Abstract
Bone can adapt its structure in response to mechanical stimuli. At the cellular level, this involves changes in chromatin organization, gene expression, and differentiation, but the underlying mechanisms are poorly understood. Here we report on the involvement of RUNX2, a bone-related transcription factor, in this process. Fluid flow shear stress loading of preosteoblasts stimulated translocation of extracellular signal-regulated kinase (ERK)/mitogen-activated protein kinase (MAPK) to the nucleus where it phosphorylated RUNX2 on the chromatin of target genes, and increased histone acetylation and gene expression. MAPK signaling and two RUNX2 phosphoacceptor sites, S301 and S319, were critical for this response. Similarly, in vivo loading of mouse ulnae dramatically increased ERK and RUNX2 phosphorylation as well as expression of osteoblast-related genes. These findings establish ERK/MAPK-mediated phosphorylation of RUNX2 as a critical step in the response of preosteoblasts to dynamic loading and define a novel mechanism to explain how mechanical signals induce gene expression in bone.
Keywords: MECHANOTRANSDUCTION, BONE, MAP KINASE, RUNX2
Introduction
Bone has the unique ability to modify its structure in response to changes in skeletal loading. Classic examples are the increase in skeletal mass associated with weight-bearing exercise,(1) elevated bone mass in loaded bones of elite athletes,(2) and conversely, dramatic losses in bone density associated with bed rest and space flight.(3,4) The anabolic effects of mechanical loading are comparable to or greater than humoral factors such as parathyroid hormone, yet remain poorly understood.
Loading of bone causes movement of interstitial fluid through channels within the bone structure (canaliculi) and marrow spaces, thereby generating fluid flow shear stress (FFSS)(4) across the surface of bone cells(for reviews, see Robling and colleagues(5) and Rubin and colleagues(6)). All cells in the osteoblast lineage (mesenchymal stem cells/marrow stromal cells, preosteoblasts, osteoblasts, and osteocytes) respond to FFSS in vitro, and may contribute to the overall anabolic response of bone during in vivo loading.(6) FFSS stimulates integrin-mediated activation of focal adhesion kinase (FAK), a component of the focal adhesions that link cells with their extracellular matrix.(7–9) Focal adhesions are proposed to function as primary mechanosensors, linking shear forces induced by fluid flow with cytoplasmic signaling pathways. FAK activates extracellular signal-regulated kinase (ERK), p38, and c-Jun N-terminal kinase (JNK) mitogen-activated protein kinase (MAPK) as well as the phosphatidylinositol 3-kinase (PI3K)/protein kinase B (AKT) pathways,(10,11) and is necessary for subsequent induction of downstream mechanoresponsive genes such as secreted phosphoprotein 1 (Spp-1), c-fos, and cyclooxygenase-2 (COX-2).(9) Moreover, conditional deletion of FAK in osteoblasts renders mice resistant to the anabolic effects of local mechanical loading.(12)
Early FFSS-induced membrane changes ultimately stimulate the differentiation of osteoblasts from mesenchymal precursors, leading to new bone formation.(13,14) An outstanding question relates to how these early signals translate into the global changes in nuclear activity, chromatin structure, and gene expression necessary for new bone formation. The RUNX2 transcription factor is ideally positioned to link cell surface signals with the nucleus. RUNX2 is required for osteoblast-specific gene expression and differentiation of mesenchymal progenitor cells to osteoblasts during development as well as in adult life.(15) It functions by associating with specific enhancers in the regulatory regions of target genes such as osteocalcin (Bglap2), bone sialoprotein (Ibsp), and osteopontin (SPP-1),(16,17) where it recruits other nuclear components to the transcriptional apparatus.(18) RUNX2 functions broadly throughout osteoblast development and is tightly controlled by both posttranslational modifications and interactions with other nuclear proteins.(19) Notably, RUNX2 is regulated by ERK1/2 and p38 MAPK-mediated phosphorylation during osteoblast differentiation, making it an attractive candidate to mediate effects of FFSS on transcription.(20–22) In vivo studies also suggest possible involvement of RUNX2 in bone mechanoresponsiveness, in that Runx2 heterozygous-null mice are resistant to the bone loss associated with skeletal unloading.(23)
Here we identify a mechanism to explain how mechanical loading regulates gene expression in bone. As we show, exposure of preosteoblast cells to FFSS or in vivo loading of mouse ulna stimulates ERK/MAPK-dependent phosphorylation of RUNX2 at specific serine residues. This is accomplished by nuclear translocation and docking of activated ERK to RUNX2 previously associated with the chromatin of target genes. MAPK activity and RUNX2 phosphorylation are required for subsequent changes in histone acetylation and gene expression. This mechanism provides a route for a biomechanical signal to be dispersed to RUNX2-regulated genes, resulting in the global changes in gene expression necessary for new bone formation.
Materials and Methods
Reagents
The reagents used in this study were from the following sources: tissue culture medium and fetal bovine serum from Hyclone Laboratories (Logan, UT, USA) and Invitrogen (Carlsbad, CA, USA), RUNX2 antibody from Medical & Biological Laboratories (Nagoya, Japan; catalog number D130-3), P-ERK and total ERK antibodies from Cell Signaling Technology (Beverly, MA, USA; catalog numbers 9101 and 9102), acetylated histone H3 and H4 and histone H3 phosphorylated at serine 10 from Millipore (Billerica, MA, USA; catalog numbers 17–615, 17–630, and 06–570), and sheep anti-mouse or donkey anti-rabbit immunoglobulin G (IgG) conjugated with horseradish peroxidase (HRP) from GE Healthcare (Piscataway, NJ, USA).
Generation of a phospho-RUNX2–specific antibody
An antibody that specifically detects RUNX2 phosphorylated at S319 was produced by Covance (Princeton, NJ, USA) using the following peptide as immunogen: YPSYLSQIMTS(P)PSIHSTTPL. The specificity of this antibody for RUNX2-S319-P has been described.(24) This antibody is specific for RUNX2-S319-P and does not cross-react with any other runt-related transcription factor.
Cell culture
We maintained MC3T3 subclone 42 preosteoblast cells in α-Minimal Essential Medium (α-MEM; Life Technologies, Inc., Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS; Hyclone Laboratories) and 1% penicillin/streptomycin as described.(25) These cells contain stably integrated copies of a 1.3-kb murine mOG2-luc reporter gene derived from Bglap2. C3H10T1/2 cells were obtained from the American Type Culture Collection (Manassas, VA, USA) and maintained in DMEM, 10% FBS.
Viral transduction
We prepared adenoviruses expressing wild-type RUNX2 or RUNX2 containing S301A, S319A mutations and used them to transduce C3H10T1/2 cells as described.(21) Cells were exposed to FFSS 24 hours after viral transduction.
FFSS loading
We plated the cells on glass microscope slides (75 × 38 × 1 mm) precoated with 0.005% poly-L-Lysine (Sigma, St. Louis, MO, USA) at a density of 50,000/cm2. After 12 hours, cells were exposed to oscillatory FFSS using a pulse-static waveform (0.033 Hz at 2 Pa, 15 seconds forward, 15 seconds reverse with a 0.2-second transition) for increasing times up to 1 hour using a custom-designed system similar to those described.(26) This system exposed cells to laminar flow resulting in a shear stress of 15 to 20 dynes/cm2. To maintain constant conditions before, during, and after exposure to FFSS, we took care to keep cells in the same medium throughout the experiment (no medium changes were included). For static controls, cells were maintained under identical conditions, but without FFSS.
RNA isolation and RT-PCR
We isolated total RNA using Trizol reagent (Invitrogen) with an RNeasy Mini Kit (Qiagen, Valencia, CA, USA). Two micrograms of total RNA was reverse-transcribed using TaqMan reverse transcriptase (Applied Biosystems, Inc., Foster City, CA, USA) for cDNA synthesis. Quantitative real-time polymerase chain reaction (PCR) detection of Bglap2, Ibsp, SPP-1, Runx2, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNAs was carried out, as described,(27) using predeveloped TaqMan probes and an ABI PRISM 7700 sequence detector (Applied Biosystems). The mRNA expression for each gene of interest was calculated based on a relative standard curve and normalized to GAPDH mRNA.
Western blots and immunoprecipitation
We harvested the cells in high-salt radioimmunoprecipitation assay (RIPA) lysis buffer (10 mM Tris, 5 mM EDTA, 0.25% Triton X-100, 300 mM NaCI, phenylmethylsulfonyl fluoride (PMSF), 1 × proteinase inhibitor cocktail; Sigma). Total protein concentration was measured with a Bio-Rad protein assay kit (Bio-Rad Laboratories, Hercules, CA, USA). Equal aliquots of protein were fractioned by SDS-PAGE using 4% to 20% Tris-Glycine gels. After transfer to nitrocellulose membranes, samples were incubated overnight at 4°C with the indicated antibodies: RUNX2 (1:500 dilution), P-ERK antibody (1:500 dilution), RUNX2-S319-P (1:1000 dilution), acetylated histone H3 and H4 (1:2000 dilution), and phosphorylated histone H3 S10-P (1:1000). A second antibody (sheep anti-mouse or donkey anti-rabbit conjugated with HRP was used at 1:10,000 dilution. HRP was detected by enhanced chemiluminescence (ECL) (Amersham Biosciences, Piscataway, NJ, USA). For certain experiments, P-RUNX2 was detected in cell extracts by immunoprecipitation using anti-RUNX2-S319 antibody followed by Western blot detection of total RUNX2.
Immunofluorescence
We carried out immunofluorescence as described.(27) Cells were fixed with 4% formaldehyde for 10 minutes at room temperature and then incubated overnight at 4°C with primary antibodies to Runx2 (1:100 dilution), P-ERK (1:100 dilution), P-Runx2 (1:500 dilution), acetylated histone H3 and H4 (1:500 dilution), and Serine 10 phosphorylated histone H3 (1:300 dilution). After incubation in Alexa Fluor 488 conjugated donkey anti-mouse and Alexa Fluor 555 conjugated donkey anti-rabbit (Invitrogen) antibodies, fluorescence was detected using an Olympus FluoView 500 laser scanning confocal microscope system with a 100× oil-immersion objective at room temperature (Microscopy and Image Analysis Laboratory, University of Michigan School of Medicine).
Chromatin immunoprecipitation
We performed chromatin immunoprecipitation (ChIP) assays as described.(17) Briefly, after FFSS, cells were cross-linked with 1% p-formaldehyde for 10 minutes at room temperature. Chromatin was then isolated and sheared by sonication to produce an average fragment size of 300 to 500 bp. Chromatin (10 μg DNA/assay) was precleared by incubation with protein A/G agarose beads (Santa Cruz Biotechnology, Santa Cruz, CA, USA) for 1 hour, incubated with primary antibodies, and precipitated with A/G agarose beads. After purification with a nucleic acid removal kit (Qiagen), input and precipitated DNA were analyzed by PCR (35 cycles) or by quantitative real-time PCR using TaqMan probes flanking OSE2a, OSE2b, or control transcribed regions in the Bglap2 gene. PCR primers were designed to detect OSE2a and OSE2b regions of the Bglap2 promoter (−160 to −120 bp and −620 to −580 bp, respectively), coding sequence in the first exon of Bglap2, a functional RUNX2 binding region in Ibsp (−130 to −59 bp), and a nonfunctional consensus RUNX2 binding region in Ibsp (−1300 to −1200 bp). All PCR primer sequences used for ChIP analysis were previously described: Bglap2(28) and Ibsp.(17)
In vivo mechanical loading
We used an ulnar loading model and custom equipment built at the University of Michigan School of Medicine for the in vivo mechanical loading studies. This loading system is based upon work by Lee and colleagues.(29) All studies with mice were performed in compliance with the University of Michigan Committee for the Use and Care of Animals. The left forelimbs of male C57BL6 mice (~16 weeks of age) were uniaxially loaded to a peak compressive force of −1.3 N (generates approximately − 2000 micro-strain at the ulnar mid-diaphysis), 500 cycles/d (sinusoidal waveform at 2 Hz) for 1, 2, or 3 days (n = 6 per group). Ten minutes after the start of each loading cycle, right and left limbs were harvested under sterile conditions and used for the analysis of mRNA and protein. For RNA isolation, ulnae were stripped of outer connective tissue and whole bones were ground in liquid nitrogen with a mortar and pestle. The powder then was resuspended in Trizol reagent (Invitrogen) and RNA was isolated.
For protein extraction and immunoprecipitation, whole ulnae were homogenized in high-salt RIPA buffer using a Polytron homogenizer. After centrifugation, the supernatant was directly separated on 4% to 20% SDS-PAGE or used in immunoprecipitation reactions.
Statistical analysis
Results are presented as mean ± SE. Sample size is indicated in the legend for each figure. Statistical significance was assessed using a one-way ANOVA followed by Tukey’s multiple-comparison test. All experiments were repeated at least twice.
Results
FFSS loading rapidly stimulates ERK/MAPK-mediated RUNX2 phosphorylation and transcriptional activity
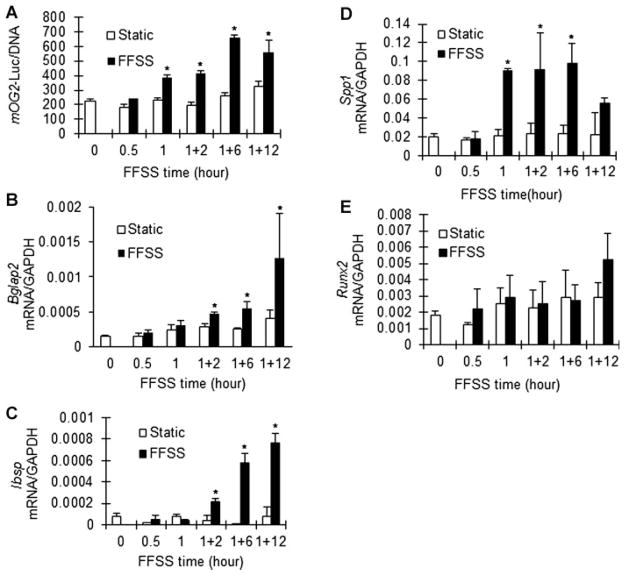
We used MC3T3-E1 clone 42 cells, a well-characterized preosteoblast cell line, to examine early effects of FFSS on osteoblast gene expression and signaling. These cells contain stably integrated copies of the murine Bglap2 promoter driving luciferase (1.3-kb mOG2-luc), making them useful both to measure transcription driven by an osteoblast-related gene promoter as well as mRNA induction. Previous studies established a good correlation between mOG2-luc activity and gene expression during in vitro osteoblast differentiation.(25) Clone 42 cells were exposed to oscillating FFSS for up to 1 hour and then cultured under static conditions for up to an additional 12 hours. FFSS rapidly stimulated transcription of the mOG2-luc reporter gene, with significant increases in luciferase activity seen after 1 hour. This FFSS stimulation persisted through the 12-hour time point (Fig. 1A). These changes were accompanied by increases in several osteoblast-related mRNAs (Bglap2, Ibsp, and SPP-1). FFSS induced Bglap2 mRNA twofold after 2 hours and this induction increased to approximately fourfold after 12 hours (Fig. 1B). Similarly, Ibsp mRNA induction was first seen after 2 hours (fourfold) and continued to increase at 6 and 12 hours (approximately 10-fold induction at both times; Fig. 1C). Spp1 mRNA was induced most rapidly, with peak levels seen after 1 and 6 hours (fivefold induction) followed by a drop 12 hours after FFSS (Fig. 1D). In contrast, Runx2 mRNA levels did not significantly change over this time interval (Fig. 1E).
Fig. 1.
FFSS induction of osteoblast gene expression. MC3T3-E1 cl42 preosteoblast cells were maintained as static cultures or exposed to FFSS for 0, 0.5, or 1 hour as described in Materials and Methods. Cells were either harvested immediately after FFSS or exposed to FFSS for 1 hour and cultured for an additional 2, 6, or 12 hours as indicated. Cells were assayed for Bglap2 promoter activity (luciferase) (A) or mRNA levels for SPP-1 (B), Bglap2 (C), Ibsp (D), and Runx2 (E). All mRNA values were normalized to GAPDH mRNA, which was not affected by FFSS. *Significantly different from time-matched static control, p <0.01, n = 3.
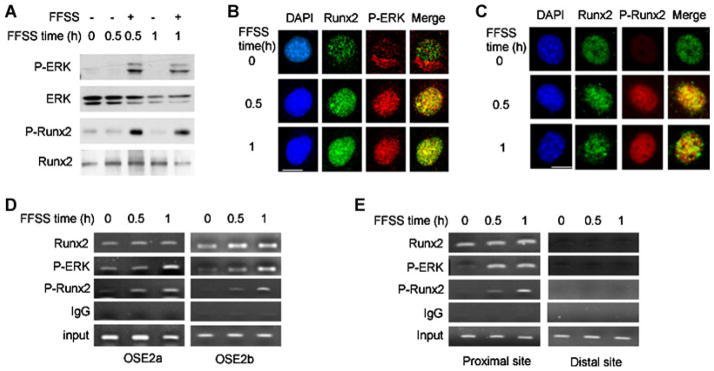
Integrin/focal adhesion kinase activation of the ERK/MAPK pathway is a well-characterized response to FFSS.(9) Furthermore, the terminal MAPK in this pathway, ERK1/2, was recently shown to stimulate RUNX2-dependent transcriptional activity by directly phosphorylating RUNX2 on specific serine residues (S43, S301, S319, and S510). Of these, both S301 and S319 were shown to be critical for transcriptional activation during osteoblast differentiation. Specifically, whereas individual S301A or S319A mutations had only a small effect on RUNX2 transcriptional activity, combined mutation of both sites almost completely blocked the ability of RUNX2 to stimulate transcription of osteoblast-related genes during in vitro osteoblast differentiation.(21) To determine if this pathway is important for FFSS induction of osteoblast gene expression, we examined ERK and RUNX2 phosphorylation (Fig. 2A and Supplementary Fig. S1). These studies used a unique antibody that specifically detects phosphorylation of RUNX2 at S319. Previous studies showed that phosphorylation at this site is well-correlated with transcriptional activity stimulated by both ERK1/2 and p38 MAPKs.(21,24) FFSS rapidly stimulated both ERK and RUNX2–S319 phosphorylation. A more detailed time course study revealed that both phosphorylation events could be detected after as little as 5 minutes of FFSS, gradually declined, and then increased again at later times (Supplementary Fig. S1). No changes in p38 phosphorylation were detected under these conditions (data not shown).
Fig. 2.
FFSS rapidly stimulates ERK1/2 activity, nuclear translocation, chromatin binding, and RUNX2 phosphorylation. (A) FFSS stimulates ERK/MAPK and RUNX2-S319 phosphorylation. Cell lysates were analyzed by probing Western blots with the indicated antibodies, except in the case of P-RUNX2 for which P-RUNX2 antibody was used for immunoprecipitation followed by Western blot detection of total RUNX2. (B,C) FFSS increases nuclear localization of P-ERK, Runx2, and P-Runx2. Immunofluorescence confocal microscopy was used to localize the indicated factors. FFSS stimulated the translocation of P-ERK from a perinuclear cytoplasmic compartment to the nucleus without affecting the nuclear localization of RUNX2 in B, and increased nuclear RUNX2-S319-P in C. Note colocalization of P-ERK and RUNX2 (in B, merged image) and P-RUNX2 with total RUNX2 (in C, merged image). Bar = 5 μm. Low-power views of multiple cells are shown in Supplementary Figs. S2 and S3. (D,E) FFSS stimulates binding of P-ERK and P-RUNX2 to proximal promoter regions of Bglap2 and Ibsp chromatin. ChIP assays were used to measure chromatin-bound RUNX2, P-RUNX2, and P-ERK1/2. Nonspecifically precipitated chromatin was measured using isotype-matched IgG. PCR primers were designed to detect OSE2a and OSE2b regions of the Bglap2 gene in D, and proximal and distal (nonfunctional) RUNX2 binding sites in Ibsp in E.
We next used immunofluorescence confocal microscopy to examine the effect of FFSS on the subcellular localization of P-ERK and RUNX2 (Fig. 2B, C shows high-magnification images of single representative cells, whereas Supplementary Figs. S2 and S3 show low-power fields). In the unstimulated state, P-ERK had a cytoplasmic/perinuclear localization whereas RUNX2 remained exclusively nuclear under all conditions. However, only very faint P-RUNX2 staining was seen in unstimulated cells. FFSS rapidly increased the nuclear localization of P-ERK without affecting total RUNX2 distribution. Furthermore, FFSS strongly stimulated nuclear P-RUNX2 immunofluorescence. Interestingly, merger of P-ERK and RUNX2 images suggests possible nuclear colocalization of these two molecules. Furthermore, extensive colocalization of P-RUNX2 with total RUNX2 was observed, as would be expected.
FFSS stimulates P-ERK binding to chromatin-associated RUNX2 and RUNX2 phosphorylation
RUNX2 regulates transcription by binding to specific enhancer sequences in the regulatory regions of target genes.(16) During osteoblast differentiation, P-ERK binds to chromatin-associated RUNX2 via a specific docking or “D site” in the C terminal portion of the runt domain.(21,27) This docking event is necessary for subsequent phosphorylation and activation of RUNX2.(21,24) On the basis of this background, studies were carried out to assess whether FFSS could stimulate P-ERK-RUNX2 binding and phosphorylation on target gene chromatin (Fig. 2D, E). ChIP assays were conducted with antibodies to total RUNX2, RUNX2-S319-P, P-ERK, and isotype-matched IgG (negative control), using PCR primers to detect association of each factor with RUNX2 enhancers in the proximal promoter regions of Bglap2 and Ibsp.(27) For Bglap2 ChIP analysis, binding near OSE2a (−130 bp) and OSE2b (−605 bp) promoter regions was separately analyzed; whereas for Ibsp, binding was examined at overlapping RUNX2 binding sites at −130 bp and at a nonfunctional site at −1300 bp (negative control). As a further control, binding to transcribed regions in the first exon of each gene was also examined and found to be negative (data not shown). In unstimulated cells, RUNX2 was bound to Bglap2 and Ibsp chromatin and this binding remained relatively constant after FFSS. In contrast, chromatin-associated P-ERK and P-RUNX2 were low to undetectable in control cells, but dramatically increased with FFSS. This is consistent with the concept that, after nuclear translocation, P-ERK binds and phosphorylates RUNX2 previously associated with target gene chromatin.
FFSS-related changes in histone acetylation and phosphorylation
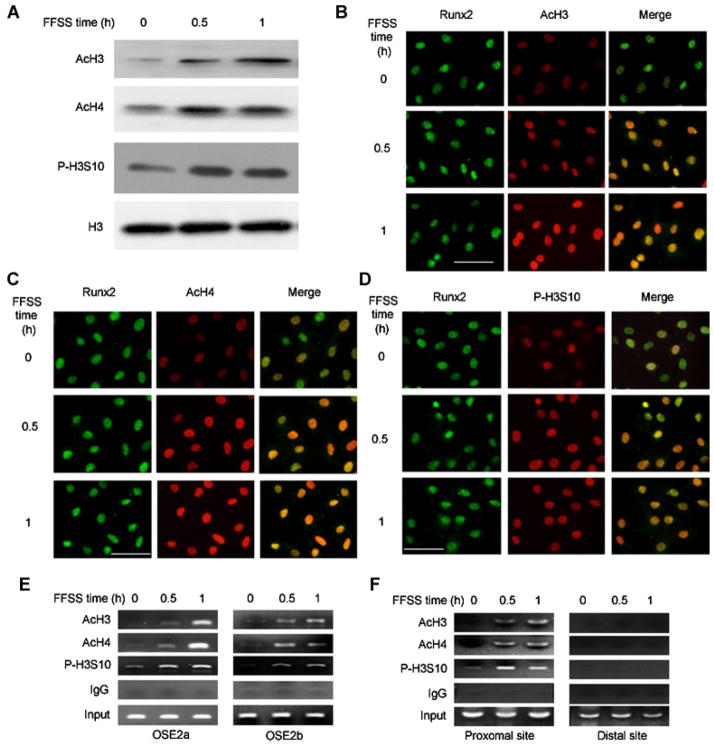
In eukaryotic cells, chromatin must progress from a condensed, inactive state to an open, active conformation before genes can be expressed.(30) The state of histone acetylation and related histone phosphorylation are major determinants of chromatin decondensation and transcriptional activity.(31) Histone H3 S10 phosphorylation was previously associated with the response to mitogenic and differentiation signals stimulated by growth factors and, in some cases, was shown to be required for subsequent histone acetyltransferase activation.(32,33) Histone H3 and H4 acetylation, particularly at K9 and K14, is selectively associated with chromatin decondensation and increased transcription.(34) As shown in Figure 3, FFSS increases both types of histone posttranslational modification. This was shown using Western blot detection of histones in whole-cell extracts (Fig. 3A), immunofluorescence (Fig. 3B–D), and ChIP analysis of RUNX2-binding regions of both Bglap2 and Ibsp (Fig. 3E, F). The immunofluorescence colocalization of RUNX2 with modified histones as well as ChIP analysis suggest that histone modifications occurred in regions of chromatin proximal to the bound RUNX2. In contrast, no histone modifications were detected using the distal Ibsp primers that amplify a region of chromatin that is approximately 1200 bp away from the nearest RUNX2 binding site (Fig. 3F).
Fig. 3.
FFSS stimulates histone acetylation and phosphorylation on chromatin of osteoblast target genes. (A) Total histone modification. Histones were extracted from nuclei and total acetylated H3 and H4 as well as S10-phosphorylated H3 were measured on Western blots. (B–D) Colocalization of acetylated/phosphorylated histones with RUNX2. Total RUNX2 and modified histones were localized by immunofluorescence confocal microscopy. (E,F) FFSS stimulates histone modification near RUNX2-binding regions of Bglap2 and Ibsp. ChIP assays were conducted using the indicated antibodies and primers to proximal promoter regions of Bglap2 and Ibsp as in Fig. 2.
FFSS-induced changes in chromatin structure and gene expression require ERK/MAPK activity and RUNX2 phosphorylation
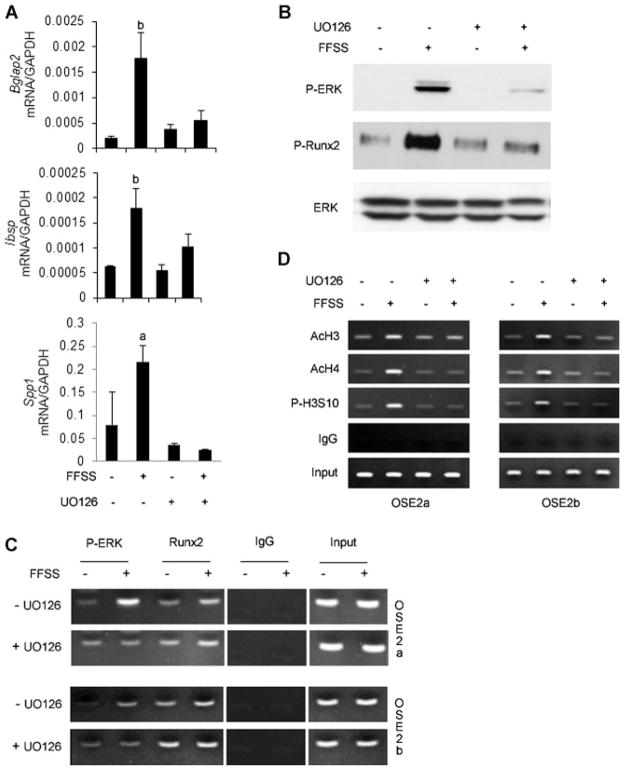
We next used chemical inhibition and genetic studies to examine whether phosphorylation of RUNX2 by ERK is necessary for the FFSS-induced changes described above. In the first study, cells were pretreated with the MAPK inhibitor, U0126, before FFSS (Fig. 4). MAPK inhibition totally blocked FFSS induction of Bglap2, Ibsp, and SPP-1 mRNAs (Fig. 4A). Furthermore, this inhibitor clearly blocked FFSS-dependent RUNX2-S319 phosphorylation (Fig. 4B), translocation of P-ERK to RUNX2 binding regions on Bglap2 chromatin (Fig. 4C), histone H3-S10 phosphorylation, and histone H3/H4 acetylation (Fig. 4D). Significantly, MAPK inhibition did not affect chromatin-bound RUNX2, indicating that RUNX2 does not need to be phosphorylated to remain chromatin-associated (Fig. 4C, right 2 lanes).
Fig. 4.
ERK1/2 MAP kinase activity is required for FFSS-induced RUNX2 phosphorylation, histone modifications, and gene expression. Cells were exposed to FFSS for 1 hour in the presence or absence of MAPK inhibitor, U0126 (10 μM), and were either immediately harvested for Western blot and ChIP analysis (B–D) or cultured for an additional 12 hours for mRNA detection (A). (A) Gene expression. The indicated mRNAs were measured by real-time RT-PCR and results were normalized to GAPDH mRNA. Significantly different from corresponding static control, ap <0.05, bp <0.01, n = 3. (B) ERK1/2 and RUNX2-S319 phosphorylation. (C,D) P-ERK binding to chromatin and histone modifications. ChIP assays were conducted as in Fig. 2 using primers to OSE2a and OSE2b regions of the mOG2 promoter.
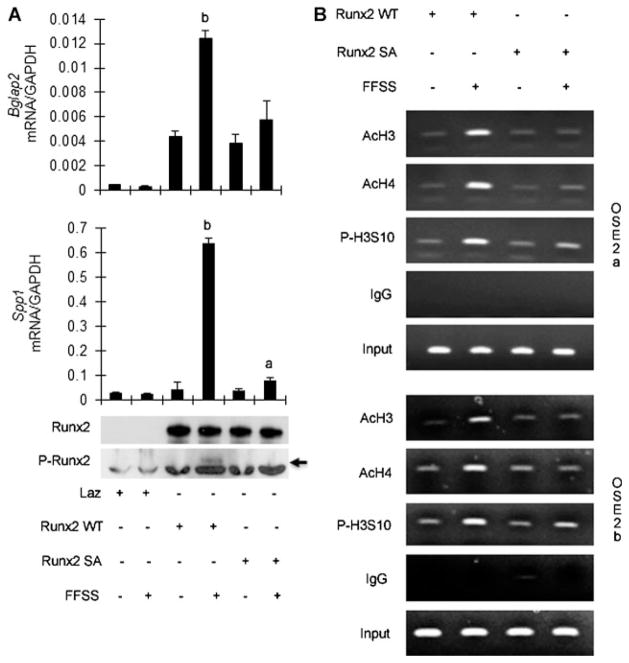
To more directly assess the specific role of RUNX phosphory-lation in the FFSS response, adenovirus expressing wild-type and S301A, S319A mutant RUNX2 were developed and used to transduce C3H10T1/2 cells (Fig. 5). As previously shown, both S301 and S319 are required for MAPK-dependent stimulation of RUNX2 transcriptional activity; mutation of both these sites blocks MAPK responsiveness whereas single mutations only slightly reduce activity.(21) The pluripotent C3H10T1/2 mesenchymal cell line normally does not express RUNX2, but undergoes osteoblast differentiation after RUNX2 transfection.(27,35) Exposure of control cells to FFSS did not affect the low basal levels of Bglap2 and SPP-1 mRNA, although it did still stimulate ERK1/2 phosphorylation (data not shown). However, transduction with wild-type RUNX2 adenovirus increased basal Bglap2 and SPP-1 mRNA in control cultures and rendered cells highly responsive to FFSS. In contrast, cells transduced with the S301A, S319A double mutant were refractory to FFSS stimulation. Note, however, that mutant RUNX2 was still able to increase basal Bglap2 and SPP-1 mRNA levels. These results could not be explained by differences in amounts of wild-type and mutant RUNX2 protein, which were expressed to the same levels. Also and as expected, FFSS stimulation of RUNX2-S319-P was only detected in cells transduced with wild-type RUNX2 (see Western blot; Fig. 5A, bottom). Cells expressing mutant RUNX2 also failed to exhibit the FFSS-induced increases in chromatin-associated histone acetylation and phosphorylation seen in wild-type cultures (Fig. 5B). This underscores the importance of RUNX2-S301/S319 phosphorylation in the response of osteoblasts to FFSS.
Fig. 5.
RUNX2 S301/S319 phosphorylation is required for downstream responses to FFSS. C3H10T1/2 cells were transduced with Lac Z control adenovirus (Laz) or adenoviruses expressing wild-type RUNX2 or RUNX2 containing S301A/S319A double mutations and, after 24 hours, exposed to FFSS for 1 hour. (A) Gene expression. Bglap2 and SPP-1 mRNAs were measured by real-time RT-PCR 12 hours after FFSS and results were normalized to GAPDH mRNA. Bottom panel shows Western blots of total and RUNX2-S319-P (arrow). Significantly different from corresponding RUNX2 static control (wild-type or mutant), ap <0.05, bp <0.01, n = 3. (B) Histone modifications. Cells were harvested for ChIP analysis immediately after FFSS. ChIP was conducted using antibodies to acetylated histone H3 and H3 and S10-phosphorylated H3 and PCR primers to OSE2a and OSE2b regions of the mOG2 promoter.
Involvement of ERK1/2 and RUNX2 phosphorylation in the in vivo response of bone to mechanical loading
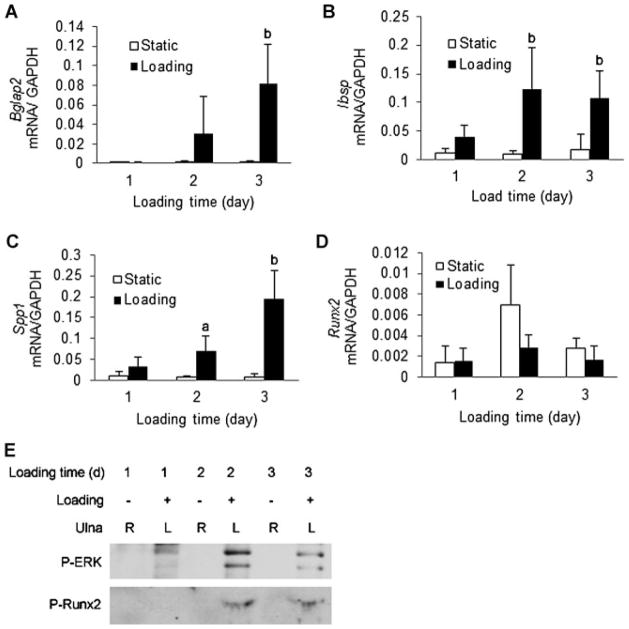
We used a mouse ulnar loading model to examine ERK and RUNX2 phosphorylation in vivo. The left forelimbs of male C57BL6 mice were exposed to cyclical uniaxial compression loading (500 cycles/d, 2 Hz) for 3 successive days. A similar loading regimen was previously shown to increase total bone formation rate after 2 weeks.(29) Immediately after each loading period, a group of animals was euthanized for isolation of total RNA and protein from whole bones. Small (twofold to threefold) increases in Bglap2, Ibsp, and Spp-1 mRNAs were detected after a single loading cycle, and the magnitude of induction continued to increase with each successive day such that fold inductions from 15- to 30-fold were seen by day 3 (Fig. 6A). Similar to FFSS results, RUNX2 mRNA levels remained relatively stable throughout the experiment. Immunoprecipitation and Western blot analysis of total bone protein extracts was used to measure ERK and RUNX2-S319 phosphorylation. After a single loading cycle, a clear increase in P-ERK was apparent. By cycle two, RUNX2-S319 phosphorylation was seen, and this persisted after the third day of loading. Because no attempt was made to fractionate bone into marrow, endosteal, or periosteal compartments, it is not possible to make any conclusions about which cell population is responsible for these changes. Nevertheless, these results clearly show that in vivo loading can induce the same changes in ERK/MAPK signaling and RUNX2 phosphorylation seen after FFSS exposure of preosteoblasts in cell culture.
Fig. 6.
In vivo mechanical loading stimulates ERK/MAPK activity, RUNX2 phosphorylation, and osteoblast gene expression. C57BL6 mice were exposed to 1, 2, or 3 days of ulnar loading. Loading was for 500 cycles at 2 Hz. Each day, bones were isolated 10 minutes after the loading cycle was completed and RNA and protein were extracted as described in Materials and Methods. (A–D) Gene expression. Bglap2, Ibsp, SPP-1, and Runx2 mRNAs were measured by real-time RT-PCR and results were normalized to GAPDH mRNA. Statistics: significantly different from time-matched unloaded control, ap <0.05, bp <0.01, n = 6. (E) Western blot detection of ERK1/2 and RUNX2 S319 phosphorylation. Whole-bone extracts were immunoprecipitated with the indicated antibodies.
Discussion
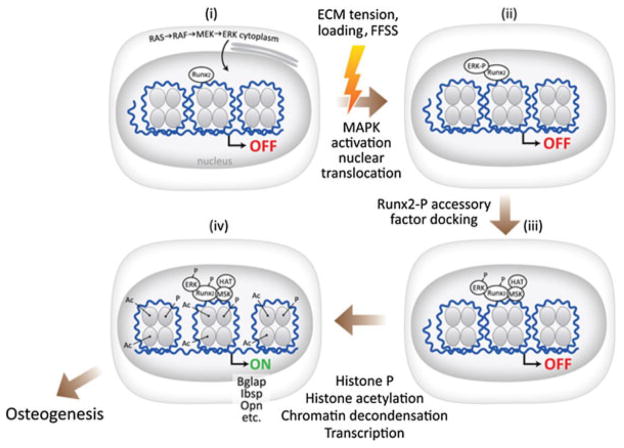
Here we describe a novel mechanism to explain how bone cells modify chromatin structure and gene expression in response to mechanical signals. Mechanical loading stimulates osteoblast precursor cells through a number of mechanisms, including FFSS. This stimulation activates the ERK/MAPK pathway, likely via activation of FAK/integrin complexes in focal adhesions. Active P-ERK1/2 translocates to the nucleus where it selectively associates with RUNX2 bound to specific enhancer regions in the regulatory regions of osteoblast target genes. P-ERK then phosphorylates RUNX2 at S301 and S319, a necessary event for subsequent changes in histone phosphorylation, acetylation, chromatin decondensation, and transcription (see Fig. 7).
Fig. 7.
Proposed mechanism for regulation of osteoblast gene expression by mechanical loading. In the unstimulated state, RUNX2 is bound to specific enhancer regions in the chromatin of target genes. Mechanical loading of bones exposes cells to fluid flow shear stress (FFSS), which activates focal adhesion kinase (FAK) in focal adhesions adjacent to extracellular matrix–cell contact points. FAK activates ERK/MAPK signaling, leading to nuclear translocation of P-ERK. In the nucleus, P-ERK docks to specific binding sites on RUNX2, where it phosphorylates this transcription factor at specific serine residues including S301 and S319. RUNX2-P recruits accessory nuclear factors, including histone acetyltransferases and possibly other protein kinases, to chromatin, leading to histone phosphorylation, acetylation, and decondensation, thereby allowing transcription of osteoblast-related genes.
Substantial evidence was presented in support of this model. Both ERK1/2 and RUNX2 S319 phosphorylation were shown to rapidly (within 5 minutes) increase after the onset of FFSS. These changes were accompanied by translocation of P-ERK from a cytoplasmic/perinuclear compartment to the nucleus where it was colocalized with RUNX2 as measured by immunofluorescence microscopy. This colocalization was also apparent on the chromatin, where P-ERK bound to RUNX2 previously associated with enhancer regions in the proximal promoters of Bglap2 and Ibsp genes. Coincident with the nuclear translocation of P-ERK was the appearance of phosphorylated RUNX2, phosphorylated histone H3, and acetylated histones H3 and H4 in chromatin regions containing RUNX2-specific enhancers. Furthermore, ERK/MAPK activity and RUNX2 S301 and S319 phosphorylation sites were required for subsequent FFSS-induced changes in histone acetylation and RUNX2-dependent transcription.
The changes in ERK and RUNX2 phosphorylation seen after FFSS in cell culture were also detected after in vivo loading of mouse ulnae. We observed that each successive period of in vivo loading increased mRNA levels as well as P-ERK and RUNX2-S319 phosphorylation. This likely indicates activation of additional osteoblasts precursor cells that will amplify the adaptive response. Bone adaptation is known to be enhanced by repeated loading periods interspersed with rest, which is due to a desensitization of bone to continuous mechanical stimuli.(36) In addition, bone will adapt little (if at all) to a single, short-term loading bout.(37) Based upon the signaling/activation mechanism we proposed, our in vivo data is consistent with previous bone adaptation studies. However, because whole ulnae were used for isolation of RNA and protein after loading, it is not possible to determine which specific cell populations respond to in vivo loading by increasing ERK and RUNX2 phosphorylation. Also, a definitive demonstration that RUNX2 phosphorylation is required for in vivo stimulation of gene expression by loading will require studies with transgenic mice containing phosphorylation-deficient RUNX2.
In order for mechanical loading to stimulate bone formation, a mechanism is required to convert a mechanical signal into biochemical changes capable of altering the pattern of gene expression in bone progenitor cells. Because osteoblast differentiation likely requires the reprogramming of a large number of genes, cells must transfer a signal to the nucleus that can be readily dispersed throughout the genome to selectively alter the expression of only those genes necessary for the osteoblast lineage. We propose that one such signal is P-ERK. Multiple tissues, including blood vessels, bone, and muscle, respond to FFSS at least in part via activation of the integrin/focal adhesion/MAPK pathway, yet each cell type displays a unique response to mechanical challenges. For example, in vascular endothelial cells, exposure to FFSS induces genes that render the endothelium resistant to proinflammatory cytokines and reduce smooth muscle proliferation, whereas in pathological conditions FFSS stimulates intimal thickening leading to atherosclerotic plaque formation.(38) A different response is initiated in bone that involves induction of a unique pattern of gene expression necessary for osteoblast differentiation and mineralization.
How might a similar FFSS-induced signal have such different effects on these two types of cells? We believe that the MAPK activation of tissue-specific transcription factors described here offers a way for mechanical loads to selectively activate only those genes associated with a specific phenotype while repressing genes involved in other lineages. This concept is illustrated in Figure 5, where exposure of RUNX2-deficient C3H10T1/2 cells to FFSS stimulated ERK1/2 phosphorylation, but had no effect on expression of Bglap2 or Ibsp mRNAs. Only after wild-type RUNX2 was transfected into cells did they acquire the ability to respond to FFSS in an osteoblast-appropriate manner (eg, induction of Bglap2 and Ibsp mRNAs). Thus, RUNX2 is necessary for P-ERK to activate an osteoblast-selective transcriptional program. Furthermore, this activation requires RUNX2 phosphorylation because the RUNX2 S301A, S319A double mutant was refractory to MAPK activation.
There is now considerable evidence in support of the concept that tissue-specific transcription factors can serve as docking sites for nuclear kinases. For example, in muscle regeneration, the tissue-specific transcription factors, MyoD and MEF2, serve as docking sites for p38α/β on the chromatin of muscle-related genes. Subsequent p38-related phosphorylation is necessary for histone acetylation, chromatin decondensation, and increased muscle-specific gene expression.(39,40) Also, in pancreatic β cells, exposure to elevated glucose concentrations stimulates the binding of P-ERK to Beta2, MafA, and PDX-1 transcription factors on the promoters of several glucose-inducible genes including insulin, where it initiates recruitment of accessory factors to stimulate transcription.(41) Of interest, studies in yeast suggest that the association of protein kinases with chromatin may be a common phenomenon in eukaryotic cells broad regulatory implications.(42) Specifically, yeast homologues of p38, ERK, and PKA all become associated with the chromatin of multiple target genes, to globally alter patterns of gene expression in response to several external stimuli such as osmotic stress, pheromones, or glucose.
In addition to stimulating osteoblast differentiation and bone formation, dynamic loading of bone can suppress other mesenchymal lineages such as adipocytes. For example, exposure of mice to low-magnitude mechanical signals preferentially increased differentiation of marrow stromal cells (MSCs) to osteoblasts and decreased adipocyte differentiation.(43) Furthermore, in vitro studies suggest that chronic exposure of MSCs to uniform biaxial strain, another type of mechanical signal that uses similar pathways to FFSS, can reduce the adipogenic potential of these cells.(44) Although the mechanistic basis for these responses is not known, nuclear P-ERK could suppress adipogenic genes while stimulating osteoblast gene expression.(45) A test of the validity of these concepts will require genomic studies to determine the extent to which P-ERK associates with RUNX2 on osteoblast-related genes as well as possible interactions with genes of adipocyte lineage.
Supplementary Material
Acknowledgments
This work was supported by NIH grant DE11723 (to RTF) and the Core Facilities of the Michigan Diabetes Research and Training Center, funded by National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) grant DK020572.
Footnotes
Authors’ roles: YL designed and performed the fluid flow experiments and assisted with in vivo studies. CG developed the anti-phospho-RUNX2 antibody and developed conditions for Western blot and IP detection of P-RUNX2. JPL, DLB, and SAG designed and performed in vivo ulna loading studies and provided advice for in vitro FFSS studies. JAR assisted with in vitro studies. RTF conceived the study, participated in data analysis, and wrote the manuscript.
Disclosures
All authors state that they have no conflicts of interest.
Additional Supporting Information may be found in the online version of this article.
References
- 1.Gleeson PB, Protas EJ, LeBlanc AD, Schneider VS, Evans HJ. Effects of weight lifting on bone mineral density in premenopausal women. J Bone Miner Res. 1990;5(2):153–8. doi: 10.1002/jbmr.5650050208. [DOI] [PubMed] [Google Scholar]
- 2.Krahl H, Michaelis U, Pieper HG, Quack G, Montag M. Stimulation of bone growth through sports. A radiologic investigation of the upper extremities in professional tennis players. Am J Sports Med. 1994;22(6):751–7. doi: 10.1177/036354659402200605. [DOI] [PubMed] [Google Scholar]
- 3.Leblanc AD, Schneider VS, Evans HJ, Engelbretson DA, Krebs JM. Bone mineral loss and recovery after 17 weeks of bed rest. J Bone Miner Res. 1990;5(8):843–50. doi: 10.1002/jbmr.5650050807. [DOI] [PubMed] [Google Scholar]
- 4.Keyak JH, Koyama AK, LeBlanc A, Lu Y, Lang TF. Reduction in proximal femoral strength due to long-duration spaceflight. Bone. 2009;44(3):449–53. doi: 10.1016/j.bone.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 5.Robling AG, Castillo AB, Turner CH. Biomechanical and molecular regulation of bone remodeling. Ann Rev Biomed Eng. 2006;8:455–98. doi: 10.1146/annurev.bioeng.8.061505.095721. [DOI] [PubMed] [Google Scholar]
- 6.Rubin J, Rubin C, Jacobs CR. Molecular pathways mediating mechanical signaling in bone. Gene. 2006;367:1–16. doi: 10.1016/j.gene.2005.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pavalko FM, Chen NX, Turner CH, Burr DB, Atkinson S, Hsieh YF, Qiu J, Duncan RL. Fluid shear-induced mechanical signaling in MC3T3-E1 osteoblasts requires cytoskeleton-integrin interactions. Am J Physiol. 1998;275(6 Pt 1):C1591–601. [PubMed] [Google Scholar]
- 8.Norvell SM, Alvarez M, Bidwell JP, Pavalko FM. Fluid shear stress induces beta-catenin signaling in osteoblasts. Calcif Tissue Int. 2004;75(5):396–404. doi: 10.1007/s00223-004-0213-y. [DOI] [PubMed] [Google Scholar]
- 9.Young SR, Gerard-O’Riley R, Kim JB, Pavalko FM. Focal adhesion kinase is important for fluid shear stress-induced mechanotransduction in osteoblasts. J Bone Miner Res. 2009;24(3):411–24. doi: 10.1359/JBMR.081102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boutahar N, Guignandon A, Vico L, Lafage-Proust MH. Mechanical strain on osteoblasts activates autophosphorylation of focal adhesion kinase and proline-rich tyrosine kinase 2 tyrosine sites involved in ERK activation. J Biol Chem. 2004;279(29):30588–99. doi: 10.1074/jbc.M313244200. [DOI] [PubMed] [Google Scholar]
- 11.Sen B, Styner M, Xie Z, Case N, Rubin CT, Rubin J. Mechanical loading regulates NFATc1 and beta-catenin signaling through a GSK3beta control node. J Biol Chem. 2009;284(50):34607–17. doi: 10.1074/jbc.M109.039453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leucht P, Kim JB, Currey JA, Brunski J, Helms JA. FAK-Mediated mechanotransduction in skeletal regeneration. PLoS One. 2007;2(4):e390. doi: 10.1371/journal.pone.0000390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li YJ, Batra NN, You L, Meier SC, Coe IA, Yellowley CE, Jacobs CR. Oscillatory fluid flow affects human marrow stromal cell proliferation and differentiation. J Orthop Res. 2004;22(6):1283–9. doi: 10.1016/j.orthres.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 14.Liu L, Shao L, Li B, Zong C, Li J, Zheng Q, Toong X, Gao C, Wang J. Extracellular signal-regulated kinase1/2 activated by fluid shear stress promotes osteogenic differentiation of human bone marrow-derived mesenchymal stem cells through novel signaling pathways. Int J Biochem Cell Biol. 2011;43:1591–601. doi: 10.1016/j.biocel.2011.07.008. [DOI] [PubMed] [Google Scholar]
- 15.Karsenty G, Wagner EF. Reaching a genetic and molecular understanding of skeletal development. Dev Cell. 2002;2(4):389–406. doi: 10.1016/s1534-5807(02)00157-0. [DOI] [PubMed] [Google Scholar]
- 16.Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89(5):747–54. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- 17.Roca H, Phimphilai M, Gopalakrishnan R, Xiao G, Franceschi RT. Cooperative interactions between RUNX2 and homeodomain protein-binding sites are critical for the osteoblast-specific expression of the bone sialoprotein gene. J Biol Chem. 2005;280(35):30845–55. doi: 10.1074/jbc.M503942200. [DOI] [PubMed] [Google Scholar]
- 18.Pratap J, Lian JB, Javed A, Barnes GL, van Wijnen AJ, Stein JL, Stein GS. Regulatory roles of Runx2 in metastatic tumor and cancer cell interactions with bone. Cancer Metastasis Rev. 2006;25(4):589–600. doi: 10.1007/s10555-006-9032-0. [DOI] [PubMed] [Google Scholar]
- 19.Franceschi R, Ge C, Xiao G, Roca H, Jiang D. Transcriptional regulation of osteoblasts. Ann N Y Acad Sci. 2007;1116:196–207. doi: 10.1196/annals.1402.081. [DOI] [PubMed] [Google Scholar]
- 20.Xiao G, Jiang D, Thomas P, Benson MD, Guan K, Karsenty G, Franceschi RT. MAPK pathways activate and phosphorylate the osteoblast-specific transcription factor, Cbfa1. J Biol Chem. 2000;275(6):4453–9. doi: 10.1074/jbc.275.6.4453. [DOI] [PubMed] [Google Scholar]
- 21.Ge C, Xiao G, Jiang D, Yang Q, Hatch NE, Roca H, Franceschi RT. Identification and functional characterization of ERK/MAPK phosphorylation sites in the Runx2 transcription factor. J Biol Chem. 2009;284(47):32533–43. doi: 10.1074/jbc.M109.040980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greenblatt MB, Shim JH, Zou W, Sitara D, Schweitzer M, Hu D, Lotinun S, Sano Y, Baron R, Park JM, Arthur S, Xie M, Schneider MD, Zhai B, Gygi S, Davis R, Glimcher LH. The p38 MAPK pathway is essential for skeletogenesis and bone homeostasis in mice. J Clin Invest. 2010;120(7):2457–73. doi: 10.1172/JCI42285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salingcarnboriboon R, Tsuji K, Komori T, Nakashima K, Ezura Y, Noda M. Runx2 is a target of mechanical unloading to alter osteoblastic activity and bone formation in vivo. Endocrinology. 2006;147(5):2296–305. doi: 10.1210/en.2005-1020. [DOI] [PubMed] [Google Scholar]
- 24.Ge C, Yang Q, Zhao G, Yu H, Kirkwood K, Franceschi RT. Interactions between extracellular signal-regulated kinase 1/2 and p38 MAP kinase pathways in the control of Runx2 phosphorylation and transcriptional activity. J Bone Miner Res. doi: 10.1002/jbmr.561. Epub 2011 Nov 9. [DOI] [Google Scholar]
- 25.Xiao G, Cui Y, Ducy P, Karsenty G, Franceschi RT. Ascorbic acid-dependent activation of the osteocalcin promoter in MC3T3-E1 preosteoblasts: requirement for collagen matrix synthesis and the presence of an intact OSE2 sequence. Mol Endocrinol. 1997;11(8):1103–13. doi: 10.1210/mend.11.8.9955. [DOI] [PubMed] [Google Scholar]
- 26.Kapur S, Baylink DJ, Lau KH. Fluid flow shear stress stimulates human osteoblast proliferation and differentiation through multiple interacting and competing signal transduction pathways. Bone. 2003;32(3):241–51. doi: 10.1016/s8756-3282(02)00979-1. [DOI] [PubMed] [Google Scholar]
- 27.Li Y, Ge C, Franceschi R. Differentiation-dependent association of phosphorylated extracellular signal-regulated kinase with the chromatin of osteoblast-related genes. J Bone Miner Res. 2010;25(1):154–68. doi: 10.1359/jbmr.090705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roca H, Franceschi RT. Analysis of transcription factor interactions in osteoblasts using competitive chromatin immunoprecipitation. Nucleic Acids Res. 2008;36(5):1723–30. doi: 10.1093/nar/gkn022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee KC, Maxwell A, Lanyon LE. Validation of a technique for studying functional adaptation of the mouse ulna in response to mechanical loading. Bone. 2002;31(3):407–12. doi: 10.1016/s8756-3282(02)00842-6. [DOI] [PubMed] [Google Scholar]
- 30.Grewal SI, Moazed D. Heterochromatin and epigenetic control of gene expression. Science. 2003;301(5634):798–802. doi: 10.1126/science.1086887. [DOI] [PubMed] [Google Scholar]
- 31.Zippo A, Serafini R, Rocchigiani M, Pennacchini S, Krepelova A, Oliviero S. Histone crosstalk between H3S10ph and H4K16ac generates a histone code that mediates transcription elongation. Cell. 2009;138(6):1122–36. doi: 10.1016/j.cell.2009.07.031. [DOI] [PubMed] [Google Scholar]
- 32.Bode AM, Dong Z. Inducible covalent posttranslational modification of histone H3. Sci STKE. 2005;2005(281):re4. doi: 10.1126/stke.2812005re4. [DOI] [PubMed] [Google Scholar]
- 33.Nowak SJ, Corces VG. Phosphorylation of histone H3: a balancing act between chromosome condensation and transcriptional activation. Trends Genet. 2004;20(4):214–20. doi: 10.1016/j.tig.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 34.Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, Zhao K. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40(7):897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang S, Wei D, Wang D, Phimphilai M, Krebsbach PH, Franceschi RT. In vitro and in vivo synergistic interactions between the Runx2/Cbfa1 transcription factor and bone morphogenetic protein-2 in stimulating osteoblast differentiation. J Bone Miner Res. 2003;18(4):705–15. doi: 10.1359/jbmr.2003.18.4.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robling AG, Burr DB, Turner CH. Recovery periods restore mechanosensitivity to dynamically loaded bone. J Exp Biol. 2001;204(Pt 19):3389–99. doi: 10.1242/jeb.204.19.3389. [DOI] [PubMed] [Google Scholar]
- 37.Forwood MR, Turner CH. The response of rat tibiae to incremental bouts of mechanical loading: a quantum concept for bone formation. Bone. 1994;15(6):603–9. doi: 10.1016/8756-3282(94)90307-7. [DOI] [PubMed] [Google Scholar]
- 38.Braddock M, Schwachtgen JL, Houston P, Dickson MC, Lee MJ, Campbell CJ. Fluid shear stress modulation of gene expression in endothelial cells. News Physiol Sci. 1998;13:241–6. doi: 10.1152/physiologyonline.1998.13.5.241. [DOI] [PubMed] [Google Scholar]
- 39.Serra C, Palacios D, Mozzetta C, Forcales SV, Morantte I, Ripani M, Jones DR, Du K, Jhala US, Simone C, Puri PL. Functional interdependence at the chromatin level between the MKK6/p38 and IGF1/PI3K/AKT pathways during muscle differentiation. Mol Cell. 2007;28(2):200–13. doi: 10.1016/j.molcel.2007.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simone C, Forcales SV, Hill DA, Imbalzano AN, Latella L, Puri PL. p38 pathway targets SWI-SNF chromatin-remodeling complex to muscle-specific loci. Nat Genet. 2004;36(7):738–43. doi: 10.1038/ng1378. [DOI] [PubMed] [Google Scholar]
- 41.Lawrence MC, McGlynn K, Shao C, Duan L, Naziruddin B, Levy MF, Cobb MH. Chromatin-bound mitogen-activated protein kinases transmit dynamic signals in transcription complexes in beta-cells. Proc Natl Acad Sci U S A. 2008;105(36):13315–20. doi: 10.1073/pnas.0806465105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pokholok DK, Zeitlinger J, Hannett NM, Reynolds DB, Young RA. Activated signal transduction kinases frequently occupy target genes. Science. 2006;313(5786):533–6. doi: 10.1126/science.1127677. [DOI] [PubMed] [Google Scholar]
- 43.Luu YK, Capilla E, Rosen CJ, Gilsanz V, Pessin JE, Judex S, Rubin CT. Mechanical stimulation of mesenchymal stem cell proliferation and differentiation promotes osteogenesis while preventing dietary-induced obesity. J Bone Miner Res. 2009;24(1):50–61. doi: 10.1359/JBMR.080817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sen B, Xie Z, Case N, Ma M, Rubin C, Rubin J. Mechanical strain inhibits adipogenesis in mesenchymal stem cells by stimulating a durable beta-catenin signal. Endocrinology. 2008;149(12):6065–75. doi: 10.1210/en.2008-0687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adams M, Reginato MJ, Shao D, Lazar MA, Chatterjee VK. Transcriptional activation by peroxisome proliferator-activated receptor gamma is inhibited by phosphorylation at a consensus mitogen-activated protein kinase site. J Biol Chem. 1997;272(8):5128–32. doi: 10.1074/jbc.272.8.5128. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.