Abstract
Mutations in the SARS-Coronavirus (SARS-CoV) can alter its clinical presentation, and the study of its mutation patterns in human populations can facilitate contact tracing. Here, we describe the development and validation of an oligonucleotide resequencing array for interrogating the entire 30-kb SARS-CoV genome in a rapid, cost-effective fashion. Using this platform, we sequenced SARS-CoV genomes from Vero cell culture isolates of 12 patients and directly from four patient tissues. The sequence obtained from the array is highly reproducible, accurate (>99.99% accuracy) and capable of identifying known and novel variants of SARS-CoV. Notably, we applied this technology to a field specimen of probable SARS and rapidly deduced its infectious source. We demonstrate that array-based resequencing-by-hybridization is a fast, reliable, and economical alternative to capillary sequencing for obtaining SARS-CoV genomic sequence on a population scale, making this an ideal platform for the global monitoring of SARS-CoV and other small-genome pathogens.
In April 2003, the SARS coronavirus (SARS-CoV), a single-stranded RNA virus, was identified as the pathogen responsible for Severe Acute Respiratory Syndrome (Drosten et al. 2003; Fouchier et al. 2003). Soon after, the consensus genome sequence for SARS-CoV was published (Marra et al. 2003; Ruan et al. 2003). Our studies have shown that the SARS-CoV mutates at a significant rate. In addition to single nucleotide changes, we observed small deletions of5–6 nucleotides in some SARS-CoV isolates. Also, we identified several recurrent sequence variants capable of distinguishing genotypes linked to strains originating in different geographical regions (Ruan et al. 2003). As mutations can alter virulence and therapeutic response ofviral pathogens, it is important to develop methods for the rapid monitoring of genetic changes in the SARS-CoV in human populations. Moreover, the rapid detection ofgenetic variants in newly confirmed SARS cases could also provide important clues to the geographic origins of infection, thus facilitating contact tracing.
Sequencing of viral genomes has historically used standard dye termination technologies. Recently, sequencing by hybridization to oligonucleotides has been described (Drmanac et al. 1992; Maskos and Southern 1992a; Southern et al. 1992; Schena et al. 1995) and commercialized (Pease et al. 1994; Nuwaysir et al. 2002). These methods have been adapted for studies of genetic diversity in disease, genes, and between species (Drmanac et al. 1998; Hacia et al. 1998; Wang et al. 1998; Hacia 1999; Vahey et al. 1999; Brenner et al. 2000; Drmanac and Drmanac 2001; Fan et al. 2002). Despite these advances, several limitations temper the enthusiasm for sequencing by hybridization, including difficulties in detection ofdeletions and cost considerations that significantly restrict array modifications and reformatting for optimization (Cutler et al. 2001; Drmanac et al. 2002; Nuwaysir et al. 2002). We have developed a DNA resequencing array containing 383,102 in situ synthesized oligonucleotide probes capable of interrogating the entire 29.7-kb SARS-CoV genome. This array can detect single-base sequence variations with respect to the consensus SARS-CoV sequence, and all known deletions and insertions reported previously in 14 SARS-CoV isolates. As the array synthesis process is maskless, it is highly flexible, allowing any new sequence variation to be rapidly included on redesigned arrays within 2 d of discovery without additional manufacturing costs.
To validate this sequencing platform, we resequenced the complete SARS-CoV genomes from Vero cell culture isolates of 12 patients and directly from four patient tissue samples. We found that the sequence obtained from the array is highly reproducible and accurate (>99.99% accuracy), and capable ofidentifying known and novel variants ofSARS-CoV. We show that the approach presented here is ideally suited for the rapid and cost-effective genotyping of SARS-CoV variants on a population scale. Finally, we applied this technology to a field specimen of probable SARS (SIN0409) and deduced its infectious source.
RESULTS
Resequencing Array Design
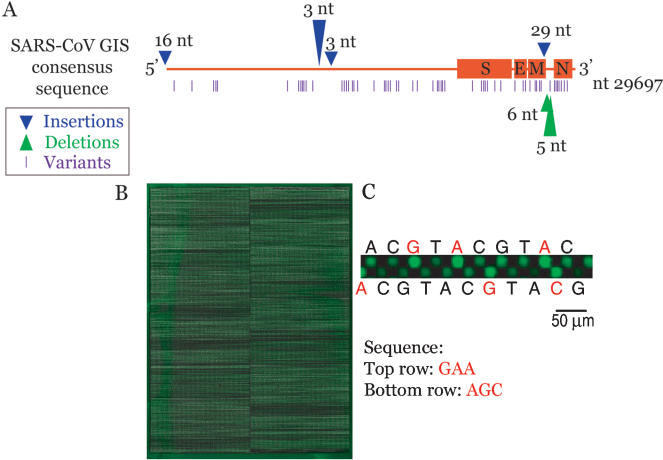
We constructed a full-genome SARS-CoV resequencing array on the basis of a consensus sequence derived from the full-length reads ofviral isolates SIN2500, SIN2677, SIN2679, SIN2748, and SIN2774 sequenced at the Genome Institute ofSingapore (Ruan et al. 2003). Additionally, we designed the array to detect all known insertions and deletions found in 14 SARS isolates published at the NCBI nucleotide database (http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi) as of April 24, 2003. These include a 6-bp deletion in SIN2677 at position 27782, a 5-bp deletion in SIN2748 at position 27810, two single base insertions in the GZ01 isolate at positions 11915, and 12021, and a 29-bp insertion in GZ01 at position 27882. Two extended 5′ ends were also included, 33 bp 5′ ofthe GIS consensus taken from the GZ01 isolate, and 26 bp taken from the Urbani isolate (Fig. 1A).
Figure 1.
SARS Resequencing array. (A) Diagram of the different sequence variants that can be detected by the array. Specific probes were designed to screen for previously published insertion and deletion sequences. (B) Resequencing array hybridized with Cy-3-labeledSARS-CoV cDNA. (C) Close-up view of oligonucleotide probes synthesized on the array. The four possible nucleotides for each position are synthesized adjacent to each other. SARS cDNA bound to perfect-match (PM) probes (in red) fluoresce with higher intensity than those bound to mismatch (MM) probes (in black).
Array-based resequencing depends on the differential hybridization of genomic fragments to short perfect-match (PM) and mismatch (MM) oligonucleotides. On our array platform, each nucleotide to be queried is located at the 15th position ofa PM 29-mer oligonucleotide. For each PM oligo, probes representing the three possible mismatch nucleotides at the 15th position were also synthesized on the array. The differences in hybridization signal intensities between sequences that bind strongly to the PM oligo and those that bind poorly to the corresponding MM oligos make it possible to discern the correct base at a given sequence position (Fig. 1B,C). Our array contains probes to resequence all but 14 bases on either end ofthe 29.7-kb viral genome in replicate fashion. As the viral target in the hybridization reaction is double-stranded cDNA, the array contains probes to interrogate both strands ofthe SARS-CoV target. Because preliminary data suggest greater sequence polymorphism at the 3′ end ofthe SARS-CoV genome, probes representing the last 18,000 bases are present in duplicate. Thus, each base is queried two to four times in a single hybridization. In all, our arrays contain 383,102 29-mer probes, with up to 4 PM probes per base position to resequence a total of48,887 bases. By hybridizing each sample on two arrays, each base is effectively queried four to eight times, similar to the standard replication level used in traditional ABI capillary sequencing (ACS). The arrays were synthesized using NimbleGen System's Maskless Array Synthesizer (MAS) technology, which uses a computer-controlled digital mirror device for the in situ light-directed synthesis ofoligonucleotides (Fig. 1B; Singh-Gasson et al. 1999; Nuwaysir et al. 2002).
Sequencing of SARS-CoV Strains
SARS-CoV RNA, extracted from each of 12 Vero E cell culture isolates and four patient tissue samples, was amplified by RT–PCR using optimized primers to generate 15 or 16 ∼2–3 kb viral genome fragments that were labeled and hybridized to the array (see Methods and Fig. 1B). Custom software was used to extract signal intensities and to compile the full genome sequence for each sample. Ofnote, genome sequences corresponding to the PCR primers were systematically censored in our analysis, as sequence variations in these regions are potentially masked by the invariant primer sequence. The number ofcensored bases ranged from 900 to 960, depending on the number of primer pairs used to amplify the sample. Conventional ABI capillary sequencing (ACS) was performed simultaneously on selected samples to allow direct sequence comparisons.
Sequence Concordance, Discordance, and Ambiguous Calls
Sequence calls were made by statistical analysis ofthe hybridization intensities and combining data from both strands using a customized version ofABACUS run at its default thresholds (Cutler et al. 2001). For statistical confidence, we hybridized each sample onto two replicate arrays. A mean call rate (i.e., rate of confident base calls) of 99.04% ± 0.69% was achieved with duplicate hybridizations (Table 1). We found that little improvement ofthe call rate could be gained with more replication. For example, Tissue 1 was hybridized on a total ofsix arrays and had a call rate of 99.24% for the first two arrays, 99.79% for four replicate arrays, and 99.82% for six replicate arrays for a maximum gain ofonly 0.58%. Using the ACS sequence as the gold standard for comparison, we found that our resequencing array achieves an accuracy rate >99.99% for called bases. This corresponds to an average ofonly 3.8 discordant calls per 100,000 bases sequenced. Notably, this is comparable with the average error rate found in the highest quality sequence obtained by ACS using the Phred algorithm (2.2 errors per 100,000 bases; Ewing et al. 1998; Richterich 1998). The precise causes ofthese discordant calls are unclear. Whereas their occurrence appears random with respect to samples and genomic position, they are reproducible within sample replicates. For example, Tissue 1 (1–1, 1–2, and 1–3; Table 1) contains one discordant call that is reproducible in 3/3 replicates. Using primer extension assay coupled with mass spectroscopy (Sequenom), we determined that this discordant call was the result of differential detection of coinfecting strains, where ACS called the correct base ofone strain, and the resequencing chip called the correct base ofthe other (J.J. Liu, pers. commun.). This phenomenon, although having a small impact on our results, warrants further investigation.
Table 1.
Call Rate and Accuracy of SARS Resequencing Array
| Array sequence | Discordant calls | Ambiguous calls (Ns) | Call rate | Accuracy |
|---|---|---|---|---|
| SIN2500 | 3 | 495 | 98.33% | 99.989% |
| SIN2677 | 4 | 179 | 99.40% | 99.986% |
| SIN2679 | 0 | 138 | 99.53% | 100% |
| SIN2748 | 2 | 223 | 99.22% | 99.993% |
| Vero isolate 1 | 1 | 230 | 99.02% | 99.997% |
| Vero isolate 2 | 1 | 328 | 98.89% | 99.997% |
| Vero isolate 3 | 4 | 183 | 99.38% | 99.986% |
| Vero isolate 4 | 0 | 198 | 99.33% | 100% |
| Vero isolate 5 | 0 | 218 | 99.36% | 100% |
| Vero isolate 6 | 0 | 210 | 99.29% | 100% |
| Vero isolate 7 | 0 | 307 | 98.96% | 100% |
| Vero isolate 8 | 0 | 220 | 99.26% | 100% |
| Tissue 1-1 | 1 | 227 | 99.24% | 99.993% |
| Tissue 1-2 | 1 | 982 | 96.69% | 99.997% |
| Tissue 1-3 | 2 | 120 | 99.60% | 99.997% |
| Tissue 2 | 0 | 278 | 99.07% | 100% |
Discordant calls differ between array and ACS. Ambiguous calls refer to bases that lack sufficient information for high-confidence base assignment. Call rate is the percentage of genome sequence with high-confidence base calls. Accuracy is the percentage of correctly called bases (as determined by ACS) over the total number of bases called (excluding ambiguous calls). These results are based on duplicate hybridizations. Tissue 1 was hybridized on three pairs of arrays. The data for tissues 3 and 4 are not shown as ACS sequence is not available.
Approximately 1% ofbases (i.e., mean = 284 bases per genome) were ambiguous calls (N calls). A base position was scored as N ifthe ABACUS algorithm could not make a confident call, or when different calls were made for the same position on replicate arrays. We found that the ambiguous calls were often reproducible (∼51% occurred in two or more samples) and frequently occurred in runs oftwo or more Ns, suggestive ofgenomic regions with poor hybridization characteristics (Fig. 2).
Figure 2.
Distribution and frequency of ambiguous calls across the SARS-CoV genome. We observed N calls at a total of 1148 bases in this study, of which 580 occurred in more than one sample.
Poor Annealing Characteristics Lead to Ambiguous Calls
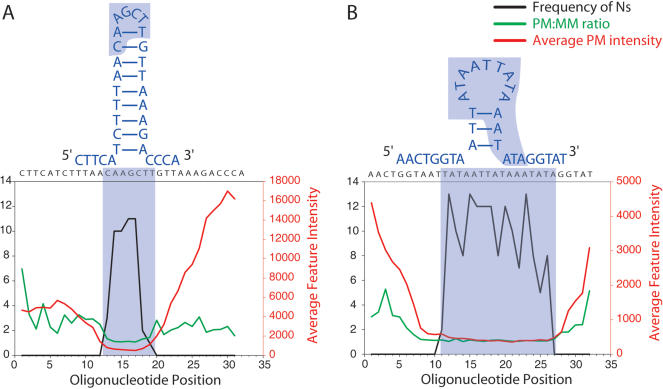
Ambiguous calls result from a lack of discrimination between the PM and MM oligo probes and/or discordance between strand calls. To determine the underlying cause ofthese N calls, we examined the extent to which they could be explained by annealing artifacts. We found that lack of discrimination between PM and MM probes were, to a large extent, caused by suboptimal probe melting temperatures. Whereas the actual Tm ofprobes attached to the surface is difficult to calculate, it is directly related to the G/C content ofeach probe. Probes with G/C content <20% or >50% had relatively poor mismatch discrimination (Fig. 3). This led to a corresponding lower call rate, particularly for probes with <20% G/C. We also observed a markedly reduced average feature intensity for probes with <20% G/C, with maximal average intensities for probes with >50% G/C. Together, these findings support the view that probes with low G/C content tend to hybridize weakly and produce insufficient signals for base calling, whereas probes with high G/C content hybridize strongly, such that a single mismatch is less likely to significantly destabilize hybridization (Fig. 3; see Methods section Data Extraction and Analysis).
Figure 3.
Stratification of probes according to %G/C and assessment of probe performance. All PM probes were binned according to %G/C, and average PM/MM ratios, call rates, and average feature intensities were calculated. G/C content <20% or >50% leads to lowest PM/MM ratios, resulting in increased rate of ambiguous calls.
We also found that probe secondary structure contributes to the ambiguous calls. In particular, probes containing regions of self-homology predicted to form stem-loop structures yielded insufficient signals for base-call discrimination. For example, we observed a string of3–5 N calls beginning at position 25955. The probe-length sequence at this position is palindromic, predicted by GeneRunner software to have a 9-bp stem and 4-bp loop (Fig. 4A). The largest recurrent run ofNs spanned 12–16 bases, and is centered around position 22785. Examination ofthe sequence in this region revealed an AT-rich region resulting in a 3-bp stem and 9-bp loop and, concurrently, had a G/C content ofonly 17% (Fig. 4B). In both cases, the frequency of ambiguous calls peaked within the predicted loop structure and coincided with low signal intensities and low PM:MM ratios. Together, we estimate that the majority ofthe recurrent N calls on a given array can be attributed to Tm (∼42%) and secondary structure (∼15%) phenomena.
Figure 4.
Effects of secondary structure on probe annealing and ambiguous calls. The most stable structure as predicted using GeneRunner software, is illustrated for two sequences with recurrent Ns; (A) bases 25953–25959, (B) bases 22781–22796. In both cases, the frequency of ambiguous calls peaks at the bases within the predicted loop structure.
Strategies being investigated to obviate these problems include (1) altering probe lengths in areas with suboptimal G/C content to reduce Tm artifacts (Southern et al. 1999), (2) modifying hybridization buffers and conditions to minimize the impact ofsecondary structure (Maskos and Southern 1992b; Nguyen and Southern 2000), and (3) improving algorithms to better discern heterozygous base calls.
Strain Determination and Identification of New Strains
To determine the impact ofambiguous calls on our ability to discern viral strains, we resequenced, by array, four SARS-CoV strains (SIN2500, SIN2677, SIN2679, and SIN2748) that were sequenced previously by ACS (Ruan et al. 2003). BLASTN analysis ofeach ofthese sequences against a database of50 published SARS-CoV genomes (which includes the ACS sequences for these four isolates) correctly identified (by highest bit score) the cognate ACS sequence (data not shown). Notably, the arrays also correctly identified the 6-bp deletion and 5-bp deletion found in SIN2677 and SIN2748, respectively (Suppl. Fig. 1 available online at www.genome.org).
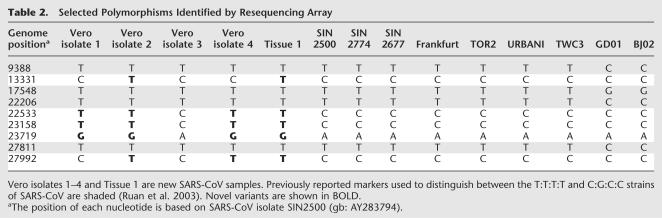
To assess the ability ofthe arrays to identify single-base variants, we compared polymorphisms found within all the SARS-CoV samples sequenced here with those found in the published data. Table 2 shows examples ofknown and novel polymorphisms identified by the arrays in five previously unsequenced samples. Subsequent ACS and Sequenom analysis confirmed the accuracy of these base calls, thus validating the ability ofthe chip to correctly identify known and novel sequence polymorphisms.
Table 2.
Selected Polymorphisms Identified by Resequencing Array
Sequence Analysis of a Field Specimen
During the writing ofthis manuscript, a lone SARS case (SIN0409) emerged in Singapore. The infected individual was a graduate student working in an infectious disease laboratory that had cultured SARS-CoV (P.L. Lim, A. Kurup, G. Gopalakrishna, K.P. Chan, C.W. Wong, L.C. Ng, S. Thoe, L. Oon, X. Bai, L.W. Stanton, et al., in prep.). Sequence analysis using the resequencing array revealed that SIN0409 was most similar to SIN2774, a SARS-CoV strain used frequently by research laboratories in Singapore. The major difference between these two samples, however, was a novel 47-bp deletion between bases 27744 and 27790 in SIN0409. Subsequent resequencing ofa vial offrozen virus, SINWNV, revealed identity with SIN0409 at the 13 signature genome positions used for SARS-CoV strain determination, including the 47-bp deletion, which was not observed in any other SARS-CoV strain sequenced to date. Thus, we concluded that the student was infected by a laboratory strain of SARS-CoV.
DISCUSSION
Efficacy of the SARS-CoV Resequencing Array
The SARS-CoV resequencing chip described here repeatedly called >99% ofthe bases and yielded highly accurate sequence (>99.99%). Of14 samples sequenced by both our resequencing array and ACS, we detected an average of1.21 discordant calls and 284 ambiguous calls per genome, ofwhich ∼51% were reproducible, and therefore predictable. In experimental samples, the array correctly identified known and novel SARS-CoV sequence variants. Under field conditions, we amplified SARS-CoV from the sputum samples of a graduate student who contracted SARS in September, 2003. Within 3 d, we were able to obtain ∼90% ofthe genome sequence using the resequencing array. With this sequence, we were able to rapidly and accurately deduce the infectious source; the student was infected by the same strain ofSARS-CoV that had contaminated his laboratory samples. These results demonstrate the efficacy and applicability ofthis platform in tracking the genetic diversity ofSARS-CoV, facilitating contact tracing and identifying the infectious source.
Resequencing Versus ACS
Capillary sequencing, unlike resequencing by hybridization, allows the direct determination ofDNA sequence. Whereas resequencing arrays are most efficacious in detecting single nucleotide changes, novel deletions and insertions can often be inferred. Clues to the presence of large (>5 bp) insertions and deletions are detected with the resequencing array by a consecutive series ofambiguous calls in a region that normally provides good sequence calls. For example, in the SIN0409 SARS case, the resequencing array identified, by virtue of a long string ofNs, the presence ofa novel ∼50-bp deletion common only to the sequences ofthe laboratory contaminant and the patient. This deletion was later confirmed by ACS and found to be 47 bp.
The advantages ofthe resequencing arrays over ACS are most apparent when multiple genomes need to be sequenced rapidly, as in the case ofan epidemiologic study ofviral genetic diversity, or for identification of infectious origin. For both methods, a time-limiting step is the amplification of all genomic PCR products from the isolated RNA. However, optimization of primers by high-specificity sequence design and empirical testing can improve PCR success rates and reduce cDNA preparation time to about 1–2 d. We have found that the optimal amount ofcDNA required for array hybridizations is ∼200 ng (i.e., 100 ng for each of two arrays). This compares favorably with the 12 μg ofcDNA required by capillary sequencing, and translates into less PCR and cDNA preparation time. Because the resequencing process relies on a previously determined consensus sequence, full-sequence attainment is achieved by simple base-call prediction rather than the more time-consuming, and computationally intensive process ofsequence assembly required ofconventional ACS. We project that one technician using the resequencing array format can process up to 50 cDNA samples (i.e., 100 arrays) in 5 d, whereas the same sample volume would require 50 days and two technicians by conventional ACS.
We find that the reagent costs of sequencing a genome the size ofSARS-CoV are approximately equal for both methods. However, the only specialized equipment required for array hybridization is a 5-μm-resolution scanner (identical to that used for 2-color DNA microarrays), and an inexpensive hybridization apparatus. Thus, the initial capital and continuing maintenance costs for the resequencing array are significantly reduced compared with that required for a capillary sequencing machine. Furthermore, the resequencing platform does not require any specialized training for sample preparation and sequence acquisition. Taken together, the resequencing array is able to obtain sequence information more rapidly, and has significantly less manpower needs and infrastructure costs than ACS. Thus, the resequencing platform is ideal for investigating viral sequence variants in a parallel and whole-genome fashion under field conditions.
Efficacy of Direct Sequencing From Patient Samples
The isolation and identification of viral pathogens from clinical samples has historically required some cell culture passaging to obtain sufficient virus for molecular manipulation. In the case of the SARS-CoV, Vero E6 (monkey kidney) cells were identified early in the epidemic as an effective vehicle for viral amplification capable ofachieving titers in the range of108 viral copies/μL. From a sequencing standpoint, this proved fortuitous, as the ability to sequence the 29.7-kb genome in rapid fashion depended on high-purity viral isolates. The drawback to this methodology, however, is that often more than 1 wk may be required to propagate and purify virus. Coupled with an additional week's time to prepare and sequence the genome, delays the identification ofthe SARS-CoV strain, which would allow for tracing ofthe infectious source. Here, we show that not only can the resequencing chip reduce the amount oftime required to obtain full sequence, but that we can bypass cell-culture passaging and obtain reliable sequence data directly from patient tissue samples. This latter innovation was achieved in large part by a systematic primer design strategy to select primers based on not only optimal annealing characteristics, but also low predicted cross-homology with human transcripts—that is, the same criteria applied to the selection ofoligo probes for expression microarrays. Through empirical testing ofselect primers, we derived the RT, first-round PCR, and nested PCR conditions necessary to amplify the entire genome from tissue specimens with approximate viral titers ofonly 2 × 105 copies/μL. Notably, in the SARS case SIN0409, we amplified the full genome sequence (with the exception of ∼2 kb) from a sputum sample and positively identified the viral strain within 3 d. In our validation studies, we also observed a consistent number ofsingle-base variants when comparing viral sequence obtained from patient tissues with sequence following amplification in Vero E cell culture (V.B. Vega, C. Lee, Y. Ruan, J.J. Liu, P. Kolatkar, E.T. Liu, L.W. Stanton, P. Long, in prep.). Thus, direct SARS-CoV sequence from primary tissues would overcome the confounding artificial mutations generated during Vero cell passage.
Conclusion
Here, we demonstrate that chip-based resequencing by hybridization is a fast and reliable method for acquiring small-genome sequences in parallel. PCR not withstanding, we were able to hybridize and sequence the SARS-CoV genomes from each of 16 clinical/laboratory samples in as little as 3 d, resulting in >475 kb of finalized sequence, from which we could correctly identify known and novel sequence variants. Novel deletions or insertions encountered by the resequencing array can be confirmed rapidly by performing capillary sequencing only on that small PCR fragment giving ambiguous calls, rather than on the entire genome. Because probes are directly synthesized on the array with great design flexibility, additional probes can be added to screen for any new deletions or insertions identified by de novo sequencing. A new array, capable of screening for these features, can be produced within 48 h, given the facile nature of the maskless array manufacturing process. In the case of pathogens in which strain coinfection is common, key SNP regions that distinguish strains could be specifically assessed by sequence technologies that can discriminate and quantitate heterozygous sequences, such as the mass-spectometry-based technology.
We conclude that it is now possible to resequence large numbers ofSARS-CoV isolates in a rapid, highly parallel, and accurate manner should the virus resurface again. Entire infected populations could be monitored effectively, even in regions that do not have high-throughput sequencing equipment. The relatively low cost ofthe resequencing array in both monetary and manpower terms, coupled with its rapid sequence turn-around time, makes this an ideal platform for the global monitoring of any small-genome pathogen. Whereas we have used this technology for resequencing the SARS-CoV genome, the iterative design flexibility and high density of probes makes this a highly attractive platform for gene expression analysis, comparative genomic hybridization, SNP discovery, and other genomic applications.
METHODS
Amplification of SARS Viral RNA
A detailed protocol is published on our Web site. Briefly, total RNA was extracted from patient lung, sputum, or fecal samples, or from Vero E cultured cells innoculated with SARS-CoV RNA. RNA was reverse-transcribed into cDNA, using 13 30-mer primers and Powerscript enzyme (Clontech). Double-stranded DNA was synthesized from this template as described previously. Tissue samples were amplified using a nested-PCR strategy (see Primer Design below). The sequences ofthese primers are listed in Supplemental Table 1. Samples from Vero E cell isolates were amplified using 30 primer pairs, as described previously (Ruan et al. 2003).
PCR Primer Design for SARS Tissue Samples
A consensus sequence (GIS consensus) for SARS Co-V was derived from the complete genome sequences of SARS Co-V isolates SIN2500, SIN2677, SIN2679, SIN2748, and SIN2774; 30-mer oligonucleotides tiling across the entire GIS consensus was generated in both strands. Using a proprietary algorithm (NimbleGen Systems), the primers were scored and ranked for uniqueness against the human transcriptome, secondary structure, and other characteristics. Upon receipt ofthe probe list, we selected 13 primers from the reverse strand with the highest possible scores for the Reverse Transcription reaction. Another 30 primers were selected to produce 15 overlapping PCR products spanning the entire SARS Co-V genome. Within each PCR product, we also selected at least one primer pair for nested PCR. For samples with low viral titer (<2 × 105 copies/μL), in addition to the process above, we searched for PCR primer pairs using Gene Runner 3.05 (http://www.generunner.com). The primer pairs found were then ranked by their ability to hybridize to perfectly matched sequence, using data from SARS resequencing arrays. Highly ranked primer pairs were then selected for nested PCR amplification ofthese low viral titer samples.
Fragmentation and Labeling of DNA
For each sample, PCR product fragments were pooled at an equimolar ratio. A total of100 ng ofpooled DNA was digested at 37°C for 2 min with 0.025 U DNase I (Invitrogen) and 10× One-Phor-All buffer (Amersham Biosciences) in a total volume of 20 μL. DNase I was inactivated by incubation at 97°C for 15 min. Sample was end-labeled with 1 μL Biotin-N6 ddATP (Perkin Elmer) and 25 U Terminal Deoxynucleotidyl Transferase (Promega) at 37°C for 90 min, and terminal transferase was inactivated by incubation at 97°C for 15 min.
Microarrays, Hybridization, and Staining
The arrays were synthesized as described previously (Singh-Gasson et al. 1999; Nuwaysir et al. 2002). A detailed protocol of the hybridization and staining procedure is published on the supplementary Web site. Briefly, the resequencing arrays were hybridized with biotinylated DNA in the presence ofresequencing hybridization buffer [100 mM MES, 2.5 M tetramethylammoniumchloride, 0.01% (v/v) Tween-20]. Before application to the array, the array was prehybridized with hybridization buffer and samples were heated to 95°C for 5 min, heated to 45°C, and centrifuged for 5 min at >12000g. After application of DNA, arrays were placed in a customized hybridization chamber and incubated at 45°C for 14–16 h in a rotisserie oven. The next morning, arrays were washed with nonstringent wash buffer [6× SSPE, 0.01% (v/v) Tween-20], followed by six 5-min washes in stringent wash buffer [100 mM MES, 0.1 M NaCl, 0.01% (v/v) Tween-20] at 50°C. The arrays were stained with a solution containing Cy3-Streptavidin conjugate (Amersham Biosciences) for 10 min, and washed again with nonstringent wash buffer. The Cy3 signal was amplified by secondary labeling of the DNA with biotinylated goat anti-streptavidin (Vector Laboratories). The secondary antibody was washed off with nonstringent wash buffer, and the array restained with the Cy3-Streptavidin solution. Finally, the stain solution was removed, and the array was washed in non-stringent wash buffer, followed by a 30-sec wash in NimbleGen Final Wash Buffer (NimbleGen Systems, Inc.). The arrays were dunked five times in 0.2× SSC, five times in ice-cold ethanol, and immediately dried down by centrifugation.
Data Extraction and Analysis
Microarrays were scanned at 5 μm resolution using the Genepix 4000b scanner (Axon Instruments). The image was interpolated and scaled up 2.5× in size using NIH Image software (http://rsb.info.nih.gov/nih-image/). Each feature on the array consists of49 pixels, and pixel intensities were extracted using NimbleScan Software (NimbleGen Systems). Sequence calls were made by statistical analysis ofthe hybridization intensities combining data from both strands using a customized version of ABACUS run at its default thresholds (Cutler et al. 2001). Briefly, ABACUS makes base calls on the basis of the differential hybridization of genomic fragments to short perfect-match (PM) and mismatch (MM) oligonucleotides. In making base calls, ABACUS uses both the mean intensity for a feature as well as the variance in the intensity ofthe feature. Generally, large PM:MM intensity ratios are readily called. However, very small PM:MM ratios can be detected ifthe variance in intensity ofthose features is small. On the other hand, very large differences, when compromised by high variance, can result in no calls. When run at the default thresholds, a typical base call usually involves an observed mean intensity (on the PM oligo) more than 10 standard errors higher than the next brightest MM oligo. A single set ofthresholds were used for this study. As samples were hybridized to at least two SARS Resequencing arrays, data from multiple arrays were integrated by combining called bases from multiple arrays. Discordant calls between arrays were designated as N. This analysis generated complete genome sequences in FASTA format and tabulated the polymorphisms identified. Visualization and multiple sequence alignment were performed using Gene Runner 3.05. BLAST analysis was performed on an internal Unix server, using a database of50 SARS Co-V sequences downloaded from NCBI Taxonomy database on August 7, 2003 (Sequences listed in Supplemental Table 2; Altschul et al. 1997).
Accession Numbers
Accession numbers have been published previously (Ruan et al. 2003) and are listed in Supplemental Table 2. Accession numbers for the newer samples which were resequenced here will be submitted and published on our Web site, once they have been confirmed and assembled by ABI capillary sequencing.
Acknowledgments
We thank Kun Yang and Melissa Riederer for technical assistance, Martin Hibberd, Yijun Ruan, Jianjun Liu, Chia Lin Wei, Patrick Ng, and Lisa Ng for providing SARS-CoV RNA and/or helpful discussion.
The publication costs ofthis article were defrayed in part by payment ofpage charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2141004.
Footnotes
[Supplemental material is available online at www.genome.org and http://www.gis.a-star.edu.sg/homepage/toolssup.jsp.]
References
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation ofprotein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner, S., Johnson, M., Bridgham, J., Golda, G., Lloyd, D.H., Johnson, D., Luo, S., McCurdy, S., Foy, M., Ewan, M., et al. 2000. Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat. Biotechnol. 18: 630–634. [DOI] [PubMed] [Google Scholar]
- Cutler, D.J., Zwick, M.E., Carrasquillo, M.M., Yohn, C.T., Tobin, K.P., Kashuk, C., Mathews, D.J., Shah, N.A., Eichler, E.E., Warrington, J.A., et al. 2001. High-throughput variation detection and genotyping using microarrays. Genome Res. 11: 1913–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drmanac, R. and Drmanac, S. 2001. Sequencing by hybridization arrays. Methods Mol. Biol. 170: 39–51. [DOI] [PubMed] [Google Scholar]
- Drmanac, R., Drmanac, S., Labat, I., Crkvenjakov, R., Vicentic, A., and Gemmell, A. 1992. Sequencing by hybridization: Towards an automated sequencing ofone million M13 clones arrayed on membranes. Electrophoresis 13: 566–573. [DOI] [PubMed] [Google Scholar]
- Drmanac, S., Kita, D., Labat, I., Hauser, B., Schmidt, C., Burczak, J.D., and Drmanac, R. 1998. Accurate sequencing by hybridization for DNA diagnostics and individual genomics. Nat. Biotechnol. 16: 54–58. [DOI] [PubMed] [Google Scholar]
- Drmanac, R., Drmanac, S., Chui, G., Diaz, R., Hou, A., Jin, H., Jin, P., Kwon, S., Lacy, S., Moeur, B., et al. 2002. Sequencing by hybridization (SBH): Advantages, achievements, and opportunities. Adv. Biochem. Eng. Biotechnol. 77: 75–101. [DOI] [PubMed] [Google Scholar]
- Drosten, C., Gunther, S., Preiser, W., van der Werf, S., Brodt, H.R., Becker, S., Rabenau, H., Panning, M., Kolesnikova, L., Fouchier, R.A., et al. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. New Engl. J. Med. 348: 1967–1976. [DOI] [PubMed] [Google Scholar]
- Ewing, B., Hillier, L., Wendl, M.C., and Green, P. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8: 175–185. [DOI] [PubMed] [Google Scholar]
- Fan, J.B., Gehl, D., Hsie, L., Shen, N., Lindblad-Toh, K., Laviolette, J.P., Robinson, E., Lipshutz, R., Wang, D., Hudson, T.J., et al. 2002. Assessing DNA sequence variations in human ESTs in a phylogenetic context using high-density oligonucleotide arrays. Genomics 80: 351–360. [DOI] [PubMed] [Google Scholar]
- Fouchier, R.A., Kuiken, T., Schutten, M., van Amerongen, G., van Doornum, G.J., van den Hoogen, B.G., Peiris, M., Lim, W., Stohr, K., and Osterhaus, A.D. 2003. Aetiology: Koch's postulates fulfilled for SARS virus. Nature 423: 240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacia, J.G. 1999. Resequencing and mutational analysis using oligonucleotide microarrays. Nat. Genet. 21: 42–47. [DOI] [PubMed] [Google Scholar]
- Hacia, J.G., Makalowski, W., Edgemon, K., Erdos, M.R., Robbins, C.M., Fodor, S.P., Brody, L.C., and Collins, F.S. 1998. Evolutionary sequence comparisons using high-density oligonucleotide arrays. Nat. Genet. 18: 155–158. [DOI] [PubMed] [Google Scholar]
- Marra, M.A., Jones, S.J., Astell, C.R., Holt, R.A., Brooks-Wilson, A., Butterfield, Y.S., Khattra, J., Asano, J.K., Barber, S.A., Chan, S.Y., et al. 2003. The Genome sequence ofthe SARS-associated coronavirus. Science 300: 1399–1404. [DOI] [PubMed] [Google Scholar]
- Maskos, U. and Southern, E.M. 1992a. Oligonucleotide hybridizations on glass supports: A novel linker for oligonucleotide synthesis and hybridization properties ofoligonucleotides synthesised in situ. Nucleic Acids Res. 20: 1679–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maskos, U. and Southern, E.M. 1992b. Parallel analysis ofoligodeoxyribonucleotide (oligonucleotide) interactions. I. Analysis of factors influencing oligonucleotide duplex formation. Nucleic Acids Res. 20: 1675–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, H.K. and Southern, E.M. 2000. Minimizing the secondary structure ofDNA targets by incorporation ofa modified deoxynucleoside: Implications for nucleic acid analysis by hybridization. Nucleic Acids Res. 28: 3904–3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuwaysir, E.F., Huang, W., Albert, T.J., Singh, J., Nuwaysir, K., Pitas, A., Richmond, T., Gorski, T., Berg, J.P., Ballin, J., et al. 2002. Gene expression analysis using oligonucleotide arrays produced by maskless photolithography. Genome Res. 12: 1749–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pease, A.C., Solas, D., Sullivan, E.J., Cronin, M.T., Holmes, C.P., and Fodor, S.P. 1994. Light-generated oligonucleotide arrays for rapid DNA sequence analysis. Proc. Natl. Acad. Sci. 91: 5022–5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richterich, P. 1998. Estimation oferrors in “raw” DNA sequences: A validation study. Genome Res. 8: 251–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan, Y.J., Wei, C.L., Ee, A.L., Vega, V.B., Thoreau, H., Su, S.T., Chia, J.M., Ng, P., Chiu, K.P., Lim, L., et al. 2003. Comparative full-length genome sequence analysis of14 SARS coronavirus isolates and common mutations associated with putative origins ofinfection. Lancet 361: 1779–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schena, M., Shalon, D., Davis, R.W., and Brown, P.O. 1995. Quantitative monitoring ofgene expression patterns with a complementary DNA microarray. Science 270: 467–470. [DOI] [PubMed] [Google Scholar]
- Singh-Gasson, S., Green, R.D., Yue, Y., Nelson, C., Blattner, F., Sussman, M.R., and Cerrina, F. 1999. Maskless fabrication of light-directed oligonucleotide microarrays using a digital micromirror array. Nat. Biotechnol. 17: 974–978. [DOI] [PubMed] [Google Scholar]
- Southern, E., Mir, K., and Shchepinov, M. 1999. Molecular interactions on microarrays. Nat. Genet. 21: 5–9. [DOI] [PubMed] [Google Scholar]
- Southern, E.M., Maskos, U., and Elder, J.K. 1992. Analyzing and comparing nucleic acid sequences by hybridization to arrays of oligonucleotides: Evaluation using experimental models. Genomics 13: 1008–1017. [DOI] [PubMed] [Google Scholar]
- Vahey, M., Nau, M.E., Barrick, S., Cooley, J.D., Sawyer, R., Sleeker, A.A., Vickerman, P., Bloor, S., Larder, B., Michael, N.L., et al. 1999. Performance of the Affymetrix GeneChip HIV PRT 440 platform for antiretroviral drug resistance genotyping ofhuman immunodeficiency virus type 1 clades and viral isolates with length polymorphisms. J. Clin. Microbiol. 37: 2533–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, D.G., Fan, J.-B., Siao, C.-J., Berno, A., Young, P., Sapolsky, R., Ghandour, G., Perkins, N., Winchester, E., Spencer, J., et al. 1998. Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science 280: 1077–1082. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi; NCBI Taxonomy database.
- http://www.generunner.com; GeneRunner v. 3.05 download page.
- http://rsb.info.nih.gov/nih-image/; NIH Image software download page.
- http://www.gis.a-star.edu.sg/homepage/toolssup.jsp; Supplemental information for this study.