Abstract
Crk and CrkL adaptors play essential neuronal positioning roles downstream of Reelininduced Dab1 tyrosine phosphorylation. Recently we identified Cin85 to be a CrkL-SH3 binding partner from embryonic murine brain while others found Cin85 binds directly to Dab1. Here using mass spectrometry, biochemical and mutational analyses we show that Dab1 suppresses Cin85 phosphorylation at Ser587. Furthermore a Cin85 Ser587 phosphomimetic disrupts the Dab1-Cin85 complex without affecting the Cin85-CapZ complex. These data provide an early glimpse into how Cin85 phosphorylation might alter the composition of its scaffolding partners to regulate its diverse roles including vesicular trafficking, receptor endocytosis and actin remodeling.
Keywords: Dab1, Cin85, Reelin, phosphorylation, CrkL, kinase
1. Introduction
During central nervous system development newly born neurons take fates dependent on instructional cues in their environments. Some instructional cues direct a neuron’s ultimate position in the mature tissue. One such master positional cue is the secreted glycoprotein Reelin. Reelin governs neuronal positioning throughout the central nervous system with its function most readily evident in the cerebellum, cerebral cortex and hippocampus. In spite of major advances toward understanding Reelin signaling [1-5], it remains only partially understood.
The canonical Reelin pathway clusters its receptors Very Low Density Lipoprotein Receptor (VLDLR) and ApoE Receptor 2 (ApoER2) found on responsive cells [6]. Bound to Reelin receptors intracellularly is the adaptor protein Disabled-1 (Dab1). Reelin receptor clustering leads to Dab1 tyrosine phosphorylation by the Src family of tyrosine kinases (SFKs) [7-10]. At the level of phosphotyrosyl-Dab1 (pY-Dab1) the pathway bifurcates with Tyr185 and Tyr198 responsible for the recruitment and activation of phosphatidylinositol 3-kinase (PI3K)-Akt signaling and Tyr220 and Tyr232leading to the recruitment of the adaptor molecules Crk and Crk-Like (CrkL) [11,12]. Genetic dissection of this bifurcation indicates that both PI3K-Akt and Crk/CrkL binding are essential in Reelin signaling [11,13]. We have identified several Crk/CrkL binding proteins that could serve as Reelin effectors in both targeted [12] and large-scale proteomic analyses [14]. We hypothesized that Crk/CrkL could recruit effector proteins to the Reelin signaling complex where they could be locally regulated by either PI3K-Akt signaling or by SFKs. Indeed we found that the Crk/CrkL binding partner C3G became tyrosine phosphorylated in response to Reelin and this led to activation of the small G protein Rap1 [12]. Among the proteins from embryonic murine brain extracts that we found bound to the CrkL-SH3 domain was the Cbl-interacting protein of 85 kDa (Cin85) [14]. Intriguingly, Sato et al. found Cin85 bound directly to the carboxyl-terminal region of Dab1 and that this interaction was disrupted when Dab1 was phosphorylated by Cyclin-dependent kinase 5 [15], a kinase that plays critical roles in brain development (reviewed in [16]).
Taken together these data suggest that Cin85 might participate in Reelin signaling in a highly regulated way and we therefore asked if Cin85 became phosphorylated at tyrosine residues or in an Akt consensus motif in a setting where Reelin-Dab1 signaling was engaged. To our surprise we found that Dab1 reduced Cin85 phosphorylation in an Akt-like motif. We identified the primary site of this regulated phosphorylation to be Ser587. Furthermore we found that a Ser587 Cin85 phosphomimetic showed dramatically reduced binding to Dab1. The implications of the regulated Cin85-Dab1 complex are discussed.
2. Materials and Methods
2.1. Plasmids and site-directed mutagenesis
The Flag-CIN85 expression construct was a gift of Dr. Ivan Dikic (Goethe University school of Medicine), the FKBP-Dab1-WT and FKBP-Dab1-5F expression constructs were gifts of Dr. Johannes Nimpf (Max Perutz Laboratories), and the Myr-Akt-HA construct was a gift of Phil Tsichlis (Tufts University Medical School). The following constructs were generated using a QuikChange site directed mutagenesis kit (Stratagene, La Jolla, CA): Flag-CIN85-ΔCT (Ser587STOP) and Flag-Cin85 Pro492Ala. DNA sequence confirmation was performed by the University of Vermont Advanced Genome Technologies Core. Flag-Cin85 Ser587Ala and Flag-Cin85 Ser587Asp were generated and sequenced-verified by Bio Basic (Markham, ON).
2.2. Mammalian cell culture, transfections, inhibitors, stimuli, and lysis
E1A-transformed Human embryonic kidney (HEK 293E) cells were grown in DMEM (Mediatech, Manassas, VA) supplemented with 5% Fetal Bovine Serum (FBS), 5% Cosmic Calf Serum (sera were from Hyclone, Logan, UT), 50 units/ml of penicillin and 50 μg/ml of streptomycin. The cells were transfected by calcium phosphate precipitation when at 75% of confluence and between six to sixteen hours after plating. Cells were washed with warm PBS four hours after transfection and returned to full growth media for 72 hours. In the case of stimulations, cells were washed once with PBS and returned to media without serum for twelve hours prior to treatments as indicated in the figure legends. Stimuli and inhibitors were used at the following concentrations and were from the following sources: Calyculin A (100 nM), Epidermal Growth Factor (EGF, 100nM), Phorbol 12-myristate 13-acetate (PMA, 100 nM) were from Cell Signaling Technology (Danvers, MA). AP20187 (200nM) was from ARIAD Pharmaceuticals (Cambridge, MA).The CaMKII inhibitor (CK-59, 500nM) was from Calbiochem (San Diego, CA). After treatments cells were washed once in ice-cold PBS and lysed in ice-cold lysis buffer (25 mM Tris pH 7.2, 137 mM NaCl, 10% glycerol, 1% Igepal, 25 mM NaF, 10 mM Na4P2O7, 1 mM Na3VO4, 1 mM PMSF, 10 μg/mL leupeptin, and 10 μg/mL pepstatin). The insoluble material was cleared using a tabletop micro-centrifuge at 13,000 rpm for 30 minutes at 4 °C and the clarified supernatant was kept for biochemical analyses.
2.3. Antibodies, Immunoprecipitations and immunoblotting
The anti-CIN85 (H-300), anti-Dab1 (H-103) and anti-CapZ (130309) antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA). The anti-Flag M2 antibody and anti-Flag M2 affinity gel were from Sigma (Saint Louis, MO). The anti-phosphotyrosine (4G10) was from Upstate Biotechnology/Milipore (Billerica, MA). Anti-phospho-Akt substrate (9611), anti-alpha tubulin (DM1A) and anti-phospho-Thr202/Tyr204-ERK1/2 (E10) antibodies were from Cell Signaling Technology (Danvers, MA). The anti-HA antibody (Clone 16B12) was from Covance (Princeton, NJ). The anti-ERK1/2 antibody was a gift of John Blenis (Harvard Medical School). Anti-rabbit IgG and anti-mouse IgG secondary antibodies conjugated to horse radish peroxidase (HRP) were obtained from Chemicon/Millipore (Billerica, MA). Immunoprecipitations were conducted using 250-500 μg of whole cell extract and 30 μl of packed anti-Flag affinity resin except for the immunoprecipitations prior to mass spectrometry where two mg of whole cell extract were used. Immunoprecipitations rocked overnight at 4 °C and were washed three times with lysis buffer prior to SDS-PAGE using 10% (37.5:1 acrylamide:bis-acrylamide) polyacrylamide gels. Immunoblotting was conducted as described previously [7].Briefly, proteins were transferred to nitrocellulose membranes and then blocked using 5% dry milk for 45 minutes in tris-buffered saline containing 0.05% Tween-20 (TBST). Primary antibodies were diluted in TBST containing 1.5% BSA and allowed to incubate on the membranes for 2-12 hours. Membranes were washed five times (5-10 minutes each) with TBST prior to incubation with HRP-conjugated secondary antibodies for 1-2 hours. After five final TBST washes the membranes were subjected to enhanced chemiluminescence (ECL) reagents (Pierce, Rockford, IL) and exposure to x-ray film (Denville scientific, Metuchen, NJ). Immunoblots were quantified in Adobe Photoshop after converting images to inverted histograms. Phosphosignals above background were normalized to anti-Flag Cin85 levels.
2.4. Mass Spectrometry
Immunopurified Flag-Cin85 was excised from the coomassie-stained polyacrylamide gel and subjected to in-gel tryptic digestion as described previously [17]. Extracted peptides were subjected to LC-MS/MS in a linear ion trap (LTQ) mass spectrometer (Thermo Electron, San Jose, CA ) as described previously [17]. Tandem mass spectra were searched using SEQUEST against a human Cin85 amino acid sequence database requiring no enzyme specificity, a 2 Da precursor mass tolerance and allowing for the following differential mass modifications: methionine oxidation (+ 16); phosphorylation on serine, threonine and tyrosine (+80); and acrylamide adduction on cysteine (+71).
3. Results
3.1. Dab1 negatively regulates Cin85 phosphorylation in an Akt-like consensus motif
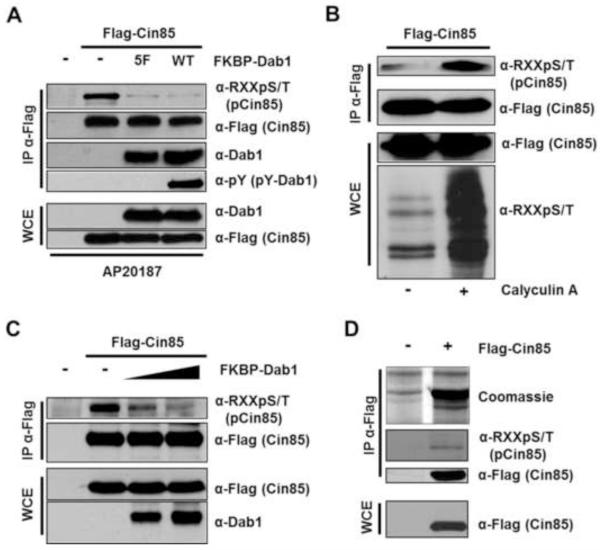
To test the hypothesis that Cin85 could be phosphorylated by SFKs or Akt when Reelin signaling is active, we used a Dab1 dimerization construct developed in the lab of J. Nimpf [6]. In this construct wildtype Dab1, or a form of Dab1 that cannot be phosphorylated at Reelininduced tyrosine phosphorylation sites (Dab15F), is fused to FKBP. The resultant proteins expressed in mammalian cells can be acutely dimerized by a bivalent small molecule (AP20187) capable of binding to FKBP [18]. This system was shown to mimic Reelin receptor clustering as dimerized wildtype Dab1-FKBP, but not Dab15F-FKBP, led to SFK-dependent Dab1 tyrosine phosphorylation and the activation of Akt in non-neuronal cell lines [6]. We co-expressed Flag-tagged Cin85 with either wildtype Dab1-FKBP or Dab15F-FKBP, induced dimerization and immunoprecipitated Flag-Cin85. The immune complexes were probed with anti-phosphotyrosine antibodies and anti-phospho-Akt substrate minimal motif (RXXS/T) antibodies. While no signal was observed in the anti-phosphotyrosine blot (not shown), we were surprised to see a decrease, not an increase, in the phosphorylation of Cin85 when using the anti-phospho-RXXS/T motif antibody (Fig. 1A). Furthermore, this decrease in Cin85 phosphorylation was independent of the phosphorylation state of Dab1 as the results were the same when co-expressing either wildtype-Dab1-FKBP or Dab15F-FKBP (Fig. 1A). We confirmed the co-immunoprecipation of Dab1 and Cin85 [15] and that only wildtype-Dab1-FKBP was tyrosine phosphorylated (Fig. 1A). We also confirmed that Cin85 exhibited reversible phosphorylation in a RXXS/T motif by a kinase endogenous to HEK293 cells. This was achieved independently of clustering by treating cells transfected with Flag-Cin85 with the serine/threonine phosphatase inhibitor Calyculin A, followed by anti-Flag immunopurification and blotting with the anti-phospho-RXXS/T motif antibody (Fig. 1B). We next demonstrated that increasing concentrations of co-transfected Dab1 led to increasingly diminished anti-phospho-RXXS/T signal on immunoprecipitated Flag-Cin85 (Fig. 1C). Toward the identification of the phosphorylated serine residue on Cin85 which was negatively regulated by Dab1 we immunopurified coomassie-stainable levels of Flag-Cin85 for mass spectrometry analysis (Fig. 1D).
Fig. 1.
Dab1 negatively regulates Cin85 phosphorylation in a RXXS/T motif. (A) The negative regulation of Cin85 RXXS/T phosphorylation by Dab1 is independent of Dab1 tyrosine phosphorylation. HEK 293 cells were transfected with the indicated constructs, starved of serum and treated with AP20187 (200 nM) for 30 minutes prior to lysis. Clarified extracts were either directly subjected to SDS-PAGE and immunoblotting with the indicated antibodies or first to anti-Flag immunopreciptation as indicated. (B) HEK 293 cells were transfected with Flag-Cin85, starved of serum and treated with or without 100 nM Calyculin A for 30 minutes prior to lysis as indicated. Clarified extracts were treated as in A. (C) HEK 293 cells were transfected with the indicated constructs. Co-transfected 10 cm dishes received 15 μg of Flag-Cin85 and either zero, 5 μg or 10 μg (as in A) of FKBP-Dab1. (D) Flag-Cin85 was immunoprecipitated from transfected cells, subjected to SDS-PAGE and stained with coomassie blue (upper panel). Whole cell extract or 10% of the immune complexes were subjected to immunoblotting with the indicated antibodies.
3.2. Serine 587 is the primary Cin85 phosphorylation site recognized by the phospho-RXXS/T motif antibody
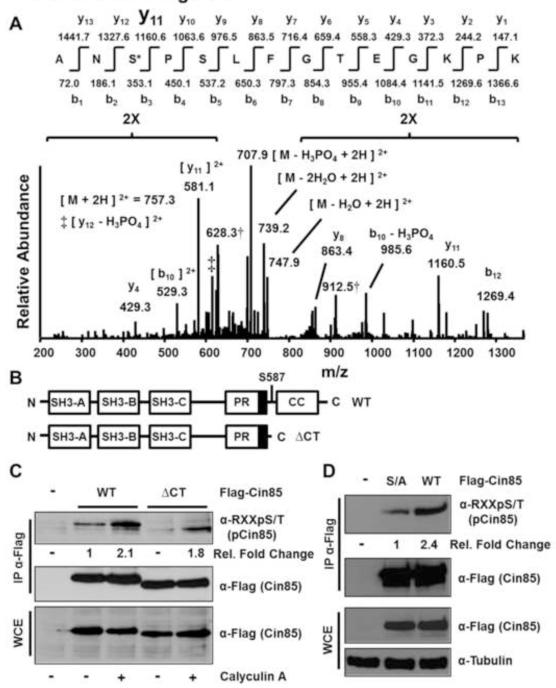
The immunopurified Flag-Cin85 from Figure 1D was cut from the SDS-PAGE gel, subjected to in-gel tryptic digestion and liquid chromatography tandem mass spectrometry in a linear ion trap mass spectrometer. One tryptic phosphopeptide was confidently identified corresponding to the CIN85 sequence R/AN(pS)PSLFGTEGKPK/(Fig. 2A) where pS indicates phosphorylated serine 587 and R/and K/indicate the sites of tryptic cleavage. Ser587 lies in a RXXS/T motif and would likely be recognized by the phospho-RXXS/T motif antibody. The peptide identification, and phosphorylation site assignment as Ser587, and not as Ser589 or Thr593 were unequivocal. This is most readily shown by the prominent y-11 daughter ion generated by fragmentation at the fragile site amino-terminal to the proline at position four, which was also observed in the spectrum of the unphosphorylated tryptic peptide harboring Ser587 (Fig. S1). In the domain structure of Cin85 Ser587 is situated between a proline-rich region and a coiled-coil domain (Fig. 2B). To determine if Ser587 was the primary phospho-RXXS/T site in Cin85 recognized by the motif-specific antibody we first compared the phospho-RXXS/T signal of immunopurified wildtype Cin85 and a Ser587Stop truncation mutant from cells with or without Calyculin A treatment following starvation of serum for twelve hours. Both the baseline phospho-RXXS/T signal from untreated cells as well as the signal following Calyculin A treatment were greatly reduced in the Ser587Stop truncation mutant compared to wildtype Cin85 (Fig. 2C). Furthermore, a Cin85 Ser587Ala mutant also showed greatly diminished anti-phospho-RXXS/T reactivity (Fig. 2D). Taken together these data suggest that Ser587 is the primary but not the only Cin85 phosphorylation site that falls within a RXXS/T motif. Indeed, phosphositeplus [19] reports the identification of four other Cin85 phosphorylation sites in addition to Ser587 that lie in a RXXS/T motif (Thr179, Ser230, Ser436 and Ser493). Phosphorylation of Ser230 has been identified in 28 studies, compared to 17 studies for Ser587, six studies for Ser436 and one study each for Thr179 and Ser493. Additionally, six other Cin85 phosphorylation sites have been identified that lie in either a KXXS or KXXT motif to which the antibody has some affinity and which could contribute to the overall phospho-RXXS/T signal.
Fig. 2.
Serine 587 is the primary Cin85 phosphorylation site recognized by the anti-phospho-RXXS/T motif antibody from growing HEK 293 cells. (A) Low energy collision-induced dissociation (CID) MS/MS spectrum identified from peptides extracted following in-gel tryptic digestion of the Flag-Cin85 band shown in Fig. 1D. Compare this spectrum to the spectrum from the unphosphorylated peptide shown in Fig. S1. (B) Schematic of the domain structure of Cin85. The three Src homology 3 (SH3) domains are depicted as well as a proline-rich (PR) region. The dark box in the PR region indicates the CapZ binding region. CC indicates the coiled-coil domain. (C) Cells were transfected with the indicated constructs and experiments were conducted as described in the legend to Fig. 1B. Fold changes in phospho-Cin85 levels relative to Flag-Cin85 levels and sample type are indicated. (D) Cells were transfected with the indicated constructs (S/A indicates Ser587Ala) and experiments were conducted as described in Fig. 1A. Fold changes in phospho-Cin85 levels relative to Flag-Cin85 levels and sample type are indicated.
3.3. Examination of potential Cin85 RXXS/T motif kinases
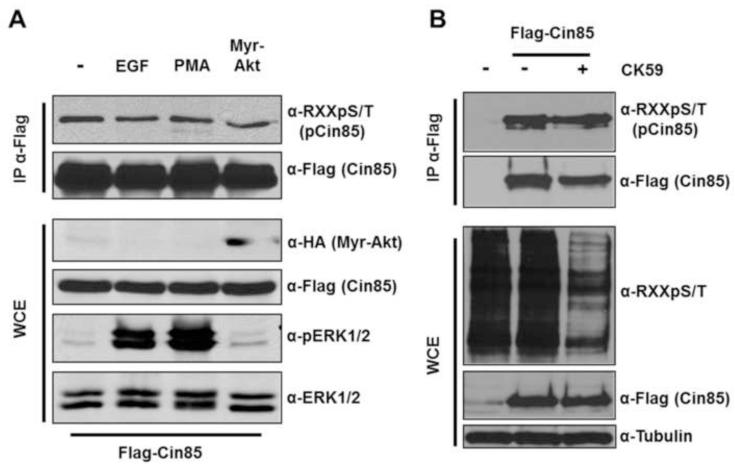
Toward the identification of the possible kinase(s) involved in the phosphorylation of Cin85 RXXS/T motifs we first tested the effect of depriving cells of serum and then stimulating with factors known to activate several basophilic kinases that target RXXS/T motifs. Stimulation with Epidermal Growth Factor (EGF) or the phorbol ester PMA variably activates several basophilic kinase families including Akt, RSK, MSK, S6K and PKC. However, stimulation with EGF or PMA did not increase Cin85 phosphorylation in a RXXS/T motif (Fig 3A). EGF and PMA were shown to be active in the system by the strong induction of activation loop phosphorylation on ERK1/2 (Fig. 3A). Furthermore, because this study began with the hypothesis that Akt might induce Cin85 phosphorylation we also specifically asked if co-transfection of Cin85 with an activated allele of Akt would increase Cin85 phosphorylation in an RXXS/T motif in unstimulated cells. It did not (Fig. 3A). One of the large-scale phosphoproteomic studies that identified Cin85 Ser587 phosphorylation suggested that this site might be phosphorylated by CamKII [20]. We therefore examined the effect of the CamKII inhibitor CK59 [21] on Cin85 phosphorylation in a RXXS/T motif. While CK59 significantly reduced phospho-RXXS/T in the whole cell extract, CK59 did not reduce Cin85 phosphorylation in a RXXS/T motif (Fig 3B). While these results reduce the list of possible Cin85 RXXS/T kinases, they remain to be identified.
Fig. 3.
Examination of potential Cin85 phospho-RXXS/T kinases. (A) HEK 293 cells were transfected with wildtype Cin85 (or co-transfected with myristoylated-Akt-HA as indicated). Cells were starved of serum for 12 hours, left untreated or treated with 100nM EGF or PMA for thirty minutes as indicated. Whole cell extracts were directly subjected to immunoblotting or first to anti-Flag immunoprecipitation as indicated. (B) HEK 293 cells were treated as in A except following serum starvation, cells were treated with or without the CamKII inhibitor CK59 (500 nM for 30 minutes).
3.4 A Cin85 Ser587Asp phosphomimetic dramatically reduces Dab1binding but not CapZ binding to Cin85
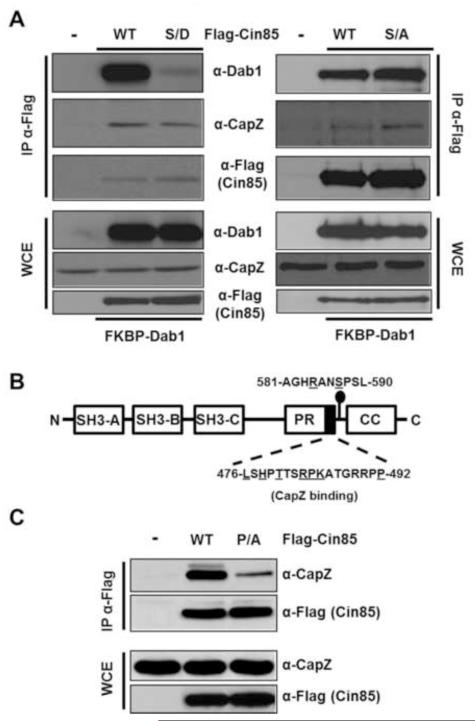
Given the precedent that Cdk5 phosphorylation of Dab1 reduces the binding of Dab1 to Cin85 [15], we asked if a Cin85 Ser587Asp phosphomimetic would exhibit reduced binding to Dab1 in reciprocal fashion. Indeed the Ser587Asp but not the Ser587Ala showed dramatically reduced binding to Dab1 without affecting the binding of Cin85 to another of its interacting partners CapZ (Fig. 4A). CapZ has previously been shown to bind to a Cin85-related protein CD2AP via a 17 residue stretch containing several conserved residues also found in Cin85 (Fig. 4B). To test the requirement of this conserved region in the binding of CapZ to Cin85 while at the same time providing contrast to lack of effect of the Ser587Asp mutation on the Cin85-CapZ interaction, we showed that a single Ser492Ala mutation in Cin85 dramatically reduced its binding to CapZ (Fig. 4C). This suggests Ser587 of Cin85 lies in a unique and functionally distinct regulatory region.
Fig. 4.
A Cin85 Ser587Asp phosphomimetic exhibits dramatically reduced binding to Dab1 but retains its binding to CapZ. (A) HEK 293 cells co-transfected with the indicated Flag-Cin85 constructs and FKBP-Dab1 were lysed and extracts were either directly subjected to SDS-PAGE and immunoblotting with the indicated antibodies or were first subjected to anti-Flag immunoprecipitations. (B) Schematic depicting the domain structure of Cin85 highlights the amino acids critical (underlined) for the binding of Cin85 to CapZ. (C) Cin85 Pro492Ala is dramatically impaired in its ability to bind CapZ. HEK 293 cells were transfected with the indicated Flag-Cin85 constructs and cell extracts were subjected directly to immunoblotting or first to anti-Flag immunoprecipitations as indicated.
4. Discussion
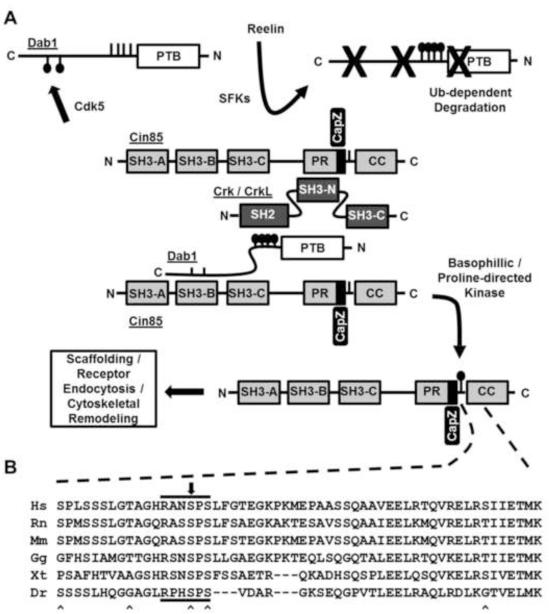
Our original hypotheses centered on the idea that Cin85 recruitment to Reelin signaling complexes would enable Cin85 to become locally phosphorylated by either active SFKs or Akt. Furthermore, we anticipated that Cin85 might be involved in the canonical neuronal positioning aspects of Reelin signaling. This latter concept was in part suggested by the fact that the interaction of Cin85 could be disrupted by Cdk5 phosphorylation of Dab1 [15] and Cdk5 deficiencies exhibit dramatic developmental defects that have similarities to Reelin deficiencies [16]. However, neither of these notions appears to be experimentally substantiated. Indeed, while our studies were underway, the role of Cin85 in the brain was explored by targeted genetic disruption of the neuronal isoforms of Cin85 [22]. In these animals no overt Reeler-like phenotype was observed, including no hypoplasia of the cerebellum and its associated loss of motor control. However, genetic disruption of Cin85 in the brain revealed Cin85 had important postsynaptic roles, particularly in the regulation of dopamine receptor trafficking [22]. This shifts the possible connection between Cin85 and Reelin signaling away from primary cytoarchitectural, developmental effects and more toward synaptic regulation. Intriguingly, both Cin85 and Reelin signaling components are localized in postsynaptic dendritic regions and Reelin has been implicated in synaptic transmission and synaptic integrity [22-24]. If so, Reelin signaling in these tissues may lead to Cin85 Ser587 phosphorylation as Reelin-dependent degradation of pY-Dab1 proceeds (Fig. 5). In this regard, it is important to note that reconstitution of Reelin signaling by synthetic clustering as shown in Fig. 1A does not induce Dab1 degradation. Alternatively, synaptic activity could increase the activity of Cin85 Ser587 kinases independently of Reelin and reduce the binding of Cin85 to Dab1, potentially freeing Cin85 for alternative roles [25].
Fig. 5.
(A) Simple model depicting phospho-dependent regulation of the Dab1-Cin85 protein complex. Dab1 phosphorylation by Cdk5 at Ser400 and Ser491disrupts its binding to Cin85. Conversely, phosphorylation of Cin85 at Ser587 disrupts its binding to Dab1. Upon Reelin stimulation Cin85 levels could increase in proximity of the Reelin signaling complex via recruitment to pY-Dab1 through the Crk/CrkL family of adaptors. Ultimately phsphotyrosyl-Dab1 is targeted for proteasome-dependent degradation and Cin85 phosphorylation at Ser587 can accumulate. It is hypothesized that phospho-Ser587 Cin85 regulates its role in receptor endocytosis or actin cytoskeletal remodeling. (B) Multiple sequence alignment shows Ser587 lies in a RXXSPS motif conserved among Cin85 orthologues in common vertebrates. The black arrow above the alignment indicates the position of Ser587. “^” markings below the alignment indicate the positions of previously identified phosphorylation sites. Letters preceding sequences indicate the first letter of both the genus and species of the Cin85 orthologues (human, rat, mouse, chicken, frog, zebrafish).
However, it is possible that the regulation of Cin85 by Dab1 reflects the effect any number of Cin85 SH3 binding proteins [25] may have including Dab2 which lacks the tyrosine phosphorylation sites of Dab1 and thus is not targeted for degradation in a similar manner. If the relative abundance of Cin85 SH3 binding proteins is high relative to Cin85 it would then be predicted that Cin85 Ser587 phosphorylation would be low. It is anticipated that Cin85 Ser587 phosphorylation plays an important role in governing the composition of proteins on, and the ultimate effect of, the Cin85 scaffold. It therefore will be important to determine which Cin85 binding proteins have altered binding to the Ser587Asp mutant. Such a line of inquiry will likely prove revealing not only to establish the function of Ser587 phosphorylation but as well for the several other Cin85 phosphorylation sites [25] that have been identified primarily via large-scale proteomic screens.
Supplementary Material
Highlights.
Dab1 prevents phosphorylation of its binding partner Cin85 at serine 587.
A Cin85 Ser587Asp mutation dramatically reduces the binding to Dab1 but not to CapZ.
Cin85 Ser587 lies in a RXXSP motif conserved in common vertebrates.
Acknowledgements
This work was supported by NSF grant IOS 1021795 (to BAB), the Vermont Genetics Network through NIH Grant P20 RR16462 from the INBRE program of the NCRR/NIGMS (support given to BAB and BKB), and NIH Grant 5 P20 RR016435 from the COBRE (neuroscience) program of the NCRR/NIGMS (support given to BAB and BKB). BKB was also supported by a NEAGAP fellowship. We thank J. Blenis, J. Nimpf, P.Tsichlis and I. Dikic for generously providing critical reagents.
Abbreviations Used
- Dab1
Disabled-1
- CIN85
Cbl-Interacting protein of 85kDa
- ApoER2
Apolipoprotein E receptor 2
- Crk
CT10-regulator of kinase
- CrkL
Crk-like
- SFK
Src family kinase
- SH3
Src homology-3 domain
- SH2
Src homology-2 domain
- PR
Proline-rich region
- CC
Coiled-coil domain
- CT
Carboxyl Terminal region
- VLDLR
Very Low Density Lipoprotein Receptor
- EGF
Epidermal Growth Factor
- EGFR
Epidermal Growth Factor Receptor
- PMA
Phorbol 12-myristates 13-acetate
- CaMKII
Calmodulin-dependent Kinase 2
- PKC
Protein Kinase C
- CK-59
CamKII inhibitor 59
- Cdk-5
Cyclin-dependent kinase 5
- CapZ
F-actin capping protein
- SH3KBP-1
SH3 Kinase binding protein-1
- RSK
90 kilodalton Ribosomal S6 Kinase
- S6K
70 kilodalton Ribosomal S6 Kinase
- MAPK
Mitogen-activated protein Kinase
- ERK
Extracellular Regulated Kinase
- MSK
Mitogen and Stress-Activated Kinase
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- WCE
whole cell extract
- IP
immunoprecipitation
- ECL
enhanced chemiluminescence
- FKBP
FK506 binding protein
- RXXS
arginine-any two amino acids-serine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Honda T, Kobayashi K, Mikoshiba K, Nakajima K. Regulation of cortical neuron migration by the Reelin signaling pathway. Neurochem Res. 2011;36:1270–9. doi: 10.1007/s11064-011-0407-4. [DOI] [PubMed] [Google Scholar]
- [2].Zhao S, Frotscher M. Go or stop? Divergent roles of Reelin in radial neuronal migration. Neuroscientist. 2010;16:421–34. doi: 10.1177/1073858410367521. [DOI] [PubMed] [Google Scholar]
- [3].Forster E, Bock HH, Herz J, Chai X, Frotscher M, Zhao S. Emerging topics in Reelin function. Eur J Neurosci. 2010;31:1511–8. doi: 10.1111/j.1460-9568.2010.07222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Katsuyama Y, Terashima T. Developmental anatomy of reeler mutant mouse. Dev Growth Differ. 2009;51:271–86. doi: 10.1111/j.1440-169X.2009.01102.x. [DOI] [PubMed] [Google Scholar]
- [5].Cooper JA. A mechanism for inside-out lamination in the neocortex. Trends Neurosci. 2008;31:113–9. doi: 10.1016/j.tins.2007.12.003. [DOI] [PubMed] [Google Scholar]
- [6].Strasser V, et al. Receptor clustering is involved in Reelin signaling. Mol Cell Biol. 2004;24:1378–86. doi: 10.1128/MCB.24.3.1378-1386.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Ballif BA, Arnaud L, Cooper JA. Tyrosine phosphorylation of Disabled-1 is essential for Reelin-stimulated activation of Akt and Src family kinases. Brain Res Mol Brain Res. 2003;117:152–9. doi: 10.1016/s0169-328x(03)00295-x. [DOI] [PubMed] [Google Scholar]
- [8].Arnaud L, Ballif BA, Forster E, Cooper JA. Fyn tyrosine kinase is a critical regulator of disabled-1 during brain development. Curr Biol. 2003;13:9–17. doi: 10.1016/s0960-9822(02)01397-0. [DOI] [PubMed] [Google Scholar]
- [9].Kuo G, Arnaud L, Kronstad-O’Brien P, Cooper JA. Absence of Fyn and Src causes a reeler-like phenotype. J Neurosci. 2005;25:8578–86. doi: 10.1523/JNEUROSCI.1656-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Bock HH, Herz J. Reelin activates SRC family tyrosine kinases in neurons. Curr Biol. 2003;13:18–26. doi: 10.1016/s0960-9822(02)01403-3. [DOI] [PubMed] [Google Scholar]
- [11].Feng L, Cooper JA. Dual functions of Dab1 during brain development. Mol Cell Biol. 2009;29:324–32. doi: 10.1128/MCB.00663-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ballif BA, Arnaud L, Arthur WT, Guris D, Imamoto A, Cooper JA. Activation of a Dab1/CrkL/C3G/Rap1 pathway in Reelin-stimulated neurons. Curr Biol. 2004;14:606–10. doi: 10.1016/j.cub.2004.03.038. [DOI] [PubMed] [Google Scholar]
- [13].Park TJ, Curran T. Crk and Crk-like play essential overlapping roles downstream of disabled-1 in the Reelin pathway. J Neurosci. 2008;28:13551–62. doi: 10.1523/JNEUROSCI.4323-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Cheerathodi M, Ballif BA. Identification of CrkL-SH3 binding proteins from embryonic murine brain: implications for Reelin signaling during brain development. J Proteome Res. 2011;10:4453–62. doi: 10.1021/pr200229a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Sato Y, et al. Regulation of the interaction of Disabled-1 with CIN85 by phosphorylation with Cyclin-dependent kinase 5. Genes Cells. 2007;12:1315–27. doi: 10.1111/j.1365-2443.2007.01139.x. [DOI] [PubMed] [Google Scholar]
- [16].Dhavan R, Tsai LH. A decade of CDK5. Nat Rev Mol Cell Biol. 2001;2:749–59. doi: 10.1038/35096019. [DOI] [PubMed] [Google Scholar]
- [17].Ballif BA, Cao Z, Schwartz D, Carraway KL, 3rd, Gygi SP. Identification of 14-3-3epsilon substrates from embryonic murine brain. J Proteome Res. 2006;5:2372–9. doi: 10.1021/pr060206k. [DOI] [PubMed] [Google Scholar]
- [18].Spencer DM, Wandless TJ, Schreiber SL, Crabtree GR. Controlling signal transduction with synthetic ligands. Science. 1993;262:1019–24. doi: 10.1126/science.7694365. [DOI] [PubMed] [Google Scholar]
- [19].Hornbeck PV, Kornhauser JM, Tkachev S, Zhang B, Skrzypek E, Murray B, Latham V, Sullivan M. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 2012;40:D261–70. doi: 10.1093/nar/gkr1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Olsen JV, et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal. 2010;3:ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- [21].Konstantopoulos N, Marcuccio S, Kyi S, Stoichevska V, Castelli LA, Ward CW, Macaulay SL. A purine analog kinase inhibitor, calcium/calmodulin-dependent protein kinase II inhibitor 59, reveals a role for calcium/calmodulin-dependent protein kinase II in insulin-stimulated glucose transport. Endocrinology. 2007;148:374–85. doi: 10.1210/en.2006-0446. [DOI] [PubMed] [Google Scholar]
- [22].Shimokawa N, et al. CIN85 regulates dopamine receptor endocytosis and governs behaviour in mice. EMBO J. 2010;29:2421–32. doi: 10.1038/emboj.2010.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Niu S, Renfro A, Quattrocchi CC, Sheldon M, D’Arcangelo G. Reelin promotes hippocampal dendrite development through the VLDLR/ApoER2-Dab1 pathway. Neuron. 2004;41:71–84. doi: 10.1016/s0896-6273(03)00819-5. [DOI] [PubMed] [Google Scholar]
- [24].Ventruti A, Kazdoba TM, Niu S, D’Arcangelo G. Reelin deficiency causes specific defects in the molecular composition of the synapses in the adult brain. Neuroscience. 2011;189:32–42. doi: 10.1016/j.neuroscience.2011.05.050. [DOI] [PubMed] [Google Scholar]
- [25].Havrylov S, Redowicz MJ, Buchman VL. Emerging roles of Ruk/CIN85 in vesicle-mediated transport, adhesion, migration and malignancy. Traffic. 2010;11:721–31. doi: 10.1111/j.1600-0854.2010.01061.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.