Abstract
The Arabidopsis (Arabidopsis thaliana) acyl-coenzyme A (CoA) desaturase-like (ADS) gene family contains nine genes encoding fatty acid desaturase-like proteins. The biological function of only one member of the family, fatty acid desaturase5 (AtADS3/FAD5, At3g15850), is known, and this gene encodes the plastidic palmitoyl-monogalactosyldiacylglycerol Δ7 desaturase. We cloned seven members of the gene family that are predicted not to have a chloroplast transit peptide and expressed them in the yeast Saccharomyces cerevisiae. All seven have previously undescribed desaturase activity on very-long-chain fatty acid (VLCFA) substrates and exhibit diverse regiospecificity, catalyzing introduction of double bonds relative to the methyl end of the molecule (n-x) at n-6 (AtADS4, At1g06350), n-7 (AtADS1.3, At1g06100 and AtADS4.2, At1g06360), n-9 (AtADS1, At1g06080 and AtADS2, At2g31360) or Δ9 (relative to the carboxyl end of the molecule) positions (AtADS1.2, At1g06090 and AtADS1.4, At1g06120). Through forward and reverse genetics it was shown that AtADS2 is involved in the synthesis of the 24:1(n-9) and 26:1(n-9) components (X:Y, where X is chain length and Y is number of double bonds) of seed lipids, sphingolipids, and the membrane phospholipids phosphatidylserine, and phosphatidylethanolamine. Plants deficient in AtADS2 expression showed no obvious phenotype when grown under normal growing conditions, but showed an almost complete loss of phosphatidylethanolamine(42:4), phosphatidylserine(42:4), dihydroxy-monohexosylceramide(42:2)-2, trihydroxy-monohexosylceramide(42:2)-3, and trihydroxy-glycosylinositolphosphoceramide(42:2)-3, lipid species that contain the VLCFA 24:1(n-9), and trihydroxy-glycosylinositolphosphoceramide(44:2)-3, a lipid containing 26:1(n-9). Acyl-CoA profiling of these plants revealed a major reduction in 24:1-CoA and a small reduction in 26:1-CoA. Overexpression of AtADS2 resulted in a substantial increase in the percentage of glycerolipid and sphingolipids species containing 24:1 and a dramatic increase in the percentage of very-long-chain monounsaturated fatty acids in the acyl-CoA pool. Plants deficient in AtADS1 expression had reduced levels of 26:1(n-9) in seed lipids, but no significant changes in leaf phospholipids or sphingolipids were observed. These findings indicate that the 24-carbon and 26-carbon monounsaturated VLCFAs of Arabidopsis result primarily from VLCFA desaturation, rather than by elongation of long chain monounsaturated fatty acids.
The ADS (for acyl-CoA desaturase-like) gene family of Arabidopsis (Arabidopsis thaliana) encodes a group of nine proteins with homology to the Δ9 acyl-lipid desaturases of cyanobacteria, the Δ9 acyl-CoA desaturases of yeast (Saccharomyces cerevisiae) and mammals (Fukuchi-Mizutani et al., 1998; Heilmann et al., 2004b) and the membrane-bound desaturases of insects (Knipple et al., 2002). Eight of these genes are located in three clusters on chromosomes I and III. The remaining gene, designated AtADS2 (At2g31360), is present on chromosome II. With the exception of Arabidopsis fatty acid desaturase5 (AtADS3/FAD5, At3g15850), which encodes the plastidic palmitoyl-monogalactosyldiacylglycerol Δ7 desaturase (Heilmann et al., 2004b), the biological role of these enzymes in Arabidopsis is currently unknown. AtADS3/FAD5 and a second closely linked homolog designated AtADS3.2 (At3g15870), are the only members of the gene family predicted to encode proteins with a plastid transit peptide.
The first study, to our knowledge, to report evidence of desaturase activity associated with an Arabidopsis ADS, AtADS1 (At1g06080), described the heterologous expression of the gene in Brassica juncea. Seeds from transformed plants contained decreased levels of saturated fatty acids and a slight increase in oleic acid content (Yao et al., 2003). Although the evidence was indirect, the results suggested that AtADS1 may encode a Δ9 desaturase. More detailed studies involving in vivo expression of AtADS1, AtADS2, and AtADS3 (without the plastid transit peptide) in yeast (Saccharomyces cerevisiae) have shown that all three enzymes can catalyze the Δ9 or Δ7 desaturation of palmitic (16:0) and stearic (18:0) acids (X:Y, where X is chain length and Y is number of double bonds), with regiospecificity being partly influenced by fatty acid substrate (Heilmann et al., 2004a). In this work, the substrate for desaturation was suggested to be a glycerolipid rather than acyl-CoA. The bifunctionality of these enzymes was further demonstrated by expression in Arabidopsis. When AtADS3 was expressed as the full-length form including the plastid transit peptide, or when AtADS1 and AtADS2 were retargeted to the plastid by the addition of a plastid transit peptide, 16:1Δ7 became the predominant monounsaturated 16-carbon (C16) fatty acid. The Arabidopsis plants used in the study were fab1/fae1 (for fatty acid elongase1) double mutant lines lacking the activity of KASII (for 3-ketoacyl-acyl-carrier protein synthase; FAB1, At1g74960) and the FAE1 condensing enzyme (At4g34520), and consequently exhibiting higher than normal levels of 16:0 and low very-long-chain unsaturated fatty acid content in the seed lipids.
Homologs of the Arabidopsis ADS enzymes have been identified in other plant species, but their catalytic activity and acyl-substrates are not well characterized. Heterologous expression of a complementary DNA (cDNA) encoding an ADS-like protein from white spruce (Picea glauca) gave evidence of Δ9 activity when expressed in yeast (Marillia et al., 2002). The lipid substrate of this desaturase was not determined and the cDNA appeared to encode an enzyme with a plastid transit peptide. The Δ5 desaturase catalyzing the synthesis of 20:1Δ5 in the seeds of Limnanthes alba is also an ADS homolog (Cahoon et al., 2000). The substrate for this reaction is thought to be the 20:0-CoA thioester (Pollard and Stumpf, 1980; Moreau et al., 1981). Δ5 desaturase activity on fatty acids with chain length longer than 18 carbons (very-long-chain fatty acids [VLCFAs]) has also been demonstrated from two ADS homologs isolated from Anemone leveillei (Sayanova et al., 2007). Indirect evidence suggesting that both enzymes utilized acyl-CoA substrates was presented based on characterization of acyl-CoA pools in developing seeds of transgenic Arabidopsis expressing the A. leveillei desaturases.
In addition to functioning in the synthesis of chloroplast lipids (AtADS3/FAD5) and VLCFAs of certain seed oils, ADS proteins have been suggested to play a role in petal senescence in roses (Rosa spp.; Fukuchi-Mizutani et al., 1995) and the expression of Arabidopsis AtADS1 and AtADS2 appears to be regulated in response to changes in temperature (Fukuchi-Mizutani et al., 1998; Byun et al., 2009). A potential role in drought tolerance has also been suggested for a member of the gene family (At1g06100; Allen et al., 2012). This group of plant enzymes therefore appears to contain members showing a diversity of lipid substrate utilization, desaturation regiospecificity and biological function that merits further investigation.
Only four members of the Arabidopsis ADS gene family have documented nomenclature. Based on the existing literature we propose a systematic nomenclature of the ADS gene family based on their chromosomal location and grouping (Table I). We chose the seven members of the gene family predicted not to have a plastid transit peptide for further characterization. Expression in yeast indicates that all seven have previously undescribed desaturase activity on VLCFA substrates. Forward and reverse genetics have revealed a role for AtADS2 in the production of very-long-chain monounsaturated fatty acids (VLCMUFAs) in seed lipids and in membrane phospholipids and sphingolipids.
Table I. The Arabidopsis ADS gene family.
| The Arabidopsis Information Resource Locus | Gene Name | Universal Protein Resource Code | References |
|---|---|---|---|
| At1g06080 | AtADS1 | O65797 | Fukuchi-Mizutani et al. (1998); Heilmann et al. (2004a, 2004b); Yao et al. (2003) |
| At1g06090 | AtADS1.2 | Q9LND9 | |
| At1g06100 | AtADS1.3 | Q9LND8 | Allen et al. (2012) |
| At1g06120 | AtADS1.4 | Q9FPD5 | |
| At2g31360 | AtADS2 | Q9SID2 | Fukuchi-Mizutani et al. (1998); Heilmann et al. (2004a, 2004b) |
| At3g15870 | AtADS3/FAD5 | Q9LVZ3 | Heilmann et al. (2004a, 2004b) |
| At3g15850 | AtADS3.2 | Q949X0 | |
| At1g06350 | AtADS4 | Q9LMI4 | |
| At1g06360 | AtADS4.2 | Q9LMI3 |
RESULTS
Comparison of Deduced Amino Acid Sequences and Subcellular Localization
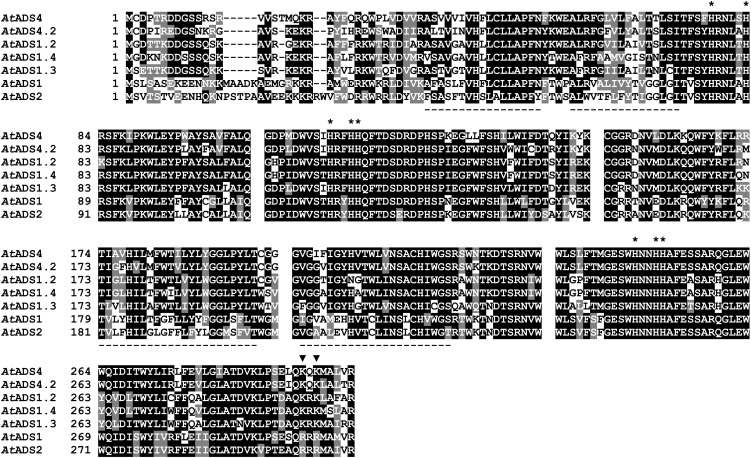
Comparison of deduced amino acid sequences (Fig. 1) showed that all of the Arabidopsis ADS proteins contained the putative Δ9 FAD-like conserved domain (cd03505; Marchler-Bauer et al., 2011) featuring the His box motifs characteristic of fatty acid desaturases (Shanklin et al., 1994; Shanklin and Cahoon, 1998). Phylogenetic analysis (Supplemental Fig. S1; Dereeper et al., 2008) indicated that AtADS1.2 and AtADS1.4 were the most highly conserved members of the ADS family with 89.6% amino acid identity, followed by AtADS4 and AtADS4.2 with 81.3% identity. AtADS1 and AtADS2 shared 75.9% identity. All ADS proteins are designated as multipass membrane proteins located in the endoplasmic reticulum (ER) membrane (http://uniprot.org). A search for potential hydrophobic membrane spanning regions using on-line analysis tools including TMHMM and PredictProtein (http://www.cbs.dtu.dk/services/TMHMM-2.0 and www.predictprotein.org) gave a consensus of four major transmembrane helices in each protein (Fig. 1) with both N and C termini located in the cytosol. None of the seven proteins contained a putative plastid transit peptide, and a search for ER localization signals using SignalP version 4.0 (http://www.cbs.dtu.dk/services/SignalP; Petersen et al., 2011) did not reveal any obvious signal peptides. AtADS1 and AtADS2 appeared to contain a putative di-Arg (-RXR-) ER retention/retrieval motif (Zerangue et al., 1999) close to the C terminus. In the other five ADS proteins this motif was replaced with Lys (-KXK-) residues.
Figure 1.
Comparison of the deduced amino acid sequences of seven Arabidopsis ADS family members. Universal Protein Resource code numbers are given in Table I. Sequences were clustered using clustalW (http://www.genome.jp/tools/clustalw/) and colored using BOXSHADE 3.21 (http://www.ch.embnet.org/software/BOX_form.html). Gaps were inserted manually to reflect polypeptide sequences encoded by individual exons. His residues conserved in membrane-bound desaturases are marked by asterisks. Black triangles mark positions of residues potentially involved in ER retention/retrieval. Underlined sequences are predicted hydrophobic membrane spanning regions (http://www.cbs.dtu.dk/services/TMHMM-2.0/; www.predictprotein.org).
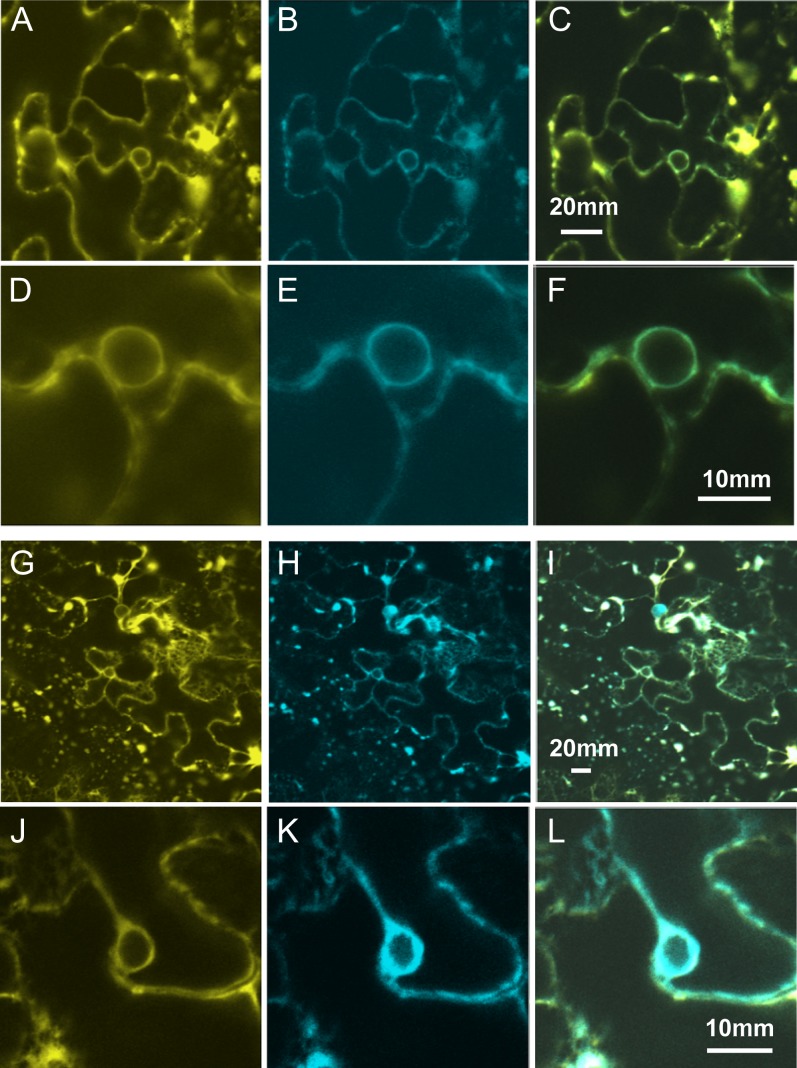
To examine the subcellular localization of AtADS1 and AtADS2, proteins were transiently expressed as yellow fluorescent protein (YFP)-fusions in Nicotiana benthamiana leaves using Agrobacterium tumefaciens infiltration. Examination of leaves by confocal microscopy revealed very low level diffuse YFP florescence when YFP was fused to the N terminus of either AtADS1 or AtADS2 (data not shown). Fusion proteins composed of YFP fused at the C terminus (Fig. 2) colocalized with the ER membrane marker CD3-953 (Nelson et al., 2007), suggesting that these proteins are components of the ER membrane system.
Figure 2.
Subcellular localization of AtADS1 and AtADS2 was investigated using transient expression in Nicotiana benthamiana leaves. A and D, Location of AtADS1-YFP; G and J, location of AtADS2-YFP; B, E, H, and K, distribution of ER marker CD3-953; C, merge of A and B; F, merge of D and E; I, merge of G and H; L, merge of J and K.
Ectopic Expression of Arabidopsis ADS cDNAs in Yeast
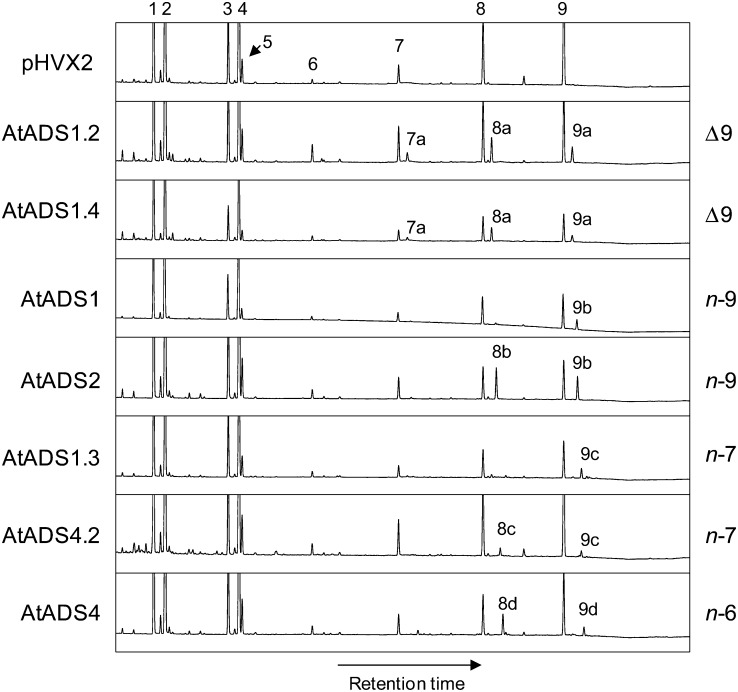
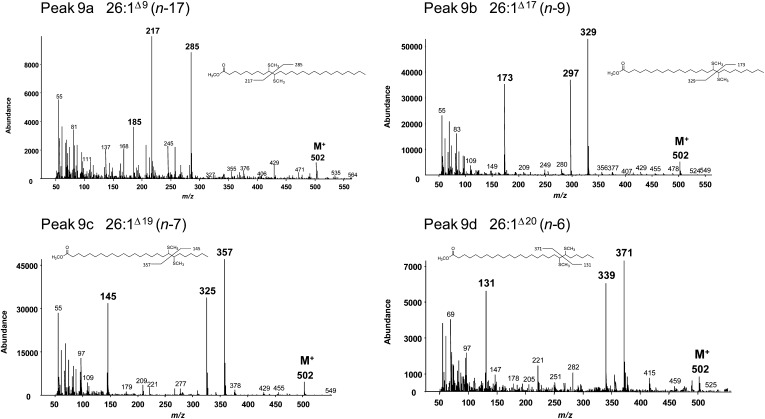
For characterization by expression in yeast, the ADS cDNAs were each cloned into an expression vector under control of a constitutive promoter. The vectors were used to transform a standard laboratory yeast strain YPH499 (Invitrogen). Analysis by gas chromatography (GC) of fatty acid methyl esters (FAMEs) prepared from total yeast lipids revealed a small amount (less than 1%) of a fatty acid identified by GC-mass spectrometry (MS) to be a monounsaturated C26 fatty acid (26:1) in cells expressing AtADS2 (data not shown). All vectors were subsequently used to transform a second yeast line (Fat1Δ) that was known to accumulate VLCFAs. Cells transformed with each the seven different vectors all showed the accumulation of VLCFAs that were not present in cells transformed with the empty vector (Fig. 3). GC-MS conducted on the FAMEs confirmed that the new VLCFAs were monounsaturated and double-bond positions were determined by GC-MS of the dimethyl disulfide (DMDS)-FAME adducts [double bond nomenclature is given as Δx relative to the carboxyl end of the molecule, or (n-x) relative to the methyl end of the molecule; Fig. 4; Supplemental Fig. S2]. Yeast cells expressing AtADS1 produced 26:1(n-9) as the predominant new fatty acid, with a very low amount of 24:1(n-9) also detected. Cells expressing AtADS2 produced both 24:1(n-9) and 26:1(n-9). These results suggested n-9 desaturase activity for AtADS1 and AtADS2. The novel VLCMUFAs in cells expressing AtADS1.2 and AtADS1.4 were identified as 22:1(n-13), 24:1(n-15), and 26:1(n-17), suggesting that these enzymes were able to catalyze desaturation at the Δ9 position. The novel VLCFAs in cells expressing AtADS4.2 were 24:1(n-7) and 26:1(n-7), whereas those in cells expressing AtADS4 were 24:1(n-6) and 26:1(n-6), suggesting n-7 and n-6 desaturase activity for these enzymes, respectively. Desaturation at the n-7 position was also observed in cells expressing AtADS1.3. These results indicated that when expressed in yeast, the ADS proteins could catalyze the formation of VLCMUFAs. Mild saponification of yeast expressing AtADS2 indicated that the VLCMUFAs were largely associated with the glycerolipid/free fatty acid fraction and were not present in significant amounts in the yeast sphingolipids (Supplemental Fig. S3). No hydroxylated C24 or C26 VLCMUFAs were observed, and when FAMEs were prepared from yeast by base catalyzed transesterification, only very low levels of VLCFAs were observed, suggesting that these fatty acids were largely present as free fatty acids.
Figure 3.
Gas chromatographs showing fatty acid profiles of yeast transformed with the empty vector pHVX2 and with the pHVX2 vector containing cDNAs encoding the ADS family members as indicated. Fatty acids: 1, 16:0; 2, 16:1Δ9; 3, 18:0; 4, 18:1Δ9; 5, 18:1Δ11; 6, 20:0; 7, 22:0; 8, 24:0; and 9, 26:0. Peaks: 7a, 22:1Δ9 (n-13); 8a, 24:1Δ9 (n-15); 9a, 26:1Δ9 (n-17); 8b, 24:1Δ15 (n-9); 9b, 26:1Δ17 (n-9); 8c, 24:1Δ17 (n-7); 9c, 26:1Δ19 (n-7); 8d, 24:1Δ18 (n-6); and 9d, 26:1Δ20 (n-6).
Figure 4.
Mass spectra of DMDS-FAMEs showing location of double bonds in the novel C26 fatty acids produced by yeast expressing the ADS family members. Diagnostic fragment ions are labeled in boldface. M+, Molecular ion.
Overexpression of AtADS1 of AtADS2 in Arabidopsis
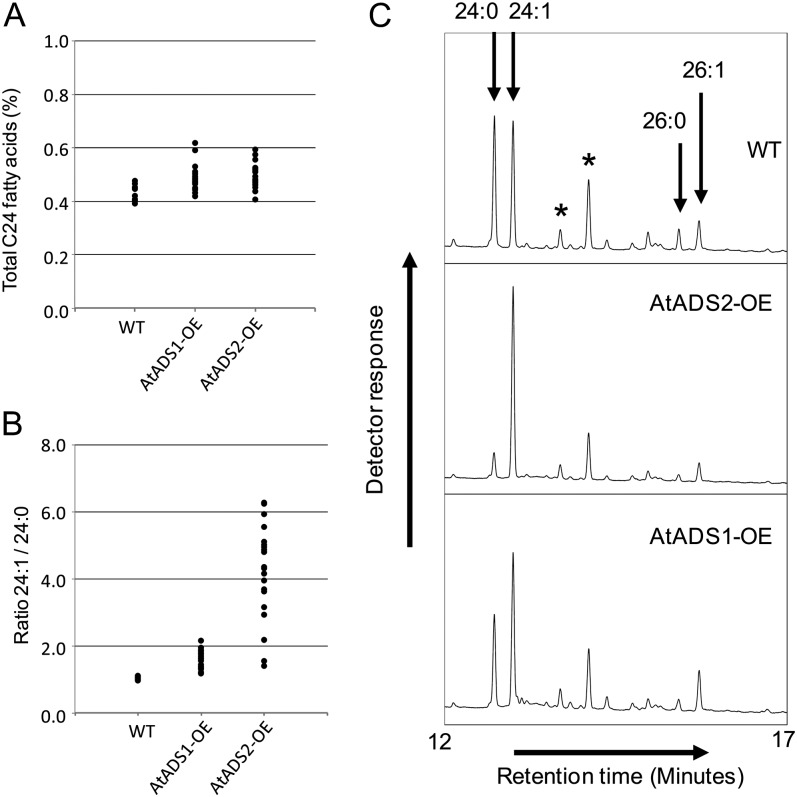
To determine whether the Arabidopsis ADS proteins could catalyze the synthesis of VLCMUFAs in a plant, Arabidopsis plants were transformed with a vector containing either AtADS1 or AtADS2 under control of the seed specific Brassica napus napin promoter. Presence of the transgene cassette was verified by PCR and transformants (T1 generation) were grown to maturity. Seed fatty acid composition was determined by GC of FAMEs prepared from total seed lipids (T2 seed). In wild-type Arabidopsis, C24 fatty acids accounted for between 0.4% and 0.5% of total seed fatty acids, with 24:0 and 24:1 being present in approximately equal amounts. The C26 fatty acids were present at levels too low for accurate quantification. Plants expressing the AtADS1 or AtADS2 cDNAs appeared to show, on average, very similar total levels of C24 fatty acids to the wild type (Fig. 5A). The ratios of 24:1 to 24:0 in lines overexpressing AtADS2 were, however, dramatically changed compared with the wild-type lines (Fig. 5B; Supplemental Fig. S4), with 24:1 becoming the predominant C24 fatty acid. Lines overexpressing AtADS1 showed a smaller change in the ratio of C24 fatty acids, but for most lines there was a clear increase in 24:1 and a reduction in 24:0. Close scrutiny of fatty acid profiles showed that in many lines overexpressing AtADS1 or AtADS2, 26:1 levels also appeared to be increased, with a corresponding decrease in 26:0 (Fig. 5C). Fatty acids with chain length less than C24 were not significantly different between the lines (Table II). Arabidopsis plants were also transformed with a construct containing AtADS2 under control of the constitutive Cauliflower mosaic virus (CaMV) 35S promoter. Expression of the transgene was verified by reverse transcription (RT)-PCR (Supplemental Fig. S5A). Analysis of seed fatty acid profiles showed changes in 24:0 and 24:1 levels similar to those when AtADS2 was expressed under control of the seed-specific promoter (Supplemental Fig. S5B). Seed fatty acid composition of line 13, used for further study, is given in Table II.
Figure 5.
Seed-specific OE of AtADS1 and AtADS2 in Arabidopsis. Total seed fatty acid composition was determined for T2 seeds from individuals of the population of untransformed (WT) plants, and plants transformed with vectors pMD123 (AtADS1-OE) and pMD124 (AtADS2-OE). A, Plot of total C24 fatty acids determined for individual plants. B, Plot of ratio of 24:1 to 24:0 determined for individual plants. C, Region of gas chromatograph showing C24 and C26 fatty acids for representative samples of the wild type (WT) and AtADS1 and AtADS2 OE lines. Peaks labeled with asterisks are unidentified components.
Table II. Seed fatty acid composition of wild-type Arabidopsis (Columbia-0), lines transformed with AtADS1 and AtADS2 under control of the napin promoter (lines MD123/5 [Napin-AtADS1 OE5] and MD124/10 [Napin-AtADS2 OE10], respectively), and a line transformed with AtADS2 under control of the CaMV 35S promoter (line CaMV 35S-AtADS2 OE13).
Data are presented as percentage ± sd (n = 5). Wild-type plants (Columbia-0) and OE lines were grown together, but in separate pots. The ads T-DNA insertion lines and wild-type 2 plants (also Columbia-0) were grown as mixed plants in randomized pots to reduce the effect of pot-to-pot variation.
| Line | Fatty Acid |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 16:0 | 16:1Δ9 | 18:0 | 18:1Δ9 | 18:1Δ11 | 18:2 | 18:3 | 20:0 | 20:1Δ11 | 20:1Δ13 | 20:2 | 20:3 | 22:0 | 22:1Δ13 | 24:0 | 24:1 | 24:1/24:0 | |
| % | |||||||||||||||||
| Wild type | 7.49 ± 0.09 | 0.25 ± 0.04 | 3.53 ± 0.05 | 11.30 ± 0.29 | 1.23 ± 0.01 | 27.28 ± 0.01 | 20.19 ± 0.16 | 2.89 ± 0.04 | 18.76 ± 0.48 | 1.56 ± 0.08 | 2.09 ± 0.04 | 0.57 ± 0.01 | 0.38 ± 0.02 | 2.02 ± 0.12 | 0.22 ± 0.02 | 0.22 ± 0.02 | 1.01 ± 0.04 |
| Napin-AtADS1 OE5 | 7.91 ± 0.36 | 0.32 ± 0.03 | 3.62 ± 0.19 | 12.14 ± 0.97 | 1.38 ± 0.06 | 26.35 ± 0.44 | 20.65 ± 0.72 | 2.72 ± 0.12 | 17.85 ± 0.69 | 1.86 ± 0.12 | 1.87 ± 0.10 | 0.54 ± 0.04 | 0.40 ± 0.02 | 1.88 ± 0.13 | 0.20 ± 0.01 | 0.32 ± 0.04 | 1.65 ± 0.12 |
| Napin-AtADS2 OE10 | 7.64 ± 0.04 | 0.26 ± 0.04 | 3.55 ± 0.04 | 11.33 ± 0.20 | 1.28 ± 0.01 | 27.36 ± 0.20 | 20.52 ± 0.16 | 2.84 ± 0.04 | 18.05 ± 0.19 | 1.56 ± 0.01 | 2.06 ± 0.02 | 0.57 ± 0.01 | 0.38 ± 0.01 | 2.01 ± 0.02 | 0.07 ± 0.01 | 0.51 ± 0.01 | 7.05 ± 0.38 |
| CaMV 35S- AtADS2 OE13 | 7.85 ± 0.02 | 0.28 ± 0.01 | 2.96 ± 0.06 | 13.17 ± 0.32 | 1.38 ± 0.02 | 26.8 ± 0.08 | 19.58 ± 0.24 | 1.86 ± 0.02 | 19.35 ± 0.13 | 1.80 ± 0.04 | 1.90 ± 0.02 | 0.51 ± 0.01 | 0.30 ± 0.01 | 1.76 ± 0.05 | 0.12 ± 0.01 | 0.37 ± 0.01 | 3.05 ± 0.21 |
| Wild type 2 | 7.96 ± 0.17 | 0.27 ± 0.02 | 2.97 ± 0.08 | 12.93 ± 1.25 | 1.33 ± 0.03 | 27.93 ± 0.70 | 19.11 ± 0.74 | 1.98 ± 0.12 | 18.93 ± 0.09 | 1.73 ± 0.07 | 1.89 ± 0.13 | 0.50 ± 0.05 | 0.31 ± 0.02 | 1.77 ± 0.15 | 0.20 ± 0.01 | 0.20 ± 0.01 | 1.00 ± 0.04 |
| ads1-1 | 7.51 ± 0.15 | 0.29 ± 0.02 | 2.74 ± 0.14 | 15.09 ± 0.60 | 1.46 ± 0.03 | 26.81 ± 0.18 | 19.60 ± 0.15 | 1.89 ± 0.14 | 18.05 ± 0.41 | 1.92 ± 0.12 | 1.64 ± 0.07 | 0.47 ± 0.02 | 0.30 ± 0.02 | 1.81 ± 0.10 | 0.21 ± 0.01 | 0.22 ± 0.02 | 1.02 ± 0.03 |
| ads1-2 | 7.83 ± 0.10 | 0.28 ± 0.01 | 2.80 ± 0.01 | 13.07 ± 0.63 | 1.33 ± 0.03 | 27.04 ± 0.16 | 19.98 ± 0.61 | 1.98 ± 0.04 | 18.91 ± 0.24 | 1.77 ± 0.02 | 1.89 ± 0.05 | 0.54 ± 0.03 | 0.31 ± 0.01 | 1.88 ± 0.04 | 0.20 ± 0.01 | 0.21 ± 0.01 | 1.04 ± 0.05 |
| ads2-1 | 7.83 ± 0.21 | 0.27 ± 0.02 | 2.99 ± 0.04 | 13.30 ± 1.02 | 1.33 ± 0.05 | 27.87 ± 0.12 | 19.09 ± 0.69 | 1.90 ± 0.09 | 19.19 ± 0.32 | 1.63 ± 0.06 | 1.83 ± 0.08 | 0.48 ± 0.04 | 0.29 ± 0.02 | 1.68 ± 0.07 | 0.27 ± 0.02 | 0.07 ± 0.01 | 0.25 ± 0.02 |
| ads2-2 | 7.86 ± 0.14 | 0.26 ± 0.02 | 2.99 ± 0.08 | 13.13 ± 0.93 | 1.30 ± 0.03 | 28.08 ± 0.15 | 19.04 ± 0.58 | 1.95 ± 0.09 | 19.04 ± 0.19 | 1.61 ± 0.07 | 1.87 ± 0.08 | 0.49 ± 0.04 | 0.30 ± 0.01 | 1.73 ± 0.11 | 0.28 ± 0.01 | 0.07 ± 0.01 | 0.24 ± 0.01 |
| ads2-3 | 7.95 ± 0.12 | 0.28 ± 0.01 | 2.96 ± 0.02 | 13.86 ± 0.92 | 1.35 ± 0.04 | 27.69 ± 0.18 | 18.83 ± 0.49 | 1.88 ± 0.07 | 18.88 ± 0.41 | 1.73 ± 0.08 | 1.77 ± 0.07 | 0.47 ± 0.03 | 0.30 ± 0.02 | 1.65 ± 0.09 | 0.15 ± 0.01 | 0.26 ± 0.01 | 1.71 ± 0.03 |
Characterization of Arabidopsis T-DNA Insertion Alleles of AtADS1 and AtADS2
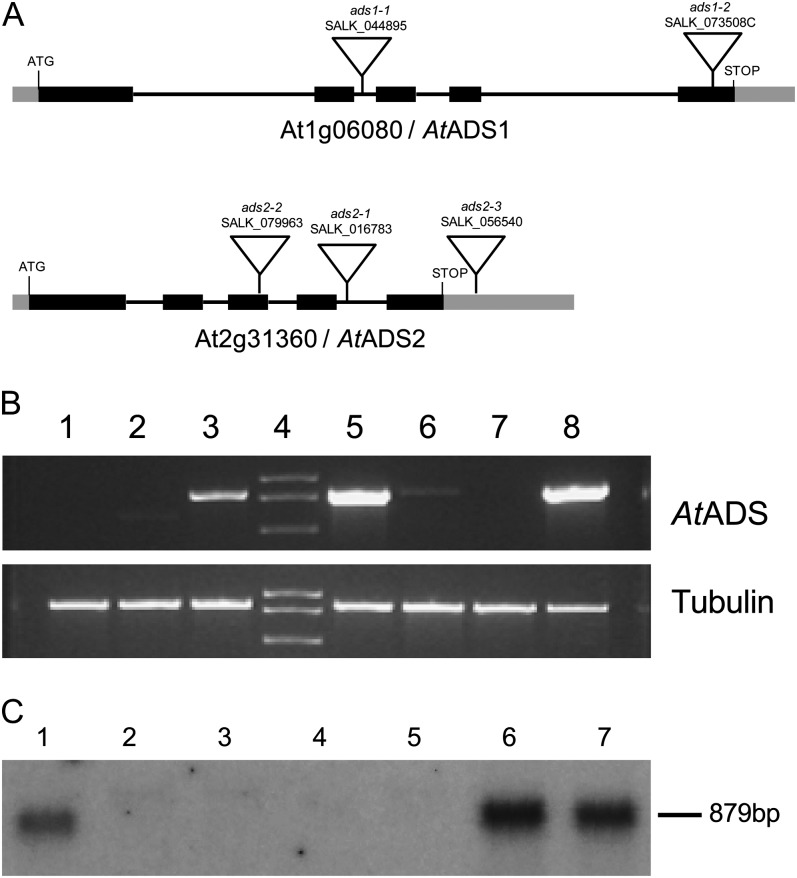
To examine the role of AtADS1 and AtADS2 by a reverse genetics approach, transfer DNA (T-DNA) insertion lines (Fig. 6a) were obtained from the Arabidopsis Biological Resource Center. Homozygous plants were identified by PCR (Supplemental Fig. S6A) and T-DNA insertion positions were verified by sequencing. For ads1-1 (SALK_044895C), the T-DNA insertion position was approximately 1,143 bp downstream of the start codon, and was located in the second intron. The second allele, ads1-2 (SALK_073508C), appeared to contain a concatemeric T-DNA insert in the fifth exon between 2,348 and 2,400 bp downstream of the start codon. Two of the three ads2 alleles also appeared to contain reversed concatemeric T-DNA insertions, allowing PCR products to be generated using the LBb1.3 primer and gene-specific primers designed to bind on either side of the T-DNA insertion site (reverse and forward primers). For ads2-1 (SALK_016783), the insertion site was located at approximately 1,060 bp downstream of the start codon, in the fourth intron. The ads2-2 (SALK_079963) allele contained an insertion toward the end of the third exon, between 811 and 831 bp downstream of the start codon. Insertion allele ads2-2 (SALK_056540) contained a T-DNA insertion in the 3′ untranslated region approximately 209 bp downstream of the stop codon. To determine whether the T-DNA insertion lines lacked ADS transcript of the correct length, RT-PCR was conducted using cDNA generated from seedling leaf total RNA (Fig. 6B; Supplemental Fig. S6B). Both AtADS1 and AtADS2 could be amplified from cDNA prepared from wild-type material, indicating that the genes were expressed in this tissue under normal growing conditions. Using primer pairs designed to amplify the entire coding region, no AtADS1 transcript was detected in the two ads1 T-DNA insertion lines, ads1-1 and ads1-2. Similarly, AtADS2 transcript could not be detected in plants homozygous for the ads2-1 and ads2-2 alleles. Results indicated that the ads2-3 allele was not a knockout allele because transcript was easily detected in the homozygous plants. In RT-PCR, using primer pairs that preceded the T-DNA insertion sites, a fragment could be amplified from all of the ads2 alleles, but no significant product was observed in the ads1 alleles (Supplemental Fig. S6B). To further characterize the ads2 alleles we therefore conducted northern-blot analysis using RNA extracted from plants homozygous for the individual ads2 alleles (Fig. 6C). Results indicated that transcript was not present at significant levels in the ads2-1 and ads2-2 alleles and also suggested that AtADS2 transcript level in the ads2-3 plants was likely higher than in wild-type plants. No obvious growth phenotype was seen for the T-DNA insertion lines when grown under continuous light at 23°C. We observed overall size and appearance of the plants, and also time to flowering and maturity.
Figure 6.
Characterization of ads1 and ads2 T-DNA insertion alleles. A, Gene structure and T-DNA insertion sites. Black boxes indicate exons, black lines indicate introns, and gray boxes indicate untranslated regions of the cDNA. B, RT-PCR showing gene expression. Template is cDNA generated from total RNA extracted from rosette leaves. Lane 1, ads1-2; lane 2, ads1-1; lanes 3 and 5, wild type; lane 6, ads2-1; lane 7, ads2-2; lane 8, ads2-3. Primer pairs: Top, RTADS1F2 and RTADS1R2 for lanes 1 to 3 and RTADS2F and RTADS2R for lanes 5 to 8. Bottom, Tubulin β-7 chain (At2g29550), primers AtTub2For and AtTub2Rev. C, Northern blot using AtADS2 cDNA as a probe. Lane 1, Wild type; lanes 2 and 3, ads2-1; lanes 4 and 5, ads2-2; lanes 6 and 7, ads2-3.
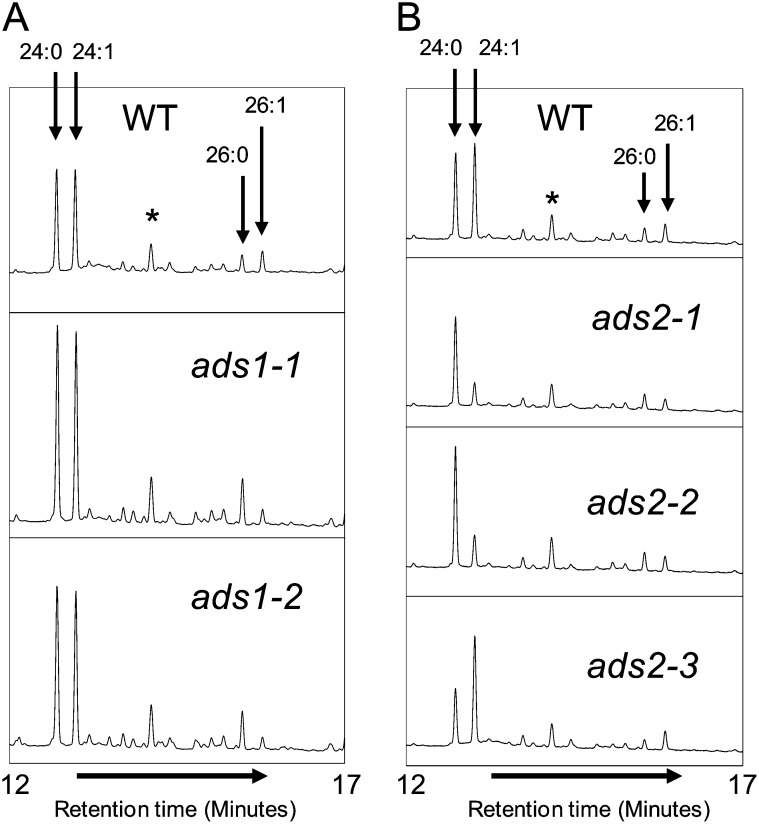
Plants homozygous for each mutant allele were grown to maturity and seed fatty acid composition was determined (Table II). As shown in detail in Figure 7B, ads2-1 and ads2-2 plants had lower levels of 24:1 in their seeds compared with control lines, with 24:0 being the predominant C24 fatty acid. Seeds from ads2-3 plants appeared to have slightly increased 24:1 levels. Examination of the ads1-1 and ads1-2 lines showed little difference in the levels or ratios of 24:0 and 24:1 compared with wild-type lines. Small differences were, however, observed in the C26 fatty acids (Fig. 7A) with the mutant lines and a change in the ratio of 26:0 and 26:1 was evident, with 26:0 becoming the predominant C26 fatty acid. Other seed fatty acids were not significantly different between the lines, with the exception of 18:1Δ9 in ads1-1, which showed a slight increase compared with the wild type. These results further implicated AtADS2 and AtADS1 in the synthesis of C24 and C26 monounsaturated fatty acids in the seed.
Figure 7.
Gas chromatographs showing C24 and C26 FAMEs prepared from mature seeds of wild-type (WT), ads1, and ads2 T-DNA insertion alleles. The peaks labeled with asterisks are unidentified components.
Lipidomic Studies
For a comparative examination of Arabidopsis leaf lipids, quantitative lipidomic analysis was conducted on total lipid extracts prepared from young rosettes. Data were collected from two separate sets of plants grown at different times. Although in experiment 1 lipids were extracted using a protocol not optimized for sphingolipid extraction, this lipid class was detected in significant amounts and data were collected and are presented.
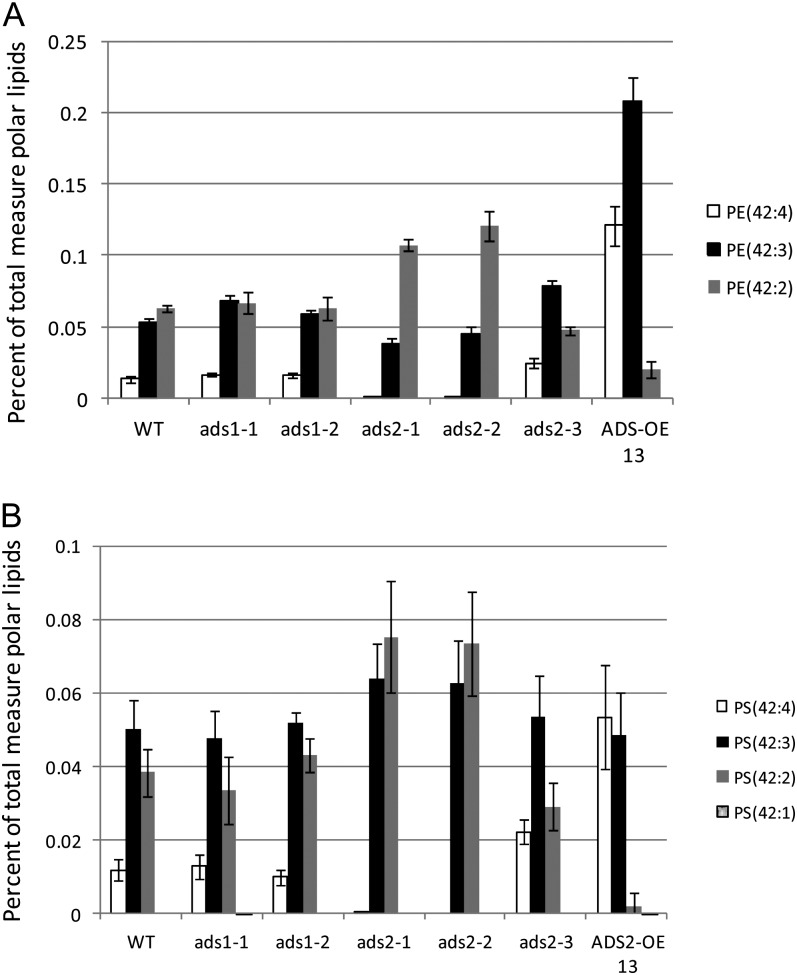
Comparison of the phospholipid molecular species in wild-type and experimental lines, within data from experiment 1, revealed significant differences in phosphatidylethanolamine (PE) and phosphatidyl-Ser (PS) species containing C24 VLCFAs in the ads2 T-DNA insertion lines and the ADS2 overexpression (OE) line (Fig. 8). No significant differences between the wild type and the ads1 T-DNA insertion lines were observed. Total PE content in wild-type plants was close to 5.2% of leaf phospholipids (equivalent to around 12.8 nmol mg–1 dry weight) with C42 PE molecular species totaling 0.13% and PE(42:2) and PE(42:3) being most abundant. The ads2-1 and ads2-2 plants contained PE(42:2) as the major C42 species (Fig. 8A), with significantly reduced levels of PE(42:3) and almost no PE(42:4). Compared with the wild type, the ads2-3 lines showed an increase in PE(42:3) and PE(42:4), with lower levels of PE(42:2). A dramatic increase in the more unsaturated C42 PE species was seen in the ADS2 OE line with PE(42:3) and PE(42:4) becoming the dominant molecular species. This line also showed a relative increase in C42 PE, with these species accounting for 0.35% of total leaf phospholipids and a slight decrease in C40 PE (0.059%–0.049%), corresponding to a reduction in PE(40:2) (Supplemental Fig. S7). A similar trend was seen when C42 PS species were examined (Fig. 8B; Supplemental Fig. S8). PS is a low abundance lipid in Arabidopsis leaf accounting for only 0.3% (or approximately 0.8 nmol mg–1 dry weight) of total leaf phospholipids in wild-type lines, with the dominant molecular species being PS(42:3) and PS(42:2). No significant increase in total PS content was seen in any of the lines examined, however the ads2-1 and ads2-2 lines showed significantly increased levels of PS(42:2) and an almost complete loss of PS(42:4). In the ads2-3 plants there was an increase in the relative amount of PS(42:4), and a corresponding decrease in percentage of PS(42:2). The AtADS2 OE line contained PS(42:4) as the dominant C42 PS species, with very much reduced PS(42:2) levels. PS composition of the ads1 T-DNA insertion lines resembled that of the wild-type plants. Data from the second experiment, comparing the wild type, ads2-1, and the ADS2 OE13 lines, showed similar trends. Changes in PS species that contain C26 fatty acid, PS(44:3) and PS(44:2), were observed (Supplemental Fig. S8), but due to the very low abundance of these lipids, significance could not be demonstrated.
Figure 8.
C42 molecular species of (A) PE and (B) PS in young rosettes of the wild type (WT), ads T-DNA insertion lines and AtADS2 OE13. The data are given as average percentage of total measured phospholipids ± sd (n = 5). Lipid nomenclature is A:B, where A is the total acyl carbons and B is total number of double bonds.
Data were also collected for the major anionic and neutral sphingolipid classes, the glycosylinositolphosphoceramides (GIPCs) and monohexosylceramides (HexCers), respectively. Experiment 1 included the wild type, both ads1 alleles, three ads2 alleles, and the ADS2 OE13 line, whereas experiment 2 was limited to the wild type, ads2-1, and ADS2 OE13. Because the extraction procedure for experiment 1 was not optimized for sphingolipid recovery, we compared relative extraction of sphingolipids between the two experiments. Comparison revealed that substantial recovery of sphingolipids had been achieved in experiment 1 (Supplemental Table S1). HexCer, for example, was measured at 150 normalized signal mg–1 dry weight (approximately equivalent to 150 nmol mg–1 dry weight) in wild-type tissue in the first extraction, compared with 107 normalized signal mg–1 dry weight in experiment 2, and GIPC was 2.6 and 3.2 normalized signal mg–1 dry weight, respectively. As data from experiment 1 cannot be considered quantitative, particularly for the less easily extracted GIPCs, results are expressed as percentages for each lipid class.
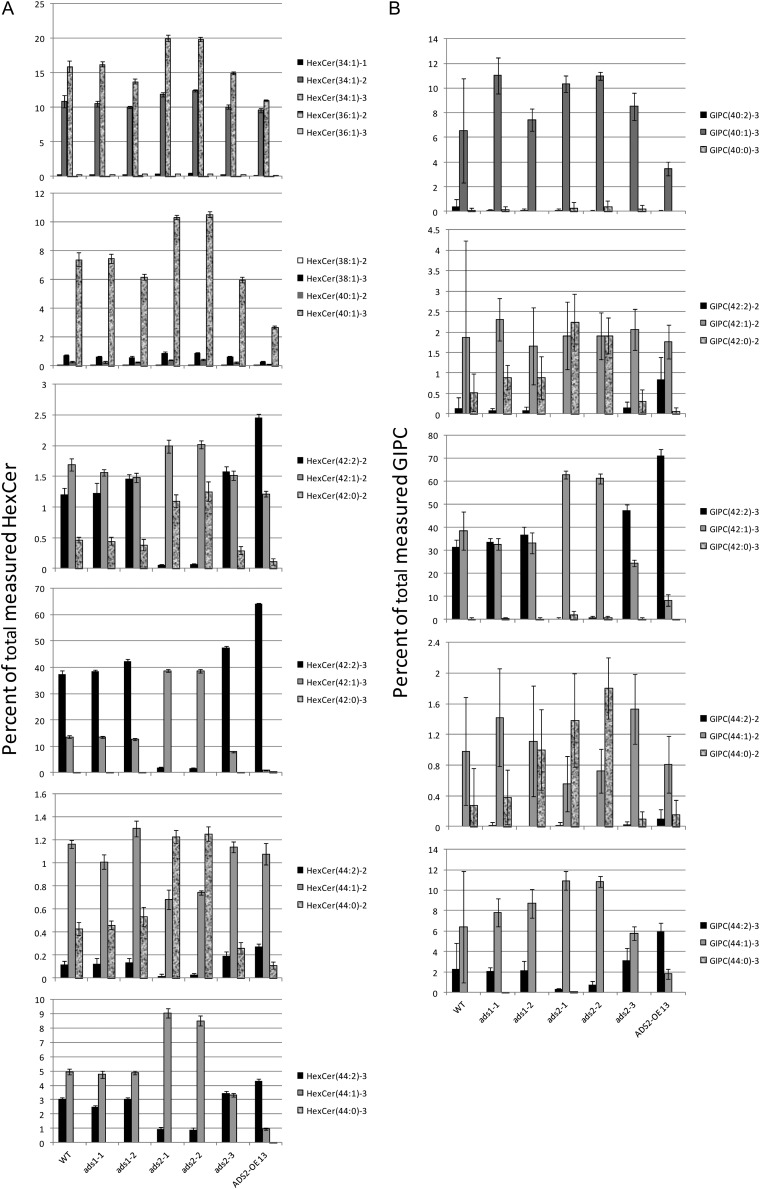
Results from the sphingolipid analysis revealed a dramatic difference in the composition of the C42 and C44 HexCers and GIPCs in the ads2 alleles and in the ADS2 OE line (Fig. 9). In contrast, the ads1 lines did not differ significantly from the wild type. For the trihydroxy C42 and C44 HexCer classes, the ads2-1 and ads2-2 plants showed a shift toward more saturated species with HexCer(42:2)-3 and HexCer(44:2)-3 being very much reduced, and HexCer(42:1)-3 and HexCer(44:1)-3 showing corresponding increases (Fig. 9A). The ads2-3 line had a composition similar to the wild-type plants but with a slightly higher percentage of (42:2)-3. ADS2 OE plants showed a shift toward a higher level of unsaturation with an increase in the percentage of (42:2)-3 species. Compared with the trihydroxy C42 and C44 HexCers, the dihydroxy species appeared enriched in fully saturated species. For the C42 HexCer, ads2-1 and ads2-2 showed an almost complete absence of (42:2)-2, a slight increase in (42:1)-2, and a substantial increase in (42:0)-2. The ads2-3 plants showed a slight increase in (42:2)-2, whereas in the over-expressing line HexCer (42:2)-2 was the most abundant lipid in the dihydroxy C42 class, comprising almost 2.5% of total HexCer compared with 1.2% in the wild type. The increase in saturated species was more obvious in the C44 HexCer class, with both (44:4)-2 and (44:1)-2 being reduced and (44:0)-2 increasing from close to 0.5% to over 1.2% in the ads2-1 and ads2-2 plants. For the GIPCs (Fig. 9B), the more abundant trihydroxy classes showed almost complete loss of (42:2)-3 and (44:2)-3 species and corresponding increases in (42:1)-3 and (44:1)-3 in the ads2-1 and ads2-2 plants. The ads2-3 and AtADS2 OE line both showed an increase in (42:2)-3 and (44:2)-3 and decrease in (42:1)-3 and (44:1)-3, with the changes being more pronounced in the AtADS2 OE line. Data from the less abundant dihydroxy GIPCs contained a large amount of variability. Clear trends in sphingolipid composition could be seen, but statistical significance could not be demonstrated. For the C42 dihydroxy GIPC class, a virtually complete loss of (42:2)-2 was seen in the ads2-1 and ads2-2 material. This was accompanied by a large increase in (42:0)-2. For the ADS2 OE line, a substantial increase in (42:2)-2 was seen. The trends seen in the data from experiment 1 were confirmed by data from the second experiment (Supplemental Fig. S9). OE, or loss of AtADS2 activity, clearly had a profound effect on the levels of C24 monounsaturated fatty acids in Arabidopsis leaf.
Figure 9.
Sphingolipid composition, as determined by electrospray ionization tandem MS, of leaves of wild-type (WT) Arabidopsis, ads alleles and AtADS2 OE13. Lipids were extracted from young rosettes and analyzed as described in “Materials and Methods.” The data are from experiment 1 and are given as an average percentage of total measured sphingolipid class ± sd (n = 5). Lipid nomenclature is A(B:C)-d, where A is the sphingolipid class, B is the total carbons in the fatty amide plus base, C is the number of double bonds in the fatty acid or base, and D is the number of free hydroxyl groups.
Acyl-CoA Profiling
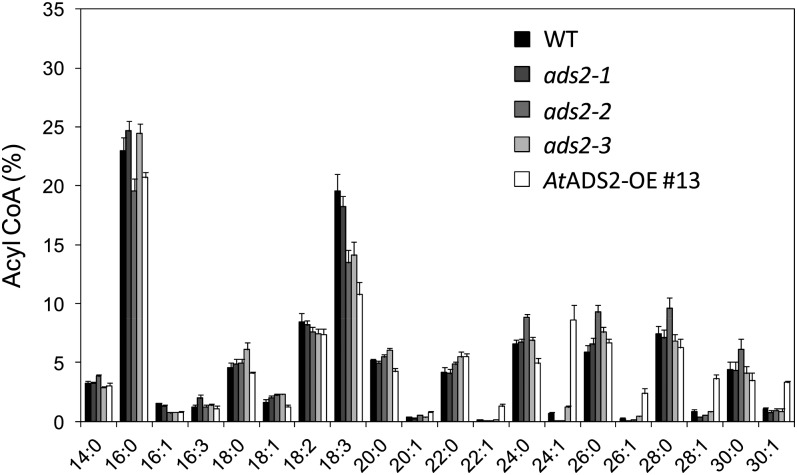
Lipidomic studies conducted on leaf glycerolipids and sphingolipids were complemented by acyl-CoA profiling (Fig. 10), using young rosettes collected 14 d after germination. In wild-type plants, acyl-CoAs with chain lengths from 14 to 30 carbons were detected, with 16:0-CoA (23%) and 18:3-CoA (20%) being the most abundant and saturated VLCFAs (VLCSFAs) dominating the VLCFA acyl-CoA profile. Although some variation was apparent in 16:0-CoA and 18:3-CoA, the most dramatic differences were observed in the VLCMUFAs with chain lengths of C22 to C30 (Fig. 10; Supplemental Fig. S10). These fatty acids were relatively minor components of the acyl-CoA pool in wild-type material, each present at levels of around 1% or less. Compared with the wild-type control, the AtADS2 OE line showed a 12-fold increase in 24:1-CoA, with levels approaching 9% of total measured leaf acyl-CoAs. A 10-fold increase in 26:1-CoA and increases in 28:1-CoA and 30:1-CoA of 4- and 3-fold, respectively, were also seen. The percentage of 22:1, below 0.1% in the wild type, was also increased in this line. The percentage of VLCSFAs in the OE line showed little correlation to the pattern observed in the VLCMUFA profile, with only 24:0 showing a modest but significant reduction. Examination of the VLCMUFA composition of the acyl-CoAs from the ads2 T-DNA insertion lines (enlarged in Supplemental Fig. S10) showed no significant change in the percentage of 22:1-CoA or 30:1-CoA. Levels of 26:1 and 28:1 were slightly reduced, compared with the wild type, in the ads2-1 and ads2-2 plants, but not in the ads2-3 line. Percentage of 24:1-CoA in the ads2-1 and ads2-2 plants was reduced from 0.7% to 0.09% and 0.06%, respectively, whereas in the ads2-3 plants the average 24:1-CoA content was increased to 1.2%. In this study, the ads2-2 plants appeared to show a small increase in levels of saturated VLCFA-CoAs when compared with the other lines.
Figure 10.
Acyl-CoA profiles of seedlings 14 d after germination from wild-type (WT) Arabidopsis, ads2 alleles, and ADS2 OE13 (AtADS2-OE #13). Results are expressed as mol % of total acyl-CoAs measured (± se), with four biological replicates for each line.
DISCUSSION
The Arabidopsis ADS gene family encodes a group of highly related proteins that have characteristic membrane bound fatty acid desaturase motifs. Their chromosomal location and homology suggests that they likely arose through a series of gene duplication events. A search of the genome of Arabidopsis lyrata uncovered close homologs of seven of the nine Arabidopsis ADS genes (Supplemental Table S2). No homologs were found for AtADS3.2 or AtADS1.4. Comparison of gene arrangement between the two species indicates that gene order of ADS1, ADS1.2, and ADS1.3 is conserved, but a homolog of AtADS1.4 is absent in A. lyrata (Supplemental Fig. S11). As AtADS1.2 and AtADS1.4 have the same catalytic activity and are the most conserved members of the gene family, it seems plausible that AtADS1.4 may have arisen from duplication of AtADS1.2 after the divergence of the two Arabidopsis species. No homologs of any ADS genes were found in the genomes of rice (Oryza sativa), Brachypodium distachyon, or maize (Zea mays), suggesting that ADS genes may be absent in monocots.
Previous studies aimed at determining enzyme activity using yeast expression gave evidence that AtADS1 and AtADS2 were capable of catalyzing Δ9 or Δ7 desaturation of C16 and C18 saturated fatty acids. Our work in yeast has revealed that significant accumulation of VLCMUFAs is seen in yeast cells expressing any of the seven Arabidopsis nonchloroplast targeted ADS desaturases. With the exception of 26:0, VLCFAs are not present at significant levels in normal laboratory yeast strains and are easily overlooked. Use of a mutant strain (Fat1Δ) that accumulates VLCSFAs was necessary to effectively characterize the activity of the ADS proteins. The Fat1Δ yeast strain has an insertion in the gene encoding the fatty acid transport protein FAT1 (YBR041W). FAT1 encodes an acyl-CoA synthetase required for both the import of long-chain fatty acids into the yeast cell, and the activation of VLCFAs (Faergeman et al., 1997; Watkins et al., 1998; Zou et al., 2002, 2003).
The VLCMUFAs produced in yeast differed in double-bond position, with AtADS1 and AtADS2 associated with n-9 fatty acids, AtADS1.3 and AtADS4.2 associated with n-7 fatty acids, AtADS4 with n-6 fatty acids, and AtADS1.2 and AtADS1.4 with Δ9 fatty acids. We were not able to directly assay desaturase activity of these enzymes on VLCSFAs in vitro, and the inability of the Fat1Δ yeast cells to effectively take up exogenous VLCSFAs did not allow for feeding studies to be carried out. The evidence, however, strongly suggests that the VLCMUFAs result from desaturase activity on VLCSFA substrates. Yeast cells do not readily elongate monounsaturated fatty acids beyond C18:1 and it is unlikely that the novel VLCMUFAs are elongation products of novel monounsaturated C16 and C18 fatty acids. For example, n-9 fatty acids are already abundant in yeast as 18:1Δ9 (n-9) is the major C18 monounsaturated fatty acid, but n-9 VLCMUFAs are not seen in the lipids of untransformed yeast. In addition, both AtADS1.2 and AtADS1.4 are associated with production of C24 and C26 Δ9 unsaturated fatty acids. If these were a result of an elongation pathway, the position of double bonds would be constant relative to the methyl end of the molecule (n-x) because elongation occurs by addition to the carboxyl end of the fatty acid. Finally, desaturase activity on VLCFAs has previously been reported for ADS like proteins. Δ5 desaturase activity on VLCSFAs was demonstrated for ADS proteins from L. alba and A. leveillei (Cahoon et al., 2000; Sayanova et al., 2007). A related desaturase from the fungus Mortierella alpina has also been reported to catalyze synthesis of 26:1 (MacKenzie et al., 2002). Positional diversity of double-bond insertion catalyzed by this class of closely related proteins offers a unique opportunity to explore the mechanisms controlling regiospecificity of membrane-bound fatty acid desaturases.
Subcellular localization studies conducted with AtADS1 and AtADS2 indicate that both proteins are located in the ER and contain a putative di-Arg retention/retrieval motif. The true subcellular location of the other five ADS proteins is uncertain because the potential di-Lys motif is atypical (-KXKXXXXX, rather than -KXKXX; Benghezal et al., 2000). The lack of localization of N-terminal tagged proteins suggests that this region may be important in protein targeting to the ER. Localization studies conducted with the plant fatty acid desaturases FAD2 (oleate 12-desaturase; At3g12120) and FAD3 (linoleate desaturase; At2g29980) have shown that both are inserted into the ER membrane cotranslationally with a noncleavable signal sequence likely contained within the first transmembrane helix (McCartney et al., 2004). In common with FAD2 and FAD3, the ADS proteins contain putative transmembrane domains close to the N terminus. Further work is required to determine the targeting and subcellular location of the ADS proteins.
To attempt to determine a function for the ADS desaturases in Arabidopsis we focused on AtADS1 and AtADS2, using Arabidopsis T-DNA insertion lines and plants engineered to overexpress AtADS2 under control of either a seed-specific promoter or a constitutive promoter. We chose three T-DNA insertion alleles of AtADS2 and discovered that while ads2-1 and ads2-2 plants lack detectable full-length AtADS2 transcript, transcript can be detected in the plants homozygous for the ads2-3 allele. The ads2-3 allele contains an insertion in the AtADS2 3′ untranslated region. The AtADS2 transcript in the ads2-3 plants appears slightly larger than in the control plants. Data from the lipidomics studies and acyl-CoA profiling also suggest that the ads2-3 allele may exhibit a slight increase in AtADS2 activity. Further work is in progress to explore these observations.
In Arabidopsis, C24 and C26 VLCFAs are a minor component of seed lipids, but are also found in the membrane PE and PS, in sphingolipids, as precursors and components of cuticular and epicuticular waxes, and in suberin (Li-Beisson et al., 2010). As previous work reported changes in seed lipid composition in Arabidopsis fab1/fae1 lines overexpressing AtADS2 (Heilmann et al., 2004a), we first examined seed lipids and observed dramatic and reciprocal alterations in 24:0 to 24:1 ratios in lines overexpressing AtADS2 and in AtADS2 T-DNA insertion alleles. These results suggested that the primary effect of AtADS2 OE or down-regulation was the alteration of VLCMUFA content. Our OE work is not directly comparable with the previous study because we used the Arabidopsis Columbia-0 line for transformation rather than the fab1/fae1 line.
More conclusive evidence for the role of AtADS2 in VLCFA metabolism came from the lipidomics studies conducted on total lipids extracted from young rosettes. Results from RT-PCR and northern blot demonstrate that there is substantial expression of AtADS2 in this tissue. From the lipidomics data it can be seen that the ads2-1 and ads2-2 plants are almost completely devoid of the PE(42:4) and PS(42:4) lipid species. The studies of Devaiah et al. (2006) have shown that the two fatty acids present in these phospholipid molecules are 24:1 and 18:3. These lipid species also show increased relative percentage in the ADS2 OE line and a moderate increase in ads2-3. Furthermore, the percentage of 42:2 lipid species containing 24:0 and 18:2 are increased in ads2-1 and ads2-2 and decreased in the OE line. The ADS2 alleles lacking transcript therefore appear deficient in 24:1, with increased 24:0, demonstrating that AtADS2 plays a role in the biosynthesis of VLCMUFAs required for Arabidopsis phospholipids.
Sphingolipids are a major component of plant cells and are estimated to account for over 40% of the lipids in the plasma membrane (Sperling et al., 2005). Arabidopsis contains a relatively simple and well-characterized complement of sphingolipids, with GIPC and HexCer species accounting for 64% and 34% of total leaf sphingolipids, respectively (Markham et al., 2006; Markham and Jaworski, 2007). The long-chain base component is predominantly trihydroxy 18:1 with a Δ8 double bond. (Markham et al., 2006). Sphingolipid fatty acids range from C16 to C26, saturated or monounsaturated, with a double bond at the n-9 position and a hydroxyl group at the α (C2) position (Imai et al., 2000). Examination of sphingolipid composition in the AtADS2 OE and ads2 plants produced results consistent with those of the phospholipid studies. In the ads2-1 and ads2-2 plants, sphingolipids containing 24:1(n-9) [HexCer(42:2)-2, HexCer(42:2)-3, GIPC(42:2)-2, and GIPC(42:2)-3], were almost completely absent, and species containing 24:0 [HexCer(42:0)-2 and GIPC(42:0)-2] were increased. For the two most abundant sphingolipid classes, GIPC(42)-3 and HexCer(42)-3, a slight change in total percentage composition was only observed for the latter, with the ads2-1 and ads2-2 lines showing around 20% reduction in C42-trihydroxy HexCer. The AtADS2 OE line showed an increase in percentage of species containing 24:1 and a reduction of sphingolipids with 24:0 as the acyl group. The yeast results indicate that AtADS2 is active on both 24:0 and 26:0 and, consistent with this, we also observed a reduction in sphingolipid species containing 26:1(n-9) [HexCer(44:2)-2, HexCer(44:2)-3, and GIPC(44:2)-3] in the ads2-1 and ads2-2 plants and an increase in the ADS2 OE line.
The most plausible explanation for the observed compositional changes in these lipids is that lack of ADS2 activity largely removes 24:1 and 26:1 from the acyl-CoA pool that would be used for sphinganine acylation, sphingolipid remodeling, and biosynthesis of the VLCFA-containing phospholipids. The data from the acyl-CoA profiling support this suggestion, in that 24:1-CoA is only present at very low levels in ads2-1 and ads2-2 plants, and the percentage of 26:1-CoA is also reduced. Furthermore, the acyl-CoA pool in the OE line is clearly greatly enriched in VLCMUFAs. Acyl-CoA profiling, however, only gives a total measurement of the acyl-CoAs present in the plant tissue at the time of extraction. This pool is considered to be highly dynamic and represents the sum of acyl-CoAs likely involved in multiple metabolic pathways. For example, the relatively high levels of C28 and C30 fatty acids, which are not a major component of phospholipids and sphingolipids, suggest that acyl-CoA intermediates of cuticular lipid biosynthesis are represented in this profile.
AtADS2 clearly plays a major role in VLCFA metabolism in Arabidopsis; the role of AtADS1, however, remains uncertain. The enzyme showed activity when expressed in yeast, and small changes were seen in seed VLCFAs when AtADS1 was overexpressed or down-regulated by T-DNA insertion. By RT-PCR we were able to detect AtADS1 expression in the rosette tissue used for the lipidomics study, but the two T-DNA insertion alleles did not show significant differences in measurable phospholipid or sphingolipid species. Although the overlap in enzyme activity suggests that enzyme redundancy may be possible, the fatty acid phenotype of the ads2 T-DNA insertion alleles indicates that AtADS1 and AtADS2 probably do not have the same functional role in Arabidopsis rosette leaves.
The acyl-lipid substrate of the Arabidopsis ADS enzymes remains to be determined, as does their ability to utilize substrates with chain length longer than C26. The ADS enzymes have been suggested to desaturate acyl-CoA, but conclusive evidence has not yet been reported. The results from the yeast expression work suggest that these enzymes likely are not active on yeast sphingolipids, as VLCMUFAs were largely absent from this lipid class. The relative abundance of VLCMUFAs in the acyl-CoA pool in Arabidopsis overexpressing AtADS2 suggests that acyl-CoA may be a substrate for desaturation, but desaturation on a glycerolipid with subsequent transfer to the acyl-CoA pool cannot be discounted. The increase in 28:1-CoA and 30:1-CoA in the acyl-CoA pool from lines overexpressing AtADS2 may indicate that this enzyme is active on saturated fatty acids longer than C26.
The Arabidopsis T-DNA insertion and AtADS OE lines studied for this work did not appear to exhibit any obvious morphological phenotype when grown under normal conditions. Changes in phospholipid fatty acid composition were considerable, especially for PS, a low abundance component of leaf membrane lipids (Devaiah et al., 2006; Yamaoka et al., 2011). Little is known about the biological role of this lipid in plants, although it has recently been reported to be required for microspore development in Arabidopsis (Yamaoka et al., 2011). Sphingolipids containing VLCFAs have been shown to play an important role in plant development and reduced levels of VLCFAs in Arabidopsis is accompanied by severe morphological disturbances (Zheng et al., 2005; Roudier et al., 2010; Markham et al., 2011). Changes in the degree of VLCFA unsaturation clearly have different consequences. Alteration to membrane lipid composition has previously been linked to temperature adaptation in plants (Burgos et al., 2011; Welti et al., 2002; for review see Upchurch, 2008) and AtADS1 and AtADS2 have both been reported to show changes in expression in response to cold treatment (Fukuchi-Mizutani et al., 1998). The authors of that study did not report any functional characterization of the desaturases but suggested, through analogy to their work with the Δ12 desaturase of cyanobacteria, that ADS2 may be involved in maintaining membrane fluidity after a downward shift in temperature. Our work supports their suggestion in that we have provided evidence that AtADS2 plays a significant role in determining the degree of unsaturation of membrane lipids, and in sphingolipids in particular. Δ8 unsaturation of the long-chain base component of sphingolipids was recently shown to be important for low temperature survival in Arabidopsis (Chen et al., 2012), illustrating the role of these lipids in temperature adaptation. In common with the ads2 plants in our study, plants lacking Δ8 unsaturation exhibited no obvious phenotype under normal growing conditions. Desaturation of the fatty acid component of sphingolipids has received little attention. The correlation of ADS expression to temperature is intriguing and is currently under detailed investigation in our laboratory.
With the exception of work by Fukuchi-Mizutani et al. (1998), no studies specifically examining ADS gene expression in Arabidopsis have been published. A survey of relative expression levels using the Arabidopsis eFP Browser (Winter et al., 2007; http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi) indicated that members of the gene families showed diverse expression patterns. For example, AtADS1.2/AtADS1.4 appears to be specifically expressed in root epidermis, and highest expression of AtADS4/AtADS4.2 was reported for the epidermis in upper regions of the Arabidopsis stem (Suh et al., 2005). These studies are slightly impeded by the lack of gene-specific probes for the ADS gene family on commercial Arabidopsis microarrays. These observations suggest that the Arabidopsis ADS gene family, in addition to being the largest desaturase family, contains members involved in diverse metabolic functions. Exploration of the roles of these enzymes will be an exciting and enlightening undertaking.
MATERIALS AND METHODS
Isolation of Full-Length cDNAs
Vectors encoding AtADS1 (C00120), AtADS1.3 (U62926), AtADS2 (U10113), and AtADS4.2 (C00098) were obtained from the Arabidopsis Biological Resource Center (http://abrc.osu.edu/). After sequencing to verify identity, cDNAs were cloned individually into the yeast (Saccharomyces cerevisiae) expression vector pHVX2 (Volschenk et al., 1997), under control of the constitutive promoter PGK1, to create vectors pMS686 (AtADS1), pMD6 (AtADS1.3), pMD5 (AtADS2), and pMD7 (AtADS4.2). To clone the three remaining genes, total RNA extracted from Arabidopsis (Arabidopsis thaliana) roots grown in liquid medium was used as a template to synthesize cDNA using Superscript II (Invitrogen) and an oligo(dT) primer. Sequences encoding AtADS1.2, AtADS1.4, and AtADS4 were amplified by PCR using Pfu Turbo polymerase (Stratagene), and resulting products were cloned into vector pCR2.1 (Invitrogen) using the TOPO cloning system. Primer pairs used for PCR amplification were: ADS12B-FP and ADS12B-RP (AtADS1.2); ADS14B-FP and ADS14B-RP (AtADS1.4); and ADS4B-FP and ADS4B-RP (AtADS4). Primer sequences are shown in Supplemental Table S3). After sequencing, the AtADS cDNAs were cloned into the yeast expression vector pHVX2 to give vectors pMD104 (AtADS1.2), pMD105 (AtADS1.4), and pMD106 (AtADS4).
Subcellular Localization by Transient Expression in Nicotiana benthamiana
To prepare the ADS cDNAs for transient expression in N. benthamiana, PCR products were prepared using primer pairs ADS1-LoF and ADS1-LoR or ADS1-LoF and ADS1-LoNSR with ADS1 cDNA as a template, and primer pairs ADS2-LoF and ADS2-LoR or ADS2-LoF and ADS2-LoNSR with ADS2 cDNA as a template. Primer sequences are given in Supplemental Table S3. PCR products were cloned into the pCR8/GW/TOPO vector (Invitrogen) and the resulting plasmids were confirmed by sequencing. The ADS coding regions were then transferred to pEarleygate 101 or 104 (Earley et al., 2006) by Gateway cloning (Invitrogen) to produce YFP fusion proteins tagged at the C or N termini. N. benthamiana plants were grown in a growth chamber at 22°C (day) and 16°C (night) with a minimum of 16 h of light. Cells from individual overnight cultures of Agrobacterium tumefaciens strain GV3101 harboring the localization vectors were washed with sterile 20 mm MgSO4 and suspended in an equal volume of infiltration buffer (10 mm MES, 10 mm MgSO4, and 100 mm acetosyringone, pH 5.2) followed by incubation at 28°C with shaking for 3 h. Cultures were mixed 1:1 with similarly prepared cultures containing the construct 35S:p19 to provide coexpression of the Tomato bushy stunt virus p19 protein, which suppressed gene silencing (Voinnet et al., 2003). The culture mixtures were infiltrated into young, nearly expanded N. benthamiana leaves (Sparkes et al., 2006) using a plastic 1-mL syringe. Transient expression of the cyan fluorescent protein-tagged ER marker CD3-953 (Nelson et al., 2007) was also conducted. After infiltration the infiltrated leaves were marked and the plants were left at room temperature for 1 h, after which they were returned to the growth chamber before imaging. At time intervals of 24, 48, and 72 h after infiltration, the abaxial leaf surface was imaged by confocal laser-scanning microscopy (LSM510 Meta; Carl Zeiss) using HCX APO L 20×/0.5 and 63×/0.9 water-dipping lenses. The excitation lines of an argon ion laser were used alternately with line switching, using the multitrack facility. Signal for YFP was obtained with 514-nm excitation with HFT 405/514, NFT 515, and BP 530–600 filters. The cyan fluorescent protein signal was obtained with 458-nm excitation with HFT 485 and BP 475–525 filters.
Expression of ADS cDNAs in Yeast
The yeast strain FAT1Δ (BY4741; MATa; his3D1; leu2D0; met15D0; ura3D0; YBR041w::kanMX4) was obtained from Invitrogen. Transformation of yeast cells was conducted as described in the manual of the pYES2.1 TOPO TA Expression Kit (Invitrogen). Transformants were selected in minimal medium without leonine. For analysis of yeast total cellular fatty acids, cells from 5-mL liquid cultures grown for 18 h at 28°C were pelleted by centrifugation and resuspended in 2 mL of 3 m HCl in methanol (Supelco) with 300 μL of hexane in screw-top Pyrex tubes. The tubes were tightly capped and heated at 80°C for 2 h. After cooling, 2 mL of 0.9% NaCl was added to each tube and FAMEs were recovered by collecting the hexane fraction. FAMEs were separated by GC using an Agilent 6890N GC equipped with a DB-23 capillary column (0.25 mm × 30 m, 0.25 mm thickness; J&W Scientific) and flame ionization detector. Double-bond position in the monounsaturated fatty acids was determined by GC-MS following conversion of FAMEs to their DMDS adducts using a method adapted from Shibahara et al. (2008). The DMDS-FAMEs were separated on a DB-5 capillary column using an Agilent 6890N gas chromatograph equipped with a 5973N mass selective detector. For mild saponification, pelleted yeast cells were resuspended in 4 mL of freshly prepared 1% KOH in methanol and heated at 80°C for 1 h. After neutralization with HCl, the suspension was extracted with hexane and the cells were collected by centrifugation. FAMEs were prepared from the unsaponified lipids by resuspending the pellet in 2 mL of 3 m HCl in methanol, with heating at 80°C for 2 h.
Verification of T-DNA Insertion Lines and Northern-Blot Analysis
To confirm the presence and location of the T-DNA insertion, genomic DNA was extracted from young rosette leaves by grinding in extraction buffer (200 mm Tris HCl, pH7.5, 250 mm NaCl, 25 mm EDTA, and 0.5% SDS). After centrifugation to remove leaf material, genomic DNA was precipitated from the supernatant using an equal volume of isopropanol and PCR was conducted using the primer pairs described in Supplemental Figure S5. PCR products were directly sequenced (ABI 3730) to identify the location of the T-DNA insertion.
To determine transcript presence or absence in the T-DNA insertion lines, seeds were geminated on one-half-strength Murashige and Skoog medium and grown to 4–5 leaf stage at 24°C under a 16 h photoperiod. Leaf RNA was extracted using an RNeasy mini kit (Qiagen) and cDNA was synthesized using Superscript II reverse transcriptase, according to the manufacturer’s protocol (Invitrogen). For each plant, PCR was conducted using the primer pairs described in Figure 6. To examine the expression of ADS2 in putative knockout lines by northern blot, the entire inflorescence was removed from wild-type and homozygous ads2 T-DNA insertion plants. Material was ground in liquid nitrogen and total RNA was extracted using the Qiagen RNeasy Plant kit. RNA was separated on an agarose gel and transferred to Hybond C membrane by capillary blotting. For hybridization, a P32-labeled DNA probe was synthesized by random priming using the coding region of ADS2, generated using primers ADS2NFP and ADS2NRP, as a template using standard procedures.
OE of AtADS1 and AtADS2 in Arabidopsis
To assemble binary vectors for Arabidopsis transformation, the coding regions of AtADS1 and AtADS2 were amplified by PCR from vectors C00120 and U10113, respectively. PCR primer pairs were ADS1-5X and ADS1-3Sac (AtADS1) and ADS2-5X and ADS2-3Sac (AtADS2; Supplemental Table S3). After restriction digest, the PCR products were cloned into the vector pBI121 (Clontech) to create vectors pMD12 (CaMV 35S-AtADS1-nopaline synthase (NOS) terminator) and pMD13 (CaMV 35S-AtADS2-NOS terminator), or pSE129 (pRD400 containing the napin promoter and NOS terminator within the T-DNA borders) to create vectors pMD123 (napin-AtADS1-NOS) and pMD124 (napin-AtADS2-NOS). The binary vectors were used for the A. tumefaciens-mediated transformation of Arabidopsis (ecotype Columbia-0) using the floral dip method (Clough and Bent, 1998). Transformants were selected by resistance to kanamycin and grown to maturity in soil at 23°C with constant light (100 μE m–2 s–1). To determine seed fatty acid composition, samples of approximately 100 seeds from each plant were placed in Pyrex tubes with 2 mL of 1 m HCl in methanol and 300 μL of hexane. Capped tubes were heated at 80°C for 2 h, and FAMEs were recovered by collecting the hexane phase after addition of 2 mL of 0.9% NaCl to the cooled tubes. FAMEs were separated by GC as described for yeast fatty acid analysis.
Growth of Plants and Lipid Extraction for Lipidomic Analysis
To collect material for lipidomic analysis, Arabidopsis seeds were planted directly into soil (Sunshine Mix #3; Sun Gro Horticulture) and grown for 25 d in constant light (approximately 100 μE m−2 s−1 at pot level) under the same conditions as described for the OE lines above. For experiment 1, lipid extraction was conducted according to the method of Welti et al. (2002). Three plants for each line were removed by cutting just below the rosette leaves, and were immediately immersed in 3 mL of isopropanol containing 0.01% butylated hydroxytoluene, preheated to 75°C. After incubation for 15 min, 1.5 mL of chloroform and 0.6 mL of water were added to each tube and samples were shaken at 22°C for 1 h. Lipid extracts were transferred to clean Pyrex tubes and five additional extractions were carried out, using 4 mL of chloroform/methanol (2:1) with 0.01% butylated hydroxytoluene each time. For each sample, the combined lipid fractions were further extracted using 1 mL of 1 m KCl and 2 mL of water, then dried under hexane and shipped on dry ice for analysis. Extracted leaf tissue was dried overnight at 105°C before weighing. The lipid profile data were acquired at the Kansas Lipidomics Research Center (http://www.k-state.edu/lipid/lipidomics/), using electrospray ionization tandem MS. Instrument acquisition and method development at the Kansas Lipidomics Research Center was supported by National Science Foundation grants MCB 0455318, MCB 0920663, and DBI 0521587, the Kansas Idea Network of Biomedical Research Excellence (National Institutes of Health grant no. P20 RR16475 from the Idea Network of Biomedical Research Excellence program of the National Center for Research Resources), National Science Foundation Office of Experimental Program to Stimulate Competitive Research grant no. EPS–0236913, Kansas Technology Enterprise Corporation, and Kansas State University. For experiment 2, plants were grown as described above, and lipids were extracted using the Plant Bligh-Dyer-Solvent H protocol (Supplemental File S1) for thorough extraction of sphingolipids. Data sets were subjected to Q-test analysis to detect discordant data points, as recommended by the Kansas Lipidomics Research Center (http://k-state.edu/lipid/lipidomics/Q-test.htm).
Acyl-CoA Analyses
To obtain plant tissue for analysis, seeds were surface sterilized, stratified for 3 d in a cold room, and plated on square plates containing one-half-strength Murashige and Skoog salt mixture in 0.9% (w/v) phytagel. Seeds were germinated and grown in a controlled environment cabinet with a photoperiod of 16 h of light (150 µE m−2 s−1) at 22°C and 8 h of dark at 18°C. Seedlings were grown for 14 d and the rosettes, with roots removed, were frozen in liquid nitrogen and stored at –80°C until further analyses. Samples were extracted according to the method described by Larson and Graham (2001) and analyzed by liquid chromatography-tandem MS with multiple reaction monitoring, operated in positive mode, essentially following the method described by Haynes et al. (2008). Liquid chromatography separation was conducted using an Agilent 1200 LC system equipped with a Gemini C18 column (2 mm i.d., 150 mm in length, with 5-mm particles) and MS was performed using an AB4000 QTRAP (Applied Biosystems) mass spectrometer (spray voltage set to 5 kV, nebulizing gas at 40 pounds per square inch [p.s.i.], focusing gas at 40 p.s.i., and curtain gas at 20 p.s.i.). The source temperature was held at 750°C. Declustering potential and collision energy were optimized on a compound-dependent basis. The acyl-CoAs were identified via multiple reaction monitoring and precursor-product ion pairs of specific acyl-CoAs were followed; for example, 24:0-CoA was monitored by the precursor-product ion pair of mass-to-charge ratio 1,118.5 and 611.36, and 24:1-CoA by the precursor-product ion pair of mass-to-charge ratio 1,116.5 and 609.5. For the purpose of identification and calibration, standard acyl-CoA esters with acyl chain lengths from C14 to C20 were purchased from Sigma as free acids or lithium salts.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phylogram illustrating relation between seven members of the Arabidopsis ADS gene family.
Supplemental Figure S2. Mass spectra of DMDS-FAMEs showing location of double bonds in all novel VLCMUFAs produced by yeast expressing the ADS family members.
Supplemental Figure S3. Distribution of VLCMUFAs in the lipids of yeast expressing AtADS2.
Supplemental Figure S4. OE of AtADS2 in Arabidopsis under control of the CaMV 35S promoter.
Supplemental Figure S5. Comparative chromatograms of two plants from a segregating line of Arabidopsis transformed with AtADS2 under control of the napin promoter.
Supplemental Figure S6. Homozygous ads1 and ads2 T-DNA alleles verified by PCR and RT-PCR probing gene expression.
Supplemental Figure S7. Lipidomics data for experiment 1. PE species as percentage of total measured polar lipids.
Supplemental Figure S8. Lipidomics data for experiment 1. PS species as percentage of total measured polar lipids.
Supplemental Figure S9. Sphingolipid analysis of wild-type Arabidopsis, ads2-1, and AtADS2 OE13.
Supplemental Figure S10. VLCMUFA-CoA profiles of seedlings 14 d after germination from wild-type Arabidopsis, ads2 alleles, and OE13.
Supplemental Figure S11. Comparison of ADS group 1 gene arrangement in the genomes of Arabidopsis and A. lyrata.
Supplemental Table S1. Comparison of sphingolipid recovery between lipidomics experiments 1 and 2.
Supplemental Table S2. ADS homologs identified in Arabidopsis lyrata.
Supplemental Table S3. Sequences of oligonucleotide primers used in this study.
Supplemental File S1. Lipid extraction procedure used in lipidomics experiment 2.
Acknowledgments
The authors acknowledge assistance of the National Research Council DNA facility for DNA sequencing and oligonucleotide primer synthesis, Drs. Jitao Zou and Pierre Fobert (National Research Council Canada) for critical review of the draft manuscript, and Dr. Jonathan Page (National Research Council Canada) and team members for providing N. benthamiana plants and vectors for localization studies. The authors would also like to thank Ruth Welti and Mary Roth of the Kansas Lipidomics Research Center, for lipidomics analysis and helpful advice.
Glossary
- VLCFA
very-long-chain fatty acid
- cDNA
complementary DNA
- VLCMUFA
very-long-chain monounsaturated fatty acid
- ER
endoplasmic reticulum
- YFP
yellow fluorescent protein
- GC
gas chromatography
- MS
mass spectrometry
- FAME
fatty acid methyl ester
- DMDS
dimethyl disulfide
- T-DNA
transfer DNA
- RT
reverse transcription
- OE
overexpression
- PE
phosphatidylethanolamine
- PS
phosphatidyl-Ser
- GIPC
glycosylinositolphosphoceramides
- HexCer
monohexosylceramine
- VLCSFA
very-long-chain saturated fatty acid
- CaMV
Cauliflower mosaic virus
- NOS
nopaline synthase
References
- Allen SM, Luck S, Mullen J, Sakai H, Tingey SV, Williams RW, inventors. February 23, 2012. Drought tolerant plants and related constructs and methods involving genes encoding fatty acid desaturase family peptides. United States Patent Application No. US2012/0047603 A1
- Benghezal M, Wasteneys GO, Jones DA. (2000) The C-terminal dilysine motif confers endoplasmic reticulum localization to type I membrane proteins in plants. Plant Cell 12: 1179–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgos A, Szymanski J, Seiwert B, Degenkolbe T, Hannah MA, Giavalisco P, Willmitzer L. (2011) Analysis of short-term changes in the Arabidopsis thaliana glycerolipidome in response to temperature and light. Plant J 66: 656–668 [DOI] [PubMed] [Google Scholar]
- Byun Y-J, Kim H-J, Lee D-H. (2009) LongSAGE analysis of the early response to cold stress in Arabidopsis leaf. Planta 229: 1181–1200 [DOI] [PubMed] [Google Scholar]
- Cahoon EB, Marillia E-F, Stecca KL, Hall SE, Taylor DC, Kinney AJ. (2000) Production of fatty acid components of meadowfoam oil in somatic soybean embryos. Plant Physiol 124: 243–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Markham JE, Cahoon EB. (2012) Sphingolipid Δ8 unsaturation is important for glucosylceramide biosynthesis and low-temperature performance in Arabidopsis. Plant J 69: 769–781 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, et al. (2008) Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36: W465–W469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah SP, Roth MR, Baughman E, Li M, Tamura P, Jeannotte R, Welti R, Wang X. (2006) Quantitative profiling of polar glycerolipid species from organs of wild-type Arabidopsis and a phospholipase Dα1 knockout mutant. Phytochemistry 67: 1907–1924 [DOI] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS. (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Faergeman NJ, DiRusso CC, Elberger A, Knudsen J, Black PN. (1997) Disruption of the Saccharomyces cerevisiae homologue to the murine fatty acid transport protein impairs uptake and growth on long-chain fatty acids. J Biol Chem 272: 8531–8538 [DOI] [PubMed] [Google Scholar]
- Fukuchi-Mizutani M, Savin K, Cornish E, Tanaka Y, Ashikari T, Kusumi T, Murata N. (1995) Senescence-induced expression of a homologue of Δ9 desaturase in rose petals. Plant Mol Biol 29: 627–635 [DOI] [PubMed] [Google Scholar]
- Fukuchi-Mizutani M, Tasaka Y, Tanaka Y, Ashikari T, Kusumi T, Murata N. (1998) Characterization of Δ9 acyl-lipid desaturase homologues from Arabidopsis thaliana. Plant Cell Physiol 39: 247–253 [DOI] [PubMed] [Google Scholar]
- Haynes CA, Allegood JC, Sims K, Wang EW, Sullards MC, Merrill AH., Jr (2008) Quantitation of fatty acyl-coenzyme As in mammalian cells by liquid chromatography-electrospray ionization tandem mass spectrometry. J Lipid Res 49: 1113–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilmann I, Pidkowich MS, Girke T, Shanklin J. (2004a) Switching desaturase enzyme specificity by alternate subcellular targeting. Proc Natl Acad Sci USA 101: 10266–10271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilmann I, Mekhedov S, King B, Browse J, Shanklin J. (2004b) Identification of the Arabidopsis palmitoyl-monogalactosyldiacylglycerol Δ7-desaturase gene FAD5, and effects of plastidial retargeting of Arabidopsis desaturases on the fad5 mutant phenotype. Plant Physiol 136: 4237–4245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai H, Yamamoto K, Shibahara A, Miyatani S, Nakayama T. (2000) Determining double-bond positions in monoenoic 2-hydroxy fatty acids of glucosylceramides by gas chromatography-mass spectrometry. Lipids 35: 233–236 [DOI] [PubMed] [Google Scholar]
- Knipple DC, Rosenfield C-L, Nielsen R, You KM, Jeong SE. (2002) Evolution of the integral membrane desaturase gene family in moths and flies. Genetics 162: 1737–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson TR, Graham IA. (2001) Technical advance: a novel technique for the sensitive quantification of acyl CoA esters from plant tissues. Plant J 25: 115–125 [DOI] [PubMed] [Google Scholar]
- Li-Beisson Y, Shorrosh B, Beisson F, Andersson MX, Arondel V, Bates PD, Baud S, Bird D, Debono A, Durrett TP, et al. (2010) Acyl-lipid metabolism. The Arabidopsis Book 8: e0133, doi/10.1199/tab.0133. [DOI] [PMC free article] [PubMed]
- MacKenzie DA, Carter AT, Wongwathanarat P, Eagles J, Salt J, Archer DB. (2002) A third fatty acid Δ9-desaturase from Mortierella alpina with a different substrate specificity to ole1p and ole2p. Microbiology 148: 1725–1735 [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, et al. (2011) CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res 39: D225–D229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marillia E-F, Giblin EM, Covello PS, Taylor DC. (2002) A desaturase-like protein from white spruce is a Δ9 desaturase. FEBS Lett 526: 49–52 [DOI] [PubMed] [Google Scholar]
- Markham JE, Li J, Cahoon EB, Jaworski JG. (2006) Separation and identification of major plant sphingolipid classes from leaves. J Biol Chem 281: 22684–22694 [DOI] [PubMed] [Google Scholar]
- Markham JE, Jaworski JG. (2007) Rapid measurement of sphingolipids from Arabidopsis thaliana by reversed-phase high-performance liquid chromatography coupled to electrospray ionization tandem mass spectrometry. Rapid Commun Mass Spectrom 21: 1304–1314 [DOI] [PubMed] [Google Scholar]
- Markham JE, Molino D, Gissot L, Bellec Y, Hématy K, Marion J, Belcram K, Palauqui J-C, Satiat-Jeunemaître B, et al. (2011) Sphingolipids containing very-long-chain fatty acids define a secretory pathway for specific polar plasma membrane protein targeting in Arabidopsis. Plant Cell 23: 2362–2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney AW, Dyer JM, Dhanoa PK, Kim PK, Andrews DW, McNew JA, Mullen RT. (2004) Membrane-bound fatty acid desaturases are inserted co-translationally into the ER and contain different ER retrieval motifs at their carboxy termini. Plant J 37: 156–173 [DOI] [PubMed] [Google Scholar]
- Moreau RA, Pollard MR, Stumpf PK. (1981) Properties of a Δ5-fatty acyl-CoA desaturase in the cotyledons of developing Limnanthes alba. Arch Biochem Biophys 209: 376–384 [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A. (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8: 785–786 [DOI] [PubMed] [Google Scholar]
- Pollard MR, Stumpf PK. (1980) Biosynthesis of C20 and C22 fatty acids by developing seeds of Limnanthes alba: chain elongation and Δ5 desaturation. Plant Physiol 66: 649–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roudier F, Gissot L, Beaudoin F, Haslam R, Michaelson L, Marion J, Molino D, Lima A, Bach L, Morin H, et al. (2010) Very-long-chain fatty acids are involved in polar auxin transport and developmental patterning in Arabidopsis. Plant Cell 22: 364–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayanova O, Haslam R, Venegas Caleron M, Napier JA. (2007) Cloning and characterization of unusual fatty acid desaturases from Anemone leveillei: identification of an acyl-coenzyme A C20 Δ5-desaturase responsible for the synthesis of sciadonic acid. Plant Physiol 144: 455–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanklin J, Cahoon EB. (1998) Desaturation and related modifications of fatty acids. Annu Rev Plant Mol Biol 49: 611–641 [DOI] [PubMed] [Google Scholar]
- Shanklin J, Whittle E, Fox BG. (1994) Eight histidine residues are catalytically essential in a membrane-associated iron enzyme, stearoyl-CoA desaturase, and are conserved in alkane hydroxylase and xylene monooxygenase. Biochemistry 33: 12787–12794 [DOI] [PubMed] [Google Scholar]
- Shibahara A, Yamamoto K, Kinoshita A, Anderson BL. (2008) An improved method for preparing dimethyl disulfide adducts for GC/MS analysis. J Am Oil Chem Soc 85: 93–94 [Google Scholar]
- Sparkes IA, Runions J, Kearns A, Hawes C. (2006) Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat Protoc 1: 2019–2025 [DOI] [PubMed] [Google Scholar]
- Sperling P, Franke S, Lüthje S, Heinz E. (2005) Are glucocerebrosides the predominant sphingolipids in plant plasma membranes? Plant Physiol Biochem 43: 1031–1038 [DOI] [PubMed] [Google Scholar]
- Suh MC, Samuels AL, Jetter R, Kunst L, Pollard M, Ohlrogge J, Beisson F. (2005) Cuticular lipid composition, surface structure, and gene expression in Arabidopsis stem epidermis. Plant Physiol 139: 1649–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upchurch RG. (2008) Fatty acid unsaturation, mobilization, and regulation in the response of plants to stress. Biotechnol Lett 30: 967–977 [DOI] [PubMed] [Google Scholar]
- Voinnet O, Rivas S, Mestre P, Baulcombe D. (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J 33: 949–956 [DOI] [PubMed] [Google Scholar]
- Volschenk H, Viljoen M, Grobler J, Petzold B, Bauer F, Subden RE, Young RA, Lonvaud A, Denayrolles M, van Vuuren HJ. (1997) Engineering pathways for malate degradation in Saccharomyces cerevisiae. Nat Biotechnol 15: 253–257 [DOI] [PubMed] [Google Scholar]
- Watkins PA, Lu J-F, Steinberg SJ, Gould SJ, Smith KD, Braiterman LT. (1998) Disruption of the Sacharomyces cerevisiae FAT1 gene decreases very-long-chain fatty acid acyl-CoA synthetase activity and elevates intracellular very-long-chain fatty acid concentration. J Biochem 273: 18210–18219 [DOI] [PubMed] [Google Scholar]
- Welti R, Li W, Li M, Sang Y, Biesiada H, Zhou HE, Rajashekar CB, Williams TD, Wang X. (2002) Profiling membrane lipids in plant stress responses. Role of phospholipase Dα in freezing-induced lipid changes in Arabidopsis. J Biol Chem 277: 31994–32002 [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. (2007) An “electronic fluorescent pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaoka Y, Yu Y, Mizoi J, Fujiki Y, Saito K, Nishijima M, Lee Y, Nishida I. (2011) PHOSPHATIDYLSERINE SYNTHASE1 is required for microspore development in Arabidopsis thaliana. Plant J 67: 648–661 [DOI] [PubMed] [Google Scholar]
- Yao K, Bacchetto RG, Lockhart KM, Friesen LJ, Potts DA, Covello PS, Taylor DC. (2003) Expression of the Arabidopsis ADS1 gene in Brassica juncea results in a decreased level of total saturated fatty acids. Plant Biotechnol J 1: 221–229 [DOI] [PubMed] [Google Scholar]
- Zerangue N, Schwappach B, Jan YN, Jan LY. (1999) A new ER trafficking signal regulates the subunit stoichiometry of plasma membrane K(ATP) channels. Neuron 22: 537–548 [DOI] [PubMed] [Google Scholar]
- Zheng H, Rowland O, Kunst L. (2005) Disruptions of the Arabidopsis enoyl-CoA reductase gene reveal an essential role for very-long-chain fatty acid synthesis in cell expansion during plant morphogenesis. Plant Cell 17: 1467–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Z, DiRusso CC, Ctrnacta V, Black PN. (2002) Fatty acid transport in Saccharomyces cerevisiae. Directed mutagenesis of FAT1 distinguishes the biochemical activities associated with Fat1p. J Biol Chem 277: 31062–31071 [DOI] [PubMed] [Google Scholar]
- Zou Z, Tong F, Faergeman NJ, Børsting C, Black PN, DiRusso CC. (2003) Vectorial acylation in Saccharomyces cerevisiae. Fat1p and fatty acyl-CoA synthetase are interacting components of a fatty acid import complex. J Biol Chem 278: 16414–16422 [DOI] [PubMed] [Google Scholar]