Abstract
Human FOXP3+CD25+CD4+ regulatory T cells (Tregs) are essential to the maintenance of immune homeostasis. Several genes are known to be important for murine Tregs, but for human Tregs the genes and underlying molecular networks controlling the suppressor function still largely remain unclear. Here, we describe a strategy to identify the key genes directly from an undirected correlation network which we reconstruct from a very high time-resolution (HTR) transcriptome during the activation of human Tregs/CD4+ T-effector cells. We show that a predicted top-ranked new key gene PLAU (the plasminogen activator urokinase) is important for the suppressor function of both human and murine Tregs. Further analysis unveils that PLAU is particularly important for memory Tregs and that PLAU mediates Treg suppressor function via STAT5 and ERK signaling pathways. Our study demonstrates the potential for identifying novel key genes for complex dynamic biological processes using a network strategy based on HTR data, and reveals a critical role for PLAU in Treg suppressor function.
Keywords: high time-resolution time series, human CD4 regulatory T cell, infer key genes from undirected gene networks, Plau knockout mice, Treg development and suppressor function
Network-based analysis of transcriptome dynamics during activation in two human T-cell subpopulations identifies key regulators, and reveals that PLAU plays a critical role in both human and murine regulatory T-cell function.
Introduction
Autoimmune diseases are currently ranked as the third most important disease category in the United States, following heart disease and cancer. Autoimmunity plays a crucial role in >80 human diseases. A subpopulation of T cells, called regulatory T cells, currently recognized as FOXP3+CD25+CD4+ regulatory T cells (Tregs), are pivotal in suppressing autoimmunity and maintaining immune homeostasis (Sakaguchi, 2004; Ochs et al, 2005; Bacchetta et al, 2007; Germain, 2008). Furthermore, Tregs suppress immune responses against allergens (Chatila, 2005), tumor cells (Knutson et al, 2007), infections (Belkaid, 2007), potentially neurodegenerative diseases (He and Balling, 2012), and other diseases (Bacchetta et al, 2007). The suppressive activity of Tregs can be beneficial or detrimental to the host. Defects in Tregs may lead to the development of autoimmunity or the failure to control the intensity of effector responses, whereas overreactive Tregs may contribute to tumor progression. Therefore, it is essential to reveal the underlying molecular mechanisms involved in Treg function.
A number of human and/or murine genes, such as FOXP3, NFAT, EOS, S1PR1, GAL1, GAL10, FGL2, and RUNX1 are important for the suppressive function and anergy of Tregs (Fontenot et al, 2003; Sakaguchi, 2005; Wu et al, 2006; Ziegler, 2006; Garin et al, 2007; Kubach et al, 2007; Ono et al, 2007; Shalev et al, 2008; Josefowicz and Rudensky, 2009; Liu et al, 2009; Pan et al, 2009; Shevach, 2009; Sakaguchi et al, 2010). Foxp3 for example has been described as a master regulator of murine Treg function and development, and a number of target genes of this transcriptional regulator have been identified by chromatin-immunoprecipitation analysis (Marson et al, 2007; Zheng et al, 2007). Although significant advances have been made in understanding murine Treg lineage commitment and function gained mainly by studying Foxp3, the dominant role of FOXP3 in human Treg cells might need to be reconsidered (Probst-Kepper et al, 2010). Differences in the role of this so-called master regulator between human and murine Tregs might indicate additional differences in the molecular networks controlling the suppressor function of human and murine Tregs. Recently, we and others have shown that GARP (LRRC32) is important for the suppressor function and the regulation of FOXP3 expression in human Tregs (Probst-Kepper et al, 2009; Wang et al, 2009). However, little has been done to systematically identify other genes involved and the underlying gene networks that control human Treg function. This is partially due to the lack of sufficient high-quality genome-wide dynamic data and the lack of a suitable strategy by which one can construct reliable gene networks and identify the key genes that regulate human Treg function (He et al, 2009).
The inference of mechanistic transcription regulatory networks in mammalian cells is an important basis to obtain insight into biological and disease processes. However, conclusions about their accuracy and the experimental validation of potential interactions that have been computationally generated are still very challenging (Walhout, 2011; Della Gatta et al, 2012; Quo et al, 2012; Stelniec-Klotz et al, 2012). Determination of multivariate dependencies of target genes, that are under the synergistic control of multiple regulators, is even more difficult and can often only be solved by approximation methods (Huynh-Thu et al, 2010; Quo et al, 2012) such as maximum entropy techniques (Margolin et al, 2010). Additional difficulties arise in validating these multiregulator-synergistic regulatory networks. Inferring gene association networks by taking into account multivariate stochastic dependencies among genes, for instance using partial correlation from static measurements is possible (also known as covariance selection) (Toh and Horimoto, 2002; de la Fuente et al, 2004; Schafer and Strimmer, 2005), which however cannot easily transfer to the cases of dynamic data. Some advanced approaches have been developed to identify synergistically interacting genes based on multivariate dependencies, which however form undirected correlation networks that therefore cannot provide direct functional regulatory evidence (Anastassiou, 2007). In the present work, we describe a simplified pairwise correlation network that nevertheless allows the identification of important network components.
Lee et al (2004) have proposed a strategy to obtain correlation linkages and constructed gene–gene correlation networks in yeast by combining different data sets under diverse conditions. Other efforts have also been made to identify correlation networks (Huttenhower et al, 2006; Langfelder and Horvath, 2008; Ruan et al, 2010). The validity of parts of such gene–gene correlation networks has been successfully demonstrated in yeast and in human B cells by small-scale experimental verification (Basso et al, 2005; Lee et al, 2007). However, the results are primarily based on the integration of static array data obtained under different conditions and thus do not take into account the dynamics of process-specific gene regulation. Furthermore, no studies attempted to identify the genes critical for a given physiological or pathological process obtained from the inferred undirected correlation networks based on gene expression data.
Here, we used a combination of high time-resolution (HTR) genome-wide gene expression analysis, systems-biology reverse engineering and small-scale biochemical, genetic, and cell-biological studies in addition to murine models to construct a correlation network and identify key genes in human Tregs during the early process of Treg activation.
Results
Generating HTR dynamic transcriptome
Based on the assumption that the key regulatory events underlying Treg activation occur very early after activation, we analyzed the dynamics of genome-wide gene expression in Tregs and CD4+ T effector cells (Teffs) during the first 6 h after stimulation.
Stable Tregs derived from sorted human CD4+CD25high T cells and alloantigen-specific Teffs derived from sorted CD4+CD25− T cells were from the same batch of Tregs and Teffs from our recent work (Probst-Kepper et al, 2009). Whereas the long-term responses of these T cells in the course of days following activation have already been characterized (Probst-Kepper et al, 2009), in the present study we employed HTR gene expression analysis with a sampling time interval of only 20 min during the first 6 h post activation of anti-CD3/-CD28/IL2 (Figure 1A). This enabled us to monitor the dynamics of immediate response of transcriptional changes, which cannot be captured by static and long time-interval measurements (He et al, 2009).
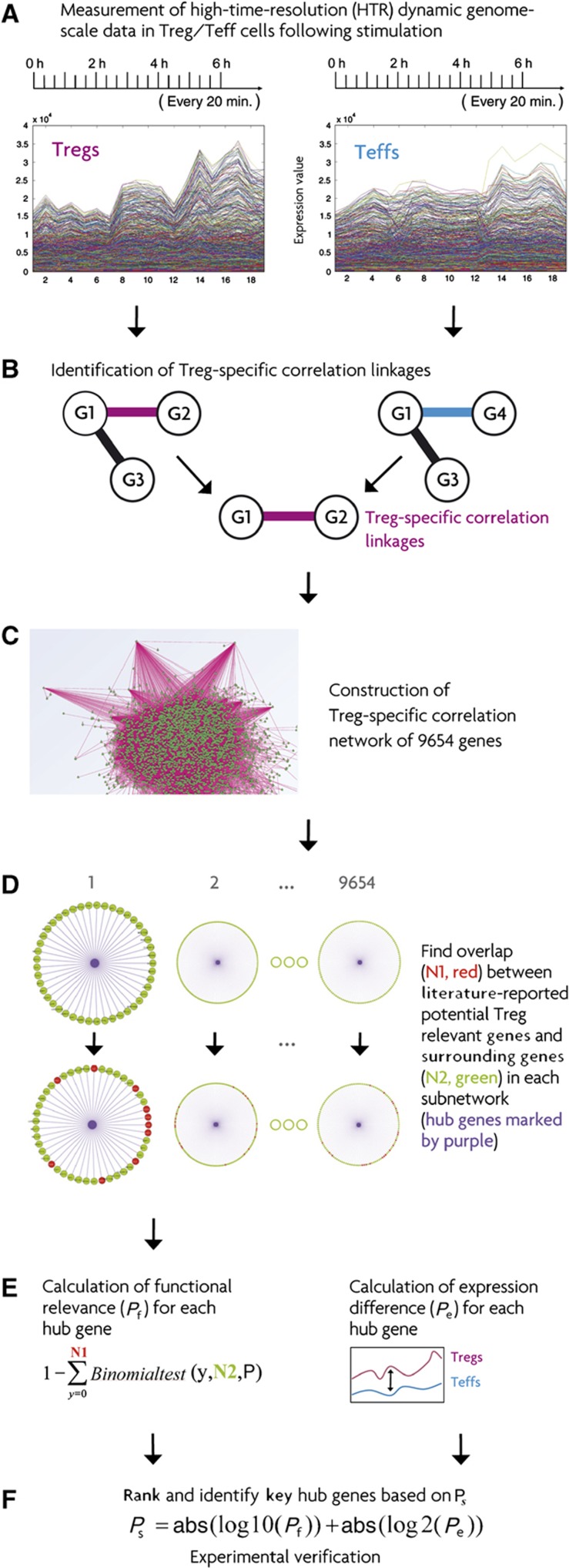
Figure 1.
Strategy for identifying novel key genes for Treg suppressor function by constructing a dynamic correlation network specific for Tregs. (A) The sampling schedule for generating high time-resolution (HTR) time-series transcriptomic data in Tregs/Teffs immediately following anti-CD28/-CD3/IL2 stimulation. (B) Construction of individual gene–gene correlation networks for Tregs/Teffs and identification of the Treg-specific correlation linkages that only exist in Tregs, but not in Teffs. (C) Construction of the Treg-specific correlation network from Treg-specific correlation linkages. (D) We first extracted the subnetwork for each given gene. The subnetwork consisted of all the genes with a correlation linkage to the given gene. We then found the overlap between the collection of literature-reported potential Treg-relevant genes and the genes with a correlation linkage to the given gene in each subnetwork. The hub gene is the purple one in the middle of each subnetwork. (E) In the left column, we inferred the functional relevance (Pf) for each hub gene by using a cumulative binomial distribution test based on the information obtained in (D). P is the overall percentage of the collection of literature-reported potential Treg genes among all the 9654 genes analyzed here. In the right column, we calculated the expression difference (Pe) between Tregs and Teffs for each hub gene. (F) We integrated the two values Pf and Pe by logarithm transforming them into a comparable range. We then identified the testable key hub candidate genes based on the ranked integrated value Ps. The sign Abs in the panel indicates the absolute value.
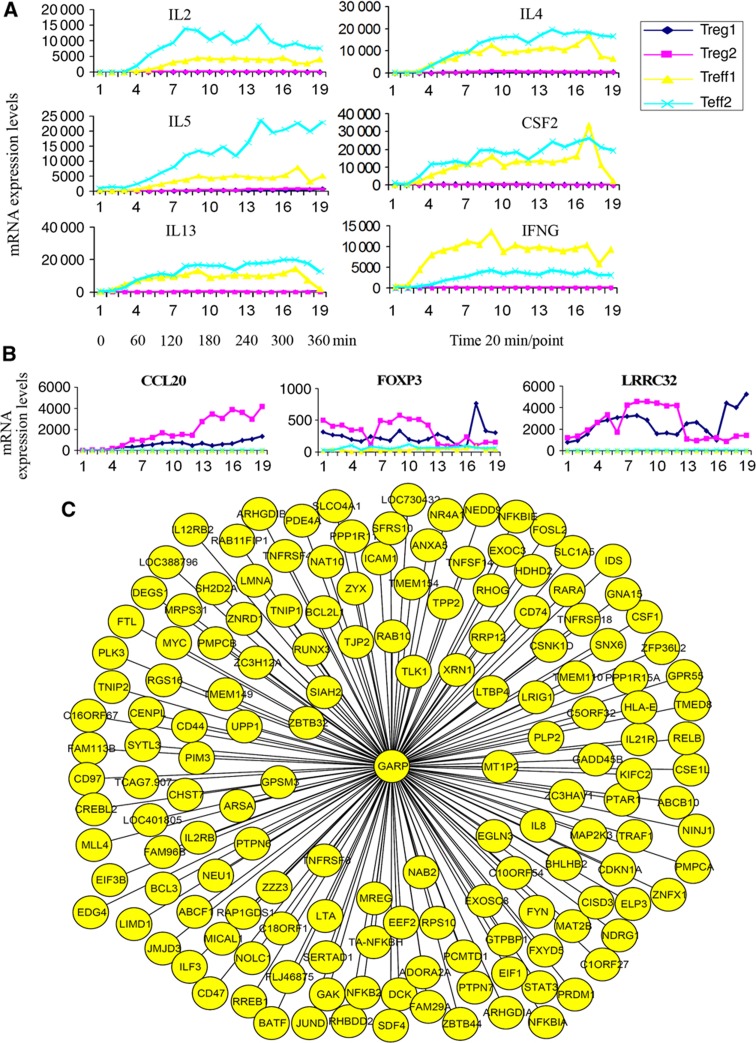
Tregs are notable for their lack of effector cytokine production in response to stimulation, as observed in long-term stimulation experiments (Sakaguchi et al, 2006). We found an almost undetectable low level of Th1 and Th2 cytokine gene expression (e.g., IL2, IL4, IL13, CSF2, and IFNG) in unstimulated Tregs and Teffs. However, surprisingly it is already within 1 h instead of days following stimulation that these cytokines were significantly upregulated in Teffs but not in Tregs (Figure 2A). An immediate-upregulation response might be partially due to the fact that most of the Tregs and Teffs become CD45RO+ memory T cells after several rounds of exposure to irradiated allogeneic Epstein-Barr virus (EBV)-transformed B cells (EBV-B cells) (Supplementary Figure S13a). Nevertheless, the demonstration of a differential expression of cytokines by Tregs and Teffs already within 1 h underscores the importance of monitoring gene expression early after activation and the necessity of sampling at a high frequency. Within 6 h, 1667 genes showed significantly higher expression in Tregs compared to Teffs, while 977 genes were expressed at lower levels in Tregs (see Materials and methods and all the related data are available on the webpage http://www.uni.lu/lcsb/publications/databases_networks_tools).
Figure 2.
Low expression of effector cytokines, high expression of CCL20, FOXP3, and GARP (LRRC32) in stimulated Tregs versus Teffs in the first 6 h and GARP subnetwork. (A) The kinetics of transcriptional expression of chosen cytokines in the first 6 h assessed by HTR time-series data. Treg1 and Treg2 (or Teff1 and Teff2) are the two independent repeated HTR time-series experiments for Tregs (or Teffs). Numbers on the x-axis indicate the corresponding sampling time points. Time point 1 is the activation starting time zero and each interval is 20 min. (B) The kinetics of CCL20 (left), FOXP3 (middle), and GARP (right) expression in the first 6 h assessed by HTR time-series data. (C) The GARP subnetwork extracted from the constructed Treg-specific correlation network based on the HTR transcription data by the proposed strategy. Each oval represents one gene. Each line between GARP and the other genes represents a correlation linkage.
The direct targets of murine Foxp3 have been reported to be significantly enriched for genes involved in the T-cell receptor signaling pathway found by chromatin immunoprecipitation-chip analysis (Marson et al, 2007). In the current study, the genes displaying a significant downregulation of expression immediately after activation were significantly enriched in the human T-cell receptor signaling pathway (Supplementary Figure S1, cumulative binomial distribution test, P-value=2.6 × 10−6). This is the first direct evidence from time-course expression results, which suggest that the human T-cell receptor signaling pathway has been significantly downregulated immediately following activation. Although the level of FOXP3 expression was much higher in Tregs versus Teffs, the expression level of FOXP3 in Tregs was not significantly changed over time after activation (Figure 2B). The HTR expression data presented here provide evidence that the role of FOXP3 in human Tregs might be overestimated and that other genes in addition to FOXP3 might play a key role in human Treg function also as indicated in a recent computational study (Fu et al, 2012).
The overlap was very significant between differentially expressed genes identified by us and those reported by other Treg-expression studies, although it could be affected by many facts, such as stimulation duration differences and the complexity of different Treg subpopulations (Feuerer et al, 2010). Approximately 57% of the genes reported by Lucas and his colleagues to be upregulated in human Tregs after stimulation for 1 day showed an overlap with the genes identified by us (Stockis et al, 2009) (P-value=6.1 × 10−6, cumulative binomial distribution test, Supplementary Table 1). Pfoertner et al (2006) have also reported upregulated and downregulated gene sets in human Tregs. Approximately 53% of their reported upregulated genes also show significantly higher expression in Tregs than in Teffs in our HTR data (P-value=1.3 × 10−9). Among the genes reported to be downregulated by them, ∼45% of the genes were also significantly downregulated in Tregs in our present work (P-value=2.8 × 10−6). In addition, Hill et al (2007) reported a so-called reliable murine Treg signature. Being aware of the differences between human and murine Tregs, ∼34% of the genes in their murine Treg signature set are also differentially expressed in human Tregs versus Teffs in the current study (P-value=5.9 × 10−3).
Constructing a Treg-specific correlation network from HTR data
On the basis of the high-quality HTR time-series data, we generated a probabilistic function-associated correlation networks, specific for Tregs. For this purpose, we used a combination of two methods, the Trend Correlation (TC) (He and Zeng, 2006) method and the Local Clustering (LC) (Qian et al, 2001) coefficient method. Both methods are able to identify dynamic correlation linkages between pairs of genes on the basis of time-series data by considering time shift and positive/inverted correlation and are based on the assumption that a robust correlation between the expression of genes over time is highly linked to the degree of functional association (Lee et al, 2004; He et al, 2009). By comparing the correlation linkages obtained between Tregs and Teffs, we identified a Treg-specific correlation network. Using stringent P-value criteria (refer to Materials and methods), we finally obtained 1 419 544 correlation linkages in the Treg-specific correlation network. In contrast, random shuffling of our data from different time points using the same methods only resulted in identification of ∼4000 correlation linkages (Supplementary Figure S2, a theoretical false positive rate of 2 × 10−3). First, we checked the overall quality of the constructed Treg-specific correlation network. Because two genes directly connected in the constructed network are expected to share the same biological process or similar function, we systematically examined the co-occurrence of two genes in the annotated same biological processes for each correlation linkage of the entire constructed correlation network. We observed that correlation linkages, in which two genes share an identical biological process, were significantly enriched for 212 out of the 860 annotated individual biological processes (Supplementary Table 2). These significantly enriched pathways include regulation of immune responses, T cell costimulation, leukocyte migration, T-cell receptor signaling, and the process involved in regulatory T-cell differentiation (Supplementary Figure S3).
Second, to evaluate the quality of the constructed Treg-specific correlation network, we examined the particular subnetworks implicated in Treg function in more detail. One of these subnetworks contains genes with a linkage to FOXP3, a known master regulator of Tregs. When we compared the genes identified in our study to a gene set composed of murine Foxp3-binding genes (Marson et al, 2007), we observed a significant enrichment by a cumulative binomial distribution test (P-value=8.9 × 10−4). A total of 18% of the genes overlapping murine Foxp3-binding genes and genes known to be involved in T-cell function were also linked to FOXP3 in our network (P-value=4.9 × 10−4). Importantly, the genes surrounding FOXP3 were significantly enriched for human genes known to be involved in autoimmune diseases (17%, P-value=2.7 × 10−6, Supplementary Table 3).
Third, we also checked the subnetwork of GARP, a gene recently identified by our group and others to be important for Treg function (Probst-Kepper et al, 2009; Wang et al, 2009; Figure 2C). As shown by the HTR data (Figure 2B), GARP was significantly upregulated immediately following activation of Tregs but not in Teffs, indicating the importance of GARP for Tregs. We identified 152 genes with a linkage to GARP (Supplementary Table 4). A total of 59 out of these 152 genes exhibited significantly differential expression in Tregs versus Teffs (Supplementary Table 4). A total of 39 genes in the GARP subnetwork overlapped with either murine Foxp3-binding genes (Marson et al, 2007), Foxp3 target genes (as reported by Marson et al, 2007), human autoimmune diseases-related genes, the murine Treg signature gene set (Hill et al, 2007) or the genes reported to be important for Treg function (Supplementary Tables 4 and 5).
Because the identification of a linkage between two genes in our correlation network might indicate either a transcriptional relationship or other functional associations, we experimentally tested whether the genes linked to GARP were also dependent on GARP expression. For this purpose, we used Teffs retrovirally overexpressing GARP as described (Probst-Kepper et al, 2009). Here, we measured and compared the expression of genes in GARP-overexpressing Teffs versus empty vector-control GFP-expressing Teffs at three time points (0, 100, and 360 min, Supplementary Table 4). Notably, 97 out of the 152 genes showed at least two-fold changes at no fewer than two measured time points (P-value=1.7 × 10−7). We found that ∼43% of the 152 genes were changed at least two-fold after stimulation at both 100 and 360 min (P-value=1.2 × 10−7). Taken together, we provide three lines of evidence confirming that the constructed Treg-specific correlation network is biologically relevant.
Identifying PLAU as a key functional hub from the constructed network
We were interested in using the constructed Treg-specific correlation network as a hypothesis generator to come up with new testable ideas concerning the function of individual genes or gene sets that might be relevant to Treg suppressor function. Using this constructed Treg-specific correlation network, we predicted and ranked key candidate genes.
Successfully extracting such information from large and highly complex gene–gene networks is still very difficult. To solve this problem, we developed a strategy to infer the functional relevance of genes from constructed undirected gene–gene networks by integrating information from subnetworks, literature mining, and expression differences.
As a starting point, we performed text mining to assemble a list of potential Treg-relevant genes to be used for predicting the functional importance of genes in the constructed network. We collected ∼400 genes mainly based on reported genomic data as described in Supplementary Table 5.
We then compared these 400 genes with those directly connected to the genes with more than a certain number of linkages in the constructed network, with 10 used here (Figure 1). Genes with >10 correlation linkages were called hubs. If the genes directly connected to a given gene were significantly enriched in the collection of 400 genes on the basis of a cumulative binomial test, then we defined this gene as a functional hub. Of note, we did not compare the 400 genes with the hubs themselves, but we inferred the importance of the hub genes according to the enrichment of the genes directly connected to them. This is based on our hypothesis that the higher the enrichment of literature-reported Treg genes directly connected to a given gene, the higher the possibility the given gene is a Treg-important gene. This method can be imagined as if one searches for a queen bee, analogous to a novel key gene, by specifically looking for those bees heavily surrounded by other bees. We call this the queen bee-surrounding principle (Figure 1).
To prioritize potential candidate hub genes for functional validation studies, we ranked functional hubs by integrating two values, which were the cumulative binomial test P-value based on the identified probabilistic functional association network indicating the potential functional importance of the hubs (Pf) and the P-value representing the degree of expression difference between Tregs and Teffs (Pe) (Figures 1E and F). Using this strategy, we expect to identify other important genes that cannot be discovered by only comparing the mRNA and/or protein expression differences that are typically not big enough to warrant further investigation.
Among the top 100 hub genes whose expression was significantly upregulated in Tregs versus Teffs, 4 hubs are known to be involved in human autoimmune diseases in OMIM database (refer to Materials and methods and Supplementary Table 6, cumulative binomial distribution, P=2.6 × 10−2). From the top 100 hub genes whose expression was significantly downregulated (refer to Materials and methods), 8 hubs were reported to be human autoimmune disease-related genes (Supplementary Table 8, P=4.6 × 10−5). 10 out of the top 100 downregulated hubs are murine Foxp3-binding genes (Supplementary Table 7, P=4.2 × 10−2). In contrast to the downregulated genes, the upregulated genes were not significantly enriched for Foxp3-binding genes among the top 100 hub genes (P=0.74), which is consistent with the report that murine Foxp3 mainly functions as a negative transcriptional regulator (Marson et al, 2007).
Surprisingly, 6 out of the top 10 ranked upregulated hub genes identified by our strategy are known to be involved in Treg suppressor function or autoimmune diseases or disorders (Table I). The top-ranked hub gene was GARP. GARP is a key receptor for Treg suppressor function, and for the regulation of FOXP3 expression in Tregs (Probst-Kepper et al, 2009; Wang et al, 2009). The second and the seventh key hub genes identified in our work were IL1R1 (CD121a) and IL1R2 (CD121b), respectively. IL1R1- and IL1R2-positive cells show significantly higher suppressive capability than IL1R1- and IL1R2-negative cells (Tran et al, 2009). We also identified CCL20 as the third-ranked gene. The receptor of CCL20, CCR6, regulates the migration of Tregs (Yamazaki et al, 2008). Though CCL20 was not expressed in murine Tregs, it was significantly upregulated in activated human Tregs but not in Teffs (Figure 2B). Another gene, ID3 (inhibitor of DNA binding 3), was ranked as the fifth key gene in our work. Id3-deficient mice develop many autoimmune disease symptoms found in primary Sjogren's syndrome patients (Li et al, 2004). Single-nucleotide polymorphisms in the neutrophil cytosolic factor 2 (NCF2) gene, the ninth-ranked hub, have been associated with systematic lupus erythematosus in a Chinese population (Yu et al, 2011).
Table 1. Top 10 ranked hub genes identified in the constructed correlation network specific for human Tregs versus Teffs.
| Gene symbol | Gene title | N1 | N2 | P f | Abs(log10(Pf)) | Abs(log2(Pe)) | Abs(log10(Pf))+Abs(log2(Pe)) | Comments |
|---|---|---|---|---|---|---|---|---|
| LRRC32 | Leucine-rich repeat containing 32 | 21 | 152 | 3.7E−07 | 6.43 | 7.81 | 14.2 | Known to be important for Treg function (Probst-Kepper et al, 2009; Wang et al, 2009) |
| IL1R1 | Interleukin 1 receptor, type I | 10 | 45 | 1.69E−06 | 5.77 | 7.12 | 12.9 | Known to be important for Treg function (Tran et al, 2009) |
| CCL20 | Chemokine (C-C motif) ligand 20 | 27 | 348 | 0.000742 | 3.13 | 7.96 | 11.1 | Its receptor CCR6 to be involved in migration of Tregs (Yamazaki et al, 2008) |
| PLAU | Plasminogen activator, urokinase | 21 | 127 | 1.38E−08 | 7.86 | 2.51 | 10.4 | Verified in this work |
| ID3 | Inhibitor of DNA binding 3, dominant-negative helix-loop-helix protein | 16 | 127 | 2.14E−05 | 4.67 | 5.44 | 10.1 | Id3-deficient mice with symptoms found in primary Sjogren's syndrome patients (Li et al, 2004) |
| SLC35F3 | Solute carrier family 35, member F3 | 37 | 407 | 3.74E−06 | 5.43 | 3.54 | 8.97 | Unknown |
| IL1R2 | Interleukin 1 receptor, type II | 27 | 307 | 9.75E−05 | 4.01 | 4.81 | 8.82 | See IL1R1 |
| FAM13A1 | Family with sequence similarity 13, member A1 | 28 | 274 | 4.64E−06 | 5.33 | 2.99 | 8.33 | Results not shown here |
| NCF2 | Neutrophil cytosolic factor 2 (65 kDa, chronic granulomatous disease, autosomal 2) | 18 | 180 | 0.000194 | 3.71 | 4.42 | 8.13 | Associated with systematic lupus erythematosus (Yu et al, 2011) |
| C16orf67 | Chromosome 16 open reading frame 67 | 33 | 371 | 1.72E−05 | 4.76 | 3.24 | 8.01 | Unknown |
Note: N1, the number of literature-reported potential Treg-relevant genes with a correlation linkage to the corresponding gene in the Treg-specific correlation network; N2, the number of genes with a correlation linkage to the corresponding gene; Pf, the P-value indicating potential functional relevance of the corresponding gene (refer to Materials and Methods); Pe, the expression difference (refer to Materials and methods).
In addition to hub genes already known to be important for Treg function or autoimmune diseases or disorders (e.g., ID3), a number of new key hub genes were identified that had so far not been reported to play a role in regulating Treg function. One of them was the plasminogen activator urokinase (PLAU) gene, ranked as the fourth hub gene in our constructed correlation network of Tregs. PLAU is a serine protease that catalyzes the conversion of plasminogen to plasmin. Two crucial regions have been defined: the N-terminal domain, which binds the receptor, and the C-terminal domain, which is endowed with catalytic activity (Mondino and Blasi, 2004). PLAU activates plasminogen into plasmin, which in turn degrades fibrin and prevents its extracellular deposition. This process might be dysregulated in several disease processes involving inflammation and tissue repair. Selective abrogation of the murine Plau-Plau-receptor interaction in vivo reveals a role for these proteins in the suppression of fibrin-associated inflammation (Connolly et al, 2010). Besides the role of PLAU in fibrinolysis, it is required for murine T lymphocyte proliferation and activation when T cells are studied as a whole population (Gyetko et al, 1999). Although the results from the whole T-cell population showed that PLAU might be required for T lymphocyte activation, it is not clear whether PLAU is involved in activation and suppressor function of Tregs, the specific subpopulation of CD4+ T cells. This is not only because the phenotype of this tiny subpopulation cannot be easily represented by the whole T lymphocytes but also because Tregs and conventional T cells (or effective T cells) play a completely opposite role and might function through different pathways.
Higher expression of PLAU in stimulated Tregs
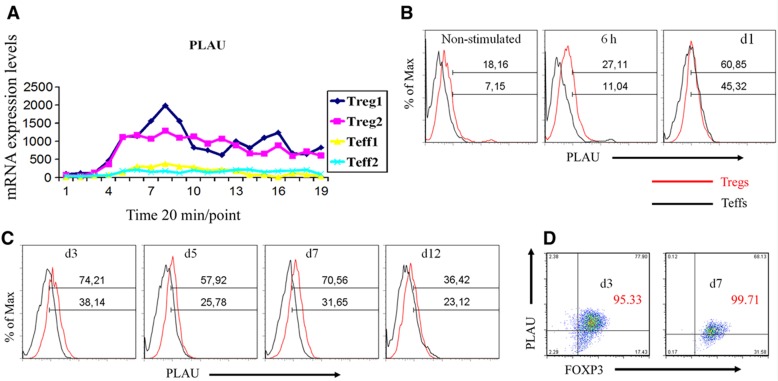
To investigate the function of PLAU in Tregs, we first characterized the kinetics of PLAU mRNA and protein expression by measuring the time course of expression of PLAU in both Tregs and Teffs following stimulation. The results show that PLAU mRNA expression was at a very low level in unstimulated Tregs and Teffs (Figure 3A). Following activation by anti-CD3/-CD28/IL2, the expression of PLAU mRNA was sharply upregulated in Tregs (Figure 3A). The expression of PLAU was also slightly upregulated in Teffs upon activation, but its expression level was significantly lower than in Tregs (Figure 3A). Such expression differences of PLAU between Tregs and Teffs were significant, but not as large as those of GARP or IL1R1 (refer to expression difference values in Table I, Figure 2B, and Supplementary Figure S13). Thus, PLAU might be overlooked by only checking expression differences. The dynamics of PLAU protein expression were measured by flow cytometry (FACS) using a PLAU-specific antibody (see Materials and methods). As shown in Figure 3B, similar to the mRNA, PLAU protein expression was higher in Tregs compared to Teffs within the first 6 h. We further monitored the protein expression of PLAU for up to 12 days (Figure 3C) following stimulation by EBV-B cells daily treated with IL2 (Probst-Kepper et al, 2009). Our results show that the protein level of PLAU in Tregs was approximately two times higher than in Teffs at different time points during the first 7 days (Figure 3C). PLAU expression declined after ∼12 days following activation (Figure 3C). We also show that the percentage of FOXP3high Treg cells was >95% at different measuring time, indicating a high purity of Tregs even after several rounds of expansion (Figure 3D). We further assessed whether PLAU was co-expressed with FOXP3. By co-staining for PLAU and FOXP3, we observed that PLAU was mainly expressed in FOXP3high Tregs following stimulation (Figure 3D). These expression data indicate that PLAU might play an important role in Treg activity.
Figure 3.
PLAU is highly expressed in Tregs. (A) mRNA expression of PLAU assessed by HTR time-series microarray in the first 6 h following anti-CD28/-CD3/IL2 stimulation. (B) Single staining of surface expression of PLAU assessed by FACS for the first day. For the first 6 h, the cells were restimulated by anti-CD28/-CD3/IL2, but for the rest of the sampling time, the cells were independently restimulated by EBV-B cells daily treated with IL2. ‘Max’, maximum. (C) Single staining of surface expression of PLAU assessed by FACS at time points from day 3 to day 12. (D) Coexpression of PLAU and FOXP3 assessed by FACS on day 3 and day 7 following EBV-B cell restimulation. Enlarged red numbers indicate the overall percentage of the two right quadrants (i.e., FOXP3+ cells). Data represent three independent experiments.
PLAU regulates FOXP3 and other important Treg genes
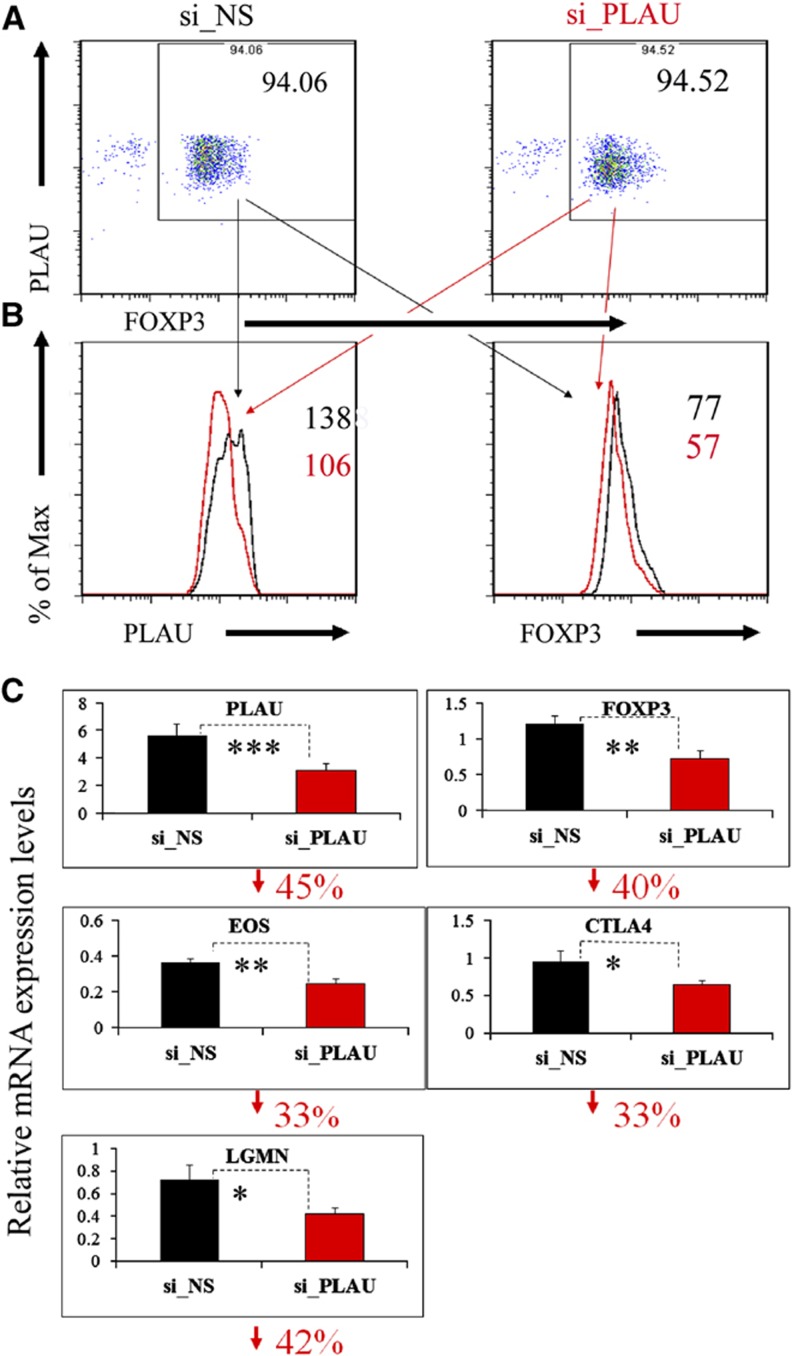
To further evaluate whether PLAU is involved in the regulation of FOXP3 expression, we assessed the regulatory effects of PLAU on FOXP3 by using small interfering RNA (siRNA) technology to knockdown PLAU. Our results show that a knockdown of PLAU significantly downregulated FOXP3 protein expression compared to a control treated with non-specific siRNA (si_NS) (Figures 4A and B). We observed a 2-day delay of FOXP3 protein downregulation compared to a decrease in PLAU protein expression. We further detected a significantly lower FOXP3 mRNA level in si_PLAU Tregs compared to control Tregs transfected with si_NS (Figure 4C). Our additional analyses show that >99.5% of the gated analyzed Tregs were DAPI-negative living cells at day 3 post siRNA transfection (Supplementary Figure S4). Together, these results demonstrate that PLAU is critical for regulating expression of FOXP3 in activated Tregs.
Figure 4.
Knockdown of PLAU downregulates FOXP3 and other known important Treg genes. (A) Knockdown of PLAU by electroporation-mediated siRNA assessed by FACS. Before performing the knockdown experiments, we first restimulated the Tregs by co-culturing them with EBV-B cells for 3 days. To remove the B cells, we collected CD4+ cells by FACS before the knockdown experiments. The Tregs were rested for another 3 days after siRNA transfection and were then analyzed by FACS. (B) Left, the gated Tregs, PLAU expression assessed by FACS; right, the gated Tregs, expression of FOXP3 protein assessed by FACS. Enlarged numbers indicate the geometric mean of fluorescence intensity of the corresponding protein. ‘Max’, maximum. (C) Downregulation of mRNA levels of PLAU, FOXP3, EOS, CTLA4, and LGMN assessed by real-time qPCR analysis. The numbers on the y axis indicate fold change relative to RPS9 (Bruder et al, 2004). The percentage under each subfigure represents the decreased percentage of the corresponding gene in Tregs transfected with specific siRNA against PLAU (si_PLAU) compared to Tregs transfected with non-silencing siRNA (si_NS). Error bars represent standard deviation (s.d.) values. The P-values indicate the results from a one-tailed Student’s t-test for three repeated measurements. P≤0.05 is indicated by *P≤0.01, by **P≤0.001, and by *** for all the figures. The qPCR results displayed here were from experiments performed on day 3 post siRNA transfection for all the genes, except for LGMN on day 2 post siRNA. Data represent two to four independent experiments.
To test whether PLAU is also involved in controlling the expression of other genes reported to be involved in or potentially important to Treg suppressor function, we measured the transcription levels of 11 specific genes (e.g., EOS, CTLA4, and LGMN; Supplementary Table 8). The measurements were performed on each of the first 3 days post siRNA treatment. A zinc-finger transcription factor of the Ikaros family, Eos mediates Foxp3-dependent gene silencing in murine Tregs by directly interacting with Foxp3 (Pan et al, 2009). Notably, our results show that knockdown of PLAU caused a significant downregulation of EOS starting from day 1 post siRNA transfection (only shown on day 3, Figure 4C). The gene coding for cytotoxic T-lymphocyte antigen 4 (CTLA4) might be involved in a central mechanism through which Tregs control the function of antigen-presenting cells by binding with CD80 and/or CD86 (Wing and Sakaguchi, 2010). We observed a significant downregulation of CTLA4 in the PLAU-knockdown Tregs on day 3 (Figure 4C). Another example of a gene important for Treg function is the endosomal cysteine protease legumain (LGMN) gene, which is significantly upregulated in Tregs relative to Teffs and potentially induces a partial Treg signature (Probst-Kepper et al, 2009). Similarly to EOS and CTLA4, the mRNA level of LGMN was significantly decreased on day 2 after the knockdown of PLAU in Tregs (Figure 4C). These results illustrate that the expression of a number of key genes involved in Treg-suppressive function is under the control of PLAU activity.
PLAU expression is positively correlated with Treg suppressor function but inversely with cytokine expression
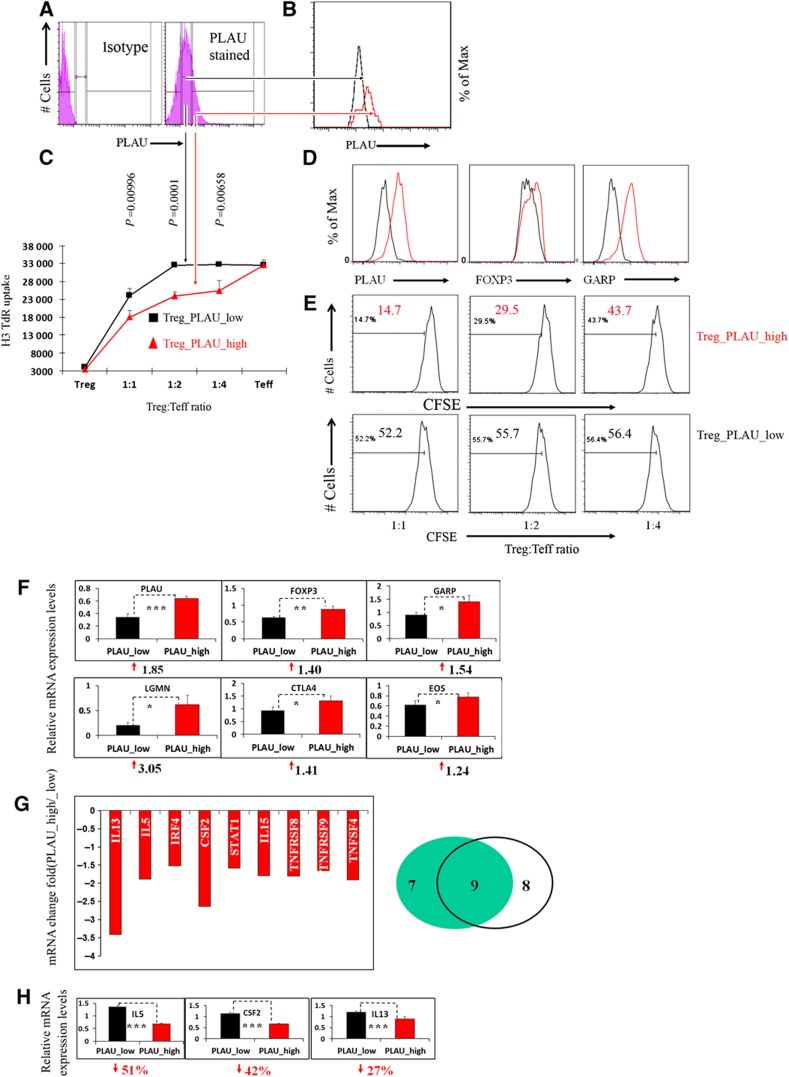
To obtain further insight into the role of PLAU in Treg function, we sorted Tregs into two subpopulations, PLAUlow and PLAUhigh Tregs, according to the expression level of PLAU protein by FACS (Figures 5A and B). To eliminate a potential inhibitory effect of the PLAU antibody on PLAU function in Tregs and to obtain more cells for further analyses, we expanded the FACS-sorted PLAUlow and PLAUhigh Tregs by re-stimulating them for at least four rounds. By co-culturing either PLAUlow or PLAUhigh Tregs with Teffs, we observed that PLAUhigh Tregs had a significantly greater potency in suppressing Teffs’ proliferation relative to PLAUlow Tregs as demonstrated by both H3 thymidine uptake experiments (Figure 5C) and CFSE dilution measurement of Teffs (Figure 5E).
Figure 5.
PLAUhigh Tregs exhibit higher potential in suppressing effector T cells and express higher levels of Treg-important genes, but they express lower levels of cytokine genes compared to PLAUlow Tregs. (A) PLAUhigh and PLAUlow Tregs were sorted by PLAU expression level (left, isotype control; the figure in the right column is divided into three parts: negative, low, and high levels of PLAU expression). (B) FACS of the PLAUhigh (red line) and PLAUlow Tregs (black line) immediately after sorting. (C) The suppression assay of different cells populations assessed by uptake of [H3]thymidine (H3-TdR) (refer to Materials and methods). Error bars represent s.d. values. Results are representative of three independent experiments. (D) Higher expression of PLAU, FOXP3, and GARP in PLAUhigh than PLAUlow Tregs assessed by FACS at day 3 post restimulation. The PLAU subsets of Tregs were expanded for at least four rounds. Of note, in this panel EBV-B cells were not sorted out before FACS analysis. Results are representative of three independent experiments. (E) The CFSE dilution measurement of the Teff proliferation co-cultured with Tregs at various ratios of PLAU_high (upper row) or PLAU_low (lower row) Tregs to Teffs as well as co-cultured EBV-B cells after 4 days. Number in each histogram represents the percentage of dividing cells from the total population (the total population indicates the gated DAPI-negative and CD4+, CFSE stained Teffs). Results are representative of three independent experiments. (F) Higher mRNA levels of FOXP3, GARP, LGMN, CTLA4, and EOS in PLAUhigh Tregs assessed by real-time qPCR analysis. The numbers on the y axis indicate fold changes relative to RPS9. The numbers under each subfigure represent the increased fold changes in PLAUhigh Tregs compared to PLAUlow Tregs. The qPCR analysis was performed after the removal of B cells by FACS before extracting RNA. The P-values represented by ‘*’ symbols indicate the results from a one-tailed Student’s t-test for three replicates. Results are representative of three independent experiments. (G) PLAUlow Tregs reversed the expression of Treg-repressed cytokines as assessed by a human Th1-Th2-Th3 cytokine PCR array. Left, numbers on the y axis indicate fold changes relative to actin. Data represent two independent experiments. The reported genes are the overlap of the two independent experiments, which show at least 1.5-fold decrease in PLAUhigh versus PLAUlow Tregs. Right, Venn diagram of the two independent experiments. The numbers specify the number of genes that decreased at least 1.5-fold. (H) Independent real-time PCR confirmed the expression of the Treg-repressed cytokines in (G), such as IL5, IL13, and CSF2. The percentage under each subfigure represents the decreased percentage of the corresponding gene in PLAUhigh versus PLAUlow Tregs. Error bars represent s.d. values. The P-values represented by ‘*’ symbols indicate the results from a one-tailed Student’s t-test for three replicates.
To further check whether low levels of PLAU expression relative to PLAUhigh Tregs have a similar effect compared to the knockdown of PLAU in Tregs versus control, we measured the expression level of the 11 genes as described for the siRNA experiments. As expected, the protein expression of PLAU and FOXP3 was significantly higher in PLAUhigh Tregs compared to PLAUlow Tregs in the expanded Treg set (Figure 5D). We observed significantly higher mRNA expression for 6 out of the 11 genes in PLAUhigh versus PLAUlow Tregs. These include genes, for example, FOXP3, PLAU, GARP, CTLA4, and EOS (Figure 5F). We also observed a higher protein expression of GARP in PLAUhigh Tregs (Figure 5D). In addition, we found a significantly higher mRNA level of LGMN in PLAUhigh Tregs (Figure 5F). These results not only further confirm a key role of PLAU in Treg function, similar to the data obtained from siRNA experiments, but also show a correlation between the expression of some known key genes and the suppressive activity of Tregs. Our results also show that PLAU is a stable surface marker for selecting human Tregs with higher suppressive potency.
We further studied the mRNA expression of a number of cytokines and related genes in PLAUhigh Tregs compared to PLAUlow Tregs using real-time PCR array. A total of 84 genes related to the three classes of CD4+ T cells (Th1, Th2, and Th3) were analyzed. An inverse relationship was found between the level of PLAU in Tregs and the expression of cytokines normally repressed in Tregs. In all, 9 out of the 84 genes were significantly downregulated in PLAUhigh Tregs (Figure 5G) though no cytokine genes were repeatedly observed to be significantly upregulated. For instance, the Th2 cytokine genes IL5 and IL13 and the Th1 cytokine colony-stimulating factor 2 (CSF2) gene were significantly upregulated in PLAUlow Tregs, while other important Treg genes were downregulated (Figures 5G and F). The genes CSF2, IL5, and IL13 (Figure 5H) are located in the same cluster on chromosome region 5q31 (Le Beau et al, 1993) and therefore might be directly or indirectly coregulated by PLAU.
We also observed a significant downregulation of interferon regulatory factor 4 (IRF4) and IL15 in PLAUhigh Tregs (Figure 5G). IRF4 promotes Th2 cytokine production in CD4+ Teffs (Honma et al, 2008), while IL15 induces type 1 and type 2 CD4+ T-cell proliferation (Niedbala et al, 2002). The expression of murine Irf4 in Tregs might depend on Foxp3 and regulate Th2 differentiation (Zheng et al, 2009). The complexity of these regulations might be worthy of further investigation. The costimulator of antigen-specific CD4+ T cells (Niedbala et al, 2002) TNFRSF9, also known as CD137 or 4-1BB, was significantly decreased in PLAUhigh Tregs. Similarly, the expression of TNFRSF8 (CD30), which is involved in NF-κB activation (Aizawa et al, 1997), was also reduced in PLAUhigh Tregs. The results about cytokines and key Treg genes indicate that the low level of PLAU in PLAUlow Tregs could be responsible for the high expression of Treg-mediated cytokines and make PLAUlow Tregs more reminiscent of the molecular characteristics of Teffs.
PLAU mediates Treg suppressor function via STAT5 and ERK signaling pathways
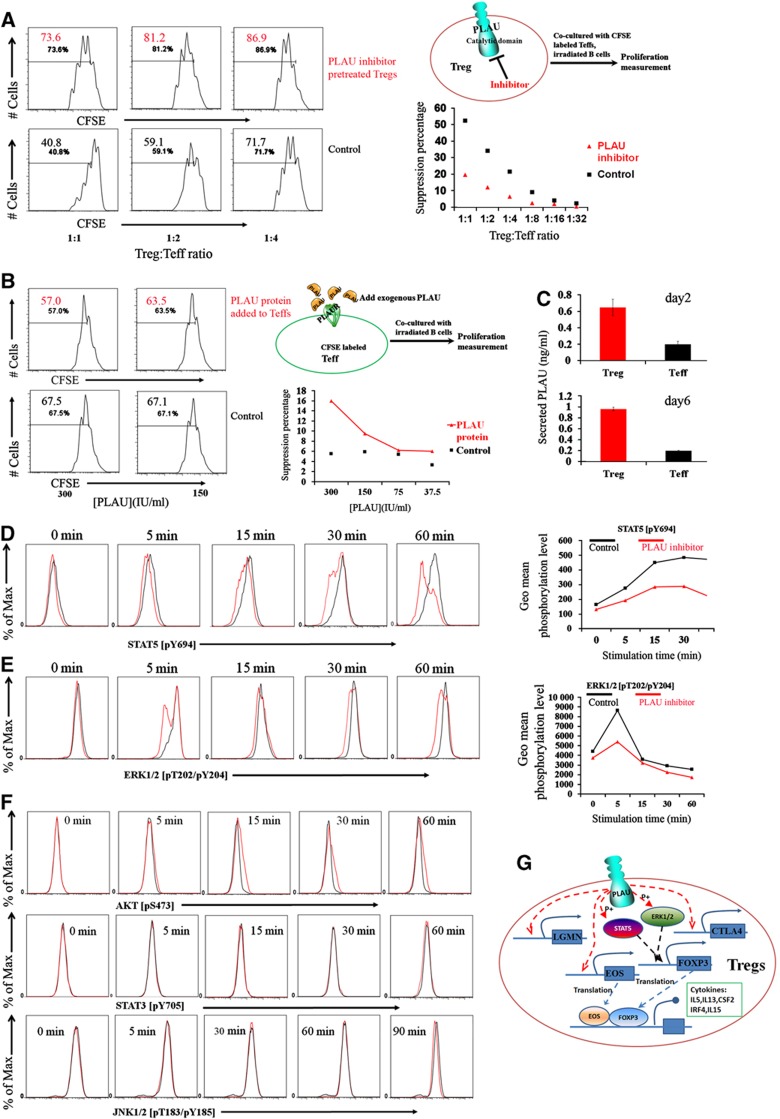
To investigate the mechanisms through which PLAU mediates Treg suppressor function, we studied the two defined domains of the PLAU protein, that is, the N-terminal domain binding the receptor as well as the C-terminal domain with catalytic activity (Mondino and Blasi, 2004). We first examined the catalytic domain using a specific inhibitory antibody. Interestingly, the suppressive capability of Tregs pretreated with the catalytic inhibitor was significantly impaired as shown by CFSE dilution measurement of co-cultured Teffs (Figure 6A) while the control IgG did not show significant effects (Supplementary Figure S5). In order to identify the signaling pathways through which the PLAU catalytic domain mediates Treg suppressor function, we assessed STAT1, STAT3, STAT5, ERK1/2, AKT, and JNK1/2 as potential PLAU targeted signaling pathways suggested by the molecular pathway databases (Nikitin et al, 2003) or by the literature (Murawski et al, 2006). Following stimulation of Tregs by anti-CD3/-CD28/IL2, we did not observe any significant difference in the phosphorylation of STAT3, AKT, and JNK1/2 in the presence or absence of the catalytic inhibitory antibody (Figure 6F). Moreover, we could not observe apparent STAT1 phosphorylation in the first 90 min of stimulation either in the presence or in the absence of the catalytic inhibitory antibody (Supplementary Figure S6). However, Tregs pretreated with the catalytic inhibitory antibody showed much less phosphorylation of STAT5 and ERK1/2 protein compared with untreated control Tregs (Figures 6D and E). It has been described that activation of STAT5 is required for upregulation of FOXP3 expression in both human and murine Tregs (Murawski et al, 2006) and that human Tregs display a greater capacity to phosphorylate ERK compared with Teffs upon TCR-mediated activation (Crellin et al, 2007). We therefore conclude that PLAU mediates Treg suppressor function via STAT5 and ERK signaling pathways.
Figure 6.
PLAU mediates Treg suppressor function via STAT5 and ERK signaling pathways. (A) The inhibition of the catalytic domain of the membrane-bound PLAU in Tregs impaired Treg suppressor function. Left, the CFSE dilution measurement of the Teff proliferation at various ratios of Tregs to Teffs at day 4 after being co-cultured with Tregs pretreated (upper row) or not (lower row) with the PLAU catalytic domain inhibitor as well as EBV-B cells. Number in each histogram represents the percentage of dividing cells from the total population (the total population indicates the gated DAPI-negative and CD4+, CFSE stained Teffs). Right, the upper figure is the experimental scheme. The lower one is the suppression percentage converted from the CFSE results in the left column. The suppression percentage is converted by comparing the proliferation percentage of Teffs at a given ratio to Tregs to the proliferation of stimulated Teffs alone without the presence of Tregs. (B) Adding exogenous PLAU protein with the specific binding capability to the PLAUR into the culture media of Teffs has only very slight suppressive effects on the proliferation of Teffs at very high doses. Left, the upper row is the CFSE measurement of the Teff proliferation at various concentrations of exogenous PLAU protein at day 4 after being co-cultured with EBV-B cells. The lower row is the control one in the absence of exogenous PLAU protein but with the same concentration of buffer. Right, the upper figure is the experimental scheme. The lower one is the suppression percentage converted from the CFSE results in the left column. Lines are a guide for the eye. (C) Very low level of secreted PLAU in the supernatant of cultured Tregs or Teffs. The upper one is the measurement from the supernatant of cultured cells at day 2 after being stimulated by anti-CD3/-CD28/IL2. The lower one is the measurement at day 6 after being re-stimulated by EBV-B cells. Error bars represent s.d. values. (D) Impaired STAT5 phosphorylation assessed by FACS in Tregs at different time points following the anti-CD3/-CD28/IL2 stimulation in the presence of the PLAU catalytic domain inhibitor. Only gated viable Tregs were displayed for all the phosphorylation results. Right, geometric (geo) mean fluorescence intensity of Tregs converted from the left column. Max, maximum. Lines are a guide for the eye. (E) Impaired ERK1/2 phosphorylation assessed by FACS in Tregs at different time points following the anti-CD3/-CD28/IL2 stimulation in the presence of the PLAU catalytic domain inhibitor. Right, geometric (geo) mean fluorescence intensity of Tregs converted from the left column. (F) Unaffected AKT, STAT3, and JNK1/2 phosphorylation assessed by FACS in Tregs at different time points following the anti-CD3/CD28/IL2 stimulation in the presence of the PLAU catalytic domain inhibitor. (G) Schematic diagram showing the mechanistic model through which PLAU mediates the suppressive function of Tregs. The red dashed arrows are the new knowledge added by this work. Data are representative of 2 (C) or 4–5 independent experiments in (A, B, D, E, F).
We next examined whether the receptor-binding domain of PLAU plays a role in controlling the suppressor function of Tregs toward Teffs. To evaluate whether PLAU protein can directly affect the proliferation of Teffs via binding to the PLAU receptor (PLAUR), which is highly expressed on human Teffs but at a very low level on Tregs (Supplementary Figure S7), we introduced purified high-molecular weight human PLAU protein, known to have specific PLAUR binding capability, into the culture media of Teffs. Addition of human exogenous PLAU protein has only slight suppressive effects on the proliferation of Teffs even at very high non-physiological concentrations (∼300 IU/ml, Figure 6B). Nevertheless, only a very low level (<0.1 IU/ml, for ng and IU conversion see Supplementary Figure S8) of secreted PLAU has been detected in the supernatant of both stimulated Treg and Teffs, respectively (Figure 6C). Since PLAU protein can only slightly affect the proliferation of Teffs, based on the minor effect, a partial blockage of PLAUR by the PLAUR antibodies is not expected to significantly change this minor effect. Indeed, a pretreatment with or without the PLAUR inhibitor did not show a significant difference on the proliferation of Teffs. Although the secreted level of PLAU is very low, the expression level of the membrane-bound PLAU of activated Tregs is quite high, raising an alternative possibility that the membrane-bound PLAU might bind to the PLAUR on Teffs and subsequently affect the proliferation of Teffs. Therefore, we investigated the alternative possibility by asking whether a blockage of PLAUR on Teffs, before a co-culture with Tregs, can alter the suppressive capability of Tregs toward Teffs. The results did not demonstrate a significant effect of the blockage (Supplementary Figure S9). This rules out the possibility that membrane-bound PLAU on Tregs promotes the suppressive function of Tregs via binding to PLAUR on Teffs. We therefore conclude that PLAU mediates the Treg suppressor function intracellularly rather than via binding to PLAUR on Teffs (Figure 6G).
Elevated Treg development but impaired suppressor function of Cd44highCd62Llow Tregs in Plau-KO mice
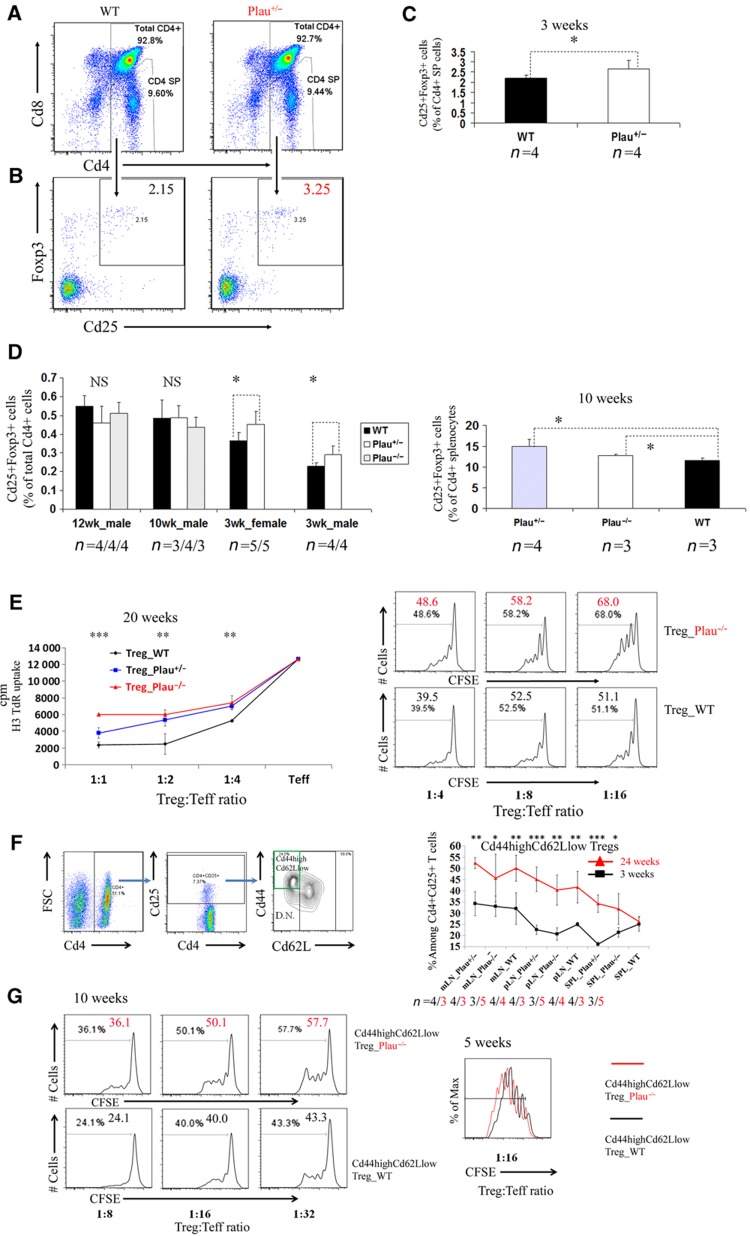
The critical role of PLAU in human Tregs motivated us to further study its role in the murine Treg system. To investigate whether murine Plau is involved in the development and suppressor function of Tregs, we analyzed Plau-knockout (Plau-KO) mice (Carmeliet et al, 1994). For this purpose, we compared Plau heterozygous and/or homozygous knockout (Plau+/− and/or Plau−/−) mice with age- and sex-matched, wild-type (WT) mice. Unexpectedly, in 3-week-old Plau+/−- mice we observed a significantly higher frequency of Cd25+Foxp3+ cells among total Cd4+ single positive (SP) T cells in the thymus compared to WT mice (Figure 7C). Further analysis shows that almost all Cd4+Cd25+Foxp3+ cells were Cd4+ SP cells in the thymus and the frequency of Cd25+Foxp3+ cells among Cd4+ SP T cells in the thymus was significantly higher in Plau+/- mice than in WT mice (Figures 7A–C). We further analyzed older mice, aged 10 and 12 weeks and found there was no significant difference in the frequency of Cd25+Foxp3+ cells among total Cd4+ T cells of thymocytes between KO mice and WT mice (Figure 7D). This might be caused by thymic involution as the number of total thymocytes in KO mice was sharply diminished in these older mice versus 3-week-old mice while WT mice had only a mild decrease (Supplementary Figure S10). We have also checked splenocytes in 10-week-old mice and found that the frequency of Cd25+Foxp3+ cells among Cd4+ splenocytes in both Plau+/− and Plau−/− mice was significantly higher than in WT mice (Figure 7D). These results of enhanced Treg development in Plau-KO mice are concomitant with the observation that sustained expression of a serine protease inhibitor can increase the population of Tregs in a specific mouse strain (Subramanian et al, 2011).
Figure 7.
Elevated thymic Foxp3+ Treg population while diminished suppressive capability of Cd44highCd66Llow Tregs in Plau-KO mice. (A) Percentages of Cd4+ SP cells among murine thymocytes isolated from wild-type (WT) control mice and Plau heterozygous knockout mice (Plau+/−) assessed by FACS. (B) Percentages of Cd25+Foxp3+ cells among murine Cd4+ SP thymocytes isolated from WT and Plau+/− mice assessed by FACS. Only gated Cd4+ SP cells from (A) are shown in (B). Data represent one of the four mice per group. (C) Comparison of percentages of Cd25+Foxp3+ cells among murine Cd4+ SP thymocytes between 3-week-old WT and Plau+/− groups. Data are representative of two independent experiments. (D) Left, comparison of percentages of Cd25+Foxp3+ cells among murine total Cd4+ thymocytes between WT and Plau+/− and/or Plau−/− groups, aged 12, 10 and 3 weeks. Error bars represent s.d. values. P-value represented by ‘*’ symbols indicate results from a one-tailed Student’s t-test for n mice per group (the corresponding number n per group is indicated under each group column). Right, comparison of percentages of Cd25+Foxp3+ cells among murine total Cd4+ splenocytes between WT, Plau+/− and Plau−/− groups in 10-week-old mice. (E) Comparison of suppressive capability of Cd4+Cd25+ Tregs between littermate WT, Plau+/−, and Plau−/− groups, age ≥20 weeks. Left, proliferation assessed by uptake of [H3]thymidine. The P-value above each ratio represented by ‘*’ symbols indicates the results from a one-tailed Student’s t-test between WT and Plau−/− groups. Right, proliferation assessed by the CFSE dilution measurement of Cd25− Ths co-cultured with Plau−/− or WT Tregs as well as irradiated feeder cells and Cd3 at various ratios of Tregs to Teffs after 3.5 days. Number in each histogram represents percentage of dividing cells from the total population (the total population indicates the gated DAPI-negative and CD4+, CFSE stained Ths). (F) Left, the gating scheme used to analyze frequencies of Cd44highCd62Llow Tregs. Right, comparison of percentages of Cd44highCd62Llow Tregs among total Tregs in spleen (SPL), peripheral LN (pLN) or mesenteric LN (mLN) from WT and Plau+/− and/or Plau−/− between groups of mice aged 24 and 3 weeks. The P-value represented by ‘*’ symbols indicates the results from a one-tailed Student’s t-test. (G) Cd44highCd62Llow Tregs from Plau–/– mice has significantly lower suppressive function than that from WT littermates. Left, proliferation assessed by the CFSE dilution measurement of Cd25− Ths co-cultured with Cd44highCd62Llow Tregs from Plau−/− or WT mice aged 10 weeks as well as irradiated feeder cells and Cd3 at various ratios of Tregs to Teffs after 3.5 days. Right, comparison of proliferation of Cd25− Ths co-cultured with Cd44highCd62Llow Tregs from littermate Plau−/− or WT mice aged 5 weeks as well as irradiated feeder cells and Cd3 at various ratios of Tregs to Teffs after 3.5 days. Max, maximum. Data are representative of 3–5 independent experiments in (E, F, G).
We further studied the suppressive capability of peripheral Tregs in Plau-KO mice and WT mice. Interestingly, our results show that the Tregs from spleens in 20-week-old WT mice had a significantly higher suppressor potential than Plau−/− mice as demonstrated by both CFSE dilution and H3 thymidine uptake measurement (Figure 7E). But the Tregs isolated from 3-week-old Plau−/− or WT mice splenocytes had no significant difference in suppressor potential (Supplementary Figure S11). We further addressed the bases of the difference between relatively younger and older mice. By co-staining Cd44, Cd62L, Cd25, and Cd4, we have found that the Cd44highCd62Llow Tregs in secondary lymphoid organs were present at significant higher percentages in older mice than in 3-week-old mice regardless of WT or Plau-KO mice (Figure 7F). This observation triggered us to check whether there were differences in the suppressive function between two Tregs subsets Cd44highCd62Llow and Cd44lowCd62Lhigh Tregs in Plau-KO and WT mice. As indicated by both division percentages and division numbers of CFSE dilution peaks, we have found that the isolated Cd44highCd62LlowCd4+Cd25+T cells in littermate control WT mice had a significantly higher suppressor potential than Plau−/− mice independent of whether they are younger (5 weeks old) or older (10 weeks old) as shown in Figure 7G. In contrast, no significant difference was observed in the suppressor capabilities of another Treg subset Cd44lowCd62Lhigh Tregs between littermate WT and Plau−/− mice (Supplementary Figure S12). Therefore, the high percentage of Cd44highCd62Llow Tregs among the total Tregs in older mice contributes to the phenotypic differences of the total Tregs between WT and Plau−/− mice. The effects of Plau on total Tregs in relatively older mice coincide very well with that in cultured human Tregs. This is due to the fact that the major fraction of Tregs in older mice are Cd44highCd62Llow Tregs. These cells are ‘memory-like’ Tregs while almost all the cultured human Tregs are CD45RO+ memory Tregs after several-round exposures to EBV-B cells. In summary, these data suggest that Plau expression impaired the development of Cd4+Cd25+Foxp3+ Tregs in murine thymus while it was critical for promoting Cd44highCd62Llow Treg suppressor function.
Discussion
In this work, we identified PLAU as a novel key gene for Treg suppressor function from an undirected correlation network instead of a transcription regulatory network of Tregs. The high predictive power of this network strategy is a result of the utilization of the queen bee-surrounding principle described here.
The proposed strategy is especially powerful in identifying key Treg genes which are not among the most significantly differentially expressed genes, but are still significantly differentially expressed between Tregs and Teffs. As shown in Table I, PLAU did not show a very-high expression difference between Tregs and Teffs compared to the other top hubs. PLAU has already been reported to be involved in fibrinolysis, tumor metastasis (Ulisse et al, 2009), neurodegenerative (Finckh et al, 2003), and other diseases (reviewed in Mondino and Blasi, 2004). However in this work, we showed a new function of PLAU, namely, an important role in human and murine Treg suppressor function. Therefore, PLAU might be important for autoimmunity. It has already been reported that murine Plau is required for T lymphocyte activation and proliferation when T cells were studied as a whole population (Gyetko et al, 1999). Our results however show that the expression level of PLAU was inversely correlated with the expression level of Th1 and Th2 cytokines and CD4+ activation markers in human Tregs. This might demonstrate a dual role of PLAU in human Teffs and Tregs. PLAU might be a mitogen (Cohen et al, 1981) for human Teffs and activator of cytokine expression of Teffs but act as an inhibitor of cytokine expression for Tregs. This dual role might be caused by Treg-specific genes and/or pathways. Treg-repressed cytokine expression was already reported to be repressed by FOXP3, EOS, and/or other important genes in Tregs.
Moreover, we assessed the regulatory effects of PLAU by siRNA-knockdown experiments, antibody inhibitory experiments, and FACS-sorted cellular experiments in human Tregs. These experiments have not only demonstrated the importance of PLAU in the expression regulation of key Treg genes, but also shown the critical role of PLAU in Treg suppressor function. Furthermore, our results from murine experiments have revealed that Plau expression decreases murine thymic Foxp3+ Treg development while promoting Cd44highCd62Llow but not Cd44lowCd62Lhigh Treg suppressor function. Our further investigation shows that PLAU mediates Treg suppressor function via STAT5 and ERK signaling pathways. The data presented here conclusively demonstrate that PLAU is important for the suppressive capability of Tregs. Thus, a new component of regulation of Treg suppressor function was discovered.
The construction of a dynamic correlation network of human Tregs provides a valid starting point for researchers who study human autoimmune diseases and/or Treg function. A key value of this human Treg functional association network is to provide subnetworks of other genes involved in the suppressor function of Tregs, such as the subnetworks of FOXP3 and GARP described here. Furthermore, since we successfully predicted and validated a vital role for PLAU in Treg suppressor function, the other top-ranked candidate hubs predicted from this work might also be very relevant. The proposed strategy for the identification of PLAU is generally applicable in discovering key candidate genes and the underlying networks associated with other human diseases.
Materials and methods
Mice
B6.129S2-Plautm1mlg/J mice have been described elsewhere (Carmeliet et al, 1994) and were purchased from the Jackson Laboratory. All of the WT, Plau+/−, and Plau−/− mice used were siblings from breeding pairs between heterozygous Plau+/− mice. All mice were kept and bred in specific pathogen-free conditions at the Animal Facility at Helmholtz Center for Infection Research in Braunschweig, Germany. All animal experiments were in accordance with protocols approved by the local authorities.
Isolation and cultivation of human Treg/Teff cells
Stable human alloantigen-specific Tregs and Teffs derived from sorted CD4+CD25high and CD4+ CD25− peripheral blood T cells were from the same batch of Tregs and Teffs from our recent work (Probst-Kepper et al, 2009). Retroviral transduction, isolation, and culture of human alloantigen-specific Teff cells with GARP coupled to an IRES-driven GFP or empty control GFP has been described (Probst-Kepper et al, 2009). IMDM (Gibco) supplemented with 10% FCS (Gibco), 100 U/ml penicillin, 100 μg/ml streptomycin (PAA) and 50 μM μ-mercaptoethanol (Sigma-Aldrich) was used for cell culture. Recombinant human IL2 (Proleukin, Novartis) was added daily to Treg culture medium at a concentration of 100 U/ml, as we previously described (Probst-Kepper et al, 2009) and in a similar way by others (Stockis et al, 2009; Tran et al, 2009). All the Tregs/Teffs were stably maintained and restimulated by EBV-B cells weekly as previously described (Probst-Kepper et al, 2009). In addition, peripheral blood CD4+CD25− Teffs (but not Tregs) freshly isolated using Invitrogen Dynabeads human regulatory CD4+CD25+ T cell kits were characterized in Supplementary Figure S13 and were used for suppression assays. Informed consent was obtained before the blood was taken.
Flow-cytometric analysis and antibodies
Human FOXP3 expression was detected with anti-FOXP3 mAbs after fixation and permeabilization of cells by the protocol provided by the manufacturer (206D, Biolegend). PLAU protein expression was analyzed by surface staining with mAbs against human PLAU B-chain (No. 394 or 3689, American Diagnostica) followed by a secondary antibody PE Goat anti-mouse IgG (minimal cross-reactivity) (Biolegend). PE or PE-Cy5 rat anti-mouse Foxp3 (FJK16s, eBioscience), FITC rat anti-mouse Cd4 (L3T4, BD), APC rat anti-mouse Cd25 (PC61, BD), and pacific-blue rat anti-mouse Cd8 (53-6.7, eBioscience) were used to analyze murine cell populations. Pacific-blue rat anti-mouse Cd4 (RM4-5, Biolegend), APC rat anti-mouse Cd25 (PC61, BD), PE-cy7 rat anti-mouse Cd44 (1M7, BD), and PE rat anti-mouse Cd62L (MEL-14, eBioscience) were used to sort Cd44highCd62Llow and Cd44lowCD62Lhigh Treg subsets. FACSCalibur (BD LSR II for murine cells) was used for data acquisition, and the data were analyzed with FlowJo software.
Cell sorting and suppression assay
To remove the EBV-B cells from human Tregs, we collected CD4+ cells by FACS before the knockdown experiments. The cultured Tregs were first stained with FITC-conjugated mAbs against CD4 (RPA-T4, BD) and then sorted with a BD Aria II. To separate Tregs with different PLAU expression levels, the Tregs were first restimulated for 3 days, stained with PLAU, and then sorted by the BD Aria II into two subpopulations: PLAUlow and PLAUhigh Tregs. For the suppression assay of human Tregs by [H3]thymidine incorporation, each of the different sets of Tregs was co-cultured with Teffs (105) with different ratios, as well as EBV-B cells (without IL2 addition). The human cells were cultured for 72 h in 96-well flat bottom plates and pulsed with [H3]thymidine (1 μCi/well, Amersham, now Perkin-Elmer) for the last 6 h. For mouse Treg suppression assays by [H3]thymidine incorporation, Cd4+Cd25+ Tregs and Cd4+Cd25− Teffs were sorted by costaining Cd4-FITC (RM4-5, BD) and Cd25-APC (PC61, BD) on murine splenocytes and peripheral lymph nodes (LNs). Cd4+Cd25+ Tregs sorted from WT or Plau+/− or Plau−/− mice were co-cultured with 105 WT Cd4+Cd25− Teffs at different ratios, together with WT irradiated splenocytes as feeder cells and 1 μg/ml CD3e (BD). The isolated cells were co-cultured in RPMI medium (Gibco) supplemented with 10% FCS (Gibco), 50 U/ml penicillin/streptomycin, 25 mM HEPES (Gibco) and 50 μM β-mercaptoethanol (Invitrogen). The murine cells were cultured for 72 h in 96-well round bottom plates and pulsed with [H3]thymidine (1 μCi/well) for the last 6 h. For the human CFSE dilution measurement, each of the different sets of Tregs was co-cultured with CFSE-labeled Teffs (105) with different ratios, as well as EBV-B cells (without IL2 addition). The human cells were cultured for 4 or 5 days in 96-well flat bottom plates and the gated DAPI (from Invitrogen) negative and CD4+, CFSE (from Invitrogen) stained Teffs were measured by FACS. CFSE at a concentration of 1 μM was used to label Teffs. For the murine CFSE dilution measurement, different sets of Tregs (total Cd4+Cd25+ Tregs, or Cd44highCd62Llow or Cd44lowCD62Lhigh Treg subsets) and Cd4+Cd25- Teffs were sorted from murine splenocytes and peripheral LNs. Different sets of Tregs sorted from WT or Plau−/− mice were co-cultured with 105 WT CFSE-labeled Cd4+Cd25− Teffs at different ratios, together with 4E5 WT irradiated splenocytes as feeder cells and 1 μg/ml CD3e. All the feeder cells were depleted with total T cells by total T-cell microbeads (CD90.2, Miltenyi Biotec) and were isolated from relatively older mice (>20 weeks) to ensure the maturation and effectiveness of antigen-presenting cells (Dakic et al, 2004). The gated DAPI-negative and CD4+, CFSE stained murine Teffs were measured by FACS after 3.5 day.
PLAU catalytic inhibitory, exogenous PLAU protein, secreted PLAU and PLAUR blockage experiments
For the human PLAU catalytic inhibitory experiments, Tregs were prestained with the specific PLAU catalytic inhibitor (394OA, American Diagnostica) at a concentration of 100 μg/ml at room temperature for 90 min and then washed twice before a following experiment, such as suppression assay or signaling pathway analysis. The normal murine IgG control without azide was from BD. For the control purpose of the experiments of introducing exogenous purified high-molecular weight human PLAU (124, American Diagnostica), we used the same concentration of blank buffer, which was used as the solvent of the purified PLAU protein. Secreted PLAU in the supernatant of cultured media was measured by ELISA kit following manufacture’s protocols (894, IMUBIND ELISA kit, American Diagnostica). For the measurement of the secreted PLAU at day 2 stimulated by anti-CD3/-CD28/IL2, 1E5 Teffs or Tregs were seeded in 200 μl media of 96-well plate; for the measurement at day 6 restimulated by EBV-B cells (with IL2), 5E5 Teffs or Tregs were seeded in 1 ml media of 24-well plate. For the PLAUR blockage experiments, Teffs were prestained with the specific PLAUR antibody (3936, American Diagnostica) at a concentration of 40 μg/ml at room temperature for 60 min and then washed twice before a following experiment.
Phosphorylation assessment by FACS
Following the human PLAU catalytic inhibitor prestaining, Tregs were stimulated by anti-CD3/-CD28/IL2. Before stimulation at 37°C, Tregs were first premixed with anti-CD3/-CD28 beads at 4°C for 20 min. Tregs were then sampled and fixed by BD cytofix buffer at various time points. The monoclonal antibodies recognizing the specific phosphorylation sites for Alex647 STAT1[pY701], PE STAT3[pY705], Alex647 STAT5[pY694], PE-Cy7 ERK1/2[pT202/pY204], and Alex647 AKT[pS473] were all BD Phosflow products. Rabbit mAB JNK1/2[pT183/pY185] and its secondary antibody Alex647 goat anti-rabbit IgG were from Invitrogen. The staining and measurement of the phosphorylation of these proteins was performed using BD Phosflow protocols.
siRNA knockdown
siRNA against human PLAU and non-silencing control siRNA were obtained from Santa Cruz Biotechnology. One hundred picomoles of siRNA was mixed with Tregs resuspended in 100 μl human T cell Nucleofector solution (Lonza Amaxa). The mixture was immediately exposed to electroporation by the Nucleofector II device (Lonza Amaxa) and placed in 37°C prewarmed IMDM with 100 U/ml IL2. Before performing the knockdown experiments, we first restimulated Tregs by co-culturing them with EBV-B cells for 3 days. To remove the B cells, we collected CD4+ cells by FACS before the knockdown experiments. Before a further analysis, the Tregs were rested for another 1 day, 2 days, or 3 days after siRNA transfection.
Real-time RT-PCR (qPCR) and pathway PCR array
RNA was isolated with the RNeasy Mini Kit (Qiagen) according to manufacturer’s protocol and additionally purified by on-column RNase-free DNase digestion (Qiagen). Purified and concentrated RNA was then reverse-transcribed using Superscript III reverse transcriptase (Invitrogen). qPCR was conducted on a LightCycler 480 (Roche) as previously described (Pfoertner et al, 2006). The following primers were finally used in our qPCR analysis. FOXP3 forward (5′–3′) ACC TAC GCC ACG CTC ATC; FOXP3 reverse (5′–3′) TCA TTG AGT GTC CGC TGC T; CTLA4 forward TGC AGC AGT TAG TTC GGG GTT GTT; CTLA4 reverse CTG GCT CTG TTG GGG GCA TTT TC; TGFB1 forward CGC AAG GAC CTC GGC TGG AAG TGG; TGFB1 reverse GAG GCG CCC GGG TTA TGC TGG TTG. The primers GARP forward GAT GGG GAA ACT GAG GCT TAG GAA, GARP reverse ACC CCC AAT CTC ACC CCA CAA ATA, LGMN forward CTC GCT CCA GGA CCT TCT TCA CAA, and LGMN reverse GCT TCC TGC TCC TCA AAA CTA ACA were from our previous work (Probst-Kepper et al, 2009). We also used qPCR primers from Qiagen for the genes RPS9, PLAU, IL1R1, S1PR1, IKZF4 (EOS), GITR, LGALS3, CD127, IL13, CSF2, IL5, and IL2. RPS9 was used as an internal standard (Bruder et al, 2004) in our qPCR analysis. Human Th1-Th2-Th3 cytokine PCR array was employed to systematically analyze the expression of cytokine gene representative of Th1, Th2, and Th3 cells by the protocol provided by the manufacturer (SABiosciences). Internal controls were included in the array, such as β-actin (chosen here). PCR array analysis was followed by the SABiosciences online tool.
Microarray sampling and RNA preparation
Human Tregs or Teffs were mixed with anti-CD3/anti-CD28-coated Dynabeads (Invitrogen) at a ratio of 1:1 and with 100 U/ml of IL2 (Proleukin, Novartis) and were distributed with 4 × 106 cells into 1.5 ml microtubes (Eppendorf) for each time point. Before the cells were cultured at 37°C, they were first stored at 4°C to allow cells and beads to settle properly. Samples were taken every 20 min during the first 6 h following stimulation, except for TeffGARP and TeffGFP cells, which were measured at time points 0, 100, and 360 min. The cells were centrifuged, and the cell pellets were lysed in RLT buffer (Qiagen) and immediately stored at −70°C. RNA was isolated with the RNeasy Mini Kit (Qiagen) according to manufacturer’s protocol and additionally purified by on-column RNase-free DNase digestion (Qiagen). RNA quality was checked by RNA 6000 Nano assay (2100 Bioanalyzer, Agilent). The comparability between stimulation results by anti-CD28/anti-CD3 beads (IL2, 100 U/ml) and EBV-B cells (IL2, 100 U/ml) is shown in Supplementary Figures S14 and S15.
Microarray measurement and data preprocessing
For oligonucleotide microarray hybridization, 3 μg RNA per sample was labeled, fragmented, and hybridized to an Affymetrix HG-U133 plus 2.0 oligonucleotide array at the Array Facility of the Helmholtz Centre for Infection Research. After the arrays were scanned, the expression value for each gene was calculated by using Affymetrix Microarray software 5.0 (MAS5). The average intensity difference values were normalized across the sample set. Probe sets that were absent in all the time-series samples, according to Affymetrix flags, were removed. Probes with intensities <50 at all the time points were excluded. In this work, probes that were not changed >2-fold among any two different time points were also excluded. We further excluded the probes with an average intensity of <50 and the largest intensity among different time points of <100. Probes were also excluded corresponding to more than one Entrez gene ID based on the HG-U133 plus 2.0 array annotation file ( http://www.affymetrix.com/, version 24, 7 March 2008). The microarray sample at time point 120 min of one Treg time series was lost during operation, so we simply interpolated the expression value for this time point by employing an intermediate linear interpolation using the values at time points 100 and 140 min.
Correlation network construction and analysis
Network construction
In this work, we demonstrated the identification of key genes by constructing and analyzing a Treg-specific correlation network. We constructed the correlation network by identifying correlation linkages among pairs of genes. The correlation linkages were identified by employing both the LC and the TC methods. Both methods are capable of discovering potential functional association between genes from time-series data by considering positive/inverted correlation and time shift. A significant correlation with time shift between two genes might indicate a transcription regulatory relationship (Bar-Joseph et al, 2012). But to obtain accurate inference, we need much additional information, a big portion of which are not available. We therefore only simply considered the pairs with this type of correlation as potential function-associated linkages in this work by not distinguishing the specific type of detailed functional association (e.g., signal transduction, transcription regulation or in the same pathways). The LC method has been widely used to identify potential functional association mainly based on a point-to-point comparison of gene expression values from time-series data as recently reviewed (Bar-Joseph et al, 2012). The TC method has also been demonstrated to be useful in identifying potential functional association from time-series data and to be complementary to the LC method based on extracting main features of the change trend and the change level of gene expression between consecutive time points (He and Zeng, 2006). Due to different principles behind the LC and the TC methods, we therefore combined both methods to complement each other in order to identify more complete potential functional associations between genes.
For this purpose, we calculated the LC score as defined by the LC method (Qian et al, 2001), the maximal matched change trend (sc) score and the correlation coefficient (cc) score for the maximal matched change trend as defined by the TC method (He and Zeng, 2006) for each pair of genes in all the time-series data sets of Tregs and Teffs. We then calculated PLC for the scores resulting from the LC method as described (Qian et al, 2001). We also calculated PTC1 for the TC method by extraction procedure I, as previously described (He and Zeng, 2006). For the TC method, we here introduced another adapted P-value (PTC2) calculation based on the reordered sc and cc distribution curve (for details, see Supplementary Figure S16). Briefly speaking, the reason to reorder the distribution table (curve) is that we found that some of the pairs, which have a very high cc score but the sc score is not very high, might still have a high chance to be potentially functionally linked. So far, we obtained three different P-values (PTC1, PTC2, and PLC) for each pair of genes in each replicate of the time-series data set generated for a given cell type (Tregs or Teffs). In the following, we set rules to extract Treg-specific correlation linkages. First, to guarantee the reproducibility of the results, we restricted the significantly correlated pairs to those repeatedly showing significant P-values in both replicates of Tregs constantly by one of the three P-values (PTC1, PTC2, and PLC). A standard P-value cutoff of 0.05, the well-established and extensively used threshold was applied. Second, in order to obtain cumulative evidence generated in both replicates and gained by both methods, we employed naive Bayesian integration (combination) (Lu et al, 2005) by multiplying different P-values from both replicates by the LC and the TC methods. This is based on the assumption of the independency of the P-values generated by two different methods due to the utilization of different principles and the independency of the results from each replicate due to the difficulty in obtaining synchronized T-cell stage before activation although all the T cells were at the resting stage. Since both PTC1 and PTC2 were caused by the TC method, we chose the minimal P-value generated by one of them. Thus, only four P-values were eventually utilized for the integration purpose. The cutoff for the integrated P-values was chosen at 1e−9 based on a more stringent but also well-established P-value threshold 0.01. The overall threshold 1e−9 was even stricter than the product of the four individual P-values ((0.01)4=1e−8) in order to avoid the cases, such as those pairs with all the individual P-values very close to the threshold of 0.01, which could be marginal. We therefore set min (PTC1,Treg1*PTC1,Treg2, PTC2,Treg1* PTC2,Treg2) *PLC,Treg1*PLC,Treg2≤1e−9, where min() indicates the minimal value of the two products of P values; Treg1/2 indicates the replicate of Treg 1 or 2. In the end, we only included the pairs satisfying both of the two criteria for the reproducibility and the integrated evidence in Tregs but the same pairs did not even meet the standards of the reproducibility in Teffs, as Treg-specific correlation linkages. The strict criteria of the reproducibility largely excluded those pairs with integrated P-values slightly higher than the integrated P-value threshold in Tregs but slightly lower than the integrated threshold in Teffs. Obviously, the pairs that cannot satisfy the basic prerequisites of the reproducibility (≤0.05) has very less chance to pass the integrated P-value threshold (based on 0.01). This has been demonstrated in Supplementary Figure 17, where >99 and 95.5% of all the Treg-specific correlation linkages show around 3- and 10-fold less P-values, respectively, in average for each of the four individual P-values in Tregs than in Teffs. For the remaining tiny proportion (<1%) of the correlation linkages, they showed a very significant correlation only in one of the two replicates of Teffs but had no significance in another one, which still causes a very small final product of P-values integrated from replicates. Apparently, we cannot consider this tiny fraction as significantly correlated pairs in Teffs without consistent results between replicates. Thus, all of linkages including the tiny fraction were Treg-specific correlation interactions with high confidence.
Threshold robustness
In addition to being rational to choose the P-value thresholds, we have also performed a robustness test in order to examine whether a reasonable variation of the P-value thresholds will cause significant changes in the resulted overall Treg-specific correlation network and in the degree of predicted functional importance of some key candidate genes. As shown in Supplementary Figure S18A, an order of magnitude variation of thresholds [0.01, 0.1] around the reproducibility threshold 0.05 has generally obtained <5% differences between the total number of correlation linkages at various thresholds and those at the threshold 0.05. The maximum difference actually only 14.6% reduction of the entire number of correlation linkages occurs at the case when changing the threshold to 0.01. Particularly for the PLAU subnetwork, the same threshold changes cause only very slight changes [6.82, 8.12] for the logarithm (−log10) of Pf (see text), which indicates the degree of potential functional importance (Supplementary Figure S18B). For comparing purpose, it is worthy to notice that the −log10Pf value is 7.86 at the threshold 0.05 (Table I). For the GARP subnetwork, the −log10Pf values have only fallen in a very narrow range [6.00, 7.12] around 6.43 at the threshold 0.05 (Supplementary Figure S18D). All these resulted −log10Pf values were still very significant and consequently caused no change in the predicted high degree of functional importance of these key candidate genes. The overlap between the identified correlation linkages with PLAU (or GARP) at the varied thresholds and those at 0.05 is also very high (≥81% and ≥88% for PLAU and GARP, respectively) (Supplementary Figures S18C and E). We also changed the integrated P-value threshold around 1e−9 and a similar observation has been obtained. We therefore concluded that our strategy is quite insensitive to the choice of P-value thresholds.
Differential expression
We also defined the differential expression between Tregs and Teffs in a combinatorial way. The expression difference over the measured continuous time points between Tregs and Teffs was first evaluated by two-tail paired Student’s t-test. Considering the difficulty in synchronizing the cell state between the replicates of T cells, we generated t-test P-values for all the possible pairs between the replicates of the two cell types, namely, PtTreg1,Teff1, PtTreg1,Teff2, PtTreg2,Teff1, and PtTreg2,Teff2. To guarantee the reproducibility of the expression difference between the two cell types for all the replicates, we only chose genes with all the four P-values of ≤0.05. For those significantly increased genes in Tregs versus Teffs, we required additional criteria as following. First, we only chose genes with at least 50% of all the measured time points (we here chose 9 out of 19 points) showing a Present/Marginal call in both replicates of Tregs to obtain more benefits as suggested (McClintick and Edenberg, 2006). Second, we only chose genes with average intensity ≥50 in both replicates of the time-series data sets of Tregs as low intensity will produce high variation (Amit et al, 2007; Ambroise et al, 2011). Third, since t-test cannot distinguish which cell type has a higher or lower expression, we only chose genes with average intensity higher in both replicates of Tregs than both replicates of Teffs by generating pair-wised comparisons as paired for the t-test. For those significantly downregulated genes in Tregs, we asked for the same three requirements for Teffs replacing Tregs as stated above.
Key gene predictions
According to our network construction strategy (Figure 1E), we calculated the functional P-value Pf for each gene using a cumulative binomial distribution test. Furthermore, considering the non-synchronization of the replicates of T cells, we calculated the average expression difference by generating all the possible pairs between the replicates of the two cell types, namely, mean(Treg1)/mean(Teff1), mean(Treg1)/mean(Teff2), mean(Treg2)/mean(Teff1), and mean(Treg2)/mean(Teff2), where mean() indicates the average intensity of a given gene over time;Teff1/2 designates the replicate of Teff 1/2. We then calculated the arithmetic mean (Pe) of the four pairs and subsequently executed a log2 transformation of the mean (log2(Pe)) to calculate average change fold for each gene between Tregs and Teffs as a log2 transformation in general has better performance (Yang et al, 2002; Ambroise et al, 2011).
In order to make the values of functional P-values (Pf) comparable to the log2-transformed expression difference values, we performed a log10 transformation on Pf. We then again used naive Bayesian integration (Lee et al, 2004) by integrating the information of the expression difference and the degree of potential functional importance of a given gene assuming both information is independent. Following the rationale above, we used abs(log10(Pf))+abs(log2(Pe)) as the integrated value to rank the potential importance of the identified functional hub candidates. Furthermore, to avoid the cases of the genes with very high expression difference but with no significant Pf values or the cases of the genes with very significant Pf values but no clear expression difference, we set up the following prerequisite conditions to be matched before ranking: Pf≤0.056 and the corresponding gene was significantly downregulated or upregulated (the upregulated ones are the main focus of this work), according to our definition above. The only reason of choosing 0.056 rather than the standard threshold 0.05 is to include FOXP3, the marginality of which has been argued in the text. Of note, the upregulated or downregulated hub genes were ranked separately.
Data availability
The complete time-series microarray data set (81 arrays) is available in the Gene Expression Omnibus (GSE11292).
Supplementary Material
Supplementary figures S1-18, Supplementary tables S1-8
Acknowledgments
This work was mainly supported by intra-mural funding from German Helmholtz-Association (Program Infection and Immunity) and partially supported by LCSB, Luxembourg. HC has obtained a PhD fellowship from the Helmholtz-China Scholarship Council program. FH was supported by German Helmholtz-Association (2008–2010) and currently by LCSB. We thank Jochen Hühn for expert advice in murine T-cell experiments and Lothar Gröbe, Petra Hagendorff, Maria Höxter, Nicolas Bonjean, and Annegrät Daujeumont for technical support; acknowledge Nikos Vlassis for advice on computational analyses. We greatly appreciate the constructive comments suggested by the anonymous reviewers.
Author contributions: FH, APZ, and RB designed the project and FH, HC, MPK, and KS designed the experiments. FH performed part of the laboratory experiments for human cells and murine experiments, drafted the manuscript, designed and performed key gene inference from the constructed network and analyzed the expression data. HC performed part of human cellular experiments and major parts of murine experiments and drafted part of the manuscript. MPK technically guided the experiments and performed part of the experiments. FH and HC analyzed the results. RG measured the microarray samples.ADS and SE annotated the network with GO terms. RB revised the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Aizawa S, Nakano H, Ishida T, Horie R, Nagai M, Ito K, Yagita H, Okumura K, Inoue J, Watanabe T (1997) Tumor necrosis factor receptor-associated factor (TRAF) 5 and TRAF2 are involved in CD30-mediated NFkappaB activation. J Biol Chem 272: 2042–2045 [DOI] [PubMed] [Google Scholar]
- Ambroise J, Bearzatto B, Robert A, Govaerts B, Macq B, Gala JL (2011) Impact of the spotted microarray preprocessing method on fold-change compression and variance stability. BMC Bioinformatics 12: 413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amit I, Citri A, Shay T, Lu Y, Katz M, Zhang F, Tarcic G, Siwak D, Lahad J, Jacob-Hirsch J, Amariglio N, Vaisman N, Segal E, Rechavi G, Alon U, Mills G, Domany E, Yarden Y (2007) A module of negative feedback regulators defines growth factor signaling. Nat Genet 39: 503–515 [DOI] [PubMed] [Google Scholar]
- Anastassiou D (2007) Computational analysis of the synergy among multiple interacting genes. Mol Syst Biol 3: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacchetta R, Gambineri E, Roncarolo MG (2007) Role of regulatory T cells and FOXP3 in human diseases. J Allergy Clin Immunol 120: 227–235quiz 236-227 [DOI] [PubMed] [Google Scholar]
- Bar-Joseph Z, Gitter A, Simon I (2012) Studying and modelling dynamic biological processes using time-series gene expression data. Nat Rev Genet 13: 552–564 [DOI] [PubMed] [Google Scholar]
- Basso K, Margolin AA, Stolovitzky G, Klein U, Dalla-Favera R, Califano A (2005) Reverse engineering of regulatory networks in human B cells. Nat Genet 37: 382–390 [DOI] [PubMed] [Google Scholar]
- Belkaid Y (2007) Regulatory T cells and infection: a dangerous necessity. Nat Rev Immunol 7: 875–888 [DOI] [PubMed] [Google Scholar]
- Bruder D, Probst-Kepper M, Westendorf AM, Geffers R, Beissert S, Loser K, von Boehmer H, Buer J, Hansen W (2004) Neuropilin-1: a surface marker of regulatory T cells. Eur J Immunol 34: 623–630 [DOI] [PubMed] [Google Scholar]
- Carmeliet P, Schoonjans L, Kieckens L, Ream B, Degen J, Bronson R, De Vos R, van den Oord JJ, Collen D, Mulligan RC (1994) Physiological consequences of loss of plasminogen activator gene function in mice. Nature 368: 419–424 [DOI] [PubMed] [Google Scholar]
- Chatila TA (2005) Role of regulatory T cells in human diseases. J Allergy Clin Immunol 116: 949–959quiz 960 [DOI] [PubMed] [Google Scholar]
- Cohen SD, Israel E, Spiess-Meier B, Wainberg MA (1981) Plasminogen activator is an apparent lymphocyte mitogen. J Immunol 126: 1415–1420 [PubMed] [Google Scholar]
- Connolly BM, Choi EY, Gardsvoll H, Bey AL, Currie BM, Chavakis T, Liu S, Molinolo A, Ploug M, Leppla SH, Bugge TH (2010) Selective abrogation of the uPA-uPAR interaction in vivo reveals a novel role in suppression of fibrin-associated inflammation. Blood 116: 1593–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crellin NK, Garcia RV, Levings MK (2007) Flow cytometry-based methods for studying signaling in human CD4+CD25+FOXP3+ T regulatory cells. J Immunol Methods 324: 92–104 [DOI] [PubMed] [Google Scholar]
- Dakic A, Shao QX, D'Amico A, O'Keeffe M, Chen WF, Shortman K, Wu L (2004) Development of the dendritic cell system during mouse ontogeny. J Immunol 172: 1018–1027 [DOI] [PubMed] [Google Scholar]
- de la Fuente A, Bing N, Hoeschele I, Mendes P (2004) Discovery of meaningful associations in genomic data using partial correlation coefficients. Bioinformatics 20: 3565–3574 [DOI] [PubMed] [Google Scholar]
- Della Gatta G, Palomero T, Perez-Garcia A, Ambesi-Impiombato A, Bansal M, Carpenter ZW, De Keersmaecker K, Sole X, Xu L, Paietta E, Racevskis J, Wiernik PH, Rowe JM, Meijerink JP, Califano A, Ferrando AA (2012) Reverse engineering of TLX oncogenic transcriptional networks identifies RUNX1 as tumor suppressor in T-ALL. Nat Med 18: 436–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuerer M, Hill JA, Kretschmer K, von Boehmer H, Mathis D, Benoist C (2010) Genomic definition of multiple ex vivo regulatory T cell subphenotypes. Proc Natl Acad Sci USA 107: 5919–5924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finckh U, van Hadeln K, Muller-Thomsen T, Alberici A, Binetti G, Hock C, Nitsch RM, Stoppe G, Reiss J, Gal A (2003) Association of late-onset Alzheimer disease with a genotype of PLAU, the gene encoding urokinase-type plasminogen activator on chromosome 10q22.2. Neurogenetics 4: 213–217 [DOI] [PubMed] [Google Scholar]
- Fontenot JD, Gavin MA, Rudensky AY (2003) Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol 4: 330–336 [DOI] [PubMed] [Google Scholar]
- Fu W, Ergun A, Lu T, Hill JA, Haxhinasto S, Fassett MS, Gazit R, Adoro S, Glimcher L, Chan S, Kastner P, Rossi D, Collins JJ, Mathis D, Benoist C (2012) A multiply redundant genetic switch 'locks in' the transcriptional signature of regulatory T cells. Nat Immunol 13: 972–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garin MI, Chu CC, Golshayan D, Cernuda-Morollon E, Wait R, Lechler RI (2007) Galectin-1: a key effector of regulation mediated by CD4+CD25+ T cells. Blood 109: 2058–2065 [DOI] [PubMed] [Google Scholar]
- Germain RN (2008) Special regulatory T-cell review: A rose by any other name: from suppressor T cells to Tregs, approbation to unbridled enthusiasm. Immunology 123: 20–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyetko MR, Libre EA, Fuller JA, Chen GH, Toews G (1999) Urokinase is required for T lymphocyte proliferation and activation in vitro. J Lab Clin Med 133: 274–288 [DOI] [PubMed] [Google Scholar]
- He F, Balling R (2012) The role of regulatory T cells in neurodegenerative diseases. Wiley Interdiscip Rev Syst Biol Med DOI doi:; DOI: 10.1002/wsbm.1187 [DOI] [PubMed] [Google Scholar]
- He F, Balling R, Zeng AP (2009) Reverse engineering and verification of gene networks: principles, assumptions, and limitations of present methods and future perspectives. J Biotechnol 144: 190–203 [DOI] [PubMed] [Google Scholar]
- He F, Zeng AP (2006) In search of functional association from time-series microarray data based on the change trend and level of gene expression. BMC Bioinformatics 7: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill JA, Feuerer M, Tash K, Haxhinasto S, Perez J, Melamed R, Mathis D, Benoist C (2007) Foxp3 transcription-factor-dependent and -independent regulation of the regulatory T cell transcriptional signature. Immunity 27: 786–800 [DOI] [PubMed] [Google Scholar]
- Honma K, Kimura D, Tominaga N, Miyakoda M, Matsuyama T, Yui K (2008) Interferon regulatory factor 4 differentially regulates the production of Th2 cytokines in naive versus effector/memory CD4+ T cells. Proc Natl Acad Sci USA 105: 15890–15895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenhower C, Hibbs M, Myers C, Troyanskaya OG (2006) A scalable method for integration and functional analysis of multiple microarray datasets. Bioinformatics 22: 2890–2897 [DOI] [PubMed] [Google Scholar]
- Huynh-Thu VA, Irrthum A, Wehenkel L, Geurts P (2010) Inferring regulatory networks from expression data using tree-based methods. PLoS ONE 5: pii: e12776.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josefowicz SZ, Rudensky A (2009) Control of regulatory T cell lineage commitment and maintenance. Immunity 30: 616–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutson KL, Disis ML, Salazar LG (2007) CD4 regulatory T cells in human cancer pathogenesis. Cancer Immunol Immunother 56: 271–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubach J, Lutter P, Bopp T, Stoll S, Becker C, Huter E, Richter C, Weingarten P, Warger T, Knop J, Mullner S, Wijdenes J, Schild H, Schmitt E, Jonuleit H (2007) Human CD4+CD25+ regulatory T cells: proteome analysis identifies galectin-10 as a novel marker essential for their anergy and suppressive function. Blood 110: 1550–1558 [DOI] [PubMed] [Google Scholar]
- Langfelder P, Horvath S (2008) WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9: 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Beau MM, Espinosa R 3rd, Neuman WL, Stock W, Roulston D, Larson RA, Keinanen M, Westbrook CA (1993) Cytogenetic and molecular delineation of the smallest commonly deleted region of chromosome 5 in malignant myeloid diseases. Proc Natl Acad Sci USA 90: 5484–5488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Date SV, Adai AT, Marcotte EM (2004) A probabilistic functional network of yeast genes. Science 306: 1555–1558 [DOI] [PubMed] [Google Scholar]
- Lee I, Li Z, Marcotte EM (2007) An improved, bias-reduced probabilistic functional gene network of Baker's Yeast, Saccharomyces cerevisiae. PLoS ONE 2: e988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Dai M, Zhuang Y (2004) A T cell intrinsic role of Id3 in a mouse model for primary Sjogren's syndrome. Immunity 21: 551–560 [DOI] [PubMed] [Google Scholar]
- Liu G, Burns S, Huang G, Boyd K, Proia RL, Flavell RA, Chi H (2009) The receptor S1P1 overrides regulatory T cell-mediated immune suppression through Akt-mTOR. Nat Immunol 10: 769–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu LJ, Xia Y, Paccanaro A, Yu H, Gerstein M (2005) Assessing the limits of genomic data integration for predicting protein networks. Genome Res 15: 945–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin AA, Wang K, Califano A, Nemenman I (2010) Multivariate dependence and genetic networks inference. IET Syst Biol 4: 428–440 [DOI] [PubMed] [Google Scholar]
- Marson A, Kretschmer K, Frampton GM, Jacobsen ES, Polansky JK, MacIsaac KD, Levine SS, Fraenkel E, von Boehmer H, Young RA (2007) Foxp3 occupancy and regulation of key target genes during T-cell stimulation. Nature 445: 931–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintick JN, Edenberg HJ (2006) Effects of filtering by Present call on analysis of microarray experiments. BMC Bioinformatics 7: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondino A, Blasi F (2004) uPA and uPAR in fibrinolysis, immunity and pathology. Trends Immunol 25: 450–455 [DOI] [PubMed] [Google Scholar]
- Murawski MR, Litherland SA, Clare-Salzler MJ, Davoodi-Semiromi A (2006) Upregulation of Foxp3 expression in mouse and human Treg is IL-2/STAT5 dependent: implications for the NOD STAT5B mutation in diabetes pathogenesis. Ann NY Acad Sci 1079: 198–204 [DOI] [PubMed] [Google Scholar]
- Niedbala W, Wei X, Liew FY (2002) IL-15 induces type 1 and type 2 CD4+ and CD8+ T cells proliferation but is unable to drive cytokine production in the absence of TCR activation or IL-12/ IL-4 stimulation in vitro. Eur J Immunol 32: 341–347 [DOI] [PubMed] [Google Scholar]
- Nikitin A, Egorov S, Daraselia N, Mazo I (2003) Pathway studio—the analysis and navigation of molecular networks. Bioinformatics 19: 2155–2157 [DOI] [PubMed] [Google Scholar]
- Ochs HD, Ziegler SF, Torgerson TR (2005) FOXP3 acts as a rheostat of the immune response. Immunol Rev 203: 156–164 [DOI] [PubMed] [Google Scholar]
- Ono M, Yaguchi H, Ohkura N, Kitabayashi I, Nagamura Y, Nomura T, Miyachi Y, Tsukada T, Sakaguchi S (2007) Foxp3 controls regulatory T-cell function by interacting with AML1/Runx1. Nature 446: 685–689 [DOI] [PubMed] [Google Scholar]
- Pan F, Yu H, Dang EV, Barbi J, Pan X, Grosso JF, Jinasena D, Sharma SM, McCadden EM, Getnet D, Drake CG, Liu JO, Ostrowski MC, Pardoll DM (2009) Eos mediates Foxp3-dependent gene silencing in CD4+ regulatory T cells. Science 325: 1142–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfoertner S, Jeron A, Probst-Kepper M, Guzman CA, Hansen W, Westendorf AM, Toepfer T, Schrader AJ, Franzke A, Buer J, Geffers R (2006) Signatures of human regulatory T cells: an encounter with old friends and new players. Genome Biol 7: R54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst-Kepper M, Balling R, Buer J (2010) FOXP3: required but not sufficient. the role of GARP (LRRC32) as a safeguard of the regulatory phenotype. Curr Mol Med 10: 533–539 [DOI] [PubMed] [Google Scholar]
- Probst-Kepper M, Geffers R, Kroger A, Viegas N, Erck C, Hecht HJ, Lunsdorf H, Roubin R, Moharregh-Khiabani D, Wagner K, Ocklenburg F, Jeron A, Garritsen H, Arstila TP, Kekalainen E, Balling R, Hauser H, Buer J, Weiss S (2009) GARP: a key receptor controlling FOXP3 in human regulatory T cells. J Cell Mol Med 13: 3343–3357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian J, Dolled-Filhart M, Lin J, Yu H, Gerstein M (2001) Beyond synexpression relationships: local clustering of time-shifted and inverted gene expression profiles identifies new, biologically relevant interactions. J Mol Biol 314: 1053–1066 [DOI] [PubMed] [Google Scholar]
- Quo CF, Kaddi C, Phan JH, Zollanvari A, Xu M, Wang MD, Alterovitz G (2012) Reverse engineering biomolecular systems using -omic data: challenges, progress and opportunities. Brief Bioinformatics 13: 430–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan J, Dean AK, Zhang W (2010) A general co-expression network-based approach to gene expression analysis: comparison and applications. BMC Syst Biol 4: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi S (2004) Naturally arising CD4+ regulatory t cells for immunologic self-tolerance and negative control of immune responses. Annu Rev Immunol 22: 531–562 [DOI] [PubMed] [Google Scholar]
- Sakaguchi S (2005) Naturally arising Foxp3-expressing CD25+CD4+ regulatory T cells in immunological tolerance to self and non-self. Nat Immunol 6: 345–352 [DOI] [PubMed] [Google Scholar]
- Sakaguchi S, Miyara M, Costantino CM, Hafler DA (2010) FOXP3+ regulatory T cells in the human immune system. Nat Rev Immunol 10: 490–500 [DOI] [PubMed] [Google Scholar]
- Sakaguchi S, Ono M, Setoguchi R, Yagi H, Hori S, Fehervari Z, Shimizu J, Takahashi T, Nomura T (2006) Foxp3+ CD25+ CD4+ natural regulatory T cells in dominant self-tolerance and autoimmune disease. Immunol Rev 212: 8–27 [DOI] [PubMed] [Google Scholar]
- Schafer J, Strimmer K (2005) An empirical Bayes approach to inferring large-scale gene association networks. Bioinformatics 21: 754–764 [DOI] [PubMed] [Google Scholar]
- Shalev I, Liu H, Koscik C, Bartczak A, Javadi M, Wong KM, Maknojia A, He W, Liu MF, Diao J, Winter E, Manuel J, McCarthy D, Cattral M, Gommerman J, Clark DA, Phillips MJ, Gorczynski RR, Zhang L, Downey G et al. (2008) Targeted deletion of fgl2 leads to impaired regulatory T cell activity and development of autoimmune glomerulonephritis. J Immunol 180: 249–260 [DOI] [PubMed] [Google Scholar]
- Shevach EM (2009) Mechanisms of foxp3+ T regulatory cell-mediated suppression. Immunity 30: 636–645 [DOI] [PubMed] [Google Scholar]
- Stelniec-Klotz I, Legewie S, Tchernitsa O, Witzel F, Klinger B, Sers C, Herzel H, Bluthgen N, Schafer R (2012) Reverse engineering a hierarchical regulatory network downstream of oncogenic KRAS. Mol Syst Biol 8: 601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockis J, Fink W, Francois V, Connerotte T, de Smet C, Knoops L, van der Bruggen P, Boon T, Coulie PG, Lucas S (2009) Comparison of stable human Treg and Th clones by transcriptional profiling. Eur J Immunol 39: 869–882 [DOI] [PubMed] [Google Scholar]
- Subramanian S, Shahaf G, Ozeri E, Miller LM, Vandenbark AA, Lewis EC, Offner H (2011) Sustained expression of circulating human alpha-1 antitrypsin reduces inflammation, increases CD4+FoxP3+ Treg cell population and prevents signs of experimental autoimmune encephalomyelitis in mice. Metab Brain Dis 26: 107–113 [DOI] [PubMed] [Google Scholar]
- Toh H, Horimoto K (2002) Inference of a genetic network by a combined approach of cluster analysis and graphical Gaussian modeling. Bioinformatics 18: 287–297 [DOI] [PubMed] [Google Scholar]
- Tran DQ, Andersson J, Hardwick D, Bebris L, Illei GG, Shevach EM (2009) Selective expression of latency-associated peptide (LAP) and IL-1 receptor type I/II (CD121a/CD121b) on activated human FOXP3+ regulatory T cells allows for their purification from expansion cultures. Blood 113: 5125–5133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulisse S, Baldini E, Sorrenti S, D'Armiento M (2009) The urokinase plasminogen activator system: a target for anti-cancer therapy. Curr Cancer Drug Targets 9: 32–71 [DOI] [PubMed] [Google Scholar]
- Walhout AJ (2011) What does biologically meaningful mean? A perspective on gene regulatory network validation. Genome Biol 12: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Kozhaya L, Mercer F, Khaitan A, Fujii H, Unutmaz D (2009) Expression of GARP selectively identifies activated human FOXP3+ regulatory T cells. Proc Natl Acad Sci USA 106: 13439–13444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wing K, Sakaguchi S (2010) Regulatory T cells exert checks and balances on self tolerance and autoimmunity. Nat Immunol 11: 7–13 [DOI] [PubMed] [Google Scholar]
- Wu Y, Borde M, Heissmeyer V, Feuerer M, Lapan AD, Stroud JC, Bates DL, Guo L, Han A, Ziegler SF, Mathis D, Benoist C, Chen L, Rao A (2006) FOXP3 controls regulatory T cell function through cooperation with NFAT. Cell 126: 375–387 [DOI] [PubMed] [Google Scholar]
- Yamazaki T, Yang XO, Chung Y, Fukunaga A, Nurieva R, Pappu B, Martin-Orozco N, Kang HS, Ma L, Panopoulos AD, Craig S, Watowich SS, Jetten AM, Tian Q, Dong C (2008) CCR6 regulates the migration of inflammatory and regulatory T cells. J Immunol 181: 8391–8401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP (2002) Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res 30: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Chen Y, Wu Q, Li P, Shao Y, Zhang J, Zhong Q, Peng X, Yang H, Hu X, Chen B, Guan M, Wan J, Zhang W (2011) The association between single-nucleotide polymorphisms of NCF2 and systemic lupus erythematosus in Chinese mainland population. Clin Rheumatol 30: 521–527 [DOI] [PubMed] [Google Scholar]
- Zheng Y, Chaudhry A, Kas A, deRoos P, Kim JM, Chu TT, Corcoran L, Treuting P, Klein U, Rudensky AY (2009) Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control T(H)2 responses. Nature 458: 351–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Josefowicz SZ, Kas A, Chu TT, Gavin MA, Rudensky AY (2007) Genome-wide analysis of Foxp3 target genes in developing and mature regulatory T cells. Nature 445: 936–940 [DOI] [PubMed] [Google Scholar]
- Ziegler SF (2006) FOXP3: of mice and men. Annu Rev Immunol 24: 209–226 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures S1-18, Supplementary tables S1-8
Data Availability Statement
The complete time-series microarray data set (81 arrays) is available in the Gene Expression Omnibus (GSE11292).