Abstract
Nodulation in legumes requires the recognition of rhizobially made Nod factors. Genetic studies have revealed that the perception of Nod factors involves LysM domain receptor-like kinases, while biochemical approaches have identified LECTIN NUCLEOTIDE PHOSPHOHYDROLASE (LNP) as a Nod factor-binding protein. Here, we show that antisense inhibition of LNP blocks nodulation in Lotus japonicus. This absence of nodulation was due to a defect in Nod factor signaling based on the observations that the early nodulation gene NODULE INCEPTION was not induced and that both Nod factor-induced perinuclear calcium spiking and calcium influx at the root hair tip were blocked. However, Nod factor did induce root hair deformation in the LNP antisense lines. LNP is also required for infection by the mycorrhizal fungus Glomus intraradices, suggesting that LNP plays a role in the common signaling pathway shared by the rhizobial and mycorrhizal symbioses. Taken together, these observations indicate that LNP acts at a novel position in the early stages of symbiosis signaling. We propose that LNP functions at the earliest stage of the common nodulation and mycorrhization symbiosis signaling pathway downstream of the Nod factor receptors; it may act either by influencing signaling via changes in external nucleotides or in conjunction with the LysM receptor-like kinases for recognition of Nod factor.
The acquisition of mineral nutrients from the environment often limits plant growth, and many plants have established symbiotic interactions with beneficial microorganisms that facilitate nutrient acquisition. The association with arbuscular mycorrhizal fungi helps in nutrient uptake, particularly phosphate, and this symbiosis is almost ubiquitous within the plant kingdom, reflecting its early establishment during the evolution of higher plants (Parniske, 2008). Several plant species also form symbioses with nitrogen-fixing bacteria such as rhizobia and Frankia spp., and these symbioses are restricted to plants in the Rosid I clade (Doyle, 1998). The establishment of both mycorrhizal and rhizobial interactions in legumes involves a molecular signal exchange between the plant and its symbiont. Legumes release strigolactones and flavonoids into the rhizosphere, and these are recognized by mycorrhizal fungi and rhizobia, respectively (Oldroyd et al., 2009). In turn, the symbionts release signals to the plant: lipochitooligosaccharide Nod factors from rhizobia (Dénarié et al., 1996) and Myc factors from mycorrhizal fungi (Kosuta et al., 2003; Oláh et al., 2005; Maillet et al., 2011). Species-specific decorations on the Nod factor backbone are recognized by the host legume, and this determines specificity in the legume-rhizobial interaction (Perret et al., 2000). In contrast, there appears to be little specificity in mycorrhizal interactions (Parniske, 2008).
Nodulation-defective legume mutants have revealed a signaling pathway that is conserved between Nod factor and mycorrhizal recognition (Wais et al., 2000; Walker et al., 2000; Kistner et al., 2005), and considering the different evolutionary histories of these two symbioses, it is legitimate to surmise that the evolution of nodulation involved the recruitment of the preexisting mycorrhizal signaling pathway for the recognition of Nod factor. At the core of this common symbiosis signaling (SYM) pathway are oscillations in nucleus-associated cytosolic calcium levels, termed calcium spiking, acting as a secondary messenger to induce gene expression. A receptor-like kinase (SYMRK/DMI2 [Endre et al., 2002; Stracke et al., 2002]), three components of the nuclear pore (NUP133, NUP85, and NENA [Kanamori et al., 2006; Saito et al., 2007; Groth et al., 2010]), and nucleus-localized cation channels (POLLUX/DMI1, CASTOR [Ané et al., 2004; Imaizumi-Anraku et al., 2005; Charpentier et al., 2008]) are all required for the induction of calcium spiking, while a calcium- and calmodulin-dependent protein kinase (CCaMK [Lévy et al., 2004; Mitra et al., 2004; Tirichine et al., 2006]) and a protein of unknown function (CYCLOPS/IPD3 [Messinese et al., 2007; Yano et al., 2008]) are necessary for perception of the calcium signal and the induction of downstream genes. In addition to the common symbiosis pathway, Nod factor signaling also involves the nodulation-specific receptor-like kinases NFR1/LYK3 and NFR5/NFP (Limpens et al., 2003; Madsen et al., 2003; Radutoiu et al., 2003; Arrighi et al., 2006) that appear to function in Nod factor perception. There are also several nodulation-specific transcription factors that act downstream of CCaMK (Libault et al., 2009).
Genetic evidence indicates that different Nod factors are differentially perceived during Nod factor signaling (Ardourel et al., 1994), with perinuclear calcium spiking associated with early gene induction (Oldroyd and Downie, 2006) and calcium influx at the root hair tip proposed to be associated with the initiation of infection (Miwa et al., 2006a). This suggests a highly specific Nod factor recognition mechanism, which can discriminate between different Nod factor structures. The LysM receptor-like kinases NFR1/LYK3 and NFR5/NFP have emerged as strong candidates to function as Nod factor receptors based on the findings that purified NFR1 and NFR5 bind Nod factors with high affinity (Broghammer et al., 2012) and that the specificity of Nod factor recognition resides within the LysM domains (Radutoiu et al., 2007; Bek et al., 2010).
Whereas the LysM receptor-like kinases were identified using nodulation-defective mutants, we isolated a Nod factor-binding lectin from the legume Dolichos biflorus (also known as Vigna unguiculata; Quinn and Etzler, 1987). This protein can hydrolyze phosphoanhydride bonds of nucleoside triphosphates and diphosphates (Etzler et al., 1999). In mammalian systems, such enzymatic activity was proposed to remove extracellular ATP, known to be toxic to cells. This LECTIN NUCLEOTIDE PHOSPHOHYDROLASE (LNP) is a peripheral membrane protein present on the surface of epidermal root hairs in the zone of the root with the capacity to nodulate and is redistributed to the tips of the root hairs upon exposure to rhizobia or Nod factors (Kalsi and Etzler, 2000). The importance of LNP for the establishment of the rhizobial symbiosis was indicated following pretreatment of the roots with antiserum against recombinant LNP that inhibited both root hair deformation and nodulation in response to rhizobia (Etzler et al., 1999; Day et al., 2000). Two soybean (Glycine max) apyrases have been characterized, the Golgi-localized endoapyrase GS50 and the plasma membrane-associated ectoapyrase GS52 (Day et al., 2000). GS52 is rapidly induced by rhizobial inoculation (Day et al., 2000), its overexpression increased both nodulation and rhizobial infection (McAlvin and Stacey 2005), and the apyrase activity was required for the enhanced nodulation (Tanaka et al., 2011). Furthermore, silencing of GS52 expression by RNA interference (RNAi) reduced nodule development and infection (Govindarajulu et al., 2009); this phenotype was partially rescued by the addition of ADP, implying that a role for the ectoapyrase may be to produce ADP from the extracellular ATP that is present around root hair tips (Kim et al., 2006). These data suggest that the role for the apyrase activity could be in the production of ADP rather than the reduction of ATP levels. The roles of apyrases and extracellular nucleotides in plant signaling have been reviewed recently (Clark and Roux, 2011).
These studies indicate that LNP binds Nod factor and is necessary for nodulation signaling. In parallel, genetic studies implicate the LysM receptor-like kinases in specific Nod factor recognition (Radutoiu et al., 2007), and high-affinity Nod factor binding to purified NFR1 and NFR5 has been demonstrated recently (Broghammer et al., 2012). In order to better understand the function that Nod factor binding to LNP plays in the establishment of symbiotic interactions, we generated transgenic lines of Lotus japonicus that have markedly reduced LNP levels. These lines are defective for both nodulation and mycorrhization, suggesting that LNP plays a role in the common symbiosis signaling pathway. Nod factor-induced responses such as gene induction, calcium influx, and calcium spiking were absent, but root hair deformation was retained, implying that LNP functions at a very early stage in the symbiosis signaling pathway.
RESULTS
Isolation of Stable Antisense Transformants Lacking LNP
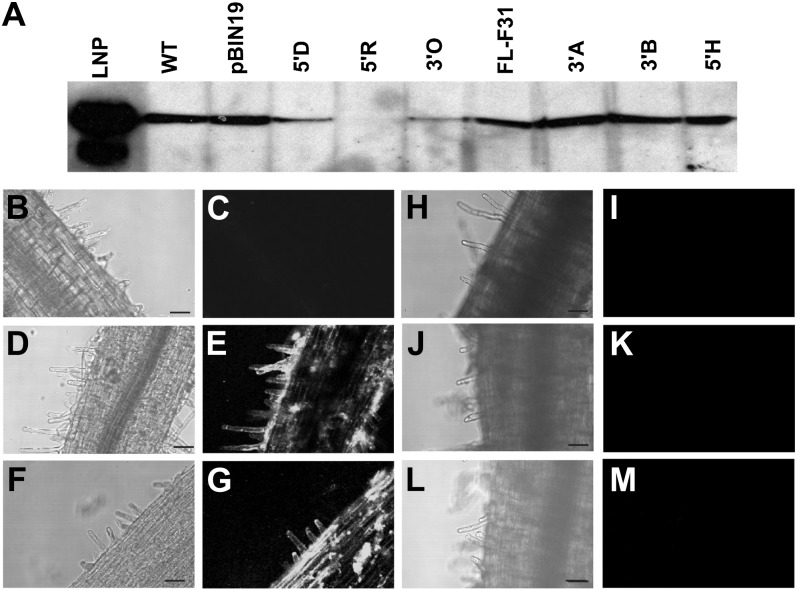
L. japonicus, ecotype Gifu, was stably transformed with three different L. japonicus LNP antisense constructs under the control of the constitutive 35S promoter. The full-length antisense construct contained the complete 1,489-nucleotide complementary DNA from L. japonicus (Roberts et al., 1999), whereas the 5′ and 3′ constructs contained nucleotides 1 to 719 and nucleotides 536 to 1,383, respectively. Seven independent transformed lines (one with the full-length construct, three with the 3′ construct, and three with the 5′ construct) were selected from independent calli based on their strong kanamycin resistance. Extracts of the uninoculated roots from the seven lines, the wild type, and the vector control were immunoblotted with antiserum to LNP. Three of the transformed antisense lines, named 5′D, 5′R, and 3′O (generated using the 5′ or 3′ construct), had substantially reduced levels of immunoreactive material compared with the wild type (Fig. 1A), so these lines along with the vector controls were propagated for seed production. The initial T0 transformed plants were selfed, and T1 seed were selected on antibiotic plates. Homozygous T1 lines were identified by testing their progeny for both kanamycin resistance (imparted by the transformed vector) and LNP protein levels using immunoblotting. If all the tested progeny from a single line were kanamycin resistant and showed substantially reduced LNP levels, we considered these lines to be homozygous. This approach was taken for 5′D, 5′R, 3′O, and the vector control lines, and homozygosity was achieved for each construct in different generations (T1, T2 or T3), and thus T2, T3, or T4 seed was used accordingly for further analysis.
Figure 1.
Immunoblot and confocal immunofluorescence microscopic analyses of LNP from wild-type (WT), vector control, and transgenic antisense lines. A, Equivalent amounts of extract from pooled roots of uninoculated 8-d-old L. japonicus lines (Table I) were subjected to SDS-urea-PAGE followed by immunoblot analysis using antiserum prepared against recombinant LNP. The single immunoreactive band corresponds to approximately 46 kD, the predicted size of LjLNP. B to M, Confocal immunofluorescence microscopy was performed using preimmunization serum (B and C) or anti-LNP serum (D–M) on whole mounts of fixed 7-d-old uninoculated roots from the following L. japonicus lines: the wild type (B–E), vector control (F and G), 5′D (H and I), 5′R (J and K), and 3′O (L and M). Bars = 50 μm.
LNP can be detected around the surface of root hairs using immunofluorescence (Kalsi and Etzler, 2000; Fig. 1E). To validate the LNP suppression in the 5′D, 5′R, and 3′O lines, we assessed the protein levels on the surface of the root. Whereas the wild type and the vector control lines showed good levels of LNP on the root surface (Fig. 1, D–G), the 5′D, 5′R, and 3′O lines showed no detectable levels of LNP (Fig. 1, H–M).
Searching the genome sequence of L. japonicus (Sato et al., 2008) revealed a second gene (chr1.CM0104.1830.r2.d) with clear similarity to LNP; the DNA and predicted protein sequences of the coding regions of this gene are 58% and 72% identical. We have called this gene LNP2, and to understand how these two genes are related to each other and to other apyrases, we generated a phylogenetic comparison of 68 plant apyrases and integrated this with an in silico prediction of their targeting sequences (Supplemental Fig. S1). From this combined analysis, there are a number of observations that can be made. Most of the sequences were predicted to contain one of three types of putative targeting sequence (secretory pathway, mitochondrial, or chloroplast). Among the proteins with predicted secretory targeting sequences, there were three clades, which could be separated into Fabaceae (including DbLNP, LjLNP, and GS52, which appear to be orthologs based on the phylogenetic analysis), Poaceae, and Solanaceae, with representatives from Brassicaceae. Within the Fabaceae clade, there are considerably more apyrases among those legumes (e.g. pea [Pisum sativum] and Medicago truncatula; Cohn et al., 2001; Navarro-Gochicoa et al., 2003) that form indeterminate nodules than those (e.g. Glycine soja and L. japonicus) that form determinate nodules. D. biflorus apyrase2 and soybean GS50 form a separate grouping within the predicted ectoapyrases. LjLNP2 falls within the clade of legume apyrases that are predicted to contain mitochondrial targeting sequences and is related to the Arabidopsis (Arabidopsis thaliana) apyrases APY1 and APY2, which appear to be involved in regulating stomatal aperture (Clark et al., 2011); the double apy1, apy2 mutant is male sterile (Steinebrunner et al., 2003).
We used quantitative reverse transcription (RT)-PCR with gene-specific primers to assay the levels of transcripts of the two LjLNP genes in the 5′D LNP antisense line and the empty vector control. We found an average 53% ± 22% reduction in the transcript levels of the LNP gene to which we made the construct and 46.5% ± 28% reduction in LNP2 transcript levels in the 5′D antisense line (from four independent biological replicates). Antisense is likely to function via both transcript stability in an RNAi-type mechanism and possibly through suppression of translation. Hence, the absolute levels of transcript abundance are unlikely to be the only cause of the dramatic reductions we observed in the level of LNP protein in roots. These results indicate that the LNP antisense construct may target both LNP genes, and this may explain why we observed relatively poor seed set and seed viability with some antisense lines. The two LNP genes are located in very close proximity (separated by only about 30 kb) on the genome, and this greatly limits the ability to generate a double mutant line. However, the key observation is that LNP is not antigenically detectable on the root surface and, therefore, is not available to function in symbiosis.
Antisense Suppression of LNP Results in a Nod− Phenotype
The homozygous 5′R (T2), 5′D (T3), and 3′O (T4) lines were tested for their ability to nodulate by inoculating 10-d-old seedlings with Mesorhizobium loti strain NZP2235. No nodules formed on the 5′D, 5′R, or 3′O plants (Table I); in comparison, the wild type and the vector control plants formed nodules (Table I; Fig. 2). When grown in the absence of added nitrate or ammonia, the nodulated plants all grew more vigorously than the nonnodulated plants (Table I), confirming that the nodules were fixing nitrogen. The fact that the three independent LNP knockdown lines were all defective for nodulation demonstrates that the loss of LNP caused the nodulation defect.
Table I. Nodulation in LNP antisense lines 4 weeks post inoculation with M. loti.
| Line | Generation | No. of Nodules per Plant | Shoot Mass | Root Mass |
|---|---|---|---|---|
| mg plant−1 | ||||
| Wild type | 23.7 ± 3.2 | 18.7 ± 4.4 | 10.2 ± 2.7 | |
| pBIN19 | T3 | 13.3 ± 2.4 | 15.0 ± 1.4 | 8.0 ± 0.4 |
| 5′D | T3 | 0.0 ± 0.0 | 8.3 ± 0.2 | 6.0 ± 0.7 |
| 5′R | T2 | 0.0 ± 0.0 | 5.9 ± 1.3 | 6.1 ± 0.2 |
| 3′O | T4 | 0.0 ± 0.0 | 4.4 ± 0.7 | 4.8 ± 0.7 |
Figure 2.
Root nodule phenotype of the L. japonicus 5′D LNP antisense line. Nodules formed in wild-type plants (A and D) and vector control (B and E) 3 weeks after inoculation with M. loti. No nodules were observed in the 5′D LNP antisense line (C and F). Bars = 10 mm (A–C) and 5 mm (D–F). [See online article for color version of this figure.]
Antisense Suppression of LNP Inhibits the Ability of Plants to Associate with Mycorrhizal Fungi
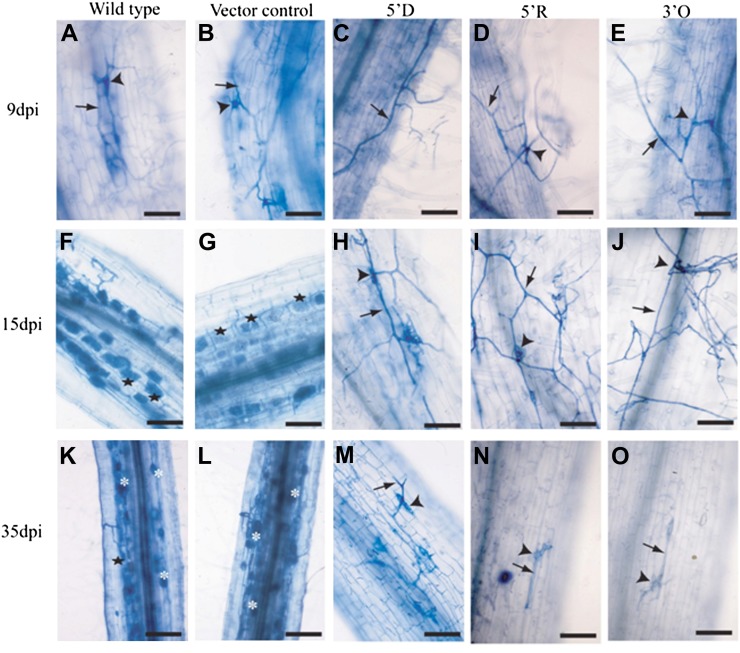
Many mutants defective for early nodulation signaling also fail to form symbioses with arbuscular mycorrhizal fungi (Parniske, 2008), and mycorrhizal fungi induce calcium spiking during initiation of the symbiosis (Kosuta et al., 2008). Mutations in NFR1 or NFR5 do not block mycorrhization, but mutations in genes of the SYM pathway do block the normal infection by mycorrhizal fungi (Parniske, 2008). We investigated the ability of the three LNP antisense lines, 5′D (T3), 5′R (T2), and 3′O (T4), to be colonized by Glomus intraradices. Ten plants were assessed at each time point, and roots were stained and analyzed for fungal colonization, with a total of 50 plants analyzed for each line. Nine days post inoculation (dpi), the attachment of fungal hyphae and appressoria formation were observed on the epidermal cell surface of all roots, and no difference was observed between the wild type, the vector control, and the three LNP antisense lines (Fig. 3, A–E). At 15 dpi, roots of both wild-type and vector control plants were heavily colonized by G. intraradices, with fungal colonization of inner cortical cells forming arbuscules (Fig. 3, F and G). No such invasion of roots by fungal hyphae was observed in the 5′D, 5′R, and 3′O lines (Fig. 3, H–J), and this lack of colonization was sustained even at 35 dpi, when wild-type and vector control roots were strongly colonized by G. intraradices (Fig. 3, K and L). Enhanced branching of fungal hyphae was observed on the epidermal cells of LNP antisense lines compared with the wild type and vector control, possibly because of the unsuccessful attempts of the fungal hyphae to gain entry into roots (Fig. 3, M–O). No balloon-like swellings of fungal hyphae or deformations were observed on the epidermal or inner cells of these three antisense lines at any time point. At 60 d after inoculation with G. intraradices, lines 5′D and 5′R showed some fungal colonization, but this was observed in only 6% of the root segments of the antisense lines compared with approximately 90% of the root segments obtained from the wild type. These results show that these three LNP antisense lines are defective for the initiation of the symbiotic association with mycorrhizal fungi, and this block is at the epidermal cell surface.
Figure 3.
Colonization of L. japonicus roots of wild-type and transgenic plants by G. intraradices. At 9 dpi (A–E), hyphal growth of G. intraradices (arrows) and appressoria formation (arrowheads) were observed on the epidermal surfaces of wild-type (A), vector control (B), 5′D (C), 5′R (D), and 3′O (E) roots. At 15 dpi (F–J) and 35 dpi (K–O), fungal hyphae formed arbuscules (stars) and vesicles (asterisk) in the cortical cells of wild-type (F and K) and vector control (G and L) roots. Only appressoria and fungal hyphal branching were observed on the epidermal surface of roots of 5′D (H and M), 5′R (I and N), and 3′O (J and O). Bars = 50 µm (A–J and M–O) and 100 µm (K and L). [See online article for color version of this figure.]
LNP Is Required for Nod Factor Induction of NODULE INCEPTION Expression
All three LNP antisense lines showed defects in rhizobial and mycorrhizal colonization, implying that LNP acts in the common symbiosis signaling pathway. To test this, we assessed features of symbiosis signaling, using gene induction and calcium responses to Nod factor as the measure. The fact that all three independent antisense lines showed defects in symbiosis (Table I; Fig. 3) strongly supports the fact that this must be the result of the suppression of LNP. From our observations, it appears that suppression of LNP may affect seed production and long-term viability of the seed, since both seed yield and seed germination levels for these lines appeared to be reduced, and long-term storage was affected. The 5′D line was frequently propagated, but the other two lines were not, and the seed of the 5′R and 3′O lines were no longer viable when it came to the analysis of the SYM pathway. Thus, it has only been possible to measure SYM pathway activities in the 5′D line.
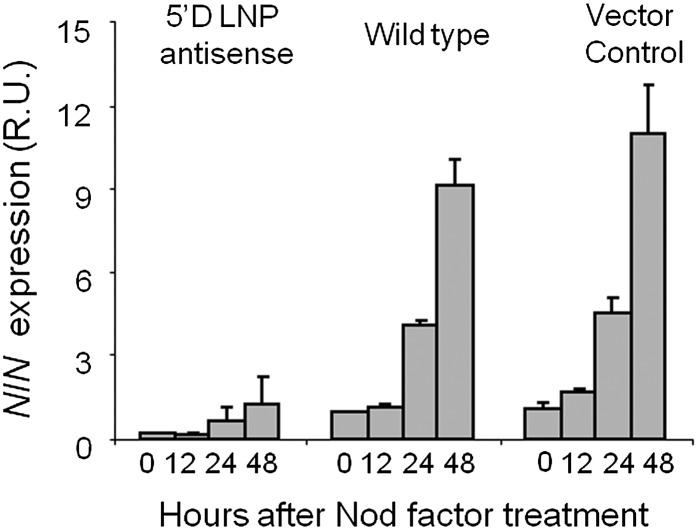
As an initial indication of the SYM pathway activity in the 5′D line, we assayed the transcription of NODULE INCEPTION (NIN), which is normally induced in L. japonicus a few hours after Nod factor treatment. The transcript abundance of this gene represents a useful indicator for the activity of Nod factor signal transduction, because mutations that block the Nod factor-induced signaling pathway in L. japonicus block NIN induction (Radutoiu et al., 2003; Imaizumi-Anraku et al., 2005; Miwa et al., 2006b; Marsh et al., 2007). NIN induction was observed in roots of wild-type plants and in the vector control line 24 and 48 h after Nod factor treatment (Fig. 4). In contrast, no significant induction of NIN transcript was seen in the 5′D LNP antisense line at any of the time points tested (Fig. 4). These data indicate that LNP is required for the full transcriptional activation of NIN and suggest that LNP functions in Nod factor signal transduction.
Figure 4.
The L. japonicus 5′D LNP antisense line does not induce NIN expression after Nod factor treatment. Quantitative RT-PCR analysis is shown for NIN expression in 5′D LNP antisense, wild-type, and vector control plants at 0, 12, 24, and 48 h after Nod factor treatment (100 nm). R.U., Relative units. POLYUBIQUITIN was used as an internal control. There were three technical replicates of three experiments for each time point. Mean values ± sd are shown.
LNP Is Required for Nod Factor-Induced Calcium Responses
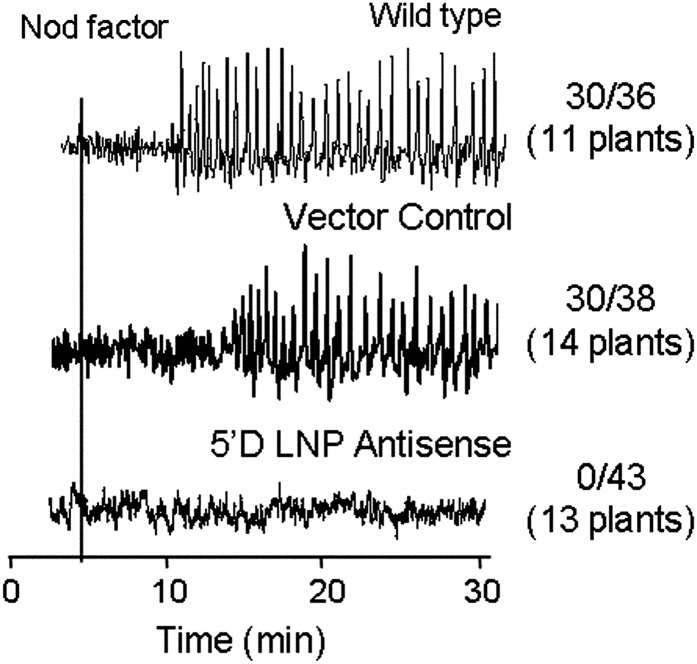
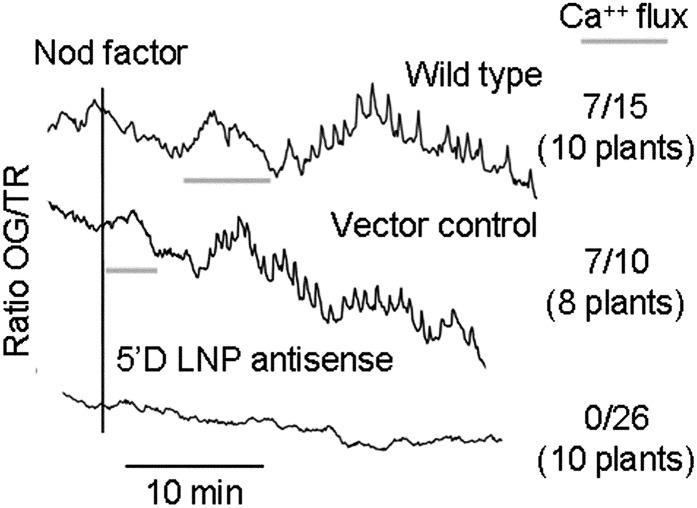
To test if LNP is required for Nod factor signaling, we measured Nod factor-induced changes in root hair calcium by dual-dye pseudoratiometric imaging following microinjection of the calcium-responsive dye Oregon Green and the reference dye Texas Red. The 5′D line was completely defective for Nod factor-induced calcium spiking: of 43 root hairs tested on 13 plants, none induced calcium spiking in response to Nod factor. In contrast, wild-type plants and the transgenic line carrying the empty vector were clearly positive for Nod factor-induced calcium spiking (Fig. 5). These data show that LNP, like NFR1, NFR5, and components of the common symbiosis signaling pathway, is required for Nod factor-induced calcium spiking.
Figure 5.
Calcium spiking is absent in the 5′D LNP antisense line. Roots of wild-type, vector control, and 5′D LNP antisense plants were treated with 100 nm M. loti Nod factor (black vertical bar). Data are presented as derivative traces representing the change in fluorescence intensity of Oregon Green/Texas Red from one point to the next (xn+1 − xn). The number of cells showing calcium spiking is shown as a fraction of the total number of cells analyzed (with the number of total plants tested in parentheses).
LNP could act as part of a Nod factor receptor (e.g. together with NFR1 and NFR5) or could be required for the common symbiosis signaling pathway. A component that discriminates between these two scenarios is a root hair tip-focused influx of calcium induced by Nod factor that appears to be independent of calcium spiking (Miwa et al., 2006a). The nfr1 and nfr5 mutants are completely defective for both Nod factor-induced calcium influx and spiking, whereas symRK, nup133, nup85, castor, and pollux mutants are defective for Nod factor-induced calcium spiking but retain the calcium flux (Miwa et al., 2006a). As the calcium influx response is localized in the tip of the root hair (Shaw and Long, 2003), we analyzed changes in intracellular calcium in the cells and in the tip of every root hair tested; only those cells that induced a significant transient increase in tip calcium compared with other cytoplasmic or nuclear regions were considered positive for the calcium influx. We observed no significant Nod factor-induced changes in cytoplasmic calcium concentration in the 5′D line (n = 26 cells from 10 plants), whereas a clear response was seen with wild-type plants and the empty vector control plants (Fig. 6). These results provide evidence that LNP is required for both Nod factor-induced calcium responses, as has been observed in nfr1 and nfr5 mutants (Miwa et al., 2006a).
Figure 6.
Nod factor-induced calcium flux is absent in the 5′D LNP antisense line. Calcium levels were monitored in individual root hairs of wild-type, vector control, and 5′D LNP antisense plants after the addition of 100 nm Nod factor (black vertical line). Ratios (arbitrary units) of the fluorescence of Oregon Green (OG; calcium-sensitive dye) and Texas Red (TX; calcium-insensitive dye) were calculated every 5 s for more than 30 min. The number of cells showing calcium influx is shown as a fraction of the total number of cells analyzed (with the total number of plants tested in parentheses).
To test the possibility that the generation of the 5′D antisense line might have caused silencing of NFR1 or NFR5 and that this could explain the observed phenotypes, we checked the expression of NFR1 and NFR5. Quantitative PCR analysis of the expression of NFR1 indicated that its expression, if anything, may be slightly increased in the 5′D antisense line (1.8-fold higher than in the vector control line), but the difference was not significantly different. NFR5 expression was also slightly increased in the 5′D antisense line (1.5-fold more than in the vector control), but again the difference was not significant. Based on these observations, we conclude that the phenotype of the 5′D antisense line is not due to an off-target effect causing silencing of NFR1 or NFR5. Since Nod factor-induced calcium influx is blocked in the 5′D antisense line, this phenotype cannot be explained by an off-target effect on any of the other known nodulation signaling genes.
LNP Is Required for Infection But Not for Root Hair Deformation
The finding that the knockdown of LNP resulted in the absence of calcium spiking, calcium influx, and NIN expression, together with the evidence showing that LNP binds Nod factor (Etzler et al., 1999), suggested that LNP could play a role in the perception of Nod factor. This would place LNP upstream of the common symbiosis signaling pathway in a position shared with NFR1 and NFR5. If LNP does indeed form a key component of Nod factor perception, the antisense line would be expected to lack all Nod factor responses. Root hair deformation is an early response to Nod factor, but it can occur in some L. japonicus mutants defective for Nod factor-induced calcium spiking (Miwa et al., 2006a). Using a root hair deformation assay with increased sensitivity (Miwa et al., 2006a), the only mutants that did not show root hair deformation responses to Nod factor were those lacking NFR1 or NFR5 (Radutoiu et al., 2003; Miwa et al., 2006a). Using this assay, the 5′D line clearly showed a significant root hair deformation (but not the shepherd’s crook structures typical of full root hair curling) in response to both M. loti inoculation and to 10 nm Nod factor (Fig. 7) and 100 nm Nod factor (P < 0.001). This result, which is somewhat different from that described previously using a less sensitive assay (Etzler and Roberts 2005), reveals that the LNP antisense line is able to perceive and respond to Nod factor, suggesting that LNP probably acts downstream of Nod factor perception by NFR1 and NFR5 or in a parallel position to these receptor-like kinases.
Figure 7.
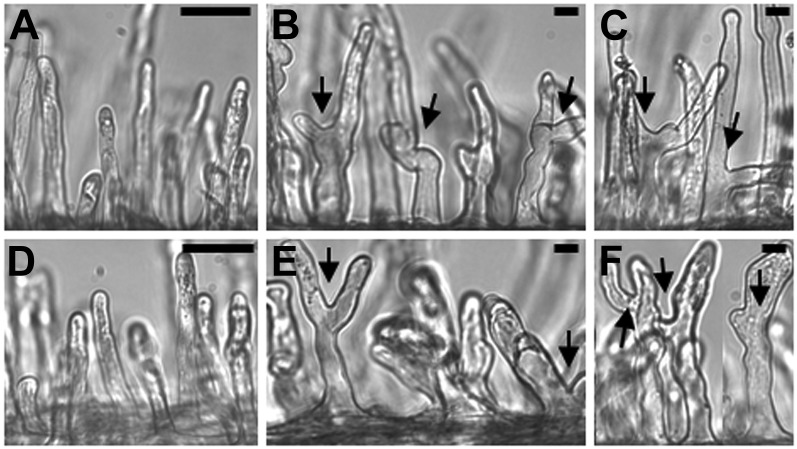
Root hair responses in vector control and 5′D antisense lines following Nod factor addition or M. loti inoculation. A to C, Vector control root hairs of uninoculated plants (A), plants treated with 10 nm Nod factor (B), and seedlings inoculated with M. loti (C). D to F, 5′D LNP antisense root hair cells of uninoculated plants (D), plants treated with 10 nm Nod factor (E), and seedlings inoculated with M. loti (F). The data on root hair deformation were analyzed using Student’s t test, and the effects of Nod factor (compared with the no-Nod factor control) were found to be significant for both the wild type (P < 0.003) and the 5′D LNP antisense line (P < 0.001). The deformation induced by M. loti was also significant (P < 0.005 for both the wild type and the 5′D LNP antisense line). Arrows indicate root hair deformation. Bars = 50 µm (A and D) and 10 µm (B, C, E, and F).
In view of the fact that we observed root hair deformation, we scored seedlings for infection foci and infection threads using lacZ-expressing M. loti by staining the roots for β-galactosidase activity. Two weeks after inoculation of wild-type plants, we detected an average of 37 ± 12 (n = 24) infection threads per seedling and 6 ± 3 (n = 24) infection foci that had not progressed to infection threads. In the 5′D antisense line, no infection threads were observed, and we observed an average of 0.5 ± 1 (n = 22) potential infection foci. In other lines in which infection is blocked, the number of infection foci is greatly increased compared with the wild type (Xie et al., 2012), so the very few potential infection foci observed suggest that even the formation of infection foci is mostly blocked.
DISCUSSION
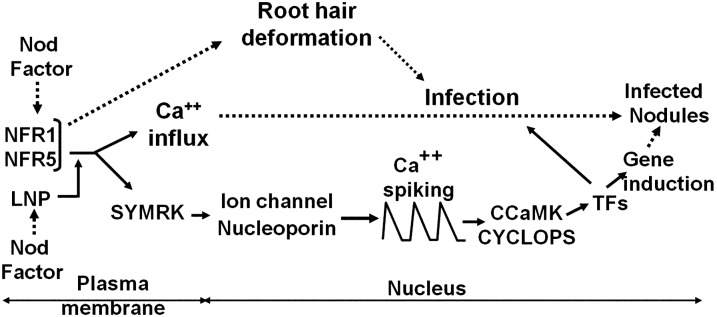
LNP was initially identified for its Nod factor-binding ability, with preferential binding to Nod factor relative to chitin (Etzler et al., 1999). The localization of LNP to legume root hair cells, its rhizobia-induced accumulation at root hair tips, the ability of LNP antiserum to interfere with nodulation (Etzler et al., 1999; Kalsi and Etzler, 2000), and the finding that silencing LNP in soybean reduces nodulation (Govindarajulu et al., 2009) all pointed to a function for LNP in nodulation signaling. In this work, we inhibited the formation of LNP and revealed that in three independent antisense lines, there was no nodulation following inoculation with M. loti. Furthermore, these three LNP antisense lines also showed defects in mycorrhizal colonization. Using one of the antisense lines, we showed that LNP is required for Nod factor-induced gene expression, calcium spiking, and calcium influx, implying a role for LNP in Nod factor signaling. Our work places LNP at a novel position, very early in the common symbiosis signaling pathway (Fig. 8).
Figure 8.
Genetic model showing the location of LNP in the nodulation signaling pathway in root hairs. Based on the lack of Nod factor-induced calcium influx in the LNP antisense line and the retention of root hair deformation by Nod factor and M. loti, LNP is proposed to act upstream of SYMRK (mutation of SYMRK did not block calcium influx) but downstream of the Nod factor receptors NFR1 and NFR5, possibly influencing their output. TFs, Transcription factors. The model is modified from that described previously (Miwa et al., 2006a); the ion channel genes are CASTOR and POLLUX, and the nucleoporin genes are NUP133, NUP8, and NENA, as described in the text. The solid lines indicate the proposed genetic pathway, and the broken lines show the induced phenotypes.
Previously characterized components of symbiosis signaling fall into four major classes: the hypothetical Nod factor receptors NFR1 and NFR5/NFP are necessary for all Nod factor responses so far measured but are not required for mycorrhization (Madsen et al., 2003; Radutoiu et al., 2003; Arrighi et al., 2006); SYMRK/DMI2, CASTOR, POLLUX/DMI1, NUP133, NUP85, and NENA are necessary for Nod factor-induced calcium spiking and gene expression as well as mycorrhization but are not required for calcium influx or root hair deformation (Wais et al., 2000; Endre et al., 2002; Stracke et al., 2002; Kanamori et al., 2006; Miwa et al., 2006a; Saito et al., 2007; Groth et al., 2010); CCaMK and CYCLOPS have similar functions but sit downstream of calcium spiking (Wais et al., 2000; Miwa et al., 2006a; Yano et al., 2008); while NSP1, NSP2, ERN, and NIN are nodulation-specific transcription factors downstream of the SYM pathway (Stracke et al., 2002; Kaló et al., 2005; Smit et al., 2005; Marsh et al., 2007; Middleton et al., 2007). From this genetics perspective, LNP represents a novel class. Like NFR1 and NFR5/NFP, LNP is required for both calcium influx and calcium spiking, but unlike NFR1 and NFR5/NFP, LNP is not required for root hair deformation and is required for mycorrhization. Such a phenotype implies a function for LNP in the common SYM pathway, but unlike LNP, components of the common SYM pathway are not required for Nod factor-induced calcium influx. Thus, the phenotype of the LNP antisense lines suggests a novel position for LNP in symbiosis signaling lying between the LysM receptor-like kinases and the common SYM pathway (Fig. 8).
NFR1 and NFR5 are Nod factor receptors based on the observations that (1) they encode receptor-like kinases with LysM domains known to function in polysaccharide binding (Madsen et al., 2003; Radutoiu et al., 2003; Arrighi et al., 2006); (2) nfr1 and nfr5 mutants show no Nod factor responses (Radutoiu et al., 2003; Miwa et al., 2006a); (3) NFR1 and NFR5 bind Nod factors with high affinity (Broghammer et al., 2012); (4) the specificity of Nod factor recognition has been shown to be a function of the LysM domains of NFR5 (Bek et al., 2010); and (5) transfer of NFR1 and NFR5 to M. truncatula transferred the specificity of the L. japonicus rhizobial interaction (Radutoiu et al., 2007). We considered the possibility that LNP might work in combination with the LysM receptor-like kinases during Nod factor perception. LNP could bind Nod factor and present it to the LysM receptor-like kinases and could even function as a complex with NFR1 and NFR5 together with Nod factor. Such a hypothesis is supported by the finding that LNP is a peripheral membrane protein (Kalsi and Etzler, 2000). However, for such a hypothesis to hold, we would have to assume that Nod factors could activate a receptor complex lacking LNP (or containing very low levels of LNP) to induce root hair deformation but not calcium spiking. Of all the Nod factor responses, root hair deformation shows the lowest specificity for the Nod factor structure (Ardourel et al., 1994; Walker and Downie, 2000), so we can surmise that root hair deformation may be activated at suboptimal levels of Nod factor perception. Thus, in this hypothesis, LNP may be required to present Nod factor to the LysM receptor-like kinases, and in its absence, Nod factor perception would be severely impaired but not eliminated. We would have to presume that LNP fulfills a comparable function in mycorrhization, and this is consistent with the recent report indicating that mycorrhizal fungi produce lipochitooligosaccharides structurally similar to Nod factors (Maillet et al., 2011).
An alternative hypothesis is that LNP is a component of the common SYM pathway. Unlike LNP, the SYM pathway is not necessary for the Nod factor-induced calcium influx response (Miwa et al., 2006a), although it has been reported that M. truncatula mutated in dmi1 or dmi2 cannot support the complete calcium influx response (Shaw and Long, 2003). Furthermore, analysis in L. japonicus mutants of SYM pathway components identified various steps of mycorrhizal colonization of roots (Novero et al., 2002; Demchenko et al., 2004; Kistner et al., 2005). In all the early L. japonicus mutants defective for arbuscular mycorrhization identified so far, fungal hyphae have been observed to enter between epidermal cells, but the intracellular penetration of epidermal or outer cell layers is blocked except in Ljsym15, in which mycorrhization is blocked at the separation of anticlinal walls of epidermal cells and arbuscule formation (Demchenko et al., 2004). In the LjsymRK mutant, fungal hyphae form appressoria and balloon-like deformations, but the subsequent fungal entry in cells of epidermis and exodermis was aborted, blocking the intracellular passage (Demchenko et al., 2004; Kistner et al., 2005). Attachment of fungal hyphae and appressorium formation were observed on the root surface of three LNP antisense lines, but intercellular or intracellular entry in the epidermal cells was not observed, implying a role for LNP in events leading to the epidermal cell entry of fungal hyphae. Thus, for both mycorrhizal signaling and Nod factor signaling, LNP appears to be acting upstream of other identified L. japonicus SYM genes. The predominant function of the SYM signaling pathway appears to be to transmit the recognition of symbiotic signaling molecules to the activation of calcium spiking and, ultimately, the transmission of this signal to transcription factors. It is difficult to ascertain the role for a second level of Nod factor binding that acts in SYM signaling but downstream of the LysM receptor-like kinases. However, one possibility is an interaction between LNP and SYMRK/DMI2, a receptor-like kinase whose function remains enigmatic.
A final scenario is one that places LNP in a parallel position to the LysM receptor-like kinases and the SYM pathway but possibly with a function required for the activation of symbiosis signaling (Fig. 8). Pertinent to this scenario is the apyrase activity of LNP. Presuming that the apyrase activity is relevant for Nod factor signaling, it is possible that either reducing the level of extracellular ATP or increasing the level of ADP or other degradation products of ATP may be important for symbiosis signaling. The mammalian extracellular apyrase NTPDase1/CD39 hydrolyzes extracellular ATP and/or ADP to AMP, which is subsequently degraded to adenosine. ATP, ADP, and adenosine serve as ligands to ligand-gated ion channel P2X receptors and G-protein-coupled P2Y and P1 receptors (Khakh and Burnstock, 2009). The addition of extracellular ADP increased legume nodulation and partially rescued the low nodule infection observed in soybean with RNAi of the expression of ectoapyrase GS52 (Govindarajulu et al., 2009), implying that ADP may be the key product of these apyrases. Alternatively, extracellular ATP may suppress symbiosis signaling. It has been suggested that extracellular ATP may act as a signal in plant-pathogen interactions (Demidchik et al., 2003), and pathogen-associated patterns such as chitin elicit increases in extracellular ATP (Kim et al., 2006). In theory, LNP could decrease ATP levels and thereby suppress plant defenses following recognition of the symbionts. However, such a role does not appear to fit well with the requirement for LNP in most aspects of symbiosis signaling. The effect of GS52 silencing in soybean (Govindarajulu et al., 2009) primarily caused the formation of uninfected nodules, and this seems more likely to be due to an effect on infection; the observation that nodule primordia are formed suggests that in the GS52 RNAi line, activation of the calcium spiking pathway can occur because this is clearly linked to nodule morphogenesis (Oldroyd and Downie, 2006). However, it is possible that the calcium influx at the root hair tip could be affected, and this has been proposed to play a role in the initiation of infection (Miwa et al., 2006a). There are two ways in which LNP might influence infection rates. First, induction of the calcium influx requires a much higher level of Nod factor than does the induction of calcium spiking (Shaw and Long, 2003, Miwa et al., 2006a), so if GS52 silencing were to cause a change in sensitivity to Nod factor, it is possible that nodule morphogenesis could be induced but infection could mostly be blocked. Second, it is possible that Nod factor activation of LNP could increase extracellular ADP levels, and extracellular ADP has been shown to induce an inwardly directed calcium flux across the plasma membrane of root epidermal cells (Demidchik et al., 2011). This fits well with the proposal that a calcium influx may promote infection by rhizobia (Miwa et al., 2006a), but it does not fit so well with the observation that the soybean GS52 apyrase shows significantly higher activity with CTP, CDP, and ADP than it does with ATP (Tanaka et al., 2011).
If a major role for LNP is in infection, as appears to be the case with GS52 in soybean, it may be significant that in legumes (pea and M. truncatula) that form indeterminate nodules, there is much greater diversification of secreted apyrases (Supplemental Fig. S1), because in indeterminate nodules, infection threads are more widely distributed and more persistent. Possibly in legumes forming indeterminate nodules, there has been selection for multiple apyrases, and this redundancy could explain why no clear nodulation phenotypes have been observed in M. truncatula. The LjLNP antisense lines analyzed here have a more severe effect than RNAi of GS52 in soybean; the resulting block of nodule morphogenesis is consistent with the lack of Nod factor-induced calcium spiking.
Whatever the role for LNP, our work highlights a novel function for a Nod factor-binding protein in both mycorrhizal and rhizobial signaling. Considering the dual role for LNP in nodulation and mycorrhization, we can presume that LNP initially evolved as a function in mycorrhizal signaling that was secondarily recruited into nodulation signaling. In this regard, it may be significant that many plants, including Poaceae and members of the Solanaceae, have ectoapyrases, but no such ectoapyrases from Brassicaceae were found within this group. This correlates with the lack of infection of Brassicaceae plants by mycorrhizal fungi and may imply a role for these ectoapyrases in mycorrhization. The work described here reveals an additional parallel mechanism in the establishment of these two symbiotic processes, implying novel commonalities in proteins that bind the microbia-derived symbiotic signals.
MATERIALS AND METHODS
Biological Materials
Lotus japonicus cv Gifu seeds were gifts from Dr. J. Webb (Institute of Grassland and Environmental Research) and Dr. K. Szczyglowski (Michigan State University-Department of Energy Plant Research Laboratory). The seeds were scarified, surface sterilized, and germinated in Hoagland medium lacking nitrate and ammonium (Hoagland and Arnold, 1950). The seedlings were grown in petri dishes containing 0.7% to 1.0% phytagar (Gibco BRL) or filter paper imbibed with the above medium for 4 to 14 d before use. Mesorhizobium loti and Bradyrhizobium sp. 24A10 (Lipha Tech) were grown in 0.5% gluconic acid, 0.1% Glu, and 0.1% yeast extract (in mineral salts vitamin base) medium (Nieuwkoop et al., 1987) and 0.5% tryptone, 0.3% yeast extract, and 6 mmol CaCl2 medium (Rosenberg et al., 1981), respectively. For gene expression, calcium, and root hair deformation analyses, seeds of L. japonicus were scarified using sand paper, soaked in 0.1% sodium hypochlorite for 15 to 20 min, washed five times in sterile water, and imbibed for 1 to 2 h in sterile water. Imbibed seeds were grown for 2 d on water agar medium in the dark and placed upside down to allow the roots to grow vertically. Subsequently, seedlings with similar root lengths (0.5–2 cm) were selected and transferred to Fahraeus nitrogen-free plant agar medium (Fahraeus, 1957). Filter paper was placed between the agar and the roots to prevent the roots growing into the agar. The roots were then covered by another filter paper to keep them moist. The petri dishes were incubated in a vertical position in a controlled environment (20°C/15°C, day/night cycles of 18/6 h), and the region of the petri dish containing the roots was covered with black plastic.
Plant Transformation
The three different LjLNP constructs described were cloned into the XbaI site of the shuttle vector pDH51 (Pietrzak et al., 1986) in the reverse orientation relative to the cauliflower mosaic virus 35S promoter and terminator. The antisense cassettes were purified from pDH51 after EcoRI digestion and ligated into the EcoRI site of the binary vector, pBIN19, which utilizes nptII as a selectable kanamycin resistance marker under the control of the Nos promoter (Bevan, 1984). Agrobacterium tumefaciens strain AGL1 (Lazo et al., 1991) was transformed with these antisense vectors and a pBIN19 vector control. The resulting strains were used to transform L. japonicus hypocotyls as described previously (Stiller et al., 1997) but with the following modifications: (1) plants were grown on regeneration medium for only 4 to 5 d; (2) no geneticin was used once the calli were placed on shoot induction medium; and (3) full-strength cefotaxime was used to control agrobacterial growth on all shoot and root media. On some lines, successful root initiation was only achieved using the higher auxin concentration described in the hairy root regeneration protocol of Stiller et al. (1997). Regenerated plants were grown under greenhouse conditions, and seeds were tested for geneticin resistance on phytagel (Sigma-Aldrich) plates containing 5 μg mL−1 geneticin, 1% Suc, and 1× Gamborg’s B5 medium (Gamborg et al., 1968), pH 5.5.
Immunoassays
Recombinant LjLNP, representing residues 1 to 414 of the mature protein (Roberts et al., 1999), was expressed in Escherichia coli using the pET-28c expression vector (Novagen). The protein was extracted from isolated inclusion bodies, further purified by ion-exchange chromatography, and used as an immunogen for the production of anti-LNP serum. Both this antiserum as well as an antiserum previously prepared against recombinant LNP from Dolichos biflorus (Etzler et al., 1999) reacted with the 46-kD LNP in immunoblots of L. japonicus roots.
Protein was extracted from L. japonicus roots, run in SDS-urea-PAGE, and subjected to immunoblot analysis as described previously (Kalsi and Etzler, 2000). Whole mounts of fixed L. japonicus roots were fixed, treated for 20 min with preimmunization serum or antiserum, washed, and then treated for 20 min with fluorescein-labeled goat anti-rabbit IgG (Sigma-Aldrich). After washing, the roots were examined with a Leica TCS NT confocal microscope (Leica, Wetzlar) using a 488-nm excitation laser line and a 520-nm barrier filter.
Nodulation Assays
Four days after germination, approximately 50 uniform seedlings from each transgenic line were transferred to pots (10 × 10 × 8 cm) containing sterile vermiculite and perlite (1:1, v/v) and placed in a growth chamber. The seedlings were grown for another 6 d and then thinned to approximately eight plants per pot to maintain uniformity. Four pots from each line were then inoculated with 50 mL per pot of a fresh overnight culture of M. loti, diluted to 105 cells mL−1 with Hoagland solution (Hoagland and Arnold, 1950) lacking nitrate and ammonium. Preliminary studies established that this dosage of rhizobia is in excess of what is required to get the maximum number of nodules per plant. An additional one pot per line was used as an uninoculated control. Three LNP− lines (5′D, 5′R, and 3′O) were retested in similar nodulation assays two more times and then tested again using 107 rhizobia mL−1. The placement of each inoculated pot was randomized in the growth chamber, and the plants were grown using a 26°C, 16-h-light (350 μmol m−2 s−1)/22°C, 8-h-dark cycle. The plants were watered daily using sterile water that was supplemented weekly with sterile Hoagland solution (Hoagland and Arnold, 1950) lacking nitrate and ammonium. Four weeks after inoculation, the plants were gently removed from the soil and washed, and the number of visible nodules was counted on each plant. The shoot and root material in each pot were separated and dried at 65°C for 2 d prior to weighing. L. japonicus wild-type plants, vector control, and 5′D LNP antisense line were grown in polystyrene K-resin pods (Phytatrey; Sigma-Aldrich) containing sterile clay granules (Seramis). Plants were grown in a growth chamber (20°C/15°C, day/night cycles of 18/6 h) and inoculated with M. loti (optical density = 0.05).
Expression Analysis
RNA was extracted from root tissue using the RNeasy Plant Mini Kit (Qiagen) according to the manufacturer’s instructions followed by DNaseI treatment (Ambion). Complementary DNA was prepared from 1 μg of RNA using the SuperScript II first-strand synthesis system for RT-PCR (Life Technologies, Invitrogen) using oligo(dT) primers according to the manufacturer’s protocol. Quantitative RT-PCR was performed using the CFX96 Real-Time System (Bio-Rad) using SYBR Green Master Mix (Sigma-Aldrich). The POLYUBIQUITIN gene was used as an internal positive control. Forward and reverse primers used were as follows: NIN, 5′-CCAAGCAGCAGTGAATGAGA-3′ and 5′-AGGAGCCCAAGTGAGTGCTA-3′; LNP, 5′-TTGGTGGGATATGGAATGGT-3′ and 5′-TGGCATTGGGTTTATTCACA-3′; LNP2, 5′-TGGGTCAAGCTTAAACGAATG-3′ and 5′-CCCTCCACCATTCCATATACC-3′; POLYUBIQUITIN, 5′-ATGCAGATCTTCGTCAAGACCTTGAC-3′ and 5′-ACCTCCCCTCAGACGAAGGA-3′ (Imaizumi-Anraku et al., 2005). In order to avoid the amplification of the antisense LNP RNA when quantifying the transcript levels, LNP primers were designed using a 3′ region that was absent from the 5′D antisense vector. Sequence data of LNP2 can be found at www.kazusa.or.jp/lotus (chr1.CM0104.100.nd). An initial denaturation step of 95°C for 4 min was followed by 40 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s. At the end of the reaction, the samples were heated at 72°C for 10 min. Results were expressed as a threshold cycle (CT) value. Gene expression was normalized to that of the reference gene, subtracting the CT value of the NIN or LNP genes from that of the POLYUBIQUITIN gene to give ΔCT. For the analysis of NIN expression, fold induction was calculated by normalizing the data for each time series to that of the untreated sample taken at 0 h. Thus, the ΔCT value for the treated sample was subtracted from the ΔCT of the untreated samples to give a ΔΔCT value. For LNP1 and LNP2 expression analysis, data were normalized to that of control vector. For the analysis of NFR1 and NFR5 expression, the primers used for amplification were as follows: for NFR1, 5-GGCCCTTTCAACACAAGATG-3′ and 5-TGTAGCCTTCGCTAGTTCCTG-3′; for NFR5, 5-TGTAACTTGCGGTTGCGCCG-3′ and 5-CTTTGACGCGCTCTGGCAACA-3′, using the POLYUBIQUITIN control as above. All quantitative PCR data were plotted as 2-ΔΔCT. For all quantitative PCR results, the data from three technical replicates of at least three biological repetitions were plotted showing sd.
Calcium Imaging
The calcium-sensitive dye Oregon Green-dextran 10,000 MW and the reference dye Texas Red-dextran 10,000 MW (Molecular Probes) were microinjected by iontophoresis into L. japonicus seedlings as described by Miwa et al. (2006a). Nod factor was added to 100 μL of liquid Fahraeus nitrogen-free plant agar medium in the chamber containing the seedling to give an estimated final concentration of 100 nm. Cells were imaged on an inverted epifluorescence microscope (model TE2000; Nikon) using a monochromator (modelOptoscan; Cairn Research) to generate specific wavelengths of light. Images were captured with a CCD camera (model ORCA-ER), and fluorescence data were analyzed using Metaflor (Molecular Devices). For calcium flux analysis, fluorescence imaged at the tip and at the shaft of the root hair were taken for each cell and compared with each other. The tip area included about 10 µm from the tip of the root hair, whereas the shaft area included part of the root hair cell protruding from the root. Only those cells showing a calcium increase in the area of the root hair cell protruding from the root as well as the root hair tip were considered positive for calcium flux. After taking a series of images, the pseudoratiometric traces were calculated by dividing Oregon Green fluorescence by that of Texas Red at each time point. Derivative traces represent the change in fluorescence intensity of Oregon Green/Texas Red from one point to the next (xn+1 − xn). Traces were generated using Microsoft Excel.
Root Hair Deformation and Infection Assays
Seedlings prepared as described previously (Miwa et al., 2006a) were assayed for root hair deformation 24 h after the addition of 10 nm Nod factor or an inoculum of freshly grown M. loti at about 105 cells mL−1. Root hairs were examined by light microscopy, and root hair deformation was scored without prior knowledge of the treatment to the seedlings. The analysis included 25 untreated seedlings, 13 treated with 10 nm Nod factor, 23 treated with 100 nm Nod factor, and 13 inoculated with M. loti. Root hair deformation was qualitatively scored on a scale of 0 to 3, and the data were analyzed using Genstat version 13 using a two-way ANOVA. Images were taken using an inverted microscope with a digital camera. Infection threads were scored as described previously using M. loti R7A marked with lacZ and stained with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside as described previously (Lombardo et al., 2006).
Mycorrhizal Colonization Assays
The ability of L. japonicus roots to be colonized by mycorrhizal fungi was tested using Glomus intraradices inocula from the International Culture Collection of Arbuscular and Vesicular-Arbuscular Mycorrhizal Fungi. The plants were grown in pots containing inoculum mixed with sterile vermiculite (1:9, v/v). Daily watering with sterile water was supplemented weekly with sterile Hoagland solution No. 2 (Hoagland and Arnold, 1950), pH 6.8, containing the standard 6 mm KNO3 and 4 mm Ca(NO3)2 but only 20 μm NH4H2PO4. The growth conditions were similar to the conditions used in the nodulation experiment. For the assay of colonization, the plants were gently removed from the soil at various time points and washed. The roots were cleared in 2.5% KOH at 90°C for 90 min, rinsed in distilled water, acidified in 1% HCl for 60 min, and stained in trypan blue at 90°C for 1 h. After destaining overnight in 50% glycerol, the roots were mounted on microscope slides in glycerol and observed with a microscope for fungal colonization. The roots were cut into approximately 1-cm segments and arranged in parallel on glass slides. The fungal colonization was analyzed by scanning across each segment once with a 2.1-mm field of view using a compound microscope. Over 100 segments were analyzed for each treatment.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phylogenetic comparison of 70 plant apyrases.
Acknowledgments
We thank Judith Murphy (University of California, Davis) for her excellent technical assistance, Dr. Vic Claassen (University of California, Davis), Dr. Eva Weigel (John Innes Centre), and Beth Thomas (International Culture Collection of Arbuscular and Vesicular-Arbuscular Mycorrhizal Fungi) for their advice with the mycorrhizae experiments, and Dr. John Harada and Dr. Biao Wu (University of California, Davis) for many helpful discussions. We also thank Dr. Bob Ludwig (University of California, San Diego) for his gift of Agrobacterium (AGL1) and Dr. Judith Webb (Institute of Grassland and Environmental Research) and Dr. K. Szczyglowski (Michigan State University) for supplying the L. japonicus seeds used in this investigation. Finally, we thank Fang Xie, Hale Tufan, Graham McGrann, Sibylle Hirsh, Jongho Sun, and Myriam Charpentier (John Innes Centre) for their scientific help, Roger Moraga (AgResearch, Ltd.) for phylogenetic analysis and advice, and Sarah Shailes (John Innes Centre) for communicating unpublished results.
Glossary
- RT
reverse transcription
- RNAi
RNA interference
- SYM
symbiosis signaling
- dpi
days post inoculation
References
- Ané JM, Kiss GB, Riely BK, Penmetsa RV, Oldroyd GE, Ayax C, Lévy J, Debellé F, Baek JM, Kalo P, et al. (2004) Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Science 303: 1364–1367 [DOI] [PubMed] [Google Scholar]
- Ardourel M, Demont N, Debellé F, Maillet F, de Billy F, Promé JC, Dénarié J, Truchet G. (1994) Rhizobium meliloti lipooligosaccharide nodulation factors: different structural requirements for bacterial entry into target root hair cells and induction of plant symbiotic developmental responses. Plant Cell 6: 1357–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrighi JF, Barre A, Ben Amor B, Bersoult A, Soriano LC, Mirabella R, de Carvalho-Niebel F, Journet EP, Ghérardi M, Huguet T, et al. (2006) The Medicago truncatula lysin [corrected] motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiol 142: 265–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bek AS, Sauer J, Thygesen MB, Duus JØ, Petersen BO, Thirup S, James E, Jensen KJ, Stougaard J, Radutoiu S. (2010) Improved characterization of nod factors and genetically based variation in LysM receptor domains identify amino acids expendable for nod factor recognition in Lotus spp. Mol Plant Microbe Interact 23: 58–66 [DOI] [PubMed] [Google Scholar]
- Bevan M. (1984) Binary Agrobacterium vectors for plant transformation. Nucleic Acids Res 12: 8711–8721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broghammer A, Krusell L, Blaise M, Sauer J, Sullivan JT, Maolanon N, Vinther M, Lorentzen A, Madsen EB, Jensen KJ, et al. (2012) Legume receptors perceive the rhizobial lipochitin oligosaccharide signal molecules by direct binding. Proc Natl Acad Sci USA 109: 13859–13864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier M, Bredemeier R, Wanner G, Takeda N, Schleiff E, Parniske M. (2008) Lotus japonicus CASTOR and POLLUX are ion channels essential for perinuclear calcium spiking in legume root endosymbiosis. Plant Cell 20: 3467–3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark G, Fraley D, Steinebrunner I, Cervantes A, Onyirimba J, Liu A, Torres J, Tang W, Kim J, Roux SJ. (2011) Extracellular nucleotides and apyrases regulate stomatal aperture in Arabidopsis. Plant Physiol 156: 1740–1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark G, Roux SJ. (2011) Apyrases, extracellular ATP and the regulation of growth. Curr Opin Plant Biol 14: 700–706 [DOI] [PubMed] [Google Scholar]
- Cohn JR, Uhm T, Ramu S, Nam YW, Kim DJ, Penmetsa RV, Wood TC, Denny RL, Young ND, Cook DR, et al. (2001) Differential regulation of a family of apyrase genes from Medicago truncatula. Plant Physiol 125: 2104–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day RB, McAlvin CB, Loh JT, Denny RL, Wood TC, Young ND, Stacey G. (2000) Differential expression of two soybean apyrases, one of which is an early nodulin. Mol Plant Microbe Interact 13: 1053–1070 [DOI] [PubMed] [Google Scholar]
- Demchenko K, Winzer T, Stougaard J, Parniske M, Pawlowski K. (2004) Distinct roles of Lotus japonicus SYMRK and SYM15 in root colonization and arbuscule formation. New Phytol 163: 381–392 [DOI] [PubMed] [Google Scholar]
- Demidchik V, Nichols C, Oliynyk M, Dark A, Glover BJ, Davies JM. (2003) Is ATP a signaling agent in plants? Plant Physiol 133: 456–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demidchik V, Shang Z, Shin R, Colaço R, Laohavisit A, Shabala S, Davies JM. (2011) Receptor-like activity evoked by extracellular ADP in Arabidopsis root epidermal plasma membrane. Plant Physiol 156: 1375–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dénarié J, Debellé F, Promé JC. (1996) Rhizobium lipo-chitooligosaccharide nodulation factors: signaling molecules mediating recognition and morphogenesis. Annu Rev Biochem 65: 503–535 [DOI] [PubMed] [Google Scholar]
- Doyle JJ. (1998) Phylogenetic perspectives on nodulation: evolving views of plants and symbiotic bacteria. Trends Plant Sci 3: 473–478 [Google Scholar]
- Endre G, Kereszt A, Kevei Z, Mihacea S, Kaló P, Kiss GB. (2002) A receptor kinase gene regulating symbiotic nodule development. Nature 417: 962–966 [DOI] [PubMed] [Google Scholar]
- Etzler ME, Kalsi G, Ewing NN, Roberts NJ, Day RB, Murphy JB. (1999) A nod factor binding lectin with apyrase activity from legume roots. Proc Natl Acad Sci USA 96: 5856–5861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etzler ME, Roberts NJ. (2005). LNP, a protein involved in the initiation of mycorrhizal infection in plants. US Patent Application No. 6,849,777
- Fahraeus G. (1957) The infection of clover root hairs by nodule bacteria studied by a simple glass slide technique. J Gen Microbiol 16: 374–381 [DOI] [PubMed] [Google Scholar]
- Gamborg OL, Miller RA, Ojima K. (1968) Nutrient requirements of suspension cultures of soybean root cells. Exp Cell Res 50: 151–158 [DOI] [PubMed] [Google Scholar]
- Govindarajulu M, Kim SY, Libault M, Berg RH, Tanaka K, Stacey G, Taylor CG. (2009) GS52 ecto-apyrase plays a critical role during soybean nodulation. Plant Physiol 149: 994–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth M, Takeda N, Perry J, Uchida H, Dräxl S, Brachmann A, Sato S, Tabata S, Kawaguchi M, Wang TL, et al. (2010) NENA, a Lotus japonicus homolog of Sec13, is required for rhizodermal infection by arbuscular mycorrhiza fungi and rhizobia but dispensable for cortical endosymbiotic development. Plant Cell 22: 2509–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoagland DR, Arnold DI, et al. (1950) The water-culture method for growing plants without soil. Calif Agric Exp Stn Circ 347: 1–39 [Google Scholar]
- Imaizumi-Anraku H, Takeda N, Charpentier M, Perry J, Miwa H, Umehara Y, Kouchi H, Murakami Y, Mulder L, Vickers K, et al. (2005) Plastid proteins crucial for symbiotic fungal and bacterial entry into plant roots. Nature 433: 527–531 [DOI] [PubMed] [Google Scholar]
- Kaló P, Gleason C, Edwards A, Marsh J, Mitra RM, Hirsch S, Jakab J, Sims S, Long SR, Rogers J, et al. (2005) Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308: 1786–1789 [DOI] [PubMed] [Google Scholar]
- Kalsi G, Etzler ME. (2000) Localization of a Nod factor-binding protein in legume roots and factors influencing its distribution and expression. Plant Physiol 124: 1039–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanamori N, Madsen LH, Radutoiu S, Frantescu M, Quistgaard EM, Miwa H, Downie JA, James EK, Felle HH, Haaning LL, et al. (2006) A nucleoporin is required for induction of Ca2+ spiking in legume nodule development and essential for rhizobial and fungal symbiosis. Proc Natl Acad Sci USA 103: 359–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakh BS, Burnstock G. (2009) The double life of ATP. Sci Amer 301: 84–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Sivaguru M, Stacey G. (2006) Extracellular ATP in plants: visualization, localization, and analysis of physiological significance in growth and signaling. Plant Physiol 142: 984–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistner C, Winzer T, Pitzschke A, Mulder L, Sato S, Kaneko T, Tabata S, Sandal N, Stougaard J, Webb KJ, et al. (2005) Seven Lotus japonicus genes required for transcriptional reprogramming of the root during fungal and bacterial symbiosis. Plant Cell 17: 2217–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosuta S, Chabaud M, Lougnon G, Gough C, Dénarié J, Barker DG, Bécard G. (2003) A diffusible factor from arbuscular mycorrhizal fungi induces symbiosis-specific MtENOD11 expression in roots of Medicago truncatula. Plant Physiol 131: 952–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosuta S, Hazledine S, Sun J, Miwa H, Morris RJ, Downie JA, Oldroyd GED. (2008) Differential and chaotic calcium signatures in the symbiosis signaling pathway of legumes. Proc Natl Acad Sci USA 105: 9823–9828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazo GR, Stein PA, Ludwig RA. (1991) A DNA transformation-competent Arabidopsis genomic library in Agrobacterium. Biotechnology (N Y) 9: 963–967 [DOI] [PubMed] [Google Scholar]
- Lévy J, Bres C, Geurts R, Chalhoub B, Kulikova O, Duc G, Journet EP, Ané JM, Lauber E, Bisseling T, et al. (2004) A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science 303: 1361–1364 [DOI] [PubMed] [Google Scholar]
- Libault M, Joshi T, Benedito VA, Xu D, Udvardi MK, Stacey G. (2009) Legume transcription factor genes: what makes legumes so special? Plant Physiol 151: 991–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limpens E, Franken C, Smit P, Willemse J, Bisseling T, Geurts R. (2003) LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science 302: 630–633 [DOI] [PubMed] [Google Scholar]
- Lombardo F, Heckmann AB, Miwa H, Perry JA, Yano K, Hayashi M, Parniske M, Wang TL, Downie JA. (2006) Identification of symbiotically defective mutants of Lotus japonicus affected in infection thread growth. Mol Plant Microbe Interact 19: 1444–1450 [DOI] [PubMed] [Google Scholar]
- Madsen EB, Madsen LH, Radutoiu S, Olbryt M, Rakwalska M, Szczyglowski K, Sato S, Kaneko T, Tabata S, Sandal N, et al. (2003) A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425: 637–640 [DOI] [PubMed] [Google Scholar]
- Maillet F, Poinsot V, André O, Puech-Pagès V, Haouy A, Gueunier M, Cromer L, Giraudet D, Formey D, Niebel A, et al. (2011) Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature 469: 58–63 [DOI] [PubMed] [Google Scholar]
- Marsh JF, Rakocevic A, Mitra RM, Brocard L, Sun J, Eschstruth A, Long SR, Schultze M, Ratet P, Oldroyd GE. (2007) Medicago truncatula NIN is essential for rhizobial-independent nodule organogenesis induced by autoactive calcium/calmodulin-dependent protein kinase. Plant Physiol 144: 324–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAlvin CB, Stacey G. (2005) Transgenic expression of the soybean apyrase in Lotus japonicus enhances nodulation. Plant Physiol 137: 1456–1462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messinese E, Mun JH, Yeun LH, Jayaraman D, Rougé P, Barre A, Lougnon G, Schornack S, Bono JJ, Cook DR, et al. (2007) A novel nuclear protein interacts with the symbiotic DMI3 calcium- and calmodulin-dependent protein kinase of Medicago truncatula. Mol Plant Microbe Interact 20: 912–921 [DOI] [PubMed] [Google Scholar]
- Middleton PH, Jakab J, Penmetsa RV, Starker CG, Doll J, Kaló P, Prabhu R, Marsh JF, Mitra RM, Kereszt A, et al. (2007) An ERF transcription factor in Medicago truncatula that is essential for Nod factor signal transduction. Plant Cell 19: 1221–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra RM, Gleason CA, Edwards A, Hadfield J, Downie JA, Oldroyd GE, Long SR. (2004) A Ca2+/calmodulin-dependent protein kinase required for symbiotic nodule development: gene identification by transcript-based cloning. Proc Natl Acad Sci USA 101: 4701–4705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa H, Sun J, Oldroyd GE, Downie JA. (2006a) Analysis of Nod-factor-induced calcium signaling in root hairs of symbiotically defective mutants of Lotus japonicus. Mol Plant Microbe Interact 19: 914–923 [DOI] [PubMed] [Google Scholar]
- Miwa H, Sun J, Oldroyd GE, Downie JA. (2006b) Analysis of calcium spiking using a cameleon calcium sensor reveals that nodulation gene expression is regulated by calcium spike number and the developmental status of the cell. Plant J 48: 883–894 [DOI] [PubMed] [Google Scholar]
- Navarro-Gochicoa MT, Camut S, Niebel A, Cullimore JV. (2003) Expression of the apyrase-like APY1 genes in roots of Medicago truncatula is induced rapidly and transiently by stress and not by Sinorhizobium meliloti or Nod factors. Plant Physiol 131: 1124–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwkoop AJ, Banfalvi Z, Deshmane N, Gerhold D, Schell MG, Sirotkin KM, Stacey G. (1987) A locus encoding host range is linked to the common nodulation genes of Bradyrhizobium japonicum. J Bacteriol 169: 2631–2638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novero M, Faccio A, Genre A, Stougaard J, Webb KJ, Mulder L, Parniske M, Bonfante P. (2002) Dual requirement of the LjSYM4 gene for mycorrhizal development in epidermal and cortical cells of Lotus japonicus roots. New Phytol 154: 741–749 [DOI] [PubMed] [Google Scholar]
- Oláh B, Brière C, Bécard G, Dénarié J, Gough C. (2005) Nod factors and a diffusible factor from arbuscular mycorrhizal fungi stimulate lateral root formation in Medicago truncatula via the DMI1/DMI2 signalling pathway. Plant J 44: 195–207 [DOI] [PubMed] [Google Scholar]
- Oldroyd GED, Downie JA. (2006) Nuclear calcium changes at the core of symbiosis signalling. Curr Opin Plant Biol 9: 351–357 [DOI] [PubMed] [Google Scholar]
- Oldroyd GED, Harrison MJ, Paszkowski U. (2009) Reprogramming plant cells for endosymbiosis. Science 324: 753–754 [DOI] [PubMed] [Google Scholar]
- Parniske M. (2008) Arbuscular mycorrhiza: the mother of plant root endosymbioses. Nat Rev Microbiol 6: 763–775 [DOI] [PubMed] [Google Scholar]
- Perret X, Staehelin C, Broughton WJ. (2000) Molecular basis of symbiotic promiscuity. Microbiol Mol Biol Rev 64: 180–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietrzak M, Shillito RD, Hohn T, Potrykus I. (1986) Expression in plants of two bacterial antibiotic resistance genes after protoplast transformation with a new plant expression vector. Nucleic Acids Res 14: 5857–5868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn JM, Etzler ME. (1987) Isolation and characterization of a lectin from the roots of Dolichos biflorus. Arch Biochem Biophys 258: 535–544 [DOI] [PubMed] [Google Scholar]
- Radutoiu S, Madsen LH, Madsen EB, Felle HH, Umehara Y, Grønlund M, Sato S, Nakamura Y, Tabata S, Sandal N, et al. et al. (2003) Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425: 585–592 [DOI] [PubMed] [Google Scholar]
- Radutoiu S, Madsen LH, Madsen EB, Jurkiewicz A, Fukai E, Quistgaard EMH, Albrektsen AS, James EK, Thirup S, Stougaard J. (2007) LysM domains mediate lipochitin-oligosaccharide recognition and Nfr genes extend the symbiotic host range. EMBO J 26: 3923–3935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts NJ, Brigham J, Wu B, Murphy JB, Volpin H, Phillips DA, Etzler ME. (1999) A Nod factor-binding lectin is a member of a distinct class of apyrases that may be unique to the legumes. Mol Gen Genet 262: 261–267 [DOI] [PubMed] [Google Scholar]
- Rosenberg C, Boistard P, Dénarié J, Casse-Delbart F. (1981) Genes controlling early and late functions in symbiosis are located on a megaplasmid in Rhizobium meliloti. Mol Gen Genet 184: 326–333 [DOI] [PubMed] [Google Scholar]
- Saito K, Yoshikawa M, Yano K, Miwa H, Uchida H, Asamizu E, Sato S, Tabata S, Imaizumi-Anraku H, Umehara Y, et al. (2007) NUCLEOPORIN85 is required for calcium spiking, fungal and bacterial symbioses, and seed production in Lotus japonicus. Plant Cell 19: 610–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S, Nakamura Y, Kaneko T, Asamizu E, Kato T, Nakao M, Sasamoto S, Watanabe A, Ono A, Kawashima K, et al. (2008) Genome structure of the legume, Lotus japonicus. DNA Res 15: 227–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw SL, Long SR. (2003) Nod factor elicits two separable calcium responses in Medicago truncatula root hair cells. Plant Physiol 131: 976–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit P, Raedts J, Portyanko V, Debellé F, Gough C, Bisseling T, Geurts R. (2005) NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 308: 1789–1791 [DOI] [PubMed] [Google Scholar]
- Steinebrunner I, Wu J, Sun Y, Corbett A, Roux SJ. (2003) Disruption of apyrases inhibits pollen germination in Arabidopsis. Plant Physiol 131: 1638–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiller J, Martirani L, Tuppale S, Chian RJ, Chiurazzi M, Gresshoff PM. (1997) High frequency transformation and regeneration of transgenic plants in the model legume Lotus japonicus. J Exp Bot 48: 1357–1365 [Google Scholar]
- Stracke S, Kistner C, Yoshida S, Mulder L, Sato S, Kaneko T, Tabata S, Sandal N, Stougaard J, Szczyglowski K, et al. (2002) A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 417: 959–962 [DOI] [PubMed] [Google Scholar]
- Tanaka K, Nguyen CT, Libault M, Cheng J, Stacey G. (2011) Enzymatic activity of the soybean ecto-apyrase GS52 is essential for stimulation of nodulation. Plant Physiol 155: 1988–1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirichine L, Imaizumi-Anraku H, Yoshida S, Murakami Y, Madsen LH, Miwa H, Nakagawa T, Sandal N, Albrektsen AS, Kawaguchi M, et al. (2006) Deregulation of a Ca2+/calmodulin-dependent kinase leads to spontaneous nodule development. Nature 441: 1153–1156 [DOI] [PubMed] [Google Scholar]
- Wais RJ, Galera C, Oldroyd G, Catoira R, Penmetsa RV, Cook D, Gough C, Denarié J, Long SR. (2000) Genetic analysis of calcium spiking responses in nodulation mutants of Medicago truncatula. Proc Natl Acad Sci USA 97: 13407–13412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker SA, Downie JA. (2000) Entry of Rhizobium leguminosarum bv. viciae into root hairs requires minimal Nod factor specificity, but subsequent infection thread growth requires nodO or nodE. Mol Plant Microbe Interact 13: 754–762 [DOI] [PubMed] [Google Scholar]
- Walker SA, Viprey V, Downie JA. (2000) Dissection of nodulation signaling using pea mutants defective for calcium spiking induced by nod factors and chitin oligomers. Proc Natl Acad Sci USA 97: 13413–13418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie F, Murray JD, Kim J, Heckmann AB, Edwards A, Oldroyd GED, Downie JA. (2012) Legume pectate lyase required for root infection by rhizobia. Proc Natl Acad Sci USA 109: 633–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano K, Yoshida S, Müller J, Singh S, Banba M, Vickers K, Markmann K, White C, Schuller B, Sato S, et al. (2008) CYCLOPS, a mediator of symbiotic intracellular accommodation. Proc Natl Acad Sci USA 105: 20540–20545 [DOI] [PMC free article] [PubMed] [Google Scholar]