Abstract
Purpose of review
In this review, we describe the ‘state-of-the- art’ in our knowledge of asthma and what gaps exists, which can be exploited in the future for effective translation of our knowledge from the bench or population studies to diagnosis and therapy.
Recent findings
The advent of microbiome research has expounded the potential role of microbes in asthma. There has been significant increase in our understanding the pathologic, genetic, cellular and molecular mechanisms of asthma. Nonetheless, the contribution of microbes to the genesis, exacerbation and treatment of asthma are poorly understood.
Summary
Asthma is a complex chronic disease of the lung whose incidence is growing at all ages despite the progress that has been made in the areas of diagnosis and treatment of asthma. The complexity is partly due to the environmental insults such allergens and microbial infections that play differential roles in the pathogenesis of childhood vis-à-vis elderly asthma. Microbes may play important roles in the exacerbation of asthma and hence in the co-morbidities due to asthma, and also in the causation of asthma.
Keywords: asthma, eosinophil, microbe, elderly, microbiota
1. Introduction
Asthma is a multifaceted disorder, which over the past few decades has increased noticeably in prevalence. [1*] It was seen that occurrence was highest amongst Puerto Rican Hispanics (14.6%) and Non-Hispanic Blacks (10.2%) as compared to the US average (7.9%). [2] In 2007, it was estimated that for asthma treatment, the direct medical expenditure was approximately $37.2 billion US dollars. [3] Outcomes could be improved via promotion of suitable medical care as well as adequate self-management instruction. [2]
There is significant discrepancy in asthma prevalence in different age groups. For example, currently the prevalence of asthma in patients >60 years of age is more than ten percent. In contrast to the general population, elderly asthmatics have an increased mortality from lung diseases, most likely respiratory tract infections. [4] About 40% of non-pneumonic lower respiratory illnesses have been due to viral infection. [5*] In the elderly, asthma is commonly incorrectly diagnosed or even missed due to an uncharacteristic appearance, underestimation of their shortness of breath, and accompanying comorbidities such as Chronic Obstructive Pulmonary Disease (COPD). [4]
In this report, we intend to review the diversity of microbes affecting asthma, examine the correlation between infection and pathogenesis of asthma, and the role of microbial infection in exacerbation of asthma. Also, it is intended to identify cellular and molecular links between microbes and asthma and discuss the potential mechanisms, which are under investigation and those that remain to be explored and elucidated.
2. Asthma Pathogenesis
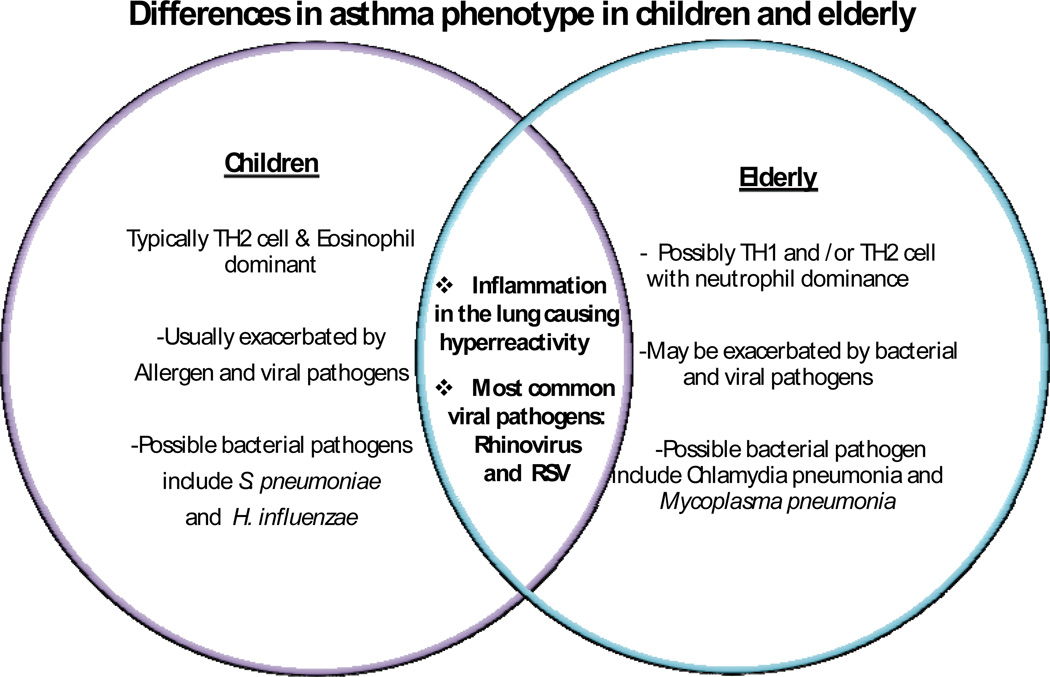
It has been suggested that mostly allergen-driven asthma in genetically predisposed children or adults is typically Th2- and eosinophil–dominant. In contrast, it has been suggested that asthma in older patients may be under epigenetic control and may be exacerbated by viral and bacterial pathogens instead of allergens, and may involve Th1 and/or Th2 cells and exhibit neutrophil dominance. A Venn diagram comparing asthma in children versus that in the elderly is depicted in Figure 1.
Figure 1.
Comparison of asthma in children and the elderly. Asthma in children is more TH2 type and eosinophillic in nature as opposed to elderly, in whom asthma is more infectious and neutrophilic [4,6,8,9]
The pathogenesis of childhood asthma consists not only of genetic tendency and environmental effects but also developmental influences since this is a period of rapid growth for not only the lung but also the immune system. [6] It is strongly connected to atopy, which is a genetic tendency to produce immunoglobulin E antibodies as a reaction to everyday exposures that is associated with Th2 responses, characterized by increased levels of Th2-cytokines such as IL-4, IL-5, IL-6 and IL-13 and increased levels of total and allergen specific antibodies. [7]
The pathogenesis of the asthma phenotype in elderly asthmatics is still poorly understood. [8] The function of eosinophil remains, but the amount of neutrophils goes up. [9] Immunosenescence, which is the enhanced vulnerability to begin or aggravate airway inflammation, has been recognized to shape the innate and adaptive immune systems with age. [4,8] For example, studies in older asthmatics have presented changes in the innate immune system via senescence of epithelial cells of the lung, augmented neutrophils in the airway, reduced function of eosinophils, and a decline in the amount of macrophages and cytotoxic natural killer cells. Adaptive immune system modifications presented were the reduction in the performance of dendritic cells and a persistent proinflammatory condition illustrated by high quantities of TNF-α, IL-1 and IL-6. [4]
CD 4+ regulatory T (Treg) cells repress the effector performance of T cell subsets, such as Th1, Th2 and Th17. Natural Treg cells depict the transcription factor forkhead box protein 3 (FoxP3) and facilitate suppression principally via cell-to-cell interaction. Various data of asthma with aging have proposed a protective role of Treg cells. Furthermore, this study determined that ageing asthmatics had decreased FoxP3 mRNA expression in peripheral blood mononuclear cells. This maintains the notion of reduced immune tolerance by Treg cells in the elderly asthmatics and therefore favors a persistent inflammatory process. [8] Another study involving mouse models stated that initial infection of the mice with the gastric pathogen Helicobacter pylori improves the amount of Treg cells in the airways and may therefore protect against the formation of asthma. Since it was seen that the amount of immature dendritic cells went up along with the amount of Treg cells in the lungs of neonatally infected mice, this study hypothesized that H. pylori-induced Treg cells could provide protection against airway inflammation by blocking the growth of dendritic cells in the lung. [10]
3. Diversity of Microbes
A general presumption is that the human microbiome is favorable to the host because of stimulation and development of immune systems, maturation of mucosal structure and function as well as delivering actual ‘colonization resistance’ against pathogen attack. [11] Typically the lung has been understood to be sterile with little to no specific microbiome in a normal person. Instead, the lung holds low quantities of bacterial communities comparable to the upper respiratory tract, which may be steady or transient. [12]
Frequent causes of acute wheezing and exacerbations of asthma are due to both respiratory infections as well as allergen exposure. [13] Potential pathogens include respiratory syncytial virus (RSV), human rhinovirus (HRV), human metapneumovirus, human parainfluenza virus (HPIV), human bocavirus, influenza viruses, adenoviruses, and enterovirus. Both RSV and HRV have been most commonly linked to the exacerbation of asthma as well as the pathogenesis. [14] Low levels of parainfluenza and influenza viruses have generally been detected in asthma exacerbations. [15]
Newer studies have begun to introduce typical bacteria such as Streptococcus pneumonia and Haemophilus influenza, as well as atypical bacteria such as Chlamydia pneumonia as being responsible for acute wheezing in children. [16] Chlamydia appears to promote innate immunity via pro-inflammatory cytokines and allergic reactions by a Th2 cellular pathway as well as adaptive immunity via antibody construction. [17] Table 1 lists all the different microbes involved in the possible causation and/or exacerbation of asthma.
Table 1.
The Microbes involved in Causation and Exacerbation of Asthma
| Genome | Causation/Exacerbation | References | |
|---|---|---|---|
| Bacteria | |||
| Streptococcus pneumoniae | Gram +, α-hemolytic | ++/− | [9,19] |
| Haemophilus influenza | Gram -, rod shaped | ++/− | [9,19] |
| Moraxella Catarrhalis | Gram -, oxidase positive | ++/− | [9,19] |
| Staphylococcus Aureus | Gram + | ++/− | [9,19] |
| Ureaplasma Urealyticum | Lack a cell wall | ++/− | [9,19] |
| Chlamydia pneumonia | Obligate intracellular pathogen | ++/++ | [9,19] |
| Mycoplasma pneumonia | Lack a cell wall | −/++ | [9,19] |
| Virus | |||
| RNA: | |||
| Picornaviruses- | ss +ve | ||
| Human Rhinovirus | −/+++ (20–80%) | [9,10,22,23,24] | |
| Paramyxovirus | ss −ve | ||
| Respiratory Syncytial Virus | ++/+++ (5–50%) | [5,9,10,22,23,24] | |
| Human Metapneumovirus | −/++(2–13%) | [9,10,22,23,24] | |
| Human Parainfluenzavirus | −/+(1–8%) | [9,10,22,23,24] | |
| Orthomyxovirus | ss −ve | ||
| Human Influenza A,B,C virus | −/+(1–9%) | [9,10,22,23,24] | |
| Coronavirus- | ss +ve | ||
| HCoV-229E HCoV-NL63 HCoV-OC43 HCoV-HKU1 |
−/+ (1–4%) | [9,10,22,23 ,24] | |
| DNA: | |||
| Adenovirus | ds linear | −/+ (<7%) | [9,10,22,23,24] |
| Bocavirus | ss circular | −/+ (6–13%) | [9,10,22,23,24] |
| Fungus | |||
| Aspergillus | Thermotolerant, ubiquitous | −/++ | [26,27] |
| Cladosporium | −/++ | [26,27] |
Abbreviations: ++, +++ represent degree of evidence showing causation and -,means no evidence.
Fungal infections have developed into a considerable health care problem in recent years and there has been a remarkable increase in the incidence of fungal respiratory tract illnesses over the past few decades. [20] Indoor fungal exposure showed an elevation in asthma exacerbations, whereas outdoor contact was more clearly linked to symptom impairment. A two-year prospective analysis, the most frequently retrieved outdoor fungi included the Aspergillus, Cladosporium, Penicillium and Alternaria genera. [21]
4. Microbiome and Asthma
The outermost sections of the nose, the nostrils, assist in filtering inhaled air and are dominated by bacteria from the classes: Staphylococcus, Corynebacterium and Propionibacterium. Studies using 16S rRNA gene clone libraries found that Actinobacteria were the most common sequences from this site. On the other hand, the oropharynx harbors bacteria from the classes Streptococcus, Haemophilus, Neisseria with minor amounts of Staphylococcus and assorted anaerobic bacteria. [22] It has been noted that children with asthma had more proteobacteria in their respiratory system, including pathogens such as influenza while non-asthmatics were seen to have more bacteroidetes, which is bacteria found in the environment such as seawater and soil. [23]
Huang et al. postulated, in a first of its kind study, that the airway maintains a multifarious group of bacteria that may add to clinical aspects of asthma among asthmatics that use inhaled corticosteroids. Also noted was that the amount of hyperresponsiveness in the bronchi demonstrated by participants was associated to community organization and the amount of particular bacterial families encompassing the microbiota in the airway. It was suggested that occupation by certain bacteria could promote inflammation, disease production, and possibly even disease heterogeneity (see Table 2). For example, bacteria in the family Comamonadaceae, which are recognized to have the ability to break down steroids, can potentially be a factor to steroid resilient asthma pathology. [24]
Table 2.
List of Bacteria from the Proteobacteria Phylum involved in bronchial hyperresponsiveness*
| Family/Taxon Representative Species | Clinical Manifestations | Reference |
|---|---|---|
| Bartonellaceae/Bartonella quintana | Trench fever, Chronic Bacteremia, Endocarditis, Bacillary Angiomatosis, Lymphadenopathy and recently correlated with bronchial hyperresponsiveness | [32] |
| Caulobacteraceae/Brevundimonas diminuta | Undetermined clinical significance, possible Nosocomial infections and recently correlated with bronchial hyperresponsiveness | [33, 34] |
| Comamonadaceae/Comamonas testosteroni | Rarely been implicated in human infections of the abdomen, bloodstream, central nervous system, and urinary tract. Recently correlated with bronchial hyperresponsiveness | [35] |
| Enterobacteriaceae/Serratia fonticola | Possible respiratory tract infections and recently correlated with bronchial hyperresponsiveness | [36, 37] |
| Nitrosomonadaceae/Nitrosomonas eutropha | Recently correlated with bronchial hyperresponsiveness | [38] |
| Nitrosomonadaceae/Nitrosomonas europaea | Recently correlated with bronchial hyperresponsiveness | [39] |
| Ralstoniaceae/Wautersia paucula | Strains have sporadically been isolated from blood, wounds, sputum, urine, eye, throat and peritoneal fluid. Recently correlated with bronchial hyperresponsiveness | [40] |
| Shewanellaceae/Shewanella sp. | Rarely implicated in cellulitis, bacteremia, otitis infections, respiratory distress, intra-abdominal infection, pneumonia, and empyema. Recently correlated with bronchial hyperresponsiveness | [41] |
| Sphingomonadaceae/Sphingomonas paucimobilis | Nosocomial infections, rarely been isolated from blood, sputum, urine, wound, bile, cerebrospinal fluid, vagina and cervix. Recently correlated with bronchial hyperresponsiveness. | [42] |
Compiled based on the report by [31]
The gastrointestinal tract of a human is considered sterile prior to birth with establishment of microbes directly after and completion by about the age of 2 years and throughout life, intestinal flora are fairly steady and hard to alter permanently. [36] Also, diet was reported to have a substantial influence on the configuration of the gut microbiota. [37*] Several studies have determined that the microbiota of allergic and non-allergic infants are distinct even before the progression of symptoms, with the most critical period being the first 6 months. [38] At one time the gut microbiota was believed to be only destructive, but now it is clear that commensal bacteria produce many favorable tasks as well, such as organizing the inflammatory reactions. [37*] Findings depicted that Staphylococcus, Enterobacteriaceae, Bacteroides and Clostridium are linked to an increased chance for atopic disease while Bifidobacterium and Lactobacillus presented a more shielding outcome. [38] For instance, children from urban cities of Sweden harbored less Bifidobacterium and Lactobacillus and more Staphylococcus and Clostridium in their bowels compared to children from Estonia, where asthma is not prevalent. Non-digestible nutrients called probiotics have been used to selectively promote an increase of bifidobacteria and lactobacilli. [39]
As an increasing trend in various parts of the world, probiotics have been encouraged as a way to control allergic diseases. It has been recognized since the beginning of the 20th century, that lactic acid bacilli in fermented milk can produce advantages in health. [36] Theoretically, probiotics could regulate TLRs leading to a Th1 response. [39] However, currently there is no hard evidence based on conducted human studies, to support an advantageous role of probiotics in the management of asthma. [36] It may be necessary to include dietary concerns in future studies since probiotics need fiber for their metabolism. It is also unclear in these prior studies if the probiotics were able to persist during their passage through the stomach. [37*]
It has been recognized for many years now that children raised on farms are less likely to develop asthma and allergies than others. This is the essence of the ‘hygiene hypothesis,’ which is the notion that the comparatively germ-free lifestyle most of us now live can disturb the maturation of the immune system. [23] These ideas presumably led to the ‘diet hypothesis’ that suggests a different idea, in which the children living on farms have a vastly different diet from those in urban environments. A noteworthy challenge is that of Japan, where there is a great amount of hygiene and development, but fewer cases of asthma than in the United States. Another example is that of the American urban lower class, which have a greater number of asthma cases as well as infectious diseases that suggests diet has more of a correlation than originally believed. [37*]
5. Links between Microbes & Asthma: What are the mechanisms?
Asthma is a chronic inflammatory disorder of the airways comprising of several cells and mediators. [15] It is a type-1 hypersensitivity reaction characterized by inflammation, recurrent bronchial obstruction, mucus hypersecretion and bronchial hyper-reactivity. The principal cells involved are eosinophils, CD4+ T cells and mast cells. [40] Studies show that Th2 cells, which are part of the innate immune response, predominate in asthma. [15] Mast cells are chief cells in the immediate reaction in asthma. After an initial exposure, that allergen or offending pathogenic antigen may trigger these cells to begin their IgE-mediated degranulation within minutes following their next contact. Granules from mast cells include proinflammatory molecules such as heparin, proteases, tumor necrosis factor and histamine. Leukotrienes and prostaglandins are also secreted from these cells, which begin the activation of basophils, eosinophils, T cells, macrophages, neutrophils and other immune cells in the lung which further cause an immediate reaction of symptoms such as bronchial spasms, edema and smooth muscle contraction. [40]
Pathogenic Link
A dynamic relationship occurs amongst the respiratory host defenses and the microbial flora, which inhabits the airway mucosal surfaces or enters via inhaled air. Each segment of the respiratory tract is predisposed to disease. The respiratory epithelium is an important barricade, which includes a coating of mucus secretions, immunoglobulins and cilia that generate mucociliary clearance. It has been noted that with age, there is a decrease of mucociliary performance due to a delayed clearance time and a decrease in the rate of ciliary beating. [17] The inhaled air then travels down the trachea towards the alveolar space while the bronchial mucosa entraps microbes, allergens and debris to either remove or react to them. The mucus and goblet cell secretions capture these particles to be driven back up the airway, while the macrophages and dendritic cells take up the other items. The host must be able to authorize discriminatory antigen tolerance otherwise it would react to every environmental antigen that was inhaled. Once the air makes its way to the respiratory bronchioles, the surfactant proteins assist in controlling inflammation via the innate immunity response. When a bacteria presents to the alveolus, it becomes opsonized and phagocytized by a macrophage possibly requiring a C3b fragment. If necessary, macrophages discharge chemokines that create inflammation and draw neutrophils. [17]
Cellular Links
Despite extensive studies, notably the questions still remain as to the patho-physiologic link concerning infection and long-term allergic disease. [41**] Numerous studies have begun to display the consequence of wheezing due to lower respiratory viral infections and the subsequent risk of asthma. [42*] The latest theory for the development of asthma is that viral infections function alongside allergy to generate the essential immune and physiologic conditions. [41**]
Eosinophils, usually thought to be key effector cells in asthma, are also considered to potential link between viral infections and asthma. Recruitment of eosinophils is considered to be a key trigger of the inflammation, which leads to bronchoconstriction, destruction of tissue and ultimately respiratory dysfunction. Lately, studies have depicted eosinophils stimulating the removal of viruses and antiviral host immunity and more specifically, decreasing the contamination of RSV in epithelial cells in vitro. Although no direct research has established a causal correlation, an extended study involving the viruses occurring during the winter and the timing of birth independently estimated the incidence of asthma. [41**]
Acute viral and bacterial infections of the lung also cause neutrophil infiltration to the lungs as part of the innate immune response. This is particularly true for non-allergic subjects, who have less circulating eosinophils. [43] There is increasing evidence showing that neutrophils are significant in the lungs of some patients with chronic asthma as well as in some patients whose death has been connected to exacerbations of asthma. One study suggested that intra-tracheal antigen challenge-induced late asthmatic response (LAR) is partially facilitated by the invasion of neutrophils but not eosinophils. This antigen stimulated neutrophilia was reliant on activated CD4+ cells that participated in the development of IL-17A, keratinocyte-derived chemokine (KC) and macrophage inflammatory protein (MIP)-2. [43] Further, during an inflammatory response, circulating neutrophils must cross the blood vessel endothelium in a luminal-to-abluminal direction to gain access to injured lung tissue. A recent report has shown that transendothelial migration does not always proceed down in one direction; neutrophils can undergo 'reverse transendothelial migration' and potentially spread the inflammatory response to other tissues. [44]
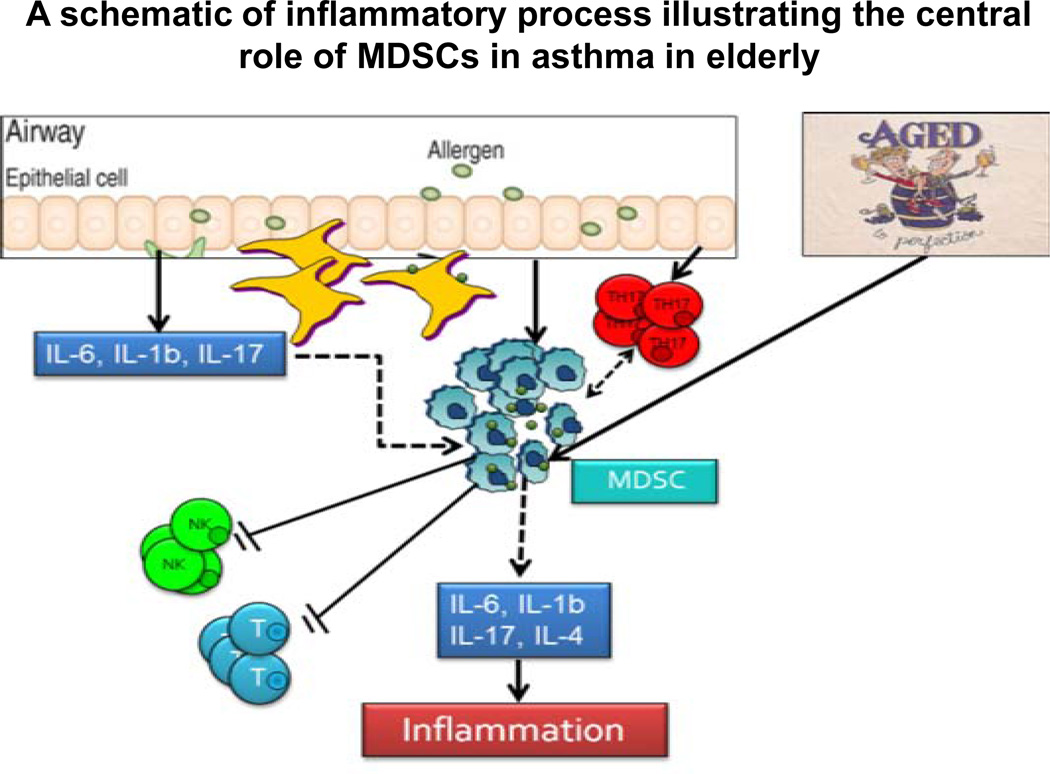
Moreover, a heterogeneous group of myeloid cells, termed myeloid derived suppressor cells (MDSC) is known to accumulate in almost all known inflammatory conditions. MDSC is comprised of pathologically activated precursors of granulocytes, macrophages and dendritic cells. They are characterized by the potent ability to inhibit T cell functions. Our unpublished studies in mice, which involved low doses of bacterial lipopolysaccharide injection or Respiratory Syncytial Virus infection showed a significant expansion and accumulation of the MDSC population. The increase was associated with proinflammatory factors like IL-7 and IL-17. Thus, MDSCs might be involved as a regulatory component, of the immune system and protect tissue from immune mediated damage. Under physiological conditions, these cells are absent and exist as immature myeloid cells. In conditions of chronic infection or inflammation as seen in asthma, there is a marked expansion of and activation of a large population of myeloid cells with immunosuppressive abilities. These cells termed MDSC accumulate in peripheral lymphoid organs and home into sites of inflammation, where they contribute to immunosuppression and are particularly relevant to the elderly asthmatics. A schematic illustration of the role of MDSCs in asthmatics is shown in Figure 2. Furthermore, some evidence suggests that MDSCs can also induce the expansion of CD 4+ regulatory T (Treg) cells. Future studies will reveal the biology of these cells in asthma and if can be considered part of a natural immune regulatory network.
Figure 2.
The respiratory system in older adults is continuously exposed to a barrage of potentially harmful external factors, leading to systemic inflammation. Inflammation involves a well-orchestrated group of cytokines and cellular events from an acute to a chronic stage. Acute inflammation, begins with the exposure of airway epithelium to environmental factors such as allergens, viral and bacterial infections, which produce a number of cytokines and chemokines including IL-6, IL-1β, IL-17 and TNF-α. These cytokines then act on monocytes and activate both the innate and adaptive immunity in the lung. The MDSCs play a central role in the process.
Molecular Links
The major molecular links between microbial infection and asthma comprise (i) activation of innate immune system of the host via a combination of innate immune cells and molecules that are necessary for the detection and removal of pathogens via activation of a number of signaling pathways that initiate antimicrobial response, (ii) a cascading inflammatory response associated with innate immune activation, and (iii) the production of cytokines and chemokines that play a role also in asthma pathogenesis. In order to invade the host cell, intracellular microbe must regulate both the innate and adaptive immune systems. [45]
The innate immune system has a crucial part in identifying viral infections and inducing primary anti-viral responses. Once the virus inhabits its host, infected non-immune cells identify it, and with the help of innate immune cells they promptly induce a response by the assembly of type I interferons (IFN) and proinflammatory cytokines. Intracellular signaling cascades monitor the expressions of these genes, which are triggered by germline-encoded pattern recognition receptors (PRRs). Nucleotide-binding oligomerization domain (NOD)-like receptor (NLR) proteins, which exist in ‘inflammasomes,’ toll-like receptors (TLRs), and retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs) are groups of innate pattern recognition receptors that recognize viral RNAs or DNAs. TLRs mainly distinguish viruses within the endosomal compartments whereas RLRs are important in the recognition of cytosolic pathogens. [46]
Bacteria may be divided into two major classes depending upon different staining characteristics of their cell walls, specifically Gram-positive and Gram-negative. The distinctive cell wall components stimulate the immune cells and serve as pathogen-associated molecular patterns (PAMPs) and are recognized by specific TLRs. [47] TLRs can detect several constituents of the bacterial cell wall, such as peptidoglycan from gram-positive bacteria and lipopolysaccharides from gram-negative bacteria. [48] Recent studies show that intracellular bacteria infect the host cells and flourish inside the cell, thus avoiding luminal defenses and therefore depend upon NLRs for detection. [47] These signals then instigate an inflammatory cascade, which assists in surrounding the infection as well as activating the adaptive immune system. [49]
The ultimate molecular mechanisms that link microbial infections to asthma include a plethora of cytokines and chemokines, which are produced in response to infection and innate and adaptive immune activation, but are also relevant to modulating asthma. Cytokines and chemokines attach to cells and conduct cell-to-cell communications among dendritic cells and lymphocytes. [47, 50] The adaptive immune system is made up of B and T cells that impart pathogen-specific immunity to the host via reordering of antigen receptor genes. [48] B cells generate pathogen-specific antibodies to counteract toxins distributed by pathogens, while T cells deliver cytokines to eliminate pathogen infected cells via their cytotoxic effects or from signals to B cells. [47] A key feature of the adaptive immune system is memory. [50] Once an antigen is presented to a naïve T cell that CD4+ cell will further differentiate into a T helper type 1 cell (Th1) or T helper type 2 cell (Th2). Th1 cells assist delayed type hypersensitivity reactions and specifically makes IL-2 and IFN-γ. The Th2 cells produce cytokines such as IL-4, IL-5, and IL-13, which stimulate B-lymphocytes to produce IgE. [40]
8. Concluding Remarks
There has been substantial progress in our understanding of the relationship between different infections and asthma. However, several significant questions remain unanswered. A major question remains is the impact of external microbial population in a specific geographic location on local and systemic immunity of humans, who live in this environment, which also may be referred to as “ecological immunity”. The microbial infections may produce differential responses based on ecological immunity. What is still unanswered about the epigenetics of asthma in the elderly and how do they contrast from those of younger adults and children? Where and how does fungus play a role in asthma? A better understanding of the qualities of a host’s genome and microbiota, and their interactions, will lead to more individualized methodologies to the prevention and management of asthma.
Further, there has been advance in understanding the link between viral infections and the development or exacerbation of asthma, but less is known on the role of bacterial infections and for both the molecular mechanism underlying the causal nature of the relationship remains to be elucidated. From the presented data, the development of asthma arises from a multifaceted communication of genetic susceptibility and environmental aspects with viral and bacterial infection likely playing a meaningful role in the effect of asthma inception. Moreover, the role of local microbiota in the nasal cavity or in the lung may significantly affect the effects of exogenous infectious agent (s). An integration of ecological immunity (combines host genetics and environmental microbial burden) and local microbiota will be required to investigate the immunologic, cellular and molecular effects of an incipient microbe. Future studies involving appropriate population and laboratory investigations, animal models and in vitro studies are necessary to achieve elucidate the links between microbial infections and either pathogenesis or protection of asthma.
Key Points.
Bacteria have now been recognized as a possible causation of asthma while fungal microbes have been identified in exacerbation cases.
Asthmatics were noted to have more proteobacteria in their respiratory system while non-asthmatics were seen to have more bacteroidetes.
The potential link between microbial infections and asthma is now thought to be innate immune cells such as eosinophils, neutrophils and he myeloid-derived suppressor cells.
Cytokines and chemokines elaborated by infections constitute the molecular mechanisms linking microbial infections to asthma.
ACKNOWLEDGEMENTS
The financial support by NIH (1RO1CA152005-01,1P30HL101265-0) and VA Merit review and RCS Award by Dept of Veteran Affairs are duly acknowledged
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest
Authors have no conflicts of interest to declare.
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
* of special interest
** of outstanding interest
- 1. Umetsu DT, DeKruyff RH. 99th Dahlem Conference on Infection, Inflammation and Chronic Inflammatory Disorders: Microbes, apoptosis and TIM-1 in the development of asthma. Clin Exp Immunol. 2010;160:125–129. doi: 10.1111/j.1365-2249.2010.04136.x.. This article provides insight as to possible reasons why current studies have indicated the Hepatitis A virus as an important atopy susceptibility gene.
- 2.Health Data Interactive. (HDI) [Accessed July 2011];CDC. National Center for Health Statistics. www.cdc.gov/nchs/hdi.htm.
- 3.Kamble S, Bharmal M. Incremental Direct Expenditure of Treating Asthma in the United States. J Asthma. 2009;46(1):73–80. doi: 10.1080/02770900802503107. [DOI] [PubMed] [Google Scholar]
- 4.Tzortzaki EG, Proklou A, Siafakas NM. Asthma in the Elderly: Can We Distinguish It from COPD? J Allergy. 2011:843543. doi: 10.1155/2011/843543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jartti L, Langen H, Soderlund-Venermo M, et al. New Respiratory Viruses and the Elderly. The Open Respiratory Medicine Journal. 2011;5:61–69. doi: 10.2174/1874306401105010061.. This review presents new viruses and their clinical importance with specificity to the elderly population.
- 6.Chung HL. Asthma in childhood: a complex, heterogeneous disease. Korean J Pediatrics. 2011;54:1–5. doi: 10.3345/kjp.2011.54.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumar RK, Hitchins MP, Foster PS. Epigenetic changes in childhood asthma. Disease Models & Mechanisms. 2009;2:549–553. doi: 10.1242/dmm.001719. [DOI] [PubMed] [Google Scholar]
- 8.Vale-Pereira S, Todo-Bom A, Geraldes L, Schmidt-Weber C, Akdis CA, Mota-Pinto A. FoxP3, GATA-3 and T-bet expression in elderly asthma. Clin Exp Allergy. 2011;41:490–496. doi: 10.1111/j.1365-2222.2010.03640.x. [DOI] [PubMed] [Google Scholar]
- 9.Reed CE. Asthma in the elderly: Diagnosis and management. J Allergy Clin Immunol. 2010;126:681–687. doi: 10.1016/j.jaci.2010.05.035. [DOI] [PubMed] [Google Scholar]
- 10.Arnold IC, Dehzad N, Reuter S, Martin H, Becher B, Taube C, Müller A. Helicobacter pylori infection prevents allergic asthma in mouse models through the induction of regulatory T cells. J Clin Invest. 2011;121(8):3088–3093. doi: 10.1172/JCI45041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blaser MJ, Falkow S. What are the consequences of the disappearing human microbiota? Nat Rev Microbiol. 2009;7:887–894. doi: 10.1038/nrmicro2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Charlson ES, Bittinger K, Haas AR, Fitzgerald AS, Frank I, Yadav A, et al. Topographical Continuity of Bacterial Populations in the Healthy Human Respiratory Tract. Am J Respir Crit Care Med. 2011 doi: 10.1164/rccm.201104-0655OC. Epub 2011 June 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kloepfer KM, Gern JE. Virus/Allergen Interactions and Exacerbations of Asthma. Immunology and Allergy Clinics of North America. 2010;30:553–563. doi: 10.1016/j.iac.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujitsuka A, Tsukagoshi H, Arakawa M, Goto-Sugai K, Ryo A, Okayama Y, et al. A molecular epidemiological study of respiratory viruses detected in Japanese children with acute wheezing illness. BMC Infectious Diseases. 2011;11:168. doi: 10.1186/1471-2334-11-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McErlean P, Greiman A, Favoreto S, Avila, Pedro C. Viral Diversity in Asthma. Immunology and Allergy Clinics of North America. 2010;30:481–495. doi: 10.1016/j.iac.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bisgaard H, Hermansen M, Bønnelykke K, Stokholm J, Baty F, Skytt NL, et al. Association of bacteria and viruses with wheezy episodes in young children: prospective nested birth cohort study. BMJ. 2010;341:c4978. doi: 10.1136/bmj.c4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reynolds HY. Modulating airway defenses against microbes. Curr Opin Pulm Med. 2002;8:154–165. doi: 10.1097/00063198-200205000-00002. [DOI] [PubMed] [Google Scholar]
- 18.Kusel MM, de Klerk NH, Kebadze T, Vohma V, Holt PG, Johnston SL, et al. Early life respiratory viral infections, atopic sensitization, and risk of subsequent development of persistent asthma. J Allergy Clin Immunol. 2007;119:1105–1110. doi: 10.1016/j.jaci.2006.12.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Paula NT, Carneiro BM, Yokosawa J, Freitas GRO, de Mattos Oliveira TF, Costa LF, et al. Human rhinovirus in the lower respiratory tract infections of young children and the possible involvement of a secondary respiratory viral agent. Mem Inst Oswaldo Cruz. 2011;106(3):316–321. doi: 10.1590/s0074-02762011000300010. [DOI] [PubMed] [Google Scholar]
- 20.Biswas D, Agarwal S, Sidhwani G, Rawat J. Fungal colonization in patients with chronic respiratory diseases from Himalayan region of India. Ann Clin Microbiol Antimicrobials. 2010;9:28. doi: 10.1186/1476-0711-9-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pongracic JA, O’Connor GT, Muilenberg ML, et al. Differential effects of outdoor versus indoor fungal spores on asthma morbidity in inner-city children. J Allergy Clin Immunol. 2010;125:593–599. doi: 10.1016/j.jaci.2009.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lemon KP, Klepac-Ceraj V, Schiffer HK, Brodie EL, Lynch SV, Kolter R. Comparative analyses of the bacterial microbiota of the human nostril and oropharynx. mBio. 2010;1(3) doi: 10.1128/mBio.00129-10. e00129-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huffnagle GB. The Microbiota and Allergies/Asthma. PLoS Pathog. 2010;6(5):e1000549. doi: 10.1371/journal.ppat.1000549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang YJ, Nelson CE, Brodie EL, et al. Airway microbiota and bronchial hyperresponsiveness in patients with suboptimally controlled asthma. J Allergy Clin Immunol. 2011;127:372–381. doi: 10.1016/j.jaci.2010.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Foucault C, Brouqui P, Raoult D. Bartonella quintana characteristics and clinical management. Emerg Infect Dis. 2006;12(2):217–223. doi: 10.3201/eid1202.050874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scotta C, Bennasar A, Moore ER, Lalucat J, Gomila M. Taxonomic characterisation of ceftazadime-resistant Brevundimonas isolates and description of Brevundimonas faecalis sp. nov. Syst Appl Microbiol. 2011;34(6):408–413. doi: 10.1016/j.syapm.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 27.Han XY, Andrade RA. Brevundimonas diminuta infections and its resistance to fluroquinolones. J Antimicrob Chemother. 2005;55(6):853–859. doi: 10.1093/jac/dki139. [DOI] [PubMed] [Google Scholar]
- 28.Tsui TL, Tsao SM, Liu KS, Chen TY, Wang YL, Teng YH, et al. Comamonas testosteroni infection in Taiwan: Reported two cases and literature review. J Microbiol Immunl Infect. 2011;44:67–71. doi: 10.1016/j.jmii.2011.01.013. [DOI] [PubMed] [Google Scholar]
- 29.Bollet C, Gainnier M, Sainty JM, Orhesser P, De Micco P. Serratia fonticola isolated from a leg abscess. J Clin Microbiol. 1991;29(4):834–835. doi: 10.1128/jcm.29.4.834-835.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Samonis G, Vouloumanou EK, Christofaki M, Dimopoulou D, Maraki S, Triantafyllou E, et al. Serratia infections in a general hospital: characteristics and outcomes. Eur J Clin Microbiol Infect Dis. 2011;30(5):653–660. doi: 10.1007/s10096-010-1135-4. [DOI] [PubMed] [Google Scholar]
- 31.Stein LY, Arp DJ, Berube PM, Chain PS, Hauser L, Jetten MS, et al. Whole-genome analysis of the ammonia-oxidizing bacterium, Nitrosomonas eutropha C91: implications for niche adaptation. Environ Microbiol. 2007;9(12):2993–3007. doi: 10.1111/j.1462-2920.2007.01409.x. [DOI] [PubMed] [Google Scholar]
- 32.Chain P, Lamerdin J, Larimer F, Regala W, Lao V, Land M, et al. Complete genome sequence of the ammonia-oxidizing bacterium and obligate chemolithoautotroph Nitrosomonas europaea. J Bacteriol. 2003;185(9):2759–2773. doi: 10.1128/JB.185.9.2759-2773.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vandamme P, Goris J, Coenye T, Hoste B, Janssens D, Kersters K, et al. Assignment of Centers for Disease Control group IVc-2 to the genus Ralstonia as Ralstonia paucula sp. nov. Int J Syst Bacteriol. 1999;49(Pt 2):663–669. doi: 10.1099/00207713-49-2-663. [DOI] [PubMed] [Google Scholar]
- 34.Hau HH, Gralnick JA. Ecology and biotechnology of the genus Shewanella. Annu Rev Microbiol. 2007;61:237–258. doi: 10.1146/annurev.micro.61.080706.093257. [DOI] [PubMed] [Google Scholar]
- 35.Toh HS, Tay HT, Kuar WK, Weng TC, Tang HJ, Tan CK. Risk factors associated with Sphingomonas paucimobilis infection. J Microbiol Immunol Infect. 2011;44:289–295. doi: 10.1016/j.jmii.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 36.Yao TC, Chang CJ, Hsu YH, Huang JL. Probiotics for allergic diseases: Realities and myths. Ped Allergy Immunol. 2010;21:900–919. doi: 10.1111/j.1399-3038.2009.00955.x. [DOI] [PubMed] [Google Scholar]
- 37. Maslowski KM, Mackay CR. Diet, gut microbiota and immune responses. Nature Immunology. 2011;12:5–9. doi: 10.1038/ni0111-5.. This article revisits the hygiene hypothesis and suggests a ‘diet hypothesis’ with connection to urbanization in reference to asthma.
- 38.Vael C, Vanheirstraeten L, Desager KN, Goossens H. Denaturing gradient gel electrophoresis of neonatal intestinal microbiota in relation to the development of asthma. BMC Microbiol. 2011;11:68. doi: 10.1186/1471-2180-11-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Michail S. The role of probiotics in allergic diseases. Allergy Asthma Clin Immunol. 2009;5:5. doi: 10.1186/1710-1492-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Madore AM, Laprise C. Immunological and genetic aspects of asthma and allergy. J Asthma Allergy. 2010;3:107–121. doi: 10.2147/JAA.S8970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Callaway Z, Kim CK. Respiratory Viruses, Eosinophilia and Their Roles in Childhood Asthma. Int Arch Allergy Immunol. 2011;155:1–11. doi: 10.1159/000319842.. This is a very interesting article making the connection between asthma and viruses to be the eosinophil.
- 42. Sly PD. The early origins of asthma: who is really at risk? Curr Opin Allergy Clin Immunol. 2011;11:24–28. doi: 10.1097/ACI.0b013e328342309d.. This review highlights the major risk factors for asthma, such as viral lower respiratory infections, and their transition into asthma.
- 43.Nabe T, Hosokawa F, Matsuya K, et al. Important role of neutrophils in the late asthmatic response in mice. Life Sci. 2011;88:1127–1135. doi: 10.1016/j.lfs.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Woodfin A, Voisin MB, Beyrau M, Colom B, Caille D, Diapouli FM, et al. The junctional adhesion molecule JAM-C regulates polarized transendothelial migration of neutrophils in vivo. Nature Immunol. 2011;12:761–769. doi: 10.1038/ni.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hartlova A, Krocova Z, Cerveny L, Stulik J. A proteomic view of the host–pathogen interaction: The host perspective. Proteomics. 2011;11:3212–3220. doi: 10.1002/pmic.201000767. [DOI] [PubMed] [Google Scholar]
- 46.Takeuchi O, Akira S. Recognition of viruses by innate immunity. Immunol Rev. 2007;220:214–224. doi: 10.1111/j.1600-065X.2007.00562.x. [DOI] [PubMed] [Google Scholar]
- 47.Yano T, Kurata S. Intracellular recognition of pathogens and autophagy as an innate immune host defence. J Biochem. 2011;150(2):143–149. doi: 10.1093/jb/mvr083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumar H, Kawai T, Akira S. Toll-like receptors and innate immunity. Biochem Biophys Res Commun. 2009;388:621–625. doi: 10.1016/j.bbrc.2009.08.062. [DOI] [PubMed] [Google Scholar]
- 49.Barton GM. A calculated response: control of inflammation by the innate immune system. J Clin Invest. 2008;118:413–420. doi: 10.1172/JCI34431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Palm NW, Medzhitov R. Pattern recognition receptors and control of adaptive immunity. Immunol Rev. 2009;227:221–233. doi: 10.1111/j.1600-065X.2008.00731.x. [DOI] [PubMed] [Google Scholar]