Abstract
Photoreceptors, especially the far-red light-absorbing phytochrome A, play a crucial role in early seedling development, triggering the transition from etiolated to photomorphogenic growth. Here, we describe the biological functions of two GRAS proteins from Arabidopsis (Arabidopsis thaliana), SCARECROW-LIKE21 (SCL21) and PHYTOCHROME A SIGNAL TRANSDUCTION1 (PAT1), which are specifically involved in phytochrome A signal transduction. Loss-of-function mutants show an elongated hypocotyl under far-red light and are impaired in other far-red high-irradiance responses. The SCL21 transcript itself is down-regulated by far-red light in a phytochrome A- and PAT1-dependent manner. Our results demonstrate that both SCL21 and PAT1 are positive regulators of phytochrome A signal transduction for several high-irradiance responses. Genetic and biochemical evidence suggest a direct interaction of the two proteins.
Light is an important environmental cue that plants exploit to monitor environmental changes and to optimize growth and photosynthesis. Using an array of photoreceptors, plants can sense different qualities and quantities of light and regulate responses such as seed germination, seedling deetiolation, expansion and elongation of all aboveground organs, and induction of flowering (Sullivan and Deng, 2003; Franklin and Quail, 2010; Kami et al., 2010). Three major classes of photoreceptors, the blue (B)/UV-A light-sensing cryptochromes, the phototropins, and the red (R)/far-red (FR) light-responsive phytochromes have been characterized in plants at the molecular level (Schäfer and Nagy, 2006). In Arabidopsis (Arabidopsis thaliana), phytochromes are encoded by a small gene family of five members, PHYA to PHYE (Clack et al., 1994). They exist as dimeric chromoproteins attached to a linear tetrapyrrole chromophore and are capable of photoreversible conformational changes between the Pr and the Pfr forms (Quail, 1997; Nagy and Schäfer, 2002). The Pfr form is considered the active form and has been shown to migrate into the nucleus (for review, see Kevei et al., 2007; Fankhauser and Chen, 2008). Phytochrome A (phyA) and phyB are the major phytochromes in plants (Smith, 1999; Quail, 2002; Chen et al., 2004; Bae and Choi, 2008). PhyB to PhyE are light stable and are found mainly in green tissues, with phyB being the main sensor of R light characterized by the R/FR reversible induction of responses. PhyB (and to a minor extent also phyD and phyE) is important for the adaptation of plants to changing R:FR ratios, caused for example by light reflected from neighboring plants or shading from canopies (Smith, 2000; Franklin, 2008; Ruberti et al., 2012).
PhyA predominates in etiolated tissues, as it is light labile and represents the primary sensor of FR light (Quail, 1997; Smith, 1999; Chen et al., 2004). It is necessary for the deetiolation process under the very low fluence response (VLFR) and the high irradiance response (HIR) of FR light and, as recently shown, also R light (Casal et al., 1998; Franklin and Whitelam, 2007). Via phyA, plants are able to react to light conditions under which other phytochromes are not active. This is crucial for the germination of buried seeds or seeds under dense canopies. Furthermore, seedlings undergo at least partial deetiolation under light perceived by phyA. Besides the more prominent phyA-dependent phenotypes observed in the seedling stage, phyA is clearly involved in plant development throughout the life cycle (Franklin and Whitelam, 2007; Kneissl et al., 2008).
Several protein intermediates have been isolated to date that are important for phyA signaling (for review, see Bae and Choi, 2008). Genetic screens have exploited, in most cases, hypocotyl elongation as a parameter for mutant selection. Only three mutants, far-red elongated hypocotyl1 (fhy1), fhy3, and phytochrome A signal transduction1 (pat1-1; Whitelam et al., 1993; Bolle et al., 2000), have been isolated in which hypocotyl elongation specifically under FR light is nearly fully abolished, similar to a phyA photoreceptor mutant. FHY1 and its homolog FHY1-LIKE1 (FHL1) have been shown to be necessary for the nuclear import of phyA (Zhou et al., 2005; Hiltbrunner et al., 2006), and FHY1 facilitates the localization of phyA to its target gene promoters and coactivating transcription (Chen et al., 2012). The transposase-derived transcription factor FHY3 and its homolog FAR-RED IMPAIRED RESPONSE1 (FAR1), appear to play roles not only in the transcription of light-regulated gene expression and circadian pathways but also in other phases of plant development (Ouyang et al., 2011; Stirnberg et al., 2012; Tang et al., 2012).
Several other mutants have been isolated that exhibit an intermediate response (long after far-red light1 [laf1], laf3, laf6, far1, fhl, long hypocotyl in far-red1 [hfr1], far-red insensitive2 [fin2], and fin219; Soh et al., 1998, 2000; Hudson et al., 1999; Fairchild et al., 2000; Fankhauser and Chory, 2000; Hsieh et al., 2000; Ballesteros et al., 2001; Møller et al., 2001; Hare et al., 2003; Zhou et al., 2005). Yet other mutants have been isolated that are hypersensitive toward FR light (empfindlicher im dunkelroten licht1 [eid1], suppressor of phyA1 (spa1), and spa4; Hoecker et al., 1998; Dieterle et al., 2001; Laubinger et al., 2004). Although most mutants have been characterized at the molecular level, the interplay of the factors is not yet fully understood.
We have previously reported the isolation of the Arabidopsis mutant pat1-1, which acts in a dominant-negative way, as the truncated PAT1 gene is still expressed (Bolle et al., 2000). PAT1 is a member of the plant-specific GRAS protein family, which plays important regulatory roles in diverse aspects of plant development (Bolle, 2004). The family name is derived from the first three members that were cloned, GIBBERELLIC ACID INSENSITIVE (GAI), REPRESSOR OF GIBBERELLIC ACID INSENSITIVE3 (RGA), and SCARECROW (SCR; Pysh et al., 1999). Some GRAS proteins are involved in such developmental processes as meristem formation and maintenance (e.g. LATERAL SUPPRESSOR, HAIRY MERISTEM [HAM]; Schumacher et al., 1999; Stuurman et al., 2002; Greb et al., 2003) or radial patterning (e.g. SCR, SHORT ROOT [SHR]; Di Laurenzio et al., 1996; Helariutta et al., 2000). Others are involved in signal transduction pathways, such as the members of the DELLA protein subbranch (GAI, RGA, RGA-LIKE1-3), which are negative regulators of gibberellin signal transduction (Peng et al., 1997; Silverstone et al., 1997; Davière et al., 2008; Schwechheimer, 2008). The GRAS protein family, which is relatively large, with at least 33 identified ORFs in the Arabidopsis genome (Bolle, 2004; Tian et al., 2004), can be organized using sequence alignment and phylogenetic analysis into several subfamilies. Four proteins in Arabidopsis are highly homologous to PAT1: SCARECROW-LIKE1 (SCL1), SCL5, SCL13, and SCL21. Therefore, we reasoned that perhaps all proteins of the PAT1 branch may be involved in light signaling pathways.
In this study, we have investigated loss-of-function lines of PAT1 and SCL21 and characterized their biological functions using genetic and molecular approaches. Both proteins are positive-acting factors specific for the phyA signal transduction pathway. Using genetic and biochemical studies, we show that they are involved in the same signaling pathway. Nevertheless, SCL21 expression is light regulated via phyA and PAT1.
RESULTS
SCL21 and PAT1 Are Members of the Same Subgroup of GRAS Proteins
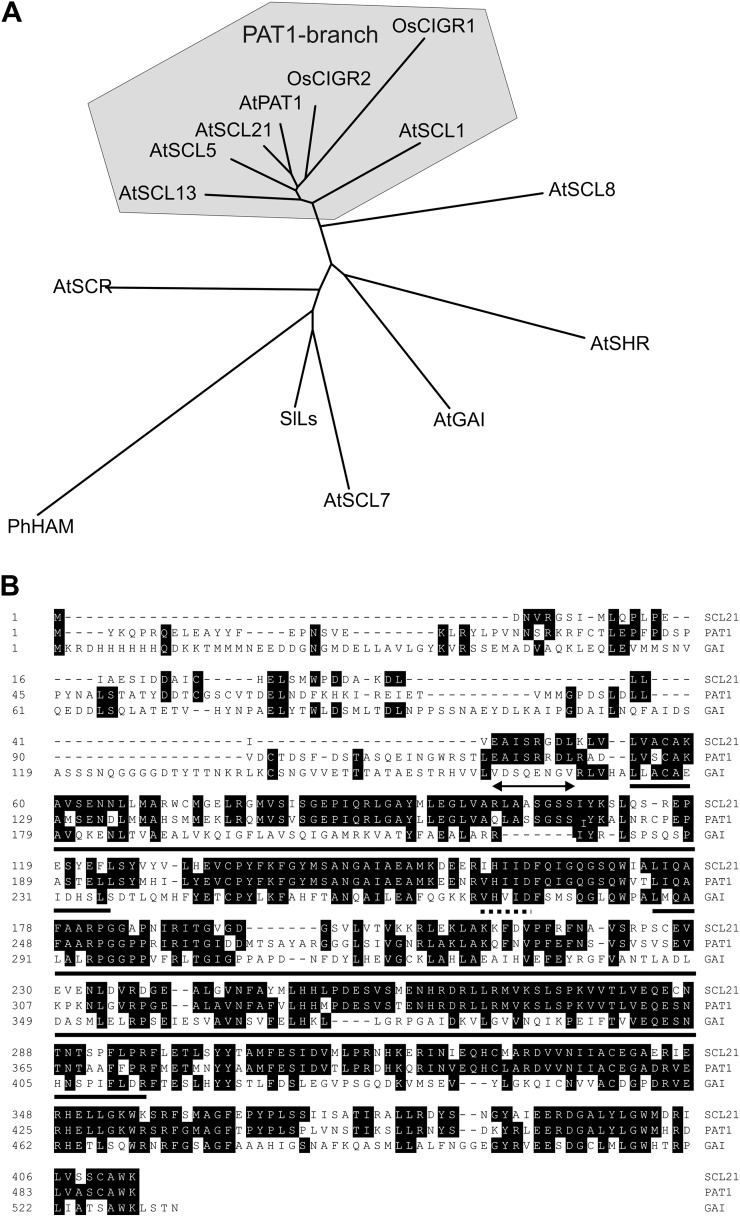
pat1-1 was isolated as a semidominant mutant that exhibits an extremely long hypocotyl under FR light, similar to the phyA loss-of-function mutant (Bolle et al., 2000). Phylogenetic analysis demonstrated that the PAT1 protein belongs to the GRAS protein family and that it is a member of one subbranch of this large protein family, which consists of PAT1, SCL1, SCL5, SCL13, and SCL21 (Fig. 1A; Bolle, 2004; Tian et al., 2004). SCL21, a 413-amino acid protein (At2g04890), is the closest homolog to PAT1 in Arabidopsis, sharing 68% identity on the protein level.
Figure 1.
SCL21 and PAT1 are members of the same subbranch of the GRAS protein family. A, Phylogenetic tree of the GRAS protein family. Representatives of all major branches of the GRAS family are shown (AtGAI, AtSCR, SlLs, AtSCL7, AtSHR, PhHAM, and AtSCL8). All Arabidopsis proteins in the PAT1 cluster are presented. The tree was generated with the PHYLIP program. At, Arabidopsis; Os, rice; Ph, Petunia hybrida; Sl, Solanum lycopersicum. B, Alignment of the amino acid sequences of Arabidopsis SCL21 with PAT1 and GAI as a more distant relative. Conserved sequences are shaded. Gaps are introduced to facilitate the alignment of conserved residues and are indicated as dashes in the sequence. The conserved VHIID domain is demarcated with a dotted line, the two Leu-rich domains with a solid line, and the EAISRRDL motif with an arrow.
All the PAT1 branch members contain the conserved signature motifs described for GRAS proteins within their C terminus (Fig. 1B): the conserved V/I HIID amino acid residues flanked by two Leu-rich domains, the PFYRE, RVER, and SAW motifs, and a putative Tyr phosphorylation site (Bolle, 2004). The biological functions of these conserved domains are still not fully understood. Although the C-terminal part of the GRAS proteins is highly conserved, their N termini vary in length and sequence. The alignment of SCL21 with PAT1 shows that SCL21 shares some conserved amino acids with PAT1 in the N-terminal part of the protein (the EAISRRDL motif), but its N terminus is much shorter than that of PAT1 (Fig. 1B; Supplemental Fig. S1). A database search was performed to find sequences of GRAS proteins homologous to the N termini of PAT1 and SCL21 in other plant species. Whereas sequences with homology to the PAT1 N terminus were observed in several other dicots, to date, protein sequences homologous to SCL21 could only be identified within the Brassicaceae (Arabidopsis lyrata; Supplemental Figs. S1 and S2). Several monocot GRAS proteins can be allocated to the PAT1 subbranch, although their sequences vary substantially within the first 46 to 90 amino acids when compared with PAT1. Among these are the rice (Oryza sativa) proteins OsCIGR1 and OsCIGR2 (for chitin-inducible gibberellin-responsive), which are inducible by the elicitor N-acetylchitooligosaccharide and exogenous gibberellins (Day et al., 2004). The closest Physcomitrella patens homologs, PAL1A and PAL1B, though, lack the complete N-terminal domain upstream of the first Leu-rich domain (122 amino acids compared with PAT1), suggesting that the N-terminal part of PAT1 and SCL21 could be a recent acquisition in evolution (Supplemental Figs. S1 and S2).
To obtain the complete coding sequence of SCL21, 5′ RACE analysis was performed, revealing a single intron within the 5′ untranslated leader sequence very similar in position to those of PAT1 and SCL13, another member of this subbranch (Torres-Galea et al., 2006). Sequence identities of the introns and the leader sequence between SCL21 and PAT1 were around 51%, whereas the identity between the coding regions was higher on the DNA level (69%).
Identification of SCL21 and PAT1 Knockdown and Knockout Lines
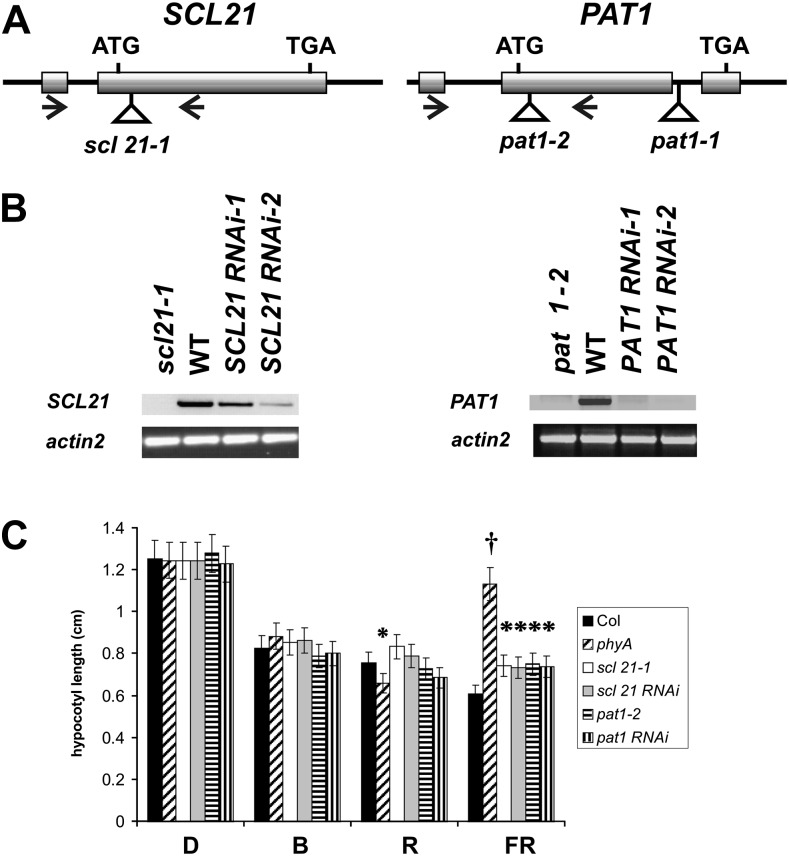
In order to evaluate the function of SCL21 in vivo and to determine whether it contributes to the phyA signaling pathway in a manner similar to PAT1, loss-of-function lines were identified (Fig. 2A). As the pat1-1 mutant itself is not a loss-of-function line, the pat1-2 insertion line from the SALK collection (SALK_064220) was isolated, in which the open reading frame is disrupted at amino acid 36. For SCL21, an insertion line was identified that carries a transfer DNA (T-DNA) insertion 119 bp after the ATG and disrupts the coding sequence at amino acid 39 (SAIL 313_G09; scl21-1).
Figure 2.
Analysis of the loss-of-function lines for SCL21 and PAT1. A, Schematic structures of the genomic SCL21 and PAT1 loci showing the positions of the T-DNA insertions and intron (line)/exon (bar) structure. The translation initiation (ATG) and termination (TAA) codons are indicated. Arrows indicate the positions of the primers used for B. B, RT-PCR was performed on total RNA from 2-week-old plants grown under continuous W light. The expression levels of SCL21 and PAT1 are shown in the wild type (WT), RNAi lines, and insertion lines. actin2 was used as a reference. C, Hypocotyl lengths of 4-d-old seedlings grown in darkness (D) or under continuous FR (0.5 μmol m−2 s−1), R (1 μmol m−2 s−1), and B (8 µmol m−2 s−1) light. Statistically significant variations from the wild type (Columbia [Col]) are indicated with asterisks (P < 0.05) or a cross (P < 0.01). Error bars indicate sd.
To complement this analysis, several independent antisense and RNA interference (RNAi) lines for SCL21 and PAT1 were generated. The reduction of the RNA levels in all lines used for the following experiments was confirmed by semiquantitative reverse transcription (RT)-PCR. RT-PCR products were obtained using complementary DNAs (cDNAs) derived from wild-type seedlings but not from RNA of loss-of-function mutants (Fig. 2B). Independent RNAi lines showed varying degrees of RNA reduction, and those lines that displayed the strongest reduction (greater than 70%), such as SCL21 RNAi-2, were chosen for further experiments (Fig. 2B). Taken together, these data demonstrate that all the isolated lines used for further physiological assessment were disrupted in normal SCL21 and PAT1 gene function.
Both PAT1 and SCL21 Are Involved in phyA-Dependent Signaling
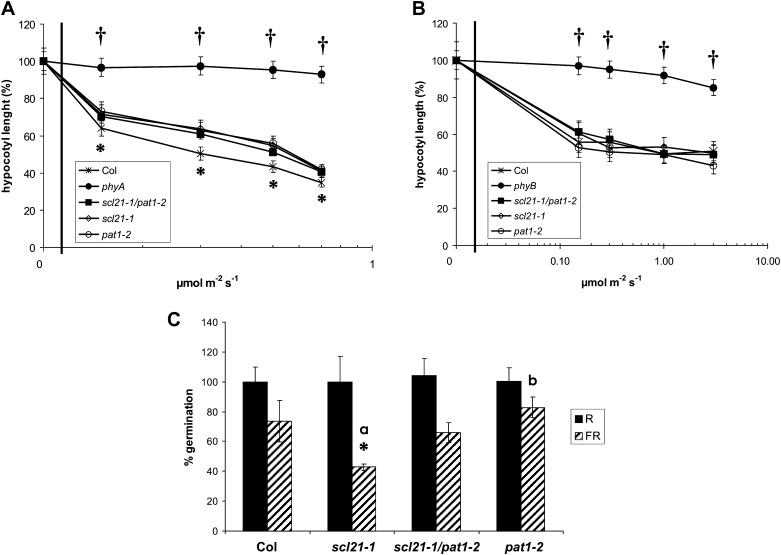
To establish whether the loss of PAT1 and SCL21 function alters the deetiolation process, we analyzed the selected lines under different light regimes (darkness and continuous R, FR, and B light). As presented in Figure 2C, all lines in which PAT1 and SCL21 gene expression was reduced or abolished had elongated hypocotyls under FR light as compared with the wild type (P < 0.05), although this elongation was not as strong as in the phyA mutant. This was also true for antisense lines (data not shown). Under R light, phyA mutants were shorter compared with the wild type, which has been described previously (P < 0.05; Hennig et al., 2001). The variability shown between scl21-1 and pat1-2 was not statistically significant (P > 0.1). No statistically significant difference could be determined under B light. When grown under darkness, the lines were indistinguishable from the wild type, indicating that the effects of the mutations are light dependent.
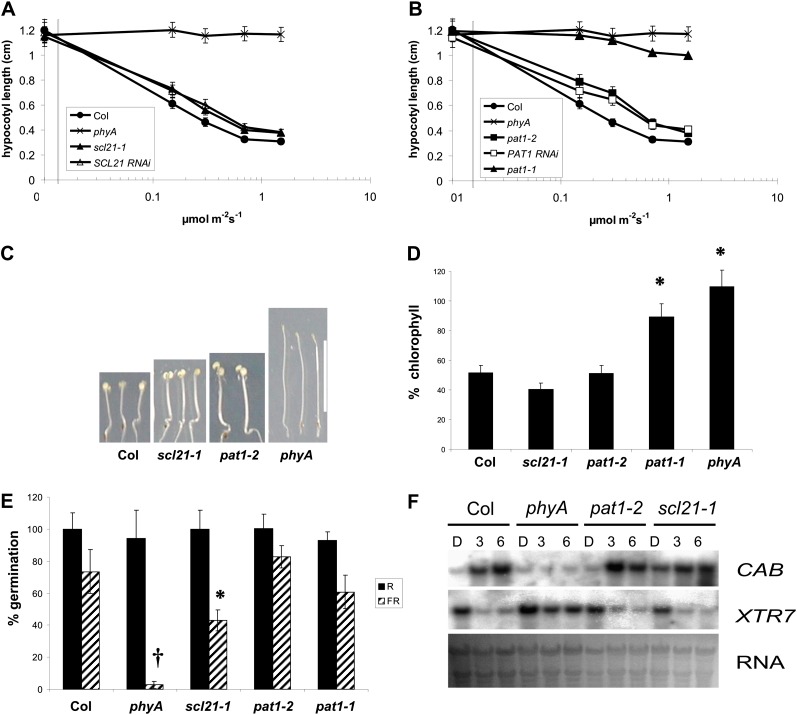
To confirm the FR light-dependent phenotype, the hypocotyl length of the different lines was analyzed under different fluence rates of FR light (Fig. 3, A–C). Figure 3, A and B, show that the hypocotyl length of a phyA mutant does not change even under higher fluence rates, whereas the hypocotyl length of wild-type seedlings is drastically reduced. The pat1-1 mutant displays a hypocotyl length similar to phyA, although slightly shorter under higher fluences. The scl21 and pat1 loss-of-function lines exhibit a slightly, but statistically significant, longer hypocotyl than the wild type (P < 0.05). The loss of inhibition of hypocotyl elongation was stronger at lower fluences but still evident under higher fluences. In contrast to hypocotyl elongation, cotyledon opening was not impaired under different fluences of FR light but was similar to the wild type (Fig. 3C).
Figure 3.
Physiological analysis of the SCL21 and PAT1 loss-of-function lines. A and B, Fluence response curves for hypocotyl elongation under FR light of the wild type (Columbia [Col]), phyA, scl21-1, pat1-1, pat1-2, and representative SCL21 and PAT1 RNAi lines. All hypocotyl values are statistically different from the respective wild-type values (P < 0.05). Error bars indicate sd. C, Phenotypes of scl21-1 and pat1-2 after 4 d of FR light. Mutant lines are compared with the wild type and phyA. Bar = 1 cm. D, Greening after prolonged FR light. Seedlings were grown for 4 d in continuous FR light (0.3 μmol m−2 s−1) and then transferred for 3 d into W light (80 μmol m−2 s−1). Chlorophyll accumulation was subsequently determined. Chlorophyll levels in phyA were set to 100%. Statistically significant variations from the wild type are indicated with asterisks (P < 0.05). Error bars indicate sd. E, Germination assays of the wild type and scl21-1, pat1-1, pat1-2, and phyA mutants. Seeds were treated with a FR light pulse after sterilization. Treated seeds were irradiated with a 10-min R light pulse (5 µmol m−2 s−1) after 3 h or a 10-min FR light pulse (1 µmol m−2 s−1) after 48 h. Germination was assessed after 7 d in darkness. Statistically significant variations from the wild type are indicated with an asterisk (P < 0.05) or a cross (P < 0.01). Error bars indicate sd. F, Northern analysis of phyA-regulated genes in the wild type, pat1-2, scl21-1, and phyA. Total RNA was harvested from 4-d-old seedlings grown in darkness without exposure to FR (D) or after 3 h (3) or 6 h (6) of irradiation with continuous FR light (1 μmol m−2 s−1). Each lane contained 5 μg of total RNA. Duplicate samples for each treatment were from seedlings grown independently under identical conditions. The same blot was probed for transcripts encoding CAB and XTR7. Ethidium bromide staining of the ribosomal RNA is shown as a loading control. [See online article for color version of this figure.]
The fact that the insertion, antisense, and RNAi lines all exhibit slightly, but statistically significant, elongated hypocotyls under FR light indicates that both PAT1 and SCL21 are involved in phyA-dependent signaling responses. Because the loss-of-function lines show a decreased responsiveness to FR light, both proteins act as positive regulators of phyA signaling. The analysis of the hypocotyl length of scl21-1/phyA or pat1-2/phyA double mutants did not indicate any additive phenotype with respect to the phyA phenotype, confirming the specificity of these GRAS proteins for the phyA signaling pathway (Supplemental Fig. S3).
Another physiological marker for phyA-dependent signaling is the FR light-dependent block of chlorophyll biosynthesis in subsequent white (W) light (van Tuinen et al., 1995; Barnes et al., 1996). Plants do not green in FR light because the committal step of chlorophyll biosynthesis needs higher light energy to be activated (Armstrong et al., 1995). Plastids exposed to FR light undergo a partial differentiation but cannot complete their development when given W light afterward. pat1-1 lines are able to green efficiently after 4 d of 1.2 µmol m−2 s−1 FR and subsequent W light treatment, similar to phyA mutants, whereas the wild type dies (Bolle et al., 2000). When given sublethal amounts of FR light (0.3 µmol m−2 s−1), wild-type plants survive but could only accumulate low amounts of chlorophyll (Fig. 3D). The PAT1 and SCL21 loss-of-function lines accumulate chlorophyll levels comparable to the wild type and, therefore, are not compromised in their sensitivity to the phyA-dependent block of greening, similar to the wild type (Fig. 3D).
Hypocotyl elongation under continuous FR light is a typical HIR. To test whether VLFRs, which are also phyA dependent, are likewise affected in the mutant lines, we performed germination assays. Germination was induced by a FR light pulse 48 h after imbibition and assayed in comparison with germination without any light. A phyA mutant is not able to germinate under these conditions (Shinomura et al., 1996; Kneissl et al., 2008). Whereas pat1-1 and pat1-2 displayed no significant reduction of germination under FR light, scl21-1 seeds germinated less efficiently (P < 0.05). The capacity to germinate after a R light pulse 3 h after imbibition, a well-characterized phyB response, was not impaired in any of the lines tested (Fig. 3E). These findings suggest that VLFRs, at least those important for germination, are not transduced via PAT1 and only in part by SCL21, indicating that these proteins are predominantly involved in HIR. Hypocotyl length under hourly pulses of FR light, another established test for VLFRs, also did not show any significant changes between the wild type and the pat1 and scl21 loss-of-function lines (data not shown). Double mutants of phyA with pat1-1, pat1-2, or scl2-1 showed the same behavior as phyA mutants under the HIR and VLFR tested, strengthening the fact that both proteins are predominantly important for the phyA signal transduction. Recently, it has been suggested that phyA is also responsible for a R-HIR (Franklin and Whitelam, 2007). Neither PAT1 nor SCL21 is involved in this pathway, as under 100 µmol m−2 s−1 R light, the hypocotyl length of pat1-1, pat1-2, and scl2-1 was similar to that of the wild type (data not shown).
pat1-1 has been shown to be essential for the appropriate expression of a subset of phyA-regulated genes (Bolle et al., 2000). To ascertain whether similar genes are affected in the SCL21 loss-of-function lines, we compared the expression levels of transcripts encoding the chlorophyll a/b-binding protein (CAB), which is induced by FR light, and a xyloglucan endotransglycosylase (XTR7), which is negatively regulated by light. Figure 3F shows that in the phyA mutant, in contrast to the wild type, CAB mRNA levels are not induced by FR light and XTR7 is not reduced. This is also true for pat1-1 (Bolle et al., 2000). The expression pattern in scl21-1 and pat1-2 resembles that in the wild type and, if at all, only subtle changes can be observed (Fig. 3F).
Light Regulates the Expression of SCL21 But Not of PAT1
The expression level of the SCL21 is very low, even lower than that observed for PAT1, and is barely detectable by northern analysis and often falling below the threshold of detection for microarrays. Digital northern analysis using data generated by AtGenExpress microarrays (Schmid et al., 2005), which compares expression levels in different developmental stages and tissues, revealed elevated levels of SCL21 only in maturing seeds as compared with seedlings and adult plants (Supplemental Fig. S4). This suggests a functional role for SCl21 in germination and the early phases of plant development. On the other hand, PAT1 is expressed at higher levels in all tissues. In seedlings, it is detectable predominantly in the hypocotyl and in adult tissue in the mature flower (especially stamen and petals) and the stem (Supplemental Fig. S4).
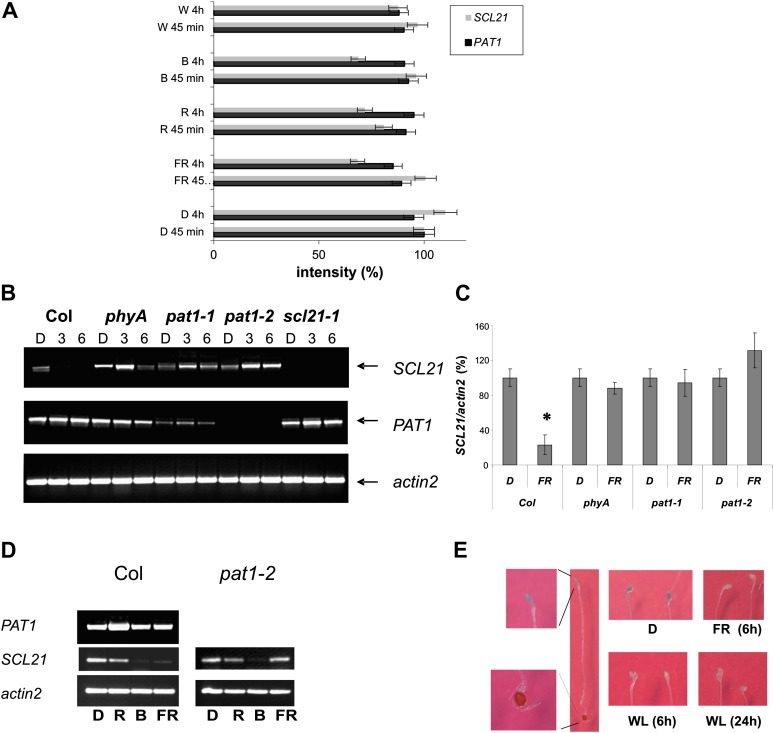
Microarray data derived from a set of light experiments during early stages of Arabidopsis seedling deetiolation (Peschke and Kretsch, 2011) indicated that SCL21, in contrast to PAT1, is negatively regulated by light. Figure 4A shows that 4-h B, R, W, and FR light treatment all down-regulated the level of SCL21 mRNA accumulation as compared with 4 h of darkness, but after 45 min, only under R light could a reduction be observed. Semiquantitative RT-PCR analysis revealed that SCL21 mRNA levels in the wild type were reduced at least 4-fold upon FR light treatment as compared with plants grown in the dark (Fig. 4, B and C). SCL21 expression in response to FR light was no longer down-regulated in the phyA mutant. These data suggest that PHYA is required for the regulation of SCL21 mRNA levels. Interestingly, the reduction of SCL21 expression is also impaired in the pat1-1 and pat1-2 mutants, also implicating PAT1 in the control of SCL21 down-regulation. The expression of PAT1, on the other hand, is not light regulated and not dependent on the presence of SCL21 or phyA. The reduced expression level of SCL21 mRNA in response to B or R light could also be confirmed by semiquantitative RT-PCR, with a stronger effect for B light in contrast to R light. This negative regulation, however, is not dependent on PAT1, as in the pat1-2 mutant, both expression levels are still reduced, whereas under FR light, expression is no longer reduced (Fig. 4D).
Figure 4.
Expression levels of SCL21 and PAT1 in different tissues and conditions. A, Expression of SCL21 and PAT1 after 45 min or 4 h of continuous W, R, B, and FR light compared with the expression in darkness (D; light treatments are described at http://arabidopsis.org/servlets/TairObject?type=expression_set&id=1007966126; Peschke and Kretsch, 2011). For better comparison, the dark value (45 min) was set at 100%. Error bars indicate sd. B, Semiquantitative RT-PCR analysis of SCL21 and PAT1 gene expression. Total RNA was harvested from 4-d-old seedlings (Columbia wild type [Col], phyA, pat1-1, pat1-2, and scl21-1) grown in darkness without exposure to FR (D) or after 3 h (3) or 6 h (6) of irradiation with continuous FR light (1 μmol m−2 s−1). As a reference, actin2 was used. C, Quantification of data from three independent semiquantitative RT-PCR analyses described in B. Expression of SCL21 was corrected with the expression of actin2, and values for the expression in the dark were set at 100% to calculate the relative expression of SCL21 after 6 h of FR light. Error bars indicate sd. Statistically significant variation from the dark level is indicated with an asterisk (P < 0.01). D, Semiquantitative RT-PCR analysis of SCL21 and PAT1 gene expression. Total RNA was harvested from 4-d-old seedlings (Columbia wild type and pat1-2) grown in darkness without exposure to light (D) or after 4 h of irradiation with R (1 μmol m−2 s−1), B (8 µmol m−2 s−1), or FR (1 μmol m−2 s−1) light. As a reference, actin2 was used. E, Light-repressed expression pattern of a SCL21::GUS reporter gene fusion construct in Arabidopsis seedlings. Four-day-old etiolated seedlings (D) were assayed for GUS activity by histochemical staining with the X-Gluc substrate. Details of the staining in the cotyledons and the root tip are shown. Additionally, 4-d-old etiolated seedlings were incubated either for 6 h in FR light (1 μmol m−2 s−1) or 6 and 24 h in W light (80 μmol m−2 s−1) and subsequently stained with X-Gluc.
To dissect the expression at a tissue-specific level, SCL21-promoter::GUS reporter gene lines were generated. In seedlings, the expression is limited to the cotyledons and the apical hook and the tip of the root. A decrease of GUS activity could also be noted when etiolated seedlings were treated for 6 or 24 h under FR or W light conditions as compared to dark-grown seedlings (Fig. 4E). In 3-week-old seedlings, however, GUS activity was distributed throughout the plant, which was not expected from the microarray data but could reflect the higher stability of the GUS protein (Supplemental Fig. S5).
Subcellular Localization of SCL21
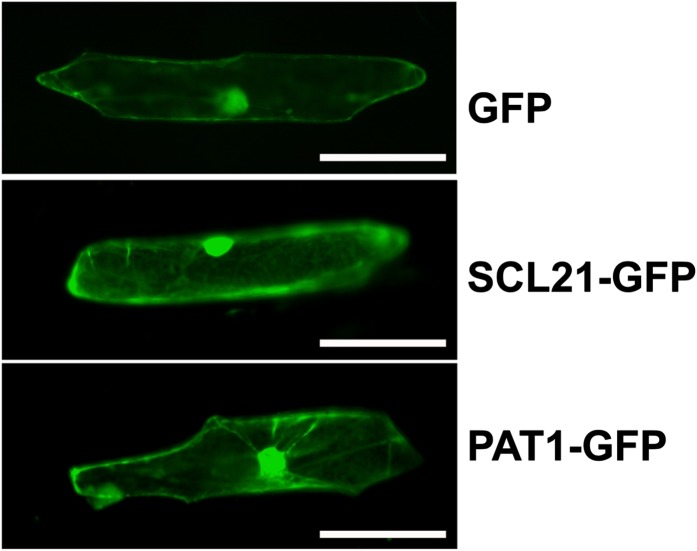
Classical nuclear localization signals could not be detected by computer analysis in either PAT1 or SCL21. To experimentally determine the subcellular localization, we fused the respective coding regions with their C termini to the GFP reporter gene and introduced these constructs transiently into onion (Allium cepa) epidermal cells by particle bombardment. As shown in Figure 5, the resulting SCL21-GFP protein was localized throughout the cytoplasm and the nucleus, similar to PAT1. The distribution was not altered by different light conditions or by the fusion of GFP to the N terminus (data not shown). These results indicate that both proteins could act in the cytoplasm as well as in the nucleus.
Figure 5.
Subcellular localization of PAT1 and SCL21. Onion epidermal cells were bombarded with 35S-GFP, 35S-SCL21-GFP, or 35S-PAT1-GFP and visualized using a fluorescence microscope. Bars = 100 µm.
Analysis of Epistasis between PAT1 and SCL21
Because both SCL21 and PAT1 are involved in phyA-dependent signaling, we wanted to investigate whether these proteins operate in the same or in divergent signaling pathways. To test the genetic relationship between SCL21 and PAT1, we generated a pat1-2/scl21-1 double mutant. Physiological analysis of hypocotyl elongation under different light regimes determined that the double mutant has an elongated hypocotyl under FR light but was wild type under all other light conditions (Fig. 6, A and B). The double mutant was statistically longer than the wild type under FR light (P < 0.05), but compared with the phenotype of the parental lines, hypocotyl elongation was not increased in the double mutant (P > 0.1), suggesting that both proteins act in the same signaling pathway. Hence, in FR light, both proteins cannot substitute for each other. The double mutant is still not as long as the pat1-1 mutant under FR light, suggesting that perhaps an additional protein from the PAT1 subbranch of the GRAS protein family is involved in FR light signaling that can partially substitute for PAT1 and SCL21.
Figure 6.
Phenotypic analysis of the scl21-1/pat1-2 double mutant. Fluence response curves for hypocotyl elongation under FR (A) and R (B) light of Columbia wild type (Col), phyA, phyB, scl21-1/pat1-2, scl21-1, and pat1-2. Hypocotyl lengths in darkness were considered as 100%. Statistically significant variations from the wild type are indicated with asterisks (P < 0.05) or crosses (P < 0.01). Error bars indicate sd. C, Germination assays of wild-type Columbia, the double mutant, and the parental lines. Seeds were treated with a FR light pulse after sterilization. Treated seeds were irradiated with a 10-min R light pulse (5 µmol m−2 s−1) after 3 h or a 10-min FR light pulse (1 µmol m−2 s−1) after 48 h. Germination was assessed after 7 d in darkness. Statistically significant variations from the wild type are indicated with an asterisk (P < 0.05) and from the double mutant with lowercase letters (aP < 0.01, bP < 0.05). Error bars indicate sd.
As we had previously determined a difference in germination efficiency after a FR light pulse between scl21-1 and pat1-2, we also tested the double mutant under these conditions. An intermediate phenotype between both parental lines could be observed (scl21-1, P < 0.01; pat1-2, P < 0.05), but no statistical difference from the wild type was found (P > 0.1; Fig. 6C). This suggests that SCL21 and PAT1 are part of mechanisms that differ between HIR and VLFR.
SCL21 and PAT1 Can Interact with Each Other
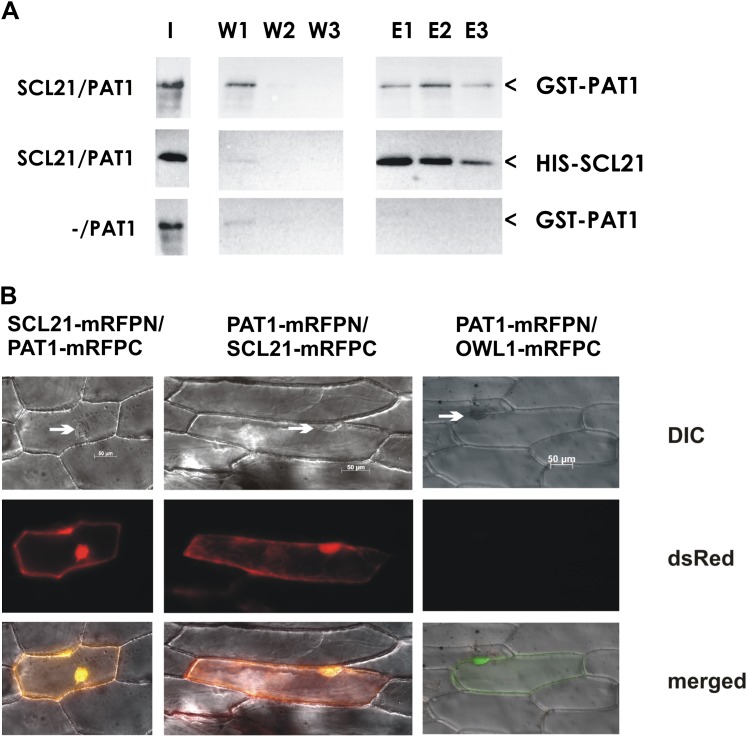
The sequence similarity and the genetic interactions between SCL21 and PAT1 prompted us to examine whether the two proteins could interact physically. For OsGAI/SLR, it has been shown that dimerization may play a role in the function of GRAS proteins (Itoh et al., 2002). In yeast (Saccharomyces cerevisiae) two-hybrid assays, we were not able to detect any dimerization between SCL21 and PAT1, as both proteins function as transcriptional activators in this system. In vitro pull-down assays, however, suggested an interaction between the two proteins (Fig. 7A). For this assay, overexpressed HIS-SCL21 and glutathione S-transferase (GST)-PAT1 fusion proteins were used. GST-PAT1 was able to interact with HIS-SCL21 bound to nickel-nitrilotriacetic acid agarose (Ni-NTA). GST-PAT1 could not bind to the Ni-NTA by itself or to Escherichia coli proteins present in the overexpressing strain. Additionally, GST alone was also not able to interact with SCL21.
Figure 7.
Physical interaction between SCL21 and PAT1. A, In vitro pull-down assays showing the interaction between PAT1 and SCL21. Recombinant HIS-SCL21 bound to Ni-NTA was used in pull-down assays with overexpressed GST-PAT1 (SCL21/PAT1). As a control, HIS-SCL21 was substituted with extracts from a not-transformed bacterial culture (−/PAT1). Samples were separated by SDS-PAGE, and the blots were probed either with anti-HIS or anti-GST antibodies. I, Input protein (unbound fraction); W, proteins eluted during wash steps 1 to 3; E, proteins eluted from the column with higher amounts of imidazole (1–3). Arrowheads mark the GST-PAT1 and HIS-SCL21 bands. Experiments were repeated three times with similar results. B, BiFC assays. Fluorescence microscopy images of onion epidermal cells coexpressing SCL21 and PAT1 fused either with the N-terminal part of mRFP (mRFPN) or the C-terminal part (mRFPC). Reconstitution of functional mRFP as detected by dsRed fluorescence occurs in the nucleus but also in the cytoplasm. Differential interference contrast (DIC) images are shown and were overlapped with the images of the dsRed channel (merged). Arrows indicate the nucleus. As a negative control, OWL1-mRFPC was used. Coexpressed GFP was detected to verify transient expression (green staining in the merged image).
To confirm this finding, we used a bimolecular fluorescence complementation (BiFC) system based on a red fluorescent protein (Zilian and Maiss, 2011). Split-monomeric red fluorescent protein (mRFP) fusions with SCL21 and PAT1 were cobombarded into onion epidermal cells and an mRFP fluorescence signal was recovered, corroborating the interaction between these two components in vivo (Fig. 7B). Moreover, transient expression of the mRFP fusions revealed that the interaction could be observed in the nucleus and the cytoplasm, corroborating the subcellular localization observed for these proteins. These results indicated that SCL21 and PAT1 can indeed directly interact with one another.
DISCUSSION
Here, we present the physiological, biochemical, and genetic analysis of two GRAS proteins, PAT1, a previously described phyA signaling intermediate (Bolle et al., 2000), and SCL21, a novel member of the PAT1 branch of the GRAS protein family. SCL21 is the closest homolog to PAT1, sharing 68% identity at the amino acid level. Not surprisingly, the two proteins differ in the composition and length of their N termini, the least conserved region of this protein family. Interestingly, SCL21, PAT1, and SCL5, also a member of the PAT1 subbranch, all share a common EAISRRDL protein motif in their N terminus that the other family members lack. Because scl21, scl5, and pat1 all exhibit a phyA-related mutant phenotype (this paper; P. Torres-Galea and C. Bolle, unpublished data), it is possible that this motif plays a functional role in mediating the phyA signal. SCL13, another close homolog that lacks this motif, is involved mainly in phyB signaling (Torres-Galea et al., 2006). Gene duplication events accompanied by the gain or deletion of sequences could account for the variations in the N-terminal domains even within a subbranch. Moreover, the subbranches of the GRAS proteins seem to have diversified prior to the divergence of the moss and flowering plant lineages (Engstrom, 2011). It is possible that the divergence of the N terminus of the GRAS proteins during evolution increased the functional capacity of the protein family, including the mediation of gibberellin and phyA signals. Sun et al. (2011) predicted that molecular recognition features within GRAS protein N termini can adopt different tertiary structures facilitating the recruitment of additional protein partners. This could suggest that the N-terminal domain determines the specificity for a distinct pathway, whereas the C-terminal so-called “GRAS domain” carries out an as yet unknown function.
Three independently generated loss-of-function lines (insertion, RNAi, and antisense lines) established that both SCL21 and PAT1 function as positive-acting components in the phyA-dependent signaling pathway. These loss-of-function mutants display a very similar phenotype, a decreased inhibition of hypocotyl elongation under FR light, but not under any other light treatment (W, B, or R light or in the dark). Although the semidominant pat1-1 mutant phenotype is stronger than the loss-of-function pat1-2 mutant, the responses observed are very similar.
To analyze whether the SCL21 and PAT1 functions overlap, a double mutant was generated. Physiological analysis of scl21-1/pat1-2 indicated that the hypocotyl length of the double mutant in response to FR light was very similar to the individual parental lines. These data suggest that both SCL21 and PAT1 are required for FR light-mediated inhibition of hypocotyl elongation. It is possible that SCL21 and PAT1 together form a heterodimeric complex that is needed for functionality in the phyA signaling pathway. This hypothesis is supported by the fact that both proteins are localized to the same subcellular compartments (cytoplasm and nucleus) and that the interaction of both proteins in these compartments can be confirmed in vivo by a BiFC assay. Furthermore, an in vitro pull-down experiment confirmed this interaction biochemically. If the interaction of SCL21 and PAT1 is necessary for their function, the loss of one partner (as in the single mutant) would compromise the function of the other protein, explaining why the phenotype of the single mutants and the double mutant are very similar. As no additive phenotypes could be observed in the double mutant under HIR conditions, we can rule out that both proteins act in different pathways or can partially substitute for each other’s function.
The observation that the pat1/scl21 double mutant phenotype is not as extreme as that observed for pat1-1 can be explained in two ways. First, the functionality of the multiprotein complex is compromised by the loss of the PAT1 C-terminal domain. This truncated form of PAT1 could negatively affect PAT1 function by altering protein stability and/or altering PAT1’s ability to interact with proteins other than SCL21 through the missing C-terminal domain, thereby changing the transfer of the signal down the cascade. It is feasible, for example, that PAT1 interacts only transiently with a protein, whereas the truncated protein forms a stable interaction, thereby not allowing a desensitization of a signaling pathway. Another possibility that cannot be ruled out at this point is that other GRAS proteins, namely SCL1 and SCL5, can compensate in part for PAT1 and SCL21, as they also seem to play a functional role in the phytochrome signaling pathway (P. Torres-Galea and C. Bolle, unpublished data). In this case, a quadruple mutant would be expected to have a similar phenotype to pat1-1.
Our data also demonstrate that both SCL21 and PAT1 regulate a specific subset of FR light-dependent responses. Although mutations in both genetic loci affect hypocotyl elongation, the unfolding of cotyledons and FR-dependent block of greening, two HIR processes, were not altered. We could not find any evidence that the pat1 single mutants, including the semidominant mutation pat1-1, affect VLFRs, indicating that only the HIRs seem to be mediated through this protein. In contrast, SCL21 seems to also play a minor role in germination after a FR pulse, a typical VLFR. Dimerization between PAT1 and SCL21 seems not to be necessary for some functions, such as for the inhibition of germination under VLFR conditions, as the scl21 single mutant is more strongly impaired than the pat1/scl21 double mutant. It could be hypothesized that in the absence of SCL21, PAT1 could homodimerize or interact with another GRAS protein and thereby inhibit germination. If SCL21 alone were responsible for the germination phenotype, than in the double mutant, we would expect a similar phenotype to that in scl21-1. The fact that the double mutant is slightly less efficient in germination compared with the pat1-2 mutant can be a first hint that, indeed, PAT1 also plays a role in VLFR-dependent germination, albeit to a lesser extent than SCL21.
Expression data also suggest that SCL21 and PAT1 have unique functions as well. The fact that SCL21 is expressed at higher levels in maturing seeds as compared with seedlings hints that SCL21 is necessary in the preliminary stages of germination. This fits with our physiological observation that SCL21 is important for germination under very low fluences. Furthermore, SCL21 mRNA levels are down-regulated upon FR, R, and B light treatment. PAT1 mRNA levels, however, are much more abundant, ubiquitously expressed, and not modulated by light treatment. These data suggest that both SCL21 and PAT1 are required in germinating seedlings to perceive FR light via phyA signal transduction, whereas PAT1 and perhaps other GRAS proteins of the PAT1 branch are important for later stages of development. Regulation on the transcriptional level can influence the presence of the individual GRAS proteins of the PAT1 branch proteins at different time points of development, thereby allowing varying heterodimerization events between these GRAS proteins that could create an additional level of regulation. Indeed, we demonstrate here that SCL21 gene expression is dependent upon the presence of both PAT1 and phyA. PAT1 and SCL21 could be acting in a hierarchical manner comparable to that observed for two other GRAS proteins, SCR and SHR, with SHR operating upstream of SCR, controlling its levels of expression (Helariutta et al., 2000).
It is interesting that the levels of SCL21 mRNA are reduced in response to light, a situation when one would hypothesize that the presence of SCL21 would be required. In fact, the expression of other genes encoding proteins involved in FR light signal transduction, such as FHY1, has been shown to behave in a similar way (Desnos et al., 2001; Zeidler et al., 2001). It is possible that these proteins are required for the initial perception of light and that, after its perception, the signal must be abrogated, a desensitization process. The mRNA levels for other components of phyA signaling, however, are unaffected by light (e.g. PAT1, FAR1; Hudson et al., 1999) or actually increased by FR light (e.g. HFR1; Fairchild et al., 2000). On the other hand, the expression of the negative regulator SPA1 is induced by FR light (Hoecker et al., 1998). Because pure FR light conditions rarely exist outside of the laboratory, a more in-depth evaluation of SCL21 expression under natural conditions should be undertaken. However, additional photoreceptors seem to be involved as well in the down-regulation of SCL21 transcript levels, as SCL21 mRNA levels are also reduced in R, B, and W light. This may reflect a point of interaction between the signal transduction pathways associated with these different photoreceptors.
In conclusion, this study provides evidence that both SCL21 and PAT1 mediate phyA-dependent light signaling, functioning in some instances in the same pathway. It seems likely that both proteins are part of the same regulatory protein complex that acts to modulate early responses to phyA-dependent light changes. Other GRAS proteins interact with a variety of different proteins; the DELLA proteins in particular have been shown to interact with transcription factors (de Lucas et al., 2008; Feng et al., 2008; Gallego-Bartolomé et al., 2010). Further studies are necessary to elucidate how PAT1 and SCL21 function at the biochemical level to mediate phyA signaling.
MATERIALS AND METHODS
Plant Material and Growth Conditions
All lines used in this study are in the Arabidopsis (Arabidopsis thaliana) Columbia background. Seeds were surface sterilized for 10 min in 30% (v/v) commercial bleach with the addition of 0.05% Triton X-100, rinsed at least three times, and sown on petri dishes (11 cm diameter) containing one-half-strength Murashige and Skoog basal medium (Sigma) and 0.8% (w/v) agar (Duchefa). To select transgenic lines, the medium was supplemented with 3% Suc and with kanamycin or Basta, respectively. Unless used for germination assays, plates were stored at 4°C for 3 d, and germination was induced by 4 h of W light followed by 22 h of darkness at 21°C. After this treatment, plates were transferred to appropriate light conditions. Light intensities were determined with spectroradiometers (W, B, and R light, model Li-1800 [LiCor]; FR light, model SKP200 with a sensor for 730 nm [Skye Instruments]). The B, R, and FR light sources were generated by light-emitting diodes using diodes with emission maxima at 469, 660, and 740 nm (Quantum Devices; PVP).
Analysis of Mutants
Insertion lines were obtained from the SAIL (scl21-1; 313_G09) and SALK (pat1-2; SALK_064220) collections. Plants were backcrossed, selfed, and grown on selective medium. Genomic DNA was extracted from resistant plants and analyzed by PCR to see whether they were homozygous for the insertion. For scl21-1 and pat1-2, primers at the 5′ end of the coding sequence (SCL21, 5′-CCCTTATCGACTTCCACCG-3′; PAT1, 5′-ATGTACAAGCAGCCTAGACAAG-3′) and from the left border of the T-DNA insertion (SAIL, 5′-GAAATGGATAAATAGCCTTGCTTCC-3′; SALK, 5′-GTTCACGTAGTGGGCCATCG-3′) were used to detect the insertion. The fragments were sequenced to confirm the insertion sites. Gene-specific primers flanking the insertion sites were used to distinguish between heterozygous and homozygous plants. Homozygous mutant plants were selfed, retested in the next generation, and used for physiological experiments. Double mutants were generated by crossing and identified in the F2 generation by PCR. F3 seeds were used for the physiological experiments.
Physiological Measurements
For fluence-response experiments, seedlings were grown under appropriate light conditions for 4 d. All lines were analyzed in parallel. The experiments were repeated at least three times, and each time a minimum of 50 seedlings were analyzed. Hypocotyl lengths and cotyledon sizes were documented using a digital camera and measured with the NIH Image software (ImageJ; National Institutes of Health).
Germination assays were performed according to Shinomura et al. (1996). Briefly, seeds were sterilized and plated on one-half-strength Murashige and Skoog medium without Suc. After plating, the seeds were pulsed with FR light (1 μmol m−2 s−1) for 10 min and transferred to dark. To test for R light responsiveness, seeds were illuminated again after 3 h with R light (5 μmol m−2 s−1) for 10 min. To test for FR light responsiveness, a FR light pulse (1 μmol m−2 s−1) of 15 min was given after 48 h. After the appropriate light pulse, the seedlings were kept in darkness for 6 d and germination was scored as positive as soon as the radicle was visible. Germination efficiency was normalized for seeds that could germinate without the second light pulse and seeds that did not germinate under W light. Experiments were repeated at least three times with seeds from different batches.
For analysis of the FR light-dependent block of greening effect, seedlings were grown for 4 d in continuous FR light (0.3 µmol m−2 s−1). Subsequently, seedlings were transferred for 3 d into W light (80 µmol m−2 s−1) before chlorophyll content was determined (Kneissl et al., 2008). Experiments were repeated at least three times.
Statistical Analysis
Where appropriate, the physiological experiments were evaluated with Student’s t tests (Excel; two-tailed distribution) between the wild type and the mutant line or, when indicated, between mutant lines. P values are given.
Transgenic Plants
To generate PAT1 and SCL21 RNAi lines, a part of each cDNA was amplified, resulting in a 665-bp-long fragment for PAT1 and an 855-bp-long one for SCL21, and cloned into the pENTR/D-TOPO cloning vector (Invitrogen). For directed cloning, a 5′-CACC extension was added to the forward primer (PAT1, 5′-CACCGACTTCAGCGTATGCTC-3′ and 5′-GCACACGAGGCAACCAAATC-3′; SCL21, 5′-CACCAACTCTCCATGTGGCCTG-3′ and 5′-GATTCGAACATTGCCGTG-3′). With the help of LR Clonase (Invitrogen), the fragments were combined into the Gateway-adapted binary vectors pK7GWIWG2(I) and pB7GWIWG2(I) (Karimi et al., 2002), which contain two tail-to-tail insertion sites separated by an intron. The expression is driven by the 35S-RNA cauliflower mosaic virus (CaMV) promoter.
To examine SCL21 promoter activity, a 1,946-bp fragment upstream of the ATG start codon was amplified with PCR (forward, 5′-CACCGCAACAAACTGAACAAG-3′; reverse, 5′-CAGCTATCTCTGGCAGTGGCTG-3′) and cloned into the pENTR/D-TOPO cloning vector (Invitrogen). The fragment was then transferred to the Gateway-adapted vector pKGWFS7 (Karimi et al., 2002), which contains the coding sequences of GFP and GUS downstream of the insertion site.
All constructs were verified by restriction and sequence analysis. Constructs were transformed into Arabidopsis plants via the Agrobacterium tumefaciens floral dip method (Clough and Bent, 1998). Transformants were selected on kanamycin or Basta-containing medium, self-fertilized, and homozygous progeny were selected.
For histochemical analysis of GUS staining, transgenic SCL21::GUS reporter gene-containing seedlings and adult plants (F2 and F3) were incubated for 24 h in 0.1 m phosphate buffer, pH 7.0, containing 0.1% Triton X-100 (v/v), 10 mm EDTA, 0.5 mm ferrocyanide, 0.5 mm ferricyanide, and 0.125 mm 5-bromo-4-chloro-3-indolyl-β-glucuronic acid (X-Gluc; Roth). Chlorophyll was removed with 70% ethanol, and the blue staining was analyzed with a microscope.
RNA Analysis
For FR light transcriptional induction experiments, seedlings were prepared as described above. After germination induction, seedlings were grown for 4 d in darkness before being transferred (0 h) to FR light (0.7 μmol m−2 s−1) for 3 or 6 h. Tissue was collected and frozen in liquid nitrogen, and RNA was extracted using the Qiagen Plant RNeasy kit according to the manufacturer’s instructions. To isolate RNA from adult plants, the TRIZOL method (Invitrogen) was used.
Ten micrograms of total RNA was separated on 1.2% formaldehyde-MOPS gels and blotted onto Hybond-N nylon membranes (GE Healthcare). Hybridizations were performed overnight at 68°C in a buffer containing 7% (w/v) SDS, 0.5 m sodium phosphate, pH 7.0, and 1 mm EDTA. PCR-generated probes were labeled by random priming using the DecaLabel DNA Labeling Kit (MBI Fermentas). Washing steps were carried out to a final stringency of 0.5× SSC and 0.1% (w/v) SDS at 65°C. The signal was quantified using a PhosphorImager (Molecular Dynamics) and the software AIDA (package version 3.25 β; Raytest) and normalized for loading using the 18S ribosomal RNA signal.
For the analysis of transcript levels of low-abundance genes, a RT reaction was performed with total RNA extracted from wild-type and mutant lines. An oligo(dT)18 primer was hybridized to 1 μg of total RNA. RT was performed with the Omniscript RT Kit (Qiagen) according to the manufacturer’s instructions. After the RT reaction, 2 μL of the cDNA was used for the PCR with gene-specific primers for SCL21 and PAT1 (PAT1, 5′-CATGGAACTTCAATAACATTCACTAC-3′ and 5′-GCACACGAGGCAACCAAATC-3′; SCL21, 5′-CCCTTATCGACTTCCACCG-3′ and 5′-GATTCGAACATTGCCGTG-3′). To avoid amplification of contaminating DNA, the 5′ forward primer was located 5′ of the intron in the leader sequence, which also prevented amplifying the RNAi constructs. PCRs were stopped after 25 and 30 cycles to avoid saturation of the reaction, and the reaction mixtures were analyzed on agarose gels. PCR products were visualized with BioDocAnalyze Digital gel documentation (Biometra) and quantified using the BioDocAnalyze software (Biometra). Each PCR was repeated at least three times. Amplification of ACTIN2 cDNA (forward, 5′-CTCTTTCTTTCCAAGCTCATAAAAAATG-3′; reverse, 5′-CAGCACAATACCGGTTGTACGAC-3′) was used as a normalization control.
To determine the 5′ transcription start site, a cDNA library, which was generated by fusion of linkers to the 5′ and 3′ ends of the cDNA (Marathon cDNA Amplification Kit; BD Biosciences Clontech), was used for RACE according to the manufacturer’s instructions.
Subcellular Localization and BiFC Assay
The SCL21 and PAT1 open reading frames were amplified by PCR from cDNA using primers containing restriction sites for XbaI and KpnI and inserted into the pGFP vector (Kost et al., 1998), generating a SCL21-GFP or PAT1-GFP fusion driven by the 35S-RNA CaMV promoter. Onion (Allium cepa) epidermal cells were bombarded with this construct using a helium biolistic gun and incubated in darkness for 12 h. To test for possible effects of light, the cells were subsequently incubated for 3 h under either FR or W light. A 35S-RNA CaMV-GFP construct was used as a control. To visualize GFP fluorescence, cells were examined using an Axioskop microscope (Carl Zeiss).
For the BiFC assay, the SCL21 and PAT1 open reading frames were amplified by PCR from cDNA using primers containing restriction sites for SalI and BamHI, cloned into pGEM-T Easy (Promega), and inserted with SalI and BamHI into both vectors, pBiFC_1 (pCB:mRFPN-GOI) and pBiFC_2 (pCB:mRFPC-GOI; Zilian and Maiss, 2011). The vectors encode the N-terminal amino acids 1 to 168 and the C-terminal amino acids 169 to 225 of the mRFP, and SCL21 and PAT1 were fused to their C terminus. Either SCL21-mRFPN/PAT1-mRFPC or PAT-mRFPN/SCL21-mRFPC was transiently coexpressed in onion epidermal cells, and interaction was observed using an Axioskop microscope (Carl Zeiss). As a negative control, OWL1, which has previously been shown not to interact with PAT1, was used (OWL1-mRFPC). To localize the transformed cells faster, pGFP was cobombarded.
In Vitro Pull-Down Assay
Full-length cDNAs of SCL21 and PAT1 were cloned from the pENTR/D-TOPO vector into the pDEST15 and pDEST17 vectors, respectively, to generate HIS-SCL21 and GST-PAT1 fusions. The constructs were transformed into Escherichia coli strain BL21(DE3)pLysS (Stratagene), and liquid cultures of single colonies were grown at 37°C to an optical density of 0.6. Expression of the fusion proteins was then induced with 1 mm isopropylthio-β-galactoside, and cells were incubated for 3 h at 37°C. The cultures were harvested, resuspended in lysis buffer (50 mm NaH2PO4, 300 mm NaCl, and 10 mm imidazole, pH 8.0), and cells were lysed under native conditions and then transferred to −80°C for 30 min. After sonification (3 × 10 s) and centrifugation (15 min, 4°C, 10,000g), the pellet with inclusion bodies was resuspended in 500 µL of 1% lithium dodecyl sulfate, 12.5% Suc, 5 mm ε-aminocaproic acid, 1 mm benzamidine, and 50 mm HEPES-KOH, pH 7.8 (adapted from Pribil et al., 2010). Subsequently, the proteins were subjected to 10 min on ice, 10 min at 90°C, 15 min at 25°C, 20 min at −20°C, thawed on ice, and kept at 25°C for 10 min. Octyl-glucopyranoside (1% [w/v] final concentration) was added, and the solution was kept on ice for 15 min. Afterward, KCl (75 mm final concentration) was added to precipitate the lithium dodecyl sulfate detergent. After centrifugation at 16,000g at 4°C for 15 min, the supernatant contained the enriched fraction of refolded HIS-SCL21 and GST-PAT1. The amount of proteins was visualized by SDS-PAGE. HIS-SCl21 was preincubated with the Ni-NTA resin for 30 min, and equal amounts of GST-PAT1 were added and incubated further for 30 min by gently shaking at 4°C. As a control, proteins isolated from a nontransformed E. coli strain were incubated with the resin and GST-PAT1 was added. After centrifugation, the unbound proteins were removed with the supernatant. The Ni-NTA resin was washed with 10 volumes of buffer (50 mm NaH2PO4, 300 mm NaCl, and 20 mm imidazole). Finally, the proteins were eluted from the resin in three steps with 1 volume of the elution buffer (50 mm NaH2PO4, 300 mm NaCl, and 250 mm imidazole, pH 8.0). Samples from all factions were separated using SDS-PAGE, and immunodetection was performed with anti-Penta-HIS (Qiagen; dilution, 1:5,000) and anti-GST (Sigma-Aldrich; 1:10,000) as primary antibodies. As secondary antibodies, anti-mouse (for anti-HIS) and anti-rabbit (for anti-GST) antibodies conjugated to horseradish peroxidase (Invitrogen), diluted 1:10,000, were applied. Blocking buffer (Tris-buffered saline plus Tween 20) was substituted with 3% nonfat milk powder. The signal was detected by chemiluminescence, and for quantification, the program BioDocAnalyze (Biometra) was used.
Sequence Analysis
Database searches were performed with BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Alignment of sequences was performed with the ClustalW program (DNAstar). The phylogenetic tree was generated with the PHYLIP program using SEQBOOT for bootstrapping (100 replicates), PROTDIST, and FITCH (Fitch and Margoliash) analysis. The phylogenetic tree was produced with DRAWTREE. Additionally, a phylogenetic tree was generated using MegAlign (setting, accurate/Gonnet) with bootstrapping (trials 1,000/seed 111) of the DNAstar program (Lasergene).
Analysis of Microarray Data
The original data published by the AtGenExpress consortium were downloaded (Expression Atlas of Arabidopsis Development; http://arabidopsis.org/servlets/TairObject?type=expression_set&id=1006710873 [described in Schmid et al., 2005] and Light Treatments; http://arabidopsis.org/servlets/TairObject?type=expression_set&id=1007966126 [described in Peschke and Kretsch, 2011]) and were evaluated according to the analysis performed at the Nottingham Arabidopsis Stock Centre (http://affymetrix.arabidopsis.info). Values not marked with P (present) were discharged. A mean value was generated from triplicates, and sd was calculated. Furthermore, the data were compared using the Genevestigator program (Zimmermann et al., 2004).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers NP_178566 (SCL21), NP_974903 (PAT1), and NP_172945.1 (GAI).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Alignment of the amino acid sequences of Arabidopsis SCL21, PAT1, and their closest homologs.
Supplemental Figure S2. Phylogenetic tree of the proteins aligned in Supplemental Figure S1.
Supplemental Figure S3. Hypocotyl lengths of 4-d-old seedlings (wild type [Columbia], phyA, phyB, double mutants of scl21-1, pat1-2, and pat1-1 with phyA or phyB) grown under continuous FR light.
Supplemental Figure S4. Expression pattern of SCL21 and PAT1 evaluated by microarray data.
Supplemental Figure S5. Expression pattern of a SCL21::GUS reporter gene fusion construct in Arabidopsis adult plants.
Acknowledgments
We thank Prof. Dr. D. Leister for his support and Christine Matzenbacher, Anina Neumann, Ingrid Duschanek, Christine Gonzales-Serrano, Corinna Hofer, and Martina Reymers for their excellent assistance in this project. Prof. Dr. Edgar Maiss is thanked for the vectors used in the BiFC assay. We are grateful to Nam-Hai Chua for helpful discussions and to Randy Foster for discussions and assistance with the manuscript. We thank the Nottingham Arabidopsis Stock Centre and Syngenta for providing mutant seed.
Glossary
- R
red
- FR
far-red
- B
blue
- W
white
- RNAi
RNA interference
- RT
reverse transcription
- VLFR
very low fluence response
- HIR
high irradiance response
- Ni-NTA
nickel-nitrilotriacetic acid agarose
- GST
glutathione S-transferase
- BiFC
bimolecular fluorescence complementation
- CaMV
cauliflower mosaic virus
- X-Gluc
5-bromo-4-chloro-3-indolyl-β-glucuronic acid
- T-DNA
transfer DNA
- cDNA
complementary DNA
References
- Armstrong GA, Runge S, Frick G, Sperling U, Apel K. (1995) Identification of NADPH:protochlorophyllide oxidoreductases A and B: a branched pathway for light-dependent chlorophyll biosynthesis in Arabidopsis thaliana. Plant Physiol 108: 1505–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae G, Choi G. (2008) Decoding of light signals by plant phytochromes and their interacting proteins. Annu Rev Plant Biol 59: 281–311 [DOI] [PubMed] [Google Scholar]
- Ballesteros ML, Bolle C, Lois LM, Moore JM, Vielle-Calzada JP, Grossniklaus U, Chua NH. (2001) LAF1, a MYB transcription activator for phytochrome A signaling. Genes Dev 15: 2613–2625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes SA, Nishizawa NK, Quaggio RB, Whitelam GC, Chua NH. (1996) Far-red light blocks greening of Arabidopsis seedlings via a phytochrome A-mediated change in plastid development. Plant Cell 8: 601–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolle C. (2004) The role of GRAS proteins in plant signal transduction and development. Planta 218: 683–692 [DOI] [PubMed] [Google Scholar]
- Bolle C, Koncz C, Chua NH. (2000) PAT1, a new member of the GRAS family, is involved in phytochrome A signal transduction. Genes Dev 14: 1269–1278 [PMC free article] [PubMed] [Google Scholar]
- Casal JJ, Cerdán PD, Staneloni RJ, Cattaneo L. (1998) Different phototransduction kinetics of phytochrome A and phytochrome B in Arabidopsis thaliana. Plant Physiol 116: 1533–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Shi X, Chen L, Dai M, Zhou Z, Shen Y, Li J, Li G, Wei N, Deng XW. (2012) Phosphorylation of FAR-RED ELONGATED HYPOCOTYL1 is a key mechanism defining signaling dynamics of phytochrome A under red and far-red light in Arabidopsis. Plant Cell 24: 1907–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Chory J, Fankhauser C. (2004) Light signal transduction in higher plants. Annu Rev Genet 38: 87–117 [DOI] [PubMed] [Google Scholar]
- Clack T, Mathews S, Sharrock RA. (1994) The phytochrome apoprotein family in Arabidopsis is encoded by five genes: the sequences and expression of PHYD and PHYE. Plant Mol Biol 25: 413–427 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [Google Scholar]
- Davière JM, de Lucas M, Prat S. (2008) Transcriptional factor interaction: a central step in DELLA function. Curr Opin Genet Dev 18: 295–303 [DOI] [PubMed] [Google Scholar]
- Day RB, Tanabe S, Koshioka M, Mitsui T, Itoh H, Ueguchi-Tanaka M, Matsuoka M, Kaku H, Shibuya N, Minami E. (2004) Two rice GRAS family genes responsive to N-acetylchitooligosaccharide elicitor are induced by phytoactive gibberellins: evidence for cross-talk between elicitor and gibberellin signaling in rice cells. Plant Mol Biol 54: 261–272 [DOI] [PubMed] [Google Scholar]
- de Lucas M, Davière JM, Rodríguez-Falcón M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blázquez MA, Titarenko E, Prat S. (2008) A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484 [DOI] [PubMed] [Google Scholar]
- Desnos T, Puente P, Whitelam GC, Harberd NP. (2001) FHY1: a phytochrome A-specific signal transducer. Genes Dev 15: 2980–2990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterle M, Zhou YC, Schäfer E, Funk M, Kretsch T. (2001) EID1, an F-box protein involved in phytochrome A-specific light signaling. Genes Dev 15: 939–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Laurenzio L, Wysocka-Diller J, Malamy JE, Pysh L, Helariutta Y, Freshour G, Hahn MG, Feldmann KA, Benfey PN. (1996) The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86: 423–433 [DOI] [PubMed] [Google Scholar]
- Engstrom EM. (2011) Major GRAS gene subfamilies arose early in land plant evolution. Plant Signal Behav 6: 850–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairchild CD, Schumaker MA, Quail PH. (2000) HFR1 encodes an atypical bHLH protein that acts in phytochrome A signal transduction. Genes Dev 14: 2377–2391 [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C, Chory J. (2000) RSF1, an Arabidopsis locus implicated in phytochrome A signaling. Plant Physiol 124: 39–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C, Chen M. (2008) Transposing phytochrome into the nucleus. Trends Plant Sci 13: 596–601 [DOI] [PubMed] [Google Scholar]
- Feng S, Martinez C, Gusmaroli G, Wang Y, Zhou J, Wang F, Chen L, Yu L, Iglesias-Pedraz JM, Kircher S, et al. (2008) Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA. (2008) Shade avoidance. New Phytol 179: 930–944 [DOI] [PubMed] [Google Scholar]
- Franklin KA, Quail PH. (2010) Phytochrome functions in Arabidopsis development. J Exp Bot 61: 11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA, Whitelam GC. (2007) Phytochrome a function in red light sensing. Plant Signal Behav 2: 383–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Bartolomé J, Minguet EG, Marín JA, Prat S, Blázquez MA, Alabadí D. (2010) Transcriptional diversification and functional conservation between DELLA proteins in Arabidopsis. Mol Biol Evol 27: 1247–1256 [DOI] [PubMed] [Google Scholar]
- Greb T, Clarenz O, Schafer E, Muller D, Herrero R, Schmitz G, Theres K. (2003) Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Genes Dev 17: 1175–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare PD, Moller SG, Huang LF, Chua NH. (2003) LAF3, a novel factor required for normal phytochrome A signaling. Plant Physiol 133: 1592–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helariutta Y, Fukaki H, Wysocka-Diller J, Nakajima K, Jung J, Sena G, Hauser MT, Benfey PN. (2000) The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101: 555–567 [DOI] [PubMed] [Google Scholar]
- Hennig L, Poppe C, Sweere U, Martin A, Schäfer E. (2001) Negative interference of endogenous phytochrome B with phytochrome A function in Arabidopsis. Plant Physiol 125: 1036–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiltbrunner A, Tscheuschler A, Viczián A, Kunkel T, Kircher S, Schäfer E. (2006) FHY1 and FHL act together to mediate nuclear accumulation of the phytochrome A photoreceptor. Plant Cell Physiol 47: 1023–1034 [DOI] [PubMed] [Google Scholar]
- Hoecker U, Xu Y, Quail PH. (1998) SPA1: a new genetic locus involved in phytochrome A-specific signal transduction. Plant Cell 10: 19–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh HL, Okamoto H, Wang M, Ang LH, Matsui M, Goodman H, Deng XW. (2000) FIN219, an auxin-regulated gene, defines a link between phytochrome A and the downstream regulator COP1 in light control of Arabidopsis development. Genes Dev 14: 1958–1970 [PMC free article] [PubMed] [Google Scholar]
- Hudson M, Ringli C, Boylan MT, Quail PH. (1999) The FAR1 locus encodes a novel nuclear protein specific to phytochrome A signaling. Genes Dev 13: 2017–2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh H, Ueguchi-Tanaka M, Sato Y, Ashikari M, Matsuoka M. (2002) The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell 14: 57–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kami C, Lorrain S, Hornitschek P, Fankhauser C. (2010) Light-regulated plant growth and development. Curr Top Dev Biol 91: 29–66 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kevei E, Schafer E, Nagy F. (2007) Light-regulated nucleo-cytoplasmic partitioning of phytochromes. J Exp Bot 58: 3113–3124 [DOI] [PubMed] [Google Scholar]
- Kneissl J, Shinomura T, Furuya M, Bolle C. (2008) A rice phytochrome A in Arabidopsis: the Role of the N-terminus under red and far-red light. Mol Plant 1: 84–102 [DOI] [PubMed] [Google Scholar]
- Kost B, Spielhofer P, Chua NH. (1998) A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. Plant J 16: 393–401 [DOI] [PubMed] [Google Scholar]
- Laubinger S, Fittinghoff K, Hoecker U. (2004) The SPA quartet: a family of WD-repeat proteins with a central role in suppression of photomorphogenesis in Arabidopsis. Plant Cell 16: 2293–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller SG, Kunkel T, Chua NH. (2001) A plastidic ABC protein involved in intercompartmental communication of light signaling. Genes Dev 15: 90–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy F, Schäfer E. (2002) Phytochromes control photomorphogenesis by differentially regulated, interacting signaling pathways in higher plants. Annu Rev Plant Biol 53: 329–355 [DOI] [PubMed] [Google Scholar]
- Ouyang X, Li J, Li G, Li B, Chen B, Shen H, Huang X, Mo X, Wan X, Lin R, et al. (2011) Genome-wide binding site analysis of FAR-RED ELONGATED HYPOCOTYL3 reveals its novel function in Arabidopsis development. Plant Cell 23: 2514–2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Carol P, Richards DE, King KE, Cowling RJ, Murphy GP, Harberd NP. (1997) The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev 11: 3194–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschke F, Kretsch T. (2011) Genome-wide analysis of light-dependent transcript accumulation patterns during early stages of Arabidopsis seedling deetiolation. Plant Physiol 155: 1353–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pribil M, Pesaresi P, Hertle A, Barbato R, Leister D. (2010) Role of plastid protein phosphatase TAP38 in LHCII dephosphorylation and thylakoid electron flow. PLoS Biol 8: e1000288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pysh LD, Wysocka-Diller JW, Camilleri C, Bouchez D, Benfey PN. (1999) The GRAS gene family in Arabidopsis: sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J 18: 111–119 [DOI] [PubMed] [Google Scholar]
- Quail PH. (1997) The phytochromes: a biochemical mechanism of signaling in sight? BioEssays 19: 571–579 [DOI] [PubMed] [Google Scholar]
- Quail PH. (2002) Phytochrome photosensory signalling networks. Nat Rev Mol Cell Biol 3: 85–93 [DOI] [PubMed] [Google Scholar]
- Ruberti I, Sessa G, Ciolfi A, Possenti M, Carabelli M, Morelli G. (2012) Plant adaptation to dynamically changing environment: the shade avoidance response. Biotechnol Adv 30: 1047–1058 [DOI] [PubMed] [Google Scholar]
- Schäfer E, Nagy F, editors (2006) Photomorphogenesis in Plants and Bacteria, Ed 3. Springer, Dordrecht, The Netherlands [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU. (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Schumacher K, Schmitt T, Rossberg M, Schmitz G, Theres K. (1999) The Lateral suppressor (Ls) gene of tomato encodes a new member of the VHIID protein family. Proc Natl Acad Sci USA 96: 290–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer C. (2008) Understanding gibberellic acid signaling: are we there yet? Curr Opin Plant Biol 11: 9–15 [DOI] [PubMed] [Google Scholar]
- Shinomura T, Nagatani A, Hanzawa H, Kubota M, Watanabe M, Furuya M. (1996) Action spectra for phytochrome A- and B-specific photoinduction of seed germination in Arabidopsis thaliana. Proc Natl Acad Sci USA 93: 8129–8133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone AL, Mak PY, Martínez EC, Sun TP. (1997) The new RGA locus encodes a negative regulator of gibberellin response in Arabidopsis thaliana. Genetics 146: 1087–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. (1999) Phytochromes: tripping the light fantastic. Nature 400: 710–711, 713 [DOI] [PubMed] [Google Scholar]
- Smith H. (2000) Phytochromes and light signal perception by plants: an emerging synthesis. Nature 407: 585–591 [DOI] [PubMed] [Google Scholar]
- Soh MS, Hong SH, Hanzawa H, Furuya M, Nam HG. (1998) Genetic identification of FIN2, a far red light-specific signaling component of Arabidopsis thaliana. Plant J 16: 411–419 [DOI] [PubMed] [Google Scholar]
- Soh MS, Kim YM, Han SJ, Song PS. (2000) REP1, a basic helix-loop-helix protein, is required for a branch pathway of phytochrome A signaling in Arabidopsis. Plant Cell 12: 2061–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirnberg P, Zhao S, Williamson L, Ward S, Leyser O. (2012) FHY3 promotes shoot branching and stress tolerance in Arabidopsis in an AXR1-dependent manner. Plant J 71: 907–920 [DOI] [PubMed] [Google Scholar]
- Stuurman J, Jäggi F, Kuhlemeier C. (2002) Shoot meristem maintenance is controlled by a GRAS-gene mediated signal from differentiating cells. Genes Dev 16: 2213–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan JA, Deng XW. (2003) From seed to seed: the role of photoreceptors in Arabidopsis development. Dev Biol 260: 289–297 [DOI] [PubMed] [Google Scholar]
- Sun X, Xue B, Jones WT, Rikkerink E, Dunker AK, Uversky VN. (2011) A functionally required unfoldome from the plant kingdom: intrinsically disordered N-terminal domains of GRAS proteins are involved in molecular recognition during plant development. Plant Mol Biol 77: 205–223 [DOI] [PubMed] [Google Scholar]
- Tang W, Wang W, Chen D, Ji Q, Jing Y, Wang H, Lin R. (2012) Transposase-derived proteins FHY3/FAR1 interact with PHYTOCHROME-INTERACTING FACTOR1 to regulate chlorophyll biosynthesis by modulating HEMB1 during deetiolation in Arabidopsis. Plant Cell 24: 1984–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian C, Wan P, Sun S, Li J, Chen M. (2004) Genome-wide analysis of the GRAS gene family in rice and Arabidopsis. Plant Mol Biol 54: 519–532 [DOI] [PubMed] [Google Scholar]
- Torres-Galea P, Huang LF, Chua NH, Bolle C. (2006) The GRAS protein SCL13 is a positive regulator of phytochrome-dependent red light signaling, but can also modulate phytochrome A responses. Mol Genet Genomics 276: 13–30 [DOI] [PubMed] [Google Scholar]
- van Tuinen A, Kerckhoffs LH, Nagatani A, Kendrick RE, Koornneef M. (1995) Far-red light-insensitive, phytochrome A-deficient mutants of tomato. Mol Gen Genet 246: 133–141 [DOI] [PubMed] [Google Scholar]
- Whitelam GC, Johnson E, Peng J, Carol P, Anderson ML, Cowl JS, Harberd NP. (1993) Phytochrome A null mutants of Arabidopsis display a wild-type phenotype in white light. Plant Cell 5: 757–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeidler M, Bolle C, Chua NH. (2001) The phytochrome A specific signaling component PAT3 is a positive regulator of Arabidopsis photomorphogenesis. Plant Cell Physiol 42: 1193–1200 [DOI] [PubMed] [Google Scholar]
- Zhou Q, Hare PD, Yang SW, Zeidler M, Huang LF, Chua NH. (2005) FHL is required for full phytochrome A signaling and shares overlapping functions with FHY1. Plant J 43: 356–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilian E, Maiss E. (2011) An optimized mRFP-based bimolecular fluorescence complementation system for the detection of protein-protein interactions in planta. J Virol Methods 174: 158–165 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]