Abstract
Potassium (K+) is an essential macronutrient for plant growth and development. Previous studies have demonstrated that Calcineurin B-Like Protein1 (CBL1) or CBL9 and CBL-Interacting Protein Kinase23 (CIPK23) regulate K+ uptake in Arabidopsis (Arabidopsis thaliana) roots by modulating K+ channel Arabidopsis K+ Transporter1. In this study, we show that the protein kinase CIPK9 interacts with the calcium sensor CBL3 and plays crucial roles in K+ homeostasis under low-K+ stress in Arabidopsis. Arabidopsis wild-type plants showed leaf chlorotic symptoms when grown for 10 d on low-K+ (100 μm) medium. Here, we show that plants lacking CIPK9 displayed a tolerant phenotype to low-K+ stress, which still maintained green leaves when the wild-type plants showed typical K+-deficient symptoms. Overexpressing lines of CIPK9 resulted in a low-K+-sensitive phenotype compared with wild-type plants. Furthermore, CBL2 and CBL3 were identified as upstream regulators of CIPK9. Both CBL2- and CBL3-overexpressing lines displayed similar low-K+-sensitive phenotypes and K+ contents to CIPK9-overexpressing lines. However, only cbl3 mutant plants, but not cbl2 mutant plants, showed the low-K+-tolerant phenotype similar to cipk9 mutants. Taken together, these results demonstrate that CIPK9 and CBL3 work together and function in K+ homeostasis under low-K+ stress in Arabidopsis.
As the most abundant cation, potassium (K+) plays crucial roles in many physiological processes in plant cells, such as enzyme activation, stomata movement, membrane potential maintenance, and osmotic regulation (Clarkson and Hanson, 1980). Unlike nitrogen and phosphorus, K+ cannot be metabolized and always remains in its ionic elementary form in plant cells to execute its physiological roles (Amtmann and Blatt, 2009). Therefore, K+ utilization efficiency is primarily dependent on K+ absorption from the environment and/or translocation in plants. It is well known that K+ absorption and translocation are mainly mediated by plant K+ transporters or channels; therefore, the functional identification of K+ transporters and channels as well as their regulators has become the focus in this field. A number of K+ transporters, channels, as well as their regulators have been functionally characterized during the last decade (Véry and Sentenac, 2003; Gierth and Mäser, 2007; Lebaudy et al., 2007; Ward et al., 2009).
In Arabidopsis (Arabidopsis thaliana), the High-Affinity K+ Transporter5 (AtHAK5) and the K+ channel Arabidopsis K+ Transporter1 (AKT1) have been identified as the two major K+ uptake components, which mediate high-affinity K+ absorption in roots (Pyo et al., 2010; Kim et al., 2012). Either lesion of AtHAK5 or AKT1 significantly impairs plant high-affinity rubidium(K+) uptake (Hirsch et al., 1998; Gierth et al., 2005; Pyo et al., 2010). Furthermore, the rubidium(K+) uptake in athak5 akt1 double mutant plants under low-K+ (LK) conditions could decrease by 85% compared with wild-type plants (Pyo et al., 2010). The K+ channels SKOR and AKT2 are identified as the major components responsible for K+ translocation between roots and shoots in Arabidopsis (Gaymard et al., 1998; Marten et al., 1999; Lacombe et al., 2000). The outward-rectifying K+ channel SKOR mediates K+ release into the xylem sap for transport toward shoots (Gaymard et al., 1998), and AKT2 conducts the K+ influx into phloem for long-distance K+ transport from sources to sinks (Marten et al., 1999; Deeken et al., 2000; Lacombe et al., 2000; Gajdanowicz et al., 2011). In addition to these transporter proteins, some regulatory components, such as the protein kinases Calcineurin B-Like Protein (CBL)-Interacting Protein Kinase23 (CIPK23) and CIPK6, the phosphatases AtPP2CA and AIP1, as well as the K+ channel subunit AtKC1, have also been identified and functionally characterized in plant K+ uptake and translocation (Chérel et al., 2002; Li et al., 2006; Xu et al., 2006; Lee et al., 2007; Duby et al., 2008; Geiger et al., 2009; Wang et al., 2010; Held et al., 2011). Notably, most of these regulatory components are characterized as the regulatory proteins for K+ channels. Among these regulatory proteins, the protein kinase CIPK23 and the calcium sensors CBL1 or CBL9 are identified as the essential K+ uptake regulators in Arabidopsis and also function in response to LK stress (Xu et al., 2006). Upon interaction with CBL1 (or CBL9), CIPK23 is recruited to the plasma membrane (PM) and activates the K+ channel AKT1 via phosphorylation, so that AKT1-mediated root K+ uptake is enhanced (Xu et al., 2006; Cheong et al., 2007). For the K+ transporter, transcriptional regulation seems more important than posttranslational regulation. It is reported that AtHAK5 is induced by LK stress in order to enhance K+ uptake under LK conditions, which is regulated by the transcription factor RAP2.11 (Kim et al., 2012).
The CBL proteins are regarded as a group of plant calcium sensors that could exclusively interact with Ser/Thr protein kinases named CIPK proteins. By forming diverse CBL/CIPK complexes, the proteins from these two families constitute a specific regulatory network of Ca2+ signaling in plant cells (Kim et al., 2000; Albrecht et al., 2001; Luan, 2009). Many CBLs and CIPKs have been reported to function in plant responses to abiotic stresses, especially to the ionic stresses (Luan, 2009). The CBL4/CIPK24 (SOS3/SOS2) complex as well as CBL10/CIPK24 are identified to participate in plant salt tolerance (Quintero et al., 2002; Kim et al., 2007; Quan et al., 2007). CBL2/CIPK11 (SCaBP1/PKS5) is reported to be involved in the regulation of pH homeostasis in Arabidopsis (Fuglsang et al., 2007). In this study, we demonstrate that CIPK9, the most similar homolog to CIPK23, interacts with CBL3, forming a CBL/CIPK complex, which regulates K+ homeostasis in Arabidopsis under LK conditions.
RESULTS
cipk9 Mutant Plants Are Tolerant to LK Stress
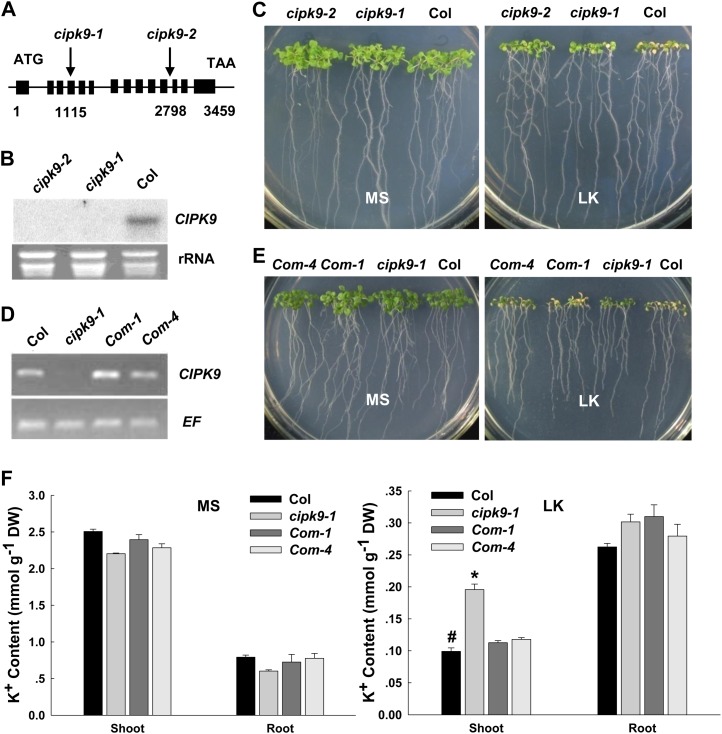
It has been reported that the CIPK23, as an important positive regulator of AKT1, is involved in K+ uptake in Arabidopsis roots (Li et al., 2006; Xu et al., 2006). Since CIPK9 is the most similar homolog to CIPK23 in this gene family, it is hypothesized that CIPK9 might be also involved in K+ uptake or K+ homeostasis in Arabidopsis, particularly under LK stress. To test this hypothesis, the transfer DNA (T-DNA) insertion mutants of CIPK9, including SALK_058629 (cipk9-1) and SALK_014699 (cipk9-2), were used to determine their K+-related phenotypes under LK (100 μm K+) conditions. The T-DNA insertion sites of cipk9-1 and cipk9-2 are located in the fourth exon (1,115 bp from ATG) and the 11th intron (2,798 bp from ATG) of the CIPK9 gene, respectively (Fig. 1A). Northern-blot analysis confirmed that the transcription of CIPK9 in these two mutant lines was disrupted (Fig. 1B). The cipk9-1 and cipk9-2 mutants did not show any significant difference from the wild-type plants under normal growth condition (Murashige and Skoog [MS] medium), but they were more tolerant to LK stress compared with wild-type plants. As shown in Figure 1C, when the leaves of cipk9 mutants remained green after growth on LK medium (100 μm K+) for 12 d, the leaves of wild-type plants had become chlorotic (Fig. 1C).
Figure 1.
Phenotype tests and K+ content measurements of cipk9 mutant plants, wild-type plants, and cipk9 complementation plants under LK stress. A, Structure of the CIPK9 gene. The boxes indicate exons and the lines represent introns. The T-DNA insertion sites in cipk9-1 and cipk9-2 are shown using arrows. B, Northern-blot analysis of CIPK9 expression levels in cipk9 mutants and wild-type plants. C, Phenotype comparison between cipk9 mutants (cipk9-1 and cipk9-2) and the wild type (Col) after growth on MS or LK medium for 12 d. D, Reverse transcription-PCR verification of CIPK9 expression levels in cipk9 complementation plants (Com-1 and Com-4). E, Phenotype comparison of wild-type plants (Col), cipk9-1 mutant plants, and complementation plants (Com-1 and Com-4) after growth on MS or LK medium for 12 d. F, K+ content measurements of various plant materials. The shoot and root K+ contents were measured after the plants were grown on MS or LK medium for 12 d. Data shown are means ± se (n = 3). Student’s t test (*P < 0.05) was used to analyze statistical significance compared with the control (#). DW, Dry weight.
To test whether the LK-tolerant phenotype of the cipk9 mutants resulted from loss of function of CIPK9, the complementation lines of cipk9 were constructed. The coding sequence of CIPK9 following its own promoter was transformed into the cipk9-1 mutant plants. Two independent complementation lines of cipk9 (Com-1 and Com-4) were obtained. The transcriptional expression levels of CIPK9 in these two complementation lines were both resumed (Fig. 1D). The phenotype test showed that both these two complementation lines displayed a similar phenotype to wild-type plants when they were grown on LK medium for 12 d (Fig. 1E). These results demonstrated that the LK-tolerant phenotype of cipk9 mutants was caused by the disruption of CIPK9.
In addition, the K+ content of various plants (ecotype Columbia [Col], cipk9-1, Com-1, and Com-4) was measured after they were grown on MS or LK medium for 12 d. Under LK conditions, cipk9-1 mutant plants accumulated more K+ in shoot tissues than wild-type plants (Fig. 1F), while the shoot K+ content of the two complementation lines was similar to that of the wild type (Fig. 1F). We observed no difference in K+ content among all these plants grown on MS medium (Fig. 1F). The results of K+ content measurements suggest that the LK-tolerant phenotype of cipk9 mutants may result from the greater K+ accumulation in the shoots of mutants under LK conditions.
Protein Interaction between CIPK9 and CBLs
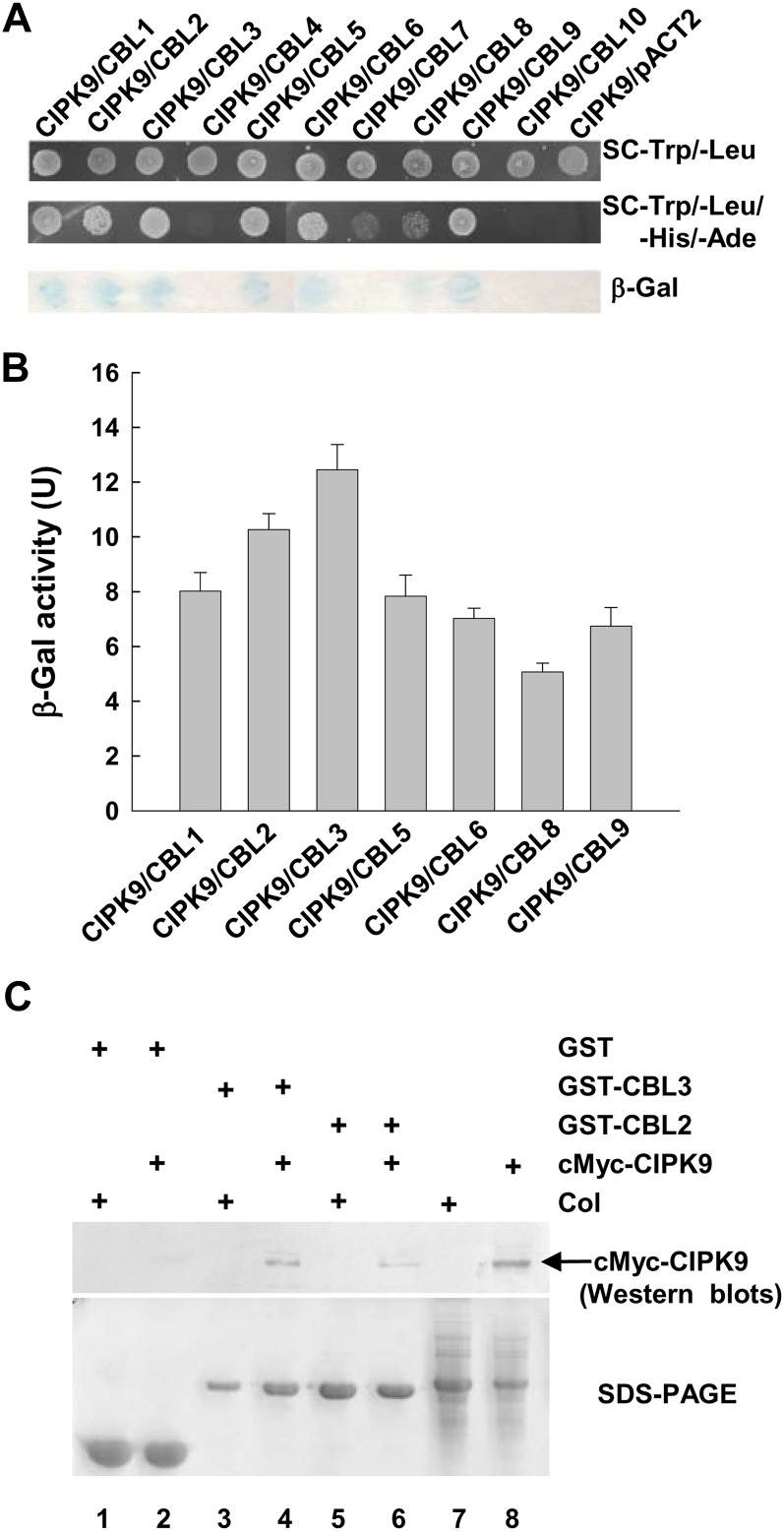
It has been reported that the activities of CIPKs are regulated by binding one or more CBL proteins (Kudla et al., 1999; Shi et al., 1999; Kim et al., 2000). To identify CBL(s) that interact with CIPK9, yeast (Saccharomyces cerevisiae) two-hybrid assays were applied to analyze the interaction between CIPK9 and each member of CBL family from Arabidopsis. The results demonstrated that seven CBLs, CBL1 to CBL3, CBL5, CBL6, CBL8, and CBL9, displayed interaction with CIPK9 (Fig. 2A). Quantitative β-galactosidase activity assays showed that CBL2 and CBL3 had the strongest interaction with CIPK9 among these seven CBLs (Fig. 2B).
Figure 2.
CIPK9 interaction with CBL2 and CBL3. A, Interaction analysis of CIPK9 with all Arabidopsis CBLs using yeast two-hybrid assays. B, Quantitative analysis of β-galactosidase activity of the positive clones containing the CIPK9/CBL complex. Data shown are means ± se (n = 3). U, Units. C, Interaction analysis of CIPK9 with CBL2 and CBL3 using pull-down assays. The GST-CBL2 and GST-CBL3 fusion proteins were used as bait to pull down from the whole plant proteins that were extracted from cMyc-CIPK9-transformed plants (cMyc-CIPK9) or wild-type plants (Col). The GST protein was used as a negative control. The top panel shows the western blot probed with anti-cMyc antibody. The arrow indicates the site of cMyc-CIPK9 protein. The bottom panel shows protein gel analysis indicating the amount of bait proteins used in pull-down assays.
To verify the interaction between CIPK9 and CBL2 or CBL3 in planta, protein pull-down assays were performed. In these assays, glutathione S-transferase (GST)-tagged CBL2 or CBL3 was used to pull down interacting proteins from the total proteins that were extracted from cMyc-CIPK9-transformed plants or wild-type plants. The cMyc antibody was used to detect cMyc-CIPK9 fusion proteins via immunoblotting. The results of pull-down assays showed that cMyc-CIPK9 proteins could be pulled down by GST-CBL2 or GST-CBL3 (Fig. 2C, lanes 6 and 4, respectively), indicating the interaction between CIPK9 and CBL2 or CBL3 in planta.
Expression Profiles of CIPK9, CBL2, and CBL3 in Arabidopsis
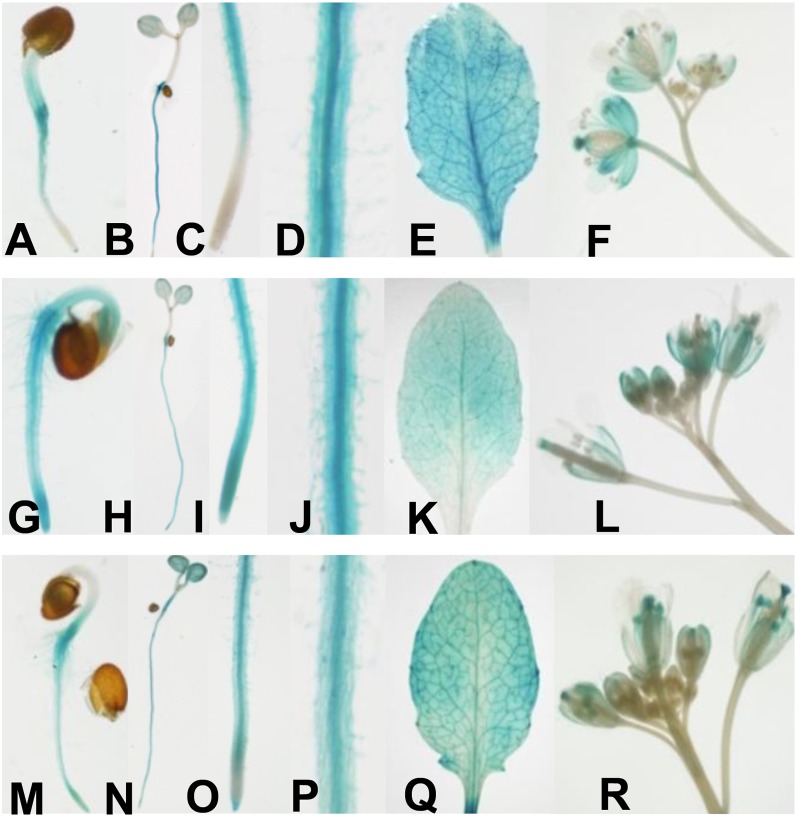
The expression profiles of CIPK9, CBL2, and CBL3 were investigated by detecting GUS activity in transgenic Arabidopsis seedlings that carried the GUS gene under the control of their own promoters (pCIPK9::GUS, pCBL2::GUS, and pCBL3::GUS). The GUS staining analysis showed that CIPK9, CBL2, and CBL3 were broadly expressed in whole seedlings at various developmental stages (Fig. 3). In root tissue, GUS activities in all transgenic seedlings were detectable (Fig. 3, B, H, and N), especially abundant in the vascular bundles (Fig. 3, D, J, and P). However, compared with CBL2 (Fig. 3I) and CBL3 (Fig. 3O), the expression of CIPK9 (Fig. 3C) could not be detected in the root tip of 5-d-old seedlings. In shoot tissue, all of them were expressed in whole leaves, particularly in the vascular tissue and hydathode (Fig. 3, E, K, and Q). These results showed that the expression profiles of CIPK9, CBL2, and CBL3 exhibited a remarkable overlap in most of the plant tissue, suggesting that CBL2 and CBL3 may function together with CIPK9 in vivo.
Figure 3.
Expression profiles of CIPK9, CBL2, and CBL3 in Arabidopsis. Expression profiles of CIPK9, CBL2, and CBL3 were shown by determining GUS activity in pCIPK9::GUS (A–F), pCBL2::GUS (G–L), and pCBL3::GUS (M–R) transgenic plants, respectively. A, G, and M, Two-day-old seedlings. B, H, and N, Five-day-old seedlings. C, I, and O, Roots of 5-d-old seedlings. D, J, and P, Root vascular tissue of 5-d-old seedlings. E, K, and Q, Leaves. F, L, and R, Flowers.
CIPK9 Interacts with CBL2 and CBL3 Mainly at the Tonoplast in Arabidopsis
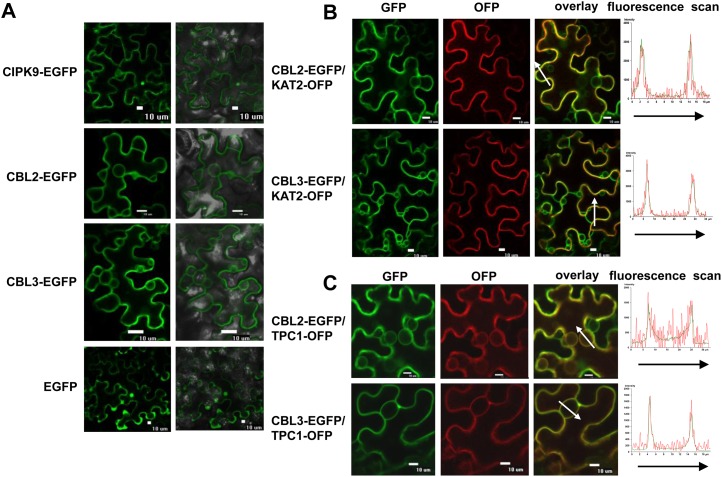
To test the subcellular localization of CIPK9, CBL2, and CBL3 in plant cells, the fusion genes CIPK9::EGFP (for enhanced GFP), CBL2::EGFP, and CBL3::EGFP were constructed and introduced into tobacco (Nicotiana benthamiana) leaves. The leaves expressing EGFP alone were used as controls. The results showed that the CIPK9-EGFP fusion protein displayed nonspecific subcellular localization (Fig. 4A). In addition to observation of the fluorescence of CBL2-EGFP and CBL3-EGFP at the tonoplast (Fig. 4A), the fluorescence of CBL2-EGFP and CBL3-EGFP was also observed to encircle the whole epidermal cell (Fig. 4A), indicating that CBL2-EGFP and CBL3-EGFP might also localize at the PM. To confirm this notion, the PM marker protein KAT2-OFP (for orange/red fluorescent protein) and the tonoplast marker protein TPC1-OFP were applied (Peiter et al., 2005; Xicluna et al., 2007). When CBL2-EGFP or CBL3-EGFP was coexpressed with KAT2-OFP or TPC1-OFP in tobacco leaves, the green fluorescence (CBL2-EGFP or CBL3-EGFP) and the red fluorescence (KAT2-OFP or TPC1-OFP) showed an obvious overlap (Fig. 4, B and C). Moreover, the CBL2-EGFP or CBL3-EGFP green fluorescence and the KAT2-OFP or TPC1-OFP red fluorescence were perfectly matched, indicating the colocalization of the two proteins (Fig. 4, B and C). These results demonstrated that CBL2 and CBL3 may locate at both the PM and tonoplast.
Figure 4.
Subcellular localization of CIPK9, CBL2, and CBL3 in tobacco leaves. A, Transient expression of CIPK9-EGFP, CBL2-EGFP, CBL3-EGFP, and EGFP. Left panels, GFP images; right panels, merge of bright-field and GFP images. Bars = 10 μm. B, Colocalization analysis of CBL2-EGFP and CBL3-EGFP with the PM marker KAT2-OFP. C, Colocalization analysis of CBL2-EGFP and CBL3-EGFP with the tonoplast marker TPC1-OFP. Left panels, GFP images; middle panels, OFP images; right panels, merge of OFP and GFP images. Bars = 10 μm. The fluorescence intensities of the arrow-covered regions were determined and are shown at right.
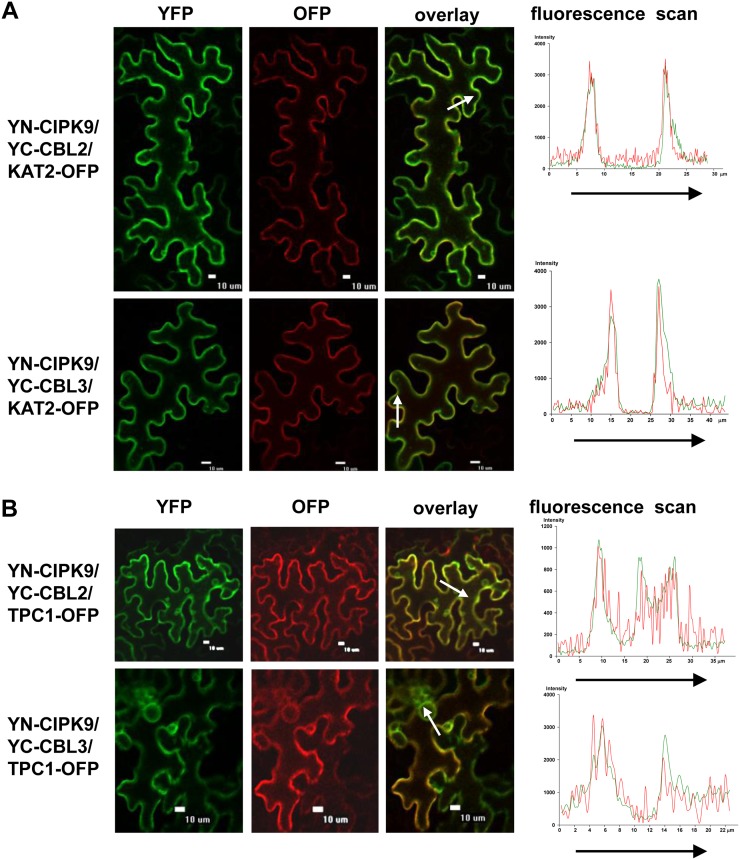
To investigate the interaction between CIPK9 and CBL2 or CBL3, bimolecular fluorescence complementation (BiFC) assays in tobacco leaves was performed (Waadt et al., 2008). CIPK9 was cloned into the split-yellow fluorescent protein (YFP) vector pSPYNE(R)173, and CBL2 or CBL3 was constructed into pSPYCE(MR) vector. The paired combinations of YN-CIPK9 and YC-CBL2 (or YC-CBL3) together with PM marker protein KAT2-OFP or the tonoplast marker protein TPC1-OFP were cotransformed into the tobacco leaves. The epidermal cells expressing CBL2/CIPK9 complex or CBL3/CIPK9 complex showed the YFP fluorescence signal at the PM and tonoplast. Moreover, the YFP fluorescence overlapped with the OFP fluorescence (Fig. 5), demonstrating that CIPK9 could interact with CBL2 and CBL3 at the PM and tonoplast in tobacco leaves.
Figure 5.
BiFC assays of CIPK9 interaction with CBL2 and CBL3 in tobacco leaves. A, KAT2-OFP was coexpressed and used as a PM marker. B, TPC1-OFP was coexpressed and used as a tonoplast marker. Left panels, YFP images; middle panels, OFP images; right panels, merge of YFP and OFP images. Bars = 10 μm. The fluorescence intensities of the arrow-covered regions were determined and are shown at right.
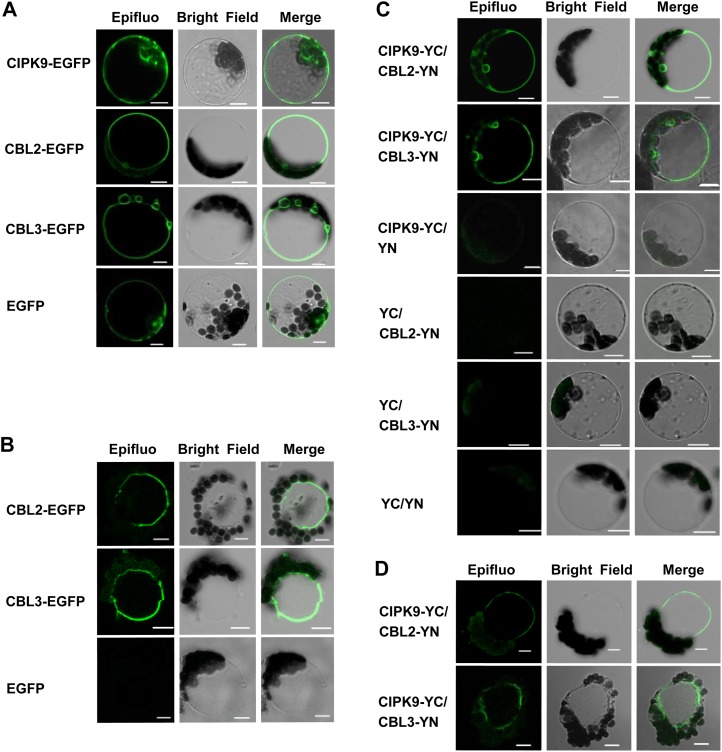
The subcellular localization of CIPK9, CBL2, and CBL3 proteins was further confirmed in Arabidopsis mesophyll protoplasts (Bracha-Drori et al., 2004; Walter et al., 2004). GFP fusion proteins of CBL2, CBL3, and CIPK9 were constructed and introduced into Arabidopsis mesophyll protoplasts. The results showed that CIPK9-EGFP did not exhibit specific localization. The fluorescence of the CBL2-EGFP or CBL3-EGFP fusion protein was mainly localized at the vacuolar membrane (Fig. 6A). To clarify the membrane identity, the transformed protoplasts were treated with hypoosmotic solution in order to release the vacuoles (Allen et al., 1998; Peiter et al., 2005). After disruption of the protoplasts expressing EGFP protein, EGFP fluorescence disappeared. CBL2-EGFP or CBL3-EGFP fluorescence was still observed at the tonoplast after the protoplasts were disrupted (Fig. 6B), suggesting the tonoplast localization of CBL2 and CBL3.
Figure 6.
The subcellular localization of CIPK9, CBL2, and CBL3 and the interaction of CIPK9 with CBL2 and CBL3 in Arabidopsis mesophyll protoplasts. A, Transient expression of CIPK9-EGFP, CBL2-EGFP, CBL3-EGFP, and EGFP in Arabidopsis mesophyll protoplasts. B, Vacuoles were isolated after bursting of the transformed protoplasts, and the images were collected immediately. C, Analysis of the interaction between CIPK9 and CBL2 or CIPK9 and CBL3 in Arabidopsis mesophyll protoplasts by BiFC assays. D, Vacuoles were isolated after bursting of the transformed protoplasts, and the images were collected immediately. Plasmid combinations are indicated on the left. Left panels, GFP fluorescence microscopy images; middle panels, bright-field microscopy images; right panels, merge of bright-field and fluorescence microscopy images. Bars = 10 μm.
For the BiFC assays in Arabidopsis mesophyll protoplasts, CIPK9 was cloned into the split-YFP vector pUC-SPYCE, and CBL2 or CBL3 was fused to pUC-SPYNE. Different combinations were cotransformed into Arabidopsis mesophyll protoplasts. In the protoplasts expressing CIPK9/CBL2 complex or CIPK9/CBL3 complex, the fluorescence signal was detected at the tonoplast (Fig. 6C). In addition, the vacuoles were released by the hypoosmotic treatment, and the fluorescence was still observed at the tonoplast (Fig. 6D). These results suggested that CIPK9 interacted with CBL2 and CBL3 mainly at the tonoplast in Arabidopsis.
Overexpressing Plants of CIPK9, CBL2, and CBL3 Are Sensitive to LK Stress
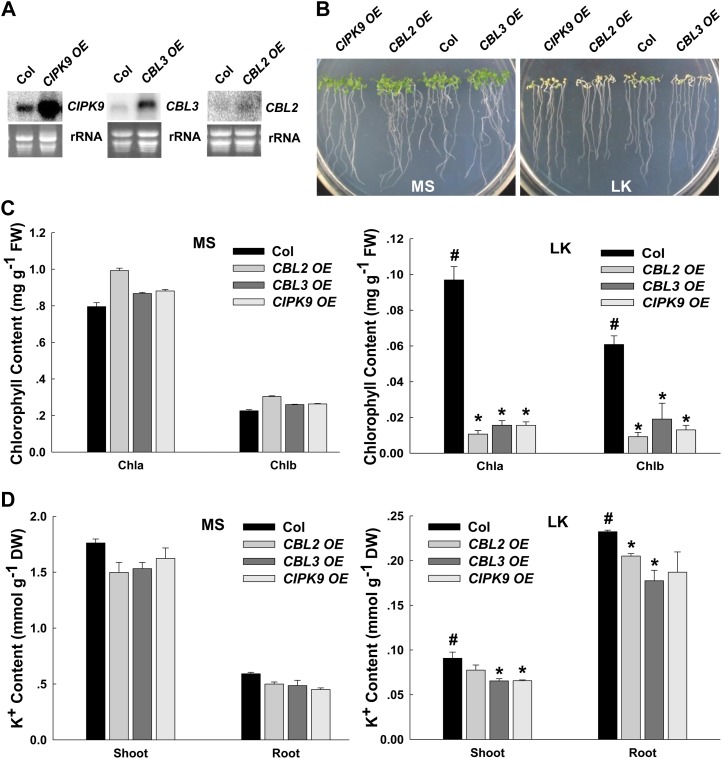
To test the hypothesis that CBL2 and/or CBL3 may play roles in plant responses to LK stress by interacting with CIPK9, the phenotypes and K+ contents of CIPK9-, CBL2-, and CBL3-overexpressing plants (CIPK9 OE, CBL2 OE, and CBL3 OE) as well as mutant plants under LK conditions were analyzed. The elevated transcriptional expression of the genes CIPK9 OE, CBL2 OE, and CBL3 OE were verified by northern blot (Fig. 7A). The phenotype test showed that all overexpressing plants exhibited more sensitive phenotypes under LK conditions (i.e. their leaves became chlorotic earlier than wild-type plants; Fig. 7B; Supplemental Figs. S3A and S5A). This sensitive phenotype was dependent on the K+ concentrations but not on NH4+ concentrations in the medium (Supplemental Figs. S1A and S2A).
Figure 7.
Phenotype tests and K+ content measurements of CIPK9-, CBL2-, and CBL3-overexpressing lines under LK stress. A, Northern-blot analysis of CIPK9, CBL2, and CBL3 expression levels in transgenic plants. B, Phenotype comparison between overexpressing lines (CIPK9 OE, CBL2 OE, and CBL3 OE) and wild-type plants after growth on MS or LK medium for 10 d. C, Chlorophyll contents of various plant materials. The samples were prepared after the plants were grown on MS or LK medium for 10 d. D, K+ content measurements of various plant materials as indicated. K+ contents of shoots and roots were measured after the plants were grown on MS or LK medium for 10 d. In C and D, data are shown as means ± se (n = 3). Student’s t test (*P < 0.05) was used to analyze statistical significance compared with the control (#). DW, Dry weight; FW, fresh weight.
To quantify the LK-sensitive phenotype, the chlorophyll a and b contents of these overexpressing plants and wild-type plants were tested. The results showed that chlorophyll contents of all the overexpressing plants were much lower than those of wild-type plants under LK conditions (Fig. 7C). In addition, the K+ contents in all three overexpressing plants were decreased compared with wild-type plants, especially under LK conditions (Fig. 7D). These results further confirmed the LK-sensitive phenotype of CIPK9-, CBL2-, and CBL3-overexpressing plants.
CBL3/CIPK9 Regulates K+ Homeostasis under LK Stress
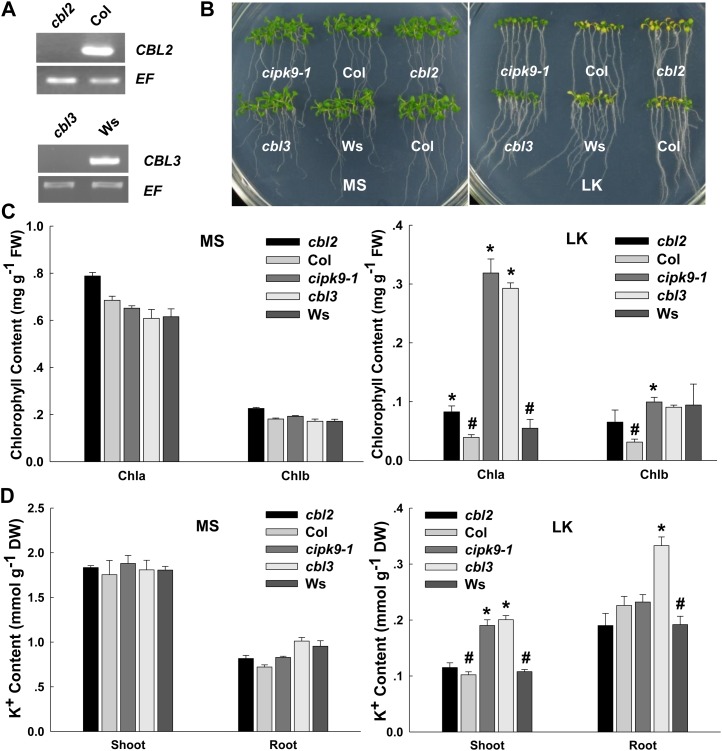
The T-DNA insertion mutants cbl2 (Col background) and cbl3 (Wassilewskija [Ws] background) were tested for their phenotypes under LK treatment. Interestingly, cbl2 mutant plants exhibited a different phenotype from cbl3 mutant plants under LK treatment, although CBL2 and CBL3 proteins showed high similarity in their amino acid sequences and both interacted with CIPK9. The cbl3 mutant plants showed a similar LK-tolerant phenotype to cipk9 mutants, whose shoots remained green after growth on LK medium for 12 d (Fig. 8B), whereas the cbl2 mutant plants and wild-type plants showed similar chlorotic symptoms (Fig. 8B; Supplemental Figs. S3B and S5B). Moreover, this tolerant phenotype of cbl3 and cipk9 mutants was also dependent on the K+ concentrations but not on NH4+ concentrations in the medium (Supplemental Figs. S1B and S2B).
Figure 8.
Comparison of phenotypes and K+ contents between cbl3 mutant plants and cipk9-1 mutant plants under LK stress. A, Reverse transcription-PCR analysis of CBL2 and CBL3 expression levels in various plants. B, Phenotype comparison between the mutant plants (cipk9-1, cbl2, and cbl3) and wild-type plants (Col and Ws) after growth on MS or LK medium for 12 d. C, Chlorophyll contents of various plant materials. The samples were prepared after the plants were grown on MS or LK medium for 12 d. D, K+ content measurements of various plant materials as indicated. K+ contents of shoots and roots were measured after the plants were grown on MS or LK medium for 12 d. In C and D, data are shown as means ± se (n = 3). Student’s t test (*P < 0.05) was used to analyze statistical significance compared with the control (#). In these assays, the data from cbl2 and cipk9 mutant plants were compared with the Col wild-type plants as a control. The data derived from Ws wild-type plants were used as the control for cbl3 mutant plants. DW, Dry weight; FW, fresh weight.
The results of chlorophyll content measurements showed that the chlorophyll a contents in cbl3 and cipk9-1 mutant plants were much higher than that in wild-type plants under LK conditions (Fig. 8C), which was consistent with their LK-tolerant phenotype. The K+ contents in shoots of cipk9 and cbl3 mutants were much higher than those of wild-type and cbl2 mutant plants after growth on LK medium for 12 d (Fig. 8D). All these results derived from phenotype tests and K+ content measurements indicated that CBL3 and CIPK9 might work together in vivo and play crucial roles in K+ homeostasis in Arabidopsis under LK conditions. Although CBL2 can physically interact with CIPK9, it may not functionally regulate CIPK9 activity. Furthermore, it is notable that the K+ content in cbl3 roots was much higher than that of any other plants, which suggested that CBL3 might also regulate other components involved in K+ uptake in Arabidopsis roots at the same time.
DISCUSSION
The Ca2+ sensor CBLs and protein kinase CIPKs work together and constitute a specific regulatory network of Ca2+ signaling in plant cells (Kim et al., 2000; Albrecht et al., 2001; Luan, 2009).
Accumulated evidence in the last decade has revealed that the CBL/CIPK regulatory pathways are mainly involved in plant responses to abiotic stresses, especially to ionic stresses, such as salt stress (Quintero et al., 2002), pH stress (Fuglsang et al., 2007), K+ deficiency (Xu et al., 2006), and nitrate deficiency (Ho et al., 2009). Because of the interaction cross talk, CBLs and CIPKs display broad functional redundancy. One CBL or CIPK protein may participate in several different physiological processes. It has been reported that CBL1 functions as a positive regulator in both salt and drought responses, and it also works as a negative regulator in the cold response in Arabidopsis (Cheong et al., 2003). In addition, CIPK23 was found to be involved in both root K+ uptake and the nitrate response (Xu et al., 2006; Ho et al., 2009). On the other hand, different CBLs or CIPKs may also work in the same pathway at the same time, so-called functional redundancy. Both CBL1 and CBL9 play roles in Arabidopsis root K+ uptake and show functional compensation (Xu et al., 2006). In this study, both CBL2- and CBL3-overexpressing lines showed the LK-sensitive phenotype and K+ contents similar to CIPK9-overexpressing lines (Fig. 7, B and D), which indicated that they may function in a CIPK9-regulated K+ deficiency response. However, the results of phenotype tests and K+ content measurements of mutant plants showed that only cbl3 and cipk9 displayed similar LK-tolerant phenotypes, but not cbl2 (Fig. 8, B and D). It is suggested that CBL3 is probably the primary upstream regulator of CIPK9 in response to K+ deficiency, while CBL2 may partially participate in this process.
Previous reports (Qi et al., 2008; Pyo et al., 2010) also tested the LK phenotypes of Arabidopsis plants on LK medium (100 μm K+), but they did not observe the typical chlorotic phenotype. However, the LK chlorotic phenotype observed in this study does not conflict with these previous reports for two main reasons. First, the NH4+ concentrations in the LK media are different. In the previous reports (Qi et al., 2008; Pyo et al., 2010), NH4+-free medium was used to test the LK phenotype, while the LK medium contained 1.25 mm NH4+ in our study. It is known that NH4+ could affect root high-affinity K+ uptake and the LK phenotype, especially under LK conditions. Second, the periods of LK treatment time are different. In the previous reports (Qi et al., 2008; Pyo et al., 2010), the LK phenotype was observed after the seedlings grew on LK medium for 6 or 7 d. One may not observe the chlorotic phenotype after 6 to 7 d of LK treatment, while the chlorotic phenotype appeared after 10 d in LK medium in our study. Thus, the time for LK treatment in our study was 10 or 12 d to test the LK-sensitive phenotype (overexpression plants) or the LK-tolerant phenotype (mutant plants), respectively.
How does the CBL3/CIPK9 complex work in plants? Our results revealed that the K+ content in all the overexpressing lines was reduced compared with wild-type plants, especially under LK conditions (Fig. 7D). According to the expression patterns of these three genes in root epidermal cells and root hairs (Fig. 3, D, J, and P), one possible hypothesis is that the overexpression of CIPK9, CBL2, and CBL3 may impair root K+ uptake from the environment and result in the reduction of plant K+ content. In addition, tissue expression profile analysis also showed that these three genes were all strongly expressed in root vascular bundles (Fig. 3, D, J, and P), which suggested a role in mineral translocation. As shown in Supplemental Figure S6, the root/shoot K+ ratio under LK conditions is increased compared with normal conditions (MS medium). We found that the root/shoot K+ ratio in cipk9 and cbl3 was much lower than that in wild-type plants under LK conditions (Supplemental Fig. S6). This result indicates that the K+ distribution (or translocation) between shoot and root is significantly affected in cipk9 and cbl3 mutants. It is hypothesized that CIPK9 might also play roles in the K+ distribution (or translocation) between shoots and roots, especially under LK conditions.
Northern-blot analysis indicated that CIPK9 might respond to LK stress (Supplemental Fig. S4). CIPK9 was slightly induced when the 7-d-old plants were transferred from MS medium to LK medium for 8 and 24 h (Supplemental Fig. S4). Since the expression of CIPK9 may impair root K+ uptake according to K+ content measurements, it seems that this induction of CIPK9 under LK conditions is not beneficial for plants to cope with LK stress. It is difficult to explain this at this point, although the increased transcription does not necessarily enhance the function of the CIPK protein. We are also confused by this result. Anyway, the transcriptional regulation of CIPK9 under LK stress is still unclear and should be further investigated in the future.
It is known that some CBL proteins could play a critical role in recruiting the interacting CIPK to the membranes to phosphorylate the membrane-associated target proteins, such as CBL1/CBL9 or SOS3 (CBL4; Xu et al., 2006; Weinl and Kudla, 2009). According to our results from subcellular localization analysis, CIPK9 was localized in the cytoplasm and nucleus in tobacco epidermal cells and CBL2 and CBL3 showed apparent localization at the tonoplast (Figs. 4C and 6B), which is consistent with the report by Batistič et al. (2010). However, by using PM marker protein, we also detected the fluorescence of CBL2-EGFP and CBL3-EGFP at the PM in tobacco epidermal cells (Fig. 4B). Some other research groups also reported the PM localization of the CBL3 protein (Oh et al., 2008; Zhou et al., 2009), suggesting their possibly diverse roles in different subcellular regions. In BiFC assays, the protein complex of CBL2/CIPK9 or CBL3/CIPK9 was also detected at the PM and tonoplast in tobacco leaves (Fig. 5) but primarily at the tonoplast when analyzed in Arabidopsis mesophyll protoplasts (Fig. 6, C and D). This implied that CBL2 or CBL3 and CIPK9 might function mainly at the tonoplast in Arabidopsis. In plants, the vacuolar membrane is required for K+ homeostasis. The vacuoles are important for the deposition of minerals. Plant growth, to a large extent, is driven by vacuolar K+ accumulation. Apart from the vacuolar deposition of minerals to provide turgor, vacuoles typically function as buffer compartments for excess nutrient uptake. During periods of deficiency, minerals such as K+ are released from the vacuole to maintain cytosolic homeostasis as well as turgor and membrane potential (Leigh and Wyn Jones, 1984; Walker et al., 1996; Latz et al., 2007), which enables plants to survive. CIPK9/CBL3 may be involved in this process to regulate K+ homeostasis under LK stress.
Until now, several CBL/CIPK regulatory pathways as well as their target proteins have been reported, and most of the target proteins of CBL/CIPK complexes are transporters (SOS1, CHL1), channels (AKT1), or proton pumps (AHA2), which may explain their important functions in plant ionic homeostasis (Quintero et al., 2002; Xu et al., 2006; Fuglsang et al., 2007; Ho et al., 2009).
Thus, it was further hypothesized that CIPK9 may regulate a K+ channel or transporter directly or indirectly. We have tested the protein interaction between CIPK9 and many K+ channels and transporters in Arabidopsis. Unfortunately, all the tested K+ channels or transporters could not interact with CIPK9, including the major two K+ uptake components AKT1 and HAK5, the K+ translocation components AKT2 and SKOR, as well as the K+ tonoplast channel TPK1 (data not shown). Therefore, the identification of target proteins of CIPK9 in the future will be very important and will provide us more clues to understand the regulatory mechanism of the CBL3/CIPK9 complex in response to LK stress.
MATERIALS AND METHODS
Plant Materials
Arabidopsis (Arabidopsis thaliana) ecotypes Col and Ws were used as wild-type plants in this study. The T-DNA insertion mutant lines of CIPK9 and CBL2 were obtained from the Arabidopsis Biological Resource Center. The cipk9 homozygous mutants (cipk9-1 and cipk9-2) were identified from SALK_058629 and SALK_014699 by the T-DNA border primer and CIPK9 gene-specific primers. The cbl2 mutant was identified from SALK_151426 by the T-DNA border primer and CBL2 gene-specific primers. The cbl3 mutant was identified from FLAG_294H06 by the T-DNA border primer and CBL3 gene-specific primers.
The overexpression of CIPK9 was constructed by fusing the CIPK9 coding sequence into pBIB vector between the enzyme sites XbaI and SacI. For CBL2 and CBL3 overexpression, the coding sequences of these two genes were cloned into the pBIB vector by using the enzyme sites SmaI and SacI, respectively. The plasmids were introduced into Agrobacterium tumefaciens GV3101 and transformed into Arabidopsis wild-type (Col) plants. For the complementation lines of cipk9, the CIPK9 coding sequence driven by its own promoter was cloned into pCAMBIA1300 vector. The construct was introduced into A. tumefaciens GV3101 and transformed into cipk9-1 mutant plants. For all experiments, T3 transgenic plants were used.
The Construction of promoter::GUS Transgenic Plants
The CIPK9 promoter fragment (1,748 bp) was amplified by using forward primer 5′-ATAAGCTTTACTAAAACGGCAAATACTAAAC-3′ and reverse primer 5′-ACGGATCCTTTCTTTTTCCGATTAAGAAA-3′ from genomic DNA and then cloned into the vector pCAMBIA1391 in front of the GUS coding sequence by using HindIII and BamHI sites. The CBL2 promoter fragment (1,546 bp) was amplified by using forward primer 5′-TTGAATTCACCCGGTTACTGGATTTGTTC-3′ and reverse primer 5′-GGGGATCCGATATATCTTAAGCAGCAAA-3′ and cloned into the vector pCAMBIA1381 between EcoRI and BamHI sites. The CBL3 promoter fragment (1,706 bp) was amplified by using forward primer 5′-AAGGATCCAAAATCAAAACAATACATGAGTCA-3′ and reverse primer 5′-AACTGCAGGATATATCTTGTAAATCAAAACTCTCC-3′ and cloned into pCAMBIA1381 vector by using the BamHI and PstI sites. All these constructs were introduced into Arabidopsis wild-type (Col) plants by using A. tumefaciens-mediated transformation. GUS activity analysis of the T3 transgenic lines at different developmental stages was performed as described (Jefferson et al., 1987).
Growth Conditions and LK Stress Treatment
The seeds were surface sterilized and germinated on MS or LK medium at 22°C under constant illumination (60 μmol m−2 s−1) for 10 or 12 d. The normal MS medium contained 0.8% (w/v) agar (Baoshi) and 3% (w/v) Suc. For LK treatment, the LK medium was modified from MS medium. Briefly, 2.99 mm CaCl2 and 1.25 mm KH2PO4 were replaced by 2.99 mm Ca(NO3)2 and 1.25 mm NH4H2PO4, 18.79 mm KNO3 and 20.6 mm NH4NO3 were removed, and 1.5 mm MgSO4 was unchanged. For making the LK medium, the actual and final K+ concentration in LK agar medium (0.8%, w/v) was adjusted to 100 ± 10 μm by adding KCl. The final K+ concentration in the LK medium for each experiment was measured and confirmed to be 100 ± 10 μm by using atomic absorption spectrophotometry.
Measurement of K+ Content
After growth on MS or LK medium for 10 or 12 d, shoots and roots of the seedlings were harvested for K+ content measurements. First, the plant samples were rinsed thoroughly with distilled water three times and dried at 80°C for 24 h. After weighing, the samples were treated in a muffle furnace at 300°C for 1 h, 575°C for 5 h, and dissolved in 0.1 n HCl. Then, the K+ concentration of the samples was measured by atomic absorption spectrophotometry (Hitachi Z5000).
Northern-Blot and Reverse Transcription-PCR Analyses
Total RNA of the plants was extracted from 10-d-old seedlings by using Trizol reagent (Invitrogen). For RNA gel-blot analysis, total RNA (30 mg for each lane) was separated by electrophoresis and then blotted onto a nylon membrane. The blot was hybridized with [α-32P]dCTP-labeled gene-specific DNA probes for CIPK9, CBL2, or CBL3. Hybridization signals were imaged by using a phosphor imager. The primers for probe amplifying were as follows: for CIPK9, forward primer 5′-AAATGGAAGAAACCGCAAAGC-3′ and reverse primer 5′-TTTCAAACCGAGTTTGAGGA-3′; for CBL2, forward primer 5′-ATGTCGCAGTGCGTTGACG-3′ and reverse primer 5′-TCAGGTATCTTCAACCTGAGAAT-3′; for CBL3, forward primer 5′-ATGTCGCAGTGCATAGACG-3′ and reverse primer 5′-TCAGGTATCTTCCACCTGCG-3′.
Yeast Two-Hybrid Assays
The coding sequence of CIPK9 was cloned into the DNA-binding domain vector (pAS2) by using the sites NdeI and XhoI. Each of the CBLs was cloned into the activation domain vector (pACT2). For yeast (Saccharomyces cerevisiae) two-hybrid assays, CIPK9-BD and each CBL-AD construct were cotransformed into the yeast strain AH109 by using the lithium acetate method (according to the TRANFOR protocol). To select the positive clones, the transformants were selected on synthetic dropout nutrient medium (SD)/−Leu−Trp medium and then streaked on selective medium (SD/−Leu−Trp−His−adenine). The β-galactosidase expression was analyzed by filter-lift assays as described (Shi et al., 1999).
For quantitative assays, the transformants were grown at 30°C to midlog phase (optical density at 600 nm = 0.5–0.8) in SD/−Leu−Trp liquid medium. The β-galactosidase activity was measured as optical density at 420 nm using o-nitrophenyl β-d-galactopyranoside as the substrate according to Shi et al. (1999) and expressed in units.
Subcellular Localization and BiFC Assays in Plant Cells
For the subcellular localization analysis, the fusion genes CIPK9::EGFP, CBL2::EGFP, and CBL3::EGFP were cloned into pBIB vector by using XbaI and SacI sites. For the generation of BiFC constructs, the coding regions of CIPK9 and CBL3 were cloned into pSPYNE(R)173 and pSPYCE(MR), respectively, by using SpeI and SalI sites. CBL2 was cloned into pSPYCE(MR) by SpeI and SmaI sites. All the plasmids were introduced into A. tumefaciens GV3101 and coinfiltrated at optical density at 600 nm = 0.6 to 0.7. After infiltration for 2 to 4 d, the fluorescence of GFP, YFP, or red fluorescent protein in transformed tobacco (Nicotiana benthamiana) leaves was observed. The images were acquired with a Nikon TE-2000 confocal microscope. Fluorescence intensity scan was performed using the quantification tool of the NIS-Elements Imaging software of Nikon ECLIPSE Ti.
For the generation of GFP and BiFC constructs expressed in Arabidopsis mesophyll protoplasts, the coding region of CIPK9 was cloned via XbaI and BamHI into pUC-EGFP and pUC-SPYCE. The coding region of CBL2 was cloned into pUC-EGFP and pUC-SPYNE by XbaI and SmaI, while the XbaI and SpeI sites were used for CBL3. Mesophyll protoplasts were isolated from leaves of about 6-week-old wild-type Arabidopsis (Col). These plasmids were purified by a Qiagen kit and transiently transformed into mesophyll protoplasts using a polyethylene glycol-mediated method according to Sheen (2001). To release the vacuoles, the protoplasts were treated in the low osmotic lysis buffer (10 mm EGTA, 250 mm sorbitol, 10 mm HEPES, pH 8.0, adjusted with KOH). Fluorescence of GFP, YFP, and red fluorescent protein was observed. The images were acquired with a LSM510 META confocal microscope after incubation at 23°C for 16 to 24 h.
GST Pull-Down Assays
The coding sequences of CBL2 and CBL3 were cloned into the pGEX-4T-2 vector by using BamHI and XhoI sites. The GST fusion constructs (GST-CBL2 and GST-CBL3) were transformed into Escherichia coli strain BL21 (DE3). The recombinant proteins were purified by glutathione Sepharose (Amersham Pharmacia) according to the manufacturer’s manual. Total proteins were extracted from 2-week-old Arabidopsis seedlings of wild-type and cMyc-CIPK9 transgenic plants by lysis buffer (50 mm Tris-HCl, pH 7.5, 100 mm NaCl, 0.2 mm CaCl2, and 0.05% Tween 20) with 1 mm phenylmethylsulfonyl fluoride and 1 mm dithiothreitol.
For pull-down assays, 200 μg of total proteins was mixed with GST-CBL2, GST-CBL3, or GST protein and then immobilized on glutathione Sepharose beads. After incubation at 4°C for 6 h, the beads were washed six times by lysis buffer. Then, the proteins were eluted by elution buffer and analyzed by western blot using anti-cMyc monoclonal antibody.
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under accession numbers At1g01140 (CIPK9), At5g55990 (CBL2), and At4g26570 (CBL2).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phenotype test of various plants under different NH4+ concentrations.
Supplemental Figure S2. Phenotype test of various plants under different K+ concentrations.
Supplemental Figure S3. Phenotype test of various plants under LK conditions on horizontal plates.
Supplemental Figure S4. Northern-blot analysis of CIPK9 in response to LK stress.
Supplemental Figure S5. Phenotype test of various plants on modified LK medium.
Supplemental Figure S6. The root/shoot K+ ratio of various plants on MS or LK medium.
Acknowledgments
We thank Dr. Jörg Kudla (Universität Münster) for providing the plasmids for BiFC assays. We are grateful to Dr. Yan Guo (China Agricultural University) for providing us the seeds of cbl3 mutant plants as well as for helpful discussions and suggestions.
Glossary
- K+
potassium
- PM
plasma membrane
- LK
low-potassium
- MS
Murashige and Skoog
- Col
ecotype Columbia
- Ws
Wassilewskija
- GST
glutathione S-transferase
- BiFC
bimolecular fluorescence complementation
- YFP
yellow fluorescent protein
- OFP
orange/red fluorescent protein
- SD
synthetic dextrose
- T-DNA
transfer DNA
References
- Albrecht V, Ritz O, Linder S, Harter K, Kudla J. (2001) The NAF domain defines a novel protein-protein interaction module conserved in Ca2+-regulated kinases. EMBO J 20: 1051–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen G, Amtmann A, Sanders D. (1998) Calcium-dependent and calcium-independent K+ mobilization channels in Vicia faba guard cell vacuoles. J Exp Bot 49: 305–318 [Google Scholar]
- Amtmann A, Blatt MR. (2009) Regulation of macronutrient transport. New Phytol 181: 35–52 [DOI] [PubMed] [Google Scholar]
- Batistič O, Waadt R, Steinhorst L, Held K, Kudla J. (2010) CBL-mediated targeting of CIPKs facilitates the decoding of calcium signals emanating from distinct cellular stores. Plant J 61: 211–222 [DOI] [PubMed] [Google Scholar]
- Bracha-Drori K, Shichrur K, Katz A, Oliva M, Angelovici R, Yalovsky S, Ohad N. (2004) Detection of protein-protein interactions in plants using bimolecular fluorescence complementation. Plant J 40: 419–427 [DOI] [PubMed] [Google Scholar]
- Cheong YH, Kim KN, Pandey GK, Gupta R, Grant JJ, Luan S. (2003) CBL1, a calcium sensor that differentially regulates salt, drought, and cold responses in Arabidopsis. Plant Cell 15: 1833–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong YH, Pandey GK, Grant JJ, Batistic O, Li L, Kim BG, Lee SC, Kudla J, Luan S. (2007) Two calcineurin B-like calcium sensors, interacting with protein kinase CIPK23, regulate leaf transpiration and root potassium uptake in Arabidopsis. Plant J 52: 223–239 [DOI] [PubMed] [Google Scholar]
- Chérel I, Michard E, Platet N, Mouline K, Alcon C, Sentenac H, Thibaud JB. (2002) Physical and functional interaction of the Arabidopsis K+ channel AKT2 and phosphatase AtPP2CA. Plant Cell 14: 1133–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson DT, Hanson JB. (1980) The mineral nutrition of higher plants. Annu Rev Plant Physiol 31: 239–298 [Google Scholar]
- Deeken R, Sanders C, Ache P, Hedrich R. (2000) Developmental and light-dependent regulation of a phloem-localised K+ channel of Arabidopsis thaliana. Plant J 23: 285–290 [DOI] [PubMed] [Google Scholar]
- Duby G, Hosy E, Fizames C, Alcon C, Costa A, Sentenac H, Thibaud JB. (2008) AtKC1, a conditionally targeted Shaker-type subunit, regulates the activity of plant K+ channels. Plant J 53: 115–123 [DOI] [PubMed] [Google Scholar]
- Fuglsang AT, Guo Y, Cuin TA, Qiu Q, Song C, Kristiansen KA, Bych K, Schulz A, Shabala S, Schumaker KS, et al. (2007) Arabidopsis protein kinase PKS5 inhibits the plasma membrane H+-ATPase by preventing interaction with 14-3-3 protein. Plant Cell 19: 1617–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajdanowicz P, Michard E, Sandmann M, Rocha M, Corrêa LGG, Ramírez-Aguilar SJ, Gomez-Porras JL, González W, Thibaud JB, van Dongen JT, et al. (2011) Potassium (K+) gradients serve as a mobile energy source in plant vascular tissues. Proc Natl Acad Sci USA 108: 864–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaymard F, Pilot G, Lacombe B, Bouchez D, Bruneau D, Boucherez J, Michaux-Ferrière N, Thibaud JB, Sentenac H. (1998) Identification and disruption of a plant Shaker-like outward channel involved in K+ release into the xylem sap. Cell 94: 647–655 [DOI] [PubMed] [Google Scholar]
- Geiger D, Becker D, Vosloh D, Gambale F, Palme K, Rehers M, Anschuetz U, Dreyer I, Kudla J, Hedrich R. (2009) Heteromeric AtKC1·AKT1 channels in Arabidopsis roots facilitate growth under K+-limiting conditions. J Biol Chem 284: 21288–21295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierth M, Mäser P. (2007) Potassium transporters in plants: involvement in K+ acquisition, redistribution and homeostasis. FEBS Lett 581: 2348–2356 [DOI] [PubMed] [Google Scholar]
- Gierth M, Mäser P, Schroeder JI. (2005) The potassium transporter AtHAK5 functions in K+ deprivation-induced high-affinity K+ uptake and AKT1 K+ channel contribution to K+ uptake kinetics in Arabidopsis roots. Plant Physiol 137: 1105–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held K, Pascaud F, Eckert C, Gajdanowicz P, Hashimoto K, Corratgé-Faillie C, Offenborn JN, Lacombe B, Dreyer I, Thibaud JB, et al. (2011) Calcium-dependent modulation and plasma membrane targeting of the AKT2 potassium channel by the CBL4/CIPK6 calcium sensor/protein kinase complex. Cell Res 21: 1116–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch RE, Lewis BD, Spalding EP, Sussman MR. (1998) A role for the AKT1 potassium channel in plant nutrition. Science 280: 918–921 [DOI] [PubMed] [Google Scholar]
- Ho CH, Lin SH, Hu HC, Tsay YF. (2009) CHL1 functions as a nitrate sensor in plants. Cell 138: 1184–1194 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BG, Waadt R, Cheong YH, Pandey GK, Dominguez-Solis JR, Schültke S, Lee SC, Kudla J, Luan S. (2007) The calcium sensor CBL10 mediates salt tolerance by regulating ion homeostasis in Arabidopsis. Plant J 52: 473–484 [DOI] [PubMed] [Google Scholar]
- Kim KN, Cheong YH, Gupta R, Luan S. (2000) Interaction specificity of Arabidopsis calcineurin B-like calcium sensors and their target kinases. Plant Physiol 124: 1844–1853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MJ, Ruzicka D, Shin R, Schachtman DP. (2012) The Arabidopsis AP2/ERF transcription factor RAP2.11 modulates plant response to low-potassium conditions. Mol Plant 5: 1042–1057 [DOI] [PubMed] [Google Scholar]
- Kudla J, Xu Q, Harter K, Gruissem W, Luan S. (1999) Genes for calcineurin B-like proteins in Arabidopsis are differentially regulated by stress signals. Proc Natl Acad Sci USA 96: 4718–4723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacombe B, Pilot G, Michard E, Gaymard F, Sentenac H, Thibaud JB. (2000) A Shaker-like K+ channel with weak rectification is expressed in both source and sink phloem tissues of Arabidopsis. Plant Cell 12: 837–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latz A, Becker D, Hekman M, Müller T, Beyhl D, Marten I, Eing C, Fischer A, Dunkel M, Bertl A, et al. (2007) TPK1, a Ca2+-regulated Arabidopsis vacuole two-pore K+ channel is activated by 14-3-3 proteins. Plant J 52: 449–459 [DOI] [PubMed] [Google Scholar]
- Lebaudy A, Véry AA, Sentenac H. (2007) K+ channel activity in plants: genes, regulations and functions. FEBS Lett 581: 2357–2366 [DOI] [PubMed] [Google Scholar]
- Lee SC, Lan WZ, Kim BG, Li L, Cheong YH, Pandey GK, Lu G, Buchanan BB, Luan S. (2007) A protein phosphorylation/dephosphorylation network regulates a plant potassium channel. Proc Natl Acad Sci USA 104: 15959–15964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh RA, Wyn Jones RG. (1984) A hypothesis relating critical potassium concentrations for growth to the distribution and functions of this ion in the plant cell. New Phytol 97: 1–13 [Google Scholar]
- Li L, Kim BG, Cheong YH, Pandey GK, Luan S. (2006) A Ca2+ signaling pathway regulates a K+ channel for low-K response in Arabidopsis. Proc Natl Acad Sci USA 103: 12625–12630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan S. (2009) The CBL-CIPK network in plant calcium signaling. Trends Plant Sci 14: 37–42 [DOI] [PubMed] [Google Scholar]
- Marten I, Hoth S, Deeken R, Ache P, Ketchum KA, Hoshi T, Hedrich R. (1999) AKT3, a phloem-localized K+ channel, is blocked by protons. Proc Natl Acad Sci USA 96: 7581–7586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SI, Park J, Yoon S, Kim Y, Park S, Ryu M, Nam MJ, Ok SH, Kim JK, Shin JS, et al. (2008) The Arabidopsis calcium sensor calcineurin B-like 3 inhibits the 5′-methylthioadenosine nucleosidase in a calcium-dependent manner. Plant Physiol 148: 1883–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiter E, Maathuis FJM, Mills LN, Knight H, Pelloux J, Hetherington AM, Sanders D. (2005) The vacuolar Ca2+-activated channel TPC1 regulates germination and stomatal movement. Nature 434: 404–408 [DOI] [PubMed] [Google Scholar]
- Pyo YJ, Gierth M, Schroeder JI, Cho MH. (2010) High-affinity K+ transport in Arabidopsis: AtHAK5 and AKT1 are vital for seedling establishment and postgermination growth under low-potassium conditions. Plant Physiol 153: 863–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Z, Hampton CR, Shin R, Barkla BJ, White PJ, Schachtman DP. (2008) The high affinity K+ transporter AtHAK5 plays a physiological role in planta at very low K+ concentrations and provides a caesium uptake pathway in Arabidopsis. J Exp Bot 59: 595–607 [DOI] [PubMed] [Google Scholar]
- Quan R, Lin H, Mendoza I, Zhang Y, Cao W, Yang Y, Shang M, Chen S, Pardo JM, Guo Y. (2007) SCABP8/CBL10, a putative calcium sensor, interacts with the protein kinase SOS2 to protect Arabidopsis shoots from salt stress. Plant Cell 19: 1415–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero FJ, Ohta M, Shi HZ, Zhu JK, Pardo JM. (2002) Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis. Proc Natl Acad Sci USA 99: 9061–9066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J. (2001) Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiol 127: 1466–1475 [PMC free article] [PubMed] [Google Scholar]
- Shi J, Kim KN, Ritz O, Albrecht V, Gupta R, Harter K, Luan S, Kudla J. (1999) Novel protein kinases associated with calcineurin B-like calcium sensors in Arabidopsis. Plant Cell 11: 2393–2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Véry AA, Sentenac H. (2003) Molecular mechanisms and regulation of K+ transport in higher plants. Annu Rev Plant Biol 54: 575–603 [DOI] [PubMed] [Google Scholar]
- Waadt R, Schmidt LK, Lohse M, Hashimoto K, Bock R, Kudla J. (2008) Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J 56: 505–516 [DOI] [PubMed] [Google Scholar]
- Walker DJ, Leigh RA, Miller AJ. (1996) Potassium homeostasis in vacuolate plant cells. Proc Natl Acad Sci USA 93: 10510–10514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schütze K, Batistic O, Weckermann K, Näke C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Wang Y, He L, Li HD, Xu J, Wu WH. (2010) Potassium channel alpha-subunit AtKC1 negatively regulates AKT1-mediated K+ uptake in Arabidopsis roots under low-K+ stress. Cell Res 20: 826–837 [DOI] [PubMed] [Google Scholar]
- Ward JM, Mäser P, Schroeder JI. (2009) Plant ion channels: gene families, physiology, and functional genomics analyses. Annu Rev Physiol 71: 59–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinl S, Kudla J. (2009) The CBL-CIPK Ca2+-decoding signaling network: function and perspectives. New Phytol 184: 517–528 [DOI] [PubMed] [Google Scholar]
- Xicluna J, Lacombe B, Dreyer I, Alcon C, Jeanguenin L, Sentenac H, Thibaud JB, Chérel I. (2007) Increased functional diversity of plant K+ channels by preferential heteromerization of the Shaker-like subunits AKT2 and KAT2. J Biol Chem 282: 486–494 [DOI] [PubMed] [Google Scholar]
- Xu J, Li HD, Chen LQ, Wang Y, Liu LL, He L, Wu WH. (2006) A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell 125: 1347–1360 [DOI] [PubMed] [Google Scholar]
- Zhou L, Fu Y, Yang Z. (2009) A genome-wide functional characterization of Arabidopsis regulatory calcium sensors in pollen tubes. J Integr Plant Biol 51: 751–761 [DOI] [PubMed] [Google Scholar]