Abstract
Little is known about the homeostasis of sulfite levels, a cytotoxic by-product of plant sulfur turnover. By employing extended dark to induce catabolic pathways, we followed key elements of the sulfite network enzymes that include adenosine-5′-phosphosulfate reductase and the sulfite scavengers sulfite oxidase (SO), sulfite reductase, UDP-sulfoquinovose synthase, and β-mercaptopyruvate sulfurtransferases. During extended dark, SO was enhanced in tomato (Solanum lycopersicum) wild-type leaves, while the other sulfite network components were down-regulated. SO RNA interference plants lacking SO activity accumulated sulfite, resulting in leaf damage and mortality. Exogenous sulfite application induced up-regulation of the sulfite scavenger activities in dark-stressed or unstressed wild-type plants, while expression of the sulfite producer, adenosine-5′-phosphosulfate reductase, was down-regulated. Unstressed or dark-stressed wild-type plants were resistant to sulfite applications, but SO RNA interference plants showed sensitivity and overaccumulation of sulfite. Hence, under extended dark stress, SO activity is necessary to cope with rising endogenous sulfite levels. However, under nonstressed conditions, the sulfite network can control sulfite levels in the absence of SO activity. The novel evidence provided by the synchronous dark-induced turnover of sulfur-containing compounds, augmented by exogenous sulfite applications, underlines the role of SO and other sulfite network components in maintaining sulfite homeostasis, where sulfite appears to act as an orchestrating signal molecule.
As a plant macronutrient, sulfur (S) is important for yield production and the quality of crops. In nature, S is mostly available in its fully oxidized anion sulfate form, which is taken up, reduced, and incorporated into Cys via sulfite and sulfide generation in the sulfate assimilation pathway (Saito, 2000). In addition to the presence of S in the amino acids Cys and Met, it is an important component of oligopeptides such as reduced glutathione (GSH), coenzymes, prosthetic groups, vitamins, secondary metabolites, and lipids (Saito, 2000; Leustek, 2002). In the latter case, the chloroplast membrane containing sulfoquinovosyldiacylglycerol (SQDG) is one of the primary S-containing components in higher plants (Shimojima, 2011).
Sulfite, a less oxidized form of sulfate, is an intermediate in the assimilation of S and a potentially cytotoxic molecule (Leustek et al., 2000) that, if not rapidly metabolized, can wreak havoc at the cellular (Sanda et al., 2001; Davidian and Kopriva, 2010) and whole-plant (Murray, 1997; Brychkova et al., 2007) levels. Roots obtain sulfate from the soil, and sulfite is generated from sulfate in the leaves by the chloroplast-localized adenosine-5′-phosphosulfate (APS) reductase (APR; Enzyme Commission [EC] 1.8.4.9; Vauclare et al., 2002). Another source of sulfite is atmospheric, originating from microbial, volcanic, or anthropogenic activities and entering the plant via the stomata or through the root system. In mammalian tissue, endogenous sulfite is thought to be formed during the degradation of the S-containing amino acids (Amy, 1988; Heber and Huve, 1998; Hänsch and Mendel, 2005); however, such a pathway has yet to be explored in plants.
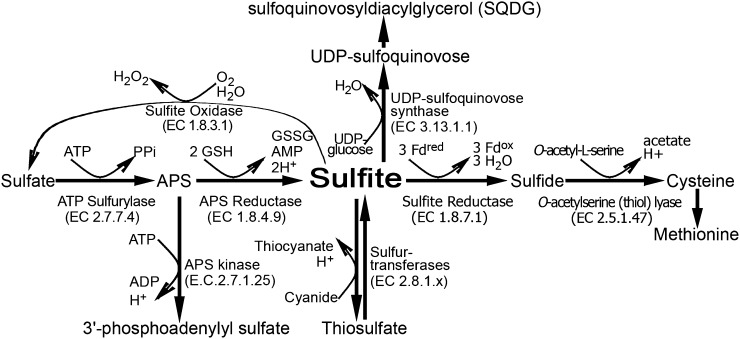
Known avenues for sulfite usage include sulfite assimilation, incorporation into metabolites, and detoxification, which together make up a potential network for the control of sulfite turnover (Fig. 1). These include ferredoxin-dependent sulfite reduction catalyzed by the chloroplast-localized sulfite reductase (SiR; EC 1.8.7.1), which yields the reduced sulfide for primary sulfate assimilation (Khan et al., 2010). Another pathway for sulfite utilization is its incorporation into sulfolipids, catalyzed by the chloroplast-localized UDP-sulfoquinovose synthase (SQD1; EC 3.13.1.1; Sanda et al., 2001). Sulfite can be reoxidized back to sulfate by the molybdenum cofactor-containing enzyme, sulfite oxidase (SO; EC 1.8.3.1), localized in the peroxisomes (Eilers et al., 2001). Alternatively, the mitochondrion- and cytosol-localized β-mercaptopyruvate sulfurtransferases (STs; EC 2.8.1.2.), MST1 and MST2, respectively, have been shown to catalyze the synthesis of the less toxic compound thiosulfate in the presence of β-mercaptopyruvate and sulfite (Papenbrock and Schmidt, 2000a; Tsakraklides et al., 2002).
Figure 1.
Schematic representation of sulfite network enzymes in plants. Sulfite is generated from sulfate in two consecutive steps. In the first step, ATP sufurylase catalyzes the adenylation of sulfate to APS, and then sulfite is produced in the chloroplast by the glutathione-dependent APR. In the chloroplast, the generated sulfite can be further reduced to sulfide by the ferredoxin-dependent SiR. The sulfide together with O-acetyl-l-Ser are the substrates for Cys biosynthesis catalyzed by OAS-TL. Alternatively, the chloroplast-localized sulfite can enter the sulfolipid reductive pathway to generate SQDG in two consecutive steps. In the first step, UDP-sulfoquinovase is catalyzed by SQD1, employing sulfite and UDP-Glc as substrates, while in the second step, SQDG is catalyzed by SQDG synthase, employing UDP-sulfoquinovose and diacylglycerols as substrates. Sulfite localized to the cytosol and mitochondria may be detoxified to thiosulfate by the STs or be generated by these enzymes from thiosulfate and cyanide (Nakamura et al., 2000; Papenbrock and Schmidt, 2000a; Tsakraklides et al., 2002). Sulfite can be oxidized to sulfate by the molybdenum cofactor-containing enzyme, peroxisomal SO. Fdox, Oxidized ferredoxin; Fdred, reduced ferredoxin; GSSG, oxidized glutathione; H2O2, hydrogen peroxide; PPi, diphosphate.
Since sulfite is cytotoxic (Leustek et al., 2000), it can be expected that its cellular levels are tightly regulated through an interplay between its production and conversion. However, virtually nothing is known about the factors regulating its homeostasis in plants and the interaction of enzymes involved in internal sulfite detoxification. In this work, extended dark stress was employed as a means to induce catabolic pathways (Keskitalo et al., 2005; Brychkova et al., 2007) that stimulate the turnover of S-containing metabolites and result in sulfite flux. We analyzed known components of the sulfite homeostasis network in tomato (Solanum lycopersicum) plants and demonstrated the essential role of SO. During dark stress, SO expression was enhanced while the expression of the other known sulfite network components, APRs, SiR, SQD1, and STs, was inhibited. In the absence of the SO activity, toxic sulfite levels accumulated in the dark and was accompanied by increased leaf damage and plant mortality. Direct sulfite application stimulated components of the sulfite network, indicating that sulfite might play an important role as a signal molecule in orchestrating sulfite homeostasis.
RESULTS
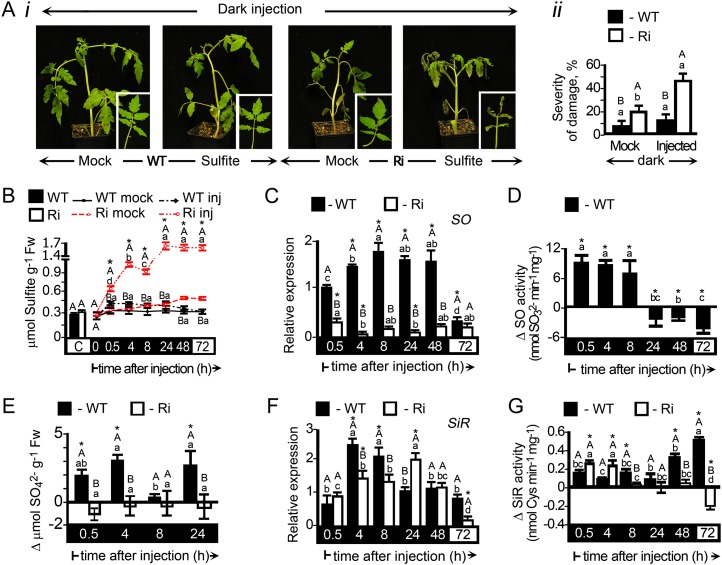
Sulfite Accumulation in Wild-Type and SO RNA Interference Plants during Extended Dark Stress
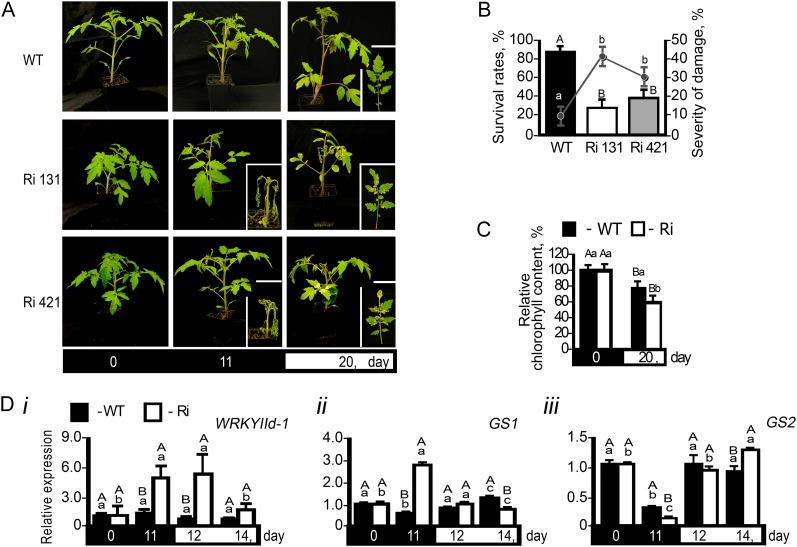
When grown under normal growth conditions, tomato and Arabidopsis (Arabidopsis thaliana) SO RNA interference (SO Ri) plants are indistinguishable from wild-type plants (Brychkova et al., 2007). However, when subjected to above-ambient concentrations of SO2, SO Ri plants showed stress symptoms that include the enhancement of senescence-associated transcripts and chlorophyll degradation (Brychkova et al., 2007). To further elucidate the role of SO in relation to other known genes that participate in sulfite turnover, we exposed tomato wild-type and SO Ri plants to extended dark (Fig. 2). Under this stress, a catabolic state is induced in the plant that results in premature senescence (Hörtensteiner and Feller, 2002; Guo and Crawford, 2005; Pruzinská et al., 2005; Brychkova et al., 2008a, 2008b). In this case, wild-type plants showed 90% survival, whereas SO Ri plants showed evidence of leaf damage after 6 d in the dark and only between 30% to 38% survival (Fig. 2, A and B) and exhibited significantly lower relative chlorophyll content than wild-type plants (Fig. 2C). Gene expression markers for senescence, WRKY IId-1 (Yang et al., 2009) and cytosolic Gln synthetase (Kawakami and Watanabe, 1988; Kamachi et al., 1991; Pageau et al., 2006), showed rapid induction only in SO Ri lines (Fig. 2, Di and Dii, respectively). Significantly, chloroplastic Gln synthetase (Kamachi et al., 1991), as a negatively expressed marker for senescence, was repressed more in SO Ri than in wild-type plants (Fig. 2Diii).
Figure 2.
The effect of 11-d dark stress on mortality and senescence-associated symptoms in tomato wild-type (WT) and SO Ri (Ri) plants. The days indicated on the black background represent dark treatment, and those on the white background represent the 16-h-light/8-h-dark regime. A, Photographs of wild-type and SO Ri plants (Ri 131 and Ri 421) after an 11-d dark treatment and subsequent 9-d recovery (total of 20 d) in a normal light/dark cycle. Closeups of mutant plants (day 11 in the dark) and leaves (20 d after treatment initiation) are presented in the insets. B, Survival rates (bars; left y axis) and damage level (line; right y axis) of wild-type and SO Ri plants evaluated at the day 9 of recovery. Error bars indicate se (n = 16). C, Leaf chlorophyll content at the day 9 of recovery. Error bars indicate se (n = 6). D, Quantitative transcript expression analysis of the senescence marker genes WRKYIId-1 (SGN-U563810; i), cytosolic Gln synthetase (GS1; SGN-U577193; ii), and chloroplastic Gln synthetase (GS2; SGN-U578319; iii). The relative expression after normalization to TFIID (SGN-U329249) is calculated by comparison with that of day 0 (set as 1.0). Error bars indicate se (n = 6). Different uppercase letters indicate significant differences between wild-type and SO Ri plants (Student’s t test; JMP 8.0 software; http://www.jmp.com/), and lowercase letters indicate significant differences within the plant species in response to treatment (Tukey-Kramer HSD test; JMP 8.0 software). The data for SO Ri plants represent means for SO Ri 131 and SO Ri 421 mutants.
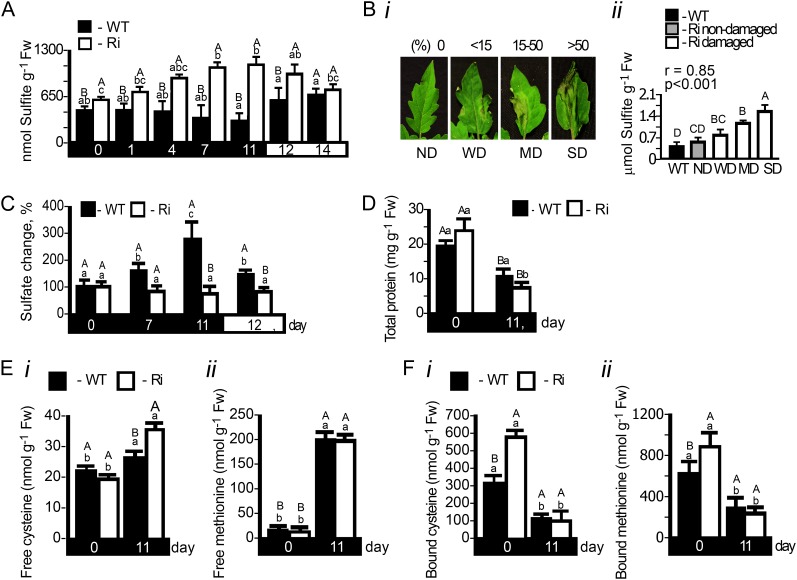
Sulfite accumulation could be a cause of the accelerated tissue damage in SO Ri plants. Indeed, under dark stress, up to a 2-fold enhancement of sulfite levels was detected in SO Ri leaves (from 0.62 to 1.2 µmol g−1 fresh weight; Fig 3A; Supplemental Table S1). The increase dissipated upon reexposure of the plants to a normal light/dark regime (Fig. 3A). In contrast, under dark conditions, a significant reduction of sulfite content was noted in wild-type leaves (from 0.52 to 0.33 µmol g−1 fresh weight; Fig 3A; Supplemental Table S1). When leaves were sampled in correlation with observed damage (days 6 and 7; Fig. 3Bi), a positive correlation between the damage and sulfite content was noted (Fig. 3Bii), further supporting the toxic role of sulfite. As sulfate is a direct product of SO activity, we examined its accumulation in wild-type and SO Ri plants. Sulfate was found to be significantly enhanced in the wild-type leaf during extended dark stress; however, its level was unchanged in the SO Ri mutants (Fig. 3C; Supplemental Table S1). The increase in sulfate could be a result of ongoing sulfate transport to the leaf or enhanced turnover of S-containing compounds in the leaves. To resolve this, the sulfate level in xylem exudate sap was determined from the stem just below the sampled leaves. In dark-stressed wild-type plants, it was more than 3-fold higher compared with the unstressed control, being more than twice higher than in SO Ri plant sap (Supplemental Table S2). These results indicate that sulfate increase in wild-type plants can originate from sulfate transport. In contrast to wild-type sap, where no dark-induced sulfite enhancement was observed, the sulfite level in the sap of SO Ri plants was enhanced. The results imply that in the SO Ri mutant, the majority of sulfite originated from sulfite transport. In normal plants, sulfite does not accumulate due to SO activity. In the absence of active SO in the SO Ri plants, sulfite, the by-product of S-containing metabolite turnover, was transported from below and accumulated, causing damage to the leaves (Supplemental Table S2).
Figure 3.
Sulfate, sulfite, total protein, and S-containing amino acid levels in tomato wild-type (WT) and SO Ri (Ri) plants as affected by extended dark stress. The days indicated on the black background represent dark treatment, and those on the white background represent the 16-h-light/8-h-dark regime. A, Sulfite accumulation. Error bars indicate se (n = 6). B, Damage level in the leaves after 6 d in the dark (i) and sulfite accumulation in the damaged area of the SO Ri leaves (ii). Nondamaged (ND; score 1), weakly damaged (WD; when damage level is less than 15%; score 2), medium damage (MD; when damage level is 15%–50%; score 3), and severely damaged (SD; when damage level is more than 50%; score 4) were defined. Error bars indicate se (n = 6). Pearson correlation analysis of the damage level in SO Ri mutants (score 1–4) versus sulfite was performed with R software (http://www.wessa.net/rwasp_correlation.wasp/). The r value is the Pearson product-moment correlation coefficient. Significance is estimated with two-sided P values. C, Sulfate accumulation in wild-type and SO Ri mutant leaves. Error bars indicate se (n = 8). D, Effect of the dark treatment on top leaf total protein content estimated by the Bradford assay. Error bars indicate se (n = 8). E and F, Free (E) and protein-bound (F) Cys (i) and Met (ii) in wild-type and SO Ri tomato plants at 0 and 11 d of dark treatment. Error bars indicate se (n = 8). Different uppercase letters indicate significant differences between wild-type and SO Ri plants (Student’s t test; JMP 8.0 software), and lowercase letters indicate significant difference within the plant species in response to treatment (Tukey-Kramer HSD test; JMP 8.0 software). All data for SO Ri plants represent means for SO Ri 131 and SO Ri 421 lines. Fw, Fresh weight.
The Levels of Free and Bound S-Containing Amino Acids in Dark Stress
The breakdown of S-containing amino acids and S-containing metabolites has been suggested as the source of endogenous sulfite appearance in mammals (Amy, 1988; Heber and Huve, 1998; Hänsch and Mendel, 2005). We explored this possibility by examining the changes in protein and S-containing amino acids during dark stress. The total protein level was found to be reduced by 24% and 63% in the dark in wild-type and SO Ri plants, respectively (Fig. 3D). Unexpectedly, in control conditions, total protein of SO Ri plants was 22% higher in comparison with the wild-type plants (Fig. 3D). While the amount of free Cys was slightly enhanced by treatment, the amount of free Met changed in wild-type or SO Ri lines by more than 13-fold (Fig. 3, Ei and Eii, respectively). The source of free Cys and Met can be attributed to the dark-induced protein degradation (Slavikova et al., 2008) detected in the tomato plants (Fig. 3D). In support of this notion, significant reductions of hydrolyzed TCA-precipitated protein-bound Cys (Fig. 3Fi) and Met (Fig. 3Fii) in both wild-type and SO Ri leaves in response to dark stress were detected. This decrease was much higher in mutant compared with wild-type leaf, being 47% versus 28% of the initial Met level and 36% versus 18% of the initial Cys level, respectively (Fig. 3, Fii and Fi, respectively). Importantly, inspection of the total S-containing amino acid content (the sum of protein-bound and free amino acids) in tomato leaves revealed a loss of 0.33 and 0.91 µmol g−1 fresh weight in wild-type and SO Ri plants, respectively (Supplemental Table S1), almost equally contributed by Cys and Met turnover (Fig. 3, E and F). These results indicate that (1) toxic endogenous sulfite accumulates in leaves during extended dark stress due to, at least in part, turnover of S-containing amino acid degradation (Fig. 3, E and F), as was shown for mammals (Amy, 1988; Heber and Huve, 1998; Hänsch and Mendel, 2005), and (2) under extended dark conditions, the presence of sulfite oxidation activity plays an essential role in sulfite detoxification by facilitating the conversion of the toxic sulfite to sulfate.
APR and SO in Dark-Stressed Wild-Type and SO Ri Plants
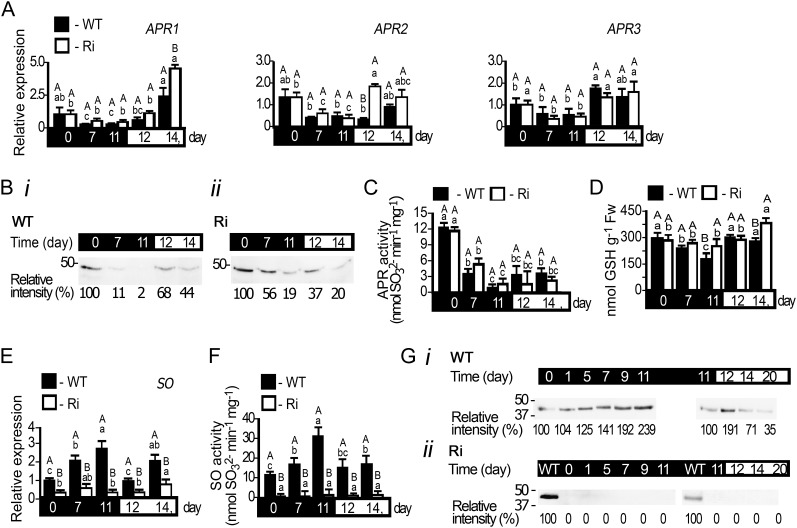
Sulfite accumulation could be a consequence of imbalance between sulfite de novo generation by APR (Kopriva et al., 2009) and its utilization by other members of the sulfite network. Inspection of APR expression revealed a reduction of all three APR transcripts in response to dark treatment and recovery of the transcript levels upon transferring the plants to light (Fig. 4A). Detection of tomato APR protein with an antibody (Brychkova et al., 2012b) revealed a decline of APR proteins with dark stress in both wild-type and SO Ri plants (Fig. 4B). Similar to APR proteins, inspection of APR activity employing APS as a substrate (Brychkova et al., 2012b) revealed that the sulfite-generating activities of the APRs in both wild-type and SO Ri plants decreased with time in the dark but were enhanced upon transfer to a normal light/dark regime thereafter (Fig. 4C). Interestingly, the SO Ri mutants displayed a tendency for higher APR activity compared with that detected in wild-type plants at days 7 and 11 in the dark, likely as a result of a higher amount of residual APR protein in SO Ri plants (Fig. 4, Bii versus Bi). Importantly, unlike the significant major decline in total Cys and Met in mutant leaves, no significant decline in GSH, the electron donor for the APR activity, was observed in SO Ri leaves compared with wild-type plants at the end of the dark stress (Fig. 4D; Supplemental Table S1).
Figure 4.
APR and SO expression in tomato wild-type (WT) and SO Ri (Ri) plants as affected by extended dark stress. The days indicated on the black background represent dark treatment, and those on the white background represent the 16-h-light/8-h-dark regime. A, Transcript analysis of APR1 (SGN-U580331), APR2 (SGN-U580235), and APR3 (SGN-U578339). The relative expression after normalization to TFIID (SGN-U329249) is calculated by comparison with that of the wild type at day 0 (set as 1.0). Error bars indicate se (n = 6). B, APR proteins extracted from leaves of wild-type (i) and SO Ri (ii) plants, fractionated by SDS-PAGE, and immunoblotted with APR-specific antiserum. Each lane contains 10 µg of soluble proteins. The data are from one of three independent experiments that yielded essentially identical results. C, Activity analysis of APR detected by sulfite appearance. Error bars indicate se (n = 6). D, Accumulation of GSH, the APR substrate, in response to dark stress. Fw, Fresh weight. E, Transcript analysis of SO (DQ853413) calculated as described in A. F, SO activity detected as sulfite disappearance. Error bars indicate se (n = 6). G, SO proteins extracted from leaves of wild-type (i) and SO Ri (ii) plants, fractionated, and immunoblotted as described in B employing SO-specific antiserum. Error bars indicate se (n = 8). Different uppercase letters indicate significant differences between wild-type and SO Ri plants (Student’s t test; JMP 8.0 software), and lowercase letters indicate significant differences within the plant species in response to treatment (Tukey-Kramer HSD test; JMP 8.0 software). All data for SO Ri plants represent means for SO Ri Ri 131 and SO Ri 421 lines. For both SO Ri lines, representative in gel activities and immunoblot analyses are presented.
As elevated sulfite in the dark was readily detected in the mutants but not in wild-type tissues, we investigated how the expression of SO responded to the dark stress. A highly significant 2.4- to 3-fold increase in the expression of wild-type SO transcript, protein, and activity was noticed (Fig. 4, E and Gi). The SO protein level and detectable activity tended to revert to lower levels when plants were returned to a normal light regime. As anticipated, SO Ri plants showed negligent amounts of SO transcript, protein, and activity (Fig. 4, E, F, and Gii).
These results indicate that (1) APR activity is reduced upon extended dark stress, although the residual amounts can still be a potential source for sulfite accumulation in SO Ri plants, and (2) SO expression enhancement in wild-type leaves paralleled the increase seen in sulfite generation in response to dark stress in SO Ri leaves, indicating that SO is essential for normal sulfite homeostasis during dark stress.
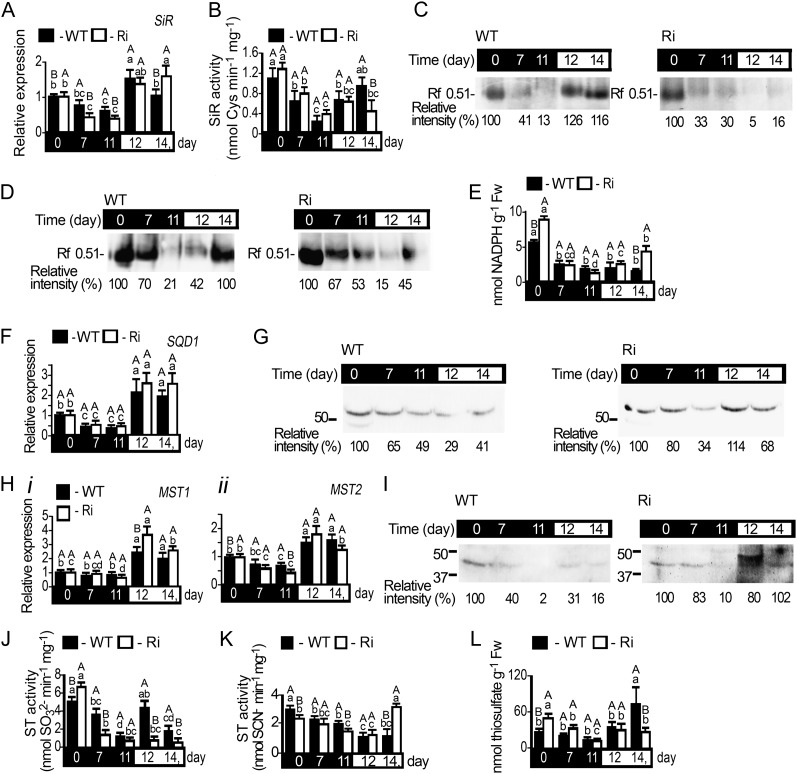
SiR Expression in Dark-Stressed Wild-Type and SO Ri Plants
SiR catalyzes the reduction of sulfite to sulfide and is likely the main route for sulfite utilization in the light (Khan et al., 2010). The relative expression of SiR transcript was found to be significantly reduced during dark stress but recovered upon transfer of the plants to a normal day/night regime in both wild-type and SO Ri plants (Fig. 5A). In order to examine SiR activity, two different activity assays were employed, a coupled O-acetyl-l-serine thiol lyase (OAS-TL)-dependent measure of SiR activity, in which the SiR activity product is detected as the final Cys product, and a direct sulfide-detecting in gel SiR activity assay (Brychkova et al., 2012c). Both the coupled SiR assay and the direct in gel assay exhibited sharp, approximately 3-fold reductions in SiR activity in wild-type and SO Ri plants. However, the activity was only partially recovered upon return to light in the SO Ri lines (Fig. 5, B and C). The reduction of in vitro SiR activity correlated with the reduced amount of detectable SiR protein (Fig. 5, compare B and C with D). As NADPH is an essential substrate for SiR activity in situ (Yonekura-Sakakibara et al., 2000; Brychkova et al., 2012c), we examined its level. As shown in Figure 5E, the NADPH level also decreased with time in the dark and recovered upon transition to the normal light/dark regime. Interestingly, upon a normal day/night regime, SiR transcript and activity, as well as NADPH, were slightly elevated in SO Ri mutants as compared with the wild-type plants (Fig. 5, A, B, and E), likely reflecting a higher need to detoxify sulfite in the SO Ri plants. The reduction in SiR protein and NADPH levels in response to dark are consistent with a dark stress-dependent decrease of SiR.
Figure 5.
SiR, SQD1, and MST1 and MST2 expression in tomato wild-type (WT) and SO Ri (Ri) plants as affected by dark stress. The days indicated on the black background represent dark treatment, and those on the white background represent the 16-h-light/8-h-dark regime. A, SiR transcript (SGN-U214723) relative expression. B and C, Kinetic activity (B) and SiR in gel assay (C) of proteins extracted from wild-type (left) and SO Ri (right) leaves. D, Immunoblot analysis of SiR protein. Protein extracted from wild-type (left) and SO Ri (right) leaves were fractionated by native PAGE and immunoblotted with SiR-specific antiserum. Each lane contains 10 µg of soluble proteins. The data are from one of three independent experiments that yielded essentially identical results. E, The level of NADPH, the reductant for SiR activity, as affected by the dark stress. Error bars indicate se (n = 6). Fw, Fresh weight. F and G, SQD1 transcript (SGN-U217001) relative expression (F) and protein immunoblot analysis (G) as described in D after protein fractionation with SDS-PAGE and immunoblotting with SQD1-specific antiserum. H, MST1 (FJ711706) and MST2 (FJ711707) relative expression analysis. All the relative expression values were calculated by comparison with that of day 0 (set as 1.0) after normalization to TFIID (SGN-U329249). Error bars indicate se (n = 6). I, MST1 protein immunoblot analysis as described in G employing MST1-specific antiserum. J, Sulfite-consuming activity of STs assayed as sulfite disappearance. K, Sulfite-producing activity of STs assayed as SCN− appearance. Error bars indicate se (n = 6). L, Thiosulfate levels, a product of sulfite-consuming activity and a substrate of sulfite-generating activity of STs, as affected by dark stress. Error bars indicate se (n = 6). Different uppercase letters indicate significant differences between wild-type and SO Ri plants (Student’s t test; JMP 8.0 software), and lowercase letters indicate significant differences within the plant species in response to treatment (Tukey-Kramer HSD test; JMP 8.0 software). All data for SO Ri plants represent means for SO Ri 131 and SO Ri 421 lines. For both SO Ri lines, representative in gel activities and immunoblot analyses are presented.
SQD1 and MSTs in Dark-Stressed Wild-Type and SO Ri Plants
Additional members of the sulfite network, SQD1, MST1, and MST2 (Supplemental Table S3), were examined for their possible contributions to sulfite regulation. The expression of chloroplast-localized SQD1 transcript decreased during dark stress and recovered upon return to light in both wild-type and SO Ri plants (Fig. 5F). Arabidopsis SQD1 is 79% identical to that of tomato, and antibodies raised against recombinant Arabidopsis SQD1 detect polypeptides of identical gel mobility (Shimojima et al., 2005; Fig. 5G). As shown in Figure 5G, the SQD1 protein decreased with dark stress, resulting in a decline in sulfolipid (SQDG) being more reduced in SO Ri than in wild-type leaves (Supplemental Table S1). The degradation of sulfolipids is an essential step in chloroplast degradation (Shimojima, 2011).
While the transcript level of MST2 was down-regulated in wild-type and SO Ri plants (Fig. 5Hii), MST1 was not affected much by dark stress (Fig. 5Hi). However, both MST transcripts were enhanced upon plant transfer to a normal light regime (Fig. 5H). Tomato STs, MST1 and MST2, exhibited strong similarity to the Arabidopsis MST1 (91% and 86% of the amino acids were identical or strongly similar to tomato MST1 and MST2 on a stretch of 253 and 329 amino acids, respectively; Supplemental Table S3). Antibody raised against the Arabidopsis MST1 protein detected tomato proteins that exhibited identical gel mobility to that shown for Arabidopsis MST1 (Papenbrock and Schmidt, 2000a) and revealed a decrease in immunoreactive polypeptide with dark stress in both wild-type and SO Ri plants (Fig. 5I).
Due to the energetically favorable equilibrium, ST activity can be measured either as sulfite generation or consumption. The detection in desalted crude protein extracts represents the sum of the activities of all the ST group members (Nakamura et al., 2000; Papenbrock and Schmidt, 2000a; Tsakraklides et al., 2002; Papenbrock et al., 2011). Sulfite-consuming activity was detected as sulfite disappearance (Tsakraklides et al., 2002) in the presence of thiocyanate (SCN−; Fig. 5J) or as SCN– disappearance (Supplemental Fig. S1A; Nakamura et al., 2000; Papenbrock and Schmidt, 2000a). To abrogate interfering SO activity, total ST activities were detected by inhibiting SO with tungstate (Brychkova et al., 2012b; Xiong et al., 2012). The reciprocal ST activity (i.e. sulfite generation) was detected indirectly as SCN– enhancement in the presence of thiosulfate and cyanide (Fig. 5K; Nakamura et al., 2000; Papenbrock and Schmidt, 2000a; Tsakraklides et al., 2002). Both ST activities (Fig. 5, J and K) as well as thiosulfate (Fig. 5L; Supplemental Fig. S1B) showed significant decreases with time in dark stress. However, upon transfer to the light, only the wild type exhibited recovered sulfite-consuming activity of STs, while sulfite-generating activity of STs was enhanced only in SO Ri plants (Fig. 5, J and K). The decreases in ST activity and protein content but not transcript level are an indication of posttranslational processes. Markedly, the potential sulfite-consuming activity of ST in wild-type plants grown under normal growth conditions as measured in vitro were about 30% to 50% the values measured for SO in vitro activity. If this is indicative of in vivo potential, it indicates a significant role for ST activity in sulfite homeostasis. The higher thiosulfate level (Fig. 5L) and ST sulfite-consuming activity (Fig. 5J) in SO Ri mutant plants in comparison with the wild type in control unstressed conditions may point to a potential role of STs in sulfite detoxification in the absence of SO activity. Due to the continuous reduction of ST activity with time in the dark, the role of ST in sulfite turnover under dark stress conditions is likely negligible. However, the increase in ST sulfite-consuming activity and enhancement of thiosulfate, the expected product of such activity, in wild-type plants after the transfer to light support the notion of ST’s potential role in sulfite homeostasis.
Sulfite Homeostasis in Normally Grown and Dark-Grown Sulfite-Injected Plants
The correlation between sulfite accumulation and leaf damage (Fig. 3Bii) indicates that the accumulated sulfite is responsible for the damaged tissue. We examined the effect of direct sulfite infiltration to the leaves by injection (Wu et al., 2011; Brychkova et al., 2012a). The three lowest leaves of 6-week-old plants were injected, and the third leaves from the bottom were used for damage evaluation and sulfite and sulfate determination. No or little damage was noted in mock or 0.5 to 5 mm sulfite-injected wild-type and SO Ri plants grown in normal light/dark conditions (Supplemental Figs. S2A and S3A). At higher concentrations (5–7.5 mm), damage severity was significant, reaching 10% and 30% in wild type and SO Ri leaves, respectively (Supplemental Fig. S2A). In contrast, SO Ri plants exposed to dark for 96 h, injected with 0.05 to 1 mm sulfite, and then left in the dark for an additional 2 d showed heightened leaf damage severity of more than 40% when injected with low levels of sulfite (0.2 mm). No damage appeared in sulfite-injected wild-type leaves exposed to similar treatment (Fig. 6A; Supplemental Fig. S2B).
Figure 6.
Damage evaluation, sulfite and sulfate levels, and expression analysis of SO and SiR in dark-stressed wild-type (WT) and SO Ri (Ri) plants as affected by 0.5 mm sulfite injection. The hours (h) indicated on the black background mean time in the dark, and those on the white background mean the 16-h-light/8-h-dark regime. A, The appearance of 6-week-old plants 72 h after sulfite or water (mock) was injected into leaves of 5-d dark-stressed plants. After injection, plants were left for an additional 48 h in the dark and then transferred to light for 24 h (i). Damaged leaf area quantification is presented as severity of damage (ii). B, Time course of sulfite level in sulfite-injected (inj) versus water-injected (mock) leaves. Error bars indicate se. C, SO transcript relative expression of wild-type and SO Ri leaf extracts. D, SO activity assayed as sulfite disappearance and presented as the difference between the activities in sulfite- and mock-injected leaves. The starting SO activities were 22.3 ± 1.35 and 2.9 ± 1.24 nmol min−1 mg−1 for the wild type and SO Ri, respectively. Error bars indicate se (n = 6). E, Sulfate levels presented as the differences between their levels in sulfite- and mock-injected leaves at each time point. Sulfate levels just before the injections were 5.92 ± 0.34 and 5.57 ± 0.53 µmol g−1 fresh weight (Fw) for wild-type and SO Ri leaves, respectively. F, SiR transcript relative expression of wild-type and SO Ri leaf extracts. G, SiR activity assayed in the coupled SiR-O-acetyl-l-serine reaction by determining Cys generation and presented as the difference between the activities in sulfite- and mock-injected leaves. The initial SiR activities before the injections were 0.15 ± 0.08 and 0.15 ± 0.11 nmol min−1 mg−1 for the wild type and SO Ri, respectively. Error bars indicate se (n = 6). Different uppercase letters indicate significant differences between wild-type and SO Ri plants (Student’s t test; JMP 8.0 software), and lowercase letters indicate significant differences within the plant species in response to treatment (Tukey-Kramer HSD test; JMP 8.0 software). Asterisks indicate significant differences between sulfite-injected and the corresponding mock-injected parameter detected. All data for SO Ri plants represent means for SO Ri 131 and SO Ri 421 lines.
For quantitative analysis, a known amount of sulfite was injected (33 µL of 0.5 mm solution) to result in an additional 0.1 µmol sulfite g−1 fresh weight. As expected, sulfite injected to dark-grown wild-type leaves resulted in an immediate sulfite enhancement to 0.4 µmol sulfite g−1 fresh weight. However, in dark-grown SO Ri leaves, similar treatment resulted in a 0.6 µmol sulfite g−1 fresh weight increase and rapidly rose to 1.6 µmol sulfite g−1 fresh weight (Fig. 6B). When similar injections were carried out in the light, much less sulfite accumulated in the SO Ri lines (Supplemental Fig. S3B). The discrepancy between SO Ri and wild-type plants and between light and dark in the SO Ri mutants can be explained by the uncoupling of sulfite production and utilization. In the absence of active SO, the residual sulfite network system was unable to cope with the injected sulfite, resulting in a cascade effect of sulfite toxicity that prevents its efficient scavenging.
Regulation of the Sulfite Network by Sulfite
To further examine the regulation of the sulfite network in response to sulfite injections, the responses of the sulfite network components were monitored. An increase in wild-type SO transcript was noted at 0.5 h (light) or 4 h (dark) after the injection and lasted 8 h in the light (Supplemental Fig. S3C) or 48 h in dark-grown plants (Fig. 6C). SO activity was 10.8 ± 0.26 nmol min−1 mg−1 in the light-grown wild type and 22.3 ± 1.35 nmol min−1 mg−1 in the dark-pretreated wild type. However, significant and rapid SO activity induction after sulfite injection was evident 0.5 h after the injection and lasted 8 h (Fig. 6D). The induction of SO activity by sulfite was higher in the dark than in light-grown plants (compare Fig. 6D with Supplemental Fig. S3D). This result demonstrates the ability of sulfite to induce SO, previously described as a constitutive enzyme (Hänsch et al., 2006; Brychkova et al., 2007; Lang et al., 2007).
Sulfate level in sulfite-injected plants compared with mock-treated controls was increased in wild-type plants but not in SO Ri plants due to the reduced SO activity (Fig. 6E; Supplemental Fig. S3E). Surprisingly, the expected increase in sulfate in wild-type leaves, especially in the dark-stressed plants, was above the amount of sulfite injected into the sampled leaves. Inspection of the xylem exudate sap, sampled from the stem below the third injected leaf 0.5 h after sulfite injection, revealed that sulfate level in dark-stressed plants was significantly increased. In contrast, sulfite level was significantly increased in plants grown under normal conditions and in dark-stressed plants (Supplemental Table S4). These results indicate that, in addition to direct oxidation of the injected sulfite by SO, sulfate accumulated due to transport from the lower parts of sulfite-injected, dark-stressed wild-type plants. Additionally, the oxidation of the transported sulfite by SO in the sampled leaf could be another significant source for sulfate enhancement (Fig. 6E; Supplemental Table S4). Thus, the presence of active SO is an important component for fast conversion of the toxic sulfite to the nontoxic sulfate in the sulfite-injected plants and for the transportation of the resultant excess sulfate to the younger leaves.
The level of SiR expression as a result of sulfite application was examined next. SiR is down-regulated by dark pretreatment itself from 0.8 ± 0.09 (wild type) and 1.09 ± 0.08 nmol min−1 mg−1 (SO Ri) in the light to 0.15 ± 0.08 (wild type) and 0.15 ± 0.11 nmol min−1 mg−1 (SO Ri) in the dark. However, the injection of sulfite rapidly induced (after 0.5 h) SiR activity in both dark- and light-grown plants (Fig. 6, F and G; Supplemental Fig. S3, F and G), where in both wild-type and SO Ri plants the highest activity levels were obtained 4 h after sulfite injection. As shown here, the induction of SiR by sulfite is in contrast to the description that it is a semiconstitutive enzyme (Leustek, 2002; Kopriva, 2006; Khan et al., 2010).
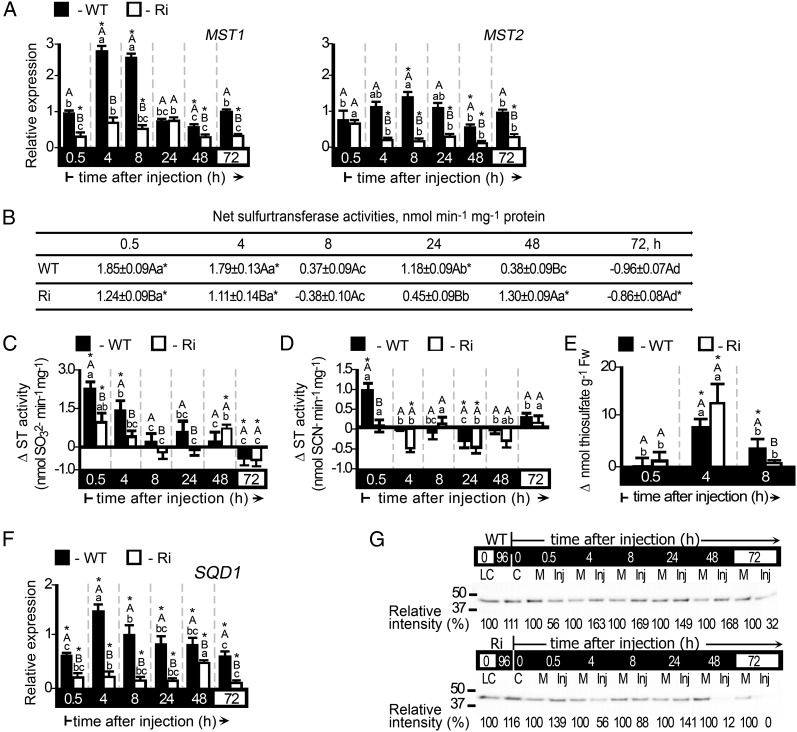
The tomato ST is a large 18-member multigene family (Supplemental Table S3) with likely varying affinities for their sulfite or thiosulfate products, such that they specialize in the catalysis of either sulfite consumption or production. Transcripts of the STs MST1 and MST2 were generally enhanced in dark-stressed plants as well as in normally grown plants in response to sulfite injection (Fig. 7A; Supplemental Fig. S4A). Net ST activity is defined as the difference between sulfite-consuming activity and sulfite-producing activity. Upon application of sulfite, the sulfite-consuming activity was enhanced, while the sulfite-producing activity of STs was either generally not affected or was down-regulated (Fig. 7, B–D; Supplemental Fig. S4, B–D). As the net ST activity is more active in the light (1.38 ± 0.14 and 4.03 ± 0.17 nmol min−1 mg−1 in wild-type and SO Ri plants, respectively) and negligible in the dark-pretreated plants (−0.13 ± 0.15 and −0.24 ± 0.08 nmol min−1 mg−1 in the wild type and SO Ri, respectively), a more drastic enhancement than in dark-stressed plants was evident after sulfite injection (compare Fig. 7B with Supplemental Fig. S4B). The sulfite-consuming ST activities would generate thiosulfate; indeed, elevated levels of thiosulfate were detected in sulfite-injected plants, being higher in the light-grown plants (Fig. 7E; Supplemental Fig. S4E).
Figure 7.
Expression analysis of the STs and SQD1 and thiosulfate levels in dark-stressed wild-type (WT) and SO Ri (Ri) plants as affected by 0.5 mm sulfite injection. The hours (h) indicated on the black background mean time in the dark, and those on the white background mean the 16-h-light/8-h-dark regime. A, MST1 (left) and MST2 (right) transcript relative expression. B, Net ST activities defined as the difference between sulfite-consuming activity and sulfite-producing activity of STs (as presented in C and D, respectively). The initial net ST activities before the injections were −0.13 ± 0.15 and −0.24 ± 0.08 nmol min−1 mg−1 for the wild type and SO Ri, respectively. Error bars indicate se (n = 6). C, Sulfite-consuming activity of STs assayed as SO32− disappearance (presented as the difference between the activities in sulfite- and mock-injected leaves). The initial activities before the injections were 0.92 ± 0.47 and 0.63 ± 0.24 nmol min−1 mg−1 for the wild type and SO Ri, respectively. Error bars indicate se (n = 6). D, Sulfite-producing activity of STs expressed as SCN− appearance in the presence of thiosulfate and cyanide. The initial activities were 1.05 ± 0.13 and 0.87 ± 0.06 nmol min−1 mg−1 for the wild type and SO Ri, respectively. Error bars indicate se (n = 6). E, Thiosulfate levels presented as the differences between their levels in sulfite- and mock-injected leaves at each time point. Thiosulfate levels in control plants were 20.7 ± 1.27 and 22.1 ± 8.25 nmol g−1 fresh weight (Fw) for wild-type and SO Ri plants, respectively. Error bars indicate se (n = 6). F, SQD1 transcript relative expression analysis. All relative expression levels after normalization to TFIID (SGN-U329249) are presented relative to the normalized expression in mock-injected leaves at each time point. Error bars indicate se (n = 3). G, SQD1 protein immunoblot analysis of wild-type (top) and SO Ri (bottom) leaf extracts. Proteins were fractionated by SDS-PAGE and immunoblotted with SQD1-specific antiserum. Each lane contains 10 µg of soluble proteins. LC, Light control; C, 96-h dark-treated control; M, mock injected; Inj, sulfite injected. The data are from one of three independent experiments that yielded essentially identical results. Different uppercase letters indicate significant differences between wild-type and SO Ri plants (Student’s t test; JMP 8.0 software), and lowercase letters indicate significant differences within the plant species in response to treatment (Tukey-Kramer HSD test; JMP 8.0 software). Asterisks indicate significant differences between sulfite-injected and the corresponding mock-injected parameter detected. All data for SO Ri plants represent means for SO Ri 131 and SO Ri 421 lines. For both SO Ri lines, representative in gel activities and immunoblot analyses are presented.
SQD1 transcript and protein levels were also significantly up-regulated in response to sulfite injection into leaves of dark-stressed wild-type plants or into wild-type and SO Ri mutant leaves grown under a natural light regime (Fig. 7, F and G; Supplemental Fig. S4, F and G). The results indicate that, in addition to SO, the other sulfite network members that assimilate sulfite, SiR, SQD1, and the STs, may have a role in sulfite homeostasis.
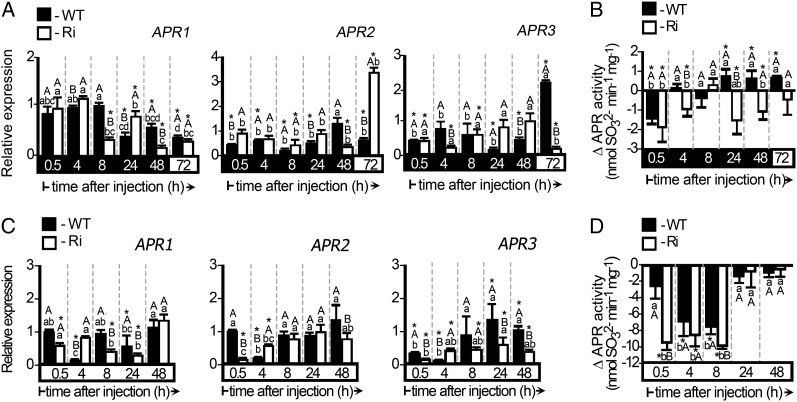
APRs generate sulfite; inspection of their transcript levels after sulfite injection revealed rapid reduction in APR2 and APR3 in both wild-type and SO Ri dark-stressed plants in the light and the dark (Fig. 8, A and C). In addition, APR activity was reduced, especially during the first 0.5 h as a result of sulfite injection (Fig. 8, B and D). As APR activity is higher in the light (14.10 ± 1.15 nmol min−1 mg−1 for the wild type and 13.36 ± 1.07 nmol min−1 mg−1 for SO Ri) compared with the dark (4.89 ± 0.32 nmol min−1 mg−1 for the wild type and 6.86 ± 0.53 nmol min−1 mg−1 for SO Ri), a more drastic reduction than in dark-stressed plants was evident after sulfite injection (Fig. 8, compare B and D). These results suggest that sulfite acts as a strong negative regulator of APRs, being able to down-regulate the APR activities not only in normally grown plants but also when these activities are already down-regulated in dark-pretreated plants.
Figure 8.
Expression and activity analysis of tomato APRs in dark-stressed and unstressed wild-type (WT) and SO Ri (Ri) plants as affected by 0.5 mm sulfite injection. The hours (h) indicated on the black background mean time in the dark, and those on the white background mean the 16-h-light/8-h-dark regime. A, Relative expression analysis of APR1, APR2, and APR3 in dark-stressed plants injected with sulfite. B, APR activity in sulfite-injected leaves of plants exposed to the dark. The results are presented as the differences between the activities in response to sulfite- and mock-injected leaves. The initial APR activities before dark injection were 4.89 ± 0.32 and 6.86 ± 0.53 nmol min−1 mg−1 for the wild type and SO Ri, respectively. Error bars indicate se (n = 6). C, Relative expression analysis of APR1, APR2, and APR3 in leaves of plants grown under normal conditions and then injected with sulfite. All the relative expression levels after normalization to TFIID (SGN-U329249) are presented relative to the normalized expression in mock-injected leaves at each time point. Error bars indicate se (n = 3). D, APR activity in sulfite-injected leaves of plants grown under normal conditions. The results are presented as in B. The initial APR activities in leaves of plants grown under normal growth conditions before the injections were 14.10 ± 1.15 and 13.36 ± 1.07 nmol min−1 mg−1 for the wild type and SO Ri, respectively. Error bars indicate se (n = 6). Different uppercase letters indicate significant differences between wild-type and SO Ri plants (Student’s t test; JMP 8.0 software), and lowercase letters indicate significant differences within the plant species in response to treatment (Tukey-Kramer HSD test; JMP 8.0 software). Asterisks indicate significant differences between sulfite-injected and the corresponding mock-injected parameter detected. All data for SO Ri plants represent means for SO Ri 131 and SO Ri 421 lines.
Although the existence of modulation events of transcriptional regulatory element(s) was not presented in this study, either because they may not exist or will be uncovered, our data indicate that sulfite may act as an orchestrating signal molecule. Here, we demonstrate, to our knowledge for the first time, the rapid sulfite-dependent induction of the sulfite network components SiR, SQD1, STs, SO, and APRs at both the transcript and activity levels. The application of sulfite to light- and dark-pretreated plants demonstrated that, in order to homeostate sulfite levels, sulfite down-regulates APR, even when already being down-regulated by the dark, up-regulates SiR, STs, and SQD1, even when already being down-regulated by the dark, and up-regulates SO, even when already being up-regulated by the dark pretreatment.
DISCUSSION
A Central Role for SO within the Sulfite Network during Extended Dark Stress
Here, we show that the sulfite network is responsive to conditions of dark stress and the presence of sulfite. The increase in sulfate levels in wild-type plants in response to sulfite injection and to the dark stress (Figs. 3C and 6E; Supplemental Fig. S3E) is consistent with a requirement for SO activity (Figs. 4F and 6D; Supplemental Fig. S3D). Thus, SO is instrumental as a detoxifying enzyme maintaining safe sulfite levels to ensure plant vitality under conditions of metabolite remobilization. This is evident from the normal phenotype of the SO Ri mutant plants, where, in the absence of active SO, additional members of the sulfite network enzymes can supplement the scavenging activity of SO under normal light conditions (Supplemental Figs. S3, F and G, and S4). However, under conditions of extended dark or application of sulfite in dark-stressed plants, where complementary sulfite network elements are repressed or when the sulfite level exceeds their assimilation capacity, the essential detoxification role of SO activity is revealed and tissue damage becomes evident (Fig. 6, A and B).
Under normal growth conditions, the other major sink for sulfite, SiR, channels it into the assimilatory reduction pathway (Leustek, 2002; Kopriva, 2006; Khan et al., 2010). However, as shown here, assimilation has a strict light requirement that is decoupled upon transfer to dark (Fig. 5, A–D). Partial absence of normal SiR activity in Arabidopsis SiR T-DNA insertion mutants resulted in a moderately higher sulfite level in mutant leaves (Khan et al., 2010). Under more extreme sulfite accumulation, such as obtained by overexpression of PaAPR in Arabidopsis and maize (Zea mays), the endogenous SiR activity and the other sulfite homeostasis components were insufficient to cope with the accumulating sulfite (Tsakraklides et al., 2002; Martin et al., 2005). Dark stress as used here accentuates the need for SO under conditions of SiR, SQD1, and ST inactivity and delineates the role of SO in maintaining sulfite levels under normal physiological conditions.
The critical point for sulfite damage during dark stress or injection of sulfite occurred when sulfite was about 3-fold higher than in normal levels (Fig. 3, A and B). Interestingly, in maize, 3-fold sulfite enhancement due to PaAPR overexpression (Martin et al., 2005) resulted in aberrant, but nonnecrotic, phenotypes, indicating differences in species-specific thresholds for sulfite. Sulfite toxicity appears to have a domino effect and rapidly damages the very agents that could contribute to its dissipation. Thus, after sulfite injection of the dark-treated plants, the sulfite network members are repressed, exacerbating damage, whereas in the light, the capacity of SiR and STs is sufficient to prevent sulfite accumulation (compare Fig. 6, A and B, with Supplemental Fig. S3, A and B). Thus, application of sulfite by injection revealed further unique aspects of the sulfite network (i.e. the role of sulfite in regulation of the sulfite network).
Subcellular Localizations of the Sulfite Network and Sulfite Homeostasis
Peroxisomal localization is ideal for the role of SO in protecting plants against sulfite toxicity. Their close vicinity to the chloroplasts (Oikawa et al., 2003) and mitochondria (Islam and Takagi, 2010) would enable the generation of a sulfite gradient facilitated by SO-dependent sulfite oxidation (Hansch and Mendel, 2008). Indeed, application by fumigation of toxic SO2 levels to normally grown Arabidopsis and tomato mutants, overexpressing or lacking SO activity, demonstrated the essential role of SO in protecting plant cell components from toxic sulfite (Brychkova et al., 2007; Lang et al., 2007; Randewig et al., 2012). By employing prolonged dark stress to induce the production of endogenous sulfite under conditions in which other sulfite network components are down-regulated, we further demonstrated that peroxisomal SO serves as the major conduit of sulfite detoxification (Fig. 2, B and C). Yet, under normal growth conditions, the activities of the other sulfite network components are sufficient to protect SO Ri plants from relatively high doses (5 mm) of toxic sulfite in the absence of SO activity (Supplemental Fig. S2A). In the chloroplast, protection against toxic levels of sulfite is afforded by the activity of the chloroplast-localized enzymes, SiR and SQD1, that have low Km for sulfite (10 μm) and are sufficiently active (Sanda et al., 2001; Khan et al., 2010). Indeed, the expression of SiR and SQD1 was rapidly up-regulated in response to sulfite application (Supplemental Figs. S3, F and G, and S4, F and G). Additionally, a possible support of potential chloroplast-localized members of the ST gene family, as shown in Arabidopsis (Papenbrock et al., 2011), should not be ignored. Thus, the battery of the chloroplast-localized sulfite-consuming enzymes could serve as a “bodyguard” to moderate sulfite generated by the chloroplast-localized APR (Kopriva, 2006). Sulfite is detoxified as well in the other cellular compartments by the cytosolic and mitochondrial localized STs (Papenbrock et al., 2011). Importantly, the in vitro sulfite-consuming activity of STs in tomato plants grown under normal growth conditions is shown here to be approximately 50% of SO activity, indicating the potential role of these enzymes, localized to all plant cell compartments, in sulfite homeostasis.
An S Balance Sheet Reveals a Critical Role of Sulfite
Supplemental Table S1 shows the distribution of S-containing compounds in the plant at the beginning and end of the dark treatment. The accounting of sulfite levels and potential pools of sulfite sources reveals that protein (designated as total S-amino acids) breakdown is largest among the identified components of dark-induced changes in S pools (Fig. 3, D–F; Supplemental Table S1). Due to the dependence of APR activity on light (Kopriva et al., 1999), the major source of dark-induced accumulation of sulfite can only be from the remobilization of internal sources of previously reduced S. These include thiol compounds, amino acids, and sulfated membrane components (Takahashi et al., 2011). SQDG has been hypothesized to act as an S storage lipid in cells (Shimojima, 2011), and its degradation in the dark would provide abundant energy for cellular processes but also release sulfite (Supplemental Table S1). As shown in Supplemental Table S1, it can be a significant source of sulfite, second to amino acid sources.
Remarkably, sulfate continues to accumulate during the dark in wild-type leaves but not in the SO Ri leaves, to an amount that is approximately 2-fold (or approximately 4.61 µmol g−1 fresh weight) higher than in control unstressed plants (Supplemental Table S1). However, when total S is measured, it continues to grow equally in both wild-type and SO Ri leaves by 8.4 and 8.18 µmol g−1 fresh weight, respectively. Thus, SO has a central function in determining the distribution of S type within the leaf but not on total leaf S uptake. This notion is further supported by the 3-fold increase in sulfate level in the xylem sap of dark-stressed wild-type plants (Supplemental Table S2). The absence of any influence of dark stress on sulfite level in the xylem sap (Supplemental Table S2) indicates that the sulfite by-product is rapidly converted into sulfate and likely to so-called “other S compounds” in wild-type leaves (compare other S compounds at 0 and 11 d in the dark in Supplemental Table S1). In contrast, sulfite increase in the xylem sap of the dark-stressed SO Ri mutant indicates that the sulfite by-product was transported from outside the leaf (Supplemental Table S2). The measured sulfite is likely an underestimate of true sulfite levels, as sulfate can also be formed by spontaneous sulfite oxidation (Hänsch et al., 2006; Brychkova et al., 2012a) and as strong nucleophile will react with other metabolites to form sulfate compounds, so-called other S compounds (King and Kaiser, 1974; Peiser et al., 1982; Hänsch and Mendel, 2005; Footitt et al., 2011). Such compounds are elevated after stress in the SO Ri mutant compared with the wild type (Supplemental Table S1). Thus, SO acts as a safety valve and converts sulfite to sulfate, resulting in less other S compounds.
Remarkably, SO Ri plants in the light contain more total S-amino acids (by 50%) but less other S compounds (Supplemental Table S1). Hence, the continuous presence of sulfite in the light increases the flux to S-containing amino acids (i.e. through SiR activity) in the SO Ri mutants. In the dark, when SiR is inactivated, the breakdown of the excess total S-amino acids in the SO Ri mutants presents a major source for the toxic sulfite (Supplemental Table S1).
Sulfite as a Regulator of S Metabolism: Sulfite Producers and Sulfite Users
The level of APR activity has been considered as the main control point in the sulfate assimilation pathway (Kopriva, 2006; Khan et al., 2010). In this work, we demonstrate its rapid down-regulation (0.5 h) after sulfite application to maintain sulfite homeostasis and avoid further toxic sulfite accumulation (Fig. 8, B and D). Hence, in addition to a role for APR in controlling the assimilation of sulfite, the level of the sulfite product is sensed to regulate both transcriptional and posttranscriptional levels of APR. We further demonstrated the regulation of SiR by sulfite, as its activity was rapidly up-regulated at the transcript and activity levels, reversing the down-regulation by the dark (Figs. 5, A–D, and 6, F and G; Supplemental Fig. S3, F and G). In addition to SiR, chloroplast-localized SQD1 exhibited sulfite-enhanced expression in response to sulfite (Fig. 7, F and G; Supplemental Fig. S4, F and G). However, the potential scavenging activities of SiR and SQD1 as measured by in vitro capacity (Sanda et al., 2001; Shimojima and Benning, 2003) is lower than SO (compare Figs. 4F and 5B as well as Fig. 6, D and G, and Supplemental Fig. S3, D and G), which explains the higher sensitivity of the SO Ri plants to sulfite (Supplemental Fig. S2). In summary, SO was demonstrated here to play an important role in sulfite consumption as a salvage housekeeping enzyme, and sulfite levels are used as a signal for rapid modification of the sulfite network.
A Potential Role for ST/Rhodanese Genes in Regulating Sulfite Levels
Rhodanese-like activity can transfer a thiol to a sulfite to generate thiosulfate in a reversible reaction. In Arabidopsis, 20 ST/rhodanese genes are localized to varied cellular positions (Papenbrock et al., 2011), and 18 members are identified in tomato (Supplemental Table S3). Their potential function, to form the less toxic thiosulfate from the toxic sulfite by STs, was previously hypothesized (Nakamura et al., 2000; Papenbrock and Schmidt, 2000b; Tsakraklides et al., 2002; Martin et al., 2005) and demonstrated using recombinant AtMST1 protein (Tsakraklides et al., 2002). However, a role of the ST members in sulfite homeostasis has not been established unambiguously. Impressively, a significant sulfite net consuming activity was demonstrated, being about 18.5% to 38% of the SO activity (compare Figs. 4F and 5, J and K). The level of thiosulfate increases in planta rapidly in response to sulfite injection (Fig. 7E; Supplemental Fig. S4E) and may represent rapid flux through ST-like activities. Conceivably, part of the generated ST products, like thiosulfate, may react with other cell components and contribute to the levels of the other S compounds. We noted differences between the levels of net sulfite-consuming activities by STs in both wild-type and mutant plants (Fig. 7B; Supplemental Fig. S4B). While these measurements are the crude sum of all the in vitro potential activities, they are likely to represent subtle changes in the populations of ST activities.
The application of extreme environmental insult to plants, such as extended darkness, has provided a system for following dramatic shifts in the redeployment of oxidized and reduced S. The turnover of S during the remobilization of cellular components is likely a necessary, albeit negative, by-product of the need for reuse of carbon skeletons as an energy source. The novel evidence provided by the synchronous dark-induced turnover of S-containing compounds augmented by exogenous sulfite applications underlines the role of SO and other sulfite network components. The network maintains sulfite homeostasis, providing a fitness component to normal physiological responses and optimizing diurnal or senescence programs in which the reutilization of existing cellular constituents takes place.
MATERIALS AND METHODS
Plant Materials, Growth Conditions, and Dark Treatment
Tomato plants (Solanum lycopersicum ‘Rheinlands Ruhm’), both wild type and SO Ri mutants (Brychkova et al., 2007), were grown in pots filled with a peat and vermiculite (4:1, v/v) mixture containing slow-release high-N Multicote 4 with microelements (0.3%, w/w; Haifa Chemicals; http://www.haifachem.com/) in a growth room under 16 h of light/8 h of dark, 22°C, 75% to 85% relative humidity, and 100 μE m−2 s−1 light, as described before (Brychkova et al., 2007, 2012a).
For dark treatment, 6-week-old tomato plants were transferred to a dark room. Samples were collected every day for 15 min, under dim light (40 μmol m−2 s−1), as a mixture of top leaves (fifth and six from the bottom) taken from five plants. After 11 d in the dark, plants were transferred back to the growth room, and the survival rate and severity of leaf damage were determined 9 d later. The average and se of the survival rate were calculated from 13 independent experiments, with at least 80 plants for each treatment.
Sulfite Injections
Infiltration of sulfite into plant leaves by injection was done as described recently (Wu et al., 2011; Brychkova et al., 2012a). The three lowest leaves of 6-week-old wild-type and SO Ri plants, normally grown or exposed to dark stress for 96 h, were injected either with buffered water (pH 5.7; mock) or 0.5 mm sulfite (pH 5.7) and left in the dark for an additional 2 d before transfer to a normal light/dark regime. The volume of the injected solutions was 33% ± 0.5% of leaf weight. The severity of damage was determined 120 h after the injection, as the mean ratio between the damaged area and the total area of the third leaves from the bottom, employing 20 leaves for each treatment and using ImageJ software (http://rsbweb.nih.gov/ij/) as described before (Brychkova et al., 2007, 2008a, 2012b). Samples for metabolite determination, gene expression, and enzyme activity analysis were collected from the third leaves from the bottom just before the injection and 0.5, 4, 8, 24, 48, and 72 h later. Plants subjected to dark stress were injected and sampled under dim light (40 μmol m−2 s−1) conditions.
Sap Exudate Collection, Protein and Chlorophyll Determination, and Quantification of Leaf Damage Levels
Exudate sap (approximately 200 μL) was collected from plants topped 1 cm below the fifth leaf. The entire root system of the tomato plants was washed with distilled water, blotted dry with filter paper, and transferred into an Arimad 2 pressure chamber (www.mrclab.com). Thereafter, the pressure was gradually increased to cause exudation. The sap (approximately 200 μL) was collected (Cramer and Lips, 1995; Hartung et al., 1998; Netting et al., 2012) under permanent pressure of 6.2 bar during 10 min and then frozen in liquid nitrogen and kept in −80°C before use for sulfate and sulfite determination. Similarly, in the case of sulfite- or water-injected plants, sap was collected from plants topped 1 cm below the third injected leaf at 0.5 h after injection. The protein content was determined according to the Bradford method using bovine serum albumin as a standard (Bradford, 1976). Total chlorophyll content was presented as remaining chlorophyll content (e.g. the ratio of chlorophyll content in a treated leaf disc [7 mm] to the untreated control leaf disc) expressed as percentage. The severity of leaf damage was estimated as a percentage of damage area to total area as described by us before (Brychkova et al., 2007, 2008a).
Preparation of RNA and Quantitative Real-Time Reverse Transcription PCR
For quantitative analysis of transcript expression, total RNA extraction, reverse transcription reaction, and quantitative real-time PCR employing specific primers (Supplemental Table S5) were performed as described before (Brychkova et al., 2007). Reactions normalized with ACTIN Tom41 (U60480), TIP1 (SGN-U321250), ELONGATION FACTOR1-α (SGN-U196120), and TFIID (SGN-U329249) as housekeeping genes (Expósito-Rodríguez et al., 2008) revealed similar results, allowing us to present results based on the TFIID.
Protein Extraction and Immunoblotting for SO, SiR, APR, SQD1, and MST1 Proteins
For immunoblot analysis of SO and SiR, proteins from leaf samples were extracted, fractionated, blotted, and subjected to immunodetection as described (Brychkova et al., 2007, 2012c). For APR, MST1, and SQD1 immunoblot detection, protein extracts were prepared under denaturing conditions (Zavgorodnyaya et al., 1997; Brychkova et al., 2012b), and aliquots of 10 μg of protein were subjected to 12.5% SDS-PAGE under denaturing conditions (Zavgorodnyaya et al., 1997; Kopriva et al., 1999; Brychkova et al., 2012b) followed by blotting to polyvinylidene difluoride membranes (Immun-Blot membranes; Bio-Rad; www.bio-rad.com/). Blotted proteins were subjected to immunodetection with specific antisera raised against recombinant APR2 as recently described for tomato (Brychkova et al., 2012b; applied in a 1:2,000 ratio; kindly supplied by Prof. Stanislav Kopriva), MST (1:1,000 ratio; kindly given by Prof. Jutta Papenbrock), or SQD1 (1:2,000 ratio; kindly gifted by Dr. Mie Shimojima), followed by 5,000-fold phosphate-buffered saline diluted secondary antibodies (anti-rabbit IgG; Sigma; http://www.sigmaaldrich.com). Protein bands were visualized by staining with the enhanced chemiluminescence SuperSignal Western Blotting System (Pierce; http://www.piercenet.com) and quantified by National Institutes of Health Image software (version 1.6).
Protein Extraction and Kinetic Assays for SO, APR, and SiR and in Gel SiR Activity
Activities for APR, SiR, and SO were extracted and assayed as described (Brychkova et al., 2012a, 2012b, 2012c). In brief, SO activity was measured as the disappearance of sulfite (Pachmayr, 1960; Lang et al., 2007; Khan et al., 2010; Teschner et al., 2010; Brychkova et al., 2012a). The desalted protein extracts were treated with 1 mm tungstate for 30 min at 4°C to inhibit SO activity (Brychkova et al., 2012a). APR activity employing APS as substrate was detected using the sulfite-specific fuchsin colorimetric detection method (Brychkova et al., 2012b). SiR activity was estimated by the coupled SiR/OAS-TL assay (Bosma et al., 1991; Khan et al., 2010) with the addition of NADPH and tungstic acid (Brychkova et al., 2012c). The resultant generated Cys was detected as described before (Gaitonde, 1967; Burandt et al., 2001; Ohtsu et al., 2010; Brychkova et al., 2012c). SiR in gel activity detection is based on the detection of sulfide, the direct product of SiR activity, reacting with lead acetate to yield lead sulfide bands (Brychkova et al., 2012c).
ST Kinetic Activity
ST activities were extracted as described previously (Nakamura et al., 2000; Papenbrock and Schmidt, 2000a; Tsakraklides et al., 2002). In brief, sulfite-generating sulfurtransferase activity was determined by colorimetric detection of SCN− formation at 460 nm as the red Fe(SCN)3 complex from cyanide and thiosulfate using acidic iron reagent (FeCl3, 50 g L−1; 65% HNO3, 200 mL L−1) as described before (Papenbrock and Schmidt, 2000a). Sulfite-consuming activity of the STs was determined as described before (Papenbrock and Schmidt, 2000a; Tsakraklides et al., 2002) with modifications. The reaction assay contained 0.1 m Tris-HCl buffer, pH 8.48, 0.1 mm Na2SO3, 5 mm β-mercaptoethanol, 50 µm NaSCN, and 80 µg mL−1 desalted protein extract treated before the assay with 1 mm tungstate for 30 min at 4°C to disrupt SO activity that consumes sulfite. The sulfite-consuming activity was measured during 15 min at 37°C and was estimated as sulfite disappearance, as described for SO activity (Pachmayr, 1960; Lang et al., 2007; Khan et al., 2010; Teschner et al., 2010; Brychkova et al., 2012a) and employing Na2SO3 as a standard in solution containing NaSCN. Sulfite-consuming activity was assayed also as SCN− disappearance by detecting Fe(SCN)3 (Papenbrock and Schmidt, 2000a). NaSCN was used as a standard in solution containing Na2SO3. Desalted protein extracts incubated with assay medium in the absence of sulfite were used as blanks. The net sulfite-generating activity was estimated as the difference between sulfite consumption activity and sulfite generation activities of STs and expressed as nmol sulfite min−1 mg−1 protein.
Extraction and Determination of NADPH, Sulfolipids, GSH, and Free and Bound Amino Acids
NADPH was extracted with 0.1 n KOH solution as described (Hajirezaei et al., 2002) and detected at 570 nm during 40 min at 30°C using the cycling assay (Matsumura and Miyachi, 1980). Total lipids were extracted in isopropanol followed by heating at 80°C for 15 min as described by Bligh and Dyer (1959). Sulfolipids were separated from the other lipids by two-dimensional thin-layer chromatography in chloroform:methanol:acetic acid:water (73:25:2:4) polar lipid separation solution, followed by quantification on Trace GC Ultra (Thermo Scientific; http://www.thermoscientific.com) as described before (Khozin et al., 1997). Free amino acids and GSH were extracted from frozen leaf samples and detected according to Matityahu et al. (2006) and Hacham et al. (2008). For protein-bound amino acid determination, total proteins were extracted from 100 mg of leaves using a standard protocol of TCA precipitation (Wang et al., 2006) followed by triple washing the SDS with 100% methanol and 80% acetone. Extracted proteins were hydrolyzed in constant boiling HCl vapors at 110°C for 22 h under nitrogen (Hacham et al., 2008). Total amino acids were determined by the AccQ·Tag method (Waters; http://www.waters.com/) using a Waters Alliance 2695 HPLC instrument. Quantification was performed using a Waters 2475 Multi 22 wavelength fluorescence detector as described previously (Hebeler et al., 2008; Dotson and Westerhoff, 2012). Reduced and oxidized forms of glutathione were determined also according to Griffith (1980) with similar results to those detected by HPLC.
Determination of Total S, Sulfate, Sulfite, and Thiosulfate
Sulfate was stabilized with 24 mm formaldehyde in 2 mm Na2CO3/0.75 mm NaHCO3 solution to prevent spontaneous sulfite oxidation (Lindgren et al., 1982) and determined as described (Brychkova et al., 2012a). Total S content was determined as described before (Busman et al., 1983). Sulfite levels were measured using sulfite detection methods based on (1) chicken SO, (2) a coupled sulfite reductase reaction linked to OAS-TL, and (3) a colorimetric fuchsin-based method as described by Brychkova et al. (2012a). Since data obtained with these three methods varied by less than 10%, only data based on chicken SO detection methods are presented. Thiosulfate content in deproteinized samples extracted in 0.1 m Tris-HCl, pH 9.0, was detected employing a method modified from Papenbrock and Schmidt (2000b). In addition to 0.058 units mL−1 bovine liver rhodanase, type II (Sigma R1756), the reaction contained 5 mm dithiothreitol in 0.1 m Tris-HCl, pH 9.0. Thiosulfate was detected as sulfide produced within 30 min at 26°C and trapped by acidified cadmium acetate (1%; pH 5.0; Murray et al., 2003) and fixed by adding 100 µL of 30 mm FeCl3 dissolved in 1.2 m HCl and 100 µL of 20 mm N,N-dimethyl-p-phenylene-diamine dissolved in 7.2 m HCl. Samples were incubated at 40°C for 10 min, and the production of methylene blue was monitored spectrophotometrically at 625 nm (Siegel, 1965; Murray et al., 2003). The calibration curves for thiosulfate without and with the addition of plant extract were linear in the tested range, 0.3 to 3.0 nmol, with correlation coefficient higher than 0.999 (Supplemental Fig S1B).
Statistical Analysis
The data for SO Ri plants represent means for SO Ri 131 and SO Ri 421 lines. Immunodetection of proteins and enzyme activities was performed on three to six independent protein preparations from different experiments. For both SO Ri lines, representative in gel activities and/or immunoblot analyses are presented. Metabolite measurements were done on six samples from two to six independent experiments. Each treatment was evaluated using ANOVA (Student’s t and Tukey-Kramer honestly significant difference [HSD] tests; JMP 8.0 software; http://www.jmp.com/). Pearson correlation analysis was performed in R statistical software (Wessa, 2012). Analysis of covariance was employed to compare linear regression line slopes (http://faculty.vassar.edu/lowry/vsancova.html).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Sulfurtransferases sulfite-consuming activity in response to dark stress and the detection of thiosulfate.
Supplemental Figure S2. Appearance and damage level of tomato leaves in response to sulfite injection.
Supplemental Figure S3. Damage evaluation, sulfite and sulfate levels, and expression analysis of sulfite oxidase and sulfite reductase in tomato plants as affected by 0.5-mm sulfite injection.
Supplemental Figure S4. Expression analysis of the sulfurtransferases, UDP-sulfoquinovose synthase, and thiosulfate level in tomato plants as affected by 0.5-mm sulfite injection.
Supplemental Table S1. Turnover of sulfur-containing metabolites in the top leaves in response to dark stress in tomato wild type and SO mutants.
Supplemental Table S2. The levels of sulfate and sulfite in the leaves and in xylem sap as affected by dark stress in tomato wild type and SO mutants.
Supplemental Table S3. Predicted representatives of the large group of tomato sulfurtransferases.
Supplemental Table S4. The levels of sulfate and sulfite in the stem xylem sap exudate in dark-stressed and normal-grown wild-type tomato plants after injections with 0.5 mm sulfite.
Supplemental Table S5. List of primers used for quantitative real-time PCR (Lycopersicon esculentum).
Acknowledgments
We thank Dr. Inna Khozin-Goldberg (Ben-Gurion University) for assistance in SQDG determination and Dr. Arye Tishbee (Weizmann Institutes of Sciences) for the total amino acid determination.
Glossary
- GSH
reduced glutathione
- SQDG
sulfoquinovosyldiacylglycerol
- APS
adenosine-5′-phosphosulfate
- APR
adenosine-5′-phosphosulfate reductase
- S
sulfur
- SiR
sulfite reductase
- SQD1
UDP-sulfoquinovose synthase
- SO
sulfite oxidase
- ST
sulfurtransferase
- SO Ri
SO RNA interference
- OAS-TL
O-acetyl-l-serine thiol lyase
- HSD
honestly significant difference
- EC
Enzyme Commission
References
- Amy NK. (1988) Effect of dietary protein and methionine on sulfite oxidase activity in rats. J Nutr 118: 941–944 [DOI] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ. (1959) A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37: 911–917 [DOI] [PubMed] [Google Scholar]
- Bosma W, Schupp R, Dekok LJ, Rennenberg H. (1991) Effect of selenate on assimilatory sulfate reduction and thiol content of spruce needles. Plant Physiol Biochem 29: 131–138 [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Brychkova G, Alikulov Z, Fluhr R, Sagi M. (2008a) A critical role for ureides in dark and senescence-induced purine remobilization is unmasked in the Atxdh1 Arabidopsis mutant. Plant J 54: 496–509 [DOI] [PubMed] [Google Scholar]
- Brychkova G, Fluhr R, Sagi M. (2008b) Formation of xanthine and the use of purine metabolites as a nitrogen source in Arabidopsis plants. Plant Signal Behav 3: 999–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brychkova G, Xia Z, Yang G, Yesbergenova Z, Zhang Z, Davydov O, Fluhr R, Sagi M. (2007) Sulfite oxidase protects plants against sulfur dioxide toxicity. Plant J 50: 696–709 [DOI] [PubMed] [Google Scholar]
- Brychkova G, Yarmolinsky D, Fluhr R, Sagi M. (2012a) The determination of sulfite levels and its oxidation in plant leaves. Plant Sci 190: 123–130 [DOI] [PubMed] [Google Scholar]
- Brychkova G, Yarmolinsky D, Sagi M. (2012b) Kinetic assays for determining in vitro APS reductase activity in plants without the use of radioactive substances. Plant Cell Physiol 53: 1648–1658 [DOI] [PubMed] [Google Scholar]
- Brychkova G, Yarmolinsky D, Ventura Y, Sagi M. (2012c) A novel in-gel assay and an improved kinetic assay for determining in vitro sulfite reductase activity in plants. Plant Cell Physiol 53: 1507–1516 [DOI] [PubMed] [Google Scholar]
- Burandt P, Schmidt A, Papenbrock J. (2001) Cysteine synthesis and cysteine desulfuration in Arabidopsis plants at different developmental stages and light conditions. Plant Physiol Biochem 39: 861–870 [Google Scholar]
- Busman LM, Dick RP, Tabatabai MA. (1983) Determination of total sulphur and chlorine in plant materials by ion chromatography. Soil Sci Soc Am J 47: 1167–1170 [Google Scholar]
- Cramer MD, Lips SH. (1995) Enriched rhizosphere CO2 concentrations can ameliorate the influence of salinity on hydroponically grown tomato plants. Physiol Plant 94: 425–432 [Google Scholar]
- Davidian J-C, Kopriva S. (2010) Regulation of sulfate uptake and assimilation: the same or not the same? Mol Plant 3: 314–325 [DOI] [PubMed] [Google Scholar]
- Dotson A, Westerhoff P. (2012) Character and treatment of organic colloids in challenging and impacted drinking water sources. J Environ Eng 138: 393–401 [Google Scholar]
- Eilers T, Schwarz G, Brinkmann H, Witt C, Richter T, Nieder J, Koch B, Hille R, Hänsch R, Mendel RR. (2001) Identification and biochemical characterization of Arabidopsis thaliana sulfite oxidase: a new player in plant sulfur metabolism. J Biol Chem 276: 46989–46994 [DOI] [PubMed] [Google Scholar]
- Expósito-Rodríguez M, Borges AA, Borges-Pérez A, Pérez JA. (2008) Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biol 8: 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Footitt EJ, Heales SJ, Mills PB, Allen GF, Oppenheim M, Clayton PT. (2011) Pyridoxal 5′-phosphate in cerebrospinal fluid: factors affecting concentration. J Inherit Metab Dis 34: 529–538 [DOI] [PubMed] [Google Scholar]
- Gaitonde MK. (1967) A spectrophotometric method for the direct determination of cysteine in the presence of other naturally occurring amino acids. Biochem J 104: 627–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith OW. (1980) Determination of glutathione and glutathione disulfide using glutathione reductase and 2-vinylpyridine. Anal Biochem 106: 207–212 [DOI] [PubMed] [Google Scholar]
- Guo FQ, Crawford NM. (2005) Arabidopsis nitric oxide synthase1 is targeted to mitochondria and protects against oxidative damage and dark-induced senescence. Plant Cell 17: 3436–3450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacham Y, Matityahu I, Schuster G, Amir R. (2008) Overexpression of mutated forms of aspartate kinase and cystathionine gamma-synthase in tobacco leaves resulted in the high accumulation of methionine and threonine. Plant J 54: 260–271 [DOI] [PubMed] [Google Scholar]
- Hajirezaei MR, Peisker M, Tschiersch H, Palatnik JF, Valle EM, Carrillo N, Sonnewald U. (2002) Small changes in the activity of chloroplastic NADP(+)-dependent ferredoxin oxidoreductase lead to impaired plant growth and restrict photosynthetic activity of transgenic tobacco plants. Plant J 29: 281–293 [DOI] [PubMed] [Google Scholar]
- Hänsch R, Lang C, Riebeseel E, Lindigkeit R, Gessler A, Rennenberg H, Mendel RR. (2006) Plant sulfite oxidase as novel producer of H2O2: combination of enzyme catalysis with a subsequent non-enzymatic reaction step. J Biol Chem 281: 6884–6888 [DOI] [PubMed] [Google Scholar]
- Hänsch R, Mendel RR. (2005) Sulfite oxidation in plant peroxisomes. Photosynth Res 86: 337–343 [DOI] [PubMed] [Google Scholar]
- Hansch R, Mendel RR. (2008) Sulfite oxidation in plants. In R Hell, C Dahl, D Knaff, T Leustek, eds, Sulfur Metabolism in Phototrophic Organisms. Springer, Dordrecht, The Netherlands, p 539
- Hartung W, Wilkinson S, Davies WJ. (1998) Factors that regulate abscisic acid concentrations at the primary site of action at the guard cell. J Exp Bot 49: 361–367 [Google Scholar]
- Hebeler R, Oeljeklaus S, Reidegeld KA, Eisenacher M, Stephan C, Sitek B, Stühler K, Meyer HE, Sturre MJG, Dijkwel PP, et al. (2008) Study of early leaf senescence in Arabidopsis thaliana by quantitative proteomics using reciprocal 14N/15N labeling and difference gel electrophoresis. Mol Cell Proteomics 7: 108–120 [DOI] [PubMed] [Google Scholar]
- Heber U, Huve K. (1998) Action of SO2 on plants and metabolic detoxification of SO2. Int Rev Cytol 177: 255–286 [Google Scholar]
- Hörtensteiner S, Feller U. (2002) Nitrogen metabolism and remobilization during senescence. J Exp Bot 53: 927–937 [DOI] [PubMed] [Google Scholar]
- Islam MS, Takagi S. (2010) Co-localization of mitochondria with chloroplasts is a light-dependent reversible response. Plant Signal Behav 5: 146–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamachi K, Yamaya T, Mae T, Ojima K. (1991) A role for glutamine synthetase in the remobilization of leaf nitrogen during natural senescence in rice leaves. Plant Physiol 96: 411–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami N, Watanabe A. (1988) Senescence-specific increase in cytosolic glutamine synthetase and its mRNA in radish cotyledons. Plant Physiol 88: 1430–1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keskitalo J, Bergquist G, Gardeström P, Jansson S. (2005) A cellular timetable of autumn senescence. Plant Physiol 139: 1635–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MS, Haas FH, Samami AA, Gholami AM, Bauer A, Fellenberg K, Reichelt M, Hänsch R, Mendel RR, Meyer AJ, et al. (2010) Sulfite reductase defines a newly discovered bottleneck for assimilatory sulfate reduction and is essential for growth and development in Arabidopsis thaliana. Plant Cell 22: 1216–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khozin I, Adlerstein D, Bigongo C, Heimer YM, Cohen Z. (1997) Elucidation of the biosynthesis of eicosapentaenoic acid in the microalga Porphyridium cruentum. II. Studies with radiolabeled precursors. Plant Physiol 114: 223–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King L-K, Kaiser ET. (1974) Nucleophilic reactions of sulfite esters in aqueous media. J Am Chem Soc 96: 1410–1418 [Google Scholar]
- Kopriva S. (2006) Regulation of sulfate assimilation in Arabidopsis and beyond. Ann Bot (Lond) 97: 479–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopriva S, Mugford SG, Matthewman C, Koprivova A. (2009) Plant sulfate assimilation genes: redundancy versus specialization. Plant Cell Rep 28: 1769–1780 [DOI] [PubMed] [Google Scholar]
- Kopriva S, Muheim R, Koprivova A, Trachsel N, Catalano C, Suter M, Brunold C. (1999) Light regulation of assimilatory sulphate reduction in Arabidopsis thaliana. Plant J 20: 37–44 [DOI] [PubMed] [Google Scholar]
- Lang C, Popko J, Wirtz M, Hell R, Herschbach C, Kreuzwieser J, Rennenberg H, Mendel RR, Hänsch R. (2007) Sulphite oxidase as key enzyme for protecting plants against sulphur dioxide. Plant Cell Environ 30: 447–455 [DOI] [PubMed] [Google Scholar]
- Leustek T. (2002) Sulfate metabolism. The Arabidopsis Book 1: e0017, doi/10.1199/tab.0017
- Leustek T, Martin MN, Bick JA, Davies JP. (2000) Pathways and regulation of sulfur metabolism revealed through molecular and genetic studies. Annu Rev Plant Physiol Plant Mol Biol 51: 141–165 [DOI] [PubMed] [Google Scholar]
- Lindgren M, Cedergren A, Lindberg J. (1982) Conditions for sulfite stabilization and determination by ion chromatography. Anal Chim Acta 141: 279–286 [Google Scholar]
- Martin MN, Tarczynski MC, Shen B, Leustek T. (2005) The role of 5′-adenylylsulfate reductase in controlling sulfate reduction in plants. Photosynth Res 86: 309–323 [DOI] [PubMed] [Google Scholar]
- Matityahu I, Kachan L, Bar Ilan I, Amir R. (2006) Transgenic tobacco plants overexpressing the Met25 gene of Saccharomyces cerevisiae exhibit enhanced levels of cysteine and glutathione and increased tolerance to oxidative stress. Amino Acids 30: 185–194 [DOI] [PubMed] [Google Scholar]
- Matsumura H, Miyachi S. (1980) Cycling assay for nicotinamide adenine dinucleotides. Methods Enzymol 69: 465–470
- Murray DB, Klevecz RR, Lloyd D. (2003) Generation and maintenance of synchrony in Saccharomyces cerevisiae continuous culture. Exp Cell Res 287: 10–15 [DOI] [PubMed] [Google Scholar]
- Murray F. (1997) Urban air pollution and health effects. In D Brune, DV Chapman, MD Gwynne, JM Pacyna, eds, The Global Environment: Science, Technology and Management. Scandinavian Science Publisher, Weinheim, Germany, pp 585–598
- Nakamura T, Yamaguchi Y, Sano H. (2000) Plant mercaptopyruvate sulfurtransferases: molecular cloning, subcellular localization and enzymatic activities. Eur J Biochem 267: 5621–5630 [DOI] [PubMed] [Google Scholar]
- Netting AG, Theobald JC, Dodd IC. (2012) Xylem sap collection and extraction methodologies to determine in vivo concentrations of ABA and its bound forms by gas chromatography-mass spectrometry (GC-MS). Plant Methods 8: 11–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsu I, Wiriyathanawudhiwong N, Morigasaki S, Nakatani T, Kadokura H, Takagi H. (2010) The L-cysteine/L-cystine shuttle system provides reducing equivalents to the periplasm in Escherichia coli. J Biol Chem 285: 17479–17487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa K, Kasahara M, Kiyosue T, Kagawa T, Suetsugu N, Takahashi F, Kanegae T, Niwa Y, Kadota A, Wada M. (2003) Chloroplast unusual positioning1 is essential for proper chloroplast positioning. Plant Cell 15: 2805–2815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachmayr F. (1960) Vorkommen und Bestimmung von Schwefelverbindungen in Mineralwasser. PhD thesis. University of Munich, Munich, Germany
- Pageau K, Reisdorf-Cren M, Morot-Gaudry J-F, Masclaux-Daubresse C. (2006) The two senescence-related markers, GS1 (cytosolic glutamine synthetase) and GDH (glutamate dehydrogenase), involved in nitrogen mobilization, are differentially regulated during pathogen attack and by stress hormones and reactive oxygen species in Nicotiana tabacum L. leaves. J Exp Bot 57: 547–557 [DOI] [PubMed] [Google Scholar]
- Papenbrock J, Guretzki S, Henne M. (2011) Latest news about the sulfurtransferase protein family of higher plants. Amino Acids 41: 43–57 [DOI] [PubMed] [Google Scholar]
- Papenbrock J, Schmidt A. (2000a) Characterization of a sulfurtransferase from Arabidopsis thaliana. Eur J Biochem 267: 145–154 [DOI] [PubMed] [Google Scholar]
- Papenbrock J, Schmidt A. (2000b) Characterization of two sulfurtransferase isozymes from Arabidopsis thaliana. Eur J Biochem 267: 5571–5579 [DOI] [PubMed] [Google Scholar]
- Peiser GD, Lizada MC, Yang SF. (1982) Sulfite-induced lipid peroxidation in chloroplasts as determined by ethane production. Plant Physiol 70: 994–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruzinská A, Tanner G, Aubry S, Anders I, Moser S, Müller T, Ongania KH, Kräutler B, Youn JY, Liljegren SJ, et al. (2005) Chlorophyll breakdown in senescent Arabidopsis leaves: characterization of chlorophyll catabolites and of chlorophyll catabolic enzymes involved in the degreening reaction. Plant Physiol 139: 52–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randewig D, Hamisch D, Herschbach C, Eiblmeier M, Gehl C, Jurgeleit J, Skerra J, Mendel RR, Rennenberg H, Hänsch R. (2012) Sulfite oxidase controls sulfur metabolism under SO2 exposure in Arabidopsis thaliana. Plant Cell Environ 35: 100–115 [DOI] [PubMed] [Google Scholar]
- Saito K. (2000) Regulation of sulfate transport and synthesis of sulfur-containing amino acids. Curr Opin Plant Biol 3: 188–195 [PubMed] [Google Scholar]
- Sanda S, Leustek T, Theisen MJ, Garavito RM, Benning C. (2001) Recombinant Arabidopsis SQD1 converts UDP-glucose and sulfite to the sulfolipid head group precursor UDP-sulfoquinovose in vitro. J Biol Chem 276: 3941–3946 [DOI] [PubMed] [Google Scholar]
- Shimojima M. (2011) Biosynthesis and functions of the plant sulfolipid. Prog Lipid Res 50: 234–239 [DOI] [PubMed] [Google Scholar]
- Shimojima M, Benning C. (2003) Native uridine 5′-diphosphate-sulfoquinovose synthase, SQD1, from spinach purifies as a 250-kDa complex. Arch Biochem Biophys 413: 123–130 [DOI] [PubMed] [Google Scholar]
- Shimojima M, Hoffmann-Benning S, Garavito RM, Benning C. (2005) Ferredoxin-dependent glutamate synthase moonlights in plant sulfolipid biosynthesis by forming a complex with SQD1. Arch Biochem Biophys 436: 206–214 [DOI] [PubMed] [Google Scholar]
- Siegel LM. (1965) A direct microdetermination for sulfide. Anal Biochem 11: 126–132 [DOI] [PubMed] [Google Scholar]
- Slavikova S, Ufaz S, Avin-Wittenberg T, Levanony H, Galili G. (2008) An autophagy-associated Atg8 protein is involved in the responses of Arabidopsis seedlings to hormonal controls and abiotic stresses. J Exp Bot 59: 4029–4043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Kopriva S, Giordano M, Saito K, Hell R. (2011) Sulfur assimilation in photosynthetic organisms: molecular functions and regulations of transporters and assimilatory enzymes. Annu Rev Plant Biol 62: 157–184 [DOI] [PubMed] [Google Scholar]
- Teschner J, Lachmann N, Schulze J, Geisler M, Selbach K, Santamaria-Araujo J, Balk J, Mendel RR, Bittner F. (2010) A novel role for Arabidopsis mitochondrial ABC transporter ATM3 in molybdenum cofactor biosynthesis. Plant Cell 22: 468–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsakraklides G, Martin M, Chalam R, Tarczynski MC, Schmidt A, Leustek T. (2002) Sulfate reduction is increased in transgenic Arabidopsis thaliana expressing 5′-adenylylsulfate reductase from Pseudomonas aeruginosa. Plant J 32: 879–889 [DOI] [PubMed] [Google Scholar]
- Vauclare P, Kopriva S, Fell D, Suter M, Sticher L, von Ballmoos P, Krähenbühl U, den Camp RO, Brunold C. (2002) Flux control of sulphate assimilation in Arabidopsis thaliana: adenosine 5′-phosphosulphate reductase is more susceptible than ATP sulphurylase to negative control by thiols. Plant J 31: 729–740 [DOI] [PubMed] [Google Scholar]
- Wang W, Vignani R, Scali M, Cresti M. (2006) A universal and rapid protocol for protein extraction from recalcitrant plant tissues for proteomic analysis. Electrophoresis 27: 2782–2786 [DOI] [PubMed] [Google Scholar]
- Wessa P. (2012) Pearson Correlation (v1.0.6) in Free Statistics Software (v1.1.23-r7). Office for Research Development and Education. http://www.wessa.net/rwasp_correlation.wasp/ (June 6, 2012)
- Wu Y, Zheng F, Ma W, Han Z, Gu Q, Shen Y, Mi H. (2011) Regulation of NAD(P)H dehydrogenase-dependent cyclic electron transport around PSI by NaHSO3 at low concentrations in tobacco chloroplasts. Plant Cell Physiol 52: 1734–1743 [DOI] [PubMed] [Google Scholar]
- Xiong J, Fu G, Yang Y, Zhu C, Tao L. (2012) Tungstate: is it really a specific nitrate reductase inhibitor in plant nitric oxide research? J Exp Bot 63: 33–41 [DOI] [PubMed] [Google Scholar]
- Yang B, Jiang Y, Rahman MH, Deyholos MK, Kav NN. (2009) Identification and expression analysis of WRKY transcription factor genes in canola (Brassica napus L.) in response to fungal pathogens and hormone treatments. BMC Plant Biol 9: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Onda Y, Ashikari T, Tanaka Y, Kusumi T, Hase T. (2000) Analysis of reductant supply systems for ferredoxin-dependent sulfite reductase in photosynthetic and nonphotosynthetic organs of maize. Plant Physiol 122: 887–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavgorodnyaya A, Papenbrock J, Grimm B. (1997) Yeast 5-aminolevulinate synthase provides additional chlorophyll precursor in transgenic tobacco. Plant J 12: 169–178 [DOI] [PubMed] [Google Scholar]