiloride analogues and sodium ions. Biochem Pharmacol. 2000;60:669–676. doi: 10.1016/s0006-2952(00)00360-9. [DOI] [PubMed] [Google Scholar]
- 36.Selent J, Sanz F, Pastor M, De Fabritiis G. Induced effects of sodium ions on dopaminergic G-protein coupled receptors. PLoS Comput Biol. 2010;6 doi: 10.1371/journal.pcbi.1000884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilson MH, Highfield HA, Limbird LE. The role of a conserved inter-transmembrane domain interface in regulating alpha(2a)-adrenergic receptor conformational stability and cell-surface turnover. Mol Pharmacol. 2001;59:929–938. doi: 10.1124/mol.59.4.929. [DOI] [PubMed] [Google Scholar]
- 38.Neve KA, et al. Modeling and mutational analysis of a putative sodium-binding pocket on the dopamine D2 receptor. Mol Pharmacol. 2001;60:373–381. doi: 10.1124/mol.60.2.373. [DOI] [PubMed] [Google Scholar]
- 39.Palczewski K, et al. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 40.Hanson MA, et al. Crystal structure of a lipid G protein-coupled receptor. Science. 2012;335:851–855. doi: 10.1126/science.1215904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lerner DJ, Chen M, Tram T, Coughlin SR. Agonist recognition by proteinase-activated receptor 2 and thrombin receptor. Importance of extracellular loop interactions for receptor function. J Biol Chem. 1996;271:13943–13947. [PubMed] [Google Scholar]
- 42.Nanevicz T, et al. Mechanisms of thrombin receptor agonist specificity. Chimeric receptors and complementary mutations identify an agonist recognition site. J Biol Chem. 1995;270:21619–21625. doi: 10.1074/jbc.270.37.21619. [DOI] [PubMed] [Google Scholar]
- 43.Blackhart BD, et al. Extracellular mutations of protease-activated receptor-1 result in differential activation by thrombin and thrombin receptor agonist peptide. Mol Pharmacol. 2000;58:1178–1187. doi: 10.1124/mol.58.6.1178. [DOI] [PubMed] [Google Scholar]
- 44.Seeley S, et al. Structural basis for thrombin activation of a protease-activated receptor: inhibition of intramolecular liganding. Chem Biol. 2003;10:1033–1041. doi: 10.1016/j.chembiol.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 45.Bahou WF, Kutok JL, Wong A, Potter CL, Coller BS. Identification of a novel thrombin receptor sequence required for activation-dependent responses. Blood. 1994;84:4195–4202. [PubMed] [Google Scholar]
- 46.Nanevicz T, Wang L, Chen M, Ishii M, Coughlin SR. Thrombin receptor activating mutations. Alteration of an extracellular agonist recognition domain causes constitutive signaling. J Biol Chem. 1996;271:702–706. doi: 10.1074/jbc.271.2.702. [DOI] [PubMed] [Google Scholar]
- 47.Shaw DE, Dror RO, Salmon JK, et al. Proceedings of the Conference on High Performance Computing Networking, Storage and Analysis. ACM; 2009. Millisecond-scale molecular dynamics simulations on Anton. in. [Google Scholar]
- 48.Rosenbaum DM, et al. GPCR engineering yields high-resolution structural insights into beta2-adrenergic receptor function. Science. 2007;318:1266–1273. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- 49.Caffrey M, Cherezov V. Crystallizing membrane proteins using lipidic mesophases. Nat Protoc. 2009;4:706–731. doi: 10.1038/nprot.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Caffrey M. Crystallizing membrane proteins for structure determination: use of lipidic mesophases. Annu Rev Biophys. 2009;38:29–51. doi: 10.1146/annurev.biophys.050708.133655. [DOI] [PubMed] [Google Scholar]
- 51.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 52.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 55.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schrodinger L. The PyMOL Molecular Graphics System. V1.5.0.4. 2010 [Google Scholar]
- 57.Cornelissen I, et al. Roles and interactions among protease-activated receptors and P2ry12 in hemostasis and thrombosis. Proc Natl Acad Sci U S A. 2010;107:18605–18610. doi: 10.1073/pnas.1013309107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dror RO, et al. Identification of two distinct inactive conformations of the beta2-adrenergic receptor reconciles structural and biochemical observations. Proc Natl Acad Sci U S A. 2009;106:4689–4694. doi: 10.1073/pnas.0811065106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fahmy K, et al. Protonation states of membrane-embedded carboxylic acid groups in rhodopsin and metarhodopsin II: a Fourier-transform infrared spectroscopy study of site-directed mutants. Proc Natl Acad Sci U S A. 1993;90:10206–10210. doi: 10.1073/pnas.90.21.10206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kräutler V, Gunsteren WFv, Hünenberger PH. A fast SHAKE algorithm to solve distance constraint equations for small molecules in molecular dynamics simulations. J. Comput. Chem. 2001;22:501–508. [Google Scholar]
- 61.Tuckerman M, Berne BJ, Martyna GJ. Reversible multiple time scale molecular dynamics. J. Chem. Phys. 1992;97:1990–2001. [Google Scholar]
- 62.Shan Y, Klepeis JL, Eastwood MP, Dror RO, Shaw DE. Gaussian split Ewald: A fast Ewald mesh method for molecular simulation. J. Chem. Phys. 2005;122:54101. doi: 10.1063/1.1839571. [DOI] [PubMed] [Google Scholar]
- 63.Dror RO, et al. Activation mechanism of the beta2-adrenergic receptor. Proc Natl Acad Sci U S A. 2011;108:18684–18689. doi: 10.1073/pnas.1110499108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dror RO, et al. Pathway and mechanism of drug binding to G-protein-coupled receptors. Proc Natl Acad Sci U S A. 2011;108:13118–13123. doi: 10.1073/pnas.1104614108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.MacKerell AD, Jr, Bashford D, Bellott M. All-Atom Empirical Potential for Molecular Modeling and Dynamics Studies of Proteins. J. Phys. Chem. 1998;B 102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 66.Mackerell AD, Jr, Feig M, Brooks CL., 3rd. Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J. Comput. Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 67.Piana S, Lindorff-Larsen K, Shaw DE. How robust are protein folding simulations with respect to force field parameterization? Biophys J. 2011;100:L47–L49. doi: 10.1016/j.bpj.2011.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Klauda JB, et al. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vanommeslaeghe K, et al. CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 2010;31:671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.