Abstract
Background
Since the detection of MRSA CC398 in pigs in 2004, it has emerged in livestock worldwide. MRSA CC398 has been found in people in contact with livestock and thus has become a public health issue. Data from a large-scale longitudinal study in two Danish and four Dutch pig herds were used to quantify MRSA CC398 transmission rates within pig herds and to identify factors affecting transmission between pigs.
Results
Sows and their offspring were sampled at varying intervals during a production cycle. Overall MRSA prevalence of sows increased from 33% before farrowing to 77% before weaning. Overall MRSA prevalence of piglets was > 60% during the entire study period. The recurrent finding of MRSA in the majority of individuals indicates true colonization or might be the result of contamination. Transmission rates were estimated using a Susceptible-Infectious-Susceptible (SIS-)model, which resulted in values of the reproduction ratio (R0) varying from 0.24 to 8.08. Transmission rates were higher in pigs treated with tetracyclins and β-lactams compared to untreated pigs implying a selective advantage of MRSA CC398 when these antimicrobials are used. Furthermore, transmission rates were higher in pre-weaning pigs compared to post-weaning pigs which might be explained by an age-related susceptibility or the presence of the sow as a primary source of MRSA CC398. Finally, transmission rates increased with the relative increase of the infection pressure within the pen compared to the total infection pressure, implying that within-pen transmission is a more important route compared to between-pen transmission and transmission through environmental exposure.
Conclusion
Our results indicate that MRSA CC398 is able to spread and persist in pig herds, resulting in an endemic situation. Transmission rates are affected by the use of selective antimicrobials and by the age of pigs.
Keywords: Methicillin resistant, Staphylococcus aureus, Transmission, Pigs, MRSA, Reproduction ratio
Background
In 2004, a distinct clone of methicillin resistant Staphylococcus aureus (MRSA CC398), referred to as livestock-associated (LA), was found in pigs and in people in contact with pigs [1]. Various observational studies have detected LA-MRSA in pig and other livestock herds worldwide, and risk factors for herds to be MRSA positive have been identified [2-7].
Antimicrobial resistant microorganisms in livestock become a public health issue when resistant organisms or resistance genes can transfer from livestock to humans. The role of animal populations in the transmission of microorganisms to humans is not only dependent on the possibility of transmission from animals to humans, but also on the possibility of transmission between animals.
The primary route of MRSA transmission between humans seems to be direct contact with individuals carrying MRSA [8,9]. However, environmental spread might be a substantially underestimated route for MRSA transmission in hospitals [10,11]. Similar mechanisms are likely for MRSA transmission between pigs. MRSA is not only isolated from pig mucosa and skin, but also from the herd environment [6,7,12], indicating that both direct and indirect transmission can occur. Little is known about MRSA transmission within pig herds and about the colonization dynamics of individual pigs over time. A single study in one pig herd assessed MRSA colonization in piglets over time and showed age-related differences in MRSA prevalence in young pigs (< 10 weeks) [13].
Transmission can be measured in longitudinal field studies and experiments, and can be expressed with the reproduction ratio (R0), which is an essential parameter in management of diseases. R0 is defined as the average number of secondary cases caused by one typical infectious individual during its entire infectious period in a completely susceptible population, and is often used as a quantitative measure of transmission [14,15]. R0 has a threshold value of 1; if R0 > 1, minor and major outbreaks can occur and an endemic situation can be established and maintained, whereas when R0 < 1 an infection does not spread and will not become endemic, i.e. the infection will fade out [15,16].
The objectives of this study were to quantify MRSA transmission rates and routes within pig herds and to identify factors affecting MRSA transmission between pigs. These objectives were obtained by a large-scale longitudinal study conducted in two Danish and four Dutch pig herds.
Methods
Selection of herds and sampling
Six farrow-to-finish herds, confirmed MRSA positive, were selected by convenience: two Danish herds (DK1, DK2) selected from a Danish pilot study [17], and four Dutch herds (NL1, NL2, NL3, NL4) selected from a cross-sectional prevalence study [7]. At each herd, one cohort of pregnant sows approaching delivery and placed in the same farrowing compartment was selected for sampling, except for herd NL4, where two cohorts of sows were included (NL4a and NL4b) with a time interval of three months.
Sows and their offspring were sampled six times during a production cycle (Table 1). Nasal swabs were taken from all pigs present at all sampling moments. Additionally, vaginal swabs were taken from the sows present at all moments, and rectal swabs were taken from new born piglets. At all sampling moments, four or five environmental wipes (Sodibox, s1 kit ringer solution, France) were taken from surfaces of each selected herd compartment.
Table 1.
Sampling moment, compartment, time in life, age group and sample type per herd
| Moment1) | Compartment | Approximate time in life | Age group |
Type of sample |
|
|---|---|---|---|---|---|
| Sows | Pigs | ||||
| 1 |
Farrowing |
1 week before farrowing |
Sows |
Nasal/vaginal |
- |
| 2 |
Farrowing |
3 days after farrowing/birth |
Sows/piglets |
Nasal/vaginal |
Nasal/rectal |
| 3 |
Farrowing |
3 wks after farrowing/birth |
Sows/piglets |
Nasal/vaginal |
Nasal |
| 4 |
Weaning2) |
6 wks after birth |
Pigs |
- |
Nasal |
| 5 |
Weaning |
10 wks after birth |
Pigs |
- |
Nasal |
| 6 | Finishing3) | 25 wks after birth | Pigs | - | Nasal |
1) Each sampling moment, 4–5 environmental wipes were taken in each compartment.
2) Pigs were moved to the weaning section approximately 4 weeks after birth.
3) Pigs were moved to the finishing section approximately 11 weeks after birth.
In total, 63 sows and their offspring were included in the study. The number of sows and piglets at each sampling moment varied depending on the number of pigs born alive and/or on movement or death of pigs. After weaning, sows returned to the breeding compartment and were omitted from further sampling. The cohorts of pigs were monitored until slaughtering time. Some pigs were lost for follow up due to sorting and mixing of pigs to other compartments. In one Dutch herd (NL1), no samples were collected at the last two sampling moments, because the pigs were moved to another location.
The study protocol was in accordance with the Dutch Law on Animal Health and Welfare and discussed with the Animal Welfare Officer of Wageningen University. The distress was considered below the European injection criterium and therefore no further approval of the Animal Welfare Commission was needed. Informed consent was obtained from each participating farmer.
Microbiological analysis
Samples were enriched using Mueller Hinton Broth with 6.5% NaCl (MHB+). Nasal, vaginal and rectal swabs were placed into 5–10 ml MHB+, environmental wipes into 100 ml MHB+. After 18 h of aerobic incubation at 37°C, a loop-full of MHB + was spread onto a chromogenic MRSA Brilliance agar (Oxoid, PO5196A, UK). One suspected colony per sample was confirmed to be MRSA CC398 by PCR [18,19].
Data analysis
At each sampling moment, prevalences of positive individuals were calculated. An individual was considered positive if either one of the swabs (nasal, rectal or vaginal) tested positive for MRSA. The exact confidence intervals (exact 95% CI) for these prevalences were calculated based on the binomial probability function (PROC FREQ) [20].
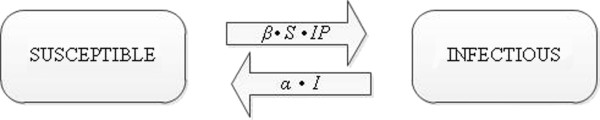
A susceptible-infectious-susceptible (SIS) model [16] was used to describe the transmission of LA-MRSA within herds. For each moment, pigs were classified infectious (I) if either one of the swabs tested positive for MRSA. Pigs were classified susceptible (S) if all swabs tested negative for MRSA. New born piglets were assumed to be MRSA negative, thus susceptible, at birth. Three sources of MRSA for a susceptible pig were allocated in our study: i) infectious individuals, including sows, within the pen; ii) infectious individuals, including sows, within the compartment (but not in the same pen); and iii) the environment of the compartment. The term total infection pressure (IP) was introduced and defined as the sum of the proportion of infectious pigs (piglets + sows) within the pen (IP within the pen), the proportion of infectious pigs (piglets + sows) within the compartment, but not in the same pen (IP other pens), and the proportion of positive environmental wipes (IP environment).
The SIS-model can then be represented in Figure 1.
Figure 1.
SIS-model used to quantify MRSA transmission rates.
In this model, β is the transmission parameter, defined as the number of secondary cases (C) out of a number of susceptible individuals (S) caused by a certain infection pressure (IP) during each time interval between samplings (Δt). The number of new cases (C) per time interval (Δt) depends directly on β, S and IP; infectious individuals (I) become susceptible again at recovery rate α. C, I, S and Δt were determined per pen. Underlying assumptions were that within a pen (1) all pigs were randomly in contact, (2) susceptible and infectious individuals were homogenous groups, i.e. all individuals were equally susceptible or infectious, and (3) infectious individuals were equally infectious over time.
The probability for each piglet to become MRSA positive during a time period Δt depends on the transmission rate β and the infectiousness present in their surroundings which we defined as IP. The probability for each piglet to become MRSA positive is therefore equal to:
From this probability, it can be shown that the number of new cases (C) in a period Δt follows a binomial distribution with parameter and index S, the number of susceptible individuals (in our case MRSA negative animals) at the start of each time period (Δt). Consequently, the relation between the expected number of cases per unit of time (E(C)) and transmission rate β, IP and the number of susceptibles is as follows (see Velthuis et al. for a detailed explanation [21]):
Our data (summarized in Table 2) were statistically analysed with SAS® 9.1 using Generalized Linear Models (described by McCullagh and Nelder [22]; PROC GENMOD [20]) with a complementary log-log link function, the term log (IP ·Δt) as offset variable, C as the number of new cases, and S as the number of trials in the binomial process. Use of such generalized linear models using an offset as explanatory variable on infectious disease data is described by Becker [23] in more detail. The relation between the expected value (E) of a number of new cases out of a number of susceptibles during a time period Δt is presented in the following basic statistical model:
Table 2.
Summarized model input per herd for piglets only used for estimation of MRSA transmission parameters
| Herd | Interval1) | Infectious (n/compartment) | Susceptible (n/compartment) | Cases (n/compartment) | Δt(days) | Pens2)(n) | Risk ab3)(n pens) | pIP(range) |
|---|---|---|---|---|---|---|---|---|
| NL1 |
I |
0 |
199 |
85 |
4 |
16 |
9 |
0.00-0.47 |
| |
II |
34 |
56 |
50 |
14 |
10 |
4 |
0.28-0.34 |
| |
III |
92 |
11 |
11 |
21 |
5 |
5 |
0.33-0.33 |
| |
IV |
- |
- |
- |
- |
- |
- |
- |
| |
V |
- |
- |
- |
- |
- |
- |
- |
| NL2 |
I |
0 |
147 |
147 |
6 |
12 |
12 |
0.33-0.33 |
| |
II |
- |
- |
- |
- |
- |
- |
- |
| |
III |
- |
- |
- |
- |
- |
- |
- |
| |
IV |
36 |
40 |
40 |
21 |
10 |
0 |
0.40-0.40 |
| |
V |
- |
- |
- |
- |
- |
- |
- |
| NL3 |
I |
0 |
90 |
86 |
5 |
8 |
8 |
0.28-0.34 |
| |
II |
17 |
4 |
4 |
14 |
2 |
0 |
0.33-0.33 |
| |
III |
42 |
1 |
1 |
21 |
1 |
0 |
0.33-0.33 |
| |
IV |
- |
- |
- |
- |
- |
- |
- |
| |
V |
- |
- |
- |
- |
- |
- |
- |
| NL4a |
I |
0 |
68 |
3 |
2 |
6 |
0 |
0.00-0.91 |
| |
II |
3 |
61 |
1 |
21 |
6 |
1 |
0.00-1.00 |
| |
III |
2 |
62 |
31 |
13 |
6 |
0 |
0.23-0.70 |
| |
IV |
23 |
30 |
23 |
21 |
5 |
0 |
0.33-0.47 |
| |
V |
16 |
2 |
1 |
80 |
2 |
0 |
0.20-0.34 |
| NL4b |
I |
0 |
79 |
1 |
4 |
6 |
0 |
0.00-1.00 |
| |
II |
- |
- |
- |
- |
- |
- |
- |
| |
III |
0 |
67 |
14 |
24 |
6 |
0 |
0.00-0.51 |
| |
IV |
14 |
53 |
7 |
15 |
6 |
0 |
0.30-0.66 |
| |
V |
4 |
19 |
8 |
94 |
2 |
0 |
0.12-0.88 |
| NL |
|
283 |
989 |
513 |
|
109 |
39 |
0.00-1.004) |
| DK1 |
I |
0 |
57 |
57 |
3 |
5 |
0 |
0.31-0.31 |
| |
II |
- |
- |
- |
- |
- |
- |
- |
| |
III |
- |
- |
- |
- |
- |
- |
- |
| |
IV |
- |
- |
- |
- |
- |
- |
- |
| |
V |
- |
- |
- |
- |
- |
- |
- |
| DK2 |
I |
0 |
142 |
97 |
4 |
14 |
14 |
0.05-0.37 |
| |
II |
27 |
41 |
34 |
18 |
7 |
0 |
0.23-0.32 |
| |
III |
86 |
17 |
17 |
22 |
7 |
1 |
0.22-0.22 |
| |
IV |
- |
- |
- |
- |
- |
- |
- |
| |
V |
- |
- |
- |
- |
- |
- |
- |
| DK | 113 | 257 | 205 | 33 | 15 | 0.05-0.374) |
NL = Dutch; DK = Danish.
1) Interval I–V = time between subsequent sampling moments starting from birth.
2) Number of pens used in statistical analysis.
3) Number of pens where > 1 pig received treatment with risk antimicrobials, i.e. tetracyclins or β-lactams.
4) NL: mean = 0.32; SD = 0.21, DK: mean = 0.27; SD = 0.08.
– : No susceptibles left.
Exponentiation of the estimated parameter log (β) gives the transmission parameter β, that denotes the transmission per day. The reproduction ratio R0 can then be calculated by multiplying β with the length of the infectious period (1/α). We assumed 17.4 days to be the length of the infectious period. This was based on the average length of the infectious period observed in a transmission experiment [24] and on the interval of sampling in this study, which ranged from 4 to 94 days.
To the basic statistical model, more explanatory variables potentially affecting transmission were added. These variables were related to antimicrobial use, age and contribution of direct and indirect transmission. Tetracyclins and β-lactam antimicrobials were defined as risk antimicrobials (ab), as these antimicrobial classes select 100% for MRSA CC398 [25] and will potentially affect its transmission. If these antimicrobials were applied on >1 pig within a pen during a time interval, then variable ‘ab’ was defined as yes, otherwise no. Age of pigs was introduced as variable, because an age-effect or presence of the sow in the farrowing compartment might affect transmission of MRSA compared to transmission in post-weaning pigs. When pigs were located in the farrowing compartment, ‘age’ was defined as pre-weaning, and after weaning when located in the weaning or finishing compartment as post-weaning. To quantify the relative effect of transmission through direct contact with pen mates compared to the total transmission through direct and indirect contact, a continuous explanatory variable was introduced. This variable (pIP) was calculated as IP within the pen divided by the total IP. A multivariable model, including the explanatory variables described above, is presented as follows:
Data from Denmark and The Netherlands were analysed separately and in combination. First, analysis was done without explanatory variables to estimate a basic transmission parameter (Table 3). Secondly, analysis including explanatory variables was done. Due to very high prevalences in new born piglets in both Danish herds, the number of susceptibles was very low after sampling moment 2, leaving no or very few cases to occur at post-weaning age. Analysis was, therefore, done on data from pre-weaning pigs only, implying that the explanatory variable ‘age’ could not be included in this analysis (Table 3). Finally, multivariable analysis was performed on Dutch data only from all pigs.
Table 3.
Models used to quantify transmission of MRSA within pig herds
| Model | Transmission parameter estimation1) | Used data |
|---|---|---|
| Basic |
TP1 = exp (log β0) |
Denmark / Netherlands |
| Bivariable |
TP2 = exp (log β0 + log β1(ab) + log β2(pIP)) |
Denmark / Netherlands |
| Multivariable | TP3 = exp (log β0 + log β1(ab) + log β2(pIP) + log β3(age)) | Netherlands |
1) In all models log (IP·Δt) was used as offset variable.
ab: use of tetracyclins or β-lactams (yes/no); age: age of pigs (pre-weaning/post-weaning); pIP: infection pressure within the pen divided by total infection pressure (continuous variable).
In bi- and multivariable analysis two-way interactions between variables were tested for significance and removed if P > 0.05. To estimate solely the herd effect, i.e. without explanatory variables, herd was included in the basic model as random effect using an exchangeable covariance structure (PROC GENMOD) [22]. Herd effect for Dutch data only accounted for 0.06% of non-explained variance, and was therefore not included in the statistical models. Herd effect for Danish data could not be estimated.
Results
MRSA prevalence
Overall MRSA prevalence of sows increased from 33.3% (exact 95% CI: 22.0-46.3%) before farrowing (moment 1) to 58.8% (exact 95% CI: 46.2-70.6%) after farrowing (moment 2) and to 77.3% (exact 95% CI: 65.3-86.7%) ~ 3 weeks later, just before weaning (moment 3). All sows in herd NL4 tested MRSA negative, whereas all sows in herd DK1 tested MRSA positive at all sampling moments (Table 4).
Table 4.
Numbers and percentages of MRSA positive sows and environmental wipes
| |
Moment 1 |
Moment 2 |
Moment 3 |
||||||
|---|---|---|---|---|---|---|---|---|---|
| |
Wipes |
Sows |
Wipes |
Sows |
Wipes |
Sows |
|||
| Herd | N pos/n total | N | % Pos | N pos/n total | N | % Pos | N pos/n total | N | % Pos |
| NL1 |
1/4 |
16 |
6.3 |
3/4 |
16 |
25.0 |
4/4 |
16 |
93.8 |
| NL2 |
4/4 |
12 |
33.3 |
4/4 |
12 |
100.0 |
4/4 |
12 |
100.0 |
| NL3 |
3/4 |
8 |
50.0 |
4/4 |
8 |
62.5 |
4/4 |
8 |
100.0 |
| NL4a |
1/4 |
6 |
0.0 |
0/4 |
6 |
0.0 |
0/4 |
4 |
0.0 |
| NL4b |
0/4 |
6 |
0.0 |
0/4 |
6 |
0.0 |
0/4 |
6 |
0.0 |
| NL |
9/20 |
48 |
18.8 |
11/20 |
48 |
43.8 |
12/20 |
46 |
76.1 |
| DK1 |
5/5 |
5 |
100.0 |
5/5 |
6 |
100.0 |
10/10 |
6 |
100.0 |
| DK2 |
5/5 |
10 |
70.0 |
4/5 |
14 |
92.9 |
5/5 |
14 |
71.4 |
| DK |
10/10 |
15 |
80.0 |
9/10 |
20 |
95.0 |
15/15 |
20 |
80.0 |
| NL + DK | 19/30 | 63 | 33.3 | 20/30 | 68 | 58.8 | 27/35 | 66 | 77.3 |
NL = Dutch; DK = Danish.
Sows were classified MRSA positive if either one of the swabs (nasal/vaginal) tested positive; from all MRSA positive samplings of sows (n = 112), 58% was classified positive based on both swabs, 39% on a positive nasal swab and 3% on a positive vaginal swab.
Pre-weaning, overall MRSA prevalence of pigs increased from 60.9% (exact 95% CI: 57.4-64.3%) in new born piglets (moment 2) to 77.8% (exact 95% CI: 74.6-80.7%) ~ 3 weeks later, just before weaning (moment 3). Post-weaning, MRSA prevalence of pigs was 79.6% (exact 95% CI: 76.2-82.8%) at moment 4 and 86.6% (exact 95% CI: 83.2-89.5%) at moment 5. In the finishing compartment, just before slaughter, MRSA prevalence was 69.6% (exact 95% CI: 64.9-74.1 (Table 5).
Table 5.
Numbers and percentages of MRSA positive pigs and environmental wipes
| |
Moment 2 |
Moment 3 |
Moment 4 |
Moment 5 |
Moment 6 |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| |
Pigs |
Pigs |
Wipes |
Pigs |
Wipes |
Pigs |
Wipes |
Pigs |
|||||
| Herd | N | % Pos | N | % Pos | N pos/N total | N | % Pos | N pos/n total | N | % Pos | N pos/n total | N | % Pos |
| NL1 |
199 |
42.7 |
186 |
93.0 |
4/4 |
125 |
100.0 |
- |
- |
- |
- |
- |
- |
| NL2 |
147 |
100.0 |
146 |
99.3 |
0/4 |
93 |
55.9 |
2/4 |
92 |
100.0 |
4/4 |
80 |
86.3 |
| NL3 |
90 |
95.6 |
90 |
98.9 |
4/4 |
87 |
100.0 |
4/4 |
86 |
100.0 |
4/4 |
82 |
63.4 |
| NL4a |
68 |
4.4 |
64 |
3.1 |
0/4 |
65 |
52.3 |
1/4 |
64 |
89.1 |
3/4 |
62 |
64.5 |
| NL4b |
79 |
1.3 |
70 |
0.0 |
1/4 |
67 |
20.9 |
0/4 |
68 |
16.2 |
0/4 |
68 |
13.2 |
| NL |
583 |
55.2 |
556 |
73.6 |
9/20 |
437 |
71.4 |
7/16 |
310 |
79.4 |
11/16 |
292 |
58.2 |
| DK1 |
57 |
100.0 |
48 |
100.0 |
10/10 |
48 |
100.0 |
7/10 |
45 |
100.0 |
5/5 |
46 |
100.0 |
| DK2 |
142 |
68.3 |
133 |
87.2 |
10/10 |
129 |
100.0 |
15/15 |
123 |
100.0 |
10/10 |
67 |
98.5 |
| DK |
199 |
77.4 |
181 |
90.6 |
20/20 |
177 |
100.0 |
22/25 |
168 |
100.0 |
15/15 |
113 |
99.1 |
| NL + DK | 782 | 60.9 | 737 | 77.8 | 29/40 | 614 | 79.6 | 29/41 | 478 | 86.6 | 26/31 | 405 | 69.6 |
NL = Dutch; DK = Danish.
Pigs were classified MRSA positive if either one of the swabs (nasal/rectal) tested positive; from all MRSA positive samplings of pigs (n = 476), 67% was classified positive based on both swabs, 31% on a positive nasal swab and 2% on a positive rectal swab.
New born piglets (moment 2) in Dutch herds were more often MRSA positive (P < 0.0001) if their dam was MRSA positive before farrowing compared to MRSA negative dams. MRSA prevalence in pigs from positive dams was 84% versus 48% from negative dams. In Danish herds, MRSA prevalence in pigs from positive dams was 78% versus 73% from negative dams. Combining data from all herds, 81% of new born pigs from MRSA positive sows were positive after birth, whereas 50% of pigs from MRSA negative sows were positive at that time (P < 0.0001).
The number of MRSA positive environmental wipes varied largely between herds and sampling moments, from no positive wipes to all wipes positive (Table 4 and 5).
Transmission quantification
Table 2 shows model input summarized per herd per sampling interval, which was used for MRSA transmission quantification. Basic R0-values including data from all pigs in the basic model, were 1.11 (95% CI: 1.00-1.22) for Dutch pigs and 1.88 (95% CI: 1.59-2.22) for Danish pigs. Including data from pre-weaning pigs only, resulted in a slightly higher R0-value for Dutch pigs (R0 = 1.96; 95% CI: 1.74-2.20), whereas the R0-value for Danish pigs did not change (R0 = 1.88; 95% CI: 1.59-2.22). Analysis on data from both countries resulted in intermediate R0-values (Table 6).
Table 6.
MRSA transmission per day and R0-values
| |
Pre-weaning pigs only |
All pigs |
||||||
|---|---|---|---|---|---|---|---|---|
| Country | C/S2) | TP1 | R0 | 95% CI | C/S2) | TP1 | R0 | 95% CI |
| NL |
377/704 |
0.112 |
1.96 |
1.74 - 2.20 |
513/989 |
0.064 |
1.11 |
1.00 - 1.22 |
| DK |
188/240 |
0.108 |
1.88 |
1.59 - 2.22 |
205/257 |
0.108 |
1.88 |
1.59 - 2.22 |
| NL + DK | 565/944 | 0.111 | 1.93 | 1.75 - 2.13 | 718/1246 | 0.071 | 1.24 | 1.14 - 1.35 |
Legend: This table shows the transmission per day from the basic model (TP1) and R0-value1) with its 95% confidence interval for Dutch (NL) and Danish (DK) herds separately and all herds (NL + DK) for pre-weaning pigs only and for all pigs.
1) To calculate R0, TP1 was multiplied with 17.4 days (= length of infectious period).
2) Number of MRSA cases (C) from total number of susceptibles (S) used in analysis.
Bivariable analysis of data from pre-weaning pigs only, showed effects of both variables: use of risk antimicrobials (ab: P < 0.0001 for Dutch data and P = 0.10 for Danish data) and the relative proportion of IP within the pen compared to the total IP (pIP: P < 0.0001 for Danish and Dutch data), and no significant interaction effect (P > 0.05). The effect of pIP was very large, especially in Danish herds. For Danish pigs, R0 was 0.02 (95% CI: 0.00-0.07) without use of risk antimicrobials and pIP at its minimum (i.e. 0), and 0.03 (95% CI: 0.00-0.16) with use of risk antimicrobials and pIP at its minimum. When pIP was at its maximum, R0-values were infinitely large both without and with use of risk antimicrobials (R0 = 121076 (95% CI: 307.31-4.77E + 07) and 1760410 (95%CI: 287.92-1.08E + 08), respectively). For Dutch pigs, R0 was 0.62 (95% CI: 0.45-0.84) without use of risk antimicrobials and pIP at its minimum, and 1.54 (95% CI: 0.84-2.83) with use of risk antimicrobials and pIP at its minimum. When pIP was at its maximum, R0-values were 3.28 (95% CI: 1.43-7.49), and 8.17 (95%CI: 2.65-25.22) for without and with use of risk antimicrobials, respectively (Table 7). Analysis on data from both countries resulted in similar results, with the lowest R0 when risk antimicrobials were not used and pIP at its minimum (R0 = 0.68; 95% CI: 0.54-0.85), and the highest R0 when risk antimicrobials were used and pIP at its maximum (R0 = 10.50; 95% CI: 4.31-25.59). Average pIP was 0.32 (SD = 0.21) for Dutch herds and 0.27 (SD = 0.08) for Danish herds (Table 2).
Table 7.
MRSA transmission parameter and R0-values
| |
|
No risk antimicrobials; pIP = 0 |
Risk antimicrobials; pIP = 0 |
No risk antimicrobials; pIP = 1 |
Risk antimicrobials; pIP = 1 |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Country | C/S4) | TP2 | R0 | 95% CI | TP2 | R0 | 95% CI | TP2 | R0 | 95% CI | TP2 | R0 | 95% CI |
| NL |
377/704 |
0.035 |
0.62 |
0.45 - 0.84 |
0.089 |
1.54 |
0.84 - 2.83 |
0.188 |
3.28 |
1.43 - 7.49 |
0.470 |
8.17 |
2.65 - 25.22 |
| DK |
188/240 |
0.001 |
0.02 |
0.00 - 0.07 |
0.002 |
0.03 |
0.00 – 0.16 |
6958 |
121076 |
307.31 - ∞ |
10117 |
176041 |
287.92 - ∞ |
| NL + DK | 565/944 | 0.039 | 0.68 | 0.54 - 0.85 | 0.076 | 1.32 | 0.84 - 2.06 | 0.309 | 5.38 | 2.74–10.56 | 0.603 | 10.50 | 4.31 - 25.59 |
Legend: this table shows the transmission per day resulting from bivariable analysis with use of risk antimcrobials1) and the relative proportion of IP compared to the total IP (pIP) included in the model2) (TP2) and R0-value3) with its 95% confidence interval for Dutch (NL) and Danish (DK) farms separately and all farms combined (NL + DK) for pre-weaning pigs only.
1) Risk antimicrobials (tetracyclins and β-lactams) in > 1 pig per pen.
2) Interaction between variables was not significant (P > 0.05).
3) To calculate R0, TP2was multiplied with 17.4 days (= length of infectious period).
4) Number of MRSA cases (C) from total number of susceptibles (S) used in analysis.
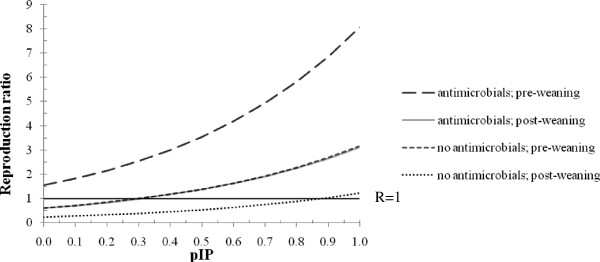
Multivariable analysis of all Dutch data showed significant effects of all three variables, ab, age and pIP (P < 0.0001); interaction effects were not significant (P > 0.05). Figure 2 shows the effect of ab and age on R0 for pIP-values between 0 and 1. R0 was lowest at 0.24 (95% CI: 0.18-0.31) when no risk antimicrobials were used in post-weaning pigs and pIP = 0. R0 increased to 0.60 (95% CI: 0.34-1.06) when risk antimicrobials were used in post-weaning pigs and a minimal pIP. R0 was above 1, though not significant (R0 = 1.56; 95% CI: 0.65-3.77), when risk antimicrobials were used in pre-weaning pigs and a minimal pIP, and below 1, though not significant, (R0 = 0.61; 95% CI: 0.34-1.10) when risk antimicrobials were used in post-weaning pigs and a minimal pIP. R0 increases with increasing pIP. Given a maximal pIP of 1, R0 was 1.22 (95% CI: 0.60-2.48) without using risk antimicrobials in post-weaning pigs and significantly above 1, i.e. 3.12 (95% CI: 1.15-8.46) when risk antimicrobials were used in this age-group. In pre-weaning pigs, R0-values were significantly above 1; R0 was 3.16 (95% CI: 1.14-8.82) when no risk antimicrobials were used, and highest with use of risk antimicrobials (R0 = 8.08; 95% CI: 2.17-30.12).
Figure 2.
Reproduction ratio for MRSA in pigs related to antimicrobial use, age and infection pressure. Reproduction ratio related to use of tetracyclins and β-lactam antimicrobials (yes, no), age of pigs (pre-weaning, post-weaning) and relative proportion of the infection pressure within the pen compared to the total infection pressure (pIP); based on 513 MRSA cases (C) from 989 susceptibles (S) from 4 herds. Note: lines for antimicrobials; post-weaning pigs and no antimicrobials; pre-weaning pigs are overlapping, thus difficult to distinguish.
Discussion
MRSA prevalences in different age groups and transmission rates between pigs were assessed longitudinally in six herds in two European countries. Sow prevalences varied widely between herds and over time. This might be explained by differences in management practices. In The Netherlands, an all in – all out system is applied in the farrowing compartment, i.e. a cohort of sows due to farrow is placed into a ‘clean’ farrowing compartment, whereas in Denmark a continuous system is practiced. Except for one Dutch herd, prevalence in sows increased in the Dutch herds during time in the farrowing compartment, which might be explained by a build-up of bacterial load, i.e. the infection pressure, during time spent in this compartment. However, given the small sample of herds studied in each country, these differences could also be due to specific management factors at the herds under study and not necessarily reflect differences between the two countries.
Prevalences in new born piglets varied from 1 to 100%. A similar explanation as given for differences in prevalences in sows might be applicable here. In one Dutch herd, prevalences in pre-weaning pigs remained low (< 5%). A rapid increase in prevalence in this herd was seen after weaning, despite the fact that no risk antimicrobials were used during that time interval and that no positive dust samples were found at the first sampling moment in the weaning compartment. In a longitudinal study on an antimicrobial-free Canadian pig farm, a similar increase in MRSA prevalence was observed around the time of weaning [13]. Co-mingling of MRSA positive and negative pigs, transmission through human handling during weaning, increased susceptibility due to stress or related to age, or a combination of these factors, might be responsible for the rapid increase in MRSA prevalence after weaning.
The recurrent finding of MRSA in the majority of sampled individuals either indicates true colonization or might be the result of contamination. To distinguish between true colonization and contamination, MRSA positive pigs should be placed in a clean environment individually for a longer time.
We used a SIS-model to describe the transmission of MRSA within herds, assuming that infectious pigs stop shedding after a while and become susceptible again. This assumption was based on the fact that most humans are intermittent carriers [26] and on data from a former experimental study [24].
The basic reproduction ratio, based on a SIS-model without explanatory variables, was significantly above one, indicating a high probability of transmission and persistence within a pig herd [27]. The reproduction ratios were calculated by multiplying estimated transmission parameters with the length of the infectious period. An infectious period is only valid for infectious individuals. Because MRSA can survive outside the host for long periods [28], the environment might also be a source of MRSA. How the contamination of the environment reflects MRSA prevalence of pigs in this environment, how long LA-MRSA persists in the environment, and how environmental contamination affects transmission, is unknown. Our method for quantification of the reproduction ratio might, therefore, be less applicable in situations where no or very few pigs are MRSA positive within a compartment, and where MRSA is present in the environment. For our calculations we used 17.4 days as the length of the infectious period, whereas the final observational time interval, i.e. the period in the finishing pig compartment, was much longer (> 10 weeks). During this period, more than one infection might have occurred in one individual, whereas our method only counts one. This implies a potential underestimation of transmission rates. Based on the prevalence in pigs just before slaughter (70%), the reproduction ratio can be estimated using R0 = 100/(100-prevalence) [27], resulting in 3.27. This is indeed higher than the estimated basic reproduction ratio based on data from all pigs in both countries (R0 = 1.24), indicating an underestimation, however, it is similar to the estimated reproduction ratio from a transmission experiment [24].
Transmission rates were higher when tetracyclins and β-lactams were used which might be explained by a selective advantage of MRSA CC398 as compared to susceptible strains present in the nasal microbiota when these antimicrobials are used. An experimental study investigating the effects of zinc and tetracycline on MRSA counts in nasal samples of pigs, showed higher counts in treated animals than in untreated animals, which seems to confirm a selective advantage of MRSA CC398 caused by both compounds [29]. The effect of zinc could not be assessed in our study as only Danish herds applied zinc in the weaning compartment where all pigs were already MRSA positive before entering.
The proportion of susceptibles and therefore, potentially new cases, was relatively high among pre-weaning pigs compared to post-weaning pigs; we actually assumed all new born piglets to be MRSA negative, and thus susceptible, before birth. MRSA prevalence in post-weaning pigs was much higher, leaving fewer susceptibles to become a case and thus estimation of transmission rates was based on less information in this age-group. Thus the power of the comparison is lower. Nevertheless, the available data indicated that transmission rates in pre-weaning pigs are significantly higher than in post-weaning pigs. This might be explained by the presence of the sow, which might be a primary source of MRSA for the new born piglets. An association between MRSA status of the sow prior to farrowing and that of the offspring just after birth was shown in our study, and was found in a longitudinal study on a Canadian pig farm as well [13]. Perinatal transmission of MRSA CC398 from sow to pigs has been demonstrated under controlled experimental conditions [30]. New born piglets might be more susceptible to acquisition of MRSA and other infectious agents due to their immature mucosal immune system and the greater impact of antimicrobials on their unbalanced microbiota [31,32].
The increased transmission rates observed in correspondence with the relative increase of the infection pressure within the pen implies that transmission through direct contact with pen mates is an important transmission route. More quantitative information on transmission rates within and between pens and the role of environmental contamination can be obtained by transmission experiments [33].
Since only 4 to 6 farrow-to-finish herds were included in the estimation of transmission rates, including some herds with very high prevalences leaving just a few trials for parameter estimation, the results might not be representative for the international pig herd population. Moreover, the observed association between explanatory variables and R0, e.g. antimicrobial use, might be confounded by other effects. Although herd effect accounted for only 0.06% of the variance in the multivariable analysis, it was not possible to distinguish the herd effect from an unconfounded estimate of the exposure effects. Prudence is therefore called for in drawing conclusions from these associations.
Conclusion
In conclusion, introduction of MRSA in a fully susceptible population most probably leads to transmission and an endemic situation where direct contact between animals is the most important route of transmission. Control programs should therefore focus on (1) prevention of introduction into a herd, and (2) prevention of transmission within a herd, e.g. by prudent antimicrobial use, all-in all-out procedures and hygiene barriers between age groups.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
MJ, EG, AG, LG, PB, JPN, PW and EB contributed to the design of the longitudinal field study. EB, PB, PW and CEG coordinated and performed the studies. EG, NV, MJ and EB analysed the data. EB, EG and CEG drafted the manuscript. All authors read, revised and approved the final manuscript.
Authors’ information
EB’s current position is at Department of Infectious Diseases and Immunology, Faculty of Veterinary Medicine, Utrecht University, The Netherlands
Contributor Information
Els M Broens, Email: e.m.broens@uu.nl.
Carmen Espinosa-Gongora, Email: ceg@life.ku.dk.
Elisabeth AM Graat, Email: lisette.graat@wur.nl.
Nadia Vendrig, Email: nadia.vendrig@wur.nl.
Peter J Van Der Wolf, Email: p.vd.wolf@gddeventer.com.
Luca Guardabassi, Email: lg@life.ku.dk.
Patrick Butaye, Email: patrick.butaye@var.fgov.be.
Jens Peter Nielsen, Email: jpni@life.ku.dk.
Mart CM De Jong, Email: mart.dejong@wur.nl.
Arjen W Van De Giessen, Email: arjen.van.de.giessen@rivm.nl.
Acknowledgements
This study was supported by the EU- HEALTH project PILGRIM of the 7th Framework Programme ( http://www.fp7-pilgrim.eu), and co-financed in Denmark by the Danish Food Industry Agency (Ministry of Food, Agriculture and Fisheries), and in The Netherlands by the INTERREG IV A Germany-Netherlands programme through the EU funding from the European Regional Development Fund (ERDF), the Ministry for Economic Affairs, Energy, Building, Housing and Transport of the State of North-Rhine Westphalia, the Ministry of Economic Affairs, Labour and Traffic of Lower Saxony, the Dutch Ministry of Economic Affairs, Agriculture and Innovation, the Product Boards for Livestock, Meat and Eggs, the Food Safety Authority in The Netherlands, and the Provinces of Friesland, Groningen, Drenthe, Overijssel, Gelderland, Noord-Brabant and Limburg, which is coordinated by the EU-region Rhine-Waal. Nol Jansen and Herman Raaijmakers are acknowledged for their assistance in taking samples on Dutch pig herds. All farmers are greatly acknowledged for their kind cooperation.
References
- Voss A, Loeffen F, Bakker J, Klaassen C, Wulf M. Methicillin-resistant Staphylococcus aureus in pig farming. Emerg Infect Dis. 2005;11:1965–1966. doi: 10.3201/eid1112.050428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TC, Male MJ, Harper AL, Kroeger JS, Tinkler GP, Moritz ED, Capuano AW, Herwaldt LA, Diekema DJ. Methicillin-resistant Staphylococcus aureus (MRSA) strain ST398 is present in midwestern U.S. swine and swine workers. PLoS One. 2009;4:e4258. doi: 10.1371/journal.pone.0004258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broens EM, Graat EAM, Van Der Wolf PJ, Van De Giessen AW, Van Duijkeren E, Wagenaar JA, Van Nes A, Mevius DJ, De Jong MCM. MRSA CC398 in the pig production chain. Prev Vet Med. 2011;98:182–189. doi: 10.1016/j.prevetmed.2010.10.010. [DOI] [PubMed] [Google Scholar]
- Graveland H, Wagenaar JA, Heesterbeek H, Mevius D, Van Duijkeren E, Heederik D. Methicillin resistant Staphylococcus aureus ST398 in veal calf farming: human MRSA carriage related with animal antimicrobial usage and farm hygiene. PLoS One. 2010;5:e10990. doi: 10.1371/journal.pone.0010990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulders MN, Haenen AP, Geenen PL, Vesseur PC, Poldervaart ES, Bosch T, Huijsdens XW, Hengeveld PD, Dam-Deisz WD, Graat EAM, Mevius D, Voss A, Van De Giessen AW. Prevalence of livestock-associated MRSA in broiler flocks and risk factors for slaughterhouse personnel in The Netherlands. Epidemiol Infect. 2010;138:743–755. doi: 10.1017/S0950268810000075. [DOI] [PubMed] [Google Scholar]
- EFSA. Analysis of the baseline-survey on the prevalence of methicillin-resistant Staphylococcus aureus (MRSA) in holdings with breeding pigs, in the EU, 2008. Part A: MRSA prevalence estimates. EFSA J. 2009;7:1376. [Google Scholar]
- Broens EM, Graat EAM, Van Der Wolf PJ, Van De Giessen AW, De Jong MCM. Prevalence and risk factor analysis of livestock associated MRSA-positive pig herds in The Netherlands. Prev Vet Med. 2011;102:41–49. doi: 10.1016/j.prevetmed.2011.06.005. [DOI] [PubMed] [Google Scholar]
- Eveillard M, Martin Y, Hidri N, Boussougant Y, Joly-Guillou ML. Carriage of methicillin-resistant Staphylococcus aureus among hospital employees: prevalence, duration, and transmission to households. Infect Contr Hosp Epidemiol. 2004;25:114–120. doi: 10.1086/502360. [DOI] [PubMed] [Google Scholar]
- Albrich WC, Harbarth S. Health-care workers: source, vector, or victim of MRSA? Lancet Infect Dis. 2008;8:289–301. doi: 10.1016/S1473-3099(08)70097-5. [DOI] [PubMed] [Google Scholar]
- Sexton T, Clarke P, O'Neill E, Dillane T, Humphreys H. Environmental reservoirs of methicillin-resistant Staphylococcus aureus in isolation rooms: correlation with patient isolates and implications for hospital hygiene. J Hosp Infect. 2006;62:187–194. doi: 10.1016/j.jhin.2005.07.017. [DOI] [PubMed] [Google Scholar]
- Boyce JM. Environmental contamination makes an important contribution to hospital infection. J Hosp Infect. 2007;65(Suppl 2):50–54. doi: 10.1016/S0195-6701(07)60015-2. [DOI] [PubMed] [Google Scholar]
- Broens EM, Graat EAM, Engel B, Van Oosterom RA, Van De Giessen AW, Van Der Wolf PJ. Comparison of sampling methods used for MRSA-classification of herds with breeding pigs. Vet Microbiol. 2011;147:440–444. doi: 10.1016/j.vetmic.2010.07.021. [DOI] [PubMed] [Google Scholar]
- Weese JS, Zwambag A, Rosendal T, Reid-Smith R, Friendship R. Longitudinal investigation of methicillin-resistant Staphylococcus aureus in piglets. Zoonoses Public Health. 2011;58:238–243. doi: 10.1111/j.1863-2378.2010.01340.x. [DOI] [PubMed] [Google Scholar]
- Kermack WO, McKendrick AG. Contributions to the mathematical theory of epidemics--I. 1927. Bull Math Biol. 1991;53:33–55. doi: 10.1007/BF02464423. [DOI] [PubMed] [Google Scholar]
- De Jong MCM, Kimman TG. Experimental quantification of vaccine-induced reduction in virus transmission. Vaccine. 1994;12:761–766. doi: 10.1016/0264-410X(94)90229-1. [DOI] [PubMed] [Google Scholar]
- Velthuis AG, Bouma A, Katsma WE, Nodelijk G, De Jong MCM. Design and analysis of small-scale transmission experiments with animals. Epidemiol Infect. 2007;135:202–217. doi: 10.1017/S095026880600673X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa-Gongora C, Larsen J, Moodley A, Nielsen JP, Skov RL, Andreasen M, Guardabassi L. Farm-specific lineages of methicillin-resistant Staphylococcus aureus clonal complex 398 in Danish pig farms. Epidemiol Infect. 2011. in press. [DOI] [PubMed]
- Stegger M, Lindsay JA, Moodley A, Skov R, Broens EM, Guardabassi L. Rapid PCR detection of Staphylococcus aureus clonal complex 398 by targeting the restriction-modification system carrying sau1-hsdS1. J Clin Microbiol. 2011;49:732–734. doi: 10.1128/JCM.01970-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Neeling AJ, Van Leeuwen WJ, Schouls LM, Schot CS, Van Veen-Rutgers A, Beunders AJ, Buiting AG, Hol C, Ligtvoet EE, Petit PL, Sabbe LJ, Van Griethuysen AJ, Van Embden JD. Resistance of staphylococci in The Netherlands: surveillance by an electronic network during 1989–1995. J Antimicrob Chemother. 1998;41:93–101. doi: 10.1093/jac/41.1.93. [DOI] [PubMed] [Google Scholar]
- SAS Institute Inc. SAS/STAT® 9.1 User's Guide. Cary, North Carolina: SAS Institute; 2004. [Google Scholar]
- Velthuis AGJ, De Jong MCM, Kamp EM, Stockhofe N, Verheijden JHM. Design and analysis of an Actinobacillus pleuropneumoniae transmission experiment. Prev Vet Med. 2003;60:53–68. doi: 10.1016/S0167-5877(03)00082-5. [DOI] [PubMed] [Google Scholar]
- McCullagh P, Nelder J. Generalized Linear Models. London: Chapman and Hall; 1989. [Google Scholar]
- Becker NG. Generalized Linear Models. Monographs on Statistics and Applied Probability: Analysis of Infectious Disease Data. London: Chapman and Hall; 2004. pp. 102–138. [Google Scholar]
- Broens EM, Graat EAM, Van De Giessen AW, Broekhuizen-Stins MJ, De Jong MCM. Quantification of transmission of livestock-associated methicillin resistant Staphylococcus aureus in pigs. Vet Microbiol. 2012;155:381–388. doi: 10.1016/j.vetmic.2011.09.010. [DOI] [PubMed] [Google Scholar]
- Kadlec K, Ehricht R, Monecke S, Steinacker U, Kaspar H, Mankertz J, Schwarz S. Diversity of antimicrobial resistance pheno- and genotypes of methicillin-resistant Staphylococcus aureus ST398 from diseased swine. J Antimicrob Chemother. 2009;64:1156–1164. doi: 10.1093/jac/dkp350. [DOI] [PubMed] [Google Scholar]
- Van Belkum A, Verkaik NJ, De Vogel CP, Boelens HA, Verveer J, Nouwen JL, Verbrugh HA, Wertheim HF. Reclassification of Staphylococcus aureus nasal carriage types. J Infect Dis. 2009;199:1820–1826. doi: 10.1086/599119. [DOI] [PubMed] [Google Scholar]
- Allen LJ, Burgin AM. Comparison of deterministic and stochastic SIS and SIR models in discrete time. Math Biosci. 2000;163:1–33. doi: 10.1016/S0025-5564(99)00047-4. [DOI] [PubMed] [Google Scholar]
- Quinn PJ, Markey BK, Carter ME, Donnelly WJ, Leonard FC. Veterinary Microbiology and Microbial disease. Oxford, United Kingdom: Blackwell Sciences Ltd; 2002. [Google Scholar]
- Moodley A, Nielsen SS, Guardabassi L. Effects of tetracycline and zinc on selection of methicillin-resistant Staphylococcus aureus Sequence Type 398 in pigs. Vet Microbiol. 2011;152:420–423. doi: 10.1016/j.vetmic.2011.05.025. [DOI] [PubMed] [Google Scholar]
- Moodley A, Latronico F, Guardabassi L. Experimental colonization of pigs with methicillin-resistant Staphylococcus aureus (MRSA): insights into the colonization and transmission of livestock-associated MRSA. Epidemiol Infect. 2010;15:1–7. doi: 10.1017/S0950268810002888. [DOI] [PubMed] [Google Scholar]
- Zoetendal EG, Cheng B, Koike S, Mackie RI. Molecular microbial ecology of the gastrointestinal tract: from phylogeny to function. Curr Issues Intest Microbiol. 2004;5:31–47. [PubMed] [Google Scholar]
- Bailey M, Haverson K, Inman C, Harris C, Jones P, Corfield G, Miller B, Stokes C. The development of the mucosal immune system pre- and post-weaning: balancing regulatory and effector function. Proc Nutr Soc. 2005;64:451–457. doi: 10.1079/PNS2005452. [DOI] [PubMed] [Google Scholar]
- Klinkenberg D, De Bree J, Laevens H, De Jong MCM. Within- and between-pen transmission of classical swine fever virus: a new method to estimate the basic reproduction ratio from transmission experiments. Epidemiol Infect. 2002;128:293–299. doi: 10.1017/s0950268801006537. [DOI] [PMC free article] [PubMed] [Google Scholar]