Abstract
CK-666 (1) is a recently discovered small molecule inhibitor of the Arp2/3 complex, a key actin cytoskeleton regulator with roles in bacterial pathogenesis and motility of cancer cells. While 1 is commercially available, the crystal structure of Arp2/3 (Actin-related protein 2/3) complex with 1 bound has not been reported, making its mechanism of action uncertain. Furthermore, its relatively low potency increases its potential for off target effects in vivo, complicating interpretation of its influence in cell biological studies and precluding its use in clinical applications. Here we report the crystal structure of 1 bound to Arp2/3 complex, which reveals that 1 binds between the Arp2 and Arp3 subunits to stabilize the inactive conformation of the complex. Based on the crystal structure, we used computational docking and free energy perturbation calculations of monosubstituted derivatives of 1 to guide optimization efforts. Biochemical assays of ten newly synthesized compounds led to the identification of compound 2, which exhibits a 3 fold increase in inhibitory activity in vitro. In addition, our computational analyses unveiled a surface groove at the interface of the Arp2 and Arp3 subunits that can be exploited for additional structure-based optimization.
Keywords: Actin, Arp2/3 complex, Amides, Cancer, Chlorine, Free Energy Perturbation, Polymers
Introduction
Many cellular processes such as motility, endocytosis, and division require the assembly of actin filaments from a pool of globular, monomeric actin.[1] Formation of the actin polymer depends on a slow nucleation step,[2] and cells utilize a handful of actin nucleating factors which are responsive to cellular signaling pathways to provide precise spatiotemporal regulation of the assembly of actin filaments.[3] Actin-related-protein 2/3 (Arp2/3) complex is a seven subunit ~225 kDa ATP-ase that is a key actin nucleator.[4] It functions by attaching itself to the side of a preexisting actin filament and nucleating a new (daughter) filament to create a Y-shaped branch. While it is already known that Arp2/3 complex is required for many cellular processes, knockouts of Arp2/3 complex subunits in multicellular organisms are lethal, so roles of the complex late in development or in adult stages remain largely undiscovered.[5] RNA interference experiments could circumvent this problem, but are relatively slow acting, irreversible, and cannot be used in some biological contexts. Therefore, fast-acting, simple, and reversible small molecule inhibitors have the potential to become powerful tools to study Arp2/3 complex in vivo. Such inhibitors may also have clinical value, since many bacterial and viral pathogens usurp host cell Arp2/3 complex for invasion or intracellular motility.[6] In addition, tumor cell migration is thought to require Arp2/3 complex, and Arp2/3 overexpression contributes to pathogenesis, growth and invasion of carcinomas.[7] Therefore, Arp2/3 complex inhibitors also hold promise as anti-tumor agents.[8]
The discovery of two related small molecule inhibitors of Arp2/3 complex, CK-636 (12) and 1 was recently reported (Figure 1).[9] Both compounds include a 2-methyl substituted indole attached at the 3 position via a short amide linker to an aromatic ring, and both inhibit Arp2/3 complex in actin polymerization assays in vitro. Importantly, these compounds are cell permeable and inhibit Listeria actin comet tail formation in infected SKOV3 cells and podosome formation in THP-1 derived monocyte cells, processes previously shown to require Arp2/3 complex.[6, 9,10] An x-ray crystal structure of 12 bound to inactive Arp2/3 complex showed that it binds at the interface between the Arp2 and Arp3 subunits to block a conformational rearrangement of these subunits required for activation.[9] Therefore, 12 represents a novel example of an allosteric interfacial inhibitor, a molecule which binds at subunit interfaces of a macromolecular assembly to block conformational dynamics critical for function.[11] Compound 1, the more potent of the two compounds, is commercially available, and has already been used in a number of in vivo studies of Arp2/3 complex function.[12] However, the IC50 (half-maximal inhibitory concentration) for 1 is in the low micromolar range and undesirably high concentrations are necessary for complete inhibition of the complex, increasing the probability of off-target effects. The potential of Arp2/3 complex inhibitors to become powerful basic research tools and the possibility of their clinical value led us to optimize parent compound 1. We first solved the crystal structure of 1 bound to Arp2/3 complex and computationally docked analogues of 1 to predict the effect of various substitutions on binding. We then synthesized and tested the analogues in an actin polymerization assay to determine their effect on Arp2/3 complex-mediated nucleation. As docking results did not correlate with experimental findings, we subsequently turned to the more sophisticated free energy perturbation calculations for binding affinity predictions, which correlated well with experimental findings. In addition to yielding an inhibitor with threefold improved inhibition, this work provides significant insights into further structure-based optimization of Arp2/3 complex inhibitors. Specifically, we identify atoms within the scaffold of 1 that cannot tolerate substitutions and reveal a position (R4) on 1, which is a promising site for future optimization efforts because it provides opportunities to exploit a groove at the interface of Arp2 and Arp3. Finally, the work presented herein provides new insights into the use of computational docking and free energy perturbation calculations to aid drug design.
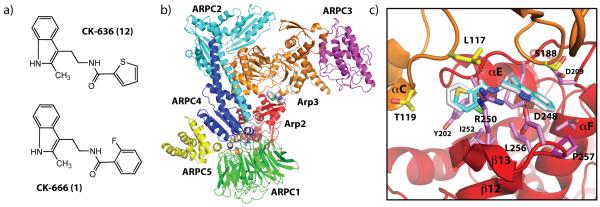
Figure 1.
(A) Previously synthesized Arp2/3 complex inhibitors 1 and 12. Compound 1 was used as a parent scaffold for substitutions indicated in Scheme 1. (B) Crystal structure of 1 bound to Bos taurus Arp2/3 complex. 1 is shown as spheres and binds at the interface of the Arp3 (orange) and Arp2 (red) subunits. The five other subunits in the complex are labeled ARPC1-5. (C). Comparison of 1 (grey carbon atoms) with 12 (turquoise carbon atoms) showing a close up of the binding pocket at the interface of Arp2 and Arp3. Coordinates for 12 for this figure were generated by overlaying Arp2 and Arp3 from 3DXK.pdb onto the structure of 1 and applying the transformation to 12.
Results
X-ray crystal structure of 1 bound to Arp2/3 complex
Crystals of the apoenzyme form of bovine Arp2/3 complex soaked in 500 μM of 1 diffracted to 2.48 Å resolution (Supplementary Table 1). Compound 1 binds to the Arp2-Arp3 interface identically to 12, except for the fluorobenzene moiety in 1, which replaces the thiophene ring in 12 (Figure 1A-C). The asymmetric distribution of density of the fluorobezene ring indicated that the fluorine is pointed towards β12 and β13 in sub domain 4 of Arp2 (Supplementary Figure 1). To verify correct assignment of the ring flip, we also modeled the ring with the fluorine pointing in the opposite direction, toward the β7/αC loop of Arp3. Refinement of this conformation followed by Fo-Fc difference electron density map generation showed the fluorine in 4.8 σ negative density, indicating that our original model was correct. The amide oxygen of 1 hydrogen bonds to Ala203 of Arp3 and the indole nitrogen forms a hydrogen bond with Asp248, a conserved residue in Arp2. Both of these polar contacts are also present in crystal structure of Arp2/3 complex with 12 bound. Compared to 12, 1 buries 16 Å2 more solvent-exposed surface area at the interface, which may account for the decreased IC50 value of 1 compared to 12 in inhibiting Arp2/3 complex in in vitro actin polymerization assays.[9] Binding of 1 causes minor conformational changes similar to those caused by 12, including rotation of Arg250Arp2 out of the binding pocket and a subtle movement of the loop connecting αE and αF in Arp2 toward the binding pocket. Since both 12 and 1 bind at an interface between Arp2 and Arp3 present only in the inactive conformation, we conclude that both inhibitors function by blocking the 25 Å movement of the Arp2 subunit thought to be required for activation of the complex.[13]
Calculation of Glide docking scores and physicochemical properties
Glide docking and scoring has been previously validated in virtual screening exercises;[14] however, we considered it necessary to test its performance with the Arp2/3 complex. We calculated Glide binding poses for inhibitors 12 and 1 and compared them to the respective inhibitor-bound Arp2/3 crystal structures. The RMSD values between non-hydrogen atoms of the Glide-calculated pose and the crystal structures of 12 or 1 bound to Arp2/3 are 0.31 and 0.42 Å, respectively, indicating close agreement between docking and the crystal structure (Supplemental Figure 2A,B). The docked poses of 12 or 1 engage in intermolecular interactions identical to those observed in the crystal structures, including hydrogen bonds between the indole amine hydrogen and the side chain of Asp248Arp2 and between the amide oxygen in the linker and the backbone NH of Ala203Arp3. One difference is that the fluorine atom of 1 is flipped in the docked pose with respect to its position in the crystal structure. However, the calculations suggest the two poses are nearly isoenergetic and we observed the fluorobenzene ring flip in a number of the Glide docked analogues.
Half of the analogues were predicted to bind better than 1 based on their docking scores (Table 1). Hydroxy and methyl substitutions at position R1 and chlorine or hydroxyl groups at R4 yielded the compounds with the lowest scores, suggesting the R1 and R4 positions are promising optimization sites. 2 had the best docking score, with a GScore 1.3 kcal/mol lower than that of 1. Calculation of the physicochemical properties for 2-11 showed that like 1, each of the analogues had predicted octanol/water partition coefficients, aqueous solubility, and Caco-2 cell permeability within the optimal range for marketed drug compounds (Supplementary Table 2).[15] In addition, all compounds comply with the Lipinski rule of five and the Jorgensen rule of three, with the exception of 3, which has a slightly lower predicted solubility (logS = −6.0) than the solubility limit of the Jorgensen rule of three (logS ≥ −5.7).[16]
Table 1.
Biochemical and computational data of compounds tested
| Compound | R1 | R2 | R3 | R4 | IC50 (μM) |
Glide 5.7 XP GScore |
ΔΔGb (kcal/mol) |
|---|---|---|---|---|---|---|---|
| 1 | H | F | H | H | 12 ± 1 | −7.10 | |
| 2 | H | F | H | OH | 4 ± 1 | −8.40 | −0.55 ± 0.15 |
| 3 | Cl | F | H | H | 18 ± 5 | −6.85 | 3.6 ± 0.16 |
| 4 | H | F | H | OCH3 | 19 ± 6 | −7.00 | −1.25 ± 0.4 |
| 5 | OH | F | H | H | 32 ± 4 | −7.97 | −1.3 ± 0.27 |
| 6 | H | F | H | Cl | 32 ± 18 | −7.04 | 0.12 ± 0.12 |
| 7 | H | H | H | Cl | 181 ± 85 | −7.92 | −0.65 ± 0.13 |
| 8 | H | F | Cl | H | 238 ± 91 | −6.95 | 1.9 ± 0.14 |
| 9 | OCH3 | F | H | H | no inhib. | −8.46 | 3.3 ± 0.5 |
| 10 | H | Cl | H | H | no inhib. | −7.66 | 3.9 ± 0.15 |
| 11 | H | H | Cl | H | no inhib. | −7.66 | 2.5± 0.17 |
| 15 | H | H | H | H | n/a | n/a | −0.53 ± 0.09 |
IC50 values were determined from the actin polymerization assay. Glide docking scores were calculated with the XP scoring function. Differences in free energies of binding, ΔΔGb, were calculated from the free energy perturbation calculations.
Chemistry
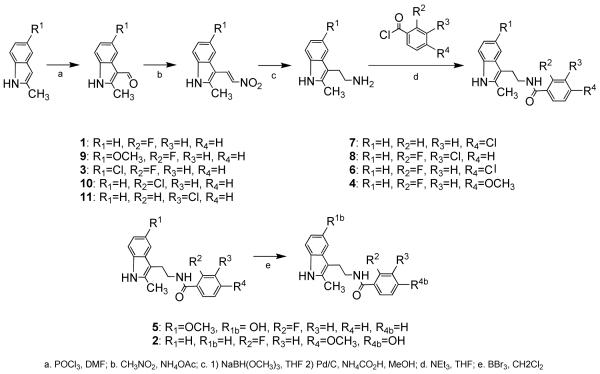
Inhibitor candidates predicted to have similar, better, or worse activity than 1 were synthesized to provide a body of data to guide analysis of structure-activity relationships and refinement of the computational studies (Scheme 1). Table 1 shows the chemical structure of all synthesized inhibitor candidates along with their experimentally determined IC50 values.
Scheme 1.
Biochemistry
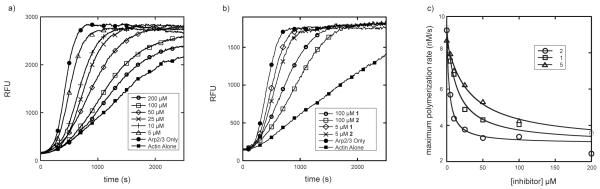
To measure the potency of the analogues, we used a standard actin polymerization assay to measure the rate of actin polymerization. In this assay, the increase in fluorescence of pyrene-labeled actin subunits upon polymerization is used to monitor the time course of filament formation.[17] Arp2/3 complex, together with its activator protein, N-WASp-VCA, increases the rate of polymer formation by nucleating branched filaments and increasing the number of elongating filament ends (Figure 2). Therefore, this assay can be used to determine the nucleation activity of Arp2/3 complex. We used Bos taurus Arp2/3 complex in our assays because it can be isolated in large quantities. Residues in the binding cleft of 1 are conserved from yeast to humans and thus, optimization of the inhibitors against the bovine complex will very likely improve IC50 values for inhibition of the complex from any species, including Homo sapiens.[9] Figure 2A shows that 1 decreased Arp2/3 complex nucleation activity in a concentration-dependent manner, with the addition of 200 μM inhibitor decreasing the maximum polymerization to a rate similar to that observed in the absence of Arp2/3 complex. The IC50 of 1 calculated under these conditions was 12 μM, in agreement with previously reported values.[9] Of the ten derivative compounds, three showed no inhibition towards Arp2/3 complex. The IC50 values of the other compounds were compared to that of 1 in Table 1: 7 and 8 showed a 15-20 fold increase in IC50, 5 and 6 showed approximately 3-fold increased IC50 values, and compounds 3 and 4 had IC50 values nearly double that of 1. 2, derivatized from 1 by replacement of a hydrogen atom at R4 with an OH group, showed a three-fold improved IC50 (Figure 2B,C).
Figure 2.
Biochemical characterization of 1 and analogues. (A) Time course of the polymerization of 30% pyrene-labeled actin in the presence of 5 nM Arp2/3 complex, 200 nM N-WASP-VCA, and either DMSO for a range of concentrations of 1. A control reaction (actin alone) lacking only Arp2/3 complex shows the intrinsic nucleation rate of actin. (B) Comparison of actin polymerization time courses in the presence of equal concentrations of 1 versus 2. Conditions are identical to panel (A). (C) Plot of maximal polymerization rate versus inhibitor concentration for 1, 2, and 5. Data were fit as described in Methods.
Free Energy Perturbation calculations
The Glide XP scores showed poor correlation with the assay data for analogs 1-14 (Table 1), indicating an inability to predict binding affinities for a series of close analogs in this system. Therefore, we initiated Free Energy Perturbation (FEP) calculations, a robust methodology for evaluating differences in free energy of binding of different ligands. The differences in free energy of binding affinity (ΔΔGb) presented in Table 1 correlate well with experimental results. In particular, all inactive analogs of 1 were correctly identified by FEP calculations, which yielded large positive differences in free energy of binding with respect to 1. Moreover, analog 2 was identified as a better binder in accordance with a 3-fold improvement in the IC50 value. For three compounds (4, 5, 7), the FEP calculations did not correctly predict the experimental results. A detailed discussion follows in the next section. Results in Table 1 are shown as ΔΔGCK-666 → ΔΔGanalog, where negative values indicate more favorable predicted binding.
Discussion
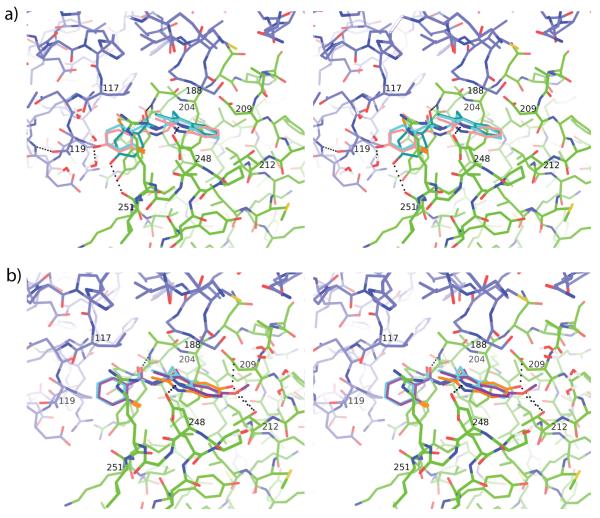
Here we present the crystal structure of 1 bound to Arp2/3 complex and show that it binds to the same pocket at the interface of Arp3 and Arp2 as 12, a previously described inhibitor, indicating that 1 and 12 use the same mechanism of inhibition.[9] Ten analogues of 1 were designed and synthesized leading to the discovery of 2, a compound with three-fold better inhibition of Arp2/3 complex than 1 and a promising physicochemical profile (Table 1 and Supplementary Table 2). Compound 2 has a hydroxyl substitution at R4, a position which allows functional groups to project into an extended groove formed at the interface of Arp2 and Arp3. Figure 3A illustrates that in the Glide docked structure of 2, the fluorobenzene ring flips and moves toward the Arp2 subunit, allowing the hydroxyl group at R4 to form a hydrogen bond with the carboxyl oxygen of Ile251 of Arp2. The movement of the ring requires a 33° deviation from planarity for the amide bond in the linker region of the inhibitor. In the FEP calculated structure the amide is within 5.4° of planarity and the R4 hydroxyl is close to the Thr119Arp3 side chain, but a hydrogen bond with the backbone amide of Asn122Arp3 locks the threonine hydroxyl in a rotamer incapable of bonding to the R4 hydroxyl. Therefore, in the FEP calculations, R4 hydroxyl hydrogen bonds only to solvent, explaining the modest decrease in calculated FEP binding energy compared to 1, which agrees more closely with the biochemical data than the Glide docking calculation. We note that examination of the crystal structure and all computational models revealed that binding requires desolvation of the nitrogen in the amide linker without a compensating hydrogen bond to the protein, suggesting that increasing the hydrophobicity of the linker may increase the net binding energy.
Figure 3.
Binding pocket of 1 with overlapping docked analogues 2, 5 and 9. (A) Stereo figure showing a comparison of 1 and the docked and FEP poses of 2. Arp3 (blue carbon atoms) and Arp2 (green carbon atoms) from the crystal structure of Arp2/3 with bound 1 (3UKR.pdb, light blue carbon atoms) are shown in line representation overlaid with the Glide docked pose (teal carbon atoms) and the FEP docked pose (pink) of 2. Selected hydrogen bonds are shown as black dotted lines. (B) Stereo figure showing a comparison of 1 and the FEP generated poses of 5 and 9. Colors are identical to panel A, except 5 is shown with orange carbon atoms and 9 is shown with purple carbon atoms.
In compound 4 a methoxy group replaces hydrogen at position R4. The Glide docked and FEP calculated poses show the methyl group making favorable van der Waals contacts with either the methyl group of Thr119Arp3 or the aliphatic portion of Arg250Arp2, respectively (Supplementary Figure 3). Accordingly, compound 4 showed a lower ΔΔG in comparison to 2 in the FEP calculations, despite the fact that it had a fivefold higher IC50 value than 2. The FEP generated pose shows the methoxy oxygen wedged in an aliphatic cleft lined by the methyl group of Thr119Arp3 and the hydrophobic part of Arg250Arp2, where it is partially shielded from solvent (Supplementary Figure 3). It hydrogen bonds to a single solvent molecule and leaves a lone pair of electrons unsatisfied. This indicates a desolvation penalty that we speculate may not be fully accounted for by the FEP calculations. Replacing the methoxy oxygen with a carbon atom would decrease the desolvation penalty and is a promising future optimization strategy.
Compound 6 has a chlorine atom in position R4 and exhibited an approximately 3-fold increase in IC50 relative to 1. Steric clash between the chlorine and the Thr119Arp3 methyl group repositions the fluorobenzene ring in both the Glide docked and FEP generated poses, though the direction of movement is the opposite in the two computations (Supplementary Figure 4). In the FEP pose, the side chain of Arg250 moves to accommodate the chlorine, whereas the side chains are fixed in the Glide docking, explaining the difference in the conformations favored for each method. These data suggest that the fluorobenzene ring can be repositioned within the pocket without a major decrease in affinity, indicating that the protein functional groups that interact with the R4 substituent may vary depending on the bulkiness of the R4 substituent. Taken together, the results for compounds 2, 4, and 6 strongly suggest that position R4 in the fluorobenzene ring is a promising site for future optimization.
Derivatives 9 and 5 featured substitutions at the R1 position and were predicted by docking to bind better than 1. 5, with a hydroxyl group at R1, exhibited an IC50 almost threefold higher than that of 1 (Table 1). The hydroxyl group at R1 was predicted to hydrogen bond to the side chains Asp209 and Thr212 of Arp2 by Glide docking and FEP calculations, but these interactions require the indole ring to distort slightly, which may outweigh energetic benefits from the predicted hydrogen bonding (Figure 3B). Replacing the R1 hydroxyl group of 1 with a methoxy group gave compound 9, which also binds with a distorted indole ring and does not hydrogen bond to the sidechain of Asp209Arp2 in the FEP pose (Figure 4B). While the oxygen of the methoxy substituent maintains one hydrogen bond with Thr212Arp2, the methyl group excludes water around the R1 site, leaving the oxygen with one unsatisfied lone pair hydrogen bond acceptor. The partial desolvation penalty of the methoxy and strain of the indole ring likely contribute to the severe decrease in binding affinity for 9, which is also predicted by the FEP calculations.
Placing a chlorine at position R1 to give 3 increased the IC50 only slightly compared to 1, despite the fact that the FEP calculation predicted this substitution was highly unfavorable (ΔΔG = +3.6 kcal/mol). This discrepancy may be due to the inability of the force field to account for the halogen bond that appears to form between the chlorine and the neighboring carbonyl of Asp209Arp2 and the side chain of Thr212Arp2. Oxygen atoms from both of these side chains are equidistant from the chlorine atom at 3.1 Å, which is close to the experimentally observed value for a halogen bond.[18]
Derivative compounds 7, 10, and 11, which featured mono-chloro substituted benzene rings in the R2, R3, and R4 positions, respectively, all showed greatly reduced or no inhibition. Substitution of chlorine at the para (7) position caused the benzene ring to tilt by 30.5° and move away from the sidechain of Thr119Arp3 to relieve steric clash (Supplementary Figure 5). The FEP calculation predicted this substitution to have little or no influence on binding. The importance of the fluorine at the ortho position is evident in 10, since its replacement with chlorine completely abolishes inhibition. The FEP calculation correctly predicted the poor binding of this compound, and the FEP calculated structures show that the bulky chlorine substituent causes a slight repositioning of the benzene ring and the amide linker (Supplementary Figure 5), that may reduce shape complementarity. Further evidence for the importance of the fluorine at the R2 position is provided by compounds 6 and 8, which have unfavorable substitutions in the para and meta positions, like 7 or 11, but due to the orthofluorine atom bind better than the monochloro compounds.
Our analysis suggests that the weak correlation between the binding affinities predicted from docking calculations and the measured inhibition constants is at least partly due to the fact that protein and water molecules are kept rigid in the docking process. FEP calculations allow for a dynamical evolution of the system, which, in several instances, resulted in changes in the positions of side chains or water molecules to accommodate the structures of the bound analogues. This induced-fit effect appeared to be a critical contributor to the ΔΔG of binding. The correlation between the FEP calculations and the measured inhibition constants demonstrate the predictive value of the FEP calculations in this system and will allow us to streamline development of improved inhibitors of Arp2/3 complex for basic research and potential clinical applications.
Experimental Section
Chemistry
The synthesis in Scheme 1 proceeds through standard protocols to produce inhibitor candidates.[19] Commercially available indoles 1 a-11 a are formylated under Vilsmeier conditions to produce aldehydes 1 b-11 b, which undergo the Henry reaction followed by condensation to yield nitroolefins 1 c-11 c. Reduction with sodium trimethoxyborohydride followed by basic aqueous workup and subsequent reduction with 10 wt.% Pd/C in the presence of ammonium formate in methanol yields amines 1 d-11 d. Acid chloride coupling completes the synthesis of 1, 3-4 and 6-11, with subsequent methoxy deprotection using BBr3 required for 2 and 5.[20] Compounds 1, 3-4, and 7-11 were all >98% pure as determined by HPLC analysis. Compound 2 was determined to be >97% pure, compound 5 was determined to be > 95% pure, and compound 6 was determined to be >96 % pure by HPLC analysis.
General Procedure for 1, 3, 4, 6, 7, 8, 9, 10, and 11
Indole amines prepared according to literature preparations were stirred in THF (1 mL/ 25 mg compound) with triethylamine (1.1 mole eq) and acid chloride (1.1 mole eq) at room temperature for 30 minutes. At the conclusion of the reaction, water was added and the resulting solution was partitioned with diethyl ether. The organic phase was washed with saturated aqueous sodium chloride, dried with MgSO4, and the solvent was removed under reduced pressure to yield crude solid product. Purification on a silica gel column using 2:1 diethyl ether:hexanes as the eluent followed by removal of solvent under reduced pressure yielded compounds in decent yields.
2-fluoro-N-(2-(2-methyl-1H-indol-3-yl)ethyl)benzamide (1)
1 (0.050 g, 42.7%) was synthesized as a white solid from 2-(2-methyl-1H-indol-3-yl)ethanamine (0.069 g, 0.40 mmol) using the general procedure described above. 1H NMR (300 MHz, (CD3)2CO): δ 9.91 (br s, 1H), 7.93 (t d, 3JHH = 7.8, 1.8 Hz, 1H), 7.64 (s, 1H), 7.59 (d, 3JHH = 6.6 Hz, 1H), 7.54 (m, 1H), 7.30 (m, 2H), 7.21 (d d d, 3JHH = 11.4, 8.1, 1.2 Hz, 1H), 7.02 (quin d, 3JHH = 6.9, 1.5 Hz, 2H), 3.68 (q, 3J = 7.2 Hz, 2H), 3.07 (t, 3JHH = 7.2 Hz, 2H), 2.42 (s, 3H).
N-(2-(5-chloro-2-methyl-1H-indol-3-yl)ethyl)-2-fluorobenzamide (3)
3 (0.040 g, 31.7%) was synthesized as a white solid from 2-(5-chloro-2-methyl-1H-indol-3-yl)ethanamine (0.080 g, 0.38 mmol) using the general procedure described above. 1H NMR (300 MHz, (CD3)2CO): δ 10.09 (br s, 1H), 7.92 (t d, 3JHH = 7.8, 1.8 Hz, 1H), 7.61 (br s, 1H), 7.59 (d, 3JHH = 2.1 Hz, 1H), 7.55 (m, 1H), 7.30 (m, 2H), 7.22 (d d d, 3JHH = 11.4, 8.4, 0.9 Hz, 1H), 7.01 (d d, 3JHH = 8.4, 2.4 Hz, 1H), 3.64 (q, 3JHH = 7.2 Hz, 2H), 3.04 (t, 3JHH = 7.2 Hz, 2H), 2.43 (s, 3H). 13C NMR (150 MHz, (CD3)2CO): δ 164.02, 161.18 (d, JCF = 246.0), 135.33, 135.21, 133.68 (d, JCF = 9.2 Hz), 132.14, 131.09, 125.48 (d, JCF = 3.5 Hz), 124.84, 123.98 (d, 2JCF = 13.2 Hz), 121.17, 118.01, 116.90 (d, 2JCF = 23.6 Hz), 112.55, 109.18, 41.37, 24.92, 11.58. FTIR (thin film): 3278, 2929, 1648, 1614, 1531, 1026 cm−1. HRMS (EI) calcd for C18H16N2OFCl (M+) 330.09352, 330.09289 found.
2-fluoro-4-methoxy-N-(2-(2-methyl-1H-indol-3-yl)ethyl)benzamide (4)
4 (0.790 g, 84.4%) was synthesized as a pink oil from 2-(2-methyl-1H-indol-3-yl)ethanamine (0.500 g, 2.87 mmol) using the general procedure described above. 1H NMR (300 MHz, (CD3)2CO): δ 9.85 (br s, 1H), 7.91 (t, 3JHH = 9 Hz, 1H), 7.56 (d d, 3JHH = 8.7, 1.8 Hz, 1H), 7.35 (br s, 1H), 7.26 (d d, 3JHH = 8.1, 1.8 Hz, 1H), 6.98 (quin d, 3JHH = 7.5, 1.5 Hz, 2H), 6.85 (d d, 3JHH = 8.7, 2.4 Hz, 1H), 6.75 (d d, 3JHH = 13.8, 2.4 Hz, 1H), 3.87 (s, 3H), 3.62 (q d, 3JHH = 7.2, 0.9 Hz, 2H), 3.02 (t, 3JHH = 7.2 Hz, 2H), 2.40 (s, 3H). 13C NMR (150 MHz, (CD3)2CO): δ 163.27 (d, JCF = 12.2 Hz), 162.59 (d, JCF = 3.5 Hz), 161.39 (d, 1JCF = 246.0 Hz), 135.86, 132.46 (d, JCF = 4.5 Hz), 132.11, 128.83, 120.28, 118.42, 117.59, 114.70 (d, 2JCF = 13.2 Hz), 110.57, 110.26, 108.09, 101.24 (d, 2JCF = 27.6 Hz), 55.40, 40.38, 24.16, 10.54. FTIR (thin film): 3402, 3294, 2920, 1620, 1499, 1270, 952, 743 cm−1. HRMS (EI) calcd for C19H19N2O2F (M+) 326.14306, 326.14214 found.
4-chloro-2-fluoro-N-(2-(2-methyl-1H-indol-3-yl)ethyl)benzamide (6)
6 (0.103 g, 7.2%) was synthesized as a white solid from 2-(2-methyl-1H-indol-3-yl)ethanamine (0.758 g, 4.35 mmol) using the general procedure described above. 1H NMR (300 MHz, (CD3)2CO): δ 9.85 (br s, 1H), 7.88 (t, 3JHH = 7.8 Hz, 1H), 7.60 (br s, 1H), 7.55 (d, 3JHH = 7.8 Hz, 1H), 7.33 (m, 2H), 7.26 (d, 3JHH = 7.8 Hz), 6.98 (quin d, 3JHH = 7.2, 1.5 Hz, 2H), 3.63 (q, 3JHH = 7.2 Hz, 2H), 3.03 (t, 3JHH = 7.2 Hz, 2H), 2.40 (s, 3H). 13C NMR (150 MHz, (CD3)2CO): δ 162.11, 159.97 (d, 1JCF = 246.0 Hz), 136.90 (d, JCF = 11 Hz), 135.85, 132.38 (d, JCF = 4.0Hz), 122.10 (d, 2JCF = 13.8 Hz) 132.16, 128.82, 124.90 (d, JCF = 3.5 Hz), 120.30, 118.44, 117.54, 116.52 (d, 2JCF = 27.6 Hz), 110.28, 107.93, 40.53, 24.03, 10.54. FTIR (thin film): 3397, 3312, 2928, 1649, 1530, 1076, 901, 742 cm−1. HRMS (EI) calcd for C18H16N2OFCl (M+) 330.09352, 330.09261 found.
4-chloro-N-(2-(2-methyl-1H-indol-3-yl)ethyl)benzamide (7)
7 (0.049 g, 54.4%) was synthesized as a white solid from 2-(2-methyl-1H-indol-3-yl)ethanamine (0.050 g, 0.29 mmol) using the general procedure described above. 1H NMR (300 MHz, (CD3)2CO): δ 9.88 (br s, 1H), 7.99 (br s, 1H), 7.90 (m, 2H), 7.50 (m, 3H), 7.27 (d, 3JHH = 7.2 Hz, 1H), 6.98 (quin d, 3JHH = 7.2, 1.2 Hz, 2H), 3.60 (q, 3JHH = 6.6 Hz, 2H), 3.01 (t, 3JHH = 6.6 Hz, 2H), 2.35 (s, 3H).
3-chloro-2-fluoro-N-(2-(2-methyl-1H-indol-3-yl)ethyl)benzamide (8)
8 (0.131 g, 39.6%) was synthesized as a white solid from 2-(2-methyl-1H-indol-3-yl)ethanamine (0.175 g, 1.00 mmol) using the general procedure described above. 1H NMR (300 MHz, (CD3)2CO): δ 9.85 (br s, 1H), 7.65 (m, 3H), 7.55 (d, 3JHH = 7.8 Hz, 1H), 7.28 (m, 2H), 6.98 (quin d, 3JHH = 7.8 Hz, 2H), 3.63 (q, 3JHH = 7.2 Hz, 2H), 3.04 (t, 3JHH = 7.2 Hz, 2H), 2.40 (s, 3H). 13C NMR (150 MHz, (CD3)2CO): δ 162.33, 155.26 (d, 1JCF = 246.0 Hz), 135.85, 132.57, 132,16, 129.42 (d, JCF = 2.3 Hz), 128.83, 125.46 (d, 2JCF = 14.3 Hz), 125.13 (d, JCF = 4.5 Hz), 120.45 (d, 2JCF = 15.8 Hz), 120.29, 118.43, 117.53, 110.27, 107.92, 40.56, 24.05, 10.55. FTIR (thin film): 3398, 3292, 1649, 1529, 1451, 1300, 1240, 745 cm−1. HRMS (EI) calcd for C 18H16N2OFCl (M+) 330.09352, 330.09398 found.
2-fluoro-N-(2-(6-methoxy-2-methyl-1H-indol-3-yl)ethyl)benzamide (9)
9 (0.103 g, 42.9%) was synthesized as a clear oil from 2-(5-methoxy-2-methyl-1H-indol-3-yl)ethanamine (0.150 g, 0.73 mmol) using the general procedure described above. 1H NMR (300 MHz, (CD3)2CO): δ 9.70 (br s, 1H), 7.90 (t d, 3JHH = 7.5, 1.8 Hz, 1H), 7.53 (m, 3H), 7.28 (t d, 3JHH = 7.5, 0.9 Hz, 1H), 7.19 (d d d, 3JHH = 11.7, 8.1, 0.9 Hz, 1H), 7.15 (d, 3JHH = 8.4 Hz, 1H), 7.09 (d, 3JHH = 2.4 Hz, 1H), 6.66 (d, 3JHH = 8.4, 2.4 Hz, 1H), 3.76 (s, 3H), 3.63 (q, 3JHH = 6.3 Hz, 2H), 3.00 (t, 3JHH = 6.3 Hz, 2H), 2.376 (s, 3H). 13C NMR (150 MHz, (CD3)2CO): δ 162.95 (d, JCF = 2.4 Hz), 160.19 (d, 1JCF = 246.0 Hz), 153.84, 132.90, 132.67 (d, JCF = 9.2 Hz), 131.16, 130.90, 129.29, 124.49, 122.99 (d, 2JCF = 13.4 Hz), 115.90 (d, 2JCF = 23.6 Hz), 110.81, 109.99, 107.97, 100.07, 54.96, 40.37, 24.13, 10.64. FTIR (thin film): 3299, 2932, 1643, 1482, 1215, 1028, 756 cm−1. HRMS (EI) calcd for C19H19N2O2F (M+) 326.14306, 326.14365 found.
2-chloro-N-(2-(2-methyl-1H-indol-3-yl)ethyl)benzamide (10)
10 (0.050 g, 55.6%) was synthesized as a clear oil from 2-(2-methyl-1H-indol-3-yl)ethanamine (0.050 g, 0.29 mmol) using the general procedure described above. 1H NMR (300 MHz, (CD3)2CO): δ 9.85 (br s, 1H), 7.55 (m, 2H), 7.37 (m, 4H), 6.99 (quin, 3JHH = 6.6 Hz, 2H), 3.61 (q, 3JHH = 7.2 Hz, 2H), 3.04 (t, 3JHH = 7.2 Hz, 2H), 2.42 (s, 3H). 13C NMR (150 MHz, (CD3)2CO): δ 166.16, 137.26, 135.83, 132.10, 130.52, 129.73, 129.16, 128.90, 126.82, 120.25, 118.42, 117.59, 110.26, 107.99, 40.29, 24.23, 10.66. FTIR (thin film): 3385, 3056, 2932, 1643, 1383, 1026, 743 cm−1. HRMS (EI) calcd for C18H17N2OCl (M+) 312.10295, 312.10365 found.
3-chloro-N-(2-(2-methyl-1H-indol-3-yl)ethyl)benzamide (11)
11 (0.042 g, 46.7%) was synthesized as a white solid from 2-(2-methyl-1H-indol-3-yl)ethanamine (0.050 g, 0.29 mmol) using the general procedure described above. 1H NMR (300 MHz, (CD3)2CO): δ 9.84 (br s, 1H), 7.96 (br s, 1H), 7.88 (t, 3JHH = 1.5 Hz, 1H), 7.82 (d t, 3JHH = 7.2, 1.5 Hz, 1H), 7.49 (m, 3H), 7.26 (d, 3JHH = 7.2 Hz), 6.98 (quin d, 3JHH = 7.5, 0.9 Hz, 2H), 3.60 (q, 3JHH = 7.2 Hz, 2H), 3.01 (t, 3JHH = 7.2 Hz, 2H), 2.36 (s, 3H). 13C NMR (150 MHz, (CD3)2CO): δ 164.98, 137.32, 135.82, 133.83, 132.09, 130.74, 130.01, 128.90, 127.16, 125.55, 120.26, 118.43, 117.52, 110.28, 108.15, 40.60, 24.08, 10.57. FTIR (thin film): 3403, 3306, 1641, 1544, 1436, 1300, 742 cm−1. HRMS (EI) calcd for C18H17N2OCl (M+) 312.10295, 312.10318 found.
General Procedure for 2 and 5
Indole amides prepared according to the general procedure described above, were stirred in CH2Cl2 (~1 mL/ 15 mg compound) under N2 at −78 °C. Boron tribromide (1M in CH2Cl2, 5 mole eq/methoxy group) was added by cannula to the stirred solutions. The reaction was allowed to warm to room temperature following addition and stirred overnight. After 18 hours, excess boron tribromide was quenched by addition of water followed by 2M sodium hydroxide until no visible reaction was evident and pH strips indicated strong basicity. The aqueous phase was acidified with 6M HCl and the phases were separated. The aqueous layer was extracted twice with 10 mL dichloromethane and the organic phases were combined. Solvent was removed by rotary evaporation, yielding an oil as crude product. Purification on a silica gel column using 6:1 diethyl ether:hexanes as eluent followed by removal of solvent under reduced pressure yielded deprotected compounds in moderate to low yields.
2-fluoro-4-hydroxy-N-(2-(2-methyl-1H-indol-3-yl)ethyl)benzamide (2)
2 (0.036 g, 9.4 %) was synthesized as a white solid from 2-fluoro-4-methoxy-N-(2-(2-methyl-1H-indol-3-yl)ethyl)benzamide (0.400 g, 1.23 mmol) using the general procedure described above. 1H NMR (300 MHz, (CD3)2 CO): δ 9.85 (br s, 1H), 9.36 (br s, 1H), 7.86 (t, 3JHH = 8.7 Hz, 1H), 7.56 (d, 3JHH = 7.2 Hz, 1H), 7.31 (br s, 1H), 7.26 (d d, 3JHH = 6.9, 1.5 Hz, 1H), 6.98 (quin d, 3JHH = 7.5, 1.5 Hz, 2H), 6.77 (d d, 3JHH = 8.7, 2.4 Hz, 1H), 6.61 (d d, 3JHH = 13.5, 2.4 Hz, 1H), 3.61 (q, 3JHH = 7.2 Hz, 2H), 3.01 (t, 3JHH = 7.2 Hz, 2H), 2.39 (s, 3H). 13C NMR (150 MHz, (CD3)2CO): δ 162.78, 161.51 (d, 1JCF = 246.0 Hz), 161.42 (d, 2JCF = 12.8 Hz), 135.86, 132.70 (d, JCF = 4.7 Hz), 132.10, 128.82, 120.28, 118.41, 117.59, 113.72 (d, 2JCF = 13.2 Hz), 111.84 (d, JCF = 2.3 Hz), 110.25, 108.10, 102.56 (d, 2JCF = 26.4 Hz), 40.35, 24.18, 10.53. FTIR (thin film): 3401, 2926, 1620, 1502, 1287, 741 cm−1. HRMS (EI) calcd for C18H17N2O2F (M+) 312.12741, 312.12653 found.
2-fluoro-N-(2-(6-hydroxy-2-methyl-1H-indol-3-yl)ethyl)benzamide (5)
5 (0.043 g, 30.9 %) was synthesized as a white solid from 2-fluoro-N-(2-(6-methoxy-2-methyl-1H-indol-3-yl)ethyl)benzamide (0.145 g, 0.44 mmol) using the general procedure described above. 1H NMR (300 MHz, (CD3)2CO): δ 9.57 (br s, 1H), 7.90 (t d, 3JHH = 7.5, 1.2 Hz, 1H), 7.55 (m, 2H), 7.29 (t d, 3JHH = 7.2, 1.2 Hz, 1H), 7.20 (d d d, 3JHH = 11.4, 8.4, 1.2 Hz, 1H), 7.08, (d, JHH = 8.4 Hz, 1H), 6.97 (d, 3JHH = 2.4 Hz, 1H), 6.60 (d d, 3JHH = 8.4, 2.4 Hz, 1H), 3.60 (q, JHH = 6.9 Hz, 2H), 2.95 (t, 3JHH = 6.9 Hz, 2H), 2.35 (s, 3H). 13C NMR (150 MHz, (CD3)2CO): δ 162.95 (d, JCF = 2.3Hz), 160.18 (d, 1JCF = 246.0 Hz), 150.65, 132.78, 132.63 (d, JCF = 8.6 Hz), 131.12, 130.52, 129.59, 124.47, 123.07 (d, 2JCF = 13.2 Hz), 115.90 (d, 2 JCF = 23.6 Hz), 110.56, 110.06, 107.33, 102.20, 40.29, 24.25, 10.62. FTIR (thin film): 3383, 3048, 2931, 1642, 1372, 1207, 1028, 756 cm−1. HRMS (EI) calcd for C18H17FN2O2 (M+) 312.12741, found 312.12790.
Protein Preparation
Bos taurus Arp2/3 complex was purified from calf thymus as previously described.[21] Rabbit skeletal muscle actin was purified from acetone powder purchased from Pel-freeze using established methods.[22] Purified actin was labeled on Cys375 with pyrene-iodoacetamide following the procedure of Pollard.[23] Human N-WASp-VCA (residues 428-505) was overexpressed in E. coli BL21(DE3) cells and purified as described by Nolen, et al.[9]
Pyrene actin polymerization assays
A solution of 2 μL 0.5 mM MgCl2 and 2 mM EGTA was added to 20 μL of 15 μM 30% pyrene-labelled actin in 2 mM Tris pH 8.0, 0.1 M ATP, 0.5 mM DTT, and 0.1 mM CaCl2. Polymerization was initiated by adding 78 μL of a solution containing Arp2/3 complex and N-WASp-VCA to bring the final buffer concentrations to 10 mM imidazole pH 7.0, 50 mM KCl, 1 mM EGTA, 1mM MgCl2 and 1 mM DTT. The fluorescence was measured every 10 seconds for 45 min using an excitation wavelength of 365 nm and an emission wavelength of 407 nm on a Tecan Safire2 plate reader. We tested each of the inhibitor analogues using a maximum concentration of 200 μM since all compounds were soluble at this concentration. The maximum polymerization rate was determined by measuring the slope of each polymerization curve at each time point and converted from RFU s−1 to nM actin s−1 assuming the total polymer concentration at equilibrium is the total actin concentration minus 0.1 μM, the critical concentration. Plots of maximum polymerization rate versus inhibitor concentration were fit using the equation below:
X-ray Crystallography
Crystals of Bos taurus Arp2/3 complex were grown by hanging drop vapor diffusion as previously described.[21] Crystals we transferred to soaking solution containing 18% PEG 8000, 50 mM Hepes pH 7.5, 100 mM potassium thiocyanate, 20 % glycerol and either 0.5 mM CK-666, 0.5 mM CK-869, or 0.5 mM CK-869 plus 2 mM ATP and 2 mM CaCl2 and soaked at 4 °C for 16 hrs. Data were collected at beamline 5.0.1 at the Advanced Light Source in Berkeley, CA. Phases were solved by molecular placement using the apo-Arp2/3 complex as a starting model (1K8K.pdb) and the structures were refined using crystallography and NMR system [24] using inhibitor parameter files generated using the prodrg server.[25] Coordinates have been deposited in the protein data bank with accession code 3UKR.
Docking Calculations
The crystal structure of Arp2/3 complexed with 1 (PDB ID: 3UKR) was employed in the docking calculations performed with Glide 5.7 (Schrödinger, LLC). The crystal structure with the substrate 12 (PDB ID: 3DXK) was also used for the validation of the docking algorithm. Both crystal structures were initially prepared within the “Protein Preparation Wizard” module of the Maestro software suite (Schrödinger, LLC). Initially, bond orders were assigned, hydrogen atoms were added, and water molecules were deleted beyond 5Å of the ligand. Missing side chains were filled in with the “Prime” module of Maestro. The hydrogen-bonding network was then optimized by reorienting atoms within hydroxyl and thiol groups, water molecules, amide groups of asparagine and glutamine, and the imidazole ring in histidine. Protonation states of histidine, aspartic acid and glutamic acid and tautomeric states of histidine were predicted. These optimizations are necessary because the orientation of atoms within these functional groups cannot be determined from the X-ray structure. Finally, the complex was submitted to a series of restrained, partial minimizations using the OPLS-AA force field.[26] The ligand structures 1-11 were drawn in ChemDraw (Cambridgesoft) and energy minimized in vacuum with the Fletcher-Powell algorithm as implemented in BOSS[27] with an energy tolerance of 0.0001 kcal/mol. All structures were then docked and scored using the Glide standard-precision (SP) mode[28] and subsequently they were re-docked and re-scored using the Glide extra-precision (XP) mode.[29] For the docking calculations, the protein structure remained fixed, while the ligand was fully flexible. In order to accommodate different-sized ligands and decrease the penalties for close ligand-protein contacts since the protein is not flexible, the van der Waals radii for non-polar ligand atoms were scaled by a factor of 0.8. Receptor atoms were not scaled. For the grid generation and ligand docking procedures, the default Glide settings were used.
Free energy perturbation calculations
FEP calculations were carried out in the context of Monte Carlo (MC) statistical mechanics simulations to predict relative free energies of binding. During a FEP calculation, a substituent on the lead compound A (e.g. fluorine) is transformed into a different substituent (e.g. chlorine) to form compound B inside the binding pocket of the protein as well as in the water phase. The difference in free energy changes for the ligands A and B in protein and in water gives the relative free energy of binding: ΔΔGb = ΔGB – ΔGA. If ΔΔGb < 0, the binding of compound B is favored with respect to A. For the FEP calculations initial structures of the analogues were generated with the ligand growing program BOMB[30] in the binding site of 1 in the Arp2/3 complex (PDB ID: 3UKR). In order to reduce computational time, a reduced model of the protein was utilized that consisted of the 215 amino acid residues closest to the binding site, which corresponds to a 23 Å radius around the binding site. A few remote side chains were neutralized so that there was no net charge for the protein. The protein-ligand and ligand-only systems were solvated in 25-Å caps and the MC/FEP calculations were executed with MCPRO.[27] The energetics for the systems were described classically with the OPLS-AA force field for the protein, OPLS/CM1A for the ligands, and TIP4P for water molecules.[26,31] After system construction, the complex was subjected to conjugate gradient optimization with MCPRO. For the MC simulations, all degrees of freedom were sampled for the ligand, while the TIP4P water molecules only translated and rotated. Bond angles and dihedral angles for protein side chains in a radius of 10 Å around the binding site were also sampled, while the backbone was kept fixed. Beyond the 10-Å radius, both the protein side chains and backbone remained fixed. All MC simulations were run at 298 K.
The FEP calculations were performed employing a simple overlap (OS) sampling scheme[32] with 11 windows as has been described previously.[33] A window refers to a MC simulation at one point along the mutation coordinate λ, which interconverts two ligands as λ goes from 0 to 1. The spacing between windows was 0.1. Each window for the unbound ligand in water consisted of 30 × 106 (30 M) configurations of MC equilibration for the first window followed by 40 M configurations of averaging. For the bound calculations, the equilibration period was 20 M configurations followed by 30 M configurations of averaging. The reported uncertainties for the free energy changes were obtained from the fluctuation in separate averages over batches of 2 M configurations. Eq 1 is used, where m is the number of batches, θi is the average of property θ for the i-th batch, and <θ> is the overall average for θ.
| Eq. 1 |
To monitor the convergence of the results, we carried out several forward and reverse mutations to ensure that hysteresis is kept minimal (i.e., for compounds 6, 7, 8, 10, 11, see Supplementary Table 3).
For compounds 7, 8, 10, 11, which bear ortho and meta substitutions, we carried out FEP calculations for both equivalent positions on the rings. In simulations within a protein environment, the finite accessible simulation time does not allow sampling of ring flips. Therefore, equivalent ortho and meta substitutions were sampled in separate simulations. For example, in Supplementary Figure 6 is shown compound 11 in the two possible orientations. We found no significant differences for mutations involving these equivalent positions with respect to the difference in free energy of binding as calculated from the FEP simulations.
Multistage FEP
For moderately large perturbations, the configurational ensembles representative of molecule A and molecule B are relatively disparate and an FEP calculation runs the risk of not converging. It is therefore recommended to break the transformation pathway into intermediate states, which ensure that the ensembles of molecule A and molecule B overlap. In this study, for mutations that introduce large changes in the overall molecular conformation, structure and characteristics of the ligand, intermediate steps were introduced, using the Multistage FEP (MFEP) method. MFEP is based on the idea of splitting a large perturbation into a series of smaller ones which are suitable for the single-stage FEP calculations. To split the perturbation, one or more intermediate molecules are constructed in addition to compound A and compound B. Since free energy is a state variable, the overall ΔG is given by the sum of the changes for each step. For the mutation of the methoxy group to hydrogen (analogues 4 and 9), we chose to initially perturb the methoxy to hydroxy, and then the hydroxy group to hydrogen For example, for compound 4, first a transformation of 4 to 2 was performed and then a transformation from 2 to 1. The values reported in Table 1 for these analogues, is the sum of these transformations. For analog 7, the ΔΔG results from a concurrent transformation of the fluorine substituent at R2 into hydrogen and of the fluorine substituent at R2 into hydrogen.
Supplementary Material
Acknowledgements
We thank the Oregon Medical Research Foundation (Grant 105DG2610189 to B.J.N.) and the National Institutes of Health (Grant 5R01GM092917-03 to B.J.N.) G.P. acknowledges a postdoctoral fellowship awarded by the Greek Ministry of Education, National Strategic Reference Framework (NSRF) 2007 – 2013 and co-funded by the European Regional Development Fund and national resources. All calculations were performed at BRFAA using an Intel Xeon cluster funded from a European Economic Area Grant. Funding for the University of Oregon Chemistry Research and Instrumentation Services has been furnished in part by the NSF (CHE-0923589).
Footnotes
Associated Content
Six figures and accompanying figure legends plus three tables.
References
- [1].Pollard TD, Cooper JA. Science. 2009;326(5957):1208–1212. doi: 10.1126/science.1175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Cooper JA, Buhle EL, Jr, Walker SB, Tsong TY, Pollard TD. Biochemistry. 1983;22(9):2193–2202. doi: 10.1021/bi00278a021. [DOI] [PubMed] [Google Scholar]
- [3].Firat-Karalar EN, Welch MD. Curr Opin Cell Biol. 2011;23(1):4–13. doi: 10.1016/j.ceb.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Goley ED, Welch MD. Nat Rev Mol Cell Biol. 2006;7(10):713–726. doi: 10.1038/nrm2026. [DOI] [PubMed] [Google Scholar]
- [5].a) Sawa M, Suetsugu S, Sugimoto A, Miki H, Yamamoto M, Takenawa T. J Cell Sci. 2003;116(Pt 8):1505–1518. doi: 10.1242/jcs.00362. [DOI] [PubMed] [Google Scholar]; b) Hudson AM, Cooley L. J Cell Biol. 2002;156(4):677–687. doi: 10.1083/jcb.200109065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Gouin E, Welch MD, Cossart P. Curr Opin Microbiol. 2005;8(1):35–45. doi: 10.1016/j.mib.2004.12.013. [DOI] [PubMed] [Google Scholar]
- [7].a) Yamazaki D, Kurisu S, Takenawa T. Cancer Sci. 2005;96(7):379–386. doi: 10.1111/j.1349-7006.2005.00062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Nurnberg A, Kitzing T, Grosse R. Nat Rev Cancer. 2011;11(3):177–187. doi: 10.1038/nrc3003. [DOI] [PubMed] [Google Scholar]; c) Zheng HC, Zheng YS, Li XH, Takahashi H, Hara T, Masuda S, Yang XH, Guan YF, Takano Y. Anticancer Res. 2008;28(4B):2225–2232. [PubMed] [Google Scholar]
- [8].To C, Shilton BH, Di Guglielmo GM. J Biol Chem. 2010;285(36):27944–27957. doi: 10.1074/jbc.M110.103036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Nolen BJ, Tomasevic N, Russell A, Pierce DW, Jia Z, McCormick CD, Hartman J, Sakowicz R, Pollard TD. Nature. 2009;460(7258):1031–1034. doi: 10.1038/nature08231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Yamaguchi H, Lorenz M, Kempiak S, Sarmiento C, Coniglio S, Symons M, Segall J, Eddy R, Miki H, Takenawa T, Condeelis J. J Cell Biol. 2005;168(3):441–452. doi: 10.1083/jcb.200407076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Pommier Y, Marchand C. Nat Rev Drug Discov. 2011;11(1):25–36. doi: 10.1038/nrd3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].a) Sun SC, Wang ZB, Xu YN, Lee SE, Cui XS, Kim NH. PLoS One. 2011;6(4):e18392. doi: 10.1371/journal.pone.0018392. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Yi K, Unruh JR, Deng M, Slaughter BD, Rubinstein B, Li R. Nat Cell Biol. 2011;13(10):1252–1258. doi: 10.1038/ncb2320. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Ilatovskaya DV, Pavlov TS, Levchenko V, Negulyaev YA, Staruschenko A. Faseb J. 2011;25(8):2688–2699. doi: 10.1096/fj.10-167262. [DOI] [PubMed] [Google Scholar]; d) Rizvi SA, Neidt EM, Cui J, Feiger Z, Skau CT, Gardel ML, Kozmin SA, Kovar DR. Chem Biol. 2009;16(11):1158–1168. doi: 10.1016/j.chembiol.2009.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Ohkawa T, Volkman LE, Welch MD. J Cell Biol. 2010;190(2):187–195. doi: 10.1083/jcb.201001162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Rouiller I, Xu XP, Amann KJ, Egile C, Nickell S, Nicastro D, Li R, Pollard TD, Volkmann N, Hanein D. J Cell Biol. 2008;180(5):887–895. doi: 10.1083/jcb.200709092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].a) Cournia Z, Leng L, Gandavadi S, Du X, Bucala R, Jorgensen WL. J Med Chem. 2009;52(2):416–424. doi: 10.1021/jm801100v. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Hare AA, Leng L, Gandavadi S, Du X, Cournia Z, Bucala R, Jorgensen WL. Bioorg Med Chem Lett. 2010;20(19):5811–5814. doi: 10.1016/j.bmcl.2010.07.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ioakimidis L, Thoukydidis L, Mirza A, Naeem S, Reynisson J. QSAR & Combinatorial Science. 2008;27(4):445–456. [Google Scholar]
- [16].a) Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Adv Drug Deliv Rev. 2001;46(1-3):3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]; b) Jorgensen WL. Acc Chem Res. 2009;42(6):724–733. doi: 10.1021/ar800236t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Cooper JA, Walker SB, Pollard TD. J Muscle Res Cell Motil. 1983;4(2):253–262. doi: 10.1007/BF00712034. [DOI] [PubMed] [Google Scholar]
- [18].Zhou P, Lv J, Zou J, Tian F, Shang Z. J Struct Biol. 2010;169(2):172–182. doi: 10.1016/j.jsb.2009.10.006. [DOI] [PubMed] [Google Scholar]
- [19].Harada H, Hirokawa Y, Suzuki K, Hiyama Y, Oue M, Kawashima H, Yoshida N, Furutani Y, Kato S. Bioorg Med Chem Lett. 2003;13(7):1301–1305. doi: 10.1016/s0960-894x(03)00073-8. [DOI] [PubMed] [Google Scholar]
- [20].Benton FL, Dillon TE. J. Am. Chem. Soc. 1942;64:1128–1129. [Google Scholar]
- [21].Nolen BJ, Littlefield RS, Pollard TD. Proc Natl Acad Sci U S A. 2004;101(44):15627–15632. doi: 10.1073/pnas.0407149101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].MacLean-Fletcher S, Pollard TD. Biochem Biophys Res Commun. 1980;96(1):18–27. doi: 10.1016/0006-291x(80)91175-4. [DOI] [PubMed] [Google Scholar]
- [23].Pollard TD. J Cell Biol. 1984;99(3):769–777. doi: 10.1083/jcb.99.3.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- [25].Schuettelkopf AW, van DMF. Aalten, Acta Crystallogr D Biol Crystallogr. 2004:D60. doi: 10.1107/S0907444904011679. in press. [DOI] [PubMed] [Google Scholar]
- [26].Jorgensen WL, Maxwell DS, TiradoRives J. J Am Chem Soc. 1996;118(45):11225–11236. [Google Scholar]
- [27].Jorgensen WL, Tirado-Rives J. J Comput Chem. 2005;26(16):1689–1700. doi: 10.1002/jcc.20297. [DOI] [PubMed] [Google Scholar]
- [28].Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE, Francis P, Shenkin PS. J Med Chem. 2004;47(7):1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- [29].Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, Sanschagrin PC, Mainz DT. J Med Chem. 2006;49(21):6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- [30].Jorgensen WL. Science. 2004;303(5665):1813–1818. doi: 10.1126/science.1096361. [DOI] [PubMed] [Google Scholar]
- [31].a) Jorgensen WL, Tirado-Rives J. Proc Natl Acad Sci U S A. 2005;102(19):6665–6670. doi: 10.1073/pnas.0408037102. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klien ML. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- [32].Jorgensen WL, Thomas LL. J Chem Theory Comput. 2008;4(6):869–876. doi: 10.1021/ct800011m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Zeevaart JG, Wang L, Thakur VV, Leung CS, Tirado-Rives J, Bailey CM, Domaoal RA, Anderson KS, Jorgensen WL. J Am Chem Soc. 2008;130(29):9492–9499. doi: 10.1021/ja8019214. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.