Abstract
Plants produce various proteinaceous inhibitors to protect themselves against microbial pathogen attack. A xyloglucan-specific endo-β-1,4-glucanase inhibitor1 gene, CaXEGIP1, was isolated and functionally characterized in pepper (Capsicum annuum) plants. CaXEGIP1 was rapidly and strongly induced in pepper leaves infected with avirulent Xanthomonas campestris pv vesicatoria, and purified CaXEGIP1 protein significantly inhibited the hydrolytic activity of the glycoside hydrolase74 family xyloglucan-specific endo-β-1,4-glucanase from Clostridium thermocellum. Soluble-modified green fluorescent protein-tagged CaXEGIP1 proteins were mainly localized to the apoplast of onion (Allium cepa) epidermal cells. Agrobacterium tumefaciens-mediated overexpression of CaXEGIP1 triggered pathogen-independent, spontaneous cell death in pepper and Nicotiana benthamiana leaves. CaXEGIP1 silencing in pepper conferred enhanced susceptibility to virulent and avirulent X. campestris pv vesicatoria, accompanied by a compromised hypersensitive response and lowered expression of defense-related genes. Overexpression of dexamethasone:CaXEGIP1 in Arabidopsis (Arabidopsis thaliana) enhanced resistance to Hyaloperonospora arabidopsidis infection. Comparative histochemical and proteomic analyses revealed that CaXEGIP1 overexpression induced a spontaneous cell death response and also increased the expression of some defense-related proteins in transgenic Arabidopsis leaves. This response was also accompanied by cell wall thickening and darkening. Together, these results suggest that pathogen-inducible CaXEGIP1 positively regulates cell death-mediated defense responses in plants.
Plant cell walls provide a physical barrier that separates challenging pathogens from the internal contents of plant cells. Additionally, the cell walls regulate cell expansion and differentiation (York et al., 2004; Flors et al., 2007; Cantu et al., 2008). Polysaccharides, such as cellulose, hemicellulose, and pectic polysaccharides, are the main components of primary cell walls. Xyloglucan (XG), the most abundant hemicellulose in the primary cell wall, plays a structural role by forming strong hydrogen bonds with cellulose microfibrils (Carpita and Gibeaut, 1993). The primary structure of XG contains a common β-(1→4)-d-glucan backbone, which is repeatedly substituted with α(1→6)-d-xylopyranosyl residues. Depolymerization of XG is proposed to play an important role during both cell wall expansion and pathogen invasion (Bourquin et al., 2002; Qin et al., 2003; Baumann et al., 2007). During cell wall expansion, plant XG endotransglycosylases cut and rejoin XG chains to allow the cellulose microfibrils to move apart. From a pathogen point of view, the carbon-rich complex represents a useful energy source for pathogen growth. To facilitate penetration into the plant tissues and to catabolize carbon sources, many plant pathogens secrete a mixture of cell wall-degrading enzymes, such as polygalacturonases, pectin methyl esterases, pectin/pecatae lyases, xylanases, and endoglucanases (Valette-Collet et al., 2003; DeBoy et al., 2008). Some microbial glycoside hydrolase (GH) family proteins, including GH5, GH12, and GH74, reportedly hydrolyze plant-derived XG (Martinez-Fleites et al., 2006; Gloster et al., 2007).
To inhibit pathogen-derived cell wall-degrading enzymes, plants secrete a mixture of inhibitor proteins into the cell wall (Qin et al., 2003; An et al., 2008; Xie et al., 2008). Some of the best characterized inhibitor proteins are polygalacturonase-inhibiting proteins (PGIPs; Albersheim and Anderson, 1971; De Lorenzo and Ferrari, 2002; Federici et al., 2006). In bean (Phaseolus vulgaris), two pairs of PGIPs, PvPGIP1/PvPGIP2 and PvPGIP3/PvPGIP4, are present in the genome. These genes may have originated from independent gene duplication events (D’Ovidio et al., 2004a). PvPGIP2 strongly inhibits polygalacturonases from Fusarium phyllophilum and Aspergillus niger via three conserved Asp residues (Spinelli et al., 2009). PGIPs reduce the hydrolytic activity of polygalacturonases to favor the accumulation of long-chain oligogalacturonides, known as elicitors of a variety of defense responses (Côté and Hahn, 1994; D’Ovidio et al., 2004b). Furthermore, transgenic expression of pear (Pyrus communis) PGIP in transgenic tomato (Solanum lycopersicum) plants limited fungal colonization, suggesting a role of PGIPs in plant defense (Powell et al., 2000).
The proteinaceous inhibitor of the cell wall-degrading enzyme xyloglucan-specific endo-β-1,4-glucanase (XEG) was identified from suspension-cultured tomatoes (Qin et al., 2003). The purified xyloglucan-specific endo-β-1,4-glucanase inhibitor protein (XEGIP) strongly inhibited XEG activity through the formation of a 1:1 protein:protein complex with XEG of Aspergillus aculeatus. More recently, two putative XEGIPs were isolated from Nicotiana benthamiana based on conserved regions found in plant XEGIP genes, and these genes were functionally characterized using virus-induced gene silencing (VIGS; Xie et al., 2008). VIGS of NbXEGIP1 strongly enhanced the wilting symptoms exhibited following infection by virulent Pseudomonas syringae pv tabaci. This finding supports the notion that NbXEGIP1 may act as an inhibitor of bacterial cell wall-degrading enzymes in N. benthamiana plants.
Programmed cell death (PCD) has been extensively characterized in plants (Lam, 2004). The hypersensitive response (HR), a well-known form of plant PCD, is one of the most efficient and immediate resistance reactions of plants. The HR is characterized by the rapid death of cells in the local region surrounding an infection site. As a result, the growth and spread of the pathogen is restricted or confined. During HR cell death development, cell wall strengthening occurs. Histochemical analyses of cells involved in melon (Cucumis melo)-powdery mildew (Podosphaera fusca) interactions demonstrate the reinforcement of the cell wall compartment as part of HR cell death-mediated resistance (Romero et al., 2008). Treatment of suspension-cultured tobacco (Nicotiana tabacum) cells with cryptogein, a 10-kD protein secreted by the oomycete Phytophthora cryptogea, induces a HR on tobacco leaves, accompanied by induced strengthening of the cell wall (Kieffer et al., 2000). However, the role of cell wall strengthening in HR cell death is poorly understood. A second type of PCD is thought to be associated with the differentiation of procambium into tracheary elements in the xylem of vascular plants (Fukuda, 2000; Lam, 2004). During the early formation of mature tracheary elements, vacuoles accumulate degradation enzymes and the cell wall is remodeled into a highly reticulated form. A similar phenomenon occurs during some plant developmental processes, including senescence and aerenchyma formation in roots (Jones, 2001).
In this study, we have isolated and functionally characterized a pepper (Capsicum annuum) xyloglucan-specific endo-β-1,4-glucanase inhibitor-protein1 gene (CaXEGIP1). Expression of CaXEGIP1 was strongly induced in pepper leaves infected with avirulent Xanthomonas campestris pv vesicatoria (Xcv) strain Bv5-4a. The purified CaXEGIP1 protein inhibited the hydrolytic activity of GH74 family XEG from the thermophilic bacterium Clostridium thermocellum. The soluble-modified GFP (smGFP)-fused CaXEGIP1 protein was localized in the external and intercellular regions of onion epidermal cells. Importantly, Agrobacterium tumefaciens-mediated transient expression of CaXEGIP1 induced the hypersensitive cell death response in pepper and N. benthamiana leaves. VIGS of CaXEGIP1 significantly enhanced the growth of virulent and avirulent Xcv in pepper leaves, accompanied by compromised HR cell death and lowered expression of CaPR1 (pathogenesis-related protein1 [PR1]) and CaDEF1 (defensin [DEF1]). We also investigated the role of CaXEGIP1 in plant cell death and defense responses using transgenic Arabidopsis (Arabidopsis thaliana) plants harboring the dexamethasone (DEX)-inducible CaXEGIP1 transgene. Overexpression of CaXEGIP1 triggered spontaneous cell death and modification of the cell wall compartment in Arabidopsis plants. Together, these results suggest that the pathogen-responsive CaXEGIP1 is involved in plant cell death-mediated defense signaling.
RESULTS
CaXEGIP1 Encodes a Putative Extracellular XEGIP
To isolate pepper genes induced during the HR, we performed macro complementary DNA (cDNA) array analysis using a cDNA library constructed from pepper leaves infected with avirulent Xcv strain Bv5-4a (Jung and Hwang, 2000; Hwang and Hwang, 2010, 2011; Hwang et al., 2011). Among the defense-related genes selected, we isolated the putative pepper CaXEGIP1 gene. This gene was strongly induced during the HR. The CaXEGIP1 cDNA (accession no. JQ673414) consists of 1,415 bp, including a 30-bp poly(A) tail, and encodes a protein of 430 amino acids with a predicted molecular mass of 47.2 kD and a pI of 9.15. The translated CaXEGIP1 amino acid sequence shared high amino acid sequence identity with various plant XEGIPs (Supplemental Fig. S1). The putative amino acid sequence encoded by the CaXEGIP1 cDNA was 45% identical to a putative XEGIP from grape (Vitis vinifera; accession no. XP_002272235) and 42% identical to a XEGIP from tomato (accession no. AAN87262; Qin et al., 2003).
CaXEGIP1 Is Strongly Induced in Leaves by Avirulent Xcv Infection
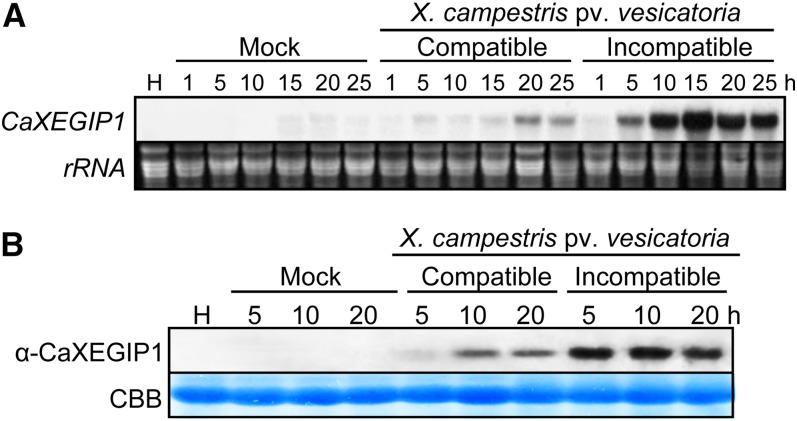
RNA gel-blot analysis was performed to investigate the expression of CaXEGIP1 in pepper leaves during compatible and incompatible interactions with Xcv (Fig. 1A). CaXEGIP1 was strongly induced in leaves inoculated with the Xcv avirulent (incompatible) strain Bv5-4a harboring avrBsT, which induces HR cell death (Kim et al., 2010; Kim and Hwang, 2011). CaXEGIP1 induction was detected as early as 5 h after Xcv avirulent Bv5-4a infection and was notably strong prior to the appearance of the HR. In contrast, CaXEGIP1 expression was low in leaves 20 and 25 h after inoculation with the virulent (compatible) strain Ds1. CaXEGIP1 transcripts were only faintly detected in mock (10 mm MgCl2)-inoculated leaves.
Figure 1.
Induction of CaXEGIP1 in pepper leaves by Xcv infection. Shown are RNA gel-blot (A) and immunoblot (B) analyses of CaXEGIP1 expression in pepper leaves at various time points after inoculation with the Xcv virulent strain Ds1 (compatible) and avirulent strain Bv5-4a (incompatible) at the six-leaf stage. H, Healthy leaves; Mock, leaves treated with 10 mm MgCl2. Equal loading of total RNA (20 μg per lane) was verified by visualizing ribosomal RNA (rRNA) on a gel stained with ethidium bromide. Protein loading was verified by Coomassie Brilliant Blue (CBB) staining. [See online article for color version of this figure.]
Immunoblot analysis using an antiserum raised against a CaXEGIP1 peptide demonstrated that infection by avirulent Xcv strain Bv5-4a strongly induced CaXEGIP1 accumulation in pepper leaves as compared with virulent Ds1 infection (Fig. 1B). As observed with the RNA gel-blot analyses, CaXEGIP1 expression was induced 5 h after avirulent Bv5-4a infection, which occurred prior to the appearance of the HR cell death. However, the induction of CaXEGIP1 was not detected in mock-inoculated leaves and was only faintly detected in leaves inoculated with virulent Xcv. This pronounced CaXEGIP1 expression during the HR cell death indicates a significant role of CaXEGIP1 in the HR-like resistance response of pepper plants to the bacterial spot pathogen Xcv.
Inhibition of XEG Activity by CaXEGIP1 Protein
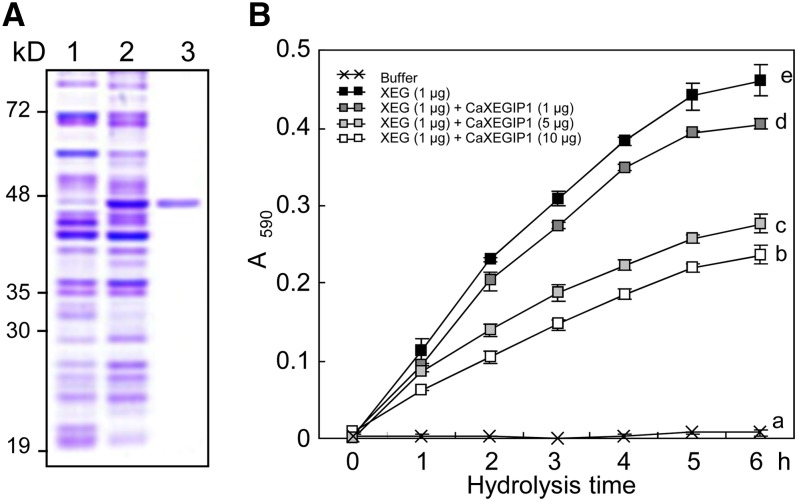
Recombinant CaXEGIP1 protein was purified from the extracts of isopropyl-β-d-thiogalactoside (IPTG)-induced Escherichia coli BL21 cells using Ni2+-nitrilotriacetic acid affinity chromatography (Fig. 2A). The homogeneity of the purified protein was confirmed by a single band on a Coomassie Brilliant Blue-stained SDS-PAGE gel (Fig. 2A, lane 3). To investigate whether recombinant CaXEGIP1 inhibits the hydrolytic activity of XEG, various amounts of the purified CaXEGIP1 were mixed with 1 μg of GH74 family XEG enzyme from C. thermocellum F7/YS (Martinez-Fleites et al., 2006). The resulting mixtures were incubated at 60°C for 1 h to permit complex formation and then added to AZO-XG solutions (Megazyme). Aliquots of each reaction mixture were taken at various time points, and the amount of reducing sugar was examined spectrophotometrically. As shown in Figure 2B, the addition of increasing amounts of purified CaXEGIP1 protein strongly inhibited XEG-catalyzed depolymerization of XG.
Figure 2.
Inhibitory effect of the recombinant CaXEGIP1 protein on XEG activity. A, Expression and purification of recombinant CaXEGIP1 in E. coli BL21 (DE3) cells. Protein loading was verified by Coomassie Brilliant Blue staining. Lane 1, uninduced E. coli cell extracts; lane 2, crude protein extracts of E. coli cells expressing the His-tagged CaXEGIP1, as induced by 1 mm IPTG; lane 3, purified His-tagged CaXEGIP1. B, Inhibition of XEG activity by CaXEGIP1. The data represent means ± sd (n = 4) from three independent experiments. Different letters indicate significant differences, as determined by Fisher’s lsd test (P < 0.05, n = 4). [See online article for color version of this figure.]
Subcellular Localization of CaXEGIP1
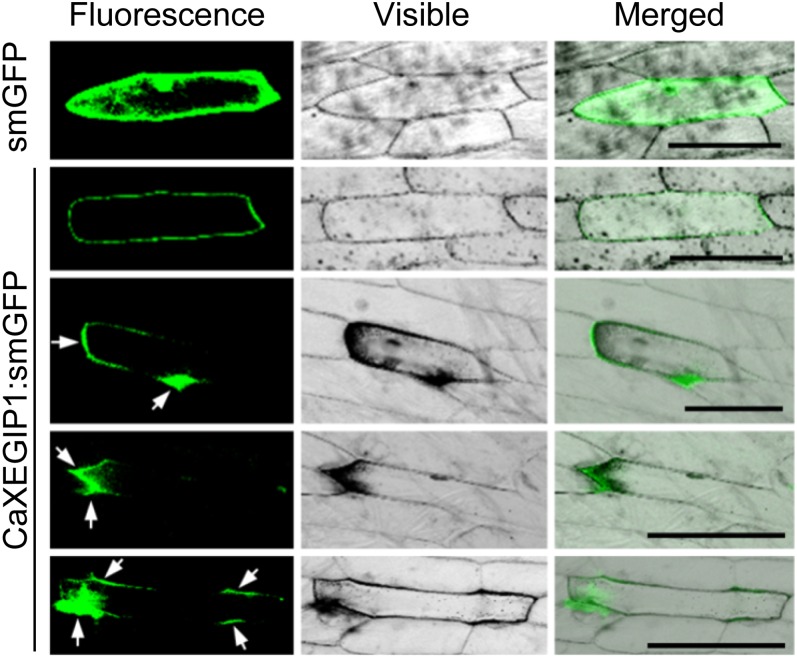
Computational analysis of the predicted protein sequence indicated that CaXEGIP1 is a secreted protein (PSORT, 82%; TargetP, 99%) and contains a signal peptide cleavage site between Ala-24 and Glu-25 (Supplemental Fig. S1). To analyze the subcellular localization of the CaXEGIP1 protein, we performed a biolistic transformation experiment. The smGFP gene was fused to the C-terminal region of CaXEGIP1. As shown in Figure 3, the CaXEGIP1:smGFP fusion protein was mainly localized in the external and intercellular regions of the onion cell, while the control smGFP protein was uniformly distributed throughout the cell. Different levels of expression of the CaXEGIP1:smGFP fusion protein were visualized based on the GFP fluorescence signaling intensity. Interestingly, the focal and strong accumulation of CaXEGIP1 proteins darkened the peripheral and apoplastic regions where the proteins localized. However, in the cells expressing lower levels of GFP-tagged CaXEGIP1, darkening of the cell peripheral region was not observed. These observations support the notion that the darkened microscopic images of onion epidermal cells resulted from the localized thickening and darkening of the cell wall by the accumulation of CaXEGIP1.
Figure 3.
Subcellular localization of CaXEGIP1 protein by transient expression of the CaXEGIP1:smGFP construct in onion epidermal cells. The smGFP gene was fused to the CaXEGIP1 3′ region. Transient expression of smGFP or CaXEGIP1:smGFP was detected by confocal laser scanning microscopy 24 h after biolistic transformation. White arrows indicate the localization of CaXEGIP1 protein in the external and intercellular regions of the onion epidermal cells. Bars = 100 µm.
Transient Expression of CaXEGIP1 Triggers Cell Death in Pepper and N. benthamiana Leaves
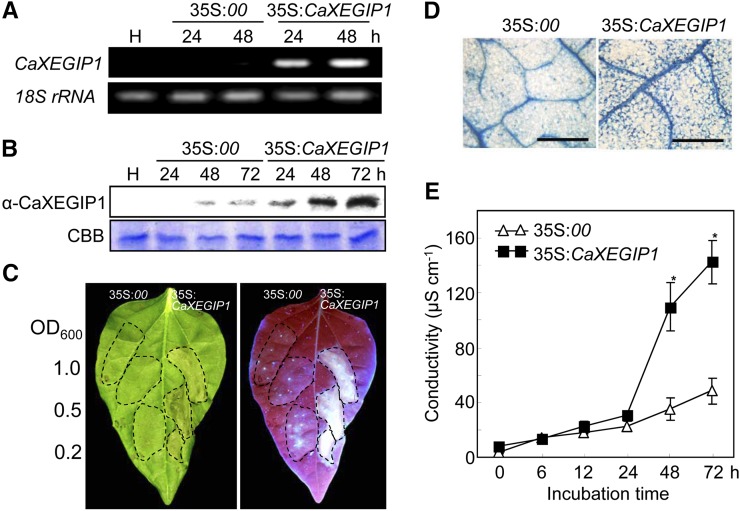
To examine the in vivo function of CaXEGIP1 in defense responses, we used A. tumefaciens-mediated transient expression in pepper and N. benthamiana plants, as described previously (Choi et al., 2009, 2011; Hwang and Hwang, 2010; Choi and Hwang, 2011; Kim and Hwang, 2011). Reverse transcription (RT)-PCR and immunoblot analyses revealed the expression of CaXEGIP1 in A. tumefaciens-infiltrated pepper leaves at both the mRNA and protein levels (Fig. 4, A and B). Transient expression of CaXEGIP1 in pepper leaves triggered a strong cell death response 48 h after agroinfiltration (Fig. 4C). This cell death induction by CaXEGIP1 transient expression was supported by the strong and differential staining of the cell death response with trypan blue and also high electrolyte leakage from the agroinfiltrated leaves (Fig. 4, D and E).
Figure 4.
Induction of the cell death response in pepper leaves by A. tumefaciens-mediated transient expression of CaXEGIP1. A and B, RT-PCR (A) and immunoblot (B) analyses of CaXEGIP1 expression in pepper leaves. Expression of pepper 18S rRNA served as a loading control. Protein loading was verified by Coomassie Brilliant Blue (CBB) staining. C, Induction of cell death in pepper leaves. The leaf areas between the lateral veins were infiltrated with A. tumefaciens at the indicated OD600 concentrations. Photographs were taken 48 h after agroinfiltration: left, visible symptoms; right, UV exposure to visualize the deposition of fluorescent pigments. D and E, Trypan blue staining (D) and electrolyte leakage measurements (E) from the leaves infiltrated by A. tumefaciens (OD600 = 0.5) strains carrying the indicated constructs. The data represent means ± sd from three independent experiments. Asterisks indicate significant differences in electrolyte leakage, as determined by the two-tailed Student’s t test (P < 0.05). Bars = 200 µm.
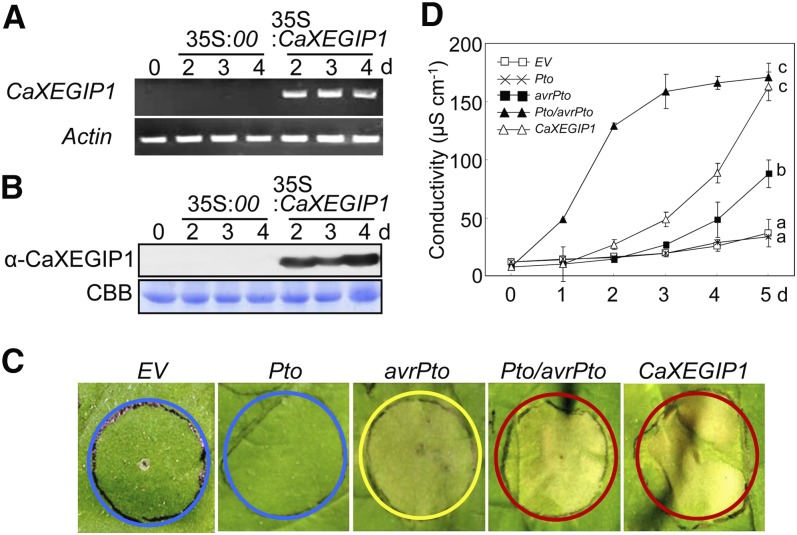
We next investigated whether transient expression of CaXEGIP1 triggers hypersensitive cell death in N. benthamiana leaves (Fig. 5). Successful transient expression of CaXEGIP1 in N. benthamiana leaves was confirmed by RT-PCR and immunoblot analyses (Fig. 5, A and B). The tomato R gene Pto and the corresponding Pseudomonas syringae pv tomato (Pst) effector gene avrPto were included as positive controls. As shown in Figure 5C, transient expression of avrPto alone triggered the chlorotic phenotype, whereas empty vector or Pto expression did not impair N. benthamiana leaves. However, transient expression of CaXEGIP1 or Pto/avrPto induced the typical hypersensitive cell death response in N. benthamiana leaves. In support of these visual cell death phenotypes, expression of CaXEGIP1 significantly enhanced electrolyte leakage from the agroinfiltrated leaves as compared with empty vector-, Pto-, or avrPto-expressing leaves (Fig. 5D). Trypan blue staining of N. benthamiana leaves also revealed CaXEGIP1-induced cell death and darkened cell wall compartments (Supplemental Fig. S2). Collectively, these results indicate that CaXEGIP1 transient expression triggers pathogen-independent hypersensitive cell death in plant tissues.
Figure 5.
Effect of A. tumefaciens-mediated transient expression of Pto, avrPto, Pto/avrPto, and CaXEGIP1 on the cell death response of N. benthamiana leaves. A and B, RT-PCR (A) and immunoblot (B) analyses of CaXEGIP1 expression in N. benthamiana leaves. Expression of the N. benthamiana Actin gene served as a control. Protein loading was verified by Coomassie Brilliant Blue (CBB) staining. C, Induction of cell death in N. benthamiana leaves. D, Measurements of electrolyte leakage from N. benthamiana leaves infiltrated with A. tumefaciens (OD600 = 0.5) carrying the indicated constructs. The blue, yellow, and red circles indicate no cell death, intermediate cell death, and full cell death, respectively. The data represent means ± sd from three independent experiments. Different letters indicate statistically significant differences, as determined by Fisher’s lsd test (P < 0.05).
Silencing of CaXEGIP1 Confers Enhanced Susceptibility to Xcv Infection
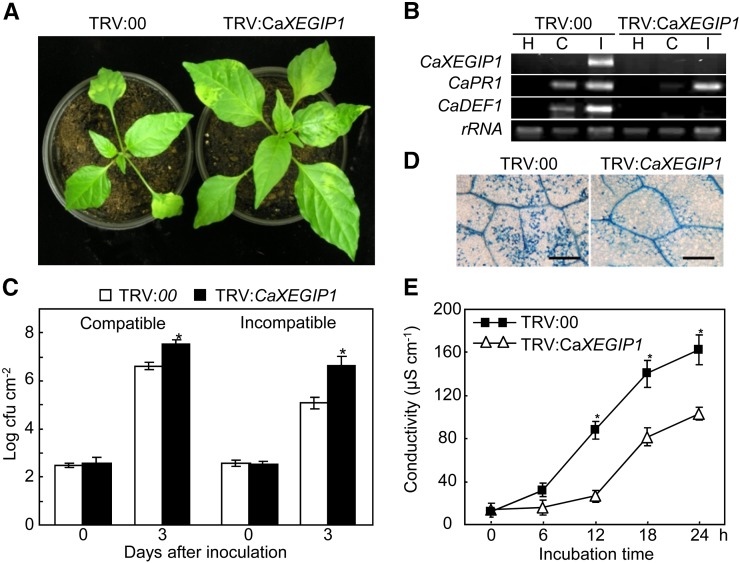
VIGS was used to investigate CaXEGIP1 loss of function in the defense responses of pepper plants (Choi et al., 2007; Hwang and Hwang, 2011; Hwang et al., 2011). Interestingly, the silencing of CaXEGIP1 enhanced the growth of the pepper plants (Fig. 6A). The efficiency of VIGS was evaluated using RT-PCR in empty vector control (TRV:00) and CaXEGIP1-silenced (TRV:CaXEGIP1) pepper leaves 12 h after inoculation with Xcv virulent and avirulent strains (Fig. 6B). Xcv infection did not induce CaXEGIP1 expression in the silenced leaves, indicating that CaXEGIP1 was fully silenced in the pepper leaves. To determine whether CaXEGIP1 silencing affects the defense response pathway in pepper, the expression of some defense-related genes was further analyzed by RT-PCR (Fig. 6B). Silencing of CaXEGIP1 greatly reduced the induction of CaPR1 in pepper leaves during virulent Xcv infection. Significantly, CaDEF1 expression was not induced in the CaXEGIP1-silenced pepper leaves inoculated with both virulent and avirulent Xcv, although CaDEF1 was strongly induced in the infected, unsilenced leaves. In the CaXEGIP1-silenced plants, Xcv growth was approximately 10-fold greater than that in the empty vector control plants (Fig. 6C), indicating that CaXEGIP1 silencing enhances Xcv growth in pepper. Trypan blue staining of avirulent Xcv-infected leaves revealed a compromised HR cell death response in CaXEGIP1-silenced pepper leaves (Fig. 6D). Furthermore, CaXEGIP1 silencing significantly reduced electrolyte leakage from avirulent Xcv-infected leaf discs (Fig. 6E). However, VIGS of empty vector (TRV:00) in pepper leaves did not affect Xcv growth and electrolyte leakage in pepper leaves compared with those in wild-type pepper leaves (Supplemental Fig. S3, A and B). Together, these results indicate that CaXEGIP1 expression is required for hypersensitive cell death signaling in pepper leaves during avirulent Xcv infection.
Figure 6.
Enhanced susceptibility of CaXEGIP1-silenced pepper plants to infection with the Xcv virulent strain Ds1 and the avirulent strain Bv5-4a. A, Enhanced growth of CaXEGIP1-silenced pepper plants. B, RT-PCR analysis of the expression of CaXEGIP1 and defense-related genes in empty vector control (TRV:00) and gene-silenced (TRV:CaXEGIP1) pepper plants 12 h after inoculation with Xcv. Expression of the pepper 18S rRNA gene served as a control. H, Uninoculated healthy leaves; C, compatible; I, incompatible. C, Bacterial growth in leaves. Asterisks indicate significant differences, as determined by the two-tailed Student’s t test (P < 0.05). D and E, Trypan blue staining (D) and electrolyte leakage measurements (E) from leaves inoculated with avirulent Xcv. The data represent means ± sd from three independent experiments. Asterisks indicate significant differences, as determined by the two-tailed t test (P < 0.05). Bars = 200 µm. [See online article for color version of this figure.]
Generation and Analysis of the DEX:CaXEGIP1 Arabidopsis Transgenic Plants
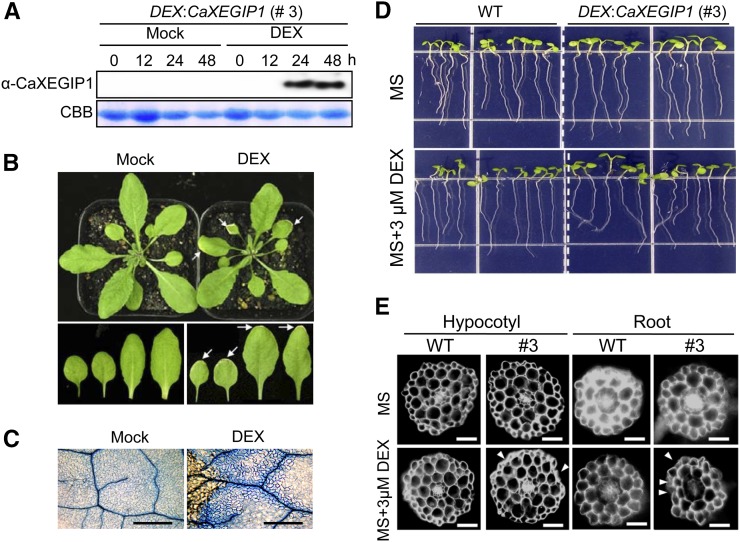
We failed to successfully generate Arabidopsis transgenic lines constitutively expressing CaXEGIP1 under the control of the cauliflower mosaic virus (CaMV) 35S promoter. Thus, the DEX-inducible promoter system was used to generate DEX:CaXEGIP1 transgenic Arabidopsis plants. DEX:CaXEGIP1 line 3 strongly expressed the CaXEGIP1 gene following induction with DEX, and this line was selected for further experiments. We did not observe any apparent phenotypic differences between wild-type and DEX:CaXEGIP1 transgenic plants (data not shown). The accumulation of CaXEGIP1 protein in the transgenic Arabidopsis leaves was detected using rabbit polyclonal antibodies raised against a CaXEGIP1 peptide (Fig. 7A). CaXEGIP1 protein greatly accumulated in the transgenic Arabidopsis leaves 24 and 48 h after DEX treatment. However, mock treatments did not induce CaXEGIP1 transcripts and protein in the DEX:CaXEGIP1 Arabidopsis leaves. The conditional induction of CaXEGIP1 by DEX treatment triggered the cell death response in transgenic Arabidopsis leaves (Fig. 7B). Visible cell death phenotypes were primarily observed in the marginal region of the DEX:CaXEGIP1 Arabidopsis leaves 2 d after DEX treatment but not in mock-treated leaves. Microscopic observations of DEX-treated leaves showed the trypan blue-stained cell death response and cell wall compartments in the DEX:CaXEGIP1 transgenic leaf tissues (Fig. 7C). However, mock and DEX treatments did not trigger any cell death response in leaves of wild-type Arabidopsis plants, as observed by visible plant growth phenotypes and trypan blue staining (Supplemental Fig. S4, A and B).
Figure 7.
CaXEGIP1-induced cell death and abnormal growth of DEX:CaXEGIP1 transgenic Arabidopsis plants after DEX treatment. A, Immunoblot analysis of CaXEGIP1 expression in leaves of transgenic plants at different time intervals after mock or 30 μm DEX treatment. Protein loading was verified by Coomassie Brilliant Blue (CBB) staining. B, CaXEGIP1-induced cell death phenotype in the leaves of DEX:CaXEGIP1 transgenic Arabidopsis plants. The visible cell death phenotype is mainly seen in the marginal region of DEX-treated leaves but not in that of mock-treated leaves, as indicated by white arrows. Photographs were taken 2 d after treatment. C, Trypan blue staining of leaves 24 h after mock or DEX treatment. Bars = 500 μm. D, Root-waving phenotype of transgenic Arabidopsis plants grown on MS medium with or without 3 μm DEX. E, Transverse sections from the roots and hypocotyls of wild-type (WT) and transgenic seedling plants (#3) grown on MS medium with or without DEX. Thin cross-sections of the roots and hypocotyls were stained with 0.1% Calcofluor (Fluorescent Brightener 28; Sigma) in water. Samples were observed with a microscope equipped with a UV lamp. Arrows indicate abnormal growth of root and hypocotyl cells of DEX:CaXEGIP1 seedling plants. [See online article for color version of this figure.]
Seven-day-old transgenic Arabidopsis seedlings were transplanted to Murashige and Skoog (MS) medium containing 3 μm DEX. Interestingly, the transgenic seedlings grown on this medium exhibited unidirectional root bending (Fig. 7D, bottom panel). However, no phenotypic differences were observed in the transgenic seedlings grown in the absence of DEX (Fig. 7D, top panel). To visualize the root bending at the cellular level, transverse sections of the root tissue were stained with 0.1% Calcofluor and observed with a microscope equipped with a UV lamp (Fig. 7E). Compared with the mock-treated wild-type and transgenic seedling roots, the DEX-treated transgenic root tissues showed shrinkage of the epidermal layers. Similar abnormal phenotypes were also observed in the epidermal layers of the hypocotyls of DEX-treated transgenic seedlings. Collectively, these results indicate that DEX-induced overexpression of CaXEGIP1 triggered the cell death response in the leaves, roots, and hypocotyls of transgenic Arabidopsis plants.
CaXEGIP1 Overexpression Confers Enhanced Cell Death But Does Not Suppress Pst Growth in Arabidopsis
We next investigated CaXEGIP1 gain of function in DEX:CaXEGIP1 transgenic Arabidopsis plants during Pst infection. Seven days after inoculation with virulent and avirulent Pst DC3000, more severe cell death developed on the leaves of DEX-treated transgenic plants compared with the mock-treated plants (Fig. 8, A and B). Interestingly, infection of the halves of leaves with avirulent Pst DC3000 (avrRpm1) induced the spreading cell death in the DEX-treated Arabidopsis plants but not in the mock-treated plants. As shown in Figure 8B, the cell death in the mock-treated plants was confined to the pathogen infection region; however, cell death spread over the infection region in the DEX-treated plants that overexpressed CaXEGIP1. In contrast to this cell death response, no significant differences in bacterial growth were detected between mock- and DEX-treated transgenic plants as well as between mock- and DEX-treated wild-type plants during both virulent and avirulent Pst DC3000 infection (Fig. 8C; Supplemental Fig. S4C). These results indicate that CaXEGIP1 overexpression enhances the cell death response but does not induce the resistance of Arabidopsis plants to Pst DC3000 infection.
Figure 8.

Cell death phenotype and bacterial growth in DEX:CaXEGIP1 transgenic Arabidopsis plants infiltrated with Pst DC3000. A and B, Disease symptoms developed on the leaves of DEX:CaXEGIP1 transgenic Arabidopsis 8 d after inoculation with virulent Pst DC3000 (A) and avirulent Pst DC3000 (avrRpm1) (B; 107 cfu mL−1). Arabidopsis plants were inoculated with Pst DC3000 24 h after mock treatment or 30 μm DEX treatment. C, Bacterial growth in DEX:CaXEGIP1 transgenic Arabidopsis leaves 0 and 3 d after inoculation with virulent Pst DC3000 and avirulent Pst DC3000 (avrRpm1) strains (105 cfu mL−1). The data represent means ± sd from three independent experiments. [See online article for color version of this figure.]
CaXEGIP1 Overexpression Confers Reduced Susceptibility to Hyaloperonospora arabidopsidis
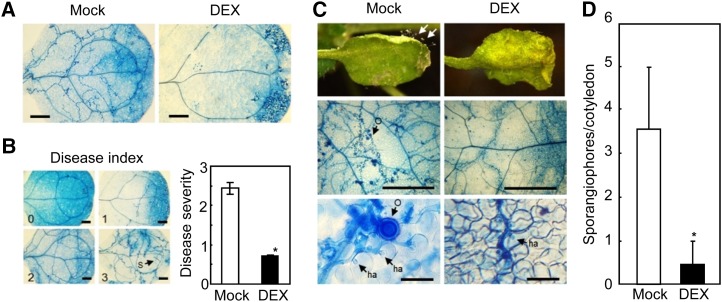
To determine whether CaXEGIP1 overexpression enhances disease resistance to a biotrophic oomycete pathogen, we inoculated DEX-treated cotyledons of 7-d-old DEX:CaXEGIP1 transgenic plants with H. arabidopsidis (Hpa) isolate Noco2. DEX-induced overexpression of CaXEGIP1 induced cell death and also significantly reduced the oomycete growth in Arabidopsis cotyledons (Fig. 9A). Three days after inoculation, the mock- or DEX-pretreated cotyledons of transgenic plants were stained with trypan blue to visualize Hpa growth. As shown in Figure 9B, DEX-treated plants showed significantly reduced disease severity compared with mock-treated plants. Trypan blue staining of infected leaves revealed a lower level of colonization with less hyphal growth and sporulation in DEX-treated transgenic plants compared with mock-treated plants (Fig. 9C). Five days after inoculation, the number of sporangiophores on over 50 cotyledons was counted with a stereomicroscope (Fig. 9D). The number of sporangiophores was significantly reduced in the DEX:CaXEGIP1 transgenic lines. These results indicate that CaXEGIP1 overexpression in Arabidopsis plants confers enhanced resistance to the obligate biotrophic oomycete pathogen Hpa.
Figure 9.
Enhanced resistance of DEX:CaXEGIP1 transgenic Arabidopsis plants to infection with Hpa isolate Noco2. A, Trypan blue staining of Hpa-infected cotyledons of untreated and DEX-treated transgenic plants 3 d after inoculation. Arabidopsis seedlings were inoculated with Hpa 24 h after mock treatment or 30 μm DEX treatment. Bars = 200 µm. B, Reduced growth of Hpa in DEX-treated transgenic Arabidopsis plants. The disease severity was rated on a 0 to 3 scale after trypan blue staining to observe fungal structure development (0, no hyphal growth; 1, minor hyphal growth; 2, severe hyphal growth but no sporangiophore formation; 3, severe hyphal growth and sporangiophore formation). S, Sporangiophore. C, Disease symptoms on the cotyledons of untreated and DEX-treated transgenic plants 5 d after spray inoculation with Hpa (5 × 104 spores mL−1). Arrows indicate sporangiophores. Trypan blue-stained fungal structures are shown at higher magnification (bottom panels). ha, Haustorium; O, oospore. Bars = 500 µm (middle row) and 50 µm (bottom row). D, The number of sporangiophores per cotyledon of untreated or DEX-treated transgenic plants 7 d after inoculation with Hpa. Asterisks indicate significant differences in disease severity, as determined by the two-tailed Student’s t test (P < 0.05).
Proteomic Analysis of DEX:CaXEGIP1 Transgenic Arabidopsis Leaves
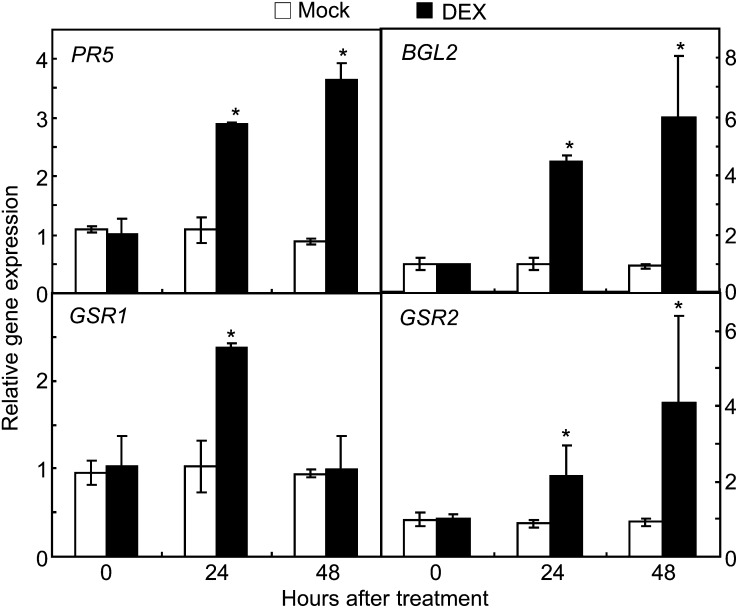
Comparative two-dimensional electrophoresis (2-DE) was used to isolate novel defense response proteins from the proteome in DEX:CaXEGIP1 transgenic plants. The entire 2-DE gel images of total proteins from mock- or DEX-treated transgenic Arabidopsis leaves are shown in Supplemental Figure S5. Among the various protein spots, four proteins newly induced in CaXEGIP1-overexpressing leaves (Supplemental Fig. S5C) were excised from the two-dimensional gels and analyzed by matrix-assisted laser-desorption ionization time of flight (Supplemental Table S1). Interestingly, two of these proteins were identified as PR proteins, PR5 (N1, thaumatin; AT1G75040) and PR2 (N2, β-1,3-glucanase2 [BGL2]; AT3G57260). The other protein spots were identified as Gln biosynthesis-related proteins, GSR2 (N3, copper ion binding/Gln-ammonia ligase; AT1G66200) and GSR1 (N4, Gln synthetase; AT5G37600). To determine whether CaXEGIP1 overexpression induces the genes encoding these four proteins, quantitative RT-PCR was performed (Fig. 10). In DEX:CaXEGIP1 transgenic leaves, none of the tested genes were significantly induced 24 and 48 h after mock treatment. In contrast, PR5, PR2, GSR1, and GSR2 were all significantly induced in DEX:CaXEGIP1 transgenic plants 24 h after DEX treatment. These results indicate that CaXEGIP1 overexpression triggers some defense response genes.
Figure 10.
Real-time RT-PCR analyses of the expression of Arabidopsis genes that are newly induced in DEX:CaXEGIP1 transgenic plants. RNA was extracted from the leaves of 4-week-old transgenic Arabidopsis plants 24 and 48 h after mock or DEX treatment. The relative amount of the gene transcript was normalized against the expression level of the UBQ gene. PR5, Pathogenesis-related gene5 (AT1G75040); BGL2, β-1,3-glucanase2 (AT3G57260); GSR1, Gln synthetase (AT5G37600); GSR2, copper-ion binding/Glu-ammonia ligase (AT1G66200). Asterisks indicate significant differences, as determined by the two-tailed Student’s t test (P < 0.05).
DISCUSSION
The HR cell death-mediated defense signaling genes of pepper were isolated from a pepper cDNA library using differential hybridization and RNA gel-blot analyses (Choi et al., 2007, 2008, 2009). One of the isolated defense response genes was identified as CaXEGIP1 based on sequence homology with tomato XEGIP1 (Qin et al., 2003). CaXEGIP1 expression is rapidly and strongly induced in pepper leaves challenged with the avirulent Xcv strain Bv5-4a. The avirulent Xcv strain Bv5-4a was recently reported to harbor the type III secretion effector protein AvrBsT, which triggers the hypersensitive cell death response and the strong induction of PR genes in pepper plants (Kim et al., 2010). In contrast, infection with virulent Xcv strain Ds1 causes susceptible symptoms accompanied by lower PR gene expression (Lee and Hwang, 2005; Choi et al., 2007). Thus, the rapid and strong induction of CaXEGIP1 in pepper leaves by infection with avirulent Xcv strain Bv5-4a strongly raises the possibility that CaXEGIP1 may be involved in the HR cell death-mediated resistance of pepper plants.
XEGIPs from tomato, tobacco, and carrot (Daucus carota) have been demonstrated to have in vitro inhibitory activity against the fungus A. niger XEG enzyme, a GH12 family XEG (Pauly et al., 1999; Qin et al., 2003; Naqvi et al., 2005; Yoshizawa et al., 2012). At the beginning of our study, we expected that the pathogen-induced CaXEGIP1 would inhibit the pathogen-derived XEG enzyme activity to reduce the pathogen virulence. However, the fungus A. niger-derived or other XEG enzymes that belong to the GH12 family were not available for us to test the inhibitory activity of CaXEGIP1. Thus, we first tested commercial XEG enzyme from the thermophilic bacterium C. thermocellum F7/YS, which belongs to a GH74 family XEG (PROZOMIX). Fortunately, we were able to detect the inhibitory effect of CaXEGIP1 on this GH74 family XEG from C. thermocellum. These results suggest that CaXEGIP1 is a functional xyloglucanase inhibitor protein. In the genome sequence of Xcv strain 85-10, the closest relative of Xcv strain Bv5-4a, 92 genes were predicted to encode putative GH family proteins (www.cazy.org). Further studies of the inhibitory effect of CaXEGIP1 on Xcv-derived GH family enzymes may determine whether CaXEGIP1 can target cell wall-degrading enzymes of invading pathogens.
Recently, structural studies using XEGIP protein from carrot and GH12 family XEG protein from A. aculeatus revealed that the two critical Arg residues from XEGIP proteins located in the inhibition loop 1 (IL-1; Arg-322) and inhibition loop 2 (IL-2; Arg-403) are conserved in XEGIP homologs from tomato and tobacco (Yoshizawa et al., 2012). All of the XEGIP homologs from carrot, tomato, and tobacco showed specific inhibitory activity on GH12 family XEG enzyme activity (Qin et al., 2003; Naqvi et al., 2005; Yoshizawa et al., 2012). However, the GH12 family XEG enzymes are structurally and mechanistically distinct from GH74 family XEG enzymes (Master et al., 2008). The inhibitory effects of plant XEGIPs on the GH74 family XEG enzymes are not yet reported. In addition, CaXEGIP1 does not have the two conserved Arg residues from IL-1 and IL-2 (Supplemental Fig. S1). Recently, Yoshizawa et al. (2011) reported the crystal structure of soybean (Glycine max) XEGIP and found that it lacks inhibitory activity on GH12 and GH11 family XEG enzymes, due to the lack of key amino acid residues, Arg-322 and Arg-403, on their IL-1 and IL-2. More importantly, the role of XEGIPs on plant cell death regulation is not fully understood. Here, we showed that overexpression of CaXEGIP1 induces pathogen-independent cell death in pepper, N. benthamiana, and Arabidopsis plants. These findings support the notion that the inhibitory mechanisms of CaXEGIP1 on GH74 family XEG may be different from those of other reported XEGIPs on GH12 family XEG enzymes. Alternatively, CaXEGIP1 may target intrinsic XG-modifying enzymes to initiate cell death signaling. However, molecular mechanisms underlying the effects of CaXEGIP1 on GH74 family XEG or the intrinsic target of CaXEGIP1 should be further elucidated.
Our topology analysis using smGFP revealed that CaXEGIP1 localizes to the cell wall and apoplast, locations where certain pathogenic microorganisms initially colonize and multiply (Fritig et al., 1998; Lee et al., 2008). Significantly, purified CaXEGIP1 protein inhibited the hydrolytic activity of GH74 family XEG from C. thermocellum F7/YS. Another important finding is that CaXEGIP1 accumulation darkens and thickens the cell wall in plant tissues, as stained by trypan blue. Interestingly, CaXEGIP1 is also ubiquitously distributed around the thickened cell walls of onion epidermal cells. Similarly, A. tumefaciens-mediated or DEX-induced overexpression of CaXEGIP1 in N. benthamiana or Arabidopsis leaves, respectively, triggered dense, pronounced staining of the cell wall compartment with trypan blue. In contrast, VIGS of CaXEGIP1 in pepper plants significantly compromised avirulent Xcv-induced HR cell death. Together, the topology and gain- or loss-of-function data support a potential functional relationship between CaXEGIP1 expression and cell wall strengthening during the HR cell death-mediated resistance response. In some plant-pathogen interactions, the resistance responses accompany rapid cell death and cell wall thickening at the site of attempted infection (Politis and Goodman, 1978; Devarenne and Martin, 2007; Jacobs, 2008). For example, infection of resistant potato (Solanum tuberosum) plants with Phytophthora infestans triggers the formation of papilla at the penetration site and thickening of the entire cell wall prior to HR-like cell death (Schmelzer, 2002). Localized cell wall apposition was also observed in tobacco leaves infected with avirulent Pseudomonas pisi (Politis and Goodman, 1978). In contrast, virulent P. syringae infection suppresses cell wall appositions in Arabidopsis leaves by secreting the type III effector proteins (Hauck et al., 2003). Our findings, along with earlier studies, suggest that CaXEGIP1 expression may strengthen the cell wall barrier, thus activating HR cell death-mediated defense responses.
Overexpression of CaXEGIP1 in pepper, N. benthamiana, and Arabidopsis leaves induced pathogen-independent, spontaneous cell death and, concomitantly, enhanced accumulation of CaXEGIP1 protein. Notably, VIGS of CaXEGIP1 in pepper plants compromised HR cell death but prompted plant growth under normal environmental conditions, as compared with empty vector control plants. These findings support the notion that pathogen-induced CaXEGIP1 up-regulates plant cell death via modification of cell wall structure. Increasing experimental evidence points to a key role for the XG family polysaccharides during various plant developmental processes, including seed germination, fruit ripening, and rapid cell wall expansion (Bourquin et al., 2002; Baumann et al., 2007; Miedes and Lorences, 2009; Anderson et al., 2010). XG is known to coat and cross-link cellulose microfibrils. Thus, the breakage of XG from the XG-cellulose network into oligosaccharides leads to cell wall loosening and cell expansion. However, the significance of xyloglucanase-mediated modification of cell wall structure during cell death signaling remains to be understood.
Silencing of CaXEGIP1 resulted in the enhanced growth of both virulent and avirulent Xcv in pepper leaves. These data suggest a potential role for CaXEGIP1 in the plant defense response to bacterial pathogens. Importantly, the high level of bacterial growth in CaXEGIP1-silenced plants was accompanied by the reduced expression of defense-related genes, such as CaPR1 (Kim and Hwang, 2000) and CaDEF1 (Do et al., 2004). These defense-related genes are strongly induced by avirulent Xcv infection. Notably, ectopic expression of CaPR1 confers tobacco (cv Xanthi) plants with a significantly enhanced resistance to a diverse range of plant pathogens, including the oomycete pathogen Phytophthora nicotianae and the bacterial pathogens Ralstonia solanacearum and P. syringae pv tabaci (Sarowar et al., 2005). Together, these findings along with other available information suggest that the CaXEGIP1 gene plays a pivotal role in the disease resistance and downstream defense signaling of pepper plants.
Unexpectedly, CaXEGIP1 overexpression did not significantly suppress the proliferation of Pst DC3000 in transgenic Arabidopsis leaves. However, the virulent and avirulent Pst DC3000 infection distinctly induced susceptible and hypersensitive cell death phenotypes in CaXEGIP1-overexpressing transgenic plants, respectively. These results suggest that CaXEGIP1 overexpression does not directly impair Pst DC3000 itself but instead positively regulates plant cell death signaling. In contrast, CaXEGIP1 overexpression did significantly suppress infection by the biotrophic oomycete pathogen Hpa by enhancing the cell death response. Comparative proteomics and quantitative PCR analyses of transgenic Arabidopsis plants identified four genes (PR2, PR5, GSR1, and GSR2) newly induced by CaXEGIP1 overexpression. Arabidopsis PR2 and PR5, which encode β-1,3-glucanase and thaumatin-like protein, respectively, have been widely used as defense molecular markers. Notably, thaumatin-like PR5 families are involved in resistance against oomycetes (van Loon et al., 2006). Reduced susceptibility of the CaXEGIP1-overexpressing transgenic Arabidopsis plants against Hpa infection may be partially due to enhanced expression of PR2 and PR5.
In summary, we have identified CaXEGIP1 as essential for the HR cell death-mediated defense response of pepper plants. Silencing of CaXEGIP1 confers pepper plants with significantly enhanced susceptibility to Xcv infection. This is accompanied by compromised HR cell death and decreased expression of CaPR1 and CaDEF1. In contrast, overexpression of CaXEGIP1 induces a pathogen-independent, spontaneous cell death response and enhances resistance to the obligate biotrophic downy mildew pathogen in Arabidopsis via cell wall modification. Together, these data suggest that extracellular and cell wall-bound CaXEGIP1 may enhance disease resistance by strengthening the host cell wall.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Pepper plants (Capsicum annuum ‘Nockwang’) and Nicotiana benthamiana were raised in a growth room at 28°C with a 16-h photoperiod at a light intensity of 100 μmol photons m−2 s−1. Arabidopsis (Arabidopsis thaliana ecotype Columbia [Col-0]) and transgenic plants were grown at 24°C under long-day conditions (16 h of light/8 h of dark) with a photosynthetic flux of 130 μmol photons m−2 s−1.
Isolation and Sequence Analysis of CaXEGIP1 cDNA
A cDNA library was constructed using poly(A+) mRNA extracted from pepper leaves inoculated with the avirulent Xanthomonas campestris pv vesicatoria strain Bv5-4a (Jung and Hwang, 2000; Choi et al., 2007). Differential hybridization screening was performed using the cDNA library, and selected clones were sequenced on the ABI 310 DNA sequencer (Applied Biosystem).
Purification and Activity Assay of CaXEGIP1
The CaXEGIP1 coding region was cloned in the pET30a vector (Novagen). Escherichia coli BL21 (DE3) (Invitrogen) was grown at 37°C until the optical density at 600 nm (OD600) reached 0.5. Gene expression was induced by adding IPTG to a final concentration of 500 μm. After 18 h of incubation at 18°C with shaking, bacterial cells were harvested by centrifugation at 3,000g for 30 min. The cells were resuspended in lysis buffer (50 mm Tris-HCl [pH 8.0], 0.3 m NaCl, and 10 mm imidazole) and lysed by sonication. Cell debris was removed by centrifugation at 15,000g for 1 h at 4°C. The soluble His-tagged CaXEGIP1 protein was purified from the supernatant by affinity chromatography using Ni2+-nitrilotriacetic acid agarose resin (Qiagen).
Recombinant XEG from Clostridium thermocellum F7/YS was purchased from PROZOMIX. The inhibitory effect of CaXEGIP1 on the enzymatic activity of XEG was assayed using AZO-XG (Megazyme) as a substrate according to the manufacturer’s instructions. Varying amounts of CaXEGIP1 were preincubated with 1 μg of XEG at 60°C for 1 h, then mixed and incubated with 0.5 mL of 10 mg mL−1 AZO-XG in 100 mm sodium acetate buffer (pH 4.5) at 60°C for 10 min. The reaction was terminated, and the high-Mr substrates were precipitated by the addition of 2 mL of 100% methanol. After centrifugation at 1,000g for 10 min, the amount of reducing sugars was measured by spectrophotometric examination of the absorbance of the supernatant solutions containing depolymerized Remazol Brilliant Blue-dyed sugars at 590 nm.
GFP Localization by Confocal Microscopy
Biolistic transformation of onion (Allium cepa) epidermal cells was performed as described previously (Choi et al., 2007). Briefly, the CaXEGIP1 coding region was cloned between the CaMV 35S promoter and the smGFP region of the binary vector p326GFP to generate a C-terminal fusion of smGFP to CaXEGIP1. The resulting plasmids were purified using the Qiagen plasmid maxi kits according to the manufacturer’s instructions. Onion epidermis was bombarded with gold particles coated with plasmids using a Bio-Rad PDS-1000/He particle-delivery system. Bombarded specimens were incubated for 24 h on 1× MS agar medium at 24°C and observed using a MRC-1024 confocal laser scanning microscope (Bio-Rad).
Pathogen Inoculation
The Xanthomonas campestris pv vesicatoria (Xcv) virulent strain Ds1 and the avirulent strain Bv5-4a were prepared as described previously (Choi et al., 2007). Briefly, bacteria were cultured overnight in YN broth (5 g of yeast extract and 8 g of nutrient broth L−1) at 28°C. Prior to inoculation, bacterial cells were collected by centrifugation and resuspended in 10 mm MgCl2 (108 colony-forming units [cfu] mL−1). Pepper plants at the six-leaf stage were inoculated by infiltrating the bacterial suspension into the abaxial side of fully expanded leaves using a syringe without a needle. The infected plants were incubated in a controlled chamber at 28°C with 100% relative humidity for 16 h.
Pseudomonas syringae pv tomato DC3000 and DC3000 (avrRpm1) were used to infect Arabidopsis. The leaves of wild-type (Col-0) and DEX:CaXEGIP1 plants were infiltrated with Pst at 107 cfu mL−1 (for disease symptom or HR observation) or 105 cfu mL−1 (for the bacterial growth assay). Bacterial growth in leaves was monitored on King’s B agar medium containing 50 μg mL−1 rifampicin and 50 μg mL−1 kanamycin.
Hyaloperonospora arabidopsidis isolate Noco2 was maintained on the susceptible Arabidopsis ecotype Col-0 (Reignault et al., 1996). One-week-old seedlings of wild-type and DEX:CaXEGIP1 plants were inoculated with a suspension of 5 × 104 conidiosporangia mL−1 in distilled tap water containing 0.05% Tween 20. Seedlings were covered with a transparent dome to maintain high humidity and were grown at 17°C with a 14-h photoperiod. Asexual sporulation was assessed by counting the sporangiophores per cotyledon 5 d after inoculation.
RNA Gel-Blot and RT-PCR Analyses
Total cellular RNA was extracted from pepper and Arabidopsis leaves using TRIzol Reagent (Invitrogen) according to the manufacturer’s instructions. For RNA gel blotting, equal quantities of RNA were separated on 1.2% formaldehyde-agarose gels in the presence of ethidium bromide and transferred to nylon membranes (Hybond N+; Amersham). To generate a gene-specific probe, the N-terminal region of CaXEGIP1 was amplified using the following primers: forward (5′-ATGGTGACATTCAAACTTCCCT-3′) and reverse (5′-CTTCCTGGTGAATCTAGGGTC-3′). A PCR-amplified fragment of 588 bp was 32P labeled using a random priming kit (Boehringer Mannheim). Agarose gel electrophoresis, RNA transfers, and hybridization with the CaXEGIP1 fragment were performed using standard procedures.
Pepper and Arabidopsis cDNA was synthesized from total RNA (2 μg) using avian myeloblastosis virus reverse transcriptase (Roche) with oligo(dT)15 as a primer (Roche) according to the manufacturer’s instructions. RT-PCR was performed using ExTaq polymerase (Takara Biomedicals). The reaction mixture contained aliquots of 1 μL of RT reaction products, 0.5 μm each of the forward and reverse primers, 1× ExTaq buffer, 250 nm deoxyribonucleotide triphosphates, and 0.5 units of ExTaq polymerase in a 50-μL solution. RT-PCR conditions were 95°C for 10 min and 30 cycles of 95°C for 30 s each, 52°C for 30 s, and 72°C for 30 s. Single bands of PCR products were confirmed on an agarose gel. The following gene-specific primers were used for RT-PCR analyses: 5′-GTGGCATCTGAAGAGTACTATATC-3′ (forward) and 5′-CTAGCCTAGAGTGGTATTATCG-3′ (reverse) for CaXEGIP1; 5′-CAGGATGCAACACTCTGGTGG-3′ (forward) and 5′-ATCAAAGGCCGGTTGGTC-3′ (reverse) for CaPR1 (accession no. AF053343); and 5′-CAGGATGCAACACTCTGGTGG-3′ (forward) and 5′-ATCAAAGGCCGGTTGGTC-3′ (reverse) for CaDEF1 (accession no. AF442388).
Agrobacterium tumefaciens-Mediated Transient Expression Assay
A. tumefaciens-mediated transient expression experiments were performed as described previously (Choi et al., 2011, 2012; Kim and Hwang, 2011). Briefly, the coding region of CaXEGIP1 was cloned between the CaMV 35S promoter and the nos terminator region of the binary vector pBIN35S. The resulting plasmid was transformed into A. tumefaciens GV3101. A. tumefaciens containing the empty vector (pBIN:00) or the pBIN35S:CaXEGIP1 construct was grown overnight at 28°C in YEP medium (10 g of yeast extract, 10 g of peptone, and 5 g of sodium chloride [Difco] L−1; pH 7.5 adjusted with NaOH) containing 50 μg mL−1 rifampicin and 50 μg mL−1 kanamycin. The bacterial cells were suspended in infiltration buffer (10 mm MgCl2, 10 mm MES [pH 5.7], and 200 μm acetosyringone) to a final OD600 of 1.0, followed by serial dilutions. Bacterial cells were infiltrated into pepper or N. benthamiana leaves. For comparison, Pto, avrPto, and Pto/avrPto were transiently expressed in N. benthamiana leaves.
VIGS
The tobacco rattle virus (TRV)-based VIGS system was used to silence CaXEGIP1 in pepper plants (Liu et al., 2002; Choi and Hwang, 2011; Lee et al., 2011). The C-terminal region of CaXEGIP1 was amplified by PCR with the following primers: 5′-GTGGCATCTGAAGAGTACTATATC-3′ (forward) and 5′-CTAGCCTAGAGTGGTATTATCG-3′ (reverse). PCR-amplified fragments containing the 558-bp 3′ region of CaXEGIP1 were cloned into the pTRV2 vector to yield pTRV2:CaXEGIP1. The resulting plasmid was transformed into A. tumefaciens strain GV3101. Transformants carrying pTRV1 or pTRV2:CaXEGIP1 were coinfiltrated into the fully expanded cotyledons of pepper plants (OD600 = 0.2). Plants were placed in a growth room at 25°C with a 16-h-light/8-h-dark photoperiod to allow viral growth and spread. Pepper plants silenced for 4 to 5 weeks were used for pathogen inoculation.
Arabidopsis Transformation
The full-length CaXEGIP1 cDNA sequence was excised from pBluescript SK− by digestion with XhoI and XbaI and subsequently cloned into pTA7002. The binary plasmids were introduced into A. tumefaciens strain GV3101 via electroporation. Transgenic Arabidopsis plants harboring the DEX-inducible CaXEGIP1 gene were generated using the floral dipping method (Clough and Bent, 1998). Transgenic plants were selected on MS agar plates containing 25 μg mL−1 hygromycin. For DEX treatment, 30 μm DEX was sprayed onto Arabidopsis leaves.
Immunoblot Analyses
After SDS-PAGE, proteins were electrotransferred to polyvinylidene difluoride membranes (GE Healthcare). Membranes were incubated with a 1:10,000 dilution of polyclonal rabbit antibodies raised against the CaXEGIP1-specific peptide NIYNPKKIDISKD (AbFrontier). Antigen-antibody complexes were detected using peroxidase-conjugated goat anti-rabbit IgG (Sigma-Aldrich).
Histochemistry
Trypan blue staining was performed as described previously (Choi et al., 2007). To visualize cell death and fungal structure, leaves were stained by boiling in lactophenol-trypan blue solution (10 mL of lactic acid, 10 mL of glycerol, 10 g of phenol, and 10 mg of trypan blue dissolved in 10 mL of distilled water) followed by destaining with chloral hydrate (2.5 g mL−1). To monitor the modification of the cell wall structure in DEX:CaXEGIP1 transgenic Arabidopsis seedlings, transverse hand sections were obtained from agarose-embedded seedlings and stained with 0.1% Fluorescent Brightener 28 (Sigma; Lin and Schiefelbein, 2001).
Electrolyte Leakage Measurement
Six leaf discs of defined area (0.6 cm in diameter) were removed and washed with distilled water following infiltration. These discs were incubated in 3 mL of distilled water at room temperature. The conductivity of the incubation medium was recorded at various time points with a conductivity meter (model sensION7; Hach; http://www.hach.com).
2-DE and Protein Staining
2-DE was performed as described previously (Choi and Hwang 2011) with minor modifications. Total soluble proteins were extracted from the leaves of mock-treated or 30 μm DEX-treated transgenic Arabidopsis plants using extraction buffer (10% [w/v] trichloroacetic acid and 0.07% [w/v] dithiothreitol [DTT] in cold acetone [−20°C]). Protein extracts (800 μg) were suspended in rehydration buffer (9 m urea, 100 mm DTT, 4% [w/v] CHAPS, 0.5% [v/v] Bio-lyte 3/10 carrier ampholytes, and 0.002% bromphenol blue) and applied to an immobilized pH gradient (IPG) strip for in-gel rehydration. Strips were rehydrated at 50 V for 24 h. Isoelectric focusing was performed using 24-cm IPG strips (Bio-Rad) with a nonlinear pH 4 to 7 gradient. Isoelectric focusing was run using a PROTEAN IEF Cell (Bio-Rad) at gradient steps of 250 V for 1 h, 500 V for 1 h, 1,000 V for 2 h, and 10,000 V for 4 h, then a final step of 10,000 V toward a total of 90 kVh. IPG strips were incubated in the equilibration buffer (50 mm Tris-HCl, pH 8.8, 6 m urea, 30% [v/v] glycerol, and 2% [w/v] SDS) containing 1% (w/v) DTT for 15 min for the first equilibration step. This was followed by incubation in 4% (w/v) iodoacetamide for the second equilibration step. For 2-DE, IPG strips were sealed on the top of the 12.5% two-dimensional gel using 1% low-melting agarose in SDS-electrophoresis buffer (25 mm Tris, 0.2 m Gly, and 0.1% SDS). SDS-PAGE was performed in an Ettan DALTsix electrophoresis unit (Amersham Biosciences) at 5 W per gel for 1 h, followed by 15 W per gel for 6 h until the bromphenol blue front reached the end of the gel.
For Coomassie Brilliant Blue staining, gels were fixed in a 40% (v/v) methanol and 7% acetic acid solution for 2 h and then stained in a Coomassie Brilliant Blue solution (0.1% [w/v] Coomassie Brilliant Blue G 250, 34% [v/v] methanol, 3% [v/v] phosphoric acid, and 17% [w/v] ammonium sulfate). Coomassie blue-stained gels were scanned with the UMAX PowerLook 1100XL scanner, and the images were analyzed using ImageMaster 2D Platinum 6.0 (Amersham Biosciences). The two-dimensional gels were compared and matched, and a quantitative determination of spot volumes was performed.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers JQ673414 (CaXEGIP1), AF053343 (CaPR1), AF442388 (CaDEF1), AT1G75040 (PR5), AT3G57260 (BGL2), AT5G37600 (GSR1), AT1G66200 (GSR2), and AT3G62250 (UBQ5).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Amino acid sequence alignment of pepper CaXEGIP1 with putative XEGIPs from grape (accession no. XP_002272235), petunia (Petunia × hybrida; accession no. ACM68432), carrot (accession no. BAA03413), tomato (accession no. AAN87262), potato (accession no. AAP84703), and Arabidopsis (accession no. AAM61574).
Supplemental Figure S2. Trypan blue staining of CaXEGIP1-induced cell death in N. benthamiana leaves.
Supplemental Figure S3. VIGS of empty vector (TRV:00) in pepper leaves does not affect Xcv growth and electrolyte leakages in pepper leaves compared with those in wild-type pepper leaves.
Supplemental Figure S4. No cell death phenotypes and no differences in bacterial growth are seen in mock- and DEX-treated wild-type Arabidopsis (Col-0) plants.
Supplemental Figure S5. Two-dimensional gel images of the proteome in mock- or DEX-treated DEX:CaXEGIP1 transgenic Arabidopsis leaves.
Supplemental Table S1. Proteins newly induced by DEX:CaXEGIP1 expression in Arabidopsis, as identified by matrix-assisted laser-desorption ionization time of flight mass spectrometry.
Acknowledgments
We thank Dr. S.P. Dinesh-Kumar (University of California, Davis) for the pTRV1 and pTRV2 vectors and Dr. U. Bonas (Martin-Luther-Universitaet) for the A. tumefaciens strain GV3101.
Glossary
- XG
xyloglucan
- XEG
xyloglucan-specific endo-β-1,4-glucanase
- VIGS
virus-induced gene silencing
- PCD
programmed cell death
- HR
hypersensitive response
- Xcv
Xanthomonas campestris pv vesicatoria
- IPTG
isopropyl-β-d-thiogalactoside
- RT
reverse transcription
- CaMV
cauliflower mosaic virus
- DEX
dexamethasone
- MS
Murashige and Skoog
- Hpa
Hyaloperonospora arabidopsidis
- 2-DE
two-dimensional electrophoresis
- IL-1
inhibition loop 1
- IL-2
inhibition loop 2
- Col-0
ecotype Columbia
- OD600
optical density at 600 nm
- cfu
colony-forming units
- DTT
dithiothreitol
- IPG
immobilized pH gradient
- cDNA
complementary DNA
- Pst
Pseudomonas syringae pv tomato
References
- Albersheim P, Anderson AJ. (1971) Proteins from plant cell walls inhibit polygalacturonases secreted by plant pathogens. Proc Natl Acad Sci USA 68: 1815–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An SH, Sohn KH, Choi HW, Hwang IS, Lee SC, Hwang BK. (2008) Pepper pectin methylesterase inhibitor protein CaPMEI1 is required for antifungal activity, basal disease resistance and abiotic stress tolerance. Planta 228: 61–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson CT, Carroll A, Akhmetova L, Somerville C. (2010) Real-time imaging of cellulose reorientation during cell wall expansion in Arabidopsis roots. Plant Physiol 152: 787–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann MJ, Eklöf JM, Michel G, Kallas AM, Teeri TT, Czjzek M, Brumer H., III (2007) Structural evidence for the evolution of xyloglucanase activity from xyloglucan endo-transglycosylases: biological implications for cell wall metabolism. Plant Cell 19: 1947–1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourquin V, Nishikubo N, Abe H, Brumer H, Denman S, Eklund M, Christiernin M, Teeri TT, Sundberg B, Mellerowicz EJ. (2002) Xyloglucan endotransglycosylases have a function during the formation of secondary cell walls of vascular tissues. Plant Cell 14: 3073–3088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantu D, Vicente AR, Labavitch JM, Bennett AB, Powell AL. (2008) Strangers in the matrix: plant cell walls and pathogen susceptibility. Trends Plant Sci 13: 610–617 [DOI] [PubMed] [Google Scholar]
- Carpita NC, Gibeaut DM. (1993) Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J 3: 1–30 [DOI] [PubMed] [Google Scholar]
- Choi HW, Kim YJ, Hwang BK. (2011) The hypersensitive induced reaction and leucine-rich repeat proteins regulate plant cell death associated with disease and plant immunity. Mol Plant Microbe Interact 24: 68–78 [DOI] [PubMed] [Google Scholar]
- Choi HW, Kim YJ, Lee SC, Hong JK, Hwang BK. (2007) Hydrogen peroxide generation by the pepper extracellular peroxidase CaPO2 activates local and systemic cell death and defense response to bacterial pathogens. Plant Physiol 145: 890–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HW, Lee BG, Kim NH, Park Y, Lim CW, Song HK, Hwang BK. (2008) A role for a menthone reductase in resistance against microbial pathogens in plants. Plant Physiol 148: 383–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HW, Lee DH, Hwang BK. (2009) The pepper calmodulin gene CaCaM1 is involved in reactive oxygen species and nitric oxide generation required for cell death and the defense response. Mol Plant Microbe Interact 22: 1389–1400 [DOI] [PubMed] [Google Scholar]
- Choi S, Hwang BK. (2011) Proteomics and functional analyses of pepper abscisic acid-responsive1 (ABR1), which is involved in cell death and defense signaling. Plant Cell 23: 823–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S, Hwang IS, Hwang BK. (2012) Requirement of the cytosolic interaction between PATHOGENESIS-RELATED PROTEIN10 and LEUCINE-RICH REPEAT PROTEIN1 for cell death and defense signaling in pepper. Plant Cell 24: 1675–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Côté F, Hahn MG. (1994) Oligosaccharins: structures and signal transduction. Plant Mol Biol 26: 1379–1411 [DOI] [PubMed] [Google Scholar]
- DeBoy RT, Mongodin EF, Fouts DE, Tailford LE, Khouri H, Emerson JB, Mohamoud Y, Watkins K, Henrissat B, Gilbert HJ, et al. (2008) Insights into plant cell wall degradation from the genome sequence of the soil bacterium Cellvibrio japonicus. J Bacteriol 190: 5455–5463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lorenzo G, Ferrari S. (2002) Polygalacturonase-inhibiting proteins in defense against phytopathogenic fungi. Curr Opin Plant Biol 5: 295–299 [DOI] [PubMed] [Google Scholar]
- Devarenne TP, Martin GB. (2007) Manipulation of plant programmed cell death pathways during plant-pathogen interactions. Plant Signal Behav 2: 188–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do HM, Lee SC, Jung HW, Sohn KH, Hwang BK. (2004) Differential expression and in situ localization of a pepper defensin (CADEF1) gene in response to pathogen infection, abiotic elicitors and environmental stress in Capsicum annuum. Plant Sci 166: 1297–1305 [Google Scholar]
- D’Ovidio R, Mattei B, Roberti S, Bellincampi D. (2004a) Polygalacturonases, polygalacturonase-inhibiting proteins and pectic oligomers in plant-pathogen interactions. Biochim Biophys Acta 1696: 237–244 [DOI] [PubMed] [Google Scholar]
- D’Ovidio R, Raiola A, Capodicasa C, Devoto A, Pontiggia D, Roberti S, Galletti R, Conti E, O’Sullivan D, De Lorenzo G. (2004b) Characterization of the complex locus of bean encoding polygalacturonase-inhibiting proteins reveals subfunctionalization for defense against fungi and insects. Plant Physiol 135: 2424–2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federici L, Di Matteo A, Fernandez-Recio J, Tsernoglou D, Cervone F. (2006) Polygalacturonase inhibiting proteins: players in plant innate immunity? Trends Plant Sci 11: 65–70 [DOI] [PubMed] [Google Scholar]
- Flors V, Leyva MdeL, Vicedo B, Finiti I, Real MD, García-Agustín P, Bennett AB, González-Bosch C. (2007) Absence of the endo-β-1,4-glucanases Cel1 and Cel2 reduces susceptibility to Botrytis cinerea in tomato. Plant J 52: 1027–1040 [DOI] [PubMed] [Google Scholar]
- Fritig B, Heitz T, Legrand M. (1998) Antimicrobial proteins in induced plant defense. Curr Opin Immunol 10: 16–22 [DOI] [PubMed] [Google Scholar]
- Fukuda H. (2000) Programmed cell death of tracheary elements as a paradigm in plants. Plant Mol Biol 44: 245–253 [DOI] [PubMed] [Google Scholar]
- Gloster TM, Ibatullin FM, Macauley K, Eklöf JM, Roberts S, Turkenburg JP, Bjørnvad ME, Jørgensen PL, Danielsen S, Johansen KS, et al. (2007) Characterization and three-dimensional structures of two distinct bacterial xyloglucanases from families GH5 and GH12. J Biol Chem 282: 19177–19189 [DOI] [PubMed] [Google Scholar]
- Hauck P, Thilmony R, He SY. (2003) A Pseudomonas syringae type III effector suppresses cell wall-based extracellular defense in susceptible Arabidopsis plants. Proc Natl Acad Sci USA 100: 8577–8582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang IS, An SH, Hwang BK. (2011) Pepper asparagine synthetase 1 (CaAS1) is required for plant nitrogen assimilation and defense responses to microbial pathogens. Plant J 67: 749–762 [DOI] [PubMed] [Google Scholar]
- Hwang IS, Hwang BK. (2010) The pepper 9-lipoxygenase gene CaLOX1 functions in defense and cell death responses to microbial pathogens. Plant Physiol 152: 948–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang IS, Hwang BK. (2011) The pepper mannose-binding lectin gene CaMBL1 is required to regulate cell death and defense responses to microbial pathogens. Plant Physiol 155: 447–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs TH. (2008) The occurrence of cell wall appositions in flag leaves of spring wheats, susceptible and partially resistant to wheat leaf rust. J Phytopathol 127: 239–249 [Google Scholar]
- Jones AM. (2001) Programmed cell death in development and defense. Plant Physiol 125: 94–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung HW, Hwang BK. (2000) Isolation, partial sequencing, and expression of pathogenesis-related cDNA genes from pepper leaves infected by Xanthomonas campestris pv. vesicatoria. Mol Plant Microbe Interact 13: 136–142 [DOI] [PubMed] [Google Scholar]
- Kieffer F, Lherminier J, Simon-Plas F, Nicole M, Paynot M, Elmayan T, Blein JP. (2000) The fungal elicitor cryptogein induces cell wall modifications on tobacco cell suspension. J Exp Bot 51: 1799–1811 [DOI] [PubMed] [Google Scholar]
- Kim DS, Hwang BK. (2011) The pepper receptor-like cytoplasmic protein kinase CaPIK1 is involved in plant signaling of defense and cell-death responses. Plant J 66: 642–655 [DOI] [PubMed] [Google Scholar]
- Kim NH, Choi HW, Hwang BK. (2010) Xanthomonas campestris pv. vesicatoria effector AvrBsT induces cell death in pepper, but suppresses defense responses in tomato. Mol Plant Microbe Interact 23: 1069–1082 [DOI] [PubMed] [Google Scholar]
- Kim YJ, Hwang BK. (2000) Pepper gene encoding a basic pathogenesis-related 1 protein is pathogen and ethylene inducible. Physiol Plant 108: 51–60 [Google Scholar]
- Lam E. (2004) Controlled cell death, plant survival and development. Nat Rev Mol Cell Biol 5: 305–315 [DOI] [PubMed] [Google Scholar]
- Lee DH, Choi HW, Hwang BK. (2011) The pepper E3 ubiquitin ligase RING1 gene, CaRING1, is required for cell death and the salicylic acid-dependent defense response. Plant Physiol 156: 2011–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SC, Hwang BK. (2005) Induction of some defense-related genes and oxidative burst is required for the establishment of systemic acquired resistance in Capsicum annuum. Planta 221: 790–800 [DOI] [PubMed] [Google Scholar]
- Lee SC, Hwang IS, Choi HW, Hwang BK. (2008) Involvement of the pepper antimicrobial protein CaAMP1 gene in broad spectrum disease resistance. Plant Physiol 148: 1004–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Schiefelbein J. (2001) Embryonic control of epidermal cell patterning in the root and hypocotyl of Arabidopsis. Development 128: 3697–3705 [DOI] [PubMed] [Google Scholar]
- Liu YL, Schiff M, Dinesh-Kumar SP. (2002) Virus-induced gene silencing in tomato. Plant J 31: 777–786 [DOI] [PubMed] [Google Scholar]
- Martinez-Fleites C, Guerreiro CI, Baumann MJ, Taylor EJ, Prates JA, Ferreira LM, Fontes CM, Brumer H, Davies GJ. (2006) Crystal structures of Clostridium thermocellum xyloglucanase, XGH74A, reveal the structural basis for xyloglucan recognition and degradation. J Biol Chem 281: 24922–24933 [DOI] [PubMed] [Google Scholar]
- Master ER, Zheng Y, Storms R, Tsang A, Powlowski J. (2008) A xyloglucan-specific family 12 glycosyl hydrolase from Aspergillus niger: recombinant expression, purification and characterization. Biochem J 411: 161–170 [DOI] [PubMed] [Google Scholar]
- Miedes E, Lorences EP. (2009) Xyloglucan endotransglucosylase/hydrolases (XTHs) during tomato fruit growth and ripening. J Plant Physiol 166: 489–498 [DOI] [PubMed] [Google Scholar]
- Naqvi SM, Harper A, Carter C, Ren G, Guirgis A, York WS, Thornburg RW. (2005) Nectarin IV, a potent endoglucanase inhibitor secreted into the nectar of ornamental tobacco plants: isolation, cloning, and characterization. Plant Physiol 139: 1389–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauly M, Andersen LN, Kauppinen S, Kofod LV, York WS, Albersheim P, Darvill A. (1999) A xyloglucan-specific endo-beta-1,4-glucanase from Aspergillus aculeatus: expression cloning in yeast, purification and characterization of the recombinant enzyme. Glycobiology 9: 93–100 [DOI] [PubMed] [Google Scholar]
- Politis DJ, Goodman RN. (1978) Localized cell wall appositions: incompatibility response of tobacco leaf cells to Pseudomonas pisi. Phytopathology 68: 309–316 [Google Scholar]
- Powell AL, van Kan J, ten Have A, Visser J, Greve LC, Bennett AB, Labavitch JM. (2000) Transgenic expression of pear PGIP in tomato limits fungal colonization. Mol Plant Microbe Interact 13: 942–950 [DOI] [PubMed] [Google Scholar]
- Qin Q, Bergmann CW, Rose JK, Saladie M, Kolli VS, Albersheim P, Darvill AG, York WS. (2003) Characterization of a tomato protein that inhibits a xyloglucan-specific endoglucanase. Plant J 34: 327–338 [DOI] [PubMed] [Google Scholar]
- Reignault P, Frost LN, Richardson H, Daniels MJ, Jones JDG, Parker JE. (1996) Four Arabidopsis RPP loci controlling resistance to the Noco2 isolate of Peronospora parasitica map to regions known to contain other RPP recognition specificities. Mol Plant Microbe Interact 9: 464–473 [DOI] [PubMed] [Google Scholar]
- Romero D, Eugenia Rivera M, Cazorla FM, Codina JC, Fernández-Ortuño D, Torés JA, Pérez-García A, de Vicente A. (2008) Comparative histochemical analyses of oxidative burst and cell wall reinforcement in compatible and incompatible melon-powdery mildew (Podosphaera fusca) interactions. J Plant Physiol 165: 1895–1905 [DOI] [PubMed] [Google Scholar]
- Sarowar S, Kim YJ, Kim EN, Kim KD, Hwang BK, Islam R, Shin JS. (2005) Overexpression of a pepper basic pathogenesis-related protein 1 gene in tobacco plants enhances resistance to heavy metal and pathogen stresses. Plant Cell Rep 24: 216–224 [DOI] [PubMed] [Google Scholar]
- Schmelzer E. (2002) Cell polarization, a crucial process in fungal defence. Trends Plant Sci 7: 411–415 [DOI] [PubMed] [Google Scholar]
- Spinelli F, Mariotti L, Mattei B, Salvi G, Cervone F, Caprari C. (2009) Three aspartic acid residues of polygalacturonase-inhibiting protein (PGIP) from Phaseolus vulgaris are critical for inhibition of Fusarium phyllophilum PG. Plant Biol (Stuttg) 11: 738–743 [DOI] [PubMed] [Google Scholar]
- Valette-Collet O, Cimerman A, Reignault P, Levis C, Boccara M. (2003) Disruption of Botrytis cinerea pectin methylesterase gene Bcpme1 reduces virulence on several host plants. Mol Plant Microbe Interact 16: 360–367 [DOI] [PubMed] [Google Scholar]
- van Loon LC, Rep M, Pieterse CM. (2006) Significance of inducible defense-related proteins in infected plants. Annu Rev Phytopathol 44: 135–162 [DOI] [PubMed] [Google Scholar]
- Xie W, Hao L, Goodwin PH. (2008) Role of a xyloglucan-specific endo-β-1,4-glucanase inhibitor in the interactions of Nicotiana benthamiana with Colletotrichum destructivum, C. orbiculare or Pseudomonas syringae pv. tabaci. Mol Plant Pathol 9: 191–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- York WS, Qin Q, Rose JK. (2004) Proteinaceous inhibitors of endo-β-glucanases. Biochim Biophys Acta 1696: 223–233 [DOI] [PubMed] [Google Scholar]
- Yoshizawa T, Shimizu T, Hirano H, Sato M, Hashimoto H. (2012) Structural basis for inhibition of xyloglucan-specific endo-β-1,4-glucanase (XEG) by XEG-protein inhibitor. J Biol Chem 287: 18710–18716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa T, Shimizu T, Yamabe M, Taichi M, Nishiuchi Y, Shichijo N, Unzai S, Hirano H, Sato M, Hashimoto H. (2011) Crystal structure of basic 7S globulin, a xyloglucan-specific endo-β-1,4-glucanase inhibitor protein-like protein from soybean lacking inhibitory activity against endo-β-glucanase. FEBS J 278: 1944–1954 [DOI] [PubMed] [Google Scholar]