Abstract
Proteomics analysis of biological samples has the potential to identify novel protein expression patterns and/or changes in protein expression patterns in different developmental or disease states. An important component of successful proteomics research, at least in its present form, is to reduce the complexity of the sample if it is derived from cells or tissues. One method to simplify complex tissues is to focus on a specific, highly purified sub-proteome. Using this approach we have developed methods to prepare highly enriched fractions of the apical plasma membrane of the syncytiotrophoblast. Through proteomics analysis of this fraction we have identified over five hundred proteins several of which were previously not known to reside in the syncytiotrophoblast. Herein, we focus on two of these, dysferlin and myoferlin. These proteins, largely known from studies of skeletal muscle, may not have been found in the human placenta were it not for discovery-based proteomics analysis. This new knowledge, acquired through a discovery-driven approach, can now be applied for the generation of hypothesis-based experimentation. Thus discovery-based and hypothesis-based research are complimentary approaches that when coupled together can hasten scientific discoveries.
Keywords: Placenta, Proteomics, Discovery-based research
1 Introduction
Protein expression patterns in cells and tissues are characteristic of their developmental, physiological, or pathological states. This being the case, it is increasingly important to determine the protein expression profile and to determine how the profile may vary from non-pathological conditions to specific states of disease. Such knowledge will lead to deeper insights into disease processes and potentially to the identification of proteins that can serve as biomarkers, providing early warning of disease paving the way to potential new therapies. The completion of the Human Genome Project coupled with advances in bioinformatics and proteomics-related technologies have enabled rapid advances in proteomics research.
The term proteomics covers a variety of methodologies and procedures that are aimed at the identification of proteins and ideally their quantification in specific biological samples. Monitoring posttranslational protein modifications that occur in many proteins which may be crucial for regulation of protein function is also a component of a complete proteomics analysis. While the Human Genome Project identified about 25,000 genes, it has been estimated that the repertoire of protein expression may be over one million [1]. This complexity provides challenges in deciphering the proteome in cells and tissues.
The human placenta is a complex and vital organ that mediates the selective transfer of solutes and gasses between mother and fetus. Additionally, the placenta produces hormones and other factors that support pregnancy and provides a barrier to the maternal immune system. The placenta employs a branching villous system to provide a large surface area for potential exchange with maternal blood that bathes these villous structures. The interface between the placenta and maternal blood is a highly specialized epithelium known as the syncytiotropholbast (STB). The apical aspect of the STB contains microvilli (MV) that further increase the absorptive surface of the placenta. The apical plasma membrane of the STB is thus of crucial importance to the function of the placenta. Nevertheless, a catalog of the full complement of proteins that reside in this membrane does not yet exist.
Proteomics analysis of the human placenta, whether normal or diseased, is at an early stage of development. There have been relatively few proteomics studies of the placenta or cell lines that are often used in place of the placenta. We have reviewed these studies recently [2]. Considering how little work has been done to date, proteomics analysis of the placenta is ripe for exploration. The discovery-based nature of much of proteomics research may not be appealing to all investigators. Those that only consider hypothesis-driven research as appropriate may regard discovery-based research in a less favorable light. We take the view that discovery-based proteomics can lead to new hypothesis-based research that may not have been conceived in the absence of the proteomics data. Proteomics analyses that yield new and novel information can thereby provide a “shortcut” to obtaining new biological insight.
2. Placental Proteomics and the Cycle of Knowledge
Scientific progress, as it applies to an individual, group, or field, may be viewed as occurring through a process akin to “punctuated equilibrium”: Periods of sluggish growth are interspersed with phases of saltatory advances. It has long been known that scientific developments are stimulated by the development of new technologies. Even in 1865, acclaimed French physiologist Claude Bernard noted: “to extend his knowledge, [mankind] has to increase the power of his organs by means of special appliances… [thereby] enabling him to penetrate inside of bodies, to dissociate them and study their hidden parts” [3]. To illustrate how technological advances may expedite scientific progress, consider the quest that Than, Bohn, and Szabo undertook to systematically characterize and catalog their seminal series of pregnancy-related proteins (PRPs). Using biochemical and immunological methods available in the era spanning the early 1970s to the early 1990s, roughly two decades were required to describe over fifty PRPs [4]. It is therefore something of a wonder to consider that large-scale mass spectrometry is now capable of identifying hundreds of proteins in a span of days to weeks. Proteomics analysis is therefore suitably poised to greatly extend this prior work, and application of this methodology may well lead to the identification of additional PRPs
The value of systematic data-gathering, such as the pursuit of PRPs, should not be undervalued. Since their discovery, PRPs have been important for both basic and clinical aspects of oncodevelopment and perinatal medicine [5]; the diagnostic and therapeutic applications of these analytes remain a source of active investigation [6-8]. However, to the defiance of reason, such discovery-based initiatives are sometimes viewed disparagingly when compared to hypothesis-driven research [9]. Insofar as scientific research necessarily places investigators at a nexus between an infirm foundation of understanding and the seemingly infinite territory of the unknown, it is unrealistic to assume that there should exist any singular approach to gaining scientific insight. Any empirical query necessitates the development of a critical reservoir of knowledge, requiring the input of large amounts of both observational and experimental data (Figure 1). It is only after the development of a suitably informed knowledge base that cogent hypotheses can be developed and subjected to rigorous testing. Proteomics seems ideal for increasing our knowledge base related to the placenta and pregnancy and simultaneously, informing the hypotheses that underpin research at a basic level. As such, proteomics offers great potential to propel forward the “cycle of knowledge” in coming years.
Figure 1.
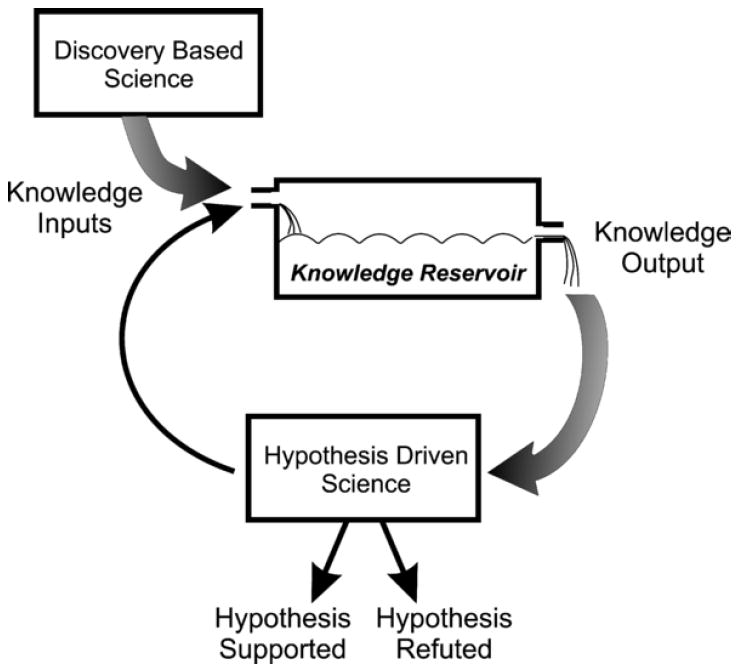
A diagram that illustrates the complementary roles of discovery-based and hypothesis-based research to the cycle of knowledge as applied to biomedical research.
3. The Importance of Sample Preparation for Proteomics Analysis
At the present time there is no single proteomics platform that can adequately obtain the proteome (identification and quantification of all of the proteins and their isoforms) of a structure as complex as an organ or an organism [10]. The dynamic range of protein expression is an important barrier that must be considered in proteomics studies. For example, in serum the protein copy number can range over 12 orders of magnitude [11] and in cells can range over 7-8 orders of magnitude [12]. Indeed this has been referred to as the “dynamic range problem” in the proteomics literature [13, 14], and becomes an important issue considering that low abundance proteins may not be well represented in the proteomics analysis. Furthermore, in many instances lower abundance proteins may be the ones of most biological and pathophysiological interest so it is crucial that such proteins are not excluded from the analysis.
How then can the problem of dynamic range of protein expression be addressed? Generally this can be accomplished by pre-fractionation of complex mixtures to enrich for a subset of the original. For example, to define the proteome of the nucleolus the nucleoli should be enriched to minimize contamination of other cellular components, as has been done [15]. Pre-fractionation has similarly yielded a high-resolution compendium of the mitochondrial proteome [16]. The complement of proteins in organelles such as the nucleolus and mitochondrion represent distinct sub-proteomes of the cell. While our own interest has been in defining the proteome of the human placenta, it remains that the placenta is a complex structure composed of multiple cell and tissue types. Moreover, the placenta contains both maternal and fetal blood which would confound a proteomics analysis of the placenta itself.
With these considerations in mind, it is reasonable to assume that analysis of the placental proteome, using currently available technology, can best be accomplished by dividing the placenta into sub-proteomes. This simplification strategy serves to increase the probability of identifying lower copy number proteins. Several methods, used individually or in combination, have been used to fractionate cells or tissues into components more suitable for proteomics study than the starting material [17-20]. We have focused our attention on the sub-proteome of the apical plasma membrane of the STB. In order to carry out meaningful proteomics analysis, a reliable method for enriching this membrane is a prerequisite. Other workers have developed methods for enriching a fraction from the apical surface of the STB. Thirteen of these studies have been reviewed and the average enrichment was ~ 20-fold with a range of 14- to 37-fold [21]. The membrane vesicles isolated in these studies were primarily used for transport studies for which this level of enrichment appears adequate. However, we realized that a greater enrichment of the apical plasma membrane of the STB would be necessary for a proteomics analysis of this membrane.
How can the apical plasma membrane of the STB be enriched more than 20-fold? A strategy, developed by Jacobson and colleagues, was to increase the density of the plasma membrane by attaching cationic colloidal silica (CCS) particles to its surface [22, 23]. This method was developed for isolating the plasma membrane of cells in culture where the plasma membrane would be easily assessable to the CCS. Following binding of the CCS particles, the cells could be disrupted and the CCS-coated membranes enriched through a series of centrifugation steps [22]. It occurred to us that if both the basal plate of the placenta was dissected away and remaining tissue was washed free of blood then the apical plasma membrane of the STB would also be accessible to CCS particles. This strategy would likely provide a suitable method for the enrichment of this specific plasma membrane.
An experiment was carried out to determine if the CCS-method of Jacobson was applicable to the human placenta. The assay used to determine if the CCS particles bind to the apical plasma membrane of the STB was electron microscopy; this could be done since the particles have inherent electron density and could be monitored by conventional transmission electron microscopy. We found that CCS particle did bind to the apical plasma membrane of the STB; however the binding was restricted to tips of the MV with very little binding to the lateral sides of the MV or to the planar aspect of this membrane at the base of MV (Fig. 2). These results suggested two possible explanations: (1) only the tips of the MV have CCS binding sites or (2) the CCS particles could not readily gain access to portions of this plasma membrane beyond the tips of the MV. In either case, this original CCS method was inadequate for isolation of the entire apical plasma membrane of the STB.
Figure 2.
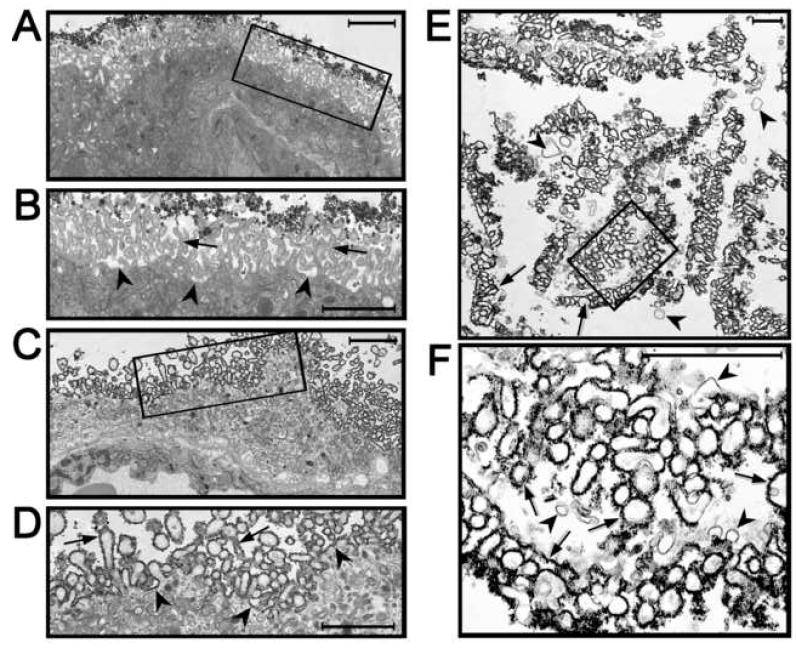
Coating of the apical plasma membrane of the STB with cationic colloidal silica and isolation of microvilli—analysis by electron microscopy. (A) Low magnification electron micrograph of placental tissue incubated with conventional CCS particles. Bar = 2 μm. (B) Higher magnification electron micrograph of the region of panel A enclosed by the rectangle. The CCS particles are present at the tips of the MV but are not present on the lateral sides of the MV (arrows) or the planar basal aspect of the apical plasma membrane (arrowheads). Bar = 2 μm. (C) Low magnification electron micrograph of placental tissue incubated with smCCS particles. Bar = 2 μm. (D) Higher magnification electron micrograph of the region of panel C enclosed by the rectangle. The smCCS particles are present at the tips of the MV and the lateral sides of the MV (arrows) as well as the planar basal aspect of the apical plasma membrane (arrowheads). Bar = 2 μm. (E) Low magnification electron micrograph of the isolated STB MV coated with smCCS. The microvillous appearance of the smCCS-coated membranes is retained (arrows). Some uncoated membrane vesicles are also present (arrowheads). Bar = 1 μm. (F) Higher magnification electron micrograph of the region of panel E in the rectangle. The smCCS particles heavily coat the extracellular face of the apical plasma membrane of the STB (arrows). Most of the membranes are coated with smCCS with only a low level of uncoated membranes being present (arrowheads). Bar = 1μm. These electron micrographs are reproduced with permission of Placenta.
To circumvent this problem, we developed an alternative procedure that utilized smaller and more uniformly sized silica particles that we have termed smCCS. We found that these particles coated the apical plasma membrane of the MV uniformly. Thus the entire apical plasma membrane of the STB has sites that will bind the cationic silica if they can gain access (Fig. 2). Once coated with smCCS, the MV could be enriched from homogenates of placenta. We have used the smCCS in our proteomics studies; however the larger CCS particles may have applicability to the placenta if one were interested in isolating the tips of the MV. Figure 3 shows the outline of the procedure used to isolate MV from the human placenta. Electron micrographs of material so isolated showed that it was highly enriched in smCCS-coated membrane with relatively little uncoated membrane contaminants (Fig. 2). In addition, the isolated MV retained morphological features reminiscent of MV in vivo.
Figure 3.
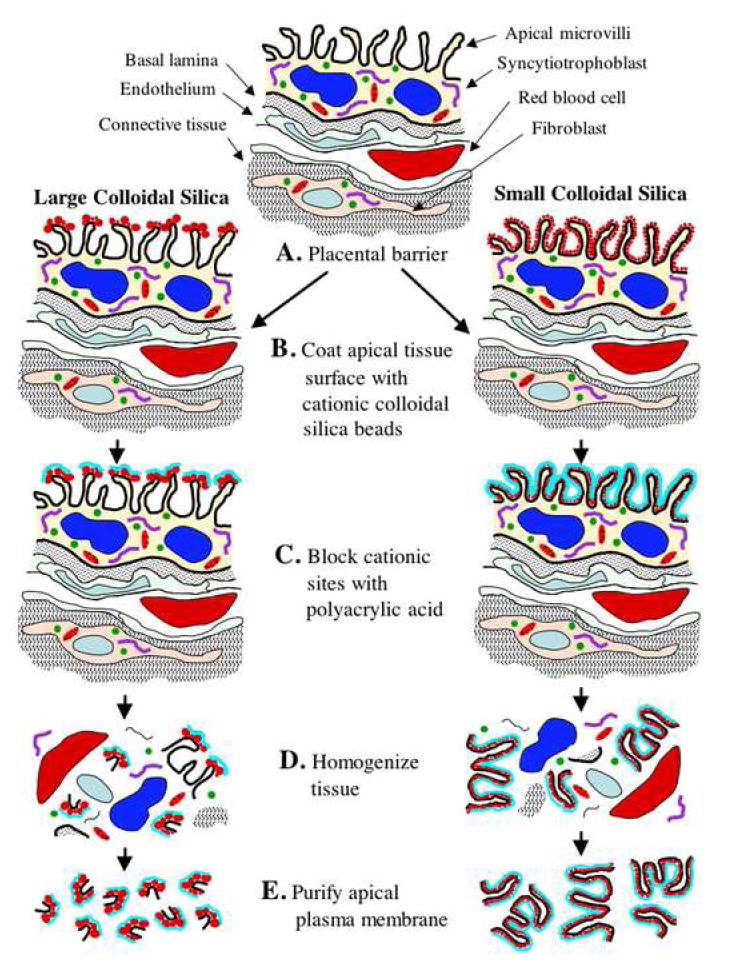
Diagram summarizing the method employed to isolate the MV fraction from the apical aspect of the STB.
Our initial characterization of the isolated MV was to compare the crude tissue homogenate and the isolated MV by 2-dimensional gel electrophoresis (2-DGE) over a broad pH range (pH 3.5 – pH 9.0). We found some dramatic differences in the spot patterns in these 2-DGE (Fig. 4). A subset of these spots were excised form the gels and identified by matrix assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF). This was a useful first step in the proteomics analysis. For example, we found that histones were significantly reduced in the MV fraction compared to the starting material. We also found enrichment for actin and ezrin in the MV fraction an expected finding since these two proteins are major components of MV. Thus the 2-DGE approach illustrated that the MV fraction was depleted of some proteins found in the starting material while others were enriched in the MV preparations. However, we abandoned the 2-DGE approaches to defining the proteome of the apical plasma membrane of the STB for several reasons. First, while numerous spots were observed in the 2-DGE preparations, they did not seem sufficiently numerous to represent the entire proteome of the MV. Secondly, to indentify these proteins one spot at a time was very labor intensive and expensive. Thirdly, we were particularly interested in identifying plasma membrane proteins and membrane proteins are not well represented in conventional 2-DGE [24]. Nevertheless these initial results suggested we were on the right track for enriching MV from the placenta. However, these results also illustrated that our MV preparations were enriched for cytoskeletal elements associated with MV. This, we reasoned, presented problems for the proteomics analysis of bona fide plasma membrane proteins.
Figure 4.
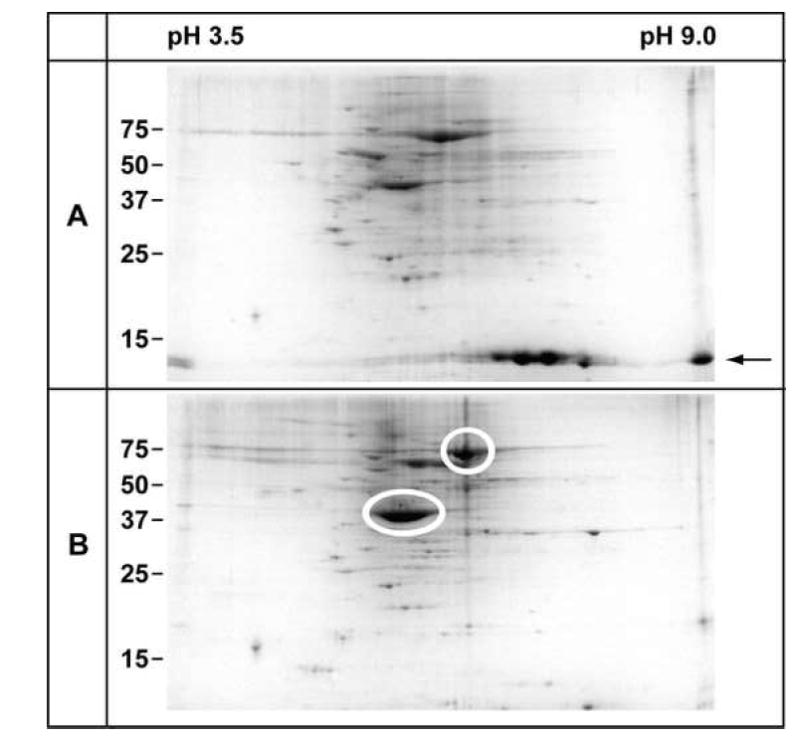
Reduction of some proteins and enrichment for others in preparation of the MV fraction. (A) A survey 2-DGE of the crude tissue homogenate. Prominent spots in the lower portion of the gel were excised, processed for MALDI-TOF mass spectrometry analysis, and identified as histones: H2A, H2B, and H4 (arrow). (B) A survey 2-DGE of the MV fraction. The histones have been largely depleted from this fraction while other proteins such as ezrin (upper circle) and actin (lower circle) have been enriched.
To assess the enrichment of the MV fraction we utilized an immunoblot procedure in which antibodies to a variety of marker proteins for different components of cells of the placenta were employed. In this way, we could compare the crude tissue homogenate and the isolated MV to assess the depletion of or enrichment for the various markers for the cytoskeleton, cellular organelles, and plasma membranes. We found that the protein placental alkaline phosphatase (PLAP), used as a marker for the apical plasma membrane of the STB, was significantly enriched in the MV fraction. The between experiment variation for enrichment of PLAP ranged from 200- to 400-fold [Robinson et al., in preparation], indicating that we had achieved a much higher level of enrichment than the ~ 20-fold enrichment reported by others (see above).
While a 200- to 400-fold enrichment for a marker of the STB plasma membrane represented a dramatic improvement, we remained concerned about the presence of potentially contaminating non-membrane proteins in the MV fraction. These proteins, such as actin which were present in high levels, might adversely affect the proteomics analysis of membrane proteins since membrane proteins are often expressed at low copy number [24]. Our strategy for depletion of non-membrane proteins from the MV fraction was to use conventional biochemical methods to disrupt protein-protein interactions. This consisted of the sequential treatment of the MV fraction to: (1) low salt; (2) high salt; (3) high pH; and (4) urea. This resulted in a further depletion of ~80% of the protein from the sample, or conversely 20% retention of the original amount of protein (Robinson et al., in preparation). When this is coupled with the original 200- to 400-fold enrichment, a final enrichment of 1,000- to 2,000-fold was achieved. It was this material that was used for the proteomics analysis.
The final extracted membrane-enriched fraction was solubilized in SDS and separated using a 1-dimensional polyacrylamide gel. Individual gel slices were excised and peptide mixtures from each gel slice were prepared for analysis using tandem mass spectrometry (MS/MS). A major advantage of MS/MS is that it offers a much higher throughput when compared to the analysis of one spot at a time when 2-DGE was coupled to MALDI-TOF analysis. Further, membrane proteins were readily resolved in the 1-dimensional SDS gel system.
4. Proteomics Analysis of the Apical Plasma Membrane of the STB
Over five hundred proteins were identified in the extracted smCCS-coated apical plasma membrane fraction derived from the STB. A full description of these results will appear in an original research publication [Vandré et al., in preparation]. In this review, a subset of the proteomics data set has been categorized to illustrate some important aspects of the proteomics results (Table 1). Reassuringly, we identified several proteins that were known from previous studies to be components of the apical plasma membrane of the STB. One of these, PLAP, is considered the standard marker protein for this membrane. Had we not identified numerous proteins known to be in this plasma membrane, our data would have been highly suspect. Most importantly, we also identified a set of proteins that had not been previously identified as components of the human placenta. In this review, we focus our attention on two of these proteins, namely, dysferlin (DYSF) and myoferlin (MYOF). A consideration of DYSF and MYOF is given below. Interestingly, we have identified a set of proteins whose function is not yet known but which were identified through the human genome project. Perhaps some of these proteins will turn out to be critically important in the placenta. The last category of proteins listed in Table 2 we have called “contaminating” non-integral membrane proteins. This subset also contains proteins that were not previously known to be expressed in the human placenta (e.g., CLIP-170; ELMO domain containing protein 2). Another interesting feature is that evidence has been presented in the literature that some members of this subset may be associated with plasma membranes; see Table 1 for specific references. We do not know as yet if any of these proteins are in fact associated with the apical plasma membrane of the STB or whether they are contaminants in our membrane fraction. If they turn out to be contaminants, it would suggest that our extraction procedure that was designed to remove non-membrane proteins is in need of further refinements.
Table 1.
An example of a classification procedure for proteins that were identified in the proteomics analysis of the apical plasma membrane of the STB.
| Characterization of proteomics data from the apical plasma membrane of the STB |
|---|
| Proteins previously know to be in the apical plasma membrane of the STB (examples) |
| PLAP |
| EGF receptor |
| Transferrin receptor |
| 5’-nucleotidase |
|
|
| Proteins previously not known to be in the apical plasma membrane of the STB (examples) |
| Dysferlin |
| Myoferlin |
| Stomatin |
| Flotillins 1 and 2 |
|
|
| Proteins known from the human genome but which have not been assigned a function (examples) |
| Hypothetical protein LOC64755 |
| Hypothetical protein LOC57003 |
| Transmembrane protein 43 |
| Transmembrane protein 85 |
|
|
| Proteins that may be “contaminating” non-integral membrane proteins (examples) |
| Dynein (cytoplasmic but also reported as membrane) [35] |
| ELMO domain containing protein 2 (cytoplasmic but also reported as membrane) [36] |
| HSP 27 (cytoplasmic but also reported as membrane) [37] |
| CLIP-170 (cytoplasmic but also reported as membrane) [38] |
The proteomics data obtained from tandem mass spectrometry analysis of tryptic digests of the proteins present in the smCCS-coated apical plasma membrane, following the extraction procedure, was in the form of a list of peptides and the identities of the proteins where such peptides are found. It is generally considered sufficient to identify two unique peptides in a protein to make definitive protein identifications. The five proteins with the greatest number of unique peptides in our data set were: DYSF, MYOF, transferrin receptor, PLAP, and assembly protein 50 with 41, 40, 35, 32, and 19 unique peptides respectively. It is interesting to note that the two proteins with the greatest number of unique peptides identified in our analysis are from the same family of proteins and were not known to be in the placenta until our proteomics analysis. These proteins, DYSF and MYOF, are in the ferlin family and are also known as the ferlin-like proteins (FerL1 and FerL3, respectively) [25]. We have shown that DYSF and MYOF primarily localize to the apical plasma membrane of the STB in immunofluorescence microscopy preparations (Fig. 5) [26]. These proteins have been shown to be of considerable important in muscle biology and pathobiology. Mutated forms of DYSF lead to the dysferlinopathies, limb girdle muscular dystrophy type 2B and Myoshi’s myopathy [27, 28]. Subsequent studies provided strong evidence for DYSF being important for the repair of damaged plasma membranes in skeletal muscle [29, 30]. More recently evidence has been presented for DYSF being important in maintenance of the plasma membranes of cardiomyocytes [31]. Myoferlin has been associated with fusion in the formation of myotubes [32]. Additionally, MYOF has also been implicated in the repair of the plasma membrane in cultured endothelial cells [33]. The next most abundant unique peptides belong to the transferrin receptor and to PLAP, the quintessential marker protein for the apical plasma membrane of the STB. Assembly protein 50 which is a component of the clathrin pit assembly complex also had numerous unique peptides identified in this analysis. Electron microscopy has shown that clathrin-coated pits are abundant in the apical plasma membrane of the STB.
Figure 5.
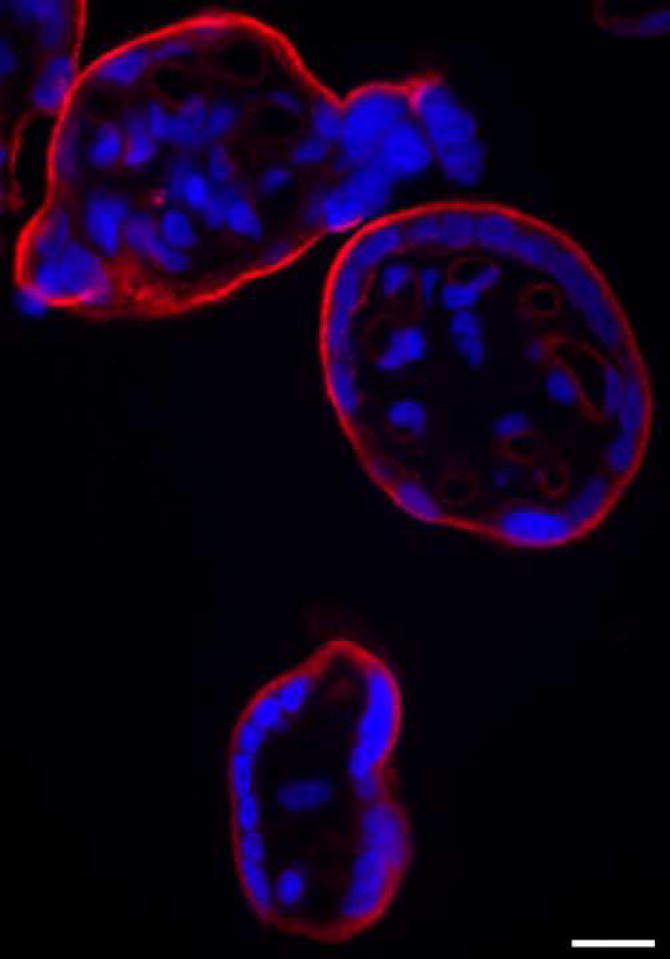
Immunofluorescence localization of DYSF in term placenta. Dysferlin (red fluorescence) is primarily localized to the apical plasma membrane of the STB. The nuclei are stained with DAPI (blue fluorescence). Bar = 20 μm.
Strong evidence has been presented for DYSF serving as a plasma membrane repair protein in skeletal muscle [29,30]; its role in placental biology is not presently known. We hypothesize that DYSF also serves as a plasma membrane repair protein in the placenta. Its location in the apical plasma membrane of the STB is consistent with that role. Experiments are currently underway to test this hypothesis. Preeclampsia is associated with shedding of membrane fragments from the apical plasma membrane of the STB. These fragments, or microparticles, contribute to the pathophysiology associated with preeclampsia [34]. This shedding of fragments is suggestive of plasma membrane instability. It is tempting to speculate that disturbances in DYSF concentration or function may contribute to this membrane instability. Thus a carefully controlled study of preeclamptic and gestational-age matched normal placentas to investigate the potential role of DYSF in this disease process is warranted.
For the purposes of this review, the identification of DYSF and MYOF in the apical plasma membrane illustrates an important point concerning the great value that proteomics can provide. When our proteomics experiments began, DYSF and MYOF were known to be in skeletal muscle [27, 28]. There would have been no logical reason to search for these proteins in the placenta using techniques suitable for identifying individual proteins, such as immunoblotting. Therefore the presence of these proteins in the human placenta may have remained unknown were it not for a proteomics analysis of a fraction derived from the apical plasma membrane of the STB from that tissue. While a proteomics screen of a biological sample may not be considered as hypothesis-driven research by some, it may nevertheless provide unique information which can lead to subsequent hypothesis-driven research. We contend that our proteomics analysis of the apical plasma membrane of the STB did precisely that; it identified these two proteins (and others not discussed in this review) that were heretofore not known to be expressed in the human placenta. Once identified as being present in the placenta, and at apparently high levels of expression, DYSF and MYOF offer the opportunity for the design of numerous hypothesis-driven experiments. Thus proteomics screens can open areas of research that would not have been contemplated in the absence of the proteomics data. This is one of the great values inherent in proteomics research.
5. Conclusions
The importance of discovery-based proteomics research and how new insights into biological processes can be obtained from such a proteomics approach form the backdrop of this paper. An equally important thread that was discussed is the importance of sample preparation in obtaining meaningful proteomics data. The discovery that DYSF and MYOF are prominent components of the apical plasma membrane of the STB may have occurred through hypothesis-based research. However, this is not certain nor is it known when that might have occurred. Proteomics analysis of a highly enriched preparation of this membrane did reveal the presence of these two related proteins. This “shortcut” proteomics method identified these proteins and now sets the stage for hypothesis-driven experiments directed toward determining the function of these ferlin molecules in the placenta in normal and diseased states.
Acknowledgments
Work form the author’s laboratories was supported in part by grants HD38764 and HD49628 from the National Institutes of Health.
Footnotes
Conflict of Interest
The author’s have no conflicts of interest related to this manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Huber LA. Is proteomics heading in the wrong direction? Nature Rev Mol Cell Biol. 2003;4:74–80. doi: 10.1038/nrm1007. [DOI] [PubMed] [Google Scholar]
- 2.Robinson JM, Ackerman WE, Kniss DA, Takizawa T, Vandré DD. Proteomics of the human placenta: promises and realities. Placenta. 2008;29:135–143. doi: 10.1016/j.placenta.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernard C, Greene HC, Henderson LJ. 1957. An Introduction to the Study of Experimental Medicine, Courier Dover Publications. [Google Scholar]
- 4.Than GN, Bohn H, Szabo DG. Informa Health Care. 1993. Advances in Pregnancy-Related Protein Research: Functional and Clinical Applications. [Google Scholar]
- 5.Than NG, Itakura A, Rao CV, Nohira T, Toth P, Mansell JP, et al. Clinical applications of pregnancy-related proteins--a workshop report. Placenta. 2003;24(Suppl A):S60–4. doi: 10.1053/plac.2002.0947. [DOI] [PubMed] [Google Scholar]
- 6.Nicolaides KH, Bindra R, Turan OM, Chefetz I, Sammar M, Meiri H, et al. A novel approach to first-trimester screening for early pre-eclampsia combining serum and Doppler ultrasound. Ultrasound Obstet Gynecol. 2006;27:13–17. doi: 10.1002/uog.2686. [DOI] [PubMed] [Google Scholar]
- 7.Spencer K, Cowans NJ, Chefetz I, Tal J, Meiri H. First-trimester maternal serum PP-13, PAPP-A and second-trimester uterine artery Doppler pulsatility index as markers of pre-eclampsia. Ultrasound Obstet Gynecol. 2007;29:128–34. doi: 10.1002/uog.3876. [DOI] [PubMed] [Google Scholar]
- 8.Romero R, Kusanovic JP, Than NG, Erez O, Gotsch F, Espinoza J, et al. First-trimester maternal serum PP13 in the risk assessment for preeclampsia. 1: Am J Obstet Gynecol. 2008;199:122.e1–122.e11. doi: 10.1016/j.ajog.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kell DB, Oliver SG. Here is the evidence, now what is the hypothesis? The complimentary roles of inductive and hypothesis-driven science in the post-genomic era. BioEssay. 2004;26:99–105. doi: 10.1002/bies.10385. [DOI] [PubMed] [Google Scholar]
- 10.Rice GE, Georgiou HM, Ahmed N, Shi G, Kruppa G. Translational proteomics: Developing a predictive capacity-a review. Placenta. 2006;27(Suppl A):S76–86. doi: 10.1016/j.placenta.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 11.Anderson NL, Anderson NG. Proteome and proteomics: new technologies, new concepts, and new ideas. Electrophoresis. 1998;19:1853–1861. doi: 10.1002/elps.1150191103. [DOI] [PubMed] [Google Scholar]
- 12.Corthals GL, Wasinger VC, Hochstrasser DF, Sanchez J-C. The dynamic range of protein expression: a challenge for proteomics research. Electrophoresis. 2000;21:1104–1115. doi: 10.1002/(SICI)1522-2683(20000401)21:6<1104::AID-ELPS1104>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 13.Wu L, Han DK. Overcoming the dynamic range problem in mass spectrometry-based shotgun proteomics. Expert Rev Proteomics. 2006;3:611–619. doi: 10.1586/14789450.3.6.611. [DOI] [PubMed] [Google Scholar]
- 14.Smalley DM, Ley K. Plasma-derived microparticles for biomarker discovery. Clin Lab. 2008;54:67–79. [PubMed] [Google Scholar]
- 15.Anderson JS, Lam YW, Leung AK, Ong SE, Lyon CE, et al. Nucleolar proteome dynamics. Nature. 2005;433:77–83. doi: 10.1038/nature03207. [DOI] [PubMed] [Google Scholar]
- 16.Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong S-E, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stasyk LF, Huber LA. Zooming in: fractionation strategies in proteomics. Proteomics. 2004;4:3704–16. doi: 10.1002/pmic.200401048. [DOI] [PubMed] [Google Scholar]
- 18.Dreger M. Subcellular proteomics. Mass Spectrometry Rev. 2003;22:27–56. doi: 10.1002/mas.10047. [DOI] [PubMed] [Google Scholar]
- 19.Righetti PG, Castagna A, Hebert B, Reymond F, Rossier JS. Prefractionation techniques in proteome analysis. Proteomics. 2003;3:1397–407. doi: 10.1002/pmic.200300472. [DOI] [PubMed] [Google Scholar]
- 20.Au CE, Bell AW, Gilchrist A, Hiding J, Nilsson T, Bergeron JJM. Cellular proteomics to create the cell map. Curr Opin Cell Biol. 2007;19:376–85. doi: 10.1016/j.ceb.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 21.Jimenez V, Henriquez M, Llanos P, Riquelme G. Isolation and purification of human plasma membrane for normal and pre-eclamptic pregnancies. Placenta. 2004;25:422–37. doi: 10.1016/j.placenta.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 22.Jacobson BS, Stolz DE, Schnitzer JE. A laboratory manual. Cold Spring Harbor Laboratory Press; 1998. Plasma membrane isolation using the cationic colloidal silica isolation technique; pp. 35.1–35.14. [Google Scholar]
- 23.Durr E, Yu J, Krasinska KM, Carver LA, Yates JR, III, Testa JE, et al. Direct proteome mapping of the lung microvascular endothelium cell surface in vivo and in cell culture. Nature Biotech. 2004;22:985–992. doi: 10.1038/nbt993. [DOI] [PubMed] [Google Scholar]
- 24.Santoni V, Molloy M, Rabilloud T. Membrane proteins and proteomics: Un amour impossible? Electrophoresis. 2000;21:1054–70. doi: 10.1002/(SICI)1522-2683(20000401)21:6<1054::AID-ELPS1054>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 25.Bansal D, Campbell KP. Dysferlin and plasma membrane repair in muscular dystrophy. Trends Cell Biol. 2004;14:206–213. doi: 10.1016/j.tcb.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 26.Vandré DD, Ackerman WE, IV, Kniss DA, Tewari AK, Mori M, Takizawa T, et al. Dysferlin is expressed in human placenta but does not associate with caveolin. Biol Reprod. 2007;77:533–42. doi: 10.1095/biolreprod.107.062190. [DOI] [PubMed] [Google Scholar]
- 27.Bashir R, Britton S, Strachan T, Keers S, Vafiadaki E, Lako M, et al. A gene related to Caenorhabditis elegans spermatogenesis factor fer-1 is mutated in limb-girdle muscular dystrophy type 2B. Nature Genet. 1998;20:37–42. doi: 10.1038/1689. [DOI] [PubMed] [Google Scholar]
- 28.Liu J, Aoki N, Illa I, Chenyan W, Fardeau M, Angelini C, et al. Dysferlin a novel skeletal muscle gene, is mutated in Miyoshi myopathy and limb girdle muscular dystrophy. Nature Genet. 1998;20:31–6. doi: 10.1038/1682. [DOI] [PubMed] [Google Scholar]
- 29.Bansal DM, Miyake K, Vogel SS, Groh S, Chen CC, Williamson R, et al. Defective membrane repair in dysferlin-deficient muscular dystrophy. Nature. 2003;423:168–72. doi: 10.1038/nature01573. [DOI] [PubMed] [Google Scholar]
- 30.Lennon NJ, Kho A, Backai BJ, Perlmutter SL, Hyman BT, Brown RH., Jr Dysferlin interacts with annexins A1 and A2 and mediates sarcolemmal wound-healing. J Biol Chem. 2003;278:50466–73. doi: 10.1074/jbc.M307247200. [DOI] [PubMed] [Google Scholar]
- 31.Han R, Bansal D, Miyake K, Muniz VP, Weiss RM, McNeil PL, et al. Dysferlin-mediated membrane repair protects the heart from stress-induced left ventricular injury. J Clin Invest. 2007;117:1805–18. doi: 10.1172/JCI30848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doherty KR, Cave A, Davis DB, Delmonte AJ, Posey A, Earley JU, Hadhazy M, McNally EM. Normal myoblast fusion requires myoferlin. Development. 2005;132:5565–75. doi: 10.1242/dev.02155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bernatchez PN, Acevedo L, Fernandez-Hernando C, Murata T, Chalouni C, Kim J, et al. Myoferlin regulates vascular endothelial growth factor receptor-2 stability and function. J Biol Chem. 2007;282:30745–30753. doi: 10.1074/jbc.M704798200. [DOI] [PubMed] [Google Scholar]
- 34.Redman CW, Sargent IL. Circulating microparticles in normal pregnancy and pre-eclampsia. Placenta. 2008;29(Suppl A):S73–7. doi: 10.1016/j.placenta.2007.11.016. [DOI] [PubMed] [Google Scholar]
- 35.Ramsden AE, Holden DW, Mota LJ. Membrane dynamics and spatial distribution of Salmonella-containing vacuoles. Trends Microbiol. 2007;15:516–24. doi: 10.1016/j.tim.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 36.Geisbrecht ER, Haralalka S, Swanson SK, Florens L, Washburn MP. Abmayr SM.Drosophila ELMO/CED-12 interacts with myoblast city to direct myoblast fusion and ommatidial organization. Dev Biol. 2008;314:137–49. doi: 10.1016/j.ydbio.2007.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fanelli MA, Montt-Guevara M, Diblasi AM, Gago FE, Tello O, Cuello-Carrion FD, et al. P-cadherin and beta-catenin are useful prognostic markers in breast cancer patients; beta-catenin interacts with heat shock protein Hsp27. Cell Stress Chaperones. 2008;13:207–20. doi: 10.1007/s12192-007-0007-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patel PC, Harrison RE. Membrane ruffles capture C3bi-opsonized particles in activated macrophages. Mol Biol Cell. 2008 doi: 10.1091/mbc.E08-02-0223. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]


