Abstract
Soybean (Glycine max) is an important crop around the world. Abiotic stress conditions, such as drought and heat, adversely affect its survival, growth, and production. The DEHYDRATION-RESPONSIVE ELEMENT-BINDING PROTEIN2 (DREB2) group includes transcription factors that contribute to drought and heat stress tolerance by activating transcription through the cis-element dehydration-responsive element (DRE) in response to these stress stimuli. Two modes of regulation, transcriptional and posttranslational, are important for the activation of gene expression by DREB2A in Arabidopsis (Arabidopsis thaliana). However, the regulatory system of DREB2 in soybean is not clear. We identified a new soybean DREB2 gene, GmDREB2A;2, that was highly induced not only by dehydration and heat but also by low temperature. GmDREB2A;2 exhibited a high transactivation activity via DRE and has a serine/threonine-rich region, which corresponds to a negative regulatory domain of DREB2A that is involved in its posttranslational regulation, including destabilization. Despite the partial similarity between these sequences, the activity and stability of the GmDREB2A;2 protein were enhanced by removal of the serine/threonine-rich region in both Arabidopsis and soybean protoplasts, suggestive of a conserved regulatory mechanism that involves the recognition of serine/threonine-rich sequences with a specific pattern. The heterologous expression of GmDREB2A;2 in Arabidopsis induced DRE-regulated stress-inducible genes and improved stress tolerance. However, there were variations in the growth phenotypes of the transgenic Arabidopsis, the induced genes, and their induction ratios between GmDREB2A;2 and DREB2A. Therefore, the basic function and regulatory machinery of DREB2 have been maintained between Arabidopsis and soybean, although differentiation has also occurred.
Soybean (Glycine max) is the most important legume crop in the world, and its production is increasing. Abiotic stress is a factor that causes a significant loss of production, and tolerance to stress conditions, such as drought, is an important agronomic trait of soybean. Additionally, drought is occasionally accompanied by heat stress in the field, and such a combination more severely affects the growth and production of plants than a single stress condition (Mittler, 2006). Therefore, increasing tolerance to multiple stresses is an important target for improving the performance of soybean in the field.
Because of their sessile growth behavior, plants have evolved complex, accurate, and precise systems to respond and adapt to ever-fluctuating environmental conditions. Many aspects of adaptation processes, which include developmental, physiological, and biochemical changes, are regulated by stress-responsive gene expression. Transcription factors play central roles in the regulation of target gene expression via specific binding to cis-acting elements in their promoters. Analyses of stress-responsive promoters have identified cis- and trans-acting elements involved in transcriptional responses to stress stimuli (Yamaguchi-Shinozaki and Shinozaki, 2006).
The dehydration-responsive element (DRE), which has a core sequence of A/GCCGAC, is a cis-acting element originally isolated from the promoter of the RD29A gene of Arabidopsis (Arabidopsis thaliana) and is involved in both cold- and dehydration-inducible gene expression via an abscisic acid (ABA)-independent pathway (Yamaguchi-Shinozaki and Shinozaki, 1994). DEHYDRATION-RESPONSIVE ELEMENT-BINDING PROTEIN2A (DREB2A) is a transcriptional activator that recognizes DRE in Arabidopsis. It is a member of the DREB subfamily within the APETALA2/ethylene responsive-element binding factor (AP2/ERF) family of transcription factors and contains a single conserved DNA-binding domain (Liu et al., 1998; Sakuma et al., 2002). Its expression is induced by dehydration, high salinity, and heat shock (Liu et al., 1998; Sakuma et al., 2006b). The transcription of DREB2A in response to dehydration and heat shock is independently regulated by different regions in the DREB2A promoter (Kim et al., 2011; Yoshida et al., 2011). The region that is responsible for dehydration inducibility contains two essential elements, a coupling element3-like sequence and an ABA-responsive element (ABRE; Kim et al., 2011), while a heat shock element in another region is necessary for the inducibility of DREB2A in response to heat shock (Yoshida et al., 2011). The inducibility in response to stress conditions is a common feature among DREB2-type transcription factors of various plants, and the promoter structure including these cis-elements is conserved among many eudicot species (Kim et al., 2011; Mizoi et al., 2012). In Poaceae (grass family), posttranscriptional control by alternative splicing works as a key regulatory mechanism of DREB2-type transcription factors, although this alternative splicing mechanism is not found in other plant families (Shen et al., 2003; Qin et al., 2007; Matsukura et al., 2010). In Arabidopsis, the accumulation of DREB2A mRNA is not sufficient for the induction of downstream genes, and posttranslational regulation is important in activating DREB2A (Liu et al., 1998; Sakuma et al., 2006a). Sakuma et al. (2006a) found that the removal of a negative regulatory domain (NRD) just downstream of the DNA-binding domain converts this protein into a constitutively active form (DREB2A CA). The overexpression of DREB2A CA in transgenic Arabidopsis resulted in the induction of dehydration- or heat shock-inducible genes and enhanced tolerance to both drought and heat shock, indicating the substantial importance of posttranslational regulation (Sakuma et al., 2006a, 2006b). The stability of DREB2A in Arabidopsis cells is increased by removal of the NRD, suggesting the involvement of the NRD in destabilizing the DREB2A protein (Sakuma et al., 2006a). The destabilization of DREB2A is mediated by the ubiquitin-proteasome pathway and is partly regulated by specific E3 ubiquitin ligases (Qin et al., 2008). However, the regulatory mechanism of stability by NRD is not clear. Whether this posttranslational regulation is common among other plant species is also unclear, although the removal of a region that corresponds to the DREB2A NRD from MtDREB2A of Medicago truncatula enhanced activity in yeast (Saccharomyces cerevisiae; Chen et al., 2009a).
Many DREB subfamily transcription factors have been isolated from various plant species other than Arabidopsis, and their involvement in stress tolerance has been proposed (Mizoi et al., 2012). In soybean, the DRE cis-element or the combination of DRE and ABRE is enriched in the promoters of dehydration stress-responsive genes, suggesting the involvement of DRE in the dehydration stress response of soybean (Maruyama et al., 2012). According to Sakuma et al. (2002), the DREB subfamily of the AP2/ERF-type proteins is further classified into A-1 to A-6 subgroups, and DREB2A is a member of the A-2 subgroup. To date, several DREB subfamily transcription factors have been isolated from soybean and have been classified into subgroups A-1 (Glycine DREB1; Chen et al., 2006), A-2 (GmDREBa and GmDREBc; Li et al., 2005), and A-5 (GmDREB1–GmDREB3 and GmDREBb; Li et al., 2005; Chen et al., 2007, 2009b). We found that the A-2 subgroup, as well as most of the other subgroups in the DREB and ERF subfamilies, had been already established in an early stage of land plant evolution, suggesting functional differentiation between these subgroups (Mizoi et al., 2012). Although GmDREBa and GmDREBc belong to the A-2 subgroup, their C-terminal activation domains are short, different from the A-2 subgroup members that have been isolated from other plants, such as ORCA1 of periwinkle (Catharanthus roseus; Menke et al., 1999), HaDREB2 of sunflower (Helianthus annuus; Díaz-Martín et al., 2005), HvDRF1 of barley (Hordeum vulgare; Xue and Loveridge, 2004), ZmDREB2A of corn (Zea mays; Qin et al., 2007), and OsDREB2B of rice (Oryza sativa; Matsukura et al., 2010). Therefore, we suspected that there may be an unidentified canonical ortholog of DREB2A that functions in the heat and dehydration stress responses of soybean.
The release of the soybean genome sequence enabled us to analyze the composition of DREB2-type transcription factors. In this study, we identified two GmDREB2A homologs that encode canonical DREB2A orthologs and were highly stress inducible. One of these homologs, GmDREB2A;2, activated transcription through the cis-element DRE, whereas GmDREBa and GmDREBc did not. The activity and stability of GmDREB2A;2 were posttranslationally regulated in both Arabidopsis and soybean cells. In addition, GmDREB2A;2 could induce the expression of DREB2A target genes and improve heat shock or drought tolerance in transgenic Arabidopsis. The results obtained in this study indicate that GmDREB2A;2 is a functional ortholog of DREB2A in soybean. However, there are differences between DREB2A and GmDREB2A;2 in the negative regulatory sequence and in the effects on gene expression in transgenic plants expressing each protein. We propose that although the basic function and regulatory machinery are conserved between Arabidopsis and soybean, specifications in the usage and regulation of DREB2 have also occurred.
RESULTS
Identification and Classification of DREB2-Type Transcription Factors in Soybean
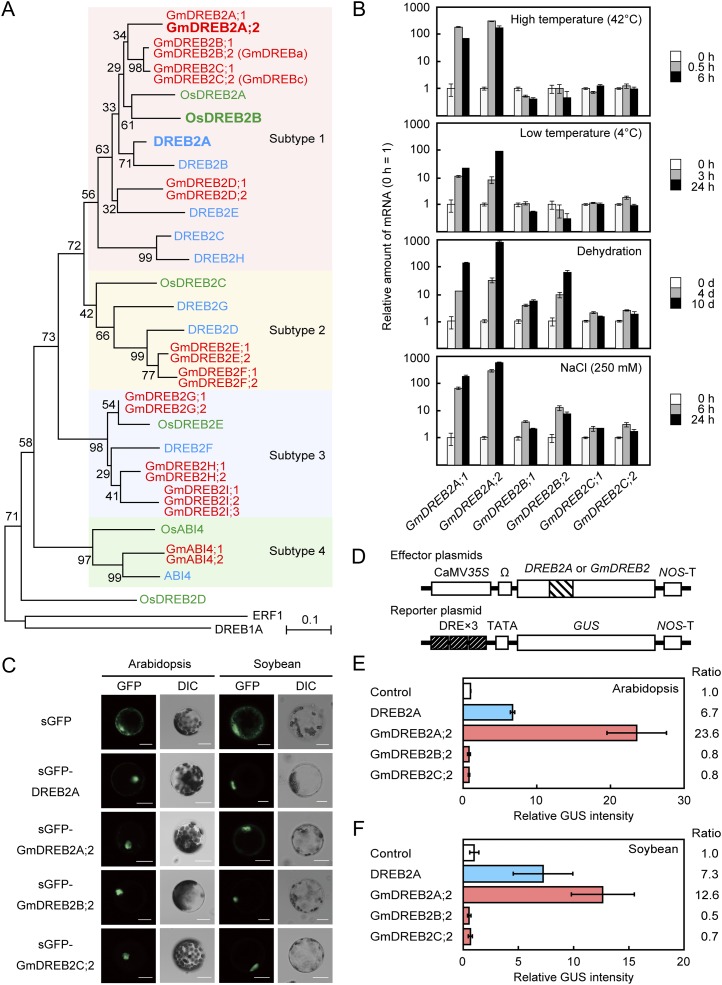
As the whole genomic sequence of soybean is now available (Schmutz et al., 2010), we searched for loci that could encode DREB2-type transcription factors in the genome. We used the peptide sequences of DREB2-type transcription factors in Arabidopsis as queries for BLASTP and TBLASTN similarity searches in the soybean genomic database at Phytozome (http://www.phytozome.net/; Goodstein et al., 2012) and identified candidate loci. Then, we deduced and classified the peptide sequences of the AP2/ERF-type DNA-binding domain for each identified candidate. As a result, 21 putative loci were classified as DREB2-type transcription factors (i.e. A-2 or A-3 subgroup; Sakuma et al., 2002), which share the common conserved motif CMIV-1 (Nakano et al., 2006; Supplemental Fig. S1; Supplemental Table S1). We reported that the DREB2-type transcription factors in Arabidopsis and rice are classified into four subtypes (Matsukura et al., 2010). Therefore, we analyzed phylogenetic relationships among the 21 candidates in soybean and all DREB2-type transcription factors in Arabidopsis and rice (Fig. 1A). We found that the 21 loci in soybean consisted of nine pairs and one group of very similar genes, which represent homologs derived from a whole-genome duplication event in the genus Glycine (Schmutz et al., 2010) and that subtypes 1, 2, 3, and 4 in soybean contain eight (four pairs), four (two pairs), seven (two pairs and one group), and two (one pair) members, respectively (Fig. 1A). We systematically named these 21 loci according to their phylogenetic relationships; GmDREB2A to GmDREB2I or GmABI4 prior to the semicolon indicate the homologous group to which each gene belongs, and the numbers following the semicolon were given according to the chromosome number on which each gene resides (Supplemental Table S1). In Arabidopsis and rice, the subtype 1 genes DREB2A and OsDREB2B have the highest stress inducibility among the DREB2 genes and have been shown to play significant roles in DRE-mediated stress responses in both plant species (Sakuma et al., 2002, 2006a, 2006b; Matsukura et al., 2010). In addition, many stress-inducible DREB2-type transcription factors that have been identified to date belong to subtype 1 (Shen et al., 2003; Xue and Loveridge, 2004; Egawa et al., 2006; Agarwal et al., 2007; Qin et al., 2007; Matsukura et al., 2010). Therefore, we focused on the eight subtype 1 genes: GmDREB2A;1, GmDREB2A;2, GmDREB2B;1, GmDREB2B;2, GmDREB2C;1, GmDREB2C;2, GmDREB2D;1, and GmDREB2D;2. GmDREB2B;2 and GmDREB2C;2 are identical to the previously described GmDREBa and GmDREBc genes, respectively (Li et al., 2005).
Figure 1.
Identification of GmDREB2A;2 as a functional ortholog of DREB2A in soybean. A, Phylogenetic tree of DREB2-type transcription factors in soybean (red), Arabidopsis (blue), and rice (green). The neighbor-joining tree was constructed based on an alignment of the peptide sequences of the N-terminal conserved region and the DNA-binding domain (Supplemental Fig. S1). The numbers on the side of each branch indicate bootstrap values from 1,000 replicates, and the scale bar indicates the substitution rate per site. Each subtype is indicated by a colored rectangle. B, Stress inducibility of GmDREB2A, GmDREB2B, and GmDREB2C homologs. The amount of each mRNA at 0 h is set to 1. The values represent the averages of three replicates, and the error bars indicate sd. C, Localization of sGFP fusion DREB2 proteins transiently expressed in Arabidopsis mesophyll protoplasts (left) or soybean stem parenchyma protoplasts (right). Confocal images of the GFP field and differential interference contrast (DIC) images are shown. Bars = 20 µm. D, Schematic diagrams of the reporter and effector constructs used in the experiments. The effector constructs contain the CaMV 35S promoter and the tobacco mosaic virus Ω sequence fused to the DREB2A or GmDREB2 coding sequences. NOS-T indicates the polyadenylation signal of a nopaline synthase gene. The striped box indicates the AP2/ERF DNA-binding domain. The reporter construct contained 75-bp fragments of the RD29A promoter tandemly repeated three times (DRE×3), the RD29A minimal promoter containing a TATA sequence, and the GUS reporter gene. E, Transactivation of the DRE×3-GUS fusion gene by GmDREB2A;2 in Arabidopsis mesophyll protoplasts. To normalize for transfection efficiency and protoplast numbers, a plasmid containing a CaMV 35S promoter-ELUC fusion gene was cotransfected in each experiment. The values indicate the ratios of normalized GUS intensity relative to that obtained with the empty effector plasmid. The bars indicate sd values from more than three replicates. F, Transactivation of the DRE×3-GUS fusion gene by GmDREB2A;2 in soybean stem parenchyma protoplasts.
GmDREB2A Homologs Share High Sequence Similarity with DREB2A
Although the complementary DNAs (cDNAs) for GmDREB2B;2/GmDREBa and GmDREB2C;2/GmDREBc have been isolated (Li et al., 2005), those for the GmDREB2A and GmDREB2D homologs have not. Therefore, we determined the cDNA sequences for these genes using the RACE method. Then, we amplified and cloned these coding regions from cDNA pools prepared from stress-treated soybean according to the deduced start and stop codons. All four homologous pairs shared high similarity with subtype 1 DREB2s of Arabidopsis in the N-terminal region, including the DNA-binding domain (Supplemental Fig. S1). Among them, the GmDREB2A homologs were most similar to DREB2A; they share the conserved motif CMIV-3 (Nakano et al., 2006) in the C-terminal activation domain as well as a Ser/Thr-rich sequence just downstream of the DNA-binding domain, which corresponds to a portion of the NRD of DREB2A (Supplemental Fig. S2). The GmDREB2D homologs had a seven-amino acid insertion in the DNA-binding domain (Supplemental Figs. S1 and S2), which is a common feature that is shared with DREB2E, which has an 8-bp insertion at the same position (Supplemental Fig. S1). In addition, each locus for GmDREB2D;1, GmDREB2D;2, and DREB2E is neighbored by a locus for a multidrug and toxic compound extrusion family transporter (Glyma07g19210, Glyma18g43740, and At2g38330, respectively), suggesting a synteny between the GmDREB2D and DREB2E loci. Therefore, the GmDREB2D homologs appear to be most related to DREB2E. The GmDREB2B and GmDREB2C homologs are truncated proteins with a short C-terminal activation domain. These proteins are notably similar to CAP2 from chickpea (Cicer arietinum; Shukla et al., 2006). Similarity searches in the Phytozome database identified only one protein of this type, which is encoded by the Medtr2g103290 locus of M. truncatula. These findings suggest that this type of protein is specific to Fabaceae (legume) plants.
These phylogenetic analyses and sequence comparisons revealed that the GmDREB2A homologs were orthologous to DREB2A. In addition, we showed that the promoter structure is conserved among DREB2A and the GmDREB2A homologs and also among DREB2-type genes of many other eudicot species (Kim et al., 2011); they share a heat shock element and ABRE or coupling element3-like sequences, which are important for transcriptional induction in response to heat and dehydration, respectively (Kim et al., 2011; Yoshida et al., 2011). Therefore, we analyzed the function of the GmDREB2A homologs in detail.
GmDREB2A Genes Are Strongly Induced in Response to Abiotic Stress Stimuli
Next, we analyzed the expression profiles of the GmDREB2A homologs under high-temperature, low-temperature, dehydration, and high-salinity conditions. Two-week-old soil-grown plants were subjected to each stress treatment, and the gene expression in whole aerial tissues was analyzed using quantitative reverse transcription (qRT)-PCR. Under all four conditions, the two GmDREB2A homologs exhibited strong inducibility (Fig. 1B). The time course of GmDREB2A;2 expression is shown in Supplemental Figure S3. We also analyzed the expression profiles of GmDREB2B;2/GmDREBa, GmDREB2C;2/GmDREBc, and their homologs. The two GmDREB2B homologs were responsive to dehydration and high salinity, whereas the two GmDREB2C homologs did not exhibit significant increases in their transcript levels under these conditions (Fig. 1B). The salt inducibility of GmDREB2B;2/GmDREBa was consistent with a previous report (Li et al., 2005), but the rate of induction was lower than that of the GmDREB2A homologs (Fig. 1B).
GmDREB2A;2 Localizes in the Nucleus and Activates Transcription through the cis-Element DRE
We then characterized the localization and activity of the GmDREB2A;2 protein. GmDREB2A;2 was selected from the pair of GmDREB2A homologs because the induction of GmDREB2A;2 was stronger than that of GmDREB2A;1, although their peptide sequences were very similar to each other (88.6% identity for full-length proteins and 100% identity for the DNA-binding domain). At first, we analyzed the subcellular localization of the GmDREB2A;2 protein by the transient expression of a synthetic GFP (sGFP; Chiu et al., 1996) fusion protein in protoplasts derived from Arabidopsis mesophyll cells or soybean stem parenchyma cells. The sGFP fusion proteins of GmDREB2A;2, GmDREB2B;2/GmDREBa, and GmDREB2C;2/GmDREBc as well as the sGFP fusion protein of DREB2A exhibited a clear signal in the nucleus in both cell types, indicating that these GmDREB2 proteins localize to the nucleus (Fig. 1C).
Next, we measured the DRE-dependent transactivation activity of the GmDREB2A;2 protein and compared it with those of the DREB2A, GmDREB2B;2, and GmDREB2C;2 proteins using the transient expression systems of Arabidopsis and soybean protoplasts. The GUS reporter plasmid contained a triplicate repeat of a 71-bp fragment from the RD29A promoter that contains a DRE sequence (Fig. 1D; Liu et al., 1998). In Arabidopsis protoplasts, GmDREB2A;2 exhibited the highest activity, which was even higher than that of DREB2A (Fig. 1E). In contrast, neither GmDREB2B;2/GmDREBa nor GmDREB2C;2/GmDREBc transactivated the reporter (Fig. 1E). Similar results were obtained for the soybean protoplasts (Fig. 1F).
The GmDREB2A homologs were similar to DREB2A in their peptide sequences, promoter sequences, and expression patterns in response to the stress treatments (Fig. 1, A and B; Kim et al., 2011). GmDREB2A;2 was targeted to the nucleus and exhibited significant transactivation activity toward the DRE-containing reporter in Arabidopsis and soybean protoplasts (Fig. 1, C–F). Collectively, these findings suggest that among the 21 DREB2-type transcription factors in soybean, the two GmDREB2A homologs are functional orthologs of DREB2A that can regulate DRE-mediated transcription in the early stages of stress responses.
The Stability of GmDREB2A;2 Is Negatively Regulated at the Posttranslational Level
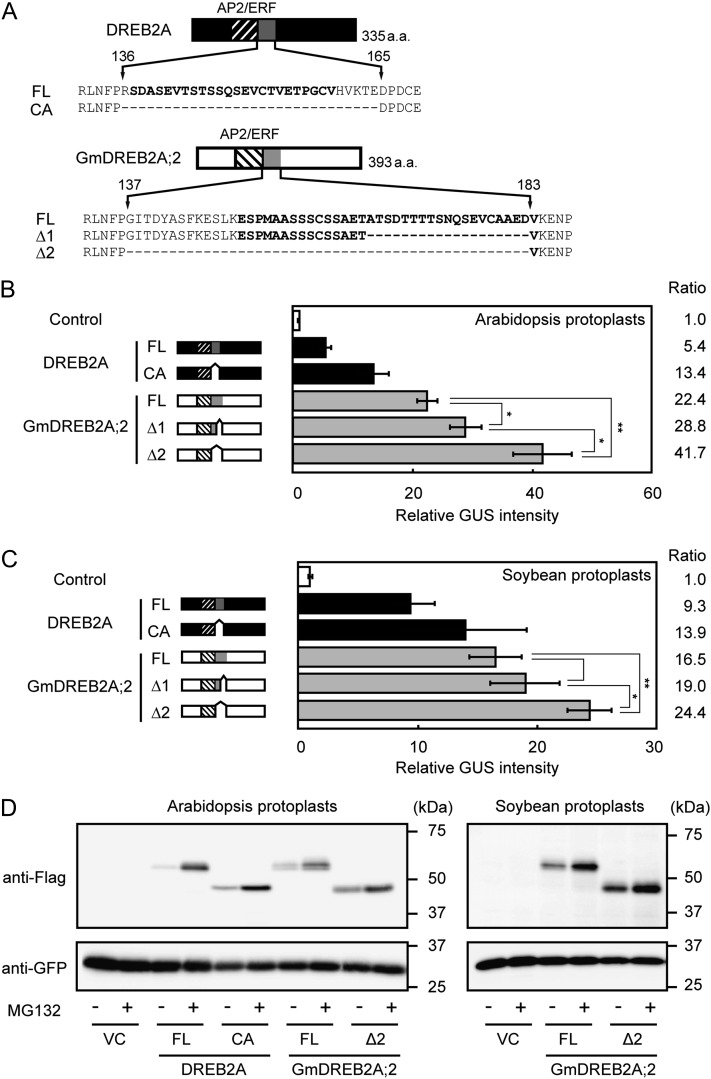
In Arabidopsis, the activity of the DREB2A protein is negatively regulated by the NRD, which is located immediately downstream of the DNA-binding domain and destabilizes the protein (Sakuma et al., 2006a). The NRD of DREB2A is a Ser/Thr-rich sequence that contains a PEST motif, which is a type of sequence identified from a number of unstable proteins (Rechsteiner and Rogers, 1996). Because GmDREB2A;2 has similarity to DREB2A in a region that corresponds to the NRD, we searched for a PEST sequence in this region using the epestfind algorithm (http://emboss.bioinformatics.nl/cgi-bin/emboss/epestfind). As a result, the region between amino acid residues 149 and 185 was identified as a potential PEST motif (PEST score = 11.46). To investigate whether the PEST-like sequence in GmDREB2A;2 can negatively regulate the activity of the protein, we generated two deletion mutants of this protein. One mutant (GmDREB2A;2 Δ1) lacks the region between Ala-165 and Asp-183, which corresponds to the region with high similarity to the NRD of DREB2A (Fig. 2A). The other mutant (GmDREB2A;2 Δ2) lacks a longer region from the residue adjacent to the DNA-binding domain (Gly-137) to Asp-183, which covers almost the full length of the potential PEST motif (Fig. 2A).
Figure 2.
Analysis of the negative regulation of the GmDREB2A;2 protein. A, Schematic diagrams of the DREB2A and GmDREB2A;2 proteins showing the NRD or PEST-like sequences. The positions of these sequences are indicated by gray boxes in the two protein models. The potential PEST sequences predicted by the epestfind algorithm are indicated by boldface letters, and the residues deleted in the mutant proteins are indicated by hyphens. The structures of the effector and reporter plasmids are the same as those in Figure 1. B and C, Effect of the deletion of GmDREB2A;2 on the transactivation of the DRE×3-GUS fusion gene in protoplasts isolated from Arabidopsis mesophyll cells (B) or soybean stem parenchyma cells (C). The schematic models of the analyzed proteins are given to the right of their names. Data represent averages and sd of four replicated samples. **P < 0.01, *P < 0.05, based on Student’s t test with the Bonferroni correction. D, Stability of the DREB2A and GmDREB2A;2 proteins in Arabidopsis (left) or soybean (right) protoplasts. The wild-type or mutant forms of the DREB2A or GmDREB2A;2 proteins were expressed as 3×Flag tag-fused proteins under the control of the CaMV 35S promoter and the tobacco mosaic virus Ω sequence. A plasmid constitutively expressing sGFP was cotransfected into the protoplasts as an internal control. Three replicates of the transfected protoplasts for each construct were combined, and the levels of the fusion proteins were analyzed by immunoblotting using an antibody against the 3×Flag tag. Protoplast suspensions were treated with MG132 for 1 h at a concentration of 50 µm. The amounts of the sGFP protein are shown as controls for equal loading. VC, Vector control.
First, we compared the transactivation activity of these mutants with the wild-type protein in Arabidopsis protoplasts using the DRE reporter (Fig. 2B). As reported by Sakuma et al. (2006a), deletion of the NRD from the full-length DREB2A protein (DREB2A FL) increased the transactivation activity (Fig. 2B; DREB2A CA). Similarly, the Δ1 and Δ2 mutants of GmDREB2A;2 exhibited higher activity than the full-length protein (GmDREB2A;2 FL), and the activity of the Δ2 mutant was significantly higher than that of the Δ1 mutant (Fig. 2B). We also measured the activity of these proteins in soybean protoplasts and obtained similar results (Fig. 2C). These results indicate that the PEST-like region in the GmDREB2A;2 protein can negatively regulate its function. In addition, although the C-terminal half of the PEST-like region that exhibits significant similarity to the NRD of DREB2A has a partial negative regulatory function, the full length of the PEST-like region appears to be necessary for full negative regulation.
The negative regulation of DREB2A by the NRD is associated with protein destabilization (Sakuma et al., 2006a). Therefore, we examined whether the high activity of GmDREB2A;2 Δ2 is associated with its stability. The full-length and deletion forms of DREB2A or GmDREB2A;2 were transiently expressed as 3× Flag-tagged proteins in Arabidopsis mesophyll protoplasts, and the levels of these proteins were analyzed by immunoblotting (Fig. 2D). sGFP was coexpressed in the protoplasts as an internal control. In the absence of the proteasome inhibitor MG132, which stabilizes DREB2A (Qin et al., 2008), DREB2A CA exhibited a stronger signal than DREB2A FL (Fig. 2D, left). Similarly, the accumulation level of GmDREB2A;2 Δ2 was higher than that of GmDREB2A;2 FL (Fig. 2D, left). In addition, similar to the DREB2A proteins, the GmDREB2A;2 proteins increased their accumulation levels in the presence of MG132 (Fig. 2D, left). To assess whether the protein level is an important determinant of the reporter activity, we varied the accumulation levels of DREB2 and GmDREB2A;2 proteins by changing the amounts of the effector plasmids (Supplemental Fig. S4). For both the wild-type and mutant proteins of DREB2A and GmDREB2A;2, the reporter activity was enhanced concurrent with an increased level of accumulation of each DREB2 protein, suggesting the importance of stability regulation for transactivation (Supplemental Fig. S4). To confirm that the enhancement of the reporter activity of GmDREB2A;2 by the Δ2 mutation is associated with the protein stability in soybean cells, we analyzed the stability of the GmDREB2A;2 protein in soybean protoplasts in a similar way and obtained a similar result (Fig. 2D, right). These results suggest that the PEST-like sequence of the GmDREB2A;2 protein can destabilize itself in a proteasome-dependent manner in soybean cells and that a posttranslational regulatory mechanism of the DREB2A orthologs is conserved between Arabidopsis and soybean.
Overexpression of an Active Form of GmDREB2A;2 Causes Growth Defects in the Germination and Seedling Stages
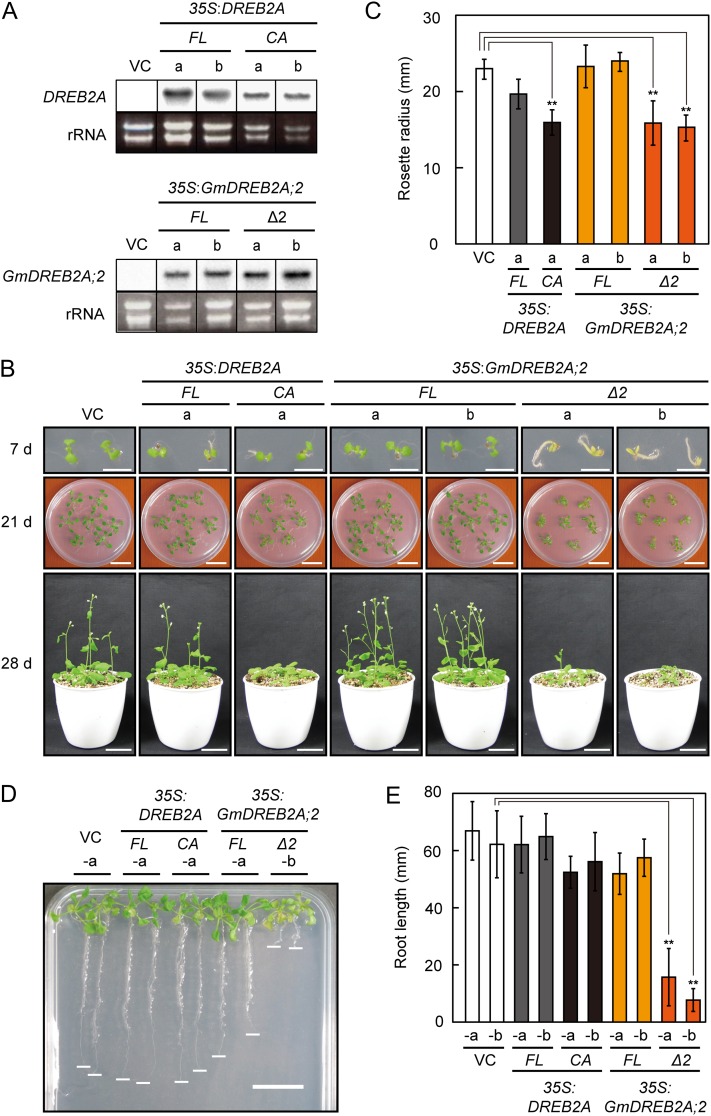
To test the function of GmDREB2A;2 in plants, we generated transgenic Arabidopsis expressing GmDREB2A;2 or GmDREB2A;2 Δ2 under the control of the cauliflower mosaic virus (CaMV) 35S promoter (35S:GmDREB2A;2 FL and 35S:GmDREB2A;2 Δ2, respectively; Fig. 3A). These transgenic plants were grown on agar plates for 3 weeks and later on soil for 1 week following transplantation. As previously reported, the overexpression of DREB2A FL did not cause any growth defects, whereas that of DREB2A CA resulted in a small rosette size and a delay in flowering (Fig. 3, B and C; 35S:DREB2A FL and 35S:DREB2A CA; Sakuma et al., 2006a). 35S:GmDREB2A;2 FL exhibited no significant morphological differences compared with the vector control plants throughout its growth (Fig. 3, B–E). In contrast, 35S:GmDREB2A;2 Δ2 exhibited a severe delay in the greening of cotyledons and hypocotyl swelling at 7 d, which was not observed in 35S:DREB2A CA (Fig. 3B). This phenotype was partially restored by reducing the Suc concentration in the medium from 3% to 0.75%, which suggests that this is a sugar-sensitive phenotype (Supplemental Fig. S5). We also compared the root growth of these transgenic plants on a vertically placed agar plate (Fig. 3D). The root growth of 35S:GmDREB2A;2 Δ2 was significantly reduced under this condition, whereas the overexpression of DREB2A CA or GmDREB2A;2 FL had only slight effects on root growth (Fig. 3, D and E). At 21 d, the rosette radii of 35S:GmDREB2A;2 Δ2 were as small as those of 35S:DREB2A CA (Fig. 3, B and C). The growth of 35S:GmDREB2A;2 Δ2 was even more severely affected than that of 35S:DREB2A CA after transplantation (Fig. 3B). The results of these growth analyses suggest that, similar to deletion of the NRD from DREB2A, deletion of the PEST-like sequence can enhance the function of GmDREB2A;2 in plants. However, the unique growth phenotype of 35S:GmDREB2A;2 Δ2 at the germination and seedling stages suggests a functional difference between the DREB2A and GmDREB2A;2 proteins.
Figure 3.
Growth of transgenic Arabidopsis plants expressing the wild-type or active form of GmDREB2A;2 under the control of the CaMV 35S promoter. A, RNA gel-blot analysis of transgenic Arabidopsis expressing the wild-type or active form of DREB2A or GmDREB2A;2. VC refers to a vector control plant, and the lowercase letters indicate two independent transgenic lines obtained using each construct. Ethidium bromide-stained images of ribosomal RNA (rRNA) are shown as loading controls. B, Growth of the generated transgenic plants. Seedlings of the generated transgenic lines were grown on agar plates for 3 weeks and then on soil for 1 week. Bars = 0.5 cm (7 d) and 2 cm (21 and 28 d). C, Rosette radii of the transgenic plants at 21 d. The radius of a rosette was defined as the radius of the smallest circle that could encompass a whole rosette. **P < 0.01 by Student’s t test with the Bonferroni correction (n = 12). D, Growth of 35S:DREB2A and 35S:GmDREB2A;2 transgenic plants on a vertically oriented medium. Seedlings were grown for 14 d under the normal condition and then grown on a vertically oriented medium for an additional 7 d. The white lines indicate the root tip. Bar = 1 cm. E, Average root lengths calculated for seedlings that were grown as shown in D. **P < 0.01 based on Student’s t test with the Bonferroni correction (n = 10).
GmDREB2A;2 Can Induce Not Only DREB2A Downstream Genes But Also Many Other Genes in Transgenic Arabidopsis
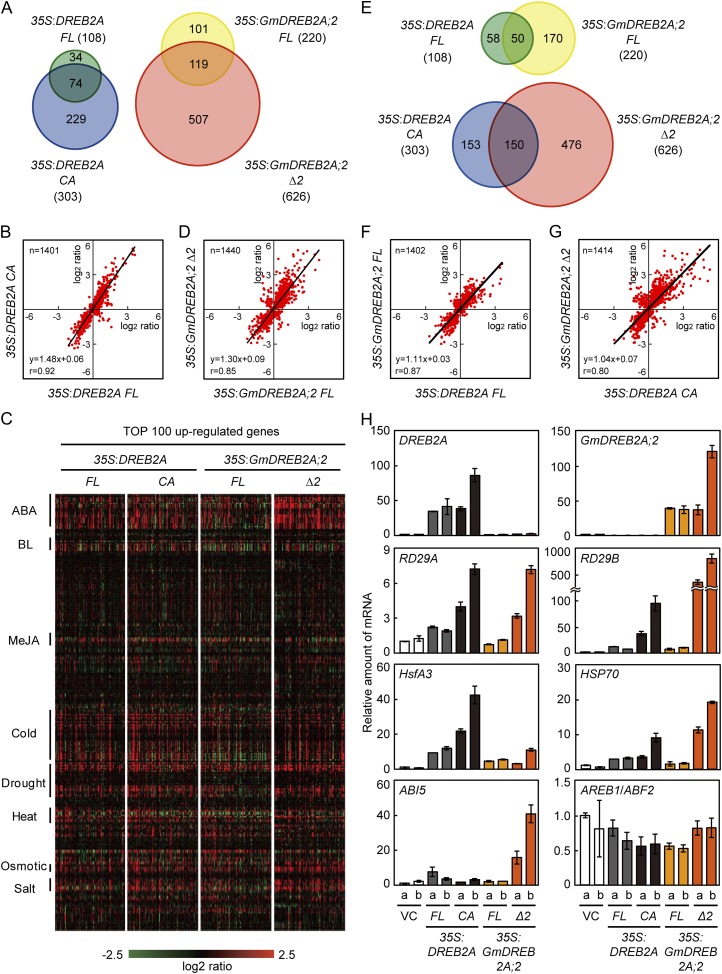
To compare the functions of GmDREB2A;2 and DREB2A in the activation of gene expression, we analyzed the gene expression profiles of the four transgenic plants. The overexpression of DREB2A FL and CA up-regulated the expression of 108 and 303 genes, respectively (fold change > 2, false discovery rate [FDR] P < 0.01, signal intensity > 200; Supplemental Tables S2 and S3), and 74 of the 108 genes (68.5%) were common between them (Fig. 4A). The expression ratios of genes that were significantly different from the vector control (FDR P < 0.01) showed a significant correlation between the two plants (r = 0.92), with the expression ratios being relatively higher in CA than in FL (regression line slope = 1.48; Fig. 4B). In addition, the expression patterns of the top 100 up-regulated genes were similar between the two plants (Fig. 4C). Therefore, deletion of the NRD from DREB2A likely did not change the target preference but merely enhanced their effects on expression levels. The overexpression of GmDREB2A;2 FL and Δ2 up-regulated the expression of 220 and 626 genes, respectively (fold change > 2, FDR P < 0.01, signal intensity > 200; Supplemental Tables S4 and S5), and 119 of the 220 genes (54.1%) were common between them (Fig. 4A). The expression levels of the downstream genes showed a correlation (r = 0.85), and the expression levels were generally higher in Δ2 than in FL (slope = 1.30; Fig. 4D). These phenomena associated with the deletion of the NRD or PEST-like sequence were similar between DREB2A and GmDREB2A;2, but 35S:GmDREB2A;2 Δ2 contained many additional downstream genes, such as those induced by ABA, compared with 35S:GmDREB2A;2 FL (Fig. 4, A and C; Supplemental Fig. S6).
Figure 4.
Comparative analysis of downstream genes of the wild-type and active forms of DREB2A and GmDREB2A;2 in transgenic Arabidopsis. A, Venn diagrams comparing the up-regulated genes between the 35S:DREB2A FL and CA plants (left) or between the 35S:GmDREB2A;2 FL and Δ2 plants (right). Genes that were significantly (FDR P < 0.01) up-regulated by more than 2-fold in these transgenic plants are shown. The total numbers of the up-regulated genes are shown in parentheses. B, Correlation analysis between the downstream genes of the wild-type and active forms of DREB2A. The induction ratios of genes that were significantly changed (FDR P < 0.01) in both of the transgenic plants are shown in the scatterplots together with the regression equation and correlation coefficient. C, Stress inducibility of the up-regulated genes in each transgenic plant. The expression ratios of the top 100 up-regulated genes in each plant (x axis, from left to right) in response to various hormone or stress treatments (y axis) are displayed as heat maps. BL, Brassinolide; MeJA, methyl jasmonate. D, Correlation analysis between the downstream genes of the wild-type and active forms of GmDREB2A;2. E, Venn diagrams comparing the up-regulated genes between the overexpressing plants of the wild-type (top) or active (bottom) forms of DREB2A and GmDREB2A;2. F and G, Correlation analysis between the wild-type (F) or active (G) forms of DREB2A and GmDREB2A;2. H, Transcript levels of the transgenes, DREB2A target genes, and ABA-related genes as analyzed by qRT-PCR. 18S ribosomal RNA was used for normalization of the data. For all genes, the amount of mRNA in the vector control plants was set to 1, with the exception that the amount of endogenous DREB2A mRNA in the vector control plants was set to 1 for GmDREBA2;2. VC refers to vector control plants, and the lowercase letters indicate the two independent transgenic lines obtained using each construct.
Next, we compared the downstream genes between the FL and active forms of DREB2A and GmDREB2A;2. More genes are up-regulated in 35S:GmDREB2A;2 FL than in 35S:DREB2A FL (Fig. 4E). Among the 108 genes up-regulated in 35S:DREB2A FL, 50 (46.3%) were shared with 35S:GmDREB2A;2 FL, and their downstream genes showed a correlation (r = 0.87; Fig. 4F). In addition, the expression patterns of the top 100 up-regulated genes were similar between these plants (Fig. 4C). A comparison between the downstream genes of the active forms of DREB2A and GmDREB2A;2 showed that, although 150 of the 303 genes (49.5%) up-regulated in 35S:DREB2A CA were shared with 35S:GmDREB2A;2 Δ2, many other genes were up-regulated in 35S:GmDREB2A;2 Δ2 (Fig. 4E; Supplemental Fig. S6). The correlation analysis suggests that the differences between the two sets of downstream genes are not caused by the overall intensity of the gene expression (slope = 1.04; Fig. 4G). Rather, this difference is attributed to a variable gene induction ratio, expressed as 35S:GmDREB2A;2 Δ2/35S:DREB2A CA (i.e. a group of genes is more highly induced in 35S:GmDREB2A;2 Δ2, whereas another group of genes show the opposite trend; Fig. 4G). This relationship is reflected by a lower correlation coefficient (r = 0.80) than those of other comparisons (Fig. 4, B, D, F, and G). These results suggest differences in the target preferences between DREB2A CA and GmDREB2A;2 Δ2.
We next confirmed the results of the microarray experiments by qRT-PCR (Fig. 4H). RD29A and RD29B are major dehydration-inducible targets of DREB2A, whereas HsfA3 and HSP70 are its major heat-inducible targets (Sakuma et al., 2006a, 2006b). The expression levels of these four genes were only slightly or moderately increased in 35S:DREB2A FL but further increased in 35S:DREB2A CA (Fig. 4H). Similarly, the effect of GmDREB2A;2 FL overexpression on the expression levels of these genes was small, whereas the overexpression of GmDREB2A;2 Δ2 resulted in marked increases in the transcript levels of the RD29A, RD29B, and HSP70 genes (Fig. 4H). Interestingly, the inducibility in 35S:DREB2A and 35S:GmDREB2A;2 transgenic plants varied for different genes. For example, RD29A was induced at similar levels in the 35S:DREB2A CA and 35S:GmDREB2A;2 Δ2 plants, whereas the induction of HsfA3 was stronger in the 35S:DREB2A plants than in the 35S:GmDREB2A;2 transgenic plants (Fig. 4H). This trend was consistent with the correlation analysis data (Fig. 4G). Because many ABA-inducible genes are specifically up-regulated in 35S:GmDREB2A;2 Δ2, we verified the expression ratios of genes that are involved in ABA-mediated gene expression and found that the expression level of the ABI5 gene was extraordinarily elevated in this plant, whereas AREB/ABF family transcription factor genes that function in vegetative tissues were not affected (Fig. 4H; Supplemental Table S5). These results confirm that GmDREB2A;2 can activate the expression of DREB2A target genes in Arabidopsis and that deletion of the PEST-like sequence from GmDREB2A;2 and deletion of the NRD from DREB2A can enhance the expression levels of target genes. Therefore, both GmDREB2A;2 and DREB2A activate gene expression in plants in a similar manner. However, functional variations are evident between the two proteins with respect to the determination of targets and their activation levels.
The DRE Core Sequence Is Enriched in the Downstream Genes of GmDREB2A;2
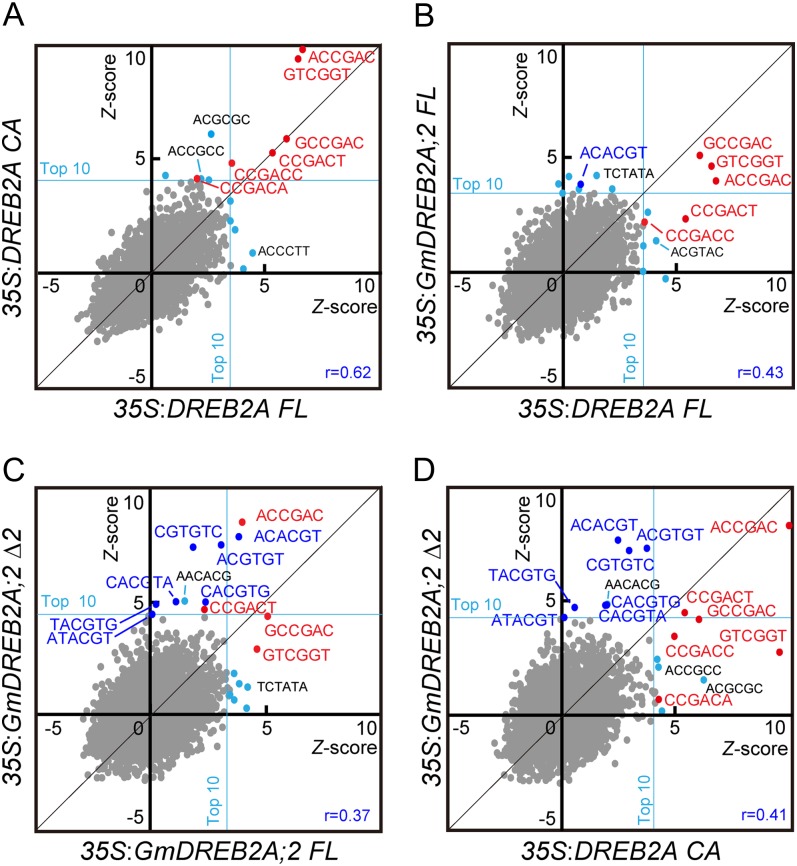
To characterize the promoters of the downstream genes, we analyzed the enrichment of hexamer motifs in the promoters of up-regulated genes in each plant. We compared the frequency of each hexamer sequence (46 = 4,096) in the promoters of the up-regulated genes with their normalized frequencies in Arabidopsis promoters according to Maruyama et al. (2012). In 35S:DREB2A CA and 35S:DREB2A FL, the most overrepresented sequence was ACCGAC, and the second was its complement sequence, GTCGGT, consistent with a previous report (Maruyama et al., 2012; Fig. 5A; Supplemental Table S6). ACCGAC is one of two forms of the DRE core sequence (A/GCCGAC) and is the most preferred target sequence of DREB2A (Sakuma et al., 2006a). The other form of DRE (GCCGAC) and partial DRE sequences (CCGACT and CCGACC) were also commonly overrepresented among the two plants (Fig. 5A; Supplemental Table S6). In 35S:GmDREB2A;2 FL, the DRE core sequences GCCGAC, GTCGGT, and ACCGAC were ranked within the top 10 (Fig. 5B; Supplemental Table S6), evidencing that the DRE core motif is the most preferred target of GmDREB2A;2 in plants. In 35S:GmDREB2A;2 Δ2, ACCGAC was the most overrepresented sequence, but many sequences related to the ABRE (PyACGTGG/TC) were ranked within the top 10 (Fig. 5, C and D; Supplemental Table S6). The difference is clearly evident in the comparison of 35S:GmDREB2A;2 Δ2 with 35S:GmDREB2A;2 CA: DRE and its related sequences appear in the top right region, whereas ABRE and its related sequences are enriched in the top middle region (Fig. 5D). A weak but similar relationship is indicated by a comparison between the plants that overexpress either of the full-length proteins (Fig. 5B). These analyses confirmed that GmDREB2A;2 prefers DRE as its binding target. The overrepresentation of ABRE in 35S:GmDREB2A;2 Δ2 appears to be indicative of a secondary effect that is associated with the possible up-regulation of an ABA-related pathway.
Figure 5.
Overrepresentation analysis of hexamer sequences in the promoters of highly up-regulated genes in the 35S:DREB2A and 35S:GmDREB2A;2 transgenic Arabidopsis plants. Promoter sequences encompassing 1,000-bp regions upstream of each transcriptional start site were obtained for the 100 most strongly up-regulated genes that had defined information on the transcriptional start sites. The quantities of all hexamer sequences in the selected 100 promoters were compared with their normalized frequencies in 100 randomly selected promoters, and the significance was evaluated using Z scores and fold enrichment values. The Z scores of all hexamers (46 = 4,096) were compared between two transgenic plants. Comparisons between 35S:DREB2A FL and CA (A), 35S:DREB2A FL and 35S:GmDREB2A;2 FL (B), 35S:GmDREB2A;2 FL and Δ2 (C), as well as 35S:DREB2A CA and 35S:GmDREB2A;2 Δ2 (D) are shown. Highly overrepresented hexamers that are related to DRE and ABRE are indicated in red and blue, respectively. The cyan dots are other hexamers that appear within the top 10. The position of the 10th hexamer is indicated by a cyan line, and the correlation coefficient is presented at the bottom right.
GmDREB2A;2 Can Improve the Heat and Drought Stress Tolerance of Arabidopsis
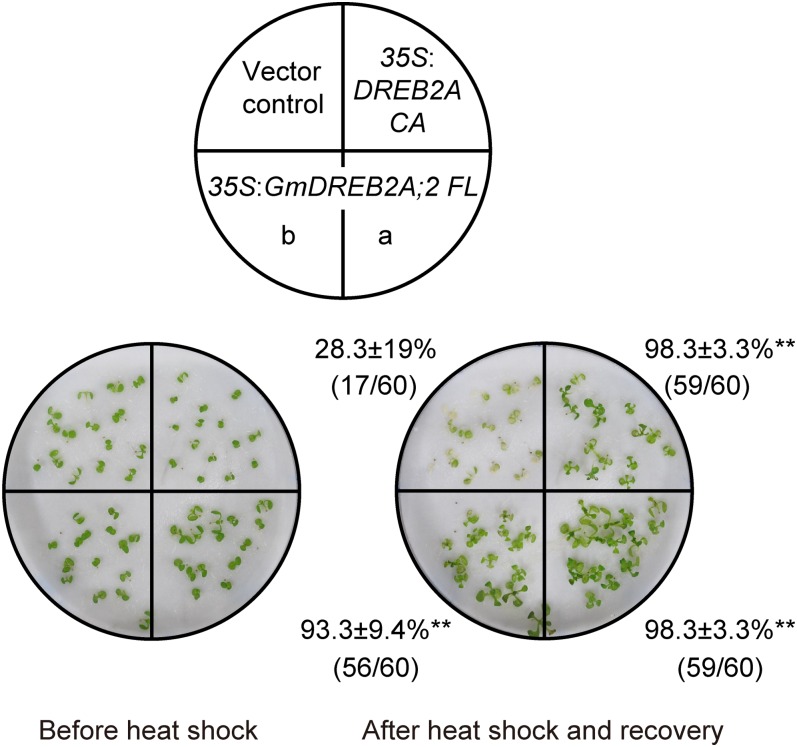
Because GmDREB2A;2 was found to enhance the expression of DREB2A downstream genes, we evaluated their function in the acquisition of heat or drought stress tolerance in plants. First, we evaluated the effect of GmDREB2A;2 overexpression on heat tolerance. Because the 35S:GmDREB2A;2 Δ2 transgenic plants showed growth defects at the germination and seedling stages (Fig. 3), we used the 35S:GmDREB2A;2 FL plants for heat stress tolerance tests. We applied a heat shock of 45°C for 60 min to 7-d-old seedlings of the vector control, 35S:DREB2A CA, and 35S:GmDREB2A;2 FL plants, which were then allowed to recover at 22°C for 6 d. While only less than 30% of the vector control plants survived, more than 90% of the 35S:DREB2A CA and 35S:GmDREB2A;2 FL transgenic plants survived (Fig. 6). This result indicates that the expression of GmDREB2A;2 can improve heat shock stress tolerance to an extent comparable to that of DREB2A CA.
Figure 6.
Improvement of heat shock tolerance by the heterologous overexpression of GmDREB2A;2. Seedlings of the vector control, 35S:DREB2A CA, and two lines of 35S:GmDREB2A;2 FL (a and b) were grown on germination medium agar plates at 22°C for 5 d and transferred onto wet filter paper. After 2 d, seedlings were treated at 45°C for 60 min. The survival rates were determined after recovery at 22°C for 7 d. Values are average survival rates and sd from four independent experiments; each experiment contained 15 seedlings of each plant. Numbers in the parentheses indicate total surviving seedlings/total tested seedlings of the four experiments. Asterisks indicate significant improvements in survival rates compared with the vector control (Student’s t test with the Bonferroni correction, **P < 0.01).
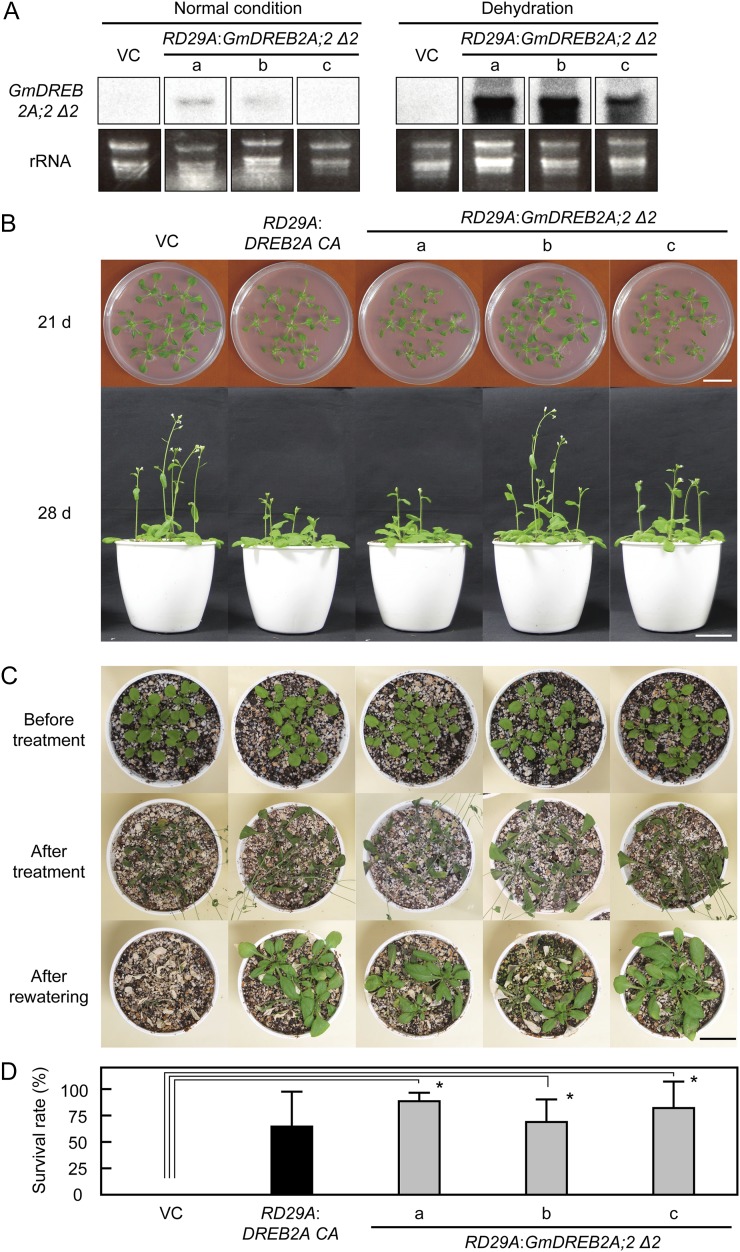
Although GmDREB2A;2 Δ2 has a higher transactivation activity and a stronger effect on the target gene expression levels in the transgenic plants than the FL protein, its overexpression severely affected plant growth (Fig. 3). Previously, we reported that the growth defects of transgenic plants overexpressing DREB1A/CBF3 or DREB2A CA can be overcome by the expression of these genes under the control of the dehydration-inducible RD29A promoter (Kasuga et al., 1999, 2004; Sakuma et al., 2006a). Therefore, we generated transgenic Arabidopsis plants that express GmDREB2A;2 Δ2 under the control of the RD29A promoter and evaluated their drought tolerance. In these transgenic plants, the expression of GmDREB2A;2 Δ2 was dehydration dependent (Fig. 7A), and the growth of these plants was almost comparable to that of the vector control plants (Fig. 7B). The vector control plants did not survive beyond the recovery following drought treatment, whereas the RD29A:GmDREB2A;2 Δ2 plants exhibited significant survival rates that were similar to those of the RD29A:DREB2A CA plants (Sakuma et al., 2006a; Fig. 7, C and D). Therefore, GmDREB2A;2 Δ2 has the ability to enhance the drought tolerance of transgenic plants at a level that is comparable to that of DREB2A CA.
Figure 7.
Improvement of drought tolerance by the dehydration-inducible expression of GmDREB2A;2. A, Generation of transgenic plants expressing GmDREB2A;2 Δ2 under the control of the dehydration-inducible RD29A promoter. The accumulation levels of GmDREB2A;2 Δ2 mRNA before (left) and after (right) dehydration are shown. VC refers to a vector control plant, and the lowercase letters indicate three independent transgenic lines. Ethidium bromide-stained images of ribosomal RNA (rRNA) are shown as loading controls. B, Growth of the generated RD29A:GmDREB2A;2 Δ2 transgenic plants. Seedlings were grown on agar medium for 21 d and later on soil for an additional 7 d. Bar = 2 cm. C and D, Drought tolerance test of the RD29A:GmDREB2A;2 Δ2 transgenic plants. Water was withheld for 2 weeks, and survival rates were determined after 1 week of recovery following rehydration. Images of the plants during the test (C; Bar = 2 cm) and survival rates (D) are shown. Asterisks in D indicate significant improvements in survival rates compared with the vector control (Student’s t tests with the Bonferroni correction, *P < 0.05; n = 3, 15 plants per test).
DISCUSSION
By leveraging genomic information, we identified 21 DREB2-type transcription factors in soybean. Among these factors, GmDREB2A;1 and GmDREB2A;2, which encode for peptides that share high sequence similarity with DREB2A (Fig. 1A; Supplemental Figs. S1 and S2), exhibited strong induction in response to heat and dehydration stresses (Fig. 1B). The rapid induction pattern of GmDREB2A;2 in response to heat shock is also characteristic of DREB2A, ZmDREB2A, and OsDREB2B (Supplemental Fig. S3; Sakuma et al., 2006b; Qin et al., 2007; Matsukura et al., 2010). The GmDREB2A genes are also induced by low temperature (Fig. 1B; Supplemental Fig. S3); this result contrasts with the lack of low-temperature inducibility of DREB2 genes in Arabidopsis (Liu et al., 1998; Sakuma et al., 2002). Although DREB1/CBF genes have important functions in DRE-mediated gene induction at low temperatures in Arabidopsis (Stockinger et al., 1997; Liu et al., 1998), the low-temperature inducibility of the GmDREB2A genes suggests that these DREB2-type transcription factors function in the DRE-mediated gene expression in response to low temperatures in soybean. Based on their expression patterns in response to stress conditions, together with their peptide sequences, the GmDREB2A homologs are considered to be critical members of the DREB2-type genes in soybean.
The GmDREB2B and GmDREB2C homologs, which include the previously isolated GmDREB2B;2/GmDREBa and GmDREB2C;2/GmDREBc (Li et al., 2005), are different from canonical subtype 1 members, as their C-terminal activation domain is short. An ortholog of these proteins was identified from chickpea (CAP2; Shukla et al., 2006), and the Phytozome database search suggested that they are specific to Fabaceae plants. Although GmDREB2B;2/GmDREBa and CAP2 can bind to the same DRE sequence that we used in the reporter plasmid and can activate transcription in yeast (Li et al., 2005; Shukla et al., 2006), we could not detect the transactivation activity of GmDREB2B;2/GmDREBa in Arabidopsis or soybean protoplasts (Fig. 1, E and F). Therefore, these Fabaceae-specific DREB2 genes are not the main transcriptional activators of the DRE-regulated pathways. However, it is interesting that these genes appear to be expressed even under normal conditions (Li et al., 2005). These unique genes may represent a Fabaceae-specific mechanism for the regulation of DRE-mediated gene transcription.
GmDREB2A;2 has a Ser/Thr-rich sequence just downstream of the DNA-binding domain, which corresponds to the NRD of DREB2A. Similar to the deletion of the NRD from DREB2A, complete deletion of this region from GmDREB2A;2 enhanced its activity and stability in both Arabidopsis and soybean protoplasts (Fig. 2). Therefore, the Ser/Thr-rich sequence of GmDREB2A;2 is functionally similar to the NRD of DREB2A with regard to its negative regulatory activity. These Ser/Thr-rich sequences contain a potential PEST motif with a core conserved sequence (167-SDTTTTSNQSEVCAAE-182 in GmDREB2A;2 and 140-SEVTSTSSQSEVCTVE-155 in DREB2A). However, compared with the NRD of DREB2A, this region in GmDREB2A;2 is long and has an additional cluster of Ser residues. The deletion analyses of the transient reporter assay systems of Arabidopsis and soybean protoplasts suggest that, despite its high similarity, the core sequence plays only a partial role and that the region containing the additional Ser cluster also has a significant function in the negative regulation of GmDREB2A;2 (Fig. 2). Because the Δ1 mutant only contains a poor PEST sequence (PEST score = 1.95), this region may have a more specific function rather than following the PEST rule. In many other DREB2A orthologs, such as ORCA1 (periwinkle; Menke et al., 1999), HaDREB2 (sunflower; Díaz-Martín et al., 2005), HvDRF1 (barley; Xue and Loveridge, 2004), and ZmDREB2A (corn; Qin et al., 2007), an additional sequence is located between the DNA-binding domain and the core conserved sequence, as is the case in GmDREB2A proteins. The mechanism underlying the negative regulation by these sequences is not clear; however, data obtained in this study and further comparisons between GmDREB2A;2 and DREB2A will provide important clues to understand the negative regulation of DREB2A orthologs in angiosperms.
The binding specificity of DREB2A has been well studied in vitro, and the DRE core sequence (A/GCCGAC) is most preferred, with A being more preferred at the first position (Sakuma et al., 2002, 2006a). However, the specificity may vary between plants, because HvDRF1 of barley shows a preference for TT/AACCGCCTT (Xue and Loveridge, 2004). In contrast to these comprehensive in vitro studies, the specificity of DREB2-type proteins in other plants has not been well studied. The specificity of DREB2A observed in vitro was confirmed in vivo by analyzing the promoter sequences of up-regulated genes in 35S:DREB2A CA transgenic Arabidopsis plants, with ACCGAC being the most significantly enriched hexamer (Maruyama et al., 2012). In this study, we applied this method to identify the preferred target sequence of GmDREB2A;2 (Fig. 5; Supplemental Table S6). In both the 35S:GmDREB2A;2 FL and Δ2 plants, ACCGAC was one of the most highly overrepresented hexamers, suggesting that ACCGAC is a preferred binding sequence of GmDREB2A;2 (Fig. 5C). A large difference between GmDREB2A;2 and DREB2A is a significant overrepresentation of ABRE-related sequences in 35S:GmDREB2A;2 Δ2 (Fig. 5D; Supplemental Table S6). A weak but similar trend was also found in 35S:GmDREB2A;2 FL (Fig. 5B; Supplemental Table S6). Because the ABRE motif is not similar to DRE, they most likely reflect a secondary activation of the ABRE-regulated pathway, as suggested by the up-regulation of ABA-inducible genes (Fig. 4C). However, this difference should initially be due to different target selectivity between the GmDREB2A; 2 and DREB2A proteins. Although we do not have specific data that can account for the difference in target selectivity, we speculate that differences in their DNA-binding specificity and/or variations in the interactions with other transcription factors or transcriptional cofactors may affect the induction patterns of downstream genes.
The overexpression of GmDREB2A;2 Δ2 resulted in severe growth defects, whereas that of GmDREB2A;2 FL did not cause any significant differences in growth compared with the vector control (Fig. 3). A similar phenomenon is associated with the overexpression of DREB2A FL or CA (Fig. 3; Sakuma et al., 2006a). However, the 35S:GmDREB2A;2 Δ2 plants differ from the 35S:DREB2A CA plants in the severe inhibition of cotyledon greening and root growth (Fig. 3, B, D, and E). Although this effect is caused by the heterologous expression under the control of the constitutive promoter, these results likely reflect a functional difference between GmDREB2A;2 and DREB2A in the recognition or activation of target promoters. From analysis of the microarray data, we found a specific increase in the transcript level of ABI5 in the 35S:GmDREB2A;2 Δ2 plants, which may account for the elevated induction of ABA-inducible genes and the sugar-sensitive developmental arrest phenotype (Figs. 2 and 4, C and H; Supplemental Fig. S5; Lopez-Molina et al., 2001; Brocard et al., 2002). Because ABI5 does not contain a DRE in the upstream region, the induction of ABI5 is not a typical case of DRE-dependent activation and, therefore, may be a side effect of heterologous expression and/or deletion. DRE showed the highest level of overrepresention in the promoters of up-regulated genes in 35S:GmDREB2A;2 Δ2 (Fig. 5D; Supplemental Table S6). In addition, there were significant overlaps and correlations between the up-regulated genes in 35S:GmDREB2A;2 Δ2 and 35S:GmDREB2A;2 FL (Fig. 4, A and D), and their expression levels, including those of typical DREB2A downstream genes, were generally higher in 35S:GmDREB2A;2 Δ2 than in 35S:GmDREB2A;2 FL (Fig. 4, D and H). Therefore, with the exception of the additional up-regulation of ABA-inducible genes, GmDREB2A;2 Δ2 can be considered to function as an active form of GmDREB2A;2 in Arabidopsis.
Despite technological advances in the transformation of soybean, the generation of transgenic soybean plants is still difficult and time consuming. To enhance the characterization of soybean genes, we used protoplasts derived from soybean cells and transgenic Arabidopsis plants. The transgenic plant expressing GmDREB2A;2 FL exhibited improved heat stress tolerance (Fig. 6). Although GmDREB2A;2 FL is not a constitutively active form, the increased expression of heat-inducible genes, including HsfA3, which is a key DREB2A target gene, can account for this phenotype (Fig. 4H). GmDREB2A;2 Δ2 has the ability to induce not only dehydration-inducible genes, including RD29A and RD29B, but also ABA-inducible genes, including ABI5, which might cause developmental arrest (Fig. 4, C and H). However, we demonstrated a positive effect of GmDREB2A;2 Δ2 on drought tolerance by stress-induced expression (Fig. 7). Therefore, similar to its Arabidopsis ortholog DREB2A, GmDREB2A;2 has the ability to improve heat and drought tolerance in plants. In soybean, GmDREB2A;2 was highly induced in response to heat or osmotic stress conditions in a pattern similar to that of DREB2A (Fig. 1B; Supplemental Fig. S3). In addition, GmDREB2A;2 can activate DRE-mediated transcription, and its activity and stability were regulated following the same rule as DREB2A in Arabidopsis (Figs. 1, D–F, and 2). Furthermore, GmDREB2A;2 has a preference for the DRE sequences in plants (Fig. 5, B–D), and DRE or DRE-ABRE combinations are enriched at least in dehydration-inducible promoters in soybean (Maruyama et al., 2012). These findings suggest that GmDREB2A;2 plays an important role in the dehydration and heat stress response in soybean as a canonical subtype 1 DREB2.
In conclusion, we identified 21 loci for DREB2-type transcription factors in soybean. Among them, the two GmDREB2A homologs encode for proteins with high sequence similarities to DREB2A and are highly induced in response to abiotic stress treatment. The GmDREB2A;2 protein exhibited high transactivation activity through DRE, and its activity was negatively regulated by the PEST-like sequence in a manner similar to that of DREB2A. Finally, the heterologous expression of GmDREB2A;2 resulted in an enhanced expression of DREB2A target stress-inducible genes and improved the heat and drought stress tolerance of transgenic Arabidopsis. These findings indicate that GmDREB2A;2 is a genuine functional ortholog of DREB2A in soybean that functions in abiotic stress responses. However, in contrast to DREB2A, the GmDREB2A homologs were induced by low temperature. Furthermore, the constitutive expression of GmDREB2A;2 was associated with unique developmental phenotypes, and variations in the induced genes and their induction ratios were observed between the transgenic plants of GmDREB2A;2 and DREB2A. These results suggest that the function of the DREB2-DRE system in the stress response has been differentiated between soybean and Arabidopsis. Due to significant advances in genome sequencing technologies, the genomic sequences of many crops are becoming increasingly available. Canonical DREB2 transcription factors are potential targets of molecular breeding because they function as positive regulators of the responses against multiple stress conditions, notably dehydration and heat shock. Although DREB2 forms a multigene family in each plant, we showed that our classification is useful for identifying a functional ortholog of DREB2A in a plant of interest. In addition, the Ser/Thr-rich sequence adjacent to the DNA-binding domain is widely found in DREB2A orthologs in angiosperms, and we showed that the function of this region is conserved between Arabidopsis and soybean, regardless of their differences in length. Therefore, we propose the use of an active form of DREB2 in combination with a promoter having the desired stress and/or tissue specificity as a solution to increase the performance of various crops against increasing droughts and/or heat associated with global warming trends.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia plants were grown on germination medium agar plates for 3 weeks at 22°C under a day/night light regime with a 16-h photoperiod at a photon flux density of 40 µmol m−2 s−1, as described previously (Yamaguchi-Shinozaki and Shinozaki, 1994). For the microarray analysis, seeds were imbibed and treated at 4°C in the dark for 2 d and were then sown on Perlite-containing soil (Dio Chemicals) in a plastic pot with a diameter of 6 cm. The plants were grown for 3 weeks under the conditions described above.
Soybean (Glycine max ‘Norin 2’) plants were grown on Perlite-containing soil in a plastic pot with a diameter of 7.5 cm in a plant incubator (CLH-301; TOMY) with a light regime of 12 h of light (25°C, 225 ± 25 µmol photons m−2 s−1)/12 h of dark (20°C). The relative humidity was 60% ± 15% during growth. For the preparation of protoplasts, plants were grown in a greenhouse set at a controlled temperature of 25°C (16 h)/20°C (8 h) and a relative humidity of 60% ± 15%.
Phylogenetic Analysis
The DREB2-type protein sequences of soybean were collected from BLASTP and TBLAST searches against protein and nucleotide databases, respectively, at Phytozome (http://www.phytozome.net/; Goodstein et al., 2012) using peptide sequences of Arabidopsis DREB2-type transcription factors as queries. Peptide or translated nucleotide sequences were aligned and clustered using ClustalX 2.1 (Larkin et al., 2007). The alignments were manually adjusted using the GeneDoc 2.6 program (http://www.nrbsc.org/gfx/genedoc/). The phylogeny of the selected sequences was constructed by the MEGA5.1 program using conserved sites (Supplemental Fig. S1; Tamura et al., 2011). The distances between branches were calculated by the neighbor-joining method based on the Jones-Taylor-Thornton model (Jones et al., 1992) with 1,000 bootstrap samples.
Stress Treatments and Expression Analysis in Soybean
Two-week-old soybean plants grown under the conditions described above were used for stress treatments. For the dehydration stress treatment, water was withheld for 10 d. For the high- and low-temperature treatments, the plants were moved to an incubator set at 42°C or a cold room set at 4°C, respectively. For the high-salinity treatment, the pots were pretreated with water for 1 d, and the water was replaced with 250 mm NaCl. For each time point, the entire aerial portions of three plants were harvested, combined, and frozen in liquid nitrogen. The samples were stored at −80°C before RNA isolation. RNA isolation, cDNA synthesis, and quantitative PCR analysis were performed as described previously (Tanaka et al., 2012).
RACE
RACE experiments were performed using the SMARTer RACE cDNA Amplification Kit (Takara Bio) according to the manufacturer’s instructions. The primers used are listed in Supplemental Table S7.
Transient Reporter Assays and Fluorescence Microscopy of Protoplasts
Effector plasmids were constructed as described in Supplemental Materials and Methods S1. A plasmid containing three tandem repeats of a 71-bp DRE-containing fragment of the RD29A promoter (DRE×3-GUS) was used as a reporter. As an internal control, a plasmid expressing a Pyrearinus termitilluminans luciferase (Emerald Luc; TOYOBO) under the control of the CaMV 35S promoter and the tobacco mosaic virus Ω sequence (pBI35SΩELUC) was constructed (Supplemental Materials and Methods S1). Plasmids expressing GFP fusion proteins of DREB2A and GmDREB2A were constructed as described in Supplemental Materials and Methods S1.
Transient expression assays using protoplasts derived from Arabidopsis mesophyll cells or soybean stem parenchyma cells were performed according to Yoo et al. (2007) with modifications as described previously (Kidokoro et al., 2009). The soybean stem parenchyma protoplasts were prepared as described in Supplemental Materials and Methods S1. Fluorescence images of protoplasts expressing GFP fusion proteins were obtained using a confocal laser scanning microscope (LSM 5 PASCAL; Carl Zeiss) as described by Yoshida et al. (2011).
Protein Stability Analysis in Protoplasts
The 3×Flag-DREB2 plasmids used in this study were constructed as described in Supplemental Materials and Methods S1. Approximately 3 × 104 protoplasts were transformed with 2.5 µg of each 3×Flag-DREB2 plasmid, 3.5 µg of DRE×3-GUS, 4 µg of pBI35SΩELUC, and 2.5 µg of pGKX-NsGFP in triplicate as described above. After a 16-h incubation at 22°C in the dark, the triplicates were combined, divided into two aliquots, and then incubated in washing and incubation solution (4 mm MES-KOH, 0.5 m mannitol, and 20 mm KCl, pH 5.7) with 50 µm MG132 or 0.25% (v/v) dimethyl sulfoxide (solvent control). After 1 h, the protoplasts were precipitated by centrifugation at 200g for 2 min at 25°C and dissolved in 40 µL of a solution containing 10 m urea, 150 mm dithiothreitol, 3 mm EDTA, 1.5% (w/v) SDS, and 75 mm Tris-HCl (pH 6.8). The extracts were denatured at 95°C for 3 min and analyzed by SDS-PAGE and immunoblotting. The 3×Flag tag-fused proteins were detected with the anti-Flag M2 antibody (Sigma-Aldrich), and sGFP was detected with a polyclonal antibody against GFP (Tanaka et al., 2012). Horseradish peroxidase-conjugated anti-rabbit or anti-mouse IgG antibodies (Pierce), SuperSignal West Dura Extended Duration Substrate (Pierce), and an ImageQuant LAS 4000 system (GE Healthcare) were used to visualize the signals.
Generation of DREB2A-Overexpressing Plants
The plasmids used in the transient reporter assays were applied to Arabidopsis transformation by the floral dip method (Clough and Bent, 1998).
Microarray Experiment and Data Processing
Genome-wide expression studies using the Arabidopsis 3 Oligo Microarray (Agilent Technologies) were performed as described below. Total RNA (200 ng) isolated from more than five seedlings with RNAiso plus (Takara Bio) was labeled with a Low RNA Input Linear Amplification/Labeling Kit (Agilent Technologies) according to the manufacturer’s instructions. Biological replication was performed by analyzing the samples obtained from two independent DREB2 transgenic lines. For each experiment, in which the vector control and one transgenic line were compared, two sets of microarrays were analyzed using a Cy3 and Cy5 dye swap. After hybridization, the microarray slides were scanned with a G2505C scanner (Agilent Technologies) and processed using Feature Extraction software (10.10.1.1; Agilent Technologies). Integration, normalization, and statistical analyses of the data were performed using GeneSpring GX11 (Agilent Technologies). The signal intensities of the spots were normalized by the Lowess method, and the significance of the expression changes was evaluated by Student’s t test. The P values were corrected by the Benjamini-Hochberg FDR method, and probes that showed a corrected P value of less than 0.01 were used for further analyses. All microarray data are available at Array Express (http://www.ebi.ac.uk/arrayexpress/) with accession number E-MEXP-3692.
Analysis of Microarray Data
A meta-profile analysis of the up-regulated genes in each transgenic plant was performed in a public microarray database (Genevestigator; https://www.genevestigator.com/gv/; Hruz et al., 2008). Overrepresentation analysis of hexamer sequences in the promoters of up-regulated genes was performed as described previously (Maruyama et al., 2012) using 1-kb upstream sequences of approximately 14,000 loci with defined transcriptional start sites to compute the normalized frequencies of all hexamers.
Stress Tolerance Test of Transgenic Arabidopsis
The drought stress and heat shock tests were performed as described by Sakuma et al. (2006a, 2006b).
The accession numbers of the cDNA sequences are as follows: GmDREB2A;1 (JX440386), GmDREB2A;2 (JX440387), GmDREB2D;1 (JX440388), and GmDREB2D;2 (JX440389). The locus identifiers of the GmDREB2 and GmABI4 genes at Phytozome are listed in Supplemental Table S1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Alignment of the DNA-binding domain and the N-terminal conserved region of DREB2-type transcription factors in soybean, Arabidopsis, and rice.
Supplemental Figure S2. Schematic models of subtype 1 DREB2-type transcription factors in soybean and Arabidopsis.
Supplemental Figure S3. Expression time course of GmDREB2A;2 in response to abiotic stress treatments.
Supplemental Figure S4. Effects of DREB2A and GmDREB2A;2 protein accumulation levels on the reporter activity.
Supplemental Figure S5. Sugar sensitivity of the 35S:GmDREB2A;2 Δ2 plants.
Supplemental Figure S6. Venn diagram showing the relationship among the significantly up-regulated genes in the 35S:DREB2A FL, 35S:DREB2A CA, 35S:GmDREB2A;2 FL, and 35S:GmDREB2A;2 Δ2 plants.
Supplemental Table S1. Loci for DREB2-type transcription factors in soybean, rice, and Arabidopsis.
Supplemental Table S2. Genes that were significantly up- or down-regulated in the 35S:DREB2A FL plants.
Supplemental Table S3. Genes that were significantly up- or down-regulated in the 35S:DREB2A CA plants.
Supplemental Table S4. Genes that were significantly up- or down-regulated in the 35S:GmDREB2A;2 FL plants.
Supplemental Table S5. Genes that were significantly up- or down-regulated in the 35S:GmDREB2A;2 Δ2 plants.
Supplemental Table S6. Overrepresentation analysis of hexamer sequences in the promoters of highly up-regulated genes in each transgenic plant.
Supplemental Table S7. Oligonucleotides used in this study.
Supplemental Materials and Methods S1. Construction of plasmid vectors.
Acknowledgments
We thank Y. Tanaka, S. Murasaki, K. Amano, and E. Kishi for providing excellent technical assistance.
Glossary
- ABA
abscisic acid
- ABRE
ABA-responsive element
- NRD
negative regulatory domain
- DRE
dehydration-responsive element
- qRT
quantitative reverse transcription
- FDR
false discovery rate
- CaMV
cauliflower mosaic virus
- cDNA
complementary DNA
- sGFP
synthetic GFP
References
- Agarwal P, Agarwal PK, Nair S, Sopory SK, Reddy MK. (2007) Stress-inducible DREB2A transcription factor from Pennisetum glaucum is a phosphoprotein and its phosphorylation negatively regulates its DNA-binding activity. Mol Genet Genomics 277: 189–198 [DOI] [PubMed] [Google Scholar]
- Brocard IM, Lynch TJ, Finkelstein RR. (2002) Regulation and role of the Arabidopsis abscisic acid-insensitive 5 gene in abscisic acid, sugar, and stress response. Plant Physiol 129: 1533–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J-R, Lü J-J, Wang T-X, Chen S-Y, Wang H-F. (2009a) Activation of a DRE-binding transcription factor from Medicago truncatula by deleting a Ser/Thr-rich region. In Vitro Cell Dev Biol Plant 45: 1–11 [Google Scholar]
- Chen M, Wang QY, Cheng XG, Xu ZS, Li LC, Ye XG, Xia LQ, Ma YZ. (2007) GmDREB2, a soybean DRE-binding transcription factor, conferred drought and high-salt tolerance in transgenic plants. Biochem Biophys Res Commun 353: 299–305 [DOI] [PubMed] [Google Scholar]
- Chen M, Xu Z, Xia L, Li L, Cheng X, Dong J, Wang Q, Ma Y. (2009b) Cold-induced modulation and functional analyses of the DRE-binding transcription factor gene, GmDREB3, in soybean (Glycine max L.). J Exp Bot 60: 121–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Chen P, de los Reyes B. (2006) Differential responses of the cultivated and wild species of soybean to dehydration stress. Crop Sci 46: 2041–2046 [Google Scholar]
- Chiu W, Niwa Y, Zeng W, Hirano T, Kobayashi H, Sheen J. (1996) Engineered GFP as a vital reporter in plants. Curr Biol 6: 325–330 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Díaz-Martín J, Almoguera C, Prieto-Dapena P, Espinosa JM, Jordano J. (2005) Functional interaction between two transcription factors involved in the developmental regulation of a small heat stress protein gene promoter. Plant Physiol 139: 1483–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egawa C, Kobayashi F, Ishibashi M, Nakamura T, Nakamura C, Takumi S. (2006) Differential regulation of transcript accumulation and alternative splicing of a DREB2 homolog under abiotic stress conditions in common wheat. Genes Genet Syst 81: 77–91 [DOI] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al. (2012) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40: D1178–D1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. (2008) Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinformat 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. (1992) The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8: 275–282 [DOI] [PubMed] [Google Scholar]
- Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. (1999) Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat Biotechnol 17: 287–291 [DOI] [PubMed] [Google Scholar]
- Kasuga M, Miura S, Shinozaki K, Yamaguchi-Shinozaki K. (2004) A combination of the Arabidopsis DREB1A gene and stress-inducible rd29A promoter improved drought- and low-temperature stress tolerance in tobacco by gene transfer. Plant Cell Physiol 45: 346–350 [DOI] [PubMed] [Google Scholar]
- Kidokoro S, Maruyama K, Nakashima K, Imura Y, Narusaka Y, Shinwari ZK, Osakabe Y, Fujita Y, Mizoi J, Shinozaki K, et al. (2009) The phytochrome-interacting factor PIF7 negatively regulates DREB1 expression under circadian control in Arabidopsis. Plant Physiol 151: 2046–2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JS, Mizoi J, Yoshida T, Fujita Y, Nakajima J, Ohori T, Todaka D, Nakashima K, Hirayama T, Shinozaki K, et al. (2011) An ABRE promoter sequence is involved in osmotic stress-responsive expression of the DREB2A gene, which encodes a transcription factor regulating drought-inducible genes in Arabidopsis. Plant Cell Physiol 52: 2136–2146 [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Li XP, Tian AG, Luo GZ, Gong ZZ, Zhang JS, Chen SY. (2005) Soybean DRE-binding transcription factors that are responsive to abiotic stresses. Theor Appl Genet 110: 1355–1362 [DOI] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. (1998) Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10: 1391–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, Chua NH. (2001) A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc Natl Acad Sci USA 98: 4782–4787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama K, Todaka D, Mizoi J, Yoshida T, Kidokoro S, Matsukura S, Takasaki H, Sakurai T, Yamamoto YY, Yoshiwara K, et al. (2012) Identification of cis-acting promoter elements in cold- and dehydration-induced transcriptional pathways in Arabidopsis, rice, and soybean. DNA Res 19: 37–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsukura S, Mizoi J, Yoshida T, Todaka D, Ito Y, Maruyama K, Shinozaki K, Yamaguchi-Shinozaki K. (2010) Comprehensive analysis of rice DREB2-type genes that encode transcription factors involved in the expression of abiotic stress-responsive genes. Mol Genet Genomics 283: 185–196 [DOI] [PubMed] [Google Scholar]
- Menke FL, Champion A, Kijne JW, Memelink J. (1999) A novel jasmonate- and elicitor-responsive element in the periwinkle secondary metabolite biosynthetic gene Str interacts with a jasmonate- and elicitor-inducible AP2-domain transcription factor, ORCA2. EMBO J 18: 4455–4463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R. (2006) Abiotic stress, the field environment and stress combination. Trends Plant Sci 11: 15–19 [DOI] [PubMed] [Google Scholar]
- Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. (2012) AP2/ERF family transcription factors in plant abiotic stress responses. Biochim Biophys Acta 1819: 86–96 [DOI] [PubMed] [Google Scholar]
- Nakano T, Suzuki K, Fujimura T, Shinshi H. (2006) Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol 140: 411–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin F, Kakimoto M, Sakuma Y, Maruyama K, Osakabe Y, Tran L-SP, Shinozaki K, Yamaguchi-Shinozaki K. (2007) Regulation and functional analysis of ZmDREB2A in response to drought and heat stresses in Zea mays L. Plant J 50: 54–69 [DOI] [PubMed] [Google Scholar]
- Qin F, Sakuma Y, Tran LSP, Maruyama K, Kidokoro S, Fujita Y, Fujita M, Umezawa T, Sawano Y, Miyazono K, et al. (2008) Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress-responsive gene expression. Plant Cell 20: 1693–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechsteiner M, Rogers SW. (1996) PEST sequences and regulation by proteolysis. Trends Biochem Sci 21: 267–271 [PubMed] [Google Scholar]
- Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K. (2002) DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem Biophys Res Commun 290: 998–1009 [DOI] [PubMed] [Google Scholar]
- Sakuma Y, Maruyama K, Osakabe Y, Qin F, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. (2006a) Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 18: 1292–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma Y, Maruyama K, Qin F, Osakabe Y, Shinozaki K, Yamaguchi-Shinozaki K. (2006b) Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. Proc Natl Acad Sci USA 103: 18822–18827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, et al. (2010) Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183 [DOI] [PubMed] [Google Scholar]
- Shen YG, Zhang WK, He SJ, Liu Q, Chen SY. (2003) An EREBP/AP2-type protein in Triticum aestivum was a DRE-binding transcription factor induced by cold, dehydration and ABA stress. Theor Appl Genet 106: 923–930 [Google Scholar]
- Shukla RK, Raha S, Tripathi V, Chattopadhyay D. (2006) Expression of CAP2, an APETALA2-family transcription factor from chickpea, enhances growth and tolerance to dehydration and salt stress in transgenic tobacco. Plant Physiol 142: 113–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger EJ, Gilmour SJ, Thomashow MF. (1997) Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci USA 94: 1035–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H, Osakabe Y, Katsura S, Mizuno S, Maruyama K, Kusakabe K, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. (2012) Abiotic stress-inducible receptor-like kinases negatively control ABA signaling in Arabidopsis. Plant J 70: 599–613 [DOI] [PubMed] [Google Scholar]
- Xue GP, Loveridge CW. (2004) HvDRF1 is involved in abscisic acid-mediated gene regulation in barley and produces two forms of AP2 transcriptional activators, interacting preferably with a CT-rich element. Plant J 37: 326–339 [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. (1994) A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6: 251–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. (2006) Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol 57: 781–803 [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Yoshida T, Ohama N, Nakajima J, Kidokoro S, Mizoi J, Nakashima K, Maruyama K, Kim JM, Seki M, Todaka D, et al. (2011) Arabidopsis HsfA1 transcription factors function as the main positive regulators in heat shock-responsive gene expression. Mol Genet Genomics 286: 321–332 [DOI] [PubMed] [Google Scholar]