Abstract
The Arabidopsis (Arabidopsis thaliana) two-component signaling system, which is composed of sensor histidine kinases, histidine phosphotransfer proteins, and response regulators, mediates the cytokinin response and various other plant responses. We have previously shown that ARABIDOPSIS HISTIDINE KINASE2 (AHK2), AHK3, and cold-inducible type A ARABIDOPSIS RESPONSE REGULATORS (ARRs) play roles in cold signaling. However, the roles of type B ARRs and ARABIDOPSIS HISTIDINE PHOSPHOTRANSFER PROTEINS (AHPs) have not been investigated in cold signaling. Here, we show that ARR1 and AHP2, AHP3, and AHP5 play positive roles in the cold-inducible expression of type A ARRs. arr1 mutants showed greatly reduced cold-responsive expression of type A ARRs compared with the wild type, whereas ARR1-overexpressing Arabidopsis exhibited the hypersensitive cold response of type A ARRs as well as enhanced freezing tolerance with cytokinin, suggesting that ARR1 functions as a positive factor of cold signaling. Transgenic Arabidopsis expressing ARR1ΔDDK:GR lacking the amino-terminal receiver domain showed wild-type expression levels of type A ARRs in response to cold, indicating that the signal receiver domain of ARR1 might be important for cold-responsive expression of type A ARRs. ahp2 ahp3 ahp5 triple mutations greatly reduced type A ARR expression in response to cold, whereas the single or double ahp mutants displayed wild-type levels of ARR expression, suggesting that AHP2, AHP3, and AHP5 are redundantly involved in cold signaling. Taken together, these results suggest that ARR1 mediates cold signal via AHP2, AHP3, or AHP5 from AHK2 and AHK3 to express type A ARRs. We further identified a cold transcriptome affected by ahk2 ahk3 mutations by microarray analysis, revealing a new cold-responsive gene network regulated downstream of AHK2 and AHK3.
Two-component systems are utilized in plant responses to hormones as well as to biotic and abiotic environmental stimuli (Schaller, 2000; Hutchison and Kieber, 2002; Hwang et al., 2002, 2012). Two-component systems were originally identified in bacteria and involve His sensor kinases and response regulators in their simplest configuration (Mizuno, 1997; Stock et al., 2000). Cytokinin signaling of Arabidopsis (Arabidopsis thaliana) utilizes a multistep phosphorelay composed of sensor His kinases, His phosphotransfer proteins, and response regulators similar to the two-component signaling system of bacterial and yeast cells (To and Kieber, 2008). The Arabidopsis cytokinin receptor family comprises the three hybrid His kinases ARABIDOPSIS HISTIDINE KINASE2 (AHK2), AHK3, and AHK4/CYTOKININ RESPONSE1/WOODEN LEG1 (Inoue et al., 2001; Suzuki et al., 2001; Ueguchi et al., 2001; Yamada et al., 2001; Kakimoto, 2003). X-ray crystal structure analysis of the AHK4 sensor domain in complex with cytokinins has revealed that cytokinin is bound to the membrane-distal period circadian protein, aryl hydrocarbon receptor nuclear translocator protein, single-minded protein domain of the cyclases, and His kinases-associated sensory extracellular domain (Hothorn et al., 2011). ARABIDOPSIS HISTIDINE PHOSPHOTRANSFER PROTEINS (AHPs) mediate the transfer of phosphoryl groups from AHKs to ARABIDOPSIS RESPONSE REGULATORS (ARRs; Hwang et al., 2002). Five AHPs act as redundant, positive regulators of cytokinin signaling (Hutchison et al., 2006). AHP6 is a pseudophosphotransfer protein that performs an inhibitory function in cytokinin signaling for protoxylem formation (Mähönen et al., 2006a).
Twenty-three ARRs are classified into type A, type B, and type C (Hwang et al., 2002; Lohrmann and Harter, 2002; Gattolin et al., 2006; Horák et al., 2008; Pils and Heyl, 2009). The type B ARRs (ARR1, ARR2, ARR10\x{2013}ARR14, and ARR18\x{2013}ARR21) are transcription factors that contain an N-terminal receiver domain and a large C-terminal region harboring a Myb-like DNA-binding domain and a Gln-rich domain and that function as positive regulators of cytokinin signaling (Mason et al., 2005; Argyros et al., 2008). The middle segments of ARR1 and ARR2 can bind to DNA in a sequence-specific manner in vitro, and their C-terminal halves function as transactivation domains in plant cells (Sakai et al., 2000). Type B ARRs directly activate the expression of type A ARRs (Hwang and Sheen, 2001; Sakai et al., 2001; Mason et al., 2005). Cytokinin-facilitated proteolysis of ARR2 has been suggested to be involved in the attenuation of signaling output in two-component circuitry (Kim et al., 2012). The type A ARRs (ARR3\x{2013}ARR9 and ARR15\x{2013}ARR17) are composed of a receiver domain and a divergent C-terminal extension. The type A ARRs are rapidly and transiently induced by cytokinin treatment, but the type B ARRs are not inducible by cytokinins. The type A ARRs function as partially redundant negative regulators of cytokinin signaling (Kiba et al., 2003; To et al., 2004; Lee et al., 2007). Negative regulation of cytokinin signaling by the type A ARRs involves phosphorylation-dependent interactions (Lee et al., 2007; To et al., 2007), and a subset of the type A ARRs are stabilized in response to cytokinin, in part, via phosphorylation (To et al., 2007). Arabidopsis genome microarray data suggest that ARR7 acts as a transcriptional repressor for a variety of early cytokinin-regulated genes in a phosphorylation-dependent manner (Lee et al., 2007, 2008). The type C ARRs have been recently defined as ARRs (ARR22 and ARR24) that have a domain structure similar to the type A ARRs, but their expression is not induced by cytokinins (Kiba et al., 2004; Horák et al., 2008; Pils and Heyl, 2009). However, the role of the type C ARRs in cytokinin signaling is unclear (Pils and Heyl, 2009).
A variety of studies have suggested that the AHKs play roles not only in organ growth and development (Higuchi et al., 2004; Nishimura et al., 2004; Riefler et al., 2006) but also in stress responses such as abscisic acid (ABA), drought, cold, and high-salinity stress signaling (Tran et al., 2007, 2010; Jeon et al., 2010; Kang et al., 2012). We have previously shown that AHK2 and AHK3 are involved in mediating the cold signal for the expression of a subset of type A ARRs, ARR5, ARR6, ARR7, and ARR15, and a type C ARR, ARR22, and that ARR7 plays a negative regulatory role in cold stress signaling (Jeon et al., 2010). Here, we investigated whether type B ARRs and AHPs are involved in Arabidopsis cold signaling. Our results demonstrate that ARR1 plays a critical role in cold signaling and that AHP2, AHP3, and AHP5 are redundantly involved in cold signaling as positive factors. Microarray analysis of the Arabidopsis cold transcriptome affected by ahk2 ahk3 mutations compared with the wild type suggested that a new cold-regulatory gene network exists downstream of AHK2 and AHK3.
RESULTS
ARR1 Performs a Primary Role as a Positive Factor in Mediating Cold for the Expression of Type A ARRs
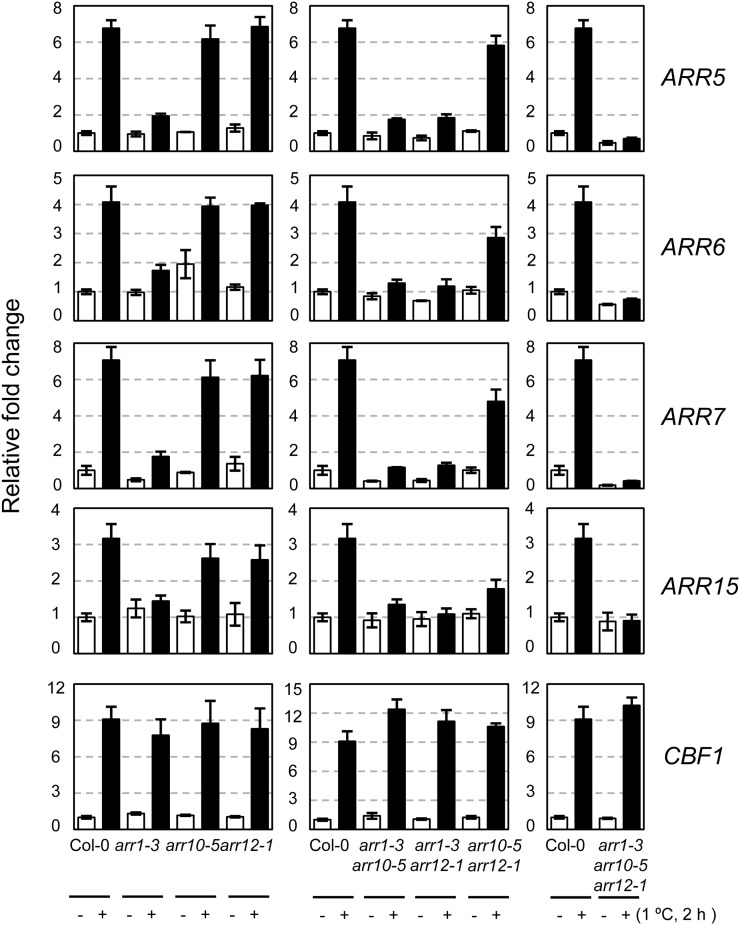
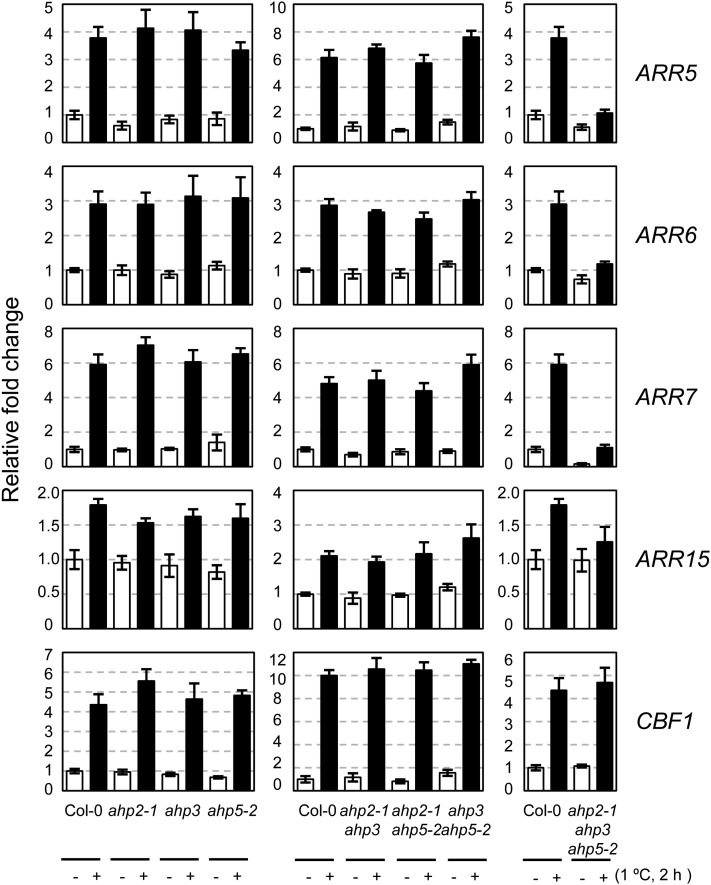
To determine whether type B ARRs are involved in cold-inducible expression of type A ARRs, we analyzed the expression of ARR5, ARR6, ARR7, and ARR15 in type B arr1, arr10, or arr12 single T-DNA insertional mutants as well as in multiple mutants in response to cold by using the quantitative reverse transcription (RT)-PCR method. Plants were treated with cold at the peak expression time of 2 h, as reported previously (Jeon et al., 2010). The arr1 single mutants showed greatly reduced transcript levels of ARR5, ARR6, ARR7, and ARR15 in response to cold compared with the wild type (Fig. 1). The type B arr double mutants, including the arr1 mutation, exhibited greatly reduced levels of these transcripts in response to cold. In contrast, arr10 arr12 double mutants showed relatively unchanged expression levels of ARR5, ARR6, and ARR7 and displayed highly reduced levels of ARR15 expression. Expression of these ARRs in arr1 arr10 arr12 triple mutants was completely unresponsive to cold. These results indicate that ARR1 is primarily involved in mediating cold signal for expressing type A ARRs and that ARR10 and ARR12 play some additive roles in this cold response. We found a similar expression pattern of those cold-inducible type A ARRs in response to cytokinin benzyladenine (BA) in arr1, arr10, and arr12 mutants, although slightly higher expression levels of ARRs were detected in BA-treated arr1 mutants compared with that in cold-treated arr1 mutants (Supplemental Fig. S1).
Figure 1.
Expression of ARR5, ARR6, ARR7, and ARR15 in response to cold in type B arr mutants compared with that in the wild-type plants. Eleven-day-old light-grown seedlings were treated for 2 h at 1°C, and total RNA isolated was subjected to quantitative RT-PCR. CBF1 was employed to verify cold-inducible gene expression. Relative fold changes were plotted after normalization to ACTIN7 RNA. Mean values and se from triplicate biological experiments are plotted.
We next asked if cold induces the expression of the type B ARRs, ARR1, ARR10, and ARR12, thereby resulting in the up-regulation of cold-responsive type A ARRs. A quantitative time-course expression analysis using real-time RT-PCR showed that cold did not significantly induce the expression of ARR1, ARR10, and ARR12 within 2 h and up to 24 h of cold treatment (Supplemental Fig. S2). As we have reproducibly observed significant cold-responsive ARR expression within 30 min and peak expression at 2 h, this result demonstrated that cold-responsive type A ARR expression is not primarily due to the increase in transcript levels of these type B ARRs.
ARR1 Overexpression in Transgenic Arabidopsis Causes Hypersensitive Cold-Responsive Expression of Type A ARRs
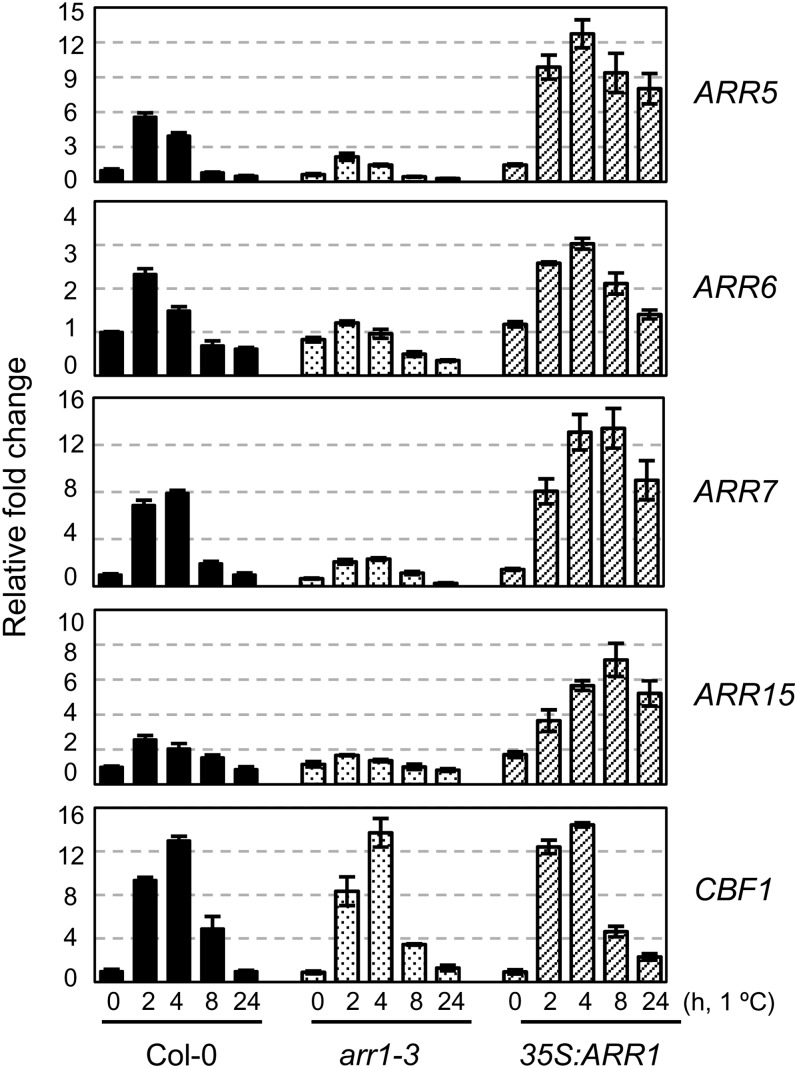
As ARR1 loss-of-function mutation caused a severely reduced cold-inducible expression of type A ARRs, we investigated whether ARR1 overexpression could superinduce the expression of type A ARRs by exposure to cold. We treated arr1 and ARR1-overexpressing transgenic plants with cold for 0, 2, 4, 8, or 24 h and assessed the expression of ARR5, ARR6, ARR7, ARR15, and C-REPEAT/DEHYDRATION-RESPONSIVE ELEMENT BINDING FACTOR1 (CBF1)/DEHYDRATION-RESPONSIVE ELEMENT BINDING FACTOR1B (DREB1B) by quantitative RT-PCR compared with that of the wild-type plants (Fig. 2). ARR1-overexpressing transgenic plants exhibited a hypersensitive response of type A ARRs to cold, whereas arr1 mutants showed reduced cold-responsive expression of type A ARRs. Expression analysis of CBF1, utilized as a cold marker gene, confirmed the effects of cold treatment with regard to the induction of gene expression. These results indicate that ARR1 plays a positive role in regulating type A ARR expression in response to cold.
Figure 2.
Expression of ARR5, ARR6, ARR7, and ARR15 in response to cold in arr1-3 and 35S:ARR1 compared with that in the wild-type plants. Eleven-day-old light-grown seedlings were treated for the indicated times with cold at 1°C. Treatment and analysis of the samples were performed as described in the Figure 1 legend. Black, dotted, and striped bars represent transcript levels from the wild-type plants, arr1-3, and 35S:ARR1 transgenic plants, respectively.
The Signal Receiver Domain of ARR1 Might Be Important in Inducing Type A ARR Expression in Response to Cold
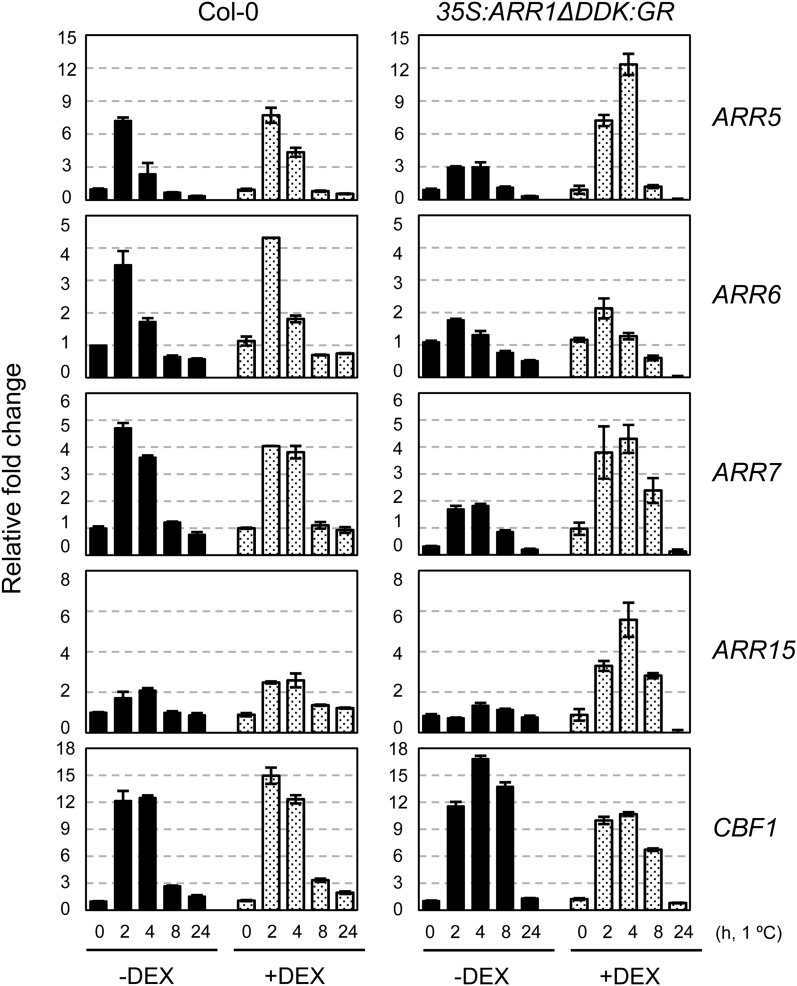
ARR1 is composed of three functional domains, the N-terminal receiver domain, the C-terminal Myb-like DNA-binding domain, and the transactivation domain (Sakai et al., 2000). Under normal growth conditions, ARR1-overexpressing transgenic plants did not show significant phenotypic change due to inhibition of the ARR1 transactivation domain by the signal receiver domain. However, ARR1-overexpressing plants showed cytokinin-related phenotypic changes upon cytokinin treatment, probably by unmasking of the ARR1 transactivation domain through a conformational change in the receiver domain (Sakai et al., 2001). To determine the role of the ARR1 N-terminal signal receiver domain during cold signaling, we analyzed the expression of cold-inducible type A ARRs in transgenic Arabidopsis overexpressing ARR1ΔDDK fused with the glucocorticoid receptor hormone-binding domain (GR; ARR1ΔDDK:GR), but lacking the ARR1 receiver domain, in a dexamethasone (DEX)-inducible manner. Expression of type A ARRs increased following DEX treatment, allowing nuclear transport of constitutively active ARR1ΔDDK, indicating that ARR1ΔDDK in transgenic plants is capable of activating ARR1 target gene expression (Supplemental Fig. S3). The expression profiles of type A ARRs in ARR1ΔDDK plants after cold treatment were obtained with or without DEX and then normalized by removing apparently increased basal expression levels of type A ARRs before cold treatment with DEX in ARR1ΔDDK:GR plants (Fig. 3). Although ARR1ΔDDK:GR plants displayed slightly increased ARR5 and ARR15 transcript levels after 4 h of cold treatment compared with the wild type, we found that ARR1ΔDDK:GR plants displayed an expression pattern of cold-inducible type A ARRs similar to that in the wild type treated with DEX in general. This result indicates that the ARR1 signal receiver domain might be important in type A ARR expression in response to cold; therefore, phosphorelay might be involved in cold signaling via the two-component signaling system. Interestingly, ARR1ΔDDK:GR plants showed reduced expression of type A ARRs in response to cold without DEX compared with that in the wild type. This result might be due to a dominant negative mutation effect of the ARR1ΔDDK:GR proteins in the cytosol, probably by a squelching mechanism.
Figure 3.
Expression levels of ARR5, ARR6, ARR7, and ARR15 after subtracting increased basal expression levels of ARR by DEX in response to cold in 35S:ARR1ΔDDK:GR compared with that in the wild-type plants. Eleven-day-old wild-type (Col-0) seedlings were incubated on MS plates with or without 10 μm DEX for an additional 2 d and then incubated at 1°C for varying periods of time. Treatment and analysis of the samples were performed as described in the Figure 1 legend.
Preincubation of ARR1-Overexpressing Transgenic Plants with Cytokinin Causes Enhanced Freezing Tolerance Compared with That in Untreated Plants
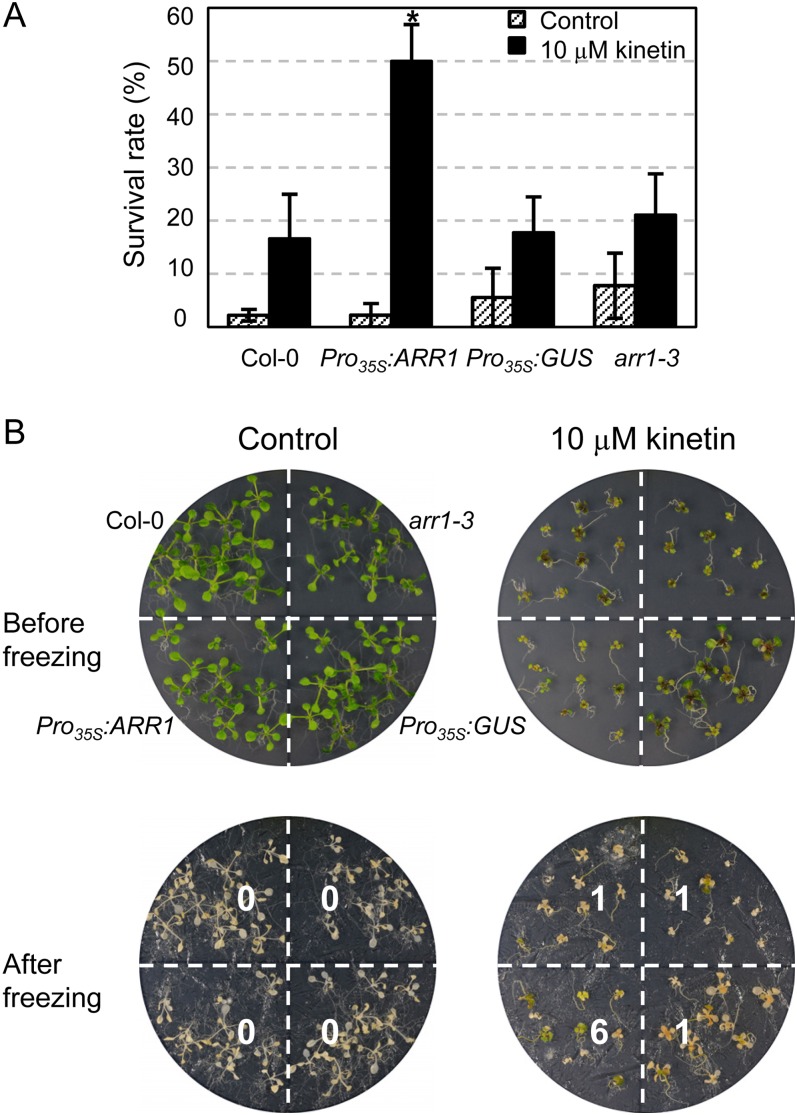
We tested the freezing tolerance of arr1 single mutants and ARR1-overexpressing transgenic plants to demonstrate that ARR1 acts as a regulator of cold stress adaptation response (Supplemental Fig. S4). However, we could not detect a significant change in freezing tolerance of ARR1-overexpressing plants compared with that in the wild type. We have shown previously that pretreatment of the wild type and ahk single mutants with cytokinin kinetin for 1 d equally enhanced the freezing tolerance of these plants, but longer treatment of kinetin, such as 7 d, induced more enhanced freezing tolerance in the ahk single mutants than in the wild-type plants (Jeon et al., 2010). We thus examined whether pretreatment with cytokinin impacts the responsiveness of arr1 mutants and ARR1-overexpressing transgenic plants to cold compared with the wild type (Fig. 4). Pretreatment with kinetin for 7 d resulted in enhanced freezing tolerance of ARR1-overexpressing transgenic plants compared with that of arr1 mutants and two other control plants (ecotype Columbia [Col-0] and Pro35S:GUS). This result indicates that ARR1 is involved in the cold stress tolerance response via cytokinin signal transduction.
Figure 4.
Effect of cytokinin preincubation on freezing tolerance of arr1-3, Pro35S:ARR1, and Pro35S:GUS plants compared with that in the wild-type plants. A, Freezing tolerance assays with or without cytokinin. arr1-3, Pro35S:ARR1, Pro35S:GUS, and wild-type plants were grown for 3 d in the light, transferred to MS plates or MS plates containing 10 µm cytokinin kinetin, and grown for an additional 7 d. These 10-d-old plants preincubated with or without cytokinin were subjected to freezing treatment at −5°C for 4 h. Plants that survived after incubation at 23°C for 2 d for recovery were counted. Experiments were conducted in triplicate, and mean values and se are plotted; n ≥ 30. Statistically significant change at P < 0.05 (Student’s t test) compared with the other samples treated with cytokinin is indicated by the asterisk. B, Example plates showing arr1-3, Pro35S:ARR1, Pro35S:GUS, and the wild-type plants preincubated with or without cytokinin for 7 d and subjected to freezing treatment. One plate (approximately 150 mm × 20 mm) contained 10 plants of each type of plant.
AHP2, AHP3, and AHP5 Are Redundantly Involved in Inducing Type A ARR Expression in Response to Cold
To evaluate the role of AHPs in cold signaling for the expression of type A ARRs, we analyzed the expression of ARR5, ARR6, ARR7, and ARR15 in ahp2, ahp3, and ahp5 single and multiple mutant backgrounds in response to cold using the quantitative RT-PCR method. ARR5, ARR6, ARR7, and ARR15 showed wild-type levels of gene expression in all single and double ahp mutants in response to cold. In contrast, ahp2 ahp3 ahp5 triple mutants showed greatly reduced transcript levels of these ARRs in response to cold compared with that of the wild type (Fig. 5). This result indicates that AHP2, AHP3, and AHP5 are redundantly involved in mediating the cold signal for the expression of the type A ARRs. The Arabidopsis electronic fluorescent pictograph browser (Schmid et al., 2005; Kilian et al., 2007; Winter et al., 2007) showed that all AHPs except for AHP6 are not inducible by cold within 24 h. Cytokinins did not affect AHP expression within 3 h. Thus, a change in the AHP transcript levels is not involved in the cold response of type A ARRs via the two-component signaling system. The result that AHPs and type B ARRs (Supplemental Fig. S2) are not transcriptionally activated by cold is consistent with a known mechanism for cytokinin signaling through a two-component phosphorelay that would not require a change in expression of the signaling components.
Figure 5.
Expression of ARR5, ARR6, ARR7, and ARR15 in response to cold in ahp mutants compared with that in the wild-type plants. Expression of ARR5, ARR6, ARR7, and ARR15 is shown in ahp single, double, and triple mutant backgrounds. Eleven-day-old light-grown seedlings were treated for 2 h at 1°C, and total RNA isolated was subjected to quantitative RT-PCR. CBF1 was employed to verify cold-inducible gene expression. Treatment and analysis of the samples were performed as described in the Figure 1 legend.
Analysis of Nuclear-Cytosolic Distribution of AHP-EGFP Fusion Proteins with or without Cold
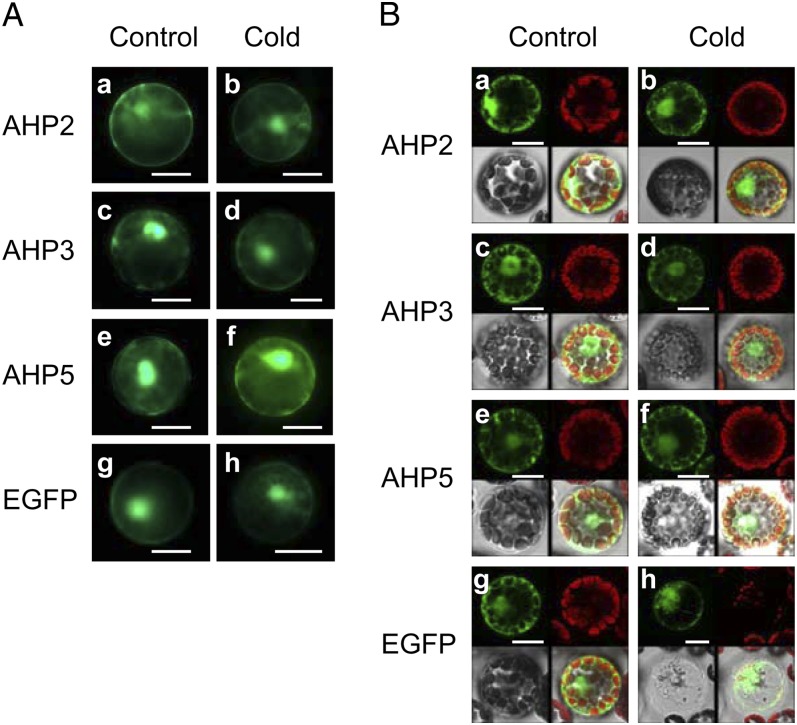
To assess whether the AHP proteins might be relocalized to the nucleus in response to cold to mediate a phosphorelay to ARRs, Arabidopsis mesophyll protoplasts were transfected with constructs expressing AHP-EGFP fusion proteins, and the subcellular distribution of AHP2-EGFP, AHP3-EGFP, and AHP5-EGFP with or without exposure to cold was determined by both confocal scanning microscopy and fluorescence microscopy (Fig. 6). We found that all of these AHP-EGFP fusion proteins were distributed in both the cytosol and the nucleus and that this distribution pattern was not altered by cold treatment. This result suggests that the flux of AHP proteins between the cytosol and nucleus might allow phosphorelay from AHKs to type B ARRs in response to cold.
Figure 6.
Analysis of the subcellular distribution of AHP2-EGFP, AHP3-EGFP, and AHP5-EGFP in the wild-type Arabidopsis mesophyll protoplasts. A, Representative fluorescence micrographs of protoplasts transfected with plasmids encoding different AHP-EGFP fusion proteins. a, c, e, and g show the protoplasts at 23°C, and b, d, f, and h show the protoplasts treated at 1°C for 3 h. Representative images are shown from three independent experiments. Bars = 10 µm. B, Representative confocal scanning laser micrographs of protoplasts transfected with plasmids encoding different AHP-EGFP fusion proteins. a, c, e, and g show the protoplasts at 23°C, and b, d, f, and h show the protoplasts treated at 1°C for 1 h. Representative images are shown from two independent experiments. The top panels show epifluorescence from EGFP and autofluorescence (chloroplast), and the bottom panels show bright-field images and merged images from left to right, respectively. Bars = 10 µm.
Microarray Analysis of the Cold Transcriptome Regulated by AHK2 and AHK3
Our previous study showed that cold induces the expression of a subset of type A ARR genes via AHK2 and AHK3 and that ahk2 ahk3 mutants exhibit enhanced freezing tolerance compared with that of the wild type (Jeon et al., 2010). A previous microarray analysis of the ahk2 ahk3 mutants compared with the wild type identified 40 or 48 genes that were up- or down-regulated with a ratio of greater than 2 in the ahk2 ahk3 mutants under unstressed conditions (Tran et al., 2007). We thought that AHK2 and AHK3 might modulate yet unidentified cold-responsive genes for the adaptation of plants to cold temperature. We conducted a microarray analysis with Affymetrix GeneChip Arabidopsis ATH1 arrays to find other cold-regulated genes downstream of AHK2 and AHK3. Both the wild type and ahk2 ahk3 mutants were treated with or without cold at 1°C for 2 h. We first verified the cold responsiveness of the wild type and ahk2 ahk3 mutants by analyzing the transcript levels of ARR5, ARR6, ARR7, ARR15, and CBF1 prior to microarray analysis (Supplemental Fig. S5). Total RNAs isolated from each sample were then subjected to microarray analysis, and the experiment was conducted in triplicate biological replications. We extracted the genes regulated by cold in the wild type with a false discovery rate (FDR) < 0.1 and isolated the genes whose cold-responsive expression was reduced or enhanced over 1.25-fold with FDR < 0.1 in ahk2 ahk3 mutants compared to that in the wild type (Supplemental Tables S1 and S2). To retrieve many meaningful cold-responsive genes affected by ahk2 ahk3 mutations, we applied a less stringent FDR value and a low fold change relative to the untreated plants as a cutoff value to isolate differentially regulated cold-responsive genes. As the relative fold change provides an interpretable cold transcriptome data set in general, we presented the impact of ahk2 ahk3 mutations on cold-responsive gene expression as the relative fold change, although it reduced the apparent impact of ahk2 ahk3 mutations on the genes exhibiting lowered basal expression levels in the ahk2 ahk3 mutants compared with the wild type. These cold-responsive genes whose expression was reduced or enhanced by ahk2 ahk3 mutations were regarded as a cold transcriptome positively or negatively regulated by AHK2 and AHK3, respectively.
We performed quantitative RT-PCR analysis for selected gene sets to confirm the expression levels of the cold transcriptome identified by the microarray analysis. We determined the transcript levels of CALMODULIN-LIKE38, GLYCINE-RICH PROTEIN, CYTOKININ RESPONSE FACTOR2 (CRF2), ARABIDOPSIS THALIANA DnaJ HOMOLOG3, and RELATED TO ABI/VP1 (RAV1), which were significantly down-regulated by ahk2 ahk3 mutations in response to cold, as representative of the genes exhibiting reduced expression by ahk2 ahk3 mutations (Supplemental Fig. S6A). Expression of these cold-regulated genes was significantly reduced by cold in ahk2 ahk3 mutants compared with that of the wild type. We also assessed the transcript levels of BON ASSOCIATION PROTEIN1, GLYCOSYLTRANSFERASE, INFLORESCENCE DEFICIENT IN ABSCISSION-LIKE, CALMODULIN-LIKE37, and CALCIUM-BINDING PROTEIN, which were enhanced by ahk2 ahk3 mutations in response to cold (Supplemental Fig. S6A). The expression of these genes was also significantly induced by cold in ahk2 ahk3 mutants. We also analyzed the expression of the CBF/DREB pathway genes CBF1, CBF2/DREB1C, and CBF3/DREB1A, three CBF3 downstream genes, PECTINESTERASE INHIBITOR PROTEIN, LIGHT-REGULATED ZINC FINGER PROTEIN1/SALT TOLERANCE HOMOLOG3 (LZF1/STH3), and AT1G70420, and MYB15, an upstream negative regulator of CBF3 (Supplemental Fig. S7). Although MYB15 transcript levels were not significantly changed in response to cold in the wild type, MYB15 expression increased over 3-fold in ahk2 ahk3 mutants in response to cold. Thus, the expression of CBF3, a target of MYB15, was reduced in ahk2 ahk3 mutants in response to cold compared with that of the wild type. The transcript levels of three CBF3 downstream genes also decreased in ahk2 ahk3 mutants in response to cold, as expected. In contrast, CBF1 and CBF2 transcript levels were not significantly changed in ahk2 ahk3 mutants in response to cold compared with that of the wild type. All these quantitative RT-PCR data are consistent with our microarray data.
Microarray analysis of the wild type treated with or without cold showed that 286 genes were up-regulated and 156 genes were down-regulated by cold (Table I). The transcript levels of CBF1, CBF2, and CBF3 increased over 55-fold and up to 128-fold in response to cold, indicating the efficacy of the microarray analysis for cold-treated Arabidopsis plants. Other cold-inducible genes, such as SALT TOLERANCE ZINC FINGER, RAV1, and EARLY RESPONSIVE TO DEHYDRATION10, were also identified by microarray analysis. Cytokinin-responsive genes such as ARR5, ARR6, ARR7, ARR15, Arabidopsis PSEUDO RESPONSE REGULATOR5 (APRR5), and CRF2 were up-regulated over 2-fold in response to cold. Microarray analysis of ahk2 ahk3 mutants treated with or without cold showed that 520 genes were up-regulated and 420 genes were down-regulated in response to cold (Table I). In general, we found similar sets of cold-responsive genes, but with different levels of expression in ahk2 ahk3 mutants compared with those in the wild type. Comparative genome-wide expression analysis of ahk2 ahk3 mutants and the wild type showed that 249 genes were up-regulated and 126 genes were down-regulated over 2-fold in ahk2 ahk3 mutants. Approximately 34%, 59%, and 55% of down-regulated genes were cold-, drought-, and osmotic-regulated genes, respectively (Table II; Supplemental Table S3). Approximately 22% or 33% of up- or down-regulated genes, respectively, were ABA-regulated genes, and the expression of significant portions of various hormone-regulated genes in addition to cytokinins such as BA or zeatin was altered in the ahk2 ahk3 mutants compared with those in the wild type (Supplemental Tables S4 and S5). These data are consistent with a previous microarray analysis of the ahk2 ahk3 mutants compared with the wild type using complementary DNA (cDNA) microarray showing that many of the 40 genes identified as up-regulated with a ratio greater than 2 in the ahk2 ahk3 mutants are stress and/or ABA responsive and also responsive to various plant hormones (Tran et al., 2007). These data indicate that there are significant networks among plant hormone signaling in Arabidopsis. Hierarchical cluster analysis of genes in which expression was modulated by ahk2 ahk3 mutations with or without cold compared with the wild type showed that the expression of several distinct sets of genes was regulated by AHK2 and AHK3 and the cold-induced expression of similar sets of genes in both the wild type and ahk2 ahk3 mutants (Supplemental Fig. S8).
Table I. Summary of differentially expressed genes by ahk2 ahk3 mutations compared with the wild type under cold at 1°C for 2 h.
| Comparison | Up | Down | Total |
|---|---|---|---|
| 1. Col-0 (−cold) versus Col-0 (+cold) | 286 | 156 | 442 |
| 2. Col-0 (−cold) versus ahk2 ahk3 (−cold) | 2,282 | 2,072 | 4,354 |
| 3. Col-0 (+cold) versus ahk2 ahk3 (+cold) | 1,484 | 1,594 | 3,078 |
| 4. ahk2 ahk3 (−cold) versus ahk2 ahk3 (+cold) | 520 | 420 | 940 |
| 5. Comparison between 1 and 4 | – | – | 1,162 |
| 6. Comparison between 1 and 4 (FDR < 0.1%) | 245 | 395 | 640 |
Table II. Stress-regulated genes differentially regulated by ahk2 ahk3 mutations compared with the wild type without cold.
FDR < 0.1 and change greater than 2-fold.
| Genes | Up-Regulated (249) |
Down-Regulated (126) |
||||
| ↑a | ↓ | Total | ↑ | ↓ | Total | |
| Cold-regulated genes | 26b (10.4%) | 8 (3.2%) | 34 (13.7%) | 21 (16.7%) | 23 (18.3%) | 44 (34.9%) |
| Drought-regulated genes | 35 (14.1%) | 13 (5.2%) | 48 (19.3%) | 48 (38.1%) | 27 (21.4%) | 75 (59.5%) |
| Osmotic-regulated genes | 57 (22.9%) | 5 (2.0%) | 62 (24.9%) | 34 (27.0%) | 36 (28.6%) | 70 (55.6%) |
| Salt-regulated genes | 17 (6.8%) | 10 (4.0%) | 27 (10.8%) | 29 (23.0%) | 23 (18.3%) | 52 (41.3%) |
Number of genes that are up-regulated (↑) or down-regulated (↓) by a given stimulus in Genevestigator.
Genes that respond to other responses in addition to a given stress were also included in the number of a given stress-regulated genes.
We conducted bioinformatic analysis of these genes and identified genes that are regulated by various stresses, including cold, drought, osmotic, and high salt, to gain insights into the functions of cold-responsive genes that are down- or up-regulated by ahk2 ahk3 mutations (Table III; Supplemental Tables S6-1, S6-2, S6-3, and S6-4). We found that 640 genes with FDR < 0.1 are stress regulated (Supplemental Tables S6-1 and S7-1). Among them, 186 genes (Supplemental Tables S6-2 and S6-4) and 57 genes (Table III; Supplemental Table S6-3) displayed cold-responsive gene expression that was decreased over 1.25- or 1.5-fold by ahk2 ahk3 mutations compared with that of the wild type, respectively. Although there were some overlaps among stress-regulated genes in terms of the number of genes that belong to each category of stress, cold-regulated genes, with a 1.25- or a 1.5-fold difference, constituted 57% (Supplemental Table S6-4) or 75% (Table III) of the stress-regulated genes, respectively. The number of genes that displayed increased cold-responsive gene expression in ahk2 ahk3 mutations compared with the wild type was much lower than that of reduced cold-responsive gene expression (72 genes with greater than 1.25-fold change [Supplemental Tables S6-2 and S6-4] and 14 genes with greater than 1.5-fold change [Table III; Supplemental Table S6-3]). Moreover, no significant bias for cold-regulated genes was observed compared with other stress-regulated genes. We also identified genes that are regulated by various phytohormones, including ABA, 1-aminocyclopropane-1-carboxylic acid (ACC), cytokinin BA, brassinosteroid, GAs, auxin indole acetic acid, methyl jasmonate, salicylic acid, and zeatin (Table IV; Supplemental Tables S7-1, S7-2, S7-3, and S7-4). Most of the phytohormones examined influenced gene expression, but ACC and the GAs did not regulate any of those genes. ABA-regulated genes with a 1.25- and a 1.5-fold difference constituted 20% (Supplemental Table S7-4) and 23% (Table IV) among the hormone-regulated genes, respectively. ABA was the most influential phytohormone for the genes differentially regulated by ahk2 ahk3 mutations.
Table III. Stress-regulated genes differentially regulated by ahk2 ahk3 mutations compared with the wild type under cold treatment at 1°C for 2 h.
FDR < 0.1 and change greater than 1.5-fold.
| Genes | Up-Regulated (14) |
Down-Regulated (57) |
||||
| ↑a | ↓ | Total | ↑ | ↓ | Total | |
| Cold-regulated genes | 5 (35.7%) | 1 (7.1%) | 6 (42.9%) | 42 (73.7%) | 1 (1.8%) | 43 (75.4%) |
| Drought-regulated genes | 4 (28.6%) | 1 (7.1%) | 5 (35.7%) | 14 (24.6%) | 8 (14.0%) | 22 (38.6%) |
| Osmotic-regulated genes | 5 (35.7%) | 0 (0.0%) | 5 (35.7%) | 21 (36.8%) | 5 (8.8%) | 26 (45.6%) |
| Salt-regulated genes | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 6 (10.5%) | 4 (7.0%) | 10 (17.5%) |
Number of genes that are up-regulated (↑) or down-regulated (↓) by a given stimulus in Genevestigator.
Table IV. Hormone-regulated genes differentially regulated by ahk2 ahk3 mutations compared with the wild type under cold treatment at 1°C for 2 h.
FDR < 0.1 and change greater than 1.5-fold.
| Genes | Up-Regulated (14) |
Down-Regulated (57) |
||||||||||
| ↑a |
↓ |
Total |
↑ |
↓ |
Total |
|||||||
| ABA-regulated genes | 2 | (14.3%) | 1 | (7.1%) | 3 | (21.4%) | 11 | (19.3%) | 2 | (3.5%) | 13 | (22.8%) |
| ACC-regulated genes | 0 | (0.0%) | 0 | (0.0%) | 0 | (0.0%) | 0 | (0.0%) | 0 | (0.0%) | 0 | (0.0%) |
| BA-regulated genes | 2 | (14.3%) | 0 | (0.0%) | 2 | (14.3%) | 5 | (8.8%) | 1 | (1.8%) | 6 | (10.5%) |
| Brassinolide-regulated genes | 0 | (0.0%) | 0 | (0.0%) | 0 | (0.0%) | 6 | (10.5%) | 1 | (1.8%) | 7 | (12.3%) |
| Ethylene-regulated genes | 0 | (0.0%) | 0 | (0.0%) | 0 | (0.0%) | 4 | (7.0%) | 1 | (1.8%) | 5 | (8.8%) |
| GA3-regulated genes | 0 | (0.0%) | 0 | (0.0%) | 0 | (0.0%) | 0 | (0.0%) | 0 | (0.0%) | 0 | (0.0%) |
| Indole acetic acid-regulated genes | 0 | (0.0%) | 1 | (7.1%) | 1 | (7.1%) | 7 | (12.3%) | 1 | (1.8%) | 8 | (14.0%) |
| Methyl jasmonate-regulated genes | 2 | (14.3%) | 1 | (7.1%) | 3 | (21.4%) | 5 | (8.8%) | 3 | (5.3%) | 8 | (14.0%) |
| Salicylic acid-regulated genes | 1 | (7.1%) | 1 | (7.1%) | 2 | (14.3%) | 3 | (5.3%) | 2 | (3.5%) | 5 | (8.8%) |
| Zeatin-regulated genes | 1 | (7.1%) | 1 | (7.1%) | 2 | (14.3%) | 4 | (7.0%) | 0 | (0.0) | 4 | (7.0) |
Number of genes that are up-regulated (↑) or down-regulated (↓) by a given stimulus in Genevestigator.
Meta-Profile and Hierarchical Cluster Analyses
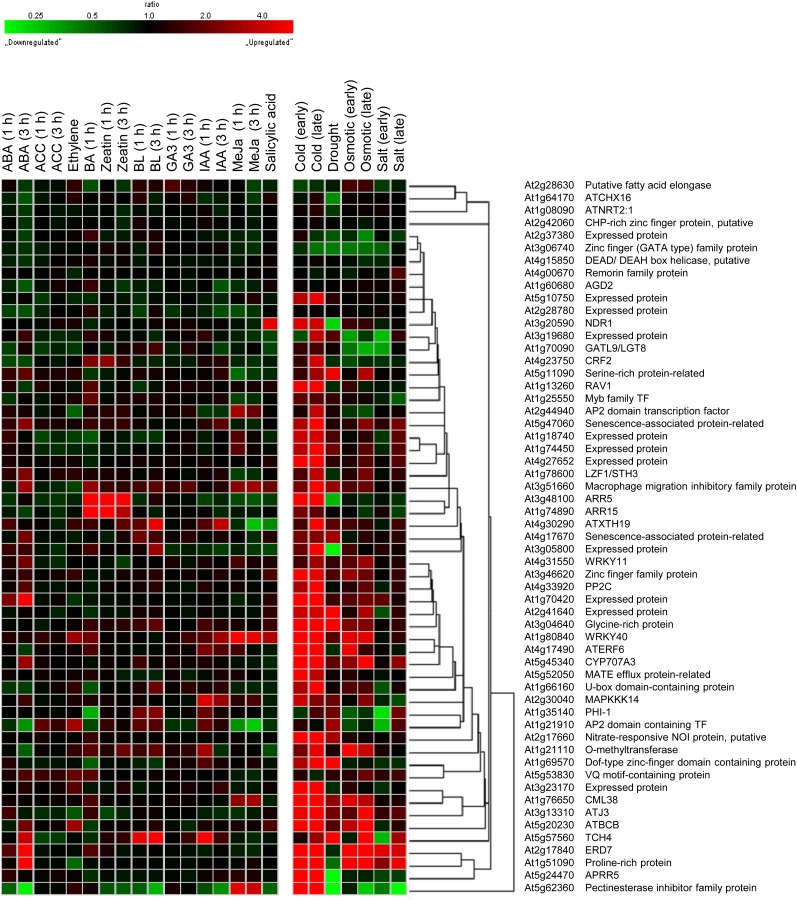
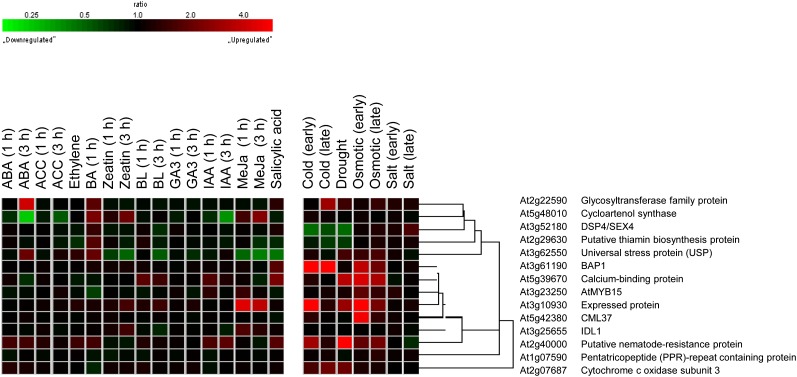
Meta-profile analysis in Genevestigator is used to summarize expression levels according to the biological contexts of the samples. We used the perturbation tool, which provides a summary of gene expression responses to a variety of perturbations such as chemicals, hormones, stresses, mutations, and others, to analyze the expression profiling of cold-responsive genes regulated by ahk2 ahk3 mutations. We first conducted a meta-profile analysis of cold-responsive genes down-regulated with a difference greater than 1.5-fold by ahk2 ahk3 mutations compared with that of the wild type, followed by hierarchical cluster analysis of the same set of genes, to group genes with a common expression pattern using a similarity search (Fig. 7). More than 80% of these genes showed a strong up-regulated response to cold. The same set of genes showed an up-regulated response to drought and osmotic stress as well, but the response was weaker than that to cold. In contrast, most of these genes hardly respond to cytokinins, except for the well-known cytokinin-response genes such as CRF2, ARR5, and ARR15. ABA up- or down-regulates some of these genes, but the ABA response is weaker than cold, drought, or osmotic stress. Other phytohormones weakly regulate these sets of genes. Figure 8 shows the meta-profile analysis and hierarchical cluster analyses of cold-responsive genes up-regulated 1.5-fold or greater by ahk2 ahk3 mutations compared with that of the wild type. The expression patterns of these genes were distinct from those of down-regulated genes. Some gene clusters showed a significant induction response to cold, drought, and osmotic stress, whereas other clusters showed a down-regulated response to these stresses. Most of these genes did not exhibit a significant response to phytohormones, except for some genes responding to ABA. Taken together, the results of these analyses suggest that a new cold-responsive gene network other than the cytokinin response is modulated by AHK2 and AHK3.
Figure 7.
Hierarchical cluster analysis of down-regulated cold-responsive genes by ahk2 ahk3 mutations compared with that of the wild type under cold treatment. A multiple testing correction was applied to the P values of the F statistics to adjust the FDRs. Genes with FDR < 0.1 were extracted. Fifty-seven genes down-regulated with a ratio of 1.5 or greater were selected. The response of ahk2 ahk3-down-regulated genes to a given stimulus was obtained from Genevestigator. BL, Brassinolide; MeJa, methyl jasmonate.
Figure 8.
Hierarchical cluster analysis of up-regulated cold-responsive genes by ahk2 ahk3 mutations compared with that of the wild type under cold treatment. A multiple testing correction was applied to the P values of the F statistics to adjust the FDRs. Genes with FDR < 0.1 were extracted. Fourteen genes up-regulated with a ratio of 1.5 or greater were selected. The response of ahk2 ahk3-down-regulated genes to a given stimulus was obtained from Genevestigator. BL, Brassinolide; MeJa, methyl jasmonate.
Bioinformatic Analysis of cis-Acting Elements in the Promoters of Cold-Regulated Genes Affected by ahk2 ahk3 Mutations
We analyzed cis-acting elements in the promoters of cold-regulated genes affected by ahk2 ahk3 mutations to gain insights into the mechanism of cold-regulated gene expression downstream of AHK2 and AHK3. The promoter regions within 1,000 bp relative to the AUG codon of the cold-responsive genes were identified by cis-acting element analysis using the ATTED-II Web tool that detects position-sensitive promoter elements regardless of their biochemical or biological roles (http://atted.jp/). ARR1-target genes have previously been identified using ARR1-overexpressing Arabidopsis (Taniguchi et al., 2007). Ten of the 57 genes identified as the cold transcriptome were found to be ARR1-target genes (Supplemental Table S8; Taniguchi et al., 2007; Heyl et al., 2008). The conserved ARR1-binding site, 5′-AGATT-3′ sequence element, was found in the promoter regions of these 10 genes. ARR5 and ARR15 have six and seven ARR1-binding sites, respectively. Five of the 57 genes identified as the cold transcriptome were CBF3 downstream genes (Supplemental Table S9; Seki et al., 2001; Maruyama et al., 2004, 2009). AT1G70420, EARLY RESPONSIVE TO DEHYDRATION7, INVERTASE/PECTIN METHYLESTERASE INHIBITOR PROTEIN, and PROLINE-RICH PROTEIN have a CRT/DRE core motif in the promoter regions, indicating that these genes might be direct targets of CBF3. However, LZF1/STH3 did not contain CRT/DRE in the promoter regions within 2,000 bp relative to the AUG codon, indicating that this might be a secondary CBF3 response gene.
Most of the genes identified as the cold transcriptome were not regulated by CBF3 or ARR1, indicating the existence of unidentified transcription factors mediating the cold response downstream of AHK2 and AHK3. We analyzed additional cis-acting elements in the promoters of the cold transcriptome, showing that the circadian responsive elements including an evening element (EE; or EE-like sequence) were highly enriched in the promoters of the cold transcriptome (Supplemental Table S10). The analysis of the cold transcriptome regulated by AHK2 and AHK3 showed that 37 genes, which constitute 65% of the cold transcriptome, contained an EE (5′-AAAATATCT-3′) or an EE-like sequence (5′-AATATCT-3′) and/or the CIRCADIAN CLOCK-ASSOCIATED1 (CCA1)-binding site (5′-AAAAATCT-3′; Supplemental Table S10).
DISCUSSION
While type B ARRs play a redundant role in Arabidopsis cytokinin signaling (Mason et al., 2005; Argyros et al., 2008; Ishida et al., 2008), each type B ARR seems to have a functional specificity in its signaling. For example, ARR2 is involved in leaf senescence, the ethylene response, and the immune response (Hass et al., 2004; Kim et al., 2006; Choi et al., 2010). This study revealed the positive role of ARR1 in cold signaling of Arabidopsis via the two-component system. Expression analysis of type A ARRs in arr1, arr10, and arr12 single, double, and triple mutants (Fig. 1) indicated that ARR1 plays a primary role in cold signaling with minimal ARR10 and ARR12 functional redundancy. It is noteworthy that arr1 mutants showed no apparent physiological phenotype, whereas arr double mutants exhibited a relatively insensitive response to cytokinins; moreover, arr triple mutants displayed a severe dwarf phenotype and complete cytokinin insensitivity similar to ahk2 ahk3 ahk4 triple mutants (Mason et al., 2005; Argyros et al., 2008; Ishida et al., 2008). Thus, while ARR1, ARR10, and ARR12 play essential and redundant roles during cytokinin signaling, ARR1 might possess an additional function during cold signaling. It is not known how ARR1 has a significant functional specificity in cold signaling compared with other type B ARRs such as ARR10 and ARR12. A selective change in ARR1 protein stability in response to cold might be one mechanism to be tested, as cytokinin-facilitated degradation of ARR2 is involved in attenuating cytokinin signaling output (Kim et al., 2012).
Our results showed that while Arabidopsis overexpressing ARR1 exhibited hypersensitive expression of type A ARRs in response to cold compared with that of the wild type, Arabidopsis expressing ARR1ΔDDK:GR lacking the receiver domain (Sakai et al., 2001) lost the hypersensitive response of type A ARRs to cold with DEX treatment (Fig. 3), indicating that the ARR1 N-terminal signal receiver domain might be important in cold-inducible type A ARR expression. The truncated ARR1 without the N-terminal signal receiver domain results in higher transactivation activity than that generated by the full-length ARR1 (Sakai et al., 2000), indicating that the signal receiver domain might partially inhibit transactivation. Transgenic Arabidopsis overexpressing ARR1 showed no phenotypic change under normal growth conditions but displayed a phenotypic change in response to cytokinins, whereas Arabidopsis expressing ARR1ΔDDK:GR lacking the receiver domain exhibited a phenotypic change under normal growth conditions, likely resulting from the constitutive transactivating function of ARR1ΔDDK (Sakai et al., 2001). Deletion of the initial 45-amino acid residues of ARR18 could also liberate the inhibitory role and induce the cytokinin response (Liang et al., 2012). Thus, the inhibition of ARR1 activity by the signal receiver domain may be derepressed by cold, leading to activation of ARR1 to promote its target gene expression. Seventeen ARR1-direct target genes have been previously identified by exploiting ARR1ΔDDK:GR, which can transactivate ARR1-direct target genes in transgenic plants in response to DEX and by analyzing the primary responses of these genes affected by arr1 mutation (Taniguchi et al., 2007). Meta-profile and hierarchical cluster analyses of these ARR1-direct target genes in response to cold and cytokinins were conducted by using Genevestigator, showing that 16 and 13 genes exhibited cytokinin response and cold response, respectively (Supplemental Fig. S9). This analysis indicates that ARR1 could regulate the majority of its cytokinin-responsive target genes in response to cold.
While loss-of-function mutation in arr1 did not show a cold phenotype, pretreatment of cytokinin induced a greater increase in enhanced freezing tolerance in the ARR1-overexpressing transgenic plants than in the two control plants and arr1 mutant plants (Fig. 4). The reason why the cold phenotype could not be observed in the arr1 mutant is not clear. The cytokinin signaling pathway may act to coregulate a subset of the cold-responsive targets. The activated type B ARRs including ARR1 could function in tandem with cold-responsive transcription factors to coregulate a set of genes. Thus, there might be a functional redundancy with other cold-responsive transcription factors. Moreover, microarray analysis indicates that unidentified transcription factors regulate many other cold-responsive genes than ARR1 downstream of AHK2 and AHK3. Additional components might coact with ARR1. Exogenous treatment of cytokinins could strengthen or potentiate the cytokinin signal transduction pathway, resulting in enhanced freezing tolerance in ARR1-overexpressing lines compared with the wild-type plants.
Single ahp mutants were indistinguishable from wild-type seedlings in cytokinin response assays, whereas various higher order mutants displayed reduced sensitivity to cytokinin in diverse cytokinin assays, indicating both a positive role for AHPs in cytokinin signaling and functional overlap among the AHPs (Hutchison et al., 2006). Our expression analysis of type A ARRs in ahp single and multiple mutant backgrounds in response to cold showed that AHP2, AHP3, and AHP5 are redundantly involved in mediating cold signaling (Fig. 5). The high degree of amino acid sequence similarity among AHP2, AHP3, and AHP5 and their overlapping expression patterns (Suzuki et al., 2000; Tanaka et al., 2004) might also be consistent with a redundant role of these AHPs in cold signaling. Previous microscopic observations had suggested a model in which the AHPs are relocalized to the nucleus in response to exogenous cytokinins (Hwang and Sheen, 2001). However, a recent study showed that the AHPs are persistently nucleocytosolic and unresponsive to exogenous cytokinins or to cytokinin signaling status (Punwani et al., 2010; Punwani and Kieber, 2010). Our analysis of the subcellular distribution of AHP2, AHP3, and AHP5 in Arabidopsis protoplasts showed that the nucleocytosolic distribution of these AHPs was not altered by cold treatment (Fig. 6). Therefore, as suggested for the cytokinin response (Punwani et al., 2010), bidirectional movement of the AHPs might shuttle the phosphoryl groups generated from AHK2 and AHK3 to the nucleus in response to cold, resulting in the activation of ARR1.
A microarray analysis has shown that CBFs regulate only approximately 12% of the cold-responsive transcriptome (Fowler and Thomashow, 2002), suggesting that other low-temperature gene networks might contribute significantly to cold response and freezing tolerance. Our meta-profile analysis and hierarchical cluster analyses of the microarray data conducted on ahk2 ahk3 mutants with or without cold compared with the wild type revealed a new cold-responsive gene network regulated downstream of AHK2 and AHK3. We found that 57 cold-responsive genes were down-regulated by the ahk2 ahk3 mutation compared with that of the wild type under cold treatment at 1°C for 2 h with a ratio of 1.5 or greater. Among them, 10 genes, including APRR5, ARR15, CRF2, and ARR5, were found to be targets of ARR1. While AHK2 and AHK3 function independently of CBFs, they modulate several CBF3 target genes via negative regulation of MYB15 (Agarwal et al., 2006), which acts as a negative regulator of CBF3, indicating that a link might exist between the CBF3 pathway via MYB15 and the AHK2/AHK3-responsive cold signaling pathway. However, most of the cold-regulated genes downstream of AHK2 and AHK3 were not regulated by CBF3 or ARR1, indicating that those cold-responsive genes are regulated by unidentified transcription factors acting downstream of AHK2 and AHK3.
CCA1, a Myb transcription factor acting as a key regulator of circadian rhythm, binds to the promoter DNA that contains the 5′-AAAAATCT-3′ or 5′-AGATTTTT-3′ element, which are highly similar to the target sequence of ARR1 (5′-AGATT-3′). An analysis of the cis-acting elements in the promoter regions of the AHK2/AHK3-responsive cold transcriptome showed overrepresentation of the EE and/or the EE-like sequence element and CCA1-binding sites (approximately 65%), as compared with 6% of the 8,000 Arabidopsis genes that exhibit circadian changes in steady-state mRNA levels (Harmer et al., 2000), indicating that circadian rhythm might be an important aspect of cold-responsive gene expression via AHK2 and AHK3. Approximately 43% (93 genes) of CBF-independent cold-responsive genes (218 genes) also contain EE motifs (Mikkelsen and Thomashow, 2009). A variety of studies have revealed the critical importance of circadian rhythm in the cold response. Clock- and cold-regulated expression of CBF-independent CONSTANS-like1 and COLD-REGULATED GENE27 is integrated through the regulatory proteins that bind to the EE and EE-like elements (Mikkelsen and Thomashow, 2009). CBF1, CBF2, and CBF3 are subject to circadian regulation, and their cold induction is gated by the circadian clock (Fowler et al., 2005). The functional implication of circadian rhythm in CBF-imparted cold tolerance has been demonstrated by the analysis of cca1 and late elongated hypocotyl double mutants exhibiting impaired freezing tolerance and diminished circadian regulation and cold induction of three CBF regulon genes (Dong et al., 2011). Overexpression of CCA1α conferred freezing tolerance in Arabidopsis, whereas that of CCA1β resulted in increased sensitivity to freezing tolerance; moreover, cold temperatures reduced CCA1β production by suppressing CCA1 alternative splicing, indicating that cold regulation of CCA1 alternative splicing contributes to freezing tolerance (Seo et al., 2012). APRR arrhythmic triple mutants exhibited enhanced freezing tolerance (Nakamichi et al., 2009), and disruption of the Arabidopsis circadian clock is responsible for extreme variation in the cold-responsive transcriptome (Bieniawska et al., 2008). It remains to be seen how the EE or EE-like motifs existing in cold-regulated gene promoters are linked to AHK2/AHK3-mediated cold regulation.
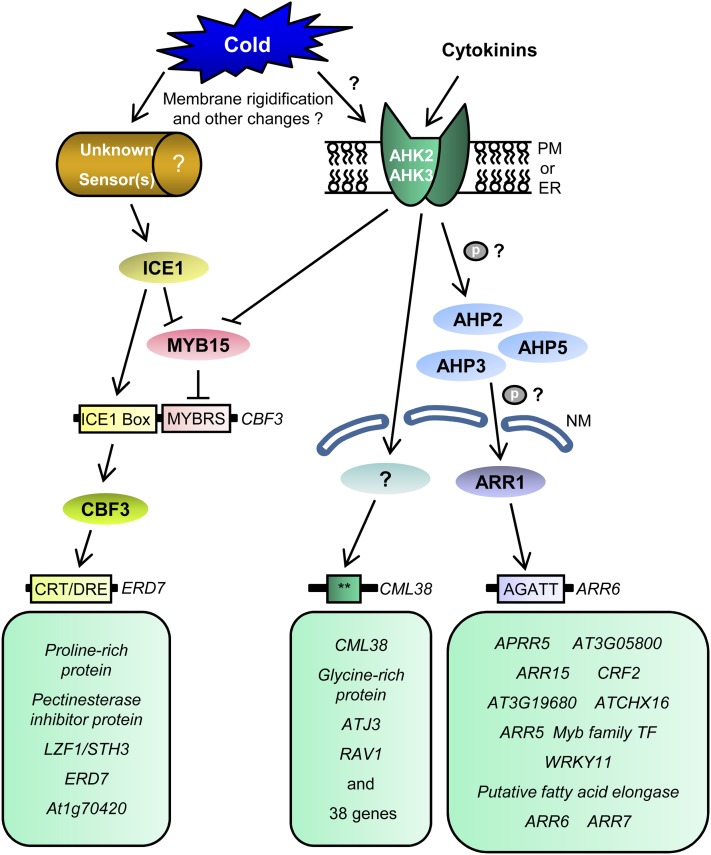
Based on gene expression studies with various cytokinin signaling mutants and microarray analysis, we propose the following model depicting a cold-responsive transcriptional network regulated downstream of AHK2 and AHK3 (Fig. 9). The cold signal can be perceived by the AHK2 and AHK3 proteins directly or, most likely, indirectly, such as through a change in membrane rigidity. AHK2 and AHK3 activated by cold stress might autophosphorylate their His residue and then transfer the phosphate group to the Asp residue in the receiver domain. The phosphate group is then transferred to AHP2, AHP3, or AHP5. These AHPs might transfer the phosphoryl group to the receiver domain of ARR1, a type B ARR, which, in turn, activates the expression of many target genes, including type A ARR genes. AHK2 and AHK3 also activate the expression of a wide variety of cold-responsive genes in ARR1-independent pathways via unidentified transcription factors. AHK2 and AHK3 modulate several CBF3 target genes via negative regulation of MYB15, a negative regulator of CBF3. While phosphorelay during cytokinin signaling has been experimentally demonstrated (Imamura et al., 1998, 2003; Inoue et al., 2001; Kim et al., 2006; Mähönen et al., 2006b; Lee et al., 2007; To et al., 2007), it remains to be shown for cold signaling via the two-component signaling system. Our data are also consistent with an alternative model in which the cytokinin signaling pathway acts to coregulate a subset of the cold-responsive targets. The activated type B ARRs including ARR1 could function in tandem with cold-responsive transcription factors to coregulate a set of genes.
Figure 9.
Diagram of the AHK2- and AHK3-mediated cold-responsive transcriptional network in Arabidopsis. Solid arrows indicate activation, and lines ending with bars represent negative regulation. Double asterisks indicate unknown cis-elements. CRT/DRE, C-repeat/dehydration-responsive element; ER, endoplasmic reticulum; ICE1 box, ICE1 binding site; MYB15, myeloblastosis15; MYBBS, MYB-binding site; NM, nuclear membrane; PM, plasma membrane. The CBF pathway was redrawn from Chinnusamy et al. (2007).
MATERIALS AND METHODS
Plant Materials
The wild-type and mutant lines of Arabidopsis (Arabidopsis thaliana) were all in Col-0. The arr1-3 (CS6971), arr10-5 (salk-098604), arr12-1 (CS6978), arr1-3 arr10-5 (CS39990), arr1-3 arr12-1 (CS6981), arr10-5 arr12-1 (CS39991), arr1-3 arr10-5 arr12-1 (CS39992), ahp2-1 (CS860144), ahp3 (CS860145), ahp5-2 (CS860148), ahp2-1 ahp3 (CS860151), ahp2-1 ahp5-2 (CS860153), ahp3 ahp5-2 (CS860155), and ahp2-1 ahp3 ahp5-2 (CS860161) seeds were obtained from the Arabidopsis Biological Resource Center. Gene-specific primers were used in combination with T-DNA-specific primers to identify and confirm T-DNA insertions by PCR (Supplemental Table S11). These primer combinations and the gene-specific primer combinations flanking the T-DNA insertion sites were used to isolate homozygous lines. The homozygous lines for T-DNA insertions were used in subsequent assays. 35S:ARR1 and 35S:ARR1ΔDDK:GR seeds and ahk2-2 3-3 seeds were provided generously by Dr. T. Aoyama (Sakai et al., 2000) and Dr. T. Kakimoto (Higuchi et al., 2004), respectively, and confirmed via genotyping prior to usage.
Plasmid Constructs
AHP2, AHP3, and AHP5 full-length DNA were PCR amplified using Pfu-X DNA polymerase (Solgent) and inserted into the Pro35S:EGFP vector (Lee et al., 2008) at the XhoI and BamHI sites as translational fusions, yielding the Pro35S:EGFP:AHP2, Pro35S:EGFP:AHP3, and Pro35S:EGFP:AHP5 constructs. PCR-amplified DNA sequences were used for subcloning after verification by DNA sequencing.
Cold and DEX Treatment
Arabidopsis was grown on germination agar plates containing 0.5× Murashige and Skoog (MS) medium with vitamins, 1.5% Suc, 2.5 mm MES, pH 5.7, and 0.8% agar at 23°C with a 16-h photoperiod and treated essentially as described previously (Jeon et al., 2010). The light-grown seedlings were incubated at 1°C for varying periods of time with white fluorescent light for cold treatment. Plants were grown on sterile filter paper on MS plates for 10 d for the DEX treatments and then transferred to MS plates containing 10 μm DEX for an additional 2 d.
RNA Isolation and Real-Time RT-PCR
The Arabidopsis plants were immediately frozen in liquid nitrogen following treatment and stored at −80°C. Total RNA was isolated from frozen Arabidopsis samples using the RNeasy Plant mini kit (Qiagen). Real-time RT-PCR was conducted using a QuantiTect SYBR Green RT-PCR kit (Qiagen) in a Rotor-Gene 2000 Real-Time Thermal Cycling System (Corbett Research). To determine the copy numbers of the transcripts in the treated samples, real-time PCR was conducted for each sample with a known quantity of the in vitro-transcribed RNA (Promega), yielding specific cycle threshold values, as described previously (Jeon et al., 2010). Real-time RT-PCR conditions and primer sequences were as described (Supplemental Table S12).
Protoplast Isolation and Analysis of Subcellular Distribution
The plasmids were purified using a Qiagen Plasmid Midi kit prior to protoplast transformation. The protoplasts from Arabidopsis mesophyll cells were prepared as described previously (Lee et al., 2008). Protoplasts isolated from the rosette leaves of 2- to 3-week-old Arabidopsis plants grown under a 16-h photoperiod were transfected with plasmid DNA by polyethylene glycol-mediated protoplast transformation and incubated for 12 to 18 h in the dark at room temperature. The subcellular localization of the EGFP:AHP fusion proteins was monitored using either confocal scanning laser microscopy or fluorescence microscopy. A TCS SP5 AOBS spectral confocal and multiphoton microscope system (Leica Microsystems) was used to analyze and capture confocal microscopic images. Confocal images of the GFP fusion proteins were acquired at the Korea Basic Science Institute. Argon (488 nm) and helium/neon (633 nm) lasers were utilized for GFP excitation and autofluorescence of chlorophyll excitation, respectively. A Leica DM2500 was used to analyze and capture fluorescence microscopic images.
Gene Expression Profiling
Wild-type (Col-0) and ahk2-2 ahk3-3 light-grown seedlings were grown for 11 d on 0.5× MS plates and incubated at 1°C for 2 h with white fluorescent light for GeneChip analysis. Affymetrix GeneChip Arabidopsis ATH1 genome arrays were used for the microarray analysis. Samples were prepared according to the manufacturer’s instructions and recommendations. Total RNA was isolated using an RNeasy Mini kit (Qiagen). RNA quality was assessed with an Agilent 2100 bioanalyzer with an RNA 6000 Nano Chip (Agilent Technologies), and quantity was determined by an ND-1000 spectrophotometer (NanoDrop Technologies). Six micrograms of RNA sample was used as input into the Affymetrix procedure as recommended by the protocol (http://www.affymetrix.com). The RNA sample was converted to double-stranded cDNA using the oligo(dT) primer incorporating a T7 promoter. Amplified RNA was generated from the double-stranded cDNA template through an in vitro transcription reaction and purified with the Affymetrix sample cleanup module. The amplified RNA was fragmented using 8 μL of 5× fragmentation buffer (sample cleanup module; Affymetrix). Fragmentation was checked on 1% agarose gels stained with ethidium bromide and hybridized to the gene containing over 22,500 probe sets, as described in the Gene Chip Expression Analysis Technical Manual (Affymetrix). The chips were stained and washed in a Genechip Fluidics Station 450 (Affymetrix) after hybridization and scanned by a Genechip Array scanner 3000 7G (Affymetrix).
Data Analysis
Affymetrix GeneChip Arabidopsis ATH1 genome oligonucleotide arrays were scanned using the Affymetrix model 3000 G7 scanner, and image data were extracted through the Affymetrix Command Console software 1.1. Expression data were generated by Affymetrix Expression Console software version 1.1. The Robust Multi Average algorithm implemented in Affymetrix Expression Console software was used for the normalization. Probe sets that were not called “present” by the MAS5 detection call in 50% or more of the samples in at least one sample group were filtered out to reduce noise for the significance analysis. An unpaired Student’s t test was performed on the Robust Multi Average expression values to determine whether genes were differentially expressed between the two groups. A multiple testing correction (Benjamini and Hochberg, 1995) was applied to the P values of the F statistics to adjust the FDR. We performed hierarchical clustering in MultiExperiment Viewer software 4.0 (http://www.tm4.org) to classify the coexpression gene group that had a similar expression pattern. The Database for Annotation, Visualization, and Integrated Discovery was used to perform the biological interpretation of differentially expressed genes (http://david.abcc.ncifcrf.gov/home.jsp). Gene ontology of these genes was classified based on the information of gene function in the Panther ontology database (http://www.pantherdb.org).
Microarray data were deposited into ArrayExpress with the accession number E-MEXP-3714 (http://www.ebi.ac.uk/at-miamexpress).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Expression of ARR5, ARR6, ARR7, and ARR15 in response to cytokinin in type B arr mutants compared with that in the wild-type plants.
Supplemental Figure S2. Quantitative analysis of type B ARR expression in response to cold.
Supplemental Figure S3. Expression of ARR5, ARR6, ARR7, and ARR15 in response to cold in 35S:ARR1ΔDDK:GR compared with that in the wild-type plants.
Supplemental Figure S4. Freezing tolerance assays of arr1-3 mutants and Pro35S:ARR1 transgenic plants compared with those in the wild-type plants.
Supplemental Figure S5. Expression analysis of ARR5, ARR6, ARR7, and ARR15 in response to cold in ahk2 ahk3 mutants compared with that in the wild-type plants prior to microarray analysis.
Supplemental Figure S6. Quantitative RT-PCR analysis of regulated genes in response to cold in ahk2 ahk3 mutants compared with the wild type.
Supplemental Figure S7. Quantitative RT-PCR analysis of the CBF cold response pathway-related genes in response to cold in ahk2 ahk3 mutants compared with that in the wild type.
Supplemental Figure S8. Hierarchical cluster analysis of cold-regulated genes affected by ahk2-2 ahk3-3 mutations compared with that in the wild type.
Supplemental Figure S9. Meta-profile and hierarchical cluster analysis of genes directly regulated by ARR1.
Supplemental Table S1. Gene list of the cold transcriptome down-regulated by ahk2 ahk3 mutations compared with that in the wild type under cold treatment.
Supplemental Table S2. Gene list of the cold transcriptome up-regulated by ahk2 ahk3 mutations compared with that in the wild type under cold treatment.
Supplemental Table S3. Stress-regulated genes differentially regulated by ahk2 ahk3 mutations compared with those in the wild type.
Supplemental Table S4. Hormone-regulated genes differentially regulated by ahk2 ahk3 mutations compared with the wild type without cold.
Supplemental Table S5. Hormone-regulated genes differentially regulated by ahk2 ahk3 mutations compared with that in the wild type.
Supplemental Table S6. Stress-regulated genes differentially regulated by ahk2 ahk3 mutations compared with that in the wild type under cold treatment.
Supplemental Table S7. Hormone-regulated genes differentially regulated by ahk2 ahk3 mutations compared with that in the wild type under cold treatment.
Supplemental Table S8. Analysis of the ARR1-binding site in the promoter of cold-responsive ARR1 target genes down-regulated by ahk2 ahk3 mutations.
Supplemental Table S9. Analysis of CBF/DREB in the promoter of cold-responsive CBF3 target genes down-regulated by ahk2 ahk3 mutations.
Supplemental Table S10. Analysis of circadian-responsive elements in the promoter of cold-responsive genes down-regulated by ahk2 ahk3 mutations.
Supplemental Table S11. Oligonucleotides and PCR conditions for genotyping.
Supplemental Table S12. Oligonucleotides and PCR conditions for quantitative RT-PCR.
Supplemental Data S1. All data of microarray analysis.
Acknowledgments
We thank Dr. T. Aoyama for providing us with the 35S:ARR1 and 35S:ARR1ΔDDK:GR seeds and Dr. T. Kakimoto for the ahk2-2 ahk3-3 mutant seeds.
Glossary
- ABA
abscisic acid
- RT
reverse transcription
- BA
benzyladenine
- DEX
dexamethasone
- Col-0
ecotype Columbia
- FDR
false discovery rate
- ACC
1-aminocyclopropane-1-carboxylic acid
- EE
evening element
- MS
Murashige and Skoog
References
- Agarwal M, Hao Y, Kapoor A, Dong CH, Fujii H, Zheng X, Zhu JK. (2006) A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J Biol Chem 281: 37636–37645 [DOI] [PubMed] [Google Scholar]
- Argyros RD, Mathews DE, Chiang YH, Palmer CM, Thibault DM, Etheridge N, Argyros DA, Mason MG, Kieber JJ, Schaller GE. (2008) Type B response regulators of Arabidopsis play key roles in cytokinin signaling and plant development. Plant Cell 20: 2102–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B 57: 289–300 [Google Scholar]
- Bieniawska Z, Espinoza C, Schlereth A, Sulpice R, Hincha DK, Hannah MA. (2008) Disruption of the Arabidopsis circadian clock is responsible for extensive variation in the cold-responsive transcriptome. Plant Physiol 147: 263–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V, Zhu J, Zhu JK. (2007) Cold stress regulation of gene expression in plants. Trends Plant Sci 12: 444–451 [DOI] [PubMed] [Google Scholar]
- Choi J, Huh SU, Kojima M, Sakakibara H, Paek KH, Hwang I. (2010) The cytokinin-activated transcription factor ARR2 promotes plant immunity via TGA3/NPR1-dependent salicylic acid signaling in Arabidopsis. Dev Cell 19: 284–295 [DOI] [PubMed] [Google Scholar]
- Dong MA, Farré EM, Thomashow MF. (2011) Circadian clock-associated 1 and late elongated hypocotyl regulate expression of the C-repeat binding factor (CBF) pathway in Arabidopsis. Proc Natl Acad Sci USA 108: 7241–7246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler S, Thomashow MF. (2002) Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14: 1675–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler SG, Cook D, Thomashow MF. (2005) Low temperature induction of Arabidopsis CBF1, 2, and 3 is gated by the circadian clock. Plant Physiol 137: 961–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattolin S, Alandete-Saez M, Elliott K, Gonzalez-Carranza Z, Naomab E, Powell C, Roberts JA. (2006) Spatial and temporal expression of the response regulators ARR22 and ARR24 in Arabidopsis thaliana. J Exp Bot 57: 4225–4233 [DOI] [PubMed] [Google Scholar]
- Harmer SL, Hogenesch JB, Straume M, Chang HS, Han B, Zhu T, Wang X, Kreps JA, Kay SA. (2000) Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290: 2110–2113 [DOI] [PubMed] [Google Scholar]
- Hass C, Lohrmann J, Albrecht V, Sweere U, Hummel F, Yoo SD, Hwang I, Zhu T, Schäfer E, Kudla J, et al. (2004) The response regulator 2 mediates ethylene signalling and hormone signal integration in Arabidopsis. EMBO J 23: 3290–3302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyl A, Ramireddy E, Brenner WG, Riefler M, Allemeersch J, Schmülling T. (2008) The transcriptional repressor ARR1-SRDX suppresses pleiotropic cytokinin activities in Arabidopsis. Plant Physiol 147: 1380–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi M, Pischke MS, Mähönen AP, Miyawaki K, Hashimoto Y, Seki M, Kobayashi M, Shinozaki K, Kato T, Tabata S, et al. (2004) In planta functions of the Arabidopsis cytokinin receptor family. Proc Natl Acad Sci USA 101: 8821–8826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horák J, Grefen C, Berendzen KW, Hahn A, Stierhof YD, Stadelhofer B, Stahl M, Koncz C, Harter K. (2008) The Arabidopsis thaliana response regulator ARR22 is a putative AHP phospho-histidine phosphatase expressed in the chalaza of developing seeds. BMC Plant Biol 8: 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hothorn M, Dabi T, Chory J. (2011) Structural basis for cytokinin recognition by Arabidopsis thaliana histidine kinase 4. Nat Chem Biol 7: 766–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison CE, Kieber JJ. (2002) Cytokinin signaling in Arabidopsis. Plant Cell (Suppl) 14: S47–S59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison CE, Li J, Argueso C, Gonzalez M, Lee E, Lewis MW, Maxwell BB, Perdue TD, Schaller GE, Alonso JM, et al. (2006) The Arabidopsis histidine phosphotransfer proteins are redundant positive regulators of cytokinin signaling. Plant Cell 18: 3073–3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Chen HC, Sheen J. (2002) Two-component signal transduction pathways in Arabidopsis. Plant Physiol 129: 500–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Sheen J. (2001) Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 413: 383–389 [DOI] [PubMed] [Google Scholar]
- Hwang I, Sheen J, Müller B. (2012) Cytokinin signaling networks. Annu Rev Plant Biol 63: 353–380 [DOI] [PubMed] [Google Scholar]
- Imamura A, Hanaki N, Umeda H, Nakamura A, Suzuki T, Ueguchi C, Mizuno T. (1998) Response regulators implicated in His-to-Asp phosphotransfer signaling in Arabidopsis. Proc Natl Acad Sci USA 95: 2691–2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura A, Kiba T, Tajima Y, Yamashino T, Mizuno T. (2003) In vivo and in vitro characterization of the ARR11 response regulator implicated in the His-to-Asp phosphorelay signal transduction in Arabidopsis thaliana. Plant Cell Physiol 44: 122–131 [DOI] [PubMed] [Google Scholar]
- Inoue T, Higuchi M, Hashimoto Y, Seki M, Kobayashi M, Kato T, Tabata S, Shinozaki K, Kakimoto T. (2001) Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature 409: 1060–1063 [DOI] [PubMed] [Google Scholar]
- Ishida K, Yamashino T, Yokoyama A, Mizuno T. (2008) Three type-B response regulators, ARR1, ARR10 and ARR12, play essential but redundant roles in cytokinin signal transduction throughout the life cycle of Arabidopsis thaliana. Plant Cell Physiol 49: 47–57 [DOI] [PubMed] [Google Scholar]
- Jeon J, Kim NY, Kim S, Kang NY, Novák O, Ku SJ, Cho C, Lee DJ, Lee EJ, Strnad M, et al. (2010) A subset of cytokinin two-component signaling system plays a role in cold temperature stress response in Arabidopsis. J Biol Chem 285: 23371–23386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakimoto T. (2003) Perception and signal transduction of cytokinins. Annu Rev Plant Biol 54: 605–627 [DOI] [PubMed] [Google Scholar]
- Kang NY, Cho C, Kim NY, Kim J. (2012) Cytokinin receptor-dependent and receptor-independent pathways in the dehydration response of Arabidopsis thaliana. J Plant Physiol 169: 1382–1391 [DOI] [PubMed] [Google Scholar]
- Kiba T, Aoki K, Sakakibara H, Mizuno T. (2004) Arabidopsis response regulator, ARR22, ectopic expression of which results in phenotypes similar to the wol cytokinin-receptor mutant. Plant Cell Physiol 45: 1063–1077 [DOI] [PubMed] [Google Scholar]
- Kiba T, Yamada H, Sato S, Kato T, Tabata S, Yamashino T, Mizuno T. (2003) The type-A response regulator, ARR15, acts as a negative regulator in the cytokinin-mediated signal transduction in Arabidopsis thaliana. Plant Cell Physiol 44: 868–874 [DOI] [PubMed] [Google Scholar]
- Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D’Angelo C, Bornberg-Bauer E, Kudla J, Harter K. (2007) The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J 50: 347–363 [DOI] [PubMed] [Google Scholar]
- Kim HJ, Ryu H, Hong SH, Woo HR, Lim PO, Lee IC, Sheen J, Nam HG, Hwang I. (2006) Cytokinin-mediated control of leaf longevity by AHK3 through phosphorylation of ARR2 in Arabidopsis. Proc Natl Acad Sci USA 103: 814–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Ryu H, Cho YH, Scacchi E, Sabatini S, Hwang I. (2012) Cytokinin-facilitated proteolysis of ARABIDOPSIS RESPONSE REGULATOR2 attenuates signaling output in two-component circuitry. Plant J 69: 934–945 [DOI] [PubMed] [Google Scholar]
- Lee DJ, Kim S, Ha YM, Kim J. (2008) Phosphorylation of Arabidopsis response regulator 7 (ARR7) at the putative phospho-accepting site is required for ARR7 to act as a negative regulator of cytokinin signaling. Planta 227: 577–587 [DOI] [PubMed] [Google Scholar]
- Lee DJ, Park JY, Ku SJ, Ha YM, Kim S, Kim MD, Oh MH, Kim J. (2007) Genome-wide expression profiling of ARABIDOPSIS RESPONSE REGULATOR 7 (ARR7) overexpression in cytokinin response. Mol Genet Genomics 277: 115–137 [DOI] [PubMed] [Google Scholar]
- Liang Y, Wang X, Hong S, Li Y, Zuo J. (2012) Deletion of the initial 45 residues of ARR18 induces cytokinin response in Arabidopsis. J Genet Genomics 39: 37–46 [DOI] [PubMed] [Google Scholar]
- Lohrmann J, Harter K. (2002) Plant two-component signaling systems and the role of response regulators. Plant Physiol 128: 363–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mähönen AP, Bishopp A, Higuchi M, Nieminen KM, Kinoshita K, Törmäkangas K, Ikeda Y, Oka A, Kakimoto T, Helariutta Y. (2006a) Cytokinin signaling and its inhibitor AHP6 regulate cell fate during vascular development. Science 311: 94–98 [DOI] [PubMed] [Google Scholar]
- Mähönen AP, Higuchi M, Törmäkangas K, Miyawaki K, Pischke MS, Sussman MR, Helariutta Y, Kakimoto T. (2006b) Cytokinins regulate a bidirectional phosphorelay network in Arabidopsis. Curr Biol 16: 1116–1122 [DOI] [PubMed] [Google Scholar]
- Maruyama K, Sakuma Y, Kasuga M, Ito Y, Seki M, Goda H, Shimada Y, Yoshida S, Shinozaki K, Yamaguchi-Shinozaki K. (2004) Identification of cold-inducible downstream genes of the Arabidopsis DREB1A/CBF3 transcriptional factor using two microarray systems. Plant J 38: 982–993 [DOI] [PubMed] [Google Scholar]
- Maruyama K, Takeda M, Kidokoro S, Yamada K, Sakuma Y, Urano K, Fujita M, Yoshiwara K, Matsukura S, Morishita Y, et al. (2009) Metabolic pathways involved in cold acclimation identified by integrated analysis of metabolites and transcripts regulated by DREB1A and DREB2A. Plant Physiol 150: 1972–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason MG, Mathews DE, Argyros DA, Maxwell BB, Kieber JJ, Alonso JM, Ecker JR, Schaller GE. (2005) Multiple type-B response regulators mediate cytokinin signal transduction in Arabidopsis. Plant Cell 17: 3007–3018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen MD, Thomashow MF. (2009) A role for circadian evening elements in cold-regulated gene expression in Arabidopsis. Plant J 60: 328–339 [DOI] [PubMed] [Google Scholar]
- Mizuno T. (1997) Compilation of all genes encoding two-component phosphotransfer signal transducers in the genome of Escherichia coli. DNA Res 4: 161–168 [DOI] [PubMed] [Google Scholar]
- Nakamichi N, Kusano M, Fukushima A, Kita M, Ito S, Yamashino T, Saito K, Sakakibara H, Mizuno T. (2009) Transcript profiling of an Arabidopsis PSEUDO RESPONSE REGULATOR arrhythmic triple mutant reveals a role for the circadian clock in cold stress response. Plant Cell Physiol 50: 447–462 [DOI] [PubMed] [Google Scholar]
- Nishimura C, Ohashi Y, Sato S, Kato T, Tabata S, Ueguchi C. (2004) Histidine kinase homologs that act as cytokinin receptors possess overlapping functions in the regulation of shoot and root growth in Arabidopsis. Plant Cell 16: 1365–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pils B, Heyl A. (2009) Unraveling the evolution of cytokinin signaling. Plant Physiol 151: 782–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punwani JA, Hutchison CE, Schaller GE, Kieber JJ. (2010) The subcellular distribution of the Arabidopsis histidine phosphotransfer proteins is independent of cytokinin signaling. Plant J 62: 473–482 [DOI] [PubMed] [Google Scholar]
- Punwani JA, Kieber JJ. (2010) Localization of the Arabidopsis histidine phosphotransfer proteins is independent of cytokinin. Plant Signal Behav 5: 896–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riefler M, Novak O, Strnad M, Schmülling T. (2006) Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell 18: 40–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai H, Aoyama T, Oka A. (2000) Arabidopsis ARR1 and ARR2 response regulators operate as transcriptional activators. Plant J 24: 703–711 [DOI] [PubMed] [Google Scholar]
- Sakai H, Honma T, Aoyama T, Sato S, Kato T, Tabata S, Oka A. (2001) ARR1, a transcription factor for genes immediately responsive to cytokinins. Science 294: 1519–1521 [DOI] [PubMed] [Google Scholar]
- Schaller GE. (2000) Histidine kinases and the role of two-component systems in plants. Adv Bot Res 32: 109–148 [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU. (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Abe H, Kasuga M, Yamaguchi-Shinozaki K, Carninci P, Hayashizaki Y, Shinozaki K. (2001) Monitoring the expression pattern of 1300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray. Plant Cell 13: 61–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo PJ, Park MJ, Lim MH, Kim SG, Lee M, Baldwin IT, Park CM. (2012) A self-regulatory circuit of CIRCADIAN CLOCK-ASSOCIATED1 underlies the circadian clock regulation of temperature responses in Arabidopsis. Plant Cell 24: 2427–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock AM, Robinson VL, Goudreau PN. (2000) Two-component signal transduction. Annu Rev Biochem 69: 183–215 [DOI] [PubMed] [Google Scholar]
- Suzuki T, Miwa K, Ishikawa K, Yamada H, Aiba H, Mizuno T. (2001) The Arabidopsis sensor His-kinase, Ahk4, can respond to cytokinins. Plant Cell Physiol 42: 107–113 [DOI] [PubMed] [Google Scholar]
- Suzuki T, Sakurai K, Imamura A, Nakamura A, Ueguchi C, Mizuno T. (2000) Compilation and characterization of histidine-containing phosphotransmitters implicated in His-to-Asp phosphorelay in plants: AHP signal transducers of Arabidopsis thaliana. Biosci Biotechnol Biochem 64: 2486–2489 [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Suzuki T, Yamashino T, Mizuno T. (2004) Comparative studies of the AHP histidine-containing phosphotransmitters implicated in His-to-Asp phosphorelay in Arabidopsis thaliana. Biosci Biotechnol Biochem 68: 462–465 [DOI] [PubMed] [Google Scholar]
- Taniguchi M, Sasaki N, Tsuge T, Aoyama T, Oka A. (2007) ARR1 directly activates cytokinin response genes that encode proteins with diverse regulatory functions. Plant Cell Physiol 48: 263–277 [DOI] [PubMed] [Google Scholar]
- To JP, Deruère J, Maxwell BB, Morris VF, Hutchison CE, Ferreira FJ, Schaller GE, Kieber JJ. (2007) Cytokinin regulates type-A Arabidopsis response regulator activity and protein stability via two-component phosphorelay. Plant Cell 19: 3901–3914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- To JP, Haberer G, Ferreira FJ, Deruère J, Mason MG, Schaller GE, Alonso JM, Ecker JR, Kieber JJ. (2004) Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell 16: 658–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- To JP, Kieber JJ. (2008) Cytokinin signaling: two-components and more. Trends Plant Sci 13: 85–92 [DOI] [PubMed] [Google Scholar]
- Tran LS, Shinozaki K, Yamaguchi-Shinozaki K. (2010) Role of cytokinin responsive two-component system in ABA and osmotic stress signalings. Plant Signal Behav 5: 148–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran LS, Urao T, Qin F, Maruyama K, Kakimoto T, Shinozaki K, Yamaguchi-Shinozaki K. (2007) Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis. Proc Natl Acad Sci USA 104: 20623–20628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi C, Sato S, Kato T, Tabata S. (2001) The AHK4 gene involved in the cytokinin-signaling pathway as a direct receptor molecule in Arabidopsis thaliana. Plant Cell Physiol 42: 751–755 [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada H, Suzuki T, Terada K, Takei K, Ishikawa K, Miwa K, Yamashino T, Mizuno T. (2001) The Arabidopsis AHK4 histidine kinase is a cytokinin-binding receptor that transduces cytokinin signals across the membrane. Plant Cell Physiol 42: 1017–1023 [DOI] [PubMed] [Google Scholar]