Abstract
The ability to precisely engineer plant genomes offers much potential for advancing basic and applied plant biology. Here, we describe methods for the targeted modification of plant genomes using transcription activator-like effector nucleases (TALENs). Methods were optimized using tobacco (Nicotiana tabacum) protoplasts and TALENs targeting the acetolactate synthase (ALS) gene. Optimal TALEN scaffolds were identified using a protoplast-based single-strand annealing assay in which TALEN cleavage creates a functional yellow fluorescent protein gene, enabling quantification of TALEN activity by flow cytometry. Single-strand annealing activity data for TALENs with different scaffolds correlated highly with their activity at endogenous targets, as measured by high-throughput DNA sequencing of polymerase chain reaction products encompassing the TALEN recognition sites. TALENs introduced targeted mutations in ALS in 30% of transformed cells, and the frequencies of targeted gene insertion approximated 14%. These efficiencies made it possible to recover genome modifications without selection or enrichment regimes: 32% of tobacco calli generated from protoplasts transformed with TALEN-encoding constructs had TALEN-induced mutations in ALS, and of 16 calli characterized in detail, all had mutations in one allele each of the duplicate ALS genes (SurA and SurB). In calli derived from cells treated with a TALEN and a 322-bp donor molecule differing by 6 bp from the ALS coding sequence, 4% showed evidence of targeted gene replacement. The optimized reagents implemented in plant protoplasts should be useful for targeted modification of cells from diverse plant species and using a variety of means for reagent delivery.
Sequence-specific nucleases are powerful reagents for creating targeted modifications to genomes in vivo (Arnould et al., 2011; Bogdanove and Voytas, 2011; Carroll, 2011). The double-strand breaks introduced at target loci by these nucleases activate the cell’s DNA-repair pathways, principally nonhomologous end joining (NHEJ) and homologous recombination. NHEJ rejoins the broken chromosomes, and because repair is often imprecise, mutations are introduced at the cut site that can disrupt gene function. Homologous recombination, on the other hand, can be harnessed to carry out gene replacement or targeted gene insertion (both referred to here as gene targeting), enabling precise modifications to genes and genomes.
In plants, the use of sequence-specific nucleases for targeted genome modification has many applications, ranging from dissecting gene function to creating plants with new traits (Curtin et al., 2012). Although still in the early stages of being deployed, sequence-specific nucleases have already been used to engineer plants with improved disease resistance, altered metabolite profiles, and tolerance to herbicides (Shukla et al., 2009; Townsend et al., 2009; Li et al., 2012). Traditional methods for introducing foreign genes into plants could also be improved using sequence-specific nucleases. For example, transgenes could be targeted to genomic regions conducive to expression, and multiple transgenes could be inserted at a single locus for linked transmission in genetic crosses.
One challenge for targeted genome modification of plants is the efficient delivery of the nucleases to cells, or in the case of gene targeting, both the nucleases and donor DNA molecules used to repair the broken chromosomes. For many plants, the efficiency of transformation of tissue explants by Agrobacterium tumefaciens or biolistic/physical means is low (Barampuram and Zhang, 2011), and as such, it is difficult to generate and analyze the large numbers of transformants needed to identify those rare plants with the desired genome modification. For a subset of row crops, vegetables, and ornamentals, however, plants can be regenerated from protoplasts derived from leaf mesophyll cells stripped of their cell walls (Davey et al., 2005). For these species, individual cells are totipotent and can be induced to grow and divide in culture to form cell clusters (calli) and ultimately differentiated into shoots and roots. Efficient means of DNA transfer into protoplasts, including electroporation and polyethylene glycol-mediated transformation, make it possible to introduce nucleases and donor molecules into large populations of cells and easily attain the requisite number of transformation events to recover targeted sequence alterations.
As a model for optimizing methods for plant genome engineering, our laboratory uses tobacco (Nicotiana tabacum), for which protoplast transformation and regeneration regimes are well established (Wright et al., 2005; Townsend et al., 2009). Our targets are tobacco’s duplicate acetolactate synthase (ALS) genes (SurA and SurB), which encode enzymes involved in branched-chain amino acid biosynthesis. Specific amino acid substitutions in ALS confer resistance to sulfonylurea and imidazolinone herbicides in a dominant fashion (Tranel and Wright, 2002). We previously demonstrated the ability of engineered zinc finger nucleases (ZFNs) to create herbicide-resistant plants through gene targeting (Townsend et al., 2009). The identification of gene-targeting events was made possible by the powerful selection conferred by herbicide resistance; however, since most targeted sequence modifications in plants do not confer selectable phenotypes, it would be desirable to achieve frequencies of site-specific mutagenesis and gene replacement sufficient to recover modifications without selection. Furthermore, despite recent advances (Sander et al., 2011b), engineering ZFNs with the desired specificity for some loci is not always possible.
In the past few years, transcription activator-like effector nucleases (TALENs) have emerged as the reagent of choice for many genome engineering applications (Bogdanove and Voytas, 2011). Much like ZFNs, TALENs are chimeric proteins made by fusing an engineered DNA-binding domain with the catalytic domain of FokI endonuclease (Christian et al., 2010; Li et al., 2011), which cleaves as a dimer. TALENs (and ZFNs), therefore, work in pairs: two monomers bind opposing strands of DNA separated by a spacer of an appropriate length, allowing FokI to dimerize and cleave DNA. One of the primary advantages of TALENs is that the DNA-binding domain can be easily engineered to recognize virtually any DNA sequence (Cermak et al., 2011; Reyon et al., 2012).
Whereas TALENs have been shown to function well as mutagens in species like zebrafish, rat, and human cells (Miller et al., 2007; Huang et al., 2011; Sander et al., 2011a; Tesson et al., 2011), only a few studies of TALENs in plants have been published to date, all of which have used TALENs to create mutations by NHEJ (Cermak et al., 2011; Mahfouz et al., 2011; Li et al., 2012). In this study, we used our tobacco model system to determine the optimal TALEN architecture, expression, and delivery methods to achieve both high-efficiency mutagenesis by NHEJ and gene targeting by homologous recombination. Our data indicate that by using TALENs, it is now possible to efficiently alter endogenous plant genes at frequencies that obviate the need for selection regimes or enrichment protocols.
RESULTS
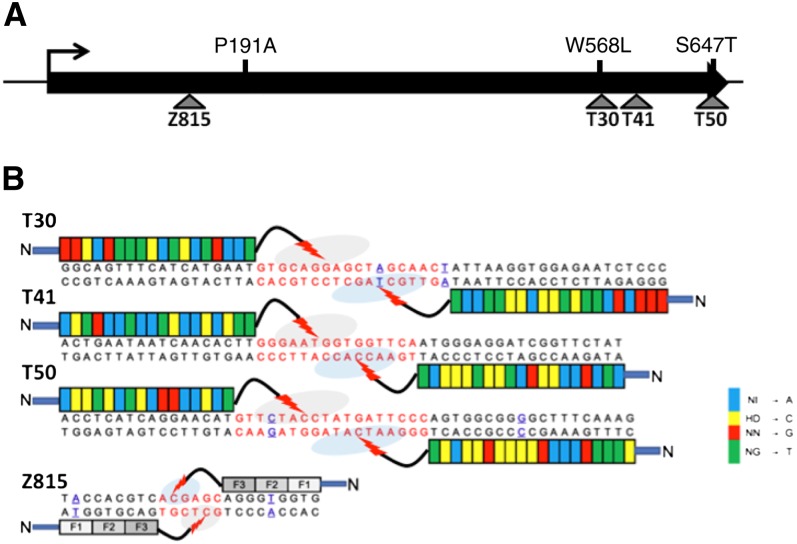
Highest efficiencies of gene targeting are achieved if DNA cleavage occurs near the desired site of modification. Accomplishing this with ZFNs can be a challenge, as it is often difficult to engineer them to recognize any given sequence. In previous work, we were able to design a total of three ZFNs that target SurB (Townsend et al., 2009). In contrast, 223 potential TALEN target sites were identified in the SurB coding sequence using our software package TALE-NT (Doyle et al., 2012). Three sites were chosen at positions in the coding sequence where amino acid substitutions are known to confer herbicide resistance (Fig. 1; Tranel and Wright, 2002; Townsend et al., 2009). For TALEN pairs targeting sites T30 and T41, the recognition sequences for both TALENs are conserved in SurA (Fig. 1B; Supplemental Fig. S1). For the pair targeting site T50, a single-nucleotide polymorphism exists in SurA in one of the two TALEN monomer-binding sites. As a control and point of comparison, we used a previously characterized ZFN (Z815), for which there are two base differences in SurA, one in each of the ZFN monomer-binding sites (Townsend et al., 2009).
Figure 1.
Targeting the tobacco ALS genes with TALENs. A, Schematic of the tobacco SurB gene, which encodes ALS, showing target sites for the TALENs and the ZFN used in this study. Amino acid substitutions that confer herbicide resistance are shown above the coding sequence. B, DNA sequences of the TALEN and ZFN recognition sites in SurB. Bases that differ in SurA are blue and underlined; sequence alignments between SurA and SurB are shown in Supplemental Figure S1. The colored boxes denote the TAL effector repeats. Each color represents a different repeat-variable diresidue, for which nucleotide specificities are given at the far right.
TAL effector repeat arrays recognizing the three SurB target sites (Fig. 1B) were constructed using our Golden Gate assembly platform (Cermak et al., 2011). The arrays were then cloned into three TALEN backbone architectures with various truncations in the N- and C-terminal regions flanking the DNA-binding domain (Miller et al., 2011; Mussolino et al., 2011; Sun et al., 2012; Supplemental Fig. S1B). All three architectures have a 152-amino acid N-terminal truncation (designated NΔ152). The C-terminal truncations vary in length and are designated by the number of amino acids after the repeat array, namely 18 (C18), 23 (C23), or 63 (C63) amino acids.
TALEN activity was tested in tobacco protoplasts using a yellow fluorescent protein (YFP) single-strand annealing (SSA) reporter (Supplemental Fig. S2). The reporter has a TALEN recognition site flanked by a 255-bp direct repeat of YFP coding sequence, a length determined to be effective for efficient recombination after cleavage and thereby reconstitution of a functional YFP gene (Supplemental Fig. S3). The YFP SSA reporter was intended to be codelivered to protoplasts along with TALEN-encoding plasmids, and YFP expression was intended to be quantified by flow cytometry to provide a readout of TALEN activity. One of the first applications of the SSA assay was to assess the effectiveness of different TALEN expression strategies. Both the T30 and T40 TALENs were tested in two configurations: (1) as tandem genes each expressed from a cauliflower mosaic virus 35S promoter, and (2) as a single expression unit in which the two TALEN coding sequences were separated by a T2A translational skipping sequence and expressed from a single 35S promoter. Both configurations resulted in comparable levels of TALEN activity as measured by the SSA assay (Supplemental Fig. S4).
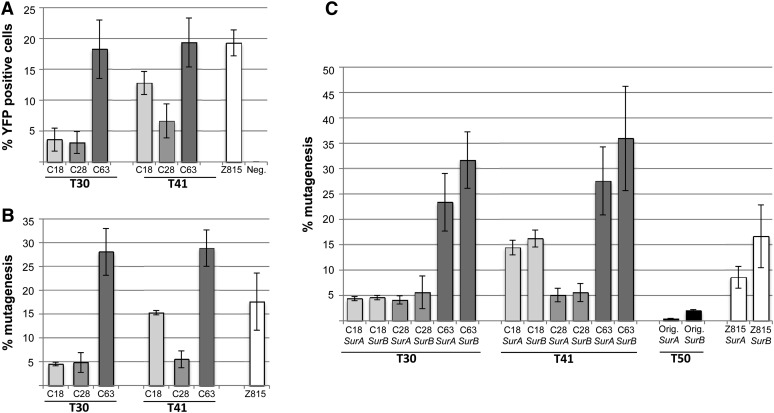
The SSA assay was then used to assess the activity of the three truncated TALEN scaffolds with the T30 and T41 DNA-binding domains (Fig. 2A). For the most active TALENs, approximately 20% of treated cells showed YFP fluorescence, a level comparable to that observed for ZFN Z815. To corroborate the tobacco SSA data, activities of the TALENs were also evaluated at their endogenous target sites in the tobacco genome. After introducing TALEN constructs into protoplasts, genomic DNA was isolated, and a 370-bp fragment encompassing the target site was amplified by PCR. The PCR amplicon was subjected to high-throughput DNA sequencing to assess the number of mutations introduced at the TALEN recognition site through imprecise repair of the break by NHEJ (Fig. 2B). The different TALEN architectures showed mutagenesis frequencies highly correlated with their activities in the protoplast SSA assay (r = 0.97, P = 4.17 × 10−05; Fig. 2A). Therefore, we conclude that the protoplast SSA assay is a reliable means of assessing TALEN activity at endogenous chromosomal target sites, barring potential differences in chromatin status that might affect activity at such sites.
Figure 2.
Gene targeting at SurA and SurB using TALENs. A, Activity of TALENs and the ZFN measured using a YFP SSA reporter in tobacco protoplasts. YFP-positive cells were quantified by flow cytometry. In the negative control, protoplasts were transformed only with the ZFN SSA reporter. B, Frequencies of NHEJ-induced mutagenesis in tobacco protoplasts by TALENs and the ZFN as revealed by high-throughput DNA sequencing. Cleavage activities are expressed as the percentage of sequencing reads with insertion/deletion mutations at the target site. Transformation efficiencies for experiments depicted in A and B were 94% and 93%, respectively (data not shown). C, The data depicted in B were parsed, and the reads derived from SurA and SurB were plotted separately. Data derived from TALEN T50 cloned in the original TALEN architecture (Orig.), described by Christian et al. (2010), was also included. The T50 target site has a single base difference in SurA in one of the TALEN binding sites. Mutagenesis for T50 at SurA (0.45% + 0.04%) was significantly lower than at SurB (1.99% ± 0.23%). In contrast, ZFN Z815 has one base difference in the target site of each of the ZFN monomers, and both SurA and SurB were targeted at frequencies that were not statistically different. Error bars in all panels represent sd of three replicates.
Because of sequence differences between SurA and SurB, it was possible to assess relative mutagenesis frequencies at these genes using the 454 sequencing data (Fig. 2C). When the target sites were identical in both genes (e.g. T30 and T41), mutagenesis frequencies were comparable. Frequencies of NHEJ-induced mutations were also obtained at the endogenous target site for TALEN T50, which was cloned in the original architecture (Christian et al., 2010; Supplemental Fig. S1B). TALEN T50 has a single nucleotide difference in SurA for one of the TALEN recognition sites, and this had a significant, negative impact, resulting in an approximately 4-fold decrease in mutagenesis activity. In comparison, mutagenesis frequencies for ZFN Z815 were statistically indistinguishable between the two loci, despite the fact that there are two base mismatches in SurA, one in each of the two DNA-binding domains. The ability to discriminate among closely related DNA sequences in plants suggests that TALENs are highly specific.
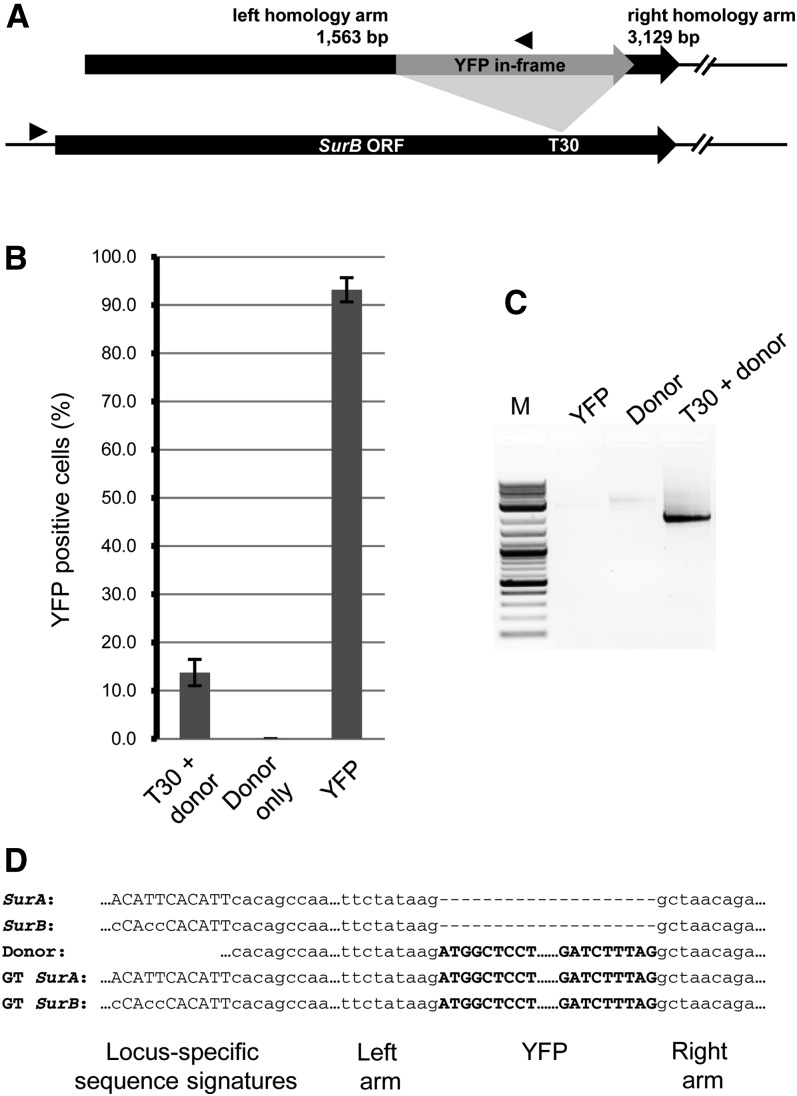
The 454 data demonstrated that the TALENs were effective in creating targeted mutations through NHEJ. We next tested the ability of TALENs to alter Sur loci through homologous recombination. We designed a donor template that would create an in-frame gene fusion between ALS and YFP (Fig. 3A). This donor template was introduced into protoplasts along with plasmids encoding the T30 NΔ152,C63 TALENs, which target an identical sequence in SurA and SurB. Targeted gene insertion was measured by quantifying YFP fluorescence by flow cytometry. Approximately 14% of protoplasts transformed with both the TALENs and the YFP donor template fluoresced (Fig. 3B), a frequency comparable to the frequency with which the T30 NΔ152,C63 TALENs induced mutations by NHEJ (Fig. 1B). Protoplasts transformed with the donor construct alone showed no fluorescence, indicating that the fluorescent cells attained in treatments with the donor and the TALEN were recombinants.
Figure 3.
Targeted YFP insertion. A, Schematic of the gene-targeting experiment. Above the diagram of SurB is the donor template that inserts the YFP coding sequence in frame. Lengths of the homology arms are indicated. Arrowheads denote PCR primers used in the molecular characterization of recombinants. B, Targeted insertion of YFP in ALS as measured by flow cytometry. Error bars represent sd of three replicates. C, Molecular evidence of gene targeting of YFP into Sur genes. Protoplasts were transformed with either TALEN T30 and the YFP donor or the donor alone. As an additional control, protoplasts were transformed with a YFP expression construct. After 24 h, DNA was prepared from total protoplasts and PCR amplified with primers complementary to a site in YFP and a site upstream of the Sur genes (as in A). Only in the presence of both the donor and TALEN T30 was a PCR product obtained. D, Representative DNA sequences obtained from clones of the PCR fragment in C. Sequences from the left and right homology arms are shown in lowercase; YFP sequences are in boldface. GT, Gene targeting. Note that both SurA and SurB loci were targeted.
DNA prepared from the transformed protoplasts was analyzed by PCR using a primer within the YFP coding sequence and a second primer that recognizes a site flanking the Sur genes and not present on the donor (Fig. 3A). Amplification products were only obtained in cells treated with the YFP donor and the nuclease, consistent with YFP fluorescence arising from recombination that placed YFP in frame with the Sur coding sequence. The DNA sequences of the PCR products confirmed this conclusion (Fig. 3D). To assess the fidelity of recombination at individual chromosomes, PCR products from two independent experiments were cloned, and six clones from each experiment were sequenced. Two clones from one experiment had either a 4- or 28-bp deletion in the T30 spacer sequence, indicating that these events arose through a combination of homologous recombination and NHEJ (Supplemental Fig. S5).
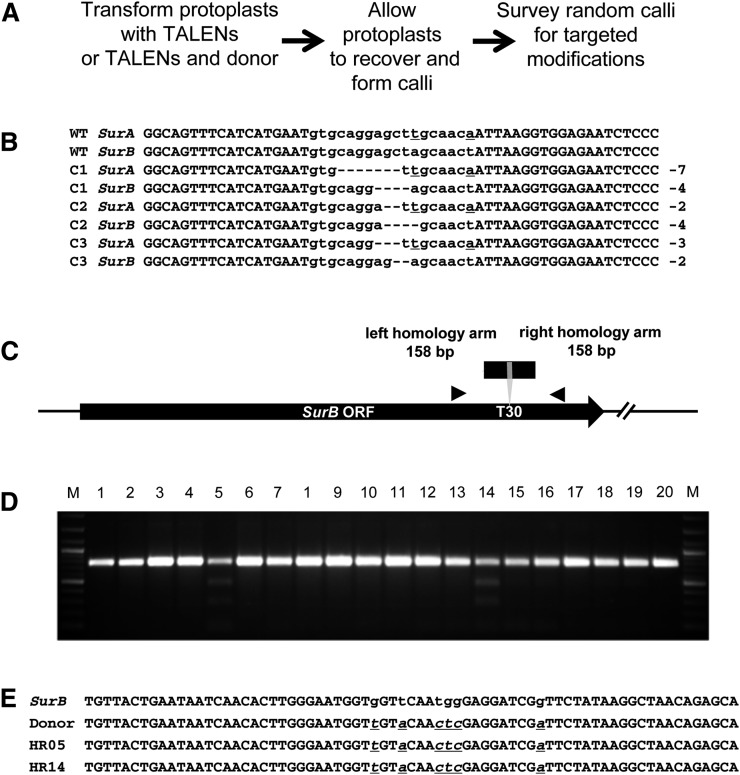
The high frequencies of targeted mutagenesis and gene replacement suggested that modifications of Sur loci could be recovered without the use of reporters or selection. Plasmids encoding the T30 TALENs in the NΔ152,C63 architecture were transformed into tobacco protoplasts, which were then grown into calli on nonselective medium (Fig. 4A). Calli from three independent experiments were randomly selected; a DNA fragment encompassing the TALEN recognition site was PCR amplified from each callus and analyzed for mutations that caused loss of a restriction enzyme site in the TALEN spacer sequence through imprecise NHEJ. In each experiment, mutations in Sur genes were identified in three of 12, 16 of 48, and four of 13 calli analyzed (weighted mean, 32%). Mutations in 16 randomly selected calli from one experiment were further analyzed to determine which Sur genes were modified and whether more then one allele of each gene was altered. All 16 calli had mutations in one allele each of SurA and SurB. Because the Sur genes encode an enzyme involved in amino acid biosynthesis, it is unlikely that mutations could be recovered in all alleles. Representative DNA sequences of altered alleles in three calli are shown in Figure 4B. In addition, a PCR survey was performed with primer sets recognizing both the 5′ and 3′ ends of the TALEN coding sequences to assess whether the circular TALEN-encoding plasmids had integrated into the genome. While the control sample using DNA prepared 24 h after transformation gave a very strong signal with both primer sets, only a very weak PCR product was obtained in two of 16 calli 30 d after transformation (Supplemental Fig. S6). This suggests that the TALEN-encoding plasmid was lost before becoming stably integrated into the genome. More rigorous tests need to be performed to fully determine whether small pieces of the plasmid were retained in the genomes of these calli.
Figure 4.
Recovery of targeted modifications without selection or enrichment. A, Flow diagram of the experimental protocol. B, DNA sequences of NHEJ-induced mutations in ALS recovered from three representative calli derived from protoplasts transformed with TALEN T30. All three calli have one allele each of SurA and SurB modified, as revealed by distinctive DNA sequence signatures (underlined residues). Sequences in lowercase denote spacers between the two TALEN binding sites. C, Schematic of the gene-targeting experiment using a donor molecule that modifies 6 bp of coding sequence; the modifications create an XhoI restriction site. Arrowheads denote PCR primers used in the molecular characterization of recombinants. ORF, Open reading frame. D, Molecular analysis of independent calli derived from treatment with TALEN T30 and the donor molecule depicted in C. A region of the coding sequence was PCR amplified (arrowheads in C) and digested with XhoI. Lanes 3 to 20 are the digestion products from 18 randomly chosen calli. Lanes 1 and 2 are products from calli treated with the donor only or TALEN T30 only, respectively. M, Molecular length markers. E, DNA sequences of recombinants from lanes 5 and 14 in D. Underlined sequences in lowercase denote base changes introduced into the coding sequence by homologous recombination.
We next determined if targeted gene replacements could also be recovered without selection. Tobacco protoplasts were transformed with the T30 TALENs in the NΔ152,C63 architecture and a 322-bp donor molecule that carried a 6-bp signature distinguishing it from both SurA and SurB (Fig. 4C). After growth on nonselective medium, DNA was prepared from calli, and PCR amplification and sequence analysis were used to determine if modifications of ALS were created by homologous recombination (Fig. 4, D and E). In three experiments, calli were surveyed, and two of 18, one of 48, and one of 48 gene-targeting events were recovered (weighted mean, 4%). All four events appeared to result from perfect homologous recombination; none of them showed evidence of repair by both homologous recombination and imprecise NHEJ. We conclude that TALEN-mediated modification of plant cells is highly efficient, and genomic alterations, including targeted mutations and gene replacements, can be readily recovered without selection regimes or high-throughput screening.
DISCUSSION
We demonstrate here that TALEN architecture, defined as the coding sequences flanking the DNA-binding domain, is an important determinant of activity in plant cells. Others, working principally in mammalian cells, have previously noted the positive impact on TALEN activity of minimizing the length of the N and C termini (Miller et al., 2011; Mussolino et al., 2011; Sun et al., 2012). Although not experimentally tested here, we speculate that removal of flanking coding sequences, particularly C-terminal residues, helps stabilize TALEN proteins or facilitates folding. Nonetheless, TALENs made using our original architecture are capable of introducing mutations (Cermak et al., 2011; Fig. 2B), and Li et al. (2012) fused the catalytic domain of FokI to a full-length TAL effector (i.e. with no truncations) and achieved efficient targeted mutagenesis in rice. Because TALENs are inherently large proteins, the truncations, in addition to improving activity, also make it easier to clone and deliver TALENs to cells. Strategies such as use of the T2A translational skipping sequence further help minimize the length of TALEN constructs by obviating the need for two promoters.
The experiments described here with TALENs warrant comparison with our previous study in which ZFNs were used to create targeted modifications of the duplicate tobacco ALS genes (Townsend et al., 2009). The most active ALS ZFN (Z815) was comparable in activity to the best TALENs, as assessed by both 454 sequencing and the SSA assay. Whereas both classes of nucleases displayed comparable activity, it is clear that TALENs are much easier to engineer to recognize novel DNA sequences: three ZFNs could be engineered to target ALS, but using conservative design parameters, 223 TALEN sites were identified. In previous studies, we have shown that more than 90% of TALENs are functional using these design criteria (Cermak et al., 2011). Reyon et al. (2012) recently showed that most of these criteria are not strictly necessary, suggesting that multiple TALENs, on average, can be targeted to every base pair in DNA. One clear advantage of TALENs over ZFNs, therefore, is their superior targeting range.
The frequency of NHEJ-induced mutagenesis, excluding possible locus-specific differences in DNA repair, is determined both by the ability of the nuclease to recognize its target site and the inherent activity of the nuclease. Frequencies of gene targeting, on the other hand, are also influenced by the donor template and the types of modifications to be introduced into the target. In previous work with Z815, a donor was used with left and right homology arms on either side of the Z815 cut site of 253 and 4,434 bp, respectively (Townsend et al., 2009). Gene targeting was achieved at frequencies ranging from 0.2% (for introducing six nucleotide substitutions 1,541 bp from the cut site) to 4.0% (for introducing eight substitutions 188 bp away). In the case of TALEN T30, a donor with left and right homology arms of 1,563 and 3,129 bp, respectively, resulted in targeted YFP insertion at a frequency of 14% 99 bp from the cleavage site. We believe that these differences in frequencies of gene targeting are not the consequence of the sequence-specific nucleases but rather the length of the homology arms and the number, type, and distribution of sequence differences between the donor and target. Remarkably, we could recover gene-targeting events in 4% of calli derived from cells treated with TALEN T30 and a 322-bp donor that incorporated six nucleotide changes. A systematic study using multiple nucleases and donor molecules with varying lengths of homology arms and types of sequence modifications to be incorporated into the target locus would help better establish best practices for gene targeting in plants. In the mean time, we typically engineer nucleases to cut as close to the site of modification as possible, something that can easily be accomplished with TALENs, and we use homology arms between 750 and 1,000 bp.
In our previous work using ZFNs to introduce sequence modifications at the Sur loci, we observed that approximately 20% of the homologous recombination events were associated with nearby mutations (Wright et al., 2005). This was consistent with the repair of DNA double-strand breaks by synthesis-dependent strand annealing, in which occasionally both homologous recombination and NHEJ are used for break repair (Orel et al., 2003; Puchta, 2005). Among 12 clones sequenced from cells with YFP insertions, we observed two in which NHEJ-induced mutations were present at the nuclease cleavage site on one side of the insertion, consistent with repair by synthesis-dependent strand annealing. Because the donor molecule had an intact TALEN recognition site, however, it is also possible that these events arose by first repairing the targeted break through homologous recombination and then cleaving the recombinant chromosome again and repairing the break imprecisely by NHEJ. A more comprehensive analysis of repair products will have to be undertaken to assess the fidelity of recombination and whether the incidence of such NHEJ-induced mutations can be reduced by altering the TALEN recognition site on the donor template.
Many plant species are genetically modified using Agrobacterium or physical methods to deliver DNA to plant tissues (Barampuram and Zhang, 2011). A recently described method for in planta gene targeting suggests that efficient recombination can be achieved by first integrating the donor molecule in the chromosome using one of these transformation strategies (Fauser et al., 2012). The donor is flanked with nuclease recognition sites such that expression of the nuclease cleaves both the target and releases the donor from the chromosome, resulting in high-frequency homologous recombination. The targeting range of TALENs makes them ideally suited for such in planta gene-targeting strategies.
For those species that can be transformed and regenerated from protoplasts, the data provided in this study suggest that it is now possible to readily recover targeted mutations, insertions, and gene replacements. Furthermore, transient expression of TALENs is sufficient for high-frequency genome modification, as many of the modified cells we analyzed did not have evidence of an integrated TALEN construct. Of course, not all plants can be regenerated from protoplasts, but the genomes of amenable species, including rice (Oryza sativa), tomato (Solanum lycopersicum), potato (Solanum tuberosum), canola (Brassica napus), and sugarcane (Saccharum officinarum), can now be altered in a variety of ways to both study gene function and engineer plants with new traits.
CONCLUSION
We report here optimal TALEN architectures and expression strategies for high-frequency gene knockout, insertion, and replacement in plants. All reagents are compatible with our previously described TALEN assembly platform (Cermak et al., 2011), thereby providing a seamless pipeline for TALEN assembly, validation, and expression in plant cells. Furthermore, we believe that our data provide a framework for the engineering of plant genomes with TALENs using Agrobacterium or the direct delivery of DNA to plant tissues.
MATERIALS AND METHODS
Plasmid Construction
TALENs were constructed using the Golden Gate Assembly method described previously (Cermak et al., 2011). The TALEN expression vectors, pTAL3 and pTAL4, are identical with the exception of the yeast selectable markers (HIS3 and LEU2, respectively). The TALEN coding sequences in both were modified to have different N- and C-terminal truncations flanking the TAL effector repeat array using standard cloning procedures. The truncations included NΔ152/C18, NΔ152/C28, and NΔ152/C63. The plasmids are listed in Supplemental Table S1. DNA sequences of the TALEN target sites T30, T41, and T50 and the TAL repeat-variable diresidue arrays are provided in Figure 1. The TALEN plasmids targeting these sites are listed in Supplemental Table S1. Sequences of the TALEN plasmids are available upon request.
A Gateway-compatible entry plasmid, pZHY013, was created using PCR8 (Invitrogen) for transient expression of TALENs in plants. This plasmid contains two heterodimeric FokI nuclease domains (Miller et al., 2007) separated by a T2A translational skipping sequence (Halpin et al., 1999). TAL arrays in the plasmids from Supplemental Table S1 are released by digestion with XbaI/BamHI: one array (left array) is first cloned into pZHY013 as an XbaI/BamHI fragment; the other array (right array) is then cloned into NheI/BglII sites, which have ends compatible with XbaI and BamHI. Lastly, a Gateway LR reaction (Life Technologies) is performed to move the TALENs into the destination vector, pZHY051, which has a 35S promoter and a NOS terminator to drive expression of the TALEN pair. The resulting plasmids are listed in Supplemental Table S2.
Plasmids for the YFP SSA assay include the SSA reporter and a positive control. The positive control plasmid, pZHY162, contains the 721-bp YFP coding region between the 35S promoter and the NOS terminator. The YFP SSA reporter, pZHY402, is a derivative of pZHY162 and has a 255-bp internal sequence duplication separated by restriction sites. These restriction sites, BglII and SpeI, allow one to clone double-stranded oligonucleotides that contain the TALEN pair binding sites with single-stranded overhangs complementary to the BglII and SpeI overhangs.
Donor plasmids were generated to measure gene targeting. One donor plasmid, pZHY417, is used for targeted insertion of the YFP coding sequence into SurA or SurB (Fig. 3A). pZHY417 is a derivative pDW1927 (Townsend et al., 2009) and contains the YFP coding sequence fused in frame with the SurB coding sequence. On either side of the YFP coding sequence are SurB homology arms of 1,563 and 3,129 bp upstream and downstream, respectively. pZHY_WL, a second donor plasmid, introduces 6 bp into the SurB coding sequence to aid in the identification of gene-targeting events (Fig. 4C). This plasmid contains a 322-bp fragment of the SurB coding sequence with the 6-bp nucleotide change identical to those in pDW1927. Sequences of these plasmids are available upon request.
TALEN Activity in Tobacco Protoplasts
The protoplast-based SSA assay was developed to test TALEN activity in plant cells. Tobacco (Nicotiana tabacum) protoplasts were isolated from young leaves of approximately 4-week-old plants (cv Xanthi) grown in a growth chamber (Conviron) at 22°C with 16 h of daylight. The protocol for the protoplast isolation procedure was based on previous work (Yoo et al., 2007). Approximately 10 fully expanded tobacco leaves were harvested and sliced into 1- to 2-mm strips with a sharp razor. Leaf strips were transferred to enzyme solution (1.0% cellulase R10, 0.25% macerozyme R10, 0.45 m mannitol, 20 mm MES, 20 mm KCl, 10 mm CaCl2, and 0.1% bovine serum albumin) and incubated 12 h at 25°C and 40 rpm in the dark. The digested product was filtered through a 100-µm cell strainer onto a 10-cm petri plate, and the filtrate was transferred into a sterile 50-mL polypropylene tube containing 10 mL of washing buffer (0.45 m mannitol + 10 mm CaCl2). The sample was centrifuged for 5 min at 100g at room temperature. The supernatant was then removed, and the protoplast pellet was resuspended in 5 mL of washing buffer and transferred to a new 15-mL tube. Protoplasts were further purified by adding 8 mL of 0.55 m Suc solution and centrifuging for 5 min at 1,000g at room temperature. The protoplasts floating on the top were transferred into a 50-mL tube containing 10 mL of wash buffer and centrifuged for another 5 min at 100g at room temperature. After removal of the supernatant, the protoplast pellet was gently resuspended in 5 mL of wash buffer. The density of living protoplasts was determined using a hemocytometer as described previously (Yoo et al., 2007). Protoplasts were collected by centrifuging for 2 min at 100g at room temperature and resuspended with 4 m mannitol, 15 mm MgCl2, and 4 mm MES to the desired density (106 mL−1).
Protoplast transformation was performed based on the protocol described previously (Yoo et al., 2007) with slight modification. A total of 200 µL of protoplast solution (containing 2 × 10−5 protoplasts) was gently mixed with 30 µL of plasmid DNA and 230 μL of 40% polyethylene glycol transformation buffer. After a 10-min incubation at room temperature, transformation was stopped by adding 900 µL of wash buffer. Protoplasts were then collected by centrifuging for 5 min at 200g at room temperature and washed one more time with 800 µL of wash buffer by centrifuging for another 5 min at 200g. Protoplasts were resuspended in 1 mL of K3/G1 medium (Elzen et al., 1985) and transferred to a six-well culture plate. The plate was placed in the dark at room temperature for 20 to 24 h prior to flow cytometry. The growth of protoplasts into calli was performed as described previously (Elzen et al., 1985). To account for the differences in size of the various TALEN architectures, different amounts of each plasmid were used to ensure equal molar concentration in the transformations. A total of 20 µg of SSA reporter or donor plasmids was cotransformed with 3 pmol of nuclease plasmids in protoplast SSA assays or gene-targeting experiments.
Flow Cytometry
YFP-positive cells were quantified by flow cytometry using a FACSCanto II (Becton Dickinson) equipped with a 488-nm solid sapphire 20-mW laser for excitation. YFP fluorescence was detected with a fluorescein isothiocyanate 530/30-nm band-pass filter, and red spectrum autofluorescence from living protoplasts (due to chlorophyll) was detected with a 670-nm long-pass filter in the PcrCP channel. The forward scatter and side light scatter detectors were set to 130 and 250 V, respectively. For each sample, 20,000 protoplasts were analyzed and gated according to YFP and red spectrum autofluorescence values. The gate boundaries were defined using negative controls (protoplasts that were transformed with a target plasmid alone). Data were analyzed by FlowJo (Tree Star).
High-Throughput DNA Sequencing
Genomic DNA was extracted from transformed protoplast populations using the DNeasy Plant Mini Kit (Qiagen). TALEN target sites were PCR amplified, and PCR conditions and sequences of primers with barcodes for sequencing are available upon request. PCR products were purified with the AMPure XP PCR purification kit (Beckman Coulter) and sequenced on a GS FLX+ System (Roche Diagnostics). Sequence reads were aligned with wild-type target DNA sequences, and those reads with deletions or insertions greater than 1 bp in the spacer region between the two TALEN binding sites were considered as TALEN-induced mutations. The mutagenesis frequency was calculated as the number of reads with deletions or insertions divided by the total number of reads. Pearson’s product-moment correlation was used to assess relationships between the frequencies of NHEJ-induced mutations and the SSA data.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Target sites and TALEN architectures used in this study.
Supplemental Figure S2. The YFP SSA assay for measuring nuclease activity in protoplasts.
Supplemental Figure S4. Strategies for expressing sequence-specific nucleases.
Supplemental Figure S5. Evidence for repair of breaks by both HR and NHEJ.
Supplemental Figure S6. Analysis of mutant calli for integration of the TALEN expression construct.
Supplemental Table S1. Golden Gate-compatible TALEN plasmids with different architectures.
Supplemental Table S2. Nuclease expression vectors targeting SurB.
Glossary
- TALEN
transcription activator-like effector nuclease
- NHEJ
nonhomologous end joining
- ALS
acetolactate synthase
- ZFN
zinc finger nuclease
- YFP
yellow fluorescent protein
- SSA
single-strand annealing
References
- Arnould S, Delenda C, Grizot S, Desseaux C, Pâques F, Silva GH, Smith J. (2011) The I-CreI meganuclease and its engineered derivatives: applications from cell modification to gene therapy. Protein Eng Des Sel 24: 27–31 [DOI] [PubMed] [Google Scholar]
- Barampuram S, Zhang ZJ. (2011) Recent advances in plant transformation. Methods Mol Biol 701: 1–35 [DOI] [PubMed] [Google Scholar]
- Bogdanove AJ, Voytas DF. (2011) TAL effectors: customizable proteins for DNA targeting. Science 333: 1843–1846 [DOI] [PubMed] [Google Scholar]
- Carroll D. (2011) Genome engineering with zinc-finger nucleases. Genetics 188: 773–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF. (2011) Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res 39: e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Bogdanove AJ, Voytas DF. (2010) Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 186: 757–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtin SJ, Voytas DF, Stupar RM. (2012) Genome engineering of crops with designer nucleases. Plant Genome 5: 42–50 [Google Scholar]
- Davey MR, Anthony P, Power JB, Lowe KC. (2005) Plant protoplasts: status and biotechnological perspectives. Biotechnol Adv 23: 131–171 [DOI] [PubMed] [Google Scholar]
- Doyle EL, Booher NJ, Standage DS, Voytas DF, Brendel VP, Vandyk JK, Bogdanove AJ. (2012) TAL Effector-Nucleotide Targeter (TALE-NT) 2.0: tools for TAL effector design and target prediction. Nucleic Acids Res 40: W117–W122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elzen PJM, Townsend JA, Lee KY, Bedbrook JR. (1985) A chimaeric hygromycin resistance gene as a selectable marker in plant cells. Plant Mol Biol 5: 299–302 [DOI] [PubMed] [Google Scholar]
- Fauser F, Roth N, Pacher M, Ilg G, Sánchez-Fernández R, Biesgen C, Puchta H. (2012) In planta gene targeting. Proc Natl Acad Sci USA 109: 7535–7540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpin C, Cooke SE, Barakate A, El Amrani A, Ryan MD. (1999) Self-processing 2A-polyproteins: a system for co-ordinate expression of multiple proteins in transgenic plants. Plant J 17: 453–459 [DOI] [PubMed] [Google Scholar]
- Huang P, Xiao A, Zhou M, Zhu Z, Lin S, Zhang B. (2011) Heritable gene targeting in zebrafish using customized TALENs. Nat Biotechnol 29: 699–700 [DOI] [PubMed] [Google Scholar]
- Li T, Huang S, Jiang WZ, Wright D, Spalding MH, Weeks DP, Yang B. (2011) TAL nucleases (TALNs): hybrid proteins composed of TAL effectors and FokI DNA-cleavage domain. Nucleic Acids Res 39: 359–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Liu B, Spalding MH, Weeks DP, Yang B. (2012) High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat Biotechnol 30: 390–392 [DOI] [PubMed] [Google Scholar]
- Mahfouz MM, Li L, Shamimuzzaman M, Wibowo A, Fang X, Zhu JK. (2011) De novo-engineered transcription activator-like effector (TALE) hybrid nuclease with novel DNA binding specificity creates double-strand breaks. Proc Natl Acad Sci USA 108: 2623–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JC, Holmes MC, Wang J, Guschin DY, Lee YL, Rupniewski I, Beausejour CM, Waite AJ, Wang NS, Kim KA, et al. (2007) An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol 25: 778–785 [DOI] [PubMed] [Google Scholar]
- Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, et al. (2011) A TALE nuclease architecture for efficient genome editing. Nat Biotechnol 29: 143–148 [DOI] [PubMed] [Google Scholar]
- Mussolino C, Morbitzer R, Lütge F, Dannemann N, Lahaye T, Cathomen T. (2011) A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res 39: 9283–9293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orel N, Kyryk A, Puchta H. (2003) Different pathways of homologous recombination are used for the repair of double-strand breaks within tandemly arranged sequences in the plant genome. Plant J 35: 604–612 [DOI] [PubMed] [Google Scholar]
- Puchta H. (2005) The repair of double-strand breaks in plants: mechanisms and consequences for genome evolution. J Exp Bot 56: 1–14 [DOI] [PubMed] [Google Scholar]
- Reyon D, Tsai SQ, Khayter C, Foden JA, Sander JD, Joung JK. (2012) FLASH assembly of TALENs for high-throughput genome editing. Nat Biotechnol 30: 460–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander JD, Cade L, Khayter C, Reyon D, Peterson RT, Joung JK, Yeh JR. (2011a) Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nat Biotechnol 29: 697–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander JD, Dahlborg EJ, Goodwin MJ, Cade L, Zhang F, Cifuentes D, Curtin SJ, Blackburn JS, Thibodeau-Beganny S, Qi Y, et al. (2011b) Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA). Nat Methods 8: 67–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla VK, Doyon Y, Miller JC, DeKelver RC, Moehle EA, Worden SE, Mitchell JC, Arnold NL, Gopalan S, Meng X, et al. (2009) Precise genome modification in the crop species Zea mays using zinc-finger nucleases. Nature 459: 437–441 [DOI] [PubMed] [Google Scholar]
- Sun N, Liang J, Abil Z, Zhao H. (2012) Optimized TAL effector nucleases (TALENs) for use in treatment of sickle cell disease. Mol Biosyst 8: 1255–1263 [DOI] [PubMed] [Google Scholar]
- Tesson L, Usal C, Ménoret S, Leung E, Niles BJ, Remy S, Santiago Y, Vincent AI, Meng X, Zhang L, et al. (2011) Knockout rats generated by embryo microinjection of TALENs. Nat Biotechnol 29: 695–696 [DOI] [PubMed] [Google Scholar]
- Townsend JA, Wright DA, Winfrey RJ, Fu F, Maeder ML, Joung JK, Voytas DF. (2009) High-frequency modification of plant genes using engineered zinc-finger nucleases. Nature 459: 442–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tranel P, Wright T. (2002) Resistance of weeds to ALS-inhibiting herbicides: what have we learned? Weed Sci 50: 700–712 [Google Scholar]
- Wright DA, Townsend JA, Winfrey RJ, Jr, Irwin PA, Rajagopal J, Lonosky PM, Hall BD, Jondle MD, Voytas DF. (2005) High-frequency homologous recombination in plants mediated by zinc-finger nucleases. Plant J 44: 693–705 [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]