Abstract
The methylesterification status of cell wall homogalacturonans, mediated through the action of pectin methylesterases (PMEs), influences the biophysical properties of plant cell walls such as elasticity and porosity, important parameters for cell elongation and water uptake. The completion of seed germination requires cell wall extensibility changes in both the radicle itself and in the micropylar tissues surrounding the radicle. In wild-type seeds of Arabidopsis (Arabidopsis thaliana), PME activities peaked around the time of testa rupture but declined just before the completion of germination (endosperm weakening and rupture). We overexpressed an Arabidopsis PME inhibitor to investigate PME involvement in seed germination. Seeds of the resultant lines showed a denser methylesterification status of their cell wall homogalacturonans, but there were no changes in the neutral sugar and uronic acid composition of the cell walls. As compared with wild-type seeds, the PME activities of the overexpressing lines were greatly reduced throughout germination, and the low steady-state levels neither increased nor decreased. The most striking phenotype was a significantly faster rate of germination, which was not connected to altered testa rupture morphology but to alterations of the micropylar endosperm cells, evident by environmental scanning electron microscopy. The transgenic seeds also exhibited an apparent reduced sensitivity to abscisic acid with respect to its inhibitory effects on germination. We speculate that PME activity contributes to the temporal regulation of radicle emergence in endospermic seeds by altering the mechanical properties of the cell walls and thereby the balance between the two opposing forces of radicle elongation and mechanical resistance of the endosperm.
Plant morphology is inevitably shaped by the cell wall, which provides mechanical support and protection at the cost of cellular motility. Plant cell growth is driven by water uptake and controlled changes in the extensibility of the cell wall; indeed, the biomechanical properties of the cell wall determine the morphology of the whole plant and its constituent tissues and organs by controlling the shape and size of cells (Cosgrove, 2005; Schopfer, 2006). Therefore, modifications that influence cell wall loosening, or conversely strengthen cell walls, are of major importance throughout the plant life cycle.
The primary plant cell wall consists mainly of a hydrated gel matrix of hemicellulosic and pectic polysaccharides, as well as cellulose microfibrils, along with proteins and aromatic substances (for review, see Fry, 2000; Cosgrove, 2005; Knox, 2008). Cell wall pectins are found either as homogalacturonans or as substituted molecules, the rhamnogalacturonans I and II as well as xylogalacturonan. Composed of a linear chain of 1,4-linked α-d-galacturonic acid (GalUA) residues, the homogalacturonans can be methylesterified at the C-6 carboxylic acid groups of the GalUA residues (Fry, 2000; Wolf et al., 2009). Depending on their position in the pectin polysaccharide network, unesterified GalUA residues can form Ca2+ cross links between the homogalacturonan molecules. The degree and pattern of pectin methylesterification are thus critical for the biomechanical properties of the cell wall, influencing various characteristics such as porosity, permeability, elasticity, and compressibility (Wolf et al., 2009; Peaucelle et al., 2012). Demethylesterification of cell wall pectins is mediated by pectin methylesterases (PMEs; EC 3.1.1.11). PME activity thus alters cell walls and, hence, mediates various physiological and biochemical processes in plants, including elongation growth and fruit ripening (for review, see Micheli, 2001; Wolf et al., 2009; Peaucelle et al., 2012). Some of the changes in the biomechanical properties of cell walls resulting from PME action are mediated through an increased susceptibility of homogalacturonans to polygalacturonases (Wakabayashi et al., 2003) and modulation of the activity of cell wall expansins through a decrease in cell wall pH (Moustacas et al., 1991; McQueen-Mason and Cosgrove, 1994).
Germination begins with imbibition and the perception of a germination-inducing signal by a nondormant seed and ends when the radicle has protruded through all covering layers (Bewley, 1997a). Mechanical restraint by the seed tissues surrounding the radicle is one potential factor that regulates the completion of germination. According to the “mechanical restraint” hypothesis, the growth potential of the radicle must exceed the mechanical resistance of the surrounding living seed tissues (e.g. the endosperm, perisperm, or megagametophyte, depending on the seed type) to permit the completion of germination (Bewley, 1997a; Müller et al., 2006). Cell wall hydrolases appear to be involved in both processes (i.e. in both tissue weakening and an increased growth potential of the embryo). Extension of the radicle may rely on the extensibility of the radicle cell walls (Bewley, 1997a). The activity of Xyloglucan Endotransglycosylase (XET), an enzyme capable of reversibly cleaving xyloglucan molecules, increases in the apical region of maize (Zea mays) seedling roots during their elongation (Wu et al., 1994), but this increase occurs after germination has been completed. Although not related to cell wall remodeling for radicle extension, a gene encoding a GA-regulated XET that is strongly expressed during germination has been implicated in the weakening of the endosperm cap of tomato (Solanum lycopersicum) seeds (Chen et al., 2002). The LeXET4 gene transcripts, which are restricted to the endosperm cap, are detected in seeds within 12 h of imbibition and reach a maximum at 24 h (Chen et al., 2002); they decline after radicle emergence despite a continued degradation of the lateral endosperm cell walls.
Other candidates for cell wall-loosening enzymes are the expansins, which do not have significant hydrolytic activity but rather possess the ability to disrupt hydrogen bonds between cell wall polymers (e.g. matrix polysaccharides and cellulose microfibrils; for review, see Cosgrove, 2005). Expression of an expansin gene in tomato (LeEXP4), specifically within the tomato endosperm cap, is correlated with weakening of this tissue during germination (Chen and Bradford, 2000). Tobacco (Nicotiana tabacum) endosperm cell walls may be weakened by β-1,3-glucanase to permit germination, since the activity of this enzyme increases within the micropylar endosperm before radicle emergence; abscisic acid (ABA) retards both enzyme accumulation and activity as well as endosperm rupture (Leubner-Metzger et al., 1995). In tomato, a germination-specific endo-β-mannanase gene is expressed in the micropylar endosperm cap of seeds (Nonogaki et al., 2000). Seeds of Datura ferox produce endo-β-mannanase and β-mannosidase in the micropylar region of the endosperm after red light stimulation, several hours before the radicle protrudes through it (Sanchez and de Miguel, 1997). In seeds of white spruce (Picea glauca), weakening of the micropylar end of the megagametophyte and nucellus precedes radicle protrusion, and this weakening is associated with endo-β-mannanase activity (Downie et al., 1997).
Very little is known about the role of PMEs in the germination of angiosperm seeds, although one study has characterized their role in the dormancy breakage and germination of conifer seeds (Ren and Kermode, 2000).
One aspect of PME regulation is the inhibition by specific proteinaceous PME inhibitors (PMEIs). The first described PMEI was that purified from kiwifruit (Actinidia chinensis; Balestrieri et al., 1990), and since then, several PMEIs from Arabidopsis (Arabidopsis thaliana) and other plant species have been characterized (Wolf et al., 2003; Raiola et al., 2004). Individual PMEIs interact with a number of different PMEs, often even over species boundaries (Giovane et al., 2004). In wheat (Triticum aestivum), PMEIs control the organ- or tissue-specific activity of specific PME isoforms (Rocchi et al., 2012).
In this study, we show the following: (1) PME activities are differentially regulated during the different phases of seed germination in Arabidopsis; (2) the degree of pectin methylesterification of the cell walls of seed tissues influences the rate of germination of the seeds; (3) changes to the degree of pectin methylesterification through overexpression (OE) of a PMEI in Arabidopsis altered the sensitivity of seed germination to ABA and changed the properties of the micropylar endosperm and radicle cells (characteristics during germination, especially endosperm rupture, and the size of cells, respectively); and (4) altered pectin methylesterification led to a number of strong morphological phenotypes during other stages of the Arabidopsis life cycle.
RESULTS
PME Activities Change during Different Phases of Arabidopsis Seed Germination
Water uptake by germinating seeds can be divided into three phases: phase I is imbibition and is characterized by rapid water uptake by the dry seed; phase II is associated with a lag, during which water uptake slows considerably and several metabolic changes take place (Bewley, 1997a; Weitbrecht et al., 2011). In Arabidopsis and related endospermic species, such as cress (Lepidium sativum) and tobacco, the completion of germination requires testa rupture (part of phase II) followed by endosperm rupture (Liu et al., 2005; Manz et al., 2005; Müller et al., 2006) and radicle elongation. Subsequently, there is further water uptake associated with the onset of seedling growth during phase III (Bewley, 1997a; Weitbrecht et al., 2011).
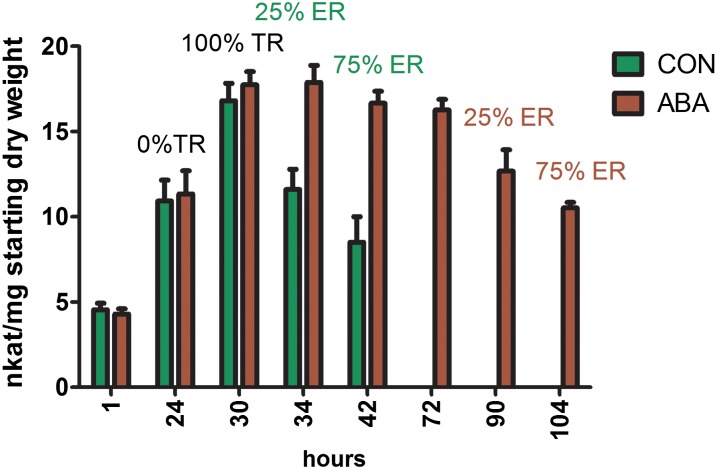
Large numbers of genes encoding pectin-modifying enzymes and associated regulators show significant differential regulation during the first 24 h of Arabidopsis seed germination (Nakabayashi et al., 2005). Based on this information, we sought to determine the temporal pattern of PME activity changes during the different stages of germination of wild-type Arabidopsis seeds (Fig. 1). PME activities increased before and shortly after testa rupture and decreased once endosperm rupture commenced and phase III set in. A principal component analysis run on Fourier transform infrared spectra of embryos from wild-type seeds at different stages of germination (Supplemental Fig. S1) supported the view that these activity changes affect the degree of pectin methylesterification. The samples were separated based on principal component 1, whose loading was strong in the region associated with ester bonds (Derbyshire et al., 2007; Pelletier et al., 2010).
Figure 1.
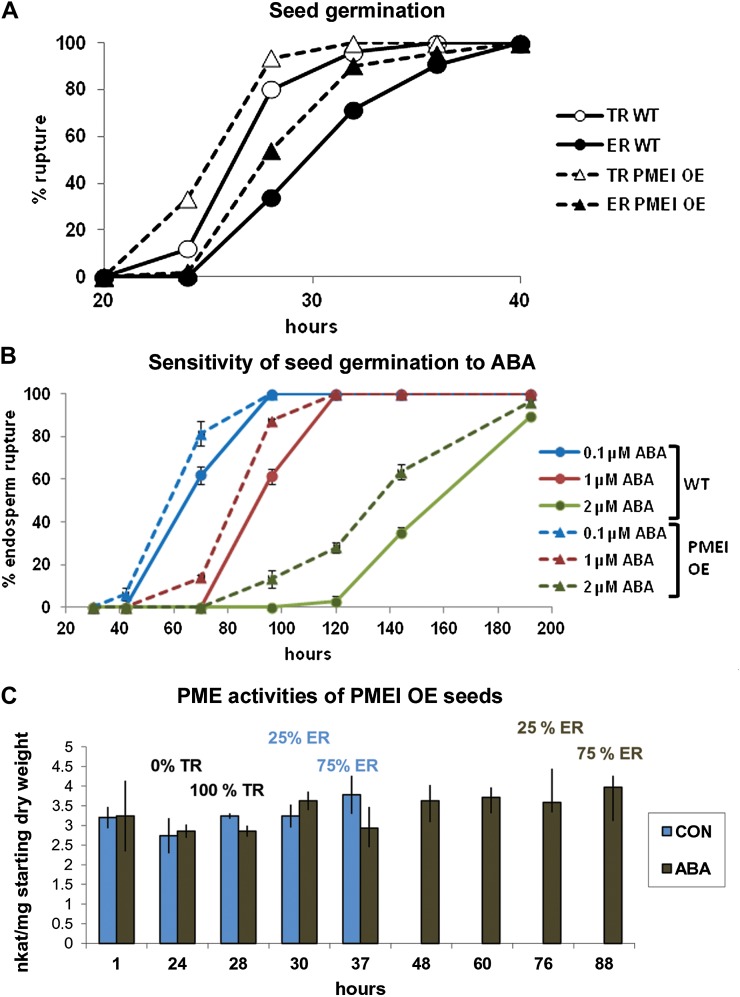
PME activities during germination of Arabidopsis wild-type seeds in the absence (control [CON]; green bars) and presence of 1 μm ABA (ABA; brown bars). Data represent averages ± se of four biological replicates of about 100 seeds each. Seeds were weighed before imbibition, and the activity shown is relative to the starting seed weight. The percentages over the bars list the proportions of seeds that had achieved testa rupture (TR) and endosperm rupture (ER). Note that both control seeds and ABA-treated seeds have reached 100% testa rupture at 32 h. Control seeds proceed to endosperm rupture, while ABA-treated seeds remain at the testa rupture stage for several days.
In order to determine whether the increase of PME activities over the first approximately 24 to 30 h of imbibition is connected to the event of testa rupture or is associated with the plateau in water uptake associated with phase II of germination, the effects of ABA were determined, as this hormone specifically prolongs phase II of germination by delaying endosperm rupture without affecting the timing of testa rupture (Müller et al., 2006; Weitbrecht et al., 2011). ABA significantly extended the period associated with higher PME activities in the seeds (e.g. in the period between 30 and 72 h; Fig. 1). The patterns mimicked those found in untreated seeds in that the activities declined once endosperm rupture was reached. Thus, we conclude that high PME activity is a feature of phase II of Arabidopsis seed germination both in the presence and in the absence of exogenous ABA and might be linked to the ABA-mediated delay in endosperm rupture and prolonged plateau phase of water uptake.
PMEI5 (At2g31430) Is a Novel PMEI Expressed in Seeds, Buds, and Mature Flowers
We chose a transgenic OE approach to determine whether changes in PME activities, and thus the degree of cell wall pectin methylesterification, would affect seed germination. PMEs and PMEIs are encoded by large gene families, with over 60 members in Arabidopsis. An examination of 20 SALK lines with T-DNA insertions in single PMEI (Supplemental Table S1) or PME (Supplemental Table S2) genes did not uncover any strong germination phenotypes, most likely due to the high functional redundancy within the groups. Therefore, in order to target a larger number of PMEs at the activity level, we generated Arabidopsis lines overexpressing At2g31430, a gene encoding a PMEI that we denoted PMEI5 (the fifth PMEI with a confirmed function in Arabidopsis; Wolf et al., 2012). PMEI5 is predicted to encode a protein of 179 amino acids with a PMEI domain (pfam PF04043) spanning almost the entire length of the protein. Two complementary DNA (cDNA) sequences are available at the National Center for Biotechnology Information (NCBI; AY327264 and AY327265). The latter shows retention of the one intron; translation of the transcripts would lead to a truncated protein due to a premature stop codon.
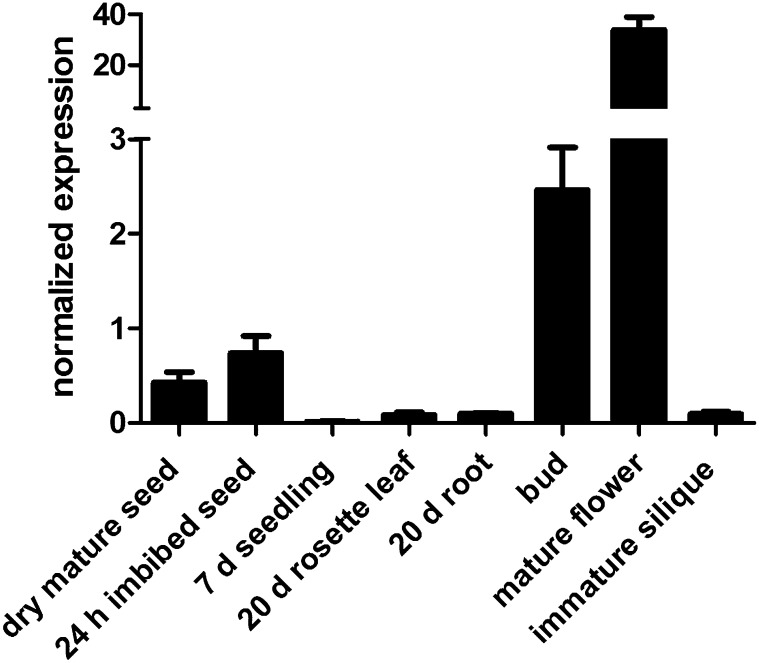
Before embarking on our PMEI5 OE work, we examined expression of the PMEI5 gene during different stages of the Arabidopsis life cycle based on the NCBI sequence AY327264 that yields the full-length protein. Transcripts encoding the full-length PMEI5 were barely detectable in seedlings, rosette leaves, roots of 20-d-old seedlings, and immature siliques (Fig. 2). Mature dry seeds and 24-h imbibed seeds showed moderate PMEI5 expression; the highest expression occurred in flower buds, particularly in mature flowers.
Figure 2.
Transcript abundance of PMEI5 in different organs and at different life cycle stages of Arabidopsis (ecotype Columbia wild type) as determined by qRT-PCR. Transcript levels were normalized to the mean of two reference genes, EF1a and ACT7. Averages of four biological replicates ± se are shown.
OE of PMEI5 Reduces PME Activities of Seeds and Leads to an Increased Degree of Cell Wall Pectin Methylesterification
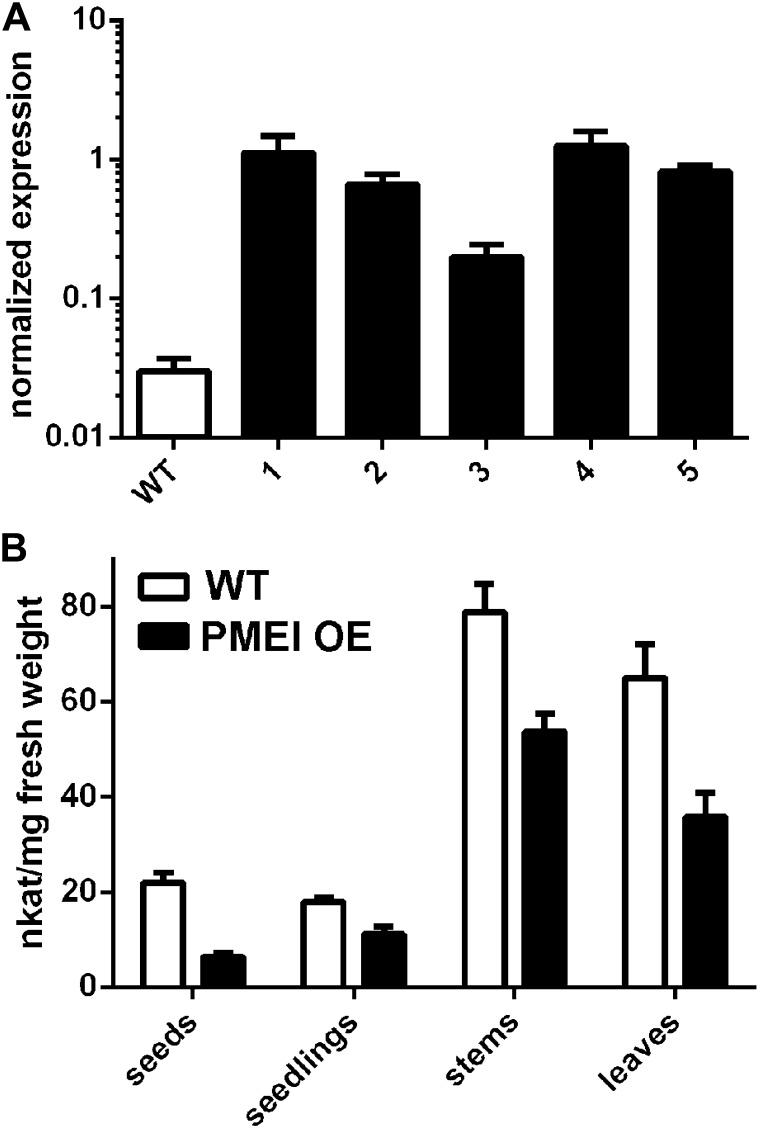
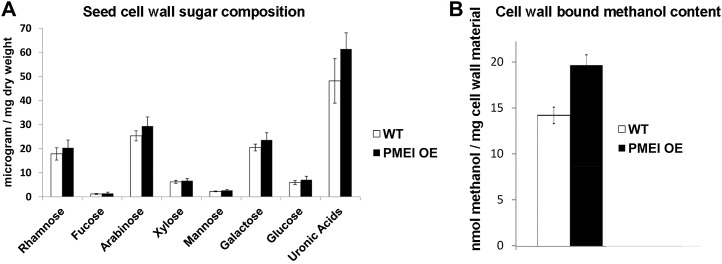
We overexpressed the PMEI5 gene by placing its full-length coding sequence under the control of a cauliflower mosaic virus 35S promoter. Several independent transgenic lines were obtained (see “Materials and Methods”), and five of the lines were examined for their PMEI5 transcript levels and for PME activity. PMEI5 transcripts in the OE lines were 10- to 50-fold higher in leaves (Fig. 3A) than in the wild-type untransformed controls. PME activities were significantly reduced in seeds (P < 0.005) and seedlings (P < 0.02) as well as in vegetative tissues of adult plants (e.g. in leaves and stems; P < 0.02; Fig. 3B). An in situ effect of the PMEI was confirmed by the incubation of protein extracts derived from the wild-type plants with extracts from PMEI OE plants. This resulted in an average reduction in PME activity of 30.3% ± 5.1%. After undergoing extensive methylesterification in the Golgi complex, homogalacturonans are secreted into the cell wall and subsequently demethylesterified by PMEs (Zhang and Staehelin, 1992; Sterling et al., 2006); inhibition of PME activity through OE of the PMEI, therefore, was expected to lead to a marked increase in the degree of pectin methylesterification in both the vegetative tissues of the plants and in seeds. To determine whether the effect on the overexpressor cell walls was specific to the process of pectin demethylesterification, water-soluble and trifluoracetic acid residue fractions of the cell walls of the seeds were examined. Indeed, an analysis of PMEI OE seed cell walls after the nonadherent mucilage was removed showed no statistically significant difference in their neutral sugar and uronic acid composition from that of wild-type seeds (Fig. 4A). However, there was a higher methanol content of the saponified cell wall fraction from the PMEI OE seeds as compared with that in the wild-type seeds (Fig. 4B), which is indicative of a greater predominance of cell wall methylesters due to PMEI5 OE.
Figure 3.
Effect of PMEI5 OE on PMEI5 transcript levels and PME activity. A, Transcript abundance of PMEI5 in the leaves of wild-type plants and in the leaves of five independent PMEI OE lines (1–5) as determined by qRT-PCR. Transcript levels were normalized to the mean of two reference genes, EF1a and ACT7. B, PME activities in different tissues of adult Arabidopsis plants and in seeds and seedlings. The leaves and stems were of plants harvested 24 d after sowing. The seeds were sampled after 16 h of imbibition (i.e. during phase II of germination), and seedlings were 5 d post germination. WT, Wild-type or nontransgenic controls. The data in B are based on average PME activities determined from five wild-type lines and five PMEI5 OE lines that represented independent insertions ± se.
Figure 4.
Cell wall sugars and methylester content. A, Sugar composition of cell walls of wild-type (WT) and PMEI OE seeds (2 h after imbibition). The data are based on averages of five replicates ± se. B, Estimated methylester content as based on methanol associated with saponified cell wall materials. The data are based on averages of three replicates ± se.
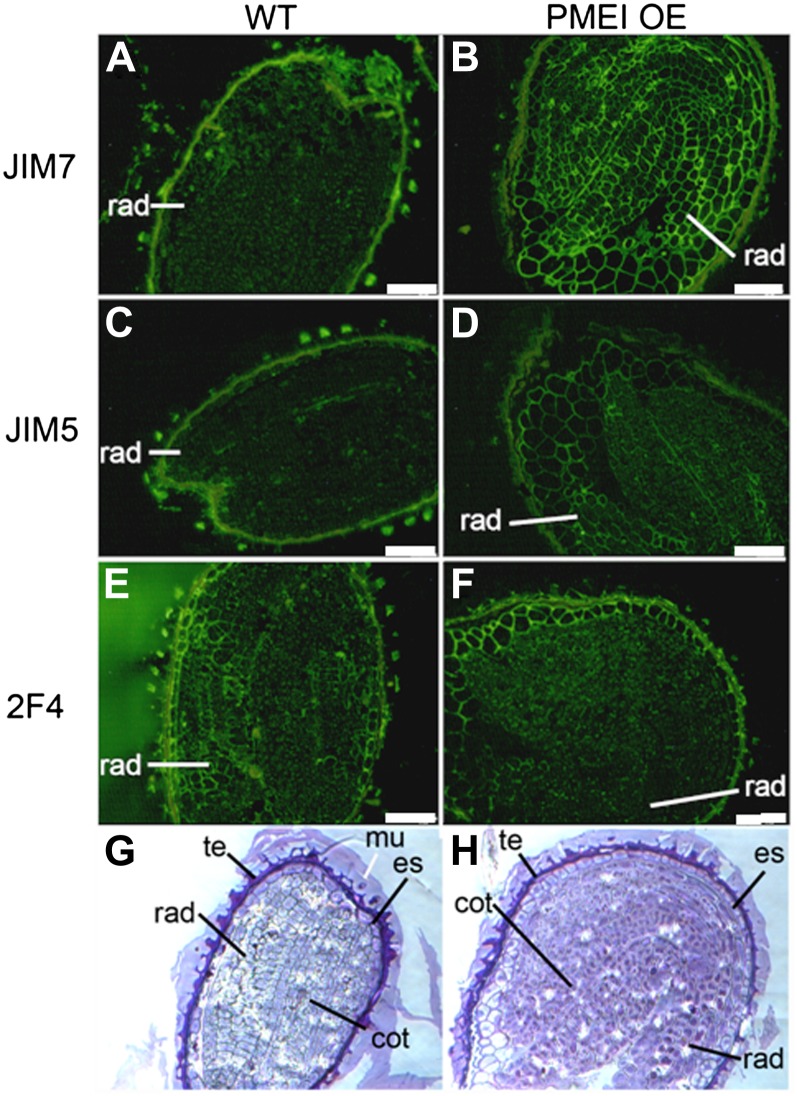
To investigate the degree of methylesterification of cell wall pectins, wild-type and PMEI OE seeds at phase II of germination were analyzed by immunofluorescence, in which they were challenged with different antibodies specific for homogalacturonans of differing degrees and patterns of methylesterification. These included a JIM7 antibody, which binds exclusively to highly methylesterified homogalacturonans with dense stretches of methylesters, and JIM5, which recognizes pectins with lower degrees of methylesterification and more sporadic methylesters. Also used was the 2F4 antibody, which recognizes homogalacturonan stretches that are largely demethylesterified and cross linked by calcium bridges (Willats et al., 2000). As expected from the reduced PME activity of the PMEI OE lines, the most dramatic differences were observed with the JIM7 antibody, whose signal was much greater in the cell walls of PMEI OE seeds as compared with the wild-type seeds (Fig. 5, A and B). This was pronounced in all parts of the embryo and to a slightly lesser extent in the endosperm; no differences were evident in the testa. While the signal was generally quite weak, the presence of JIM5 epitopes was more evident in the PMEI OE embryos (but less abundant in the testa) as compared with that of wild-type seeds (Fig. 5, C and D). The 2F4 antibody bound most tenaciously to the endosperm cells in all samples (Fig. 5, E and F) and revealed some regional differences in the OE versus wild-type seeds. Thus, the inhibition of PME activity by OE of PMEI5 caused a more dense methylesterification of cell wall homogalacturonans in the embryo and endosperm.
Figure 5.
Effects of PMEI5 OE on cell size and the degree of pectin methylesterification of the embryo, endosperm, and testa. A to F, Immunolabeling of representative sections derived from PMEI OE and wild-type (WT) seeds with the JIM7 antibody (A and B), JIM5 antibody (C and D), and 2F4 antibody (E and F). G and H, Staining of sections. All images were taken at the same magnification for the WT and PMEI OE. Note that the cells of PMEI OE seeds are larger. cot, Cotyledons; es, endosperm; mu, mucilage; rad, radical; te, testa. Bars = 65 μm.
PME Inhibition Leads to Faster Seed Germination and a Lowered Sensitivity of Seeds to the Inhibitory Effects of ABA on Germination
One key characteristic of the PMEI OE lines was that their rate of seed germination was significantly faster than that exhibited by wild-type seeds; this was the case for both testa rupture and endosperm rupture (Fig. 6A). Both fresh and after-ripened seeds of the OE lines exhibited faster germination, showing that the effect of the resultant cell wall modifications is not mediated through reduced dormancy. Germination on medium containing 1 µm ABA led to a prolonged phase II and higher PME activities in wild-type seeds throughout phase II (Fig. 1). The OE seeds showed a reduced sensitivity to ABA with respect to a delay of endosperm rupture (Fig. 6B). To more closely examine the delay of endosperm rupture in response to ABA, we compared the times required to reach 50% endosperm rupture of the OE and wild-type seed populations as a function of the concentration of exogenous ABA (0.1–2 µm). We found that with increasing ABA concentrations, the average delay of endosperm rupture within the population is less pronounced for the PMEI OE seeds than for the wild-type seeds (data not shown). Thus, the OE seeds exhibited a greater speed of completion of germination relative to the wild-type seeds as the ABA concentration was increased. While this is indicative of a lowered ABA sensitivity of the OE seeds, we recognize the limitations of these exogenous ABA studies.
Figure 6.
Effects of PMEI5 OE on seed germination characteristics. A, Testa rupture (TR) and endosperm rupture (ER) in seeds of the PMEI OE lines as compared with the wild type (WT). Data represent averages ± se of three replicates of 50 seeds each. B, Effects of different concentrations of ABA on the germination (endosperm rupture) of PMEI OE lines versus the WT. Data represent averages ± se of three replicates of 50 seeds each. C, PME activities of protein extracts derived from PMEI OE lines during germination in the absence (control [CON]; blue bars) and presence of 1 μm ABA (ABA; gray bars). Data are based on averages ± se of four biological replicates of about 100 seeds each. Seeds were weighed before imbibition, and the activity shown is relative to the starting seed weight. The percentages over the bars list the proportion of seeds that had achieved testa rupture and endosperm rupture.
We assayed PME activities in the OE lines to see if there were any changes in PME activities during phase II of the germination process despite the presence of the inhibitor in OE lines. Unlike the PME activities of wild-type seeds (Fig. 1), the PME activities of the OE lines (while reduced relative to the wild-type seeds) were maintained at a low level throughout germination, and exogenous ABA had little effect on the low steady-state levels (Fig. 6C). The faster germination and apparent decline in sensitivity to ABA of the OE seeds was thus associated with both the reduction of PME activities as well as a loss of differential regulation of PME activities during germination.
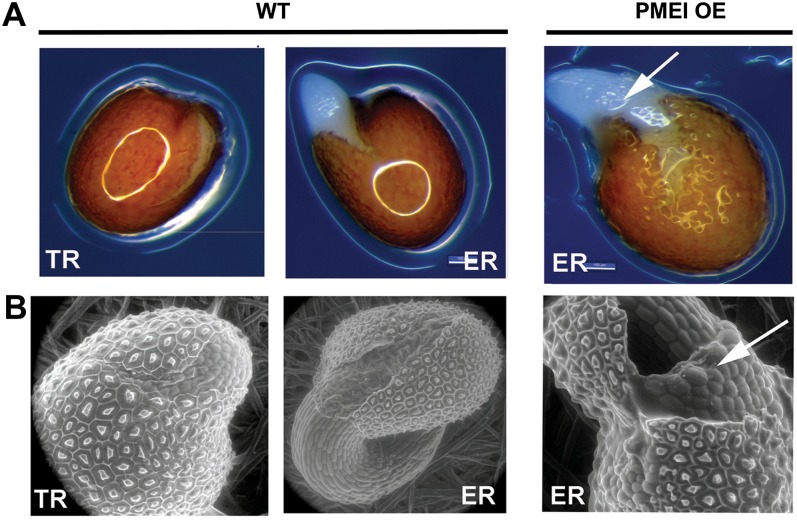
To examine the process of testa rupture and endosperm rupture in more detail, wild-type and PMEI OE seeds were examined by light microscopy (Fig. 7A) and by environmental scanning electron microscopy (eSEM; Fig. 7B). Testa rupture in the PMEI OE seeds occurred in a morphologically similar fashion to that found for the wild-type seeds (data shown only for wild-type seeds; Fig. 7). In both cases, cell separation causes testa rupture, and the outlines of the cells constituting the testa were clearly visible. Thus, any abnormal rupture of the testa does not accompany the faster germination of the PMEI5 OE seeds. The endosperm appeared to stretch more extensively over the radicle in the PMEI OE seeds during phase II; a residual collar-like structure appeared around the radicle after endosperm rupture (Fig. 7). This collar-like structure was composed of endosperm cells and was particularly evident in the eSEM images; the rupture point of the endosperm was unaffected. In order to see if these changes in testa morphology also led to changes in the mucilage that is secreted upon imbibition, imbibed seeds were stained with ruthenium red (Supplemental Fig. S2). There were no obvious differences in the mucilage volume or density when the seeds were stained with solutions containing water or other agents. The other agents included EDTA, which chelates calcium ions of the cell wall and thereby destroys calcium bridges, extending the mucilage volume, and CaCl2, which contracts the mucilage due to the formation of calcium bridges. A comparison of the two sets of seeds (OE and wild type) stained in the presence of these agents showed that they exhibited similar mucilage profiles (Supplemental Fig. S2).
Figure 7.
Testa rupture (TR) and endosperm rupture (ER) of wild-type (WT) and PMEI OE seeds. A, Light microscopy images of WT and PMEI OE seeds. B, eSEM images. Arrows show how the endosperm of PMEI OE seeds is stretched more extensively and retained as a collar-like structure of cells that clings to the radicle after endosperm rupture. Note the clear rupture lines of the testa shown for WT seeds in B.
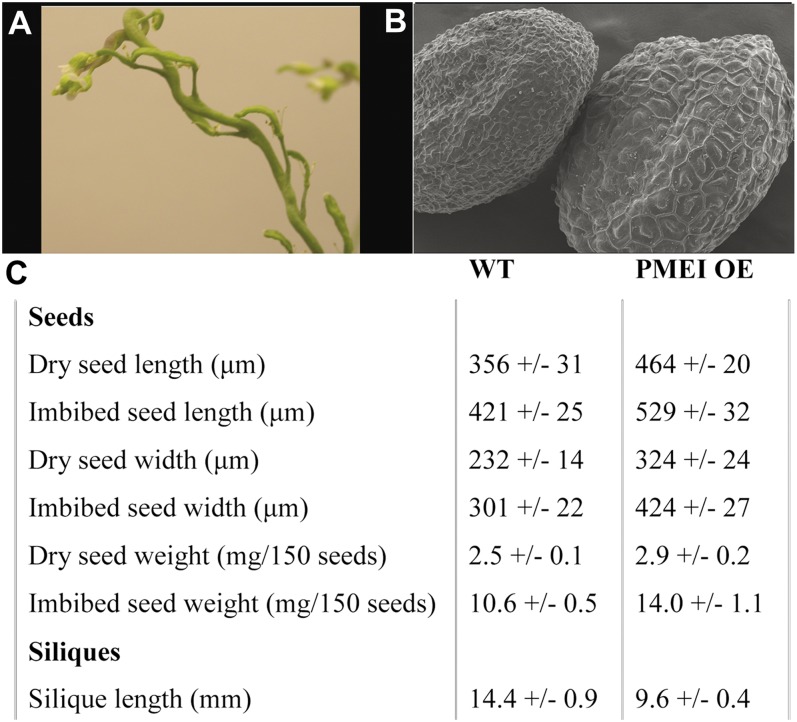
Another feature of the PMEI OE seeds that was evident was the larger cell size; larger cells made up the testa, endosperm, and embryo of the PMEI OE seeds as compared with wild-type seeds (Figs. 5 and 8B). In accordance with the larger cell size but similar cell numbers, both dry and imbibed seeds of the PMEI OE lines were larger and heavier than dry and imbibed wild-type seeds, respectively (Fig. 8C). This change could be due to the altered cell wall properties or to the fact that PMEI OE plants showed a marked reduction of seed production, with often only one or two seeds per silique. This could perhaps be due to greater resource allocation from the parent plant (because of the lower seed number). In any event, the siliques were shortened and wrinkled (Fig. 8A).
Figure 8.
Morphological phenotypes of PMEI OE seeds and siliques. A, Siliques of PMEI OE plants are wrinkled and shorter than those of wild-type plants. Also note the twisted growth of the stem. B, eSEM image of dry seeds of the wild type (WT; left) and PMEI OE (right). Note the larger size of PMEI OE testa cells. C, Quantitative differences between the characteristics of seeds, siliques, and plant growth of wild-type and PMEI OE lines.
In relation to the causes of the lowered seed numbers per silique, reciprocal hand crosses with wild-type plants and even between two PMEI OE parents demonstrated that this was not exclusively due to defective pollen (e.g. with an inability to fertilize the egg cell) or a mechanical problem with the silique shape, as the artificial transfer of the pollen led to a significant increase in seed numbers. Hand crosses between PMEI OE parents led to 30 ± 2 seeds per silique, PMEI OE pollen on wild-type stigma (PMEI OE × wild type) developed 36 ± 2 seeds per silique, and wild type × PMEI OE generated 44 ± 3 seeds per silique. These seed numbers are still below that characteristic of the wild type (56 ± 5 seeds per silique). A closer visual examination of the reproductive organs of the PMEI OE lines showed that filament elongation is likely not sufficiently coordinated with style elongation, as the anthers often seemed too short for the pollen to reach the stigma.
In addition to the effects on seed germination and other seed characteristics, the OE of PMEI5 led to several marked morphological phenotypes of seedlings and adult plants (Supplemental Fig. S3).
DISCUSSION
OE of a Novel PMEI as a System to Study PME Function
Functional redundancy within the large PME family is problematic for determining the effects of the degree of cell wall pectin methylesterification on plant development and morphology by a PME gene knockout strategy. We circumvented this problem by expressing the PME inhibitor PMEI5 ectopically. The inhibitory activity of PMEI5 on PME activity was evident in the transgenic lines, and we were able to show that there were resultant changes (increases) in the degree of pectin methylesterification in the seed tissues of the PMEI OE lines as compared with the wild type. Four PMEIs have been functionally characterized in Arabidopsis to date. PMEI1 and PMEI2 (Wolf et al., 2003) are highly expressed in flowers; PMEI1 expression is specific to pollen, while the expression of PMEI3 is particularly high in the apical meristem (Peaucelle et al., 2008). PMEI4 is up-regulated during growth acceleration in skotomorphic hypocotyls (Pelletier et al., 2010). PMEI5 is not represented on the most commonly used microarray chips; thus, no prior information was available about its expression patterns. Our finding that PMEI5 is most highly expressed in flowers shows that its spatial expression bears similarity to PMEI1 and PMEI2. We confirmed the information from the NCBI that some cDNAs of PMEI5 show intron retention. In wheat, regulation of the PMEI genes themselves is mediated in part by intron retention. Processed and unprocessed transcripts of two PMEI genes (Tdpmei2.1 and Tdpmei2.2) accumulate in several organs of wheat, but anthers contain exclusively mature transcripts. PMEI3 of wheat lacks introns, and its transcript accumulates mainly in stem internodes (Rocchi et al., 2012).
PMEs show differential expression during different stages of the Arabidopsis life cycle (Electronic Fluorescent Pictograph browser [http://www.bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi]; Winter et al., 2007). The phenotypes affected by PMEI5 OE (Supplemental Fig. S3) are likely indicative of this inhibitor interacting with a number of different PMEs, similar to the findings of others in which PMEIs show cross-species and even cross-family interactions (Giovane et al., 2004). Pelletier et al. (2010) overexpressed the PMEI3 gene in Arabidopsis to study the role of pectin methylesterification in dark-induced hypocotyl elongation. Interestingly, they did not observe several of the morphological phenotypes evident in our PMEI5 OE line, including the looping stem growth (Supplemental Fig. S3). This demonstrates that the PMEIs may have selective effects despite interacting with several PMEs.
PME Activity Plays a Role in Arabidopsis Seed Germination
Biochemical changes in the cell wall that cause cell wall loosening in seed micropylar tissues are an important aspect of germination. In order for a seed to complete germination, the growth potential of the radicle must be greater than the resistance of the tissues enveloping it (Bewley, 1997b; Müller et al., 2006). Hydrolase-mediated changes in the mechanical properties of the cell walls, particularly in the radicle itself (to promote its growth potential) and in the seed micropylar tissues surrounding the radicle tip (to promote mechanical weakening), are thus crucial to this process. These extensibility changes obviously must be subject to strict spatial and temporal control. Expansins and xyloglucan endotransglucosylases are expressed in the endosperm cap of germinating tomato seeds (Chen et al., 2002), and differential regulation of the expression and activity of the hydrolases β-1,3-glucanase and β-1,4-mannanase in the endosperm accompanies seed germination in tobacco (Leubner-Metzger et al., 1995) and tomato (Nonogaki et al., 2000), respectively. OE of a β-1,3-glucanase gene in tobacco alters dormancy but does not affect the germination process per se (Leubner-Metzger, 2002).
In this study, we identified a role for PME activity in seed germination and found a significant effect of PME inhibition on increasing the rate of germination; dormancy was unaffected. PME activities increased in wild-type seeds over the first 24 to 30 h of germination, a period associated with several metabolic changes (e.g. the translation of newly transcribed mRNAs, mitochondrial repair and biogenesis, and the mobilization of soluble sugars to prepare the seed for the completion of germination) and testa rupture (Weitbrecht et al., 2011). The activities declined thereafter during the period associated with the completion of germination (endosperm rupture and radicle protrusion).
As noted in the introduction, higher PME activity (and thus cell wall pectin demethylesterification) influences cell wall biomechanical properties. The proposed mechanisms of PME action in these processes can be summarized as follows: (1) creating an acidic environment within the cell wall as a result of deesterification of pectins, thus promoting cell wall extension or growth, perhaps through the modulation of expansins (see above); (2) facilitating the hydrolysis of polygalacturonic chains by polygalacturonases (Wakabayashi et al., 2003); and (3) promoting the formation of calcium cross linkages (through demethylation of pectins) that ultimately change the state of the pectin matrix by generating free carboxyl groups that are able to bind Ca2+ (Micheli, 2001; Peaucelle et al., 2012).
In yellow cedar (Callitropsis nootkatensis) seeds, the micropylar megagametophyte decreases in mechanical strength following a dormancy-breaking treatment, and during germination, the cells of the megagametophyte in the area immediately surrounding the radicle exhibit a loss of their internal structure, which would represent significant weakening to allow radicle emergence (Ren and Kermode, 1999). Concurrently, the embryo exhibits increased turgor and a reduced sensitivity to low osmotic potentials; both the weakening and strengthening processes are positively associated with PME activities (Ren and Kermode, 2000).
Thus, decreasing the extent of cell wall pectin demethylesterification (e.g. through PME inhibition) could lead to either enhanced or reduced elongation growth, implying an increased or decreased wall extensibility, respectively. For example, reduced elongation is the case for the growth of hypocotyls and pollen tubes (Bosch and Hepler, 2005; Derbyshire et al., 2007; Pelletier et al., 2010). As far as germination is concerned, in this study, the overexpressing PMEI lines exhibited a higher degree of cell wall pectin methylesterification, and the seeds completed germination faster. We would need to conduct studies on the resultant changes in the mechanical strength of the different seed tissues (especially those of the micropylar endosperm and radicle) to determine the precise effects of the PME inhibition of cell wall extensibility of these tissues. These experiments are very difficult to perform in Arabidopsis seed tissues. We speculate that PME inhibition led to faster endosperm rupture and completion of germination by improving the capacity of the radicle to break through this tissue; this improved growth potential of the embryo could be related to an increased rate of water uptake. Accordingly, the reduced PME activities associated with seeds of the overexpressing lines could have made their cell walls more permeable to water and more extensible, possibly through altering the ability of cell wall homogalacturonans to form Ca2+ bridges, a process requiring their demethylesterification. Indirect evidence in support of this comes from a study by Rautengarten et al. (2008), in which increased PME activity in a subtilase-deficient mutant, atsbt1.7, was associated with a decrease in germination capability under water-limiting conditions induced by polyethylene glycol. It is possible that the mechanical resistance of the micropylar endosperm was also altered; our morphological studies (Fig. 7) indicate that this is the case. Changes in mechanical resistance could be caused directly by the increased pectin methylesterification changing the biophysical properties of the cell wall. It is also possible that the faster radicle emergence of the OE seeds is due to the larger size of cells of the PMEI OE seeds; thus, cell size-related changes in cell wall mechanical properties may be operative. In any case, the endosperm cells may be weakened in relation to the opposing force of the growing radicle. A stronger turgor-driven expansion of the radicle cells is also possible.
Wolf et al. (2012) recently demonstrated a link between altered cell wall pectin methylesterification and brassinosteroid signaling. They uncovered an up-regulation of brassinosteroid signaling pathways as a result of the inhibition of pectin methylesterase activity via genetic and pharmacological means, including PMEI5 OE. The authors propose that this up-regulation of the brassinosteroid pathway is likely the result of a feedback (“compensatory”) mechanism that the plant uses to restore cell wall homeostasis. In this way, the potential for catastrophic changes in cell wall integrity can be avoided. It is possible that the faster germination of PMEI OE seeds in our study is due to this up-regulation of brassinosteroid signaling, as these pathways can act to promote Arabidopsis seed germination (Steber and McCourt, 2001).
Microarray data certainly support the contention that changes in the pectin matrix occur during germination; transcripts encoding a large number of pectin-modifying enzymes change during germination phases I and II, and the regulation of these genes appears to be highly complex (Nakabayashi et al., 2005). PME activity increases in a tissue-specific manner during seed dormancy release and germination of yellow cedar seeds (Ren and Kermode, 2000), where pectin is the predominant polysaccharide in the cell walls of the megagametophyte (Ren and Kermode, 1999). In the seeds of this highly dormant conifer species, the addition of ABA to intact seeds after dormancy breakage suppresses the activity of two PME isoforms (Ren and Kermode, 2000).
Increased Cell Wall Pectin Methylesterification Leads to an Apparent Reduced Sensitivity of Seed Germination to ABA
In seeds of several species, ABA specifically delays endosperm rupture without having a major effect on the timing of testa rupture (Müller et al., 2006). In Arabidopsis wild-type seeds, the delay in the onset of endosperm rupture caused by exogenous ABA was associated with a prolonged period of higher PME activities. It is still unclear how the increase in the duration of phase II is brought about by the presence of ABA. In wild-type seeds, PME activities peaked in phase II of seed germination; ABA prolongs the period of high PME activities, such that they do not decline until just around the onset of endosperm rupture. As with the control seeds, in the ABA-treated seeds, the PME activities stay declined once 75% of the seeds had reached endosperm rupture. Thus, changes in the cell wall pectin characteristics might play a role in the mechanism by which ABA delays endosperm rupture. In cress seeds, ABA leads to a delay in endosperm weakening (Müller et al., 2006), which is part of the process that allows the radicle to emerge through this tissue. In accordance with the hypothesis that PME activity contributes to ABA inhibition of endosperm rupture, ABA delayed the endosperm rupture of PMEI OE seeds to a lesser degree than that of wild-type seeds. More information is needed to support the contention that ABA specifically delays endosperm rupture via changes in cell wall pectin characteristics. While we did not address any role of endogenous ABA, this factor has been linked to cell wall composition changes. For example, leaves of the ABA-deficient sitiens mutant of tomato have lower levels of GalUA in their cell walls and are less susceptible to degradation by pectinases than those of the wild-type leaves (Curvers et al., 2010). The authors conclude that the GalUA residues within the cell walls of the sitiens leaf cells, while lower in amount, nonetheless have a higher degree of pectin methylesterification, which in turn renders them more resistant to pectinolysis. While this effect is on cell wall composition and therefore differs from our study, it is interesting that endogenous ABA can influence cell wall properties.
CONCLUSION
The properties of the pectin matrix are dynamically changed during the plant life cycle as the plant reacts to its environment and undergoes developmental programs that require cell wall extensibility changes. Our findings support the view that pectins and their degree of methylesterification, as influenced by PME activities, play a role in seed germination. We speculate that PME activity contributes to the temporal regulation of radicle emergence in endospermic seeds by altering the mechanical properties of the cell walls and, thereby, the balance between the two opposing forces of radicle elongation and the mechanical resistance of the endosperm.
MATERIALS AND METHODS
PME Activity Assays
Soluble protein extracts were generated from whole seeds, stems, leaves, and seedlings of Arabidopsis (Arabidopsis thaliana). The tissue was ground in liquid nitrogen and added to twice the fresh weight (w/v) of extraction buffer (100 mm Tris-HCl, pH 7.5, 500 mm NaCl containing protease inhibitor cocktail composed of 100 mm phenylmethylsulfonyl fluoride, 2 mm bestatin, 0.3 mm pepstatin A, and 0.3 mm E-64 [ABMGood; www.abmgood.com]). Extracts were then rotated at 4°C for 30 min and centrifuged at 11,500g at 4°C for 20 min. Fresh supernatants were used immediately for all enzyme assays. A coupled enzymatic assay was performed as described by Grsic-Rausch and Rausch (2004) using a spectrophotometric plate reader (SpectraMax M2e; Molecular Devices; www.moleculardevices.com). A Student’s t test of the data sets was run in Graphpad Prism 6.0 (www.graphpad.com).
RNA Extraction and cDNA Synthesis
Arabidopsis parts were ground in liquid nitrogen, and total RNA was extracted as described by Chang et al. (1993) with the following modifications: after the addition of buffer (2% hexadecyl trimethyl-ammonium bromide, 2% polyvinylpyrrolidone [molecular weight = 40,000], 100 mm Tris-HCl, pH 8.0, 25 mm EDTA, pH 8.0, 2 m NaCl, and 2% β-mercaptoethanol), the extracts were kept at 65°C for 10 min. All chloroform-isoamylalcohol extractions were repeated once. RNA was treated with DNase-I (Fermentas; www.thermoscientificbio.com/fermentas/) to remove remaining genomic DNA, and the quantity and purity of the RNA were determined with a Nanodrop spectrophotometer (ND-2000C; Thermo Scientific; www.thermoscientific.com). One microgram of RNA was reverse transcribed using the EasyScript Plus kit (ABMGood; www.abmgood.com) with a mixture of random hexamers and oligo(dT) primers. cDNA from four biological replicate RNA samples was used for quantitative reverse transcription (qRT)-PCR.
qRT-PCR
qRT-PCR was run in 15-μL reactions on an ABI7900HT machine (Applied Biosystems; www.appliedbiosystems.com) using PerfeCTa Sybr Green Supermix with ROX (Quanta Biosciences; www.quantabio.com). Primers were designed with the primer3 (Rozen and Skaletsky, 2000) tool in Geneious 4.8.5 and based on sequence AY327264 from the NCBI (forward primer 5′-TTGCGATAACGCAGTCAAAAATG-3′ and reverse primer 5′-GAAGTCCAAGTTCCCAAGCTG-3′). Actin7 (ACT7) and Elongation Factor1α (EF1α) were used as reference genes. The reaction mixture consisted of 150 ng of cDNA (RNA equivalent), 7.5 μL of Supermix, and 140 nm of each primer. The mix was subjected to a temperature regime of 3 min at 95°C and 40 cycles of 15 s at 95°C and 1 min at 60°C. A dissociation curve was run after each quantitative PCR to validate that only one product had been amplified in each well. The efficiency (E) of the primer pairs was calculated as the average of the efficiencies of the individual reactions by using raw fluorescence data with the publicly available PCR Miner tool (http://www.miner.ewindup.info/version2; Zhao and Fernald, 2005). The efficiency was then used to calculate transcript abundance for the individual samples as (1 + E) ^ (−CT) as described by Graeber et al. (2011). No-template controls were included for each primer pair to check for contamination of the reaction solutions, and no-reverse transcription controls were used to check for genomic DNA contamination in the RNA.
Generation of Transgenic Arabidopsis Lines
The open reading frame of At2g31430 was amplified from genomic Arabidopsis DNA using 31430_fw primer (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTGatggccacaatgctaataaaccac-3′) and 31430_rv primer (5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCttaggtcacaagcttgttgaaaataag-3′), including Gateway (Invitrogen; www.invitrogen.com) attB1 and attB2 sites, respectively. The PCR product was then inserted into the pDONR207 donor plasmid, sequenced, and recombined with the binary destination vector pB2GW7 (Karimi et al., 2002). Arabidopsis plants were transformed by the floral dip method (Clough and Bent, 1998). The transgenic progeny were selected by spraying BASTA (120 mg L−1) on seedlings grown in soil. Experiments were conducted on the T1 and T2 generations, as later generations were characterized by gene silencing that generally led to loss of phenotypes over about four to five generations. A total of five selections were performed on T1 seedlings at different times, yielding 10 to 25 independent transgenic lines per selection, all of which displayed the described phenotypes.
Germination Testing
Arabidopsis plants (wild-type ecotype Columbia and lines overexpressing the PMEI5 gene [At2g31430]) were grown in soil in pots maintained in a growth chamber at 22°C with a 16-h photoperiod (long days). Mature dry seeds were either used immediately for germination testing (“fresh seeds”) or were monitored for their germination after first undergoing after-ripening at room temperature for 3 months. For germination tests, seeds were surface sterilized with 70% ethanol, washed with 100% ethanol, and air dried. Three sets of 50 seeds were sown on one-half-strength Murashige and Skoog medium (pH 6.5) solidified with 1% agar. Germination was scored with a Zeiss dissection microscope. Seeds in which the endosperm was visible through a crack in the testa were considered to have reached testa rupture; seeds in which the radicle tip had emerged through the endosperm were considered to have reached endosperm rupture. The effects of ABA on germination were also determined. For these studies, (±)-ABA (Sigma; www.sigmaaldrich.com) at various concentrations as indicated was added to the germination medium.
Immunofluorescence Studies and Histological Examination
Seeds at 35 h after imbibition were prepared for high-pressure freezing, freeze substitution, resin embedding, and sectioning as described by Rensing et al. (2002). Two percent (w/v) osmium tetroxide and 8% (v/v) dimethoxypropane in acetone were used for general histological examination of sections, and 0.25% (v/v) glutaraldehyde and 2% (w/v) uranyl acetate in 8% (v/v) dimethoxypropane in acetone were used for immunolabeling. The samples were embedded in Spurr’s resin (Spurr, 1969), and sections (0.5 μm) were stained with toluidine blue for general examination. For immunolabeling, the samples were embedded in LR White (London Resin; www.2spi.com), and sections were incubated in a blocking solution (1% bovine serum albumin [Sigma] in 20 mm Tris, 500 mm NaCl, and 0.2% Tween 20). After rinsing in buffer (20 mm Tris, 500 mm NaCl, and 0.2% Tween 20), the sections were incubated in the primary antibodies JIM7, JIM5, and 2F4 (Knox et al., 1990; 1:10 dilutions; Plant Probes; www.plantprobes.net). Following incubation in the secondary antibody (Alexa Fluor 488 goat anti-rat IgG [Invitrogen A11006], used at a dilution of 1:100 for JIM5 and JIM7; Alexa Fluor 488 goat anti-mouse IgG [Invitrogen A11001], used at a dilution of 1:100 for 2F4; www.invitrogen.com), the sections were examined via epifluorescence using a DMR light microscope (Leica Microsystems; www.leica.com).
Quantification of Methylesters in Seed Cell Walls
Seeds (100–120) of PMEI OE and wild-type plants were imbibed for 2 h at 22°C in petri dishes containing filter paper and water. The seeds were then ground in liquid nitrogen, and 200 μL of methanol was added. The ground seed materials were extracted four times with a 1:1 (v/v) methanol:chloroform mixture, washed once with acetone, and dried overnight at room temperature. The weight of the dried cell wall materials was determined, and 0.5 to 1.0 mg was washed with 2 mL of water. To release the methylesters, the cell wall materials were incubated for 1 h at room temperature with 100 µL of 0.5 m NaOH. After neutralization with 50 µL of 1 m HCl, the samples were centrifuged at 2,000g for 10 min. Fifty microliters of the supernatant was used to quantify the methanol released during saponification (Klavons and Bennett, 1986). In brief, the released methanol was oxidized for 15 min at room temperature on a shaker using 0.03 units of alcohol oxidase (Sigma-Aldrich; www.sigmaaldrich.com) in 50 µL of 20 mm phosphate buffer, pH 7.5. The resultant extract was then developed for 15 min at 60°C in 20 mm acetyl acetone, 50 mm acetic acid, and 2 m ammonium acetate. Absorbance was measured at 412 nm using a Spectramax M2e microplate reader (Molecular Devices; www.moleculardevices.com) and compared with a standard curve generated with a methanol dilution series. A Student’s t test of the data sets was run in Graphpad Prism 6.0 (www.graphpad.com).
eSEM
All images were taken with a Quanta 250 FEG environmental scanning microscope (FEI; www.fei.com). An accelerating voltage of 10 kV was used for all images; the stage was cooled to approximately 3°C for all samples, and a disk of wetted filter paper was used to keep the samples hydrated. The relative humidity was maintained at 80% to 100% during imaging.
Cell Wall Composition Analysis
Water-soluble mucilage was extracted from 6 to 8 mg of seeds by shaking them in 1 mL of distilled water for 2 h at 37°C. The remaining pellets were ground in a ball mill (Retsch; www.retsch.de) to a fine powder, and seed cell wall material was prepared by washing with 1 mL of 70% (v/v) ethanol, three times with 1 mL of chloroform:methanol (1:1), and twice with acetone before drying. Starch was removed from the pellet by incubation with α-amylase and amyloglucosidase overnight before hydrolysis to monosaccharides. Hydrolysis was performed by incubation with 2 m trifluoracetic acid for 1 h at 121°C. The content of uronic acids was determined by the m-hydroxybiphenyl colorimetric uronic acid assay (Blumenkrantz and Asboe-Hansen, 1973). The content of cell wall monosaccharides was determined by gas chromatography-mass spectrometry analysis of the respective alditol acetates, as described previously (Foster et al., 2010).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Principal component analyses of FTIR spectra recorded from embryos of Arabidopsis wild-type seeds at different times of germination.
Supplemental Figure S2. Ruthenium red staining of seeds to examine the mucilage associated with wild-type seeds and with PMEI OE seeds.
Supplemental Figure S3. Effects of PMEI5 overexpression on morphology at various stages of the Arabidopsis life cycle.
Supplemental Table S1. SALK lines representing single PMEI gene knockouts that show no germination- or visible growth- phenotypes.
Supplemental Table S2. SALK lines representing single PME gene knockouts that show no germination- or visible growth- phenotypes.
Acknowledgments
We thank Dr. Kazumi Nakabayashi (Max Planck Institute for Plant Breeding Research) for providing expression data and Dr. Ralf Thomann (Freiburg Material Research Centre) for help with the eSEM. We are also grateful to Dr. Karen Tanino (University of Saskatoon) and Dr. Ferenc Borondics (Canadian Lightsource Synchrotron) for their help with the Fourier transform infrared studies.
Glossary
- PME
pectin methylesterase
- ABA
abscisic acid
- PMEI
PME inhibitor
- NCBI
National Center for Biotechnology Information
- OE
overexpression
- eSEM
environmental scanning electron microscopy
- qRT
quantitative reverse transcription
References
- Balestrieri C, Castaldo D, Giovane A, Quagliuolo L, Servillo L. (1990) A glycoprotein inhibitor of pectin methylesterase in kiwi fruit (Actinidia chinensis). Eur J Biochem 193: 183–187 [DOI] [PubMed] [Google Scholar]
- Bewley JD. (1997a) Seed germination and dormancy. Plant Cell 9: 1055–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley JD. (1997b) Breaking down the walls: a role for endo-β-mannanase in release from seed dormancy? Trends Plant Sci 2: 464–469 [Google Scholar]
- Blumenkrantz N, Asboe-Hansen G. (1973) New method for quantitative determination of uronic acids. Anal Biochem 54: 484–489 [DOI] [PubMed] [Google Scholar]
- Bosch M, Hepler PK. (2005) Pectin methylesterases and pectin dynamics in pollen tubes. Plant Cell 17: 3219–3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Puryear J, Cairney J. (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep 11: 113–116 [Google Scholar]
- Chen F, Bradford KJ. (2000) Expression of an expansin is associated with endosperm weakening during tomato seed germination. Plant Physiol 124: 1265–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Nonogaki H, Bradford KJ. (2002) A gibberellin-regulated xyloglucan endotransglycosylase gene is expressed in the endosperm cap during tomato seed germination. J Exp Bot 53: 215–223 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. (2005) Growth of the plant cell wall. Nat Rev Mol Cell Biol 6: 850–861 [DOI] [PubMed] [Google Scholar]
- Curvers K, Seifi H, Mouille G, de Rycke R, Asselbergh B, Van Hecke A, Vanderschaeghe D, Höfte H, Callewaert N, Van Breusegem F, et al. (2010) Abscisic acid deficiency causes changes in cuticle permeability and pectin composition that influence tomato resistance to Botrytis cinerea. Plant Physiol 154: 847–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbyshire P, McCann MC, Roberts K. (2007) Restricted cell elongation in Arabidopsis hypocotyls is associated with a reduced average pectin esterification level. BMC Plant Biol 7: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downie B, Hilhorst HWM, Bewley JD. (1997) Endo-β-mannanase activity during dormancy alleviation and germination of white spruce (Picea glauca) seeds. Physiol Plant 101: 405–415 [Google Scholar]
- Foster CE, Martin TM, Pauly M. (2010) Comprehensive compositional analysis of plant cell walls (lignocellulosic biomass). Part II. Carbohydrates. J Vis Exp 37: e1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry SC. (2000) The Growing Plant Cell Wall: Chemical and Metabolic Analysis. Blackburn Press, Caldwell, NJ [Google Scholar]
- Giovane A, Servillo L, Balestrieri C, Raiola A, D’Avino R, Tamburrini M, Ciardiello MA, Camardella L. (2004) Pectin methylesterase inhibitor. Biochim Biophys Acta 1696: 245–252 [DOI] [PubMed] [Google Scholar]
- Graeber K, Linkies A, Wood ATA, Leubner-Metzger G. (2011) A guideline to family-wide comparative state-of-the-art quantitative RT-PCR analysis exemplified with a Brassicaceae cross-species seed germination case study. Plant Cell 23: 2045–2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grsic-Rausch S, Rausch T. (2004) A coupled spectrophotometric enzyme assay for the determination of pectin methylesterase activity and its inhibition by proteinaceous inhibitors. Anal Biochem 333: 14–18 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. (2002) Gateway vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Klavons JA, Bennett RD. (1986) Determination of methanol using alcohol oxidase and its application to methyl ester content of pectins. J Agric Food Chem 34: 597–599 [Google Scholar]
- Knox JP. (2008) Revealing the structural and functional diversity of plant cell walls. Curr Opin Plant Biol 11: 308–313 [DOI] [PubMed] [Google Scholar]
- Knox JP, Linstead PJ, King J, Cooper C, Roberts K. (1990) Pectin esterification is spatially regulated both within cell walls and between developing tissues of root apices. Planta 181: 512–521 [DOI] [PubMed] [Google Scholar]
- Leubner-Metzger G. (2002) Seed after-ripening and over-expression of class I β-1,3-glucanase confer maternal effects on tobacco testa rupture and dormancy release. Planta 215: 959–968 [DOI] [PubMed] [Google Scholar]
- Leubner-Metzger G, Fründt C, Vögeli-Lange R, Meins F., Jr (1995) Class I β-1,3-glucanases in the endosperm of tobacco during germination. Plant Physiol 109: 751–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P-P, Koizuka N, Homrichhausen TM, Hewitt JR, Martin RC, Nonogaki H. (2005) Large-scale screening of Arabidopsis enhancer-trap lines for seed germination-associated genes. Plant J 41: 936–944 [DOI] [PubMed] [Google Scholar]
- Manz B, Müller K, Kucera B, Volke F, Leubner-Metzger G. (2005) Water uptake and distribution in germinating tobacco seeds investigated in vivo by nuclear magnetic resonance imaging. Plant Physiol 138: 1538–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQueen-Mason S, Cosgrove DJ. (1994) Disruption of hydrogen bonding between plant cell wall polymers by proteins that induce wall extension. Proc Natl Acad Sci USA 91: 6574–6578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micheli F. (2001) Pectin methylesterases: cell wall enzymes with important roles in plant physiology. Trends Plant Sci 6: 414–419 [DOI] [PubMed] [Google Scholar]
- Moustacas AM, Nari J, Borel M, Noat G, Ricard J. (1991) Pectin methylesterase, metal ions and plant cell-wall extension: the role of metal ions in plant cell-wall extension. Biochem J 279: 351–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller K, Tintelnot S, Leubner-Metzger G. (2006) Endosperm-limited Brassicaceae seed germination: abscisic acid inhibits embryo-induced endosperm weakening of Lepidium sativum (cress) and endosperm rupture of cress and Arabidopsis thaliana. Plant Cell Physiol 47: 864–877 [DOI] [PubMed] [Google Scholar]
- Nakabayashi K, Okamoto M, Koshiba T, Kamiya Y, Nambara E. (2005) Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: epigenetic and genetic regulation of transcription in seed. Plant J 41: 697–709 [DOI] [PubMed] [Google Scholar]
- Nonogaki H, Gee OH, Bradford KJ. (2000) A germination-specific endo-β-mannanase gene is expressed in the micropylar endosperm cap of tomato seeds. Plant Physiol 123: 1235–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peaucelle A, Braybrook SA, Höfte H. (2012) Cell wall mechanics and growth control in plants: the role of pectins revisited. Front Plant Sci 3: 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peaucelle A, Louvet R, Johansen JN, Höfte H, Laufs P, Pelloux J, Mouille G. (2008) Arabidopsis phyllotaxis is controlled by the methyl-esterification status of cell-wall pectins. Curr Biol 18: 1943–1948 [DOI] [PubMed] [Google Scholar]
- Pelletier S, Van Orden J, Wolf S, Vissenberg K, Delacourt J, Ndong YA, Pelloux J, Bischoff V, Urbain A, Mouille G, et al. (2010) A role for pectin de-methylesterification in a developmentally regulated growth acceleration in dark-grown Arabidopsis hypocotyls. New Phytol 188: 726–739 [DOI] [PubMed] [Google Scholar]
- Raiola A, Camardella L, Giovane A, Mattei B, De Lorenzo G, Cervone F, Bellincampi D. (2004) Two Arabidopsis thaliana genes encode functional pectin methylesterase inhibitors. FEBS Lett 557: 199–203 [DOI] [PubMed] [Google Scholar]
- Rautengarten C, Usadel B, Neumetzler L, Hartmann J, Büssis D, Altmann T. (2008) A subtilisin-like serine protease essential for mucilage release from Arabidopsis seed coats. Plant J 54: 466–480 [DOI] [PubMed] [Google Scholar]
- Ren C, Kermode AR. (1999) Analyses to determine the role of the megagametophyte and other seed tissues in dormancy maintenance of yellow cedar (Chamaecyparis nootkatensis) seeds: morphological, cellular and physiological changes following moist chilling and during germination. J Exp Bot 50: 1403–1419 [Google Scholar]
- Ren C, Kermode AR. (2000) An increase in pectin methyl esterase activity accompanies dormancy breakage and germination of yellow cedar seeds. Plant Physiol 124: 231–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing KH, Samuels AL, Savidge RA. (2002) Ultrastructure of vascular cambial cell cytokinesis in pine seedlings preserved by cryofixation and substitution. Protoplasma 220: 39–49 [DOI] [PubMed] [Google Scholar]
- Rocchi V, Janni M, Bellincampi D, Giardina T, D’Ovidio R. (2012) Intron retention regulates the expression of pectin methyl esterase inhibitor (Pmei) genes during wheat growth and development. Plant Biol (Stuttg) 14: 365–373 [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ. (2000) Primer3 on the WWW for general users and for biologist programmers. In S Krawetz, S Misener, eds, Bioinformatics Methods and Protocols: Methods in Molecular Biology. Humana Press, Totowa, NJ, pp 365–386 [Google Scholar]
- Sanchez RA, de Miguel L. (1997) Phytochrome promotion of mannan-degrading enzymes in the micropylar endosperm of Datura ferox L. seeds and its relationship with germination. J Exp Bot 37: 1574–1580 [Google Scholar]
- Schopfer P. (2006) Biomechanics of plant growth. Am J Bot 93: 1415–1425 [DOI] [PubMed] [Google Scholar]
- Spurr AR. (1969) A low-viscosity epoxy resin embedding medium for electron microscopy. J Ultrastruct Res 26: 31–43 [DOI] [PubMed] [Google Scholar]
- Steber CM, McCourt P. (2001) A role for brassinosteroids in germination in Arabidopsis. Plant Physiol 125: 763–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterling JD, Atmodjo MA, Inwood SE, Kumar Kolli VS, Quigley HF, Hahn MG, Mohnen D. (2006) Functional identification of an Arabidopsis pectin biosynthetic homogalacturonan galacturonosyltransferase. Proc Natl Acad Sci USA 103: 5236–5241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi K, Hoson T, Huber DJ. (2003) Methyl de-esterification as a major factor regulating the extent of pectin depolymerization during fruit ripening: a comparison of the action of avocado (Persea americana) and tomato (Lycopersicon esculentum) polygalacturonases. J Plant Physiol 160: 667–673 [DOI] [PubMed] [Google Scholar]
- Weitbrecht K, Müller K, Leubner-Metzger G. (2011) First off the mark: early seed germination. J Exp Bot 62: 3289–3309 [DOI] [PubMed] [Google Scholar]
- Willats WG, Limberg G, Buchholt HC, van Alebeek GJ, Benen J, Christensen TM, Visser J, Voragen AA, Mikkelsen JD, Knox JP. (2000) Analysis of pectic epitopes recognised by hybridoma and phage display monoclonal antibodies using defined oligosaccharides, polysaccharides, and enzymatic degradation. Carbohydr Res 327: 309–320 [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf S, Grsic-Rausch S, Rausch T, Greiner S. (2003) Identification of pollen-expressed pectin methylesterase inhibitors in Arabidopsis. FEBS Lett 555: 551–555 [DOI] [PubMed] [Google Scholar]
- Wolf S, Mouille G, Pelloux J. (2009) Homogalacturonan methyl-esterification and plant development. Mol Plant 2: 851–860 [DOI] [PubMed] [Google Scholar]
- Wolf S, Mravec J, Greiner S, Mouille G, Höfte H. (2012) Plant cell wall homeostasis is mediated by brassinosteroid feedback signaling. Curr Biol 22: 1732–1737 [DOI] [PubMed] [Google Scholar]
- Wu Y, Spollen WG, Sharp RE, Hetherington PR, Fry SC. (1994) Root growth maintenance at low water potentials: increased activity of xyloglucan endotransglycosylase and its possible regulation by abscisic acid. Plant Physiol 106: 607–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang GF, Staehelin LA. (1992) Functional compartmentation of the Golgi apparatus of plant cells: immunocytochemical analysis of high-pressure frozen- and freeze-substituted sycamore maple suspension culture cells. Plant Physiol 99: 1070–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Fernald RD. (2005) Comprehensive algorithm for quantitative real-time polymerase chain reaction. J Comput Biol 12: 1047–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]