Structure of a 20-amino-acid peptide of AHNAK bound asymmetrically to the AnxA2–S100A10A heterotetramer (1:2:2 symmetry) provides insights into the atomic level interactions that govern this membrane-repair scaffolding complex.
Keywords: annexin A2, S100A10, AHNAK, calcium-binding proteins, membrane repair, 2:2:1 protein complex
Abstract
AHNAK, a large 629 kDa protein, has been implicated in membrane repair, and the annexin A2–S100A10 heterotetramer [(p11)2(AnxA2)2)] has high affinity for several regions of its 1002-amino-acid C-terminal domain. (p11)2(AnxA2)2 is often localized near the plasma membrane, and this C2-symmetric platform is proposed to be involved in the bridging of membrane vesicles and trafficking of proteins to the plasma membrane. All three proteins co-localize at the intracellular face of the plasma membrane in a Ca2+-dependent manner. The binding of AHNAK to (p11)2(AnxA2)2 has been studied previously, and a minimal binding motif has been mapped to a 20-amino-acid peptide corresponding to residues 5654–5673 of the AHNAK C-terminal domain. Here, the 2.5 Å resolution crystal structure of this 20-amino-acid peptide of AHNAK bound to the AnxA2–S100A10 heterotetramer (1:2:2 symmetry) is presented, which confirms the asymmetric arrangement first described by Rezvanpour and coworkers and explains why the binding motif has high affinity for (p11)2(AnxA2)2. Binding of AHNAK to the surface of (p11)2(AnxA2)2 is governed by several hydrophobic interactions between side chains of AHNAK and pockets on S100A10. The pockets are large enough to accommodate a variety of hydrophobic side chains, allowing the consensus sequence to be more general. Additionally, the various hydrogen bonds formed between the AHNAK peptide and (p11)2(AnxA2)2 most often involve backbone atoms of AHNAK; as a result, the side chains, particularly those that point away from S100A10/AnxA2 towards the solvent, are largely interchangeable. While the structure-based consensus sequence allows interactions with various stretches of the AHNAK C-terminal domain, comparison with other S100 structures reveals that the sequence has been optimized for binding to S100A10. This model adds new insight to the understanding of the specific interactions that occur in this membrane-repair scaffold.
1. Background
Regulation of membrane dynamics is crucial for normal cellular processes as well as the survival of tumor cells, and the process involves the activity of an array of intracellular proteins (Gerke & Moss, 1997 ▶; Borgonovo et al., 2002 ▶; Tekpli et al., 2011 ▶; Epp et al., 2011 ▶; Cheng & Lane, 2010 ▶; Saarikangas et al., 2010 ▶). When the integrity of the plasma membrane is compromised, intracellular proteins involved in membrane repair localize to the site of damage and in many cases are activated by the influx in Ca2+ ions from the extracellular matrix (Draeger et al., 2011 ▶; Bischofberger et al., 2009 ▶). AHNAK, a large 629 kDa protein, has been implicated in membrane repair, and the annexin A2–S100A10 heterotetramer [(p11)2(AnxA2)2] has high affinity for several regions of the long (1002-amino-acid) C-terminal domain of AHNAK (Shtivelman & Bishop, 1993 ▶; Benaud et al., 2004 ▶; Huang et al., 2007 ▶; Lennon et al., 2003 ▶; De Seranno et al., 2006 ▶; Rezvanpour et al., 2011 ▶).
Annexin A2 (AnxA2) belongs to a family of Ca2+-binding and acidic phospholipid-binding proteins characterized by a core domain of four repeats (eight in the case of annexin A6) of five α-helices and a variable N-terminal domain (Gerke & Moss, 2002 ▶; Burger et al., 1996 ▶; Rosengarth & Luecke, 2004 ▶). The protein has been implicated in numerous cellular processes; for example, aggregation of phospholipid vesicles (Blackwood & Ernst, 1990 ▶; Ayala-Sanmartin et al., 2001 ▶), G-actin polymerization (Ozorowski et al., 2012 ▶; Hayes et al., 2006 ▶), F-actin bundling (Filipenko & Waisman, 2001 ▶), apical lumen formation (Martin-Belmonte et al., 2007 ▶), endosome biogenesis (Morel et al., 2009 ▶), angiogenesis (Semov et al., 2005 ▶) and exocytosis (Lorusso et al., 2006 ▶; Umbrecht-Jenck et al., 2010 ▶). Additionally, several unique and diverse binding partners and ligands of AnxA2 have been identified, such as PCSK9 (Mayer et al., 2008 ▶), phosphatidylinositol 4,5-bisphosphate (Rescher et al., 2004 ▶), heparin (Shao et al., 2006 ▶), chlorotoxin (Kesavan et al., 2010 ▶) and progastrin (Singh et al., 2006 ▶). Most commonly, AnxA2 exists either as a monomer or in complex with a dimer of S100A10 (also called p11), forming a heterotetramer [(p11)2(AnxA2)2] (Gerke & Moss, 2002 ▶; Rescher & Gerke, 2008 ▶; Rezvanpour & Shaw, 2009 ▶; Gerke & Weber, 1984 ▶). The S100 family of proteins is comprised of approximately 10 kDa proteins that contain EF-hand calcium-binding sites and generally form dimers. The binding of calcium results in protein activation by exposing hydrophobic pockets, which then become binding sites of binding partners. S100A10 (p11) is unique among the S100 proteins in that it has lost its calcium-binding ability and is permanently activated (Rescher & Gerke, 2008 ▶; Rezvanpour & Shaw, 2009 ▶). The N-terminus of AnxA2 binds into each of the two hydrophobic pockets of S100A10, forming a symmetric 2:2 complex of two AnxA2 molecules bound to a dimer of S100A10 (Réty et al., 1999 ▶). A requirement for this strong interaction is post-translational modification of AnxA2, namely the removal of Met1 and acetylation of Ser2 at the new N-terminus (Nazmi et al., 2012 ▶; Becker et al., 1990 ▶). (p11)2(AnxA2)2 is often localized near the plasma membrane, and this symmetric platform is proposed to be involved in the bridging of membrane vesicles and trafficking of proteins to the plasma membrane (van de Graaf et al., 2003 ▶; Umbrecht-Jenck et al., 2010 ▶). A crystal structure of the S100A10 dimer in complex with N-terminal peptides of AnxA2 has previously been determined (Réty et al., 1999 ▶). A similar complex is believed to form between the N-terminal domain of AnxA2 and S100A4, although detailed structural data are not available for this heterotetramer. Different from (p11)2(AnxA2)2, this interaction occurs on the extracellular surface of endothelial cells and accelerates plasmin formation, contributing to tumor-induced angiogenesis (Semov et al., 2005 ▶).
AHNAK, first described in 1992 and named after the Hebrew word for ‘giant’ (Shtivelman et al., 1992 ▶), was originally found in the nucleus and noted for the repression of its expression in human neuroblastoma cells (Shtivelman & Bishop, 1993 ▶). The C-terminus of AHNAK has been shown to be responsible for translocation between nucleus and cytoplasm (Nie et al., 2000 ▶) and the induction of actin bundling (Haase et al., 2004 ▶), and AHNAK is a major component of enlargosomes, which are vesicles for rapid calcium-dependent exocytosis (Borgonovo et al., 2002 ▶). The C-terminal domain of AHNAK interacts with (p11)2(AnxA2)2 and all three proteins co-localize at the intracellular face of the plasma membrane in a Ca2+-dependent manner. Depletion of AnxA2 by siRNA causes release of AHNAK into the cytoplasm, suggesting that (p11)2(AnxA2)2 may recruit AHNAK to the plasma membrane and act as a scaffold, locating it nearby for cell membrane-repair activities and possibly regulation of membrane cyto-architecture (Benaud et al., 2004 ▶). Both AHNAK and annexins A1 and A2 have been shown to interact with dysferlin, a 230 kDa muscle membrane protein with roles in skeletal muscle regeneration and wound healing (Huang et al., 2007 ▶; Lennon et al., 2003 ▶; Cacciottolo et al., 2011 ▶). Furthermore, AHNAK-deficient mice were highly susceptible to Leishmania major infection owing to the proposed role of AHNAK in T-cell Ca2+ signaling mediated by Cav1 channels based on loss-of-function experiments, the expression characteristics of AHNAK in T cells and the requirement of AHNAK for the expression of L-type calcium channels (Matza et al., 2008 ▶). AHNAK has also been described as a specific binding partner for S100B, and the highest affinity of S100B was measured for peptides corresponding to repeat regions of the internal domain of AHNAK, which showed high specificity for S100B in a Ca2+-dependent and Zn2+-dependent manner (Gentil et al., 2001 ▶).
The binding interaction of AHNAK with (p11)2(AnxA2)2 has been studied more extensively, and a minimal binding motif has been mapped to a 20-amino-acid peptide corresponding to residues 5654–5673 of the AHNAK C-terminal domain (De Seranno et al., 2006 ▶; Rezvanpour et al., 2011 ▶). Various biophysical studies have also shown that the AnxA2 N-terminal domain is necessary for high-affinity binding of S100A10 to AHNAK. Most recently, NMR spectroscopy determined that the likely binding site of the AHNAK peptide is on the surface of (p11)2(AnxA2)2 and is formed by helix IV of each S100A10 monomer and the C-terminal regions of the AnxA2 peptides corresponding to the N-terminal domain (residues 2–16; Rezvanpour et al., 2011 ▶), but did not explain the specific interactions between AHNAK and (p11)2(AnxA2)2. A dissociation constant (K d) value of about 30 nM was reported by Rezvanpour and coworkers between the AHNAK peptide and an S100A10–AnxA2 hybrid construct using both fluorescence spectroscopy and isothermal calorimetry, and the former method suggested a poor interaction between the AHNAK peptide and S100A10 alone, as shown by a weak change in fluorescence. Here, we present the 2.5 Å resolution crystal structure of the 20-amino-acid peptide of AHNAK bound to the AnxA2–S100A10 heterotetramer (1:2:2 symmetry), which confirms the asymmetric platform first described by Rezvanpour and coworkers and explains why the minimal binding motif has high affinity for S100A10. Additionally, we compare this binding interface with those of other members of the S100 family.
2. Materials and methods
2.1. Expression of S100A10
S100A10 (p11) was expressed in Escherichia coli BL21 (DE3) cells and grown in Lennox broth medium to an OD600 of 0.6 at 310 K. Recombinant protein expression was then induced by adding isopropyl β-d-1-thiogalactopyranoside (IPTG) to a final concentration of 1 mM and bacteria were grown for a further 4 h. The cells were harvested by centrifugation at 5000g for 15 min at 277 K and resuspended in lysis buffer (50 mM Tris–HCl pH 8.5, 300 mM NaCl, 10 mM MgCl2, 2 mM EGTA, 1 mM PMSF and one Complete protease-inhibitor tablet) at a ratio of 3.0 ml lysis buffer per gram of wet pellet. The sample was subsequently lysed by sonication and the lysate was centrifuged at 100 000g for 60 min at 277 K. The soluble fraction was loaded onto a HiTrap Q column (GE Healthcare). The flowthrough was collected, dialyzed overnight against SP buffer and loaded onto a 5 ml HiTrap SP HP cation-exchange column (GE Healthcare). The column was subjected to a 50–1000 mM NaCl gradient and p11 eluted between 300 and 400 mM NaCl. The protein was dialyzed against 20 mM Tris pH 7.5, 50 mM NaCl, 1 mM DTT and was concentrated to 20 mg ml−1 using a 10K molecular-weight cutoff centrifugal concentrator unit (Millipore).
2.2. Peptide synthesis
Side-chain-protected Fmoc amino acids, Fmoc-PAL-PEG-polystyrene support and O-(benzotriazol-1-yl)-N,N,N′,N′-tetramethyluronium tetrafluoroborate (TBTU) were obtained from Applied Biosystems Inc. N,N-Diisopropylethylamine (DIEA), thioanisole, ethanedithiol and anisole were obtained from Aldrich Chemical Co. Inc. and trifluoroacetic acid (TFA) was obtained from Advanced ChemTech Inc. All solvents were of HPLC grade and all chemicals were of analytical (AnalaR) grade.
Automatic synthesis was performed by a batchwise method on a CS336X (C S Bio Inc.) synthesizer, employing Fmoc/t-butyl tactics consisting of TBTU/DIEA as a coupling reagent for 1 h and 2% piperidine, 2% 1,8-diazabicyclo[5.4.0]undec-7-ene (DBU) in DMF as a deprotection reagent for 7 min. At the end of the synthesis, the peptides were acetylated using 20% acetic anhydride in DMF for 30 min. Cleavage of the peptide from the resin support and concomitant deprotection of the amino-acid side chains was achieved in Reagent R (TFA:thioanisole:ethanedithiol:anisole at 90:5:3:2) using a Discover microwave solid-phase synthesis system (CEM). This was followed by removal of the exhausted resin using filtration and precipitation of the peptide product in cold (277 K) anhydrous ether. The precipitate was allowed to settle overnight at 253 K and was then washed three times with cold ether and dried under high vacuum.
Preparative reversed-phase high-performance liquid chromatography (RP-HPLC) was performed on a semi-preparative scale using a Waters Delta Prep 4000, a Waters 486 detector and a Vydac 259 VHP1522 polymer column (Grace) and a flow rate of 8 ml min−1. Crude peptide was loaded in formic acid and gradient-eluted (5–95% buffer B, 60 min) with 0.1% TFA/H2O (buffer A) and 0.1% TFA/acetonitrile (buffer B). The center cut from the preparative run was frozen in liquid nitrogen immediately upon collection and lyophilized under high vacuum.
The masses of the purified peptides were confirmed by ESI-MS using a Waters Micromass LCT Premier mass spectrometer. Lyophilized peptides were resuspended in 50 mM Tris–HCl pH 7.5, 1 mM DTT to a final concentration of 3 mg ml−1.
2.3. Crystallization and data collection
S100A10 was incubated with an equimolar amount of AnxA2 peptide in 50 mM Tris–HCl pH 7.5, 1 mM DTT for 1 h at room temperature. An equimolar amount of AHNAK was subsequently added (1:1:1 ratio of S100A10:AnxA2:AHNAK) and the mixture was incubated at room temperature for 1 h followed by overnight dialysis against 50 mM Tris–HCl pH 7.5, 1 mM DTT at 277 K. The complex was concentrated to 18 mg ml−1 (using the extinction coefficient of S100A10 for calculation) using a 10K molecular-weight cutoff centrifugal concentrator unit (Millipore).
Crystals were grown using the hanging-drop vapor-diffusion method by mixing 2 µl protein complex with 2 µl crystallization buffer [20%(w/v) PEG 8000, 100 mM Tris–HCl pH 8.5, 10%(v/v) 2-propanol] and incubating drops over a 750 µl volume of crystallization buffer at 290 K. Crystals appeared after about one week. Crystals were harvested and cooled in liquid nitrogen using 25% glycerol as a cryoprotectant. Data were collected at 100 K as 180 frames with 1° rotation each on beamline 8.2.1 at the Advanced Light Source (ALS; Berkeley, California, USA). Data were processed using HKL-2000 (Otwinowski & Minor, 1997 ▶; see Table 1 ▶ for crystallographic statistics). Our data belonged to the same space group (C2) with similar (<1.5 Å difference) unit-cell parameters as the previously reported S100A10–AnxA2 structure (PDB entry 1bt6; Réty et al., 1999 ▶); phasing was therefore achieved using REFMAC5 (Murshudov et al., 2011 ▶) with native S100A10–AnxA2 (Réty et al., 2000 ▶) as the model. Further refinement and model building were performed using PHENIX (Adams et al., 2010 ▶) and Coot (Emsley et al., 2010 ▶). The AHNAK peptide was built into the electron density by constructing a polyalanine chain and visually searching for amino-acid side chains with strong 2F o − F c electron density. Phe5 and Phe13 were initially modeled as reference residues with strong electron density, and the remaining residues were introduced based on the AHNAK peptide sequence. Additionally, the AnxA2 peptides were extended at the C-terminal end from those in the initial model owing to strong electron density. Three 2-propanol molecules were also modeled as the electron density was too large to support a water molecule.
Table 1. X-ray diffraction data-reduction and refinement statistics.
Values in parentheses are for the highest resolution shell.
| Data collection | |
| Beamline | 8.2.1, ALS |
| Wavelength (Å) | 0.979 |
| Space group | C2 |
| Unit-cell parameters (Å, °) | a = 79.07, b = 55.22, c = 63.14, α = γ = 90.0, β = 111.7 |
| Molecules in asymmetric unit | 1 |
| Resolution range (Å) | 50.00–2.50 |
| Mosaicity range (°) | 0.45–0.52 |
| Total observations | 31704 |
| Unique reflections | 8685 |
| Multiplicity | 3.7 (3.1) |
| Completeness (%) | 98.5 (96.9) |
| R merge (%) | 6.5 (42.0) |
| Average I/σ(I) | 22.38 (1.93) |
| Data-processing program | HKL-2000 |
| Refinement | |
| Refinement programs | REFMAC5, PHENIX |
| Resolution range (Å) | 29.33–2.51 |
| R factor (%) | 18.1 |
| R free † (%) | 25.1 |
| R.m.s.d. stereochemistry‡ | |
| Bond lengths (Å) | 0.009 |
| Bond angles (°) | 1.223 |
| Solvent content (%) | 46.4 |
| No. of atoms | |
| Protein | 1795 |
| Waters | 8 |
| 2-Propanol | 12 |
| Average B (Å2) | |
| Protein | 52.4 |
| Waters | 55.0 |
| 2-Propanol | 54.4 |
| Ramachandran plot§ (%) | |
| Preferred | 92.3 |
| Allowed | 7.7 |
| Outliers | 0.0 |
| PDB code | 4ftg |
R free based on a test-set size of 5% of all structure factors.
R.m.s.d. stereochemistry is the deviation from target values.
Ramachandran analysis was carried out using the program PROCHECK (Laskowski et al., 1993 ▶).
Images for figures were generated using VMD 1.9 (Humphrey et al., 1996 ▶) or PyMOL v.1.3r1 (Schrödinger). Buried surface area was calculated using the protein interfaces, surfaces and assemblies service PISA at the European Bioinformatics Institute (http://www.ebi.ac.uk/pdbe/prot_int/pistart.html; Krissinel & Henrick, 2007 ▶). Secondary-structure predictions were performed using YASPIN (Lin et al., 2005 ▶) and BCL::Jufo (http://www.meilerlab.org). Ramachandran analysis was carried out using PROCHECK (Laskowski et al., 1993 ▶). The structural model and crystallographic data have been deposited in the Protein Data Bank under PDB code 4ftg.
3. Results
3.1. Interaction of AHNAK peptide with (p11)2(AnxA2)2
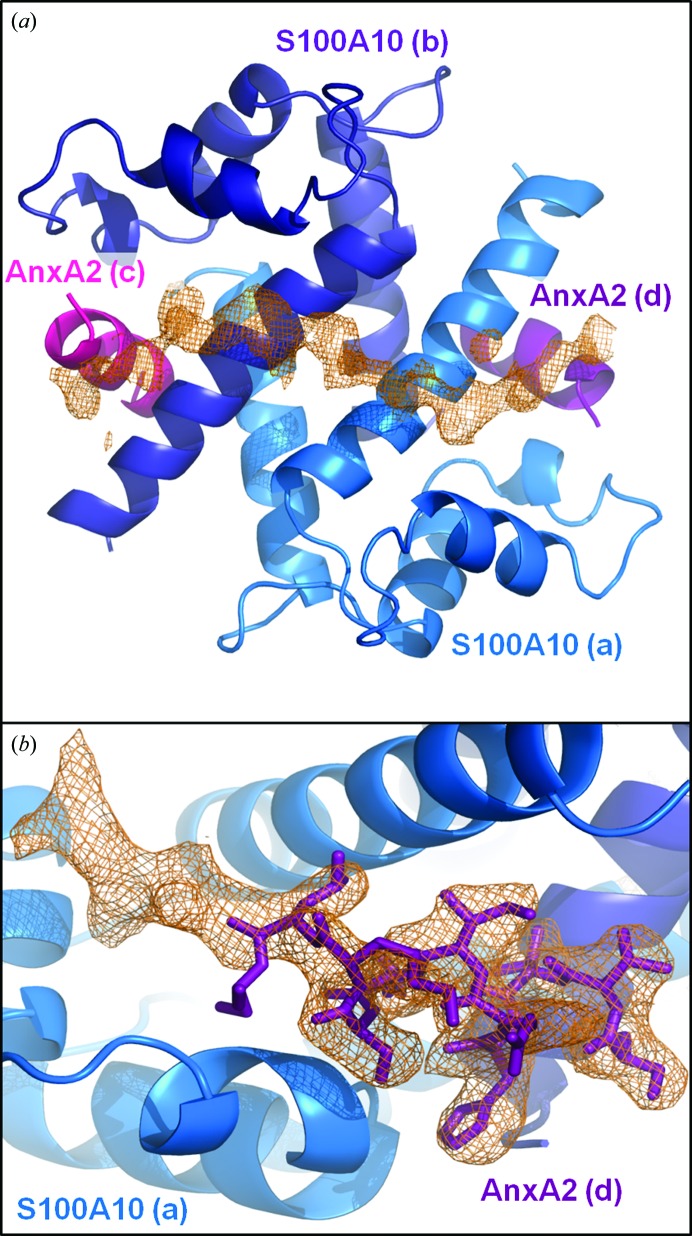
Crystals of the AHNAK–(p11)2(AnxA2)2 complex grew under conditions very similar to those used for the previously reported structure of the AnxA2–S100A10 complex (Réty et al., 1999 ▶) and diffracted to 2.5 Å resolution (Table 1 ▶). The space group (C2) is identical and the unit-cell parameters deviate by less than 1.5 Å between our structure and the previously reported AnxA2–S100A10 complex (PDB entry 1bt6). As a result, phasing was achieved using the structure of Réty and coworkers as an initial model, and both 2F o − F c and F o − F c maps revealed a long continuous chain of unassigned density spanning from the C-terminal region of one AnxA2 peptide (chain D), across the S100A10 dimer interface (chains A and B) to the C-terminal end of the second AnxA2 peptide (chain C) (Fig. 1 ▶ a). Although the crystallization drops contained an excess of AHNAK peptide (2:1 ratio of AHNAK peptide to AnxA2–S100A10 heterotetramer), the density only supports the binding of one AHNAK peptide to the heterotetramer, validating the results of Rezvanpour and coworkers that the complex is formed at a 1:2:2 ratio of AHNAK:S100A10:AnxA2. Furthermore, additional density is visible for the C-terminal residues of AnxA2 when compared with the structure of Rety and coworkers, and a change in orientation is apparent beginning with Ser12 on both AnxA2 peptides (chains C and D; Fig. 1 ▶ b). The lack of tight crystal packing near this interacting surface allowed the accommodation of the 20-amino-acid peptide (the distance between symmetry atoms is about 20 Å), explaining the nearly identical unit cells and space groups of the apo and AHNAK-bound (p11)2(AnxA2)2 structures.
Figure 1.
Electron density of AHNAK peptide and extended AnxA2 peptide. (a) A continuous chain of unassigned electron density is clearly visible in the F o − F c map after initial refinement using the crystal structure of the apo S100A10–AnxA2 tetramer as a starting model (PDB entry 1bt6). S100A10 (marine and dark blue) and AnxA2 (magenta and purple) are shown as cartoon representations. The unassigned electron density stretches from one AnxA2 peptide, over the dimer interface of S100A10, to the second AnxA2 peptide. The map is contoured at 2σ. (b) Electron density seen in the 2F o − F c map after initial refinement supports an extension of the AnxA2 at the C-terminal end and a change of direction beginning with Ser12. Monomer A of S100A10 is depicted as a cartoon (marine) and the AnxA2 peptide (chain D) is shown as sticks (purple). The map is contoured at 1σ. Lower-case letters represent chain IDs.
Binding of the AHNAK peptide to the S100A10–AnxA2 heterotetramer does not cause a significant conformational change of the S100A10 dimer. The Cα root-mean-square deviation (r.m.s.d.) between S100A10 monomer A of apo and AHNAK-bound (p11)2(AnxA2)2 is 0.40 Å, and for monomer B this value increases only slightly to 0.53 Å. When a specific chain is not used to anchor the superposition, the r.m.s.d. between apo and AHNAK-bound (p11)2(AnxA2)2 is 0.60 Å. While both AnxA2 peptides adopt a canonical n + 4 α-helix for the first eight residues, the AHNAK peptide contains no specific secondary-structure elements. Several interactions between the AHNAK peptide and both the S100A10 dimer and AnxA2 peptides are evident and will be described in detail.
The crystal structure of AHNAK in complex with the AnxA2–S100A10 heterotetramer reveals a platform that is asymmetric (apparently induced by AHNAK binding) and a binding surface composed of many residues described by NMR studies as having the largest chemical shifts upon binding of the AHNAK peptide (Rezvanpour et al., 2011 ▶; Fig. 2 ▶). Electron density is visible for the first 16 residues of the 20-amino-acid AHNAK peptide (corresponding to residues 5654–5673; UniProt Q09666). Interactions between the AHNAK peptide and the (p11)2(AnxA2)2 heterotetramer mainly involve hydrogen bonding, particularly between backbone atoms, and hydrophobic interactions of side chains with grooves and pockets on the (p11)2(AnxA2)2 surface (Fig. 2 ▶, lower insets). Several hydrogen bonds occur between the backbone atoms of AHNAK and either S100A10 or annexin A2 backbone atoms, supporting the observation that interaction between the large AHNAK protein and the (p11)2(AnxA2)2 scaffolding complex is not limited to a specific sequence but a more general consensus sequence that has previously been shown to occur in multiple regions of the 1002-amino-acid C-terminal domain of AHNAK.
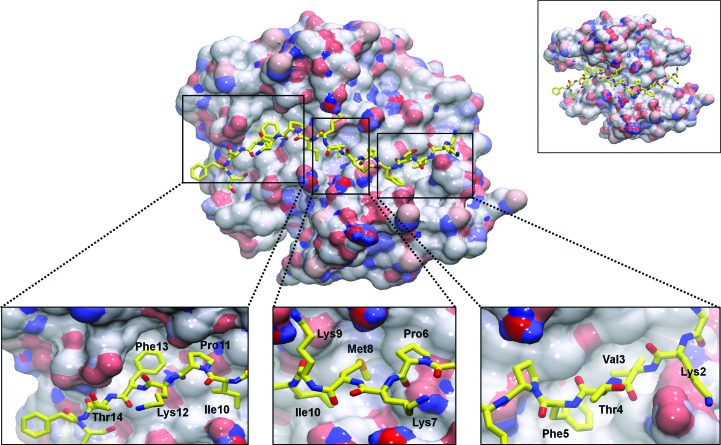
Figure 2.
AHNAK binds to various pockets on the surface of the S100A10–AnxA2 tetramer. The binding of AnxA2 to S100A10 increases the surface area of the largely hydrophobic surface formed across the dimeric interface of helix IV of S100A10 chains A and B by 363 Å2, allowing more interactions with AHNAK (yellow sticks). Removal of AnxA2 leaves the N- and C-terminal portions of AHNAK without a binding surface (upper right inset). Several hydrophobic residues of AHNAK insert themselves into hydrophobic pockets on the surface of S100A10–AnxA2, while the side chains of polar and charged residues mainly point towards the solvent (lower insets). The surface of S100A10–AnxA2 is colored by electrostatic potential, with red representing electronegative regions and blue representing electropositive regions.
It has previously been established that the affinity of AHNAK for S100A10 is much greater when AnxA2 is bound (De Seranno et al., 2006 ▶). Indeed, the crystal structure reveals direct interactions between the AnxA2 and AHNAK peptides. The backbone carbonyl of Gly1 (AHNAK) accepts a hydrogen bond from the backbone amine of Ser12 of AnxA2 (chain D; distance of 3.28 Å). The interaction is further strengthened by the backbone amide of Val3 (AHNAK), which donates a hydrogen bond to the backbone carbonyl of Ser12 of AnxA2 (chain D; 2.90 Å; Fig. 3 ▶ a). Weak electron density also suggests that a water molecule may act as a bridge between the carbonyl of Lys2 (AHNAK) and the side-chain carboxyl of Asp84 (S100A10 chain A), with an average distance of 3 Å between the center of this weak density and the neighboring atoms. However, a molecule of 2-propanol is observed very close to this region in our model, making the inclusion of a water molecule unfavorable as the O atom of water would be in very close contact (∼1.6 Å) with a methyl group of 2-propanol. This is likely to be an artifact from the use of 10%(v/v) 2-propanol as an additive in crystallization and may be excluding water molecules that would normally localize in the region in vivo. Furthermore, Asp84 (S100A10) and Ser12 (AnxA2) did show large differences in chemical shift upon AHNAK binding in the NMR experiments of Rezvanpour and coworkers.
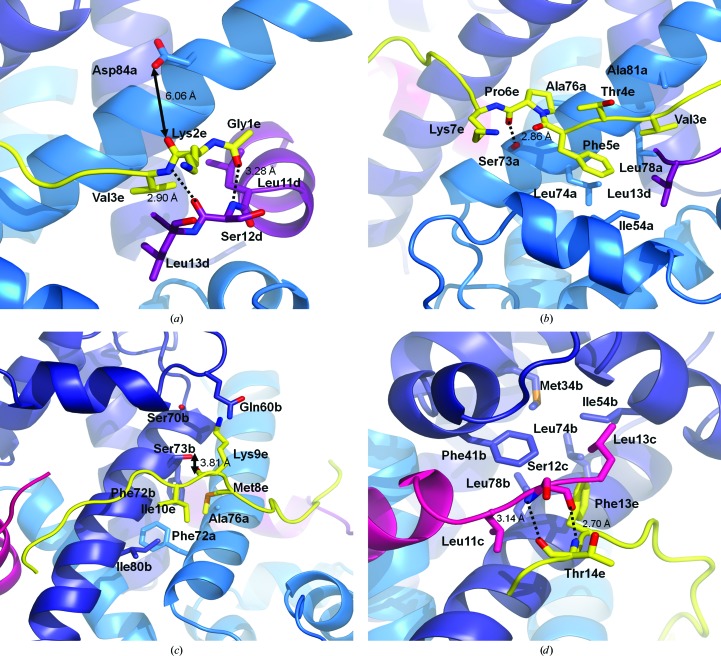
Figure 3.
AHNAK binds to S100A10–AnxA2 through a series of hydrogen bonds and hydrophobic interactions. (a) At the N-terminal end of the AHNAK peptide (yellow), the backbones of Gly1 and Val3 form hydrogen bonds to the main-chain atoms of Ser12 located at the C-terminal end of the AnxA2 peptide (chain D; purple). Hydrogen bonds are represented as dashed lines, while distances are shown for solid arrows. Weak electron density halfway between Asp84 of S100A10 (marine) and Lys2 of AHNAK suggests that a water molecule may act as a bridge between the two residues, but the model contains a nearby 2-propanol molecule whose methyl group would clash with the water O atom if modeled into the density (not shown). (b) A series of hydrophobic interactions are formed between Val3 and Phe5 of AHNAK (yellow), Leu13 of AnxA2 (chain D; purple) and Ile54, Leu74, Leu78 and Ala81 of S100A10 (chain A; marine). The main-chain carbonyl of Pro6 (AHNAK) accepts a hydrogen bond from the main-chain amide of Ala76 (S100A10, chain A). (c) The middle region of the peptide contains more hydrophobic residues which interact with hydrophobic pockets on S100A10. Met8 and Ile10 of AHNAK (yellow) face several hydrophobic residues near the S100A10 dimer interface (chain A, marine; chain B, dark blue). Additionally, the side-chain hydroxyl of Ser73 (S100A10, chain B) donates a hydrogen bond to the main-chain carbonyl of Lys9 (AHNAK). (d) The main chain of Thr14 (AHNAK; yellow) accepts and donates a hydrogen bond from Ser12 of AnxA2 (chain C; magenta). Additionally, the region contains hydrophobic interactions between Phe13 of AHNAK and several hydrophobic residues of S100A10 (chain B; dark blue) and Leu13 of AnxA2 (chain C). Lower-case letters represent chain IDs.
These hydrogen-bonding interactions are followed by a series of nonpolar residues embedded into a hydrophobic patch on the surface of (p11)2(AnxA2)2 (Fig. 3 ▶ b). The side chain of Val3 (AHNAK) faces a pocket containing Leu78 and Ala81 of S100A10 (chain A) and Leu13 of AnxA2 (chain D). While Thr4 of AHNAK is rotated towards the solvent, Phe5 points towards the hydrophobic side chains of Ile54, Leu74, and Leu78 of S100A10 (chain A) and Leu13 of AnxA2 (chain D). Finally, the side chain of Pro6 of AHNAK is in proximity to Ala76 of S100A10 (chain A). The backbone carbonyl of this proline also accepts a hydrogen bond from the side-chain hydroxyl of Ser73 of S100A10 (chain A; 2.86 Å), a residue that also showed a large chemical shift in NMR studies (Rezvanpour et al., 2011 ▶).
Met8 and Ile10 of AHNAK are buried in a hydrophobic region near the S100A10 dimer interface formed by Ala76 and Phe72 of S100A10 chain A and Phe72 and Ile80 of chain B (Fig. 3 ▶ c). Although electron density is weak for the side chain of Lys9 of AHNAK, it points into a region surrounded by Ser70, Ser73 and Gln60 of S100A10 (chain B), suggesting that a polar interaction is possible (both Ser73 and Gln60 show chemical shifts in NMR). The perturbation of Ser73 may be a result of the hydroxyl group donating a hydrogen bond to the backbone carbonyl of Lys9 (AHNAK), although the rotamer of Ser73 best supported by density has its hydroxyl group pointing away from the carbonyl.
Density is strong for the entire side chain of Phe13 of AHNAK and it is packed against a surface composed of several hydrophobic residues from chain B of S100A10 (Met34, Phe41, Ile54, Leu74 and Leu78), along with Leu11 and Leu13 of AnxA2 chain C (Fig. 3 ▶ d). AnxA2 is of further importance, as the backbone amide of Thr14 (AHNAK) donates a hydrogen bond to the main-chain carbonyl of Ser12 of AnxA2 (chain C; 2.70 Å), while the backbone amide of Ser12 donates a hydrogen bond to the main-chain carbonyl of Thr14 (AHNAK; 3.14 Å). Electron density is absent beyond Ser16 of AHNAK, suggesting that the stabilizing interactions are confined to the series of hydrogen bonds and hydrophobic patches observed in the first 16 residues. The buried surface area between the AHNAK peptide and (p11)2(AnxA2)2, calculated using PDBePISA (Krissinel & Henrick, 2007 ▶), is 1238 Å2, of which 29% (363 Å2) is contributed by the AnxA2 peptide.
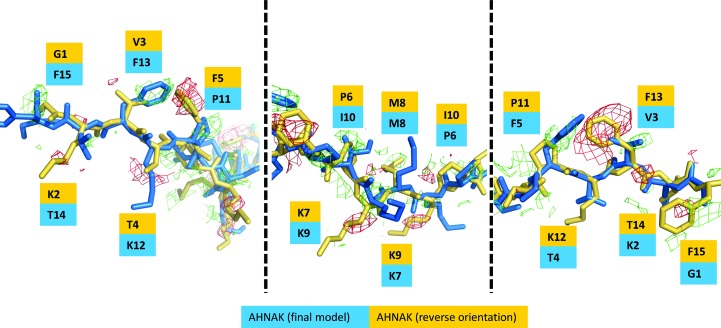
The observation that the AHNAK peptide binds to (p11)2(AnxA2)2 in only one direction led us to examine which factors contribute to this asymmetry. The superimposition of S100A10 chain B onto chain A in our model resulted in a Cα backbone r.m.s.d. value of 0.47 Å, yet significant structural changes between the two monomers were not apparent. Examination of the two monomers alone did not suggest key conformational changes that would favor one AHNAK peptide orientation over the other. The reverse orientation of the peptide was then modeled, followed by another step of refinement. F o − F c difference maps revealed missing side chains, particularly for the phenylalanine residues that have strong electron density in our model (Fig. 4 ▶). Additionally, the map revealed negative density for phenylalanine-residue side chains in the ‘reverse’ model. While the map does not support this alternate conformation, there is little evidence that the alternate conformation of the AHNAK peptide would result in steric clashes with either the S100A10 dimer or the AnxA2 peptides.
Figure 4.
Comparison of opposite orientations of the AHNAK peptide when bound to (p11)2(AnxA2)2. The orientation supported by electron density and part of the final model is represented in blue, while the hypothetical reverse orientation is colored yellow. An F o − F c difference map after refinement with the reverse orientation is shown in green (contoured at +2.5σ) and red (contoured at −2.5σ).
During sample preparation, peptides were incubated with the S100A10 dimer for several hours before crystallization attempts. It is conceivable and likely that the AHNAK peptide binds to the S100A10–AnxA2 heterotetramer in either of two conformations. Electron-density maps support the presence of only one conformation in the crystal structure, suggesting that crystal-packing effects may favor one conformation over the other and not structural differences on the (p11)2(AnxA2)2 platform. An examination of the symmetry elements does not provide a clear verdict as to how crystal packing contributes to one conformation being favored over the other, but it may be that the disordered residues 17–20 of the AHNAK peptide that are not observed in the crystal structure are responsible and more space is available for the full peptide in one orientation compared with the other.
While this manuscript was in revision, a 3.5 Å resolution crystal structure with density for only ten residues of the AHNAK peptide appeared (Dempsey et al., 2012 ▶). The coordinates had not been released at the time of writing, so a direct comparison was not available. However, the 3.5 Å resolution crystal structure contained a fusion protein in which a mutated S100A10 was covalently attached to the AnxA2 peptide via a linker, while this 2.5 Å resolution crystal structure does not have any artificial constraints that may affect complex formation. In our model, Met8 of the AHNAK peptide is centrally located at the S100A10 dimer interface, while Dempsey and coworkers place Ile10 and Pro11 in this region. Overall, the positioning of the peptide in the 3.5 Å resolution structure appears to be offset by about three residues towards the N-terminus when compared with our 2.5 Å resolution structure.
3.2. Analysis of the consensus sequence
Rezvanpour and coworkers performed a peptide-array analysis to deduce a consensus sequence based on the regions of the AHNAK C-terminal domain that bound to (p11)2(AnxA2)2 with high affinity. These sequences are presented in Fig. 5 ▶, truncated to correspond to the 20-amino-acid peptide used in this study (sequence e). Although these eight peptides contain the calculated consensus sequence, they showed different binding affinities, with the best being sequences b and e (colored green) and the weakest being sequences a, d and g (colored red).
Figure 5.

Comparison of AHNAK C-terminal peptides that display a range of binding affinities to the S100A10–AnxA2 tetramer. The 20-mer AHNAK peptide used in the cocrystallization experiment is highlighted in yellow. Phe5 and Phe13, both of which have strong electron density for their side chains, are shown in bold along with corresponding residues in the other peptides. Sequences colored in green show the tightest binding in previous studies, while those in red showed weak binding (Rezvanpour et al., 2011 ▶). The original consensus sequence (1) is listed below the peptide sequences and a revised consensus sequence (2; outlined) based on the crystal structure is also included. ϕ, hydrophobic; +, positively charged; X, variable; Z, charged/polar.
To examine whether there is a structural relationship between peptides that show high affinity for (p11)2(AnxA2)2 and those that show weaker binding, sequence a was compared with the peptide used in this study (sequence e). Sequence a contains several differences from e, but the first two (G1A and K2P) are likely to be nonconsequential, as there is room for a methyl group of alanine and the side chain of lysine points towards the solvent and does not interact with (p11)2(AnxA2)2, although the conformational rigidity of proline would be likely to distort the backbone conformation. Additionally, the substitution of a valine for aspartate at position 3 would result in unfavorable electrostatics, as the negatively charged side chain would point into a groove composed of hydrophobic residues. Thr4 of the AHNAK peptide, which has a solvent-accessible side chain, is replaced by a hydrophobic valine residue in sequence a. Additionally, the substitution of Phe5 by an aspartate would also place a charged residue in a mainly hydrophobic groove on S100A10. The remaining sequence of peptide a features more conservative substitutions that are likely to allow binding in the consensus region, but a lack of favorable interactions on the N-terminal side of this sequence is likely to explain why this peptide showed weaker binding. A similar issue occurs with sequence d, in which a Val-to-Ala substitution at position 3 results in a shorter side chain that does not fit as deeply into the hydrophobic groove on S100A10, while key hydrophobic interactions are lost by the mutation of Phe5 and Ile10 to glycine residues. An opposite effect is observed in sequence g, in which a hydrophobic residue mutation, V3P, results in a steric clash, as the side chain of the phenylalanine does not fit into the pocket occupied by a valine in sequence e. A Pro6-to-Phe substitution would be predicted to lead to further steric clashes, and a hydrophobic interaction is lost with the substitution of Met8 by serine. Favorable interactions are diminished further by the substitution of the hydrophobic Ile10 by the charged Lys10 in a very hydrophobic groove and similarly by the mutation of Phe13 to His.
Sequence b, which also showed a high binding affinity, does not have any major mutations in the first five residues that would affect binding. In fact, the consensus sequence is kept mainly intact, but it is worthwhile noting that Ile10 is mutated to Lys, as is also observed in sequence g, which shows low binding affinity. The introduction of Lys is not likely to cause a steric clash as this pocket is deep and the consensus sequence does list this position as variable. However, the residues that make up this pocket are mainly hydrophobic and this may result in peptide e being more optimal for binding to the (p11)2(AnxA2)2 portion of the AHNAK binding site than peptide b.
The crystal structure also allows a closer analysis of the computer-generated consensus sequence based on previous peptide-array experiments (Rezvanpour et al., 2011 ▶; Rezvanpour & Shaw, 2009 ▶). Of particular concern is that strong electron density and multiple interactions with (p11)2(AnxA2)2 are observed for the first five residues of the AHNAK peptide in the crystal structure, yet the consensus sequence begins at position 6. The strong electron density of the Phe5 side chain resulting from a stabilizing effect of hydrophobic residues from S100A10 and AnxA2 surrounding Phe5 suggests that this position is of significant importance for the binding affinity. A hydrophobic residue, preferably one with a relatively bulky side chain, would contribute to strong interactions between the peptide and the protein complex. A similar argument could be made for the third position, occupied by Val3 in this structure, owing to its interaction with a hydrophobic pocket. While the first two residues do contribute hydrogen bonds, these are through backbone atoms, and specific side chains are not required for interaction. Despite the variability, an argument can be made that having residues at those positions, as long as they do not cause a steric clash with neighboring atoms, is still important because it is these residues that interact with AnxA2, which, as described previously, increases the affinity of AHNAK for S100A10. Since positions 2 and 4 are solvent-exposed in this model, they are probably less important and should be occupied by polar or charged residues. In summary, the revised consensus sequence for the first five positions, which did not receive assignments in the original consensus sequence, is XZϕZϕ (where X represents variable, Z represents polar/charged and ϕ represents hydrophobic residues).
Continuing to the original consensus sequence, a hydrophobic residue is expected at position 6 and this is fulfilled by Pro6 in this structure, and the positively charged residue expected at position 7 is Lys7, the side chain of which may be able to form a salt bridge to Asp57 of S100A10 (chain A), although this is not visible in the crystal structure as very little density is observed for the side chain of Lys7. However, the crystal structure reveals that position 8, which is listed as variable in the original consensus sequence, should be changed to a hydrophobic residue (Met8 in this structure) as its side chain is inserted into a largely hydrophobic pocket. A positively charged residue at the next position is supported by our structure, and Ile10, which is listed as variable in the consensus sequence, could potentially be replaced by other residues as its side chain is not as deeply placed into the hydrophobic surface of S100A10 as Met8. Phe13, which is listed as a hydrophobic residue in the consensus sequence, is well defined in the electron-density maps, supporting that this position is well suited for a hydrophobic residue; considering that sequence b, which also shows high binding affinity, contains a phenylalanine at this position, it appears justified to adjust the consensus sequence to reflect this specificity. The side chain of Thr14 is accessible to the solvent without clear interactions. Finally, the consensus sequence suggests a hydrophobic residue at position 15 (Phe15 in this structure), but electron density is not observed for this side chain nor is there justifiable evidence of an interaction with any hydrophobic residues at the interface. Therefore, based on our structure, the consensus sequence should be adjusted to include positions 1–14 as follows: XZϕZϕϕ+ϕ+XPKFZ (where X represents variable, Z represents polar/charged, ϕ represents hydrophobic and + represents positively charged residues; Fig. 5 ▶).
3.3. Comparison with other S100 structures
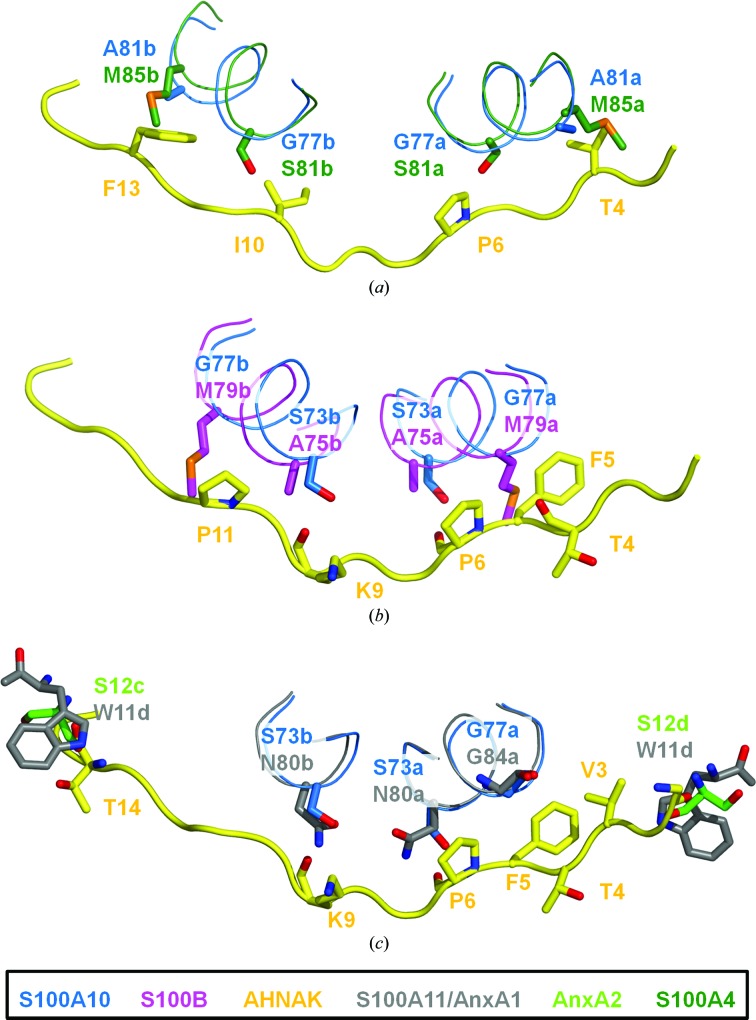
Several structures of S100 proteins in complexes with their respective ligands have been published, including S100B in complex with a CapZ peptide (TRTK12; Charpentier et al., 2010 ▶), S100A11 in complex with the annexin A1 N-terminus (Réty et al., 2000 ▶) and S100A4 in complex with myosin IIA (Kiss et al., 2012 ▶). While these complexes generally have a quaternary structure similar to that of (p11)2(AnxA2)2, namely a peptide bound in each hydrophobic pocket of the activated S100 dimer (1:1 ratio of S100 protein:ligand), the structure of S100A4 in complex with a myosin IIA peptide was the first to reveal a asymmetric binding motif in which one peptide binds to the S100A4 dimer (1:2). This structure shares similarities with the AHNAK–(p11)2(AnxA2)2 structure insofar as the peptide binds the S100 dimer in a similar region. However, while the hydrophobic pockets of AHNAK–(p11)2(AnxA2)2 are filled with AnxA2 N-terminal peptides, the myosin IIA peptide wraps itself into both pockets on the N- and C-terminal ends. Additionally, several sequence differences between S100A10 and S100A4 would cause steric clashes with the AHNAK peptide. For example, Ala81 of S100A10 is mutated to Met85 in S100A4, resulting in a steric clash with Val3 of AHNAK (Fig. 6 ▶ a). The myosin IIA peptide has a smaller alanine residue near this region, allowing a better fit. Gly77 of S100A10 is replaced by Cys81 in S100A4 (mutated to serine in the crystal structure with myosin IIA; PDB entry 3zwh; Kiss et al., 2012 ▶) and the sulfhydryl of this residue would clash (distance 1.94 Å) with the CD atom of Pro6 (AHNAK). Both S100A10 and S100A4 share a similar cleft in which the side chain of Met8 (AHNAK) is buried. Indeed, when the two structures are superimposed the myosin IIA peptide also contains a methionine about 2 Å away (Cα—Cα distance) from Met8 of AHNAK. The weak hydrogen bond observed between the main-chain carbonyl of Lys9 (AHNAK) and the side-chain hydroxyl of Ser73 (S100A10 chain B) would not be possible in S100A4, as Ser73 is a valine (Val77). Additionally, the change of Gly77 to Cys81, which would cause a clash with chain A of S100A4, presents a similar mismatch in chain B, as the CG2 atom of Ile10 (AHNAK) is within 1.59 Å of the side-chain sulfhydryl of Cys73. Phe13 of AHNAK, which fits tightly into a pocket surrounded by hydrophobic residues, now has a major clash with Met85 of S100A4 chain B (Ala81 in S100A10). Without the presence of AnxA2, key hydrogen bonds are lost at both the N- and C-terminal ends of the AHNAK peptide. It is evident that this consensus sequence of the AHNAK C-terminal domain has specificity for the S100A10–AnxA2 heterotetramer. While several AHNAK residues could potentially bind to S100A4, the lack of AnxA2 would prevent the formation of stabilizing hydrogen bonds. AnxA2 is a known binding partner of S100A4 on the extracellular surface of endothelial cells, but even if the complex were accessible to AHNAK, differences between S100A10 and S100A4, some of which cause steric hindrance in both chains, would prevent the AHNAK peptide from binding with high affinity, explaining its specificity for S100A10.
Figure 6.
Comparison of the AHNAK–S100A10–AnxA2 binding interface with other S100 proteins. (a) Superposition of S100A4–myosin IIA (PDB entry 3zwh) onto AHNAK–(p11)2(AnxA2)2 (r.m.s.d. 1.48 Å). Key differences between S100A10 (blue) and S100A4 (dark green) are thought to create steric clashes with the AHNAK peptide (yellow). Ala81 is substituted by Met85 in S100A4, resulting in a clash with Thr4 of AHNAK on chain A of S100A4 and with Phe13 of AHNAK on chain B. Additionally, the substitution of Gly77 by Ser81 in S100A4 (Cys81 in the native sequence) is predicted to result in a steric clash on both chains as the hydroxyl groups of serine comes into close contact with Pro6 and Ile10 (AHNAK). (b) Superposition of S100B–TRTK12 (PDB entry 3rm1) and AHNAK–(p11)2(AnxA2)2 (r.m.s.d. 1.49 Å). Although S100B is a known binding partner of AHNAK, the peptide which shows high affinity for S100A10–AnxA2 is not likely to be the same sequence that binds to the S100B dimer. Two mutations, Gly77 to Met79 and Ser73 to Ala75, cause steric clashes and a loss of hydrogen bonding between both chains of S100B (magenta) and the AHNAK peptide (yellow). (c) Superposition of S100A11–AnxA1 (PDB entry 1qls) onto AHNAK–(p11)2(AnxA2)2 (r.m.s.d. 1.19 Å). The AHNAK binding surface on S100A10–AnxA2 shows high similarity to this surface on S100A11–AnxA1 (gray). The hydrogen bond between Gly1 of AHNAK (yellow) and Ser12 of AnxA2 (lime green) could be supplemented by a hydrogen bond to Trp11 of AnxA1 instead. Unlike S100A4 and S100B, which do not conserve Gly77 of S100A10, S100A11 retains this residue (Gly84) and no steric clashes are predicted to occur. The only notable change is Ser73 to Asn80, which disrupts hydrogen bonds between both chains of S100A11 and AHNAK. However, the side-chain amide of Asn80 could play the role of the side-chain hydroxyl of Ser73 and donate hydrogen bonds to main-chain atoms on AHNAK. Lower-case letters represent chain IDs.
There are no published data to support binding of AHNAK to S100A4, and this is not unexpected after a structural comparison between S100A4 and S100A10 revealed key differences. However, it has been reported that AHNAK binds to S100B (Gentil et al., 2001 ▶). A similar analysis was performed using the S100B structure to determine whether the same consensus sequence recognizes both S100A10 and S100B. As established previously, AnxA2 is necessary for the high-affinity binding of AHNAK to S100A10. However, S100B is not a reported binding partner of AnxA2. Therefore, the key hydrogen bonds adding to the stability of the complex made possible by the presence of AnxA2 should not exist. S100B does bind the CapZ peptide (TRTK12) and a crystal structure of the complex is available (PDB entry 3iqq; Charpentier et al., 2010 ▶). Superimposition of the S100B–CapZ structure onto AHNAK–(p11)2(AnxA2)2 suggests that the hydrogen bond between the carbonyl of Gly1 (AHNAK) and the amide of Ser12 (AnxA2 chain D) could potentially occur with the amide of Asp6 (TRTK12) instead, as the backbone atoms of Ser12 (AnxA2) and Asp6 (TRTK12) are aligned. However, the remaining C-terminus of the TRTK12 peptide clashes heavily with the AHNAK peptide, most noticeably with the backbone atoms. As a result, one must consider only the S100B dimer, and without the added stability of AnxA2 the binding of this particular AHNAK peptide would be weakened. Gly77 of S100A10 is replaced by Met79 of S100B, resulting in a major steric clash between chain A of S100B and the side chain of Phe5 of AHNAK (Fig. 6 ▶ b). Additionally, the backbone carbonyl of Thr4 (AHNAK) is within 1.56 Å of the CG atom of Met79, resulting in loss of hydrophobic interactions and proper alignment of AHNAK within the grooves of the S100B pocket. Ser73 of S100A10 is Ala75 in S100B, preventing a hydrogen-bond interaction between the main-chain carbonyl of Pro6 (AHNAK) and the side chain of the serine residue. Met8 of AHNAK is packed into a pocket formed by the dimer interface, and this is similar to S100A10 and S100A4. The Ser73 to Ala75 change removes another hydrogen bond to chain B of S100B, this time to the backbone carbonyl of Lys9 (AHNAK). Also in chain B, the change of Ala77 to Met79 causes a major steric clash between the side chain of methionine and the ring of Pro11 (AHNAK). It is therefore unlikely that this same sequence of AHNAK binds to S100B dimers. The consensus sequence has clear specificity for S100A10, and the binding of S100B must occur elsewhere on the AHNAK protein.
S100A11 in complex with a peptide from the annexin A1 (AnxA1) N-terminus is the most closely related published structure (PDB entry 1qls; Réty et al., 2000 ▶) to the S100A10–AnxA2 heterotetramer. The S100A11–AnxA1 complex was superimposed onto the AHNAK–(p11)2(AnxA2)2 structure. While the C-terminal region of the AnxA1 peptide does not superimpose well with the AnxA2 peptide and is at a further distance away from the AHNAK peptide, a weak hydrogen bond between the carbonyl of Gly1 (AHNAK) and the backbone amide of Trp11 (AnxA1 chain D; distance 3.49 Å) is possible. Gly77 of S100A10 remains a glycine (Gly84) in S100A11 and no steric clashes are introduced that could affect the region of Val3, Thr4 or Phe5 of AHNAK (Fig. 6 ▶ c). Ser73 is mutated to Asn80 in S100A11, disrupting the hydrogen bond between the side-chain hydroxyl of serine and the main-chain carbonyl of Pro6 on AHNAK. However, it is feasible that the side-chain amide of Asn80 donates a hydrogen bond to this carbonyl (distance of 3.60 Å). Met8 of AHNAK once again fits into a groove formed by hydrophobic residues, although changes result in less bulky hydrophobic side chains in S100A11, namely the changes of Phe72 to Leu79 and of Ala76 to Gly83 at the symmetrical dimer interface. Within chain B, a Ser73-to-Asn80 mutation also affects the hydrogen bond to the main-chain carbonyl of Lys9, although it could be substituted by the side-chain amide of Asn80 acting as a donor to the carbonyl. The hydrogen bonds between the backbone atoms of Ser12 of AnxA2 chain C and Thr14 of AHNAK may be affected by the large indole ring of Trp11 of AnxA1, as it pushes the two peptides apart. No major clashes between the AHNAK peptide and the S100A11–AnxA1 heterotetramer are obvious, so it is feasible that this interaction occurs. However, interactions will be governed by cellular compartmentalization, localization and expression levels.
4. Discussion
The crystal structure of an AHNAK peptide in complex with the S100A10–AnxA2 tetramer gives a clearer understanding of the consensus sequence and provides a structural explanation as to why several regions of the AHNAK C-terminal domain show varying levels of binding affinity to (p11)2(AnxA2)2. The binding of AHNAK to the surface of (p11)2(AnxA2)2 is governed by several hydrophobic interactions between side chains of AHNAK and pockets on S100A10. A certain amount of substitution by other hydrophobic residues is tolerated, allowing the consensus sequence to be more general. Additionally, the various hydrogen bonds formed between the AHNAK peptide and (p11)2(AnxA2)2 most often involve backbone atoms of AHNAK; as a result, the side chains, particularly those that point away from S100A10–AnxA2 towards the solvent, are largely interchangeable.
Binding of the AHNAK peptide leads to a break in twofold symmetry between the S100A10 protomers, yet the disruption is not pronounced near the dimer interface. In fact, Met8 of AHNAK is at this interface and the flanking residues (Lys7 and Lys9) both have side chains near Gln60 of the nearest S100A10 chain. The symmetry breakage occurs most in the region of residues Ser73–Thr79 of S100A10, as the interacting residues of AHNAK near these regions are Pro6 and Ile10. This results in a slight conformational change between the two monomers of S100A10, in which helix IV is moved closer to helix III in chain A relative to chain B by an average distance of 0.26 Å between helix IV Cα atoms of Ser73–Thr79 with respect to a fixed position (Leu58 Cα) on helix III. This is further affected by the N-terminal portion of AHNAK, with the side chains of Val3 and Phe5 projecting into this region on chain A, while the C-terminal region produces less steric constraint by introducing only one hydrophobic side chain (Phe13) into the corresponding region on chain B of S100A10.
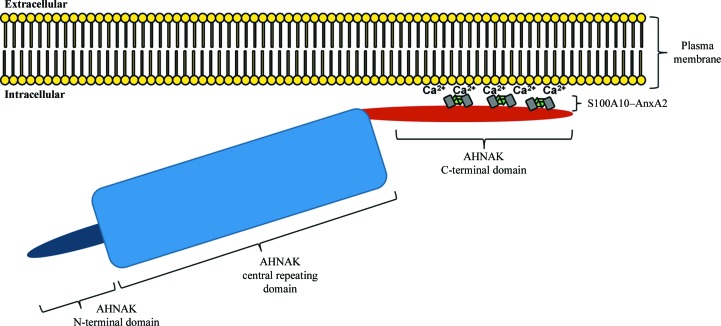
It is evident that (p11)2(AnxA2)2 can bind to various regions of the AHNAK C-terminal domain, as long as the consensus sequence is conserved, but the affinity will vary depending on amino-acid optimization in both the consensus sequence and the regions flanking it, particularly on the N-terminal side. Considering the massive size of AHNAK in relation to (p11)2(AnxA2)2, we propose a model in which multiple (p11)2(AnxA2)2 complexes bind to the C-terminal domain of AHNAK, anchoring it to the plasma membrane for roles in membrane repair (Fig. 7 ▶). While this consensus sequence allows interactions along various stretches of the AHNAK C-terminal domain, comparison with other S100 structures reveals that the sequence has been optimized for binding to S100A10 and several clashes would make binding of this specific AHNAK peptide to S100B and S100A4 unfavorable. There is strong conservation between the binding surfaces of S100A10–AnxA2 and S100A11–AnxA1, making the latter a potential AHNAK interaction partner. The possibility of this interaction should be investigated further. Our model adds new insight to our understanding of the specific interactions that occur in this membrane-repair scaffold.
Figure 7.
Proposed schematic of AHNAK, S100A10 and AnxA2 co-localization near the intracellular surface of the plasma membrane. Multiple (p11)2(AnxA2)2 complexes may bind to the C-terminal domain of AHNAK. AnxA2 binds to acidic phospholipids on the plasma membrane in a Ca2+-dependent manner, pulling AHNAK near to the plasma membrane.
Supplementary Material
PDB reference: AHNAK peptide bound to AnxA2–S100A10 heterotetramer, 4ftg
Acknowledgments
This work was supported by NIH grant R01AI78000 and the UC Irvine Center for Biomembrane Systems (HL). GO was supported by training grant T15LM07443 from the National Library of Medicine. The Advanced Light Source is supported by the Director of the US Department of Energy (Office of Science, Office of Basic Energy Sciences) under Contract DE-AC02-05CH11231.
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Ayala-Sanmartin, J., Henry, J.-P. & Pradel, L.-A. (2001). Biochim. Biophys. Acta, 1510, 18–28. [DOI] [PubMed]
- Becker, T., Weber, K. & Johnsson, N. (1990). EMBO J. 9, 4207–4213. [DOI] [PMC free article] [PubMed]
- Benaud, C., Gentil, B. J., Assard, N., Court, M., Garin, J., Delphin, C. & Baudier, J. (2004). J. Cell Biol. 164, 133–144. [DOI] [PMC free article] [PubMed]
- Bischofberger, M., Gonzalez, M. R. & van der Goot, F. G. (2009). Curr. Opin. Cell Biol. 21, 589–595. [DOI] [PubMed]
- Blackwood, R. A. & Ernst, J. D. (1990). Biochem. J. 266, 195–200. [DOI] [PMC free article] [PubMed]
- Borgonovo, B., Cocucci, E., Racchetti, G., Podini, P., Bachi, A. & Meldolesi, J. (2002). Nature Cell Biol. 4, 955–962. [DOI] [PubMed]
- Burger, A., Berendes, R., Liemann, S., Benz, J., Hofmann, A., Göttig, P., Huber, R., Gerke, V., Thiel, C., Römisch, J. & Weber, K. (1996). J. Mol. Biol. 257, 839–847. [DOI] [PubMed]
- Cacciottolo, M., Belcastro, V., Laval, S., Bushby, K., di Bernardo, D. & Nigro, V. (2011). J. Biol. Chem. 286, 5404–5413. [DOI] [PMC free article] [PubMed]
- Charpentier, T. H., Thompson, L. E., Liriano, M. A., Varney, K. M., Wilder, P. T., Pozharski, E., Toth, E. A. & Weber, D. J. (2010). J. Mol. Biol. 396, 1227–1243. [DOI] [PMC free article] [PubMed]
- Cheng, J. P. X. & Lane, J. D. (2010). Histol. Histopathol. 25, 1457–1472. [DOI] [PubMed]
- Dempsey, B. R., Rezvanpour, A., Lee, T.-W., Barber, K. R., Junop, M. S. & Shaw, G. S. (2012). Structure, 20, 1737–1745. [DOI] [PubMed]
- De Seranno, S., Benaud, C., Assard, N., Khediri, S., Gerke, V., Baudier, J. & Delphin, C. (2006). J. Biol. Chem. 281, 35030–35038. [DOI] [PubMed]
- Draeger, A., Monastyrskaya, K. & Babiychuk, E. B. (2011). Biochem. Pharmacol. 81, 703–712. [DOI] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Epp, N., Rethmeier, R., Krämer, L. & Ungermann, C. (2011). Eur. J. Cell Biol. 90, 779–785. [DOI] [PubMed]
- Filipenko, N. R. & Waisman, D. M. (2001). J. Biol. Chem. 276, 5310–5315. [DOI] [PubMed]
- Gentil, B. J., Delphin, C., Mbele, G. O., Deloulme, J. C., Ferro, M., Garin, J. & Baudier, J. (2001). J. Biol. Chem. 276, 23253–23261. [DOI] [PubMed]
- Gerke, V. & Moss, S. E. (1997). Biochim. Biophys. Acta, 1357, 129–154. [DOI] [PubMed]
- Gerke, V. & Moss, S. E. (2002). Physiol. Rev. 82, 331–371. [DOI] [PubMed]
- Gerke, V. & Weber, K. (1984). EMBO J. 3, 227–233. [DOI] [PMC free article] [PubMed]
- Graaf, S. F. van de, Hoenderop, J. G., Gkika, D., Lamers, D., Prenen, J., Rescher, U., Gerke, V., Staub, O., Nilius, B. & Bindels, R. J. (2003). EMBO J. 22, 1478–1487. [DOI] [PMC free article] [PubMed]
- Haase, H., Pagel, I., Khalina, Y., Zacharzowsky, U., Person, V., Lutsch, G., Petzhold, D., Kott, M., Schaper, J. & Morano, I. (2004). FASEB J. 18, 839–841. [DOI] [PubMed]
- Hayes, M. J., Shao, D., Bailly, M. & Moss, S. E. (2006). EMBO J. 25, 1816–1826. [DOI] [PMC free article] [PubMed]
- Huang, Y., Laval, S. H., van Remoortere, A., Baudier, J., Benaud, C., Anderson, L. V., Straub, V., Deelder, A., Frants, R. R., den Dunnen, J. T., Bushby, K. & van der Maarel, S. M. (2007). FASEB J. 21, 732–742. [DOI] [PubMed]
- Humphrey, W., Dalke, A. & Schulten, K. (1996). J. Mol. Graph. 14, 33–38. [DOI] [PubMed]
- Kesavan, K., Ratliff, J., Johnson, E. W., Dahlberg, W., Asara, J. M., Misra, P., Frangioni, J. V. & Jacoby, D. B. (2010). J. Biol. Chem. 285, 4366–4374. [DOI] [PMC free article] [PubMed]
- Kiss, B., Duelli, A., Radnai, L., Kékesi, K. A., Katona, G. & Nyitray, L. (2012). Proc. Natl Acad. Sci. USA, 109, 6048–6053. [DOI] [PMC free article] [PubMed]
- Krissinel, E. & Henrick, K. (2007). J. Mol. Biol. 372, 774–797. [DOI] [PubMed]
- Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993). J. Appl. Cryst. 26, 283–291.
- Lennon, N. J., Kho, A., Bacskai, B. J., Perlmutter, S. L., Hyman, B. T. & Brown, R. H. (2003). J. Biol. Chem. 278, 50466–50473. [DOI] [PubMed]
- Lin, K., Simossis, V. A., Taylor, W. R. & Heringa, J. (2005). Bioinformatics, 21, 152–159. [DOI] [PubMed]
- Lorusso, A., Covino, C., Priori, G., Bachi, A., Meldolesi, J. & Chieregatti, E. (2006). EMBO J. 25, 5443–5456. [DOI] [PMC free article] [PubMed]
- Martin-Belmonte, F., Gassama, A., Datta, A., Yu, W., Rescher, U., Gerke, V. & Mostov, K. (2007). Cell, 128, 383–397. [DOI] [PMC free article] [PubMed]
- Matza, D., Badou, A., Kobayashi, K. S., Goldsmith-Pestana, K., Masuda, Y., Komuro, A., McMahon-Pratt, D., Marchesi, V. T. & Flavell, R. A. (2008). Immunity, 28, 64–74. [DOI] [PMC free article] [PubMed]
- Mayer, G., Poirier, S. & Seidah, N. G. (2008). J. Biol. Chem. 283, 31791–31801. [DOI] [PubMed]
- Morel, E., Parton, R. G. & Gruenberg, J. (2009). Dev. Cell, 16, 445–457. [DOI] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Nazmi, A. R., Ozorowski, G., Pejic, M., Whitelegge, J. P., Gerke, V. & Luecke, H. (2012). Biol. Chem. 393, 1141–1150. [DOI] [PubMed]
- Nie, Z., Ning, W., Amagai, M. & Hashimoto, T. (2000). J. Invest. Dermatol. 114, 1044–1049. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Ozorowski, G., Ryan, C. M., Whitelegge, J. P. & Luecke, H. (2012). Biol. Chem. 393, 1151–1163. [DOI] [PubMed]
- Rescher, U. & Gerke, V. (2008). Eur. J. Physiol. 455, 575–582. [DOI] [PubMed]
- Rescher, U., Ruhe, D., Ludwig, C., Zobiack, N. & Gerke, V. (2004). J. Cell Sci. 117, 3473–3480. [DOI] [PubMed]
- Réty, S., Osterloh, D., Arié, J.-P., Tabaries, S., Seeman, J., Russo-Marie, F., Gerke, V. & Lewit-Bentley, A. (2000). Structure, 8, 175–184. [DOI] [PubMed]
- Réty, S., Sopkova, J., Renouard, M., Osterloh, D., Gerke, V., Tabaries, S., Russo-Marie, F. & Lewit-Bentley, A. (1999). Nature Struct. Mol. Biol. 6, 89–95. [DOI] [PubMed]
- Rezvanpour, A., Santamaria-Kisiel, L. & Shaw, G. S. (2011). J. Biol. Chem. 286, 40174–40183. [DOI] [PMC free article] [PubMed]
- Rezvanpour, A. & Shaw, G. S. (2009). Gen. Physiol. Biophys. 28, F39–F46. [PubMed]
- Rosengarth, A. & Luecke, H. (2004). Annexins, 1, 129–136.
- Saarikangas, J., Zhao, H. & Lappalainen, P. (2010). Physiol. Rev. 90, 259–289. [DOI] [PubMed]
- Semov, A., Moreno, M. J., Onichtchenko, A., Abulrob, A., Ball, M., Ekiel, I., Pietrzynski, G., Stanimirovic, D. & Alakhov, V. (2005). J. Biol. Chem. 280, 20833–20841. [DOI] [PubMed]
- Shao, C., Zhang, F., Kemp, M. M., Linhardt, R. J., Waisman, D. M., Head, J. F. & Seaton, B. A. (2006). J. Biol. Chem. 281, 31689–31695. [DOI] [PMC free article] [PubMed]
- Shtivelman, E. & Bishop, J. M. (1993). J. Cell Biol. 120, 625–630. [DOI] [PMC free article] [PubMed]
- Shtivelman, E., Cohen, F. E. & Bishop, J. M. (1992). Proc. Natl Acad. Sci. USA, 89, 5472–5476. [DOI] [PMC free article] [PubMed]
- Singh, P., Wu, H., Clark, C. & Owlia, A. (2006). Oncogene, 26, 425–440. [DOI] [PubMed]
- Tekpli, X., Holme, J. A., Sergent, O. & Lagadic-Gossmann, D. (2011). Recent Pat. Anticancer Drug Discov. 6, 347–353. [DOI] [PubMed]
- Umbrecht-Jenck, E., Demais, V., Calco, V., Bailly, Y., Bader, M.-F. & Chasserot-Golaz, S. (2010). Traffic, 11, 958–971. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: AHNAK peptide bound to AnxA2–S100A10 heterotetramer, 4ftg