Abstract
Asp299Gly (D299G) and, to a lesser extent, Thr399Ile (T399I) TLR4 polymorphisms have been associated with Gram negative sepsis and other infectious diseases, but the mechanisms by which they affect TLR4 signaling are unclear. In this study, we determined the impact of the D299G and T399I polymorphisms on TLR4 expression, interactions with myeloid differentiation factor 2 (MD2), LPS binding, and LPS-mediated activation of the MyD88- and TIR domain-containing adapter inducing IFN-β (TRIF) signaling pathways. Complementation of human embryonic kidney 293/CD14/MD2 transfectants with wild-type (WT) or mutant yellow fluorescent protein (YFP)-tagged TLR4 variants revealed comparable total TLR4 expression, TLR4-MD-2 interactions, and LPS binding. FACS analyses with anti-TLR4 Ab showed only minimal changes in the cell surface levels of the D299G TLR4. Cells transfected with D299G TLR4 exhibited impaired LPS-induced phosphorylation of p38 and TANK binding kinase-1, activation of NF-κB and IFN regulatory factor 3, and induction of IL-8 and IFN-β mRNA, while T399I TLR4 did not cause statistically significant inhibition. In contrast to WT TLR4, expression of the D299G mutants in TLR4−/− mouse macrophages failed to elicit LPS-mediated induction of TNF-α and IFN-β mRNA. Co-immunoprecipitation revealed diminished LPS-driven interaction of MyD88 and TRIF with the D299G TLR4 species, in contrast to robust adapter recruitment exhibited by WT TLR4. Thus, the D299G polymorphism compromises recruitment of MyD88 and TRIF to TLR4 without affecting TLR4 expression, TLR4-MD-2 interaction, or LPS binding, suggesting that it interferes with TLR4 dimerization and assembly of intracellular docking platforms for adapter recruitment.
INTRODUCTION
Host immune cells detect microbial pathogens, endogenous “danger” molecules, and environmental cues by a set of germ line-encoded pattern recognition receptors (PRRs)3 to initiate immediate and early innate immune responses and prime adaptive immunity (1). PRRs include membrane-associated TLRs expressed on the plasma membrane, Golgi, endoplasmic reticulum and endosomes, cytosolic nucleotide binding domain and leucine rich repeat (LRR)-containing receptors, retinoic acid-inducible gene-I-like helicases, and sensors of cytosolic DNA (2–5). TLRs are type I transmembrane glycoproteins expressed on the cell surface (TLR1, TLR2, TLR4-TLR6, TLR10, TLR11) or in endosomal compartments (TLR3, TLR7-TLR9) that share a common structural organization. All TLRs express an N-terminal ectodomain that contains LRRs involved in ligand recognition and co-receptor interaction, a transmembrane region, and an intracellular region containing a Toll-IL-1R resistance [TIR] signaling domain (2). TLR2 cooperates with TLR1 or TLR6 to recognize tri- or di-acylated lipoproteins from Gram positive bacteria, mycobacteria, or mycoplasma (6). TLR4 is the main receptor for Gram negative bacterial LPS (7), but also senses mannan, the fusion (F) protein of respiratory syncytial virus (RSV), and chlamydial heat shock proteins (Hsp) (8–10), and others. TLR5 detects bacterial flagellin (11, 12), while mouse TLR11 is important for protection against uropathogenic Escherichia coli and recognizes a profilin-like molecule from Toxoplasma gondii (13, 14). Endosomal TLRs sense viral dsRNA (TLR3) (15), ssRNA (TLR7 and TLR8) (16, 17), and hypomethylated CpG motifs present in bacterial, viral, and fungal DNA (TLR9) (2, 18). Sensing of conserved pathogen-associated molecular patterns (e.g., LPS) via the LRR-containing ectodomains leads to TLR dimerization that brings their TIR signaling domains into close proximity, forming intracellular docking platforms that enable recruitment of adapter proteins and kinases (2, 19). All TLRs except TLR3 use the signaling adapter MyD88 that associates with TLRs via TIR-TIR domain interactions and recruits IL-1R-associated kinases (IRAK) 4 and IRAK1 that signal downstream to activate NF-κB, MAP kinases, and inflammatory cytokines (20). TLR3 solely engages the TIR domain-containing adapter inducing IFN-β (TRIF) to activate inflammatory cytokines and type I IFN, while TLR4 also uses TRIF to signal expression of co-stimulatory molecules and type I IFNs via activation of TANK-binding kinase (TBK) 1 and IFN regulatory factors (IRF) 3, and IRF7 (2, 20). TLR2 and TLR4 require the “bridging” adapter TIR domain-containing adapter protein (TIRAP) (21), also called MyD88 adapter-like (Mal) (22), to recruit MyD88 to TLRs, whereas TLR4 also requires a “bridging” adapter TRIF-related adapter molecule (TRAM) that recruits TRIF to the TLR4 receptor complex (23, 24).
Two single nucleotide polymorphisms (SNPs) that encode mutations in the ectodomain of TLR4, D299G and T399I, occur in humans with a frequency of approximately 6–10% and have been associated with blunted responsiveness to inhaled LPS, decreased LPS-mediated production of cytokines by airway epithelial cells and macrophages, Gram-negative bacteremia, and sepsis (25, 26). The D299G polymorphism has also been linked to Boutonneuse fever (27), RSV infections in infants and young children (28, 29), HIV-associated tuberculosis (30, 31), Crohn’s disease (32), ulcerative colitis (33), endometriosis (34), and tonsillitis (35). Human subjects carrying the T399I polymorphism either exhibit a milder LPS-hyporesponsive phenotype or do not manifest it at all (36). In many instances, associations between these TLR4 SNPs and predisposition to disease have been shown to depend on ethnic backgrounds, gender, and/or prior immune deficiencies (reviewed in (36)). While the D299G and T399I TLR4 polymorphisms have been associated with an increased risk of infectious diseases, they have been found to protect against inflammatory diseases such as atherogenesis and rheumatoid arthritis (37).
The molecular mechanisms by which the TLR4 SNPs affect receptor functions are largely unknown. Compromised LPS responses elicited by the D299G variant were originally associated with decreased expression of the mutant TLR4 species in human airway epithelial cells (25). In contrast, others have reported that D299G or T399I TLR4 mediate diminished NF-κB activation and cytokine gene expression in response to LPS, RSV F protein, or chlamydial Hsp60 under conditions of equal total and cell surface expression of transfected TLR4 species in HEK293T cells (29, 38). In this study, we sought to determine the impact of the D299G and T399I polymorphisms on TLR4 expression, interactions with MD2, LPS binding to the TLR4/CD14/MD2 complex, recruitment of signaling adapter proteins, and activation of the MyD88 and TRIF pathways. Our data indicate that the D299G polymorphism compromises LPS-inducible recruitment of MyD88 and TRIF to TLR4 and activation of MyD88- and TRIF-dependent signaling pathways, while not significantly affecting total and cell surface TLR4 expression, its interaction with MD2, or LPS binding to the TLR4-MD2-CD14 receptor complex.
MATERIALS AND METHODS
Reagents and Cell Culture
pcDNA3-YFP-TLR4, pcDNA3-CD14, pEFBOS-Flag- or haemagglutinin (HA)-MD2, pcDNA3-AU1-MyD88, pEFBOS-Flag-TRIF, pELAM-luciferase (Luc), p125-Luc and pTK-Renilla-Luc were described (38–40). Plasmids encoding the D299G and T399I polymorphisms were engineered by site-directed mutagenesis of pcDNA3-YFP-TLR4, using QuickChange kit (Stratagene, Kirkland, WA) and the following primers: D299G, 5′-GCATACTTAGACTACTACCTCGATGGTATTATTGACTTATTTAATTGTTTGA-3′; T399I TLR4, 5′-TGCTGTTCTCAAAGTGATTTTGGGACAATCAGCCTAAAGTATTT-3′. The presence of the mutations was verified by sequencing with the Big Dye Terminator Cycle Sequencing Kit (Applied Biosystems, Carlsbad, CA) (41). Biotinylated LPS and S-[2,3-bis(palmitoyloxy)-(2-RS)-propyl]-N-palmitoyl-(R)-Cys-Ser-Lys4-OH (Pam3Cys) were purchased from Invivogen (San Diego, CA), streptavidin-allophycocyanin (SA-APC) and human TNF-α were from BioLegend (San Diego, CA). The following Abs were used: anti-green fluorescent protein (GFP) (Invitrogen, Carlsbad, CA), anti-Flag-HRP (Sigma-Aldrich, St. Louis, MI), anti-phospho (p) and anti-total p38 (Promega, Madison, WI), anti-p- and anti-total-TBK1 (Cell Signaling, Danvers, MA), anti-TLR4, anti-tubulin, anti-p- and anti-total IRF-3, anti-HA-HRP (Santa Cruz, Santa Cruz, CA), anti-HLA (BD Biosciences, San Jose, CA), and anti-AU1 (Axxora, LLC, San Diego, CA) Abs. Highly purified, lipoprotein-free E. coli K235 LPS was described previously (42). Human embryonic kidney (HEK) 293T and HEK293 cells (ATCC, Manassas, VA) were maintained in DMEM containing 10% FBS (Sigma-Aldrich), 2 mM L-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin (Invitrogen) (complete (c) DMEM). WT and TLR4−/− J2 retrovirus-immortalized (i) mouse bone marrow-derived macrophages (BMDMs) were kindly provided by Drs. Douglas Golenbock and Katherine Fitzgerald (University of Massachusetts Medical School, Worcester, MA) and maintained in cDMEM.
Transient and stable transfections
For reporter assays, HEK293T cells were plated in 24-well plates and transfected with plasmids pcDNA3-CD14, pEFBOS-Flag-MD2 (50 ng each), pcDNA3-YFP-TLR4 (150 ng), pTK-Renilla-Luc (10 ng), pELAM-Luc or p125-Luc (300 ng each), with the total amount of plasmid DNA adjusted to 1 μg with pcDNA3. For gene expression studies and Western blotting, cells were plated in 6-well plates, and transfected with pcDNA3-CD14 (0.5 μg), pEFBOS-MD2 (0.5 μg), pcDNA3-YFP-TLR4 (1.0 μg) or pcDNA3 (2.0 μg) (all amounts are per well). For immunoprecipitation, HEK293T cells were plated in 100 mm tissue culture dishes, and transfected with pcDNA3-CD14 (2.0 μg), pEFBOS-MD2 (2.0 μg), pcDNA3-YFP-TLR4 (6.0 μg) (all amounts are per dish). Transient transfection of HEK293T cells was performed, using SuperFect transfection reagent (Qiagen, Valencia, CA) per manufacturer’s recommendations. iBMDMs were transfected with plasmids encoding human YFP-TLR4 species, using Lipofectamine 2000 (Invitrogen), as recommended by the manufacturer. To obtain stable cell lines, TLR4-transfected HEK293 cells were selected in cDMEM containing 1 mg/ml G418 (Sigma-Alrdich).
Animal Procedures
Six- to 8-wk-old WT C57BL/6 mice were purchased from the Charles River Laboratories (Wilmington, MA), and TLR4−/− mice were bred and maintained within the accredited animal facility at the University of Maryland School of Medicine. All animal experiments were conducted with institutional approval. Bone marrow cells were isolated from femurs and tibiae of mice and cultured in complete RPMI in tissue culture dishes. To obtain primary BMDMs, bone marrow cells were differentiated for seven days in the presence of 25% (v/v) of L929 cell-conditioned, M-CSF-containing supernatant, as described (43). BMDMs were detached and used for nucleofection.
LPS Binding and FACS
We adapted a previously reported LPS binding protocol (44). In brief, HEK293T cells were treated for 30 min at 37° C with medium or 1 μg/ml biotinylated LPS, washed with HBSS, and incubated with or without 1 μg/ml SA-APC on ice for 30 min. To examine TLR4 cell surface expression, HEK293/CD14/MD2 transfectants expressing YFP-TLR4 variants were stained for 30 min on ice with HTA125-PE (Santa Cruz), anti-HLA-A, B, C-PE (positive control), or isotype control IgG-PE (BD Biosciences) Abs. After incubation, cells were washed with ice-cold PBS, fixed for 15 min in 2% paraformaldehyde, and analyzed on a LSRII Flow Cytometer (BD Biosciences) to measure YFP (total YFP-TLR4 expression), PE (cell surface TLR4 expression), or APC (LPS binding) fluorescence. The data were analyzed using the FlowJo software (Tree Star, Ashland, OR).
Preparation of cell extracts, immunoprecipitation and Western blot analyses
Cell extracts were prepared as described (45, 46) using a lysis buffer containing 20 mM HEPES (pH 7.4), 1 mM PMSF, 1 mM DTT, 1 mM sodium orthovanadate, 50 mM NaF, 2 mM EDTA, 150 mM NaCl, 1% Triton X-100, 20 mM β-glycerol phosphate and protease inhibitor cocktail (Roche Applied Science, Indianapolis, IN). For immunoprecipitation, cell lysates (0.5–1 mg total protein) were pre-cleared by incubation for 2 h at 4° C with 20 μl of 50% slurry of protein G-agarose (Roche Applied Science, Indianapolis, IN). Pre-cleared lysates were incubated for 20 h at 4° C with the respective Abs, followed by addition of protein G-agarose (45 μl of 50% slurry/sample) and incubation for 4 h at 4° C under rotation. After extensive washing with ice-cold lysis buffer, beads were re-suspended in Laemmli sample buffer (Bio-Rad), boiled for 10 min, and supernatants were analyzed by immunoblotting, as reported previously (40). Densitometric analysis of intensities of the corresponding bands was performed using the Quantity One software (Bio-Rad) as described previously (45).
RNA Isolation, Reverse Transcription, and Real-Time (RT) Quantitative (q) PCR
RNA was isolated with Trizol (Invitrogen), treated for 1 h at 37° C with DNase (Promega), re-purified, cDNA was prepared using reverse transcription system (Promega), and analyzed by RT-qPCR using 5 μl of cDNA, 0.3 μM gene-specific primers, and SYBR Green Supermix (Bio-Rad) on a MyIQ Real-Time PCR machine (Bio-Rad). The following primers were used: human HPRT, forward, 5′-GCTGACCTGCTGGATTACATT-3′; reverse, 5′-GTTGAGAGATCATCTCCACCA; human IL-8, forward, 5′-CACCGGAAGGAACCATCTC ACT-3′; reverse, 5′-TGCACCTTCACACAGAGCTGC-3′; human IFN-β, forward, 5′-ACTGCCTCAAGGACAGGATG-3, reverse, 5′-AGCCAGGAGGTTCTCAACAA-3′; mouse TNF-α, forward, 5′-CCCAGGCAGTCAGATCATCTTC-3′, reverse, 5′-GCTTGAGGGTTTGC TACAACA TG-3′; mouse HPRT, forward, 5′-GCTGACCTGCTGGATTACATT-3′, reverse, 5′-GTTGAGAGATCATCTCCACCA-3′. Melting curve analysis was performed to ensure specific amplification, and data were processed using 2−ΔΔCT method (47).
Nucleofection
pcDNA3-eGFP and pcDNA3-YFP-TLR4 vectors encoding WT or D299G variants were introduced into WT and TLR4−/− BMDMs by nucleofection, using the Nucleofector I device and the mouse macrophage nucleofection kit (Lonza, Basel, Switzerland), as recommended by the manufacturer. After recovery for 20 h, cells were treated with medium, LPS, TNF-α, RNA was isolated, reverse transcribed, and analyzed by RT-qPCR.
Reporter Assays
Stably (HEK293) or transiently transfected (HEK293T) cells expressing WT, D299G, or T399I YPF-TLR4, along with CD14 and Flag-MD2, were treated for 5 h at 37° C with medium, LPS, or TNF-α (100 ng/ml each) in a 5% CO2 atmosphere. After treatment, cells were lysed with passive lysis buffer (Promega), and firefly and Renilla luciferase activities were measured, using the Dual Luciferase Reporter Assay System (Promega) on a Lumat LB 9507 luminometer (Berthold Technologies). Firefly luciferase activity was normalized to Renilla luciferase activity, and values in agonist-treated cells were normalized to those detected in medium-treated cells, and expressed as fold induction.
Confocal Microscopy
Cells were seeded (4 × 105 cells) on poly-L-lysine-coated coverslips (Fischer Scientific, Pittsburg, PA) in phenol-free cDMEM, and cultured for 20 h. Transfection was performed using SuperFect transfection reagent, followed by gentle washing and recovery for 48 h. Cells were fixed with 2% paraformaldehyde and mounted onto glass slides using a DABCO-based anti-fade fluorescent mounting media. Images were acquired using an Olympus Fluoview 500 laser scanning confocal microscope (Olympus, Melville, NY) at excitation wavelength 515 nm and standard emission filters for YFP detection (YFP-TLR4).
Statistical analysis
Data were analyzed using the GraphPad Prism 5 program for Windows (GraphPad Software Inc.), and statistical differences were evaluated by the Student’s t-test with the level of significance set at p<0.05. Values are expressed as mean ± SD.
RESULTS
The D299G polymorphism impairs LPS-induced p38 and NF-κB activation, but does not affect total TLR4 levels and minimally impacts TLR4 cell surface expression
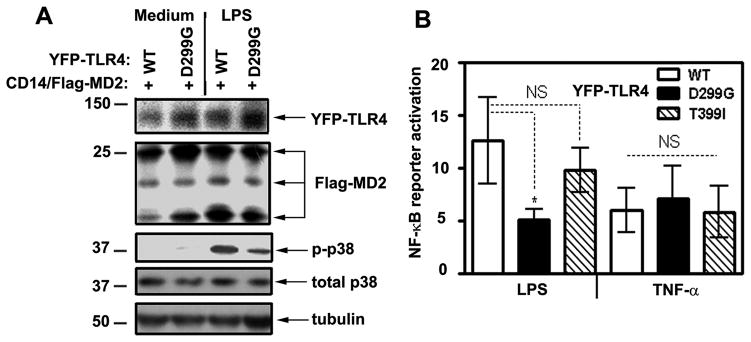
Since the magnitude of LPS responses depends on TLR4 expression (48, 49), we studied the impact of the D299G and T399I polymorphisms on TLR4 levels and signaling in TLR4-deficient HEK293 cells (50) transfected with engineered WT or mutant YFP-TLR4 species, CD14 and MD2 (to impart LPS sensitivity) (51–53). Fusion of YFP with TLR4 at the C-terminus allows for improved detection of TLR4, while not affecting its localization and functions (44, 54). Because TLR4 activates MAP kinases and NF-κB via MyD88- and TRIF-dependent pathways (24, 55), we measured expression of these integral parameters to examine receptor signaling. Since phosphorylation of p38 closely correlates with activation of its kinase activity (56), LPS-mediated p38 phosphorylation was used as a read-out. LPS induced robust phosphorylation of p38 in 293T/CD14/MD2 transfectants expressing WT YFP-TLR4, whereas a blunted response was observed in cells transfected with the D299G YFP-TLR4 under conditions of comparable expression of YFP-TLR4, Flag-MD2, p38 and tubulin (Fig. 1A). These data indicate that deficient LPS-induced p38 phosphorylation via D299G TLR4 was not due to lower expression of receptor components, differences in total p38 levels, or unequal protein loading. Compared to WT TLR4, D299G species exhibited 59% decreased LPS-induced activation of the NF-κB-dependent pELAM-Luc reporter, T399I TLR4 showed no statistically significant differences, while TNF-α similarly stimulated NF-κB in cells expressing WT or mutant TLR4 (Fig. 1B).
Figure 1. The presence of the D299G polymorphism impairs the ability of TLR4 to mediate LPS-induced p38 phosphorylation and NF-κB activation.
A, HEK293 cells stably expressing WT or D299G YFP-TLR4 were co-transfected with pcDNA3-CD14 and pEFBOS-Flag-MD2. Cells were treated for 30 min with medium or 100 ng/ml LPS, and cell lysates were analyzed by immunoblotting with Abs against GFP (YFP-TLR4), Flag (Flag-MD2), p-p38, total p38, and tubulin. The data of a representative (n=3) experiment are shown. B, HEK293T cells transiently transfected with WT, D299G or T399I YFP-TLR4 species, along with pcDNA3-CD14 and pEFBOS-Flag-MD2 were co-transfected with pELAM-Luc and pTK-Renilla-Luc. Cells were treated for 5 h with medium, 100 ng/ml LPS or TNF-α, and cell lysates were analyzed for firefly and Renilla Luc activities. The summary of five experiments (mean ± SD) is depicted. *p<0.05; NS: non-significant.
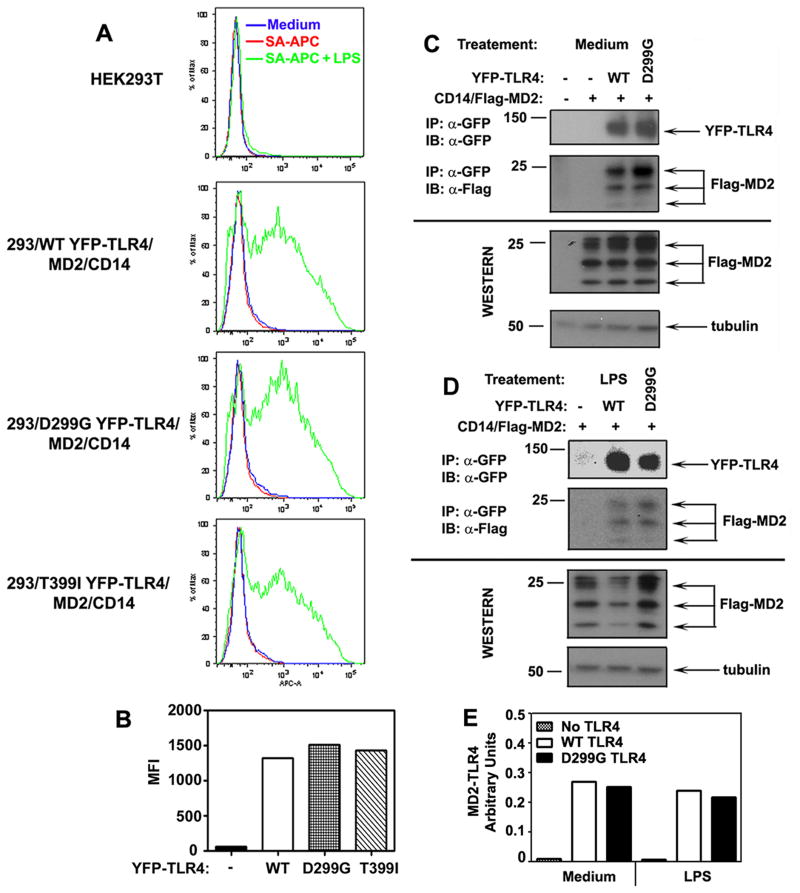
Total YFP-TLR4 expression was determined based on YFP fluorescence of expressed TLR4 species and their immunoreactivity with anti-GFP Ab that cross-reacts with YFP (44, 54, 57). Confocal imaging and FACS showed comparable fluorescence intensities of WT, D299G, and T399I YFP-TLR4 in 293/CD14/MD2/TLR4 stable transfectants (Fig. 2A and B), and immunoblot analyses revealed similar amounts of the WT vs. D299G YFP-TLR4 and Flag-MD2 proteins (Fig. 2C). To determine TLR4 cell surface expression, we stained 293/MD2/CD14 tranfsectants stably expressing WT or mutant YFP-TLR4 with PE-conjugated anti-TLR4 (HTA125) or isotype control Abs, followed by FACS analyses. Since the D299G polymorphism most profoundly inhibits TLR4 signaling (Fig. 1 and (25, 29, 38), we compared cell surface expression of WT vs. D299G TLR4. In contrast to TLR4-deficient HEK293 cells, TLR4-expressing 293/CD14/ MD2 transfectants stained with HTA125-PE showed a shift to the right in the PE fluorescence compared to the isotype control Ab staining (Supplemental Fig. 1), indicating specific detection of TLR4. HTA125 staining of 293/CD14/MD2 cells expressing WT and D299G YFP-TLR4 showed identical peaks and only marginal differences in the shoulder patterns (Fig. 2B). Thus, the D299G and T399I polymorphisms do not affect total and minimally impact cell surface TLR4 expression.
Figure 2. The impact of the D299G and T399I polymorphisms on total and cell surface expression of TLR4.
HEK293 cells stably transfected with expression vectors encoding WT, D299G or T399I YFP-TLR4 were co-transfected with pcDNA3-CD14 and pEFBOS-Flag-MD2. Cells transfected with pcDNA3, designated as “(−)”, were used as a negative control. After recovery, total TLR4 expression was determined by confocal microscopy (A) and FACS analysis (B, top panel), using the YFP fluorescence intensity (FITC channel) as a readout. TLR4 cell surface expression was determined by staining of cells with HTA125-PE (anti-TLR4 Ab) or isotype control IgG-PE, followed by FACS analyses of PE fluorescence (B, bottom panel). C: Whole cell lysates from the respective cell transfectants were subjected to Western blot analyses, using antibodies against GFP, Flag, and tubulin. The results of a representative (n=3) experiment are shown.
The D299G and T399I polymorphisms do not affect LPS binding to the TLR4/CD14/MD2 complex and TLR4 interactions with MD2
Since the D299G and T399I polymorphisms are located in the TLR4 ectodomain (25) that is involved in ligand sensing and co-receptor interactions (2, 20, 58), they could affect LPS binding to the TLR4/CD14/MD2 complex or TLR4-MD2 assembly. To test this hypothesis, 293/CD14/MD2 transfectants expressing WT or mutant YFP-TLR4 were incubated with medium or biotinylated LPS, washed, and bound LPS was detected by SA-APC staining followed by FACS analysis. In the absence of biotinylated LPS, no differences in SA-APC binding to the analyzed cell lines were observed, which was comparable to the autofluorescence profiles in medium-treated cells (Fig. 3A). Cells incubated with biotinylated LPS and SA-APC showed mean fluorescent intensity values (MFI) of 1318, 1514, and 1432 for 293/CD14/MD2 transfectants expressing WT, D299G, and T399I YFP-TLR4, respectively, significantly exceeding 58.7 MFI detected in pcDNA3-transfected cells (Fig. 3A, B). Thus, the D299G and T399I polymorphisms do not affect LPS binding to the TLR4/MD2/CD14 receptor complex.
Figure 3. LPS binding to the TLR4/CD14/MD2 complex and TLR4-MD2 interactions in HEK293/CD14/MD2 cells expressing WT, D299G or T399I YFP-TLR4 species.
A, HEK293 cells stably expressing WT, D299G or T399I YFP-TLR4 species were transiently transfected with pcDNA3-CD14 and pEFBOS-Flag-MD2. After recovery for 20 h, cells were incubated for 30 min at 37° C with medium or biotinylated LPS (1μg/ml), washed, stained with SA-APC (1μg/ml), and analyzed by FACS to detect LPS binding (APC fluorescence). B: Quantification of FACS results (mean fluorescence intensity, MFI) shown in (A), using the FlowJo program. HEK293T cells were transiently transfected with vectors encoding WT or D299G YFP-TLR4 species, along with pcDNA3-CD14/pEFBOS-Flag-MD2. After recovery, cells were treated with medium (C) or stimulated with 100 ng/ml LPS (D), cell lysates were immunoprecipitated with anti-GFP antibody (YFP-TLR4 pull-down), and analyzed by immunoblotting with anti-Flag Ab (to determine Flag-MD2 interactions with YFP-TLR4s). Tubulin immunoblot was used to control protein loading. The data of a representative experiment (n=3) are shown. E: Densotometric quantification of the data shown in (C, D). The results are presented as arbitrary units representing ratios of normalized densitometric values of MD2-TLR4 complexes detected in samples transfected with pcDNA3 (background), or vectors expressing WT or D299G YFP-TLR4. Normalization of densitometric values was calculated as a/b/c/d, where a, b, c and d correspond to the densitometric values of MD2-TLR4, total TLR4, total MD2 and tubulin, respectively.
To determine the impact of the D299G polymorphism on TLR4-MD2 interactions, transfected WT or D299G YFP-TLR4 species were immunoprecipitated with anti-GFP Ab, and the amount of TLR4-interacting Flag-MD2 was determined by immunoblotting of YFP immune complexes with anti-Flag Ab. Immunoblot analyses and densitometric quantification of WT vs. D299G YFP-TLR4 proteins immunoprecipitated from cells treated with medium or LPS revealed comparable intensities of protein bands immunoreactive with anti-GFP and anti-Flag Abs (Fig. 3C, D, top panels, and Fig. 3E). Whole cell lysates obtained from 293T/CD14/Flag-MD2 cells complemented with WT or D299G YFP-TLR4 showed comparable total levels of Flag-MD2 proteins (Fig. 3C, 2nd panel from bottom). Thus, similar amounts of Flag-MD2 interact with WT or D299G YFP-TLR4 variants under conditions of similar total expression of the interacting proteins, indicating that the D299G polymorphism does not affect TLR4-MD2 interactions.
The D299G polymorphism impairs the ability of TLR4 to recruit MyD88 and activate MyD88-dependent cytokine genes
Recruitment of MyD88 to TLR4 is one of the earliest events of TLR4 signaling (2, 20). To determine the impact of the D299G TLR4 polymorphism on this process, we used co-immunoprecipitation to study LPS-inducible association of transfected AU1-MyD88 with WT or D299G YFP-TLR4 in 293/YFP-TLR4 stable cell lines expressing the respective TLR4s, CD14 and HA-MD2. LPS induced robust association of WT YFP-TLR4 with AU1-MyD88 (3.2–14.4-fold increase over medium controls), while the D299G YFP-TLR4 had a reduced (0.98–1.8 fold increase) capacity to recruit AU1-MyD88, leading to 32–82% inhibition (Fig. 4A–D). Comparable total levels of HA-MD2 (Fig. 4A), YFP-TLR4 species, and AU1-MyD88 were seen (Fig. 4A and B), indicating that differences in MyD88 recruitment to WT vs. D299G TLR4 were not due to variations in total expression of TLR4, MD2, or MyD88.
Figure 4. Compromised LPS-mediated recruitment of MyD88 to D299G TLR4.
HEK293 cells transiently (A) or stably (B) expressing WT or D299G YFP-TLR4 were co-transfected with pcDNA3-CD14, pEFBOS-HA-MD2, and either pcDNA3-AU1 or pcDNA3. After recovery, cells were treated with medium or 100 ng/ml LPS for 1 min (A) or for the indicated time course (B), cell lysates were prepared, and YFP-TLR4 proteins were immunoprecipitated with anti-GFP Ab or isotype control IgG, followed by immunoblot analyses of the immune complexes with anti-GFP or anti-AU1 Abs. Whole cell lysates were examined by immunoblotting, using the indicated antibodies. C and D: densitometirc quantification of the data shown in A and B. Densities of the bands representing MyD88 associated with TLR4 were normalized by total TLR4 and by total MD2 values and were expressed as arbitrary units. Numbers indicate inhibition (%) of LPS-inducible MyD88-TLR4 values in D299G YFP-TLR4-transfected cells compared to WT YFP-TLR4 transfectants. Shown are the results of a representative (n=3) experiment.
Next, we studied the impact of the D299G and T399I polymorphisms on transcription of MyD88-dependent cytokine genes IL-8 and TNF-α (59–61). LPS increased the levels of IL-8 mRNA in 293T/CD14/MD2 cells transiently and stably transfected with WT YFP-TLR4 by 4.7- and 7.5-fold, respectively, whereas cells expressing the D299G variant showed 45–54% inhibition in LPS-mediated IL-8 mRNA induction (Fig. 5A and B). Transfected T399I YFP-TLR4 elicited responses that showed no statistically significant differences compared to those observed in WT YFP-TLR4-expressing transfectants (Fig. 5A and B). Since HEK293 cells have a limited repertoire of LPS-inducible genes and lack many macrophage characteristics (e.g., phagocytosis) (40, 44, 50, 62), we sought to confirm our results in WT and TLR4−/− BMDMs. Consistent with the literature (61), LPS failed to up-regulate TNF-α mRNA in TLR4−/− macrophages, whereas TLR4+/+ cells expressing endogenous mouse TLR4 responded to LPS with a 12.5-fold increase in the levels of TNF-α mRNA (Fig. 5C). Nucleofection of TLR4+/+ BMDMs with WT or D299G TLR4 had no statistically significant impact on LPS-induced TNF-α gene expression compared to the response of GFP-nucleofected TLR4+/+ cells (Fig. 5C). TLR4−/− cells complemented with human WT YFP-TLR4 responded to LPS with a 6.2-fold increase in the expression of TNF-α mRNA, while nucleofection of the D299G TLR4 failed to impart LPS responsiveness (Fig. 5C). TLR4−/− BMDMs complemented with WT vs. D299G YFP-TLR4 variants showed similar levels of transfected TLR4 species, as evidenced by their equivalent TLR4 mRNA levels (Fig. 5D). These results indicate that the D299G TLR4 variant is compromised in its ability to elicit LPS-mediated recruitment of MyD88 and expression of MyD88-dependent cytokine genes.
Figure 5. The impact of the D299G and T399I TLR4 polymorphisms on LPS-induced expression of MyD88-dependent cytokine genes.
HEK293 transient (A) and stable (B) transfectants expressing WT, D299G or T399I YFP-TLR4 were co-transfected with pcDNA3-CD14/pEFBOS-Flag-MD2. C, C57BL/6 (TLR4+/+) and TLR4−/− primary BMDMs were nucleofected with plasmids encoding eGFP (a negative control), WT or D299G YFP-TLR4, as indicated. After recovery for 24 h. cells were treated for 3 h with medium, LPS or TNF-α (100 ng/ml each), RNA was isolated, reverse-transcribed, and analyzed by real-time qPCR with primers specific for human HPRT and IL-8 (A, B); mouse HPRT and TNF-α (C), and human TLR4 (D). The results of a representative experiment (n = 3) are depicted. *p<0.05; ns: not statistically significant.
The D299G polymorphism inhibits TLR4-mediated activation of the TRIF pathway
To discern whether the D299G and T399I polymorphisms affect the TRIF pathway, we examined LPS-induced recruitment of TRIF to TLR4 and activation of the TBK1-IRF3-IFN-β signaling axis. LPS induced recruitment of transfected Flag-TRIF to WT YFP-TLR4 within 15–30 min post-stimulation, whereas no TRIF-associated TLR4 was detected in 293/CD14/MD2 cells expressing the D299G YFP-TLR4 variant (Fig. 6, top panel). Comparable total expression of WT and D299G TLR4, as well as Flag-TRIF proteins, was detected (Fig. 6, 2nd and 3rd panels from the top, respectively), indicating that deficient TRIF recruitment to the D299G TLR4 was not due to lower levels of the interacting proteins. In line with the requirement of TRIF recruitment to TLR4 for activation of TBK1 and IRF3 (23, 24), LPS induced markedly reduced phosphorylation of TBK1 and IRF3 in 293T/CD14/MD2 cells transfected with D299G YFP-TLR4, whereas robust responses were seen in WT YFP-TLR4-expressing cells (Fig. 7A and B). Comparable intensities of TLR4, MD2, TBK1, and IRF3 bands were detected (Fig. 7A and B), indicating that differences in TBK1 and IRF3 phosphorylation were not due to variations in expression of the receptor components or TBK1/IRF-3 total levels. To determine the impact of the D299G polymorphism on IRF3 transactivation, LPS-mediated induction of TRIF-controlled, IRF3-dependent, IFN-β promoter-driven luciferase reporter (p125-Luc) and transcription of the IFN-β gene were analyzed. LPS treatment of 293T/CD14/MD2 cells transiently or stably expressing WT TLR4 led to 3.2- and 4.1-fold induction of p125-Luc, respectively (Fig. 7C and D), and increased the levels of IFN-β mRNA by 2.8-fold (Fig. 7E). Cells expressing the D299G TLR4 showed significantly decreased LPS-driven activation of IFN-β-promoter reporter (Fig. 7C, D) and IFN-β gene expression (Fig. 7E) compared to the responses observed in WT TLR4-transfected cells. 293/CD14/MD2 cells transfected with WT or T399I TLR4 showed no statistically significant differences in LPS-mediated p125-Luc reporter activation and IFN-β gene expression (Fig. 7C–E). Comparable induction of the p125-Luc reporter and IFN-β gene expression were seen in TNF-α-treated 293T/CD14/MD2 cells expressing WT, D299G, or T399I TLR4 (Fig. 7C–E), indicating that the TLR4 SNPs do not affect TLR-independent responses.
Figure 6. The D299G polymorphism impairs the ability of TLR4 to recruit TRIF in response to LPS stimulation.
293/TLR4 stable cell lines expressing WT or D299G YFP-TLR4 species were co-transfected with pcDNA3-CD14, pEFBOS-HA-MD2, and pEFBOS-Flag-TRIF. After recovery for 20 h, cells were treated with medium or stimulated for the indicated times with 100 ng/ml LPS, cell lysates were prepared and immunoprecipitated with anti-GFP or anti-Flag Abs. Immune complexes were analyzed by immunoblotting with anti-GFP, anti-Flag, or isotype control IgG Abs to determine total TLR4 expression and amounts of TRIF recruited to TLR4 proteins. The data of a representative (n=3) experiments are shown.
Figure 7. LPS-mediated phosphorylaiton of TBK-1 and IRF3, activation of p125-Luc and induction of IFN-β mRNA in cells expressing WT or D299G YFP-TLR4.
A, HEK293T cells were transiently transfected with plasmids encoding WT or D299G YFP-TLR4, along with pcDNA3-CD14 and pEFBOS-Flag-MD2. After recovery, cells were treated with 100 ng/ml LPS for 30 min and lysates were analyzed by immunoblotting with anti-GFP (YFP-TLR4), anti-Flag (Flag-MD2), anti-p-TBK-1, anti-total TBK-1, and anti-tubulin Abs. B, HEK293 cells stably expressing YFP-TLR4 WT or D299G were co-transfected with pcDNA3-CD14 and pEFBOS-HA-MD2. After recovery, cells were treated for with 100 ng/ml LPS for 15 min, and cell lysates were prepared and analyzed by immunoblotting with Abs against GFP (YFP-TLR4), p-IRF3 and total IRF3, and anti-HA-HRP Ab (HA-MD2). C, HEK293T cells were transiently transfected with WT, D299G or T399I YFP-TLR4 species, along with pcDNA3-CD14 and pEFBOS-Flag-MD2. In addition, cells were co-transfected with p125-Luc and pTK-Renilla-Luc. D, HEK293 cells stably expressing WT or D299G YFP-TLR4 species were co-transfected with pcDNA3-CD14 and pEFBOS-Flag-MD2. After recovery, cells were stimulated for 5 h with LPS or TNF-α (100 ng/ml each). Cell lysates were analyzed for firefly and Renilla luciferase activities. E, HEK293T cells were transiently transfected with WT, D299G or T399I YFP-TLR4 species, along with pcDNA3-CD14 and pEFBOS-Flag-MD2. F, WT (TLR4+/+) and TLR4−/− iBMDMs were transfected as shown. After recovery, cells were stimulated for 3 h with 100 ng/ml LPS. RNA was isolated, reverse-transcribed and examined by real-time qPCR with primers specific for human HPRT and IFN-β (E) or mouse HPRT and IFN-β (F). Shown are results (mean ± SD) of the summary of three independent experiments (C and D) and a representative experiment (n=3, A, B, E and F). *p<0.05;**p<0.01; ns: not significant.
To confirm our data in cells with the macrophage phenotype, we assessed TLR4-inducible induction of IFN-β mRNA in iBMDM cell lines that have been reported to replicate LPS responses of primary macrophages (63). LPS treatment of iBMDMs complemented with WT YFP-TLR4 increased expression of IFN-β mRNA by 6-fold, while pcDNA3-transfected cells did not show IFN-β mRNA induction (Fig. 7F). In contrast, complementation of TLR4−/− iBMDMs with D299G YFP-TLR4 led to only a 2-fold increase in IFN-β mRNA in response to LPS (Fig. 7F), accounting for a 65% inhibition of the WT response. Similar relative levels of human TLR4 mRNA in iBMDMs complemented with WT vs. D299G TLR4 were observed (Supplemental Fig. 2), suggesting comparable expression of transfected YFP-TLR4 species. These data indicate that D299G polymorphism compromises the ability of TLR4 to elicit LPS-induced recruitment of TRIF to TLR4, phosphorylation of TBK1 and IRF3, activation of IRF3-driven reporter and IFN-β mRNA.
DISCUSSION
The D299G and T399I polymorphisms in the TLR4 ectodomain have been linked to sepsis, Gram-negative infections, RSV bronchiolitis, Mediterranean spotted fever, tuberculosis in HIV-infected patients, and S. pyogenes-associated tonsillitis (25–31, 35, 36). The D299G TLR4 variant exhibits blunted responses to LPS, F protein from RSV, and chlamydial Hsp60, whereas the T399I polymorphism has either a significantly reduced or no inhibitory impact on TLR4 functions (25–31, 35, 36, 38). The molecular mechanisms responsible for the observed signaling deficiencies are unclear, and controversial data have been reported regarding the impact of these polymorphisms on TLR4 expression (38, 64).
Association of the ectodomain of TLR4 with chaperones gp96 and CNPY3 regulate TLR4 trafficking and expression (65–67), critical parameters defining the magnitude of LPS responses (48). Since the D299G and T399I polymorphisms are located in the ectodomain of TLR4 (25), it is possible that they could affect TLR4 cellular localization and/or expression levels by impacting TLR4 assembly with chaperones. However, the data provided in this report do not support this hypothesis and show comparable total expression of transfected WT vs. polymorphic D299G and T399I YFP-TLR4 variants, as demonstrated by confocal microscopy, FACS, and immunoblot analyses, and using both transient and stable transfectants. Furthermore, FACS analysis revealed only minimal differences in cell surface expression of WT and mutant TLR4 species, and confocal imaging showed their comparable localization on the plasma membrane and in intracellular compartments, reported to be the Golgi apparatus and endoplasmic reticulum (44, 54). Our results support similar data reported with transfected untagged TLR4 (38) and indicate that the D229G and T399I polymorphisms do not significantly alter TLR4 expression, suggesting that signaling deficiencies of the mutant TLR4 species are not due to changes in their expression levels.
MD2 is the principal co-receptor that confers LPS responsiveness by binding LPS and presenting it to TLR4 (51). Mice with a targeted mutation in the gene encoding MD2 exhibit a phenotype identical to TLR4 knockout mice, with both knockouts lacking LPS responsiveness (61, 68). Although the D299G and T399I polymorphisms are located outside of the F440, F463, and L444 MD2-interaction interface of TLR4 (58), it could be theorized that they distantly impose conformational changes, affecting TLR4-MD2 interactions, as previously suggested (38). To address this possibility, we studied whether the D299G polymorphism, associated with the strongest inhibition of TLR4 signaling (25, 27, 29, 38), affects the ability of transfected YFP-TLR4 to interact with Flag-MD2 under basal state and upon LPS stimulation. Co-immunoprecipitation experiments revealed no measurable differences in TLR4-MD2 interactions regardless of whether or WT or D299G TLR4s were present, indicating that compromised signaling elicited by D299G TLR4 can not be accounted for by its deficient association with MD2.
Five acyl chains of lipid A bind to a hydrophobic pocket within MD2, imposing conformational changes and re-orientation of the critical F126 residue in MD2 that presents the remaining acyl chain to the neighboring TLR4-MD2 complex, promoting TLR4 homo-dimerization (58). Hydrophobic interactions of lipid A with the TLR4 residues F440, 463, and L444, and interactions of the two phosphate groups of lipid A with positively charged K388, R264, K341, K362 residues within TLR4 are important for promoting receptor homo-dimerization (58). Since the D299G and T399I polymorphisms are located relatively close to the R264, K341, K362 residues, they could influence LPS binding to the receptor complex TLR4/MD2/CD14. To address this possibility, we examined binding of biotinylated LPS to 293/CD14/MD2 transfectants expressing WT or mutant TLR4 variants, as revealed by FACS analysis of streptavidin-APC bound to cell-associated biotinylated LPS. Similar LPS binding was detected in 293/CD14/MD2 cells expressing WT, D299G, or T399I YFP-TLR4 species under conditions of comparable TLR4 expression, confirming a recent publication demonstrating comparable affinities of interactions of radioactively labeled LOS with WT, D299G, or T399I TLR4 (64). However, this cannot exclude the possibility that despite comparable LPS binding to the TLR4/MD2/CD14 complex in cells expressing WT vs. mutant versions of TLR4, the presence of the D299G polymorphism could affect intricate TLR4 interactions with MD2-presented LPS, resulting in altered TLR4 engagement and homodimerization. Structural studies of lipid A in complex with MD2-associated WT or mutant TLR4 are required to dissect the impact of the D299G and T399I polymorphisms on LPS interactions with MD2/TLR4 and TLR4/MD2 dimerization states.
LPS triggers TLR4 homo-dimerization that brings together intracellular TIR domains, forming docking platforms to enable recruitment of adapters TIRAP/Mal and MyD88 to the plasma membrane-associated TLR4, and TRIF and TRAM to TLR4 translocated to endosomes (2, 69). These events initiate TLR4 signaling pathways responsible for expression of pro-inflammatory cytokines, chemokines (MyD88-dependent) and type I IFNs and type I IFN-dependent genes (TRIF-dependent) (2, 20, 69). To the best of our knowledge, we show for the first time that impaired recruitment of MyD88 and TRIF to the D299G TLR4 species represents a principal mechanism by which this polymorphism alters receptor signaling. Consistent with a key role for Myddosome assembly for initiation of downstream signaling (58), deficient MyD88 recruitment to D299G TLR4 was associated with impaired LPS-mediated induction of IL-8 mRNA in HEK293 transfection system, and the failure of this mutant to impart LPS-mediated activation of TNF-α gene expression upon nucleofection-based complementation of TLR4−/− BMDMs. Our results provide a mechanistic explanation for deficiencies of the D299G TLR4 variant in eliciting MyD88-dependent responses to LPS (this paper and (25, 38, 64)), F protein of RSV, and chlamydial HSP60 (29), and other TLR4-dependent ligands. Furthermore, to the best of our knowledge, this paper is the first report demonstrating deficient activation of the TRIF signaling pathway via the D299G TLR4. In line with the proposed impact of the D299G polymorphism on TLR4 dimerization that is likely to compromise assembly of docking sites within the TIR domains critical for adapter recruitment, we found impaired LPS-inducible recruitment of TRIF to D299G TLR4. Deficient LPS-inducible TRIF recruitment to D299G TLR4 translated into decreased phosphorylation of TBK1 in 293T/CD14/MD2 cells transfected with D299G TLR4, compared to a robust response observed in cells complemented with WT TLR4. Consistent with the requirement for TBK1 kinase activity for IRF3 phosphorylation and activation (70), we showed compromised TLR4-elicited phosphorylation of IRF3 and IRF3-dependent activation of IFN-β promoter-driven luciferase reporter and induction of IFN-β mRNA. Since HEK293 transfectants are limited in TLR4-mediated responses (40, 44, 50, 62), we confirmed our data on the impact of the D299G TLR4 species on LPS-driven activation of the TRIF pathway in mouse iBMDMs that behave similarly to primary macrophages (63). Transfection-based complementation of TLR4−/− iBMDMs with human WT TLR4 imparted LPS-inducible IFN-β mRNA expression, whereas transfection of the D299G TLR4 species led to very little, if any, activation of this TRIF-dependent response. In contrast, cells expressing the T399I TLR4 variants exhibited no statistically significant deficiencies in LPS-induced MyD88- or TRIF-dependent responses, in line with the reported absence or relatively mild inhibitory effects of this polymorphism on TLR4 functions (29, 38, 64). Since TIRAP/Mal and TRAM are bridging adapters required for recruitment of signaling adapters MyD88 and TRIF to TLR4 (22–24, 69), deficient MyD88 and TRIF recruitment to D299G TLR4 could be due to compromised interactions of TIRAP/Mal and TRAM with the mutant receptor. This interference could result from conformational changes imposed by the mutation that would compromise MD2-assisted LPS presentation within TLR4 complexes (even though total LPS binding and TLR4-MD2 interactions are preserved), affecting TLR4 dimerization and assembly of docking platforms for adapter recruitment. Further studies are needed to address these questions.
Activation of p38 MAP kinase and the transcription factor NF-κB is governed by the MyD88-dependent (early responses) and TRIF-dependent (delayed activation) pathways. The impact of the D299G and T399I polymorphisms on LPS-driven p38 and NF-κB activation was studied based on phosphorylation of p38 and induction of the NF-κB-dependent pELAM-Luc reporter. To the best of our knowledge, this is the first demonstration of compromised LPS-mediated activation of p38 MAP kinase as a consequence of the presence of the D299G mutation in TLR4. In keeping with previous results (29, 38), we show that complementation of 293/CD14/MD2 transfectants with D299G TLR4 reduced LPS-mediated activation of the NF-κB reporter. This reduction was specific for TLR4, as TNF-α elicited similar activation of NF-κB regardless of whether WT or mutant TLR4 species were expressed. Because the D299G polymorphism affects TLR4 signaling at the level of MyD88 and TRIF recruitment, it is likely that deficient recruitment and activation of proximal adapter/kinase models is responsible for deficiencies in downstream signaling events. Consistent with defective activation of p38 and NF-κB, and in line with their importance in the transcriptional control of cytokine expression (2, 20), we observed impaired production of pro-inflammatory cytokines TNF-α and IL-8.
In summary, our data show comparable total expression of transfected WT, D299G, and T399I TLR4 species, and minimal effects of the polymorphisms on TLR4 cell surface expression and LPS binding to the TLR4/MD2/CD14 complex. This paper demonstrates for the first time that deficient recruitment of signaling adapters MyD88 and TRIF to TLR4 is a mechanistic basis for D299G-mediated impairment of TLR4-elicited, LPS-induced activation of MyD88-dependent IL-8 and TNF-α genes, deficient TRIF-dependent phosphorylation of TBK1 and IRF-3, transactivation of IRF-3 and expression of IFN-β mRNA. TLR4-driven activation of NF-κB and p38 dependent on both pathways was also impacted by the D299G polymorphism. It will be important to extend our results to cells from human patients expressing endogenous TLR4 SNPs and to conduct studies in knock-in mice expressing humanized version of TLR4 proteins to define the impact of these mutations on susceptibility to infectious and inflammatory diseases.
Supplementary Material
Acknowledgments
We are grateful to Drs. Katherine A. Fitzgerald and Douglas T. Golenbock (University of Massachusetts Medical School, Worcester, MA) for kindly providing iBMDM cell lines, Drs. Andrei Chapoval and Svetlana Chapoval (University of Maryland School of Medicine, Baltimore, MD) for help with stable transfections and FACS analyses, and Dr. Adam Puche (University of Maryland School of Medicine, Baltimore, MD) for guidance and advice with confocal studies.
Footnotes
This work was supported by NIH grants RO1AI18797 (to SNV), RO1AI059524 and R21AI067468 (to AEM).
Abbreviations used in this paper: PRRs, pattern recognition receptors; LRR, leucine-rich repeat; TIR, Toll-IL-1R resistance; F, fusion, RSV, respiratory syncytial virus; Hsp, heat shock protein; IRAK, IL-1R-associated kinase; TRIF, TIR domain-containing adapter inducing IFN-β; TBK, TANK-binding kinase; IRF, IFN regulatory factor; Mal, MyD88 adapter-like; TIRAP, TIR domain-containing adapter protein; TRAM, TRIF-related adapter molecule; MD2, myeloid differentiation factor 2; SA-APC, streptavidin-allophycocyanin; GFP, green fluorescent protein; p, phospho; HEK, human embryonic kidney; BMDMs, bone marrow-derived macrophages; c, complete; i, J2 retrovirus-immortalized; BMDMs, bone marrow-derived; RT, real-time; q, quantitative.
References
- 1.Medzhitov R, Preston-Hurlburt P, Janeway CA., Jr A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature. 1997;388:394–397. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- 2.Beutler B. Microbe sensing, positive feedback loops, and the pathogenesis of inflammatory diseases. Immunol Rev. 2009;227:248–263. doi: 10.1111/j.1600-065X.2008.00733.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schattgen SA, Fitzgerald KA. The PYHIN protein family as mediators of host defenses. Immunol Rev. 2011;243:109–118. doi: 10.1111/j.1600-065X.2011.01053.x. [DOI] [PubMed] [Google Scholar]
- 4.Davis BK, Wen H, Ting JP. The inflammasome NLRs in immunity, inflammation, and associated diseases. Annu Rev Immunol. 2011;29:707–735. doi: 10.1146/annurev-immunol-031210-101405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Onoguchi K, Yoneyama M, Fujita T. Retinoic acid-inducible gene-I-like receptors. J Interferon Cytokine Res. 2011;31:27–31. doi: 10.1089/jir.2010.0057. [DOI] [PubMed] [Google Scholar]
- 6.Lien E, Sellati TJ, Yoshimura A, Flo TH, Rawadi G, Finberg RW, Carroll JD, Espevik T, Ingalls RR, Radolf JD, Golenbock DT. Toll-like receptor 2 functions as a pattern recognition receptor for diverse bacterial products. J Biol Chem. 1999;274:33419–33425. doi: 10.1074/jbc.274.47.33419. [DOI] [PubMed] [Google Scholar]
- 7.Poltorak A, He X, Smirnova I, Liu MY, Van Huffel C, Du X, Birdwell D, Alejos E, Silva M, Galanos C, Freudenberg M, Ricciardi-Castagnoli P, Layton B, Beutler B. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 8.Bulut Y, Faure E, Thomas L, Karahashi H, Michelsen KS, Equils O, Morrison SG, Arditi M. Chlamydial heat shock protein 60 activates macrophages and endothelial cells through Toll-like receptor 4 and MD2 in a MyD88-dependent pathway. J Immunol. 2002;168:1435–1440. doi: 10.4049/jimmunol.168.3.1435. [DOI] [PubMed] [Google Scholar]
- 9.Flo TH, Ryan L, Latz E, Takeuchi O, Monks BG, Lien E, Halaas Ø, Akira S, Skjåk-Braek G, Golenbock DT, Espevik T. Involvement of toll-like receptor (TLR) 2 and TLR4 in cell activation by mannuronic acid polymers. J Biol Chem. 2002;277:35489–35495. doi: 10.1074/jbc.M201366200. [DOI] [PubMed] [Google Scholar]
- 10.Kurt-Jones EA, Popova L, Kwinn L, Haynes LM, Jones LP, Tripp RA, Walsh EE, Freeman MW, Golenbock DT, Anderson LJ, Finberg RW. Pattern recognition receptors TLR4 and CD14 mediate response to respiratory syncytial virus. Nat Immunol. 2000;1:398–401. doi: 10.1038/80833. [DOI] [PubMed] [Google Scholar]
- 11.Gewirtz AT, Navas TA, Lyons S, Godowski PJ, Madara JL. Cutting edge: bacterial flagellin activates basolaterally expressed TLR5 to induce epithelial proinflammatory gene expression. J Immunol. 2001;167:1882–1885. doi: 10.4049/jimmunol.167.4.1882. [DOI] [PubMed] [Google Scholar]
- 12.Mizel SB, Snipes JA. Gram-negative flagellin-induced self-tolerance is associated with a block in interleukin-1 receptor-associated kinase release from toll-like receptor 5. J Biol Chem. 2002;277:22414–22420. doi: 10.1074/jbc.M201762200. [DOI] [PubMed] [Google Scholar]
- 13.Zhang D, Zhang G, Hayden MS, Greenblatt MB, Bussey C, Flavell RA, Ghosh S. A toll-like receptor that prevents infection by uropathogenic bacteria. Science. 2004;303:1522–1526. doi: 10.1126/science.1094351. [DOI] [PubMed] [Google Scholar]
- 14.Yarovinsky F, Zhang D, Andersen JF, Bannenberg GL, Serhan CN, Hayden MS, Hieny S, Sutterwala FS, Flavell RA, Ghosh S, Sher A. TLR11 activation of dendritic cells by a protozoan profilin-like protein. Science. 2005;308:1626–1629. doi: 10.1126/science.1109893. [DOI] [PubMed] [Google Scholar]
- 15.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NFκB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 16.Diebold SS, Kaisho T, Hemmi H, Akira S, Reis e Sousa C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303:1529–1531. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- 17.Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, Lipford G, Wagner H, Bauer S. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303:1526–1529. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- 18.Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, Matsumoto M, Hoshino K, Wagner H, Takeda K, Akira S. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 19.Vogel SN, Fitzgerald KA, Fenton MJ. TLRs: differential adapter utilization by toll-like receptors mediates TLR-specific patterns of gene expression. Mol Interv. 2003;3:466–477. doi: 10.1124/mi.3.8.466. [DOI] [PubMed] [Google Scholar]
- 20.Doyle SL, O’Neill LA. Toll-like receptors: from the discovery of NF-κB to new insights into transcriptional regulations in innate immunity. Biochem Pharmacol. 2006;72:1102–1113. doi: 10.1016/j.bcp.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 21.Horng T, Barton GM, Medzhitov R. TIRAP: an adapter molecule in the Toll signaling pathway. Nat Immunol. 2001;2:835–841. doi: 10.1038/ni0901-835. [DOI] [PubMed] [Google Scholar]
- 22.Fitzgerald KA, Palsson-McDermott EM, Bowie AG, Jefferies CA, Mansell AS, Brady G, Brint E, Dunne A, Gray P, Harte MT, McMurray D, Smith DE, Sims JE, Bird TA, O’Neill LA. Mal (MyD88-adapter-like) is required for Toll-like receptor-4 signal transduction. Nature. 2001;413:78–83. doi: 10.1038/35092578. [DOI] [PubMed] [Google Scholar]
- 23.Fitzgerald KA, Rowe DC, Barnes BJ, Caffrey DR, Visintin A, Latz E, Monks B, Pitha PM, Golenbock DT. LPS-TLR4 signaling to IRF-3/7 and NF-kappaB involves the toll adapters TRAM and TRIF. J Exp Med. 2003;198:1043–1055. doi: 10.1084/jem.20031023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamamoto M, Sato S, Hemmi H, Hoshino K, Kaisho T, Sanjo H, Takeuchi O, Sugiyama M, Okabe M, Takeda K, Akira S. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science. 2003;301:640–643. doi: 10.1126/science.1087262. [DOI] [PubMed] [Google Scholar]
- 25.Arbour NC, Lorenz E, Schutte BC, Zabner J, Kline JN, Jones M, Frees K, Watt JL, Schwartz DA. TLR4 mutations are associated with endotoxin hyporesponsiveness in humans. Nat Genet. 2000;25:187–191. doi: 10.1038/76048. [DOI] [PubMed] [Google Scholar]
- 26.Lorenz E, Mira JP, Frees KL, Schwartz DA. Relevance of mutations in the TLR4 receptor in patients with Gram-negative septic shock. Arch Intern Med. 2002;162:1028–1032. doi: 10.1001/archinte.162.9.1028. [DOI] [PubMed] [Google Scholar]
- 27.Balistreri CR, Candore G, Lio D, Colonna-Romano G, Di Lorenzo G, Mansueto P, Rini G, Mansueto S, Cillari E, Franceschi C, Caruso C. Role of TLR4 receptor polymorphisms in Boutonneuse fever. Int J Immunopathol Pharmacol. 2005;18:655–660. doi: 10.1177/039463200501800406. [DOI] [PubMed] [Google Scholar]
- 28.Tal G, Mandelberg A, Dalal I, Cesar K, Somekh E, Tal A, Oron A, Itskovich S, Ballin A, Houri S, Beigelman A, Lider O, Rechavi G, Amariglio N. Association between common Toll-like receptor 4 mutations and severe respiratory syncytial virus disease. J Infect Dis. 2004;189:2057–2063. doi: 10.1086/420830. [DOI] [PubMed] [Google Scholar]
- 29.Awomoyi AA, Rallabhandi P, Pollin TI, Lorenz E, Sztein MB, Boukhvalova MS, Hemming VG, Blanco JC, Vogel SN. Association of TLR4 polymorphisms with symptomatic respiratory syncytial virus infection in high-risk infants and young children. J Immunol. 2007;179:3171–3177. doi: 10.4049/jimmunol.179.5.3171. [DOI] [PubMed] [Google Scholar]
- 30.Ferwerda B, Kibiki GS, Netea MG, Dolmans WM, van der Ven AJ. The toll-like receptor 4 Asp299Gly variant and tuberculosis susceptibility in HIV-infected patients in Tanzania. AIDS. 2007;21:1375–1377. doi: 10.1097/QAD.0b013e32814e6b2d. [DOI] [PubMed] [Google Scholar]
- 31.Pulido I, Leal M, Genebat M, Pacheco YM, Sáez ME, Soriano-Sarabia N. The TLR4 ASP299GLY polymorphism is a risk factor for active tuberculosis in Caucasian HIV-infected patients. Curr HIV Res. 2010;8:253–258. doi: 10.2174/157016210791111052. [DOI] [PubMed] [Google Scholar]
- 32.Brand S, Staudinger T, Schnitzler F, Pfennig S, Hofbauer K, Dambacher J, Seiderer J, Tillack C, Konrad A, Crispin A, Göke B, Lohse P, Ochsenkühn T. The role of Toll-like receptor 4 Asp299Gly and Thr399Ile polymorphisms and CARD15/NOD2 mutations in the susceptibility and phenotype of Crohn’s disease. Inflamm Bowel Dis. 2005;11:645–652. doi: 10.1097/01.mib.0000168372.94907.d2. [DOI] [PubMed] [Google Scholar]
- 33.Franchimont D, Vermeire S, El Housni H, Pierik M, Van Steen K, Gustot T, Quertinmont E, Abramowicz M, Van Gossum A, Deviere J, Rutgeerts P. Deficient host-bacteria interactions in inflammatory bowel disease? The toll-like receptor (TLR)-4 Asp299Gly polymorphism is associated with Crohn’s disease and ulcerative colitis. Gut. 2004;53:987–992. doi: 10.1136/gut.2003.030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Latha M, Vaidya S, Movva S, Chava S, Govindan S, Govatati S, Banoori M, Hasan Q, Kodati VL. Molecular pathogenesis of endometriosis; Toll-like receptor-4 A896G (D299G) polymorphism: a novel explanation. Genet Test Mol Biomarkers. 2011;15:181–184. doi: 10.1089/gtmb.2010.0178. [DOI] [PubMed] [Google Scholar]
- 35.Liadaki K, Petinaki E, Skoulakis C, Tsirevelou P, Klapsa D, Germenis AE, Speletas M. Toll-like receptor 4 gene (TLR4), but not TLR2, polymorphisms modify the risk of tonsillar disease due to Streptococcus pyogenes and Haemophilus influenzae. Clin Vaccine Immunol. 2011;18:217–222. doi: 10.1128/CVI.00460-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vogel SN, Awomoyi AA, Rallabhandi P, Medvedev AE. Mutations in TLR4 signaling that lead to increased susceptibility to infection in humans: an overview. J Endotoxin Res. 2005;11:333–339. doi: 10.1179/096805105X58724. [DOI] [PubMed] [Google Scholar]
- 37.Radstake TR, Franke B, Hanssen S, Netea MG, Welsing P, Barrera P, Joosten LA, van Riel PL, van den Berg WB. The Toll-like receptor 4 Asp299Gly functional variant is associated with decreased rheumatoid arthritis disease susceptibility but does not influence disease severity and/or outcome. Arthritis Rheum. 2004;50:999–1001. doi: 10.1002/art.20114. [DOI] [PubMed] [Google Scholar]
- 38.Rallabhandi P, Bell J, Boukhvalova MS, Medvedev A, Lorenz E, Arditi M, Hemming VG, Blanco JC, Segal DM, Vogel SN. Analysis of TLR4 polymorphic variants: new insights into TLR4/MD-2/CD14 stoichiometry, structure, and signaling. J Immunol. 2006;177:322–332. doi: 10.4049/jimmunol.177.1.322. [DOI] [PubMed] [Google Scholar]
- 39.Medvedev AE, Thomas K, Awomoyi A, Kuhns DB, Gallin JI, Li X, Vogel SN. Cutting edge: expression of IL-1 receptor-associated kinase-4 (IRAK-4) proteins with mutations identified in a patient with recurrent bacterial infections alters normal IRAK-4 interaction with components of the IL-1 receptor complex. J Immunol. 2005;174:6587–6591. doi: 10.4049/jimmunol.174.11.6587. [DOI] [PubMed] [Google Scholar]
- 40.Piao W, Song C, Chen H, Wahl LM, Fitzgerald KA, O’Neill LA, Medvedev AE. Tyrosine phosphorylation of MyD88 adapter-like (Mal) is critical for signal transduction and blocked in endotoxin tolerance. J Biol Chem. 2008;283:3109–3119. doi: 10.1074/jbc.M707400200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Medvedev AE, Lentschat A, Kuhns DB, Blanco JC, Salkowski C, Zhang S, Arditi M, Gallin JI, Vogel SN. Distinct mutations in IRAK-4 confer hyporesponsiveness to lipopolysaccharide and interleukin-1 in a patient with recurrent bacterial infections. J Exp Med. 2003;198:521–531. doi: 10.1084/jem.20030701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McIntire FC, Sievert HW, Barlow GH, Finley RA, Lee AY. Chemical, physical, biological properties of a lipopolysaccharide from Escherichia coli K-235. Biochemistry. 1967;6:2363–2372. doi: 10.1021/bi00860a011. [DOI] [PubMed] [Google Scholar]
- 43.Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Visintin A, Latz E, Monks BG, Espevik T, Golenbock DT. Lysines 128 and 132 enable lipopolysaccharide binding to MD-2, leading to Toll-like receptor-4 aggregation and signal transduction. J Biol Chem. 2003;278:48313–48320. doi: 10.1074/jbc.M306802200. [DOI] [PubMed] [Google Scholar]
- 45.Chen H, Cowan MJ, Hasday JD, Vogel SN, Medvedev AE. Tobacco smoking inhibits expression of proinflammatory cytokines and activation of IL-1R-associated kinase, p38, and NF-kappaB in alveolar macrophages stimulated with TLR2 and TLR4 agonists. J Immunol. 2007;179:6097–6106. doi: 10.4049/jimmunol.179.9.6097. [DOI] [PubMed] [Google Scholar]
- 46.Piao W, Song C, Chen H, Diaz MQ, Wahl LM, Fitzgerald KA, Li L, Medvedev AE. Endotoxin tolerance dysregulates MyD88- and Toll/IL-1R domain-containing adapter inducing IFN-β-dependent pathways and increases expression of negative regulators of TLR signaling. J Leukoc Biol. 2009;86:863–875. doi: 10.1189/jlb.0309189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 48.Du X, Poltorak A, Silva M, Beutler B. Analysis of Tlr4-mediated LPS signal transduction in macrophages by mutational modification of the receptor. Blood Cells Mol Dis. 1999;25:328–338. doi: 10.1006/bcmd.1999.0262. [DOI] [PubMed] [Google Scholar]
- 49.Kalis C, Kanzler B, Lembo A, Poltorak A, Galanos C, Freudenberg MA. Toll-like receptor 4 expression levels determine the degree of LPS-susceptibility in mice. Eur J Immunol. 2003;33:798–805. doi: 10.1002/eji.200323431. [DOI] [PubMed] [Google Scholar]
- 50.Chow JC, Young DW, Golenbock DT, Christ WJ, Gusovsky F. Toll-like receptor-4 mediates lipopolysaccharide-induced signal transduction. J Biol Chem. 1999;274:10689–10692. doi: 10.1074/jbc.274.16.10689. [DOI] [PubMed] [Google Scholar]
- 51.Shimazu R, Akashi S, Ogata H, Nagai Y, Fukudome K, Miyake K, Kimoto M. MD-2, a molecule that confers lipopolysaccharide responsiveness on Toll-like receptor 4. J Exp Med. 1999;189:1777–1782. doi: 10.1084/jem.189.11.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zanoni I, Ostuni R, Marek LR, Barresi S, Barbalat R, Barton GM, Granucci F, Kagan JC. CD14 controls the LPS-induced endocytosis of Toll-like receptor 4. Cell. 2011;147:868–880. doi: 10.1016/j.cell.2011.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wright SD, Ramos RA, Tobias PS, Ulevitch RJ, Mathison JC. CD14, a receptor for complexes of lipopolysaccharide (LPS) and LPS binding protein. Science. 1990;249:1431–1433. doi: 10.1126/science.1698311. [DOI] [PubMed] [Google Scholar]
- 54.Latz E, Visintin A, Lien E, Fitzgerald KA, Monks BG, Kurt-Jones EA, Golenbock DT, Espevik T. Lipopolysaccharide rapidly traffics to and from the Golgi apparatus with the toll-like receptor 4-MD-2-CD14 complex in a process that is distinct from the initiation of signal transduction. J Biol Chem. 2002;277:47834–47843. doi: 10.1074/jbc.M207873200. [DOI] [PubMed] [Google Scholar]
- 55.Kawai T, Adachi O, Ogawa T, Takeda K, Akira S. Unresponsiveness of MyD88-deficient mice to endotoxin. Immunity. 1999;11:115–122. doi: 10.1016/s1074-7613(00)80086-2. [DOI] [PubMed] [Google Scholar]
- 56.Raingeaud J, Gupta S, Rogers JS, Dickens M, Han J, Ulevitch RJ, Davis RJ. Pro-inflammatory cytokines and environmental stress cause p38 mitogen-activated protein kinase activation by dual phosphorylation on tyrosine and threonine. J Biol Chem. 1995;270:7420–7426. doi: 10.1074/jbc.270.13.7420. [DOI] [PubMed] [Google Scholar]
- 57.Tsien RY. The green fluorescent protein. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 58.Park BS, Song DH, Kim HM, Choi BS, Lee H, Lee JO. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature. 2009;458:1191–1195. doi: 10.1038/nature07830. [DOI] [PubMed] [Google Scholar]
- 59.Godaly G, Young DB. Mycobacterium bovis bacille Calmette Guerin infection of human neutrophils induces CXCL8 secretion by MyD88-dependent TLR2 and TLR4 activation. Cell Microbiol. 2005;7:591–601. doi: 10.1111/j.1462-5822.2004.00489.x. [DOI] [PubMed] [Google Scholar]
- 60.Walsh DE, Greene CM, Carroll TP, Taggart CC, Gallagher PM, O’Neill SJ, McElvaney NG. Interleukin-8 up-regulation by neutrophil elastase is mediated by MyD88/IRAK/TRAF-6 in human bronchial epithelium. J Biol Chem. 2001;276:35494–35499. doi: 10.1074/jbc.M103543200. [DOI] [PubMed] [Google Scholar]
- 61.Hoshino K, Takeuchi O, Kawai T, Sanjo H, Ogawa T, Takeda Y, Takeda K, Akira S. Cutting edge: Toll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipopolysaccharide: evidence for TLR4 as the Lps gene product. J Immunol. 1999;162:3749–3752. [PubMed] [Google Scholar]
- 62.Medvedev AE, Vogel SN. Overexpression of CD14, TLR4, and MD-2 in HEK293T cells does not prevent induction of in vitro endotoxin tolerance. J Endotoxin Res. 2003;9:60–64. doi: 10.1179/096805103125001360. [DOI] [PubMed] [Google Scholar]
- 63.Nagpal K, Plantinga TS, Wong J, Monks BG, Gay NJ, Netea MG, Fitzgerald KA, Golenbock DT. A TIR domain variant of MyD88 adapter-like (Mal)/TIRAP results in loss of MyD88 binding and reduced TLR2/TLR4 signaling. J Biol Chem. 2009;284:25742–25748. doi: 10.1074/jbc.M109.014886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Prohinar P, Rallabhandi P, Weiss JP, Gioannini TL. Expression of functional D299G.T399I polymorphic variant of TLR4 depends more on coexpression of MD-2 than does wild-type TLR4. J Immunol. 2010;184:4362–4367. doi: 10.4049/jimmunol.0903142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang Y, Liu B, Dai J, Srivastava PK, Zammit DJ, Lefrançois L, Li Z. Heat shock protein gp96 is a master chaperone for toll-like receptors and is important in the innate function of macrophages. Immunity. 2007;26:215–226. doi: 10.1016/j.immuni.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu B, Yang Y, Qiu Z, Staron M, Hong F, Li Y, Wu S, Li Y, Hao B, Bona R, Han D, Li Z. Folding of Toll-like receptors by the HSP90 paralogue gp96 requires a substrate-specific cochaperone. Nat Commun. 2010;1:1–10. doi: 10.1038/ncomms1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wakabayashi Y, Kobayashi M, Akashi-Takamura S, Tanimura N, Konno K, Takahashi K, Ishii T, Mizutani T, Iba H, Kouro T, Takaki S, Takatsu K, Oda Y, Ishihama Y, Saitoh S, Miyake K. A protein associated with toll-like receptor 4 (PRAT4A) regulates cell surface expression of TLR4. J Immunol. 2006;177:1772–1779. doi: 10.4049/jimmunol.177.3.1772. [DOI] [PubMed] [Google Scholar]
- 68.Nagai Y, Akashi S, Nagafuku M, Ogata M, Iwakura Y, Akira S, Kitamura T, Kosugi A, Kimoto M, Miyake K. Essential role of MD-2 in LPS responsiveness and TLR4 distribution. Nat Immunol. 2002;3:667–672. doi: 10.1038/ni809. [DOI] [PubMed] [Google Scholar]
- 69.Kagan JC, Su T, Horng T, Chow A, Akira S, Medzhitov R. TRAM couples endocytosis of Toll-like receptor 4 to the induction of interferon-beta. Nat Immun. 2008;9:361–368. doi: 10.1038/ni1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, Coyle AJ, Liao SM, Maniatis T. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol. 2003;4:491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.