Abstract
Comparative sequence analysis has evolved as an essential technique for identifying functional coding and noncoding elements conserved throughout evolution. Here, we introduce zPicture, an interactive Web-based sequence alignment and visualization tool for dynamically generating conservation profiles and identifying evolutionarily conserved regions (ECRs). zPicture is highly flexible, because critical parameters can be modified interactively, allowing users to differentially predict ECRs in comparisons of sequences of different phylogenetic distances and evolutionary rates. We demonstrate the application of this module to identify a known regulatory element in the HOXD locus, in which functional ECRs are difficult to discern against the highly conserved genomic background. zPicture also facilitates transcription factor binding-site analysis via the rVista tool portal. We present an example of the HBB complex when zPicture/rVista combination specifically pinpoints to two ECRs containing GATA-1, NF-E2, and TAL1/E47 binding sites that were identified previously as transcriptional enhancers. In addition, zPicture is linked to the UCSC Genome Browser, allowing users to automatically extract sequences and gene annotations for any recorded locus. Finally, we describe howthis tool can be efficiently applied to the analysis of nonvertebrate genomes, including those of microbial organisms.
The availability of DNA sequence information from several complete genomes has created new opportunities for formulating and testing hypotheses on the basis of phylogeny. Systematic comparisons of related genomes now permit the deduction of clear evolutionary histories and the characterization of sequence conservation profiles. Several studies have shown that sequence elements with critical biological roles are resistant to accumulating mutations and can be distinguished from the neutrally evolving background in genomic alignments (Elnitski et al. 2003). PipMaker (Schwartz et al. 2000; http://bio.cse.psu.edu/pipmaker/) and VISTA (Mayor et al. 2000; http://www-gsd.lbl.gov/VISTA/VistaInput.html) are two alignment and visualization tools used extensively to perform comparative genomic analysis to identify sequences with key biological roles. These tools have been instrumental for the sequence-based discovery of novel genes (Pennacchio et al. 2001) and gene regulatory elements (Oeltjen et al. 1997; Loots et al. 2000).
As the genome community moves toward sampling DNA from organisms on far-reaching branches of the evolutionary tree, and as a large number of whole-genome shotgun sequencing projects reach completion, comparative sequence analysis will be vital for identifying functional sequences. In particular, the vast diversity of genomes being sequenced and the increasing sophistication of the scientific questions addressed require flexible analytical tools. We have developed novel ways to visualize genomic alignments that allow the user to actively modify conservation parameters, data retrieval, and output formats. These features have been incorporated into an automated alignment and visualization tool, zPicture (http://zpicture.dcode.org), to generate reliable, highly sensitive single or multiple pairwise sequence alignments and provide the results in a visually compact, user-friendly, and interactive manner. The tool can be applied to the analysis of large genomic regions of any length from microbes to human. Threshold levels of conservation can be adjusted dynamically to optimize the detection of conserved regions in alignments, independent of the evolutionary distances separating the underlying sequences. zPicture is also capable of analyzing alignments for the presence of conserved transcription factor-binding sites via the rVista tool portal (Loots et al. 2002; http://rvista.dcode.org). Finally, this analytical tool is cross-referenced with several genome browsers and allows for automatic downloads of genomic sequences and annotation files. In this study, we describe this new server and illustrate its versatility through several examples from mammals and microbes.
RESULTS AND METHODS
Generating and Visualizing Alignments
zPicture uses BLASTZ (Schwartz et al. 2003) to generate sequence alignments between the reference sequence and one or more ortholgous sequences. Alignments are generated in <1 min for sequences up to 2 Mb in length. The `Fast Alignment' option (“T = 2 H = 2500 Y = 3400” parameters) efficiently aligns complete microbial genomes, several megabases in length, without affecting sensitivity.
At the zPicture main page, the user can submit the sequence data by choosing from several available options as follows: (1) paste in or upload sequence files from the user's computer, (2) automatically download sequence files from the UCSC Genome Browser (Kent et al. 2002; Karolchik et al. 2003; http://genome.ucsc.edu) or the NCBI database (http://www.ncbi.nlm.nih.gov/). All input sequences must be entered in the FASTA format. To download sequence and annotation data from the UCSC Genome Browser Gateway, users need to select the `UPLOAD sequence and gene annotation' button on the zPicture server home page, which will redirect the user to a new window. To use this feature, the following information has to be specified by the user: (1) the organism the sequence is derived from, (2) the genome assembly, (3) the type of annotation tables, and (4) the genome coordinates for which the data should be retrieved. The genome coordinates can be copied directly and pasted from the UCSC browser (chromosome:from-to format; for example: chr17: 1000–2000). After the first sequence and its corresponding annotation files have been uploaded, zPicture acknowledges the successful upload of the data, and informs the user to proceed with providing information for the subsequent sequence. To download sequence files from the NCBI database, the user needs to select the `NCBI accession #' option and type in the accession number.
zPicture allows for customized real-time processing of sequence alignment data by promptly returning a set of output alignment files in the same browser window where the user submitted the input sequences. These files include (1) a dot plot, (2) a dynamically interactive visualization module, (3) modifiable annotation files, (4) a transcription factor binding site analysis interface, and (5) a set of static sequence, alignment and annotation files.
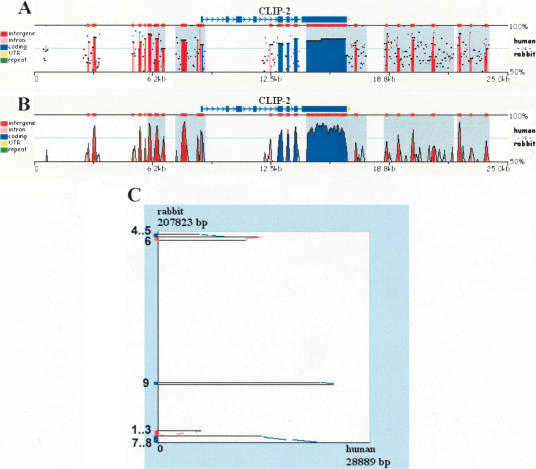
By following the visualization link, the user is directed to a conservation profile plot that can be actively modified over the Web to optimize the computational analysis. Alignments are visualized either as standard percent identity plots (PIP; Schwartz et al. 2000) or as Vista-like (Dubchak et al. 2000) smooth graphs (Fig. 1). These visualization layouts can be interchanged with a single button click. We have aligned a 260-kb human region containing the CLIP2 gene to a 208-kb unordered/unoriented rabbit draft BAC sequence (NCBI Acc. No. AC145542.1). The human genomic sequence was supplied by using the “UPLOAD” feature and providing the UCSC Genome Browser coordinates (chr19:19380000–19640000; July 2003 assembly), and the genes were mapped using RefSeq annotation files. The alignment was visualized either as a PIP- (Fig. 1A) or a smooth- (Fig. 1B) conservation plot. BLASTZ identifies matches independent of their linear organization in the input sequences; therefore, zPicture comparative analysis can be performed efficiently on draft sequences. This tool can be applied to order and orient contigs on the basis of homology to the available assembled human and mouse genomes. Contigs can be ordered and oriented using the dot-plot display, where alignments on the forward strand are depicted in red and on the reverse strand in blue (Fig. 1C). Orientation can also be determined visually from the conservation plot itself, as inverted regions have a gray-shaded background (Fig. 1A,B).
Figure 1.
Pip (A), smooth-graph (B), and Dot-plots (C) constructed by zPicture for a 29-kb region from human chromosome 19 (chr19:19501332–19530220) and an orthologous rabbit BAC (NCBI Acc# AC145542.1). The automatic sequence upload feature extracted the human sequences and the RefSeq annotation files from the UCSC database. Default parameters (>100 bp/>70% ID) were used to highlight intronic (pink) and intragenic (red) conserved elements. Untranslated Regions (UTRs) are colored in yellow, exons are in blue and repeats are in green. Inverted regions are shaded in gray. Dot-plots (C) can be used to order and orient draft sequence contigs; positive strand alignments are in red and negative strand in blue.
Dynamic Analysis of Conservation Profiles
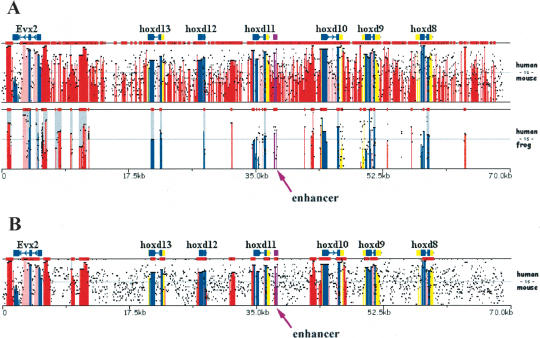
In general, there are no optimal fixed parameters for identifying functional conserved elements in any pair of sequences, as the degree of conservation varies, not only due to the evolutionary distance between species, but also due to highly variable regional mutation rates within a genome (Hardison et al. 2003). Because of these regional variations, the conservation criteria have to be adjusted in order to reflect the local interplay between purifying pressure and background noise. For several reasons, the ECR criteria of ≥100 bp and ≥70 percent identity (%ID) have been suggested as reasonable parameters for identifying functional human/mouse noncoding elements (Loots et al. 2000). However, these parameters are not always adequate for identifying the most informative conservation patterns. For example, in an alignment of 140 kb mouse and human sequences from the HOXD cluster (Fig. 2A), these standard parameters fail to detect a reasonable number of conserved elements and highlight potential regulatory elements (Gerard et al. 1997), as a result of the high degree of local similarity between humans and rodents in this particular genomic interval. Because mouse/frog alignments of the same region yield more informative alignments (Fig. 2B), we can dynamically adjust the ECR parameters accordingly (y-axis: 65%–100%; ECR: ≥500 bp/≥85%ID) and amplify the signal to noise ratio in the mouse/human alignment to yield a manageable number of conserved noncoding elements with good correspondence to conserved noncoding elements that have been identified as regulatory elements (Fig. 2B; Gerard et al. 1997). The position of this known regulatory element can be documented in the annotation file as a region of `Other' properties (OTH), and is highlighted in purple in the visual display.
Figure 2.
Dynamically modifying ECR parameters to optimize regulatory element detection. (A) Overlaid human/mouse and human/frog alignments for the HoxD cluster analyzed using default parameters (≥100 bp, ≥70%). (B) Human/mouse alignments analyzed using stringent conservation criteria (≥500 bp, ≥85%). A known enhancer was annotated as `other' feature (purple) and is pointed to by an arrow. x-axis size in kilobases; y-axis percent identity 50%–100% (A); 65%–100% (B). Frog sequence NCBI accession number AC145806.1; human and mouse sequences were downloaded from UCSC database, coordinates: human-chr2:177140000–177210000 and mouse-chr2:75814790–75951489.
This example illustrates the most critical feature of the zPicture program, the ability to actively modify the evolutionary criteria to reflect the appropriate phylogenetic relationship for the analyzed sequences. The dynamic visualization module allows for selection of (1) the minimum length and the minimum percent identity in a sliding window as a threshold for detecting ECRs by scanning BLASTZ alignments; (2) the sequence that will be displayed at a given time as the reference sequence (using the `Base-top' switch button); (3) the bottom cut-off value for percent identity (y-axis); (4) the picture resolution to either compact or zoom-into the alignments; and (5) the length of base sequence to be displayed per alignment layer. The zPicture visualization tool is capable of dynamically replotting, rescaling, and modifying the ECR-detection criteria and base sequence instantly without resubmitting the data. In contrast, other available visualization programs only provide static displays for alignments.
Annotation
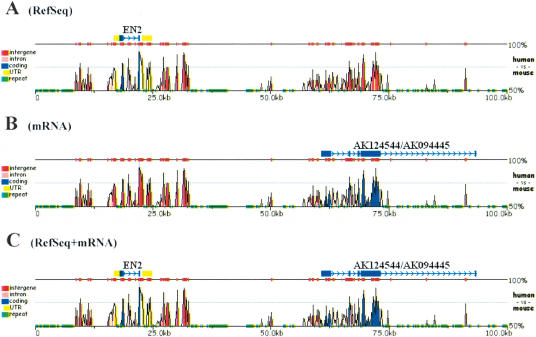
Users can optionally supply zPicture with (1) gene coordinates or annotation files and (2) repetitive DNA information. One of the most challenging tasks in genome biology is the ability to identify all of the protein-coding genes in a process called gene annotation. The ability to predict genes in otherwise anonymous sequences has been improving steadily, with the development of several ab initio gene-prediction tools coupled with experimental evidence stemming from ESTs and mRNA sequencing efforts. Several different databases store gene information and genome annotations that have been biologically validated. However, as the process of gene discovery is dynamic, no single database hosts the most comprehensive gene collection. To allow biologists to build on and improve the available annotations provided by genome projects, we have incorporated two key features into the zPicture tool. First, the user has the option to automatically download sequences and annotation data directly from the UCSC Genome Browser by indicating the organism, the assembly, the type of annotation files, and the genome coordinates to be used for extracting the desired data. Second, the user can actively improve the available annotations by interactively editing the annotation files (Fig. 3).
Figure 3.
Expanding annotation in alignments. Human (chr7:154600000–155050000) and mouse (chr5:25950131–27951930) 420-kb alignment for the Sonic Hedgehog/Engrailed region. Genomic sequences and RefSeq (A) or mRNA (B) annotation files were uploaded from the UCSC genome database. Using mRNA data, the RefSeq annotation was dynamically edited to include the most complete set of exons (C).
To edit annotation files, users have to go to the `Update annotation' section on the results page, and select the sequence for which amendments are being made. To annotate a contiguous region, the starting position, the ending position, and the type of sequence feature have to be indicated [coding exon (blue) are indicated by `CDS', untranslated regions (yellow) by `UTR', and for all other types or elements (purple) by `OTH']. To annotate a transcript, on the first line, users must indicate the direction of the gene by < or >, followed by the start, the end position, and the desired gene name. On succeeding lines, the same format should be followed as described above for contiguous regions.
Detailed gene annotations also play an essential role in distinguishing coding from noncoding conserved elements. On the basis of the resulting conservation plot, users can interactively modify sequence annotations to reflect new discoveries based on the detected shared homology of the underlying sequence data. Using this feature, gene coordinates can be edited to include for example, alternatively spliced exons, new genes, regulatory elements, or other available experimental data. The dynamic visualization interface immediately incorporates these changes without having to resubmit sequence data or recompute the alignments.
Repeat content can be annotated either by distinguishing repeats (lowercase letters) from nonredundant sequences (uppercase letters), or by running the locally installed RepeatMasker program (http://repeatmasker.genome.washington.edu/). If sequences are provided by loading data from the UCSC browser, these sequences have been preprocessed for repeats, and the first option `repeats are identified by lowercase' should be selected. In this case, annotation files are automatically extracted and pasted into the annotation window. If sequences are supplied by other means, the user can choose to mask repetitive elements by selecting the `mask repetitive elements' option and indicating the organism of choice. In this case, annotation files are not automatically provided, and the user has the option to supply their own annotation files, either by uploading a file from the user's computer, or pasting in the gene coordinates in the suggested format.
Aligning Microbial Genomes
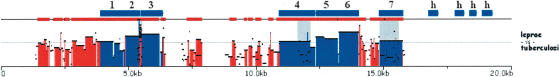
Considerable resources have been devoted to sequencing the genomes of single celled organisms, particularly of microbes that affect human health. As the available collection expands to include several closely related species, comparative genomic approaches can be applied to understand microbial pathogenesis. To illustrate how the zPicture tool can be used for the analysis of two closely related, fully sequenced microbial genomes, we aligned and analyzed the genomes of Mycobacterium leprae (NC_002677) and that of Mycobacterium tuberculosis (NC_002755; 3.3 Mb and 4.4 Mb in length, respectively; Fig. 4). Initial characterization of the Mycobacterium leprae genome identified a total of 1604 transcripts, 152 of which are putative genes (hypothetical proteins), whereas the rest have some experimental evidence of coding for proteins (including similarity to other known proteins). We addressed whether some of these hypothetical genes are also present in the Mycobacterium tuberculosis genome by analyzing the zPicture conservation profile of these two genomes. The majority of nonhypothetical genes (∼97%; 1402/1452; ≥100 bp and ≥70%ID) are highly conserved (Fig. 4), whereas only ∼20% of the hypothetical genes are conserved (30/152) in the second species. This example illustrates how the zPicture tool can easily be applied to the analysis of microbial genomes, particularly to increase the confidence of newly predicted putative transcripts through sequence conservation. Such analysis will also be very useful for obtaining insights into the molecular characteristics of individual microbial genomes to identify commonly shared traits, as well as unique sequence signatures.
Figure 4.
Comparing microbial genomes – Mycobacterium leprae vs. Mycobacterium tuberculosis. An alignment of 20 kb of the Mycobacterium leprae containing 11 genes is displayed. Genes are identified in blue and marked as (1) possible phosphatidate cytidylyltransferase; (2) ribosome recycling factor; (3) possible uridylate kinase; (4) possible amidase; (5) elongation factor EF-Ts; (6) 30S ribosomal protein S2; (7) integrase/recombinase; (h) hypothetical protein.
Transcription Factor Binding-Site Analysis
Modulation of gene expression is achieved through the complex interaction of transcription factors (TF) and DNA-binding motifs. Characterizing patterns of TF binding is a critical step for sequences-based discovery of noncoding regulatory elements. zPicture allows regulatory element analysis and transcription factor-binding sites (TFBS) visualization though the rVista tool portal (Loots et al. 2002) available at the results page. The rVista tool combines TFBS motif recognition, orthologous sequence alignments, and TFBS cluster analysis to overcome some of the limitations associated with TFBS predictions of sequences derived from a single organism. The analysis proceeds in four steps as follows: (1) identification of TFBS matches in the individual sequences, (2) identification of locally aligned noncoding TFBSs, (3) calculation of local conservation extending upstream and downstream from each orthologous TFBS, and (4) visualization of individual or clustered noncoding TFBSs. Precomputed matrices imported from the TRANSFAC database (Wingender et al. 2001; http://www.biobase.de) or user-defined consensus sequences can be used to search for TFBS motifs in the zPicture sequence alignment, and the annotation files are used to identify TFBSs present in noncoding DNA and to calculate the degree of DNA conservation encompassing each TFBS. rVista analysis provides an additional criteria for computationally defining the character of conserved noncoding sequences to possibly enrich for elements with regulatory potential.
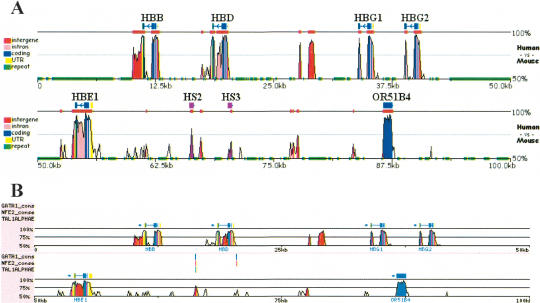
As an example of this application, the aligned human and mouse sequences containing the HBB gene complex were searched for conserved matches to binding sites for GATA-1, NF-E2, and TAL1/E47 transcription factors (Fig. 5). Only one short region (∼100 bp) within the 100 kb of aligned sequences had conserved hits to all three binding sites. This short region is also known as hypersensitive site (HS) 2 of the β-globin locus control region, a well-known erythroid enhancer (Talbot and Grosveld 1991). rVista also identifies a second conserved element in this region containing two GATA-1 and one NF-E2 binding site. This element corresponds to the previously characterized HS3 enhancer. Thus, the application of zPicture and rVista uniquely identified known transcriptional enhancer elements.
Figure 5.
zPicture and rVista analysis for the HBB gene complex. Human (chr11:5199997–5300000) and mouse (chr7:92235754–92356764) genomic sequences for the HBB locus were downloaded from UCSC browser, aligned, and the conservation was analyzed (>100 bp/>70% ID) using zPicture (A). Refseq annotation was edited by providing coordinates for the functionally characterized regulatory elements, hypersensitive site 2 and 3 (purple; HS2 and HS3). Alignments were analyzed for the presence of GATA1, NFE2, and Tal1 putative transcription factor-binding sites (TFBS). Clusters of conserved TFBS (three sites/100 bp) were identified by rVista, and correspond to known regulatory elements HS2 and HS3.
DISCUSSION
Our main objective in developing zPicture has been to create an alignment analysis tool that is dynamically Web interactive, fast, easy to use, and capable of generating multiple pairwise alignments that can be concurrently manipulated. Upon submitting the alignment request, the data is returned rapidly on the same Web page. Similar to PipMaker, zPicture can handle sequences of any length; alignments of sequences ≤1 Mb will be generated in less than 1 min; 2–3 Mb requests will be processed under 5 min, and jobs ≥5 Mb will require ≥30 min. We do not limit the size of input sequences; therefore, this tool can be used for comparing large genomic intervals or even complete bacterial genomes. If sequences are acquired from the UCSC database, zPicture analysis eliminates the need to mask repetitive elements prior to generating alignments, a step that accelerates the alignment process >100-fold, generating 1 Mb alignments in ∼30 sec. Also, the ability to extract sequence and annotation data from the UCSC Genome Browser is a unique feature that eliminates the need to manually create annotation files and expedites the process of comparative analysis. Conserved sequences can be retrieved interactively by clicking on the zPicture conservation profiles, an option unavailable for other comparative sequence analysis tools. Both zPicture and PipMaker provide users with dot-plots that present an overview of the evolutionary rearrangements in the sequences being compared. Conserved features within the dot plots can also be accessed and viewed as sequence alignments with a single mouse-click on the image. Because BLASTZ is a local aligner, zPicture identifies homologous regions independent of their location and orientation in the second sequence; therefore, this tool can efficiently be applied to the analysis of unfinished draft sequences to find overlaps between contigs and assist during assembly. The local alignment algorithm also provides the maximum efficiency in aligning distantly related genomes, such as those of mammals and fishes, in which gene order and orientation have not been faithfully preserved. Most importantly, zPicture has been designed to allow for interactive fine tuning of the conservation data to optimize the evolutionary thresholds required to extract the most significant biological data.
Comparative genomic tools have been implemented successfully for prioritizing candidate regions to be tested in functional assays, and as these tools evolve, they have the potential to be applied for the de novo identification of functional coding and noncoding sequences. The zPicture tools can be used as a reverse-engineering approach for understanding the modular structure of DNA through cross-species comparisons, and provides a theoretical solution for decrypting the sequence of genomes.
Acknowledgments
The work was performed under the auspices of the U.S. Department of Energy by the University of California, Lawrence Livermore National Laboratory Contract No. W-7405-Eng-48. Additional support was from NHGRI grant HG02238 (W.M. and R.H.)
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2129504.
References
- Dubchak, I., Brudno, M., Loots, G.G., Pachter, L., Mayor, C., Rubin, E.M., and Frazer, K.A. 2000. Active conservation of noncoding sequences revealed by three-way species comparisons. Genome Res. 10: 1304–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elnitski, L., Hardison, R.C., Li, J., Yang, S., Kolbe, D., Eswara, P., O'Connor, M.J., Schwartz, S., Miller, W., and Chiaromonte, F. 2003. Distinguishing regulatory DNA from neutral sites. Genome Res. 13: 64–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerard, M., Zakany, J., and Duboule, D. 1997. Interspecies exchange of a Hoxd enhancer in vivo induces premature transcription and anterior shift of the sacrum. Dev. Biol. 190: 32–40. [DOI] [PubMed] [Google Scholar]
- Hardison, R.C., Roskin, K.M., Yang, S., Diekhans, M., Kent, W.J., Weber, R., Elnitski, L., Li, J., O'Connor, M., Kolbe, D., et al. 2003. Covariation in frequencies of substitution, deletion, transposition, and recombination during eutherian evolution. Genome Res. 13: 13–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik, D., Baertsch, R., Diekhans, M., Furey, T.S., Hinrichs, A., Lu, Y.T., Roskin, K.M., Schwartz, M., Sugnet, C.W., Thomas, D.J., et al. 2003. The UCSC Genome Browser Database. Nucleic Acids Res. 31: 51–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent, W.J., Sugnet, C.W., Furey, T.S., Roskin, K.M., Pringle, T.H., Zahler, A.M., and Haussler, D. 2002. The human genome browser at UCSC. Genome Res. 12: 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loots, G.G., Locksley, R.M., Blankespoor, C.M., Wang, Z.E., Miller, W., Rubin, E.M., and Frazer, K.A. 2000. Identification of a coordinate regulator of interleukins 4, 13, and 5 by cross-species sequence comparisons. Science 288: 136–140. [DOI] [PubMed] [Google Scholar]
- Loots, G.G., Ovcharenko, I., Pachter, L., Dubchak, I., and Rubin, E.M. 2002. rVista for comparative sequence-based discovery of functional transcription factor binding sites. Genome Res. 12: 832–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor, C., Brudno, M., Schwartz, J.R., Poliakov, A., Rubin, E.M., Frazer, K.A., Pachter, L.S., and Dubchak, I. 2000. VISTA: Visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 16: 1046–1047. [DOI] [PubMed] [Google Scholar]
- Oeltjen, J.C., Malley, T.M., Muzny, D.M., Miller, W., Gibbs, R.A., and Belmont, J.W. 1997. Large-scale comparative sequence analysis of the human and murine Bruton's tyrosine kinase loci reveals conserved regulatory domains. Genome Res. 7: 315–329. [DOI] [PubMed] [Google Scholar]
- Pennacchio, L.A., Olivier, M., Hubacek, J.A., Cohen, J.C., Cox, D.R., Fruchart, J.C., Krauss, R.M., and Rubin, E.M. 2001. An apolipoprotein influencing triglycerides in humans and mice revealed by comparative sequencing. Science 294: 169–173. [DOI] [PubMed] [Google Scholar]
- Schwartz, S., Zhang, Z., Frazer, K.A., Smit, A., Riemer, C., Bouck, J., Gibbs, R., Hardison, R., and Miller, W. 2000. PipMaker—A Web server for aligning two genomic DNA sequences. Genome Res. 10: 577–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, S., Kent, W.J., Smit, A., Zhang, Z., Baertsch, R., Hardison, R.C., Haussler, D., and Miller, W. 2003. Human–mouse alignments with BLASTZ. Genome Res. 13: 103–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot, D. and Grosveld, F. 1991. The 5′HS2 of the globin locus control region enhances transcription through the interaction of a multimeric complex binding at two functionally distinct NF-E2 binding sites. EMBO J. 10: 1391–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingender, E., Chen, X., Fricke, E., Geffers, R., Hehl, R., Liebich, I., Krull, M., Matys, V., Michael, H., Ohnhauser, R., et al. 2001. The TRANSFAC system on gene expression regulation. Nucleic Acids Res. 29: 281–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://genome.ucsc.edu/; Human Genome Browser at UCSC.
- http://www.ncbi.nlm.nih.gov/; NCBI Database.
- http://bio.cse.psu.edu/pipmaker/; PipMaker.
- http://rvista.dcode.org/; rVista.
- http://www.biobase.de/; Transfac Database.
- http://www-gsd.lbl.gov/VISTA/VistaInput.html; Vista.
- http://repeatmasker.genome.washington.edu/; RepeatMasker.
- http://zpicture.dcode.org/; zPicture.