Abstract
In this report, we have achieved a richer view of the transcriptome for Chromosomes 21 and 22 by using high-density oligonucleotide arrays on cytosolic poly(A)+ RNA. Conservatively, only 31.4% of the observed transcribed nucleotides correspond to well-annotated genes, whereas an additional 4.8% and 14.7% correspond to mRNAs and ESTs, respectively. Approximately 85% of the known exons were detected, and up to 21% of known genes have only a single isoform based on exon-skipping alternative expression. Overall, the expression of the well-characterized exons falls predominately into two categories, uniquely or ubiquitously expressed with an identifiable proportion of antisense transcripts. The remaining observed transcription (49.0%) was outside of any known annotation. These novel transcripts appear to be more cell-line-specific and have lower and less variation in expression than the well-characterized genes. Novel transcripts were further characterized based on their distance to annotations, transcript size, coding capacity, and identification as antisense to intronic sequences. By RT-PCR, 126 novel transcripts were independently verified, resulting in a 65% verification rate. These observations strongly support the argument for a re-evaluation of the total number of human genes and an alternative term for “gene” to encompass these growing, novel classes of RNA transcripts in the human genome.
The working draft of the human genome has led to several estimates of the total number of encoded genes ranging from 30,000 to 120,000 (Ewing and Green 2000; Liang et al. 2000; Lander et al. 2001; Venter et al. 2001), indicating that the correct number of genes remains a controversial issue. Although 30,000 to 40,000 is presently the most favorable estimate, emerging data based on a variety of computational and experimental approaches indicate that these estimates need to be re-evaluated (Chen et al. 2002; Okazaki et al. 2002; Saha et al. 2002; Guigo et al. 2003; Nekrutenko et al. 2003; Rinn et al. 2003). We recently reported the results of an unbiased analysis of human Chromosomes 21 and 22 that systematically interrogated the entire nonrepetitive regions of these chromosomes for the location of transcription using high-density oligonucleotide arrays (Fodor et al. 1991, 1993; Pease et al. 1994; Kapranov et al. 2002). Transcription for 11 different cell lines (A-375, CCRF-CEM, COLO 205, FHs 738Lu, HepG2, Jurkat, NCCIT, NIH:OVCAR-3, PC-3, SK-N-AS, U-87 MG) was mapped using uniformly spaced oligonucleotide probes (25-mers) that interrogate, on average, every 35 bp of human Chromosomes 21 and 22. An unexpected result was that the vast majority of identified transcribed nucleotides (∼90%) were outside of the annotated regions (defined as exons of RefSeq and Sanger annotations as well as GenBank mRNAs with complete coding regions, as of October 2000). Based on these observations, we concluded that as much as an order of magnitude more of genomic sequence was transcribed into RNA than is accounted for by well-characterized human exons.
Following this study, other investigators have estimated the amounts of transcription emanating from human and mouse genomes using a variety of experimental approaches, such as serial analysis of gene expression (SAGE; Chen et al. 2002), long-SAGE (Saha et al. 2002), spotted DNA microarrays (Rinn et al. 2003), and large-scale EST sequencing (Okazaki et al. 2002). Each of these studies independently indicated that there is a larger proportion of the human and mouse genomes transcribed than previously estimated. Taken together, the observations all point to a large and poorly understood population of discrete RNA transcripts comprising the respective transcriptomes.
In this study, we describe an in-depth analysis of the poly(A)+ cytosolic RNA transcription data reported earlier (Kapranov et al. 2002) and how we constructed a novel view of the transcriptome from Chromosomes 21 and 22 by increasing the verification of the identified novel transcripts, using alternative statistically rigorous analyses of these data, and implementing strategies to construct contiguous transcriptional units. The resulting transcriptome map contains more information regarding well-characterized exons' differential expression and the novel array-detected transcripts size, coding capacity, strandedness, and location in the genome.
RESULTS
Generation of Transcription Maps Based on Collective Behavior of Neighboring Probes
In an earlier analysis of poly(A)+ cytosolic RNA transcripts from Chromosomes 21 and 22 (Kapranov et al. 2002), maps were constructed based on the behavior of each individual probe pair. A probe pair with a background-subtracted perfect match (PM) intensity and mismatch (MM) intensity was positive if the ratio (PM/MM) and difference (PM–MM) exceeded a threshold corresponding to a particular false-positive rate calculated from the negative control probe pairs designed against bacterial sequences. Although this analysis method was shown to be accurate and informative using independent biochemical methods, it was likely that such an analytical approach could suffer from probe sequence-specific effects (e.g., self-structure) and may also be influenced by nonspecific cross-hybridization events that would increase the rate of false-positive determinations. To alleviate these potential shortcomings, we used a robust method that makes use of neighboring-probe intensities within a predefined window to determine whether each probe pair was positive. The local expression level was estimated by calculating the “pseudo-median” or Hodges-Lehmann estimator (Hollander and Wolfe 1999) of all PM–MM values of all probe pairs that lie within the sliding window. More specifically, for probe pair i at genomic coordinate Pi, the expression level Ei is the median of all n(n + 1)/2 pairwise averages (Zj + Zk)/2, where Zj = PMj – MMj and j ≤ k is the set of all probe pairs whose genomic coordinates lie within [Pk – BW, Pk + BW], where BW is the bandwidth and the resulting widow size is given by (2 × BW) + 1. The determination of a suitable window size is constrained by two facts: the spacing of the probes along the chromosomes (approximately every 35 bp) and the median size of an exon on these chromosomes (137 bp). We used a bandwidth of 50 bp, which approximately corresponds to three probe pairs. Multiple bandwidths (comprising a specific number of bases) were initially tested for their ability to achieve the greatest detection sensitivity and specificity based on spiked-in quantitative bacterial RNA transcripts. This approach resulted in greater sensitivity for any given false-positive rate than by using the results of the performance of a single probe pair (data not shown). One fact to be mindful of when using such a window-based approach is that although the false-positive call rate was reduced in identifying the sites of transcription, the performance of the probe pairs was smoothed, making strict determination of the transcription boundaries possibly problematic.
Using this analysis approach, the average of the overall total positive probes detected from the poly(A)+ cytosolic RNA fraction in each cell line tested was 10.3% of the mapped probe pairs on the arrays (Supplemental Table S1 available online at www.genome.org and http://transcriptome.affymetrix.com/download/genome_res_data). This number is increased significantly to 25.8% if one considers all positive probes in the cumulative map of all 11 cell lines (i.e., the “1 of 11” map). These are consistent with our previous analysis (Kapranov et al. 2002).
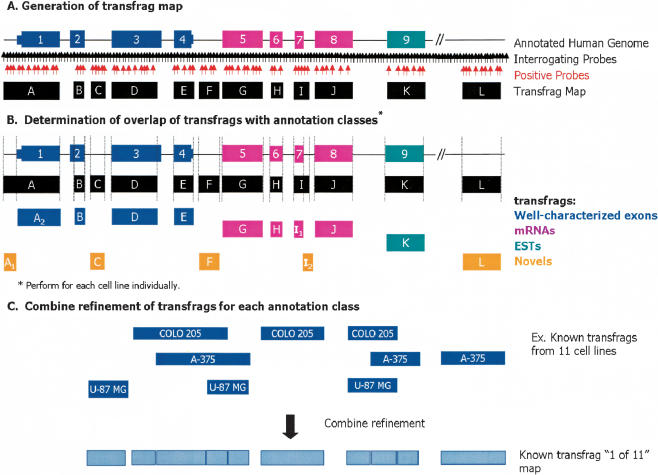
Following the determination of which of the probe pairs were likely to be detecting transcription, the behavior of neighboring probe pairs was evaluated to assemble contiguous fragments of transcribed units (i.e., transfrags). Transfrag maps were constructed by implementing a fixed intensity threshold (threshold = 150), a maximum gap between positive probe pairs (maxgap = 40), and a minimum length of adjacent positive probe pairs (minrun = 90; Fig. 1A). Transfrag maps contain all transfrags that meet these criteria and are thus a chromosome-wide summary of transcriptional fragments. A threshold of 150 gives a median false-positive rate of 2.9% from the negative bacterial controls on all arrays (Supplemental Table S2). These parameters were chosen such that a very conservative transfrag map, low in false-positive calls, would result. Transfrags in such maps would contain no negative probe pairs. This analysis allows for the formation of continuous blocks of transcription as well as further minimizing probe-specific effects. Transfrag maps were generated for each cell line individually as well as a “1 of 11” map, which contains transfrags detected in at least one of the 11 cell lines. The average total number of transfrags found in each cell line was 2965, and the average length of a transfrag was 153 bp (Supplemental Table S3). The number of transfrags increases significantly to 9001 in the “1 of 11” map, yet the average length of a transfrag remains approximately the same, 154 bp, suggesting a considerable tendency toward cell-line-specific transfrags. Other evidence discussed below supports this conclusion.
Figure 1.
Schematic representation of tiled probe pairs and generation of transfrags from human Chromosomes 21 and 22 oligonucleotide arrays. Schematic representation of oligonucleotide arrays interrogating the entire nonrepetitive regions of human Chromosomes 21 and 22 with probes regularly spaced at ∼35-bp intervals. (A) Generation of transfrag map. At each probe position, a bandwidth of 50 was used to determine positive probes above a threshold of 150 (indicated in red) wherein the pseudo-median values for each position are recomputed as a pseudo-median of all probes in the window using the Hodges-Lehmann estimator. Fragments of contiguously transcribed elements termed transfrags were generated by joining positive probes that were separated by a certain distance (maxgap = 40) and whose length was less than a particular size (minrun = 90). (B) Portions of transfrags based on annotations. The transfrags were delineated into the following classes based on their overlap with a predefined set of annotations: (1) well-characterized exons (dark blue); (2) mRNAs (pink); and (3) ESTs (green). Any observed transcription outside of these annotation classes was considered novel (orange). The vertical lines indicate the boundaries of the transfrags. (C) Combined refinement of transfrags for each annotation class. Following this classification, all transfrags that belong to a particular class are combined to form a comprehensive nonoverlapping union of transfrags termed a “1 of 11” map.
It is important to note that alterations in any of these analysis strategies and their corresponding parameters (i.e., window size, minrun, maxgap, etc.), results in different but overlapping maps. By implementing this transfrag approach, a more stringent set of maps can be generated because each probe pair must meet the collective minrun, maxgap, and threshold criteria. The choice of what values to choose for the threshold, minrun, and maxgap criteria ultimately involves a tradeoff between the desired false-positive and sensitivity rates.
Association of Detected Transfrags to Chromosome-Wide Annotations
Following the generation of transfrag maps, regions of observed transcription were classified based on the overlap of base pairs in transfrags to elements in a collection of annotation classes (Fig. 1B). The annotation classes used in this study were (1) known or well-characterized exons, (2) mRNAs from GenBank that do not overlap class 1, and (3) all publicly available ESTs that are not represented in classes 1 or 2. Any observed transcription outside of these annotation classes was considered novel. The well-characterized known class, compiled by UCSC (Kent et al. 2002; Karolchik et al. 2003) and viewable using their genome browser, is based on protein-coding genes from SWISS-PROT, TrEMBL, and TrEMBL-NEW and their corresponding mRNAs from GenBank. Transfrags overlapping with the set of Sanger annotated pseudogenes on Chromosome 22 (Collins et al. 2003) were removed from our analyses because it is difficult to attribute the genomic origin of transcription in these cases. In addition, sequences within 50 bp of annotations were masked to account for the smoothing effect of using a window-based approach (see above). Figure 1B illustrates the identification and delineation of transfrags into these annotation classes. It is important to note that the relationship of neighboring transfrags (i.e., determining if they are deemed to be part of the same transcript) cannot be inferred without additional experimental information such as isolation of contiguous cDNAs containing the identified transfrags.
In many instances, transfrags that overlapped annotations extended well beyond the annotation bounds. Such extensions could be the result of overlapping transcription from an adjacent gene, extensions of exons or UTRs, or antisense transcription at such loci. To differentiate transfrags that are synonymous with annotations versus transfrags that are extensions of known annotations, transfrags that overlapped and extended any annotation were fragmented to derive a unique set of novel transfrags (Fig. 1B). Following this classification, a “1 of 11 known” map and a “1 of 11 novel” map were compiled to represent the nonoverlapping union of all known or novel transfrags from all cell lines using a combine refinement approach (Fig. 1C).
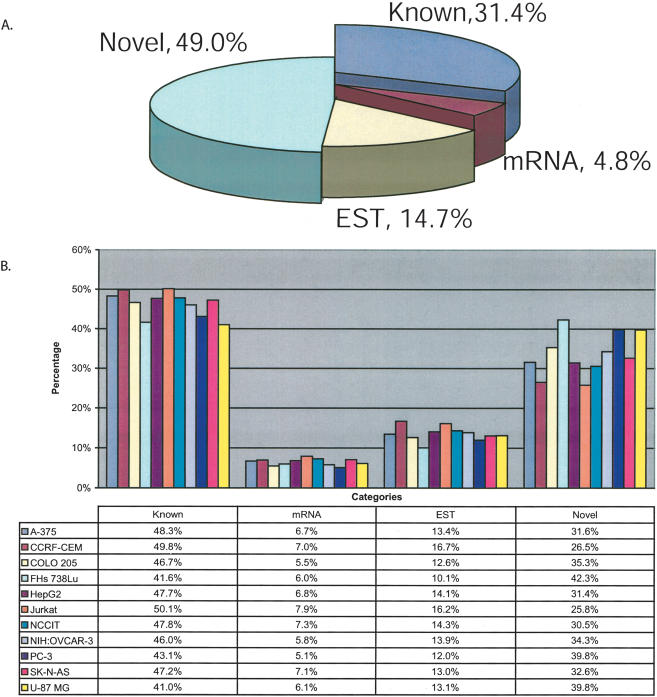
Approximately half (49.0%) of the base pairs within transfrags mapped along Chromosomes 21 and 22 do not overlap with any well-characterized exon, mRNA, or EST, in at least “1 of 11” cell lines (Fig. 2A). Only 31.4% of the observed transcription aligned within known exons. An additional 4.8% and 14.7% of the transcription were seen within mRNAs and ESTs, respectively. For each cell line, the proportion of detected transcription within the well-characterized exons ranges from 41.0%–50.1%, 5.1%–7.9% in mRNAs, 10.1%–16.7% in ESTs, and 26.5%–42.3% of the observed transcription is novel (Fig. 2B). Interestingly, if one considers only the probe pairs without the generation of transfrags, ∼52% of the positive probes are novel, 11% overlap known exons, 13% overlap mRNAs, and 24% overlap ESTs (data not shown). A similar proportion of novel transcription was seen in human fetal brain poly(A)+ RNA (data not shown), indicating that the observed abundant novel transcription was not restricted to the cultured cell lines but can also be observed in a normal tissue.
Figure 2.
Characterization of all transfrags based on annotations. Transfrags from all 11 cell lines were classified based on their base pair overlap with annotations. The annotations are (1) Known, overlapping with known annotations (compiled by UCSC Genome Browser based on protein-coding genes from SWISS-PROT, TrEMBL, and TrEMBL-NEW); (2) mRNA, overlapping with mRNAs and not known; (3) ESTs, overlapping with ESTs and not known or mRNAs; and (4) Novel, not overlapping known exons, mRNAs, or ESTs. (A) Pie chart representing all transfrags from “1 of 11” map. (B) Bar graph shows the percentage of all transfrags from each of the 11 cell lines by annotation class. An overlap of even a single base pair is considered as an intersect between the transfrag and annotation.
Relating Transfrags to Genes
Using these alternative analyses approaches, transcripts mapping to well-characterized exons of known genes were also readily detectable. On Chromosomes 21 and 22, there are 990 well-characterized genes that are composed of 6463 exons (Kent et al. 2002). Given the median size of an exon on these chromosomes (137 bp) and the 35-bp resolution of the arrays, a total of 92.8% (5995/6463) of the exons have at least one interrogating probe pair. A single probe pair interrogates 15% (899/5995) of these 5995 exons on the array. The exons that were not represented on the array are either small in size (i.e., <50 bp), enriched in repeat sequence (which are eliminated during probe selection), or contain sequences that would be problematic to interrogate (e.g., G-quartets or palindromes that were penalized against during probe selection and therefore unlikely to be included on the array). An exon was considered present when at least 30% of the interrogating probe pairs were positive (Supplemental Fig. S1). The average number of probe pairs in known exons is ∼7.5. In the “1 of 11” map, 84.5% (5068/5995) of the known exons were detected and 70.5% (27,088/38,407) of the probe pairs within these exons are positive (Supplemental Fig. S1). The total percentage of known exons detected in each cell line ranged from 40.8% to 63.8%, with 27.6%–47.1% of the probes pairs within these exons positive (Supplemental Fig. S1).
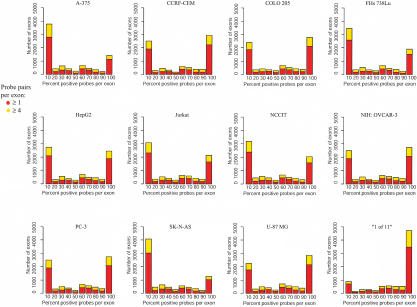
The binning of positive probes in exons resulted in a bimodal distribution indicating that the majority of exons fell into two cases: exons that have no or few positive probes (<10%) and exons that have all or mostly all (>90%) positive probes (Fig. 3). A similar trend resulted when using all exons or only exons that contain ≥4 probe pairs (yellow vs. red profiles, Fig. 3). Additionally, in the “1 of 11” map, an overwhelming population of exons appear to have >90% positive probes, indicating there was a significant amount of differential expression of exons. Exons with only a portion of the interrogating probes positive (10%–90%) imply shorter alternative exon isoforms. However, two factors make unambiguous interpretation of these results challenging: (1) probes denoting expression of a particular exon can come from either strand; and (2) any observed expression proximal to an annotated exon may result from a longer alternative isoform of the exon. Thus, larger or shorter exon isoforms and overlapping transcripts limit the comprehensive characterization of alternative splicing isoforms.
Figure 3.
Distribution of positive probes in exons. The distribution of the percent of positive probes in exons is plotted by cell line as well as for the “1 of 11” map. The red bars represent all interrogated exons, and the yellow bars represent exons that contain four more interrogating probe pairs.
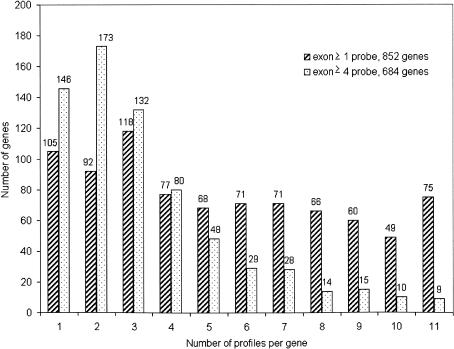
By generating an “on/off” profile for each exon on the array (5995) in all genes (990) in all cell lines, we estimated the degree of differential expression in terms of exon skipping. The collection of genes selected for this analysis came from the UCSC collection of known genes (Kent et al. 2002; Karolchik et al. 2003). For example, a two-exon gene (AF073799, galanin receptor GALR3) could contain at most four profiles: on–on, on–off, off–on, and off–off. In the case of the galanin receptor gene, only two patterns were observed, off–off and off–on (Supplemental Table S4). An exon was “on” if at least 30% of the interrogating probe pairs were positive. Figure 4 shows the count of profiles for each gene in all cell lines. The maximum number of profiles observed for any gene was 11 because there were 11 cell lines tested and only one pattern per cell line was used. In this case, a particular gene with 11 patterns had a different usage of exons in each cell line tested. Similarly, the minimal number of profiles was 1, indicating that a particular gene had the same on/off exon pattern in all 11 cell lines in which it was expressed. The blue bars represent all exons, and the red bars represent exons that contain four or more interrogating probe pairs. An on/off profile for each exon of a gene in every cell line showed that ∼12%–21% (105/852 of genes with all exons, 146/684 of genes with exons having ≥4 probes) of the genes displaying a single profile or have only one isoform in terms of exon skipping. This estimation of alternative splicing of known genes is likely to be an underestimate because it only accounts for exon-skipping events as opposed to alterative splicing of exons resulting in truncation or extensions of exons. Furthermore, the plot contains only genes with more than one exon. An important caveat is that exons with only a single or few interrogating probe pairs might be misleading because a single probe pair determines the overall expression of the exon and thus the on/off call. This might lead to a higher number of profiles that are potentially an overestimate of exon-skipping isoforms (Fig. 4, blue bars). Needless to say, this analysis allows for the determination of the usage of each exon for each gene. Thus, all alternatively spliced forms for each gene on Chromosomes 21 and 22 in each cell line tested denoted by exon-swapping as the differential splicing motif has be constructed (Supplemental Table S4).
Figure 4.
Distribution of genes by isoforms. An “on/off” profile was determined for each exon in all genes on the array. An exon was “on” if at least 30% of the interrogating probe pairs were positive. The histogram shows the count of profiles for each gene in all cell lines. The maximum number of profiles was 11, indicating that a particular gene has a different “on/off” pattern of exons. The minimal number of profiles was 1, indicating that a particular gene has the same “on/off” exon pattern in all 11 cell lines tested. The blue bars represent all exons with at least 30% of the interrogating probe pairs positive. The red bars represent exons that contain four or more interrogating probe pairs with at least 30% of the interrogating probe pairs positive. The numbers above the bars indicate the number of genes that contain the specified number of profiles. Of 852 genes, 75 genes have 11 profiles when considering all exons, and nine genes out of 684 have 11 profiles when considering exons with at least four probes.
Differential Expression of Novel Transfrags Between Cell Lines
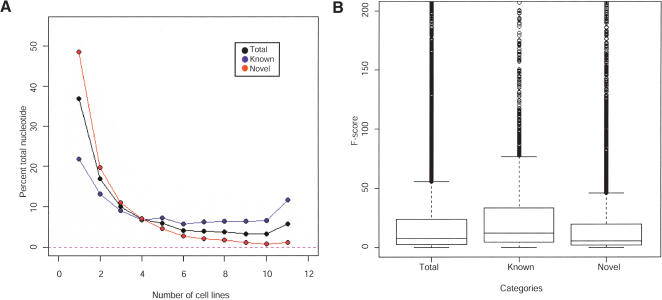
The proportion of transcription increases in the “1 of 11” map compared with individual cell lines, implying that many of the known and novel transfrags are cell-line-specific (Figs. 2 and 3). This observation can be seen as consistent with the fact that the cell types used in this study are of different developmental origins and therefore have unique expression profiles. The percentage of total nucleotides within known or novel transfrags was plotted against the number of cell lines expressing that transfrag (Fig. 5A). On average, a transfrag corresponding to a well-characterized exon was observed in approximately five cell lines, whereas any novel transfrag was found in approximately three cell lines on average. Upon closer inspection, two different and distinctly segregated populations of transfrags overlap well-characterized exons. The larger population of well-characterized transcription (30.8%) was found within transfrags that were expressed in one or two cell lines. The second and noticeably significant population of known transcription was ubiquitously expressed (11.5%). Conversely, 48.5% of the observed novel transcription was limited to a single cell line (Fig. 5A).
Figure 5.
Analysis of the expression and variance of known and novel transcription across all 11 cell lines. (A) Expression of known and novel transcription from 11 cell lines by the percentage of total nucleotides within the total (black), known (blue), and novel (red) transfrags according to the number of cell lines expressing that transfrag. (B) Estimation of the degree of differential expression across the 11 cell lines. An F-statistic was calculated for each transfrag by the variance of the average pseudo-median value in each transfrag between cell lines divided by the average of the within cell line variation of the pseudo-median value in that transfrag.
Analysis of the degree of differential expression across the 11 cell lines by an ANOVA test evaluated the variance of expression within each transfrag and between cell lines. The ANOVA F-statistic for each transfrag was defined by the variance of average pseudo-median values in each transfrag between cell lines divided by the average of the within cell line variation of pseudo-median values in that transfrag. Figure 5B illustrates the distribution of ANOVA F-statistics for the total, known, and novel transfrags. By comparing the population of ANOVA F-statistics between the known and novel transfrags, the known transfrags displayed a significantly greater differential expression across the 11 cell lines than the novel transfrags. The smaller variation in the novel transfrags was likely a result of their lower expression levels, expression in fewer cell lines, and consequently expression levels that are closer to the background making any variation difficult to measure. On average, the intensity of the positive probes of transfrags that align to known exons compared with the novel transfrags is higher, 323 compared with 187.
Characterization of Novel Transfrags
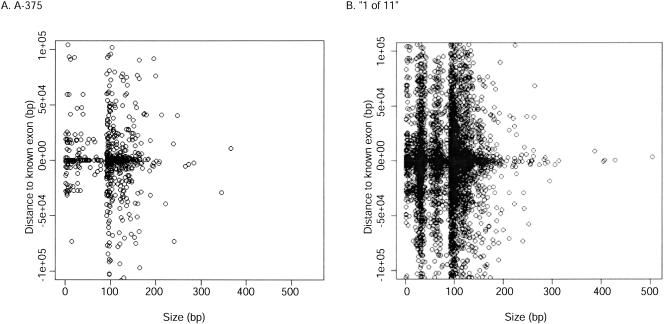
A total of 610,570 bp of sequence corresponded to novel transfrags in the “1 of 11” map. Figure 6 shows the distribution of the size of the novel transfrags relative to the distance to any well-characterized exon of a representative cell line, A375, as well as the “1 of 11” map. It shows that novel transfrags were not confined to regions proximal to annotations, with many novel transfrags identified >10 kb from a well-characterized exon (Fig. 6). Supplemental Figure S2 shows this distribution for all 11 cell lines individually. The population of novel transfrags that are <100 bp results from the fragmentation of transfrags that overlap exons but are larger than the exon (refer to Fig. 1C). These novel transfrags may represent extensions of 5′- and 3′-UTRs of the annotated genes or portions of other overlapping sense or antisense transcripts. The distal novel transfrags are often >100 bp and could represent parts of novel protein-coding genes or noncoding RNAs (ncRNAs) or small single-exon transcripts. An analysis of the coding capacity of all novel transfrags found that ∼24% have at least one open reading frame (ORF; as determined by no stop codons and minimum length of 75 bp), indicating that a majority of the observed novel transcription may be noncoding.
Figure 6.
Fragment size versus distance to annotation of novel transfrags. (A) The distribution of the size of the novel transfrags from A375 relative to the genome annotations (exons, mRNAs, ESTs). Location is determined by the distance to the nearest known exon in either the 5′ (–) or 3′ (+) direction. (B) Distribution of novel transfrags from the “1 of 11” map. The incremental pattern of transfag sizes reflects the 35 bp spacing of probes.
Strand Determination of Transfrags
All transcription maps described above were generated using non-strand-specific double-stranded cDNA made with random primers on cytosolic poly(A)+ RNA. Because one of the interesting subclasses of the novel transfrags (36% of total) is those located within the bounds of an annotated gene, that is, overlapping an intron, determination of the strand becomes important. To investigate whether these novel transfrags represent alternative exon isoforms (novel exons, extension of exons, or 5′- or 3′-UTRs) or antisense transcripts, the data obtained using the cDNA labeling assay were supplemented with a strand-specific RNA assay to assign strand information to the cDNA-derived transfrags. We used a novel direct RNA end-labeling method (K. Cole, V. Truong, D. Barone, and G. McGall, unpubl.) as opposed to using first-strand cDNA synthesis, to avoid the potential for unintended second-strand synthesis or any spurious priming events.
End-labeled RNA targets from two cell lines (A375 and Jurkat) were hybridized to the arrays representing each strand of Chromosomes 21 and 22 separately, and transfrag maps were generated using the following parameters (threshold = FPR 5%, maxgap = 40 bp, and minrun = 90 bp). The transfrag maps for each strand were combined into a nonoverlapping map (“1 of 2” [+] strand map and “1 of 2” [–] strand map) and used to assess the total amount of stranded transfrags in the cDNA-derived data by comparing them with a “1 of 2” double-stranded cDNA map obtained from the A375 and Jurkat cell lines. This “1 of 2” double-stranded cDNA-derived map resulted in 3747 (496.5 kb) transfrags that could be compared with the 1392 (159.8 kb) and 1112 (131.0 kb) transfrags in the “1 of 2” (+) strand and “1 of 2” (–) strand maps, respectively (Table 1A).
Table 1.
Strand Determination and Antisense Transcription
| A. Number of transfrags Transfragsa | No. transfrags | Base pair coverage | |||
|---|---|---|---|---|---|
| cDNA (1 of 2) | 3747 | 496,537 | |||
| + strand (1 of 2) | 1392 | 159,778 | |||
| - strand (1 of 2) | 1112 | 130,966 | |||
| B. Characterization based on annotations | |||||
| Base pair coverage
|
|||||
| Overlapping data setsb | Total known, mRNA, ESTc transfrags | Total novel transfrags | |||
| + strand | 15,440 | 8277 | |||
| - strand | 41,283 | 8529 | |||
| Totals | 56,723 | 16,806 | |||
| C. Antisense transcription | |||||
| Base pair coverage | |||||
| Overlapping data setsb | Total known, mRNA, ESTc transfrags | ||||
| + strand | 1948 | ||||
| - strand | 4187 | ||||
| Percent of antisense detectedd | 10.8% | ||||
| D. Novel transcriptione | |||||
| Base pair coverage
|
|||||
| Overlapping data setsb | Total novel intronic transfrags | Same strand | Antisense | ||
| + strand | 2596 | 1506 | 1090 | ||
| - strand | 2672 | 1071 | 1601 | ||
| Percent of novel transcription | 48.9% | 51.1% | |||
Transfrags cDNA (1 of 2) = A375 and Jurkat cDNA data combined into a nonoverlapping map. + strand (1 of 2) = A375 and Jurkat (+) strand specific data combined into a nonoverlapping map. - strand (1 of 2) = A375 and Jurkat (-) strand specific data combined into a nonoverlapping map.
A set of transfrags that overlap between the cDNA and RNA direct-labeling assays was generated. The resulting set was then compared to the indicated annotations for each chromosome and strand.
Only ESTs of confident strandedness were used, where strandedness evidence comes from splicing and/or having a polyadenylation site, possibly with associated signal.
Percent base-pair coverage in B that is antisense to the annotations.
Distribution of the base-pair coverage of the novel intronic transfrags based on the strand of the overlapping annotation.
Although the hybridization targets and conditions are quite different between the cDNA and RNA direct-labeling assays and the cDNA assay is more sensitive than the RNA assay (data not shown), ∼35% of the positive probe pairs between the cDNA and RNA end-labeled data are identical. On a transfrag level, 73.5 kb out of the 496.5 kb (14.8%) of the cDNA data overlap with the RNA end-labeled data indicating strand. Of these, ∼57 kb (77%) of the cDNA transfrags corresponds to known exons, mRNAs, or ESTs, whereas the remaining 23% (16.8 kb) corresponds to novel transcribed regions (Table 1B). Approximately 11% of the base pairs in strand-specific transfrags that overlap a known exon, mRNA, or EST were antisense (Table 1C). Of the 5.3 kb of the novel transcription (31%) that is intronic or overlaps intronic regions from well-characterized genes, 2.7 kb (51%) was antisense (Table 1D). These data indicate that a significant proportion of the observed transcription is antisense to well-characterized exons, introns, mRNAs, or ESTs.
Supplemental Figure S3 displays several examples of antisense and novel transcription observed in this study for which strandedness was determined. The stranded transcript in the 3′-UTR of SEC14L2 on Chromosome 22 represents an example of an antisense transcript of an annotated gene (Supplemental Fig. S3A). The novel (–) strand-specific transfrag adjacent to an exon in the C21orf66 gene shows an example of a possible exon extension (Supplemental Fig. S3B). The gap between the novel transfrag and the exon of C21orf66 is caused by the use of a 50-bp masked region proximal to exons to account for the smoothing effect of using a window-based approach. Although adjacent transfrags identified on the same strand are not necessarily part of a transcript, it is tempting to speculate that the two novel transfrags shown in Supplemental Figure S3C that overlap a portion of a Genscan predicted gene represent a gene. These are but a few of the observed antisense transcripts (12% of the total strand-specific transcription identified).
Additional Experimental Validation of Novel Transcripts
A total of 193 novel transcribed regions (loci) of Chromosomes 21 and 22 were selected for experimental verification and characterization using RT-PCR and Northern hybridization based on positive probes and/or transfrags. These 193 include the 63 regions previously reported by Kapranov et al. (2002) and will not be discussed further. The regions were chosen to be distal (minimal distance >5 kb) from any known annotations including ESTs because these examples would most likely be the result of false-positive array determinations based on their low expression levels, large numbers, and small lengths (Figs. 5 and 6). Several pairs of PCR primers (typically 2–15) interrogated each region. Regions that contained at least one RT-PCR product were scored positive.
Overall, 82/130 (63%) of the new regions have been successfully cloned and/or sequence-verified (Supplemental Table S5). Interestingly, sequence analysis of the PCR products showed little evidence of coding capacity for the majority of the sequences, indicating that many of these novel transfrags are likely to be noncoding transcripts (see Supplemental Table S5). Together with the regions reported previously (Kapranov et al. 2002), the rate of experimental verification is 126/193, or ∼65%.
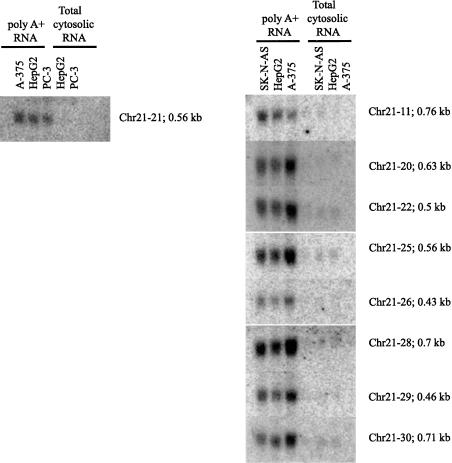
In addition, several cloned PCR products were used as probes on Northern blots with cytosolic total and poly(A)+ fraction RNAs (Fig. 7). Approximately 30% of PCR products detected a specific RNA transcript on the Northern blots. All detected RNAs were enriched in the poly(A)+ fraction, indicating that they have poly(A)+ segments. All of the detected transcripts appear to be at very low abundance, requiring 6–12 μg of poly(A)+ RNA/lane and prolonged exposures (1 wk on phosphor-screen) to be detected. This indicates that ∼70% of the regions fell below the level of detection of Northern blot technology. Nonetheless, the vast majority of the RT-PCR-detected transcripts had discrete sizes, many in the relatively small range (400–700 bases; Fig. 7). These clones are available for distribution and have been submitted to dbEST (CF798425–CF798506).
Figure 7.
Northern blot analysis of novel regions. For Northern blots, 12 μg of cytosolic RNA and the poly(A)+ fraction from each of the specified cell lines was loaded on the gel. The filters were hybridized with radiolabeled DNA probes corresponding to the cloned RT-PCR products derived from the novel array-predicted transcribed regions described in both the present and previous reports (Kapranov et al. 2002).
DISCUSSION
An in-depth analysis and characterization of the transcription identified using high-density arrays along Chromosomes 21 and 22 (Kapranov et al. 2002) have been carried out using algorithmic approaches that are less dependent on individual oligonucleotide hybridization results and that incorporate the hybridization results of neighboring probes to assist in constructing continuous regions of transcription (transfrags). These analyses provide greater statistical rigor by reducing the potential number of false-positive calls. Although these new analyses have resulted in very similar conclusions to our earlier studies, that a significant number of novel transcripts from Chromosomes 21 and 22 can be identified as RNAs transported into the cytoplasm as poly(A)+ transcripts (Kapranov et al. 2002), they also provide a less fragmented map of the transcribed regions of Chromosomes 21 and 22. Each of the detected transfrag elements has properties similar to those of gene-exon structures.
Our earlier results indicated that the number of transcribed base pairs located within any of the well-characterized exons of Chromosomes 21 and 22 was as much as an order of magnitude less than that observed outside these annotations. In our present analyses, by joining together neighboring positive probe pairs into transfrags based on a stringent set of parameters, and by overlaying the locations of transcribed regions to a more complete set of annotations including mRNAs and ESTs, almost half of the observed transcription was observed to be outside of any annotation in 1 of 11 cell lines, with two-thirds found outside of well-characterized exons (Fig. 2A). For any of the individual cell lines studied, almost two-thirds of the mapped transcription is located within one of these annotated regions, whereas novel transcripts make up only one-third of the observed transcription (Fig. 2B). Using this conservative analysis approach, the proportion of novel transcription is slightly smaller then previously reported. However, these estimates are likely to represent an underestimate of the amount of novel transcription, especially because a sliding-window-based analysis on single positive probe levels indicated that ∼90% of transcription is outside of well-characterized exons (data not shown).
Although only a third of the observed transcription corresponds to well-characterized genes, the distribution of positive probes in exons found that many exons are either all “off” or all “on” (Supplemental Fig. S1; Fig. 3). By generating differential expression profiles for all the exons within a gene, we have specifically identified a large population of alternative gene expression based on a single mode used by the splicing machinery, exon skipping (Fig. 4). Whereas our observations indicate that 12%–21% of the genes on Chromosomes 21 and 22 have a single isoform, the remaining 79%–88% have multiple forms. This estimate is considerably higher than previously noted by the analysis of the public databases (Mironov et al. 1999; Kan et al. 2001; Lander et al. 2001; Modrek et al. 2001). Our estimate of the percent of genes that exhibit alternatively spliced forms is conservative because our analysis only detected alternatively spliced forms that are the result of exon skipping. Based on these data from Chromosomes 21 and 22, there is a distinct possibility that nearly all of the coding genes of the genome exhibit alternatively spliced forms. The observed novel transcription (49.0%) appears to represent transcripts of lower abundance, as seen by their expression in only one cell line (Fig. 5A) and their lower pseudo-median value variances (Fig. 5B). Nonetheless, non-array-based biochemical verification of sites of novel transcription confirms that they represent bona fide transcripts in the cell. The 65% rate of verification (Supplemental Table S5; Fig. 7) is likely to be an underestimate given the low abundance of these transcripts and lack of knowledge of transcript structure in the novel regions.
RNA transcription of protein-coding mRNAs observed from both strands of a locus (Labrador et al. 2001), intron-containing genes (Levinson et al. 1992; Conrad et al. 2002), and transcripts with elongated or shortened exons coding for proteins with different functions (Rahman et al. 2002) have all been previously observed. Consistent with these examples, the observed novel transcription found proximal to annotations may represent alternative exon isoforms (novel exons, extension of exons, or extensions of 5′- or 3′-UTRs) or distinct transcriptional units encoded on either the same strand or antisense strand to the characterized annotation. Our strand-specific analysis of the novel transfrag data showed that approximately half of the observed novel intronic transcription emanated from the same strand as the adjacent exons and the remaining half of the transcription was antisense to well-characterized introns (Table 1; Supplemental Fig. S3).
Approximately 11% of the observed transcription of base pairs in exons, mRNAs, and EST was also found to be antisense (Table 1C; Supplemental Fig. S3). Although the strand-specific data are informative, they are considerably less sensitive than the cDNA-based maps. By extrapolation, this implies that at least 20% of the total base pairs on Chromosomes 21 and 22 constitute antisense transcription. Such wide-spread antisense transcription in human and other eukaryotic genomes is becoming increasingly evident (Lehner et al. 2002; Shendure and Church 2002; Yelin et al. 2003). Several recent studies have used computational approaches to identify and estimate the amount of antisense transcription in the human genome, with ranges from ∼2% to >8% of genes having a sense–antisense gene pair (Lehner et al. 2002; Shendure and Church 2002; Yelin et al. 2003). Although our estimate of antisense transcription is comparable, it is not directly comparable because we are reporting 20% of total base pairs are antisense and not 20% of genes. However, this report is the first chromosome-wide experimental identification of antisense transcription. Furthermore, in a companion report, we observed global antisense transcription as a common feature of RNA transcription in human cells based on mapping the locations of binding sites for three common transcription factors (Sp1, cMyc, and p53) using our high-density oligonucleotide arrays along human Chromosomes 21 and 22 (Cawley et al. 2004).
The observed novel transcription found distal to well-characterized exons is likely to represent novel (coding and noncoding) transcripts (Fig. 6). Because these novel transfrags are a significant distance to any annotation, their inclusion as interrogated sites of the genome using other types of array platforms is unlikely, given that such arrays do not interrogate the genome on such a scale or in an unbiased manner (Chen et al. 2002; Okazaki et al. 2002; Saha et al. 2002; Guigo et al. 2003; Rinn et al. 2003).
Finally, the possibility that the observed novel transcription was due to cross-hybridization to other sequence-related regions in the genome was investigated and shown not to be a major contributing factor (A. Piccolboni, S. Cawley, S. Bekiranov, and T.R. Gingeras, unpubl.). These data indicated that neither gene families, pseudogenes, nor partial duplication of probe sequences contribute significantly to the hybridization signals observed for probe pairs interrogating novel transcribed regions.
A biological function for some portion of these novel transcripts, both proximal and distal to well-characterized annotations, is supported by their evolutionary sequence conservation when compared with the mouse genome (∼20% of the novel transcribed regions using BLAST p-score cutoff of 5e–50; data not shown). This percentage of conservation is consistent with another previous estimate (Dermitzakis et al. 2002). Additionally, a small proportion (24%) of the novel transfrags had an ORF >75 bp in at least a single reading frame, indicating that the majority of the novel transfrags may be ncRNAs. However, because transfrags represent only a portion of a transcript, it is difficult to fully evaluate both mouse sequence conservation and coding capacity analyses of these novel transfrag data. Recently, a large proportion of novel noncoding mouse transcripts (Okazaki et al. 2002) and nongenic conserved blocks between mouse and human (Dermitzakis et al. 2002) have been identified supporting these conclusions. Nevertheless, Nekrutenko et al. (2003) identified a large population (14,000) of potential protein-coding exons using an evolutionary comparative genomics approach between mouse and human, with 111 of those on Chromosome 22, and found 89 (∼80%) are expressed in our transcription data (Nekrutenko et al. 2003). Additional evolutionary sequence comparisons of the expressed transfrag regions to the genomes of species closer to human are in progress and may indicate a greater degree of conservation.
With the increasing number of large-scale comparative genomic studies and the completion of a working draft of the human genome sequence, our comprehension of the organization, size, and structure of the genome continues to increase. The present estimate of 30,000–40,000 genes in the human genome (Lander et al. 2001; Venter et al. 2001) does not account for many ncRNAs. The recent identification of several types of ncRNAs, such as small nucleolar RNAs, microRNAs, guide RNAs, and antisense RNAs (Kiss 2002; Pasquinelli and Ruvkun 2002), in the gene count would significantly expand the complexity of the human genome (Storz 2002). Our own studies will likely add even more to this list of distinct RNA transcripts. As stated earlier, these novel cytosolic polyadenylated transcripts are likely to be both coding and noncoding transcripts. Many of these novel transcripts have been shown to be spliced, differentially expressed, antisense to well-characterized genes, and homologous to mouse sequences. In a separate set of studies we report that they are also associated with several common transcription factors located at their 5′ termini.
The accumulation of this groundswell of recent reports indicating a greater amount of transcription than previously determined offers two possible ideas for consideration. First, the use of the term “gene” to identify all the transcribed units in the genome may need reconsideration, given the fact that this is a term that was coined to denote a genetic concept and not necessarily a physical and measurable entity. With respect to the efforts to enumerate all functional transcribed units, it may be helpful to consider using the term “transcript(s)” in place of gene. This suggestion has the attraction of allowing for the enumeration of each of the isoforms for all presently annotated genes as well as any distinct novel transcript that possesses some but not all of the properties of well-characterized coding transcripts. Thus, the fact that a transcript possesses coding capabilities would only be one of its possible features, not the most important one. Following on this last consideration, a second possible consideration stemming from the growing list of transcribed regions of the genome is the likelihood that the present efforts in estimating the total number of genes in the genome is misguided and at the very least miscalculated. These efforts are misguided given the discussion presented previously that a more useful entity to be counted is the number of transcripts. They are also miscalculated because such estimates are biased strongly in favor of protein-coding transcripts. Although we believe it is premature to arrive at an accurate estimate of the total number of transcripts that a living cell could synthesize, based on our own studies it is likely to be a much larger number than the 30,000–40,000 present estimates.
METHODS
Cell Culture, Nucleic Acid Purification, cDNA Synthesis, Fragmentation, Labeling, and Hybridization
See Kapranov et al. (2002).
Determination of Thresholds and Estimation of Sensitivity/Specificity
By fixing the pseudo-median threshold, the corresponding false-positive rate (FPR), sensitivity (Sn), and specificity (Sp) were determined by using the present bacterial segments and the bacterial regions outside the spiked-in bacterial controls (Kapranov et al. 2002). A threshold of 150 corresponds to a median FPR of 2.9% for the 11 cell lines (Supplemental Table S2).
Construction of Transcription Maps for the Human Chromosomes 21 and 22
Arrays were quantile-normalized within replicate groups (Bolstad et al. 2003) and then scaled to have a median feature intensity of 700. The (PM, MM) intensity pairs were mapped to the genome using exact 25-mer matching. For each genomic position to which a probe pair mapped, a data set was generated consisting of all (PM, MM) pairs mapping within a window of ±50 bp. The Hodges-Lehmann estimator or pseudo-median was calculated using PM–MM data from the 3 arrays within the local data set as an estimator of the expression level at each genomic position (as described in the Results section). The pseudo-median was calculated in a sliding window across the genome. Pseudo-medians above a threshold of 150 were called positive. Contiguously transcribed elements termed transfrags were generated by joining positive probes that were separated by less than a certain distance (maxgap=40) and whose length was less than a particular size (minrun=90).
Direct RNA End-Labeling Assay
Direct labeling of RNA using T4 RNA ligase was performed as described by K. Cole, V. Truong, D. Barone, and G. McGall (unpubl.). Briefly, A375 and Jurkat poly(A)+ RNA was mixed with spiked in bacterial poly(A)+ RNA transcripts. RNA was fragmented and dephosphorylated by incubation with 2 U of shrimp alkaline phosphatase (United States Biochemicals) and Escherichia coli RNase III (0.1 U/μg RNA; New England BioLabs) in 10 mM Tris-HCl, 10 mM MgCl2, 50 mM NaCl, 1 mM DTT (pH 7.9) for 35 min at 37°C, followed by inactivation of SIP for 20 min at 65°C. The fragmented, dephosphorylated RNA was then directly labeled with T4 RNA ligase (20 U), pCp-biotin donor compound (5′-pCpteg-biotin-3′), and 12% PEG in 50 mM Tris-HCl, 10 mM MgCl2, 10 mM DTT, 1 mM ATP (pH 7.8) for 2 h at 37°C. The labeled RNA is then directly added to the hybridization solution containing 30 pM control oligo B2 and control oligo 213B (Affymetrix), 1× eukaryotic hybridization controls (Affymetrix), 0.1 mg/mLherring sperm DNA (Invitrogen), 0.5 mg/mLacetylated BSA (Invitrogen), 5% formamide (Sigma), 30 mM MES (Sigma), 74 mM MES · Na (Sigma), 0.01% Triton X-100, 0.885 M NaCl, and 20 mM EDTA. Then 5 μg of RNA per Chromosome 21–22 array was hybridized for 18 h at 50°C. The arrays were washed and stained as previously described (Kapranov et al. 2002).
RT-PCR and Sequencing of Cloned PCR Products
The RT-PCR procedure was carried out using two major approaches: (1) direct amplification from RNA with gene-specific primers using the C. therm. Polymerase One-Step RT-PCR System (Roche) following the manufacturer's protocols; (2) first converting RNA into cDNA with random hexamers as described previously (Kapranov et al. 2002) and then using gene-specific primers and TaqGold polymerase (Applied Biosystems) as suggested by the manufacturer. In both cases, the starting material was cytosolic poly(A)+ RNA treated with DNase I (Roche). Approximately 100 ng of RNA or cDNA was used per PCR reaction. Minus RT controls were run for the second method. A combination of nested primers was used for each region. The sequences of primers are available upon request. Typically, two rounds of 40 cycles of PCR were required to amplify the targets. In a few cases, the substrate for amplification was plasmid cDNA libraries (200 ng per reaction) in combination with TaqGold polymerase. PCR products were cloned into pCRScript (Strategene) and/or sequenced directly using the ABI Big Dye v 3.0 sequencing kit. Sequences were aligned back to the genome to confirm identity of the PCR products. Northern blots were performed with cloned PCR products as probes as described (Kapranov et al. 2002) on both poly(A)+ and total cytosolic RNAs from indicated cell lines.
Acknowledgments
The authors thank Victor Sementchenko, Alan Williams, and Kevin Struhl for helpful discussions during data analysis; Sandeep Patel, Ray Wheeler, and Harley Gorrell for technical assistance; Kyle Cole and Vivi Truong for advice concerning direct-RNA labeling methodology; and Robert L. Strausberg for encouragement and support. Support for this work was provided in part by NCI contracts (21XS019A and 21XS019B) and No1-LO-12400 and Affymetrix, Inc.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2094104.
Footnotes
[Supplemental material is available online at www.genome.org. All novel, sequence-verified transcripts (Supplemental Table S4) have been submitted to dbEST (CF798425–CF798506). The following individuals kindly provided unpublished information as indicated in the paper: K. Cole, V. Truong, D. Barone, G. McGall, H.H. Ng, E.A. Sekinger, A.J. Williams, R. Wheeler, B. Wong, and K. Struhl.]
References
- Bolstad, B.M., Irizarry, R.A., Astrand, M., and Speed, T.P. 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19: 185–193. [DOI] [PubMed] [Google Scholar]
- Cawley, S., Bekiranov, S., Ng, H., Kapranov, P., Sekinger, E.A., Kampa, D., Piccolboni, A., Sementchenko, V., Cheng, J., Williams, A., et al. 2004. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 point to widespread regulation of non-coding RNAs. Cell (in press). [DOI] [PubMed]
- Chen, J., Sun, M., Lee, S., Zhou, G., Rowley, J.D., and Wang, S.M. 2002. Identifying novel transcripts and novel genes in the human genome by using novel SAGE tags. Proc. Natl. Acad. Sci. 99: 12257–12262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, J.E., Goward, M.E., Cole, C.G., Smink, L.J., Huckle, E.J., Knowles, S., Bye, J.M., Beare, D.M., and Dunham, I. 2003. Reevaluating human gene annotation: A second generation analysis of human Chromosome 22. Genome Res. 13: 27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad, C., Vianna, C., Freeman, M., and Davies, P. 2002. A polymorphic gene nested within an intron of the τ gene: Implications for Alzheimer's disease. Proc. Natl. Acad. Sci. 99: 7751–7756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermitzakis, E.T., Reymond, A., Lyle, R., Scamuffa, N., Ucla, C., Deutsch, S., Stevenson, B.J., Flegel, V., Bucher, P., Jongeneel, C.V., et al. 2002. Numerous potentially functional but non-genic conserved sequences on human Chromosome 21. Nature 420: 578–582. [DOI] [PubMed] [Google Scholar]
- Ewing, B. and Green, P. 2000. Analysis of expressed sequence tags indicates 35,000 human genes. Nat. Genet. 25: 232–234. [DOI] [PubMed] [Google Scholar]
- Fodor, S.P., Read, J.L., Pirrung, M.C., Stryer, L., Lu, A.T., and Solas, D. 1991. Light-directed, spatially addressable parallel chemical synthesis. Science 251: 767–773. [DOI] [PubMed] [Google Scholar]
- Fodor, S.P., Rava, R.P., Huang, X.C., Pease, A.C., Holmes, C.P., and Adams, C.L. 1993. Multiplexed biochemical assays with biological chips. Nature 364: 555–556. [DOI] [PubMed] [Google Scholar]
- Guigo, R., Dermitzakis, E.T., Agarwal, P., Ponting, C.P., Parra, G., Reymond, A., Abril, J.F., Keibler, E., Lyle, R., Ucla, C., et al. 2003. Comparison of mouse and human genomes followed by experimental verification yields an estimated 1,019 additional genes. Proc. Natl. Acad. Sci. 100: 1140–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollander, M. and Wolfe, D.A. 1999. Nonparametric statistical methods, 2nd ed. John Wiley and Sons, Inc., New York.
- Kan, Z., Rouchka, E.C., Gish, W.R., and States, D.J. 2001. Gene structure prediction and alternative splicing analysis using genomically aligned ESTs. Genome Res. 11: 889–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov, P., Cawley, S.E., Drenkow, J., Bekiranov, S., Strausberg, R.L., Fodor, S.P., and Gingeras, T.R. 2002. Large-scale transcriptional activity in chromosomes 21 and 22. Science 296: 916–919. [DOI] [PubMed] [Google Scholar]
- Karolchik, D., Baertsch, R., Diekhans, M., Furey, T.S., Hinrichs, A., Lu, Y.T., Roskin, K.M., Schwartz, M., Sugnet, C.W., Thomas, D.J., et al. 2003. The UCSC Genome Browser Database. Nucleic Acids Res. 31: 51–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent, W.J., Sugnet, C.W., Furey, T.S., Roskin, K.M., Pringle, T.H., Zahler, A.M., and Haussler, D. 2002. The human genome browser at UCSC. Genome Res. 12: 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss, T. 2002. Small nucleolar RNAs: An abundant group of noncoding RNAs with diverse cellular functions. Cell 109: 145–148. [DOI] [PubMed] [Google Scholar]
- Labrador, M., Mongelard, F., Plata-Rengifo, P., Baxter, E.M., Corces, V.G., and Gerasimova, T.I. 2001. Protein encoding by both DNA strands. Nature 409: 1000. [DOI] [PubMed] [Google Scholar]
- Lander, E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921. [DOI] [PubMed] [Google Scholar]
- Lehner, B., Williams, G., Campbell, R.D., and Sanderson, C.M. 2002. Antisense transcripts in the human genome. Trends Genet. 18: 63–65. [DOI] [PubMed] [Google Scholar]
- Levinson, B., Kenwrick, S., Gamel, P., Fisher, K., and Gitschier, J. 1992. Evidence for a third transcript from the human factor VIII gene. Genomics 14: 585–589. [DOI] [PubMed] [Google Scholar]
- Liang, F., Holt, I., Pertea, G., Karamycheva, S., Salzberg, S.L., and Quackenbush, J. 2000. Gene index analysis of the human genome estimates approximately 120,000 genes. Nat. Genet. 25: 239–240. [DOI] [PubMed] [Google Scholar]
- Mironov, A.A., Fickett, J.W., and Gelfand, M.S. 1999. Frequent alternative splicing of human genes. Genome Res. 9: 1288–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrek, B., Resch, A., Grasso, C., and Lee, C. 2001. Genome-wide detection of alternative splicing in expressed sequences of human genes. Nucleic Acids Res. 29: 2850–2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrutenko, A., Chung, W.Y., and Li, W.H. 2003. An evolutionary approach reveals a high protein-coding capacity of the human genome. Trends Genet. 19: 306–310. [DOI] [PubMed] [Google Scholar]
- Okazaki, Y., Furuno, M., Kasukawa, T., Adachi, J., Bono, H., Kondo, S., Nikaido, I., Osato, N., Saito, R., Suzuki, H., et al. 2002. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 420: 563–573. [DOI] [PubMed] [Google Scholar]
- Pasquinelli, A.E. and Ruvkun, G. 2002. Control of developmental timing by microRNAs and their targets. Annu. Rev. Cell Dev. Biol. 18: 495–513. [DOI] [PubMed] [Google Scholar]
- Pease, A.C., Solas, D., Sullivan, E.J., Cronin, M.T., Holmes, C.P., and Fodor, S.P. 1994. Light-generated oligonucleotide arrays for rapid DNA sequence analysis. Proc. Natl. Acad. Sci. 91: 5022–5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman, L., Bliskovski, V., Reinhold, W., and Zajac-Kaye, M. 2002. Alternative splicing of brain-specific PTB defines a tissue-specific isoform pattern that predicts distinct functional roles. Genomics 80: 245–249. [DOI] [PubMed] [Google Scholar]
- Rinn, J.L., Euskirchen, G., Bertone, P., Martone, R., Luscombe, N.M., Hartman, S., Harrison, P.M., Nelson, F.K., Miller, P., Gerstein, M., et al. 2003. The transcriptional activity of human Chromosome 22. Genes & Dev. 17: 529–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha, S., Sparks, A.B., Rago, C., Akmaev, V., Wang, C.J., Vogelstein, B., Kinzler, K.W., and Velculescu, V.E. 2002. Using the transcriptome to annotate the genome. Nat. Biotechnol. 20: 508–512. [DOI] [PubMed] [Google Scholar]
- Shendure, J. and Church, G.M. 2002. Computational discovery of sense-antisense transcription in the human and mouse genomes. Genome Biol. 3: RESEARCH0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz, G. 2002. An expanding universe of noncoding RNAs. Science 296: 1260–1263. [DOI] [PubMed] [Google Scholar]
- Venter, J.C., Adams, M.D., Myers, E.W., Li, P.W., Mural, R.J., Sutton, G.G., Smith, H.O., Yandell, M., Evans, C.A., Holt, R.A., et al. 2001. The sequence of the human genome. Science 291: 1304–1351. [DOI] [PubMed] [Google Scholar]
- Yelin, R., Dahary, D., Sorek, R., Levanon, E.Y., Goldstein, O., Shoshan, A., Diber, A., Biton, S., Tamir, Y., Khosravi, R., et al. 2003. Widespread occurrence of antisense transcription in the human genome. Nat. Biotechnol. 21: 379–386. [DOI] [PubMed] [Google Scholar]