Abstract
The expression of genes that control iron (Fe) uptake and distribution (i.e. Fe utilization-related genes) is tightly regulated. Fe deficiency strongly induces Fe utilization-related gene expression; however, little is known about the mechanisms that regulate this response in plants. Transcriptome analysis of an Arabidopsis (Arabidopsis thaliana) mutant defective in RNA polymerase II C-terminal domain-phosphatase-like1 (CPL1) revealed significant up-regulation of Fe utilization-related genes (e.g. IRON-REGULATED TRANSPORTER1), suggesting the importance of RNA metabolism in Fe signaling. An analysis using multiple cpl1 alleles established that cpl1 mutations enhanced specific transcriptional responses to low Fe availability. Changes in protein level were less prominent than those in transcript level, indicating that cpl1-2 mainly affects the Fe deficiency response at the transcriptional level. However, Fe content was significantly increased in the roots and decreased in the shoots of cpl1-2 plants, indicating that the cpl1 mutations do indeed affect Fe homeostasis. Furthermore, root growth of cpl1-2 showed improved tolerance to Fe deficiency and cadmium (Cd) toxicity. cpl1-2 plants accumulated more Cd in the shoots, suggesting that Cd toxicity in the roots of this mutant is averted by the transport of excess Cd to the shoots. Genetic data indicate that cpl1-2 likely activates Fe deficiency responses upstream of both FE-DEFICIENCY-INDUCED TRANSCRIPTION FACTOR-dependent and -independent signaling pathways. Interestingly, various osmotic stress/abscisic acid (ABA)-inducible genes were up-regulated in cpl1-2, and the expression of some ABA-inducible genes was controlled by Fe availability. We propose that the cpl1 mutations enhance Fe deficiency signaling and promote cross talk with a branch of the osmotic stress/ABA signaling pathway.
Iron (Fe) is an essential metal element for nearly all organisms. In plants, Fe is present as a cofactor in many metalloproteins and is found in the active sites of photosynthetic and respiratory Fe-sulfur clusters. Fe is also required for RNA and hormone biosynthesis, nitrogen fixation, sulfate assimilation, and chlorophyll biosynthesis (Broadley et al., 2012). On the other hand, Fe is highly reactive, and excess Fe can produce reactive oxygen species via Fenton reactions (Møller et al., 2007). In soil, the predominant form of Fe is Fe(III), which is abundantly present as insoluble ferric oxides and ferric hydroxides in aerobic environments (Guerinot and Yi, 1994; Palmer and Guerinot, 2009). The solubility of Fe(III) in aqueous solution is 1 order of magnitude less than the Fe concentration needed for survival; therefore, a large portion of agricultural land is Fe deficient (Guerinot and Yi, 1994). Fe deficiency in plants induces intercostal/interveinal leaf chlorosis due to limited chlorophyll biosynthesis and results in significant yield loss of crops.
Plants have developed two distinct mechanisms, strategy I (reduction strategy) and strategy II (chelation strategy), to mobilize insoluble Fe(III) in the rhizosphere and transport it through the plasma membrane (Romheld, 1987; Welch, 1995; Schmidt, 1999; Gross et al., 2003; Grotz and Guerinot, 2006; Puig et al., 2007; Conte and Walker, 2011; Schmidt and Buckhout, 2011; Ivanov et al., 2012; White, 2012). Arabidopsis (Arabidopsis thaliana) and other dicots rely on strategy I. In this strategy, the rhizosphere is first acidified by a plasma membrane-localized Arabidopsis H+-ATPase2 (AHA2; Santi and Schmidt, 2009). Then, FERRIC CHELATE REDUCTASE2 (FRO2) reduces Fe(III) to soluble Fe(II) (Yi and Guerinot, 1996; Robinson et al., 1999). Finally, the reduced Fe is taken up by a high-affinity transporter, IRON-REGULATED TRANSPORTER1 (IRT1; Eide et al., 1996; Henriques et al., 2002; Varotto et al., 2002; Vert et al., 2002), which is strongly expressed in root epidermal cells and is localized to the plasma membrane (Eide et al., 1996; Vert et al., 2002). Once the Fe(II) is absorbed, various transporters and chemicals mobilize it. Fe(II) can be chelated by nicotianamine and transported intercellularly by YELLOW STRIPE-LIKE transporters (DiDonato et al., 2004; Waters et al., 2006; Waters and Grusak, 2008; Chu et al., 2010). In the vasculature, citrate, which is exuded by a FERRIC REDUCTASE DEFECTIVE3 (FRD3) transporter (Durrett et al., 2007), forms a tri-Fe(III), tricitrate complex for long-distance transport (Rogers and Guerinot, 2002; Green and Rogers, 2004; Rellán-Alvarez et al., 2010). In addition, OLIGOPEPTIDE TRANSPORTER3 (OPT3) may also function as a transporter of Fe chelates (Wintz et al., 2003; Stacey et al., 2008). In contrast, excess Fe is sequestered in plastids by Fe-binding proteins named ferritins (Waldo et al., 1995). Here, we collectively refer to genes that are involved in Fe uptake, mobilization, and signaling as “Fe utilization-related genes,” according to Kobayashi et al. (2009).
The expression levels of genes encoding components of the Fe acquisition mechanism, such as FRO2 and IRT1, are under transcriptional regulation in Fe-limited conditions, as their mRNA levels increase in response to Fe deficiency (Eide et al., 1996; Robinson et al., 1999; Connolly et al., 2002; Vert et al., 2002). Central to this regulation are basic helix-loop-helix (bHLH) transcription factors, FER in tomato (Solanum lycopersicum; Ling et al., 2002) and FER-LIKE IRON DEFICIENCY-INDUCED TRANSCRIPTION FACTOR (FIT) in Arabidopsis (Colangelo and Guerinot, 2004; Jakoby et al., 2004; Yuan et al., 2005; Bauer et al., 2007). In Arabidopsis, FIT regulates various Fe utilization-related genes, including IRT1, FRO2, and NICOTINAMINE SYNTHASE1 (Colangelo and Guerinot, 2004; Jakoby et al., 2004). FIT dimerizes with group Ib bHLH family transcription factors, bHLH38 or bHLH39, and directly activates the transcription of IRT1 and FRO2 (Yuan et al., 2008). While the induction of FIT expression during the Fe deficiency response is moderate (Bauer et al., 2004; Colangelo and Guerinot, 2004; Jakoby et al., 2004), bHLH38, bHLH39, bHLH100, and bHLH101 are strongly up-regulated (Wang et al., 2007). It has been proposed that dimerization of FIT with different bHLHs determines the target specificity of Fe deficiency-induced transcriptional activation (Yuan et al., 2008). To date, little is known about the upstream mechanisms that sense cellular Fe levels and regulate the early signals that lead to the expression of FIT and group Ib bHLH genes. In addition to the central pathway, a recent study reported the existence of a cell-specific regulatory mechanism mediated by the bHLH transcription factors POPEYE and IAA-LEU RESISTANT3 and the E3 ligase BRUTUS that operates in the root pericycle Fe response and controls root development (Long et al., 2010). Repression of FERRITIN1 (FER1) genes in the absence of excess Fe is mediated by the cis-element iron-dependent regulatory sequence (Petit et al., 2001). These regulators control specific branches of Fe deficiency signaling; however, the mechanisms that sense Fe availability and fine-tune the signal throughput of individual pathways have yet to be determined.
A wide range of physiological and molecular signals influence Fe signaling in Arabidopsis. Ethylene regulates the expression of IRT1, FRO2, and FIT, perhaps via the activity of EIN3 and EIL1, which stabilize FIT (Lingam et al., 2011). In contrast, cytokinin and abscisic acid (ABA) repress the expression of IRT1, FRO2, and FIT (Séguéla et al., 2008). Environmental stresses, such as salt/osmotic stress, attenuate Fe deficiency responses (Séguéla et al., 2008), whereas phosphate starvation triggers the Fe deficiency responses (Hirsch et al., 2006; Thibaud et al., 2010). Moreover, volatiles emitted by soil microbes can up-regulate the expression of Fe utilization-related genes (Zhang et al., 2009). The molecular mechanisms that facilitate multiple inputs into Fe signaling have not been determined; however, the plasticity of Fe signaling implies the presence of diverse regulatory components that are coordinated to achieve proper levels of cellular Fe.
Among the various genes that affect the expression of Fe utilization-related genes, TRIDENT (TDT) is unique as it encodes a subunit of an RNA-decapping enzyme that is involved in RNA metabolism (Goeres et al., 2007). The RNA metabolic pathway is a major mechanism in the cotranscriptional/posttranscriptional regulation of diverse developmental and environmental responses (Kuhn and Schroeder, 2003; Gregory et al., 2008; Nakaminami et al., 2012). While ABA signaling is a prototypical target of RNA metabolism-mediated regulation, many transcripts are regulated by small RNAs, such as microRNAs and small interfering RNAs (Ramachandran and Chen, 2008). The tdt mutation has been shown to repress the expression of Fe utilization genes; however, whether this is due to severe growth defects in the mutant or to specific Fe signaling defects has not been elucidated (Goeres et al., 2007).
Here, we identify an isoform of RNA metabolism regulators, CPL1 (for RNA polymerase II C-terminal domain [pol II CTD] phosphatase-like1), as a novel regulator of the Fe deficiency responses. CPL family proteins are known to dephosphorylate pol II CTD (Koiwa et al., 2004), which is the regulatory domain of polymerase II. Of the more than 20 CPL isoforms present in the Arabidopsis genome, CPL1 is a negative regulator of stress-responsive gene expression under various osmotic stresses (Koiwa et al., 2002; Xiong et al., 2002). In this report, we determined that CPL1 also functions in the plant Fe deficiency response. Microarray analysis of the cpl1 transcriptome found that both the osmotic stress/ABA response and the Fe deficiency response are activated in cpl1. cpl1 mutant plants exhibited various hallmarks of the Fe deficiency response, such as altered metal profiles, and increased tolerance to Fe deficiency and cadmium (Cd) toxicity. These results suggest that CPL1 is a previously uncharacterized regulator of the Fe deficiency response. In addition, our data suggest that a subset of ABA/osmotic stress-induced genes are coregulated by Fe deficiency signals and are targets of CPL1 regulation.
RESULTS
The Identification of Genes Uniquely Regulated in cpl1
Arabidopsis CPL1 was previously shown to regulate gene expression under osmotic stress conditions through ABA signaling (Koiwa et al., 2002; Xiong et al., 2002). To obtain insight into the regulatory networks modulated by CPL1, the global transcript profile of cpl1-2 was determined using an Affymetrix ATH1 Gene Chip. To obtain unbiased gene expression profiles, copy RNA probes were prepared from total RNA extracted from unstressed young (10-d-old) seedlings of the wild-type C24 or cpl1-2 lines grown on medium containing one-quarter-strength Murashige and Skoog (MS) salts, 0.5% Suc, and 0.8% agar. After probe-level data normalization using the Robust Multiarray Analysis algorithm (Supplemental Fig. S1A), hybridization intensity data for 19,597 out of 22,810 probes, which were above the 20th percentile in at least one out of six hybridizations, were selected for further analysis. Next, consistency between the C24 and cpl1-2 replicate data sets was confirmed by principal component analysis Triplicate data sets of each genotype clustered together in a three-dimensional scatterplot (Supplemental Fig. S1), whereas there was a clear separation of the C24 and cpl1-2 data sets. This is indicative of data consistency among replicate data sets for each genotype as well as of genotype differentiation. Differentially expressed genes were then determined by evaluating fold change (2-fold or greater was considered as the cutoff) and one-way ANOVA (P < 0.05 was considered significant), followed by the Tukey’s honestly significant difference (HSD) test and Benjamini-Hochberg multiple corrections. A total of 114 and 132 genes were significantly up- and down-regulated by more than 2-fold, respectively, in the cpl1-2 mutant data sets relative to those of the control (Supplemental Data Set S1). To confirm the microarray data, the expression of 41 up-regulated and 22 down-regulated genes was determined by reverse transcription quantitative PCR (RT-qPCR) analysis. The up-regulation of 36 genes and down-regulation of 15 genes was confirmed (i.e. 81% of the genes tested by RT-qPCR), indicating that the microarray results were generally reliable (Supplemental Data Set S1). Genes that could not be confirmed as being differentially expressed were excluded in subsequent analyses. We refer to the resulting 109 genes that were up-regulated in cpl1-2 as CUTs (for cpl1-Up Transcripts).
CUTs Encode ABA/Osmotic Stress- and Fe Deficiency-Responsive Genes
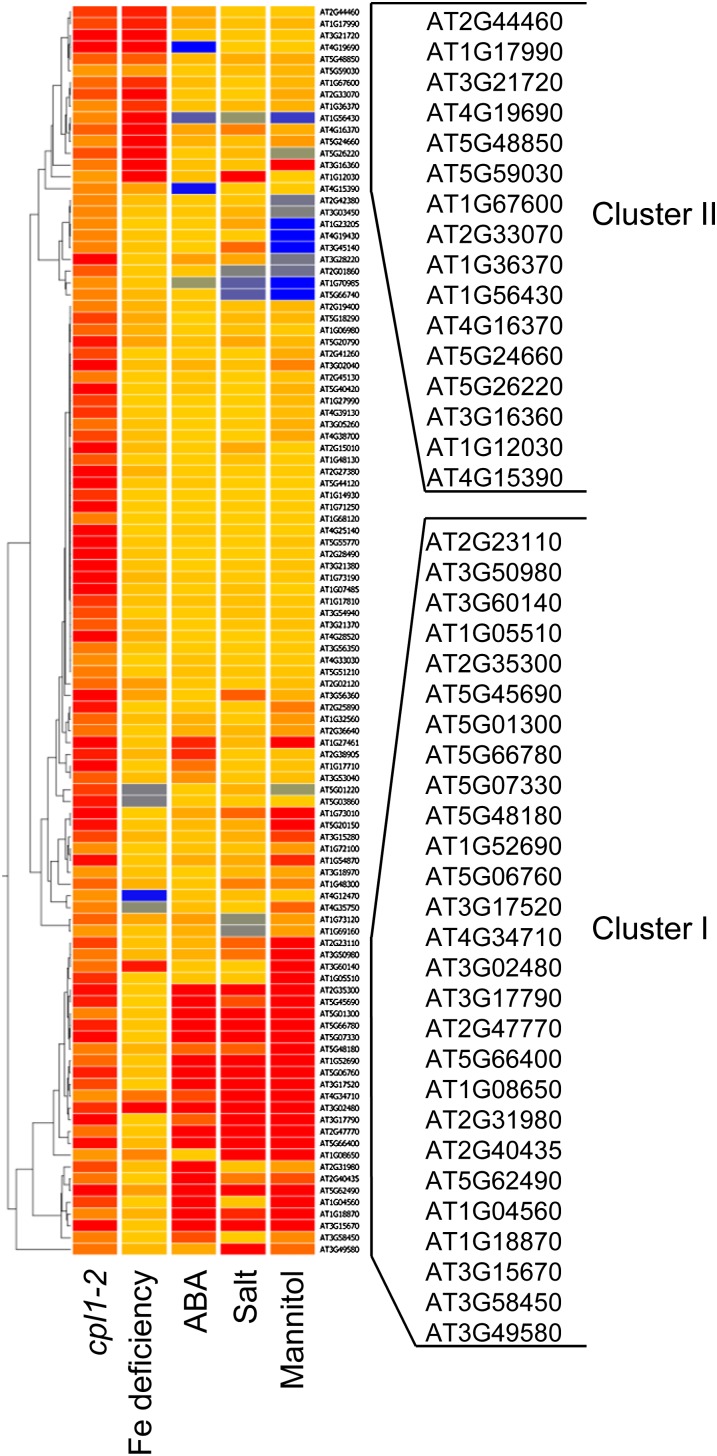
To determine the signaling pathway in which CPL1 is involved, we screened gene expression data in the AtGenExpress and Gene Expression Omnibus databases using the list of CUTs. The CUT expression profiles of four abiotic stress/ABA treatments showed overlap with that of cpl1-2 and were subjected to hierarchical clustering using Pearson correlation and the average linkage rule. Clustering results showed that no single treatment profile displayed overall similarity with the cpl1-2 profile; however, two clusters of genes were similarly up-regulated in cpl1-2 and in plants exposed to abiotic stresses and ABA (Fig. 1). Genes in cluster I (27 genes) were up-regulated by hyperosmotic stress and ABA. Cluster II contains 16 genes that were strongly induced under Fe deficiency stress, including At4g19690 (IRT1), a major Fe transporter. The differential regulation of clusters I and II by abiotic stresses was confirmed by Gene Set Enrichment Analysis (GSEA) using GeneTrail (Backes et al., 2007; Schuler et al., 2011). As shown in Supplemental Table S1, the genes in cluster I were significantly enriched by osmotic stress and ABA (P < 0.05) but not by Fe deficiency. Conversely, those in cluster II were enriched only by Fe deficiency (P < 0.05).
Figure 1.
Hierarchical clustering of CUTs based on their differential expression during abiotic stress. The expression levels of 109 CUTs in cpl1-2 plants and in wild-type plants subjected to Fe deficiency (Fe-free nutrient solution for 24 h; Gene Expression Omnibus accession GSE15189), ABA (1 µm ABA for 3 h; NASCArray accession 176), salt (130 mm NaCl for 6 h; NASCArray accession 139), or mannitol (300 mm mannitol for 12 h; NASCArray accession 140) were subjected to hierarchical clustering using the Pearson correlation distance and average linkage rule.
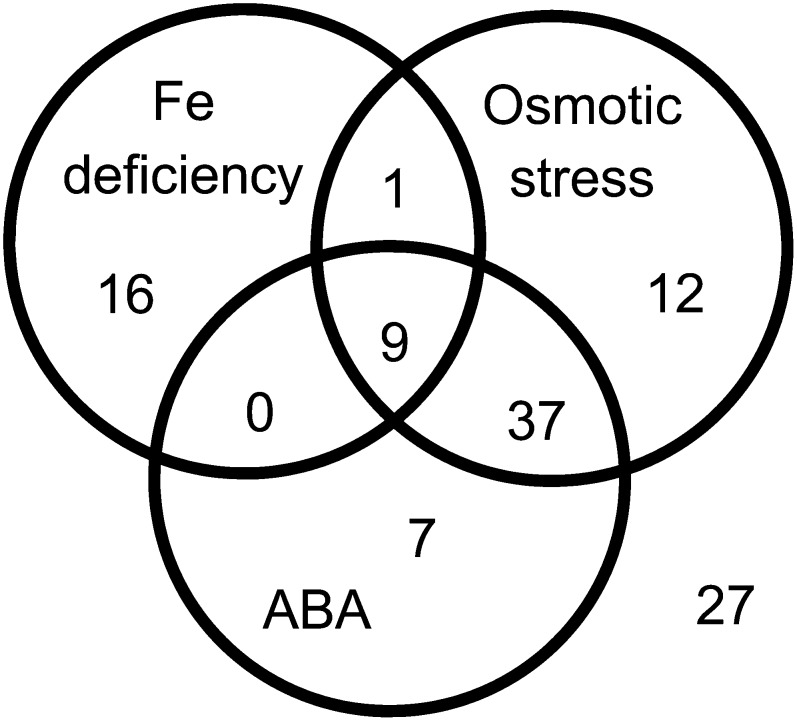
To obtain deeper insight into the regulation of CUTs by abiotic stresses, we examined the expression levels of CUTs in public microarray data sets designed to analyze responses to Fe deficiency (Colangelo and Guerinot, 2004; Dinneny et al., 2008; Buckhout et al., 2009; Long et al., 2010; Yang et al., 2010; Schuler et al., 2011; Ivanov et al., 2012), ABA treatment (Seki et al., 2002a; Nishimura et al., 2007; Goda et al., 2008; Mizoguchi et al., 2010), and osmotic stress treatment (mannitol or drought; Kreps et al., 2002; Seki et al., 2002b; Li et al., 2006; Kilian et al., 2007). Affymetrix CEL files for each experiment were processed as described above, and transcripts significantly up-regulated by 2-fold or more (P < 0.05) were identified for each microarray experiment. Genes up-regulated in both cpl1-2 and wild-type plants subjected to these abiotic stress treatments were identified and grouped (Fig. 2; Supplemental Table S2). In addition, the Fe-regulated genes described by Ivanov et al. (2012) were examined for overlaps. We identified 26 and 59 CUTs that were up-regulated in the Fe deficiency and osmotic stress data sets, respectively (P < 0.05 by GSEA; Supplemental Table S3). Of these, 10 CUTs were regulated both in Fe and osmotic stress data sets. Some of the osmotically regulated CUTs, including nine CUTs regulated by Fe deficiency, were also up-regulated in ABA data sets, implying that ABA signaling plays a role in the dual regulation of some CUTs by osmotic stress and Fe deficiency. The remaining 27 CUTs were not up-regulated in either the Fe deficiency or osmotic stress experiment.
Figure 2.
Venn diagram representing the differential regulation of CUTs by abiotic stresses. The expression levels of CUTs were examined in public microarray data sets for Fe deficiency, ABA, and osmotic stress (mannitol or drought) treatments (see text and Supplemental Table S3).
The IRT1 transcript showed the highest level of up-regulation (54.6-fold) in cpl1-2 microarray data sets, even though the plants used in the analysis were grown under Fe-sufficient conditions. Other CUTs in Fe-stress data sets include Fe utilization-related genes, such as OPT3, NICOTIANAMINE SYNTHASE4, and ISOCITRATE LYASE. To confirm that the observed gene expression changes in cpl1-2 were due to the mutation in the CPL1 locus and not to a potential second-site mutation, the expression levels of select CUTs in both cpl1-1 and cpl1-2 alleles were determined by RT-qPCR. Of 26 CUTs tested, we confirmed the significant up-regulation of 19 Fe- and/or ABA/osmotic stress-regulated CUTs in both cpl1-1 and cpl1-2 plants. In addition, the expression of five more CUTs was significantly up-regulated in at least one cpl1 allele (Table I).
Table I. Select CUTs confirmed as being up-regulated in cpl1-1 and cpl1-2.
RT-qPCR values are ±se. Asterisks (P < 0.05) indicate Student’s t test between mean values of cpl1 and C24. N/D, Not determined.
| Arabidopsis Genome Initiative No. | Classa | Gene |
cpl1-2/C24 |
cpl1-1/C24 |
|
|---|---|---|---|---|---|
| Microarray | RT-qPCR | RT-qPCR | |||
| AT4G19690 | F | Iron-responsive transporter1 (IRT1) | 54.57 | 57.68 ± 0.88* | 37.79 ± 0.56* |
| AT3G21720 | F | Isocitrate lyase, putative (ICL) | 8.99 | 5.28 ± 0.79* | 5.98 ± 0.34* |
| AT2G44460 | F | Glycosyl hydrolase family1 protein (BGLU28) | 3.92 | 7.73 ± 0.47* | 7.36 ± 0.34* |
| AT5G26220 | F | ChaC-like family protein | 3.46 | 2.64 ± 0.17* | 2.71 ± 0.06* |
| AT4G16370 | F | Oligopeptide transporter family protein (OPT3) | 3.18 | 3.25 ± 0.23* | 4.08 ± 0.10* |
| AT3G16360 | F | HPT phosphotransmitter4 (AHP4) | 2.60 | 2.64 ± 0.32* | 2.81 ± 0.14* |
| AT1G56430 | F | Nicotianamine synthase4 (NAS4) | 2.35 | 1.87 ± 0.08* | 1.79 ± 0.26 |
| AT5G24660 | F | Response to low sulfur2 (LSU2) | 2.22 | 1.87 ± 0.43* | 2.44 ± 0.40* |
| AT1G08650 | F | Phosphoenolpyruvate carboxylase kinase1 (PPCK1) | 2.03 | 3.43 ± 0.37* | N/D |
| AT1G17990 | F/O | 12-Oxophytodienoate reductase, putative (OPR) | 3.64 | 3.81 ± 0.21* | 6.87 ± 0.40* |
| AT5G62490 | F/A/O | ABA-responsive protein (HVA22b) | 6.91 | 15.67 ± 0.81* | 6.32 ± 0.50* |
| AT3G15670 | F/A/O | Late embryogenesis abundant protein (LEA) family protein | 6.32 | 8.89 ± 0.11* | 4.29 ± 0.08* |
| AT2G35300 | F/A/O | Late embryogenesis abundant protein4-2 (LEA4-2/LEA18) | 4.82 | 5.28 ± 0.52* | 3.29 ± 0.32* |
| AT5G06760 | F/A/O | Late embryogenesis abundant protein4-5 (LEA4-5) | 4.44 | 4.20 ± 0.38* | 4.00 ± 0.04* |
| AT3G02480 | F/A/O | ABA-responsive protein-related/LEA family protein (ABAR) | 4.11 | 3.43 ± 0.40* | 2.08 ± 0.07* |
| AT1G67600 | F/A/O | Acid phosphatase/vanadium-dependent haloperoxidase-related protein | 2.99 | 2.62 ± 0.32* | 1.89 ± 0.02 |
| AT3G60140 | F/A/O | β-Glucosidase30, dark inducible2, senescence-related gene2 (BGLU30/DIN2/SRG2) | 2.81 | 2.35 ± 0.28* | N/D |
| AT4G34710 | F/A/O | Arg decarboxylase2 (SPE2/ADC2) | 2.19 | 3.73 ± 0.27* | 3.46 ± 0.10* |
| AT3G17790 | A/O | Purple acid phosphatase17 (PAP17) | 5.38 | 7.89 ± 0.04* | 7.16 ± 0.24* |
| AT5G07330 | A/O | Unknown protein | 4.93 | 4.26 ± 0.13* | 2.68 ± 0.04* |
| AT5G45690 | A/O | Unknown protein | 4.49 | 5.74 ± 0.50* | 4.63 ± 0.23* |
| AT5G66780 | A/O | Unknown protein | 4.41 | 3.46 ± 0.41* | 2.06 ± 0.04* |
| AT1G05510 | A/O | Unknown protein | 4.11 | 8.40 ± 0.56* | 5.90 ± 0.37* |
| AT3G58450 | A/O | Universal stress protein (USP) family protein/adenine nucleotide α-hydrolase-like superfamily protein | 2.55 | 6.73 ± 0.62* | 5.86 ± 0.17* |
CPL1 Is Involved in Fe Stress Responses
Elevated basal expression levels of Fe utilization-related genes in cpl1-2 were unexpected, since cpl1-2 was isolated based on altered osmotic stress responses, and cpl1-2 does not exhibit the typical Fe deficiency symptoms (e.g. leaf chlorosis) seen in the irt1 and fit mutants (Connolly et al., 2002; Vert et al., 2002; Colangelo and Guerinot, 2004; Jakoby et al., 2004). To establish the causal link between cpl1 mutations and enhanced Fe deficiency responses, we tested the basal expression levels of Fe utilization-related genes (IRT1, FRO2, FIT, and group Ib bHLHs) in several independent cpl1 mutants. In addition to cpl1-1 and cpl1-2 in the C24 background, two transfer DNA (T-DNA) insertion lines, cpl1-5 and cpl1-6, in the ecotype Columbia (Col-0) background, were tested. In cpl1-5 and cpl1-6, T-DNAs were inserted at loci 11512171 and 11514092 of chromosome 4 (Supplemental Fig. S2, A and B), respectively, and the production of intact CPL1 mRNA was abolished (Supplemental Fig. S2C). The basal expression of Fe utilization-related genes was elevated in all alleles, indicating that the cpl1 mutations caused the gene expression phenotypes observed in these plants (Supplemental Table S4). The phenotype was determined to be recessive, because F1 plants produced by crossing cpl1-2 and the wild type showed a wild-type level of gene expression (Supplemental Fig. S3). To compare the involvement of different CPL paralogs in this phenotype, we tested the gene expression levels in cpl2-2 and cpl3-1. The expression levels of IRT1, FRO2, and FIT (hereafter referred to as [IRT1, FIT, FRO2]) in cpl2-2 and cpl3-1 were similar to those of the wild type (Supplemental Table S4), indicating that CPL1 is uniquely associated with regulating Fe deficiency signaling.
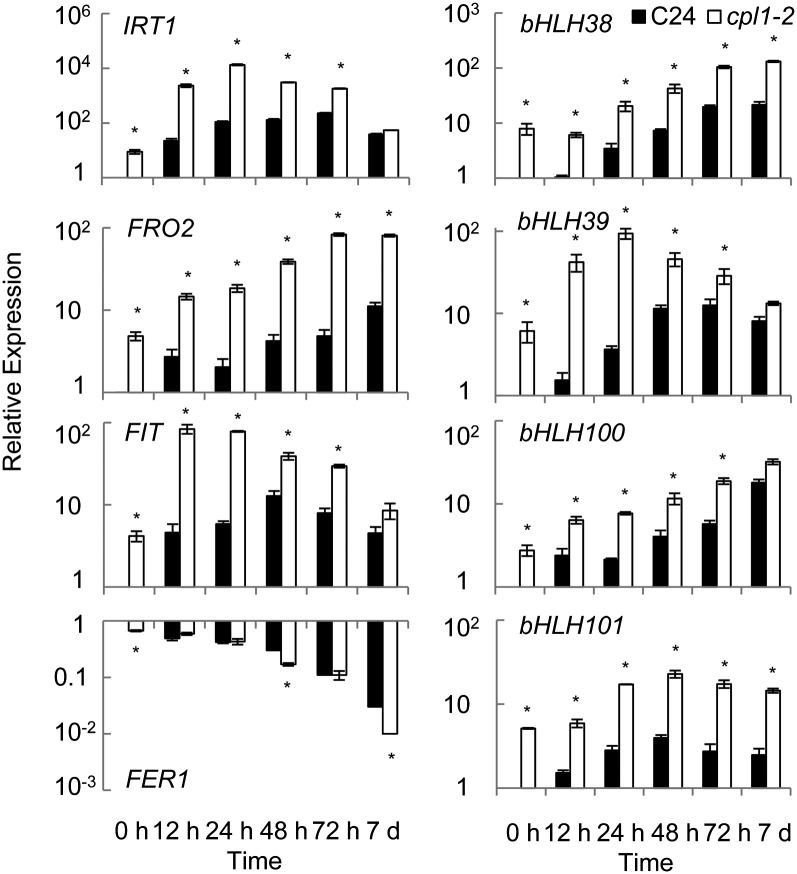
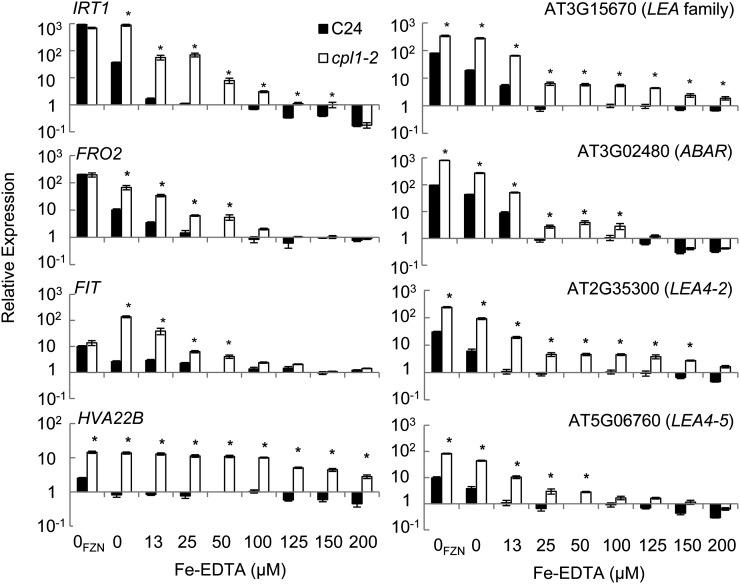
The above results established that the basal expression of Fe utilization-related genes was elevated in cpl1 mutants. Next, we systematically analyzed the Fe deficiency responses of cpl1-2. For this purpose, we established Fe-sufficient conditions using basal growth medium containing one-quarter-strength MS salts (modified to 50 µm Fe-EDTA) and 0.5% Suc (Fig. 3). Fe deficiency stress was applied by transferring plants to the same medium lacking Fe-EDTA but containing 300 µm ferrozine, which chelates Fe in the medium and makes it unavailable to the plant (Colangelo and Guerinot, 2004). An increase in Fe-EDTA from 25 to 50 µm moderately decreased the basal expression level of Fe utilization-related genes in cpl1-2. Upon transfer to Fe-deficient medium, cpl1-2 showed more rapid and greater expression of [IRT1, FIT, FRO2] and bHLH39 than did the wild type as early as 12 h after transfer. The expression of bHLH38, bHLH100, and bHLH101 was greater in cpl1-2 than in the wild type, but the response kinetics of these genes in the mutant were similar to those of the wild type. FER1 was repressed in both cpl1-2 and the wild type, albeit cpl1-2 showed lower basal expression levels. Similar genotype differences were observed when 1× MS medium was used as the basal medium (Supplemental Fig. S4). Further RT-qPCR analyses confirmed the enhanced basal and/or induced expression levels of many but not all Fe-regulated/Fe utilization-related genes in cpl1-2 (Supplemental Fig. S5). Overall, the rapid and strong induction of [IRT1, FIT, FRO2] suggests that the transcriptional response of cpl1-2 is more sensitive to Fe deficiency stress. To test this hypothesis, we determined if elevated external Fe could suppress the cpl1-2 phenotype. Basal [IRT1, FIT, FRO2] expression levels were determined in plants growing on medium containing various concentrations of Fe (Fig. 4). cpl1-2 maintained higher levels of [IRT1, FIT, FRO2], with the greatest difference between genotypes being observed at 25 µm; however, higher Fe concentrations repressed the overall expression of [IRT1, FIT, FRO2] in both the wild type and in cpl1-2. On medium containing more than 100 µm Fe, the wild type and cpl1-2 showed similar, low-level expression of [IRT1, FIT, FRO2]. Together, these results support the hypothesis that the Fe deficiency response in cpl1-2 is more sensitive than that in the wild type but is not constitutive.
Figure 3.
Time course of expression levels of Fe utilization-related genes in the roots of cpl1-2 and C24 under Fe deficiency. Plants were grown on basal medium for 7 d and then transferred to Fe-deficient basal medium containing 300 µm ferrozine (see “Materials and Methods”). Root samples were collected at the time of transfer (0) or at 12, 24, 48, or 72 h or 7 d after the transfer. The presented expression levels (relative to untreated C24 samples) are mean values of three biological replicates analyzed in duplicate. Error bars indicate se of biological replicates. *P < 0.05 by Student’s t test between mean values of cpl1-2 and C24 for the same conditions.
Figure 4.
Basal expression levels of IRT1, FIT, FRO2, and LEA family proteins under different Fe concentrations. Plants were grown for 10 d on medium containing one-quarter-strength MS salts adjusted to the indicated concentrations of Fe-EDTA, 0.5% Suc, and 1.5% agar. 0FZN indicates medium without Fe-EDTA but containing 300 µm ferrozine. Total RNA was extracted from whole plants. The presented expression levels (relative to C24 samples collected from medium containing 50 µm Fe-EDTA) are mean values of three biological replicates analyzed in duplicate. Error bars indicate the se of biological replicates. *P < 0.05 by Student’s t test between mean values of cpl1-2 and C24 for the same conditions.
The cpl1-2 Mutation Affects Fe Homeostasis
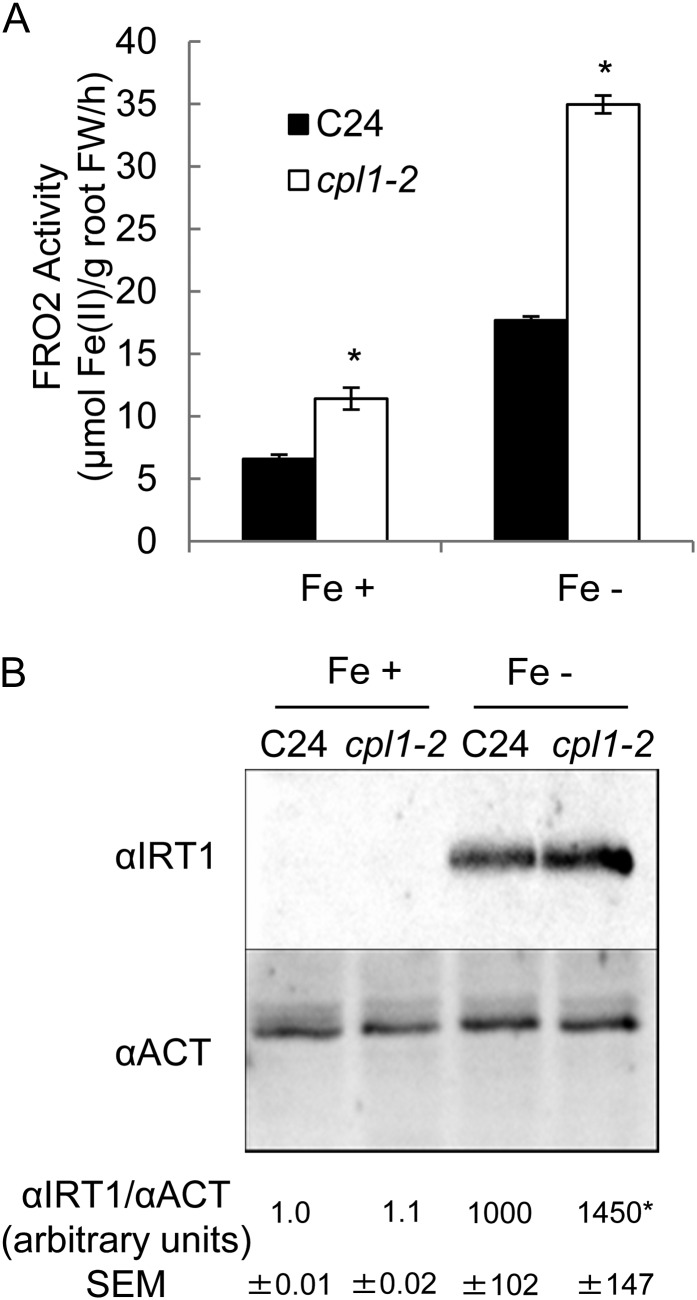
To determine whether elevated IRT1 and FRO2 transcript levels result in an increase of the corresponding proteins in the cpl1-2 mutant, we tested the levels of root FRO2 activity and IRT1 protein accumulation. Consistent with the transcript levels, FRO2 activity was 1.7- and 2-fold higher than in the wild type in Fe-sufficient and -deficient conditions, respectively (Fig. 5A). IRT1 proteins were below the level of detection under Fe-sufficient conditions in both genotypes; however, upon exposure to Fe deficiency (0 µm Fe-EDTA + 300 µm ferrozine for 3 d), cpl1-2 accumulated a slightly but significantly higher level of IRT1 than did the wild type (1.5- ± 0.24-fold; P < 0.05; Fig. 5B). The small difference in IRT1 protein level between the wild-type and mutant plants is consistent with the tight regulation of IRT1 by ubiquitin-mediated protein turnover (Kerkeb et al., 2008; Barberon et al., 2011).
Figure 5.
FRO activity and IRT1 protein accumulation in roots of cpl1-2 and C24 under Fe-sufficient or -deficient conditions. Plants were grown on basal medium for 7 d and then transferred to Fe-sufficient (Fe+; 50 µm Fe-EDTA) or Fe-deficient (Fe−; 0 µm Fe-EDTA + 300 µm ferrozine) basal medium. A, FRO2 activity after 3 d of treatment. The presented values are means of three biological replicates, each consisting of six technical repeats. Bars indicate the se of biological replicates. FW, Fresh weight. B, IRT1 protein accumulation after 3 d of treatment. Twenty micrograms of total protein was analyzed by immunoblot using anti-IRT1 antibodies (αIRT1). An anti-actin antibody (αACT [A2066; Sigma]) was used as the loading control. Average band intensities of three experiments (±se) are given in arbitrary units. *P < 0.05 by Student’s t test between mean values of cpl1-2 and C24 for the same conditions.
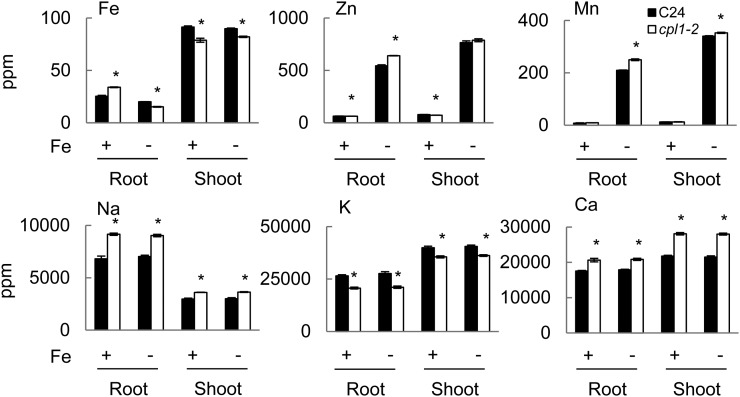
To determine the physiological consequences of the molecular changes in cpl1-2, elemental analyses were conducted for plants grown under the same conditions as described above. Roots and shoots were harvested separately, and metal contents were determined by inductively coupled plasma mass spectrometry (ICP-MS). As shown in Figure 6, in Fe-sufficient conditions, cpl1-2 accumulated 34% more Fe in the roots than did the wild type but 14% less Fe in the shoots. This suggests that although the IRT1 protein level was below our detection limit, higher basal IRT1 expression likely contributes to enhanced Fe acquisition in cpl1-2. It also suggests that translocation of Fe from the roots to the shoots is impaired in cpl1-2. As reported previously, exposure to Fe-deficient medium induced the accumulation of zinc (Zn) and manganese (Mn) in wild-type plants (Baxter et al., 2008). Interestingly, cpl1-2 roots accumulated 17.6% and 19.2% higher levels of Zn and Mn, respectively, under Fe-deficient conditions than did wild-type roots. Since IRT1 is likely responsible for increases in Zn and Mn uptake during Fe deficiency (Korshunova et al., 1999; Vert et al., 2002), the elevated IRT1 transcript and protein levels in cpl1-2 are likely to be biologically relevant. In addition, we noted that the sodium (Na) and calcium levels in the roots of cpl1-2 plants grown under Fe-sufficient conditions were elevated by 35% and 17.2%, respectively, compared with the wild type, and that potassium (K) levels were reduced by 22%. Overall, the cpl1-2 ion profile was consistent with the transcript profile of cpl1 mutants and was distinct from that of prototypical Fe homeostasis mutants such as frd3, which showed constitutive Fe deficiency stress responses and accumulated Zn and Mn even under Fe-sufficient conditions (Rogers and Guerinot, 2002).
Figure 6.
Metal contents of cpl1-2 and C24 roots and shoots under Fe-sufficient or -deficient conditions. Plants were grown on basal medium for 7 d and then transferred to Fe-sufficient (Fe+; 50 µm Fe-EDTA) or Fe-deficient (Fe−; 0 µm Fe-EDTA + 300 µm ferrozine) basal medium. After 3 d, root and shoot tissues were collected separately and dried at 65°C for 48 h, and elemental levels were determined from 100 mg of dried tissues by ICP-MS analysis. The presented elemental levels are mean values of three biological replicates analyzed in triplicate. Error bars indicate the se of biological replicates. *P < 0.05 by Student’s t test between mean values of cpl1-2 and C24 for the same conditions.
To determine if the distribution of Fe in roots is affected by the cpl1-2 mutation, Fe in root tissues was visualized by Perls staining (Supplemental Fig. S6). Unlike the previously characterized frd3-1 mutant, which accumulates Fe in vascular tissues (Green and Rogers, 2004), the root Fe profile of cpl1-2 was indistinguishable from wild-type C24 and stained predominantly root hairs.
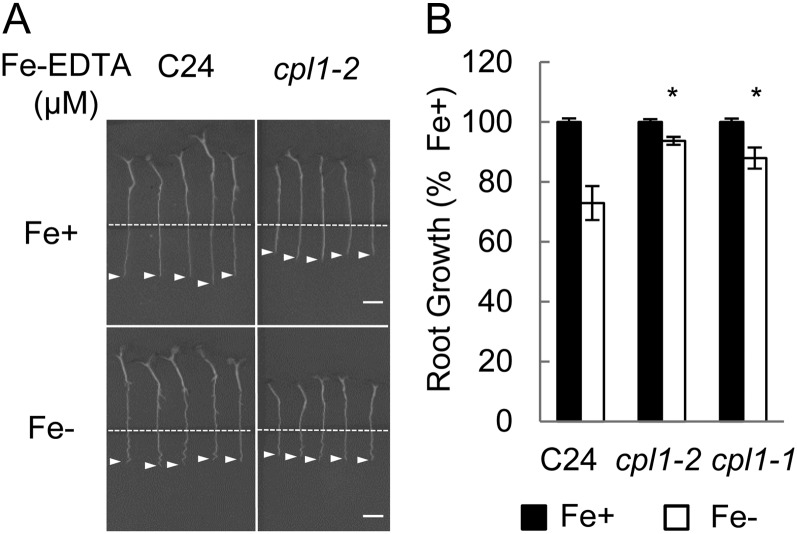
The root growth response of the cpl1 mutants to Fe deficiency stress was analyzed in vitro (Fig. 7). Plants were grown for 4 d on one-quarter-strength MS medium containing 50 µm Fe-EDTA and for another 5 d on Fe-deficient medium. Primary root growth of the wild type was inhibited by 28% ± 3% upon transfer to Fe-deficient medium, whereas that of cpl1-1 and cpl1-2 was inhibited by only 13% ± 4% and 6% ± 2%, respectively. This indicates that the cpl1 mutants are more tolerant to Fe deficiency than is the wild type. The intermediate tolerance in cpl1-1 may be due to the leaky expression of functional CPL1 transcript in this allele, which contains the T-DNA insertion in the third intron (Koiwa et al., 2002).
Figure 7.
Primary root growth of C24 and cpl1 mutants under different levels of available Fe. Four-day-old seedlings of C24, cpl1-1, and cpl1-2 were subjected to Fe deficiency as described in “Materials and Methods.” A, Root growth after 5 d of treatment. Root tip positions are marked by arrowheads. Dashed lines show the positions of the roots at the time of transfer. Bars = 10 mm. B, Root length was quantified to calculate relative root growth of the wild type and the cpl1 mutants. Error bars indicate the se of three biological replicates, each consisting of 20 seedling measurements. *P < 0.05 by Student’s t test between mean values of C24 and each mutant under Fe deficiency conditions. Fe+, 50 µm Fe-EDTA; Fe−, 0 µm Fe-EDTA + 300 µm ferrozine.
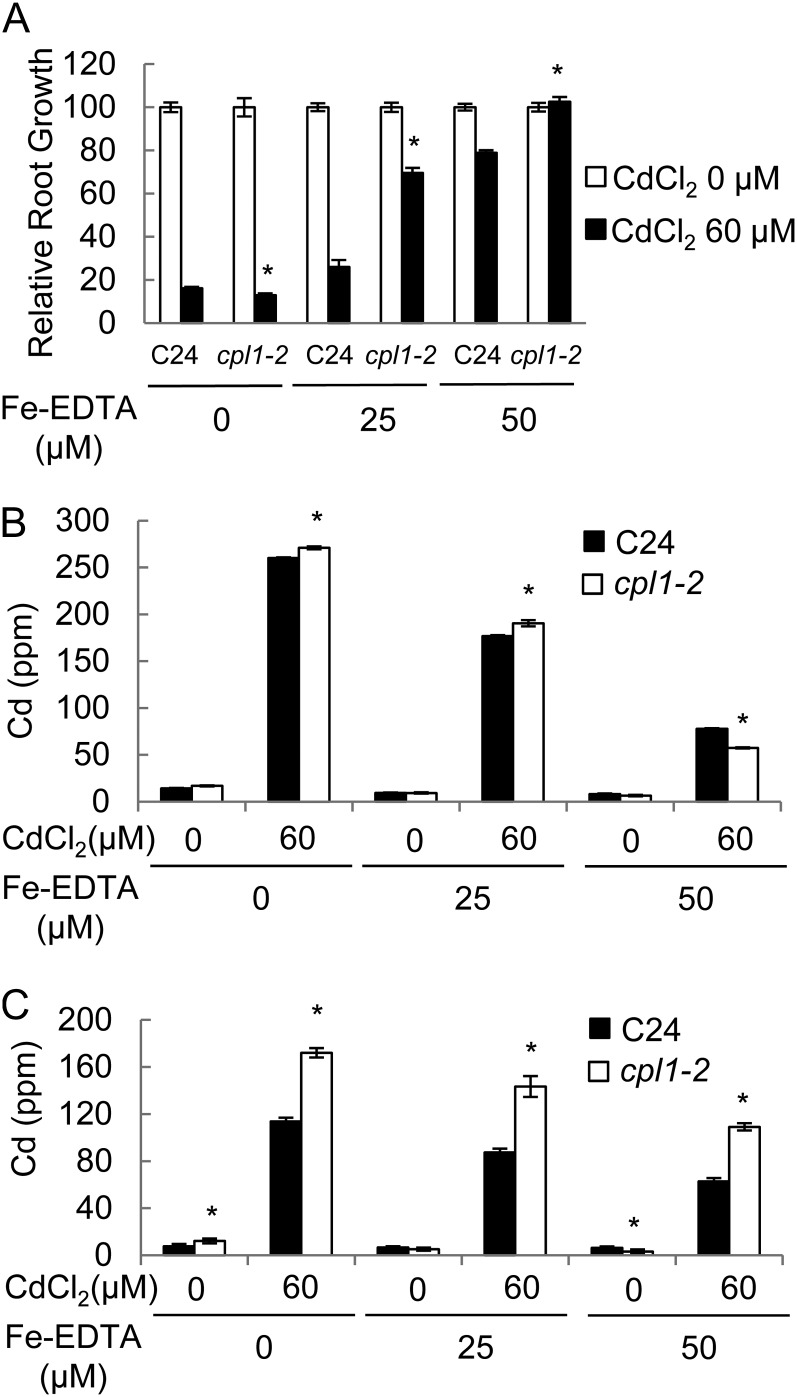
Since Cd ions can enter plant cells through Fe-uptake systems (Clemens, 2006), the cpl1-2 mutation may affect plant tolerance to Cd. To test this possibility, cpl1-2 and the C24 wild type were grown for 14 d on medium containing 60 µm CdCl2 and various concentrations of Fe. In the absence of Fe, growth of both the wild type and cpl1-2 was severely inhibited by the presence of Cd (Fig. 8A). With increasing amounts of Fe in the medium, both genotypes showed recovery; however, cpl1-2 plants showed greater tolerance to Cd on medium containing 25 or 50 µm Fe-EDTA. In the presence of 50 µm Fe-EDTA, wild-type plants showed substantial tolerance to Cd, albeit the tolerance was weaker than that of cpl1-2. Root Cd contents of wild-type and cpl1-2 plants were similar for all treatments, indicating that the basis for increased Cd tolerance in cpl1-2 does not likely involve exclusion of Cd from the roots (Fig. 8B). Surprisingly, the shoots of cpl1-2 plants accumulated 54.2% to 73% more Cd than did those of the wild type, suggesting that the long-distance transport of Cd was enhanced in cpl1-2 plants (Fig. 8C).
Figure 8.
Cd resistance of cpl1-2. Primary root growth (A) and Cd levels in the roots (B) and shoots (C) are shown for C24 and cpl1-2 growing on medium containing various levels of Fe and Cd. Seeds were germinated and grown for 10 d on one-quarter-strength MS medium adjusted to the indicated concentrations of Fe-EDTA and CdCl2. The presented root lengths are means of three biological replicates, each consisting of 20 seedlings. The presented Cd levels are means of three biological replicates analyzed in triplicate. Error bars indicate the se of biological replicates. *P < 0.05 by Student’s t test between mean values of cpl1-2 and C24 for the same conditions.
Genetic Dissection of Fe Signaling Perturbation in cpl1
It has been shown that the bHLH transcription factor FIT is a central regulator of Fe deficiency signaling and that FIT regulates its own Fe deficiency-induced expression (Colangelo and Guerinot, 2004; Jakoby et al., 2004; Wang et al., 2007). To determine whether the up-regulation of Fe utilization-related genes in the cpl1 mutants is dependent on FIT activity, the expression of these genes was analyzed in the fit cpl1 double mutant. Since a well-characterized fit-2 mutant is in the Col-0 genetic background (Colangelo and Guerinot, 2004), cpl1-6 in the Col-0 background was used in this analysis. Furthermore, to quantify FIT expression in the fit mutant background, lines with a FIT-LUC reporter gene were prepared in the wild type (Col-0), cpl1-6 and fit-2 single mutants, and the fit-2 cpl1-6 double mutant by genetic crossing (Supplemental Fig. S7).
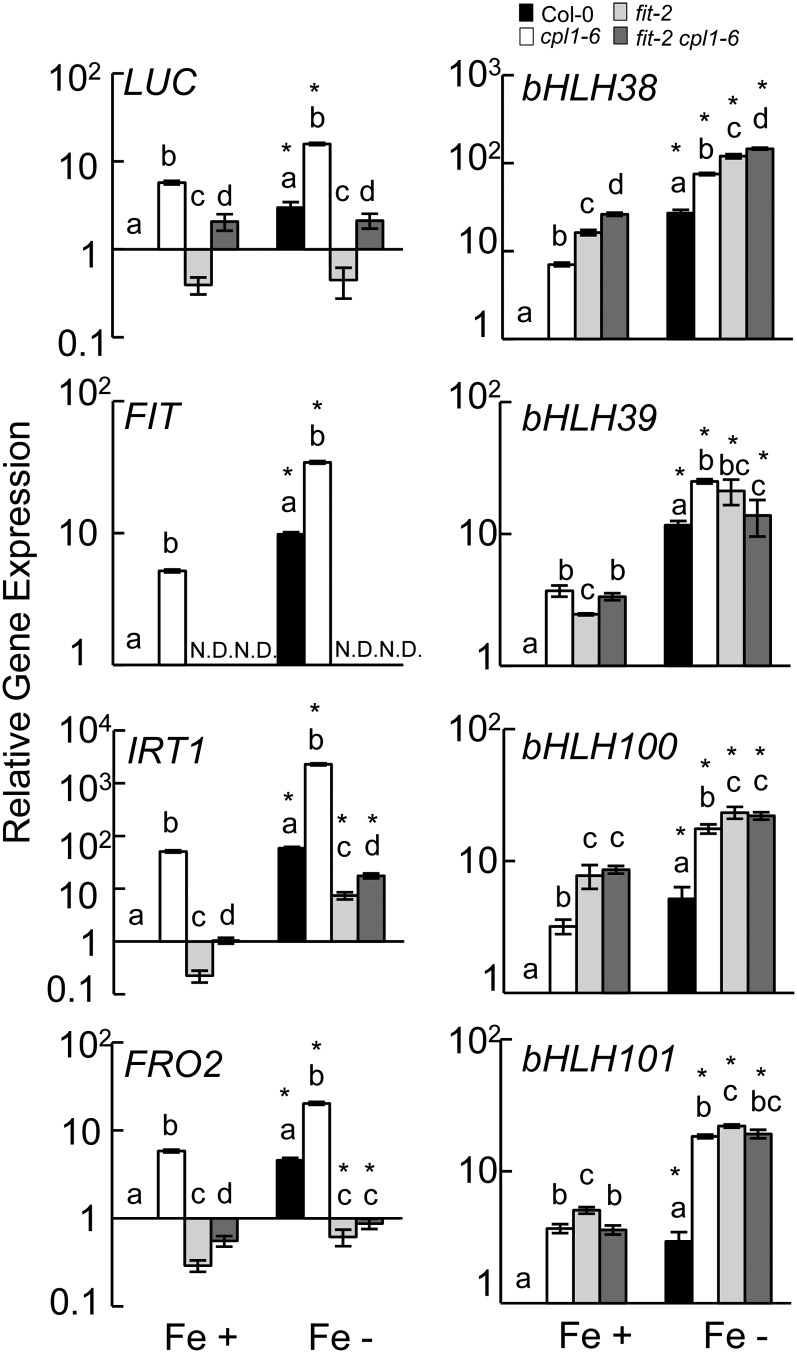
As shown in Figure 9, FIT-LUC and endogenous FIT expression were both enhanced in the cpl1-6 mutant. On the other hand, the basal expression level of FIT-LUC in fit-2 plants was 40% that of the wild-type level. In fit-2 cpl1-6, however, the level of FIT-LUC expression was similar to that of the wild type and lower than that of the cpl1-6 single mutant. This suggests that the cpl1-6 mutation can activate the FIT-independent signal to promote basal FIT expression. Furthermore, positive feedback by the FIT autoactivation mechanism likely amplifies FIT expression. Interestingly, Fe deficiency treatment did not induce the expression of FIT-LUC in the absence of functional FIT. This may indicate that FIT induction by Fe deficiency is entirely dependent on a FIT autoregulation mechanism. However, a FIT-independent component may be activated in the fit-2 background even during growth on Fe-sufficient medium, due to the constitutive Fe-deficient status of the mutants, resulting in the lack of further activation by Fe deficiency treatment.
Figure 9.
Expression levels of FIT-LUC, FIT, IRT1, FRO2, and group Ib bHLH transcription factors in Col-0, cpl1-6, fit-2, and fit-2 cpl1-6 plants containing the FIT-LUC reporter gene. Plants were grown on basal medium for 7 d and transferred to Fe-sufficient (Fe+; 50 µm Fe-EDTA) or Fe-deficient (Fe−; 0 µm Fe-EDTA + 300 µm ferrozine) basal medium. Total RNA was extracted from root tissue after 3 d of treatment. The presented expression levels (relative to untreated Col-0 samples) are mean values of biological duplicates analyzed in duplicate. Error bars indicate the se of biological replicates. Different letters show significant differences between genotypes under Fe+ and Fe− conditions (P < 0.05 by one-way ANOVA followed by Tukey’s HSD post hoc test). *P < 0.05 by Student’s t test between mean values of Fe+ and Fe− for the same genotype.
The expression profiles of genes downstream of FIT were similar to that of FIT-LUC. Under Fe-sufficient conditions, IRT1 and FRO2 levels were 22% and 29% lower in the fit-2 mutant than in the wild type, respectively; however, the expression of these genes was recovered (95% and 54% of wild-type expression, respectively) in fit-2 cpl1-6 plants. IRT1 and FRO2 expression levels increased in response to Fe deficiency treatment even in the fit-2 background, indicating that a FIT-independent pathway positively regulates the expression of these genes; however, similar to FIT-LUC, FIT is required for the full induction of IRT1 and FRO2. In contrast to [IRT1, FIT-LUC, FRO2], the basal and induced levels of group Ib bHLH expression were higher in all mutant lines tested, and their expression levels were largely similar among mutant lines other than bHLH38. It appears that both the enhanced Fe deficiency signals in cpl1-6 plants and the Fe-deficient status caused by the fit-2 mutation activate group Ib bHLH expression by the same FIT-independent mechanism. Together, these results indicate that cpl1-2 likely activates Fe deficiency responses upstream of both FIT-dependent and -independent signaling pathways.
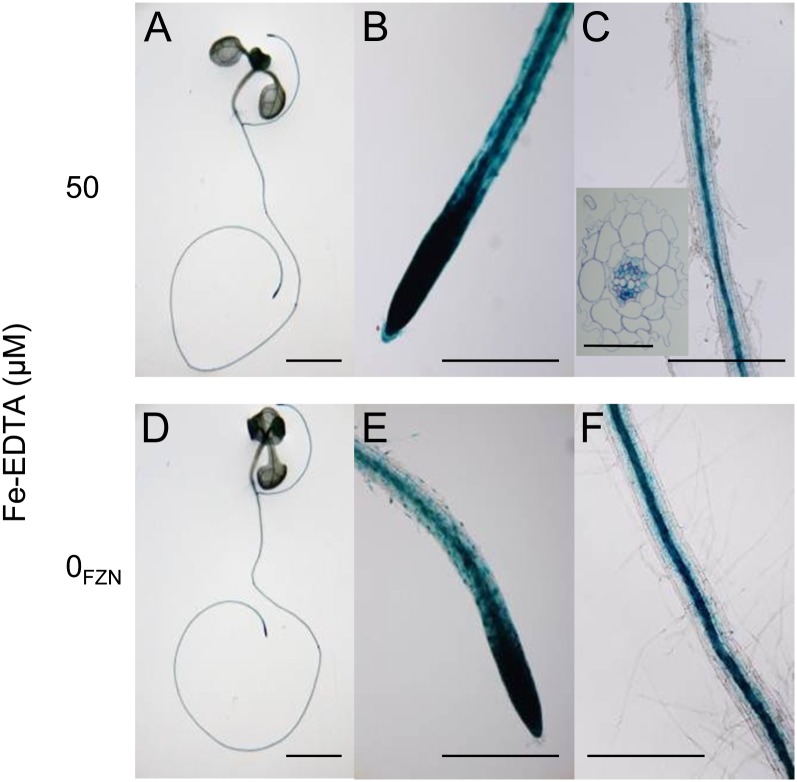
FIT is specifically expressed in the root epidermis (Colangelo and Guerinot, 2004); therefore, the direct regulation of FIT by CPL1 would require CPL1 expression in these cells. The tissue specificity of CPL1 expression was analyzed using a CPL1-GUS translational fusion construct. The GUS open reading frame was inserted in an 8.4-kb CPL1 genomic fragment immediately before the stop codon of the CPL1 open reading frame, whose expression was regulated by its native regulatory sequences. The expression of the reporter gene was monitored in the roots of transgenic Arabidopsis plants (Fig. 10). The expression profile of CPL1-GUS was similar in Fe-sufficient or -deficient conditions. CPL1-GUS expression was high in root tips; however, in the mature part of roots, CPL1-GUS expression was confined to the stele, particularly in the phloem, and virtually no activity was observed in the outer layers of root cells. Overall, except for a limited area of the root tip, CPL1 and FIT showed distinct tissue-specific expression patterns, suggesting that the regulation of FIT by CPL1 is likely indirect.
Figure 10.
CPL1 was expressed in the root tip and stele. Plants expressing the CPL1-GUS fusion protein were grown on basal medium for 7 d and transferred to Fe-sufficient (50 µm Fe-EDTA; A–C) or Fe-deficient (0 µm Fe-EDTA + 300 µm ferrozine; D–F) basal medium. After 3 d, GUS activity was visualized and documented. Bars = 10 mm in A and D; 2 mm in B, C, E, and F; and 0.05 mm in C, inset.
The cpl1-2 Mutation Promotes the Fe Deficiency-Mediated Expression of ABA/Osmotic Stress-Regulated Genes
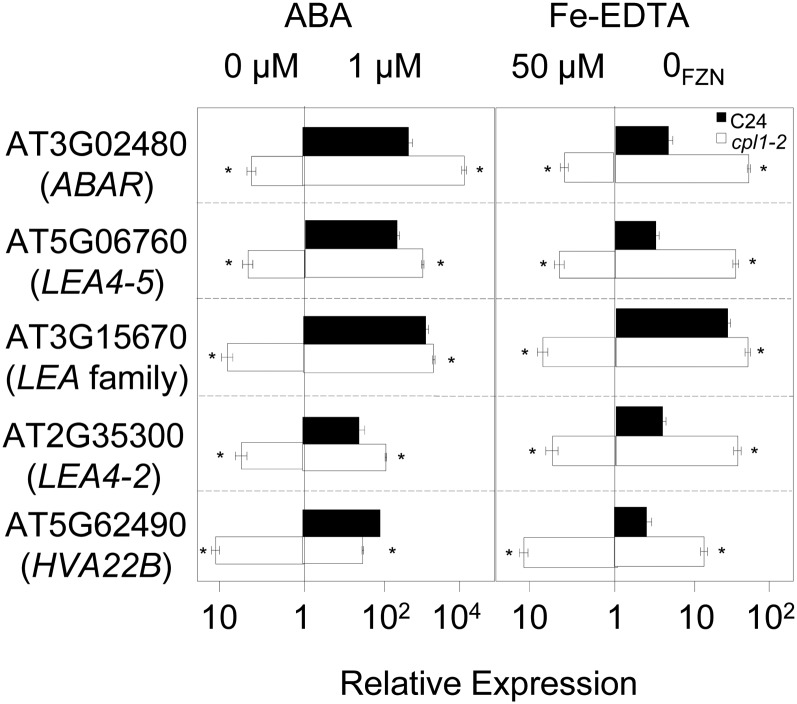
Our finding that cpl1-2 simultaneously enhances the expression of both osmotic stress/ABA-responsive and Fe deficiency-responsive genes is somewhat contradictory to the previous report that ABA and osmotic stress inhibit Fe deficiency signaling (Séguéla et al., 2008). However, an observation similar to ours was reported in rice (Oryza sativa) overexpressing IDEF1 (for iron deficiency element [IDE1]-binding factor1; Kobayashi et al., 2009). Because of the similarity between IDE1 (CATGA) and the ABA signaling cis-element (RY motif [CATGCATG]; Kobayashi et al., 2009), we hypothesized that CPL1 regulates a branch of Fe deficiency and osmotic stress/ABA signaling through similar cis-regulatory elements. While ABA treatment inhibited [IRT1, FRO2, FIT] expression in both the wild type and cpl1-2 (data not shown), the expression of genes inducible by both Fe deficiency stress and ABA treatment was substantially enhanced in cpl1-2 (Fig. 11). This indicates that some ABA-responsive genes are also regulated by Fe through a common CPL1-repressed pathway. Consistently, the presence of higher concentrations of external Fe could repress not only the expression of prototypical Fe-regulated genes [IRT1, FRO2, FIT] but also the enhanced expression of LATE EMBRYONIC ABUNDANT (LEA) in cpl1-2 (Fig. 4). Interestingly, the repression of LEA expression by excess Fe (greater than 50 µm) was less pronounced in cpl1-2 than in the wild type, and HVA22 expression was largely unaffected by increased Fe, suggesting that additional Fe-independent mechanisms up-regulate LEA expression in cpl1-2. To test the state of ABA signaling in cpl1, the expression levels of several ABA signaling components were determined. The expression of ABI1, ABI3, and AREB1/ABF2 was up-regulated 4.0-, 3.9-, and 5.4-fold in cpl1-2 under normal growth conditions. This result was consistent with the observation that cpl1-2 was hyperresponsive to ABA treatment (Fig. 11). Together, these results suggest that CPL1 regulates the signaling pathway upstream of the cross talk between ABA and Fe signals.
Figure 11.
The expression of LEA family transcripts in response to ABA or Fe deficiency treatments. Plants were grown on basal medium for 7 d and transferred to basal medium containing 1 µm ABA or to Fe-deficient basal medium (0FZN; 0 µm Fe-EDTA + 300 µm ferrozine). The duration of treatment was 1 h for ABA and 72 h for Fe deficiency. Total RNA was extracted from roots. The presented expression levels (relative to untreated C24 samples) are mean values of biological triplicates analyzed in duplicate. Error bars indicate the se of biological replicates. *P < 0.05 by Student’s t test between mean values of cpl1-2 and C24 for the same conditions.
DISCUSSION
RNA metabolism regulates diverse developmental and stress response signaling pathways in plants. This study shows that an upstream component of the Fe deficiency response was regulated by an RNA metabolism factor, CPL1. Although the Fe deficiency response signaling pathway is regulated by multiple mechanisms, such as protein-protein interactions (Yuan et al., 2008), differential promoter activation (Yuan et al., 2008; Long et al., 2010), protein ubiquitination (Kerkeb et al., 2008; Li and Schmidt, 2010), and proteasomal and nonproteasomal protein degradation (Barberon et al., 2011; Lingam et al., 2011; Sivitz et al., 2011), regulation at the level of RNA metabolism has not hitherto been established. CPL1 is a plant-specific isoform of the universal pol II CTD phosphatase family of proteins, which harbor double-stranded RNA-binding motifs and can specifically dephosphorylate pol II CTD at Ser-5-PO4 in vitro (Koiwa et al., 2002, 2004; Xiong et al., 2002). We have shown that CPL1 localizes to the root stele and negatively regulates Fe signaling. cpl1 mutations result in enhanced expression of FIT and group Ib bHLH genes, which are essential for Fe deficiency sensing/signaling and affect metal homeostasis. This finding is consistent with a previous report showing that Fe utilization-related genes are down-regulated in the RNA decapping-deficient mutant, tdt (Goeres et al., 2007).
Since alteration of the polymerase II phosphorylation status can potentially trigger alterations in global transcription and RNA maturation, it could be argued that the observed up-regulation of Fe utilization-related genes resulted from a pleiotropic effect of the cpl1-2 mutation, similar to the down-regulation of Fe utilization-related genes in the tdt mutation (Goeres et al., 2007). However, several observations argue against the involvement of pleiotropic effects. First, in contrast to the tdt mutant, the cpl1 mutants do not exhibit any major defects in growth and development. Second, cpl1-2 affects the levels of metals associated with Fe homeostasis (i.e. Fe, Zn, Mn, and Cd). Third, cpl1-2 affects multiple regulons in Fe deficiency signaling (i.e. FIT-dependent and -independent Fe deficiency responses and repression of the basal level of FER1). Fourth, the effect of cpl1-2 could be suppressed by increased Fe concentrations in the medium. Finally, the effect of cpl1-2 on the expression of [IRT1, FIT, FRO2] likely involves intercellular communication. These findings indicate that the cpl1 mutations affect Fe homeostasis and activate upstream signaling component(s) rather than cause the misexpression of FIT, regardless of the presence or absence of Fe signals. On the other hand, levels of IRT1, which are regulated posttranslationally by Fe status, are affected only moderately by cpl1 mutations under Fe deficiency stress. This suggests that CPL1 either specifically regulates branch(es) of Fe deficiency signaling or that higher thresholds of Fe deficiency signals are required to activate posttranslational Fe deficiency responses. Alternatively, the levels of IRT1 at the plasma membrane may be regulated by apoplastic Fe rather than by CPL1-mediated signaling.
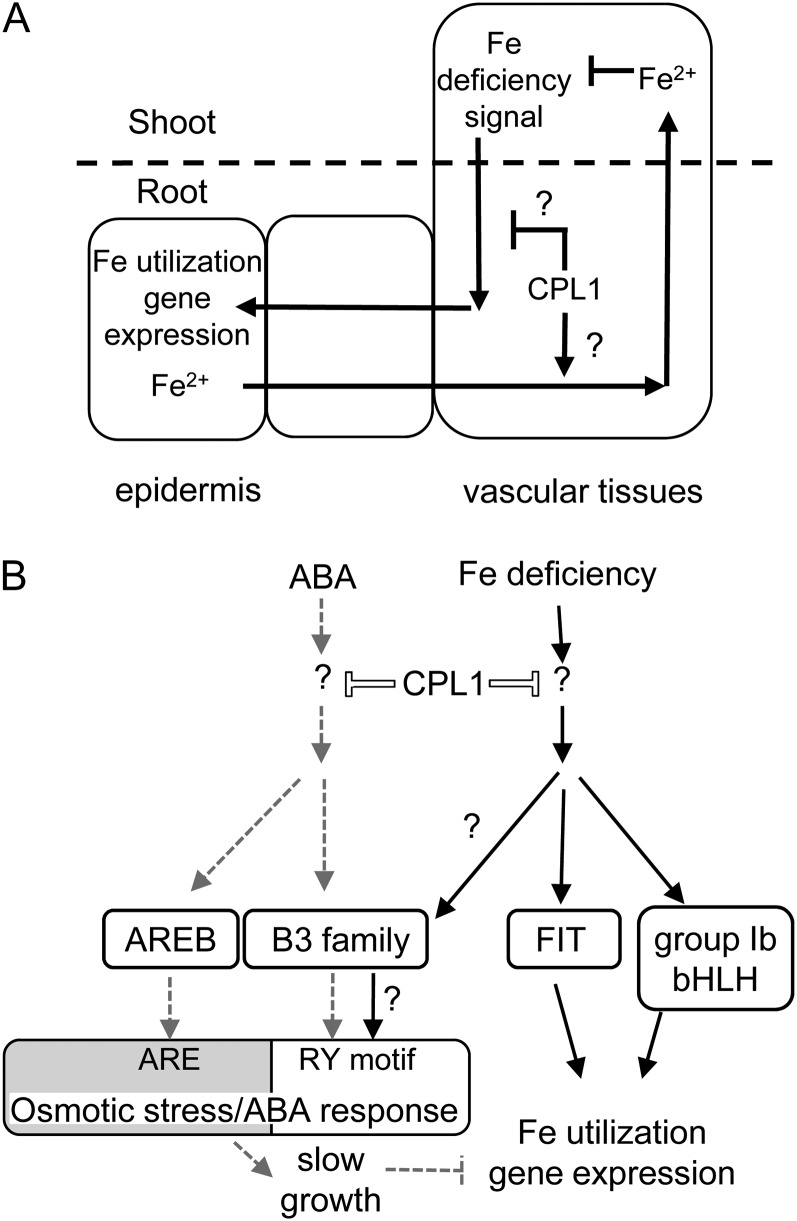
How does CPL1 regulate the Fe status and deficiency responses? Our data indicate that CPL1 is strongly expressed in the root stele and not at all in the epidermis, where FIT is expressed. It is also noteworthy that cpl1-2 induced the expression of [IRT1, FIT, FRO2] under Fe-sufficient conditions, even in the presence of elevated levels of root Fe, but repressed [IRT1, FIT, FRO2] expression when additional Fe was supplied exogenously. One possibility is that CPL1 functions at the organ level to keep the root Fe level low by repressing [IRT1, FIT, FRO2] and promoting Fe loading into the xylem (Fig. 12A). This could be achieved by promoting Fe transport from the epidermis to the xylem, by repressing sequestration/compartmentalization of Fe in the root cells, or by repressing xylem unloading of Fe in the roots. Upon Fe deficiency, Fe starvation signals generated in the shoot overcome the repression of [IRT1, FIT, FRO2] by CPL1. Upon mutation of CPL1, both the repression of [IRT1, FIT, FRO2] and the promotion of xylem loading are impaired, resulting in elevated expression of [IRT1, FIT, FRO2] and accumulation of Fe in the roots. At the same time, Fe in the shoots is reduced due to decreased root-to-shoot Fe transport, which in turn generates systemic Fe deficiency signals and promotes [IRT1, FIT, FRO2] expression in the roots. This mutant phenotype can be ameliorated by the addition of Fe, because a basal level of xylem loading still occurs in cpl1 mutants. The relatively large decrease in root Fe content in the cpl1-2 mutant during Fe deficiency may be due to the continued growth of cpl1-2 roots diluting the acquired Fe and/or to the greater mobilization of stored Fe by enhanced Fe deficiency signals. While the expression levels of FRD3 and IRON REGULATED1 (IREG1), which likely load citrate and Fe to the xylem, respectively, do not substantially differ between the wild type and cpl1-2, there is a moderate increase in the expression of some plastid Fe uptake genes in cpl1-2. This implies that CPL1 attenuates Fe compartmentalization rather than regulates xylem-loading activity (Supplemental Fig. S5B). However, we cannot exclude the possibility that cpl1-2 regulates the expression of other xylem-loading Fe transporters, since IREG1 is likely not the sole/major xylem-loading Fe transporter (Morrissey et al., 2009). Furthermore, the mode of CPL1 function in this process remains to be determined. Because the physicochemical nature of intercellular Fe deficiency signaling between the shoots and roots and the root stele and epidermal tissues remains elusive, it is difficult to address this issue. Currently, we are not able to detect significant changes in overall CTD phosphorylation status during Fe deficiency, suggesting that bulk pol II CTD phosphorylation is not involved (data not shown). The presence of the double-stranded RNA-binding motif in CPL1 suggests that CPL1 may regulate small RNA production and/or function. It is interesting that CPL1 is strongly expressed in the stele, likely in phloem companion cells, where the expression of small RNA production machinery is enriched (Mustroph et al., 2009). Indeed, several microRNAs are regulated by the Fe signal (Kong and Yang, 2010), and the potential effect of cpl1-2 on the mobility of small RNAs may explain the systemic nature of this mutation as well as its impact on multiple pathways.
Figure 12.
Model for the role of CPL1. A, Role of CPL1 in root-shoot Fe distribution. CPL1 in the stele likely promotes the root-to-shoot transport of Fe and attenuates the Fe deficiency signal in shoots. Communication between different cell types in shoot tissues is omitted in this model. B, Coregulation of a subset of osmotic stress/ABA response genes by ABA and Fe deficiency signals and its attenuation by CPL1. Solid and broken arrows indicate pathways that operate during Fe deficiency and in the presence of ABA, respectively. White bars indicate the repression activity of CPL1, which is absent in cpl1 mutants. CPL1 attenuates the Fe deficiency signaling pathway upstream of FIT and group Ib bHLH. Direct targets of CPL1 in the pathways are currently unknown. B3 family transcription factors up-regulate the expression of a subset of osmotic stress/ABA-responsive genes through the RY motif in response to Fe deficiency and ABA signaling. ABA also promotes gene expression via the ABA-responsive element. When ABA levels are elevated, the enhanced expression of ABA-responsive genes causes slow growth, which inhibits the expression of Fe utilization genes.
We initially expected that the enhanced levels of IRT1 in cpl1-2 would result in Cd oversensitivity, because IRT1 is responsible for Cd influx into root cells (Connolly et al., 2002). Instead, cpl1-2 showed Cd tolerance, and Cd levels in cpl1-2 roots were not greater than those in wild-type roots. Rather, cpl1-2 showed a substantial increase in shoot Cd levels, indicating an increase in the root-to-shoot transport of Cd. Since Cd transport and tolerance involve multiple mechanisms and have not been fully elucidated, there is no simple explanation for how cpl1-2 confers Cd tolerance. However, several genes that are up-regulated in cpl1-2 are related to Cd tolerance. For example, the elevated expression of Fe acquisition genes such as [IRT1, FIT, FRO2] and group Ib bHLH, and the resulting increase in root Fe levels, can ameliorate the toxicity of cellular Cd (Wu et al., 2012). In addition, Wu et al. (2012) also reported that coexpression of FIT and bHLH38/39 activates HMA3, MTP3, IREG2, and IRT2, which can enhance the sequestration of Cd into vacuoles in the root. In addition, OPT3 may mediate Cd tolerance in a similar manner to the yeast homolog, ScOPT2, which transports Cd into the vacuoles (Aouida et al., 2009). However, these mechanisms in general promote the sequestration of Cd into vacuoles, resulting in higher levels of Cd in the roots. In contrast, the “overflow protection mechanism” proposed for phytochelatin-mediated long-distance Cd transport from roots to shoots (Gong et al., 2003) may better explain the Cd tolerance of cpl1-2. Similar to cpl1 mutants, transgenic plants with root-specific expression of phytochelatin synthase have elevated levels of Cd in the shoots but normal levels of Cd in the roots (Gong et al., 2003). However, since phytochelatin is not likely the sole chelator that mediates long-distance Cd transport (Clemens, 2006), we cannot exclude the contribution of other mechanisms.
The simultaneous up-regulation of Fe deficiency- and ABA/osmotic stress-regulated transcripts is a unique signature of the cpl1-2 transcriptome. This may indicate that the cpl1-2 mutation activates two distinct stress signaling pathways and/or that these two pathways exhibit specific cross talk. The first possibility is supported by our finding that both Fe-specific and ABA/osmotic stress-specific genes are up-regulated in cpl1. The increase of Na (30%) and decrease of K (23%) in cpl1 are further indicators that cpl1 mutants indeed experience osmotic (salt) stress under normal growth conditions. While previous studies indicate that Fe deficiency and ABA/osmotic stress pathways exhibit cross talk (Dinneny et al., 2008; Séguéla et al., 2008; Kobayashi et al., 2009), the positive coregulation of gene expression by ABA/osmotic stress and Fe deficiency has not been studied in dicots to date. In monocots, however, specific cross talk between ABA/osmotic stress and Fe deficiency signaling occurs, as shown with rice overexpressing IDEF1, a B3 family transcription factor (Kobayashi et al., 2009). Although an obvious IDEF1 ortholog is not present in the Arabidopsis genome, IDE1 sequences are found in many Fe-responsive gene promoters, including those from Arabidopsis (Kobayashi et al., 2003, 2005). Interestingly, the closest homolog of IDEF1 in Arabidopsis is ABI3, and the expression of this gene in cpl1-2 was elevated 4-fold over wild-type levels. Furthermore, the recognition sequence of IDEF1 (CATGC) overlaps with that of ABI3/VP1-type B3 family transcription factors (CATGCA; Kobayashi et al., 2009, 2010). Therefore, coregulation of genes by ABA and Fe deficiency in cpl1-2 may be caused by a yet unidentified B3 family transcription factor(s) (Fig. 12B). Another notable change in ABA signaling in cpl1-2 was the 5.4-fold up-regulation of AREB1/ABF2. Interestingly, plants overexpressing either ABI3 or an activated AREB1-ΔQT variant show enhanced expression of the LEA genes identified in our cpl1-2 microarray, and AREB1-ΔQT overexpression causes the up-regulation of bHLH38 and bHLH39 as well (Fujita et al., 2005; Nakashima et al., 2006). It is tempting to speculate that B3 family transcription factor(s) function as an ancient form of IDEF1 in dicots and regulate both ABA and Fe deficiency signaling. However, higher level integration of ABA and Fe signaling pathways may exist. Whereas the ABA signaling pathway and ABA-responsive promoter elements/transcription factors are well established (Hubbard et al., 2010), detailed information on specific Fe-responsive cis-elements in the dicot Fe deficiency-induced promoters and on the Fe signaling components that function upstream of FIT or group Ib bHLH is lacking. In addition, the role of LEA proteins in the Fe deficiency response has not been established, but some LEA proteins appear to bind metal ions and may function in metal transport (Kruger et al., 2002). Further genetic and molecular analyses are required to determine the role and the mode of action of the ABA-Fe deficiency signal cross talk, which occurs in both strategy I and strategy II plants.
MATERIALS AND METHODS
Chemicals and Primer Information
All chemicals were obtained from Sigma. Sigma agar E was used for all plant growth experiments. Primer sequences used in this study are shown in Supplemental Table S5.
Plant Materials
The Arabidopsis (Arabidopsis thaliana) ecotypes Col-0 and C24 were used in this study. The cpl1-1 and cpl1-2 lines were described previously (Koiwa et al., 2002; Xiong et al., 2002). cpl1-5 (GK-590C07) and cpl1-6 (GK-165H09) were obtained from the Nottingham Arabidopsis Stock Center (NASC), and frd3-1 was obtained from the Arabidopsis Biological Resource Center. fit-2 seeds were provided by Dr. Paul Paré.
For general growth and microarray analyses, seeds were sown on medium containing one-quarter-strength MS salts, 0.5% Suc, and 0.8% agar. After stratification for 2 d at 4°C, the plates were kept in a growth incubator under a long-day photoperiod (16 h of light, 8 h of darkness) at 25°C for 10 d.
RNA Extraction
Total RNA was isolated using TRIzol reagent (Chomczynski and Sacchi, 1987) and treated with DNase I (Promega) to remove genomic DNA contamination.
Microarray Analyses
Total RNA was extracted from 10-d-old wild-type C24 and cpl1-2 mutant seedlings. Quality tests of the RNA samples, copy RNA synthesis, biotin labeling, hybridization to Affymetrix ATH1 GeneChips, and scanning were performed by the Affymetrix Service at the NASC. After chip hybridization and scanning, triplicate data were obtained for each genotype, and data were processed using GeneSpring GX 11.0.2 software (Agilent). Raw intensity values, computed from CEL files, were processed first by Robust Multiarray Analysis (Irizarry et al., 2003). Filtering on expression levels and fold changes (2-fold or greater) was performed using the GeneSpring package, and differentially expressed genes were determined. Statistical analyses were performed using one-way ANOVA at P < 0.05 (with asymptotic P value computation) followed by the Tukey’s HSD post hoc test and Benjamini-Hochberg false discovery rate multiple testing correction. A fold change of at least 2-fold was considered indicative of differential expression, where a P value of 0.05 or less was considered indicative of significant alteration in expression. All microarray data from this study were deposited in the NASC database under accession number 611.
For hierarchical clustering of up-regulated genes, raw gene expression data for abiotic stresses and hormone treatments were first obtained from the AtGenExpress project (http://www.arabidopsis.org/portals/expression/microarray/ATGenExpress.jsp). The CEL file names are NASCarray 176 (ABA), 140 (salt), and 139 (mannitol). The CEL file for the Fe deficiency experiment (GSE15189) was obtained from the Gene Expression Omnibus database (Buckhout et al., 2009). All data were clustered by Pearson correlation distance and the average linkage rule (Eisen et al., 1998).
Gene Set Enrichment Analysis
GSEA was performed using GeneTrail (Backes et al., 2007; Schuler et al., 2011). Briefly, 22,811 probes on ATH1 microarray data sets obtained from public databases were ranked and sorted according to fold change from the most induced to the most suppressed by each stress treatment. Subsequently, GSEA analyses were performed for each sorted data set using gene sets created from an analysis of the cpl1-2 microarray. As a reference, gene sets consisting of constitutively expressed genes in Arabidopsis were analyzed (Czechowski et al., 2005). False discovery rate was used as the P value adjustment (Benjamini and Hochberg, 1995).
RT-qPCR Analysis
Total RNA samples (2 µg) were reverse transcribed using random hexamers and SuperScript III Reverse Transcriptase (Life Technologies) in a total volume of 20 µL. One-twentieth of the reverse transcription products were analyzed using ABI 7900 Sequence Detectors and SYBR Green Master Mix (Life Technologies). The amplification reaction and data analysis were performed as described (Salzman et al., 2005). Each reaction was run in technical duplicate, and the melting curves were analyzed by SDS2.2.2 software (Life Technologies) to verify that only a single product was amplified. TUB8 (At5g23860) was used as an internal control for the normalization of data.
Preparation and Analysis of the FIT-LUC Reporter Gene
A 1,324-bp genomic DNA fragment of the FIT promoter was amplified by PCR using a primer pair (1047, 1048) and bacterial artificial chromosome (BAC) clone F24D13 as template. The PCR product was digested by HindIII and EcoRV and ligated into pEnEL2Ω-LUC (GenBank accession no. JN570503) digested with HindIII and SnaBI. A Gateway LR reaction was performed according to the manufacturer’s instructions (Life Technologies) using pBSVirHygGW as the destination vector (Ueda et al., 2008). Transformation of Agrobacterium tumefaciens GV3101(pMP90RK), floral transformation of cpl1-6, and hygromycin selection of transformants were performed as described previously (Ueda et al., 2008). The homozygous cpl1-6 FIT-LUC line was crossed with the Col-0 wild type and fit-2, and homozygous Col-0 FIT-LUC, fit-2 FIT-LUC, and cpl1-6 fit-2 FIT-LUC lines were identified in the segregating F2 population and confirmed in the F3 generation. The expression level of LUC in each line was determined by RT-qPCR.
Preparation and Analysis of the CPL1-GUS Reporter Gene
A genomic fragment (8.1 kb) of the CPL1 locus encoded by an 8.4-kb BlpI fragment of BAC clone F17L22 was cloned in SmaI-digested pENTR2B (Life Technologies). Subsequently, a GUS-coding sequence of pBI101 was inserted immediately before the stop codon of CPL1. The resulting entry clone was then recombined with pBSVirHygGW binary plasmid (Ueda et al., 2008) using Gateway LR recombinase (Life Technologies) to produce pBSVirHygCPL1-GUS, which was introduced into A. tumefaciens GV3101(pMP90RK) and then used for flower transformation of Col-0 plants (Koiwa et al., 2002). Hygromycin-resistant transgenic plants were selected on medium containing one-quarter-strength MS salts, 30 µg mL−1 hygromycin, 100 µg mL−1 cefotaxime, and 0.8% agar.
Five-day-old Col-0 CPL-GUS plants germinated on basal medium were grown for an additional 3 d on Fe-sufficient or -deficient basal medium (see below). For high-resolution GUS staining, plants were fixed in ice-cold 90% acetone for 15 min and then incubated at 37°C for 4 h in a solution containing 2 mm 5-bromo-4-chloro-3-indolyl glucuronide, 5 mm K3Fe(CN)6, 5 mm K4Fe(CN)6, 100 mm sodium phosphate buffer (pH 7.0), and 0.1% Triton X-100. After incubation, the samples were rinsed with 100 mm phosphate buffer.
For embedding, GUS-stained plants were fixed in 2% glutaraldehyde using cold microwave technology in a BioWave microwave with a 6-min vacuum cycle (2 min on/2 min off/2 min on) at 200 W. After washing in 100 mm sodium phosphate buffer (pH 7.0), the sample was dehydrated in a methanol/water graded series at 10% methanol increments (from 50% to 90%) for 1 min (30 s on/30 s off) at 200 W followed by three cycles in 100% methanol. Samples were rinsed in propylene oxide, and then 2 mL of fresh propylene oxide was added to each sample. Samples were infiltrated with Quetol 651 low-viscosity resin by the stepwise addition of 10% (v/v) resin to each sample (Ellis, 2006). After each addition of resin, samples were allowed to infiltrate for several hours to overnight before additional resin was added. After two changes of 100% resin, samples were again vacuum infiltrated for 6 min (2 min on/2 min off/2 min on) at 200 W and then embedded. Five-micrometer sections were prepared and mounted onto slides and then stained with toluidine blue before observation.
Stress Treatments
For testing plant responses to various Fe regimens, seeds were sown on basal medium containing one-quarter-strength MS salts, 50 µm Fe-EDTA, 0.5% Suc, and 1.5% agar. Fe deficiency was applied by transferring 7-d-old seedlings to basal medium without Fe-EDTA but containing 300 µm ferrozine [3-(2-pyridyl)-5,6-diphenyl-1,2,4-triazine sulfonate; 0FZN basal medium]. Total RNA was isolated at the indicated times. For growth analysis, 4-d-old seedlings grown on basal medium were transferred to the 0FZN basal medium, and plants were photographed 5 d after the transfer. Primary root lengths were analyzed using ImageJ software.
To determine Cd sensitivity, seedlings of the C24 wild type and the cpl1-2 mutant were grown on basal medium adjusted to the indicated concentrations of Fe-EDTA and in the presence or absence of 60 µm CdCl2. Seedlings were photographed after 11 d of growth, and primary root lengths were analyzed.
For ABA treatment, 7-d-old plants grown on basal medium were transferred to basal medium with or without 1 µm ABA.
Determination of Ferric Chelate Reductase Activity
Root ferric chelate reductase activity was measured spectrophotometrically as described (Yi and Guerinot, 1996). The assay solution was composed of 0.1 mm Fe(III)-EDTA and 0.3 mm ferrozine in distilled water. Roots of five plants were soaked in assay solution for 30 min in darkness, and then the absorbance of the assay solution was determined. An identical assay solution without any plants was used as a blank. Purple-colored Fe(II)-ferrozine complex formation was quantified using a molar extinction coefficient of 28.6 mm−1 cm−1 at 562 nm. The experimental results were means of three biological repeats with six technical replicates each.
IRT1 Accumulation
Total root protein was extracted from both C24 wild-type and cpl1-2 mutant plants grown either in Fe-sufficient or Fe-deficient conditions. Extracts were prepared by grinding tissues in extraction buffer on ice followed by centrifugation at 4°C for 15 min at 14,000g. Twenty micrograms of total protein extracts was resolved on a 10% SDS-polyacrylamide gel and electroblotted onto a polyvinylidene fluoride membrane (Millipore). After blocking with 6% skim milk in Tris-buffered saline, the membrane was incubated with affinity-purified rabbit anti-IRT1 antibodies (diluted 1:5,000 in Tris-Tween-buffered saline) prepared as described previously (Vert et al., 2002; Genscript) and then goat anti-rabbit IgG-horseradish peroxidase conjugate (Thermo Scientific) diluted 1:100,000 in Tris-Tween-buffered saline. The blot was developed using a Supersignal West Femto Kit (Thermo Scientific) and documented using a Cascade II CCD camera (Photometrics). Anti-actin antibody (A2066; Sigma) was used as the loading control.
Determination of Metal Content
Tissue element analysis of Na, K, calcium, Fe, Mn, Zn, and Cd using ICP-MS was performed as described (Baxter et al., 2007). Briefly, shoots were washed thoroughly with distilled water. Roots were washed first in 2 mm CaSO4 and 10 mm EDTA for 10 min and then rinsed twice in distilled water. Then, tissues were blot dried, divided into three replicates of about 100 mg fresh weight, dried in a 65°C oven for 48 h, and reweighed. The dried samples were digested completely using 0.6 mL of concentrated ultrapure-grade HNO3 (JT Baker) at 110°C for 4 h. Each sample was diluted to 6 mL with nanopure water (18.2 MΩ) and analyzed on a Perkin-Elmer NexION 300D ICP-MS device in the reaction mode. Indium (EMD Millipore) was used as an internal standard. National Institute of Standards and Technology traceable calibration standards (Alfa Aesar) were used for the calibration. Each sample was read five times in the pulse detector mode. Three biological replicates were performed per analysis.
Perls Staining of Fe
Perls staining of Arabidopsis seedlings was performed as described (Stacey et al., 2008) with slight modifications. Ten-day-old wild-type C24 and cpl1-2 mutant seedlings were first washed with 10 mm EDTA (pH 8.0) solution for 5 min and then with distilled water three times. Next, seedlings were vacuum infiltrated with equal volumes of 4% (v/v) HCl and 4% (w/v) potassium ferrocyanide (Perls solution) for 30 min at room temperature and then incubated for 30 min at 53°C. Seedlings were then rinsed several times with distilled water and observed with a BX51 microscope (Olympus).
Statistical Analysis
Statistical analyses were performed with MINITAB 16 software (Minitab). The statistical significances of differences between mean values were determined using Student’s t test or one-way ANOVA followed by Tukey’s HSD post hoc test. Differences were considered significant when the P value was less than 0.05.
Sequence data from this article can be found in the Arabidopsis Genome Initiative or the GenBank/EMBL data libraries under the following accession numbers: CPL1 (AT4G21670), CPL2 (AT5G01270), CPL3 (AT2G33540), IRT1 (AT4G19690), FIT (AT2G28160), FRO2 (AT1G01580), BHLH38 (AT3G56970), BHLH39 (AT3G56980), BHLH100 (AT2G41240), BHLH101 (AT5G04150), FER1 (AT5G01600), ICL (AT3G21720), BGLU29 (AT2G44460), OPT3 (AT4G16370), AHP4 (AT3G16360), NAS4 (AT1G56430), LSU2 (AT5G24660), PPCK1 (AT1G08650), OPR (AT1G17990), HVA22b (AT5G62490), LEA4-2/LEA18 (AT2G35300), LEA4-5 (AT5G06760), BGLU30 (AT3G60140), ADC2 (AT4G34710), PAP17 (AT3G17790), TUB8 (AT5G23860), ACT2 (AT3G18780), AT5G26220, AT3G15670, AT3G02480, AT1G67600, AT5G07330, AT5G45690, AT5G66780, AT1G05510, AT3G58450, AT5G48850, AT5G59030, AT2G33070, AT1G36370, AT1G12030, AT4G15390, AT2G23110, AT3G50980, AT5G01300, AT5G48180, AT1G52690, AT3G17520, AT2G47770, AT5G66400, AT2G31980, AT2G40435, AT1G04560, AT1G18870, and AT3G49580.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Normalization and principal component analysis of the cpl1-2 and C24 microarray.
Supplemental Figure S2. Molecular characterization of the cpl1-5 and cpl1-6 mutants.
Supplemental Figure S3. The expression levels of IRT1 and FIT in F1 plants produced after crossing cpl1-2 with the wild type.
Supplemental Figure S4. Time-course analysis of expression levels of Fe utilization-related genes in the roots of cpl1-2 and C24 under Fe deficiency.
Supplemental Figure S5. The expression of Fe utilization-related genes in roots of cpl1-2 and C24 under Fe deficiency.
Supplemental Figure S6. Fe deposition in cpl1-2 and C24 roots.
Supplemental Figure S7. Molecular characterization of fit-2 and cpl1-6 mutations and the FIT-LUC transgene.
Supplemental Table S1. GSEA of cluster I and cluster II gene sets.
Supplemental Table S2. Categories of up-regulated genes in the cpl1-2 mutant, and comparison with relevant published microarray data sets.
Supplemental Table S3. GSEA of the gene sets presented in Figure 2.
Supplemental Table S4. RT-qPCR analysis of Fe utilization-related gene expression levels in different cpl1 alleles, cpl2-2 and cpl3-1.
Supplemental Table S5. List of primers used in the study for genotyping of mutant and transgenic lines and RT-qPCR.
Acknowledgments
We thank the Purdue Ionomics Information Management System for initial determination of the cpl1 ionome. We thank Dr. William D. James for assistance in ICP-MS analyses at the Texas A&M University Elemental Analysis Laboratory. We thank Dr. Greg Cobb and Ms. E. Ann Ellis for assistance in microscopy. We thank Dr. Paul M. Hasegawa for helpful discussions and critical reading of the manuscript and Dr. Xiaoqian Wu for technical assistance. We thank Dr. Paul Paré and Dr. Mary Lou Guerinot for providing fit-2. We thank NASC/GABI-kat and the Arabidopsis Biological Resource Center for providing T-DNA lines and a BAC clone. We thank Dr. Catherine Curie for anti-IRT1 antibodies, which were used earlier in this study.
Glossary
- Fe
iron
- ABA
abscisic acid
- pol II CTD
RNA polymerase II C-terminal domain
- Cd
cadmium
- MS
Murashige and Skoog
- HSD
honestly significant difference
- RT-qPCR
reverse transcription quantitative PCR
- GSEA
Gene Set Enrichment Analysis
- Col-0
ecotype Columbia
- ICP-MS
inductively coupled plasma mass spectrometry
- Zn
zinc
- Mn
manganese
- Na
sodium
- K
potassium
- NASC
Nottingham Arabidopsis Stock Center
- BAC
bacterial artificial chromosome
- T-DNA
transfer DNA
References
- Aouida M, Khodami-Pour A, Ramotar D. (2009) Novel role for the Saccharomyces cerevisiae oligopeptide transporter Opt2 in drug detoxification. Biochem Cell Biol 87: 653–661 [DOI] [PubMed] [Google Scholar]
- Backes C, Keller A, Kuentzer J, Kneissl B, Comtesse N, Elnakady YA, Muller R, Meese E, Lenhof HP. (2007) GeneTrail: advanced gene set enrichment analysis. Nucleic Acids Res 35: W186–W192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberon M, Zelazny E, Robert S, Conejero G, Curie C, Friml J, Vert G. (2011) Monoubiquitin-dependent endocytosis of the IRON-REGULATED TRANSPORTER 1 (IRT1) transporter controls iron uptake in plants. Proc Natl Acad Sci USA 108: 12985–12986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer P, Ling HQ, Guerinot ML. (2007) FIT, the FER-LIKE IRON DEFICIENCY INDUCED TRANSCRIPTION FACTOR in Arabidopsis. Plant Physiol Biochem 45: 260–261 [DOI] [PubMed] [Google Scholar]
- Bauer P, Thiel T, Klatte M, Bereczky Z, Brumbarova T, Hell R, Grosse I. (2004) Analysis of sequence, map position, and gene expression reveals conserved essential genes for iron uptake in Arabidopsis and tomato. Plant Physiol 136: 4169–4183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter I, Ouzzani M, Orcun S, Kennedy B, Jandhyala SS, Salt DE. (2007) Purdue Ionomics Information Management System: an integrated functional genomics platform. Plant Physiol 143: 600–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter IR, Vitek O, Lahner B, Muthukumar B, Borghi M, Morrissey J, Guerinot ML, Salt DE. (2008) The leaf ionome as a multivariable system to detect a plant’s physiological status. Proc Natl Acad Sci USA 105: 12081–12086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57: 289–300 [Google Scholar]
- Broadley M, Brown P, Cakmak I, Rengel Z, Zhao F. (2012) Function of nutrients: micronutrients. In P Marschner, ed, Marschner’s Mineral Nutrition of Higher Plants, Ed 3. Academic Press, London, pp 191–248. [Google Scholar]
- Buckhout TJ, Yang TJ, Schmidt W. (2009) Early iron-deficiency-induced transcriptional changes in Arabidopsis roots as revealed by microarray analyses. BMC Genomics 10: 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162: 156–159 [DOI] [PubMed] [Google Scholar]
- Chu HH, Chiecko J, Punshon T, Lanzirotti A, Lahner B, Salt DE, Walker EL. (2010) Successful reproduction requires the function of Arabidopsis Yellow Stripe-Like1 and Yellow Stripe-Like3 metal-nicotianamine transporters in both vegetative and reproductive structures. Plant Physiol 154: 197–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens S. (2006) Toxic metal accumulation, responses to exposure and mechanisms of tolerance in plants. Biochimie 88: 1707–1719 [DOI] [PubMed] [Google Scholar]
- Colangelo EP, Guerinot ML. (2004) The essential basic helix-loop-helix protein FIT1 is required for the iron deficiency response. Plant Cell 16: 3400–3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly EL, Fett JP, Guerinot ML. (2002) Expression of the IRT1 metal transporter is controlled by metals at the levels of transcript and protein accumulation. Plant Cell 14: 1347–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conte SS, Walker EL. (2011) Transporters contributing to iron trafficking in plants. Mol Plant 4: 464–476 [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiDonato RJ, Jr, Roberts LA, Sanderson T, Eisley RB, Walker EL. (2004) Arabidopsis Yellow Stripe-Like2 (YSL2): a metal-regulated gene encoding a plasma membrane transporter of nicotianamine-metal complexes. Plant J 39: 403–414 [DOI] [PubMed] [Google Scholar]
- Dinneny JR, Long TA, Wang JY, Jung JW, Mace D, Pointer S, Barron C, Brady SM, Schiefelbein J, Benfey PN. (2008) Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science 320: 942–945 [DOI] [PubMed] [Google Scholar]
- Durrett TP, Gassmann W, Rogers EE. (2007) The FRD3-mediated efflux of citrate into the root vasculature is necessary for efficient iron translocation. Plant Physiol 144: 197–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eide D, Broderius M, Fett J, Guerinot ML. (1996) A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc Natl Acad Sci USA 93: 5624–5628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis E. (2006) Corrected formulation for Spurr low viscosity embedding medium using the replacement epoxide ERL 4221. Microsc Microanal 12: 288–289 [Google Scholar]
- Fujita Y, Fujita M, Satoh R, Maruyama K, Parvez MM, Seki M, Hiratsu K, Ohme-Takagi M, Shinozaki K, Yamaguchi-Shinozaki K. (2005) AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 17: 3470–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda H, Sasaki E, Akiyama K, Maruyama-Nakashita A, Nakabayashi K, Li WQ, Ogawa M, Yamauchi Y, Preston J, Aoki K, et al. (2008) The AtGenExpress hormone and chemical treatment data set: experimental design, data evaluation, model data analysis and data access. Plant J 55: 526–542 [DOI] [PubMed] [Google Scholar]
- Goeres DC, Van Norman JM, Zhang W, Fauver NA, Spencer ML, Sieburth LE. (2007) Components of the Arabidopsis mRNA decapping complex are required for early seedling development. Plant Cell 19: 1549–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong JM, Lee DA, Schroeder JI. (2003) Long-distance root-to-shoot transport of phytochelatins and cadmium in Arabidopsis. Proc Natl Acad Sci USA 100: 10118–10123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green LS, Rogers EE. (2004) FRD3 controls iron localization in Arabidopsis. Plant Physiol 136: 2523–2531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory BD, O’Malley RC, Lister R, Urich MA, Tonti-Filippini J, Chen H, Millar AH, Ecker JR. (2008) A link between RNA metabolism and silencing affecting Arabidopsis development. Dev Cell 14: 854–866 [DOI] [PubMed] [Google Scholar]
- Gross J, Stein RJ, Fett-Neto AG, Fett JP. (2003) Iron homeostasis related genes in rice. Genet Mol Biol 26: 477–497 [Google Scholar]
- Grotz N, Guerinot ML. (2006) Molecular aspects of Cu, Fe and Zn homeostasis in plants. Biochim Biophys Acta 1763: 595–608 [DOI] [PubMed] [Google Scholar]
- Guerinot ML, Yi Y. (1994) Iron: nutritious, noxious, and not readily available. Plant Physiol 104: 815–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques R, Jásik J, Klein M, Martinoia E, Feller U, Schell J, Pais MS, Koncz C. (2002) Knock-out of Arabidopsis metal transporter gene IRT1 results in iron deficiency accompanied by cell differentiation defects. Plant Mol Biol 50: 587–597 [DOI] [PubMed] [Google Scholar]
- Hirsch J, Marin E, Floriani M, Chiarenza S, Richaud P, Nussaume L, Thibaud MC. (2006) Phosphate deficiency promotes modification of iron distribution in Arabidopsis plants. Biochimie 88: 1767–1771 [DOI] [PubMed] [Google Scholar]
- Hubbard KE, Nishimura N, Hitomi K, Getzoff ED, Schroeder JI. (2010) Early abscisic acid signal transduction mechanisms: newly discovered components and newly emerging questions. Genes Dev 24: 1695–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. (2003) Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 31: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov R, Brumbarova T, Bauer P. (2012) Fitting into the harsh reality: regulation of iron-deficiency responses in dicotyledonous plants. Mol Plant 5: 27–42 [DOI] [PubMed] [Google Scholar]
- Jakoby M, Wang HY, Reidt W, Weisshaar B, Bauer P. (2004) FRU (BHLH029) is required for induction of iron mobilization genes in Arabidopsis thaliana. FEBS Lett 577: 528–534 [DOI] [PubMed] [Google Scholar]
- Kerkeb L, Mukherjee I, Chatterjee I, Lahner B, Salt DE, Connolly EL. (2008) Iron-induced turnover of the Arabidopsis IRON-REGULATED TRANSPORTER1 metal transporter requires lysine residues. Plant Physiol 146: 1964–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D’Angelo C, Bornberg-Bauer E, Kudla J, Harter K. (2007) The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J 50: 347–363 [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Itai RN, Ogo Y, Kakei Y, Nakanishi H, Takahashi M, Nishizawa NK. (2009) The rice transcription factor IDEF1 is essential for the early response to iron deficiency, and induces vegetative expression of late embryogenesis abundant genes. Plant J 60: 948–961 [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Nakanishi H, Nishizawa NK. (2010) Dual regulation of iron deficiency response mediated by the transcription factor IDEF1. Plant Signal Behav 5: 157–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Nakayama Y, Itai RN, Nakanishi H, Yoshihara T, Mori S, Nishizawa NK. (2003) Identification of novel cis-acting elements, IDE1 and IDE2, of the barley IDS2 gene promoter conferring iron-deficiency-inducible, root-specific expression in heterogeneous tobacco plants. Plant J 36: 780–793 [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Suzuki M, Inoue H, Itai RN, Takahashi M, Nakanishi H, Mori S, Nishizawa NK. (2005) Expression of iron-acquisition-related genes in iron-deficient rice is co-ordinately induced by partially conserved iron-deficiency-responsive elements. J Exp Bot 56: 1305–1316 [DOI] [PubMed] [Google Scholar]
- Koiwa H, Barb AW, Xiong L, Li F, McCully MG, Lee BH, Sokolchik I, Zhu J, Gong Z, Reddy M, et al. (2002) C-terminal domain phosphatase-like family members (AtCPLs) differentially regulate Arabidopsis thaliana abiotic stress signaling, growth, and development. Proc Natl Acad Sci USA 99: 10893–10898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koiwa H, Hausmann S, Bang WY, Ueda A, Kondo N, Hiraguri A, Fukuhara T, Bahk JD, Yun DJ, Bressan RA, et al. (2004) Arabidopsis C-terminal domain phosphatase-like 1 and 2 are essential Ser-5-specific C-terminal domain phosphatases. Proc Natl Acad Sci USA 101: 14539–14544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong WW, Yang ZM. (2010) Identification of iron-deficiency responsive microRNA genes and cis-elements in Arabidopsis. Plant Physiol Biochem 48: 153–159 [DOI] [PubMed] [Google Scholar]
- Korshunova YO, Eide D, Clark WG, Guerinot ML, Pakrasi HB. (1999) The IRT1 protein from Arabidopsis thaliana is a metal transporter with a broad substrate range. Plant Mol Biol 40: 37–44 [DOI] [PubMed] [Google Scholar]
- Kreps JA, Wu YJ, Chang HS, Zhu T, Wang X, Harper JF. (2002) Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol 130: 2129–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger C, Berkowitz O, Stephan UW, Hell R. (2002) A metal-binding member of the late embryogenesis abundant protein family transports iron in the phloem of Ricinus communis L. J Biol Chem 277: 25062–25069 [DOI] [PubMed] [Google Scholar]
- Kuhn JM, Schroeder JI. (2003) Impacts of altered RNA metabolism on abscisic acid signaling. Curr Opin Plant Biol 6: 463–469 [DOI] [PubMed] [Google Scholar]
- Li W, Schmidt W. (2010) A lysine-63-linked ubiquitin chain-forming conjugase, UBC13, promotes the developmental responses to iron deficiency in Arabidopsis roots. Plant J 62: 330–343 [DOI] [PubMed] [Google Scholar]
- Li YH, Lee KK, Walsh S, Smith C, Hadingham S, Sorefan K, Cawley G, Bevan MW. (2006) Establishing glucose- and ABA-regulated transcription networks in Arabidopsis by microarray analysis and promoter classification using a Relevance Vector Machine. Genome Res 16: 414–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling HQ, Bauer P, Bereczky Z, Keller B, Ganal M. (2002) The tomato fer gene encoding a bHLH protein controls iron-uptake responses in roots. Proc Natl Acad Sci USA 99: 13938–13943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingam S, Mohrbacher J, Brumbarova T, Potuschak T, Fink-Straube C, Blondet E, Genschik P, Bauer P. (2011) Interaction between the bHLH transcription factor FIT and ETHYLENE INSENSITIVE3/ETHYLENE INSENSITIVE3-LIKE1 reveals molecular linkage between the regulation of iron acquisition and ethylene signaling in Arabidopsis. Plant Cell 23: 1815–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long TA, Tsukagoshi H, Busch W, Lahner B, Salt DE, Benfey PN. (2010) The bHLH transcription factor POPEYE regulates response to iron deficiency in Arabidopsis roots. Plant Cell 22: 2219–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi M, Umezawa T, Nakashima K, Kidokoro S, Takasaki H, Fujita Y, Yamaguchi-Shinozaki K, Shinozaki K. (2010) Two closely related subclass II SnRK2 protein kinases cooperatively regulate drought-inducible gene expression. Plant Cell Physiol 51: 842–847 [DOI] [PubMed] [Google Scholar]
- Møller IM, Jensen PE, Hansson A. (2007) Oxidative modifications to cellular components in plants. Annu Rev Plant Biol 58: 459–481 [DOI] [PubMed] [Google Scholar]
- Morrissey J, Baxter IR, Lee J, Li L, Lahner B, Grotz N, Kaplan J, Salt DE, Guerinot ML. (2009) The ferroportin metal efflux proteins function in iron and cobalt homeostasis in Arabidopsis. Plant Cell 21: 3326–3338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustroph A, Zanetti ME, Jang CJ, Holtan HE, Repetti PP, Galbraith DW, Girke T, Bailey-Serres J. (2009) Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc Natl Acad Sci USA 106: 18843–18848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaminami K, Matsui A, Shinozaki K, Seki M. (2012) RNA regulation in plant abiotic stress responses. Biochim Biophys Acta 1819: 149–153 [DOI] [PubMed] [Google Scholar]
- Nakashima K, Fujita Y, Katsura K, Maruyama K, Narusaka Y, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. (2006) Transcriptional regulation of ABI3- and ABA-responsive genes including RD29B and RD29A in seeds, germinating embryos, and seedlings of Arabidopsis. Plant Mol Biol 60: 51–68 [DOI] [PubMed] [Google Scholar]
- Nishimura N, Yoshida T, Kitahata N, Asami T, Shinozaki K, Hirayama T. (2007) ABA-Hypersensitive Germination1 encodes a protein phosphatase 2C, an essential component of abscisic acid signaling in Arabidopsis seed. Plant J 50: 935–949 [DOI] [PubMed] [Google Scholar]
- Palmer CM, Guerinot ML. (2009) Facing the challenges of Cu, Fe and Zn homeostasis in plants. Nat Chem Biol 5: 333–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit JM, van Wuytswinkel O, Briat JF, Lobréaux S. (2001) Characterization of an iron-dependent regulatory sequence involved in the transcriptional control of AtFer1 and ZmFer1 plant ferritin genes by iron. J Biol Chem 276: 5584–5590 [DOI] [PubMed] [Google Scholar]
- Puig S, Andrés-Colás N, García-Molina A, Peñarrubia L. (2007) Copper and iron homeostasis in Arabidopsis: responses to metal deficiencies, interactions and biotechnological applications. Plant Cell Environ 30: 271–290 [DOI] [PubMed] [Google Scholar]
- Ramachandran V, Chen X. (2008) Small RNA metabolism in Arabidopsis. Trends Plant Sci 13: 368–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rellán-Alvarez R, Giner-Martínez-Sierra J, Orduna J, Orera I, Rodríguez-Castrillón JA, García-Alonso JI, Abadía J, Alvarez-Fernández A. (2010) Identification of a tri-iron(III), tri-citrate complex in the xylem sap of iron-deficient tomato resupplied with iron: new insights into plant iron long-distance transport. Plant Cell Physiol 51: 91–102 [DOI] [PubMed] [Google Scholar]
- Robinson NJ, Procter CM, Connolly EL, Guerinot ML. (1999) A ferric-chelate reductase for iron uptake from soils. Nature 397: 694–697 [DOI] [PubMed] [Google Scholar]
- Rogers EE, Guerinot ML. (2002) FRD3, a member of the multidrug and toxin efflux family, controls iron deficiency responses in Arabidopsis. Plant Cell 14: 1787–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romheld V. (1987) Different strategies for iron acquisition in higher-plants. Physiol Plant 70: 231–234 [Google Scholar]
- Salzman RA, Brady JA, Finlayson SA, Buchanan CD, Summer EJ, Sun F, Klein PE, Klein RR, Pratt LH, Cordonnier-Pratt MM, et al. (2005) Transcriptional profiling of sorghum induced by methyl jasmonate, salicylic acid, and aminocyclopropane carboxylic acid reveals cooperative regulation and novel gene responses. Plant Physiol 138: 352–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santi S, Schmidt W. (2009) Dissecting iron deficiency-induced proton extrusion in Arabidopsis roots. New Phytol 183: 1072–1084 [DOI] [PubMed] [Google Scholar]
- Schmidt W. (1999) Mechanisms and regulation of reduction-based iron uptake in plants. New Phytol 141: 1–26 [Google Scholar]
- Schmidt W, Buckhout TJ. (2011) A hitchhiker’s guide to the Arabidopsis ferrome. Plant Physiol Biochem 49: 462–470 [DOI] [PubMed] [Google Scholar]
- Schuler M, Keller A, Backes C, Philippar K, Lenhof HP, Bauer P. (2011) Transcriptome analysis by GeneTrail revealed regulation of functional categories in response to alterations of iron homeostasis in Arabidopsis thaliana. BMC Plant Biol 11: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Séguéla M, Briat JF, Vert G, Curie C. (2008) Cytokinins negatively regulate the root iron uptake machinery in Arabidopsis through a growth-dependent pathway. Plant J 55: 289–300 [DOI] [PubMed] [Google Scholar]
- Seki M, Ishida J, Narusaka M, Fujita M, Nanjo T, Umezawa T, Kamiya A, Nakajima M, Enju A, Sakurai T, et al. (2002a) Monitoring the expression pattern of around 7,000 Arabidopsis genes under ABA treatments using a full-length cDNA microarray. Funct Integr Genomics 2: 282–291 [DOI] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Ishida J, Nanjo T, Fujita M, Oono Y, Kamiya A, Nakajima M, Enju A, Sakurai T, et al. (2002b) Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J 31: 279–292 [DOI] [PubMed] [Google Scholar]
- Sivitz A, Grinvalds C, Barberon M, Curie C, Vert G. (2011) Proteasome-mediated turnover of the transcriptional activator FIT is required for plant iron-deficiency responses. Plant J 66: 1044–1052 [DOI] [PubMed] [Google Scholar]
- Stacey MG, Patel A, McClain WE, Mathieu M, Remley M, Rogers EE, Gassmann W, Blevins DG, Stacey G. (2008) The Arabidopsis AtOPT3 protein functions in metal homeostasis and movement of iron to developing seeds. Plant Physiol 146: 589–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibaud MC, Arrighi JF, Bayle V, Chiarenza S, Creff A, Bustos R, Paz-Ares J, Poirier Y, Nussaume L. (2010) Dissection of local and systemic transcriptional responses to phosphate starvation in Arabidopsis. Plant J 64: 775–789 [DOI] [PubMed] [Google Scholar]
- Ueda A, Li P, Feng Y, Vikram M, Kim S, Kang CH, Kang JS, Bahk JD, Lee SY, Fukuhara T, et al. (2008) The Arabidopsis thaliana carboxyl-terminal domain phosphatase-like 2 regulates plant growth, stress and auxin responses. Plant Mol Biol 67: 683–697 [DOI] [PubMed] [Google Scholar]
- Varotto C, Maiwald D, Pesaresi P, Jahns P, Salamini F, Leister D. (2002) The metal ion transporter IRT1 is necessary for iron homeostasis and efficient photosynthesis in Arabidopsis thaliana. Plant J 31: 589–599 [DOI] [PubMed] [Google Scholar]
- Vert G, Grotz N, Dédaldéchamp F, Gaymard F, Guerinot ML, Briat JF, Curie C. (2002) IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 14: 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldo GS, Wright E, Whang ZH, Briat JF, Theil EC, Sayers DE. (1995) Formation of the ferritin iron mineral occurs in plastids. Plant Physiol 109: 797–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HY, Klatte M, Jakoby M, Bäumlein H, Weisshaar B, Bauer P. (2007) Iron deficiency-mediated stress regulation of four subgroup Ib BHLH genes in Arabidopsis thaliana. Planta 226: 897–908 [DOI] [PubMed] [Google Scholar]
- Waters BM, Chu HH, Didonato RJ, Roberts LA, Eisley RB, Lahner B, Salt DE, Walker EL. (2006) Mutations in Arabidopsis yellow stripe-like1 and yellow stripe-like3 reveal their roles in metal ion homeostasis and loading of metal ions in seeds. Plant Physiol 141: 1446–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters BM, Grusak MA. (2008) Whole-plant mineral partitioning throughout the life cycle in Arabidopsis thaliana ecotypes Columbia, Landsberg erecta, Cape Verde Islands, and the mutant line ysl1ysl3. New Phytol 177: 389–405 [DOI] [PubMed] [Google Scholar]
- Welch RM. (1995) Micronutrient nutrition of plants. Crit Rev Plant Sci 14: 49–82 [Google Scholar]
- White JP. (2012) Ion uptake mechanisms of individual cells and roots: short-distance transport. In P Marschner, ed, Marschner’s Mineral Nutrition of Higher Plants, Ed 3. Academic Press, London, pp 7–47. [Google Scholar]
- Wintz H, Fox T, Wu YY, Feng V, Chen W, Chang HS, Zhu T, Vulpe C. (2003) Expression profiles of Arabidopsis thaliana in mineral deficiencies reveal novel transporters involved in metal homeostasis. J Biol Chem 278: 47644–47653 [DOI] [PubMed] [Google Scholar]
- Wu H, Chen C, Du J, Liu H, Cui Y, Zhang Y, He Y, Wang Y, Chu C, Feng Z, et al. (2012) Co-overexpression FIT with AtbHLH38 or AtbHLH39 in Arabidopsis-enhanced cadmium tolerance via increased cadmium sequestration in roots and improved iron homeostasis of shoots. Plant Physiol 158: 790–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Lee H, Ishitani M, Tanaka Y, Stevenson B, Koiwa H, Bressan RA, Hasegawa PM, Zhu J-K. (2002) Repression of stress-responsive genes by FIERY2, a novel transcriptional regulator in Arabidopsis. Proc Natl Acad Sci USA 99: 10899–10904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang TJW, Lin WD, Schmidt W. (2010) Transcriptional profiling of the Arabidopsis iron deficiency response reveals conserved transition metal homeostasis networks. Plant Physiol 152: 2130–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi Y, Guerinot ML. (1996) Genetic evidence that induction of root Fe(III) chelate reductase activity is necessary for iron uptake under iron deficiency. Plant J 10: 835–844 [DOI] [PubMed] [Google Scholar]
- Yuan Y, Wu H, Wang N, Li J, Zhao W, Du J, Wang D, Ling HQ. (2008) FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Res 18: 385–397 [DOI] [PubMed] [Google Scholar]
- Yuan YX, Zhang J, Wang DW, Ling HQ. (2005) AtbHLH29 of Arabidopsis thaliana is a functional ortholog of tomato FER involved in controlling iron acquisition in strategy I plants. Cell Res 15: 613–621 [DOI] [PubMed] [Google Scholar]
- Zhang H, Sun Y, Xie X, Kim MS, Dowd SE, Paré PW. (2009) A soil bacterium regulates plant acquisition of iron via deficiency-inducible mechanisms. Plant J 58: 568–577 [DOI] [PubMed] [Google Scholar]