Abstract
Eukaryotic algae have long been known to live in anoxic environments, but interest in their anaerobic energy metabolism has only recently gained momentum, largely due to their utility in biofuel production. Chlamydomonas reinhardtii figures remarkably in this respect, because it efficiently produces hydrogen and its genome harbors many genes for anaerobic metabolic routes. Central to anaerobic energy metabolism in many unicellular eukaryotes (protists) is pyruvate:ferredoxin oxidoreductase (PFO), which decarboxylates pyruvate and forms acetyl-coenzyme A with concomitant reduction of low-potential ferredoxins or flavodoxins. Here, we report the biochemical properties of the homodimeric PFO of C. reinhardtii expressed in Escherichia coli. Electron paramagnetic resonance spectroscopy of the recombinant enzyme (Cr-rPFO) showed three distinct [4Fe-4S] iron-sulfur clusters and a thiamine pyrophosphate radical upon reduction by pyruvate. Purified Cr-rPFO exhibits a specific decarboxylase activity of 12 µmol pyruvate min−1 mg−1 protein using benzyl viologen as electron acceptor. Despite the fact that the enzyme is very oxygen sensitive, it localizes to the chloroplast. Among the six known chloroplast ferredoxins (FDX1–FDX6) in C. reinhardtii, FDX1 and FDX2 were the most efficient electron acceptors from Cr-rPFO, with comparable apparent Km values of approximately 4 µm. As revealed by immunoblotting, anaerobic conditions that lead to the induction of CrPFO did not increase levels of either FDX1 or FDX2. FDX1, being by far the most abundant ferredoxin, is thus likely the partner of PFO in C. reinhardtii. This finding postulates a direct link between CrPFO and hydrogenase and provides new opportunities to better study and engineer hydrogen production in this protist.
Unicellular protists, including photosynthetic algae, commonly experience periods of hypoxia or anoxia as a result of low light exposure and/or biotic activities in their habitats, typically soils, fresh water, and marine water, during which their survival relies on anaerobic energy metabolism (Atteia et al., 2012; Müller et al., 2012). Surveys of genome sequences are uncovering the diversity of metabolic routes among microalgae (Atteia et al., 2006, 2012). The alga Chlamydomonas reinhardtii, a model organism for studying photosynthesis and chloroplast biogenesis (Rochaix, 2002; Eberhard et al., 2008) as well as the impact of micronutrient deficiency (Merchant et al., 2006; Page et al., 2012), has the most diverse set of anaerobic metabolic routes known, not only among algae (Atteia et al., 2006, 2012; Grossman et al., 2007, 2011) but among eukaryotes in general (Müller et al., 2012). In addition to anaerobic metabolic routes typical of plants and mammals that involve ethanol and lactate fermentations and/or Ala accumulation (de Sousa and Sodek, 2003; Bailey-Serres et al., 2012; Catalanotti et al., 2012), C. reinhardtii also has an assortment of enzymes that were long thought to be specific to anaerobic, nonphotosynthetic protists.
Pyruvate plays a central role in both aerobic and anaerobic metabolism. The metabolic fate of pyruvate is regulated in large part by the cell’s redox state. In the absence of oxygen, pyruvate can be converted into acetyl-CoA by two distinct enzymes: (1) pyruvate formate-lyase (PFL; EC 2.3.1.54) and (2) pyruvate:ferredoxin oxidoreductase (PFO [also named PFOR or PFR]; EC 1.2.7.1). PFL is a nonredox enzyme that converts pyruvate into acetyl-CoA and formate through a radical-based mechanism (Knappe and Sawers, 1990). Inactive PFL is converted to the active form by PFL-activating enzyme (EC 1.97.1.4), which introduces a free radical on the ultimate Gly residue of PFL in an S-adenosyl-l-Met- and flavodoxin-dependent reaction (Knappe et al., 1969; Wagner et al., 1992). Molecular oxygen irreversibly inactivates the radical-active form of PFL (Wagner et al., 1992). Pyruvate oxidation by PFO generates CO2, acetyl-CoA, and two electrons, which are typically delivered to a small low-redox-potential protein, either a ferredoxin (FDX) or a flavodoxin. PFO contains multiple [4Fe-4S] iron-sulfur cluster centers and thiamine pyrophosphate (TPP) as cofactors. Three types of PFO that differ in their subunit composition have been described to date. Depending on the organism, PFO can be homodimeric, heterodimeric, or multimeric (Kletzin and Adams, 1996; Hug et al., 2010). In some eukaryotes, an alternative form of PFO has been found that consists of a PFO domain fused with a NADPH-cytochrome (cyt) P450 reductase-type flavoprotein at its C terminus (Rotte et al., 2001; Lantsman et al., 2008). This pyruvate:NADP+ oxidoreductase (PNO) decarboxylates pyruvate into acetyl-CoA and requires NADP+ as electron acceptor instead of a proteinaceous electron carrier. All PFOs, with the exception of those of the Desulfovibrio spp. (Pieulle et al., 1995; Vita et al., 2008), are irreversibly inactivated by oxygen. PFL and PFO (or PNO) are crucial enzymes in the anaerobic metabolism of a great variety of prokaryotes and of a number of eukaryotes (Akhmanova et al., 1999; Horner et al., 1999; Gelius-Dietrich and Henze, 2004; Hug et al., 2010; Stairs et al., 2011). The occurrence of PFL or PFO in microalgae has been mostly inferred from genome surveys. The concomitant presence of PFL and PFO is not common among prokaryotes, and among eukaryotes, only a few microalgae, notably C. reinhardtii and the diatom Thalassiosira pseudonana (Atteia et al., 2012), appear to have both enzymes for the anaerobic conversion of pyruvate into acetyl-CoA.
In C. reinhardtii, PFL has been the subject of several studies (Atteia et al., 2006; Hemschemeier et al., 2008). PFL is dually targeted to the chloroplast and the mitochondria (Atteia et al., 2006; Terashima et al., 2010), but it is not clear whether the enzyme is functional in both cell compartments, because the intracellular localization of its activase is currently unknown. PFL is required for anaerobic metabolism in the dark, as evidenced by the production of formate from pyruvate that derives from the degradation of starch or other carbon stores (Kreuzberg, 1984; Mus et al., 2007). In the absence of PFL, fermentative metabolism is rerouted toward lactate and ethanol production (Philipps et al., 2011; Catalanotti et al., 2012). Evidence for the existence of a PFO in C. reinhardtii was first obtained from its genome sequence (Atteia et al., 2006; Grossman et al., 2007), but discrepancies between gene models are noted. Pfo transcript levels in the algal cells have been found to increase after exposure to dark anoxia (Mus et al., 2007), sulfur deprivation (Hemschemeier et al., 2008), or copper deficiency (Castruita et al., 2011). So far, physiological and biochemical studies are lacking to follow up and rationalize the transcriptomic data. Also, the intracellular localization of PFO in the photosynthetic alga is not clearly established. In parasitic anaerobic eukaryotes, PFO is located in the cytosol or the hydrogenosomes, which are hydrogen-producing mitochondria-like organelles (Müller et al., 2012). PNO is found in the mitochondria of the euglenophyte Euglena gracilis (Inui et al., 1984) and in the hydrogenosomes of the parasite Blastocystis spp. (Lantsman et al., 2008). We proposed earlier that C. reinhardtii PFO (CrPFO) is targeted to the chloroplast stroma, where its activity might be coupled to that of the iron-only hydrogenase (HYDA) via a ferredoxin under anaerobic conditions (Atteia et al., 2006). Support for its chloroplast localization came out recently from a comparative proteomic analysis of chloroplast and mitochondrial fractions of anaerobic algal cells (Terashima et al., 2010).
This study aimed at expanding our understanding of the anaerobic metabolism in C. reinhardtii and unicellular algae in general. We focused on PFO, a metalloenzyme that typically catalyzes anaerobic oxidative decarboxylation of pyruvate in a variety of bacteria and parasites but that has never been investigated in a photosynthetic eukaryote. Here, we have identified CrPFO and established its intracellular localization. The in-depth characterization of CrPFO was carried out on the purified recombinant enzyme (Cr-rPFO) that was functionally expressed in Escherichia coli. Finally, the study of the electron transfer between Cr-rPFO and different C. reinhardtii FDXs provided insight into its physiological partners. Our data on CrPFO open the way to investigate the respective roles of the two routes for anaerobic interconversion of pyruvate and acetyl-CoA.
RESULTS
The C. reinhardtii Genome Encodes a Typical Homodimeric PFO
Considering the discrepancies between the C. reinhardtii Pfo gene models available on the Joint Genome Institute (JGI) and phytozome Web portals, in particular at the 5′ and 3′ ends, reconstitution of the CrPFO coding sequence was necessary. To achieve this, we carried out a series of PCR amplifications using a C. reinhardtii complementary DNA (cDNA) library as template and different pairs of primers designed from the sequence data available on the JGI Web site. The sequences of the PCR products were assembled into a cDNA sequence of 4,338 nucleotides, which exhibits an open reading frame of 3,984 nucleotides flanked by noncoding regions of 141 bp on the 5′ end and 213 bp on the 3′ end. With a (G+C) content of 68%, the C. reinhardtii Pfo cDNA is close to the average (G+C) content of C. reinhardtii nuclear genes of about 65% (Grossman et al., 2003). The Pfo cDNA sequence relates best to the sequence on the phytozome Web portal (version 8: Cre11.g473950.t1.1), with 99% sequence identity; the differences between the two sequences lie in the absence of a 42-bp stretch in the Pfo cDNA sequence (positions 2,817–2,859 in the phytozome gene sequence) and in changes at seven nucleotide positions. The sequences found on the JGI genome portal (assemblies version 2.0, version 3.0, or version 4.0) differ by several gaps and/or insertions from the Pfo cDNA sequence determined here and are likely due to erroneous intron-exon predictions.
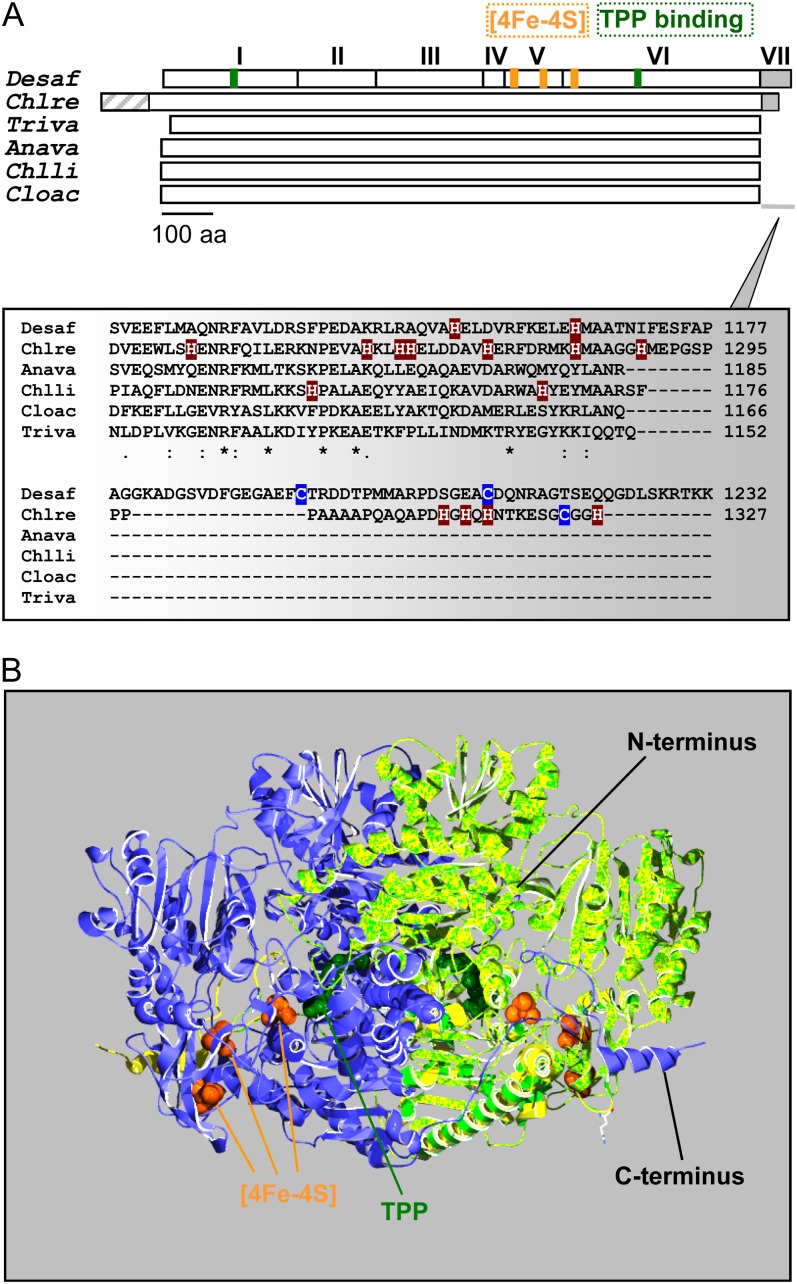
The C. reinhardtii Pfo cDNA encodes a protein of 1,327 amino acids that shows extensive sequence identity to homodimeric PFOs (Supplemental Figs. S1 and S2). CrPFO shares 52% amino acid sequence identity with its well-characterized counterparts in the heterotrophic anaerobic eukaryotes Trichomonas vaginalis and Entamoeba histolytica. The highest sequence identity with bacterial PFOs is found in deltaproteobacteria (Desulfovibrio spp.) and firmicutes (Clostridium spp., Heliobacterium spp., and Moorella spp.; 52%–54%). Homology with cyanobacterial enzymes is somewhat lower, with 42% to 45% sequence identity. CrPFO exhibits two typical ferredoxin-type sequences (Cys-X2-Cys-X2-Cys-X3-Cys-Pro) at positions 804 to 815 and 860 to 871 (Fig. 1; Supplemental Fig. S1), which are assumed to ligate the median and distal [4Fe-4S] clusters, respectively. The conserved Cys residues at positions 925, 928, 953, and 1,191 (Supplemental Fig. S2) are likely to serve as ligands for the [4Fe-4S] cluster proximal to the TPP. CrPFO also exhibits the sequence motif Gly-Gly-Asp-Gly-X3-Asp-Ile-Gly for TPP binding (Hawkins et al., 1989; Kletzin and Adams, 1996; Supplemental Fig. S1).
Figure 1.
Schematic representation and predicted structure of C. reinhardtii PFO. A, Top, the monomer of the homodimeric DaPFO has been described as being composed of seven domains (I–VII). Domains I, II, and IV are involved in dimer formation, and domain VII confers oxygen stability to the enzyme via disulfide bond formation (Pieulle et al., 1995; Chabrière et al., 1999). Bottom, as compared with bacterial and eukaryotic PFOs, the algal enzyme exhibits an extension at its N terminus that likely serves as an intracellular targeting peptide. The C-terminal extension of CrPFO differs from that of DaPFO in both length and amino acid (aa) composition, in particular in their contents of His and Cys residues. Anava, Anabaena variabilis (cyanobacterium); Chlli, Chlorobium limicola (chlorobia); Chlre, C. reinhardtii (photosynthetic eukaryote); Cloac, C. acetobutylicum (firmicute); Desaf, D. africanus (deltaproteobacterium); Triva, T. vaginalis (heterotrophic eukaryote). B, Overlay of the predicted structure of a CrPFO monomer (light green; Pro-96–His-1292) with the use of the DaPFO monomer (yellow) as a template shown in interaction with another DaPFO monomer (blue) to form the typical PFO homodimer. The 2.3-Å crystal structure of DaPFO has revealed the arrangement of the three [4Fe-4S] clusters (orange) and the TPP cofactor (dark green) within the enzyme (Chabrière et al., 1999; Charon et al., 1999). TPP is deeply buried in the protein, and its closest cluster (or proximal cluster) is approximately 13 Å away. The other two clusters are arranged successively up to the surface of the dimer, with cluster-to-cluster distances of about 12 to 15 Å. In DaPFO, the C terminus (domain VII) extends over the other monomer (Chabrière et al., 1999).
Compared with bacterial and parasitic eukaryote PFOs, the CrPFO exhibits extensions at its N terminus (approximately 95 residues) and C terminus (approximately 36 residues; Fig. 1; Supplemental Fig. S2). The N-terminal extension is suggestive of organelle targeting; however, it contains a stretch of nine Thr residues, which is uncommon among chloroplast- or mitochondria-targeting peptides (Tardif et al., 2012). The C-terminal extension comprises a hydrophobic region rich in Pro and Ala residues followed by a hydrophilic region that contains five His residues (Fig. 1A). This extension is clearly distinct from that found in the enzymes of Desulfovibrio spp. (domain VII) and that protects the proximal [4Fe-4S] cluster from oxygen via disulfide bridge formation (Cys-1195 and Cys-1212; Fig. 1; Chabrière et al., 1999; Vita et al., 2008). Because the C terminus of CrPFO lacks the pair of Cys residues, it is unlikely to protect the enzyme from oxidative damage. Modeling CrPFO (lacking the first 95 residues and the last 36 residues) on the three-dimensional structure of the Desulfovibrio africanus PFO (DaPFO) was possible due to a high similarity between the two sequences (approximately 52% amino acid identity), and as observed in Figure 1B, overlay was almost complete.
CrPFO Is a Soluble Chloroplast Protein
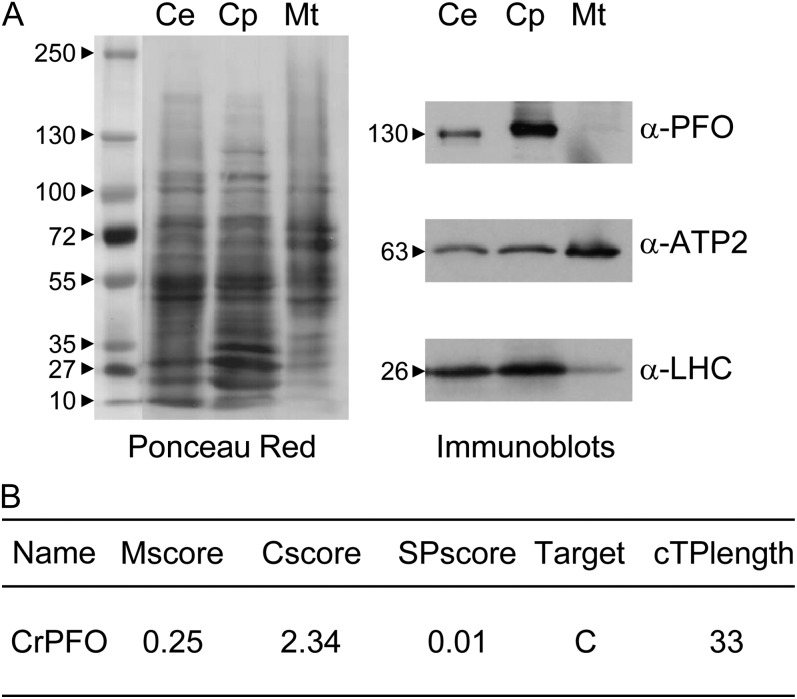
The subcellular localization of CrPFO was investigated by immunoblot analysis, thereby circumventing possible oxygen sensitivity problems in activity measurements. Chloroplasts and mitochondria were isolated from C. reinhardtii cells kept for a few hours under dark anoxic conditions and further purified by density gradient centrifugation following a standard protocol (van Lis et al., 2005). The relative purity of the organelle fractions was assessed by immunoblotting using antibodies against the proteins of the chloroplast light-harvesting complex (LHC) and the β-subunit of the mitochondrial F0F1-ATPase (ATP2). As shown in Figure 2A, mitochondrial fractions were virtually devoid of chloroplast contamination, whereas chloroplasts were slightly contaminated by mitochondria, a frequent situation (Atteia et al., 2006; Page et al., 2012). PFO antiserum detected CrPFO solely in the chloroplast fraction with an apparent molecular mass of 130 kD (Fig. 2A). The cellular CrPFO concentration was estimated from different protein blots to be in the range of 300 ng mg−1 total cell protein.
Figure 2.
Subcellular localization of PFO in C. reinhardtii. A, Analysis of chloroplasts (Cp) and mitochondria (Mt) isolated from cells (strain 10-6C) incubated in anaerobic TAP medium in the dark (Ce). Proteins (30 µg) were separated by urea/SDS-PAGE and transferred onto a nitrocellulose membrane. The nitrocellulose membrane was then stained with Ponceau red. Immunoblots show the distribution of LHCs, ATP2, and CrPFO in whole cells and isolated organelles. B, CrPFO intracellular sorting as predicted by PredAlgo (Tardif et al., 2012).
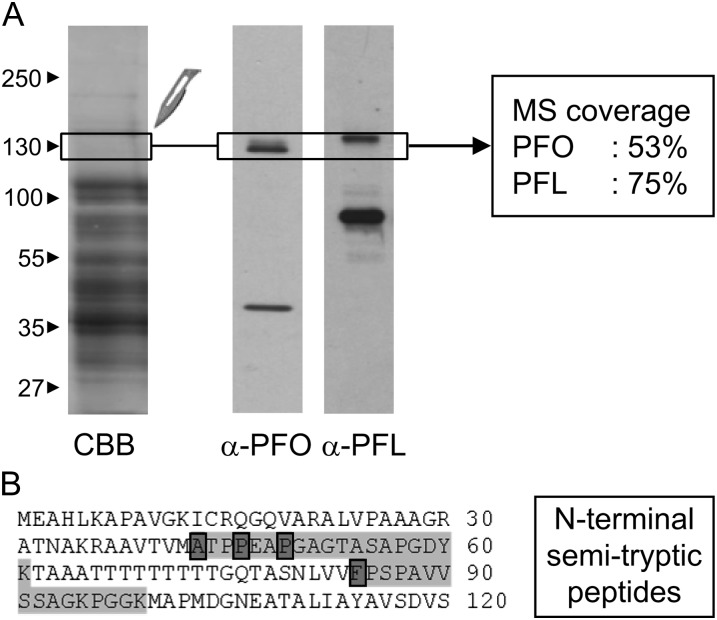
C. reinhardtii chloroplasts were broken by two cycles of freezing/thawing and fractionated by ultracentrifugation. CrPFO, present in the soluble fraction, was further purified by anion-exchange chromatography (all steps under a nitrogen atmosphere). The resultant CrPFO-enriched fraction was loaded on SDS-PAGE, and a gel band covering the 130-kD region was analyzed by mass spectrometry (MS; Fig. 3A). CrPFO was identified with a tryptic peptide coverage over 50% (Supplemental Table S2). In the same analysis, PFL was also identified (Supplemental Table S2). Immunoblot with a PFL antiserum showed that the protein at a molecular mass of approximately 130 kD represented a minor fraction of the total PFL present in this sample, which migrates at approximately 75 kD (Fig. 3A). The 130-kD PFL likely corresponds to the undissociated dimer.
Figure 3.
Identification of chloroplast CrPFO by MS. A, Proteins in a chloroplast fraction enriched in CrPFO were separated by urea/SDS-PAGE. Proteins were either stained with Coomassie brilliant blue (CBB) or transferred to nitrocellulose for immunoblot analysis with antibodies against PFO or PFL. The gel piece in which tryptic peptides for CrPFO and PFL were identified is indicated. B, CrPFO N-terminal sequence with identified semitryptic peptides. The N-terminal residue of each of the identified semitryptic peptides is boxed in dark gray. The complete list of (semi)tryptic peptides is given in Supplemental Table S2.
MS data allowed us to solve a few issues related to the divergent protein sequences predicted from gene models and the Pfo cDNA. The seven nucleotides differing in the Pfo sequence in the phytozome database and the Pfo cDNA sequence (see above) translate into four amino acid changes (D390N, E745K, F849L, and T894A; positions in CrPFO), none of which are in conserved positions or anywhere near the cofactor-binding sites (Supplemental Fig. S2). MS analysis of CrPFO identified two peptides (VIVLMGSGSAAVEEAVNYLNAQGK and LDIPASWSSLPTHVVNPNPPAK) that support the phytozome sequence(Supplemental Fig. S3). It is unclear whether these two divergent positions in the Pfo cDNA sequence result from variations in the nucleotide sequences between the strains used for cDNA synthesis and genome sequencing or from errors introduced by the DNA polymerase during nucleic acid amplification. In the 130-kD gel band, the tryptic peptide 911-GSQLQPPLMEFSGACEGCGETPYVK-935 was also identified. This peptide matches the sequences derived from the Pfo cDNA and the phytozome sequences but not that from the JGI genome portal (Supplemental Fig. S3). In the latter sequence, the last two residues (VK) are replaced by seven residues (WVLFCAR). Following the tryptic peptide indicated above, the phytozome sequence shows, as compared to the Pfo cDNA sequence, an insertion of 14 residues that shares seven residues with the JGI sequence. While these insertions may be due to an inaccurate prediction for intron/exon (exon 8), it cannot be excluded that different isoforms of CrPFO exist. Lastly, the presence of the C-terminal extension in the native enzyme (Fig. 1B) was confirmed through the identification of the tryptic peptide 1,286-HMAAGGHMEPGSPPPPAAAAPQAQAPDHGHQHNTK-1,331 (Supplemental Fig. S3).
PredAlgo, a newly released targeting prediction software dedicated to green algae (Tardif et al., 2012), gave a convincing score of 2.34 for CrPFO being targeted to the chloroplast, with a predicted chloroplast-targeting peptide length of 33 amino acids (Fig. 2B). A search for peptides with a nontryptic N-terminal site (“semitryptic peptides”) identified four peptides that match the N-terminal part of CrPFO, indicating four candidate positions for the mature N terminus of the algal enzyme (Ala-43, Pro-47, Pro-49, and Phe-84; Fig. 3B). Several hypotheses could explain the identification of multiple N-terminal peptides: (1) cleavage of the CrPFO chloroplast-targeting peptide occurs at more than one position, as described for C. reinhardtii ATP2, with mature N termini differing by one amino acid (Tardif et al., 2012); (2) processing of CrPFO proceeds through sequential steps; or (3) specific and partial proteolytic events at the N-terminal end. To discriminate between these hypotheses, further studies on the native enzyme will be needed. Of note, two proteins close together on SDS-PAGE are often immunodetected (Fig. 3A); these proteins might either be isoforms of CrPFO or enzymes with distinct N termini.
In the CrPFO-enriched fraction from anion-exchange chromatography, oxidative pyruvate decarboxylation was negligible. Rather than drastically upscaling C. reinhardtii cultures and working on the purification procedure to try to improve the yield and purity of CrPFO (and remove PFL, in particular), it appeared more practical to produce the enzyme in a heterologous system to obtain sufficient amounts of protein for enzymatic and spectroscopic characterization and shorten the purification procedure.
Expression of CrPFO in E. coli and Purification of the Recombinant Enzyme
No singular N terminus for mature C. reinhardtii PFO could be inferred from the MS data. With the objective of expressing CrPFO, we thus chose as a starting residue Pro-96, which corresponds to the N terminus of prokaryotic PFOs (Supplemental Fig. S2). Specific primers were designed to amplify the region coding for Pro-96 to His-1327. For as yet unclear reasons, amplification of that complete region using the cDNA library as template did not work. This DNA region, however, could be amplified in two parts that were then cloned into the expression vector pET24a (for details, see “Materials and Methods”). C. reinhardtii C-terminal 6-His-tagged recombinant PFO (Cr-rPFO) was produced in E. coli grown aerobically at 37°C, although at low levels (less than 1 mg protein L−1 culture). Expression under anaerobic conditions led to negligible amounts of expressed Cr-rPFO and was thus abandoned. Attempts to grow the cells at lower temperature (25°C) resulted in the degradation of Cr-rPFO with concomitant cell death, probably because this redox enzyme interferes with the metabolism of the bacterial host by competing for pyruvate. Either this effect is more problematic for the E. coli cells at lower temperatures or it is caused by a higher expression of Cr-rPFO, leading to a proteolytic response. Expression of C. reinhardtii recombinant PFO with an N-terminal 6-His tag led to a higher level of protein (by a factor 3), which, however, was mainly insoluble.
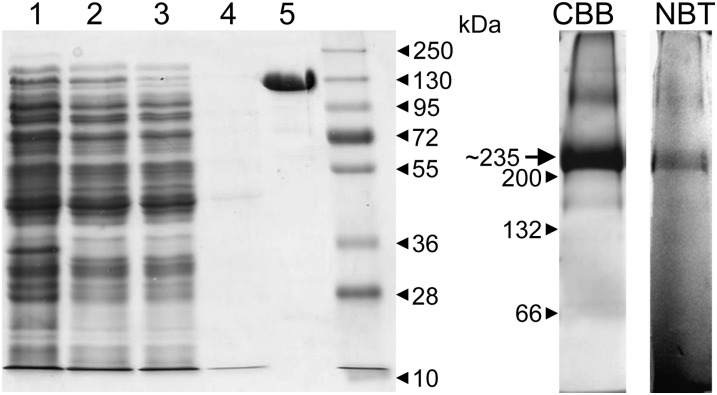
Purification of Cr-rPFO was done under a nitrogen atmosphere in a glove box. Key steps of the purification protocol were (1) cell breakage by two freeze/thaw cycles and (2) incubation with the nickel-affinity gel in the presence of 30 mm imidazole to reduce unspecific interactions. The recombinant protein showed a higher than usual affinity for the particular nickel resin used (HIS-SelectNickel Affinity Gel), as it still binds in the presence of 30 mm imidazole. A likely explanation for this strong affinity is that CrPFO itself interacts with the resin via a few His residues at its C terminus, located at the surface of the dimeric enzyme (Fig. 1). As judged by SDS-PAGE, purified Cr-rPFO was essentially homogeneous (Fig. 4, lane 5), and SDS-PAGE analysis of several preparations revealed negligible contamination. Cr-rPFO has a molecular mass of 130 kD (Fig. 4), equal to that of the native enzyme (Fig. 2). On blue native-PAGE, run in the glove box, Cr-rPFO migrated as a main band with an estimated molecular mass of approximately 235 kD (Fig. 4, lane CBB), consistent with the molecular mass of the dimer. This same band was able to reduce nitroblue tetrazolium in the presence of pyruvate and CoA (Fig. 4, lane NBT).
Figure 4.
Purification of Cr-rPFO on HIS-SelectNickel Affinity Gel. Left panel, SDS-PAGE (10% acrylamide gel) analysis of the purification of Cr-rPFO from E. coli cells. The gel was stained with Coomassie Brilliant Blue. Lane 1, E. coli cells expressing Cr-rPFO; lane 2, E. coli soluble fraction; lane 3, proteins that do not bind on the HIS-Select resin; lane 4, wash with 30 mm imidazole; lane 5, Cr-rPFO elution with 100 mm imidazole (6 µg). Right panel, Cr-rPFO (40 µg) analyzed by blue native-PAGE (5%–15% acrylamide gel) run under an N2 atmosphere. CBB, Gel lane stained with Coomassie Brilliant Blue; NBT, gel lane stained for PFO activity in an assay mixture containing 10 mm nitroblue tetrazolium as electron acceptor.
Assessment of Redox Cofactors
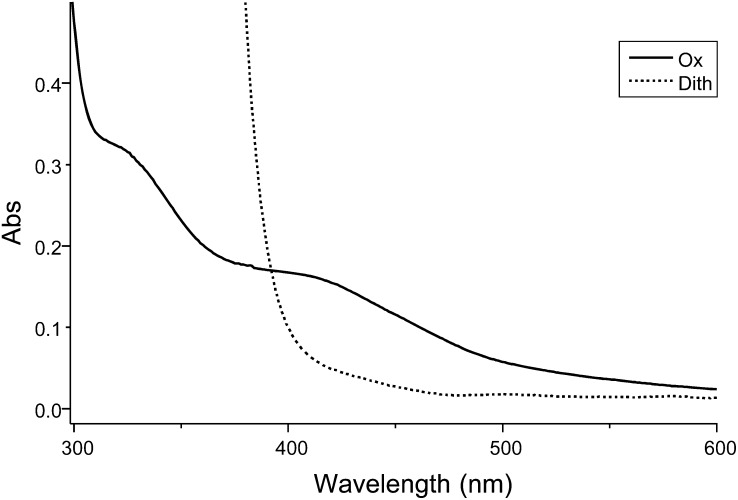
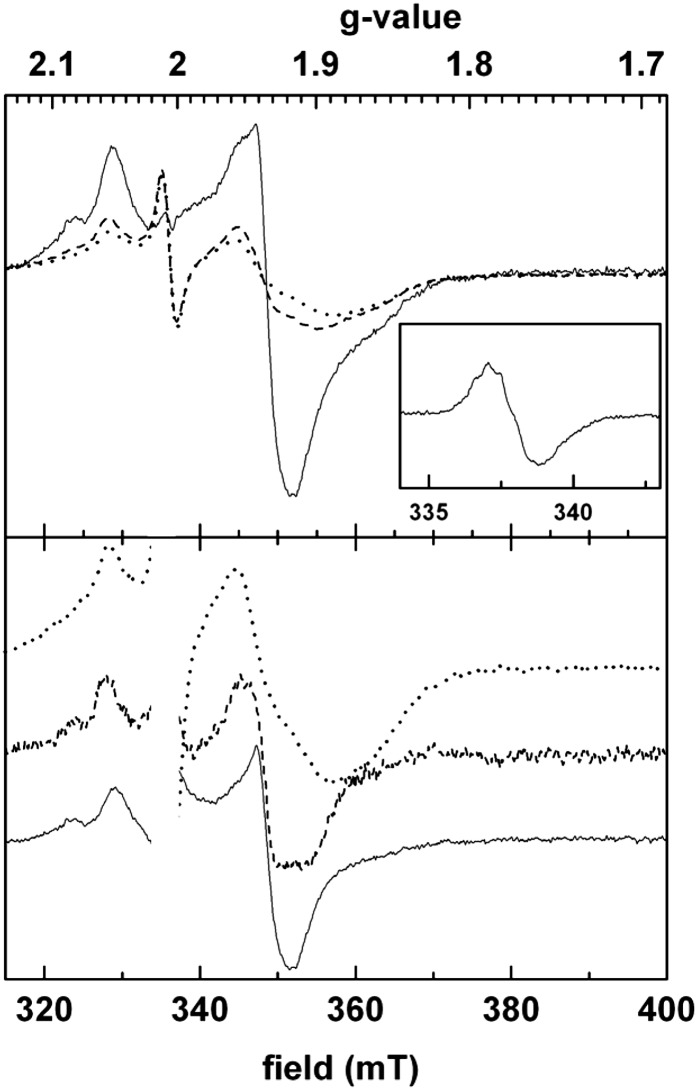
From the sequence analysis, it is inferred that all of the Cys residues ligating the three cubane [4Fe-4S] clusters in the D. africanus structure (Pieulle et al., 1997; Chabrière et al., 1999) are conserved in the PFO of the green alga (Fig. 1; Supplemental Fig. S2). Iron contents of 8 to 11 mol iron mol−1 monomeric Cr-rPFO were found by inductively coupled plasma-MS. This iron content is somewhat lower than expected (12 mol iron mol−1 Cr-rPFO monomer), likely because of an incomplete maturation of the Cr-rPFO and/or a partial degradation of the iron-sulfur clusters due to trace oxygen. The UV/visible spectrum of Cr-rPFO shows features characteristic of iron-sulfur clusters, with a broad absorption band around 420 nm that decreases upon dithionite addition (Fig. 5). To further confirm the identification of dithionite-reducible [4Fe-4S] clusters, we performed electron paramagnetic resonance (EPR) spectroscopy on the anaerobically purified Cr-rPFO. Figure 6 (top panel) shows corresponding spectra recorded on samples treated with subequimolar amounts of dithionite (dotted line), excess amounts of dithionite (dashed line), and reduced by the addition of its substrates pyruvate and CoA (solid line). The bottom panel represents normalized difference spectra together with the least-reduced spectrum recorded in the presence of low dithionite concentration. All three spectra are substantially distinct from each other and thus provide evidence for the presence of three distinct [4Fe-4S] clusters. When Cr-rPFO was incubated in the presence of pyruvate but in the absence of CoA, the EPR feature of the TPP radical (hyperfine structure) was observed (Fig. 6, inset), which very much resembles that reported for native PFO from D. africanus (Pieulle et al., 1995) and Moorella thermoacetica (Menon and Ragsdale, 1997).
Figure 5.
UV/visible spectra of purified Cr-rPFO. The spectra were recorded in an anaerobic cuvette containing 10 µm enzyme in 100 mm Gly buffer at pH 10.5 in order to ensure full reduction by dithionite. Solid line, Cr-rPFO as purified (Ox); dotted line, Cr-rPFO after reduction by dithionite (Dith).
Figure 6.
Selected EPR spectra of Cr-rPFO. Top panel, spectra recorded on only slightly reduced enzyme (dotted line), on a sample that was treated with an excess of dithionite (dashed line), and on enzyme reduced by its substrates pyruvate and CoA. Bottom panel, redox difference spectra (continuous line, substrate reduced minus excess dithionite reduced; dashed line, excess dithionite reduced minus weakly reduced) normalized to approximately equivalent signal amplitudes to facilitate comparison. The dotted line corresponds to the spectrum of weakly reduced enzyme as in the top panel. The spectra show values given to characterize paramagnetic species (gz, gy, and gx lines) in the spectral range typical for iron-sulfur clusters: that is, around 2.05 (peaks), 1.92 to 1.94 (derivatives), and 1.85 to 1.92 (troughs). The pronounced shift of the gx troughs and, to a lesser extent, the shifts of the gy lines indicate the presence of at least three different spectral species. The satellite gz peaks at 2.08 in the higher reduction states are most probably due to paramagnetic interaction between adjacent clusters. Inset, TPP radical signal. Cr-rPFO (20 µm) in 50 mm Tricine, pH 8.0, 0.1 mm TPP, and 0.1 mm MgCl2 was reduced by 10 mm pyruvate (5 min at room temperature). Instrument settings were as follows: temperature, 15 K; microwave frequency, 9.44 GHz; microwave power, 6.3 mW; modulation amplitude, 1.6 millitesla (mT).
Pyruvate Decarboxylase Activity of Cr-rPFO
The ability of Cr-rPFO to catalyze the oxidative decarboxylation of pyruvate was assessed spectrophotometrically following the reduction of different electron acceptors (Table I) The best activities were obtained with the artificial acceptor benzyl viologen (BV; midpoint redox potential = −375 mV), with an average specific activity of 12 µmol pyruvate min−1 mg−1 protein. The specific activities with methyl viologen (−450 mV) and with methylene blue (+10 mV) were, respectively, three to four times and 2.5 to three times lower than that with BV. Neither NAD+ nor NADP+ could be reduced by Cr-rPFO (Table I). As stated above, ferredoxin is a typical physiological electron acceptor of homodimeric PFOs. Therefore, as a first assessment, petF from C. reinhardtii (FDX1) and spinach (Spinacia oleracea; Fd1) were tested. Ferredoxin reduction by Cr-rPFO was followed in an indirect assay that consists of following the reduction of horse heart cyt c by PFO-reduced ferredoxins (Pieulle et al., 2004). Pyruvate decarboxylation in the presence of these [2Fe-2S] ferredoxins was achieved with rates about one-half those measured with BV, while direct reduction of cyt c by Cr-rPFO was marginal (Table I).
Table I. Electron acceptor specificity of Cr-rPFO.
The assay contained 25 mm potassium phosphate buffer (pH 7.0), 0.1 mm CoA, 5 mm pyruvate, 0.2 mm TPP, and 1 mm MgCl2 plus 2 to 4 µg of Cr-rPFO per mL. Measurements were carried out at 25°C.
Apparent Km values for the natural PFO substrates CoA and pyruvate were estimated to be 0.16 mm for pyruvate and 5.2 µm for CoA (Table II), both being of the same order of magnitude as for other characterized PFOs.
Table II. Apparent Km values for CoA and pyruvate.
The Km values were determined using 1 mm BV as electron acceptor. The Km for pyruvate was measured with 10 µm CoA; the Km for CoA was measured with 5 mm pyruvate.
| Substrate | Km |
|---|---|
| µm | |
| CoA | 5.2 ± 1.3 |
| Pyruvate | 160 ± 3.4 |
TPP is an essential cofactor for the PFO decarboxylase activity. Its required addition in the reaction mixture (Table III) indicates that this cofactor was lost during the purification of Cr-rPFO. To observe the stimulating effect of TPP, preincubation was not required. Upon addition of Mg2+, an ion that stabilizes TPP, the enzymatic activity almost doubled (Table III). The addition of 0.3 m NaCl to the standard assay mixture caused a significant inhibition (up to 80%) of BV reduction by Cr-rPFO. The presence of NaCl during the purification allowed obtaining pure Cr-rPFO in a single step, and as a consequence, the exposure of the enzyme to this salt had to remain as short as possible. The enzyme activity was unaffected by imidazole even at concentrations up to 100 mm. Dithiothreitol (DTT) was found to have no influence on the activity catalyzed by the recombinant enzyme (Table III), in contrast to the stimulative effect observed on the activity of PFOs from Desulfovibrio spp. (Vita et al., 2008). When Cr-rPFO (1.5 mg mL−1 in 50 mm Tricine, pH 8.0) was exposed to air for 1 h on ice, a drastic loss of activity was observed (Table III), indicating that the algal enzyme is oxygen sensitive, as most PFOs are. The pH optimum of Cr-rPFO in 25 mm potassium phosphate was at pH 7.0 to 7.5, being comparable to that reported for the homodimeric PFOs of the methanogenic archaeon Methanosarcina barkeri (Bock et al., 1996) and Clostridium acetobutylicum (Meinecke et al., 1989) but quite different from the pH optimum of 9 reported for DaPFO (Pieulle et al., 1995; Cavazza et al., 2006). The temperature curve maximum was at 44°C (data not shown). Generally, even under a nitrogen atmosphere, the enzyme was found to be sensitive to desalting, dilution, and concentration steps, resulting in variable loss of protein and specific activity.
Table III. Effect of diverse constituents of the assay on Cr-rPFO decarboxylase activity.
Identification of the Physiological Partner(s) of CrPFO
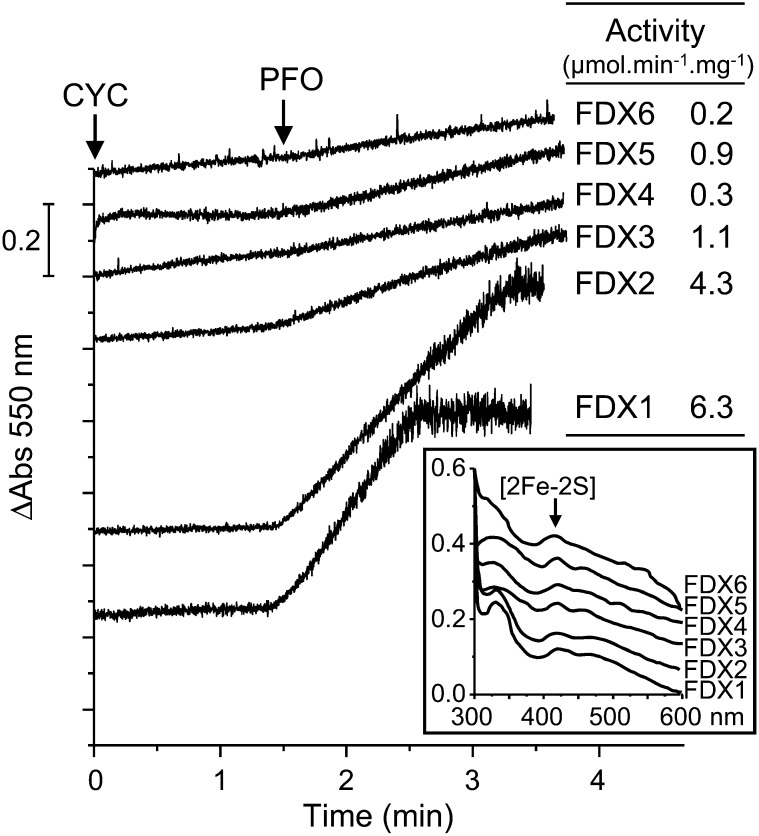
The distal [4Fe-4S] cluster near the surface of the dimer (Fig. 1B) delivers electrons to an acceptor, typically a ferredoxin or a flavodoxin. The localization of CrPFO to the chloroplasts (see above) allowed us to constrain the investigation of its physiological acceptor(s). Of the various ferredoxins encoded by the alga, FDX1 to FDX6 (all of the [2Fe-2S] type) localize to the chloroplast (Jacobs et al., 2009; Terauchi et al., 2009). No genetic or biochemical evidence exists for the presence of flavodoxin(s) in C. reinhardtii. The ability of Cr-rPFO to reduce each of the six FDXs, produced in E. coli and purified, was tested in an indirect assay using cyt c, as described above. The concentrations of holoFDXs were adjusted in the reaction mixture (Fig. 7, inset). Our data showed that FDX reduction by Cr-rPFO was far faster with FDX1 and FDX2 than with the other four (Fig. 7). The reduction rate was low with FDX3 and FDX5 and nearly undetectable with FDX4 and FDX6 (Fig. 7). In independent measurements, it was confirmed that the electron transfer from reduced FDXs to cyt c was likely not a limiting factor in the observed reduction rates (Supplemental Table S3). Apparent Km values for FDX1 and FDX2 were determined to be in the same range (approximately 4 µm; Table IV).
Figure 7.
Efficiency of the different chloroplast FDXs as electron acceptors from Cr-rPFO. PFO decarboxylase activity was assayed in the presence of 20 µm each FDX, and their reduction rate was measured as cyt c reduction at 550 nm. The values for the FDXs were corrected for direct reduction of cyt c by Cr-rPFO. Inset, FDX contents were evaluated by the absorption signal at 420 nm of the oxidized [2Fe-2S] cluster.
Table IV. Apparent Km values for FDX1 and FDX2.
The Km was determined using 50 µm cyt c in a coupled assay with ferredoxin.
| Electron Acceptor | Km |
|---|---|
| µm | |
| FDX1 | 4.6 ± 0.6 |
| FDX2 | 3.7 ± 0.9 |
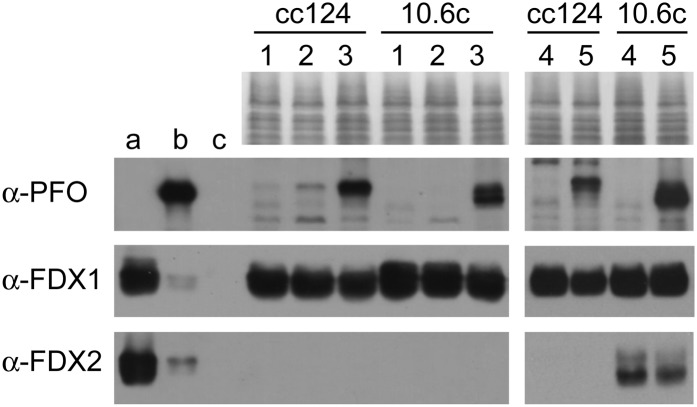
To gain insights into the significance of FDX1 and FDX2 as physiological partners of CrPFO, their intracellular levels were followed by immunoblotting. Protein levels were assessed in wild-type strain cc124 and mutant strain 10-6C, which lacks Rubisco activity (Spreitzer and Mets, 1980). For PFO induction, the cells were maintained for a few hours in dark anoxia, either in Tris-acetate-phosphate (TAP) medium or in anaerobic induction buffer (AIB), a reference medium for anaerobic studies on the green alga (Mus et al., 2007; Magneschi et al., 2012). As observed in Figure 8, the highest levels of CrPFO were immunodetected in both cells when incubated in anaerobic TAP medium (lanes 3). In AIB, CrPFO was found in trace amounts in cc124 cells but was not detectable in 10-6C cells (Fig. 8, lanes 2). FDX1 levels were high under all conditions, although slightly diminished in cells accumulating CrPFO (Fig. 8, lanes 3). By contrast, FDX2 was not detected in any of these samples, which is in line with its reported function being specific to nitrate metabolism (Terauchi et al., 2009). The algal cells were also grown in the presence of nitrate instead of ammonium and then incubated in anaerobic TAP medium (containing nitrate) to induce the expression of CrPFO (Fig. 8, lanes 5). In strain 10-6C, FDX2 levels were elevated in aerated cultures (Fig. 8, lane 4) but slightly lower in cells after anaerobic incubation (Fig. 8, lane 5). The situation with strain cc124 was different, as FDX2 was detected neither in aerobic nor in anaerobic conditions (Fig. 8, lanes 4 and 5). While these observations suggest a deviant nitrate metabolism in cc124, the fact that CrPFO is induced without concomitant FDX2 accumulation, or accompanies the lowering of existing FDX2 pools, made clear that in the alga FDX2 is not dedicated to CrPFO. Taken together, our data indicate no obvious correlation between the accumulation of FDX1 or FDX2 and CrPFO. Because FDX1 is (by far) the most abundant isoform in C. reinhardtii, it is highly likely that this ferredoxin is the physiological partner of CrPFO.
Figure 8.
Cellular levels of CrPFO, FDX1, and FDX2 under different conditions. Proteins in extracts from strains cc124 and 10-6C (25 µg) were separated by SDS-PAGE and transferred to nitrocellulose membrane for immunoblot analysis with α-PFO, α-FDX1, and α-FDX2. Lane 1, TAP light aerobic; lane 2, dark anaerobiosis in AIB medium; lane 3, dark anaerobiosis in TAP medium; lane 4, TAP-NO3− light aerobic; 5, dark anaerobiosis in nitrate containing TAP medium. a, b, and c for FDXs are 200, 40, and 5 ng of control ferredoxin; b for PFO is 40 ng of control Cr-rPFO protein. Immunodetection limits for recombinant proteins are 40 ng for both FDX1 and FDX2 and 5 ng for CrPFO (data not shown).
DISCUSSION
PFOs are found in all three domains of life (Kletzin and Adams, 1996; Müller et al., 2012). In eukaryotes, PFO activity was first discovered in the hydrogenosomes of the anaerobic flagellate Tritrichomonas foetus (Lindmark and Müller, 1973) and later found in various parasite lineages (Horner et al., 1999; Hug et al., 2010). Being a key enzyme of anaerobic metabolism of some parasites, intensive research has been devoted to the use of PFO as a medical target. The release of the C. reinhardtii genome sequence uncovered a gene for a PFO, thus suggesting that this metabolic enzyme was not restricted to anaerobic heterotrophic lineages. Here, we provide evidence for a functional PFO in the photosynthetic alga C. reinhardtii.
Chloroplast CrPFO
The Pfo cDNA sequence as well as MS data on the native CrPFO have indicated differences in reference gene models on the JGI and the phytozome portals, thereby emphasizing the need to confirm sequences prior to in silico studies, such as structure predictions or phylogenetic studies. CrPFO is a new member of the large family of homodimeric PFOs, which occur in mesophilic bacteria, such as Clostridium spp. (Uyeda and Rabinowitz, 1971; Meinecke et al., 1989) and Desulfovibrio spp. (Pieulle et al., 1995), in cyanobacteria, and in anaerobic eukaryotes (Williams et al., 1987; Hrdý and Müller 1995; Townson et al., 1996). There exist two other types of PFOs, which differ from the homodimeric enzymes in their subunit composition: the heterodimeric PFOs in archaeal halobacteria (Kerscher and Oesterhelt, 1981; Plaga et al., 1992) and the multimeric PFOs (containing four or five distinct subunits) found in anaerobic hyperthermophiles, comprising both bacteria such as Thermotoga maritima (Blamey and Adams, 1994) and archaea such as Pyrococcus furiosus (Blamey and Adams, 1993). The only type of PFO encountered in eukaryotes so far belongs to the homodimeric family. In phylogenetic trees, the eukaryotic enzymes form a completely separate clade wedged between two clades comprising bacterial representatives (Atteia et al., 2006; Hug et al., 2010). PFO was present in the eukaryote common ancestor (Embley and Martin, 2006; Müller et al., 2012), which is reflected in the high homology to PFOs from heterotrophic eukaryotes. Also, since CrPFO is more related to deltaproteobacteria and firmicutes than to cyanobacteria, a cyanobacterial origin for the algal PFO can be excluded.
C. reinhardtii PFO differs from (most of) its counterparts in bacteria and eukaryotes by the presence of an extension at its C terminus. The reported high sensitivity of the recombinant algal enzyme to oxygen rules out the possibility that this extension protects the enzyme catalytic center from oxidative damage, as is the case for the C terminus of Desulfovibrio spp. PFOs (Vita et al., 2008). Currently, we are left with the question of what the function of this short extension could be. It is worth noting that Ala stretches, as found at the C terminus of CrPFO (Fig. 1A), are also present at the C terminus of the PFO from the free-living flagellate Trimastix pyriformis (Supplemental Fig. S2). This is also true for the C termini of several other enzymes of C. reinhardtii (anaerobic) metabolism (i.e. phosphotransacetylases PTA1 and PTA2 and acetate kinase ACK2). The role of these stretches may constitute a topic for future investigation into the function and/or regulation of eukaryotic anaerobic enzymes.
Thus far, eukaryote PFO and its alternative form PNO have been found in the cytosol, the hydrogenosomes, or the mitochondrion (Müller et al., 2012). Our data provide experimental evidence for the localization of the CrPFO in the chloroplast. Therefore, PFO can function in any cell compartment. Once the evolutionary history of the eukaryote PFOs is resolved, PFO might become an interesting model to study recompartmentalization (i.e. the evolutionary move of an enzyme from one cell compartment to another; Martin, 2010).
In contrast to PFL, which was found in both the chloroplast and mitochondria of C. reinhardtii (Atteia et al., 2006; Terashima et al., 2010), CrPFO appears to be restricted to a single cell compartment. After HYDA (Happe et al., 1994) and PFL, PFO is the third highly oxygen-sensitive enzyme shown to populate the chloroplast in the alga, suggesting that there must be an underlying principle explaining this occurrence. In its natural habitats, the photosynthetic alga encounters fermentation products such as acetate, which result from the anaerobic metabolic activity of other organisms as well as its own. It was observed that acetate inhibits C. reinhardtii oxygen-producing photosynthetic activity (Heifetz et al., 2000); thus, acetate may constitute a turn-on switch for anaerobic metabolism and its oxygen-sensitive enzymes.
CrPFO was not (immuno)detected in cells growing under aerobic conditions, in contrast to PFL (Atteia et al., 2006; Magneschi et al., 2012). It is assumed that the detected PFL is in its inactivated form, which is not sensitive to oxygen. Since cellular energy is comparatively low in an anaerobic environment, it may be good housekeeping to maintain a constant level of PFL and only up-regulate the expression of the small PFL activase protein upon anoxia. In contrast, the synthesis and maturation of CrPFO seem to occur only under anoxic conditions.
Spectroscopic and Enzymatic Properties of Heterologously Expressed CrPFO
Our study of the native CrPFO indicated its relatively low intracellular levels in the culture conditions investigated, resulting in a negligible activity detected in chloroplast-soluble fractions. Hence, for the characterization of CrPFO, we pursued heterologous expression of the enzyme in E. coli. This was deemed feasible since a pyruvate:flavodoxin oxidoreductase (akin to PFO) is also part of the enzymatic arsenal of this bacterium (Blaschkowski et al., 1982). Expression of a functional Cr-rPFO was achieved by growing E. coli cells aerobically in an autoinducible rich medium, in contrast to the expression of DaPFO, which was achieved after manual induction with isopropylthio-β-galactoside under anaerobic conditions (Pieulle et al., 1997). Since under physiological conditions the cytoplasm of E. coli is reducing, anaerobic expression of anaerobic enzymes in the bacterium may not be necessary, especially when the recombinant enzyme is confined to the cytoplasm and the expression of required chaperones or the production of cofactors is not dependent on anaerobic culture conditions. Since these prerequisites were met for PFO, it seemed likely that Cr-rPFO could be properly expressed under aerobic culture conditions, as was observed. It is known that E. coli PFO is usually expressed only at low levels, playing a role in PFL activation via flavodoxin (Blaschkowski et al., 1982); the observed low heterologous expression of Cr-rPFO in E. coli cells might be explained when higher PFO levels would lead to disruptive oxidative pyruvate conversion, upsetting the redox balance and damaging or killing bacterial cells. For example, this effect is known for E. coli expressing high levels of cyt c-type proteins (R. van Lis, unpublished data).
C. reinhardtii PFO produced in E. coli is functional. We showed here that the recombinant enzyme contains the full set of redox cofactors (i.e. the catalytic TPP and three [4Fe-4S] centers), as found for most PFOs from various bacterial species and likely from heterotrophic eukaryotes, although, to our knowledge, no EPR spectra have been published. The decarboxylase activity of Cr-rPFO using the artificial acceptor BV (12 µmol pyruvate min−1 mg−1 protein) is in the activity range reported for native PFOs from the heterotrophic eukaryote T. vaginalis (10–20 µmol pyruvate min−1 mg−1 protein; Williams et al., 1987), the firmicute C. acetobutylicum (25 µmol pyruvate min−1 mg−1 protein; Meinecke et al., 1989), and the methanogenic archaeon M. barkeri (22 µmol pyruvate min−1 mg−1 protein; Bock et al., 1996). However, it is significantly smaller than the activities reported for the homodimeric PFO from D. africanus (70 µmol pyruvate min−1 mg−1 protein; Pieulle et al., 1995) or the multimeric PFO from the hyperthermophilic sulfate-reducing archaeon Archaeoglobus fulgidus (74 µmol pyruvate min−1 mg−1 protein; Kunow et al., 1995). This may have to do with the more robust heat-resistant configuration of PFOs from hyperthermophiles and the oxygen-protective C terminus found in the DaPFO, protecting these types of PFOs from structural and oxidative damage. In this regard, it is noted that the iron-sulfur cluster content of Cr-rPFO was to some extent too low, which should be kept in mind when considering its specific decarboxylating activities. Also, it cannot be excluded that the few amino acids in Cr-rPFO that differ from the native CrPFO (Supplemental Fig. S3), although they are not conserved and removed from the catalytic center(s), may have some effect on the kinetic parameters.
Addition of TPP was required for Cr-rPFO decarboxylase activity, likely because this cofactor was lost during purification. The loss of TPP is not specific for the algal enzyme. Indeed, it has been reported in various studies, such as those on the native PFOs from Klebsiella pneumoniae, M. thermoaceticum (Wahl and Orme-Johnson, 1987), and T. vaginalis (Williams et al., 1987). Similar to most homodimeric PFOs, that of the green alga shows a high sensitivity to oxygen. This sensitivity is currently explained by the degradation of the [4Fe-4S] cluster proximal to the TPP, which is in most species exposed to the solvent (Fig. 1; Pieulle et al., 1995; Chabrière et al., 1999; Vita et al., 2008).
Catalytic and Molecular Insights into the Interaction between Cr-rPFO and Ferredoxins
The oxidation of pyruvate by PFO generates low-potential electron transfer to small soluble proteins, typically ferredoxins or flavodoxins. As far as small soluble electron carrier proteins go in C. reinhardtii, no flavodoxin could be found, so we were left with six ferredoxins (FDX1–FDX6) that have been identified in the chloroplast and that were shown to be differentially regulated (Terauchi et al., 2009). So far, specific physiological roles have only been assigned to FDX1 and FDX2. FDX1 (petF), the most abundant isoform in the alga (Schmitter et al., 1988; Terauchi et al., 2009), is the electron acceptor to ferredoxin:NADP+ oxidoreductase and the best donor to HYDA (Winkler et al., 2009). FDX2 is involved in nitrate metabolism: (1) it specifically accumulates in algal cells grown on nitrate (as opposed to ammonium-grown cells), and (2) it is a better electron donor to nitrite reductase than FDX1 (Terauchi et al., 2009). A role for FDX5 in anaerobic hydrogen production was initially considered because its transcript and protein levels increase significantly upon exposure to anaerobic conditions (Mus et al., 2007; Jacobs et al., 2009). This hypothesis was later ruled out, as no hydrogen evolution could be measured via in vitro activity assays using (heterologously expressed) FDX5 (Jacobs et al., 2009).
The efficiency of the chloroplast FDXs in accepting electrons from Cr-rPFO was here evaluated in in vitro assays. FDX1 was found to be the best electron acceptor, with a maximum decarboxylase activity of 6 µmol pyruvate min−1 mg−1 protein, followed by FDX2 with an activity of 4 µmol pyruvate min−1 mg−1 protein. The other FDXs were found to be poor acceptors. FDX1 and FDX2 are related: (1) they share 68% amino acid sequence identity (mature proteins), (2) they exhibit acidic pI values in the range of approximately 4.1 to 4.3, and (3) they migrate as a dimer on SDS-PAGE (data not shown). However, as shown by Terauchi et al. (2009), FDX2 differs from FDX1 in its redox potential (which is 80 mV more positive) as well as in its surface charge in the vicinity of the [2Fe-2S] cluster (which is more negative). Our data showed that the electron transfer from Cr-rPFO and the Km values for FDX1 and FDX2 are of the same order of magnitude, thus indicating some flexibility in the interaction between Cr-rPFO and its ferredoxin partner. The low electron transfer efficiency observed with FDX3 to FDX6 as compared with FDX1 (and FDX2) might be explained by the fact that former FDXs exhibit extensions at their C termini, eight residues for FDX4 and FDX5 and 33 to 34 residues for FDX3 and FDX6. These C-terminal extensions might interfere with the interaction with CrPFO, an interaction believed to occur primarily through electrostatic forces (Pieulle et al., 2004). This assumption is comforted by the fact that no Cr-rPFO activity could be measured when FDX1 was expressed with a C-terminal 6-His tag, which adds a stretch of eight amino acids (LEHHHHHH) to the protein. It is also noted that the reduction of FDXs by ferredoxin:NADP+ oxidoreductase, using NADPH as electron donor, follows the same trend as with CrPFO (i.e. FDX1 and FDX2 are highly efficient electron acceptors but FDX3–FDX6 are not; R. van Lis, unpublished data).
Immunoblot experiments showed that the levels of FDX1 and FDX2 do not strictly correlate with the levels of PFO in the algal cells. In strain cc124, anaerobic accumulation of CrPFO in nitrate-containing culture medium occurs in the absence of any detectable FDX2. As shown here, between the two FDXs that were found to work efficiently with Cr-rPFO, FDX1 is by far the most abundant isoform and is thus very likely the main acceptor for CrPFO.
In most anaerobic eukaryotes and bacteria, PFO is a catabolic enzyme, involved in a metabolic route that can lead to hydrogen production (Verhaart et al., 2010; Rydzak et al., 2011; Müller et al., 2012). With the identification of FDX1 as the best electron acceptor from Cr-rPFO (this study) and the best donor to the hydrogenase HYDA1 (Winkler et al., 2009), the involvement of PFO in hydrogen production in the alga may be considered very likely, especially in low light or in the absence of light as a strong FDX reductant. As said before, FDX2 can connect anaerobic pyruvate decarboxylation to nitrogen metabolism via nitrite reductase (Terauchi et al., 2009). Via either FDX1 or FDX2, CrPFO could also provide reducing power for amino acid synthesis (via Glu synthase) and sulfur metabolism (via sulfite reductase; Winkler et al., 2010) under dark anoxia. In a number of autotrophic bacteria, PFO has also been shown to function as pyruvate synthase (Fuchs, 2011). The equilibrium dynamics of the enzymatic reaction catalyzed by PFO in C. reinhardtii remains to be established in further studies.
CONCLUSION
C. reinhardtii exhibits a substantial repertoire of fermentative enzymes that provide the alga with metabolic plasticity during periods of anoxia, thereby enhancing its survival (Atteia et al., 2006, 2012; Grossman et al., 2007). Our study shows that the photosynthetic alga harbors a PFO that structurally and functionally relates to homodimeric PFOs and that uses FDX1 as electron carrier. Studies of the metabolic switches and interplay between the anaerobic metabolic routes in C. reinhardtii remain limited. Especially the seemingly redundant capacity for the interconversion of pyruvate and acetyl-CoA by both PFL and PFO warrants further study. Expression and regulation studies of PFO and other anaerobic enzymes as well as intracellular and extracellular metabolic product analyses under various oxygen-excluding conditions should provide more insights into how this enzyme fits into the anaerobic metabolism of green protists.
MATERIALS AND METHODS
Algal Strains and Culture Conditions
Chlamydomonas reinhardtii strain cc124 was obtained from the Chlamydomonas Culture Collection (Duke University). Strain 10-6C (Spreitzer and Mets, 1980) was obtained from the collection of photosynthesis mutants at the Laboratoire de Physiologie Membranaire et Moléculaire du Chloroplaste (Institut de Biologie Physico-Chimique). Algae were maintained on TAP medium (Harris, 1989) solidified with 1.5% agar (w/v). For strain 10-6C, TAP medium was supplemented with 100 mg L−1 Arg. For growth on nitrate as the sole nitrogen source, ammonium in the TAP medium was replaced by nitrate (4 mm NaNO3). Liquid cultures were grown at 24°C with constant shaking (125 rpm) in dim light.
Anaerobic Induction of Cell Suspensions
Exponentially growing C. reinhardtii cultures were harvested and then resuspended either in TAP medium or in AIB containing 50 mm potassium phosphate (pH 7.0) and 3 mm MgCl2 (Ghirardi et al., 1997) to a cell concentration of 1 to 1.5 × 107 cells mL−1. Cells were then placed in an anaerobic jar containing an AnaerocultA cartridge (Merck) and incubated under agitation in the dark for 5 to 6 h at 24°C. Following anaerobic incubation, the cells were harvested by centrifugation at 2,000g for 5 min and flash frozen. Cell pellets were stored at −20°C until used.
Organelle Isolation and Partial Purification of Native CrPFO
Cells (strain 10-6C) grown to late exponential phase were harvested and then resuspended in one-tenth volume of TAP medium supplemented with 100 mg L−1 Arg. The cell suspension was transferred to a bottle and later placed in a jar containing an AnaerocultA cartridge. After 5 h of anaerobic incubation in the dark, the cells were harvested and used to isolate organelles as described (van Lis et al., 2005). Organelles were flash frozen and stored at −20°C.
For partial purification of CrPFO, all steps except ultracentrifugation were carried out in an anaerobic glove box (Jacomex; oxygen concentration less than 5 ppm). Chloroplasts, resuspended in 50 mm HEPES (pH 7.5) and supplemented with protease inhibitors (0.1 mm phenylmethylsulfonyl fluoride, 0.5 mm benzamidine, and 1 mm aminocaproic acid) to a concentration of 3 mg proteins mL−1, were broken by two cycles of freezing (in a freezing ethanol bath) and thawing at room temperature. Fractionation of the chloroplasts into their membrane and soluble constituents was achieved by ultracentrifugation (45,000 rpm for 45 min; 70Ti rotor) at 4°C. Soluble chloroplast proteins were applied to an anion-exchange chromatography column (DE52) preequilibrated with 50 mm HEPES-NaOH, pH 7.5, and then eluted by a stepwise NaCl gradient (50 mm steps) in the same buffer. CrPFO was eluted with 100 mm NaCl.
Cloning of the C. reinhardtii Pfo Gene
From the Pfo gene sequences obtained from the JGI (assemblies version 2.0, version 3.0, and version 4.0), specific primers were designed. These primers were then used in PCR to obtain the sequence of the Pfo coding region and of its flanking regions, using as DNA template a λZAPII phagemid cDNA library (Stratagene) made with mRNAs isolated from cells grown in light with 5% (v/v) CO2 (a gift from J. Davies). Among the various PCR products obtained, the longest of 3,917 bp (amplified with primers P1and P2; Supplemental Table S1) was used as a DNA template to amplify the sequence corresponding to the predicted mature Pfo coding region. Because all our attempts to amplify this region failed, we decided to amplify and clone the sequence for the mature CrPFO in two parts. A silent XhoI site was introduced for religating the two pieces after having separately cloned them. We obtained a 5′ end fragment of 598 bp using a forward primer (P3) that includes an NdeI restriction site and a reverse primer (P4) containing an XhoI site. The 3′ end fragment of 3,116 bp was obtained by using a forward primer (P5) containing an XhoI site and a reverse primer (P6) containing a HindIII site. The 5′ end fragment was digested with NdeI and XhoI, whereas the 3′ end fragment was digested with XhoI, dephosphorylated, and then cut with HindIII. The resulting fragments were ligated for 30 min with the pET24a plasmid (Merck-Novagen) predigested with NdeI and HindIII. The sequence of the 3,705-bp insert (encoding 1,235 amino acids) in the Pfo-pET24a construct thus obtained was verified by sequencing at GATC Biotech. The C. reinhardtii Pfo cDNA sequence has been deposited in GenBank under accession number FR848330.
Cr-rPFO Expression and Purification
The Pfo-pET24a construct was cotransformed with the pRKISC plasmid (Nakamura et al., 1999) into Escherichia coli BL21(DE3) strain for expression. Cells were grown overnight at 37°C in autoinducible ZYM5052 medium (Studier, 2005) supplemented with 1 mm Cys under aerobic conditions. The cells were harvested, washed in 50 mm sodium phosphate (pH 8.0) containing 300 mm NaCl (buffer 1), and flash frozen. Cell pellets were stored at −20°C until use. Cr-rPFO was purified in nitrogen atmosphere in a glove box. Pellets from a 1-L culture were resuspended in 20 mL of buffer 1 supplemented with lysozyme (0.5 mg mL−1), DNase (crystals), and the protease inhibitors described above. Cells were disrupted by two cycles of freezing and thawing. The soluble fraction, containing Cr-rPFO, was incubated with HIS-SelectNickel Affinity Gel (Sigma) in the presence of 30 mm imidazole and 2.5 mm DTT. After a 20-min anaerobic incubation at 4°C under gentle agitation, the resin was washed with 5 volumes of cold buffer 1 supplemented with 30 mm imidazole and 2.5 mm DTT. Cr-rPFO was then eluted from the resin with 100 mm imidazole in buffer 1. The eluted enzyme was immediately passed through a PD10 desalting column (GE Healthcare) preequilibrated with 50 mm Tricine, pH 8.0, and finally concentrated to approximately 1.0 to 1.5 mg mL−1. Purified enzyme was stored in liquid nitrogen.
Production of Polyclonal Anti-PFO Antibodies
Polyclonal antibodies were produced by immunization of a rabbit with a truncated CrPFO (Met-350–His-1232; Cr-tPFO). Cr-tPfo was amplified by PCR using as template the cDNA library and the primer pair P7 and P8, which contain a BamHI site and a HindIII site, respectively (Supplemental Table S1), and further cloned into the pET24a vector. E. coli BL21(DE3) cells were transformed with Cr-tPfo-pET24a to produce Cr-tPFO. The protein was then purified under denaturing conditions using HIS-SelectNickel Affinity Gel, as recommended by the supplier. Antibodies were produced by Eurogentec.
Expression and Purification of Recombinant Chloroplast Ferredoxins FDX1 to FDX6
The gene regions coding for C. reinhardtii mature chloroplast ferredoxins FDX1 to FDX6 (Jacobs et al., 2009; Terauchi et al., 2009) were amplified by PCR (including the stop codon) using primers P9 to P20 listed in Supplemental Table S1 and cloned into expression vector pET24a. Expression was done in E. coli strain BL21(DE3) as for Cr-rPFO, except that the induction was done at 25°C with 20 µm isopropylthio-β-galactoside instead of lactose. E. coli cell pellets from 2-L cultures were resuspended in 50 mm HEPES, pH 7.5, in the presence of lysozyme, DNase (crystals), and the protease inhibitors and further broken by French press. Insoluble material was removed by ultracentrifugation (244,000g for 45 min) at 4°C. For all FDXs, the soluble fraction was loaded on a DE52 anion-exchange chromatography column equilibrated with 50 mm HEPES, pH 7.5; their elution was achieved with NaCl concentrations in the range of 200 to 300 mm. Using ammonium sulfate precipitation steps at 55%, 75%, and 90%, FDX1 precipitated at 90% whereas FDX2 did not. The precipitated FDX1 was dissolved in 50 mm Tricine, pH 8.0, and desalted, whereas FDX2 was dialyzed against the same buffer and concentrated. Following DE52 chromatography steps, FDX3, FDX4, and FDX6 were further purified on a DEAE Biogel A and a hydroxyapatite column; all three FDXs eluted at 50 mm sodium phosphate. FDX5, prone to aggregation upon oxygen exposure (Jacobs et al., 2009), was purified in a glove box. FDX5-containing cells were broken by two cycles of freezing and thawing. The purification steps for FDX5 included DE52 column chromatography (elution with 250 mm NaCl), precipitation with NH4SO4 (65% saturation), and DEAE Biogel A column chromatography (Bio-Rad). FDX5 eluted from the latter column with 100 mm NaCl and then was desalted and stored in liquid nitrogen.
Protein Sample Preparation, Gel Electrophoresis, and Immunoblotting
Protein concentration was routinely determined using the bicinchoninic acid kit (Pierce), with bovine serum albumin as standard. When protein concentrations of whole cells were determined, proteins were first precipitated by CHCl3/methanol (Wessel and Flügge, 1984) and then resuspended in 2% (w/v) SDS. For protein gels, cells or cell fractions were solubilized in the presence of 2.5% SDS (w/v) and 1% β-mercaptoethanol, boiled for 2 min, and then centrifuged for 1 min at 12,000g to remove insoluble material. Proteins were separated by SDS-PAGE on a 10% polyacrylamide gel, a 5% to 12% acrylamide gel containing 6 m urea (PFO), or a 12% to 18% acrylamide gel (FDXs). Molecular masses were estimated using Prestained Protein Ladder Plus (Euromedex). On urea/SDS gel gradients, the 75- and 100-kD markers comigrate; the 75-kD protein probably runs anomalously. After electrophoresis, proteins were either stained with Coomassie Brilliant Blue R250 or transferred onto nitrocellulose membranes (Roti-NC; Roth). Primary antibodies were used at the following dilutions: anti-PFO, 1:5,000; anti-PFL, 1:15,000 (Atteia et al., 2006); anti-ATP2, 1:50,000 (Atteia et al., 2006), anti-LHC, 1:50,000; anti-FDX1, 1:40,000; and anti-FDX2, 1:2,000. Secondary antibody was horseradish peroxidase-conjugated goat anti-rabbit IgG (Sigma) used at 1:10,000. Immunodetection was done by a homemade enhanced chemiluminescence system (Durrant, 1990). Blue native-PAGE was prepared as described previously (Schägger, 2001).
MS and Bioinformatics Analyses
The gel band in the 130-kD region was manually excised and cut in pieces before being washed by six successive 15-min incubations in 25 mm NH4HCO3 and 25 mm NH4HCO3 containing 50% (v/v) acetonitrile. Gel pieces were then dehydrated with 100% acetonitrile and incubated for 45 min at 53°C with 10 mm DTT in 25 mm NH4HCO3 and for 35 min in the dark with 55 mm iodoacetamide in 25 mm NH4HCO3. Alkylation was stopped by adding 10 mm DTT in 25 mm NH4HCO3 and mixing for 10 min. Gel pieces were then washed again by incubation in 25 mm NH4HCO3. Before dehydration with 100% acetonitrile, 0.15 µg of modified trypsin (Promega; sequencing grade) in 25 mm NH4HCO3 was added to the dehydrated gel pieces for an overnight incubation at 37°C. Peptides were then extracted in three sequential extraction steps (15 min each) in 30 µL of 50% acetonitrile, 30 µL of 5% formic acid, and finally 30 µL of 100% acetonitrile. The pooled supernatants were then dried under vacuum.
The dried extracted peptides were resuspended in 5% acetonitrile and 0.1% trifluoroacetic acid and analyzed by online nano-liquid chromatography-MS/MS (Ultimate 3000 [Dionex] and LTQ-Orbitrap Velos pro [Thermo Fisher Scientific]). The nano-liquid chromatography method consisted of a 30-min gradient ranging from 5% to 45% acetronitrile in 0.1% formic acid at a flow rate of 300 nL min−1. Peptides were applied to a 300-µm × 5-mm PepMap C18 reverse-phase precolumn and then separated on a 75-µm × 250-mm C18 reverse-phase column (PepMap; Dionex). MS and MS/MS (up to 20 MS/MS acquisitions for each MS spectrum) data were acquired using Xcalibur (Thermo Fisher Scientific) and processed automatically using Mascot Daemon software (version 2.3; Matrix Science). Searches against a custom polypeptide sequence database (Nguyen et al., 2011) and a contaminants databases (34,618 sequences in total for these two databases) and the corresponding reversed databases were performed using Mascot (version 2.4). ESI-TRAP was chosen as the instrument, trypsin/Pro as the enzyme, and two missed cleavages were allowed. Precursor and fragment mass error tolerances were set at 10 ppm and 0.6 D, respectively. Peptide modifications allowed during the search were as follows: carbamidomethyl (Cys, fixed), acetyl (N terminus, variable), oxidation (Met, variable), and deamidation (Asn and Glu, variable). The IRMa software (Dupierris et al., 2009; version 1.30.4) was used to filter the results (false discovery rate < 1% for peptide identification, minimum of one unique peptide and two peptide sequences per protein). Protein hits from the contaminants database were discarded from the list of identified proteins.
Cr-rPFO and Ferredoxin-Dependent Enzymatic Assays
All assays were performed in a nitrogen atmosphere in a glove box at 25°C using a Varian Cary50 spectrophotometer with a probe of 1 cm optical length. Standard assay mixture (1 mL) for Cr-rPFO activity measurements contained 25 mm potassium phosphate (pH 7.0), 0.1 mm CoA, 5 mm sodium pyruvate, 0.2 mm TPP, and 1 mm MgCl2. The reaction was started by addition of the enzyme (approximately 2–4 µg of protein). For the pH dependency curve, 25 mm potassium phosphate was used, with increments of 0.5 pH units from pH 5.0 to pH 9.0. The rate of BV reduction was determined at 600 nm (molar absorption coefficient [ε] = 7.4 mm−1 cm−1), the rate of reduction of methyl viologen was determined at 604 nm (ε = 13.9 mm−1 cm−1), and the rate of methylene blue reduction was determined at 600 nm (ε = 30.6 mm−1 cm−1). Specific activity was defined as µmol pyruvate decarboxylated min–1 mg–1 protein at 25°C. One catalytic unit corresponds to 1 µmol pyruvate decarboxylated min–1 mg–1 protein.
Cr-rPFO activity in the presence of FDX was assessed in a coupled assay, whereby the reduction of horse heart cyt c by reduced ferredoxin was followed spectrophotometrically at 550 nm (ε = 20 mm−1 cm−1). The assay buffer was as described above except that the artificial electron acceptor was replaced by 20 µm ferredoxin and 50 µm cyt c. To determine the amount of FDX present, absorption spectra of FDXs in the reaction mixture were recorded prior to adding cyt c and Cr-rPFO. Km values were calculated with the help of the program Hyper32 for hyperbolic regression analysis (available at http://homepage.ntlworld.com/john.easterby/hyper32.html).
EPR Spectroscopy
Purified Cr-rPFO was transferred into EPR tubes in the anaerobic glove box, where they were also frozen and sealed. Additions of dithionite, pyruvate, and/or CoA were done prior to transfer into the tubes. EPR spectra were acquired on a Bruker ElecSys X-band spectrometer (Bruker) fitted with a liquid helium cryostat and temperature control system (Oxford Instruments).
Modeling of CrPFO
Using the crystal structure of DaPFO (Protein Data Bank no. 2C42) as the basis, the amino acid sequence of CrPFO was used to construct a predicted three-dimensional structure with SwissModel. This structure was overlaid with that of DaPFO using PDB Viewer. The resulting image was then imported into the program POV-Rey for an enhanced-depth view of the structures.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers FR848330 (nucleotide) and CCA61743 (protein).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Amino acid sequence of CrPFO.
Supplemental Figure S2. Multiple sequence alignment of homodimeric PFOs from different bacterial and eukaryote sources.
Supplemental Figure S3. CrPFO sequence with the tryptic peptides identified by MS.
Supplemental Table S1. Primers used to generate Pfo cDNA and protein expression constructs.
Supplemental Table S2. Proteins identified by MS in the 130-kD gel band that contained immunodetected CrPFO.
Supplemental Table S3. Reduction rate of horse heart cyt c by the different FDXs, reduced either enzymatically by Cr-rPFO in the pyruvate decarboxylase reaction assay or chemically by dithionite.
Acknowledgments
We thank Sabeeha Merchant for the gift of antibodies against FDX1 and FDX2, Olivier Vallon for antibodies against the LHC proteins, Alessandro Aliverti for the gift of the spinach (Spinacia oleracea) Fd1 expression construct, and Florence Chaspoul for the inductively coupled plasma-MS measurements. We also thank Profs. William F. Martin and Samuel I. Beale for helpful comments on the manuscript.
Glossary
- EPR
electron paramagnetic resonance
- PFL
pyruvate formate-lyase
- PFO
pyruvate:ferredoxin oxidoreductase
- TPP
thiamine pyrophosphate
- cyt
cytochrome
- PNO
pyruvate:NADP+ oxidoreductase
- JGI
Joint Genome Institute
- LHC
light-harvesting complex
- MS
mass spectrometry
- BV
benzyl viologen
- DTT
dithiothreitol
- AIB
anaerobic induction buffer
- TAP
Tris-acetate-phosphate
- cDNA
complementary DNA
- EC
Enzyme Commission
References
- Akhmanova A, Voncken FG, Hosea KM, Harhangi H, Keltjens JT, op den Camp HJ, Vogels GD, Hackstein JH. (1999) A hydrogenosome with pyruvate formate-lyase: anaerobic chytrid fungi use an alternative route for pyruvate catabolism. Mol Microbiol 32: 1103–1114 [DOI] [PubMed] [Google Scholar]
- Atteia A, van Lis R, Gelius-Dietrich G, Adrait A, Garin J, Joyard J, Rolland N, Martin W. (2006) Pyruvate formate-lyase and a novel route of eukaryotic ATP synthesis in Chlamydomonas mitochondria. J Biol Chem 281: 9909–9918 [DOI] [PubMed] [Google Scholar]
- Atteia A, van Lis R, Tielens AGM, Martin WF. (2012) Anaerobic energy metabolism in unicellular photosynthetic eukaryotes. Biochim Biophys Acta (in press) [DOI] [PubMed] [Google Scholar]
- Bailey-Serres J, Fukao T, Gibbs DJ, Holdsworth MJ, Lee SC, Licausi F, Perata P, Voesenek LA, van Dongen JT. (2012) Making sense of low oxygen sensing. Trends Plant Sci 17: 129–138 [DOI] [PubMed] [Google Scholar]
- Blamey JM, Adams MW. (1993) Purification and characterization of pyruvate ferredoxin oxidoreductase from the hyperthermophilic archaeon Pyrococcus furiosus. Biochim Biophys Acta 1161: 19–27 [DOI] [PubMed] [Google Scholar]
- Blamey JM, Adams MW. (1994) Characterization of an ancestral type of pyruvate ferredoxin oxidoreductase from the hyperthermophilic bacterium, Thermotoga maritima. Biochemistry 33: 1000–1007 [DOI] [PubMed] [Google Scholar]
- Blaschkowski HP, Neuer G, Ludwig-Festl M, Knappe J. (1982) Routes of flavodoxin and ferredoxin reduction in Escherichia coli: CoA-acylating pyruvate:flavodoxin and NADPH:flavodoxin oxidoreductases participating in the activation of pyruvate formate-lyase. Eur J Biochem 123: 563–569 [PubMed] [Google Scholar]
- Bock AK, Kunow J, Glasemacher J, Schönheit P. (1996) Catalytic properties, molecular composition and sequence alignments of pyruvate:ferredoxin oxidoreductase from the methanogenic archaeon Methanosarcina barkeri (strain Fusaro). Eur J Biochem 237: 35–44 [DOI] [PubMed] [Google Scholar]
- Castruita M, Casero D, Karpowicz SJ, Kropat J, Vieler A, Hsieh SI, Yan W, Cokus S, Loo JA, Benning C, et al. (2011) Systems biology approach in Chlamydomonas reveals connections between copper nutrition and multiple metabolic steps. Plant Cell 23: 1273–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalanotti C, Dubini A, Subramanian V, Yang W, Magneschi L, Mus F, Seibert M, Posewitz MC, Grossman AR. (2012) Altered fermentative metabolism in Chlamydomonas reinhardtii mutants lacking pyruvate formate lyase and both pyruvate formate lyase and alcohol dehydrogenase. Plant Cell 24: 692–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavazza C, Contreras-Martel C, Pieulle L, Chabrière E, Hatchikian EC, Fontecilla-Camps JC. (2006) Flexibility of thiamine diphosphate revealed by kinetic crystallographic studies of the reaction of pyruvate-ferredoxin oxidoreductase with pyruvate. Structure 14: 217–224 [DOI] [PubMed] [Google Scholar]
- Chabrière E, Charon MH, Volbeda A, Pieulle L, Hatchikian EC, Fontecilla-Camps JC. (1999) Crystal structures of the key anaerobic enzyme pyruvate:ferredoxin oxidoreductase, free and in complex with pyruvate. Nat Struct Biol 6: 182–190 [DOI] [PubMed] [Google Scholar]
- Charon MH, Volbeda A, Chabriere E, Pieulle L, Fontecilla-Camps JC. (1999) Structure and electron transfer mechanism of pyruvate:ferredoxin oxidoreductase. Curr Opin Struct Biol 9: 663–669 [DOI] [PubMed] [Google Scholar]
- de Sousa CAF, Sodek L. (2003) Alanine metabolism and alanine aminotransferase activity in soybean (Glycine max) during hypoxia of the root system and subsequent return to normoxia. Environ Exp Bot 50: 1–8 [Google Scholar]
- Dupierris V, Masselon C, Court M, Kieffer-Jaquinod S, Bruley C. (2009) A toolbox for validation of mass spectrometry peptides identification and generation of database: IRMa. Bioinformatics 25: 1980–1981 [DOI] [PubMed] [Google Scholar]
- Durrant I. (1990) Light-based detection of biomolecules. Nature 346:297–298 [DOI] [PubMed] [Google Scholar]
- Eberhard S, Finazzi G, Wollman FA. (2008) The dynamics of photosynthesis. Annu Rev Genet 42: 463–515 [DOI] [PubMed] [Google Scholar]
- Embley TM, Martin W. (2006) Eukaryotic evolution, changes and challenges. Nature 440: 623–630 [DOI] [PubMed] [Google Scholar]
- Fuchs G. (2011) Alternative pathways of carbon dioxide fixation: insights into the early evolution of life? Annu Rev Microbiol 65: 631–658 [DOI] [PubMed] [Google Scholar]
- Gelius-Dietrich G, Henze K. (2004) Pyruvate formate lyase (PFL) and PFL activating enzyme in the chytrid fungus Neocallimastix frontalis: a free-radical enzyme system conserved across divergent eukaryotic lineages. J Eukaryot Microbiol 51: 456–463 [DOI] [PubMed] [Google Scholar]
- Ghirardi ML, Togasaki RK, Seibert M. (1997) Oxygen sensitivity of algal H2-production. Appl Biochem Biotechnol 63-65: 141–151 [DOI] [PubMed] [Google Scholar]
- Grossman AR, Catalanotti C, Yang W, Dubini A, Magneschi L, Subramanian V, Posewitz MC, Seibert M. (2011) Multiple facets of anoxic metabolism and hydrogen production in the unicellular green alga Chlamydomonas reinhardtii. New Phytol 190: 279–288 [DOI] [PubMed] [Google Scholar]
- Grossman AR, Croft M, Gladyshev VN, Merchant SS, Posewitz MC, Prochnik S, Spalding MH. (2007) Novel metabolism in Chlamydomonas through the lens of genomics. Curr Opin Plant Biol 10: 190–198 [DOI] [PubMed] [Google Scholar]
- Grossman AR, Harris EE, Hauser C, Lefebvre PA, Martinez D, Rokhsar D, Shrager J, Silflow CD, Stern D, Vallon O, et al. (2003) Chlamydomonas reinhardtii at the crossroads of genomics. Eukaryot Cell 2: 1137–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Happe T, Mosler B, Naber JD. (1994) Induction, localization and metal content of hydrogenase in the green alga Chlamydomonas reinhardtii. Eur J Biochem 222: 769–774 [DOI] [PubMed] [Google Scholar]
- Harris E. (1989) The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use. Academic Press, San Diego. [DOI] [PubMed] [Google Scholar]
- Hawkins CF, Borges A, Perham RN. (1989) A common structural motif in thiamin pyrophosphate-binding enzymes. FEBS Lett 255: 77–82 [DOI] [PubMed] [Google Scholar]
- Heifetz PB, Förster B, Osmond CB, Giles LJ, Boynton JE. (2000) Effects of acetate on facultative autotrophy in Chlamydomonas reinhardtii assessed by photosynthetic measurements and stable isotope analyses. Plant Physiol 122: 1439–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemschemeier A, Jacobs J, Happe T. (2008) Biochemical and physiological characterization of the pyruvate formate-lyase Pfl1 of Chlamydomonas reinhardtii, a typically bacterial enzyme in a eukaryotic alga. Eukaryot Cell 7: 518–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner DS, Hirt RP, Embley TM. (1999) A single eubacterial origin of eukaryotic pyruvate:ferredoxin oxidoreductase genes: implications for the evolution of anaerobic eukaryotes. Mol Biol Evol 16: 1280–1291 [DOI] [PubMed] [Google Scholar]
- Hrdý I, Müller M. (1995) Primary structure and eubacterial relationships of the pyruvate:ferredoxin oxidoreductase of the amitochondriate eukaryote Trichomonas vaginalis. J Mol Evol 41: 388–396 [PubMed] [Google Scholar]
- Hug LA, Stechmann A, Roger AJ. (2010) Phylogenetic distributions and histories of proteins involved in anaerobic pyruvate metabolism in eukaryotes. Mol Biol Evol 27: 311–324 [DOI] [PubMed] [Google Scholar]
- Inui H, Miyatake K, Nakano Y, Kitaoka S. (1984) Occurrence of oxygen-sensitive, NADP+-dependent pyruvate dehydrogenase in mitochondria of Euglena gracilis. J Biochem 96: 931–934 [DOI] [PubMed] [Google Scholar]
- Jacobs J, Pudollek S, Hemschemeier A, Happe T. (2009) A novel, anaerobically induced ferredoxin in Chlamydomonas reinhardtii. FEBS Lett 583: 325–329 [DOI] [PubMed] [Google Scholar]
- Kerscher L, Oesterhelt D. (1981) Purification and properties of two 2-oxoacid:ferredoxin oxidoreductases from Halobacterium halobium. Eur J Biochem 116: 587–594 [DOI] [PubMed] [Google Scholar]
- Kletzin A, Adams MW. (1996) Molecular and phylogenetic characterization of pyruvate and 2-ketoisovalerate ferredoxin oxidoreductases from Pyrococcus furiosus and pyruvate ferredoxin oxidoreductase from Thermotoga maritima. J Bacteriol 178: 248–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knappe J, Sawers G. (1990) A radical-chemical route to acetyl-CoA: the anaerobically induced pyruvate formate-lyase system of Escherichia coli. FEMS Microbiol Rev 6: 383–398 [DOI] [PubMed] [Google Scholar]
- Knappe J, Schacht J, Möckel W, Höpner T, Vetter H, Jr, Edenharder R. (1969) Pyruvate formate-lyase reaction in Escherichia coli: the enzymatic system converting an inactive form of the lyase into the catalytically active enzyme. Eur J Biochem 11: 316–327 [DOI] [PubMed] [Google Scholar]
- Kreuzberg K. (1984) Starch fermentation via a formate producing pathway in Chlamydomonas reinhardtii, Chlorogonium elongatum and Chlorella fusca. Physiol Plant 61: 87–94 [Google Scholar]
- Kunow J, Linder D, Thauer RK. (1995) Pyruvate:ferredoxin oxidoreductase from the sulfate-reducing Archaeoglobus fulgidus: molecular composition, catalytic properties, and sequence alignments. Arch Microbiol 163: 21–28 [DOI] [PubMed] [Google Scholar]
- Lantsman Y, Tan KSW, Morada M, Yarlett N. (2008) Biochemical characterization of a mitochondrial-like organelle from Blastocystis sp. subtype 7. Microbiology 154: 2757–2766 [DOI] [PubMed] [Google Scholar]
- Lindmark DG, Müller M. (1973) Hydrogenosome, a cytoplasmic organelle of the anaerobic flagellate Tritrichomonas foetus, and its role in pyruvate metabolism. J Biol Chem 248: 7724–7728 [PubMed] [Google Scholar]
- Magneschi L, Catalanotti C, Subramanian V, Dubini A, Yang W, Mus F, Posewitz MC, Seibert M, Perata P, Grossman AR. (2012) A mutant in the ADH1 gene of Chlamydomonas reinhardtii elicits metabolic restructuring during anaerobiosis. Plant Physiol 158: 1293–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W. (2010) Evolutionary origins of metabolic compartmentalization in eukaryotes. Philos Trans R Soc Lond B Biol Sci 365: 847–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinecke B, Bertram J, Gottschalk G. (1989) Purification and characterization of the pyruvate-ferredoxin oxidoreductase from Clostridium acetobutylicum. Arch Microbiol 152: 244–250 [DOI] [PubMed] [Google Scholar]
- Menon S, Ragsdale SW. (1997) Mechanism of the Clostridium thermoaceticum pyruvate:ferredoxin oxidoreductase: evidence for the common catalytic intermediacy of the hydroxyethylthiamine pyropyrosphate radical. Biochemistry 36: 8484–8494 [DOI] [PubMed] [Google Scholar]
- Merchant SS, Allen MD, Kropat J, Moseley JL, Long JC, Tottey S, Terauchi AM. (2006) Between a rock and a hard place: trace element nutrition in Chlamydomonas. Biochim Biophys Acta 1763: 578–594 [DOI] [PubMed] [Google Scholar]
- Müller M, Mentel M, van Hellemond JJ, Henze K, Woehle C, Gould SB, Yu RY, van der Giezen M, Tielens AG, Martin WF. (2012) Biochemistry and evolution of anaerobic energy metabolism in eukaryotes. Microbiol Mol Biol Rev 76: 444–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mus F, Dubini A, Seibert M, Posewitz MC, Grossman AR. (2007) Anaerobic acclimation in Chlamydomonas reinhardtii: anoxic gene expression, hydrogenase induction, and metabolic pathways. J Biol Chem 282: 25475–25486 [DOI] [PubMed] [Google Scholar]
- Nakamura M, Saeki K, Takahashi Y. (1999) Hyperproduction of recombinant ferredoxins in Escherichia coli by coexpression of the ORF1-ORF2-iscS-iscU-iscA-hscB-hs cA-fdx-ORF3 gene cluster. J Biochem 126: 10–18 [DOI] [PubMed] [Google Scholar]
- Nguyen HM, Baudet M, Cuiné S, Adriano JM, Barthe D, Billon E, Bruley C, Beisson F, Peltier G, Ferro M, et al. (2011) Proteomic profiling of oil bodies isolated from the unicellular green microalga Chlamydomonas reinhardtii: with focus on proteins involved in lipid metabolism. Proteomics 11: 4266–4273 [DOI] [PubMed] [Google Scholar]
- Page MD, Allen MD, Kropat J, Urzica EI, Karpowicz SJ, Hsieh SI, Loo JA, Merchant SS. (2012) Fe sparing and Fe recycling contribute to increased superoxide dismutase capacity in iron-starved Chlamydomonas reinhardtii. Plant Cell 24: 2649–2665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philipps G, Krawietz D, Hemschemeier A, Happe T. (2011) A pyruvate formate lyase-deficient Chlamydomonas reinhardtii strain provides evidence for a link between fermentation and hydrogen production in green algae. Plant J 66: 330–340 [DOI] [PubMed] [Google Scholar]
- Pieulle L, Guigliarelli B, Asso M, Dole F, Bernadac A, Hatchikian EC. (1995) Isolation and characterization of the pyruvate-ferredoxin oxidoreductase from the sulfate-reducing bacterium Desulfovibrio africanus. Biochim Biophys Acta 1250: 49–59 [DOI] [PubMed] [Google Scholar]
- Pieulle L, Magro V, Hatchikian EC. (1997) Isolation and analysis of the gene encoding the pyruvate-ferredoxin oxidoreductase of Desulfovibrio africanus, production of the recombinant enzyme in Escherichia coli, and effect of carboxy-terminal deletions on its stability. J Bacteriol 179: 5684–5692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieulle L, Nouailler M, Morelli X, Cavazza C, Gallice P, Blanchet S, Bianco P, Guerlesquin F, Hatchikian EC. (2004) Multiple orientations in a physiological complex: the pyruvate-ferredoxin oxidoreductase-ferredoxin system. Biochemistry 43: 15480–15493 [DOI] [PubMed] [Google Scholar]
- Plaga W, Lottspeich F, Oesterhelt D. (1992) Improved purification, crystallization and primary structure of pyruvate:ferredoxin oxidoreductase from Halobacterium halobium. Eur J Biochem 205: 391–397 [DOI] [PubMed] [Google Scholar]
- Rochaix JD. (2002) The three genomes of Chlamydomonas. Photosynth Res 73: 285–293 [DOI] [PubMed] [Google Scholar]
- Rotte C, Stejskal F, Zhu G, Keithly JS, Martin W. (2001) Pyruvate:NADP+ oxidoreductase from the mitochondrion of Euglena gracilis and from the apicomplexan Cryptosporidium parvum: a biochemical relic linking pyruvate metabolism in mitochondriate and amitochondriate protists. Mol Biol Evol 18: 710–720 [DOI] [PubMed] [Google Scholar]
- Rydzak T, Levin DB, Cicek N, Sparling R. (2011) End-product induced metabolic shifts in Clostridium thermocellum ATCC 27405. Appl Microbiol Biotechnol 92: 199–209 [DOI] [PubMed] [Google Scholar]
- Schägger H. (2001) Blue-native gels to isolate protein complexes from mitochondria. Methods Cell Biol 65: 231–244 [DOI] [PubMed] [Google Scholar]
- Schmitter JM, Jacquot JP, de Lamotte-Guéry F, Beauvallet C, Dutka S, Gadal P, Decottignies P. (1988) Purification, properties and complete amino acid sequence of the ferredoxin from a green alga, Chlamydomonas reinhardtii. Eur J Biochem 172: 405–412 [DOI] [PubMed] [Google Scholar]
- Spreitzer RJ, Mets LJ. (1980) Non-mendelian mutation affecting ribulose-1,5-biphosphate carboxylase structure and activity. Nature 285: 114–115 [Google Scholar]
- Stairs CW, Roger AJ, Hampl V. (2011) Eukaryotic pyruvate formate lyase and its activating enzyme were acquired laterally from a Firmicute. Mol Biol Evol 28: 2087–2099 [DOI] [PubMed] [Google Scholar]
- Studier FW. (2005) Protein production by auto-induction in high density shaking cultures. Protein Expr Purif 41: 207–234 [DOI] [PubMed] [Google Scholar]
- Tardif M, Atteia A, Specht M, Cogne G, Rolland N, Brugière S, Hippler M, Ferro M, Bruley C, Peltier G, et al. (2012) PredAlgo: a new subcellular localization prediction tool dedicated to green algae. Mol Biol Evol 29: 3625–3639 [DOI] [PubMed] [Google Scholar]
- Terashima M, Specht M, Naumann B, Hippler M. (2010) Characterizing the anaerobic response of Chlamydomonas reinhardtii by quantitative proteomics. Mol Cell Proteomics 9: 1514–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terauchi AM, Lu SF, Zaffagnini M, Tappa S, Hirasawa M, Tripathy JN, Knaff DB, Farmer PJ, Lemaire SD, Hase T, et al. (2009) Pattern of expression and substrate specificity of chloroplast ferredoxins from Chlamydomonas reinhardtii. J Biol Chem 284: 25867–25878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townson SM, Upcroft JA, Upcroft P. (1996) Characterisation and purification of pyruvate:ferredoxin oxidoreductase from Giardia duodenalis. Mol Biochem Parasitol 79: 183–193 [DOI] [PubMed] [Google Scholar]
- Uyeda K, Rabinowitz JC. (1971) Pyruvate-ferredoxin oxidoreductase. 3. Purification and properties of the enzyme. J Biol Chem 246: 3111–3119 [PubMed] [Google Scholar]
- van Lis R, Atteia A, Nogaj LA, Beale SI. (2005) Subcellular localization and light-regulated expression of protoporphyrinogen IX oxidase and ferrochelatase in Chlamydomonas reinhardtii. Plant Physiol 139: 1946–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaart MR, Bielen AA, van der Oost J, Stams AJ, Kengen SW. (2010) Hydrogen production by hyperthermophilic and extremely thermophilic bacteria and archaea: mechanisms for reductant disposal. Environ Technol 31: 993–1003 [DOI] [PubMed] [Google Scholar]
- Vita N, Hatchikian EC, Nouailler M, Dolla A, Pieulle L. (2008) Disulfide bond-dependent mechanism of protection against oxidative stress in pyruvate-ferredoxin oxidoreductase of anaerobic Desulfovibrio bacteria. Biochemistry 47: 957–964 [DOI] [PubMed] [Google Scholar]
- Wagner AF, Frey M, Neugebauer FA, Schäfer W, Knappe J. (1992) The free radical in pyruvate formate-lyase is located on glycine-734. Proc Natl Acad Sci USA 89: 996–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl RC, Orme-Johnson WH. (1987) Clostridial pyruvate oxidoreductase and the pyruvate-oxidizing enzyme specific to nitrogen fixation in Klebsiella pneumoniae are similar enzymes. J Biol Chem 262: 10489–10496 [PubMed] [Google Scholar]
- Wessel D, Flügge UI. (1984) A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem 138: 141–143 [DOI] [PubMed] [Google Scholar]
- Williams K, Lowe PN, Leadlay PF. (1987) Purification and characterization of pyruvate:ferredoxin oxidoreductase from the anaerobic protozoon Trichomonas vaginalis. Biochem J 246: 529–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler M, Hemschemeier A, Jacobs J, Stripp S, Happe T. (2010) Multiple ferredoxin isoforms in Chlamydomonas reinhardtii: their role under stress conditions and biotechnological implications. Eur J Cell Biol 89: 998–1004 [DOI] [PubMed] [Google Scholar]
- Winkler M, Kuhlgert S, Hippler M, Happe T. (2009) Characterization of the key step for light-driven hydrogen evolution in green algae. J Biol Chem 284: 36620–36627 [DOI] [PMC free article] [PubMed] [Google Scholar]