Abstract
The multifunctional movement protein (MP) of Tomato mosaic tobamovirus (ToMV) is involved in viral cell-to-cell movement, symptom development, and resistance gene recognition. However, it remains to be elucidated how ToMV MP plays such diverse roles in plants. Here, we show that ToMV MP interacts with the Rubisco small subunit (RbCS) of Nicotiana benthamiana in vitro and in vivo. In susceptible N. benthamiana plants, silencing of NbRbCS enabled ToMV to induce necrosis in inoculated leaves, thus enhancing virus local infectivity. However, the development of systemic viral symptoms was delayed. In transgenic N. benthamiana plants harboring Tobacco mosaic virus resistance-22 (Tm-22), which mediates extreme resistance to ToMV, silencing of NbRbCS compromised Tm-22-dependent resistance. ToMV was able to establish efficient local infection but was not able to move systemically. These findings suggest that NbRbCS plays a vital role in tobamovirus movement and plant antiviral defenses.
Plant viruses use at least one movement protein (MP) to facilitate viral spread between plant cells via plasmodesmata (PD; Lucas and Gilbertson, 1994; Ghoshroy et al., 1997). Among viral MPs, the MP of tobamoviruses, such as Tobacco mosaic virus (TMV) and its close relative Tomato mosaic virus (ToMV), is the best characterized. TMV MP specifically accumulates in PD and modifies the plasmodesmatal size exclusion limit in mature source leaves or tissues (Wolf et al., 1989; Deom et al., 1990; Ding et al., 1992). TMV MP and viral genomic RNA form a mobile ribonucleoprotein complex that is essential for cell-to-cell movement of viral infection (Watanabe et al., 1984; Deom et al., 1987; Citovsky et al., 1990, 1992; Kiselyova et al., 2001; Kawakami et al., 2004; Waigmann et al., 2007). TMV MP also enhances intercellular RNA silencing (Vogler et al., 2008) and affects viral symptom development, host range, and host susceptibility to virus (Dardick et al., 2000; Bazzini et al., 2007). Furthermore, ToMV MP is identified as an avirulence factor that is recognized by tomato (Solanum lycopersicum) resistance proteins Tobacco mosaic virus resistance-2 (Tm-2) and Tm-22 (Meshi et al., 1989; Lanfermeijer et al., 2004). Indeed, tomato Tm-22 confers extreme resistance against TMV and ToMV in tomato plants and even in heterologous tobacco (Nicotiana tabacum) plants (Lanfermeijer et al., 2003, 2004).
To date, several host factors that interact with TMV MP have been identified. These TMV MP-binding host factors include cell wall-associated proteins such as pectin methylesterase (Chen et al., 2000), calreticulin (Meshi et al., 1989), ANK1 (Ueki et al., 2010), and the cellular DnaJ-like protein MPIP1 (Shimizu et al., 2009). Many cytoskeletal components such as actin filaments (McLean et al., 1995), microtubules (Heinlein et al., 1995), and the microtubule-associated proteins MPB2C (Kragler et al., 2003) and EB1a (Brandner et al., 2008) also interact with TMV MP. Most of these factors are involved in TMV cell-to-cell movement.
Rubisco catalyzes the first step of CO2 assimilation in photosynthesis and photorespiration. The Rubisco holoenzyme is a heteropolymer consisting of eight large subunits (RbCLs) and eight small subunits (RbCSs). RbCL was reported to interact with the coat protein of Potato virus Y (Feki et al., 2005). Both RbCS and RbCL were reported to interact with the P3 proteins encoded by several potyviruses, including Shallot yellow stripe virus, Onion yellow dwarf virus, Soybean mosaic virus, and Turnip mosaic virus (Lin et al., 2011). Proteomic analysis of the plant-virus interactome revealed that RbCS participates in the formation of virus complexes of Rice yellow mottle virus (Brizard et al., 2006). However, the biological function of Rubisco in viral infection remains unknown.
In this study, we show that RbCS plays an essential role in virus movement, host susceptibility, and Tm-22-mediated extreme resistance in the ToMV-host plant interaction.
RESULTS
NbRbCS Interacts with ToMV MP
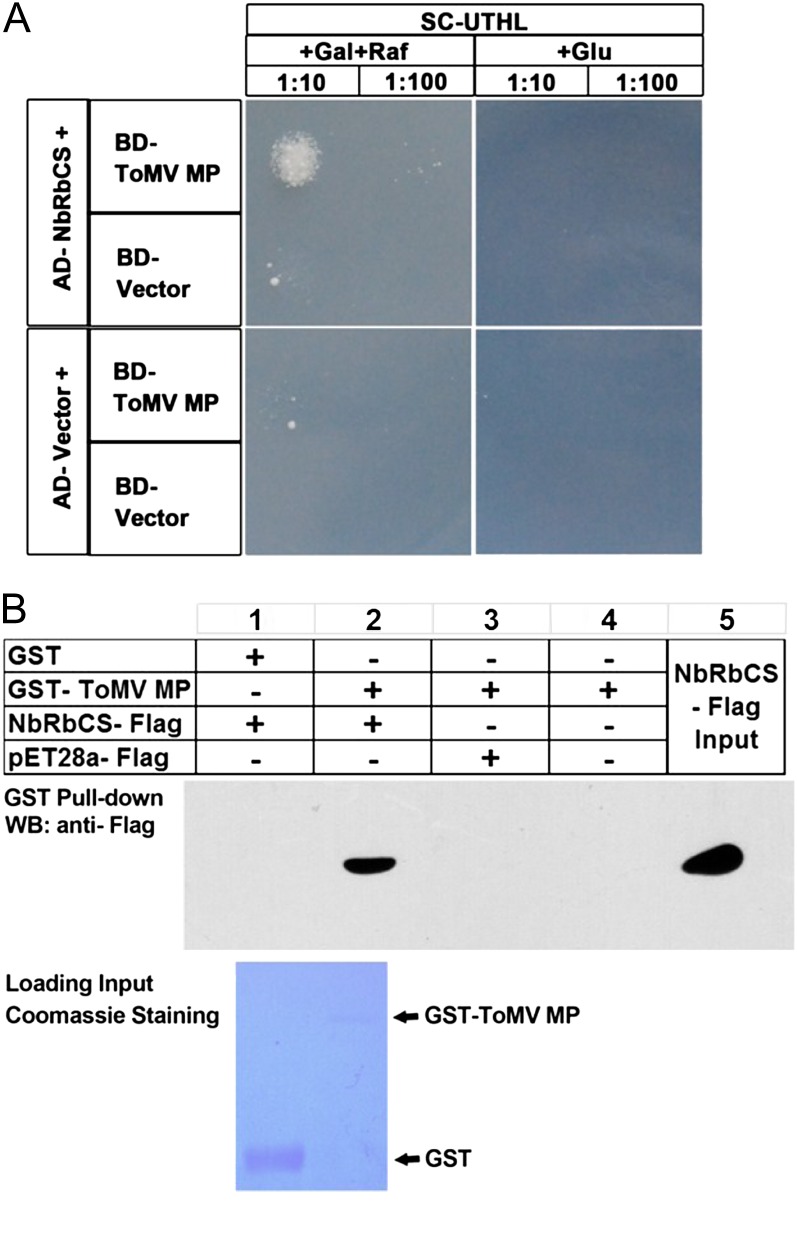
To identify ToMV MP-interacting proteins, we used ToMV MP as bait and conducted yeast (Saccharomyces cerevisiae) two-hybrid (Y2H) screens of a tomato complementary DNA (cDNA) library previously constructed in the Y2H prey vector (Liu et al., 2002b). From the screens, we obtained a number of clones that encode proteins that may interact with ToMV MP. Three of these candidates were identified as tomato RbCS cDNAs. To test whether Nicotiana benthamiana RbCS (NbRbCS) also interacts with ToMV MP, we cloned the full-length NbRbCS cDNA (N. benthamiana tentative consensus annotator no. TC23245) in the Y2H activation domain (AD) vector to produce AD::NbRbCS. Indeed, BD::ToMV MP interacted specifically with AD::NbRbCS, as indicated by the growth of yeast on Leu– plates containing Gal (Fig. 1A). However, as the controls, AD::NbRbCS and bait LexA binding domain (BD; BD vector), AD vector and BD::ToMV MP, as well as AD vector and BD vector showed no interaction (Fig. 1A).
Figure 1.
ToMV MP and NbRbCS interacted in yeast and in vitro. A, Growth of yeast strains containing NLS-LexA BD (control) or NLS-LexA BD ToMV MP baits transformed with AD-NbRbCS or AD (control) on Leu-deficient medium containing Gal and raffinose (Raf; left) or Glu (right) at 28°C for 3 d. Yeast cells were plated at 10- and 100-fold dilutions. B, GST pull-down assay to confirm the interaction between ToMV MP and NbRbCS in vitro. The total soluble proteins of E. coli expressing the NbRbCS-3×FLAG-6×His fusion protein or an empty vector 3×FLAG-6×His were incubated with purified GST-ToMV MP or GST immobilized on glutathione-Sepharose beads. Beads were washed and analyzed by SDS-PAGE and western-blot (WB) assays using anti-FLAG antibody (top panel). The bottom panel shows inputs of purified fusion proteins in pull-down assays. Equal aliquots of glutathione beads loaded with GST-ToMV MP or GST were separated by SDS-PAGE and stained with Coomassie Brilliant Blue. Black arrows indicate the bands corresponding to GST-ToMV MP and GST. [See online article for color version of this figure.]
The ToMV MP-NbRbCS interaction was further confirmed by glutathione S-transferase (GST) pull-down assays. GST-ToMV MP and NbRbCS-3×FLAG-6×His fusion proteins were separately expressed in Escherichia coli. The GST-ToMV MP fusion protein was purified using glutathione-Sepharose beads and then mixed with total soluble proteins of E. coli expressing the NbRbCS-3×FLAG-6×His fusion protein or 3×FLAG-6×His control. Western-blot analysis using an anti-FLAG antibody showed that only GST-ToMV MP specifically pulled down NbRbCS-3×FLAG-6×His, demonstrating that ToMV MP was able to bind to NbRbCS (Fig. 1B).
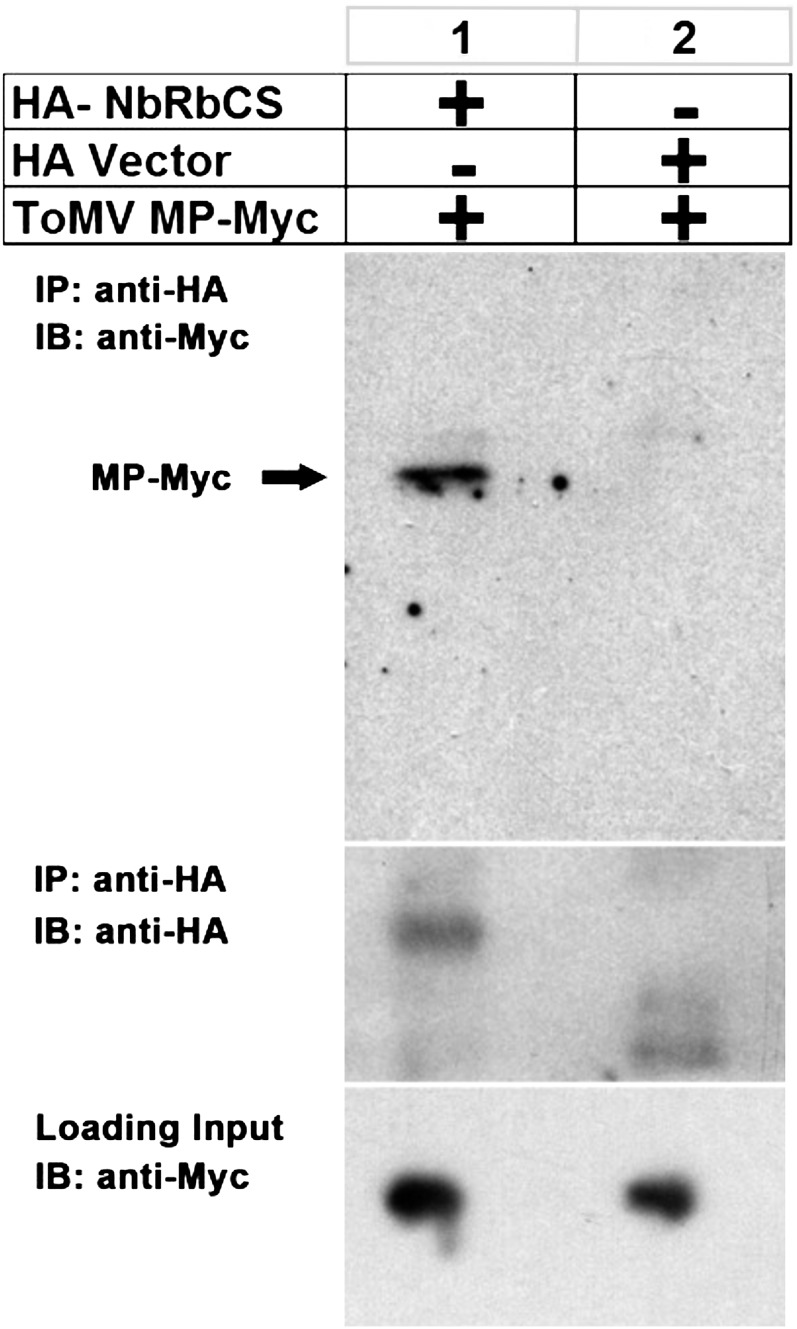
In vivo pull-down assays were performed to test if ToMV MP directly interacts with NbRbCS in planta. ToMV MP tagged with a Myc epitope (ToMV MP-Myc) and NbRbCS tagged with a hemagglutinin (HA) peptide (HA-NbRbCS) were expressed in plants under the control of Cauliflower mosaic virus 35S promoter. Expression of ToMV MP-Myc was confirmed by western-blot analysis with anti-Myc antibody (Fig. 2, bottom). Total proteins extracted from leaves coinfiltrated with Agrobacterium tumefaciens carrying the ToMV MP-Myc expression cassette together with A. tumefaciens carrying either HA vector or HA-NbRbCS expression cassettes were immunoprecipitated using an anti-HA antibody. The resulting precipitates were analyzed by western blot using an anti-Myc antibody. ToMV MP-Myc was only detected in the immunoprecipitations with HA-NbRbCS but not in immunoprecipitations with the control HA (Fig. 2, top), although the anti-HA antibody could pull down both HA-NbRbCS and control HA proteins (Fig. 2, middle). These experiments clearly demonstrate that ToMV MP interacts with NbRbCS in plant cells.
Figure 2.
In vivo pull-down assay for the interaction between ToMV MP and NbRbCS. Immunoprecipitation (IP) was performed on extracts containing transiently expressed 3×HA-NbRbCS and ToMV MP-4×Myc or extracts containing transiently expressed ToMV MP-4×Myc and 3×HA by using protein A/G agarose beads incubated with anti-HA antibody. After immunoprecipitation, precipitates were analyzed by western blot using anti-Myc (top panel) or anti-HA (middle panel) antibody. The expression of ToMV MP-4×Myc was confirmed by western-blot analysis with anti-Myc antibody (bottom panel). IB, Immunoblot.
N-Terminal and Middle Domains of ToMV MP Are Responsible for Its Interaction with NbRbCS
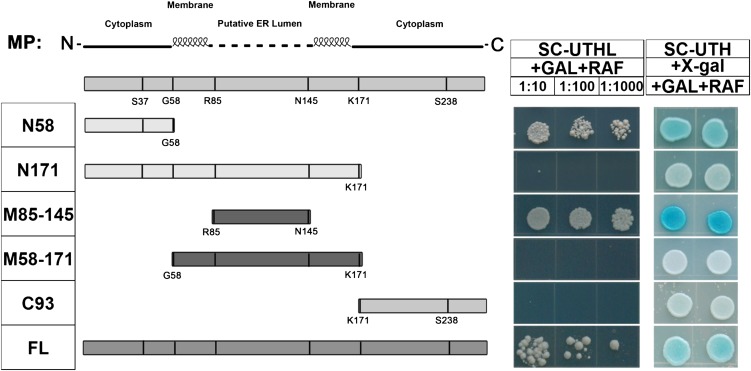
TMV MP can be divided into N-terminal, middle, and C-terminal regions joined by two putative α-helical transmembrane domains (Brill et al., 2000). The N-terminal and C-terminal regions are thought to be cytoplasmic, and the middle region of MP is presumed to fold in the endoplasmic reticulum (ER) lumen in plant cells (Brill et al., 2000, 2004). Moreover, the N-terminal and middle regions of MPs are conserved among tobamoviruses (Supplemental Fig. S1). According to this structural model, we generated a series of ToMV MP-deletion mutants in order to determine which amino acid domain(s) is responsible for ToMV MP binding to NbRbCS. The Y2H assays demonstrated that the N-terminal region of ToMV MP (amino acid residues 1–58) interacted with NbRbCS, as indicated by growth of yeast on Leu– plates containing Gal and raffinose and blue yeast colonies on plates containing 5-bromo-4-chloro-3-indolyl β-d-galactopyranoside. The middle region (amino acid residues 85–145) also interacted with NbRbCS (Fig. 3). By contrast, no interaction was observed between NbRbCS and the C-terminal fragment (amino acid residues 171–264) of ToMV MP. Surprisingly, there was also no interaction between NbRbCS and a longer N-terminal polypeptide (amino acid residues 1–171) or a longer middle amino acid section (amino acid residues 58–171). These two polypeptides contain the dual transmembrane domains, which may disturb the NbRbCS-binding activity of ToMV MP. As expected, the full-length ToMV MP protein did interact with NbRbCS (Fig. 3). These results suggest that the N-terminal and middle domains of ToMV MP are responsible for its interaction with RbCS.
Figure 3.
Analysis of NbRbCS interaction with ToMV MP deletions. The ToMV MP deletion derivatives used in Y2H assays are depicted in the left diagram. Boxes in the top bar represent the coding region, and the amino acid residues and relative positions in ToMV MP are indicated under each construct. The bars on the bottom indicate the segmental and full-length derivatives used in each assay. For each hybrid, dilutions of yeast cultures at 10−1, 10−2, and 10−3 were spotted onto Leu-deficient (left) or 5-bromo-4-chloro-3-indolyl β-d-galactopyranoside (X-gal)-containing (right) plates and grown for 3 d at 28°C. SC-UTH, Synthetic complete medium lacking uracil, Trp, and His; SC-UTHL, synthetic complete medium lacking uracil, Trp, His and Leu; RAF, raffinose.
Subcellular Localization of the NbRbCS-ToMV MP Interaction
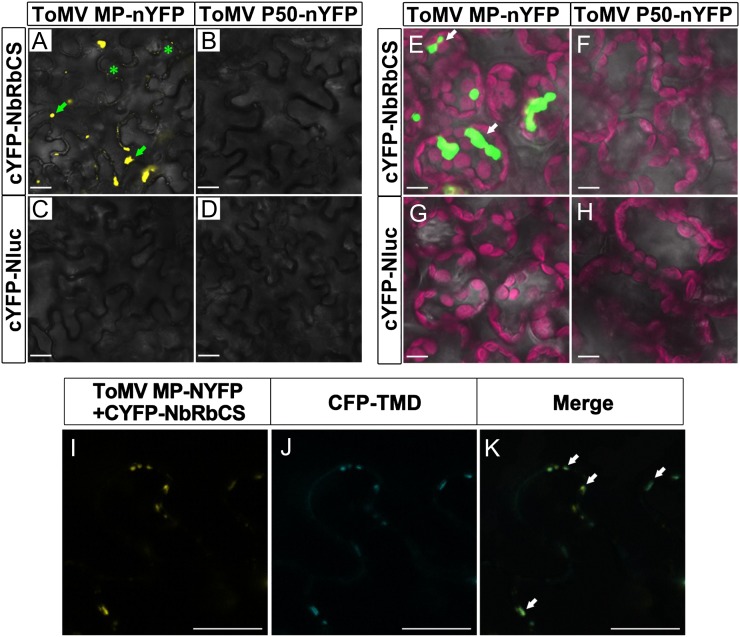
The subcellular localization of the NbRbCS-ToMV MP interaction was studied by citrine yellow fluorescent protein (YFP)-based bimolecular fluorescence complementation (BiFC) assays, where YFP was split into N-terminal (nYFP) and C-terminal (cYFP) fragments for protein fusion (Burch-Smith et al., 2007). When cYFP-NbRbCS was transiently coexpressed with ToMV MP-nYFP in N. benthamiana, bright fluorescent signals were observed in mesophyll and epidermal cells. The YFP fluorescence was observed mainly in amorphous aggresomes in the cytoplasm (Fig. 4, A and E, arrows), while some fluorescence was also found in punctate structures that were likely to be PD in the cell walls of the epidermal cells (Fig. 4A, asterisks). Indeed, the punctate-localized YFP fluorescence from the ToMV MP-RbCS interaction coincided with the PD regions labeled by the cyan fluorescent protein (CFP)-tagged PD marker CFP-TMD (for transmembrane domain of Arabidopsis plasmodesmata-located protein1a [PDLP1a]; Thomas et al., 2008; Fig. 4, I–K). Interestingly, the subcellular localization of the NbRbCS-ToMV MP interaction was similar to that of ToMV MP itself. ToMV MP-YFP accumulated primarily in PD along cell walls and partially in cytosolic inclusion bodies (referred to as virus replication complexes [VRCs]; Supplemental Fig. S2), as described previously (Kahn et al., 1998; Ashby et al., 2006). RbCS is a nucleus-encoded chloroplast protein (Chua and Schmidt, 1978; Smith and Ellis, 1979); however, no signal for NbRbCS-ToMV MP interaction was detected in chloroplasts (Fig. 4G). Moreover, no interaction signals were observed for cYFP-NbRbCS with nYFP-ToMV P50 (Fig. 4, B and F) or for various control combinations (Fig. 4, C, D, G, and H). These results demonstrate that the interaction of ToMV MP with NbRbCS may occur at PD and VRCs.
Figure 4.
In vivo subcellular localization of the NbRbCS-ToMV MP interaction by BiFC assay. A to H, NbRbCS and control N-terminal luciferase (Nluc) were tagged at the N terminus with cYFP (cYFP-NbRbCS and cYFP-nLUC) and were coexpressed transiently with ToMV MP or control ToMV P50 tagged at the C terminus with nYFP (ToMV MP-nYFP and ToMV P50-nYFP) in N. benthamiana leaves, and any fluorescence was observed in epidermal (A–D) and mesophyll (E–H) cells by confocal microscopy at 2 dpi. The NbRbCS interaction with ToMV MP was observed in cytoplasmic aggregates (indicated with arrows) and in discrete punctate sites (indicated with asterisks) along the cell edges. I to K, BiFC constructs and the PD marker CFP-tagged TMD (CFP-TMD) were coexpressed in N. benthamiana leaf epidermal cells. YFP channel (I), CFP channel (J), and merged images (K) show that good colocalization existed between the subcellular localization of the ToMV MP-NbRbCS interaction (YFP) and CFP-TMD in discrete punctate sites along the cell border. Green fluorescence shows sites with the two YFP and CFP fluorescence merged. Arrows indicate the positions of overlain YFP and CFP fluorescence. Bars = 20 μm.
Silencing of NbRbCS Enables ToMV to Induce Local Necrosis But Reduces Virus Systemic Movement
To examine the biological relevance of the NbRbCS-ToMV MP interaction to ToMV infection of plants, we used the Tobacco rattle virus (TRV) virus-induced gene silencing (VIGS) vector (Liu et al., 2002a) to silence NbRbCS in N. benthamiana. Western-blot assays showed that silencing of NbRbCS dramatically reduced NbRbCS protein levels (Fig. 5A). We also noted that silencing of NbRbCS caused leaf chlorosis and upward leaf curling (Fig. 5B, top panels).
Figure 5.
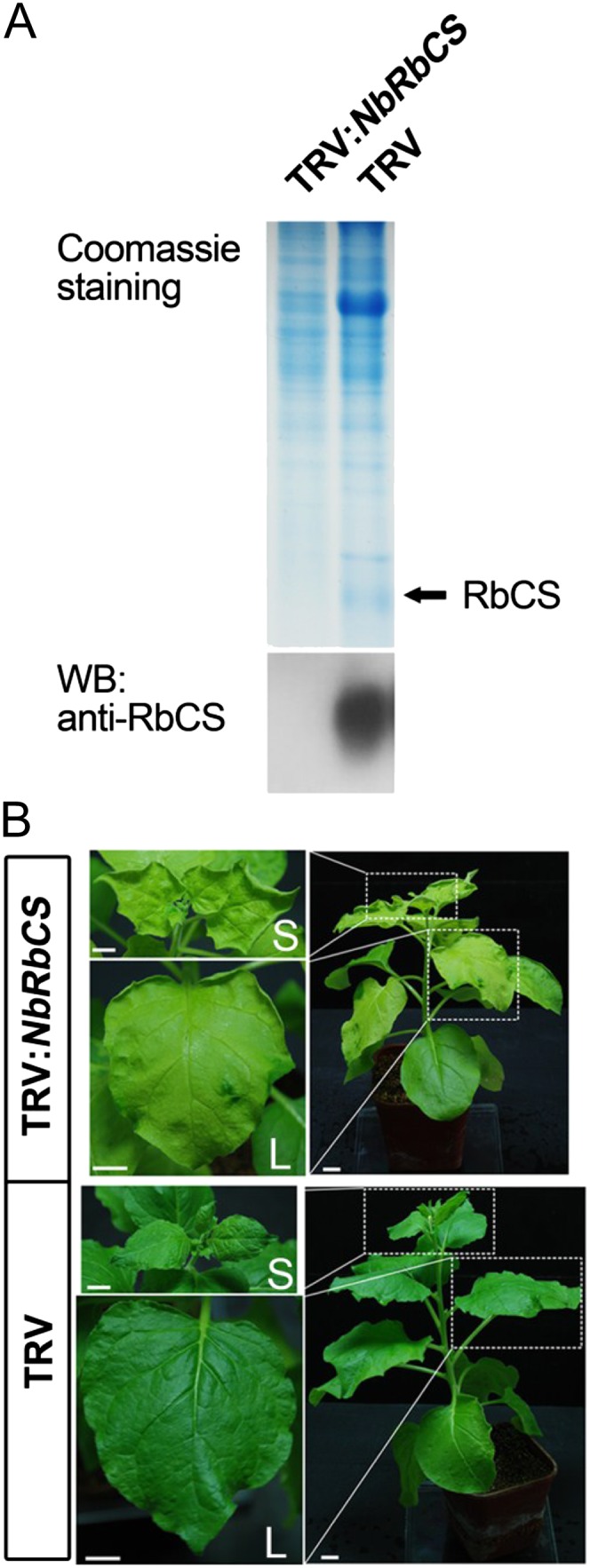
Silencing of NbRbCS induces leaf chlorosis in N. benthamiana plants. A, Western blot (WB) to confirm the silencing of NbRbCS at the protein level. Total protein from equal weights of NbRbCS-silenced leaves and nonsilenced TRV control leaves was separated by SDS-PAGE and stained with Coomassie Brilliant Blue (top panel). Parallel samples were detected by western blotting using anti-RbCS antibodies (bottom panel). For each sample, protein fractionation and immunoblotting were performed with three replications. B, Silencing of NbRbCS resulted in leaf chlorosis. Expanded upper leaves (L) and shoot apices (S) within the dotted rectangles are shown in magnification on the left. Photographs were taken 3 weeks after VIGS infiltration. Bars = 1 cm.
To test the role of RbCS in virus infection, when NbRbCS-silencing phenotypes had fully established in N. benthamiana plants, the plants were mechanically inoculated with ToMV and monitored for viral spread and symptom development. In the NbRbCS-silenced plants, ToMV induced necrosis in the inoculated leaves at approximately 60 h post infection (Fig. 6, top row, red arrows). However, the noninfective plant sap control (Supplemental Fig. S3A) and TMV-GFP (Supplemental Fig. S4A, top row) did not induce any necrosis in the NbRbCS-silenced plants, suggesting that ToMV specifically induced necrosis. Moreover, necrosis spread out along proximate veins and resulted in the collapse of large leaf patches or even the entire leaf lamina. However, no necrosis was observed in NbRbCS-nonsilenced TRV control plants (Fig. 6, bottom row). In addition, NbRbCS silencing increased the number of TMV-GFP infection foci in the inoculated leaves and the percentage of total infected leaf area (Supplemental Fig. S4), although it reduced the size of a single TMV-GFP focus (Supplemental Fig. S4). These results suggest that silencing of NbRbCS may enhance host susceptibility to tobamoviruses.
Figure 6.
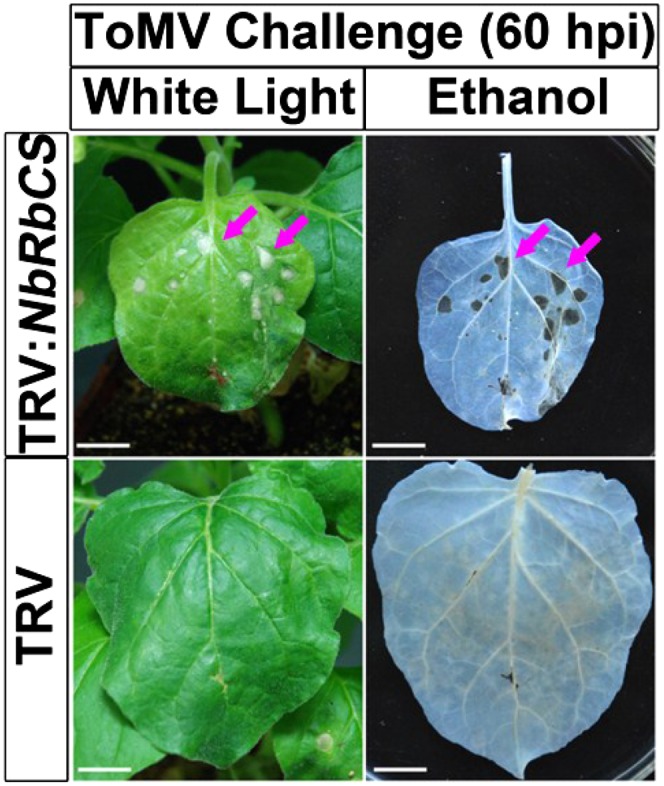
Silencing of NbRbCS enables ToMV to induce local necrosis. ToMV induced necrosis in the inoculated leaves of NbRbCS-silenced N. benthamiana plants. Leaf photographs were taken at 60 h post infection (hpi) when necrosis appeared on NbRbCS-silenced plants (left column). After being cleared with ethanol, leaves were photographed again (right column). Bars = 1 cm.
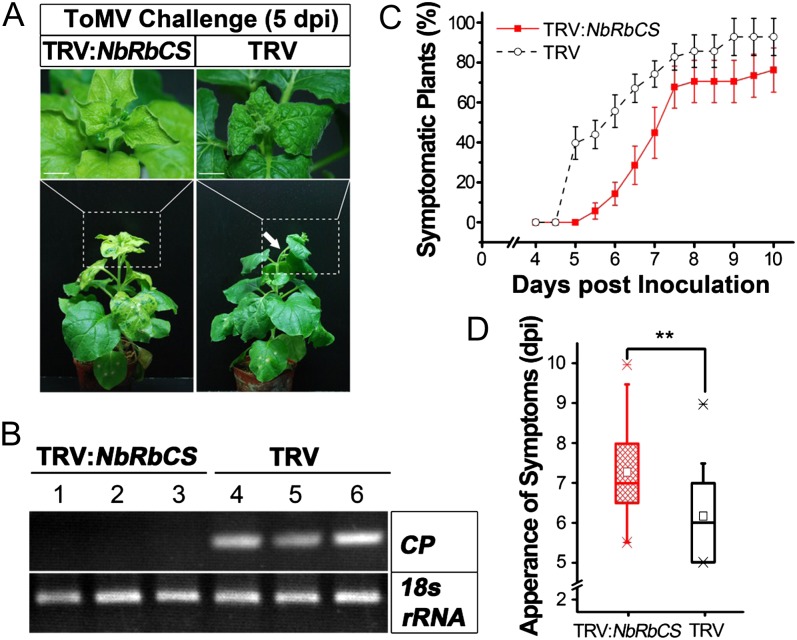
To follow up on the local infection results, we next examined the effect of NbRbCS silencing on systemic trafficking of ToMV. Viral systemic symptom development can be used as an indicator to monitor long-distance trafficking of ToMV and other plant viruses (Leisner et al., 1993; Li et al., 2005; Lewis and Lazarowitz, 2010). In N. benthamiana, ToMV systemic infection resulted in curved apical shoots, crinkled apical leaves, chlorotic mottling of newly emerged leaves, and backward winding of young leaves (Fig. 7A, right column). Surprisingly, we found that NbRbCS silencing delayed the appearance of ToMV systemic symptoms. Systemic symptoms were first observed in approximately 5% of NbRbCS-silenced plants at 5.5 d post infection (dpi), but more than 40% of nonsilenced TRV control plants had systemic ToMV infection at 5.5 dpi (Fig. 7, C and D). Furthermore, ToMV RNA can be detected by reverse transcription (RT)-PCR at 5 dpi in the upper uninoculated leaves of TRV control plants with viral symptoms but not in the upper uninoculated leaves of NbRbCS-silenced plants without viral symptoms (Fig. 7B), supporting the idea that the viral symptoms can be used as an indicator of the presence of ToMV. Systemic symptoms appeared at 7 dpi in almost 80% of nonsilenced control plants compared with approximately 45% of NbRbCS-silenced plants. Furthermore, more than 20% of NbRbCS-silenced plants remained symptomless, but all NbRbCS nonsilencing plants became systemically infected with ToMV at 10 dpi (Fig. 7C). The general tendency was that systemic infection occurred 1 to 2 d later in NbRbCS-silenced plants than in nonsilenced plants (Fig. 7D).
Figure 7.
Silencing of NbRbCS delays systemic infection by ToMV. A, Silencing of NbRbCS delays the systemic symptoms induced by ToMV. Plants were photographed at 5 dpi (bottom row). The white arrow points out the shoot curling caused by ToMV systemic infection. Shoot apices are shown in partially enlarged images in the top row. Bars = 1 cm. B, RT-PCR detection of ToMV RNA from TRV control plants with viral symptoms and NbRbCS-silenced plants without viral symptoms at 5 dpi. Representative gels show RT-PCR products (32 cycles of amplification) corresponding to fragments of ToMV CP and the control 18s rRNA. C, Percentage of plants that displayed ToMV-induced systemic symptoms. All values depicted with symbols and bars in line charts are means ± sd from six independent experiments. D, Box plots depict dpi for ToMV systemic infection. Significantly slower ToMV systemic infection was observed in NbRbCS-silenced plants compared with that in nonsilenced TRV control plants. Asterisks above the bars denote the significance of differences between NbRbCS-silenced plants and nonsilenced control plants according to two-sample Student’s t test (**P < 0.01; n = 51). Whiskers represent 5% to 95% intervals of each grouped data set, and crosses represent maximal and minimal observations, respectively.
Taken together, these findings suggest that silencing of NbRbCS enhances the local susceptibility of N. benthamiana to ToMV infection but reduces ToMV long-distance movement.
Silencing of NbRbCS Compromised Tm-22-Mediated Extreme Resistance against ToMV
Enhanced host susceptibility to viruses in NbRbCS-silenced plants suggests that NbRbCS may have a role in plant antiviral defense. To test this hypothesis, we used RT-PCR to examine the effect of silencing NbRbCS on the expression of the defense-related PR1a gene. As seen in Supplemental Figure S5, PR1a RNA levels were reduced in the NbRbCS-silenced plants compared with the nonsilenced TRV control plants agroinfiltrated with TRV empty VIGS vector. However, the level of 18S rRNA was not affected by NbRbCS gene expression. These results suggest that NbRbCS may participate in antiviral defenses in plants.
To further test this idea, we investigated whether the ToMV MP-NbRbCS interaction has a role in Tm-22 gene-mediated resistance against ToMV infection. This experiment was performed in transgenic Tm-22 N. benthamiana line T1-4, which is extremely resistant to both ToMV and TMV (J. Zhao and Y. Liu, unpublished data; Supplemental Materials and Methods S1). It should be noted that ToMV MP is the corresponding avirulence protein that is recognized by the Tm-22 gene product. As expected, the NbRbCS-nonsilenced T1-4 plants were extremely resistant to ToMV infection, and no hypersensitive response (HR) was observed in the inoculated leaves (Fig. 8A, bottom row). However, ToMV induced HR necrosis in the inoculated leaves of NbRbCS-silenced T1-4 plants at 4 to 5 dpi (Fig. 8A, top row, arrows). In addition, the noninfective plant sap control did not induce any necrosis in the NbRbCS-silenced T1-4 plants (Supplemental Fig. S3B). Furthermore, ToMV RNA was readily detectable by RT-PCR at 4 dpi in the inoculated leaves of NbRbCS-silenced plants but not in nonsilenced T1-4 plants (Fig. 8B). Moreover, we consistently failed to detect any ToMV RNA in the upper noninoculated leaves of both NbRbCS-silenced and control plants even at 6 weeks post ToMV infection (data not shown). These data suggest that NbRbCS is required for Tm-22-mediated local extreme resistance.
Figure 8.
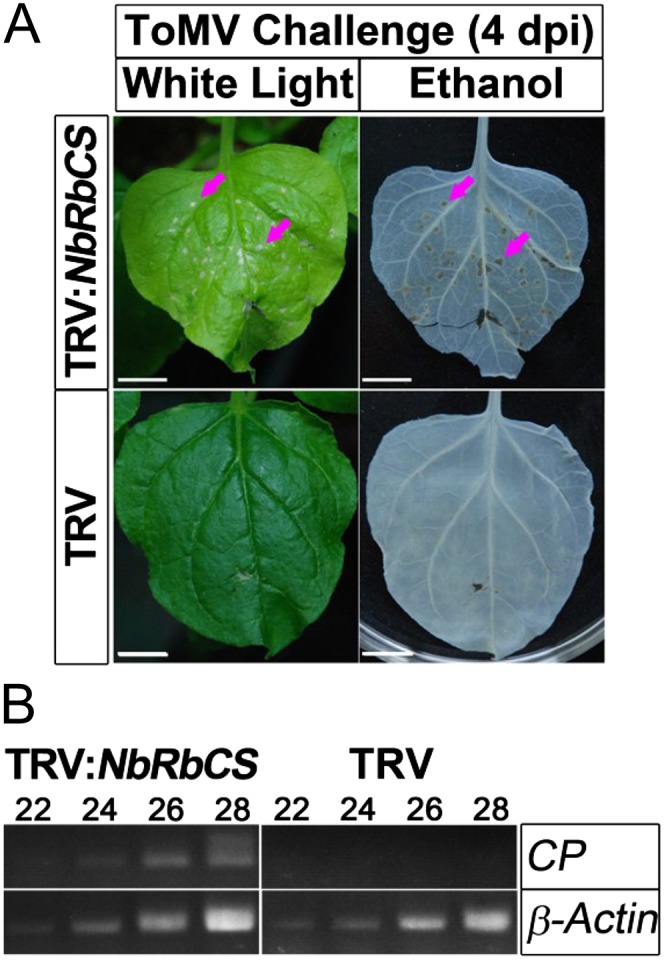
Silencing of NbRbCS compromised Tm-22-mediated local resistance against ToMV. ToMV triggered the HR in inoculated leaves in NbRbCS-silenced but not in nonsilenced control Tm-22 transgenic plants. A, Photographs were taken at 4 d post ToMV infection (left column), and then the excised leaves were decolored in ethanol and photographed again (right column). Red arrows indicate the HR lesions. Bars = 1 cm. B, RT-PCR detection of ToMV CP transcripts in inoculated leaves from NbRbCS-silenced and control Tm-22 transgenic plants. The number of PCR cycles is shown above each lane. β-Actin was included as a loading control, and photographs were typical results from three repeated experiments.
DISCUSSION
RbCS May Interact with MPs from Many Tobamoviruses
Through a range of in vitro and in vivo protein-protein interaction assays, we have identified NbRbCS as a new ToMV MP-interacting host protein. NbRbCS interacted with the N-terminal and middle domains of ToMV MP. These domains are conserved among tobamovirus MPs, suggesting that RbCS may commonly interact with MPs from tobamoviruses. Indeed, we found that NbRbCS associated and interacted with TMV MP in planta (Supplemental Figs. S6 and S7; Supplemental Results S1). RbCS is translated in the cytoplasm and then imported into chloroplasts (Keegstra et al., 1995; Schatz and Dobberstein, 1996). However, we found that NbRbCS interacts with ToMV and TMV MPs in the cytoplasm but not in the chloroplast. It is possible that both ToMV and TMV MPs may form a protein complex with NbRbCS and, consequently, hijack this protein prior to its entry into the chloroplast to facilitate virus infection of plants.
Role of RbCS in Host Susceptibility to Virus Infection and in Plant Defense
RbCS is expressed in mesophyll cells but not in nonphotosynthetic epidermal cells (Lu et al., 2002). Removal of the lower RbCS-free epidermis from plant leaves reduces infection by TMV or Tobacco necrosis virus (Wieringabrants, 1981), suggesting that RbSC may have a negative impact on these viruses. Consistent with this hypothesis, NbRbCS silencing enabled ToMV to establish severe local infections and produce necrotic lesions in inoculated leaves. Furthermore, silencing of NbRbCS caused a significant increase in the numbers of local TMV infection foci (Supplemental Fig. S4B), demonstrating that RbCS negatively affects host susceptibility to tobamoviruses. This conclusion is further supported by the finding that NbRbCS silencing reduced defense-related gene expression and compromised Tm-22 gene-mediated local extreme resistance against ToMV (Fig. 8) and TMV-GFP (Supplemental Fig. S8; Supplemental Results S1). Strikingly, NbRbCS silencing only compromised Tm-22 gene-mediated local extreme resistance but not systemic resistance against ToMV. It is possible that silencing of NbRbCS completely disrupted Tm-22 gene-mediated systemic resistance against ToMV, but the virus cannot move systemically, because the systemic movement is prevented by the lack of NbRbCS. However, we cannot rule out the possibility that Tm-22 gene-mediated local and systemic resistance can be functionally separated.
In several cases, resistance gene-mediated HR and virus resistance have been found to require light (Chandra-Shekara et al., 2006), and the expression of Rubisco genes, including RbSC, is light dependent. It is well known that dark-treated plants are more susceptible to TMV infection and that light treatments decrease plant susceptibility to TMV infection (Helms and McIntyre, 1967). These findings suggest that RbCS may play a role in plant defense. In addition, NbRbCS silencing could have a general effect on plant metabolism, and this effect may reduce plant defenses and further cause an enhanced susceptibility to viral infections. It is possible that healthy plants have sufficient levels of RbCS and are thus less susceptible to infection by tobamoviruses. However, ToMV MP may interact with RbCS and disturb its function to optimize cellular environments that are suitable for the establishment of ToMV infection. On the other hand, virus infection could be effectively established in cells in which levels of RbCS are reduced as in NbRbCS-silenced plants.
Besides RbCS, the 33-kD subunit of the oxygen-evolving complex (OEC) and Rab GDP dissociation inhibitor (GDI) are involved in TMV infection (Abbink et al., 2002; Kramer et al., 2011). Both the OEC 33-kD subunit and GDI interact with TMV 126-kD replicase and negatively regulate host susceptibility. The OEC 33-kD subunit was suggested to have a general role in defense, and GDI was suggested to participate in vesicle trafficking to enhance the establishment of an infection (Abbink et al., 2002). TMV MP was also suggested to function as an RNA silencing enhancer to influence host cell gene expression and thus to enhance host susceptibility to TMV infection (Vogler et al., 2008; Amari et al., 2012). Whether RbCS works together with the OEC 33-kD subunit and/or GDI to affect virus infection and whether RbCS participates in RNA silencing need to be further investigated.
Role of RbCS in Virus Movement
Both RbCS and RbCL were reported to interact with the P3 proteins encoded by different potyviruses (Lin et al., 2011). Furthermore, potyvirus P3 localizes to PD and acts in intercellular virus movement (Wei et al., 2010; Wen and Hajimorad, 2010). These reports are consistent with our finding that NbRbCS interacts with MPs of ToMV and TMV, which are involved in viral cell-to-cell movement. Indeed, silencing of NbRbCS reduced the size of TMV infection foci (Supplemental Fig. S4, A and D; Supplemental Results S1), suggesting that MP-binding NbRbCS participates in cell-to-cell movement of TMV. Viral infection induces the formation of irregularly shaped cytoplasmic inclusion bodies, termed X-bodies, viroplasms, or VRCs in plants (Heinlein et al., 1998; Más and Beachy, 1999; Szecsi et al., 1999). TMV-induced VRCs are ER-derived aggregates that contain viral RNA, 126- and 183-kD replicase, MP, Coat Protein (CP), and host factors (Más and Beachy, 1999). MP-binding NbRbCS seems to be one of these host factors, since we found that the NbRbCS interaction with MPs of ToMV and TMV mainly localized to irregularly shaped cytoplasmic inclusion bodies (Fig. 4; Supplemental Fig. S7) and NbRbCS could only form irregularly shaped cytoplasmic inclusion bodies when ToMV MP was coexpressed (Supplemental Fig. S9). Although the formation of inclusion bodies from ER in infected cells might be dispensable for viral replication, VRCs may exist to regulate TMV RNA movement efficiency (Boyko et al., 2000; Waigmann et al., 2007). NbRbCS may participate in viral cell-to-cell movement by regulating the function of MP within VRCs.
We found that silencing of NbRbCS reduces the systemic movement of ToMV and TMV (Fig. 7; Supplemental Fig. S10; Supplemental Results S1). This reduction may be caused by reduced virus cell-to-cell movement in NbRbCS-silenced plants, because silencing of NbRbCS reduces the size of TMV infection foci (Supplemental Fig. S4, A and D). However, there is also a possibility that NbRbCS is involved in ToMV systemic movement through the change in flow of photoassimilates in NbRbCS-silenced plants. It was proposed that plant viruses, paralleling the flow of photoassimilates, spread systemically through the vascular system (Wintermantel et al., 1997; Susi et al., 1999). Silencing of NbRbCS may cause a change in the flow of photoassimilates because of leaf chlorosis and reduced photosynthesis. Consistent with this hypothesis, TMV MP does affect carbohydrate metabolism and photoassimilate translocation during TMV infection. Transgenic overexpression of TMV MP resulted in pale-green leaves, elevated carbohydrate accumulation, and reduced rates of photoassimilate export in source leaves (Lucas et al., 1993; Olesinski et al., 1995, 1996; Almon et al., 1997). Furthermore, TMV MP induced alterations in carbon partitioning in the mesophyll and is involved in regulating photosynthesis. However, this process is independent of change in the PD size-exclusion limit (Balachandran et al., 1995; Olesinski et al., 1995). TMV MP may also attenuate photosynthesis and carbon metabolism by affecting certain mesophyllic factors (Wolf and Millatiner, 2000). Along with a decrease of Rubisco during TMV infection (Itaya et al., 2002) and tobamovirus MP-RbCS physical interactions, it is possible that RbCS may act as a potential node linking MP with photoassimilate allocation and photosynthesis. Thus, reduction of functional RbCS levels may account for the inability of tobamovirus to traffic over long distances.
MATERIALS AND METHODS
Plant Materials and Plasmids
Transgenic line T1-4 (also called TM#1) is a Tm-22-transformed Nicotiana benthamiana line that shows extreme resistance against ToMV and TMV (J. Zhao and Y. Liu, unpublished data; Supplemental Materials and Methods S1). Vectors pTRV1 and pTRV2-LIC were described previously (Liu et al., 2002a; Dong et al., 2007); pTRV2-NbRbCS was generated by cloning PCR products of NbRbCS cDNA into pTRV2-LIC. DNA fragments of TMV MP-nLUC, ToMV MP-nLUC, cLUC-NbRbCS, cYFP-NbRbCS, ToMV MP-nYFP, ToMV p50-nYFP, and CFP-TMD were obtained by overlapping PCR. The resulting PCR products were cloned between the duplicated Cauliflower mosaic virus 35S promoter and Nos terminator of pJG045, a pCAMBIA1300-based T-DNA vector (J. Zhao and Y. Liu, unpublished data). Full-length ToMV MP was cloned into pGEX4T-1 vector to express GST-tagged fusion proteins in Escherichia coli. The NbRbCS-3×FLAG-6×His fusion DNA fragment was cloned into pET28a to express double-tagged RbCS-3×FLAG-6×His in E. coli.
Y2H Screen and Interaction Assays
The full-length ToMV MP gene was PCR amplified and cloned into pYL302, in which the nuclear localization signal sequence of the LexA DNA BD is under the control of a GAL10 promoter (Y. Liu and S.P. Dinesh-Kumar, unpublished data), to generate the LexA DNA BD containing bait vector BD-ToMV MP. The full-length NbRbCS gene was PCR amplified and cloned into pJG4-5 (Kolonin et al., 2000) to produce the B42 DNA AD-containing prey vector AD-NbRbCS. The Y2H prey library containing tomato (Solanum lycopersicum) cDNA (Liu et al., 2002b) was used to screen ToMV MP-binding proteins. The Y2H screen and interaction assays were performed as described (Liu et al., 2002b).
Transient Expression by Agroinfiltration and VIGS
All plants were grown in pots at 22°C in a growth chamber under a 16-h-light/8-h-dark cycle with 120 µmol m–2 s–1 white light illumination. For Agrobacterium tumefaciens-mediated transient expression studies, GV2260 strains containing the relevant expression vector were grown overnight, pelleted, resuspended in infiltration buffer (10 mm MgCl2, 10 mm MES, and 200 μm acetosyringone, pH 5.6), and kept at room temperature for 4 h. A. tumefaciens cultures (optical density at 600 nm = 1.0) were infiltrated into N. benthamiana leaves. The infiltrated leaves were detached at 60 to 72 h post infiltration for the corresponding assays. For coexpression, equal amounts of A. tumefaciens cultures were mixed and used for infiltration. VIGS was performed as described (Liu et al., 2002a).
RNA Extraction, RT, and RT-PCR
Total RNA was extracted using TRIzol reagent (Tiangen) according to the manufacturer’s protocol. First-strand cDNA was synthesized from 2 μg of RNA using a 21-nucleotide [oligo(dT) plus two anchoring nucleotides] or gene-specific primer and Moloney murine leukemia virus reverse transcriptase (Tiangen). DNA fragments corresponding to 18S rRNA, Actin, NbRbCS, ToMV MP, CP, and GFP were PCR amplified using the respective primers. The sequences of the primers used in PCR are available upon request.
Monitoring Protein Expression Levels and in Vivo Pull-Down Assays
Total proteins from N. benthamiana leaves were extracted in buffer containing 150 mm Tris-HCl, 150 mm NaCl, 25 mm sodium fluoride, 0.1% Triton X-100, pH 7.5, 2 mm dithiothreitol, 0.5 mm phenylmethanesulfonyl fluoride (PMSF), and plant protease inhibitor cocktail (Sigma-Aldrich) as described (Serino et al., 1999). Protein extracts were separated by 10% SDS-PAGE and transferred to Immobilon-P polyvinylidene difluoride membranes (Millipore) for western-blot analysis using anti-RbCS, anti-HA, or anti-Myc (Santa Cruz) primary antibody. For in vivo pull-down assays, protein extracts were incubated with the anti-HA antibody (1:200 diluted) for 4 h at 4°C following overnight incubation with protein A/G plus agarose beads (AbMART) equilibrated with the extraction buffer. The beads were washed three times with the extraction buffer. Immunoprecipitated samples were used for western-blot analysis as described above using the anti-Myc primary antibody.
GST Pull-Down Assays
GST-ToMV MP and NbRbCS-3×FLAG-6×His fusion proteins were produced in BL21 (DE3) cells (Stratagene) and purified using glutathione-Sepharose beads (GE Healthcare) and nickel-nitrilotriacetic acid agarose resins (Qiagen) according to the manufacturer’s instructions. About 1 µg of purified GST fusion proteins or GST was incubated for 3 h with purified NbRbCS-3×FLAG-6×His or 3×FLAG-6×His at 4°C in 0.2 mL of buffer A (100 mm NaCl, 50 mm Tris-Cl, pH 7.5, 0.1 mm EDTA, 0.1 mm EGTA, 50 mm NaF, 0.2% Triton X-100, 0.1% β-mercaptoethanol, 1 mm PMSF, and complete protease inhibitor). The beads were washed three times with ice-cold buffer B (100 mm NaCl, 50 mm HEPES, pH 7.5, 0.1 mm EDTA, 1 mm PMSF, and complete protease inhibitor) at 4°C. The washed beads were boiled in SDS sample buffer, and proteins were separated by SDS-PAGE for western-blot assays using the anti-FLAG antibody.
BiFC and Fluorescence Microscopy
Citrine YFP-based BiFC was performed as described (Burch-Smith et al., 2007). Live plant imaging was performed on a Zeiss LSM710 confocal microscope. Enhanced GFP- or citrine YFP-derived fluorescence was acquired using excitation 488-nm laser and emission 493- to 598-nm filters and excitation 514-nm laser and emission 519- to 587-nm filters, respectively. CFP fluorescence was excited with a 405-nm laser line, and the emissions were captured at 430 to 460 nm. For colocalization assays, two fluorescent channels were separately monitored in two tracks, and signals were pseudocolored in digital images as described in the figure legends. Twelve-bit confocal images were acquired with an EC Plan-Neofluar 10×/0.30 M27 objective for 10× magnification and a Plan-Apochromat 40×/0.95 Korr M27 objective for 40× magnification. Images were analyzed with ZEN 2009 Light Edition and Image-Pro Plus 6.0 software.
Infection and Statistical Analysis of ToMV
Plants were rub inoculated with ToMV. Viral spread and symptoms were monitored for at least 2 weeks. All experiments were performed at least three times, and at least three plants were used per construct each time. OriginPro 8.1 and SPSS 19.0 were used to conduct statistical analyses and to generate graphs.
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: X02144 (ToMV); AY179605 (Actin); JN247448 (PR1a); AF536201 (Tm22); AAF62891 (GFP); AY818369 (YFP); GU734654 (CFP); AT5G43980 (TMD); and P03583 (TMV MP).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Amino acid sequence alignment of tobamovirus MPs.
Supplemental Figure S2. Subcellular localization of ToMV MP.
Supplemental Figure S3. Mock inoculation does not induce local necrosis in NbRbCS-silenced plants.
Supplemental Figure S4. Silencing of NbRbCS enhances local multiplication but reduces cell-to-cell movement of TMV-GFP.
Supplemental Figure S5. Silencing of NbRbCS reduces PR1a expression.
Supplemental Figure S6. TMV MP interacts with NbRbCS in plants.
Supplemental Figure S7. BiFC visualization of subcellular localization of the TMV MP-NbRbCS interaction.
Supplemental Figure S8. Silencing of NbRbCS compromised Tm-22-mediated extreme resistance against TMV-GFP.
Supplemental Figure S9. Coexpression of ToMV MP resulted in the formation of NbRbCS in cytosolic aggresomes and punctae along the cell wall.
Supplemental Figure S10. Suppression of NbRbCS delays TMV-GFP systemic infection in N. benthamiana.
Acknowledgments
We thank Jian-Min Zhou for providing the luciferase complementation imaging assay vectors.
Glossary
- MP
movement protein
- PD
plasmodesmata
- TMV
Tobacco mosaic virus
- ToMV
Tomato mosaic virus
- Y2H
yeast two-hybrid
- AD
activation domain
- BD
binding domain
- GST
glutathione S-transferase
- HA
hemagglutinin
- ER
endoplasmic reticulum
- BiFC
bimolecular fluorescence complementation
- YFP
yellow fluorescent protein
- nYFP
N-terminal yellow fluorescent protein
- cYFP
C-terminal yellow fluorescent protein
- CFP
cyan fluorescent protein
- VRCs
virus replication complexes
- VIGS
virus-induced gene silencing
- dpi
days post infection
- RT
reverse transcription
- HR
hypersensitive response
- OEC
oxygen-evolving complex
- GDI
GDP dissociation inhibitor
- PMSF
phenylmethanesulfonyl fluoride
- cDNA
complementary DNA
- TRV
Tobacco rattle virus
References
- Abbink TE, Peart JR, Mos TN, Baulcombe DC, Bol JF, Linthorst HJ. (2002) Silencing of a gene encoding a protein component of the oxygen-evolving complex of photosystem II enhances virus replication in plants. Virology 295: 307–319 [DOI] [PubMed] [Google Scholar]
- Almon E, Horowitz M, Wang HL, Lucas WJ, Zamski E, Wolf S. (1997) Phloem-specific expression of the tobacco mosaic virus movement protein alters carbon metabolism and partitioning in transgenic potato plants. Plant Physiol 115: 1599–1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amari K, Vazquez F, Heinlein M. (2012) Manipulation of plant host susceptibility: an emerging role for viral movement proteins? Front Plant Sci 3: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashby J, Boutant E, Seemanpillai M, Groner A, Sambade A, Ritzenthaler C, Heinlein M. (2006) Tobacco mosaic virus movement protein functions as a structural microtubule-associated protein. J Virol 80: 8329–8344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balachandran S, Hull RJ, Vaadia Y, Wolf S, Lucas WJ. (1995) Alteration in carbon partitioning induced by the movement protein of tobacco mosaic virus originates in the mesophyll and is independent of change in the plasmodesmal size exclusion limit. Plant Cell Environ 18: 1301–1310 [Google Scholar]
- Bazzini AA, Hopp HE, Beachy RN, Asurmendi S. (2007) Infection and coaccumulation of tobacco mosaic virus proteins alter microRNA levels, correlating with symptom and plant development. Proc Natl Acad Sci USA 104: 12157–12162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyko V, van der Laak J, Ferralli J, Suslova E, Kwon MO, Heinlein M. (2000) Cellular targets of functional and dysfunctional mutants of tobacco mosaic virus movement protein fused to green fluorescent protein. J Virol 74: 11339–11346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandner K, Sambade A, Boutant E, Didier P, Mély Y, Ritzenthaler C, Heinlein M. (2008) Tobacco mosaic virus movement protein interacts with green fluorescent protein-tagged microtubule end-binding protein 1. Plant Physiol 147: 611–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brill LM, Dechongkit S, DeLaBarre B, Stroebel J, Beachy RN, Yeager M. (2004) Dimerization of recombinant tobacco mosaic virus movement protein. J Virol 78: 3372–3377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brill LM, Nunn RS, Kahn TW, Yeager M, Beachy RN. (2000) Recombinant tobacco mosaic virus movement protein is an RNA-binding, alpha-helical membrane protein. Proc Natl Acad Sci USA 97: 7112–7117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brizard JP, Carapito C, Delalande F, Van Dorsselaer A, Brugidou C. (2006) Proteome analysis of plant-virus interactome: comprehensive data for virus multiplication inside their hosts. Mol Cell Proteomics 5: 2279–2297 [DOI] [PubMed] [Google Scholar]
- Burch-Smith TM, Schiff M, Caplan JL, Tsao J, Czymmek K, Dinesh-Kumar SP. (2007) A novel role for the TIR domain in association with pathogen-derived elicitors. PLoS Biol 5: e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra-Shekara AC, Gupte M, Navarre D, Raina S, Raina R, Klessig D, Kachroo P. (2006) Light-dependent hypersensitive response and resistance signaling against Turnip crinkle virus in Arabidopsis. Plant J 45: 320–334 [DOI] [PubMed] [Google Scholar]
- Chen MH, Sheng J, Hind G, Handa AK, Citovsky V. (2000) Interaction between the tobacco mosaic virus movement protein and host cell pectin methylesterases is required for viral cell-to-cell movement. EMBO J 19: 913–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua NH, Schmidt GW. (1978) Post-translational transport into intact chloroplasts of a precursor to the small subunit of ribulose-1,5-bisphosphate carboxylase. Proc Natl Acad Sci USA 75: 6110–6114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citovsky V, Knorr D, Schuster G, Zambryski P. (1990) The P30 movement protein of tobacco mosaic virus is a single-strand nucleic acid binding protein. Cell 60: 637–647 [DOI] [PubMed] [Google Scholar]
- Citovsky V, Wong ML, Shaw AL, Prasad BV, Zambryski P. (1992) Visualization and characterization of tobacco mosaic virus movement protein binding to single-stranded nucleic acids. Plant Cell 4: 397–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dardick CD, Golem S, Culver JN. (2000) Susceptibility and symptom development in Arabidopsis thaliana to Tobacco mosaic virus is influenced by virus cell-to-cell movement. Mol Plant Microbe Interact 13: 1139–1144 [DOI] [PubMed] [Google Scholar]
- Deom CM, Oliver MJ, Beachy RN. (1987) The 30-kilodalton gene product of tobacco mosaic virus potentiates virus movement. Science 237: 389–394 [DOI] [PubMed] [Google Scholar]
- Deom CM, Schubert KR, Wolf S, Holt CA, Lucas WJ, Beachy RN. (1990) Molecular characterization and biological function of the movement protein of tobacco mosaic virus in transgenic plants. Proc Natl Acad Sci USA 87: 3284–3288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding B, Haudenshield JS, Hull RJ, Wolf S, Beachy RN, Lucas WJ. (1992) Secondary plasmodesmata are specific sites of localization of the tobacco mosaic virus movement protein in transgenic tobacco plants. Plant Cell 4: 915–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Burch-Smith TM, Liu Y, Mamillapalli P, Dinesh-Kumar SP. (2007) A ligation-independent cloning tobacco rattle virus vector for high-throughput virus-induced gene silencing identifies roles for NbMADS4-1 and -2 in floral development. Plant Physiol 145: 1161–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feki S, Loukili MJ, Triki-Marrakchi R, Karimova G, Old I, Ounouna H, Nato A, Nato F, Guesdon JL, Lafaye P, et al. (2005) Interaction between tobacco ribulose-1,5-biphosphate carboxylase/oxygenase large subunit (Rubisco-LSU) and the PVY coat protein (PVY-CP). Eur J Plant Pathol 112: 221–234 [Google Scholar]
- Ghoshroy S, Lartey R, Sheng J, Citovsky V. (1997) Transport of proteins and nucleic acids through plasmodesmata. Annu Rev Plant Physiol Plant Mol Biol 48: 27–50 [DOI] [PubMed] [Google Scholar]
- Heinlein M, Epel BL, Padgett HS, Beachy RN. (1995) Interaction of tobamovirus movement proteins with the plant cytoskeleton. Science 270: 1983–1985 [DOI] [PubMed] [Google Scholar]
- Heinlein M, Padgett HS, Gens JS, Pickard BG, Casper SJ, Epel BL, Beachy RN. (1998) Changing patterns of localization of the tobacco mosaic virus movement protein and replicase to the endoplasmic reticulum and microtubules during infection. Plant Cell 10: 1107–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms K, McIntyre GA. (1967) Light-induced susceptibility of Phaseolus vulgaris L. to tobacco mosaic virus infection. I. Effects of light intensity, temperature, and the length of the preinoculation dark period. Virology 31: 191–196 [DOI] [PubMed] [Google Scholar]
- Itaya A, Matsuda Y, Gonzales RA, Nelson RS, Ding B. (2002) Potato spindle tuber viroid strains of different pathogenicity induces and suppresses expression of common and unique genes in infected tomato. Mol Plant Microbe Interact 15: 990–999 [DOI] [PubMed] [Google Scholar]
- Kahn TW, Lapidot M, Heinlein M, Reichel C, Cooper B, Gafny R, Beachy RN. (1998) Domains of the TMV movement protein involved in subcellular localization. Plant J 15: 15–25 [DOI] [PubMed] [Google Scholar]
- Kawakami S, Watanabe Y, Beachy RN. (2004) Tobacco mosaic virus infection spreads cell to cell as intact replication complexes. Proc Natl Acad Sci USA 101: 6291–6296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegstra K, Bruce B, Hurley M, Li HM, Perry S. (1995) Targeting of proteins into chloroplasts. Physiol Plant 93: 157–162 [Google Scholar]
- Kiselyova OI, Yaminsky IV, Karger EM, Frolova OY, Dorokhov YL, Atabekov JG. (2001) Visualization by atomic force microscopy of tobacco mosaic virus movement protein-RNA complexes formed in vitro. J Gen Virol 82: 1503–1508 [DOI] [PubMed] [Google Scholar]
- Kolonin MG, Zhong J, Finley RL. (2000) Interaction mating methods in two-hybrid systems. Methods Enzymol 328: 26–46 [DOI] [PubMed] [Google Scholar]
- Kragler F, Curin M, Trutnyeva K, Gansch A, Waigmann E. (2003) MPB2C, a microtubule-associated plant protein binds to and interferes with cell-to-cell transport of tobacco mosaic virus movement protein. Plant Physiol 132: 1870–1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer SR, Goregaoker SP, Culver JN. (2011) Association of the Tobacco mosaic virus 126kDa replication protein with a GDI protein affects host susceptibility. Virology 414: 110–118 [DOI] [PubMed] [Google Scholar]
- Lanfermeijer FC, Dijkhuis J, Sturre MJ, de Haan P, Hille J. (2003) Cloning and characterization of the durable tomato mosaic virus resistance gene Tm-2(2) from Lycopersicon esculentum. Plant Mol Biol 52: 1037–1049 [DOI] [PubMed] [Google Scholar]
- Lanfermeijer FC, Jiang GY, Ferwerda MA, Dijkhuis J, de Haan P, Yang RC, Hille J. (2004) The durable resistance gene Tm-2(2) from tomato confers resistance against ToMV in tobacco and preserves its viral specificity. Plant Sci 167: 687–692 [Google Scholar]
- Leisner SM, Turgeon R, Howell SH. (1993) Effects of host plant development and genetic determinants on the long-distance movement of cauliflower mosaic virus in Arabidopsis. Plant Cell 5: 191–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JD, Lazarowitz SG. (2010) Arabidopsis synaptotagmin SYTA regulates endocytosis and virus movement protein cell-to-cell transport. Proc Natl Acad Sci USA 107: 2491–2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wu MY, Song HH, Hu X, Qiu BS. (2005) Identification of a tobacco protein interacting with tomato mosaic virus coat protein and facilitating long-distance movement of virus. Arch Virol 150: 1993–2008 [DOI] [PubMed] [Google Scholar]
- Lin L, Luo Z, Yan F, Lu Y, Zheng H, Chen J. (2011) Interaction between potyvirus P3 and ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) of host plants. Virus Genes 43: 90–92 [DOI] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Marathe R, Dinesh-Kumar SP. (2002a) Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J 30: 415–429 [DOI] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Serino G, Deng XW, Dinesh-Kumar SP. (2002b) Role of SCF ubiquitin-ligase and the COP9 signalosome in the N gene-mediated resistance response to Tobacco mosaic virus. Plant Cell 14: 1483–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Koroleva OA, Farrar JF, Gallagher J, Pollock CJ, Tomos AD. (2002) Rubisco small subunit, chlorophyll a/b-binding protein and sucrose:fructan-6-fructosyl transferase gene expression and sugar status in single barley leaf cells in situ: cell type specificity and induction by light. Plant Physiol 130: 1335–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas WJ, Gilbertson RL. (1994) Plasmodesmata in relation to viral movement within leaf tissue. Annu Rev Phytopathol 32: 387–411 [Google Scholar]
- Lucas WJ, Olesinski A, Hull RJ, Haudenshield JS, Deom CM, Beachy RN, Wolf S. (1993) Influence of the tobacco mosaic-virus 30-kda movement protein on carbon metabolism and photosynthate partitioning in transgenic tobacco plants. Planta 190: 88–96 [Google Scholar]
- Más P, Beachy RN. (1999) Replication of tobacco mosaic virus on endoplasmic reticulum and role of the cytoskeleton and virus movement protein in intracellular distribution of viral RNA. J Cell Biol 147: 945–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean BG, Zupan J, Zambryski PC. (1995) Tobacco mosaic virus movement protein associates with the cytoskeleton in tobacco cells. Plant Cell 7: 2101–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshi T, Motoyoshi F, Maeda T, Yoshiwoka S, Watanabe H, Okada Y. (1989) Mutations in the tobacco mosaic virus 30-kD protein gene overcome Tm-2 resistance in tomato. Plant Cell 1: 515–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olesinski AA, Almon E, Navot N, Perl A, Galun E, Lucas WJ, Wolf S. (1996) Tissue-specific expression of the tobacco mosaic virus movement protein in transgenic potato plants alters plasmodesmal function and carbohydrate partitioning. Plant Physiol 111: 541–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olesinski AA, Lucas WJ, Galun E, Wolf S. (1995) Pleiotropic effects of tobacco-mosaic-virus movement protein on carbon metabolism in transgenic tobacco plants. Planta 197: 118–126 [Google Scholar]
- Schatz G, Dobberstein B. (1996) Common principles of protein translocation across membranes. Science 271: 1519–1526 [DOI] [PubMed] [Google Scholar]
- Serino G, Tsuge T, Kwok S, Matsui M, Wei N, Deng XW. (1999) Arabidopsis cop8 and fus4 mutations define the same gene that encodes subunit 4 of the COP9 signalosome. Plant Cell 11: 1967–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T, Yoshii A, Sakurai K, Hamada K, Yamaji Y, Suzuki M, Namba S, Hibi T. (2009) Identification of a novel tobacco DnaJ-like protein that interacts with the movement protein of tobacco mosaic virus. Arch Virol 154: 959–967 [DOI] [PubMed] [Google Scholar]
- Smith SM, Ellis RJ. (1979) Processing of small subunit precursor of ribulose bisphosphate carboxylase and its assembly into whole enzyme are stromal events. Nature 278: 662–664 [Google Scholar]
- Susi P, Pehu E, Lehto K. (1999) Replication in the phloem is not necessary for efficient vascular transport of tobacco mosaic tobamovirus. FEBS Lett 447: 121–123 [DOI] [PubMed] [Google Scholar]
- Szecsi J, Ding XS, Lim CO, Bendahmane M, Cho MJ, Nelson RS, Beachy RN. (1999) Development of tobacco mosaic virus infection sites in Nicotiana benthamiana. Mol Plant Microbe Interact 12: 143–152 [Google Scholar]
- Thomas CL, Bayer EM, Ritzenthaler C, Fernandez-Calvino L, Maule AJ. (2008) Specific targeting of a plasmodesmal protein affecting cell-to-cell communication. PLoS Biol 6: e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueki S, Spektor R, Natale DM, Citovsky V. (2010) ANK, a host cytoplasmic receptor for the Tobacco mosaic virus cell-to-cell movement protein, facilitates intercellular transport through plasmodesmata. PLoS Pathog 6: e1001201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogler H, Kwon MO, Dang V, Sambade A, Fasler M, Ashby J, Heinlein M. (2008) Tobacco mosaic virus movement protein enhances the spread of RNA silencing. PLoS Pathog 4: e1000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waigmann E, Curin M, Heinlein M. (2007) Tobacco mosaic virus: a model for macromolecular cell-to-cell spread. In E Waigmann, M Heinlein, eds, Viral Transport in Plants, Vol 7. Springer-Verlag, Heidelberg/Berlin, pp 29–62
- Watanabe Y, Emori Y, Ooshika I, Meshi T, Ohno T, Okada Y. (1984) Synthesis of TMV-specific RNAs and proteins at the early stage of infection in tobacco protoplasts: transient expression of the 30K protein and its mRNA. Virology 133: 18–24 [DOI] [PubMed] [Google Scholar]
- Wei T, Zhang C, Hong J, Xiong R, Kasschau KD, Zhou X, Carrington JC, Wang A. (2010) Formation of complexes at plasmodesmata for potyvirus intercellular movement is mediated by the viral protein P3N-PIPO. PLoS Pathog 6: e1000962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen RH, Hajimorad MR. (2010) Mutational analysis of the putative pipo of soybean mosaic virus suggests disruption of PIPO protein impedes movement. Virology 400: 1–7 [DOI] [PubMed] [Google Scholar]
- Wieringabrants DH. (1981) The role of the epidermis in virus-induced local lesions on cowpea and tobacco leaves. J Gen Virol 54: 209–212 [Google Scholar]
- Wintermantel WM, Banerjee N, Oliver JC, Paolillo DJ, Zaitlin M. (1997) Cucumber mosaic virus is restricted from entering minor veins in transgenic tobacco exhibiting replicase-mediated resistance. Virology 231: 248–257 [DOI] [PubMed] [Google Scholar]
- Wolf S, Deom CM, Beachy RN, Lucas WJ. (1989) Movement protein of tobacco mosaic virus modifies plasmodesmatal size exclusion limit. Science 246: 377–379 [DOI] [PubMed] [Google Scholar]
- Wolf S, Millatiner A. (2000) Effect of tobacco mosaic virus movement protein on photosynthesis in transgenic tobacco plants. J Plant Physiol 156: 253–258 [Google Scholar]