Abstract
Tobacco mosaic virus (TMV) forms dense cytoplasmic bodies containing replication-associated proteins (virus replication complexes [VRCs]) upon infection. To identify host proteins that interact with individual viral components of VRCs or VRCs in toto, we isolated viral replicase- and VRC-enriched fractions from TMV-infected Nicotiana tabacum plants. Two host proteins in enriched fractions, ATP-synthase γ-subunit (AtpC) and Rubisco activase (RCA) were identified by matrix-assisted laser-desorption ionization time-of-flight mass spectrometry or liquid chromatography-tandem mass spectrometry. Through pull-down analysis, RCA bound predominantly to the region between the methyltransferase and helicase domains of the TMV replicase. Tobamovirus, but not Cucumber mosaic virus or Potato virus X, infection of N. tabacum plants resulted in 50% reductions in Rca and AtpC messenger RNA levels. To investigate the role of these host proteins in TMV accumulation and plant defense, we used a Tobacco rattle virus vector to silence these genes in Nicotiana benthamiana plants prior to challenge with TMV expressing green fluorescent protein. TMV-induced fluorescent lesions on Rca- or AtpC-silenced leaves were, respectively, similar or twice the size of those on leaves expressing these genes. Silencing Rca and AtpC did not influence the spread of Tomato bushy stunt virus and Potato virus X. In AtpC- and Rca-silenced leaves TMV accumulation and pathogenicity were greatly enhanced, suggesting a role of both host-encoded proteins in a defense response against TMV. In addition, silencing these host genes altered the phenotype of the TMV infection foci and VRCs, yielding foci with concentric fluorescent rings and dramatically more but smaller VRCs. The concentric rings occurred through renewed virus accumulation internal to the infection front.
As obligate intracellular organisms, plant viruses must interact with various host factors essential for their accumulation and intercellular movement. These virus-host interactions are often complex and difficult to understand mechanistically, but for some of them considerable progress has been made in recent years (Ahlquist et al., 2003; Boevink and Oparka, 2005; Benitez-Alfonso et al., 2010; Harries et al., 2010; Ishibashi et al., 2010; Laliberté and Sanfaçon, 2010; Verchot-Lubicz et al., 2010; Le Gall et al., 2011; Nagy and Pogany, 2011; Niehl and Heinlein, 2011; Schoelz et al., 2011). Progress also has been made toward understanding virus-host interactions involved in the plant immune response (Kang et al., 2005; Padmanabhan and Dinesh-Kumar, 2010; Cournoyer and Dinesh-Kumar, 2011). There are other interactions that are not essential for virus accumulation and movement and that do not function in the immune response, but which also can significantly disrupt host and virus physiology and cause disease (Culver and Padmanabhan, 2007). Those involved in defense are often referred to as nonhost resistance and are not cultivar specific (Mysore and Ryu 2004; Maule et al., 2007). These interactions, whether to inhibit or aid virus accumulation, and the mechanism(s) by which they influence host physiology and disease induction are less understood than those involving essential or immune response interactions. Considering the serious effect these nonessential interactions can have on host development and disease, they require further study to devise new strategies to limit virus-mediated economic damage (Culver and Padmanabhan, 2007).
We have used the model Nicotiana spp.-Tobacco mosaic virus (TMV) system to further identify host factors that may participate in virus replication and elucidate their role in the accumulation, movement, and disease-invoking ability of a virus. TMV is a positive-sense single-stranded RNA virus that is the type member of the well-studied genus, Tobamovirus. The full-length RNA encodes the 126- and 183-kD replication-associated proteins, the latter translated by a read through of an amber termination codon at the end of the126-kD protein open reading frame (Pelham, 1978). Together they are often referred to as components of the TMV replicase. The 30-kD movement protein (MP) and the 17.5-kD coat protein (CP) are translated from the subgenomic mRNAs (Zaitlin, 1999). The 126- and 183-kD proteins are of central interest in this study. These proteins contain domains with sequences or activities similar to those of methyltransferases (MTs), helicases (HELs), and RNA-dependent RNA polymerases (POLs; Evans et al., 1985; Dunigan and Zaitlin, 1990; Koonin, 1991). In addition to their enhancement of, or necessity for, virus accumulation, respectively, the 126- and 183-kD proteins of TMV and related viruses are involved in other aspects of the viral infection. These include inducing mosaic symptoms in Nicotiana tabacum (Bao et al., 1996; Shintaku et al., 1996), mediating a necrotic resistance response through interaction with a specific host resistance gene in N. tabacum (Padgett and Beachy, 1993; Whitham et al., 1994; Abbink et al., 1998; Erickson et al., 1999), supporting intercellular virus movement in N. tabacum (Goregaoker et al., 2001; Hirashima and Watanabe, 2001, 2003), and suppressing gene silencing in N. tabacum and Nicotiana benthamiana (Ding et al., 2004). The determinants of some of these activities have been mapped to various regions within these viral proteins: chlorosis and gene silencing suppression determinants to the MT, HEL, and nonconserved domain between the MT and HEL domains, necrosis determinants to the HEL domain, and intercellular movement determinants to both the nonconserved and HEL domains (Bao et al., 1996; Padgett et al., 1997; Abbink et al., 1998; Hirashima and Watanabe, 2003; Ding et al., 2004; Wang et al., 2012).
Plant proteins associate with the 126- and/or 183-kD proteins and some of these interactions influence virus accumulation and disease development. The RNA binding subunit of host translation initiation factor eIF-3 was shown to associate with the RNA-dependent RNA POL complex of a TMV that infects tomato (Solanum lycopersicum; Osman and Buck, 1997). Another translation factor, elongation factor1A (EF1α), interacts directly with the MT domain of the TMV 126-/183-kD proteins and is necessary for normal virus accumulation (Yamaji et al., 2006, 2010). Other host factors such as membrane proteins encoded by the Tobamovirus multiplication (TOM) genes that interact with the 126-kD protein or its ortholog and an associated host GTP-binding protein, ARL8, are essential for tobamovirus replication in Arabidopsis (Arabidopsis thaliana) and N. tabacum (Yamanaka et al., 2000; Yamanaka et al., 2002; Asano et al., 2005; Nishikiori et al., 2011). Also, the tomato resistance gene, Tm-1, was shown to interact with the homologs of the 126- and 183-kD proteins from the tobamovirus Tomato mosaic virus to prevent its replication (Ishibashi et al., 2007). The allelic gene of Tm-1, tm-1, which fails to interact with the replication proteins from or provide resistance to Tomato mosaic virus, does bind the replication proteins from and inhibit the replication of Tobacco mild green mosaic virus and Pepper mild mottle virus in vitro (Ishibashi et al., 2009). In regard to disease development, the plant P58IPK, an inhibitor of a double-stranded RNA-activated protein kinase, mediates interaction between the HEL domain of TMV replicase and the N. tabacum N protein for the normal development of disease symptoms (Bilgin et al., 2003). The HEL domain of the TMV 126-kD protein also interacts with a subset of the auxin/indole-3-acetic acid protein family, and this interaction is correlated with the disruption of auxin/indole-3-acetic acid targeting, increased virus accumulation in mature tissue, and the production of a disease phenotype (Padmanabhan et al., 2005, 2006, 2008). Lastly, through a direct or indirect interaction, microfilaments were determined to be necessary for the intracellular trafficking of the TMV 126-kD protein fused with GFP and sustained intercellular movement of TMV (Liu et al., 2005; Harries et al., 2009).
In addition to interacting with host proteins, the 126- and 183-kD proteins are present in dense cytoplasmic bodies (also referred to as VRCs for TMV) within the host cell during infection. These VRCs contain other virus-encoded proteins and associate with various host cell components such as ribosomes, endoplasmic reticulum, and cytoskeleton (Shalla, 1964; Hills et al., 1987; Heinlein et al., 1998; Más and Beachy, 1999). The VRCs also are very dynamic, changing location and content over time (Szécsi et al., 1999). However, the full composition of the VRCs and their function during infection is still not understood. Also, considering that most plant viruses induce the formation of cytoplasmic bodies and the increasing experimental support that they are correlated with disease induction (Shalla et al., 1980; Liu et al., 2005; Liu et al., 2006), it is important to understand their composition and function during virus infection for practical purposes.
Here we describe the isolation and purification of complexes containing the TMV 126-/183-kD proteins from TMV-infected N. tabacum leaves and the use of proteomics to identify host proteins associated with them. Two nuclear-encoded chloroplast proteins, ATP synthase-γ subunit (AtpC, encoded by AtpC) and Rubisco activase (RCA) copurified with enriched VRCs and/or viral POL. Their influence on virus accumulation, virus spread, and disease induction was investigated through gene knockdown in N. benthamiana plants. In addition, the influence of virus infection on their transcript accumulation and presence in VRCs was assessed. These proteins were found to specifically inhibit tobamovirus spread and/or accumulation, possibly through transient interaction with the 126-/183-kD proteins or indirectly through an unknown mechanism.
RESULTS
Identification of Host Proteins Enriched in the TMV Replicase Complex
The TMV replication-associated proteins associate with different plant factors during virus replication and movement within the host. To identify more host proteins, TMV replicase complexes were isolated from N. tabacum leaves 4 d post inoculation (dpi) with the U1 strain of TMV (Shintaku et al., 1996). The fractions containing the highest TMV 126-/183-kD protein levels and RNA POL activities (Supplemental Fig. S1) were profiled by SDS-PAGE and compared with the healthy controls processed in parallel (Fig. 1A). The proteins that were present in higher levels in the extracts from TMV-inoculated leaves when compared with healthy control extracts were subjected to in-gel trypsin digestions and matrix-assisted laser-desorption ionization time-of-flight mass spectrometry (MALDI-TofMS) or nano-liquid chromatography-tandem mass spectrometry (LC-MS/MS) analyses. Resultant mass spectral data were used in Mascot (Perkins et al., 1999) to search against the National Center for Biotechnology Information nonredundant database with a restriction to the Viridiplantae. The protein identifications were based upon significant molecular weight search (MOWSE) scores and at least two peptide matches. In addition to the 183- and 126-kD replication-associated proteins and 17-kD TMV CP, we identified two host chloroplast proteins, AtpC (GenBank accession no. X63606) and RCA (EMBL accession no. Z14980). AtpC is one of the subunits of the chloroplast ATP synthase and is encoded by a nuclear gene (AtpC) as a precursor protein (41 kD) that is transported from the cytoplasm into the chloroplast stroma where it is processed into its mature form (36 kD; Larsson et al., 1992). RCA is encoded by a nuclear gene (Rca) as an immature protein (approximately 49 kD) that also is transported from the cytoplasm into the chloroplast where it is processed into its mature form (approximately 42 kD) to activate Rubisco by releasing the bound sugar and phosphates from the active site in an ATP-dependent manner (Portis and Salvucci, 2002; Spreitzer and Salvucci, 2002). RCA is also a member of the AAA+ protein family (ATPases associated with diverse cellular activities; Portis et al., 2008). For AtpC and RCA, respectively, we obtained six peptide matches covering 16% of the apparent precursor protein (Mr estimate from gel approximately 41 kD) and five peptide matches covering 19% of the apparent precursor protein (Mr estimate from gel approximately 47 kD). To further investigate the presence of host factors in TMV VRCs, we performed a crude purification of the VRCs from N. tabacum using a virus expressing an MP-GFP fusion (TMV.MP-GFP.CP). The MP-GFP fusion is present in VRCs at early stages of infection (Heinlein et al., 1998; Liu et al., 2005). It is important to note that these fractions enriched for MP-GFP and 183- and 126-kD protein were also enriched with both precursor (49 kD) and mature (42 kD) forms of RCA (six peptide matches with a MOWSE score of 422 covering 21% of the precursor protein, and thirteen peptide matches with a MOWSE score of 1,402 covering 33% of the mature protein; Fig. 1B). Amino acid composition of the peptide fragments also matched those of the predicted RCA fragments. Although by position during SDS-PAGE migration both the POL-enriched and VRC-enriched fractions contained the precursor forms of AtpC and RCA (Fig. 1, A and B), no expected transit peptide fragments were identified through MALDI-TofMS or LC-MS/MS analysis.
Figure 1.
Enriched levels of plant AtpC and RCA in fractions enriched for TMV POL activity or TMV VRCs and the interaction of RCA with TMV replicase domains. A, Fractions enriched in TMV POL activity were obtained from extracts of N. tabacum leaves infected with TMV U1 strain by differential centrifugation and anion-exchange chromatography coupled with assays for viral RNA POL activity. B, Fractions enriched for TMV VRCs were isolated by differential centrifugation of extracts from N. tabacum plants infected with TMV.MP-GFP.CP. Enriched fractions from A and B (TMV) were profiled using precast 4% to 15% SDS-polyacrylamide gels and compared with the mock-inoculated controls (Mock) processed in parallel. Protein identifications were performed using in-gel digestion followed by MALDI-TofMS (A) and nano-LC-MS/MS (B), peptide mass mapping, and database searching. C, Two-hundred microliters of N. tabacum S30 fractions was incubated with replicase domain proteins fused with GST or GST alone and pulled down with glutathione sepharose 4B beads. Proteins pulled down with the beads were immunodetected with antibody against RCA (arrows). POL, POL domain; HEL, HEL domain; I, II, nonconserved domain; MT, MT domain. [See online article for color version of this figure.]
Because RCA was found in fractions enriched in both TMV POL activity and VRCs, we further analyzed the specific domains of the TMV replicase proteins required for the interaction with RCA by in vitro pull-down assays using glutathione-S-transferase (GST). The domains of 126-/183-kD proteins were fused to the C-terminal end of GST. Equal amounts of fusion proteins bound to glutathione sepharose beads were incubated with the S30 fraction of N. tabacum leaves and the pulled-down extracts were assayed by immunoblotting with anti-RCA. RCA bound to the GST-I, II nonconserved domain, located between the MT and HEL domains, and, to a lesser extent, the GST-MT and GST-POL domains (Fig. 1C), indicating that RCA interacts most specifically with the I, II domain of the TMV-encoded replicase proteins.
Considering these results, experiments were initiated to determine if the AtpC and RCA proteins would colocalize extensively in vivo with the TMV 126-kD GFP fusion protein during ectopic expression or VRC of TMV during infection. After agroinfiltration of N. benthamiana epidermal leaves with fusions of AtpC or RCA with the mCherry reporter gene, we did not observe significant relocalization of these proteins to the VRCs induced by TMV.MP-GFP or to the inclusions induced during ectopic expression of the 126-kD GFP protein fusion (Supplemental Figs. S2 and S3).
Tobamovirus Infection Alters the Expression Levels of AtpC and Rca mRNAs
Because AtpC and RCA were enriched in replicase and/or VRC-enriched fractions from TMV-inoculated leaves when compared with the uninfected extracts, we wanted to determine the effect of virus infection on the accumulation of their respective encoding mRNAs. To this end, N. tabacum leaves were challenged with TMV U1, and the levels of AtpC and Rca mRNAs were determined by quantitative reverse transcription-PCR (RT-PCR) in the infected leaves. Both AtpC (Fig. 2A) and Rca (Fig. 2B) mRNAs were reduced by 50% in TMV-infected leaves compared with mock-inoculated leaves at 4 dpi. RCA levels in chloroplasts were also reduced by 70% after TMV infection (data not shown). In addition to TMV, N. tabacum leaves were challenged with Turnip vein clearing virus (TVCV), Cucumber mosaic virus (CMV), or Potato virus X (PVX). Infection by TVCV, a member of the Tobamovirus genus, like TMV, also reduced the levels of AtpC and Rca mRNA by 50% compared with mock-inoculated leaves (Fig. 2, A and B, respectively). It is important to note that infections with viruses belonging to different genera, the Cucumovirus CMV, and the Potexvirus PVX, modestly reduced AtpC mRNA levels and had no effect on Rca mRNA levels. The influence of tobamoviruses on AtpC and Rca mRNA levels was not due to a general down-regulation of host mRNA levels as EF1α and ubiquitin mRNA levels were induced equally by TMV, TVCV, PVX, and CMV (Supplemental Fig. S4). In addition, the influence of the tobamoviruses on AtpC and Rca mRNA levels was not due to a greater titer of these viruses compared with PVX and CMV because there was no correlation between virus titer and down-regulation of the targeted host mRNAs (compare results shown in Supplemental Fig. S5 and Fig. 2).
Figure 2.
Effect of virus infection on AtpC and Rca mRNA levels in N. tabacum plants. N. tabacum plants were mock-inoculated (Mock) or inoculated with TMV-U1 (TMV), TVCV, CMV, or PVX. Quantitative RT-PCR was used to determine the relative accumulation of AtpC (A) and Rca (B) mRNAs in mock- and virus-inoculated leaves. The expression of AtpC and Rca was normalized with EF1α mRNA. Bars represent means ± se for at least three replicates per treatment. ANOVA followed, as necessary, by an lsd calculation was used to determine significant differences between treatments. Treatment means with different letters above their bars indicate significant differences between each at the P = 0.05 level. Note that the experiments with TMV were conducted and analyzed separately from those with the other viruses.
AtpC and RCA Influence Accumulation and/or Spread of TMV, But Not PVX or Tomato bushy stunt virus
To investigate the possible roles of AtpC and Rca during TMV infection, we used Tobacco rattle virus (TRV)-mediated virus-induced gene silencing (VIGS; Liu et al., 2002) to silence these genes in N. benthamiana. By 10 dpi, the plants infiltrated with TRV expressing a fragment of the AtpC or Rca genes (TRV-AtpC or TRV-Rca) showed stunted growth with light-green areas on systemically silenced leaves compared with the plants infiltrated with TRV vector alone (Supplemental Fig. S6, A and B). Quantitative RT-PCR analyses confirmed the silencing of AtpC and Rca genes (greater than 90% decrease in mRNA levels; Fig. 3, A and B).
Figure 3.
The relative mRNA levels of AtpC (A) and Rca (B) in TRV-infiltrated (TRV) and TRV-AtpC- (A) and TRV-Rca-infiltrated (B) plants by quantitative RT-PCR. For these studies, analyzed leaves were those showing the visual silencing phenotype (Supplemental Fig. S4). The data represent mean +/− the se from five replicate experiments.
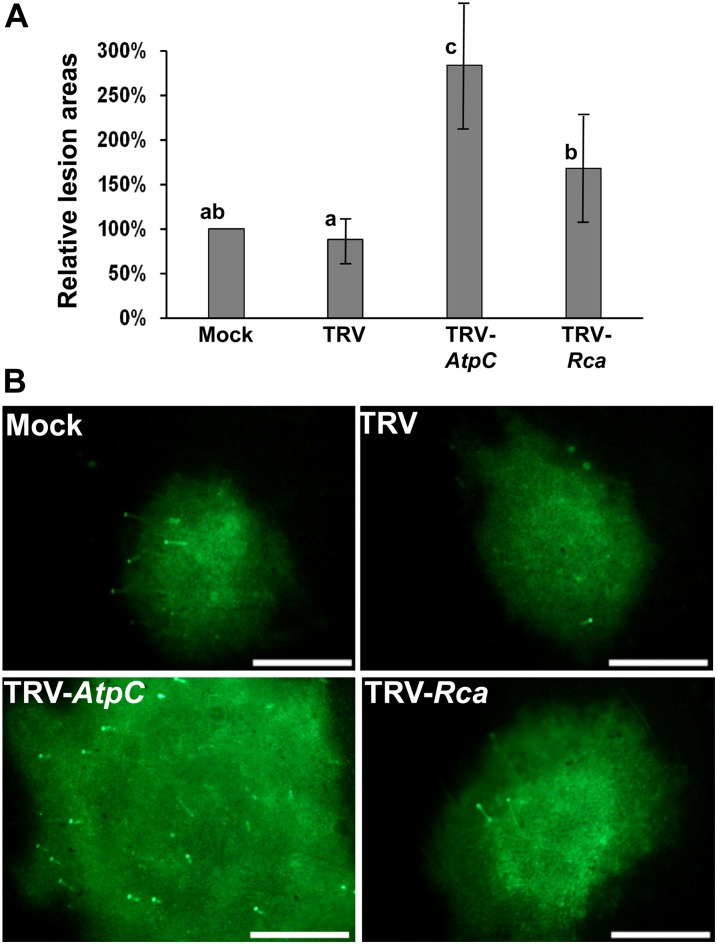
Fourteen days after infiltration with the VIGS vectors we inoculated silenced leaves with TMV.MP.GFP.CP, PVX-GFP, and Tomato bushy stunt virus (TBSV)-GFP infectious transcripts, all expressing unfused GFP. Virus intercellular movement on the TRV-AtpC/Rca, mock, and TRV controls was quantified by imaging GFP fluorescence at 4 dpi (Fig. 4A; Supplemental Fig. S7A). TMV.MP.GFP.CP-induced lesions on AtpC-silenced leaves were twice as large as those on TRV-infiltrated and mock-infiltrated leaves (Fig. 4B). It is surprising that AtpC silencing had the opposite effect on TBSV-GFP and PVX-GFP intercellular spread, resulting in generally smaller lesions compared with those on mock- and TRV-infiltrated leaves (Supplemental Fig. S7, B and C). Silencing Rca did not significantly alter the intercellular movement of TMV.MP.GFP.CP (Fig. 4B). However, lesions induced by TBSV-GFP (Supplemental Fig. S7B) and PVX-GFP (Supplemental Fig. S7C) were generally smaller on Rca-silenced leaves than on the mock- or TRV-inoculated leaves.
Figure 4.
AtpC- and Rca-silencing enhances the intercellular movement and/or accumulation of TMV.MP.GFP.CP. A, Representative images of fluorescent local lesions induced by TMV.MP.GFP.CP inoculated onto silenced leaves from TRV-AtpC- or TRV-Rca-infiltrated N. benthamiana plants. Plants infiltrated with buffer (Mock) or TRV without insert (TRV) were controls. Images were taken at 4 dpi on leaves showing both visual and biochemical evidence of host gene silencing. Bars = 1 mm. B, Relative lesion areas of TMV.MP.GFP.CP were determined at 4 dpi with the percentages standardized against those from mock-infiltrated samples. C, TMV.MP.GFP.CP accumulation in standardized areas encompassing the lesions at 4 dpi, as measured by ELISA. In both B and C, each bar represents the grand mean +/− sd of two experimental means. In B, no deviation is shown about the mock treatment grand mean because each mock treatment experimental mean was normalized to 100% for comparison with other treatment values. Each experimental mean contained at least six replicates in B and at least three replicates in C. The standard deviations about the experimental mock treatment means utilized in B averaged 28%. Different letters above the bars for each treatment within an experiment indicate significant differences between those grand means at the P = 0.05 level as determined by ANOVA and lsd. Data summarized in C was transformed for statistical analysis to correct for nonhomogenous variance.
In an effort to determine the effect of AtpC and Rca silencing on the accumulation of TMV.MP.GFP.CP at 4 dpi, we utilized an ELISA to quantify TMV CP levels per lesion. The ELISA data showed that silencing Rca caused a 7-fold increase in TMV.MP.GFP.CP accumulation per cell and silencing AtpC caused a 4-fold increase in TMV accumulation, a portion of which could be accounted for by the increased intercellular spread (Fig. 4C, and for AtpC, compare with value in Fig. 4B). It is important to note that silencing of Rca and AtpC enhanced the TMV U1 pathogenicity in N. benthamiana plants with symptoms progressing more quickly in Rca- and AtpC-silenced plants (Fig. 5). By 14 dpi, TMV U1 infection resulted in stem necrosis and apical death in plants infiltrated with TRV-Rca (Fig. 5, A and C) and TRV-AtpC (Fig. 5, B and D) compared with mock-infiltrated or TRV-infiltrated plants (Fig. 5, C and D).
Figure 5.
Systemic necrosis induced by TMV in Rca- or AtpC-silenced N. benthamiana plants. Stem necrosis and lodging of the apical regions (indicated by white arrows) of the (A) Rca-silenced and (B) AtpC-silenced N. benthamiana plants at 14 dpi with TMV (U1 strain). Range and comparison of symptoms in mock-infiltrated, TRV-infiltrated, and (C) TRV-Rca-silenced or (D) TRV-AtpC-silenced N. benthamiana plants at 14 d post challenge with TMV U1.
Silencing of AtpC and Rca Alters the TMV.MP-GFP Infection Foci and VRC Phenotypes
To further study the effect of silencing AtpC and Rca on TMV accumulation and movement, we used TMV.MP-GFP (Heinlein et al., 1995), where GFP was fused with MP and the fusion was driven by the MP subgenomic promoter. N. benthamiana leaves silenced for AtpC and Rca expression were inoculated with TMV.MP-GFP, and the spread of GFP fluorescence was monitored at 4 dpi. As observed with TMV.MP.GFP.CP, which expressed an unfused GFP (Fig. 4), lesions induced by TMV.MP-GFP on leaves silenced for AtpC were significantly larger than those on control plants (Fig. 6A). Also, for both the AtpC- and Rca-silenced leaves, we observed infection foci consisting of concentric fluorescent rings with alternating dark areas unlike those on mock-infiltrated and TRV-infiltrated controls (Fig. 6B). At least 90% of the total number of the TMV.MP-GFP foci present on AtpC-silenced leaves and 50% of those on Rca-silenced leaves had these concentric fluorescent rings.
Figure 6.
AtpC and Rca silencing influences the intercellular movement and/or visual phenotype of TMV.MP-GFP infection foci in N. benthamiana. A, Relative lesion areas were determined at 4 dpi with TMV.MP-GFP on silenced leaves from TRV-AtpC- or TRV-Rca-infiltrated plants with the percentages standardized against those from buffer-infiltrated (Mock) samples. Plants infiltrated with buffer or TRV without insert (TRV) were controls. Each bar represents a grand mean +/− sd of three experimental means. No deviation is shown for the mock treatment grand mean because each mock treatment experimental mean was normalized to 100% for comparison with other treatment values. Each experimental mean contained at least 12 replicates. The standard deviations for the experimental mock treatment means averaged 31%. Different letters above bars for each treatment indicate significant differences between those grand means at the P = 0.05 level as determined by ANOVA and lsd. B, Representative images of TMV.MP-GFP-induced infection foci at 4 dpi on mock-, TRV-, TRV-AtpC-, or TRV-Rca-infiltrated N. benthamiana plants. Bars = 1 mm.
We proposed two models to explain the formation of concentric fluorescent rings within infection foci. In the first model, we hypothesized that a reinitiation of MP-GFP accumulation either from the center of the infection foci or from the internal edge of the spreading infection occurs over time, resulting in successive rings as fluorescence spreads (Fig. 7A1). In the second model, we hypothesized that the initial infection ring spreads radially, inducing a defense response that inhibits virus accumulation as it spreads with time (through RNA interference or other inducible defense mechanisms). As the defense response proceeds, the virus, which is the inducer of the response, decreases in accumulation resulting in a dark area. At some point, the induction of the defense response becomes so low that virus again is able to accumulate, creating a second fluorescent ring outside the first. This ripple effect would continue over time radiating outward from the infection center (Fig. 7A2). To determine which of these hypotheses was correct, we compared the size of the single lesion ring at 2 dpi to those of the internal infection ring at 4 dpi with TMV.MP-GFP on AtpC-silenced leaves (compare size of ring at 2 dpi with those of internal ring at 4 dpi in Fig. 7, A and B). If the internal ring at 4 dpi was smaller than the ring at 2 dpi, the first model was supported. The internal infection rings at 4 dpi were significantly smaller than the infection rings at 2 dpi, suggesting that the appearance of concentric fluorescent rings within infection foci was by the reinitiation of TMV.MP-GFP accumulation from the center of the infection site or the internal edge of the spreading infection (Fig. 7B and model 1 in Fig. 7A).
Figure 7.
Analysis of the mechanism by which concentric rings form in AtpC-silenced leaves after TMV.MP-GFP inoculation. A, Models to explain the visual phenotype of TMV.MP-GFP lesions on AtpC-silenced N. benthamiana leaves. The first involves reinitiation of infection either from the center of the infection locus or from the internal edge of the spreading infection, and the second involves repeated lowering and increasing over time of virus accumulation at the spreading front. B, Standardized lesion areas of virus-induced single rings at 2 dpi and middle rings at 4 dpi on AtpC-silenced N. benthamiana leaves. Each bar represents the mean of 24 replicates ± se.
To examine the VRC phenotype in the concentrically ringed infection foci, the MP-GFP fluorescence at 4 dpi was observed under a higher magnification. The morphology of the TMV VRCs at the front of the infection ring induced in the AtpC- (Fig. 8C) and Rca-silenced (Fig. 8D) leaves were much smaller compared with those in tissue where these genes were not silenced (Fig. 8, A and B). In addition, a significant increase in the number and decrease in size of VRCs in the AtpC- and Rca-silenced leaves was observed compared with the controls (Fig. 8E). The huge increase in the number of VRCs in AtpC- and Rca-silenced leaves correlated with the increase in GFP accumulation in the individual lesions at 4 dpi (Fig. 8F).
Figure 8.
Silencing AtpC or Rca in N. benthamiana leaves modifies the VRC phenotype and enhances the accumulation of TMV.MP-GFP. Observation of TMV.MP-GFP induced VRCs at 4 dpi on systemic leaves from (A) mock-, (B) TRV-, (C) TRV-AtpC-, or (D) TRV-Rca-infiltrated N. benthamiana plants. Tissue inoculated with TMV was shown to be silenced for the target gene. The projected images are 15 to 20 optical sections obtained at 0.5-µm intervals. Bars = 20 µm. E, VRC numbers and sizes in cells from individual TMV.MP-GFP lesions in TRV-, TRV-AtpC-, or TRV-Rca-infiltrated N. benthamiana plants. Values represent means +/− sd from fields representing three biological replicates, subsampled a minimum of two times. Within subsamples, there was one (VRC number) or 10 (VRC size) observations. Within experiments determining VRC number or size, different letters associated with mean treatment values indicate significant differences between these means at the P = 0.001 level for VRC number or size. F, Western blot of proteins isolated from three individual TMV.MP-GFP lesions probed with anti-GFP antibodies that detects the MP-GFP fusion (top; see arrow). The bottom visualizes input protein levels (Input) through Coomassie brilliant blue staining of gel used for the western blot.
Intercellular Movement of TVCV Is Facilitated by Silencing AtpC and Rca
Because silencing of AtpC and Rca did not facilitate the spread of TBSV and PVX, both members of different genera from TMV, we wanted to know the effect of silencing these genes on the intercellular movement of a virus in the same genus as TMV. Using TVCV expressing an unfused GFP (TVCV-GFP), we determined that similar to TMV, its spread at 4 dpi was significantly enhanced in N. benthamiana leaves silenced for AtpC (Fig. 9A). TVCV-GFP spread in leaves silenced for Rca expression resulted in, at most, a modest increase in lesion size compared with the mock-inoculated and TRV-inoculated controls (Fig. 9A). This result was more similar to those observed after TMV infection than those observed after TBSV or PVX infection (compare Fig. 4 and Supplemental Fig. S7 with Fig. 9). Taken together, these data indicate that silencing AtpC in N. benthamiana increases the spread of tobamoviruses specifically.
Figure 9.
The influence of silencing AtpC or Rca on the intercellular movement of TVCV-GFP in N. benthamiana. A, Relative lesion areas were determined at 4 dpi on silenced leaves from TRV-AtpC- and TRV-Rca-infiltrated plants with the percentages standardized against those from buffer-infiltrated (Mock) samples. Plants infiltrated with buffer or TRV without insert (TRV) were controls. Each bar represents a grand mean +/− sd of three experimental means. No deviation is shown about the mock treatment grand mean because each mock treatment experimental mean was normalized to 100% for comparison with other treatment values. Each experimental mean contained at least six replicates. The standard deviations about the experimental mock treatment means averaged 20%. Different letters above bars for each treatment indicate significant differences between those grand means at the P = 0.05 level as determined by ANOVA and lsd. B, Representative images of TVCV-GFP-induced infection foci at 4 dpi on mock-, TRV-, TRV-AtpC-, or TRV-Rca-infiltrated N. benthamiana plants. Bars = 1 mm.
DISCUSSION
Host proteins play a critical role in the life cycle of an invading virus. However, host proteins that influence virus accumulation and symptoms, but are not essential for virus accumulation or identified as participants in an active defense response, are poorly understood. Identifying these proteins and determining the mechanism by which these interactions translate to less virus or host transcript accumulation could lead to novel methods to achieve virus-resistant plants.
Nuclear-Encoded Chloroplast Proteins Associated with TMV Complexes Are Influenced by Tobamovirus Infection and, in Turn, Influence Tobamovirus Accumulation
We have identified two chloroplast proteins, AtpC and RCA, which are enriched in the TMV replicase complex fractions and for RCA, also in VRC fractions, upon extraction (Fig. 1). It is interesting to note that tobamovirus infections and not those of viruses from other genera inhibited the steady-state levels of mRNAs encoding these proteins (Fig. 2). In what may be a related finding, silencing AtpC and Rca released tobamoviruses to accumulate to high levels and in AtpC-silenced tissue, to spread more quickly (Fig. 4, 6, and 8). This high accumulation (Rca- and AtpC-silenced tissue) and more rapid spread (AtpC-silenced tissue) was associated with severe disease symptoms (Fig. 5). These nuclear-encoded chloroplast proteins (Larsson et al., 1992; Portis and Salvucci, 2002) therefore appear to have a function in defending against tobamovirus infections and tobamoviruses possibly evolved to specifically inhibit the expression of these genes to enhance their own accumulation.
Considering that tobamoviruses replicate in the cytoplasm of infected cells, it may seem surprising that two nuclear-encoded chloroplast proteins would be influenced by tobamovirus accumulation and themselves influence virus accumulation. However, infections with TMV and other viruses are known to alter the transcript levels of nuclear genes encoding chloroplast-localized proteins (Itaya et al., 2002). In addition, modifying levels of particular host chloroplast proteins alters the accumulation of some viruses. The chloroplast protein, phosphoglycerate kinase, is required for the accumulation of Bamboo mosaic virus, a virus related to PVX (Lin et al., 2007). Also, chloroplast proteins are implicated in defending against infections. NRIP1, a chloroplast-targeted host protein, is important for pathogen recognition through its ability to interact with both the plant N immune receptor and TMV 126-kD replicase protein (Caplan et al., 2008). Increased accumulation of TMV was observed upon silencing the genes encoding ferredoxin I and the 33K subunit of the oxygen-evolving complex (OEC) of PSII and increased accumulation of Plum pox virus (PPV) was observed upon silencing the gene encoding PSI subunit K (PSI-K) protein (Abbink et al., 2002; Jiménez et al., 2006; Ma et al., 2008). All of these proteins therefore appear to have a role in defending the host against infection. It is interesting to note that for both the 33K subunit and PSI-K proteins, infection by TMV or PPV inhibits their respective mRNA levels (Abbink et al., 2002; Jiménez et al., 2006). Both the 33K subunit and the PSI-K proteins were identified through yeast (Saccharomyces cerevisiae) two-hybrid screens with the helicase domain of the 126-kD protein of TMV and the cylindrical inclusion protein of PPV, respectively, as bait (Abbink et al., 2002; Jiménez et al., 2006). Thus, our findings with AtpC and RCA have striking similarities to findings by others for nuclear-encoded chloroplast proteins unrelated to AtpC or RCA. The challenge from all of these studies is to further understand mechanistically how these interactions influence virus accumulation.
Toward a Mechanistic Understanding of AtpC and RCA Influence on Virus Accumulation
One way to begin a mechanistic understanding is to consider the specificity of the interaction for a particular virus. This would provide some indication of the required protein structures to elicit the response. Abbink and associates (2002) determined that silencing the 33K OEC subunit mRNA, in addition to allowing TMV overaccumulation, resulted in a significant severalfold increase in accumulation of viruses from different genera, PVX and Alfalfa mosaic virus. Although the possibility exists that proteins encoded by these disparate viruses have similar motifs that also interact with the 33K OEC subunit, it seems more likely that the down-regulation of the 33K OEC subunit accumulation, either by silencing or TMV infection, results in a general nonspecific plant response that supports accumulation of multiple viruses. Perhaps supporting this conclusion is the finding that infection of N. tabacum with Alfalfa mosaic virus did not decrease the mRNA levels of the 33K OEC subunit (Abbink et al., 2002). In our study, we determined that silencing AtpC and Rca caused a significant increase in the spread (in AtpC- silenced tissue) and accumulation (in AtpC and Rca-silenced tissue) of the tobamoviruses, TMV and TVCV, but not viruses of other genera, TBSV and PVX. These findings suggest that the modified activities of these host proteins caused by the tobamoviruses do not cause a general plant metabolic response that would aid the accumulation of all viruses. Further support for this conclusion comes from our finding that the levels of virus accumulation in infected tissues for each virus were not correlated with the level of AtpC and Rca transcript levels in these tissues (Fig. 2; Supplemental Fig. S5).
The different infection phenotypes displayed in plants silenced for AtpC and RCA indicates a further specificity in the effect of these proteins on tobamovirus physiology. That silencing of Rca mRNA resulted in only an increase in TMV accumulation, whereas silencing of AtpC resulted in predominantly an increase in virus spread indicated that different aspects of the TMV life cycle were influenced by silencing these respective genes. The main known function of RCA in plants is to activate Rubisco by releasing the bound sugar and phosphates from the active site in an ATP-dependent manner (Portis and Salvucci, 2002). Possibly of more interest, RCA is a member of the AAA+ family of proteins (Portis and Salvucci, 2002; Portis, 2003). These proteins often assemble as hexameric ring complexes and utilize a conserved ATPase domain as an energy generator to remodel macromolecules during divergent processes such as protein unfolding and degradation, organelle biogenesis, and DNA recombination and repair (Ogura and Wilkinson, 2001; Snider et al., 2008). In the study by Abbink and colleagues (2002), in addition to the previously described 33K OEC subunit, a protein belonging to the AAA family but with unknown function was identified that also interacted with a portion of the TMV replicase-associated proteins (Abbink et al., 2002). Unlike our findings with RCA, however, they found that TMV infection did not modify the transcript levels of this protein and silencing this AAA gene resulted in a decrease in TMV accumulation (Abbink et al., 2002). Additionally, we found that RCA had its strongest interaction with domain I, II between the MT and HEL domains of the TMV replicase-associated proteins whereas the AAA with unknown function interacted with the helicase domain (Fig. 1C; Abbink et al., 2002). AAA family members have a conserved domain necessary for ATPase activity that is within the core of the multimer complex, with less conserved domains which undertake the distinct activities for each family member often to the perimeter of this core region. It may be that the common domain of the AAA proteins interacts with the TMV replicase-associated domains because multiple AAA proteins have been identified that interact with the TMV 126-/183-kD proteins (Fig. 1; Abbink et al., 2002; a plant representative of the FtsH gene family [S. Bhat and R.S. Nelson, unpublished data]). However, the common domain in these AAA proteins is not sufficient to explain the opposite effects observed for RCA and the unknown AAA protein on virus accumulation: one limiting and one aiding virus accumulation. The unique domains on these AAA proteins or the unique binding sites for these proteins within the 126-/183-kD proteins may control these responses.
Regarding the response of TMV to RCA specifically, we hypothesized that RCA was retargeted to VRCs in the cytoplasm instead of chloroplasts by the 126-/183-kD proteins because putative precursor protein identified by size was present in SDS-PAGE (Fig. 1, A and B), possibly leading to the premature degradation of the replication complex. Our finding that RCA interacts in vitro with domains of 126- and 183-kD replicase, particularly with the domain I, II, is consistent with this hypothesis (Fig. 1C). An example of a relocation of another host protein by a viral protein is the observation that the TBSV P19 protein interacts in vitro with the Arabidopsis ALY2 protein involved in RNA transport and relocalizes ALY2 from its normal nuclear location to the cytoplasm (Uhrig et al., 2004). However, in our study, after agroinfiltration with fusions of AtpC or RCA with the mCherry reporter gene, we did not observe significant relocalization of these proteins to the VRCs induced by TMV.MP-GFP or to the inclusions induced during ectopic expression of the 126-GFP protein fusion (Supplemental Figs. S2 and S3). Also, no relocalization of RCA to VRCs was observed by immunofluorescence microscopy on fixed tissue incubated with rhodamine-conjugated secondary antibodies (data not shown). The interactions between host proteins and TMV VRCs may be transient and in immunofluorescence studies, disrupted during cell fixation. Another possibility is that only a small percentage of the total RCA protein and/or replicase have the correct structure(s) to bind one another and thus not at a level that could be detected by fluorescence colocalization studies. Support for this hypothesis comes from the pull-down results where only a small percentage of the input RCA was bound by the various replicase domains (Fig. 1C). However, we cannot be certain whether what was bound reflected a structure in minor amounts or was limited by the absence of the virus holoprotein or the binding capacity of the assay. It is also possible that the interaction between AtpC or RCA with the 126-kD protein is present only on extraction. The influence of these host proteins on virus accumulation then would be necessarily indirect, for example by supporting host activities whose products then inhibit virus accumulation. Assuming lower photosynthesis rates in the absence of RCA and AtpC (in particular under normal CO2 levels; Somerville et al., 1982; Rott et al., 2011), it is interesting that the tobamoviruses would overaccumulate under these conditions because previous studies indicate that TMV accumulation is indirectly supported when photosynthesis is unimpeded (Kano, 1985). Thus, it likely would be other functions of these proteins, beyond supporting photosynthesis, responsible for inhibiting tobamovirus accumulation.
AtpC and RCA host proteins could have a role in plant defense against TMV infection by manipulating signaling pathways. It is intriguing to note that the proteolytic fragments of AtpCs referred to as inceptins play an important role in defending legume plants against insect attack by inducing various phytohormone signaling pathways including salicylic acid, jasmonic acid, and ethylene (Schmelz et al., 2006, 2007). Applying this information to our findings, perhaps the reduction in AtpC mRNA induced by TMV or free AtpC due to an interaction with the 126-/183-kD protein(s) results in fewer proteolytic fragments of AtpC. This would result in less salicylic acid, and lower salicylic acid accumulation allows greater spread and accumulation of TMV (Chivasa et al., 1997; Murphy and Carr, 2002). Further work is necessary to test the efficacy of this model.
Two characteristics of the tobamovirus infection phenotypes in both Rca- and AtpC-silenced plants is the appearance of concentric fluorescent rings and the overproduction of multiple small VRCs (Figs. 6B and 8, C and D). It is likely that the increased accumulation of TMV in the AtpC-silenced plants rather than its increased spread in these plants is represented by these phenotypes because they are displayed to varying degrees during virus infection in Rca-silenced plants where only increased virus accumulation was observed (Fig. 6, B and D). Under normal conditions with no gene silencing, TMV.MP-GFP produces a single fluorescent ring due to completion of the virus life cycle and the degradation of MP-GFP in the center of the infection foci (Szécsi et al., 1999). In an effort to further understand the cause of the concentric fluorescent rings in Rca- and AtpC-silenced plants, we determined that the additional ring of fluorescence initiated either from the center of the infection foci or from the internal edge of the spreading infection rather than through waves of increased and decreased virus accumulation at the infection front (Fig. 7). This finding is consistent with a model where there is renewing of virus accumulation in tissue normally finished with this activity (Fig. 7A, model 1). It is possible that the formation of the smaller and more numerous VRCs and viruses in plants silenced for AtpC and RCA accumulation is mechanistically related to the concentric ring phenotype (Figs. 8 and 6). The appearance of these numerous and bright granules may reflect overaccumulation of virus or virus products that would lead over time to an inducible host defense (e.g. RNA interference) at the center of the lesion.
The concentric fluorescent ring phenotype displayed by TMV in the AtpC- and RCA-silenced plants resembles the concentric necrotic rings observed during infections with ringspot viruses. The development of concentric ring spots of Tomato ringspot virus in N. benthamiana induced the accumulation of hydrogen peroxide in the ring spots (Jovel et al., 2007). However, there was no correlation in virus accumulation levels between necrotic and nonnecrotic tissue in systemically infected leaves (Jovel et al., 2007). Although it is necessary to verify this finding within necrotic and nonnecrotic tissue from necrotic ringspots themselves (the more closely related system to our work), this result is different from our findings where the presence of alternating fluorescence intensities is a direct measure of alternating levels of accumulation of a virally encoded protein. In addition, the cause of the concentric rings themselves requires further study. We have indicated that the fluorescent rings may be due to a loss of a defense response internal to the infection front that in some manner is associated with AtpC or RCA silencing. Both these genes are more active in light (e.g. Kusnetsov et al., 1999; Wang et al., 2003) and thus ring formation may be due to a diurnal decrease in the efficiency in silencing in this tissue. Impaired silencing, however, would likely influence accumulation at the infection front, in addition to an influence internal to the front, leading to multiple rings appearing externally and internally as the infection spreads (i.e. model 2). Our results do not support this mechanism of ring formation. In addition, if silencing efficiency was the cause of the ring pattern, similar rings might be observed even in nonsilenced tissue due to the diurnal pattern of AtpC and RCA expression. Such a ring pattern is absent in nonsilenced tissue (Fig. 6; Szécsi et al., 1999). Although our results support the conclusion that a weakened defense response, related to AtpC and Rca silencing, allows reinitiation of infection internal to the spreading infection, the intracellular mechanism responsible for the concentric fluorescent ring phenomenon and its relationship to necrotic ringspot formation requires more study.
CONCLUSION
Taken together, our results demonstrate that the host chloroplast proteins AtpC and RCA interact with TMV replication-associated proteins and VRCs in vitro and play a role in plant defense specifically against tobamoviruses. Although both proteins limit tobamovirus infections, they have different effects on TMV spread, indicating that although they interact with similar TMV complexes, their influences are not always on the same step of the virus life cycle. A practical outcome of this work is that overexpression of these proteins in plants may result in an increased resistance to tobamoviruses. Further studies of these host proteins during TMV infection will tell much about how host factors involved in “passive” resistance to virus infection function.
MATERIALS AND METHODS
Plant Materials, Virus Strains, and Virus Inoculations
Nicotiana tabacum and Nicotiana benthamiana were grown as described (Harries et al., 2008). Purified virus (TMV) U1 strain, (Shintaku et al., 1996)] or virus from plant extracts TVCV, CMV, and PVX (Yang et al., 2004) were used in some studies. Plant extracts containing TVCV, CMV, or PVX were obtained by grinding infected leaf tissue in liquid nitrogen followed by addition of approximately 2 volumes of 0.1 m sodium phosphate buffer (pH 7). Crude extracts or purified TMV U1 strain diluted with phosphate buffer, then were inoculated directly to plants dusted with carborundum (330 grit, Fisher), as described (Ding et al., 1998). Additional studies were conducted using plasmids encoding TMV.MP.GFP.CP (previously referred to as TMV-MP-GFP-CP), TMV.MP-GFP (previously referred to as TMV-MP:GFP), and TMV.MP-GFP.CP (previously referred to as TMV-MP:GFP-CP; Cheng et al., 2000; Liu et al., 2005), and TVCV-GFP, TBSV-GFP, and PVX-GFP (sources described in Harries et al., 2009). Viral infectious transcripts were obtained from 1 µg linearized plasmid DNA using appropriate T7/T3/SP6 mMessage mMachine in vitro transcription kit (Ambion). Plant leaves were rub-inoculated mechanically with one-half of each transcript reaction using carborundum as an abrasive. Virus-inoculated plants were either placed in the growth chamber or greenhouse. Growth chamber conditions were 23°C +/− 1°C with 16 h of light (approximately 155 µmol m–2 s−1) and 8 h of dark. Greenhouse conditions were 24°C +/− 2°C with 16 h supplemental lighting (400 µmol m–2 s−1) and 60% humidity.
Isolation and Purification of TMV Replicase Complex
The TMV RNA replicase-containing fractions were purified from infected N. tabacum ‘Xanthi-nn’ as described (Osman and Buck, 1997). Leaves from N. tabacum plants inoculated with TMV (U1 strain) were harvested at 4 dpi and homogenized in buffer B. The TMV replicase complex was further isolated by differential centrifugation to give a 30,000g pellet, which was further subjected to a Suc density gradient (20% to 60% [wt/wt] in Tris-EDTA-dithiothreitol (TED) buffer) centrifugation. TED buffer consisted of 50 mM Tris-HCl (pH 8.0), 10 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol and 5% (vol/vol) glycerol. Suc density gradient fractions were analyzed for RNA POL activity as described by Osman and Buck (1997) with some modifications discussed below. Active fractions (fractions 4 and 5 from the top) were chosen for further purification. Those were diluted 10-fold with TED buffer, centrifuged at 40,000g, and the resulting pellet containing crude membrane-bound RNA POL was solubilized with sodium taurodeoxycholate and subjected to anion-exchange chromatography on DEAE-Bio-Gel A (Bio-Rad) and High Q (Bio-Rad) using an HPLC system (Pharmacia) as described previously (Osman and Buck, 1997). Collected fractions were then assayed for RNA POL activity. The reactions were performed using 50 µL of RNA POL preparation added to 50 µL 2× buffer B containing 2 mm ATP, 2 mm GTP, 2 mm CTP, 20 µm UTP, 10 µCi [α-32P]UTP, and bentonite (4.8 mg/mL). Reactions were carried out for 1 h at 30°C. Reaction products were isolated by phenol extraction and ethanol precipitation and resuspended in 10 µL of Tris-EDTA buffer (10 mm Tris-HCl, pH 8.0, 1 mm EDTA) and analyzed by PAGE containing 8M urea followed by autoradiography. Fractions showing highest RNA POL activity were analyzed for protein composition using 4% to 15% SDS-PAGE, followed by protein staining with the Bio-Rad Silver Stain Plus kit. Similar procedures were performed using tissue from uninfected plants to obtain control samples. Excision, in-gel tryptic digests, and mass spectrometry analyses of individual protein bands were conducted as described (Watson et al., 2003).
Crude VRC fractions were obtained as described (Covey and Hull, 1981) with the following modifications. Tissue was extracted at 4 dpi with TMV (U1 strain) expressing an MP-GFP fusion and CP when the virus was in maximum replication phase and the MP-GFP was present in the VRC, thus providing a marker for this complex (Liu et al., 2005). Leaves were ground in liquid nitrogen, followed by addition of buffer A (50 mm Tris-HCl, pH 7.6, 60 mm KCl, 6mm 2-mercaptoethanol, and complete EDTA-free protease inhibitor [Roche]) at 4°C. Extract was then filtered through eight layers of cheese cloth. The filtrate was subjected to centrifugation at 2,000g for 10 min at 4°C. The pellet fraction was resuspended in buffer A containing Triton X-100 (1%) and EDTA (5 mm) and repelleted by centrifugation at 2,000g for 10 min. The centrifugation process above was repeated two times and the final pellet resuspended in buffer A containing glycerol (15%). The purified TMV VRCs were analyzed for fluorescence from GFP through confocal microscopy. Proteins within fractions enriched for GFP, were separated by SDS-PAGE (Laemmli, 1970) and visualized through silver staining as described (Blum et al., 1987). Excision, in-gel tryptic digests, and nano-LC-MS/MS analyses of individual protein bands were conducted as described (Lei et al., 2005). Protein identification was performed by searching against the National Center for Biotechnology Information nonredundant protein database.
VIGS Constructs and Agroinfiltration
Full-length AtpC and RCA complementary DNA (cDNA) sequences from N. tabacum were amplified by RT-PCR using Moloney murine leukemia virus reverse transcriptase (Promega), exTaq POL (Takara), oligo dT, and gene specific primers (Supplemental Table S1). The full-length cDNAs were cloned into pCR-Blunt II-TOPO as instructed (Invitrogen). A 425-bp fragment from the open reading frame of AtpC and RCA were amplified by PCR using exTaq POL and gene specific primers (Supplemental Table S1), sequenced, and cloned into the silencing vector, pTRV2 (Liu et al., 2002), using Gateway cloning technology (Invitrogen). pTRV1 and pTRV2 constructs in Agrobacterium tumefaciens were grown and infiltrated into N. benthamiana leaves as described (Ding et al., 2004). Gene silencing was confirmed by standard RNA extraction and quantitative RT-PCR analysis.
Total RNA Extraction and Quantitative RT-PCR Analysis
Total RNA from leaf tissues were extracted using an RNeasy plant mini kit (Qiagen) and treated with DNase I. First-strand cDNA was synthesized using a 12- to 18-base oligo(dT) primer (Invitrogen) and Moloney murine leukemia virus reverse transcriptase (Promega). Two microliters of 20-fold diluted cDNA, gene specific primers, and the Power SYBR Green master mix (Applied Biosystems) were used for quantitative RT-PCR analyses of AtpC and Rca mRNA levels with an ABI Prism 7900 HT sequence detection system (Applied Biosystems). To normalize the mRNA levels of target genes between samples, relative EF1α mRNA levels were determined using EF1α-specific primers and a relative quantification method (Pfaffl, 2001). The influence of virus infection on EF1α and ubiquitin mRNA levels also was determined through this method. Additionally, virus levels in tissues were determined through reverse transcription and quantitative PCR with virus-specific primers. To create standard curves used to quantify virus levels, virus RNA was isolated from infected tissue through virus-specific procedures: TMV and TVCV (Bruening et al., 1976; Gooding and Hebert, 1967), PVX (AbouHaidar et al., 1998; Francki and McLean, 1968), and CMV (Roossinck and White, 1998). The viral RNA was quantified by spectrophotometry. Viral RNA levels from treatment tissue were quantified by comparing their cycle threshold values obtained during quantitative RT-PCR with cycle threshold values obtained from mock-inoculated tissue spiked with known amounts of viral RNA. Reverse-strand synthesis utilized SSRT III (Invitrogen). Primers for all quantitative RT-PCR reactions are provided (Supplemental Table S1).
Statistical analysis of VRC numbers and sizes in AtpC- and Rca-silenced compared with mock- and TRV-infected control plants utilized generalized linear or linear mixed models (number: GENMOD procedure, size: MIXED procedure) with repeated measurements in SAS 9.3 (SAS Institute, Inc.). As necessary, the data were transformed to meet assumptions for equal variances between means. Individual treatment mean comparisons utilized the Tukey-Kramer adjustment of P values.
ELISAs
For ELISA analyses, discs of equal size containing tissue from outside the perimeter of the largest green fluorescent lesion were harvested at a particular dpi, as described in the results, and frozen in liquid nitrogen. Frozen tissues were ground in 100 μL of phosphate-buffered saline (PBS) buffer (0.14 m NaCl, 3 mm KCl, 10 mm phosphate buffer, pH 7.0), and ELISAs were performed for TMV CP accumulation as described (Derrick et al., 1997).
GST Constructs and Pull-Down Assay
The cDNA fragments of MT (nucleotides 69–959), I, II (nucleotides 956–2487), HEL (nucleotides 2483–3416), and POL (nucleotides 3421–4916) domains of 126-/183-kD proteins were cloned into pGEX-5X-2 (GE Lifesciences) and expressed in Escherichia coli. Expressed GST or GST-fused MT, I-II, HEL, and POL domains were bound to 50 μl of glutathione sepharose 4B as instructed (GE Lifesciences). Supernatant fraction (S30) from N. tabacum was prepared as described previously (Yamaji et al., 2006). Tubes containing approximately equal amounts of GST or GST-virus gene fusions bound to sepharose beads, as determined through SDS-PAGE analysis of the bound beads, were incubated with 200 μL of S30 fraction in protein interaction buffer (PBS containing 1% Triton X-100, 0.5% NP-40, 0.1% SDS, and 1mm phenylmethylsulfonyl fluoride) for 1 h at 4°C. The beads were washed three times, twice with PBS containing 1% Triton X-100 and once in PBS. Unbound fractions were removed and the bound beads were heated in SDS-PAGE sample buffer at 100°C for 10 min and centrifuged at 18,000g for 5 min. The supernatant was resolved on 4% to 10% SDS-PAGE and subjected to immunoblotting.
Total Protein Extraction and Western-Blot Assays
Plant tissue frozen in liquid nitrogen was ground to fine powder and thawed in protein extraction buffer (5 mm Tris-HCl, pH 8.0, 150 mm NaCl, 2 mm MgCl2, 50 mm KCl, 1% Triton X-100, 0.5 mm dithiothreitol) containing protease inhibitor (cOmplete EDTA-free, Roche; 1 tablet/50 mL). Samples were placed on ice for 30 min and centrifuged at 18,000g for 10 min. Fifteen micrograms of each protein sample was boiled in SDS-PAGE sample buffer at 100°C for 10 min and centrifuged at 18,000g for 5 min. The supernatant was resolved on 4% to 10% SDS-PAGE and electroblotted to polyvinylidene difluoride membranes (Immun-Blot, Bio-Rad). Blots were blocked with Tris-buffered saline containing 5% skim milk and probed with the appropriate primary antibodies from rabbits, anti-RCA (1:7,000 dilution), or anti-GFP (0.5 μg/mL; BioVision Research Products). They were then washed in Tris-buffered saline buffer containing 0.05% Tween 20 and probed with anti-rabbit alkaline phosphatase-conjugated secondary antibody (Promega). The immunoprobed proteins were detected by colorimetric reaction using nitroblue tetrazolium and 5-bromo-4-chloro-3-indolylphosphate-p-toluidine (Promega).
Microscopy
Fluorescent viral lesion images were captured with an Olympus SZX-12 fluorescent stereomicroscope (Olympus). Lesion areas (pixels) were measured using the MetaMorph 4.5 program (Universal Imaging). TMV.MP-GFP VRCs were imaged on a Bio-Rad model 1024ES. Images were processed using Adobe Photoshop. GFP was excited using a 488-nm line from a krypton/argon laser and emissions were captured at 522 nm.
Subcellular Localization of AtpC and RCA
Full length AtpC and Rca open reading frames were cloned into 5G-mCherry C1 plasmid, obtained from Dr. Elison Blancaflor (Noble Foundation) and introduced into A. tumefaciens GV2260 by electroporation. For studies on the interaction of AtpC and RCA with ectopically expressed 126-kD protein-GFP fusion, A. tumefaciens containing the 126-kD protein-GFP fusion was coinfiltrated with A. tumefaciens containing AtpC-mCherry or Rca-mCherry using a needleless syringe and observed 2 dpi. For studies involving later challenge with the virus, N. benthamiana leaves were syringe-infiltrated with A. tumefaciens containing AtpC-mCherry or Rca-mCherry and placed in the growth chamber for 2 d. Leaves expressing AtpC-mCherry or RCA-mCherry then were inoculated with TMV.MP-GFP, and 4 dpi, the infected cells were imaged using a Perkin-Elmer UltraView ERS spinning-disc confocal system coupled to a Zeiss Observer D1 inverted microscope. The cells were observed with a 63× water-immersion objective. GFP was excited at 488 nm, and emission was detected at 510 nm. Chloroplast autofluorescence was detected by excitation at 647 nm, and emission was detected at 680 nm. mCherry was excited at 587 nm, and emission was detected at 610 nm.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers X63606 and Z14980.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Fraction analysis for TMV 126- and 183-kD proteins and RNA POL activity.
Supplemental Figure S2. Lack of AtpC- or RCA-mCherry colocalization with TMV.MP-GFP in epidermal cells of N. benthamiana.
Supplemental Figure S3. Lack of AtpC- or RCA-mCherry colocalization with a 126-kD protein-GFP fusion in epidermal cells of N. benthamiana.
Supplemental Figure S4. Host EF1α and ubiquitin mRNA levels in N. tabacum challenged with TVCV, CMV, and PVX.
Supplemental Figure S5. Virus accumulation in extracts from challenged plants analyzed for AtpC and Rca mRNA levels in Figure 2.
Supplemental Figure S6. AtpC and Rca mRNA silencing phenotypes in N. benthamiana plants.
Supplemental Figure S7. Influence of AtpC or Rca silencing on intercellular movement of TBSV-GFP and PVX-GFP.
Supplemental Table S1. Primers for VIGS constructs and quantitative RT-PCR analyses.
Acknowledgments
The authors thank S.P. Dinesh-Kumar for providing the TRV silencing vector, Michael Salvucci for RCA antibodies, and Elison Blancaflor for the mCherry construct. We thank Aline Valster for microscopy assistance, Xin Shun Ding for reagents, Frank Coker and Vicki Barrett for greenhouse assistance, Stacy Allen for assistance with quantitative RT-PCR, Stephen Webb for assistance in statistical analysis of VRC number and size, and Elison Blancaflor and Rujin Chen for critically reading the manuscript. This work was supported by the Samuel Roberts Noble Foundation, Inc.
Glossary
- TMV
Tobacco mosaic virus
- VRC
virus replication complex
- dpi
days post inoculation
- RT-PCR
reverse transcription-PCR
- TVCV
Turnip vein clearing virus
- CMV
Cucumber mosaic virus
- PVX
Potato virus X
- TRV
Tobacco rattle virus
- VIGS
virus-induced gene silencing
- TBSV
Tomato bushy stunt virus
- OEC
oxygen-evolving complex
- PPV
Plum pox virus
- cDNA
complementary DNA
- PBS
phosphate-buffered saline
- POL
polymerase
- MT
methyltransferase
- CP
coat protein
- MP
movement protein
- HEL
helicase
- MALDI-TofMS
matrix-assisted laser-desorption ionization time-of-flight mass spectrometry
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- GST
glutathione-S-transferase
References
- Abbink TE, Peart JR, Mos TN, Baulcombe DC, Bol JF, Linthorst HJ. (2002) Silencing of a gene encoding a protein component of the oxygen-evolving complex of photosystem II enhances virus replication in plants. Virology 295: 307–319 [DOI] [PubMed] [Google Scholar]
- Abbink TEM, Tjernberg PA, Bol JF, Linthorst HJM. (1998) Tobacco mosaic virus helicase domain induces necrosis in N gene-carrying tobacco in the absence of virus replication. Mol Plant Microbe Interact 11: 1242–1246 [Google Scholar]
- AbouHaidar MG, Huimin X, Hefferon KL. (1998) Potexvirus isolation and RNA extraction. Meth Molec Biol 81: 131–143 [DOI] [PubMed] [Google Scholar]
- Ahlquist P, Noueiry AO, Lee WM, Kushner DB, Dye BT. (2003) Host factors in positive-strand RNA virus genome replication. J Virol 77: 8181–8186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano M, Satoh R, Mochizuki A, Tsuda S, Yamanaka T, Nishiguchi M, Hirai K, Meshi T, Naito S, Ishikawa M. (2005) Tobamovirus-resistant tobacco generated by RNA interference directed against host genes. FEBS Lett 579: 4479–4484 [DOI] [PubMed] [Google Scholar]
- Bao Y, Carter SA, Nelson RS. (1996) The 126- and 183-kilodalton proteins of tobacco mosaic virus, and not their common nucleotide sequence, control mosaic symptom formation in tobacco. J Virol 70: 6378–6383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benitez-Alfonso Y, Faulkner C, Ritzenthaler C, Maule AJ. (2010) Plasmodesmata: gateways to local and systemic virus infection. Mol Plant Microbe Interact 23: 1403–1412 [DOI] [PubMed] [Google Scholar]
- Bilgin DD, Liu Y, Schiff M, Dinesh-Kumar SP. (2003) P58(IPK), a plant ortholog of double-stranded RNA-dependent protein kinase PKR inhibitor, functions in viral pathogenesis. Dev Cell 4: 651–661 [DOI] [PubMed] [Google Scholar]
- Blum H, Beier H, Gross HJ. (1987) Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis 8: 93–99 [Google Scholar]
- Boevink P, Oparka KJ. (2005) Virus-host interactions during movement processes. Plant Physiol 138: 1815–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruening G, Beachy RN, Scalla R, Zaitlin M. (1976) In vitro and in vivo translation of the ribonucleic acids of a cowpea strain of tobacco mosaic virus. Virology 71: 498–517 [DOI] [PubMed] [Google Scholar]
- Caplan JL, Mamillapalli P, Burch-Smith TM, Czymmek K, Dinesh-Kumar SP. (2008) Chloroplastic protein NRIP1 mediates innate immune receptor recognition of a viral effector. Cell 132: 449–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng NH, Su CL, Carter SA, Nelson RS. (2000) Vascular invasion routes and systemic accumulation patterns of tobacco mosaic virus in Nicotiana benthamiana. Plant J 23: 349–362 [DOI] [PubMed] [Google Scholar]
- Chivasa S, Murphy AM, Naylor M, Carr JP. (1997) Salicylic acid interferes with tobacco mosaic virus replication via a novel salicylhydroxamic acid-sensitive mechanism. Plant Cell 9: 547–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cournoyer P, Dinesh-Kumar SP. (2011) NB-LRR immune receptors in plant virus defence. In C Caranta, MA Aranda, M Tepfer, JJ Lopez-Moya, eds, Recent Advances in Plant Virology. Caister Academic Press, Norfolk, UK, pp 149–176
- Covey SN, Hull R. (1981) Transcription of cauliflower mosaic virus DNA. Detection of transcripts, properties, and location of the gene encoding the virus inclusion body protein. Virology 111: 463–474 [DOI] [PubMed] [Google Scholar]
- Culver JN, Padmanabhan MS. (2007) Virus-induced disease: altering host physiology one interaction at a time. Annu Rev Phytopathol 45: 221–243 [DOI] [PubMed] [Google Scholar]
- Derrick PM, Carter SA, Nelson RS. (1997) Mutation of the tobacco mosaic tobamovirus 126-and 183-kDa proteins: effects on phloem-dependent virus accumulation and synthesis of viral proteins. Mol Plant Microbe Interact 10: 589–596 [Google Scholar]
- Ding XS, Carter SA, Deom CM, Nelson RS. (1998) Tobamovirus and potyvirus accumulation in minor veins of inoculated leaves from representatives of the Solanaceae and Fabaceae. Plant Physiol 116: 125–136 [Google Scholar]
- Ding XS, Liu J, Cheng NH, Folimonov A, Hou YM, Bao Y, Katagi C, Carter SA, Nelson RS. (2004) The Tobacco mosaic virus 126-kDa protein associated with virus replication and movement suppresses RNA silencing. Mol Plant Microbe Interact 17: 583–592 [DOI] [PubMed] [Google Scholar]
- Dunigan DD, Zaitlin M. (1990) Capping of tobacco mosaic virus RNA. Analysis of viral-coded guanylyltransferase-like activity. J Biol Chem 265: 7779–7786 [PubMed] [Google Scholar]
- Erickson FL, Holzberg S, Calderon-Urrea A, Handley V, Axtell M, Corr C, Baker B. (1999) The helicase domain of the TMV replicase proteins induces the N-mediated defence response in tobacco. Plant J 18: 67–75 [DOI] [PubMed] [Google Scholar]
- Evans RK, Haley BE, Roth DA. (1985) Photoaffinity labeling of a viral induced protein from tobacco. Characterization of nucleotide-binding properties. J Biol Chem 260: 7800–7804 [PubMed] [Google Scholar]
- Francki RIB, McLean GD. (1968) Purification of potato virus X and preparation of infectious ribonucleic acid by degradation with lithium chloride. Aust J Biol Sci 21: 1311–1318 [DOI] [PubMed] [Google Scholar]
- Gooding GV, Jr, Hebert TT. (1967) A simple technique for purification of tobacco mosaic virus in large quantities. Phytopathology 57: 1285. [PubMed] [Google Scholar]
- Goregaoker SP, Lewandowski DJ, Culver JN. (2001) Identification and functional analysis of an interaction between domains of the 126/183-kDa replicase-associated proteins of tobacco mosaic virus. Virology 282: 320–328 [DOI] [PubMed] [Google Scholar]
- Harries PA, Palanichelvam K, Bhat S, Nelson RS. (2008) Tobacco mosaic virus 126-kDa protein increases the susceptibility of Nicotiana tabacum to other viruses and its dosage affects virus-induced gene silencing. Mol Plant Microbe Interact 21: 1539–1548 [DOI] [PubMed] [Google Scholar]
- Harries PA, Park J-W, Sasaki N, Ballard KD, Maule AJ, Nelson RS. (2009) Differing requirements for actin and myosin by plant viruses for sustained intercellular movement. Proc Natl Acad Sci USA 106: 17594–17599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harries PA, Schoelz JE, Nelson RS. (2010) Intracellular transport of viruses and their components: utilizing the cytoskeleton and membrane highways. Mol Plant Microbe Interact 23: 1381–1393 [DOI] [PubMed] [Google Scholar]
- Heinlein M, Epel BL, Padgett HS, Beachy RN. (1995) Interaction of tobamovirus movement proteins with the plant cytoskeleton. Science 270: 1983–1985 [DOI] [PubMed] [Google Scholar]
- Heinlein M, Padgett HS, Gens JS, Pickard BG, Casper SJ, Epel BL, Beachy RN. (1998) Changing patterns of localization of the tobacco mosaic virus movement protein and replicase to the endoplasmic reticulum and microtubules during infection. Plant Cell 10: 1107–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hills GJ, Plaskitt KA, Young ND, Dunigan DD, Watts JW, Wilson TM, Zaitlin M. (1987) Immunogold localization of the intracellular sites of structural and nonstructural tobacco mosaic virus proteins. Virology 161: 488–496 [DOI] [PubMed] [Google Scholar]
- Hirashima K, Watanabe Y. (2001) Tobamovirus replicase coding region is involved in cell-to-cell movement. J Virol 75: 8831–8836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirashima K, Watanabe Y. (2003) RNA helicase domain of tobamovirus replicase executes cell-to-cell movement possibly through collaboration with its nonconserved region. J Virol 77: 12357–12362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi K, Masuda K, Naito S, Meshi T, Ishikawa M. (2007) An inhibitor of viral RNA replication is encoded by a plant resistance gene. Proc Natl Acad Sci USA 104: 13833–13838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi K, Naito S, Meshi T, Ishikawa M. (2009) An inhibitory interaction between viral and cellular proteins underlies the resistance of tomato to nonadapted tobamoviruses. Proc Natl Acad Sci USA 106: 8778–8783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi K, Nishikiori M, Ishikawa M. (2010) Interactions between tobamovirus replication proteins and cellular factors: their impacts on virus multiplication. Mol Plant Microbe Interact 23: 1413–1419 [DOI] [PubMed] [Google Scholar]
- Itaya A, Matsuda Y, Gonzales RA, Nelson RS, Ding B. (2002) Potato spindle tuber viroid strains of different pathogenicity induces and suppresses expression of common and unique genes in infected tomato. Mol Plant Microbe Interact 15: 990–999 [DOI] [PubMed] [Google Scholar]
- Jiménez I, López L, Alamillo JM, Valli A, García JA. (2006) Identification of a plum pox virus CI-interacting protein from chloroplast that has a negative effect in virus infection. Mol Plant Microbe Interact 19: 350–358 [DOI] [PubMed] [Google Scholar]
- Jovel J, Walker M, Sanfaçon H. (2007) Recovery of Nicotiana benthamiana plants from a necrotic response induced by a nepovirus is associated with RNA silencing but not with reduced virus titer. J Virol 81: 12285–12297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang B-C, Yeam I, Jahn MM. (2005) Genetics of plant virus resistance. Annu Rev Phytopathol 43: 581–621 [DOI] [PubMed] [Google Scholar]
- Kano H. (1985) Effects of light and inhibitors of photosynthesis and respiration on the multiplication of tobacco mosaic virus in tobacco protoplasts. Plant Cell Physiol 26: 1241–1249 [Google Scholar]
- Koonin EV. (1991) The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J Gen Virol 72: 2197–2206 [DOI] [PubMed] [Google Scholar]
- Kusnetsov V, Landsberger M, Meurer J, Oelmüller R. (1999) The assembly of the CAAT-box binding complex at a photosynthesis gene promoter is regulated by light, cytokinin, and the stage of the plastids. J Biol Chem 274: 36009–36014 [DOI] [PubMed] [Google Scholar]
- Laemmli UK. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Laliberté J-F, Sanfaçon H. (2010) Cellular remodeling during plant virus infection. Annu Rev Phytopathol 48: 69–91 [DOI] [PubMed] [Google Scholar]
- Larsson KH, Napier JA, Gray JC. (1992) Import and processing of the precursor form of the gamma subunit of the chloroplast ATP synthase from tobacco. Plant Mol Biol 19: 343–349 [DOI] [PubMed] [Google Scholar]
- Le Gall O, Aranda MA, Caranta C. (2011) Plant resistance to viruses mediated by translation initiation factors. In C Caranta, MA Aranda, M Tepfer, JJ Lopez-Moya, eds, Recent Advances in Plant Virology. Caister Academic Press, Norfolk, UK, pp 177–194
- Lei Z, Elmer AM, Watson BS, Dixon RA, Mendes PJ, Sumner LW. (2005) A two-dimensional electrophoresis proteomic reference map and systematic identification of 1367 proteins from a cell suspension culture of the model legume Medicago truncatula. Mol Cell Proteomics 4: 1812–1825 [DOI] [PubMed] [Google Scholar]
- Lin J-W, Ding M-P, Hsu Y-H, Tsai C-H. (2007) Chloroplast phosphoglycerate kinase, a gluconeogenetic enzyme, is required for efficient accumulation of Bamboo mosaic virus. Nucleic Acids Res 35: 424–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JZ, Blancaflor EB, Nelson RS. (2005) The tobacco mosaic virus 126-kilodalton protein, a constituent of the virus replication complex, alone or within the complex aligns with and traffics along microfilaments. Plant Physiol 138: 1853–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JZ, Blancaflor EB, Nelson RS. (2006) The structure of the Tobacco mosaic virus replication complex is modulated by the 126-kDa protein and complex and protein traffic along microfilaments. In F Sánchez, C Quinto, IM Lopez-Lara, O Geiger, eds, Biology of Molecular Plant-Microbe Interactions, Vol 5. pp 410–415
- Liu Y, Schiff M, Dinesh-Kumar SP. (2002) Virus-induced gene silencing in tomato. Plant J 31: 777–786 [DOI] [PubMed] [Google Scholar]
- Ma Y, Zhou T, Hong Y, Fan Z, Li H. (2008) Decreased level of ferredoxin I in Tobacco mosaic virus-infected tobacco is associated with development of the mosaic symptom. Physiol Mol Plant Pathol 72: 39–45 [Google Scholar]
- Más P, Beachy RN. (1999) Replication of tobacco mosaic virus on endoplasmic reticulum and role of the cytoskeleton and virus movement protein in intracellular distribution of viral RNA. J Cell Biol 147: 945–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maule AJ, Caranta C, Boulton MI. (2007) Sources of natural resistance to plant viruses: status and prospects. Mol Plant Pathol 8: 223–231 [DOI] [PubMed] [Google Scholar]
- Murphy AM, Carr JP. (2002) Salicylic acid has cell-specific effects on tobacco mosaic virus replication and cell-to-cell movement. Plant Physiol 128: 552–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mysore KS, Ryu C-M. (2004) Nonhost resistance: how much do we know? Trends Plant Sci 9: 97–104 [DOI] [PubMed] [Google Scholar]
- Nagy P, Pogany J. (2011) Replication of plant RNA viruses. In C Caranta, MA Aranda, M Tepfer, JJ Lopez-Moya, eds, Recent Advances in Plant Virology. Caister Academic Press, Norfolk, UK, pp 19–46
- Niehl A, Heinlein M. (2011) Cellular pathways for viral transport through plasmodesmata. Protoplasma 248: 75–99 [DOI] [PubMed] [Google Scholar]
- Nishikiori M, Mori M, Dohi K, Okamura H, Katoh E, Naito S, Meshi T, Ishikawa M. (2011) A host small GTP-binding protein ARL8 plays crucial roles in tobamovirus RNA replication. PLoS Path 7: e1002409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura T, Wilkinson AJ. (2001) AAA+ superfamily ATPases: common structure—diverse function. Genes Cells 6: 575–597 [DOI] [PubMed] [Google Scholar]
- Osman TA, Buck KW. (1997) The tobacco mosaic virus RNA polymerase complex contains a plant protein related to the RNA-binding subunit of yeast eIF-3. J Virol 71: 6075–6082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett HS, Beachy RN. (1993) Analysis of a tobacco mosaic virus strain capable of overcoming N gene-mediated resistance. Plant Cell 5: 577–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett HS, Watanabe Y, Beachy RN. (1997) Identification of the TMV replicase sequence that activates the N gene-mediated hypersensitive response. Mol Plant Microbe Interact 10: 709–715 [Google Scholar]
- Padmanabhan MS, Dinesh-Kumar SP. (2010) All hands on deck—the role of chloroplasts, endoplasmic reticulum, and the nucleus in driving plant innate immunity. Mol Plant Microbe Interact 23: 1368–1380 [DOI] [PubMed] [Google Scholar]
- Padmanabhan MS, Goregaoker SP, Golem S, Shiferaw H, Culver JN. (2005) Interaction of the tobacco mosaic virus replicase protein with the Aux/IAA protein PAP1/IAA26 is associated with disease development. J Virol 79: 2549–2558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan MS, Kramer SR, Wang X, Culver JN. (2008) Tobacco mosaic virus replicase-auxin/indole acetic acid protein interactions: reprogramming the auxin response pathway to enhance virus infection. J Virol 82: 2477–2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan MS, Shiferaw H, Culver JN. (2006) The Tobacco mosaic virus replicase protein disrupts the localization and function of interacting Aux/IAA proteins. Mol Plant Microbe Interact 19: 864–873 [DOI] [PubMed] [Google Scholar]
- Pelham HR. (1978) Leaky UAG termination codon in tobacco mosaic virus RNA. Nature 272: 469–471 [DOI] [PubMed] [Google Scholar]
- Perkins DN, Pappin DJC, Creasy DM, Cottrell JS. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20: 3551–3567 [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portis AR., Jr (2003) Rubisco activase—Rubisco’s catalytic chaperone. Photosynth Res 75: 11–27 [DOI] [PubMed] [Google Scholar]
- Portis AR, Jr, Li C, Wang D, Salvucci ME. (2008) Regulation of Rubisco activase and its interaction with Rubisco. J Exp Bot 59: 1597–1604 [DOI] [PubMed] [Google Scholar]
- Portis AR, Jr, Salvucci ME. (2002) The discovery of Rubisco activase—yet another story of serendipity. Photosynth Res 73: 257–264 [DOI] [PubMed] [Google Scholar]
- Roossinck MJ, White PS. (1998) Cucumovirus isolation and RNA extraction. Methods Mol Biol 81: 189–196 [DOI] [PubMed] [Google Scholar]
- Rott M, Martins NF, Thiele W, Lein W, Bock R, Kramer DM, Schöttler MA. (2011) ATP synthase repression in tobacco restricts photosynthetic electron transport, CO2 assimilation, and plant growth by overacidification of the thylakoid lumen. Plant Cell 23: 304–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelz EA, Carroll MJ, LeClere S, Phipps SM, Meredith J, Chourey PS, Alborn HT, Teal PEA. (2006) Fragments of ATP synthase mediate plant perception of insect attack. Proc Natl Acad Sci USA 103: 8894–8899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelz EA, LeClere S, Carroll MJ, Alborn HT, Teal PEA. (2007) Cowpea chloroplastic ATP synthase is the source of multiple plant defense elicitors during insect herbivory. Plant Physiol 144: 793–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoelz JE, Harries PA, Nelson RS. (2011) Intracellular transport of plant viruses: finding the door out of the cell. Mol Plant 4: 813–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalla TA. (1964) Assembly and aggregation of Tobacco mosaic virus in tomato leaflets. J Cell Biol 21: 253–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalla TA, Shepherd RJ, Petersen LJ. (1980) Comparative cytology of nine isolates of cauliflower mosaic virus. Virology 102: 381–388 [DOI] [PubMed] [Google Scholar]
- Shintaku MH, Carter SA, Bao Y, Nelson RS. (1996) Mapping nucleotides in the 126-kDa protein gene that control the differential symptoms induced by two strains of tobacco mosaic virus. Virology 221: 218–225 [DOI] [PubMed] [Google Scholar]
- Snider J, Thibault G, Houry WA. (2008) The AAA+ superfamily of functionally diverse proteins. Genome Biol 9: 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville CR, Portis AR, Jr, Ogren WL. (1982) A mutant of Arabidopsis thaliana which lacks activation of RuBP carboxylase in vivo. Plant Physiol 70: 381–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spreitzer RJ, Salvucci ME. (2002) Rubisco: structure, regulatory interactions, and possibilities for a better enzyme. Annu Rev Plant Biol 53: 449–475 [DOI] [PubMed] [Google Scholar]
- Szécsi J, Ding XS, Lim CO, Bendahmane M, Cho MJ, Nelson RS, Beachy RN. (1999) Development of tobacco mosaic virus infection sites in Nicotiana benthamiana. Mol Plant Microbe Interact 12: 143–152 [Google Scholar]
- Uhrig JF, Canto T, Marshall D, MacFarlane SA. (2004) Relocalization of nuclear ALY proteins to the cytoplasm by the tomato bushy stunt virus P19 pathogenicity protein. Plant Physiol 135: 2411–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verchot-Lubicz J, Torrance L, Solovyev AG, Morozov SY, Jackson AO, Gilmer D. (2010) Varied movement strategies employed by triple gene block-encoding viruses. Mol Plant Microbe Interact 23: 1231–1247 [DOI] [PubMed] [Google Scholar]
- Wang L-Y, Lin S-S, Hung T-H, Li T-K, Lin N-C, Shen T-L. (2012) Multiple domains of the tobacco mosaic virus p126 protein can independently suppress local and systemic RNA silencing. Mol Plant Microbe Interact 25: 648–657 [DOI] [PubMed] [Google Scholar]
- Wang N-Y, Jiang D-A, Hong J, Zhang F, Weng X-Y. (2003) Diurnal changes of rubisco and RCA activities and their cellular localization in rice. Acta Bot Sin 45: 1076–1083 [Google Scholar]
- Watson BS, Asirvatham VS, Wang L, Sumner LW. (2003) Mapping the proteome of barrel medic (Medicago truncatula). Plant Physiol 131: 1104–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitham S, Dinesh-Kumar SP, Choi D, Hehl R, Corr C, Baker B. (1994) The product of the tobacco mosaic virus resistance gene N: similarity to toll and the interleukin-1 receptor. Cell 78: 1101–1115 [DOI] [PubMed] [Google Scholar]
- Yamaji Y, Kobayashi T, Hamada K, Sakurai K, Yoshii A, Suzuki M, Namba S, Hibi T. (2006) In vivo interaction between Tobacco mosaic virus RNA-dependent RNA polymerase and host translation elongation factor 1A. Virology 347: 100–108 [DOI] [PubMed] [Google Scholar]
- Yamaji Y, Sakurai K, Hamada K, Komatsu K, Ozeki J, Yoshida A, Yoshii A, Shimizu T, Namba S, Hibi T. (2010) Significance of eukaryotic translation elongation factor 1A in tobacco mosaic virus infection. Arch Virol 155: 263–268 [DOI] [PubMed] [Google Scholar]
- Yamanaka T, Imai T, Satoh R, Kawashima A, Takahashi M, Tomita K, Kubota K, Meshi T, Naito S, Ishikawa M. (2002) Complete inhibition of tobamovirus multiplication by simultaneous mutations in two homologous host genes. J Virol 76: 2491–2497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka T, Ohta T, Takahashi M, Meshi T, Schmidt R, Dean C, Naito S, Ishikawa M. (2000) TOM1, an Arabidopsis gene required for efficient multiplication of a tobamovirus, encodes a putative transmembrane protein. Proc Natl Acad Sci USA 97: 10107–10112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SJ, Carter SA, Cole AB, Cheng NH, Nelson RS. (2004) A natural variant of a host RNA-dependent RNA polymerase is associated with increased susceptibility to viruses by Nicotiana benthamiana. Proc Natl Acad Sci USA 101: 6297–6302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaitlin M. (1999) Elucidation of the genome organization of tobacco mosaic virus. Philos Trans R Soc Lond B Biol Sci 354: 587–591 [DOI] [PMC free article] [PubMed] [Google Scholar]