Abstract
The signalling molecule DIF-1 is required for normal cell fate choice and patterning in Dictyostelium. To understand how these developmental processes are regulated will require knowledge of how cells receive and respond to the DIF-1 signal. Previously, we have described a bZIP transcription factor, DimA, which is required for cells to respond to DIF-1. However, it was unknown whether DimA activity is required to activate the DIF response pathway in certain cells or is a component of the response pathway itself. In this study, we describe the identification of a DimA-related bZIP transcription factor, DimB. Rapid changes in the subcellular localisation of both DimA and DimB in response to DIF-1 suggest that they are directly downstream of the DIF-1 signal. Genetic and biochemical interactions between DimA and DimB provides evidence that their ability to regulate diverse targets in response to DIF-1 is partly due to their ability to form homo- and heterodimeric complexes. DimA and DimB are therefore direct regulators of cellular responses to DIF-1.
Keywords: Dictyostelium, DIF-1, bZIP, DimB
INTRODUCTION
The social amoeba Dictyostelium discoideum provides a system with which to address issues of cell type differentiation and pattern formation during development. Although Dictyostelium cells grow as single celled amoebae, a multicellular cycle is triggered in response to starvation (Kessin, 2001). Individual cells aggregate using cAMP as a chemoattractant to form a mound of cells. Within the mound, prestalk and prespore cell types differentiate intermingled with one another (Thompson et al., 2004b; Williams et al., 1989). Each cell type then sorts out into distinct regions, so that at the slug stage of development, the major prestalk and prespore cell types are arranged along the anteroposterior axis. For example, the pstA cells occupy the most anterior region constituting the slug tip, while the pstO population lies just posterior and abuts the prespore zone (Early et al., 1993; Jermyn et al., 1989; Maeda et al., 2003; Maruo et al., 2004; Yamada et al., 2005). Finally, in response to the appropriate signals, prestalk and prespore cells terminally differentiate and a fruiting body is formed, consisting of a ball of spores supported by dead stalk cells (Kessin, 2001).
In order to understand this pattern-forming process, efforts have focused on the discovery of the underlying molecules and signalling pathways. The chlorinated alkyl phenone DIF-1 is central to most ideas. DIF-1 was identified as a molecule made by developing Dictyostelium cells that induces isolated amoebae to differentiate as stalk cells in cell culture (Morris et al., 1987). DIF-1 treatment also results in prestalk marker gene induction, together with the repression of prespore markers and spore cell differentiation (Kay et al., 1999). These studies were performed on cells in culture and have been instrumental for our understanding of DIF-1 action. Furthermore, much progress has now been made in defining the role of DIF-1 during multicellular development. A consensus has now emerged in which DIF-1 is synthesized by prespore cells (Kay and Thompson, 2001) and plays a role at least in the normal differentiation of the pstO cell population (Kimmel and Firtel, 2004; Strmecki et al., 2005). The identification of DIF-1 signalling mutants has been central to this. For example, in a mutant engineered to be defective in DIF-1 biosynthesis (dmtA−), pstO cell differentiation is compromised, although pstA and prespore cells differentiate apparently normally (Maeda et al., 2003; Maruo et al., 2004; Thompson and Kay, 2000). The dmtA− mutant defines hallmark DIF-1 signalling morphological defects: the slugs are extremely long and thin compared with wild type, while culmination is clearly aberrant. These defects are due to the absence of DIF-1 as they can be rescued by addition of exogenous DIF-1. Further support for these ideas came from the identification of a mutant (dimA−) that is unable to respond to DIF-1 under all tested conditions (Thompson et al., 2004a). For example, when dimA− cells are treated with DIF-1, prestalk markers are not induced and prespore markers are not repressed. The dimA− mutant also exhibits morphological and cell type differentiation defects that phenocopy the dmtA− mutant. However, unlike the dmtA− mutant, and consistent with a role in regulating DIF responses rather than DIF-1 biosynthesis, the defects of the dimA− mutant are cell-autonomous (Foster et al., 2004).
To date, only one other DIF signalling component, STATc, has been identified (Fukuzawa et al., 2001). Like other DIF signalling mutants, the STATc mutant exhibits aberrant pstO cell differentiation. In this case, however, it is a failure to repress pstA markers in this cell type rather than any detectable defect in pstO marker induction. STATc encodes a member of the STAT family of transcription factors. Importantly, STATc is also generally accepted to be directly downstream of the DIF-1 signal because STATc exhibits DIF dependent tyrosine phosphorylation together with rapid nuclear accumulation in response to DIF-1 (Fukuzawa et al., 2001).
The disrupted gene in the dimA− mutant has been cloned and also encodes a transcription factor, although in this case of the bZIP family (Thompson et al., 2004a). It has therefore been proposed that DimA is a direct regulator of DIF-1 target gene expression. However, one problem with this idea is paradoxically due to the similarity of the dimA− and dmtA− developmental phenotypes. DimA would therefore appear to regulate the expression of most, if not all, DIF-1 target genes. Furthermore, DimA functions as both an activator of prestalk gene expression and repressor of prespore gene expression. In order to explain how DimA could have such diverse activities, two hypotheses have been put forward (1) DimA is a permissive factor required to set up cellular competence to respond to DIF-1 (Kimmel and Firtel, 2004; Strmecki et al., 2005). In this model, it is proposed that DimA is not downstream of the DIF-1 signal. Instead of directly regulating the expression of DIF-1 target genes, DimA would be required for the activation of genes that permit cells to respond to DIF-1, such as the DIF-1 signal transduction machinery. (2) DimA activity is regulated by heterodimerisation with other factors (Thompson et al., 2004a). As bZIP transcription factors not only bind DNA as homodimers, but also as heterodimeric complexes, the formation of heterodimers with other bZIP family members can greatly expand their regulatory potential (Hurst, 1995). Heterodimerisation can even turn an activating factor into a repressor (Chinenov and Kerppola, 2001). In this way, heterodimerisation could explain the ability of DimA to regulate diverse DIF-1 responses.
Most lines of evidence indicate that DimA plays a key role in the regulation of DIF responses. Conflicting views of its mode of action illustrate that in order to understand the DIF-1 signal transduction pathway, it will be vital to determine whether DimA plays an active or permissive role in its regulation. We have therefore set out to investigate how DimA activity is regulated. First, we have examined the possible role of heterodimerisation in DimA-regulated gene expression. We report the identification of a second bZIP transcription factor (DimB) that can directly interact with DimA in vitro. Consistent with the idea that DimB also regulates DIF responses, dimB− mutant cells exhibit similar but not identical phenotypes to the dimA− mutant. These observations provide support for the idea that interactions between DimA and DimB serve to regulate their activity. Second, we have investigated whether DimA and DimB are directly downstream of the DIF-1 signal or are required to set up the conditions that would allow DIF-1 responses. One way to distinguish between these possibilities would be to establish whether DIF-1 treatment elicits rapid changes in the activity of DimA or DimB, as has been reported for STATc (Fukuzawa et al., 2001). Here, we report that upon DIF-1 stimulation, DimA and DimB rapidly accumulate in the nucleus. Finally, we find that nuclear accumulation of DimA is dependent on DimB activity, which we interpret as further evidence of an in vivo interaction between these transcription factors.
MATERIALS AND METHODS
Strains, culture and maintenance
Dictyostelium strains were grown and maintained in association with Klebsiella aerogenes or in HL5 axenic medium (Sussman, 1987). Transformants were selected in 10 μg/ml blasticidin or 10 μg/ml G418. For development, cells in exponential growth phase were harvested and washed free of medium before plating at a density of 6.4×105 cells/cm2 on KK2 (16.1 mM KH2PO4, 3.7 mM K2HPO4) plates in 1.5% purified agar. Whole-mount lacZ staining was performed as described (Dingermann et al., 1989).
Monolayer assays
Stalk and spore monolayer assays, in addition to lacZ activity measurements were performed as described (Thompson et al., 2004a). For induction of prestalk and prespore markers in monolayers, AX4 and mutant cells were resuspended in stalk medium containing cAMP (2 mM NaCl, 10 mM KCl, 1 mM CaCl2, 10 mM MES (pH 6.2), 10 μg/ml streptomycin sulphate, 10 unit/ml penicillin, 37.5 μM cerulenin and 5 mM cAMP) at a density of 2.5×106/ml. Cell suspensions (10 ml) were incubated in 10 cm tissue culture dishes at 22°C for 9 hours. Each strain was further incubated with or without the addition of 100 nM DIF-1, for 1-3 hours. Samples were harvested and total RNA extracted using TRI-reagent (Sigma). Genomic DNA was removed by DNAseI (Roche) treatment followed by phenol extraction and ethanol precipitation.
mRNA measurement by quantitative PCR
Total RNA was reverse transcribed using Mu-MLV reverse transcriptase (Eurogentec). Primers were designed to flank short genomic sequences (100-200 bp) of the ecmA, ecmB and cotB genes. IG7 was used as a normalizing gene to eliminate variation in cDNA concentration between the samples. Standard PCR reactions, including addition of the nucleic acid dye SYBR Green I (Molecular Probes) at 1× concentration, were then set up in 96-well plates and cycled in an Opticon 2 Quantitative PCR machine (MJ Research). Cycle threshold (Ct) values were then obtained and differences in gene transcript levels between DIF-1 treated, and untreated samples were calculated. Values were normalised using the IG7 Ct values for each sample to give ΔΔCt values, using the following calculation: ΔΔCt= (1+Etarget)Δct target/(1+Enorm)Δct norm where Δ Ct is the difference in Ct values between the control and DIF treated samples, and E is the efficiency of the reaction, normally taken as 1.
GFP construct generation
A 3.8 kb dimA genomic fragment and a 1.8 kb dimB genomic fragment were amplified by PCR (dimA primers, 5′-CGCGGATCCATGGACTCAGATAATTGG-3 and 5′-CGCCTCGAGAATATTAGGGGTCTTATAACT-3′; dimB primers, 5′-CGCGGATCCATGAATCAATTTTATCAATCTACC-3′ and 5′-CGCCTCGAGTTATTGTCTCGAAGGTTGTTG-3′. Each fragment was cloned as a translational fusion into the BamHI and XhoI sites of pTX-GFP (Levi et al., 2000). Clonal transgenic Dictyostelium lines with equally strong GFP fluorescence were selected for further analysis. For simultaneous detection of DAPI and GFP, cells were fixed with 3.7% paraformaldehyde in PBS and permeabilised with 0.1% NP-40 before mounting in Vectashield containing DAPI (Molecular Probes).
In vitro protein interaction
Expression vectors for Glutathione S-transferase (GST) fusion proteins of DimA and DimB were constructed by cloning a 393 nucleotide fragment of the dimA or dimB gene encoding the DNA-binding domain and the dimerization domain into the pGEX4T2 (DimA) or pGEX4T1 (DimB) vectors (Amersham Biosciences). His-tag fusion proteins were generated by cloning the same fragments into pET-DEST42 (Invitrogen). All recombinant proteins were expressed in E. coli BL21 star cells. GST fusion proteins were purified with Glutathione Sepharose beads (Amersham Biosciences) according to the manufacturer's instructions. His-tag fusion proteins were purified with Talon metal affinity resins (BD Biosciences). Eluted proteins were dialyzed against 20 mM Tris-HCl (pH 7.4), 50 mM NaCl and 10% glycerol and protein concentrations were estimated using the Bradford reagent (BioRad). For GST pull-down assay, 3 μg of His-tag fusion proteins were mixed with equal amount of GST alone or GST fusion proteins in binding buffer [20 mM Tris-HCl, pH 8.0, 150 mM NaCl and 0.2% NP40 supplemented with a cocktail of protease inhibitors (Roche)]. The mixture was kept at 4°C for 1 hour before 100 μl of 50% glutathione-Sepharose beads slurry was added. After 18 hours of incubation at 4°C on a rocker, the beads were washed three times in 20 mM Tris-HCl (pH 8.0), 150 mM NaCl and 0.2% Triton X-100. Both the supernatants and the pellets were mixed with equal volumes of 2×SDS sample buffer, boiled for 5 minutes and separated by SDS-12%PAGE prior to transfer to BA85 nitrocellulose membrane (Schleicher and Schuell). The filter was first probed with rabbit anti-V5 antibody (Novus Biologicals) followed by goat anti-Rabbit-IgG antibody conjugated to alkaline phosphatase (AP).
RESULTS
Identification of a DimA interacting protein
Heterodimerisation regulates the activity of bZIP transcription factors in diverse systems (Chinenov and Kerppola, 2001; Hurst, 1995). We investigated whether this could also represent a mode of regulation of the Dictyostelium bZIP transcription factor DimA. We made four predictions for a putative DimA heterodimerisation partner. It should (1) exhibit significant sequence homology to DimA within the leucine zipper dimerisation domain; (2) directly interact with DimA in vitro; (3) be expressed at the same time as DimA; and (4) be expressed in the same cells as DimA during development.
The Dictyostelium genome contains 18 additional putative bZIP transcription factor encoding genes (Fig. 1A) (Huang and Shaulsky, 2005). We used the computational method of Fong et al. to predict which of the putative bZIP transcription factors might dimerise with DimA (Fong et al., 2004). We found eight bZIPs, including one we named DimB, that were as likely to form heterodimers with DimA as DimA was to homodimerise (data not shown). We then aligned all 19 putative leucine zipper domains and found that DimB was the most similar to DimA (Fig. 1B). These findings implicate DimB as a potential heterodimerisation partner for DimA. We therefore tested whether DimA and DimB could interact in vitro. The DNA-binding domains of DimA or DimB were expressed as either 6-His or GST fusion proteins. Pull-down assays with purified proteins showed that DimA and DimB can form homodimeric and heterodimeric complexes in vitro (Fig. 1C).
Fig. 1. Identification of a DimA-interacting protein.
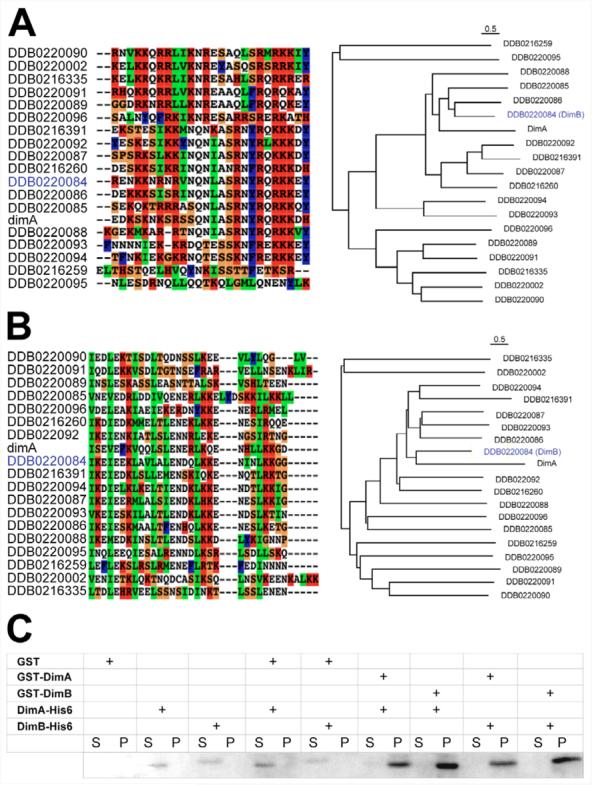
The Dictyostelium genome contains 19 potential bZIP genes. Alignments and phylogenetic trees of the DimA basic DNA-binding domain (A) and leucine zipper dimerisation domain (B) are shown. Identifiers from the Dictyostelium database (www.dictybase.org) are shown with DimB highlighted in blue. Basic residues are shown in red, hydrophobic in blue, small and polar in white, and secondary structure breakers in orange. The basic regions were defined as the first 26 amino acids from the basic DNA-binding domain, and all 19 sequences were aligned using CLUSTALX (default setting) and a phylogenetic neighbour-joining tree was plotted using NJ-Plot. Scale bars above the alignment indicate 0.5 estimated amino acid substitutions per site. Multiple sequence alignment and phylogenetic analysis were performed based on the dimerization domain, represented by the first four hepatads from the leucine zipper region of each protein. The results indicated a greater sequence identity between DimA and DimB bZIP domain. (C) DimA and DimB form homo- and heterodimers in vitro. Fusion proteins were expressed in E. coli and purified by affinity chromatography. DimA or DimB GST fusion proteins were mixed with His tagged versions before incubating in the presence of glutathione sepharose beads. Presence of His fusion proteins was monitored in the supernatant (S, unbound) and pellet (P, bound).
In order for two proteins to interact in vivo, their expression must overlap spatially and temporally. We tested whether the expression of DimB was consistent with the possibility of heterodimerisation with DimA. The levels of dimA and dimB transcripts were determined throughout development by quantitative RT-PCR. Consistent with previous northern blots (Thompson et al., 2004a), we found that dimA transcription was developmentally regulated with levels peaking at culmination (Fig. 2A). Similarly, dimB transcription was also developmentally regulated and exhibited an overlapping profile of developmental regulation (Fig. 2A). Most importantly, both genes were expressed at about 8 hours of development, the time of cell type divergence when DIF-1 is believed to act. We also tested the cell-type specificity of dimB expression by quantitative RT-PCR from RNA samples made from separated prespore and prestalk cells (Van Driessche et al., 2002). We found that dimB was present in both prespore and prestalk cells with some enrichment in the prespore cells (Fig. 2B). This pattern is essentially identical to that of dimA (Thompson et al., 2004a). The overlap in gene expression suggests that the DimA and DimB proteins are also likely to be co-expressed, allowing heterodimerisation.
Fig. 2. Developmental regulation and cell type expression of dimB RNA.
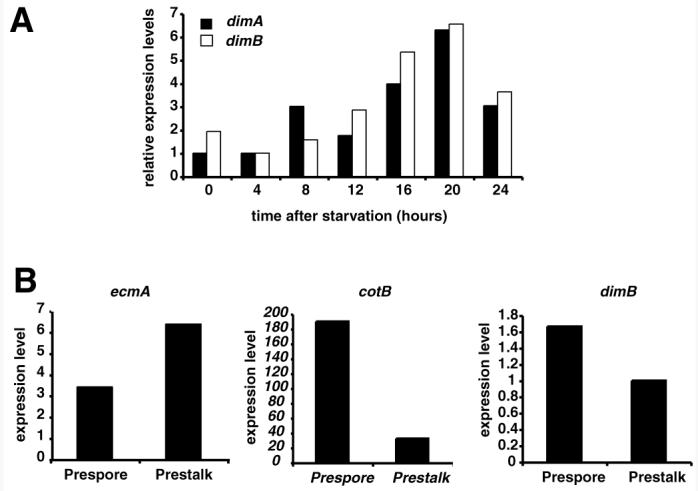
Cells were developed on agar for the times indicated (hours) and RNA was harvested. After reverse transcription, levels of each transcript were determined by quantitative PCR and normalized to IG7. (A) Developmental regulation of dimB compared with dimA. Expression of the two genes is upregulated during development. (B) RNA from prespore and prestalk cells was analyzed by quantitative RT-PCR with oligonucleotides specific to the dimB mRNA, indicating expression in both cell types with some prespore enrichment. Primers against prestalk (ecmA) and prespore (cotB) specific genes were used as controls. Expression levels are given in arbitrary units.
dimA− and dimB− mutants exhibit similar developmental defects
If protein-protein interactions between DimA and DimB are required to regulate their normal activity, then mutants in each gene would be expected to exhibit similar morphological phenotypes. A dimB insertion vector was generated by in vitro transposition (Abe et al., 2003) (Fig. 3A). Mutants in the dimB locus were generated by homologous recombination and verified by PCR (not shown). dimB− mutant cells grew normally in axenic medium. However, following starvation, development proceeded normally only until the finger stage, when the dimB− mutant produced extremely long and thin fingers/slugs compared with the wild type (Fig. 3B,C). When these slugs were allowed to migrate, they were also often found to break up. dimB− mutant slugs therefore show indistinguishable morphological defects from those previously described in the dimA− and dmtA− mutants (Thompson et al., 2004a). One simple explanation for this behaviour would be that dimA is not expressed in the dimB− mutant or vice versa. This is not so because dimA and dimB transcripts were detected in the dimB− mutant and dimA− mutant, respectively (not shown).
Fig. 3. The dimB− mutant exhibits DIF-1 signalling morphological defects.
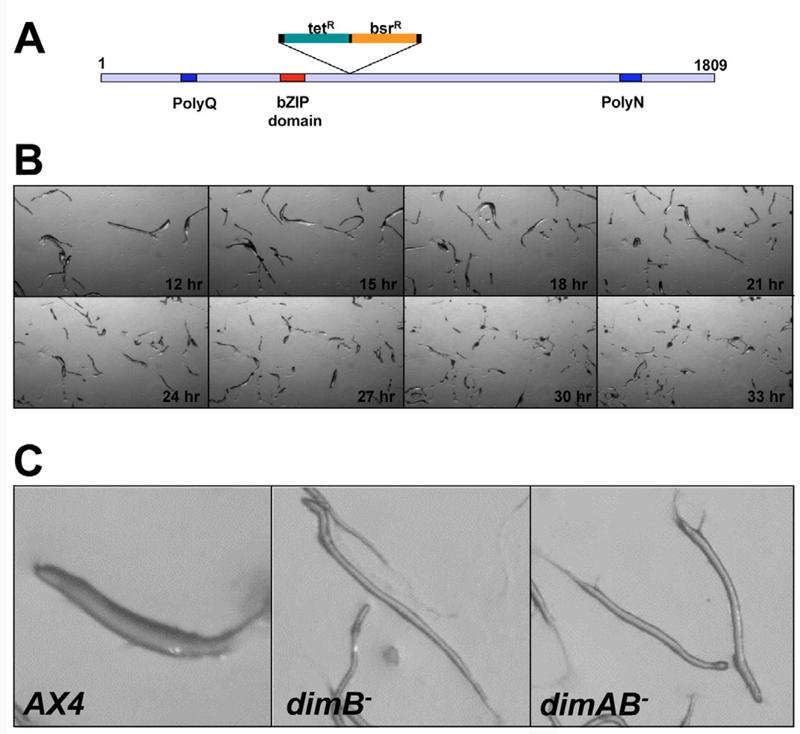
(A) Schematic of the dimB gene and disruption vector. The predicted DimB protein contains 1809 amino acids. Regions of low complexity [poly glutamine (Q) and poly asparagine (N) tracts] and the basic-leucine zipper (bZIP) domain are highlighted. The dimB disruption vector was generated by in vitro transposition of sequences containing the tetracycline resistance gene (for selection in bacteria) and the blasticidin resistance gene into the dimB-coding sequence. (B) Developmental morphology of dimB− mutant cells. (C) dimB− and dimAB− double-mutant cells form long thin slugs compared with wild type (AX4).
The dimB− allele we generated did not delete any of the coding region, so it might not be a null allele (Fig. 3A). Nevertheless, the phenotypes we found were almost identical to those described in the accompanying paper (Zhukovskaya et al., 2006). In addition, both insertions are in similar positions (nucleotide 600 and 696 with respect to the ATG). These researchers used antibodies against DimB to show that the protein was undetectable in their insertional dimB− mutant strain. We therefore conclude that our dimB− allele is essentially null. A double mutant where both dimA and dimB were disrupted, had an identical phenotype to the single gene mutations (Fig. 3C). It is therefore also unlikely that DimA and DimB play functionally redundant roles in Dictyostelium development. These observations suggest that DimB is also required for DIF-1 signalling.
The dimA− and dmtA− mutants exhibit similar defects in developmental gene expression in addition to the morphological defects (Thompson et al., 2004a). Most notably, both mutants show a reduced zone of expression of the prestalk marker ecmAO-lacZ and an expanded zone of expression of prespore-lacZ markers. Similarly, we found that in dimB− mutant slugs the relative length of the ecmAO-lacZ labelled prestalk zone was considerably shorter than in wild type (Fig. 4A). In addition, we found that the prespore zone of dimB− mutant slugs was expanded (especially when stained for long periods) (Fig. 4A). However, this was generally not seen to the same extent as in dimA− mutant slugs.
Fig. 4. dimB− exhibits gene expression and cell-autonomous defects.
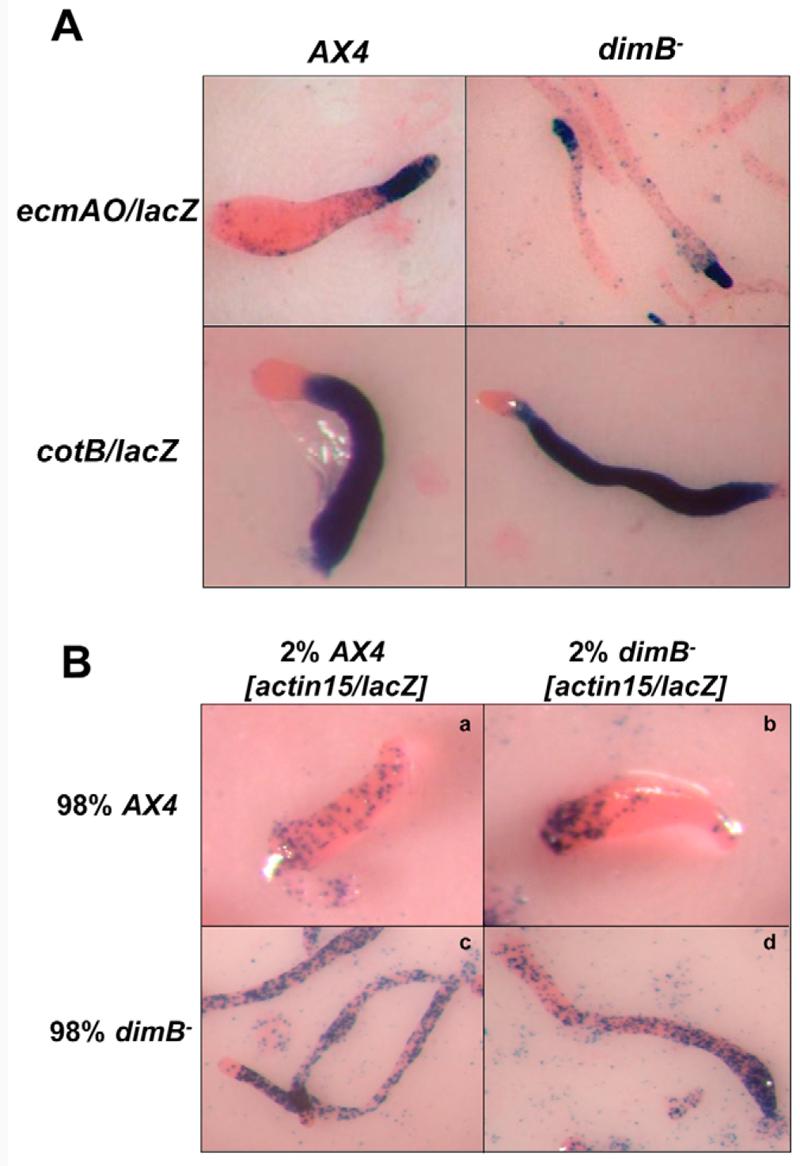
(A) Expression pattern of representative prestalk and prespore markers at the slug stage of development. The prestalk marker ecmAO/lacZ shows a reduced zone of expression compared with wild type. The prespore marker cotB/lacZ shows an expanded zone of expression, although expression is weaker in the expanded region than in the prespore zone proper. (B) The dimB− mutant exhibits cell-autonomous defects when mixed with wild-type cells. A minority (2%) of wild-type or dimB− mutant cells were labelled by constitutive expression of lacZ and mixed with unlabelled cells. Wild-type and dimB− mutant cells are evenly distributed in homotypic control chimaeras (a and d). dimB− mutant cells preferentially localize to the rear of the prespore zone when mixed with an excess of wild-type cells (b). Wild-type cells localize to the pstO and prespore zone when mixed with dimB− mutant cells (c).
In addition to sharing morphological and gene expression defects, if DimB is required to integrate responses to DIF-1, any defects would be predicted to be cell-autonomous. Consistent with this idea, we found that the morphological defects of the dimB− mutant could not be rescued by addition of DIF-1 to the agar (not shown). Second, dimB− mutant cells exhibited cell-autonomous defects in chimaeras. Wild-type or dimB− mutant cells were labelled by constitutive expression of lacZ and their position followed in chimaeric slugs when mixed with an excess of unlabelled cells. In homotypic control chimaerae, lacZ-labelled cells were distributed evenly (Fig. 4B). However, when labelled dimB− mutant cells were mixed with wild type, the mutant cells were enriched in the prespore zone. It is also noteworthy that their predominance within the rear of the prespore zone was reminiscent of the distribution of dimA− cells in similar chimaerae (Foster et al., 2004). Furthermore, in the reciprocal experiment, labelled wild-type cells were enriched in the prestalk zone and anterior prespore zone of chimaeric slugs. Taken together, the similarities between the phenotypes displayed by the dimA− and dimB− mutants strongly suggest that DimB plays a similar role to that of DimA in the regulation of DIF-1 responses.
DimB is required for normal responses to DIF-1
In order to directly test whether DimB is required for DIF-1 responses, we examined the behaviour of dimB− mutant cells in the cAMP removal monolayer assay. In this assay, cells are initially induced by cAMP to become competent to respond to DIF-1. After removal of cAMP and addition of DIF-1, wild-type cells differentiate as stalk cells. The DIF-1 non-responsive mutant, dimA−, does not produce stalk cells in response to DIF-1 in this assay. Similarly, we found that dimB− and dimA−/dimB− mutant cells also failed to respond to DIF-1 (Fig. 5A). Instead, the mutants cells remained as amoebae, demonstrating that DimB, like DimA, is required for normal responses to DIF-1.
Fig. 5. dimB− mutant cells fail to induce prestalk markers and stalk cells in response to DIF-1.
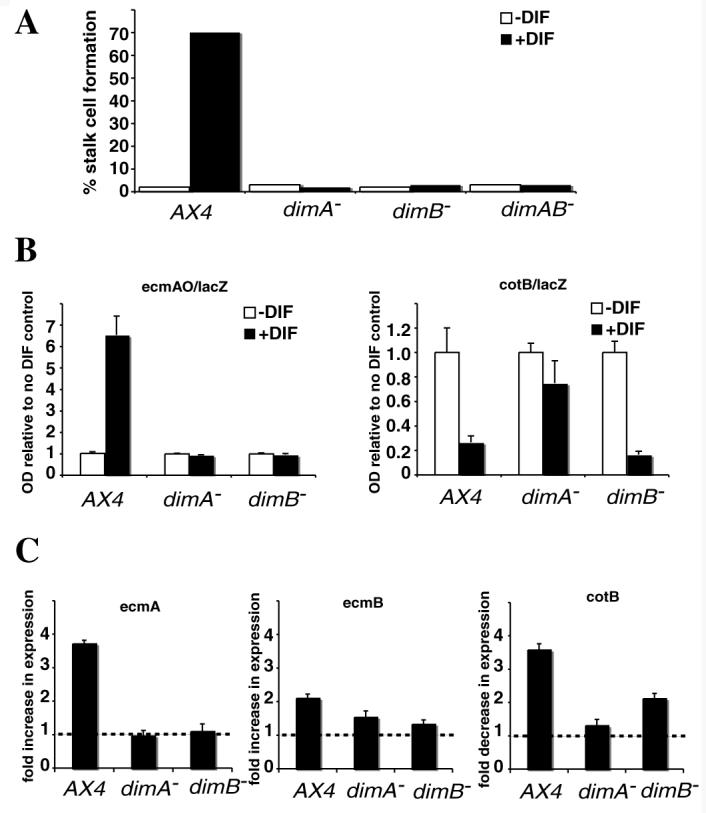
(A) Stalk cell induction was measured in response to DIF-1 in the cAMP removal assay. The assay was repeated at least three times and a representative example is shown. Wild-type cells (AX4) differentiated as stalk cells, whereas mutants defective either in dimA or in dimB, or in both (dimAB−) failed to do so. (B) In the dimB− mutant, a prestalk reporter (ecmAO/lacZ) was non responsive to DIF-1, whereas a prespore marker (cotB/lacZ) was repressed as in the wild type. The results are means of three experiments where each assay was performed in triplicate. (C) The dimB− mutant did not show rapid induction of prestalk gene expression (ecmA or ecmB) in culture, although the prespore gene (cotB) was repressed. After treatment with cAMP for 9 hours, samples were incubated with or without DIF-1 for 3 hours. Expression levels were normalized to IG7 and compared with or without DIF-1. In the case of ecmA and ecmB, fold increase is shown, and for cotB fold decrease. Broken line indicates no change.
The cAMP removal assay uses a terminal differentiation phenotype (stalk cell formation) as a measure of DIF-1 responsiveness. We therefore employed another monolayer assay that provides a more direct transcriptional readout in order to further characterize the defective DIF-1 response in the dimB− mutant. In this assay, cells do not undergo terminal differentiation owing to the continued presence of cAMP. Previously, we have demonstrated that DimA is required for both the induction of prestalk markers and repression of prespore markers under these conditions (Thompson et al., 2004a). We compared the behaviour of dimB− cells using strains transformed with the prestalk marker ecmAO-lacZ and the prespore marker cotB-lacZ. We found that dimB− mutant cells showed little or no induction of ecmAO-lacZ over a 24-hour period in the presence of DIF-1 when compared with wild type (Fig. 5B). Despite this, we were surprised to find that repression of the prespore marker cotB-lacZ by DIF-1 was unaffected (Fig. 5B). This is in marked contrast to the behaviour of dimA− mutant cells and suggests that, despite other similarities, DimA and DimB do not play identical roles. To confirm these findings, we used a second assay that gives a more rapid measure of DIF responses. In this assay, cells are initially brought to competence to respond to DIF-1 by cAMP treatment followed by a short DIF-1 treatment. Expression levels of representative prestalk and prespore transcripts were measured by quantitative real time RT-PCR. Using this assay, the dimA− mutant showed no responses to DIF-1. By contrast, although dimB− cells also exhibited defects, they were distinct from those observed in the dimA− mutant. No induction of the prestalk markers ecmA and ecmB was detected after 3 hours of DIF treatment (Fig. 5C). Despite this defect in prestalk marker induction, dimB− mutant cells exhibited clear DIF-1 dependent repression of the prespore marker cotB, although the magnitude was less than that seen in wild type (Fig. 5C).
DimB is therefore required for normal DIF responses. Unlike dimA−, however, the major defect in the dimB− mutant is in the induction of prestalk gene expression and stalk cell fate, as opposed to the repression of prespore markers. These results demonstrate that although DimA and DimB functions overlap, each plays a unique role.
DimB is required to repress autophagy independent cell death
As DIF-1 treatment in cAMP monolayer assays results in the repression of prespore gene expression in the dimB− mutant, this suggested that DimB might not play a role in DIF-dependent repression of the prespore/spore fate. To further test this idea, we employed the 8-Br-cAMP monolayer assay, in which wild-type cells efficiently differentiate into viable spores in the absence of DIF-1 (Kay, 1989). However, when DIF-1 is added, spore formation is repressed and wild-type cells differentiate as dead stalk cells or remain as detergent sensitive amoebae. This response was exploited to identify the dimA− mutant, which fails to respond to DIF-1 and therefore forms spores in the presence of DIF-1 (Thompson et al., 2004a). We examined the behaviour of dimB− cells in this assay and found that dimB− mutant cells efficiently make spores in the absence of DIF-1, illustrating that they are able to undergo terminal differentiation (Fig. 6A). As predicted from the cAMP assays, spore cell formation was repressed by DIF-1 in dimB− mutant cells as efficiently as in the wild type (Fig. 6A). This finding further supports the idea that DimB is not required for the repression of the prespore/spore fate in response to DIF-1.
Fig. 6. dimB is required for stalk cell induction and repression of non-vacuolar cell death in response to DIF-1.
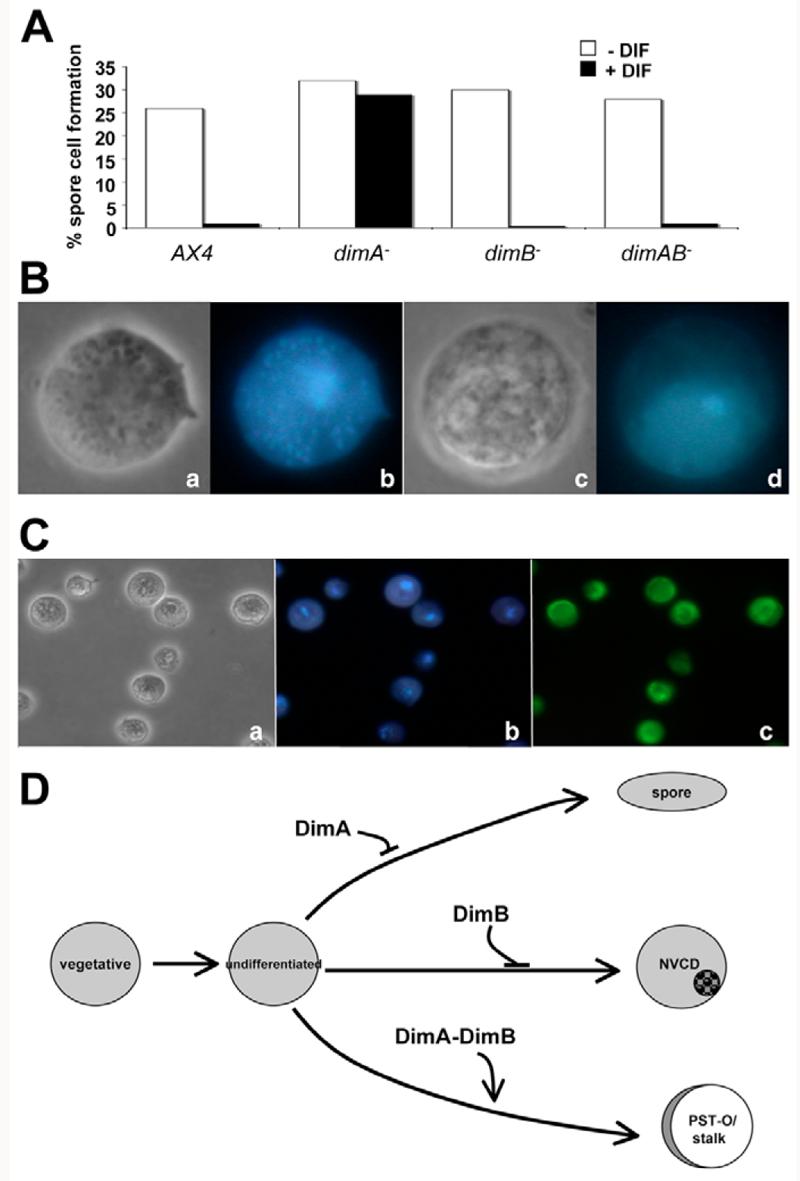
(A) Repression of spore cell formation by DIF-1 was measured in an 8-Br-cAMP assay. In the absence of DIF-1, all strains produced similar numbers of spores. Only dimA− mutant cells produced spore cells in the presence of DIF-1. Each assay was performed at least three times and the results of a representative assay are shown. dimAB− represents the double mutant dimA−dimB− and AX4 is the wild type. (B) dimB− cells were stained with DAPI without DIF-1 treatment (a,b) or after DIF-1 treatment (c,d). Micrographs were taken with phase contrast (a,c) or fluorescence microscopy (b,d). (C) dimB− mutants produce cells with identical characteristics to those that have undergone non-vacuolar cell death. (a) Phase-contrast image showing cell morphology. Cells appear rounded and contain a dense/granular region. (b) Staining of DNA with DAPI. (c) F-actin distribution detected with phalloidin. (D) Model of DIF-1 responses based on the epistatic relationship between DimA and DimB in the 8-Br-cAMP assay.
If dimB− mutant cells do not make spores in the presence of DIF-1, then what do they become? As predicted by the cAMP monolayer assays, dimB− cells still failed to make stalk cells (Fig. 6B). Surprisingly, however, despite the fact that DIF-1-treated dimB− cells made neither spores nor stalk cells, they did not remain as amoebae. Instead the dimB− mutant cells exhibited an unusual morphology, rarely seen in monolayer assays (Fig. 6B). These cells appeared to exhibit a similar morphology to DIF-1-treated atg1− cells, which are unable to undergo autophagy and do not make stalk cells in response to DIF-1 (Kosta et al., 2004). Instead, these cells exhibit non-vacuolar cell death (NVCD). We found that this similarity extended beyond morphology. Other shared features include loss of cell viability, the collapse of the cytoplasm and the organelles (mitochondria) into the perinuclear compartment (Fig. 6B), and the concentration of F-actin to the cell periphery, surrounding the central condensation of organelles (Fig. 6C).
One explanation for these observations is that stalk cell formation is a dependent sequence of events. The first step would require DimA to trigger NVCD, whereas subsequent vacuolisation requires DimB. This scheme predicts that a dimA−/dimB− double mutant should behave like a dimA− mutant in a 8-Br-cAMP monolayer assay. Instead, we found that the double mutant phenocopied the dimB− mutant, suggesting that dimB is epistatic to dimA (Fig. 6A). In order to explain this observation, we propose that there are at least three distinct routes that can be taken in response to DIF-1 in an 8-Br-cAMP assay (Fig. 6D): (1) vacuolised stalk cell formation requires both DimA and DimB; (2) NVCD is repressed by DimB; and (3) spore cell formation is repressed by DimA. These results further illustrate that DimA and DimB participate in distinct and overlapping pathways.
DIF-1 induces rapid nuclear accumulation of DimA and DimB
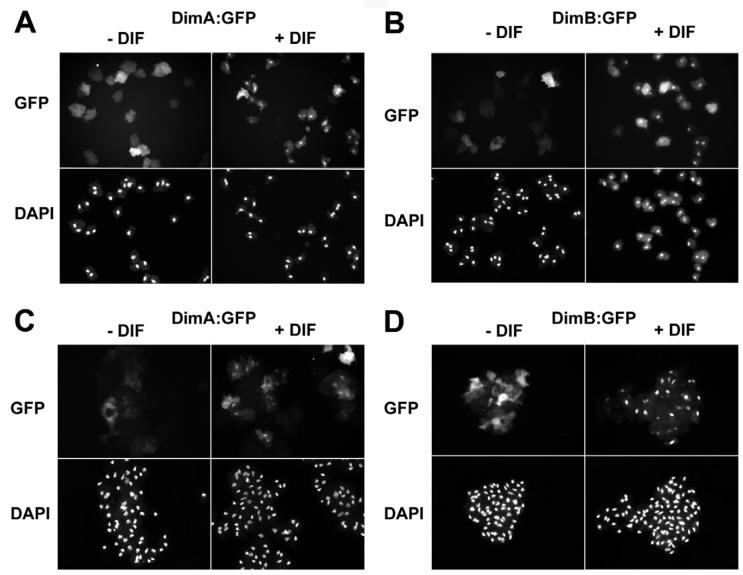
Homo and heterodimerisation of DimA and DimB goes some way to explain how a small number of transcription factors could directly regulate many different DIF-1 responses. However, testing whether DimA or DimB function downstream of the DIF-1 signal requires some change in their activity to be measured immediately after exposure to DIF-1. For example, STATc exhibits rapid nuclear accumulation and tyrosine phosphorylation in response to DIF-1 (Fukuzawa et al., 2001). Interestingly, it has also been reported that a variety of stimuli regulate the subcellular localization of bZIP transcription factors in other organisms (Kircher et al., 1999; Kuge et al., 1997). We therefore tested whether DIF-1 treatment affects the localization of the bZIP transcription factors DimA or DimB. DimA:GFP and DimB:GFP fusion proteins were expressed under the control of a constitutive promoter. Both fusion proteins were functional because DimA:GFP expression in the dimA− mutant or DimB-GFP expression in the dimB− mutant rescued stalk cell formation (not shown). In order to follow subcellular localization, cells were shaken in buffer for 4 hours before the addition of 50 nM DIF-1. In the absence of DIF-1, GFP was fairly uniformly distributed throughout the cells. However, after DIF-1 treatment, both DimA:GFP and DimB:GFP exhibited nuclear accumulation (Fig. 7A,B). In mock-treated control cells, no change in distribution was observed. DIF-1 normally accumulates to measurable levels after cell aggregation. We therefore tested whether the nuclear accumulation of the proteins could also be observed in cells that have been developed for 10 hours. The results in Fig. 7C,D show DIF-1 induced nuclear accumulation of both bZIPs. The similarity between the responses at 4 hours and 10 hours of development suggests that DimA and DimB are the initial components in the cellular response to DIF-1.
Fig. 7. DIF-1-dependent nuclear accumulation of DimA and DimB.
DimA and DimB rapidly accumulate in the nucleus of DIF-1-treated cells. DimA and DimB were constitutively expressed as GFP fusion proteins in cells starved for 4 hours (A,B) or 10 hours (C,D). After DIF-1 treatment, the number of cells with even staining in cytoplasm and nucleus rapidly decreased with a concomitant increase in the number of cells with stronger staining in the nucleus compared with cytoplasm. No change in distribution was observed in untreated control cells. Nuclear accumulation was verified by comparison with DAPI staining. In the pictures, 10 minutes DIF-1 treatment is shown. In the samples starved for 10 hours (C,D), clumping of cells can be seen. In addition, an increase in the number of cells with increased nuclear fluorescence is observed in control samples, presumably owing to endogenous DIF-1 production at this time point.
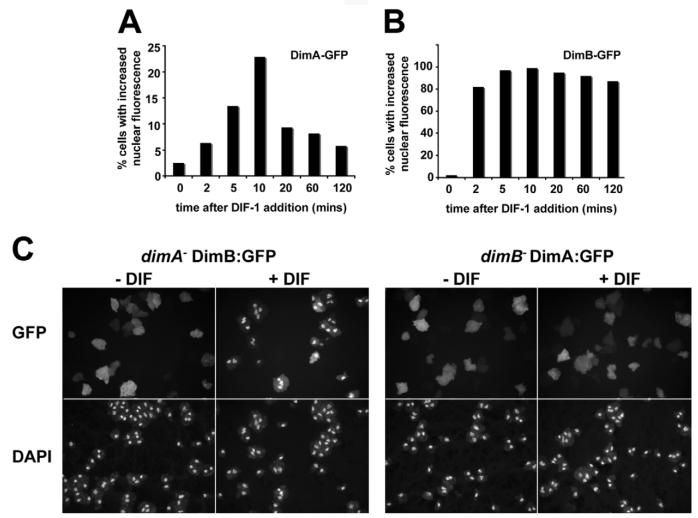
We next determined the kinetics of nuclear accumulation. We used cells that had been starved for 4 hours because they exhibit lower levels of nuclear accumulation in control samples (Fig. 7A,C). In both DimA-GFP and DimB-GFP, accumulation was transient, with maximal accumulation seen after 10 minutes (Fig. 8A,B). Remarkably, significant accumulation could even be measured after only 2 minutes. The kinetics of DIF-1 induced nuclear accumulation therefore strongly suggests that the bZIP transcription factor activities are directly affected by DIF-1 and therefore downstream of the signal. The rapid kinetics and the sharp compartmentalization of the proteins upon DIF-1 stimulation suggest that the fluorescent signal faithfully reflected the localization of the bZIP proteins. This argument is also supported by staining with antibodies directed against DimB in the accompanying paper (Zhukovskaya et al., 2006).
Fig. 8. Quantification of the nuclear accumulation kinetics.
(A,B) DimA and DimB exhibit rapid nuclear accumulation kinetics. Cells with higher levels of nuclear fluorescence than cytoplasmic fluorescence were scored as positive. (C) Nuclear accumulation of DimA-GFP is dependent on DimB. DimB-GFP was expressed in dimA− cells and DimA-GFP was expressed in dimB− cells. Nuclear accumulation of DimB-GFP was unaffected, whereas no DIF-1-induced nuclear accumulation of DimA-GFP could be detected.
Nuclear accumulation of DimA:GFP is dependent on DimB
We also tested whether nuclear accumulation of DimA:GFP or DimB:GFP was dependent on DimB or DimA, respectively. In control experiments, normal accumulation was observed. When DimA:GFP was expressed in dimA− cells, DIF-1 treatment triggered nuclear accumulation. Nuclear accumulation of DimB:GFP was also observed when expressed in dimB− cells (not shown). Similarly, when DimB:GFP was expressed in the dimA− mutant, DIF-1 induced nuclear accumulation of DimB:GFP with normal kinetics (Fig. 8C). By contrast, nuclear accumulation of DimA-GFP was not induced by DIF-1 when expressed in the dimB− mutant (Fig. 8C). This failure was not due to altered accumulation kinetics. No accumulation could be detected at any time point between 1 minute and 2 hours after DIF-1 addition (not shown). These results demonstrate that nuclear accumulation of DimA in response to DIF-1 is dependent on DimB. It should be noted, however, that nuclear DimA:GFP is still detectable in dimB− mutant cells, because the protein is uniformly distributed throughout the cell. As any nuclear DimA could be considered functional, it is therefore possible that some DimA activity remains. This provides a likely explanation for how DimA-dependent repression of prespore markers can take place in the DimB mutant (Fig. 5B,C).
DISCUSSION
DimB interacts with DimA and is required for normal DIF-1 responses
This work describes the identification of a DimA-related bZIP transcription factor, DimB. DimB interacts with DimA in vitro and the genetic data suggests that this interaction occurs in vivo and is required to integrate normal DIF-1 responses.
(1) The dimB− mutant exhibits hallmark DIF signalling morphological defects when developed. The long, thin slugs produced by the dimB− mutant are indistinguishable from those seen in the DIF-1 signalling mutants dimA− or dmtA−. These defects are due to a failure to respond normally to DIF-1, rather than DIF-1 production, as they are cell autonomous.
(2) dimB− mutant cells exhibit defective responses to DIF-1. DIF-1 treatment of dimB− mutant cells in monolayer assays does not induce prestalk marker gene expression and dimB− mutant cells fail to produce stalk cells in response to DIF-1.
(3) DimA and DimB rapidly accumulate in the nucleus of DIF-1-treated cells when developed in shaken suspension. Nuclear accumulation of DimA is dependent on DimB as it does not occur in the dimB− mutant. As DimA and DimB interact in vitro, a simple interpretation of this finding is that interactions with DimB regulate the nuclear accumulation of DimA. For example, it is possible that interactions with DimB are required to carry DimA into the nucleus. Alternatively, interactions between DimA and DNA bound DimB may serve to tether DimA protein once translocated into the nucleus. Analysis of the GFP-fusion proteins in slugs did not provide sufficient resolution to determine whether the shaken suspension development assays reflect the interaction in multicellular structures.
DimA and DimB also play independent roles
Some DIF-1 responses, such as the induction of prestalk marker gene expression, require the activity of both DimA and DimB. As they readily interact in vitro, they may function as a heterodimeric complex to regulate these responses. However, a number of DIF responses also appear to occur independently of either DimA or DimB. For example, DIF-1-mediated prespore gene repression is dependent on DimA but independent of DimB. By contrast, repression of NVCD is dependent on DimB but independent of DimA. The most simple explanation is that DimA and DimB operate as homodimers in these processes. However, as 17 other bZIP transcription factors are probably encoded by the Dictyostelium genome, the formation of additional heterodimeric complexes could play regulatory roles. For example, in mammalian cells, although Fos and Jun can form a heterodimeric complex, a network of interactions with other bZIPs vastly increase their regulatory repertoire (Chinenov and Kerppola, 2001). In order to test this possibility, we have knocked out eight of the remaining bZIPs and begun to compare their phenotypes to those displayed by the dimA− and dimB− mutants (E.H, G.S. and C.R.L.T., unpublished). Interestingly, although developmental and DIF-1 response defects have been detected, none is identical to those seen in the dimA− or dimB− mutants. It is therefore possible that other bZIPs could play a role in regulating the DimA- or DimB-specific responses reported here, or even in the regulation of currently unidentified responses to DIF-1.
DIF signalling in cell culture versus development
The morphological phenotype of the dimA− or dimB− mutants appears identical at the slug stage of development, but their behaviour in monolayer assays suggests they also play distinct roles. We do not, however, believe that these ideas are mutually exclusive. For example, the phenotype of each mutant also appears to be the same in a cAMP removal assay (failure to make stalk cells). Nevertheless, closer inspection reveals that in the dimB− mutant, a typical prespore marker is repressed in response to DIF-1, whereas expression levels remain unchanged in the dimA− mutant. We believe that a more detailed analysis of the phenotype of each mutant during development could also reveal differences, although such tools are not currently available. Such studies will undoubtedly be aided by the identification of the repertoire of genes regulated by DimA and DimB, either together or independently.
DimA and DimB are downstream of the DIF-1 response pathway
The results described here strongly suggest that DimA and DimB are direct regulators of gene expression in response to DIF rather than required to set up DIF responses. First, both DimA and DimB exhibit rapid nuclear accumulation in response to DIF-1. As significant accumulation occurs after as little as 2 minutes, it is probably due to direct post-translational modification of DimA or DimB, or indirect modification of a regulatory factor. Second, if DimA or DimB were required for the activation of permissive factors for DIF-1 responsiveness, the mutants would be expected to be completely indifferent to DIF-1, but this is not the case. For example, DIF-1 treatment induced nuclear accumulation of DimB in the dimA− mutant while prespore gene repression and induction of NVCD still occurred in the dimB− mutant. Finally, DIF-1 responses could even be measured in the dimA−/dimB− mutant, resulting in NVCD in 8-Br-cAMP monolayer assays. These results therefore demonstrate that DimA and DimB play an active role in the regulation of both overlapping and distinct aspects of DIF-1 signal transduction and gene regulation. Understanding how each factor is regulated in response to DIF-1 and the nature of genes regulated by each factor provides a challenge for future studies.
Acknowledgments
C.R.L.T. was supported by a Wellcome Trust project grant and a MRC Career Establishment Grant. G.S. was supported by grants from the NIH/NICHD P01 HD39691 and National Science Foundation EF-0328455. E.H. was supported in part by a training fellowship from the W. M. Keck Foundation of the Gulf Coast Consortia through the Keck Center for Computational and Structural Biology. The authors thank Dr Denis Headon for critical reading of the manuscript, Dr Rob Kay for advice on monolayer induction assays, and members of the Kuspa laboratory and Manchester Developmental Biology groups for stimulating discussion.
References
- Abe T, Langenick J, Williams JG. Rapid generation of gene disruption constructs by in vitro transposition and identification of a Dictyostelium protein kinase that regulates its rate of growth and development. Nucleic Acids Res. 2003;31:107–111. doi: 10.1093/nar/gng095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinenov Y, Kerppola T. Close encounters of many kinds: Fos-Jun interactions that mediate transcription regulatory specificity. Oncogene. 2001;20:2438–2452. doi: 10.1038/sj.onc.1204385. [DOI] [PubMed] [Google Scholar]
- Dingermann T, Reindl N, Werner H, Hildebrandt M, Nellen W, Harwood A, Williams J, Nerke K. Optimization and in situ detection of Escherichia coli beta-galactosidase gene expression in Dictyostelium discoideum. Gene. 1989;85:353–362. doi: 10.1016/0378-1119(89)90428-9. [DOI] [PubMed] [Google Scholar]
- Early AE, Gaskell MJ, Traynor D, Williams JG. Two distinct populations of prestalk cells within the tip of the migratory Dictyostelium slug with differing fates at culmination. Development. 1993;118:353–362. doi: 10.1242/dev.118.2.353. [DOI] [PubMed] [Google Scholar]
- Fong JH, Keating AE, Singh M. Predicting specificity in bZIP coiled-coil protein interactions. Genome Biol. 2004;5:R11. doi: 10.1186/gb-2004-5-2-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster KR, Shaulsky G, Strassmann JE, Queller DC, Thompson CR. Pleiotropy as a mechanism to stabilize cooperation. Nature. 2004;431:693–696. doi: 10.1038/nature02894. [DOI] [PubMed] [Google Scholar]
- Fukuzawa M, Araki T, Adrian I, Williams JG. Tyrosine phosphorylation-independent nuclear translocation of a Dictyostelium STAT in response to DIF signaling. Mol. Cell. 2001;7:779–788. doi: 10.1016/s1097-2765(01)00222-2. [DOI] [PubMed] [Google Scholar]
- Huang E, Shaulsky G. Components of the dictyostelium gene expression regulatory machinery. In: Loomis WF, Kuspa A, editors. Dictyostelium Genomics. Horizon Scientific Press; Norwich, UK: 2005. [Google Scholar]
- Hurst H. Transcription factors 1, bZIP proteins. Protein Profile. 1995;2:101–168. [PubMed] [Google Scholar]
- Jermyn KA, Duffy KTI, Williams JG. A new anatomy of the prestalk zone in Dictyostelium. Nature. 1989;340:144–146. doi: 10.1038/340144a0. [DOI] [PubMed] [Google Scholar]
- Kay RR. Evidence that elevated intracellular cyclic AMP triggers spore maturation in Dictyostelium. Development. 1989;105:753–759. [Google Scholar]
- Kay RR, Thompson CRL. Cross-induction of cell types in Dictyostelium: evidence that DIF-1 is made by prespore cells. Development. 2001;128:4959–4966. doi: 10.1242/dev.128.24.4959. [DOI] [PubMed] [Google Scholar]
- Kay RR, Flatman P, Thompson CRL. DIF signalling and cell fate. Semin. Cell Dev. Biol. 1999;10:577–585. doi: 10.1006/scdb.1999.0341. [DOI] [PubMed] [Google Scholar]
- Kessin RH. Dictyostelium. Cambridge University Press; Cambridge: 2001. [Google Scholar]
- Kimmel A, Firtel RA. Breaking symmetries: regulation of Dictyostelium development through chemoattractant and morphogen signal-response. Curr. Opin. Genet. Dev. 2004;14:540–549. doi: 10.1016/j.gde.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Kircher S, Wellmer F, Nick P, Rügner A, Schäfer E, Harter K. Nuclear Import of the Parsley bZIP Transcription Factor CPRF2 Is Regulated by Phytochrome Photoreceptors. J. Cell Biol. 1999;144:201–211. doi: 10.1083/jcb.144.2.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosta A, Roisin-Bouffay C, Luciani M-F, Otto GP, Kessin RH, Golstein P. Autophagy Gene Disruption Reveals a Non-vacuolar Cell Death Pathway in Dictyostelium. J. Biol. Chem. 2004;279:48404–48409. doi: 10.1074/jbc.M408924200. [DOI] [PubMed] [Google Scholar]
- Kuge S, Jones N, Nomoto A. Regulation of yAP-1 nuclear localization in response to oxidative stress. EMBO J. 1997;16:1710–1720. doi: 10.1093/emboj/16.7.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levi S, Polyakov M, Egelhoff TT. Green fluorescent protein and epitope tag fusion vectors for Dictyostelium discoideum. Plasmid. 2000;44:231–238. doi: 10.1006/plas.2000.1487. [DOI] [PubMed] [Google Scholar]
- Maeda M, Haruyo S, Maruo T, Ogihara S, Iranfar N, Fuller D, Morio T, Urushihara H, Tanaka T, Loomis WF. Changing patterns of gene expression in prestalk cell subtypes of Dictyostelium recognised by in situ hybridisation with genes from microarray analyses. Eukaryot. Cell. 2003;2:627–637. doi: 10.1128/EC.2.3.627-637.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruo T, Sakamoto H, Iranfar N, Fuller D, Morio T, Urushihara H, Tanaka Y, Maeda M, Loomis WF. Control of cell type proportioning in Dictyostelium discoideum by differentiation-inducing factor as determined by in situ hybridization. Eukaryot. Cell. 2004;3:124–128. doi: 10.1128/EC.3.5.1241-1248.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris HR, Taylor GW, Masento MS, Jermyn KA, Kay RR. Chemical structure of the morphogen differentiation inducing factor from Dictyostelium discoideum. Nature. 1987;328:811–814. doi: 10.1038/328811a0. [DOI] [PubMed] [Google Scholar]
- Strmecki L, Greene DM, Pears C. Developmental decisions in Dictyostelium discoideum. Dev. Biol. 2005;284:25–36. doi: 10.1016/j.ydbio.2005.05.011. [DOI] [PubMed] [Google Scholar]
- Sussman M. Cultivation and synchronous morphogenesis of Dictyostelium under controlled experimental conditions. Methods Cell Biol. 1987;28:9–29. doi: 10.1016/s0091-679x(08)61635-0. [DOI] [PubMed] [Google Scholar]
- Thompson CRL, Kay RR. The role of DIF-1 signaling in Dictyostelium development. Mol. Cell. 2000;6:1509–1514. doi: 10.1016/s1097-2765(00)00147-7. [DOI] [PubMed] [Google Scholar]
- Thompson CRL, Fu Q, Buhay C, Kay RR, Shaulsky G. A bZIP/bRLZ transcription factor required for DIF signaling in Dictyostelium. Development. 2004a;131:513–523. doi: 10.1242/dev.00939. [DOI] [PubMed] [Google Scholar]
- Thompson CRL, Reichelt S, Kay RR. A demonstration of pattern formation without positional information in Dictyostelium. Dev. Growth Differ. 2004b;46:363–369. doi: 10.1111/j.1440-169x.2004.00753.x. [DOI] [PubMed] [Google Scholar]
- Van Driessche N, Shaw C, Katoh M, Morio T, Sucgang R, Ibarra M, Kuwayama H, Saito T, Urushihara H, Maeda M, et al. A transcriptional profile of multicellular development in Dictyostelium discoideum. Development. 2002;129:1543–1552. doi: 10.1242/dev.129.7.1543. [DOI] [PubMed] [Google Scholar]
- Williams JG, Duffy KT, Lane DP, Mcrobbie SJ, Harwood AJ, Traynor D, Kay RR, Jermyn KA. Origins of the prestalk-prespore pattern in Dictyostelium development. Cell. 1989;59:1157–1163. doi: 10.1016/0092-8674(89)90771-x. [DOI] [PubMed] [Google Scholar]
- Yamada Y, Sakamoto S, Ogihara S, Maeda Y. Novel patterns of the gene expression regulation in the prestalk region along the antero-posterior axis during multicellular development of Dictyostelium. Gene Expr. Patterns. 2005;6:63–68. doi: 10.1016/j.modgep.2005.04.017. [DOI] [PubMed] [Google Scholar]
- Zhukovskaya N, Fukuzawa M, Yamada Y, Araki T, Williams JG. The Dictyostelium bZIP transcription factor DimB regulates prestalk-specific gene expression. Development. 2006;133:439–448. doi: 10.1242/dev.02190. [DOI] [PubMed] [Google Scholar]