Abstract
Following sequencing and assembly of the human genome, the preferred methods for identification of new drug targets have changed dramatically. Modern tactics such as genome-wide association studies (GWAS) and deep sequencing are fundamentally different from the pharmacology-guided approaches used previously, in which knowledge of small molecule ligands acting at their cellular targets was the primary discovery engine. A consequence of the “target-first, pharmacology-second” strategy is that many predicted drug targets are non-enzymes, such as scaffolding, regulatory or structural proteins, and their activities are often dependent on protein-protein interactions (PPIs). These types of targets create unique challenges to drug discovery efforts because enzymatic turnover cannot be used as a convenient surrogate for compound potency. Moreover, it is often challenging to predict how ligand binding to non-enzymes might affect changes in protein function and/or pathobiology. Thus, in the post-genomic era, targets might be strongly implicated by molecular biology-based methods, yet they often later earn the designation of “undruggable.” Can the scope of available targets be widened to include these promising, but challenging, non-enzymes? In this review, we discuss advances in high throughput screening technology and chemical library design that are emerging to deal with these challenges.
Challenges Associated with Non-Enzyme Targets
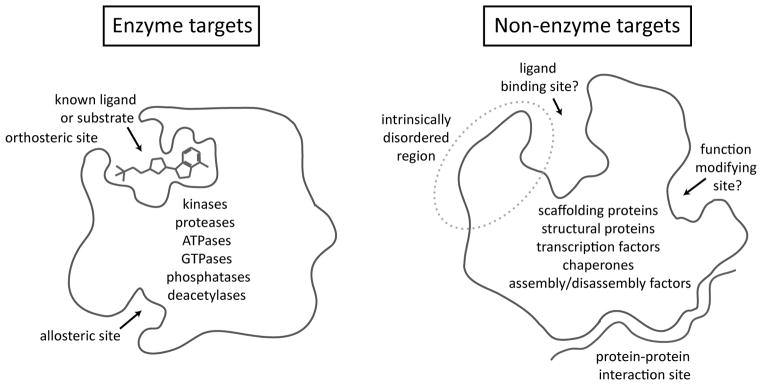
The majority of current drug targets are G-protein coupled receptors, nuclear receptors, ion channels or enzymes (e.g. kinases, proteases, deacetylases, etc.) [1, 2]. Many of these targets were historically identified based on their pharmacology: agonists or antagonists were used to probe the biology of the target, followed by progression to therapeutic candidates. As a consequence, many of these proteins, by definition, contain deep grooves that are amenable to binding by low molecular weight, “drug-like” small molecules. In contrast, the modern shift towards molecular biology- and genomics-based target identification has often implicated other types of targets, including non-enzymes (Figure 1). Non-enzymes make up a majority of the human proteome and they include proteins involved in organizing signaling pathways, maintaining structural integrity, assembly/disassembly of protein complexes, chaperoning, subcellular transport, transcription, translation and other critical functions. Rather than using enzymatic turnover to carry out their biology, most non-enzymes use protein-protein interactions (PPIs), either transient or stable contacts that form the backbone of all major cellular pathways [3]. In turn, the challenges of targeting PPIs have been well documented [4–6].
Figure 1.
Nonenzyme targets present unique challenges to drug discovery. Classic enzyme targets have well-defined active sites and many have clear allosteric sites, which make attractive binding regions for orthosteric and allosteric inhibitors. In contrast, most non-enzymes lack obvious binding pockets or they are involved in protein-protein interactions that involve larger, more diffuse contact areas.
Modern drug discovery approaches, such as high-throughput screening (HTS), typically rely on the measurement of enzymatic turnover to drive discovery of potential clinical leads; thus, non-enzymes pose a particular challenge. Rather, known “inhibitors” of non-enzymes typically bind to the target and either block binding to other proteins or otherwise alter structure-function (e.g. change oligomerization, alter protein stability, etc.). It is often difficult to predict what will happen to biological pathways in response to these changes and it is more difficult to envision HTS platforms that will rapidly identify potential ligands. To make matters worse, non-enzymes often lack natural ligands or even ligand binding sites, posing a further hurdle to drug discovery campaigns. Finally, many non-enzymes are either structurally uncharacterized or intractable for structural biology (i.e. they contain regions of intrinsic disorder), which often precludes the use of most structure-guided design methods.
Despite these significant challenges, the prominent role of non-enzymes in biology and pathobiology is certain, so what can be done to expand the number of “druggable” targets to include these proteins? What HTS methods can be adapted for use against non-enzymes? What strategies are amenable to hit identification in the absence of structural information? Is it possible to identify a ligand binding site de novo? If so, how can one predict whether or not such a site is “druggable”? In this review, we provide an update on the methods being used to identify molecules that bind to non-canonical targets and categorize them into affinity-, computational- and stability-based techniques. We also discuss how chemical library design is evolving to meet the specific challenges of post-genomic drug discovery.
Affinity-Based Techniques
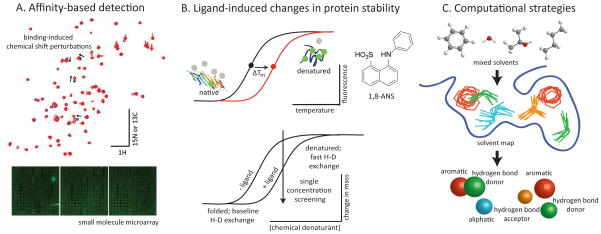
A major problem in many potential drug discovery campaigns involving non-enzymes is that it is difficult to identify molecules that bind the target. In the absence of an enzymatic function, there is no convenient surrogate for ligand binding, so the interaction must be directly measured. Nuclear magnetic resonance (NMR)-based screening has proven to be particularly amenable to the label-free, affinity-based selection of ligands that bind a target of interest, including non-enzymes [7]. The most information-rich platform for NMR-based screening uses a two-dimensional experiment (HSQC or TROSY) and observes 15N or 13C isotopically labeled protein. In these experiments, mixtures of library compounds, generally low molecular weight fragments, are added to a solution of the protein. Hits result in binding-induced perturbations of the chemical shifts associated with N-H or C-H bonds (Figure 2A) and, if the NMR spectrum is assigned to the protein’s primary sequence, then the ligand binding site may be directly determined from this experiment. The binding site is often used to prioritize hits and the screen may be carried out in the presence of a competitive orthosteric ligand to favor the identification of second-site binders. False positive rates are typically low, and nonspecific binding is often readily recognizable. However, protein-observed NMR screening requires that the protein be highly soluble, stable, and homogenous at high concentrations (50 to 500 μM [8]), able to be recombinantly expressed in isotopically enriched media, and relatively small (less than ~80 kDa, though this limit depends on the type of labeling and experiment used [9]). Thus, to complement the protein-observed experiments, a number of one-dimensional, ligand-observed experiments may also be used, including saturation transfer [10, 11] and diffusion based experiments [12, 13]. These approaches are selection-based, meaning that only the ligands interacting with the target protein are identified from mixtures. Ligand-observed experiments require relatively low concentrations of protein (typically 1 to 10 μM), and the protein need not be isotopically labeled. In addition, they have higher throughput and lower experimental cost than protein-observed experiments. However, ligand-observed methods do not distinguish between specific and nonspecific interactions, they offer no information on the binding site, and they suffer from higher false positive rates (though the combination of several ligand-observed experiments may increase reliability [14]). For both ligand- and protein-observed NMR experiments, relatively weak interactions (KD values between 0.1 μM to 10 mM) can be measured, but stronger interactions can give false negatives [9].
Figure 2.
Selected biophysical methods for ligand discovery. A: Ligand-induced changes in chemical shifts of a 1H, 15N HSQC spectrum of a protein target suitable for NMR-based screening indicate binding. Fluorescent spots on a small molecule microarray indicate the presence of a fluorescently labeled protein bound to the immobilized ligands. B: Differential scanning fluorimetry measures changes in the melting temperature (Tm) of a protein target induced by ligand binding. Similarly, hydrogen-deuterium exchange can measure changes in stability to chemical denaturation due to small molecule binding. C: The mixed-solvent molecular dynamics method may be used for both binding site identification and the construction of a pharmacophore.
In addition to its utility as a screening strategy, NMR can be extremely valuable for de novo binding site identification in targets for which no orthosteric site is known or for which an allosteric site is desired [15–19]. These methods might even reveal sites that are not obvious from available crystal structures because NMR is solution-based. Because of the reliability of NMR in identifying binding sites for small molecules, hit rates from fragment-based NMR screens are often used to categorize a protein target for its potential “druggability” [20]. The theory in this approach is that higher hit rates are suggestive of more and deeper binding sites. For example, Hajduk and colleagues observed a correlation between high experimental NMR hit rates (>0.2%) and the success of medicinal chemistry campaigns to develop molecules with high affinity (<300 nM) among a set of 23 protein targets [20]. This approach might be particularly useful in targeting PPIs, because of the notoriously shallow contact surfaces involved and the advantages of using allostery to disrupt these interfaces [21, 22].
To illustrate the potential of NMR-based screening campaigns, it is useful to consider the specific example of survivin. Survivin is a cell cycle regulator and inhibitor of apoptosis that is upregulated in most tumor cell types but absent in most other adult tissues [23]. High levels of survivin have been associated with poor prognosis in patients [24] and antisense oligonucleotides and siRNA against survivin decrease proliferation in a number of cancer cell lines [25, 26]. Survivin has no enzymatic activity or known endogenous small molecule regulators, and, accordingly, no robust biochemical assay of survivin function has been established. Wendt and colleagues at Abbott Laboratories chose to employ two complementary affinity-based screening methods, NMR-based screening and affinity selection mass spectrometry (AS-MS) to pursue lead generation of molecules that bind survivin [7]. AS-MS experiments start with the incubation of a mixture of ligands with the protein target of interest, followed by a separation step to remove unbound molecules and mass spectrometry-based identification of eluted compound(s) [27]. These methods are highly sensitive and allow for the evaluation of large chemical libraries (up to 108 to date, [28]) without the need to add labels [27, 29]. However, because it is prone to false positives, this method is complemented by protein-based NMR screening. Thus, the Abbott group used these two methods in combination to discover a novel small molecule-binding site on the dimer interface of survivin. They also used the relative hit rates from the screening campaign to evaluate the relative druggability of this new site, concluding that the dimer interface may be particularly promising (0.35% relative to 0.01% for a known peptide-binding interface). One lead series was developed into a class of compounds with nanomolar affinity for survivin [7]. Although it is not yet clear how interactions with this binding site impact survivin biology, the lead compounds from this campaign are expected to be powerful probes for target validation.
Surface plasmon resonance (SPR) is a label-free platform for the detection of direct binding interactions. Briefly, the target protein is typically immobilized to a gold chip, and potential ligands are introduced to this surface. Real-time association and dissociation rates of the interaction are measured, giving useful information about binding kinetics. The well-known nutlin class of MDM2-p53 protein-protein inhibitors originated from a competition SPR screen, in which the ability of molecules to disrupt this PPI was monitored [30]. The throughput of SPR experiments is lower than that of other affinity-based techniques, but these rates are increasing with newer generations of the technology; the latest instrument from GE Healthcare, the Biacore 4000, handles up to 4,800 samples per day [31]. However, this technique is still more widely applied to the evaluation of small, focused libraries during lead optimization. Some improvement in throughput can be obtained using biolayer interferometry (BLI). In commercialized BLI platforms, pins with immobilized ligand are dipped into wells of 96- or 384-well microtiter plates containing solutions of analytes, and the association and dissociation rates are measured in real-time. Using this approach, the OctetRed384 (ForteBio) can process up to 7000 samples per day [32]. Both SPR and BLI are flexible platforms that are well suited to the study of non-canonical targets because no structural information is required, no ligand binding site needs to be identified and no enzymatic activity is necessary.
Microarray techniques facilitate the discovery of new ligands via binding of a target to arrays of immobilized compounds. In this approach, small molecules or peptides are covalently attached to modified glass microscope slides, followed by incubation with the protein target of interest that is either directly labeled with a fluorophore or detected using a fluorescent antibody (Figure 2A). This approach has been used to discover new ligands for non-enzymes, including the yeast transcription factor Hap3p [33] and the extracellular signaling protein Sonic hedgehog [34]. In the Hap3p campaign, a collection of 12,400 immobilized compounds was screened, leading to the discovery of haptamide B. Haptamide B binds Hap3p and inhibits its transcriptional activity [33], likely by blocking PPIs in the transcription complex. In a similar strategy, Stanton and colleagues screened 10,000 immobilized compounds and identified robotnikinin, which binds the N-terminus of Sonic hedgehog and inhibits signaling [34, 35]. In another recent adaptation of this technology, Landry and colleagues combined microarrays with ellipsometry to obtain affinity values for binding to 104 immobilized small molecules [36, 37]. The oblique-incidence reflectivity difference microscope that was constructed for this use is not yet commercialized [38], but it has the potential to accelerate affinity-based lead discovery by microarrays by facilitating rank-ordering of potential ligands.
Stability-Based Methods
Monitoring ligand-induced changes in protein stability is another way to discover potential ligands for non-enzymes. Historically, the drug discovery applications of ligand-induced stability were pioneered in attempts to develop “pharmacological chaperones”, or molecules that stabilize the folded form of a mutated or damaged protein. Pharmacological chaperones have been successfully used to correct disease phenotypes in a number of disorders caused by a loss of protein stability [39], including phenylketonuria [40, 41], Gaucher disease [42, 43], Tay-Sachs disease [44], cystic fibrosis [45, 46], and transthyretin amyloidosis [47, 48]. One molecule, tafamidis, has been approved in Europe for the treatment of a form of transthyretin amyloidosis, familial amyloid polyneuropathy [49]. Tafamidis kinetically stabilizes the tetrameric conformation of transthyretin, increasing the activation barrier of dissociation of the tetramer to an unstable monomer [47]. Similarly, a recently discovered peptide inhibitor of caspase-6 acts by stabilizing an inactive, tetrameric conformation of the protein [50]. Recent work suggests that even some classic ligands might, in fact, use a pharmacological chaperone mechanism; for example, nicotine appears to exert its effects by thermodynamically stabilizing a specific conformation of the acetylcholine receptor [51, 52]. There are a number of methods available for discovering ligands that bind and stabilize targets and, because these methods do not rely on enzymatic turnover, they are particularly versatile tools for discovery in a post-genomic era.
Differential scanning fluorimetry (DSF) is one technique for measuring the ligand-induced changes in the thermal stability of a protein [53]. In these experiments, a protein solution is heated, leading to thermal denaturation. This unfolding is monitored using an environmentally sensitive fluorophore, such as 1-anilinonapthalene-8-sulfonic acid (1,8-ANS) (Figure 2B) and ligands are identified by their ability to shift the apparent melting transition (ΔTm). DSF experiments can be miniaturized for use in 384-well microtiter plates [54–57], permitting the screening of chemical libraries.
One illustrative example of a DSF campaign was reported for the transcription factor p53, which is a tumor suppressor that normally functions to regulate cell cycle arrest and apoptosis. Knockout mice (p53−/−) have high rates of spontaneous tumors, and p53 null or mutant tumors are associated with poor prognosis and resistance to chemotherapy in a number of human cancers [58]. The suppressor oncogenes MDM2 and HDM2 engage in PPIs that activate p53, leading to cell cycle arrest and apoptosis. These observations suggested that inhibitors of the p53-MDM2/HDM2 interactions might be promising anti-tumor agents, yet the drug target was clearly a non-enzyme, PPI interface. A team at Johnson & Johnson used DSF to screen a focused collection of 22,000 1,4-benzodiazepine-2,5-diones for affinity to HDM2 [59, 60]. The screening hits were then evaluated for inhibition of the p53-HDM2 PPI by a competitive fluorescence polarization (FP) assay, resulting in the development of inhibitors with nanomolar potency in cancer cell lines [58]. In this example, the candidate molecules, discovered by DSF, appear to bind HDM2 and stabilize a conformation that prevents the p53 interaction. DSF has more recently been applied to an HTS campaign against the F508Δ mutant of the cystic fibrosis transmembrane conductance regulator (CFTR) [61]. This point mutant is responsible for most cases of cystic fibrosis and it is known to destabilize the protein, causing F508Δ CFTR to be aberrantly retained in the ER and degraded rather than trafficked to the plasma membrane, where it normally functions as a chloride channel. DSF was used to prioritize hits from a cell-based primary screen and it was found that the most promising molecules bind to the first nucleotide-binding domain of the CFTR, helping to restore the folding free energy (ΔG) lost by the mutant. These efforts resulted in the identification of a phenylhydrazone, RDR1, which acts as a pharmacological chaperone for the misfolded F508Δ CFTR mutant [61]. Finally, several variations of DSF experiments have been reported. For example, intrinsic fluorophores, such as tryptophan or a cofactor, can be used in place of an extrinsic dye [62]; cysteine residues can be used in combination with thiol-specific fluorochromes in the same manner [63].
Hydrogen-deuterium exchange (HDX) coupled with NMR or mass spectrometry can be a powerful method for the detection of ligand-induced changes in protein stability. When a folded protein is placed in a buffer containing deuterated water, exchangeable protons on amide nitrogens and side chain heteroatoms are replaced with deuterons at a rate that is proportional to their relative solvent accessibility. Upon unfolding of the protein, internal protons become exchangeable [64], thus ligands can be detected by their ability to delay or prevent deuteration (Figure 2B) [65]. This technique has been developed for HTS by the Fitzgerald laboratory using the prolyl isomerase cyclophilin A as a model system [66–68]. In this example, a library of 104 compounds was screened at a single timepoint and a single denaturant concentration, with a throughput of ~100,000 compounds per day.
In Silico Methods
Another tool in the discovery of ligands for non-enzyme targets is de novo binding site identification, which uses geometrical, energy-based, evolutionary, or probe mapping techniques to scan for sites that may be deep enough to accommodate small molecules with good binding affinity [69, 70]. This approach is often used as a prelude to the development of pharmacophores that might bind the new site, which enriches subsequent HTS campaigns with predicted inhibitors. Most of the available de novo site prediction methods search for potential sites by identifying concave ‘pockets’ on the surface of a rigid protein structure [69]. Alternatives include energy-based approaches, which use a 3D potential grid to identify contiguous regions of predicted low energy interactions [71] and evolutionary (or genomic) methods, which consider the degree of conservation of amino acids on a protein’s surface [70, 72]. Lastly, probe mapping techniques coat the surface of the protein with small organic molecules and calculate the interaction energies between the probes and the surface to predict likely sites [69]. These four strategies may be used alone or in combination [69].
One significant limitation of the current de novo methods is that they are generally used with a rigid protein structure, which makes them fast but inaccurate for flexible binding sites [70]. However, one recent advance is based on the multiple-solvent crystal structure (MSCS) approach [73]. In the MSCS experiment, a target protein is crystallized and placed in solutions containing organic solvent. The organic probes displace water and they tend to accumulate at sites where favorable interactions may be possible. When multiple solvents are used, the contributions of aromatic, aliphatic, and hydrogen bonding interactions are identified (Figure 2C). The computational equivalent of the MSCS approach is mixed-solvent molecular dynamics [74], which employs an ensemble of protein structures from multiple crystal or NMR experiments in a virtual box of mixed aqueous and organic solvent molecules (e.g. benzene + propane + water). This system is minimized in a molecular dynamics (MD) simulation to build pharmacophore models of potential binding sites [74]. While the incorporation of explicit solvents and protein flexibility represent an improvement, the predictive power of any de novo method remains to be demonstrated for any non-enzyme.
Another possible contribution of in silico methods is that, once binding sites are identified, they may be computationally assessed for potential druggability. Though this subfield is in its early stages, a number of interesting studies have been reported [75–78]. Briefly, these methods use a combination of physical and physicochemical parameters, including the shape, size, hydrophobicity, and hydrogen bonding capability of the pocket, and they compare these values to training sets of known ligand-protein pairs. Cheng et al. developed one such method for predicting maximal affinity using a scoring system based on the hydrophobicity of the solvent accessible surface area (SASA) and the shape (curvature) of the ligand binding sites [75]. This method was able to confirm approximately 60 known protein-ligand maximal affinities. More importantly, they also carried out pilot screens of 11,000 compounds against two target enzymes, one of which was predicted to be “druggable” (i.e. good maximal affinities) by their computational method and the other “difficult” (i.e. weak maximal affinities). These screens gave hit rates of 1.8% and 0.15%, respectively, consistent with the prediction. Moreover, additional optimization at Pfizer produced eleven sub-micromolar potency leads from the “druggable” target project, but none for the “difficult” one. Further development of these methods may yield an important tool for non-enzymes.
Application to Members of Highly Similar Enzyme Families
While not the major focus of this review, another class of “undruggable” targets includes certain members of highly similar enzymes (e.g. kinase families). The reason these targets are “undruggable” is that the key amino acids populating the enzyme active site are identical or highly conserved, making it difficult to acquire selectivity. These observations have led many groups to pursue secondary sites, allosteric pockets or PPIs to differentiate between members of these families (recently reviewed in [79]). Often, the regions outside the active site cleft are not subject to the same evolutionary pressure and these surfaces can be distinct among family members. Accordingly, the discovery methods described here, such as NMR and SPR, may be applicable to these targets. In some of these approaches, it might even be beneficial to saturate the enzyme active site to preclude or discourage discovery of substrate-competitive inhibitors [80].
What Is the Appropriate Chemical Space for Libraries that Target Non-Enzymes?
One theory to describe the apparent “un-druggability” of a non-enzyme target is that the types of molecules being used in most HTS campaigns do not sample the appropriate chemical space [81, 82]. For example, commercial chemical libraries appear to be ill suited for the discovery of inhibitors that bind PPIs [82, 83]. Inhibitors of PPIs tend to have higher molecular mass and more complex topology (e.g. macrocycles, high number of chiral centers) than inhibitors of traditional, enzyme targets (recently reviewed in [84]). Thus, the success of HTS for non-canonical targets may be critically dependent on the selection of the appropriate chemical library and seemingly failed screens for non-enzymes may, in fact, have arisen from poor sampling of chemical space. Consequently, creative construction of new chemical libraries is a vibrant and important area of research that is likely to expand our definition of “druggable” targets.
Diversity-oriented synthesis (DOS) is one approach to expand the chemical space sampled by synthetic chemical libraries. Many current HTS libraries consist of molecules representing a relatively small number of chemical scaffolds, with physicochemical properties resembling existing drugs [81]. DOS approaches rely on divergent synthetic steps, in which the product of one complexity-generating transformation is a substrate in a second, and so on [85–87] (Figure 3A). Thus, in contrast to target-oriented synthesis or medicinal chemistry, DOS methods tend to access structures with increased scaffold complexity and variety in a limited number of synthetic steps.
Figure 3.
A: Diversity-oriented synthesis uses sequences of modular, complexity-generating reactions to build compound libraries of diverse scaffolds (figure adapted from [87]). B: Focused libraries of natural product-inspired scaffolds and cyclic peptides may be useful for lead generation against non-enzymes and protein-protein interactions. C: Fragment-based screening enables the evolution of low-affinity, high-efficiency binders into high affinity leads.
Natural products provided some of the original inspiration for DOS libraries [88, 89], because these natural compounds tend to be more structurally complex, with more chiral centers, a higher proportion of carbon, hydrogen, and oxygen atoms and fewer nitrogen atoms than synthetic compounds (reviewed in [90]). They also tend to be larger (> 500 Da) and frequently more water-soluble [91]. These compounds have evolved to be bioactive; thus, they tend to have relatively favorable pharmacokinetic properties and high affinity and specificity [91]. Unsurprisingly, a large proportion (>60%) of FDA-approved drugs are natural products or natural product derivatives [92–94]. Inspired by these favorable properties, libraries assembled based on privileged core natural products have been constructed around a number of scaffolds, including carbohydrates, steroids and sterols, fatty acid derivatives, polyketides, linear and cyclic peptides, terpenoids, flavonoids, alkaloids, macrolactones and macrolactams, and many others [90] (Figure 3B). In a related concept, Hopkins and Groom presented the idea that the majority of drugs compete against endogenous small molecule regulators for binding sites on proteins [95]. This concept has led to the use of metabolomic profiling as a way to identify druggable binding sites, and to the development of metabolite and cofactor mimetic libraries [96].
Another interesting property of natural products is that they can sometimes inhibit otherwise intractable classes of drug targets, such as PPIs [97]. For example, we recently screened a small library of plant-derived natural products and successfully identified inhibitors of the challenging PPI between the anti-bacterial targets DnaK and DnaJ [98, 99]. The difficulty of targeting PPIs using commercial libraries is thought to result, in part, from incompatible physicochemical properties [22, 83]. For example, a 2010 analysis compared 66 PPI inhibitors with a diverse set of 557 typical drugs, using 1,666 molecular descriptors [83]. The study concluded that PPI inhibitors are larger, more lipophilic, and have more aromatic rings and fused ring systems [83]. Thus, natural products may be especially suitable for targeting PPIs, as many natural products overlap with this region of chemical space [81, 100].
Ribosomal and non-ribosomal peptides are natural products that exhibit a wide range of biological activities. Synthetic peptides are often assembled by solid phase synthesis, using functionalized polystyrene resin beads as solid support. Natural and unnatural amino acids may be modularly incorporated to rapidly assemble a large amount of diversity using split-and-pool methods. In one-bead-one-compound combinatorial libraries, each solid-support resin bead is coated with a homogenous population of a unique peptide or peptoid [101–103]. Such libraries can then be incubated with a fluorescently labeled target of interest to find binding partners. Like other affinity-based selection techniques, such as phage display [104], this platform can be applied to any type of target molecule, even non-enzymes [102]. Linear peptides generally have poor pharmacokinetic properties (poor absorption and susceptibility to rapid degradation by proteases), but this can be circumvented using a number of well-established strategies [105]. For example, synthetic biological agents such as stapled peptides with covalently constrained secondary structure may be cell-permeable and resistant to cellular proteases [106, 107]. Moreover, the conformational restriction imposed by the covalent stabilization of secondary structure can efficiently mimic the binding surface of a protein (most notably α-helices), resulting in tight and productive binding. Stapled peptides have been successfully developed to modulate a number of noncanonical targets such as transcription factors as well as PPIs [108–110]. Cyclic peptides are another class of natural products suitable for use in targeting non-enzymes. The reduced conformational flexibility of cyclized peptides is advantageous for target binding, membrane permeability, and stabilization against digestion by endoproteases [111]. Until recently, the one-bead-one-compound technique was limited to the screening of linear peptide or peptoid libraries, because Edman degradation sequencing requires a free amino terminus. However, Liu et al. developed a clever strategy to circumvent this obstacle [111], including both linear and cyclic versions of peptides embedded on either the inner or outer layers of a polymeric resin support (Figure 3B) [111]. In a proof-of-principle study, a library of 107 cyclic peptides was generated and screened against the human prolactin receptor, resulting in molecules with low micromolar affinity for an allosteric site on the receptor [111]. Similarly, a screen of a focused library of over 106 cyclic peptides designed to competitively inhibit the calcineurin-NFAT PPI resulted in the discovery of several ligands with low micromolar potency [112].
DNA-encoded libraries (DELs) are another way to select for small molecules with affinity for a target of interest [29, 113, 114]. Analogous to phage-display, DELs link small molecule selection with unique, covalently attached DNA “bar codes” [114]. Molecules with affinity for the target are identified by PCR amplification and sequencing of the DNA tag. DELs may be synthesized using split-and-pool combinatorial assembly or DNA-templated synthetic methods [113]. For example, Wrenn et al. synthesized and screened 108 DNA-encoded 8-mer peptoids for binding to the N-terminal SH3 domain of the proto-oncogene Crk (p38) [28], which successfully resulted in the identification of several peptoids with low- to mid-micromolar affinity for Crk. One drawback of this approach is that synthetic transformations used in library construction must be DNA-compatible, but a relatively wide range of orthogonal reactions have been reported [115]. Compound discovery by DELs may be applied to any type of target class, and is relatively inexpensive after the initial investment of library construction.
Fragment-based screening utilizes chemical libraries consisting of low-molecular weight, low complexity compounds (“fragments”), which are tested for binding to a target protein by NMR, x-ray crystallography, or SPR. Low-affinity hits (generally with KD values between 0.1 and 10 mM [13]) are then evolved into higher-affinity binders though structure-based design and medicinal chemistry (Figure 3C). Fragment-based screening has gained widespread application over the past decade [116], and provides several strategic advantages. First, fragment-based screening libraries exhibit high sampling efficiency; they offer greater coverage of chemical space with a smaller number of library members [117]. In fact, a library of 103 fragments represents the same chemical space as 1013 drug-like molecules [117]. Second, fragment based screening produces weak but high-quality binders. Absolute binding affinities range from micromolar to millimolar, but ligand efficiency (or binding energy per non-hydrogen atom [118]), is comparable or stronger than HTS hits [119]. The reason for this observation is that molecules binding to their target must overcome the entropic cost of the interaction, estimated for a rigid body to be ~15–20 kJ/mol [118]. As a result, a fragment that binds with 100 μM affinity actually contributes over half of the binding energy to an optimized, nanomolar KD molecule [118], as long as the incorporated fragment still takes advantage of the same binding interactions. Lastly, fragment hits have favorable physicochemical properties as starting points for pharmaceutical design. As compared to typical HTS hits, fragment hits are much lower in molecular weight, less lipophilic, and more soluble [120].
It may be the case that “failure of a [well-designed] screen to identify a chemical starting point can be simplified to one of two factors: the target itself is un-druggable (unable to be modulated appropriately by a small molecule), or the screen did not test the correct compounds (yet)” [121]. The expansion of screening collections may therefore increase the number of targets that are considered “druggable.”
The Concept of “Druggability” is Evolving
There is a fundamental dilemma associated with categorizing “druggable” targets and “drug-like” molecules on the basis of past success stories [121]. If we categorize “druggable” targets as only those that resemble successfully drugged targets, and “drug-like” small molecules as only those that resemble current FDA-approved compounds, then we discourage innovation and exclude the possibility that either target space or drug space might be expanded by new technology. In other words, until we try – and fail – it is not clear that any target is “undruggable” and, even then, it is only “undruggable” under the current paradigm. Accordingly, we have focused this review on high-throughput methods for selecting ligands with affinity for non-canonical targets. In many cases, these methods have been used against targets that lack enzymatic activity, structural information, existing ligands, or known ligand binding sites. Combined with efforts to expand chemical space and enrich for modulators of non-canonical targets, these advances are helping to expand the definition of “druggable”. However, it is unlikely that these examples represent the final word on drug discovery for post-genomic targets. The real lesson is that no target is “undruggable”.
Acknowledgments
The authors thank members of the Gestwicki Lab for helpful conversations. Our work on protein-protein interactions is funded by NIH grant NS059690 and NSF grant MCB-0844512. L.N.M. is funded by an NIH pre-doctoral fellowship (GM007767) and the American Foundation for Pharmaceutical Education.
References
- 1.Overington JP, Al-Lazikani B, Hopkins AL. Opinion - How many drug targets are there? Nature Reviews Drug Discovery. 2006;5(12):993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 2.Imming P, Sinning C, Meyer A. Opinion - Drugs, their targets and the nature and number of drug targets. Nature Reviews Drug Discovery. 2006;5(10):821–834. doi: 10.1038/nrd2132. [DOI] [PubMed] [Google Scholar]
- 3.Rual JF, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437(7062):1173–8. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 4.Arkin MR, Whitty A. The road less traveled: modulating signal transduction enzymes by inhibiting their protein-protein interactions. Curr Opin Chem Biol. 2009;13(3):284–90. doi: 10.1016/j.cbpa.2009.05.125. [DOI] [PubMed] [Google Scholar]
- 5.Smith MC, Gestwicki JE. Features of protein-protein interactions that translate into potent inhibitors: topology, surface area and affinity. Expert Rev Mol Med. 2012;14:e16. doi: 10.1017/erm.2012.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thompson AD, et al. Fine-tuning multiprotein complexes using small molecules. ACS Chemical Biology. 2012;7(8):1311–1320. doi: 10.1021/cb300255p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wendt MD, et al. Discovery of a novel small molecule binding site of human survivin. Bioorg Med Chem Lett. 2007;17(11):3122–9. doi: 10.1016/j.bmcl.2007.03.042. [DOI] [PubMed] [Google Scholar]
- 8.Renaud JP, Delsuc MA. Biophysical techniques for ligand screening and drug design. Curr Opin Pharmacol. 2009;9(5):622–8. doi: 10.1016/j.coph.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 9.Holdgate GA, et al. Affinity-based, biophysical methods to detect and analyze ligand binding to recombinant proteins: matching high information content with high throughput. J Struct Biol. 2010;172(1):142–57. doi: 10.1016/j.jsb.2010.06.024. [DOI] [PubMed] [Google Scholar]
- 10.Dalvit C, et al. WaterLOGSY as a method for primary NMR screening: practical aspects and range of applicability. J Biomol NMR. 2001;21(4):349–59. doi: 10.1023/a:1013302231549. [DOI] [PubMed] [Google Scholar]
- 11.Mayer M, Meyer B. Characterization of ligand binding by saturation transfer difference NMR spectroscopy. Angewandte Chemie-International Edition. 1999;38(12):1784–1788. doi: 10.1002/(SICI)1521-3773(19990614)38:12<1784::AID-ANIE1784>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 12.Hajduk PJ, Olejniczak ET, Fesik SW. One-dimensional relaxation- and diffusion-edited NMR methods for screening compounds that bind to macromolecules. Journal of the American Chemical Society. 1997;119(50):12257–12261. [Google Scholar]
- 13.Scott DE, et al. Fragment-Based Approaches in Drug Discovery and Chemical Biology. Biochemistry. 2012 doi: 10.1021/bi3005126. [DOI] [PubMed] [Google Scholar]
- 14.Brough PA, et al. Combining hit identification strategies: fragment-based and in silico approaches to orally active 2-aminothieno[2,3-d]pyrimidine inhibitors of the Hsp90 molecular chaperone. J Med Chem. 2009;52(15):4794–809. doi: 10.1021/jm900357y. [DOI] [PubMed] [Google Scholar]
- 15.Stockman BJ, et al. Identification of allosteric PIF-pocket ligands for PDK1 using NMR-based fragment screening and 1H-15N TROSY experiments. Chem Biol Drug Des. 2009;73(2):179–88. doi: 10.1111/j.1747-0285.2008.00768.x. [DOI] [PubMed] [Google Scholar]
- 16.Jahnke W, et al. Strategies for the NMR-based identification and optimization of allosteric protein kinase inhibitors. Chembiochem. 2005;6(9):1607–10. doi: 10.1002/cbic.200500100. [DOI] [PubMed] [Google Scholar]
- 17.Kristiansen M, et al. Identification, synthesis, and characterization of new glycogen phosphorylase inhibitors binding to the allosteric AMP site. J Med Chem. 2004;47(14):3537–45. doi: 10.1021/jm031121n. [DOI] [PubMed] [Google Scholar]
- 18.Krimm I, Lancelin JM, Praly JP. Binding evaluation of fragment-based scaffolds for probing allosteric enzymes. J Med Chem. 2012;55(3):1287–95. doi: 10.1021/jm201439b. [DOI] [PubMed] [Google Scholar]
- 19.Jahnke W, et al. Allosteric non-bisphosphonate FPPS inhibitors identified by fragment-based discovery. Nat Chem Biol. 2010;6(9):660–6. doi: 10.1038/nchembio.421. [DOI] [PubMed] [Google Scholar]
- 20.Hajduk PJ, Huth JR, Fesik SW. Druggability indices for protein targets derived from NMR-based screening data. J Med Chem. 2005;48(7):2518–25. doi: 10.1021/jm049131r. [DOI] [PubMed] [Google Scholar]
- 21.Arkin MR, Wells JA. Small-molecule inhibitors of protein-protein interactions: progressing towards the dream. Nat Rev Drug Discov. 2004;3(4):301–17. doi: 10.1038/nrd1343. [DOI] [PubMed] [Google Scholar]
- 22.Pagliaro L, et al. Emerging classes of protein-protein interaction inhibitors and new tools for their development. Curr Opin Chem Biol. 2004;8(4):442–9. doi: 10.1016/j.cbpa.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 23.Altieri DC. Survivin, cancer networks and pathway-directed drug discovery. Nat Rev Cancer. 2008;8(1):61–70. doi: 10.1038/nrc2293. [DOI] [PubMed] [Google Scholar]
- 24.Altieri DC. Targeting survivin in cancer. Cancer Lett. 2012 doi: 10.1016/j.canlet.2012.03.005. http://dx.doi.org/10.1016/j.canlet.2012.03.005. [DOI] [PMC free article] [PubMed]
- 25.Zaffaroni N, Pennati M, Daidone MG. Survivin as a target for new anticancer interventions. J Cell Mol Med. 2005;9(2):360–72. doi: 10.1111/j.1582-4934.2005.tb00361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carrasco RA, et al. Antisense inhibition of survivin expression as a cancer therapeutic. Mol Cancer Ther. 2011;10(2):221–32. doi: 10.1158/1535-7163.MCT-10-0756. [DOI] [PubMed] [Google Scholar]
- 27.Jonker N, et al. Recent developments in protein-ligand affinity mass spectrometry. Anal Bioanal Chem. 2011;399(8):2669–81. doi: 10.1007/s00216-010-4350-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wrenn SJ, et al. Synthetic ligands discovered by in vitro selection. J Am Chem Soc. 2007;129(43):13137–43. doi: 10.1021/ja073993a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clark MA. Selecting chemicals: the emerging utility of DNA-encoded libraries. Curr Opin Chem Biol. 2010;14(3):396–403. doi: 10.1016/j.cbpa.2010.02.017. [DOI] [PubMed] [Google Scholar]
- 30.Vassilev LT, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303(5659):844–8. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 31.GE Healthcare Biacore 4000 Systems Overview. http://www.biacore.com/lifesciences/products/systems_overview/Biacore_4000/System-Information/index.html.
- 32.ForteBio OctetRed384 System Specifications. http://www.fortebio.com/octet_384_specs.html.
- 33.Koehler AN, Shamji AF, Schreiber SL. Discovery of an inhibitor of a transcription factor using small molecule microarrays and diversity-oriented synthesis. J Am Chem Soc. 2003;125(28):8420–1. doi: 10.1021/ja0352698. [DOI] [PubMed] [Google Scholar]
- 34.Stanton BZ, et al. A small molecule that binds Hedgehog and blocks its signaling in human cells. Nat Chem Biol. 2009;5(3):154–6. doi: 10.1038/nchembio.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peng LF, et al. Syntheses of aminoalcohol-derived macrocycles leading to a small-molecule binder to and inhibitor of Sonic Hedgehog. Bioorg Med Chem Lett. 2009;19(22):6319–25. doi: 10.1016/j.bmcl.2009.09.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Landry JP, Fei Y, Zhu XD. High Throughput, Label-free Screening Small Molecule Compound Libraries for Protein-Ligands using Combination of Small Molecule Microarrays and a Special Ellipsometry-based Optical Scanner. Int Drug Discov. 2011:8–13. [PMC free article] [PubMed] [Google Scholar]
- 37.Landry JP, Fei Y, Zhu X. Simultaneous Measurement of 10,000 Protein-Ligand Affinity Constants Using Microarray-Based Kinetic Constant Assays. Assay Drug Dev Technol. 2012;10(3):250–259. doi: 10.1089/adt.2011.0406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fei YY, et al. A novel high-throughput scanning microscope for label-free detection of protein and small-molecule chemical microarrays. Rev Sci Instrum. 2008;79(1):013708. doi: 10.1063/1.2830286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ulloa-Aguirre A, et al. Pharmacologic rescue of conformationally-defective proteins: implications for the treatment of human disease. Traffic. 2004;5(11):821–37. doi: 10.1111/j.1600-0854.2004.00232.x. [DOI] [PubMed] [Google Scholar]
- 40.Kozarich JW. The biochemistry of disease: desperately seeking syzygy. Annu Rev Biochem. 2009;78:55–63. doi: 10.1146/annurev-biochem-120108-082254. [DOI] [PubMed] [Google Scholar]
- 41.Pey AL, et al. Identification of pharmacological chaperones as potential therapeutic agents to treat phenylketonuria. J Clin Invest. 2008;118(8):2858–67. doi: 10.1172/JCI34355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sawkar AR, D’Haeze W, Kelly JW. Therapeutic strategies to ameliorate lysosomal storage disorders--a focus on Gaucher disease. Cell Mol Life Sci. 2006;63(10):1179–92. doi: 10.1007/s00018-005-5437-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu Z, Sawkar AR, Kelly JW. Pharmacologic chaperoning as a strategy to treat Gaucher disease. FEBS J. 2007;274(19):4944–50. doi: 10.1111/j.1742-4658.2007.06042.x. [DOI] [PubMed] [Google Scholar]
- 44.Tropak MB, et al. High-throughput screening for human lysosomal beta-N-Acetyl hexosaminidase inhibitors acting as pharmacological chaperones. Chem Biol. 2007;14(2):153–64. doi: 10.1016/j.chembiol.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Y, et al. Additive effect of multiple pharmacological chaperones on maturation of CFTR processing mutants. Biochem J. 2007;406(2):257–63. doi: 10.1042/BJ20070478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Y, et al. Modulating the folding of P-glycoprotein and cystic fibrosis transmembrane conductance regulator truncation mutants with pharmacological chaperones. Mol Pharmacol. 2007;71(3):751–8. doi: 10.1124/mol.106.029926. [DOI] [PubMed] [Google Scholar]
- 47.Hammarstrom P, et al. Prevention of transthyretin amyloid disease by changing protein misfolding energetics. Science. 2003;299(5607):713–6. doi: 10.1126/science.1079589. [DOI] [PubMed] [Google Scholar]
- 48.Johnson SM, et al. Native state kinetic stabilization as a strategy to ameliorate protein misfolding diseases: a focus on the transthyretin amyloidoses. Acc Chem Res. 2005;38(12):911–21. doi: 10.1021/ar020073i. [DOI] [PubMed] [Google Scholar]
- 49.Johnson SM, et al. The Transthyretin Amyloidoses: From Delineating the Molecular Mechanism of Aggregation Linked to Pathology to a Regulatory-Agency-Approved Drug. J Mol Biol. 2012;421(2–3):185–203. doi: 10.1016/j.jmb.2011.12.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stanger K, et al. Allosteric peptides bind a caspase zymogen and mediate caspase tetramerization. Nat Chem Biol. 2012;8(7):655–660. doi: 10.1038/nchembio.967. [DOI] [PubMed] [Google Scholar]
- 51.Kuryatov A, et al. Nicotine acts as a pharmacological chaperone to up-regulate human alpha4beta2 acetylcholine receptors. Mol Pharmacol. 2005;68(6):1839–51. doi: 10.1124/mol.105.012419. [DOI] [PubMed] [Google Scholar]
- 52.Lester HA, et al. Nicotine is a selective pharmacological chaperone of acetylcholine receptor number and stoichiometry. Implications for drug discovery. AAPS J. 2009;11(1):167–77. doi: 10.1208/s12248-009-9090-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Holdgate GA, Ward WH. Measurements of binding thermodynamics in drug discovery. Drug Discov Today. 2005;10(22):1543–50. doi: 10.1016/S1359-6446(05)03610-X. [DOI] [PubMed] [Google Scholar]
- 54.Cummings MD, Farnum MA, Nelen MI. Universal screening methods and applications of ThermoFluor. J Biomol Screen. 2006;11(7):854–63. doi: 10.1177/1087057106292746. [DOI] [PubMed] [Google Scholar]
- 55.Niesen FH, Berglund H, Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat Protoc. 2007;2(9):2212–21. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 56.Lo MC, et al. Evaluation of fluorescence-based thermal shift assays for hit identification in drug discovery. Anal Biochem. 2004;332(1):153–9. doi: 10.1016/j.ab.2004.04.031. [DOI] [PubMed] [Google Scholar]
- 57.DeSantis K, et al. Use of differential scanning fluorimetry as a high-throughput assay to identify nuclear receptor ligands. Nucl Recept Signal. 2012;10:e002. doi: 10.1621/nrs.10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koblish HK, et al. Benzodiazepinedione inhibitors of the Hdm2:p53 complex suppress human tumor cell proliferation in vitro and sensitize tumors to doxorubicin in vivo. Mol Cancer Ther. 2006;5(1):160–9. doi: 10.1158/1535-7163.MCT-05-0199. [DOI] [PubMed] [Google Scholar]
- 59.Grasberger BL, et al. Discovery and cocrystal structure of benzodiazepinedione HDM2 antagonists that activate p53 in cells. J Med Chem. 2005;48(4):909–12. doi: 10.1021/jm049137g. [DOI] [PubMed] [Google Scholar]
- 60.Parks DJ, et al. 1,4-Benzodiazepine-2,5-diones as small molecule antagonists of the HDM2-p53 interaction: discovery and SAR. Bioorg Med Chem Lett. 2005;15(3):765–70. doi: 10.1016/j.bmcl.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 61.Sampson HM, et al. Identification of a NBD1-binding pharmacological chaperone that corrects the trafficking defect of F508del-CFTR. Chem Biol. 2011;18(2):231–42. doi: 10.1016/j.chembiol.2010.11.016. [DOI] [PubMed] [Google Scholar]
- 62.Forneris F, et al. ThermoFAD, a Thermofluor-adapted flavin ad hoc detection system for protein folding and ligand binding. FEBS J. 2009;276(10):2833–40. doi: 10.1111/j.1742-4658.2009.07006.x. [DOI] [PubMed] [Google Scholar]
- 63.Isom DG, et al. A miniaturized technique for assessing protein thermodynamics and function using fast determination of quantitative cysteine reactivity. Proteins. 2010;79(4):1034–1047. doi: 10.1002/prot.22932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ghaemmaghami S, Fitzgerald MC, Oas TG. A quantitative, high-throughput screen for protein stability. Proc Natl Acad Sci U S A. 2000;97(15):8296–301. doi: 10.1073/pnas.140111397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tang L, et al. H/D exchange- and mass spectrometry-based strategy for the thermodynamic analysis of protein-ligand binding. Anal Chem. 2007;79(15):5869–77. doi: 10.1021/ac0700777. [DOI] [PubMed] [Google Scholar]
- 66.Powell KD, Fitzgerald MC. High-throughput screening assay for the tunable selection of protein ligands. J Comb Chem. 2004;6(2):262–9. doi: 10.1021/cc034051e. [DOI] [PubMed] [Google Scholar]
- 67.Hopper ED, et al. Throughput and efficiency of a mass spectrometry-based screening assay for protein-ligand binding detection. J Am Soc Mass Spectrom. 2008;19(9):1303–11. doi: 10.1016/j.jasms.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dearmond PD, et al. Discovery of novel cyclophilin A ligands using an H/D exchange- and mass spectrometry-based strategy. J Biomol Screen. 2010;15(9):1051–62. doi: 10.1177/1087057110382775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Perot S, et al. Druggable pockets and binding site centric chemical space: a paradigm shift in drug discovery. Drug Discov Today. 2010;15(15–16):656–67. doi: 10.1016/j.drudis.2010.05.015. [DOI] [PubMed] [Google Scholar]
- 70.Henrich S, et al. Computational approaches to identifying and characterizing protein binding sites for ligand design. J Mol Recognit. 2010;23(2):209–19. doi: 10.1002/jmr.984. [DOI] [PubMed] [Google Scholar]
- 71.An JH, Totrov M, Abagyan R. Pocketome via comprehensive identification and classification of ligand binding envelopes. Molecular & Cellular Proteomics. 2005;4(6):752–761. doi: 10.1074/mcp.M400159-MCP200. [DOI] [PubMed] [Google Scholar]
- 72.Kozakov D, et al. Structural conservation of druggable hot spots in protein-protein interfaces. Proc Natl Acad Sci U S A. 2011;108(33):13528–33. doi: 10.1073/pnas.1101835108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mattos C, Ringe D. Locating and characterizing binding sites on proteins. Nat Biotechnol. 1996;14(5):595–9. doi: 10.1038/nbt0596-595. [DOI] [PubMed] [Google Scholar]
- 74.Lexa KW, Carlson HA. Full Protein Flexibility Is Essential for Proper Hot-Spot Mapping. J Am Chem Soc. 2010;133(2):200–202. doi: 10.1021/ja1079332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cheng AC, et al. Structure-based maximal affinity model predicts small-molecule druggability. Nat Biotechnol. 2007;25(1):71–5. doi: 10.1038/nbt1273. [DOI] [PubMed] [Google Scholar]
- 76.Halgren TA. Identifying and characterizing binding sites and assessing druggability. J Chem Inf Model. 2009;49(2):377–89. doi: 10.1021/ci800324m. [DOI] [PubMed] [Google Scholar]
- 77.Zhong S, MacKerell AD., Jr Binding response: a descriptor for selecting ligand binding site on protein surfaces. J Chem Inf Model. 2007;47(6):2303–15. doi: 10.1021/ci700149k. [DOI] [PubMed] [Google Scholar]
- 78.Nayal M, Honig B. On the nature of cavities on protein surfaces: application to the identification of drug-binding sites. Proteins. 2006;63(4):892–906. doi: 10.1002/prot.20897. [DOI] [PubMed] [Google Scholar]
- 79.Dar AC, Shokat KM. The evolution of protein kinase inhibitors from antagonists to agonists of cellular signaling. Annu Rev Biochem. 2011;80:769–95. doi: 10.1146/annurev-biochem-090308-173656. [DOI] [PubMed] [Google Scholar]
- 80.Chang L, et al. Chemical screens against a reconstituted multiprotein complex: myricetin blocks DnaJ regulation of DnaK through an allosteric mechanism. Chemical Biology. 2011;18(2):210–221. doi: 10.1016/j.chembiol.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bauer RA, Wurst JM, Tan DS. Expanding the range of ‘druggable’ targets with natural product-based libraries: an academic perspective. Curr Opin Chem Biol. 2010;14(3):308–14. doi: 10.1016/j.cbpa.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dandapani S, Marcaurelle LA. Grand challenge commentary: Accessing new chemical space for ‘undruggable’ targets. Nat Chem Biol. 2010;6(12):861–3. doi: 10.1038/nchembio.479. [DOI] [PubMed] [Google Scholar]
- 83.Sperandio O, et al. Rationalizing the chemical space of protein-protein interaction inhibitors. Drug Discov Today. 2010;15(5–6):220–9. doi: 10.1016/j.drudis.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 84.Smith MC, Gestwicki JE. Features of protein-protein interactions that translate into potent inhibitors: topology, surface area and affinity. Expert Reviews in Molecular Medicine. 2012 doi: 10.1017/erm.2012.10. http://dx.doi.org/10.1017/erm.2012.10. [DOI] [PMC free article] [PubMed]
- 85.Schreiber SL. Organic chemistry: Molecular diversity by design. Nature. 2009;457(7226):153–4. doi: 10.1038/457153a. [DOI] [PubMed] [Google Scholar]
- 86.Galloway WR, Isidro-Llobet A, Spring DR. Diversity-oriented synthesis as a tool for the discovery of novel biologically active small molecules. Nat Commun. 2010;1:80. doi: 10.1038/ncomms1081. [DOI] [PubMed] [Google Scholar]
- 87.Burke MD, Schreiber SL. A planning strategy for diversity-oriented synthesis. Angew Chem Int Ed Engl. 2004;43(1):46–58. doi: 10.1002/anie.200300626. [DOI] [PubMed] [Google Scholar]
- 88.Isidro-Llobet A, et al. Diversity-oriented synthesis of macrocyclic peptidomimetics. Proc Natl Acad Sci U S A. 2011;108(17):6793–8. doi: 10.1073/pnas.1015267108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Renner S, et al. Recent trends and observations in the design of high-quality screening collections. Future Med Chem. 2011;3(6):751–66. doi: 10.4155/fmc.11.15. [DOI] [PubMed] [Google Scholar]
- 90.Boldi AM. Libraries from natural product-like scaffolds. Curr Opin Chem Biol. 2004;8(3):281–6. doi: 10.1016/j.cbpa.2004.04.010. [DOI] [PubMed] [Google Scholar]
- 91.Clardy J, Walsh C. Lessons from natural molecules. Nature. 2004;432(7019):829–37. doi: 10.1038/nature03194. [DOI] [PubMed] [Google Scholar]
- 92.Harvey AL. Natural products as a screening resource. Curr Opin Chem Biol. 2007;11(5):480–4. doi: 10.1016/j.cbpa.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 93.Harvey AL. Natural products in drug discovery. Drug Discov Today. 2008;13(19–20):894–901. doi: 10.1016/j.drudis.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 94.Bottcher T, Pitscheider M, Sieber SA. Natural products and their biological targets: proteomic and metabolomic labeling strategies. Angew Chem Int Ed Engl. 2010;49(15):2680–98. doi: 10.1002/anie.200905352. [DOI] [PubMed] [Google Scholar]
- 95.Hopkins AL, Groom CR. The druggable genome. Nat Rev Drug Discov. 2002;1(9):727–30. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 96.Zhang L, et al. High-throughput synergy screening identifies microbial metabolites as combination agents for the treatment of fungal infections. Proc Natl Acad Sci U S A. 2007;104(11):4606–11. doi: 10.1073/pnas.0609370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hegde NS, et al. The transcription factor FOXM1 is a cellular target of the natural product thiostrepton. Nat Chem. 2011;3(9):725–31. doi: 10.1038/nchem.1114. [DOI] [PubMed] [Google Scholar]
- 98.Chang L, et al. Chemical screens against a reconstituted multiprotein complex: myricetin blocks DnaJ regulation of DnaK through an allosteric mechanism. Chem Biol. 2011;18(2):210–21. doi: 10.1016/j.chembiol.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Evans CG, Chang L, Gestwicki JE. Heat shock protein 70 (hsp70) as an emerging drug target. J Med Chem. 2010;53(12):4585–602. doi: 10.1021/jm100054f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Koehn FE, Carter GT. The evolving role of natural products in drug discovery. Nat Rev Drug Discov. 2005;4(3):206–20. doi: 10.1038/nrd1657. [DOI] [PubMed] [Google Scholar]
- 101.Lam KS, Lebl M, Krchnak V. The “One-Bead-One-Compound” Combinatorial Library Method. Chem Rev. 1997;97(2):411–448. doi: 10.1021/cr9600114. [DOI] [PubMed] [Google Scholar]
- 102.Chen X, et al. Expanded polyglutamine-binding peptoid as a novel therapeutic agent for treatment of Huntington’s disease. Chem Biol. 2011;18(9):1113–25. doi: 10.1016/j.chembiol.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zuckermann RN, Kodadek T. Peptoids as potential therapeutics. Current Opinion in Molecular Therapeutics. 2009;11(3):299–307. [PubMed] [Google Scholar]
- 104.Pande J, Szewczyk MM, Grover AK. Phage display: concept, innovations, applications and future. Biotechnol Adv. 2010;28(6):849–58. doi: 10.1016/j.biotechadv.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 105.Audie J, Boyd C. The synergistic use of computation, chemistry and biology to discover novel peptide-based drugs: the time is right. Curr Pharm Des. 2010;16(5):567–82. doi: 10.2174/138161210790361425. [DOI] [PubMed] [Google Scholar]
- 106.Harrison RS, et al. Downsizing human, bacterial, and viral proteins to short water-stable alpha helices that maintain biological potency. Proc Natl Acad Sci U S A. 2010;107(26):11686–91. doi: 10.1073/pnas.1002498107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Verdine GL, Hilinski GJ. All-hydrocarbon stapled peptides as Synthetic Cell-Accessible Mini-Proteins. Drug Discov Today. 2012;9(1):e41–e47. doi: 10.1016/j.ddtec.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 108.Kawamoto SA, et al. Design of triazole-stapled BCL9 alpha-helical peptides to target the beta-catenin/B-cell CLL/lymphoma 9 (BCL9) protein-protein interaction. J Med Chem. 2012;55(3):1137–46. doi: 10.1021/jm201125d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Madden MM, et al. Synthesis of cell-permeable stapled peptide dual inhibitors of the p53-Mdm2/Mdmx interactions via photoinduced cycloaddition. Bioorg Med Chem Lett. 2011;21(5):1472–5. doi: 10.1016/j.bmcl.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Moellering RE, et al. Direct inhibition of the NOTCH transcription factor complex. Nature. 2009;462(7270):182–8. doi: 10.1038/nature08543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Liu T, et al. Synthesis and screening of a cyclic peptide library: discovery of small-molecule ligands against human prolactin receptor. Bioorg Med Chem. 2009;17(3):1026–33. doi: 10.1016/j.bmc.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Liu T, et al. High-throughput screening of one-bead-one-compound libraries: identification of cyclic peptidyl inhibitors against calcineurin/NFAT interaction. ACS Comb Sci. 2011;13(5):537–46. doi: 10.1021/co200101w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kleiner RE, Dumelin CE, Liu DR. Small-molecule discovery from DNA-encoded chemical libraries. Chem Soc Rev. 2011;40(12):5707–17. doi: 10.1039/c1cs15076f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mannocci L, et al. 20 years of DNA-encoded chemical libraries. Chem Commun (Camb) 2011;47(48):12747–53. doi: 10.1039/c1cc15634a. [DOI] [PubMed] [Google Scholar]
- 115.Gartner ZJ, Kanan MW, Liu DR. Expanding the reaction scope of DNA-templated synthesis. Angew Chem Int Ed Engl. 2002;41(10):1796–800. doi: 10.1002/1521-3773(20020517)41:10<1796::aid-anie1796>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 116.Leach AR, Hann MM. Molecular complexity and fragment-based drug discovery: ten years on. Curr Opin Chem Biol. 2011;15(4):489–96. doi: 10.1016/j.cbpa.2011.05.008. [DOI] [PubMed] [Google Scholar]
- 117.Edfeldt FN, Folmer RH, Breeze AL. Fragment screening to predict druggability (ligandability) and lead discovery success. Drug Discov Today. 2011;16(7–8):284–7. doi: 10.1016/j.drudis.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 118.Murray CW, Rees DC. The rise of fragment-based drug discovery. Nat Chem. 2009;1(3):187–92. doi: 10.1038/nchem.217. [DOI] [PubMed] [Google Scholar]
- 119.Carr RA, et al. Fragment-based lead discovery: leads by design. Drug Discov Today. 2005;10(14):987–92. doi: 10.1016/S1359-6446(05)03511-7. [DOI] [PubMed] [Google Scholar]
- 120.Keseru GM, Makara GM. The influence of lead discovery strategies on the properties of drug candidates. Nat Rev Drug Discov. 2009;8(3):203–12. doi: 10.1038/nrd2796. [DOI] [PubMed] [Google Scholar]
- 121.Drewry DH, Macarron R. Enhancements of screening collections to address areas of unmet medical need: an industry perspective. Curr Opin Chem Biol. 2010;14(3):289–98.0. doi: 10.1016/j.cbpa.2010.03.024. [DOI] [PubMed] [Google Scholar]