Abstract
The deep sequencing of an mRNA population, RNA-seq, is a very successful application of next-generation sequencing technologies (NGSTs). RNA-seq takes advantage of two key NGST features: (1) samples can be mixtures of different DNA pieces, and (2) sequencing provides both qualitative and quantitative information about each DNA piece analyzed. We recently used RNA-seq to study the transcriptome of Aspergillus fumigatus, a deadly human fungal pathogen. Analysis of the RNA-seq data indicates that there are likely tens of unannotated and hundreds of novel genes in the A. fumigates transcriptome, mostly encoding for small proteins. Inspection of transcriptome-wide variation between two isolates reveals thousands of single nucleotide polymorphisms. Finally, comparison of the transcriptome profiles of one isolate in two different growth conditions identified thousands of differentially-expressed genes. These results demonstrate the utility and potential of RNA-seq for functional genomics studies in A. fumigatus and other fungal human pathogens.
Keywords: novel genes, annotation, population structure, differential expression, transcriptome profiling
Introduction
Recent technological advances in genome science have enabled researchers to routinely generate unprecedented amounts of sequence data from almost any species, opening the floodgates for the study of the genome content and function in non-model organisms.1,2 The main catalyst for these changes has been the development of several different so called next-generation sequencing technologies (NGSTs).3 Astonishingly, the amount of sequence data that a single NGST machine can currently produce in a few days is larger than the total amount of sequence data ever collected and deposited in sequence databases by individual users through traditional methods.4 Importantly, NGST technologies yield not only qualitative information about the sequence of every DNA fragment analyzed, but also quantitative information about the relative abundance of each DNA fragment in the library sequenced.1
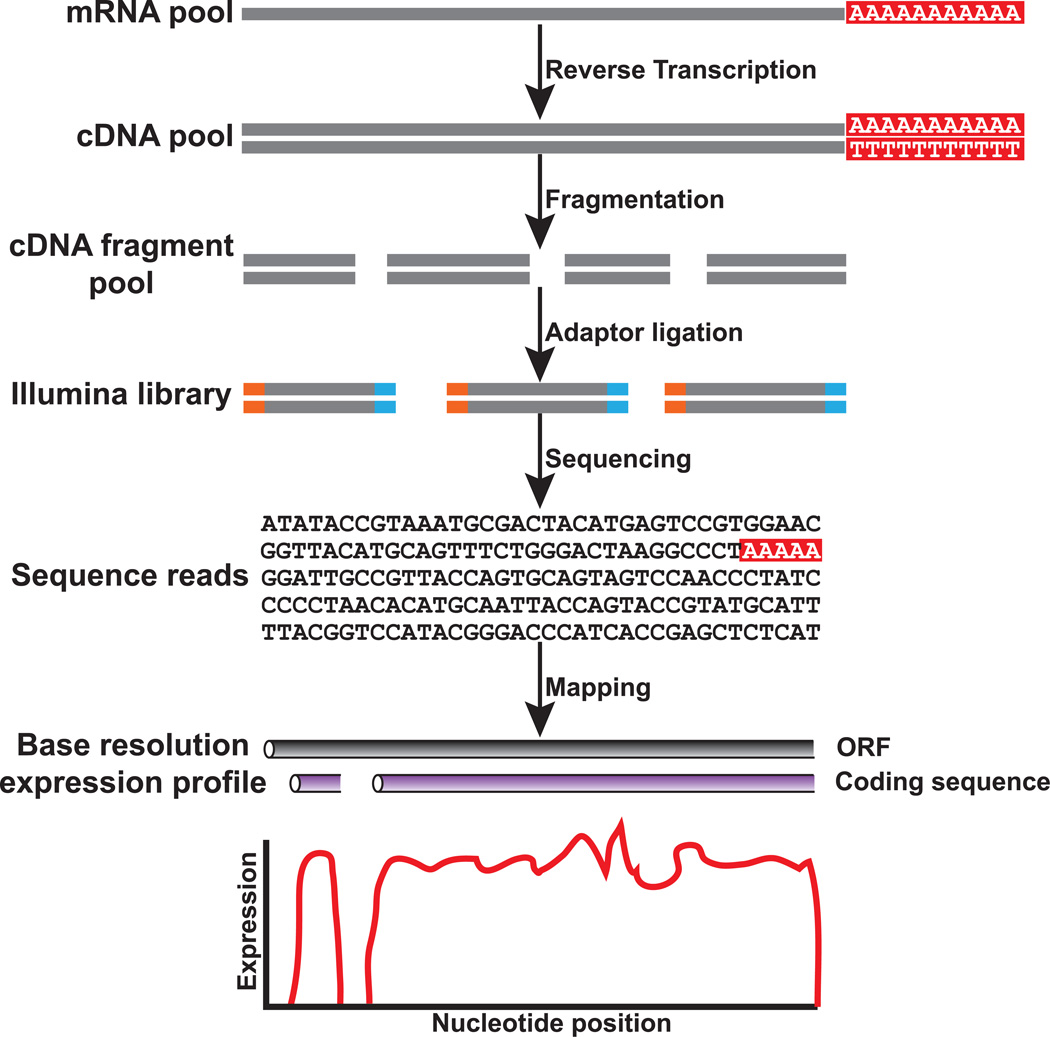
The abundance of NGST-produced data, their qualitative and quantitative nature, and their applicability to any organism for which fresh DNA or RNA is available, has enabled researchers to tailor NGSTs for a variety of different questions beyond the sequencing of genomes.1,5 One of the most powerful such applications is RNA-seq, the employment of NGSTs for transcriptome profiling.6,7 A typical RNA-seq experiment begins with the isolation of mRNA, its conversion into cDNA, followed by fragmentation and addition of adaptors to each DNA fragment’s ends (Fig. 1). Sequencing the library of fragments in a high-throughput fashion returns as many as billions of sequence reads that can vary in length and in their characteristics (e.g., single-end or paired-end), depending on the NGST technology being employed and the experiment being conducted3. Once sequence reads have been obtained, they can be used for a wide variety of functional analyses, including but not limited to the study of alternative splicing, gene expression, allele-specific expression, identification of transcription start sites, identification of gene fusion,6,8,9 as well as for a variety of evolutionary analyses.2,10 For example, in what is perhaps its most frequent application, when RNA-seq is performed on an organism whose genome and annotation is already characterized, one can directly map the sequence reads to the reference genome or transcriptome, thus simultaneously calculating its abundance as well as its sequence (Fig. 1).
Figure 1.
The workflow of a typical RNA-seq experiment. Briefly, following isolation, the mRNA pool is converted to a cDNA pool and is then fragmented. Next, NGST adaptors are added to each cDNA fragment, the resulting library of fragments is sequenced, and the sequence of each fragment is read using NGST. Once sequence reads have been obtained, they can be used for a variety of analyses. For example, if the mRNA pool is from an organism whose genome and annotation is known, the sequence reads can be aligned or mapped to the reference genome or transcriptome and the sequence of the entire transcript as well as its relative expression can be calculated. Thus, RNA-seq technology is simultaneously qualitative (i.e., it can determine the sequences of different sequence fragments in a pool) and quantitative (i.e., it can determine the relative abundance of different sequence fragments in a pool).
The filamentous fungal genus Aspergillus contains approximately 250 species and spans over 200 million years of evolutionary history.11 Several species in the genus can cause a range of frequently deadly diseases, which are collectively known as aspergillosis.12,13 Aspergillosis usually affects individuals that have compromised immune defences and is established following inhalation of Aspergillus spores. The great majority of Aspergillus-induced infections is caused by A. fumigatus,12,14,15 a very abundant and widely distributed species. A. fumigatus is one of the most common species found in decaying vegetation, and a prolific spore producer.16–19 When A. fumigatus establishes an infection in the human lung, it usually forms a dense colony of filaments embedded in a polymeric extracellular matrix.20,21 To identify candidate genes involved in this colony or biofilm-like growth (COG), we previously used RNA-seq to compare the transcriptomes of COG and liquid planktonic growth (PLG) conditions.22 Here, we use this data to highlight the multitude of utilities of RNA-seq technology for functional genomics studies of A. fumigatus, by focusing specifically on three applications: characterizing the structure of the A. fumigatus transcriptome; measuring transcriptome-wide levels of variation between isolates; and, finally, comparing the transcriptome-wide expression profile of A. fumigatus during different non-nutritional growth conditions.
RNA-seq for annotation: characterizing transcriptome structure
Our RNA-seq data provided very good coverage of the A. fumigatus Af293 reference transcriptome. Specifically, 27,236,154 sequence reads, each 42 bp long, generated by RNA-seq from growth of the ATCC46645 isolate in the COG and PLG conditions, were mapped against the Af293 reference transcriptome. Sequence reads mapped to 90.5% (8,952/9,887) of reference transcripts, recovering 77% of the sequence of the reference transcriptome (11.1/14.4 Mb). Approximately 73% of the sequence of the average transcript was recovered but for over 60% of the transcripts more than 90% of their sequence was recovered.
To identify the extent of unannotated and novel genes in the A. fumigatus genome, we further mapped the RNA-seq generated sequence reads to the A. fumigatus Af293 reference genome, using the reference gene models23 as guides. Briefly, gene models are hypotheses about the structure of transcripts produced by the set of genes in a genome. Although the majority of gene models constructed through the annotation of the reference Af293 isolate is of high quality and supported by a variety of evidence (e.g., expressed sequence tags, high similarity scores to other genes, etc.), annotation is a very challenging process and examples of misannotated genes or genes omitted from an annotation are present in any eukaryotic genome.24 After the mapping step, we assembled the mapped reads into gene models using two different state-of-the-art programs, Cufflinks25 and Scripture.26 Interestingly, whereas the Cufflinks program is designed to maximize precision, the Scripture programis designed to maximize sensitivity, resulting in significantly different annotations from the same set of data, especially for lower expressed genes.9 We then compared the annotation produced by the two programs to the reference annotation and on the basis of the results classified, the gene models constructed by the two programs as annotated, unannotated or novel. Gene models whose sequence overlapped with that from any reference gene and whose location was on the same strand as their reference counterpart were considered to be “annotated.” For remaining gene models, we used their protein sequence products in BLAST similarity searches against the NCBI nr database for the presence of homologs in the genome of any organism. Gene models with significant hit(s) in the nr database were considered “unannotated,” whereas gene models without any hits in the same database were considered “novel.”
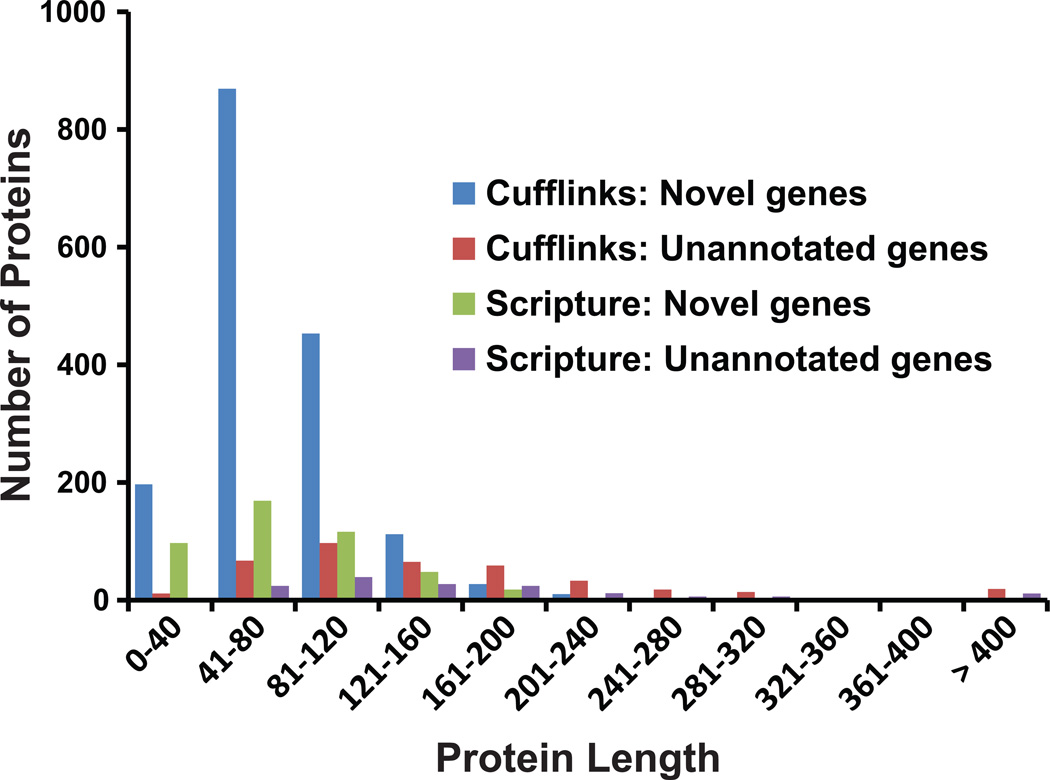
Examination of the annotation produced from the Cufflinks and Scripture programs using the RNA-seq data identified hundreds of unannotated and thousands of novel gene models (Table 1). In line with previous analyses,9 the numbers of unannotated and novel gene models differed greatly between the two programs. Interestingly, the great majority of novel genes predicted by the two programs encoded for small proteins (Fig. 2), with 91% (1,519/1,673) and 83% (382/460) of the protein products of novel gene models constructed by Cufflinks and Scripture, respectively, being equal or shorter than 120 amino acids (aa). In contrast, the percentages for unannotated genes were 45% (Cufflinks) and 43% (Scripture). Similarly, the median lengths of novel proteins were 68 aa (Cufflinks) and 72 aa (Scripture), whereas the median lengths of unannotated proteins were 132 aa (Cufflinks) and 136 aa (Scripture). Although these analyses are preliminary, the RNA-seq data suggest that, conservatively, there are likely tens of previously unannotated and hundreds of novel gene models in A. fumigatus, the great majority of which encode for small proteins. Determining the function of these small proteins as well as their role in pathogenicity, if any, represents a very interesting research challenge and opportunity for future functional genomics research in A. fumigatus.
Table 1.
Summary of unannotated and novel genes reconstructed using RNA-seq data
| Software | Annotated genes | Unannotated genes |
Novel genes | |
|---|---|---|---|---|
| Protein- coding |
RNA-coding | |||
| Cufflinks | 7,764 (8663) | 390 (411) | 24 (25) | 1,673 (1,703) |
| Scripture | 4,285 (6852) | 156 (214) | 23 (54) | 462 (653) |
Number in bracket indicates the total number of transcripts in each category. Some loci are included in more than one category.
Figure 2.
The majority of putative novel genes identified by RNA-seq in A. fumigatus encodes for small proteins. ATCC46645 sequence reads were mapped against the A. fumigatus af293 reference genome with the TopHat software,43 using the reference gene models23 as guides and not allowing for introns > 1,000 bp. Mapped reads were assembled into transcripts using the Cufflinks25 and Scripture26 programs, and their gene predictions were compared to the reference gene models. ORFs overlapping with exons of any reference gene model were classified as “annotated” genes. For the remaining ORFs, their protein products were searched against the NCBI nr database; these loci were classified either as “unannotated,” if the encoded protein had at least one homolog in the NCBInr database, or as “novel” if the encoded protein had no homologs in the NCBInr database. The x-axis corresponds to groups of protein lengths encoded by putative novel or unannotated genes identified by the Cufflinks and Scripture programs (indicated by bars of different colors). The y-axis corresponds to the number of genes belonging to each group.
RNA-seq for population genetics: characterizing transcriptome variation between isolates
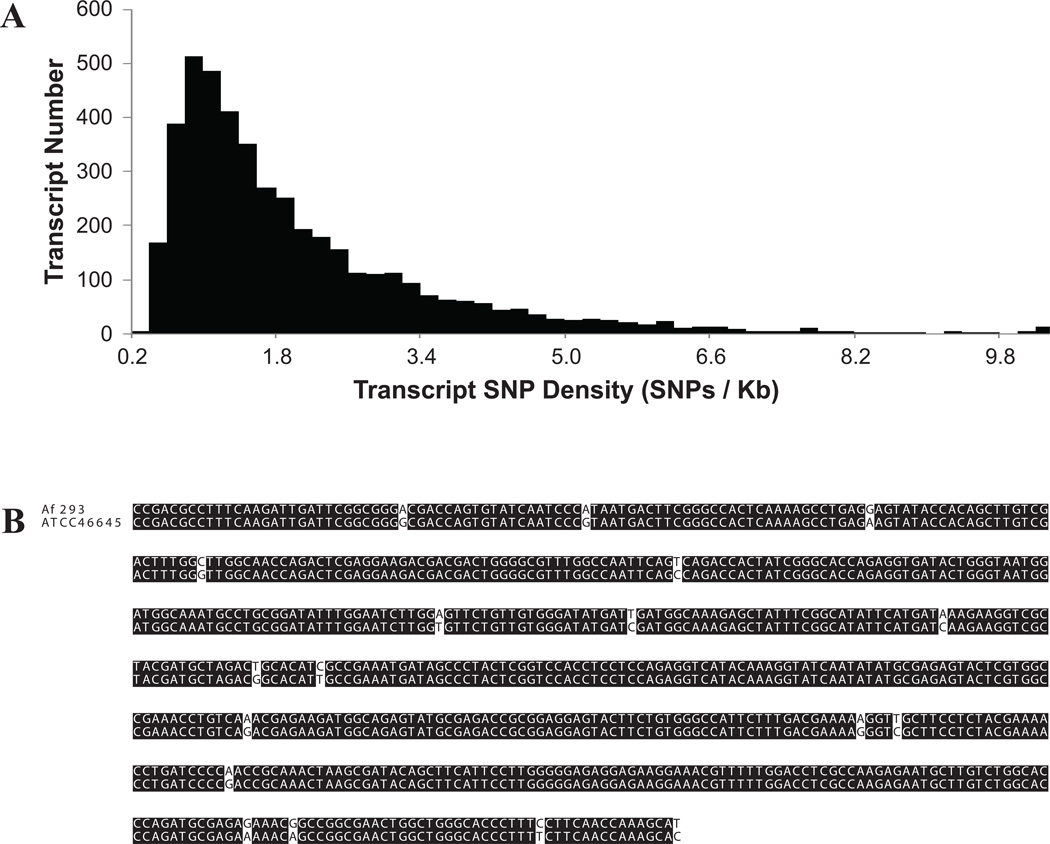
Although many fungal species, including several human pathogens, show populationstructure,27–29 it was thought that A. fumigatus lacks populationstructure.30–32 However, two recent multilocus studies, one using isolates from around the world31 and the other using isolates from the Netherlands,33 identified genetically distinct lineages within A. fumigatus, suggesting that the absence of population structure in older studies could be due to the use of fewer and less informative markers. To evaluate the potential of RNA-seq to provide novel markers for population genetic and, more generally, evolutionary analysis we compared the transcriptomes of ATCC46645 (reconstructed through mapping to the Af293 reference transcriptome) and Af293 isolates. Examination of sequence alignments between the two isolates from 8,952 genes identified 12,872 single nucleotide polymorphisms (SNPs) in 4,923 genes, representing nearly 50% (4,923/9,887) of A. fumigatus reference genes. The average number of SNPs in variable genes was 2.6, with nearly two-thirds of the genes containing 1 or 2 SNPs, 12 containing more than 10 SNPs, and two containing 40 SNPs. Per kilobase of transcriptome sequence, the average SNP density was 1.2 (12,872 SNPs / 11,109,536 recovered nucleotides; Fig. 3A), with 12 genes showing SNP densities greater than 10 (Table 2 and Fig. 3B). Finally, 51% of SNPs were nonsynonymous substitutions, with the remaining 49% being synonymous ones.
Figure 3.
Transcriptome-wide variation between A. fumigatus strains ATCC46645 and Af293. (A) Histogram plot of transcript SNP density. The x-axis corresponds to groups of different transcript SNP densities (number of SNPs/Kb). The y-axis corresponds to the number of transcripts in each SNP density group. (B) Partial nucleotide sequence alignment between Af293 and ATCC46645 strains for a putative protein kinase (Afu3g03740), one of the genes with the highest SNP density in our comparison (see also Table 2). High quality SNPs were identified by filtering for variable sites with coverage values ≥ 5 and an average quality scores ≥ 20 using the cns2snp script in the Maq software.44 Pairwise nucleotide diversity was calculated as π = n/N; where n = the number of differences between sequences and N = the total number of sites examined. SNP density per Kb was calculated as π * 1,000.
Table 2.
Transcripts with SNP densities greater than 10 SNPs per Kilobase
| Gene Name | Function | SNP Density (total SNPs) |
|---|---|---|
| Afu2g01890 | CAT5 protein | 10.4 (8) |
| Afu2g17900 | conserved hypothetical protein | 14.3 (7) |
| Afu3g01500 | hypothetical protein | 10.3 (7) |
| Afu3g03740 | putative protein kinase | 27.0 (18) |
| Afu3g07310 | conserved hypothetical protein | 14.5 (12) |
| Afu4g00580 | hypothetical protein | 10.1 (10) |
| Afu5g00890 | hypothetical protein | 10.2 (2) |
| Afu5g01650 | putative bZIP transcription factor (JlbA) | 10.4 (7) |
| Afu6g03420 | clock-controlled gene-9 protein | 11.4 (7) |
| Afu7g08410 | putative transposase | 19.3 (18) |
| Afu7g08470 | peroxisomal copper amine oxidase | 11.0 (18) |
| Afu8g06430 | hypothetical protein | 12.2 (3) |
The identification of thousands of SNPs between the two isolates, suggests that in addition to whole-genome sequencing, RNA-seq is a powerful tool for the study of genetic differentiation in A. fumigatus. Importantly, because the A. fumigatus transcriptome is approximately 50% of the genome and because transcripts are grossly unevenly abundant (varying over several orders of magnitude22), even shallow NGST sequencing should provide in depth sampling of a few hundred loci, and of hundreds if not thousands of SNPs, simply by sequencing transcripts in proportion to their representation in the library.10 Thus, RNA-seq is a powerful alternative to the standard multilocus sequence typing currently used for the study of isolate identification and population structure in A. fumigatus, and in filamentous fungi in general.34
RNA-seq for functional genomics: characterizing global transcriptome changes between different growth conditions
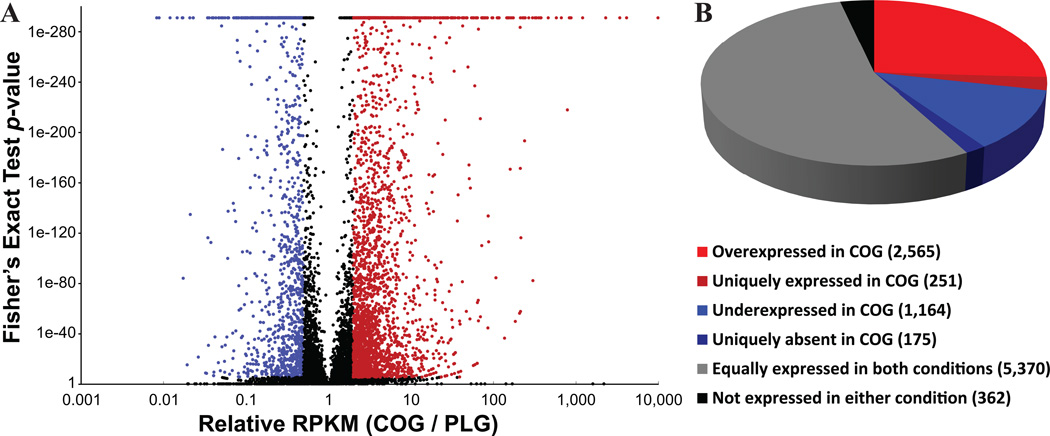
The most common application of RNA-seq is for the identification of genes that show differential regulation under certain conditions. Examination of gene expression using our RNA-seq data obtained from A. fumigatus growth in the COG and PLG conditions revealed that 92% ofreference transcripts (9,099/9,887) were expressed in both conditions and 4.3% (426/9,887) were uniquely expressed in either condition. By considering differentially expressed genes as only those that exhibited a 2-fold biological difference in relative gene expression between conditions and a statistically significant P value below 5.5e–06, we identified 2,861 genes that were either significantly upregulated in COG relative to PLG or uniquely expressed in COG, and 1,339 that were either significantly downregulated in the same comparisonor uniquely expressed in PLG (Fig. 4A). The remaining genes either showed uniform expression in the two conditions (5,370) or were not expressed in either condition (362; Fig. 4B). Remarkably, the range of expression values in both samples ranged seven orders of magnitude.
Figure 4.
RNA-seq identifies thousands of differentially expressed genes from A. fumigatus ATCC46645 grown in the colony (COG) and plankton (PLG) growth conditions. (A) Volcano plot of the differentially regulated genes between COG and PLG conditions. For each gene, the rRPKM value (RPKM(COG) / RPKM(PLG)) was plotted against its respective Fisher’s exact test P value. P values smaller than 1e−290 were reported as 1e−290. The dotted line running parallel to the x-axis indicates the statistical cutoff (P < 5.5e−6), whereas the dotted line running parallel to the y-axis indicates the biological cutoff (2-fold difference in RPKM between COG and PLG). The red-colored and blue-colored dots correspond to upregulated and downregulated genes between COG and PLG, respectively. (B) Pie chart showing the partitioning of the 9,887 A. fumigatus genes with respect to their expression profile in the two growth conditions.
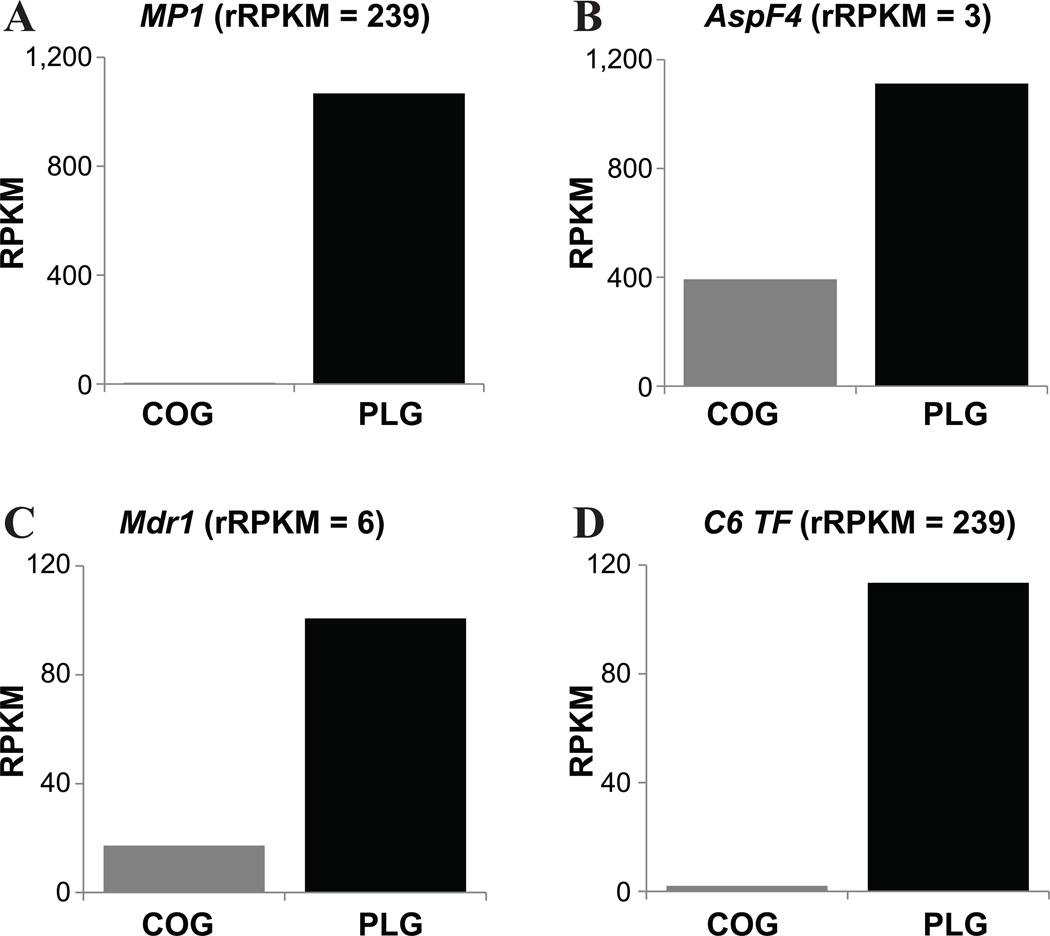
Upregulated and downregulated genes were non-randomly distributed across the genome and showed strong association with specific functional categories (Fig. 5).22 Some of the strongest associations were for cell wall genes (out of 409 genes, 169 were significantly upregulated and only 41 were significantly downregulated), for pump and transporter genes (out of 319 genes, 146 were upregulated and 16 were downregulated), and for allergens (out of 81 genes, 41 were upregulated and 13 were downregulated). Thus, the application of RNA-seq to study a single difference in non-nutritional environmental conditions identified thousands of differentially expressed genes. Considering that the breadth and sensitivity of other technologies for measuring macromolecule abundance differences, such as microarrays and 2-D gel electrophoresis, are much narrower,35 RNA-seq appears to be the most powerful tool for genome-wide functional comparisons of fungal growth to date.
Figure 5.
Examples of differentially regulated genes from specific functional categories that constitute candidates for the observed pathobiological and morphological differences between the two conditions. (A) The cell wall galactomannoprotein MP1(Afu4g03240). (B) The allergen AspF4 (Afu2g03830). (C) The ABC multidrug transporter Mdr1 (Afu5g06070). (D) The C6 transcription factor of the fumitremorgin secondary metabolism gene cluster (Afu8g00420). In each graph, the x-axis corresponds to the two growth conditions (COG in black and PLG in gray) and the y-axis to the RPKM expression value of corresponding gene in each of the two conditions. The rRPKM (RPKM (COG) /RPKM (PLG)) value for each comparison is reported in parentheses next to each gene’s name.
Finally, it important to emphasize that the gene expression values measured by RNA-seq do not show significant variation when replicated. Biological and technical replicates are not yet standard in the RNA-seq literature, for the simple reason that the RNA-seq technique is much more accurate than microarrays,36 although the inclusion of technical and biological replicates offers additional power.37 We performed both biological and technical replicates for a subset of our RNA-seq experiments to verify that our RNA-seq experiments worked as expected.22 We observed a very high degree of replicability of our results at both the biological and the technical level in the subset tested. For example, the correlation values between three biological PLG replicates performed on three different A. fumigatus strains as well as on one technical BFG replicate are extremely high (r > 0.91),22 on par with similar studies in the literature.38 These data suggest that not only are our results unaffected by biological or technical replication issues, but also that they hold across different A. fumigatus strains.
Designing and executing a RNA-seq experiment
RNA-seq is a very versatile tool that can be used to address a wide variety of basic and applied science questions, from increasing the genomic depth of the tree of life10 to characterizing the genetic makeup of cancer in the human body.39 Consequently, the design of a RNA-seq experiment will vary depending on the question asked and the nature of the investigation, which will in turn determine the acquisition of RNA-seq data, its handling and analysis, as well as the pursuit of follow-up experiments.
In contrast to many other types of bioinformatics analyses where the tools (e.g., the BLAST algorithm40 is the near universal choice for examining the identity of a sequence) and databases (e.g., the PFAM database41 is one of a few standard protein domain databases) are well established, the toolkit for the analysis of RNA-seq data is far less well defined, largely because both the technology and its bioinformatics tools are not only very new but also rapidly changing. Although this pace of change in technology and software makes the design of benchmark studies challenging, a very useful resource for RNA-seq and NGSTs in general is the http://seqanswers.com/ website that aims to provide “an information resource and user-driven community focused on all aspects of next-generation genomics,” and which routinely hosts discussions on a variety of topics such as analysis practices ands oftware choice or performance. Although RNA-seq, and NGSTs in general, are touted as “cheap” technologies, it should be emphasized that the most important challenge to applying this technology is the cost associated with the bioinformatics analysis of the data.
Because of the large-scale nature of RNA-seq experiments, considerable attention is also required to choosing the appropriate experimental design. Fang and Cui42 recently described a number of experimental design principles that require careful consideration, some of which were discussed in the paragraphs above, including randomization of samples, technical and biological replication of the experiment, the depth and type of sequencing that needs to be performed, as well as whether validation of the results is required by an independent approach, such as qRT-PCR.
Conclusions
Even though NGSTs and RNA-seq are less than a decade old, it is abundantly clear that they have dramatically altered the landscape of functional genomics studies in non-model organisms. Analysis of the data produced by a single experiment in A. fumigatus has uncovered tens of putative unannotated and hundreds of novel small genes, thousands of SNPs, and hundreds of candidates for downstream functional experiments to identify the molecular basis of colony growth and its potential role in the establishment of some forms of aspergillosis. In the near future, we anticipate that RNA-seq applications will not only lead to far greater understanding of the parts, structure, and function of the A. fumigatus genome, but will also identify the key differences between in vitro and in vivo models of the disease, as well as define the molecular interactions between the human host and the fungal pathogen during infection.
Acknowledgments
This work was conducted in part using the resources of the Advanced Computing Center for Research and Education at Vanderbilt University. J.G.G. is funded by the graduate program in biological sciences at Vanderbilt University and the National Institute of Allergy and Infectious Diseases, National Institutes of Health (NIH, NIAID: F31AI091343-01). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIAID or the NIH. Work in J.-P.L.’s Aspergillus lab is partly funded by the ESF Grant Fuminomics and the ALLFUN FP7 project. Research in A.R.’s lab is supported by the Searle Scholars Program and the National Science Foundation (DEB-0844968).
Footnotes
Conflicts of interest
The authors declare no conflicts of interest.
References
- 1.Rokas A, Abbot P. Harnessing genomics for evolutionary insights. Trends Ecol. Evol. 2009;24:192–200. doi: 10.1016/j.tree.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 2.Gibbons JG, et al. Benchmarking next-generation transcriptome sequencing for functional and evolutionary genomics. Mol. Biol. Evol. 2009;26:2731–2744. doi: 10.1093/molbev/msp188. [DOI] [PubMed] [Google Scholar]
- 3.Glenn TC. Field guide to next-generation DNA sequencers. Mol. Ecol. Resour. 2011;11:759–769. doi: 10.1111/j.1755-0998.2011.03024.x. [DOI] [PubMed] [Google Scholar]
- 4.Gilad Y, Pritchard JK, Thornton K. Characterizing natural variation using next-generation sequencing technologies. Trends Genet. 2009;25:463–471. doi: 10.1016/j.tig.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kahvejian A, Quackenbush J, Thompson JF. What would you do if you could sequence everything? Nat. Biotechnol. 2008;26:1125–1133. doi: 10.1038/nbt1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Z, Gerstein M, Snyder M. RNA-seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wold B, Myers RM. Sequence census methods for functional genomics. Nat. Meth. 2008;5:19–21. doi: 10.1038/nmeth1157. [DOI] [PubMed] [Google Scholar]
- 8.Ozsolak F, Milos PM. RNA sequencing: advances, challenges and opportunities. Nat. Rev. Genet. 2011;12:87–98. doi: 10.1038/nrg2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garber M, Grabherr MG, Guttman M, Trapnell C. Computational methods for transcriptome annotation and quantification using RNA-seq. Nat. Meth. 2011;8:469–477. doi: 10.1038/nmeth.1613. [DOI] [PubMed] [Google Scholar]
- 10.Hittinger CT, Johnston M, Tossberg JT, Rokas A. Leveraging skewed transcript abundance by RNA-seq to increase the genomic depth of the tree of life. Proc. Natl. Acad. Sci. USA. 2010;107:1476–1481. doi: 10.1073/pnas.0910449107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geiser DM, et al. The current status of species recognition and identification in Aspergillus. Stud. Mycol. 2007;59:1–10. doi: 10.3114/sim.2007.59.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Denning DW. Invasive aspergillosis. Clin. Infect. Dis. 1998;26:781–803. doi: 10.1086/513943. [DOI] [PubMed] [Google Scholar]
- 13.Latge JP. Aspergillus fumigatus and aspergillosis. Clin. Microbiol. Rev. 1999;12:310–350. doi: 10.1128/cmr.12.2.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morgan J, et al. Incidence of invasive aspergillosis following hematopoietic stem cell and solid organ transplantation: interim results of a prospective multicenter surveillance program. Med. Mycol. 2005;43(Suppl 1):S49–S58. doi: 10.1080/13693780400020113. [DOI] [PubMed] [Google Scholar]
- 15.Schmitt HJ, Blevins A, Sobeck K, Armstrong D. Aspergillus species from hospital air and from patients. Mycoses. 1990;33:539–541. doi: 10.1111/myc.1990.33.11-12.539. [DOI] [PubMed] [Google Scholar]
- 16.Klich MA. Biogeography of Aspergillus species in soil and litter. Mycologia. 2002;94:21–27. [PubMed] [Google Scholar]
- 17.Shelton BG, Kirkland KH, Flanders WD, Morris GK. Profiles of airborne fungi in buildings and outdoor environments in the United States. Appl. Environ. Microbiol. 2002;68:1743–1753. doi: 10.1128/AEM.68.4.1743-1753.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klich MA. Health effects of Aspergillus in food and air. Toxicol. Ind. Health. 2009;25:657–667. doi: 10.1177/0748233709348271. [DOI] [PubMed] [Google Scholar]
- 19.Raper KB, Fennell DI. The Genus Aspergillus. Baltimore: Williams & Wilkins; 1965. [Google Scholar]
- 20.Loussert C, et al. In vivo biofilm composition of Aspergillus fumigatus. Cell. Microbiol. 2010;12:405–410. doi: 10.1111/j.1462-5822.2009.01409.x. [DOI] [PubMed] [Google Scholar]
- 21.Beauvais A, et al. An extracellular matrix glues together the aerial-grown hyphae of Aspergillus fumigatus. Cell. Microbiol. 2007;9:1588–1600. doi: 10.1111/j.1462-5822.2007.00895.x. [DOI] [PubMed] [Google Scholar]
- 22.Gibbons JG, et al. Global transcriptome changes underlying colony growth in the opportunistic human pathogen Aspergillus fumigatus. Eukaryot. Cell. 2012;11:68–78. doi: 10.1128/EC.05102-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nierman WC, et al. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature. 2005;438:1151–1156. doi: 10.1038/nature04332. [DOI] [PubMed] [Google Scholar]
- 24.Yandell M, Ence D. A beginner's guide to eukaryotic genome annotation. Nat. Rev. Genet. 2012;13:329–342. doi: 10.1038/nrg3174. [DOI] [PubMed] [Google Scholar]
- 25.Trapnell C, et al. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guttman M, et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat. Biotechnol. 2010;28:503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hittinger CT, et al. Remarkably ancient balanced polymorphisms in a multi-locus gene network. Nature. 2010;464:54–58. doi: 10.1038/nature08791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Milgroom MG. Recombination and the multilocus structure of fungal populations. Annu. Rev. Phytopathol. 1996;34:457–477. doi: 10.1146/annurev.phyto.34.1.457. [DOI] [PubMed] [Google Scholar]
- 29.Taylor JW, Turner E, Townsend JP, Dettman JR, Jacobson D. Eukaryotic microbes, species recognition and the geographic limits of species: examples from the kingdom Fungi. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2006;361:1947–1963. doi: 10.1098/rstb.2006.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Debeaupuis JP, Sarfati J, Chazalet V, Latge JP. Genetic diversity among clinical and environmental isolates of Aspergillus fumigatus. Infect. Immun. 1997;65:3080–3085. doi: 10.1128/iai.65.8.3080-3085.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pringle A, et al. Cryptic speciation in the cosmopolitan and clonal human pathogenic fungus Aspergillus fumigatus. Evolution. 2005;59:1886–1899. [PubMed] [Google Scholar]
- 32.Rydholm C, Szakacs G, Lutzoni F. Low genetic variation and no detectable population structure in Aspergillus fumigatus compared to closely related Neosartorya species. Eukaryot. Cell. 2006;5:650–657. doi: 10.1128/EC.5.4.650-657.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klaassen CH, Gibbons JG, Fedorova N, Meis JF, Rokas A. Evidence for genetic differentiation and variable recombination rates among Dutch populations of the opportunistic human pathogen Aspergillus fumigatus. Mol. Ecol. 2012;21:57–70. doi: 10.1111/j.1365-294X.2011.05364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klaassen CH. MLST versus microsatellites for typing Aspergillus fumigatus isolates. Med. Mycol. 2009;47(Suppl 1):S27–S33. doi: 10.1080/13693780802382244. [DOI] [PubMed] [Google Scholar]
- 35.Bruns S, et al. Functional genomic profiling of Aspergillus fumigatus biofilm reveals enhanced production of the mycotoxin gliotoxin. Proteomics. 2010;10:3097–3107. doi: 10.1002/pmic.201000129. [DOI] [PubMed] [Google Scholar]
- 36.Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Auer PL, Doerge RW. Statistical design and analysis of RNA sequencing data. Genetics. 2010;185:405–416. doi: 10.1534/genetics.110.114983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bruno VM, et al. Comprehensive annotation of the transcriptome of the human fungal pathogen Candida albicans using RNA-seq. Genome Res. 2010;20:1451–1458. doi: 10.1101/gr.109553.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maher CA, et al. Transcriptome sequencing to detect gene fusions in cancer. Nature. 2009;458:97–101. doi: 10.1038/nature07638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Finn RD, et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fang Z, Cui X. Design and validation issues in RNA-seq experiments. Brief. Bioinform. 2011;12:280–287. doi: 10.1093/bib/bbr004. [DOI] [PubMed] [Google Scholar]
- 43.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]