Abstract
Background
Plant resistance genes, which encode R-proteins, constitute one of the most important and widely investigated gene families. Thanks to the use of both genetic and molecular approaches, more than 100 R genes have been cloned so far. Analysis of resistance proteins and investigation of domain properties may afford insights into their role and function. Moreover, genomic experiments and availability of high-throughput sequence data are very useful for discovering new R genes and establish hypotheses about R-genes architecture.
Result
We surveyed the PRGdb dataset to provide valuable information about hidden R-protein features. Through an in silico approach 4409 putative R-proteins belonging to 33 plant organisms were analysed for domain associations frequency. The proteins showed common domain associations as well as previously unknown classes. Interestingly, the number of proteins falling into each class was found inversely related to domain arrangement complexity. Out of 31 possible theoretical domain combinations, only 22 were found. Proteins retrieved were filtered to highlight, through the visualization of a Venn diagram, candidate classes able to exert resistance function. Detailed analyses performed on conserved profiles of those strong putative R proteins revealed interesting domain features. Finally, several atypical domain associations were identified.
Conclusion
The effort made in this study allowed us to approach the R-domains arrangement issue from a different point of view, sorting through the vast diversity of R proteins. Overall, many protein features were revealed and interesting new domain associations were found. In addition, insights on domain associations meaning and R domains modelling were provided.
Keywords: Disease resistance gene, Plant UniGene, Domain arrangements, Bioinformatics analyses
Background
During their life plants are continuously under pathogen attack. Due to their nature, namely the lack of mobility, plants have developed molecular and chemical features to withstand biotic stresses. The plant immune system is based on receptors that recognise broadly conserved molecules associated to a wide range of pathogens. Resistance gene products (R proteins) are thought to recognise signal molecules produced by the pathogen and to respond by initiating rapid changes in host cell physiology and metabolism so as to directly inhibit pathogen growth.
To date, more than 100 R genes have been cloned (http://www.prgdb.org). Five typical protein structures were recognised as involved in the resistance process: the TIR-NBS-LRR (TNLs; e.g. N gene) [1], the CC-NBS-LRR (CNLs; e.g. I2 gene) [2], the receptor-like kinase (RLKs; e.g. FLS2) [3], the receptor-like protein (RLPs; e.g. CF4 gene) [4] and the kinase-like protein (e.g. PTO gene) [5]. The five R-protein types share common features: two of the five classes (RLK and RLP) contain a transmembrane domain (TM) that anchors them into the membrane, and four of them contain a leucine-rich repeat region (LRR) [6]. Classes TNL and CNL, lacking clear membrane anchor domains, operate mainly in the cytoplasm. Both contain a Nucleotide-Binding Site (NBS) and an LRR domain [7]. The TNL class has, additionally, a N-terminal domain with homology to the animal Toll-Interleukin Receptor (TIR). By contrast, the CNL class lacks the TIR domain and may include a C-terminal Coiled-Coil region (CC). Several RLK and RLP proteins confer resistance to biotic stresses. However, their function should be tested experimentally, because these proteins are involved also in other cellular mechanisms not related to resistance. RLKs consist of an intracellular serine kinase domain (KIN) and extracellular leucine-rich repeat region (eLRR) of 25-38 amino acids (AA) that confer a broad interaction surface, well suited to interact with multiple ligands [8]. The eLRR domain plays a recognising role, while the kinase triggers the downstream activation cascades [9]. RLKs can either function as homodimers [10] or require heteromeric interactions with other proteins to initiate a defence response [11,12]. Moreover, those genes can have multi-functionality activity [13]. Similar in function and structure, the RLP family consists of a serine/threonine receptor containing a leucine-rich region (KIN-LRR), a transmembrane region of ~25 AA, and a short cytoplasmic region, with no kinase domain [14]. The RLP extracellular leucine-rich repeat (eLRR) shows homology with the eLRR of the RLKs. Moreover, RLPs can be involved in other cellular mechanisms, like RLKs do [14]. Finally, proteins containing only a kinase (KIN) domain, like the tomato PTO gene [5] that confers resistance to Pseudomonas syringae, completes the panorama of R proteins. In addition to these well-studied five R-classes, many other resistance proteins (Oth-R), which exert their function in different ways, have been discovered. Sometimes they share conserved domains with the classified R proteins, but their functional mechanisms are usually so different that they cannot be simply classified [15,16]. In this class fall the Hordeum vulgare MLO[17] and the Arabidopsis thaliana RPW8 genes [18], that confer resistance against the powdery mildew caused by Blumeria graminis and Golovinomyces cichoracearum, respectively. The study of this class of proteins may be of great interest to gain insights into the plant immune system overall [19].
For a long time R proteins were thought to recognize specific pathogen proteins using ill-defined mechanisms. Many models have been proposed to explain the way R proteins act, including the guard hypothesis, the zig-zag model and the switch model [20,21]. The most widely endorsed model connects various actors, assuming a collaborative role among PTI (PAMP-triggered immunity) proteins and resistance proteins [22]. By using domain architecture comparisons and domain property investigation, the role and function of R proteins and the ways for generating novelty may be better appraised. Genomic experiments, availability of sequenced genomes and high-throughput analysis can be very useful to discover new R-proteins and lay down new hypotheses concerning the domain reorganization process.
Recently, the plant resistance gene database (PRGdb, http://www.prgdb.org) has been developed. It is a specific resource collecting all functional R-genes and many putative sequences predicted by UNIGENE and NCBI nucleotide datasets. Prediction analysis using such data shows that plant genomes not only code for R proteins with known domain arrangements. but also for proteins with new resistance domain associations [23].
The aim of this paper is to revisit the information generated until now on R-proteins, analysing in depth the largest plant R UniGene dataset. We first analysed the frequency of domain associations to provide data on R-domain distribution and to discover new putative R-protein models. Then, after a manual curated data filtration, we highlighted features and levels of conservation among R-classes. Finally, we explored sequences similar to the neglected “other” R class (Oth-R) and with atypical R-domain associations (Aty-R).
Results
Analysis of R-domain associations in the UniGene PRG dataset
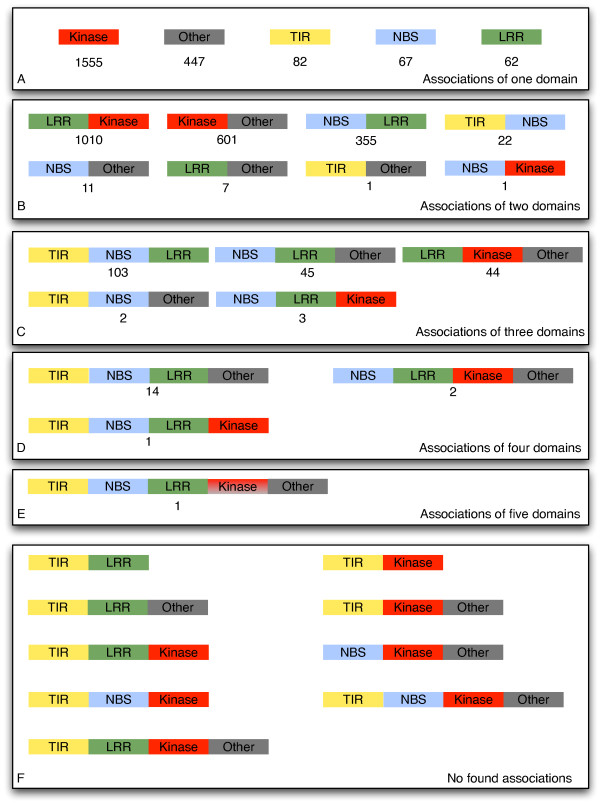
The PRGdb full dataset was surveyed to identify UniGenes starting with proper initial codon (methionine). Of 10,463 translated proteins belonging to 33 organisms (UniGene eudicotyledons), 4409 were selected and classified according to their conserved domains. A contingency table (data not shown) allowed us to divide proteins into 22 subfamilies according with their domain composition. As shown in Figure 1, a considerable number of proteins are composed by a single domain (40%): 1555 proteins showed a kinase domain, 82 a TIR domain, 67 a NBS domain, 62 a LRR domain and 447 have domains classified as other (Oth-R). Associations comprising from two to five domains were also found. Few proteins showed a complex domain association comprising more than three domains. Most of the domain associations described here (42% of the total) were already reported in the literature. In particular, the already described classes are subdivided into kinases (KIN) (35.2%), transmembrane receptors (RLP or RLK) (22.9%), and cytoplasmic proteins (CNL and TNL) (9.2%). The protein domain arrangements not yet described ranged from plausible highly represented classes (e.g Kinase-OthR, 13%) to the non-ordinary class TIR-NBS-LRR-KIN-Oth-R (less than 1%). Looking at R-domain occurrence in the full dataset, the NBS domain was found in 13 classes, followed by the LRR domain in 12, the KIN domain in 9, and finally TIR domains in 8 classes. Preferential associations were observed between LRR-NBS and LRR-KIN domains that are present in 8 and 6 classes, respectively. Out of all possible combinations, nine were not found in our analysis. All of them, except one, contain a TIR domain (Figure 1F).
Figure 1.
R-domain associations found in the UniGene PRG dataset. Predicted sequences are grouped according to the number of domains identified (A-E). Box F reports the associations not found in our dataset. LRR = Leucine-rich repeat; NBS = Nucleotide-binding site; TIR = Toll-interleukin like receptor; KIN = kinase.
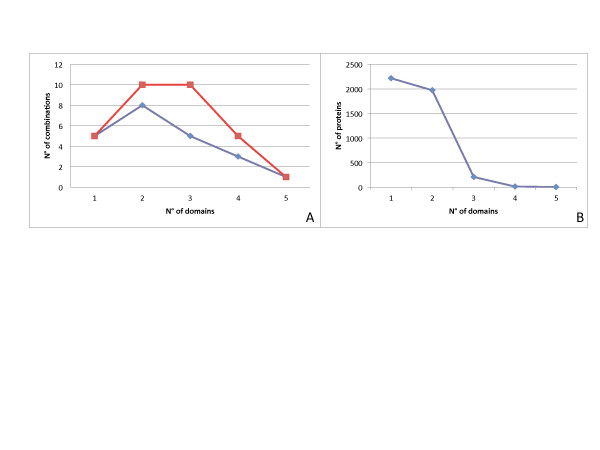
To assess the biological relevance of the domain associations, we compared the theoretical and observed domain association distributions (Figure 2A). Overall, 31 hypothetical domain associations were displayed. Our theoretical model calculated by a binomial formula showed that proteins composed by three domains and two domains were the most numerous (33%), followed by one and four-domain associations (16%). Arrangements up to five domains should be rare. In our processed dataset only 22 out of 31 possible combinations were found. Five classes showed only one domain (20%), 8 classes two domains (37%), 8 classes three domains (25%), 3 classes four domains (11%), and 1 class five domains (2.7%). The distribution displayed by our sample, in comparisons with the theoretical distribution of proteins domain composition, indicates that R-domain combinations are not random in nature. Indeed, the number of proteins in each class is inversely related to domain arrangement complexity (Figure 2B).
Figure 2.
A) Distribution of theoretical (red) and observed (blue) domain associations. B) R proteins arranged in ascending order with corresponding observed frequencies.
Data filtering
To perform a reliable R-proteins classification, data obtained from PRGdb has been filtered according to specific R-protein features. Our specific goal was to deeply annotate putative resistance genes and to find signatures that could support a resistance function. For the most important resistance class (NBS-LRR proteins) a coiled-coil prediction analysis, allowed us to divid into two sub classes: NBS-LRR (45 proteins) and CNL (310 proteins). Using the same procedure, proteins containing only a NBS domain have been divided into CC-NBS (30 proteins) and NBS (37 protein). Instead a transmembrane motif prediction allowed us to divide transmembrane receptors into two classes: RLK (transmembrane motif in the middle of protein) and RLP (transmembrane protein at the C terminal of protein). In order to extract receptors putatively involved in resistance processes RLP proteins have been filtered according to the Fritz-Laylin method. Proteins containing only a LRR domain (LRR-Oth-R, LRR-KIN-Oth-R) have been deleted and kinases have been filtered to select only PTO-like proteins. A total of 107 RLP proteins close related to previous cloned R proteins and 17 proteins containing the PTO-like domain were identified (PTHR24420:SF785 or PTHR23258:SF418, Panther database). In addition, proteins containing an Oth-R domain have been filtered to select only MLO-like and RPW8-like proteins. Seventy-five proteins containing the MLO-like domain (PF03094, Pfam database) and 7 proteins possessing the RPW8 domain (PF05659, Pfam database) were selected. RPW8-like proteins have been divided in two sub-classes: proteins containing only the RPW8 domain and proteins containing a RPW8 domain associated with an NBS-LRR profile (see “Atypical association paragraph”). The 75 MLO-like proteins have been phylogenetically analysed to extract 11 MLO-like proteins that cluster in the same clade of the Arabidopsis and tomato MLOs functional genes [17]. Finally, to confirm the cellular localization of R proteins, a transmembrane region prediction was performed on all the selected candidates. Through a combined in silico approach, using Phobius, TMHMM v2.0 and Interproscan tools, each protein was scanned for the presence of transmembrane domains (TM). Interestingly, transmembrane signatures have been found in cytoplasmic classes (Table 1).
Table 1.
Proteins containing putative transmembrane motifs
| R-domain associations | n. TM | % TM | 1 TM | 2 TM | More TM |
|---|---|---|---|---|---|
| CNL |
20 |
6.4% |
17 |
3 |
0 |
| RLP |
107 |
100% |
63 |
41 |
3 |
| TNL |
17 |
16.5% |
12 |
3 |
2 |
| TIR |
9 |
10.9% |
6 |
2 |
1 |
| NBS-LRR |
1 |
2.2% |
1 |
0 |
0 |
| NBS |
3 |
8.1% |
8 |
0 |
0 |
| TIR-NBS |
2 |
9% |
0 |
2 |
0 |
| NBS-OthR |
1 |
8.3% |
1 |
0 |
0 |
| PTO-like |
9 |
56.2% |
1 |
8 |
0 |
| MLO-like |
11 |
100% |
0 |
4 |
7 |
| TNL-OthR | 1 | 7.1% | 0 | 1 | 0 |
For each class we report the number of sequences with a TM signature (n.), the percentage of sequences with a TM motif on the total (%) and the number of sequences with one, two or more TM domains.
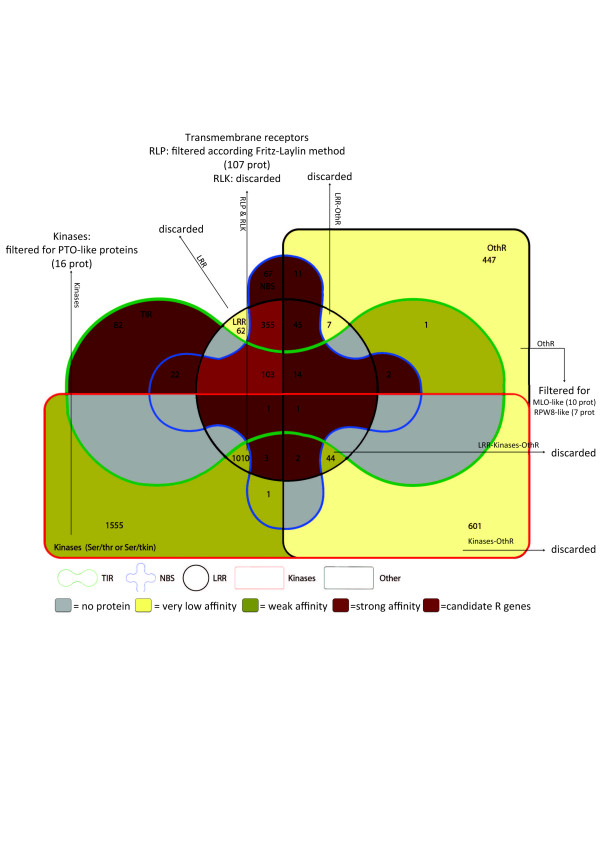
To better appreciate the overall filtering process, an Edwards Venn diagram has been drawn where proteins are divided according to our annotation. Using information about the predicted function of each domain, different groups have been obtained (Figure 3). Out of 4409 proteins, 817 strong R-candidates have been selected. Proteins coloured with darker colours showed a high probability to exert merely a resistance function. Purple red has been used for the CNL and TNL families that are the best candidates to exert a resistance function. The families that could exert a resistance function, but have not been described yet in the literature as functional classes, were brown coloured. Green and yellow colours have been used for families that need further analysis to validate their putative resistance function. Among them, 76 proteins with new domains or with a new domain association were found. Grey areas showed classes that were not found in our dataset.
Figure 3.
Edwards Venn diagram in which proteins are grouped according to their interpro-scan profile. The edge colors represent the five domains [LRR, Kinase, NBS, Other (OthR) and TIR] reported in legend. The group’s intersections showed all possible domain combinations. The colour filling intensity indicated the affinity of a determinate group to exert a resistance function. Stronger colours showed a high probability to exert only a resistance function; Purple red has been used for the CNL and TNL families. The families that could exert a resistance function, but have been not yet described in the literature, were brown coloured. Green and yellow colours have been used for families that need further analysis to validate their putative resistance function. Grey areas showed classes not found in our dataset. Numbers indicated proteins belonging to a specific group. Arrows were used to highlight the analyses performed for a given group in order to select further R-protein candidates.
Resistance families’ comparison
Classes with more than 10 sequences were analysed for their level of similarity in order to reveal domain features and the level of conservation within members belonging to the same class (Table 2). The length of proteins ranged from 1146 AA of TNL protein to 226 AA of proteins with only a TIR domain. The number of members belonging to each class ranged from 310 (CNL) to 10 (MLO-like proteins). Class identity ranged from 14.1% to 55.5%. Identical amino acid sites were not found in RLP class. Comparisons of multiple alignments of random samples, supported by an ANOVA test, revealed that identity is unaffected by number and length of aligned sequences.
Table 2.
Multiple alignment comparison of 10 R protein groups composed by typical resistance domains
| Class Name | n. of sequences | Average sequence length | Alignment length | Identity | Identical sites |
|---|---|---|---|---|---|
| CNL |
310 |
1014 |
5050 |
16.3% |
0.3% |
| RLP |
107 |
651 |
1734 |
17.1% |
0.0% |
| TNL |
103 |
1146 |
3717 |
25.6% |
0.1% |
| TIR |
82 |
226 |
801 |
29.3% |
0.5% |
| NBS-LRR |
45 |
831 |
2750 |
15.4% |
0.4% |
| NBS |
37 |
361 |
1070 |
14.5% |
0.1% |
| CC-NBS |
30 |
395 |
804 |
16.4% |
0.1% |
| TIR-NBS |
22 |
502 |
833 |
31.8% |
1.0% |
| PTO-like |
16 |
561 |
921 |
35.7% |
6.0% |
| MLO-like | 10 | 427 | 622 | 55.5% | 14.1% |
Groups were ordered according to the number of retrieved sequences. Average sequence lengths, percentage of identity and percentage of identical sites are reported.
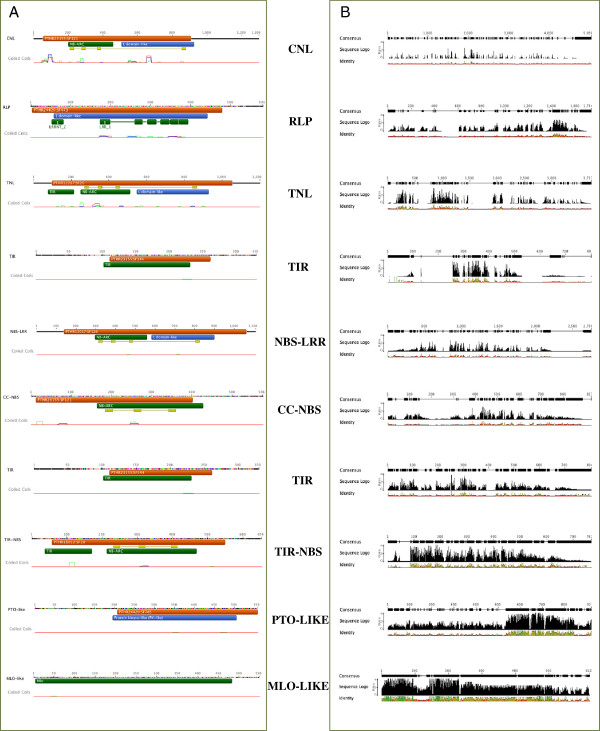
InterProScan protein signature check allowed us to underline conserved domains along the proteins (Figure 4). The pattern of conservation within each group provided evidence that the most conserved classes are the single protein PTO-like and MLO-like, followed by those in which a TIR domain is present: TNL, TIR, TIR-NBS. By contrast, the less conserved are those in which the LRR domain is present. In general, the LRR profile showed a high variability in typology and number of repetitions among proteins. Moreover, in RLPs the LRR is positioned at the N-terminal, while in CNL and TNL at the C-terminal. RLP proteins showed a consensus starting motif MALS(L) n and a high level of conservation at the kinase level. The NBS domain is more conserved in TNLs than in CNLs. TIR is the most conserved R domain, with the peak of conservation in N-terminal that reveal a common starting motif MA(S)n and a core conserved domain region. Also for this domain differences among single classes can be evidenced. The TIR domain is more conserved in the TIR-NBS class and in proteins not characterized by other signatures. Moreover, proteins composed by only one TIR domain showed differences in the last part of the profile (Figure 4).
Figure 4.
A) Interproscan results of consensus sequences of each analysed class B) Pattern of conservation along consensus sequence among members of each class.
R domain atypical associations (Aty-R)
Interproscan annotation of putative resistance proteins evidenced the presence of proteins containing unreported domains associated with a R domain. Table 3 shows the most interesting associations found in our dataset. In Arabidopsis the RPW8 domain, known to be involved in the resistance against powdery mildew, was found associated with NBS and LRR domains. Moreover, some of the “Aty-R” domains identified in this work overlap R domains, indicating that they could play a similar role. An example is the TIRAP domain, a Toll/Interleukin-1 Receptor domain containing an adaptor protein, present in Arabidopsis thaliana, Vitis vinifera and Gossypium hirsutum, which showed a very similar sequence to the TIR domain. It has not been found yet in the resistance gene families. Interestingly, some associations were found only in certain species, whereas others were ubiquitous. The association between NBS and “Aty-R” only occurred in Oryza sativa. Retrotransposon elements in association with the R-gene domain were also found. In particular, seven proteins of Oryza sativa, four of Populus trichocarpa and one of Arabidopsis thaliana were evidenced (Table 4).
Table 3.
Atypical R-domain combinations found in the UniGene PRGdb dataset
| Unigene ID | Resistance Class | Aty-R domain(s) | Species |
|---|---|---|---|
| At.43365 |
NBS-LRR-AtyR |
Mob1 |
Arabidopsis thaliana |
| Os.24417 |
NBS-LRR-AtyR |
Zinc finger, ZZ-type |
Oryza sativa |
| Os.25168 |
NBS-AtyR |
GTPase Containing Family |
Oryza sativa |
| Os.25222 |
NBS-AtyR |
Phosphatase 2C |
Oryza sativa |
| Os.25268 |
NBS-AtyR |
WRKY transcription factor; Gag-Pol-related Retrotransposon |
Oryza sativa |
| Os.27097 |
NBS-AtyR |
WRKY transcription factor |
Oryza sativa |
| Os.78619 |
NBS-AtyR |
Zinc finger, CCHC-type |
Oryza sativa |
| Os.79969 |
NBS-AtyR |
DUF1979 |
Oryza sativa |
| Os.93921 |
NBS-AtyR |
Cecropin; Origin replication binding protein |
Oryza sativa |
| Pth.15498 |
NBS-AtyR |
Gag-Pol-related Retrotransposon |
Populus trichocarpa |
| Pth.15636 |
NBS-AtyR |
Zinc finger, BED-type |
Populus trichocarpa |
| At.3076 |
TIR-LRR-NBS-KIN-Serthr-AtyR |
Phenylalanine Hydroxylase (PAH); WRKY transcription factor |
Arabidopsis thaliana |
| At.38115 |
TIR-AtyR |
DUF541 |
Arabidopsis thaliana |
| At.51652 |
TIR-AtyR |
Toll-IL-1 receptor domain-containing adapter protein (TIRAP) |
Arabidopsis thaliana |
| Ghi.9199 |
TIR-AtyR |
Toll-IL-1 receptor domain-containing adapter protein (TIRAP) |
Gossypium hirsutum |
| Gma.3221 |
TIR-AtyR |
Helix-loop-helix structural domain (EF-HAND 2) |
Glycine max |
| Vvi.2456 |
TIR-AtyR |
Toll-IL-1 receptor domain-containing adapter protein (TIRAP) |
Vitis vinifera |
| At.46853 |
TNL-AtyR |
Pleckstrin homology domain (PH); Regulator of chromosome condensation |
Arabidopsis thaliana |
| Pth.16040 |
TNL-AtyR |
CALCINEURIN B |
Populus trichocarpa |
| Pth.16041 |
TNL-AtyR |
DNA-Directed RNA Polymerase II |
Populus trichocarpa |
| Pth.16058 |
TNL-AtyR |
Gag-Pol-related Retrotransposon |
Populus trichocarpa |
| Han.34 |
TIR-NBS-AtyR |
Steroid Binding Protein |
Helianthus annuus |
| Psi.4721 | TIR-NBS-AtyR | Toll-IL-1 receptor domain-containing adapter protein (TIRAP) | Picea sitchensis |
Sequences are divided according to the R domain and sorted by species.
Table 4.
Candidate R genes showing a transposon insertion
| Gene Name | Species | Class or domain |
|---|---|---|
|
Os.25268 |
Oryza sativa |
CNL-WRKY |
|
Os.53813 |
Oryza sativa |
CNL |
|
Os.78767 |
Oryza sativa |
Ser/thr |
|
Os.79795 |
Oryza sativa |
CNL |
|
Os.79928 |
Oryza sativa |
RLP |
|
Os.79975 |
Oryza sativa |
LRR |
|
Os.83660 |
Oryza sativa |
RLP |
|
Pth.15498 |
Populus trichocarpa |
NBS |
|
Pth.16071 |
Populus trichocarpa |
TNL-TIR |
|
Pth.16077 |
Populus trichocarpa |
TNL-TIR |
| Pth.8196 | Populus trichocarpa | TNL |
Sequences are divided according to species and resistance classes. Gene names correspond to UniGene Id.
Discussion
The analysis performed in this paper sheds light on the complex panorama of resistance proteins, highlighting the “underground” information of this family. Although R-proteins are an important and useful family in plant species, some of their characteristics have not been elucidated yet [6,24]. With the advent of the genomic era, the classification of R-proteins into five families is now at odds with the latest discoveries in this field. From our data a possible new scenario emerges, where a broader repertoire of proteins might be involved in the resistance process. In the PRG UniGene dataset, using semi-automated prediction analysis, we detected proteins that were similar to functional R proteins. By choosing only UniGene sequences (set of tailed transcript sequences from the same locus), we avoided selecting pseudo-genes or predicted sequences derived from annotation errors. To ensure that our sampling was sufficiently accurate, the analysis was made more rigorous, selecting a subset of 4409 UniGene homologues to R-proteins and starting with a methionine. The use of a specific R-proteins prediction tool allowed us to place a large number of sequences in known R classes. However, numerous sequences similar to R proteins but with unknown domain arrangements were identified, including new associations among known R domains, proteins with a R domain repetition and sequences containing just one R domain.
Since protein domains are major evolutionary units, the identification of domain loss, transfer, duplication and combination with other domains to form new proteins is important [25]. The distribution of domain associations could be affected by natural pressure that somehow acts to select the most favourable associations to achieve a given task. Domains are considered to be the basic unit of proteins, and reorganizing these blocks may lead to significant changes in the physical structure as well as the biochemical activity of the corresponding proteins [26]. In our data, out of 31 theoretical combinations, only 22 associations were found. Interestingly, the observed data lack TIR associations. Domain shuffling was found to have an important role in the evolution of innate immune systems in both vertebrates and invertebrates [27]. In our study, a high number of proteins with one or two domains were found. R proteins could exert their function associated in a multi-protein complex or alone [28]. Proteins with multi-domains should be able to offer all specific needs for resistance (recognition, signal transduction and energy sourcing), while single domain proteins could change conformation more easily to be able to work in a protein complex. It may be more advantageous for living organisms to produce a higher number of proteins that allow flexible associations. Indeed, recent data suggest that the R domains need to be separated to exert their function [29-31]. Moreover, in our dataset, TIR-LRR, TIR-Ser/thr and more complex derived combinations were not found. These findings suggest that non-detected combinations may not be advantageous. Each domain has a specific function: LRR is involved in recognition and intramolecular interactions [21], kinase in signal transduction [32], NBS in ATP binding [33] and TIR in signalling and molecular interaction [34,35]. Described R domains seem to be essential to initiate a defence response in different patho-systems, but they can be associated in different ways. The associations evidenced in this work offer the opportunity to explore the full panorama of R proteins and understand the rationale of domain association.
In order to further characterize a subset of strong putative R-proteins, a filtering process was conducted. The most difficult part of the process was to find an efficient way to select good candidates to exert resistance function, loosing as few sequences as possible. The construction of a Venn diagram for visualizing the probability of proteins to exert resistance function based on the presence of “strong putative domain” and “weak putative domain” was very useful. Following this approach we were able to highlight putative disease resistance proteins. More detailed analyses were conducted only on proteins showing at least one domain homologous to domains identified in proteins with undisputed resistance function as reported elsewhere [8,24].
Proteins localization could affect domain function, activity, protein structure and affinity for other proteins. Hence, to outline the localization and conformations of putative R-proteins, we performed a transmembrane prediction. Some typical cytosolic classes like the CNL and TNL evidenced transmembrane domains. In order to verify evinced attributes more detailed studies should be performed.
Focusing on single protein class conservation pattern we evidenced some peculiar features. Indeed, little alterations of motifs could have a considerable effect on the functional specificities of the corresponding domains. The LRR domain was the most variable R domain in terms of number of leucine repetitions, length and conservation. The LRR domain is a common motif in more than 2000 proteins. At least four different families, LRR_1, LRR_2, LRR_3, FNIP [36-39], have been found. Differences in number and motifs composition among plant R proteins have been already reported [6]. Looking at R proteins, it is important to underline that the localization of this domain in the CNL/TNL classes and the RLP class is different. In the first case LRR repetitions are positioned at the C-terminal of the proteins, while in the second one at the N-terminal. These data suggest the occurrence of a different evolutionary process for the CNL/TNL and RLP classes, even if they share a common domain. NBS and TIR showed a high percentage of conservation to preserve their function as well as the RLP kinase domains. The NBS domain, associated with the TIR domain (TNL and TIR-NBS classes), is more conserved than the NBS found alone and the NBS present in the CNL proteins. In an Arabidopsis survey, the NBS domain of TNL and CNL are clearly distinguished in different phylogenetic branches [7]. Moreover, the NBS domain of TNL is reported to contain an additional loop [40]. Interestingly, the conservation profile of proteins characterized only by the TIR domain showed some specific peculiarities at the C-terminal part [16].
Finally, many sequences evidenced associations of R domains with domains involved in other processes or domains with unknown functions. Novel identified proteins were collected in a catalogue termed Aty-R (atypical resistance proteins). Several sequences often have a R domain with an additional motif. Interestingly, proteins with a WRKY motif (a motif found in zinc-finger, transcription factor and present also in the RRS1 R-protein, [41]) were found. Aty-R domain associations could have occurred to improve specificity of the protein without changing its structure (TIRAP domain similar with TIR, STRUMBLING receptor similar to Ser/thr) or could have become established to enhance protein expression and stability through domains like WRKY, Zinc finger and EF-Hand [41].
The discoveries of domain associations and the presence of R-domains integrated in transposon elements enhance the possible organizations of R genes, adding new information on the feature of this family. Interestingly, a peculiarity for each species was found, namely the presence of transposon elements in the Oryza and Populus dataset. In Oryza sativa a transposon insertion in genes involved in the resistance process has already been found [42]. Besides transposon insertions, an association composed by TNL-TIR (repetition of TIR domain in the final part of the proteins) was found in Populus trichocarpa. A TIR-TIR interaction between the N and Ntr genes was revealed in Nicotiana, suggesting that two TIR domain interactions could increase resistance ability [43]. Overall, many protein features were revealed and interesting new domain associations were found.
The analysis performed in this study paves the way to understand how plant resistance domain associations are originated. Insights on R domains modelling were also provided. The panorama of R candidate proteins emerging from this analysis makes the current R-protein classification too restrictive. In addition, the recent increasing number of functional R-proteins found, difficult to classify, is a clear indication that a revision is needed [8,37].
Conclusions
The purposes of our study were to investigate the domain architecture of translated expressed sequences similar to R-proteins and to develop approaches to identify candidates for functional studies. We believe that this work is the starting point to explore the panorama of resistance proteins within a different perspective. From our data it emerges that there are several aspects that merit an in-depth study. Tools should be developed to better discriminate general plant receptors from receptors involved in resistance process, to visualize new domain arrangements, to analyse possible 3D domain interactions and to provide models of action. Within the complex R-proteins scenario, our data pose new questions concerning the absence of some domain combinations, the role of sequences containing single domains, the possible involvement of new classes in the resistance process, the role of tandem R domains and the role of transposons in the functionality and expression of R genes. All analysed proteins and all produced datasets were available in a special section of the PRGdb (http://www.prgdb.org), with downloadable data, in-depth studies and advanced search method to extract specific proteins of interest.
Methods
Data selection
Overall, we inspected 10463 UniGene sequences, similar to proteins that exert resistance function, annotated through a specific R-protein prediction pipeline. The dataset was selected by the full NCBI UniGene plant dataset of 600,000 sequences translated by Estscan v.3.0.2 and analysed by the DRAGO pipeline [23]. From PRGdb with ad hoc queries, 4409 proteins starting with a methionine were extrapolated from the entire set and divided, according to their domains, into different classes.
Through an exhaustive data filtering system, a total of 817 proteins have been selected as strong putative resistance proteins. Ubiquitous proteins involved also in other cellular processes (like LRR, RLK, LRR-OthR, LRR-KIN-OthR and KIN-OthR classes) have been excluded from our dataset and stored in separate files. OthR, RLP and kinases classes have been filtered with specific phylogenetic and interproscan analyses.
Protein analysis
Sequences were analysed with InterProScan 4.8 stand-alone version with the last update (spring 2011) of all 13 integrated databases (PROSITE, PRINTS, Pfam, ProDom, SMART, TIGRFAMs, PIR super family, SUPERFAMILY Gene3D, PANTHER and HAMAP) [44]. The output of each sequence was semi-manually checked for conserved domains and 4409 proteins were divided according to their conserved features. To classify putative R proteins in accordance with domain occurrence, a contingency table was obtained using R statistical software [45]. The total set of proteins was examined for the presence of transmembrane domains using Phobius [46] and TMHMM Geneious tools [47] while the coiled-coil prediction was performed by the coiled-coil tool of Geneious [47]. Proteins with new domain associations or containing domains involved in the resistance mechanism but not specific for it were manually inspected for discovering new R protein features. Data were recorded and used for further investigations.
Domain associations
All possible R-domain combinations were calculated using the following binomial formula:
in which n is the number of different domains and k the number of domains that can be found in a single protein. The distribution of theoretical R-domain associations was used to perform a comparison with our dataset. In this study the number of domain (n) is 5 and the number of domains that can be found in a single protein (k) is between 1 and 5.
RLPs filtering
RLP reference resistance proteins (downloaded from PRG selecting “RLP class reference set”), RLPs predicted from our previous analysis and RLP not involved in resistance process were aligned with Muscle v3.6 [48] using a maximum number of iteration of 32. The transmembrane C3-F domains [14] was extracted and the alignment refined. This aligned region was used for a phylogenetic analysis with PHYML v3.0, using the JTT substitution model, transition/transversion model estimated, proportion of invariable site estimated, gamma distribution estimate and number of substitution for categories equal to 4. A tree/length/branch optimization has been obtained and accuracy has been calculated with aLRT statistics method [49]. This approach allowed us to separate RLPs homologues to reference resistance RLPs from others.
MLO filtering
A phylogenetic analysis was performed on the predicted MLO-like proteins to select MLO- proteins that can confer resistance. The MLO reference resistance gene (http://prgdb.crg.eu/gene.php?id=35723&type=ref) [17] and three Arabidosis MLO-like proteins phylogenetically closed to it [50] were downloaded. The 75 MLO-like proteins predicted with our pipeline were aligned with references genes [50] for performing a phylogenetic analysis, following the procedure described in previous paragraph. Proteins belonging to same clade of MLO reference resistance gene have been selected.
Pairwise identity and ANOVA test
Associations of known R domains consisting of more than 10 sequences were grouped and analysed for identity. The alignments were performed with MUSCLE v.3.6 with a maximum of 16 iterations. A total of ten groups of aligned proteins were obtained. Alignments were manually checked and unaligned regions were discarded. ANOVA analysis at 0.05 and 0.01 level of significance was performed on identity results obtained on 10 random batches of 10 sequences collected within each class and compared with results obtained using the total number of proteins belonging to each class. The conservation profile of each group was obtained and examined.
Competing interests
The authors have declared that no competing interests exist.
Authors’ contributions
Conceived and designed the experiments: MRE and WS; Performed data analysis: WS; Analysed results: MRE; Wrote the paper: MRE and WS. Both authors read and approved the final manuscript.
Contributor Information
Walter Sanseverino, Email: walter.sanseverino@unina.it.
Maria Raffaella Ercolano, Email: ercolano@unina.it.
Acknowledgements
We thank Dr. L. Tardella for his help in R software analyses, Dr. F. Giannino and Dr. G. Incerti for statistical support, Dr. A. Ferrigno for mathematical support and M. Walters for editing the manuscript and Prof. D. Carputo for reading our manuscript, for providing suggestions. Contribution no. from the DISSPAPA.
Funding
Ministry of Education, University and Research (GenoPOM-PRO). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- Erickson FL, Holzberg S, Calderon-Urrea A, Handley V, Axtell M, Corr C, Baker B. The helicase domain of the TMV replicase proteins induces the N-mediated defence response in tobacco. Plant J. 1999;18(1):67–75. doi: 10.1046/j.1365-313X.1999.00426.x. [DOI] [PubMed] [Google Scholar]
- Ori N, Eshed Y, Paran I, Presting G, Aviv D, Tanksley S, Zamir D, Fluhr R. The I2C family from the wilt disease resistance locus I2 belongs to the nucleotide binding, leucine-rich repeat superfamily of plant resistance genes. Plant Cell. 1997;9(4):521–532. doi: 10.1105/tpc.9.4.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Gomez L, Boller T. FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell. 2000;5(6):1003–1011. doi: 10.1016/S1097-2765(00)80265-8. [DOI] [PubMed] [Google Scholar]
- Thomas C, Jones D, Parniske M, Harrison K, Balint-Kurti P, Hatzixanthis K, Jones J. Characterization of the tomato Cf-4 gene for resistance to Cladosporium fulvum identifies sequences that determine recognitional specificity in Cf-4 and Cf-9. The Plant Cell Online. 1997;9(12):2209. doi: 10.1105/tpc.9.12.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G, Brommonschenkel S, Chunwongse J, Frary A, Ganal M, Spivey R, Wu T, Earle E, Tanksley S. Map-based cloning of a protein kinase gene conferring disease resistance in tomato. Science. 1993;262(5138):1432–1436. doi: 10.1126/science.7902614. [DOI] [PubMed] [Google Scholar]
- van Ooijen G, van den Burg HA, Cornelissen BJC, Takken FLW. Structure and function of resistance proteins in solanaceous plants. Annu Rev Phytopathol. 2007;45:43–72. doi: 10.1146/annurev.phyto.45.062806.094430. [DOI] [PubMed] [Google Scholar]
- Meyers B, Kozik A, Griego A, Kuang H, Michelmore R. Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell. 2003;15(4):809. doi: 10.1105/tpc.009308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu SH, Bleecker AB. Plant receptor-like kinase gene family: diversity, function, and signaling. Sci STKE. 2001;2001(113):re22. doi: 10.1126/stke.2001.113.re22. [DOI] [PubMed] [Google Scholar]
- Bent A. Plant disease resistance genes: function meets structure. Plant Cell. 1996;8(10):1757–1771. doi: 10.1105/tpc.8.10.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morillo S, Tax F. Functional analysis of receptor-like kinases in monocots and dicots. Curr Opin Plant Biol. 2006;9(5):460–469. doi: 10.1016/j.pbi.2006.07.009. [DOI] [PubMed] [Google Scholar]
- Weber ANR, Moncrieffe MC, Gangloff M, Imler J-L, Gay NJ. Ligand-receptor and receptor-receptor interactions act in concert to activate signaling in the Drosophila toll pathway. J Biol Chem. 2005;280(24):22793–22799. doi: 10.1074/jbc.M502074200. [DOI] [PubMed] [Google Scholar]
- Karlova R, Boeren S, Russinova E, Aker J, Vervoort J, de Vries S. The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE1 protein complex includes BRASSINOSTEROID-INSENSITIVE1. Plant Cell. 2006;18(3):626–638. doi: 10.1105/tpc.105.039412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masle J, Gilmore S, Farquhar G. The ERECTA gene regulates plant transpiration efficiency in Arabidopsis. Nature. 2005;436(7052):866–870. doi: 10.1038/nature03835. [DOI] [PubMed] [Google Scholar]
- Fritz-Laylin L, Krishnamurthy N, Tor M, Sjolander K, Jones J. Phylogenomic analysis of the receptor-like proteins of rice and Arabidopsis. Plant Physiol. 2005;138(2):611–623. doi: 10.1104/pp.104.054452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandwagt BF, Mesbah LA, Takken FL, Laurent PL, Kneppers TJ, Hille J, Nijkamp HJ. A longevity assurance gene homolog of tomato mediates resistance to Alternaria alternata f. sp. lycopersici toxins and fumonisin B1. Proc Natl Acad Sci USA. 2000;97(9):4961–4966. doi: 10.1073/pnas.97.9.4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romer P, Hahn S, Jordan T, Strauss T, Bonas U, Lahaye T. Plant pathogen recognition mediated by promoter activation of the pepper Bs3 resistance gene. Science. 2007;318(5850):645–648. doi: 10.1126/science.1144958. [DOI] [PubMed] [Google Scholar]
- Buschges R, Hollricher K, Panstruga R, Simons G, Wolter M, Frijters A, van Daelen R, van der Lee T, Diergaarde P, Groenendijk J. et al. The barley Mlo gene: a novel control element of plant pathogen resistance. Cell. 1997;88(5):695–705. doi: 10.1016/S0092-8674(00)81912-1. [DOI] [PubMed] [Google Scholar]
- Xiao S, Ellwood S, Calis O, Patrick E, Li T, Coleman M, Turner JG. Broad-spectrum mildew resistance in Arabidopsis thaliana mediated by RPW8. Science. 2001;291(5501):118–120. doi: 10.1126/science.291.5501.118. [DOI] [PubMed] [Google Scholar]
- Jones J, Dangl J. The plant immune system. Nature. 2006;444(7117):323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Dangl JL, Jones JD. Plant pathogens and integrated defence responses to infection. Nature. 2001;411(6839):826–833. doi: 10.1038/35081161. [DOI] [PubMed] [Google Scholar]
- Takken FLW, Tameling WIL. To nibble at plant resistance proteins. Science. 2009;324(5928):744–746. doi: 10.1126/science.1171666. [DOI] [PubMed] [Google Scholar]
- Boller T, He SY. Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science. 2009;324(5928):742–744. doi: 10.1126/science.1171647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanseverino W, Roma G, De Simone M, Faino L, Melito S, Stupka E, Frusciante L, Ercolano MR. PRGdb: a bioinformatics platform for plant resistance gene analysis. Nucleic Acids Res. 2010;38(Database issue):D814–821. doi: 10.1093/nar/gkp978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GB, Bogdanove AJ, Sessa G. Understanding the functions of plant disease resistance proteins. Annu Rev Plant Biol. 2003;54:23–61. doi: 10.1146/annurev.arplant.54.031902.135035. [DOI] [PubMed] [Google Scholar]
- Yang S, Bourne PE. The evolutionary history of protein domains viewed by species phylogeny. PLoS One. 2009;4(12):e8378. doi: 10.1371/journal.pone.0008378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Zmasek CM, Dishaw LJ, Mueller MG, Ye Y, Litman GW, Godzik A. Novel genes dramatically alter regulatory network topology in amphioxus. Genome Biol. 2008;9:R123. doi: 10.1186/gb-2008-9-8-r123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue JX, Meyers BC, Chen JQ, Tian D, Yang S. Tracing the origin and evolutionary history of plant nucleotide-binding site-leucine-rich repeat (NBS-LRR) genes. New Phytol. 2012;193(4):1049–1063. doi: 10.1111/j.1469-8137.2011.04006.x. [DOI] [PubMed] [Google Scholar]
- Jones DA, Takemoto D. Plant innate immunity - direct and indirect recognition of general and specific pathogen-associated molecules. Curr Opin Immunol. 2004;16(1):48–62. doi: 10.1016/j.coi.2003.11.016. [DOI] [PubMed] [Google Scholar]
- Moffett P, Farnham G, Peart J, Baulcombe D. Interaction between domains of a plant NBS-LRR protein in disease resistance-related cell death. EMBO J. 2002;21(17):4511. doi: 10.1093/emboj/cdf453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M, Shao F, Innes RW, Dixon JE, Xu Z. The crystal structure of Pseudomonas avirulence protein AvrPphB: a papain-like fold with a distinct substrate-binding site. Proc Natl Acad Sci USA. 2004;101(1):302–307. doi: 10.1073/pnas.2036536100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney HC, Van't Klooster JW, Van der Hoorn RA, Joosten MH, Jones JD, De Wit PJ. Cladosporium Avr2 inhibits tomato Rcr3 protease required for Cf-2-dependent disease resistance. Science. 2005;308(5729):1783–1786. doi: 10.1126/science.1111404. [DOI] [PubMed] [Google Scholar]
- Stone JM, Walker JC. Plant protein kinase families and signal transduction. Plant Physiol. 1995;108(2):451–457. doi: 10.1104/pp.108.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tameling WI, Elzinga SD, Darmin PS, Vossen JH, Takken FL, Haring MA, Cornelissen BJ. The tomato R gene products I-2 and MI-1 are functional ATP binding proteins with ATPase activity. Plant Cell. 2002;14(11):2929–2939. doi: 10.1105/tpc.005793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp EB, Medzhitov R. The Toll-receptor family and control of innate immunity. Curr Opin Immunol. 1999;11(1):13–18. doi: 10.1016/S0952-7915(99)80003-X. [DOI] [PubMed] [Google Scholar]
- Collier SM, Moffett P. NB-LRRs work a "bait and switch" on pathogens. Trends Plant Sci. 2009;14(10):521–529. doi: 10.1016/j.tplants.2009.08.001. [DOI] [PubMed] [Google Scholar]
- Dievart A, Clark SE. LRR-containing receptors regulating plant development and defense. Development. 2004;131(2):251–261. doi: 10.1242/dev.00998. [DOI] [PubMed] [Google Scholar]
- Panstruga R. Discovery of novel conserved peptide domains by ortholog comparison within plant multi-protein families. Plant Mol Biol. 2005;59(3):485–500. doi: 10.1007/s11103-005-0353-0. [DOI] [PubMed] [Google Scholar]
- Kobe B, Deisenhofer J. Crystal structure of porcine ribonuclease inhibitor, a protein with leucine-rich repeats. Nature. 1993;366(6457):751–756. doi: 10.1038/366751a0. [DOI] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R. et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34(Database issue):D247–251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHale L, Tan X, Koehl P, Michelmore RW. Plant NBS-LRR proteins: adaptable guards. Genome Biol. 2006;7(4):212. doi: 10.1186/gb-2006-7-4-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslandes L, Olivier J, Theulieres F, Hirsch J, Feng DX, Bittner-Eddy P, Beynon J, Marco Y. Resistance to Ralstonia solanacearum in Arabidopsis thaliana is conferred by the recessive RRS1-R gene, a member of a novel family of resistance genes. Proc Natl Acad Sci USA. 2002;99(4):2404–2409. doi: 10.1073/pnas.032485099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Ramakrishna W. Retrotransposon insertion polymorphisms in six rice genes and their evolutionary history. Gene. 2008;412(1–2):50–58. doi: 10.1016/j.gene.2008.01.012. [DOI] [PubMed] [Google Scholar]
- Stange C, Matus JT, Dominguez C, Perez-Acle T, Arce-Johnson P. The N-homologue LRR domain adopts a folding which explains the TMV-Cg-induced HR-like response in sensitive tobacco plants. J Mol Graph Model. 2008;26(5):850–860. doi: 10.1016/j.jmgm.2007.05.006. [DOI] [PubMed] [Google Scholar]
- Hunter S, Apweiler R, Attwood T, Bairoch A, Bateman A, Binns D, Bork P, Das U, Daugherty L, Duquenne L. et al. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37(Database issue):D211–215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihaka RG R. R: A language for data analysis and graphics. J Comput Graph Stat. 1996;5:15. [Google Scholar]
- Kall L, Krogh A, Sonnhammer E. Advantages of combined transmembrane topology and signal peptide prediction--the Phobius web server. Nucleic Acids Res. 2007;35(Web Server issue):W429–432. doi: 10.1093/nar/gkm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305(3):567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisimova M, Gascuel O. Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst Biol. 2006;55(4):539–552. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
- Bai Y, Pavan S, Zheng Z, Zappel NF, Reinstadler A, Lotti C, De Giovanni C, Ricciardi L, Lindhout P, Visser R. et al. Naturally occurring broad-spectrum powdery mildew resistance in a Central American tomato accession is caused by loss of mlo function. Mol Plant Microbe Interact. 2008;21(1):30–39. doi: 10.1094/MPMI-21-1-0030. [DOI] [PubMed] [Google Scholar]