Abstract
Recent evidence of abundant transcript variation (e.g., alternative splicing, alternative initiation, alternative polyadenylation) in complex genomes indicates that cataloging the complete set of transcripts from an organism is an important project. One challenge is the fact that most high-throughput experimental methods for characterizing transcripts (such as EST sequencing) give highly detailed information about short fragments of transcripts or protein products, instead of a complete characterization of a full-length form. We analyze this “multiassembly problem”—reconstructing the most likely set of full-length isoform sequences from a mixture of EST fragment data—and present a graph-based algorithm for solving it. In a variety of tests, we demonstrate that this algorithm deals appropriately with coupling of distinct alternative splicing events, increasing fragmentation of the input data and different types of transcript variation (such as alternative splicing, initiation, polyadenylation, and intron retention). To test the method's performance on pure fragment (EST) data, we removed all mRNA sequences, and found it produced no errors in 40 cases tested. Using this algorithm, we have constructed an Alternatively Spliced Proteins database (ASP) from analysis of human expressed and genomic sequences, consisting of 13,384 protein isoforms of 4422 genes, yielding an average of 3.0 protein isoforms per gene.
There is now great interest in measuring and cataloging transcriptomes and proteomes. mRNA processing can produce different protein-coding sequences from a single gene, which we will refer to as isoforms. Recent genomics studies found that 30%–60% of human genes have multiple isoforms produced by alternative splicing (Mironov et al. 1999; Brett et al. 2000; Croft et al. 2000; International Human Genome Sequencing Consortium 2001; Kan et al. 2001; Modrek et al. 2001), expanding the diversity of the human proteome. In addition to alternative splicing, alternative initiation and termination also contribute to the diversity of the human proteome. A recent computational analysis identified 68,645 first exons in human genome, indicating that alternative initiation might happen in a considerable portion of human genes (Davuluri et al. 2001).
There has been a tremendous amount of valuable work on the cataloging and construction of full-length transcript sequences, both by experimental and bioinformatics methods. For example, experimental construction and sequencing of libraries containing full-length cDNA clones by the Mammalian Gene Collection (MGC) has produced 14,462 mRNA sequences for 10,718 human genes (as of 2/2003), an average of 1.3 isoforms per gene (Strausberg et al. 1999). The RefSeq database has cataloged these transcripts (Pruitt et al. 2000). Another major source of transcript information is EST fragment data. However, these data require software assembly to reconstruct full-length transcript sequences. There are ∼4 million human EST sequences in public databases. Clustering (Schuler 1997; Christoffels et al. 2001), indexing (Liang et al. 2000a,b; Quackenbush et al. 2001), and mapping (Zhuo et al. 2001) of these data onto the genome has made a major contribution to genomics research. One central resource has been the construction of a consensus sequence for each group of ESTs (Quackenbush et al. 2000).
A particularly exciting aspect of this research has been development of software methods to discern multiple full-length isoform sequences (e.g., representing alternative splice forms) from EST fragment data (Burke et al. 1998; Harrison et al. 2002; Kan et al. 2002). An innovative method has been described for constructing alternative transcript sequences, by comparing ESTs with a full-length mRNA sequence adopted as “the reference sequence” for a gene (Kan et al. 2002). This ingeniously avoids the problem of assembly by simply assuming that the reference sequence already provides a complete assembly to which the ESTs can be compared one by one.
Although this method has made a vital contribution, we believe additional problems need to be solved. First, in 82% of human UniGene clusters, there is no full-length mRNA, and thus the “reference sequence” approach is not possible. And in other organisms, with less intensive mRNA sequencing efforts, the situation is worse. More fundamentally, assuming that we should use a single sequence as the master template for all isoforms of a gene neglects a basic question about how the totality of transcript evidence should be properly weighed and integrated. In working with real data, many questions arise. What if there are two mRNA sequences for a gene that disagree? What if a large majority of EST evidence disagrees with the “reference sequence” at one splice: should the reference sequence's anomalous splice really be used as the default template? Such questions point out the need both for a global consideration of all the evidence simultaneously, and for a probabilistic approach that identifies the most likely set of isoforms given all this evidence.
Complete analysis of the human transcriptome and proteome faces several challenges. First, there are many different sources of variation producing multiple isoforms. Not only alternative splicing, but alternative initiation, polyadenylation, intron retention, nonsense-mediated decay, and RNA editing contribute different kinds of changes to the transcriptome. Second, these mRNA processing differences are superimposed on a broad variety of genetic polymorphisms, from which they must be distinguished, and with which they can interact (e.g., SNPs that change alternative splicing as in tau exon 10; D'Souza and Schellenberg 2000). Third, most of the experimental data (e.g., ESTs) consist of small fragments, and 86% of all alternative splices can only be detected using ESTs (as opposed to mRNAs; see Table 1), yet these splice forms are only interpretable if we know their full-length transcript sequences. Fourth, alternative splices at different points in a transcript can be strongly coupled (e.g., in mutually exclusive exon selection), complicating the analysis. Fifth, the data contain a high level of experimental error. For example, ESTs may contain genomic or other contaminations, incomplete mRNA processing, or library construction artifacts such as chimeric sequences (for a review, see Modrek and Lee 2002). Because there is a substantial probability of error in any given EST, a probabilistic assembly method is required, to compute the most likely full-length isoforms given the set of EST observations. Finally, to produce the most biologically meaningful results, this analysis must integrate information from many different data types, including genomic sequence, full-length mRNAs, ESTs, and protein open reading frames (ORFs).
Table 1.
Detection of Alternative Splicing in Human
| Number of | Dec-00 | Jan-02 |
|---|---|---|
| Splices | 39,862 | 133,369 |
| Splices in mRNA | 24,934 (62.6%) | 114,708 (86.0%) |
| Alternative splice relationships | 6201 | 30,793 |
| Alternative splice relationships detected in mRNAs | 815 (13.1%) | 4289 (13.9%) |
We will refer to these challenges as the “multiassembly problem.” It is analogous to the well-known problem of assembling a consensus transcript sequence from a set of fragments (Liang et al. 2000a,b; Quackenbush et al. 2000, 2001). However, we can no longer assume there exists a single consensus, and instead must assume a hidden mixture of isoforms. We describe the mixture as “hidden” because we know neither the number of isoforms it contains, nor their interrelationships, nor which observations (expressed sequence fragments) came from which isoforms.
In this paper, we describe a multiassembly method based on several principles:
It is essential to use as complete information as possible, including genomic sequence, mRNAs, ESTs, protein open-reading frame information, and so on.
The method should make no assumptions of a single consensus or reference sequence for isoform construction.
All of the above challenges reflect the presence of complex structure in the experimental data that cannot be represented adequately as a string (the typical representation of a sequence), but rather as a partial order alignment graph (Irizarry et al. 2000; Lee et al. 2002; Lee 2003). Specifically, we have represented alternative splice forms as distinct paths through a “splicing graph” whose nodes are exons and whose edges are splices (Modrek et al. 2001; Heber et al. 2002; Lee et al. 2002), considering all of the different forms of isoform variation (not just alternative splicing) and coupling.
The method should use a maximum likelihood approach to sort out strong versus weak evidence, to predict the most likely set of isoforms given the observations.
We adopt a simplified approach of first constructing isoforms based on observational evidence, and second filtering out experimental artifacts using prior knowledge (e.g., length distribution of ORFs). In the first stage, all evidence is used, and nothing is thrown away. In the second stage, evaluation of the isoforms takes into account their global properties (e.g., protein translation) to filter out those that are likely to be artifacts.
We present the detailed algorithm, statistical analysis of its accuracy against a variety of validation tests, and its results for 29,204 human transcript isoforms.
METHODS
Data Sources
The results in this paper are based on human genomic sequence assemblies (International Human Genome Sequencing Consortium 2001; downloaded from ftp://ftp.ncbi.nih.gov/genomes/H_sapiens January 2002) and human EST and mRNA sequences from UniGene (Schuler 1997; downloaded from ftp://ftp.ncbi.nih.gov/repository/UniGene January 2002).
Algorithm Overview
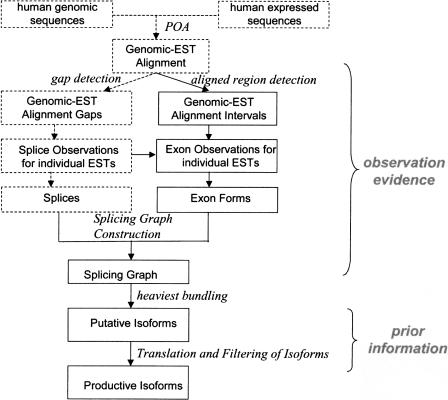
An overview of the isoform generation procedure is shown in Figure 1. It has two stages: first, construction of isoforms based on observational evidence (ESTs and mRNAs, which we shall refer to collectively as “expressed sequences”); second, filtering of the isoforms using prior statistical information that distinguishes “productive isoforms” from EST artifacts (e.g., rules for finding a good ORF). The first phase itself divides into two parts: construction of a splice graph (Heber et al. 2002; for an example, see Fig. 3 below) showing how all the different exons in the gene can be spliced together; followed by traversal of the graph using the heaviest bundling algorithm to extract the most likely set of assembled isoforms consistent with these observational data. Throughout our analysis of the observational evidence, we follow the rules of parsimony (constructing the simplest possible gene model consistent with the observational evidence) and of maximum likelihood (constructing the model that maximizes the likelihood of the observations).
Figure 1.
Flowchart of isoform generation. The alignment of genomic sequence, mRNAs, and ESTs is analyzed on two separate paths: first, to produce splice information; second, to produce exon form information. Together, these constitute a “splice graph” (see Fig. 3). The heaviest bundle algorithm analyzes this graph to predict the most likely assembled isoforms, which finally are filtered by protein-coding criteria.
Figure 3.
Exon-form generation and splice graph construction. Based on the alignment of ESTs (shown as horizontal lines with gaps, and vertical lines indicating their 5′ and 3′-ends) to the genomic sequence (top), a set of exon forms are constructed (boxes, below). The exon forms constitute the nodes of a graph, connected by edges representing the splices observed between them in the ESTs.
Alternative Splicing Analysis
Our algorithm begins with partial order alignment of expressed sequences to genomic sequence using the program POA, and identification of splices observed as gaps within each expressed sequence (splice_obs) as described previously (Irizarry et al. 2000; Modrek et al. 2001; Lee et al. 2002; dashed boxes in Fig. 1). Briefly, EST and mRNA sequences for an individual UniGene cluster were analyzed by the Bayesian ReOrienter (BRO; Irizarry et al. 2000), which finds matches between all the sequences, considering all possible orientations, and infers a global orientation solution. A Bayesian statistical model is used to infer missing EST orientation information and to correct errors in reported EST orientations. Next, reoriented EST and mRNA sequences were aligned by Partial Order Alignment to the genomic sequence, using full dynamic programming local alignment and a gap penalty truncated beyond 16 bp to allow for introns (Modrek et al. 2001). We excluded expressed sequences with nonconsensus splices (lacking canonical splice site sequences at the putative splice junctions) entirely from our isoform construction process. We allowed both U1/U2 splice sites (GT– AG) and U11/U12 splice sites (AT–AC).
Assembling Observational Evidence for Individual Exons
To identify exons in each expressed sequence, the algorithm first searches the alignment of each expressed sequence to the genomic sequence for genomic-expressed sequence alignment intervals, defined as contiguous blocks of alignment without gaps. It is common for an individual expressed sequence to have small insertions or deletions within one exon. Therefore, the algorithm merges adjacent intervals that appear to constitute a single exon: adjacent genomic-expressed sequence alignment intervals are merged if the insertion/deletion between them is (1) not an insertion (in the expressed sequence, relative to the genomic sequence) of >6 bp, and (2) not a deletion of >11 bp. After all possible merges are performed, we term the resulting merged interval an exon observation (exon_obs), because it represents the observational evidence for a single exon, from a single expressed sequence. At each end of the exon observation, we also record whether it has a splice (observed continuation to another exon).
Defining the Minimal Set of Exon Forms Consistent With the Exon Observations
It is important to distinguish a wide variety of effects that can alter exons and their interconnections, including alternative 3′- and 5′-splicing, alternative initiation and termination (polyadenylation), and intron retention. In addition to this complex mixture of effects, we must also consider the effects of random fragmentation and coverage bias [e.g., far more sequencing from the 3′-end(s) of a gene] typical in ESTs. Sorting this out requires some care. For example, we cannot make the common assumption that overlapping exon observations from different expressed sequences should be merged as a single exon. Only in the absence of alternative 3′/5′-splicing, alternative polyadenylation, and intron retention, would this assumption be valid.
Thus, it is necessary to introduce a new definition, exon form, as an interval of genomic sequence with fixed endpoints, which differs from the standard notion of an exon in that multiple, distinct exon forms can overlap in the genomic sequence. This definition preserves our ability to distinguish splice variants that differ by alternative 3′-splicing, or other such effects, that would otherwise be obscured. In Figure 3, for example, exon 2 has three different exon forms. At each end of the exon form, we must also record whether it has a splice (continuation to another exon). We require that splices are only allowed at consensus splice junction sequences.
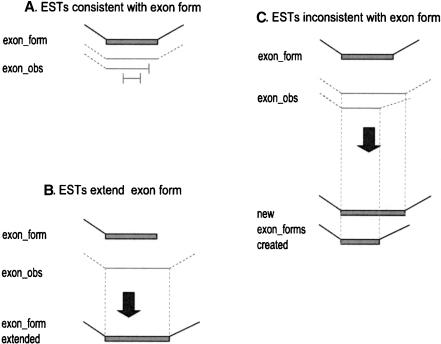
Our algorithm constructs exon forms from the exon observations by parsimony: it only adds a new exon form if there is no other way to explain the exon observations. The key principle is the rule of consistency of an exon observation to an exon form: If a given end of an exon observation has a splice, it must match the splice at that end of the exon form (Fig. 2A); if the exon_obs end has no splice, it must simply be contained within the interval of the exon form (Fig. 2A). If a given end of an exon form has a splice, it cannot be extended (Fig. 2C). However, if it has no splice, then it can be extended to match an exon_obs that extends past its end (Fig. 2B).
Figure 2.
Exon-form extension rules. Exon forms are shown as filled boxes; splices are shown as angled lines; exon observations (exon_obs) from individual ESTs are shown as horizontal lines; the 5′- and 3′-ends of ESTs are shown as vertical lines. (A) An exon_obs is consistent with an exon form if it matches exactly at both ends of the exon form, or its only mismatch is that the EST ends within the exon_form. (B) An exon form with no splice site on one side can be extended on that side by exon_obs that extend the alignment interval. (C) An exon form with defined splice sites cannot explain exon_obs with different splice sites; new exon forms must be created to explain these exon_obs.
It should be emphasized that under these rules a given exon_obs may be consistent with multiple exon forms. For example, a very short EST that is entirely contained within a region that is shared between two exon forms (e.g., alternative 3′-variants of the same exon), is consistent with both. (For an example, see the right exon_obs of the second EST from the top in Fig. 3. It is consistent with both the top and bottom exon forms of that exon.) Therefore, after constructing all the exon forms, we check whether each exon_obs is consistent with each exon form, and record a variety of information: whether this is an exact match, a strong match (the exon_obs interval is contained within the exon form interval and matches it to within 10 bp distance at both ends), or a weak match (the exon_obs interval is contained within the exon form interval but ends more than 10 bp away from at least one end of the exon form). If a given exon_obs is consistent with only one form, it is recorded as being unique evidence for that exon form.
Assembling the Splice Graph
The splice graph (Fig. 3) integrates the following information: exon forms are represented as nodes in the graph; splices are represented as weighted, directed edges in the graph. Our algorithm adds edges to the graph as follows: each splice_obs in a given expressed sequence connects a pair of exon_obs in that expressed sequence; we map this pair of exon_obs to the exon forms that they are consistent with, and add an edge between the corresponding pairs of exon forms. We process all splice_obs to add edges to the graph, disallowing redundant edges (each pair of exon forms will be connected by at most one edge). Initially, each edge is assigned a weight equal to the number of expressed sequences whose splice_obs crosses that edge.
Generation of Putative Isoforms by Heaviest Bundling
We used a graph algorithm, Heaviest Bundling (HB; Lee 2003), to assemble multiple isoforms from the splice graph, using the isoform generation mode of the algorithm. HB finds the consensus traversal of the splice graph that maximizes the probability of the observations. Each edge in the graph is assigned a weight equal to the amount of evidence (number of expressed sequences) for that edge, and HB uses dynamic programming to find the path across the graph that has maximum weight. We simply used the number of expressed sequence observations as the edge weight factor for heaviest bundling. However, this method could easily incorporate additional weighting factors such as statistical confidence (e.g., mRNA quality, or PHRED quality scores), intron length, or splice site strength. For each node, it selects the incoming edge with maximum edge weight; to resolve a tie, it selects the edge whose predecessor node has the large node score. After recording the best incoming edge, it saves the score for this node, equal to the sum of the selected incoming edge weight, plus the score of that edge's predecessor node (Fig. 5A below). After performing heaviest bundling on the whole graph, the algorithm selects the end node with maximum score, and backtracks the saved path, which constitutes the Maximum Likelihood isoform based on this splice graph.
Figure 5.
Heaviest bundling of Hs.2012. (A) Heaviest bundling algorithm (see text). (B) Heaviest bundling produces four maximum likelihood traversals of the Hs.2012 splice graph, each highlighted in red. Numbered nodes represent the exon forms; arrows represents splices between exons. The starting template sequence used for each heaviest bundle is indicated by thick outlines on the nodes that constitute its exons (see text).
Heaviest bundling's “isoform generation mode” guarantees that any set of alternative splicing events that were observed together in an expressed sequence will be represented together in at least one assembled isoform. This is achieved by two rules: iteration (HB isoform generation is repeated until all expressed sequences are explained by [consistent with] at least one generated isoform); and templating (during each round, HB up-weights the expressed sequence spanning the largest number of exon forms but not explained by any isoform generated so far). This template corresponds to the longest mRNA/EST in the data set that is not represented by any of the isoforms yet constructed. Increasing its edge weight contribution to 1000 times that of other expressed sequences causes it to dominate in the region of the gene it covers; however, if the template is incomplete (covers only part of the gene, as is typical for ESTs), HB will extend it by following the maximum likelihood path indicated by the other expressed sequences. Full details of the HB algorithm and template weighting are given in Lee (2003).
We check each expressed sequence for consistency with each isoform by the following rule: every exon_obs and splice_ obs in the expressed sequence must be consistent with the exon forms and splices in the isoform. We record the number of consistent/unique expressed sequence evidence for each isoform. The isoform with the largest number of consistent expressed sequence support in a cluster is classified as the “major” form and the others as “minor” forms.
Translation and Filtering of Putative Isoforms
Each isoform's transcript sequence is constructed by assembling the genomic sequence intervals that constitute its exons. To generate the protein data in this paper, we simply searched for the longest ORF in each transcript.
Because EST data are fragmentary, it is necessary to evaluate the isoform transcript sequences for completeness and potential artifacts. First, we seek to distinguish “productive isoforms” that give rise to a real protein product, from those that probably reflect EST artifacts. Second, we try to screen for full-length isoforms by checking that the sequences are complete enough to give at least the full protein sequence. We apply the following simple filters to assess each isoform transcript sequence: (1) the longest ORF for each transcript must begin with a start codon and end with a stop codon. This can help identify transcripts that are clearly incomplete, but of course does not guarantee full length. (2) Each transcript's inferred translation must exceed 50 amino acids. This eliminates many EST artifacts, but is by no means a guarantee of valid protein forms. (3) The translation of each minor transcript must match at least 50% of the length of the major protein isoform. Large truncations of protein products induce nonsense-mediated decay (NMD) mechanisms that degrade “nonproductive” mRNAs (Maquat and Carmichael 2001). This rule seeks to eliminate EST artifacts such as intron retention, but is not a rigorous rule. It has been shown that less severe truncations can cause NMD.
These filters are merely a set of heuristic guidelines for eliminating many incomplete, artifactual, or nonproductive isoforms, for the convenience of researchers who wish to concentrate on productive isoforms. However, they are only a heuristic, designed to eliminate many false positives (incomplete or nonproductive isoforms) without incurring an excessively large false-negative rate (failure to report isoforms that may be of real biological interest). The larger question of how to distinguish productive isoforms from nonproductive isoforms is a complex topic that is separate from the primary concern of this paper (how to construct isoform sequences from a fragment mixture). We applied these filters purely as a convenience to biologists who wish to use our protein data set, but wish to emphasize that more rigorous work is needed.
Validation of Protein Isoforms
We randomly selected 80 UniGene clusters from our database with corresponding SWISS-PROT entries, by matching the gene symbol of the UniGene cluster to that reported in the SWISS-PROT entry. We applied several criteria for deciding whether each cluster could be used as a “Gold Standard” for comparison (i.e., where any mismatch to the SWISS-PROT sequence can automatically be assumed to be an error): (1) We required that the SWISS-PROT sequence be a direct translation of a complete ORF. Specifically, we excluded “conceptual translations,” and those lacking a start codon (methionine). (2) We required that there is no nonconsensus splice in the gene structure for the UniGene cluster. (3) We required that the alignment of the UniGene cluster to genomic sequence indicate a reliable/unambiguous genomic sequence (because our procedure uses the genomic sequence as the basis for constructing isoforms). Specifically, we excluded clusters with stretches of uncalled bases (NNNNN) in the genomic sequence, or strong evidence of insertion/deletion between the genomic sequence and the mRNA(s). In total, 57 clusters passed all these criteria, and were used as our test set (see Results).
As a second test, we validated both our major and minor protein isoform sequences by comparing them with a set of well-characterized genes to estimate the rate of false positives in our data set. The validation procedure was based on an exhaustive literature search. We randomly selected 20 human genes whose mRNA and protein expression patterns have been described in the literature, and whose protein isoform sequences have been experimentally verified by Western blot, and/or shown to demonstrate distinct and different functional behavior based on biochemical analysis. We compared the amino acid sequences of the experimentally verified isoforms against the computational isoforms generated by our methods. We also identified alteration of the protein product caused by alternative splicing, and checked to see if it matched that reported in the literature. We used the ExPASy Proteomic Tool (http://www.expasy.org/tools/pi_tool.html) to calculate the molecular weight of the protein forms that were used to validate our results. We used the CAI program in the EMBOSS suite v.2.5.1 (Rice et al. 2000) to calculate codon adaptation index.
RESULTS
Isoform Generation for Transcobalamin I (TCN1)
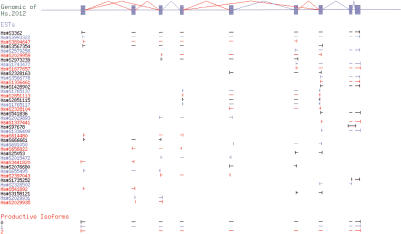
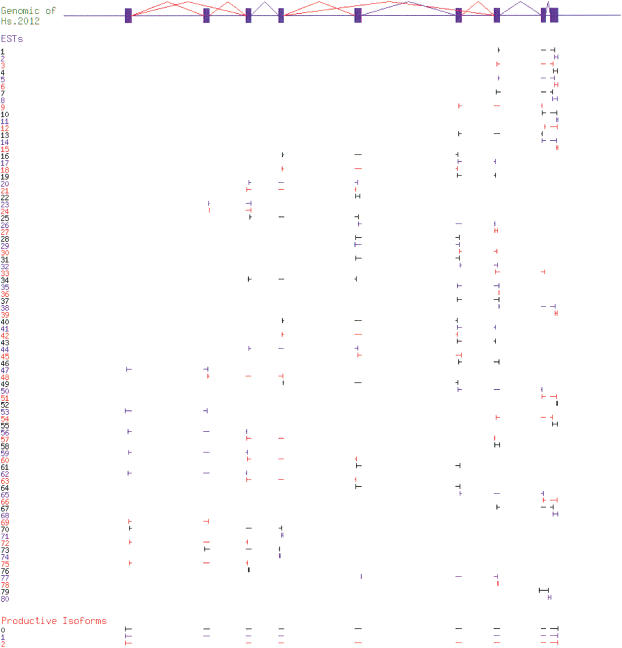
We used the TCN1 gene (UniGene cluster Hs.2012) as an example to show the isoform construction process. Figure 4 summarizes the raw input data: alignment of all the mRNA and EST sequences in Hs.2012 to its genomic region; and the isoforms we constructed from the alignment. Analysis of the alignment produced Exon Obs and Splice Obs for each sequence, resulting in a splice graph of 10 Exon Forms (Fig. 5B). Exon Form 8 was a singleton observed in only one EST (Hs#S97678). This EST appears to be intron inclusion or genomic contamination (see Fig. 4). Note that Exon Form 8 overlaps both Exon Forms 9 and 10 in the alignment, but because no splice is ever observed connecting it to another exon, it is an isolated node in the splice graph (see Fig. 5B). This ability to distinguish overlapping, potentially confusing exon forms, and keep their relationships clear is an important property of the splice graph data structure.
Figure 4.
Isoform generation for Hs.2012. Genomic sequence (top) is shown as exons (filled boxes) and splices (angled lines; alternative splices are colored red). The alignment of mRNAs and ESTs is shown schematically by alignment intervals (horizontal lines); the ends of each expressed sequence are indicated with vertical lines. The isoforms produced from the alignment data by the algorithm are shown at the bottom. Adjacent ESTs (and isoforms) are colored differently to make it easier to distinguish them.
Heaviest bundling produced four isoforms as traversals of this graph, starting from the templates Hs#S3362 (mRNA), Hs#S3567354 (EST), Hs#S614480 (EST), and Hs#S97678 (EST), respectively. The first three isoforms span the complete gene structure of Hs.2012, whereas the last isoform only contains Exon Form 8. We then constructed the transcript sequence for each isoform using the genomic sequence of each Exon Form, and translated the transcript sequence into protein sequence. The algorithm identified the first isoform as the “major” isoform because it was supported by the largest amount of expressed sequence evidence. The last isoform was rejected as an “invalid” isoform because its protein sequence (48 amino acids, 9% identity to major isoform) fell below the filtering criteria.
The three other isoforms represent alternative splice forms of the TCN1 transcript. The major isoform matches an existing mRNA sequence, but the two additional isoforms appear to be novel (i.e., no known mRNA), either skipping exon 2, or skipping exons 5 and 6. Because no expressed sequence was observed containing both alternative splice events, they were constructed as separate isoforms.
Detection of Coupled Alternative Splicing
One fundamental benefit of our algorithm is that it guarantees that any set of alternative splicing events that were observed together in an expressed sequence will be represented together in at least one assembled isoform. Alternative splicing events in different parts of a gene will be reported as coupled in one isoform if and only if they were observed together in at least one expressed sequence. For example, in nitric oxide synthase 2A (Hs.193788), the algorithm generated two isoforms (Fig. 6A). The major isoform, containing 27 exons, was supported by eight mRNAs. The minor isoform, skipping exons 24 and 26, was supported by two independent ESTs. These two exon skips were combined in a single isoform because they were always observed to be coupled in the EST data. If these two exon skips were observed in unlinked ESTs, they would each have given rise to a different isoform (producing a total of three isoforms, as in Hs.2012; see Fig. 5). Another form of coupling we commonly observed is mutually exclusive exon usage (e.g., Hs.117572; data not shown).
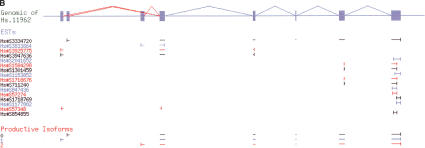
Figure 6.
Coupled alternative splicing detected by heaviest bundling. Genomic sequence (top) is shown as exons (filled boxes) and splices (angled lines; alternative splices are colored red). The alignment of mRNAs and ESTs is shown schematically by alignment intervals (horizontal lines); the ends of each expressed sequence are indicated with vertical lines. The isoforms produced from the alignment data by the algorithm are shown at the bottom. Adjacent ESTs (and isoforms) are colored differently to make it easier to distinguish them. (A) In Hs.193788, two nonadjacent alternative splices (skipping of exons 24 and 26) were always observed together (in two ESTs), thus, the algorithm coupled both events in a single isoform (isoform 1). (B) An example of alternative initiation.
The Heaviest Bundling algorithm was also able to detect a wide variety of isoform variations in addition to true alternative splicing (competition between splice sites), such as alternative initiation (Fig. 6B), alternative polyadenylation (data not shown), and intron retention (e.g., Hs#S97678 in Fig. 4). For example, in the apoptosis regulator gene BCL-G (Hs.11962), the algorithm reported three isoforms differing only in their choice of first exon (Fig. 6B). One of these corresponds to the mRNA sequence deposited in GenBank for the gene. The other two are novel isoforms detected in some 5′-ESTs revealing two alternative first exons. Our algorithm combined these alternative first exons with the major isoform path through the rest of the gene structure.
Validation Tests
We first tested the robustness of our multiassembly algorithm by directly testing its sensitivity to increasing fragmentation. We artificially fragmented the mRNA and EST sequences in Hs.2012 into small pieces and reran the isoform construction algorithm. Each mRNA or EST sequence was chopped into fragments of 300 bp or less (Fig. 7). The isoform generation program still produced the same three valid isoforms from those tiny fragmentary sequences. The only difference in the calculation occurred in the processing of the singleton Exon Form 8 (based on the single EST Hs#S97678; see Fig. 4). This EST was chopped into two pieces that still cannot be connected with any other exon forms; thus, this EST gave rise to two isoforms (each consisting of a single-Exon Form) that were excluded as invalid proteins by our filter criteria.
Figure 7.
Effect of fragmentation on isoform generation for Hs.2012. All mRNAs and ESTs in Hs.2012 were broken into fragments of 300 nt or less. The isoforms produced from these data by the algorithm (indicated at the bottom) are identical to the results prior to fragmentation (see Fig. 4). Genomic sequence (top) is shown as exons (filled boxes) and splices (angled lines; alternative splices are colored red). The alignment of mRNAs and ESTs is shown schematically by alignment intervals (horizontal lines); the ends of each expressed sequence are indicated with vertical lines. Adjacent ESTs (and isoforms) are colored differently to make it easier to distinguish them.
In practice, the real question is whether the algorithm can reliably reconstruct full-length transcript sequences from EST data alone. We therefore performed a practical test on a random sample of clusters (40 isoforms), by removing mRNA sequences from each cluster to see whether the algorithm would produce identical results from the EST data alone (see below). In all cases (32 isoforms) in which the ESTs covered the full gene structure, the algorithm produced the same isoforms from the ESTs alone. In eight cases we observed data coverage errors, in which the EST data were missing exons or splices observed only in the mRNAs. In 13% of cases (5/40 isoforms tested), an isoform was missed because the ESTs were missing a constitutive exon, and in 8% of cases (3/40 isoforms tested), because the ESTs were missing an alternative splice. Overall, we saw no signs of algorithmic error; when no essential data were missing, the algorithm reliably produced correct isoforms.
To assess the reliability of the protein products of our isoform generation algorithm, we generated a random sample of 57 UniGene clusters that could be mapped to a unique entry in the SWISS-PROT database (see Methods for criteria). We then compared the SWISS-PROT sequences mapped to these 57 clusters with our protein isoform sequences. In 52 clusters, we found a match of >99% identity (measured over the full length of the SWISS-PROT sequence) to our protein isoforms. In 50 out of 52 match cases, the SWISS-PROT sequence matched our major isoform sequence, most likely because only a small fraction of SWISS-PROT entries represent alternative splice forms. In a single cluster (Hs.96), we found an algorithmic error. The SWISS-PROT sequence (APR_HUMAN) matched not the longest ORF but the second longest ORF in the mRNA. Thus, although our algorithm generated a correct mRNA isoform, its translation (generated by finding the longest ORF) did not match the SWISS-PROT entry. This indicates that our method can be unreliable for very short proteins (APR_HUMAN is only 54 amino acids in length), in which the longest ORF is not a reliable predictor. Because all of the input data were correct, we consider this to be an algorithmic error. Of the remaining clusters, four revealed data coverage problems. In three of these clusters, the generated protein isoform sequence matched the SWISS-PROT sequence perfectly but was not as long, owing to lack of EST/mRNA coverage in part of the gene; in another cluster, because of lack of coverage, we are unable to generate any productive isoform. Thus, the overall rate of algorithmic error for protein isoform generation in this data set was 1.8% (1/57 clusters), and the rate of data coverage errors was 7.0% (4/57 clusters).
Construction of an Alternatively Spliced Proteins (ASP) Sequence Database
We have applied our algorithm to the subset of the human genome that produces multiple splice forms, to produce a database of Alternatively Spliced Proteins (Table 2). Beginning with the draft human genome sequence and UniGene expressed sequence data, we identified 17,581 multiexon genes that produced a total of 61,508 isoforms, an average of 3.5 isoforms per gene. After applying our isoform filters (require a complete ORF, disallow gross truncations of the predicted protein product, etc.), a total of 29,204 isoforms in 13,608 genes passed all our criteria, including 22,232 isoforms in 6636 genes with at least two productive isoforms per gene. In all, 80% of these isoforms produced an altered protein product, resulting in 17,742 distinct protein isoforms. Finally, by requiring that each major–minor isoform pair must share an alternative splicing relationship (excluding possibility of EST artifacts, i.e., genomic contamination), we generated an Alternatively Spliced Proteins database (ASP), with 13,384 protein isoforms for 4422 human genes, on average 3.0 protein isoforms per gene.
Table 2.
Analysis of the Human Transcriptome by Isoform Assembly
| UniGene | cluster | Isoforms | ||
|---|---|---|---|---|
| Total clusters | 96,109 | |||
| Mapped to genome | 68,032 | 71% | ||
| Detected consensus splices | 18,173 | 27% | ||
| Produced putative isoforms | 17,581 | 97% | 61,508 | |
| Productive mRNA isoforms | 13,608 | 77% | 29,204 | 47% |
| Multiple productive isoforms per gene | 6636 | 49% | 22,232 | 76% |
| DIstinct protein isoforms | 17,742 | 80% | ||
| ASP (alternatively spliced proteins) database | 4422 | 13,384 | ||
ASP significantly expands available protein isoform data for the human proteome. Over half of ASP consists of novel isoforms not matching any mRNA or protein sequence deposited in GenBank. At present, SWISS-PROT (Bairoch and Apweiler 1998) provides annotated alternative protein isoforms for 967 human genes via its VARSPLIC feature. Adding the ASP data expands the data set of alternatively spliced protein forms to a total of 5021 human genes (after removing overlaps between ASP and SWISS-PROT), a five-fold increase. All of the ASP data will be made freely available on the Web upon publication (http://www.bioinformatics.ucla.edu/ASAP).
Validation of Alternative Protein Isoforms
One of the principal difficulties in analyzing the proteome-wide impact of alternative splicing is the relative lack of experimental data for directly validating alternatively spliced protein products. Whereas most biochemical studies focus on the major protein isoform, experimental evidence concerning alternative isoforms tends to be at the transcript (mRNA) level. Standard methods for identifying isoforms (hybridization-based methods like RT-PCR or cDNA sequencing) are not applicable to direct detection of protein isoforms. Whereas high-throughput genomics methods have generated large data sets of transcript sequence (4 million human expressed sequences), there is so far little high-throughput experimental data (such as mass spectrometry) surveying alternative splicing throughout the proteome. Usually, evidence of alternative splice forms is of mRNAs, rather than detection of the protein products themselves, and actual protein sequencing data are very rare. In this context, it is not easy to determine which genes can be considered a “gold standard” for protein isoform validation; that is, can we assume all mRNA and protein isoforms of these genes have been fully and correctly reported in the published literature?
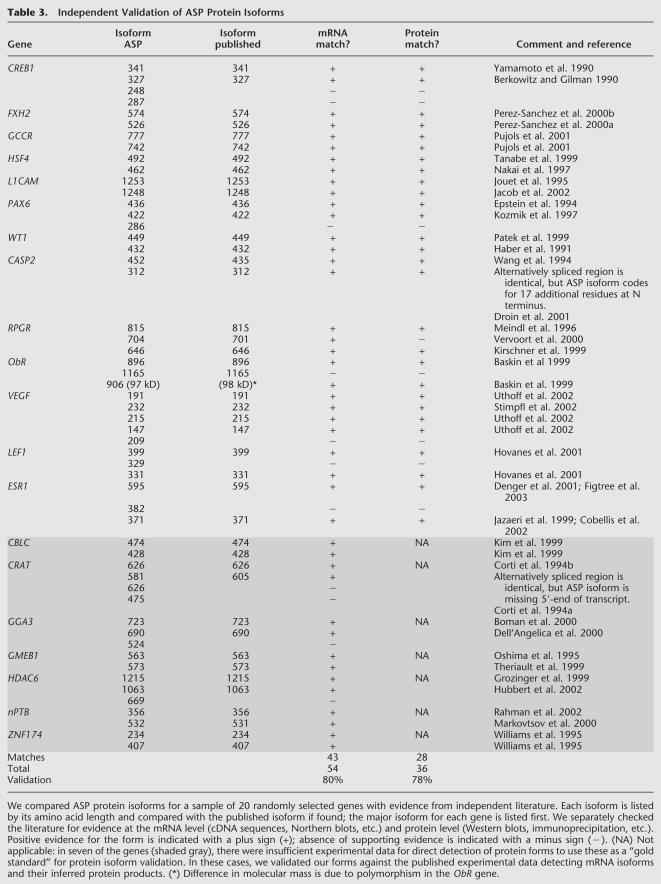
Nonetheless, we have sought to assess the accuracy of the transcript and protein isoform sequences in the ASP database, using a variety of independent experimental data. First, 59% of all major isoform sequences and 27% of all minor isoforms in the ASP database are supported by at least one mRNA sequence deposited in GenBank. Thus, 41% of ASP matches previously published, human-curated sequences. To evaluate our minor isoform results in more detail, we examined a random sample of 20 well-characterized genes (Table 3). Of the ASP mRNA isoforms for these genes, 80% were validated by published experimental data (sequencing, Northern blots, etc.). For minor ASP isoforms, 68% were validated. Turning to published protein isoform evidence (Western blots, etc.), we were able to validate 78% of the ASP protein sequences, and 65% of the ASP minor protein isoform sequences. These data indicate that most of the protein isoforms in ASP can be confirmed when appropriate experimental studies are performed.
Table 3.
Independent Validation of ASP Protein Isoforms
It should be emphasized that these assessments do not test the accuracy of our algorithm, so much as the accuracy of the input data (ESTs), and the completeness of identification of all splice forms in the experimental literature for these genes. It is possible that a splice form found in ESTs does not actually constitute a sizeable fraction of the transcripts of that gene in any tissue (and which we might consider to be a false positive in the EST data). It is also possible that a given splice form is specific to a particular cell type or activation state that has not yet been analyzed in the experimental literature (a false negative in the published literature). Our tests indicate that the sum of both false positives in the EST data and false negatives in the published literature constitutes ∼32% of minor isoforms, but give no information as to how many are false positives versus how many are false negatives.
Statistical Analysis of ASP Protein Isoforms
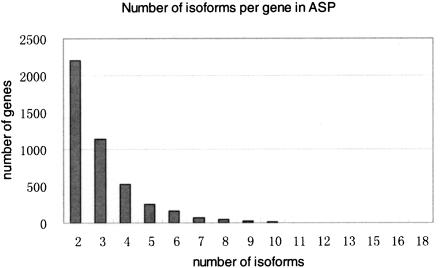
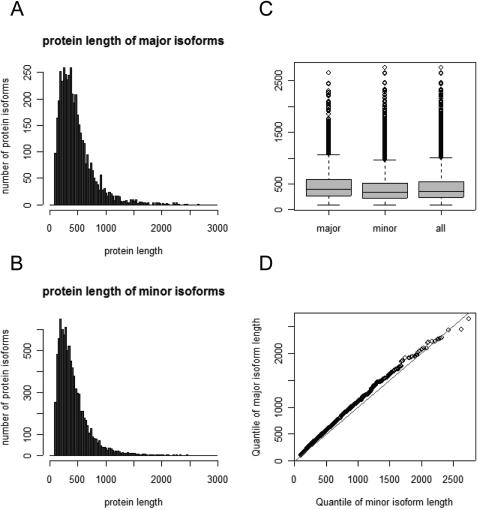
We have performed a series of statistical analyses of the ASP database, both to assess it versus previous studies of alternative splicing, and to test whether the ASP alternative splice forms (i.e., minor forms) show evidence of aberrant characteristics. About half of the genes in ASP were represented by a single major protein isoform and a single minor isoform, with the remaining genes expressing from three to 18 distinct protein isoforms (Fig. 8). The average length of major protein isoforms is 471 amino acids, and the average length of minor protein isoforms is shorter (420 amino acids; Fig. 9). This length difference (50 amino acids) is analogous to the skipping of a single exon (the average length of an internal exon is 150 nt; Modrek et al. 2001), and is consistent with a previous study on Chromosome 22 reporting that alternative exon usage usually produces a minor isoform product that is shorter than the major isoform, because of exon skipping (Hide et al. 2001).
Figure 8.
Number of isoforms per gene in ASP.
Figure 9.
Comparison of protein length distributions for major versus minor isoforms. Histograms of protein length for (A) major isoforms; (B) minor isoforms. (C) Comparison of the mean and standard deviation ranges; (D) quantile–quantile plot.
Next, we assessed whether ASP minor transcript isoforms showed evidence of aberrant protein coding characteristics, by calculating the codon adaptation index (CAI) of the ORF in each minor ASP isoform. As a group, minor isoforms in ASP had high CAI scores, identical to those of major isoforms (Fig. 10). Both the mean value and distribution of CAI were identical between major versus alternatively spliced (minor) isoforms. As another test of unusual functional characteristics, we tested for sequences that might be subject to nonsense-mediated decay (NMD). NMD can occur if the translation stop codon is >50 nt upstream of the last exon–exon junction site (Maquat 2002). It has recently been reported that a third of alternative splice forms detected in ESTs might be NMD candidates by this definition (Lewis et al. 2003). In a test set of 18,714 multiexon human mRNA sequences, we found only 3.9% were NMD candidates by this criterion (Table 4). Among ASP major isoforms, we found a similarly low fraction (3.7%). In ASP minor isoforms, we found a significant increase in NMD candidates (11.1%), but far lower than that expected in EST alternative splice forms as a whole (33%). This indicates that our filtering steps have largely removed alternative splice forms that have aberrant characteristics or may not actually code for a protein product.
Figure 10.
Comparison of codon adaptation index for major versus minor isoforms. Histograms of codon adaptation index for (A) major isoforms; (B) minor isoforms. (C) Comparison of the mean and standard deviation ranges; (D) quantile–quantile plot.
Table 4.
Analysis of Nonsense-Mediated Decay (NMD) Candidates in ASP
| mRNA | Major ASP isoforms | Minor ASP isoforms | |
|---|---|---|---|
| Normal | 17,985 | 4257 | 7966 |
| NMD Targets | 729 | 165 | 996 |
| NMD% | 3.90% | 3.73% | 11.11% |
Finally, we have tested whether ASP minor isoforms might be due to spliceosomal errors caused by occasionally selecting a weak splice site. This has been observed at very low frequency in some genes (Skandalis et al. 2002). We evaluated whether minor isoforms contained suboptimal splice sites by calculating an HMM splice site score (Fairbrother et al. 2002) for splice sites in both major and minor isoforms (Fig. 11). We trained the splice site HMM on 89,506 constitutive human splice sites. The distribution of splice site scores for major versus minor isoforms was virtually identical (as shown by the quantile–quantile plot), indicating that minor isoforms show no evidence of being spliceosomal error products associated with poor splice sites. We obtained similar results comparing minor splice sites versus constitutive splice sites (data not shown).
Figure 11.
Comparison of splice site strength for major versus minor isoforms. Histograms of splice site strength for (A) major isoforms; (B) minor isoforms. (C) Quantile–quantile plot.
DISCUSSION
The work described in this paper is a minor extension of a large body of previous research on EST indexing and consensus construction. Previous research has elucidated the construction of EST clusters (Schuler 1997; Christoffels et al. 2001), alignments, gene indexes (Liang et al. 2000a,b; Quackenbush et al. 2001), consensus sequences (Quackenbush et al. 2000), and identification of sequence variants (Burke et al. 1998; Fasulo et al. 2002; Harrison et al. 2002; Kan et al. 2002). Much work has also been devoted to the mapping of expressed sequence data (Zhuo et al. 2001), analysis, and visualization (Haas et al. 2000; Muilu et al. 2001) of the interrelationships among these sequences. It should be emphasized that the work described in this paper does not address the prior problem of EST clustering, but simply assembles isoforms for a given EST cluster.
The problem of fragment multiassembly highlighted in this paper is likely to remain important for the analysis of high-throughput transcriptome and proteome data in the foreseeable future. For example, despite very significant efforts to generate full-length transcript sequences for human genes (e.g., MGC), the percentage of alternative splice events that were detected in the context of a full-length mRNA sequence remains low (14%). The reason for this is that high-throughput sequencing of EST fragments is much larger, and has grown as rapidly. This bias toward fragment data is not unique to ESTs. Other high-throughput methods such as DNA microarrays and mass spectrometry also give primarily “local” information about part of a transcript or protein, rather than “global” information about the full-length isoform. For example, although probes specific enough (25–70 nt in length) to distinguish different splice forms can be incorporated in a microarray, they only detect one splice or exon at a time. Conceptually, the hybridization intensities from such a “splicing array” are equivalent to sequencing a very large number of short (25–70 nt) EST fragments. Inferring the mix of full-length isoforms from such data is equivalent to the multiassembly problem outlined in this paper. Unfortunately, because the resulting protein isoforms depend on knowing the exact full-length transcript sequences, this problem must be considered.
Our tests indicate that Heaviest Bundling is an effective multiassembly algorithm, which can reconstruct full-length transcripts from fragmentary EST mixtures without relying on the presence of full-length sequences. Unless the full-length sequences contain unique information, fragmentation or even removal of the mRNAs does not greatly affect the algorithm's results. If the algorithm is supplied full-length mRNA sequences, it makes use of them, and guarantees that any observed coupling of alternative splice events will be preserved in the constructed isoforms. In the absence of full-length mRNAs, the algorithm will produce a maximum likelihood solution using all the fragment data. Our tests show that unless the ESTs are missing specific exons or splices, this produces identical results.
Our approach has many defects. For example, there are circumstances in which transcript assembly from ESTs is not appropriate. In general, for applications that require validated transcript sequences, EST assembly is not reliable enough, because of the likelihood of false positives. For example, projects such as the human annotation workshop (HAWK) have elected not to attempt this. Second, our filtering procedure may remove some biologically important transcript sequences, such as RNAs that perform regulatory functions rather than encoding proteins. It may be interesting to analyze the subset of sequences that were filtered out, for indications of such regulatory functions.
ASP offers biologists a useful resource for experimental testing and functional characterization of novel alternative splice forms. The ASP transcript sequences provide a data set of good candidate isoforms for experimental validation efforts, and enable automated design of PCR primers for detecting these splice forms. ASP can also be incorporated into high-throughput mass spectrometry to assist experimental identification of alternative splice forms. Analysis of how alternative splicing alters functional regions in the ASP protein isoforms (e.g., disruption of domain architecture, protein localization signals) can indicate likely functional effects, and guide further experiments.
Acknowledgments
We thank C. Grasso, B. Modrek, M. Gorlick, Q. Xu, and L. Atanelovfor their helpful discussions and comments on this work. This work was supported by the National Institute of Mental Health and the National Institute of Neurological Disorders and Stroke (Grant MH65166), National Science Foundation grant IIS-0082964, and Department of Energy grant DEFG0387ER60615.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1304504. Article published online before print in February 2004.
References
- Bairoch, A. and Apweiler, R. 1998. The SWISS-PROT protein sequence data bank and its supplement TrEMBL in 1998. Nucleic Acids Res. 26: 38–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin, D.G., Schwartz, M.W., Seeley, R.J., Woods, S.C., Porte Jr., D., Breininger, J.F., Jonak, Z., Schaefer, J., Krouse, M., Burghardt, C., et al. 1999. Leptin receptor long-form splice-variant protein expression in neuron cell bodies of the brain and co-localization with neuropeptide Y mRNA in the arcuate nucleus. J. Histochem. Cytochem. 47: 353–362. [DOI] [PubMed] [Google Scholar]
- Berkowitz, L.A. and Gilman, M.Z. 1990. Two distinct forms of active transcription factor CREB (cAMP response element binding protein). Proc. Natl. Acad. Sci. 87: 5258–5262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boman, A.L., Zhang, C., Zhu, X., and Kahn, R.A. 2000. A family of ADP-ribosylation factor effectors that can alter membrane transport through the trans-Golgi. Mol. Biol. Cell 11: 1241–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brett, D., Hanke, J., Lehmann, G., Haase, S., Delbruck, S., Krueger, S., Reich, J., and Bork, P. 2000. EST comparison indicates 38% of human mRNAs contain possible alternative splice forms. FEBS Lett. 474: 83–86. [DOI] [PubMed] [Google Scholar]
- Burke, J., Wang, H., Hide, W., and Davison, D.B. 1998. Alternative gene form discovery and candidate gene selection from gene indexing projects. Genome Res. 8: 276–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoffels, A., van Gelder, A., Greyling, G., Miller, R., Hide, T., and Hide, W. 2001. STACK: Sequence Tag Alignment and Consensus Knowledgebase. Nucleic Acids Res. 29: 234–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobellis, L., Reis, F.M., Driul, L., Vultaggio, G., Ferretti, I., Villa, E., and Petraglia, F. 2002. Estrogen receptor α mRNA variant lacking exon 5 is co-expressed with the wild-type in endometrial adenocarcinoma. Eur. J. Obstet. Gynecol. Reprod. Biol. 102: 92–95. [DOI] [PubMed] [Google Scholar]
- Corti, O., DiDonato, S., and Finocchiaro, G. 1994a. Divergent sequences in the 5′ region of cDNA suggest alternative splicing as a mechanism for the generation of carnitine acetyltransferases with different subcellular localizations. Biochem. J. 303: 27–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti, O., Finocchiaro, G., Rossi, E., Zuffardi, O., and DiDonato, S. 1994b. Molecular cloning of cDNAs encoding human carnitine acetyltransferase and mapping of the corresponding gene to chromosome 9q34.1. Genomics 23: 94–99. [DOI] [PubMed] [Google Scholar]
- Croft, L., Schandorff, S., Clark, F., Burrage, K., Arctander, P., and Mattick, J.S. 2000. ISIS, the intron information system, reveals the high frequency of alternative splicing in the human genome. Nat. Genet. 24: 340–341. [DOI] [PubMed] [Google Scholar]
- Davuluri, R.V., Grosse, I., and Zhang, M.Q. 2001. Computational identification of promoters and first exons in the human genome. Nat. Genet. 29: 412–417. [DOI] [PubMed] [Google Scholar]
- Dell'Angelica, E.C., Puertollano, R., Mullins, C., Aguilar, R.C., Vargas, J.D., Hartnell, L.M., and Bonifacino, J.S. 2000. GGAs: A family of ADP ribosylation factor-binding proteins related to adaptors and associated with the Golgi complex. J. Cell Biol. 149: 81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denger, S., Reid, G., Kos, M., Flouriot, G., Parsch, D., Brand, H., Korach, K.S., Sonntag-Buck, V., and Gannon, F. 2001. ERα gene expression in human primary osteoblasts: Evidence for the expression of two receptor proteins. Mol. Endocrinol. 15: 2064–2077. [DOI] [PubMed] [Google Scholar]
- Droin, N., Rebe, C., Bichat, F., Hammann, A., Bertrand, R., and Solary, E. 2001. Modulation of apoptosis by procaspase-2 short isoform: Selective inhibition of chromatin condensation, apoptotic body formation and phosphatidylserine externalization. Oncogene 20: 260–269. [DOI] [PubMed] [Google Scholar]
- D'Souza, I. and Schellenberg, G.D. 2000. Determinants of 4-repeat τ expression: Coordination between enhancing and inhibitory splicing sequences for exon 10 inclusion. J. Biol. Chem. 275: 17700–17709. [DOI] [PubMed] [Google Scholar]
- Epstein, J.A., Glaser, T., Cai, J., Jepeal, L., Walton, D.S., and Maas, R.L. 1994. Two independent and interactive DNA-binding subdomains of the Pax6 paired domain are regulated by alternative splicing. Genes & Dev. 8: 2022–2034. [DOI] [PubMed] [Google Scholar]
- Fairbrother, W.G., Yeh, R.F., Sharp, P.A., and Burge, C.B. 2002. Predictive identification of exonic splicing enhancers in human genes. Science 297: 1007–1013. [DOI] [PubMed] [Google Scholar]
- Fasulo, D., Halpern, A., Dew, I., and Mobarry, C. 2002. Efficiently detecting polymorphisms during the fragment assembly process. Bioinformatics 18 Suppl 1: S294–S302. [DOI] [PubMed] [Google Scholar]
- Figtree, G.A., McDonald, D., Watkins, H., and Channon, K.M. 2003. Truncated estrogen receptor α 46-kDa isoform in human endothelial cells: Relationship to acute activation of nitric oxide synthase. Circulation 107: 120–126. [DOI] [PubMed] [Google Scholar]
- Grozinger, C.M., Hassig, C.A., and Schreiber, S.L. 1999. Three proteins define a class of human histone deacetylases related to yeast Hda1p. Proc. Natl. Acad. Sci. 96: 4868–4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas, S.A., Beissbarth, T., Rivals, E., Krause, A., and Vingron, M. 2000. GeneNest: Automated generation and visualization of gene indices. Trends Genet. 16: 521–523. [DOI] [PubMed] [Google Scholar]
- Haber, D.A., Sohn, R.L., Buckler, A.J., Pelletier, J., Call, K.M., and Housman, D.E. 1991. Alternative splicing and genomic structure of the Wilms tumor gene WT1. Proc. Natl. Acad. Sci. 88: 9618–9622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison, P.M., Kumar, A., Lang, N., Snyder, M., and Gerstein, M. 2002. A question of size: The eukaryotic proteome and the problems in defining it. Nucleic Acids Res. 30: 1083–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heber, S., Alekseyev, M., Sze, S.H., Tang, H., and Pevzner, P.A. 2002. Splicing graphs and EST assembly problem. Bioinformatics 18 Suppl. 1: S181–S188. [DOI] [PubMed] [Google Scholar]
- Hide, W.A., Babenko, V.N., van Heusden, P.A., Seoighe, C., and Kelso, J.F. 2001. The contribution of exon-skipping events on chromosome 22 to protein coding diversity. Genome Res. 11: 1848–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovanes, K., Li, T.W., Munguia, J.E., Truong, T., Milovanovic, T., Lawrence Marsh, J., Holcombe, R.F., and Waterman, M.L. 2001. β-Catenin-sensitive isoforms of lymphoid enhancer factor-1 are selectively expressed in colon cancer. Nat. Genet. 28: 53–57. [DOI] [PubMed] [Google Scholar]
- Hubbert, C., Guardiola, A., Shao, R., Kawaguchi, Y., Ito, A., Nixon, A., Yoshida, M., Wang, X.F., and Yao, T.P. 2002. HDAC6 is a microtubule-associated deacetylase. Nature 417: 455–458. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921. [DOI] [PubMed] [Google Scholar]
- Irizarry, K., Kustanovich, V., Li, C., Brown, N., Nelson, S., Wong, W., and Lee, C. 2000. Genome-wide analysis of single-nucleotide polymorphisms in human expressed sequences. Nat. Genet. 26: 233–236. [DOI] [PubMed] [Google Scholar]
- Jacob, J., Haspel, J., Kane-Goldsmith, N., and Grumet, M. 2002. L1 mediated homophilic binding and neurite outgrowth are modulated by alternative splicing of exon 2. J. Neurobiol. 51: 177–189. [DOI] [PubMed] [Google Scholar]
- Jazaeri, O., Shupnik, M.A., Jazaeri, A.A., and Rice, L.W. 1999. Expression of estrogen receptor α mRNA and protein variants in human endometrial carcinoma. Gynecol. Oncol. 74: 38–47. [DOI] [PubMed] [Google Scholar]
- Jouet, M., Rosenthal, A., and Kenwrick, S. 1995. Exon 2 of the gene for neural cell adhesion molecule L1 is alternatively spliced in B cells. Brain Res. Mol. Brain Res. 30: 378–380. [DOI] [PubMed] [Google Scholar]
- Kan, Z., Rouchka, E.C., Gish, W.R., and States, D.J. 2001. Gene structure prediction and alternative splicing analysis using genomically aligned ESTs. Genome Res. 11: 889–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan, Z., States, D., and Gish, W. 2002. Selecting for functional alternative splices in ESTs. Genome Res. 12: 1837–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, M., Tezuka, T., Suziki, Y., Sugano, S., Hirai, M., and Yamamoto, T. 1999. Molecular cloning and characterization of a novel cbl-family gene, cbl-c. Gene 239: 145–154. [DOI] [PubMed] [Google Scholar]
- Kirschner, R., Rosenberg, T., Schultz-Heienbrok, R., Lenzner, S., Feil, S., Roepman, R., Cremers, F.P., Ropers, H.H., and Berger, W. 1999. RPGR transcription studies in mouse and human tissues reveal a retina-specific isoform that is disrupted in a patient with X-linked retinitis pigmentosa. Hum. Mol. Genet. 8: 1571–1578. [DOI] [PubMed] [Google Scholar]
- Kozmik, Z., Czerny, T., and Busslinger, M. 1997. Alternatively spliced insertions in the paired domain restrict the DNA sequence specificity of Pax6 and Pax8. EMBO J. 16: 6793–6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, C. 2003. Generating consensus sequences from partial order multiple sequence alignment graphs. Bioinformatics 19: 999–1008. [DOI] [PubMed] [Google Scholar]
- Lee, C., Grasso, C., and Sharlow, M. 2002. Multiple sequence alignment using partial order graphs. Bioinformatics 18: 452–464. [DOI] [PubMed] [Google Scholar]
- Lewis, B.P., Green, R.E., and Brenner, S.E. 2003. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc. Natl. Acad. Sci. 100: 189–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, F., Holt, I., Pertea, G., Karamycheva, S., Salzberg, S.L., and Quackenbush, J. 2000a. Gene Index analysis of the human genome estimates approximately 120,000 genes. Nat. Genet. 25: 239–240. [DOI] [PubMed] [Google Scholar]
- Liang, F., Holt, I., Pertea, G., Karamycheva, S., Salzberg, S.L., and Quackenbush, J. 2000b. An optimized protocol for analysis of EST sequences. Nucleic Acids Res. 28: 3657–3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat, L.E. 2002. Nonsense-mediated mRNA decay. Curr. Biol. 12: R196–R197. [DOI] [PubMed] [Google Scholar]
- Maquat, L.E. and Carmichael, G.G. 2001. Quality control of mRNA function. Cell 104: 173–176. [DOI] [PubMed] [Google Scholar]
- Markovtsov, V., Nikolic, J.M., Goldman, J.A., Turck, C.W., Chou, M.Y., and Black, D.L. 2000. Cooperative assembly of an hnRNP complex induced by a tissue-specific homolog of polypyrimidine tract binding protein. Mol. Cell. Biol. 20: 7463–7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meindl, A., Dry, K., Herrmann, K., Manson, F., Ciccodicola, A., Edgar, A., Carvalho, M.R., Achatz, H., Hellebrand, H., Lennon, A., et al. 1996. A gene (RPGR) with homology to the RCC1 guanine nucleotide exchange factor is mutated in X-linked retinitis pigmentosa (RP3). Nat. Genet. 13: 35–42. [DOI] [PubMed] [Google Scholar]
- Mironov, A.A., Fickett, J.W., and Gelfand, M.S. 1999. Frequent alternative splicing of human genes. Genome Res. 9: 1288–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrek, B. and Lee, C. 2002. A genomic view of alternative splicing. Nat. Genet. 30: 13–19. [DOI] [PubMed] [Google Scholar]
- Modrek, B., Resch, A., Grasso, C., and Lee, C. 2001. Genome-wide analysis of alternative splicing using human expressed sequence data. Nucleic Acids Res. 29: 2850–2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muilu, J., Rodriguez-Tome, P., and Robinson, A. 2001. GBuilder—An application for the visualization and integration of EST cluster data. Genome Res. 11: 179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai, A., Tanabe, M., Kawazoe, Y., Inazawa, J., Morimoto, R.I., and Nagata, K. 1997. HSF4, a new member of the human heat shock factor family which lacks properties of a transcriptional activator. Mol. Cell. Biol. 17: 469–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima, H., Szapary, D., and Simons Jr., S.S. 1995. The factor binding to the glucocorticoid modulatory element of the tyrosine aminotransferase gene is a novel and ubiquitous heteromeric complex. J. Biol. Chem. 270: 21893–21901. [DOI] [PubMed] [Google Scholar]
- Patek, C.E., Little, M.H., Fleming, S., Miles, C., Charlieu, J.P., Clarke, A.R., Miyagawa, K., Christie, S., Doig, J., Harrison, D.J., et al. 1999. A zinc finger truncation of murine WT1 results in the characteristic urogenital abnormalities of Denys-Drash syndrome. Proc. Natl. Acad. Sci. 96: 2931–2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Sanchez, C., Arias-de-la-Fuente, C., Gomez-Ferreria, M.A., Granadino, B., and Rey-Campos, J. 2000a. FHX.L and FHX.S, two isoforms of the human fork-head factor FHX (FOXJ2) with differential activity. J. Mol. Biol. 301: 795–806. [DOI] [PubMed] [Google Scholar]
- Perez-Sanchez, C., Gomez-Ferreria, M.A., de La Fuente, C.A., Granadino, B., Velasco, G., Esteban-Gamboa, A., and Rey-Campos, J. 2000b. FHX, a novel fork head factor with a dual DNA binding specificity. J. Biol. Chem. 275: 12909–12916. [DOI] [PubMed] [Google Scholar]
- Pruitt, K.D., Katz, K.S., Sicotte, H., and Maglott, D.R. 2000. Introducing RefSeq and LocusLink: Curated human genome resources at the NCBI. Trends Genet. 16: 44–47. [DOI] [PubMed] [Google Scholar]
- Pujols, L., Mullol, J., Perez, M., Roca-Ferrer, J., Juan, M., Xaubet, A., Cidlowski, J.A., and Picado, C. 2001. Expression of the human glucocorticoid receptor α and β isoforms in human respiratory epithelial cells and their regulation by dexamethasone. Am. J. Respir. Cell Mol. Biol. 24: 49–57. [DOI] [PubMed] [Google Scholar]
- Quackenbush, J., Liang, F., Holt, I., Pertea, G., and Upton, J. 2000. The TIGR Gene Indices: Reconstruction and representation of expressed gene sequences. Nucleic Acids Res. 28: 141–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quackenbush, J., Cho, J., Lee, D., Liang, F., Holt, I., Karamycheva, S., Parvizi, B., Pertea, G., Sultana, R., and White, J. 2001. The TIGR Gene Indices: Analysis of gene transcript sequences in highly sampled eukaryotic species. Nucleic Acids Res. 29: 159–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman, L., Bliskovski, V., Reinhold, W., and Zajac-Kaye, M. 2002. Alternative splicing of brain-specific PTB defines a tissue-specific isoform pattern that predicts distinct functional roles. Genomics 80: 245–249. [DOI] [PubMed] [Google Scholar]
- Rice, P., Longden, I., and Bleasby, A. 2000. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 16: 276–277. [DOI] [PubMed] [Google Scholar]
- Schuler, G. 1997. Pieces of the puzzle: Expressed sequence tags and the catalog of human genes. J. Mol. Med. 75: 694–698. [DOI] [PubMed] [Google Scholar]
- Skandalis, A., Ninniss, P.J., McCormac, D., and Newton, L. 2002. Spontaneous frequency of exon skipping in the human HPRT gene. Mut. Res. 501: 37–44. [DOI] [PubMed] [Google Scholar]
- Stimpfl, M., Tong, D., Fasching, B., Schuster, E., Obermair, A., Leodolter, S., and Zeillinger, R. 2002. Vascular endothelial growth factor splice variants and their prognostic value in breast and ovarian cancer. Clin. Cancer Res. 8: 2253–2259. [PubMed] [Google Scholar]
- Strausberg, R.L., Feingold, E.A., Klausner, R.D., and Collins, F.S. 1999. The mammalian gene collection. Science 286: 455–457. [DOI] [PubMed] [Google Scholar]
- Tanabe, M., Sasai, N., Nagata, K., Liu, X.D., Liu, P.C., Thiele, D.J., and Nakai, A. 1999. The mammalian HSF4 gene generates both an activator and a repressor of heat shock genes by alternative splicing. J. Biol. Chem. 274: 27845–27856. [DOI] [PubMed] [Google Scholar]
- Theriault, J.R., Charette, S.J., Lambert, H., and Landry, J. 1999. Cloning and characterization of hGMEB1, a novel glucocorticoid modulatory element binding protein. FEBS Lett. 452: 170–176. [DOI] [PubMed] [Google Scholar]
- Uthoff, S.M., Duchrow, M., Schmidt, M.H., Broll, R., Bruch, H.P., Strik, M.W., and Galandiuk, S. 2002. VEGF isoforms and mutations in human colorectal cancer. Int. J. Cancer 101: 32–36. [DOI] [PubMed] [Google Scholar]
- Vervoort, R., Lennon, A., Bird, A.C., Tulloch, B., Axton, R., Miano, M.G., Meindl, A., Meitinger, T., Ciccodicola, A., and Wright, A.F. 2000. Mutational hot spot within a new RPGR exon in X-linked retinitis pigmentosa. Nat. Genet. 25: 462–466. [DOI] [PubMed] [Google Scholar]
- Wang, L., Miura, M., Bergeron, L., Zhu, H., and Yuan, J. 1994. Ich-1, an Ice/ced-3-related gene, encodes both positive and negative regulators of programmed cell death. Cell 78: 739–750. [DOI] [PubMed] [Google Scholar]
- Williams, A.J., Khachigian, L.M., Shows, T., and Collins, T. 1995. Isolation and characterization of a novel zinc-finger protein with transcription repressor activity. J. Biol. Chem. 270: 22143–22152. [DOI] [PubMed] [Google Scholar]
- Yamamoto, K.K., Gonzalez, G.A., Menzel, P., Rivier, J., and Montminy, M.R. 1990. Characterization of a bipartite activator domain in transcription factor CREB. Cell 60: 611–617. [DOI] [PubMed] [Google Scholar]
- Zhuo, D., Zhao, W.D., Wright, F.A., Yang, H.Y., Wang, J.P., Sears, R., Baer, T., Kwon, D.H., Gordon, D., Gibbs, S., et al. 2001. Assembly, annotation, and integration of UNIGENE clusters into the human genome draft. Genome Res. 11: 904–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- ftp://ftp.ncbi.nih.gov/genomes/H_sapiens; human genome sequence, downloaded from January 2002.
- ftp://ftp.ncbi.nih.gov/repository/UniGene; human EST and mRNA sequences from UniGene, downloaded from January 2002.
- http://www.expasy.org/tools/pi_tool.html; ExPASy Proteomic Tool.
- http://www.bioinformatics.ucla.edu/ASAP; Alternative Splicing Annotation Project.