Abstract
Background
Recombinant DNA technology has been extensively employed to generate a variety of products from genetically modified organisms (GMOs) over the last decade, and the development of technologies capable of analyzing these products is crucial to understanding gene expression patterns. Liquid chromatography coupled with mass spectrometry is a powerful tool for analyzing protein contents and possible expression modifications in GMOs. Specifically, the NanoUPLC-MSE technique provides rapid protein analyses of complex mixtures with supported steps for high sample throughput, identification and quantization using low sample quantities with outstanding repeatability. Here, we present an assessment of the peptide and protein identification and quantification of soybean seed EMBRAPA BR16 cultivar contents using NanoUPLC-MSE and provide a comparison to the theoretical tryptic digestion of soybean sequences from Uniprot database.
Results
The NanoUPLC-MSE peptide analysis resulted in 3,400 identified peptides, 58% of which were identified to have no miscleavages. The experiment revealed that 13% of the peptides underwent in-source fragmentation, and 82% of the peptides were identified with a mass measurement accuracy of less than 5 ppm. More than 75% of the identified proteins have at least 10 matched peptides, 88% of the identified proteins have greater than 30% of coverage, and 87% of the identified proteins occur in all four replicates. 78% of the identified proteins correspond to all glycinin and beta-conglycinin chains.
The theoretical Uniprot peptide database has 723,749 entries, and 548,336 peptides have molecular weights of greater than 500 Da. Seed proteins represent 0.86% of the protein database entries. At the peptide level, trypsin-digested seed proteins represent only 0.3% of the theoretical Uniprot peptide database. A total of 22% of all database peptides have a pI value of less than 5, and 25% of them have a pI value between 5 and 8. Based on the detection range of typical NanoUPLC-MSE experiments, i.e., 500 to 5000 Da, 64 proteins will not be identified.
Conclusions
NanoUPLC-MSE experiments provide good protein coverage within a peptide error of 5 ppm and a wide MW detection range from 500 to 5000 Da. A second digestion enzyme should be used depending on the tissue or proteins to be analyzed. In the case of seed tissue, trypsin protein digestion results offer good databank coverage. The Uniprot database has many duplicate entries that may result in false protein homolog associations when using NanoUPLC-MSE analysis. The proteomic profile of the EMBRAPA BR-16 seed lacks certain described proteins relative to the profiles of transgenic soybeans reported in other works.
Keywords: Soybean, Seed proteomics, NanoUPLC-MSE, Uniprot database
Background
Soybean Glycine max (L) Merrill] is one of the most important leguminous crops in the world with a vital importance to the economies of many countries. Brazil is responsible for 27% of the world soybean production and is second only to the U.S., which produces 35% [1]. Soybean seed products are used in a variety of industrial goods derived from oil (58%) and protein (68%) and are used to feed both humans and animals [1].
In the last decade, efforts have been undertaken to improve soybean crop yields. To this end, genetic engineering has been extensively used to develop soybean plants with abiotic and biotic resistance or tolerance [2]. However, both the quantity of grain produced and the nutritional content of the grain are critical; therefore, the production of highly nutritional seeds of many important crops is currently a focus of research [3-5]. Furthermore, the soybean is also a viable platform for the production of recombinant pharmaceutical molecules, such as human growth hormone [6] and coagulation factor IX [7], for several reasons: the soybean can undergo long-term storage at ambient temperatures [8,9], can provide an appropriate biochemical environment for protein stability through the creation of specialized storage compartments [9,10], is not contaminated by human or animal pathogens [8,11], its desiccation characteristics prevent it from undergoing non-enzymatic hydrolysis or protease degradation [11], and it does not carry harmful substances that are present in certain plant leaves, which is important for downstream processing [11,12].
To enhance protein content analysis efforts, the use of technologies that permit the analysis of protein expression patterns has become a necessity in evaluating the genetic modification of these plants [5,13-15]. The seed, leaf and root proteins of a variety of cultivars have been well documented [15,16]. Two-dimensional gel electrophoresis (2DE) is the most commonly used technique in proteomic analysis, and many types of proteomic studies based on 2DE have been reported [17]; however, 2DE is an extremely time-consuming technique. High throughput protein identification via 2DE requires the use of replicate gels as well as gel excision and digestion procedures [18]; these steps can be complicated and slow. Database comparisons are typically performed using peptide mass fingerprinting [19,20], and quantization is performed by gel image intensity evaluation or by protein tagging [21,22]. All of these stages of 2DE are time-consuming and can produce inconsistent results.
The coupling of liquid chromatography with mass spectrometry, as in NanoUPLC-MSE procedures, provides more robust throughput sample analysis capabilities than other techniques. Complex samples may be prepared in single vials, and all processes associated with chromatography, MS and MS/MS acquisition and database searching can be performed in a few steps [23]. These experiments have led to significant innovations, such as the ability to obtain linear sequence structural information at the femtomole level [24], small surface areas and minimal dead volumes, which minimize analyte losses due to surface adsorption, as well as low flow rates, which minimize the required analyte dilution [25]. Low-abundance analytes can be separated with a high recovery rate when they are associated with a high dynamic range and a high-quality MS detection system [26]. In this present study, we used MSE, which is a data-independent acquisition method that uses low and high collision energies without precursor selection, unlike other methods such as data-dependent acquisition (DDA) [27]. Ion detection, clustering and the normalization of data-independent, alternate scanning LC-MSE data have been explained in detail elsewhere [27,28].
Here, we present a statistical assessment of soybean seeds using NanoUPLC-MSE proteomic experiments and provide a comparison with the theoretical tryptic digestion of sequences from the Uniprot [29,30] soybean database.
Results and discussion
NanoUPLC-MSE proteomics
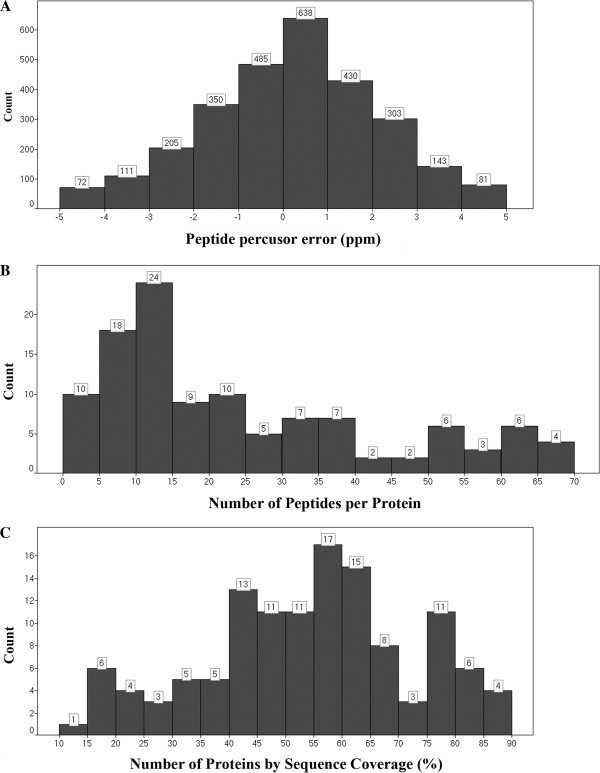
The resulting soybean seed NanoUPLC-MSE peptide data generated by the PLGS process are shown in Figure 1A. The experiment resulted in 3,400 identified peptides; 58% of these peptides were obtained from peptide match type data in the first pass, and 6% were obtained in the second pass [31]. A total of 17% of the peptides were identified by a missed trypsin cleavage, whereas an in-source fragmentation rate of 13% was expected for the Synapt G2 data. Figure 2A shows the peptide parts per million error (ppm) indicating that 82% of the peptides were detected with an error of less than 5 ppm. As shown in Figure 2B, 75% of the identified proteins have at least 10 matched peptides, and 88% of the identified proteins have greater than 30% coverage (Figure 2C). The experiment revealed 113 proteins, of which 87% were replicated 4 times, as shown in Figure 1B and Table 1. These results far exceed the minimum protein identification quality compared to other proteomic data, such as those obtained from the 2DE technique, in which only 10 to 20% of the identified proteins exhibit a coverage greater than 30% [14,20].
Figure 1.

Peptide detection type, repetition rate, and protein function chart.A) On peptide match type, PepFrag1 and Pepfrag2 correspond to the peptide matches when compared to database by PLGS, VarMod corresponds to variable modifications, InSource corresponds to fragmentation that occurred on ionization source, MissedCleavage indicates the missed cleavage performed by trypsin and Neutral loss H2O and NH3 correspond to water and ammonia precursor losses; B) Repeat rate indicates the number of times that an identified protein apears on the replicas; C) Protein function of the identified proteins clustered in storage, defense, energy processing, embryogenesis, seed maturation or other functions.
Figure 2.
Experiment PPM error at the peptide level, number of identified peptides per protein and experimental protein sequence coverage.A) Indicates the number of identified peptides in a 5ppm error range; B) Indicates the number of identified peptides per protein; C) indicates the number of proteins with sequence coverage from 10 to 90%.
Table 1.
Protein identification table
| Uniprot code | Description | Protein MW | ScoreAVG | ProductsAVG | PeptidesAVG | Fmol Covariance | NgramAVG | Repeate Rate | % of TSP |
|---|---|---|---|---|---|---|---|---|---|
|
P04776 |
Glycinin G1 OS Glycine max GN GY1 PE 1 S |
56333.71 |
147104.4 |
523.25 |
38.5 |
0.02 |
85.28 |
4 |
17.68 |
|
Q549Z4 |
Proglycinin A2B1 OS Glycine max PE 2 SV |
54961.11 |
69391.44 |
427.25 |
38.25 |
0.67 |
55.32 |
4 |
11.47 |
|
Q4LER5 |
Beta conglycinin alpha subunit Fragment |
70348.96 |
105347.27 |
301.75 |
54.25 |
0.75 |
32.68 |
4 |
6.78 |
|
Q3V5S6 |
Beta conglycinin alpha subunit OS Glycin |
70569.34 |
104417.56 |
286.5 |
53.75 |
0.72 |
30.91 |
4 |
6.41 |
|
P04405 |
Glycinin G2 OS Glycine max GN Gy2 PE 1 S |
54961.11 |
63666.28 |
370 |
36.5 |
1.97 |
19.12 |
4 |
3.96 |
|
Q7XXT2 |
Prepro beta conglycinin alpha prime subu |
72532.23 |
28368.36 |
295 |
66 |
0.67 |
18.17 |
4 |
3.77 |
|
P04347 |
Glycinin OS Glycine max PE 1 SV 1 |
58412.39 |
23431.64 |
191.5 |
53 |
0.22 |
15.99 |
4 |
3.31 |
|
P02858 |
Glycinin G4 OS Glycine max GN GY4 PE 1 S |
64043.42 |
31521.33 |
290.75 |
60 |
0.26 |
14.25 |
4 |
2.95 |
|
Q9SB12 |
Glycinin OS Glycine max PE 3 SV 1 |
58685.70 |
29853.17 |
251 |
68.75 |
0.48 |
13.30 |
4 |
2.76 |
|
Q9S9D0 |
Glycinin G4 subunit OS Glycine max PE 3 |
64135.62 |
32357.33 |
309 |
60.25 |
0.31 |
11.44 |
4 |
2.37 |
|
Q04672 |
Sucrose binding protein OS Glycine max G |
60921.87 |
11679.68 |
186.25 |
33.5 |
0.02 |
11.37 |
4 |
2.36 |
|
Q9ATY1 |
Kunitz trypsin inhibitor OS Glycine max |
24434.59 |
35543.40 |
112.5 |
13.75 |
0.17 |
11.34 |
4 |
2.35 |
|
Q948Y0 |
Beta conglycinin alpha prime subunit OS |
72423.19 |
17151.17 |
223.5 |
65.75 |
0.75 |
10.95 |
4 |
2.27 |
|
Q948X9 |
Beta conglycinin alpha subunit OS Glycin |
72760.60 |
15765.20 |
186.25 |
44.25 |
0.75 |
9.25 |
4 |
1.92 |
|
P05046 |
Lectin OS Glycine max GN LE1 PE 1 SV 1 |
30928.02 |
10795.06 |
63.75 |
10.25 |
0.01 |
7.95 |
4 |
1.65 |
|
Q94LX2 |
Beta conglycinin alpha subunit OS Glycin |
70591.40 |
113386.25 |
365.5 |
51.75 |
1.01 |
7.57 |
4 |
1.57 |
|
P00489 |
GLYCOGEN PHOSPHORYLASE MUSCLE FORM EC |
97671.64 |
3801.66 |
161.5 |
37.5 |
0.00 |
7.33 |
4 |
1.52 |
|
P11827 |
Beta conglycinin alpha chain OS Glycine |
74610.54 |
11508.88 |
185.25 |
65 |
0.86 |
7.31 |
4 |
1.52 |
|
C6SWW4 |
Putative uncharacterized protein OS Glyc |
22731.98 |
17601.57 |
100.5 |
12.25 |
0.07 |
7.12 |
4 |
1.48 |
|
P25974 |
Beta conglycinin beta chain OS Glycine m |
50609.06 |
19512.84 |
186.25 |
34 |
0.71 |
7.10 |
4 |
1.47 |
|
Q39898 |
Kunitz trypsin inhibitor OS Glycine max |
24361.54 |
54750.93 |
144.25 |
14.5 |
1.16 |
6.72 |
4 |
1.39 |
|
C6T9L1 |
Putative uncharacterized protein OS Glyc |
50621.07 |
18189.66 |
182.5 |
33.25 |
0.96 |
6.30 |
4 |
1.31 |
|
P93708 |
Glycinin OS Glycine max GN Gly A3B4 PE 2 |
58691.62 |
39274.08 |
237.25 |
62.5 |
1.18 |
5.91 |
4 |
1.23 |
|
P13916 |
Beta conglycinin alpha chain OS Glycine |
70578.31 |
113406.84 |
362 |
49.5 |
0.76 |
5.75 |
4 |
1.19 |
|
B3TDK4 |
Lipoxygenase OS Glycine max PE 3 SV 1 |
94639.64 |
4359.21 |
144.25 |
35.75 |
0.03 |
5.63 |
4 |
1.17 |
|
Q93VL9 |
Beta conglycinin beta subunit OS Glycine |
50580.97 |
17965.49 |
159.75 |
32 |
0.72 |
5.29 |
4 |
1.10 |
|
Q852U5 |
Glycinin A1bB2 445 OS Glycine max PE 2 S |
54845.14 |
19226.30 |
97 |
16 |
0.33 |
4.59 |
4 |
0.95 |
|
P13917 |
Basic 7S globulin OS Glycine max GN BG P |
47134.49 |
4532.62 |
62.5 |
14 |
0.03 |
3.51 |
4 |
0.73 |
|
P01070 |
Trypsin inhibitor A OS Glycine max GN KT |
24290.46 |
40525.39 |
125 |
14 |
1.97 |
3.36 |
4 |
0.70 |
|
Q70EM0 |
Dehydrin OS Glycine max GN lea D 11 PE 3 |
23787.84 |
11066.56 |
101.5 |
19 |
0.02 |
3.24 |
4 |
0.67 |
|
Q0MUU5 |
Beta conglycinin alpha subunit OS Glycin |
70172.28 |
24742.62 |
233.5 |
63.25 |
2.00 |
3.14 |
4 |
0.65 |
|
C0KG62 |
Mutant glycinin A3B4 OS Glycine max PE 2 |
60494.82 |
42247.79 |
242.5 |
62.25 |
2.00 |
2.46 |
4 |
0.51 |
|
Q7GC77 |
Glycinin A3B4 subunit OS Glycine max PE |
58643.62 |
50693.51 |
331 |
57.5 |
2.00 |
2.39 |
4 |
0.49 |
|
Q53WV6 |
Napin type 2S albumin 3 OS Glycine max P |
19030.31 |
21340.11 |
82.75 |
13 |
1.17 |
2.19 |
4 |
0.45 |
|
Q43452 |
Glycinin OS Glycine max GN Gy4 PE 3 SV 1 |
64389.76 |
32891.71 |
289.25 |
50.5 |
2.00 |
2.19 |
4 |
0.45 |
|
Q852U4 |
Glycinin A1bB2 784 OS Glycine max PE 2 S |
54868.22 |
18987.27 |
81.75 |
13.5 |
1.30 |
1.90 |
4 |
0.39 |
|
P10538 |
Beta amylase OS Glycine max GN BMY1 PE 1 |
56485.05 |
1358.99 |
57 |
23.5 |
0.72 |
1.87 |
4 |
0.39 |
|
Q8RVH5 |
Basic 7S globulin 2 OS Glycine max PE 1 |
47889.37 |
2086.68 |
37.25 |
11.75 |
0.38 |
1.87 |
4 |
0.39 |
|
Q39853 |
Soybean beta conglycinin alpha subunit F |
24453.59 |
88244.12 |
88.5 |
9.25 |
1.89 |
1.77 |
4 |
0.37 |
|
C6SYA7 |
Putative uncharacterized protein OS Glyc |
19027.27 |
23176.74 |
87 |
15 |
1.19 |
1.75 |
4 |
0.36 |
|
Q39871 |
Late embryongenesis abundant protein OS |
50643.91 |
919.20 |
40.5 |
14 |
0.17 |
1.65 |
2 |
0.34 |
|
C6T7B0 |
Putative uncharacterized protein Fragmen |
48939.81 |
23743.45 |
190.25 |
54.25 |
1.16 |
1.61 |
4 |
0.33 |
|
C0J370 |
Ribulose bisphosphate carboxylase large |
52835.80 |
2789.04 |
34.25 |
9.25 |
0.75 |
1.60 |
4 |
0.33 |
|
C6TKH0 |
Putative uncharacterized protein OS Glyc |
32117.31 |
1246.92 |
38.75 |
10 |
0.03 |
1.54 |
4 |
0.32 |
|
Q9SB11 |
Glycinin OS Glycine max GN A5A4B3 PE 2 S |
64253.61 |
38444.74 |
381.5 |
49 |
1.98 |
1.24 |
4 |
0.26 |
|
Q39858 |
Soybean glycinin A3 B4 subunit Fragment |
27467.17 |
26110.47 |
58.5 |
10.25 |
2.00 |
1.20 |
4 |
0.25 |
|
Q39816 |
7S storage protein alpha subunit OS Glyc |
27538.85 |
8464.43 |
117.75 |
29 |
1.19 |
0.97 |
4 |
0.20 |
|
Q39805 |
Dehydrin like protein OS Glycine max PE |
23717.75 |
10257.65 |
91.5 |
17 |
1.61 |
0.96 |
4 |
0.20 |
|
P19594 |
2S albumin OS Glycine max PE 1 SV 2 |
19030.31 |
20001.72 |
82 |
14.75 |
1.74 |
0.93 |
4 |
0.19 |
|
P11828 |
Glycinin G3 OS Glycine max GN GY3 PE 3 S |
54869.09 |
20193.73 |
96 |
16.5 |
0.83 |
0.86 |
4 |
0.18 |
|
C6TDF5 |
Putative uncharacterized protein OS Glyc |
42166.14 |
2011.92 |
45 |
12.5 |
0.61 |
0.81 |
2 |
0.17 |
|
Q39876 |
Maturation associated protein OS Glycine |
23713.76 |
11376.82 |
107.75 |
19.25 |
1.99 |
0.80 |
4 |
0.17 |
|
Q42795 |
Beta amylase OS Glycine max PE 1 SV 1 |
56413.93 |
1363.11 |
54.75 |
22.5 |
1.99 |
0.73 |
4 |
0.15 |
|
Q42447 |
Maturation protein OS Glycine max GN MAT |
25658.99 |
3286.16 |
26.25 |
6.25 |
0.73 |
0.64 |
4 |
0.13 |
|
Q9SEK9 |
Seed maturation protein PM25 OS Glycine |
25842.80 |
1323.54 |
30.33 |
7.66 |
0.37 |
0.63 |
3 |
0.13 |
|
Q9SP11 |
Sucrose binding protein homolog S 64 OS |
56176.59 |
2088.61 |
62.5 |
24 |
0.69 |
0.62 |
4 |
0.13 |
|
Q9XET0 |
Putative uncharacterized protein OS Glyc |
15154.43 |
2553.76 |
27.75 |
5.75 |
0.03 |
0.57 |
4 |
0.12 |
|
Q9ATY0 |
Truncated Kunitz trypsin inhibitor OS Gl |
16134.39 |
18524.95 |
49.5 |
7.25 |
2.00 |
0.57 |
4 |
0.12 |
|
C6T1Q7 |
Putative uncharacterized protein OS Glyc |
18394.94 |
4148.8 |
41.25 |
10.75 |
0.67 |
0.55 |
4 |
0.11 |
|
Q9LLQ6 |
Seed maturation protein PM34 OS Glycine |
32052.25 |
1013.75 |
24.5 |
10 |
0.39 |
0.55 |
2 |
0.11 |
|
P01064 |
Bowman Birk type proteinase inhibitor D |
10323.16 |
4799.89 |
22.25 |
4.5 |
0.68 |
0.54 |
4 |
0.11 |
|
P25273 |
Kunitz type trypsin inhibitor KTI2 OS Gl |
23085.45 |
2073.41 |
19.75 |
7.25 |
0.87 |
0.51 |
4 |
0.11 |
|
C6T588 |
Putative uncharacterized protein OS Glyc |
16817.85 |
2001.68 |
23 |
6.25 |
0.14 |
0.49 |
4 |
0.10 |
|
A1KR24 |
Dehydrin OS Glycine max GN LEA 2 D11 PE |
25384.72 |
3672.93 |
34.25 |
8.75 |
0.44 |
0.46 |
4 |
0.10 |
|
P09439 |
Seed lipoxygenase 2 OS Glycine max GN LO |
97430.94 |
763.76 |
45 |
27 |
1.41 |
0.40 |
2 |
0.08 |
|
C6TB67 |
Putative uncharacterized protein OS Glyc |
23271.56 |
1060.30 |
16 |
5.33 |
0.03 |
0.40 |
3 |
0.08 |
|
P27066 |
Ribulose bisphosphate carboxylase large |
53066.07 |
2905.08 |
31.75 |
8.75 |
2.00 |
0.37 |
4 |
0.08 |
|
P01063 |
Bowman Birk type proteinase inhibitor C |
9999.68 |
5359.28 |
33.25 |
4.25 |
0.10 |
0.34 |
4 |
0.07 |
|
B3TDK5 |
Lipoxygenase OS Glycine max PE 3 SV 1 |
97068.50 |
753.71 |
44 |
26.5 |
1.41 |
0.33 |
2 |
0.07 |
|
Q39875 |
Soybean lipoxygenase 1 Fragment OS Glyci |
36808.29 |
1111.12 |
23 |
12.333333 |
1.65 |
0.31 |
3 |
0.06 |
|
Q9SBA9 |
Bowman Birk proteinase inhibitor Fragmen |
5100.41 |
8774.56 |
15 |
1 |
0.03 |
0.31 |
4 |
0.06 |
|
Q76B18 |
Kunitz trypsin inhibitor OS Glycine max |
24433.60 |
24408.10 |
91 |
12.5 |
2.00 |
0.30 |
4 |
0.06 |
|
O23957 |
Dehydrin OS Glycine max GN GmPM12 PE 2 S |
17319.94 |
1858.29 |
13.5 |
3 |
1.41 |
0.28 |
2 |
0.06 |
|
C6T9Z5 |
Putative uncharacterized protein OS Glyc |
43109.03 |
1650.38 |
28 |
11.5 |
1.41 |
0.27 |
2 |
0.06 |
|
Q84V19 |
Sucrose binding protein 2 OS Glycine max |
56117.52 |
2226.42 |
65.75 |
23.25 |
1.93 |
0.25 |
4 |
0.05 |
|
O22121 |
Beta subunit of beta conglycinin Fragmen |
47975.72 |
20300.60 |
200.5 |
32.75 |
2.00 |
0.25 |
4 |
0.05 |
|
P08170 |
Seed lipoxygenase 1 OS Glycine max GN LO |
94597.61 |
3028.73 |
90.75 |
35.75 |
1.18 |
0.21 |
4 |
0.04 |
|
Q53B72 |
Putative chalcone isomerase 4 OS Glycine |
23495.71 |
1460.83 |
11.5 |
2.5 |
0.01 |
0.20 |
2 |
0.04 |
|
Q43709 |
Bowman Birk proteinase isoinhibitor D II |
12351.73 |
5458.41 |
27 |
4.5 |
2.00 |
0.20 |
4 |
0.04 |
|
Q9ZNZ4 |
Napin type 2S albumin 1 OS Glycine max P |
18404.98 |
4001.44 |
39.75 |
10.5 |
2.00 |
0.18 |
4 |
0.04 |
|
Q94IA1 |
Kunitz trypsin inhibitor OS Glycine max |
24333.52 |
34662.18 |
107 |
12 |
2.00 |
0.18 |
4 |
0.04 |
|
Q588Z3 |
Beta amylase OS Glycine max GN Gm BamyDa |
56441.98 |
1367.07 |
58.5 |
22.75 |
2.00 |
0.16 |
4 |
0.03 |
|
C7EA91 |
Mutant glycinin subunit A1aB1b OS Glycin |
44092.82 |
71745.98 |
326.25 |
33 |
0.42 |
0.15 |
4 |
0.03 |
|
Q70EL8 |
Dehydrin OS Glycine max GN lea D 11 PE 4 |
23733.70 |
9455.43 |
82.25 |
16.5 |
2.00 |
0.11 |
4 |
0.02 |
|
P93165 |
Em protein OS Glycine max PE 4 SV 1 |
11491.34 |
3266.78 |
22.25 |
6 |
0.06 |
0.11 |
4 |
0.02 |
|
C6TBB3 |
Putative uncharacterized protein OS Glyc |
12345.51 |
1829.78 |
14.75 |
4.5 |
0.21 |
0.10 |
4 |
0.02 |
|
Q4LER6 |
Beta conglycinin alpha prime subunit OS |
72513.18 |
26588.66 |
264.25 |
63.5 |
2.00 |
0.09 |
4 |
0.02 |
|
Q7M1N5 |
Glycinin A1aB1b Fragments OS Glycine max |
6315.28 |
13404.83 |
14.5 |
3.5 |
1.09 |
0.07 |
4 |
0.01 |
|
P25272 |
Kunitz type trypsin inhibitor KTI1 OS Gl |
22831.11 |
11727.02 |
70.75 |
12.5 |
0.16 |
0.06 |
4 |
0.01 |
|
C7EA92 |
Mutant glycinin subunit A1aB1b OS Glycin |
43990.73 |
65306.21 |
275.25 |
30 |
0.70 |
0.05 |
4 |
0.01 |
|
C6T7Y4 |
Putative uncharacterized protein OS Glyc |
33364.68 |
1386.46 |
32.5 |
13.5 |
2.00 |
0.04 |
4 |
0.01 |
|
Q5K3Q9 |
Putative dehydrin Fragment OS Glycine ma |
20395.17 |
7625.21 |
70.5 |
15.75 |
2.00 |
0.00 |
4 |
0.00 |
|
P01071 |
Trypsin inhibitor B OS Glycine max PE 1 |
20268.75 |
17312.94 |
48.75 |
7.5 |
2.00 |
0.00 |
4 |
0.00 |
|
Q9FZP9 |
Alpha subunit of beta conglycinin Fragme |
65199.76 |
25899.30 |
239.25 |
55.5 |
|
0.00 |
4 |
0.00 |
|
O22120 |
Alpha subunit of beta conglycinin Fragme |
63221.91 |
105305.79 |
299.75 |
42.5 |
|
0.00 |
4 |
0.00 |
|
Q588Z5 |
Beta amylase OS Glycine max GN Gm BamyKz |
56419.97 |
1193.51 |
46.25 |
20.75 |
|
0.00 |
4 |
0.00 |
|
Q588Z6 |
Beta amylase OS Glycine max GN Gm BamyTk |
56499.03 |
945.94 |
42 |
20 |
|
0.00 |
4 |
0.00 |
|
Q588Z4 |
Beta amylase OS Glycine max GN Gm BamyTk |
56426.96 |
1290.96 |
50.5 |
23.25 |
|
0.00 |
4 |
0.00 |
|
Q45UE7 |
Beta amylase OS Glycine max PE 2 SV 1 |
56412.98 |
1420.14 |
54.25 |
21.25 |
|
0.00 |
4 |
0.00 |
|
Q84UB3 |
Beta conglycinin alpha subunit Fragment |
45018.62 |
24184.62 |
193.75 |
39.75 |
|
0.00 |
4 |
0.00 |
|
Q50JD8 |
Beta conglycinin beta subunit Fragment O |
48387.22 |
22348.6 |
219 |
28.25 |
|
0.00 |
4 |
0.00 |
|
Q70EL7 |
Dehydrin OS Glycine max GN lea D 11 PE 3 |
25275.54 |
3943.08 |
30 |
8 |
|
0.00 |
3 |
0.00 |
|
Q70EL9 |
Dehydrin OS Glycine max GN lea D 11 PE 3 |
25551.97 |
3591.29 |
24 |
5.6666665 |
|
0.00 |
3 |
0.00 |
|
Q7M211 |
Glycinin A3B4 Plasmid pSPGD41 Fragment O |
21495.15 |
29529.21 |
86 |
16.75 |
|
0.00 |
4 |
0.00 |
|
Q7M210 |
Glycinin A3B4 Plasmid pSPGL1 Fragment OS |
27293.87 |
30284.32 |
105.25 |
23.75 |
|
0.00 |
4 |
0.00 |
|
Q6LBP7 |
Glycinin B 1b subunit 15 AA Fragment OS |
1698.00 |
8521.25 |
7.6666665 |
2.3333333 |
|
0.00 |
3 |
0.00 |
|
P93707 |
Glycinin OS Glycine max GN Gly A3B4 PE 2 |
58633.58 |
45602.53 |
283 |
58.75 |
|
0.00 |
4 |
0.00 |
|
Q9LD16 |
Kunitz trypsin inhibitor 3 OS Glycine ma |
24295.44 |
27398.43 |
66.25 |
9.25 |
|
0.00 |
4 |
0.00 |
|
Q39899 |
Kunitz trypsin inhibitor OS Glycine max |
24308.45 |
17312.92 |
48.75 |
7.5 |
|
0.00 |
4 |
0.00 |
|
Q7XAW0 |
Lea protein OS Glycine max GN ZLDE 2 PE |
25368.76 |
3593.48 |
33.75 |
8.5 |
|
0.00 |
4 |
0.00 |
|
Q39870 |
Lipoxygenase OS Glycine max GN lox2 PE 2 |
97551.04 |
754.83 |
39 |
25.5 |
|
0.00 |
2 |
0.00 |
|
C6T488 |
Putative uncharacterized protein OS Glyc |
24403.62 |
35506 |
113.5 |
13.5 |
|
0.00 |
4 |
0.00 |
| C6SX26 | Putative uncharacterized protein OS Glyc | 12914.35 | 3157.77 | 23.5 | 3.5 | 0.00 | 4 | 0.00 |
The ScoreAVG is the average PLGS score for each hit. ProductsAVG is the average fragment ion products for a protein hit. PeptideAVG is the average of the peptide hits per identified protein. FmolCovariance is the covariance at the femtomole detection level for each protein in the replicate analyses. NgramAVG is the average (in nanograms) of each protein load on the column. The repetition rate is the repeatability of each protein in the replicates. % of TSP is the percentage of each protein in the total soluble mixture relative to the total protein load (in nanograms) on the column.
Figure 3 shows the results obtained from dynamic range detection, indicating that 95 proteins were quantified. A 3-log range and a good detection distribution of high and low molecular weights were obtained, as indicated by the size of the squares. The 10 least abundant proteins includes B3TDK5_SOYBN-Lipoxygenase, C7EA91_SOYBN-Mutant glycinin subunit A1aB1b, Q588Z3_SOYBN-Beta amylase, KTI1_SOYBN-Kunitz type trypsin inhibitor, LOX1_SOYBN-Seed lipoxygenase 1, Q4LER6_SOYBN-Beta conglycinin alpha prime, C7EA92_SOYBN-Mutant glycinin subunit A1aB1b, C6T7Y4_SOYBN-Putative uncharacterized protein, Q5K3Q9_SOYBN-Putative dehydrin Fragment and ITRB_SOYBN-Trypsin inhibitor B. The 10 most abundant proteins are composed of GLYG1_SOYBN-Glycinin G1, Q549Z4_SOYBN-Proglycinin A2B1, Q4LER5_SOYBN-Beta conglycinin alpha subunit, Q9ATY1_SOYBN-Kunitz trypsin inhibitor, Q3V5S6_SOYBN-Beta conglycinin alpha subunit, GLYG2_SOYBN-Glycinin G2, C6SWW4_SOYBN-Putative uncharacterized protein, Q39898_SOYBN-Kunitz trypsin inhibitor, GLYG5_SOYBN-Glycinin and LEC_SOYBN-Lectin. A detailed description of each protein is presented in Table 1. A comparison of our results with other proteomic data reveals that there is a discrepancy in the number of proteins that were identified. Barbosa et al. [14] described 192 identified proteins, although these 192 2DE spots likely correspond to a lower number of proteins because many of the identifications are associated with the same protein with a pI shift. The same trend can be observed in the work presented by Mooney et al. [20], which described 96 identifications of 150 spots detected via 2DE. Sakata et al. [32] described more than 500 spots in gels from cotyledons but reported only 34 identified proteins. Our results mainly identify single proteins. However, there were some exceptions, especially for proteins that possess subunits with similar amino acid sequences, such as glycinin and beta-conglycinin, but are identified with a different accession number in the Uniprot database.
Figure 3.
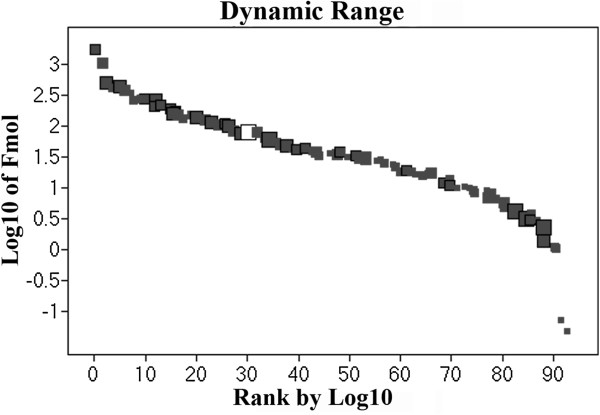
Detection dynamic range of the experiment. White square represent rabbit phosphorylase B as an external standard. Dark squares indicate proteins identified in the experiment. The size of each square represents the molecular weight of the identified protein.
We also compared the identified proteins and correlated them to their function. All identified proteins having at least two replicates are shown in Table 1. A total of 42% of these data correspond to storage proteins, as shown in Figure 1C. From TSP, 78% of the identified species correspond to glycinin and beta-conglycinin chains: GLYG1_SOYBN-Glycinin and Q549Z4_SOYBN-Proglycinin A2B1 correspond to 18% and 12% of TSP, respectively [33]; 13% correspond to inhibitors, including Q9ATY1_SOYBN kunitz type (2.35% of TSP) and Q9SBA9_SOYBN Bowman Birk proteinase inhibitor (0.06% of TSP) [34]; 16% correspond to energy-related proteins, such as C0J370_SOYBN Ribolose (0.33% TSP), Q42795_SOYBN beta-amylases (0.57%) and B3TDK4_SOYBN lipoxygenase (1.14% TSP); and 17% are associated with abiotic stress, including Q7XAW0_SOYBN dehydrin (1% TSP) and LEA proteins (<0.01 % TSP) [35]. An additional 13% correspond to putative uncharacterized proteins. Late embryogenesis proteins and maturation proteins represent 7% of the proteins, including Q39871_SOYBN (0.34% TSP), P93165_SOYBN Em protein (0.2% TSP) and Q9LLQ6_SOYBN seed maturation protein (0.13% TSP) [36,37]. These results are in agreement with those of other studies [15,17,20,32].
Uniprot data assessment
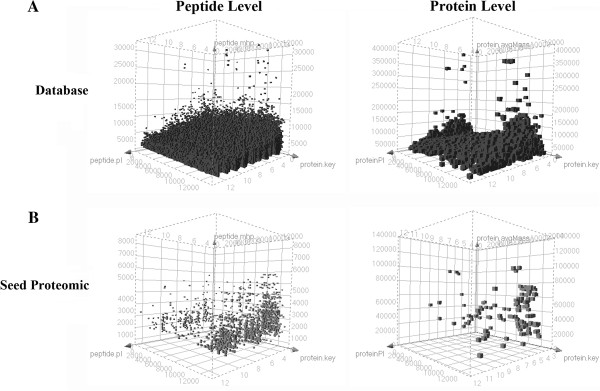
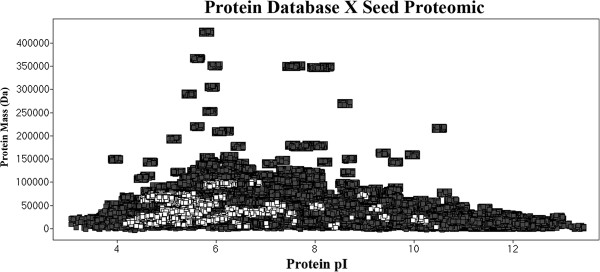
There are 13,117 soybean sequence entries in the Uniprot database. The theoretical tryptic digestion results show 368,435 peptides. Assuming one missed cleavage, the theoretical peptide database has 723,749 entries, 548,336 of which possess a molecular weight greater than 500 Da. These results and a comparison with proteomic data are presented in Figure 4. Seed proteins represent 0.86% of the protein database entries. At the peptide level, trypsin-digested seed proteins represent only 0.3% of the theoretical peptide database, including missed cleavage proteins, which are responsible for only 0.08% of the identified data. Of the seed proteins detected in our experiments, 78% have a pI value between 4.2 and 6. This result is presented in Figure 5, which shows that seed proteins have acidic characteristics. This characteristic was also reported by Robic et al. [38]. At the peptide level (Figure 6), 22% of all database peptides have a pI of less than 5, and 25% of them have a pI between 5 and 8. Figure 6 also shows that 43% of peptides resulting from the experiments have a pI value of less than 5. This pattern is characteristic of tryptic digestion and LC-ESI-MS experiments because the method favors charged peptides.
Figure 4.
Distribution of peptides and proteins in the Uniprot database and seed proteomics by NanoUPLC-MSE.A) Corresponds to the database proteins; B) Indicates the proteins identified in this work.
Figure 5.
Distribution of seed proteins identified within the Uniprot protein database by the isoelectric point. Data shown in dark squares correspond to database proteins, and data shown in white squares are hits identified by this work.
Figure 6.
Distribution of peptide isoelectric points for the database and identified peptides. Dark squares corresponds to database peptides. White squares represents peptides identified using NanoUPLC-MSE.
In Table 2, we present the number of proteins that are not detected within a particular peptide molecular mass detection range. When assuming the minimum and maximum peptide detection levels found using NanoUPLC-MSE experiments, i.e., 500 to 5000 Da, 64 proteins do not have detectable peptides after trypsin digestion (Table 2 and 3). The majority of these proteins correspond to putative and uncharacterized proteins, although NU6C_SOYBN NAD(P)H-quinone oxidoreductase is within the detection range and is not related to seed proteins. Assuming 1 peptide at a threshold of 5,000 Da (Table 3), a few seed proteins are not detected by NanoUPLC-MSE: ACT6_SOYBN Actin-6, ACT7_SOYBN Actin-7, ALL50_SOYBN Major Gly 50 kDa allergen and Q7M212_SOYBN Water-soluble 35K protein. With 2 peptides at an upper threshold of 5,000 Da (Table 3), several putative proteins are not detected, including Q3HM31_SOYBN Hydrophobic seed protein and Q692Y3_SOYBN Glycinin gy1 (Fragment). With 3 peptides at an upper threshold of 5,000 Da (Table 3), the not detected protein list is mainly composed of putative and uncharacterized proteins and other protein fragments that have short amino acid sequences.
Table 2.
Number of proteins with 0 peptides over a given peptide detection range
| Peptide MW range | Number of proteins that has 0 peptides |
|---|---|
|
500<x<1000 |
379 |
|
500<x<2000 |
139 |
|
500<x<3000 |
101 |
|
500<x<4000 |
73 |
|
500<x<5000 |
64 |
|
1000<x<2000 |
311 |
|
2000<x<3000 |
700 |
|
3000<x<4000 |
2163 |
|
4000<x<5000 |
4820 |
|
5000<x<6000 |
7953 |
| >6000 | 9006 |
Table 3.
Uniprot access codes for proteins containing 0, 1, 2 or 3 peptides for a given database peptide detection threshold
|
Nº |
500<x<5000 MW cutoff |
500<x<3000 MW cutoff |
||||||
|---|---|---|---|---|---|---|---|---|
| 0.peptides | 1.peptides | 2.peptides | 3.peptides | 0 peptides | 1.peptides | 2.peptides | 3.peptides | |
|
1 |
C6T0N1 |
A8ICX6 |
B3TDL0 |
C6SWN8 |
C6SZL6 |
A8ICX6 |
B3TDL0 |
B4XH43 |
|
2 |
C6T0N3 |
A8IFI6 |
C6SV77 |
C6SYE8 |
C6T029 |
A8IFI6 |
C6SV77 |
C6SVW9 |
|
3 |
C6T0N7 |
C6SV73 |
C6SVQ7 |
C6SYW9 |
C6T0N1 |
C6SV73 |
C6SVK4 |
C6SVX8 |
|
4 |
C6T0R2 |
C6SW74 |
C6SWW3 |
C6SZA0 |
C6T0N3 |
C6SW74 |
C6SVQ7 |
C6SZD7 |
|
5 |
C6T0R6 |
C6SXX1 |
C6SXB8 |
C6T0K8 |
C6T0N7 |
C6SWU9 |
C6SWN8 |
C6SZQ4 |
|
6 |
C6T0S2 |
C6SY49 |
C6SY76 |
C6T0W0 |
C6T0Q2 |
C6SXX1 |
C6SWW3 |
C6SZT3 |
|
7 |
C6T0S5 |
C6SYJ1 |
C6SYR8 |
C6T0Y8 |
C6T0R2 |
C6SY49 |
C6SX62 |
C6SZT6 |
|
8 |
C6T0T0 |
C6T079 |
C6SYR9 |
C6T2N3 |
C6T0R6 |
C6SYJ1 |
C6SXB8 |
C6T013 |
|
9 |
C6T0T6 |
C6T0X6 |
C6SZK8 |
C6T4B2 |
C6T0S2 |
C6SYR8 |
C6SXL1 |
C6T022 |
|
10 |
C6T0U0 |
C6T113 |
C6SZV3 |
C6T5Y8 |
C6T0S5 |
C6SYW9 |
C6SY37 |
C6T0H4 |
|
11 |
C6T0U6 |
C6T128 |
C6T0P3 |
C6T604 |
C6T0T0 |
C6SZA0 |
C6SY76 |
C6T0W0 |
|
12 |
C6T0U8 |
C6T145 |
C6T0P7 |
C6T695 |
C6T0T6 |
C6SZK8 |
C6SYE8 |
C6T0Y8 |
|
13 |
C6T0V0 |
C6T149 |
C6T0Q2 |
C6T697 |
C6T0U0 |
C6SZM6 |
C6SYI5 |
C6T196 |
|
14 |
C6T0V2 |
C6T5D7 |
C6T0R0 |
C6T6A1 |
C6T0U6 |
C6T079 |
C6SYL8 |
C6T1C2 |
|
15 |
C6T0V4 |
C6T5W7 |
C6T0R4 |
C6T6D9 |
C6T0U8 |
C6T0K8 |
C6SYR9 |
C6T1C3 |
|
16 |
C6T0V8 |
C6T614 |
C6T0R8 |
C6T6I0 |
C6T0V0 |
C6T0X6 |
C6SZV3 |
C6T1N5 |
|
17 |
C6T0W6 |
C6T635 |
C6T0S8 |
C6T6I2 |
C6T0V2 |
C6T115 |
C6T0P3 |
C6T272 |
|
18 |
C6T0W8 |
C6T656 |
C6T0T4 |
C6T6J6 |
C6T0V4 |
C6T145 |
C6T0P7 |
C6T2A4 |
|
19 |
C6T0Y0 |
C6T676 |
C6T0W2 |
C6T6Q9 |
C6T0V8 |
C6T149 |
C6T0R0 |
C6T343 |
|
20 |
C6T108 |
C6T6A8 |
C6T0W4 |
C6T6S9 |
C6T0W6 |
C6T173 |
C6T0R4 |
C6T382 |
|
21 |
C6T110 |
C6T6C5 |
C6T0X2 |
C6T6T3 |
C6T0W8 |
C6T1V9 |
C6T0R8 |
C6T384 |
|
22 |
C6T124 |
C6T6D5 |
C6T0X4 |
C6T6U7 |
C6T0X2 |
C6T3J3 |
C6T0S8 |
C6T400 |
|
23 |
C6T130 |
C6T6F4 |
C6T0Y2 |
C6T6U8 |
C6T0Y0 |
C6T426 |
C6T0T4 |
C6T4D2 |
|
24 |
C6T132 |
C6T6H2 |
C6T0Y4 |
C6T7A3 |
C6T108 |
C6T4U9 |
C6T0V6 |
C6T4E9 |
|
25 |
C6T137 |
C6T6L2 |
C6T0Z0 |
C6TDF1 |
C6T110 |
C6T5D7 |
C6T0W2 |
C6T4I4 |
|
26 |
C6T141 |
C6T6L9 |
C6T0Z5 |
C6TE34 |
C6T113 |
C6T5Y5 |
C6T0W4 |
C6T4U1 |
|
27 |
C6T5Q9 |
C6T6R9 |
C6T0Z7 |
C6TFK5 |
C6T124 |
C6T614 |
C6T0X4 |
C6T5Y9 |
|
28 |
C6T5V9 |
C6T6V2 |
C6T0Z9 |
C6TFK6 |
C6T128 |
C6T635 |
C6T0Y2 |
C6T5Z3 |
|
29 |
C6T5W3 |
C6T6X9 |
C6T101 |
C6TFL5 |
C6T130 |
C6T656 |
C6T0Y4 |
C6T604 |
|
30 |
C6T602 |
C6T6Y2 |
C6T122 |
C6TFS1 |
C6T132 |
C6T676 |
C6T0Z0 |
C6T661 |
|
31 |
C6T606 |
C6T7V8 |
C6T143 |
C6TFT9 |
C6T137 |
C6T6A8 |
C6T0Z5 |
C6T695 |
|
32 |
C6T618 |
C6TC31 |
C6T1V9 |
C6TGK1 |
C6T141 |
C6T6C5 |
C6T0Z7 |
C6T6A1 |
|
33 |
C6T620 |
C6TE40 |
C6T2B3 |
C6THI2 |
C6T2D4 |
C6T6F4 |
C6T0Z9 |
C6T6B5 |
|
34 |
C6T643 |
C6TG02 |
C6T2D4 |
C6TIA8 |
C6T2Q5 |
C6T6H2 |
C6T101 |
C6T6I9 |
|
35 |
C6T664 |
C6TGX9 |
C6T2Q5 |
C6TIG4 |
C6T5N2 |
C6T6L2 |
C6T122 |
C6T6K7 |
|
36 |
C6T666 |
C6TIG9 |
C6T2Z6 |
C6TKN2 |
C6T5Q9 |
C6T6L9 |
C6T143 |
C6T6M6 |
|
37 |
C6T684 |
C6TKR3 |
C6T3C3 |
C6TMR8 |
C6T5U2 |
C6T6R9 |
C6T144 |
C6T6Q9 |
|
38 |
C6T688 |
C6TLP1 |
C6T5U2 |
C6TNC0 |
C6T5U9 |
C6T6V2 |
C6T233 |
C6T6T3 |
|
39 |
C6T6C7 |
C6TN83 |
C6T5U9 |
C6TNW8 |
C6T5V9 |
C6T6Y2 |
C6T269 |
C6T6U7 |
|
40 |
C6T6E5 |
O49223 |
C6T5V1 |
O49225 |
C6T5W3 |
C6T774 |
C6T2B3 |
C6T6U8 |
|
41 |
C6T6F5 |
P13993 |
C6T5X3 |
P08012 |
C6T5W7 |
C6T7U7 |
C6T2N3 |
C6T6W7 |
|
42 |
C6T6G1 |
P15986 |
C6T5Y1 |
P08297 |
C6T5Y1 |
C6TA13 |
C6T2Z6 |
C6T6X5 |
|
43 |
C6T6H7 |
P15987 |
C6T5Z8 |
P49159 |
C6T5Z8 |
C6TB42 |
C6T3A7 |
C6T743 |
|
44 |
C6T6J2 |
P55960 |
C6T608 |
Q2PMQ7 |
C6T602 |
C6TD14 |
C6T3C3 |
C6T7A3 |
|
45 |
C6T6J4 |
P82947 |
C6T622 |
Q2PMR6 |
C6T606 |
C6TD15 |
C6T404 |
C6T7H3 |
|
46 |
C6T6J8 |
Q6JC67 |
C6T629 |
Q2PMR9 |
C6T618 |
C6TDF1 |
C6T411 |
C6T8U0 |
|
47 |
C6T6J9 |
Q6JC68 |
C6T631 |
Q2PMS0 |
C6T620 |
C6TDT6 |
C6T4B2 |
C6T9A8 |
|
48 |
C6T6K2 |
Q7M212 |
C6T633 |
Q2PMT7 |
C6T643 |
C6TE71 |
C6T5V1 |
C6T9K4 |
|
49 |
C6T6L0 |
Q84U84 |
C6T639 |
Q39829 |
C6T652 |
C6TG02 |
C6T5X3 |
C6TA51 |
|
50 |
C6T6L7 |
Q8S3C5 |
C6T645 |
Q4W663 |
C6T664 |
C6TGS5 |
C6T5Y8 |
C6TAK0 |
|
51 |
C6T6N6 |
Q8S3C6 |
C6T649 |
Q6LBP7 |
C6T666 |
C6TIG4 |
C6T608 |
C6TBL2 |
|
52 |
C6T6Q0 |
Q8W238 |
C6T652 |
Q6Q0T0 |
C6T671 |
C6TIG9 |
C6T622 |
C6TDM5 |
|
53 |
C6T6Q2 |
Q9JMW3 |
C6T659 |
Q6X0N6 |
C6T678 |
C6TKR3 |
C6T629 |
C6TDP4 |
|
54 |
C6T6Q6 |
Q9S8F2 |
C6T671 |
Q7M1K4 |
C6T684 |
C6TL18 |
C6T631 |
C6TDP6 |
|
55 |
C6T6S4 |
Q9S8F3 |
C6T678 |
Q9S8R7 |
C6T688 |
C6TL51 |
C6T633 |
C6TDR0 |
|
56 |
C6T6T1 |
Q9S8K3 |
C6T682 |
Q9S8X5 |
C6T6C7 |
C6TLP1 |
C6T639 |
C6TDR8 |
|
57 |
C6T6T7 |
Q9S904 |
C6T686 |
Q9S926 |
C6T6D5 |
O49223 |
C6T645 |
C6TEN4 |
|
58 |
C6T6T9 |
Q9S905 |
C6T690 |
Q9SBB0 |
C6T6E5 |
P13993 |
C6T649 |
C6TF71 |
|
59 |
C6T6W6 |
Q9S929 |
C6T693 |
|
C6T6F5 |
P15986 |
C6T659 |
C6TFK5 |
|
60 |
C6T6W8 |
|
|
|
C6T6G1 |
P15987 |
C6T682 |
C6TFS1 |
|
61 |
C6T837 |
|
|
|
C6T6H7 |
P49159 |
C6T686 |
C6TFT9 |
|
62 |
C6TDZ9 |
|
|
|
C6T6I0 |
P55960 |
C6T690 |
C6TFY8 |
|
63 |
C6TN18 |
|
|
|
C6T6J2 |
P82947 |
C6T693 |
C6TGK1 |
|
64 |
Q2PMN5 |
|
|
|
C6T6J4 |
Q2PMQ7 |
C6T697 |
C6TGZ6 |
|
65 |
|
|
|
|
C6T6J8 |
Q2PMS0 |
C6T698 |
C6TIK8 |
|
66 |
|
|
|
|
C6T6J9 |
Q2PMS6 |
C6T6B0 |
C6TJX4 |
|
67 |
|
|
|
|
C6T6K2 |
Q2PMT6 |
C6T6B7 |
C6TKH6 |
|
68 |
|
|
|
|
C6T6K4 |
Q2PMT7 |
C6T6D9 |
C6TKN2 |
|
69 |
|
|
|
|
C6T6L0 |
Q2PMU0 |
C6T6F3 |
C6TLE1 |
|
70 |
|
|
|
|
C6T6L7 |
Q39806 |
C6T6F7 |
C6TLT6 |
|
71 |
|
|
|
|
C6T6N6 |
Q4W663 |
C6T6H9 |
C6TLV5 |
|
72 |
|
|
|
|
C6T6P1 |
Q6JC67 |
C6T6I2 |
C6TM33 |
|
73 |
|
|
|
|
C6T6Q0 |
Q6JC68 |
C6T6J6 |
C6TM52 |
|
74 |
|
|
|
|
C6T6Q2 |
Q6X0N6 |
C6T6K0 |
C7S8C3 |
|
75 |
|
|
|
|
C6T6Q6 |
Q7M212 |
C6T6K6 |
O49225 |
|
76 |
|
|
|
|
C6T6S4 |
Q84U84 |
C6T6R3 |
O65110 |
|
77 |
|
|
|
|
C6T6T1 |
Q8S3C6 |
C6T6S9 |
P08012 |
|
78 |
|
|
|
|
C6T6T7 |
Q8W238 |
C6T6W1 |
P69421 |
|
79 |
|
|
|
|
C6T6T9 |
Q9JMW3 |
C6T737 |
Q2PMQ8 |
|
80 |
|
|
|
|
C6T6W6 |
Q9S8K3 |
C6T7G8 |
Q2PMR4 |
|
81 |
|
|
|
|
C6T6W8 |
Q9S904 |
C6T7P1 |
Q2PMS7 |
|
82 |
|
|
|
|
C6T6X9 |
Q9S905 |
C6T7Y0 |
Q39829 |
|
83 |
|
|
|
|
C6T7V8 |
Q9S929 |
C6T9H9 |
Q39863 |
|
84 |
|
|
|
|
C6T837 |
|
C6TBB6 |
Q41267 |
|
85 |
|
|
|
|
C6TC31 |
|
C6TBR9 |
Q4W666 |
|
86 |
|
|
|
|
C6TCV3 |
|
C6TBS2 |
Q6J5U8 |
|
87 |
|
|
|
|
C6TDZ9 |
|
C6TBV4 |
Q6LBP7 |
|
88 |
|
|
|
|
C6TE40 |
|
C6TD51 |
Q6LED6 |
|
89 |
|
|
|
|
C6TFL5 |
|
C6TD69 |
Q7M1K4 |
|
90 |
|
|
|
|
C6TGX9 |
|
C6TE34 |
Q9S8R7 |
|
91 |
|
|
|
|
C6TLD5 |
|
C6TEN7 |
Q9S8X5 |
|
92 |
|
|
|
|
C6TLV0 |
|
C6TF61 |
Q9S926 |
|
93 |
|
|
|
|
C6TN00 |
|
C6TF86 |
Q9S9H5 |
|
94 |
|
|
|
|
C6TN18 |
|
C6TFK6 |
|
|
95 |
|
|
|
|
C6TN83 |
|
C6TFW2 |
|
|
96 |
|
|
|
|
Q2PMN5 |
|
C6TFY6 |
|
|
97 |
|
|
|
|
Q3HM31 |
|
C6TH40 |
|
|
98 |
|
|
|
|
Q8S3C5 |
|
C6THI2 |
|
|
99 |
|
|
|
|
Q9S8F2 |
|
C6TIA8 |
|
|
100 |
|
|
|
|
Q9S8F3 |
|
C6TJW9 |
|
|
101 |
|
|
|
|
Q9SBB0 |
|
C6TK75 |
|
|
102 |
|
|
|
|
|
|
C6TKY5 |
|
|
103 |
|
|
|
|
|
|
C6TLF7 |
|
|
104 |
|
|
|
|
|
|
C6TMR8 |
|
|
105 |
|
|
|
|
|
|
C6TNC0 |
|
|
106 |
|
|
|
|
|
|
C6TNG5 |
|
|
107 |
|
|
|
|
|
|
C6TNW8 |
|
|
108 |
|
|
|
|
|
|
O65109 |
|
|
109 |
|
|
|
|
|
|
P08297 |
|
|
110 |
|
|
|
|
|
|
P69195 |
|
|
111 |
|
|
|
|
|
|
Q0GPJ4 |
|
|
112 |
|
|
|
|
|
|
Q2PMR5 |
|
|
113 |
|
|
|
|
|
|
Q2PMR6 |
|
|
114 |
|
|
|
|
|
|
Q2PMR9 |
|
|
115 |
|
|
|
|
|
|
Q2TI80 |
|
|
116 |
|
|
|
|
|
|
Q4W664 |
|
|
117 |
|
|
|
|
|
|
Q692Y3 |
|
|
118 |
|
|
|
|
|
|
Q6J5X9 |
|
|
119 |
|
|
|
|
|
|
Q6Q0T0 |
|
|
120 |
|
|
|
|
|
|
Q75NI2 |
|
|
121 |
|
|
|
|
|
|
Q7M285 |
|
|
122 |
|
|
|
|
|
|
Q9S8H6 |
|
|
123 |
|
|
|
|
|
|
Q9S8P7 |
|
|
124 |
|
|
|
|
|
|
Q9S8X4 |
|
| 125 | Q9S8X6 | |||||||
Many of these undetected proteins have been found in soybean seeds and described in other studies [15,17,20,32]. An analysis of the undetected proteins in the database shows that the majority of the sequences are composed of short amino acid sequences with at most 20 residues. This observation may explain the level of missed detection in the NanoUPLC experiments. Other proteins that are not described in this work, such as glyceraldehyde 3-phosphate (Q2I0H4_SOYBN), Malate dehydrogenase (B0M1B0_SOYBN), Glutathione S-transferase (C6ZQJ7_SOYBN), Isoflavone reductase (Q9SDZ0_SOYBN), Alcohol dehydrogenase 1 (Q8LJR2_SOYBN) and In2-1 protein (Q9FQ95_SOYBN), have been described as soybean seed proteins. These proteins have been described in a previous study on the proteomics of transgenic soybean seeds expressing CTAG recombinant proteins [23]. Further experiments must be performed to clarify this issue.
We hypothesize that environmental stress may have altered the seed expression profiles because the EMBRAPA BR-16 seeds were cultivated in the field, and the transgenic seeds were grown in a greenhouse. For example, Barbosa et al. [14] and Brandão et al. [22] reported different expression levels of enzymes in the transgenic soybean proteome of Monsanto Roundup-ready seeds. The authors state that the genetic modification itself could be a stress factor and may produce alterations in the seed proteome. A comparison between the results of this work and our previous study provides evidence in support of this hypothesis, indicating the need for further experiments to confirm possible proteome alterations due to genetic modification. Nevertheless, highly hydrophobic or insoluble proteins will not be detected due to the necessity for in-solution protease digestion; special protocols are needed for the digestion of these types of protein.
Conclusions
NanoUPLC-MSE experiments are a viable choice as a proteomic pipeline for soybean protein detection. NanoUPLC-MSE provides good protein coverage with a 5 ppm peptide error, reduced sample manipulation relative to other techniques and detection of a wide range of peptide MWs, i.e., from 500 to 5000 Da. Because not all proteins from the Uniprot database are covered, the use of a second digestion enzyme is recommended depending on the tissue to be analyzed. In the case of seed tissue, trypsin protein digestion results in good database coverage. The Uniprot database has many duplicate entries that may result in false protein homolog association and must be formatted prior to use or the use of the reviewed sequences only. It also has many fragment entries that are not suitable for NanoUPLC-MSE analysis but may be used in other techniques. The proteomic profile of EMBRAPA BR-16 seed lacks certain described proteins relative to transgenic soybean profiles reported in other studies. This discrepancy demonstrates the need for further transgenic and nontransgenic proteome analyses.
Methods
Extraction of total soluble protein from soybean seeds
Seeds from the EMBRAPA BR-16 cultivar were used in this work. The soybean seeds were ground to a fine powder using a coffee grinder. A 100 mg sample of powder was weighed and placed in a 2 mL capped centrifuge tube. Petroleum ether (1 mL) was added, and the sample was gently agitated for 15 min. The supernatant was discarded, and this step was repeated twice. The petroleum ether was evaporated for 10 min, and 1 mL of 20 mM Tris–HCl pH 8.3, 1.5 mM KCl, 10 mM DTT, 1 mM PMSF and 0.1% V/V SDS was added. The sample was slowly vortexed at room temperature for 10 min and centrifuged for 5 min at 10000g at 4°C. The supernatant was then transferred to a new centrifuge tube. For each 200 μL of sample, 800 μL of cold acetone was added to the centrifuge tube. The sample was vortexed thoroughly and incubated at −20°C for 1 h with vortexing performed every 15 min. The sample was then centrifuged for 10min at 15700g. The supernatant was discarded, and the pellet was dried at room temperature for 30min. The pellet was carefully dissolved in 500 μL of 50 mM ammonium bicarbonate and quantified using a Quant-iT™ Protein Assay Kit (Invitrogen, USA). The sample was finally diluted with 50 mM ammonium bicarbonate to a protein concentration of 1 μg.μL-1.
Sample preparation for NanoUPLC-MSE acquisition
A 50 μL aliquot of the 1 μg.μL-1 sample was added to 10 μL of 50 mM ammonium bicarbonate in a microcentrifuge tube. Then, 25 μL of RapiGEST™ (Waters, USA) (0.2% v/v) was added, and the sample was vortexed and incubated in a dry bath at 80°C for 15 min. The sample was briefly centrifuged, and 2.5 μL of 100 mM DTT was added. The sample was vortexed gently and incubated at 60°C for 30 min followed by centrifugation. Iodoacetamide (2.5 μL of a 300 mM solution) was added, and the sample was briefly vortexed and incubated in the dark at room temperature for 30 min. Then, 10 μL of trypsin (with 400 μL of 50 mM ammonium bicarbonate added per 20 μg vial of trypsin) was added, and the sample was briefly vortexed. The sample was digested at 37°C in a dry bath overnight. To cleave and precipitate the RapiGEST™, 10 μL of a 5% TFA solution was added, and the sample was vortexed, incubated for 90 min at 37°C in a dry bath, and centrifuged at 18000g at 6°C for 30 min. The supernatant was transferred to a Waters Total Recovery vial (Waters, USA), and 5 μL of Rabbit Phosphorylase B (Waters, part number 186002326) (with 1 mL of 3% acetonitrile and 0.1% formic acid) and 85 μL of a 3% acetonitrile and 0.1% formic acid solution were added. The final concentration of the protein was 250 ng.μL-1, and the final concentration of Phosphorylase B was 25 fmol.μL-1. The final volume was 200 μL.
NanoUPLC-MSE acquisition
The nanoscale LC separation of tryptic peptides from TSP was performed using a nanoACQUITY™ system (Waters Corp., USA) equipped with a Symmetry C18 5μm, 5mm x 300μm precolumn and a nanoEase™ BEH130 C18 1.7 μm, 100 μm x 100 mm analytical reversed-phase column (Waters, USA). The samples were initially transferred to the pre-column using an aqueous 0.1% formic acid solution with a flow rate of 5 μL.min-1 for 2 min. Mobile phase A consisted of 0.1% formic acid in water, and mobile phase B consisted of 0.1% formic acid in acetonitrile. The peptides were separated using a gradient of 3-40% mobile phase B for 200 min with a flow rate of 600 ηL.min-1 followed by a 10 min rinse with 85% of mobile phase B. The column was re-equilibrated to the initial conditions for 20 min. The column temperature was maintained at 35°C. The lock mass was delivered from the fluidics system of a SynaptG2 pump using a constant flow rate of 400 ηL.min-1 at a concentration of 200 fmol of GFP to the reference sprayer of the NanoLockSpray source of the mass spectrometer. All samples were analyzed in four replicates.
The tryptic peptides were analyzed using a Synapt G2 HDMS™ mass spectrometer (Waters, Manchester, UK) with a hybrid quadrupole/ion mobility/orthogonal acceleration time-of-flight (oa-TOF) geometry. For all measurements, the mass spectrometer was operated in the sensitive mode of analysis with a typical resolving power of at least 10000 full-width half-maximum (FWHM). All analyses were performed using a positive nanoelectrospray ion mode (nanoESI +). The time-of-flight analyzer of the mass spectrometer was externally calibrated with GFP b+ and y+ ions from 50 to 1990 m/z with the data post acquisition lock mass corrected using the GFP double charged precursor ion [M + 2H]2+ = 785.8426. The reference sprayer was sampled at a frequency of 30 s. The exact mass retention time (EMRT) [28] nanoLC-MSE data were collected in an alternating low energy and elevated energy acquisition mode. The continuum spectra acquisition time in each mode was 1.5 s with a 0.1 s interscan delay. In the low-energy MS mode, data were collected at constant collision energy of 3 eV. In the elevated-energy MS mode, the collision energy was increased from 12 to 45 eV during each 1.5 s spectrum. The radiofrequency that was applied to the quadrupole mass analyzer was adjusted such that the ions from 50 to 2000 m/z were efficiently transmitted, which ensured that any ions less than 50 m/z observed in the LC-MS data were only derived from dissociations in the TRAP T-wave collision cell.
Data processing and protein identification
The MS data that were obtained from the LC-MSE analysis were processed and searched using the ProteinLynx Global Server (PLGS) version 2.5 (Waters, Manchester, UK). Proteins were identified using the software’s embedded ion accounting algorithm and a search of the Glycine max database with MassPREP digestion standards (MPDS) UniProtKB/Swiss-Prot sequences (Phosphorylase - P00489 - PHS2_RABIT, Bovine Hemoglobin - P02070 - HBB_BOVIN, ADH - P00330 - ADH1_YEAST, BSA - P02769 - ALBU_BOVIN) that were appended to the database. Identifications and quantitative data packaging were performed using dedicated algorithms [28,31] and a search against a soybean Uniprot database. The ion detection, clustering, and log-scale parametric normalizations were performed in PLGS with an ExpressionE license installed. The intensity measurements were typically adjusted for these components, i.e., the deisotoped and charge state-reduced EMRTs that were replicated throughout the entire experiment for the analysis at the EMRT cluster level. The fixed modification of carbamidomethyl-C was specified, and the included variable modifications were acetylation of the N-terminus, deamidation of N, deamidation of Q and oxidation of M. Components were typically clustered with a 10ppm mass precision and a 0.25 min time tolerance against the database-generated theoretical peptide ion masses with a minimum of one matched peptide. The alignment of elevated-energy ions with low-energy precursor peptide ions was performed with an approximate precision of 0.05 min. One missed cleavage site was allowed. The precursor and fragment ion tolerances were determined automatically. The protein identification criteria also included the detection of at least three fragment ions per peptide, 6 fragments per protein and the determination of at least one peptide per protein; the identification of the protein was allowed with a maximum 4% false positive discovery rate in at least four technical replicate injections. Using protein identification replication as a filter, the false positive rate was minimized because false positive protein identifications, i.e., chemical noise, have a random nature and do not tend to replicate across injections. For the analysis of the protein identification and quantification level, the observed intensity measurements were normalized to the intensity measurement of the identified peptides of the digested internal standard. Protein tables generated by PLGS were merged, and the dynamic range of the experiment was calculated using the in-house software program MassPivot by setting the minimum repeat rate for each protein in all replicates to 2.
Uniprot soybean database digestion and experiment analysis
Glycine max protein sequences were obtained from Uniprot ( http://www.uniprot.org), and the theoretical tryptic digestion was performed using the in-house software Digestion tool. The digestion was performed allowing 1 missed cleavage, and the molecular mass and isoelectric point of all peptides and proteins were calculated. The peptide and protein tables from PLGS were compared with the database digestion table using the Spotfire software ( http://spotfire.tibco.com/), suitable graphics were generated for all data. Microsoft Excel (Microsoft, USA) was used for table manipulations.
Authors’ contributions
All authors contributed equally to this work. All authors read and approved the final manuscript.
Contributor Information
Andre M Murad, Email: andre.murad@embrapa.br.
Elibio L Rech, Email: elibio.rech@embrapa.br.
Acknowledgements
This work was supported by the Brazilian Agricultural Research Corporation (EMBRAPA), the National Council for Scientific and Technological Development (CNPq) and the Fundação de Apoio a Pesquisa-DF (FAP-DF). The authors acknowledge support from C. Bloch at the Mass Spectrometry Laboratory-EMBRAPA.
References
- Soystats. www.soystats.com.
- BASF and Embrapa’s Cultivance soybeans receive approval for commercial cultivation in Brazil. http://www.basf.com/group/pressrelease/P-10-148.
- Aluru MR, Rodermel SR, Reddy MB. Genetic modification of low phytic Acid 1–1 maize to enhance iron content and bioavailability. J Agric Food Chem. 2011;59(24):12954–12962. doi: 10.1021/jf203485a. [DOI] [PubMed] [Google Scholar]
- Drakakaki G, Marcel S, Glahn RP, Lund EK, Pariagh S, Fischer R, Christou P, Stoger E. Endosperm-specific co-expression of recombinant soybean ferritin and Aspergillus phytase in maize results in significant increases in the levels of bioavailable iron. Plant Mol Biol. 2005;59(6):869–880. doi: 10.1007/s11103-005-1537-3. [DOI] [PubMed] [Google Scholar]
- Herman EM, Helm RM, Jung R, Kinney AJ. Genetic modification removes an immunodominant allergen from soybean. Plant Physiol. 2003;132(1):36–43. doi: 10.1104/pp.103.021865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha NB, Murad AM, Cipriano TM, Araujo ACG, Aragao FJL, Leite A, Vianna GR, McPhee TR, Souza GHMF, Waters MJ. et al. Expression of functional recombinant human growth hormone in transgenic soybean seeds. Transgenic Res. 2010;20(4):811–826. doi: 10.1007/s11248-010-9460-z. [DOI] [PubMed] [Google Scholar]
- Cunha NB, Murad AM, Ramos GL, Maranhao AQ, Brıgido MM, Araujo ACG, Lacorte C, Aragao FJL, Covas DT, Fontes AM. et al. Accumulation of functional recombinant human coagulation factor IX in transgenic soybean seeds. Transgenic Res. 2010;20(4):841–855. doi: 10.1007/s11248-010-9461-y. [DOI] [PubMed] [Google Scholar]
- Boothe J, Nykiforuk C, Shen Y, Zaplachinski S, Szarka S, Kuhlman P, Murray E, Morck D, Moloney MM. Seed-based expression systems for plant molecular farming. Plant Biotechnol J. 2010;8:588–606. doi: 10.1111/j.1467-7652.2010.00511.x. [DOI] [PubMed] [Google Scholar]
- Cunha NB, Araújo ACG, Leite A, Murad AM, Vianna GR, Rech EL. Correct targeting of proinsulin in protein storage vacuoles of transgenic soybean seeds. Genet Mol Res. 2010;9(2):1163–1170. doi: 10.4238/vol9-2gmr849. [DOI] [PubMed] [Google Scholar]
- Jolliffe NA, Craddock CP, Frigerio L. Pathways for protein transport to seed storage vacuoles. Biochem Soc Trans. 2005;33:1016–1018. doi: 10.1042/BST20051016. [DOI] [PubMed] [Google Scholar]
- Ma JK-C, Drake PMW, Christou P. The production of recombinant pharmaceutical proteins in plants. Nat Rev Genet. 2003;4:794–805. doi: 10.1038/nrg1177. [DOI] [PubMed] [Google Scholar]
- Tremblay R, Wang D, Jevnikar AM, Ma S. Tobacco, a highly efficient green bioreactor for production of therapeutic proteins. Biotechnol Adv. 2010;28:214–221. doi: 10.1016/j.biotechadv.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y-H, Choi SJ, Lee H-A, Moon TW. Quantitation of CP4 5-Enolpyruvylshikimate-3-Phosphate synthase in soybean by two-dimensional gel electrophoresis. J Microbiol Biotechnol. 2006;16(1):25–31. [Google Scholar]
- Barbosa HS, Arruda SC, Azevedo RA, Arruda MA. New insights on proteomics of transgenic soybean seeds: evaluation of differential expressions of enzymes and proteins. Anal Bioanal Chem. 2012;402(1):299–314. doi: 10.1007/s00216-011-5409-1. [DOI] [PubMed] [Google Scholar]
- Natarajan SS, Xu C, Bae H, Caperna TJ, Garrett WM. Characterization of storage proteins in wild (Glycine soja) and cultivated (Glycine max) soybean seeds using proteomic analysis. J Agric Food Chem. 2006;54(8):3114–3120. doi: 10.1021/jf052954k. [DOI] [PubMed] [Google Scholar]
- Aghaei K, Ehsanpour AA, Shah AH, Komatsu S. Proteome analysis of soybean hypocotyl and root under salt stress. Amino Acids. 2009;36(1):91–98. doi: 10.1007/s00726-008-0036-7. [DOI] [PubMed] [Google Scholar]
- Komatsu S, Ahsan N. Soybean proteomics and its application to functional analysis. J Proteomics. 2009;72(3):325–336. doi: 10.1016/j.jprot.2008.10.001. [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc. 2006;1(6):2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- Yang Y, Zhang S, Howe K, Wilson DB, Moser F, Irwin D, Thannhauser TW. A comparison of nLC-ESI-MS/MS and nLC-MALDI-MS/MS for GeLC-based protein identification and iTRAQ-based shotgun quantitative proteomics. J Biomol Tech. 2007;18:226–237. [PMC free article] [PubMed] [Google Scholar]
- Mooney BP, Krishnan HB, Thelen JJ. High-throughput peptide mass fingerprinting of soybean seed proteins: automated workflow and utility of UniGene expressed sequence tag databases for protein identification. Phytochemistry. 2004;65(12):1733–1744. doi: 10.1016/j.phytochem.2004.04.011. [DOI] [PubMed] [Google Scholar]
- Nogueira SB, Labate CA, Gozzo FC, Pilau EJ, Lajolo FM, Oliveira do Nascimento JR. Proteomic analysis of papaya fruit ripening using 2DE-DIGE. J Proteomics. 2012;75(4):1428–1439. doi: 10.1016/j.jprot.2011.11.015. [DOI] [PubMed] [Google Scholar]
- Brandao AR, Barbosa HS, Arruda MA. Image analysis of two-dimensional gel electrophoresis for comparative proteomics of transgenic and non-transgenic soybean seeds. J Proteomics. 2010;73(8):1433–1440. doi: 10.1016/j.jprot.2010.01.009. [DOI] [PubMed] [Google Scholar]
- Murad AM, Souza GH, Garcia JS, Rech EL. Detection and expression analysis of recombinant proteins in plant-derived complex mixtures using nanoUPLC-MS(E) J Sep Sci. 2011;34(19):2618–2630. doi: 10.1002/jssc.201100238. [DOI] [PubMed] [Google Scholar]
- Shen Y, Zhao R, Berger SJ, Anderson GA, Rodriguez N, Smith RD. High-efficiency nanoscale liquid chromatography coupled on-line with mass spectrometry using nanoelectrospray ionization for proteomics. Anal Chem. 2002;74:4235–4249. doi: 10.1021/ac0202280. [DOI] [PubMed] [Google Scholar]
- Liu H, Finch JW, Lavallee MJ, Collamati RA, Benevides CC, Gebler JC. Effects of column length, particle size, gradient length and flow rate on peak capacity of nano-scale liquid chromatography for peptide separations. J Chromatogr A. 2007;1147(1):30–36. doi: 10.1016/j.chroma.2007.02.016. [DOI] [PubMed] [Google Scholar]
- Levin Y, Wang L, Ingudomnukul E, Schwarz E, Baron-Cohen S, Palotás A, Bahn S. Real-time evaluation of experimental variation in large-scale LC–MS/MS-based quantitative proteomics of complex samples. J Chromatogr B. 2009;877:1299–1305. doi: 10.1016/j.jchromb.2008.11.007. [DOI] [PubMed] [Google Scholar]
- Geromanos SJ, Vissers JPC, Silva JC, Dorschel CA, Li G-Z, Gorenstein MV, Bateman RH, Langridge JI. The detection, correlation, and comparison of peptide precursor and product ions from data independent LC-MS with data dependant LC-MS/MS. Proteomics. 2009;9(6):1683–1695. doi: 10.1002/pmic.200800562. [DOI] [PubMed] [Google Scholar]
- Silva JC, Gorenstein MV, Li G-Z, Vissers JPC, Geromanos SJ. Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Mol Cell Proteomics. 2005;5(1):144–156. doi: 10.1074/mcp.M500230-MCP200. [DOI] [PubMed] [Google Scholar]
- Bairoch A, Apweiler R, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M. et al. The universal protein resource (UniProt) Nucleic Acids Res. 2005;33(Database issue):D154–D159. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leinonen R, Diez FG, Binns D, Fleischmann W, Lopez R, Apweiler R. UniProt archive. Bioinformatics. 2004;20(17):3236–3237. doi: 10.1093/bioinformatics/bth191. [DOI] [PubMed] [Google Scholar]
- Li G-Z, Vissers JPC, Silva JC, Golick D, Gorenstein MV, Geromanos SJ. Database searching and accounting of multiplexed precursor and product ion spectra from the data independent analysis of simple and complex peptide mixtures. Proteomics. 2009;9:1696–1719. doi: 10.1002/pmic.200800564. [DOI] [PubMed] [Google Scholar]
- Sakata K, Ohyanagi H, Nobori H, Nakamura T, Hashiguchi A, Nanjo Y, Mikami Y, Yunokawa H, Komatsu S. Soybean proteome database: a data resource for plant differential omics. J Proteome Res. 2009;8(7):3539–3548. doi: 10.1021/pr900229k. [DOI] [PubMed] [Google Scholar]
- Shutov AD, Kakhovskaya IA, Bastrygina AS, Bulmaga VP, Horstmann C, Muntz K. Limited proteolysis of beta-conglycinin and glycinin, the 7S and 11S storage globulins from soybean [Glycine max (L.) Merr.]. Structural and evolutionary implications. Eur J Biochem. 1996;241(1):221–228. doi: 10.1111/j.1432-1033.1996.0221t.x. [DOI] [PubMed] [Google Scholar]
- Lee KJ, Kim JB, Ha BK, Kim SH, Kang SY, Lee BM, Kim DS. Proteomic characterization of Kunitz trypsin inhibitor variants, Tia and Tib, in soybean [Glycine max (L.) Merrill] Amino Acids. 2011;43(1):379–388. doi: 10.1007/s00726-011-1092-y. [DOI] [PubMed] [Google Scholar]
- Wang W, Vinocur B, Altman A. Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta. 2003;218(1):1–14. doi: 10.1007/s00425-003-1105-5. [DOI] [PubMed] [Google Scholar]
- Hill JE, Breidenbach RW. Proteins of soybean seeds: II. Accumulation of the major protein components during seed development and maturation. Plant Physiol. 1974;53(5):747–751. doi: 10.1104/pp.53.5.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsing YC, Tsou CH, Hsu TF, Chen ZY, Hsieh KL, Hsieh JS, Chow TY. Tissue- and stage-specific expression of a soybean (Glycine max L.) seed-maturation, biotinylated protein. Plant Mol Biol. 1998;38(3):481–490. doi: 10.1023/A:1006079926339. [DOI] [PubMed] [Google Scholar]
- Robic G, Farinas CS, Rech EL, Miranda EA. Transgenic soybean seed as protein expression system: aqueous extraction of recombinant beta-glucuronidase. Appl Biochem Biotechnol. 2010;160(4):1157–1167. doi: 10.1007/s12010-009-8637-5. [DOI] [PubMed] [Google Scholar]