Abstract
A three year field study (2007–2009) of the diversity and numbers of the total and metabolically active free-living diazotophic bacteria and total bacterial communities in organic and conventionally managed agricultural soil was conducted using the Nafferton Factorial Systems Comparison (NFSC) study, in northeast England. Fertility management appeared to have little impact on both diazotrophic and total bacterial communities. However, copy numbers of the nifH gene did appear to be negatively impacted by conventional crop protection measures across all years suggesting diazotrophs may be particularly sensitive to pesticides. Impacts of crop management were greatly overshadowed by the influence of temporal effects with diazotrophic communities changing on a year by year basis and from season to season. Quantitative analyses using qPCR of each community indicated that metabolically active diazotrophs were highest in year 1 but the population significantly declined in year 2 before recovering somewhat in the final year. The total bacterial population in contrast increased significantly each year. It appeared that the dominant drivers of qualitative and quantitative changes in both communities were annual and seasonal effects. Moreover, regression analyses showed activity of both communities was significantly affected by soil temperature and climatic conditions.
Introduction
Yields of arable crops depend on sufficient reservoirs of plant available nitrogen in agricultural soils. However, as conventional fertility management using inorganic nitrogen fertiliser is becoming increasingly expensive and is recognised as having significant detrimental effects on the environment [1], there is growing interest in more sustainable systems that can exploit biologically fixed nitrogen or use inorganic nitrogen as efficiently as possible.
One microbiological approach to improve sustainability is to enhance the activity of the nitrogen fixing bacteria in soil [2] and to optimise the effects of land use [3], crop management [4], [5], N management [4], and seasonal variations [6] on N fixation processes, especially by free-living diazotrophs.
Fertility management, crop protection and crop rotation have all been shown to exert significant effects on the soil microbial communities present in organically and conventionally managed agricultural soils. Previous studies that looked at the impact of farm management on the function and diversity of these communities report the most significant factor affecting soil microbial communities is the fertility management regime [7], [8]. However, results are equivocal and studies have mostly focussed on comparing farming systems over a single season. Here we extend these studies by exploring the effects of different organic and conventional farm management practices on the total bacterial and free-living N fixing community using a factorial design that allows us to investigate the individual effects of crop protection practices and fertility management over several seasons. In general, organic fertility management systems, that include the application of farmyard manure, and the use of diverse crop rotations have been shown to have a positive effect on microbial biomass, diversity and activity [9], [10], [11], [12], when compared with conventional systems. These differences are mainly attributed to; the increased organic C added as manure; lower background levels of readily-available nitrogen, and pH values that are, on average, closer to neutral in organically managed soils [13], [14]. As nitrogen fixation is energy-expensive, it is reliant on carbon sources that are more abundant and are retained longer in organically managed compared to conventional soils [15]. Therefore, organic soils are more likely to offer optimal conditions for nitrogen fixation and it is perhaps unsurprising that increases in soil organic carbon have been shown to stimulate nitrogen fixation, although results have been inconsistent [16], [17], [18], [2]. Additional drivers of the activity of the diazotrophic community have been identified as the soil microbial biomass and total nitrogen [3] both of which are typically higher in organic systems.
Other secondary effects of fertility management could also be significant, in particular, changes in soil pH, which is considered a predictor of soil microbial community composition [19]. Hallin et al, [20] found that pH affected total bacterial community composition among soils treated with different fertilizers. Phosphorus can also stimulate nitrogen fixation as it is required for microbial energy production. Reed et al, [21] observed doubling of nitrogen fixation in response to the addition of phosphorus. Most studies discussing free-living N fixing bacteria have described results from a single season (e.g. [22], [23]. Since free-living N fixers are known to be very sensitive to environmental conditions, it is important to establish whether crop management effects are consistent across dates within a given year, and over several growing seasons.
Crop protection measures could also potentially affect the soil microbial community. Conventional farmers can use a complex mixture of pesticides to protect their crops [24]; whereas, organic farmers rely on the diversification of crops in the field (intercropping) and over time (crop rotation), use of resistant varieties, optimal timing of tillage for weed control, and use of a limited range of organically approved pesticides [25]. While conventional crop protection measures include the use of chemicals that are toxic to specific organisms, the majority of studies into the effects of chemical pesticides on the soil microbial community have found that they do not significantly affect microbial populations when used at the correct dose [26], [27], [28]. Nitrogen fixing bacteria are thought to be especially sensitive to pesticides [29]. However, most of the work looking at the effects of pesticides has been carried out on symbiotic diazotrophs. For example, it was shown both in vitro and in vivo that around 30 different pesticides have a negative effect on the relationship between S. meliloti and alfalfa probably due to a disruption in the chemical signalling between the bacteria and its host [30], [31].
In this study we utilise an existing factorial field trial that enables comparisons between elements of organic and conventional systems, including fertility management, crop protection and crop rotation, to be analysed over several years. Previously, we have used this trial to demonstrate how organic and conventional crop rotations affect the bulk soil microbial community with emphasis on free-living diazotrophs [23] within a single growing season. In this paper the effects of fertility management and crop protection as well as sample date and sample year, on the general total bacterial and diazotrophic communities over three years are reported.
Materials and Methods
Soil Sampling
The soil (sandy loam; Stagnogley) used in this study was taken from the Nafferton Factorial Systems Comparison (NFSC) study, a field trial based at Nafferton Farm in the Tyne Valley, northeast England. The NFSC was established in 2001 and consists of a series of four field experiments established within four replicate blocks. Each experiment is a split split-plot design with three factors. The main factor is crop rotation: an eight year, conventional cereal intensive rotation is compared to an eight year, diverse legume intensive organic crop rotation. Each crop rotation main plot is split to compare two levels of crop protection: organic (ORG CP; according to Soil Association organic farming standards [32]) and conventional (CON CP; following British Farm Assured practice). Each crop protection subplot is further split into two fertility management sub-subplots: organic (ORG FM) and conventional (CON FM). Each of the four field experiments has the same design, but was begun in a different year to allow a diversity of crops to be grown in the trial in any given year. In this study soils were taken from potato plots (6 x 24 m in size) grown in 2007, 2008 and 2009 on three dates in each year (March, July, September). Soil samples in March were taken from bare soil which had been mouldboard ploughed the previous autumn, prior to the application of any fertility or crop protection treatments. Before samples were taken in June, the soil had undergone secondary tillage and potato planting, as well as frequent ridging for weed control. Pesticides had been applied to CON CP treatments and mineral fertilisers to CON FM treatments. Compost was applied to ORG FM treatments in April prior to potato planting. Final samples were taken post harvest. Prior to potato harvest CON CP treatments were treated with a chemical defoliant, while ORG CP plots were mechanically defoliated. All potato crops followed a winter cereal the previous year; however, in 2007, the potatoes were in a conventional crop rotation following a crop of winter barley that followed two previous years of winter wheat following a grass/clover ley. In contrast, both the 2008 and 2009 potato crops were grown in an organic crop rotation following a preceding crop of winter wheat that followed a grass/clover ley.
Full details of the organic and conventional fertility management and crop protection practices used in the potato crop and for the preceding year are shown in Table 1.
Table 1. Crop protection protocols and fertility management used in the NFSC experiments for 2006, 2007, 2008 and 2009 under organic crop protection (ORG CP) or conventional crop protection (CON CP) and organic fertility management (ORG FM) or conventional fertility management (CON FM).
| Current crop | |
| Potatoes (2007–9) | |
| ORG CP | mechanical weeding (ridging); copper-oxychlorideb (23 kg/ha) |
| CON CP | aldicarbd (33.5 kg/ha); linurona (3.5 L/ha); fluazinamc (1.5 L/ha); mancozeb and metalaxyl-Mc (4.7 kg/ha); oiquate (2 L/ha) |
| ORG FM | composted cattle manure (equivalent to 180 kg total N/ha with 2–9% of total N in plant available forms; 2–17 kg total P2O5/ha; 5–149 kg total K2O/ha) |
| CON FM | 0∶20:30 (134 kg P2O5/ha; 200 kg K2O/ha); Nitram (180 kg N/ha) |
| Previous crop | |
| Winter barley (2006) | |
| ORG CP | mechanical weeding (finger weeder) |
| CON CP | Pendimethalina (2.5 L/ha); isoproturona (1.5 L/ha); Duplosana (1 L/ha); Acantob (0.4 L/ha); Prolineb (0.4 L/ha); Corbelb (0.5 L/ha); Fluroxypyrb (0.75L/ha); Amistarb (0.25 L/ha); Bravo 500b (0.5 L/ha); Cleancrop EPXb (0.4 L/ha) |
| ORG FM | no amendment |
| CON FM | 0∶20:30 (64 kg P2O5/ha; 96 kg K2O/ha); Nitram (170 kg N/ha) |
| Winter Wheat (2007–2008) | |
| ORG CP | mechanical weeding (finger weeder) |
| CON CP | isoproturona (6 L/ha); Opticaa (1 L/ha); Pendimethalina (1.5 L/ha); Corbelb (0.2 L/ha); Cleancrop EPXb (1.25 L/ha); Bravo 500b (1.75 L/ha); chlormequatc (2.3 L/ha); Ternb (0.15 L/ha); Twistb (0.25 L/ha) |
| ORG FM | no amendment |
| CON FM | 0∶20:30 (64 kg P2O5/ha; 96 kg K2O/ha); Nitram (210 kg N/ha) |
herbicide;
fungicide;
growth regulator;
nematicide;
desiccant.
Soils were sampled and a standard set of soil properties (pH, soil organic C, soil total N, NO3-N, NH4-N, soil basal respiration (SBR) and Mehlich 3-extractable P, K, and Fe) were analysed as described in Orr et al, [23]. Weather conditions at the experimental site were monitored on an hourly basis using a Delta-T GP1 datalogger with sensors. Mean results for soil temperature and rainfall in the 14 days prior to each soil sampling occasion are shown in Table S1.
Nucleic Acid Extraction and PCR
RNA was extracted from 0.25 g of soil with the UltraClean microbial RNA isolation kit (MoBio) and reverse transcribed with the Superscript II reverse transcriptase kit (Invitrogen). DNA was extracted from 0.25 g of soil using the UltraClean Soil DNA extraction kit (MoBio). The nifH gene was amplified using an adapted method first described by Wartiainen et al [33]. Initially a 360 bp fragment is amplified using using PolF and PolR primers [34] followed by nesting with PolFI and AQER-GC30 primers [33]. To amplify the total bacterial community, the V3 variable region of 16S rRNA was amplified using V3FC and V3R primers [35]. Full PCR conditions and primer sequences are described in Orr et al, [23].
DGGE
DGGE was carried out using the D-Code system (Bio-Rad Laboratories) as described by Baxter & Cummings [35]. PCR products were electrophoresed through gels containing 35–55% denaturing gradient at 200 V for either 6 (nifH) or 4.5 (16S rRNA) hours. Bands were identified and relative intensities were calculated with Quantity One software (Bio-Rad). Shannon’s diversity index (H′) was calculated by the formula H′ = -Σpiln(pi), where pi is the ratio of intensity of band i compared with the total intensity of the lane.
qPCR
Reactions were set up using SYBR green (Thermo Fisher Scientific) according to Orr et al, [23] with the Rotor-Gene RG 3000 (Corbett Research). PolF and PolR primers were used for nifH qPCR, and Eub338 and V3R were used for total bacteria qPCR. A standard curve was set up using 10-fold dilutions of pGEM-T Easy vector plasmid DNA containing either the nifH gene of Rhizobium sp. strain IRBG74 bacterium [36] or the 16S rRNA gene of Pseudomonas aeruginosa NCTC10662. Each soil extraction, no-template control, and standard curve dilution was replicated three times. Average copy number was converted into copies of the gene per g of oven dry soil.
Standard deviation was determined (by the Rotor-Gene 6 software [Corbett Research]) on the replicate threshold cycle (CT) scores. qPCR was repeated if the deviation was above 0.4. Samples were considered to be below reasonable limits of detection if the CT score was above 30 [37]. In the system used in this study, this would equate to results below 1.0×104 copies per g of soil being rejected. All no-template control results fell below this threshold (35.4±2.8). The standard curve produced was linear (r 2 = 0.98), and the PCR efficiency was 0.9.
Sequencing
All sequencing was carried out on a 3130 genetic analyzer (Applied Biosystems). DGGE bands of interest were excised from the gel. DNA was eluted into 10 µl of sterile water and used as the template in the nifH PCR. The process was repeated until the band of interest was isolated. The PCR product was then cleaned up using ExoSAP-IT. PCR products were then purified using ethanol precipitation. Sequence data was analyzed using the NCBI BLAST tool.
Statistical Analysis
In all tests, significant effects/interactions were those with a P value of 0.05. Treatment effects were analyzed by analysis of variance of a linear mixed effects model, using the lme function in R version 2.6.1 [38] with the maximum likelihood method and the random error term (block/year/date/crop protection) specified to reflect the nested structure of the design [39]. The combined data for all years were analyzed first, and where interaction terms were significant, further analyses were conducted at each level of the interacting factor. Where analysis at a given level of a factor was carried out, that factor was removed from the random error term. The normality of the residuals of all models was tested with QQ plots, and data were log-transformed when necessary to meet the criteria of normal data distribution. Differences between main effects were tested by analysis of variance (ANOVA). Differences between years or sample dates (both across years or within a year) were tested with Tukey contrasts in the general linear hypothesis testing (glht) function of the multcomp package in R. A linear mixed effects model was used for the Tukey contrasts, containing a year or sample date main effect with the random error term specified as described above.
Step-wise regression was carried out in Minitab [40] using the results over the three years and three sample dates for qPCR and Shannon’s diversity index data as response variables and the measured soil parameters listed above (pH, NO3 −, NH4 +, soil basal respiration, total N and organic C) as well as environmental variables (average soil temperature and total rainfall in the 14 days prior to sampling) as explanatory variables.
DGGE data were analyzed by detrended correspondence analysis (DCA) on relative intensities followed by direct ordination with Monte Carlo permutation testing. Direct ordination was either by canonical correspondence analysis (CCA) or redundancy discriminate analysis (RDA), depending on the length of the DCA axis (where an axis of >3.5 = CCA and an axis of <3.5 = RDA) using CANOCO 4.5 and CANODRAW for Windows [41].
Results
Diversity and Expression of nifH and 16S mRNA Transcripts and Genes
A single band of 360 bp, corresponding to the expected nifH mRNA transcript, was successfully amplified from RNA extracted from all 2007 and 2009 plots. However, the nifH mRNA transcript could not be detected in certain plots in September 2008. When using the qPCR approach the CT score for the nifH mRNA transcript was below the reasonable limits of detection for all plots at all sample dates in 2008. In contrast, acceptable copy numbers of the 16S mRNA transcript were successfully amplified from all 2008 samples suggesting that the nifH gene was not being expressed at certain dates in 2008 rather than a problem with the extraction and amplification protocol.
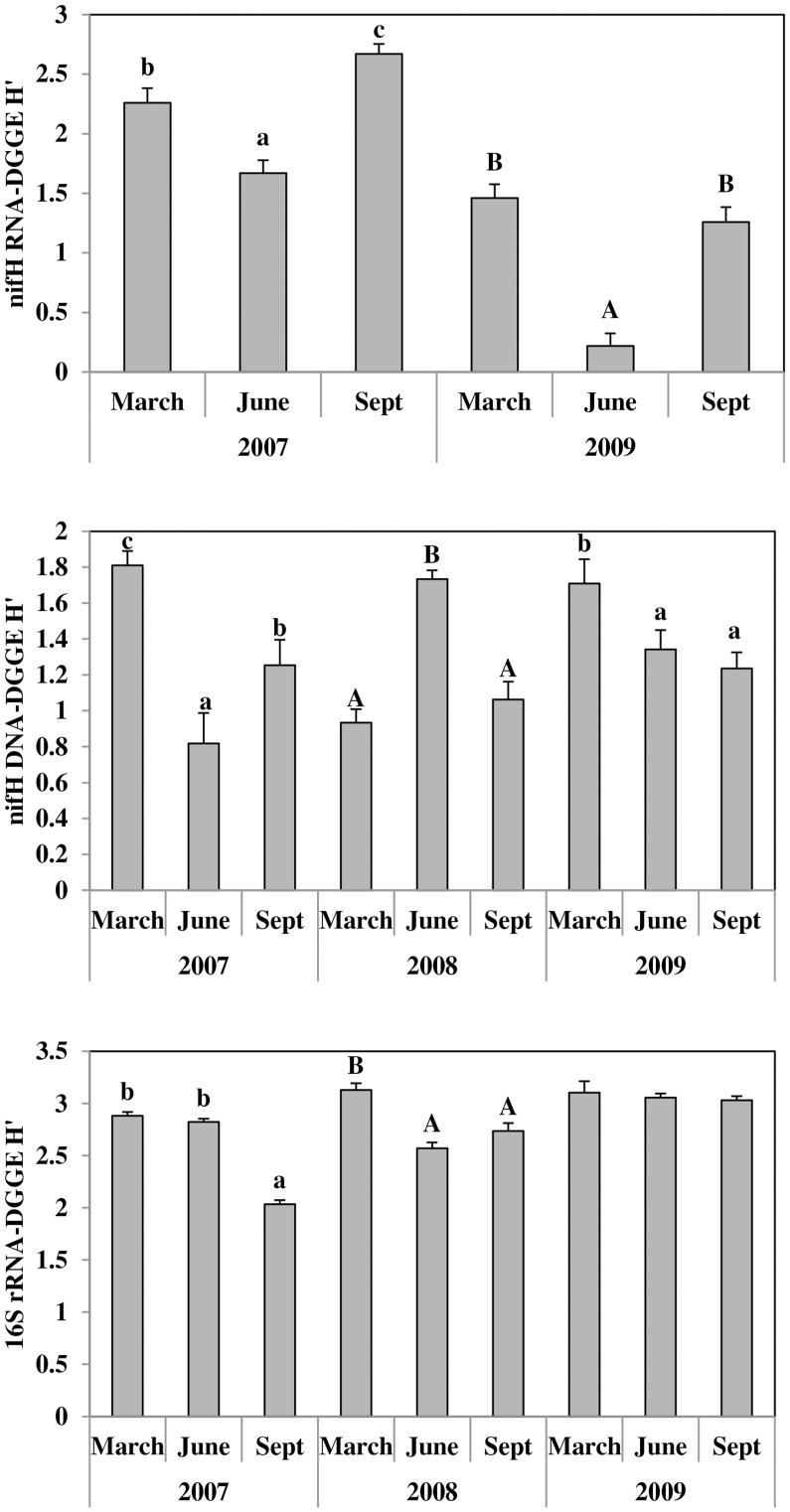
When nifH RNA diversity indices (H′) from 2007 and 2009 were analyzed (Table 2), the year, sample date and sample date × year interaction terms were all significant, while crop protection and fertility management factors did not contribute to a significant proportion of the variation in results. H′ was significantly higher overall in 2007 and generally, the June sample date had the lowest diversity across the three years. However, when separate analyses were conducted for each year, sample date was highly significant for both 2007 and 2009 (P = 0.002 in both years). In both years June samples had the lowest nifH mRNA transcript diversity, although H′ values for June 2009 were extremely low compared with June 2007 (Fig. 1). In addition, September 2007 nifH mRNA transcript diversity was significantly higher than the other two sample dates in that year, whereas in 2009, there was no difference in nifH mRNA transcript diversity between March and September sample dates.
Table 2. Summary of Shannon diversity analysis of all DGGE results from all sample years and nucleic acid types.
| H′ for nifH DGGE (RNA) band data (mean+SE) | H′ for nifH DGGE band data (DNA) (mean+SE) | H′ for 16S rRNA DGGE band data (mean+SE) | |
| Year (Y) | |||
| 2007 | 2.20±0.08 a | 1.29±0.10 a | 2.58±0.06 a |
| 2008 | 1.24±0.07 a | 2.81±0.05 a | |
| 2009 | 0.98±0.10 b | 1.43±0.07 a | 3.06±0.04 b |
| Sample Date (SD) | |||
| March | 1.86±0.11 b | 1.48±0.081a | 3.04±0.05 b |
| June | 0.95±0.15 a | 1.30±0.09 a | 2.82±0.04 a |
| September | 1.97±0.15 b | 1.18±0.07 a | 2.60±0.07 a |
| Crop protection (CP) | |||
| ORG | 1.62±0.13 a | 1.37±0.06 a | 2.82±0.05 a |
| CON | 1.56±0.13 a | 1.27±0.07 a | 2.82±0.05 a |
| Fertility management (FM) | |||
| ORG | 1.57±0.13 a | 1.28±0.06 a | 2.79±0.05 a |
| CON | 1.61±0.13 a | 1.37±0.07 a | 2.85±0.05 a |
| ANOVA P -values | |||
| Y | 0.001 | <0.001 | |
| SD | <0.001 | 0.012 | <0.001 |
| Y*SD | 0.040 | <0.001 | <0.001 |
| CP*FM | 0.006 | ||
Although date was a significant factor in the ANOVA, means comparison tests did not indicate any significant differences among dates.
P-values are only shown for terms with P<0.05. Means followed by the same letter for a given factor are not significantly different (P<0.05; Tukey’s HSD test where there are more than two treatment levels).
Figure 1. Showing Shannon’s diversity indices of the metabolically active and total diazotrophic bacteria and the total bacterial communities derived from the DGGE analyses.
The nifH mRNA transcripts are represented by the top, the nifH gene by the middle and the 16S mRNA transcript by the bottom panels respectively. Bars labelled with the same letter in the same year are not different (P = 0.05). Bars are standard errors (n = 16).
In contrast to the RNA results, analysis of the nifH DNA-DGGE diversity results showed that year was not a significant factor but there was a significant interaction between sample date and year (Table 2); again, crop protection and fertility management were not significant factors in the model. Since the year × sample date term was significant, a separate analysis was conducted for each year for both RNA and DNA extractions (Fig. 1). This showed that the ranking of dates for DNA-DGGE diversity was not the same in each year. In 2007 and 2009 H′ was highest for the March sampling date, while in 2008 it was highest in June.
The diversity of the active bacterial community was also analyzed (Table 2). ANOVA indicated that overall diversity was highest in 2009, and within a given year was greatest in March; however, there were significant year by date interactions. These are illustrated in Fig. 1 which shows that sample date had no effect on bacterial community diversity in 2009, while on the other two dates there were some differences among sample dates.
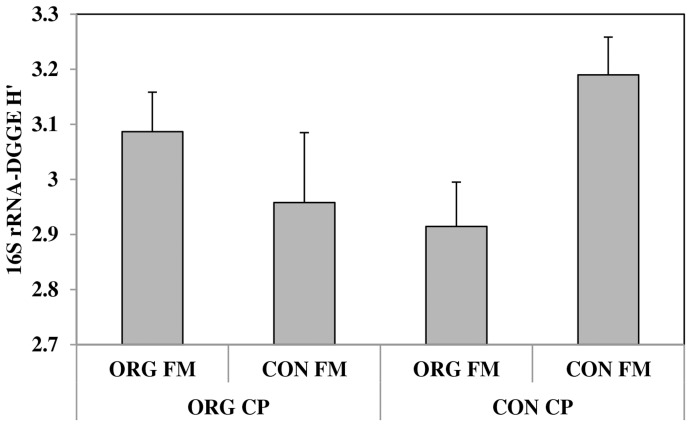
In contrast to nifH community diversity, the overall bacterial community diversity was affected by crop management practices. There was also a significant interaction between sample date, fertility management and crop protection. When only the March samples were analyzed across all three years, there was a significant crop protection by fertility management interaction (P = 0.007), although neither factor had a significant main effect. Fig. 2 shows that in March of every year, highest bacterial diversity was measured in the fully conventionally managed plots. However, on the other two sampling dates, crop management had no effect on bacterial community diversity and year was the only significant factor in the model. For all sample dates, highest bacterial community diversity was measured in 2009.
Figure 2. The interaction between organic and conventional crop protection (ORG CP, CON CP) and organic and conventional fertility management (ORG FM, CON FM) on March sample dates only for three years (2007, 2008 and 2009) for Shannon’s diversity index of the 16S mRNA transcript.
Bars are standard errors (n = 12).
The diazotrophic and total bacterial community composition were further analysed using constrained ordination, for each sample date, to determine how soil biochemical properties measured on the sample dates and environmental conditions in the two weeks prior to sampling may be driving community composition. The importance of the fertility management and crop protection treatments as drivers of community composition were also investigated in the constrained ordinations. For nifH, although fertility management and crop protection did not represent a significant portion of the variance on any of the sample dates, factors that were significantly affected by fertility management on all sample dates did contribute significantly to the variation in nifH community structure. Specifically, soil basal respiration (greater under organic FM, P<0.001) and nitrate and ammonium (both greater under conventional FM, P<0.001 and = 0.003 respectively) were correlated with changes in nifH diversity at certain dates. Factors associated with fertility management also contributed to a much greater proportion of the variance when analysed as separate factors rather than grouped as one management factor (Table 3). The constrained ordination did, however show that crop management significantly affected total bacterial community composition in June 2007 and June 2009 and that fertility management significantly affected total bacterial community composition in September 2008. The significant effect of fertility management in 2008 coincides with pH also significantly affecting total bacterial community composition (Table 3).
Table 3. Summary of CCA and RDA analysis of RNA DGGE results showing significant variables.
| Gene of interest | Sample date | Variables tested | Significant variables selected by forward selection | Variance of DGGE data explained by the model (%) | ||||
| 2007 | 2008 | 2009 | 2007 | 2008 | 2009 | |||
| nifH | March | FM | 8.0 | 4.8 | ||||
| CP | 5.8 | 5.1 | ||||||
| Associated variables1 | 12.7 | 13.2 | ||||||
| Associated variables1, FM, CP | 14.4 | 15.6 | ||||||
| June | FM | 6.1 | 8.0 | |||||
| CP | 6.4 | 10.1 | ||||||
| Associated variables1 | NH4 + | 11.1 | 23.6 | |||||
| Associated variables1, FM, CP | SBR | NH4 + | 23.4 | 36 | ||||
| September | FM | 7.2 | 9.1 | |||||
| CP | 5.2 | 5.5 | ||||||
| Associated variables1 | NO3 −, NH4 + | 14.3 | 20.6 | |||||
| Associated variables1, FM, CP | NO3 −, NH4 + | 15.2 | 22.6 | |||||
| 16S rRNA | March | FM | 3.1 | 5.8 | 4.7 | |||
| CP | 6.2 | 6.1 | 5.9 | |||||
| Associated variables1 | 9.9 | 4.5 | 4.2 | |||||
| Associated variables1, FM, CP | 10 | 8.8 | 9.1 | |||||
| June | FM | 6.2 | 4.5 | 4.4 | ||||
| CP | CP | CP | 14.1 | 8.0 | 11.7 | |||
| Associated variables1 | 9.1 | 9.2 | 6.8 | |||||
| Associated variables1, FM, CP | CP | CP | 17.8 | 10.5 | 13.5 | |||
| September | FM | FM | 11.3 | 9.3 | 5.8 | |||
| CP | 4.7 | 3.9 | 5.3 | |||||
| Associated variables1 | pH | NO3 − | 11.2 | 12.9 | 11.2 | |||
| Associated variables1, FM, CP | FM, pH | NO3 − | 22.1 | 18.5 | 12.9 | |||
FM = fertility management, CP = crop protection.
Associated variables measured at the time of sampling pH, soil basal respiration (SBR), ammonium (NH4 +) and nitrate (NO3 −). Soil basal respiration was measured in June samples only and pH was measured in September samples only.
For the nifH community attempts were made to sequence all bands on the gels. This resulted in 22 bands being sequenced and identified from the DGGE gels. The sequences were around 200 bp in length and enabled gross taxonomic resolution but were too short for higher level phylogenetic affiliation to be determined. Sequence data is available at the GenBank database under accession numbers JQ618105–JQ618126. Table S2 shows the closest match from the NCBI database. Of the 22 sequences, 17 were from uncultured taxa; 10 sequences belonged to Alpha-Proteobacteria, 9 belonged to Beta-Proteobacteria, 2 belonged to Gamma-Proteobacteria and 1 belonged to the order Clostridia. The remaining 3 bands were identified as belonging to Rhizobium huautlense. By analyzing the relative intensities of the sequenced bands using ANOVA (data not shown) it was found that management type did not significantly affect the presence or intensity of any of the sequenced bands.
Quantification of the nifH and 16S mRNA Transcripts and Genes with qPCR
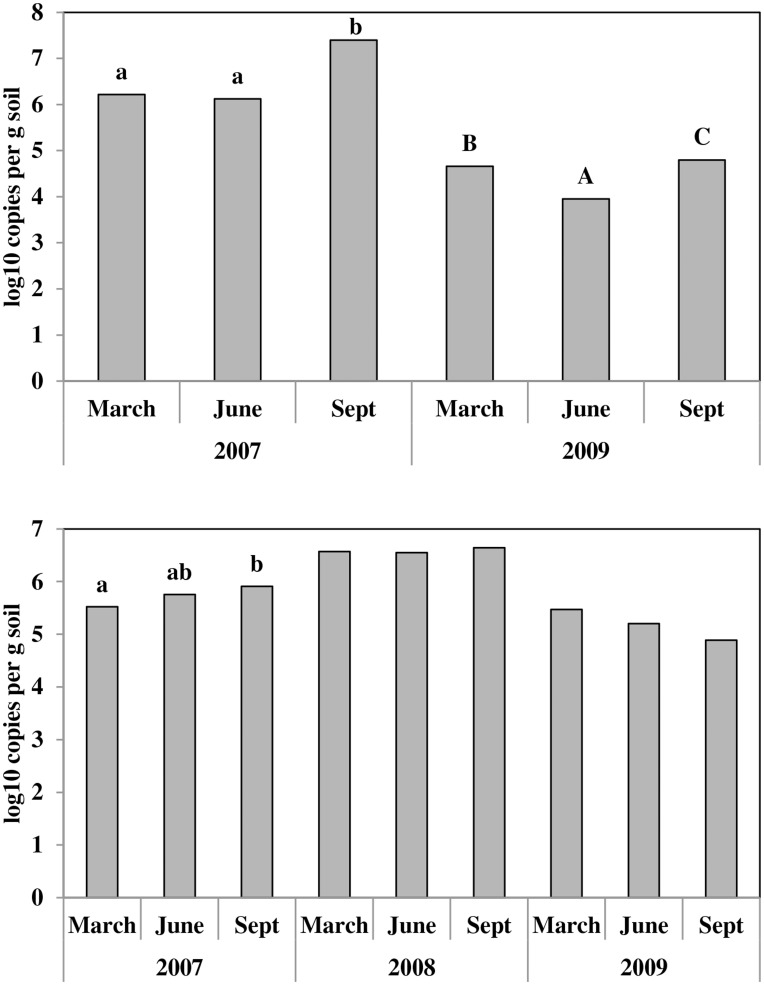
The predominant factors affecting nifH mRNA transcripts and DNA copy numbers and the 16S ribosomal mRNA transcript copy numbers were Year and Sample date, although in some cases crop management practices also affected these parameters (Table 4). On average there were significantly more copies of the nifH mRNA transcript detected in 2007 compared with 2009. September samples also had more copies of nifH mRNA; however, there were strong interactions between Year and Sample date, and Year and Fertility Management. For this reason a separate year by year analysis was conducted. In both years Sample date was the predominant factor affecting nifH mRNA transcript copy numbers. Highest numbers were detected in September, although in 2009 significantly lower numbers were detected in June while in 2007 March and June results did not differ (Fig. 3). In both years the use of organic fertility management always resulted in higher levels of nifH gene expression than conventional fertilisation.
Table 4. Summary of qPCR analysis across all years and sample dates for all genes and nucleic acid types.
| qPCR average copy numbers for nifH RNA data set (mean ± SE) | qPCR average copy numbers fornifH DNA data set (mean ± SE) | qPCR average copy numbers for 16SrRNA gene data set (mean ± SE) | |
| year (Y) | |||
| 2007 | 9.29×106±2.85×106b | 5.69×105±6.81×104b | 8.05×107±1.17×107a |
| 2008 | 3.89×106±2.62×105c | 5.26×107±2.30×107a | |
| 2009 | 3.92×104±5.37×103a | 1.77×105±6.34×104a | 2.96×108±1.29×108a |
| sample date (SD) | |||
| March | 8.46×105±4.99×105b | 1.45×106±2.65×105a | 2.99×108±1.29×108 |
| June | 6.62×105±2.33×105b | 1.42×106±2.51×105a | 4.31×107±7.29×106a |
| September | 1.25×107±4.16×106a | 1.76×106±3.45×105a | 8.66×107±2.41×107a |
| Crop protection (CP) | |||
| org | 6.25×106±2.72×105a | 1.74×106±2.44×105a | 1.06×107±5.20×107a |
| con | 3.07×106±1.18×106a | 1.35×106±2.27×105b | 1.81×108±8.13×107a |
| fertility management | |||
| org | 4.97×106±2.59×106a | 1.64×106±2.31×105a | 1.23×108±3.92×107a |
| con | 4.35×106±1.53×106a | 1.45×106±2.42×105 | 1.63×108±7.99×107a |
| ANOVA P values | |||
| Y | <0.001 | <0.001 | |
| SD | <0.001 | 0.032 | |
| CP | 0.013 | ||
| Y*SD | 0.005 | <0.001 | |
| Y*FM | 0.001 | 0.032 | |
| CP*FM | 0.037 | ||
| Y*SD*FM | 0.022 |
P-values are only shown for terms with P<0.05; all data were log-transformed before analysis. Means followed by the same letter for a given factor are not significantly different (P<0.05; Tukey’s HSD test where there are more than two treatment levels).
Figure 3. Showing copy numbers of the nifH mRNA transcript and the nifH gene.
nifH mRNA transcripts shown in the top and nifH gene in the bottom panels respectively. Bars labelled with the same letter in the same year are not different (P = 0.05). Unlabelled bars in the same year are not significantly different.
Quantities of the nifH gene were also strongly affected by Year with highest copy numbers observed in 2008, but in contrast to the nifH mRNA transcript, crop protection practices were also significant. The use of organic crop protection practices resulted in significantly higher quantities of the nifH gene compared with conventional crop protection (Table 4). Year by year analysis of the nifH gene shows that sample date is only a significant factor in 2007 with nifH gene copy number increasing throughout the year (Table 5). Year by year analysis also shows that in 2007 organic fertility management results in increased nifH gene copy number (Table 5).
Table 5. Average copy numbers for nifH gene amplified from DNA and reverse transcribed RNA for each year of the trial.
| Copies of nifH RNA/g soil (mean±SE) | Copies of nifH DNA/g soil(mean±SE) | Copies of 16S rRNA gene/g soil(mean±SE) | ||||||
| 2007 | 2009 | 2007 | 2008 | 2009 | 2007 | 2008 | 2009 | |
| Sample date (SD) | ||||||||
| March | 1.65×106± | 4.56×104± | 3.31×105± | 3.72×106± | 2.94×105± | 6.19×107± | 1.66×107± | 8.20×108± |
| 9.71×105b | 1.16×104b | 1.08×105a | 3.16×105a | 1.78×105a | 1.88×107a | 4.24×106a | 3.58×108b | |
| June | 1.32×106± | 8.97×103± | 5.65×105± | 3.54×106± | 1.59×105± | 7.21×107± | 1.52×107± | 4.20×107± |
| 4.10×105b | 1.13×102a | 1.20×105ab | 3.46×105a | 6.35×104a | 1.54×107a | 3.09×107a | 1.20×107a | |
| September | 2.49×107± | 6.28×104± | 8.10×105± | 4.39×106± | 7.70×104± | 1.08×108± | 1.26×108± | 2.62×107± |
| 7.15×106a | 5.95×103c | 1.00×105b | 6.31×105a | 2.47×104a | 2.50×107a | 6.65×107b | 4.49×106a | |
| Crop protection (CP) | ||||||||
| ORG | 1.01×107± | 3.42×104± | 5.75×105± | 4.37×106± | 2.81×105± | 7.68×107± | 3.12×107± | 2.08×108± |
| 5.00×106a | 6.79×103a | 9.46×104a | 2.73×105a | 1.23×105a | 1.41×107a | 8.73×106a | 1.07×108a | |
| CON | 8.50×106± | 4.41×104± | 5.62×105± | 3.40×106± | 7.26×104± | 8.43×107± | 7.39×107± | 3.83×108± |
| 2.86×106a | 8.33×103a | 1.00×105a | 4.30×105a | 1.94×104a | 1.90×107a | 4.53×107a | 2.36×108a | |
| Fertility mgt (FM) | ||||||||
| ORG | 9.91×106± | 4.67×104± | 7.62×105± | 4.03×106± | 1.26×105± | 8.75×107± | 7.33×107± | 2.08×108± |
| 2.76×106a | 8.08×103a | 1.07×105a | 3.12×105a | 4.32×104a | 1.76×107a | 4.53×107a | 1.04×108a | |
| CON | 8.62×106± | 3.16×104± | 3.76×105± | 3.74×106±a | 2.27×105± | 7.36×107± | 3.18×107± | 3.83×108± |
| 4.929×106b | 6.75×103b | 6.44×104b | 4.25×105 | 1.20×105a | 1.57×107a | 8.70×106a | 2.37×108a | |
| ANOVA P -values | ||||||||
| SD | 0.001 | <0.001 | 0.050 | 0.011 | 0.009 | |||
| FM | 0.026 | 0.016 | 0.005 | |||||
| SD*FM | 0.036 | |||||||
| SD*CP*FM | 0.048 | |||||||
P-values are only shown for terms with P<0.05; data for nifH RNA 2007 and 2009, nifH DNA 2009 only, and 16S rRNA all years, were log-transformed before analysis. Means followed by the same letter for a given factor are not significantly different (P<0.05; Tukey’s HSD test where there are more than two treatment levels).
Although the ANOVA results indicated that sample date had a significant effect on copies of the 16S mRNA transcript (Table 4) there were no significant differences among the months identified using Tukey’s HSD test. Year was not a significant factor affecting numbers of 16S mRNA transcript but a significant Year by Sample date interaction was observed. When each year was analysed individually (Table 5) it was found that Sample date was a significant factor in 2008, where highest numbers of 16S mRNA transcript were observed in September, and in 2009, where highest numbers were observed in March. Stepwise regression was used to determine how soil biochemical properties may be driving nifH and 16S RNA transcript and gene activity (qPCR) and diversity (DGGE H′) (Table S3). This analysis indicated that soil temperature had a slightly negative effect on nifH diversity (for both transcript and gene) and a positive effect on nifH transcript activity. Rainfall was negatively correlated with nifH transcript diversity (RNA) and positively related to nifH gene diversity. In addition, the diversity of the nifH mRNA transcript was negatively related to soil C and soil basal respiration. Whereas for activity of the nifH gene measured using DNA extracts, pH had a positive effect while soil basal respiration was negatively correlated with expression (Table S3). In general there was a positive correlation between nifH RNA diversity and copy number and likewise a positive correlation between 16S rRNA diversity and copy number. For the 16S rRNA gene, copy numbers were also negatively correlated with rainfall. Negative correlations were observed between the DGGE H′ data set and average soil temperature with average rainfall positively correlated with both nifH and 16S DNA DGGE H′. 16S rRNA DGGE H′ was also affected by available nitrate; total carbon and available ammonium. These correlations to environmental parameters are distinct from those of 16S expression, suggesting nifH expression did not simply mirror the response of the broader bacterial community (Table S3).
Discussion
The NFSC trial enables studies to be conducted with spatial and temporal replication of each system of interest allowing for robust analyses of the impact of management and environmental effects on the microbial communities [42]. Previously we have shown that soils in a conventional crop rotation had a significantly greater diversity and number of free-living diazotrophic bacteria than those within an organic rotation [23]. Here we compared the effect of organic versus conventional fertility management and crop protection activities on the total and free-living N fixing bacterial communities in three different years, on three sampling dates in each year. We hypothesised that the predominant factor affecting diazotrophic and total bacterial diversity, biomass, activity and community structure would be enhanced under organic fertility management, as a result of increased levels of organic carbon, phosphorus and higher soil pH, as has been previously observed [10], [43]–[47].
However, although overall activity of soil organisms was enhanced under ORG FM (e.g. higher soil basal respiration 1.14 mg CO2 kg−1 h−1 for CON FM versus 1.38 mg CO2 kg−1 h−1 for ORG FM), and pH was significantly reduced in conventional fertility management (6.35 for CON FM versus 6.58 for ORG FM on average) while the availability of P, nitrate and ammonium was increased; (Table S4) our data demonstrated that the most significant explanatory variables for quantitative changes in both the diversity and numbers of free-living diazotrophic and total bacterial populations in agricultural soil in a multiple year study were temporal and seasonal effects. These observations contrast with previous work, where fertility source (farmyard manure versus mineral or no fertilizer) was the dominant factor driving bacterial community structure [8], [11], indicating that an increase in organic carbon, associated with organic fertility management activities, had a positive correlation with bacterial soil diversity [48], [49]. However, other studies that have more resonance with our data, indicate that changes to bacterial structure and diversity due to management practices are often subtle [50] and seasonal and plant growth effects often have a greater influence than those due to management processes [51]. One explanation for our findings may be that, although the NFSC trial has been ongoing since 2001, there were no significant differences in soil organic C or total N between the soil management treatments in any of the study years.
Although overall fertility management had no effect on the diversity of the diazotrophs, the factors soil basal respiration and available nitrate were associated with changes in nifH diversity and activity (Table 3). There are very few studies on the impact of organic farming on the free-living diazotrophic communities in agricultural soil. DeLuca et al, [22] compared the use of cattle manure and urea fertilizers and found that both fertilizer types inhibited nitrogen fixation (measured by acetylene reduction assay) and that pH was correlated with nitrogen fixation ability. Previous studies, looking at the effect of individual attributes of farm management on the rhizospheric nitrogen fixing community, suggested that the application of increased amounts of nitrogen fertilizer (normally associated with conventional fertility management) would result in decreased diazotrophic diversity and activity [4], [52]. Rather our data suggests that many different factors affect the nitrogen fixing community (Table 3 and S3). Although our results were not as conclusive as previous studies, organic fertility management was observed to correlate with increased nifH mRNA transcripts in 2007 and 2009, and increased nifH gene copy number in 2007 (Table 5). Soil nitrate levels were also negatively correlated with nifH qPCR data (Table S3), which corresponds to other studies which have reported inhibition of nitrogenase activity in free-living N2 fixing bacteria [53]–[56]. The interacting effects of nitrogen level, carbon availability and crop protection practices, make it difficult to recommend one suite of management practices that can be expected to enhance N fixation by diazotrophs in agricultural soils.
It was hypothesised that conventional crop protection would have a negative effect on nifH diversity, and expression, as studies into the environmental impacts of pesticides have shown that they can significantly affect the bacterial community as a whole and that diazotrophs could be particularly affected [57], [58], [51]. Our results suggest that conventional crop protection did in fact exert a negative effect on the diazotrophic activity when nifH copy numbers derived from the DNA data set were considered (Table 4) but appeared not to impact on the diversity or structure of the community. The DNA results suggest that the size of the nifH population in plots under conventional crop protection has been significantly reduced due to the long-term application of pesticides. That levels of nifH expression (RNA-qPCR results) did not mirror the DNA-qPCR results suggests that activity of the diazotrophic community is not limited by its size, but rather by other controlling factors. A range of pesticides are applied to the potato crops in the NFSC experiment (Table 1) some of which have been shown to have some inhibitory effect on diazotrophs at high concentrations [57], [59], [60]. Many previous studies looking at the effect of pesticides on the diazotrophic community have focussed on nitrogen fixers which are symbiotic with legumes. Bradyrhizobium japonicum, for example, has been found to be particularly susceptible to the effects of glyphosphate due to the sensitivity of its phosphate synthase enzyme [61], [62]. Other studies have found that herbicides will affect nitrogenase activity, nodule formation, nodule biomass and leghaemoglobin concentrations [61]–[63]. However, it is unclear whether this is due to direct changes in the rhizobia, indirect physiological changes in the plant, or both and does not explain why we see significant changes in the free-living nitrogen fixing community [64], [61].
In contrast crop protection strategy had no significant effect on the activity of the total bacterial population, although, it was a significant driver of community structure in June of both 2007 and 2009. To our knowledge this is the first study which fully investigates the effects of crop protection protocols in the field on the activity of both diazotrophic and total bacterial communities.
The temporal effects observed on both diversity and number of the diazotrophic, and total bacterial communities, were primarily affected by the recent environmental conditions. On most occasions, rainfall and soil temperature were significant factors affecting activity and diversity according to stepwise regression (Table S3), although the effects were not always positive. Diversity tended to be higher in March for both nifH (mRNA transcripts and genes) and 16S mRNA transcripts. Optimum temperature for growth and activity of diazotrophs is between 10 and 25°C (similar to the temperature range in the field between June and September) [65], [66]. Activities of the general bacterial community were largely unaffected by sample date, suggesting that this community included species with a wide range of optimal temperatures that were able to adapt to the environmental conditions throughout the growing season. As expected, diversity and copy number of the 16S rRNA gene were always higher than diversity and copy number of the nifH gene. Ratios of the nifH gene to the 16S rRNA gene (∼1 copies of nifH gene: 50 copies of 16S rRNA gene) were similar to ratios seen between the 16S rRNA gene and genes used in nitrogen cycling found in other studies [67], [68].
Seasonal effects observed in this study may also be related to the crop management practices that occur throughout the year. Samples taken in March are from a relatively undisturbed soil with no plant cover. June samples may be affected by frequent cultivations for weed control, especially in the organic crop protection plots. This makes it difficult to separate soil temperature and moisture effects in this study from the effects of seasonal management practices. Stage of crop growth can also influence microbial community composition. Certain members of the soil bacterial community, particularly Acidobacteria, Bacteroidetes and Alpha-, Beta-, and Gammaproteobacteria, have previously been observed to be diminished in summer in crop land [49]. It has been demonstrated that growth stage and seasonal effects significantly affect diversity in soil under potato and maize [69]. For example, when culture dependent and independent (cloning and DGGE) methods were used to assess bacterial diversity in bulk and rhizosphere soil in 3 species of potato, bacterial communities were observed to change as the plant developed. Higher diversity was observed around 25 days after planting, compared to growth 65 and 140 days after planting [70]. Similarly in maize, bacterial activity, as measured by PLFA and BIOLOG, changed as maize went through five leaf stage, flowering and maturity [71]. It is assumed that these observations reflect changes in the amount and quality of root exudates as the plant reaches maturity [47].
We found that management activity, temporal and seasonal factors appeared to exert no significant effect on the most abundant diazotrophs identified by sequencing the DGGE bands (Table S2). A follow up study is currently underway using pyrosequencing to more thoroughly resolve the taxonomic structure of the diazotrophic communities in these soils. Previous work looking at the impact of differing levels of nitrogen fertilization on the diazotrophic communities of soil showed that the predominant taxa were present in all soils regardless of the amounts of nitrogen fertilizer used [52], [72]. It has been suggested that the predominant taxa remain unaffected by the level of N fertilization, whereas the minor members of this community are more sensitive to such changes [73]. In conclusion we found the dominant factors affecting the diversity and numbers of both the nitrogen fixing and the total bacterial community are temporal. The only exception was the impact of conventional crop protection protocols that seemed to reduce the number of diazotrophs within the soils but not their activity. Fertility management appeared to have little effect on the diversity of both the nitrogen fixing and the total bacterial community, although soil parameters, particularly pH and the concentrations of nitrate and ammonium, were significant factors in determining community structures. The combination of our study and the work of others suggests that rather than the bacterial communities being affected directly by the nature of the fertilizers applied they are more likely to respond to changes in carbon and nitrogen levels in the soil [10], [43], [44], [74]. Although crop management practices were found to impact on the activity and function of soil bacteria, the overriding factor was consistently the year and date of sampling.
Supporting Information
Summary of environmental conditions measured in the experimental field during the 14 days prior to each sample date.
(DOCX)
The closest matches for the 22 sequenced bands derived from the NCBI database.
(DOC)
Significant explanatory variables for nifH and 16S rRNA gene activity (qPCR) and diversity (DGGE H’) determined by stepwise regression.
(DOC)
The impact of farm management and year of sampling on environmental variables measured in each soil
(DOC)
Funding Statement
This work was funded by the following: European Community financial participation under the Seventh Framework Programme for Research, Technological Development and Demonstration Activities, for the Integrated Project NUE CROPS EU-FP7 222–645. The Yorkshire Agricultural Society. www.yas.co.uk. Grant NPD/JMD/08/72. Nafferton Ecological Farming Group. http://research.ncl.ac.uk/nefg. Institutional funding was provided by Northumbria University Research Development Fund. http://www.northumbria.ac.uk. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Kibblewhite MG, Ritz K, Swift MJ (2008) Soil health in agricultural systems. Phil Trans Roy Soc B-Biol Sci 363: 685–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hsu SF, Buckley DH (2009) Evidence for the functional significance of diazotroph community structure in soil. ISME 3: 124–136. [DOI] [PubMed] [Google Scholar]
- 3. Hayden HL, Drake J, Imhof M, Oxley APA, Norng S, et al. (2010) The abundance of nitrogen cycle genes amoA and nifH depends on land-uses and soil types in South-Eastern Australia. Soil Biol Biochem 42: 1774–1783. [Google Scholar]
- 4. Coelho MRR, Marriel IE, Jenkins SN, Lanyon CV, Seldin L, et al. (2009) Molecular detection and quantification of nifH gene sequences in the rhizosphere of sorghum (Sorghum bicolor) sown with two levels of nitrogen fertilizer. Appl Soil Ecol 42: 48–53. [Google Scholar]
- 5. Hauggaard-Nielsen H, Mundus S, Jensen E (2009) Nitrogen dynamics following grain legumes and subsequent catch crops and the effects on succeeding cereal crops. Nut Cycl Agroecosyst 84: 281–291. [Google Scholar]
- 6. Gamble M, Bagwell C, LaRocque J, Bergholz P, Lovell C (2010) Seasonal Variability of Diazotroph Assemblages Associated with the Rhizosphere of the Salt Marsh Cordgrass, Spartina alterniflora . Microb Ecol 59: 253–265. [DOI] [PubMed] [Google Scholar]
- 7. Hartmann M, Fliessbach A, Oberholzer HR, Widmer F (2006) Ranking the magnitude of crop and farming system effects on soil microbial biomass and genetic structure of bacterial communities. FEMS Microbiol Ecol 57: 378–388. [DOI] [PubMed] [Google Scholar]
- 8. Esperschütz J, Gattinger A, Mäder P, Schloter M, Fließbach A (2007) Response of soil microbial biomass and community structures to conventional and organic farming systems under identical crop rotations. FEMS Microbiol Ecol 61: 26–37. [DOI] [PubMed] [Google Scholar]
- 9. Bossio DA, Scow KM, Gunapala N, Graham KJ (1998) Determinants of soil microbial communities: effects of agricultural management, season, and soil type on phospholipid fatty acid profiles. Microb Ecol 36: 1–12. [DOI] [PubMed] [Google Scholar]
- 10. van Diepeningen AD, de Vos OJ, Korthals GW, van Bruggen AHC (2006) Effects of organic versus conventional management on chemical and biological parameters in agricultural soils. Appl Soil Ecol 31: 120–135. [Google Scholar]
- 11. Widmer F, Rasche F, Hartmann M, Fließbach A (2006) Community structures and substrate utilization of bacteria in soils from organic and conventional farming systems of the DOK long-term field experiment. Appl Soil Ecol 33: 294–307. [Google Scholar]
- 12. Wessén E, Hallin S, Philippot L (2010) Differential responses of bacterial and archaeal groups at high taxonomical ranks to soil management. Soil Biol Biochem 42: 1759–1765. [Google Scholar]
- 13. Fernández-Calviño D, Bååth E (2010) Growth response of the bacterial community to pH in soils differing in pH. FEMS Microbiol Ecol 73: 149–156. [DOI] [PubMed] [Google Scholar]
- 14. Mäder P, Fließbach A, Dubois D, Gunst L, Fried P, et al. (2002) Soil Fertility and Biodiversity in Organic Farming. Science 296: 1694–1697. [DOI] [PubMed] [Google Scholar]
- 15. Pimentel D, Hepperly P, Hanson J, Douds D, Seidel R (2005) Environmental, Energetic, and Economic Comparisons of Organic and Conventional Farming Systems. BioSci 55: 573–582. [Google Scholar]
- 16. Keeling AA, Cook JA, Wilcox A (1998) Effects of carbohydrate application on diazotroph populations and nitrogen availability in grass swards established in garden waste compost. Biores Technol 66: 89–97. [Google Scholar]
- 17. Hartley AE, Schlesinger WH (2002) Potential environmental controls on nitrogenase activity in biological crusts of the northern Chihuahuan Desert. J Arid Environ 52: 293–304. [Google Scholar]
- 18. Bürgmann H, Meier S, Bunge M, Widmer F, Zeyer J (2005) Effects of model root exudates on structure and activity of a soil diazotroph community. Environ Microbiol 7: 1711–1724. [DOI] [PubMed] [Google Scholar]
- 19. Fierer N, Jackson RB (2006) The diversity and biogeography of soil bacterial communities. PNAS 103: 626–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hallin S, Jones CM, Schloter M, Philippot L (2009) Relationship between N-cycling communities and ecosystem functioning in a 50-year-old fertilization experiment. ISME J 3: 597–605. [DOI] [PubMed] [Google Scholar]
- 21. Reed SC, Seastedt TR, Mann CM, Suding KN, Townsend AR, et al. (2007) Phosphorus fertilization stimulates nitrogen fixation and increases inorganic nitrogen concentrations in a restored prairie. Appl Soil Ecol 36: 238–242. [Google Scholar]
- 22. DeLuca TH, Drinkwater LE, Wiefling BA, DeNicola DM (1996) Free-living nitrogen-fixing bacteria in temperate cropping systems: Influence of nitrogen source. Biol Fert Soil 23: 140–144. [Google Scholar]
- 23. Orr CH, James A, Leifert C, Cooper JM, Cummings SP (2011) Diversity and Activity of Free-Living Nitrogen Fixing Bacteria and Total Bacteria in Organic and Conventionally Managed Soils. Appl Env Microbiol 77: 911–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hussain S, Siddique T, Saleem M, Arshad M, Khalid A, et al.. (2009) Impact of Pesticides on Soil Microbial Diversity, Enzymes, and Biochemical Reactions. In: Advances in Agronomy. New York. USA: Academic Press. 159–200. [Google Scholar]
- 25.Cooper J, Niggli U, Leifert C (2007) Handbook of organic food safety and quality. Cambridge, UK: Woodhead Publishing Limited. 521 p. [Google Scholar]
- 26. Bending GD, Rodríguez-Cruz MS, Lincoln SD (2007) Fungicide impacts on microbial communities in soils with contrasting management histories. Chemosphere 69: 82–88. [DOI] [PubMed] [Google Scholar]
- 27. Cycoń M, Piotrowska-Seget Z (2009) Changes in bacterial diversity and community structure following pesticides addition to soil estimated by cultivation technique. Ecotoxicol 18: 632–642. [DOI] [PubMed] [Google Scholar]
- 28. Spyrou I, Karpouzas D, Menkissoglu-Spiroudi U (2009) Do botanical pesticides alter the structure of the soil microbial community? Microb Ecol 58: 715–727. [DOI] [PubMed] [Google Scholar]
- 29. Omar SA, Abd-Alla MH (1992) Effect of pesticides on growth, respiration and nitrogenase activity of Azotobacter and Azospirillum . World J Microbiol Biotech 8: 326–328. [DOI] [PubMed] [Google Scholar]
- 30. Fox JE, Gulledge J, Engelhaupt E, Burow ME, McLachlan JA (2007) Pesticides reduce symbiotic efficiency of nitrogen-fixing rhizobia and host plants. PNAS 104: 10282–10287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fox JE, Starcevic M, Jones PE, Burrow ME, McLachlan JA (2004) Phytoestrogen signalling and Symbiotic Gene Activation Are Disrupted by Endocrine-Disrupting Chemicals. Environ Health Persp 112: 672–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soil Association (2005). Soil Association organic standards, Bristol, UK. [Google Scholar]
- 33. Wartiainen I, Eriksson T, Zheng W, Rasmussen U (2008) Variation in the active diazotrophic community in rice paddy-nifH PCR-DGGE analysis of rhizosphere and bulk soil. Appl Soil Ecol 39: 65–75. [Google Scholar]
- 34. Poly F, Monrozier LJ, Bally R (2001) Improvement in the RFLP procedure for studying the diversity of nifH genes in communities of nitrogen fixers in soil. Res Microbiol 152: 95–103. [DOI] [PubMed] [Google Scholar]
- 35. Baxter J, Cummings SP (2008) The degradation of the herbicide bromoxynil and its impact on bacterial diversity in a top soil. J Appl Microbiol 104: 1605–1616. [DOI] [PubMed] [Google Scholar]
- 36. Cummings SP, Gyaneshwar P, Vinuesa P, Farruggia FT, Andrews M, et al. (2009) Nodulation of Sesbania species by Rhizobium (Agrobacterium) strain IRBG74 and other rhizobia. Environ Microbiol 11: 2510–2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Karlen Y, McNair A, Perseguers S, Mazza C, Mermod N (2007) Statistical significance of quantitative PCR. BMC Bioinform 8: 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.R Development Core Team (2006) R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- 39.Venables WN, Ripley BD (2002) Modern applied statistics with S, 4th ed. New York, USA: Springer. [Google Scholar]
- 40.Minitab (2006) Minitab Statistical Software. Release 15 for Windows. State College, Pennsylvania, Minitab® is a registered trademark of Minitab Inc. [Google Scholar]
- 41. Lindström ES, Bergström AK (2005) Community composition of bacterioplankton and cell transport in lakes in two different drainage areas. Aquatic Sci 67: 210–219. [Google Scholar]
- 42. Prosser JI (2010) Replicate or lie. Environ Microbiol 12: 1806–1810. [DOI] [PubMed] [Google Scholar]
- 43. Postma J, Schilder MT, Bloem J, van Leeuwen-Haagsma WK (2008) Soil suppressiveness and functional diversity of the soil microflora in organic farming systems. Soil Biol Biochem 40: 2394–2406. [Google Scholar]
- 44. Toljander JF, Santos-González JC, Tehler A, Finlay RD (2008) Community analysis of arbuscular mycorrhizal fungi and bacteria in the maize mycorrhizosphere in a long-term fertilization trial. FEMS Microbiol Ecol 65: 323–338. [DOI] [PubMed] [Google Scholar]
- 45. Birkhofer K, Bezemer TM, Bloem J, Bonkowski M, Christensen S, et al. (2008) Long-term organic farming fosters below and aboveground biota: Implications for soil quality, biological control and productivity. Soil Biol Biochem 40: 2297–2308. [Google Scholar]
- 46. Tamm L, Thürig B, Bruns C, Fuchs J, Köpke U, et al. (2010) Soil type, management history, and soil amendments influence the development of soil-borne (Rhizoctonia solani, Pythium ultimum) and air-borne (Phytophthora infestans, Hyaloperonospora parasitica) diseases. Eur J Plant Path 127: 465–481. [Google Scholar]
- 47. Ngosong C, Jarosch M, Raupp J, Neumann E, Ruess L (2010) The impact of farming practice on soil microorganisms and arbuscular mycorrhizal fungi: Crop type versus long-term mineral and organic fertilization. Appl Soil Ecol 46: 134–142. [Google Scholar]
- 48. Lejon DPH, Chaussod R, Ranger J, Ranjard L (2005) Microbial community structure and density under different tree species in an acid forest soil (Morvan, France). Microb Ecol 50: 614–625. [DOI] [PubMed] [Google Scholar]
- 49. Jangid K, Williams MA, Franzluebbers AJ, Sanderlin JS, Reeves JH, et al. (2008) Relative impacts of land-use, management intensity and fertilization upon soil microbial community structure in agricultural systems. Soil Biol Biochem 40: 2843–2853. [Google Scholar]
- 50. Drenovsky RE, Vo D, Graham KJ, Scow KM (2004) Soil Water Content and Organic Carbon Availability Are Major Determinants of Soil Microbial Community Composition. Microb Ecol 48: 424–430. [DOI] [PubMed] [Google Scholar]
- 51. Moreno B, Garcia-Rodriguez S, Cañizares R, Castro J, Benítez E (2009) Rainfed olive farming in south-eastern Spain: Long-term effect of soil management on biological indicators of soil quality. Agri Ecosys Environ 131: 333–339. [Google Scholar]
- 52. Coelho MRR, de Vos M, Carneiro NP, Marriel IE, Paiva E, et al. (2008) Diversity of nifH gene pools in the rhizosphere of two cultivars of sorghum Sorghum bicolor treated with contrasting levels of nitrogen fertilizer. FEMS Microbiol Letts 279: 15–22. [DOI] [PubMed] [Google Scholar]
- 53. Knowles R, Denike D (1974) Effect of ammonium-nitrogen, nitrite-nitrogen and nitrate-nitrogen on anaerobic nitrogenase activity in soil. Soil Biol Biochem 6: 353–358. [Google Scholar]
- 54. Chapin DM, Bliss LC, Bledsoe LI (1991) Environmental-regulation of nitrogen-fixation in a high arctic lowland ecosystem. Can J Bot 69: 2744–2755. [Google Scholar]
- 55. Kitoh S, Shiomi N (1991) Effect of mineral nutrients and combined nitrogen-sources in the medium on growth and nitrogen-fixation of the azolla-anabaena association. Soil Sci Plant Nutr 37: 419–426. [Google Scholar]
- 56. Roper MM, Turpin JE, Thompson JP (1994) Nitrogenase activity (C2H2 reduction) by free-living bacteria in soil in a long term tillage and stubble management experiment on a vertisol. Soil Biol Biochem 26: 1087–1091. [Google Scholar]
- 57. Doneche B, Seguin G, Ribereau-Gayon P (1983) Mancozeb Effect on Soil Microorganisms and Its Degradation in Soils. Soil Sci 135: 361–366. [Google Scholar]
- 58. Cycoń M, Piotrowska-Seget Z (2007) Effect of selected pesticides on soil microflora involved in organic matter and nitrogen transformations: pot experiment. Pol J Ecol 55: 207–220. [Google Scholar]
- 59. Sturz AV, Kimpinski J (1999) Effects of fosthiazate and aldicarb on population of plant-growth-promoting bacteria, root lesion nematodes and bacteria-feeding nematodes in the root zome of potatoes. Plant Pathol 48: 26–32. [Google Scholar]
- 60. Miloševiã NA, Govedarica MM (2002) Effect of herbicides on microbiological properties of soil. Proceedings for Natural Sciences 102: 5–21. [Google Scholar]
- 61. Zablotowicz RM, Reddy KN (2007) Nitrogenase activity, nitrogen content, and yield responses to glyphosate in glyphosate-resistant soybean. Crop Prot 26: 370–376. [Google Scholar]
- 62. Bohm GMB, Alves BJR, Urquiaga S, Boddey RM, Xavier GR, et al. (2009) Glyphosate- and imazethapyr-induced effects on yield, nodule mass and biological nitrogen fixation in field-grown glyphosate resistant soybean. Soil Biol Biochem 41: 420–422. [Google Scholar]
- 63. Reddy KN, Zablotowicz RM (2002) Glyphosate-resistant soybean response to various salts of glyphosate and glyphosate accumulation in soybean nodules. Weed Sci 51: 496–502. [Google Scholar]
- 64. Vieira R, Silva C, Silveira A (2007) Soil microbial biomass C and symbiotic processes associated with soybean after sulfentrazone herbicide application. Plant Soil 300: 95–103. [Google Scholar]
- 65. Beauchamp CJ, Lévesque G, Prévost D, Chalifour FP (2006) Isolation of free-living dinitrogen-fixing bacteria and their activity in compost containing de-inking paper sludge. Biores Technol 97: 1002–1011. [DOI] [PubMed] [Google Scholar]
- 66. Eckford R, Cook FD, Saul D, Aislabie J, Foght J (2002) Free-living Heterotrophic Nitrogen-fixing Bacteria Isolated from Fuel-Contaminated Antarctic Soils. Appl Environ Microbiol 68: 5181–5185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kandeler E, Deiglmayr K, Tscherko D, Bru D, Philippot L (2006) Abundance of narG, nirS, nirK, and nosZ Genes of Denitrifying Bacteria during Primary Successions of a Glacier Foreland. Appl Environ Microbiol 72: 5957–5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Morales SE, Cosart T, Holben WE (2010) Bacterial gene abundances as indicators of greenhouse gas emission in soils. ISME J 4: 799–808. [DOI] [PubMed] [Google Scholar]
- 69. Diallo S, Crépin A, Barbey C, Orange N, Burini JF, et al. (2010) Mechanisms and recent advances in biological control mediated through the potato rhizosphere. FEMS Microbiol Ecol 75: 351–364. [DOI] [PubMed] [Google Scholar]
- 70. van Overbeek L, van Elsas JD (2008) Effects of plant genotype and growth stage on the structure of bacterial communities associated with potato (Solanum tuberosum L.) FEMS Microbiol Ecol. 64: 283–296. [DOI] [PubMed] [Google Scholar]
- 71. Griffiths BS, Caul S, Thompson J, Birch ANE, Scrimgeour C, et al. (2006) Soil Microbial and Faunal Community Responses to Maize and Insecticide in Two Soils. J Environ Qual 35: 734–741. [DOI] [PubMed] [Google Scholar]
- 72. Ogilvie L, Hirsch P, Johnston A (2008) Bacterial Diversity of the Broadbalk ‘Classical’ Winter Wheat Experiment in Relation to Long-Term Fertilizer Inputs. Microbial Ecol 56: 525–537. [DOI] [PubMed] [Google Scholar]
- 73. Knauth S, Hurek T, Brar D, Reinhold-Hurek B (2005) Influence of different Oryza cultivars on expression of nifH gene pools in roots of rice. Environ Microbiol 7: 1725–1733. [DOI] [PubMed] [Google Scholar]
- 74. Campbell BJ, Polson SW, Hanson TE, Mack MC, Schuur EAG (2010) The effect of nutrient deposition on bacterial communities in Arctic tundra soil. Environ Microbiol 12: 1842–1854. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Summary of environmental conditions measured in the experimental field during the 14 days prior to each sample date.
(DOCX)
The closest matches for the 22 sequenced bands derived from the NCBI database.
(DOC)
Significant explanatory variables for nifH and 16S rRNA gene activity (qPCR) and diversity (DGGE H’) determined by stepwise regression.
(DOC)
The impact of farm management and year of sampling on environmental variables measured in each soil
(DOC)