Abstract
Medicago truncatula is one of the most studied model plants. Nevertheless, the genome of this legume remains incompletely determined. We used RNA-Seq to characterize the transcriptome during the early organogenesis of the nodule and during its functioning. We detected 37,333 expressed transcription units; to our knowledge, 1,670 had never been described before and were functionally annotated. We identified 7,595 new transcribed regions, mostly corresponding to 5′ and 3′ untranslated region extensions and new exons associated with 5,264 previously annotated genes. We also inferred 23,165 putative transcript isoforms from 6,587 genes and measured the abundance of transcripts for each isoform, which suggests an important role for alternative splicing in the generation of proteome diversity in M. truncatula. Finally, we carried out a differential expression analysis, which provided a comprehensive view of transcriptional reprogramming during nodulation. In particular, depletion of nitric oxide in roots inoculated with Sinorhizobium meliloti greatly increased our understanding of the role of this reactive species in the optimal establishment of the symbiotic interaction, revealing differential patterns of expression for 2,030 genes and pointing to the inhibition of the expression of defense genes.
Legumes (Fabaceae) are of considerable socioeconomic importance, accounting for one-third of the world’s primary crop production (Graham and Vance, 2003). These plants are characterized principally by their ability to reduce atmospheric nitrogen through the establishment of a symbiotic association with bacteria called rhizobia. This ability to fix atmospheric nitrogen allows these plants to grow on soils that are deficient in nitrogen, decreasing both the need for costly nitrogen fertilizers and the water pollution they may cause (Graham and Vance, 2003).
A new organ, the root nodule, is formed to house this symbiosis. Within this organ, there is an exchange of nutrients: the bacteria provide the plant with ammonia, and the plant provides the bacteria with carbohydrates. Root nodule formation in the symbiotic relationship involving the model plant Medicago truncatula and its bacterial symbiont Sinorhizobium meliloti involves several steps. An exchange of signals induces cell division in the inner root cortex and the formation of a nodule primordium (Long, 2001). In parallel, rhizobia infect the root via infection threads initially formed in root hairs (Jones et al., 2007). The rhizobia are then taken up by endocytosis, forming organelle-like structures called symbiosomes and differentiating into nitrogen-fixing bacteroids. On histological examination, the final nodule can be seen to consist of several different zones: a meristematic zone I, an infection zone II, a fixation zone III, and, in older nodules, a senescence zone IV (Oldroyd and Downie, 2008). Many factors have been shown to control the nodulation process (Oldroyd and Downie, 2008), but nitric oxide (NO), which was initially reported to be involved in plant-pathogen interactions (Delledonne et al., 1998; Durner et al., 1998), seems to play an important role (Shimoda et al., 2005; del Giudice et al., 2011). NO depletion has been shown to cause the down-regulation of plant genes involved in nodule development and to delay nodule formation (del Giudice et al., 2011).
Over the past 10 years, hundreds of plant and bacterial genes displaying differential expression during the nodulation process have been identified by transcriptome analyses in M. truncatula and S. meliloti (Becker et al., 2004; El Yahyaoui et al., 2004; Capela et al., 2006; Starker et al., 2006; Benedito et al., 2008; Jones et al., 2008; Maunoury et al., 2010; Moreau et al., 2011). Studies of bacterial and plant mutants have led to the identification of plant genes involved in early stages of the symbiotic interaction (Mitra and Long, 2004). In one study, complementary DNA (cDNA) microarrays carrying 2,366 single genes were used to analyze M. truncatula nodule formation (Maunoury et al., 2010). This study demonstrated that a two-stage reprogramming of gene expression takes place during nodule formation. More recently, more than 3,400 differentially regulated genes and associated regulators have been identified in studies with a 16.4 K 70-mer oligonucleotide microarray (Moreau et al., 2011); this analysis led to the definition of four different stages of transcription reprogramming during the course of nodulation. Additional resources relevant to M. truncatula functional genomics include the M. truncatula Gene Expression Atlas (http://mtgea.noble.org/v3/), which provides developmental expression data for most of the genes of M. truncatula (Benedito et al., 2008). However, the picture remains incomplete despite the availability of all these data, because all the studies to date have been carried out with microarrays based on an incomplete genome. The genome sequence of M. truncatula is currently being annotated by the International Medicago Genome Annotation Group, which recently reported a draft sequence of the M. truncatula euchromatin based on a bacterial artificial chromosome assembly supplemented with an Illumina shotgun sequence. On the basis of alignments with ESTs, the authors estimated that about 94% of all M. truncatula genes have now been captured (Young et al., 2011). In total, 64,388 gene loci were described in version 3.5 of the genome sequence, in which 14,322 of the predicted genes were annotated as transposons (Young et al., 2011). However, the annotation of the M. truncatula genome remains incomplete.
The development of high-throughput DNA sequencing technologies has provided new methods for the characterization and quantification of transcriptomes (RNA-Seq) at the single-nucleotide level. Here, we used the RNA-Seq approach for transcriptome characterization during early nodule organogenesis and nodule functioning. Our data significantly improve the annotation of the M. truncatula genome by identifying 7,595 new transcribed regions in the radius of known genes, 139 new genes from previously annotated intergenic regions, and 1,670 new transcription units (TUs) not mapped on the reference genome. We also paid particular attention to alternative splicing (AS) and to the role of NO in the optimal establishment of symbiosis.
RESULTS
High-Throughput Sequencing and Mapping of the M. truncatula Transcriptome
We profiled the nodulation transcriptome by isolating mRNA from noninoculated nitrogen-starved roots (referred to hereafter as “noninoculated roots”), roots 4 d after inoculation (“inoculated roots”), and 21-d-old nodules (“nodules”). We also investigated the role of NO in modulating transcriptional reprogramming during the establishment of symbiosis by isolating mRNA from inoculated roots treated with the NO scavenger 2-(4-carboxyphenyl)-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide (cPTIO). We harvested only the 2-cm region of the roots in which most of the nodule primordia appeared (see “Materials and Methods”; Supplemental Fig. S1). For each set of conditions, we used two biological replicates to generate independent libraries for RNA-Seq that were sequenced separately with an Illumina genome analyzer II.
We generated approximately 135.5 million high-quality 36-bp reads (Supplemental Table S1), which were then aligned with the M. truncatula genome sequence (Mt3.0 version) and with sequences from a custom splice-junction database, for the detection of transcribed regions and splicing sites. This provided information about the accuracy of gene predictions, the extent and pervasiveness of genome transcription, and the frequency of AS events. We identified a unique map position on the reference genome for 54.6% of the 135.5 million reads (Table I). About 8% of these reads mapped to known alternatively spliced junctions. We also analyzed the distribution with respect to genes of the 74 million uniquely mapped reads. We found that most (78.8%) mapped to protein-coding genes (Table I). The others mapped to introns (1.25%), untranslated regions (UTRs; 12%), and intergenic regions (6.2%). The proportion of reads mapping to UTRs and new exons was lower for inoculated roots and nodules than for noninoculated roots, whereas no such difference was observed in cPTIO-treated inoculated roots.
Table I. Genomic distribution of the reads and base coverage for the different conditions.
| Distribution | No. of Mapped Reads | Reads | bp Covered |
|---|---|---|---|
| All map reads (no known junctions) | % | ||
| Noninoculated roots | 9,630,799 | 100 | 28,124,194 |
| Inoculated roots | 24,971,580 | 100 | 31,231,171 |
| cPTIO-treated inoculated roots | 24,548,808 | 100 | 37,288,391 |
| Nodules | 15,220,330 | 100 | 29,672,799 |
| All | 74,371,517 | 100 | 48,478,409 |
| Coding sequences | |||
| Noninoculated roots | 7,448,707 | 77.34 | 17,145,704 |
| Inoculated roots | 19,982,322 | 80.02 | 18,749,601 |
| cPTIO-treated inoculated roots | 18,838,610 | 76.74 | 19,480,139 |
| Nodules | 12,349,557 | 81.13 | 17,447,569 |
| All | 58,619,196 | 78.82 | 22,131,430 |
| UTRs | |||
| Noninoculated roots | 1,299,977 | 13.50 | 3,711,333 |
| Inoculated roots | 2,790,178 | 11.17 | 3,959,526 |
| cPTIO-treated inoculated roots | 3,174,530 | 12.93 | 4,671,251 |
| Nodules | 1,570,623 | 10.32 | 3,741,933 |
| All | 8,835,308 | 11.88 | 5,502,782 |
| Introns | |||
| Noninoculated roots | 146,403 | 1.52 | 3,205,597 |
| Inoculated roots | 234,802 | 0.94 | 3,781,346 |
| cPTIO-treated inoculated roots | 364,144 | 1.48 | 6,427,895 |
| Nodules | 188,121 | 1.23 | 3,487,953 |
| All | 933,471 | 1.25 | 10,395,202 |
| New coding sequences on annotated genes | |||
| Noninoculated roots | 159,973 | 1.66 | 1,225,276 |
| Inoculated roots | 285,762 | 1.14 | 1,338,164 |
| cPTIO-treated inoculated roots | 387,905 | 1.58 | 1,666,726 |
| Nodules | 208,173 | 1.36 | 1,272,683 |
| All | 1,041,813 | 1.40 | 1,989,813 |
| New TUs | |||
| Noninoculated roots | 33,501 | 0.35 | 57,548 |
| Inoculated roots | 126,469 | 0.50 | 63,179 |
| cPTIO-treated inoculated roots | 118,831 | 0.48 | 73,094 |
| Nodules | 52,004 | 0.34 | 75,795 |
| All | 330,805 | 0.44 | 103,984 |
| Intergenic | |||
| Noninoculated roots | 542,238 | 5.63 | 2,778,736 |
| Inoculated roots | 1,552,047 | 6.21 | 3,339,355 |
| cPTIO-treated inoculated roots | 1,664,788 | 6.78 | 4,969,286 |
| Nodules | 851,852 | 5.59 | 3,646,866 |
| All | 4,610,925 | 6.20 | 8,355,198 |
| Known junctions | |||
| Noninoculated roots | 862,939 | 8.96 | |
| Inoculated roots | 2,299,765 | 9.21 | |
| cPTIO-treated inoculated roots | 1,643,062 | 6.69 | |
| Nodules | 1,578,057 | 10.36 | |
| All | 6,383,823 | 8.58 |
New TUs and AS
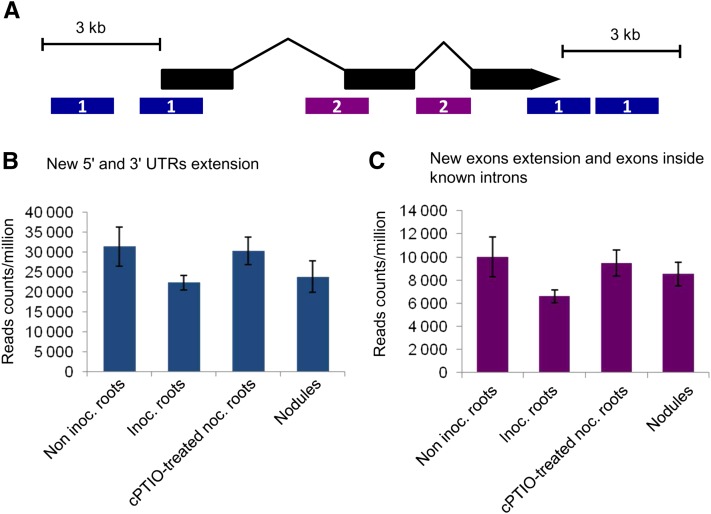
For improvement of the current annotation of the M. truncatula genome, we first clustered the reads mapping to nonannotated regions into 7,875 putative new transcribed regions. Most of these regions (7,595) were within 3,000 nucleotides of 5,264 known genes (Fig. 1; Supplemental Table S2). The remaining newly identified transcribed regions were used to define 139 new TUs (Supplemental Table S3). Functional annotations were obtained for 61 of these TUs. Differences in the number of reads corresponding to these new transcribed regions were observed between conditions (Fig. 1, B and C; Supplemental Table S2), with almost 30% fewer reads mapping to new 5′ and 3′ UTR extensions in inoculated roots and nodules than in noninoculated roots. No such difference was observed in cPTIO-treated inoculated roots (Fig. 1B).
Figure 1.
New transcribed regions identified on the annotated Mt3.0 version of the M. truncatula genome. A, Most of these putative new transcribed regions (7,595) fell within a radius of 3,000 nucleotides of already known genes (5,092 genes). These new transcribed regions can be divided in two classes: (1) 4,107 regions (3,393 genes) that represent extended new 5′ and 3′ UTR extensions (in blue); and (2) 3,488 regions (2,544 genes) that represent exon extensions and exons inside known introns (in purple). B, Sum of all the read counts per million pertaining to new 5′ and 3′ UTR extensions in the four conditions analyzed. C, Sum of all the read counts per million mapping on new exon extensions and exons inside known introns in the different conditions.
A large proportion of the reads (38%) could not be mapped to the Mt3.0 version of the M. truncatula reference genome, probably due to incomplete genome coverage in this version of the reference sequence. We assembled these unmapped reads into 16,335 TUs of 101 to 5,984 nucleotides in size. Most (12,071) matched with M. truncatula Gene Index sequences (tentative consensus, full-length cDNA sequences, and singleton ESTs from The Institute for Genomic Research database) and clustered into 7,122 TUs. We obtained a final set of 11,434 TUs that were also mapped against the new RNA-Seq data accompanying the Mt3.5 version of the M. truncatula genome (Young et al., 2011). Finally, 1,670 TUs could not be mapped to any known M. truncatula Gene Index sequence, Medicago Affymetrix probe set, or RNA-Seq data (Mt3.5). These 1,670 TUs without any correspondence in public databases were considered to be new TUs. Functional annotations were obtained by BLAST2go interrogation (http://www.blast2go.com) of National Center for Biotechnology Information nonredundant databases for only 881 of these new TUs (E value ≤ 1 × 10−8).
We analyzed splice variants with Cufflinks (Trapnell et al., 2010), which is not restricted by prior gene annotation and therefore can be used to identify completely new isoform transcripts and to determine their abundance. Despite limitations due to the lack of paired reads, we identified 6,587 genes with a minimum of two isoforms, giving a total of 23,165 different transcripts (Supplemental Table S4). Of these, 8,557 were known isoforms referenced in the available M. truncatula transcriptome annotation (Mt3.5), whereas 14,608 were new. In addition, 2,457 of these newly identified isoforms presented a reference intron fully covered by the reads, indicating intron retention events. The relative abundance of each of the isoforms, reported in expected fragments per kilobase of transcript per million fragments sequenced, revealed that the multiple gene isoforms were often coexpressed in the same organ but regulated differently during nodulation (Supplemental Tables S5 and S6). The frequency of statistically significant splicing events in cPTIO-treated inoculated roots was more than twice that in untreated inoculated roots (Supplemental Table S7).
Global Analysis of the M. truncatula Transcriptome during Symbiotic Interaction
On a single-gene basis, more than 48% of the genes and almost 68% of the exons for which we had evidence of transcription in at least one condition were covered by reads over more than 90% of their length. We defined 37,333 TUs supported by at least two reads in at least one of the four conditions assayed (Table II). The application of a more stringent criterion, a minimum of five reads, decreased this number slightly, to 34,515 TUs, with 29,227 for noninoculated roots and 32,663 for inoculated roots treated with cPTIO. Moreover, in order to assess the agreement between biological replicates for each condition, we calculated the percentage of TUs detected in both replicates at different reads per kilobase per million mapped reads (RPKM) thresholds relative to the total number of detected TUs in either replicate. The results showed a high agreement between replicates, with overlap values ranging from 87.9% to 93.2% at an RPKM threshold of 1. Similar results were found also for higher thresholds (Supplemental Fig. S2).
Table II. Number of TUs identified in the different conditions according to the number of reads and with a minimum expression level of 2.77 RPKM.
We investigated whether the differences in gene coverage between RNA sources were due to differences in sample size in terms of mapped reads (ranging from 16.4 million [7.24 + 9.16] for root control to 45.4 million [22.84 + 22.57] for inoculated roots) or to real differences in transcriptome complexity by subsampling and inferring the transcript discovery rate as a function of the number of uniquely mapped reads (Supplemental Fig. S3). The graphs clearly show that, at the depth of sequencing achieved, we have almost reached a plateau for each sample. Furthermore, Spearman’s rank correlation analysis on crude counts showed a strong correlation between the two replicates of each condition: 0.90, 0.83, 0.99, and 0.86 for control, inoculated roots, cPTIO-treated inoculated roots, and nodules, respectively.
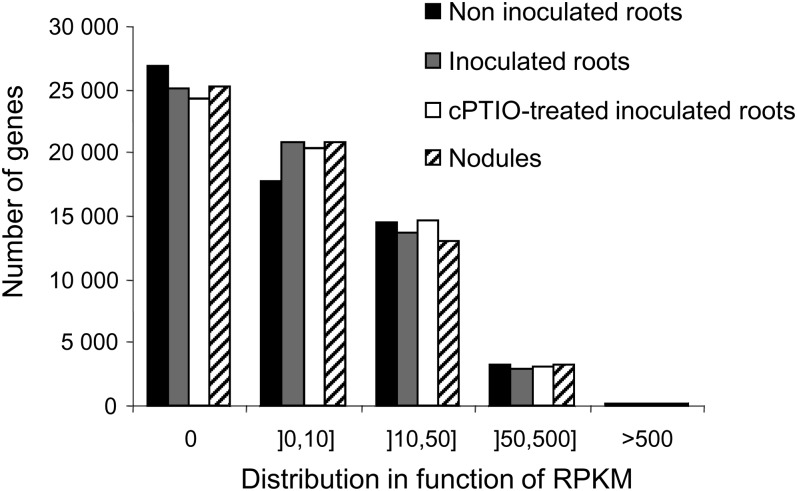
Expression levels of the 50,962 TUs predicted from version Mt3.0 of the M. truncatula genome and of the 11,573 TUs identified (new TUs aligned with the reference genome and all nonaligned TUs) were determined with ERANGE (Mortazavi et al., 2008) and are expressed in RPKM. The distribution of genes on the basis of expression level was similar in the four samples (Fig. 2). In total, 28,744 TUs were identified in at least one condition, with a cutoff value of 2.77 RPKM corresponding to a confidence level of 95% (Table II; Supplemental Table S8). Most of the TUs (76.5%; 21,967) were common to all four conditions, consistent with previous microarray analyses (Benedito et al., 2008). The expression patterns of a set of genes identified in our RNA-Seq analysis were validated by quantitative real-time (qRT)-PCR (consistent results for 74% of 50 tested genes; Supplemental Table S9). It must be underlined here that the qRT-PCR experiments were performed with the two biological replicates used for the RNA-Seq analysis together with three other biological replicates.
Figure 2.
Distribution of genes based on the expression level in RPKM. Five classes have been defined from nonexpressed (0 RPKM) to highly expressed (more than 500 RPKM).
Dynamic Reprogramming of the M. truncatula Transcriptome during Nodulation
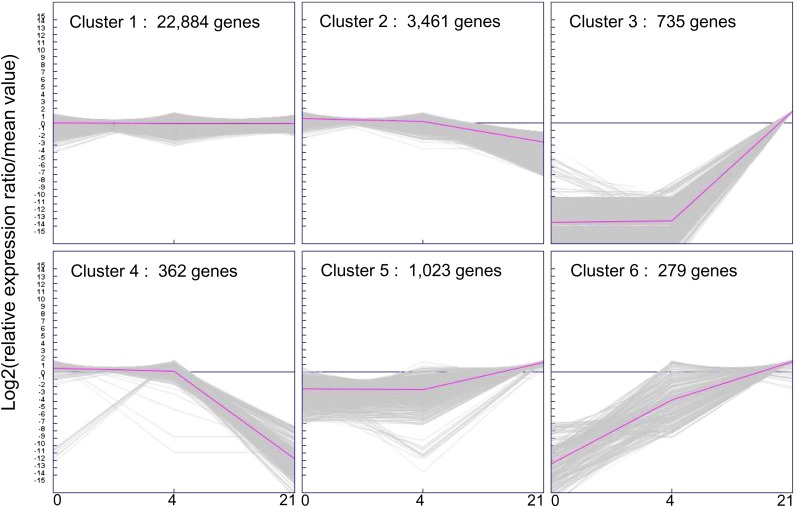
The 28,744 TUs were grouped by K-means analysis (Sturn et al., 2002) into six clusters differing in terms of their expression dynamics during nodulation (Fig. 3). These clusters differed considerably in size: clusters 4 and 6 each contained only 1% of all the TUs, whereas cluster 1, which contained all the genes stably transcribed in the various conditions, accounted for 80% of the TUs. Genes down-regulated or not expressed in nodules were grouped into clusters 2 (3,461 TUs) and 4 (362 TUs), whereas most of the genes induced in nodules were grouped into clusters 5 (1,023 TUs) and 3 (735 TUs). The last cluster (cluster 6) was composed of 279 TUs induced in both inoculated roots and mature nodules (Supplemental Table S8).
Figure 3.
Cluster analysis by K-means method of the expression profiles in the three stages sequenced. Cluster analysis was made on the 28,744 genes detected in roots, inoculated roots, and nodules. Prior to K-means analysis, the expression data (in RPKM) were transformed with the Expression Profiler software from EBI (www.ebi.ac.uk) around the gene’s average value. The K-means analysis performed with the Genesis bioinformatics tool (Sturn et al., 2002; http://genome.tugraz.at) allowed us to discover six main patterns of expression occurring during these three stages. In the scale at bottom, the numbers 0, 4, and 21 correspond to the three conditions sequenced: roots (before infection), inoculated roots (4 d post infection), and nodules (21 d post infection), respectively. [See online article for color version of this figure.]
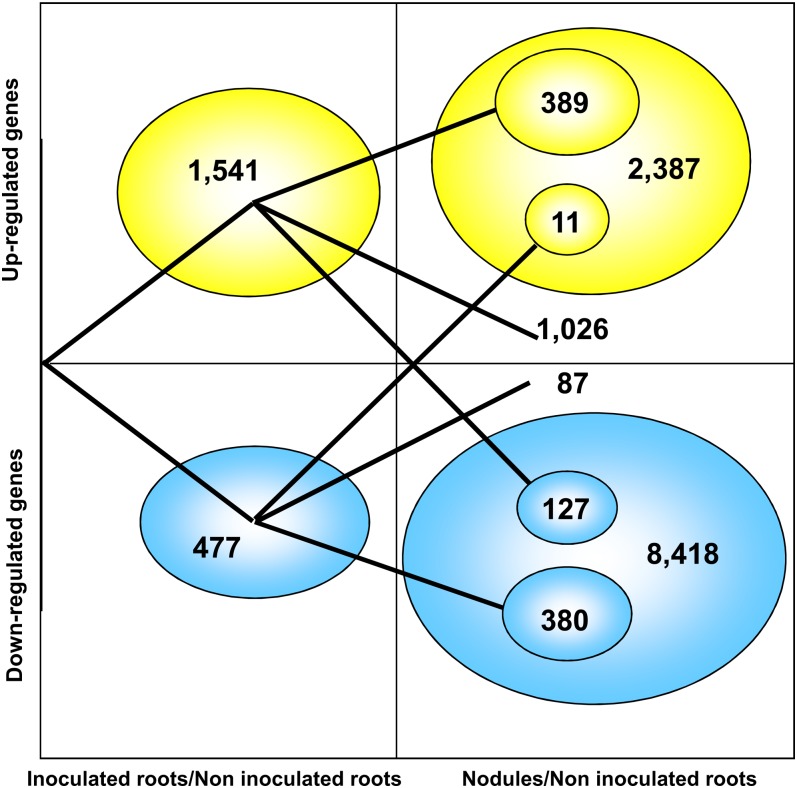
We further explored transcriptional reprogramming during early nodule formation and functioning by calculating Student’s t statistics, with a significance threshold of P < 0.001 and a false discovery rate (FDR) of 0.001. In this analysis, in which we selected only transcripts for which expression levels differed by a factor of at least 2 between two conditions (Table III), we identified 2,018 TUs displaying differential expression in inoculated and noninoculated roots and 10,805 TUs displaying differential expression in nodules and noninoculated roots (Supplemental Tables S10 and S11). In the early stage of the interaction (inoculated roots), most (1,541) of the 2,018 differentially expressed TUs were up-regulated (Fig. 4; Supplemental Table S10), indicating the occurrence of substantial changes during the development of a new organ from the root tissue. Only one-quarter (389) of these TUs continued to be up-regulated in nodules (Fig. 4). By contrast, most (380) of the 477 TUs down-regulated in inoculated roots continued to be down-regulated in nodules. By contrast to what was observed for inoculated roots, most (8,418) of the 10,805 TUs differentially expressed in nodules were down-regulated (Fig. 4; Supplemental Table S11).
Table III. Differential expression analysis.
Shown are the number of TUs for which expression levels differed by a factor of at least 2 between two conditions with a significantly different level based on Student’s t test and a FDR obtained with the R package qvalue (Dabney et al., 2010) threshold of q < 0.001. In parentheses, the number of up-regulated TUs is given.
| Parameter | Inoculated Roots versus Noninoculated Roots | cPTIO-Treated Inoculated Roots versus Inoculated Roots | Common cPTIO-Treated and Inoculated Roots | Nodules versus Noninoculated Roots |
|---|---|---|---|---|
| No. of TUs | 2,018 | 2,030 | 415 | 10,805 |
| Up-regulated | 1,541 | 783 | 170 | 2,387 |
| Down-regulated | 477 | 1,242 | 245 | 8,418 |
| With Affymetrix probe seta | 1,276 | 1,293 | 267 | 7,506 |
| Identified in Affymetrix GeneChipb | 449 | 4,704 | ||
| Without protein homologyc | 305 | 317 | 84 | 1,325 |
| Hypothetical and unknown proteind | 346 | 430 | 70 | 1,807 |
| TFs | 113 (87) | 95 (32) | 11 | 530 (158) |
Number of Affymetrix probe sets corresponding to the TUs identified.
TUs that have been identified as differentially expressed in an Affymetrix study (Benedito et al., 2008).
Results of BLAST against public protein databases.
From the annotation in the Mt3.0 M. truncatula genome.
Figure 4.
Dynamics of the differentially expressed genes in the inoculated roots and the nodules compared with the roots. Only genes showing at least a 2-fold change with a significant difference based on Student’s t test and a FDR obtained with the R package qvalue (Dabney et al., 2010) threshold of q < 0.001 are represented. A total of 2,018 and 10,806 TUs were identified in inoculated roots and nodules compared with the roots, respectively. The expression of 1,026 and 87 genes was found to be modified only in inoculated roots. [See online article for color version of this figure.]
We compared our RNA-Seq data with data generated in a previous genome-wide analysis of M. truncatula gene expression based upon Affymetrix chip technology (Benedito et al., 2008). We identified a set of 4,697 genes found to be differentially regulated in nodules by both techniques; 1,292 of the 2,387 up-regulated genes were described as up-regulated in 14- and 28-d-old nodules in the M. truncatula Gene Expression Atlas, while 3,405 TUs were found to be down-regulated by both techniques. It should be noted that a low median of expression (<2 RPKM) was observed for the 5,013 TUs not identified as down-regulated in the M. truncatula Gene Expression Atlas. Of the 2,387 up-regulated TUs, 1,998 were identified as induced only at this late stage of symbiosis. We also identified a number of new differentially expressed genes not represented on the current Affymetrix Medicago GeneChip: 37.4% and 30.8% of the 2,018 and 10,805 genes modulated in inoculated roots and nodules, respectively, including several new nodule-specific Cys-rich peptides (annotated as late nodulins: 82 of 252 genes up-regulated in nodules) and transcription factor (TF; 95 of 530 genes induced in nodules) genes. This highlights the greater power of RNA-Seq than of microarrays in terms of sensitivity and ability to discover new genes.
We then analyzed the data set for inoculated roots treated with cPTIO, a scavenger of NO. The efficiency of cPTIO treatment was confirmed by the observed regulation of three genes known to be regulated by NO: those encode a Glu-Cys ligase (Medtr5g010350), which is involved in reduced glutathione synthesis (Innocenti et al., 2007), a putative lipoxygenase (Contig3307), which is involved in the jasmonate pathway (Palmieri et al., 2008), and class I hemoglobin (Contig525), all of which were repressed by cPTIO. The NO-dependent up-regulation of a class I hemoglobin was also observed in Lotus japonicus during the early stage of nodulation (Shimoda et al., 2005). cPTIO treatment resulted in changes in the expression of 2,030 TUs with respect to the pattern observed in inoculated roots (P < 0.001, FDR = 0.001; Table IV). The most striking finding was the large number (1,242) of TUs down-regulated by this treatment. In total, we identified 415 TUs displaying differential regulation in inoculated versus noninoculated roots and in cPTIO-treated inoculated roots versus inoculated roots (Supplemental Table S13). For 245 of these TUs, the up-regulation observed in inoculated roots was prevented by NO depletion (Supplemental Table S13). Among the remaining 170 TUs, of which the down-regulation upon inoculation was prevented by NO depletion, genes related to plant defenses were well represented (53 TUs; Supplemental Table S14), indicating that, in contrast to pathogenic interactions, NO repressed defense reactions.
Table IV. List of TFs and associated regulators of gene expression differentially expressed in nodules compared with inoculated roots.
| Bin Code | Name | Genes | No. of TFs Induced in Nodules | Known |
|---|---|---|---|---|
| 27.3.99 | Unclassified | 50 | 15 | |
| 27.3.6 | bHLH, basic helix-loop-helix family | 67 | 7 | |
| 27.3.60 | NIN-like bZIP-related family | 2 | 2 | |
| 27.3.22 | HB, homeobox TF family | 33 | 7 | |
| 27.3.11 | C2H2 zinc finger family | 38 | 14 | |
| 27.3.66 | Psudo-ARR TF family | 3 | 3 | |
| 27.3.14 | CCAAT box-binding factor family, HAP2 | 4 | 4 | HAP2.1; HAP2.3 |
| 27.3.35 | bZIP TF family | 19 | 8 | |
| 28.1.3 | DNA synthesis/chromatin structure, histone | 10 | 2 | |
| 27.3.44 | Chromatin-remodeling factors | 4 | 2 | |
| 27.1.20 | RNA processing, degradation dicer | 1 | 1 | |
| 27.3.54 | Histone acetyltransferases | 2 | 1 | |
| 27.3.1 | ABI3/VP1-related B3 domain-containing TF family | 2 | 1 | |
| 27.3.7 | C2C2(Zn) CO-like, constans-like zinc finger family | 11 | 1 | |
| 27.3.34 | Orphan family | 2 | 1 | |
| 27.3.4 | ARF, auxin response factor family | 8 | 0 | |
| 27.3.52 | Global TF group | 1 | 1 | |
| 27.3.63 | PHD finger TF | 2 | 1 | |
| 27.3.18 | E2F/DP TF family | 1 | 1 | |
| 27.3.32 | WRKY domain TF family | 34 | 7 | |
| 27.3.3 | AP2/EREBP, APETALA2/ethylene-responsive factor | 42 | 12 | |
| 27.3.26 | MYB-related TF family | 10 | 3 | |
| 27.3.57 | JUMONJI family | 1 | 1 | |
| 27.3.19 | EIN3-like (EIL) TF family | 1 | 1 | |
| 27.3.25 | MYB domain TF family | 52 | 4 | |
| 27.3.40 | Auxin/indole-3-acetic acid family | 15 | 0 | |
| 27.3.69 | SET-domain transcriptional regulator family | 9 | 0 | |
| 27.3.23 | HSF, heat shock TF family | 3 | 0 | |
| 27.3.15 | CCAAT box-binding factor family, HAP3 | 4 | 2 | |
| 27.3.65 | Polycomb group (PcG) | 1 | 0 | |
| 27.3.5 | ARR | 3 | 1 | RR4 |
| 17.3.2.2 | Hormone metabolism, brassinosteroid, signal transduction, BZR | 1 | 0 | |
| 27.3.9 | C2C2(Zn) GATA TF family | 9 | 1 | |
| 27.3.12 | C3H zinc finger family | 25 | 8 | |
| 27.3.37 | AS2, lateral organ boundaries gene family | 15 | 5 | |
| 27.3.62 | Nucleosome/chromatin assembly factor group | 1 | 0 | |
| 27.3.39 | AtSR TF family | 1 | 0 | |
| 27.3.20 | G2-like TF family, GARP | 11 | 4 | |
| 27.3.46 | DNA methyltransferases | 5 | 0 | |
| 27.3.24 | MADS box TF family | 10 | 1 | |
| 27.3.67 | Putative transcription regulator | 19 | 6 | |
| 27.3.27 | NAC domain TF family | 11 | 5 | |
| 27.3.16 | CCAAT box-binding factor family, HAP5 | 2 | 2 | HAP5.2 |
| 27.3.31 | TUB TF family | 1 | 0 | |
| 27.3.49 | GeBP like | 1 | 0 | |
| 27.3.64 | PHOR1 | 6 | 0 | |
| 27.3.50 | General transcription | 2 | 0 | |
| 27.3.41 | B3 TF family | 7 | 0 | |
| 27.3.30 | Trihelix, triple-helix TF family | 8 | 2 | |
| 27.3.8 | C2C2(Zn) DOF zinc finger family | 12 | 1 | |
| 27.3.29 | TCP TF family | 6 | 3 | |
| 27.3.17 | CPP(Zn), CPP1-related TF family | 1 | 0 | |
| 27.3.21 | GRAS TF family | 9 | 5 |
To assess the specificity of the cPTIO effects, the expression levels of defense genes responding to the cPTIO treatment were tested via qRT-PCR in the presence of compounds that are known to be inhibitors of NO synthesis: tungstate, which inhibits nitrate reductase, and l-NG-nitro-arginine methyl ester (l-NAME), which inhibits NO synthase. The obtained results (Supplemental Fig. S4) well validated again the RNA-Seq results and showed that the cPTIO effect could be mimicked by tungstate for a majority of the genes tested, while l-NAME appeared to be ineffective. This indicates that the observed cPTIO effects are linked, at least partially, to NO levels. Furthermore, analyses on transgenic roots with promoter-reporter gene fusions (promoters of the Medtr1g124600 and Medtr7g071380 genes, encoding a glutathione transferase and a chalcone synthase, respectively) confirmed the effect of tungstate on the transcriptional regulation of genes identified as regulated by cPTIO (Supplemental Fig. S5). The MapMan representation tool (http://mapman.gabipd.org) provided an overview of the biotic stress pathway, leading to the identification of major NO-dependent modifications (Supplemental Fig. S6), clearly suggesting that NO has a beneficial effect, repressing the defense reactions of the plant to favor establishment of the symbiotic interaction.
DISCUSSION
This work describes the transcriptome landscape of M. truncatula and its expression dynamics at two important stages of the symbiotic interaction with S. meliloti: early organogenesis of the nodules and fully differentiated and functioning nodules.
We used RNA-Seq techniques to generate about 5 Gb of sequence information, corresponding to about 10 times the size of the M. truncatula genome (estimated size of 471–583 Mb [Medicago Genome Sequence Consortium, 2007]). After alignment with the reference genome, the data set of the 74 million uniquely mapped reads provided a rich resource for improving gene annotation across the M. truncatula genome. We identified 7,595 new transcribed regions associated with 5,264 annotated genes. These new transcribed regions contained numerous new exons within introns, new 5′ and 3′ UTRs, and extensions of known 5′ and 3′ UTRs, making it possible to better define gene boundaries. Extended 5′ UTRs may reflect alternative transcription start sites, as demonstrated in Schizosaccharomyces pombe, in which more than 20 genes display dynamic changes in 5′ UTR length during the course of sexual differentiation (Wilhelm et al., 2008). The extension of 3′ UTRs may have an impact on gene expression regulation (e.g. if providing microRNA binding sites). We also identified 139 new TUs in intergenic regions that were not described in version Mt3.0 of the M. truncatula genome, more than one-half of which may correspond to noncoding RNAs, as no functional annotation was found in public protein databases (National Center for Biotechnology Information nonredundant database). Finally, we assembled a large number of reads that did not map onto the reference genome into 11,435 TUs, 1,670 of which had not been identified before. These data are currently used, in addition to others, to gain a more comprehensive view of the expressed genome in various organs and physiological conditions and thereby to improve the M. truncatula genome annotation.
Our analysis of gene expression during the symbiotic interaction identified 28,744 genes expressed, in at least one condition, above the selected cutoff value of 2.77 RPKM and confirmed the power of RNA-Seq for the in-depth quantitative analyses of transcriptomes. A K-means clustering analysis showed that 80% of the expressed genes were transcribed to similar levels in the different conditions and revealed major transcriptional reprogramming in fully formed and functional nodules (clusters 2–5). Therefore, we used a stringent Student’s t test analysis (P < 0.001, FDR = 0.001) to investigate, in depth, the two steps in the symbiotic interaction studied here (Table IV). A comparison of inoculated and noninoculated roots showed that 2,018 genes were differentially regulated in these two conditions (Supplemental Table S10), most (1,541) being up-regulated during symbiosis. Most of these genes (1,026) did not appear to be differentially regulated in mature nodules (Fig. 4). Our analysis of the genes up-regulated in inoculated roots highlights the importance of (1) genes encoding extensins (root nodule extensins), which are abundant in the infection thread lumen (Brewin, 2004); (2) cyclin-like and ribosomal protein-encoding genes, which are involved in the cell cycle and development; and (3) genes involved in redox regulation, consistent with the role of reactive oxygen species and antioxidant defenses in key stages of nodule formation, such as infection thread development and nodule meristem formation (Chang et al., 2009). This analysis also revealed changes in the level of expression of genes not previously identified as involved in nodulation that (1) are involved in protein processing and degradation, highlighting the reorganization of the protein pool occurring during nodulation; (2) encode cytochrome P450s, which may be involved in flavonoid synthesis (Zhang et al., 2007); and (3) encode putative proteins similar to arginase (Supplemental Fig. S7A), an enzyme that has been reported to hydrolyze Arg to generate urea and Orn in higher plants, Orn being a precursor of polyamines (Chen et al., 2004). The 389 genes that continued to be up-regulated in nodules included many encoding nodulins and early nodulins, such as MtNIN, ENOD40, ENOD20, HAP2.1, MtN1, MtN2, and MtN6 (Crespi et al., 1994; Gamas et al., 1996; Combier et al., 2006; Marsh et al., 2007), highlighting the role of these compounds in nodule development and function. A similar expression profile was found for genes encoding formins, which play an important role in cytokinesis and cell expansion (Blanchoin and Staiger, 2010), and genes encoding proteins involved in steroid metabolism. These findings suggest that cardenolides (Bauer et al., 2010), in addition to their possible role in the synthesis of brassinosteroids, which have been shown to be involved in nodulation (Ferguson et al., 2005), are synthesized from the earliest stages of the interaction.
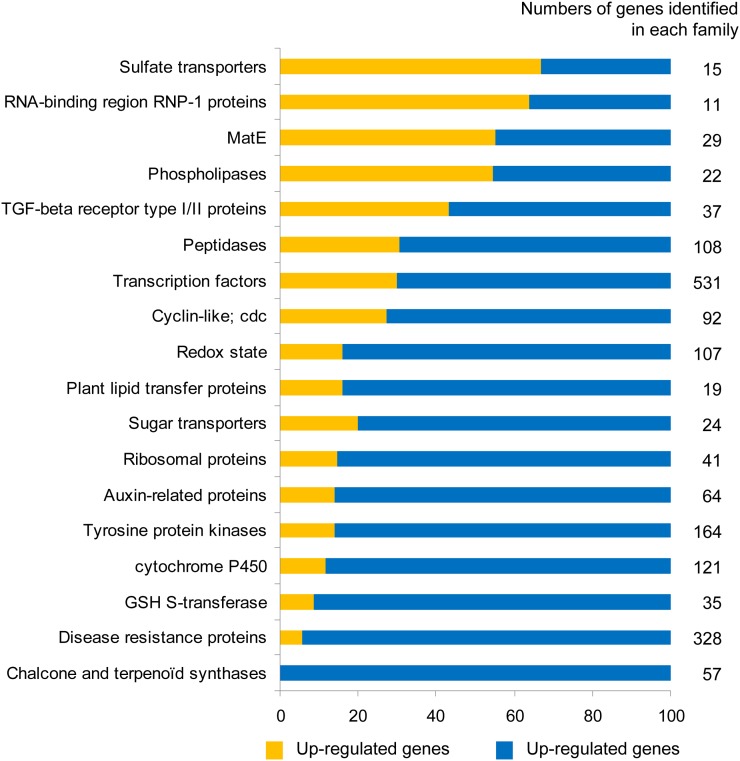
A large number of genes (10,805) displayed differential expression in nodules and noninoculated roots (Supplemental Table S11), highlighting the major reprogramming triggered by the establishment of a functional symbiosis. However, most (78%) of these genes were down-regulated in nodules. One of the most striking results of the global analysis of the gene families represented (Fig. 5) is the repression of genes encoding chalcone and terpenoid synthases (57 down-regulated TUs) and the repression of 95% of the disease resistance genes (310 of 328). Furthermore, consistent with the extensive down-regulation of genes observed at this stage, approximately 70% of the 598 genes encoding TFs and associated regulators of gene expression, the largest group of genes identified, were found to be down-regulated or even switched off in nodules (Table IV). More than 80% of the genes related to auxin, redox state, and sugar transporters (Fig. 5) were also down-regulated. This suggests that hormone and redox balance may be modified in the functioning nodule and indicates that dicarboxylic acids replace sugars as the carbon metabolites transported in this organ (White et al., 2007; Benedito et al., 2010). The large number of genes down-regulated is of particular importance, highlighting the high degree of functional specialization of the nodule.
Figure 5.
Transcriptional regulation of gene families in nodules. The number of genes identified in each family is indicated at right and corresponds to 100% in each family. The rate of genes belonging to the different classes is represented according to the total number of genes identified in each family (in percentage). The classification of each gene in the families is based on the protein function indicated in the Mt3.0 annotation. cdc, Cell division control; GSH, glutathione; MatE, multiantimicrobial extrusion protein; Redox, protein related to the redox state. [See online article for color version of this figure.]
By contrast, the 22% of induced genes belonged to a large number of gene families. The genes displaying the greatest differences in transcript abundance included those encoding leghemoglobins MtLb1 and MtLb2, nodulins, late nodulins and nodule-specific Cys-rich genes, and many genes involved in carbon and nitrogen metabolism, such as Suc, Gln, and Asn synthetases. Many genes encoding sulfate transporters, multiantimicrobial extrusion proteins, and TGF-β receptors type I/II (including nitrate transporters, oligonucleotide transporters, etc.), probably important for exchanges between the two partners, were up-regulated in nodules, confirming the importance of transport functions in nodule development and function. The observed up-regulation of nitrate transporters is of particular interest and may reflect the existence of a “nitrate respiration” process (Horchani et al., 2011). Finally, a large number of TF-encoding genes not identified in previous analyses were identified here: these genes included those encoding two CCAAT-binding factors (two HAP3-like), one NIN-like, 17 WRKY, and no less than 92 zinc finger proteins (Supplemental Table S11). Overall, about 35% of the genes identified by RNA-Seq as displaying differential regulation between nodules or inoculated roots and noninoculated roots are not present on the Affymetrix Medicago GeneChip. Consequently, additional genetic determinants specific to nodule development and functioning, that are reported here to our knowledge for the first time, can now be deciphered.
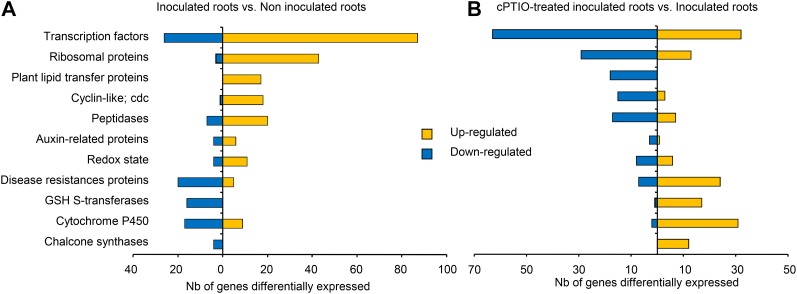
NO is required for optimal establishment of the M. truncatula-S. meliloti symbiotic interaction (del Giudice et al., 2011). Therefore, we characterized the effects of NO depletion on the nodulation transcriptome during the early phases of the symbiosis by carrying out RNA-Seq analysis on inoculated roots treated with the NO scavenger cPTIO. Surprisingly, NO depletion decreased the number of reads pertaining to 5′ and 3′ UTRs and new exons from that observed in inoculated roots and nodules to that for noninoculated roots, indicating a probable role for NO in decreasing transcriptome complexity during nodulation that merits further investigation (Fig. 1; Supplemental Table S2). A comparison of NO-depleted inoculated roots and inoculated roots revealed differential patterns of expression for 2,030 genes (Table III; Supplemental Table S12). The most striking finding was the large number of genes from several gene families for which NO depletion prevented the up-regulation induced by inoculation with the microsymbiont (Fig. 6). These genes included many encoding TFs and several genes encoding proteins involved in nodule development, such as cyclin-like proteins, peptidase, or ribosomal protein families, consistent with the reported effect of NO on the nodulation process (del Giudice et al., 2011). Another interesting finding is the failure of NO-depleted M. truncatula roots to trigger the expression of 11 of the 14 members of the plant lipid transfer protein gene family induced early in nodulation, as the proteins encoded by these genes may participate in the control of bacterial infection and the autoinhibitory regulation of nodule numbers by the host plant (Pii et al., 2009). NO depletion also triggered major changes in hormone signaling, as suggested by the lack of induction of several genes encoding auxin-responsive proteins (Supplemental Fig. S6), and resulted in a lack of activation of genes encoding proteins involved in the cell cycle and protein synthesis, both of which are required for the dedifferentiation of cortical cells and the induction of cell division during nodule formation. Thus, a large number of transcriptional regulators appear to be down-regulated by cPTIO treatment (Supplemental Table S12). One HAP3-like (Medtr1g099630.1) gene appeared to be regulated by NO, possibly accounting in part for the impairment of nodulation by cPTIO (del Giudice et al., 2011). Other TF families known to be involved in nodulation (such as the NIN-like proteins) were not affected, highlighting the specificity of the effect of NO on TF regulation. The genes for which down-regulation in inoculated roots was prevented by NO depletion (Supplemental Table S13) included the gene encoding the NADPH oxidase RBOHC and a large number of genes encoding Tyr kinases, PR proteins, glutathione S-transferases and chalcone synthases, and genes encoding proteins involved in terpene, flavonoid, and phenylpropanoid pathways (Fig. 6; Supplemental Fig. S6C). These findings clearly suggest that NO plays a crucial role in repressing the defense reaction in symbiotic conditions, thereby favoring establishment of the beneficial plant-microbe interaction. This action differs markedly from the signaling functions of NO in pathogenic interactions, in which NO cooperates with reactive oxygen species to induce hypersensitive cell death and functions independently of such intermediates in the induction of defense-related genes (Delledonne et al., 1998). This hypothesis was confirmed, at least partly, by the experiments performed with tungstate; it must be noted here that l-NAME, which inhibits NO synthase-like activity, did not affect the expression level of the defense genes. Taken together, these results shed new light on the possible role of nitrate reductase in the synthesis of NO during the first steps of the nodulation process.
Figure 6.
Comparison of the transcriptional regulation of gene families in inoculated roots treated or not with cPTIO. Only genes showing at least a 2-fold change with a significant difference based on Student’s t test and a FDR obtained with the R package qvalue (Dabney et al., 2010) threshold of q < 0.001 are represented. A, Transcriptional regulation of gene families in inoculated roots compared with roots. B, Transcriptional regulation of gene families in inoculated roots treated for 8 h with 1 mm cPTIO compared with inoculated roots. The classification of each gene in the families is based on the protein function indicated in the Mt3.0 annotation. cdc, Cell division control; GSH, glutathione; Redox, protein related to the redox state. [See online article for color version of this figure.]
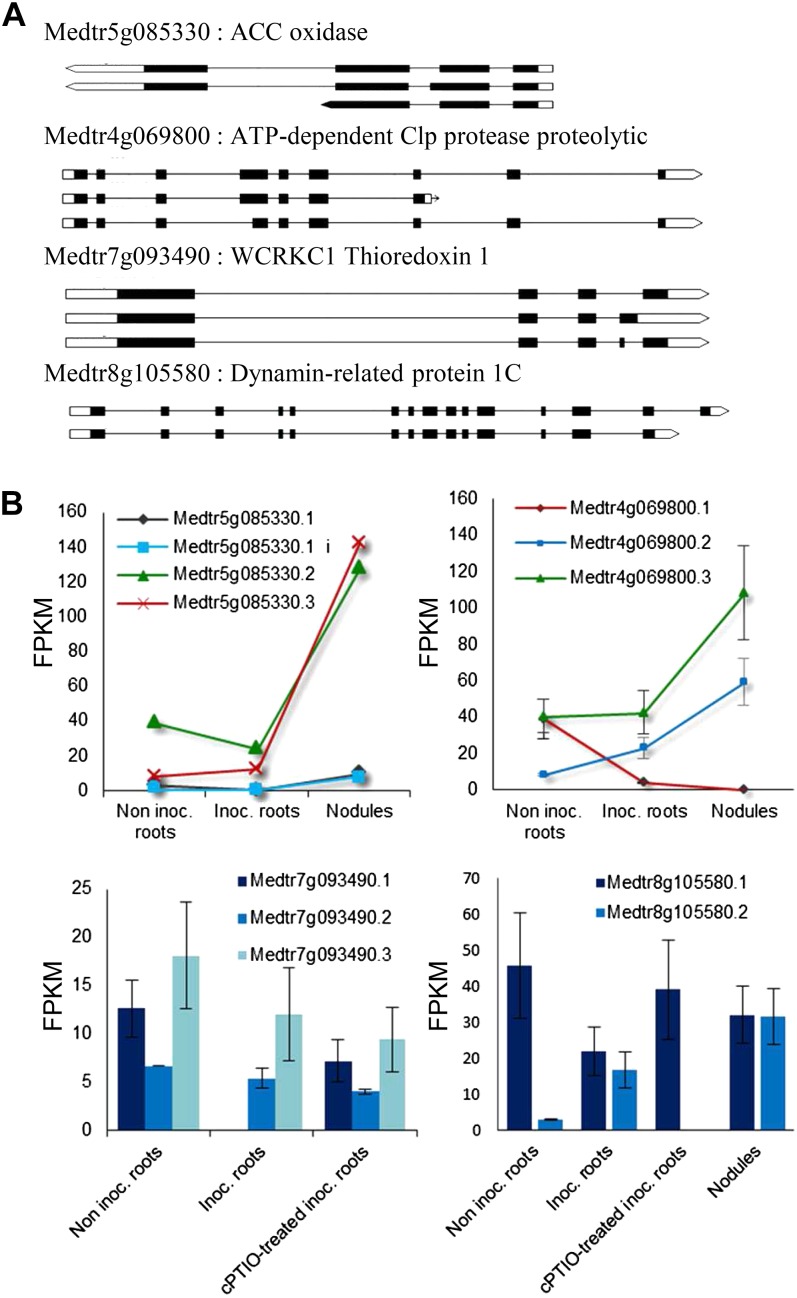
Finally, we assessed the complexity in the nodulation transcriptome by determining the frequency of the various alternatively spliced forms. We identified 23,164 different transcripts derived from 6,587 genes. Although originally thought to be less frequent in plants than in animals, AS is now known to be widespread in plants (Reddy et al., 2012). A recent analysis using RNA-Seq has revealed that over 40% of intron-containing genes in Arabidopsis (Arabidopsis thaliana; Filichkin et al., 2010) and about 48% in rice (Oryza sativa; Lu et al., 2010) undergo AS. Concerning legumes, the only data available are based on EST alignments: the observed frequency of alternatively spliced genes was described to be lower in M. truncatula (approximately 10%) and L. japonicus (approximately 3%) than in Arabidopsis and rice; however, AS frequencies appeared comparable in all four species when EST levels were normalized (Wang et al., 2008). Although it is not known how much of AS is due to noise in the splicing process (Melamud and Moult, 2009), the estimated extent of AS frequency in M. truncatula obtained in this study (6,587 genes) appears significantly higher than the one (1,107 genes) reported previously, based on EST alignments (Wang et al., 2008). This underscores the superior detection capability of the RNA-Seq method over the conventional EST approach, especially for low-abundance transcript variants (Filichkin et al., 2010). Therefore, we concluded that AS may play an important role in increasing the diversity of the M. truncatula proteome. Comparisons of inoculated roots or 21-d-old nodules with noninoculated roots led to the identification of 63 and 65 genes, respectively, 29 of which were common to the two comparisons. Expression analyses showed that several AS events were dynamically modulated, indicating the association of AS events with a specific phase of the nodulation process, confirming recent reports that many splice variants are differentially expressed in a development-specific manner or in response to developmental cues (Reddy et al., 2012). The alternatively spliced forms included four isoforms from a gene encoding a potential 1-aminocyclopropane-1-carboxylate oxidase (Medtr5g085330) involved in ethylene biosynthesis. Three of these isoforms were already known, and the fourth corresponded to an intron retention event. The major isoforms expressed during the nodulation were isoforms 2 and 3 rather than isoform 1 (Fig. 7). For the ATP-dependent Clp protease (Medtr4g069800), isoform 1, which was expressed in noninoculated roots, was totally repressed during symbiotic interaction, whereas the other two isoforms were induced (Fig. 7). Many switches between major and minor transcript isoforms were observed in our analysis, suggesting a probable role of AS in the adaptive changes observed during nodulation. Moreover, cPTIO treatment enhanced splicing (148 genes). For example, changes to the major isoform of dynamin-related protein1C were blocked in NO-depleted inoculated roots (Fig. 7). cPTIO treatment seemed to block the repression of isoform 1 selectively, without affecting the other isoforms, in the case of a gene encoding a thioredoxin-like protein (Medtr7g093490; Fig. 7). These two examples suggest that NO may exert fine control over transcriptome complexity and expression dynamics.
Figure 7.
Transcript abundances of multiple isoform genes regulated during the nodulation process and/or the cPTIO treatment. A, Gene models from the M. truncatula reference genome (version Mt3.5) showing the structure of the isoforms with the positions of the exons, introns, and UTR sequences. B, Relative abundance of transcript isoforms measured as a function of the condition in RPKM. In the case of the 1-aminocyclopropane-1-carboxylate (ACC) oxidase, we observed one unknown isoform (Medtr5g085330.1 i) in which part of an intron is conserved. Inoc., Inoculated roots; Inoc + cPTIO, inoculated roots treated for 8 h with cPTIO.
CONCLUSION
In conclusion, although many genes differentially expressed during nodulation have been identified in previous studies, this investigation of the transcriptome of M. truncatula during establishment of the symbiotic interaction has considerably refined and expanded our knowledge of the basic building blocks required for the colonization and correct functioning of the nodule. The resulting view of the transcriptome, at single-base resolution, markedly improves our understanding of expression dynamics during the symbiotic process and has major biological implications that can now be evaluated experimentally.
MATERIALS AND METHODS
Growth Conditions and Isolation of Total RNA
Sinorhizobium meliloti strain RCR2011 (Rosenberg et al., 1981) was grown in Luria-Bertani medium supplemented with 2.5 mm CaCl2 and 2.5 mm MgSO4. Streptomycin at 100 to 300 μg mL−1 was added when required. Seeds of Medicago truncatula ‘Jemalong A17’ were scarified with H2SO4, surface sterilized in a bleach solution, rinsed with sterile distilled water, germinated on agar plates for 3 d in the dark, and allowed to grow on nitrogen-free Fahraeus medium plates (covered with pouch paper) during 8 or 21 d in a culture room (22°C–25°C) with a light/dark photoperiod of 16 h/8 h (http://www.noble.org/MedicagoHandbook/). Four days after the germinated seeds were transferred onto medium plates, the plants were inoculated with 200 µL per root of S. meliloti grown in liquid Luria-Bertani medium supplemented with 2.5 mm CaCl2 and 2.5 mm MgSO4, pelleted at 4,500 rpm, washed twice with sterile distilled water, and resuspended in sterile distilled water to a final optical density at 600 nm of 0.01 (corresponding to an inoculum concentration of approximately 105 to 106 bacteria cells per mL). The control condition was inoculation with 200 µL of sterile distilled water. Four days post inoculation, some of the plants were treated with 250 µL of a 1 mm solution of cPTIO (Sigma) prepared in 10 mm Tris-KCl buffer that was added along the whole length of the roots. Plants were allowed to stay in contact with cPTIO for 8 h. For the inoculated root condition, roots were treated with 250 µL of 10 mm Tris-KCl buffer for 8 h. Treatments with l-NAME (Sigma) and tungstate (Fluka) were done following the same protocol using 5 and 2 mm, respectively, of each inhibitor of NO synthesis. Furthermore, in order to obtain a higher concentration of RNA from nodule primordia, a 2-cm region of the root in which most of the nodule primordia appear was harvested and immediately frozen in liquid nitrogen. To localize the root infection zone, we marked the position of the primary root apex on the day of the inoculation. Four days later, the infection zone was mainly located around this mark. For the nodule condition, plants grew on a medium plate for 21 d. Nodules were then harvested and immediately frozen in liquid nitrogen. A total of 100 mg of plant material was ground in liquid nitrogen, and total RNA was isolated using QIAshredder and RNeasy Mini kit columns (Qiagen; www.qiagen.com). The optional DNase treatment step was added to the protocol using the RNase-free DNase set (Qiagen). The integrity of total RNA was checked on a 2100 Bioanalyzer (Agilent; www.agilent.com), and its quantity as well as purity were determined with a Nanodrop 2000 apparatus (Thermo Scientific; www.thermofisher.com).
qRT-PCR Validation
For the qRT-PCR validation, cDNAs were made from the RNA isolated either by RNeasy Mini kit columns (as described previously) or Trizol reagent (Invitrogen; www.invitrogen.com). A total of 100 to 200 mg of plant material (root) was ground in liquid nitrogen, and total RNA was isolated using Trizol reagent. The integrity of total RNA was checked on agarose gels, and its quantity as well as purity were determined spectrophotometrically. A total of 500 to 1,500 ng of RNA was used as a template for reverse transcription reaction in a 20-µL reaction volume using the Omniscript RT Kit (Qiagen; www.qiagen.com). Real-time qRT-PCR was carried out using the qPCR Mastermix Plus for SYBR Green I reagent (Eurogentec; www.eurogentec.com). Reactions were run on the Chromo4 Real-Time PCR Detection System (Bio-Rad; www.bio-rad.com), and quantification was performed with the Opticon Monitor analysis software version 3.1 (Bio-Rad). Data were quantified using Opticon Monitor 2 (MJ Research) and analyzed with RqPCRBase, an R package working on the R computing environment for analysis of quantitative real-time PCR data (T. Tran and F. Hilliou, unpublished data). The mRNA levels were normalized against three constitutively expressed endogenous genes, Mtc27 (TC106535; Van de Velde et al., 2006) and two genes (Mtr.10324.1.S1_at and Mtr.31250.1.S1_at) selected as reference genes (del Giudice et al., 2011). PCR for each biological replicate was performed in three technical replicates. For each reaction, 5 µL of 100-fold-diluted cDNA and 0.3 µm primers were used. The initial denaturing time was 10 min, followed by 40 PCR cycles at 95°C for 10 s and 60°C for 1 min. The specificity of the amplification was confirmed by a single peak in a dissociation curve at the end of the PCR procedure. For each experiment, the stability of the reference genes across samples was tested using geNorm software (Vandesompele et al., 2002). The absence of genomic DNA contaminations in the RNA samples was tested by PCR analysis of all samples using oligonucleotides bordering an intron in the glutathione synthetase gene of M. truncatula.
The gene-specific primers used are listed in Supplemental Table S15.
Library Preparation and Sequencing
Poly(A) mRNA was isolated from the extracted RNA to prepare a nondirectional Illumina RNA-Seq library with the mRNA-SEquation 8 sample prep kit (Illumina). We modified the gel extraction step by dissolving excised gel slices at room temperature to avoid underrepresentation of AT-rich sequences (Quail et al., 2008). Library quality control and quantification were performed with a Bioanalyzer Chip DNA 1000 series II (Agilent). Each library had an insert size of 200 bp, and 36- to 44-bp sequences were generated on an Illumina genome analyzer II. The reads were distributed within the four conditions as follows: 16.4 million for the root control, 45.4 million for inoculated roots, 46.6 million for inoculated roots treated with cPTIO, and 27 million for nodules (Supplemental Table S1).
Mapping and Analysis of Illumina Reads
M. truncatula genome and splice database sequence alignments were generated with Bowtie (Langmead et al., 2009). Bowtie supports up to three mismatches, reports reads aligning to multiple locations in the genome, is able to assess the quality of the sequences generated, and its output can be directly parsed by the ERANGE package (Mortazavi et al., 2008) that was used for differential expression analysis. For our analysis, we allowed up to two mismatches (parameter “-v 2”). The sequences were discarded if they aligned on more than 10 different loci (parameter “-m 10”). The program reported all the matches it was able to find for the remaining reads (parameter “-k 11”). Alignment of the reads was made on the Mt3.0 version of the M. truncatula genome sequence (www.medicago.org), and the output was processed using the software suite BedTools (http://code.google.com/p/bedtools/; Quinlan and Hall, 2010) in order to assign each read to an exon, intron, UTR, or intergenic region. Reads mapped onto external exons fell within a 3-kb catchment from both ends of a gene, promoting the investigation of putative undiscovered exons. Intergenic reads represented those sequence reads that fell outside this catchment. The program ERANGE defined potentially novel clusters of expression on the basis of their alignment; they were categorized as novel sections (exons/UTRs) of a known gene if they fell inside a radius of 3,000 bp from them (average gene density/2). The remaining expressed clusters were marked as potential new genes.
In order to identify potential new isoforms of known genes, we remapped all reads against the M. truncatula genome using TopHat (Trapnell et al., 2010), with a segment length of 16 due to the short length of our reads, and defined the new isoforms of known genes by performing a Cufflinks analysis on each sample, with standard parameters, followed by an analysis with Cuffcompare to merge transcripts identified on different samples (Trapnell et al., 2010). We used the latest genome sequence and annotations provided by the M. truncatula research community (Mt3.5; http://www.medicago.org/). The Mt3.0 version consists of essentially the same sequence data found in the more recent Mt3.5 version, except that the assembly of Mt3.5 was based on newer optical map results (Branca et al., 2011; Young et al., 2011). Therefore, we used the Mt3.5 version only for the identification of AS events.
Clustering
The expression data of the 28,744 genes expressed (in RPKM) were transformed with the software Expression Profiler from the European Bioinformatics Institute (www.ebi.ac.uk) to get a relative expression ratio (in log2 base) around the gene’s average value. The clustering was made following the K-means method with Pearson’s correlation distance using the Genesis bioinformatic tool (Sturn et al., 2002; http://genome.tugraz.at). Different numbers of clusters were tested to identify the minimum number of clusters to represent all the separated expression profiles.
Identification of Novel Transcribed Regions
We used the reads that had not been mapped against the Mt3.0 sequence from every sample to assemble separately our contigs, using the Velvet program (Zerbino and Birney, 2008), using a sensitive hash length of 29 for the reads with a length of 44 bp and of 21 for the rest. The contigs were subsequently clustered together using the software CAP3 (Huang and Madan, 1999), with a minimum overlap of 90%, requiring an overlap identity of 80%. Contigs mapping against the reference genome with identity of 90% or greater and coverage of 90% or greater after BLAT alignment were discarded from further analysis. All the contigs were also mapped against the accompanying RNA-Seq data of the Mt3.5 version with GMAP (version 2012-04-21). The contigs, with an alignment coverage on the sequence length of 90% or greater and on the identity of 90% or greater, were merged together using the program mergeBed from the BEDTools suite (http://code.google.com/p/bedtools/).
Gene Expression Analysis
The evaluation of gene expression was performed with the ERANGE software (version 3.1) available at http://woldlab.caltech.edu/RNA-Seq (Mortazavi et al., 2008). ERANGE requires Cistematic 2.5 to execute RunStandardanalysis.sh. Therefore, a Python script (medicago3.py) was developed to import the M. truncatula reference sequence (Mt3.0) and gff annotation into Cistematics Genomes, and a Perl script (gff2knowngene.pl; Zenoni et al., 2010) was used to convert the gff annotation file to the knowngene.txt file used by RunStandardanalysis.sh. ERANGE reports the number of mapped reads per kilobase of exon per million mapped reads, measuring the transcriptional activity for each gene. To obtain an accurate measure of gene expression not biased by reads mapping to splice junctions in genes with many introns, ERANGE considers both reads mapping to the genome or to the custom splice junctions database. ERANGE was preferred over commercial packages such as CASAVA and the GenomeStudio platform from Illumina because of its open nature. This allowed us to adapt and reuse code for our own analysis with greater flexibility than a comparable closed-source commercial package.
Differential Gene Expression Statistics for RNA-Seq
ERANGE software computes the normalized gene locus expression level (named RPKM) by assigning reads to their site of origin and counting them. In the case of reads that match equally well to several sites, ERANGE assigns them proportionally to their most likely site(s) of origin (Mortazavi et al., 2008). The number of reads falling in a given gene locus can be estimated from the RPKM value as follows: n = RPKM × L × NTot × 10−9, where n = number of mapping reads at a given gene locus, L = estimated length (bp) of the gene locus, NTot = number of total mapping reads, and RPKM = gene locus RPKM value. The null hypothesis of no differential gene expression for each gene was tested using the R package qvalue (Storey, 2002; Storey and Tibshirani, 2003; Dabney et al., 2010) on the R working environment. FDRs were calculated based on P values obtained running Student’s t test on the raw read counts using the basic R package stats.
The threshold value for the FDR was 0.001, and genes were first selected using this filter. Differentially expressed genes were then filtered again based on a fold change greater than 2.
Construction of Promoter Fusion, and GUS Detection in Planta
To analyze promoter activity, 1,567 and 1,330 bp of the genomic region upstream from the coding regions of glutathione S-transferase (Medtr7g071380) and chalcone synthase (Medtr1g124600), respectively, were amplified by PCR with the following primers: 5′-AAAAAGCAGGCTTCCGTGACTTATCCCATTTATGTG-3′ (GST F), 5′-AGAAAGCTGGGTGCTTCAATTCTTAATCAAACAACTG-3′ (GST R), 5′-AAAAAGCAGGCTTCGCACCAACACTTCTAATTGA-3′ (CHS F), and 5′-GAAAGCTGGGTGGGTAGCAAATGAAAGTCTCAG-3′ (CHS R). The amplified fragments were reamplified with the Gateway adapter primers 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCT-3′ and 5′-GGGGACCACTTTGTACAAGAAAGCTGGGT-3′, cloned into the pDONR207 Gateway vector (Invitrogen), and then transferred to the GUS::GFP-containing binary vector pKGWFS7 (Karimi et al., 2002), generating the plasmids ProGST::GUS and ProCHS::GUS.
For GUS detection, roots transformed with ProGST::GUS and ProCHS::GUS were incubated for 1 h in 90% acetone (diluted in a phosphate buffer: 0.1 m Na2HPO4 and 0.1 m NaH2PO4, pH 7.4) at −20°C. Roots were then washed twice in the phosphate buffer and incubated at room temperature, protected from light, in phosphate buffer containing potassium ferricyanure (0.5 mm), potassium ferrocyanure (0.5 mm), and 5-bromo-4-chloro-3-indolyl-β-glucuronic acid (0.5 ng mL−1) for 3 and 16 h. The roots were observed by optical microscopy (Axioplan imagin2; Zeiss). Photographs were taken with an AxioCam (Zeiss) and acquired with the corresponding software.
All Illumina sequence data have been deposited in the National Center for Biotechnology Information short-read archive, and Sanger-sequenced PCR products have been deposited in GenBank (accession no. SRA048731). Assembled contigs longer than 200 bp have been deposited at Transcriptome Shotgun Assembly (accession nos. JR366937–JR375780). Coverage data are available at http://ddlab.sci.univr.it/cgi-bin/gbrowse/medicago/.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Photographs of the harvested plants at 4 d post inoculation.
Supplemental Figure S2. Percentage of TUs detected in both replicates at different RPKM thresholds.
Supplemental Figure S3. Number of genes detected with five or more uniquely mapped reads as a function of the sequencing depth.
Supplemental Figure S4. Comparison of cPTIO and two different NO synthesis inhibitors treatments on the transcript level on six genes.
Supplemental Figure S5. Histochemical localization of GUS activity in transgenic roots expressing ProGST::GUS and ProCHS::GUS fusion.
Supplemental Figure S6. Overview of biotic stress pathway regulated during the early organogenesis and by the cPTIO treatment using MapMan.
Supplemental Figure S7. Overview of general metabolism regulated by the cPTIO treatment during the early organogenesis using MapMan.
Supplemental Table S1. Number of reads obtained for each sample in the experiment.
Supplemental Table S2. List of new transcribed regions identified.
Supplemental Table S3. TUs newly identified mapped on the Mt3.0 genome version (named FAR).
Supplemental Table S4. Cuff tracking data of all transcripts from Cufflinks analysis.
Supplemental Table S5. Isoforms.fpkm.tracking data of noninoculated roots versus inoculated roots.
Supplemental Table S6. Isoforms.fpkm.tracking data of noninoculated roots versus nodules.
Supplemental Table S7. Isoforms.fpkm.tracking data of inoculated roots versus cPTIO-treated inoculated roots.
Supplemental Table S8. TUs expressed in the different condition above 2.77 RPKM.
Supplemental Table S9. Validation of RNA-Seq expression results by qRT-PCR.
Supplemental Table S10. TUs differentially expressed in inoculated roots compared with noninoculated roots.
Supplemental Table S11. TUs differentially expressed in nodules compared with noninoculated roots.
Supplemental Table S12. TUs differentially expressed in cPTIO-treated inoculated roots compared with inoculated roots.
Supplemental Table S13. TUs differentially expressed either by inoculation or cPTIO treatment.
Supplemental Table S14. Differential expression analysis of defense genes families affected by cPTIO treatment.
Supplemental Table S15. Primer sequences used for real-time qPCR analysis.
Acknowledgments
We thank Pascal Gamas for useful comments on the manuscript and Sébastien Gucciardo for his dedicated technical assistance. We also thank Giovanni Malerba for support with statistical analysis and William Rhamey for English proofreading of the manuscript.
Glossary
- NO
nitric oxide
- TUs
transcription units
- cPTIO
2-(4-carboxyphenyl)-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide
- UTR
untranslated region
- RPKM
reads per kilobase per million mapped reads
- qRT
quantitative reverse transcription
- FDR
false discovery rate
- l-NAME
l-NG-nitro-arginine methyl ester
- TF
transcription factor
- AS
alternative splicing
- qRT
quantitative real-time
References
- Bauer P, Munkert J, Brydziun M, Burda E, Müller-Uri F, Gröger H, Muller YA, Kreis W. (2010) Highly conserved progesterone 5 beta-reductase genes (P5 beta R) from 5 beta-cardenolide-free and 5 beta-cardenolide-producing angiosperms. Phytochemistry 71: 1495–1505 [DOI] [PubMed] [Google Scholar]
- Becker A, Bergès H, Krol E, Bruand C, Rüberg S, Capela D, Lauber E, Meilhoc E, Ampe F, de Bruijn FJ, et al. (2004) Global changes in gene expression in Sinorhizobium meliloti 1021 under microoxic and symbiotic conditions. Mol Plant Microbe Interact 17: 292–303 [DOI] [PubMed] [Google Scholar]
- Benedito VA, Li H, Dai X, Wandrey M, He J, Kaundal R, Torres-Jerez I, Gomez SK, Harrison MJ, Tang Y, et al. (2010) Genomic inventory and transcriptional analysis of Medicago truncatula transporters. Plant Physiol 152: 1716–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedito VA, Torres-Jerez I, Murray JD, Andriankaja A, Allen S, Kakar K, Wandrey M, Verdier J, Zuber H, Ott T, et al. (2008) A gene expression atlas of the model legume Medicago truncatula. Plant J 55: 504–513 [DOI] [PubMed] [Google Scholar]
- Blanchoin L, Staiger CJ. (2010) Plant formins: diverse isoforms and unique molecular mechanism. Biochim Biophys Acta 1803: 201–206 [DOI] [PubMed] [Google Scholar]
- Branca A, Paape TD, Zhou P, Briskine R, Farmer AD, Mudge J, Bharti AK, Woodward JE, May GD, Gentzbittel L, et al. (2011) Whole-genome nucleotide diversity, recombination, and linkage disequilibrium in the model legume Medicago truncatula. Proc Natl Acad Sci USA 108: E864–E870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewin NJ. (2004) Plant cell wall remodelling in the rhizobium-legume symbiosis. Crit Rev Plant Sci 23: 293–316 [Google Scholar]
- Capela D, Filipe C, Bobik C, Batut J, Bruand C. (2006) Sinorhizobium meliloti differentiation during symbiosis with alfalfa: a transcriptomic dissection. Mol Plant Microbe Interact 19: 363–372 [DOI] [PubMed] [Google Scholar]
- Chang C, Damiani I, Puppo A, Frendo P. (2009) Redox changes during the legume-rhizobium symbiosis. Mol Plant 2: 370–377 [DOI] [PubMed] [Google Scholar]
- Chen H, McCaig BC, Melotto M, He SY, Howe GA. (2004) Regulation of plant arginase by wounding, jasmonate, and the phytotoxin coronatine. J Biol Chem 279: 45998–46007 [DOI] [PubMed] [Google Scholar]
- Combier JP, Frugier F, de Billy F, Boualem A, El-Yahyaoui F, Moreau S, Vernié T, Ott T, Gamas P, Crespi M, et al. (2006) MtHAP2-1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula. Genes Dev 20: 3084–3088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespi MD, Jurkevitch E, Poiret M, d’Aubenton-Carafa Y, Petrovics G, Kondorosi E, Kondorosi A. (1994) enod40, a gene expressed during nodule organogenesis, codes for a non-translatable RNA involved in plant growth. EMBO J 13: 5099–5112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabney A, Storey JD, Warnes GR. (2010) qvalue: Q-value estimation for false discovery rate control. R package version 1.24.0. http://CRAN.R-project.org/package=qvalue (October 26, 2010)
- del Giudice J, Cam Y, Damiani I, Fung-Chat F, Meilhoc E, Bruand C, Brouquisse R, Puppo A, Boscari A. (2011) Nitric oxide is required for an optimal establishment of the Medicago truncatula-Sinorhizobium meliloti symbiosis. New Phytol 191: 405–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delledonne M, Xia Y, Dixon RA, Lamb C. (1998) Nitric oxide functions as a signal in plant disease resistance. Nature 394: 585–588 [DOI] [PubMed] [Google Scholar]
- Durner J, Wendehenne D, Klessig DF. (1998) Defense gene induction in tobacco by nitric oxide, cyclic GMP, and cyclic ADP-ribose. Proc Natl Acad Sci USA 95: 10328–10333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Yahyaoui F, Küster H, Ben Amor B, Hohnjec N, Pühler A, Becker A, Gouzy J, Vernié T, Gough C, Niebel A, et al. (2004) Expression profiling in Medicago truncatula identifies more than 750 genes differentially expressed during nodulation, including many potential regulators of the symbiotic program. Plant Physiol 136: 3159–3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson BJ, Ross JJ, Reid JB. (2005) Nodulation phenotypes of gibberellin and brassinosteroid mutants of pea. Plant Physiol 138: 2396–2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filichkin SA, Priest HD, Givan SA, Shen R, Bryant DW, Fox SE, Wong WK, Mockler TC. (2010) Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res 20: 45–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamas P, Niebel FdeC, Lescure N, Cullimore J. (1996) Use of a subtractive hybridization approach to identify new Medicago truncatula genes induced during root nodule development. Mol Plant Microbe Interact 9: 233–242 [DOI] [PubMed] [Google Scholar]
- Graham PH, Vance CP. (2003) Legumes: importance and constraints to greater use. Plant Physiol 131: 872–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horchani F, Prévot M, Boscari A, Evangelisti E, Meilhoc E, Bruand C, Raymond P, Boncompagni E, Aschi-Smiti S, Puppo A, et al. (2011) Both plant and bacterial nitrate reductases contribute to nitric oxide production in Medicago truncatula nitrogen-fixing nodules. Plant Physiol 155: 1023–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Madan A. (1999) CAP3: a DNA sequence assembly program. Genome Res 9: 868–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innocenti G, Pucciariello C, Le Gleuher M, Hopkins J, de Stefano M, Delledonne M, Puppo A, Baudouin E, Frendo P. (2007) Glutathione synthesis is regulated by nitric oxide in Medicago truncatula roots. Planta 225: 1597–1602 [DOI] [PubMed] [Google Scholar]
- Jones KM, Kobayashi H, Davies BW, Taga ME, Walker GC. (2007) How rhizobial symbionts invade plants: the Sinorhizobium-Medicago model. Nat Rev Microbiol 5: 619–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KM, Sharopova N, Lohar DP, Zhang JQ, VandenBosch KA, Walker GC. (2008) Differential response of the plant Medicago truncatula to its symbiont Sinorhizobium meliloti or an exopolysaccharide-deficient mutant. Proc Natl Acad Sci USA 105: 704–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. (2002) Gateway vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long SR. (2001) Genes and signals in the rhizobium-legume symbiosis. Plant Physiol 125: 69–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu TT, Lu GJ, Fan DL, Zhu CR, Li W, Zhao QA, Feng Q, Zhao Y, Guo YL, Li WJ, et al. (2010) Function annotation of the rice transcriptome at single-nucleotide resolution by RNA-seq. Genome Res 20: 1238–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh JF, Rakocevic A, Mitra RM, Brocard L, Sun J, Eschstruth A, Long SR, Schultze M, Ratet P, Oldroyd GED. (2007) Medicago truncatula NIN is essential for rhizobial-independent nodule organogenesis induced by autoactive calcium/calmodulin-dependent protein kinase. Plant Physiol 144: 324–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maunoury N, Redondo-Nieto M, Bourcy M, Van de Velde W, Alunni B, Laporte P, Durand P, Agier N, Marisa L, Vaubert D, et al. (2010) Differentiation of symbiotic cells and endosymbionts in Medicago truncatula nodulation are coupled to two transcriptome-switches. PLoS ONE 5: e9519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medicago Genome Sequence Consortium. (2007) Medicago truncatula genome “Mt2.0” release whitepaper. README: prerelease of Medicago truncatula genome sequences. http://medicago.org/genome/downloads/Mt2 (August 10, 2007) [Google Scholar]
- Melamud E, Moult J. (2009) Stochastic noise in splicing machinery. Nucleic Acids Res 37: 4873–4886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra RM, Long SR. (2004) Plant and bacterial symbiotic mutants define three transcriptionally distinct stages in the development of the Medicago truncatula/Sinorhizobium meliloti symbiosis. Plant Physiol 134: 595–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau S, Verdenaud M, Ott T, Letort S, de Billy F, Niebel A, Gouzy J, de Carvalho-Niebel F, Gamas P. (2011) Transcription reprogramming during root nodule development in Medicago truncatula. PLoS ONE 6: e16463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Oldroyd GE, Downie JA. (2008) Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu Rev Plant Biol 59: 519–546 [DOI] [PubMed] [Google Scholar]
- Palmieri MC, Sell S, Huang X, Scherf M, Werner T, Durner J, Lindermayr C. (2008) Nitric oxide-responsive genes and promoters in Arabidopsis thaliana: a bioinformatics approach. J Exp Bot 59: 177–186 [DOI] [PubMed] [Google Scholar]
- Pii Y, Astegno A, Peroni E, Zaccardelli M, Pandolfini T, Crimi M. (2009) The Medicago truncatula N5 gene encoding a root-specific lipid transfer protein is required for the symbiotic interaction with Sinorhizobium meliloti. Mol Plant Microbe Interact 22: 1577–1587 [DOI] [PubMed] [Google Scholar]
- Quail MA, Kozarewa I, Smith F, Scally A, Stephens PJ, Durbin R, Swerdlow H, Turner DJ. (2008) A large genome center’s improvements to the Illumina sequencing system. Nat Methods 5: 1005–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy AS, Rogers MF, Richardson DN, Hamilton M, Ben-Hur A. (2012) Deciphering the plant splicing code: experimental and computational approaches for predicting alternative splicing and splicing regulatory elements. Front Plant Sci 3: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg C, Boistard P, Dénarié J, Casse-Delbart F. (1981) Genes controlling early and late functions in symbiosis are located on a megaplasmid in Rhizobium meliloti. Mol Gen Genet 184: 326–333 [DOI] [PubMed] [Google Scholar]
- Shimoda Y, Nagata M, Suzuki A, Abe M, Sato S, Kato T, Tabata S, Higashi S, Uchiumi T. (2005) Symbiotic rhizobium and nitric oxide induce gene expression of non-symbiotic hemoglobin in Lotus japonicus. Plant Cell Physiol 46: 99–107 [DOI] [PubMed] [Google Scholar]
- Starker CG, Parra-Colmenares AL, Smith L, Mitra RM, Long SR. (2006) Nitrogen fixation mutants of Medicago truncatula fail to support plant and bacterial symbiotic gene expression. Plant Physiol 140: 671–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD. (2002) A direct approach to false discovery rates. J R Stat Soc Ser B Stat Methodol 64: 479–498 [Google Scholar]
- Storey JD, Tibshirani R. (2003) Statistical significance for genomewide studies. Proc Natl Acad Sci USA 100: 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturn A, Quackenbush J, Trajanoski Z. (2002) Genesis: cluster analysis of microarray data. Bioinformatics 18: 207–208 [DOI] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Velde W, Guerra JC, De Keyser A, De Rycke R, Rombauts S, Maunoury N, Mergaert P, Kondorosi E, Holsters M, Goormachtig S. (2006) Aging in legume symbiosis: a molecular view on nodule senescence in Medicago truncatula. Plant Physiol 141: 711–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang BB, O’Toole M, Brendel V, Young ND. (2008) Cross-species EST alignments reveal novel and conserved alternative splicing events in legumes. BMC Plant Biol 8: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J, Prell J, James EK, Poole P. (2007) Nutrient sharing between symbionts. Plant Physiol 144: 604–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm BT, Marguerat S, Watt S, Schubert F, Wood V, Goodhead I, Penkett CJ, Rogers J, Bähler J. (2008) Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature 453: 1239–1243 [DOI] [PubMed] [Google Scholar]
- Young ND, Debellé F, Oldroyd GED, Geurts R, Cannon SB, Udvardi MK, Benedito VA, Mayer KFX, Gouzy J, Schoof H, et al. (2011) The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480: 520–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenoni S, Ferrarini A, Giacomelli E, Xumerle L, Fasoli M, Malerba G, Bellin D, Pezzotti M, Delledonne M. (2010) Characterization of transcriptional complexity during berry development in Vitis vinifera using RNA-Seq. Plant Physiol 152: 1787–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. (2008) Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18: 821–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Subramanian S, Zhang Y, Yu O. (2007) Flavone synthases from Medicago truncatula are flavanone-2-hydroxylases and are important for nodulation. Plant Physiol 144: 741–751 [DOI] [PMC free article] [PubMed] [Google Scholar]