Abstract
Dysfunctional mitochondria are central in the pathogenesis of diabetic cardiomyopathy. Mitochondrial proteomic alterations resulting from diabetes mellitus have been reported although the mechanisms driving changes in proteomic signatures are unknown. microRNAs (miRNAs) have been considered as potential regulators of proteins. The goal of this study was to determine whether miRNAs play a role in diabetes-induced mitochondrial proteomic alterations. Quanitative RT-PCR miRNA screening in diabetic mice, 5 wk following multiple low-dose streptozotocin treatment was associated with alteration in the expression of 29 miRNAs in the diabetic heart compared with control. Among those miRNAs upregulated in the diabetic heart was miR-141 (P < 0.002). miRNA target prediction analyses identified miR-141 as a potential regulator of the inner mitochondrial membrane phosphate transporter, solute carrier family 25 member 3 (Slc25a3), which provides inorganic phosphate to the mitochondrial matrix and is essential for ATP production. With the use of a luciferase reporter construct with a Slc25a3 3′-untranslated region (UTR) target sequence, overexpression of miR-141 downregulated luciferase activity levels confirming miR-141/Slc25a3 3′-UTR binding. miR-141 overexpression in HL-1 cells elicited a decrease in Slc25a3 protein content, ATP production and a decrease in ATP synthase activity, similar to the diabetic phenotype (P < 0.05, for both). Diabetic interfibrillar mitochondria (IFM) displayed decreased Slc25a3 protein content, which was inversely correlated with increased miR-141 expression. Further, diabetic IFM ATP synthase activity was also decreased (P < 0.05). Together these results indicate that miR-141 can regulate Slc25a3 protein expression in the diabetic heart. Further, diabetes-induced miRNA changes may influence mitochondrial proteomes and functional processes such as mitochondrial ATP production.
Keywords: mitochondria, diabetes mellitus, miRNA
cardiac complications, including diabetic cardiomyopathy, are the leading cause of morbidity and mortality among type 1 diabetic patients. Mitochondrial dysfunction plays a key role in cardiac abnormalities associated with the type 1 diabetic heart (11, 15, 35, 39). Cardiac mitochondria are characterized as spatially distinct subpopulations, including those located beneath the sarcolemma, termed subsarcolemmal mitochondria (SSM), and those located between the myofibrils, termed interfibrillar mitochondria (IFM). Mounting evidence suggests that mitochondrial subpopulations are differentially affected in pathological settings (4, 11, 12, 22, 23, 32, 37, 44). We previously reported that IFM are dysfunctional in type 1 diabetic cardiac tissue through mechanisms that include enhanced oxidative stress, decreased oxidative phosphorylation, decreased cardiolipin content, and decreased nuclear-encoded mitochondrial protein import (4, 11). Further, we have observed alterations in type 1 diabetic mitochondrial proteomes, although the mechanisms accounting for the changes are not entirely clear (7, 17, 39, 40). A large proportion of dysregulated proteins reside within the inner mitochondrial membrane (IMM) and may be associated with disruption of essential mitochondrial functions such as oxidative phosphorylation and protein import.
One particular protein of interest is the IMM phosphate transporter solute carrier family 25 member 3 protein (Slc25a3), which has been shown to be significantly decreased in abundance during a type 1 diabetic insult (4). This transmembrane protein is essential for ATP production as it acts as a conduit for inorganic phosphate in its movement from the cytoplasm into the matrix, providing a substrate to fuel the F0F1-ATPase (20). Decrements to Slc25a3 have been shown to exert deleterious effects on ATP production and cellular viability (2, 30, 34). Clinically, patients with a homozygous mutation in the Slc25a3 gene display exercise intolerance, proximal muscle weakness, and cardiomyopathy (29). Currently, the regulatory mechanisms accounting for reduced Slc25a3 abundance during type 1 diabetes mellitus have yet to be explored.
One potential set of regulatory mechanisms that may account for alterations to protein expression are microRNAs (miRNA), which are 21–23 nucleotide noncoding RNAs capable of posttranscriptional regulation. Originally described in Caenorhabditis elegans, miRNAs have been identified in many species including all vertebrates (43). With the help of the RNA-induced silencing complex, miRNAs influence protein abundance by degrading or inhibiting the translation of the target mRNA (24). The mode of action is thought to depend largely on the binding efficiency of the miRNA to the target mRNA. miRNAs exist in all parts of the body and have been implicated in many cellular processes (38). miRNAs have been extensively studied in the context of cancer, heart failure, and cellular homeostasis (8, 41). As an example, the miR-200 family (miR-200a, 200b, 200c, 141, and 429) has been shown to modulate E-cadherin transcriptional repressors ZEB1 and SIP1, critical factors in epithelial to mesenchymal transition and tumor metastasis (16). Interestingly, miR-141 and miR-200c were shown to be upregulated in an acute hindlimb ischemic model of hypoxia, presumably through a mechanism of enhanced reactive oxygen species production (26).
In this study, we evaluated the impact of type 1 diabetes mellitus on miRNA regulation in the heart. We performed broad scale analyses on 376 of the most abundant mouse miRNAs and found that miR-141 was the most significantly increased miRNA within the diabetic heart. Through miRNA target prediction analyses, we identified Slc25a3 as a potential target of miR-141. We then hypothesized that enhanced expression of miR-141 could negatively regulate Slc25a3 within cardiac mitochondria during diabetes mellitus and that a decrease in Slc25a3 would lead to diminished mitochondrial ATP synthesis rates.
MATERIALS AND METHODS
Experimental animals and diabetes induction.
The animal experiments in this study conformed to National Institutes of Health Guidelines for the Care and Use of Laboratory Animals and were approved by the West Virginia University Animal Care and Use Committee. Male FVB mice (Jackson Laboratory, Bar Harbor, ME) were housed in the West Virginia University Health Sciences Center animal facility. Mice were given unlimited access to a rodent diet and water. Type 1 diabetes mellitus was induced in 5-wk-old mice following the protocol of the Animal Models of Diabetic Complications Consortium using multiple low-dose streptozotocin (STZ; Sigma, St. Louis, MO) injections as previously described (4, 11). Briefly, injections of 50 mg/kg body wt STZ dissolved in sodium citrate buffer (pH 4.5) were performed daily for 5 consecutive days after 6 h of fasting. Mice that served as vehicle controls were given the same volume per body weight of sodium citrate buffer. One week postinjections, hyperglycemia was confirmed by measuring fasting blood glucose levels using a commercially available kit (Bayer, Mishiwaka, IN). Blood glucose levels >250 mg/dl were considered diabetic. Five weeks posthyperglycemia onset, animals were killed for further experimentation.
Preparation of individual mitochondrial subpopulations.
At 5 wk posthyperglycemia onset, FVB mice were killed and their hearts were excised. Hearts were rinsed in PBS (pH 7.4) and then blotted dry. SSM and IFM were isolated as previously described following the methods of Palmer et al. (31) with minor modifications (4, 10–12, 44). Mitochondrial subpopulation pellets were resuspended in sucrose-based SEM buffer (250 mM sucrose, 1 mM EDTA, and 10 mM MOPS, pH 7.2) or mitochondrial extraction buffer (Biovision, Mountain View, CA) depending on the assay to be performed. Mitochondrial protein concentrations were determined using the Bradford method and BSA as a standard (5).
Western blot analyses.
SDS-PAGE was run on 4–12% gradient gels as described previously (21), with equal amounts of protein loaded. Relative amounts of Slc25a3 were quantified in isolated mitochondrial subpopulations and HL-1 cells (a kind gift from Dr. William Claycomb) using an anti-Slc25a3 rabbit antibody (product no. 15855-AP; Proteintech, Chicago, IL) and anti-Slc25a3 goat antibody (product no. SC161226; Santa Cruz, Santa Cruz, CA), respectively. The secondary antibodies used in the analyses were a goat anti-rabbit IgG horseradish peroxidase conjugate (product no. 31463; Pierce Biotechnology, Rockford, IL) and a donkey anti-goat IgG horseradish peroxidase conjugate (product no. sc2020; Santa Cruz). Detection and quantitation of chemiluminescent signals were performed using a G:BOX (Syngene, Frederick, MD), and the data were expressed as arbitrary optical density units. Control for protein loading in isolated mitochondria was confirmed using a cyclooxygenase IV rabbit antibody (product no. ab16056; Abcam, Cambridge, MA), while control for protein loading in HL-1 cells was confirmed using an anti-GAPDH mouse antibody (product no. ab8245; Abcam) in conjunction with the secondary mouse antibody described above.
RNA isolation.
Hearts were rinsed in PBS (pH 7.4) and then snap frozen in liquid nitrogen. Total RNA (small and large) was isolated from heart (25 mg) tissue using a miRNeasy mini kit per manufacturer's protocol (Qiagen, Valencia, CA). Total RNA quantification and purity were assessed using a Nanodrop ND-1000 (Thermo Fisher Scientific, Wilmington, DE). Samples were then stored at −80°C until further evaluation.
miRNA quantitative RT-PCR array.
For miRNA quantitative RT-PCR (RTQ-PCR) analyses, enriched miRNA was converted to cDNA using a RT2 miRNA First Strand kit (SABiosciences, Frederick, MD). cDNA samples were then used for the RT2 miRNA PCR Whole Genome Array system (SABiosciences, Frederick, MD), allowing for simultaneous detection of 376 mouse miRNAs, which represented functional miRNAs, housekeeping genes (Rnu6, snoRNA, and PPC), and RNA quality controls (SABiosciences, Frederick, MD). The analyses were performed according to the manufacturer's protocol using a Taqman 7900HT system (Applied Biosystems, Foster City, CA) utilizing sequence detection systems software version 2.3 (SDS 2.3). Threshold cycles (Ct) were used to determine miRNA copy number and levels of respective miRNAs. Relative quantitative (q)PCR expression was normalized to the expression of housekeeping genes using ΔΔCt methodology (25).
Cell culture and plasmid transfection.
Cells were grown at 37°C in a humidified atmosphere of 5% CO2-95% air and maintained in DMEM (Cellgro, Manassas, VA; HEK293) or Claycomb media (Sigma Aldrich, St. Louis, MO; HL-1) with 10% FBS (Sigma). miRNA expression plasmids (pCMV-MIR) housing the mouse precursor sequence of miR-141 (5′-GGGUCCAUCUUCCAGUGCAGUGUUGGAUGGUUGAAGUAUGAAGCUCCUAACACUGUCUGGUAAAGAUGGCCC-3′, mature sequence in bold and underlined) and human miR-200c precursor sequence (5′- CCCUCGUCUUACCCAGCAGUGUUUGGGUGCGGUUGGGAGUCUCUAAUACUGCCGGGUAAUGAUGGAGG-3′, mature sequence in bold and underlined) were utilized for transfection studies (Origene, Rockville, MD). Two micrograms of plasmid DNA were transfected using a FuGENE 6 transfection kit per the manufacturer's protocol (Promega, Madison, WI) in six-well culture plates. An Empty pCMV-MIR plasmid was used as a negative control (Origene, Rockville, MD). Seventy-two hours posttransfection, cells were washed with PBS and harvested in either 1× RIPA buffer (Sigma) for protein-based experimentation or RNA lysis buffer (Origene) for RNA isolation.
miRNA targeting luciferase assay.
The mouse Slc25a3 3′-untranslated region (UTR) 5′-TTTAAACTAGCGG CCGCTAGTCTTTATCTGCTTGTTGATCAGTGTTGTATATATTCTAGA-3′ (target sequence in bold and underlined) was duplexed (Integrated DNA Technologies, Coralville, IA), cut by Pme1/Xba1, and ligated into the PmirGLO Dual-Luciferase miRNA Target Expression Vector for miRNA binding analyses. Then, 500 ng of Slc25a3 PmirGLO vector were cotransfected as described above with 500 ng of mouse miR-141, human miR-200c, or empty pCMV-MIR in triplicate in 12-well culture plates. Twenty-four hours posttransfection, luciferase activities were assayed with the Dual-Luciferase Reporter Assay System (Promega, Madison, WI) per manufacturer's protocol using a Flexstation 3 Luminometer (Molecular Devices, Sunnyvale, CA). Luciferase activity was quantified and reported as the ratio of firefly luciferase to Renilla luciferase.
RTQ-PCR analyses.
Isolated total RNA from HL-1 cells was converted to cDNA using the miRNA First Strand cDNA synthesis kit (Origene) per the manufacturer's protocol. Briefly, 15 ng of cDNA sample, 0.5 μM of human miR-141 primer pairs (Origene), and 2× SYBR Green I qPCR Master Mix (Origene) were used to perform RTQ-PCR analysis using a Taqman 7900HT system (Applied Biosystems). RTQ-PCR was performed as follows: activation, 50°C for 2 min; presoak, 95°C for 10 min, followed by 42 cycles: denaturation, 95°C for 15 s; annealing 55°C for 10 s; and extension 72°C for 30 s. Threshold cycles (Ct) were used to determine miRNA copy number and levels of miR-141. Relative qPCR expression of miR-141 and Slc25a3 was normalized to the expression of GAPDH using the ΔΔCt method as above (25).
Mitochondrial ATP synthase activity.
ATP synthase activity was measured in mitochondrial subpopulations and cellular lysate as oligomycin-sensitive ATPase activity using an assay coupled with pyruvate kinase, which converts the ADP to ATP and produces pyruvate from phosphoenolpyruvate as previously described (10, 14, 33, 36). Protein content was assessed as described above (5) with final values expressed as nanomoles consumed per minute per milligram of protein, which was equal to the nanomoles of NADH oxidized per minute per milligram of protein.
ATP content.
Bioluminescent determination of ATP was performed as per the manufacturer's instructions, using a kit (Invitrogen, Carlsbad, CA). Briefly, cellular ATP was determined on a Molecular Devices luminescence plate reader and normalized to protein content.
Statistics.
The means ± SE were calculated for all data sets. Data were analyzed with a one-way ANOVA method to evaluate the main treatment effect, diabetes induction (GraphPad Software, La Jolla, CA). Fisher's least significant difference post hoc tests were performed to determine the significant differences among means. When appropriate, a Student's t-test was employed. P < 0.05 was considered significant.
RESULTS
miRNA alterations in the diabetic heart.
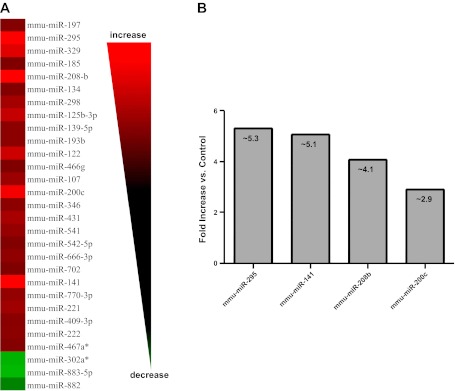
To gain insight into how type 1 diabetes mellitus influences miRNA contents within the heart, we performed a broad scale RTQ-PCR analysis of 376 mouse miRNAs. Diabetic hearts displayed significant increases in 26 miRNAs and significant decreases in 3 miRNAs compared with control (Fig. 1A). Among the most highly upregulated miRNAs within the diabetic hearts were miR-295 (∼5.3-fold, P < 0.03), miR-141 (∼5.1-fold, P < 0.002), miR-208b (∼4.1-fold, P < 0.0004), and miR-200c (∼2.9-fold, P < 0.05; Fig. 1B).
Fig. 1.
MicroRNA (miRNA) alterations in type 1 diabetic heart. A: heat map of quantitative RT-PCR analyses identifying miRNAs that are significantly altered in type 1 diabetic mouse hearts compared with controls. Alterations are expressed using a log2 scale (−ΔΔCt). Changes in green indicate downregulation while changes in red indicate upregulation; n = 4 per group. B: graphical representation of the four miRNAs displaying the greatest fold increases in the diabetic heart compared with control. miR-295, P < 0.02; miR-141, P < 0.002; miR-208b, P < 0.0004; and miR-200c, P < 0.05.
Slc25a3 is a target of mir-141.
Because miR-141 elicited the greatest combination of fold change and statistical significance in the diabetic heart compared with control, we focused upon genes that were potentially regulated by this specific miRNA. Using miRNA target predictor programs (MirBase, Target Scan, Microcosm), we were able to identify the mitochondrial phosphate carrier, Slc25a3, as a potential target of miR-141. Figure 2A highlights the putative recognition site of miR-141 and the Slc25a3 3′-UTR. Further, the miR-141 seeding sequence to the Slc25a3 3′-UTR was highly conserved across multiple vertebrate species including mouse, rat, human, and guinea pig (Fig. 2B). High sequence homology and conserved vertebrate seeding regions are both positive indicators of potential miR-141 targeting and Slc25a3 regulation.
Fig. 2.
miR-141 binding to solute carrier family 25 member 3 (Slc25a3). A: sequence homology between Slc25a3 3′-UTR (top) and miR-141 (bottom). Black lines represent perfect match, and grey lines represent G-U wobbles. B: miR-141 recognition sequence (seeding region) of Slc25a3 gene in mouse (Mus musculus), rat (Rattus norvegicus), human (Homo sapiens), and guinea pig (Cavia porcellus) as indicted by Target Scan (www.TargetScan.org). Bold underlined characters represent the seed sequence for miR-141.
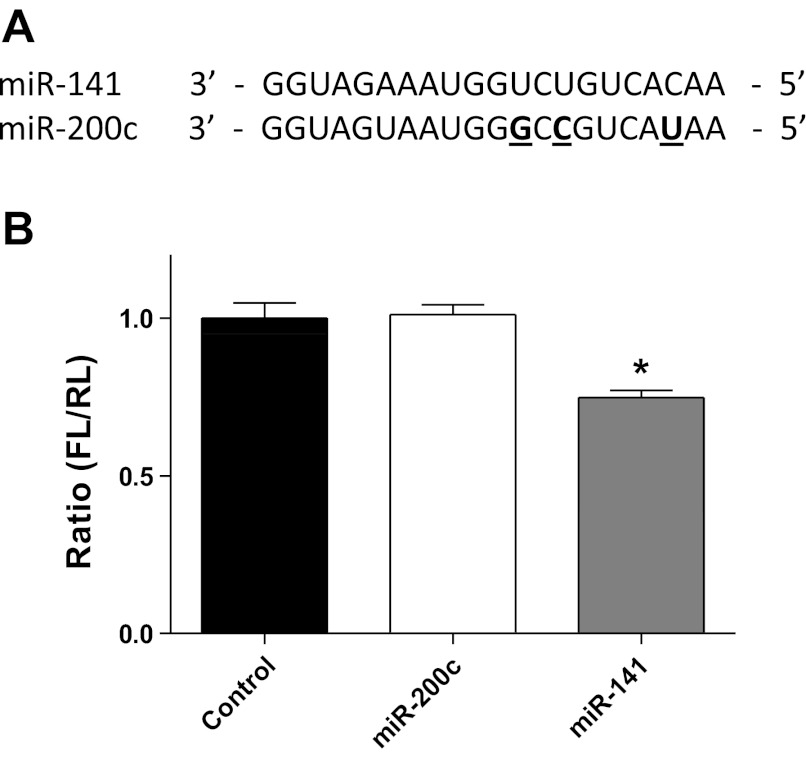
To determine whether miR-141 was able to effectively bind to the 3′-UTR of Slc25a3, we performed a cell-based luciferase reporter assay in which the Slc25a3 3′-UTR of miR-141 was cloned downstream of the firefly luciferase gene in the pMIR-GLO plasmid. To determine specificity of miR-141, the luciferase reporter plasmid was cotransfected into HEK293 cells with miR-141, miR-200c, or pCMV-MIR control. miR-200c contains a slightly different seeding region than miR-141 (Fig. 3A); therefore, it is not expected to bind with the 3′-UTR of Slc25a3. As shown in Fig. 3B, miR-141 was able to significantly reduce luciferase activity by 30%, whereas the control miRNA plasmid (pCMV-MIR) and miR-200c could not (Fig. 3B). These results indicate that Slc25a3 is a potential target of miR-141. Further, miR-141 recognizes the putative recognition site in the 3′-UTR of Slc25a3.
Fig. 3.
miR-141 binding to the Slc25a3 3′-UTR target sequence. A: sequence homology between miR-141 and family member miR-200c. Bold underlined characters indicate differences in miR-200c nucleotide sequence compared with miR-141. B: relative luciferase activities of Slc25a3 3′-UTR reporter coexpressed with miR-141, miR-200c, or plasmid (pCMV-MIR) control in HEK293 cells 24 h post transfection. Firefly luciferase (FL) activity was normalized to Renilla luciferase (RL) activity. Values are represented as means ± SE; n = 4 per group. *P < 0.05 for miR-141 vs. all other groups.
Slc25a3 is decreased in type 1 diabetic IFM.
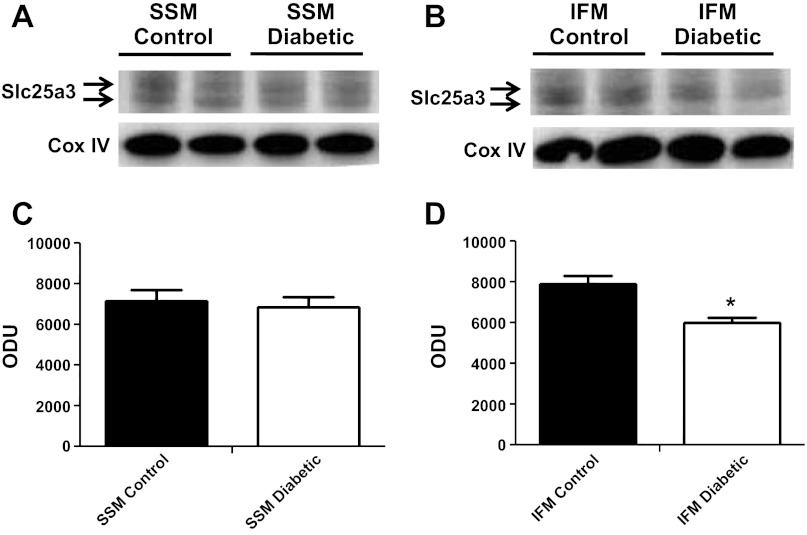
We next analyzed Slc25a3 mRNA levels and protein contents from control and diabetic hearts to determine if miR-141 upregulation correlated with decreased Slc25a3 mRNA levels and/or protein contents. Interestingly, mRNA levels of Slc25a3 were not significantly changed by diabetes mellitus compared with controls (Fig. 4). Western blots analyses of mitochondrial subpopulations revealed a significant decrease in Slc25a3 protein content within diabetic IFM compared with control IFM (Fig. 5, B and D) with no significant change in the SSM subpopulation (Fig. 5, A and C). The results suggest that if miR-141 regulates Slc25a3, it is through a mechanism of translational repression and not transcriptional degradation. Further, the effect seems to impact the IFM subpopulation specifically.
Fig. 4.
Slc25a3 mRNA expression levels. Relative mRNA levels of Slc25a3 in control and diabetic heart tissue. GAPDH was used as an internal control. Values are calculated as the ratio of Slc25a3 mRNA levels to GAPDH mRNA levels. Data are represented as means ± SE; n = 4 per group.
Fig. 5.
Slc25a3 protein contents. Western blots for Slc25a3 protein expression in subsarcolemmal mitochondria (SSM; A) and interfibrillar mitochondria (IFM; B) from control and diabetic hearts (n = 4). Densitometry analysis of Western blots for Slc25a3 protein expression in SSM (C) and IFM (D). Control for protein loading was confirmed by cyclooxygenase IV (Cox IV) Western blot. Values are calculated as arbitrary optical density units (ODU). Data are represented as means ± SE; n = 4 per group. *P < 0.05 for IFM diabetic vs. IFM control.
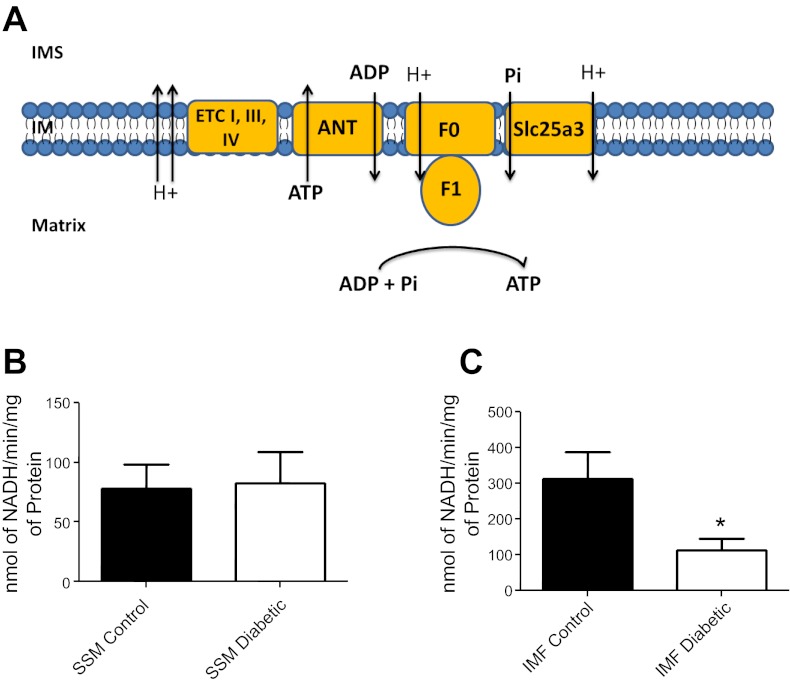
We examined whether loss of Slc25a3 would influence mitochondrial functionally through analysis of ATP synthase activity. The ATP synthase is able to create ATP by utilizing essential substrates ADP and phosphate, which are translocated into the mitochondrial matrix through adenine nucleotide transporter (ANT) and Slc25a3, respectively (Fig. 6A). ATP synthase activity within the diabetic IFM was significantly decreased compared with control, correlating with decreased Slc25a3 levels (Fig. 6C). Diabetic SSM ATP synthase activity was not significantly changed compared with control (Fig. 6B).
Fig. 6.
Mitochondrial subpopulation ATP synthase activity. A: general schematic of how ATP synthase produces ATP with the aid of Slc25a3 (phosphate transport) and adenine nucleotide transporter (ANT; ADP transport) within the mitochondrion (IM, inner membrane; IMS, inner membrane space; F0 and F1, ATPase subunits). ATP synthase activity was assessed in SSM (B) and IFM (C) from hearts of control and diabetic animals using an assay coupled with pyruvate kinase which converts ADP to ATP and produces pyruvate from phosphoenolpyruvate. Final values are expressed as nanomoles of NADH per minutes per milligrams of protein ± SE; n = 4 for each group. ETC, electron transport chain. *P < 0.05 for IFM diabetic vs. IFM control.
miR-141 directly regulates Slc25a3.
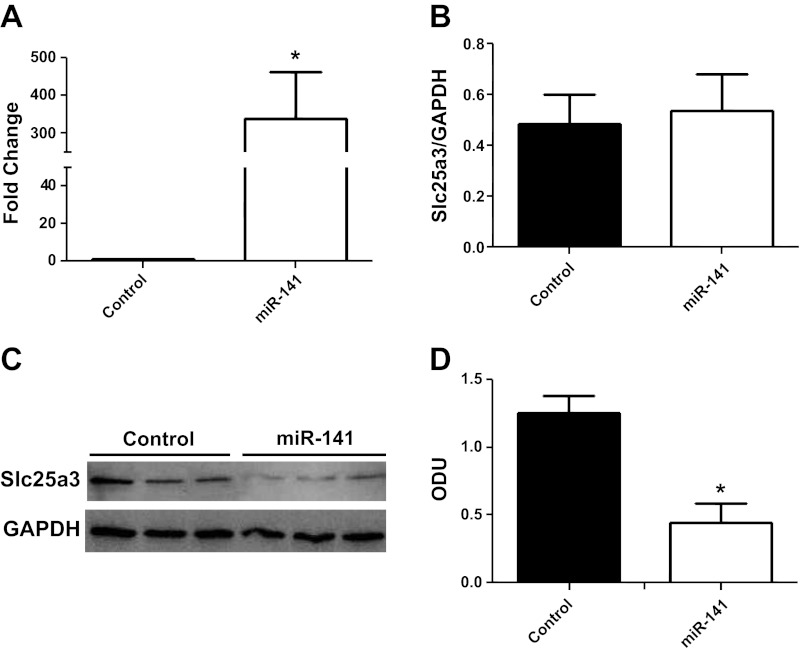
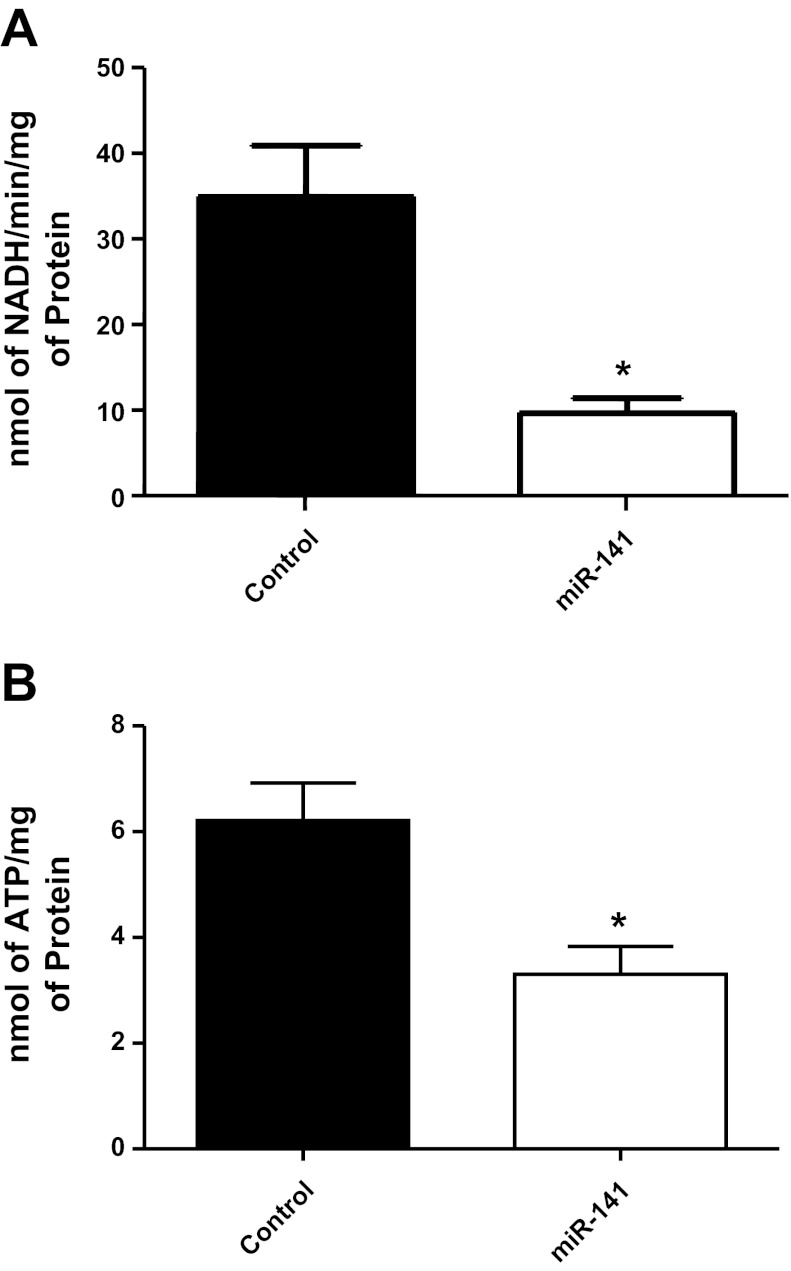
To determine whether miR-141 directly regulates Slc25a3 expression, we performed miR-141 overexpression studies using HL-1 cells. The HL-1 cell line is derived from a cardiac muscle cell line and retains a phenotype that is similar to adult cardiomyocytes (9). Transient transfections increased miR-141 mRNA expression by ∼300 fold compared with control in HL-1 cells (Fig. 7A). miR-141 overexpression did not significantly change Slc25a3 mRNA expression levels compared with control (Fig. 7B). However, protein expression of Slc25a3 was significantly decreased (70%) in miR-141 HL-1 cells 72 h posttransfection compared with control (Fig. 7, C and D). These results corroborate the findings observed in the diabetic heart and indicate that miR-141 action is through translational repression. Functionally, ATP synthase activity and ATP contents were significantly decreased within miR-141 overexpressing cells (Fig. 8, A and B) indicating that overexpression of miR-141 has functional implications for mitochondrial ATP production through a mechanism of decreased inorganic phosphate transport.
Fig. 7.
miR-141 overexpression. A: fold increase of miR-141 in HL-1 cells 72 h posttransfection with control (pCMV-MIR) or miR-141 expression plasmids. B: relative mRNA levels of Slc25a3 in HL-1 control cells and miR-141 cells. GAPDH was used as an internal control. Values are calculated as the ratio of Slc25a3 mRNA levels to GAPDH mRNA levels. Western blots (C) and densitometry analysis (D) of Slc25a3 protein expression in HL-1 control and miR-141 cells. GAPDH was used as an internal control. Values are represented as means ± SE; n = 4 per group. *P < 0.05 for miR-141 vs. control.
Fig. 8.
miRNA-141 overexpression and ATP synthase activity. A: ATP synthase activity was assessed in HL-1 control cells and miR-141 cells using an assay coupled with pyruvate kinase which converts ADP to ATP and produces pyruvate from phosphoenolpyruvate. Final values are expressed as nanomoles of NADH per minutes per milligrams of protein ± SE. B: ATP content was assessed in HL-1 control cells and miR-141 cells using a bioluminescent assay. Final values are expressed as nanomoles of ATP/ per milligrams of protein ± SE; n = 4 for each group. *P < 0.05 for miR-141 vs. control.
DISCUSSION
Mitochondrial dysfunction is known to play a central role in the pathogenesis of diabetic cardiomyopathy. Proteomic dysregulation is associated with mitochondrial and subsequently, cardiac dysfunction observed in type 1 diabetic models (11). Nevertheless, identification of the mechanisms involved remains limited. miRNAs are capable of posttranscriptional and translational repression, making them attractive for testing hypothesized mechanisms of proteomic dysregulation. Therefore, the goal of this study was to determine the impact of diabetes mellitus on miRNA contents in the heart. Further, we wanted to identify specific mitochondrial proteomic targets whose contents were influenced during diabetes mellitus and could be influenced by miRNAs. To accomplish this goal, we performed a broad scale RTQ-PCR miRNA analysis from the hearts of control and STZ-treated mice to evaluate miRNA dysregulation resulting from diabetes mellitus.
Because miRNAs have the ability to modulate physiological processes through the regulation of specific target genes, studies have begun to elucidate how miRNAs regulate essential processes within the heart during pathologies (13). RTQ-PCR analysis of 376 of the most abundant miRNAs revealed increases in 26 of the 29 significantly altered miRNAs in the diabetic heart. Four miRNAs that displayed the highest fold change increases were miR-295, miR-141, miR-208b, and miR-200c. The miR-208 family has been identified as regulators of cardiac contractility through the regulation of β-myosin heavy chain expression within the heart (42). Interestingly, cardiac myosin heavy chain switching from α to β has been shown in multiple type 1 and type 2 diabetic models implicating miR-208b as a potential key regulator of heart function during diabetic cardiomyopathy (3). Further research into the impact of miR-208 expression on cardiac myosin heavy chain switching during diabetes mellitus is warranted.
miRNA family members miR-141 and miR-200c, which are located on chromosome 6 in the mouse genome (19), were significantly increased in the diabetic heart compared with control by approximately fivefold and threefold, respectively. Because miR-141 displayed the greatest combination of fold change and significant difference in the diabetic heart compared with control, we focused on identifying nuclear-encoded mitochondrial genes that could potentially be regulated by this miRNA. Through the use of multiple miRNA targeting databases (MirBase, Target Scan, Microcosm), the IMM phosphate carrier Slc25a3 was identified as a potential target due to its perfect seeding region match and high sequence homology between vertebrate species. Luciferase inhibition after coincubation of the Slc25a3 3′-UTR with miR-141 confirmed binding between Slc25a3 mRNA and miR-141. Recently, miR-141 and miR-200c have been shown to be activated during conditions of enhanced oxidative stress (26, 27). Exposing both human umbilical vein endothelial cells and C2C12 myoblasts to hydrogen peroxide, Magenta et. al. (26) noted increases in both miR-141 and miR-200c expression (26). Additional evidence is provided by an acute hindlimb ischemia model of oxidative stress, which was also associated with elevated miR-141 and miR-200c levels. Our laboratory and others have shown that diabetic mitochondria display increased reactive oxygen species levels and oxidative damage, suggesting a potential mechanism accounting for the elevation of miR-141 during type 1 diabetic insult (11, 17, 35).
miR-141 overexpression studies were carried out to validate target gene (Slc25a3) repression. As expected, enhanced levels of miR-141 significantly decreased Slc25a3 protein content by 50% and decreased mitochondrial ATP synthesis rates. Multiple studies have linked detriments to Slc25a3 with decreased ATP synthesis in the mitochondrion, highlighting the critical role phosphate transport plays in mitochondrial energy production (2, 30, 34). Most notably, patients with mitochondrial phosphate-carrier deficiency display decreased ATP synthesis in muscle mitochondria, which correlates with hypertrophic cardiomyopathy and diminished cardiac output (28). It should be noted that changes in proteins other than Slc25a3, such as ANT, which mediates the exchange of ADP and ATP across the IMM, can influence ATP levels in the cell. Mice possess three ANT isoforms, ANT1, which is expressed primarily in heart and skeletal muscle, ANT2, which is expressed in most tissues, and ANT4, which is located primarily in the testis (6). Previous proteomic analyses from our laboratory indicate that the major cardiac ANT isoforms (ANT1 and ANT2) are not significantly impacted as a result of type 1 diabetic insult (4).
Cardiac and skeletal muscle contain two spatially distinct mitochondrial subpopulations, SSM located beneath the sarcolemma and IFM situated between the myofibrils (31). Interestingly, miR-141 overexpression within the diabetic heart correlated with decreased protein content of Slc25a3 as well as decreased ATP synthase activity only within cardiac IFM. These results were somewhat unexpected as our assumption was that Slc25a3 would be decreased within both mitochondrial subpopulations. It should be noted that mitochondrial isolation procedures, in general, can result in some selection of more/less viable mitochondria, which represent a potential limitation of these analyses. In addition, because physiological and pathological states have been associated with differential proteomic responses of mitochondrial subpopulations in cardiac and skeletal muscle tissue (1, 4, 11, 18, 23, 44), mechanisms other than miRNAs need to be considered, such as altered nuclear-encoded mitochondrial protein import or enhanced posttranslational modifications as previously reported (4). Nevertheless, future studies dedicated to elucidating the regulatory role of miRNAs in the context of individual mitochondrial subpopulations is warranted.
In conclusion, these findings indicate that pathologies, such as diabetic cardiomyopathy, have the ability to alter mitochondrial specific proteins through differential expression of miRNAs. Specifically, we report for the first time that miR-141 is significantly upregulated within the diabetic heart and is capable of regulating the mitochondrial phosphate carrier Slc25a3. Perturbation of mitochondrial energy production via miR-141 could be a potential mechanism accounting for mitochondrial dysfunction and the pathogenesis of the diabetic heart.
GRANTS
This work was supported by National Institutes of Diabetes and Digestive and Kidney Diseases Grant DP2-DK-083095. This work was also supported by a Grant-In-Aid from the American Heart Association Grant 0855484D. E. Dabkowski is a recipient of an American Heart Association Predoctoral Fellowship (0815406D). W. Baseler is a recipient of an American Heart Association Predoctoral Fellowship (10PRE3420006). W. Baseler and T. Croston are recipients of National Institutes of Health Predoctoral Fellowships (T32HL090610).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: W.A.B., E.R.D., and J.M.H. conception and design of research; W.A.B., D.T., R.J., E.R.D., and T.L.C. performed experiments; W.A.B., D.T., R.J., and J.M.H. analyzed data; W.A.B., R.J., and J.M.H. interpreted results of experiments; W.A.B., D.T., R.J., and J.M.H. prepared figures; W.A.B. and J.M.H. drafted manuscript; W.A.B., D.T., R.J., T.L.C., and J.M.H. edited and revised manuscript; W.A.B., D.T., R.J., E.R.D., T.L.C., and J.M.H. approved final version of manuscript.
ACKNOWLEDGMENTS
We would like to acknowledge Dr. William Claycomb for the HL-1 cells utilized in the cell culture experiments.
REFERENCES
- 1. Adhihetty PJ, Ljubicic V, Menzies KJ, Hood DA. Differential susceptibility of subsarcolemmal and intermyofibrillar mitochondria to apoptotic stimuli. Am J Physiol Cell Physiol 289: C994–C1001, 2005 [DOI] [PubMed] [Google Scholar]
- 2. Alcala S, Klee M, Fernandez J, Fleischer A, Pimentel-Muinos FX. A high-throughput screening for mammalian cell death effectors identifies the mitochondrial phosphate carrier as a regulator of cytochrome c release. Oncogene 27: 44–54, 2008 [DOI] [PubMed] [Google Scholar]
- 3. Aragno M, Mastrocola R, Medana C, Catalano MG, Vercellinatto I, Danni O, Boccuzzi G. Oxidative stress-dependent impairment of cardiac-specific transcription factors in experimental diabetes. Endocrinology 147: 5967–5974, 2006 [DOI] [PubMed] [Google Scholar]
- 4. Baseler WA, Dabkowski ER, Williamson CL, Croston TL, Thapa D, Powell MJ, Razunguzwa TT, Hollander JM. Proteomic alterations of distinct mitochondrial subpopulations in the type 1 diabetic heart: contribution of protein import dysfunction. Am J Physiol Regul Integr Comp Physiol 300: R186–R200, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254, 1976 [DOI] [PubMed] [Google Scholar]
- 6. Brower JV, Lim CH, Han C, Hankowski KE, Hamazaki T, Terada N. Differential CpG island methylation of murine adenine nucleotide translocase genes. Biochim Biophys Acta 1789: 198–203, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bugger H, Chen D, Riehle C, Soto J, Theobald HA, Hu XX, Ganesan B, Weimer BC, Abel ED. Tissue-specific remodeling of the mitochondrial proteome in type 1 diabetic akita mice. Diabetes 58: 1986–1997, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer 6: 857–866, 2006 [DOI] [PubMed] [Google Scholar]
- 9. Claycomb WC, Lanson NA, Jr, Stallworth BS, Egeland DB, Delcarpio JB, Bahinski A, Izzo NJ., Jr HL-1 cells: a cardiac muscle cell line that contracts and retains phenotypic characteristics of the adult cardiomyocyte. Proc Natl Acad Sci USA 95: 2979–2984, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dabkowski ER, Baseler WA, Williamson CL, Powell M, Razunguzwa TT, Frisbee JC, Hollander JM. Mitochondrial dysfunction in the type 2 diabetic heart is associated with alterations in spatially distinct mitochondrial proteomes. Am J Physiol Heart Circ Physiol 299: H529–H540, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dabkowski ER, Williamson CL, Bukowski VC, Chapman RS, Leonard SS, Peer CJ, Callery PS, Hollander JM. Diabetic cardiomyopathy-associated dysfunction in spatially distinct mitochondrial subpopulations. Am J Physiol Heart Circ Physiol 296: H359–H369, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dabkowski ER, Williamson CL, Hollander JM. Mitochondria-specific transgenic overexpression of phospholipid hydroperoxide glutathione peroxidase (GPx4) attenuates ischemia/reperfusion-associated cardiac dysfunction. Free Radic Biol Med 45: 855–865, 2008 [DOI] [PubMed] [Google Scholar]
- 13. Feng B, Chen S, George B, Feng Q, Chakrabarti S. miR133a regulates cardiomyocyte hypertrophy in diabetes. Diabetes Metab Res Rev 26: 40–49, 2010 [DOI] [PubMed] [Google Scholar]
- 14. Feniouk BA, Suzuki T, Yoshida M. Regulatory interplay between proton motive force, ADP, phosphate, and subunit epsilon in bacterial ATP synthase. J Biol Chem 282: 764–772, 2007 [DOI] [PubMed] [Google Scholar]
- 15. Flarsheim CE, Grupp IL, Matlib MA. Mitochondrial dysfunction accompanies diastolic dysfunction in diabetic rat heart. Am J Physiol Heart Circ Physiol 271: H192–H202, 1996 [DOI] [PubMed] [Google Scholar]
- 16. Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol 10: 593–601, 2008 [DOI] [PubMed] [Google Scholar]
- 17. Hamblin M, Friedman DB, Hill S, Caprioli RM, Smith HM, Hill MF. Alterations in the diabetic myocardial proteome coupled with increased myocardial oxidative stress underlies diabetic cardiomyopathy. J Mol Cell Cardiol 42: 884–895, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kelley DE, He J, Menshikova EV, Ritov VB. Dysfunction of mitochondria in human skeletal muscle in type 2 diabetes. Diabetes 51: 2944–2950, 2002 [DOI] [PubMed] [Google Scholar]
- 19. Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem 283: 14910–14914, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kramer R. Structural and functional aspects of the phosphate carrier from mitochondria. Kidney Int 49: 947–952, 1996 [DOI] [PubMed] [Google Scholar]
- 21. Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685, 1970 [DOI] [PubMed] [Google Scholar]
- 22. Lesnefsky EJ, Gudz TI, Moghaddas S, Migita CT, Ikeda-Saito M, Turkaly PJ, Hoppel CL. Aging decreases electron transport complex III activity in heart interfibrillar mitochondria by alteration of the cytochrome c binding site. J Mol Cell Cardiol 33: 37–47, 2001 [DOI] [PubMed] [Google Scholar]
- 23. Lesnefsky EJ, Slabe TJ, Stoll MS, Minkler PE, Hoppel CL. Myocardial ischemia selectively depletes cardiolipin in rabbit heart subsarcolemmal mitochondria. Am J Physiol Heart Circ Physiol 280: H2770–H2778, 2001 [DOI] [PubMed] [Google Scholar]
- 24. Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433: 769–773, 2005 [DOI] [PubMed] [Google Scholar]
- 25. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2[-delta delta C(T)] method. Methods 25: 402–408, 2001 [DOI] [PubMed] [Google Scholar]
- 26. Magenta A, Cencioni C, Fasanaro P, Zaccagnini G, Greco S, Sarra-Ferraris G, Antonini A, Martelli F, Capogrossi MC. miR-200c is upregulated by oxidative stress and induces endothelial cell apoptosis and senescence via ZEB1 inhibition. Cell Death Differ 18: 1628–1639, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mateescu B, Batista L, Cardon M, Gruosso T, de Feraudy Y, Mariani O, Nicolas A, Meyniel JP, Cottu P, Sastre-Garau X, Mechta-Grigoriou F. miR-141 and miR-200a act on ovarian tumorigenesis by controlling oxidative stress response. Nat Med 17: 1627–1635, 2011 [DOI] [PubMed] [Google Scholar]
- 28. Mayr JA, Merkel O, Kohlwein SD, Gebhardt BR, Bohles H, Fotschl U, Koch J, Jaksch M, Lochmuller H, Horvath R, Freisinger P, Sperl W. Mitochondrial phosphate-carrier deficiency: a novel disorder of oxidative phosphorylation. Am J Hum Genet 80: 478–484, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mayr JA, Zimmermann FA, Horvath R, Schneider HC, Schoser B, Holinski-Feder E, Czermin B, Freisinger P, Sperl W. Deficiency of the mitochondrial phosphate carrier presenting as myopathy and cardiomyopathy in a family with three affected children. Neuromuscul Disord 21: 803–808, 2011 [DOI] [PubMed] [Google Scholar]
- 30. Nishi Y, Fujimoto S, Sasaki M, Mukai E, Sato H, Sato Y, Tahara Y, Nakamura Y, Inagaki N. Role of mitochondrial phosphate carrier in metabolism-secretion coupling in rat insulinoma cell line INS-1. Biochem J 435: 421–430, 2011 [DOI] [PubMed] [Google Scholar]
- 31. Palmer JW, Tandler B, Hoppel CL. Biochemical properties of subsarcolemmal and interfibrillar mitochondria isolated from rat cardiac muscle. J Biol Chem 252: 8731–8739, 1977 [PubMed] [Google Scholar]
- 32. Palmer JW, Tandler B, Hoppel CL. Heterogeneous response of subsarcolemmal heart mitochondria to calcium. Am J Physiol Heart Circ Physiol 250: H741–H748, 1986 [DOI] [PubMed] [Google Scholar]
- 33. Paradies G, Ruggiero FM, Petrosillo G, Quagliariello E. Enhanced cytochrome oxidase activity and modification of lipids in heart mitochondria from hyperthyroid rats. Biochim Biophys Acta 1225: 165–170, 1994 [DOI] [PubMed] [Google Scholar]
- 34. Poncet D, Pauleau AL, Szabadkai G, Vozza A, Scholz SR, Le Bras M, Briere JJ, Jalil A, Le Moigne R, Brenner C, Hahn G, Wittig I, Schagger H, Lemaire C, Bianchi K, Souquere S, Pierron G, Rustin P, Goldmacher VS, Rizzuto R, Palmieri F, Kroemer G. Cytopathic effects of the cytomegalovirus-encoded apoptosis inhibitory protein vMIA. J Cell Biol 174: 985–996, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rolo AP, Palmeira CM. Diabetes and mitochondrial function: role of hyperglycemia and oxidative stress. Toxicol Appl Pharmacol 212: 167–178, 2006 [DOI] [PubMed] [Google Scholar]
- 36. Rosca MG, Okere IA, Sharma N, Stanley WC, Recchia FA, Hoppel CL. Altered expression of the adenine nucleotide translocase isoforms and decreased ATP synthase activity in skeletal muscle mitochondria in heart failure. J Mol Cell Cardiol 46: 927–935, 2009 [DOI] [PubMed] [Google Scholar]
- 37. Rosca MG, Vazquez EJ, Kerner J, Parland W, Chandler MP, Stanley W, Sabbah HN, Hoppel CL. Cardiac mitochondria in heart failure: decrease in respirasomes and oxidative phosphorylation. Cardiovasc Res 80: 30–39, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sen CK. MicroRNAs as new maestro conducting the expanding symphony orchestra of regenerative and reparative medicine. Physiol Genomics 43: 517–520, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shen X, Zheng S, Thongboonkerd V, Xu M, Pierce WM, Jr, Klein JB, Epstein PN. Cardiac mitochondrial damage and biogenesis in a chronic model of type 1 diabetes. Am J Physiol Endocrinol Metab 287: E896–E905, 2004 [DOI] [PubMed] [Google Scholar]
- 40. Turko IV, Murad F. Quantitative protein profiling in heart mitochondria from diabetic rats. J Biol Chem 278: 35844–35849, 2003 [DOI] [PubMed] [Google Scholar]
- 41. van Rooij E, Olson EN. MicroRNAs: powerful new regulators of heart disease and provocative therapeutic targets. J Clin Invest 117: 2369–2376, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science 316: 575–579, 2007 [DOI] [PubMed] [Google Scholar]
- 43. Walker MD. Role of MicroRNA in pancreatic beta-cells: where more is less. Diabetes 57: 2567–2568, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Williamson CL, Dabkowski ER, Baseler WA, Croston TL, Alway SE, Hollander JM. Enhanced apoptotic propensity in diabetic cardiac mitochondria: influence of subcellular spatial location. Am J Physiol Heart Circ Physiol 298: H633–H642, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]