Abstract
Arabidopsis (Arabidopsis thaliana) UV RESISTANCE LOCUS8 (UVR8) is a photoreceptor that specifically mediates photomorphogenic responses to ultraviolet (UV)-B in plants. UV-B photoreception induces the conversion of the UVR8 dimer into a monomer that interacts with the CONSTITUTIVELY PHOTOMORPHOGENIC1 (COP1) protein to regulate gene expression. However, it is not known how the dimeric photoreceptor is regenerated in plants. Here, we show, by using inhibitors of protein synthesis and degradation via the proteasome, that the UVR8 dimer is not regenerated by rapid de novo synthesis following destruction of the monomer. Rather, regeneration occurs by reversion from the monomer to the dimer. However, regeneration of dimeric UVR8 in darkness following UV-B exposure occurs much more rapidly in vivo than in vitro with illuminated plant extracts or purified UVR8, indicating that rapid regeneration requires intact cells. Rapid dimer regeneration in vivo requires protein synthesis, the presence of a carboxyl-terminal 27-amino acid region of UVR8, and the presence of COP1, which is known to interact with the carboxyl-terminal region. However, none of these factors can account fully for the difference in regeneration kinetics in vivo and in vitro, indicating that additional proteins or processes are involved in UVR8 dimer regeneration in vivo.
UV-B wavelengths (280–315 nm) act as a key regulatory signal to initiate photomorphogenic responses in plants (Frohnmeyer and Staiger, 2003; Ulm and Nagy, 2005; Jenkins, 2009; Heijde and Ulm, 2012). These responses include the suppression of hypocotyl extension (Ballaré et al., 1995; Kim et al., 1998; Suesslin and Frohnmeyer, 2003; Shinkle et al., 2004), regulation of plant morphology (Hectors et al., 2007; Wargent et al., 2009), and the production of UV-absorbing phenolic compounds that act as a sunscreen in the outer tissues (Li et al., 1993; Stapleton and Walbot, 1994; Jordan, 1996; Rozema et al., 1997). Photomorphogenic responses to UV-B are mediated by the photoreceptor UV RESISTANCE LOCUS8 (UVR8; Kliebenstein et al., 2002; Brown et al., 2005; Brown and Jenkins, 2008; Favory et al., 2009; Rizzini et al., 2011). UVR8 specifically regulates the expression of numerous genes that underpin photomorphogenic responses to UV-B (Brown et al., 2005; Favory et al., 2009). Some of these genes encode proteins that help to prevent or repair damage by UV-B (Ulm et al., 2004; Brown et al., 2005; Favory et al., 2009; Stracke et al., 2010). Hence, uvr8 mutant Arabidopsis (Arabidopsis thaliana) plants, which fail to express these genes, are highly sensitive to elevated levels of UV-B (Kliebenstein et al., 2002; Brown et al., 2005).
Among the genes regulated by UVR8 is that encoding the ELONGATED HYPOCOTYL5 (HY5) transcription factor (Brown et al., 2005; Favory et al., 2009). HY5, sometimes acting redundantly with ELONGATED HYPOCOTYL5 HOMOLOG, mediates many gene expression responses initiated by UVR8 (Brown et al., 2005; Brown and Jenkins, 2008). UVR8 binds to histones in chromatin, including at the HY5 gene (Brown et al., 2005; Cloix and Jenkins, 2008), raising the possibility that UVR8 regulates transcription by promoting the recruitment or activation of transcription factors and/or chromatin-remodeling proteins. However, the processes involved in transcriptional regulation by UVR8 are not understood.
Map-based cloning of the UVR8 gene revealed that it encodes a seven-bladed β-propeller protein (Kliebenstein et al., 2002). The UVR8 protein exists as a homodimer in plants and in vitro, but it rapidly dissociates into monomers in response to UV-B treatment (Rizzini et al., 2011, Christie et al., 2012; Wu et al., 2012). The recently determined crystal structure of UVR8 reveals that the dimer is maintained by salt-bridge interactions between charged amino acids at the dimeric interface (Christie et al., 2012; Wu et al., 2012). UV-B photoreception causes the disruption of these salt bridges and hence monomerization. UVR8 absorbs UV-B via specific Trp amino acids that act as chromophores for the photoreceptor and that are adjacent to salt-bridging Arg residues (Christie et al., 2012; Wu et al., 2012).
Photoreception leads to both the rapid nuclear accumulation of UVR8 (Kaiserli and Jenkins, 2007) and interaction with the CONSTITUTIVELY PHOTOMORPHOGENIC1 (COP1) protein (Favory et al., 2009; Rizzini et al., 2011; Cloix et al., 2012). COP1 is a positive regulator of UVR8-mediated gene expression (Oravecz et al., 2006), in contrast to its function as a negative regulator of photomorphogenesis in dark-grown seedlings (Osterlund et al., 2000; Yi and Deng, 2005). Whereas COP1 acts as an E3 ubiquitin ligase to degrade positive regulators such as HY5 in dark-grown seedlings, there is no evidence that it functions as an E3 ubiquitin ligase in UV-B photomorphogenic responses. The REPRESSOR OF UV-B PHOTOMORPHOGENESIS1 (RUP1) and RUP2 proteins negatively regulate UVR8-mediated photomorphogenic responses and interact with UVR8 (Gruber et al., 2010). The RUP proteins are expressed in response to UV-B, and both UVR8 and COP1 are required for their UV-B-induced expression (Gruber et al., 2010).
An important issue is how the functional UVR8 photoreceptor is regenerated following photoreception. Since UV-B photoreception converts the UVR8 dimer into the monomer, how is the dimeric photoreceptor restored? In principle, there are two possible mechanisms. First, the monomer could be degraded after it functions and protein synthesis could replace the dimer in the cell. This would entail quite rapid and continual turnover of the UVR8 protein. Second, the monomer could revert to the dimer to reconstitute the functional photoreceptor without any requirement for synthesis and degradation. Experiments with the homogeneous purified protein show that the monomer can revert to the dimer and that the reformed dimer is functional in UV-B photoreception (Christie et al., 2012; Wu et al., 2012). However, the kinetics of reversion in vitro are slow; although some dimer is reformed within a few hours, complete reversion takes 24 to 48 h. Hence, our aim in this study was to investigate the kinetics and mechanism of regeneration of the UVR8 photoreceptor in plants.
RESULTS
Regeneration of the UVR8 Dimer Is Rapid in Intact Plants
To examine whether UVR8 is in the dimeric or monomeric form, we used a SDS-PAGE assay developed by Rizzini et al. (2011). In this assay, protein samples for electrophoresis are prepared using sample buffer containing SDS, but they are not boiled prior to loading on a SDS-PAGE gel. Interactions that maintain the dimer are sufficiently strong to resist denaturation by SDS under these conditions. Hence, the UVR8 dimer and monomer are clearly resolved on the gels and are visualized by incubating an immunoblot of the gel with an anti-UVR8 antibody.
We used this assay to monitor the relative amounts of UVR8 dimer and monomer following UV-B treatment of intact wild-type Arabidopsis plants. As shown in Figure 1A, essentially all the UVR8 protein is present as a dimer before UV-B exposure, and it is then converted to the monomer in response to a 3-h UV-B treatment. When plants are subsequently transferred to darkness, a decrease in the amount of monomer and a concomitant increase in the dimer are seen within 15 min. After 1 h in darkness, virtually all the UVR8 protein is present as the dimer. The total amount of UVR8 does not appear to change significantly over the time course, at least up to 2 h following the end of illumination.
Figure 1.
Regeneration of the UVR8 dimer after UV-B exposure is much more rapid in vivo than in vitro. A, Immunoblot of whole cell extracts from wild-type Ler plants probed with anti-UVR8 antibody. Plants were exposed to 2.5 μmol m−2 s−1 UV-B for 3 h (UV-B +) and then transferred to darkness for the indicated time periods before extracts were prepared. Extract samples were prepared for electrophoresis without boiling and resolved on a 7.5% SDS-PAGE gel prior to immunoblotting. The UVR8 dimer (D) and monomer (M) are indicated. Ponceau staining of Rubisco large subunit (rbcL) is shown as a loading control. B, Coomassie-stained SDS-PAGE gel of purified UVR8 protein exposed to 1.5 μmol m−2 s−1 UV-B for 1 h (UV-B +) and then transferred to darkness at room temperature for the indicated times. Samples were analyzed without boiling on a 7.5% SDS-PAGE gel. A non-UV-B-treated boiled sample is shown as a control. C, Quantification of the loss of monomer in darkness following UV-B exposure for the wild type in vivo (solid line) and purified UVR8 in vitro (solid line with points). Dotted and dashed lines show 95% confidence limits of the best fit curves from three replicates. D, Immunoblot of UV-B-treated whole cell extract from wild-type Ler plants probed with anti-UVR8 antibody. The extract was exposed to 1.5 μmol m−2 s−1 UV-B for 1 h (UV-B +) and then transferred to darkness at room temperature for the indicated times. Samples were analyzed without boiling on a 7.5% SDS-PAGE gel prior to immunoblotting. Ponceau staining of Rubisco large subunit is shown as a loading control.
In contrast, reappearance of the dimer in darkness following UV-B treatment of purified UVR8 protein shows much slower kinetics (Fig. 1B). Although reversion to the dimer is detectable 3 h after transfer to darkness, most of the protein is still in the monomeric form 6 h after the end of UV-B illumination, and approximately 30 h are required to see nearly complete dimer regeneration.
In an attempt to quantify the kinetics of dimer regeneration, the percentage of UVR8 in the monomeric form was determined by measuring band intensities on western blots from several independent experiments. As shown in Figure 1C, in vivo, the monomer declines exponentially in darkness following UV-B exposure, whereas in vitro, the rate is much slower. The mean time required for 50% loss of the monomer was calculated from the graphical data with 95% confidence limits (Table I). In vivo, 50% of the monomer is lost within approximately 18 min, whereas in vitro, it takes about 15 h.
Table I. Quantification and statistical analysis of the lifetime of the UVR8 monomer in different experimental conditions.
UVR8 protein bands were quantified on three representative western blots for each type of experiment using ImageJ software. The value for the monomer at each time point was normalized against that after UV-B illumination, taken as 100%. Percentage values of monomer with time were plotted (Fig. 1C; Supplemental Fig. S1) and fitted using the Curve Fitting Toolbox in MATLAB (version 7.12.0). The best fit was chosen, and the time point at which the monomer reached 50% is shown ± the 95% confidence values at that point. The R2 value indicates how well the line fits the data points, where 1.0 would represent a perfect fit.
| Source of UVR8 | Treatment | In Vivo/In Vitro | 50% Monomer | R2 |
|---|---|---|---|---|
| Wild type | In vivo | 18 min (+4/−3) | 0.94 | |
| Wild type | Protein extract | In vitro | 15 h (+/−2) | 0.85 |
| cop1-4 | In vivo | 75 min (+15/−9) | 0.87 | |
| Wild type | CHX | In vivo | 60 min (+/−6) | 0.98 |
| Wild type | DMSO (CHX control) | In vivo | 14 min (+2/−3) | 0.99 |
| Wild type | MG132 | In vivo | 17 min (+6/−4) | 0.95 |
| Wild type | DMSO (MG132 control) | In vivo | 15 min (+/−2) | 0.98 |
| uvr8-1/GFP-UVR8 | In vivo | 60 min (+/−15) | 0.94 | |
| uvr8-1/GFP-ΔC27UVR8 | In vivo | 5.5 h (+/−1) | 0.80 | |
| Purified UVR8 | In vitro | 15 h (+/−1) | 0.98 | |
| Purified UVR8 | Trypsin treated | In vitro | 14.5 h (+1.5/−2) | 0.95 |
In the introduction, we raised two possible mechanisms of dimer regeneration following monomerization: (1) reversion of monomer to dimer, and (2) dimerization of newly synthesized monomer to replace degraded monomer. From the above data, we conclude that if the UVR8 dimer is regenerated by reversion, the process occurs much more quickly in intact plants than in vitro. However, if there is monomer degradation and resynthesis, both must occur rapidly in vivo and must be coordinated to maintain a constant amount of UVR8.
Rapid Regeneration of the UVR8 Dimer Requires Intact Cells
To further explore the mechanism of regeneration, we monitored the amounts of UVR8 dimer and monomer following UV-B treatment of plant extracts. UV-B exposure of Arabidopsis extracts in vitro initiates monomerization of UVR8 (Fig. 1D). However, reappearance of the dimer is much slower in extracts than in intact plants, and the kinetics are very similar to those seen for reversion of the purified protein (compare Fig. 1, B and D), with most of the UVR8 protein being in the monomeric form 12 h after the end of UV-B illumination. Quantification of the loss of monomer (Supplemental Fig. S1A) further demonstrates the similarity in kinetics for the purified protein and plant cell extract in vitro (Table I). We conclude that rapid regeneration of the UVR8 dimer requires intact cells, implying the involvement of physiological processes.
Protein Synthesis Is Required for Rapid Regeneration of the Dimer
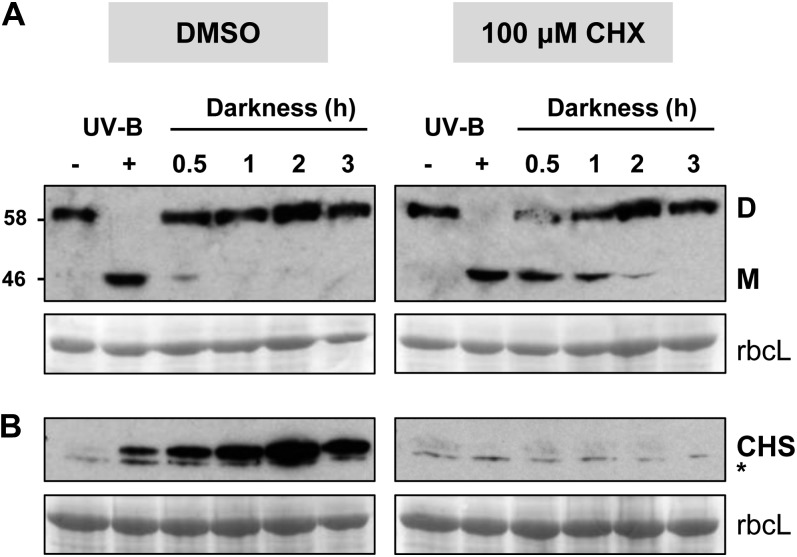
To examine whether protein synthesis is important in dimer regeneration, we treated plants with cycloheximide (CHX). Plants were transferred to liquid medium containing CHX 1 h before the start of UV-B exposure to ensure that the chemical entered the cells. To test the effectiveness of the CHX treatment, we monitored the UV-B induction of CHALCONE SYNTHASE (CHS) expression, which is inhibited by CHX (Christie and Jenkins, 1996). Control plants were treated in exactly the same way except that CHX was omitted. As shown in Figure 2A, CHX treatment did not impair UV-B-induced conversion of the UVR8 dimer to the monomer. Nevertheless, CHX was active in the tissue because CHS expression, detected by a specific antibody, was prevented (Fig. 1B).
Figure 2.
Protein synthesis is required to maximize the rate of dimer regeneration. A, Immunoblots of whole cell extracts from wild-type Ler plants probed with anti-UVR8 antibody. Plants were placed in medium containing 0.1% DMSO with or without CHX 1 h before exposure to 2.5 μmol m−2 s−1 UV-B for 3 h (UV-B +) and then transferred to darkness for the indicated times before extracts were prepared. Samples were prepared for electrophoresis without boiling and resolved on a 7.5% SDS-PAGE gel prior to immunoblotting. The UVR8 dimer (D) and monomer (M) are indicated. B, Immunoblots from the experiment shown in A probed with anti-CHS antibody. The asterisk indicates a nonspecific band recognized by the antibody. Ponceau staining of Rubisco large subunit (rbcL) is shown as a loading control.
We reasoned that if protein synthesis is required to replenish the UVR8 dimer following hypothetical rapid degradation of the monomer, then the amount of UVR8 should decrease substantially following CHX treatment. However, as shown in Figure 2A, CHX treatment did not affect the total amount of UVR8 up to at least 3 h following the end of UV-B exposure, by which time UVR8 is in the dimeric form. This result indicates that synthesis of the UVR8 protein is not required for dimer regeneration following UV-B exposure. Nevertheless, Figure 2A shows that the rate of dimer reappearance is slower in the plants treated with CHX; whereas very little monomer remains in control plants 30 min after transfer to darkness following UV-B treatment, in the CHX-treated plants, a substantial amount of monomer remains after 1 h and is still detectable after 2 h of darkness. Quantification of the loss of monomer (Supplemental Fig. S1B) reveals the slower kinetics following CHX treatment (Table I). We conclude that protein synthesis is required to maximize the rate of reversion from monomer to dimer.
No Evidence of Targeted Proteolysis of UVR8 via the Proteasome
To complement the experiments with CHX, we examined the effect of MG132, which inhibits protein degradation by the proteasome, on the amount of UVR8 following UV-B exposure. Since several previous studies employed quite long preincubations with MG132 to see inhibition of the proteasome (Jang et al., 2005; Yang et al., 2005; Dong et al., 2006), plants were transferred to liquid medium containing MG132 11 h before the start of UV-B illumination. MG132 did not impair UV-B-induced conversion of the UVR8 dimer to the monomer (Fig. 3A). Furthermore, there was no effect on regeneration of the dimer following transfer to darkness (Fig. 3A) and no quantitative difference in the kinetics for monomer loss in MG132-treated and control plants (Table I; Supplemental Fig. S1C). To check that MG132 had entered the tissue, an immunoblot was incubated with an antibody to ubiquitin. Inhibition of proteasomal degradation should lead to the accumulation of polyubiquitylated proteins in the cell, and it is evident that increased amounts of such proteins are present in the plants treated with the inhibitor compared with the control (Fig. 3B). The key observation in this experiment is that the total amount of UVR8 did not change over the time course of illumination and dimer regeneration in darkness (Fig. 3, A and C). Therefore, we conclude that UVR8 is not subject to proteasomal degradation following UV-B exposure.
Figure 3.
Monomeric UVR8 is not degraded via the proteasome. A, Immunoblots of whole cell extracts from wild-type Ler plants probed with anti-UVR8 antibody. Plants were placed in medium containing 0.1% DMSO with or without MG132 11 h before exposure to 2.5 μmol m−2 s−1 UV-B for 3 h (UV-B +) and then transferred to darkness for the indicated times before extracts were prepared. Samples were prepared for electrophoresis without boiling and resolved on a 7.5% SDS-PAGE gel prior to immunoblotting. The UVR8 dimer (D) and monomer (M) are indicated. Ponceau staining of Rubisco large subunit (rbcL) is shown as a loading control. B, Immunoblots from the experiment shown in A probed with anti-ubiquitin antibody. C, Immunoblots prepared as in A probed with anti-UVR8 antibody but with samples of whole cell extract boiled prior to electrophoresis on a 7.5% SDS-PAGE gel.
The C Terminus of UVR8 Is Required for Rapid in Vivo Regeneration of the Photoreceptor
To further study regeneration of the UVR8 dimer, we examined the importance of the C terminus of the protein. A 27-amino acid region in the C terminus from residues 397 to 423 (termed C27) is both necessary and sufficient for interaction with the WD40 region of the COP1 protein and can also interact with other WD40 proteins (Cloix et al., 2012). As shown in Figure 4A, the uvr8-2 mutant, which lacks the C-terminal 40 amino acids including the C27 region (Brown et al., 2005; Cloix et al., 2012), shows normal UV-B induction of monomerization but slower regeneration of the dimer in subsequent darkness compared with the wild type; the monomer is still detectable 4 h after the end of illumination. A similar observation is seen with a UVR8 mutant protein lacking specifically the C27 region (Fig. 4B). This experiment was undertaken with a transgenic uvr8-1 line expressing UVR8 lacking the C27 region fused to GFP at the N terminus (uvr8-1/GFP-ΔC27UVR8; Cloix et al., 2012); control plants expressed wild-type UVR8 fused to GFP (uvr8-1/GFP-UVR8; Kaiserli and Jenkins, 2007). Both lines show UV-B-induced monomerization, but regeneration of the dimer was much slower in the uvr8-1/GFP-ΔC27UVR8 plants compared with the uvr8-1/GFP-UVR8 control (Fig. 4B). Quantification (Table I; Supplemental Fig. S1D) shows the slower kinetics of the loss of monomer in the uvr8-1/GFP-ΔC27UVR8 plants. We conclude that the C27 region is required to maximize the rate of UVR8 dimer regeneration in vivo.
Figure 4.
The C terminus of UVR8 is required for rapid regeneration of the dimer in vivo. A, Immunoblots of whole cell extracts from wild-type Ler (WT) and uvr8-2 plants probed with anti-UVR8 antibody. Plants were exposed to 2.5 μmol m−2 s−1 UV-B for 3 h (UV-B +) and then transferred to darkness for the indicated times before extracts were prepared. Extract samples were prepared for electrophoresis without boiling and resolved on a 7.5% SDS-PAGE gel prior to immunoblotting. The UVR8 dimer (D) and monomer (M) are indicated. Ponceau staining of Rubisco large subunit (rbcL) is shown as a loading control. B, Immunoblots of whole cell extracts from uvr8-1/GFP-UVR8 and uvr8-1/GFP-ΔC27UVR8 plants probed with anti-GFP antibody. Plants were exposed to UV-B and transferred to darkness, and samples were analyzed by electrophoresis as in A. The asterisk indicates a nonspecific band recognized by the anti-GFP antibody. C, Coomassie-stained SDS-PAGE of purified UVR8 protein digested with trypsin, exposed to 1.5 μmol m−2 s−1 UV-B for 1 h (UV-B +), and then transferred to darkness for the indicated times. Samples were analyzed without boiling on a 7.5% SDS-PAGE gel. A non-UV-B-treated boiled sample is shown as a control. D, Immunoblot of whole cell extract from cop1-4 plants probed with anti-UVR8 antibody. Plants were exposed to UV-B and transferred to darkness, and samples were analyzed by electrophoresis as in A.
To test whether the C terminus affected the rate of dimer regeneration in vitro, purified UVR8 was subjected to a mild trypsin treatment, which removes 40 amino acids from the C terminus (Christie et al., 2012). This C-terminally truncated protein undergoes normal UV-B-induced dimer-to-monomer conversion, as reported previously (Christie et al., 2012). However, in contrast to the in vivo situation, the rate of dimer regeneration in darkness following UV-B exposure was not slower for C-terminally truncated UVR8 compared with the wild-type protein; the kinetics of regeneration were indistinguishable for the two proteins (compare Figs. 1B and 4C). The same conclusion can be drawn from measurements of monomer loss (Table I; Supplemental Fig. S1E). This finding indicates that the absence of the C27 region only slows regeneration in intact cells, suggesting that interaction of one or more proteins with the C27 region may be required to maximize the rate of regeneration.
COP1 Is Required for Rapid Dimer Regeneration in Vivo
Since the C27 region interacts with COP1, we reasoned that this interaction might be important for dimer regeneration. The kinetics of regeneration, therefore, was examined in the cop1-4 mutant. As shown in Figure 4D, cop1-4 plants show normal UV-B-induced UVR8 monomerization, but the rate of dimer regeneration in subsequent darkness is slower than in wild-type plants; whereas the monomer is no longer detectable in wild-type plants after 1 h of darkness, monomer is present in cop1-4 plants after at least 2 h of darkness (compare Fig. 4D with Figs. 1A and 4A). Quantification of monomer loss reveals the slower kinetics for cop1-4 plants compared with the wild type (Table I; Supplemental Fig. S1F). Thus, COP1 is required to maximize the rate of dimer regeneration in vivo.
DISCUSSION
No Evidence for Rapid Turnover of UVR8 in Intact Plants
In intact plants, exposure to UV-B converts dimeric UVR8 into the monomer, and the dimer is then totally regenerated in less than 1 h of darkness. Furthermore, there is no change in the total amount of UVR8 over the time course of monomerization and regeneration. The question we addressed here is how the dimer is regenerated. We considered the possibility that the monomer is degraded following its formation and that the dimer is replaced following the synthesis of new monomer. If this were the case, the processes would have to be rapid and closely coordinated to maintain a constant amount of UVR8. The experiments presented in Figures 2 and 3 do not provide any evidence of such rapid turnover.
The preincubation with CHX was evidently effective because it prevented the accumulation of CHS in response to UV-B treatment. We previously reported that CHX inhibits CHS transcript accumulation in response to UV-B without generally affecting gene expression (Christie and Jenkins, 1996). Hence, the CHX treatment would be expected to inhibit the synthesis of UVR8 following UV-B treatment if it was required to replace the dimer. However, CHX treatment did not prevent the regeneration of the dimer, and there was no detectable change in the total amount of UVR8 over the duration of the experiment. The fact that the dimer reappeared completely in the presence of CHX indicates that UVR8 protein synthesis is not required for dimer regeneration.
MG132 has been used widely to inhibit proteasomal activity (Osterlund et al., 2000; Yang et al., 2005; Dong et al., 2006; Yu et al., 2007). The presence of MG132 in cells causes the general accumulation of polyubiquitylated proteins, which can be seen in our experiment using an anti-ubiquitin antibody. Although MG132 evidently entered the tissue, there was no effect either on the regeneration of the dimer or on the total amount of UVR8. Clearly, if the monomer was rapidly degraded via the proteasome following UV-B exposure, MG132 would have inhibited its disappearance. Therefore, we conclude that the monomer is not subject to significant proteasomal degradation. Although it is not possible to rule out monomer degradation by other types of proteolysis, the fact that the total amount of UVR8 remained constant throughout all the experiments presented in this study indicates that the protein is not subject to rapid turnover. This conclusion is consistent with previous studies showing that UVR8 is essentially constitutively expressed; the protein is present in all plant tissues analyzed to date (Rizzini et al., 2011), and its abundance is not affected by different light qualities (Kaiserli and Jenkins, 2007).
We conclude from the above experiments that the UVR8 dimer is regenerated by reversion of the monomer to the dimer. Since the photoreceptor appears to be relatively stable, we anticipate that, in vivo, UVR8 will cycle between the dimeric and monomeric forms according to the prevailing ambient level of UV-B. Thus, a UVR8 dimer/monomer photoequilibrium may be established analogous to the balance between the inactive Pr and active Pfr forms of phytochromes in daylight. The stability of the UVR8 photoreceptor is similar to that of the light-stable phytochromes and cry1 but contrasts with that of the phyA and cry2 photoreceptors, both of which are subject to proteasomal degradation (Seo et al., 2004; Yu et al., 2007).
Protein Synthesis Is Required to Maximize the Rate of Dimer Regeneration
Regeneration of the UVR8 dimer following UV-B exposure occurs much more slowly with the homogeneous purified protein than it does in vivo. The purified protein is very stable in vitro, and it monomerizes in response to UV-B even after 48 h of incubation in darkness at room temperature (Christie et al., 2012). In addition, dimer regeneration occurs with the same slow kinetics following exposure of plant extracts to UV-B. Therefore, it appears that a normal cellular environment is required to maximize the rate of regeneration, presumably because cellular compartmentation or particular physiological processes are needed.
The CHX experiment indicates that protein synthesis following UV-B exposure is required to facilitate the rapid reversion from monomer to dimer. CHX treatment did not affect the total amount of UVR8 or prevent dimer regeneration, but it did slow the kinetics of monomer disappearance and dimer accumulation. A likely scenario is that one or more proteins synthesized in response to UV-B facilitate reversion of the monomer. Nonetheless, dimer formation still occurred in the presence of CHX, so protein synthesis was evidently not essential for regeneration. Moreover, the rate of dimer formation in vivo in the presence of CHX was still a lot faster than that in vitro, so clearly, protein synthesis is not the only factor required for rapid dimer regeneration.
The C Terminus of UVR8 Is Required for Rapid Regeneration in Vivo
Deletion of the C-terminal 40 amino acids of UVR8, in the uvr8-2 allele, and specifically the C27 region, in the GFP-ΔC27UVR8 fusion, reduced the rate of dimer regeneration in vivo. To address the possibility that removal of the C-terminal region impaired reversion to the dimer because of an adverse effect on UVR8 structure, we examined the regeneration of trypsin-treated purified UVR8, which is equivalent to the uvr8-2 allele because it lacks the C-terminal 40 amino acids. Since there was no difference in the kinetics of dimer regeneration for the wild-type and trypsin-treated UVR8 proteins in vitro, we conclude that truncation of the C terminus does not impair regeneration for structural reasons. That the absence of the C-terminal region only affects regeneration in vivo suggests that this part of UVR8 may interact with proteins that facilitate reversion to the dimer. This could include proteins synthesized in response to UV-B, as discussed above. The presence of preexisting proteins is not sufficient to maximize the rate of reversion, at least in vitro, because there is no difference in reversion kinetics between purified UVR8 and UV-B-treated whole cell extracts. Nevertheless, the absence of COP1, which is present prior to UV-B exposure, does diminish the rate of dimer regeneration, indicating that it is involved, most likely with other proteins, in facilitating reversion of the monomer.
We reported recently that COP1 interacts with the C27 region of UVR8 to initiate signal transduction (Cloix et al., 2012). It is possible that this interaction is required to promote the recruitment of other proteins that facilitate reversion to the dimer. In principle, the C terminus could interact with a range of proteins; it is known to interact with the WD40 domain proteins COP1, RUP1, and RUP2 (Cloix et al., 2012), but there are numerous WD40 domain proteins in plants, and other types of proteins might also interact. Moreover, COP1 is required for the UV-B induction of many UVR8-regulated genes, so the absence of COP1 in the cop1-4 mutant may impair the synthesis of one or more components needed for rapid regeneration. Therefore, we do not know whether the role of COP1 in dimer regeneration is direct, via its ability to interact with the C terminus of UVR8, or indirect, via its requirement to synthesize one or more other proteins that can interact with the C terminus. It should be noted, however, that regeneration of the dimer in cop1-4 plants and in UVR8 with C-terminal deletions in vivo is still faster than in vitro, so additional factors are likely to maximize the rate of dimer regeneration.
CONCLUSION
In summary, this study demonstrates that the dimeric UVR8 photoreceptor is regenerated rapidly in vivo and that this is accomplished by reversion of the monomer to the dimer rather than by a mechanism involving rapid turnover of the protein. This process is crucial, as it will enable the photoreceptor to respond rapidly and sensitively to changes in ambient UV-B levels in sunlight to regulate photomorphogenic responses. The data presented suggest that the process of reversion from monomer to dimer is complex and is facilitated by several factors: the presence of intact cells, protein synthesis in response to UV-B, and interaction of the C-terminal region of UVR8 with proteins, including COP1. Therefore, further research is required to identify the components and processes involved.
MATERIALS AND METHODS
Plant Materials and Treatments
Seeds of wild-type Arabidopsis (Arabidopsis thaliana) ecotype Landsberg erecta (Ler) and the cop1-4 mutant were obtained from the Nottingham Arabidopsis Stock Centre. The uvr8-2 mutant (Ler background) was isolated by Brown et al. (2005) and further characterized by Cloix et al. (2012). Transgenic lines expressing the GFP-UVR8 and GFP-ΔC27UVR8 (UVR8 lacking amino acids 397–423) fusions under the control of the UVR8 promoter were made in the uvr8-1 background, as described by Kaiserli and Jenkins (2007) and Cloix et al. (2012), respectively.
Plants were grown on agar plates containing one-half-strength Murashige and Skoog (MS) salts under 100 µmol m−2 s−1 constant white light (warm white fluorescent tubes; Osram) at 21°C for 10 d and then placed in darkness for 16 h. Plants were exposed to narrow-band UV-B (Philips TL20W/01RS; spectrum shown in Christie et al., 2012) as indicated in the figure legends and subsequently transferred to darkness for the durations shown in the figures before the preparation of protein extracts.
For CHX and MG132 treatments, plants were transferred to liquid one-half-strength MS medium containing CHX (100 μm, dissolved in 0.1% dimethyl sulfoxide [DMSO]; Sigma) or MG132 (100 μm, dissolved in 0.1% DMSO; Calbiochem) and incubated for 1 or 11 h, respectively, prior to the UV-B treatment. Control plants were transferred to one-half-strength MS medium containing 0.1% DMSO alone.
UVR8 Dimer/Monomer Analysis
Arabidopsis whole cell extracts were prepared as described by Kaiserli and Jenkins (2007). The dimer/monomer status of UVR8 in whole cell extracts was examined essentially as described by Rizzini et al. (2011). Four times loading buffer (250 mm Tris-HCl, pH 6.8, 2% SDS, 20% β-mercaptoethanol, 40% glycerol, and 0.5% bromphenol blue) was added to the samples, and the proteins were loaded on a 7.5% SDS-PAGE gel without boiling (unless indicated otherwise). Immunoblots were incubated with an anti-UVR8 antibody directed against the C terminus of the protein (Kaiserli and Jenkins, 2007), except for the uvr8-2 experiment, when an antibody recognizing the N terminus was used (Cloix and Jenkins, 2008), or anti-GFP antibody (Clontech) in the case of the GFP fusions.
For direct UV-B illumination of extracts (Fig. 1C), exposure to 1.5 µmol m−2 s−1 narrow-band UV-B for 1 h was carried out on ice (control extracts were not illuminated), and the extracts were then transferred to darkness at room temperature.
Preparation of purified UVR8 protein as well as trypsin treatment (N-tosyl-l-phenylalanyl chloromethyl ketone treated; Sigma) were performed as described by Christie et al. (2012). Purified proteins were exposed to 1.5 µmol m−2 s−1 narrow-band UV-B on ice for 1 h and transferred to darkness at room temperature for the indicated times. Protein samples were prepared for electrophoresis without boiling (unless indicated otherwise) and loaded on a 7.5% SDS-PAGE gel. Gels were stained with Coomassie blue.
For the analysis of CHS and polyubiquitylated proteins, protein samples were boiled prior to electrophoresis on a 7.5% SDS-PAGE gel. Immunoblots were probed with anti-CHS (Santa Cruz) or anti-ubiquitin (Agrisera) antibodies.
Immunoblots were stained with Ponceau S to reveal the Rubisco large subunit, which was used as a loading control. The data shown are representative of at least three independent experiments.
Quantification of UVR8 monomer loss in darkness following UV-B exposure was undertaken for representative western blots from three independent experiments. The immunodetected UVR8 bands were quantified using ImageJ software. Data were corrected for background and normalized against the value of the monomer after UV-B illumination, taken as 100%. Points were plotted and fitted using the Curve Fitting Toolbox in MATLAB (version 7.12.0). The best fit was chosen, and a 95% confidence level of the fit is shown. To facilitate comparison between treatments and genotypes, the time taken for loss of 50% of the monomer was calculated.
The Arabidopsis Genome Initiative locus identifier for UVR8 is At5g63860 and that for COP1 is At2g32950.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Kinetics of the loss of UVR8 monomer in darkness following UV-B exposure under the experimental conditions used in this study.
Acknowledgments
We are particularly grateful to John M. Christie for advice with experiments, discussion and suggestions, and comments on the manuscript. We also thank Brian O. Smith, Catherine Cloix, and all members of the Jenkins and Christie laboratories for discussion of the research and Tobias Nagel for help with statistical analysis. We are grateful to Ari Sadanandom for the anti-ubiquitin antibody.
Glossary
- CHX
cycloheximide
- Ler
Landsberg erecta
- MS
Murashige and Skoog
- DMSO
dimethyl sulfoxide
References
- Ballaré CL, Barnes PW, Flint SD. (1995) Inhibition of hypocotyl elongation by ultraviolet-B radiation in de-etiolating tomato seedlings. 1. The photoreceptor. Physiol Plant 93: 584–592 [Google Scholar]
- Brown BA, Cloix C, Jiang GH, Kaiserli E, Herzyk P, Kliebenstein DJ, Jenkins GI. (2005) A UV-B-specific signaling component orchestrates plant UV protection. Proc Natl Acad Sci USA 102: 18225–18230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown BA, Jenkins GI. (2008) UV-B signaling pathways with different fluence-rate response profiles are distinguished in mature Arabidopsis leaf tissue by requirement for UVR8, HY5, and HYH. Plant Physiol 146: 576–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie JM, Arvai AS, Baxter KJ, Heilmann M, Pratt AJ, O’Hara A, Kelly SM, Hothorn M, Smith BO, Hitomi K, et al. (2012) Plant UVR8 photoreceptor senses UV-B by tryptophan-mediated disruption of cross-dimer salt bridges. Science 335: 1492–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie JM, Jenkins GI. (1996) Distinct UV-B and UV-A/blue light signal transduction pathways induce chalcone synthase gene expression in Arabidopsis cells. Plant Cell 8: 1555–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloix C, Jenkins GI. (2008) Interaction of the Arabidopsis UV-B-specific signaling component UVR8 with chromatin. Mol Plant 1: 118–128 [DOI] [PubMed] [Google Scholar]
- Cloix C, Kaiserli E, Heilmann M, Baxter KJ, Brown BA, O’Hara A, Smith BO, Christie JM, Jenkins GI. (2012) C-terminal region of the UV-B photoreceptor UVR8 initiates signaling through interaction with the COP1 protein. Proc Natl Acad Sci USA 109: 16366–16370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong CH, Agarwal M, Zhang Y, Xie Q, Zhu JK. (2006) The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc Natl Acad Sci USA 103: 8281–8286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favory JJ, Stec A, Gruber H, Rizzini L, Oravecz A, Funk M, Albert A, Cloix C, Jenkins GI, Oakeley EJ, et al. (2009) Interaction of COP1 and UVR8 regulates UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. EMBO J 28: 591–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohnmeyer H, Staiger D. (2003) Ultraviolet-B radiation-mediated responses in plants: balancing damage and protection. Plant Physiol 133: 1420–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber H, Heijde M, Heller W, Albert A, Seidlitz HK, Ulm R. (2010) Negative feedback regulation of UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. Proc Natl Acad Sci USA 107: 20132–20137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hectors K, Prinsen E, De Coen W, Jansen MAK, Guisez Y. (2007) Arabidopsis thaliana plants acclimated to low dose rates of ultraviolet B radiation show specific changes in morphology and gene expression in the absence of stress symptoms. New Phytol 175: 255–270 [DOI] [PubMed] [Google Scholar]
- Heijde M, Ulm R. (2012) UV-B photoreceptor-mediated signalling in plants. Trends Plant Sci 17: 230–237 [DOI] [PubMed] [Google Scholar]
- Jang I-C, Yang J-Y, Seo HS, Chua N-H. (2005) HFR1 is targeted by COP1 E3 ligase for post-translational proteolysis during phytochrome A signaling. Genes Dev 19: 593–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins GI. (2009) Signal transduction in responses to UV-B radiation. Annu Rev Plant Biol 60: 407–431 [DOI] [PubMed] [Google Scholar]
- Jordan BR. (1996) The effects of ultraviolet-B radiation on plants: a molecular perspective. Adv Bot Res 22: 97–162 [Google Scholar]
- Kaiserli E, Jenkins GI. (2007) UV-B promotes rapid nuclear translocation of the Arabidopsis UV-B specific signaling component UVR8 and activates its function in the nucleus. Plant Cell 19: 2662–2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BC, Tennessen DJ, Last RL. (1998) UV-B-induced photomorphogenesis in Arabidopsis thaliana. Plant J 15: 667–674 [DOI] [PubMed] [Google Scholar]
- Kliebenstein DJ, Lim JE, Landry LG, Last RL. (2002) Arabidopsis UVR8 regulates ultraviolet-B signal transduction and tolerance and contains sequence similarity to human regulator of chromatin condensation 1. Plant Physiol 130: 234–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JY, Ou-Lee TM, Raba R, Amundson RG, Last RL. (1993) Arabidopsis flavonoid mutants are hypersensitive to UV-B irradiation. Plant Cell 5: 171–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oravecz A, Baumann A, Máté Z, Brzezinska A, Molinier J, Oakeley EJ, Adám É, Schäfer E, Nagy F, Ulm R. (2006) CONSTITUTIVELY PHOTOMORPHOGENIC1 is required for the UV-B response in Arabidopsis. Plant Cell 18: 1975–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund MT, Hardtke CS, Wei N, Deng XW. (2000) Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405: 462–466 [DOI] [PubMed] [Google Scholar]
- Rizzini L, Favory J-J, Cloix C, Faggionato D, O’Hara A, Kaiserli E, Baumeister R, Schäfer E, Nagy F, Jenkins GI, et al. (2011) Perception of UV-B by the Arabidopsis UVR8 protein. Science 332: 103–106 [DOI] [PubMed] [Google Scholar]
- Rozema J, van de Staaij J, Björn LO, Caldwell M. (1997) UV-B as an environmental factor in plant life: stress and regulation. Trends Ecol Evol 12: 22–28 [DOI] [PubMed] [Google Scholar]
- Seo HS, Watanabe E, Tokutomi S, Nagatani A, Chua N-H. (2004) Photoreceptor ubiquitination by COP1 E3 ligase desensitizes phytochrome A signaling. Genes Dev 18: 617–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinkle JR, Atkins AK, Humphrey EE, Rodgers CW, Wheeler SL, Barnes PW. (2004) Growth and morphological responses to different UV wavebands in cucumber (Cucumis sativum) and other dicotyledonous seedlings. Physiol Plant 120: 240–248 [DOI] [PubMed] [Google Scholar]
- Stapleton AE, Walbot V. (1994) Flavonoids can protect maize DNA from the induction of ultraviolet radiation damage. Plant Physiol 105: 881–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R, Favory J-J, Gruber H, Bartelniewoehner L, Bartels S, Binkert M, Funk M, Weisshaar B, Ulm R. (2010) The Arabidopsis bZIP transcription factor HY5 regulates expression of the PFG1/MYB12 gene in response to light and ultraviolet-B radiation. Plant Cell Environ 33: 88–103 [DOI] [PubMed] [Google Scholar]
- Suesslin C, Frohnmeyer H. (2003) An Arabidopsis mutant defective in UV-B light-mediated responses. Plant J 33: 591–601 [DOI] [PubMed] [Google Scholar]
- Ulm R, Baumann A, Oravecz A, Máté Z, Adám E, Oakeley EJ, Schäfer E, Nagy F. (2004) Genome-wide analysis of gene expression reveals function of the bZIP transcription factor HY5 in the UV-B response of Arabidopsis. Proc Natl Acad Sci USA 101: 1397–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulm R, Nagy F. (2005) Signalling and gene regulation in response to ultraviolet light. Curr Opin Plant Biol 8: 477–482 [DOI] [PubMed] [Google Scholar]
- Wargent JJ, Gegas VC, Jenkins GI, Doonan JH, Paul ND. (2009) UVR8 in Arabidopsis thaliana regulates multiple aspects of cellular differentiation during leaf development in response to ultraviolet B radiation. New Phytol 183: 315–326 [DOI] [PubMed] [Google Scholar]
- Wu D, Hu Q, Yan Z, Chen W, Yan C, Huang X, Zhang J, Yang P, Deng H, Wang J, et al. (2012) Structural basis of ultraviolet-B perception by UVR8. Nature 484: 214–219 [DOI] [PubMed] [Google Scholar]
- Yang J, Lin R, Sullivan J, Hoecker U, Liu B, Xu L, Deng XW, Wang H. (2005) Light regulates COP1-mediated degradation of HFR1, a transcription factor essential for light signaling in Arabidopsis. Plant Cell 17: 804–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi C, Deng XW. (2005) COP1: from plant photomorphogenesis to mammalian tumorigenesis. Trends Cell Biol 15: 618–625 [DOI] [PubMed] [Google Scholar]
- Yu X, Klejnot J, Zhao X, Shalitin D, Maymon M, Yang H, Lee J, Liu X, Lopez J, Lin C. (2007) Arabidopsis cryptochrome 2 completes its posttranslational life cycle in the nucleus. Plant Cell 19: 3146–3156 [DOI] [PMC free article] [PubMed] [Google Scholar]