Abstract
The endoplasmic reticulum (ER) body, a large compartment specific to the Brassicales, accumulates β-glucosidase and possibly plays a role in the defense against pathogens and herbivores. Although the ER body is a subdomain of the ER, it is unclear whether any ER body-specific membrane protein exists. In this study, we identified two integral membrane proteins of the ER body in Arabidopsis (Arabidopsis thaliana) and termed them MEMBRANE PROTEIN OF ENDOPLASMIC RETICULUM BODY1 (MEB1) and MEB2. In Arabidopsis, a basic helix-loop-helix transcription factor, NAI1, and an ER body component, NAI2, regulate ER body formation. The expression profiles of MEB1 and MEB2 are similar to those of NAI1, NAI2, and ER body β-glucosidase PYK10 in Arabidopsis. The expression of MEB1 and MEB2 was reduced in the nai1 mutant, indicating that NAI1 regulates the expression of MEB1 and MEB2 genes. MEB1 and MEB2 proteins localize to the ER body membrane but not to the ER network, suggesting that these proteins are specifically recruited to the ER body membrane. MEB1 and MEB2 physically interacted with ER body component NAI2, and they were diffused throughout the ER network in the nai2 mutant, which has no ER body. Heterologous expression of MEB1 and MEB2 in yeast (Saccharomyces cerevisiae) suppresses iron and manganese toxicity, suggesting that MEB1 and MEB2 are metal transporters. These results indicate that the membrane of ER bodies has specific membrane proteins and suggest that the ER body is involved in defense against metal stress as well as pathogens and herbivores.
Proteins entering the secretory pathway are synthesized on the rough endoplasmic reticulum (ER). COPII vesicles, which are approximately 50 nm in diameter, represent the majority of transport vesicles moving from the ER (Alberts et al., 2002). However, plants differ from animals in that they also produce different types of ER-derived vesicles, and these are involved in the accumulation of proteins (Galili, 2004; Hara-Nishimura et al., 2004; Herman and Schmidt, 2004; Herman, 2008). Protein bodies (PBs) store seed storage proteins in the endosperm of maize (Zea mays) and rice (Oryza sativa; Herman and Larkins, 1999; Satoh-Cruz et al., 2010; Nagamine et al., 2011). Precursor-accumulating vesicles accumulate seed protein precursors in the maturing cotyledons of pumpkin (Cucurbita maxima; Hara-Nishimura et al., 1998). Similar structures occur in the seeds of maigo mutants that have a defect in the transport of seed storage proteins between the ER and Golgi in Arabidopsis (Arabidopsis thaliana; Takahashi et al., 2010). KDEL-tailed proteinase-accumulating vesicles and ricinosomes accumulate a papain-type proteinase in dying tissues in mung bean (Vigna mungo) and castor bean (Ricinus communis) seedlings (Toyooka et al., 2000; Schmid et al., 2001). Interestingly, these unique vesicles are larger (approximately 500 nm) than COPII vesicles and appear at specific stages of the plant life cycle.
In Arabidopsis, several research groups identified another type of ER structure, designated the ER body/fusiform body (Gunning, 1998; Ridge et al., 1999; Matsushima et al., 2003a). The spindle-shaped, 5- to 10-µm-long ER bodies can be easily detected in Arabidopsis expressing an ER-targeted GFP (Hayashi et al., 2001). Electron microscopy revealed that ER bodies have a fibrous structure, have a connection with the ER network, and are surrounded by a single ribosome-bearing membrane (Gunning, 1998; Hayashi et al., 2001). In young seedlings, ER bodies are uniformly distributed throughout the epidermis of cotyledons and hypocotyls but subsequently disappear as the plant grows (Matsushima et al., 2002). In contrast, most root tissues constitutively express ER bodies (Matsushima et al., 2003a). Interestingly, wounding or treatment with the wound hormone jasmonate induces ER body accumulation in adult leaves (Matsushima et al., 2002), suggesting that the ER body is involved in pest/pathogen resistance in Arabidopsis (Hara-Nishimura and Matsushima, 2003; Yamada et al., 2011). In Arabidopsis seedlings, PYK10, a β-glucosidase with the ER-retention signal KDEL, is the major component of the ER body (Matsushima et al., 2003b). Recently, PYK10 was identified as a β-glucosidase that cleaves scopolin, a major glucoside in the root (Ahn et al., 2010). PYK10 is also involved in resistance against the mutualistic root fungus Piriformospora indica (Sherameti et al., 2008). PYK10 deficiency causes hyperinfection by the fungus, which ends the mutualistic interaction and suggests that the ER body is involved in resistance to fungal infection. Structures similar to the ER bodies of Arabidopsis have been reported as dilated ER cisternae in cells of various organs in other Brassicales (Iversen, 1970a, 1970b; Behnke and Eschlbeck, 1978).
PYK10 is the major component of the ER body in the seedlings and roots of Arabidopsis (Matsushima et al., 2003b). Wound-induced ER bodies accumulate BGLU18, a β-glucosidase with the ER-retention signal REEL (Ogasawara et al., 2009). Structural changes to PYK10 via the mutation or down-regulation of PYK10 and BGLU21 (the closest homolog of PYK10) results in the elongation of the ER body (Nagano et al., 2009). These findings suggest that ER body contents regulate ER body morphology. Most ER-soluble proteins contain a C-terminal ER-retention signal that is required for retention in the ER. The presence of KDEL on PYK10 and of REEL on BGLU18 suggests that ER-retention mechanisms are used to localize these proteins in the ER body.
In a previous study, Arabidopsis nai1 and nai2 mutants were found to lack ER bodies; NAI1 was found to encode a basic helix-loop-helix-type transcription factor, while NAI2 was found to encode a novel ER body protein (Matsushima et al., 2004; Yamada et al., 2008). NAI1 regulates the expression of NAI2 and PYK10. NAI2 is an ER body component involved in ER body formation and PYK10 accumulation (Yamada et al., 2008, 2009, 2011). NAI1 also regulates the expression of JACALIN-RELATED LECTIN genes (JAL22, JAL23, JAL31, JAL33, and PBP1/JAL30) and GDSL-LIPASE-LIKE PROTEIN genes (GLL23 and GLL25; Matsushima et al., 2004; Nagano et al., 2005, 2008). PBP1 localizes to the cytosol, while PYK10 localizes to the ER body (Nagano et al., 2005). When cells are broken, PYK10 forms large complexes with JALs and GLLs that regulate the size of the PYK10 complex and may regulate substrate specificity (Nagano et al., 2008). Although soluble ER body components have been identified, including NAI2, PYK10, and BGLU18, the components of the ER body membrane remain unknown. This study focuses on two novel ER body membrane proteins, the localization of which is regulated by the ER body component NAI2.
RESULTS
Identification of MEB1 and MEB2 by in Silico Analysis
The failure of Arabidopsis nai1 mutants to form ER bodies suggests that the transcription factor NAI1 regulates the expression of ER body genes. Microarray analysis of a nai1-1 mutant showed that the expression of several genes was reduced compared with wild-type plants (Nagano et al., 2008), including two homologous genes, At4g27860 and At5g24290, that encode integral membrane proteins (Table I). The ATTED-II database showed that the expression profile of At4g27860 and At5g24290 was similar to that of PYK10, NAI1, and NAI2 (Table I), suggesting that these genes encode proteins related to the ER body. We named these genes MEMBRANE PROTEIN OF ENDOPLASMIC RETICULUM BODY1 (MEB1; At4g27860) and MEB2 (At5g24290). Cell fractionation and proteomic analysis showed that the MEB2 protein cofractionates with NAI2, but the localization of both proteins could not be clearly resolved (Dunkley et al., 2006). These findings suggest that MEB1 and MEB2 localize to the ER body, which includes NAI2 in Arabidopsis.
Table I. Expression levels of MEB1 and MEB2 in the nai1 mutant and correlation with the expression levels of NAI1, NAI2, and PYK10.
Fold indicates fold change determined by microarray analysis of Arabidopsis nai1-1 and the Columbia wild type. Cor indicates Pearson’s correlation coefficient for gene expression patterns. These correlation coefficients are listed in the ATTED-II database (http://atted.jp; Obayashi et al., 2011). Rank orders of correlation coefficients for each gene are shown in parentheses.
| Gene | Locus | Fold (nai1/Wild Type) | Cor (NAI1) | Cor (NAI2) | Cor (PYK10) |
|---|---|---|---|---|---|
| NAI1 | At2g22770 | – | 1.00 (–) | 0.68 (12) | 0.59 (24) |
| NAI2 | At3g15950 | 0.13 | 0.68 (5) | 1.00 (–) | 0.89 (4) |
| PYK10 | At3g09260 | 0.23 | 0.59 (19) | 0.89 (6) | 1.00 (–) |
| MEB1 | At4g27860 | 0.31 | 0.66 (3) | 0.69 (13) | 0.63 (15) |
| MEB2 | At5g24290 | 0.18 | 0.73 (1) | 0.64 (21) | 0.53 (66) |
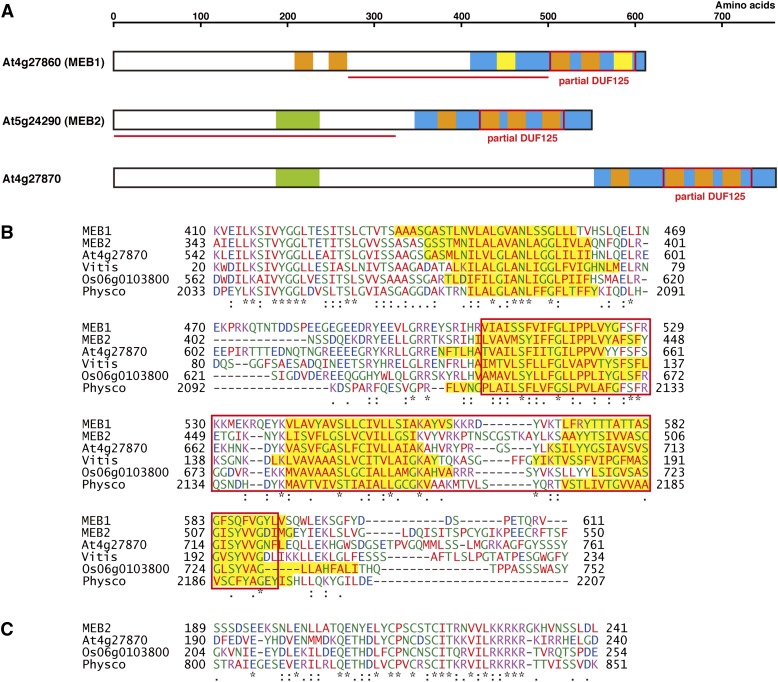
MEB1 and MEB2 have homologous C-terminal regions designated as DOMAIN OF UNKNOWN FUNCTION125 (DUF125) in the Pfam database (Finn et al., 2010). MEB1 and MEB2 also contain four hypothetical transmembrane domains (Fig. 1A). It is possible that the two transmembrane domains of MEB1 are not actual transmembrane regions, because they are not assigned as transmembrane domains with some transmembrane prediction tools. There is another homolog, At4g27870 in Arabidopsis. Proteins with homology to MEB1/MEB2 exist in various plants, including grape (Vitis vinifera; CAO17728), rice (Oryza sativa; Os06g0103800), and moss (Physcomitrella patens; XP_001760792; Fig. 1B). The N-terminal regions of these proteins are enriched in hydrophilic amino acids, but their amino acid sequences diverged highly. Interestingly, MEB1 has two characteristic hydrophobic N-terminal regions, presumably transmembrane domains, which are absent in MEB2, At4g27870, CAO17728, Os06g0103800, and XP_001760792 (Fig. 1A). Instead of these hydrophobic regions, MEB2, At4g27870, Os06g0103800, and XP_001760792 have a short homologous region that we have named the MEB2 domain (Fig. 1C). GenBank/EMBL/DNA Data Bank of Japan was queried using the unique N-terminal regions of MEB1 and MEB2 to identify MEB1-like and MEB2-like proteins, respectively. The potential homologs of MEB1 were found only in the Brassicaceae. In Arabidopsis lyrata, a single protein (EFH45796) appeared to be a homolog of MEB1. In Brassica napus, a tentative consensus sequence (TC11332) was assembled from ESTs (http://compbio.dfci.harvard.edu/tgi/plant.html) that appeared to encode a homolog of MEB1 (Supplemental Fig. S1A). These genes encode proteins that have two transmembrane regions in their N-terminal regions, which is a characteristic feature of MEB1 (Supplemental Fig. S1B). These findings suggest that MEB1-like proteins diverged during the evolution of Brassicales, which have ER bodies. Several proteins with both the MEB2 domain and a partial DUF125 were identified in various plant species (Fig. 1, B and C), suggesting that MEB2-like proteins are widely distributed.
Figure 1.
Structures of MEB1, MEB2, and At4g27870 proteins. A, Diagram showing the structural regions of MEB1, MEB2, and At4g27870. Orange and yellow boxes indicate putative transmembrane regions with high and low probability, respectively. Red boxes indicate partial DUF125 sequences. Blue boxes show homologous C-terminal regions. Green boxes show MEB2 domains. Sequences underlined in red indicate regions used as antigens for the production of specific antibodies. B, Alignment of the C-terminal regions (blue boxes in A) of MEB1- and MEB2-related proteins. Homologous regions of MEB1, MEB2, At4g27870, grape CAO17728, rice Os06g0103800 (NP_001056545), and moss XP_001760792 were aligned using ClustalW2. Numbers indicate amino acids. Red boxes show partial DUF125 regions. Yellow boxes show putative transmembrane regions. C, Alignment of the MEB2 domain (green boxes in A) of MEB2-related proteins. Homologous regions of MEB2, At4g27870, Os06g0103800, and XP_001760792 were aligned using ClustalW2. Numbers indicate amino acids.
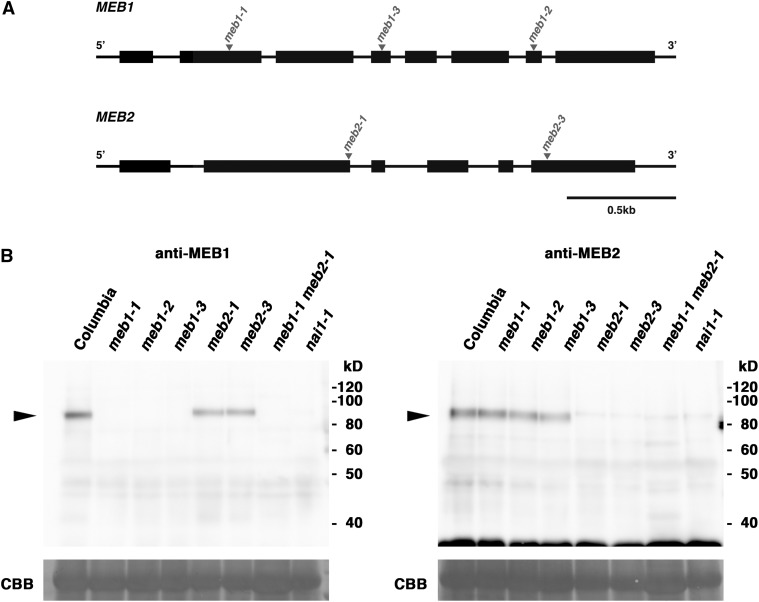
Antibodies were generated against the unique N-terminal regions of MEB1 and MEB2 (Fig. 1A). T-DNA insertion mutants of MEB1 and MEB2 were obtained (Fig. 2A). All mutants have a T-DNA insertion in the protein-coding region and therefore may produce truncated proteins. The anti-MEB1 antibody recognized an approximately 84-kD polypeptide absent in all meb1 mutants, and the anti-MEB2 antibody recognized an approximately 82-kD polypeptide absent in all meb2 mutants (Fig. 2B), indicating that these bands represent the MEB1 and MEB2 polypeptides and showing that full-length MEB1 and MEB2 are expressed in Arabidopsis.
Figure 2.
Detection of MEB1 and MEB2 proteins. A, Exon/intron structures of MEB1 (top) and MEB2 (bottom) genes. Black boxes indicate exons. Triangles indicate T-DNA insertion sites in each mutant. B, Immunoblot analysis of 7-d-old seedlings from the strains indicated was performed using antibodies against MEB1 (left) or MEB2 (right). Arrowheads indicate bands corresponding to MEB1 and MEB2 polypeptides. Coomassie blue staining (CBB) shows the Rubisco large subunit, which served as a loading control.
NAI1 Regulates the Expression of MEB1 and MEB2
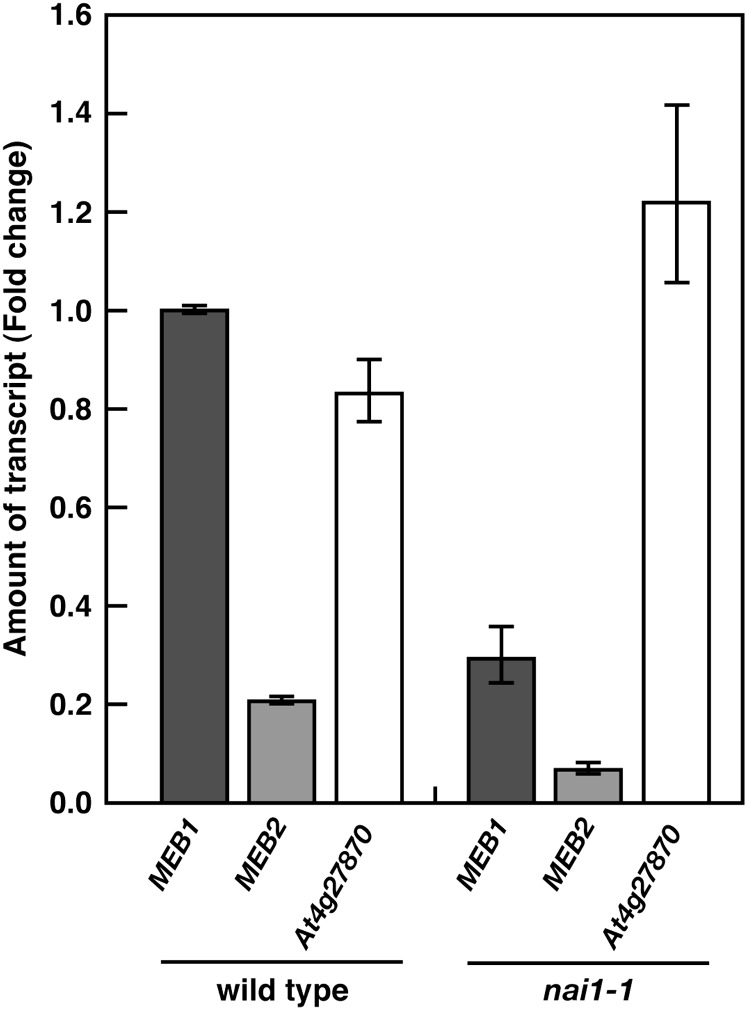
NAI1 is a basic helix-loop-helix transcription factor that regulates the expression of ER body-related genes in Arabidopsis (Nagano et al., 2008). The microarray data showed that the expression of MEB1 and MEB2 is reduced in the nai1 mutant (Table I). To determine the expression level of MEB1, MEB2, and At4g27870 in Arabidopsis, quantitative reverse transcription (qRT)-PCR was performed in nai1 mutant and wild-type Arabidopsis seedlings (Fig. 3). The expression of MEB1 and MEB2 was reduced by approximately two-thirds in nai1 mutant seedlings compared with wild-type seedlings, indicating that these genes are regulated by NAI1. This result is consistent with the absence of MEB1 and MEB2 protein in the nai1 mutant (Fig. 2B). The expression of At4g27870 is not reduced by the nai1 mutation, suggesting that NAI1 is not a transcription factor for At4g27870.
Figure 3.
MEB1, MEB2, and At4g27870 mRNA levels in the wild type and the nai1 mutant. Total RNA from 7-d-old seedlings was subjected to qRT-PCR analysis. The data were normalized to UBIQUITIN10 mRNA levels. The relative abundance of each mRNA was calibrated against the amount of MEB1 mRNA in wild-type plants. Relative quantities were calculated using the 2−ΔΔCt method (Livak and Schmittgen, 2001). Error bars indicate se of threshold cycle (Ct) values, which are calculated using the equation 2−ΔΔCt ± se. These data represent the results of three independent biological replicates.
MEB1 and MEB2 Are ER Body Integral Membrane Proteins
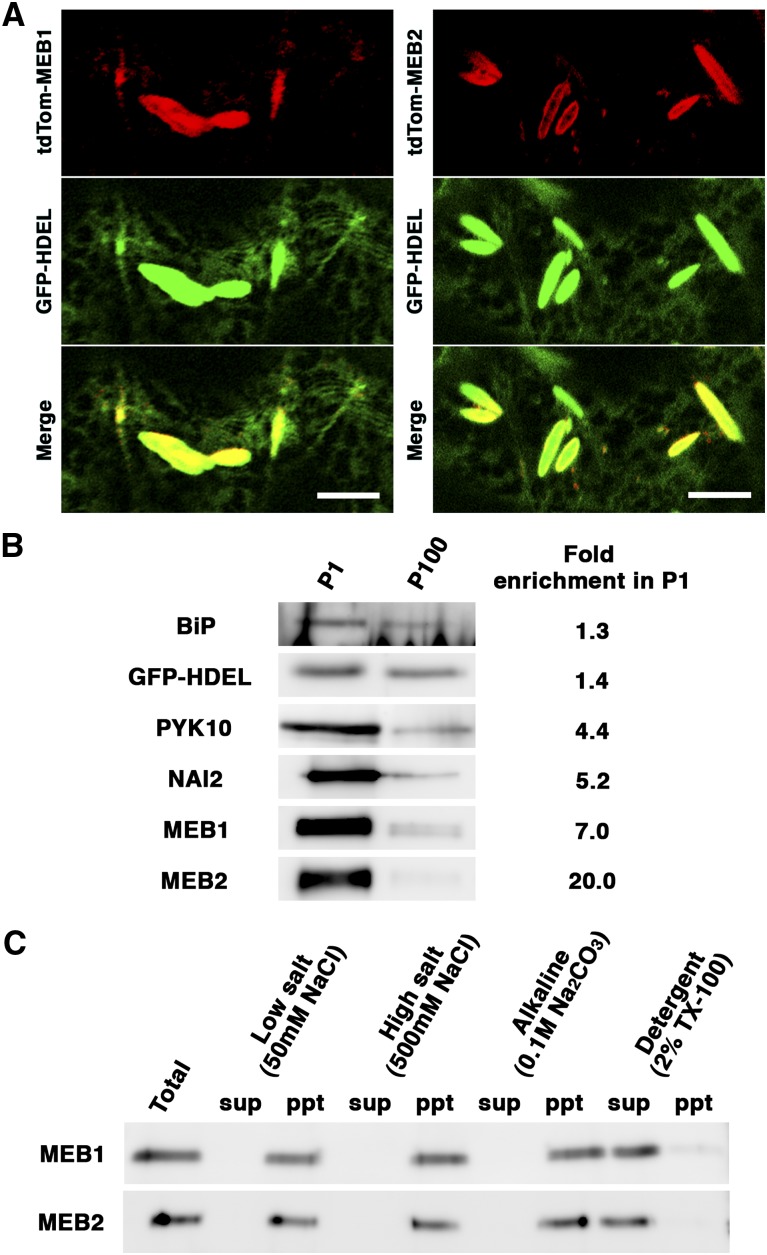
To examine the localization of MEB1 and MEB2, we generated expression vectors encoding fusion proteins between the fluorescent protein tdTomato (tdTOM) and MEB1 or MEB2. These proteins were transiently expressed in the cotyledons of transgenic Arabidopsis (GFP-h) in which the ER was marked by GFP (Fig. 4A). The signal from tdTOM-MEB1 and tdTOM-MEB2 was observed in the peripheral regions of the ER bodies (Fig. 4A; Supplemental Fig. S2), indicating that these fusion proteins localize to the ER body membranes.
Figure 4.
MEB1 and MEB2 localize to the ER body membrane. A, Fluorescence images of cotyledon epidermal cells transiently expressing tdTom-MEB1 (left) or tdTom-MEB2 (right) fusion proteins in 7-d-old GFP-h plants. Bar = 10 µm. B, Immunoblot analysis of subcellular fractions enriched for ER bodies (P1) or ER networks (P100) using antibodies against BiP, GFP (GFP-HDEL), NAI2, MEB1, and MEB2. The same sample volume was loaded in each lane. Fold enrichment in P1 indicates the ratio of P1 to P100 band intensities. C, Immunoblot analysis of the P1 fraction shown in B following buffer treatment and ultracentrifugation to separate soluble and membrane fractions. Low-salt (50 mm NaCl) buffer extracts soluble proteins, high-salt (500 mm NaCl) and alkaline buffers extract peripheral membrane proteins, and detergent (2% [v/v] Triton X-100) buffer extracts integral membrane proteins. ppt, Precipitate; sup, supernatant.
To confirm the localization of MEB1 and MEB2, ER body-rich (P1) and ER network-rich (P100) fractions were isolated using a subcellular fractionation method established previously (Matsushima et al., 2003b). These fractions were subjected to immunoblot analysis using specific antibodies against MEB1 and MEB2. Binding protein (BiP) and GFP-HDEL were detected in both the P1 and P100 fractions (Fig. 4B), indicating that these proteins accumulate in both the ER and the ER body. By contrast, MEB1 and MEB2 were concentrated in the P1 fraction along with the ER body resident proteins NAI2 and PYK10. These data confirm that MEB1 and MEB2 are ER body proteins.
Since MEB1 and MEB2 contain putative transmembrane regions (Fig. 1), we investigated whether they were integral membrane proteins. The P1 fractions were treated with different buffers: a low-salt buffer to extract soluble proteins, a high-salt buffer and alkaline buffer to extract peripheral membrane proteins, or a detergent buffer to extract integral membrane proteins. Following centrifugation, the supernatants and precipitates were subjected to immunoblot analysis (Fig. 4C). MEB1 and MEB2 were not extracted in the low-salt, high-salt, and alkaline buffers but were extracted in the detergent buffer. These data show that MEB1 and MEB2 are integral membrane proteins of the ER body.
Accumulation of MEB1 and MEB2 on ER Bodies Produced by NAI2
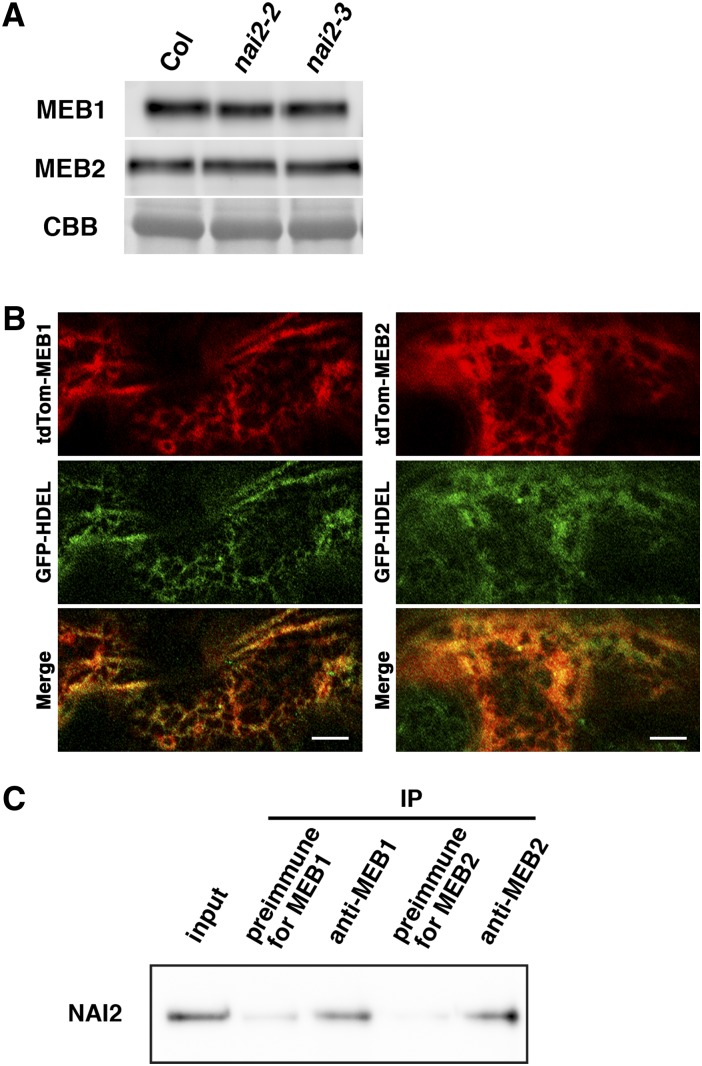
In the Arabidopsis nai2 mutant, which has no ER body, PYK10 protein levels are reduced and PYK10 expression is diffuse throughout the ER network (Yamada et al., 2008). To examine the effect of NAI2 depletion on MEB1 and MEB2 protein levels, we performed immunoblot analysis of MEB1 and MEB2 with the nai2 mutants. Immunoblot analysis showed no reduction in the levels of MEB1 or MEB2 in the Arabidopsis nai2 mutants (Fig. 5A). Next, the localization of MEB1 and MEB2 was assessed in the nai2 mutant by the transient expression of tdTOM-MEB1 and tdTOM-MEB2. The fusion proteins colocalized with the ER GFP signal (Fig. 5B), indicating that NAI2 depletion results in the dispersion of MEB1 and MEB2 to the ER network. This result suggests that MEB1 and MEB2 accumulate on ER bodies produced by NAI2. Since NAI2 is an ER body protein, it is possible that NAI2 forms a complex with MEB1 and MEB2 on the ER body membrane. To examine this, proteins were extracted from Arabidopsis seedlings, and MEB1- and MEB2-containing protein complexes were immunoprecipitated using specific antibodies. NAI2 was detected in MEB1- and MEB2-containing protein complexes (Fig. 5C), indicating that NAI2 and MEB1 or MEB2 are located in the same complexes. These results suggest that NAI2 interacts directly or indirectly with MEB1 and MEB2 on the ER body membrane.
Figure 5.
ER localization of MEB1 and MEB2 in the nai2 mutant. A, MEB1 and MEB2 levels in nai2 mutants. Total protein extract from 7-d-old seedlings was subjected to immunoblot analysis using antibodies against MEB1 or MEB2. Coomassie Brilliant Blue staining (CBB) shows the Rubisco large subunit as a loading control. B, Fluorescence images of cotyledon epidermal cells from 7-d-old seedlings transiently expressing tdTOM-MEB1 or tdTOM-MEB2. A nai2-2 mutant constitutively expressing ER-targeted GFP (GFP-HDEL) was bombarded with gold particles coated with the appropriate plasmids and germinated for 1 d. Bars = 10 µm. C, MEB1 and MEB2 proteins were immunoprecipitated (IP) from Arabidopsis seedlings with anti-MEB1 and anti-MEB2 antibodies. Complexes containing NAI2 protein were detected with biotinylated anti-NAI2 antibody. Sera from preimmunized rabbits were used as negative controls. Input shows samples used for immunoprecipitation (1:100 dilution).
MEB1 and MEB2 Are Functional Homologs of the Yeast Iron Transporter CCC1
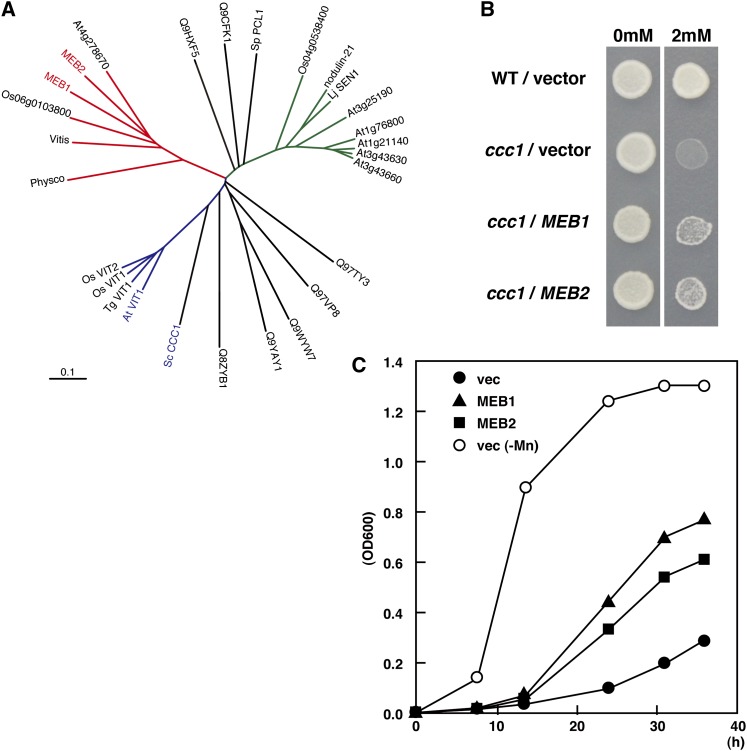
MEB1 and MEB2 have DUF125 sequence. The proteins containing a DUF125 sequence are assigned as the VACUOLAR IRON TRANSPORTER (VIT) family, including CCC1 in yeast (Saccharomyces cerevisiae), AtVIT1 in Arabidopsis, TgVIT1 in tulip (Tulipa gesneriana), NODULIN21 in soybean (Glycine max), and LjSEN1 in Lotus japonicus (Delauney et al., 1990; Li et al., 2001; Kim et al., 2006; Momonoi et al., 2009; Hakoyama et al., 2012). The VIT family includes the iron/manganese uptake transporter. Although the protein structures of MEB1 and MEB2 are different from canonical VITs such as CCC1 in yeast and VIT1 in Arabidopsis, the DUF125 sequence of those proteins could be aligned during phylogenetic tree analysis (Fig. 6A). There are three subfamilies of plant DUF125 proteins: the VIT1 subfamily represented by AtVIT1 (Kim et al., 2006), the SEN1/NODULIN21 subfamily represented by LjSEN1 (Hakoyama et al., 2012), and the MEB1/MEB2 subfamily. In yeast, CCC1 sequesters excess cytosolic iron to the vacuole, and a defect of CCC1 increases iron sensitivity by disrupting the regulation of cytosolic iron concentration (Li et al., 2001; Lin et al., 2011). Interestingly, overexpression of MEB1 or MEB2 in the ccc1 mutant weakly suppressed iron toxicity. The yeast ccc1 mutant could not grow on synthetic Gal medium supplemented with 2 mm iron, whereas the wild type grew on medium with and without iron. The ccc1 mutant transformed with MEB1 or MEB2 could grow on medium containing 2 mm iron, although growth was reduced (Fig. 6B). This indicates that MEB1 or MEB2 partially reduces the iron toxicity in the yeast ccc1 mutant. CCC1 transports manganese, and overexpression of CCC1 suppresses the toxicity resulting from high manganese concentrations in wild-type yeast (Lapinskas et al., 1996). Overproduction of MEB1 or MEB2 also suppresses the growth retardation caused by high concentrations of manganese in yeast (Fig. 6C). These results show that MEB1 and MEB2 serve as functional homologs of CCC1, which is an iron/manganese transporter. In yeast, MEB1 and MEB2 may localize in the ER and sequester excess cytosolic iron and manganese into the ER to reduce metal ion toxicity.
Figure 6.
Functional analysis of MEB1 and MEB2 in yeast. A, Phylogenetic tree of DUF125 domains. DUF125 regions were extracted, and the dendrogram was drawn with ClustalW2 and iTOL. Plant DUF125-containing proteins group into three subfamilies. The MEB1/MEB2 subfamily (red branch) contains MEB1, MEB2, At4g27870, grape CAO17728 (Vitis), rice Os06g0103800, and moss XP_001760792 (Physco). The VIT1 subfamily (blue branch) contains Arabidopsis VIT1 (At2g01770), tulip VIT1 (TgVIT1; BAH98154), and rice VIT1 (Os04g0463400) and VIT2 (Os09g0396900). The SEN1/NODULIN21 subfamily (green branch) contains L. japonicus SEN1 (BAL46698), soybean NODULIN21 (P16313), rice Os04g0538400, and Arabidopsis At3g25190, At1g76800, At1g21140, At3g43630, and At3g43660. The dendrogram also includes DUF125 domain-containing proteins from yeast (S. cerevisiae [CCC1] and Schizosaccharomyces pombe [PCL1]) and unicellular bacteria ([Sulfolobus solfataricus [Q97VP8 and Q97TY3], Thermotoga maritima [Q9WYW7], Aeropyrum pernix [Q9YAY1], Pyrobaculum aerophilum [Q8ZYB1], Pseudomonas aeruginosa [Q9HXF5], and Lactococcus lactis [Q9CFK1]). MEB1 and MEB2 are highlighted in red. ScCCC1 and AtVIT1, which have iron/manganese transporter activities, are highlighted in blue. The horizontal scale represents the evolutionary distance expressed as the number of substitutions per amino acid. B, MEB1 and MEB2 suppress the toxicity of iron in the ccc1 mutant. Four yeast strains, the wild type harboring empty vector (WT/vector), ccc1 mutant harboring empty vector (ccc1/vector), MEB1 (ccc1/MEB1), and MEB2 (ccc1/MEB2), were grown on synthetic minimal medium with (left) or without (right) 2 mm iron. The same amount of cells were spotted and germinated for 4 d. C, Three yeast strains, the wild type harboring empty vector (vec; black circles), MEB1 (black triangles), and MEB2 (black squares) were germinated in liquid medium containing 15 mm MnCl2, and the growth curves were plotted. White circles indicate the growth curve of yeast without manganese.
MEB1 and MEB2 Are Dispensable for ER Body Formation
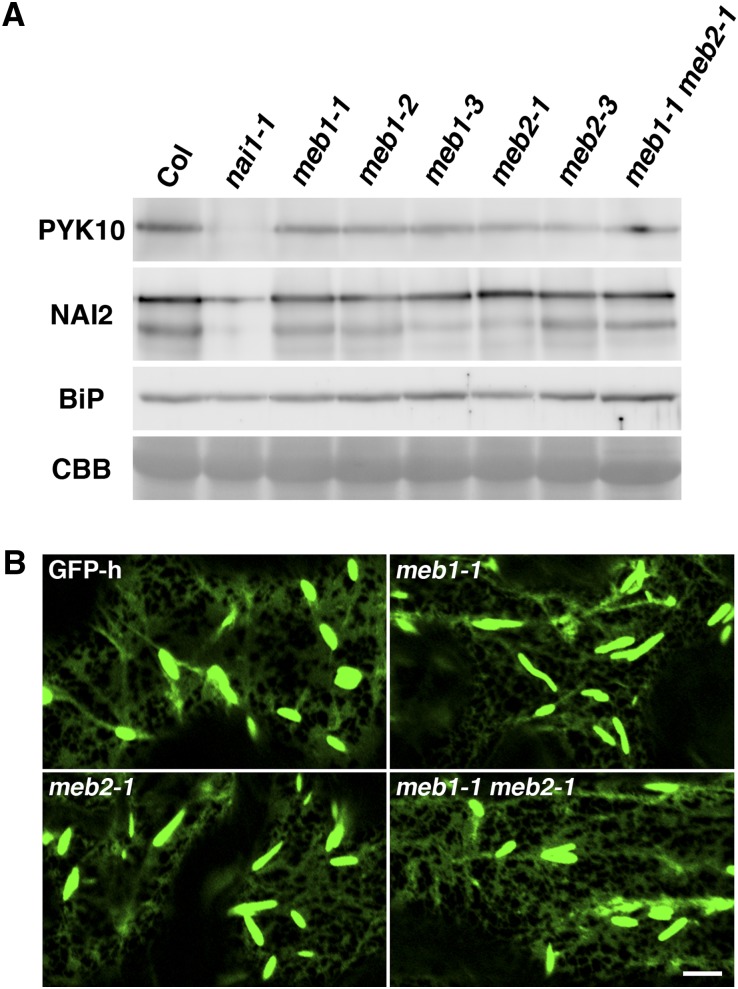
To examine the effects of MEB1 and MEB2 on ER body formation, knockout mutants of these proteins were examined. The meb1 and meb2 mutants were isolated, and the meb1 meb2 double mutant was generated. These mutants were viable and similar to wild-type plants. No differences in PYK10 and NAI2 levels were detected between wild-type plants and any of the three mutants (Fig. 7A), suggesting that MEB1 and MEB2 are not essential for the accumulation of ER body components in Arabidopsis. Mutant and wild-type plants showed a similar amount of ER bodies, with no observable differences in ER body morphology (Fig. 7B), suggesting that MEB1 and MEB2 are not essential for ER body formation or PYK10 accumulation in ER bodies.
Figure 7.
PYK10 accumulation and ER body morphology are unchanged in meb1 and meb2 mutants. A, PYK10, NAI2, and BiP levels in meb1, meb2, and meb1 meb2 mutants. Total protein extract from 7-d-old seedlings was subjected to immunoblot analysis using antibodies against PYK10, NAI2, or BiP. Coomassie Brilliant Blue staining (CBB) shows the Rubisco large subunit as a loading control. Col, Columbia accession. B, Fluorescence images of cotyledon epidermal cells from 7-d-old seedlings in the wild type (GFP-h) and meb1-1, meb2-1, and meb1-1 meb2-1 mutants expressing ER-targeted GFP (GFP-HDEL). Bar = 10 µm.
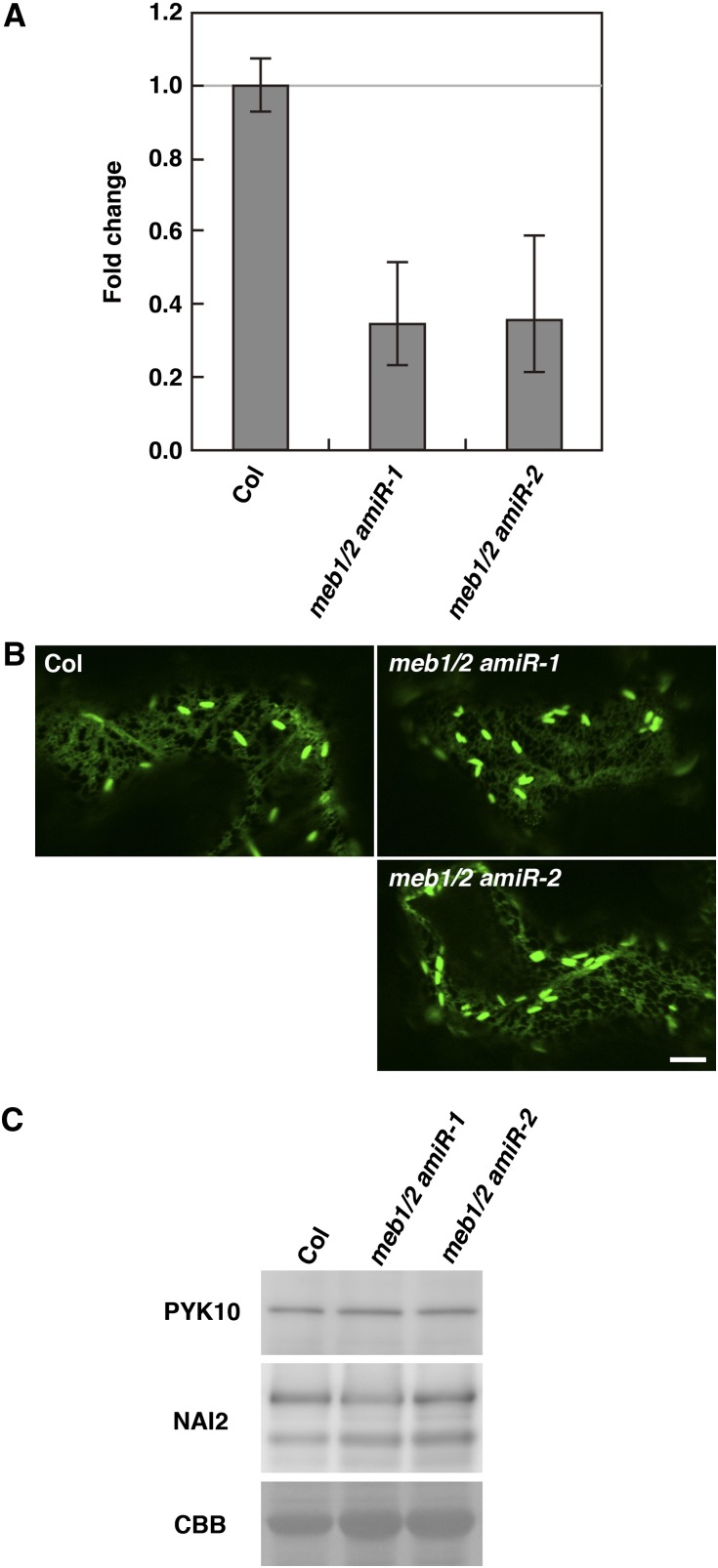
At4g27870 is a homolog of MEB1 and MEB2 expressed in seedlings (Fig. 3). Functional redundancy between MEB1, MEB2, and At4g27870 might explain the lack of phenotype in the meb1 meb2 double mutant. Since the proximity of the At4g27870 and meb1 (At4g27860) genes precludes the generation of a meb1 at4g27870 double mutant, two independent artificial microRNA (amiR) genes were constructed for the At4g27870 gene and introduced into the meb1 meb2 double mutant. Two independent lines decreased expression of the At4g27870 gene (Fig. 8A). ER body formation was unchanged in these meb1 meb2 at4g27870 triple mutants (Fig. 8B), suggesting that these membrane proteins are dispensable for ER body formation. Similarly, no differences were detected in PYK10 or NAI2 levels between wild-type plants and meb1 meb2 at4g27870 triple mutants (Fig. 8C), suggesting that these membrane proteins are not essential for the accumulation of ER body components in Arabidopsis.
Figure 8.
PYK10 accumulation and ER body morphology are unchanged in meb1 meb2 at4g27870 triple knockdown/knockout mutants. A, At4g27870 mRNA levels in meb1-1 meb2-1 at4g27870-1 (meb1/2 amiR-1) and meb1-1 meb2-1 at4g27870-2 (meb1/2 amiR-2) triple knockdown/knockout mutants. meb1/2 amiR-1 and meb1/2 amiR-2 plants harbor different constructs to knockdown gene At4g27870 (see “Materials and Methods”). Total RNA from 10-d-old seedlings was subjected to qRT-PCR analysis. The data were normalized to UBIQUITIN10 mRNA levels. The relative quantity of each mRNA was calibrated to the signal observed in wild-type plants (accession Columbia [Col]). The relative quantity was calculated by the 2−ΔΔCt method (Livak and Schmittgen, 2001). Error bars indicate se of the threshold cycle (Ct) values, calculated using the equation 2−ΔΔCt ± se. The data represent the results of three independent biological replications. B, Fluorescence images of cotyledon epidermal cells from seedlings transiently expressing ER-targeted GFP. Six-day-old wild-type or meb1 meb2 at4g27870 mutant plants were bombarded with gold particles coated with GFP-HDEL plasmids and germinated for 1 d before inspection. Bar = 10 µm. C, Total protein extract from 10-d-old seedlings was subjected to immunoblot analysis using antibodies against PYK10 and NAI2. Coomassie brilliant blue staining (CBB) shows the Rubisco large subunit as a loading control.
The ER body is believed to be involved in defense against fungal infection (Hara-Nishimura and Matsushima, 2003; Matsushima et al., 2003a; Sherameti et al., 2008; Yamada et al., 2011). We examined whether pathogen sensitivity is altered in the meb1 meb2 double mutant compared with wild-type plants. We examined Alternaria brassicicola, Fusarium oxysporum, Pythium sylvaticum, and Pseudomonas syringae pv glycinea, which are pathogenic to Arabidopsis accession Columbia. These pathogens gradually damaged wild-type plants. The sensitivity of wild-type and mutant plants to A. brassicicola, F. oxysporum, and P. sylvaticum was not significantly different. In addition, the growth of P. syringae pv glycinea was similar in mutant and wild-type plants (data not shown). These results suggest that MEB1 and MEB2 are not involved in defense against these pathogens.
DISCUSSION
The ER Body Membrane Is Distinct from ER Network Membranes and Contains MEB1 and MEB2
The ER body is a dilated ER subdomain that accumulates PYK10 and NAI2 (Matsushima et al., 2003b; Yamada et al., 2008, 2009). The specific components of the ER body membrane remain unknown. We found that MEB1 and MEB2 contain transmembrane regions and localize to the ER body membrane. This indicates that the ER body membrane is distinct from membranes in the ER network. In cereal plants, PBs are ER-derived structures that accumulate specific seed proteins. In rice, prolamin mRNAs accumulate on the prolamin PB membrane (Choi et al., 2000), suggesting that this membrane is distinct from the ER network. In maize, FLOURY1 (FL1) is a PB membrane protein involved in PB formation (Holding et al., 2007). Although MEB1 and MEB2 are structurally unrelated to FL1, these findings suggest that other ER-derived structures, such as ricinosomes and KDEL-tailed proteinase-accumulating vesicles (Toyooka et al., 2000; Schmid et al., 2001), may contain integral membrane proteins that distinguish them from the ER network.
The lengths of MEB1 and MEB2 are 611 and 550 amino acids, respectively, and their predicted molecular masses are 68.2 and 61.0 kD, respectively. In SDS-PAGE, the observed molecular masses of MEB1 and MEB2 were approximately 84 and 82 kD, respectively. This difference between predicted and observed molecular masses may be attributable to the large number of hydrophobic residues, which affect the behavior of proteins in SDS-PAGE analysis (Rath et al., 2009).
MEB1 and MEB2 Accumulate ER Bodies Produced by NAI2
NAI2 is an ER body component that regulates ER body formation. MEB1 and MEB2 were diffusely distributed throughout the ER network in the nai2 mutant. Because there is no ER body in the nai2 mutant, MEB1 and MEB2 cannot localize to the ER body, which resulted in a diffuse ER network in the nai2 mutant. This suggests that NAI2 function is to make an ER body and ER body membrane that acts as a scaffold for MEB1 and MEB2. Alternatively, it is possible that NAI2 itself is a scaffold for MEB1 and MEB2, because NAI2 and MEB1 or MEB2 form a protein complex. We propose that ER body components, such as PYK10 and NAI2, induce an ER body and then trap MEB1 and MEB2 during ER body formation.
Possible Functions of MEB1 and MEB2
Our results indicate that MEB1, MEB2, and At4g27870 are not involved in PYK10 accumulation. Furthermore, MEB1, MEB2, and At4g27870 appear dispensable for ER body formation in Arabidopsis. This implies that MEB1 and MEB2 are mainly involved in ER body function rather than ER body formation.
The C-terminal region of MEB1 and MEB2 contains a DUF125 sequence. Proteins containing a DUF125 sequence are assigned to the VIT family, including CCC1 in yeast, AtVIT1 in Arabidopsis, and OsVIT1 and OsVIT2 in rice. CCC1 is a vacuolar iron/manganese transporter that is responsible for the regulation of cytosolic iron homeostasis (Li et al., 2001). The Arabidopsis protein AtVIT1, a close homolog of CCC1, transports both iron and manganese, but only iron homeostasis is altered following mutation of vit1 in seeds (Kim et al., 2006; Roschzttardtz et al., 2009). OsVIT1 and OsVIT2 transport iron and zinc and are responsible for the accumulation of these metals (Zhang et al., 2012). We found that MEB1 and MEB2 partially reduced the iron sensitivity of the ccc1 mutant and enhanced manganese resistance in yeast. These results suggest that the DUF125 sequence of these proteins underlies their ability to transport metal ion. This also suggests that ER bodies containing MEB1 and MEB2 are involved in metal ion homeostasis. The ER body is constitutively observed in roots (Hara-Nishimura and Matsushima, 2003; Matsushima et al., 2003a, 2003b). Root tissues may be exposed to high concentrations of metal ions. Therefore, it is reasonable to expect that roots have detoxifying mechanisms for metals, which in part may be mediated by ER bodies. Another example of a new function of the ER body was recently revealed. Takahashi et al. (2012) showed that the chlorophyll-binding protein BoWSCP accumulates in the ER body of Brussels sprout (Brassica oleracea). When cells are disrupted, BoWSCP is released from the ER body and then immediately scavenges chlorophyll located in the damaged thylakoid membranes to suppress the production of reactive oxygen species (Takahashi et al., 2012). These observations led to the proposal that the ER body is involved in defense against other environmental stresses in addition to pathogens/herbivores.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana accession Columbia) and transgenic Arabidopsis expressing GFP in the ER and ER bodies (GFP-h) were used as the wild-type background (Yamada et al., 2008). Arabidopsis nai1-1 and nai2-2 mutants were also used (Matsushima et al., 2004; Yamada et al., 2008). nai2-2 was transformed with the Agrobacterium tumefaciens strain C58C1Rif, which contains a pBI121 binary vector harboring the SP-GFP-HDEL gene (Yamada et al., 2008). T-DNA insertion mutants of MEB1 and MEB2, identified by the Salk Institute Genomic Analysis Laboratory or Syngenta, were obtained from the Arabidopsis Biological Resource Center. The T-DNA insertion was confirmed by genome PCR using a gene-specific primer and a T-DNA primer (Supplemental Table S1). The following mutants were used: SALK_011418 (meb1-1), SALK_024453 (meb1-2), SALK_107203 (meb1-3), CS810089 (meb2-1), and SALK_098787 (meb2-3; McElver et al., 2001; Alonso et al., 2003). These mutants were crossed with a GFP-h line to obtain transgenic plants expressing GFP in the ER and ER bodies. All plants were germinated aseptically at 22°C under continuous light (approximately 100 mE s−1 m−2) on Murashige and Skoog plates containing 0.4% Gellan Gum (Wako), 0.5% (w/v) MES-KOH buffer (pH 5.7), and 1× Murashige and Skoog salts mixture (Wako).
To produce at4g27870 knockdown plants in meb1 meb2 double mutants, two amiRs specific to At4g27870 were constructed. The method was described previously (Schwab et al., 2006). Two amiR candidate DNA sequences (amiRat4g27870-1 and amiRat4g27870-2) were calculated using the WMD3 program (http://wmd3.weigelworld.org/cgi-bin/webapp.cgi). Specific primers were generated (Supplemental Table S1), and DNA fragments were amplified by PCR using primers amiR oligo A and B (Schwab et al., 2006) and the template amiR vectors in pRS300 (a kind gift from D. Weigel). The DNA fragments were digested with EcoRI/NotI and introduced into the EcoRI/NotI site of the Gateway entry vector pENTR1a (Invitrogen). The entire amiR coding regions were transferred into the binary vector pMDC7 (Curtis and Grossniklaus, 2003) to generate pMDC7/amiRat4g27870-1 and pMDC7/amiRat4g27870-2. These plasmids encode the amiR gene under the control of the estradiol-inducible promoter. The plasmids were introduced into GV3101 (pMP90RK) and then transformed into meb1-1 meb2-1 double mutants. Transgenic homozygote plants were germinated on a plate containing 5 µm 17β-estradiol and analyzed (Zuo et al., 2002).
Full-Length cDNAs for MEB1 and MEB2
Complementary DNA (cDNA) fragments containing the MEB1 and MEB2 genes were PCR amplified using gene-specific primer sets (Supplemental Table S1) and EST clone templates (U21078 for MEB1 and U23495 for MEB2). The fragments were introduced into the Gateway entry vector pENTR/SD/d-TOPO (Invitrogen) to produce pENTR/MEB1 and pENTR/MEB2. The EST clones were Salk/Stanford/Plant Gene Expression Center Consortium full-length cDNA/open reading frame clones obtained from the Arabidopsis Biological Resource Center.
qRT-PCR
qRT-PCR was performed as reported previously (Yamada et al., 2008). Total RNAs were isolated from 20 7-d-old wild-type or mutant line seedlings using 700 µL of Isogen (Nippon Gene). RNA was dissolved in 175 µL of distilled water, and first-strand cDNA was synthesized from 5 µL of the RNA solution using Ready-to-Go RT-PCR beads (GE Healthcare Life Science) and random oligomers. Real-time PCR was performed using the 7500 Fast Real-Time PCR System (Applied Biosystems) and a TaqMan Gene Expression Assay Kit (Applied Biosystems) according to the manufacturer’s instructions. The assay identifiers were At02300790_g1 for MEB1, At02185786_g1 for MEB2, At02300797_g1 for At4g27870, and At02358313_s1 for UBIQUITIN10. The 2−ΔΔCt method was used for relative quantification (Livak and Schmittgen, 2001).
Microscopy
A confocal laser scanning microscope (LSM510; Carl Zeiss) was used to observe fluorescent proteins. An argon laser (488 nm) and a 505/530-nm band-pass filter were used to observe GFP, whereas a helium-neon laser (543 nm) and a 560/615-nm band-pass filter were used for tdTOM.
Particle Bombardment
The entire protein-coding region of pENTR/MEB1 and pENTR/MEB2 was transferred to vector ptdGW (a kind gift from S. Mano) to generate ptd/MEB1 and ptd/MEB2. These plasmids carry Pro-35S:tdTom-MEB1 or Pro-35S:tdTom-MEB2, which encode fusion proteins of tdTOM (Shaner et al., 2004) with MEB1 or MEB2. Arabidopsis plants were germinated aseptically on Murashige and Skoog plates. Plasmid DNAs were bombarded into 5-d-old seedlings using the Biolistic Particle Delivery System (Bio-Rad Laboratories) according to the manufacturer’s instructions (Yamada et al., 2007).
Antibody Preparation
PCR was used to amplify two partial cDNA fragments containing MEB1 and MEB2, which encode amino acids 272 to 502 (MEB1/C) and 1 to 325 (MEB2/ΔTM), respectively. PCR was performed using gene-specific primer sets (Supplemental Table S1) and EST clones. The amplified fragments were cloned initially into pENTR/SD/d-TOPO. The protein-coding regions were subsequently transferred into pDEST17 (Invitrogen) for the expression of His-tagged fusion proteins in Escherichia coli (BL21-AI; Invitrogen). Total E. coli proteins were dissolved in 50 mm sodium phosphate buffer (pH 7.2) containing 0.3 m NaCl and 50 mm imidazole, and then recombinant MEB1/C and MEB2/ΔTM proteins were purified by nickel-column chromatography. The purified proteins were injected into rabbits to raise antibodies (Scrum). Anti-PYK10/IM (Matsushima et al., 2003b), anti-BiP (Hara-Nishimura et al., 1998), anti-NAI2/ΔSP (Yamada et al., 2008), and anti-GFP (Mitsuhashi et al., 2000) antibodies were also used.
Immunoblot Analysis
The protein extraction method was described previously (Yamada et al., 2008). Proteins were separated by SDS-PAGE, transferred to a nylon membrane, and subjected to immunoblot analysis using anti-MEB1/C (1:6,000 dilution), anti-MEB2/ΔTM (1:12,000 dilution), anti-PYK10/IM (1:10,000 dilution), anti-BiP (1:5,000 dilution), anti-NAI2/ΔSP (1:2,000 dilution), and anti-GFP (1:5,000 dilution) antibodies. Can Get Signal (Toyobo) was used for immunoblots with anti-MEB1, anti-MEB2, or anti-PYK10/IM. Alternatively, proteins were stained with Coomassie Brilliant Blue R-250 to detect levels of Rubisco for use as a loading control. The band intensity was determined using the Gels tool in ImageJ software.
Subcellular Fractionation and Extraction of Membrane Protein
The subcellular fractionation method was described previously (Yamada et al., 2008). Briefly, 0.83 g of 8-d-old seedlings was chopped on ice in 2.5 mL of chopping buffer that contained 50 mm HEPES-NaOH, pH 7.5, 5 mm EDTA, 0.4 m Suc, and protease inhibitor cocktail (one tablet per 50 mL; Boehringer-Mannheim). The homogenate was filtered through cheesecloth and then centrifuged at 1,000g at 4°C for 20 min. The pellet was designated the P1 fraction. The supernatant was centrifuged once at 8,000g at 4°C for 20 min and then again at 100,000g at 4°C for 1 h. The pellet after ultracentrifugation was designated the P100 fraction. The P1 and P100 fractions were resuspended in the same volume of chopping buffer and subjected to immunoblot analysis. For the separation of soluble and membrane components, the P1 fractions obtained from 8-d-old seedlings (2 g) were treated with different buffers: a low-salt buffer (10 mm HEPES-NaOH [pH 7.5], 1 mm EDTA, and 50 mm NaCl), a high-salt buffer (10 mm HEPES-NaOH [pH 7.5], 1 mm EDTA, and 500 mm NaCl), an alkaline buffer (0.1 m Na2CO3 [pH 11]), or a detergent buffer (10 mm HEPES-NaOH [pH 7.5] and 2% [v/v] Triton X-100). The mixtures were then centrifuged at 100,000g for 30 min and separated into soluble (supernatant) and precipitate fractions. The precipitate was resuspended in the buffer of equal volume to supernatant and subjected to immunoblot analysis.
Immunoprecipitation
Total protein was extracted from 20 8-d-old Arabidopsis seedlings using 350 µL of Tris-buffered saline (TBS; 50 mm Tris-HCl [pH 7.5] and 150 mm NaCl) and protease inhibitor cocktail (one tablet per 50 mL; Boehringer-Mannheim). The homogenate was centrifuged at 9,000g for 3 min at 4°C to remove cell debris. Protein extract (100 µL) was mixed with 900 µL of TBS and anti-MEB1 serum (2.5 µL) or anti-MEB2 serum (0.5 µL) and then incubated overnight at 4°C. Following the addition of 20 µL of Protein G Sepharose (GE Healthcare Life Science) beads, the mixture was incubated for 3 h. The Protein G Sepharose beads were recovered by centrifugation and washed several times with TBS. Bound proteins were eluted with 30 µL of 2× SDS-PAGE sample buffer (20 mm Tris-HCl [pH 6.8], 2% [w/v] SDS, 2% [v/v] 2-mercaptoethanol, and 40% [v/v] glycerol).
Analysis of Protein Structure
Several transmembrane prediction tools on the ExPASy Web site (http://au.expasy.org/tools/), including the DAS, HMMTOP, SOSUI, Tmpred, and TMHMM programs, were used to identify transmembrane regions. We also referred to Aramemnone (http://aramemnon.botanik.uni-koeln.de/) for the transmembrane prediction. The ClustalW2 program (http://www.ebi.ac.uk/Tools/msa/clustalw2/) was used to align amino acid sequences. The program iTOL (http://itol.embl.de/index.shtml) was used for dendrogram analysis (Letunic and Bork, 2011).
Yeast Strains and Analysis
Yeast (Saccharomyces cerevisiae) strains BY4741 (wild type; MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) and ccc1 (BY4741 Δccc1::KanMX) were used (these strains were kind gifts from Y. Kamada and Y. Ohsumi, respectively). The entire protein-coding region of pENTR/MEB1 and pENTR/MEB2 was transferred to vector p415 GAL1-GW to generate p415/MEB1 and p415/MEB2. These plasmids carry a Gal-inducible promoter. The Gateway destination vector p415 GAL1-GW was derived from p415 GAL1 (Mumberg et al., 1994), which had been modified into a Gateway vector by using the Gateway Vector Conversion System (Invitrogen). Cassette B from the Gateway Vector Conversion System was inserted in the SmaI restriction site of p415 GAL1. Yeast strains harboring a control vector (p415 GAL1), p415/MEB1, or p415/MEB2 were grown in synthetic minimal medium containing 2% (w/v) Gal (SG) supplemented with His, Met, and uracil but not Leu. For the iron toxicity assay, 5 µL of cell suspension (optimal density at 600 nm = 0.1) was spotted on an SG plate containing 2% (w/v) agar supplemented with His, Met, and uracil, with or without 2 mm FeSO4, and subsequently cultured at 30°C for 4 d. For the manganese toxicity assay, 10 µL of cell suspension (optimal density at 600 nm = 1) was applied to 5 mL of SG containing 2% (w/v) Suc supplemented with His, Met, and uracil, with or without 15 mm MnCl2, and subsequently cultured at 30°C and 300 rpm. The optimal density at 600 nm was monitored at various time intervals.
Pathogen Infection Experiment
Alternaria brassicicola (MAFF726527) was obtained from the National Institute of Agrobiological Sciences gene bank. A. brassicicola, Fusarium oxysporum strain NDF9 (a kind gift from M. Nozue), and Pythium sylvaticum strain DSM2322 (a kind gift from P. Schulze-Lefert and P. Bednarek) were maintained in potato dextrose agar (BD Difco). Pseudomonas syringae pv glycinea (a kind gift from D. Takemoto) was maintained in Plusgrow medium (Nacalai Tesque). Conidia of A. brassicicola were suspended in 0.5% (w/v) Triton X-100 and placed on the cotyledons of 8-d-old seedlings, and symptoms were observed for 2 weeks. Mycelia of F. oxysporum or P. sylvaticum were placed close to aseptically grown, 1-week-old plants, and symptoms were observed for 1 week. P. syringae cells were suspended in a solution containing 10 mm MgCl2 and 0.01% (w/v) Triton X-100, and seedlings were placed in the solution for 1 h. After several days, the bacteria were extracted from cotyledons by homogenization, and the number of colony-forming units was counted.
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: MEB1/At4g27860 (NM_001203926), MEB2/At5g24290 (NM_180736), At4g27870 (NM_118925), NAI1/At2g22770 (NM_179700), NAI2/At3g15950 (NM_112466), PYK10/At3g09260 (NM_111760), and Arabidopsis lyrata MEB1 homolog (EFH45796).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Structures of MEB1 homologs.
Supplemental Figure S2. Fluorescence intensities of GFP and tdTOM in Figure 4A.
Supplemental Table S1. Nucleotide sequences of oligonucleotide primers used in this study.
Acknowledgments
We thank Detlef Weigel (Max Planck Institute for Developmental Biology), Shoji Mano (National Institute for Basic Biology), Yoshinori Ohsumi (Tokyo Institute for Technology), Yoshiaki Kamada (National Institute for Basic Biology), Masayuki Nozue (Shinshu University), Paul Schulze-Lefert (Max Planck Institute for Plant Breeding Research), Paweł Bednarek (Max Planck Institute for Plant Breeding Research), and Daigo Takemoto (Nagoya University) for providing plant pathogens, yeast strains, and vectors. We also thank the National Institute for Basic Biology Center for Analytical Instruments for instrumentation.
Glossary
- ER
endoplasmic reticulum
- PB
protein body
- qRT
quantitative reverse transcription
- amiR
artificial microRNA
- TBS
Tris-buffered saline
- SG
synthetic minimal medium containing 2% (w/v) Gal
- cDNA
complementary DNA
References
- Ahn YO, Shimizu B, Sakata K, Gantulga D, Zhou C, Bevan DR, Esen A. (2010) Scopolin-hydrolyzing β-glucosidases in roots of Arabidopsis. Plant Cell Physiol 51: 132–143 [DOI] [PubMed] [Google Scholar]
- Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. (2002) Intracellular vesicular traffic. In B Alberts, A Johnson, J Lewis, M Raff, K Roberts, P Walter, eds, Molecular Biology of the Cell, Ed 4. Garland Science, New York, pp 711–766
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Behnke H-D, Eschlbeck G. (1978) Dilated cisternae in Capparales: an attempt towards the characterization of a specific endoplasmic reticulum. Protoplasma 97: 351–363 [Google Scholar]
- Choi S-B, Wang C, Muench DG, Ozawa K, Franceschi VR, Wu Y, Okita TW. (2000) Messenger RNA targeting of rice seed storage proteins to specific ER subdomains. Nature 407: 765–767 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. (2003) A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delauney AJ, Cheon C-I, Snyder PJ, Verma DPS. (1990) A nodule-specific sequence encoding a methionine-rich polypeptide, nodulin-21. Plant Mol Biol 14: 449–451 [DOI] [PubMed] [Google Scholar]
- Dunkley TP, Hester S, Shadforth IP, Runions J, Weimar T, Hanton SL, Griffin JL, Bessant C, Brandizzi F, Hawes C, et al. (2006) Mapping the Arabidopsis organelle proteome. Proc Natl Acad Sci USA 103: 6518–6523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Tate J, Coggill PC, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. (2010) The Pfam protein families database. Nucleic Acids Res 38: D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galili G. (2004) ER-derived compartments are formed by highly regulated processes and have special functions in plants. Plant Physiol 136: 3411–3413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunning BES. (1998) The identity of mystery organelles in Arabidopsis plants expressing GFP. Trends Plant Sci 3: 417. [DOI] [PubMed] [Google Scholar]
- Hakoyama T, Niimi K, Yamamoto T, Isobe S, Sato S, Nakamura Y, Tabata S, Kumagai H, Umehara Y, Brossuleit K, et al. (2012) The integral membrane protein SEN1 is required for symbiotic nitrogen fixation in Lotus japonicus nodules. Plant Cell Physiol 53: 225–236 [DOI] [PubMed] [Google Scholar]
- Hara-Nishimura I, Matsushima R. (2003) A wound-inducible organelle derived from endoplasmic reticulum: a plant strategy against environmental stresses? Curr Opin Plant Biol 6: 583–588 [DOI] [PubMed] [Google Scholar]
- Hara-Nishimura I, Matsushima R, Shimada T, Nishimura M. (2004) Diversity and formation of endoplasmic reticulum-derived compartments in plants: are these compartments specific to plant cells? Plant Physiol 136: 3435–3439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara-Nishimura I, Shimada T, Hatano K, Takeuchi Y, Nishimura M. (1998) Transport of storage proteins to protein storage vacuoles is mediated by large precursor-accumulating vesicles. Plant Cell 10: 825–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Y, Yamada K, Shimada T, Matsushima R, Nishizawa NK, Nishimura M, Hara-Nishimura I. (2001) A proteinase-storing body that prepares for cell death or stresses in the epidermal cells of Arabidopsis. Plant Cell Physiol 42: 894–899 [DOI] [PubMed] [Google Scholar]
- Herman EM. (2008) Endoplasmic reticulum bodies: solving the insoluble. Curr Opin Plant Biol 11: 672–679 [DOI] [PubMed] [Google Scholar]
- Herman EM, Larkins BA. (1999) Protein storage bodies and vacuoles. Plant Cell 11: 601–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman EM, Schmidt M. (2004) Endoplasmic reticulum to vacuole trafficking of endoplasmic reticulum bodies provides an alternate pathway for protein transfer to the vacuole. Plant Physiol 136: 3440–3446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holding DR, Otegui MS, Li B, Meeley RB, Dam T, Hunter BG, Jung R, Larkins BA. (2007) The maize Floury1 gene encodes a novel endoplasmic reticulum protein involved in zein protein body formation. Plant Cell 19: 2569–2582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iversen T-H. (1970a) Cytochemical localization of myrosinase (β-thioglucosidase) in root tips of Sinapis alba. Protoplasma 71: 451–466 [Google Scholar]
- Iversen T-H. (1970b) The morphology, occurrence, and distribution of dilated cisternae of the endoplasmic reticulum in tissues of plants of the Cruciferae. Protoplasma 71: 467–477 [Google Scholar]
- Kim SA, Punshon T, Lanzirotti A, Li L, Alonso JM, Ecker JR, Kaplan J, Guerinot ML. (2006) Localization of iron in Arabidopsis seed requires the vacuolar membrane transporter VIT1. Science 314: 1295–1298 [DOI] [PubMed] [Google Scholar]
- Lapinskas PJ, Lin SJ, Culotta VC. (1996) The role of the Saccharomyces cerevisiae CCC1 gene in the homeostasis of manganese ions. Mol Microbiol 21: 519–528 [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P. (2011) Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res 39: W475–W478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Chen OS, McVey Ward D, Kaplan J. (2001) CCC1 is a transporter that mediates vacuolar iron storage in yeast. J Biol Chem 276: 29515–29519 [DOI] [PubMed] [Google Scholar]
- Lin H, Li L, Jia X, Ward DM, Kaplan J. (2011) Genetic and biochemical analysis of high iron toxicity in yeast: iron toxicity is due to the accumulation of cytosolic iron and occurs under both aerobic and anaerobic conditions. J Biol Chem 286: 3851–3862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Matsushima R, Fukao Y, Nishimura M, Hara-Nishimura I. (2004) NAI1 gene encodes a basic-helix-loop-helix-type putative transcription factor that regulates the formation of an endoplasmic reticulum-derived structure, the ER body. Plant Cell 16: 1536–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima R, Hayashi Y, Kondo M, Shimada T, Nishimura M, Hara-Nishimura I. (2002) An endoplasmic reticulum-derived structure that is induced under stress conditions in Arabidopsis. Plant Physiol 130: 1807–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima R, Hayashi Y, Yamada K, Shimada T, Nishimura M, Hara-Nishimura I. (2003a) The ER body, a novel endoplasmic reticulum-derived structure in Arabidopsis. Plant Cell Physiol 44: 661–666 [DOI] [PubMed] [Google Scholar]
- Matsushima R, Kondo M, Nishimura M, Hara-Nishimura I. (2003b) A novel ER-derived compartment, the ER body, selectively accumulates a β-glucosidase with an ER-retention signal in Arabidopsis. Plant J 33: 493–502 [DOI] [PubMed] [Google Scholar]
- McElver J, Tzafrir I, Aux G, Rogers R, Ashby C, Smith K, Thomas C, Schetter A, Zhou Q, Cushman MA, et al. (2001) Insertional mutagenesis of genes required for seed development in Arabidopsis thaliana. Genetics 159: 1751–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuhashi N, Shimada T, Mano S, Nishimura M, Hara-Nishimura I. (2000) Characterization of organelles in the vacuolar-sorting pathway by visualization with GFP in tobacco BY-2 cells. Plant Cell Physiol 41: 993–1001 [DOI] [PubMed] [Google Scholar]
- Momonoi K, Yoshida K, Mano S, Takahashi H, Nakamori C, Shoji K, Nitta A, Nishimura M. (2009) A vacuolar iron transporter in tulip, TgVit1, is responsible for blue coloration in petal cells through iron accumulation. Plant J 59: 437–447 [DOI] [PubMed] [Google Scholar]
- Mumberg D, Müller R, Funk M. (1994) Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res 22: 5767–5768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamine A, Matsusaka H, Ushijima T, Kawagoe Y, Ogawa M, Okita TW, Kumamaru T. (2011) A role for the cysteine-rich 10 kDa prolamin in protein body I formation in rice. Plant Cell Physiol 52: 1003–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano AJ, Fukao Y, Fujiwara M, Nishimura M, Hara-Nishimura I. (2008) Antagonistic jacalin-related lectins regulate the size of ER body-type β-glucosidase complexes in Arabidopsis thaliana. Plant Cell Physiol 49: 969–980 [DOI] [PubMed] [Google Scholar]
- Nagano AJ, Maekawa A, Nakano RT, Miyahara M, Higaki T, Kutsuna N, Hasezawa S, Hara-Nishimura I. (2009) Quantitative analysis of ER body morphology in an Arabidopsis mutant. Plant Cell Physiol 50: 2015–2022 [DOI] [PubMed] [Google Scholar]
- Nagano AJ, Matsushima R, Hara-Nishimura I. (2005) Activation of an ER-body-localized β-glucosidase via a cytosolic binding partner in damaged tissues of Arabidopsis thaliana. Plant Cell Physiol 46: 1140–1148 [DOI] [PubMed] [Google Scholar]
- Obayashi T, Nishida K, Kasahara K, Kinoshita K. (2011) ATTED-II updates: condition-specific gene coexpression to extend coexpression analyses and applications to a broad range of flowering plants. Plant Cell Physiol 52: 213–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogasawara K, Yamada K, Christeller JT, Kondo M, Hatsugai N, Hara-Nishimura I, Nishimura M. (2009) Constitutive and inducible ER bodies of Arabidopsis thaliana accumulate distinct β-glucosidases. Plant Cell Physiol 50: 480–488 [DOI] [PubMed] [Google Scholar]
- Rath A, Glibowicka M, Nadeau VG, Chen G, Deber CM. (2009) Detergent binding explains anomalous SDS-PAGE migration of membrane proteins. Proc Natl Acad Sci USA 106: 1760–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridge RW, Uozumi Y, Plazinski J, Hurley UA, Williamson RE. (1999) Developmental transitions and dynamics of the cortical ER of Arabidopsis cells seen with green fluorescent protein. Plant Cell Physiol 40: 1253–1261 [DOI] [PubMed] [Google Scholar]
- Roschzttardtz H, Conéjéro G, Curie C, Mari S. (2009) Identification of the endodermal vacuole as the iron storage compartment in the Arabidopsis embryo. Plant Physiol 151: 1329–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh-Cruz M, Crofts AJ, Takemoto-Kuno Y, Sugino A, Washida H, Crofts N, Okita TW, Ogawa M, Satoh H, Kumamaru T. (2010) Protein disulfide isomerase like 1-1 participates in the maturation of proglutelin within the endoplasmic reticulum in rice endosperm. Plant Cell Physiol 51: 1581–1593 [DOI] [PubMed] [Google Scholar]
- Schmid M, Simpson DJ, Sarioglu H, Lottspeich F, Gietl C. (2001) The ricinosomes of senescing plant tissue bud from the endoplasmic reticulum. Proc Natl Acad Sci USA 98: 5353–5358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. (2006) Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18: 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. (2004) Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol 22: 1567–1572 [DOI] [PubMed] [Google Scholar]
- Sherameti I, Venus Y, Drzewiecki C, Tripathi S, Dan VM, Nitz I, Varma A, Grundler FM, Oelmüller R. (2008) PYK10, a β-glucosidase located in the endoplasmatic reticulum, is crucial for the beneficial interaction between Arabidopsis thaliana and the endophytic fungus Piriformospora indica. Plant J 54: 428–439 [DOI] [PubMed] [Google Scholar]
- Takahashi H, Tamura K, Takagi J, Koumoto Y, Hara-Nishimura I, Shimada T. (2010) MAG4/Atp115 is a Golgi-localized tethering factor that mediates efficient anterograde transport in Arabidopsis. Plant Cell Physiol 51: 1777–1787 [DOI] [PubMed] [Google Scholar]
- Takahashi S, Yanai H, Nakamaru Y, Uchida A, Nakayama K, Satoh H. (2012) Molecular cloning, characterization and analysis of the intracellular localization of a water-soluble Chl-binding protein from Brussels sprouts (Brassica oleracea var. gemmifera). Plant Cell Physiol 53: 879–891 [DOI] [PubMed] [Google Scholar]
- Toyooka K, Okamoto T, Minamikawa T. (2000) Mass transport of proform of a KDEL-tailed cysteine proteinase (SH-EP) to protein storage vacuoles by endoplasmic reticulum-derived vesicle is involved in protein mobilization in germinating seeds. J Cell Biol 148: 453–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Fukao Y, Hayashi M, Fukazawa M, Suzuki I, Nishimura M. (2007) Cytosolic HSP90 regulates the heat shock response that is responsible for heat acclimation in Arabidopsis thaliana. J Biol Chem 282: 37794–37804 [DOI] [PubMed] [Google Scholar]
- Yamada K, Hara-Nishimura I, Nishimura M. (2011) Unique defense strategy by the endoplasmic reticulum body in plants. Plant Cell Physiol 52: 2039–2049 [DOI] [PubMed] [Google Scholar]
- Yamada K, Nagano AJ, Nishina M, Hara-Nishimura I, Nishimura M. (2008) NAI2 is an endoplasmic reticulum body component that enables ER body formation in Arabidopsis thaliana. Plant Cell 20: 2529–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Nagano AJ, Ogasawara K, Hara-Nishimura I, Nishimura M. (2009) The ER body, a new organelle in Arabidopsis thaliana, requires NAI2 for its formation and accumulates specific β-glucosidases. Plant Signal Behav 4: 849–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Xu YH, Yi HY, Gong JM. (2012) Vacuolar membrane transporters OsVIT1 and OsVIT2 modulate iron translocation between flag leaves and seeds in rice. Plant J 72: 400–410 [DOI] [PubMed] [Google Scholar]
- Zuo J, Niu QW, Frugis G, Chua NH. (2002) The WUSCHEL gene promotes vegetative-to-embryonic transition in Arabidopsis. Plant J 30: 349–359 [DOI] [PubMed] [Google Scholar]