Abstract
High-throughput sequencing has drastically changed the research of genes responsible for genetic disorders and is now gradually introduced as an additional genetic diagnostic testing in clinical practice. The current debates on the emerging technical, medical and ethical issues as well as the potential optimum use of the available technology are discussed.
Key Words: Diagnosis, Ethical aspects, Genetic disorders, Next-generation sequencing, Prenatal diagnosis
Mendelian or monogenic genetic disorders are not caused by an external factor but by high-penetrance pathogenic variants of the individual's genome; the possibilities of curative treatment are currently limited, and each one separately is rare.
Rare disorders have currently gained considerable attention and are recognized as an important issue of public health [Donaldson, 2009]. The European Commission defines as rare disorder any disorder with a frequency of less than 1/2,000, further stating that 6–8% of the population suffers from a rare disorder which means 27 to 36 million people [Li et al., 2010]. The European Organization for Rare Diseases (EURORDIS; http://www.eurordis.org/) estimates that at least 80% of them have a genetic origin, and their considerable burden in the health system has been discussed in a few studies. Kumar et al. [2001] estimate that approximately 20% of the visits in the pediatric emergencies are from patients suffering or suspected of being suffering from genetic disorders. Of these patients only 3% had a definitive genetic disorder, whereas the other 97% were not accurately diagnosed. The burden of genetic disorders is also demonstrated in the study by Yoon et al. [1997] showing that 9–12% of all the hospitalizations in pediatric hospitals were related to birth defects and genetic anomalies and those children stayed longer in the hospital and accounted for 184% more hospital charges. Additionally, Stevenson and Carey [2004] studied the reason of death of >500 children in a pediatric hospital and showed that 31% of deaths in children of all ages and 51% of deaths in children under 1 year were due to genetic disorders.
The contradiction between the above data and the relatively poor literature for the vast majority of these diseases can easily be explained by the difficulty to study them. It is of notice that the gene for cystic fibrosis, one of the most common autosomal recessive Mendelian disorders with an incidence of 1 per 2–3,000 [Scotet et al., 2008], has been identified only in 1989 [Riordan et al., 1989]. The discovery of genes responsible for rare monogenic disorders has accelerated since the first draft of the human genome sequence in 2001 [Lander et al., 2001; Venter et al., 2001] was published, but the identification of the responsible genes remained laborious and often impossible [Antonarakis and Beckmann, 2006]. Traditional genetic techniques used to pinpoint the candidate regions, e.g. linkage analysis, require either single large families or many small families with genetic homogeneity of the responsible gene and could only restrict the analysis to a few Mb. These usually contain from dozens to hundreds of genes and other functional genomic elements, which then should be individually sequenced and studied.
Rare genetic disorders did not benefit from the explosion of genome-wide association studies performed in the last 10 years, which were more suited for the study of frequent disorders and tested the ‘common variant, common disease’ hypothesis. Furthermore, it has been shown that many factors made the study of rare disorders by genome-wide association studies impossible. Sample sizes are usually very small, and the pathogenic variants are usually heterogeneous precluding their tagging by common SNPs (single-nucleotide polymorphisms) [Emond et al., 2012].
The laboratory capabilities have drastically changed when high-throughput sequencing (HTS) technologies appeared a couple of years ago. Genetic laboratories have now the opportunity to sequence much larger areas of the genome for a small fraction of the cost of the traditional Sanger sequencing. The study of Kircher and Kelso [2010] shows the capabilities of HTS both in terms of output and low cost. By the traditionally used methods (Sanger sequencing) a laboratory can produce 6 Mb of sequence per day at a cost of 500 dollars per Mb. In contrast, Illumina and ABI instruments (the 2 technologies tested in the study) could produce at the time of the study 5,000 Mb/day at a cost of 0.5 dollars/Mb; that is 830 times more data at 1/1,000th of the cost.
The use of HTS has introduced new types of expertise in the research laboratories. In contrast to Sanger sequencing, only 1 allele is read at a time and read length (how long the DNA fragments are read by the sequencer), the total number of reads, and depth of coverage (how many times each nucleotide is read by the sequencer) have been introduced and characterize each experiment. The depth of sequence is arguably the most important factor that determines the sensitivity and specificity of the nucleotide calls.
The large number of data generated by the modern sequencers, where every experiment can produce up to several hundreds of Gb (table 1) of information, has also made the development of substantial bioinformatics/biocomputing expertise by the laboratories necessary, and analysis of the data is now the longest part of every sequencing project.
Table 1.
Comparative table of the maximum output, read length and time/run for different commercially available HTS platforms
| Maximum output | Read length | Time/run | Reference | |
|---|---|---|---|---|
| HiSeq 2500 (illumina®) | 600 Gb | 2 × 100 bp | 11 days | www.illumina.com |
| Miseq (illumina®) | 8.5 Gb | 2 × 250 bp | 39 h | www.illumina.com |
| 5500xl (Applied Biosystems®) | 360 Gb | 50 bp | 12 days | www.appliedbiosystems.com |
| Titanium XL+ (Roche®) | 700 Mb | 700 bp | 23 h | www.454.com |
| Ion Proton (Ion Torrent®) | 10 Gb | 200 bp | 2–4 h | www.iontorrent.com |
The information was retrieved in August 2012 from the respective websites of the companies.
Additional new technologies have also been developed in order to target more efficiently the sequencing of genomic areas of interest. Commercially available technologies are now dominated by liquid capture which allow for the selection of the quasi-totality of the RefSeq coding exons and a large part of the 5′ and 3′ UTR, constantly expanding and including more regions of potential interest (i.e. non-coding RNAs). In addition, it is also possible to design custom libraries in order to focus on the specific areas of interest of the genome.
The power of HTS was first shown in a proof of concept study [Ng et al., 2009] which ‘rediscovered’ pathogenic variants in the MYH3 gene as the cause of Freeman-Sheldon syndrome. Only 4 unrelated affected individuals participated in this study and the discovery was not based on family studies (of course the success of this example depended on the genetic homogeneity of the syndrome studied). This was followed by many other reports which have expanded our knowledge of genetic defects that cause Mendelian disorders. In published exome data, the number of genes harboring novel heterozygous rare variants is usually reported to be 500 per individual [Bamshad et al., 2012] and several strategies are used in order to identify the pathogenic variants among them. In a recent review, Gilissen et al. [2012] have summarized the different ways that can be used in order to take advantage of HTS in the discovery of pathogenic variants, namely: classic linkage [Krawitz et al., 2010; Züchner et al., 2011], the identification of genes with 2 pathogenic mutations in compound heterozygosity in the absence of consanguinity [Gilissen et al., 2010], the search for pathogenic mutations in a common gene in a well-characterized small cohort of patients with a presumed dominant disorder [Hoischen et al., 2010; Ng et al., 2010], the search for de novo mutations in patients with presumed dominant disorders [Vissers et al., 2010; O'Roak et al., 2011], a candidate gene approach [Haack et al., 2010], and finally, homozygosity mapping for autosomal recessive disorders in selected families of consanguineous matings [Becker et al., 2011; Hamamy et al., 2011]. Especially for the latter case there is evidence that the sequencing of 2 affected sibs instead of 1 drastically reduces the number of candidate variants [Pippucci et al., 2011]. A search of the OMIM database in August 2012 with the terms ‘exome’ and ‘allelic variants’ resulted in 148 different entries, the vast majority of which are new gene-disease associations for Mendelian disorders discovered by HTS strategies.
HTS and the Diagnosis of Rare Disorders
The power of HTS has renewed the interest for rare genetic disorders in the research laboratories, and is slowly beginning to be used in the routine clinical genetics.
Current Laboratory Diagnostic Practices
Rare genetic diseases are severely underdiagnosed; Kumar et al. [2001] estimated that only 3% of the patients in their study of pediatric emergency genetic disorders had a definite genetic diagnosis. In addition, a considerable fraction of the dysmorphology cases do not enjoy a specific molecular diagnosis, and thus, the clinical characterization is only descriptive at the phenotypic level.
One reason for the underdiagnosis of rare genetic disorders is the fact that they are indeed rare and thus systematically overlooked or are unknown in the everyday clinical practice. As an indicative comparison, 12% of women will develop breast cancer in their lifetimes (http://seer.cancer.gov/csr/1975 2009 pops09/) while the frequency of cystic fibrosis, one of the commonest autosomal recessive disorders in the European population, has an incidence of approximately 1 per 2–3,000 [Scotet et al., 2008]. The practical outcome for the patients with Mendelian genetic disorders is a dysfunctional medical care, since these patients are in contact with health professionals for many years without an exact diagnosis and are thus subjected to numerous unnecessary or expensive diagnostic procedures and sometimes dubious treatments.
A series of equally important factors contribute to the limited use of genetic diagnostic tests that unless all are fully addressed, genetic testing will remain a rarity compared to other diagnostic tests despite the substantial number of patients with Mendelian genetic disorders.
The technical aspects of diagnoses of Mendelian genetic disorders are difficult. Standardized methods exist for only a handful of genes, while for the large majority, each diagnostic laboratory is performing tests designed in house without a large scale and communal validation. Significant progress has been achieved both in terms of control and accreditation by various national and international bodies, but a significant improvement is desirable and necessary. For example, the rate of genotype errors in diagnostic laboratories may range from 1.1 to 5.2% [Seneca et al., 2008].
The difficulties for setting up a specific genetic testing for each Mendelian disorder led to the fact that the vast majority of laboratories are restricted to testing a few dozen genes among those who cause the 2,840 (OMIM, August 2012) known Mendelian (and mostly rare) genetic disorders. It is a common practice for clinical geneticists to actively search the world for a suitable diagnostic or even research laboratory capable of offering the desired test at a reasonable turnaround time. The lack of availability of diagnostic tests in combination with long turnaround times, which for some rare and technically challenging tests can even be in the range of years, severely prevents the wide use of diagnostic genetic tests.
An equally important aspect is the interpretation of genetic tests. The identification of a novel or rare variant raises questions of its pathogenicity, particularly in cases of reduced penetrance. An additional problem is that of genetic heterogeneity in which all genes involved in the particular phenotype need to be tested. For example, for a complete molecular work-up of a phenotype such as retinitis pigmentosa, more than 100 genes [Nishiguchi and Rivolta, 2012] need to be tested, and this number increases with the addition of new knowledge.
Use of HTS for Diagnosis of Genetic Disorders
From a purely technical point of view, the protocols for the performance of the HTS experiments are well-established and are used by research laboratories for more than 4 years; thus, there is no apparent difficulty to transfer the necessary know-how to experienced diagnostic laboratories. The analysis of the results from the output of the sequencers to the actual list of the variants is still not completely optimized and automated, but the necessary tools and technical expertise already exist.
There are 2 general strategies to employ candidate gene HTS for diagnostic purposes: One is to select and sequence all the protein-coding genes and then focus on candidate genes by computational means [Rosewich et al., 2012]; the other is to select and sequence only a list of candidate genes [Redin et al., 2012]. Obviously, there are advantages and disadvantages for each of these options, but in both cases only the genes prescribed by the referring physician will be tested and analyzed, and each laboratory could choose the one or the other option depending on cost and turnaround time.
Concerning the clinical utility of genetic tests, HTS can drastically improve the specificity and sensitivity of genetic testing in cases of disorders with genetic heterogeneity by testing all the candidate genes [Dixon-Salazar et al., 2012]. Examples of such use would be the use of panels of genes for the hundreds of genes related to intellectual disability [van Bokhoven, 2011], the already mentioned retinitis pigmentosa [Nishiguchi and Rivolta, 2012], the 71 genes related to non-syndromic hearing loss (http://hereditaryhearingloss.org), the 25 genes related to Charcot-Marie-Tooth syndrome [Pareyson and Marchesi, 2009], and the 14 genes related to familial hypertrophic cardiomyopathy (http://www.ncbi.nlm.nih.gov/books/NBK1768/). In addition, HTS could easily test the extremely long genes, i.e. TTN (titin), that were problematic in the past.
The problem of interpretation of individual variants is not new and the literature on the variants of unknown significance particularly in the BRCA1 and BRCA2 testing is extensive [Akbari et al., 2011]. Thanks to large HTS projects, however, such as the Exome Variant Server (http://evs.gs.washington.edu/EVS/) and the 1,000 Genomes Project (2010), there is for the first time the possibility to have accurate measurements of the frequency of even extremely rare variants (minimum allele frequency <0.1%) in the human population and construct a frequency argument for a potential pathogenicity of a given genomic variant.
Additionally, several computer algorithms have been developed for the likely pathogenicity of non-synonymous and splicing variants that take into account not only the biochemical consequence of the variant but also the evolutionary conservation of the reference allele [Thusberg et al., 2011].
HTS is likely to replace rapidly every other diagnostic sequencing test (particularly as the costs are dropping), but technical limitations of the present HTS technologies dictate that alternative methods will still be used. For example, one of the most frequently performed tests in molecular diagnostics, the search for the CGG expansion in the FMR1 gene causing fragile-X syndrome (and all the other trinucleotide expansion disorders), will continue to be performed by alternative methods due to the limitation of the read's length by the current HTS methods. Another problem is the difficulty to accurately detect large deletions and insertions which are still diagnosed by alternative methods.
HTS and Family Planning/Prenatal Diagnosis
HTS will likely have a profound effect in fetal diagnosis and family planning. A few years ago, Chiu et al. [2008] demonstrated the possibility to diagnose the presence of aneuploidies in the embryo by examining a maternal blood sample. The method is based on the measurement of the relative frequency of the reads of each chromosome in the cell-free DNA circulating in the mother's blood, a significant portion of which comes from the embryo, and has since been refined, and it is now offered as a clinical service by private companies (see for example http://www.sequenomcmm.com/home/health-care-professionals/trisomy-21/). Currently, only trisomy 21 is tested, but it is expected that soon enough all possible gross chromosomal aberrations will be detected offering a safe, non-invasive diagnosis to the majority of pregnant women during the first trimester [Chiu et al., 2011]. After clinical validation in large cohorts of patients and a further reduction of the sequencing costs, the offer of this test may become a standard during pregnancy. The issue is controversial, since this test has the possibility of eliminating completely the common chromosomal abnormalities, should the couples decide for the termination of the affected pregnancy.
Additional technological advances that could revolutionize prenatal diagnostics is the successful genomic sequencing of the fetal genome by sampling the mother's blood [Fan et al., 2012; Kitzman et al., 2012] and in the case of preimplantation diagnosis the sequencing of the genome of a single cell [Hou et al., 2012; Xu et al., 2012].
Another test that is currently in the phase of development is the detection of recessive alleles in consanguineous couples. The increased risk of consanguineous couples to have offspring with autosomal recessive disorders is well-documented because of the sharing of alleles by descent [Tadmouri et al., 2009]. In these couples, HTS sequencing of each of the parents will reveal the shared pathogenic variants in heterozygosity; thus, a catalog of risk genes for the couple could be established and informed decisions could be taken. The difficulty in these analyses is the discovery of rare or private variants of unknown significance in candidate recessive genes that complicate the genetic counseling.
Ethical Aspects
There has been a considerable discussion about the ethical aspects of HTS sequencing, but it is our opinion that the use of HTS as an additional diagnostic tool will not raise issues that are new to the medical community. One key aspect, however, is that in using ‘diagnostic’ HTS the line that separates routine diagnosis from research is not always well-defined.
The most important issue is that of incidental findings, i.e. the identification of likely pathogenic variants that are not related to the clinical question asked; the problem in this case is the disclosure of the incidental findings to the patient or the health professional. There is a growing literature concerning the attitudes towards these findings particularly in the research setting [Wolf et al., 2012]. Common recommendations are that research results on incidental findings may be disclosed if the genetic information reveals significant risk for a condition which has substantial health implications or is life-threatening, or can be used for either a treatment or intervention in order to modify the outcome. These genomic variants are commonly referred to as ‘actionable variants’ [Wolf et al., 2012]. The content of the pre-test informed consent is of paramount importance and the research subject needs to be well-informed of the issues regarding incidental findings and disclosure options. A well-thought-out and debated informed consent preserves the right to know or not-to-know of specific genetic information.
There is also the uncertainty of what exactly is an ‘actionable variant’. A useful definition is ‘when specific strategies contingent upon the result in question have been documented to ameliorate expected morbidity and mortality’ [Evans and Rothschild, 2012]. However, this narrow definition is not satisfactory to all, since genomic variants likely to influence life decisions could be included in the definition of ‘actionable variants’. It is interesting to note that even without an exact definition in mind in a study by Green et al. [2012] clinical geneticists have had a very high degree of concordance concerning the results that should be disclosed to the patients.
Another notable issue is that for many of the predicted damaging genomic variants there is insufficient clinical validation; this, coupled with the reduced penetrance issue, greatly diminishes the ability to provide a sound genetic counseling for the majority of genomic variants.
Conclusion
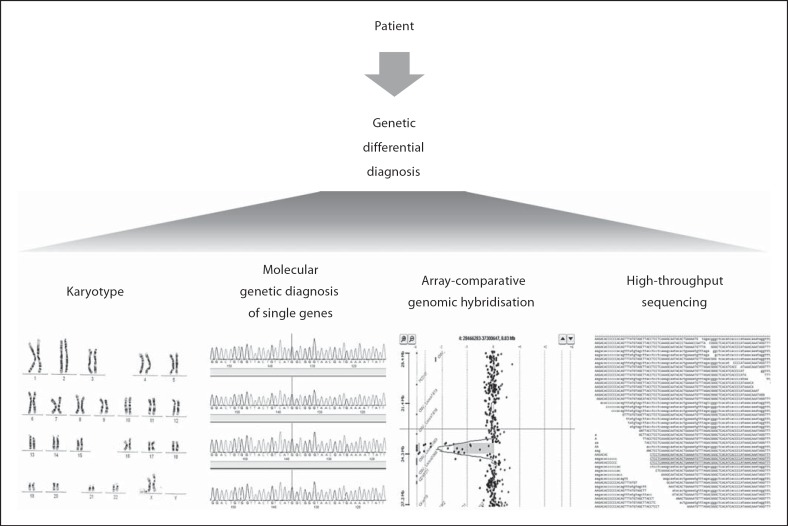
HTS technologies that have appeared less than 10 years ago have already revolutionized the research for the identification of pathogenic genes for rare genetic disorders and revitalized the interest about them. These technologies are gradually finding their place (fig. 1) in the clinical diagnosis providing novel capabilities for the diagnosis of genetic disorders which will help the millions of affected patients and their families, and those who until now did not even have access to the diagnosis of their disorder. The implementation of genomic medicine is proceeding carefully, with active debates on the emerging issues and the optimum use of the available technology.
Fig. 1.
High-throughput sequencing is slowly being introduced as an additional genetic diagnostic test and in the future it will replace most if not all the other genetic tests.
Acknowledgements
P.M. is supported by a grant of the Bodossaki Foundation.
References
- Akbari MR, Zhang S, Fan I, Royer R, Li S, et al. Clinical impact of unclassified variants of the BRCA1 and BRCA2 genes. J Med Genet. 2011;48:783–786. doi: 10.1136/jmedgenet-2011-100305. [DOI] [PubMed] [Google Scholar]
- Antonarakis SE, Beckmann JS. Mendelian disorders deserve more attention. Nat Rev Genet. 2006;7:277–282. doi: 10.1038/nrg1826. [DOI] [PubMed] [Google Scholar]
- Bamshad MJ, Shendure JA, Valle D, Hamosh A, Lupski JR, et al. The Centers for Mendelian Genomics: a new large-scale initiative to identify the genes underlying rare Mendelian conditions. Am J Med Genet A. 2012;158A:1523–1525. doi: 10.1002/ajmg.a.35470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker J, Semler O, Gilissen C, Li Y, Bolz HJ, et al. Exome sequencing identifies truncating mutations in human SERPINF1 in autosomal-recessive osteogenesis imperfecta. Am J Hum Genet. 2011;88:362–371. doi: 10.1016/j.ajhg.2011.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu RW, Chan KC, Gao Y, Lau VY, Zheng W, et al. Noninvasive prenatal diagnosis of fetal chromosomal aneuploidy by massively parallel genomic sequencing of DNA in maternal plasma. Proc Natl Acad Sci USA. 2008;105:20458–20463. doi: 10.1073/pnas.0810641105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu RW, Akolekar R, Zheng YW, Leung TY, Sun H, et al. Non-invasive prenatal assessment of trisomy 21 by multiplexed maternal plasma DNA sequencing: large scale validity study. BMJ. 2011;342:c7401. doi: 10.1136/bmj.c7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon-Salazar TJ, Silhavy JL, Udpa N, Schroth J, Bielas S, et al. Exome sequencing can improve diagnosis and alter patient management. Sci Transl Med. 2012;4:138–178. doi: 10.1126/scitranslmed.3003544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson L. On the State of the Public Health: Annual Report of the Chief Medical Officer 2009. UK: Department of Health; 2009. [Google Scholar]
- Emond MJ, Louie T, Emerson J, Zhao W, Mathias RA, et al. Exome sequencing of extreme phenotypes identifies DCTN4 as a modifier of chronic Pseudomonas aeruginosa infection in cystic fibrosis. Nat Genet. 2012;44:886–889. doi: 10.1038/ng.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans JP, Rothschild BB. Return of results: not that complicated? Genet Med. 2012;14:358–360. doi: 10.1038/gim.2012.8. [DOI] [PubMed] [Google Scholar]
- Fan HC, Gu W, Wang J, Blumenfeld YJ, El-Sayed YY, Quake SR. Non-invasive prenatal measurement of the fetal genome. Nature. 2012;487:320–324. doi: 10.1038/nature11251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1000 Genomes Project Consortium: A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilissen C, Arts HH, Hoischen A, Spruijt L, Mans DA, et al. Exome sequencing identifies WDR35 variants involved in Sensenbrenner syndrome. Am J Hum Genet. 2010;87:418–423. doi: 10.1016/j.ajhg.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilissen C, Hoischen A, Brunner HG, Veltman JA. Disease gene identification strategies for exome sequencing. Eur J Hum Genet. 2012;20:490–497. doi: 10.1038/ejhg.2011.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RC, Berg JS, Berry GT, Biesecker LG, Dimmock DP, et al. Exploring concordance and discordance for return of incidental findings from clinical sequencing. Genet Med. 2012;14:405–410. doi: 10.1038/gim.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haack TB, Danhauser K, Haberberger B, Hoser J, Strecker V, et al. Exome sequencing identifies ACAD9 mutations as a cause of complex I deficiency. Nat Genet. 2010;42:1131–1134. doi: 10.1038/ng.706. [DOI] [PubMed] [Google Scholar]
- Hamamy H, Antonarakis SE, Cavalli-Sforza LL, Temtamy S, Romeo G, et al. Consanguineous marriages, pearls and perils: Geneva International Consanguinity Workshop Report. Genet Med. 2011;13:841–847. doi: 10.1097/GIM.0b013e318217477f. [DOI] [PubMed] [Google Scholar]
- Hoischen A, van Bon BW, Gilissen C, Arts P, van Lier B, et al. De novo mutations of SETBP1 cause Schinzel-Giedion syndrome. Nat Genet. 2010;42:483–485. doi: 10.1038/ng.581. [DOI] [PubMed] [Google Scholar]
- Hou Y, Song L, Zhu P, Zhang B, Tao Y, et al. Single-cell exome sequencing and monoclonal evolution of a JAK2-negative myeloproliferative neoplasm. Cell. 2012;148:873–885. doi: 10.1016/j.cell.2012.02.028. [DOI] [PubMed] [Google Scholar]
- Kircher M, Kelso J. High-throughput DNA sequencing – concepts and limitations. Bioessays. 2010;32:524–536. doi: 10.1002/bies.200900181. [DOI] [PubMed] [Google Scholar]
- Kitzman JO, Snyder MW, Ventura M, Lewis AP, Qiu R, et al. Noninvasive whole-genome sequencing of a human fetus. Sci Transl Med. 2012;4:137–176. doi: 10.1126/scitranslmed.3004323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawitz PM, Schweiger MR, Rödelsperger C, Marcelis C, Kölsch U, et al. Identity-by-descent filtering of exome sequence data identifies PIGV mutations in hyperphosphatasia mental retardation syndrome. Nat Genet. 2010;42:827–829. doi: 10.1038/ng.653. [DOI] [PubMed] [Google Scholar]
- Kumar P, Radhakrishnan J, Chowdhary MA, Giampietro PF. Prevalence and patterns of presentation of genetic disorders in a pediatric emergency department. Mayo Clin Proc. 2001;76:777–783. doi: 10.1016/S0025-6196(11)63220-5. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Li Y, Pawlik B, Elcioglu N, Aglan M, Kayserili H, et al. LRP4 mutations alter Wnt/beta-catenin signaling and cause limb and kidney malformations in Cenani-Lenz syndrome. Am J Hum Genet. 2010;86:696–706. doi: 10.1016/j.ajhg.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SB, Turner EH, Robertson PD, Flygare SD, Bigham AW, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SB, Bigham AW, Buckingham KJ, Hannibal MC, McMillin MJ, et al. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat Genet. 2010;42:790–793. doi: 10.1038/ng.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiguchi KM, Rivolta C. Genes associated with retinitis pigmentosa and allied diseases are frequently mutated in the general population. PLoS One. 2012;7:e41902. doi: 10.1371/journal.pone.0041902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Roak BJ, Deriziotis P, Lee C, Vives L, Schwartz JJ, et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat Genet. 2011;43:585–589. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pareyson D, Marchesi C. Diagnosis, natural history, and management of Charcot-Marie-Tooth disease. Lancet Neurol. 2009;8:654–667. doi: 10.1016/S1474-4422(09)70110-3. [DOI] [PubMed] [Google Scholar]
- Pippucci T, Benelli M, Magi A, Martelli PL, Magini P, et al. EX-HOM (EXome HOMozygosity): a proof of principle. Hum Hered. 2011;72:45–53. doi: 10.1159/000330164. [DOI] [PubMed] [Google Scholar]
- Redin C, Le Gras S, Mhamdi O, Geoffroy V, Stoetzel C, et al. Targeted high-throughput sequencing for diagnosis of genetically heterogeneous diseases: efficient mutation detection in Bardet-Biedl and Alstrom Syndromes. J Med Genet. 2012;49:502–512. doi: 10.1136/jmedgenet-2012-100875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riordan JR, Rommens JM, Kerem B, Alon N, Rozmahel R, et al. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science. 1989;245:1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- Rosewich H, Thiele H, Ohlenbusch A, Maschke U, Altmüller J, et al. Heterozygous de-novo mutations in ATP1A3 in patients with alternating hemiplegia of childhood: a whole-exome sequencing gene-identification study. Lancet Neurol. 2012;11:764–773. doi: 10.1016/S1474-4422(12)70182-5. [DOI] [PubMed] [Google Scholar]
- Scotet V, Assael BM, Duguépéroux I, Tamanini A, Audrézet MP, et al. Time trends in birth incidence of cystic fibrosis in two European areas: data from newborn screening programs. J Pediatr. 2008;152:25–32. doi: 10.1016/j.jpeds.2007.07.046. [DOI] [PubMed] [Google Scholar]
- Seneca S, Morris MA, Patton S, Elles R, Sequeiros J. Experience and outcome of 3 years of a European EQA scheme for genetic testing of the spinocerebellar ataxias. Eur J Hum Genet. 2008;16:913–920. doi: 10.1038/ejhg.2008.29. [DOI] [PubMed] [Google Scholar]
- Stevenson DA, Carey JC. Contribution of malformations and genetic disorders to mortality in a children's hospital. Am J Med Genet A. 2004;126A:393–397. doi: 10.1002/ajmg.a.20409. [DOI] [PubMed] [Google Scholar]
- Tadmouri GO, Nair P, Obeid T, Al Ali MT, Al Khaja N, Hamamy HA. Consanguinity and reproductive health among Arabs. Reprod Health. 2009;6:17. doi: 10.1186/1742-4755-6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thusberg J, Olatubosun A, Vihinen M. Performance of mutation pathogenicity prediction methods on missense variants. Hum Mutat. 2011;32:358–368. doi: 10.1002/humu.21445. [DOI] [PubMed] [Google Scholar]
- van Bokhoven H. Genetic and epigenetic networks in intellectual disabilities. Annu Rev Genet. 2011;45:81–104. doi: 10.1146/annurev-genet-110410-132512. [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Vissers LE, de Ligt J, Gilissen C, Janssen I, Steehouwer M, et al. A de novo paradigm for mental retardation. Nat Genet. 2010;42:1109–1112. doi: 10.1038/ng.712. [DOI] [PubMed] [Google Scholar]
- Wolf SM, Crock BN, Van Ness B, Lawrenz F, Kahn JP, et al. Managing incidental findings and research results in genomic research involving biobanks and archived data sets. Genet Med. 2012;14:361–384. doi: 10.1038/gim.2012.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Hou Y, Yin X, Bao L, Tang A, et al. Single-cell exome sequencing reveals single-nucleotide mutation characteristics of a kidney tumor. Cell. 2012;148:886–895. doi: 10.1016/j.cell.2012.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon PW, Olney RS, Khoury MJ, Sappenfield WM, Chavez GF, Taylor D. Contribution of birth defects and genetic diseases to pediatric hospitalizations. A population-based study. Arch Pediatr Adolesc Med. 1997;151:1096–1103. doi: 10.1001/archpedi.1997.02170480026004. [DOI] [PubMed] [Google Scholar]
- Züchner S, Dallman J, Wen R, Beecham G, Naj A, et al. Whole-exome sequencing links a variant in DHDDS to retinitis pigmentosa. Am J Hum Genet. 2011;88:201–206. doi: 10.1016/j.ajhg.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]