Abstract
Comparative genomics is a promising approach to the challenging problem of eukaryotic regulatory element identification, because functional noncoding sequences may be conserved across species from evolutionary constraints. We systematically analyzed known human and Saccharomyces cerevisiae regulatory elements and discovered that human regulatory elements are more conserved between human and mouse than are background sequences. Although S. cerevisiae regulatory elements do not appear to be more conserved by comparison of S. cerevisiae to Schizosaccharomyces pombe, they are more conserved when compared with multiple other yeast genomes (Saccharomyces paradoxus, Saccharomyces mikatae, and Saccharomyces bayanus). Based on these analyses, we developed a sequence-motif-finding algorithm called CompareProspector, which extends Gibbs sampling by biasing the search in regions conserved across species. Using human–mouse comparison, CompareProspector identified known motifs for transcription factors Mef2, Myf, Srf, and Sp1 from a set of human-muscle-specific genes. It also discovered the NFAT motif from genes up-regulated by CD28 stimulation in T-cells, which implies the direct involvement of NFAT in mediating the CD28 stimulatory signal. Using Caenorhabditis elegans–Caenorhabditis briggsae comparison, CompareProspector found the PHA-4 motif and the UNC-86 motif. CompareProspector outperformed many other computational motif-finding programs, demonstrating the power of comparative genomics-based biased sampling in eukaryotic regulatory element identification.
Regulatory elements are short DNA sequences that determine the timing, location, and level of gene expression (Pennacchio and Rubin 2001). Although often only 5 to 20 bp in length, they are critical for understanding gene regulation. Experimental procedures for regulatory element discovery such as electrophoretic mobility shift assays and nuclease protection assays typically verify one element at a time. Therefore, computational methods have been developed to predict regulatory elements (characterized as sequence motifs) and their locations in a high-throughput manner.
A widely used computational strategy for regulatory element identification is to search for common sequence patterns in the promoters of genes from a single species that are known or hypothesized to be coregulated (e.g., coexpressed genes from microarray experiments). Methods such as Consensus (Hertz et al. 1990), MEME (Bailey and Elkan 1994), and Gibbs Motif Sampler (Liu et al. 1995) [and its variations AlignACE (Roth et al. 1998) and BioProspector (Liu et al. 2001)] have successfully applied this strategy in finding regulatory elements from lower organisms such as bacteria and yeast. In higher eukaryotes, however, regulatory elements tend to be shorter and dispersed among long intergenic sequences, and their identification is significantly more difficult. Existing methods not only take longer to converge, but also often converge on sequence motifs that are not biologically relevant.
As the complete sequences of >130 microbial and 20 eukaryotic genomes have become publicly available (http://www.ncbi.nlm.nih.gov/Genomes/index.html), a comparative genomics strategy has been proposed to aid regulatory element identification by examining orthologous sequences from multiple species. In bacteria, transcription-factor-binding motifs have been discovered simply by comparing orthologous promoters from multiple species (McGuire et al. 2000; McCue et al. 2001; Qin et al. 2003). In higher eukaryotes, comparison of orthologous promoters from multiple species has been helpful in identifying the regulatory elements of a single gene (Blanchette and Tompa 2002). However, when finding motifs from groups of genes hypothesized to be coregulated, simple inclusion often adds more noise than signal. Hardison (2000) proposed searching for functional noncoding sequences from genomic regions that are highly conserved across species, which usually are under stronger evolutionary constraints than nonfunctional (“background”) DNA. When 81% of the least conserved upstream sequences between human and mouse were filtered out, the Gibbs Motif Sampler was able to find three out of the six known motifs from 28 human-muscle-specific genes (Wasserman et al. 2000).
Choosing a good conservation threshold is critical to balancing sensitivity and specificity in the above approach. To find a good pair of species to compare and good sequence conservation threshold, we performed systematic analyses of sequence conservation of known human regulatory elements (compared with mouse) and Saccharomyces cerevisiae elements (compared with Schizosaccharomyces pombe using pairwise alignment, Saccharomyces paradoxus, Saccharomyces mikatae, and Saccharomyces bayanus using multiple alignment). We chose to study the regulatory elements in human and S. cerevisiae because the complete genomes as well as a significant number of experimentally determined regulatory elements are available for these organisms.
Based on the results of our conservation analyses, we designed CompareProspector, aimed to take advantage of comparative genomics information to aid sequence motif-finding in higher eukaryotes. Given the upstream sequences of a group of genes that are known or predicted to be coregulated in a given species, and local sequence conservation (represented as window percent identity values, or WPIDs) calculated based on alignments with orthologous sequences, CompareProspector uses a Gibbs sampling approach to search for motifs in the input sequences, biasing the search toward conserved regions by integrating sequence conservation into the posterior probability in the sampling process. We tested CompareProspector on two data sets from humans using human–mouse comparisons and two data sets from Caenorhabditis elegans using C. elegans–Caenorhabditis briggsae comparisons, and compared its performance with other motif-finding programs.
RESULTS
Conservation of Known Human Regulatory Elements Between Human and Mouse
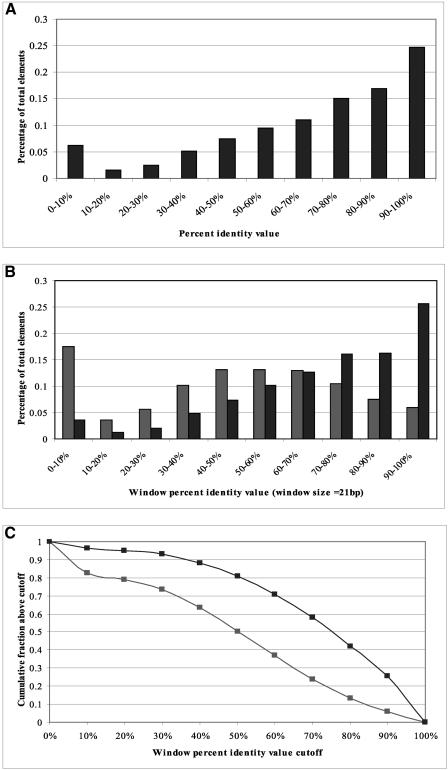
Among all the human regulatory elements documented in TRANSFAC (Wingender et al. 2000), we selected 467 elements upstream of 127 human genes (with RefSeq sequences) whose orthologous mouse genes can be retrieved from LocusLink. The genomic sequences upstream (5000 bp upstream of RefSeqs) of each orthologous gene pair were retrieved from the respective genome and aligned by a global alignment program, LAGAN (Brudno et al. 2003). We mapped the known regulatory elements onto the human–mouse alignments and calculated their percent identity values, finding that 81% of the elements are ≥50% conserved between human and mouse, and the average percent identity value of all the regulatory elements is 69.5% (Fig. 1A).
Figure 1.
Conservation of known human regulatory elements between human and mouse. (A) Histogram of percent identity values of known human regulatory elements between human and mouse. (B) Histogram of WPID values (21-bp window) of known human regulatory elements (dark) and background sequences (light) within 1000 bp upstream of TSS. In general, the 21-bp windows around known human regulatory elements are more conserved between human and mouse than those around background sequences. (C) Cumulative distribution of WPID values of known human regulatory elements and background sequences. WPID value cutoffs around 0.5 to 0.8 seem to give the biggest fraction difference between regulatory elements and background.
In practical motif-finding problems, the width of a regulatory element is often unknown and variable with different transcription factors. A 21-bp window, a span wider than most of the known eukaryotic regulatory elements, can be used to characterize the local sequence context around most elements. Therefore, we studied the sequence conservation using a 21-bp sliding window along the human–mouse alignment. We focused on 333 known regulatory elements that are within 1000 bp upstream of the transcription start site (TSS), and “background sequences”—all 21-bp windows from the same upstream regions that do not overlap with a known element. Known elements have a different window percent identity (WPID) distribution from background sequences, with an average WPID value of 75% compared with 53% (Fig. 1B). These differences indicate that we can use the WPID value to guide regulatory element identification, as sequences with high WPID values have a higher regulatory element signal-versus-noise ratio. The cumulative distributions of known regulatory elements and background (Fig. 1C) also indicate that cutoffs around 0.5 to 0.8 achieve the best balance between element enrichment and background elimination.
Conservation of Known S. cerevisiae Regulatory Elements Among Yeasts
We collected 274 known regulatory elements in S. cerevisiae from SCPD (Zhu and Zhang 1999) and 107 pairs of orthologous genes between S. cerevisiae and S. pombe. The sequences 1000 bp upstream of every orthologous gene pair were aligned with LAGAN, and the WPID values (window size = 21bp) of known regulatory elements and background sequences were calculated. The average WPID value between S. cerevisiae and S. pombe is 42% for known regulatory elements and 43% for the background sequences (Fig. 2A,B). S. cerevisiae and S. pombe diverged 300–1144 million years ago (Wood et al. 2002). Therefore, the regulatory elements in S. cerevisiae and S. pombe may have diverged to such an extent as to be indistinguishable from background sequences by comparison of the two genomes. The sequences of three other yeast species (S. paradoxus, S. mikatae, and S. bayanus) have recently become available (Kellis et al. 2003). We aligned the orthologous upstream regions from S. cerevisiae and these three species using MLAGAN, a multiple sequence alignment program (Brudno et al. 2003), and calculated WPIDs for both known regulatory elements and background sequences (Fig. 2C,D). In this case, known regulatory elements are more conserved than background sequences, but the two distributions do not differ as much as those of the human–mouse comparisons.
Figure 2.
Conservation of known S. cerevisiae regulatory elements in yeasts. (A) Histogram of WPID values (21-bp window) of known S. cerevisiae regulatory elements (dark) and background sequences (light) within 1000 bp upstream of the translation start site between S. cerevisiae and S. pombe. (B) Cumulative distribution of WPID values. (C) Histogram of WPID values (21-bp window) of known S. cerevisiae regulatory elements and background sequences among S. cerevisiae, S. paradoxus, S. mikatae, and S. bayanus. (D) Cumulative distribution of the WPID values among the four yeast species. The conservation distributions of known elements and background are very similar between S. cerevisiae and S. pombe, without the good separation observed in the human–mouse comparisons. The distributions of known elements and background among the four yeasts are more differentiable than those of S. cerevisiae and S. pombe, indicating that sequences from multiple species can help identify regulatory elements.
The above analyses indicate that the identification of human regulatory elements is likely to benefit from genome comparison with mouse, and that of S. cerevisiae regulatory elements will benefit from comparison with multiple related species, but not from comparison with S. pombe alone. Although at present there is not enough genome sequence and regulatory element data to establish the most suitable range of evolutionary distances for cross-genome comparison, a general rule of thumb is that the evolutionary distance between the species should be neither too far nor too close. C. elegans and C. briggsae diverged 25–50 million years ago (Kent and Zahler 2000), which is comparable to the human/mouse divergence age of 75 million years (Waterston et al. 2002). Therefore, we expect C. elegans–C. briggsae comparison to be beneficial to C. elegans regulatory element identification.
CompareProspector
We developed CompareProspector to take advantage of comparative genomics information to aid sequence motif finding. CompareProspector is built upon BioProspector (Liu et al. 2001), which is an extension of the original Gibbs Sampler (Liu et al. 1995) with improved flexibility and performance. CompareProspector takes as input a list of sequences from one species that is predicted to share common regulatory element(s). Such sequences can be obtained from high-throughput genomics techniques such as gene expression profile clustering or chromatin immunoprecipitation followed by microarray (ChIP-chip). It also takes as input an array of WPID for each input sequence (when its ortholog is available). We calculate the WPIDs based on the LAGAN alignment of each input sequence with its ortholog from another species. In the Gibbs sampling iterations, CompareProspector biases the motif finding toward sequences conserved across species. First of all, the user can specify two WPID thresholds, Tch (high conservation threshold) and Tcl (low conservation threshold). In BioProspector, a site score Ax is calculated for every site x in the input sequence as the ratio of the probability of generating x from the motif model over the probability of generating x from the background distribution. A new site is sampled with probability proportional to Ax. In CompareProspector, during initial iterations of Gibbs sampling, only positions whose WPID values are above Tch are sampled. Subsequently, the WPID cutoff is gradually decreased from Tch to Tcl to allow sampling of less conserved positions. The new site score A′x is weighted by sequence conservation (A′x = Ax × WPIDx, WPIDx being the WPID of site x) to favor sampling of more conserved sequences. Sequences without orthologs are assigned Tcl as the WPIDx for all x, so they only participate in sampling in later iterations. Finally, in the original BioProspector, sites with a high enough score Ax are automatically added to the motif without sampling. CompareProspector restricts autoatic additions to only sites whose WPIDs are above Tch. This step further down weighs the influence of divergent sites and sequences without orthologs. The output of CompareProspector includes a list of highest-scoring motifs as position-specific probability matrices, the individual sites used to construct each motif, and the locations of the sites on the input sequences.
Based on the results of the above conservation analysis of known human regulatory elements, we used 0.8 as Tch and 0.5 as Tcl for human–mouse comparisons. Although the divergence of C. elegans and C. briggsae is more recent than that of human and mouse, we chose 0.5 as Tch and 0.3 as Tcl for C. elegans–C. briggsae comparison because of their shorter generation time. We applied CompareProspector on two human and two C. elegans data sets.
Results of CompareProspector on Human Data Sets
We first tested CompareProspector on 28 pairs of human–mouse orthologous genes up-regulated in skeletal muscles, a data set with detailed annotation of experimentally verified regulatory elements (Wasserman et al. 2000). Known regulatory elements in this data set include binding sites for transcription factors Mef2, Myf, Srf, Sp1, Tef, and Nvl. We applied CompareProspector, using human–mouse comparisons, to the upstream sequences of the 28 human genes to search for motifs (summarized results in Table 1). When the motif width was set to 8 bp, the top 15 motifs identified by CompareProspector all have the consensus RACAGSTG, which agrees with the known MYF consensus RRCAGSTG. Nine out of the 20 individual sites of this motif reported by CompareProspector have experimental evidence of function. Using conservation filtering, the Gibbs Motif Sampler (Wasserman et al. 2000) also identified the MYF motif with 19 sites, six of which are known sites. When the width was set to 10 bp, all of the top 15 motifs found by CompareProspector share the MEF2 motif consensus. Seven out of the 13 sites reported by CompareProspector are known sites, whereas five out of the eight reported by Wasserman et al. are known to be functional. To find other motifs, we masked out all the sites matching the MYF and MEF2 consensus and ran CompareProspector again. At motif width 10, the best motif identified was the SP1 motif, with one out of the 13 reported sites being a known SP1 site. The SRF motif was also identified, with three out of the 15 sites being known sites. CompareProspector failed to identify the NVL motif, because the eight NVL binding sites vary too much in sequence to construct a discernable consensus. It also failed to find the TEF motif, which has only five known sites in the 28 genes.
Table 1.
Motifs Identified by CompareProspector From the Human Skeletal Muscle Data Set
In summary, all the motifs identified by CompareProspector were known motifs. The conservation filtering approach by Wasserman et al. (2000) identified three motifs (MEF2, MYF, and SRF), whereas CompareProspector identified four (MEF2, MYF, SRF, and SP1). For the three motifs identified by both approaches, CompareProspector improved the individual element prediction sensitivity from 34% (Wasserman et al. 2000) to 41% while maintaining the same false-positive rate of 63%. It is worth noting that the false-positive rate is an overestimation, because some predicted sites may, indeed, have biological function. When other motif-finding programs were applied to this data set, BioProspector (Liu et al. 2001) failed to identify any known motifs; Consensus (Hertz et al. 1990) identified the SP1 motif; AlignACE (Roth et al. 1998) and MEME (Bailey and Elkan 1994) each identified the SP1 and MEF2 motifs.
The second human data set contained 11 genes shown to be up-regulated by the activation of CD28, a surface receptor on T-cells (Diehn et al. 2002). The genomic sequences of 10 genes could be reliably identified, and the orthologous mouse sequences of eight genes were retrieved using LocusLink. Because the 5′-ends of RefSeqs often do not extend to TSS, sequences 3000 bp upstream of the start of each RefSeq were retrieved. Diehn et al. (2002) hypothesized that the nuclear factor of activated T-cells (NFAT) plays a key role in mediating the CD28 signal based on the enrichment of independently confirmed NFAT targets among the CD28 responsive genes (TNF, CD69, SCYA3, and EGR2). Whereas the consensus of NFAT was known to be TGGAAA (Rao et al. 1997), none of the de novo computational methods, including AlignACE, BioProspector, Consensus, and MEME, were able to find this motif using the human sequences as input. When we applied CompareProspector to this data set, the top motif discovered was TTGGAAA, which matches the known NFAT consensus. CompareProspector identified nine sites for this motif, one in each of the upstream regions of the CD28-responsive genes except the gene TNFRSF11A. Our findings support the claim that NFAT may play a key role in mediating the CD28 costimulatory signal. They also indicate that the nine genes with NFAT-binding sites are directly regulated by NFAT, rather than being regulated by factors downstream from NFAT.
Results of CompareProspector on C. elegans Data Sets
Our first C. elegans data set had 240 genes obtained from microarray as up-regulated by PHA-4, a transcription factor that specifies organ identity for C. elegans pharyngeal cells (Gaudet and Mango 2002). Of these, 211 genes have identifiable genomic sequences from WormBase (http://www.wormbase.org), and among them 122 have C. briggsae orthologs. We searched for motifs in the 1000 bp upstream of the translation start site of the genes, as the transcription start sites of many C. elegans genes are yet to be determined. All the 10 top-ranking motifs identified by CompareProspector have the consensus TGTKTGC (Table 2), which agrees with the known PHA-4 binding consensus TRTT-KRY (Overdier et al. 1994). As expected, the predicted sites are conserved between C. elegans and C. briggsae: among the 138 individual sites predicted in the best motif, 49 are upstream of C. elegans genes with C. briggsae orthologs. Of these 49 sites, 17 (35%) are 100% conserved between C. elegans and C. briggsae, and seven differ by only one nucleotide. AlignACE and Consensus failed to find this motif. MEME reported the PHA-4 motif as the fifth-ranked motif, but took 9 h to run on a 400-MHz Sun Sparc II workstation, compared with 54 min by CompareProspector. BioProspector took 63 min to run and identified the PHA-4 motif as the eighth-ranked motif. When all sites matching the PHA-4 motif consensus were removed from the data set, CompareProspector discovered a motif with consensus AGAGACGCAG. This motif was also identified from genes involved in acute ethanol exposure in worms, and mutation of this motif led to disruption of the ethanol response (14th International C. elegans Conference abstract 1113C). The function of this motif in pharyngeal cells remains to be explored.
Table 2.
Motifs Discovered by CompareProspector on the C. elegans PHA-4 Data Set
Our second data set contained three C. elegans genes that were known to bind to UNC-86, a transcription factor necessary for the production and differentiation of touch cells (Duggan et al. 1998). DNase I footprinting experiments were conducted on mec-3 (Xue et al. 1992) and mec-7 (Duggan et al. 1998), and the UNC-86-binding consensus was determined to be AAATKCAT. We searched the 1000-bp region upstream of the translation start of the genes for motifs, using C. elegans–C. briggsae conservation information. All top three motifs reported by CompareProspector have consensus CAATGCAT, which resembles the known binding consensus. In addition, the sites identified upstream of the mec-3 and mec-7 genes both lay in the region known to be protected by UNC-86 in DNase I protection assays. When using BioProspector, the CAATGCAT motif was also found, but only as the fourth-ranked motif. AlignACE, Consensus, and MEME failed to identity this motif.
Comparison Between Conservation Filtering and Biased Sampling
One simple approach to integrate comparative genomics in existing motif-finding programs is to filter out sequences not conserved across species (Wasserman et al. 2000). The drawback of this approach is the difficulty in choosing a good conservation cutoff and dealing with input sequences that do not have identifiable orthologs. For each of our four data sets, we compiled new input sequences by filtering out sequences without orthologs and nucleotides whose conservation level is below Tch (0.8 for human and 0.5 for C. elegans). AlignACE, BioProspector, Consensus, and MEME all have improved performance— BioProspector identified the same known motifs as CompareProspector from the four data sets; AlignACE, Consensus, and MEME all found the UNC-86 motif, but failed to find the NFAT motif from the CD28 data set. If we only filter out nucleotides whose conservation level was below Tcl (0.5 for human and 0.3 for C. elegans), most of the improvements from these algorithms diminished. This indicates that in order for conservation filtering to improve motif finding, the conservation cutoff needs to be set high. However, at this high cutoff, functional sites that are not conserved enough will be excluded from consideration. CompareProspector starts motif finding from highly conserved positions, then gradually lowers the threshold to sample more positions. CompareProspector identified 138 potential PHA-4 binding sites in 138 genes, whereas BioProspector with Tch conservation-filtered input identified only 83 sites in 83 genes. At least one gene, ZK816.4, predicted to contain a PHA-4 site by CompareProspector but missed by BioProspector, is known to be a true PHA-4 target (Gaudet and Mango 2002). Even when we lowered the Tcl to be 0.01 for both human and C. elegans, CompareProspector still managed to find the same known motifs as before in all the data sets.
DISCUSSION
Comparative genomics is widely regarded as a promising approach for identification of eukaryotic regulatory elements. In this paper, we presented a systematic analysis of the conservation of known regulatory elements in human and S. cerevisiae by comparison with other species. It would be desirable to repeat these comparisons for worms (C. elegans and C. briggsae) and diptera (Drosophila melanogaster and Anopheles gambiae, the malaria mosquito), but information on their regulatory elements was too limited to allow for any systematic analysis. Human–mouse comparisons were helpful in differentiating regulatory elements from background sequences, but S. cerevisiae–S. pombe pairwise comparisons were not. Comparison of sequences from multiple species showed considerable promise in differentiating S. cerevisiae known regulatory elements from background sequences. However, the difference between the two distributions is not as significant as the one seen in human–mouse comparisons. For future work, two strategies can be used to improve the separation. One is to use more sophisticated statistics, such as those used by Elnitski et al. (2003), to maximize the separation between known regulatory elements and background sequences. The other strategy is to include more species (Cliften et al. 2003; Kellis et al. 2003). Once known regulatory elements can be sufficiently separated from background, we can extend CompareProspector from pairwise species to multiple species. This can readily be done by using sequence similarity measures calculated from multiple alignments rather than pairwise alignments.
CompareProspector identifies regulatory elements using information from both intraspecies pattern enrichment (e.g., coregulated genes from the same species) and interspecies sequence conservation. This distinguishes it from other phylogenetic footprinting programs that use orthologous promoters of a single gene from multiple species to identify regulatory elements. Although many more eukaryotic genomes are being sequenced, the number of whole eukaryotic genome sequences available will remain limited because of the high cost and long duration of such genome sequencing projects. Before enough eukaryotic genomes are completely sequenced to support the single-gene, multiple-species phylogenetic footprinting approach, our multiple-gene, multiple-species approach provides a better alternative.
Both global aligners (such as LAGAN) and local aligners (such as BLASTZ; Schwartz et al. 2003) can be used to calculate the local sequence context around a site. D. Pollard and colleagues simulated sequence divergence with interspersed blocks of constraint and evaluated the quality of alignments in the blocks of constraint using both LAGAN and BLASTZ. They found that for sequences with low divergence (<1 substitution per unconstrained site), LAGAN and BLASTZ give similar results. However, for medium to highly divergent sequences (>1 substitution per unconstrained site), BLASTZ has poor alignment coverage in constrained blocks, whereas LAGAN provides complete alignment coverage and maintains high alignment accuracy (D. Pollard and C. Bergman, pers. comm.).
METHODS
We collected known human regulatory elements from the Transcription Factor Database (TRANSFAC 6.2; Wingender et al. 2000) and known S. cerevisiae regulatory elements from the S. cerevisiae Promoter Database (SCPD; Zhu and Zhang 1999), respectively. To ensure data quality, we selected regulatory elements according to the following criteria: (1) Its location relative to the gene is known. (2) The documented element sequence is found in genomic sequences extracted from the genome assembly. (3) The regulated gene has a known orthologous gene in mouse (for human elements) or S. pombe (for S. cerevisiae elements). Given the accession number of a gene, we retrieved its gene sequence from the above Web sites and mapped its location on the genome using BLAT (Kent 2002). With the location of the gene, we then retrieved its upstream sequences from the genome for analysis.
We identified the orthologous gene pairs between human and mouse with LocusLink (Pruitt and Maglott 2001). For S. cerevisiae and S. pombe, the orthologous gene pairs were identified by BLASTP using the “reciprocal best hit” criteria (Altschul et al. 1997). Orthologous gene pairs for S. cerevisiae, S. paradoxus, S. mikatae, and S. bayanus were obtained from the Saccharomyces Genome Database (http://www.yeastgenome.org). As the proteome of C. briggsae is not yet available, the orthologous gene pairs between C. elegans and C. briggsae were identified using TBLASTN (Altschul et al. 1997). We chose the top hit from the C. briggsae genome as the C. elegans ortholog if the expectation value was <10–15 and the TBLASTN hit started within the first 30 amino acids of the C. elegans query. For some C. elegans genes that are organized in operons (Blumenthal et al. 2002), we also included the first gene of the operon in our data set.
To align the genomic sequence of orthologous genes, we used the global alignment program LAGAN and MLAGAN with default parameters (Brudno et al. 2003). For pairwise alignment, the percent identity value of a nucleotide was assigned 1 if the nucleotide is the same in the pairwise alignment and 0 otherwise. For multiple alignment, the percent identity value was 1 if the nucleotide is completely conserved in all sequences and 0 otherwise. The percent identity value of a regulatory element was calculated as the average percent identity over the length of the element. The window percent identify (WPID) value of a nucleotide was calculated as the average percent identity value of the 21-bp window centered at that nucleotide.
Built on top of BioProspector (Liu et al. 2001), CompareProspector also uses Gibbs sampling for motif finding but biases toward regions that are conserved across species. The inputs of CompareProspector are:
A set of sequences that may share the same regulatory element(s) (e.g., the promoter sequences of genes in the same expression profile cluster).
An array of WPIDs associated with each sequence based on the LAGAN alignment of the input sequence with its ortholog.
A high (Tch) and a low (Tcl) conservation threshold for window percent identity values. From our experiences, cutoff values within a reasonable range give similar results.
The width of the motif to search for. When motif width is unknown, different widths from 6 bp up to 15 bp are recommended for testing. The width should be specified shorter if top motifs have very degenerate positions at the two ends, and specified longer if the consensuses of several top motifs overlap and there are conserved nonoverlapping positions at either ends.
A file containing probabilities that characterize the background nucleotide distribution. When this is not available, CompareProspector estimates the background nucleotide distribution from all the input sequences.
CompareProspector outputs a list of highest-scoring motifs as position-specific probability matrices, the individual sites used to construct each motif, and the locations of the sites on the input sequences. The same motif or similar motifs being reported repeatedly is an indication of the motif's statistical significance.
Acknowledgments
We thank Cristian Castillo-Davis, Max Diehn, Josh Stuart, James Lund, Mike Brudno, Jason Lieb, Stuart Kim, and Douglas Brutlag for their insight and help during the preparation of this manuscript. We also thank the three anonymous reviewers for their constructive comments. We acknowledge the support of NIH grant GM61374 (R.B.A. and Y.L.), NIH grant HG003162 (Y.L. and S.B.), and a Stanford Graduate Fellowship (Y.L.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1327604.
Footnotes
[Supplemental data are available at www.genome.org and at http://compareprospector.stanford.edu. The program CompareProspector is available at http://compareprospector.stanford.edu.]
References
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey, T.L. and Elkan, C. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2: 28–36. [PubMed] [Google Scholar]
- Blanchette, M. and Tompa, M. 2002. Discovery of regulatory elements by a computational method for phylogenetic footprinting. Genome Res. 12: 739–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal, T., Evans, D., Link, C.D., Guffanti, A., Lawson, D., Thierry-Mieg, J., Thierry-Mieg, D., Chiu, W.L., Duke, K., Kiraly, M., et al. 2002. A global analysis of Caenorhabditis elegans operons. Nature 417: 851–854. [DOI] [PubMed] [Google Scholar]
- Brudno, M., Do, C.B., Cooper, G.M., Kim, M.F., Davydov, E., Green, E.D., Sidow, A., and Batzoglou, S. 2003. Lagan and Multi-Lagan: Efficient tools for large-scale multiple alignment of genomic DNA. Genome Res. 13: 721–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliften, P., Sudarsanam, P., Desikan, A., Fulton, L., Fulton, B., Majors, J., Waterston, R., Cohen, B.A., and Johnston, M. 2003. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science 301: 71–76. [DOI] [PubMed] [Google Scholar]
- Diehn, M., Alizadeh, A.A., Rando, O.J., Liu, C.L., Stankunas, K., Botstein, D., Crabtree, G.R., and Brown, P.O. 2002. Genomic expression programs and the integration of the Cd28 costimulatory signal in T cell activation. Proc. Natl. Acad. Sci. 99: 11796–11801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggan, A., Ma, C., and Chalfie, M. 1998. Regulation of touch receptor differentiation by the Caenorhabditis elegans Mec-3 and Unc-86 genes. Development 125: 4107–4119. [DOI] [PubMed] [Google Scholar]
- Elnitski, L., Hardison, R.C., Li, J., Yang, S., Kolbe, D., Eswara, P., O'Connor, M.J., Schwartz, S., Miller, W., and Chiaromonte, F. 2003. Distinguishing regulatory DNA from neutral sites. Genome Res. 13: 64–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet, J. and Mango, S.E. 2002. Regulation of organogenesis by the Caenorhabditis elegans Foxa protein Pha-4. Science 295: 821–825. [DOI] [PubMed] [Google Scholar]
- Hardison, R.C. 2000. Conserved noncoding sequences are reliable guides to regulatory elements. Trends Genet. 16: 369–372. [DOI] [PubMed] [Google Scholar]
- Hertz, G.Z., Hartzell III, G.W., and Stormo, G.D. 1990. Identification of consensus patterns in unaligned DNA sequences known to be functionally related. Comput. Appl. Biosci. 6: 81–92. [DOI] [PubMed] [Google Scholar]
- Kellis, M., Patterson, N., Endrizzi, M., Birren, B., and Lander, E.S. 2003. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423: 241–254. [DOI] [PubMed] [Google Scholar]
- Kent, W.J. 2002. BLAT—The BLAST-like alignment tool. Genome Res. 12: 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent, W.J. and Zahler, A.M. 2000. Conservation, regulation, synteny, and introns in a large-scale C. briggsae–C. elegans genomic alignment. Genome Res. 10: 1115–1125. [DOI] [PubMed] [Google Scholar]
- Liu, J.S., Neuwald, A.F., and Lawrence, C.E. 1995. Bayesian Models for multiple local sequence alignment and Gibbs sampling strategies. J. Am. Stat. Assoc. 90: 1156–1170. [Google Scholar]
- Liu, X., Brutlag, D.L., and Liu, J.S. 2001. Bioprospector: Discovering conserved DNA motifs in upstream regulatory regions of co-expressed genes. Pac. Symp. Biocomput. 127–138. [PubMed]
- McCue, L., Thompson, W., Carmack, C., Ryan, M.P., Liu, J.S., Derbyshire, V., and Lawrence, C.E. 2001. Phylogenetic footprinting of transcription factor binding sites in proteobacterial genomes. Nucleic Acids Res. 29: 774–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire, A.M., Hughes, J.D., and Church, G.M. 2000. Conservation of DNA regulatory motifs and discovery of new motifs in microbial genomes. Genome Res. 10: 744–757. [DOI] [PubMed] [Google Scholar]
- Overdier, D.G., Porcella, A., and Costa, R.H. 1994. The DNA-binding specificity of the hepatocyte nuclear factor 3/Forkhead domain is influenced by amino-acid residues adjacent to the recognition helix. Mol. Cell. Biol. 14: 2755–2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennacchio, L.A. and Rubin, E.M. 2001. Genomic strategies to identify mammalian regulatory sequences. Nat. Rev. Genet. 2: 100–109. [DOI] [PubMed] [Google Scholar]
- Pruitt, K.D. and Maglott, D.R. 2001. RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res. 29: 137–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, Z.S., McCue, L.A., Thompson, W., Mayerhofer, L., Lawrence, C.E., and Liu, J.S. 2003. Identification of co-regulated genes through Bayesian clustering of predicted regulatory binding sites. Nat. Biotechnol. 21: 435–439. [DOI] [PubMed] [Google Scholar]
- Rao, A., Luo, C., and Hogan, P.G. 1997. Transcription factors of the NFAT family: Regulation and function. Annu. Rev. Immunol. 15: 707–747. [DOI] [PubMed] [Google Scholar]
- Roth, F.P., Hughes, J.D., Estep, P.W., and Church, G.M. 1998. Finding DNA regulatory motifs within unaligned noncoding sequences clustered by whole-genome mRNA quantitation. Nat. Biotechnol. 16: 939–945. [DOI] [PubMed] [Google Scholar]
- Schwartz, S., Kent, W.J., Smit, A., Zhang, Z., Baertsch, R., Hardison, R.C., Haussler, D., and Miller, W. 2003. Human–mouse alignments with BLASTZ. Genome Res. 13: 103–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman, W.W., Palumbo, M., Thompson, W., Fickett, J.W., and Lawrence, C.E. 2000. Human–mouse genome comparisons to locate regulatory sites. Nat. Genet. 26: 225–228. [DOI] [PubMed] [Google Scholar]
- Waterston, R.H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J.F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562. [DOI] [PubMed] [Google Scholar]
- Wingender, E., Chen, X., Hehl, R., Karas, H., Liebich, I., Matys, V., Meinhardt, T., Pruss, M., Reuter, I., and Schacherer, F. 2000. Transfac: An integrated system for gene expression regulation. Nucleic Acids Res. 28: 316–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood, V., Gwilliam, R., Rajandream, M.A., Lyne, M., Lyne, R., Stewart, A., Sgouros, J., Peat, N., Hayles, J., Baker, S., et al. 2002. The genome sequence of Schizosaccharomyces pombe. Nature 415: 871–880. [DOI] [PubMed] [Google Scholar]
- Xue, D., Finney, M., Ruvkun, G., and Chalfie, M. 1992. Regulation of the Mec-3 gene by the C. elegans homeoproteins Unc-86 and Mec-3. EMBO J. 11: 4969–4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J. and Zhang, M.Q. 1999. SCPD: A promoter database of the yeast Saccharomyces cerevisiae. Bioinformatics 15: 607–611. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- ftp://genome-ftp.stanford.edu/pub/yeast/genome_seq/all_fasta/; Saccharomyces Genome Database (S. cerevisiae).
- http://compareprospector.stanford.edu; CompareProspector.
- http://genome.ucsc.edu/; UCSC Genome Browser (human, June 2002 assembly).
- http://genome.ucsc.edu/; UCSC Genome Browser (mouse, Feb. 2002 assembly).
- http://www.genome.wustl.edu/projects/cbriggsae/; C. briggsae sequencing at GSC (C. briggsae).
- http://www.ncbi.nlm.nih.gov/Genomes/index.html; microbial and eukaryotic genomes.
- http://www.sanger.ac.uk/Projects/S_pombe/; The S. pombe Genome Project (S. pombe).
- http://www.wormbase.org/; WormBase (C. elegans).
- http://www.yeastgenome.org/; Saccharomyces Genome Database (S. paradoxus, S. mikatae, and S. bayanus).