Abstract
The Arabidopsis (Arabidopsis thaliana) genome is the most well-annotated plant genome. However, transcriptome sequencing in Arabidopsis continues to suggest the presence of polyadenylated (polyA) transcripts originating from presumed intergenic regions. It is not clear whether these transcripts represent novel noncoding or protein-coding genes. To understand the nature of intergenic polyA transcription, we first assessed its abundance using multiple messenger RNA sequencing data sets. We found 6,545 intergenic transcribed fragments (ITFs) occupying 3.6% of Arabidopsis intergenic space. In contrast to transcribed fragments that map to protein-coding and RNA genes, most ITFs are significantly shorter, are expressed at significantly lower levels, and tend to be more data set specific. A surprisingly large number of ITFs (32.1%) may be protein coding based on evidence of translation. However, our results indicate that these “translated” ITFs tend to be close to and are likely associated with known genes. To investigate if ITFs are under selection and are functional, we assessed ITF conservation through cross-species as well as within-species comparisons. Our analysis reveals that 237 ITFs, including 49 with translation evidence, are under strong selective constraint and relatively distant from annotated features. These ITFs are likely parts of novel genes. However, the selective pressure imposed on most ITFs is similar to that of randomly selected, untranscribed intergenic sequences. Our findings indicate that despite the prevalence of ITFs, apart from the possibility of genomic contamination, many may be background or noisy transcripts derived from “junk” DNA, whose production may be inherent to the process of transcription and which, on rare occasions, may act as catalysts for the creation of novel genes.
The advent of tiling arrays and high-throughput sequencing has led to the discovery of a complex transcriptional landscape in eukaryotic genomes. Studies in yeast (Saccharomyces cerevisiae; David et al., 2006), animals (Bertone et al., 2004; Carninci et al., 2005), and plants (Yamada et al., 2003; Li et al., 2007; Matsui et al., 2008) have revealed the presence of a large number of unannotated, novel transcripts. These novel transcripts may represent alternatively spliced forms of known genes (Filichkin et al., 2010), products of antisense (Yamada et al., 2003) or bidirectional transcription (Xu et al., 2009), retained introns (Ner-Gaon et al., 2004; Filichkin et al., 2010), transcript fusions (Ruan et al., 2007), or intergenic transcriptional units (referred to hereafter as intergenic transcribed fragments [ITFs]). Among these novel transcripts, ITFs are unique in that they do not overlap with known genomic features and may represent novel genic sequences. The prevalence of intergenic transcription raises the possibility that there are many more functional genes yet to be discovered. However, there are two outstanding questions regarding ITFs. First, it is not clear what proportion of ITFs code for proteins. Second, whether most ITFs are functional is under debate (Mattick, 2009; Ponting and Belgard, 2010).
After ITFs are identified with whole-genome tiling arrays or high-throughput sequencing, computational methods are used to determine if they display characteristics of noncoding RNA (ncRNA; Fahlgren et al., 2007; Li et al., 2007; Gregory et al., 2008). These methods rely on secondary structure prediction, similarity to known ncRNAs, and conservation between species. The protein-coding potential of ITFs, on the other hand, is determined based on ab initio gene prediction, open reading frame (ORF) length, evolutionary conservation measures, pairwise alignment scores, predicted secondary structure, and entropy (Nekrutenko et al., 2002; Liu et al., 2006; Dinger et al., 2008). For example, in a global gene expression study in Arabidopsis (Arabidopsis thaliana), a 50-amino acid length threshold was used to define potential protein-coding intergenic transcripts (Stolc et al., 2005). Similarly, the Functional Annotation of the Mammalian Genome consortium defined putative protein-coding mRNAs using an ORF length cutoff of 300 nucleotides (Okazaki et al., 2002). Reliance on length cutoffs can result in longer random ORFs being falsely annotated as protein coding and can also lead to the exclusion of true small ORFs such as those that have been identified in yeast, humans, and Arabidopsis (Basrai et al., 1997; Hanada et al., 2007; Pruitt et al., 2007). Proteomics and polyribosome immunoprecipitation (Zanetti et al., 2005; Sparkes et al., 2006) allow more direct identification of potentially protein-coding ITFs than computational approaches. Currently, there has yet to be a systematic assessment of ITF protein-coding potential based on a combination of computational and experimental approaches.
In addition to the question of whether ITFs code for proteins, the functional relevance of intergenic transcription is not well understood. One hypothesis is that most transcripts simply represent transcriptional noise. For example, based on the genome-wide distribution of RNA polymerase II and TATA-binding protein in yeast, approximately 90% of RNA polymerase II transcriptional initiation events were estimated to be the result of low polymerase fidelity and may represent transcriptional noise (Struhl, 2007). Consistent with the “noise” hypothesis, several studies have shown that ITFs tend to have significantly higher evolutionary rates than known genes. For example, the Encyclopedia of DNA Elements (ENCODE) consortium found that 93% of the unannotated transcribed regions in the human genome show no clear evidence of evolutionary constraint (Birney et al., 2007). The alternative hypothesis is that most ITFs are functional (Dinger et al., 2009). Differential expression, alternative splicing, and/or association with chromatin modification marks have been cited as evidence for ITF functionality (Guttman et al., 2009; Hiller et al., 2009). In addition, the functions of a growing number of novel transcripts have been experimentally determined. Examples include Xist, RepA, Air, and Hotair, which regulate the recruitment of Polycomb proteins onto DNA (Mercer et al., 2009), as well as a recently discovered long noncoding RNA called COLDAIR shown to be important in regulating vernalization responses in Arabidopsis (Heo and Sung, 2011). Based on these studies, it is clear that some ITFs are functional. The main question concerns the abundance of functional ITFs relative to those derived from noisy transcription.
To date, most studies of intergenic transcription have focused on the presumably noncoding fraction of the transcriptome. In addition, currently, there is no published study assessing the evolutionary significance of plant intergenic transcription. In this study, we focused on intergenic polyadenylated (polyA) RNA transcripts to gain more insight into the nature of plant intergenic transcription by RNA polymerase II. We first analyzed eight different Arabidopsis mRNA sequencing (mRNA-seq) data sets from this study and two other sources (Filichkin et al., 2010; Jiao and Meyerowitz, 2010) to determine the extent of intergenic polyA transcription. We then investigated whether ITFs are likely protein coding using (1) ribosome immunoprecipitation data generated in this study as well as public data sets (Jiao and Meyerowitz, 2010), (2) proteomics data (Baerenfaller et al., 2008; Castellana et al., 2008), and (3) fusion protein expression studies on selected targets. Finally, making use of the polymorphism data from 80 different Arabidopsis accessions (Cao et al., 2011) and protein-coding genes and genome sequences of other plants, we explored whether ITFs, especially those that may code for proteins, are likely functional based on within- and cross-species conservation.
RESULTS AND DISCUSSION
Defining Transcribed Regions in the Arabidopsis Genome
To explore the functional significance of intergenic transcription further, a rigorous definition of transcribed regions within the Arabidopsis genome is necessary. To this end, we analyzed mRNA-seq data from three different sources: (1) 7-d-old seedlings generated in this study, (2) whole flower (Jiao and Meyerowitz, 2010), and (3) 12-d-old seedlings grown under six environmental conditions (Filichkin et al., 2010; Supplemental Table S1). We assembled transcript fragments (TxFrags) using two approaches (see “Materials and Methods”). In the first approach, contiguous regions in Arabidopsis occupied by mapped mRNA-seq reads were defined as expressed (Set 1 TxFrags). In the second, more stringent approach, we assembled TxFrags using the transcript assembly program Cufflinks (Set 2 TxFrags).
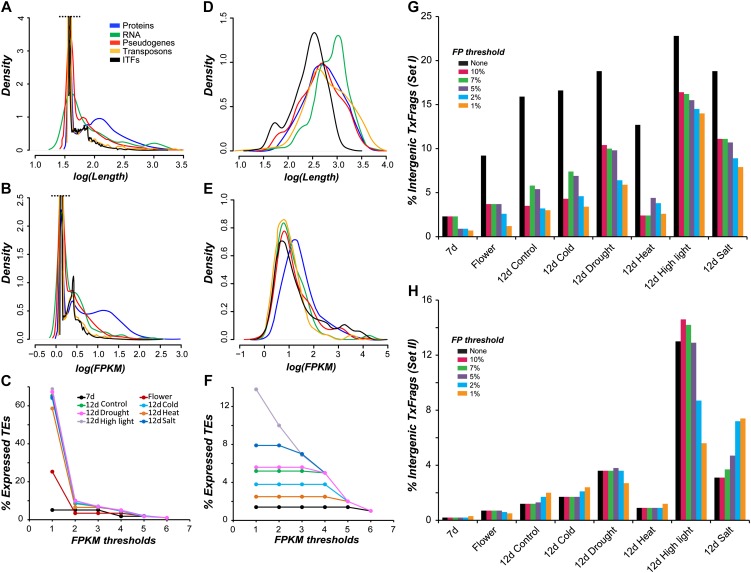
We first compared the characteristics of Set 1 TxFrags among annotated features including protein-coding genes, RNA genes, pseudogenes, and transposons. Regardless of the genomic feature and data set, the Set 1 TxFrag length distributions are bimodal with the first peaks located near the mRNA-seq single read length, indicating that most Set 1 TxFrags consist of a single read (Fig. 1A; Supplemental Fig. S1). Next, the fragments per kilobase of exon model per million mapped reads (FPKM) measure was used to assess Set 1 TxFrag expression level. Similar to length distributions, the Set 1 TxFrag FPKM distributions are bimodal with the first peaks at very low FPKM, mostly consisting of single-read TxFrags (Fig. 1B; Supplemental Fig. S2). The likely sources of low-FPKM TxFrags are (1) genes with very low-level or highly specific expression, (2) “transcriptional noise” representing background genome transcription (Struhl, 2007), or (3) low-level genomic DNA contamination in the sequenced mRNA sample. If the presence of one or more Set 1 TxFrags is considered evidence of expression, 78% to 94% of protein-coding genes are expressed. However, 19% to 68% of pseudogenes and 5% to 69% of transposons would be considered expressed based on the same criterion (Supplemental File S1A). Given that Arabidopsis transposons have been documented to be underexpressed (Schmid et al., 2005) and subject to strong posttranscriptional silencing through DNA methylation (Zhang et al., 2006; Zilberman et al., 2007), transposon expression was used as a conservative error estimate of expression calls.
Figure 1.
Characteristics of Set 1 and Set 2 TxFrags. A and B, Length (A) and expression level distributions (B) of various genomic features, proteins (blue), RNA (green), pseudogenes (red), transposons (orange), and ITFs (black), based on Set 1 TxFrags identified across all eight RNA-seq data sets. Both axes are logarithmically scaled with base 10. To emphasize the lower peaks, curves beyond the black dashed line are truncated. C, Percentage of transposons considered expressed based on Set 1 TxFrags identified from eight data sets at various FPKM thresholds. For details of other features, see Supplemental Table S1. D and E, Length (D) and expression level distributions (E) for Set 2 TxFrags. F, Percentage of transposons considered expressed based on Set 2 TxFrags. G and H, Percentage of TxFrags defined as intergenic at different FPKM thresholds among data sets. Set 1 TxFrags (G) and Set 2 TxFrags (H) were identified as intergenic without an FPKM threshold (black) and at progressively more stringent FPKM thresholds according to transposon-based false-positive (FP) rates of 1%, 2%, 5%, 7%, and 10%. The x axis indicates the data sets used to identify TxFrags. The y axis represents the percentage of true-positive TxFrags that are intergenic at each FP threshold. Note that the percentage did not monotonically decrease because some TxFrags overlapping with annotated features also were filtered out when false-positive thresholds were applied.
To stringently control for false positives arising from background transcription and/or low-level genomic contamination, we applied multiple FPKM thresholds defined according to the percentage of transposon TxFrags considered expressed (Fig. 1C; Supplemental File S1A). Comparing percentage of transposons expressed, we found that the FPKM thresholds have significantly different impacts on data sets (Fig. 1C). For example, an FPKM threshold based on the 90th percentile of the transposon expression distribution results in a 57.4% reduction of transposon expression in the 12-d seedling drought stress data set compared with no FPKM threshold but causes no reduction in the 7-d data (Fig. 1C). This difference in the degree of transposon expression due to FPKM threshold choice is not simply due to differences in sequencing depth, as the numbers of mapped reads are both approximately 4.8 × 106 (Supplemental Table S1). In addition, this difference cannot be attributed to stress treatments, as degrees of transposon expression in the stress treatment and control samples are similar regardless of FPKM thresholds (Fig. 1C, green). We note that only 22% to 38% of reads from the 12-d data sets can be mapped to the Arabidopsis genome compared with 71% and 75% for 7-d and flower data, respectively. Thus, data quality may significantly impact gene expression calls, even after quality filtering and mapping the reads to the genome.
For comparison, we applied a second, more stringent transcript assembly approach using Cufflinks with bias corrections of transcript models based on sequences, positions, and abundance (Trapnell et al., 2010) to generate Set 2 TxFrags. Compared with Set 1 TxFrags, Set 2 TxFrags are significantly longer (Fig. 1D; Kolmogrov-Smirnov [KS] test, P < 2.2e-16) and have significantly higher FPKM values (Fig. 1E; KS test, P < 2.2e-16). In addition, Set 2 TxFrags length and coverage distributions overlap with the right tails of Set 1 TxFrags (Fig. 1, A, B, D, and E), indicating that the main difference between these two sets is enrichment for longer and more abundant transcripts in Set 2 TxFrags. Increasingly stringent FPKM thresholds still have a significant effect on the numbers of transposons considered expressed for several 12-d data sets (Fig. 1F). Nonetheless, the second approach allows for better control in calling transposon expression, which we considered to be mostly false positive, than the first, simpler approach.
Pervasiveness of Transcription in Arabidopsis Intergenic Regions
Previous microarray-based studies in Arabidopsis have shown that a large number of polyA transcripts are produced from the intergenic regions of the genome (Yamada et al., 2003; Matsui et al., 2008). Considering the advantages of RNA-seq over microarrays for expression studies (Agarwal et al., 2010), we reassessed the preponderance of intergenic transcription using RNA-seq data sets. Here, TxFrags located within intergenic regions are referred to as ITFs. We found that the analysis method (Set 1 versus Set 2), FPKM threshold, and data set significantly influence estimates of ITF abundance (Fig. 1, G and H; Supplemental File S1). For example, 9.2% of Set 1 TxFrags from the flower data are considered ITFs when no FPKM threshold is applied, but this proportion drops to 3.7% with an FPKM threshold of 1.33, which corresponds to a 10% false-positive rate (Fig. 1G). Comparing between data sets by allowing a 10% false-positive rate, ITF estimates differ by 7-fold (2.3%–16.4%) and 73-fold (0.2%–14.6%) based on Set 1 TxFrags and Set 2 TxFrags, respectively (Supplemental File S1). Despite these differences, there are two consistent characteristics among data sets that separate Set 1 and Set 2 ITFs. Set 1 ITFs tend to be significantly shorter than Set 2 ITFs (Fig. 1, A and D; KS test, P < 2.2e-16). In addition, Set 1 ITF expression levels are not significantly different from those of Set 1 TxFrags (Fig. 1B; KS test, P = 0.28) but are significantly lower than protein-coding gene TxFrags (KS test, P < 2.2e-16). Set 2 ITFs have significantly lower expression levels than protein-coding gene TxFrags as well (Fig. 1E; KS test, P < 1e-2), although the pattern is not as pronounced as for Set 1 ITFs, presumably due to the bias corrections applied on the data set by Cufflinks. Our findings are consistent with earlier studies in Arabidopsis (Hanada et al., 2007; Matsui et al., 2008) and mammals (Wang et al., 2004; van Bakel et al., 2010), which found that intergenic sequences tend to be lowly expressed.
We next focused on Set 2 TxFrags, which represent a more stringently defined set of transcripts. Across data sets, 0.2% to 14.6% of TxFrags are potentially derived from intergenic transcription based on a 5% false-positive rate (Fig. 1H; Supplemental File S1B). This proportion corresponds to 10,511 ITFs across eight RNA-seq data sets, together representing 6,545 nonoverlapping intergenic transcribed genomic regions and spanning 3.6% of the assembled intergenic region in Arabidopsis. Our ITF estimate is comparable to an earlier tiling array-based study in Arabidopsis, where 7,719 unannotated transcriptional units were defined as novel, non-protein-coding RNAs (Matsui et al., 2008). Other studies have provided more conservative estimates of Arabidopsis intergenic expressed regions, from 104 (Stolc et al., 2005) to 2,397 (Yamada et al., 2003). In mammals, however, the ENCODE project as well as other studies have reported significantly more pervasive intergenic transcription (Bertone et al., 2004; Birney et al., 2007; Kapranov et al., 2007). The ENCODE project reported that 488,906 (22.6%) TxFrags lie in intergenic regions and that 93% of the ENCODE bases have transcription evidence (Birney et al., 2007). Compared with Arabidopsis (3.6%), a significantly larger proportion of the ENCODE region is transcribed, even if we consider Set 1 TxFrags (13.7% at a 5% false-positive rate) that are not as rigorously defined as Set 2 TxFrags.
There are several possible explanations for the differences in ITF pervasiveness between plants and humans. First, the ENCODE study analyzed transcripts obtained from 31 different cell lines and tissues, which represents a much broader sampling of the transcriptome than our study. Second, known issues with tiling arrays used in the ENCODE study, particularly cross hybridization (Agarwal et al., 2010; van Bakel et al., 2010), may lead to an overestimation of ITFs. Consistent with this possibility, a previous RNA-seq study of human 293T cell total RNA found that only approximately 4% of reads were intergenic (van Bakel et al., 2011), similar to our Arabidopsis estimate. Third, the intergenic space in Arabidopsis constitutes only approximately 40% of the genome, compared with approximately 99% in the human genome. If intergenic transcripts are largely derived from noisy transcription or genomic contamination, species with larger genomes may have more RNA-seq reads from intergenic space. The fourth reason may be that larger genomes have more functional elements. However, variation in genome size can be due to extreme proliferation of transposable elements (Hawkins et al., 2006; Piegu et al., 2006). Thus larger genomes do not necessarily contain more genes. Finally, elements of our experimental design, such as the use of tissue samples with multiple cell types or insufficient coverage, may lead to an underestimate of ITFs. To address some of the issues concerning our study design, we analyzed cell type-specific transcriptome data obtained using directional Illumina sequencing.
Factors Affecting ITF Estimates
The data sets we analyzed have the following limitations that may affect estimates of ITF abundance (Clark et al., 2011). First, all data sets were generated using complex tissue samples that may render cell type-specific ITFs undetectable. Second, the sequencing was performed using single reads without directionality information, which may result in misassembly of ITFs. Third, the read length and coverage may be insufficient for detecting ITFs expressed at low levels. To address these issues, we directionally sequenced polyA-selected RNA from T87 suspension culture cells with longer reads (72 bp) and greater depth (two to nine times more sequenced bases; approximately 2.3 Gb, approximately 3 × 107 reads). We found that 0.9% of reads and 4.2% of TxFrags (identified using the same criteria as Set 2 TxFrags) from the suspension culture data are intergenic (Supplemental Fig. S3), consistent with the proportions of intergenic reads and TxFrags identified from more complicated tissues (Supplemental Table S1). In addition, in the suspension cell data set, 3,052 (9.8%) TxFrags and 170 (13.1%) ITFs overlap with one or more other TxFrags and ITFs, respectively, that are in the opposite orientation. Thus, lack of read directionality information and misassembly can lead to an approximately 13% underestimate of ITFs that overlap in opposite orientations.
Another factor affecting the estimate of ITFs is that the data sets we have analyzed so far are derived from polyA RNA. Nonpolyadenylated (nonpolyA) RNA may constitute the bulk of the transcriptome and significantly contribute to intergenic expression (Cheng et al., 2005; Armour et al., 2009; Xu et al., 2010). However, an earlier study focusing on both polyA and nonpolyA RNAs in Arabidopsis found that 3.5% of reads are intergenic (Lister et al., 2008), which is comparable to the 0.4% to 8.2% reads that are intergenic in the mRNA-seq data sets we analyzed (Supplemental Table S1). In addition, a study of human 293T cell ribosomal RNA-depleted total RNA revealed that approximately 4% of reads were intergenic (van Bakel et al., 2011). This suggests that our estimates of intergenic transcription in Arabidopsis based on polyA RNA sequencing are reasonable. Nonetheless, detailed studies of nonpolyA ITFs will be necessary to estimate the contribution of nonpolyA transcripts to intergenic transcription.
Taken together, we have identified 6,545 ITFs (5% false-positive rate) that are likely novel transcriptional units not previously defined in the Arabidopsis genome. Two outstanding questions remain. First, because these ITFs are derived from polyA RNAs, are they parts of novel protein-coding or noncoding RNA genes? Second, do some of these ITFs have clear evidence of selection, therefore suggesting their functionality? To address the first question, we assessed the protein-coding potential of ITFs by analyzing ribosome-associated transcripts and shotgun proteomics data sets.
Distinguishing Coding from Noncoding Intergenic Transcripts Based on Ribosome Association
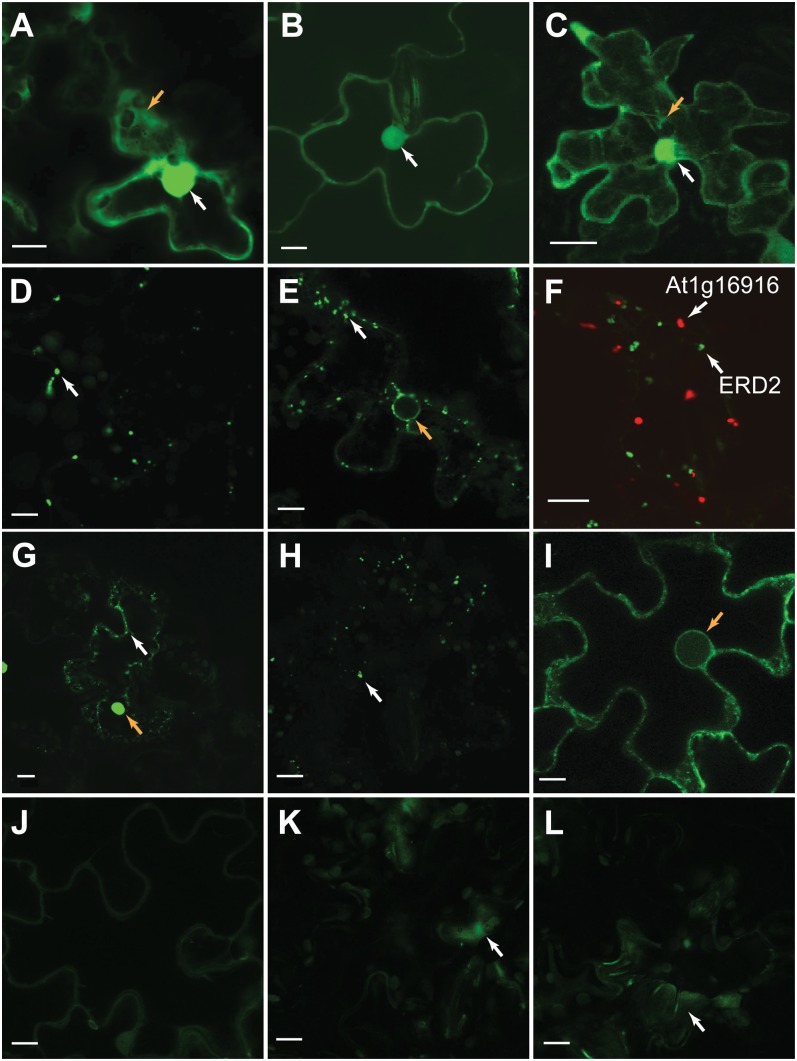
Translation initiation is the rate-limiting step in protein translation; therefore, transcripts associated with the ribosome are more likely to be translated (Kawaguchi and Bailey-Serres, 2002; Bailey-Serres et al., 2009). Studies in Arabidopsis (Branco-Price et al., 2008; Jiao and Meyerowitz, 2010), mouse (Doyle et al., 2008), and yeast (Ingolia et al., 2009) have taken advantage of this property to globally investigate translational regulation. To assess whether ribosome association of intergenic transcripts is a good measure of their translation potential, we first sequenced ribosome-associated transcripts from 7-d-old seedlings. After identifying the ribosome-associated transcript fragments (R-TxFrags), we selected eight genomic regions with evidence of ribosome association and seven without for in vivo translation studies (Supplemental Table S2). These regions overlap with putative small open reading frame (sORF) genes that were originally computationally predicted from intergenic regions (Hanada et al., 2007). Several of these regions have since been annotated solely based on computational predictions and/or complementary DNA (cDNA) evidence. The 5′ untranslated regions (UTRs) and coding sequences of the sORFs were fused in frame to a yellow fluorescent protein (YFP) reporter that lacks a translational start codon, and the translation of these sequences in transiently transformed tobacco (Nicotiana tabacum) leaf epidermal cells was evaluated (see “Materials and Methods”; Fig. 2).
Figure 2.
In vivo translation of predicted protein-coding sequences in transiently transformed tobacco leaf epidermal cells. A, Enhanced YFP (EYFP) is localized to the cytoplasm (orange arrow) and nucleus (white arrow). B and C, AT_3|+|1|14212973-14213269-EYFP (B) and AT_1|-|2|20126281-20126376-EYFP (C) have similar localization patterns. Nuclei are indicated by white arrows. In C, a series of 18 slices (1 µm each) was merged to highlight cytoplasmic strands (orange arrow). D and E, AT_1|-|2|5786755-5786853-EYFP appears to be vesicle localized (white arrow; D), similar to endoplasmic reticulum/Golgi marker ERD2-GFP (white arrow; E). F, AT_1|-|2|5786755-5786853-EYFP (recently annotated as At1g16916; red) does not colocalize with ERD2-GFP (green). G and H, AT_3|-|0|3663786-3663977-EYFP (G) and AT_3|+|2|4574607-4574900-EYFP (H) also have punctate expression patterns (white arrows). The orange arrow in G indicates potential aggregation of the AT_3|-|0|3663786-3663977-EYFP fusion protein. I, AT_1|+|1|11469497-11469754-EYFP appears to localize to the endoplasmic reticulum and nuclear envelope (orange arrow), similar to ERD2 (orange arrow in E). J to L, sORFs in a known noncoding small nucleolar RNA At1g12013 (J) and in an intron of a protein-coding gene, At1g43560 (K), are not translated based on a signal similar to a leaf infiltrated with A. tumefaciens lacking a fusion protein construct (L). Signal observed in K and L (white arrows) is likely due to cell damage. Bars = 10 µm. Names of all protein-coding sequences are as previously published (Hanada et al., 2007).
Of the eight genomic regions with R-TxFrag evidence, five were translated in tobacco, while only one of the seven regions without R-TxFrag support was translated (Fig. 2; Supplemental Table S2). Thus, there was a significant enrichment of sORFs with R-TxFrag evidence among those translated in vivo (Fisher’s exact test, P < 0.05). The observed localization patterns of the protein fusions were largely consistent with signal peptide predictions (Fig. 2; Supplemental Table S2), indicating that the fusion proteins were likely correctly translated and targeted in tobacco. However, three sORFs with ribosome association evidence do not appear to be translated in the transient expression assay. These sORFs may not be translated, or this may be an artifact due to the use of a heterologous system (tobacco). One translated sORF is an annotated “other RNA” gene (At1g31935; Fig. 2I; Supplemental Table S2). In addition, a number of annotated other RNA genes have either ribosome association or proteomics evidence, which highlights the importance of experimentally evaluating protein-coding potential. Overall, based on the findings of our in vivo translation assays, we conclude that features with evidence of ribosome association are more likely to be translated than those without.
Translation Evidence for ITFs
Given that ribosome association is a good indicator of the translation potential of intergenic sequences, we further analyzed R-TxFrags from the 7-d-old seedling data to estimate the proportion of ITFs likely to be parts of coding genes. To address potential issues due to sequencing coverage or tissue-specific expression and translation, R-TxFrags were also identified using ribosome-associated transcript data of whole flowers and specific floral domains expressing three homeotic genes (Jiao and Meyerowitz, 2010). For comparison, we also incorporated shotgun proteomics data from two studies examining protein expression in multiple tissues and developmental stages (Baerenfaller et al., 2008; Castellana et al., 2008). These data are collectively referred to as “translation data sets” and are summarized in Supplemental Table S3.
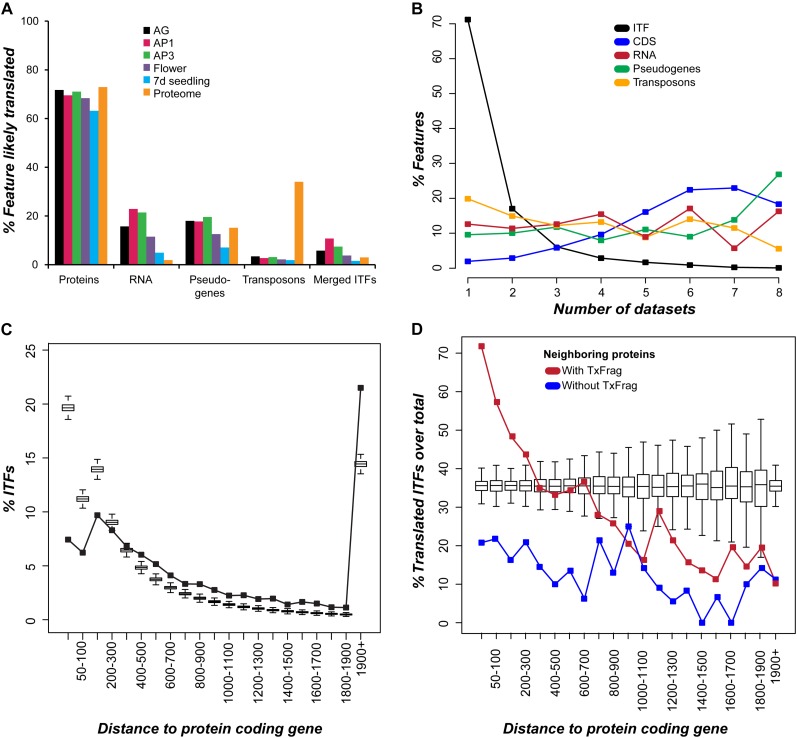
As with mRNA-seq, most R-TxFrags and proteomics tags mapped to previously annotated regions in the genome, particularly protein-coding genes. Among ribosome immunoprecipitation data sets, 63% to 73% of protein-coding genes have one or more R-TxFrags (Fig. 3A). Similarly, 74% of protein-coding genes have one or more proteomics tags (Fig. 3A). In addition, 62% to 67% of the R-TxFrags overlap with one or more proteomic tags. These findings demonstrate that ribosome-associated transcripts tend to be translated, consistent with our in vivo translation studies (Fig. 2). On the other hand, 5% to 23% of annotated ncRNA genes and 7% to 15% of pseudogenes have uniquely mapped R-TxFrags and/or proteomics tags. If all annotated RNA genes are truly noncoding, calling a feature translated based on a corresponding R-TxFrag and/or proteomics tag can have a 5% to 23% false-positive rate depending on the data set (Fig. 3A, RNA). One anomaly is that 34.0% of transposons have proteomics tags, although only 1.9% to 3.4% have R-TxFrags (Fig. 3A). This discrepancy is in sharp contrast to our finding that the proportions of protein-coding genes possessing R-TxFrags and proteomics tags are both approximately 70% (Fig. 3A). This observation, also noted in the original study (Castellana et al., 2008), is inconsistent with studies demonstrating reduced transcription of transposons (Schmid et al., 2005) and their extensive methylation (Zhang et al., 2006; Zilberman et al., 2007). Using the number of proteomics tags as a proxy for protein expression level, transposons with proteomics evidence tend to have significantly fewer tags than protein-coding genes (Supplemental Fig. S5; KS test, P < 2.2e-16). In addition, 67.6% of transposons with proteomics evidence have only one tag compared with 26.6% of protein-coding genes, suggesting that if the transposons are expressed and translated, it happens at significantly lower levels than protein-coding genes.
Figure 3.
Translation evidence, breadth, and distance of ITFs from neighboring genes. A, Percentage of features with overlapping translation evidence was calculated for protein-coding genes, RNA genes (excluding other RNA), pseudogenes, transposons, and ITFs obtained from the 7-d seedling and flower transcriptomes. Ribosome immunoprecipitation data are for AGAMOUS (AG), APETALA1 (AP1), AP3, flower, and 7-d seedling. Proteomics data are combined data from two studies. Only uniquely mapping R-TxFrags and proteomics tags were used as evidence. B, Breadth of expression (as indicated by the number of data sets where a feature can be found) of ITFs (black) and TxFrags mapped to protein-coding genes (blue), RNA genes (red), pseudogenes (green), and transposons (orange). CDS, Coding sequence. C, Distance distribution of ITFs to their nearest protein-coding genes. The box plots depict distance distributions between 10,000 sets of randomly sampled intergenic sequences and their nearest protein-coding genes. D, Percentage of translated ITFs over all ITFs in the same distance bin is shown as a function of distance to the nearest protein-coding gene. ITFs neighboring proteins with and without transcript evidence are represented by red and blue lines, respectively. Box plots represent the randomly expected proportions in each distance bin obtained by permuting the association between distance and presence/absence of translation evidence. The medians of random expectations are approximately 35%, because approximately 35% of ITFs have one or more pieces of translation evidence.
How many ITFs have evidence of translation? Among the 6,545 nonoverlapping ITFs identified from eight mRNA-seq data sets, 2,107 (32.2%) have one or more R-TxFrags from one or more of the translation data sets analyzed (Fig. 3A; Supplemental Fig. S4). Unlike protein-coding genes, there is substantially stronger support for ITF translation from ribosome association than from proteomics data (Fig. 3A). Some of the ribosome-associated ITFs may contain protein-coding regions even though there is no proteomics support, partly due to the fact that proteomics data tend to be biased toward more abundantly translated proteins (Baerenfaller et al., 2008). It is also likely that a significant number of ribosome-associated ITFs are derived from the UTRs of protein-coding transcripts. Taken together, even if the false-positive rate is 23%, approximately 1,622 ITFs are likely parts of transcripts destined to be translated after eliminating potential false positives. Thus, a significant number of intergenic transcripts may be part of larger protein-coding genes, either as coding sequences or as UTRs. Our finding highlights the importance of assessing the translational potential of polyA intergenic transcripts before defining them as sequences that function solely at the RNA level.
Relationship between ITFs and Neighboring, Annotated Genes
Based on an analysis of mRNA sequencing data, we uncovered thousands of short, low-abundance transcripts from intergenic regions. In addition, many of these ITFs are supported by translation evidence. One immediate question is whether these ITFs, translated or not, are extensions of previously annotated or novel protein-coding genes. To address this question, we assessed whether there is a significant bias in where ITFs are located within the Arabidopsis genome by calculating the distance between each ITF and its closest annotated protein-coding gene. We found that although a substantial number of ITFs are closer to genes, they are not any closer than intergenic sequences sampled randomly based on ITF number and size (Fig. 3C). This is contrary to the expectation that ITFs are predominantly extensions of existing genes.
Given that ITFs in general are not closer to neighboring genes than randomly selected intergenic sequences, do ITFs with translation evidence behave similarly? First, we found that translation evidence (proteomics tags and R-TxFrags) tends to lie farther away from protein-coding genes than random expectation (Supplemental Fig. S6A). However, ITFs with translation evidence tend to lie closer to genes than ITFs without translation evidence (Supplemental Fig. S6B), suggesting that most ITFs with translation evidence may be parts of neighboring protein-coding genes. If translated ITFs are indeed missing parts of annotated genes, ITFs closer to genes with transcription evidence should be enriched in the translated set compared with ITFs closer to nontranscribed genes. Consistent with this expectation, among the 4,942 ITFs closest to a transcribed annotated protein, 37.9% have translation evidence, while among the 563 ITFs closest to a nontranscribed annotated protein, only 14.2% have translation evidence (Fisher’s exact test, P < 2.2e-16; Fig. 3D). These observations suggest that most ITFs with translation evidence that are close to annotated genes may be missing parts of those genes or associated with the transcription of those genes via an unknown mechanism.
Taken together, we have demonstrated the presence of ITFs from more than 6,000 intergenic regions in Arabidopsis from multiple RNA sequencing data sets. More than 20% of these ITFs are likely translated or are part of protein-coding transcripts. Among the 6,545 ITFs, 59.4% are located more than 300 bp away from an annotated gene. Of these, 847 (21.7%) have translation evidence. Considering that 300 bp is approximately the 90th percentile of both Arabidopsis intron and UTR lengths, these relatively distant ITFs may be parts of novel transcriptional units. However, ITFs, in general, tend to be significantly shorter and expressed at lower levels than protein-coding genes. We also find that ITFs, in general, tend to be expressed narrowly, in a data set-specific manner, while TxFrags corresponding to annotated features are present in multiple data sets (Fig. 3B). The translation of approximately 32.2% of ITFs is supported by one or more ribosome immunoprecipitation and/or proteomics data sets, compared with 88.0%, 44.6%, and 36.9% for protein-coding genes, pseudogenes, and transposons, respectively. In terms of translation, ITFs behave similarly to pseudogenes and transposons. Previous studies have suggested that the breadth of expression as well as the level of expression can be considered as proxy indicators of functionality (Nuzhdin et al., 2004; Subramanian and Kumar, 2004; Movahedi et al., 2011). However, genes can have highly specific expression and/or low expression levels. Thus, one remaining question is whether these ITFs are parts of functional sequences with clear evidence of selection.
Evidence of Natural Selection on ITFs at the Nucleotide Level
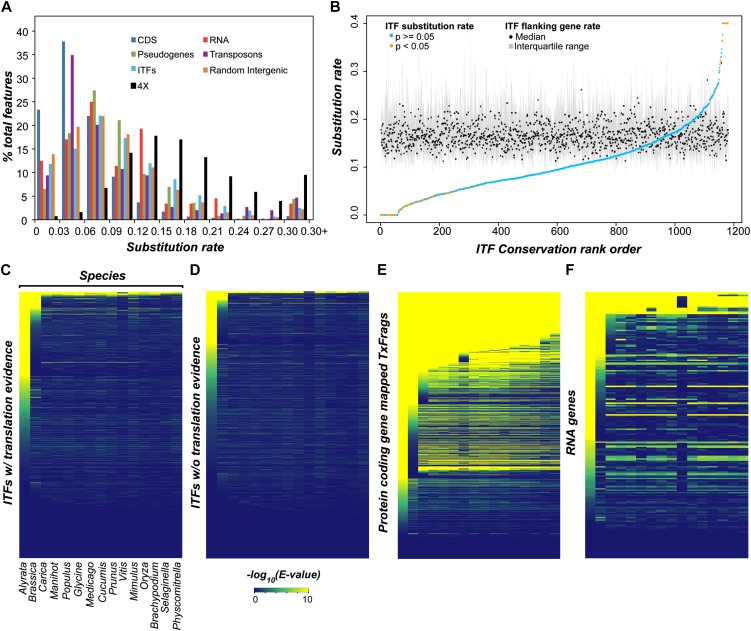
Intergenic transcripts that are independent transcriptional units may be derived from noisy, background transcription or unannotated genes that are functional (Struhl, 2007; Dinger et al., 2009; van Bakel et al., 2010, 2011; Clark et al., 2011). Transcripts not important to cellular function are expected to accumulate mutations much like neutrally evolving sequences. In contrast, functional ITFs should be selected for and show signs of nonneutral evolution. To assess whether there is a clear signature of natural selection that is indicative of functionality, the ITF nucleotide substitution rates were estimated using syntenic genomic regions of Arabidopsis and Arabidopsis lyrata, which diverged from their common ancestor approximately 10 million years ago (Hu et al., 2011). We also estimated the 4-fold degenerate site substitution rates of protein-coding orthologs as proxies for neutral evolution rates. Substitution rates for protein-coding genes, RNA genes, and randomly chosen intergenic regions not overlapping with ITFs were also estimated for comparison.
Among 6,545 ITFs, only 1,238 (18.9%) have identifiable syntenic regions for substitution rate estimation between the two Arabidopsis species. The proportion of syntenic ITFs is significantly lower than those of protein-coding genes (90.9%; Fisher’s exact test, P < 2.2e-16), RNA genes (35.7%; P < 9.1e-10), and pseudogenes (33.4%; P < 2.2e-16) but significantly higher than transposons (10.9%; P < 1.5e-16). Thus, many “orphan” ITFs without putative orthologs likely evolved rapidly with little or no selective constraint. For ITFs found within syntenic regions, substitution rates are significantly higher than those of annotated protein-coding genes (KS test, P < 2.2e-16; Fig. 4A). On the other hand, ITF substitution rates in general are significantly lower than those of 4-fold degenerate sites (KS test, P < 2.2e-16; Fig. 4A). These observations suggest that ITFs may constitute a mixed population, with the first population under strong selective constraint and the second one evolving neutrally. Using the 5th percentile of the 4-fold degenerate site rate distribution (rate = 0.07) as a threshold, only 6.4% of the 6,545 ITFs are likely under strong purifying selection. The remaining 93.6% are likely under little or no purifying selection. To control for local rate variation, we compared the rate of each ITF with the 4-fold site rates of neighboring genes. Based on this approach, a much smaller percentage, 2.7%, of ITFs were found to be under selection (Fig. 4B).
Figure 4.
Evolutionary conservation of ITF sequences. A, Between-species nucleotide substitution rate distributions of different features and 4-fold degenerate sites (4x). CDS, Coding sequence. B, Substitution rates of ITFs compared with local substitution rates of 4x sites. 4x sites of up to 60 neighboring protein-coding genes were used to determine the distributions of local substitution rates. Black circles indicate medians of the distributions, gray lines define the interquartile ranges, and each orange or blue circle indicates the substitution rate of the ITF in the given region. The ITFs are arranged from low to high z scores. An orange circle indicates a significant z score at P < 0.05, while a blue circle indicates P ≥ 0.05. C to F, Heat maps indicating degree of cross-species similarity of ITFs with translation evidence (C), ITFs without translation evidence (D), 10,000 randomly selected TxFrags mapped to annotated protein-coding genes (E), and all TxFrags mapped to annotated RNAs (F). TxFrags mapping to proteins and annotated RNAs were chosen based on the size distribution of the ITFs. Each row represents a feature, and each column represents the subject species for similarity search. The expect (E) values were converted to a negative logarithmic scale and adjusted to be between 0 and 10, with 0 (blue) indicating E ≥ 1 and 10 (yellow) indicating E ≤ 1e-10.
One issue in comparing any sequence feature with 4-fold sites is that there can be significant alignment bias. This is because a sequence feature (e.g. an ITF) is aligned to its putative ortholog at the nucleotide level, whereas 4-fold sites are identified from nucleotide sites originally aligned based on protein sequences. Given that the alignment process involves finding an alignment with the best score, regardless of whether the sites are homologous or not, it will tend to make a nucleotide-based alignment look more similar than it really is. Thus, the lower substitution rate among sequence features compared with 4-fold sites can simply be due to this artifact. To account for this, we selected random intergenic regions that have no evidence of expression and calculated their substitution rates. Similar to ITFs (6.4%), 7.5% of random intergenic region samples are under strong purifying selection. More importantly, there is little, but statistically significant, difference in the substitution rate distributions between ITFs and random intergenic sequence (median rates, 0.09 and 0.08, respectively; KS test, P < 2e-05). Therefore, after accounting for potential alignment bias, potentially even fewer than 6.4% of ITFs are under significant selective constraints. The implication is that, based on cross-species comparison, the majority of ITFs appear to evolve in a way similar to presumably nonfunctional, nonexpressed random intergenic regions.
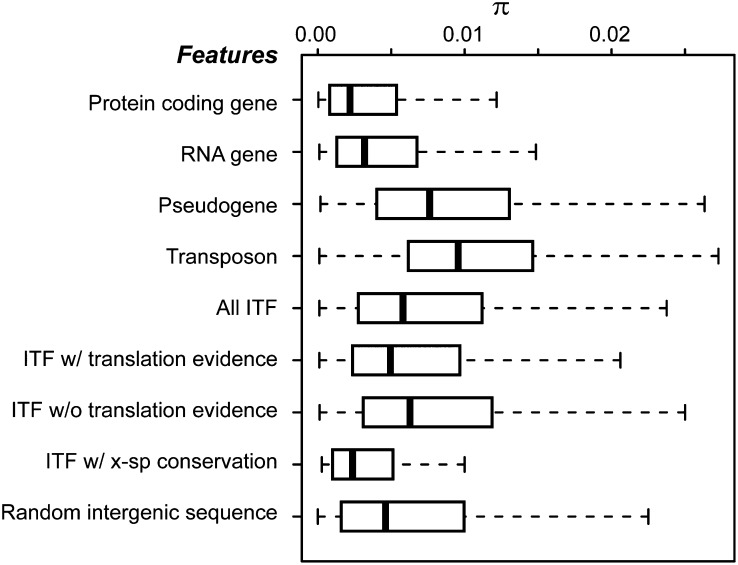
To assess the possibility that some ITFs may have a species-specific function in Arabidopsis, making the signature of selection only obvious at the intraspecific level, we analyzed genomic sequences of 80 accessions of Arabidopsis (Cao et al., 2011) and estimated the nucleotide diversity (π) for ITFs, annotated sequence features, and randomly selected intergenic sequences not overlapping with ITFs. The π value allows us to assess the genetic variability of different genomic features among Arabidopsis populations (Li, 1997). Our findings suggest that ITFs have π values significantly higher than coding sequences and RNA genes (KS tests, both P < 2.2e-16) but similar to random intergenic sequences not overlapping with ITFs (Fig. 5). We also estimated Tajima’s D, Fu and Li’s D, and Fay and Wu’s H statistics based on site-frequency spectrum to assess if ITFs are under selection. The distributions of all three statistics were comparable between ITFs and randomly sampled, unexpressed intergenic sequences (KS tests, all P > 0.1) but significantly different from those of protein-coding genes and RNA genes (KS tests, all P < 0.001; Supplemental Fig. S7). These findings suggest that there is much more relaxed selection within species on ITFs compared with protein-coding genes. Furthermore, the intensity of selection on ITFs is similar to that on random intergenic regions, which are likely largely nonfunctional and evolve neutrally.
Figure 5.
Distribution of π values for genomic features. The π values were calculated using population genomic data of 80 Arabidopsis accessions. x-sp, Cross species. Random intergenic sequences were selected from regions without transcript support.
Selection on ITFs with Translation Evidence
Our findings indicate that some ITFs are under strong selective constraint and may be functional. However, a much larger number of ITFs do not have clear signatures of selection. One immediate question is whether ITFs under strong selective constraint tend to be those that are translated, given that ITFs with translation evidence tend to be located closer to neighboring genes. We performed a similarity search between ITF sequences and the genomes of 15 land plants ranging from a bryophyte to angiosperms. For comparison and to address potential annotation issues, we also analyzed TxFrags mapping to protein-coding genes and RNA genes. ITFs with evidence of translation were slightly more conserved over ITFs with no evidence of translation (compare Fig. 4, C and D), but most ITFs have significant similarities only between Arabidopsis and A. lyrata, with sequence similarity rapidly declining beyond the Arabidopsis genus. On the other hand, there is a significantly higher degree of cross-species similarity between protein-coding genes: 6,895 of the 10,000 randomly selected protein sequences had E values of less than 1e-5 in more than one species (Fig. 4E). Even RNA genes, which are not expected to be translated, have higher sequence similarities than ITFs (Fig. 4F). Thus, at both the nucleotide and amino acid sequence levels, relatively few ITFs are under selection based on cross-species comparisons.
Of the 847 ITFs with translation evidence that are located more than 300 bp away from genes, 799 did not show significant conservation. Considering that these ITFs tend to be expressed at low levels, such sequences may represent translational noise. But we cannot rule out the possibility that they are lineage-specific coding sequences. Of the 49 ITFs that did show conservation, 16 had similar sequences present in more than one species at the amino acid level (E value < 1e-5), and 10 showed overlap with computationally predicted sORFs with high protein-coding potential (Hanada et al., 2010). The π value distribution of these 49 ITFs is statistically indistinguishable from that of TxFrags mapping to protein-coding sequences (KS test, P = 0.2; Fig. 5). These ITFs may thus represent novel functional genes. Nonetheless, only approximately 5% of ITFs with translational evidence are subject to strong purifying selection among Arabidopsis accessions, reinforcing the notion that most of them are products of noisy transcription.
CONCLUSION
In this study, we analyzed the intergenic polyA transcriptome of Arabidopsis to address the issues of abundance, coding/noncoding nature, and functional relevance of intergenic polyA transcripts. Our results indicate that approximately 5% of the TxFrags in the Arabidopsis transcriptome can be reliably called intergenic. One limitation of our analyses, as we have noted before, is our focus on the polyA fraction of the transcriptome. It is likely that the nonpolyA fraction of the transcriptome may harbor additional novel noncoding genes that need to be further investigated. Another limitation is that the read lengths of the RNA-seq data we used are short. It is possible that some ITFs belong to the same transcriptional units, making the number of ITFs an overestimate.
Our results indicate that approximately 3.6% of the intergenic space in Arabidopsis is transcribed by RNA polymerase II, and approximately 40% of what is transcribed tends to lie within 300 bp of annotated genes. Around one-third of ITFs have translation evidence, and we find a significant bias in their distribution; they tend to be closer to transcribed protein-coding genes, raising the possibility that some ITFs may in fact be unannotated extensions of known genes. Our primary sequence-level evolutionary analysis indicates that a relatively low fraction (approximately 5%) of the ITFs have experienced strong purifying selection either within species or between species. We should emphasize that our criteria for evaluating selection is stringent. Furthermore, some ITFs may be more strongly constrained at the secondary structural level, similar to noncoding RNA genes (Washietl et al., 2005). In addition, some long noncoding RNAs such as Air and Xist are poorly conserved (Pang et al., 2006; Ponting et al., 2009), indicating that a lack of conservation may not always mean lack of function. Thus, it is likely we will miss some ITFs that are under selection or are functional. However, most ITFs are short, and unlike the long noncoding RNAs, tend to be data set specific and expressed at a very low level compared with annotated genes. In addition, most ITFs have characteristics more similar to pseudogenes and transposons than to protein-coding and RNA genes. Taken together, most ITFs bear the hallmarks of neutrally evolving sequences, suggesting that they are products of noisy transcription, as proposed earlier (Struhl, 2007).
The idea of transcriptional noise has been intensely debated over the past few years. Some studies support the theory that the transcriptional machinery might be error prone and that many transcripts may be the result of false starts and/or stops (Li et al., 2007; Struhl, 2007; Xu et al., 2009; van Bakel et al., 2010). Such errors may occur because it is not possible to regulate any biological process to the point that there is no error; noisy transcription may exist simply because it incurs little fitness cost. Considering that the vast majority of mutations are neutral or nearly neutral (Ohta, 1992), this paradigm for gene evolution may also apply to other molecular events, including transcription. As has been postulated before, the target of natural selection may be the effects of error-prone transcription rather than the transcriptional process itself (Hurst, 2009). Another possibility is that the effect of genetic drift, particularly on organisms with smaller effective population sizes, may render selection against erroneous transcript production ineffective. In either case, the hypothesis is that transcriptional errors may not always be subjected to purifying selection. Based on our findings, it would seem that much of the intergenic transcription falls into this category. We should emphasize that, for most ITFs, there is little or no evidence to reject the null hypothesis that ITFs are nonfunctional. The reason to consider nonfunctionality as null is simply because only functionality can be experimentally tested (van Bakel et al., 2010).
In a recently published series of papers by the ENCODE consortium, 80.4% of the human genome was found to have evidence of functionality (Dunham et al., 2012). Functionality of a sequence, in this case, was defined at the biochemical level. That is, a functional sequence is presumed to have at least one RNA- and/or chromatin-associated event, such as transcription factor binding, nucleosome binding, or DNA methylation, in at least one cell type. However, it remains unclear to what extent these biochemically functional sequences may have physiological function, given that these biochemical events can also be due to noise. As an example, among the novel long intergenic polyA TxFrags obtained in the ENCODE study, only 4% of the bases were conserved between humans and macaques. Among the 96% of bases not conserved, only 6% to 11% showed evidence of lineage-specific constraint in humans, comparable to what we found in Arabidopsis (Ward and Kellis, 2012). In addition, the ITFs found in this study were found to be present at less than 0.1 copy per cell (Djebali et al., 2012), consistent with our finding that most of the Arabidopsis ITFs have a very low expression level.
Considering the dramatically increased sensitivity and throughput in sequencing, noisy transcription and even contaminating sequences, such as trace amounts of genomic DNA, can be readily detected. Thus, a transcription event as detected by sequencing may not be considered functional by default. Evolutionary constraint acting on novel sequences or other evidence should be demonstrated prior to their annotation. The increasing availability of population-wide polymorphism data sets and genome sequences of related species provide more robust tools for such evolutionary studies, especially those focusing on lineage-specific selection (Ward and Kellis, 2012). In addition to the question of functionality, we show that a significant number of ITFs are associated with ribosomes and a smaller fraction of them have proteomics tags. Thus, novel transcripts should not be regarded as noncoding by default without rigorous experimental analysis of their coding potential. Our results also suggest the need to have a clearer understanding of the mechanistic aspects of RNA polymerase action on how noisy transcription may arise. Use of an integrated approach to validate novel RNA predictions and their functionality would be important in this regard. For example, a previous study in mouse used an array of approaches, including the identification of conserved histone modification marks, evolutionary analyses of promoter regions, gene set enrichment analysis, transcription factor chromatin immunoprecipitation, and RNA interference assays, to identify putative functional long ncRNAs (Guttman et al., 2009). We surmise that such an approach will allow us to explore more deeply the mechanistic and evolutionary aspects of the transcriptional process in plants.
MATERIALS AND METHODS
Plant Material and RNA Isolation
For transcriptome and ribosome immunoprecipitation studies, transgenic Arabidopsis (Arabidopsis thaliana) Columbia seeds expressing a His6FLAG-tagged version of the ribosomal large subunit protein L18B (35S:HF-RPL18B) were surface sterilized, stratified for 3 d, and sown on 0.5× Murashige and Skoog medium containing 1% (w/v) Suc and 0.4% phytagel. Seedlings were grown vertically under a 16-h-day (125 μE m−2 s−1 photosynthetically active radiation)/8-h-night cycle for 7 d as described previously (Branco-Price et al., 2008). Seven-day-old seedlings were harvested at the end of the light period. Total RNA extraction and ribosome immunoprecipitation were done as described previously (Branco-Price et al., 2008) for three biological replicates, except that the RNeasy Plant Mini Kit purification step was omitted. Total RNA and ribosome-immunoprecipitated RNA were quantified using a Nanodrop spectrophotometer (Nanodrop Technologies), and RNA quality was assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies).
Illumina RNA-seq and Data Analysis
For each in-house (7-d seedlings) RNA sample, cDNA libraries were constructed by first isolating polyA RNA from 3 µg of total or ribosome-associated RNA. Libraries for RNA-seq were prepared using the Illumina mRNA-seq Sample Prep Kit. Briefly, polyA RNA was fragmented and reverse transcribed using random primers. Adapters were ligated to double-stranded cDNA, and fragments from 175 to 225 bp were gel purified. After PCR amplification, the cDNA libraries were sequenced on an Illumina Genome Analyzer. Each library was loaded onto at least two lanes; however, usable sequence was only obtained for one polyA RNA library and two ribosome-associated RNA libraries (four lanes total). Three lanes of ribosome-associated RNA sequencing, corresponding to two biological replicates, were combined to give 19,818,643 reads, and one lane of polyA RNA yielding 7,028,772 36-bp reads was obtained. The original sequencing reads were deposited in the National Center for Biotechnology Information Short Read Archive under accession number SRA053376 (http://www.ncbi.nlm.nih.gov/Traces/sra). All public data sets (Supplemental Table S1; Supplemental Fig. S2) were downloaded from the National Center for Biotechnology Information Short Read Archive. The following procedure was commonly performed on both the in-house and public data sets.
The short reads, after quality trimming, were mapped to The Arabidopsis Information Resource Arabidopsis genome release 10 using Bowtie version 0.12.7 (Langmead et al., 2009) and TopHat version 1.2.0 (Trapnell et al., 2009). The default settings were used except that the maximum combined intron size was set at 5,000 bp. The mapped reads were assembled with two approaches. In the first approach, reads with overlapping genomic locations were merged into TxFrags (Set 1 TxFrags) without considering the possibility that neighboring TxFrags may be derived from the same transcriptional units. In the second approach, Cufflinks 0.9.3 (Trapnell et al., 2010) was used with default parameters, except that a maximum combined intron size was set at 5,000 bp (Set 2 TxFrags). All TxFrags overlapping with annotated features by 1 bp or more, including those in introns or UTRs, were flagged as genic transcripts.
Estimating Expression Level and Breadth of Expression of Features
For estimating expression level, the FPKM measure was used. Since Set 1 TxFrags represent a set of unique, nonoverlapping TxFrags, the entire TxFrag was considered an exon for the purpose of FPKM estimation. For Set 2 TxFrags, FPKM values were estimated by Cufflinks. The breadth of expression was calculated for the 6,545 merged ITFs. For comparison, we also measured the expression breadth of TxFrags mapping to annotated features. We used the number of data sets in which a particular feature had expression evidence (one or more overlapping TxFrag) as a measure of the breadth of expression of that feature.
5′ RACE and Transient Expression of YFP Fusion Proteins in Tobacco
The 5′ UTRs of putative coding sORF sequences were identified from publicly available cDNA sequences (Aubourg et al., 2007) or were amplified by 5′ RACE (RLM-RACE kit [Ambion] or SMART RACE cDNA amplification kit [Clontech]). The 5′ UTRs and coding sequences of each sORF were amplified from genomic DNA and cloned into the TOPO-TA entry vector (Invitrogen). The sequences were then transferred by recombination mediated by LR Clonase (Invitrogen) into a modified pMDC83 destination vector (Curtis and Grossniklaus, 2003), containing the enhanced YFP sequence (Clontech) and lacking a translational start codon, under the control of the 35S promoter. Constructs containing sORFs fused in frame with YFP were transformed into Agrobacterium tumefaciens GV3101. Transient transformation was performed to express sORF-YFP fusions in tobacco (Nicotiana tabacum) cells (Sparkes et al., 2006). Transgenic A. tumefaciens cells were cultivated overnight, and 200 μL of the culture (optical density A600 approximately 1–2) was pelleted and resuspended with sterile water to 0.1 optical density. A. tumefaciens cells were infiltrated into tobacco leaves, and the infiltrated tobacco was kept under constant light for 72 h. Infiltrated areas of tobacco leaves were detached and observed with an inverted laser scanning confocal microscope (Olympus Spectral FV 1000). YFP signals were detected with the 514-nm argon laser excitation line with a band-pass emission filter of 517.5 to 542.5 nm. For visualization of AT_1|-|2|5786755-5786853 (Hanada et al., 2007) and Endoplasmic Reticulum Retention Defective2 (ERD2) colocalization, equal volumes of A. tumefaciens cultures were mixed prior to infiltration. Fluorescence was visualized after 3 d with a Meta Zeiss confocal microscope using the argon laser excitation lines of 458 and 514 nm and band-pass emission filters of 475 to 525 nm and 530 to 600 nm for blue-shifted GFP and YFP, respectively.
Evolutionary Conservation Analyses
To identify ITFs under selection at the nucleotide level, the Arabidopsis ITFs were first mapped to the Arabidopsis lyrata genome using GMAP version 2007-09-28 (Wu and Watanabe, 2005) with default settings. Putative ITF orthologs were defined as pairs of similar sequences (80% or greater coverage, 80% or greater identity, 40 bp or greater match length) between Arabidopsis and A. lyrata flanked by one or more putative orthologous genes among 10 protein-coding genes on either side of the ITF. Putative orthologs between these two species were identified based on reciprocal best-match and synteny information. The orthologous ITFs were aligned using Clustal 2.1 (Thompson et al., 1994), and the nucleotide substitution rate was calculated using baseml with the HKY substitution model in PAML (Yang, 2007). ITFs with a substitution rate lower than the 95th percentile (HKY distance ≤ 0.07) of the 4-fold degenerate site substitution rates of all protein-coding orthologs were deemed to be evolving under strong purifying selection. To control for genome-wide variation in local substitution rates, the 4-fold degenerate site substitution rates of up to 60 protein-coding genes in the vicinity of the ITFs were used to determine the 5% significance level using a z test. We did not conduct a nonsynonymous substitutions per nonsynonymous site (Ka)/synonymous substitutions per synonymous site (Ks) analysis for ITFs because (1) most ITFs are short, so the variance of Ka and Ks estimates for short sequences tend to be high, and (2) it is not clear what the correct reading frame is, if these ITFs are translated. Instead, to compare the levels of conservation at the coding sequence level, we performed a translated BLAST search between ITF/TxFrag sequences and the draft assemblies of 14 plant species in Phytozome 5.0 (http://www.phytozome.org/). The negative logarithm of the E value of the top match in each species was used to plot a heat map. All negative log values of 10 or greater or 0 or less were set to 10 and 0, respectively.
For conservation analyses within species, we used polymorphism data in the form of a genome matrix file from 80 different Arabidopsis accessions (Cao et al., 2011). For each genomic feature type, we reconstructed the aligned sequences based on the genome matrix file. The aligned sequences were analyzed for π, Tajima’s D, and Fu and Li’s D using Variscan (Vilella et al., 2005) with the following parameters: RefPos = 1, Outgroup = none, RunMode = 12, UseMuts = 0, CompleteDeletion = 0, FixNum = 1, NumNuc = 60. For Fay and Wu’s H, we used the orthologs in A. lyrata as outgroups with RunMode = 22. For comparison, π values for 10,000 randomly chosen protein-coding genes, RNA genes, transposons, and pseudogenes were also calculated. For features with fewer than 10,000 sequences, bootstrap samples were used. To determine the background π values, 10,000 random intergenic sequences were sampled based on the size distribution of ITFs. Only those intergenic sequences not overlapping with any TxFrags were used for analysis. For each sequence in each feature type, a π value was estimated. The π distributions were then compared statistically.
The presence of ambiguous nucleotides or the short size of the ITFs can affect the error margins associated with π estimates. To assess whether these factors influenced our findings, we conducted additional analysis by changing the minimum number of sites analyzed (MinLength) and the proportion of the aligned length with nonambiguous bases (coverage). We sampled a range of MinLength (0, 50, 100, 150, and 200) at no coverage threshold (Supplemental Fig. S8) and a range of coverage (0, 0.25, 0.50, 0.75, and 1) at no MinLength threshold (Supplemental Fig. S9). Our analyses suggested that the trend observed in Figure 5 is not affected by the presence of ambiguous nucleotides or the short length of the ITF.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number SRA053376.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Size distributions of TxFrags in different data sets.
Supplemental Figure S2. Expression level distributions of TxFrags in different data sets.
Supplemental Figure S3. Directional sequencing of mRNA from T87 cells.
Supplemental Figure S4. Pair-wise proportions of ITFs with translation evidence across different data sets.
Supplemental Figure S5. Number of peptides per feature.
Supplemental Figure S6. Distances of translation evidence from annotated genes.
Supplemental Figure S7. Population-level tests of neutrality for different features.
Supplemental Figure S8. Changing MinLength thresholds does not impact π distributions.
Supplemental Figure S9. Changing coverage thresholds does not impact π distributions.
Supplemental Table S1. Description of input data sets.
Supplemental Table S2. Sequences tested for coding potential.
Supplemental Table S3. Description of data sets used for translation analyses.
Supplemental File S1. Percentages of features considered expressed at different false-positive rate thresholds.
Acknowledgments
We are grateful to Kevin Carr, Shari Tjugum-Holland, and Jeff Landgraff for help with sequencing and data preprocessing, Melinda Frame for help with confocal laser scanning microscopy, John Johnston for computational support, Kelian Sun for help with cultivating T87 cells, and Andy Lin for his contributions toward preliminary studies.
Glossary
- polyA
polyadenylated
- nonpolyA
nonpolyadenylated
- ITF
intergenic transcribed fragment
- ncRNA
noncoding RNA
- ORF
open reading frame
- mRNA-seq
mRNA sequencing
- TxFrags
transcript fragments
- FPKM
fragments per kilobase of exon model per million mapped reads
- KS
Kolmogrov-Smirnov
- R-TxFrags
ribosome-associated transcript fragments
- sORF
small open reading frame
- YFP
yellow fluorescent protein
- UTR
untranslated region
- π
nucleotide diversity
- cDNA
complementary DNA
- ENCODE
Encyclopedia of DNA Elements
References
- Agarwal A, Koppstein D, Rozowsky J, Sboner A, Habegger L, Hillier LW, Sasidharan R, Reinke V, Waterston RH, Gerstein M. (2010) Comparison and calibration of transcriptome data from RNA-Seq and tiling arrays. BMC Genomics 11: 383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armour CD, Castle JC, Chen R, Babak T, Loerch P, Jackson S, Shah JK, Dey J, Rohl CA, Johnson JM, et al. (2009) Digital transcriptome profiling using selective hexamer priming for cDNA synthesis. Nat Methods 6: 647–649 [DOI] [PubMed] [Google Scholar]
- Aubourg S, Martin-Magniette M-L, Brunaud V, Taconnat L, Bitton F, Balzergue S, Jullien PE, Ingouff M, Thareau V, Schiex T, et al. (2007) Analysis of CATMA transcriptome data identifies hundreds of novel functional genes and improves gene models in the Arabidopsis genome. BMC Genomics 8: 401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baerenfaller K, Grossmann J, Grobei MA, Hull R, Hirsch-Hoffmann M, Yalovsky S, Zimmermann P, Grossniklaus U, Gruissem W, Baginsky S. (2008) Genome-scale proteomics reveals Arabidopsis thaliana gene models and proteome dynamics. Science 320: 938–941 [DOI] [PubMed] [Google Scholar]
- Bailey-Serres J, Sorenson R, Juntawong P. (2009) Getting the message across: cytoplasmic ribonucleoprotein complexes. Trends Plant Sci 14: 443–453 [DOI] [PubMed] [Google Scholar]
- Basrai MA, Hieter P, Boeke JD. (1997) Small open reading frames: beautiful needles in the haystack. Genome Res 7: 768–771 [DOI] [PubMed] [Google Scholar]
- Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, Epstein CB, Frietze S, Harrow J, Kaul R, et al. (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertone P, Stolc V, Royce TE, Rozowsky JS, Urban AE, Zhu X, Rinn JL, Tongprasit W, Samanta M, Weissman S, et al. (2004) Global identification of human transcribed sequences with genome tiling arrays. Science 306: 2242–2246 [DOI] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. (2007) Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branco-Price C, Kaiser KA, Jang CJH, Larive CK, Bailey-Serres J. (2008) Selective mRNA translation coordinates energetic and metabolic adjustments to cellular oxygen deprivation and reoxygenation in Arabidopsis thaliana. Plant J 56: 743–755 [DOI] [PubMed] [Google Scholar]
- Cao J, Schneeberger K, Ossowski S, Günther T, Bender S, Fitz J, Koenig D, Lanz C, Stegle O, Lippert C, et al. (2011) Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat Genet 43: 956–963 [DOI] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. (2005) The transcriptional landscape of the mammalian genome. Science 309: 1559–1563 [DOI] [PubMed] [Google Scholar]
- Castellana NE, Payne SH, Shen Z, Stanke M, Bafna V, Briggs SP. (2008) Discovery and revision of Arabidopsis genes by proteogenomics. Proc Natl Acad Sci USA 105: 21034–21038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, Long J, Stern D, Tammana H, Helt G, et al. (2005) Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science 308: 1149–1154 [DOI] [PubMed] [Google Scholar]
- Clark MB, Amaral PP, Schlesinger FJ, Dinger ME, Taft RJ, Rinn JL, Ponting CP, Stadler PF, Morris KV, Morillon A, et al. (2011) The reality of pervasive transcription. PLoS Biol 9: e1000625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David L, Huber W, Granovskaia M, Toedling J, Palm CJ, Bofkin L, Jones T, Davis RW, Steinmetz LM. (2006) A high-resolution map of transcription in the yeast genome. Proc Natl Acad Sci USA 103: 5320–5325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinger ME, Amaral PP, Mercer TR, Mattick JS. (2009) Pervasive transcription of the eukaryotic genome: functional indices and conceptual implications. Brief Funct Genomics Proteomics 8: 407–423 [DOI] [PubMed] [Google Scholar]
- Dinger ME, Pang KC, Mercer TR, Mattick JS. (2008) Differentiating protein-coding and noncoding RNA: challenges and ambiguities. PLoS Comput Biol 4: e1000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. (2012) Landscape of transcription in human cells. Nature 489: 101–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JP, Dougherty JD, Heiman M, Schmidt EF, Stevens TR, Ma G, Bupp S, Shrestha P, Shah RD, Doughty ML, et al. (2008) Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell 135: 749–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren N, Howell MD, Kasschau KD, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Law TF, Grant SR, Dangl JL, et al. (2007) High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS ONE 2: e219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filichkin SA, Priest HD, Givan SA, Shen R, Bryant DW, Fox SE, Wong WK, Mockler TC. (2010) Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res 20: 45–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory BD, O’Malley RC, Lister R, Urich MA, Tonti-Filippini J, Chen H, Millar AH, Ecker JR. (2008) A link between RNA metabolism and silencing affecting Arabidopsis development. Dev Cell 14: 854–866 [DOI] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. (2009) Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458: 223–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K, Akiyama K, Sakurai T, Toyoda T, Shinozaki K, Shiu S-H. (2010) sORF finder: a program package to identify small open reading frames with high coding potential. Bioinformatics 26: 399–400 [DOI] [PubMed] [Google Scholar]
- Hanada K, Zhang X, Borevitz JO, Li W-H, Shiu S-H. (2007) A large number of novel coding small open reading frames in the intergenic regions of the Arabidopsis thaliana genome are transcribed and/or under purifying selection. Genome Res 17: 632–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins JS, Kim H, Nason JD, Wing RA, Wendel JF. (2006) Differential lineage-specific amplification of transposable elements is responsible for genome size variation in Gossypium. Genome Res 16: 1252–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo JB, Sung S. (2011) Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331: 76–79 [DOI] [PubMed] [Google Scholar]
- Hiller M, Findeiss S, Lein S, Marz M, Nickel C, Rose D, Schulz C, Backofen R, Prohaska SJ, Reuter G, et al. (2009) Conserved introns reveal novel transcripts in Drosophila melanogaster. Genome Res 19: 1289–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu TT, Pattyn P, Bakker EG, Cao J, Cheng J-F, Clark RM, Fahlgren N, Fawcett JA, Grimwood J, Gundlach H, et al. (2011) The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat Genet 43: 476–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst LD. (2009) Evolutionary genomics and the reach of selection. J Biol 8: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JRS, Weissman JS. (2009) Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324: 218–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Meyerowitz EM. (2010) Cell-type specific analysis of translating RNAs in developing flowers reveals new levels of control. Mol Syst Biol 6: 419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermüller J, Hofacker IL, et al. (2007) RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316: 1484–1488 [DOI] [PubMed] [Google Scholar]
- Kawaguchi R, Bailey-Serres J. (2002) Regulation of translational initiation in plants. Curr Opin Plant Biol 5: 460–465 [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Wang X, Sasidharan R, Stolc V, Deng W, He H, Korbel J, Chen X, Tongprasit W, Ronald P, et al. (2007) Global identification and characterization of transcriptionally active regions in the rice genome. PLoS ONE 2: e294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W-H. (1997) Molecular Evolution. Sinauer Associates, Sunderland, MA
- Lister R, O’Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR. (2008) Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133: 523–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Gough J, Rost B. (2006) Distinguishing protein-coding from non-coding RNAs through support vector machines. PLoS Genet 2: e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui A, Ishida J, Morosawa T, Mochizuki Y, Kaminuma E, Endo TA, Okamoto M, Nambara E, Nakajima M, Kawashima M, et al. (2008) Arabidopsis transcriptome analysis under drought, cold, high-salinity and ABA treatment conditions using a tiling array. Plant Cell Physiol 49: 1135–1149 [DOI] [PubMed] [Google Scholar]
- Mattick JS. (2009) The genetic signatures of noncoding RNAs. PLoS Genet 5: e1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mattick JS. (2009) Long non-coding RNAs: insights into functions. Nat Rev Genet 10: 155–159 [DOI] [PubMed] [Google Scholar]
- Movahedi S, Van de Peer Y, Vandepoele K. (2011) Comparative network analysis reveals that tissue specificity and gene function are important factors influencing the mode of expression evolution in Arabidopsis and rice. Plant Physiol 156: 1316–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrutenko A, Makova KD, Li W-H. (2002) The K(A)/K(S) ratio test for assessing the protein-coding potential of genomic regions: an empirical and simulation study. Genome Res 12: 198–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ner-Gaon H, Halachmi R, Savaldi-Goldstein S, Rubin E, Ophir R, Fluhr R. (2004) Intron retention is a major phenomenon in alternative splicing in Arabidopsis. Plant J 39: 877–885 [DOI] [PubMed] [Google Scholar]
- Nuzhdin SV, Wayne ML, Harmon KL, McIntyre LM. (2004) Common pattern of evolution of gene expression level and protein sequence in Drosophila. Mol Biol Evol 21: 1308–1317 [DOI] [PubMed] [Google Scholar]
- Ohta T. (1992) The nearly neutral theory of molecular evolution. Annu Rev Ecol Syst 23: 263–286 [Google Scholar]
- Okazaki Y, Furuno M, Kasukawa T, Adachi J, Bono H, Kondo S, Nikaido I, Osato N, Saito R, Suzuki H, et al. (2002) Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 420: 563–573 [DOI] [PubMed] [Google Scholar]
- Pang KC, Frith MC, Mattick JS. (2006) Rapid evolution of noncoding RNAs: lack of conservation does not mean lack of function. Trends Genet 22: 1–5 [DOI] [PubMed] [Google Scholar]
- Piegu B, Guyot R, Picault N, Roulin A, Sanyal A, Kim H, Collura K, Brar DS, Jackson S, Wing RA, et al. (2006) Doubling genome size without polyploidization: dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice. Genome Res 16: 1262–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP, Belgard TG. (2010) Transcribed dark matter: meaning or myth? Hum Mol Genet 19: R162–R168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP, Oliver PL, Reik W. (2009) Evolution and functions of long noncoding RNAs. Cell 136: 629–641 [DOI] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Maglott DR. (2007) NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res 35: D61–D65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Y, Ooi HS, Choo SW, Chiu KP, Zhao XD, Srinivasan KG, Yao F, Choo CY, Liu J, Ariyaratne P, et al. (2007) Fusion transcripts and transcribed retrotransposed loci discovered through comprehensive transcriptome analysis using Paired-End diTags (PETs). Genome Res 17: 828–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU. (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Sparkes IA, Runions J, Kearns A, Hawes C. (2006) Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat Protoc 1: 2019–2025 [DOI] [PubMed] [Google Scholar]
- Stolc V, Samanta MP, Tongprasit W, Sethi H, Liang S, Nelson DC, Hegeman A, Nelson C, Rancour D, Bednarek S, et al. (2005) Identification of transcribed sequences in Arabidopsis thaliana by using high-resolution genome tiling arrays. Proc Natl Acad Sci USA 102: 4453–4458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. (2007) Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat Struct Mol Biol 14: 103–105 [DOI] [PubMed] [Google Scholar]
- Subramanian S, Kumar S. (2004) Gene expression intensity shapes evolutionary rates of the proteins encoded by the vertebrate genome. Genetics 168: 373–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bakel H, Nislow C, Blencowe BJ, Hughes TR. (2010) Most “dark matter” transcripts are associated with known genes. PLoS Biol 8: e1000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bakel H, Nislow C, Blencowe BJ, Hughes TR. (2011) Response to “The Reality of Pervasive Transcription.” PLoS Biol 9: e1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilella AJ, Blanco-Garcia A, Hutter S, Rozas J. (2005) VariScan: analysis of evolutionary patterns from large-scale DNA sequence polymorphism data. Bioinformatics 21: 2791–2793 [DOI] [PubMed] [Google Scholar]
- Wang J, Zhang J, Zheng H, Li J, Liu D, Li H, Samudrala R, Yu J, Wong GK-S. (2004) Mouse transcriptome: neutral evolution of ‘non-coding’ complementary DNAs. Nature 431: 1. [PubMed] [Google Scholar]
- Ward LD, Kellis M. (2012) Evidence of abundant purifying selection in humans for recently acquired regulatory functions. Science 337: 1675–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washietl S, Hofacker IL, Lukasser M, Hüttenhofer A, Stadler PF. (2005) Mapping of conserved RNA secondary structures predicts thousands of functional noncoding RNAs in the human genome. Nat Biotechnol 23: 1383–1390 [DOI] [PubMed] [Google Scholar]
- Wu TD, Watanabe CK. (2005) GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21: 1859–1875 [DOI] [PubMed] [Google Scholar]
- Xu AG, He L, Li Z, Xu Y, Li M, Fu X, Yan Z, Yuan Y, Menzel C, Li N, et al. (2010) Intergenic and repeat transcription in human, chimpanzee and macaque brains measured by RNA-Seq. PLoS Comput Biol 6: e1000843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Münster S, Camblong J, Guffanti E, Stutz F, Huber W, Steinmetz LM. (2009) Bidirectional promoters generate pervasive transcription in yeast. Nature 457: 1033–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Lim J, Dale JM, Chen H, Shinn P, Palm CJ, Southwick AM, Wu HC, Kim C, Nguyen M, et al. (2003) Empirical analysis of transcriptional activity in the Arabidopsis genome. Science 302: 842–846 [DOI] [PubMed] [Google Scholar]
- Yang Z. (2007) PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24: 1586–1591 [DOI] [PubMed] [Google Scholar]
- Zanetti ME, Chang I-F, Gong F, Galbraith DW, Bailey-Serres J. (2005) Immunopurification of polyribosomal complexes of Arabidopsis for global analysis of gene expression. Plant Physiol 138: 624–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW-L, Chen H, Henderson IR, Shinn P, Pellegrini M, Jacobsen SE, et al. (2006) Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 126: 1189–1201 [DOI] [PubMed] [Google Scholar]
- Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. (2007) Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet 39: 61–69 [DOI] [PubMed] [Google Scholar]