Abstract
Many plant proteins are modified with N-linked oligosaccharides at asparagine-X-serine/threonine sites during transit through the endoplasmic reticulum and the Golgi. We have identified a number of Arabidopsis (Arabidopsis thaliana) proteins with modifications consisting of an N-linked N-acetyl-d-glucosamine monosaccharide (N-GlcNAc). Electron transfer dissociation mass spectrometry analysis of peptides bearing this modification mapped the modification to asparagine-X-serine/threonine sites on proteins that are predicted to transit through the endoplasmic reticulum and Golgi. A mass labeling method was developed and used to study N-GlcNAc modification of two thioglucoside glucohydrolases (myrosinases), TGG1 and TGG2 (for thioglucoside glucohydrolase). These myrosinases are also modified with high-mannose (Man)-type glycans. We found that N-GlcNAc and high-Man-type glycans can occur at the same site. It has been hypothesized that N-GlcNAc modifications are generated when endo-β-N-acetylglucosaminidase (ENGase) cleaves N-linked glycans. We examined the effects of mutations affecting the two known Arabidopsis ENGases on N-GlcNAc modification of myrosinase and found that modification of TGG2 was greatly reduced in one of the single mutants and absent in the double mutant. Surprisingly, N-GlcNAc modification of TGG1 was not affected in any of the mutants. These data support the hypothesis that ENGases hydrolyze high-Man glycans to produce some of the N-GlcNAc modifications but also suggest that some N-GlcNAc modifications are generated by another mechanism. Since N-GlcNAc modification was detected at only one site on each myrosinase, the production of the N-GlcNAc modification may be regulated.
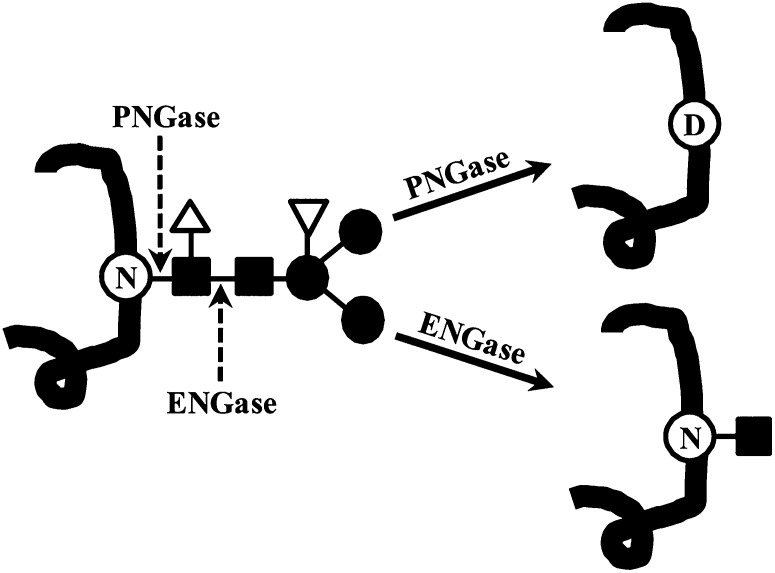
Many plant proteins are modified with N-linked (Glc)3(Man)9(GlcNAc)2 glycan(s) at the Asn of the Asn-X-Ser/Thr sequon, where X is any amino acid except Pro, as they enter the endoplasmic reticulum (ER; Pattison and Amtmann, 2009). These N-glycans are then modified by the removal and addition of sugars as the protein transits through the secretory system. Through this process, a diverse array of glycan structures can be generated. These glycan structures have been divided into four categories (Lerouge et al., 1998). High-Man-type glycans are produced in the ER. Complex-, paucimannosidic-, and hybrid-type glycans, which are produced in the Golgi, have Xyl and Fuc attached to the protein-proximal Man and GlcNAc of their (Man)3(GlcNAc)2 common core (Fig. 1).
Figure 1.
PNGase and ENGase reactions. PNGase and ENGase cleave the common core of N-linked glycans, which is composed of GlcNAc (closed squares) and Man (closed circles), at the sites indicated. In plants, the common core can be modified with Fuc (open triangle), which inhibits PNGase F, and Xyl (inverted triangle). When a glycan is removed by PNGase F, the modified Asn (N) is converted to an Asp (D).
N-Glycans influence protein conformation, stability, and activity. They play a major role in the quality-control mechanisms that (1) retain misfolded proteins in the ER until they become properly folded and (2) target terminally misfolded proteins for destruction by the endoplasmic reticulum-associated protein degradation (ERAD) pathway (Vembar and Brodsky, 2008). During ERAD, misfolded proteins are exported to the cytosol, the glycans are removed and recycled, and the proteins are degraded by the 26S proteasome. There is also precedence for glycosylation status regulating the activity of plant proteins. Concanavalin A (Con A), a lectin from jackbean (Canavalia ensiformis), is synthesized as an inactive high-Man precursor protein that is deglycosylated as part of the processing that converts it to an active lectin (Min et al., 1992; Sheldon and Bowles, 1992).
Modification of the Asn of the Asn-X-Ser/Thr sequon with an N-linked N-acetyl-d-glucosamine monosaccharide (N-GlcNAc) occurs in fungi (Hase et al., 1982) and has recently been discovered in animals (Chalkley et al., 2009; Wang et al., 2010). N-GlcNAc modification also occurs in plants. The Asn-X-Ser/Thr sequon of ribosome-inactivating proteins from sponge gourd (Luffa cylindrica) and pokeweed (Phytolacca americana) is modified with N-GlcNAc (Islam et al., 1991; Zeng et al., 2003). Matrix-assisted laser-desorption ionization time-of-flight analysis of trypsin fragments derived from antibodies expressed in transgenic maize (Zea mays) and tobacco (Nicotiana tabacum) detects fragments with masses consistent with N-GlcNAc modification (Bakker et al., 2006; Rademacher et al., 2008).
Several mechanisms for the origin of this modification have been proposed. It has been proposed that cytosolic endo-β-N-actetylglucosaminidase (ENGase) produces the N-GlcNAc modification when it removes larger N-linked glycans (Fig. 1). However, ENGase is more active toward free oligosaccharides than glycoproteins (Chantret and Moore, 2008), and it is unclear how cytosol-localized ENGase gains access to these glycoproteins, which are not generally localized in the cytosol. One possibility is that ENGase is acting on misfolded proteins that have been exported to the cytosol for degradation by the ERAD pathway. Alternatively, it is possible that N-GlcNAc modifications are an artifact that is generated during sample preparation. Thus, it remains an open question how N-GlcNAc modifications are produced and if they have a role in the cell.
Here, we report the discovery of a number of Arabidopsis (Arabidopsis thaliana) proteins bearing N-GlcNAc modifications that are attached to the Asn of the Asn-X-Ser/Thr sites. We present evidence that these modifications occur in planta. An analysis of mutants indicates that, in at least one case, the modification is produced by Arabidopsis ENGases, but it also suggests that some N-GlcNAc modifications are generated by another mechanism.
RESULTS
Arabidopsis Proteins Can Be Modified with an N-GlcNAc
During the course of experiments to identify O-GlcNAc-modified proteins of Arabidopsis, we characterized the glycans with terminal GlcNAc that remain after removal of the complex and hybrid glycans, which are abundant and commonly terminate with GlcNAc (Rayon et al., 1999). Peptide:N-glycosidase F (PNGase F) removes many types of N-linked glycans and in the process converts the modified Asn to Asp (Fig. 1). However, plant complex and hybrid glycans are resistant to removal by PNGase F because the protein-proximal GlcNAc is modified with Fuc. This modification does not occur in the fucta/fuctb/xylt triple mutant; thus, PNGase F removes the complex and hybrid glycans on proteins from this mutant (Strasser et al., 2004; Supplemental Fig. S1). Using an assay that detects terminal GlcNAc by capping it with [3H]Gal, we found that many proteins still had glycans terminating with GlcNAc after PNGase F treatment (Fig. 2). β-Elimination chemistry was used to determine if these glycans were N or O linked; O-linked glycans are removed by this chemistry. Most of the modifications were resistant to β-elimination, suggesting that they are N linked (Fig. 2).
Figure 2.
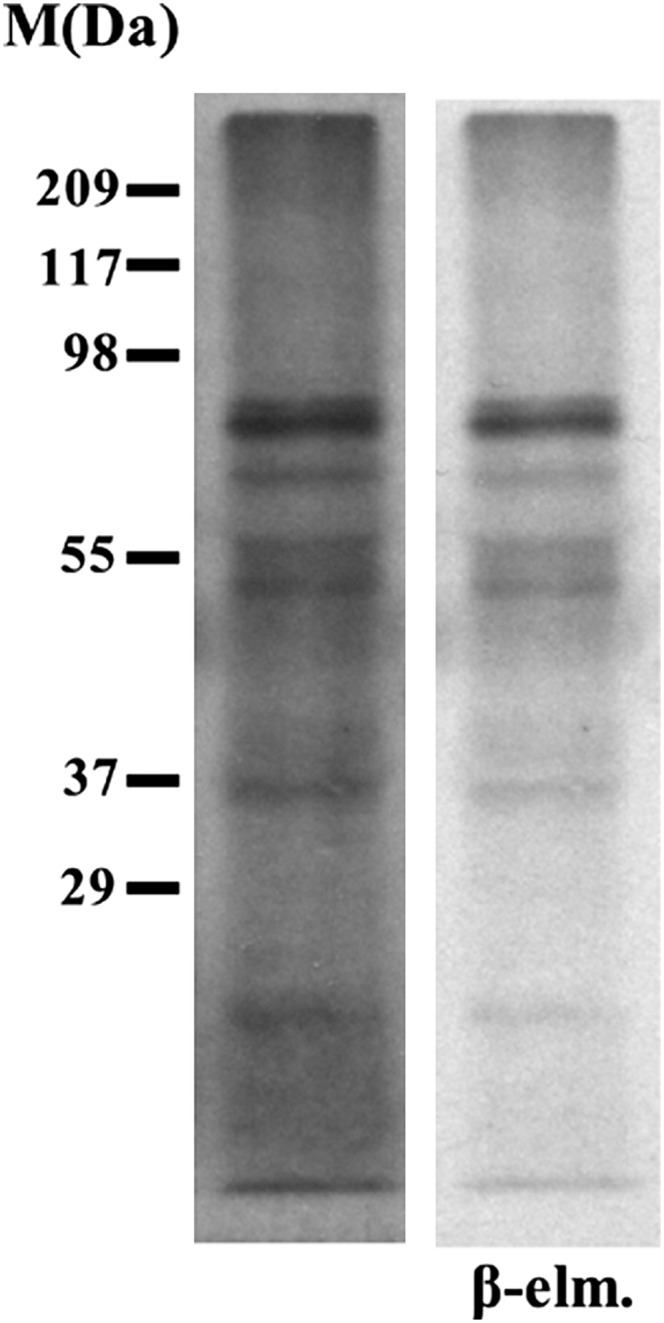
PNGase F-resistant glycans with terminal GlcNAc on fucta/fuctb/xylt triple mutant proteins are resistant to β-elimination chemistry. Glycans with terminal GlcNAc on a blot containing PNGase F-treated fucta/fuctb/xylt triple mutant proteins were detected by labeling with [3H]Gal. The lane on the left shows the blot prior to β-elimination, and the right lane (β-elm.) shows the same blot after subjecting it to β-elimination chemistry, which removes O-linked glycans.
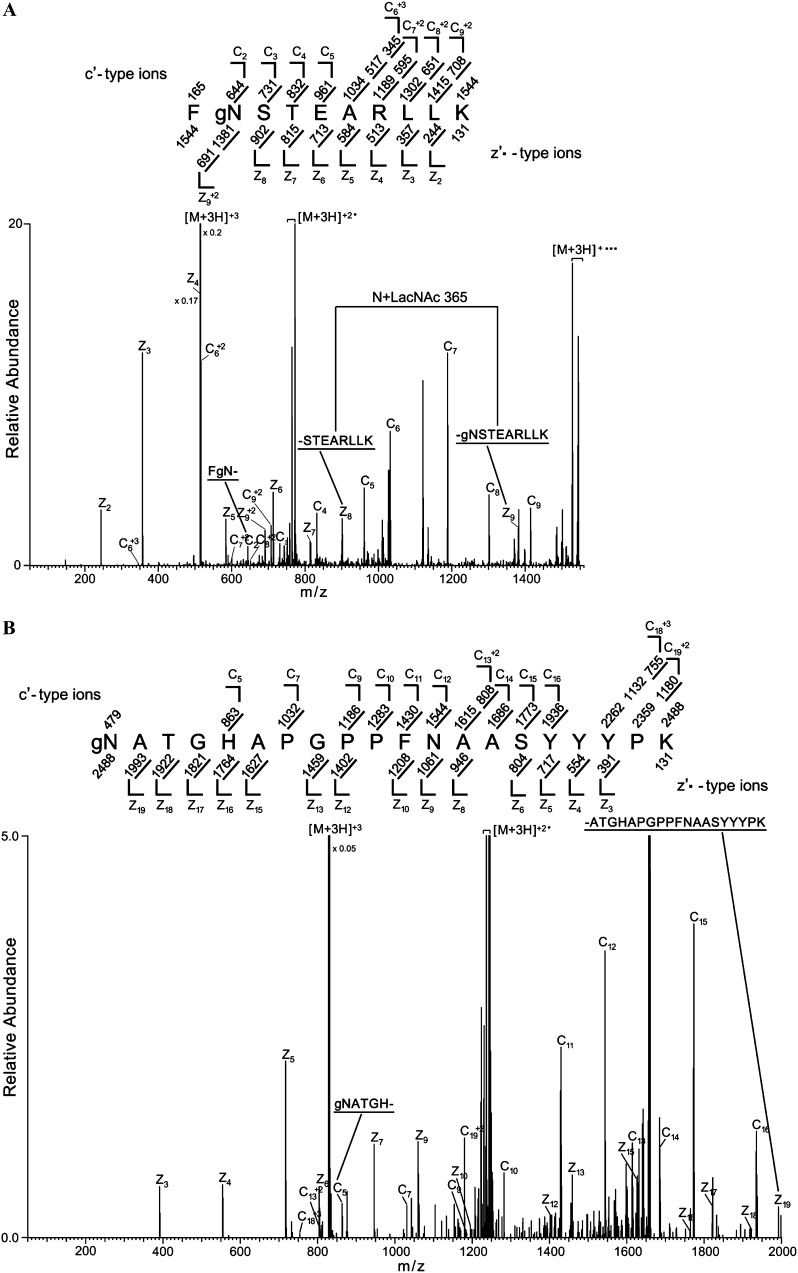
The identities of the proteins bearing modifications with terminal GlcNAc and the nature of the modifications were investigated using mass spectrometry. Proteins from the fucta/fuctb/xylt triple mutant were treated with PNGase F and digested with endo-Lys-C or trypsin. Peptides bearing modifications with terminal GlcNAc were then enriched by lectin affinity chromatography (Kim et al., 2011). In this procedure, terminal GlcNAc is enzymatically capped with Gal using bovine β-1,4-galactosyltransferase, creating the disaccharide N-acetyl-d-lactosamine (LacNAc). Peptides with LacNAc modifications are then enriched by chromatography on a Ricinus Communis Agglutinin I lectin (RCA I) column (Supplemental Fig. S2). Electron transfer dissociation (ETD) tandem mass spectrometry (MS/MS) analysis identified peptides from 14 different proteins with precursor ion masses 365 D (the mass of one LacNAc) greater than the unmodified peptide that produced tandem mass spectra consistent with them having modifications consisting of a single N-linked LacNAc. For example, Figure 3A shows the tandem mass spectrum for a triply charged precursor ion 515.2663 mass-to-charge ratio (m/z), which supports a match to the thioglucoside glucohydrolase2 (TGG2) peptide FNSTEARLLK with an N-linked LacNAc modification at the Asn residue (Asn-340 of full-length TGG2). Consistent with amino acids 3 to 10 of the peptide being unmodified, the masses of fragment ions z2 to z8 are the same as the theoretically unmodified fragment ion m/z values. In contrast, and consistent with the N being modified, the mass of the next larger z ion (z9) is larger than the previous ion by the mass of N+LacNAc. This is further supported because all of the detected c ions have masses 365 D greater than theoretically unmodified fragment ions. This conclusion is further supported because the singly, doubly, and triply charged full-length peptide ions (labeled [M+3H]+, [M+3H]+2, and [M+3H]+3 in the figure) have masses that are 365 D greater than the unmodified peptide. Figure 3B shows the tandem mass spectrum for a triply charged precursor ion 830.0481 m/z, which supports a match to TGG1 peptide NATGHAPGPPFNAASYYYPK with a LacNAc modification at the first Asn (Asn-379 [N379] of TGG1). Fragment ions c5, c7, c9, c10, and c11 all have masses 365 D larger than the theoretically unmodified fragment ions, which is consistent with a LacNAc modification within the first four amino acids. The masses of all of the observed z ions, including z19, which encompasses amino acids 2 to 20, are consistent with the ions being unmodified, suggesting that the first amino acid of the peptide is modified. This conclusion is supported because the doubly and triply charged full-length peptide (labeled [M+3H]+2 and [M+3H]+3 in the figure) have masses 365 D larger than the unmodified peptide. All spectra (Table I; Supplemental Fig. S3) were manually inspected for verification of fragment ions that support localization of the LacNAc-modified amino acids; the quality of the ETD MS/MS spectra in Supplemental Figure S3 are similar to the quality shown in Figure 3. Subsequent experiments identified 34 additional N-LacNAc-modified proteins (Supplemental Table S1). All of the N-LacNAc modifications occur on the Asn of the canonical Asn-X-Ser/Thr N-glycosylation site. The presence of the LacNAc on these peptides is consistent with them being modified with N-linked GlcNAc in planta.
Figure 3.
TGG1 and TGG2 are N-GlcNAc modified. Proteins from the fucta/fuctb/xylt triple mutant were digested with Lys-C (or trypsin) and treated with PNGase F. Glycans with terminal GlcNAc were capped with Gal, and GlcNAc-modified peptides were enriched by RCA I lectin affinity chromatography (Supplemental Fig. S2). The enriched peptides were then analyzed by mass spectrometry. A, ETD MS/MS spectrum recorded on [M+3H]+3 ions (m/z 515.2663) from the LacNAc (365.1322)-modified TGG2 peptide FgNSTEARLLK. B, [M+3H]+3 ions (m/z 830.0481) corresponding to the LacNAc-modified TGG1 peptide gNATGHAPGPPFNAASYYYPK. Predicted c′- and z′·-type ions are listed above and below the peptide sequence, respectively. Singly and doubly charged fragment ions are listed as monoisotopic masses. Ions observed and labeled in the spectrum are underlined. Ions corresponding to charge-reduced species and those resulting from neutral losses are bracketed. Modified residues are preceded by “g” to signify modification by a single LacNAc moiety.
Table I. List of single N-linked GlcNAc-modified peptides for MS/MS analysis.
| Arabidopsis Genome Initiative Code | Peptide Sequence | Observed z | Observed m/z | Calculated m/z | ppm |
|---|---|---|---|---|---|
| At1g01300 | gNVTHAPRPGGFSSSVVSGLSQGSGEYFTR | +4 | 837.4022 | 837.4014 | 1.0 |
| At1g02816 | STGQFHAYFgNK | +3 | 555.5859 | 555.5865 | −0.1 |
| At1g55260 | IEDNYgNSTSPTQIHK | +3 | 704.6583 | 704.6590 | −0.1 |
| At1g62660 | IgNRTGISLIYDTTDFK | +3 | 741.3726 | 741.3743 | −2.4 |
| At1g78680 | LSgNATDASSIAASYVK | +3 | 654.9868 | 654.9849 | 3.8 |
| At2g17120 | VPIHCSCSgNGTGVSNR | +3 | 703.9774 | 703.9777 | 0.3 |
| gNATTLRNIQTLFAVK | +3 | 685.7048 | 685.7041 | 1.0 | |
| At2g28470 | AgNVTYDHRALVIDGK | +3 | 679.6774 | 679.6763 | 1.2 |
| At3g05180 | TARSgNGTIITAK | +3 | 533.2801 | 533.2812 | −2.0 |
| At3g15950 | AgNDSTGDKDDDEHVAR | +4 | 528.2223 | 528.2231 | −0.4 |
| At4g16660 | TEgNTTKEEEQSK | +3 | 596.9335 | 596.9342 | −1.1 |
| At5g20630 | KNPDQVTENDFAFTGLGTAGgNTSNIIK | +3 | 1,073.1876 | 1,073.1846 | 2.7 |
| At5g25980 | FgNSTEARLLK | +3 | 515.2662 | 515.2669 | −1.3 |
| At5g26000 | gNATGHAPGPPFNAASYYYPK | +3 | 830.0481 | 830.0486 | −0.6 |
| At5g67360 | RgNYTCDPSK | +3 | 502.5519 | 502.5527 | −0.5 |
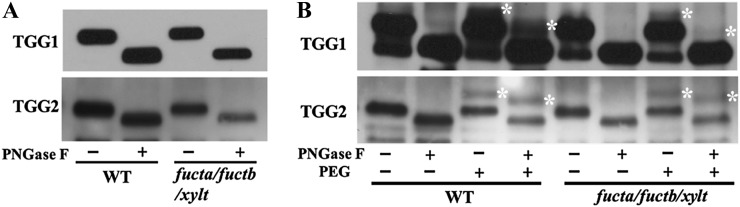
Because antibodies were available against two of the putatively N-GlcNAc-modified proteins, TGG1 and TGG2 (Ueda et al., 2006), we were able to investigate the modifications on them further. TGGs are thioglucoside hydrolases (Enzyme Commission 3.2.1.147) that are also called myrosinases. TGGs play a role in plant defense by hydrolyzing glucosinolates to produce toxic compounds. TGGs and glucosinolates are located in different cells but can come into contact with each other when microbes or herbivores damage cells. In addition to the N-GlcNAc modification reported here, TGG1 and TGG2 are also modified with N-linked high-Man-type glycans that retard their migration during SDS-PAGE (Ueda et al., 2006). Since, even in wild-type plants, high-Man-type glycans are not modified with Fuc, PNGase F removes them. The migration of TGG1 and TGG2 from the triple mutant was similar to proteins from wild-type plants, and treatment with PNGase F affected the migration similarly, suggesting that glycosylation is unaffected in the triple mutant (Fig. 4A).
Figure 4.
TGG1 and TGG2 from wild-type plants are N-GlcNAc modified. A, Immunoblot detection of TGG1 and TGG2 from wild-type (WT) and triple mutant plants following treatment with or without PNGase F. PNGase F treatment removes high-Man glycans, which increases the rate of TGG1 and TGG2 migration during electrophoresis (Ueda et al., 2006). B, Immunoblots showing the effect of PEGylating wild-type and triple mutant proteins after treatment with or without PNGase F. Asterisks indicate the PEGylated proteins.
We developed a mass-labeling strategy for the detection of terminal GlcNAc that is conceptually similar to but uses different chemistry from that described by Rexach et al. (2010). In this strategy, N-azidogalactosamine (GalNAz) is enzymatically added to terminal GlcNAc, and then one molecule of polyethylene glycol (PEG) 5000 (PEGylation) is added to the GalNAz using Click chemistry. Since PEG retards the mobility of the protein during SDS-PAGE (Supplemental Fig. S4), it is possible to monitor both the fraction of protein that is modified as well as the number of modifications. When modification of TGG1 and TGG2 was studied using this method, a fraction of both became PEGylated (Fig. 4B), and the magnitude of the mobility shift of the PEGylated form was consistent with the addition of only one molecule of PEG. The amount and number of PEGylations to TGG1 and TGG2 from the fucta/fuctb/xylt triple mutant and the wild type are similar, indicating that N-GlcNAc modification is unaffected in the mutant. Since it has been reported that PNGase F can remove N-GlcNAc from some peptides at a low rate (Fan and Lee, 1997), it is important to note that, under the conditions used here, treatment with PNGase F had no detectable effect on the fraction of either TGG that was PEGylated. Therefore, the N-GlcNAc modifications are not affected by the PNGase F treatment.
N-GlcNAc- and PNGase F-Sensitive Glycans Occur at the Same Site on TGG1
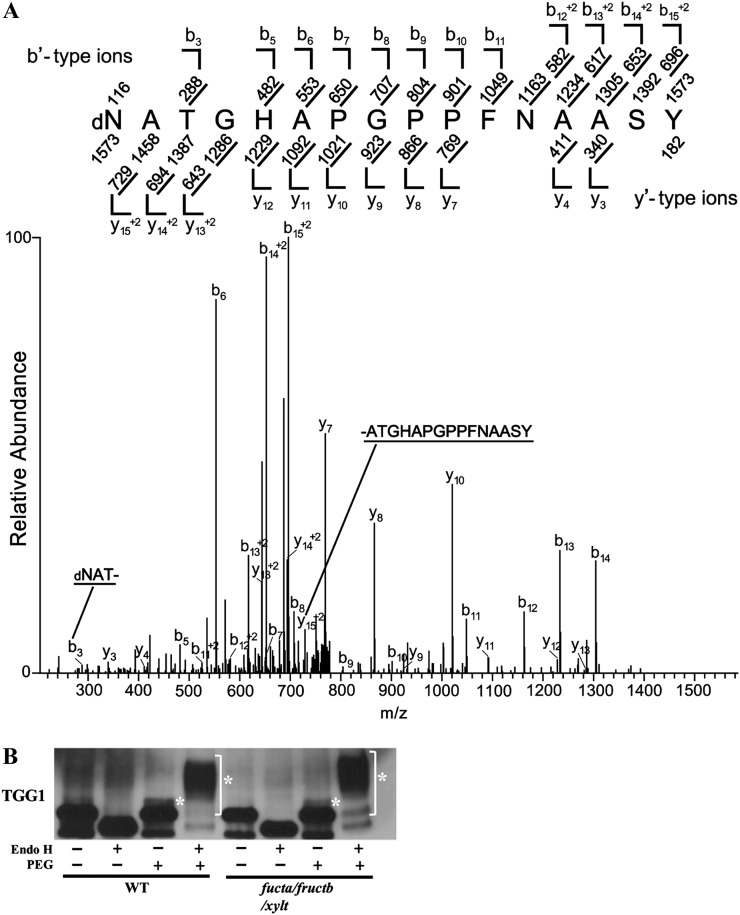
Removal of high-Man-type glycans by PNGase F results in deamidation of the modified Asn, which converts it to an Asp (Fig. 1). In contrast, endo-β-N-acetylglucosaminidase H (Endo H) creates an N-GlcNAc modification when it removes high-Man-type modifications (Fig. 1). The possibility that N-GlcNAc and high-Man-type glycans occur at the same site on TGG1 was investigated by determining if PNGase F treatment caused deamidation at N379, the site of N-GlcNAc modification. TGG1 was enriched by Con A lectin affinity chromatography (Supplemental Fig. S5A), incubated with PNGase F, Endo H, or buffer alone, and then purified by two-dimensional (2D)-PAGE. The region of the gel containing TGG1 was excised, proteins on the gel were digested with trypsin, and the resulting peptides were analyzed by collision-induced dissociation (CID) MS/MS (Fig. 5A). Deamidation of N379 was detected in PNGase F-treated preparations but not in control or Endo H-treated preparations, suggesting that, in addition to being modified with N-GlcNAc, N379 is also modified with high-Man-type glycans. Surprisingly, N-GlcNAc modification of the trypsin fragment containing N379 was not detected in the control or PNGase F-treated samples but was detected in the Endo H-treated samples (Supplemental Fig. S5B). Therefore, it is likely that N-GlcNAc modification of N379 on untreated TGG1 is not sufficiently abundant for detection by CID MS/MS and that the action of Endo H on high-Man-type glycans increases the abundance of this modification sufficiently for detection, thus providing further evidence that both N-GlcNAc and high-Man-type glycans can occur at this site.
Figure 5.
High-Man-type glycans occur at multiple sites on TGG1, including N379, the site of N-GlcNAc modification. A, TGG1 was enriched by Con A-Sepharose affinity chromatography, treated with PNGase F, and further purified by 2D PAGE. TGG1 was cut out from a 2D gel, digested with trypsin, and analyzed by MS/MS. The CID MS/MS spectrum was recorded on [M+2H]+2 ions (m/z 786.8574) corresponding to the peptide dNATGHAPGPPFNAASYYYPK deamidated at Asn (N379). Predicted b′- and y′-type ions are listed above and below the peptide sequence, respectively. Singly and doubly charged fragment ions are listed as monoisotopic masses. Ions observed and labeled in the spectrum are underlined. The modified residue is preceded by “d” to signify deamidated Asn. B, An immunoblot showing that extensive PEGylation of TGG1 occurs after treatment with Endo H. Asterisks indicate the PEGylated proteins.
Identification of Four Additional Sites Where TGG1 Is Modified with High-Man-Type Glycans
TGG1 has nine potential Asn-X-Ser/Thr N-modification sites. In the course of determining if N379 is modified with high-Man-type glycans, we found that four additional Asn-X-Ser/Thr sites (Asn-33, Asn-108, Asn-493, and Asn-512) were deamidated following treatment with PNGase F (Supplemental Table S2; Supplemental Fig. S6) and had detectable N-GlcNAc modifications only after treatment with Endo H (Supplemental Fig. S7), suggesting that these sites are modified with high-Man-type glycans. In addition, PEGylation following treatment with Endo H caused a large reduction in the migration of TGG1 during SDS-PAGE, consistent with most of the TGG1 being modified with multiple high-Man-type glycans (Fig. 5B).
ENGase Activity Is Required for GlcNAc Modification of TGG2
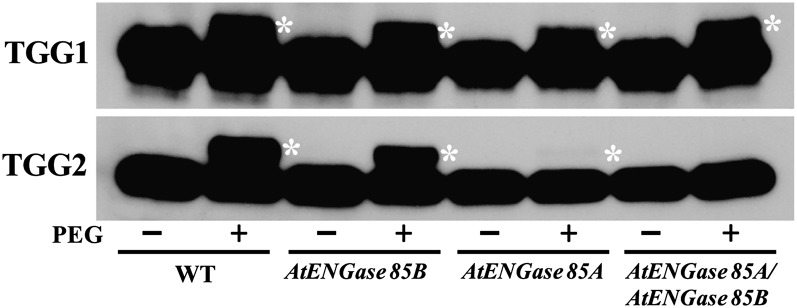
It has been proposed that endogenous ENGase produces the N-GlcNAc modifications found on some animal proteins (Wang et al., 2010), but to our knowledge, this hypothesis has not been tested. Arabidopsis has two functional ENGase genes, At3g11040 (AtENGase85B) and At5g05460 (AtENGase85A), and a pseudogene. Previous studies have found that free oligosaccharides with one GlcNAc at the reducing end are not detectable in AtENGase85A AtENGase85B double mutants, suggesting that the double mutant lacks all ENGase activity (Fischl et al., 2011; Kimura et al., 2011). We tested the hypothesis that Arabidopsis ENGase is responsible for the production of N-GlcNAc by determining if the abundance of this modification on TGG1 and TGG2 is affected in ENGase single and double mutants. Lines homozygous for transfer DNA (T-DNA) mutations affecting AtENGase85B and AtENGase85A were identified and then used to construct a double mutant. TGG1 and TGG2 from the single and double mutant lines were tested for defects in N-GlcNAc modification. While N-GlcNAc modification of TGG1 was unaffected in the mutants, modification of TGG2 was greatly reduced in the AtENGase85A single mutant and undetectable in the double mutant (Fig. 6). These results suggest that N-GlcNAc modification of TGG2 is produced when the ENGases remove high-Man-type glycans and that the N-GlcNAc modification on TGG1 is produced by another mechanism.
Figure 6.
ENGase is required for N-GlcNAc modification of TGG2. Immunoblots show PEGylation of TGG1 and TGG2 from wild-type (WT), AtENGase85BSalk_031210, AtENGase85ASail_714_D09, and AtENGase85BSalk_031210 AtENGase85ASail_714_D09 double mutant plants. Asterisks indicate PEGylated TGG1 and TGG2.
DISCUSSION
We have identified 48 Arabidopsis proteins bearing posttranslational modifications consisting of an N-GlcNAc. Several properties of the modification suggest that it is derived from larger N-linked glycans. The modification occurs at the sequon Asn-X-Ser/Thr, which is also where N-linked oligosaccharides occur (Pattison and Amtmann, 2009). All of the modified proteins contain signal sequences or noncleavable signal sequences (Supplemental Table S3), suggesting that they transit through the secretory system. PNGase F treatment deamidated the N-GlcNAc modification site (N379) of TGG1, suggesting that this site is also modified with high-Man-type glycans. Finally, as discussed below, this modification is not present on TGG2 from the AtENGase85A AtENGase85B double mutant, suggesting that it is produced by the action of ENGase on high-Man-type glycans.
Although many of the N-GlcNAc-modified proteins have multiple Asn-X-Ser/Thr sequons (Supplemental Table S3), the modification was detected at only one or two sites on each protein. The failure to detect the modification at multiple sites could reflect limitations in the enrichment and mass spectrometry methods or could indicate that production of the modification is regulated. Although both TGG1 and TGG2 have multiple Asn-X-Ser/Thr sites and are multiply glycosylated (Fig. 5B), proteins with more than one N-GlcNAc were not detected using a mass-labeling assay that allows the number of modifications with terminal GlcNAc on a protein to be determined (Fig. 4B). We analyzed the regions surrounding the N-GlcNAc modifications for evidence of a consensus modification but found no consensus beyond the Asn-X-Ser/Thr sequon, suggesting that, if the location of the modification is regulated, it is not determined by the primary sequence surrounding the site. Among the proteins with multiple Asn-X-Ser/Thr sequons, there was no consistent pattern for which site was N-GlcNAc modified. Therefore, it remains an open question whether the site of N-GlcNAc modifications is regulated and, if so, what determines the location of the modification.
Results from the mass-labeling assay showed that the majority of TGG1 and TGG2 was not N-GlcNAc modified (Fig. 4B). Although the N-GlcNAc modification was discovered on PNGase F-treated proteins isolated from the fucta/fuctb/xylt triple mutant, the abundance of the modification was unaffected by PNGase F and was similar in the wild type and the fucta/fuctb/xylt triple mutant, indicating that the modification was not produced by PNGase F or the triple mutant. Although it cannot be completely ruled out, it is unlikely that N-GlcNAc modifications are an artifact that is produced during protein purification. We prepared protein samples by several different methods, including direct extraction from leaves into SDS followed by immediate boiling or aqueous extraction and purification by lectin affinity chromatography, and the extraction method had no detectable effect on the fraction of TGG1 or TGG2 that was modified. In addition, TGG2 from the AtENGase85A AtENGase85B double mutant was not detectably modified, while modification of TGG1 was not affected.
It has been proposed that N-GlcNAc modifications on animal proteins are produced by ENGase acting on glycoproteins that are being degraded by the ERAD pathway (Wang et al., 2010); however, the role of ENGase in this process is unclear (Chantret et al., 2010). Free oligosaccharides with one GlcNAc at the reducing end are found in animals and plants (Suzuki and Funakoshi, 2006; Chantret and Moore, 2008; Funakoshi and Suzuki, 2009), but these could arise from ENGase acting on glycoproteins or the oligosaccharides after they have been released from the protein. Arabidopsis mutants lacking ENGase activity have a large increase in free oligosaccharides with two GlcNAc at the reducing end (Fischl et al., 2011; Kimura et al., 2011), suggesting that ENGase acts primarily on free oligosaccharides. Our finding that TGG2 from the AtENGase85B AtENGase85A double mutant is not N-GlcNAc modified (Fig. 6) strongly supports the hypothesis that ENGases can produce some of the N-GlcNAc modifications. However, the result that N-GlcNAc modification of TGG1 is unaffected in the double mutant suggests that some modifications can be produced by another mechanism. In the future, it will be important to learn the nature of this other mechanism as well as the relative importance of the different mechanisms.
Our results also suggest that AtENGase85B and AtENGase85A have specialized functions. While N-GlcNAc modification of TGG2 was greatly reduced in AtENGase85A and undetectable in double mutant plants, modification of TGG1 was unaffected in these mutants. Modification of TGG1 and TGG2 was largely unaffected in AtENGase85B plants. These results are consistent with the model that AtENGase85A is the enzyme mostly responsible for the modification on TGG2 and that AtENGase85B has a much smaller role. Interestingly, neither ENGase seems to have a role in producing N-GlcNAc modifications on TGG1. Since Arabidopsis is believed to have just two ENGases (Fischl et al., 2011; Kimura et al., 2011), these results suggest that multiple mechanisms exist for the production of N-GlcNAc modifications. Bacterial N-linked glycosyltransferases that glycosylated Asn-X-Ser/Thr sites using either UDP-Glc or UDP-Gal have recently been identified (Choi et al., 2010; Grass et al., 2010), but we were unable to identify a gene encoding a similar enzyme in the Arabidopsis genome. The Arabidopsis genome also contains a predicted ENGase pseudogene, At3g61010, which is unlikely to encode a functional ENGase (Fischl et al., 2011). It is also possible that one or both alleles used in the double mutant are not null alleles, but this is unlikely because the AtENGase85B allele used here was also used previously to create an ENGase double mutant that had no detectable ENGase activity (Fischl et al., 2011; Kimura et al., 2011). Although we employed a different AtENGase85A allele than was used in other studies and the T-DNA is inserted in an intron, TGG2 modification was almost undetectable in single mutants with this allele, suggesting that the allele encodes little or no functional ENGase.
It is an open question whether N-GlcNAc modification occurs as an intermediate step in the degradation of misfolded proteins or if the modification has a function such as regulating protein localization or activity. Consistent with N-GlcNAc modification being produced during the degradation of misfolded proteins by the ERAD pathway, AtENGase85A and AtENGase85B are localized in the cytosol when transiently expressed in Nicotiana benthamiana (Fischl et al., 2011). However, several other lines of evidence suggest that the production of N-GlcNAc modification may not be generally associated with protein turnover. The modification to TGG1 and TGG2 is relatively abundant, which suggests that the proteins are rapidly turning over or that the modified protein is relatively stable. Second, rather than occurring at all positions that are modified with N-glycans, it seems to occur only at a subset of these positions. Possible roles for the modification include modifying protein-protein interactions, protein localization, or regulation of activity. We did not detect any difference in myrosinase activity in wild-type and AtENGase85B or AtENGase85A mutant plants (Supplemental Fig. S8). However, the lack of effect of the AtENGase85B mutation on myrosinase activity is not surprising, because N-GlcNAc modification of TGG1 is unaffected in this mutant and TGG1 is more active than TGG2 (Barth and Jander, 2006). It will be interesting to learn if myrosinase activity is affected when modification to both enzymes is blocked.
MATERIALS AND METHODS
Plant Materials and Cytosolic Protein Extraction
Wild-type and fucta/fuctb/xylt triple mutant Arabidopsis (Arabidopsis thaliana) plants in the Columbia background were grown in soil at 22°C under fluorescent lights (16 h of light/8 h of dark) for 2 weeks, at which time shoots were harvested, flash frozen in liquid nitrogen, and stored at −80°C until being used for protein extraction. Preparation of cytosolic proteins was carried out as described by Calikowski and Meier (2006). T-DNA alleles affecting AtENGase85A (stock no. CS876187; Sail_714_D09) and AtENGase85B (Salk_031210) were obtained from the Arabidopsis Biological Resource Center. Homozygous AtENGase85ASail_714_D09 and AtENGase85BSalk_031210 single mutants were identified and used to create an AtENGase85ASail_714_D09 and AtENGase85BSalk_031210 double mutant.
Recombinant Protein Preparation and Detection of GlcNAcylated Proteins on Blot
A construct expressing CT-5 was produced by cloning the portion of the Arabidopsis gene encoding amino acids 685 to 1,041 of GIGANTEA into pET32. O-GlcNAc-modified CT-5 was prepared by coexpressing it with SECRET AGENT as described previously (Kim et al., 2011). Detection of GlcNAcylated proteins on the blots was performed as described (Kim et al., 2011). β-Elimination of O-GlcNAc on the protein blots was performed as described previously (Duk et al., 1997).
Enrichment of GlcNAc-Modified Peptides by Lectin Affinity Chromatography
Details of RCA I affinity chromatography procedures are described (Kim et al., 2011). Protein extraction for the purification of TGG1 and TGG2 by Con A affinity chromatography was performed as described previously (Pessina et al., 1990) and according to the Con A resin manufacturer’s protocol (Pharmacia Biotech).
Deglycosylation and PEGylation of Proteins
Proteins were treated with PNGase F (New England Biolabs) and Endo H (Promega) according to the manufacturers’ protocols. PEGylation, the addition of PEG to terminal GlcNAc modifications, proceeds in two steps: enzymatic labeling of GlcNAc with GalNAz, followed by attaching PEG-alkyne via a copper-catalyzed azide-alkyne Huisgen cycloaddition reaction. Enzymatic labeling was done with the Click-iT O-GlcNAc Enzymatic Labeling System (Invitrogen) following the manufacturer’s instructions. After the labeling reaction, the sample was methanol-chloroform precipitated as in the instructions, but the GalNAz-labeled proteins were resuspended in 100 mm sodium phosphate (pH 8.0) with 1% SDS to an estimated concentration of 25 μg μL−1. Samples were boiled for 5 min and cooled on ice for 2 min followed by a brief spin. The protocol for the Click reaction was modified from the procedure of Deiters et al. (2004). Conditions for the Click reaction were 100 mm sodium phosphate (pH 8.0), 0.5% SDS, 1 mg mL−1 PEG-alkyne (Creative PEGWorks), 1 mm CuSO4, 1 mm Tris[(1-benzyl-1H1,2,3-triazol-4-yl)methyl]amine (Chan et al., 2004), and 2 mm Tris(carboxyethyl)phosphine. Tris[(1-benzyl-1H1,2,3-triazol-4-yl)methyl]amine was used as a 10 mm stock solution in dimethyl sulfoxide. The reaction was run overnight at room temperature, after which the proteins were resolved by SDS-PAGE.
SDS-PAGE, 2D-PAGE, and Immunoblotting
For one-dimensional SDS-PAGE, 25 µg of plant protein was loaded in each lane, subjected to electrophoresis, and transferred to an Immobilon-P membrane by standard methods. Immunoblot analysis was performed as described (Kim et al., 2011). For 2D-PAGE, 50-µg aliquots of Con A-purified plant protein were either untreated or treated with 500 units of PNGase F or Endo H and precipitated. The protein samples were dissolved in 250 µL of immobilized pH gradient running buffer (7 m urea, 2 m thiourea, 2% CHAPS [GE Healthcare], 1% n-dodecyl β-d-maltoside [Sigma], trace bromphenol blue, 0.5% [v/v] ampholyte buffer pH 3–10 NL [GE Healthcare], 12 µL mL−1 Destreak reagent [GE Healthcare], and 10 mm dithiothreitol). The sample was allowed to rehydrate into a 13-cm Immobiline DryStrip pH 3–10 NL (GE Healthcare) overnight under low current (30 V for 7 h, followed by 60 V for 7 h). After overnight rehydration, the Immobiline DryStrip sample was resolved with a voltage ramp of 500 V for 0.5 kVh, 1,000 V for 0.8 kVh, and 8,000 V for 17.3 kVh in an Ettan IPGphor isoelectric focusing apparatus (Amersham) until a total 19.255 kVh was reached. The isoelectric focusing strip was equilibrated for 0.5 h in SDS equilibration buffer (50 mm Tris, pH 8.8, 8 m urea, 30% glycerol, 4% SDS, and 1% dithiothreitol) followed by separation on a 10% to 20% Tris-HCl SDS-PAGE Criterion gel (Bio-Rad). After SDS-PAGE, the 2D gel was stained with Deep Purple total protein stain (GE Healthcare) according to the manufacturer’s instructions. The gel image was visualized with the Typhoon 8610 scanner (GE Healthcare) set at a photo multiplier tube voltage of 600 V, 532-nm excitation, and 610BP30 emission filter.
Nanoflow Liquid Chromatography-MS/MS and Analysis of MS/MS Data
Reverse-phase nano-HPLC online with electrospray ionization mass spectrometry using a LTQ-OrbitrapXL equipped with an ETD source (Thermo Scientific) was performed as described (Bandhakavi et al., 2009) with a few exceptions. Peptides were eluted using an acetonitrile gradient of 2% to 40% over 60 min. Survey scans from 360 to 1,800 m/z were acquired using the Orbitrap analyzer at 60,000 resolution. The data-dependent settings included selection of the top five most abundant ions in each survey scan for MS/MS, excluding 1+ or undetermined charge states; dynamic exclusion was enabled for 20 s. Fragment ions were detected in the linear ion trap for both CID and ETD activation modes. For experiments using ETD fragmentation, the scan parameters were as follows: precursor ion isolation window of 3 m/z units; precursor ion automatic gain control of 2 × 104 charges; precursor injection time of 100 ms; fluoranthene reagent ion; reagent ion automatic gain control of 400,000; reagent ion injection time of 50 ms; reagent ion reaction time of 100 ms. Fragmentation with CID was performed as described previously (Bandhakavi et al., 2009) with 35% normalized collision energy. CID and ETD data were analyzed using Scaffold version 3 (www.proteomesoftware.com) and OMSSA (version 2.1.7), respectively, and confirmed via manual inspection.
Prior to database searching, raw CID data were extracted and converted to the mzXML format using MS Convert software from ProteoWizard. Data were searched using SEQUEST version 27, revision 12, against the National Center for Biotechnology Information-derived reference sequence Arabidopsis database from September 2009, which included common contaminants from http://www.thegpm.org/crap/index.htm, totaling 33,440 entries. Precursor and fragment ion tolerances for database searches were set at 10 ppm and 0.8 atomic mass units (amu). Semitryptic specificity was selected with up to two missed cleavage sites. Since the samples had been alkylated, the addition of 57.02146 amu to Cys was set as a fixed modification, and variable modifications included the addition of 15.9949 amu to Met, to allow for the oxidation of Met, which is a common spontaneous modification. In addition, variable modifications of 203.0794 and 0.9848 amu to Asn were allowed to detect N-GlcNAc modifications and the conversion of Asn to Asp, which occurs when PNGase F removes N-GlcNAc, respectively. The probabilities that peptide candidate identifications were correct (Keller et al., 2002) were calculated through the use of Scaffold version 3. Protein identifications were filtered using the following filter criteria: 10 ppm precursor mass tolerance, more than 95% peptide probability, and full trypsin specificity prior to confirmation by manual inspection.
When ETD data were analyzed using OMSSA, .dta files were generated using the DTA generator (http://www.chem.wisc.edu/~coon/), and the MS/MS spectra were searched against all Arabidopsis proteins in the National Center for Biotechnology Information nonredundant database. The parameters for the search were the same as described above with the addition of allowing variable modification of 365.1322 amu to Asn, Ser, and Thr to account for possible N- and O-linked LacNAc modifications.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. PNGase F removes few of the glycans with terminal GlcNAc from the wild type but removes many of these glycans from fucta/fuctb/xylt triple mutant proteins.
Supplemental Figure S2. Enrichment of GlcNAc-modified fucta/fuctb/xylt triple mutant peptides by RCA I affinity chromatography.
Supplemental Figure S3. Mapping single N-linked GlcNAc sites on Arabidopsis peptides.
Supplemental Figure S4. PEGylation of a control O-GlcNAc-modified protein.
Supplemental Figure S5. Western analysis of TGG1 purification by Con A-Sepharose affinity chromatography.
Supplemental Figure S6. Mapping PNGase F-deamidated sites on TGG1 using liquid chromatography-MS/MS.
Supplemental Figure S7. After Endo H treatment, N-GlcNAc was mapped using liquid chromatography-MS/MS.
Supplemental Figure S8. Loss of ENGase activity does not affect myrosinase activity.
Supplemental Table S1. List of detected single N-linked GlcNAc-modified peptides.
Supplemental Table S2. Location of deamidated and N-GlcNAc-modified Asn on the TGG1 protein.
Supplemental Table S3. List of detected single N-linked GlcNAc-modified peptides.
Acknowledgments
We thank Ikuko Hara-Nishimura for providing antibodies against TGG1 and TGG2, Georg Jander for providing tgg mutant seed, and Cory Livingston for isolating the AtENGase85B and AtENGase85A single mutants and creating the double mutant. We also thank LeeAnn Higgins for assistance with the manuscript and Kerry Sokol for critical reading of the manuscript.
Glossary
- ER
endoplasmic reticulum
- ERAD
endoplasmic reticulum-associated protein degradation
- N-GlcNAc
N-linked N-acetyl-d-glucosamine monosaccharide
- ENGase
endo-β-N-actetylglucosaminidase
- PNGase F
peptide:N-glycosidase F
- LacNAc
N-acetyl-d-lactosamine
- MS/MS
tandem mass spectrometry
- m/z
mass-to-charge ratio
- GalNAz
N-azidogalactosamine
- PEG
polyethylene glycol
- Endo H
endo-β-N-acetylglucosaminidase H
- 2D
two-dimensional
- CID
collision-induced dissociation
- N379
Asn-379
- Con A
concanavalin A
- amu
atomic mass units
- T-DNA
transfer DNA
- ETD
electron transfer dissociation
References
- Bakker H, Rouwendal GJ, Karnoup AS, Florack DE, Stoopen GM, Helsper JP, van Ree R, van Die I, Bosch D. (2006) An antibody produced in tobacco expressing a hybrid beta-1,4-galactosyltransferase is essentially devoid of plant carbohydrate epitopes. Proc Natl Acad Sci USA 103: 7577–7582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandhakavi S, Stone MD, Onsongo G, Van Riper SK, Griffin TJ. (2009) A dynamic range compression and three-dimensional peptide fractionation analysis platform expands proteome coverage and the diagnostic potential of whole saliva. J Proteome Res 8: 5590–5600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth C, Jander G. (2006) Arabidopsis myrosinases TGG1 and TGG2 have redundant function in glucosinolate breakdown and insect defense. Plant J 46: 549–562 [DOI] [PubMed] [Google Scholar]
- Calikowski TT, Meier I. (2006) Isolation of nuclear proteins. Methods Mol Biol 323: 393–402 [DOI] [PubMed] [Google Scholar]
- Chalkley RJ, Thalhammer A, Schoepfer R, Burlingame AL. (2009) Identification of protein O-GlcNAcylation sites using electron transfer dissociation mass spectrometry on native peptides. Proc Natl Acad Sci USA 106: 8894–8899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan TR, Hilgraf R, Sharpless KB, Fokin VV. (2004) Polytriazoles as copper(I)-stabilizing ligands in catalysis. Org Lett 6: 2853–2855 [DOI] [PubMed] [Google Scholar]
- Chantret I, Fasseu M, Zaoui K, Le Bizec C, Yayé HS, Dupré T, Moore SE. (2010) Identification of roles for peptide:N-glycanase and endo-beta-N-acetylglucosaminidase (Engase1p) during protein N-glycosylation in human HepG2 cells. PLoS ONE 5: e11734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chantret I, Moore SE. (2008) Free oligosaccharide regulation during mammalian protein N-glycosylation. Glycobiology 18: 210–224 [DOI] [PubMed] [Google Scholar]
- Choi KJ, Grass S, Paek S, St Geme JW, III, Yeo HJ. (2010) The Actinobacillus pleuropneumoniae HMW1C-like glycosyltransferase mediates N-linked glycosylation of the Haemophilus influenzae HMW1 adhesin. PLoS ONE 5: e15888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deiters A, Cropp TA, Summerer D, Mukherji M, Schultz PG. (2004) Site-specific PEGylation of proteins containing unnatural amino acids. Bioorg Med Chem Lett 14: 5743–5745 [DOI] [PubMed] [Google Scholar]
- Duk M, Ugorski M, Lisowska E. (1997) β-Elimination of O-glycans from glycoproteins transferred to Immobilon P membranes: method and some applications. Anal Biochem 253: 98–102 [DOI] [PubMed] [Google Scholar]
- Fan JQ, Lee YC. (1997) Detailed studies on substrate structure requirements of glycoamidases A and F. J Biol Chem 272: 27058–27064 [DOI] [PubMed] [Google Scholar]
- Fischl RM, Stadlmann J, Grass J, Altmann F, Léonard R. (2011) The two endo-β-N-acetylglucosaminidase genes from Arabidopsis thaliana encode cytoplasmic enzymes controlling free N-glycan levels. Plant Mol Biol 77: 275–284 [DOI] [PubMed] [Google Scholar]
- Funakoshi Y, Suzuki T. (2009) Glycobiology in the cytosol: the bitter side of a sweet world. Biochim Biophys Acta 1790: 81–94 [DOI] [PubMed] [Google Scholar]
- Grass S, Lichti CF, Townsend RR, Gross J, St Geme JW., III (2010) The Haemophilus influenzae HMW1C protein is a glycosyltransferase that transfers hexose residues to asparagine sites in the HMW1 adhesin. PLoS Pathog 6: e1000919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hase S, Fujimura K, Kanoh M, Ikenaka T. (1982) Studies on heterogeneity of Taka-amylase A: isolation of an amylase having one N-acetylglucosamine residue as the sugar chain. J Biochem 92: 265–270 [DOI] [PubMed] [Google Scholar]
- Islam MR, Kung SS, Kimura Y, Funatsu G. (1991) N-Acetyl-D-glucosamine-asparagine structure in ribosome-inactivating proteins from the seeds of Luffa cylindrica and Phytolacca americana. Agric Biol Chem 55: 1375–1381 [PubMed] [Google Scholar]
- Keller A, Nesvizhskii AI, Kolker E, Aebersold R. (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem 74: 5383–5392 [DOI] [PubMed] [Google Scholar]
- Kim YC, Udeshi ND, Balsbaugh JL, Shabanowitz J, Hunt DF, Olszewski NE. (2011) O-GlcNAcylation of the Plum pox virus capsid protein catalyzed by SECRET AGENT: characterization of O-GlcNAc sites by electron transfer dissociation mass spectrometry. Amino Acids 40: 869–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura Y, Takeoka Y, Inoue M, Maeda M, Fujiyama K. (2011) Double-knockout of putative endo-β-N-acetylglucosaminidase (ENGase) genes in Arabidopsis thaliana: loss of ENGase activity induced accumulation of high-mannose type free N-glycans bearing N,N′-acetylchitobiosyl unit. Biosci Biotechnol Biochem 75: 1019–1021 [DOI] [PubMed] [Google Scholar]
- Lerouge P, Cabanes-Macheteau M, Rayon C, Fischette-Lainé AC, Gomord V, Faye L. (1998) N-Glycoprotein biosynthesis in plants: recent developments and future trends. Plant Mol Biol 38: 31–48 [PubMed] [Google Scholar]
- Min W, Dunn AJ, Jones DH. (1992) Non-glycosylated recombinant pro-concanavalin A is active without polypeptide cleavage. EMBO J 11: 1303–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattison RJ, Amtmann A. (2009) N-Glycan production in the endoplasmic reticulum of plants. Trends Plant Sci 14: 92–99 [DOI] [PubMed] [Google Scholar]
- Pessina A, Thomas RM, Palmieri S, Luisi PL. (1990) An improved method for the purification of myrosinase and its physicochemical characterization. Arch Biochem Biophys 280: 383–389 [DOI] [PubMed] [Google Scholar]
- Rademacher T, Sack M, Arcalis E, Stadlmann J, Balzer S, Altmann F, Quendler H, Stiegler G, Kunert R, Fischer R, et al. (2008) Recombinant antibody 2G12 produced in maize endosperm efficiently neutralizes HIV-1 and contains predominantly single-GlcNAc N-glycans. Plant Biotechnol J 6: 189–201 [DOI] [PubMed] [Google Scholar]
- Rayon C, Cabanes-Macheteau M, Loutelier-Bourhis C, Salliot-Maire I, Lemoine J, Reiter WD, Lerouge P, Faye L. (1999) Characterization of N-glycans from Arabidopsis: application to a fucose-deficient mutant. Plant Physiol 119: 725–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rexach JE, Rogers CJ, Yu SH, Tao J, Sun YE, Hsieh-Wilson LC. (2010) Quantification of O-glycosylation stoichiometry and dynamics using resolvable mass tags. Nat Chem Biol 6: 645–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon PS, Bowles DJ. (1992) The glycoprotein precursor of concanavalin A is converted to an active lectin by deglycosylation. EMBO J 11: 1297–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser R, Altmann F, Mach L, Glössl J, Steinkellner H. (2004) Generation of Arabidopsis thaliana plants with complex N-glycans lacking beta1,2-linked xylose and core alpha1,3-linked fucose. FEBS Lett 561: 132–136 [DOI] [PubMed] [Google Scholar]
- Suzuki T, Funakoshi Y. (2006) Free N-linked oligosaccharide chains: formation and degradation. Glycoconj J 23: 291–302 [DOI] [PubMed] [Google Scholar]
- Ueda H, Nishiyama C, Shimada T, Koumoto Y, Hayashi Y, Kondo M, Takahashi T, Ohtomo I, Nishimura M, Hara-Nishimura I. (2006) AtVAM3 is required for normal specification of idioblasts, myrosin cells. Plant Cell Physiol 47: 164–175 [DOI] [PubMed] [Google Scholar]
- Vembar SS, Brodsky JL. (2008) One step at a time: endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol 9: 944–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Udeshi ND, Slawson C, Compton PD, Sakabe K, Cheung WD, Shabanowitz J, Hunt DF, Hart GW. (2010) Extensive crosstalk between O-GlcNAcylation and phosphorylation regulates cytokinesis. Sci Signal 3: ra2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng ZH, He XL, Li HM, Hu Z, Wang DC. (2003) Crystal structure of pokeweed antiviral protein with well-defined sugars from seeds at 1.8A resolution. J Struct Biol 141: 171–178 [DOI] [PubMed] [Google Scholar]