Abstract
Functional modules are considered the primary building blocks of biomolecular systems. Here we study to what extent functional modules behave cohesively across genomes:That is, are functional modules also evolutionary modules? We probe this question by analyzing for a large collection of functional modules the phyletic patterns of their genes across 110 genomes. The majority of functional modules display limited evolutionary modularity. This result confirms certain comparative genome analyses, but is in contrast to implicit assumptions in the systems analysis of functional genomics data. We show that this apparent interspecies flexibility in the organization of functional modules depends more on functional differentiation within orthologous groups of genes, than on noise in the functional module definitions. When filtering out these sources of nonmodularity, even though very few functional modules behave perfectly modular in evolution, about half behave at least significantly more modular than a random set of genes. There are substantial differences in the evolutionary modularity between individual functional modules as well as between collections of functional modules, partly corresponding to conceptual differences in the functional module definition, which make comparisons between functional module collections biologically difficult to interpret. Analysis within one collection does not suffer from such differences, and we show that within the EcoCyc metabolic pathway database, biosynthetic pathways are evolutionarily more modular than catabolic pathways.
With the sequencing of complete genomes there has been a shift from determining the function of individual proteins towards determining how these proteins interact with each other to form functional modules such as protein complexes and metabolic pathways (Ihmels et al. 2002; Milo et al. 2002; Ravasz et al. 2002; Snel et al. 2002; von Mering et al. 2002; Wang et al. 2002; Rives and Galitski 2003; Wolf and Arkin 2003). Here we ask the question: To what extent do the components of such functional modules behave cohesively in evolution and can therefore also be considered evolutionary modules? Genome sequences actually provide data to measure the evolutionary modularity of functional modules by assessing whether the orthologs of the proteins in a specific module occur together across genomes. Although evolutionary modularity of functional modules has been assumed in the analysis of functional genomics data (Rives and Galitski 2003), and transcriptional modules have been suggested to be evolutionarily conserved modules (Wang et al. 2002), thus far analyses based on this measure have given conflicting reports on the evolutionary conservation of functional modules. On the one hand, metabolic pathways display considerable variation across complete genomes (Huynen et al. 1999; Peregrin-Alvarez et al. 2003), and only a few known regulons can be predicted using similarity of phyletic patterns (Manson McGuire and Church 2000). On the other hand, there should be some evolutionary modularity of functional modules, as the similarity in the phyletic patterns of genes has been shown to indicate a functional relation between their proteins (Huynen and Bork 1998; Pellegrini et al. 1999; Tatusov et al. 2001; Ramani and Marcotte 2003; von Mering et al. 2003), and genes in operons tend to have a tendency to have similar phyletic patterns (Moreno-Hagelsieb et al. 2001; Moreno-Hagelsieb and Collado-Vides 2002). However, the studies using phylogenetic profiles for function prediction tend to omit how many interactions they fail to predict.
In measuring the evolutionary modularity of functional modules one is confronted with a number of conceptual, biological, and technical issues that have largely been ignored in genome-scale analyses published thus far. The first set of issues revolves around the definition of what constitutes a functional module and how it is different from an evolutionary module: Are functional modules just protein complexes or metabolic pathways, or do we also include sets of coregulated proteins (Winther 2001)? And, if we do include different types of functional modules, do we observe differences in their evolutionary modularity? Furthermore, do we include modules derived from genomics data, which are very noisy in nature? The second issue is, when we measure modularity by the similarity in the phyletic patterns of orthologous groups, how do we account for functional differentiation within orthologous groups (Galperin and Koonin 2000; Sonnhammer and Koonin 2002), which basically reflects flexibility at a lower level than that of the composition of modules? A third issue is how to quantify modularity. How modular do we consider the evolution of a functional module when the `same' module in another species is partly composed of different proteins and/or of fewer proteins? Or, in other words, one might not expect a functional module to be a perfectly evolutionary module as well, but is it at least more modular than a random set of genes? Given the large number of arguable answers to the above questions, one cannot obtain a simple unequivocal answer to the question of evolutionary modularity of functional modules. What we can do, however, is identify trends that are independent of the various functional module definitions and orthology measures, and develop a scoring system that compares the observed level of evolutionary modularity with the level expected for a random set of proteins. Using this scoring system we quantify the evolutionary modularity of a collection of 1387 functional modules through the presence of their proteins in 110 genomes. This means that we propose an evolutionary (as opposed to functional) module to be a group of genes that tends to be present and absent together. To account for the issues mentioned above, we (1) measure the effect of noise in the functional module definition by comparing and filtering the data sets, (2) estimate the impact of functional differentiation within an orthologous group by filtering for gene duplication, and (3) account for the type of functional module by comparing a diverse set of biomolecular systems, including metabolic pathways, protein complexes, and transcriptional modules.
With this systematic approach a comprehensive view of functional module evolution is obtained. The majority of functional modules display a large degree of flexibility. We show that this flexibility depends more on the functional differentiation within orthologous groups of genes than on a noisy module definition. Furthermore, some data sets are evolutionarily more modular than others, and, within metabolic pathways, biosynthetic pathways are evolutionarily more modular than catabolic pathways.
RESULTS
Measuring Modularity
We survey the evolutionary modularity of nine different collections of functional modules (Table 1). These systems vary in how they are obtained (manually curated, results of high-throughput genomics, bioinformatic analyses of genomics data) and what `type' of biomolecular system they represent (metabolic pathways, protein complexes, transcriptional modules). Such a wide selection of data sets allows us to draw conclusions on the propensity of functional modules to also represent evolutionary modules without depending on idiosyncratic properties of any particular data set. At the same time, the use of various types of data allows a comparison of evolutionary modularity with the biological type and (experimental) source of each set of data.
Table 1.
Overview and General Properties of the Data Sets
| Name of data seta | Type of functional modules | Method by which functional modules were determined | No. of modules | Average module size |
|---|---|---|---|---|
| Known complexes in Yeast (Mewes et al. 2002; von Mering et al. 2002) | Protein complexes | Manually curated | 126 | 4.36 |
| Known operons in E. coli (Salgado et al. 2001) | Transcriptional modules | Manually curated | 149 | 3.15 |
| EcoCyc metabolic pathways (Karp et al. 2002) | Metabolic pathways | Manually curated | 103 | 4.08 |
| HMS-PCI (Ho et al. 2002) | Protein complexes | High-throughput genomics data | 375 | 5.35 |
| TAP (Gavin et al. 2002) | Protein complexes | High-throughput genomics data | 190 | 7.39 |
| Genes sharing TFB sites (Lee et al. 2002) | Transcriptional modules | High-throughput genomics data | 175 | 5.41 |
| Transcriptional clusters (Ihmels et al. 2002) | Transcriptional modules | Bioinformatics analysis of genomics data | 84 | 33.79 |
| KEGG maps (Ogata et al. 1999) | Metabolic pathways; Protein complexes | Manually curated | 61 | 31.62 |
| Predicted regulons in E. coli (van Nimwegen et al. 2002) | Transcriptional modules | Bioinformatics analysis of genomics data | 124 | 10.02 |
The data sets were obtained from the Web sites given in their respective publications: See Methods for details.
The complete data set consists of 1387 functional modules. We track the evolutionary distribution of their constituent genes by the presence and absence of orthologous genes in the genomes of 110 species (16 archaea, 85 bacteria and eight eukarya, obtained from the SWISS-PROT Proteome [Pruess et al. 2003] project). Although homology has also been used to determine the phylogenetic distribution of a gene (Pellegrini et al. 1999; Peregrin-Alvarez et al. 2003), orthology is better suited here, because orthologs are much more likely to have equivalent functions than homologs (Tatusov et al. 1997; Sonnhammer and Koonin 2002). Orthologies were assigned using the clusters of orthologous groups (COG) database (Tatusov et al. 1997, 2001).
When a functional module would behave as an evolutionary module, one expects to observe either a large fraction (the module is present) or a small fraction (the module is absent) of its proteins in a given species. Thus, to measure evolutionary modularity for a functional module, we take the sum of the deviation of the number of components of the functional module for each genome to the average number of module components per genome, that is, the sum of the deviations to the average (Fig. 1; Methods). In order to normalize this observed deviation between 1 (perfectly modular) and 0 (behaving like a random set of genes), we compute (A) the maximal deviation of an ideal evolutionary module (i.e., the module is always completely present or completely absent), and (B) the expected random deviation and its variation in any species under a null model of random gene distribution (absence of evolutionary modularity). Any choice for the random deviation under a null model is subject to our (limited) knowledge of the evolution of gene content: that is, the presence/absence pattern of any set of orthologous groups over a set of genomes is decidedly nonrandom, and depends on the evolutionary distance between the species and their genome size (Snel et al. 1999). To counter this uncertainty, we choose two score baselines. The first one ignores the phylogenetic and size patterns in the distribution of genomes. It calculates the expected fraction of genes and its variation by redistributing orthologs of the proteins in the module randomly across all species. We refer to this randomization as `random shuffling.' Our second baseline does take into account inherent signals in shared gene content such as phylogenetic distance and genome size. It achieves this by sampling other genes from the species in which the functional module is defined that have the same frequency as the genes from the functional module, thereby conserving any intrinsic signals in the presence of genes. We refer to this randomization as `random sampling.'
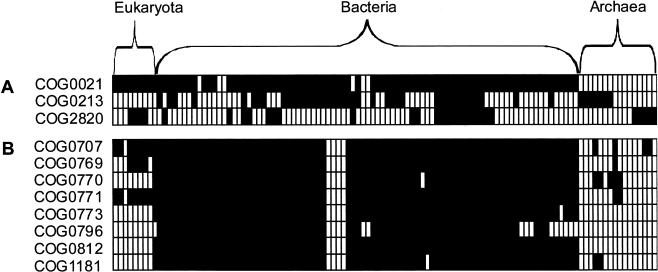
Figure 1.
Phyletic patterns of the components of two functional modules. Each row is an orthologous group of genes, and each column is a species. Filled squares indicate that the orthologous group is present; blank squares indicate absence. Panel (A) shows the (deoxy) ribose phosphate metabolism pathway from EcoCyc. It has three components that evolve very flexibly: The observed deviation from the average number of module components per species is 0.682, whereas the random deviation is 0.677 for random shuffling and 0.802 for random sampling. The deviation when the module would behave evolutionarily perfectly modular would be 1.50. The modularity score of this pathway is thus 0.006 (random shuffling) or –0.172 (random sampling; see Methods). As the SD's of both types of random are respectively 0.033 and 0.074, this pathway falls within one SD of random and is thus as flexible as a random evolving group of genes would be. (B) The peptidoglycan biosynthesis pathway from EcoCyc. The modularity score of this pathway is 0.82 for random shuffling and 0.71 for random sampling; both are significantly more than random (more than 2 SD).
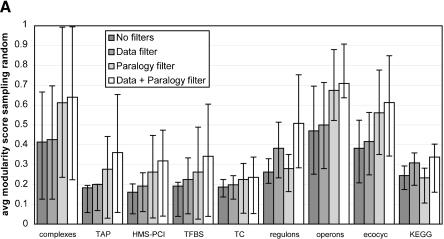
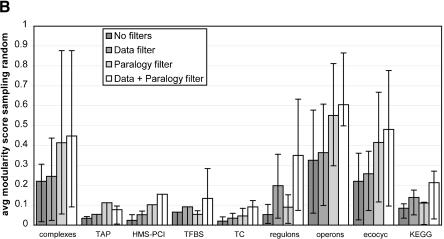
Surveying all 1387 functional modules with this approach reveals that, although the average functional module is closer to being as flexible as a random system would be (i.e., closer to the null model) than to being as modular as an ideal evolutionary module (Fig. 2A), most functional modules are significantly more modular than random (more than two standard deviations, Table 2). The tendency to limited but significant modularity is observed across all collections of biomolecular systems (Fig. 2) and thus does not depend on any collection in particular. Interestingly, the average score drops substantially when we compare against the second baseline that is the expected presence of genes based on phylogeny and genome size (Fig. 2B). In fact, using this baseline for the score reveals many functional modules that are not significantly more modular than a randomly evolving set of genes would be (Table 2). This means that a substantial portion of the observed evolutionary modularity of the components cannot be discriminated from phylogenetic signals and convergent pressure on gene content through genome size effects.
Figure 2.
Distribution of the average scores for various filters for all data sets. The error bars reveal the spread of the modularity score within a single data set. The length of the bars represents the lower quartile and upper quartile respectively. Note that this allows for asymmetric bars. Very asymmetric bars suggest a huge gap between mean and median. (A) The average scores when the random baseline is obtained by random shuffling. This randomization ignores the phylogenetic and size patterns in the distribution of genomes, and is obtained by redistributing orthologs of the proteins in the module randomly across all species. (B) The average scores when the random baseline is obtained by random sampling. This baseline does take into account inherent signals in shared gene content such as phylogenetic distance and genome size. It achieves this by sampling other genes from the species in which the functional module is defined that have the same frequency as the genes from the functional module, thereby conserving any intrinsic signals in the presence of genes.
Table 2.
Fraction of Functional Modules More Than Two Standard Deviations Modular Than Random
|
No data filter by cross-comparison
|
Data filter by cross-comparison
|
|||
|---|---|---|---|---|
| No paralogy filter | Random shuffling | Random sampling | Random shuffling | Random sampling |
| Known complexes in yeast | 0.89 | 0.54 | 0.91 | 0.56 |
| TAP protein complexes | 0.58 | 0.17 | 0.63 | 0.22 |
| HMS-PCI protein complexes | 0.64 | 0.17 | 0.71 | 0.25 |
| Genes sharing TFBS | 0.61 | 0.25 | 0.67 | 0.28 |
| Transcriptional clusters | 0.30 | 0.00 | 0.37 | 0.01 |
| Predicted regulons in E. coli | 0.91 | 0.05 | 0.95 | 0.41 |
| Known operons in E. Coli | 0.95 | 0.68 | 0.97 | 0.73 |
| EcoCyc metabolic pathways | 0.93 | 0.50 | 0.97 | 0.59 |
| KEGG maps | 0.48 | 0.03 | 0.85 | 0.13 |
| Paralogy filter | Random shuffling | Random sampling | Random shuffling | Random sampling |
| Known complexes in yeast | 0.95 | 0.68 | 0.94 | 0.69 |
| TAP protein complexes | 0.67 | 0.37 | 0.74 | 0.43 |
| HMS-PCI protein complexes | 0.66 | 0.35 | 0.80 | 0.38 |
| Genes sharing TFBS | 0.72 | 0.28 | 0.72 | 0.40 |
| Transcriptional clusters | 0.60 | 0.14 | 0.67 | 0.13 |
| Predicted regulons in E. coli | 0.95 | 0.20 | 0.97 | 0.67 |
| Known operons in E. coli | 1.00 | 0.91 | 0.97 | 0.92 |
| EcoCyc metabolic pathways | 0.98 | 0.71 | 1.00 | 0.74 |
| KEGG maps | 0.60 | 0.17 | 0.88 | 0.35 |
Filtering for Nonmodular Processes
The on-average low level of evolutionary modularity we observe can be the result of a host of conceptual, methodological, and biological issues (see above). As we are interested in the biological flexibility on the module level, that is, intrinsic flexibility, we try to remove the two extrinsic sources of nonmodular behavior that were most frequently observed in a manual survey of the results: noise in the definition of functional modules, and genuine evolutionary flexibility at a lower level, that of the functions of the proteins within an orthologous group (Sonnhammer and Koonin 2002). We independently correct for both sources of evolutionary nonmodular behavior and measure their relative effect.
We correct for noise in the functional module definition by overlaying the nine different data sets with each other: That is, for each functional module we only retain those proteins that are linked in a single module in at least one of the other data sets (see Methods). Such a cross-comparison filter should increase the reliability of the functional module definition, because it has been shown that the overlap between functional genomics data sets that predict interactions between proteins has a drastically higher accuracy than any of the individual data sets (von Mering et al. 2002). The cross-filtered functional modules show a limited increase in the average score (Fig. 2), and a larger increase in the fraction of modules whose evolution now is significantly more modular than random (Table 2). Some of the initially observed flexibility is thus indeed due to noise in the original functional module definition.
We can correct for the effect of functional differentiation within orthologous groups by filtering out duplicated genes. Filtering in such a way might be regarded as noise reduction. However, the original definition of orthology of genes being related by speciation allows for gene duplications after the speciation event, so-called inparalogs. Inparalogs allow (recent) gene duplications to belong to the same orthologous groups, although they likely have undergone functional differentiation (Sonnhammer and Koonin 2002). The presence of a functionally differentiated paralog (the copy) can lead to lower modularity scores. For example, when the members of the original functional module disappear the original will also be deleted, but the copy (and thus the orthologous group) will be retained due to its newly acquired function, and the tendency for cohesive evolution in that species will seem disrupted. Filtering for functional differentiation at the level of individual proteins in this manner indeed results in a substantial increase in average scores (Fig. 2) and in the fraction of modules that behaves significantly modular in evolution (Table 2). We filter out orthologous groups for gene duplications such that we still retain the least duplicated half of the orthologous groups for further analysis (at a gene species ratio threshold of less than 1.33, only one species in three is allowed to have two representatives of the orthologous group). The evolutionary modularity of the genes that are thereby kept is markedly increased. These results stress the importance of high-quality function definition and suggest that part of the observed variation in the presence of pathways in previous studies was due to the usage of an even lower level of resolution at the level of protein function: that is, homologs rather than orthologs (Peregrin-Alvarez et al. 2003). Surprisingly for most data sets, the effect of filtering for paralogy leads to a bigger shift towards evolutionary modularity than filtering the data sets by cross-comparison. The paralogy effect reflects real biological flexibility, albeit at a lower level than that of complete modules, whereas the cross-filtering of data sets is noise reduction. Thus our initial observation that the evolution of functional modules is flexible appears to be caused in a larger part by inherent biological flexibility at the level of protein function within orthologous groups, except for the cases of the (predicted) regulons and the KEGG maps, which display a larger effect of data set filtering.
Applying both corrections naturally gives the largest shift from flexibility to modularity (Fig. 2). The average score for a functional module varies between 0.077 and 0.709 depending on data set and baseline. Using random shuffling as baseline, the evolution of almost all functional modules can now be said to be more modular than random, and even for the sampling baseline the evolution of the majority of functional modules is now more cohesive than a random set of genes. This result thereby expands on what has already been found to a certain extent for metabolic pathways and reveals it to be the case as well for protein complexes and transcriptional module: Functional modules evolve cohesively to some extent, but even when taking into account systematic sources of nonmodularity, they are rarely perfectly modular.
Comparing Biomolecular Systems
Certain data sets show a higher level of evolutionary modularity than others, and also within each data set the individual functional modules reveal tremendous variation (Fig. 2). The data set of known protein complexes is evolutionarily more modular than the transcriptional modules of genes sharing transcription factor binding sites (TFBSs) obtained by high-throughput genomics experiments (Lee et al. 2002) across all combinations of filters (Fig. 2). Both KEGG and EcoCyc (metabolic) pathways are manually curated, yet EcoCyc pathways are evolutionarily more modular (Table 2), probably because of conceptual differences between pathways (Eco-Cyc) and pathway maps (KEGG), which is also reflected in the average size of their modules (Table 1). The operons from regulon DB behave evolutionarily exceptionally modular compared to other transcriptional modules and in fact compared to all other data sets. This confirms findings from studies that attempt to predict operons in genomic sequences (Moreno-Hagelsieb et al. 2001; Moreno-Hagelsieb and Collado-Vides 2002). However in addition to being transcriptional units, genes in operons by definition also constitute evolutionary units, because evolutionary processes that affect gene content, such as deletions, duplications, and horizontal transfers often affect chromosomal segments rather than individual genes (Lawrence and Roth 1996). Indeed the set of predicted regulons (groups of coregulated genes and operons) from Escherichia coli reveals less modularity in its evolution than the operons (Table 2), as was already suggested by the inability of phyletic patterns to predict regulons (Manson McGuire and Church 2000). Thus the observed high average score of the operons is caused by functional as well as genome structural reasons.
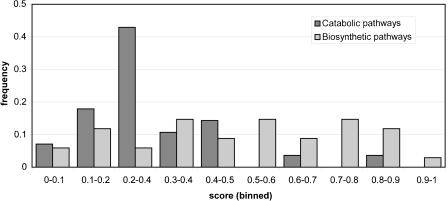
Analysis of classes of functional modules within a data set does not suffer from differences in quality and concept of their definition, and allows a ceteris paribus comparison. The differences in evolutionary modularity within each data set are thus likely to reflect real biological flexibility. Such an analysis within the EcoCyc database (Karp et al. 2002) reveals that biosynthetic pathways are evolutionarily significantly more modular than catabolic pathways (Fig. 3; P < 0.001 Wilcoxon rank-sum test). Catabolic pathways thus show a greater flexibility in evolution. Apparently there is more variation in the ways of breaking down compounds than in synthesizing them. This difference in evolutionary mode could reflect a difference in possible paths on the underlying biochemical landscape, for example, it is conceivable that there are more thermodynamic or biochemical constraints to synthesizing metabolites than to breaking them down, offering less alternative feasible routes.
Figure 3.
Distribution of modularity scores in catabolic versus biosynthetic pathways. Histogram of the distribution of modularity scores among catabolic versus biosynthetic metabolic pathways in EcoCyc (Karp et al. 2002).
DISCUSSION
Analyses of complete genome data have given conflicting results with regards to the evolutionary modularity of functional modules. Metabolic pathways display considerable variation across genomes (Huynen et al. 1999; Peregrin-Alvarez et al. 2003), while at the same time the similarity in the distribution of orthologous groups has been shown to indicate a functional relation (Huynen and Bork 1998; Pellegrini et al. 1999; Tatusov et al. 2001; Ramani and Marcotte 2003; von Mering et al. 2003). This paradox seems to depend on what is being measured: the general behavior of pathways versus the justifiable focus on a minimal number of false positives for a certain set of co-occurrence predictions. In any case, the direct test of evolutionary modularity of functional modules presented here reveals substantial flexibility. This flexibility delimits the potential of using the co-occurrence of genes (i.e., phylogenetic profiles) for the prediction of function relations (Huynen and Bork 1998; Pellegrini et al. 1999), at least over large collections of genomes.
Whether the evolutionary behavior of the functional modules that we observe can be called modular or not, is of course in the eye of the beholder: Most groups of genes which have been proposed to be functional modules display considerable variation in evolution, but about half of the functional modules do tend to evolve more cohesively than random when the randomization considers inherent phylogenetic and genome size signals in the presence of genes. This intermediate level of modularity is close to the estimate in a study on Pyrococci, where 40%–50% of the gene gains and losses were found to be modular (Ettema et al. 2001). All in all, functional modules tend to correlate with evolutionary modules, but at the same time there are many biological processes that result in flexibility. More importantly, certain trends can be delineated by filtering and by comparing biomolecular systems. Some data sets (KEGG, predicted regulons in E. coli) show a substantial increase in their score when their functional modules are filtered by cross-confirmation to reduce noise. Yet, this increase due to noise removal is small compared to the differences in modularity between the data sets that are the result of conceptual and biological issues. One biological issue that we explicitly test, gene duplication, alone has a bigger impact on the score than the noise reduction in the definition of the data sets by cross-filtering. Paralogy likely adversely affects the evolutionary modularity score, because functionally differentiated copies can be retained while the functional module is codeleted with its module members (see above). More generally, the results are consistent with a view in which genes that are not duplicated are not able to evolve new functions and are evolutionarily `trapped' in modules.
The limited modularity in certain data sets also raises issues by itself. In sharp contrast to what has been proposed (Wang et al. 2002), all of the yeast transcriptional module-like data sets have an especially limited modularity compared to the other types of data sets. In one data set in particular (the transcriptional clusters), the fraction of functional modules that is significantly more modular than sampling random remains as low as 13% after applying both the paralogy and cross-confirmation filter. However, there are analyses that suggest that the yeast transcription data are not even modular on a functional level (Rung et al. 2002), which would explain the observed limited evolutionary modularity. The flexibility in the high-throughput-determined protein complex data hints at noise in the functional module definition. Yet, filtering these data sets by cross-comparisons has a very limited effect on modularity. Another explanation is that many shared components exist between complexes (Gavin et al. 2002), and that the observed evolutionary flexibility actually reflects the functional flexibility. Generally the differing modularity scores between functional modules within a data set and among data sets are likely to be caused by a myriad of evolutionary and biological reasons, which cannot all be captured by our set of filters. Flexibility can also reside in analogous displacement of enzymes (Morett et al. 2003) or reflect a core-periphery organization of functional modules (Rives and Galitski 2003).
In general, the observed flexibility is in contrast to expectations in the current literature on systems analysis of high-throughput genomics data (Milo et al. 2002; Wang et al. 2002; Rives and Galitski 2003), but it is consistent with the conventional view that genes are the primary agents of function. Genes by definition constitute ideal functional modules, and some of the highest-scoring functional modules consist of only two orthologous groups of genes. If many larger collections of genes would be ideal functional modules, we might also have seen more gene fusions than are actually observed (Snel et al. 2000). Thus when studying genome evolution globally, a system-level description of genomes cannot consist solely of a genome as a collection of modules; it still needs a proper description of the genes and their relations (Marcotte 2001). Modularity might however still be very relevant on shorter time scales, but it is eroded by subsequent deletion and addition of components. For example, the duplication of the complete hox gene cluster in vertebrates is an example of a modular evolutionary event, but also of the subsequent divergence by independent loss of genes in each cluster.
The flexibility in the make-up of functional modules throughout the tree of life might also reveal something about the origin of complex biomolecular systems. We observe many, to the human eye, `partial' or incomplete functional modules. All these instances are likely to be functional or at the very least not be deleterious for the organisms in which they occur. Although we cannot directly reconstruct how functional modules such as protein complexes came into existence, we can conclude that gradual growth is possible because partial modules are viable.
METHODS
Genomes and Orthology
We used the SWISS-PROT Proteome (Pruess et al. 2003) set of March 12, containing the 110 complete proteomes of 85 bacteria, 16 archaea, and eight eukarya. Each proteome is the complete complement of (predicted) protein coding sequences of a genome. For comparative genomics we need to define equivalent genes across genomes. Although homology is frequently used to determine the phylogenetic distribution of a gene (Pellegrini et al. 1999; Peregrin-Alvarez et al. 2003), orthology is better suited because of its evolutionary definition: Whereas homologies between species might have already been present as multiple genes at the time of speciation between genomes, orthologs stem from the same gene at the time of speciation between genomes and are thus more likely to have equivalent functions (Fitch 1970). Orthologies were assigned using the clusters of orthologous groups (COG) database (Tatusov et al. 1997, 2001). Proteomes unassigned by the COG database were assigned using an in-house COGnitor perl script using Smith-Waterman searches against the COG database (Tatusov et al. 1997; von Mering et al. 2003). To be able to perform comparative genome analysis across the tree of life (i.e., between eukaryotes and prokaryotes) in the current COG setup, which has separate orthologies for eukaryotes and unicellular organisms, we assigned eukaryotic orthologous groups to COGs through best bidirectional hit sequence comparisons when appropriate. After assigning, we scored the gene/species ratio of each COG as a measure of paralogy within each orthologous group.
Biomolecular Systems Data Sets
The data sets were obtained from the Web sites given in their respective publications: overlapping transcriptional clusters (Ihmels et al. 2002), genes sharing transcription factor binding sites (TFBSs; Lee et al. 2002), EcoCyc metabolic pathways (Karp et al. 2002), KEGG maps (Ogata et al. 1999), known operons from E. coli (Salgado et al. 2001), and the predicted regulons in E. coli (van Nimwegen et al. 2002). Note that regulons are different from operons in the sense that a regulon encompasses a set of operons and sometimes single genes that are regulated by the same transcriptional regulator. The compilation of known protein complexes in yeast from MIPS (Mewes et al. 2002), which was used as reference data sets for the comparative assessment of large-scale data sets of protein-protein interactions (von Mering et al. 2002) was obtained from Christian von Mering (EMBL). The purified protein complexes data sets obtained using the TAP protocol (Gavin et al. 2002) and the HMS-PCI protocol (Ho et al. 2002) were downloaded from the MIPS server (Mewes et al. 2002). For each data set, we removed from further analysis a functional module when another functional module as defined in that same data set was a subset of the functional module. We could unfortunately not survey the modules from the metabolic network studied by Ravasz et al. (2002), because in that work the modules consisted of metabolites rather than proteins.
Modularity
We defined modularity in the evolution of a functional module by the deviation in the presence of orthologous genes of the proteins in a species from the average, and thus expected, number of orthologs per species (Fig. 1). When a functional module is an evolutionary module, we suppose that either a majority (the module is present) or minority (the module is absent) of its orthologs is present in a given species. To quantify this we first obtained the observed deviation of the number of module genes for each species from the average number of module genes per species. We also need to know this deviation from the average number of module genes under a null model. Here we used two score baselines. The first one ignores the phylogenetic and size patterns in the distribution of genomes (Snel et al. 1999) and simply calculates the expected fraction of genes and its variation by redistributing orthologs of the proteins in the module randomly across all species 100 times. Our second baseline does take into account inherent signals in gene content evolution such as phylogenetic signals and genome size signals. It achieves this by random sampling 100 times other genes from the species where the functional module is defined that have the same frequency as the genes from the functional module, thereby conserving any intrinsic signals in the presence of genes. The random baseline then is zero in our score and to obtain a 1, the maximum possible raw score for a module, the deviation from expected is computed when the module is always completely present or completely absent (i.e., it evolves perfectly modular). So our score is then computed by ([observed deviation from expected – random deviation from expected] / [maximum deviation from expected – random deviation from expected]). Subsequently, by checking the variation around the (shuffling and sampling) random deviation, we can use the standard deviation (SD) of this randomization to check whether the observed deviation from expected of a module is distinguishable from a randomly evolving module. Two standard deviations was chosen as a reasonable approximation of a significant deviation, as visual inspection of both a number of random shufflings and a number of random drawings revealed a Gaussian distribution. The main aim of this statistic is to quantify how nonrandom or modular the evolution of the functional modules in a data set is, in addition to the average modularity score.
Filtering by Cross-Comparison
To enhance the original functional module definition, we compared functional modules from all data sets to each other. The rationale here is that it has been shown that taking the overlap between data sets that predict interactions between proteins drastically increases the reliability of the predictions (von Mering et al. 2002). Specifically, we marked all proteins in a functional module as `connected' in its data set, and then tried to rebuild via single linkage each functional module using connections from all other data sets but not from the data set in which the module was originally defined. If only subsets of the proteins within one functional module could be connected to each other via other data sets, we chose the largest subset to represent the original functional module. The phyletic pattern of all genes of all functional modules of all data sets under all four filtering conditions are available as Supplemental material.
Acknowledgments
We thank Christian von Mering for allowing us to use the reference set of known protein complexes as compiled from MIPS. We also thank the anonymous referees for their useful comments. This work was supported in part by a grant from the Netherlands organization for scientific research (NWO).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1969504.
Footnotes
[Supplemental material is available online at www.genome.org.]
References
- Ettema, T., van der Oost, J., and Huynen, M. 2001. Modularity in the gain and loss of genes: Applications for function prediction. Trends Genet. 17: 485–487. [DOI] [PubMed] [Google Scholar]
- Fitch, W.M. 1970. Distinguishing homologous from analogous proteins. Syst. Zool. 19: 99–113. [PubMed] [Google Scholar]
- Galperin, M.Y. and Koonin, E.V. 2000. Who's your neighbor? New computational approaches for functional genomics. Nat. Biotechnol. 18: 609–613. [DOI] [PubMed] [Google Scholar]
- Gavin, A.C., Bosche, M., Krause, R., Grandi, P., Marzioch, M., Bauer, A., Schultz, J., Rick, J.M., Michon, A.M., Cruciat, C.M., et al. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415: 141–147. [DOI] [PubMed] [Google Scholar]
- Ho, Y., Gruhler, A., Heilbut, A., Bader, G.D., Moore, L., Adams, S.L., Millar, A., Taylor, P., Bennett, K., Boutilier, K., et al. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415: 180–183. [DOI] [PubMed] [Google Scholar]
- Huynen, M.A. and Bork, P. 1998. Measuring genome evolution. Proc. Natl. Acad. Sci. 95: 5849–5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynen, M.A., Dandekar, T., and Bork, P. 1999. Variation and evolution of the citric-acid cycle: A genomic perspective. Trends Microbiol. 7: 281–291. [DOI] [PubMed] [Google Scholar]
- Ihmels, J., Friedlander, G., Bergmann, S., Sarig, O., Ziv, Y., and Barkai, N. 2002. Revealing modular organization in the yeast transcriptional network. Nat. Genet. 31: 370–377. [DOI] [PubMed] [Google Scholar]
- Karp, P.D., Riley, M., Saier, M., Paulsen, I.T., Collado-Vides, J., Paley, S.M., Pellegrini-Toole, A., Bonavides, C., and Gama-Castro, S. 2002. The EcoCyc Database. Nucleic Acids Res. 30: 56–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence, J.G. and Roth, J.R. 1996. Selfish operons: Horizontal transfer may drive the evolution of gene clusters. Genetics 143: 1843–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, T.I., Rinaldi, N.J., Robert, F., Odom, D.T., Bar-Joseph, Z., Gerber, G.K., Hannett, N.M., Harbison, C.T., Thompson, C.M., Simon, I., et al. 2002. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298: 799–804. [DOI] [PubMed] [Google Scholar]
- Manson McGuire, A. and Church, G.M. 2000. Predicting regulons and their cis-regulatory motifs by comparative genomics. Nucleic Acids Res. 28: 4523–4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotte, E.M. 2001. The path not taken. Nat. Biotechnol. 19: 626–627. [DOI] [PubMed] [Google Scholar]
- Mewes, H.W., Frishman, D., Guldener, U., Mannhaupt, G., Mayer, K., Mokrejs, M., Morgenstern, B., Munsterkotter, M., Rudd, S., and Weil, B. 2002. MIPS: A database for genomes and protein sequences. Nucleic Acids Res. 30: 31–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milo, R., Shen-Orr, S., Itzkovitz, S., Kashtan, N., Chklovskii, D., and Alon, U. 2002. Network motifs: Simple building blocks of complex networks. Science 298: 824–827. [DOI] [PubMed] [Google Scholar]
- Moreno-Hagelsieb, G. and Collado-Vides, J. 2002. A powerful non-homology method for the prediction of operons in prokaryotes. Bioinformatics (Suppl. 1) 18: S329–336. [DOI] [PubMed] [Google Scholar]
- Moreno-Hagelsieb, G., Trevino, V., Perez-Rueda, E., Smith, T.F., and Collado-Vides, J. 2001. Transcription unit conservation in the three domains of life: A perspective from Escherichia coli. Trends Genet. 17: 175–177. [DOI] [PubMed] [Google Scholar]
- Morett, E., Korbel, J.O., Rajan, E., Saab-Rincon, G., Olvera, L., Olvera, M., Schmidt, S., Snel, B., and Bork, P. 2003. Systematic discovery of analogous enzymes in thiamin biosynthesis. Nat. Biotechnol. 21: 790–795. [DOI] [PubMed] [Google Scholar]
- Ogata, H., Goto, S., Sato, K., Fujibuchi, W., Bono, H., and Kanehisa, M. 1999. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 27: 29–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini, M., Marcotte, E.M., Thompson, M.J., Eisenberg, D., and Yeates, T.O. 1999. Assigning protein functions by comparative genome analysis: Protein phylogenetic profiles. Proc. Natl. Acad. Sci. 96: 4285–4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peregrin-Alvarez, J.M., Tsoka, S., and Ouzounis, C.A. 2003. The phylogenetic extent of metabolic enzymes and pathways. Genome Res. 13: 422–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruess, M., Fleischmann, W., Kanapin, A., Karavidopoulou, Y., Kersey, P., Kriventseva, E., Mittard, V., Mulder, N., Phan, I., Servant, F., et al. 2003. The Proteome Analysis database: A tool for the in silico analysis of whole proteomes. Nucleic Acids Res. 31: 414–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramani, A.K. and Marcotte, E.M. 2003. Exploiting the co-evolution of interacting proteins to discover interaction specificity. J. Mol. Biol. 327: 273–284. [DOI] [PubMed] [Google Scholar]
- Ravasz, E., Somera, A.L., Mongru, D.A., Oltvai, Z.N., and Barabasi, A.L. 2002. Hierarchical organization of modularity in metabolic networks. Science 297: 1551–1555. [DOI] [PubMed] [Google Scholar]
- Rives, A.W. and Galitski, T. 2003. Modular organization of cellular networks. Proc. Natl. Acad. Sci. 100: 1128–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rung, J., Schlitt, T., Brazma, A., Freivalds, K., and Vilo, J. 2002. Building and analysing genome-wide gene disruption networks. Bioinformatics (Suppl. 2) 18: S202–210. [DOI] [PubMed] [Google Scholar]
- Salgado, H., Santos-Zavaleta, A., Gama-Castro, S., Millan-Zarate, D., Diaz-Peredo, E., Sanchez-Solano, F., Perez-Rueda, E., Bonavides-Martinez, C., and Collado-Vides, J. 2001. RegulonDB (version 3.2): Transcriptional regulation and operon organization in Escherichia coli K-12. Nucleic Acids Res. 29: 72–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snel, B., Bork, P., and Huynen, M.A. 1999. Genome phylogeny based on gene content. Nat. Genet. 21: 108–110. [DOI] [PubMed] [Google Scholar]
- Snel, B., Bork, P., and Huynen, M.A. 2000. Genome evolution. Gene fusion versus gene fission. Trends Genet. 16: 9–11. [DOI] [PubMed] [Google Scholar]
- Snel, B., Bork, P., and Huynen, M.A. 2002. The identification of functional modules from the genomic association of genes. Proc. Natl. Acad. Sci. 99: 5890–5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer, E.L. and Koonin, E.V. 2002. Orthology, paralogy and proposed classification for paralog subtypes. Trends Genet. 18: 619–620. [DOI] [PubMed] [Google Scholar]
- Tatusov, R.L., Koonin, E.V., and Lipman, D.J. 1997. A genomic perspective on protein families. Science 278: 631–637. [DOI] [PubMed] [Google Scholar]
- Tatusov, R.L., Natale, D.A., Garkavtsev, I.V., Tatusova, T.A., Shankavaram, U.T., Rao, B.S., Kiryutin, B., Galperin, M.Y., Fedorova, N.D., and Koonin, E.V. 2001. The COG database: New developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 29: 22–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nimwegen, E., Zavolan, M., Rajewsky, N., and Siggia, E.D. 2002. Probabilistic clustering of sequences: Inferring new bacterial regulons by comparative genomics. Proc. Natl. Acad. Sci. 99: 7323–7328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Mering, C., Krause, R., Snel, B., Cornell, M., Oliver, S.G., Fields, S., and Bork, P. 2002. Comparative assessment of large-scale data sets of protein-protein interactions. Nature 417: 399–403. [DOI] [PubMed] [Google Scholar]
- von Mering, C., Huynen, M., Jaeggi, D., Schmidt, S., Bork, P., and Snel, B. 2003. STRING: A database of predicted functional associations between proteins. Nucleic Acids Res. 31: 258–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W., Cherry, J.M., Botstein, D., and Li, H. 2002. A systematic approach to reconstructing transcription networks in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 99: 16893–16898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winther, R.G. 2001. Varieties of modules: Kinds, levels, origins, and behaviors. J. Exp. Zool. 291: 116–129. [DOI] [PubMed] [Google Scholar]
- Wolf, D.M. and Arkin, A.P. 2003. Motifs, modules and games in bacteria. Curr. Opin. Microbiol. 6: 125–134. [DOI] [PubMed] [Google Scholar]