Abstract
When females of Drosophila melanogaster and males of Drosophila simulans are mated, the male progeny are inviable, whereas the female progeny display manifold malformations and are sterile. These abnormalities result from genetic incompatibilities accumulated since the time the lineages of the species diverged, and may have their origin in aberrant gene transcription. Because compensatory changes within species may obscure differences at the regulatory level in conventional comparisons of the expression profile between species, we have compared the gene-expression profile of hybrid females with those of females of the parental species in order to identify regulatory incompatibilities. In the hybrid females, we find abnormal levels of messenger RNA for a large fraction of the Drosophila transcriptome. These include a gross underexpression of genes preferentially expressed in females, accompanying gonadal atrophy. The hybrid females also show significant overexpression of male-biased genes, which we attribute to incompatibilities in the regulatory mechanisms that normally act to control the expression of these genes in females. The net result of the multiple incompatibilities is that the gene-expression profiles of the parental females are more similar to each other than either is to that of the hybrid.
Experiments with hybrids (Dickinson 1980; Dickinson et al. 1984; Nielsen et al. 2000) offer an unparalleled opportunity to uncover regulatory incompatibilities that may be overlooked in conventional comparisons of the gene-expression profile between species, owing to compensatory changes that may obscure the existing interspecific differences (Ranz et al. 2003; Rifkin et al. 2003). The reason for this is that different genetic changes, fixed independently in the lineages of the species, may result in a qualitatively and quantitatively similar modification of the level of gene expression relative to the ancestral state, and hence, to one another. In the genetic background of interspecific hybrids, however, these changes may no longer compensate, and the lack of genetic coadaptation will be observed as anomalies in the levels of gene expression.
Interspecific hybrids in Drosophila display diverse types of abnormalities affecting primarily the reproductive system rather than the soma, which usually result in complete sterility (Sturtevant 1920; Bonnier 1924; Kerkis 1933; Civetta and Singh 1998; Hollocher et al. 2000). These abnormalities reflect deleterious genetic interactions as a consequence of independent, noncoordinated changes (Ludwig et al. 2000; Shaw et al. 2002) accumulated between the two gene pools in the absence of gene flow (Dobzhansky 1936; Muller 1942). Some of these changes affect gene regulation (Orr et al. 1997; Skaer and Simpson 2000; Barbash et al. 2003) and may have played a major role in the species evolution (Dickinson and Carson 1979; Dickinson 1980; Kopp et al. 2000; Sucena and Stern 2000; Wittkopp et al. 2002; Sucena et al. 2003). In the hybrids, both allometric differences relative to the parental species and divergence in gene regulation will appear as anomalous patterns of gene expression, which will be reflected as aberrant levels for the affected mRNA species (Dickinson et al. 1984; Skaer and Simpson 2000; Barbash et al. 2003).
Naturally occurring hybrids between Drosophila melanogaster and Drosophila simulans have been reported (Sperlich 1962), although the lineages that led to these species diverged in Africa ∼2.5 million years ago (Mya; Lachaise 1988). The females of these two species are externally indistinguishable, whereas the hybrid females display minor morphological defects in addition to severely reduced gonads (Sturtevant 1919, 1920; Hollocher et al. 2000), whose absence is compensated by hypertrophy of the fat body (Dickinson et al. 1984). In this study, we have used cDNA microarrays to determine how the abnormalities in the hybrid females that arise as a result of interspecific genetic incompatibilities are reflected in the gene-expression profile of the hybrids versus those of the parents. We have found that a remarkable fraction of the Drosophila transcriptome shows a significant effect.
On the other hand, many of the regulatory aberrations observed in the interspecific hybrids can be related to genes that show a sex bias in their expression. An estimated 30%–50% of the Drosophila transcriptome shows sex-biased expression (Jin et al. 2001; Arbeitman et al. 2002; Parisi et al. 2003; Ranz et al. 2003), and sex-biased genes (especially male-biased genes) evolve exceptionally rapidly (Carson 1985; Coulthart and Singh 1988; Civetta and Singh 1999; Wu 2001; Meiklejohn et al. 2003; Michalak and Noor 2003; Ranz et al. 2003). Population theory and experimental work predict that, as the regulation of male-biased genes evolves so rapidly, interspecific hybrids might be expected to exhibit greater regulatory incompatibilities for those genes than for other classes of genes. In the interspecific hybrid females, we have found that male-biased genes, as compared with genes that are not sex-biased, are preferentially overexpressed in comparison with the parental species.
RESULTS AND DISCUSSION
We compared the gene-expression profile for the whole body of the three female genotypes through 21 competitive mRNA hybridizations, and controlled for the effects due to atrophied gonads with 12 additional competitive hybridizations using head mRNA (Fig. 1). Relative levels of gene expression were estimated by means of a Bayesian procedure (Townsend and Hartl 2002; Supplemental Tables 1 and 2 available online at www.genome.org), and the robustness of the results tested in parallel with a mixed-model ANOVA (analysis of variance; Wolfinger et al. 2001; see Methods). Substantial differences in gene expression among the three female genotypes were found both for the whole-body experiments and for those using heads only (Fig. 2). Altogether, ∼89% (3965/4450) of the elements in the whole-body comparisons and ∼54% (2507/4448) of the elements in the head comparisons were significantly different in at least one pairwise comparison. Various factors contribute to the greater number of differences detected with whole-body mRNA as compared with head mRNA. First, the comparison of the expression profile using whole-body mRNA is affected more by the allometric differences between the hybrid females and the females of D. melanogaster and D. simulans than the experiments using head mRNA only (see below). Second, the level of replication and some methodological aspects are different (Methods). Overall, the remarkable number of significant differences in mRNA abundance emphasizes the breakdown of regulatory mechanisms resulting from incompatibilities acquired during the ∼2.5 Mya divergence between the transcriptomes of the females of D. melanogaster and D. simulans.
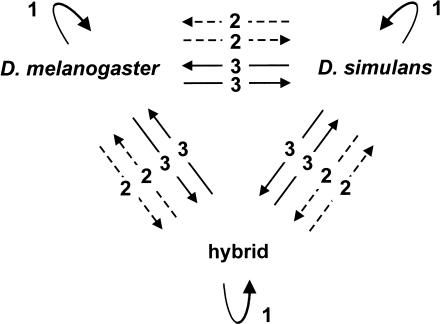
Figure 1.
Set of competitive hybridizations onto cDNA microarrays performed to compare the expression profile among D. melanogaster, D. simulans, and hybrid females. Twenty-one hybridizations were carried out with mRNA from whole flies (solid arrows) and 12 with head-specific amplified mRNA (dashed arrows). Arrowhead, sample labeled with Cy5; base of arrow, sample labeled with Cy3. Each comparison was replicated six times for whole-fly mRNA and four times for head mRNA. Three self-self hybridizations were also carried out for the whole-body experiment. The resulting experimental design was balanced for dyes and samples.
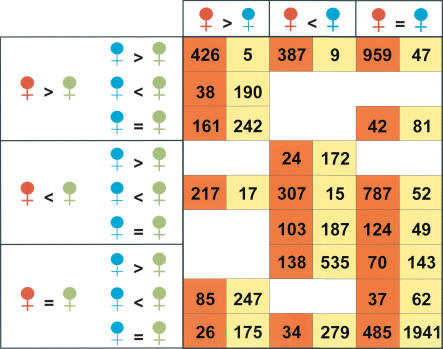
Figure 2.
Classification of the cDNAs assayed according to the relationships at expression level among D. melanogaster (red), D. simulans (blue), and hybrid (green) in whole-body (orange cells) and heads only (yellow cells) experiments. Only the 4450 array elements regarded as consistent with the Bayesian methodology (Townsend and Hartl 2002) in the whole-body analysis have been included (Methods); in the case of the experiments with head mRNA, no reliable estimate was obtained by Bagel for two of the 4450 array elements considered. The logical operators (=, <, >) denote the relationship among the 95% credible intervals of the mean expression levels obtained for the three kind of females. The number that appears in a particular cell shows the total number of genes on our cDNA microarray that meet the three different conditions under which each gene can be classified. The eight empty cells correspond to mutually incompatible requirements. See Supplemental Figure 2 for numerical code of the cells in orange.
In contrast to the expression profiles of the heads, in which the hybrids are often intermediate between the parental species (Fig. 2), pairwise comparisons for the whole body indicate that the parental species are often more similar to each other than either is to the hybrid. Females of the parental species show significant differences for 43.7% (1946 array elements) of their transcriptome, whereas the number of differences for the comparisons D. melanogaster-hybrid and D. simulans-hybrid is 79.5% (3573 elements) and 78.1% (3475 elements), respectively. Two main patterns of misregulation in hybrid females are evident in the whole-body experiments (Fig. 3). The first involves a reduced abundance of transcript in the hybrid for 1772 elements, and the second involves an overabundance of the transcript in the hybrid for 1311 elements. Illustrative examples are shown in Figure 4. In both cases, we find that the magnitude of the difference in mRNA abundance is greater than twofold for approximately one-third of the genes that show statistical significance (Suppl. Fig. 1). Overall, 69.3% of the assayed transcriptome of D. melanogaster and D. simulans displays misregulation in the hybrid females, resulting in either overexpression or underexpression relative to the females of both parental species. An additional 10.9% (487 elements) displays simple dominance, in which the level of expression equals that of one of the parental species. In this case, there is a slight but nonsignificant tendency for the D. simulans level of expression to be dominant (G = 3.5, d.f. = 1, P = 0.06). Unexpectedly, only 1.4% of the genes (62 elements) show an intermediate level of expression relative to those in the parental species.
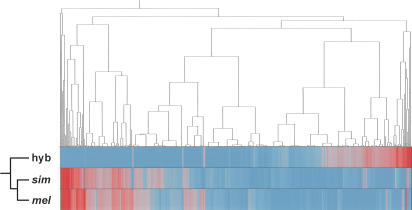
Figure 3.
Hierarchical clustering of the mean levels of expressions estimated using a Bayesian methodology (BAGEL) for the whole-body experiments. Underexpressed genes (blue); overexpressed genes (red). Ward's minimum variance was used as a distance metric. The dendrogram includes 3965 genes that showed significantly different expression levels in at least one of the three pairwise comparisons between females. (mel) D. melanogaster; (sim) D. simulans; (hyb) hybrid.
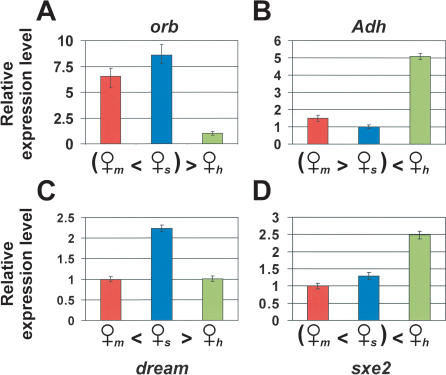
Figure 4.
Patterns of misregulation between hybrids and females of D. melanogaster and D. simulans in whole-body experiments. (A) orb, a gene underexpressed in the hybrid and required for correct oogenesis in a normal female. (B) Adh, a gene overexpressed in the hybrid that is highly expressed in the fat body (Dickinson et al. 1984). (C) dream, a gene for which the allele of D. melanogaster is dominant over that of D. simulans, which involves a higher level of expression. (D) sxe2, a gene overexpressed in the hybrid that is expressed in the brain of males only (Fujii and Amrein 2002). Female symbol subscripts indicate the genotype; (m) D. melanogaster; (s) D. simulans; (h) hybrid. Error bars indicate the 95% credible intervals around the estimated mean expression level. Three pairwise comparisons are possible among the three female genotypes. Differences in level of gene expression are regarded to be statistically significant if the 95% credible intervals of the estimates failed to overlap. (=) No significant difference; (>) significant overexpression.
Using the currently available functional annotation of the Drosophila genome, we performed a systematic analysis of the underexpressed and overexpressed genes to group the misregulated genes into major functional categories. Underexpression of genes in the hybrid is highly correlated with the process of oogenesis (Table 1 for some examples and Suppl. Table 3 for detailed list), which is consistent with the profound reduction of germ-line tissues in the atrophied ovaries of the hybrid females (Sturtevant 1920; Hollocher et al. 2000). Because most female-biased genes are inferred to be expressed preferentially in the gonads (Arbeitman et al. 2002; Parisi et al. 2003), it is expected that the majority of female-biased genes will be underexpressed in their hybrids. Accordingly, we examined those genes classified previously as female-biased in gene expression both in D. melanogaster and D. simulans using the same array platform (Ranz et al. 2003). Among 1194 genes considered female-biased under conservative criteria (Table 2), 84.7% show underexpression in the hybrid females, 0.7% overexpression, and 14.6% other patterns (Gadj = 1052, d.f. = 2, P = 3.2 × 10–229). In good agreement with the expectation, only 1.2% of the female-biased genes that are significantly underexpressed in the whole-body comparisons are also significantly underexpressed in the heads.
Table 1.
Examples of Functional Categories Found to be Transcriptionally Enriched in the Samples of Genes Under- and Overexpressed in Hybrid Females of D. melanogaster and D. simulans
| Functional category (GMRC terma, P valueb) | |
|---|---|
| Underexpressed Genes | |
| Metabolic and Regulatory Pathway | Transcription factors (dme03022; P = 2.9 × 10-3) |
| Molecular Function | Nucleic acid binding activity (GO:0003676; P = 6.6 × 10-13) |
| RNA binding activity (GO:0003723; P = 4.6 × 10-4) | |
| Helicase activity (GO:0004386; P = 3.3 × 10-2) | |
| Biological Process | DNA metabolism (GO:0006259; P = 1.1 × 10-9) |
| Cell growth and/or maintenance (GO:0008151; P = 8.8 × 10-7) | |
| Transcription (GO:0006350; P = 4.1 × 10-5) | |
| Cellular Component | Nucleus (GO:0005634; P = 2.0 × 10-25) |
| Replication fork (GO:0005657; P = 5.0 × 10-6) | |
| Transcription factor complex (GO:0005667; P = 2.8 × 10-4) | |
| Overexpressed Genes | |
| Metabolic and Regulatory Pathway | Oxidative phosphorylation (dme00190; P = 1.6 × 10-7) |
| Fatty acid metabolism (dme00071; P = 4.6 × 10-6) | |
| Glycolysis/Gluconeogenesis (dme00010; P = 6.3 × 10-6) | |
| Molecular Function | Oxireductase activity (GO:0016491; P = 2.8 × 10-10) |
| Serine-type peptidase activity (GO:0008236; P = 1.3 × 10-7) | |
| Hydrolase activity (GO:0016787; P = 2.8 × 10-2) | |
| Biological Process | Ion transporter (GO:0006811; P = 4.5 × 10-4) |
| Defense response (GO:0006952; P = 5.8 × 10-3) | |
| Pathways of carbohydrate metabolism (GO:0006092; P = 7.7 × 10-3) | |
| Cellular Component | Membrane (GO:0016020; P = 8.2 × 10-12) |
| Microsome (GO:0005792; P = 3.2 × 10-6) | |
| Mitochondrial electron transport chain (GO:0005746; P = 4.7 × 10-3) |
GMRC, Gene Merge Class (see Methods).
After Bonferroni correction.
Table 2.
Relationship Between Sex Bias in Gene Expression in Normal Individuals and Misregulation in Hybrids
| Misregulation in the hybrid | % female-biased | Sex bias in gene expression % male-biased | % nonsex-biased |
|---|---|---|---|
| Underexpression | 84.5 (+45)/93.9 (+54.4) | 4.7 (-19.2)/1.8 (-22) | 10.8 (-25.9)/4.3 (-32.3) |
| Overexpression | 1.1 (-38.4)/0.3 (-39.1) | 54.6 (+30.9)/58.3 (+34.5) | 44.3 (+7.6)/41.3 (+4.6) |
Before the slash, percentage of genes regardless of the level of misregulation in the hybrid in relation to the parental species; after the slash, percentage of misregulated genes with a difference in gene expression >2. In parenthesis, difference between the observed and the expected percentage of genes in each category according to its reported representation on the array (Ranz et al. 2003): female-biased (39.5%); male-biased (23.8%); nonsex-biased (36.7%). Classification in one particular category involves that the same tendency was found for both D. melanogaster and D. simulans. Differences in expression between the hybrid and the parental species must be significant at P < 0.01, both with BAGEL and with the mixed-model ANOVA, for a gene to be included in the analysis. Also, only genes from Ranz et al. (2003) regarded here as consistent (see Methods) are included in the analysis. Number of underexpressed genes = 1194; number of overexpressed genes = 948; number of underexpressed genes above twofold = 603; number of overexpressed genes above twofold = 383.
Among overexpressed genes, the functional classes that stand out include genes involved in diverse metabolic pathways (Table 1 for some examples, and Suppl. Table 4 for detailed list), some fraction of which reflects the hypertrophy of different tissues, the fat body in particular, which allometrically compensates for the minute gonads (Dickinson et al. 1984). The fat body serves as a nutritional reservoir and also acts as an endocrine organ involved in the innate immune response in insects (Miller et al. 2002), which is also consistent with certain functional classes of genes that are found to be transcriptionally enriched. On the other hand, overexpressed genes exhibit a nonuniform association with the reported patterns of gene expression by sex (Jin et al. 2001; Arbeitman et al. 2002; Parisi et al. 2003; Ranz et al. 2003). In particular, a disproportionate number of overexpressed genes in hybrid females are male-biased in expression in both D. melanogaster and D. simulans, relative to genes without sex bias (Ranz et al. 2003). Accordingly, we found an excess over expectation of four times more male-biased genes than nonsex-biased genes, and this ratio increases to eight when only genes with a difference in expression above twofold are considered (Table 2). Among 938 genes classified as overexpressed under conservative criteria (Table 2), 518 are male-biased in their expression in both parental species as against a random expectation of 367, whereas 420 show no sex bias in the parental species as against a random expectation of 571 (Gadj = 98.9, d.f. = 1, P = 2.6 × 10–23). Examples of this tendency are sxe2 (Fig. 4D) and to, which are genes preferentially expressed in the brain of males, and have an important role in sexspecific behavior (Dauwalder et al. 2002; Fujii and Amrein 2002).
Although additional, systematic tissue-specific analysis would be informative, even experiments with whole-body mRNA compared with head mRNA demonstrate variation in the breakdown of the expression network across tissues. Many of the underexpressed genes in whole-body mRNA experiments can be attributed to allometric differences associated with the atrophy of the ovaries, which may result from the early failure in development of a few key genes. Nevertheless, in the case of the overexpressed genes in the hybrids, allometric differences alone are not enough to account for the disproportionate fraction of overexpressed genes in body tissues that are male-biased in their expression. Two, not mutually exclusive causes can explain the observed pattern for this class of genes. First, there could be a general breakdown of the regulatory mechanisms that normally act to repress excess transcription of male-biased genes in females. Alternatively, some regulatory factors may have a dominant, collateral enhancement on the expression of genes from the other species.
Unless hybrid-rescue mutant strains are used (Hutter 1997), the lethality of hybrid males between D. melanogaster and D. simulans wild types precludes examination of the expression pattern of male-biased genes in hybrid males. Yet, the significant excess of male-biased genes showing overexpression in the F1 progeny of intraspecific crosses in D. melanogaster suggests that hybrid males may also show this phenomenon (G. Gibson, R. Riley-Berger, L. Harshman, A. Kopp, S. Vacha, S. Nuzhdin, M. Wayne, unpubl.). So far in Drosophila, information relative to the expression profile in hybrid males has been obtained for the pairs of species D. simulans–D. mauritiana (Michalak and Noor 2003) and D. pseudoobscura–D. persimilis (Reiland and Noor 2002). For the first pair, hundreds of genes were found to be misregulated in the hybrid males relative to the parental species. In that group of genes, those related with male reproduction appear to be more common, and in contrast with the results of the present findings in females, underexpression seems to be rule. This same pattern of misregulation was also found for one male-specific gene in the hybrids of D. pseudoobscura–D. persimilis (Reiland and Noor 2002). The fact that females were analyzed in the present study, that all the species pairs examined represent different phylogenetic distances, and that there are some methodological differences in the comparisons might account for the somewhat different patterns of misregulation found in the different studies.
The present results between D. melanogaster and D. simulans agree with the results obtained in hybrids of more recently diverged species (Reiland and Noor 2002; Michalak and Noor 2003), supporting the inference that male-biased genes are preferentially affected by regulatory meltdown resulting from genetic incompatibilities between species. These results serve to emphasize the importance of rapid evolution of male-biased genes in the divergence between species and the acquisition of reproductive isolation (Singh and Kulathinal 2000; Wu 2001).
METHODS
Flies
D. melanogaster Canton S and D. simulans Sim-1 inbred strains were maintained at 25°C on glucose–cornmeal–yeast medium. Males and virgin females were collected for the interspecific and conspecific crosses. All crosses were performed at 18°C using seven to eight individuals from each sex. Virgin females from the parental species, as well as hybrid females were stored separately for 5–6 d at room temperature and then snap frozen in liquid nitrogen invariably at the same time of the day in a time window of 2 h. For each cross, two independent cohorts were used as the source of mRNA.
cDNA Microarrays
Microarrays based on the Drosophila Gene Collection version 1.0 of expressed sequence tags (Rubin et al. 2000) were constructed as described in Ranz et al. (2003). A total of 4776 clones were appropriate for analysis. The suitability of the D. melanogaster cDNA microarrays to compare the expression profile of closely related species of the melanogaster species subgroup has been demonstrated by competitive hybridizations using genomic DNA (Ranz et al. 2003), which is in good agreement with the absence of a significant bias in the number of genes preferentially overexpressed in D. melanogaster versus D. simulans (Ranz et al. 2003; this study).
mRNA Extraction, Hybridizations, and Imaging
The mRNA extractions were carried out from adult female progeny of two independent sets of crosses grown and collected at different times in order to incorporate among-culture variation into the hybridizations. The extraction procedure was as described in Ranz et al. (2003). For head-specific mRNA, we used an amplification technique resulting in linear increase in the amount of each mRNA in the sample (Baugh et al. 2001). Several amplifications using one or two heads each were carried out and pooled. Apart from the extraction technique, the only difference between the experiments was the amount of mRNA hybridized, 2 μg in the case of mRNA from the whole body and 3 μg in the case of mRNA from the amplification of heads. Hybridizations and imaging were done as in Ranz et al. (2003). Raw microarray data is available at the Gene Expression Omnibus database (Edgar et. al. 2002) under accession nos. GSM14975–GSM15007.
Data Analysis
Relative level of expression of each gene in each competitive hybridization was estimated using the ratio between the fluorescence intensity of the channels for the corresponding array element. A Bayesian method (BAGEL, software version 2.4; Townsend and Hartl 2002) was used to estimate the mean expression level and 95% credible interval for each gene. BAGEL assumes that the ratio of normalized expression levels in any hybridization is distributed as the ratio of two suitably truncated normal distributions. A difference in level of gene expression was judged to be statistically significant if the 95% credible intervals of the estimates failed to overlap. Ten independent Markov chain Monte Carlo trajectories were simulated per gene. A comparison of gene-expression levels was regarded as statistically significant only if the 95% credible intervals were nonoverlapping in at least 9 of the 10 trajectories. Among the 4450 elements meeting this criterion, 99.8% of the significant differences showed nonoverlapping credible intervals in all 10 trajectories (those referred to as consistent in Suppl. Table 1). To estimate the rate of false positives, we created 10 independent data sets by randomizing with replacement the expression ratios for each gene across hybridizations. These randomized data sets were analyzed with BAGEL. On average, among 4745 elements tested, 248.2 ± 15.2 (5.2% ± 0.3%) were statistically significant under our criterion of nonoverlapping 95% credible intervals.
To evaluate the robustness of the analysis, we also carried out a two-step mixed-model analysis of variance (Jin et al. 2001; Wolfinger et al. 2001) and obtained similar results. In the first step, log2-transformed raw fluorescent intensities were centered to the mean of each hybridization by capturing the residuals after fitting dye, array, and array by dye interaction effects. These relative fluorescence intensities were then subject to gene-specific models with Genotype (melanogaster, simulans, or hybrid) and Dye as fixed terms, and Array as a normally distributed random effect (to control for correlation between dyes on each array). Pairwise differences among parental species and hybrids were computed with the Diffs option in PROC MIXED in SAS Version 8.0 (Cary, NC).
Comparison of the classification of the genes at different significant threshold probabilities indicated that the number of spots with exactly the same relationships at expression level among the three female genotypes, and therefore occupying the same cell in Figure 2, was 65.2% at P < 0.05, 67.5% at P < 0.01, and 69.5% at P < 0.001 (for example, see Suppl. Table 1 for P < 0.01). For those spots that differ in their classification between methodologies, most differences were between adjacent cells in Figure 2, that is two of the three pairwise relationships among the different females were coincidental. Dramatic changes, such as being classified as overexpressed by one methodology and as underexpressed by the other were infrequent. For example, at P < 0.01, and according to the number of coincidences in the way a gene is classified for the three comparisons performed, the results are 3003 array elements with three coincidences, 1118 elements with two coincidences, 319 elements with one coincidence, and 10 elements with no coincidence (the number of coincidences is corresponded to the nomenclature used for classifying the elements on the array in Suppl. Table 1, namely, “identical”, “similar”, and “discordant”, respectively). The disparities found may stem from the different underlying assumptions of the statistical frameworks. Whereas both methods model the variance of each gene independently, the Bayesian methodology used does not necessarily assume that the variance at the level of gene expression is the same for all of the nodes (samples) under comparison, whereas the mixed-model analysis of variance does so and uses a centering step to control for dye and array effects. Yet, the relative number of array elements found to exhibit one particular pattern of expression among the three female genotypes is not affected by those few disparities. Accordingly, the product-moment correlation coefficient between both statistical frameworks in the number of array elements assigned to each of the categories in Figure 2 was r = 0.97, P = 1.0 × 10–11 at P < 0.05; r = 0.98, P = 3.9 × 10–13 at P < 0.01; and r = 0.98, P = 1.5 × 10–13 at P < 0.001.
Functional Analysis
Significant overrepresentation of particular functional categories among differentially expressed genes was determined using the software GeneMerge with corrections for multiple tests (Castillo-Davis and Hartl 2003). The functional categories scrutinized are molecular function, biological process, and cellular component as defined in the Gene Ontology Consortium database (http://www.geneontology.org), as well as regulatory pathway as defined in the Kyoto Encyclopaedia of Genes and Genomes database (http://www.genome.ad.jp/kegg). GO and Kegg terms are denominated as GeneMeRGe terms (GMRG) in the outputs of GeneMerge (Suppl. Tables 3 and 4). Given the found rate of false positives, we only considered genes whose under- or overexpression in the hybrid females in relation to both parental species was significant at P < 0.01 by both the Bayesian methodology (Townsend and Hartl 2002) and the mixed-model ANOVA (Wolfinger et al. 2001), that is, 1419 underexpressed genes and 1214 overexpressed genes after removing redundancy from duplicated clones, controls, and few clones with no FlyBase qualifier.
Acknowledgments
We thank the staff of Bauer Center for Genomic Research of Harvard University for material and technical help and M. Ashburner, A. Berry, C. Machado, M. Noor, J. Roote, one anonymous reviewer, and members from Hartl and Wakeley laboratories for valuable comments on the manuscript. Work funded by a post-doctoral fellowship from the Ministerio de Ciencia y Tecnología (J.M.R.) and a National Institute of Health grant (D.L.H.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2019804. Article published online before print in February 2004.
Footnotes
References
- Arbeitman, M.N., Furlong, E.E., Iman, F., Johnson, E., Null, B.H., Baker, B.S., Krasnow, M.A., Scott, M.P., and White, K.P. 2002. Gene expression during the life cycle of Drosophila melanogaster. Science 297: 2270–2275. [DOI] [PubMed] [Google Scholar]
- Barbash, D.A., Siino, D.F., Tarone, A.M., and Roote, J. 2003. A rapidly evolving MYB-related protein causes species isolation in Drosophila. Proc. Natl. Acad. Sci. 100: 5302–5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh, L.R., Hill, A.A., Brown, E.L., and Hunter, C.P. 2001. Quantitative analysis of mRNA amplification by in vitro transcription. Nucleic Acids Res. 29: E29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnier, G. 1924. Contributions to the knowledge of intra- and interspecific relationships in Drosophila. Acta Zool. 5: 1–122. [Google Scholar]
- Carson, H.L. 1985. Unification of speciation theory in plants and animals. Syst. Bot. 10: 380–390. [Google Scholar]
- Castillo-Davis, C.I. and Hartl, D.L. 2003. GeneMerge—Post-genomic analysis, data mining, and hypothesis testing. Bioinformatics 19: 891–892. [DOI] [PubMed] [Google Scholar]
- Civetta, A. and Singh, R.S. 1998. Sex and speciation: Genetic architecture and evolutionary potential of sexual versus nonsexual traits in the sibling species of the Drosophila melanogaster complex. Evolution 52: 1080–1092. [DOI] [PubMed] [Google Scholar]
- Civetta, A. and Singh, R.S. 1999. Broad-sense sexual selection, sex gene pool evolution, and speciation. Genome 42: 1033–1041. [PubMed] [Google Scholar]
- Coulthart, M.B. and Singh, R.S. 1988. High level of divergence of male-reproductive tract proteins, between Drosophila melanogaster and its sibling species, D. simulans. Mol. Biol. Evol. 5: 182–191. [DOI] [PubMed] [Google Scholar]
- Dauwalder, B., Tsujimoto, S., Moss, J., and Mattox, W. 2002. The Drosophila takeout gene is regulated by the somatic sex-determination pathway and affects male courtship behavior. Genes & Dev. 16: 2879–2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson, W.J. 1980. Regulation of the tissue specificity of enzyme expression by diffusible factors: Evidence in Drosophila hybrids. Science 207: 995–997. [DOI] [PubMed] [Google Scholar]
- Dickinson, W.J. and Carson, H.L. 1979. Regulation of the tissue specificity of an enzyme by a cis-acting genetic element: Evidence from interspecific Drosophila hybrids. Proc. Nat. Acad. Sci. 76: 4559–4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson, W.J., Rowan, R.G., and Brennan, M.D. 1984. Regulatory gene evolution: Adaptive differences in expression of alcohol dehydrogenase in Drosophila melanogaster and Drosophila simulans. Heredity 52: 215–225. [DOI] [PubMed] [Google Scholar]
- Dobzhansky, T. 1936. Studies on hybrid sterility. II. Localization of sterility factors in Drosophila pseudoobscura hybrids. Genetics 21: 113–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar, R., Domrachev, M., and Lash, A.E. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30: 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii, S. and Amrein, H. 2002. Genes expressed in the Drosophila head reveal a role for fat cells in sex-specific physiology. EMBO J. 21: 5353–5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollocher, H., Agopian, K., Waterbury, J., O'Neill, R.W., and Davis, A.W. 2000. Characterization of defects in adult germline development and oogenesis of sterile and rescued female hybrids in crosses between Drosophila simulans and Drosophila melanogaster. J. Exp. Zool. 288: 205–218. [DOI] [PubMed] [Google Scholar]
- Hutter, P. 1997. Genetics of hybrid inviability in Drosophila. Adv. Genet. 36: 157–185. [DOI] [PubMed] [Google Scholar]
- Jin, W., Riley, R.M., Wolfinger, R.D., White, K.P., Passador-Gurgel, G., and Gibson, G. 2001. The contributions of sex, genotype and age to transcriptional variance in Drosophila melanogaster. Nat. Genet. 29: 389–395. [DOI] [PubMed] [Google Scholar]
- Kerkis, J. 1933. Development of gonads in hybrids between Drosophila melanogaster and Drosophila simulans. J. Exp. Zool. 66: 477–509. [Google Scholar]
- Kopp, A., Duncan, I., Godt, D., and Carroll, S.B. 2000. Genetic control and evolution of sexually dimorphic characters in Drosophila. Nature 408: 553–559. [DOI] [PubMed] [Google Scholar]
- Lachaise, D., Cariou, M.-L., David, J.R., Lemeunier, F., Tsacas, L., and Ashburner, M. 1988. Historical biogeography of the Drosophila melanogaster subgroup. Evol. Biol. 22: 159–225. [Google Scholar]
- Ludwig, M.Z., Bergman, C., Patel, N.H., and Kreitman, M. 2000. Evidence for stabilizing selection in a eukaryotic enhancer element. Nature 403: 564–567. [DOI] [PubMed] [Google Scholar]
- Meiklejohn, C.D., Parsch, J., Ranz, J.M., and Hartl, D.L. 2003. Rapid evolution of male-biased gene expression in Drosophila. Proc. Natl. Acad. Sci. 100: 9894–9899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalak, P. and Noor, M.A. 2003. Genome-wide patterns of expression in Drosophila pure species and hybrid males. Mol. Biol. Evol. 20: 1070–1076. [DOI] [PubMed] [Google Scholar]
- Miller, J.M., Oligino, T., Pazdera, M., López, A.J., and Hoshizaki, D.K. 2002. Identification of fat-cell enhancer regions in Drosophila melanogaster. Insect Mol. Biol. 11: 67–77. [DOI] [PubMed] [Google Scholar]
- Muller, H.J. 1942. Isolating mechanisms, evolution, and temperature. Biol. Symp. 6: 71–125. [Google Scholar]
- Nielsen, M.G., Wilson, K.A., Raff, E.C., and Raff, R.A. 2000. Novel gene expression patterns in hybrid embryos between species with different modes of development. Evol Dev. 2: 133–144. [DOI] [PubMed] [Google Scholar]
- Orr, H.A., Madden, L.D., Coyne, J.A., Goodwin, R., and Hawley, R.S. 1997. The developmental genetics of hybrid inviability: A mitotic defect in Drosophila hybrids. Genetics 145: 1031–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi, M., Nuttall, R., Naiman, D., Bouffard, G., Malley, J., Andrews, J., Eastman, S., and Oliver, B. 2003. Paucity of genes on the Drosophila X chromosome showing male-biased expression. Science 299: 697–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranz, J.M., Castillo-Davis, C.I., Meikeljohn, C.D., and Hartl, D.L. 2003. Sex-dependent gene expression and evolution of the Drosophila transcriptome. Science 300: 1742–1745. [DOI] [PubMed] [Google Scholar]
- Reiland, J. and Noor, M.A.F. 2002. Little qualitative RNA misexpression in sterile male F1 hybrids of Drosophila pseudoobscura and D. persimilis. BMC Evol. Biol. 2: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rifkin, S.A, Kim, J., and White, K.P. 2003. Evolution of gene expression in the Drosophila melanogaster subgroup. Nat. Genet. 33: 138–144. [DOI] [PubMed] [Google Scholar]
- Rubin, G.M., Hong, L., Brokstein, P., Evans-Holm, M., Frise, E., Stapleton, M., and Harvey, D.A. 2000. A Drosophila complementary DNA resource. Science 287: 2222–2224. [DOI] [PubMed] [Google Scholar]
- Shaw, P.J., Wratten, N.S., McGregor, A.P., and Dover, G.A. 2002. Coevolution in bicoid-dependent promoters and the inception of regulatory incompatibilities among species of higher Diptera. Evol. Dev. 4: 265–277. [DOI] [PubMed] [Google Scholar]
- Singh, R.S. and Kulathinal, R.J. 2000. Sex gene pool evolution and speciation: A new paradigm. Genes Genet. Syst. 75: 119–130. [DOI] [PubMed] [Google Scholar]
- Skaer, N. and Simpson, P. 2000. Genetic analysis of bristle loss in hybrids between Drosophila melanogaster and D. simulans provides evidence for divergence of cis-regulatory sequences in the achaete-scute gene complex. Dev. Biol. 221: 148–167. [DOI] [PubMed] [Google Scholar]
- Sperlich, D. 1962. Hybrids between Drosophila melanogaster and D. simulans in nature. Drosoph. Inf. Serv. 36: 118. [Google Scholar]
- Sturtevant, A.H. 1919. A new species closely resembling Drosophila melanogaster. Psyche 26: 153–155. [Google Scholar]
- Sturtevant, A.H. 1920. Genetic studies on Drosophila simulans. I. Introduction: Hybrids with Drosophila melanogaster. Genetics 5: 488–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sucena, E. and Stern, D.L. 2000. Divergence of larval morphology between Drosophila sechellia and its sibling species caused by cis-regulatory evolution of ovo/shaven-baby. Proc. Natl. Acad. Sci. 97: 4530–4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sucena, E., Delon, I., Jones, I., Payre, F., and Stern, D.L. 2003. Regulatory evolution of shavenbaby/ovo underlies multiple cases of morphological parallelism. Nature 424: 935–938. [DOI] [PubMed] [Google Scholar]
- Townsend, J.P. and Hartl, D.L. 2002. Bayesian analysis of gene expression levels: Statistical quantification of relative mRNA level across multiple strains or treatments. Genome Biol. 3: RESEARCH0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp, P.J., Vaccaro, K., and Carroll, S.B. 2002. Evolution of yellow gene regulation and pigmentation in Drosophila. Curr. Biol. 12: 1547–1556. [DOI] [PubMed] [Google Scholar]
- Wolfinger, R.D., Gibson, G., Wolfinger, E.D., Bennett, L., Hamadeh, H., Bushel, P., Afshari, C., and Paules, R.S. 2001. Assessing gene significance from cDNA microarray expression data via mixed models. J. Comput. Biol. 8: 625–637. [DOI] [PubMed] [Google Scholar]
- Wu, C.-I. 2001. Genes and speciation. J. Evol. Biol. 6: 899–891. [Google Scholar]
WEB SITE REFERENCES
- http://www.geneontology.org; Gene Ontology Consortium database.
- http://www.genome.ad.jp/kegg; the Kyoto Encyclopaedia of Genes and Genomes database.