Abstract
The molecular basis of primary wall extension endures as one of the central enigmas in plant cell morphogenesis. Classical cell wall models suggest that xyloglucan endo-transglycosylase activity is the primary catalyst (together with expansins) of controlled cell wall loosening through the transient cleavage and religation of xyloglucan-cellulose cross links. The genome of Arabidopsis (Arabidopsis thaliana) contains 33 phylogenetically diverse XYLOGLUCAN ENDO-TRANSGLYCOSYLASE/HYDROLASE (XTH) gene products, two of which were predicted to be predominant xyloglucan endohydrolases due to clustering into group III-A. Enzyme kinetic analysis of recombinant AtXTH31 confirmed this prediction and indicated that this enzyme had similar catalytic properties to the nasturtium (Tropaeolum majus) xyloglucanase1 responsible for storage xyloglucan hydrolysis during germination. Global analysis of Genevestigator data indicated that AtXTH31 and the paralogous AtXTH32 were abundantly expressed in expanding tissues. Microscopy analysis, utilizing the resorufin β-glycoside of the xyloglucan oligosaccharide XXXG as an in situ probe, indicated significant xyloglucan endohydrolase activity in specific regions of both roots and hypocotyls, in good correlation with transcriptomic data. Moreover, this hydrolytic activity was essentially completely eliminated in AtXTH31/AtXTH32 double knockout lines. However, single and double knockout lines, as well as individual overexpressing lines, of AtXTH31 and AtXTH32 did not demonstrate significant growth or developmental phenotypes. These results suggest that although xyloglucan polysaccharide hydrolysis occurs in parallel with primary wall expansion, morphological effects are subtle or may be compensated by other mechanisms. We hypothesize that there is likely to be an interplay between these xyloglucan endohydrolases and recently discovered apoplastic exo-glycosidases in the hydrolytic modification of matrix xyloglucans.
The molecular basis of primary wall extension endures as one of the central enigmas in plant cell morphogenesis (Cosgrove, 2005; Geitmann and Ortega, 2009; Jarvis, 2009; Torres et al., 2009). Classical models of the primary cell wall in land plants describe the wall as a composite structure composed of load-bearing, semicrystalline cellulose fibrils surrounded by an amorphous matrix of cross-linking glycans (hemicelluloses and pectins), structural (glyco)proteins, and, in some cases, polyphenolics (Carpita and Gibeaut, 1993; Carpita and McCann, 2000; Cosgrove, 2005; Jarvis, 2009). Moreover, primary plant cell walls have a high water content (approximately 80% in primary walls; Jarvis, 2009), which is responsible for maintaining the structure in a dynamic, hydrogel-like state (Ha et al., 1997; Zwieniecki et al., 2001; Jarvis, 2009). Contemporary structural analysis, using increasingly sophisticated probes, is refining our understanding of the exquisite spatial localization of cell wall polymers (Knox, 2008; Yarbrough et al., 2009). Genetic and biochemical approaches are at present uncovering the individual enzymes responsible for building these macromolecules, and models of coordinated supramolecular assembly at the plasma membrane continue to evolve (Geisler et al., 2008; Liepman et al., 2010). Lastly, there is a growing interest in the roles of endogenous glycoside hydrolases in primary plant cell wall remodeling (Vicente et al., 2007; Lopez-Casado et al., 2008; Minic, 2008). Nonetheless, we are far from having a holistic model of cell wall development for even a single wall or tissue type (Somerville et al., 2004).
The pectins notwithstanding, the dominant matrix glycans in the primary cell walls of the majority of land plants (dicots, noncommelinoid monocots, and gymnosperms) are the xyloglucans (XGs), which may constitute up to one-fourth of the wall dry weight. In contrast, the commelinoid monocots, grasses in particular, contain comparatively low levels of XG (1%–5% in the primary wall), which is substituted by glucuronoarabinoxylans and mixed linkage glucans (Carpita and Gibeaut, 1993; Carpita and McCann, 2000; Vogel, 2008; Hsieh and Harris, 2009). The broad distribution of XGs in the plant kingdom (Carpita and McCann, 2000) has spurred a large body of research on the taxonomy of XG fine structures (Hoffman et al., 2005; Hsieh and Harris, 2009) and the nature of the cellulose-XG interaction (Zhou et al., 2007; Zhang et al., 2011, and refs. therein). Indeed, many contemporary models implicate XGs as the main hemicelluloses, which contribute to the strength of the primary wall by cross linking cellulose fibrils (Pauly et al., 1999; Carpita and McCann, 2000; Cosgrove, 2005), although the proposed distance over which this cross linking is thought to occur has recently been called into question (Cosgrove and Jarvis, 2012; Park and Cosgrove, 2012).
Primary cell wall extension necessarily requires relaxation of the cellulose-matrix glycan/pectin network under turgor-induced strain. Xyloglucan endo-transglycosylases (XETs; EC 2.4.1.207) are believed to play a key role in this process, due to the ability of these enzymes to catalyze the nonhydrolytic cleavage and rearrangement of XGs (Rose et al., 2002). XETs have thus been suggested to mediate transient wall creep by breaking XG cross links under tension, followed by chain ligation to generate a new cross link (Rose et al., 2002). The validity of this dogmatic model, however, has been questioned due to a lack of experimental evidence (Cosgrove, 2000, 2005; Kutschera, 2001). Indeed, XET activity has been directly observed in myriad plant tissues, and both wall-strengthening and wall-loosening effects have been ascribed (Cosgrove, 2000, 2005; Eklöf and Brumer, 2010, and refs. therein).
Since the landmark discovery of XET activity in the early 1990s (Farkas et al., 1992; Fry et al., 1992; Nishitani and Tominaga, 1992), these enzymes are now known to be encoded by large XYLOGLUCAN ENDO-TRANSGLYCOSYLASE/HYDROLASE (XTH) gene families present in diverse land plant species, including both monocots and dicots (Eklöf and Brumer, 2010). As the name suggests, a small number of XTH gene products have demonstrable xyloglucan endo-hydrolase (XEH; EC 3.2.1.151) activity (for review, see Eklöf and Brumer, 2010). Based on detailed enzyme structure-function and protein phylogenetic analyses, we have previously suggested that predominant XEHs are segregated into a subdivision of the historical phylogenetic group III, called group III-A (Baumann et al., 2007). In particular, members of group III-A include both the archetypal nasturtium (Tropaeolum majus) xyloglucanase1 (TmNXG1), which catalyzes the hydrolysis of storage XG during seed germination, and proteins encoded by ESTs during the ripening of various fruits (Baumann et al., 2007). Intriguingly, surveys of available genomes revealed the presence of at least one XTH group III-A member in all higher land plants, including both dicots and cereal grasses (Del Bem and Vincentz, 2010; Eklöf and Brumer, 2010). The potential functions of these predicted enzymes remain enigmatic, especially in those species that lack predominant storage xyloglucans or fleshy fruits.
In this study, we have elaborated on this previous work through the functional characterization of two Arabidopsis (Arabidopsis thaliana) group III-A homologs, AtXTH31 and AtXTH32, which are expressed in expanding vegetative tissues. Heterologous production and detailed kinetic analysis confirmed that the AtXTH31 gene product was indeed a predominant XEH, in keeping with the phylogenetic prediction of enzyme activity. Although single and double knockout lines, as well as overexpressing lines, of AtXTH31 and AtXTH32 failed to demonstrate significant growth phenotypes, XEH activity was severely reduced in diverse tissues by gene disruption. These findings suggest that matrix xyloglucan hydrolysis occurs in parallel with primary cell wall expansion and that specific XEHs may have integral roles in wall assembly and disassembly.
RESULTS
AtXTH31 and AtXTH32 Encode the Two Group III-A Proteins of Arabidopsis
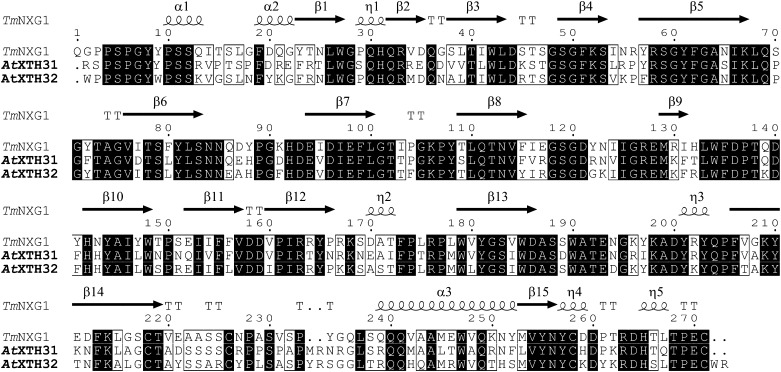
Thirty-three members of the XTH gene family have been found previously (Yokoyama and Nishitani, 2001) in the genome of Arabidopsis (Arabidopsis Genome Initiative, 2000). Of these, only two, AtXTH31 (locus At3g44990) and AtXTH32 (locus At2g36870), encode proteins belonging to group III-A (Eklöf and Brumer, 2010) in the updated phylogeny proposed by Baumann et al. (2007). Protein sequence alignment of the AtXTH31 and AtXTH32 gene products with TmNXG1, whose three-dimensional structure has been solved (Protein Data Bank identifier 2uwa; Baumann et al., 2007), clearly demonstrates the structural similarity of these group III-A members (Fig. 1). AtXTH31 and AtXTH32 have 66% and 69% identities with TmNXG1, respectively (82% and 83% similarities, respectively), whereas their identities with the group I representative, Populus tremula × Populus tremuloides XET16-34 (Johansson et al., 2004; Kallas et al., 2005), drop to 35% and 37% identities, respectively (50% and 52% similarities, respectively).
Figure 1.
Protein sequence alignment of TmNXG1 (GenBank accession no. CAA48324), AtXTH31 (GenBank accession nos. AAL07012 and At3g44990), and AtXTH32 (GenBank accession nos. AAD31572 and At2g36870) of group III-A. Identical residues are indicated by black boxes, and similar residues are indicated by white boxes. Secondary structural elements from the experimentally determined tertiary structure of TmNXG1 (Baumann et al., 2007) are indicated above the alignment. The figure was produced with ESPript (Gouet et al., 2003).
AtXTH31 and AtXTH32 were thus predicted to encode proteins with predominant XEH (EC 3.2.1.151; i.e. endoxyloglucanase) activity. In particular, both Arabidopsis homologs possess the long variant of loop 3 previously observed in TmNXG1, which is typical of group III-A members and which has been shown to increase the hydrolysis-to-transglycosylation ratio in these enzymes (Baumann et al., 2007).
Heterologous Expression of AtXTH31 and AtXTH32
Recombinant expression of AtXTH31 in the yeast Pichia pastoris SMD1168H yielded soluble protein secreted to the medium under the guidance of the yeast α-factor secretion signal peptide, which replaced the predicted native signal peptide (the full construct sequence is given in Supplemental Fig. S1). Purification on a xyloglucan oligosaccharide affinity column yielded apparently pure protein with a molar mass of approximately 34 kD, as judged by SDS-PAGE (Supplemental Fig. S2A). Western-blot analysis using an anti-His6 IgG indicated that the full-length protein was produced, which contained the c-myc and hexa-His tags in series at the C terminus (Supplemental Fig. S2B). However, high-resolution intact protein mass spectrometry (Supplemental Fig. S3) indicated that a significant amount of cleavage occurred at the Kex2 protease site located between the c-myc and His6 tags; the preparation was thus a mixture of two predominant protein forms (compare with Supplemental Fig. S1; EAEA-AtXTH31-c-myc-His6, average mass calculated at 34,490.2 D, observed at 3,4491 D, 50% base peak intensity; EAEA-AtXTH31-LEQKLISEE, average mass calculated at 32,952.7 D, observed at 32,954 D, base peak). Like all other group III-A members, AtXTH31 lacks a consensus N-glycosylation site (Eklöf and Brumer, 2010) and, therefore, was not N-glycosylated, as revealed by the protein mass spectrum; O-glycosylation was similarly not indicated.
Despite several attempts, including the use of codon-optimized complementary DNA (cDNA), recombinant production of AtXTH32 in P. pastoris was unsuccessful. mRNA levels were comparable to TmNXG1-producing control strains (data not shown), thus hinting toward a problem with translation or protein folding. Similarly, we have previously observed an approximately 50% success rate for the heterologous expression of diverse XTH genes in P. pastoris (Kaewthai et al., 2010). Transient expression of AtXTH32 in Nicotiana benthamiana (Sainsbury et al., 2009) was likewise unsuccessful.
Enzyme Kinetic Analysis of AtXTH31
The bicinchoninic acid (BCA) reducing-sugar assay indicated that AtXTH31 cleaved 1 g L–1 tamarind (Tamarindus indica) seed xyloglucan (York et al., 1993) with a specific activity of 4.4 ± 0.1 mol min–1 mol–1 protein at the pH optimum of 4.75 (pH rate data not shown). In comparison, our previous work indicated that the nasturtium seed xyloglucanase TmNXG1 exhibited a specific activity toward tamarind xyloglucan of 5.5 ± 0.5 mol min–1 mol–1 protein (substrate concentration [S] = 1 g L–1; pH optimum of 4.8; Baumann et al., 2007). High-performance size-exclusion chromatography with evaporative light-scattering detection (HPSEC-ELS) analysis (data not shown) demonstrated that AtXTH31 is an endo-acting enzyme, which significantly decreased the molar mass of xyloglucan prior to the appearance of the limit digestion products XXXG, XLXG, XXLG, and XLLG (for high-performance anion-exchange chromatography with pulsed amperometric detection [HPAEC-PAD], see Supplemental Fig. S4; for oligosaccharide nomenclature, see Fry et al. [1993]). AtXTH31 had no detectable activity toward carboxymethyl cellulose, medium-viscosity barley (Hordeum vulgare) β-glucan, wheat (Triticum aestivum) arabinoxylan, carob (Ceratonia siliqua) galactomannan, lupin (Lupinus albus) galactan, or hydroxyethyl cellulose (all at 1 g L–1) in overnight assays.
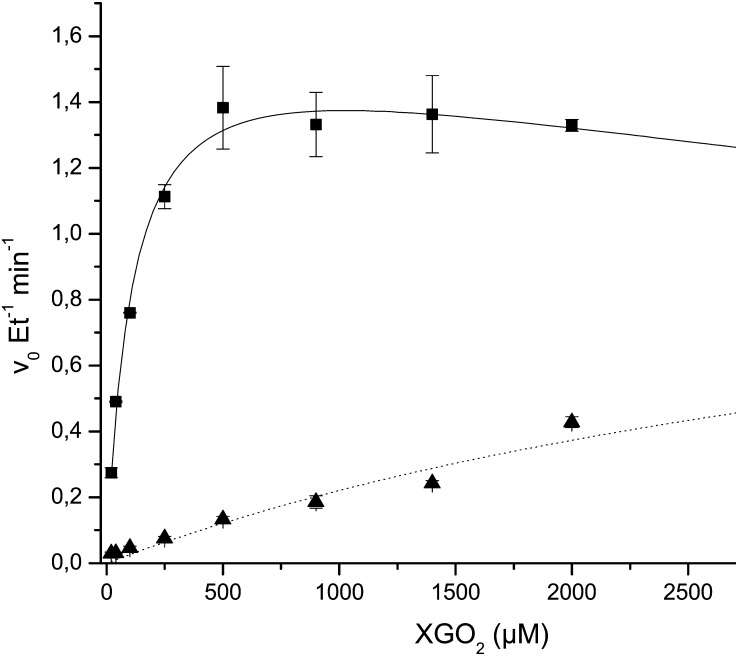
Initial rates of hydrolysis and transglycosylation by AtXTH31 were determined with a previously established HPLC assay (Baumann et al., 2007) using minimal oligosaccharide donor substrates based on a XXXGXXXG core structure. As prepared from tamarind xyloglucan by limited hydrolysis, “XGO2” is a mixture of variably galactosylated oligosaccharides. Endohydrolysis of XGO2 occurs at the central, unbranched Glc to yield two molecules of XGO1. Endo-transglycosylation, in contrast, yields one molecule each of XGO3 and XGO1, which are quantified simultaneously by HPLC in the stopped assay.
As shown in Figure 2, AtXTH31 exhibited predominant XEH activity (EC 3.2.1.151) across a broad range of substrate concentrations up to and including the maximum tested, 3 mm. In competition with water, the nonreducing end of the XXXGXXXG-based donor substrates are also capable of acting as glycosyl acceptors in the breakdown of the covalent glycosyl enzyme intermediate (Eklöf and Brumer, 2010). Thus, as the XGO2 concentration approached millimolar levels, an increase in the transglycosylation rate was observed, with a concomitant slight decrease in the hydrolytic rate (Fig. 2).
Figure 2.
Initial rate kinetics of hydrolysis and transglycosylation catalyzed by AtXTH31. Squares indicate the specific hydrolytic rate of XGO2 (variably galactosylated XXXGXXXG-based oligosaccharides) to produce two XGO1 molecules, and triangles indicate the specific transglycosylation rate of XGO2 to produce XGO3 plus XGO1.
Regrettably, our inability to heterologously express AtXTH32 precluded the experimental determination of enzymatic activity. However, AtXTH32 has high protein sequence identity with both AtXTH31 and TmNXG1 (approximately 70% identity and 80% similarity for both; compare with Fig. 1), which suggests that AtXTH32 may be predicted to likewise exhibit predominant hydrolytic (XEH) activity.
Expression of AtXTH31 and AtXTH32 in Arabidopsis Ecotype Columbia Tissues
Organ-specific and temporal expression patterns of all 33 Arabidopsis (ecotype Columbia [Col-0]) XTH genes from Genevestigator data (Zimmermann et al., 2004) have previously been analyzed in detail, in connection with extensive reporter gene fusion data (Becnel et al., 2006). In this study, Genevestigator expression levels of AtXTH31 (At3g44990) and AtXTH32 (At2g36870) were reevaluated to take advantage of the current larger data set. Across the lifetime of the plant (Supplemental Fig. S5A), AtXTH31 exhibits the strongest expression in the first 14 d of growth, from germination through the seedling stage, after which time expression drops dramatically. In these early stages, transcript levels of AtXTH31 are notably higher than those of AtXTH32, which generally appears to be constitutively expressed, with a slight up-regulation in more mature stages.
At the tissue level, AtXTH31 and AtXTH32 also exhibit distinct expression patterns (Supplemental Fig. S5B). In keeping with the general low- to medium-level expression observed in temporal studies, AtXTH32 only exhibits very-high-level expression in two tissues, namely stems and the shoot apical meristem. AtXTH31, on the other hand, appears to be highly to very highly expressed in tissues undergoing elongation: hypocotyl, radicle, root elongation zone, and lateral root. In support of these data, AtXTH31::GUS transgenic Arabidopsis plant lines have previously been shown to exhibit prominent staining in roots, whereas AtXTH32::GUS lines were stained prominently in the shoot apex of older seedlings (Becnel et al., 2006).
To verify the observation that AtXTH31 and AtXTH32 are expressed in tissues undergoing rapid expansion, semiquantitative PCR was performed in this study (Supplemental Fig. S6). AtXTH31 was expressed at notably higher levels than AtXTH32 in root, hypocotyl, and etiolated whole seedlings. In our hands, AtXTH31 and AtXTH32 also exhibited contrasting tissue-specific expression patterns in Arabidopsis Col-0. Whereas AtXTH31 was highly expressed in the root and hypocotyls, expression of AtXTH32 appeared to be predominantly localized to cotyledons, in agreement with Genevestigator data. Furthermore, analysis of protein levels in 5- and 11-d-old hypocotyls by mass spectrometry has likewise recently indicated that the AtXTH31 gene product is in significantly higher abundance than AtXTH32 in this tissue (Jamet et al., 2009).
Reverse Genetics Analysis of AtXTH31 and AtXTH32
To investigate the effect of the AtXTH31 and AtXTH32 gene products on plant growth and development, we isolated plant lines homozygous for T-DNA insertions in these genes (designated xth31-1 and xth32-1, respectively) as well as a double knockout line (xth31-1/xth32-1). A complete lack of the corresponding transcripts was verified in all knockout lines by reverse transcription (RT)-PCR (data not shown). In addition, lines that individually contained single insertions of 35S::AtXTH31 and 35S::AtXTH32 constructs were prepared to test the effects of ectopic overexpression in Arabidopsis Col-0.
Observation of the general growth patterns of the xth31-1, xth32-1, and xth31-1/xth32-1 knockout lines did not reveal any obvious phenotypic differences with respect to the wild type. Germination, growth rate, and organ development (leaf size, bolt extension, flowering time, flower and seed pod morphology, etc.) were all essentially identical by visual surveys (data not shown). Detailed analysis of root and etiolated hypocotyl extension also failed to demonstrate any significant phenotype in each single mutant and the double mutant line. In particular, total root length after 7 d in short-day conditions was statistically identical for all lines, and the root growth between days 8 and 11 was indistinguishable in all plant lines at approximately 0.5 cm d−1 (data not shown). The dark-grown hypocotyl length of all lines was also statistically indistinguishable at days 5 and 11 under dark conditions. Likewise, no significant growth phenotypes were observed in 35S::AtXTH31 and 35S::AtXTH32 overexpression lines.
Genevestigator data indicated that AtXTH31 and AtXTH32 are 2.6- and 1.67-fold up-regulated in response to Suc, respectively. Nevertheless, reducing the sugar content in the medium from 1% to 0.5% did not affect the phenotype of the knockout lines relative to the wild type (data not shown). We also examined whether AtXTH31 and AtXTH32 could be involved more specifically in the auxin-mediated increase in hypocotyl elongation at higher temperature (Vandenbussche et al., 2005). Wild-type seedlings grown at 22°C for 2 d post germination and subsequently shifted to 28°C for 3 d demonstrated an increase in hypocotyl length of approximately 10%, which was statistically identical in the xth31-1/xth32-1 double knockout line (data not shown).
Despite extensive analysis, the only observable developmental phenotype was a very mild effect on hypocotyl extension immediately post germination. Quantitative kinematic analysis over 96 h post germination revealed a slight, but statistically significant, reduction in hypocotyl extension in both 35S::AtXTH31 and 35S::AtXTH32 at 24 and 48 h, which converged with the wild type at 72 to 96 h (Supplemental Fig. S8). On the other hand, kinematic analysis of the xth31-1/xth32-1 line over the first 27 h indicated that no statistically significant difference in hypocotyl extension was observed versus the wild type (data not shown).
In Situ Visualization of XET and XEH Activities in the Wild-Type and Double Knockout Lines
In light of the minor effects of AtXTH31 and AtXTH32 on Arabidopsis development, we directed our attention to the analysis of potential changes in XET and XEH enzyme activities in situ using specific fluorescent (Vissenberg et al., 2000) and fluorogenic (Ibatullin et al., 2009) probes, respectively. Based on the close homology of AtXTH31 and AtXTH32 as group III-A members, the double knockout line was examined directly to avoid potential complications due to functional compensation.
The resorufin β-glycoside of XXXG (XXXG-Res) has previously been demonstrated to be a substrate for the predominant XEH from nasturtium, TmNXG1, which catalyzes the release of the fluorescent resorufinyl anion in vitro and allows the detection of activity in vivo (Ibatullin et al., 2009). At the pH value of 5.5 used for real-time in situ assays, the specific activity of AtXTH31 at 1 mm XXXG-Res was 0.068 ± 0.008 mol min−1 mol−1 AtXTH31, while at the optimum pH of 4.75, the specific activity was 0.28 ± 0.01 mol min−1 mol−1 AtXTH31 ([S] = 1 mm), as determined in a stopped assay. It should be noted that in the in situ assays, which follow fluorescence development continuously, the resorufin aglycone will be 33% ionized at pH 5.5 (pKa = 5.8, ε = 62,000 m−1 cm−1; Po and Senozan, 2001). This pH was selected as a compromise between maximum enzyme activity and maximum fluorescence detection sensitivity. Thus, XEH activity was revealed in both the roots and etiolated hypocotyls of Arabidopsis Col-0 and xth31-1/xth32-1 plants by monitoring the release of the resorufinyl anion by confocal fluorescence microscopy.
Roots
Examination of wild-type Col-0 (Supplemental Fig. S9B) and the xth31-1/xth32-1 mutant (Supplemental Fig. S9B) using the Complex Carbohydrate Research Center-Monoclonal1 (CCRC-M1) antibody, which recognizes α(1-2)-linked fucosyl residues present in xyloglucan and rhamnogalacturonan-I (Puhlmann et al., 1994), revealed no significant differences in microscopic cell wall composition in the mutant line. CCRC-M1 staining was observed throughout the roots, including the root hairs, except at the root tip, similar to earlier reports (Freshour et al., 1996). As observed previously for the wild type (McCartney et al., 2003), staining of rhamnogalacturonan-I-associated (1→4)-β-d-galactan epitope by the Leeds Monoclonal5 (LM5) antibody was restricted to the rapid expansion zone in both lines (Supplemental Fig.S9, C and D), which served as a marker for this region.
In Situ XET Activity.
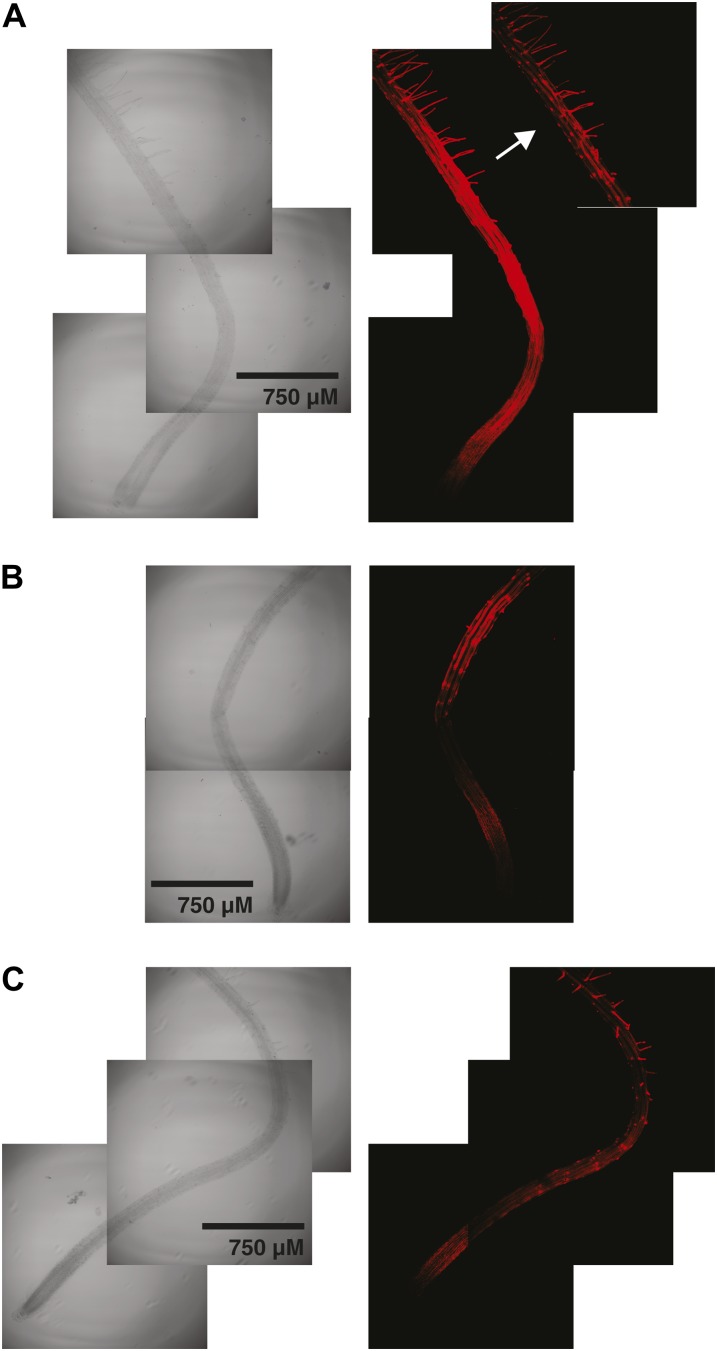
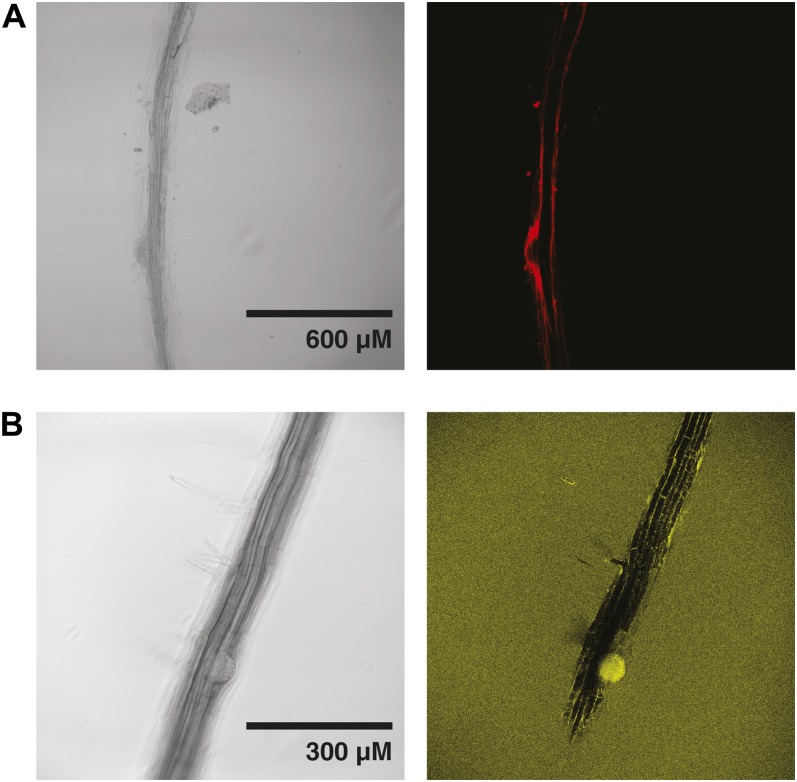
For comparison with previous studies, XET activity was visualized in Col-0 and xth31-1/xth32-1 roots by monitoring the incorporation of the xyloglucan oligosaccharide-sulforhodamine conjugate XXXG-SR into the cell wall by confocal fluorescence microscopy. In this stopped assay, XXXG-SR functions as an alternate glycosyl acceptor for enzymes with XET activity, thus forming covalent xyloglucan-XXXG-SR conjugates at sites where the polysaccharide and enzyme are colocalized (Vissenberg et al., 2000; Nishikubo et al., 2007). As shown in Figure 3, the localization of XET activity was essentially identical in wild-type (Fig. 3A) and xth31-1/xth32-1 (Fig. 3, B and C) plants, although mutant plants in general exhibited weaker signal. In both lines, XET activity was observed in the root elongation zone (compare with Supplemental Fig. S9) and at sites of root hair initiation, precisely as in previous studies (Vissenberg et al., 2000, 2001).
Figure 3.
Localization of XET activity in 6-d-old Arabidopsis roots using XXXG-SR. A, Wild-type Col-0. The inset (arrow) with detector attenuation highlights root hair initiation and root hair staining. B, xth31-1/xth32-1 double knockout, showing root hair initiation and elongation zone staining. C, xth31-1/xth32-1 double knockout, a second example showing root hair, root hair initiation, and elongation zone staining. Representative images from the analysis of approximately 10 plants from each line are shown. [See online article for color version of this figure.]
In Situ XEH Activity.
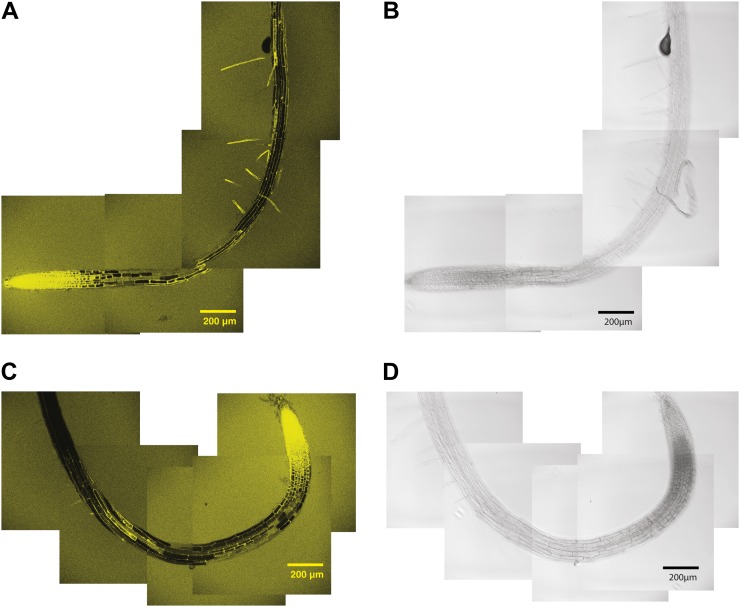
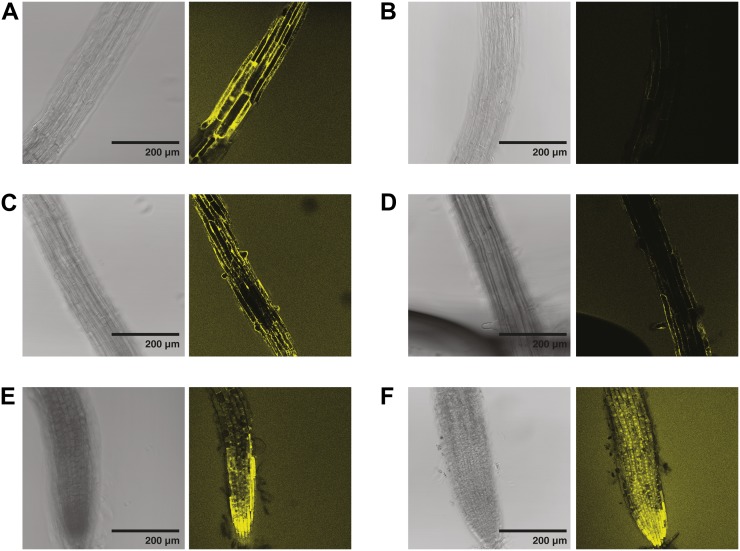
XEH activity, as revealed by the hydrolysis of XXXG-Res, was highest in the cells of the growing root tips of both wild-type and xth31-1/xth32-1 plants, to the extent that significant diffusion of the resorufinyl anion into the medium was observed (Fig. 4). Closer inspection of the root tip indicated that the two lines exhibited similar XEH activity in this region and that significant amounts of resorufinyl anion were contained in internal compartments (Fig. 5, E and F), perhaps due to the uptake and sequential hydrolysis of XXXG-Res by endogenous α-xylosidase and β-glucosidase activity (Iglesias et al., 2006).
Figure 4.
Localization of XEH activity in 6-d-old Arabidopsis roots using XXXG-Res. A and B, Confocal fluorescence (A) and bright-field (B) images of wild-type Col-0. C and D, Confocal fluorescence (C) and bright-field (D) images of xth31-1/xth32-1 double knockout. Representative images from the analysis of approximately 10 plants from each line are shown. [See online article for color version of this figure.]
Figure 5.
Detailed views from the localization of XEH activity in 6-d-old Arabidopsis roots using XXXG-Res. A, Col-0, the region corresponding to the top portion of Figure 4A (approximately 2 mm from the root tip). B, xth31-1/xth32-1 double knockout, the region corresponding to the top portion of Figure 4B (approximately 2 mm from the root tip). C, Col-0, emerging root hairs. D, xth31-1/xth32-1 double knockout, emerging root hairs. E, Col-0, root tip. F, xth31-1/xth32-1 double knockout, root tip. As in Figure 4, representative images from the analysis of approximately 10 plants from each line are shown. [See online article for color version of this figure.]
XEH activity, however, was distinctly lower in specific regions distal to the root tip of xth31-1/xth32-1 knockout plants than in the wild type. Specifically, XEH activity was high in the root hairs of wild-type plants and was essentially absent in the root hairs of xth31-1/xth32-1 knockout plants (Fig. 4, A and C). In the root hair elongation zone, XEH activity was high and clearly observable in the cell wall, whereas significantly less wall-associated XEH activity was present in the xth31-1/xth32-1 double mutant (Fig. 5, A and B). Similarly, XEH activity in root hair initiation zones was significantly reduced in the double mutant (Fig. 5D) compared with the wild type (Fig. 5C), although no gross alterations in root hair morphology were observed.
Notably, a localized increase in XEH activity was also clearly observed in Col-0 plants during lateral root initiation (Fig. 6). Strong XET activity was also observed in lateral root initiation zones, as has been demonstrated previously in Arabidopsis (Vissenberg et al., 2000). XEH activity was also high in wild-type cell walls in the junction region between root and hypocotyls, also known as the collet, which contains early emerging root hairs (Lin and Schiefelbein, 2001; Scheres et al., 2002; Supplemental Fig. S10), and was strongly reduced in the cell walls of xth31-1/xth32-1 double knockout plants, especially in the primary root (Supplemental Fig. S10, C and D).
Figure 6.
Localization of XET and XEH activity in wild-type Arabidopsis Col-0 during lateral root initiation. A, XET activity detected with XXXG-SR. B, XEH activity detected with XXXG-Res. Representative images from the analysis of approximately 10 plants from each line are shown. [See online article for color version of this figure.]
Etiolated Hypocotyls
In Situ XET Activity.
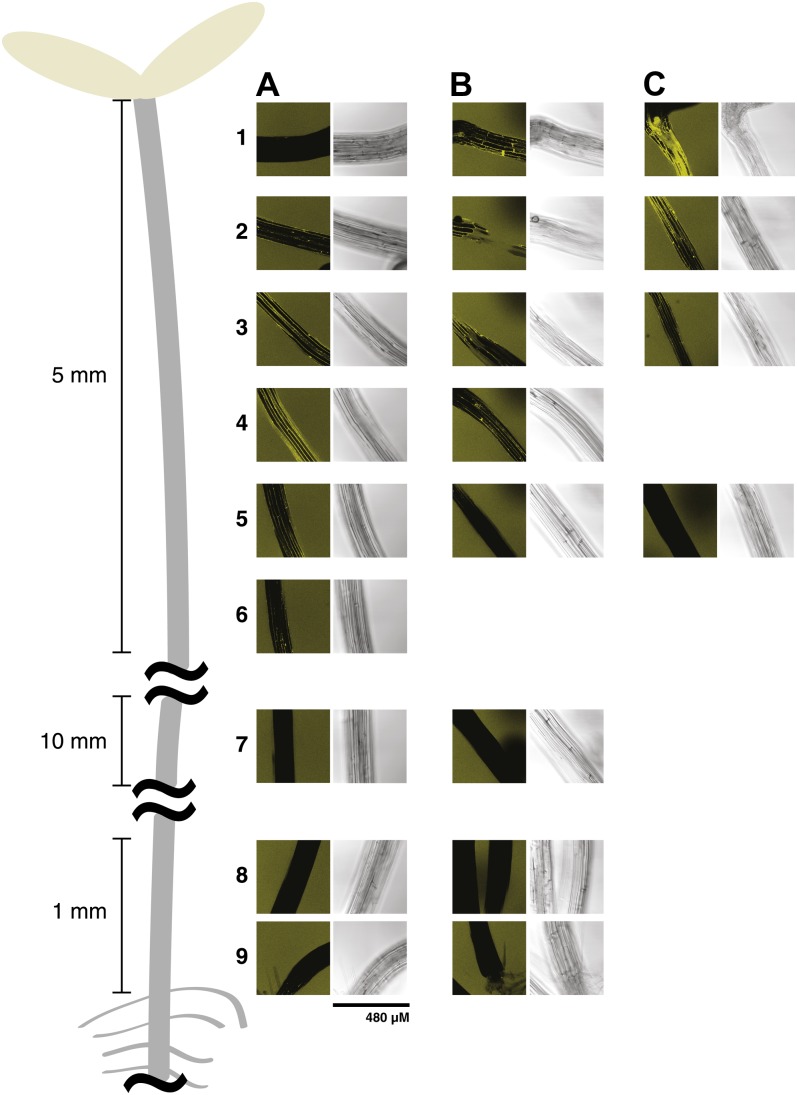
To the best of our knowledge, XET activity has previously not been spatially localized in Arabidopsis hypocotyls, although whole-tissue measurements of activity have been performed in soybean (Glycine max; Wu et al., 2005). Thus, to allow comparison with in situ XEH activity analysis (see below), XET activity was highlighted across the entire dark-grown hypocotyl length by the incorporation of XXXG-SR (Supplemental Fig. S11). In the Col-0 wild-type line, XET activity started approximately 0.5 mm below the cotyledons and extended down 1 to 2 mm (Supplemental Fig. S11A, zones 2 and 3), but it was conspicuously absent along the majority of the hypocotyl (Supplemental Fig. S11A, zones 4–8; typified by the zone 7 image). This pattern suggests that XET activity is highest in the growing region of the hypocotyl (Gendreau et al., 1997; compare with Fig. 2d in Wolf et al., 2012). Slight XET activity was also observed in the cotyledons (Supplemental Fig. S11A, zone 1), and high activity was apparent in the hypocotyl-root junction (Supplemental Fig. S11A, zone 9). A very similar pattern of XET activity was observed in the xth31-1/xth32-1 mutant line, with the notable exception that XXXG-SR incorporation was much higher in the region immediately below the cotyledons, with activity persisting much farther along the hypocotyl length (Supplemental Fig. S11B, zones 1–4). Interestingly, the high XET activity observed at the cotyledon-hypocotyl junction in the double mutant mirrored observations of high XEH activity in this zone (see below).
In Situ XEH Activity.
XEH activity in hypocotyls, as revealed by XXXG-Res hydrolysis, was observed from the apical hook to 5 mm down the hypocotyl of the wild-type plant (Fig. 7A, zones 2–5) but was clearly absent in the lower two-thirds, including the root junction zone, in both the wild type and xth31-1/xth32-1 mutant lines. As for XET activity (see above), XEH activity appeared to reach a maximum in the growing region of the hypocotyl (compare with Fig. 2d in Wolf et al., 2012). Moreover, the double knockout line demonstrated significantly reduced XEH activity in the expanding zone (Fig. 7, B and C, zones 3–5). Curiously, one-third of the xth31-1/xth32-1 mutant plants surveyed had greatly increased XEH activity directly below the cotyledon-hypocotyl junction, which decreased in a gradient to approximately 10 mm down the hypocotyl (Fig. 7C, zones 1 and 2). This corresponds to the same region in which XET activity appeared to be increased in the mutants (Fig. 7D, zone 1).
Figure 7.
Localization of XEH activity in 5-d-old Arabidopsis etiolated hypocotyls using XXXG-Res. A, Wild-type Col-0. B, xth31-1/xth32-1 double knockout. C, A second example of xth31-1/xth32-1 double knockout. Numbered zones show the approximate positions at which images were taken along the length of the hypocotyl and correspond to those in Supplemental Figure S11. The composite image was produced from representative images from the analysis of approximately 20 plants from each line. [See online article for color version of this figure.]
DISCUSSION
The importance of xyloglucan-cellulose cross links in modulating the strength and extensibility of the primary plant cell wall is a key feature of classical models of this composite structure (Carpita and McCann, 2000; Cosgrove, 2005; Mellerowicz et al., 2008). Although the molecular mechanisms of cell wall extension are largely unknown, a key tenet is transient wall loosening effected by the cleavage and religation of matrix xyloglucan by XET (EC 2.4.1.207). Indeed, since the discovery of XET activity in the early 1990s, a strong focus on xyloglucan transglycosylation in the context of wall extension and remodeling continues to be sustained (Takeda et al., 2002; Nishikubo et al., 2007, 2011; Baba et al., 2009; Hernández-Nistal et al., 2010; Lee et al., 2010; Miedes et al., 2010, 2011; Opazo et al., 2010; Stratilová et al., 2010).
The observation that the XTH gene family of Arabidopsis also encodes two proteins homologous to the nasturtium seed xyloglucanase TmNXG1 (Fig. 1; Baumann et al., 2007) suggested the possibility that xyloglucan hydrolysis might also play a role in primary cell wall extension. In particular, Arabidopsis is not known to utilize xyloglucan as a major seed storage polysaccharide (compare with nasturtium) and does not undergo fleshy fruit softening (compare with tomato [Solanum lycopersicum]), which might be expected to require specific endoxyloglucanases (Edwards et al., 1986; Vicente et al., 2007). Indeed, ESTs of TmNXG1 homologs, which comprise XTH phylogenetic group III-A, are often found in ripening fruits (Baumann et al., 2007). And whereas nasturtium and tamarind have abundant xyloglucan in the seed endosperm (Marcus et al., 2008; von Schantz et al., 2009), xyloglucan has thus far only been localized in the columella (Western et al., 2000) of the Arabidopsis seed coat epidermis (J.P. Knox, unpublished data, available at http://www.personal.leeds.ac.uk/~bmbjpk/gal6.htm). Nonetheless, Genevestigator data analysis and semiquantitative PCR analysis in this study revealed that the group III-A homologs AtXTH31 and AtXTH32 were in fact expressed in diverse tissues of the Arabidopsis Col-0, including those undergoing significant expansion.
Detailed enzyme kinetic analysis upheld our prediction that AtXTH31 was a predominant XEH (EC 3.2.1.151), in keeping with its group III-A membership (Baumann et al., 2007). AtXTH31 hydrolyzed high-molar-mass xyloglucan and the fluorogenic substrate XXXG-Res at rates comparable to those of TmNXG1. In a simultaneous assay on minimal XGO2 substrates, substrate hydrolysis clearly dominated over transglycosylation, even at donor/acceptor substrate concentrations as high as 3 mm (Fig. 2). Our inability to recombinantly express AtXTH32 cDNA in the yeast P. pastoris, as well as in the plant N. benthamiana, mirrored previous results with certain XTH genes, in which no sequence-specific correlation with the lack of expression could be found (Kaewthai et al., 2010). The high degree of protein sequence similarity between AtXTH32 and both AtXTH31 and TmNXG1 (Fig. 1) nonetheless allows the inference, with a high degree of confidence, that AtXTH32 also possesses predominant XEH activity.
Remarkably, single and double knockout lines of AtXTH31 and AtXTH32 exhibited no significant growth phenotypes versus the wild type, despite the strong reduction of XEH activity in specific tissues of the double mutant, as revealed by in situ enzyme analysis (discussed further below). Likewise, individual AtXTH31 and AtXTH32 overexpression lines exhibited only a marginal decrease in hypocotyl growth rate immediately post germination. Keeping in mind that these differences are subtle, they are nonetheless difficult to rationalize in terms of current models of cell wall expansion, which would predict exactly the opposite effect: increased hydrolytic cleavage of xyloglucan due to overexpression of these XEHs should cause wall loosening and, therefore, increase the expansion/growth rate.
One explanation for these apparently counterintuitive observations is that altering XEH activity directly affects the levels of small xyloglucan oligosaccharides in the apoplast, which may act as signaling molecules affecting growth (York et al., 1984; McDougall and Fry, 1989; Vargas-Rechia et al., 1998; Salamoni et al., 2008). Here, AtXTH31 may be the dominant player in Col-0, as it is significantly up-regulated (Supplemental Fig. S5B) in both imbibed seeds (absolute expression value [AEV] 8,300) and the radicle (AEV 11,000), whereas AtXTH32 has very low-level expression (AEV 880 and 590, respectively, as supported by promoter-GUS analysis [Becnel et al., 2006]).
Other possible explanations for the lack of directly observable growth phenotypes are related to the ambiguous structural role xyloglucans play in the cell wall. Although fucogalactoxyloglucan may constitute up to 20% to 25% of the primary cell wall dry weight in dicots (Vogel, 2008), the polysaccharide is, remarkably, dispensable: Arabidopsis plants that completely lack xyloglucan, due to the disruption of two essential xylosyltransferases, have an essentially normal growth habit (Cavalier et al., 2008). This suggests a significant ability to compensate for even drastic effects on xyloglucan composition and quantity. Likewise, recent elegant work from Park and Cosgrove (2012), using ex vivo cucumber (Cucumis sativus) and Arabidopsis cell wall systems, has provided strong evidence that extensive cleavage of bulk matrix xyloglucan chains does not induce wall creep. Rather, these authors suggest that wall extensibility is dominated by comparatively few intimately associated chains of cellulose and xyloglucan, which require mixed-function cellulose/xyloglucanase enzymes to disrupt. Although untested at present, AtXTH31 and/or AtXTH32 may not have the capacity to access such regions. Collectively, these observations may explain why XTH-related wall phenotypes are often slight in those cases where they are observed (Cosgrove, 2000, 2005; Osato et al., 2006; Miedes et al., 2011; Nishikubo et al., 2011).
It is also worthwhile to note that plant XEHs, including AtXTH31 and TmNXG1, are catalytically feeble when compared with, for example, microbial XEHs (endoxyloglucanases or xyloglucan-specific endoglucanases), which can have specific activities toward the polysaccharide of up to 3 orders of magnitude higher (Ibatullin et al., 2008). Therefore, AtXTH31 and AtXTH32, and, by extension, their homologs in higher plants may only play subtle roles in xyloglucan editing in the larger context of the cell wall complex.
Despite the lack of striking morphological phenotypes, a combination of reverse genetics and chemical biology clearly highlighted that XEH activity due to AtXTH31 and/or AtXTH32 is nonetheless widespread in developing tissues. We had previously presented XXXG-Res as a new probe for the real-time visualization of XEH activity in situ (Ibatullin et al., 2009). In connection with the xth31/xth32 double knockout line, comparative analysis versus the Col-0 wild type indicated suppression of XEH in distinct zones of the root and hypocotyl of the mutant. Notably, XET activity also appeared to be slightly reduced in the double knockout, perhaps indicating a compensatory mechanism or secondary effects leading to altered transcript levels of other XTH family members. In the absence of a detailed global analysis, it is tempting to speculate that a reduction in xyloglucan oligosaccharide release from the apoplast may affect a wall-sensing signal mechanism (York et al., 1984; McDougall and Fry, 1989; Vargas-Rechia et al., 1998; Salamoni et al., 2008).
Moving from the root apical meristem, the elongation zone was clearly delineated by LM5 staining (Supplemental Fig. S9) as well as XET activity staining (Fig. 3), with both the AtXTH31/AtXTH32 knockout and wild-type lines exhibiting similar patterns, which were equivalent to those observed previously for the wild type (Vissenberg et al., 2000, 2001; McCartney et al., 2003). AtXTH31 is highly expressed in this zone (AEV 12,000 versus 870 for AtXTH32); however, the lack of differential signal from XXXG-Res hydrolysis implied that either sequential α-xylosidase and β-glucosidase activity (Iglesias et al., 2006), or one or more other enzyme(s) with XEH activity, is active here (Fig. 4). However, XEH activity was notably diminished in double knockout plants in the region between the expansion zone and the zone of differentiation. Signal was completely abolished in the walls of many cells, including the root hairs (Figs. 4C and 5, B and D). Therefore, it was somewhat surprising that there were no obvious growth phenotypes, despite such distinct reductions in XEH activity.
Remarkably, XEH activity is clearly observed in root hair bulges in the wild type, where XET activity has previously been demonstrated (compare Fig. 5C with Fig. 3A; Vissenberg et al., 2001). In that previous study, root hair-deficient mutants failed to show a localized increase in XET activity; however, the explicit requirement of XET activity for root hair initiation has not been proven, to our knowledge. Despite the elimination of XEH activity in initiating root hairs in the xth31/xth32 mutant (Fig. 5D), root hair initiation nonetheless appeared to be grossly unaffected. Furthermore, neither AtXTH31 (AEV 970) nor AtXTH32 (AEV 270) is significantly expressed in the root hair zone (Supplemental Fig. S5B). Thus, it is concluded that xyloglucan endohydrolysis is a latent activity in the root cell wall remaining from the period of cell elongation (where, in particular, AtXTH31 transcript levels are high), which is not strictly essential for root hair initiation.
Likewise, both XET and XEH activity were clearly observed during lateral root initiation in the wild type (Fig. 6). Given the similarities between embryonic and lateral root development (Malamy and Benfey, 1997), it is tempting to speculate that similar wall polysaccharide hydrolases may be involved. AtXTH31 is indeed tremendously up-regulated in lateral roots (AEV 18,000 versus 1,300 for AtXTH32), as it is in the root elongation zone. In the roots examined, lateral root initiation was a rare event; thus, little can be surmised from the inability to covisualize XEH activity and initiation in the xth31/xth32 mutant. A more focused study including the use of lateral root mutants (Malamy and Benfey, 1997) together with the XXXG-Res probe would clearly be warranted to further elucidate the importance of XEH activity in this tissue.
The observation of XEH activity in etiolated hypocotyls likewise highlighted xyloglucan degradation during cell wall remodeling and expansion. As was observed in various root cells, knocking out the homologs AtXTH31 and AtXTH32 significantly diminished signal from XXXG-Res hydrolysis in the expanding zone (Fig. 7, image row 4). Here, the majority of the effect is likely to be due to loss of the AtXTH31 gene product (AEV 20,000), although AtXTH32 was also significantly expressed in the hypocotyl (AEV 3,900). The lack of activity in the lower two-thirds of the hypocotyl indicated a cessation of matrix xyloglucan hydrolysis due to enzyme inactivation in, or elimination from, the apoplast/cell wall. XEH activity was clearly not persistent in regions where cell wall expansion had ceased, as was true for XET activity (Supplemental Fig. S11), thus potentially implicating a combined role of these two activities.
The observation that approximately one-third of xth31/xth32 plants demonstrated elevated levels of XXXG-Res hydrolysis in the region of the hypocotyl abutting the cotyledons was indeed odd (Fig. 7C). Likewise, some plants also exhibited increased levels of XET activity in this same region (Supplemental Fig. S11). It was surmised that the increased signal in both assays is due to compensation by at least one unidentified enzyme with both XEH and XET activity, or one or more predominant XEH and one or more XET. In this context, it should be highlighted that a drawback of the XET in situ assay is that incorporation of XXXG-SR into high-molar-mass xyloglucan may detect predominant xyloglucan hydrolases with only minor transglycosylation activity, since only one possible product of the enzyme is detected. Furthermore, both the XET and XEH in situ assays are limited by the fact that they cannot be directly correlated to individual enzymes. XXXG-Res can potentially be broken down by other enzymes with XEH side or main activities, for example, members of GH5, GH16 (i.e. other XTH gene products), or other glycoside hydrolases encoded by the Arabidopsis genome (Henrissat et al., 2001), in which xyloglucanase activity has yet to be identified.
CONCLUSION
This study has highlighted that XEH activity is specifically localized in a range of Arabidopsis tissues, especially those undergoing cell wall expansion, and that this activity is due to group III-A XTH gene products. These observations have potentially important implications for present models of primary plant cell wall biosynthesis and maturation, in which transglycosylation has been considered to be the preeminent xyloglucan-remodeling activity (Rose et al., 2002; Eklöf and Brumer, 2010). Although general endoglucanase (cellulase) activity has long been postulated to have a role in wall loosening (Fry, 1989; Rose and Bennett, 1999; Urbanowicz et al., 2007; Vicente et al., 2007), our results directly implicate the action of endogenous xyloglucan-specific hydrolases from glycoside hydrolase family 16 in the apoplastic arena. However, these results do not support XEHs as major actors in controlling cell wall extension. The XEHs encoded by AtXTH31 and AtXTH32 may instead be the key enzymes in a xyloglucan-recycling pathway composed of recently discovered Arabidopsis α-l-fucosidases, β-galactosidases, α-xylosidases, β-glucosidases, and their homologs, which are similarly up-regulated in cells undergoing wall extension and/or remodeling (Iglesias et al., 2006; Sampedro et al., 2010, 2012; Günl et al., 2011). In light of our current work and these recent analyses, continued scrutiny of the role of polysaccharide hydrolases in the context of cell wall morphogenesis is clearly warranted.
MATERIALS AND METHODS
Recombinant Expression of AtXTH31 and AtXTH32 in Pichia pastoris SMD1168H
A synthetic gene corresponding to the nucleotide sequence of AtXTH31 (locus At3g44990) and wild-type and codon-optimized versions of AtXTH32 (locus At2g36870) were obtained from GENEART. PCR amplification using AtXTH31-PF and AtXTH31-PR (Supplemental Table S1) was used to generate XhoI and XbaI restriction sites to facilitate cloning into the pPICZ α-C vector containing the alcohol oxidase promoter (Invitrogen). In the pPICZ α-C-AtXTH31 construct, the sequence encoding the predicted native signal peptide was replaced by that encoding the yeast α-factor secretion signal peptide. Competent SMD1168H cells were transformed with 5 to 10 µg of plasmid DNA, which had previously been linearized with SacI as described by the manufacturer (Invitrogen; Pichia Expression Kit manual). After transformation, the cells were placed on yeast extract-peptone-dextrose-sorbitol agar plates containing zeocin (100 mg mL−1) and incubated at 30°C for 3 to 5 d. The colonies from yeast extract-peptone-dextrose-sorbitol + zeocin plates were further grown on yeast extract-peptone-dextrose containing zeocin (100 mg mL−1) for small-scale expression.
The purified clones were expressed in a small scale to identify positive clones, by inoculating 5 mL of buffered glycerol/complex medium in a BD Falcon 50-mL conical tube. The cultures were grown overnight at 30°C and 180 to 220 rpm, at which time the medium was changed to buffered methanol/complex medium. The cultures were induced for 4 to 5 d at 22°C, with methanol at a final concentration of 0.5% (v/v) on the first day and 1% (v/v) for the remainder of the cultivation time. On day 4, optical density at 600 nm values were measured, the cells were centrifuged at 3,400g for 10 min, and the supernatant was used for screening protein expression using the Bradford protein assay and SDS-PAGE (NuPAGE; Invitrogen), essentially as described (Kaewthai et al., 2010). For SDS-PAGE analysis, 20 µL of supernatant was added to SDS-PAGE loading buffer. Western-blot analyses were performed using 0.45-µm nitrocellulose blotting membranes (Bio-Rad). The AtXTH31 protein fused with poly-His tag was detected with Hisprobe-HRP (Thermo Scientific; catalog no. 15165), nickel (Ni2+)-activated derivative conjugated to horseradish peroxidase, and visualization by ECL (ECL Advance Western Blotting Detection Kit; Amersham; product code no. RPN2135), according to the manufacturer’s directions. (The same procedure was applied for the recombinant expression of wild-type and codon-optimized AtXTH32, but these experiments were consistently unsuccessful.)
Large-scale production of AtXTH31 and subsequent affinity purification were performed according to the methods described by Baumann et al. (2007). Protein electrospray mass spectrometry was performed as described by Sundqvist et al. (2007).
Biochemical Characterization of Recombinant AtXTH31
pH Dependence of Xyloglucan Hydrolysis
The reducing ends produced by the hydrolysis of xyloglucan by AtXTH31 at different pH values were quantified using the BCA method relative to a Glc standard (2.5–50 µm). A total assay volume of 250 µL containing 1 g L–1 xyloglucan, 50 mm buffer (NaOAc, pH 4–5.5; sodium citrate, pH 5.5–6), and 0.15 µm AtXTH31 was incubated for 20 min and stopped by the addition of 250 µL of the BCA reagent (McFeeters, 1980). Color was developed at 80°C for 20 min, and the A560 was recorded on a Cary 50 Bio UV/Vis spectrophotometer (Varian).
Analysis of Xyloglucan Hydrolysis Products
Samples (100 µL) containing XG (1 g L–1) and AtXTH31 (0.3 µm) in 70 mm NH4OAc, pH 5.5, were incubated for 0, 105, 240, or 600 min and overnight at 30°C. The reactions were stopped by heating in a metal block at 95°C for 10 min and were subsequently frozen and lyophilized. Products were dissolved in 350 µL of dimethyl sulfoxide for HPSEC-ELS. HPSEC-ELS was performed with a Gynkotek M480G pump (Optimize Technologies), a PL-ELS 1000 ELS detector (Polymer Laboratories; Varian), and two 300- × 7.5-mm PLgel 10-μm MIXED-B columns (Polymer Laboratories) coupled in series with a matching 50-mm guard column; the columns were maintained at 70°C. The eluent was 100% dimethyl sulfoxide (1 mL min–1), and the injection volume was 100 μL. Polymer Laboratories pullulan standards were used for molar mass calibration (Mr 738–1,660,000).
The end products of AtXTH31-catalyzed xyloglucan hydrolysis were determined by incubating 1 g L–1 xyloglucan, 100 mm NaOAc, pH 4.75, and 1.5 µm AtXTH31 overnight at 30°C. The reaction was subsequently diluted six times and analyzed by HPAEC-PAD according to the method described by Maris et al. (2011).
Substrate Specificity
The hydrolysis of various β-1,4-linked polysaccharides by AtXTH31 was quantified using the BCA method. Polysaccharides were assayed at 1 g L–1 in 50 mm NaOAc, pH 4.75, with 0.15 µm AtXTH31. The assayed polysaccharides were tamarind (Tamarindus indica) xyloglucan, carboxymethyl cellulose, medium-viscosity barley (Hordeum vulgare) β-glucan, wheat (Triticum aestivum) arabinoxylan, carob (Ceratonia siliqua) galactomannan, galactan from lupin (Lupinus albus; all from Megazyme), and hydroxyethyl cellulose (Fluka).
Initial Rate Kinetics with a Minimal Xyloglucooligosaccharide Substrate
The rates of hydrolysis and transglycosylation were measured by HPAEC-PAD with a mixture of Glc8-based xyloglucooligosaccharides essentially as described by Baumann et al. (2007) with the gradient and column described by Maris et al. (2011). Assays (12.5 μL) contained 50 mm NaOAc, pH 4.75, 0.77 µm AtXTH31, and Glc8-based xyloglucooligosaccharides in concentrations ranging from 20 to 3,000 µm. Assays were incubated at 30°C for 10 min prior to the addition of 1 m NaOH (5 μL). Products were quantified against standards containing Glc4- and Glc12-based xyloglucooligosaccharides from 1 to 50 µm prepared according to Martinez-Fleites et al. (2006).
Activity of Heterologously Expressed AtXTH31 on XXXG-Res
XXXG-Res was prepared according to Ibatullin et al. (2009). The AtXTH31-catalyzed hydrolysis of XXXG-Res was determined by measuring the release of the resorufin anion (λmax = 571 nm, ε = 62,000 m−1 cm−1) in duplicate assays using a Cary 50 Bio UV/Vis spectrophotometer (Varian). A total assay volume of 50 µL, containing 1 mm XXXG-Res, 75 mm buffer (NaOAc, pH 4.75, or MES, pH 5.5), and 0.27 µm AtXTH31, was incubated at 30°C for 120 or 285 min. The A571 was measured immediately after the addition of an equal volume of 0.35 m Na2CO3. The background hydrolysis rate was determined by incubating XXXG-Res in identical conditions without enzyme.
Generation of Mutant Plant Lines and Genotyping
Plant Materials and Growth Conditions
Seeds of Arabidopsis (Arabidopsis thaliana) Col-0, AtXTH31 and AtXTH32 mutants, as well as xth31-1/xth32-1 were sown and grown on soil or surface sterilized, germinated, and grown on solid (0.7% agar) Murashige and Skoog medium supplemented with 1% Suc (except where otherwise indicated) at 22°C under a 16-h-light/8-h-dark illumination regime at 117 µmol m−2 s−1 in a growth chamber (Sanyo MLR-351).
Analysis of AtXTH31 and AtXTH32 Gene Transcript Levels in Different Tissues of Arabidopsis Col-0
Seedlings of the wild type were grown under light conditions for 7 d for collection of cotyledons, hypocotyls, and roots. Etiolated seedlings were grown for 5 d for collection of whole seedlings. RNA extraction was performed using Trizol (Invitrogen), and cDNA was synthesized using SuperScript II reverse transcriptase (Invitrogen). Semiquantitative RT-PCR of AtXTH31 and AtXTH32 in tissues was performed using primers (Supplemental Table S1) specific to AtXTH31 (AtXTH31-RT-F and AtXTH31-RT-R) and AtXTH32 (AtXTH32-RT-F and AtXTH32-RT-R). Primers for the housekeeping gene clathrin were used for controls.
Isolation and Characterization of T-DNA Insertion Lines
Genomic DNA of plant lines was extracted using Quick DNA Prep (Weigel and Glazebrook, 2002) for all genotyping and T-DNA insertion analysis; primers used for genotyping are described in Supplemental Table S1. Four T-DNA insertion lines were obtained from the Salk Institute Genomic Analysis Laboratory: SALK_114136 (AtXTH31; insertion in first exon), SALK_129686 (AtXTH31; insertion in third exon), SAIL_672_D08 (AtXTH32; insertion in third exon), and SALK_131432 (AtXTH32; insertion in third intron). Of these, homozygotes were directly identified in M2 plants for SALK_129686 and SALK_131432 (Supplemental Fig. S7); thus, the SALK_114136 and SAIL_672_D08 lines were not examined further. The SALK_129686 and SALK_131432 alleles were designated xth31-1 and xth32-1, respectively. Each line was backcrossed to wild-type plants two times, and the genotypes of the F1 and F2 progeny were verified by PCR, using primers specific to AtXTH31 and AtXTH32 and the T-DNA insert (data not shown). PCR fragments were also sequenced and confirmed the predicted T-DNA insertions in each gene. Clean homozygous single-mutant plant lines were thus obtained for use in all further experiments. Homozygous double mutants were generated by crossing pollen of the homozygous xth31-1 and xth32-1 lines. To check expression levels, RNA from Col-0, xth31-1, xth32-1, and xth31-1/xth32-1 at 5 and 11 d were extracted using the RNeasy Plant Mini Kit (Qiagen; catalog no. 69104). RT-PCR was performed using the TITANIUM One Step RT-PCR kit (Clontech; catalog no. 639504) and the gene-specific primer pairs AtXTH31-Forward/AtXTH31-Reverse and AtXTH32-Forward/AtXTH32-Reverse (Supplemental Table S1).
Generation of AtXTH31 and AtXTH32 Ectopic Overexpression Lines
AtXTH31 and AtXTH32 were amplified by PCR from 5-d-old dark-grown hypocotyl genomic DNA, cloned into the pENTR entry vector (Invitrogen), and transferred into pK2GW7 in frame with the 35S promoter. Arabidopsis Col-0 was then transformed with either the 35S::AtXTH31 or 35S::AtXTH32 construct. Monoinsertional homozygote lines were selected based on their kanamycin resistance (five lines for each construct).
Phenotyping
General
A Sony DSC-W55 camera was used for photography of plant development. Images were analyzed using the ImageJ software program (http://rsbweb.nih.gov/ij/) to quantify growth. Development of all mutants was observed by following the stages suggested by Boyes et al. (2001) over this period.
Root Growth Analyses
The root growth rate of all mutants was analyzed as described by Weigel and Glazebrook (2002) on seedlings grown with or without 1% Suc. After 2 d of vernalization, plates were placed in a growth chamber at 22°C to 23°C in a 16-h/8-h day/night cycle. After roots grew to a length of approximately 1.5 cm, healthy seedlings with approximately equal root lengths were chosen to monitor growth at 24-h intervals for 4 d by marking the position of the root tip prior to photography. To measure the root length of 7-d-old seedlings, plates were prepared in the same manner as described for root growth rate analysis.
Hypocotyl Growth Analyses
After 2 d of vernalization, germination was induced by exposure to light for 6 h. Plates were subsequently placed in the growth chamber and maintained in the dark for 5 or 11 d prior to photography. For the experiment on the effect of endogenous auxin on hypocotyl length, plates were maintained in the dark at 22°C for 2 d and then transferred to 28°C for 3 d. Control plates were maintained in the dark at 22°C for 5 d.
For kinematic analysis of hypocotyl growth, after vernalization, plates were oriented vertically in infrared light (880-nm light-emitting diodes) at 22°C. Hypocotyl elongation was monitored every 2 h for 5 d by a Canon EOS 50D. Hypocotyl length at 24, 48, 72, and 96 h after germination was measured on 10 seedlings, and this process was repeated three times; growth curves thus represent the average of 30 seedlings.
Immunolocalization of Fucogalactoxyloglucan and Rhamnogalacturonan-I
Six-day-old seedlings were fixed for 1 h in 4% paraformaldehyde in phosphate-buffered saline (PBS) and rinsed twice in PBS. Five percent bovine serum albumin in PBS was used as blocking solution for 1 h prior to the addition of 1/100 LM5 antibody (Plantprobes) and 1/100 CCRC-M1 antibody (CarboSource) in 2.5% bovine serum albumin. After 1 h of incubation and five washes with PBS, secondary antibodies were applied: 1/200 anti-rat CY5 and 1/200 anti-mouse tetramethyl rhodamine isothiocyanate, respectively.
In Situ XET and XEH Assays
In Situ XET Assay.
The in situ detection of XET activity was based on the method described by Vissenberg et al. (2000) with minor modifications. Hypocotyls from 5-d-old plants and roots from 6-d-old plants were collected in 25 mm MES buffer and subsequently incubated with 6.5 µm XXXG-SR (synthesized as described by Nishikubo et al. [2007]) in 25 mm MES buffer, pH 5.5, in the dark at room temperature for 2 h. Hypocotyls or roots were intensively washed in ethanol:formic acid:water (15:1:4, v/v) for 10 min and subsequently washed with 5% formic acid for 3 h to remove unincorporated XXXG-SR. The tissues were then examined by confocal laser scanning microscopy (Leica Microsystems) with a 561-nm laser, and the signal was detected between 566 and 650 nm. Negative controls were prepared in the same manner as the sample sections, except that samples were heated at 95°C for 1 h to denature enzymes.
In Situ XEH Assay.
In situ detection of XEH activity employed the method devised by Ibatullin et al. (2009) with minor modifications. Hypocotyls from 5-d-old plants and roots from 6-d-old plants were collected in 25 mm MES buffer, pH 5.5. The solution was replaced by 25 mm MES, pH 5.5, containing 9.3 × 10−4 m XXXG-Res, prepared according to Ibatullin et al. (2009). The reaction was allowed to proceeded in the dark for 1 to 1.5 h for hypocotyls and 1.5 h for roots. The samples were then examined by confocal laser scanning microscopy (Leica Microsystems) using 561-nm laser excitation and monitoring emission at 566 to 650 nm. Negative controls were prepared in the same manner as the sample sections, except that samples were heated at 95°C for 1 h to denature enzymes.
Sequence data from this article can be found in the GenBank data libraries under accession numbers TmNXG1 (CAA48324), AtXTH31 (AAL07012 and At3g44990), and AtXTH32 (AAD31572 and At2g36870), and in the Arabidopsis Information Resource (CS67890 [xth31-1], CS67891 [xth32-1], and CS67892 [xth31-1 xth32-1]).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. AtXTH31 expression construct sequences.
Supplemental Figure S2. SDS-PAGE and western-blot analyses of the AtXTH31 expression product.
Supplemental Figure S3. Reconstructed mass spectrum of recombinant AtXTH31.
Supplemental Figure S4. HPAEC-PAD analysis of AtXTH31 tamarind seed xyloglucan limit digest products.
Supplemental Figure S5. Expression profiles of AtXTH31 (At3g44990) and AtXTH32 (At2g36870) from the Genevestigator data set (Zimmermann et al., 2004).
Supplemental Figure S6. Semiquantitative PCR of AtXTH31 and AtXTH32 in different tissues grown under long-day (16 h) light conditions at 7 d post germination versus clathrin as a control.
Supplemental Figure S7. Homozygous Arabidopsis T-DNA insertion lines obtained for AtXTH31 (SALK_129686) and AtXTH32 (SALK_131432).
Supplemental Figure S8. Dark-grown hypocotyl length post germination.
Supplemental Figure S9. Localization of polysaccharides in roots.
Supplemental Figure S10. Localization of xyloglucan endohydrolase activity in the root-hypocotyl junction zone of 6-d-old Arabidopsis using XXXG-Res.
Supplemental Figure S11. Localization of xyloglucan endo-transglycosylase activity in 5-d-old Arabidopsis etiolated hypocotyls using XXXG-SR.
Supplemental Table S1. PCR primers.
Acknowledgments
We thank Prof. Malcolm Bennett (University of Nottingham) for helpful discussions on growth kinematic profiling.
Glossary
- XG
xyloglucan
- XET
xyloglucan endo-transglycosylase
- XEH
xyloglucan endo-hydrolase
- BCA
bicinchoninic acid
- HPSEC-ELS
high-performance size-exclusion chromatography with evaporative light-scattering detection
- HPAEC-PAD
high-performance anion-exchange chromatography with pulsed amperometric detection
- Col-0
ecotype Columbia
- RT
reverse transcription
- AEV
absolute expression value
- PBS
phosphate-buffered saline
References
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Baba K, Park YW, Kaku T, Kaida R, Takeuchi M, Yoshida M, Hosoo Y, Ojio Y, Okuyama T, Taniguchi T, et al. (2009) Xyloglucan for generating tensile stress to bend tree stem. Mol Plant 2: 893–903 [DOI] [PubMed] [Google Scholar]
- Baumann MJ, Eklöf JM, Michel G, Kallas AM, Teeri TT, Czjzek M, Brumer H., III (2007) Structural evidence for the evolution of xyloglucanase activity from xyloglucan endo-transglycosylases: biological implications for cell wall metabolism. Plant Cell 19: 1947–1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becnel J, Natarajan M, Kipp A, Braam J. (2006) Developmental expression patterns of Arabidopsis XTH genes reported by transgenes and Genevestigator. Plant Mol Biol 61: 451–467 [DOI] [PubMed] [Google Scholar]
- Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Görlach J. (2001) Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell 13: 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpita N, McCann M. (2000). The cell wall. In B Buchanan, W Gruissem, R Jones, eds, Biochemistry and Molecular Biology of Plants. John Wiley & Sons, Somerset, NJ, pp 52–108
- Carpita NC, Gibeaut DM. (1993) Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J 3: 1–30 [DOI] [PubMed] [Google Scholar]
- Cavalier DM, Lerouxel O, Neumetzler L, Yamauchi K, Reinecke A, Freshour G, Zabotina OA, Hahn MG, Burgert I, Pauly M, et al. (2008) Disrupting two Arabidopsis thaliana xylosyltransferase genes results in plants deficient in xyloglucan, a major primary cell wall component. Plant Cell 20: 1519–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ. (2000) Expansive growth of plant cell walls. Plant Physiol Biochem 38: 109–124 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. (2005) Growth of the plant cell wall. Nat Rev Mol Cell Biol 6: 850–861 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ, Jarvis MC. (2012) Comparative structure and biomechanics of plant primary and secondary cell walls. Front Plant Sci 3: 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Bem LE, Vincentz MG. (2010) Evolution of xyloglucan-related genes in green plants. BMC Evol Biol 10: 341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards M, Dea ICM, Bulpin PV, Reid JSG. (1986) Purification and properties of a novel xyloglucan-specific endo-(1-4)-β-D-glucanase from germinated nasturtium seeds (Tropaeolum majus L.). J Biol Chem 261: 9489–9494 [PubMed] [Google Scholar]
- Eklöf JM, Brumer H. (2010) The XTH gene family: an update on enzyme structure, function, and phylogeny in xyloglucan remodeling. Plant Physiol 153: 456–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas V, Sulova Z, Stratilova E, Hanna R, Maclachlan G. (1992) Cleavage of xyloglucan by nasturtium seed xyloglucanase and transglycosylation to xyloglucan subunit oligosaccharides. Arch Biochem Biophys 298: 365–370 [DOI] [PubMed] [Google Scholar]
- Freshour G, Clay RP, Fuller MS, Albersheim P, Darvill AG, Hahn MG. (1996) Developmental and tissue-specific structural alterations of the cell-wall polysaccharides of Arabidopsis thaliana roots. Plant Physiol 110: 1413–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry SC. (1989) Cellulases, hemicelluloses and auxin-stimulated growth: a possible relationship. Physiol Plant 75: 532–536 [Google Scholar]
- Fry SC, Smith RC, Renwick KF, Martin DJ, Hodge SK, Matthews KJ. (1992) Xyloglucan endotransglycosylase, a new wall-loosening enzyme activity from plants. Biochem J 282: 821–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry SC, York WS, Albersheim P, Darvill A, Hayashi T, Joseleau JP, Kato Y, Lorences EP, Maclachlan GA, McNeil M, et al. (1993) An unambiguous nomenclature for xyloglucan-derived oligosaccharides. Physiol Plant 89: 1–3 [Google Scholar]
- Geisler DA, Sampathkumar A, Mutwil M, Persson S. (2008) Laying down the bricks: logistic aspects of cell wall biosynthesis. Curr Opin Plant Biol 11: 647–652 [DOI] [PubMed] [Google Scholar]
- Geitmann A, Ortega JKE. (2009) Mechanics and modeling of plant cell growth. Trends Plant Sci 14: 467–478 [DOI] [PubMed] [Google Scholar]
- Gendreau E, Traas J, Desnos T, Grandjean O, Caboche M, Höfte H. (1997) Cellular basis of hypocotyl growth in Arabidopsis thaliana. Plant Physiol 114: 295–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouet P, Robert X, Courcelle E. (2003) ESPript/ENDscript: extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res 31: 3320–3323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günl M, Neumetzler L, Kraemer F, de Souza A, Schultink A, Pena M, York WS, Pauly M. (2011) AXY8 encodes an α-fucosidase, underscoring the importance of apoplastic metabolism on the fine structure of Arabidopsis cell wall polysaccharides. Plant Cell 23: 4025–4040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha MA, Apperley DC, Jarvis MC. (1997) Molecular rigidity in dry and hydrated onion cell walls. Plant Physiol 115: 593–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrissat B, Coutinho PM, Davies GJ. (2001) A census of carbohydrate-active enzymes in the genome of Arabidopsis thaliana. Plant Mol Biol 47: 55–72 [PubMed] [Google Scholar]
- Hernández-Nistal J, Martín I, Labrador E, Dopico B. (2010) The immunolocation of XTH1 in embryonic axes during chickpea germination and seedling growth confirms its function in cell elongation and vascular differentiation. J Exp Bot 61: 4231–4238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman M, Jia ZH, Peña MJ, Cash M, Harper A, Blackburn AR, II, Darvill A, York WS. (2005) Structural analysis of xyloglucans in the primary cell walls of plants in the subclass Asteridae. Carbohydr Res 340: 1826–1840 [DOI] [PubMed] [Google Scholar]
- Hsieh YSY, Harris PJ. (2009) Xyloglucans of monocotyledons have diverse structures. Mol Plant 2: 943–965 [DOI] [PubMed] [Google Scholar]
- Ibatullin FM, Banasiak A, Baumann MJ, Greffe L, Takahashi J, Mellerowicz EJ, Brumer H. (2009) A real-time fluorogenic assay for the visualization of glycoside hydrolase activity in planta. Plant Physiol 151: 1741–1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibatullin FM, Baumann MJ, Greffe L, Brumer H. (2008) Kinetic analyses of retaining endo-(xylo)glucanases from plant and microbial sources using new chromogenic xylogluco-oligosaccharide aryl glycosides. Biochemistry 47: 7762–7769 [DOI] [PubMed] [Google Scholar]
- Iglesias N, Abelenda JA, Rodiño M, Sampedro J, Revilla G, Zarra I. (2006) Apoplastic glycosidases active against xyloglucan oligosaccharides of Arabidopsis thaliana. Plant Cell Physiol 47: 55–63 [DOI] [PubMed] [Google Scholar]
- Jamet E, Roujol D, San-Clemente H, Irshad M, Soubigou-Taconnat L, Renou JP, Pont-Lezica R. (2009) Cell wall biogenesis of Arabidopsis thaliana elongating cells: transcriptomics complements proteomics. BMC Genomics 10: 505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis MC. (2009) Plant cell walls: supramolecular assembly, signalling and stress. Struct Chem 20: 245–253 [Google Scholar]
- Johansson P, Brumer H, III, Baumann MJ, Kallas ÅM, Henriksson H, Denman SE, Teeri TT, Jones TA. (2004) Crystal structures of a poplar xyloglucan endotransglycosylase reveal details of transglycosylation acceptor binding. Plant Cell 16: 874–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaewthai N, Harvey AJ, Hrmova M, Brumer H, Ezcurra I, Teeri TT, Fincher GB. (2010) Heterologous expression of diverse barley XTH genes in the yeast Pichia pastoris. Plant Biotechnol 27: 251–258 [Google Scholar]
- Kallas ÅM, Piens K, Denman SE, Henriksson H, Fäldt J, Johansson P, Brumer H, Teeri TT. (2005) Enzymatic properties of native and deglycosylated hybrid aspen (Populus tremula × tremuloides) xyloglucan endotransglycosylase 16A expressed in Pichia pastoris. Biochem J 390: 105–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox JP. (2008) Revealing the structural and functional diversity of plant cell walls. Curr Opin Plant Biol 11: 308–313 [DOI] [PubMed] [Google Scholar]
- Kutschera U. (2001) Stem elongation and cell wall proteins in flowering plants. Plant Biol 3: 466–480 [Google Scholar]
- Lee J, Burns TH, Light G, Sun Y, Fokar M, Kasukabe Y, Fujisawa K, Maekawa Y, Allen RD. (2010) Xyloglucan endotransglycosylase/hydrolase genes in cotton and their role in fiber elongation. Planta 232: 1191–1205 [DOI] [PubMed] [Google Scholar]
- Liepman AH, Wightman R, Geshi N, Turner SR, Scheller HV. (2010) Arabidopsis: a powerful model system for plant cell wall research. Plant J 61: 1107–1121 [DOI] [PubMed] [Google Scholar]
- Lin Y, Schiefelbein J. (2001) Embryonic control of epidermal cell patterning in the root and hypocotyl of Arabidopsis. Development 128: 3697–3705 [DOI] [PubMed] [Google Scholar]
- Lopez-Casado G, Urbanowicz BR, Damasceno CMB, Rose JKC. (2008) Plant glycosyl hydrolases and biofuels: a natural marriage. Curr Opin Plant Biol 11: 329–337 [DOI] [PubMed] [Google Scholar]
- Malamy JE, Benfey PN. (1997) Down and out in Arabidopsis: the formation of lateral roots. Trends Plant Sci 2: 390–396 [Google Scholar]
- Marcus SE, Verhertbruggen Y, Herve C, Ordaz-Ortiz JJ, Farkas V, Pedersen HL, Willats WGT, Knox JP. (2008) Pectic homogalacturonan masks abundant sets of xyloglucan epitopes in plant cell walls. BMC Plant Biol 8: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maris A, Kaewthai N, Eklöf JM, Miller JG, Brumer H, Fry SC, Verbelen J-P, Vissenberg K. (2011) Differences in enzymic properties of five recombinant xyloglucan endotransglucosylase/hydrolase (XTH) proteins of Arabidopsis thaliana. J Exp Bot 62: 261–271 [DOI] [PubMed] [Google Scholar]
- Martinez-Fleites C, Guerreiro CI, Baumann MJ, Taylor EJ, Prates JAM, Ferreira LMA, Fontes CM, Brumer H, Davies GJ. (2006) Crystal structures of Clostridium thermocellum xyloglucanase, XGH74A, reveal the structural basis for xyloglucan recognition and degradation. J Biol Chem 281: 24922–24933 [DOI] [PubMed] [Google Scholar]
- McCartney L, Steele-King CG, Jordan E, Knox JP. (2003) Cell wall pectic (1→4)-beta-d-galactan marks the acceleration of cell elongation in the Arabidopsis seedling root meristem. Plant J 33: 447–454 [DOI] [PubMed] [Google Scholar]
- McDougall GJ, Fry SC. (1989) Structure-activity relationships for xyloglucan oligosaccharides with antiauxin activity. Plant Physiol 89: 883–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFeeters RF. (1980) A manual method for reducing sugar determinations with 2,2′-bicinchoninate reagent. Anal Biochem 103: 302–306 [DOI] [PubMed] [Google Scholar]
- Mellerowicz EJ, Immerzeel P, Hayashi T. (2008) Xyloglucan: the molecular muscle of trees. Ann Bot (Lond) 102: 659–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miedes E, Herbers K, Sonnewald U, Lorences EP. (2010) Overexpression of a cell wall enzyme reduces xyloglucan depolymerization and softening of transgenic tomato fruits. J Agric Food Chem 58: 5708–5713 [DOI] [PubMed] [Google Scholar]
- Miedes E, Zarra I, Hoson T, Herbers K, Sonnewald U, Lorences EP. (2011) Xyloglucan endotransglucosylase and cell wall extensibility. J Plant Physiol 168: 196–203 [DOI] [PubMed] [Google Scholar]
- Minic Z. (2008) Physiological roles of plant glycoside hydrolases. Planta 227: 723–740 [DOI] [PubMed] [Google Scholar]
- Nishikubo N, Awano T, Banasiak A, Bourquin V, Ibatullin F, Funada R, Brumer H, Teeri TT, Hayashi T, Sundberg B, et al. (2007) Xyloglucan endo-transglycosylase (XET) functions in gelatinous layers of tension wood fibers in poplar: a glimpse into the mechanism of the balancing act of trees. Plant Cell Physiol 48: 843–855 [DOI] [PubMed] [Google Scholar]
- Nishikubo N, Takahashi J, Roos AA, Derba-Maceluch M, Piens K, Brumer H, Teeri TT, Stålbrand H, Mellerowicz EJ. (2011) Xyloglucan endo-transglycosylase-mediated xyloglucan rearrangements in developing wood of hybrid aspen (Populus tremula × tremuloides). Plant Physiol 155: 399–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitani K, Tominaga R. (1992) Endo-xyloglucan transferase, a novel class of glycosyltransferase that catalyzes transfer of a segment of xyloglucan molecule to another xyloglucan molecule. J Biol Chem 267: 21058–21064 [PubMed] [Google Scholar]
- Opazo MC, Figueroa CR, Henríquez J, Herrera R, Bruno C, Valenzuela PDT, Moya-León MA. (2010) Characterization of two divergent cDNAs encoding xyloglucan endotransglycosylase/hydrolase (XTH) expressed in Fragaria chiloensis fruit. Plant Sci 179: 479–488 [DOI] [PubMed] [Google Scholar]
- Osato Y, Yokoyama R, Nishitani K. (2006) A principal role for AtXTH18 in Arabidopsis thaliana root growth: a functional analysis using RNAi plants. J Plant Res 119: 153–162 [DOI] [PubMed] [Google Scholar]
- Park YB, Cosgrove DJ. (2012) A revised architecture of primary cell walls based on biomechanical changes induced by substrate-specific endoglucanases. Plant Physiol 158: 1933–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauly M, Albersheim P, Darvill A, York WS. (1999) Molecular domains of the cellulose/xyloglucan network in the cell walls of higher plants. Plant J 20: 629–639 [DOI] [PubMed] [Google Scholar]
- Po HN, Senozan NM. (2001) The Henderson-Hasselbalch equation: its history and limitations. J Chem Educ 78: 1499–1503 [Google Scholar]
- Puhlmann J, Bucheli E, Swain MJ, Dunning N, Albersheim P, Darvill AG, Hahn MG. (1994) Generation of monoclonal antibodies against plant cell-wall polysaccharides. I. Characterization of a monoclonal antibody to a terminal α-(1→2)-linked fucosyl-containing epitope. Plant Physiol 104: 699–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose JKC, Bennett AB. (1999) Cooperative disassembly of the cellulose-xyloglucan network of plant cell walls: parallels between cell expansion and fruit ripening. Trends Plant Sci 4: 176–183 [DOI] [PubMed] [Google Scholar]
- Rose JKC, Braam J, Fry SC, Nishitani K. (2002) The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: current perspectives and a new unifying nomenclature. Plant Cell Physiol 43: 1421–1435 [DOI] [PubMed] [Google Scholar]
- Sainsbury F, Thuenemann EC, Lomonossoff GP. (2009) pEAQ: versatile expression vectors for easy and quick transient expression of heterologous proteins in plants. Plant Biotechnol J 7: 682–693 [DOI] [PubMed] [Google Scholar]
- Salamoni AT, Sierakowski MR, Boeira JP, Neto OAR, Quoirin M. (2008) Effect of seed xyloglucans and derivates on the growth of Arabidopsis thaliana. Ciencia Florestal 18: 315–320 [Google Scholar]
- Sampedro J, Gianzo C, Iglesias N, Guitián E, Revilla G, Zarra I. (2012) AtBGAL10 is the main xyloglucan β-galactosidase in Arabidopsis, and its absence results in unusual xyloglucan subunits and growth defects. Plant Physiol 158: 1146–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampedro J, Pardo B, Gianzo C, Guitián E, Revilla G, Zarra I. (2010) Lack of α-xylosidase activity in Arabidopsis alters xyloglucan composition and results in growth defects. Plant Physiol 154: 1105–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres B, Benfey P, Dolan L. (2002). Root development. The Arabidopsis Book 1: e0101, doi/10.1199/tab.0101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville C, Bauer S, Brininstool G, Facette M, Hamann T, Milne J, Osborne E, Paredez A, Persson S, Raab T, et al. (2004) Toward a systems approach to understanding plant cell walls. Science 306: 2206–2211 [DOI] [PubMed] [Google Scholar]
- Stratilová E, Ait-Mohand F, Rehulka P, Garajová S, Flodrová D, Rehulková H, Farkas V. (2010) Xyloglucan endotransglycosylases (XETs) from germinating nasturtium (Tropaeolum majus) seeds: isolation and characterization of the major form. Plant Physiol Biochem 48: 207–215 [DOI] [PubMed] [Google Scholar]
- Sundqvist G, Stenvall M, Berglund H, Ottosson J, Brumer H. (2007) A general, robust method for the quality control of intact proteins using LC-ESI-MS. J Chromatogr B Analyt Technol Biomed Life Sci 852: 188–194 [DOI] [PubMed] [Google Scholar]
- Takeda T, Furuta Y, Awano T, Mizuno K, Mitsuishi Y, Hayashi T. (2002) Suppression and acceleration of cell elongation by integration of xyloglucans in pea stem segments. Proc Natl Acad Sci USA 99: 9055–9060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres FG, Troncoso OP, Lopez D, Grande C, Gomez CM. (2009) Reversible stress softening and stress recovery of cellulose networks. Soft Matter 5: 4185–4190 [Google Scholar]
- Urbanowicz BR, Bennett AB, Del Campillo E, Catalá C, Hayashi T, Henrissat B, Höfte H, McQueen-Mason SJ, Patterson SE, Shoseyov O, et al. (2007) Structural organization and a standardized nomenclature for plant endo-1,4-β-glucanases (cellulases) of glycosyl hydrolase family 9. Plant Physiol 144: 1693–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche F, Verbelen JP, Van Der Straeten D. (2005) Of light and length: regulation of hypocotyl growth in Arabidopsis. Bioessays 27: 275–284 [DOI] [PubMed] [Google Scholar]
- Vargas-Rechia C, Reicher F, Sierakowski MR, Heyraud A, Driguez H, Linart Y. (1998) Xyloglucan octasaccharide XXLGol derived from the seeds of Hymenaea courbaril acts as a signaling molecule. Plant Physiol 116: 1013–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente AR, Saladie M, Rose JKC, Labavitch JM. (2007) The linkage between cell wall metabolism and fruit softening: looking to the future. J Sci Food Agric 87: 1435–1448 [Google Scholar]
- Vissenberg K, Fry SC, Verbelen JP. (2001) Root hair initiation is coupled to a highly localized increase of xyloglucan endotransglycosylase action in Arabidopsis roots. Plant Physiol 127: 1125–1135 [PMC free article] [PubMed] [Google Scholar]
- Vissenberg K, Martinez-Vilchez IM, Verbelen JP, Miller JG, Fry SC. (2000) In vivo colocalization of xyloglucan endotransglycosylase activity and its donor substrate in the elongation zone of Arabidopsis roots. Plant Cell 12: 1229–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J. (2008) Unique aspects of the grass cell wall. Curr Opin Plant Biol 11: 301–307 [DOI] [PubMed] [Google Scholar]
- von Schantz L, Gullfot F, Scheer S, Filonova L, Gunnarsson LC, Flint JE, Daniel G, Nordberg-Karlsson E, Brumer H, Ohlin M. (2009) Affinity maturation generates greatly improved xyloglucan-specific carbohydrate binding modules. BMC Biotechnol 9: 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Glazebrook J. (2002). Arabidopsis: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Western TL, Skinner DJ, Haughn GW. (2000) Differentiation of mucilage secretory cells of the Arabidopsis seed coat. Plant Physiol 122: 345–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf S, Hématy K, Höfte H. (2012) Growth control and cell wall signaling in plants. Annu Rev Plant Biol 63: 381–407 [DOI] [PubMed] [Google Scholar]
- Wu YJ, Jeong BR, Fry SC, Boyer JS. (2005) Change in XET activities, cell wall extensibility and hypocotyl elongation of soybean seedlings at low water potential. Planta 220: 593–601 [DOI] [PubMed] [Google Scholar]
- Yarbrough JM, Himmel ME, Ding SY. (2009) Plant cell wall characterization using scanning probe microscopy techniques. Biotechnol Biofuels 2: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama R, Nishitani K. (2001) A comprehensive expression analysis of all members of a gene family encoding cell-wall enzymes allowed us to predict cis-regulatory regions involved in cell-wall construction in specific organs of Arabidopsis. Plant Cell Physiol 42: 1025–1033 [DOI] [PubMed] [Google Scholar]
- York WS, Darvill AG, Albersheim P. (1984) Inhibition of 2,4-dichlorophenoxyacetic acid-stimulated elongation of pea stem segments by a xyloglucan oligosaccharide. Plant Physiol 75: 295–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- York WS, Harvey LK, Guillen R, Albersheim P, Darvill AG. (1993) The structure of plant cell walls. 36. Structural analysis of tamarind seed xyloglucan oligosaccharides using beta-galactosidase digestion and spectroscopic methods. Carbohydr Res 248: 285–301 [DOI] [PubMed] [Google Scholar]
- Zhang Q, Brumer H, Ågren H, Tu YQ. (2011) The adsorption of xyloglucan on cellulose: effects of explicit water and side chain variation. Carbohydr Res 346: 2595–2602 [DOI] [PubMed] [Google Scholar]
- Zhou Q, Rutland MW, Teeri TT, Brumer H. (2007) Xyloglucan in cellulose modification. Cellulose 14: 625–641 [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwieniecki MA, Melcher PJ, Holbrook NM. (2001) Hydrogel control of xylem hydraulic resistance in plants. Science 291: 1059–1062 [DOI] [PubMed] [Google Scholar]