Abstract
Insertional mutagenesis is a powerful tool for determining gene function in both model and crop plant species. Tnt1, the transposable element of tobacco (Nicotiana tabacum) cell type 1, is a retrotransposon that replicates via an RNA copy that is reverse transcribed and integrated elsewhere in the plant genome. Based on studies in a variety of plants, Tnt1 appears to be inactive in normal plant tissue but can be reactivated by tissue culture. Our goal was to evaluate the utility of the Tnt1 retrotransposon as a mutagenesis strategy in soybean (Glycine max). Experiments showed that the Tnt1 element was stably transformed into soybean plants by Agrobacterium tumefaciens-mediated transformation. Twenty-seven independent transgenic lines carrying Tnt1 insertions were generated. Southern-blot analysis revealed that the copy number of transposed Tnt1 elements ranged from four to 19 insertions, with an average of approximately eight copies per line. These insertions showed Mendelian segregation and did not transpose under normal growth conditions. Analysis of 99 Tnt1 flanking sequences revealed insertions into 62 (62%) annotated genes, indicating that the element preferentially inserts into protein-coding regions. Tnt1 insertions were found in all 20 soybean chromosomes, indicating that Tnt1 transposed throughout the soybean genome. Furthermore, fluorescence in situ hybridization experiments validated that Tnt1 inserted into multiple chromosomes. Passage of transgenic lines through two different tissue culture treatments resulted in Tnt1 transposition, significantly increasing the number of insertions per line. Thus, our data demonstrate the Tnt1 retrotransposon to be a powerful system that can be used for effective large-scale insertional mutagenesis in soybean.
Soybean (Glycine max) is a major commodity crop that offers a wealth of resources, including proteins, oils, mineral nutrients, and natural products, that impact human health and nutrition. The products of soybean are widely used as vegetable oil and protein sources for human consumption and are valuable feedstock for the livestock industry (Gepts et al., 2005; O’Brian and Vance, 2007). Research on soybean is driven by its importance as a food crop worldwide. In recent years, considerable progress has been made in developing genomic resources for soybean, including the complete sequencing of the genome, which predicts 46,430 high-confidence protein-encoding genes (Schmutz et al., 2010). Utilizing the Illumina Solexa sequencing platform, a gene expression atlas of the soybean genome was developed that documented the transcription of up to 55,616 annotated genes (Libault et al., 2010). One remaining major challenge is the elucidation of the function of these genes, especially those encoding important agronomic traits. This challenge can be met, in part, by the development of insertional mutagenesis tools to investigate soybean gene function.
Insertional mutagenesis is an effective method for functional genomics studies. Mutagenesis can modulate gene expression and create very useful loss-of-function mutants, whose phenotypes can validate and explore gene function. Insertional mutagenesis has been successfully used to study gene function in both model and crop plant species (Cowperthwaite et al., 2002; Alonso et al., 2003; An et al., 2003; Fladung et al., 2004; Tadege et al., 2008; Mathieu et al., 2009). A clear example is the use of T-DNA tagging to create large mutant populations of Arabidopsis (Arabidopsis thaliana; Alonso et al., 2003). However, although the development of a T-DNA insertional mutant repository in soybean is technically possible, it would require a tremendous amount of labor, since each mutant line would require an independent transformation event. Thus, a transposon-tagging strategy, where many mutations could be derived from one primary transformation event, is an attractive approach for a plant such as soybean in which transformation requires a much longer time frame (roughly 1 year, from seed to seed; Parrott and Clemente, 2004; Mathieu et al., 2009).
Transposon tagging has been used successfully in soybean. For example, Mathieu et al. (2009) utilized the well-characterized maize (Zea mays) transposon, Ac/Ds, to identify a soybean male-sterile line. Ac/Ds is a class II transposon, which transposes into new locations in plant genomes via a “cut-and-paste” mechanism (Wessler, 2006). However, similar to the situation with T-DNA insertions, the use of this transposon requires many independent transformation events to create a library sufficiently large to target the entire soybean genome, because it tends to transpose to linked sites (Jones et al., 1990; Ito et al., 1999; Parinov and Sundaresan, 2000). Perhaps a more promising alternative is mPing, a class II element originally isolated in rice (Oryza sativa; Jiang et al., 2003; Kikuchi et al., 2003; Nakazaki et al., 2003), where it transposes at a high frequency and can reach a high copy number in some cultivars (Naito et al., 2006). Recently, Hancock et al. (2011) reported that mPing can successfully transpose in soybean and generated stable, heritable insertions. However, one possible limitation to the utility of the mPing element is that the element continues to transpose, even under normal plant growth conditions, thereby creating somatic mutations that could complicate both phenotypic and genetic analyses.
Compared with the class II transposons, the class I retrotransposons present some advantages for use as an insertional mutagenic tool (Kumar and Hirochika, 2001). Retrotransposons transpose in a “copy-and-paste” manner via an RNA intermediate (Kumar and Bennetzen, 1999). Several retrotransposons have already been used effectively as mutagens in plants. For example, Tos17, an endogenous retrotransposon of rice, is active in the rice genome during tissue culture and has been used for gene tagging in rice (Piffanelli et al., 2007). LORE1, an exon-targeting endogenous retrotransposon in Lotus japonicus, was recently used to develop a medium-sized mutagenized population composed of 2,450 plant lines (Fukai et al., 2012). The Tto1 element, from tobacco (Nicotiana tabacum), has also been used for mutagenesis in Arabidopsis (Okamoto and Hirochika, 2000) and rice (Hirochika et al., 1996). Tnt1, originally isolated from tobacco, has been successfully used in several heterologous hosts, including Medicago truncatula (d’Erfurth et al., 2003; Tadege et al., 2005, 2008; Iantcheva et al., 2009), Arabidopsis (Lucas et al., 1995; Courtial et al., 2001), and lettuce (Lactuca sativa; Mazier et al., 2007). Collectively, these studies demonstrate that retrotransposons transpose preferentially into gene-rich regions, thus making them highly mutagenic. While retrotransposons are activated in tissue culture, they appear to be stable in mature, transgenic plants. Therefore, relatively few primary transgenic lines can lead to large populations of mutants by repeated transfer through tissue culture. Indeed, the retrotransposon Tnt1 has been used successfully in the model legume plant M. truncatula to build useful mutant populations (d’Erfurth et al., 2003; Tadege et al., 2005, 2008; Iantcheva et al., 2009). The published M. truncatula Tnt1 population contains nearly 12,000 insertion lines, representing over 300,000 insertions, and has been used successfully in both forward and reverse genetics studies (Tadege et al., 2008; Cheng et al., 2011). However, reactivation of retrotransposon transposition does not occur in every plant species examined. For example, Ishizaki and Kato (2005) failed to detected tissue culture reactivation of the Tto1 retrotransposon in transgenic potato (Solanum tuberosum) plants.
The goal of our study thus was to explore the utility of the Tnt1 retrotransposon as a mutagenesis strategy in soybean. Our findings demonstrate that Tnt1 is an attractive and efficient system that can now be used for large-scale insertion mutagenesis in soybean.
RESULTS AND DISCUSSION
Generation of Tnt1 Retrotransposon-Containing Soybean Lines
Although Tnt1 transposes very efficiently in the legume model plant M. truncatula and several other plants, including tobacco, Arabidopsis, and lettuce (Courtial et al., 2001; d’Erfurth et al., 2003; Tadege et al., 2005, 2008; Mazier et al., 2007; Iantcheva et al., 2009), it was important to evaluate its utility in soybean for two reasons. First, due to the economic importance of soybean, it is critical to develop improved gene-discovery tools. Second, it is known that some retrotransposons are genotype specific for transposition or exhibit high efficiency only on specific genotypes. For example, the Tto1 retrotransposon from tobacco transposes in tobacco, Arabidopsis, and rice (Hirochika, 1993; Hirochika et al., 1996; Okamoto and Hirochika 2000) but does not transpose in potato tissue culture (Ishizaki and Kato, 2005). In M. truncatula, the reactivation protocol of Tnt1 optimized to cv R108 is not applicable for cv Jemalong (Iantcheva et al., 2009). Therefore, it is necessary to determine if Tnt1 transposes in soybean and also to optimize the methodology to induce its transposition.
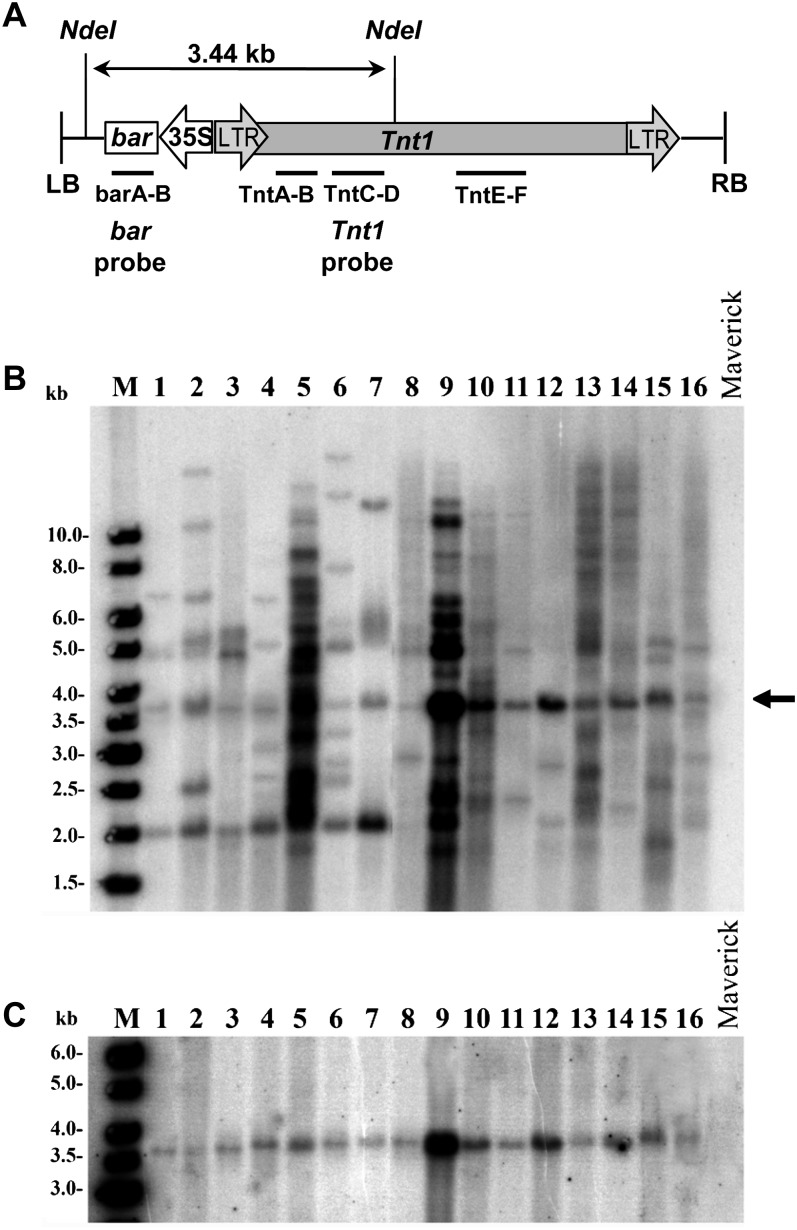
To investigate whether the Tnt1 element can transpose in soybean during tissue culture, Agrobacterium tumefaciens-mediated transformation was performed using a modified soybean cotyledonary node transformation protocol (Zeng et al., 2004). The plasmid pSH-Tnt1 containing the Tnt1 element was constructed by inserting the Tnt1 DNA into the binary vector pZY101, which carries a selectable bar gene marker for glufosinate resistance (Fig. 1A). Transformations were performed in cv Maverick, a genotype that is susceptible to glufosinate. cv Maverick is an elite soybean genotype that is resistant to stress conditions and shows a consistently higher transformation frequency when compared with other genotypes (Z.Y. Zhang, unpublished data). Twenty-seven independent glufosinate resistance plants (verified by leaf-painting assay) were generated by this approach. To determine if these regenerated plants harbored Tnt1, PCR experiments were performed using three primer pairs specific for Tnt1 and one primer pair specific for the bar gene, as indicated in Figure 1A. All 27 lines gave positive PCR amplifications for all Tnt1- and bar-specific primer pairs (data not shown).
Figure 1.
A, Diagram of plasmid pSH-Tnt1 containing the Tnt1 element in the binary vector pZY101. LB and RB, Left and right borders, respectively; bar, gene conferring glufosinate resistance; 35S, promoter 35S; LTR, long terminal repeat. NdeI restriction sites and PCR fragments for bar (barA-B) and Tnt1 (TntA-B, TntC-D and TntE-F) amplification are shown. PCR fragment barA-B or TntC-D was used in Southern-blot analysis to probe for bar or Tnt1, respectively. B, Southern-blot analysis of Tnt1 primary transgenic lines to identify Tnt1-hybridizing bands. Chromosomal DNA (15–20 µg) from each transgenic line was digested with NdeI and probed with the TntC-D PCR fragment. The arrow indicates hybridization bands representing nontransposed Tnt1 (i.e. T-DNA associated). M, Molecular weight markers. Lanes 1 to 16 show Tnt1 mutants BS2-5, BS2-6, BS3-5, BS5-4, BS5-6, BS5-10, BS5-12, BS5-13, BS6-19, BS6-20, BS7-5, BS7-7, BS7-8, BS7-10, BS8-5, and BS8-7, respectively. cv Maverick is the parent line. C, Southern-blot analysis of Tnt1 transgenic lines to identify bar-hybridizing bands. The blot used in B was stripped and rehybridized using the barA-B PCR fragment as a probe.
Tnt1 Transposes in Regenerated Soybean Plants
To verify the PCR results and to determine if Tnt1 integrated into the soybean genome and transposed during tissue culture, we performed Southern-blot analysis on all 27 lines that were positive by PCR. Chromosomal DNA of these plants was extracted and digested with restriction enzyme NdeI. Southern-blot analysis was performed using a 755-bp Tnt1 internal fragment as the probe, which corresponds to bases 1,067 to 1,822 of the retrotransposon. The same blot was then stripped and reprobed with a 480-bp bar internal fragment. The NdeI sites and probe locations are shown in Figure 1A. NdeI cuts the Tnt1 DNA once at position 1,983 and also cuts once within the T-DNA region (near the left border). Therefore, a line carrying Tnt1 associated with a T-DNA should show a 3.44-kb band when the above-mentioned probe is used, while most other hybridization bands would represent transposed Tnt1 copies. Southern-blot analysis of 16 Tnt1-containing plants and the parent line cv Maverick using the Tnt1 probe is shown in Figure 1B. As expected, a 3.44-kb band (indicated by the arrow) was present, representing nontransposed Tnt1 (i.e. T-DNA associated) in all 27 lines. This band also hybridized with the bar probe (Fig. 1C), further confirming that it is T-DNA associated. We detected no plant carrying a Tnt1 element without the T-DNA, as has been reported in M. truncatula, where 11.2% of the regenerated plants carried only the retrotransposon (d’Erfurth et al., 2003). In addition to the 3.44-kb band, multiple Tnt1-hybridizing bands, which did not hybridize to the bar probe, were detected in all plants tested, indicating that Tnt1 is able to transpose in soybean during the tissue culture associated with transformation (Fig. 1B). Transposed Tnt1 copy numbers ranged from four to 19, with an average of approximately eight copies per line. Thus, the results of these experiments confirm that the Tnt1 element was stably transformed in soybean plants by A. tumefaciens-mediated transformation. Since the plants we analyzed were derived directly from the tissue culture of primary transformation, the observed transposition events likely occurred during A. tumefaciens-mediated transformation. The copy numbers of transposed Tnt1 elements in soybean were similarly, perhaps slightly less, than that reported for M. truncatula (ranging from four to more than 30 insertions), Arabidopsis (ranging from zero to 26 insertions), or lettuce (more than 30 copies; Courtial et al., 2001; d’Erfurth et al., 2003; Mazier et al., 2007). Further optimization of the transformation method may permit the generation of lines with significantly more Tnt1 insertion events.
Southern-blot analysis revealed that all Tnt1-harboring soybean plants contained transposed Tnt1 elements. No plant contained just a single copy of T-DNA. This result is similar to M. truncatula but contrasts with reports in Arabidopsis, where several regenerated plants contained no transposed Tnt1 copy, or in lettuce, where four different regenerated lettuce plants were found to contain only a truncated version of the T-DNA but no transposed copies of Tnt1.
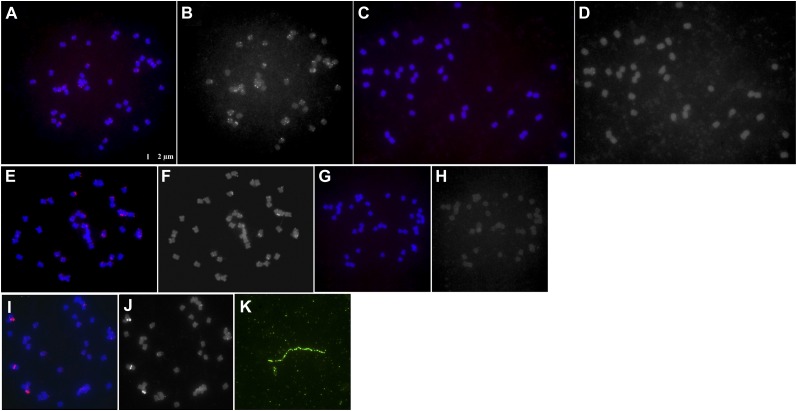
In order to be useful for large-scale mutagenesis in soybean, it is critical that the Tnt1 insertion pattern does not exclude any chromosomes. As one method to determine the Tnt1 transposition pattern, two Tnt1-containing plants were analyzed using fluorescence in situ hybridization (FISH). The Tnt1-containing line BS5-6 chromosomes were hybridized with Texas Red-labeled pSH-Tnt1 plasmid DNA. Chromosomes of untransformed cv Maverick served as controls. As shown in Figure 2A, the hybridization signals (red dots) were detected on most BS5-6 chromosomes, whereas no signal was detected in the parental control line (Fig. 2C). Moreover, multiple hybridization signals were observed in several chromosomes (Fig. 2, A and B). Based on the Southern-blot analysis, line BS5-6 possesses approximately 19 transposed copies of Tnt1 (Fig. 1B). The FISH data showing multiple Tnt1-hybridizing regions are consistent with these results. We should note that although the probe DNA used for FISH experiments contained the entire Tnt1 and T-DNA, the Southern-blot analysis revealed that most of the hybridization signals observed in the chromosomes of BS5-6 were transposed Tnt1 elements. To further clarify this, we cloned the 5.3-kb Tnt1 DNA into vector pBluescript SK+ and used the resulting plasmid construct (pBS-Tnt1) as a probe for FISH experiments to examine line BS5-6. We found that hybridization signals observed with the pBS-Tnt1 probe (Fig. 2, E and F) were comparable to those obtained with pSH-Tnt1 (Fig. 2, A and B). No hybridization signals were detected when the empty pBluescript SK+ vector was used as a probe (data not shown). Furthermore, a FISH experiment was also performed on another Tnt1 line, BS6-19, using pBS-Tnt1 as a probe. The results (Fig. 2, I and J) revealed very strong hybridization signals on several chromosomes. In addition, several weaker signals were also detected on other chromosomes. As expected, no signal was detected in the parental control line (Fig. 2, G and H). Southern-blot analysis predicted 15 Tnt1 insertions in line BS6-19. To examine the nature of the stronger FISH hybridizing signals, we performed fiber-FISH to estimate the size of the hybridizing region. When a pBluescript SK+ plasmid harboring Tnt1 was labeled with biotin and used as a probe (Fig. 2K), a strong signal was detected in line BS6-19 over an estimated length of approximately 34 kb. A plausible explanation for this length is that multiple copies of Tnt1 DNA inserted into the same position on one chromosome in this line. This would suggest that the weaker FISH hybridizing signals likely represent one or at most a few inserted Tnt1 elements, whereas the stronger hybridizing bands likely represent tandemly arrayed, multiple copies of Tnt1.
Figure 2.
FISH-based characterization of Tnt1 mutant lines BS5-6 and BS6-19. A and C, Chromosomes of line BS5-6 or the parent line cv Maverick were hybridized with Texas Red-labeled pSH-Tnt1 plasmid DNA, respectively. E and G, Chromosomes of line BS5-6 or cv Maverick were hybridized with Texas Red-labeled pBS-Tnt1 plasmid DNA, respectively. I, Chromosomes of line BS6-19 were hybridized with pBS-Tnt1 probe. B, D, F, H, and J, Grayscale images of the chromosomes in A, C, E, G, and I, respectively. K, Fiber-FISH shows an approximately 34-kb Tnt1 DNA fiber in line BS6-19.
Tnt1 Efficiently Transposes into Coding Regions
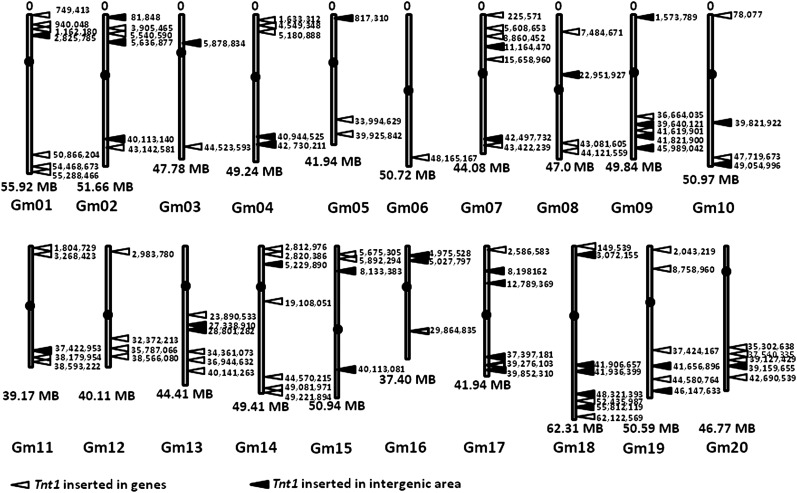
To identify Tnt1 insertion sites, we performed thermal asymmetric interlaced (TAIL)-PCR (Ratet et al., 2006) on 18 independent transgenic lines to recover Tnt1 flanking sequences. Of the 99 Tnt1 insertion sites identified, 62 were located in annotated genes (Table I). Moreover, Tnt1 insertions were found in all 20 soybean chromosomes (Fig. 3), as indicated by mapping the flanking sequences to the published soybean genome sequence (Schmutz et al., 2010). Therefore, consistent with the results of the FISH analysis, Tnt1 appears to transpose throughout the soybean genome.
Table I. Tnt1 hit genes identified by TAIL-PCR.
| Gene | Gene Location | Tnt1 Location |
|---|---|---|
| Glyma01g01090 | Gm01:746970.0.750314 | 749,413 |
| Glyma01g01300 | Gm01:937365.0.939890 | 940,048 |
| Glyma01g01560 | Gm01:1160245.0.1164111 | 1,162,180 |
| Glyma01g38870 | Gm01:50865317.0.50867505 | 50,866,204 |
| Glyma01g43460 | Gm01:54468406.0.54469790 | 54,468,673 |
| Glyma01g44730 | Gm01:55288549.0.55291102 | 55,288,466 |
| Glyma02g04770 | Gm02:3902268.0.3906661 | 3,905,465 |
| Glyma02g06890 | Gm02:5533736.0.5541825 | 5,540,590 |
| Glyma02g37830 | Gm02:43141981.0.43145206 | 43,142,581 |
| Glyma03g38120 | Gm03:44522550.0.44526793 | 44,523,593 |
| Glyma04g02350 | Gm04:1631557.0.1634732 | 1,633,312 |
| Glyma04g05960 | Gm04:4543997.0.4549817 | 4,549,348 |
| Glyma04g06710 | Gm04:5178279.0.5181655 | 5,180,888 |
| Glyma05g28120 | Gm05:33994151.0.33996925 | 33,994,629 |
| Glyma05g36000 | Gm05:39924225.0.39927013 | 39,925,842 |
| Glyma06g45450 | Gm06:48160130.0.48165969 | 48,165,167 |
| Glyma07g00500 | Gm07:224177.0.230842 | 225,571 |
| Glyma07g06950 | Gm07:5607639.0.5610266 | 5,608,653 |
| Glyma07g10570 | Gm07:8857596.0.8860631 | 8,860,452 |
| Glyma07g15940 | Gm07:15658959.0.15662391 | 15,658,960 |
| Glyma07g38720 | Gm07:43421544.0.43422862 | 43,422,239 |
| Glyma08g10350 | Gm08:7484480.0.7485874 | 7,484,671 |
| Glyma08g43240 | Gm08:43081527.0.43082006 | 43,081,605 |
| Glyma08g45860 | Gm08:45121245.0.45131117 | 45,121,559 |
| Glyma09g29810 | Gm09:36663252.0.36666676 | 36,664,035 |
| Glyma09g35670 | Gm09:41618503.0.41620799 | 41619901 |
| Glyma10g00260 | Gm10:75667.0.78986 | 78,077 |
| Glyma10g40240 | Gm10:47715528.0.47720018 | 47,719,673 |
| Glyma11g02780 | Gm11:1804302.0.1806201 | 1,804,729 |
| Glyma11g04750 | Gm11:3266510.0.3268970 | 3,268,423 |
| Glyma11g36880 | Gm11:38179557.0.38181293 | 38,179,954 |
| Glyma11g37500 | Gm11:38592441.0.38597663 | 38,593,222 |
| Glyma12g04480 | Gm12:2982711.0.2987078 | 2,983,780 |
| Glyma12g28980 | Gm12:32372131.0.32373204 | 32,372,213 |
| Glyma12g32280 | Gm12:35785488.0.35787769 | 35,787,066 |
| Glyma12g35440 | Gm12:38564240.0.38567572 | 38,566,080 |
| Glyma13g20420 | Gm13:23889695.0.23897572 | 23,890,533 |
| Glyma13g32090 | Gm13:34359721.0.34361227 | 34,361,073 |
| Glyma13g35520 | Gm13:36943648.0.36950770 | 36,944,632 |
| Glyma13g39540 | Gm13:40114057.0.40114640 | 40,114,263 |
| Glyma14g04190 | Gm14:2812976.0.2815950 | 2,812,976 |
| Glyma14g04200 | Gm14:2818106.0.2821324 | 2,820,386 |
| Glyma14g17330 | Gm14:19105307.0.19109002 | 19,108,051 |
| Glyma14g35580 | Gm14:44570215.0.44571936 | 44,570,215 |
| Glyma14g40090 | Gm14:49077959.0.49084565 | 49,081,971 |
| Glyma14g40250 | Gm14:49220022.0.49221998 | 49,221,894 |
| Glyma15g08070 | Gm15:5675190.0.5675401 | 5,675,305 |
| Glyma15g08350 | Gm15:5892318.0.5898492 | 5,892,294 |
| Glyma16g25770 | Gm16:29864908.0.29872224 | 29,864,835 |
| Glyma17g03940 | Gm17:2586560.0.2590026 | 2,586,583 |
| Glyma17g35300 | Gm17:39275298.0.39276346 | 39,276,103 |
| Glyma18g00440 | Gm18:148373.0.150712 | 149,539 |
| Glyma18g43100 | Gm18:52435074.0.52438457 | 52,435,987 |
| Glyma18g53850 | Gm18:62121982.0.62123472 | 62,122,569 |
| Glyma19g02320 | Gm19:2042249.0.2046314 | 2,043,219 |
| Glyma19g07410 | Gm19:8755326.0.8759630 | 8,758,960 |
| Glyma19g29690 | Gm19:37423341.0.37424387 | 37,424,167 |
| Glyma19g37430 | Gm19:44580492.0.44583090 | 44,580,764 |
| Glyma20g25660 | Gm20:35302630.0.35306802 | 35,302,638 |
| Glyma20g28620 | Gm20:37540008.0.37541602 | 37,540,335 |
| Glyma20g30490 | Gm20:39122592.0.39129208 | 39,127,429 |
| Glyma20g34300 | Gm20:42690498.0.42692467 | 42,690,537 |
Figure 3.
Locations of Tnt1 insertion sites in the soybean genome. Tnt1 flanking sequences were identified in 18 Tnt1 lines by TAIL-PCR. White arrowheads, Tnt1 inserted in coding regions; black arrowheads, Tnt1 inserted in intergenic regions; black circles, centromeres.
To obtain efficient mutagenesis in plants such as soybean, which has a relatively large genome, it will be important to use a transposon system with an insertional preference for coding regions rather than intergenic regions. One of the main advantages of using retrotransposons for mutagenesis is that they have been documented to transpose preferentially into gene-rich regions. Tnt1 does transpose preferentially into gene-rich regions in M. truncatula, Arabidopsis, and lettuce (Courtial et al., 2001; d’Erfurth et al., 2003; Mazier et al., 2007). Our analysis of 99 Tnt1 flanking sequences revealed that the element inserted into 62 (62%) annotated genes. If Tnt1 insertion into the soybean genome had occurred randomly, the tagging efficiency should have been 9.8% (46,430 genes of 2 kb per 950-Mb genome; Schmutz et al., 2010). Therefore, our results suggest that Tnt1 preferentially inserts into protein-coding regions in soybean.
Tnt1 Insertions Are Stable and Heritable in Soybean
We examined the expression of Tnt1 transposase using reverse transcription (RT)-PCR in young leaves of progeny lines derived from three independent transgenic events. As shown in Figure 4, expression of the Tnt1 was detected in the tissues of the Tnt1 transgenic plants but not in leaves of the parent line cv Maverick. Comparison of three different transgenic events showed that the level of transposase expression was variable, which may be due to positional effects at the various Tnt1 insertion sites.
Figure 4.
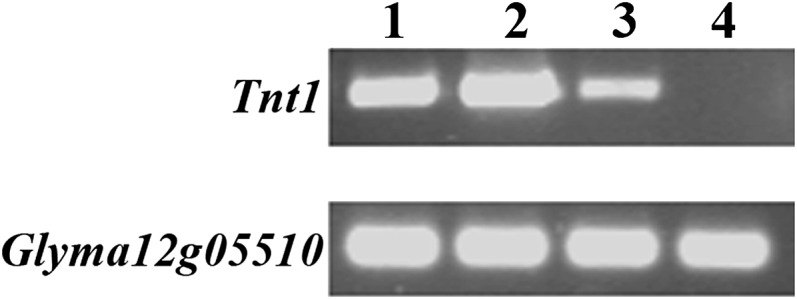
Analysis of Tnt1 expression in young leaves of Tnt1 transgenic plants by RT-PCR. As a control, expression of the constitutively expressed Glyma12g05510 gene (Libault et al., 2010) was also determined. Lane 1, BS5-12-8R; lane 2, BS5-12-12C; lane 3, BS6-19; lane 4, cv Maverick (parent line).
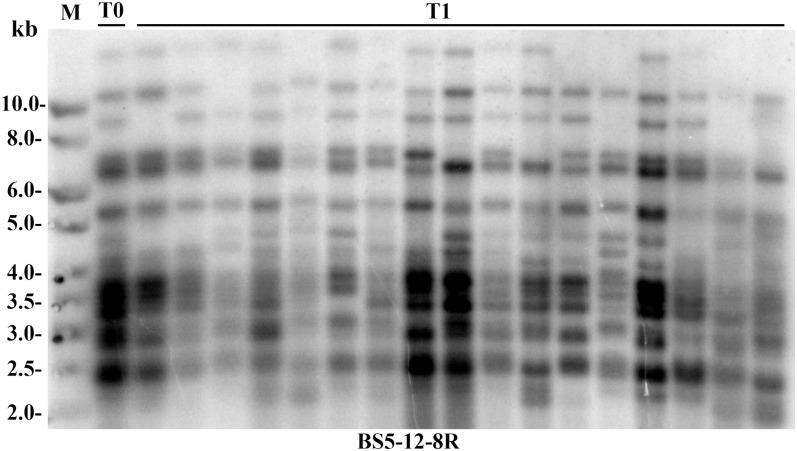
To determine if Tnt1 insertions are active in self-fertilized progeny plants, the original T0 Tnt1 soybean transgenic events were allowed to self-fertilize, and the locations of the Tnt1 insertions in progeny of six lines were examined by Southern-blot analysis. The Southern-blot analysis results of the T1 progeny of Tnt1 lines BS5-12-8R, BS6-19, BS5-6, BS5-12, BS12-7, and BS5-12-12C, as well as T2 progeny of line BS6-19, using the Tnt1 probe are shown in Figure 5 and Supplemental Figure S1. The locations of the Tnt1 insertions were found to be stable in the progeny lines because no new band was observed, indicating an absence of additional germinal or somatic transpositions. Because the Southern-blot analysis method was not accurate enough to resolve all of the different insertions in a transgenic event, the segregation of individual Tnt1 insertions from one event (BS5-12-8R) was examined using PCR amplification of selected flanking regions and scored as present, absent, or heterozygous in each of the progeny lines (Table II). For three of four tested loci, the segregation pattern was close to a ratio of 1:2:1 (25% wild type, 50% heterozygous, and 25% homozygous for a given Tnt1 insertion). These results indicate that Tnt1 insertions do follow Mendelian segregation. The segregation results for insertion 3 (Table II) indicated that no homozygous mutant locus was detected, which could be attributable to lethality associated with insertion or that the population tested (18 plants) was too small.
Figure 5.
Southern-blot analysis of the Tnt1 line BS5-12-8R and its T1 progeny obtained by self-pollination. Fifteen micrograms of chromosomal DNA from different plants was digested with NdeI and hybridized with Tnt1 probe. M, Molecular weight markers; T0, BS5-12-8R; T1, progeny of BS5-12-8R.
Table II. Segregation of Tnt1 insertions in line BS5-12-8R.
PCR analysis results are shown for the segregation of Tnt1 insertions in the progeny of line BS5-12-8R. Insertions 1, 2, 3, and 4 were inserted into Glyma20g34300, Glyma17g35300, Glyma01g01300, and Glyma19g07410, respectively. +/+, Wild-type homozygous plants; +/−, heterozygous plants; −/−, homozygous plants for a given Tnt1 insertion.
| BS5-12-8R | Insertion No. 1 | Insertion No. 2 | Insertion No. 3 | Insertion No. 4 |
|---|---|---|---|---|
| T0 | +/− | +/− | +/− | +/− |
| Plant 1 | −/− | +/− | +/− | +/− |
| Plant 2 | +/− | +/− | +/+ | +/− |
| Plant 3 | +/− | +/+ | +/− | +/+ |
| Plant 4 | +/− | +/+ | +/− | +/− |
| Plant 5 | +/− | +/− | +/− | −/− |
| Plant 6 | +/− | +/− | +/+ | +/− |
| Plant 7 | +/− | −/− | +/− | +/− |
| Plant 8 | −/− | +/+ | +/− | +/− |
| Plant 9 | −/− | +/− | +/− | +/− |
| Plant 10 | +/+ | +/+ | +/− | +/− |
| Plant 11 | +/− | +/− | +/− | +/− |
| Plant 12 | +/+ | +/− | +/− | −/− |
| Plant 13 | +/− | +/− | +/+ | +/− |
| Plant 14 | +/− | +/− | +/− | +/+ |
| Plant 15 | +/− | +/+ | +/− | +/− |
| Plant 16 | +/− | +/− | +/− | +/− |
| Plant 17 | −/− | −/− | +/− | +/− |
| Plant 18 | +/+ | +/+ | +/− | +/− |
| cv Maverick | +/+ | +/+ | +/+ | +/+ |
| Ratio | ||||
| +/+ | 3 | 6 | 3 | 2 |
| +/− | 11 | 10 | 15 | 14 |
| −/− | 4 | 2 | 0 | 2 |
In order to be useful as a mutagen, Tnt1 insertions should remain inactive during normal plant growth and exhibit segregation consistent with a single locus. It is important for mutant analysis that new rounds of transposition do not occur in subsequent generations. Similar to our findings in soybean, studies have shown that Tnt1 insertions are genetically independent and follow Mendelian segregation in Arabidopsis, M. truncatula, and lettuce (Courtial et al., 2001; d’Erfurth et al., 2003; Mazier et al., 2007). In Arabidopsis and M. truncatula, while RT products of Tnt1 are detectable, they did not result in the integration of new germinal Tnt1 copies in the progeny of transformed plants (Courtial et al., 2001; d’Erfurth et al., 2003). Although expression of the Tnt1 transposase could be detected in the vegetative tissue of transgenic plants, there was no evidence that the Tnt1 element was able to transpose in mature plants and in subsequent generations under normal growth conditions. These stable insertion events were heritable and segregated in a Mendelian fashion.
Tnt1 Transposition Can Be Reactivated in Soybean by in Vitro Culture
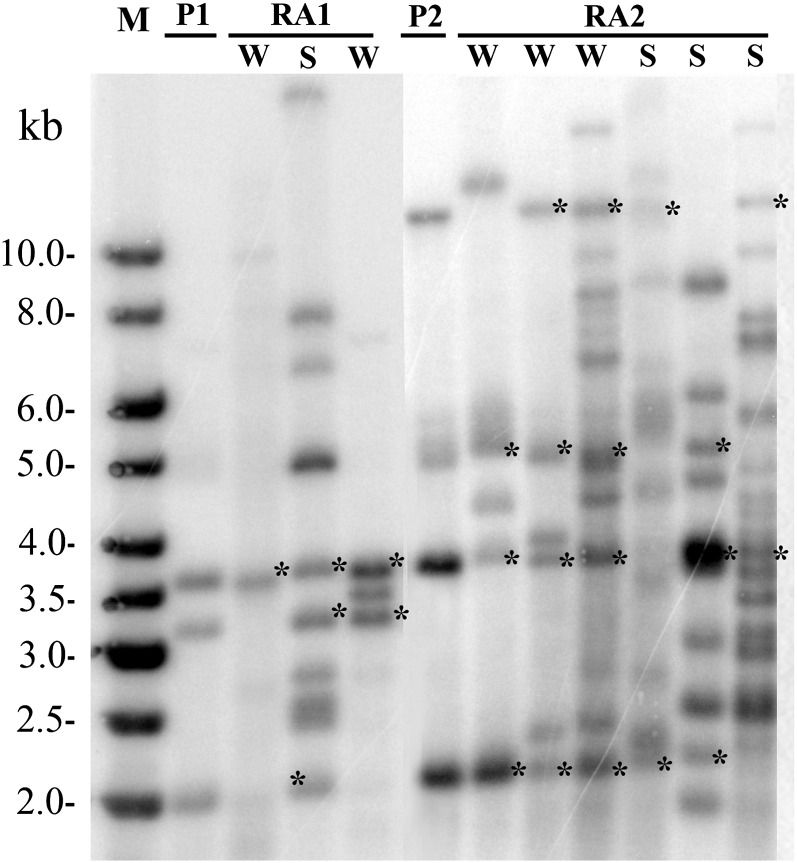
Given the time needed to produce independent soybean transgenic lines, practical use of the Tnt1 transposon in soybean would require that a few initial transgenic lines be used to reactivate the transposon through tissue culture in order to generate populations with large numbers of independent insertions. To investigate the feasibility of this approach, we tested the two published methods for soybean regeneration to gauge their ability to reactivate Tnt1 transposition. The cotyledons of the Tnt1-containing T1 plant seeds were used as explants for the first approach (Zeng et al., 2004). The explants were treated by wounding or wounding followed by 12 h of shaking in a 1 m Suc solution. The latter treatment was tested since it was reported to significantly increase the frequency of Tnt1 transposition in M. truncatula ‘Jemalong’ (Iantcheva et al., 2009). Over 40 plants were regenerated from wounded cotyledons of seven Tnt1 T0 lines with or without Suc treatment. Those plants were examined by Southern-blot analysis. The results (Fig. 6) revealed that one line, BS5-12, showed a significant number of new Tnt1 transposition events (up to 20 copies). The original T0 parental line contained only four Tnt1 insertion sites. The regenerated plants from other lines tested by this approach produced zero to five new Tnt1 insertions in the genome. However, the Suc treatment did not enhance the frequency of Tnt1 transposition in these experiments.
Figure 6.
Remobilization of Tnt1 transposition by tissue culture using cotyledons as explants with wound or wounding plus Suc treatment. Southern-blot analysis was performed by using NdeI-digested chromosomal DNA of regenerated plants from Tnt1 lines BS5-14 and BS5-12 with the Tnt1 probe. The hybridization bands that presented in parent lines are marked with asterisks, and the unmarked bands were potential novel insertions in regenerated plants. M, molecular weight standards; P1 and P2, Tnt1 primary transgenic lines BS5-14 and BS5-12, respectively; RA1, plants regenerated from line BS5-14; RA2, plants regenerated from line BS5-12; W, plants regenerated from wound-treated cotyledons; S, plants regenerated from wound plus 1 m Suc-treated explants.
In order to verify that the new bands observed were novel Tnt1 insertion sites, TAIL-PCR was performed to recover the flanking soybean sequence from four plants generated from reactivation of event BS5-12. Twenty Tnt1 flanking sequences were obtained from those plants. Ten specific primers were designed from the Tnt1 flanking sequences of reactivated line BS5-12-8R, and five primers were designed from the Tnt1 flanking sequences of reactivated line BS5-12-12C. Those primers were paired with a Tnt1-specific primer, LTR7, and used for PCR. PCR results revealed that all of the BS5-12-8R primers (paired with the LTR7 primer) produced PCR products with BS5-12-8R chromosomal DNA. Similarly, all five BS5-12-12C primers produced PCR products with genomic DNA of BS5-12-12C. No PCR product was produced using the genomic DNA of the T0 plant BS5-12 or the parent plant as template. These results confirm that the new hybridization bands observed by Southern-blot analysis were indeed novel Tnt1 insertions. Thus, our results clearly demonstrate that the cotyledon approach does reactivate Tnt1 transposition and generate additional insertion sites. However, the fact that only one line exhibited a high frequency of transposition suggests that the original site of Tnt1 insertion may affect the ability to transpose. These results are similar to the case of M. truncatula, where only a few lines were shown to transpose at a high frequency by repeated transfer in tissue culture. However, these “starter lines” were sufficient to generate a large insertional mutant population (d’Erfurth et al., 2003; Iantcheva et al., 2009). In the case of soybean, our results suggest that an extended period in tissue culture, perhaps with repeated wounding, enhanced the frequency of transposition in the BS5-12 line. Consistent with the previous results, analysis of the Tnt1 flanking sequences obtained from the reactivated plants showed that Tnt1 inserted preferentially into annotated genes in 12 (60%) of the isolated integration sites.
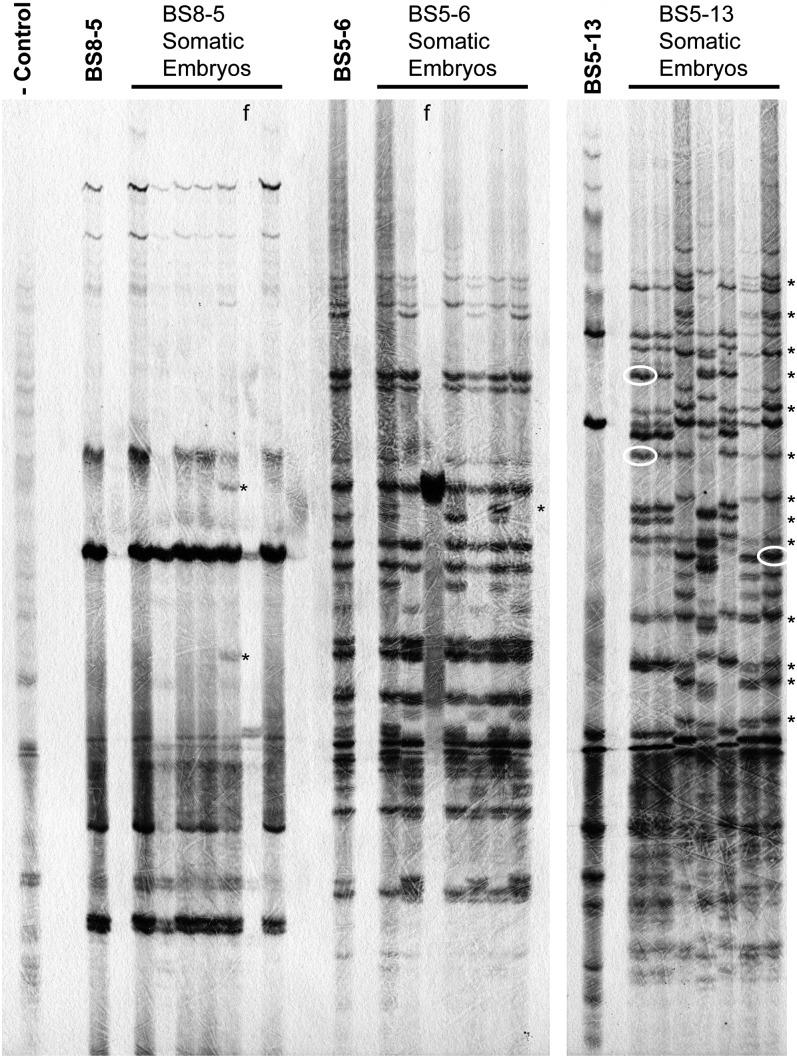
In addition to the use of cotyledonary nodes, soybean can also be regenerated from somatic embryos (Trick et al., 1997). Somatic embryos were generated from immature embryos collected from Tnt1-transformed T1 plants. Individual plants from five independent Tnt1-containing lines were selected for passage through somatic embryogenesis. During the tissue culture treatment, seven mature embryos were selected at the end of a 5-week histodifferentiation step for transposon display analysis (Van den Broeck et al., 1998; Hancock et al., 2011) using Tnt1-specific primers. This allowed for comparison of the Tnt1 insertions in the original plant and the resulting somatic embryos. As expected, the Tnt1 insertions present in the parent plant were found to segregate in a Mendelian fashion in the somatic embryos. In addition, the somatic embryos of four of the genotypes tested showed a small number (one to five) of novel bands that were not present in the parent (i.e. BS8-5 and BS5-6; Fig. 7). However, the somatic embryos produced from the BS5-13 line showed a large increase in the total number of novel bands (up to 20) in the somatic embryos (Fig. 7). Some of these novel insertions were shared between embryos, indicating that they occurred early in the production of embryogenic tissue. Some bands were also unique to single embryos, suggesting that they occurred later in embryo development. We should note that Tnt1-containing line BS5-12 was also tested for reactivation by this approach but showed only limited reactivation. Similarly, reactivation experiments performed using the cotyledon-node approach with event BS5-13 produced only one or two new Tnt1 copies (data not shown). These results suggest that the efficiency of reactivation approaches are related to the genotypes of the original Tnt1-containing lines used.
Figure 7.
Autoradiograph of Tnt1 transposon display analysis of somatic embryos produced from three Tnt1-containing lines. Potential novel insertions (not present in the parent plant) are marked with asterisks. White ovals indicate the bands that were excised from the gel for sequence analysis. Untransformed soybean DNA was used as the negative control. f, Failed lanes.
To verify that novel insertion sites arose during somatic embryogenesis, we excised three novel bands from the transposon display gel, performed PCR amplification with appropriate primers, and sequenced the PCR products. This resulted in four sequences that included both the end of the Tnt1 element and soybean genomic sequence. A homology search allowed for the insertion sites to be located in the soybean genome (Table III). Using primers that flank the Tnt1 insertion sites, we were able to verify the presence of three of these Tnt1 insertions (data not shown). Of these, one was found to be present in the original BS5-13 plant, but the remaining two Tnt1 insertions were confirmed to be novel insertions that occurred during tissue culture treatment. This analysis confirms that the majority of the bands observed in transposon display represent true transposition events.
Table III. Tnt1 insertions identified in somatic embryos.
Asterisks indicate insertions that could be verified by PCR.
| Tnt1 Location | Type of Insertion |
|---|---|
| GM10:48222768 | Intergenic |
| GM08:16425209 | Potential promoter region* |
| GM03:40433849 | Exon* |
| GM09:895535 | Approximately 2.5-kb downstream of a coding region* (present in BS5-13) |
In summary, we compared the ability of two different tissue culture methods to reactivate Tnt1 transposition. In both methods, the majority of the lines tested showed modest transposition, but in each case, a single line showed a much higher frequency of transposition. An interesting finding was that different lines were optimal for the two methods: event BS5-12 showed a higher frequency of transposition using the cotyledonary node approach, while line BS5-13 showed higher transposition during somatic embryogenesis. Therefore, these two lines represent promising starter lines for the construction of large mutant populations of soybean.
Tnt1: An Insertion Mutagen in Soybean?
Practical use of Tnt1 for mutagenesis in soybean requires the generation of several initial transgenic lines for subsequent reactivation by repeated tissue culture regeneration. This approach is especially well suited for a plant such as soybean, in which generation of the original transgenic events is laborious and time consuming. In this strategy, a higher number of insertions per line allows for a lower number of individual plants to be maintained in order to create a population suitable for mutant screening. The flanking sequences in this population can be readily identified using high-throughput sequencing methods to create a searchable database of insertion sites, comparable to those currently available for model species (Williams-Carrier et al., 2010; Urbański et al., 2012). Clearly, this approach has advantages over T-DNA or Ac/Ds mutagensis of soybean, in which a large number of independent transgenic lines would be needed (Scholte et al., 2002; Wessler, 2006; Mathieu et al., 2009). The Ac/Ds system has the added limitation that most insertions occur within a short distance (within a few centimorgan) of the original insertion site (Jones et al., 1990; Ito et al., 1999; Parinov and Sundaresan, 2000). Unlike the mPing transposable element (Hancock et al., 2011), Tnt1 appears to be stable in mature plants with no evidence of additional germinal or somatic insertions. Consistent with findings in other plant species, such as M. truncatula, Arabidopsis, and lettuce (Courtial et al., 2001; d’Erfurth et al., 2003; Mazier et al., 2007), Tnt1 transposition in soybean targets gene-rich regions preferentially, which makes it highly effective for mutagenic gene-function studies. Our data revealed that Tnt1 transposition generates from four to 20 insertions per plant in soybean. These insertions are stable during the life cycle of soybean, and they are genetically independent and can be separated by recombination. Therefore, unwanted insertions can be removed through serial backcrossing to the parental line. If one wants to work with a line with a clean single Tnt1 insertion, a couple of rounds of backcrossing will be required.
Soybean is an ancient tetraploid whose genome has undergone at least two rounds of whole-genome duplication (Schmutz et al., 2010). This raises the possibility that gene functional redundancy due to the presence of homeologous gene copies could limit the ability to obtain informative phenotypes for single transposon insertions. However, clearly, phenotypes can be obtained by chemical or radiation mutagenesis (Cooper et al., 2008; Bolon et al., 2011). Moreover, we previously identified a male-sterile mutant of soybean using Ac/Ds mutagenesis (Mathieu et al., 2009). Even in the model plant Arabidopsis, the presence of multigene families can limit the ability to obtain phenotypes by mutating a single member of the family (Stacey et al., 2006). Hence, it remains to be seen whether the paleotetraploid nature of the soybean genome would create any significant limitations to the use of large-scale transposon mutagenesis for gene functional studies.
CONCLUSION
We successfully introduced the Tnt1 retrotransposon into stably transformed soybean plants by A. tumefaciens-mediated transformation. The inserted Tnt1 elements appear to be inactive in somatic plant tissues and were inherited in a Mendelian fashion. However, the activity of these elements could be reactivated by two different tissue culture treatments. Analysis of the sequences flanking the Tnt1 insertion sites showed that the element preferentially inserts into protein-coding regions. Two Tnt1 lines, originally containing only a few copies of the Tnt1 element, were shown to be highly efficient for transposition upon passage through tissue culture; therefore, they represent highly promising lines for the development of large, mutant populations in soybean. The development and characterization of such a population would create an extremely useful resource for both basic and applied studies of this important crop plant.
MATERIALS AND METHODS
Plant Material and Plant Growth Conditions
Soybean (Glycine max ‘Maverick’) was used for all plant transformation experiments. Soybean plants were grown in soil in the greenhouse and watered alternatively with deionized water and a nutrient solution (Miracle-Gro) with a cycle of 18 h of light at 29°C and 6 h of dark at 24°C.
Bacterial Strains and T-DNA Vectors
The Escherichia coli strain DH5α (Sambrook et al., 1989) was used for cloning and the propagation of the different vectors. Agrobacterium tumefaciens strain AGL1 was used in all plant transformation experiments. Plasmids were introduced into AGL1 by direct DNA transfer (An et al., 1988). An EcoRI fragment containing the entire Tnt1 element from plasmid pHLV4909 (a gift from Helene Lucas) was cloned into the binary vector pZY101 (Vega et al., 2008) to yield pSH-Tnt1. The vector pZY101 carries the bar gene for glufosinate resistance. The resulting plasmid pSH-Tnt1 was used for all transformations. The A. tumefaciens strain was grown in yeast extract peptone medium containing rifampicin (30 mg L−1) and spectinomycin (100 mg L−1) and kept at 250-rpm shaking overnight at 28°C. Cotyledonary explants derived from 5-d-old seedlings of genotype cv Maverick were used for the cocultivation.
Plant Transformation and Selection
All T0 transgenic soybean events were developed following the protocol as described previously (Zeng et al., 2004), except that antioxidants dithiothreitol and sodium thiosulfate were added to the cocultivation medium at the concentrations of 3.3 and 1.0 mm, respectively (Olhoft et al., 2003); also, 0, 10, and 5 mg L−1 glufosinate was added to the first and second shoot induction media as well as the shoot elongation medium, respectively. Each regenerated plant was screened three times from plantlet to plant stage using herbicide leaf painting to assess the functional expression of the bar gene. All of the plant transformations were performed at the University of Missouri Plant Transformation Core facility.
Examining the Occurrence of the Tnt1 Element and T-DNA in Transformed Plants
Chromosomal DNA was isolated from plants according to Dellaporta et al. (1983). PCR experiments were performed to examine the Tnt1 insertions and T-DNA in regenerated plants. Three pairs of Tnt1-specific primers and one pair of bar-specific primers (Fig. 1) were used: TntA, 5′-TGGTATCAGAGCACAGGTTCTGCT-3′; TntB, 5-AAATGTGACAAAAAATTCGTACCT-3′; TntC, 5′-AACGGACTAATCACACAGCTTGCC-3′; TntD, 5′-ATAACTCTCGTATCCATCTCGGTC-3′; TntE, 5′-TTGATTTTGACGAAATTTTCTCCC-3′; TntF, 5′-CCTGCCATATCAGCATCTGTATAG; barA, 5-TACCATGAGCCCAGAACGCCC-3′; and barB, GGCTGAAGTCCAGCTGCCAGAAAG-3′.
Molecular Analysis
Standard procedures were used in the isolation of plasmid DNA, gel electrophoresis, PCR, DNA ligation, transformation, and electroporation (Sambrook et al., 1989). Restriction and modification enzymes were obtained from Promega. Soybean plant chromosomal DNA was extracted from young leaves according to the procedures described by Dellaporta et al. (1983). Fifteen micrograms of RNase A-treated genomic DNA for each line was digested with NdeI and separated on a 0.8% agarose Tris-acetate EDTA gel running at 30 V overnight. DNAs were transferred to Zeta Probe GT Nylon membrane (Bio-Rad Laboratories) and used for Southern-blot analysis. Southern-blot hybridizations were carried out following the procedures of Klein-Lankhorst et al. (1991). A 755-bp Tnt1 internal fragment corresponding to bases 1,067 to 1,822 of the retrotransposon was used as a probe (Fig. 1A). The Prime-a-Gene DNA labeling system (Promega) was used for labeling DNA probes. The [α-32P]dATP (3,000 Ci mol−1)-labeled probes were used for hybridization. After hybridizing with the Tnt1 probe, the blots were stripped according to the instructions of the manufacturer and reprobed with a 480-bp bar internal fragment. After washing, the membrane was exposed to a phosphor imager screen and then visualized using the FujiFilm Fluorescent Imager Analyzer FLA 3000.
RNA Isolation and RT-PCR
Total RNA from young leaves was isolated using Trizol Reagent according to the manufacturer’s instructions (Invitrogen). The isolated RNA was further purified and treated with DNase TURBO DNA-free according to the manufacturer’s instructions (Ambion) The first-strand complementary DNA was synthesized using avian myeloblastosis virus reverse transcriptase (Promega) and used as input in PCR using Taq polymerase (Promega) according to the manufacturer’s instructions with the following PCR conditions: 94°C for 5 min, then 35 cycles of 94°C for 30 s, 57°C for 1 min, and 72°C for 2 min, followed by 72°C for 5 min. The Tnt1 gene-specific forward and reverse primers used were as follows: 5′-TGGTATCAGAGCACAGGTTCTGCT-3′ (forward primer) and 5′-AAATGTGACAAAAAATTCGTACCT-3′ (reverse primer). The Cons 6 primers (Libault et al., 2010; 5′-AGATAGGGAAATTGTGCAGGT-3′ [forward primer] and 5′-CTAATGGCAATTGCAGCTCTC-3′ [reverse primer]), designed from the sequence of gene Glyma12g05510, were used as internal controls.
FISH and Fiber-FISH Analyses
Sample preparation, FISH and fiber-FISH experiments, and image processing were performed precisely as described by Gill et al. (2009). The plasmid DNAs of pSH-Tnt1 or pBS-Tnt1 (Tnt1 DNA clone into pBluescript SK+) were labeled with Texas Red and used as probes for FISH experiments. The biotin-labeled pBS-Tnt1 DNA was used as a probe for fiber-FISH experiments.
Genetic Analysis
Genomic DNA was isolated from plants according to Dellaporta et al. (1983). The segregation of different Tnt1 insertions in a randomly chosen line, BS5-12-8R, was examined by PCR using the following gene-specific forward and reverse primers on genomic DNA: 5′-CGAACATTACACCACTAAGATGTC-3′ (Glyma20g34300F) and 5′-TGACATCTCAAATTACTTTCATTG-3′ (Glyma20g34300R); 5′-TAAGGTCGTCAGCTAATGCCGATC-3′ (Glyma17g35300F) and 5′-TCAATTCTTCCCGATCGTTTACAC-3′ (Glyma17g35300R); 5′-ACCAAGCTTTGTGACTGCATCCAC-3′ (Glyma01g01300F) and 5′-TATATCTTCTTGTGGACTACAAGG-3′ (Glyma01g01300R); 5′-GCCAAGCTTGATTCCAGGGAGATA-3′ (Glyma19g07410F) and 5′-TGTTTCTGTATGGTCAGACATAAC-3′ (Glyma19g07410R); in combination with the Tnt1 right border primer 5′-TATTATTCCGCTTTATTACCGTGA-3′ (LTR7). PCR was conducted using ExTaq Polymerase (Takara) under the following conditions: 94°C for 5 min, 35 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 2 min, followed by 72°C for 5 min.
Tnt1 Flanking Sequence Isolation and Sequencing
The Tnt1 flanking sequences were recovered by TAIL-PCR as described by Ratet et al. (2006). The arbitrary primers used were AD1 [5′-NTCGA(G/C)T(A/T)T(G/C)G(A/T)GTT-3′], AD2 [5′-NGTCGA(G/C)(A/T_GANA(A/T)GAA-3′], and AD3 [5′-(A/T)GTGNAG(A/T)ANCANAGA-3′]. Three Tnt1-specific primers, Tntail3 (5′-TCTGGATGAATGAGACTGGAGG-3′, corresponding to bases 4,696–4,717 of Tnt1), LTR4 (5′-TACCGTATCTCGGTGCTACA-3′, corresponding to bases 534–553/5,258–5,277 of Tnt1), and LTR7 (5′-TATTATTCCGCTTTATTACCGTGA-3′, corresponding to bases 555–578/5,279–5,302 of Tnt1) were used for primary, secondary, and tertiary PCR, respectively. The PCR products were cloned into pGem-T Easy vector (Promega) and sequenced. DNA sequencing was performed at the DNA Core Facility of the University of Missouri.
Homology Searching
The flanking sequences of the tagged loci were compared with the sequences of the database using the BLAST program at http://blast.ncbi.nlm.nih.gov, http://www.phytozome.net/soybean.php (Phytozome), and http://soybase.org (Soybase).
Reactivation of Tnt1 Transposition
The T0 transgenic Tnt1 events were reactivated using two different tissue culture approaches. The first approach used cotyledons as explants through organogenesis-based in vitro tissue culture. All the steps and media followed the protocol described by Zeng et al. (2004) with modifications, and no A. tumefaciens inoculation was involved. The major modifications included the replacement of Murashige and Skoog-based medium (Murashige and Skoog, 1962) with B5-based medium (Gamborg et al., 1968) for all culture stages, the use of 0.2 mg L−1 indoleacetic acid and 2 mg L−1 zeatin riboside for the shoot elongation stage, as well as the deployment of a step-up selection strategy. Briefly, seeds of primary transgenic Tnt1 events were germinated for 5 d on B5-based germination medium. The cotyledonary node explants were prepared by wounding with a razor blade with or without Suc solution treatment. Suc treatment was performed by shaking (120 rpm) the wounded explants in 1 m Suc solution for 12 h. Treated explants were then cultured on B5-based shoot induction medium for the first 2 weeks and followed by an additional 2 weeks of subculture on the same fresh medium amended with 5 mg L−1 glufosinate. Explants were transferred biweekly onto fresh B5-based shoot elongation medium amended with 10 mg L−1 glufosinate. Shoots longer than 3 cm were excised and cultured in B5-based rooting medium without glufosinate selection. Each plantlet (with a shoot and roots) was transferred to Metro-mix 200 soil (Hummert International) in a Jiffy pot inside a Magenta culture vessel for acclimatization. Hardened plantlets were transferred to 3-gallon pots containing Promix soil mixed with Peters 20-20-20 (Hummert International) in a greenhouse. Plants were watered as needed. Each event was screened three times from plantlet to plant stage using herbicide leaf painting for the functional expression of the bar gene.
For the second approach, seeds of Tnt1-containing T0 plants were germinated and grown in the greenhouse. Somatic embryogenesis and plant regeneration were performed on the immature embryos collected from these plants. The production of somatic embryos was performed as described previously (Trick et al., 1997), excluding bombardment and antibiotic selection. DNA purification of the parent plant and differentiated embryos was performed using the cetyl-trimethyl-ammonium bromide method (Murray and Thompson, 1980). The transposon display protocol was essentially the same as described by Hancock et al. (2011) except using Tnt1-specific primers (Tnt1 P3 [primary amplification], 5′-CCAACCAAACCAAGTCAACA-3′; Tnt1 P4 [secondary amplification], 5′-GGTTGGCTACCAAACCAAAG-3′). Excised transposon display bands were PCR amplified with the appropriate primers and cloned into pJET1.2 (Fermentas) for sequencing.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Southern-blot analysis of the T1 progeny of Tnt1 lines BS6-19, BS5-6, BS5-12, BS12-7, and BS5-12-12C, as well as the T2 progeny of line BS6-19, obtained by self-pollination.
Acknowledgments
We are grateful to Helene Lucas and Pascal Ratet (Institut National de la Recherche Agronomique, Versailles) for providing the Tnt1 plasmid pHLV4909. Thanks to Neng Wan (University of Missouri) for taking care of the transgenic plants in the greenhouse.
Glossary
- FISH
fluorescence in situ hybridization
- TAIL
thermal asymmetric interlaced
- RT
reverse transcription
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- An G, Ebert PR, Mitra A, Ha SB. (1988) Binary vectors. In SB Gelvin, RA Schilperoot, eds, Plant Molecular Biology Manual, Vol 3. Martinus Nijhoff, Dordrecht, The Netherlands, pp 1–19
- An S, Park S, Jeong D-H, Lee D-Y, Kang H-G, Yu J-H, Hur J, Kim S-R, Kim Y-H, Lee M, et al. (2003) Generation and analysis of end sequence database for T-DNA tagging lines in rice. Plant Physiol 133: 2040–2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolon YT, Haun WJ, Xu WW, Grant D, Stacey MG, Nelson RT, Gerhardt DJ, Jeddeloh JA, Stacey G, Muehlbauer GJ, et al. (2011) Phenotypic and genomic analyses of a fast neutron mutant population resource in soybean. Plant Physiol 156: 240–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Wen J, Tadege M, Ratet P, Mysore KS. (2011) Reverse genetics in Medicago truncatula using Tnt1 insertion mutants. Methods Mol Biol 678: 179–190 [DOI] [PubMed] [Google Scholar]
- Cooper JL, Till BJ, Laport RG, Darlow MC, Kleffner JM, Jamai A, El-Mellouki T, Liu S, Ritchie R, Nielsen N, et al. (2008) TILLING to detect induced mutations in soybean. BMC Plant Biol 8: 8–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtial B, Feuerbach F, Eberhard S, Rohmer L, Chiapello H, Camilleri C, Lucas H. (2001) Tnt1 transposition events are induced by in vitro transformation of Arabidopsis thaliana, and transposed copies integrate into genes. Mol Genet Genomics 265: 32–42 [DOI] [PubMed] [Google Scholar]
- Cowperthwaite M, Park W, Xu Z, Yan X, Maurais SC, Dooner HK. (2002) Use of the transposon Ac as a gene-searching engine in the maize genome. Plant Cell 14: 713–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaporta SL, Wood J, Hicks JB. (1983) A plant DNA minipreparation: version II. Plant Mol Biol 1: 19–21 [Google Scholar]
- d’Erfurth I, Cosson V, Eschstruth A, Lucas H, Kondorosi A, Ratet P. (2003) Efficient transposition of the Tnt1 tobacco retrotransposon in the model legume Medicago truncatula. Plant J 34: 95–106 [DOI] [PubMed] [Google Scholar]
- Fladung M, Deutsch F, Hönicka H, Kumar S. (2004) T-DNA and transposon tagging in aspen. Plant Biol (Stuttg) 6: 5–11 [DOI] [PubMed] [Google Scholar]
- Fukai E, Soyano T, Umehara Y, Nakayama S, Hirakawa H, Tabata S, Sato S, Hayashi M. (2012) Establishment of a Lotus japonicus gene tagging population using the exon-targeting endogenous retrotransposon LORE1. Plant J 69: 720–730 [DOI] [PubMed] [Google Scholar]
- Gamborg OL, Miller RA, Ojima K. (1968) Nutrient requirements of suspension cultures of soybean root cells. Exp Cell Res 50: 151–158 [DOI] [PubMed] [Google Scholar]
- Gepts P, Beavis WD, Brummer EC, Shoemaker RC, Stalker HT, Weeden NF, Young ND. (2005) Legumes as a model plant family: genomics for food and feed report of the cross-legume advances through genomics conference. Plant Physiol 137: 1228–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill N, Findley S, Walling JG, Hans C, Ma J, Doyle J, Stacey G, Jackson SA. (2009) Molecular and chromosomal evidence for allopolyploidy in soybean. Plant Physiol 151: 1167–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock CN, Zhang F, Floyd K, Richardson AO, Lafayette P, Tucker D, Wessler SR, Parrott WA. (2011) The rice miniature inverted repeat transposable element mPing is an effective insertional mutagen in soybean. Plant Physiol 157: 552–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirochika H. (1993) Activation of tobacco retrotransposons during tissue culture. EMBO J 12: 2521–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirochika H, Otsuki H, Yoshikawa M, Otsuki Y, Sugimoto K, Takeda S. (1996) Autonomous transposition of the tobacco retrotransposon Tto1 in rice. Plant Cell 8: 725–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iantcheva A, Chabaud M, Cosson V, Barascud M, Schutz B, Primard-Brisset C, Durand P, Barker DG, Vlahova M, Ratet P. (2009) Osmotic shock improves Tnt1 transposition frequency in Medicago truncatula cv Jemalong during in vitro regeneration. Plant Cell Rep 28: 1563–1572 [DOI] [PubMed] [Google Scholar]
- Ishizaki T, Kato A. (2005) Introduction of the tobacco retrotransposon Tto1 into diploid potato. Plant Cell Rep 24: 52–58 [DOI] [PubMed] [Google Scholar]
- Ito T, Seki M, Hayashida N, Shibata D, Shinozaki K. (1999) Regional insertional mutagenesis of genes on Arabidopsis thaliana chromosome V using the Ac/Ds transposon in combination with a cDNA scanning method. Plant J 17: 433–444 [DOI] [PubMed] [Google Scholar]
- Jiang N, Bao ZR, Zhang XY, Hirochika H, Eddy SR, McCouch SR, Wessler SR. (2003) An active DNA transposon family in rice. Nature 421: 163–167 [DOI] [PubMed] [Google Scholar]
- Jones JD, Carland F, Lim E, Ralston E, Dooner HK. (1990) Preferential transposition of the maize element Activator to linked chromosomal locations in tobacco. Plant Cell 2: 701–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K, Terauchi K, Wada M, Hirano HY. (2003) The plant MITE mPing is mobilized in anther culture. Nature 421: 167–170 [DOI] [PubMed] [Google Scholar]
- Klein-Lankhorst RM, Rietveld P, Machiels B, Verkerk R, Weide R, Gebhardt C, Koornneef M, Zabel P. (1991) RFLP markers linked to the root knot nematode resistance gene Mi in tomato. Theor Appl Genet 81: 661–667 [DOI] [PubMed] [Google Scholar]
- Kumar A, Bennetzen JL. (1999) Plant retrotransposons. Annu Rev Genet 33: 479–532 [DOI] [PubMed] [Google Scholar]
- Kumar A, Hirochika H. (2001) Applications of retrotransposons as genetic tools in plant biology. Trends Plant Sci 6: 127–134 [DOI] [PubMed] [Google Scholar]
- Libault M, Farmer A, Joshi T, Takahashi K, Langley RJ, Franklin LD, He J, Xu D, May G, Stacey G. (2010) An integrated transcriptome atlas of the crop model Glycine max, and its use in comparative analyses in plants. Plant J 63: 86–99 [DOI] [PubMed] [Google Scholar]
- Lucas H, Feuerbach F, Kunert K, Grandbastien MA, Caboche M. (1995) RNA-mediated transposition of the tobacco retrotransposon Tnt1 in Arabidopsis thaliana. EMBO J 14: 2364–2373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu M, Winters EK, Kong F, Wan J, Wang S, Eckert H, Luth D, Paz M, Donovan C, Zhang Z, et al. (2009) Establishment of a soybean (Glycine max Merr. L) transposon-based mutagenesis repository. Planta 229: 279–289 [DOI] [PubMed] [Google Scholar]
- Mazier M, Botton E, Flamain F, Bouchet J-P, Courtial B, Chupeau M-C, Chupeau Y, Maisonneuve B, Lucas H. (2007) Successful gene tagging in lettuce using the Tnt1 retrotransposon from tobacco. Plant Physiol 144: 18–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Murray MG, Thompson WF. (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8: 4321–4325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito K, Cho E, Yang GJ, Campbell MA, Yano K, Okumoto Y, Tanisaka T, Wessler SR. (2006) Dramatic amplification of a rice transposable element during recent domestication. Proc Natl Acad Sci USA 103: 17620–17625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazaki T, Okumoto Y, Horibata A, Yamahira S, Teraishi M, Nishida H, Inoue H, Tanisaka T. (2003) Mobilization of a transposon in the rice genome. Nature 421: 170–172 [DOI] [PubMed] [Google Scholar]
- O’Brian MR, Vance CP. (2007) Legume biology: sequence to seeds. Plant Physiol 144: 537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto H, Hirochika H. (2000) Efficient insertion mutagenesis of Arabidopsis by tissue culture-induced activation of the tobacco retrotransposon Tto1. Plant J 23: 291–304 [DOI] [PubMed] [Google Scholar]
- Olhoft PM, Flagel LE, Donovan CM, Somers DA. (2003) Efficient soybean transformation using hygromycin B selection in the cotyledonary-node method. Planta 216: 723–735 [DOI] [PubMed] [Google Scholar]
- Parinov S, Sundaresan V. (2000) Functional genomics in Arabidopsis: large-scale insertional mutagenesis complements the genome sequencing project. Curr Opin Biotechnol 11: 157–161 [DOI] [PubMed] [Google Scholar]
- Parrott WA, Clemente TE. (2004). Transgenic soybean. In JE Specht, HR Boerma, eds, Soybeans: Improvement, Production, and Uses. ASA-CSA-SSSA, Madison, WI, pp 265–302
- Piffanelli P, Droc G, Mieulet D, Lanau N, Bès M, Bourgeois E, Rouvière C, Gavory F, Cruaud C, Ghesquière A, et al. (2007) Large-scale characterization of Tos17 insertion sites in a rice T-DNA mutant library. Plant Mol Biol 65: 587–601 [DOI] [PubMed] [Google Scholar]
- Ratet P, Porcedu A, Tadege M, Mysore KS. (2006) Insertional mutagenesis in Medicago truncatula using Tnt1 retrotransposon. In U Mathesius, EP Journet, LW Sumner, eds, The Medicago truncatula handbook. http://www.noble.org/MedicagoHandbook/ (December 3, 2012)
- Sambrook J, Fritsch EF, Maniatis T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, et al. (2010) Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183 [DOI] [PubMed] [Google Scholar]
- Scholte M, d’Erfurth I, Ripa S. (2002) T-DNA tagging in the model legume Medicago truncatula allows efficient gene discovery. Mol Breed 10: 203–215 [Google Scholar]
- Stacey MG, Osawa H, Patel A, Gassmann W, Stacey G. (2006) Expression analyses of Arabidopsis oligopeptide transporters during seed germination, vegetative growth and reproduction. Planta 223: 291–305 [DOI] [PubMed] [Google Scholar]
- Tadege M, Ratet P, Mysore KS. (2005) Insertional mutagenesis: a Swiss Army knife for functional genomics of Medicago truncatula. Trends Plant Sci 10: 229–235 [DOI] [PubMed] [Google Scholar]
- Tadege M, Wen J, He J, Tu H, Kwak Y, Eschstruth A, Cayrel A, Endre G, Zhao PX, Chabaud M, et al. (2008) Large-scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula. Plant J 54: 335–347 [DOI] [PubMed] [Google Scholar]
- Trick HN, Dinkins RD, Santarem ER, Di R, Samoylov V, Meurer C, Walker D, Parrott WA, Finer JJ, Collins GB. (1997) Recent advances in soybean transformation. Plant Tissue Cult Biotechnol 3: 9–26 [Google Scholar]
- Urbański DF, Małolepszy A, Stougaard J, Andersen SU. (2012) Genome-wide LORE1 retrotransposon mutagenesis and high-throughput insertion detection in Lotus japonicus. Plant J 69: 731–741 [DOI] [PubMed] [Google Scholar]
- Van den Broeck D, Maes T, Sauer M, Zethof J, De Keukeleire P, D’hauw M, Van Montagu M, Gerats T. (1998) Transposon display identifies individual transposable elements in high copy number lines. Plant J 13: 121–129 [DOI] [PubMed] [Google Scholar]
- Vega JM, Yu W, Kennon AR, Chen X, Zhang ZJ. (2008) Improvement of Agrobacterium-mediated transformation in Hi-II maize (Zea mays) using standard binary vectors. Plant Cell Rep 27: 297–305 [DOI] [PubMed] [Google Scholar]
- Wessler SR. (2006) Transposable elements and the evolution of eukaryotic genomes. Proc Natl Acad Sci USA 103: 17600–17601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams-Carrier R, Stiffler N, Belcher S, Kroeger T, Stern DB, Monde RA, Coalter R, Barkan A. (2010) Use of Illumina sequencing to identify transposon insertions underlying mutant phenotypes in high-copy Mutator lines of maize. Plant J 63: 167–177 [DOI] [PubMed] [Google Scholar]
- Zeng P, Vadnais DA, Zhang Z, Polacco JC. (2004) Refined glufosinate selection in Agrobacterium-mediated transformation of soybean [Glycine max (L.) Merrill]. Plant Cell Rep 22: 478–482 [DOI] [PubMed] [Google Scholar]